Abstract
Genomic deletions provide a powerful loss-of-function model in noncoding regions to assess the role of purifying selection on genetic variation. Regulatory element function is characterized by nonuniform tissue and cell type activity, necessarily linking the study of fitness consequences from regulatory variants to their corresponding cellular activity. We generated a callset of deletions from genomes in the Alzheimer's Disease Neuroimaging Initiative (ADNI) and used deletions from The 1000 Genomes Project Consortium (1000GP) in order to examine whether purifying selection preserves noncoding sites of chromatin accessibility marked by DNase I hypersensitivity (DHS), histone modification (enhancer, transcribed, Polycomb-repressed, heterochromatin), and chromatin loop anchors. To examine this in a cellular activity-aware manner, we developed a statistical method, pleiotropy ratio score (PlyRS), which calculates a correlation-adjusted count of “cellular pleiotropy” for each noncoding base pair by analyzing shared regulatory annotations across tissues and cell types. By comparing real deletion PlyRS values to simulations in a length-matched framework and by using genomic covariates in analyses, we found that purifying selection acts to preserve both DHS and enhancer noncoding sites. However, we did not find evidence of purifying selection for noncoding transcribed, Polycomb-repressed, or heterochromatin sites beyond that of the noncoding background. Additionally, we found evidence that purifying selection is acting on chromatin loop integrity by preserving colocalized CTCF binding sites. At regions of DHS, enhancer, and CTCF within chromatin loop anchors, we found evidence that both sites of activity specific to a particular tissue or cell type and sites of cellularly pleiotropic activity are preserved by selection.
Large-scale sequencing studies have provided tremendous insight into biological function and human disease, with statistical signatures of natural selection serving as a primary identifying feature. The classic example is the analysis of selective constraints on protein-coding genes evident from the depletion of missense or nonsense genetic variants. These advances, however, are not directly translatable to the analysis of noncoding DNA, which has increasingly become a focus of human genetics research. Genomic studies have revealed numerous regions of regulatory activity marked by chromatin accessibility or histone modification (The ENCODE Project Consortium 2012; Roadmap Epigenomics Consortium et al. 2015). Association signals for common human phenotypes are enriched in these regulatory regions of the genome (Maurano et al. 2012; Trynka et al. 2013; Gusev et al. 2014; Finucane et al. 2015), showcasing the importance of specialized cellular function. In contrast to protein-coding sequences, the function of regulatory sequences is not determined by triplet codon structure, thereby providing no obvious analog to protein-truncating single-nucleotide variants (SNVs) to identify loss of function. This lack of knowledge of the mutational consequences of individual nucleotides within regulatory sequences complicates the ability to study their function through the lens of purifying natural selection. Previous work focusing on SNVs within noncoding regions developed sophisticated genetic models that relied on functional proxies such as transcription factor binding sites, nucleotide conservation across species, or machine learning (Kircher et al. 2014; Ritchie et al. 2014; Lee et al. 2015; Quang et al. 2015; Zhou and Troyanskaya 2015; Ionita-Laza et al. 2016; Huang et al. 2017; Rojano et al. 2019). However, it is difficult to clearly interpret these findings in terms of selection against the disruption of regulation. In contrast to SNVs, deletions are a class of variation that provide a direct loss of normal regulatory function at a locus by physically removing the sequence of a regulatory element in at least a heterozygous manner. This logic underlies experimental studies of regulatory function using CRISPR-Cas9 systems (Zhu et al. 2016; Liu et al. 2017). Yet, natural population genetic variation provides a more unbiased genome-wide view of the action of selection on deletions. Work performed by sequencing consortia has shown the reduction of deletion variation in various categories of regulatory sequences (Sudmant et al. 2015a,b; Abel et al. 2020).
The hallmark of human regulatory loci is their nonuniform activity across tissues and cell types. Here, we offer a population genetic analysis of natural deletions in light of variable regulatory activity across tissues and cell types (collectively called “biosamples”). Deletions that remove sites of genomic regulation with pleiotropic cellular effects (what we term “cellular pleiotropy”; i.e., the same regulatory locus is active in more than one biosample) might be expected to be, on average, more deleterious (i.e., fitness-reducing) than deletions that remove sites with activity specific to a particular biosample, because any changes at the DNA level to the cellularly pleiotropic regulatory loci potentially affect multiple biosamples simultaneously. Another possibility is that because particular biosample regulation is what enables widespread cellular diversity, these regulatory sites must be under strong selective constraint to preserve their specialized biological function. These two potential modes of selection that preserve the regulation of cellular activity are not mutually exclusive, as selection may be operating to remove overlapping deletions to preserve the utility of both types of regulatory loci. Prior work has provided suggestive evidence that biosample activity count is a contributor to selective constraint in regulatory sequences (Cheng et al. 2014; Huang et al. 2017; Quiver and Lachance 2018; Abel et al. 2020; Xu et al. 2020). Studying purifying selection on noncoding deletions is thus inherently tied to the cellular activity of corresponding deleted regulatory sequences. These previous studies, although providing great contributions to the field, do not fully grant clarity of interpretation for examining these questions, however, because biosample counts were inflated owing to a high correlation of the cellular activity among the tissues and cell types analyzed. Thus, the method used to numerically count affected cellular activity influences interpretation of results. To address this, we have developed a statistical method, pleiotropy ratio score (PlyRS), to quantify the amount of biosample activity (i.e., cellular pleiotropy) for individual nucleotides in light of the hierarchical developmental structure of human biosamples, while controlling for their correlation rather than using a simple biosample count. We then analyzed separately several diverse epigenomic features (open chromatin, histone modifications, and chromatin loop anchors), taking into account nonindependence of these individual annotations across biosamples by using our PlyRS values. In this way, we assessed the effect of purifying selection on millions of nucleotide positions in the human noncoding genome by examining patterns of PlyRS values within naturally occurring deletion sequences.
The reduction of genetic variation and a shift in the allele frequency (AF) spectrum (AFS) toward rare variants are two key signatures of purifying selection. If selection is operating on the removal of deleterious deletions overlapping regulatory regions, we would expect to see both a reduction in deletion variation overlapping the important regulatory features and a shift in the AFS of remaining overlapping deletions toward rarer alleles, relative to neutral expectations. These conditions on segregating deletions should be simultaneously present to confidently conclude that purifying natural selection is acting to preserve a particular regulatory epigenomic feature, as either reduced deletion counts or a shift in the deletion AFS alone may indicate deletion-calling artifacts or confounding genomic covariates. Both of these signatures are prone to misspecification from various technical or biological confounders, particularly for structural variation. For example, the accuracy of deletion calls is influenced by their length and AF (Huddleston and Eichler 2016). Longer deletions have more prevalent missing coverage, and common deletions are observed more often in the population, so these types of deletions are more likely to be correctly identified using current methods based on analyzing short-read sequencing data. Variant calling accuracy also depends on the mappability of the sequence (Treangen and Salzberg 2012). Therefore, the observed negative correlation of deletion length and AF (Mills et al. 2011; Sudmant et al. 2015a) could be owing to these deletion-caller algorithm biases, underlying biology, or both.
In addition to technical confounders, biological factors unrelated to the direct pressure of selection may affect the number of segregating mutations and the AFS. For example, the amount of variation is linearly proportional to mutation rate; however, the deletion mutation rate at fine-scale is still unknown and could be influenced by sequence GC content and other local genomic properties. In contrast to the overall amount of variation, AFS does not depend on mutation rate when examined in the form of the distribution of relative proportions of variants with a given frequency. In this representation, for small populations in which there is only one mutation event per segregating site, as relevant for this work, AFS depends on the genealogical history of the site and does not depend on mutation rate. This is important for our analysis, because the AFS test shows that the depletion of deletions is not owing to reduction of mutation rate. Together with amount of variation, however, AFS could be influenced by complex mechanisms like background selection (Cvijović et al. 2018).
To address the technical and population genetic confounding effects described above, we simulated length-matched positions of each real deletion while keeping the original AF label and took into account relevant genomic confounding variables co-occurring with the same deletion. We examined PlyRS values within deletion coordinates, checking for a reduction in observed PlyRS values compared with simulations that would be indicative of purifying selection. We also checked for a shift in the AFS of overlapping deletions toward rarer alleles while examining the magnitude of PlyRS depletion compared with simulations. By using this analysis framework of PlyRS values, we assessed the potential of purifying selection to preserve noncoding epigenomic features by comparing the observed diversity and AFS of real deletions to the expectations based on simulations.
