Abstract
DNA methylation is an important factor regulating gene expression in organisms. However, whether DNA methylation plays a key role in adaptive evolution is unknown. Here, we show evidence of naturally selected DNA methylation in Arabidopsis thaliana. In comparison with single nucleotide polymorphisms, three types of methylation—methylated CGs (mCGs), mCHGs, and mCHHs—contributed highly to variable gene expression levels among an A. thaliana population. Such variably expressed genes largely affect a large variation of specialized metabolic quantities. Among the three types of methylations, only mCGs located in promoter regions of genes associated with specialized metabolites show a selective sweep signature in the A. thaliana population. Thus, naturally selected mCGs appear to be key mutations that cause the expressional diversity associated with specialized metabolites during plant evolution.
All living organisms use DNA as a heritable material. The heritable DNA-based information depends on the order of four chemical bases: adenine (A), guanine (G), cytosine (C), and thymine (T). Among the four bases, C can undergo an enzyme-mediated chemical modification called DNA methylation. DNA methylation significantly affects transcriptional regulation. Consequently, DNA methylation plays important roles in development, cell differentiation, reprogramming, and stress responses in both plants and mammals (Feng et al. 2010). There are three types of DNA methylation: CG, CHG, and CHH—where H = A, T, or C. Each type of DNA methylation is maintained by different systems in plants (Kawashima and Berger 2014; Zhang et al. 2018). Among methylated CG (mCG), methylated CHG (mCHG), and methylated CHH (mCHH), only the mCG is inherited from parents (Iwasaki and Paszkowski 2014). Especially, the inheritance of mCGs in plant genomes is stable over many generations compared to animal genomes (Feng et al. 2010; Zemach et al. 2010; Takuno et al. 2016). Therefore, it is of interest to examine the adaptive evolution triggered by mutated mCGs as the other heritable material. However, mCHG and mCHH tend to be modified in specific developmental stages through trans effects (Bouyer et al. 2017; Kawakatsu et al. 2017; Kawakatsu and Ecker 2019). Inherited mCGs cause phenotypic changes in Arabidopsis, and the changes may be advantageous for a particular condition, indicating that mCGs contribute to adaptive evolution in Arabidopsis (Dubin et al. 2015; Williams and Gehring 2017; He et al. 2018). Nevertheless, there are no reports of inherited mCGs undergoing positive selection.
It is unclear what kinds of traits triggered by mCGs have been positively selected in an Arabidopsis population. There is a large variety of specialized metabolites produced by plants, and the accumulated specialized metabolites are highly diverse within a single species (Wink 2008; Pichersky and Lewinsohn 2011; Weigel 2012; Carreno-Quintero et al. 2013; Alseekh et al. 2015; Matsuda et al. 2015; Pichersky and Raguso 2018; Tohge et al. 2018). Some of the specialized metabolites have various functions related to reproduction and responses to abiotic/biotic stress (Pichersky and Lewinsohn 2011; Pichersky and Raguso 2018; Tohge et al. 2018). The differences in the accumulated specialized metabolites strongly affect fitness in the natural environment (Kerwin et al. 2015). Among the A. thaliana accessions, differentially methylated regions are concentrated in genes related to specialized metabolites (Kawakatsu et al. 2016). DNA methylation in the promoter region of a gene may alter the production of specialized metabolites in A. thaliana (Kooke et al. 2019). Thus, specialized metabolite diversity may result from the diversity of DNA methylation in Arabidopsis (Kawakatsu et al. 2016; Kooke et al. 2019). However, at the genomic level, gene expression is rarely controlled by DNA methylation, except that of transposable elements (TEs), in comparison with nucleotide mutations (Matzke et al. 2015; Meng et al. 2016; Zhang et al. 2018). Therefore, the contribution of DNA methylation to specialized metabolite production remains unclear at the genomic level.
Results
Association between gene expression and DNA methylation
We obtained 1,397,934 single nucleotide polymorphisms (SNPs) and 4,448,076 single methylation polymorphisms (SMPs) (mCGs: 1,653,070, mCHGs: 814,938, and mCHHs: 1,980,068) from 620 accessions of A. thaliana in the 1001 Genomes Consortium and Kawakatsu et al. (Supplemental Table S1; The 1001 Genomes Consortium 2016; Kawakatsu et al. 2016). Among the 24,030 genes, all the SMP types were concentrated in the promoter regions, from 3 kb upstream of the transcriptional start sites to the termination sites (Fig. 1A). The expression levels of the 24,030 genes in the 620 accessions were obtained from the RNA-seq data presented in Kawakatsu et al. (2016) (Supplemental Table S2). To examine the range of cis-regulatory effects of SMPs on gene expression, we identified SMPs having significant correlations with gene expression using a linear mixed model and 1000 randomly selected genes. The associated SMPs were concentrated from 1 kb upstream of the transcriptional start sites to l kb downstream from the termination sites (Fig. 1B). To address the cis-regulatory effects, we focused on this region in each gene.
Figure 1.
Distribution of DNA methylation associated with gene expression. (A) The distributions of mCGs, mCHGs, and mCHHs in 24,320 genes. The x-axis represents SMP positions relative to the transcriptional start and termination sites in 24,320 genes from 620 A. thaliana accessions. The blue line represents the distribution of the mCGs, mCHGs, and mCHHs. (B) The distributions of mCGs, mCHGs, and mCHHs associated with gene expression in a cis-regulatory manner in 1000 randomly selected genes. The x-axis represents SMP positions relative to the transcriptional start and termination sites in 1000 randomly selected genes from 620 A. thaliana accessions. (C) The numbers of genes with significant associations between expression and mCGs, mCHGs, mCHHs, or SNPs.
To compare the cis-regulatory effects between SMPs (mCGs, mCHGs, and mCHHs) and SNPs in each of the 24,030 genes, we identified mCGs, mCHGs, mCHHs, and SNPs that were significantly associated with gene expression levels among 620 accessions (FDR-corrected P values < 0.05) (Table 1; Supplemental Tables S3–S6). The numbers of genes with at least one significantly associated mCG, mCHG, mCHH, and SNP were 6690, 3319, 3463, and 21,924, respectively (Fig. 1C), indicating that SNPs have much larger cis-regulatory effects than SMPs. There were 8425 genes that had significantly associated SMPs in the mCGs, mCHGs, or mCHHs. Only 405 of 8425 genes did not have any associated SNPs (Fig. 1C). Out of 21,924 genes with significantly associated SNPs, 13,904 genes did not have associated SMPs in the mCGs, mCHHs, or mCHHs (Fig. 1C). These results indicate that, at the genomic level, SNPs are a major factor regulating gene expression in view of the number of associated genes. This result is supported by previous reports (Matzke et al. 2015; Meng et al. 2016; Zhang et al. 2018). On the other hand, genes associated with SMPs in the mCGs or mCHGs tend to have higher r2 than genes associated with SNPs (Supplemental Fig. S1). This result indicates that each mCG or mCHG has a large effect on gene expression compared to SNPs.
