Abstract
Centromeres play an essential function in cell division by specifying the site of kinetochore formation on each chromosome for mitotic spindle attachment. Centromeres are defined epigenetically by the histone H3 variant Centromere Protein A (Cenpa). Cenpa nucleosomes maintain the centromere by designating the site for new Cenpa assembly after dilution by replication. Vertebrate centromeres assemble on tandem arrays of repetitive sequences, but the function of repeat DNA in centromere formation has been challenging to dissect due to the difficulty in manipulating centromeres in cells. Xenopus laevis egg extracts assemble centromeres in vitro, providing a system for studying centromeric DNA functions. However, centromeric sequences in Xenopus laevis have not been extensively characterized. In this study, we combine Cenpa ChIP-seq with a k-mer based analysis approach to identify the Xenopus laevis centromere repeat sequences. By in situ hybridization, we show that Xenopus laevis centromeres contain diverse repeat sequences, and we map the centromere position on each Xenopus laevis chromosome using the distribution of centromere-enriched k-mers. Our identification of Xenopus laevis centromere sequences enables previously unapproachable centromere genomic studies. Our approach should be broadly applicable for the analysis of centromere and other repetitive sequences in any organism.
Accurate chromosome segregation during cell division requires the centromere, a nucleoprotein complex assembled on each chromosome that is essential for chromosome segregation. Centromeres provide the assembly site for the mitotic kinetochore that mediates microtubule attachment and error correction during mitosis (Foley and Kapoor 2013). Centromeres are defined epigenetically by the histone H3 variant, Centromere Protein A (Cenpa), the presence of which is both necessary and sufficient for centromere formation (Musacchio and Desai 2017). Unlike histone H3.1 nucleosomes, that are assembled as chromosomes replicate in S-phase, Cenpa nucleosomes are replenished after replication during the next G1 phase of the cell cycle. Cenpa nucleosomes in chromatin appear to epigenetically dictate the sites of new Cenpa incorporation, thereby providing a mechanism for self-maintenance (Zasadzińska and Foltz 2017).
In humans, centromeres form on tandem repeats of an ∼171-bp DNA sequence termed ɑ-satellite. Each 171-bp monomer shares ∼60% sequence homology with other monomers. Tandem arrays of monomers are repeated in blocks of higher order repeats (HORs), resulting in long stretches of virtually identical repeat sequences (Willard and Waye 1987; Rudd et al. 2003; McNulty and Sullivan 2018). Investigation into the genetic features required to form stable human artificial chromosomes (HACs) identified repetitive ɑ-satellite DNA as sufficient for de novo centromere formation when introduced into human cells (Harrington et al. 1997; Ohzeki et al. 2015). These studies demonstrated that repetitive DNA promotes centromere formation in vertebrates.
Perturbing centromere function in cells often leads to cell death; thus, cell-free systems using budding yeast and Xenopus laevis egg extracts have been invaluable for studying centromere and kinetochore assembly (Ng and Carbon 1987; Hyman et al. 1992; Sorger et al. 1994; Desai et al. 1997; Akiyoshi et al. 2010; Guse et al. 2011; Moree et al. 2011). Budding yeast centromeres are defined by a single 125-bp DNA sequence that is sufficient to recruit much of the centromere and kinetochore in cell extracts. As in humans, Xenopus laevis builds its centromeres on repetitive sequences (Edwards and Murray 2005), and thus Xenopus egg extract provides a unique system to study the functions of repetitive DNA in driving centromere formation. A 174-bp centromeric repeat has been previously identified in Xenopus laevis by chromatin immunoprecipitation of Cenpa followed by cloning and sequencing (Edwards and Murray 2005). This repeat sequence termed Frog centromere repeat 1 (Fcr1) forms large repetitive arrays and is AT rich, as are centromeric repeats from other vertebrates (Manuelidis 1978; McDermid et al. 1986; Melters et al. 2013; Sullivan et al. 2017). Fcr1 is detected on only 60%–70% of Xenopus laevis centromeres, suggesting that there must be other sequence elements that comprise Xenopus laevis centromeres. Xenopus laevis is an allotetraploid species: the genome is composed of two related subgenomes named the long (L) and short (S) based on the length of the homoeologous chromosomes (Session et al. 2016). Whether there is conservation of centromeric repeats within each subgenome or between homoeologous chromosomes remains unknown.
In this study, we identified and characterized Cenpa-associated sequences in Xenopus laevis. We utilized a k-mer-based method that does not depend on an assembled reference genome and thus provides an unbiased approach to identify sequence motifs present at the centromere. Our results demonstrate the sequence diversity at active Xenopus laevis centromeres and enable future studies of the function of repetitive elements in centromere formation and function.
Results
Cenpa-associated sequences are composed of diverse but related repetitive sequences
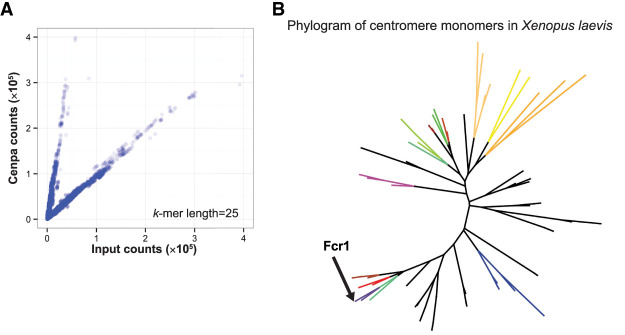
To characterize centromeric sequences in Xenopus laevis, we prepared libraries for high-throughput sequencing from solubilized mononucleosomal fractions of total genomic DNA, henceforth referred to as “input DNA,” as well as from a Cenpa immunoprecipitation (ChIP-seq) (Supplemental Fig. S1A). Because Xenopus laevis centromeric DNA is repetitive (Edwards and Murray 2005), we performed an alignment-independent analysis based on k-mer counting (Hayden and Willard 2012). We identified repeat sequences enriched for Cenpa by generating 25-bp-long k-mers from each read set and comparing the k-mer composition of the Cenpa immunoprecipitate to the input fraction (Supplemental Fig. S1B). For each k-mer, we calculated a centromere enrichment score by dividing the number of times this k-mer was found in the Cenpa ChIP data by the number of times it was found in the input data, normalized to the size of the sequencing libraries. We identified k-mers that were: (1) abundant in both the Cenpa ChIP and input samples (found at least 1000 times), and (2) enriched in the Cenpa immunoprecipitate compared to the input based on the centromere enrichment score. This approach identified a population of k-mers that are more prevalent in the Cenpa ChIP than input DNA (Fig. 1A). Similar analysis performed with different k-mer lengths yielded the same conclusions (Supplemental Fig. S1C).
Figure 1.
Identification of Cenpa-associated sequences by k-mer analysis. (A) Scatterplot of 25-bp k-mer counts normalized to sequencing depth found in input and Cenpa ChIP-seq libraries. (B) Phylogram of representative Cenpa-associated sequences that contained a minimum of 80 enriched 25-bp k-mers identified as most abundant after clustering by sequence similarity. FCR monomers chosen for FISH experiments are colored and Fcr1 identified by Edwards and Murray (2005) is labeled.
