Summary
Colorectal cancer (CRC) progression is a complex process that is not well understood. We describe an in vitro organ-on-chip model that emulates in vivo tissue structure and the tumor microenvironment (TME) to better understand intravasation, an early step in metastasis. The CRC-on-chip incorporates fluid flow and peristalsis-like cyclic stretching and consists of endothelial and epithelial compartments, separated by a porous membrane. On-chip imaging and effluent analyses are used to interrogate CRC progression and the resulting cellular heterogeneity. Mass spectrometry-based metabolite profiles are indicative of a CRC disease state. Tumor cells intravasate from the epithelial channel to the endothelial channel, revealing differences in invasion between aggressive and non-aggressive tumor cells. Tuning the TME by peristalsis-like mechanical forces, the epithelial:endothelial interface, and the addition of fibroblasts influences the invasive capabilities of tumor cells. The CRC-on-chip is a tunable human-relevant model system and a valuable tool to study early invasive events in cancer.
Subject areas: Classification Description, Bioengineering, Tissue engineering, Cancer, Biomedical materials
Graphical abstract
Highlights
-
•
A human colorectal cancer chip recapitulates aspects of CRC biology
-
•
On-chip imaging and metabolomic effluent analyses offer insight into progression
-
•
Organ-on-chip conditions induce phenotypic heterogeneity compared to 2D cultures
-
•
Stromal cell cross talk and mechanical forces increase tumor cell intravasation
Classification Description: Bioengineering; Tissue engineering; Cancer; Biomedical materials
Introduction
Colorectal cancer (CRC) is one of the deadliest cancers worldwide with over 900,000 people dying from the disease each year (Siegel et al., 2021). In the United States, the 5-year survival rate for patients with metastatic CRC is less than 15% (Siegel et al., 2020). To address this dismal outcome, there is an urgent need to better understand and ultimately control aspects of cancer progression. Tumor metastasis is an elaborate cascade of events whereby cells from the primary tumor invade the surrounding tissue and intravasate into the blood or lymphatic vessels, extravasate into a distant organ, evade the immune system, and grow into secondary, metastatic tumors (Batista et al., 2019). Genetically engineered mouse models and patient-derived xenografts have been critical in advancing the tumor metastasis field by providing an in vivo system that can model tumor spread from primary to distant organ sites through the vascular network (Bürtin et al., 2020; Walrath et al., 2010). However, limitations remain with traditional preclinical cancer models including a lack of in vitro model systems that imitate human physiology and the inability of in vivo animal experiments to recapitulate and tune human organ microenvironments (Gould et al., 2015; Ledford, 2011; Mak et al., 2014). Recent advances in in vitro culture systems have overcome some of these limitations. In particular, 3D microfluidic organ-on-chip (Organ Chip) systems incorporate unique features to better model in vivo cell-cell and cell-extracellular matrix (ECM) interactions (Bhatia and Ingber, 2014; Sarvestani et al., 2020), which can support novel interrogations of cancer progression.
Organ chips are designed to model normal or diseased organ-level structure and function by incorporating tissue compartments and physical forces that mimic in vivo cyclic strain (i.e., peristalsis-like motions) and fluid shear stress (Basson, 2007; Gayer and Basson, 2009). The microfluidic nature of these systems sustains longer-term experiments and allows for continuous effluent collection to monitor byproducts as an indirect measure of tissue function and viability (Bai et al., 2015; Jang et al., 2019; McAleer et al., 2019; Pavesi et al., 2017). When combined with imaging-based approaches (Lee et al., 2018; Pavesi et al., 2017; Ying et al., 2015), organ chips support dynamic cell phenotyping in a noninvasive manner. Organ chip models across a variety of cancer types (e.g., breast, lung, colon, and pancreatic cancers) have been engineered to interrogate important biological processes in cancer, such as angiogenesis, epithelial-mesenchymal transition, cancer cell metastasis, and therapeutic response (Caballero et al., 2017; Sontheimer-Phelps et al., 2019).
Our goal was to develop a CRC-on-chip, integrating multiple in vivo-relevant cell types and physical forces, to reveal how diverse tumor microenvironment (TME) cues work in concert to influence the spread of a primary colon tumor. We introduced aspects of the TME in a stepwise fashion to support the study of tumor-TME interactions in a tunable, physiologically relevant system. Specifically, we expanded upon previous organ-on-chip models by incorporating physical forces to mimic peristalsis, an important factor in colon physiology (Gayer and Basson, 2009) and host-microbe interactions in the healthy intestine (Grassart et al., 2019; Kim et al., 2016). In addition, we integrated aspects of the stromal TME, such as cancer-associated fibroblasts (CAFs), which have been implicated in metastatic spread (Quail and Joyce, 2013). This resulted in a heterocellular tumor compartment interfaced with a blood vessel compartment to study CRC as a diseased organ. We demonstrate that this model is suitable to investigate early stages of the CRC metastatic process, mimicking the intravasation of tumor cells into a blood vessel, which can be monitored via on-chip imaging and mass spectrometry-based metabolomics. As depicted in the graphical abstract, the CRC-on-Chip model can be used to interrogate tumor cell behavior in a “multiplexed” fashion. From a single chip, tumor cell morphology, growth rate, and invasion dynamics can be monitored via confocal microscopy and frequent analyses of the effluent, such as metabolomics or cell shedding, can be performed.
Results
Development and characterization of CRC-on-chip
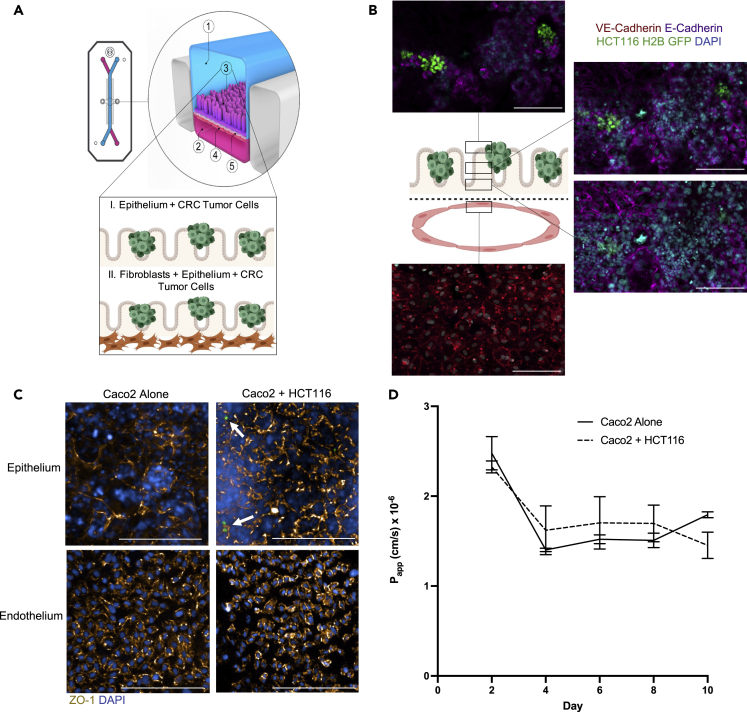
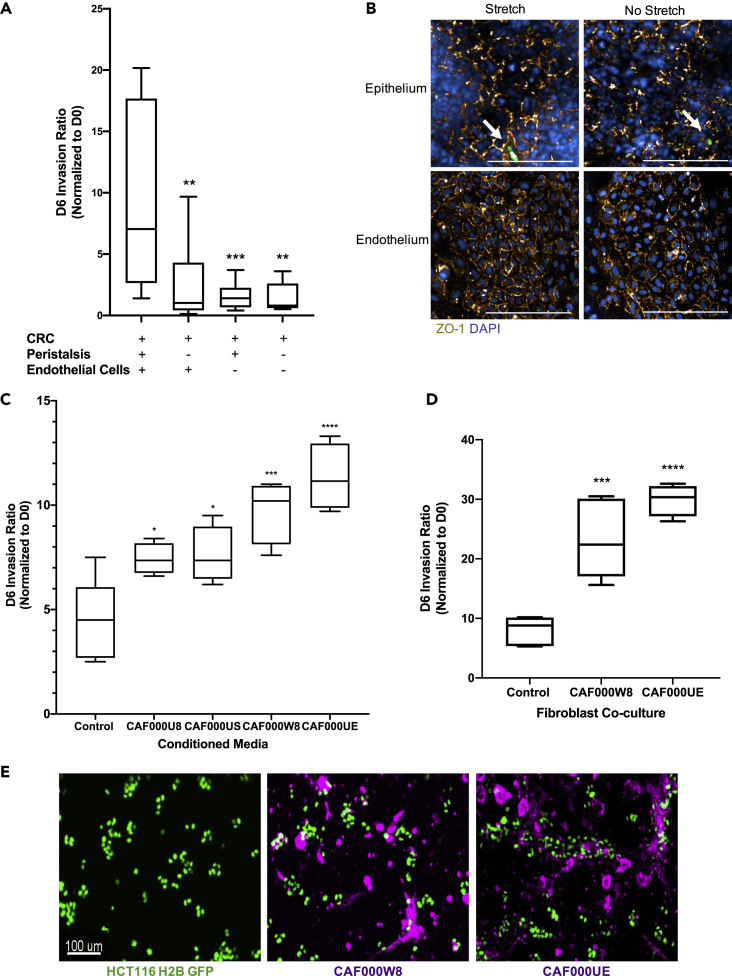
An Intestine Chip, consisting of intestinal epithelial cells (Caco2 C2BBe1) and endothelial cells (human umbilical vein endothelial cells; HUVECs) seeded in an ECM-coated chip, has been previously developed to model the intestine (Jalili-Firoozinezhad et al., 2018; Kim et al., 2012, 2016). On chip, the Caco2 C2bbe1 clone more closely resembles the normal human colon due to the formation of a polarized monolayer of epithelial cells displaying an apical brush border (Peterson and Mooseker, 1992). With fluid flow and cyclic strain, the Caco2 C2BBe1 cells form 3D-like architecture and differentiate into the four main intestinal cell lineages (goblet, enteroendocrine, Paneth, and enterocytes) (Figure S1). The Intestine Chip emulates tissue function by displaying an intact intestinal barrier with the endothelial cells forming vessel-like structures along the bottom channel (Jalili-Firoozinezhad et al., 2018; Kim et al., 2012, 2016). We modified the Caco2 Intestine Chip to model CRC by introducing epithelial and stromal cells (tumor cell lines or patient-derived tumor organoids and patient-derived CAFs) into the top channel (Figure 1A) and endothelial cells (HUVEC) into the bottom channel. ECM composition, cell types, on-chip locations, and fluorescent labels used to distinguish cell types are outlined in Table 1. Fluid flow and cyclic, peristalsis-like mechanical deformations were introduced to complete the physiologically relevant epithelial:endothelial tissue:tissue interface and create a CRC-on-Chip system.
