Abstract
Background
Fibroblast (FGFs) and insulin (IGF) growth factor pathways are among 10 most recurrently altered genomic pathways in pancreatic ductal adenocarcinoma (PDAC). However, the prognostic and therapeutic relevance of FGF and IGF pathways in PDAC is largely unknown.
Methods
We investigated the relationship between fibroblast and insulin pathway gene expression and clinicopathological features in three independent transcriptomic cohorts of 532 PDAC patients. Furthermore, we have examined the coexpressed genes specific to the prognostic marker identified from these cohorts. Statistical tests including Fisher-exact\Chi-square, Kaplan–Meier, Pearson Correlation and cox regression analyses were performed. Additionally, pathway analysis of gene-specific co-expressed genes was also performed.
Results
The dysregulation of six genes including FGF9, FGF14, FGFR1, FGFR4, IGF2BP2 and IGF2BP3 were significantly associated with different clinical characteristics (including grade, stage, recurrence and nodes) in PDAC cohorts. 11 genes (including FGF9, FGF13, FGF14, FGF17, FGFR1, FGFRL1, FGFBP3, IGFBP3, IGF2BP2, IGF2BP3 and IGFBPL1) showed association with overall survival in different PDAC cohorts. Interestingly, overexpression of FGF14 was found associated with better overall survival (OS) in all three cohorts. Of note, multivariate analysis also revealed FGF14 as an independent prognostic marker for better OS in all three cohorts. Furthermore, FMN2 and PGR were among the top genes that correlated with FGF14 in all 3 cohorts. Of note, overexpression of FMN2 and PGR was found significantly associated with good overall survival in PDAC patients, suggesting FMN2 and PGR can also act as potential markers for the prediction of prognosis in PDAC patients.
Conclusion
FGF14 may define a distinct subset of PDAC patients with better prognosis. Moreover, FGF14-based sub-classification of PDAC suggests that FMN2 and PGR can be employed as good prognostic markers in PDAC and this classification may lead to new therapeutic approaches.
Introduction
Pancreatic adenocarcinoma (PDAC) is the fourth most fatal cancer with increasing mortality rate [1]. Interestingly, PDAC accounts for more than 95% of exocrine pancreatic cancers [2]. PDAC imposes great mortality risk due to its rapid spread, poor prognosis, scarcity of available treatment options, late detection, invulnerability towards chemotherapy and lower overall survival rate [3]. The sensitivity profiles exhibited by PDAC towards chemotherapy and radiotherapy remain very low [4, 5]. As PDAC shows very high resistance to treatment, it is ranked among the most aggressive tumor types [6]. The major non-genetic risk factors of PDAC are chronic pancreatitis, diabetes, and obesity [7]. Among all these risk factors, diabetes is the most prominent factor, as pancreatic cancer and diabetes both affect the same organ [8]. In addition, both of these diseases also share common risk factors [9]. Keeping in view the aggressive nature and low therapeutic options available for PDAC, proper screening and identification of suitable therapeutic targets is urgently required [10].
The aggressiveness exhibited by PDAC is not only due to environmental or specific molecular traits but also due to genetically altered pathways [11]. Importantly, two growth factor receptor pathways including Fibroblast growth factors (FGFs) and insulin growth factors (IGF) are frequently reported in PDAC [12]. For instance, altered expression of FGFR1 and FGFR2 receptors is reported in PDAC pathogenesis [13, 14]. Very importantly, Motoda et al. revealed that FGF19 (specific ligand of FGFR4) is involved in suppressing PDAC progression by stimulating FGFR4 expression i.e. the overexpression of FGFR4 correlates with good survival outcomes in PDAC [15]. Furthermore, studies also reported that dysregulation of genes of Insulin growth factor pathway also lead to excessive growth stimulation in human pancreatic cancer [16–18]. Particularly, IGF-I is considered as potential therapeutic target for pancreatic tumors (along with IGF-IR) [19].
In this study, we aimed to investigate integrated role of FGFs and IGF family genes on the prognosis of PDAC patients. In addition, we also examined gene-specific co-expressed genes with the independent prognostic marker revealed in the first half of analyses and investigated their significance in PDAC patients. For these purposes, microarray and RNA-seq data of 532 PDAC patients were analyzed in PDAC patients.
Materials and methods
Data collection and processing
The overall study design is presented in S1 Fig in S1 File. This study is certified and approved by COMSATS University review board (CIIT/Bio/ERB/16/21). Transcriptomic profiling of FGF and IGF pathway genes in 532 (65+179+288) PDAC patients was performed. The discovery cohort 1 (GSE62452) was retrieved from Gene Expression Omnibus (GEO) database [20]. Robust multiarray average (RMA) normalization was performed to generate mRNA expression. The expression data was divided into high and low expression groups based on their median values. The clinicopathological parameters of cohort 1 included grade, stage and survival status, details are summarized in S1 Table in S1 File. The discovery cohort 2 of 179 PDAC patients was obtained from public cBioPortal database (https://www.cbioportal.org/). Similarly, reads per kilobase million (RPKM) expression values were divided into high and low expression groups. The clinicopathological parameters of cohort 2 included age, gender, stage, grade, T-stage, nodes, metastasis, residual, survival and disease-free survival (DFS) status, type-2 diabetes and alcohol consumption history (S2 Table in S1 File). For validation of PDAC results, expression data of 288 PDAC patients was retrieved from ArrayExpress (E-MTAB-6134) database and RMA processing was done to generate mRNA expression. The cohort 3 included classification system based on tumor features and includes clinicopathological parameters such as gender, nodes, survival status and t-stage. The details of clinicopathological features of cohort 3 are described in S3 Table in S1 File. R-script was used for the modification and pre-processing of all the cohorts under study.