Table 1.
Numbers of SMPs and SNPs in 620 global A. thaliana accessions

Association between expression variation and DNA methylation
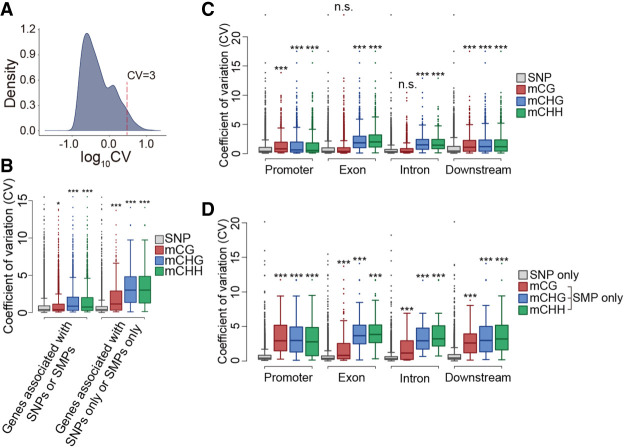
We examined the variation in gene expression within a single species. It is possible that SMPs have different effects on expressional variations compared to SNPs. Therefore, we evaluated the degree of expressional variation among the 620 accessions using the coefficients of variation (CVs). Approximately 95% of genes had low levels of expressional variation (CV < 3), and only ∼5% had large levels of expressional variation (CV ≥ 3) (Fig. 2A). In addition, the distribution of CVs has two peaks (Fig. 2A). The higher peak is largely contributed by TE genes (median of CV = 3.18) compared to the other genes (median of CV = 0.41) (Supplemental Figs. S2, S3). We then compared the expressional variations between genes associated with SMPs and SNPs. Genes associated with mCHGs and mCHHs tended to have higher CVs than genes associated with SNPs (median of CV in SNPs = 0.4, median of CV in mCGs = 0.41, median of CV in mCHGs = 0.87, median of CV in mCHHs = 0.75; mCG: P = 1.51 × 10−2, mCHG: P < 2.20 × 10−16, and mCHH: P < 2.20 × 10−16; Wilcoxon rank-sum test) (Fig. 2B). Furthermore, for all SMP types, genes associated with only SMPs and not SNPs tended to have much higher CVs than genes associated with only SNPs and not SMPs (median of CV in SNPs = 0.4, median of CV in mCGs = 1.16, median of CV in mCHGs = 3.03, median of CV in mCHHs = 3.03; mCG: P < 2.20 × 10−16, mCHG: P < 2.20 × 10−16, and mCHH: P < 2.20 × 10−16; Wilcoxon rank-sum test) (Fig. 2B). This trend is shown in both TE genes and the other genes (Supplemental Figs. S4–S7). These results indicate that SMPs are a main factor in the control of highly variable gene expression within a single species.
Figure 2.
Variation in the expression of genes associated with SMPs or SNPs. (A) Coefficients of variation (CVs) for 24,320 genes from 620 accessions. (B) CVs of genes associated with SNPs, mCGs, mCHGs, and mCHHs. (C) CVs of genes associated with SNPs, mCGs, mCHGs, and mCHHs located in promoter, exon, intron, or downstream regions. (D) CVs of genes associated with only SNPs, only mCGs, only mCHGs, and only mCHHs located in promoter, exon, intron, or downstream regions. In each box plot, the box represents the 25%–75% range, the middle line represents the median, the dotted line represents the 1%–99% range, and the outer circles represent outliers. The significant differences in CVs among genes associated with SNPs and the SMP groups were evaluated using the Wilcoxon rank-sum test; (***) P < 0.001, (*) P < 0.05, (ns) not significant.
DNA methylation has different effects on gene expression depending on its location (Zhang et al. 2018). The DNA methylation of TE regions causes the TE to be silenced, and the DNA methylation of promoters mainly causes transcriptional repression. The DNA methylation of a gene body is correlated with constitutive expression, not repression (Zhang et al. 2006; Kawakatsu et al. 2016). To address which types of SMPs control highly variable gene expression within a single species, SMPs and SNPs were classified into promoter (1-kb region upstream of the transcriptional start site to the transcriptional start site), exon, intron, and downstream (transcriptional termination site to l kb downstream from the transcriptional termination site). For genes associated with mCGs, mCHGs, mCHHs, and SNPs, CVs were compared among these regions. The genes associated with most of the SMPs tended to have significantly higher CVs than genes associated with SNPs, except for mCGs located on exon or intron regions (Fig. 2C). This trend was clearer for genes associated with only SMPs and not SNPs (Fig. 2D). Genes with mCHGs and mCHHs located on exon regions, compared to the other regions, tended to have higher CVs (P < 0.05; Wilcoxon rank-sum test) (Supplemental Tables S7, S8). Genes with mCGs located on promoter and downstream regions, compared to exon and intron regions, tended to have higher CVs (P < 0.05; Wilcoxon rank-sum test) (Fig. 2C,D; Supplemental Tables S7, S8). These results indicate that mCHGs and mCHHs located on exon regions associated with highly variable gene expression, but only mCGs located on promoter and downstream regions are associated with highly variable gene expression. Thus, there is a difference in how CG methylation and non-CG methylation regulates gene expression.
Association between DNA methylation and specialized metabolite diversity
To understand the functional categories having higher expressional variation levels, we examined Gene Ontology (GO) terms enriched for genes having the 5% highest and lowest CV values. These GO terms were associated with specialized metabolism and ubiquitous categories, respectively (Fig. 3A,B). Thus, genes associated with specialized metabolism tended to have high levels of expressional variation. We further examined the GO terms enriched for genes associated with only SMPs. The genes associated with only mCG, mCHG, or mCHH tended to be enriched in GO terms related to specialized metabolism (Supplemental Table S9). Thus, mCG, mCHG, and mCHH are highly correlated with the expressional variation of metabolome quantitative trait genes (mQTGs).
Figure 3.
Relationship between DNA methylation and mQTG expression. (A) Top 10 enriched GO terms in genes with high CVs (top 5%). (B) Top 10 enriched GO terms in genes with low CVs (bottom 5%). The false discovery rate (FDR) was calculated from GO enrichment analyses using the χ2 test. (C) The rates of mQTGs having high CVs (top 5%) associated with SNPs, mCGs, mCHGs, and mCHHs in 620 A. thaliana accessions. The rates represent the numbers of mQTGs with high CVs (top 5%) associated with SNPs, mCGs, mCHGs, and mCHHs each divided by the number of total genes associated with SNPs, mCGs, mCHGs, and mCHHs, respectively. (D) The rates of mQTGs with high CVs (top 5%) associated with only SNPs, only mCGs, only mCHGs, and only mCHHs in the 620 accessions. The rates represent the numbers of mQTGs with high CVs (top 5%) associated with only SNPs, only mCGs, only mCHGs, and only mCHHs each divided by the number of genes associated with only mCGs, only mCHGs, and only mCHHs, respectively. (E) The rates of mQTGs with high CVs (top 5%) associated with SNPs, mCGs, mCHGs, and mCHHs in 28 accessions. (F) The rates of mQTGs with high CVs (top 5%) associated with only SNPs, only mCGs, only mCHGs, and only mCHHs in 28 accessions. The differences in the rates between SNPs and each of the three kinds of SMPs (mCGs, mCHGs, and mCHHs) were evaluated using the χ2 test; (***) P < 0.001, (**) P < 0.01.