Figure 1.
CRC-on-Chip tissue structure
(A) The organ-on-chip platform (schematic courtesy of Emulate, Inc.) consists of an epithelial channel (1) comprising epithelial and cancerous cells (3) and an endothelial channel (2) comprising HUVEC cells (4) separated by a porous membrane (5). To model cell-cell interactions in the TME, the CRC-on-Chip was modified to include layers of different cell types in the epithelial channel. CRC tumor cells were seeded on top of the epithelial cells. A stromal layer, comprised of CAFs, can be incorporated into the epithelial channel.
(B) Confocal fluorescence images of a chip cross-section spanning 106 μm from the top of the endothelial channel into the epithelial channel, highlighting the endothelial:epithelial tissue:tissue interface. HUVEC cells are labeled with anti-VE cadherin (red). Caco2 C2BBe1 cells labeled with anti-E-Cadherin (purple) form 3D-like structures in the top epithelial channel. HCT116 H2B-GFP cells grow in clusters on top of the Caco2 cells. Nuclei are labeled with DAPI (blue). Scale bar is 100 μm.
(C) Representative confocal immunofluorescent images of the epithelial (top) and endothelial (bottom) channels of an Intestine Chip (left) and CRC-on-Chip (right) stained for ZO-1 (gold) on day 6. DAPI (blue) labels the nuclei of the Caco2 C2BBe1 cells in the epithelial channel and HUVECs in the endothelial channel. White arrows designate HCT116 (green) in the epithelial channel of the CRC-on-Chip. Scale bars represent 200 μm. Images are maximum projections that span a 15 μm Z-height in the epithelial channel and a 10 μm Z-height in the endothelial channel with a 5 μm step size.
(D) The apparent permeability (Papp) of the intestinal epithelial cells in the top channel was not changed when HCT116 tumor cells were added to the CRC Chips. The concentration of inulin-FITC that diffused from the epithelial channel to the endothelial channel was used to calculate Papp (N = 3 Chips). Data are represented as mean ± SEM and analyzed using a two-way ANOVA; p > 0.05.
Table 1.
Chip locations specified for cell types and ECM composition used in the CRC-on-Chip
| Channel | Cell type | Cell | Fluorescent tag | ECM |
|---|---|---|---|---|
| Bottom | Endothelial | HUVEC | RFP | Matrigel and Collagen I |
| Top | Epithelial | Caco2 C2BBe1 | Unlabeled | Matrigel and Collagen I |
| Top | Stroma (CAF) | 000UE, 000UK, 000U8, 000US | Cell Tracker Deep Red | Collagen IV, Matrigel and Collagen I overlay |
| Top | Cancer Cell Line | HCT116 or HT29 | H2B-GFP | Matrigel and Collagen I |
| Top | CRC Organoid | ORG000US | H2B-GFP | Matrigel |
To generate the CRC-on-Chip, we first allowed the epithelium to form a monolayer, develop villi-like structures, and establish a complete, functional barrier (approximately 2-3 days (Kim et al., 2012; Kim et al., 2016)) before seeding CRC tumor cells. CRC tumor cells were seeded at a low density relative to the Caco2 C2BBe1 epithelial cells (1:5 tumor:epithelial cell seeding) in order to mimic cancer development within the colon epithelium. After the addition of tumor cells, the chips were placed under constant flow (30 μL hr −1) and stretch conditions (10% deformation; 0.2 Hz) for up to two weeks. The epithelial:endothelial tissue layers were visualized by immunofluorescence staining of the endothelial cells (VE-cadherin) in the bottom compartment and the Caco2 C2BBe1 (E-cadherin) and cancerous (H2B-GFP) epithelial cells in the top compartment (Figure 1B). Clusters of HCT116 tumor cells were observed on top of the 3D structures formed by the Caco2 C2BBe1 cells. We found the addition of CRC tumor cell lines did not noticeably impact the formation of tight junctions, as shown by immunofluorescence staining of ZO-1 in the epithelial and endothelial channel (Figure 1C). Large-scale images show strong ZO-1 expression across the length of the epithelial channel in both the Caco2 Intestine Chips and the CRC-on-Chips seeded with HCT116 tumor cells (HCT116-CRC-on-Chip) (Figures S2A and S2B). In addition, the presence of the HCT116 tumor cells did not significantly change the ability of the Caco2 C2BBe1 cells to form a stable intestinal barrier over the course of the experiments (Figure 1D). We modeled the development of CRC “hot spots” along the length of the colon. On chips, the tumor cells grew in 3D clusters extending into the lumen rather than the monolayer morphology seen on plastic (Figures S2C and S2D), suggesting the chip structure can be used to mimic the progression of colon cancer in which a polyp forms in the colonic crypts before eventually evolving into a cancerous lesion that grows into the intestinal lumen (Dekker et al., 2019; Humphries and Wright, 2008).
Metabolic comparison of Intestine Chips versus CRC chips
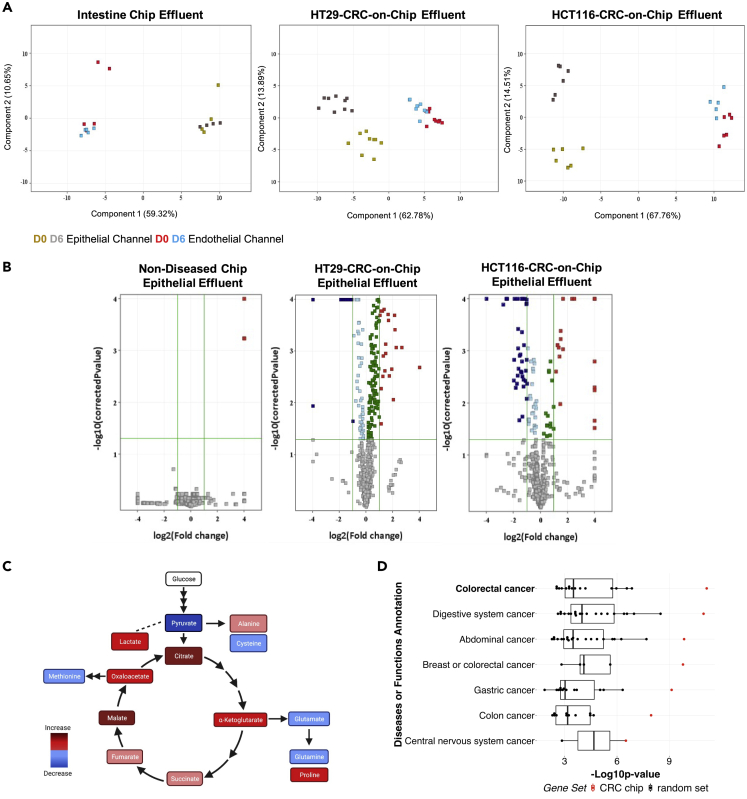
The microfluidic nature of the CRC-on-Chip system supports dynamic measurements of the effluent. To determine whether our CRC-on-Chip model mimics important aspects of CRC biology, we performed mass spectrometry-based metabolomics. Metabolite extracts of inlet and outlet media from the top epithelial channel and the bottom endothelial channel were analyzed from Intestine Chips and diseased CRC-on-Chips on days 0 (D0) and 6 (D6). In order to better understand the metabolomic profiles across different stages of CRC aggressiveness, we performed experiments with diseased CRC-on-Chips seeded with HCT116 (HCT116-CRC-on-Chip) and HT29 (HT29-CRC-on-Chip) cell lines (Tables S1, S2, S3, and S4). A principal component analysis (PCA) of the metabolite intensities shows clear separation between the epithelial and the endothelial channel effluents for both the Intestine Chips and the CRC-on-Chips (Figure 2A). When evaluating the metabolites in the epithelial channel, there was significant overlap in the Intestine Chip between D0 and D6, resulting in only a few differential metabolites, while there were several differential metabolites detected in the HT29-CRC-on-Chip, and a much larger number of differential metabolites detected in the HCT116-CRC-on-Chip between days (Figure 2B). Furthermore, metabolic profiles between the Intestine Chips and the HCT116-CRC-on-Chip on D6 yielded differentially expressed metabolites (Table S5). The differential metabolites that were identified using our in-house library between the Intestine Chip and the HCT116-CRC-on-Chip were mapped to pathways using Ingenuity Pathway Analysis (IPA) to identify the most affected pathways in the HCT116-CRC-on-Chips (Table S6). The TCA cycle and several amino acid metabolism pathways were the most significantly altered pathways, as highlighted in Figure 2C.
