Abstract
Background
Primary Graft Dysfunction (PGD) is a risk factor for Chronic Lung Allograft Dysfunction (CLAD). However, the association between PGD and degree of allograft injury remains poorly defined. In this study, we leverage a novel biomarker for allograft injury, percentage donor-derived cell-free DNA (%ddcfDNA), to study the association between PGD, degree of allograft injury, and the development of CLAD.
Methods
This prospective cohort study recruited 99 lung transplant recipients and collected plasma samples on days 1, 3 and 7 for %ddcfDNA measurements. Clinical data on day 3 was used to adjudicate for PGD. %ddcfDNA levels were compared between PGD grades. In PGD patients, %ddcfDNA was compared between those who developed CLAD and those who did not.
Results
On post-transplant day 3, %ddcfDNA was higher in PGD than in non PGD patients (median (IQR): 12.2% (8.2, 22.0) vs 8.5% (5.6, 13.2) p = 0.01). %ddcfDNA correlated with the severity grade of PGD (r = 0.24, p = 0.02). Within the PGD group, higher levels of %ddcfDNA correlated with increased risk of developing CLAD (log OR(SE) 1.38 (0.53), p=0.009). PGD patients who developed CLAD showed ~2 times higher %ddcfDNA levels than patients who did not develop CLAD (median (IQR): 22.4% (11.8, 27.6) vs. 9.9% (6.7, 14.9), p = 0.007).
Conclusion
PGD patients demonstrated increased early post-transplant allograft injury, as measured by %ddcfDNA, in comparison to non PGD patients, and these high %ddcfDNA levels were associated with subsequent development of CLAD. This study suggests that %ddcfDNA identifies PGD patients at greater risk of CLAD than PGD alone.
Introduction
Primary Graft Dysfunction (PGD) is characterized by acute hypoxemic respiratory failure and allograft specific pulmonary infiltrates without an alternative identifiable cause that develops within 72 hours after lung transplantation [1]. PGD remains a leading cause of early mortality after lung transplantation and has been identified as an independent risk factor for the development of Chronic Lung Allograft Dysfunction (CLAD)[2, 3]. Furthermore, the severity of PGD correlates with the risk of developing CLAD [2]. Given the associated morbidity and mortality in patients with PGD, these patients may benefit from further risk stratification to identify patients at highest risk of long-term complications. Patients deemed at higher risk of long-term complications may benefit from more intensive monitoring in the post-transplant setting.
Given that the diagnosis and severity of PGD are currently based on clinical criteria alone, it may be difficult to discern the degree of allograft injury. A more direct, quantitative marker of allograft injury may therefore be helpful. Plasma Donor Derived Cell Free DNA (%ddcfDNA) has emerged as a noninvasive biomarker for allograft injury following solid organ transplant [4–6]. Following lung transplantation, levels of plasma %ddcfDNA increase in the setting of acute cellular rejection (ACR) and antibody mediated rejection (AMR) [6, 7]. Mean levels of plasma %ddcfDNA in the months following lung transplantation also serve as a predictor of long term outcomes, including CLAD[7]. Similarly, levels of plasma %ddcfDNA may provide further insight into the degree of early allograft injury in patients with PGD, and may provide an adjunctive tool for quantitatively assessing the severity of PGD and for identifying patients at risk for development of long-term complications. In this study, we investigated the relationship between the development and severity of PGD with levels of plasma %ddcfDNA, as well as the association between levels of plasma %ddcfDNA and subsequent development of CLAD.
Methods
Study Design
We conducted an observational analysis that included subjects from two ongoing prospective cohort studies. The first study, Genome Transplant Dynamics (GTD) (NCT01985412), is a single center study that began in 2010 at Stanford University Hospital to evaluate the utility of plasma %ddcfDNA to monitor for acute rejection. The second study, Genomic Research Alliance for Transplantation (GRAfT) (NCT0243070), began recruitment in 2015 at three centers (the Johns Hopkins Hospital and University of Maryland Medical Center, and Inova Fairfax Hospital). Both studies recruited subjects who were at least 18 years of age and awaiting lung transplantation. Subjects were enrolled between June 1, 2015 and November 1, 2017. After transplantation, patients underwent routine post-transplant care including regular clinic visits, serial spirometry and surveillance bronchoscopy. Arterial blood gas (ABG) measurements and chest X-rays were obtained on day 3 after transplantation to adjudicate for PGD grade. Plasma samples were serially collected post-transplant and analyzed for %ddcfDNA. Since the diagnosis of PGD was made at the 72-hour mark post-transplant, we only included patients with available day 3 plasma samples for %ddcfDNA measurements in this study. This study was approved by the Institutional Review Board at each center.
