Abstract
Germ cells in Drosophila melanogaster are specified maternally shortly after fertilization and are transcriptionally quiescent until their zygotic genome is activated to sustain further development. To understand the molecular basis of this process, we analyzed the progressing transcriptomes of early male and female germ cells at the single-cell level between germline specification and coalescence with somatic gonadal cells. Our data comprehensively cover zygotic activation in the germline genome, and analyses on genes that exhibit germline-restricted expression reveal that polymerase pausing and differential RNA stability are important mechanisms that establish gene expression differences between the germline and soma. In addition, we observe an immediate bifurcation between the male and female germ cells as zygotic transcription begins. The main difference between the two sexes is an elevation in X Chromosome expression in females relative to males, signifying incomplete dosage compensation, with a few select genes exhibiting even higher expression increases. These indicate that the male program is the default mode in the germline that is driven to female development with a second X Chromosome.
The germline-soma dichotomy is established as germ cells are specified and is central to the unique ability of germline to transition to the totipotent state of embryos after fertilization (Johnson and Alberio 2015). In mice, WNT and BMP signaling induce germline formation via the expression of multiple transcriptional regulators (Sybirna et al. 2019). In Drosophila melanogaster, germ plasm loaded in the posterior of oocytes contains RNAs and proteins that are important for germline specification and early germline characteristics such as repression of transcription and migration (Santos and Lehmann 2004). The phase of transcriptional quiescence is thought to prevent erratic expression of somatic genes (Hanyu-Nakamura et al. 2008; Asaoka et al. 2019). However, it is unclear what occurs downstream of specification that embodies the germline program to drive development of this lineage.
One important developmental choice germ cells must make early on is to determine their sex. Sex determination is controlled by select genes whose activities reflect sex chromosome dosage and content. The genetics underlying this process can be distinct between the soma and germline and is much better understood in the soma of most species. For fruit flies, successful establishment of germline sex requires matching somatic input and intrinsic decision, but the genes involved are not the same (Murray et al. 2010), and it is not yet known which genes read the sex chromosomal content autonomously in the germline. Sex-specific germ cell differentiation is first observed in late embryogenesis, and these events include the division and stratification of male germ cells, whereas the female ones remain quiescent (Wawersik et al. 2005; Le Bras and Van Doren 2006). Divergence in gene expression that underlies these differences most likely arises earlier.
Determination of sex chromosome composition also dictates whether there is a need for dosage compensation, a process encompassing a variety of mechanisms across species designed to equalize expression of X Chromosome genes between the two sexes, as females have two doses compared to just one in males. In the mammalian female germline, both X Chromosomes are active, unlike in somatic cells in which one X is inactivated (Sangrithi et al. 2017; Chitiashvili et al. 2020). X Chromosome expression also appears to be sex- and stage-dependent, which is thought to be important in accommodating specific biology needed during major developmental transitions, including lineage specification and entry to meiosis (Turner 2015; Sangrithi et al. 2017; Chitiashvili et al. 2020). In fruit flies, male somatic cells increase their X Chromosome gene expression twofold to match expression in the female soma via the dosage compensation complex (Conrad and Akhtar 2012). However, this mechanism is not used in germ cells (Rastelli and Kuroda 1998), and X Chromosome expression has not been evaluated in the earliest germ cells that undertake key developmental changes.
The emergence of single-cell RNA-sequencing (scRNA-seq) techniques has opened a new window to investigating critical early germline developmental events, especially because germ cells exist in small numbers at these stages. Multiple studies have analyzed transcriptomes of germ cells at the single-cell level but mostly at later stages of development (Li et al. 2017; Jevitt et al. 2020; Niu and Spradling 2020; Rust et al. 2020; Slaidina et al. 2020; Mahadevaraju et al. 2021). Here, we profile the transcriptomes of fruit fly germ cells immediately after specification and before they have coalesced with somatic cells, and we perform this survey for both male and female germ cells. By examining the earliest stages of Drosophila germline development, we aim to reveal the events and molecular characteristics that define the germline lineage.
Results
Purification of early germ cells for single-cell transcriptome profiling
To obtain germ cells after specification and before gonad coalescence for scRNA-seq, we FACS-purified germ cells from 0- to 8-h embryos based on germline-specific GFP expression using the vas-GFP transgene (Fig. 1A,B; Supplemental Fig. S1A–D,I). We also performed a second round of scRNA-seq for which we processed 5- to 8-h female and male germ cells separately to achieve greater resolution between the sexes (Supplemental Fig. S1E–H). This was made possible by the addition of an Sxl-GFP transgene which is highly expressed in female but not male 5- to 8-h embryos (Supplemental Fig. S1J). These two female and male samples contained some somatic cells due to GFP expression from the Sxl-GFP transgene (Supplemental Fig. S1K), but this did not pose a problem for bioinformatics analyses as there are well-established markers that can help us unequivocally identify germ cell populations. The numbers of cells sequenced as well as the mean reads and median genes detected per cell in all three samples are as summarized (Supplemental Fig. S2A).
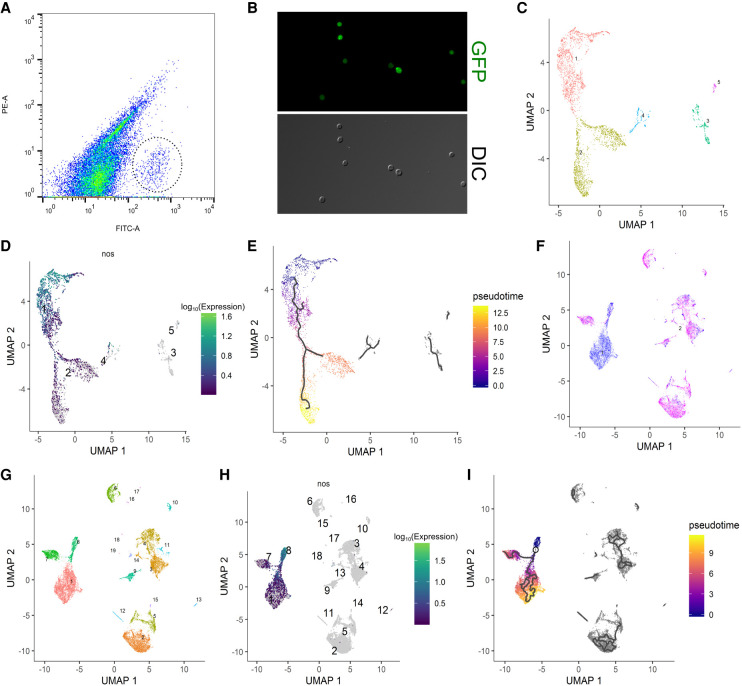
Figure 1.
Clustering and pseudotime analysis of single-cell germline transcriptomes. (A) FACS plot for sorting germ cells from embryonic homogenates of 0- to 8-h embryos with the GFP+ germ cells highlighted by a dotted circle. x-axis is the green channel. (B) Morphology by DIC and GFP profiles of sorted 0- to 8-h germ cells in the unsexed sample. Note that virtually all cells recovered are GFP-positive cells whose morphology resembles that of germline. (C) Clustering analysis of the unsexed sample. Each dot represents a cell in the data set, and individual clusters are numbered and colored differently. (D) Expression profile of nos of the unsexed sample showing that clusters 1 and 2 make up the main germline cluster. Each dot represents a cell in the data set, the numbers indicate individual clusters, and the color code for expression levels is indicated on the right. (E) Pseudotime analysis of the germline cluster from the unsexed sample. The black lines within the clusters indicate pseudotime trajectories. The legend on the right explains the color code of pseudotime. (F) The sexed data set is plotted after dimension reduction. Each blue and magenta dot represents a cell from the male and female data set, respectively. (G) Clustering analysis of the sexed sample. Each dot represents a cell in the data set, and individual clusters are numbered and colored differently. (H) Expression profile of nos after clustering analysis of the sexed data set indicating that clusters 1, 7, and 8 contain germ cells. Details of the graph are the same as in D. (I) Pseudotime analysis of the germline cluster from the sexed data set. Black lines indicate pseudotime trajectories and the color codes of pseudotime are indicated on the right.
Clustering and pseudotime analyses reveal bifurcation between early male and female germline
The primary bioinformatics tool we used for scRNA-seq data analysis was Monocle 3, which enables both clustering and pseudotime analyses (Cao et al. 2019). Clustering analysis of the sequencing data from 0- to 8-h unsexed cells resulted in a major Y-shaped germline cluster based on the expression profiles of known germline marker genes such as nanos (nos) and vasa (vas) (Fig. 1C,D; Supplemental Fig. S3A,B). The female and male samples, when combined (herein referred to as the “sexed sample/data set”), also produced a Y-shaped germline cluster as evidenced by the profiles of nos and vas expression (Fig. 1G,H; Supplemental Fig. S3E,F). In the unsexed sample, cells in cluster 4 also express germline markers (Supplemental Fig. S3A–D). However, they contain a much higher proportion of mitochondrial reads compared to cells from the main germline cluster (cluster 2) and are likely germ cells dying during sample preparation (Supplemental Fig. S2C). No mitochondrial reads were detected in the sexed sample (Supplemental Fig. S2B).