Figure 2.
Metabolic analyses validate use of CRC-on-Chip to model CRC progression
(A) Epithelial and endothelial effluent was collected from the Intestine Chip and the HT29-and HCT116-CRC-on-Chips on days 0 and 6 of the experiment and mass spectrometry-based metabolomics was performed. The principal component analysis (PCA) on the differential metabolites demonstrates the clustering of samples corresponding to the effluent compartment (epithelium or endothelium) and the different time points.
(B) Volcano plots comparing the metabolites from the top epithelial channel on day 0 and day 6 for the Intestine Chip and the HT29-and HCT116-CRC-on-Chips. Each point represents a metabolite. Analytes with p values <0.05 and fold change >2 were regarded as statistically significant (colored red and blue upregulated and downregulated, respectively).
(C) Differential metabolites between the Intestine Chip and the HCT116-CRC-on-Chip from the epithelial effluent showed altered TCA cycle and amino acid metabolism via Ingenuity Pathway Analysis (IPA).
(D) The 50 differential metabolites that matched to our internal database from the epithelial channel of the HCT116-CRC-on-Chip mapped to colorectal cancer with the highest significance (highest –Log10p value) using IPA. Each group is ranked by p value and colored based on the 50 differentially expressed metabolites from our dataset, termed “CRC Chip” (red; asterisk denotes significant outliers) or 20 permutations of 50 randomly selected metabolites, termed “random set” (black box plots).Wilcoxon signed rank test compared the CRC Chips to the random selection sets for the “colorectal cancer” disease state, p = 0.0005.
Separate clusters were observed between the time points in the HCT116-CRC-on-Chip data set (Figure 2B), with the identification of 50 significantly differentially expressed metabolites in the epithelial effluent indicating CRC tumor cell growth. Differential metabolites (p < 0.05 and with a fold change greater than 2) in the epithelial effluent of the HCT116-CRC-on-Chip mapped to ‘CRC’ disease state with the highest significance using IPA (p = 7.19 × 10−12), while those from the HT29-CRC-on-Chip (p < 0.05 and with a fold change greater than 1) mapped to CRC to a less significant degree (p = 2.31 × 10−9). To confirm the specificity of this finding, we performed 20 permutations in which we selected 50 random metabolites from our library of identified compounds from the HCT116-CRC-on-Chip experiments and these “random sets” were evaluated with the same pathway analysis. The mapping of our HCT116-CRC-on-Chip data to ‘CRC’ and other related disease states is not based on chance, suggesting that our CRC-on-Chip is a good model system to further study CRC progression (Figure 2D).
Examination of tumor cell intravasation
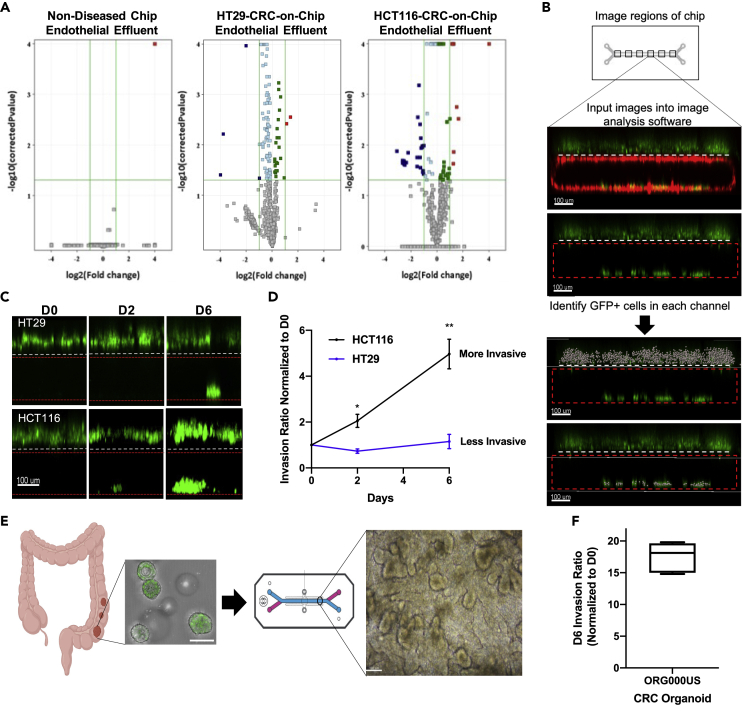
When we examined the PCA for the endothelial channel effluent (Figure 2A), we observed a similar pattern to what was detected in the epithelial channel. Separate clusters of metabolites between D0 and D6 were identified in the HCT116-CRC-on-Chip, minimal separation was seen in the HT29-CRC-on-Chip, and no separation was found in the Intestine Chip. We detected over 20 differential metabolites in the HCT116-CRC-on-Chip endothelial effluent and only 5 differential metabolites in the HT29-CRC-on-Chip (Figure 3A), suggesting metabolic changes within the endothelial channel occur over time only when CRC tumor cells are present in the model. When we analyzed the differentially expressed metabolites from the endothelial compartment in the HCT116-and HT29-CRC-on-Chips using IPA, we found that several pathways were significantly altered in the HCT116-CRC-on-Chips (glycine betaine degradation, alanine metabolism, HIF1α signaling, and adenine and adenosine salvage pathways) (Table S7) while no pathways were identified as significantly altered in the HT29-CRC-on-Chips.
Figure 3.
Validation of CRC tumor cell invasion from an epithelial to endothelial compartment, mimicking intravasation
(A) Volcano plots comparing the metabolites from the bottom endothelial channel on day 0 and day 6 for the Intestine Chip and the HT29-and HCT116-CRC-on-Chips. Each point represents a metabolite. Analytes with p values < 0.05 and fold change >2 were regarded as statistically significant (colored red and blue).
(B) 6 regions of the chip were imaged via confocal microscopy and input into 3-D reconstruction software for GFP + cell quantification. An invasion ratio was calculated based on the number of GFP + cells in the bottom channel compared to the top channel and normalized by the day 0 counts. Scale bars represent 100 μm.
(C and D) Tumor cell (HCT116 or HT29) invasion was monitored over time by imaging the same chip regions at various time points, days 0, 2, 6. Representative images show different invasion behavior for each tumor cell (C) and quantification is also depicted (D). N = 6 Chips. Data are represented as mean ± SEM and analyzed using a two-way ANOVA; ∗p < 0.05; ∗∗p < 0.01). Scale bars represent 100 μm.
(E) CRC organoids (H2B GFP labeled) from patient 000US were dissociated and fragments were seeded onto the ECM-coated epithelial channel. Brightfield image of the 000US organoids seeded in the epithelial channel of the chip showing the CRC tissue architecture on day 6 (D6). Scale bars represent 100 μm. Chip schematic courtesy of Emulate, Inc.
(F) Invasion of ORG000US was measured on D6 of the experiment (N = 6 Chips). An invasion ratio was calculated based on the number of GFP + cells in the bottom channel compared to the top channel and normalized by the day 0 counts. Data are represented using a boxplot.
Based on the metabolic changes we observed within the endothelial compartment when CRC tumor cells of various aggressiveness were present on-chip, we hypothesized that the CRC Chip could help address important questions related to tumor metastasis, in particular early tumor cell dissemination. Specifically, tumor cells were monitored via confocal microscopy to visualize and quantify the invasion of cells from the top epithelial channel into the bottom endothelial channel through an ECM coating and a porous membrane. Given that the tumor cells traversed the epithelium and ECM to invade through the porous membrane and into the endothelium channel, we termed this an “invasion assay”. We calculated a ratio of the number of invaded cells (tumor cells in the bottom channel) to the number of non-invaded cells (tumor cells in the top channel) over time to determine an invasion rate (Figure 3B). Data were normalized to D0 to account for any initial cell seeding variability between chips (Figure S3A). While the epithelial Caco2 C2BBe1 cells showed minimal invasion (Figure S3A), the invasion rate differed between CRC tumor cell lines, with the HT29 H2B-GFP cells having a significantly lower invasion rate as compared with the HCT116 H2B-GFP (Figures 3C and 3D). Specifically, the average day 6 invasion ratio for all three cell lines was: HCT116 = 4.97; HT29 = 1.15; Caco2 C2BBe1 = 0.37. This finding mimics the aggressiveness of the tumors from which these cells were derived and is supported by extensive in vitro and in vivo data in which HCT116 cells are highly aggressive and known to develop liver metastases in vivo, while the HT29 cells possess less efficient metastatic capabilities (Olejniczak et al., 2018). This trend of the HCT116 cells invading more than the HT29 cells corroborates the metabolic changes seen in the endothelial compartment in the CRC-on-Chips (discussed above), suggesting the differential metabolites and altered metabolic pathways may be a result of CRC tumor cells invading into the endothelium.