To determine whether DNA methylation was associated with the diversity of specialized metabolites, we identified 13,425 mQTGs in Arabidopsis using our previous study (Shirai et al. 2017). We focused on 415 of 13,425 mQTGs that had CVs > 2.978 (top 5%). Although all of genes with the 5% highest CVs include a high rate of TE genes (370/1209 = 31%), mQTGs with the 5% highest CVs include a low rate of TE genes (22/415 = 5.30%). Moreover, mQTGs with the 1% highest CVs associated with mCGs do not include any TE genes. Therefore, it is unlikely that TE genes may be a main determinant for specialized metabolism. Of the genes associated with mCGs, mCHGs, mCHHs, and SNPs, 124, 106, 102, and 319 genes, respectively, were mQTGs with the 5% highest CVs (mCGs: 124/6690 = 1.85%, mCHGs: 106/3319 = 3.19%, mCHHs: 102/3463 = 2.95%, SNPs: 319/21,924 = 1.46%) (Fig. 3C). The proportions of mQTGs having the 5% highest CVs among the genes associated with all SMP types were significantly greater than that of the SNPs (mCGs: P = 6.49 × 10−3, mCHGs: P = 6.00 × 10−17, mCHHs: P = 2.40 × 10−13; χ2 test) (Fig. 3C). Thus, SMPs may be correlated with mQTGs having large expressional variations. These tendencies are not dependent on the locations of the SMPs (Supplemental Figs. S8, S9). Similar trends were also shown for genes associated with either only SMPs or SNPs. Out of 262, 195, 188, and 13,904 genes associated with mCGs, mCHGs, mCHHs, and SNPs, 18, 18, 14, and 171 genes, respectively, were mQTGs having the 5% highest CVs (mCGs: 18/262 = 6.87%, mCHGs: 18/195 = 9.23%, mCHHs: 14/188 = 7.45%, and SNPs: 171/13,904 = 1.23%) (Fig. 3D). The proportions of mQTGs having the 5% highest CVs among genes associated with all the SMP types were significantly greater than that of the SNPs (mCGs: P = 1.20 × 10−16, mCHGs: P = 3.78 × 10−24, mCHHs: P = 1.04 × 10−14; chi-square test) (Fig. 3D).
To validate the associations between DNA methylation and mQTGs with high CVs, we prepared another type of data set. We focused on only 28 characteristic accessions selected from a microarray profile of 21,957 genes in 75 A. thaliana accessions (Methods; Supplemental Tables S10, S11; Shirai et al. 2017). This data set tended to have greater expressional variation than random selections (Supplemental Fig. S10). For the selected 28 accessions, we obtained 1,911,086 SMPs (mCGs: 638,352, mCHGs: 383,377, and mCHHs: 889,357) and 1,269,078 SNPs. The distributions of the SMPs were quite similar to those in the 620 accessions (Supplemental Fig. S11). We focused on SMPs and SNPs located on gene bodies (exon and intron) and 1-kb regions on either side of the genes (Supplemental Table S12). As in the former analysis, we performed an association analysis between the SMPs (or the SNPs) and gene expression by the same method (Methods; Supplemental Tables S13–S16). Out of 854, 312, 541, and 5881 genes associated with mCGs, mCHGs, mCHHs, and SNPs, 103, 48, 78, and 323 genes, respectively, were mQTGs having the 5% highest CVs (mCG: 103/854 = 12.06%, mCHGs: 48/312 = 15.38%, mCHHs: 78/541 = 14.42%, and SNPs: 323/5881 = 5.49%) (Fig. 3E). The proportions of mQTGs having the 5% highest CVs among the genes associated with all SMP types were significantly greater than that of the SNPs (mCGs: P = 3.59 × 10−17, mCHGs: P = 1.73 × 10−14, and mCHHs: P = 8.08 × 10−20; χ2 test) (Fig. 3E). Similar trends were also shown in the analysis of the genes associated with either only SMPs or SNPs. Out of 444, 150, 274, and 5216 genes associated with mCGs, mCHGs, mCHHs, and SNPs, 49, 16, 32, and 233 genes, respectively, were mQTGs having the 5% highest CVs (mCG: 49/444 = 11.04%, mCHGs: 16/150 = 10.67%, mCHHs: 32/274 = 11.68%, and SNPs: 233/5216 = 4.47%) (Fig. 3F). The proportions of mQTGs having the 5% highest CVs among the genes associated with all SMP types were significantly greater than that of the SNPs (mCGs: P = 2.08 × 10−11, mCHGs: P = 2.37 × 10−4, and mCHHs: P = 7.53 × 10−9; χ2 test) (Fig. 3F). In summary, association analyses of the global 620 accessions and the 28 characteristic accessions indicated that all the SMP types tended to be associated with mQTGs having large expressional variations.
Selective sweep of DNA methylation
Specialized metabolite diversity contributes to local adaptation. Therefore, the DNA methylation associated with specialized metabolites may have been positively selected among the 620 A. thaliana accessions. We first focused on three kinds of DNA methylation associated with mQTGs having the 5% highest CVs (1892 mCGs, 1761 mCHGs, and 1949 mCHHs) and evaluated the selection pressure using Tajima's D test. If a SMP had undergone directional selection that indicated either positive or purifying selection, then accessions harboring the SMP would have lower Tajima's D values than accessions lacking the SMP in the genomic region. Therefore, we compared Tajima's D values in a 10-kb region (5-kb upstream of and 5-kb downstream from the chosen DNA methylation site) between accessions with and without DNA methylation (Supplemental Tables S17, S18). Accessions with either mCGs located on any regions, mCHGs located on any regions, or mCHH located on only promoter region tended to have significantly lower Tajima's D values than accessions lacking the DNA methylation (P < 1.00 × 10−3; Wilcoxon rank-sum test) (Supplemental Fig. S12A; Supplemental Table S19). This tendency became much stronger when mQTGs having the 1% highest CVs (330 mCGs, 214 mCHGs, and 188 mCHHs) were analyzed using the same procedure (P < 1.00 × 10−3; Wilcoxon rank-sum test) (Fig 4A; Supplemental Table S19). We calculated the ratios of median of Tajima's D in accessions with either mCGs or mCHGs to the median of Tajima's D in accessions lacking the DNA methylation in either the 1% or 5% highest CVs. (Supplemental Table S20). The ratios in mQTGs having the 1% highest CVs are higher than those in mQTGs having the 5% highest CVs (the 1% highest CVs, mCGs: 1.55–3.03, mCHGs: 1.33–1.96; the 5% highest CVs, mCGs: 1.50–1.87, mCHGs: 1.13–1.41). Thus, mCGs and mCHGs may be subjected to directional selection. However, most of the mCHHs are unlikely to be controlled by directional selection. The differences of Tajima's D might be caused by population structure. However, accessions with DNA methylation essentially had the same subpopulation classified by geographic distributions in comparison with those without DNA methylation (Supplemental Fig. S13). Therefore, it is unlikely that the differences of population structure significantly affect Tajima's D between accessions with and without DNA methylation. In addition, Tajima's D shows significantly different trends among the locations (promoter, exon, intron, downstream) or types of SMPs (mCG, mCHG, mCHH) (Fig 4A). Furthermore, we inferred Tajima's D values under a neutral process based on the expected demographic history of 620 accessions (Supplemental Methods; Supplemental Fig. S14; Supplemental Tables S21, S22; Hudson 2002; Ossowski et al. 2010; Excoffier and Foll 2011). The inferred Tajima's D values are significantly higher than those in any types/locations of SMPs (FDR < 0.05; Wilcoxon rank-sum test) (Supplemental Fig. S15; Supplemental Table S23). These results suggest that the time-course change of population size is also not a main determinant for Tajima's D between accessions with and without DNA methylation.