Results
Pleiotropy ratio score
We score deletions with respect to their effect on regulatory function by considering deletion overlap in the context of both the number of removed regulatory sites and the activity of each site across biosamples. In contrast to SNVs, a noncoding deletion can potentially remove regulatory function at a genomic locus along two distinct “axes” (for a cartoon, see Fig. 1A). The horizontal axis corresponds to the amount of regulatory space removed by the deletion irrespective of its biosample activity. The vertical axis corresponds to the combined amount of regulatory activity across biosamples of each base pair (i.e., the cellular pleiotropy of a regulatory coordinate). Thus, for any deletion overlapping regulatory sequences, there will be a simultaneous removal at that locus along both axes, which we quantify by a counting score for each axis.
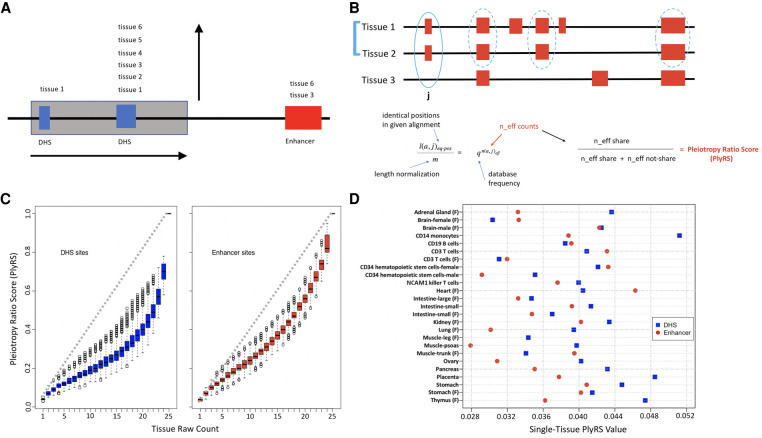
Figure 1.
Pleiotropy ratio score (PlyRS). (A) Purifying selection against deletions can operate along two “axes” at a genomic locus. A deletion (here shaded gray) can remove putative genomic function along a horizontal axis by overlapping one or more regulatory sites (here overlapping two DHSs [hotspot; blue] but missing an enhancer [H3K4me1; red]). The regulatory sites overlapped can have annotated activity specific to a particular biosample (as the leftmost DHS) or cellularly pleiotropic activity (as the rightmost DHS), thus removing putative genomic function along a vertical axis. (B) Schematic of derivation of PlyRS. By using biosamples sharing an annotation at a given base pair, we compute effective number (n_eff) of biosamples by equating the observed sharing fraction to that under the expectation of independent annotations. PlyRS is given by n_eff values normalized across biosamples. (C) Comparison of PlyRS to simple biosample counts. PlyRS can range from zero, representing no regulatory activity at a particular base pair in any biosample, to one, representing regulatory activity in all biosamples analyzed. Because PlyRS accounts for the positive regulatory activity correlation between biosamples analyzed, PlyRS (y-axis) will always fall at, or below, the diagonal versus a simple count (x-axis). Each regulatory feature will display a different PlyRS distribution (e.g., the enhancer feature [H3K4me1; right] has a PlyRS distribution that lies closer to the diagonal than for the DHS feature [left]), based on the activity covariance of the biosamples that are analyzed of that regulatory feature. Boxplot midlines correspond to the median PlyRS value, with the box delimiting the second and third quartile range and the whiskers extending from box edges out up to 1.5 times the box range or the furthest PlyRS value if within that bound. (D) Comparison of PlyRS values for sites of activity specific to a particular biosample. When regulatory activity at a base pair is annotated as specific to a particular biosample, each biosample will have a PlyRS value corresponding to it that may be below, at, or above an otherwise simple count of 0.04 (i.e., one biosample divided by 25 total possible biosamples analyzed). This is owing to PlyRS up-weighting biosamples that have relatively rare activity genome-wide and down-weighting biosamples with relatively common activity. Because of the highly correlated nature of DHS biosamples (blue squares) to one another (or enhancer biosamples [red circles] to one another) among the biosamples that we analyzed, many biosamples fall below a count of 0.04 when regulatory activity at a base pair is specific to a particular biosample.
A single deletion, depending on its length, may remove one or more adjacent regulatory elements. For the horizontal axis, we calculate the amount of deleted regulatory space on a per-annotation basis. We examine annotations at the base pair level rather than element level because there is a mismatch between the start and end coordinates of most pleiotropic regulatory elements when compared across biosamples (Roadmap Epigenomics Consortium et al. 2015). The base pair annotation does not require fixing precise coordinate boundaries between each biosample, enabling regulatory site annotations to be equally compared across biosamples. We do not require the removal of an entire regulatory element for the horizontal count because even a partial deletion of a regulatory element might render it inoperable (Ibn-Salem et al. 2014). For example, if 15 regulatory base pairs are deleted, this might correspond to roughly 1/10 of the regulatory element being deleted (using an average regulatory element coordinate length of 150 bp) (John et al. 2011), therefore serving as a scaled proxy of real regulatory element removal. Consequently, for a deletion overlapping regulatory space, the horizontal axis count score can range from as low as one (only a single regulatory base pair deleted in any biosample) to as high as the length of the deletion (all base pairs along the deletion length overlap a regulatory annotation).
The vertical axis measures the breadth of cellular activity deleted at a particular genomic locus. A simple numerical count of the number of biosamples where regulatory space has activity is not sufficient for properly specifying cellular pleiotropy; this count can be heavily influenced by the cellular diversity of the particular biosamples included in the analysis. This would be particularly true in the case of a subset of highly correlated biosamples, such as blood cells, dominating a data set. For example, a count of three in an analysis performed with heart tissue, lung tissue, and 10 blood cell types would not have the same interpretation as a count of three in an analysis performed with heart tissue, lung tissue, and only one blood cell type. In the former, it could be that the count of three comes from three highly correlated blood cell types, but in the latter, the count of three would have to come from the more developmentally diverse set of all three tissues. Therefore, in recognition of these issues, we developed a statistical method, called pleiotropy ratio score (PlyRS; pronounced “ply-ers” as in the pliers hand tool), which calculates a correlation-adjusted count of cellular pleiotropy among the biosamples analyzed. This count is calculated per base pair for each regulatory annotation separately.
To construct PlyRS, we adapted the PSIC method (Sunyaev et al. 1999), which was originally developed to assess independent observations when looking at a multiple sequence alignment of amino acid substitutions. Figure 1B provides a helpful schematic of the derivation of PlyRS. For a set of biosamples analyzed (three in Fig. 1B), we find the biosamples that share a regulatory annotation at a particular base pair (in Fig. 1B, position j has Tissue 1 and Tissue 2 sharing a regulatory annotation). We compare the genomic fraction of positions with shared annotation to that expected if biosamples were independent (presented as independent Bernoulli experiments with the corresponding annotation frequencies, q). We then calculate effective number of independent biosamples (“n_eff share”) as the number of independent biosamples that would have the same shared fraction of annotation as the observed data. This gives us an adjusted count based on the underlying correlation between the biosamples analyzed. We similarly compute effective number of biosamples that are not annotated in this position (“n_eff not-share” in Fig. 1B; position j has Tissue 3 not sharing a regulatory annotation). The effective number in either case will not be very informative by itself, because it would scale differently at each base pair depending on the set of biosamples sharing a regulatory annotation at that position, especially when the number of biosamples analyzed is large. Therefore, from the two effective numbers (n_eff share and n_eff not-share), we derive the PlyRS by taking the n_eff counts from sharing and divide by the total n_eff counts from both sharing and not sharing.
At any base pair coordinate within a deletion, the PlyRS value scale can range from zero to a maximum of one. A PlyRS value of zero corresponds to base pair for which there is no annotated regulatory activity at that genomic position in any biosample analyzed in the total set of biosamples. Conversely, a PlyRS value of one corresponds to base pair for which there is annotated regulatory activity at that genomic position in all biosamples analyzed. Between these extreme bounds, the counts of regulatory sites active in biosamples with common activity will be down-weighted, whereas the counts of sites active in biosamples with rare activity will be up-weighted. Figure 1C illustrates how PlyRS corresponds to the simple biosample counts. Similarly, for each base pair that has activity specific to a particular biosample, the PlyRS value of that base pair will be different depending on which particular biosample has activity and how its activity covaries across the genome with the other biosamples being analyzed (see Fig. 1D). For our purposes, we define a deletion at a regulatory locus as having both horizontal and vertical axis components, even if that deletion overlaps a regulatory annotation with activity only specific to a particular biosample.