Traditionally, in vitro tumor cell invasion has been studied using transwell assays. We performed modified transwell experiments by culturing HUVEC cells on the bottom of the transwell membrane and co-culturing Caco2 C2BBe1 and CRC tumor cells on top of the membrane to mimic the culturing of multiple cell types in the chip system. HT29 cells were less invasive than HCT116 cells in the transwell system; however, when the number of invaded cells is compared between transwell and on-chip experiments at a similar time point (day 2 for both experiments), the on-chip invasion assay indicated less invasion (Figure S3B). While the pore size and the cells seeded per culture area were the same between the two different model systems, the presence of mechanical cyclic stretching and fluid flow in the chip could explain these differences. This suggests that the chip system more closely models in vivo intravasation as a rare event compared to traditional in vitro assays (Deryugina and Kiosses, 2017; Hapach et al., 2019; Wyckoff et al., 2000).
Previous studies have combined the organ-on-chip system with organoids derived from healthy small and large intestine tissues and demonstrated more in vivo-like composition of intestinal cell types (Kasendra et al., 2018), mucus physiology (Sontheimer-Phelps et al., 2020) and drug response (Kasendra et al., 2020). We adapted these published methods to seed organoids derived from human colon tumors in the top channel of the chips (without the presence of the Caco2 C2BBe1 cells) (Figure 3E). We confirmed that tumor organoid cells were capable of invading into the endothelial channel under the same fluid and mechanical forces as applied in the Intestine Chip (Figure 3F).
CRC cells exhibit phenotypic heterogeneity during intravasation
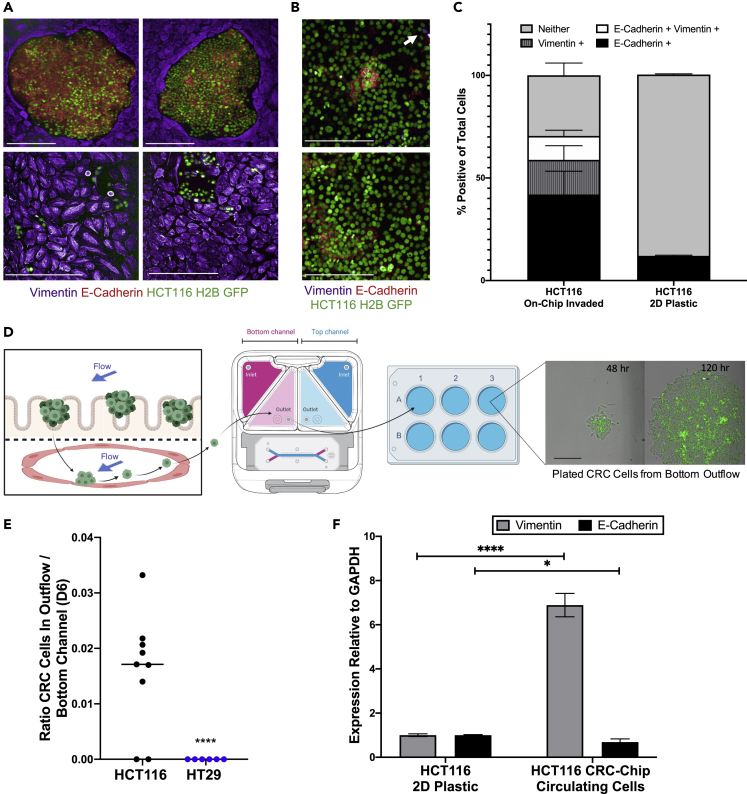
Given the differential invasive phenotypes of HT29 or HCT116 cells, we sought to identify changes occurring during intravasation in our CRC Chips. First, we investigated the expression of epithelial or mesenchymal markers in CRC Chips cultured with HT29 or HCT116 tumor cells, which differ in their EMT programming (Pino et al., 2010). HT29 tumor cells in the top, epithelial channel formed tight clusters that stained strongly for the adhesion molecule E-cadherin and weakly for vimentin, suggesting these cells are epithelial-like (Figure S4, left panels). In contrast, staining of HCT116 tumor cells showed decreased E-cadherin expression toward the center of the tumor clusters and stronger vimentin expression at the edges of these clusters (Figure S4, right panels), suggesting that the HCT116 tumor cells are losing epithelial features and gaining a more mesenchymal-like phenotype.
We further investigated HCT116 cells that invaded and adhered to the bottom, endothelial channel and discovered heterogeneity in the expression of epithelial or mesenchymal markers. Some tumor cells stained strongly for E-cadherin and were negative for vimentin, forming large clusters bordered by HUVECs (Figure 4A, top panels). While in other regions of the same chip, there were invaded HCT116 cells that did not express E-cadherin, but stained positive for vimentin and showed a mesenchymal-like morphology (Figure 4A, bottom panels). When we examined HCT116 cells cultured in traditional cell culture conditions (plastic, 2D), we found less E-cadherin-positive and vimentin-positive cells (Figure 4B). When we quantitated the percentage of cells identified as positive or negative for E-cadherin or vimentin expression, we discovered that there was greater heterogeneity in the invaded HCT116 cells on-chip compared to plastic (Figure 4C). The percentage of E-cadherin+, E-cadherin+ and vimentin+, and vimentin + HCT116 cells increased on-chip, while the percentage of cells that were negative for both E-cadherin and vimentin decreased on-chip as compared to HCT116 cultured on plastic. Taken together, these results suggest that HCT116 tumor cells are more heterogeneous on-chip, recently corroborated by another tumor-on-a-chip model system (Hachey et al., 2021).
Figure 4.
CRC cell heterogeneity during intravasation on-chip
(A) Representative images depict the phenotypic heterogeneity of invaded HCT116 cells adhered to endothelial cells in the bottom chip channel. HUVECs uniformly express vimentin (purple), while clusters of HCT116 cells (green) grow in tight aggregates and highly express E-Cadherin (red) (top panels) or grow in more disperse colonies with higher vimentin (purple) expression (bottom panels). Scale bar represents 200 μm and images are from day 6 of the experiment.
(B) Representative images depict the heterogeneity of HCT116 cells cultured on plastic. HCT116 cells (green) show moderate expression of E-Cadherin (red), with relatively few vimentin-positive cells (purple) (white arrow). Cells were grown to 70% confluency before fixation, immunofluorescent staining, and imaging. Scale bar represents 200 μm.
(C) HCT116 heterogeneity from (A) and (B) was quantitated using PerkinElmer Harmony software. GFP + tumor cells were segmented and classified as E-Cadherin + or vimentin + based on intensity thresholds (N = 5 Chips for chip experiments, N = 4 replicates for 2D plastic experiments). Data are represented as mean ± SEM.
(D) Invaded CTCs are found in the endothelial effluent where they are collected and cultured for down-stream analyses. Scale bar represents 200 μm.
(E) Viable tumor cells were collected from the effluent of the bottom endothelial channel reservoir on day 6. Cells were plated and counted via HCS imaging system once the cells had attached to the plate (6-10 hr later) (N = 6 Chips for HT29 experiments, N = 9 Chips for HCT116 experiments; ∗∗∗∗p < 0.0001).
(F) RT-qPCR results show invaded HCT116 cells have reduced E-Cadherin and increased vimentin expression compared to HCT116 cells grown on plastic tissue culture dishes (N = 6 replicates). Data are represented as mean ± SEM and analyzed using multiple unpaired t-tests; ∗∗∗∗p < 0.0001, ∗p < 0.05).
Moreover, the microfluidic design of the chips allows for interrogation of any viable, circulating cells that have perfused through the channel and accumulate in the outlet reservoir (Figure 4D). We collected media from the outflow of the bottom channel every two days. HT29 cells did not appear in the bottom channel effluent, however we were able to isolate, quantify, and relate the number of circulating HCT116 cells in the bottom channel outflow to the number of invaded HCT116 cells still adhered to the endothelial channel (Figure 4E). These invaded, circulating cells had lower E-cadherin and higher vimentin expression than HCT116s on plastic, indicating these cells had acquired mesenchymal-like phenotypes (Figure 4F).
Mechanical and biochemical cues from the TME impact CRC invasion
It is understood that cancer cells respond to mechanical cues present in the body (e.g., shear force from fluid flow) (Follain et al., 2020), however many of the mechanistic details remain unknown as these processes are difficult to study. A major advantage of the CRC-on-Chip model over other organs-on-chip technologies is the ability to mimic peristalsis, a physiological process of muscle contraction and relaxation that naturally occurs in the colon. We showed that in the presence of peristalsis (10% strain; 0.2 Hz), HCT116 tumor cell invasion increased dramatically compared to static conditions (>3-fold) (Figure 5A). In addition, CRC-on-Chips with HT29 tumor cells showed increased invasion in the presence of peristalsis (2-fold) (Figure S5). Tight junction formation between epithelial or endothelial cells was not noticeably affected by the presence of cyclic stretching, as measured by immunofluorescent staining for ZO-1 on stretched and not stretched CRC-on-Chips (Figures 5B and S6). The increased invasive phenotype in response to mechanical forces requires further study, including alternative mechanisms beyond epithelial barrier disfunction.
Figure 5.
The TME influences tumor cell invasion
(A) CRC-on-Chips were cultured in the presence of cyclic peristalsis-like mechanical strain and HUVECs (N = 12 Chips), without cyclic strain and with HUVECs (N = 12 Chips), with cyclic strain and without HUVEC cells (N = 12 Chips) or without cyclic strain or HUVECs (N = 9 Chips). The invasion ratio of the HCT116 cells was determined via microscopy. Data are represented as boxplots and analyzed using a 1-way ANOVA with multiple comparisons; ∗∗p < 0.01, ∗∗∗p < 0.001.