Figure 4.

Selective sweep of DNA methylation associated with mQTG expression. (A) Tajima's D values near SMPs associated with mQTG expression. The x-axis represents SMP or SNP locations, and the y-axis represents Tajima's D values. The boxes represent Tajima's D values of accessions with no DNA methylation or accessions with DNA methylation at SMP sites associated with mQTGs having high CVs (top 1%). (B) The differences in nucleotide diversity (π) near SMPs associated with mQTG expression. The x-axis represents the SMP or SNP locations, whereas the y-axis represents the differences in π. Gray boxes represent the π differences of neutral SNPs. The other boxes represent the π differences of each type of SMP associated with mQTGs having high CVs (top 1%). The differences between the neutral SMPs and the SNPs associated with mQTGs were analyzed at each location using a Wilcoxon rank-sum test. In each box plot, the box represents the 25%–75% range, the middle line represents the median, the dotted line represents the 1%–99% range, and the outer circles represent outliers. The differences between the two groups were determined at each location using a Wilcoxon rank-sum test; (***) P < 0.001, (**) P < 0.01.
However, SNP density in genomic regions may affect Tajima's D extensively. We then recalculated Tajima's D values in 100 SNPs windows (upstream 50 SNPs and downstream 50 SNPs of the chosen DNA methylation site) instead of 10-kb windows and performed the same analyses (Supplemental Methods). Consequently, only accessions with mCGs located on promoter regions tended to have significantly lower Tajima's D values than accessions lacking the DNA methylation (P < 1.00 × 10−2; Wilcoxon rank-sum test) (Supplemental Fig. S16) as shown in the 10-kb window analysis (Fig. 4). This result indicates that mCGs located on promoter regions have been controlled by strong directional selection in comparison to either the other types/locations of SMPs. We also inferred Tajima's D values in 100 SNPs window analysis under a neutral process based on the expected demographic history. The inferred Tajima's D values are significantly higher than those in any types/locations of SMPs (FDR < 0.05; Wilcoxon rank-sum test) (Supplemental Fig. S17; Supplemental Table S24). Thus, these results support the outputs of the10-kb window analysis.
We evaluated the selection pressure of DNA methylation by analyzing nucleotide diversity (π). The π near DNA methylation sites is decreased by a selective sweep when genes controlled by DNA methylation are under a strong positive selection pressure (Maynared Smith and Haigh 1974). To assess the selective sweeps near the DNA methylation sites in 620 A. thaliana accessions, we calculated differences in π values between accessions with and without DNA methylation at sites associated with mQTGs. As a negative control, we prepared neutral SNPs and calculated identified differences in π values between accessions with major alleles and those with minor alleles (Supplemental Table S25). We first focused on 1892 mCGs, 1761 mCHGs, and 1949 mCHHs associated with mQTGs having the 5% highest CVs. However, differences in π values were not identified between these DNA methylation sites and neutral SNPs (Supplemental Fig. S12B; Supplemental Table S26). We then focused on 330 mCGs, 214 mCHGs, and 188 mCHHs associated with mQTGs having the 1% highest CVs. Only mCGs located on promoter regions had larger differences in π than neutral SNPs (P = 7.88 × 10−3; Wilcoxon rank-sum test) (Fig. 4B; Supplemental Table S26). In addition, accessions with mCGs located on promoter regions tend to have lower π values than neutral SNPs (Supplemental Fig. S18). These results indicate that the mCGs on promoter regions had undergone positive selective sweeps in the mQTGs having highly varied expression (1% highest CVs) among the 620 A. thaliana accessions. Although differences in π were not identified between mCHGs and neutral SNPs, mCHGs were subjected to directional selection as determined by Tajima's D analysis. This suggests that mCHGs were subjected to purifying selection rather than positive selection. For mCHHs, the Tajima's D and π analyses suggested that mCHHs have not undergone strong selection.
To further support the selective sweep to mCGs on promoter regions, we calculated Z-transformed FST (ZFST) between accessions with and without selected mCGs at mCG sites associated with mQTGs (Supplemental Methods; Axelsson et al. 2013). As a result, ZFST values tend to be higher than those of the other genomic regions (Supplemental Fig. S19). These results strongly support that the mCGs on promoter regions had undergone selective sweeps.
Discussion
Using genome, methylome, and transcriptome data from 620 accessions of A. thaliana, we identified 13,462 cis-regulatory SMPs (6690 mCGs, 3319 mCHGs, and 3453 mCHHs) and 21,924 cis-regulatory SNPs that were associated with the expression of 24,030 genes. Thus, SNPs tended to have much greater cis-regulatory effects than SMPs recognized by mCG, mCHG, and mCHH. However, these mutations only led to a low level of diversity in gene expression because most of the genes did not have large expressional variations among the 620 accessions (Fig. 2A–D). Therefore, we examined which mutations led to a high level of diversity in gene expression. The presence of the three kinds of SMPs was largely associated with a higher diversity in gene expression than the presence of SNPs. Furthermore, specialized metabolites tended to be regulated by genes, leading to a high level of diversity in gene expression among accessions (Fig. 3A–E). This trend was validated by our genome, methylome, and transcriptome data from 28 accessions. Thus, the present analysis proposes that DNA methylation causes a large variation in gene expression, which contributes to the diversity of specialized metabolites in A. thaliana.
Our results suggest that DNA methylation contributes to local adaptation by promoting specialized metabolite diversity in A. thaliana. Recent studies have also suggested a relationship between DNA methylation and local adaptation in plants (Dubin et al. 2015; Gardiner et al. 2018; He et al. 2018; Schmid et al. 2018). However, there are few reports of naturally selected DNA methylation. Therefore, we examined the positive selective sweep of DNA methylation using π, Tajima's D, and ZFST values. Our results showed that naturally selected mCGs on promoter regions are key mutations resulting in the expressional diversity associated with specialized metabolites during plant evolution.