Construction of deletion and regulatory data sets
To examine selective constraints on deletions within regulatory regions, we needed fine-resolution of genomic coordinates for both deletions and regulatory regions as well as high-confidence deletion AFs from population data. For this, we compiled deletion data from two callsets and regulatory data from seven callsets, and we applied additional filters relevant to our analysis. For additional criteria used to ensure high-quality data sets, see Methods.
We used deletions that we called and genotyped (see Supplemental Note S1; Supplemental Figs. S2–S4; Supplemental Tables S13, S14) across 752 individuals sequenced as part of the Alzheimer's Disease Neuroimaging Initiative (ADNI) (Petersen et al. 2010), using the CNV algorithm Genome STRiP (Handsaker et al. 2011). We additionally used deletion data (Supplemental Note S2) from The 1000 Genomes Project Consortium (1000GP) phase 3 callset of breakpoint-resolved deletions that had been genotyped in 2504 individuals from 26 modern human populations (Sudmant et al. 2015a). We restricted our analysis to noncoding deletions. As expected, the bulk (>80%) of deletions in our data sets remaining after filtering were rare (≤1% AF).
To analyze genomic deletions within regulatory regions, we used biochemical data associated with regulatory activity from the NIH Roadmap Epigenomics Consortium (REC) (Roadmap Epigenomics Consortium et al. 2015). In particular, we used two callsets of chromatin accessibility data (DNase I hypersensitivity [DHS]) and four callsets of histone modification data (H3K4me1 “enhancer,” H3K36me3 “transcribed,” H3K27me3 “Polycomb-repressed,” and H3K9me3 “heterochromatin”). Two sets of DHS annotation (“hotspot” and “MACS”) were used to check for consistency in the analyses. DHS annotations are typically associated with sites of open chromatin, allowing accessibility for regulator binding, and histone annotations are typically associated with sites of specific regulatory activity, as noted. Although histone annotations from REC callsets may be located widely throughout the genome, activity marks such as H3K4me1 (enhancer) are often located within DHS sites, although not exclusively. Even though we used high-probability regulatory annotations (Supplemental Note S3) to define regulatory loci, the loci identified should be considered as containing candidate regulatory elements because of the nature of the annotations. For example, histone modification may spread beyond the regulatory element at which it initiates, and we did not attempt to define the core regulatory element in order to be conservative in our downstream analyses. We additionally used data that demarcate chromatin loop anchors (Rao et al. 2014), which are associated as defining local genomic regions of physically interacting DNA.
Depletion of variation at DHS or enhancer sites
We first tested whether there was evidence of the depletion of noncoding deletion variation overlapping chromatin accessibility or histone modification. The depletion should be measured with respect to a background distribution of deletions along the genome in the absence of selection. We constructed this background distribution using simulations (Supplemental Note S4). For each real deletion in both the 1000GP and ADNI data sets, we randomly placed 1000 deletions of the same length to occur on the same chromosome and same noncoding genomic compartment space (intronic or intergenic). We confined to uniquely mappable sequence coordinates for both real deletions and simulated deletions. This procedure corrects for the confounding effects of mappability, deletion length, and AF. We summed the PlyRS values calculated per base pair along the length of every deletion. This sum, denoted PlyRSsum (Supplemental Note S5), corresponds to the total cellular pleiotropy (for a specific regulatory feature) of the deletion, encompassing both the horizontal and vertical axes along which purifying selection may be operating on the deletion. To evaluate depletion, we first compared the PlyRSsum of each real deletion to its own simulated counterparts by computing an empirical P-value. We then summed logarithms of the empirical P-values of all the individual comparisons and estimated the overall depletion across the deletion set by comparing this sum to the background expectation using Cohen's d-statistic. For more detail on our procedure of generating length-matched simulations and the calculation of depletion, see Supplemental Notes S4.2 and S6, respectively.
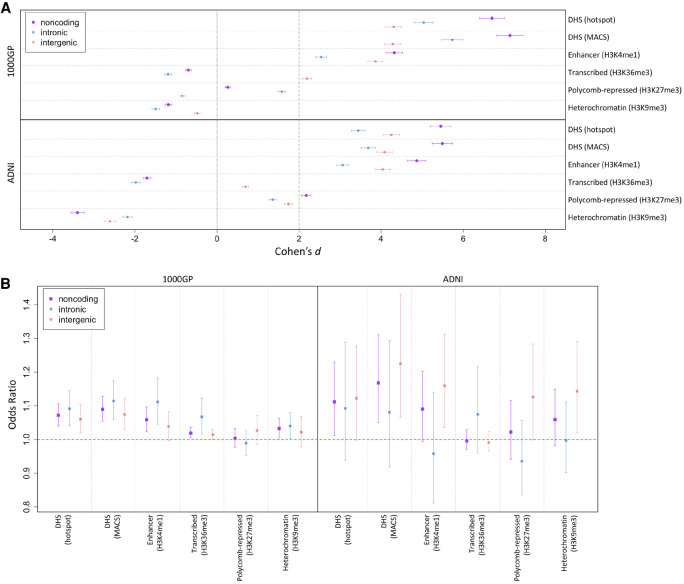
Figure 2A shows PlyRSsum effect sizes from comparing real data to simulations and indicates significant depletion of deletions (Cohen's d > 2, corresponding to 2 SD) overlapping noncoding DHS or enhancer regions. The depletion of deletions overlapping DHS or enhancer sites was significant not only in the full deletion sets but also in both the intronic and intergenic genomic compartments. Additionally, we found concordance between effect sizes in the 1000GP and ADNI data sets for DHS or enhancer deletion depletions, suggesting reliable capture of biological information from deletion callsets with differing characteristics. These results suggest that purifying selection may be operating broadly on deletions to preserve DHS and enhancer epigenomic features. We did not detect a significant depletion for deletions overlapping noncoding transcribed, Polycomb-repressed, or heterochromatin epigenomic features. For this analysis, Cohen's d corresponding to zero indicates a level of depletion that is consistent with that of the noncoding background for the nonrepetitive coordinates that we allow (see Supplemental Note S4.1). A negative Cohen's d suggests that a regulatory feature may be enriched for overlapping deletions compared with expectation; however, given the restricted set of noncoding coordinates we allow, our analysis methodology is not designed to provide indications of enrichment, as a different set of allowable coordinates may be more appropriate in those situations. Supplemental Table S1 lists the effect sizes found in the depletion simulations.
Figure 2.
Depletion of deletions and shift of deletion allele frequency (AF) spectrum (AFS) overlapping regulatory sites. (A) We calculated PlyRSsum for every deletion to quantify overlap with sites of chromatin accessibility or histone modification. We plot the degree of reduction in the PlyRSsum for real deletions relative to simulation. This reduction is measured using Cohen's d, which is the effect size of a t-test on PlyRSsum values in units of SD (plotted with 95% confidence intervals [CIs] showing the uncertainty owing to the finite number of simulations). Two units of effect size (Cohen's d = 2) approximately correspond to the 95% CI of significance in depletion. Higher values of Cohen's d indicate larger depletion within those sets compared with simulation. In presence of the true effect, there is a sample-size dependence on the underlying t-test, and the expected value of Cohen's d would be higher for larger data sets. (B) For each deletion, we determined the magnitude of PlyRSsum depletion, calculated as a ratio between its PlyRSsum and the average PlyRSsum of its length-matched simulated counterparts, for sites of chromatin accessibility or histone modification. We tested whether PlyRSsum depletion magnitude depends on AF (deletions categorized as rare [AF ≤ 1%] or common), using multivariate logistic regression in the presence of genomic covariates. We plot the regression odds ratio with 95% profile likelihood-based CIs. Results above one indicate a positive correlation of the magnitude of PlyRSsum depletion with AF. This corresponds to an excess of rare alleles overlapping the regulatory feature in the real data set compared with simulation, which is the expected result for features being preserved by the action of purifying selection against overlapping deletions.
Shift in AFS at DHS or enhancer sites
We next tested whether there was a shift in the AFS of noncoding deletions overlapping the chromatin accessibility or histone modification epigenomic features. The analysis of AF distribution is important because the overall amount of variation can be confounded by mutation rate (unlike SNVs, we do not have good models for mutation rate along the genome for deletions) (Kloosterman et al. 2015). The AF distribution, when examined using proportions rather than mutation counts, does not depend on mutation rate for relatively small populations (within the limits of the infinite sites approximation), but owing to the recent explosive growth of the human population, this assumption may break down for extremely large sample sizes, at which point recent recurrent mutations become relevant. However, for the sample sizes analyzed here, the AF distribution can be assumed to be independent of mutation rate (Li 1997) because the chance of recurrent mutations is extremely small for deletions, which would require the start and end coordinates to be identical. Therefore, a shift in the AFS of real deletions in our data sets compared with simulated deletions would likely reflect the action of purifying selection at an extent greater than that of the rest of the noncoding genome that we analyze.