Clinical Endpoints
PGD was defined as the development of pulmonary infiltrates in the transplanted lung(s) consistent with non-cardiogenic pulmonary edema 72 hours post-transplant and graded according to the severity of hypoxemia as reflected by the ratio of partial pressure of arterial oxygen to fraction of inspired oxygen (P:F). The radiographic criteria for PGD was established for each patient through the independent review of their chest radiographs at 72 hours by an adjudication committee member at each clinical site. The adjudication committees were comprised of thoracic surgeons, transplant pulmonologists and thoracic radiologists. Patients were then assigned a PGD grade according to the International Society for Heart and Lung Transplantation (ISHLT) criteria [1]. CLAD was defined according to ISHLT criteria as a > 20% decline in FEV1 +/- FVC from baseline to at least 3 months post-transplant, that persisted on separate measurements > 3 weeks apart [8]. Infection was classified as the average number of respiratory pathogens isolated by BAL per 100 days.
Measurement of %ddcfDNA
Plasma %ddcfDNA was measured using an automated shotgun sequencing method. This approach has been described previously [9]. In summary, donor and recipient pre-transplant genomic DNA was isolated and genotyped. The data were then compared to identify single-nucleotide polymorphisms (SNPs). After transplantation, recipient plasma cell free DNA (cfDNA) was isolated in order to create a DNA library for paired end shotgun sequencing. The cfDNA sequence reads were then evaluated for the presence of donor and recipient SNPs and %ddcfDNA was calculated as the percentage of donor SNPs to total (recipient and donor) SNPs. The value for %ddcfDNA in single lung transplant recipients was adjusted by multiplying the initial value by 2[6, 7, 10].
Statistical Analysis
Categorical variables were summarized as counts and percentages, and compared using Chi-square tests or Fisher exact tests. Continuous variables were summarized as mean (SD) or median (IQR) and were compared using t-tests or Wilcoxon rank sum tests. Linear mixed models were used to analyze repeated measures of continuous variables. Standard residual diagnostics were used to check model assumptions. For each analysis, %ddcfDNA was log-transformed unless non-parametric methods were used. Categorical outcomes were analyzed in multivariable logistic regression models to adjust for potential confounders, which were selected by univariate analysis (p < 0.1), including gender, race, type of transplant, donor-recipient race mismatch and PGD (Supplementary Table 1). All analyses were performed using SAS version 9.4, and p-values were 2-sided with a value of < 0.05 indicating significance.
Results
Cohort Description
Ninety-nine subjects met criteria for inclusion in our analysis; however, 13 of these patients did not have available day 3 CXR or ABG data necessary to adjudicate for PGD. 86 patients were therefore included in our final analysis (Supplementary Figure 1). The mean (SD) age at transplantation was 54.3 (14.4) years and the mean (SD) Lung Allocation Score (LAS) was 44.9 (12.1). Interstitial lung disease was the most common indication for transplant (46.4%) and 70.2% of patients underwent bilateral lung transplant. The mean (SD) donor age was 36.3 (14.1) years old (Table 1). Mean post-transplant follow-up was 24.0 +/- 10.5 months. Following transplantation, 39 (45.4%) patients did not develop PGD, 19 (22.1%) patients developed Grade 1 PGD, 12 (14.0) patients developed Grade 2 PGD and 16 (18.6) patients developed Grade 3 PGD. Eight patients lacked data to adjudicate for CLAD. 22/78 (28.2) patients in our cohort developed CLAD. This included 11 patients with CLAD who had PGD and 11 patients with CLAD who did not have PGD.