(B) Representative confocal immunofluorescent images of the epithelial (top) and endothelial (bottom) channels of the CRC-on-Chips in the presence (left) or absence (right) of peristalsis stained for ZO-1 (gold) on day 6. DAPI (blue) labels the nuclei of Caco2 C2BBe1 cells in the epithelial channel and HUVECs in the endothelial channel. White arrows indicate HCT116 cells. Scale bars represent 200 μm. Images are maximum projections that span a 15 μm Z-height in the epithelial channel and a 10 μm Z-height in the endothelial channel with a 5 μm step size.
(C) Conditioned media from CAFs derived from four patients (N = 4 Chips for each patient-derived CAF) was flowed through the epithelial channel for the duration of the experiment. Differences in HCT116 cell invasion ratio quantification is depicted. Data are represented as boxplots and analyzed using 1-way ANOVA with multiple comparisons; ∗p < 0.05; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001.
(D) Representative CAFs derived from two patients were labeled with Cell Tracker Deep Red and seeded in the top channel prior to epithelial cell and HCT116 cell seeding (N = 4 replicates for each patient-derived CAF) and the invasion ratio was quantified on day 6. Data are represented as boxplots and analyzed using a 1-way ANOVA with multiple comparisons; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001.
(E) Representative images of CAF000W8 and CAF000UE on day 0 (one day after tumor cell seeding) illustrate heterocellular interactions on chip. Scale bar represents 100 μm.
In addition to the physical forces present in the TME, heterocellular interactions can also be examined using this model. We showed that cancer-endothelial cross talk is important for driving an invasive phenotype. In the presence of HUVECs, the HCT116 invasion rate was significantly higher than when HUVECs were not seeded in the CRC-on-Chip (Figure 5A). There was no difference in HCT116 invasion ratio in the presence or absence of cyclic strain when the HUVECs were absent, suggesting an important role of the endothelial cells in the invasive phenotype in the presence of peristalsis. Our data support published evidence that the tumor:blood vessel tissue:tissue interface is a critical modulator of cancer progression (Amos and Choi, 2021; Choi and Moon, 2018; Nguyen et al., 2019).
Furthermore, CAFs, the most abundant cell type in the cancer stroma, have been implicated in promoting invasion (Karnoub et al., 2007; Orimo et al., 2005) and cancer metastasis (Sahai et al., 2020). In order to interrogate the role of CAFs in CRC invasion in our system, we examined the effects of CAF-derived secreted factors using conditioned media (CM), as well as the physical presence of CAFs seeded on the chips. For the secreted factor analysis, we perfused CAF CM through the epithelial channel of the chip for the duration of the experiment (6 days). CM from four different patient-derived CAF lines significantly increased HCT116 invasion (Figure 5C). We measured the cytokines secreted in the CM and, while some cytokines overlapped between the CAFs, there was significant patient heterogeneity in the cytokine profiles (Figure S7 and Tables S8 and S9). While the continual exposure of tumor cells to CAF secreted factors through microfluidics is an exciting advancement over current methods, the physical interaction between CAFs and tumor cells has been shown to be important for invasion (Gaggioli et al., 2007). We introduced a layer of CAFs to the CRC Chip after modification of the ECM in the top channel to ensure CAF attachment and viability (set-up ii of Figure 1A, Table 1), leaving the endothelial channel unchanged. The CAFs were initially seeded as a monolayer on the chip membrane coated with collagen IV but transitioned to form extended networks after seeding the Caco2 C2BBe1 layer and the HCT116 tumor cells two days later. Similar to the CM results, the presence of the CAF000UE and CAF000W8 significantly increased invasion (Figure 5D). Notably, confocal microscopy revealed that the physical presence of the CAFs influenced how the HCT116 cells seeded onto the epithelium. The HCT116 cells clustered around and on top of the CAF networks (Figure 5E). While the invasion rates were increased approximately 2-fold in the presence of CAF CM or the physical CAFs (CAF000W8 and CAF000UE), the HCT116 seeding morphology suggests the physical interaction between CAFs and tumor cells is important and requires further study.
Discussion
We expanded upon a previously characterized Intestine Chip (Kim et al., 2012) to generate a CRC-on-Chip that encapsulates cancer-specific biomimetic microenvironments to advance our knowledge in the CRC research domain. Some of the results presented here corroborate findings from previous cancer progression studies as a metric to validate our model system. However, we also demonstrate for the first-time a CRC-on-Chip that recapitulates many aspects of the complex TME that are difficult to reconstruct in other model systems. This has led to the discovery of novel preliminary findings, including the impact of peristalsis and the epithelial:endothelial tissue:tissue interface on tumor cell intravasation. While further mechanistic studies are needed to explore the impact of these findings, we demonstrate the utility of the microfluidics-based CRC-on-Chip for the study of CRC progression.
The current chip design can be used to mimic the progression of colon cancer in which a polyp forms in the colonic crypts before eventually evolving into a cancerous lesion that grows into the intestinal lumen (Dekker et al., 2019; Humphries and Wright, 2008). When we introduced CRC tumor cells into the model, the presence of the epithelium-tumor boundary resulted in 3D “hot spots” of tumor cells. There was also a measurable shift in metabolite profiles from the collected effluent between the Intestine Chip and the CRC Chip, which mapped to the CRC disease state. Metabolic reprogramming has been extensively studied in cancer and is a hallmark of cancer progression (Faubert et al., 2020). Specifically, our results are corroborated by published metabolomics datasets that found alterations in the TCA cycle, urea cycle, and amino acid metabolism in patients with CRC when compared to healthy controls (Farshidfar et al., 2016; Tan et al., 2013). Although additional studies are needed to address the metabolic reprogramming that occurred in the CRC Chip, the microfluidic attributes of the chip technology will aid in identifying metabolic phenotypes that link to disease progression and metabolic vulnerabilities that may be targeted in metastatic tumor cells.
In addition to effluent-based metabolomics studies confirming CRC pathway-specific metabolites in the chip, it also revealed tumor cell intravasation as a feature of the model. This finding was based on dynamic metabolite changes in the endothelial channel indicative of tumor cell presence. This prompted us to further explore the capacity of our system to interrogate the invasive potential of CRC tumor cells. An advantage of this model is the transparent nature of the chip material, which supports dynamic confocal imaging of cell phenotypes. We were able to quantitate substantial differences in invasion into the vascular channel between aggressive and non-aggressive CRC tumor cells and identify a metabolic profile for an aggressive, invasive CRC tumor. More work is needed to understand whether these metabolic changes are influenced by tumor-endothelial cross talk or solely the presence of invaded tumor cells. In addition, we were able to visualize cellular heterogeneity as tumor cells intravasated. However, questions emerge as to why some invaded tumor cells remain as tightly adhered clusters to the endothelial wall, whereas, others end up in circulation and presumably are the mediators of distant metastases. Carefully interrogating the role of dynamic processes such as heterogeneity in cancer progression has proved challenging due to the lack of appropriate models (Brabletz et al., 2018), however our preliminary work suggests the CRC-on-Chip is a useful tool in elucidating cellular plasticity, invasiveness, and cancer progression.
Furthermore, we demonstrated we can tune mechanical and biochemical cues within the TME to better understand early tumor cell dissemination. Previously, the role of mechanical forces on cancer progression has been technically challenging to study in the laboratory. The introduction of microfluidic-based systems has expanded our understanding of how fluid shear forces influence tumor cell (particularly circulating tumor cells) survival in circulation and extravasation at the metastatic site(s) (reviewed extensively in (Follain et al., 2020)). Previous work studying metastasis in a zebrafish model suggests that tumor cells which form stable adhesions with the endothelium are more likely to extravasate (Follain et al., 2018; Osmani et al., 2019). The CRC-on-Chip is an advantageous model to better understand and potentially target the metastatic capabilities of the tumor cells that adhere to the endothelial channel. Future work will investigate whether the tumor cells in the endothelial channel have a propensity to extravasate and seed distant organs, such as the liver, given its frequency as a metastatic site for CRC. Other researchers have designed microfluidic devices to study these later steps in the metastatic cascade. For example, one group connected a colon cancer chamber to organ-specific epithelial cell chambers (i.e. lung or liver) to study metastatic organotropism (Aleman and Skardal, 2019; Skardal et al., 2016).
While microfluidic systems have offered insights into the role of shear forces in tumor biology, few microfluidic organ chips incorporate mechanical rhythmic deformations. We hypothesize that the increased invasive capabilities of CRC tumor cells in the presence of peristalsis may be mediated by endothelial-tumor cross talk. The mechanobiology of endothelial cells has been well studied, with several reports describing how endothelial cells change cell orientation, proliferation, and cell surface markers in response to cyclic strain (Thodeti et al., 2009, Yamashiro and Yanagisawa, 2020), however the role of mechanotransduction in CRC requires further study. A non-small-cell lung cancer (NSCLC) organ chip model demonstrated that rhythmic mechanical forces mimicking breathing decreased NSCLC cell invasive behavior (Hassell et al., 2017). However, in our CRC model, we show that peristalsis-like motions increase tumor cell invasion. This discrepancy may be explained by different organs responding differentially to biophysical cues. Recent evidence suggests that these mechanical forces may accelerate cancer progression rather than act as a passive bystander (Ciasca et al., 2016) and can alter the stromal milieu, which in turn influences tumor cell behavior (Huang et al., 2013). In the context of CRC, it has been shown that increased pressure in the tumor promotes tumor cell proliferation and adhesion through Src and FAK signaling (Basson et al., 2000). While these reports investigated how various physical forces influenced cancer progression, another recent study showed that CRC tumor cell lines representing degrees of metastatic potential were distinguished based on cell deformability when exposed to shear stress, indicating there may be prognostic value in understanding the mechanobiology of CRC progression (Armistead et al., 2020). There remains a dearth of information on the importance of mechanical forces during cancer progression; however, organ chip technologies are primed to fill this scientific void.