The Y-shaped nature of the germline clusters suggests that the early germ cells we collected are on a transcriptional continuum that diverges once during this time period. When we examined known maternally contributed germline genes such as polar granule component (pgc) and germ cell-less (gcl) (Jongens et al. 1992; Nakamura et al. 1996), we found that their transcript levels are highest in cells at the end of the Y-stems and gradually decrease toward the branches of the germline clusters (Supplemental Fig. S3C,D,G,H). This indicates that the earliest germ cells are at the base of the Y-stems, a position assigned as the “root” of the clusters for pseudotime analyses, which subsequently gave rise to trajectories along pseudotime which start at the base of Y-shape, branch in the middle, and progress toward the tips of the two prongs (Fig. 1E,I). In the sexed sample, the stem contains germline from both sexes whereas the branches are comprised largely of either male or female germline (Fig. 1F,I). This indicates that sexual differentiation has begun in this early time point prior to gonad coalescence.
Distinct germline expression patterns along pseudotime
The presence of distinct expression patterns common to many genes is suggestive of developmental changes and transitions between physiological states. Of the multiple expression modules determined for the germline clusters, several exhibit a clear and continuous trend over pseudotime and are shared between the unsexed and sexed samples (Supplemental Fig. S4A,B).
One category, exemplified by module 3 of the unsexed sample and module 10 of the sexed sample, represents genes that are mainly expressed maternally (Fig. 2A,D). Another type portrays increased gene expression along pseudotime, reflecting zygotic activation of the germline genome and includes module 2 from the unsexed sample and module 3 from the sexed sample (Fig. 2B,E). Moreover, there are also modules that show higher expression in one branch than the other (module 6 from the unsexed sample and module 5 from the sexed sample) (Fig. 2C,F), and we know from the sexed sample that the branch with higher expression is comprised of female germ cells (Fig. 1F). In contrast, there are no modules with the reverse trend.
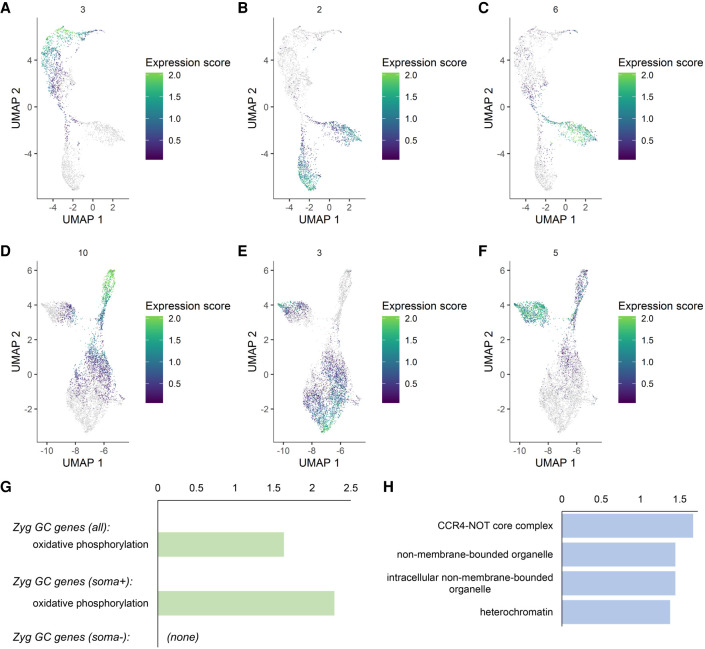
Figure 2.
Expression trends in the germline clusters and activation of the zygotic germline genome. (A–F) Expression modules in the unsexed (A–C) and sexed (D–F) samples indicating patterns of maternal contribution (A,D), zygotic activation (B,E), and female-biased expression (C,F). Module numbers and color codes are on the top and to the right of each graph, respectively. (G) KEGG pathway enrichment results for the genes activated in the zygotic germline. The upper bar shows the enrichment term found for all zygotic genes together and the lower bar is for those that are soma-positive. There were no terms found to be enriched for the soma-negative genes. (H) GO-Term: Cellular Components enrichment analysis for the zygotic soma-negative germline genes. (G,H) The x-axes indicate −log(p) values.
We next determined “marker genes” for the germline population from the unsexed sample before and after the bifurcation point to find genes that are maternally deposited versus zygotically produced (Supplemental Table S1, “Maternal all” and “Zygotic all”). We also picked out a subset of markers that are highly germline-enriched by removing those that showed substantial somatic expression as determined by the somatic cells included in the sexed sample (Supplemental Table S1, “Maternal soma-negative,” “Zygotic soma-negative”).
The gene lists were subsequently validated with the embryonic in situ hybridization database of the Berkeley Drosophila Genome Project (BDGP, https://insitu.fruitfly.org/cgi-bin/ex/insitu.pl) for all genes from the “Maternal soma-negative” and the “Zygotic soma-negative” groups. We found that, for both lists, the in situ patterns of most genes that have a clear expression pattern in the BDGP database match the expectations based on our sequencing results (Supplemental Fig. S5A). We compiled example images of one gene each from the maternal and zygotic lists to demonstrate the similarities between the BDGP results and ours (Supplemental Fig. S5B–G). We also examined the in situ patterns of two zygotically activated candidates not profiled by BDGP, one belonging to the soma-negative group (Heterochromatin protein 6, HP6) and the other to the soma-positive group (P32). We observed that their expression indeed gradually increases in the embryonic germline as indicated in our results (cf. Supplemental Fig. S5H vs. S5I–L). Overall, there is very good accordance between our two rounds of scRNA-seq and the BDGP in situ patterns, indicating that our transcriptome results are robust.
Activation of the zygotic germline genome
Of the genes activated zygotically in the germline, some are highly germline-enriched, but the majority are also expressed in the soma, albeit at lower levels. When we looked at the molecular functions of these two types of genes, those that are also expressed somatically are mostly “housekeeping” genes (Fig. 2G; Supplemental Table S2). The enrichment terms with the highest significance for zygotic germline genes were mitochondrial components (Fig. 2G), an observation also made for the embryonic human germline (Li et al. 2017). This may reflect a conserved requirement for energy consumption in the early germline. In comparison, those that are more germline-specific include germline-enriched features such as RNA granules as well as components that regulate gene expression, which may be important for germline lineage development (Fig. 2H).
To investigate the genes that may delineate the germline program, we parsed our data sets for germline-specific genes known to or that potentially encode RNA- and DNA-binding proteins, as they can mediate germline-specific expression via transcriptional activation, elongation, RNA stabilization, or translational control (Supplemental Table S3). Those that are maternally contributed are expressed prior to zygotic activation and are much more likely to be involved in regulating germline-specific zygotic expression. The zygotic ones may contribute to germline-specific gene expression or functions at later stages. There are previous reports of maternally contributed germline genes that are needed for proper expression of vas and nos (Yatsu et al. 2008; Nakamura et al. 2019); in our data sets, these genes exhibit varying degrees of expression in the soma in addition to the germline, implying that genes with roles in the early germline do not need to be highly germline-specific (Supplemental Fig. S6A).
To explore whether additional factors regulate zygotic activation, we looked for enrichment of known transcription factor (TF) binding sites and also performed de novo discovery of enriched sequence motifs in the promoter regions of zygotic germline genes (Fig. 3A; Supplemental Fig. S6B). The distribution of the motifs identified by both strategies is concentrated close to transcription start sites and conforms to what is commonly observed for TF binding sites (Supplemental Fig. S6C). In addition, their presence on zygotic soma-negative and -positive germline genes are similar (Fig. 3B), suggesting that these two groups of genes are activated similarly. To compare, we examined TF binding site enrichment in marker genes of three somatic clusters in our sexed data set (clusters 2–4 in Fig. 1G). We found that, although the motifs enriched in germline genes are also present in somatic markers (Fig. 3B), their frequencies are reduced and the TF binding sites most significantly enriched in the soma are distinct from those in the germ cells (Fig. 3A).