As an additional example of selective sweep to mCGs, we focused on a mCG located on the promoter region of AT1G14800 (Chromosome 1: 5,098,964). The mCG is associated with mQTGs having the 1% highest CVs. The genomic region near the mCG site has the 1% highest difference of π, the 1% highest ZFST, and negative Tajima's D. For the mCG, we applied SweeD, a software for detecting selective sweep from site frequency spectrum (Supplemental Methods; Supplemental Fig. S20; Pavlidis et al. 2013). As a result, the composite likelihood ratio (CLR) near the mCG site is significantly more elevated than the neighboring genomic regions (Supplemental Fig. S21; Supplemental Table S27). On the other hand, the elevation of CLR near the mCG site is not observed in accessions lacking the mCG (Supplemental Fig. S21; Supplemental Table S28). Thus, we show a clear evidence for selective sweep to mCGs on promoter regions.
We identified the evidence of a positive selective sweep for only mCGs, not for mCHGs and mCHHs. This difference in selection pressure may be caused by the difference in mCG, mCHG, and mCHH maintenance. The CG methylation patterns are maintained by METHYLTRANSFERASE 1 during DNA replication (Iwasaki and Paszkowski 2014; Kawashima and Berger 2014; Zhang et al. 2018), and the mutated mCGs are transmitted to the next generation (Iwasaki and Paszkowski 2014; Kawashima and Berger 2014). Therefore, the variation in mCG contributes to adaptive evolution, similarly to SNPs, in plants. However, the CHG and CHH methylation patterns are not transmitted through generations because of their maintenance mechanisms (Iwasaki and Paszkowski 2014; Kawashima and Berger 2014; Zhang et al. 2018). In addition, the CHG and CHH methylation patterns are highly dependent on trans-regulation (Greaves et al. 2016; Zhang et al. 2016). Therefore, positive selection has limited direct effects on mCHGs and mCHHs.
We detected a positive selective sweep of mCGs in the promoter region. The DNA methylation of promoter regions strongly suppresses gene expression in plants (Zhang et al. 2006, 2018; Yang et al. 2015) by inhibiting the binding of transcription factors (Kawakatsu et al. 2016; O'Malley et al. 2016). Here, mCGs of promoters suppressed the expression of specialized metabolite genes (Supplemental Fig. S22; Supplemental Table S29). Thus, suppressing gene expression through CG methylation induces specialized metabolite diversity in some of A. thaliana accessions, which may contribute to their adaptability.
The mCGs in exon and intron regions are also naturally selected but are not subject to selective sweep. mCGs in exon and intron regions significantly affect splicing patterns (Zhang et al. 2006; Kawakatsu et al. 2016). In addition, mCGs in exon region are likely to prohibit TE insertion (Regulski et al. 2013). Thus, mCGs in exon and intron regions contribute to functional stability of expressed genes. Therefore, mCGs in exon and intron regions may be maintained by purifying selection rather than selective sweep.
Although mCHGs are rarely subjected to a selective sweep, they appear to be associated with the highly diverse expression of specialized metabolic genes that are under strong purifying selection. This selection may be related to the characteristic functions and maintenance mechanisms of mCHGs. The mCHGs are maintained by feedback loops with the dimethylation of lysine 9 in histone H3 (H3K9me2) (Iwasaki and Paszkowski 2014; Kawashima and Berger 2014; Zhang et al. 2018). H3K9me2, like DNA methylation, is also associated with the repression of gene expression and the silencing of TEs (Zhang et al. 2018). Moreover, mCHGs and H3K9me2 are associated with imprinted paternally expressed genes (Klosinska et al. 2016; Moreno-Romero et al. 2019), and they both tend to repress the maternal alleles of these genes (Klosinska et al. 2016; Moreno-Romero et al. 2019). Some of the imprinted genes regulate specialized metabolism (Roy 2016). Thus, the functional importance of imprinting may be controlled by strong purifying selection through mCHGs.
The Tajima's D analysis also indicated that only mCHHs located in introns were affected by directional selection. Some introns of plant genes contain several TEs (Zhang et al. 2018). These regions are highly methylated to avoid alternative splicing (Ong-Abdullah et al. 2015). The directional selection toward mCHHs in introns may be correlated with these expressional regulations. However, selection pressure toward mCHHs is clearly weak compared to those of mCG and mCHG. This weak selection may be caused by the instability of mCHHs. Methylation levels at CHH sites change greatly throughout a plant's life (Bouyer et al. 2017; Kawakatsu et al. 2017; Kawakatsu and Ecker 2019). In addition, mCHHs have no mechanisms for stable maintenance compared with mCGs and mCHGs. Thus, mCHHs may not play important roles in plant evolution.
The mCGs located in promoters are subjected to positive selective sweeps. However, the tendency is mild compared to that of SNPs (Supplemental Fig. S23; Supplemental Table S26). Thus, the mild selective sweep may be correlated with the mutational rates at the mCG sites. Methylated cytosines frequently mutate to thymines through deamination. This causes the high mutational rate of methylated cytosine (Duncan and Miller 1980; Ossowski et al. 2010; Gardiner et al. 2018). In fact, in the present study, methylated cytosine tended to mutate more to thymine compared to unmethylated cytosine (Supplemental Fig. S24; Supplemental Table S30). Thus, the instability of methylated sites may result in their limited contribution to adaptive evolution compared with SNPs.
In summary, the present study revealed that all types of DNA methylation were associated with genes, leading to a high level of gene expressional diversity among 620 A. thaliana accessions. The genes associated with DNA methylation were frequently related to specialized metabolite diversity. Although all types of DNA methylation are similarly associated with gene expression, the selection pressure is quite different among mCGs, mCHGs, and mCHHs. To examine evidence of naturally selected DNA methylation in A. thaliana accessions, we focused on previously identified mQTGs for 1335 specialized metabolites (Shirai et al. 2017). In genes related to specialized metabolite diversity, the mCGs and mCHGs tended to be affected by strong directional selection. In particular, mCGs located in promoter regions tended to be associated with positive selective sweeps. Thus, the present study shows that mCGs contribute to adaptive evolution in plants.
Methods
SNPs and SMPs in 620 A. thaliana accessions
We obtained SNPs, SMPs, and expression profiles of 620 accessions of A. thaliana from the 1001 Genomes Consortium and Kawakatsu et al. (The 1001 Genomes Consortium 2016; Kawakatsu et al. 2016). We removed SNPs and SMPs called in less than half of the accessions. The remaining 1,397,934 SNPs and 4,448,076 SMPs (mCGs: 1,653,070, mCHGs: 814,938, and mCHHs: 1,980,068) with allele frequencies > 5% were used in further analyses. The locations of the SNPs and SMPs were annotated using the SnpEff program (Cingolani et al. 2012). The expression data of 24,030 genes were also collected from Kawakatsu et al. (2016) (NCBI Gene Expression Omnibus [GEO; https://www.ncbi.nlm.nih.gov/geo/]) under accession number GSE80744 (Supplemental Table S2).