Table 1:
Patient Demographics
| Recipient Age (Years) | 54.3 (14.4) |
| Donor Age (Years) | 36.3 (14.1) |
| Male Recipient (%) | 42.7% |
| LAS Score | 44.9 (12.1) |
| Bilateral Transplant (%) | 70.2% |
| Diagnosis | |
| COPD | 17.9% |
| Cystic Fibrosis | 16.7% |
| Interstitial Lung Disease | 46.4% |
| Pulmonary Arterial Hypertension | 4.8% |
| Other | 8.2% |
| Re-transplantation | 6% |
| Race | |
| White | 81.7% |
| Black | 15.9% |
| Asian | 2.4% |
| Recipient-Donor Race Mismatch | 26.3% |
LAS: Lung Allocation Score, COPD: Chronic Obstructive Pulmonary Disease
Levels of %ddcfDNA in patients with PGD vs no PGD
A total of 269 plasma samples were assessed for %ddcfDNA by shotgun sequencing. Duplicated reads and low mapping quality (MAPQ < 30) reads were filtered out for estimation of %ddcfDNA [9], after which an average of 67% of total reads were used for computing %ddcfDNA. The median day 1 level of %ddcfDNA for all patients was 26.2% (IQR: 18.4 – 36.4%). Levels then declined following two phase logarithmic decay kinetics to 10.7% on day 3 (IQR: 6.6 – 17.3%), 4.2% on day 7 (IQR: 2.6 – 6.8%), and 1.4% on day 30 (IQR: 1.0 – 2.7%).
Figure 1a demonstrates the trends in median values of %ddcfDNA over the first 7 days post-transplant for both PGD and non-PGD patients. %ddcfDNA on Day 3 was higher in patients with PGD than in patients without PGD (median (IQR): 12.2% (8.2, 22.0) vs 8.5% (5.6, 13.2), p = 0.01) (Figure 1b). Even before the diagnosis of PGD was made on Day 3, levels of %ddcfDNA on Day 1 were higher in PGD patients than non-PGD patients (32.8% (22.2, 40.2) vs 22.0% (16.7, 28.1), p = 0.01). The difference between values on Day 1 and 3 was similar (p=0.94 for interaction, p=0.006 for the average PGD effect). On Day 