Moreover, tumor-stromal cell interactions are important to recapitulate in preclinical cancer model systems, in particular, CAFs given their abundance in the TME. It is well documented that CAFs can promote invasion (Karnoub et al., 2007; Orimo et al., 2005), which we demonstrated in our CRC Chip model. Some studies have shown adhesions between E-cadherin and N-cadherin on tumor cells and CAFs, respectively, enable CAFs to physically guide tumor cells through the invasion process (Labernadie et al., 2017). One limitation of the current chip model is the inability to perform time-lapse imaging without sacrificing continual and controlled fluid flow and cyclic stretching. Future iterations of this system may support the imaging demands required to capture the leader-follower phenomenon during intravasation. In addition to CAFs, the CRC-on-Chip is amenable to inclusion of other cell types to more closely model the complete TME. The addition of immune cells and the microbiome as previously explored in the Intestine Chip (Kim et al., 2016) would be extremely relevant given the importance of the immune system in cancer progression (Gonzalez et al., 2018) and the intriguing microbiome-CRC cross talk (Geller et al., 2017).
The organ chip field is expanding in popularity and demonstrated utility; however, it is still in its infancy especially in the context of cancer. Conversely, organoids have revolutionized the cancer field because they are thought to better replicate organ complexity and function and have been shown to predict patient anticancer drug response in early in vitro drug screens (Driehuis et al., 2019; Drost and Clevers, 2018; Ooft et al., 2019; Vlachogiannis et al., 2018; Yao et al., 2019). Nevertheless, the organoid model lacks aspects of the TME discussed here, such as stromal-tumor interactions, tumor:endothelial tissue:tissue interfaces, and physical forces. We demonstrated the utility of combining organoids and the organ chip technology to study tumor cell progression as evidenced by patient-derived tumor organoids invading into the endothelial channel using our CRC-on-Chip. Expanding this work to encompass organoids from various patients with CRC will give us greater insights into inter-tumor heterogeneity. In addition, the tissue-tissue interface of the Organ Chips also supports the performance of drug treatment studies in a physiological manner by flowing drugs through the endothelial channel and measuring tumor cell response in the epithelial channel as seen in previous reports (Carvalho et al., 2019). Further development of the patient-derived CRC-on-Chip to include autologous normal and tumor organoids will pave the way for studying cancer progression in a patient-specific manner and may enhance precision medicine approaches (Ramzy et al., 2020).
In conclusion, the CRC-on-Chip provides a human-relevant model system to examine CRC progression within the TME milieu. The ability to monitor key steps in cancer metastasis is unparalleled to other preclinical cancer models. This system will be critical to better understand the mechanisms surrounding CRC early metastatic spread and potentially elucidate novel therapeutic targets within the TME.
Limitations of the study
We use a combination of primary, patient-derived cells and immortalized cell lines to build the CRC Chip. In this case, we believe the increased complexity of the microfluidic organ-on-chip system more closely resembles in vivo tissue structure and function and will greatly advance CRC mechanistic studies; however we acknowledge that the use of cell lines is a limitation of this study, especially the use of the C2BBe1 clone of the Caco2 cell line. Although this cell line has been selected because it is more representative of the human colon, we are working to include patient-derived normal colon epithelial cells to this model system. Patient-derived cell populations require further optimizations and, as such, the patient-derived organoid model we describe here is exclusively cancer organoids. Future work will include additional primary cell types from diverse patient populations to better represent the heterogeneity of human cancer biology. In addition, since our Organ Chip platform is not currently compatible with live, dynamic imaging, the use of static time points in this study is a limitation. As the technology improves, we will be able to monitor cell behavior in real time on-chip to better understand cancer progression. Furthermore, while we suggest our CRC-on-Chip is capable of establishing an intact barrier based on ZO-1 expression and permeability assays, an important caveat to this finding is the lack of transepithelial/transendothelial electrical resistance (TEER) measurements of the tight junction integrity.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-E Cadherin antibody [HECD-1] - Intercellular Junction Marker | Abcam | Cat# ab1416; RRID:AB_300946 |
| Recombinant Anti-Vimentin antibody [EPR3776] – Cytoskeleton Marker | Abcam | Cat# ab92547; RRID:AB_10562134 |
| Goat anti-Rabbit IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa fluor 555 | Molecular Probes, Invitrogen | Cat# A21428; RRID:AB_141784 |
| Goat anti-Mouse IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa fluor 647 | Molecular Probes, Invitrogen | Cat# A21235; RRID:AB_2535804 |
| Rabbit Anti-Human VE Cadherin Polyclonal Antibody, Unconjugated | Abcam | Cat# ab33168; RRID:AB_870662 |
| ZO-1 Monoclonal Antibody (ZO1-1A12), Alexa Fluor 594 | Thermo-Fisher | Cat# 339194 RRID:AB_2532188 |
| Bacterial and virus strains | ||
| LentiBrite Histone-H2B-GFP Lentiviral Biosensor | Millipore | Cat#17-10229 |
| Biological samples | ||
| Patient-derived sample: CAF000UE | Shannon M. Mumenthaler This paper |
N/A |
| Patient-derived sample: CAF000U8 | Shannon M. Mumenthaler This paper |
N/A |
| Patient-derived sample: CAF000W8 | Shannon M. Mumenthaler This paper |
N/A |
| Patient-derived sample: CAF000US | Shannon M. Mumenthaler This paper |
N/A |
| Patient-derived sample: ORG000US | Shannon M. Mumenthaler This paper |
N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Basement membrane extract (BME) | Cultrex | Cat#3533-005-02 |
| Noggin | Tonbo | Cat#21-7075-U500 |
| Epidermal growth factor (EGF) | Life Technologies | Cat#PHG0313 |
| SB202190 | Sigma Aldrich | Cat#S7067 |
| TGF-β RI Kinase Inhibitor IV (A83-01) | Millipore | Cat#616454-2MG |
| Nicotinamide | Sigma Aldrich | Cat#N0636 |
| B27 | Gibco | Cat#17504-001 |
| N-Acetyl-L-cysteine | Sigma Aldrich | Cat#A9165 |
| N2 | Gibco | Cat#17502-048 |
| HEPES | Gibco | Cat#15630-080 |
| GlutaMax | Gibco | Cat#35050-061 |
| 16% paraformaldehyde | Electron Microscopy Sciences | Cat#15710 |
| Inulin-Fluorescein isothiocyanate (FITC, 2-5kDa) | Sigma Aldrich | Cat# F3272 |
| Collagenase, Type II | Millipore | Cat#234155-100MG |
| Bovine hyaluronidase | MP Biomedicals | Cat#0210074080 |
| Hoechst 33342 (nuclear dye) | Invitrogen | Cat# H1399 |
| Saponin | Sigma Aldrich | Cat#84510 |
| Bovine Serum Albumin | Millipore | Cat#260-500GM |
| InfinityLab Deactivator Additive | Agilent | Cat#5191-4506 |
| EGM-2 Endothelial Cell Growth Medium-2 BulletKit | Lonza | Cat# CC-3162 |
| DMEM | Gibco | Cat#10569-010 |
| Eagle’s Minimum Essential Medium | Corning | Cat#10-009-CV |
| Advanced DMEM/F-12 | Gibco | Cat#12634-010 |
| Rat tail collagen type I | Corning | Cat# C354249 |
| Matrigel | Corning | Cat#356231 |
| ER-1 | Emulate, Inc | Cat# Basic Research Kit |
| ER-2 | Emulate, Inc | Cat# Basic Research Kit |
| Rat tail collagen IV | Sigma Aldrich | Cat# C5533 |
| Cell Tracker Deep Red | Invitrogen | Cat# C34565 |
| Gentle Cell Dissociation Reagent | STEMCELL Technologies | Cat#07174 |
| TrypLE | Gibco | Cat#12605-028 |
| LY-27632 | Millipore | Cat#5.09228.0001 |
| McCoy’s 5A (Modified) Media | Gibco | Cat#16600-082 |
| DAPI | Sigma Aldrich | Cat# D9542 |
| Critical commercial assays | ||
| RNAspin Mini RNA Isolation Kit | GE Healthcare | Cat#25-0500-71 |
| Proteome Profiler Human XL Cytokine Array Kit | R&D Systems | Cat# ARY022B |
| Metabolomics QC Standard 2 | Cambridge Isotope Laboratories, Inc | Cat# MSK-QC2 |
| Metabolomics QC Standard 1 | Cambridge Isotope Laboratories, Inc | Cat# MSK-QC1 |
| iScript Reverse Transcription Supermix | Bio-Rad | Cat#1708841 |
| iQ SYBR Green Master | Bio-Rad | Cat#1708880 |
| Organ-on-Chip | Emulate, Inc | Basic Research Kit |
| Experimental models: cell lines | ||
| Human: RFP expressing Human umbilical vein endothelial cells | Angio-Proteomie | Cat# cAP-0001RFP |
| Human: Caco2 C2BBe1 | ATCC | Cat# CRL-2102 |
| Human: HCT116 | ATCC | Cat# CCL-247 |
| Human: HT29 | ATCC | Cat# HTB-38 |
| Human: CCD18Co | ATCC | Cat# CRL-1459 |
| Oligonucleotides | ||
| Primers for analysis of gene-expression changes, see Table S11 | Integrated DNA Technologies (IDT) | N/A |
| Software and algorithms | ||
| R statistical environment | https://www.R-project.org/ | v4.0.2 |
| VennDiagram R package | R statistical environment | v1.6.20 |
| Harmony High-Content Imaging and Analysis Software | Perkin Elmer | N/A |
| Imaris image analysis | Imaris | N/A |
| Profinder | Agilent Technologies | N/A |
| Mass Profiler Professional (MPP) | Agilent Technologies | N/A |
| Ingenuity Pathway Analysis (IPA) | digitalinsights.qiagen. com | N/A |
| GraphPad Prism 8 | GraphPad Software, Inc. | https://www.graphpad.com/ |
| Mass Hunter | Agilent Technologies | N/A |
| Biorender | Biorender | https://biorender.com/ |
| Other | ||
| Fluoroblok™ transwell inserts with PET membranes | Corning | #351152 |
| Zoë ™ Culture Module | Emulate, Inc. | N/A |
| Orb™ Hub Module | Emulate, Inc. | N/A |
| Perkin Elmer Operetta High Content Screening (HCS) platform | Perkin Elmer | N/A |
| Perkin Elmer Operetta CLS High Content System | Perkin Elmer | N/A |
| Spectramax | Molecular Devices | N/A |
| Olympus FV3000 | Olympus | N/A |
| Ultra high performance liquid chromatography (UHPLC) system 1290 | Agilent Technologies | N/A |
| Quadrupole time of flight (Q-TOF 6545) mass spectrometer | Agilent Technologies | N/A |
| InfinityLab Poroshell 120 HILIC | Agilent Technologies | 675775-924 |
| ZORBAX RRHD Eclipse Plus C18 | Agilent Technologies | 959757-902 |
Resource availability
Lead contact
Requests for resources and additional information should be directed to and will be fulfilled by the Lead Contact, Shannon M. Mumenthaler (smumenth@usc.edu).