To test whether the magnitude of PlyRSsum depletion depends on AF, we used logistic regression. Binarized AF is the outcome variable (deletions categorized as rare [AF ≤ 1%] or common) and PlyRSsum as predictor (Supplemental Note S7). To measure the magnitude of PlyRSsum depletion for each deletion, we calculated a ratio between its PlyRSsum and the average PlyRSsum of its length-matched simulated deletions. If purifying selection is, in fact, acting against deletions overlapping regulatory features, we would expect the largest PlyRSsum depletions to be found in common deletions. Still, the AF distribution can be affected by a number of variables unrelated to selective pressure. We accounted for relevant genomic covariates by including them into the multivariate logistic regression model. To take into account the potential effect of background selection, we controlled for regional (50 kb ± deletion coordinates) SNV nucleotide diversity and recombination rate, as well as the distance to the nearest transcription start site. We additionally controlled for regional GC content. Because of technical confounders, AF is expected to be influenced by deletion length, so we also controlled for length explicitly.
Figure 2B shows that for deletions overlapping DHS sites, the odds ratio (OR) significantly (confidence interval [CI] 95%) exceeded one in both data sets. This indicates the action of purifying selection by PlyRSsum depletion magnitude showing positive correlation with AF. Additionally, for deletions overlapping enhancer sites, the OR significantly exceeded one in the 1000GP data set, whereas the lower CI boundary of the OR was nearly significant, at 0.995, in the ADNI data set. All intronic and intergenic genomic compartment sets for DHS or enhancer features had mean OR > 1 (except ADNI intronic enhancers at 0.96). Supplemental Table S2 lists the ORs found in the logistic regressions. These results suggest that purifying selection may be preserving noncoding DHS and enhancer epigenomic features by reducing AFs of overlapping deletions. On the other hand, there is a lack of consistent AF shift for genomic compartment sets for transcribed, Polycomb-repressed, and heterochromatin features in both deletion data sets, with the mean OR sometimes falling below one and the OR CI often extending below one. This indicates that any negative selection against deletions overlapping these features would be of a strength comparable to, or less than, that of the noncoding background. In light of the insufficient evidence across the data sets for an excess of rare alleles for these features, combined with the lack of reduction in variation described above, we focused the analysis below on noncoding DHS or enhancer epigenomic features that showed statistical significance of both key signatures of broad selection against overlapping deletions.
Differential selection on preserving cellular activity
The results described above have indicated that purifying selection is acting against the total cellular pleiotropic burden (PlyRSsum) of noncoding deletions, preserving both DHS and enhancer regulatory sites. However, these analyses do not clarify if purifying selection preserves DHS or enhancer sites of both activity specific to a particular biosample and cellularly pleiotropic activity. One possibility is that deletions removing regulatory sites active in multiple biosamples incur a greater fitness cost. Another possibility is that because sites with activity specific to a particular biosample are vital to organismal development, deletions removing them are subject to a stronger selective effect. It could also be the case that purifying selective pressure on deletions is acting to preserve both types of regulatory sites simultaneously. To distinguish between these scenarios, we calculated two additional PlyRS measures, PlyRSsum-mono and PlyRSsum-pleio (Supplemental Note S5). PlyRSsum-mono included the sum of PlyRS values of deleted base pairs for those only associated with regulatory activity specific to a particular biosample. PlyRSsum-pleio included the sum of PlyRS values of deleted base pairs for those associated with cellularly pleiotropic regulatory activity. The sum of these two components across the length of a deletion's coordinates is the original measure of total cellular pleiotropic burden, PlyRSsum. With these additional PlyRS measures, we performed the same analyses as above to examine both a potential reduction in variation and a shift in AF, now applied separately to each component of PlyRSsum. This allowed us to determine, within the same sets of real deletions, which scenario of regulatory activity preservation was contributing to the signal of depletion in variation and shift in the AFS as found above.
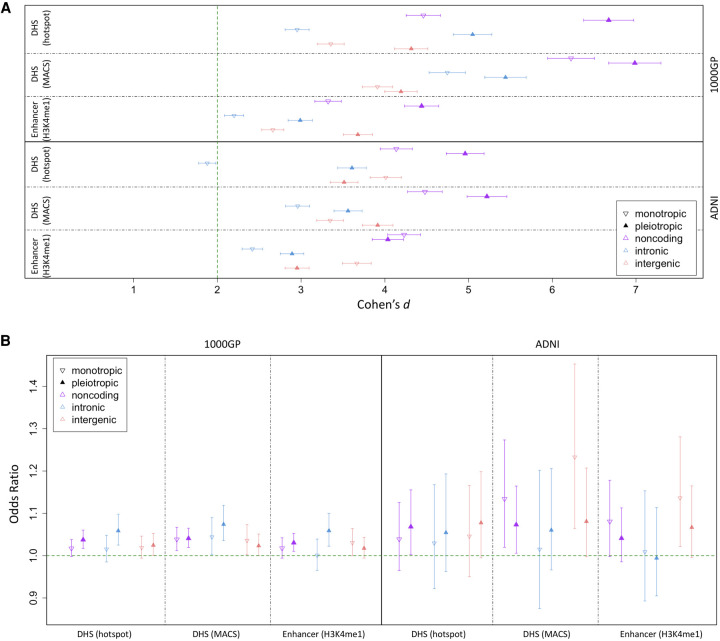
Figure 3A shows a significant depletion of variation for DHS or enhancer sites corresponding to both sites of activity specific to a particular biosample and cellularly pleiotropic activity in both the 1000GP and ADNI data sets. The effect size of this reduction in variation for PlyRSsum-mono or PlyRSsum-pleio was greater for PlyRSsum-pleio for both noncoding regulatory features, except for enhancer sites in ADNI deletions, where the effect size was comparable (error bars overlapping). Supplemental Tables S3 and S4 list the effect sizes found in the depletion simulations, including those for intronic and intergenic compartments where depletion values did not consistently favor greater reduction of PlyRSsum-pleio. Figure 3B shows that the magnitude of deletion depletion overlapping DHS or enhancer sites leads to a significantly shifted AFS at both sites of activity specific to a particular biosample and cellularly pleiotropic activity. For DHS or enhancer sites in all genomic compartments, the mean ORs of the magnitude of depletion for PlyRSsum-mono or PlyRSsum-pleio in association to AF were greater than one in both deletion data sets (except ADNI intronic enhancers) and were comparable between PlyRSsum-mono and PlyRSsum-pleio. Supplemental Tables S5 and S6 list the ORs found in the logistic regressions. These results collectively indicate that purifying selection is acting to preserve DHS or enhancer sites of activity specific to a particular biosample as well as cellularly pleiotropic activity.
Figure 3.
Depletion of deletions and shift of AFS overlapping DHS or enhancer sites of variable cellular activity. (A) We calculated PlyRSsum-mono (monotropic) and PlyRSsum-pleio (pleiotropic) for every deletion to quantify overlap with DHS or enhancer sites. We plot the degree of reduction in PlyRSsum-mono (or PlyRSsum-pleio) for real deletions relative to simulation measured using Cohen's d (with 95% CIs showing the uncertainty owing to the finite number of simulations). (B) For each deletion, we determined the magnitude of PlyRSsum-mono (monotropic) and PlyRSsum-pleio (pleiotropic) depletion, calculated as a ratio between its PlyRSsum-mono (or PlyRSsum-pleio) and the average PlyRSsum-mono (or PlyRSsum-pleio) of its length-matched simulated counterparts, for DHS or enhancer sites. We tested whether PlyRSsum-mono (or PlyRSsum-pleio) depletion magnitude depends on AF (deletions categorized as rare [AF ≤ 1%] or common), using multivariate logistic regression in the presence of genomic covariates. We plot the regression odds ratio with 95% profile likelihood-based CIs.
Purifying selection on CTCF sites within chromatin loop anchors
We also investigated whether there was evidence of depletion of variation and a shift in the AFS of deletions overlapping chromatin loop anchors. Chromatin loops are large regions of self-interacting DNA that facilitate cis-regulatory effects at a wider scale than that of individual regulators (Lupiáñez et al. 2015; Schoenfelder and Fraser 2019), and so, deletions removing a loop anchor (i.e., side endpoint of the chromatin loop) may be under strong purifying natural selection to preserve the loop integrity. The distance between loop anchors is greater than the longest deletions in our data sets, so deletions can only overlap, at most, one loop anchor. Additionally, the loop data are less precise than chromatin accessibility or histone modification annotations, so the number of base pairs of a deletion overlapping a loop anchor may not reflect actual deleteriousness of the mutation but rather correspond to imprecise annotations on the edges. These characteristics of loop annotation mean that using PlyRSsum to define the total cellular pleiotropy of overlapping deletions can propagate a potential bias in the measure. To avoid this and still test whether purifying selection may be operating on deletions overlapping loop anchors, we measured overlap both as a binary variable and by calculating the maximal PlyRS value (PlyRSmax) (Supplemental Note S5) along the length of an overlapping deletion. We performed the same analyses as for the chromatin accessibility or histone modification annotations.
