Abstract
Background
Temozolomide (TMZ) resistance in glioblastoma multiforme (GBM) is mediated by the DNA repair protein O6-methylguanine DNA methyltransferase (MGMT). MGMT promoter methylation (occurs in about 40% of patients) is associated with loss of MGMT expression (MGMT−) that compromises DNA repair, leading to a favorable response to TMZ therapy. The 60% of patients with unmethylated MGMT (MGMT+) GBM experience resistance to TMZ; in these patients, understanding the mechanism of MGMT-mediated repair and modulating MGMT activity may lead to enhanced TMZ activity. Here, we report a novel mode of regulation of MGMT protein activity by poly(ADP-ribose) polymerase (PARP).
Methods
MGMT-PARP interaction was detected by co-immunoprecipitation. PARylation of MGMT and PARP was detected by co-immunoprecipitation with anti-PAR antibody. O6-methylguanine (O6-MetG) adducts were quantified by immunofluorescence assay. In vivo studies were conducted in mice to determine the effectiveness of PARP inhibition in sensitizing GBM to TMZ.
Results
We demonstrated that PARP physically binds with MGMT and PARylates MGMT in response to TMZ treatment. In addition, PARylation of MGMT by PARP is required for MGMT binding to chromatin to enhance the removal of O6-MetG adducts from DNA after TMZ treatment. PARP inhibitors reduced PARP-MGMT binding and MGMT PARylation, silencing MGMT activity to repair O6-MetG. PARP inhibition restored TMZ sensitivity in vivo in MGMT-expressing GBM.
Conclusion
This study demonstrated that PARylation of MGMT by PARP is critical for repairing TMZ-induced O6-MetG, and inhibition of PARylation by PARP inhibitor reduces MGMT function rendering sensitization to TMZ, providing a rationale for combining PARP inhibitors to sensitize TMZ in MGMT-unmethylated GBM.
Keywords: DNA damage repair, MGMT PARylation, PARP, TMZ resistance
Key Points.
PARP PARylates MGMT and regulates its activity in MGMT+ GBM.
All PARP inhibitors regulate MGMT activity, irrespective of their trapping function.
Combining PARP inhibitors with TMZ showed therapeutic benefit in MGMT+ GSCs.
Importance of the Study.
Increasing evidence supports the notion that DNA damage repair signaling plays an important role in inducing TMZ resistance; hence, it has emerged as a molecular target for therapeutic development. PARP is an enzyme involved in base excision repair, and PARP inhibitors are being developed as sensitizing agents to improve TMZ efficacy. Here, we report a novel mode of regulation of MGMT activity by PARP. PARP acts as a double-edged sword in unmethylated (MGMT+) glioma: PARP physically interacts with and PARylates MGMT to remove O6-MetG adducts in damaged DNA, independent of base excision repair, in response to TMZ treatment; second, PARP acts as a sensor to elicit response pathways involved in base excision repair. PARP inhibitors suppress PARP-MGMT binding and abolish MGMT function. In this study, we identified MGMT as a novel PARylation substrate for PARP, providing a rationale for using PARP inhibitors as sensitizers to modulate TMZ therapy in MGMT+ GBM.
Glioblastoma multiforme (GBM) is a lethal primary brain tumor with limited treatment options. The standard therapy is surgical resection, followed by adjuvant radiotherapy and the treatment with the DNA alkylating agent temozolomide (TMZ). Although TMZ displays antitumor activity and limited toxicity, its survival benefit is merely 2.5 months because of the rapid occurrence of resistance and tumor relapse. Further, the median overall survival among O6-methylguanine DNA methyltransferase (MGMT) methylated patients was 18.2 months as compared to 12.2 months among those without methylation. Inhibition of TMZ-induced DNA damage repair response represents an attractive strategy for potentiating the cytotoxic effects of TMZ and improving the outcome of GBM treatment.
The cytotoxicity of TMZ is mediated by the addition of methyl groups at N7 and O6 sites on guanines and the N3 site on adenines (N7-MetG, O6-MetG, and N3-MetA, respectively) in genomic DNA. O6-MetG is the most toxic and mutagenic DNA modification produced by TMZ; it is quickly repaired by the enzyme O6-metG DNA methyl transferase (MGMT). The methylation status of the MGMT promoter, which silences MGMT expression, has been reported to be a prognostic predictor of TMZ chemotherapy.1,2 In MGMT-unmethylated GBM tumors (approximately 60% of patients), TMZ-induced O6-MetG adducts are rapidly removed by MGMT, conferring resistance to TMZ. Understanding the underlying mechanism of MGMT-mediated repair and modulating MGMT activity in the unmethylated GBM group will be critical to enhancing the TMZ mediated response to chemotherapy.
On the other hand, N3-MetA and N7-MetG repair are mediated by the base excision repair (BER) machinery in a process that involves poly(ADP-ribose) polymerase (PARP), which ADP-ribosylates DNA and proteins. PARP proteins create a docking site for the incorporation of other components of the BER machinery, when acting on DNA, which complete the repair process.
Poly(ADP-ribose) polymerase 1 (PARP1) is well known as an ADP-ribosylating enzyme involved in a number of cellular processes, including DNA repair, genomic stability, and programmed cell death. PARP activity is triggered by various stress stimuli; among which, DNA strand breaks are the best characterized. In particular, PARP1 acts as a primary “DNA nick sensor” that physically recognizes DNA lesions to elicit the intervention of several DNA damage response pathways, including the BER/single-strand break repair (BER/SSBR) pathway. Recognition of DNA strand breaks is followed by PARP1 activation and auto-ribosylation.3 The extensive branching network of poly(ADP-ribose) (PAR) on PARP1 attracts, and assists in assembling, multiprotein complexes that are involved in chromatin remodeling, DNA repair, and damage checkpoint signaling.
PARP1-mediated BER/SSBR is critical for the repair of N7-MetG and N3-MetA, and the combination of PARP inhibitors with TMZ increases TMZ-induced cytotoxicity by disrupting N7-MetG and N3-MetA repair. PARP inhibitors have potentiated TMZ efficacy in numerous preclinical models,4–7 providing a rationale for their clinical development to potentiate TMZ therapy in GBM. Various studies have shown that PARP inhibitors are more efficient at sensitizing TMZ in MGMT-unmethylated cells than in methylated cells8,9; however, the mechanism of how PARP inhibitors are more effective in unmethylated GBM remains to be explored, and this preferential sensitization cannot be explained by PARP-mediated N7-MetG and N3-MetA repair. To explore the mechanism mediated by PARP inhibitors in sensitizing glioma sphere-forming cells (GSCs) harboring unmethylated MGMT promoter to TMZ, in our current study, we showed that PARP interacts with MGMT and poly(ADP-ribosyl)ates (PARylates) MGMT, and inhibition of MGMT PARylation by PARP inhibitors suppressed MGMT function to repair O6-MetG, rendering sensitization to TMZ.