Material availability
This study did not generate new unique reagents.
Data and code availability
All relevant data are available from the Lead Contact upon request. The published article includes all datasets generated or analyzed during this study.
Experimental model and subject details
Cell culture
Commercially available cell lines
Human umbilical vein endothelial cells (HUVEC) expressing Red Fluorescent Protein (RFP) (Angio-Proteomie, #cAP-0001RFP) were expanded in EBM-2 media with EGM-2 SingleQuots Supplements (2% FBS, 1% Pen-Strep, Hydrocortisone, hFGF-B, VEGF, R3-IGF-1, Ascorbic Acid, hEGF, and heparin in proprietary concentrations) (Lonza #CC-3162; supplemented with 1% Penicillin-Streptomycin (Pen-Strep) in lieu of Gentamicin). Caco2 C2BBe1 cells (ATCC, #CRL-2102) were grown in DMEM (Gibco, #10569-010) with 10% fetal bovine serum (FBS) and 1% Pen-Strep. HCT116 and HT29 cells (ATCC #CCL-247 and #HTB-38) were grown in McCoy’s 5A media (Gibco, #16600-082) with 10% FBS and 1% Pen-Strep, labeled with LentiBrite Histone-H2B-GFP Lentiviral Biosensor (Millipore, #17-10229), and sorted to achieve a pure fluorescent population. CCD18Co (ATCC, #CRL-1459) cells were grown in Eagle’s Minimum Essential Medium (Corning, #10-009-CV) with 10% FBS and 1% Pen-Strep. All cells were cultured under standard laboratory conditions (5% CO2, 37°C).
Patient-derived samples
Tissue resections were received from the USC Norris Comprehensive Cancer Center following Institutional Review Board (IRB) approval (Protocol HS-06-00678; approval date 08-02-2019) and patient consent. Tumor profiles, including known tumor mutations, sex, and treatment information, are detailed in Table S10. Human primary fibroblasts and organoids were derived from CRC tumors via a previously described method (Sato et al., 2009, 2011). Briefly, tumor pieces were minced and enzymatically digested using 1.5 mg mL-1 collagenase (Millipore, #234155), 10 μM LY27632 (Millipore, #5.09228.0001) and 20 μg mL-1 hyaluronidase (MP Biomedicals, #0210074080) for 30 minutes at 37°C. The resulting cell mixture was either plated in basement membrane extract (BME; Cultrex, #3533-005-02) to derive organoids or on plastic tissue culture plates to select for fibroblasts. Organoids and fibroblasts were cultured in a defined colon media (ADMEM/F12 (Gibco, #12634-010), 10% FBS, 1% Pen-Strep, supplemented with 100 ng mL-1 Noggin (Tonbo, #21-7075-U500), 50 ng mL-1 epidermal growth factor (EGF) (Life Technologies, #PHG0313), 10 μM SB202190 (Sigma Aldrich, #S7067), 500 nM TGF-β RI Kinase Inhibitor IV (A83-01) (Millipore, #616454-2MG), 10 mM Nicotinamide (Sigma Aldrich, #N0636), 1 x B27 (Gibco, #17504-001), 1mM N-acetylcysteine (Sigma Aldrich, #A9165), 1 x N2 (Gibco, #17502-048), 1 x HEPES (Gibco, #15630-080), 1 x GlutaMax (Gibco, #35050-061)). After establishment and expansion of organoids, they were subsequently labeled with H2B-GFP lentivirus. Cells cultured on plastic tissue culture plates were confirmed to be cancer-associated fibroblasts (CAFs) based on morphology and gene expression signatures of vimentin (Vim), alpha smooth muscle actin (Acta2), and fibronectin (Fn1), as measured via RT-qPCR (Figure S8). Primary CAFs were used for experiments between passage 3 and passage 7.
Method details
RT-qPCR
To confirm the identity of CAFs and to interrogate epithelial or mesenchymal marker expression in invaded tumor cells from the CRC-on-Chips, we performed RT-qPCR analysis on cultured primary cells and cultured HCT116 collected from the chip effluent. Cellular RNA was extracted using RNAspin Mini RNA Isolation Kit (GE Healthcare; #25-0500-71) and cDNA was reverse transcribed using iScript Reverse Transcription Supermix (Bio-Rad, #1708841) following manufacturer’s instructions. The cDNA was then amplified using iScript SYBR Green Master (Bio-Rad; #1708880). The sequences for PCR primers are listed in Table S11. Results were normalized to GAPDH expression for all experiments.
CAF conditioned media
CAFs were seeded into a 6-well plate and allowed to grow until they reached 70% confluency at which point media was exchanged for fresh colon media. After 72 hours, the media was collected, centrifuged at 900 rpm (200 rcf) for 5 minutes, and the supernatant was removed and stored at -20°C. CAF CM was diluted 1:1 with DMEM, 10% FBS, 1% Pen-Strep for all experiments.
CAF secretome analysis
CAFs were seeded into a 6-well plate to collect CM as described above. When cells reached 70% confluency, one well of the 6-well plate was cultured in un-supplemented colon media (ADMEM/F12) without FBS or Pen-Strep for 72 hours. This media was collected, spun down to remove debris, aliquoted, and stored at -80°C. Frozen CM was thawed and analyzed using a cytokine array (R&D Systems, #ARY022B) following manufacturer’s instructions. Analyses of over- and under-expressed cytokines were performed in the R statistical environment (v4.0.2). Z-scores were generated by gene-wise scaling and were assessed for overlap at thresholds of 0.5. Visualization of overlaps was facilitated by the VennDiagram package (v1.6.20) (Chen and Boutros, 2011).
Tumor cell transwell invasion assay
Fluoroblok™ transwell inserts with PET membranes (6.5 mm membrane diameter, 8 μm membrane pore size, 0.3 cm2 cell culture area, Corning, #351152) were coated with 30 μg mL-1 type I collagen (Corning, #C354249) and 100 μg mL-1 Matrigel (Corning, #356231) for 2 hours at 37°C before the ECM was gently aspirated from the insert. For transwell experiments with endothelial cells, the inserts were inverted and seeded with HUVECs (2.1x105 cells in 35 μL; 7x105 cells cm-2) and incubated at 37°C for 2 hours. The inserts were flipped over, placed in wells with 500 μL endothelial media in the bottom chamber, and Caco2 C2BBe1 cells were then seeded on top (1.1x105 cells in 200 μL; 3.7x105 cells cm-2). CRC tumor cell lines (HCT116 H2B GFP or HT29 H2B GFP) were added (3.6x104 cells in 100μL; 1.2x105 cells cm-2) 48 hours later. Cells were incubated in the same cell culture media as organ-chip experiments in order to facilitate comparisons between experiments. Caco2 C2BBe1 and CRC tumor cells in the top chamber were maintained in DMEM with 10% FBS and 1% Pen-Strep. Fully supplemented EBM-2 media (2% FBS, 1% Pen-Strep, Hydrocortisone, hFGF-B, VEGF, R3-IGF-1, Ascorbic Acid, hEGF, and heparin in proprietary concentrations) was placed in the bottom of the wells. The transwells were imaged the day after tumor cell seeding (D0) and 48 hours later (D2) using the Perkin Elmer Operetta High Content Screening (HCS) platform. The number of GFP+ cells in the bottom chamber were quantified using Perkin Elmer Harmony software.