SNPs and SMPs in 28 accessions
We used our previous microarray expression data for 26,995 genes in 75 A. thaliana accessions (GEO, GSE89805) (Shirai et al. 2017). From the 26,995 genes, we selected 21,957 genes that had no SNPs in the microarray probe sites (Supplemental Table S11). Using the expression patterns, we performed a clustering analysis of the 75 accessions using the ward.D2 method in R package “hclust” (Supplemental Fig. S25; R Core Team 2015). On the basis of the clustering results, we manually chose 28 accessions with characteristic expression patterns (Supplemental Fig. S25; Supplemental Table S10). We extracted genomic DNA from two-wk-old seedlings of the 28 accessions. The extracted DNAs were fragmented using a Covaris sonicator (Covaris). Genomic libraries were constructed using an Illumina TruSeq Sample Preparation kit following the manufacturer's instructions. The libraries were treated with sodium bisulfite using an EpiTect Bisulfite Kit (Qiagen) following the manufacturer's instructions. The bisulfite-converted libraries were PCR-amplified using Taq hot-start polymerase (TaKaRa Bio). The library quality was monitored using an Agilent 2100 Bioanalyzer. We performed paired-end sequencing (2 × 100 bp) of the bisulfite-treated and nontreated libraries on an Illumina HiSeq 2000 platform (see Data access). The reads were adapter-trimmed and quality-filtered using Trimmomatic software (Bolger et al. 2014). The reads from nontreated DNA were mapped to the TAIR10 genome (https://www.arabidopsis.org) using BWA software (Li and Durbin 2009). PCR duplicates were removed using the Picard package (version 1.07) (http://broadinstitute.github.io/picard/). SNP calling and quality control were performed using GATK 3.4 software (McKenna et al. 2010). We removed the sites with a missing rate > 0.5. The remaining SNPs with allele frequencies > 5% were used for analyses. The reads from bisulfite-treated DNA were mapped and PCR duplicates removed using Bismark v0.14.5 (Krueger and Andrews 2011). The methylation levels were detected using the same tool. SMP sites were detected by binomial tests (false discovery rate [FDR] < 0.05) following the methods of Lister et al. (2008). SMPs called in less than half of the accessions were removed. The remaining SMPs with allele frequencies > 5% were used for analyses.
Metabolome quantitative trait genes
We performed a metabolite–transcriptome correlation analysis to detect mQTGs having low false positive rates (Shirai et al. 2017). This analysis enabled us to compare the effects on gene expression between SNPs and SMPs. Our previous study detected mQTGs by applying this method to 1335 specialized metabolites. From the candidate mQTGs, we selected 13,425 genes having significant correlations with more than five metabolites as mQTGs for the present study (Supplemental Table S31).
Association analysis
To examine the relationships between SMPs (or SNPs) and targeted gene expression, we focused on SMPs (or SNPs) located in the targeted gene body (exon and intron) and 1 kb on both sides of the targeted gene. To examine the association between the SMPs (or SNPs) and the gene expression levels, we applied a linear mixed model using the R package “lme4′(R Core Team 2015). In the model, the predictive variables are SMPs (or SNPs). The response variables are expression levels. First, we performed an association analysis between SMPs (or SNPs) and expression data from the RNA-seq of 620 A. thaliana accessions. To determine population structure, we used an ADMIXTURE analysis of the 620 accessions (Alexander et al. 2009). As a result, the 620 accessions were divided into 13 groups on the basis of their ancestry (Supplemental Fig. S26; Supplemental Table S1). The differences among the groups were used as the random effects in the linear mixed model. Then, we performed an association analysis between each SMP (or SNP) and expression using the microarray profile of each gene in the 28 accessions. The differences among accessions were used as the random effects when considering the population structure in the linear mixed model. For each association analysis, we used FDR < 0.05 as the threshold. We calculated the false positive rates of the models. For the calculation, we prepared negative control data that included 5000 sets of randomly selected genes and randomly selected SMPs (or SNPs). The numbers of the selected SMPs (or SNPs) at each gene were determined by the average numbers of SMPs (or SNPs) at the gene in each data set (Supplemental Table S32). We applied each model to the negative control data. From the results, we counted the number of false positive associations and true negative associations. The false positive rates were calculated using the following equation: False positive rate = False positives/(False positives + True negatives). For each model, the false positive rate of the analysis was less than 0.1 (Supplemental Fig. S27).
Detection of selective sweeps
To detect selection pressure for DNA methylation related to specialized metabolite diversity, we calculated Tajima's D values (Tajima 1989). At each SMP site associated with the expression of an mQTG, we divided the 620 accessions into two groups, with and without DNA methylation at the SMP site. In each group, we calculated the Tajima's D value in a 10-kb region (5-kb upstream of and 5-kb downstream from the SMP site) using the VCFtools program (Danecek et al. 2011). The distributions of Tajima's D values were compared using a Wilcoxon rank-sum test.
To detect selective sweeps for DNA methylation, we calculated π in accordance with our previously published methods (Shirai et al. 2017). At each SMP site associated with the expression of an mQTG, we divided the 620 accessions into two groups, with and without DNA methylation at the SMP site. In each group, we calculated π in a 10-kb region (5-kb upstream of and 5-kb downstream from the SMP site) using the VCFtools program (Danecek et al. 2011). The differences in π were calculated using the following equation: The differences in π = |log10 (the π of the accessions with DNA methylation/the π of accessions without DNA methylation)|. Similarly, we analyzed π at neutral SNP sites. We randomly selected 4500 SNP sites from the 1,397,934 SNP sites (Supplemental Table S25). We excluded SNPs with strongly biased frequencies (allele frequency > 0.8 or allele frequency < 0.2) that were potentially affected by selection pressure (Supplemental Fig. S28). We defined the remaining SNPs as neutral SNPs. At each neutral SNP site, we divided the 620 accessions into two groups on the basis of the allele (minor or major allele). In each allele group, we calculated π in the 10-kb region. The difference in π was calculated using the following equation: The differences in π = |log10 (the π of the accessions with minor alleles/the π of accessions with major alleles)|. The distributions of the differences in π were compared using a Wilcoxon rank-sum test.
Data access
The raw sequencing data generated in this study have been submitted to the DNA Data Bank of Japan (DDBJ; https://www.ddbj.nig.ac.jp) under accession number DRA003230.
Supplementary Material
Acknowledgments
We thank Lesley Benyon, PhD, from the Edanz Group (https://en-author-services.edanzgroup.com/ac) for editing a draft of this manuscript. We also thank the National Institute of Genetics of the Research Organization of Information and Systems for providing excellent supercomputer services. This study was supported by Grants-in-Aid for Scientific Research (20H03317, 20H05905, 20H05906, 25710017, 18KK0176, 19H05348, 18H02420, 19K22313 to K.H.) and Asahi Glass Foundation (to K.H.).