To elucidate the diversity of centromeric sequences in Xenopus laevis, we analyzed 150-bp single-end reads from our Cenpa ChIP-seq libraries that contained at least one Cenpa-enriched k-mer. As expected, a larger proportion of Cenpa ChIP-seq reads contained Cenpa-enriched k-mers compared to input (Supplemental Fig. S1D). We then hierarchically grouped these Cenpa ChIP-seq reads by sequence similarity and identified a representative read for each cluster. A phylogram of these enriched 150-bp representative reads illustrates the diversity of Cenpa-associated repeat sequences (Fig. 1B). The previously identified Fcr1 sequence (Edwards and Murray 2005) was present in one clade of the tree validating both the experimental approach and the k-mer-based analysis used to identify repetitive elements. All of these sequences were homologous to Fcr1, thus we refer to these as frog centromere repeat (FCR) monomers (Supplemental Fig. S2A). The FCR monomer sequences are 150 bp long due to the sequencing length and MNase digestion, and thus the center of each read reflects the core position of the dyad axis of the Cenpa nucleosome on the ∼174-bp-long centromeric monomer. On average, the FCR monomers were 39.5% GC (Supplemental Fig. S2B), similar to Fcr1 (Edwards and Murray 2005). In addition to identifying FCR monomers from sequencing reads, we used Tandem Repeat Finder to search the Xenopus laevis genome for satellite-sized repeat monomers (Benson 1999). Monomers identified in centromeric regions were extracted and clustered, revealing a similar phylogram of Fcr1-related sequences (Supplemental Fig. S2C). Using RepeatMasker to identify repetitive sequences from Cenpa ChIP as well as from input libraries identified other repeat classes as enriched in the Cenpa data set compared to input, but k-mers from these repeats were not identified using the k-mer counting method (Supplemental Fig. S2D). We suggest that this discrepancy arises because RepeatMasker groups together related repeats with variation in sequence composition that the strict k-mer counting method would treat separately. Overall, our data demonstrate that the repetitive sequences associated with Xenopus laevis Cenpa are related to the previously identified Fcr1 but that they comprise a diverse, previously uncharacterized family.
FCR monomers vary in their abundance and chromosome-specific localization
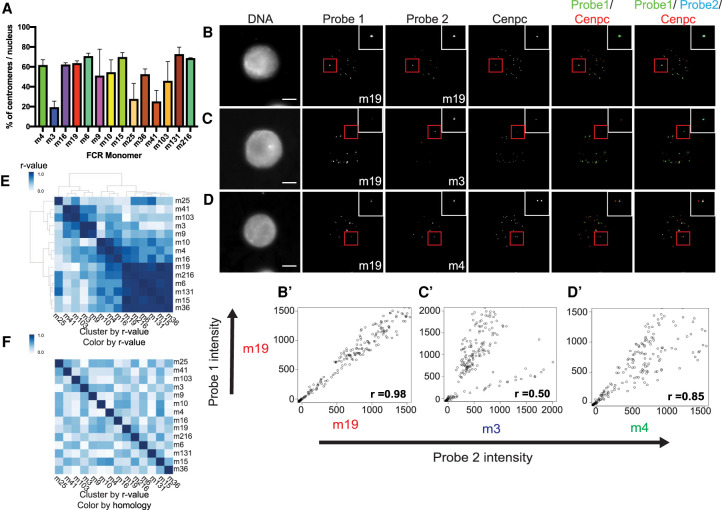
To validate the centromeric localization of the FCR sequences, we performed fluorescence in situ hybridization (FISH) combined with immunofluorescence for the constitutive centromere-associated protein, Cenpc. Using Xenopus laevis sperm nuclei incubated in Xenopus laevis egg extract, we identified the percentage of centromeres to which each FCR monomer localized (Fig. 2A). We confirmed that the previously identified Fcr1 sequence, a member of FCR monomer 16 subfamily, localized to ∼60% of centromeres (Edwards and Murray 2005). Several other FCR monomers also displayed a similar localization to 60% of centromeres. However, some FCR monomers were present at fewer centromeres, suggesting that these may be FCR variant sequences that are specific to a subset of chromosomes (Fig. 2A). Most FCR monomer FISH signals were centromeric, with some localization expanded beyond the Cenpc signal but with very little off-centromere localization, indicating that FCR monomers are found almost exclusively at centromeric regions of chromosomes.
Figure 2.
FCR monomers exhibit distinct centromeric localization independent of sequence similarity. (A) Bar plot of the percentage of centromeres per nucleus that are positive for a given FCR monomer. Bar color corresponds to color on phylogram. Averages of two independent experiments are shown with standard error displayed. (B–D) Maximum projection images of two-color FISH with immunofluorescence for the centromere marker, Cenpc. (B) FCR monomer 19 versus FCR monomer 19, (C) FCR monomer 19 versus FCR monomer 3, (D) FCR monomer 19 versus FCR monomer 4. Scale bar, 10 µM. (B′,C′,D′) Scatterplots of background subtracted probe intensities for each centromere from two-color FISH experiments. Pearson coefficients are displayed in the bottom right corner. (E) Clustered heat map of FCR monomer Pearson correlation to other FCR monomers as determined by two-color FISH. (F) Heat map ordered based on FISH Pearson correlation clustering; color map displays sequence similarity between FCR monomers.
We hypothesized that the distinct branches on the phylogram of Cenpa-associated sequences (Fig. 1B) might have originated from centromeric sequences found on the parental subgenomes that gave rise to the allotetraploid Xenopus laevis genome. To test this hypothesis, we performed two-color FISH combined with immunofluorescence in order to determine whether the different FCR monomers are present on distinct sets of chromosomes. We performed pairwise localization for 14 FCR monomers. Labeling the same FCR monomer sequence in two different colors resulted in overlapping FISH signals with similar intensities (Fig. 2B,B′). In contrast, colocalization of different FCR monomers resulted in variable localization and intensities. Some probes appeared to be mutually exclusive such that, if a centromere was positive for one probe, it was not positive for the other (Fig. 2C,C′). These likely correspond to FCR monomers occupying distinct chromosomes. Other pairs of FCR monomers were observed on a common set of chromosomes but to a different degree (Fig. 2D,D′). In these cases, some chromosomes appeared positive for both probes but with a stronger signal for one of them. Additionally, some FCR monomers appeared equally abundant on individual centromeres (Supplemental Fig. S3). To investigate the degree to which different FCR monomers colocalized, we calculated Pearson correlation coefficients for the intensity of each set of probes at each centromere (Fig. 2B′,C′,D′, bottom right) and clustered these correlations. We observed a set of highly correlated probes (Fig. 2E) all of which are found at ∼60% of centromeres (Fig. 2A). Therefore, FCR monomers that localize to the majority of chromosomes are likely to be found together on the same centromeres.
To test if the colocalization of different monomers was due to sequence similarity between monomers or due to the localization of distinct monomers on the same chromosome, we colored the heat map of FISH probes by sequence similarity but maintained the clustering by colocalization (Fig. 2F). This resulted in a loss of the clustered structure, indicating that FCR monomers that colocalize are not necessarily closely related at the sequence level. The lack of sequence similarity among FCR monomers that were correlated by FISH indicates that the branches on the phylogram do not predict colocalization. Thus, pairwise FISH analysis of related sequences that all share similarity with the originally identified Fcr1 reveals that not all FCR monomers are found on the same number of chromosomes and that different chromosomes have distinct centromeric repetitive arrays.