Functional analysis of gene-specific coexpressed genes
The gene-specific coexpressed genes obtained through the analysis were subjected to pathway analysis using the ClusterProfiler program [21] implemented in R software version 4.0.3. Only genes that were common in cohort 1, 2 and 3 were subjected to this analysis. The analysis was conducted under the specific parameters including statistical Fisher’s exact test, along with Benjamini correction and the cut-off value set for the analysis was adjusted P-value <0.05.
Statistical analysis
IBM SPSS® software version 20.0 (Armonk, NY, USA) and R software version 4.0.3 were used for the statistical evaluation of the data. Statistical correlation of gene expression was performed against clinical parameters available for each cohort. The association was determined using Chi-square and Fisher Exact tests. We generated Kaplan Meier plots to determine the association with overall survival and disease-free survival. Furthermore, to predict independent prognostic markers for PDAC, cox regression univariate and multivariate analysis was performed. The probability value less than 0.05 was considered statistically significant. To examine gene-specific coexpressed genes, Pearson’s correlation using the gene of interest was employed on all the three cohorts of PDAC patients using R-script. VennDiagram [22] package implemented in R was used to generate customizable venn diagrams to identify common significantly associated genes among the cohorts used in the analysis.
Results
Association of genes with clinical parameters in cohort 1
The dichotomized expression values of FGF and IGF pathway genes were evaluated against the clinical features available. According to the results, most of the genes showed significant (*p ≤ 0.05) association with early tumor grade and stage. The information regarding significant association of genes with clinical parameters is summarized in S4 Table in S1 File. The overexpression of FGF6, FGF9, FGF14, FGFR1 and FGFR4 genes was correlated with early tumor grade (i.e., grade stage < = 2) (S4 Table in S1 File). In addition, overexpression of FGFR1 and IGFBP4 genes was found significantly associated with early tumor stage (i.e., tumor stage < = 2). However, the overexpression of FGFR1OP2, IGF2BP2, IGF2BP3, IGFL2 and IRS1 genes was significant in advance stages of grade (S4 Table in S1 File).
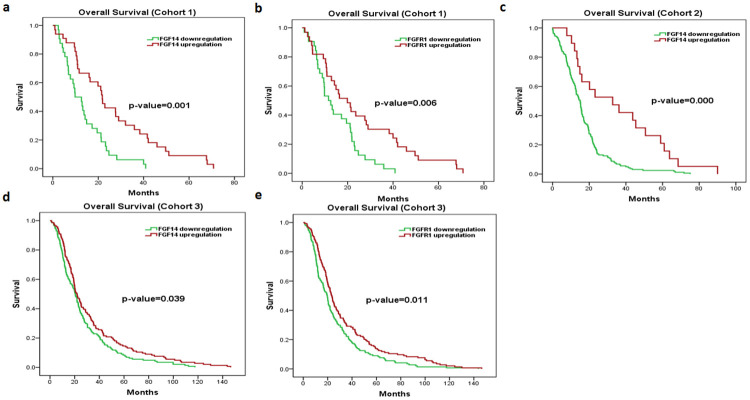
Kaplan-Meier survival analysis showed significantly lower overall survival (OS) of IGF2BP2 gene and IGF2BP3 overexpression, with Mantel-cox p-value of 0.046 and 0.021, respectively. (S2 Fig in S1 File) Interestingly, the overexpression of FGFR1 and FGF14 genes showed good overall survival (OS), with Mantel-cox p-value of 0.006 and 0.001, respectively (Fig 1).
Fig 1. Kaplan–Meier survival analyses of FGF14 and FGFR1 in PDAC cohorts 1, 2 and 3.
Red represents expression ≥ the median, while green represents expression < the median. Overall survival plots of FGF14 (a), FGFR1 (b) in cohort 1. Overall survival plots of FGF14 (c) in cohort 2. Overall survival plots of FGF14 (d), FGFR1 (e) in cohort 3. (* p ≤ 0.05).
Next, univariate/multivariate cox regression survival analysis was performed to identify potential prognostic markers for overall survival. According to univariate results, overall survival was significantly associated with FGF14 (HR 0.398, 95% CI 0.232–0.682, P = 0.001), FGFR1 (HR 0.475, 95% CI 0.277–0.817, P = 0.007), IGF2BP2 (HR 1.667, 95% CI 1.003–2.771, P = 0.049), IGF2BP3 (HR 1.836, 95% CI 1.089–3.094, P = 0.023) and Grade (HR 0.504, 95% CI 0.295–0.859, P = 0.012) (Table 1). Of note, multivariate analysis revealed FGF14 gene as the only independent prognostic marker for better overall survival (i.e., HR < 1) of PDAC patients (Table 1).