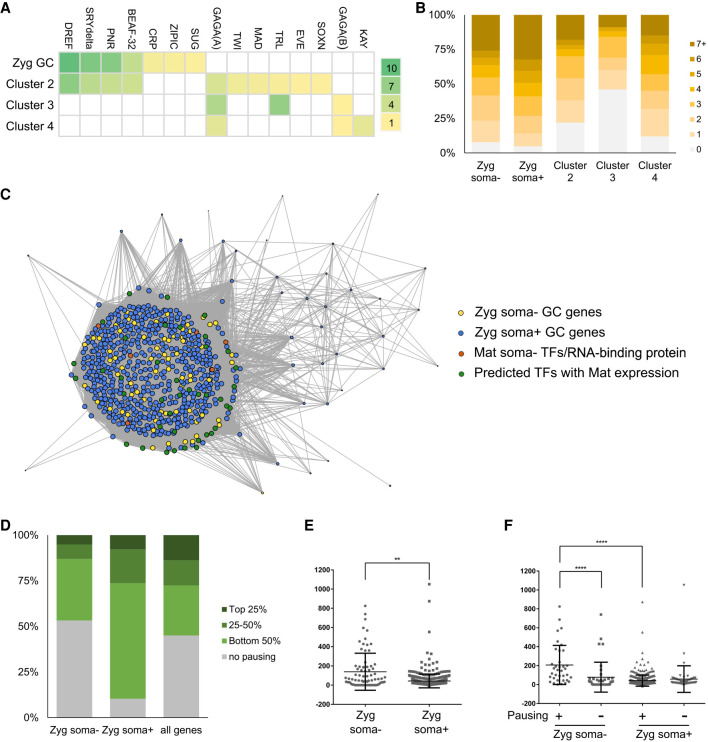
Figure 3.
Regulation of early germline-specific expression. (A) Heat map display of TF binding site enrichment analysis for all zygotic germline genes and markers of somatic clusters 2–4. The names of TFs are listed on the top and the scale on the right indicates the –log(p) values for each entry. (B) Numbers of binding sites found in the promoter regions of zygotic germline genes or markers of somatic clusters for TFs enriched in the germline. (C) Network view of pairwise expression correlation values in the germline cluster of the unsexed data set between genes of the following four groups with one another: zygotic soma-negative germline genes (yellow dots), zygotic soma-positive germline genes (blue dots), maternally deposited soma-negative TFs and RNA-binding protein genes (orange dots), and TFs identified through our enrichment analyses with detectable maternal expression in germ cells (green dots). Only correlations >0.5 are displayed, the sizes of dots indicate the numbers of correlations each gene has above the 0.5 cutoff, and the lengths of the connection lines inversely reflect the correlation values between a pair of genes. (D) Polymerase pausing of zygotic germline genes in the early embryonic soma based on a study from Saunders et al. (2013). Genes are categorized based on their pausing index into top 25%, 25%–50%, bottom 50%, or no pausing groups. We present the comparisons between soma-negative zygotic germline genes, soma-positive zygotic germline genes, and all genes in the genome. (E) Test of RNA stability in the soma using ratios of nascent RNA levels in the gene body regions to steady-state RNA levels of various genes. The middle horizontal lines indicate the means calculated for the soma-negative and soma-positive groups of genes, whereas the top and bottom horizontal lines mark the standard deviations. (**) P < 0.005. (F) Comparisons of RNA stability between the soma-negative and -positive zygotic germline genes that exhibit pausing and not. The values were calculated as in E. (****) P < 0.0001.
To further examine the potential significance of the TFs highlighted above in regulating zygotic germline expression, we carried out gene regulatory network (GRN) analyses between zygotically activated genes and a subset of the aforementioned TFs—those that exhibit clear expression prior to germline zygotic activation in the unsexed data set. There are several observations we made from these network analyses. First, expression of the majority of zygotically activated genes are highly correlated (Fig. 3C). This suggests that zygotic activation is comprised of parallel expression of many genes, likely the result of the block of RNA polymerase II being relieved, which enables genes to be transactivated by TFs present in germ cells at this stage. Second, there is no clear separation between zygotic soma-negative and -positive genes in the network (Fig. 3C). This is consistent with our TF binding analyses suggesting that activation of these genes is similar. Third, multiple TFs also exhibit strong correlation with the bulk of zygotically activated germline genes (Fig. 3C; Supplemental Fig. S6D), indicating that these TFs act concurrently at the start of zygotic activation rather than in a hierarchical fashion. Furthermore, germline-specific TFs (Supplemental Table S3; orange dots in Fig. 3C) are not more strongly correlated with zygotically activated germline genes than those identified through TF binding site analyses which are not germline-restricted (green dots in Fig. 3C). Overall, these results support the idea that multiple TFs are involved in zygotic germline transcription, and the actions of many such TFs are not germline-restricted.
Polymerase pausing and regulated RNA stability contribute to germline-specific expression
If germline-enriched expression is not the result of cell type–specific transcriptional activation, there would need to be additional mechanisms post-transcription to confer germline-specific gene expression, such as inhibition of transcription elongation and regulation of transcript stability. Here, we took advantage of a genome-wide nascent RNA-sequencing (GRO-seq) study in the soma of early Drosophila embryos carried out by Saunders et al. (2013) to investigate the possibilities mentioned above. In the GRO-seq study, pausing of RNA polymerase II after initiation of transcription was observed for more than half of all genes in somatic cells, and this phenomenon was suggested as a strategy to accommodate greater flexibility in gene expression control during development.
By referencing their results, we found that a subset of zygotic germline genes, both soma-negative and -positive, exhibit promoter-proximal polymerase pausing (Fig. 3D). Although there is currently no data on whether pausing also occurs in the germ cells, this analysis does lend support to the idea that differential polymerase pausing after similar transcriptional activation between the soma and germline contributes to how certain genes achieve germline-enriched expression.
We further used the GRO-seq data to investigate whether differential RNA stability also regulates expression of germline genes in the soma. We determined RNA stability by calculating the ratio of nascent RNA levels in the gene body of various genes from the GRO-seq study to steady-state RNA levels detected in the somatic cells profiled in our data set. Increases in this ratio would indicate greater RNA instability. We found that RNA turnover in soma-negative zygotic germline genes is greater than that in soma-positive ones, and the same trend also holds true when just the genes that exhibit polymerase pausing were examined (Fig. 3E,F). This suggests that faster turnover rates of transcripts are in part why RNAs of soma-negative genes fail to accumulate despite being transcribed in the soma. Moreover, transcripts of genes that exhibit polymerase pausing are less stable than those that do not pause (Fig. 3F). These findings suggest that promoter-proximal pausing and regulation of RNA stability are important mechanisms that mediate germline-specific gene expression and contribute to establishing the distinct programs between germline and soma.
X Chromosome expression in the embryonic germline is partially dosage compensated
Our clustering and pseudotime analyses indicate that, as zygotic transcription is activated, the transcriptomes of female and male germ cells diverge (Fig. 1E,I). In addition, there are only expression modules in which female expression is substantially higher than that of males but not the reverse (Fig. 2C,F; Supplemental Fig. S4). When the chromosomal locations of such female-enriched genes were examined, we found almost all of them to be positioned on the X Chromosome and distributed across the entire X Chromosome (Fig. 4A,B). The observation that expression of many X Chromosome genes is higher in female germ cells than males brought to our mind the issue of dosage compensation. Drosophila germ cells do not utilize the dosage compensation complex present in somatic cells to perform dosage compensation (Rastelli and Kuroda 1998), and results from multiple studies examining larval and adult male germline have suggested X Chromosome dosage to be dynamic (Deng et al. 2011; Mikhaylova and Nurminsky 2011; Meiklejohn and Presgraves 2012; Vibranovski et al. 2012; Shi et al. 2020; Mahadevaraju et al. 2021). This issue has not been addressed in the embryonic germline that can have different gene expression requirements, as these cells undergo developmental events not encountered by germ cells in later stages.
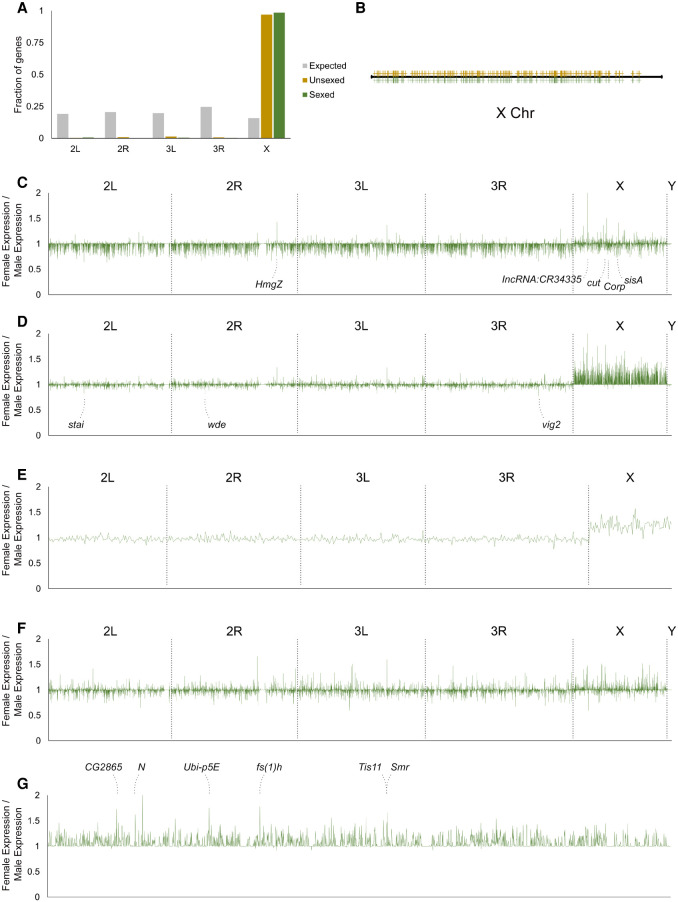
Figure 4.