Identification of centromeric repeat arrays on each Xenopus laevis chromosome
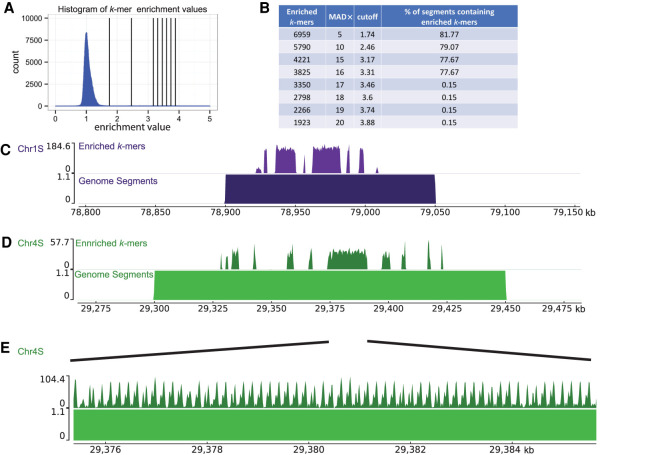
We next identified the location of the centromeres on each chromosome by identifying the regions in the genome that contained the most Cenpa-associated k-mers. The histogram of centromere enrichment scores has a median near one with a long tail of enrichment values above 1 (Fig. 3A). Using an updated version of the Xenopus laevis genome that has a single contig per chromosome (Xenopus laevis genome v10.1 is available at NCBI's Nucleotide database [https://www.ncbi.nlm.nih.gov/nuccore] under accession number JAGEVR000000000), we partitioned each chromosome into non-overlapping 50-kb segments and identified the 50-kb segments that contained Cenpa-enriched k-mers. We selected k-mers for alignment to the genome by increasing the threshold for inclusion based on the magnitude of the centromere enrichment scores. At low enrichment scores, we observed that most genome segments contained at least one k-mer (Fig. 3A,B). Increasing the centromere enrichment scores we used as a cutoff resulted in a steady decrease in the percentage of genome segments containing an enriched k-mer (Fig. 3B). Upon increasing the threshold for inclusion from an enrichment value of 3.31 to 3.46, the number of genome segments containing an enriched k-mer dropped from 77.67% of all segments to 0.15% of segments. This dramatic reduction in genome segments containing enriched k-mers arose from a modest reduction in the total number of k-mers from 3825 to 3350 enriched k-mers. Further increasing the stringency caused no change in the percentage of genome segments with enriched k-mers, yielding 84 50-kb segments that represent the centromere repeat-containing segments of the Xenopus laevis genome.
Figure 3.
Identification of centromeres on Xenopus laevis chromosomes. (A) Histogram of centromere enrichment scores for 25-bp k-mers. Enrichment scores are the ratio of normalized k-mer counts for the Cenpa data set over the input data set. Vertical lines display stringency cutoffs of (1, 2, 5, 10, 15, and 20) median absolute deviations away from the median enrichment value. (B) Table displaying the number of enriched 25-bp k-mers, the median absolute deviations (MAD×) away from the median used as the cutoff value, the enrichment value cutoff, and the percentage of genome segments containing an enriched k-mer. (C–E) Representative genome browser images with aligned enriched k-mers (top) and aligned genome segments (bottom). E is a zoom-in on a region in D.
We mapped these 84 segments onto the Xenopus laevis genome and found that almost all chromosomes contained a single locus of centromere repetitive arrays (Supplemental Fig. S4A). On each chromosome, the location of 50-kb genome segments containing Cenpa-enriched k-mers were frequently contiguous (Fig. 3C,D). Within individual 50-kb segments, we found gaps between regions containing Cenpa-enriched k-mers (Supplemental Fig. S4B). Both of these observations were consistent across chromosomes, including the Chr 9_10 homoeologous pair, which arose from a chromosome fusion event. In human centromeres, highly homogeneous repetitive arrays at the core of the centromere become less homogeneous in flanking genomic regions (Miga et al. 2014). Similarly, when we examined the edges of the repetitive array, we observed a lower density of enriched k-mers (Fig. 3C,D). Local alignment of the enriched k-mers in each 50-kb segment revealed peaks and valleys that span the genome segments (Fig. 3E). The distance between peaks is ∼170 bp, similar to the monomer size of human centromeric repeats and the size of the originally reported Fcr1 (174 bp). Not all monomer sequences in the repetitive array possessed k-mer peaks of an identical shape, further suggesting that these tandem arrays are not composed of identical repeated monomers but of diverse monomers that potentially form higher-order repeats. We estimate that the size of the repetitive array on each chromosome can vary from as little as ∼20 kb on Chr 7S up to ∼1 Mb on Chr 1L (Supplemental Fig. S4C). These estimates are based on the distance between the first and last base pair where a Cenpa-enriched k-mer aligned after manual selection of the centromeric region based on 50-kb genome segments. These estimates include the gaps between repetitive arrays on each chromosome. It is important to note that centromeres based on these estimates are only partially covered by Cenpa-enriched k-mers (Supplemental Fig. S4D). Whether these gaps also contain Cenpa nucleosomes not identified by k-mer analysis because the sequences are not repetitive is unknown. It is possible that centromere length calculations are underestimated because repetitive array lengths on genome assemblies could be limited by the long-read sequencing data from which they are made. However, these centromere size estimates are similar to CENPA-containing arrays observed in humans (Sullivan et al. 2011; Miga et al. 2014). Unlike human centromeric repeats, regions with Cenpa-enriched k-mers in the Xenopus laevis genome were not enriched for the human CENPB box (Supplemental Fig. S5A).
When aligning Cenpa-enriched k-mers to genomic regions (Fig. 3C,D,E), we allowed each individual k-mer to align multiple times. Thus, regions where Cenpa-enriched k-mers align to the genome do not represent all the locations of Cenpa nucleosomes but instead show the genomic coordinates that contain sequence features enriched in the Cenpa ChIP-seq data set. We cannot determine what fraction of these repetitive regions are occupied by Cenpa nucleosomes on any given chromosome. Some chromosomes (e.g., Chr 2S) contain other repetitive sequences nearby and interspersed with Cenpa-enriched k-mers, indicating that not all repeats are enriched for Cenpa nucleosomes (Supplemental Fig. S5B). In contrast, other, more homogeneous chromosomes only contain repeats that are composed of Cenpa-enriched k-mers, surrounded by unique sequence (Supplemental Fig. S5C).