Materials and Methods
Cell Lines and Reagents
The GSC lines were established by isolating neurosphere-forming cells from fresh surgical specimens of human GBM tissue obtained at The University of Texas MD Anderson Cancer Center (Houston, Texas) from 2005 through 2008, as described previously.10 Thirteen GSC cells were cultured in DMEM/F12 medium containing B27 supplement (Invitrogen), basic fibroblast growth factor, and epidermal growth factor (20 ng/mL each). Short tandem repeats using the Applied Biosystems AmpFISTR Identifier kit (Foster City, CA) were used to authenticate cells. The last authentication test was performed in July 2017. Talazoparib was from WuXi AppTec (Wuxi, Jiangsu, China). TMZ was from Sigma-Aldrich; veliparib, olaparib, pamiparib, and (O6-benzylguanine [O6BG]) were from Selleck. For in vitro use, all inhibitors were dissolved in dimethyl sulfoxide (Sigma-Aldrich). All cell lines were tested negative for Mycoplasma contamination using the MycoAlert Detection Kit (Lonza).
Cell Viability Assay and Sphere Formation Assay
For the cell viability assay, 5000 cells per well were plated in 96-well plates and treated in triplicate for 5 days with serially diluted TMZ, combined with the indicated doses of talazoparib. Cell proliferation was estimated using the CellTiter-Glo (Promega, Madison, WI) viability assay kit. The IC50 values were calculated as the mean drug concentration required to inhibit cell proliferation by 50% compared with vehicle-treated controls. For the sphere formation assay, cells were seeded in 96-well plates (1 cell/well) by flow cytometry sorting and treated with TMZ (5 μM), talazoparib (5 nM), or a combination for 3 weeks. Wells with sphere formation were counted and recorded using the Cell3 iMager CC-5000 System (SCREEN Holdings, Kyoto, Japan). The effect of the drug combination was determined by the Bliss independence model.11
Bliss Independence Model
Bliss independence model is based on the probability theory for independent events. The Bliss model was used to compute the expected combined effects of two drugs as the product of their individual effects on the basis of the assumption that there was no drug-drug interaction effect. The expected combination effect was calculated using Effect(a+b) = E(a) + E(b) − E(a) E(b). The drug combinations were synergistic if their observed effects were higher than the expected combined effects, antagonistic if they were lower, and additive if they were equal. With the excess over Bliss (EOB) score, the drug combination effect is determined at each combination dose.
Immunoblotting Analysis
Cells were harvested in lysis solution, and the extracted proteins were subjected to immunoblotting, as described previously,12 using the following primary antibodies: anti-PARP1, anti-MGMT (Cell Signaling, Boston, MA), anti-gamma H2AX (phosphor-S139) antibody (Abcam) and anti-PAR (Trevigen). Anti-β-actin antibody was purchased from Sigma and used as a loading control.
Immunocytochemical Assay for O6-MetG
An immunocytochemical assay was performed as described previously to quantify O6-MetG adducts.13,14 Cells were plated onto Lab-Tek II tissue culture slides (Thermo Fisher) and treated with TMZ and PARP inhibitor for 72 hours. O6-MetG adducts were stained with monoclonal mouse anti-(O6-MetG) antibody EM-2-3 (1:6000) for 16 hours at 4°C. Binding of the primary antibody was visualized by subsequent staining with a secondary antibody (Alexa Fluor 488 goat anti-mouse IgG; Invitrogen) for 1 hour. The cells were counterstained with Vecta shield sealant containing 4′,6-diamidino-2-phenylindole (Vector Laboratories), and images were obtained with Olympus FV1000 confocal microscope. The percentage of cells displaying foci was quantified by counting 5 random fields.
Apoptosis Analysis by Annexin V Staining
Cell apoptosis was detected using the Annexin-V-Fluor staining kit (Roche, San Francisco, CA) according to the manufacturer’s instructions. In brief, 1 × 106 cells were collected and resuspended in 100 µL of binding buffer. FITC annexin V (5 µL) and 7-amino-actinomycin D (7-AAD) (5 µL) were added, and cells were incubated for 15 minutes in the dark at ambient temperature. Samples were tested using BD FACS Celesta (BD Biosciences, Franklin Lakes, NJ) and analyzed using FlowJo software version 10.3.
Immunoprecipitation
Cells were lysed in lysis buffer (50 mM Tris-HCl [pH 8.0], 5 mM EDTA, 150 mM NaCl, 0.5% NP-40, and 1 mM phenylmethylsulfonyl fluoride). Cell lysate (200-1000 µg of protein) was immunoprecipitated with anti-PARP antibody (Cell Signaling, Boston, MA), anti-MGMT antibody (Sigma), or anti-PAR antibody (Trevigen) for 18 hours at 4°C. Normal preimmune mouse and rabbit IgG (Cell Signaling Technology, Boston, MA) was also used as negative controls. The immunoprecipitates were eluted by the 2× SDS sample buffer and then detected by Western blot analysis.
Subcellular Fractionation Assay
To detect chromatin-bound MGMT, cells were collected and cytoplasmic, nuclear soluble, and chromatin-bound proteins were fractionated using a subcellular protein fractionation kit from Thermo Scientific (#78840), following the manufacturer’s instructions.15 Immunoblotting was carried out using standard procedures. Actin was blotted for the cytoplasmic fraction marker, Lamin B for the nuclear soluble marker, and histone H3 for the chromatin-bound marker.
Animal Studies
All animal studies were conducted in the veterinary facilities of MD Anderson in accordance with institutional rules. All the animals used in this study were 4- to 6-week-old male nu/nu mice. To create the subcutaneous tumor model, luciferase-tagged GSC23 or GSC272 cells (5 × 106) were implanted into the hind flanks of nu/nu mice. For intracranial tumor model, the luciferase-tagged GSC23 or GSC272 cells (5 × 105) was implanted intracranially into nude mice using a previously described guide-screw system.16 Two cycles of sequential treatment were administrated. For each cycle, TMZ (50 mg/kg/day for 5 days) was given by oral gavage; PARP inhibitor treatment (talazoparib at 0.33 mg/kg/day for 5 days/week or olaparib at 25 mg/kg/day for 5 days/week) was given the following week for 3 weeks. Tumor growth was visualized and quantified using the IVIS Spectrum in vivo imaging system. Mice were monitored daily and euthanized when they became moribund. Body weight was monitored at every 2-week interval. Whole tumors were extracted, rapidly frozen in liquid nitrogen, and stored at −70°C.
Statistical Analysis
The statistical comparison was performed using Student’s t-test or Fisher’s exact test, as appropriate. The results are presented as the mean of at least 3 independent experiments. All tests were two-sided. Statistical analyses were carried out using GraphPad Prism software. Survival curves were plotted using the Kaplan-Meier method, and log-rank tests were used to compare survival curves between groups. A P value of <.05 was considered statistically significant.
Study Approval
The animal study was approved by the institutional review board of The University of Texas MD Anderson Cancer Center.