Analyses of sex differences in the early germline. (A) Chromosomal distribution of genes from the female-enriched expression modules. The fraction of genes belonging to each chromosome is plotted. Gray bars, expected distribution based on number of genes on each chromosome; yellow bars, distribution of genes from the unsexed sample; green bars, distribution of genes from the sexed sample. (B) Distribution of X Chromosome genes from female-enriched expression modules along the X Chromosome. The yellow and green marks designated genes from the unsexed and sexed data sets, respectively. (C) Differences in expression levels of all genes between female and male germ cells prior to the bifurcation point of the germline cluster from the adjusted sexed data set in which male reads were reduced randomly by 25%. The x- and y-axes are the same for C–F: the x-axes plot the positions on individual chromosomes as indicated and the y-axes plot the fold differences of expression of female germ cells over male germ cells. The identities of the most female-biased genes are indicated. (D) Differences in expression levels of all genes between female and male germ cells after they bifurcate in the germline cluster from the adjusted sexed data set. The gene names of the most male-biased peaks are indicated. (E) Expression differences of the 647 zygotically activated germline genes between the two sexes based on the adjusted sexed data set. (F) Female-to-male expression ratio of all genes of somatic cells included in the adjusted sexed sample. (G) Female-to-male expression ratios of X Chromosome genes of germ cells after bifurcation from the adjusted sexed data set. The identities of the highest peaks are indicated.
To investigate dosage compensation in the embryonic germline, we compared sex differences in autosomal and sex chromosomal expression in the germline before and after the bifurcation point so as to track changes that may occur as a result of zygotic transcription and possible dosage compensation. By plotting the ratio of female-to-male gene expression on different chromosomes, we observed several patterns. In the sexed data set, X Chromosome gene expression in the females is on average higher than in males, and this phenomenon is specific to the X Chromosome (Supplemental Fig. S7A). Female-to-male expression ratios for X Chromosome genes between one and two indicate partial dosage compensation, as complete dosage compensation would result in average ratios of one, and a complete lack of compensation would give rise to ratios on average of two. For autosomes, the average female-to-male expression ratios were slightly <1 (Supplemental Fig. S7A). This was also the case for genes across all chromosomes when comparing female and male somatic cells from the sexed data set even though the expectation was that these cells would exhibit full dosage compensation and give rise to average sex expression ratios of one (Supplemental Fig. S7B). We reasoned that this may arise from our male sample being sequenced to 25% greater depth than the female sample, thereby resulting in higher expression values. To correct for this effect, we randomly removed 25% of reads from the male data set and reanalyzed sex expression ratios in this adjusted sexed data set. We found that the higher expression levels in males promptly disappeared and that sex expression ratios of autosomal genes in the germline are now on average one (Fig. 4D). Likewise, sex expression ratios for all genes in the somatic cells now also averaged one, in line with the expected complete dosage compensation (Fig. 4F). In comparison, the higher female expression in X Chromosomes of female germ cells compared to males was preserved (Fig. 4D). Consistent with these observations, there was also an X Chromosome-specific expression increase in females compared to males in the unsexed data set (Supplemental Fig. S8A).
Another trend we observed was that the sex difference in X Chromosome expression occurred only after zygotic transcription begins (Fig. 4C,D). However, these X Chromosome genes could be expressed maternally and/or zygotically; therefore, it is necessary to focus on those that are only zygotically activated for a more accurate analysis of the possible effects of dosage compensation. We replotted the female-to-male expression ratios for the 647 zygotically activated genes (Supplemental Table S1) and found that the average expression on the X in females is clearly higher than that in males (Fig. 4E; Supplemental Fig. S8B). Taken together, our results demonstrate that there is partial, incomplete dosage compensation for the X Chromosome in the early germline.
Candidate genes that control germline sex determination
Our single-cell transcriptome profiles have captured the earliest stages of germline sexual differentiation, and this data enabled us to look for candidate genes of germline sex determination on the X Chromosome. From the germline sex expression ratio plots, one can identify individual peaks that are particularly high, and they correspond to greater female-to-male expression ratios (Fig. 4G; Supplemental Fig. S8C). When the unsexed and sexed samples were compared, the identities of their higher peaks matched very well (Fig. 4G; Supplemental Fig. S8C). This suggests that these genes are regulated differently from most other genes on the X Chromosome, and their higher levels of expression are likely to be functionally significant for sex-specific germline development. There are also a few higher peaks in the sex expression ratio plot of germline prior to bifurcation (Fig. 4C). Among those, two are X-encoded transcription factors cut (Ct) and sisterless A (SisA), the latter being a known X-dosage sensor in the soma and recently shown to regulate germline expression of Sex lethal (Sxl), a known regulator of female germline sexual development (Cline and Meyer 1996; Goyal et al. 2019). These genes may also contribute to germline sex determination. In contrast, the peaks that exhibit male-biased expression are not as distinct (Fig. 4D).
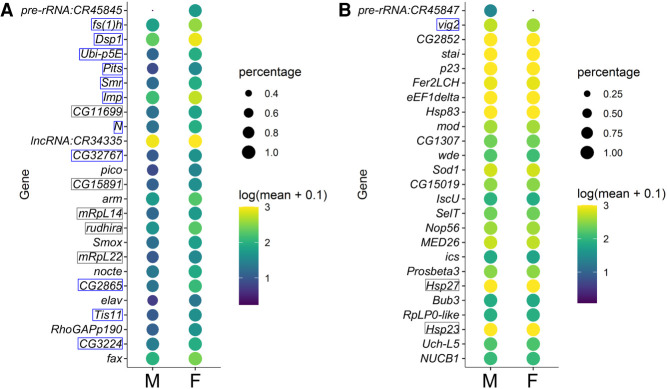
Another method for identifying sex markers is to call top marker genes for the female and male germ cells past the bifurcation point. We conducted this analysis without taking into consideration transcriptomes of earlier germ cells or somatic cells, as germline sex-determining factors would not necessarily have to be germline- or stage-specific. For the sexed data set, we used the original data set in which the male read depth was greater to obtain maximum information and resolution. For female germline markers, we focused on the few with the most stringent significance values, as many others will show female-enriched expression due to incomplete dosage compensation (Supplemental Table S4). Of the top 25 female markers, 16 are common between the unsexed and sexed samples and overlap with the genes that exhibit the highest female-to-male expression ratios (Figs. 4G, 5A; Supplemental Figs. S8C, S9A). Furthermore, several of those, such as female sterile (1) homeotic (fs(1)h), Smrter (Smr), and Protein interacting with Ttk69 and Sin3A (Pits), encode transcriptional regulators or contain nucleic acid binding domains (Fig. 5A; Supplemental Fig. S9A). These genes are prime candidates for controlling germline sex.
Figure 5.
Top sex markers from the sexed data set. (A) Expression profiles of the top 25 female markers in the male and female germline clusters after zygotic activation are plotted, with the colors reflecting mean expression as indicated on the right; the size of the dots reflects the fraction of cells in the clusters with detectable expression of each gene. The gene names boxed in gray or blue are those also determined as top female markers in the unsexed data set (Supplemental Fig. S9); the ones predicted to be transcription- or chromatin-associated factors are marked with blue boxes. (B) Expression patterns of the top 25 male markers plotted in the same way as in A.
We also looked for male germline markers and found that not only is this list much shorter than that for female markers, the extent of increased expression compared to female germ cells is also smaller (Figs. 4D, 5B, 6A–D; Supplemental Figs. S8A, S9B). This is possibly due to the 8-h male germline being at the cusp of differentiation and thus showing limited male-specific gene activation. There are also other genes known to have important sex-specific germline functions or expression patterns in embryogenesis, with the most notable examples being Sxl and PHD finger protein 7 (Phf7) which have been shown to trigger germline sex reversal (Oliver et al. 1987; Pauli et al. 1993; Casper and Van Doren 2009; Hashiyama et al. 2011; Yang et al. 2012). In our data sets, Sxl exhibits a female bias in expression smaller than the top female markers we identified, whereas Phf7 is expressed at low levels with a weak female bias, likely due to it being on the X Chromosome (Supplemental Fig. S10A–C). The other genes do not show sex-biased expression in our data sets, possibly because their sex-enriched expression occurs later (Supplemental Fig. S10C).