Chromosome-specific assignment of centromere sequences
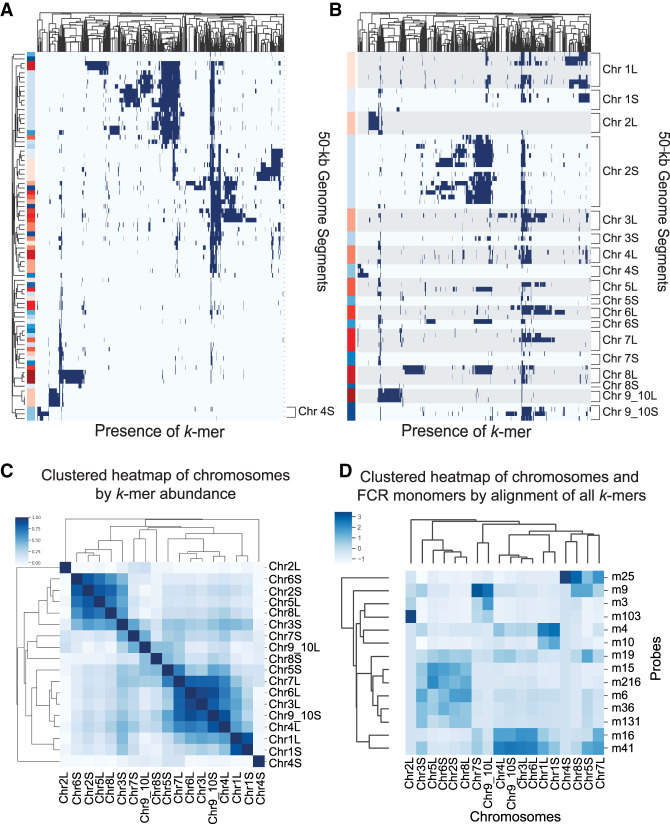
To determine which k-mers were present in each centromeric region, we clustered each 50-kb genome region by the similarity of their Cenpa-enriched k-mers (Fig. 4A). We observed that some groups of k-mers were only found on genome segments from the same chromosome (Chr 4S), whereas other groups of k-mers, seen as vertical stripes on the heat map, were found on genomic regions from several different chromosomes. No strong correlations could be made for subsets of k-mers localizing to one ancestral subgenome or the other. To illustrate the relationship between the k-mer content on the two subgenomes, we reordered the genome segments (rows) by their chromosome of origin while maintaining the clustering by k-mer similarity (Fig. 4B). This shows that some homoeologous chromosomes can have very similar k-mer spectra (e.g., Chr 1L and 1S), whereas other homoeologous chromosome pairs have a distinct centromeric makeup (Chr 2L and 2S, Chr 4L and 4S, and Chr 9_10L and 9_10S) (Fig. 4B).
Figure 4.
Assignment of FCR monomers to chromosomes by k-mer content. (A) Clustered heat map showing the presence (blue) or absence (white) of individual enriched k-mers on each centromeric genome segment. Both rows and columns are clustered to show k-mers and segments that display similar distributions. Genome segments, on the y-axis, are labeled on the left side indicating the L subgenome (blue), S subgenome (red). (B) Similar to A, but the genome segments, y-axis, are not ordered based on similar k-mer content and are instead listed by chromosome. L subgenome chromosomes are shaded with gray for clarity. (C) Clustered heat map of chromosomes by abundance of Cenpa-enriched k-mers. By combining 50-kb genome segments from each chromosome, an array of counts for each k-mer was used to generate a Euclidean distance between chromosomes used for clustering. Coloring of heat map is (1 − Euclidean distance). (D) Clustered heat map of counts reported from Bowtie of the number of times any k-mer from each FCR monomer aligns to each chromosomal contig.
No clear trends emerged for the similarity of k-mer signatures between homoeologous chromosomes or between chromosomes from the same ancestral genome, suggesting a diverse evolutionary history of Xenopus centromeres. To evaluate the similarity of centromeres between homoeologous chromosomes, we clustered the chromosomes by the frequency with which shared k-mers were found between chromosomes (Fig. 4C). Only one homoeologous pair (Chr 1L and Chr 1S) cluster together based on shared k-mer abundance. Chromosomes from both the L and S subgenomes cluster together, and in some cases chromosomes with distinct k-mer content do not cluster with any other chromosomes (Chr 2L and Chr 4S). Chromosomes with fewer unique k-mers (Supplemental Fig. S6A) do not neatly cluster with other sets of chromosomes (Chr 7S, Chr 8S, Chr 9_10L). The clustering of these chromosomes may be determined by the absence of shared k-mers rather than the k-mers they contain. L subgenome chromosomes have more unique centromeric k-mers, with the exception of Chr 2 and Chr 9_10 pairs, which may reflect a higher centromeric mutation rate in that subgenome before the allotetraploidization of Xenopus laevis.
Chr 7S and Chr 8S had the smallest regions identified by Cenpa-enriched k-mers. In order to investigate the possibility that Cenpa nucleosomes might be positioned at nonrepetitive sequences on these chromosomes, we aligned reads using Bowtie (Langmead et al. 2009) and only kept reads with single alignments (-m1). Cenpa ChIP-seq reads align outside the region where Cenpa-enriched k-mers are present on Chr 7S and Chr 8S. On Chr 7S, reads align at the site of Cenpa-enriched k-mers and extend beyond this region on one side (Supplemental Fig. S6B). On Chr 8S, Cenpa ChIP reads align ∼1.8 Mb away from the position of Cenpa-enriched k-mers (Supplemental Fig. S6C). This is in contrast to centromeres with larger repetitive arrays, which do not exhibit enrichment of uniquely mapping reads adjacent to regions with Cenpa-enriched k-mers (Supplemental Fig. S6D). Importantly, the region on Chr 8S where Cenpa ChIP reads align has no repeat characteristics, unlike the regions with Cenpa-enriched k-mers on Chr 7S and Chr 8S (Supplemental Fig. S6E). This indicates that, for this Xenopus laevis chromosome, the centromere can be assembled and maintained on nonrepetitive DNA.
We have evaluated the k-mer content of each Xenopus laevis chromosome; however, to understand which 174-bp monomer sequences make up the centromeric arrays on each chromosome requires mapping the individual monomers to specific chromosomes. To assign FCR monomer sequences to specific chromosomes, we extracted k-mers from the monomers that we localized by FISH (Fig. 2) and aligned just those k-mers to individual Xenopus laevis chromosomes. Using Bowtie, we allowed sequences to align as many times as possible without mismatches. By measuring the number of times k-mers from an FCR monomer aligned to each chromosome, we assessed from which chromosome each FCR monomer was likely derived (Fig. 4D). Stronger signal in the heat map indicates the chromosomes to which the FCR monomers would be predicted to have the strongest hybridization. The clustering of FCR monomers in this heat map is similar to the clustering from the two-color FISH experiments (Fig. 2E). Most notably, the dominant cluster by FISH containing FCR monomers 216, 131, 36, 19, 15, and 6 is recapitulated by this method. Additionally, by FISH, FCR monomer 25 clustered by itself and by this alignment-based analysis, FCR monomer 25 k-mers aligned most to Chr 4S, indicating that Chr 4S is the chromosome detected by FCR monomer 25 in the FISH experiment. Chr 8S also has k-mers from FCR monomer 25, but the repetitive array on this chromosome is short, <60 kb (Supplemental Fig. S7), which may explain why only one centromere stains strongly for FCR monomer 25 by FISH. Aligning only 25-bp k-mers that are unique to each 150-bp monomer and not shared between them generated a very similar heat map to the alignment of all k-mers (Supplemental Fig. S6B), supporting that the differences between FCR monomers drives the observed signal of chromosomal localizations. Our ability to assign specific FCR monomers to individual chromosomes makes genomic analysis of individual chromosomal centromeres in Xenopus laevis possible.