Results
PARP Inhibitor Potentiated TMZ Response in MGMT-Unmethylated GSCs
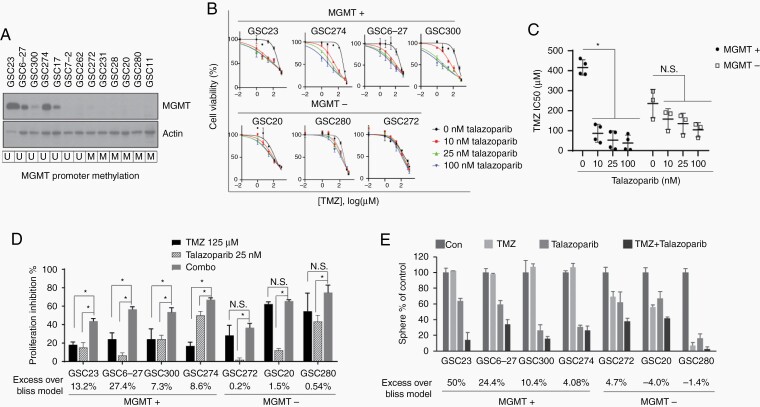
MGMT promoter methylation and MGMT expression were evaluated in 13 GSC lines. As detected by methylation-specific PCR, MGMT promoter, was methylated in 6 of 13 GSC cell lines (46%) (Figure 1A), consistent with previous clinical data showing that 40%-45% of GBMs have MGMT promoter methylation.1,2 Of 7 unmethylated GSCs, 5 showed MGMT protein expression (Figure 1A).
Fig. 1.
PARP inhibitor potentiated TMZ response in MGMT+ GSCs. A, MGMT expression was detected in 13 GSC cell lines by western blot, MGMT promoter methylation status determined by sequencing was shown in the bottom, U for unmethylated, M for methylated. B-D, MGMT+ and MGMT− GSCs were treated with serial diluted TMZ with or without talazoparib at indicated concentration. Dose-response curves were plotted (B), IC50 of TMZ calculated by GraphPad (C), Graph shows cell proliferation inhibition by TMZ, talazoparib, and combinational treatment. Synergistic effect for each cell line as calculated by bliss model was shown in the bottom (D). E, Effect of combination of TMZ and talazoparib on sphere formation in MGMT+ and MGMT− GSCs. Synergistic effect for each cell line as calculated by bliss model was shown in the bottom. Abbreviations: GSCs, glioma sphere-forming cells; MGMT, O6-methylguanine DNA methyltransferase; PARP, poly(ADP-ribose) polymerase; TMZ, temozolomide.
To assess the ability of the PARP inhibitor to synergize with TMZ in GSCs, we treated 4 MGMT-unmethylated/MGMT expression (MGMT+) and 3 MGMT methylated/no MGMT expression (MGMT−) cell lines with TMZ and PARP inhibitor talazoparib and measured cell viability by the CellTiter-Glo assay (Figure 1B). As expected, MGMT+ cells were resistant to TMZ monotherapy, as indicated by less inhibition of cell proliferation and a higher IC50 compared to that of MGMT− cells (Figure 1C and D). However, 25 nM talazoparib significantly enhanced TMZ-induced inhibition of proliferation in MGMT+ GSCs but not in MGMT− GSCs (Figure 1C).
It is noteworthy that although we used a very low concentration of talazoparib, GSCs showed varying sensitivity to single-agent talazoparib. To account for this bias, we used a bliss independence model to calculate the synergistic effect and determine the combination effect in GSCs. The EOB additivity was as high as 27.4% in MGMT+ cells, indicating that the combination had a significant synergistic effect in MGMT+ cells, while the EOB was close to 0% in MGMT− cells, indicating no synergistic effect (Figure 1D, Supplementary Figure S1A).
We depleted MGMT in MGMT+ GSC23 using O6BG—a competitive inhibitor of MGMT. We show that treatment of MGMT-expressing GSC 23 with O6BG to deplete MGMT was able to reverse the sensitizing effect of PARP inhibitor talazoparib, thereby showing that MGMT is the specific target of PARP inhibition for the TMZ and talazoparib synergistic activity (Supplementary Figure S1B).
We also analyzed the sphere-forming capability of both MGMT+ and MGMT− cells in the presence of either TMZ alone or combination with PARP inhibitors. MGMT+ cells were resistant to TMZ monotherapy, as shown by the no inhibition of sphere formation by TMZ, whereas TMZ suppressed sphere formation in MGMT− GSCs. The addition of talazoparib remarkably potentiated the TMZ-induced inhibition of sphere formation in MGMT+ cells, while no significant potentiation was seen in MGMT− cells (Figure 1E).
PARP Physically Binds With MGMT and PARylates MGMT
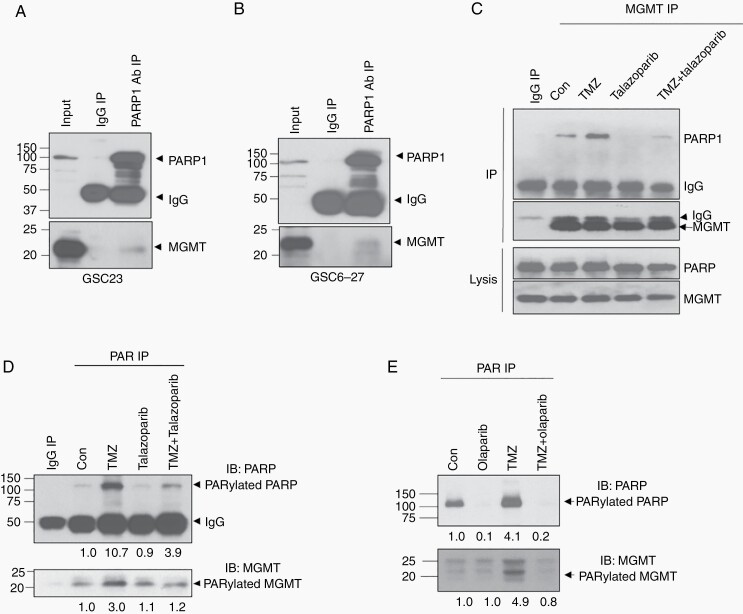
As PARP inhibitors potentiated TMZ-induced cytotoxicity in MGMT+ GSCs but not in MGMT− GSCs, we hypothesized that PARP regulates MGMT function. We first determined if there is an interaction between PARP and MGMT. In MGMT+ GSC23 cells, we showed that the endogenous MGMT was co-immunoprecipitated along with PARP by PARP1-specific antibody, suggesting a physical interaction between MGMT and PARP. IgG antibody failed to pull down MGMT, ruling out nonspecific immunoprecipitation (Figure 2A). The interaction between PARP and MGMT was validated in another MGMT+ cell line GSC6-27 (Figure 2B). The interaction between MGMT and PARP was moderate, as only a partial fraction of MGMT was pulled down by anti-PARP antibody (Figure 2A and B). We reasoned that the basal DNA damage level is low. We then asked if TMZ can induce increased PARP-MGMT interaction. Indeed, the baseline MGMT-PARP interaction was weak, as only a partial fraction of PARP was co-immunoprecipitated along with MGMT by anti-MGMT antibody; however, after TMZ treatment, a significant amount of PARP was co-immunoprecipitated, suggesting that the MGMT-PARP interaction was increased in response to TMZ-induced DNA damage (Figure 2C). TMZ-induced PARP-MGMT interaction was suppressed by the PARP inhibitor talazoparib (Figure 2C), suggesting that PARP activity is crucial for MGMT-PARP interaction.