Table 1. Univariate and multivariate analysis of genes for overall survival in cohort 1.
| Factors | Univariate Analysis | Multivariable analysis | ||||||
|---|---|---|---|---|---|---|---|---|
| P-value | HR | 95.0% CI | P-value | 95.0% CI | ||||
| Lower | Upper | HR | Lower | Upper | ||||
| FGF14 | 0.001 | 0.398 | 0.232 | 0.682 | 0.045 | 0.545 | 0.301 | 0.987 |
| FGFR1 | 0.007 | 0.475 | 0.277 | 0.817 | ||||
| IGF2BP2 | 0.049 | 1.667 | 1.003 | 2.771 | ||||
| IGF2BP3 | 0.023 | 1.836 | 1.089 | 3.094 | ||||
| Grade | 0.012 | 0.504 | 0.295 | 0.859 | ||||
Association of genes with clinical parameters in cohort 2
In cohort 2, several genes under study have shown significant (*p ≤ 0.05) association with multiple clinicopathological features including age, gender, grade, stage, tumor recurrence, residual, nodes, survival, DFS status, diabetes and alcohol consumption history. The detailed information of significant association of genes with clinicopathological parameters is summarized in S5 Table in S1 File. For instance, overexpression of FGF4 and IGF2BP3 genes was associated with age < 50. The overexpression of IRS2 genes was significantly associated with alcoholic PDAC patients. Interestingly, overexpression of FGFR2, FGF7 genes and low expression of FGFR4 gene was found significantly associated with diabetic PDAC patients. The overexpression FGF7, FGF9, FGF14, FGF18 and IGF1 genes was found significant in early tumor grades. However, only overexpression of FGFBP1 gene showed association with advanced tumor grade. Furthermore, overexpression of FGF12, FGF14, FGF17, FGFR1, FGFBP3 and IGFBPL1 genes was found in early tumor stage, while, overexpression of IGF1R, IGF2BP2 and INSRR was found in advanced tumor stages. In addition, overexpression of FGF1, FGF9 and FGF14 genes were found significant in node negative status of PDAC patients (S5 Table in S1 File).
According to Kaplan Meier plots for overall survival, the overexpression of FGFRL1 (p-value = 0.033), IGFBP3 (p-value = 0.037) and IGFL1 (p-value = 0.001) showed poor overall survival, while, overexpression of FGF9 (p-value = 0.007), FGF13 (p-value = 0.015), FGF17 (p-value = 0.000), FGFBP3 (p-value = 0.000) and IGFBPL1 (p-value = 0.002) genes was associated with good overall survival of PDAC patients (S3 Fig in S1 File). Of note, FGF14 (p-value = 0.000) in overexpressed state correlated with good OS of PDAC patients (Fig 1). According to Kaplan Meier plots for disease-free survival (DFS), overexpression of FGF9 (p-value = 0.012), FGF12 (p-value = 0.001), FGF13 (p-value = 0.014), FGF14 (p-value = 0.000), FGF17 (p-value = 0.000), FGFBP3 (p-value = 0.000) and IGFBPL1 (p-value = 0.000) genes showed increased DFS, while, overexpression of FGF10 (p-value = 0.018), FGFRL1 (p-value = 0.031) and IGFL1 (p-value = 0.001) genes reduced DFS of PDAC patients (S4 Fig in S1 File).
Moreover, univariate analysis of discovery cohort 2 showed significant association of FGF9 (HR 0.541, 95% CI 0.342–0.855, P = 0.008), FGF13 (HR 0.510, 95% CI 0.293–0.887, P = 0.017), FGF14 (HR 0.397, 95% CI 0.240–0.657, P = 0.000), FGF17 (HR 0.244, 95% CI 0.106–0.560, P = 0.001), FGFBP3 (HR 0.272, 95% CI 0.137–0.540, P = 0.000), FGFRL1 (HR 1.658, 95% CI 1.035–2.656, P = 0.035), IGFBP3 (HR 1.558, 95% CI 1.022–2.375, P = 0.039), IGFBPL1 (HR 0.235, 95% CI 0.085–0.646, P = 0.005), Nodes (HR 0.714, 95% CI 0.513–0.995, P = 0.047) and Residual tumor (HR 0.673, 95% CI 0.486–0.932, P = 0.017) with hazard ratio and 95% CI presented in Table 2. Consistent with the results in cohort 1, FGF14 (HR 0.531, 95% CI 0.303–0.930, P = 0.027) was found as independent prognostic marker in multivariate analysis.
Table 2. Univariate and multivariate analysis of genes for overall survival in cohort 2.
| Factors | Univariate Analysis | Multivariable analysis | ||||||
|---|---|---|---|---|---|---|---|---|
| P-value | HR | 95.0% CI | P-value | HR | 95.0% CI | |||
| Lower | Upper | Lower | Upper | |||||
| FGF9 | 0.008 | 0.541 | 0.342 | 0.855 | ||||
| FGF13 | 0.017 | 0.510 | 0.293 | 0.887 | ||||
| FGF14 | 0.000 | 0.397 | 0.240 | 0.657 | 0.027 | 0.531 | 0.303 | 0.930 |
| FGF17 | 0.001 | 0.244 | 0.106 | 0.560 | ||||
| FGFBP3 | 0.000 | 0.272 | 0.137 | 0.540 | ||||
| FGFRL1 | 0.035 | 1.658 | 1.035 | 2.656 | ||||
| IGFBP3 | 0.039 | 1.558 | 1.022 | 2.375 | ||||
| IGFBPL1 | 0.005 | 0.235 | 0.085 | 0.646 | ||||
| Nodes | 0.047 | 0.714 | 0.513 | 0.995 | ||||
| Residual | 0.017 | 0.673 | 0.486 | 0.932 | 0.014 | 0.654 | 0.466 | 0.918 |
Of note, we have also evaluated the association of independent prognostic marker FGF14 with IGF pathway genes. Interestingly, FGF14 inversely correlated with important IGF pathway genes i.e. IGF2BP1, IGF2BP2, IGF2BP3, IGFBP6, IGFL1, IGFL2 and IGFL3 in cohort 1 and 2 of PDAC patients (S6 Fig in S1 File).