Discussion
In this study, we characterized the active centromeric regions in Xenopus laevis using native MNase Cenpa ChIP-seq. We utilized a k-mer-based strategy to functionally define the active centromeric repeat DNA on each chromosome in Xenopus laevis. The k-mer counting approach we developed can be applied to study any repeats present in a genome using ChIP-seq or analogous data sets. We found that, in Xenopus laevis centromeres, the primary sequences associated with Cenpa are a diverse set of repeat sequences related to Fcr1 (Edwards and Murray 2005). However, two chromosomes from the S subgenome were identified which had centromeres at nonrepetitive regions, despite containing short repetitive regions composed of Cenpa-enriched k-mers. We used sequence mapping and in situ hybridization to show that groups of these FCR monomers form repeat arrays that can be either (1) unique to individual chromosomes, (2) shared between subsets of chromosomes with different levels of abundance, or (3) mutually exclusive when compared between chromosomes. These observations lead to several different models for how centromeric sequences are established and maintained in Xenopus laevis. Homoeologous chromosomes that possess similar k-mer content (e.g., Chr 1L and 1S) suggest that the ancestral chromosome before divergence may have contained this same repetitive centromeric array and that both homoeologs maintained this HOR. Alternatively, some homoeologous chromosomes have distinct centromeric k-mer spectra (e.g., Chr 4L and 4S). As Chr 4L shares k-mers with other chromosomes, including Chr 1L and 1S, an ancestral centromeric repeat may have been shared between these chromosomes. Chr 4S may have once harbored this same ancestral repeat but acquired a distinct centromeric repeat that became multimerized and fixed over time. Transposable elements or intrachromosomal recombination may also generate the diversity observed between pairs of homoeologs and within each subgenome. The presence of diverse sequences at centromeres suggests that multiple sequences have the capacity for retaining Cenpa and maintaining centromeres.
Although diverse sequences compose centromeres in Xenopus laevis, these sequences could have common properties that allow them to be competent for centromere establishment. Studies from fission yeast (Ngan and Clarke 1997), fly (Peacock et al. 1974; Sun et al. 1997, 2003), and human (Hayden et al. 2013) suggest that multiple distinct repetitive units have the capacity to harbor active centromeres and may be able to form centromeres de novo. These studies demonstrate the capacity of repetitive DNA to establish an active centromere in eukaryotic systems. Although the role of CENPA in defining centromeres is undisputed, these investigations highlight the potential function of the underlying centromeric DNA in specification and establishment of centromeres.
In humans, alpha satellite DNA can promote CENPA assembly in part by providing a binding site for the CENPB protein (Ohzeki et al. 2002). Xenopus laevis appears to lack a centromere binding CENPB homolog, yet sequence features of Xenopus laevis centromeric DNA may facilitate centromere function, as has been suggested for other species (Kasinathan and Henikoff 2018). A recent study demonstrated that certain nonrepetitive chromosomal fragments contain the ability to retain CENPA after transient targeting of HJURP to deposit CENPA (Logsdon et al. 2019), suggesting that some nonalphoid sequences are competent for CENPA retention. These studies challenge the notion that centromeres are specified purely by epigenetic factors and motivate the investigation of DNA sequence contributions to centromere maintenance in diverse model organisms.
In this study, we have characterized Xenopus laevis centromeric repeats, enabling dissection of the genetic determinants of Xenopus laevis centromeres. By assigning repeat monomers to specific chromosomes, we allow further study of Xenopus laevis centromeres using genomic techniques. How DNA elements synergize with centromere assembly factors that epigenetically promote Cenpa nucleosome formation is a key question in centromere formation and inheritance. The approach that we apply to Xenopus laevis centromeres should be broadly applicable to the study of centromere or other repeat sequences in any organism.
Methods
MNase ChIP-seq library preparation
Adult J-strain Xenopus laevis were anesthetized and sacrificed before blood was drawn. For each frog, 6–8 aliquots of ∼300 µL of blood were washed three times by centrifugation for 5 min at 1400g with 1 mL of Buffer 4 (15 mM sodium citrate, 150 mM NaCl) to prevent clotting. Two additional washes were performed with 1 mL each of Buffer 1 (2.5 mM EDTA, 0.5 M EGTA, 15 mM Tris-HCl pH 7.4, 15 mM NaCl, 60 mM KCl, 15 mM sodium citrate 0.5 mM spermidine, 0.15 mM spermine, 340 mM sucrose, supplemented with 0.1 mM PMSF). Cells were resuspended in Buffer 1, pooled, and lysed by dounce homogenization with Wheaton pestle B (30–50×). Cells were checked on a hemocytometer to confirm complete lysis of cell membrane and intact nuclei. After lysis, cells were washed two times with Buffer 3 (15 mM Tris-HCl pH 7.4, 15 mM NaCl, 60 mM KCl, 0.5 mM spermidine, 0.15 mM spermine, 340 mM sucrose, supplemented with 0.1 mM PMSF), and resuspended in 500 µL Buffer 3. CaCl2 was added to 5 mM. Chromatin was digested with 300 U MNase 30 min at room temperature. Digestion was quenched with a final concentration of 5 mM EDTA and 10 mM EGTA. To lyse nuclei, 10% IGEPAL CA-630 was added to a final concentration of 0.05%. Samples were incubated on ice for 10 min followed by sedimentation of the chromatin at 5 min 1500g, 4°C and removal of the supernatant. Chromatin was resuspended in 500 µL Buffer 3 supplemented with 200 mM NaCl. The chromatin was extracted by overnight rotation at 4°C. The chromatin was pelleted at ∼16,000g for 10 min at 4°C. The supernatant was collected, and a sample was processed to confirm digestion to mostly mononucleosomes. This supernatant is the ChIP input. The input was precleared by rotation at 4°C for 4 h to overnight with 100 µL Protein A dynabeads prewashed with TBST (0.1% Triton X-100). For immunoprecipitation, 5 µg of antibody (Xenopus laevis Cenpa) (Milks et al. 2009) was coupled to 20 µL of Protein A dynabeads that had been washed three times with 400 µL TBST by rotation at 4°C in a final volume of 200 µL TBST. Antibody-bound beads were washed three times with TBST, and beads were collected. Precleared beads were collected, and the precleared input was split evenly between antibody-bound beads. A sample was also taken for input library preparation. Samples were rotated overnight at 4°C to bind nucleosomes to beads. After overnight rotation, beads were collected and washed three times with 400 µL TBST. Beads were then resuspended in 40 µL TE (10 mM Tris-HCl pH 8.0, 1 mM EDTA) supplemented with 0.1% SDS. Proteinase K was added to 0.25 mg/mL (0.5 µL of 20 mg/mL), and samples were incubated at 65°C with 850 rpm shaking from 4 h to overnight. Beads were collected, and ChIP samples were transferred to a new tube.
AMPure XP beads were used to isolate the mononucleosomal fraction. Briefly, 1.6× sample volume of beads were mixed with the sample and incubated for 5 min at room temperature. Beads were collected, and supernatant was removed. Beads were washed two times with EtOH and allowed to air dry on a magnet for 5 min. Beads were then eluted with 27 µL 10 mM Tris-HCl pH 8.0. ChIP eluates and input were assessed by high-sensitivity Qubit and Agilent Bioanalyzer. Sequencing libraries were prepared with the NEBNext Ultra II DNA Library prep kit with up to 1 µg of input or ChIP eluate DNA following the manufacturer's protocol. Two replicates of each sample were sequenced on MiSeq sequencer and one replicate on a HiSeq Illumina NGS sequencer.
DNA FISH and IF protocol
Xenopus laevis CSF-arrested egg extract was prepared as described and supplemented with Xenopus laevis sperm nuclei. The extracts were released into interphase for 75–90 min before being fixed with 2% formaldehyde, sedimented onto poly-lysine-coated coverslips and processed for immunofluorescence (French et al. 2017). Briefly, coverslips were washed quickly with PBS, and Antibody Dilution Buffer (AbDil) (150 mM NaCl, 20 mM Tris-HCl pH 7.4, 0.1% Triton X-100, 2% BSA, 0.1% sodium azide), before blocking in AbDil for 30 min. Samples were incubated in primary antibody for 30 min at room temperature (1 µg/mL rabbit anti-Xenopus laevis Cenpc antibody [Milks et al. 2009] diluted in AbDil), washed quickly three times in AbDil, and incubated in secondary antibody (donkey anti rabbit-Alexa 647 [Thermo Fisher Scientific A-31573]) diluted 1:1000 in AbDil for 30 min at room temperature. Samples were washed again in AbDil three times.