Figure 6.
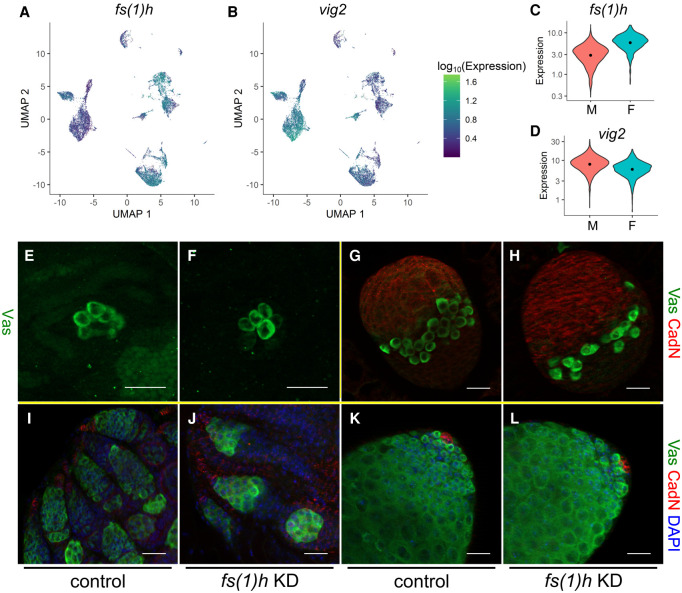
Female-specific involvement of fs(1)h in germline development. (A,B) Expression profiles of fs(1)h (A) and vig2 (B) in the sexed data set. Color codes of the expression levels are indicated to the right. (C,D) Violin plots of the female marker fs(1)h (C) and male marker vig2 (D). Orange and turquoise populations indicate male and female expression levels, respectively. (E–L) RNAi knockdown phenotypes of fs(1)h in the germline of ovaries at embryonic stage 17 (E,F), third-instar larva (G,H), newly eclosed adult females (I,J), and testes from newly eclosed males (K,L). (E,G,I,K) Controls (nos-Gal4/+); (F,H,J,L) Germline knockdown of fs(1)h (HMS02723/+;nos-Gal4/+). (E,F) Stained with a Vasa antibody (green); (G,H) stained with Vasa (green) and Cadherin-N (red) antibodies; and (I–L) stained in addition with DAPI (blue). Scale bars, 20 µm.
A top female marker, fs(1)h, regulates female-specific germline development
To investigate whether candidate genes exhibiting sex-enriched expression are important for germline sexual development, we performed functional tests for one of the genes that has consistently ranked highly in our tests of female marker genes, fs(1)h. This gene is located on the X Chromosome and encodes a bromodomain protein known to cause female sterility. Many studies have demonstrated its functions in gene expression regulation in various contexts (Digan et al. 1986; Chang et al. 2007; Florence and Faller 2008), but its possible roles in germline development have not been reported. We tested the functions of fs(1)h in female-specific germline development by germline-specific RNAi knockdown (nos-Gal4, UAS-fs(1)h RNAi). Immunofluorescence staining revealed that at stage 17, a stage shortly after female-biased expression of fs(1)h is detected, there are no differences in the female germline when fs(1)h is knocked down compared to controls, but this is a time point during which female germ cells are quiescent (Fig. 6E,F). In comparison, in third-instar larval ovaries in which the germ cells start to divide and develop, there are strong defects in both germ cell number and organization compared to controls (Fig. 6G,H). This defect persists to adulthood whereas the undifferentiated male germline is mostly normal (Fig. 6I–L). These results indicate that fs(1)h is indeed important for female-specific germline development. Furthermore, this functional validation indicates that the higher expression of female marker genes is physiologically important and is likely established with unique mechanisms.
Discussion
Our single-cell transcriptome profiling of the embryonic Drosophila germline provides a comprehensive coverage of the molecular signatures at this critical developmental stage. It demonstrates the power of using single-cell techniques to build trajectories that can dissect and reveal characteristics of important lineages that exist in small cell numbers.
Regulation of germline zygotic activation
Here, we investigated regulations of zygotic activation at the transcriptional and post-transcriptional steps and looked into how germline-specific expression may be achieved as it is an essential aspect for securing the germline identity. Our results show that, during zygotic activation, a large wave of transcription occurs through the actions of different TFs present in but not exclusive to the embryonic germline. We attempted to decipher the network of TFs that regulate germ cell gene expression via GRN analysis. However, it appears that the cells we have collected span up to a very limited time window after zygotic activation, and therefore there is insufficient information and resolution to construct potential hierarchies of TFs that act within the embryonic germline.
Our data further suggest that post-transcriptional mechanisms contribute substantially to germline-specific gene expression at the time of zygotic activation. Cross-analyses with a nascent RNA-sequencing study of somatic cells from early embryos (Saunders et al. 2013) clearly indicate that polymerase pausing and transcriptional readthrough occurs on a substantial number of germline genes in the soma. These suggest that regulation of transcription elongation and RNA stability are important for establishing germline-restricted gene expression. The time windows profiled in the Saunders study are a few hours earlier than the somatic cells we analyzed (5–8 h), but assuming that expression profiles do not change abruptly in the soma within these time frames, we can obtain insightful comparisons between the GRO-seq data set and our own. In fact, our analyses here may yet underestimate the contribution of polymerase pausing and differential RNA stability to gene expression differences between the soma and germline as the Saunders study was a profile on the entire soma rather than the subset that most resembles germ cells in their usage of transcription factors. Future studies that directly compare promoter-proximal polymerase pausing and RNA stability between germ cells and specific somatic cells will clarify the extent to which these processes are involved in establishing the distinction between the germline and somatic programs. Among the RNA-binding protein-encoding genes that are expressed maternally in the germline, three are germline-specific and already noted for their functions in regulating early germline development: nos, oskar (osk), and bruno 1 (bru1) (Lehmann and Nüsslein-Volhard 1986; Kim-Ha et al. 1995; Asaoka et al. 2019). There is already an example of nos regulating stability of a transcript in the embryonic germline (Sugimori et al. 2018). It is quite possible that their roles in germline biology are more extensive than we currently appreciate.
Male versus female early development
Germ cells need a mechanism to read their sex chromosome composition to determine their sex. In our trajectories, we observed a bifurcation between male and female germ cells at zygotic activation, indicating that germline sex determination has occurred at this early developmental stage. Our data sets also reveal multiple transcription factors and chromatin-associated factors that reproducibly exhibit close to twofold higher expression in the female germline than in males. This could be a parallel to how sex determination occurs in the soma in which the combinatorial activity of multiple transcription factors regulates expression of the sex switch gene Sxl. Functional testing of one of the female-enriched germline factors, fs(1)h, showed that it is indeed important for female-specific germline development. Further studies will reveal whether Fs(1)h is indeed part of the machinery that determines germline sex and if other factors that we also identified as being female-biased at this early stage act cooperatively to read out the X Chromosomal dose in germ cells.
An important consequence of the determination of sex chromosomal content is the activation of dosage compensation. A recent study based on scRNA-seq data in larval and adult germline indicated that the male X Chromosome dose is largely compensated in spermatogonia (Mahadevaraju et al. 2021). In our data sets, we directly compared expression levels of each gene in the female and male embryonic germline of equivalent stages and clearly demonstrated partial compensation of X Chromosome genes in the germline once the zygotic genome has been activated. Whether the incompleteness of X dosage compensation is a snapshot of a transitory state toward full compensation or representative of spermatogonia in general will require examination of the full spectrum of germline development. Furthermore, the mechanism responsible for germline dosage compensation bypasses certain genes, leaving them to be expressed at higher female-to-male ratios, and we presume those genes to be the most likely candidates in counting sex chromosomes in the germline.
We noticed that some genes also exhibit higher sex expression ratios in the germline population prior to the bifurcation point of the sexed data set. To explain how their mRNA levels would become sex-dependent prior to zygotic transcription, one would need to invoke special scenarios such as somatic induction, leaky transcriptional repression in this early phase, or paternal contribution. However, the sex expression differences are modest and stem from a single data set; thus, independent validation of such differences would be needed for future investigations.
In the developmental trajectories, the male branches appear to extend further along pseudotime than the female branches (Fig. 1E,I). This suggests some degree of development present in the male germline at 8 h of embryogenesis and is in line with our knowledge that male germline development precedes that of their female counterparts. Nonetheless, none of our various attempts to find genes that are male-enriched past the bifurcation point resulted in candidates with sufficient confidence beyond vig2 and heat shock protein genes, as described earlier. We hypothesize that the male germline at this time point is at the very start of development, with vig2 involved in modifying chromatin and heat-shock chaperones being produced to assist folding of the upcoming wave of new proteins.