3, log10 (%ddcfDNA) levels correlated with severity of PGD grade (Pearson correlation: r = 0.24, p = 0.02), levels were ≈ 1.5 times higher for PGD grade 3 compared to PGD 1+2 and ≈ 2 times higher for PGD grade 3 compared to non-PGD patients (8.5% (5.6, 13.2) vs 10.9% (8.20, 19.3) vs 17.6% (8.1, 22.8), p = 0.03, Kruskal-Wallis Test) (Supplementary Figure 2).
Figure 1.
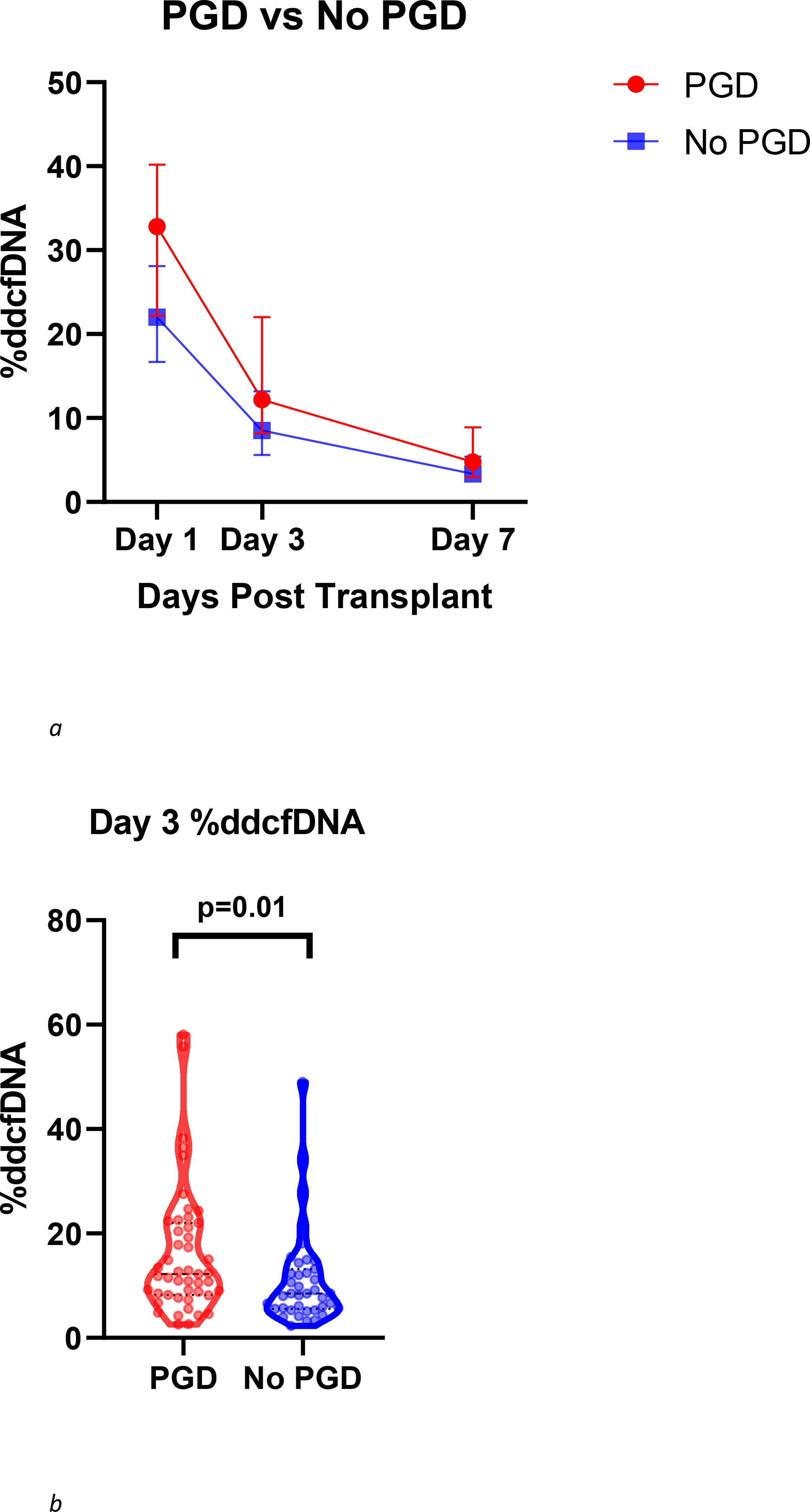
(a) Trend in %ddcfDNA over the first post-transplant week in PGD and non-PGD patients. Each point represents median values with interquartile range. Levels of %ddcfDNA were higher in patients with PGD than in patients without PGD on days 1, 3 and 7 (Day 7 values, median (IQR): 4.7% (3.0,8.9) vs 3.3% (2.4, 5.4), p=0.046) (b) Comparison of day 3 %ddcfDNA between PGD and non-PGD patients displayed with violin plots including all individual data points, median and IQR values PGD patients demonstrated higher levels of %ddcfDNA than non-PGD patients (median (IQR): 12.2% (8.2, 22.0) vs 8.5% (5.6, 13.2), p = 0.01).
Levels of %ddcfDNA in PGD patients who develop CLAD
In an effort to further characterize the risk of developing CLAD in patients with PGD, we analyzed differences in allograft injury as assessed by %ddcfDNA between PGD patients who developed CLAD (PGD+/CLAD+) compared to PGD patients who did not develop CLAD (PGD+/CLAD−). The decay in %ddcfDNA over the first post-transplant week for PGD+/CLAD+ and PGD+/CLAD−is shown in Figure 2a. At diagnosis of PGD on Day 3, PGD patients who subsequently developed CLAD (PGD+/CLAD+) demonstrated over 2-fold higher levels of %ddcfDNA than PGD patients that did not develop CLAD (PGD+/CLAD−) (Day 3 median (IQR): 22.4% (11.8, 27.6) vs. 9.9% (6.7, 14.9), p = 0.007) (Figure 2b).
Figure 2.