After immunofluorescence, samples were fixed again in 2.5% formaldehyde diluted in PBS for 10 min and washed three times in PBS. Samples were treated with 100 µg/mL RNase A in PBS for 30 min and washed again in PBS for 30 min. Samples were then dehydrated with an ethanol series for 1–2 min in 70%, 80%, 95%, and 100% EtOH before being allowed to air dry for 2 min. Probes were diluted and denatured at 75°C for 5 min before being spotted onto coverslips. The coverslips were then inverted onto slides and incubated on a heat block at 80°C for 10 min. Slides were then transferred to a humid chamber and hybridized at 37°C overnight. Coverslips were floated and inverted off of slides with 4× SSC (0.6 M NaCl, 60 mM sodium citrate) and washed with 4× SSC. Coverslips were then washed three times with 2× SSC prewarmed to 37°C with 50% formamide for 5 min each, three times with 2× SSC prewarmed to 37°C for 5 min each, one time with room temperature 1× SSC for 10 min, and one time with room temperature 4× SSC for 5 min. After washing, coverslips were stained with 10 µg/mL Hoechst-33342 diluted in AbDil for 10 min and washed one time with PBS containing 0.1% Triton X-100 and one time with PBS before being mounted (20 mM Tris-HCl pH 8.8, 0.5% p-Phenylenediamine, 90% glycerol) on slides and sealed with nail polish.
Imaging was performed on an IX70 Olympus microscope with a DeltaVision system (Applied Precision), a Sedat quad-pass filter set (Semrock), and monochromatic solid-state illuminators, controlled via softWoRx 4.1.0 software (Applied Precision). Images of sperm nuclei were acquired using a 60× 1.4 NA Plan Apochromat oil immersion lens (Olympus). Images were acquired with a charge-coupled device camera (CoolSNAP HQ; Photometrics) and digitized to 16 bits. Z-sections were taken at 0.2-µm intervals. Displayed images of sperm nuclei are maximum intensity projections of z-stacks.
FISH probes were generated using random hexamer priming. To generate FISH probes, 150-bp FCR monomer sequences were ordered as GeneBlocks from IDT. GeneBlocks were blunt-ligated into the pJET1.2 vector. PCR products containing the FCR monomers were amplified using the pJET1.2 forward and reverse sequencing primers. One microgram of PCR product was mixed with 5 µL of 25 µM random hexamer primer, and water was added up to 38 µL. The PCR product was denatured at 95°C for 10 min and then snap-cooled on ice. During denaturation, 2.5 µL of 1 mM dA,C,G, 2.5 µL of 1 mM Alexa fluorophore conjugated dUTP, 5 µL of 10× NEB Buffer 2, and 2 µL of Klenow (exo-) polymerase were premixed. Both Alexa 488 and 568 dUTP conjugated fluorophores were used in these experiments. Nucleotide and polymerase mix were then mixed with denatured template and primers and incubated, protected from light, at 37°C overnight. The reaction was quenched with 2 µL of 10 mM EDTA and desalted with a Microbio-spin 6 column (Bio-Rad) to remove unincorporated nucleotides. Probes were then precipitated by adding 10 µL of 10 mg/mL salmon sperm DNA, 6 µL of 3 M sodium acetate, and 120 µL 100% EtOH. Probes were vortexed and precipitated at −80°C for at least 30 min. Samples were spun at ∼16,000g at 4°C for 10 min to pellet probes. Supernatant was removed, and the pellet was resuspended in 1 mL 70% EtOH and spun again to wash. Supernatant was removed again, and the pellet was allowed to air dry. Probes were then resuspended in 50 µL hybridization buffer (65% formamide, 5× SSC, 5× Denhardt's Buffer [0.1% Ficoll-400, 0.1% Polyvinylpyrrolidone], with 150 µg/mL yeast tRNA, and 0.5 mg/mL salmon sperm DNA). Probes were incubated at 37°C for 15 min to allow complete solubilization and stored at −20°C for 2 to 3 mo. When used for single-color FISH experiments, 4 µL of probe was mixed with 4 µL of hybridization buffer, and for two-color FISH experiments, 4 µL of each probe were diluted together.
Phylogram generation
All Cenpa-associated sequences with at least 20 enriched k-mers were isolated. Enriched k-mers were defined as those with a centromere enrichment score above 25 median absolute deviations away from the median. These sequences were then entered into sequential rounds of cluster generation based on sequence similarity using cd-hit-est, first clustering sequences together that were 98% identical, then 95%, and finally 90% identical by sequence (Fu et al. 2012). This generated a list of representative sequences. The top 50 most abundant sequences were then used to generate the phylogram using Geneious (7.1.4) Tree Builder with the following settings: Genetic Distance Model = Tamura-Nei, Tree building method = Neighbor-joining, Outgroup = No outgroup, Alignment Type = Global alignment, Cost Matrix = 93% similarity. Colors were manually added to branches that contain FCR monomers used for validation by FISH. The originally identified Fcr1 monomer is labeled with an arrow (Fig. 1B). Tandem Repeat Finder was used to identify repeat monomers within the centromeric regions of the genome (Benson 1999). Repeat monomers of similar size to Fcr1 were extracted manually and clustered using cd-hit-est as described above. A phylogram of monomers identified in the genome was generated as described above. GC content of the top 50 FCR monomers and input reads were plotted using FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Alignment of FCR monomers to Fcr1 was performed using a Geneious map to reference tool with default settings.
FISH data analysis probe correlations and heat map generation
Axial projections of images were analyzed as previously described (Moree et al. 2011). Cenpc immunofluorescence signal was used as the centromere fiducial marker. Image-specific background values were subtracted from FISH intensities at each centromere. For quantification of percentage of centromeres per nucleus positive for a FISH probe, a cutoff background subtracted intensity of 200 AU was used to call a centromere positive or negative. A custom bash script (Supplemental Code) was used to quantify the percentage of centromeres per nucleus that were FISH-positive. Quantifications for single-color FISH are from three or four experimental replicates with 10–12 nuclei quantified per experiment. For two-color FISH experiments, background subtracted centromere intensities were plotted on a scatterplot and Pearson correlations were calculated in R (R Core Team 2018). Two-color FISH experiments were performed in duplicate with at least 200 centromeres quantified per slide per experiment. Results shown are from one representative experiment.