Validation of FGF14 as independent prognostic marker of PDAC
Interestingly, univariate analysis of validation cohort also showed significant association of FGF14 (HR 0.782, 95% CI 0.619–0.988, P = 0.039), FGFR1 (HR 0.740, 95% CI 0.586–0.935, P = 0.000) and nodes (HR 1.668, 95% CI 1.270–2.191, P = 0.000) with overall survival of PDAC patients. (Table 3) (Fig 1) Furthermore, multivariate analysis also revealed FGFR1 and FGF14 as independent prognostic markers for better overall survival in pancreatic cancer patients (Table 3). In addition, we have also evaluated the association of FGF14 with IGF pathway genes in the validation cohort. FGF14 inversely correlated with IGF2BP1, IGF2BP2, IGF2BP3, IGFBP6 and IGFL2 (S6 Fig in S1 File). Intriguingly, these results were consistent with the findings of cohort 1 and 2 of PDAC patients. Of note, inverse correlation of FGF14 with IGF2BP1, IGF2BP2, IGF2BP3, IGFBP6 and IGFL2 was found in all three cohorts of PDAC patients under study.
Table 3. Univariate and multivariate analysis of genes for overall survival in cohort 3.
| Factors | Univariate Analysis | Multivariable analysis | ||||||
|---|---|---|---|---|---|---|---|---|
| P-value | HR | 95.0% CI | P-value | HR | 95.0% CI | |||
| Lower | Upper | Lower | Upper | |||||
| FGF14 | 0.039 | 0.782 | 0.619 | 0.988 | 0.050 | 0.791 | 0.626 | 1.000 |
| FGFR1 | 0.011 | 0.740 | 0.586 | 0.935 | 0.011 | 0.738 | 0.583 | 0.934 |
| Nodes | 0.000 | 1.668 | 1.270 | 2.191 | 0.000 | 1.707 | 1.299 | 2.243 |
Hence, according to our results FGF14 is the predictor for overall survival in all three PDAC cohorts, while, FGFR1 is the predictor for overall survival in two out of three PDAC cohorts. Similarly, FGF14 inversely correlated with oncogenic IGF pathway genes in all three cohorts. Furthermore, common clinicopathological associations with FGF and IGF pathway genes in cohort 1 and 2 are summarized in (S6 Table in S1 File).
Identification of FGF14 co-expressed genes
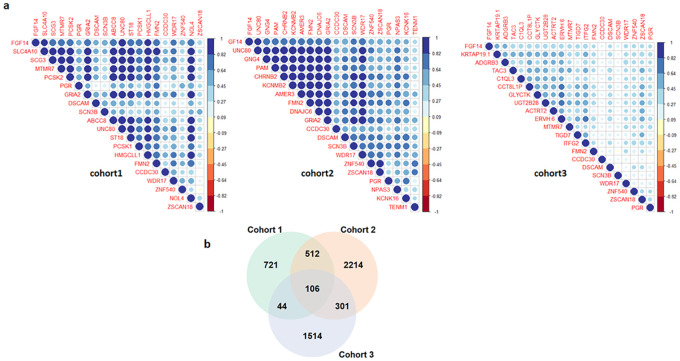
The correlation of expression profile of FGF14 gene was examined with the expression profiles of approximately 20,000 available genes across all the three independent cohorts of PDAC patients. The correlation employed on cohort 1, 2 and 3 revealed a large number of genes that are positively correlated with FGF14 expression in these datasets. Top positively correlated genes are shown in Fig 2a. The common (106) top positively correlated genes of FGF14 across all the three cohorts were used for further analysis (Fig 2b).
Fig 2. (a) Top positively correlated genes associated with FGF14 at expression level in three independent cohorts of PDAC patients (b) Venn diagram illustrating top positively correlated genes of FGF14 at expression level common among the three independent cohorts of PDAC patients.
Functional specification of FGF14-coexpressed genes
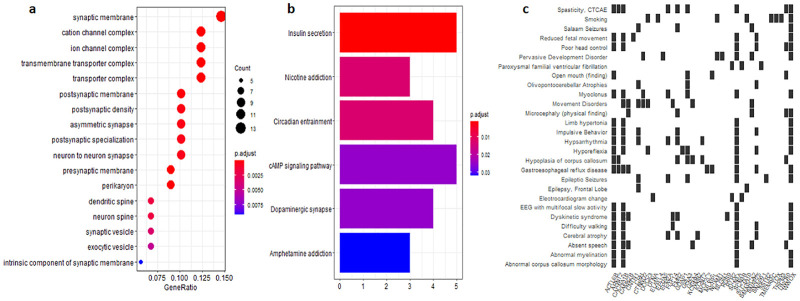
According to functional analysis results, co-expressed genes common in all three cohorts clustered into several enriched Gene ontology (GO) terms (Fig 3a) including intrinsic component of synaptic membrane, exocytic vesicles, synaptic vesicles, ion channel complex, transporter complex etc. The GO terms suggest that nerves related membranes and vesicles might play an important role in PDAC. Additionally, the analysis revealed that these FGF14-specific coexpressed genes are significantly involved in several Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways (Fig 3b) such as ‘Amphetamine addiction’, ‘Dopaminergic synapse’, ‘cAMP signalling pathway’, ‘Insulin secretion’, etc. Furthermore, different diseases associated with FGF14-coexpressed genes were also identified (Fig 3c).