Fig. 2.
PARP physically binds with MGMT and PARylates MGMT. A-B, Endogenous physical interaction of PARP and MGMT was detected by PARP antibody IP in GSC23 (A) and GSC6-27 (B). C, GSC23 was treated with 100 μM TMZ, 100 nM talazoparib, or combination for 3 days, and MGMT-PARP interaction was detected by MGMT antibody IP. D, GSC23 was treated with TMZ, talazoparib or combination, IP was performed with anti-PAR antibody to purify all PARylated proteins and followed by western blot with PARP and MGMT antibodies to detect PARylated PARP and PARylated MGMT. Quantification of PARylated PARP and PARylated MGMT was shown in the bottom. E, GSC23 was treated with TMZ, olaparib, or combination, IP was performed with anti-PAR antibody to purify all PARylated proteins and followed by western blot with PARP and MGMT antibodies to detect PARylated PARP and PARylated MGMT. Quantification of PARylated PARP and PARylated MGMT was shown in the bottom. Abbreviations: GSCs, glioma sphere-forming cells; IP, immunoprecipitation; MGMT, O6-methylguanine DNA methyltransferase; PARP, poly(ADP-ribose) polymerase; TMZ, temozolomide.
The main role of PARP1 is to catalyze the polymerization of ADP-ribose units, resulting in the attachment of either linear or branched PAR polymers to themselves or to other target proteins that are involved in DNA damage repair or chromatin remodeling. Therefore, we further determined whether MGMT is a PARylation target of PARP. We used anti-PAR antibody to immunoprecipitate all PARylated proteins in GSC23 cells, followed by immunoblotting with PARP antibody to detect PARylated PARP and anti-MGMT antibody to detect PARylated MGMT. We showed that PARP itself was PARylated; the PARylation of PARP was increased in response to TMZ treatment (Figure 2D and E), suggesting that PARP was activated by TMZ-induced DNA damage. As expected, the PARP inhibitor talazoparib suppressed TMZ-induced PARylation of PARP. Interestingly, we found that MGMT was also PARylated and that the PARylation was induced by TMZ treatment. More interestingly, the TMZ-induced PARylation of MGMT was suppressed by the PARP inhibitors talazoparib (Figure 2D) and olaparib (Figure 2E).
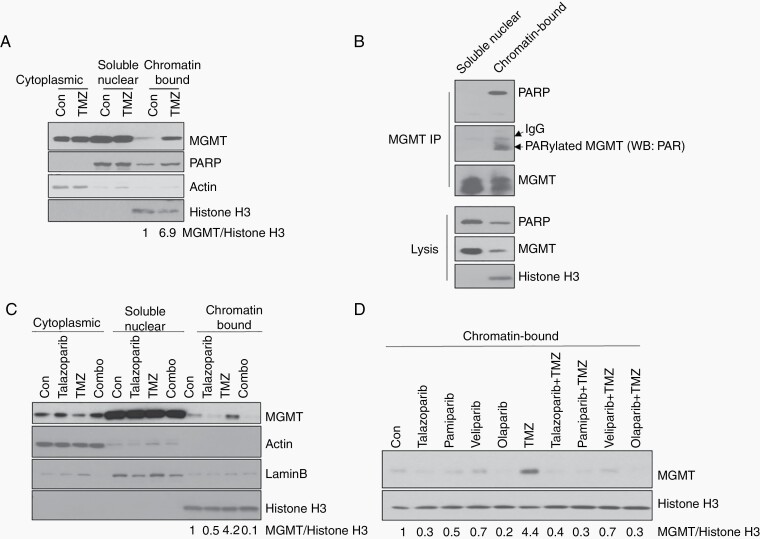
PARylated MGMT Binds to Chromatin and PARP Inhibitors Suppressed MGMT-Chromatin DNA Binding
It is well known that the PARylation of chromatin remodeling proteins or DNA repair proteins by PARP enhances accessibility to chromatin; therefore, we determined whether the PARylation of MGMT improves the DNA-binding capacity of MGMT. We first fractionated the nuclear protein into a soluble nuclear fraction and chromatin-bound fraction. As shown in Figure 3A, a small fraction of MGMT was detected in the chromatin-bound fraction compared to in the soluble nuclear fraction, indicating that the basal level of MGMT-DNA binding is very low. However, after TMZ treatment, the levels of both chromatin-bound PARP and chromatin-bound MGMT were increased, indicating that TMZ induced the binding of PARP to damaged DNA and that PARP recruited MGMT to chromatin.
Fig. 3.
PARylated MGMT binds to chromatin and PARP inhibitors suppressed MGMT-Chromatin DNA binding. A, GSC23 was treated with 100 μM TMZ for 3 days. Cytoplasmic, nuclear soluble, and chromatin-bound fraction were analyzed for MGMT expression by western blot. Actin, Lamin B, and histone H3 were used as makers for cytoplasm, nuclear and chromatin-bound fraction, respectively. B, Nuclear soluble and chromatin-bound fractions from TMZ-treated GSC23 were analyzed by IP with anti-MGMT antibody, followed by western blot with PARP antibody to detect MGMT-PARP interaction, and PAR antibody to detect PARylated MGMT. C, GSC23 was treated with TMZ and talazoaprib, western blot to show MGMT expression in subcellular fraction. D, GSC23 was treated with TMZ and different PARP inhibitors, Chromain-bound subcellular fraction was purified and MGMT expression was analyzed by western blot. Abbreviations: IP, immunoprecipitation; MGMT, O6-methylguanine DNA methyltransferase; PARP, poly(ADP-ribose) polymerase; TMZ, temozolomide.
To further demonstrate that PARP recruited MGMT and that PARylation of MGMT is coupled with its chromatin translocation, we performed immunoprecipitation in the soluble nuclear and chromatin-bound nuclear fraction. Although there was an abundance of MGMT and PARP in soluble nuclear fraction lysate, PARP was not co-precipitated with MGMT by anti-MGMT antibody, suggesting that no MGMT-PARP interaction was detected in the soluble nuclear fraction (Figure 3B). In contrast, despite the low expression of MGMT and PARP in the chromatin-bound nuclear fraction, a large portion of PARP was co-precipitated with MGMT using anti-MGMT antibody, indicating that the PARP-MGMT interaction specifically occurred in the chromatin fraction. More interestingly, only chromatin-bound MGMT was PARylated by PARP (Figure 3B), suggesting that PARylation of MGMT is critical for MGMT binding to DNA.