Genome segment analysis
In order to identify regions on each chromosome that contain Cenpa-enriched k-mers, an updated version of the Xenopus laevis genome (v10.1; https://www.ncbi.nlm.nih.gov/nuccore/JAGEVR000000000.1) was first separated into 50-kb segments using a custom Python script (Supplemental Code). The bbduk program was then used to extract genome segments that had enriched k-mers. Enriched k-mers were defined by the ratio of normalized k-mer abundance in the Cenpa and input libraries. Cutoffs were established by variable multiples of median absolute deviation from the median enrichment ratio. A stringent cutoff of 17 median absolute deviations from the median enrichment ration of all k-mers was chosen for downstream analysis. Genome segments with a minimal k-mer density were used to characterize centromeres on each chromosome by its k-mer content. Eighty-four total 50-kb genome segments were identified as containing a high abundance of Cenpa-enriched k-mers. A binary matrix was generated indicating the presence or absence of each enriched k-mer on each genome segment. This matrix was then plotted as a heat map, clustering both axes (method = “jaccard”) to show both k-mers and genome segments that are found together, or without clustering by genome segment to preserve the ordering of source chromosomes from each segment. Additionally, chromosomes were clustered by the abundance of each Cenpa-enriched k-mer (method = “euclidean”), after collapsing 50-kb genome segments by chromosome. For genome visualization, Cenpa-enriched k-mers and genome segments with Cenpa-enriched k-mers were aligned to the updated Xenopus laevis genome using Bowtie 2 (Langmead and Salzberg 2012) allowing for multiple alignment of k-mers. Genome alignments were then visualized using the pygenometracks Python module (Ramírez et al. 2018). To estimate repeat array size, bedGraphs with 1-bp resolution were created using deepTools2 (Ramírez et al. 2016). From this bedGraph, the centromeric region on each chromosome was selected manually based on the 50-kb genome segment analysis, and then the centromere length was defined as the distance between the first and last base pair that had a Cenpa-enriched k-mer aligned. We also report the total base pairs within the centromere on each chromosome that have Cenpa-enriched k-mers aligned and the fraction of the centromere with a Cenpa-enriched k-mer.
Bowtie analysis
FCR monomers (150 bp) were initially split into 25-bp k-mers (126 total) using KMC (Kokot et al. 2017). k-mers from each monomer were then aligned to the Xenopus laevis genome 10.2 provided by Jessen Bredeson and Dan Rokhsar using Bowtie (Langmead et al. 2009) allowing for no mismatches and for alignment as many times as possible. Alignment files were then used to count the number of times k-mers from each FCR monomer aligned to each chromosome. This produced a table of counts of the number of times k-mers from each FCR monomer aligned to each chromosome. This table was then plotted as a heat map, normalizing the intensity to the highest count on each chromosome. This same analysis was also performed with k-mers that were specific to individual FCR monomers. Alignment counts were normalized by the number of k-mers that were specific to each FCR monomer to account for differences in the number of specific k-mers for each FCR monomer.
To investigate mapping to nonrepetitive regions, Bowtie was used to align Cenpa ChIP and input single-end 150-bp reads using (-m1) to only return reads with single matches. Using deepTools2, alignment files from each library were converted to bigWig using bamCoverage with 25-bp window size and then bamCompare to calculate a log2 ratio of Cenpa ChIP/input signal. These alignments were plotted similar to alignments of k-mers described above with the pygenometracks Python module.
Dotplot analysis
Self dot plots were generated using FlexiDot v1.6 using word size of 150 bp or 50 bp and did not allow for wobble or substitution (Seibt et al. 2018). Sequences used for this analysis were selected based on the presence of Cenpa-enriched k-mers or unique mapping of Cenpa reads. GFF files corresponding to Chr 2S and Chr 4S were used to annotate regions that contain Cenpa-enriched k-mers in cyan and regions without Cenpa-enriched kmers in red boxes.
CENPB box analysis
Instances of the CENPB box “NTTCGNNNNANNCGGGN” were identified in the Xenopus laevis v10.2 genome and the hg38 human genome using FIMO (Grant et al. 2011) with thresh = 0.001 and ‐‐max-stored-scores = 100000000. An enrichment analysis was then performed using GAT (Heger et al. 2013), with ‐‐num-samples = 1000, to determine if the instances of CENPB box motif for each genome were enriched within the centromeric regions. For Xenopus laevis v10.2, centromeric regions were defined by the presence of Cenpa-enriched k-mers. For human hg38, centromeric regions were defined based on UCSC annotations for centromere models.
RepeatMasker analysis
RepeatMasker 4.0.9 was used to identify repeat classes in 20 M reads from Cenpa and Input sequencing libraries using the giri Repbase library for Xenopus repeats (Smit et al. 2013–2015). Counts for each repeat class were summarized, and an enrichment score was calculated for each class which was reported as a bar blot.
Data access
All raw and processed sequencing data generated in this study have been submitted to the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE153058. Our k-mer analysis pipeline is available at GitHub (https://github.com/straightlab/xenla-cen-dna-paper) and as Supplemental Code.
Supplementary Material
Acknowledgments
We thank Magdalena Strzelecka and Andrew Grenfell for discussion. We thank members of the Straight laboratory for comments on the manuscript. We thank Jessen Bredeson and Daniel Rokhsar for providing early access to Xenopus laevis genome release 10.2. Some of the computing for this project was performed on the Sherlock cluster. We also thank Stanford University and the Stanford Research Computing Center for providing computational resources and support that contributed to these research results. This work was supported by National Institutes of Health (NIH), National Institute of General Medical Sciences (NIGMS) R01 GM074728 to A.F.S. and NIH (NIGMS) R35 GM118183 to R.H. O.K.S. was supported by NIH (NIGMS) T32 GM113854-02 and a National Science Foundation Graduate Research Fellowships Program.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at https://www.genome.org/cgi/doi/10.1101/gr.267781.120.
Competing interest statement
The authors declare no competing interests.
References
- Akiyoshi B, Sarangapani KK, Powers AF, Nelson CR, Reichow SL, Arellano-Santoyo H, Gonen T, Ranish JA, Asbury CL, Biggins S. 2010. Tension directly stabilizes reconstituted kinetochore-microtubule attachments. Nature 468: 576–579. 10.1038/nature09594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson G. 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res 27: 573–580. 10.1093/nar/27.2.573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai A, Deacon HW, Walczak CE, Mitchison TJ. 1997. A method that allows the assembly of kinetochore components onto chromosomes condensed in clarified Xenopus egg extracts. Proc Natl Acad Sci 94: 12378–12383. 10.1073/pnas.94.23.12378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards NS, Murray AW. 2005. Identification of Xenopus CENP-A and an associated centromeric DNA repeat. Mol Biol Cell 16: 1800–1810. 10.1091/mbc.e04-09-0788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley EA, Kapoor TM. 2013. Microtubule attachment and spindle assembly checkpoint signalling at the kinetochore. Nat Rev Mol Cell Biol 14: 25–37. 10.1038/nrm3494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- French BT, Westhorpe FG, Limouse C, Straight AF. 2017. Xenopus laevis M18BP1 directly binds existing CENP-A nucleosomes to promote centromeric chromatin assembly. Dev Cell 42: 190–199.e10. 10.1016/j.devcel.2017.06.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu L, Niu B, Zhu Z, Wu S, Li W. 2012. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28: 3150–3152. 10.1093/bioinformatics/bts565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant CE, Bailey TL, Noble WS. 2011. FIMO: scanning for occurrences of a given motif. Bioinformatics 27: 1017–1018. 10.1093/bioinformatics/btr064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guse A, Carroll CW, Moree B, Fuller CJ, Straight AF. 2011. In vitro centromere and kinetochore assembly on defined chromatin templates. Nature 477: 354–358. 10.1038/nature10379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington JJ, Van Bokkelen G, Mays RW, Gustashaw K, Willard HF. 1997. Formation of de novo centromeres and construction of first-generation human artificial microchromosomes. Nat Genet 15: 345–355. 10.1038/ng0497-345 [DOI] [PubMed] [Google Scholar]