Fig 3. (a) Enriched GO terms among the FGF14-specific coexpressed genes common in all three cohorts of PDAC patients (b) Pathway analysis of FGF14-specific coexpressed genes common in all three cohorts of PDAC patients (c) Gene-disease associations of FGF14-specific coexpressed genes common in all three cohorts of PDAC patients.
Prognostic profiling of FGF14-specific coexpressed genes
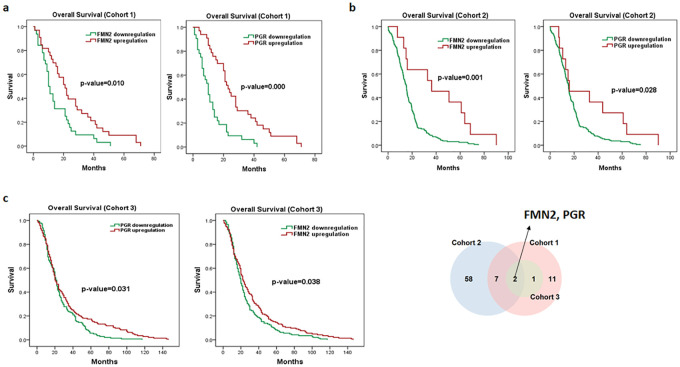
According to Kaplan Meier plots for OS, several genes out of 106 FGF14-specific coexpressed genes showed significant association with the OS of PDAC patients in cohort 1 and 2 (S7 and S8 Tables in S1 File). Predominantly, the overexpression of CCDC30, DSCAM, FMN2, PGR, SCN3B, WDR17, ZNF540 and ZSCAN18 from FGF14-specific coexpressed genes showed significant association with good OS in cohort 1 and 2 (S5 Fig in S1 File). Next the association of these FGF14-specific coexpressed genes that correlated with OS in PDAC patients was examined in cohort 3 of PDAC patients. Notably, the overexpression of FMN2 and PGR genes showed association with good OS in all three cohorts of PDAC patients, suggesting the potential to formulate a FGF14-specific signature (Fig 4).
Fig 4. Kaplan–Meier survival analyses of FMN2 and PGR (top common genes of FGF14) in PDAC cohorts 1, 2 and 3.
Red represents expression ≥ the median, while green represents expression < the median. Overall survival plots of FMN2, PGR (a) in cohort 1. Overall survival plots of FMN2, PGR (b) in cohort 2. Overall survival plots of FMN2, PGR (c) in cohort 3. (* p ≤ 0.05).
Discussion
For FGF pathway, the downregulation of FGF9, FGF14 and FGFR1 genes was associated with advanced clinical features, suggesting its potential tumor suppressive role (S6 Table in S1 File). Particularly, FGF14 expression, was identified as an independent prognostic marker in all three PDAC cohorts. To the best of our knowledge this is the first study demonstrating the linkage of FGF14 dysregulation with PDAC patients. In addition, we suggest that overexpression of FMN2 and PGR is correlated with good OS in PDAC. The main findings of our study suggest that mRNA expression of FGF14, FMN2, PGR and FGFR1 genes define a potentially new molecular subtype of PDAC.
Earlier, reduced FGFR1 was reported in multiple cancers including lung, pancreas, and breast cancers [23–25]. In addition, downregulation of FGF9 was reported to regulate tumorigenesis in lungs cancer [26]. Interestingly, prediction of FGF14 as a novel biomarker of survival is of immense clinical significance. Previously, the dysregulation of FGF14 using pancreatic cancer cell lines was reported [27]. Consistently, Liu et al. also proposed that dysregulation of FGF14 (homologue of FGF13) is involved in progression of cervical cancer [28]. Very recently, Tianhong Su and colleagues extensively investigated the role of FGF14 in colorectal cancer cell lines. Interestingly, they have suggested the tumor suppressor role of FGF14 in colorectal cancer [29]. Of note, a recent study reported that FGF12 (the homologue of FGF14 [30]) have a malignancy- inhibitor effect on pancreatic cancer cell lines [31]. Two most recent studies reported that FGF14 overexpression leads to tumor suppressive effects in lung adenocarcinoma and nasopharyngeal carcinoma [32, 33]. In line with the previous in-vitro and in-vivo studies, our in-silico data also suggests that inhibitors targeting FGF14 expression would prove to be a promising therapeutic option against solid tumors.
For IGF pathway, IGF2BP1, IGF2BP2, IGF2BP3, IGFBP6 and IGFL2 inversely correlated with FGF14. In addition, IGF2BP2 was found associated with advanced clinical features in PDAC patients. Consistent with previous studies, up-regulation of IGF2BP2 promoted liver, colorectal and breast tumorigenesis [34–36]. Similarly, studies have also reported the tumor-promoting role of IGF2BP1, IGF2BP3, IGFBP6 and IGFL2 in several cancers such as hepatocellular carcinoma, pancreatic and prostate cancers [37–40]. The potential association of FGF14 with IGF pathways genes, if validated by additional studies, may serve as a pivotal to understand the existing associations between growth factor pathways that results in pathogenesis of PDAC.