To further confirm that MGMT PARylation is mediated by PARP1, we treated cells with TMZ, talazoparib, or combination. The results showed that chromatin-bound MGMT was decreased in talazoparib compared to the control group, suggesting that talazoparib inhibited the basal level of MGMT binding to chromatin (Figure 3C). More importantly, chromatin-bound MGMT was decreased remarkably after combination treatment compared to TMZ treatment alone, suggesting that talazoparib suppressed TMZ-induced MGMT-DNA binding (Figure 3C). Since talazoparib has PARP trapping activity, we determined whether this inhibition of MGMT-DNA binding was due to the trapping activity of the PARP inhibitor. For that, we used other PARP inhibitors (veliparib, olaparib, and pamiparib) with different PARP trapping abilities. Both trapping and non-trapping PARP inhibitors suppressed TMZ-induced MGMT-DNA binding (Figure 3D).
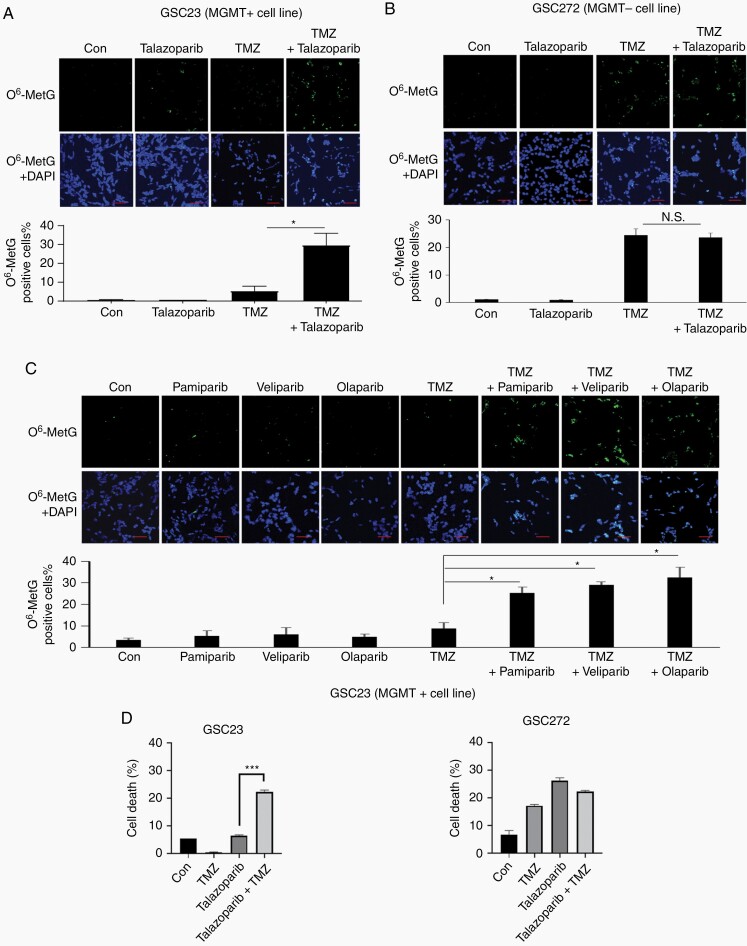
PARP Inhibition Potentiated TMZ-Induced O6-MetG Accumulation and Apoptosis in MGMT+ Cells
Based on the data that PARP inhibition blocked MGMT binding to DNA, we hypothesized that PARP inhibition suppressed the repair function of MGMT to remove TMZ-induced O6-MetG. To test this, we treated the MGMT+ cell line GSC23 and MGMT− cell line GSC272 with the PARP inhibitor talazoparib and TMZ; O6-MetG was detected by immunocytochemical analysis. In the MGMT+ GSC23, TMZ alone induced a weak accumulation of O6-MetG level indicating that MGMT actively removed TMZ-induced O6-MetG; the addition of talazoparib significantly enhanced TMZ-induced O6-MetG (Figure 4A). This suggests that PARP inhibition suppressed MGMT PARylation and subsequent MGMT-DNA binding, leading to decreased MGMT activity.
Fig. 4.
PARP inhibition potentiated TMZ-induced O6-MetG accumulation in MGMT+ cells. GSC23 (A) and GSC272 (B) were treated with 100 µM TMZ, 100 nM talazoparib, or combination for 3 days, cells were stained with O6-MetG antibody and DAPI. C, GSC23 cells were treated with TMZ, pamiparib, veliparib, olaparib, or combination, cells were stained with O6-MetG antibody and DAPI. Representative fields were shown. and O6-MetG-positive cells were quantified by counting 3 fields of each treatment. Scale bars, 50 microns. D, GSC23 and GSC272 cells were treated with 100 μM TMZ, 100 nM talazoparib, or combination for 3 days. Cells were stained with FITC annexin V and 7-amino-actinomycin D and cell death was measured by flow cytometry. Abbreviations: DAPI, 4′,6-diamidino-2-phenylindole; GSCs, glioma sphere-forming cells; MGMT, O6-methylguanine DNA methyltransferase; PARP, poly(ADP-ribose) polymerase; TMZ, temozolomide.
In contrast, TMZ alone induced strong O6-MetG accumulation in MGMT− GSC272 cells; the addition of talazoparib failed to enhance TMZ-induced O6-MetG in GSC272 cells (Figure 4B), suggesting that the observed effect of PARP inhibition on O6-MetG is MGMT dependent. Further, we showed that other PARP inhibitors pamiparib, veliparib, and olaparib also potentiated TMZ-induced O6-MetG accumulation in MGMT+ GSC23 cells (Figure 4C), suggesting that all PARP inhibitors, regardless of their trapping capability, suppressed MGMT activity.
To show that increased O6-MetG accumulation leads to increased double-strand break (DSB) and subsequent cell death, we did apoptosis analysis using annexin V staining. Results showed that in MGMT+ GSC23, TMZ combined with PARP inhibitor induced apoptosis as shown by increased annexin V staining in comparison to TMZ or PARP inhibitor alone. However, the induction of apoptosis was not observed in MGMT− GSC 272 (Figure 4D). We further showed that increased apoptosis in MGMT+ GSC23 was due to increased DSB accumulation shown by elevated γH2AX expression in TMZ and PARP inhibitor treatment groups (Supplementary Figure S2).