- Hayden KE, Willard HF. 2012. Composition and organization of active centromere sequences in complex genomes. BMC Genomics 13: 324. 10.1186/1471-2164-13-324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden KE, Strome ED, Merrett SL, Lee HR, Rudd MK, Willard HF. 2013. Sequences associated with centromere competency in the human genome. Mol Cell Biol 33: 763–772. 10.1128/MCB.01198-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heger A, Webber C, Goodson M, Ponting CP, Lunter G. 2013. GAT: a simulation framework for testing the association of genomic intervals. Bioinformatics 29: 2046–2048. 10.1093/bioinformatics/btt343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman AA, Middleton K, Centola M, Mitchison TJ, Carbon J. 1992. Microtubule-motor activity of a yeast centromere-binding protein complex. Nature 359: 533–536. 10.1038/359533a0 [DOI] [PubMed] [Google Scholar]
- Kasinathan S, Henikoff S. 2018. Non-B-form DNA is enriched at centromeres. Mol Biol Evol 35: 949–962. 10.1093/molbev/msy010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokot M, Długosz M, Deorowicz S. 2017. KMC 3: counting and manipulating k-mer statistics. Bioinformatics 33: 2759–2761. 10.1093/bioinformatics/btx304 [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logsdon GA, Gambogi CW, Liskovykh MA, Barrey EJ, Larionov V, Miga KH, Heun P, Black BE. 2019. Human artificial chromosomes that bypass centromeric DNA. Cell 178: 624–639.e19. 10.1016/j.cell.2019.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manuelidis L. 1978. Complex and simple sequences in human repeated DNAs. Chromosoma 66: 1–21. 10.1007/BF00285812 [DOI] [PubMed] [Google Scholar]
- McDermid HE, Duncan AM, Higgins MJ, Hamerton JL, Rector E, Brasch KR, White BN. 1986. Isolation and characterization of an α-satellite repeated sequence from human chromosome 22. Chromosoma 94: 228–234. 10.1007/BF00288497 [DOI] [PubMed] [Google Scholar]
- McNulty SM, Sullivan BA. 2018. α satellite DNA biology: finding function in the recesses of the genome. Chromosome Res 26: 115–138. 10.1007/s10577-018-9582-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melters DP, Bradnam KR, Young HA, Telis N, May MR, Ruby JG, Sebra R, Peluso P, Eid J, Rank D, et al. 2013. Comparative analysis of tandem repeats from hundreds of species reveals unique insights into centromere evolution. Genome Biol 14: R10. 10.1186/gb-2013-14-1-r10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miga KH, Newton Y, Jain M, Altemose N, Willard HF, Kent WJ. 2014. Centromere reference models for human chromosomes X and Y satellite arrays. Genome Res 24: 697–707. 10.1101/gr.159624.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milks KJ, Moree B, Straight AF. 2009. Dissection of CENP-C-directed centromere and kinetochore assembly. Mol Biol Cell 20: 4246–4255. 10.1091/mbc.e09-05-0378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moree B, Meyer CB, Fuller CJ, Straight AF. 2011. CENP-C recruits M18BP1 to centromeres to promote CENP-A chromatin assembly. J Cell Biol 194: 855–871. 10.1083/jcb.201106079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musacchio A, Desai A. 2017. A molecular view of kinetochore assembly and function. Biology 6: 5. 10.3390/biology6010005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng R, Carbon J. 1987. Mutational and in vitro protein-binding studies on centromere DNA from Saccharomyces cerevisiae. Mol Cell Biol 7: 4522–4534. 10.1128/MCB.7.12.4522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngan VK, Clarke L. 1997. The centromere enhancer mediates centromere activation in Schizosaccharomyces pombe. Mol Cell Biol 17: 3305–3314. 10.1128/MCB.17.6.3305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohzeki J, Nakano M, Okada T, Masumoto H. 2002. CENP-B box is required for de novo centromere chromatin assembly on human alphoid DNA. J Cell Biol 159: 765–775. 10.1083/jcb.200207112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohzeki J, Larionov V, Earnshaw WC, Masumoto H. 2015. Genetic and epigenetic regulation of centromeres: a look at HAC formation. Chromosome Res 23: 87–103. 10.1007/s10577-015-9470-z [DOI] [PubMed] [Google Scholar]
- Peacock WJ, Brutlag D, Goldring E, Appels R, Hinton CW, Lindsey DL. 1974. The organization of highly repeated DNA sequences in Drosophila melanogaster chromosomes. Cold Spring Harb Symp Quant Biol 38: 405–416. 10.1101/SQB.1974.038.01.043 [DOI] [PubMed] [Google Scholar]
- Ramírez F, Ryan DP, Grüning B, Bhardwaj V, Kilpert F, Richter AS, Heyne S, Dündar F, Manke T. 2016. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res 44: W160–W165. 10.1093/nar/gkw257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez F, Bhardwaj V, Arrigoni L, Lam KC, Grüning BA, Villaveces J, Habermann B, Akhtar A, Manke T. 2018. High-resolution TADs reveal DNA sequences underlying genome organization in flies. Nat Commun 9: 189. 10.1038/s41467-017-02525-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2018. R: language and environment for statistical computing. R Foundation for Statistical Computing, Vienna. https://www.R-project.org/. [Google Scholar]
- Rudd MK, Schueler MG, Willard HF. 2003. Sequence organization and functional annotation of human centromeres. Cold Spring Harb Symp Quant Biol 68: 141–150. 10.1101/sqb.2003.68.141 [DOI] [PubMed] [Google Scholar]
- Seibt KM, Schmidt T, Heitkam T. 2018. FlexiDot: highly customizable, ambiguity-aware dotplots for visual sequence analyses. Bioinformatics 34: 3575–3577. 10.1093/bioinformatics/bty395 [DOI] [PubMed] [Google Scholar]
- Session AM, Uno Y, Kwon T, Chapman JA, Toyoda A, Takahashi S, Fukui A, Hikosaka A, Suzuki A, Kondo M, et al. 2016. Genome evolution in the allotetraploid frog Xenopus laevis. Nature 538: 336–343. 10.1038/nature19840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit AFA, Hubley R, Green P. 2013–2015. RepeatMasker Open version-4.0. http://www.repeatmasker.org.
- Sorger PK, Severin FF, Hyman AA. 1994. Factors required for the binding of reassembled yeast kinetochores to microtubules in vitro. J Cell Biol 127: 995–1008. 10.1083/jcb.127.4.995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan LL, Boivin CD, Mravinac B, Song IY, Sullivan BA. 2011. Genomic size of CENP-A domain is proportional to total alpha satellite array size at human centromeres and expands in cancer cells. Chromosome Res 19: 457–470. 10.1007/s10577-011-9208-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan LL, Chew K, Sullivan BA. 2017. Α satellite DNA variation and function of the human centromere. Nucleus 8: 331–339. 10.1080/19491034.2017.1308989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Wahlstrom J, Karpen G. 1997. Molecular structure of a functional Drosophila centromere. Cell 91: 1007–1019. 10.1016/S0092-8674(00)80491-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Le HD, Wahlstrom JM, Karpen GH. 2003. Sequence analysis of a functional Drosophila centromere. Genome Res 13: 182–194. 10.1101/gr.681703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willard HF, Waye JS. 1987. Hierarchical order in chromosome-specific human α-satellite DNA. Trends Genet 3: 192–198. 10.1016/0168-9525(87)90232-0 [DOI] [Google Scholar]
- Zasadzińska E, Foltz DR. 2017. Orchestrating the specific assembly of centromeric nucleosomes. Prog Mol Subcell Biol 56: 165–192. 10.1007/978-3-319-58592-5_7 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.