Moreover, we also evaluated the prognostic relevance of FGF14-specific coexpressed genes. The top genes CCDC30, DSCAM, FMN2, PGR, SCN3B, WDR17, ZNF540 and ZSCAN18 which correlated with FGF14 were found highly associated with good OS in two cohorts. Overexpression of FMN2 and PGR was correlated with good OS in all three cohorts of PDAC patients, implying their relevance as potential prognostic markers. In a very recent study, findings show that ZNF540 and PGR were among top 267 down-regulated genes in the squamous cell lung cancer tissues compared to the adjacent normal tissues [41]. PGR is reported as a mediator for anti-tumor activities exerted by Tamoxifen drug in ER+-breast cancer tissue [42]. The expression of FMN2 is predominantly observed in tumor suppressor pathway [43]. Moreover, the dysregulation of FMN2 is identified in large cohort of colorectal cancer patients, and could serve as early diagnostic marker of colorectal cancer [44]. The current study extensively demonstrated the potential link of FGF14 overexpression with the better prognosis of PADC patients, followed by FGF14 based subtyping and strong literature support. This is the first study demonstrating the role of FGF14 and its associated genes in PDAC, extensively elucidating the underlying molecular mechanism. Therefore, we suggest that FGF14 based classification is helpful in categorizing PDAC patients into good and poor prognosis groups. Thus, this study can pave way to design FGF14 specific inhibitors with good efficacy.
Furthermore, FGFR1 showed promising results in our study along with good correlation profiles with FGF14 (S7 Fig in S1 File). Our results of FGFR1 expression are in agreement with our recent study in which we established the clinical significance of FGFR1 through analysis of 313 pancreatic cancer patients along with the cross-validation done through Immunohistochemistry (IHC) of FGFR1 protein. Consistently, FGFR1 overexpression was found significantly correlated with the better overall survival in pancreatic cancer patients [45].
Altogether, we conclude that FGF14 overexpression may be used as prognostic biomarker for better overall survival in PDAC patients. In addition, our data suggests that FMN2 and PGR overexpression can also act as good prognostic marker for the diagnosis of PDAC. In future, preclinical studies are required to establish the therapeutic relevance of FGF14 in pancreatic cancer. The findings of this study may provide a deeper understanding of molecular mechanism involved in the onset and progression of PDAC.
Supporting information
(PDF)
Acknowledgments
We are grateful to Higher Education Commission (HEC) of Pakistan and COMSATS University Islamabad for facilitating this research.
Abbreviations
- CI
Confidence interval
- DFS
Disease-free survival
- FGF
Fibroblast growth factor
- GEO
Gene Expression Omnibus
- GO
Gene-ontology
- IGF
Insulin growth factor
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- OS
Overall survival
- PDAC
Pancreatic ductal adenocarcinoma
- HR
Hazard ratio
- RMA
Robust multiarray average
- RPKM
Reads per kilobase million
Data Availability
All relevant data are within the manuscript and its Supporting information files.
Funding Statement
The authors received no specific funding for this work.
References
- 1.Goggins M. Molecular Markers of Early Pancreatic Cancer. JCO. 2005;23: 4524–4531. 10.1200/JCO.2005.19.711 [DOI] [PubMed] [Google Scholar]
- 2.Pour PM, Pandey KK, Batra SK. What is the origin of pancreatic adenocarcinoma? Molecular Cancer. 2003;2: 13. 10.1186/1476-4598-2-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bardeesy N, DePinho RA. Pancreatic cancer biology and genetics. Nature Reviews Cancer. 2002;2: 897–909. 10.1038/nrc949 [DOI] [PubMed] [Google Scholar]
- 4.Long J, Luo G, Xiao Z, Liu Z, Guo M, Liu L, et al. Cancer statistics: Current diagnosis and treatment of pancreatic cancer in Shanghai, China. Cancer Letters. 2014;346: 273–277. 10.1016/j.canlet.2014.01.004 [DOI] [PubMed] [Google Scholar]
- 5.Provenzano PP, Cuevas C, Chang AE, Goel VK, Von Hoff DD, Hingorani SR. Enzymatic Targeting of the Stroma Ablates Physical Barriers to Treatment of Pancreatic Ductal Adenocarcinoma. Cancer Cell. 2012;21: 418–429. 10.1016/j.ccr.2012.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oberstein PE, Olive KP. Pancreatic cancer: why is it so hard to treat? Therap Adv Gastroenterol. 2013;6: 321–337. 10.1177/1756283X13478680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Becker AE, Hernandez YG, Frucht H, Lucas AL. Pancreatic ductal adenocarcinoma: Risk factors, screening, and early detection. World Journal of Gastroenterology. 2014;20: 11182–11198. 10.3748/wjg.v20.i32.11182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kukreja M. Immunosignaturing Microarrays Distinguish Antibody Profiles of Related Pancreatic Diseases. Journal of Proteomics & Bioinformatics. 2013;01. 10.4172/jpb.S6-001 [DOI] [Google Scholar]