PARP Inhibition Increased TMZ Sensitivity in MGMT+ Tumors In Vivo
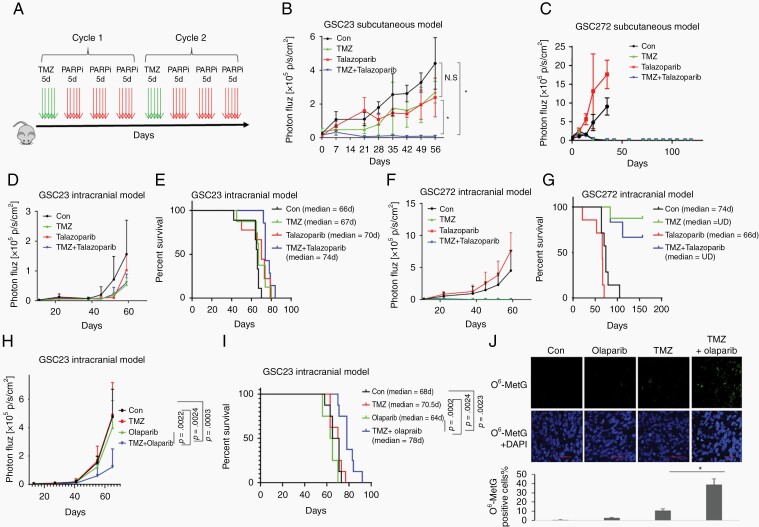
To determine whether PARP inhibition can restore TMZ sensitivity in vivo, we first studied the combination effect in subcutaneous mouse models (Figure 5A). In the MGMT+ GSC23 model (Figure 5B), TMZ treatment alone did not significantly inhibit tumor progression, which is consistent with our in vitro data and the results of clinical reports1,2,9 that showed that MGMT expression confers resistance to TMZ therapy. Consistent with the results of our previous report,17 talazoparib alone did not significantly inhibit tumor growth in GSC23. However, sequential combination dosing of TMZ and talazoparib significantly suppressed tumor growth (Figure 5B). In MGMT− GSC272 cells, TMZ single-agent treatment markedly reduced tumor growth, and the addition of talazoparib did not show an additive effect (Figure 5C). We next determined whether talazoparib can potentiate TMZ therapy in an intracranial model. Similar to the subcutaneous model, TMZ treatment alone showed no significant survival benefit in MGMT+ GSC23 mice (Figure 5D and E); however, we observed increased survival in MGMT− GSC272 mice (Figure 5F and G). The addition, talazoparib did not potentiate TMZ therapy with regard to tumor suppression or survival extension in either cell line, probably because of limited BBB (blood-brain barrier) penetration of talazoparib, as described previously.17
Fig. 5.
PARP inhibition restored TMZ sensitivity in MGMT+ tumor in vivo. A, A graphic scheme for sequential combination of TMZ with PARP inhibitors, TMZ 50 mg/kg/day; talazoparib 0.33 mg/kg/day, olaparib 25 mg/kg/day. B and C, MGMT+ GSC23 (B) and MGMT− GSC272 (C) were implanted in nude mice to establish subcutaneous models. Mice were administrated with TMZ, talazoparib, or sequential combination. Tumor size was monitored by bioluminescence imaging. D-G, Mice bearing GSC23 (D-E) and GSC272 (F-G) intracranial tumors were administrated with TMZ, talazoparib, or sequential combination. Tumor growth was evaluated by bioluminescence imaging (D and F), survival curves were compared using Kaplan-Meier survival plots (E and G). H-J, Mice bearing GSC23 intracranial tumors were administrated with TMZ, olaparib, or sequential combination. Tumor growth was evaluated by bioluminescence imaging (H), survival curves were compared using Kaplan-Meier survival plots (I). The tissue sections were incubated with antibodies against O6-MetG and quantified by Image-Pro Plus (J). Scale bars: 50 microns. Abbreviations: GSCs, glioma sphere-forming cells; MGMT, O6-methylguanine DNA methyltransferase; PARP, poly(ADP-ribose) polymerase; TMZ, temozolomide.
Further studies were performed with other BBB-penetrant PARP inhibitors, olaparib,18 veliparib, and niraparib to study the sensitizing effects on TMZ -treatment. Olaparib, veliparib, and niraparib alone did not suppress tumor progression or extend animal survival in a GSC23 intracranial model (Figure 5H-J and Supplementary Figure S3). GSC23 tumors were resistant to TMZ monotherapy as TMZ did not suppress tumor progression, nor did it extend animal survival. However, combination of olaparib (Figure 5H), veliparib, and niraparib (Supplementary Figure S3) with TMZ significantly inhibited tumor growth (Figure 5H; P = .0003 for combination vs control, P = .0022 for combination vs olaparib alone, P = .0024 for combination vs TMZ alone) and extended animal survival (Figure 5I, P = .0024 for combination vs TMZ alone, P = .0002 for combination vs olaparib alone, and P = .0023 for combination vs control, log-rank test). The toxicity was evaluated in nude mice for 9 weeks. Animals receiving the regimens involving TMZ lost 7%-10% body weight, however, they recovered promptly after completion of treatment. Combining PARP inhibitors with TMZ did not confer additional toxicity as the body weights of TMZ and combination groups were comparable. Analysis of tumor tissue showed that TMZ alone failed to introduce O6-MetG accumulation; addition of olaparib significantly enhanced TMZ-induced O6-MetG (Figure 5J). Taken together, our data showed that PARP inhibition restored TMZ sensitivity in MGMT+ tumors in vivo.
Discussion
This study shows that PARP inhibitors increase TMZ sensitivity and selectivity in MGMT-unmethylated GBM by regulating MGMT protein activity by a mechanism independent of BER. PARP inhibitor-mediated inhibition of BER activity is considered to be the likely mechanism by which TMZ-resistant GBM tumors are sensitized to TMZ. It is imperative to understand the mechanisms that trigger the potentiation effect of PARP inhibitors to identify molecularly defined GBM patients who will benefit from such combinations.18
In this study, we showed that PARP inhibitors restored TMZ cytotoxicity in MGMT+ GSCs, both in vitro and in vivo, by a mechanism independent of the classic BER function of PARP inhibitors. We showed that PARP regulates MGMT activity by PARylating MGMT and enhances MGMT/DNA binding in response to TMZ treatment and that PARP inhibition suppresses MGMT PARylation, abolishes O6-MetG repair, and sensitizes MGMT+ GSCs to TMZ. Our results demonstrate that PARP inhibitors synergize with TMZ in MGMT+ GSCs but not in MGMT− cells.
PARP plays a key role in the BER/SSBR pathway, which repairs oxidative stress and other stimuli induced SSBs. Given the role of PARP in SSBR and BER in repairing N3-methyladenine and N7-methylguanine lesions, PARP inhibitor-mediated potentiation of TMZ sensitivity has been attributed to be due to inhibition of BER. However, recent studies showed that PARP inhibitor-mediated restoration of TMZ sensitivity could be independent of BER, as genetic and pharmacologic blockage of BER pathway enzymes and XRCC1 knockdown failed to recapitulate this phenotype.19 Consistent with this finding, we showed that PARP regulates MGMT activity by PARylating MGMT in response to TMZ treatment. Further, PARP inhibitors suppressed MGMT PARylation, MGMT-DNA binding, and O6-MetG repair, rendering cells sensitive to TMZ. These findings are supported by observations from a previous report8 that showed that MGMT expression provides resistance to TMZ in a PARP-dependent manner. Interestingly, that study also showed that PARP inhibitors did not sensitize cells to TMZ when MGMT activity was inhibited,8 suggesting that MGMT is required for this function. Our study conclusively demonstrated that PARP physically interacted with MGMT and PARylated MGMT and that PARP inhibitors inhibited the PARP-mediated PARylation of MGMT, providing a unique mechanistic explanation for predicting PARP inhibitors potentiation to TMZ. These results suggest that PARP inhibition suppressed MGMT PARylation and subsequent MGMT-DNA binding, leading to decreased MGMT activity and increased O6-MetG accumulation. This increased accumulation of O6-MetG resulted in increased DNA damage in MGMT+ GSCs and sensitization to TMZ, leading to cell death.