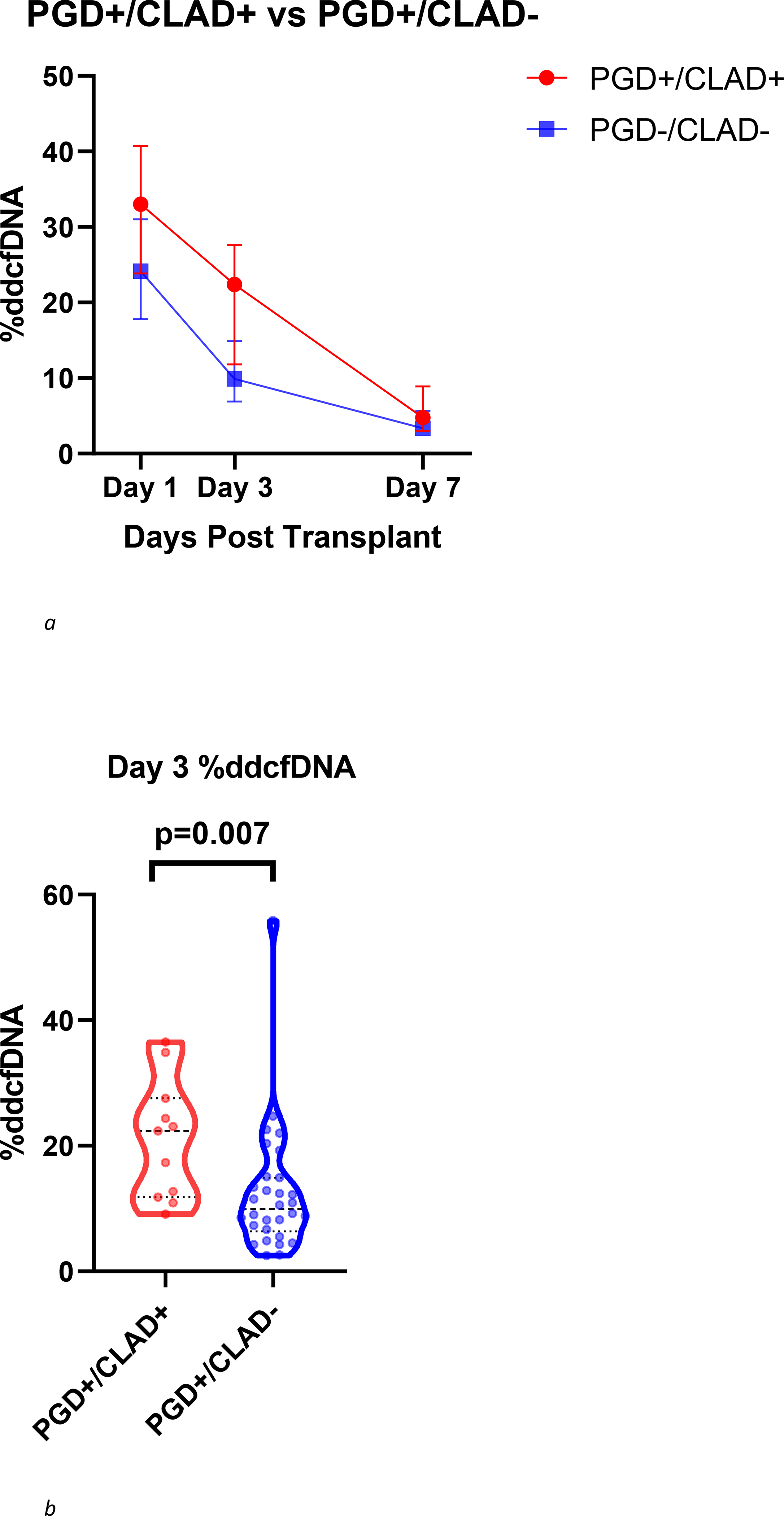
(a) Trend in %ddcfDNA over the first week post-transplant in PGD+/CLAD+ patients and PGD+/CLAD− patients. Each point represents median values with interquartile range. (b) Comparison of day 3 %ddcfDNA between PGD+/CLAD+ patients vs PGD+/CLAD− patients displayed with violin plots including all individual data points, median and IQR values. PGD+/CLAD+ patients demonstrated higher levels of %ddcfDNA than PGD+/CLAD− patients (Day 3 median (IQR): 22.4% (11.8, 27.6) vs. 9.9% (6.9, 14.9), p = 0.007).
Multivariate logistic regression analysis demonstrated that the relationship between levels of %ddcfDNA and the development of CLAD was different between the PGD patients and the non-PGD patients (p=0.004), after adjusting for race (White vs non-White) based on our univariate analysis (Supplementary Table 1). In patients who experienced PGD, higher levels of %ddcfDNA correlated with increased risk of developing CLAD (logOR(SE) 1.38 (0.53), p=0.009). A similar relationship was not observed in non-PGD patients (the logOR(SE) -0.64 (0.45), p=0.16). Given that ACR and infection may serve as risk factors for CLAD, we next added these covariates to our multivariate model. Accounting for infection and the number of episodes of ACR, higher levels of %ddcfDNA in patients with PGD still correlated with an increased risk of developing CLAD (logOR(SE) 1.66 (0.70), p = 0.02). An ROC curve of a model incorporating PGD and Race alone demonstrated an AUC of 0.58, however, the AUC increased to 0.77 when adding %ddcfDNA to the model and further increased to 0.86 with the addition of infection and ACR. (Supplementary Figures 3a, 3b and 3c).