- 9.Ben Q, Xu M, Ning X, Liu J, Hong S, Huang W, et al. Diabetes mellitus and risk of pancreatic cancer: A meta-analysis of cohort studies. European Journal of Cancer. 2011;47: 1928–1937. 10.1016/j.ejca.2011.03.003 [DOI] [PubMed] [Google Scholar]
- 10.Stathis A, Moore MJ. Advanced pancreatic carcinoma: current treatment and future challenges. Nat Rev Clin Oncol. 2010;7: 163–172. 10.1038/nrclinonc.2009.236 [DOI] [PubMed] [Google Scholar]
- 11.Reiter JG, Iacobuzio-Donahue CA. Pancreatic cancer: Pancreatic carcinogenesis—several small steps or one giant leap? Nat Rev Gastroenterol Hepatol. 2016;14: 7–8. 10.1038/nrgastro.2016.190 [DOI] [PubMed] [Google Scholar]
- 12.Ozawa F, Friess H, Tempia‐Caliera A, Kleeff J, Büchler MW. Growth factors and their receptors in pancreatic cancer. Teratogenesis, Carcinogenesis, and Mutagenesis. 2001;21: 27–44. [DOI] [PubMed] [Google Scholar]
- 13.Lehnen NC, von Mässenhausen A, Kalthoff H, Zhou H, Glowka T, Schütte U, et al. Fibroblast growth factor receptor 1 gene amplification in pancreatic ductal adenocarcinoma. Histopathology. 2013;63: 157–166. 10.1111/his.12115 [DOI] [PubMed] [Google Scholar]
- 14.Vescarelli E, Ceccarelli S, Angeloni A. Role of Fibroblast Growth Factor Receptor 2 in Pancreatic Cancer: Potential Target for New Therapeutic Approach? Pancreatic Disorders & Therapy. 2015;5: 1–4. 10.4172/2165-7092.1000164 [DOI] [Google Scholar]
- 15.Motoda N, Matsuda Y, Onda M, Ishiwata T, Uchida E, Naito Z. Overexpression of fibroblast growth factor receptor 4 in high-grade pancreatic intraepithelial neoplasia and pancreatic ductal adenocarcinoma. Int J Oncol. 2011;38: 133–143. [PubMed] [Google Scholar]
- 16.Neid M, Datta K, Stephan S, Khanna I, Pal S, Shaw L, et al. Role of Insulin Receptor Substrates and Protein Kinase C-ζ in Vascular Permeability Factor/Vascular Endothelial Growth Factor Expression in Pancreatic Cancer Cells. J Biol Chem. 2004;279: 3941–3948. 10.1074/jbc.M303975200 [DOI] [PubMed] [Google Scholar]
- 17.Samovski D, Dhule P, Pietka T, Jacome-Sosa M, Penrose E, Son N-H, et al. Regulation of Insulin Receptor Pathway and Glucose Metabolism by CD36 Signaling. Diabetes. 2018;67: 1272–1284. 10.2337/db17-1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brännmark C, Nyman E, Fagerholm S, Bergenholm L, Ekstrand E-M, Cedersund G, et al. Insulin Signaling in Type 2 Diabetes. J Biol Chem. 2013;288: 9867–9880. 10.1074/jbc.M112.432062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kopantzev EP, Grankina EV, Kopantseva MR, Sverdlov ED. The IGF-I/IGF-IR Signaling System and Pancreatic Cancer. Mol Genet Microbiol Virol. 2017;32: 131–136. 10.3103/S0891416817030041 [DOI] [Google Scholar]
- 20.Yang S, He P, Wang J, Schetter A, Tang W, Funamizu N, et al. A Novel MIF Signaling Pathway Drives the Malignant Character of Pancreatic Cancer by Targeting NR3C2. Cancer Res. 2016;76: 3838–3850. 10.1158/0008-5472.CAN-15-2841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu G, Wang L-G, Han Y, He Q-Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16: 284–287. 10.1089/omi.2011.0118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen H, Boutros PC. VennDiagram: a package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinformatics. 2011;12: 35. 10.1186/1471-2105-12-35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lehnen NC, von Mässenhausen A, Kalthoff H, Zhou H, Glowka T, Schütte U, et al. Fibroblast growth factor receptor 1 gene amplification in pancreatic ductal adenocarcinoma. Histopathology. 2013;63: 157–166. 10.1111/his.12115 [DOI] [PubMed] [Google Scholar]
- 24.Weiss J, Sos ML, Seidel D, Peifer M, Zander T, Heuckmann JM, et al. Frequent and Focal FGFR1 Amplification Associates with Therapeutically Tractable FGFR1 Dependency in Squamous Cell Lung Cancer. Science Translational Medicine. 2010;2: 62ra93–62ra93. 10.1126/scitranslmed.3001451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Turner N, Pearson A, Sharpe R, Lambros M, Geyer F, Lopez-Garcia MA, et al. FGFR1 Amplification Drives Endocrine Therapy Resistance and Is a Therapeutic Target in Breast Cancer. Cancer Res. 2010;70: 2085–2094. 10.1158/0008-5472.CAN-09-3746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Q, Liu S, Zhao X, Wang Y, Tian D, Jiang W. MiR-372-3p promotes cell growth and metastasis by targeting FGF9 in lung squamous cell carcinoma. Cancer Medicine. 2017;6: 1323–1330. 10.1002/cam4.1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hagihara A, Miyamoto K, Furuta J, Hiraoka N, Wakazono K, Seki S, et al. Identification of 27 5′ CpG islands aberrantly methylated and 13 genes silenced in human pancreatic cancers. Oncogene. 2004;23: 8705–8710. 10.1038/sj.onc.1207783 [DOI] [PubMed] [Google Scholar]