Our scRNA-seq results portray the early sex-developmental sequence of D. melanogaster germline as such: when zygotic transcription is derepressed, higher expression of select X Chromosome genes due to a double dose of X Chromosomes drives the germline toward the female program, away from the default male fate. Male germ cells which exhibit lower expression of these female-determining genes initiate development. In contrast, establishment of the female fate would result in the mitotic and developmental quiescence of the germ cells until later larval stages. This is a new model that paves the way for future functional studies on the central germline sex-determining factors.
Methods
Fly strains
Fly strains used in this study include vas-GFP (Shigenobu et al. 2006) and Sxl-Pe-EGFP.G G78b (abbreviated as Sxl-GFP, Bloomington Stock Center). Sxl-GFP was used for sexing of embryos. In the first round of scRNA-seq, we used flies that contained four copies of vas-GFP. The second round of sequencing was carried out with flies homozygous for the Sxl-GFP and vas-GFP transgenes, both of which are on Chromosome 2. RNAi knockdown of fs(1)h in germ cells was performed with flies carrying the P{TRiP.HMS02723}attP40 RNAi construct (Bloomington Stock Center) and nos-Gal4 transgene (Van Doren et al. 1998) raised at 29°C.
Isolation of germ cells
Embryos of the desired age and genotype were collected on grape juice plates, dechorionated, and homogenized with the loose pestle in a Dounce homogenizer for 6–7 strokes. We found that this step alone could release sufficient single germ cells, thus we chose to forgo further enzymatic treatments. The lysates were filtered twice through 40-µm mesh, centrifuged at 850g for 2 min, and FACS-sorted (FACSAria, BD) to obtain GFP+ germ cells. A small fraction of the sorted cells was examined on a fluorescence microscope (AxioSkop, Zeiss) to document the integrity, purity, and cell number of the resulting samples before being used for library construction on the 10x Genomics single-cell RNA-seq platform and high-throughput sequencing on the NovaSeq 6000 System (Illumina).
We performed two rounds of scRNA-seq experiments. In the first round, we collected 0- to 4-h and 4- to 8-h vas-GFP germ cells separately, as the respective yields for germ cells were quite different (Supplemental Fig. S1A–D). The purified cells were then mixed in equal numbers and processed via the 10x Genomics Drop-seq pipeline (Single Cell 3′ v2). In the second round, we performed scRNA-seq (Single Cell 3′ v3) on sexed 5- to 8-h germ cells, obtained by FACS-sorting GFP+ cells from 5- to 8-h Sxl-GFP, vas-GFP embryos; 5- to 8-h female embryos show clear Sxl-GFP signals whereas male ones do not, and we hand-separated male and female embryos under fluorescent stereoscopes (Supplemental Fig. S1J). We validated our sex assignment by examining the sexes of hundreds of sorted embryos after their development to adulthood (100% correct, more than 700 flies scored). Embryos that have been separated by sex were subsequently homogenized as described above and sorted by FACS. Compared to the first round (Supplemental Fig. S1A,C), gating for live cells was further restricted to a subset enriched for germ cells (Supplemental Fig. S1E,G) to reduce inclusion of somatic cells that may also express GFP from the Sxl-GFP transgene (Supplemental Fig. S1F,H). The purity of the first unsexed sample was close to 100% germ cells as estimated by examining a fraction of the sorted cells by fluorescence microscopy (Fig. 1B); the female and male samples were a mix of germ cells and somatic cells (Supplemental Fig. S1K).
Analysis of scRNA-seq data
For the unsexed sample, we obtained sequencing results for 3810 cells that passed through quality control with the CellRanger software (version 3.0.1, 10x Genomics) with the mean reads per cells being 33,487 and the median genes detected per cell being 3166. For the sexed samples, 11,001 and 7222 cells from the female and male samples, respectively, passed through quality control. The overall cell count for the female sample was higher than that of the males due to the female sample cell count being underestimated at the time of sample collection. The mean reads per cell were 22,241 (female) and 29,231 (male), and the median genes detected per cell were 2045 (female) and 3482 (male); 84.9%, 80.1%, and 83.9% of all reads from the three samples, unsexed, female, and male, respectively, mapped to the D. melanogaster transcriptome (BDGP6.28).
Sequencing results were analyzed with the Monocle 3 package to determine clusters and construct a pseudotime that signifies the developmental trajectory of early germ cells (Cao et al. 2019). The data underwent preprocessing (number of dimensions set to 100), dimension reduction using the UMAP method (McInnes et al. 2018), and clustering with the Leiden algorithm (Traag et al. 2019). This gave rise to Y-shaped clusters for germ cells which were determined by expression of two germline markers, nos and vas. To order cells in the germline clusters, we chose the “root” to be at the end of the stem of the Y-shaped clusters based on the expression patterns of pgc and gcl (Supplemental Fig. S3C,D), which consequently enabled assignment of pseudotime in the germline clusters.
To investigate expression trends, we determined expression modules using default parameters except for the resolution being set to 0.0001. To identify markers of designated clusters, we utilized the top_markers function to find top genes that delineate various cell populations and used q values or marker scores in addition to prioritize candidate genes and adjust stringency. To identify germline genes with either predominant maternal or zygotic expression, we examined genes called as marker genes for the germ cells prior to or after the bifurcation point in the main germline cluster from the unsexed data set. From the corresponding lists of top 2000 markers (Supplemental Table S5), a cutoff of 0.3 in marker score was chosen. We noticed a few genes in the marker lists that overlap between the maternal and zygotic germline marker lists. When we examined the expression profiles of duplicated entries, most of them had relatively high expression throughout pseudotime. These genes were a minority on the lists and were removed from the list of zygotically activated genes. To generate the subsets of maternal or zygotic markers that are also highly germline-enriched, we utilized our sexed data set and eliminated genes whose average expression in all somatic cells we profiled was greater than 0.03.
To examine expression progression of specific genes, we selected the germ cells in the stem of the Y-shaped clusters as well as in the male branch, as these cells together represented a linear developmental progression along pseudotime. This allowed us to graph changes in expression of individual genes as a function of pseudotime.
Analyses of marker gene characteristics
To identify pathways that are enriched in the zygotically activated germline genes, we performed ordered queries for KEGG Pathway and GO Term: Cellular Components analyses using the g:Profiler platform (biit.cs.ut.ee/gprofiler/gost) (Raudvere et al. 2019) with false discovery rate thresholds of 0.05.
To look for enrichment of transcription factor binding sites in the zygotic germline genes, we used the g:Profiler interface to search the 1-kb region upstream of and downstream from the transcription start sites (TSSs) of candidates for binding sites cataloged in the TRANSFAC database (Release 2019.3, classes: v2) with P < 0.05. For de novo identification of sequence motifs enriched in the promoter regions of zygotic germline genes, we extracted the 1-kb region upstream of TSSs of all soma-positive and -negative zygotic germline genes from the Ensembl BioMart database and used the MEME-suite for motif discovery, using the 0-order model for background correction and with the statistical significance (E-value) cutoff of 0.0001 (Bailey et al. 2009). To examine the distribution of motifs in markers of different clusters, we used the FIMO tool for motif scanning of match sites in the 1-kb region upstream of TSSs of germline genes or top 100 markers of clusters 2–4 with P < 0.0001 (Grant et al. 2011).
To examine the extent of polymerase pausing in the embryonic soma, we referenced pausing indices from the Saunders study that performed GRO-seq for 2- to 2.5-h embryos (NCBI Gene Expression Omnibus [GEO; https://www.ncbi.nlm.nih.gov/geo/] accession number GSE41611) (Saunders et al. 2013). To determine transcript stability based on the Saunders study, we referenced a reanalysis of the 3- to 3.5-h embryo data which were mapped to the newer Drosophila genome assembly (dm6, GSM3281693, GSM3281694) to calculate mapped reads per bin (MRPB) in the gene body regions defined as being from +100 to the end of each gene. This was to avoid reads that reflect promoter-proximal polymerase pausing rather than transcriptional readthrough. MRPB values for gene body regions were determined by subtracting the MRPB values in the first 100 bps downstream from TSSs from those for the entire length of genes, all computed with multiBigwigSummary (version 3.3.2.0.0) from the deepTools2 package via the Galaxy platform and corrected for their relative lengths (Goecks et al. 2010; Ramírez et al. 2014). The lowest MRPB values were used in subsequent analyses for genes with more than one isoform. The gene body MRPB values were further divided by the average steady-state expression levels calculated for all somatic cells combined, based on our own scRNA-seq data, to obtain indices that reflect RNA stability. The Wilcoxon rank-sum test was used to determine statistical significance comparing RNA stability of zygotic germline soma-negative and -positive genes. The Kruskal–Wallis test followed by Dunn's multiple comparison test was used to compare RNA stability of soma-negative and -positive germline genes that exhibit polymerase pausing and not.