Discussion
This study reports two important findings supporting the use of %ddcfDNA in patients with PGD. First, from Day 1 of transplantation, %ddcfDNA correlated with the presence and severity of PGD. Second, in patients with PGD, %ddcfDNA correlated with the subsequent risk of developing CLAD. Tanaka et al. previously reported an inverse correlation between %ddcfDNA and oxygenation in a cohort of 15 patients undergoing living donor-lobar lung transplantation as well as higher levels of %ddcfDNA in patients with acute rejection [11]. Our study builds on these findings in a larger cohort of cadaveric lung transplant patients, further extending the relationship of %ddcfDNA and PGD to the subsequent development of CLAD.
These findings have several implications. %ddcfDNA is emerging as a useful biomarker of allograft injury in solid organ transplants, and is primarily used to detect acute rejection [12–14]. The relationship between levels of %ddcfDNA and the severity of PGD lends further credence to the hypothesis that %ddcfDNA is a molecular marker for allograft injury, and our results now extend this concept to the early post-transplant setting. The current diagnostic criteria of PGD relies on a combination of radiographic findings and arterial blood gas measurements in the absence of other identifiable etiologies. Incorporation of %ddcfDNA into the diagnosis may allow for a more specific and quantitative marker of allograft injury to increase diagnostic certainty and more adequately define PGD severity.
The association of early post-transplant allograft injury to long term outcomes such as CLAD has been reported in prior investigations. For example, our previous analysis showed that %ddcfDNA within the first 3 months post-transplant served as a predictor for the development of CLAD [7]. Fiser and colleagues previously demonstrated that ischemia-reperfusion injury, characterized by radiographic infiltrates and hypoxemia within the first 24 hours of transplantation, was an independent risk factor for the development and progression of bronchiolitis obliterans syndrome (BOS) [15]. Daud and colleagues later corroborated these findings by establishing PGD as an independent risk factor for the development of BOS, with the risk of BOS increasing with respect to the severity of PGD [3]. This study further strengthens these observations by demonstrating the association between the degree of allograft injury at PGD diagnosis with developing CLAD.
The mechanism by which early allograft injury contributes to the development of CLAD remains undefined. However, accumulating evidence suggests that an increase in allograft immunogenicity may play a substantial role. Early ischemia-reperfusion injury after kidney transplantation results in recruitment of recipient MHC Class-II leukocytes to the allograft [16]. In animal models, early ischemia reperfusion injury after lung transplant caused increased expression of donor MHC Class II antigen[17]. Furthermore, in adult lung transplant patients, an increase in pro-inflammatory mediators during PGD has been associated with the subsequent development of anti-HLA II alloantibodies, and patients with CLAD exhibit higher levels of early post-transplant inflammatory mediators and increased anti-HLA Class II alloantibodies[18, 19]. More recently, it has been postulated that early allograft injury may result in the release of exosomes expressing HLA antigen and lung associated self-antigens (SAags) to K alpha 1 tubulin and collagen-V. This results in the development of alloantibodies to these antigens which are upregulated in patients with CLAD, providing further evidence for the role of early allograft injury-induced immunogenicity [20–24].
Currently, clinicians are unable to use early clinical parameters to accurately predict long term complications after lung transplantation. Accurate risk stratification in the early post-transplant setting would help guide subsequent monitoring and surveillance required for routine follow up, and could potentially alter future diagnostic and therapeutic decision making. Our findings suggest that the combination of the presence of PGD on day 3 and levels of %ddcfDNA may be a better predictor of CLAD than PGD alone. Future studies focusing on the development of predictive models for CLAD may benefit from incorporating %ddcfDNA into proposed algorithms.
In this study we did not find an association between %ddcfDNA and development of CLAD in non-PGD patients. Our prior report in a larger population of patients examined %ddcfDNA at a different early post-transplant period (Day 14 to 90) and showed correlation of %ddcfDNA to CLAD in PGD and non-PGD patients[7]. Future studies with a larger cohort size may be needed to fully examine the association of Day 3 %ddcfDNA and CLAD in non-PGD patients. These future studies should also examine PGD diagnosed at an earlier post-transplant period, as it is possible that some Day 3 %ddcfDNA values were sampled from patients who had resolving lung injury after experiencing PGD earlier than Day 3. In addition, these future studies should also compare the ddcfDNA tissue of origin within the allograft between PGD and non-PGD patients. Nonetheless, our results suggest that %ddcfDNA on Day 3 has a bigger effect size in distinguishing PGD than non-PGD patients for their risk of CLAD.