Rao and Huntley et al. (2014) identified that a large majority (∼86%) of chromatin loop anchor loci had binding from the insulator protein CTCF, which ensures integrity of DNA looping and, consequently, chromatin loop fidelity (Guo et al. 2015; Nora et al. 2017). Given this critical function of CTCF and its presence within most loop anchors as specific binding points, we suspected that deletions that overlap loop anchors might be under stronger purifying selection if a deletion also simultaneously overlaps a CTCF site within the loop anchor, thereby removing a vital binding point. To elucidate this, in addition to identifying the full set of deletions overlapping chromatin loop anchors (loops), we further refined deletions into two subsets: deletions overlapping the loop anchor but not simultaneously overlapping a CTCF binding site (loopsnoCTCF) and deletions overlapping a loop anchor while simultaneously overlapping a CTCF binding site (loopsCTCF). Only ∼1% of all deletions in our data sets overlapped loopsCTCF, so we ignored intronic and intergenic designations in the analysis (but maintained them in simulations).
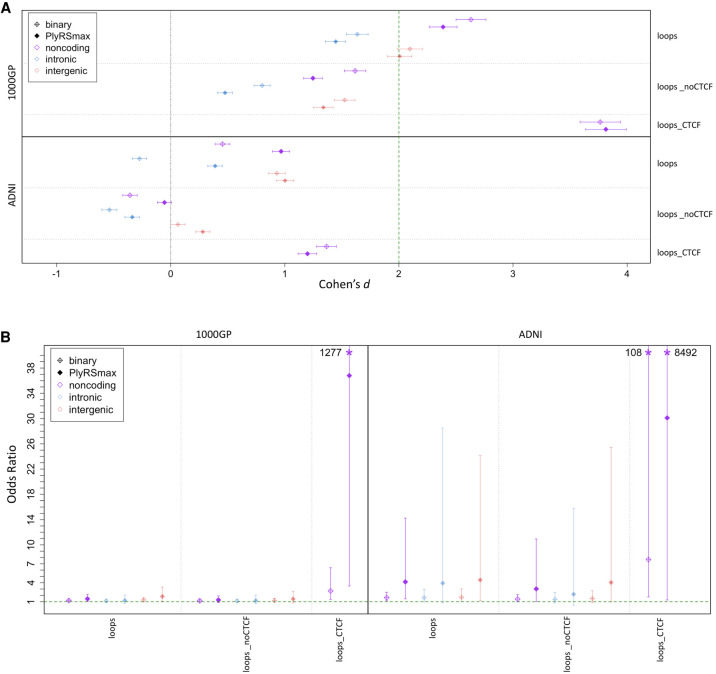
Figure 4A shows the effect sizes of binary overlap or PlyRSmax overlap from comparing real deletions to simulations and indicates that, with respect to the full set of loop anchors being overlapped (irrespective of whether CTCF sites are simultaneously overlapped), there was minimal depletion of deletion variation, if any. However, as also seen in Figure 4A, separation into the loopsnoCTCF and loopsCTCF subsets revealed that a signal of depletion was evident only for deletions overlapping loopsCTCF. Deletions in the ADNI data set showed the same characteristic pattern of greater reduction in variation in loopsCTCF versus loopsnoCTCF as was seen in the 1000GP data set; however, the reduction seen in ADNI deletions overlapping loopsCTCF was not statistically significant. We did not find any difference between the effect size of depletion for binary overlap compared with the PlyRSmax overlap measure, suggesting that there may not be stronger selection against deletions overlapping the most cellularly pleiotropic loopsCTCF. Supplemental Tables S7 and S8 list the effect sizes found in the chromatin loop anchor depletion simulations.
Figure 4.
Depletion of deletions and shift of AFS overlapping chromatin loop anchor sites. (A) We calculated a binary variable and PlyRSmax for every deletion to quantify overlap with loop sites. We plot the degree of reduction in the binary variable (or PlyRSmax) for real deletions relative to simulation measured using Cohen's d (with 95% CIs showing the uncertainty owing to the finite number of simulations). (B) For each deletion, we determined the magnitude of binary variable (or PlyRSmax) depletion, calculated as the difference between the binary variable (or PlyRSmax) and the average binary variable (or PlyRSmax) of its length-matched simulated counterparts, for loop sites. We tested whether binary variable (or PlyRSmax) depletion magnitude depends on AF (deletions categorized as rare [AF ≤ 1%] or common), using multivariate logistic regression in the presence of genomic covariates. We plot the regression odds ratio with 95% profile likelihood-based CIs. Asterisks denote odds ratio CIs that extend above the plotted y-axis, with values as indicated.
We also examined whether the depletion magnitude of binary overlap or PlyRSmax overlap at loop anchor loci showed dependence on AF using the same logistic regression framework as above for chromatin accessibility or histone modification annotations. Figure 4B shows evidence of a shift in the deletion AFS based on the magnitude of depletion at loopsCTCF, for which the mean OR estimate for binary overlap in 1000GP was 2.70 (minimum [min] 95% CI: 1.35) and in ADNI was 7.67 (min CI: 1.76). The mean OR estimate for PlyRSmax overlap of loopsCTCF in 1000GP was 36.80 (min CI: 3.49) and in ADNI was 30.11 (min CI: 1.27). The excess of rare alleles overlapping loopsCTCF exceeded the shift for loopsnoCTCF, which displayed only a modest effect in the ADNI data set (min CI: 1.02) and was not significant in the 1000GP data set. These results collectively suggest that purifying selection may be acting to preserve chromatin loop integrity by specifically preserving CTCF binding motifs within loop anchors. Supplemental Tables S9 and S10 list the ORs found in the chromatin loop anchor logistic regressions.
Discussion
By using the clarity of genomic deletions to identify loss of noncoding regulatory function, we used a novel data set of genomic deletions and a consortium-released deletion data set to determine whether purifying selection is operating to preserve noncoding regulatory loci. We examined sites of chromatin accessibility (DHS), histone modification (enhancer, transcribed, Polycomb-repressed, and heterochromatin), and chromatin loop anchors. Analysis of selection in the noncoding genome is motivated by prior findings in human genetics from genome-wide association studies that conclude most of heritability is owing to relatively common noncoding alleles within regulatory annotations (Maurano et al. 2012; Trynka et al. 2013; Gusev et al. 2014; Finucane et al. 2015). Initially, these findings appeared inconsistent with the expectation that disease-associated alleles are under pressure from purifying selection. However, recent studies showed that complex trait effect sizes are negatively correlated with AF, hinting at the action of purifying selection (Gazal et al. 2018; Zeng et al. 2018; Schoech et al. 2019). These observations put the question of the effect of noncoding regulatory alleles on function and fitness at the forefront of genomic studies ranging from basic evolutionary genetics to the allelic architecture of common human traits.
Broadly, our results are consistent with prior studies of SNV variation that indicated that certain putative regulatory sites are selectively constrained (Vernot et al. 2012; Ward and Kellis 2012; Huang et al. 2017). Recent analyses of structural variation (Abel et al. 2020; Xu et al. 2020) similarly underscored the importance of regulatory sequence for specifying critical cellular regulation. Our work characterizes purifying selection against complete removal of putative regulatory sites in the context of cellular activity, bringing together different aspects of previous work on regulatory selection accounting for dependency among biosamples while correcting for genomic confounders. Because the majority of deletion alleles in our data sets are rare, we make inferences regarding purifying selection on heterozygous mutations as we are underpowered to detect recessive selection.
Because a principal characteristic of human regulatory element function is their nonuniform activity across biosamples, interpreting fitness consequences from genetic variants in noncoding regions is inherently linked to corresponding regulatory site cellular activity, and proper quantification of this activity is critical for reliable interpretation of results. To incorporate this defining feature into the study of noncoding purifying selection, we developed a statistical method, PlyRS, which quantifies the extent of abundance of cellularly pleiotropic activity for individual base pairs. By use of the PlyRS method, our results indicate that purifying selection acts on both DHS and enhancer sites, as evident by both the depletion of deletions overlapping these annotations and a shift in the AFS of overlapping deletions toward rare alleles.