Author contributions: R.N. prepared samples and performed Illumina library preparation. M.S. and Y.S. performed Illumina sequencing. M.P.S. and K.S. collected genome and methylome data. K.S. analyzed the data. K.S. and K.H. wrote the manuscript. The research was directed by K.S. and K.H.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at https://www.genome.org/cgi/doi/10.1101/gr.271726.120.
Competing interest statement
The authors declare no competing interests.
References
- The 1001 Genomes Consortium. 2016. 1,135 genomes reveal the global pattern of polymorphism in Arabidopsis thaliana. Cell 166: 481–491. 10.1016/j.cell.2016.05.063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander DH, Novembre J, Lange K. 2009. Fast model-based estimation of ancestry in unrelated individuals. Genome Res 19: 1655–1664. 10.1101/gr.094052.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alseekh S, Tohge T, Wendenberg R, Scossa F, Omranian N, Li J, Kleessen S, Giavalisco P, Pleban T, Mueller-Roeber B, et al. 2015. Identification and mode of inheritance of quantitative trait loci for secondary metabolite abundance in tomato. Plant Cell 27: 485–512. 10.1105/tpc.114.132266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axelsson E, Ratnakumar A, Arendt ML, Maqbool K, Webster MT, Perloski M, Liberg O, Arnemo JM, Hedhammar Å, Lindblad-Toh K. 2013. The genomic signature of dog domestication reveals adaptation to a starch-rich diet. Nature 495: 360–364. 10.1038/nature11837 [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouyer D, Kramdi A, Kassam M, Heese M, Schnittger A, Roudier F, Colot V. 2017. DNA methylation dynamics during early plant life. Genome Biol 18: 179. 10.1186/s13059-017-1313-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carreno-Quintero N, Bouwmeester HJ, Keurentjes JJB. 2013. Genetic analysis of metabolome-phenotype interactions: from model to crop species. Trends Genet 29: 41–50. 10.1016/j.tig.2012.09.006 [DOI] [PubMed] [Google Scholar]
- Cingolani P, Platts A, Wang LL, Coon M, Nguyen T, Wang L, Land SJ, Lu X, Ruden DM. 2012. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 6: 80–92. 10.4161/fly.19695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, et al. 2011. The variant call format and VCFtools. Bioinformatics 27: 2156–2158. 10.1093/bioinformatics/btr330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubin MJ, Zhang P, Meng D, Remigereau M-S, Osborne EJ, Paolo Casale F, Drewe P, Kahles A, Jean G, Vilhjálmsson B, et al. 2015. DNA methylation in Arabidopsis has a genetic basis and shows evidence of local adaptation. eLife 4: e05255. 10.7554/eLife.05255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan BK, Miller JH. 1980. Mutagenic deamination of cytosine residues in DNA. Nature 287: 560–561. 10.1038/287560a0 [DOI] [PubMed] [Google Scholar]
- Excoffier L, Foll M. 2011. fastsimcoal: a continuous-time coalescent simulator of genomic diversity under arbitrarily complex evolutionary scenarios. Bioinformatics 27: 1332–1334. 10.1093/bioinformatics/btr124 [DOI] [PubMed] [Google Scholar]
- Feng S, Jacobsen SE, Reik W. 2010. Epigenetic reprogramming in plant and animal development. Science 330: 622–627. 10.1126/science.1190614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner LJ, Joynson R, Omony J, Rusholme-Pilcher R, Olohan L, Lang D, Bai C, Hawkesford M, Salt D, Spannagl M, et al. 2018. Hidden variation in polyploid wheat drives local adaptation. Genome Res 28: 1319–1332. 10.1101/gr.233551.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaves IK, Eichten SR, Groszmann M, Wang A, Ying H, Peacock WJ, Dennis ES. 2016. Twenty-four–nucleotide siRNAs produce heritable trans-chromosomal methylation in F1 Arabidopsis hybrids. Proc Natl Acad Sci 113: E6895–E6902. 10.1073/pnas.1613623113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Wu W, Zinta G, Yang L, Wang D, Liu R, Zhang H, Zheng Z, Huang H, Zhang Q, et al. 2018. A naturally occurring epiallele associates with leaf senescence and local climate adaptation in Arabidopsis accessions. Nat Commun 9: 460. 10.1038/s41467-018-02839-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson RR. 2002. Generating samples under a Wright–Fisher neutral model of genetic variation. Bioinformatics 18: 337–338. 10.1093/bioinformatics/18.2.337 [DOI] [PubMed] [Google Scholar]
- Iwasaki M, Paszkowski J. 2014. Epigenetic memory in plants. EMBO J 33: 1987–1998. 10.15252/embj.201488883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakatsu T, Ecker JR. 2019. Diversity and dynamics of DNA methylation: epigenomic resources and tools for crop breeding. Breed Sci 69: 191–204. 10.1270/jsbbs.19005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakatsu T, Huang SC, Jupe F, Sasaki E, Schmitz RJJ, Urich MAA, Castanon R, Nery JRR, Barragan C, He Y, et al. 2016. Epigenomic diversity in a global collection of Arabidopsis thaliana accessions. Cell 166: 492–505. 10.1016/j.cell.2016.06.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakatsu T, Nery JR, Castanon R, Ecker JR. 2017. Dynamic DNA methylation reconfiguration during seed development and germination. Genome Biol 18: 171. 10.1186/s13059-017-1251-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima T, Berger F. 2014. Epigenetic reprogramming in plant sexual reproduction. Nat Rev Genet 15: 613–624. 10.1038/nrg3685 [DOI] [PubMed] [Google Scholar]
- Kerwin R, Feusier J, Corwin J, Rubin M, Lin C, Muok A, Larson B, Li B, Joseph B, Francisco M, et al. 2015. Natural genetic variation in Arabidopsis thaliana defense metabolism genes modulates field fitness. eLife 4: e05604. 10.7554/eLife.05604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klosinska M, Picard CL, Gehring M. 2016. Conserved imprinting associated with unique epigenetic signatures in the Arabidopsis genus. Nat Plants 2: 16145. 10.1038/nplants.2016.145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooke R, Morgado L, Becker F, Van Eekelen H, Hazarika R, Zheng Q, De Vos RCH, Johannes F, Keurentjes JJB. 2019. Epigenetic mapping of the Arabidopsis metabolome reveals mediators of the epigenotype-phenotype map. Genome Res 29: 96–106. 10.1101/gr.232371.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger F, Andrews SR. 2011. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 27: 1571–1572. 10.1093/bioinformatics/btr167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25: 1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, O'Malley RC, Tonti-Filippini J, Gregory BD, Berry CC, Millar AH, Ecker JR. 2008. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell 133: 523–536. 10.1016/j.cell.2008.03.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda F, Nakabayashi R, Yang Z, Okazaki Y, Yonemaru JI, Ebana K, Yano M, Saito K. 2015. Metabolome-genome-wide association study dissects genetic architecture for generating natural variation in rice secondary metabolism. Plant J 81: 13–23. 10.1111/tpj.12681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke MA, Kanno T, Matzke AJM. 2015. RNA-directed DNA methylation: the evolution of a complex epigenetic pathway in flowering plants. Annu Rev Plant Biol 66: 243–267. 10.1146/annurev-arplant-043014-114633 [DOI] [PubMed] [Google Scholar]