Our findings are limited by the observational study design, missing data, and relatively small sample size. In addition, this study focuses on a phenotype of PGD characterized by persistent allograft dysfunction at 72 hours post-transplant [25]. While this excludes patients with PGD that resolves within 72 hours, it focuses on a phenotype of patients at higher risk for poor outcomes [25]. To the best of our knowledge, this study offers the largest analysis of %ddcfDNA levels concurrent with PGD diagnosis. We were unable to obtain data regarding the use of cardiopulmonary bypass or pulmonary artery pressures at the time of transplant, both of which may increase the risk of PGD and potentially increase %ddcfDNA levels. While the number of re-transplantation procedures was low in our cohort (6%), future studies should aim to assess the impact of re-transplantation on early %ddcfDNA levels. Lastly, our pre-specified analysis used the incidence of CLAD as an outcome, however, time to CLAD may represent a more robust outcome measure in future studies using a larger cohort of patients. Validation of these findings in a larger independent cohort is needed. As such, these findings should be viewed as hypothesis generating.
In conclusion, this study demonstrates an association between plasma %ddcfDNA and the diagnosis and severity of PGD, as well as a correlation between %ddcfDNA levels in patients with PGD and the subsequent development of CLAD. This work lays the foundation for future studies evaluating the role of early %ddcfDNA as a predictive biomarker for long term outcomes after lung transplantation.
Supplementary Material
Acknowledgement
We would like to acknowledge the National Cancer Institute for genotyping as well the Adjudication committee for PGD: Errol Bush (Johns Hopkins), Pali Shah (Johns Hopkins), Joshua Diamond (University of Pennsylvania), Anne Brown (Inova Fairfax). We acknowledge Dr. Hannah Valantine for guidance and for reviewing the manuscript.
Funding
The Genome Transplant Dynamics Study (NCT01985412) is funded by the National Institutes of Health, Grant RC4AI092673. The Genomic Research Alliance for Transplantation Study (NCT02423070) is funded by the Division of Intramural Research of the National Heart, Lung, and Blood Institute. Funders had no role in study design, data collection, data analysis, interpretation, writing of the report. Dr. Sean Agbor-Enoh receives funding from the Cystic Fibrosis Foundation, the National Institute of Health Distinguished Scholar Program and the National Heart, Lung and Blood Institute Intramural Research division.
Author Financial Disclosures
Dr. Errol Bush has received consulting fees from VeraMedica Institute for occupational/environmental exposure litigation. Dr. Kiran Khush reports receiving grants and personal fees as a scientific advisor and participant on a speaker’s bureau from CareDx Inc. outside of the scope of the submitted work.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Snell GI, et al. , Report of the ISHLT Working Group on Primary Lung Graft Dysfunction, part I: Definition and grading-A 2016 Consensus Group statement of the International Society for Heart and Lung Transplantation. J Heart Lung Transplant, 2017. 36(10): p. 1097–1103. [DOI] [PubMed] [Google Scholar]
- 2.Kreisel D, et al. , Short- and long-term outcomes of 1000 adult lung transplant recipients at a single center. J Thorac Cardiovasc Surg, 2011. 141(1): p. 215–22. [DOI] [PubMed] [Google Scholar]
- 3.Daud SA, et al. , Impact of immediate primary lung allograft dysfunction on bronchiolitis obliterans syndrome. Am J Respir Crit Care Med, 2007. 175(5): p. 507–13. [DOI] [PubMed] [Google Scholar]
- 4.Gielis EM, et al. , Cell-Free DNA: An Upcoming Biomarker in Transplantation. Am J Transplant, 2015. 15(10): p. 2541–51. [DOI] [PubMed] [Google Scholar]