- 28.Liu M-Y, Zhang H, Hu Y-J, Chen Y-W, Zhao X-N. Identification of key genes associated with cervical cancer by comprehensive analysis of transcriptome microarray and methylation microarray. Oncology Letters. 2016;12: 473–478. 10.3892/ol.2016.4658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Su T, Zhang N, Huang L, Wei G, Peng S, Zeng Z. IDDF2019-ABS-0113 FGF14 is a functional tumor suppressor through inhibiting PI3K/AKT/mTOR pathway in colorectal cancer. Gut. 2019;68: A15–A15. 10.1136/gutjnl-2019-IDDFAbstracts.28 [DOI] [Google Scholar]
- 30.Khalid A, Javaid MA. Fibroblast Growth Factors and their Emerging Cancer-Related Aspects. Journal of Cancer Science & Therapy. 2016;08. 10.4172/1948-5956.1000413 [DOI] [Google Scholar]
- 31.Kawano M, Miura T, Fujita M, Koike S, Imadome K, Ishikawa A, et al. The FGF1/CPP-C chimera protein protects against intestinal adverse effects of C-ion radiotherapy without exacerbating pancreatic carcinoma. Clinical and Translational Radiation Oncology. 2019;14: 8–16. 10.1016/j.ctro.2018.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Turkowski K, Herzberg F, Günther S, Brunn D, Weigert A, Meister M, et al. Fibroblast Growth Factor—14 Acts as Tumor Suppressor in Lung Adenocarcinomas. Cells. 2020;9: 1755. 10.3390/cells9081755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang P, He Q, Lei Y, Li Y, Wen X, Hong M, et al. m6A-mediated ZNF750 repression facilitates nasopharyngeal carcinoma progression. Cell Death Dis. 2018;9. 10.1038/s41419-018-1224-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu F-y, Zhou S -j, Deng Y -l, Zhang Z -y, Zhang E -l, Wu Z -b, et al. MiR-216b is involved in pathogenesis and progression of hepatocellular carcinoma through HBx-miR-216b-IGF2BP2 signaling pathway. Cell Death & Disease. 2015;6: e1670. 10.1038/cddis.2015.46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li X, Li Y, Lu H. miR-1193 Suppresses Proliferation and Invasion of Human Breast Cancer Cells Through Directly Targeting IGF2BP2. Oncology Research Featuring Preclinical and Clinical Cancer Therapeutics. 2017;25: 579–585. 10.3727/97818823455816X14760504645779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ye S, Song W, Xu X, Zhao X, Yang L. IGF2BP2 promotes colorectal cancer cell proliferation and survival through interfering with RAF-1 degradation by miR-195. FEBS Letters. 2016;590: 1641–1650. 10.1002/1873-3468.12205 [DOI] [PubMed] [Google Scholar]
- 37.Glaß M, Michl P, Hüttelmaier S. RNA Binding Proteins as Drivers and Therapeutic Target Candidates in Pancreatic Ductal Adenocarcinoma. International Journal of Molecular Sciences. 2020;21: 4190. 10.3390/ijms21114190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Taniuchi K, Furihata M, Hanazaki K, Saito M, Saibara T. IGF2BP3-mediated translation in cell protrusions promotes cell invasiveness and metastasis of pancreatic cancer. Oncotarget. 2014;5: 6832–6845. 10.18632/oncotarget.2257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bach LA. Recent insights into the actions of IGFBP-6. J Cell Commun Signal. 2015;9: 189–200. 10.1007/s12079-015-0288-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zou B, Liu X, Gong Y, Cai C, Li P, Xing S, et al. A novel 12-marker panel of cancer-associated fibroblasts involved in progression of hepatocellular carcinoma. Cancer Manag Res. 2018;10: 5303–5311. 10.2147/CMAR.S176152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu Z, Wang Y-M, Dai Y, Chen L-A. POLE2 Serves as a Prognostic Biomarker and Is Associated with Immune Infiltration in Squamous Cell Lung Cancer. Med Sci Monit. 2020;26: e921430-1-e921430-11. 10.12659/MSM.921430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fang Q, Yao S, Luo G, Zhang X. Identification of differentially expressed genes in human breast cancer cells induced by 4-hydroxyltamoxifen and elucidation of their pathophysiological relevance and mechanisms. Oncotarget. 2017;9: 2475–2501. 10.18632/oncotarget.23504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yamada K, Ono M, Perkins ND, Rocha S, Lamond AI. Identification and functional characterization of FMN2, a regulator of the cyclin-dependent kinase inhibitor p21. Mol Cell. 2013;49: 922–933. 10.1016/j.molcel.2012.12.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li D-J, Feng Z-C, Li X-R, Hu G. Involvement of methylation-associated silencing of formin 2 in colorectal carcinogenesis. World J Gastroenterol. 2018;24: 5013–5024. 10.3748/wjg.v24.i44.5013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Haq F, Sung Y-N, Park I, Kayani MA, Yousuf F, Hong S-M, et al. FGFR1 expression defines clinically distinct subtypes in pancreatic cancer. Journal of Translational Medicine. 2018;16: 374. 10.1186/s12967-018-1743-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
Data Availability Statement
All relevant data are within the manuscript and its Supporting information files.