- Maynared Smith J, Haigh J. 1974. The hitch-hiking effect of a favourable gene. Genet Res 23: 23–35. 10.1017/S0016672300014634 [DOI] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20: 1297–1303. 10.1101/gr.107524.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng D, Dubin M, Zhang P, Osborne EJ, Stegle O, Clark RM, Nordborg M. 2016. Limited contribution of DNA methylation variation to expression regulation in Arabidopsis thaliana. PLoS Genet 12: e1006141. 10.1371/journal.pgen.1006141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Romero J, Del Toro-De León G, Yadav VK, Santos-González J, Köhler C. 2019. Epigenetic signatures associated with imprinted paternally expressed genes in the Arabidopsis endosperm. Genome Biol 20: 41. 10.1186/s13059-019-1652-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Malley RC, Huang SSC, Song L, Lewsey MG, Bartlett A, Nery JR, Galli M, Gallavotti A, Ecker JR. 2016. Cistrome and epicistrome features shape the regulatory DNA landscape. Cell 165: 1280–1292. 10.1016/j.cell.2016.04.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong-Abdullah M, Ordway JM, Jiang N, Ooi SE, Kok SY, Sarpan N, Azimi N, Hashim AT, Ishak Z, Rosli SK, et al. 2015. Loss of Karma transposon methylation underlies the mantled somaclonal variant of oil palm. Nature 525: 533–537. 10.1038/nature15365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossowski S, Schneeberger K, Lucas-Lledó JI, Warthmann N, Clark RM, Shaw RG, Weigel D, Lynch M. 2010. The rate and molecular spectrum of spontaneous mutations in Arabidopsis thaliana. Science 327: 92–94. 10.1126/science.1180677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlidis P, Živković D, Stamatakis A, Alachiotis N. 2013. SweeD: likelihood-based detection of selective sweeps in thousands of genomes. Mol Biol Evol 30: 2224–2234. 10.1093/molbev/mst112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichersky E, Lewinsohn E. 2011. Convergent evolution in plant specialized metabolism. Annu Rev Plant Biol 62: 549–566. 10.1146/annurev-arplant-042110-103814 [DOI] [PubMed] [Google Scholar]
- Pichersky E, Raguso RA. 2018. Why do plants produce so many terpenoid compounds? New Phytol 220: 692–702. 10.1111/nph.14178 [DOI] [PubMed] [Google Scholar]
- R Core Team. 2015. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna. https://www.R-project.org/. [Google Scholar]
- Regulski M, Lu Z, Kendall J, Donoghue MTA, Reinders J, Llaca V, Deschamps S, Smith A, Levy D, McCombie WR, et al. 2013. The maize methylome influences mRNA splice sites and reveals widespread paramutation-like switches guided by small RNA. Genome Res 23: 1651–1662. 10.1101/gr.153510.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S. 2016. Function of MYB domain transcription factors in abiotic stress and epigenetic control of stress response in plant genome. Plant Signal Behav 11: e1117723. 10.1080/15592324.2015.1117723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid MW, Heichinger C, Coman Schmid D, Guthörl D, Gagliardini V, Bruggmann R, Aluri S, Aquino C, Schmid B, Turnbull LA, et al. 2018. Contribution of epigenetic variation to adaptation in Arabidopsis. Nat Commun 9: 4446. 10.1038/s41467-018-06932-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirai K, Matsuda F, Nakabayashi R, Okamoto M, Tanaka M, Fujimoto A, Shimizu M, Shinozaki K, Seki M, Saito K, et al. 2017. A highly specific genome-wide association study integrated with transcriptome data reveals the contribution of copy number variations to specialized metabolites in Arabidopsis thaliana accessions. Mol Biol Evol 34: 3111–3122. 10.1093/molbev/msx234 [DOI] [PubMed] [Google Scholar]
- Tajima F. 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123: 585–595. 10.1093/genetics/123.3.585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takuno S, Ran JH, Gaut BS. 2016. Evolutionary patterns of genic DNA methylation vary across land plants. Nat Plants 2: 15222. 10.1038/nplants.2015.222 [DOI] [PubMed] [Google Scholar]
- Tohge T, Perez de Souza L, Fernie AR. 2018. On the natural diversity of phenylacylated-flavonoid and their in planta function under conditions of stress. Phytochem Rev 17: 279–290. 10.1007/s11101-017-9531-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D. 2012. Natural variation in Arabidopsis: from molecular genetics to ecological genomics. Plant Physiol 158: 2–22. 10.1104/pp.111.189845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams BP, Gehring M. 2017. Stable transgenerational epigenetic inheritance requires a DNA methylation-sensing circuit. Nat Commun 8: 2124. 10.1038/s41467-017-02219-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wink M. 2008. Plant secondary metabolism: diversity, function and its evolution. Nat Prod Commun 3: 1205–1216. 10.1177/1934578X0800300801 [DOI] [Google Scholar]
- Yang H, Chang F, You C, Cui J, Zhu G, Wang L, Zheng Y, Qi J, Ma H. 2015. Whole-genome DNA methylation patterns and complex associations with gene structure and expression during flower development in Arabidopsis. Plant J 81: 268–281. 10.1111/tpj.12726 [DOI] [PubMed] [Google Scholar]
- Zemach A, McDaniel IE, Silva P, Zilberman D. 2010. Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science 328: 916–919. 10.1126/science.1186366 [DOI] [PubMed] [Google Scholar]
- Zhang X, Yazaki J, Sundaresan A, Cokus S, Chan SWL, Chen H, Henderson IR, Shinn P, Pellegrini M, Jacobsen SE, et al. 2006. Genome-wide high-resolution mapping and functional analysis of DNA methylation in Arabidopsis. Cell 126: 1189–1201. 10.1016/j.cell.2006.08.003 [DOI] [PubMed] [Google Scholar]
- Zhang Q, Wang D, Lang Z, He L, Yang L, Zeng L, Li Y, Zhao C, Huang H, Zhang H, et al. 2016. Methylation interactions in Arabidopsis hybrids require RNA-directed DNA methylation and are influenced by genetic variation. Proc Natl Acad Sci 113: E4248–E4256. 10.1073/pnas.1607851113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Lang Z, Zhu JK. 2018. Dynamics and function of DNA methylation in plants. Nat Rev Mol Cell Biol 19: 489–506. 10.1038/s41580-018-0016-z [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.