To calculate gene-by-gene female-to-male expression ratios in the sexed data set, we used the average expression of each gene in all germ cells in the male sample, all germ cells in the female sample, all somatic cells in the male sample, and all somatic cells in the female sample. Counts normalized by log-transformation were used as expression values for each gene. For the unsexed data set, the same ratios were calculated for cells in the two postbifurcation branches of the germline cluster.
To perform gene regulatory network analyses, pseudotime values for each cell in the germ cell clusters along with normalized log values of gene expression were extracted and input into the LEAP package to calculate pairwise expression correlation values between all genes of the following four categories (Specht and Li 2017): zygotic soma-negative germline genes, zygotic soma-positive germline genes, maternally deposited soma-negative germline genes that encode transcription factors or RNA-binding proteins (nine factors) (Supplemental Table S3), and TFs with predicted significance. The last group refers to TFs identified through binding site enrichment or motif discovery; they also have to exhibit an average expression of 0.1 in the germ cells prior to the bifurcation point in the unsexed data set. This last group includes 31 factors, 25 of which are E-box motif-binding TFs. Network visualization was performed with igraph with correlations greater than 0.5 depicted (Pemberton 1975).
In situ hybridization and immunofluorescence staining
In situ hybridization chain reactions (HCR v3.0, Molecular Instruments) were performed on 0- to 16-h (25°C) embryos to validate scRNA-seq results. Embryos were dechorionated with 100% bleach for 2 min prior to fixation in 4.5% formaldehyde and clearing with xylene substitute (Sigma-Aldrich) to minimize autofluorescence. Subsequently, samples were hybridized with 2 pmol of split-initiator probes overnight at 37°C to detect mRNA targets, then incubated with 6 pmol of hairpins labeled with various fluorophores overnight at room temperature to generate fluorescent amplification polymers. The embryos were then stained with a rabbit-α-Vasa antibody (1:250, d-260, sc-30210, Santa Cruz Biotechnology) followed by an Alexa Flour 488-conjugated goat-α-rabbit secondary antibody (1:1000, 111-545-003, Jackson ImmunoResearch) to mark the germ cells. Confocal images were taken on a LSM780 (Zeiss).
Immunofluorescence of embryonic, larval, and adult gonads was performed as previously described (Yang et al. 2012). Primary antibodies used were rabbit-α-Vasa and rat-α-Cadherin-N (1:20, DN-Ex #8, DSHB); goat secondary antibodies used were conjugated with Alexa Flours 488 and 594 (1:1000, 111-545-003, 112-585-143, Jackson ImmunoResearch). Samples were also stained with DAPI (1 µg/mL) to mark nuclei before imaging on Apotome.2 (Zeiss).
Data access
All raw and processed sequencing data generated in this study have been submitted to the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE150568.
Supplementary Material
Acknowledgments
The authors thank Li Bing Lin for technical assistance, and Mark Van Doren and the lab of Brian Oliver for discussions. Services of scRNA-seq and demultiplexing of the sequencing reads were provided by BioTools. Flow cytometry and confocal microscopy were performed at the Core Facility and Imaging Center at Chang Gung University. This work was funded by a Ministry of Science and Technology grant from Taiwan (108-2628-B-182-007) to S.Y.Y. and Chang Gung Memorial Hospital grants to S.Y.Y. (CMRPD1K0231) and S.D.F. (CMRPD1J0251).
Author contributions: Conceptualization, S.Y.Y; data acquisition, H.-C.C., H.W.L, H.H.H., and S.Y.Y.; data analysis, S.Y.Y., Y.-R.L., and S.D.F.; writing, S.Y.Y., S.D.F., and Y.-R.L.; funding, S.Y.Y. and S.D.F.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at https://www.genome.org/cgi/doi/10.1101/gr.271148.120.
Competing interest statement
The authors declare no competing interests.
References
- Asaoka M, Hanyu-Nakamura K, Nakamura A, Kobayashi S. 2019. Maternal Nanos inhibits Importin-α2/Pendulin-dependent nuclear import to prevent somatic gene expression in the Drosophila germline. PLoS Genet 15: e1008090. 10.1371/journal.pgen.1008090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. 2009. MEME Suite: tools for motif discovery and searching. Nucleic Acids Res 37: W202–W208. 10.1093/nar/gkp335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J, Spielmann M, Qiu X, Huang X, Ibrahim DM, Hill AJ, Zhang F, Mundlos S, Christiansen L, Steemers FJ, et al. 2019. The single-cell transcriptional landscape of mammalian organogenesis. Nature 566: 496–502. 10.1038/s41586-019-0969-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casper AL, Van Doren M. 2009. The establishment of sexual identity in the Drosophila germline. Development 136: 3821–3830. 10.1242/dev.042374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y-L, King B, Lin S-C, Kennison JA, Huang D-H. 2007. A double-bromodomain protein, FSH-S, activates the homeotic gene ultrabithorax through a critical promoter-proximal region. Mol Cell Biol 27: 5486–5498. 10.1128/MCB.00692-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitiashvili T, Dror I, Kim R, Hsu FM, Chaudhari R, Pandolfi E, Chen D, Liebscher S, Schenke-Layland K, Plath K, et al. 2020. Female human primordial germ cells display X-chromosome dosage compensation despite the absence of X-inactivation. Nat Cell Biol 22: 1436–1446. 10.1038/s41556-020-00607-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline TW, Meyer BJ. 1996. Vive la différence: males vs females in flies vs worms. Annu Rev Genet 30: 637–702. 10.1146/annurev.genet.30.1.637 [DOI] [PubMed] [Google Scholar]
- Conrad T, Akhtar A. 2012. Dosage compensation in Drosophila melanogaster: epigenetic fine-tuning of chromosome-wide transcription. Nat Rev Genet 13: 123–134. 10.1038/nrg3124 [DOI] [PubMed] [Google Scholar]
- Deng X, Hiatt JB, Nguyen DK, Ercan S, Sturgill D, Hillier LW, Schlesinger F, Davis CA, Reinke VJ, Gingeras TR, et al. 2011. Evidence for compensatory upregulation of expressed X-linked genes in mammals, Caenorhabditis elegans and Drosophila melanogaster. Nat Genet 43: 1179–1185. 10.1038/ng.948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Digan ME, Haynes SR, Mozer BA, Dawid IB, Forquignon F, Gans M. 1986. Genetic and molecular analysis of fs(1)h, a maternal effect homeotic gene in Drosophila. Dev Biol 114: 161–169. 10.1016/0012-1606(86)90392-1 [DOI] [PubMed] [Google Scholar]
- Florence BL, Faller DV. 2008. Drosophila female sterile (1) homeotic is a multifunctional transcriptional regulator that is modulated by Ras signaling. Dev Dyn 237: 554–564. 10.1002/dvdy.21432 [DOI] [PubMed] [Google Scholar]
- Goecks J, Nekrutenko A, Taylor J. 2010. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol 11: R86. 10.1186/gb-2010-11-8-r86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyal R, Baxter E, Van Doren M. 2019. sisterless A is required for the activation of Sex lethal in the germline. bioRxiv 10.1101/2019.12.17.880070 [DOI]
- Grant CE, Bailey TL, Noble WS. 2011. FIMO: scanning for occurrences of a given motif. Bioinformatics 27: 1017–1018. 10.1093/bioinformatics/btr064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanyu-Nakamura K, Sonobe-Nojima H, Tanigawa A, Lasko P, Nakamura A. 2008. Drosophila Pgc protein inhibits P-TEFb recruitment to chromatin in primordial germ cells. Nature 451: 730–733. 10.1038/nature06498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashiyama K, Hayashi Y, Kobayashi S. 2011. Drosophila Sex lethal gene initiates female development in germline progenitors. Science 333: 885–888. 10.1126/science.1208146 [DOI] [PubMed] [Google Scholar]
- Jevitt A, Chatterjee D, Xie G, Wang X-F, Otwell T, Huang Y-C, Deng W-M. 2020. A single-cell atlas of adult Drosophila ovary identifies transcriptional programs and somatic cell lineage regulating oogenesis. PLoS Biol 18: e3000538. 10.1371/journal.pbio.3000538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson AD, Alberio R. 2015. Primordial germ cells: the first cell lineage or the last cells standing? Development 142: 2730–2739. 10.1242/dev.113993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jongens TA, Hay B, Jan LY, Jan YN. 1992. The germ cell-less gene product: a posteriorly localized component necessary for germ cell development in Drosophila. Cell 70: 569–584. 10.1016/0092-8674(92)90427-E [DOI] [PubMed] [Google Scholar]
- Kim-Ha J, Kerr K, Macdonald PM. 1995. Translational regulation of oskar mRNA by Bruno, an ovarian RNA-binding protein, is essential. Cell 81: 403–412. 10.1016/0092-8674(95)90393-3 [DOI] [PubMed] [Google Scholar]
- Le Bras S, Van Doren M. 2006. Development of the male germline stem cell niche in Drosophila. Dev Biol 294: 92–103. 10.1016/j.ydbio.2006.02.030 [DOI] [PubMed] [Google Scholar]
- Lehmann R, Nüsslein-Volhard C. 1986. Abdominal segmentation, pole cell formation, and embryonic polarity require the localized activity of oskar, a maternal gene in Drosophila. Cell 47: 141–152. 10.1016/0092-8674(86)90375-2 [DOI] [PubMed] [Google Scholar]
- Li L, Dong J, Yan L, Yong J, Liu X, Hu Y, Fan X, Wu X, Guo H, Wang X, et al. 2017. Single-cell RNA-seq analysis maps development of human germline cells and gonadal niche interactions. Cell Stem Cell 20: 858–873.e4. 10.1016/j.stem.2017.03.007 [DOI] [PubMed] [Google Scholar]
- Mahadevaraju S, Fear JM, Akeju M, Galletta BJ, Pinheiro MMLS, Avelino CC, Cabral-de-Mello DC, Conlon K, Dell'Orso S, Demere Z, et al. 2021. Dynamic sex chromosome expression in Drosophila male germ cells. Nat Commun 12: 892. 10.1038/s41467-021-20897-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInnes L, Healy J, Melville J. 2018. UMAP: Uniform Manifold Approximation and Projection for dimension reduction. arXiv:1802.03426 [stat.ML].