By using simulated deletions in a length-matched framework and covariate-aware analyses, we found statistically significant evidence at noncoding DHS or enhancer regions that both sites of activity specific to particular biosample activity and cellularly pleiotropic activity are preserved by selection. This finding establishes not only that some individual DHS or enhancer single-activity regulatory sites are selectively preserved but also that these single-activity biosamples are preserved as a class, indicating that selective preservation for these is likely the general rule rather than the exception. We found some indication that cellularly pleiotropic variants may be subject to a stronger reduction in variation than variants specific to a particular biosample, although the difference in AFS shifts is not significant. Because ADNI deletions are rarer, on average, than 1000GP deletions and because the AFS test we used collapses all rare deletions into a single class, it may be expected that real associations might appear statistically weaker in the ADNI data set for this test format. Additional analysis on larger data sets would be needed to accurately quantify the relative contributions of selection on sites of variable regulatory activity. For all analyses involving DHS or histone modification regulatory features, we excluded deletions (and genomic space) overlapping chromatin loop anchor base pairs, as deletions disrupting chromatin loop integrity may already be under purifying selection owing to the potentially resulting cis-regulatory effects. In this way, we ensure reliable interpretation of selective effects on deletions disrupting chromatin accessibility or histone modification, without introducing potential confounding from selective pressure from chromatin loop anchor disruption, which we analyzed separately.
In contrast to the findings above, we did not find evidence of purifying selection acting on other epigenomic annotations such as noncoding transcribed, Polycomb-repressed, or heterochromatin sites, consistent with previously reported findings (Sudmant et al. 2015a; Abel et al. 2020). In the absence of statistical confirmation, we can conclude that, notwithstanding any specific regulatory locus potentially being under selective constraint, these classes of epigenomic annotations as a whole are not selectively preserved at a greater extent than that of the noncoding background. Most methods to detect natural selection from DNA polymorphism data compare characteristics of genetic variation to expectation under neutrality. We compare degree of diversity and AFs of deletions overlapping genomic annotations to simulated deletions randomly placed in the uniquely mappable human genome. Although studies have assumed that most of the noncoding human genome evolves neutrally and that random unannotated sequence represents a “neutral standard,” estimates of the size of the “functional” genome vary widely (Ponting and Hardison 2011; The ENCODE Project Consortium 2012; Rands et al. 2014). It is difficult to judge the validity of the assumption that randomly placed deletions are truly neutral. Previous work examining selection on indels has used ancestral repeats as a proxy for neutral mutations (Barton and Zeng 2019); however, the deletions in our data sets are much longer, and we do not assume a neutral set of deletions in uniquely mappable regions. Therefore, we interpret our results as indication that DHS and enhancer sequences are under stronger purifying selection than the noncoding genome on average. For transcribed, Polycomb-repressed, and heterochromatin regions, we conclude that we do not find evidence of purifying selection being greater than that of the uniquely-mappable noncoding sequence background we analyze. Cohen's d is negative for heterochromatic regions, and it is difficult to unequivocally interpret this finding. One possibility is that these regions are less depleted in deletions than the noncoding genome as a whole. However, neither of these annotations shows a corresponding AF shift toward common alleles. Alternatively, the mutation rate of deletions may be elevated in heterochromatic regions because dense chromatin interferes with double-strand break repair (Watts 2016). It is also possible that this is a technical artifact of removal of repetitive sequence in our analysis.
Our results give indication to support the hypothesis that long deletions are under stronger selection (Girirajan et al. 2011; Sudmant et al. 2015a). Supplemental Figure S1 shows that there appears to be a qualitative trend of greater depletion of deletions longer than the median length in our data sets, which is likely because longer deletions overlap multiple DHS or enhancer sites. This trend is especially pronounced for those deletions that overlap cellularly pleiotropic sites.
Driven by human genetics examples (Lupiáñez et al. 2015; Akdemir et al. 2020), there is a considerable interest in the effect of deletions on TAD-loops, many of which are demarcated by chromatin loop anchors (Rao et al. 2014). We did not find statistical evidence that selection is acting against deletions overlapping loop anchors without simultaneous removal of CTCF sites. However, purifying selection is indeed operating to preserve chromatin loop integrity by preserving colocalized CTCF binding sites within chromatin loop anchors. These findings are in agreement with other studies (Fudenberg and Pollard 2019; Kentepozidou et al. 2020). In both 1000GP and ADNI, deletions of CTCF binding sites within loop anchors show a significant excess of rare alleles. These deletions are also significantly depleted in 1000GP, with the ADNI data set showing a similar qualitative but not significant effect. The difference in significance for the depletion signal between the two data sets may simply be owing to the difference in power to see this effect, as there are approximately four times the number of deletions in 1000GP in comparison to ADNI.
We did not find statistically significant evidence in either data set that the loop CTCF binding sites of highest cellular pleiotropy show additional signal for purifying selection beyond that for sites with activity specific to a particular biosample. We cannot exclude that this equivalence is owing to lack of power: either five biosamples in chromatin loop anchor analysis are not numerous enough to see a difference (compared with the 25 biosamples used in the analysis of chromatin accessibility or histone modification features) or deletions overlapping cellularly pleiotropic loop anchors are already so few in number that power is limited (only ∼4% [1000GP] or ∼8% [ADNI] of all deletions in our data sets). An additional limitation on power comes from sparsity of loop anchor annotations overlapping ∼10% of deletions. Constrained by this sparsity, we analyzed chromatin loop anchor depletion using the otherwise full genomic coordinates allowed without excluding colocalizing regulatory sequences.
Methods
We used deletions from two data sets, a callset we generated from the genomes of participants in ADNI (Petersen et al. 2010) and a callset filtered from 1000GP (Sudmant et al. 2015a), to examine selective constraint within regulatory regions. The two deletion data sets have different callset properties, enabling robustness of the analysis. ADNI consists of deletions derived from high-coverage WGS data that are on average longer and rarer, using genotypes from the subset of individuals that we determined were of European ancestry as identified by principal components analysis. An extended description of the ADNI data set construction process is given in Supplemental Note S1. 1000GP consists of deletions derived from low-coverage whole-genome sequencing (WGS) that span a wider length range and are genotyped from individuals of diverse demographic histories (see Supplemental Note S2.1). For both deletion data sets, we restricted our analyses to noncoding deletions by removing any deletion that overlapped any exon or UTR by ≥1 bp, as exonic deletions have been previously shown to be under strong purifying selection because of their protein-altering effects (Conrad et al. 2010). We also examined only deletions occurring on autosomes because sex-chromosome functional elements may involve complex sex-biased regulation (Khramtsova et al. 2019), which might be subject to unique selective properties. To mitigate nonuniform (i.e., biased) deletion callability in the noncoding genome (Supplemental Fig. S5), which might distort the AFS of the remaining set of deletions, we additionally excluded deletions overlapping any regions of low mappability, segmental duplications, centromeres, and reference assembly gaps. Additional details on the deletion filtering criteria are given in Supplemental Note S2.2. Specific characteristics of the filtered deletion data sets are shown in Supplemental Table S11.
We used regulatory data from the NIH REC for definition of regulatory breakpoints as well as uniform processing across multiple biosamples (Roadmap Epigenomics Consortium et al. 2015). We used annotation data for sites of chromatin accessibility (DHS) and histone modification (H3K4me1 “enhancer,” H3K36me3 “transcribed,” H3K27me3 “polycomb-repressed,” and H3K9me3 “heterochromatin”). Two sets of DHS annotation (hotspot and MACS) were used to check for consistency. Because regulatory element boundaries are not perfectly aligned between biosamples, it could be the case that a partial deletion of an element observed in one particular biosample may correspond to a complete deletion of the element observed in another distinct biosample. Therefore, we assume we cannot confidently determine which coordinates are the exact peak signal for a cellularly pleiotropic regulatory element. Because of this, we use the specific base pair annotation from narrow regions of enrichment (P ≤ 0.01) for histone modification data and DHS data (MACS peak caller, NarrowPeak) (Zhang et al. 2008) and additionally the specific base pair annotation from general-sized regions of DNA accessibility (P ≤ 0.01) for DHS data (hotspot algorithm, BroadPeak) (John et al. 2011). We used all 25 primary biosamples for which data were available across all six callsets for each biosample. We additionally used chromatin loop anchor data consisting of a callset of five human noncancerous biosamples (Rao et al. 2014). Additional details on the regulatory data sets are given in Supplemental Note S3. Identity of the biosamples analyzed from REC is shown in Supplemental Table S12.