It has been well documented that PARP plays an important role in recognizing DNA lesions and initiating BER, NER, HR, and NHEJ pathways by recruiting repair machinery to the site of DNA damage.20,21 PARP functions by modifying directly (covalently) or indirectly (noncovalently) by adding poly(ADP-ribosyl)ation to repair proteins. PARP catalyzes the transfer of ADP-ribose polymers to substrates, including numerous DNA repair enzymes to sense DNA lesions, activates DNA damage responses, and facilitates DNA damage repairs.22 Our study further showed that PARP recognizes TMZ-induced DNA damage and auto-modified PARP directs PARylation of MGMT. Inhibition of PARP enzymatic activity by PARP inhibitors suppressed MGMT PARylation, MGMT-DNA binding, and O6-MetG repair, suggesting that MGMT PARylation induces the conformational changes in MGMT that are needed for its easy accessibility to the alkylated base. PARP inhibitors have also been reported to sensitize mismatch repair (MMR)-defective GBM cells to TMZ.19,23,24 Mechanistically, this PARP inhibitor-mediated synthetic phenotype is independent of BER blockage remains to be investigated.19 As TMZ-induced O6-MetG is repaired by MGMT or MMR, MMR−GBM tumors rely on MGMT to repair O6-MetG. Meanwhile, MGMT expression has been found to be negatively correlated with MMR complex expression: GBM cells with a very low MMR expression level had a high MGMT expression level.25 Thus, it is likely that restoration of TMZ sensitivity in those MMR−GBM tumors was mediated by PARP inhibitor-repressed MGMT activity.
Several PARP inhibitors have been developed in preclinical and clinical studies for various tumors, including GBM.26 Combinations of the PARP inhibitors olaparib and veliparib with TMZ have been studied in a phase I trial and phase II/III trials, respectively.27,28 However, excessive toxicity has limited the efficacy of TMZ combination therapy and has led to clinical trial failure. It is important to determine strategies and identify predictive biomarkers to select GBM patients who will most likely benefit from PARP inhibitors. We tested various PARP inhibitors, including olaparib, pamiparib, and veliparib, in addition to talazoparib, and found that irrespective of their DNA trapping function, all PARP inhibitors sensitized MGMT+ GSCs to TMZ. Therefore, the trapping function of PARP inhibitors may not play a part in this sensitization, as olaparib and veliparib have low trapping ability compared to talazoparib and pamiparib but had similar effects in sensitizing MGMT+ cells to TMZ. Therefore, the catalytic activity of PARP inhibitors is required for this function, as shown by both in vitro and in vivo models using these PARP inhibitors. A recent study in Ewings Sarcoma also confirmed that PARP1/MGMT interaction may enhance the repair of TMZ-induced DNA damage.29
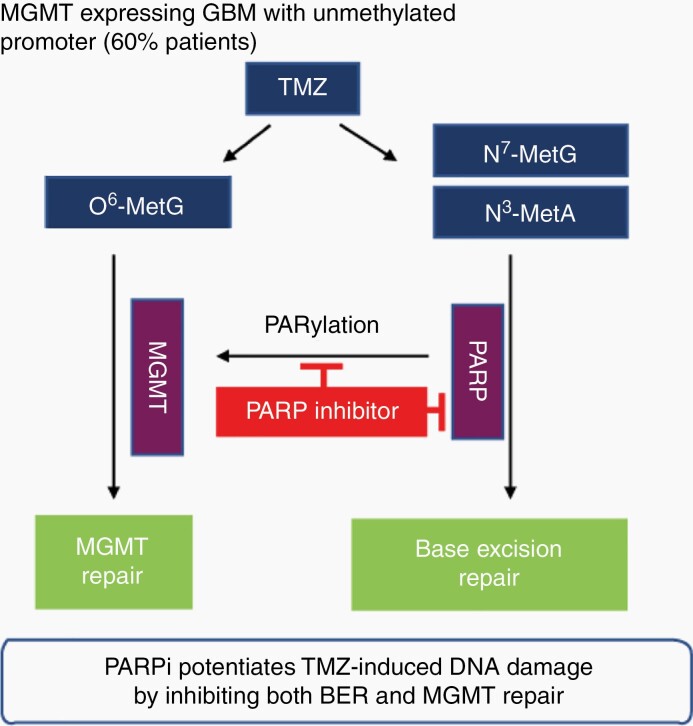
We propose that PARP inhibition by PARP inhibitors acts as a double-edged sword in MGMT-expressing GBM, first by blocking the BER/SSBR pathway to repair TMZ-induced N7-MetG and N3-MetA and second, more importantly, by suppressing MGMT activity to repair O6-MetG, resulting in augmented cytotoxicity (Figure 6). We described the mechanistic relationship of PARP-MGMT binding and further clarified the mechanism of MGMT-mediated repair of TMZ-induced O6-MetG, providing a rationale for sensitizing TMZ using PARP inhibitors in patients with MGMT-unmethylated GBM.
Fig. 6.
Model summarizing the proposed mechanism. PARP physically interacts with and PARylates MGMT to remove O6-MetG adducts in damaged DNA, independent of base excision repair, in response to TMZ treatment. In addition, PARP acts as a sensor to elicit response pathways in base excision repair. PARP inhibitors suppress PARP-MGMT binding and abolish MGMT function. Therefore, treatment with TMZ and PARP inhibitors inhibits both BER- and MGMT-mediated repair and results in an increased antitumor effect. Abbreviations: BER, base excision repair; MGMT, O6-methylguanine DNA methyltransferase; PARP, poly(ADP-ribose) polymerase; TMZ, temozolomide.
Supplementary Material
Acknowledgments
The authors thank Verlene Henry and Tiara Collier for performing animal studies and Ann Sutton in the Scientific Publications department for manuscript editing.
Funding
This study was funded by Cancer Center support grant (CA016672) from Defeat GBM Research Collaborative, a subsidiary of the National Brain Tumor Society to W.K.A.Y. and J.F.G.
Conflict of interest statement. W.K.A.Y. discloses a conflict of interest as a consultant with DNATrix. The rest of the authors have no conflicts of interest to disclose.
Author contributions. D.K., W.K.A.Y., and S.W. designed the study. S.W., F.G., and X.L. conducted the experiment and collected the data. S.W. performed the statistical analysis. S.W. and D.K. prepared the manuscript. D.K. and W.K.A.Y. revised the manuscript. All authors read and approved the final manuscript.