- 5.Bloom RD, et al. , Cell-Free DNA and Active Rejection in Kidney Allografts. J Am Soc Nephrol, 2017. 28(7): p. 2221–2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Vlaminck I, et al. , Noninvasive monitoring of infection and rejection after lung transplantation. Proc Natl Acad Sci U S A, 2015. 112(43): p. 13336–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Agbor-Enoh S, et al. , Donor-derived cell-free DNA predicts allograft failure and mortality after lung transplantation. EBioMedicine, 2019. 40: p. 541–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Verleden GM, et al. , Chronic lung allograft dysfunction: Definition, diagnostic criteria, and approaches to treatment-A consensus report from the Pulmonary Council of the ISHLT. J Heart Lung Transplant, 2019. 38(5): p. 493–503. [DOI] [PubMed] [Google Scholar]
- 9.Agbor-Enoh S, et al. , Applying rigor and reproducibility standards to assay donor-derived cell-free DNA as a non-invasive method for detection of acute rejection and graft injury after heart transplantation. J Heart Lung Transplant, 2017. 36(9): p. 1004–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Agbor-Enoh S, et al. , Late manifestation of alloantibody-associated injury and clinical pulmonary antibody-mediated rejection: Evidence from cell-free DNA analysis. J Heart Lung Transplant, 2018. 37(7): p. 925–932. [DOI] [PubMed] [Google Scholar]
- 11.Tanaka S, et al. , Donor-derived cell-free DNA is associated with acute rejection and decreased oxygenation in primary graft dysfunction after living donor-lobar lung transplantation. Sci Rep, 2018. 8(1): p. 15366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Knight SR, Thorne A, and Lo Faro ML, Donor-specific Cell-free DNA as a Biomarker in Solid Organ Transplantation. A Systematic Review. Transplantation, 2019. 103(2): p. 273–283. [DOI] [PubMed] [Google Scholar]
- 13.Gadi VK, et al. , Soluble donor DNA concentrations in recipient serum correlate with pancreas-kidney rejection. Clin Chem, 2006. 52(3): p. 379–82. [DOI] [PubMed] [Google Scholar]
- 14.Garcia Moreira V, et al. , Cell-free DNA as a noninvasive acute rejection marker in renal transplantation. Clin Chem, 2009. 55(11): p. 1958–66. [DOI] [PubMed] [Google Scholar]
- 15.Fiser SM, et al. , Ischemia-reperfusion injury after lung transplantation increases risk of late bronchiolitis obliterans syndrome. Ann Thorac Surg, 2002. 73(4): p. 1041–7; discussion 1047–8. [DOI] [PubMed] [Google Scholar]
- 16.Penfield JG, et al. , Transplant surgery injury recruits recipient MHC class II-positive leukocytes into the kidney. Kidney Int, 1999. 56(5): p. 1759–69. [DOI] [PubMed] [Google Scholar]
- 17.Waddell TK, et al. , Major histocompatibility complex expression and lung ischemia-reperfusion in rats. Ann Thorac Surg, 1996. 62(3): p. 866–72. [DOI] [PubMed] [Google Scholar]
- 18.Bharat A, et al. , Early posttransplant inflammation promotes the development of alloimmunity and chronic human lung allograft rejection. Transplantation, 2007. 83(2): p. 150–8. [DOI] [PubMed] [Google Scholar]
- 19.Bharat A, et al. , Immunological link between primary graft dysfunction and chronic lung allograft rejection. Ann Thorac Surg, 2008. 86(1): p. 189–95; discussion 196–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goers TA, et al. , De novo production of K-alpha1 tubulin-specific antibodies: role in chronic lung allograft rejection. J Immunol, 2008. 180(7): p. 4487–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gunasekaran M, et al. , Donor-Derived Exosomes With Lung Self-Antigens in Human Lung Allograft Rejection. Am J Transplant, 2017. 17(2): p. 474–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mohanakumar T, et al. , A novel mechanism for immune regulation after human lung transplantation. J Thorac Cardiovasc Surg, 2019. 157(5): p. 2096–2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zaffiri L, et al. , Collagen type-V is a danger signal associated with primary graft dysfunction in lung transplantation. Transpl Immunol, 2019. 56: p. 101224. [DOI] [PubMed] [Google Scholar]
- 24.Iwata T, et al. , Anti-type V collagen humoral immunity in lung transplant primary graft dysfunction. J Immunol, 2008. 181(8): p. 5738–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shah RJ, et al. , Latent class analysis identifies distinct phenotypes of primary graft dysfunction after lung transplantation. Chest, 2013. 144(2): p. 616–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