To determine if the action of purifying selection is occurring against deletions overlapping regulatory sites, we required the identification of two key signatures: reduction of genetic variation overlapping the sites and a shift in the AFS toward rare variants of the remaining alleles overlapping the sites. These signatures were assessed in light of results from deletion simulations (see Supplemental Note S4). Identification of both signatures would indicate selective pressure to preserve the corresponding regulatory feature(s). A description of the significance calculation of reduction in variation is given in Supplemental Note S6 (see also Supplemental Fig. S6). Descriptions of the procedure involving multivariate regression on deletion genomic covariates and significance calculation of shift in AFS are given in Supplemental Note S7. To examine potential purifying selection against deletions to preserve regulatory features, we examined deletion overlap in the context of regulatory biosample activity. To properly “count” biosample activity removed by deletions overlapping regulatory features, we developed a statistical method called pleiotropy ratio score (PlyRS), which calculates a correlation-adjusted count of cellular pleiotropy for each base pair in the noncoding genome. A description of the derived PlyRS measures calculated for deletions is given in Supplemental Note S5. The PlyRS method is flexible and easily allows for the addition of new and larger regulatory data sets as they become available for medical or evolutionary applications.
Data access
In accordance with the ADNI Consortium data policy, ADNI-related data files that we have made available (see Supplemental Note S1.6) may be obtained from the Laboratory of Neuro Imaging (LONI) Image and Data Archive (IDA) hosted at the University of Southern California, Los Angeles, California. All files obtained from the LONI IDA require that investigators download, review, sign, and submit the ADNI WGS data use agreement and be a registered user of ADNI data. More information on obtaining ADNI data access can be found at http://adni.loni.usc.edu/data-samples/access-data/. Once registered and logged in at the site above, data files for this project may be located by browsing for author name in the data portal section. Source code of PlyRS calculation is made available at the repository on GitHub (https://github.com/davidwradke/PlyRS) and main scripts are included in Supplemental Note S8.
Supplementary Material
Acknowledgments
We thank members of the Sunyaev laboratory and Matt Maurano for helpful discussion on the topic. We thank Sheila Sutti Diamond for administrative help related to the ADNI data used and generated in this project. We also thank Sung Chun, Nikolaos Patsopoulos, Deb Farlow, and Ruth McCole for helpful discussion related to the ADNI work. We thank the ADNI individuals for their participation in the consortium. This material is based upon work supported by the National Science Foundation Graduate Research Fellowship Program under grant no. DGE1144152 (D.W.R.). Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the National Science Foundation. D.W.R. was also supported by the National Institutes of Health (NIH) under the Ruth L. Kirschstein National Research Service Awards 4T32HG002295-14 and 5T15LM007092-27 and supported by NIH grant 5R01HG010372. S.R.S. is currently supported by NIH grants R35GM127131, R01MH101244, and U01HG006500. R.C.G. is currently supported by NIH grants U01-AG24904, R01-AG047866, R01-HL143295, and U01-TR003201; the Snite Foundation; and the Franca Sozzanni Fund. Data collection and sharing for this project were funded by the ADNI (NIH grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, by the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie, Alzheimer's Association, Alzheimer's Drug Discovery Foundation, Araclon Biotech, BioClinica, Biogen, Bristol-Myers Squibb Company, CereSpir, Cogstate, Eisai, Elan Pharmaceuticals, Eli Lilly and Company, EuroImmun, F. Hoffmann-La Roche and its affiliated company Genentech, Fujirebio, GE Healthcare, IXICO, Janssen Alzheimer Immunotherapy Research & Development, Johnson & Johnson Pharmaceutical Research & Development, Lumosity, Lundbeck, Merck & Co., Meso Scale Diagnostics, NeuroRx Research, Neurotrack Technologies, Novartis Pharmaceuticals Corporation, Pfizer, Piramal Imaging, Servier, Takeda Pharmaceutical Company, and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer's Therapeutic Research Institute at the University of Southern California. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at https://www.genome.org/cgi/doi/10.1101/gr.275263.121.
Competing interest statement
R.C.G. has received compensation for advising AIA, Genomic Life, Grail, OptumLabs, Verily, Vibrent Health, and Wamberg and is cofounder of Genome Medical. The remaining authors declare no competing interests.
References
- Abel HJ, Larson DE, Regier AA, Chiang C, Das I, Kanchi KL, Layer RM, Neale BM, Salerno WJ, Reeves C, et al. 2020. Mapping and characterization of structural variation in 17,795 human genomes. Nature 583: 83–89. 10.1038/s41586-020-2371-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akdemir KC, Le VT, Chandran S, Li Y, Verhaak RG, Beroukhim R, Campbell PJ, Chin L, Dixon JR, Futreal PA, et al. 2020. Disruption of chromatin folding domains by somatic genomic rearrangements in human cancer. Nat Genet 52: 294–305. 10.1038/s41588-019-0564-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton HJ, Zeng K. 2019. The impact of natural selection on short insertion and deletion variation in the great tit genome. Genome Biol Evol 11: 1514–1524. 10.1093/gbe/evz068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Ma Z, Kim BH, Wu W, Cayting P, Boyle AP, Sundaram V, Xing X, Dogan N, Li J, et al. 2014. Principles of regulatory information conservation between mouse and human. Nature 515: 371–375. 10.1038/nature13985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad DF, Pinto D, Redon R, Feuk L, Gokcumen O, Zhang Y, Aerts J, Andrews TD, Barnes C, Campbell P, et al. 2010. Origins and functional impact of copy number variation in the human genome. Nature 464: 704–712. 10.1038/nature08516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvijović I, Good BH, Desai MM. 2018. The effect of strong purifying selection on genetic diversity. Genetics 209: 1235–1278. 10.1534/genetics.118.301058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ENCODE Project Consortium. 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489: 57–74. 10.1038/nature11247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finucane HK, Bulik-Sullivan B, Gusev A, Trynka G, Reshef Y, Loh PR, Anttila V, Xu H, Zang C, Farh K, et al. 2015. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat Genet 47: 1228–1235. 10.1038/ng.3404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fudenberg G, Pollard KS. 2019. Chromatin features constrain structural variation across evolutionary timescales. Proc Nat Acad Sci 116: 2175–2180. 10.1073/pnas.1808631116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazal S, Loh PR, Finucane HK, Ganna A, Schoech A, Sunyaev S, Price AL. 2018. Functional architecture of low-frequency variants highlights strength of negative selection across coding and non-coding annotations. Nat Genet 50: 1600–1607. 10.1038/s41588-018-0231-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girirajan S, Brkanac Z, Coe BP, Baker C, Vives L, Vu TH, Shafer N, Bernier R, Ferrero GB, Silengo M, et al. 2011. Relative burden of large CNVs on a range of neurodevelopmental phenotypes. PLoS Genet 7: e1002334. 10.1371/journal.pgen.1002334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Xu Q, Canzio D, Shou J, Li J, Gorkin DU, Jung I, Wu H, Zhai Y, Tang Y, et al. 2015. CRISPR inversion of CTCF sites alters genome topology and enhancer/promoter function. Cell 162: 900–910. 10.1016/j.cell.2015.07.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusev A, Lee SH, Trynka G, Finucane H, Vilhjálmsson BJ, Xu H, Zang C, Ripke S, Bulik-Sullivan B, Stahl E, et al. 2014. Partitioning heritability of regulatory and cell-type-specific variants across 11 common diseases. Am J Hum Genet 95: 535–552. 10.1016/j.ajhg.2014.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handsaker RE, Korn JM, Nemesh J, McCarroll SA. 2011. Discovery and genotyping of genome structural polymorphism by sequencing on a population scale. Nat Genet 43: 269–276. 10.1038/ng.768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YF, Gulko B, Siepel A. 2017. Fast, scalable prediction of deleterious noncoding variants from functional and population genomic data. Nat Genet 49: 618–624. 10.1038/ng.3810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huddleston J, Eichler EE. 2016. An incomplete understanding of human genetic variation. Genetics 202: 1251–1254. 10.1534/genetics.115.180539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibn-Salem J, Köhler S, Love MI, Chung HR, Huang N, Hurles ME, Haendel M, Washington NL, Smedley D, Mungall CJ, et al. 2014. Deletions of chromosomal regulatory boundaries are associated with congenital disease. Genome Biol 15: 423. 10.1186/s13059-014-0423-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ionita-Laza I, McCallum K, Xu B, Buxbaum JD. 2016. A spectral approach integrating functional genomic annotations for coding and noncoding variants. Nat Genet 48: 214–220. 10.1038/ng.3477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- John S, Sabo PJ, Thurman RE, Sung MH, Biddie SC, Johnson TA, Hager GL, Stamatoyannopoulos JA. 2011. Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nat Genet 43: 264–268. 10.1038/ng.759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kentepozidou E, Aitken SJ, Feig C, Stefflova K, Ibarra-Soria X, Odom DT, Roller M, Flicek P. 2020. Clustered CTCF binding is an evolutionary mechanism to maintain topologically associating domains. Genome Biol 21: 5. 10.1186/s13059-019-1894-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khramtsova EA, Davis LK, Stranger BE. 2019. The role of sex in the genomics of human complex traits. Nat Rev Genet 20: 173–190. 10.1038/s41576-018-0083-1 [DOI] [PubMed] [Google Scholar]