- Meiklejohn CD, Presgraves DC. 2012. Little evidence for demasculinization of the Drosophila X chromosome among genes expressed in the male germline. Genome Biol Evol 4: 1007–1016. 10.1093/gbe/evs077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikhaylova LM, Nurminsky DI. 2011. Lack of global meiotic sex chromosome inactivation, and paucity of tissue-specific gene expression on the Drosophila X chromosome. BMC Biol 9: 29. 10.1186/1741-7007-9-29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray SM, Yang SY, Van Doren M. 2010. Germ cell sex determination: a collaboration between soma and germline. Curr Opin Cell Biol 22: 722–729. 10.1016/j.ceb.2010.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura A, Amikura R, Mukai M, Kobayashi S, Lasko PF. 1996. Requirement for a noncoding RNA in Drosophila polar granules for germ cell establishment. Science 274: 2075–2079. 10.1126/science.274.5295.2075 [DOI] [PubMed] [Google Scholar]
- Nakamura S, Hira S, Fujiwara M, Miyagata N, Tsuji T, Kondo A, Kimura H, Shinozuka Y, Hayashi M, Kobayashi S, et al. 2019. A truncated form of a transcription factor Mamo activates vasa in Drosophila embryos. Commun Biol 2: 422. 10.1038/s42003-019-0663-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu W, Spradling AC. 2020. Two distinct pathways of pregranulosa cell differentiation support follicle formation in the mouse ovary. Proc Natl Acad Sci 117: 20015–20026. 10.1073/pnas.2005570117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver B, Perrimon N, Mahowald AP. 1987. The ovo locus is required for sex-specific germ line maintenance in Drosophila. Genes Dev 1: 913–923. 10.1101/gad.1.9.913 [DOI] [PubMed] [Google Scholar]
- Pauli D, Oliver B, Mahowald AP. 1993. The role of the ovarian tumor locus in Drosophila melanogaster germ line sex determination. Development 119: 123–134. [DOI] [PubMed] [Google Scholar]
- Pemberton JR. 1975. Retention of mercurial preservatives in desiccated biological products. J Clin Microbiol 2: 549–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez F, Dündar F, Diehl S, Grüning BA, Manke T. 2014. deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res 42: W187–W191. 10.1093/nar/gku365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastelli L, Kuroda MI. 1998. An analysis of maleless and histone H4 acetylation in Drosophila melanogaster spermatogenesis. Mech Dev 71: 107–117. 10.1016/S0925-4773(98)00009-4 [DOI] [PubMed] [Google Scholar]
- Raudvere U, Kolberg L, Kuzmin I, Arak T, Adler P, Peterson H, Vilo J. 2019. g:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res 47: W191–W198. 10.1093/nar/gkz369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rust K, Byrnes LE, Yu KS, Park JS, Sneddon JB, Tward AD, Nystul TG. 2020. A single-cell atlas and lineage analysis of the adult Drosophila ovary. Nat Commun 11: 5628. 10.1038/s41467-020-19361-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangrithi MN, Royo H, Mahadevaiah SK, Ojarikre O, Bhaw L, Sesay A, Peters AHFM, Stadler M, Turner JMA. 2017. Non-canonical and sexually dimorphic X dosage compensation states in the mouse and human germline. Dev Cell 40: 289–301.e3. 10.1016/j.devcel.2016.12.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos AC, Lehmann R. 2004. Germ cell specification and migration in Drosophila and beyond. Curr Biol 14: R578–R589. 10.1016/j.cub.2004.07.018 [DOI] [PubMed] [Google Scholar]
- Saunders A, Core LJ, Sutcliffe C, Lis JT, Ashe HL. 2013. Extensive polymerase pausing during Drosophila axis patterning enables high-level and pliable transcription. Genes Dev 27: 1146–1158. 10.1101/gad.215459.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Z, Lim C, Tran V, Cui K, Zhao K, Chen X. 2020. Single-cyst transcriptome analysis of Drosophila male germline stem cell lineage. Development 147: dev184259. 10.1242/dev.184259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigenobu S, Arita K, Kitadate Y, Noda C, Kobayashi S. 2006. Isolation of germline cells from Drosophila embryos by flow cytometry. Dev Growth Differ 48: 49–57. 10.1111/j.1440-169X.2006.00845.x [DOI] [PubMed] [Google Scholar]
- Slaidina M, Banisch TU, Gupta S, Lehmann R. 2020. A single-cell atlas of the developing Drosophila ovary identifies follicle stem cell progenitors. Genes Dev 34: 239–249. 10.1101/gad.330464.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Specht AT, Li J. 2017. LEAP: constructing gene co-expression networks for single-cell RNA-sequencing data using pseudotime ordering. Bioinformatics 33: 764–766. 10.1093/bioinformatics/btw729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimori S, Kumata Y, Kobayashi S. 2018. Maternal Nanos-dependent RNA stabilization in the primordial germ cells of Drosophila embryos. Dev Growth Differ 60: 63–75. 10.1111/dgd.12414 [DOI] [PubMed] [Google Scholar]
- Sybirna A, Wong FCK, Surani MA. 2019. Genetic basis for primordial germ cells specification in mouse and human: conserved and divergent roles of PRDM and SOX transcription factors. Curr Top Dev Biol 135: 35–89. 10.1016/bs.ctdb.2019.04.004 [DOI] [PubMed] [Google Scholar]
- Traag VA, Waltman L, van Eck NJ. 2019. From Louvain to Leiden: guaranteeing well-connected communities. Sci Rep 9: 5233. 10.1038/s41598-019-41695-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner JMA. 2015. Meiotic silencing in mammals. Annu Rev Genet 49: 395–412. 10.1146/annurev-genet-112414-055145 [DOI] [PubMed] [Google Scholar]
- Van Doren M, Williamson AL, Lehmann R. 1998. Regulation of zygotic gene expression in Drosophila primordial germ cells. Curr Biol 8: 243–246. 10.1016/S0960-9822(98)70091-0 [DOI] [PubMed] [Google Scholar]
- Vibranovski MD, Zhang YE, Kemkemer C, Lopes HF, Karr TL, Long M. 2012. Re-analysis of the larval testis data on meiotic sex chromosome inactivation revealed evidence for tissue-specific gene expression related to the Drosophila X chromosome. BMC Biol 10: 49. 10.1186/1741-7007-10-49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wawersik M, Milutinovich A, Casper AL, Matunis E, Williams B, Van Doren M. 2005. Somatic control of germline sexual development is mediated by the JAK/STAT pathway. Nature 436: 563–567. 10.1038/nature03849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SY, Baxter EM, Van Doren M. 2012. Phf7 controls male sex determination in the Drosophila germline. Dev Cell 22: 1041–1051. 10.1016/j.devcel.2012.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatsu J, Hayashi M, Mukai M, Arita K, Shigenobu S, Kobayashi S. 2008. Maternal RNAs encoding transcription factors for germline-specific gene expression in Drosophila embryos. Int J Dev Biol 52: 913–923. 10.1387/ijdb.082576jy [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.