References
- 1. Hegi ME, Diserens AC, Gorlia T, et al. . MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352(10):997–1003. [DOI] [PubMed] [Google Scholar]
- 2. Esteller M, Garcia-Foncillas J, Andion E, et al. . Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents. N Engl J Med. 2000;343(19):1350–1354. [DOI] [PubMed] [Google Scholar]
- 3. Ko HL, Ren EC. Functional aspects of PARP1 in DNA repair and transcription. Biomolecules. 2012;2(4):524–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Palma JP, Rodriguez LE, Bontcheva-Diaz VD, et al. . The PARP inhibitor, ABT-888 potentiates temozolomide: correlation with drug levels and reduction in PARP activity in vivo. Anticancer Res. 2008;28(5A): 2625–2635. [PubMed] [Google Scholar]
- 5. Horton TM, Jenkins G, Pati D, et al. . Poly(ADP-ribose) polymerase inhibitor ABT-888 potentiates the cytotoxic activity of temozolomide in leukemia cells: influence of mismatch repair status and O6-methylguanine-DNA methyltransferase activity. Mol Cancer Ther. 2009;8(8):2232–2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tentori L, Ricci-Vitiani L, Muzi A, et al. . Pharmacological inhibition of poly(ADP-ribose) polymerase-1 modulates resistance of human glioblastoma stem cells to temozolomide. BMC Cancer. 2014;14:151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gupta SK, Smith EJ, Mladek AC, et al. . PARP inhibitors for sensitization of alkylation chemotherapy in glioblastoma: impact of blood-brain barrier and molecular heterogeneity. Front Oncol. 2018;8:670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Erice O, Smith MP, White R, et al. . MGMT expression predicts PARP-mediated resistance to temozolomide. Mol Cancer Ther. 2015;14(5):1236–1246. [DOI] [PubMed] [Google Scholar]
- 9. Mansouri A, Hachem LD, Mansouri S, et al. . MGMT promoter methylation status testing to guide therapy for glioblastoma: refining the approach based on emerging evidence and current challenges. Neuro Oncol. 2019;21(2):167–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Saito N, Fu J, Zheng S, et al. . A high Notch pathway activation predicts response to γ secretase inhibitors in proneural subtype of glioma tumor-initiating cells. Stem Cells. 2014;32(1):301–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Greco WR, Bravo G, Parsons JC. The search for synergy: a critical review from a response surface perspective. Pharmacol Rev. 1995;47(2):331–385. [PubMed] [Google Scholar]
- 12. Wu S, Wang S, Zheng S, Verhaak R, Koul D, Yung WK. MSK1-mediated β-catenin phosphorylation confers resistance to PI3K/mTOR inhibitors in glioblastoma. Mol Cancer Ther. 2016;15(7):1656–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Seiler F, Kirstein U, Eberle G, Hochleitner K, Rajewsky MF. Quantification of specific DNA O-alkylation products in individual cells by monoclonal antibodies and digital imaging of intensified nuclear fluorescence. Carcinogenesis. 1993;14(9):1907–1913. [DOI] [PubMed] [Google Scholar]
- 14. Thomale J, Seiler F, Müller MR, Seeber S, Rajewsky MF. Repair of O6-alkylguanines in the nuclear DNA of human lymphocytes and leukaemic cells: analysis at the single-cell level. Br J Cancer. 1994;69(4): 698–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Murai J, Huang SY, Renaud A, et al. . Stereospecific PARP trapping by BMN 673 and comparison with olaparib and rucaparib. Mol Cancer Ther. 2014;13(2):433–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lal S, Lacroix M, Tofilon P, Fuller GN, Sawaya R, Lang FF. An implantable guide-screw system for brain tumor studies in small animals. J Neurosurg. 2000;92(2):326–333. [DOI] [PubMed] [Google Scholar]
- 17. Wu S, Gao F, Zheng S, et al. . EGFR amplification induces increased DNA damage response and renders selective sensitivity to talazoparib (PARP inhibitor) in glioblastoma. Clin Cancer Res. 2020;26(6):1395–1407. [DOI] [PubMed] [Google Scholar]
- 18. Hanna C, Kurian KM, Williams K, et al. . Pharmacokinetics, safety, and tolerability of olaparib and temozolomide for recurrent glioblastoma: results of the phase I OPARATIC trial. Neuro Oncol. 2020;22(12):1840–1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Higuchi F, Nagashima H, Ning J, Koerner MVA, Wakimoto H, Cahill DP. Restoration of temozolomide sensitivity by PARP inhibitors in mismatch repair deficient glioblastoma is independent of base excision repair. Clin Cancer Res. 2020;26(7):1690–1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ray Chaudhuri A, Nussenzweig A. The multifaceted roles of PARP1 in DNA repair and chromatin remodelling. Nat Rev Mol Cell Biol. 2017;18(10):610–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li M, Yu X. The role of poly(ADP-ribosyl)ation in DNA damage response and cancer chemotherapy. Oncogene. 2015;34(26):3349–3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yoshimoto K, Mizoguchi M, Hata N, et al. . Complex DNA repair pathways as possible therapeutic targets to overcome temozolomide resistance in glioblastoma. Front Oncol. 2012;2:186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yuan AL, Ricks CB, Bohm AK, et al. . ABT-888 restores sensitivity in temozolomide resistant glioma cells and xenografts. PLoS One. 2018;13(8):e0202860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cheng CL, Johnson SP, Keir ST, et al. . Poly(ADP-ribose) polymerase-1 inhibition reverses temozolomide resistance in a DNA mismatch repair-deficient malignant glioma xenograft. Mol Cancer Ther. 2005;4(9):1364–1368. [DOI] [PubMed] [Google Scholar]
- 25. Perazzoli G, Prados J, Ortiz R, et al. . Temozolomide resistance in glioblastoma cell lines: implication of MGMT, MMR, P-glycoprotein and CD133 expression. PLoS One. 2015;10(10):e0140131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ganguly B, Dolfi SC, Rodriguez-Rodriguez L, Ganesan S, Hirshfield KM. Role of biomarkers in the development of PARP inhibitors. Biomark Cancer. 2016;8(Suppl 1):15–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lesueur P, Lequesne J, Grellard JM, et al. . Phase I/IIa study of concomitant radiotherapy with olaparib and temozolomide in unresectable or partially resectable glioblastoma: OLA-TMZ-RTE-01 trial protocol. BMC Cancer. 2019;19(1):198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Baxter PA, Su JM, Onar-Thomas A, et al. . A phase I/II study of veliparib (ABT-888) with radiation and temozolomide in newly diagnosed diffuse pontine glioma: a Pediatric Brain Tumor Consortium study. Neuro Oncol. 2020;22(6):875–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dauren A, Jodie C, Rostislav L, et al. . PARP1 and MGMT interaction-based sensitivity to DNA damage in Ewing sarcoma. bioRxiv. 2020.01.26.920405. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.