- Kircher M, Witten DM, Jain P, O'Roak BJ, Cooper GM, Shendure J. 2014. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet 46: 310–315. 10.1038/ng.2892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloosterman WP, Francioli LC, Hormozdiari F, Marschall T, Hehir-Kwa JY, Abdellaoui A, Lameijer EW, Moed MH, Koval V, Renkens I, et al. 2015. Characteristics of de novo structural changes in the human genome. Genome Res 25: 792–801. 10.1101/gr.185041.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D, Gorkin DU, Baker M, Strober BJ, Asoni AL, McCallion AS, Beer MA. 2015. A method to predict the impact of regulatory variants from DNA sequence. Nat Genet 47: 955–961. 10.1038/ng.3331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li WH. 1997. Molecular evolution. Sinauer Associates, Sunderland, MA. [Google Scholar]
- Liu SJ, Horlbeck MA, Cho SW, Birk HS, Malatesta M, He D, Attenello FJ, Villalta JE, Cho MY, Chen Y, et al. 2017. CRISPRi-based genome-scale identification of functional long noncoding RNA loci in human cells. Science 355: eaah7111. 10.1126/science.aah7111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupiáñez DG, Kraft K, Heinrich V, Krawitz P, Brancati F, Klopocki E, Horn D, Kayserili H, Opitz JM, Laxova R, et al. 2015. Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell 161: 1012–1025. 10.1016/j.cell.2015.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurano MT, Humbert R, Rynes E, Thurman RE, Haugen E, Wang H, Reynolds AP, Sandstrom R, Qu H, Brody J, et al. 2012. Systematic localization of common disease-associated variation in regulatory DNA. Science 337: 1190–1195. 10.1126/science.1222794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills RE, Walter K, Stewart C, Handsaker RE, Chen K, Alkan C, Abyzov A, Yoon SC, Ye K, Cheetham RK et al. 2011. Mapping copy number variation by population-scale genome sequencing. Nature 470: 59–65. 10.1038/nature09708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nora EP, Goloborodko A, Valton AL, Gibcus JH, Uebersohn A, Abdennur N, Dekker J, Mirny LA, Bruneau BG. 2017. Targeted degradation of CTCF decouples local insulation of chromosome domains from genomic compartmentalization. Cell 169: 930–944.e22. 10.1016/j.cell.2017.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen RC, Aisen PS, Beckett LA, Donohue MC, Gamst AC, Harvey DJ, Jack CR, Jagust WJ, Shaw LM, Toga AW, et al. 2010. Alzheimer's Disease Neuroimaging Initiative (ADNI): clinical characterization. Neurology 74: 201–209. 10.1212/WNL.0b013e3181cb3e25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting CP, Hardison RC. 2011. What fraction of the human genome is functional? Genome Res 21: 1769–1776. 10.1101/gr.116814.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quang D, Chen Y, Xie X. 2015. DANN: a deep learning approach for annotating the pathogenicity of genetic variants. Bioinformatics 31: 761–763. 10.1093/bioinformatics/btu703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiver MH, Lachance J. 2018. Adaptive eQTLs reveal the evolutionary impacts of pleiotropy and tissue-specificity, while contributing to health and disease in human populations. bioRxiv 10.1101/444737 [DOI] [PMC free article] [PubMed]
- Rands CM, Meader S, Ponting CP, Lunter G. 2014. 8.2% of the human genome is constrained: variation in rates of turnover across functional element classes in the human lineage. PLoS Genet 10: e1004525. 10.1371/journal.pgen.1004525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao SSP, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, Sanborn AL, Machol I, Omer AD, Lander ES, et al. 2014. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159: 1665–1680. 10.1016/j.cell.2014.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie GRS, Dunham I, Zeggini E, Flicek P. 2014. Functional annotation of noncoding sequence variants. Nat Methods 11: 294–296. 10.1038/nmeth.2832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roadmap Epigenomics Consortium, Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, Kheradpour P, Zhang Z, Wang J, et al. 2015. Integrative analysis of 111 reference human epigenomes. Nature 518: 317–330. 10.1038/nature14248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojano E, Seoane P, Ranea JAG, Perkins JR. 2019. Regulatory variants: from detection to predicting impact. Briefings Bioinform 20: 1639–1654. 10.1093/bib/bby039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoech AP, Jordan DM, Loh PR, Gazal S, O'Connor LJ, Balick DJ, Palamara PF, Finucane HK, Sunyaev SR, Price AL. 2019. Quantification of frequency-dependent genetic architectures in 25 UK Biobank traits reveals action of negative selection. Nat Commun 10: 790. 10.1038/s41467-019-08424-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenfelder S, Fraser P. 2019. Long-range enhancer–promoter contacts in gene expression control. Nat Rev Genet 20: 437–455. 10.1038/s41576-019-0128-0 [DOI] [PubMed] [Google Scholar]
- Sudmant PH, Rausch T, Gardner EJ, Handsaker RE, Abyzov A, Huddleston J, Zhang Y, Ye K, Jun G, Fritz MHY, et al. 2015a. An integrated map of structural variation in 2,504 human genomes. Nature 526: 75–81. 10.1038/nature15394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudmant PH, Mallick S, Nelson BJ, Hormozdiari F, Krumm N, Huddleston J, Coe BP, Baker C, Nordenfelt S, Bamshad M, et al. 2015b. Global diversity, population stratification, and selection of human copy-number variation. Science 349: aab3761. 10.1126/science.aab3761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunyaev SR, Eisenhaber F, Rodchenkov IV, Eisenhaber B, Tumanyan VG, Kuznetsov EN. 1999. PSIC: profile extraction from sequence alignments with position-specific counts of independent observations. Protein Eng 12: 387–394. 10.1093/protein/12.5.387 [DOI] [PubMed] [Google Scholar]
- Treangen TJ, Salzberg SL. 2012. Repetitive DNA and next-generation sequencing: computational challenges and solutions. Nat Rev Genet 13: 36–46. 10.1038/nrg3117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trynka G, Sandor C, Han B, Xu H, Stranger BE, Liu XS, Raychaudhuri S. 2013. Chromatin marks identify critical cell types for fine mapping complex trait variants. Nat Genet 45: 124–130. 10.1038/ng.2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernot B, Stergachis AB, Maurano MT, Vierstra J, Neph S, Thurman RE, Stamatoyannopoulos JA, Akey JM. 2012. Personal and population genomics of human regulatory variation. Genome Res 22: 1689–1697. 10.1101/gr.134890.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward LD, Kellis M. 2012. Evidence of abundant purifying selection in humans for recently acquired regulatory functions. Science 337: 1675–1678. 10.1126/science.1225057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts FZ. 2016. Repair of DNA double-strand breaks in heterochromatin. Biomolecules 6: 47. 10.3390/biom6040047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Gokcumen O, Khurana E. 2020. Loss-of-function tolerance of enhancers in the human genome. PLoS Genet 16: e1008663. 10.1371/journal.pgen.1008663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng J, de Vlaming R, Wu Y, Robinson MR, Lloyd-Jones LR, Yengo L, Yap CX, Xue A, Sidorenko J, McRae AF, et al. 2018. Signatures of negative selection in the genetic architecture of human complex traits. Nat Genet 50: 746–753. 10.1038/s41588-018-0101-4 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. 2008. Model-based Analysis of ChiP-Seq (MACS). Genome Biol 9: R137. 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Troyanskaya OG. 2015. Predicting effects of noncoding variants with deep learning-based sequence model. Nat Methods 12: 931–934. 10.1038/nmeth.3547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S, Li W, Liu J, Chen CH, Liao Q, Xu P, Xu H, Xiao T, Cao Z, Peng J, et al. 2016. Genome-scale deletion screening of human long non-coding RNAs using a paired-guide RNA CRISPR-Cas9 library. Nat Biotechnol 34: 1279–1286. 10.1038/nbt.3715 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.