Tumor cell immunofluorescence in 2D
HCT116 H2B GFP or HT29 H2B GFP cells were seeded on 24-well cell culture plates and allowed to grow to 70% confluency. Cells were washed with PBS and fixed with 4% paraformaldehyde (Electron Microscopy Sciences, #15710), incubated for 15 minutes, and permeabilized with 1% saponin (Sigma Aldrich, #84510). Blocking buffer of 2% bovine serum albumin (BSA) (Millipore, #260-500GM) and primary antibodies were incubated overnight at 4°C before a 2-hour incubation with secondary antibodies (1:100, Molecular Probes, Invitrogen, #A21428 and #A21235) diluted in blocking buffer. The primary antibodies used for these studies were anti-E-cadherin (1:25; Abcam, #ab1416) and anti-vimentin (1:50; Abcam, #ab92547). Cells were imaged using the Perkin Elmer Operetta CLS High Content System. Quantitation of E-Cadherin+ and Vimentin+ tumor cells (identified by GFP) was performed using the Perkin Elmer Harmony software.
Microfluidic organ-chip design and culture
Chips were acquired from Emulate, Inc. The fabrication methods have been previously described (Huh et al., 2013). In brief, the chips are made of transparent elastomeric polymer (polydimethylsiloxane, PDMS). They are divided into upper (1 mm high x 1 mm wide) and lower (0.2 mm high x 1 mm wide) microfluidic compartments separated by a thin porous membrane (50 μm thick with 7 μm diameter pores; 17.1 mm2 co-culture region). The upper compartment hosts epithelial cells (plus additional cell types), and the lower compartment hosts the endothelial cells. Each compartment is coated with a tissue-specific ECM prior to cell seeding. The chips are attached to a Pod™ portable module that encloses the chips to control sterility, holds inlet cell culture media and effluent, allows for monitoring via microscopy, and is designed to ensure no pressure differentials between channels. The chips and pod are then housed in an automated culture module instrument (Zoë™ culture module and Orb™ hub module, Emulate, Inc.) that controls the fluid flow and stretching forces while inside an incubator.
Caco2 Intestine Chip
Methods for establishing the Intestine Chip cell culture have been previously described (Kim et al., 2012). Briefly, the chip PDMS membranes were activated by proprietary Emulate Reagents 1 and 2 (Emulate, Inc, ER-1 and ER-2) under UV light for 20 minutes. The epithelial and endothelial channels were coated with a mixture of 30 μg mL-1 type I collagen (Corning, #354249) and 100 μg mL-1 Matrigel (Corning, #356231) for 2 hours at 37°C before washing with PBS. RFP-labeled HUVEC cells were seeded into the bottom channel (1.2 x 105 cells in 20 μL; 7 x 105 cells cm-2). The chips were inverted and incubated at 37°C for 2 hours. The density of HUVEC cells was optimized to ensure the cells would self-assemble a tube along the channel. After HUVEC attachment, Caco2 C2BBe1 cells were seeded into the top channel (62,500 cells in 50 μL; 3.7 x 105 cells cm-2) and the chips were incubated overnight at 37°C. The chips were perfused with DMEM, 10% FBS, 1% Pen-Strep in the top channel and endothelial media (EBM-2 fully supplemented with 2% FBS, 1% Pen-Strep, Hydrocortisone, hFGF-B, VEGF, R3-IGF-1, Ascorbic Acid, hEGF, and heparin in proprietary concentrations) in the bottom channel at 30 μL hour -1 (0.02 dyne cm-2) starting the day after cell seeding. Cyclic, peristalsis-like membrane deformations (10% strain, 0.2 Hz) were also initiated the day after cell seeding using an electronic vacuum pump system (Emulate, Inc).
Caco2 + CRC cell lines
To modify the Intestine Chip to model CRC, we introduced CRC cells on top of the Caco2 C2BBe1 epithelial cell layer. Forty-eight hours after Caco2 cell seeding, CRC cells (HCT116 H2B-GFP or HT29 H2B-GFP) were seeded in the top channel (2x104 cells in 50 μL; 1.2x105 cells cm-2). The chips were incubated for 2 hours at 37°C after which 30 μL hour -1 (0.02 dyne cm-2) flow was re-established with DMEM, 10% FBS, 1% Pen-Strep in the top channel and endothelial media (EBM-2 fully supplemented with 2% FBS, 1% Pen-Strep, Hydrocortisone, hFGF-B, VEGF, R3-IGF-1, Ascorbic Acid, hEGF, and heparin in proprietary concentrations) in the bottom channel. Cyclic, peristalsis-like membrane deformations (10% strain, 0.2 Hz) were initiated the day after HCT116 seeding.
Caco2 + CRC cell lines + patient-derived fibroblasts
Fibroblasts were introduced to the epithelial channel based on methods developed by Emulate, Inc. Briefly, the top channel was coated with 200 μg mL-1 type IV collagen (Sigma Aldrich, #C5533) and the bottom channel was coated with 100 μg mL-1 Matrigel (Corning, #356231) and 30 μg mL-1 type I collagen (Corning, #354249). HUVECs were seeded on the bottom channel as described above. Fibroblasts were labeled with Cell Tracker Deep Red (1:1000; Invitrogen, #C34565) and seeded onto the top channel (7.5x104 cells in 50 μL), incubated for at least 4 hours, and then overlaid with a 100 μg mL-1 Matrigel (Corning, #356231) and 30 μg mL-1 type I collagen (Corning, #354249) layer and incubated for an additional 2 hours. The Caco2 C2BBe1 and HCT116 cells were then seeded as described above. Chips were perfused with 1:1 Colon media: DMEM with 10% FBS and 1% Pen-Strep in the top channel and endothelial media (EBM-2 fully supplemented with 2% FBS, 1% Pen-Strep, Hydrocortisone, hFGF-B, VEGF, R3-IGF-1, Ascorbic Acid, hEGF, and heparin in proprietary concentrations) in the bottom channel.
Patient-derived colon cancer organoids
The above CRC-on-Chip protocol was amended by introducing human colon tumor-derived organoids to the top channel. Briefly, the top channel was coated with 250 μg mL-1 Matrigel (Corning, #356231) and the bottom channel was coated with 100 μg mL-1 Matrigel (Corning, #356231) and 30 μg mL-1 type I collagen (Corning, #354249). RFP-labeled HUVECs were seeded on the bottom channel as described above. Organoids were incubated in Gentle Cell Dissociation Reagent (STEMCELL Technologies, #07174) on ice before enzymatically dissociated using 50% TrypLE (Gibco; 12605-028) supplemented with 10 mM LY-27632 (Millipore, #5.09228.0001) and seeded on the top channel as described previously (Kasendra et al., 2020). Chips were incubated overnight before starting flow and mechanical deformations as noted above. Chips were perfused with fully supplemented Colon media in the top channel and endothelial media (EBM-2 fully supplemented with 2% FBS, 1% Pen-Strep, Hydrocortisone, hFGF-B, VEGF, R3-IGF-1, Ascorbic Acid, hEGF, and heparin in proprietary concentrations) in the bottom channel.
On-chip permeability assay
The barrier function of the intestinal epithelium was measured by adding Inulin-Fluorescein isothiocyanate (FITC, 2-5kDa; Sigma Aldrich, #F3272) to the epithelial channel at the start of fluid flow and maintained for the duration of the experiment. The effluent from the bottom channel was collected every two days and the fluorescence was measured using a plate reader (Spectramax). Apparent permeability (Papp) was calculated using the following formula:
where Coutput is the concentration of Inulin-FITC in the endothelial outflow, A is the seeded area, and Cinput is the concentration of Inulin-FITC flowed into the epithelial channel. Duplicate or triplicate chips per condition were performed for a set of three separate experiments.
CRC-on-chip immunofluorescence
CRC Organ Chips were manually washed by flowing PBS through the endothelial and epithelial channels. The chips were fixed with 4% paraformaldehyde (Electron Microscopy Sciences, #15710), incubated for 15 minutes, and permeabilized with 1% saponin. Blocking buffer of 2% bovine serum albumin (BSA) and primary antibodies were incubated overnight at 4°C before a 2 hour incubation with secondary antibodies (1:100, Molecular Probes, Invitrogen, #A21428 and #A21235) diluted in blocking buffer. The primary antibodies used for these studies were anti-E-cadherin (1:25; Abcam, #ab1416), anti-VE-cadherin (1:25; Abcam, #ab33168), anti-vimentin (1:50; Abcam, #ab93547), anti-ZO-1 (1:100; Thermo-Fisher, #339194). DAPI (Sigma-Aldrich, D9542) was used to label all nuclei. Chips were imaged using the Perkin Elmer Operetta CLS High Content System. Quantitation of E-Cadherin+ and Vimentin+ (tumor cells identified by GFP) was performed using the Perkin Elmer Harmony software based on intensity thresholds.
Tumor cell invasion assay on-chip
CRC-Chips were prepared as described above. Tumor cell behavior was monitored via live cell imaging using a confocal laser-scanning microscope (Olympus FV3000). Imaging began the day after CRC cell seeding (day 0) and the same regions were imaged on subsequent imaging days. Images (10x magnification) were acquired at six separate regions across the epithelial channel (in the xy-direction), starting from the bottom of the endothelial channel up to the top of the epithelial channel (roughly 400 μm) at 5 μm increments (in the z-direction). Image z-stacks were imported into Imaris image analysis software for quantification of invasion. Images were manually divided into top (above the membrane) and bottom (below the membrane) channel regions. The Imaris cell detection algorithm was used to detect the number of GFP+ cells in each region. An invasion ratio was calculated based on the total number of GFP+ cells in the bottom channel divided by the total number of GFP+ cells in the top channel at specified time points. Raw invasion ratios were normalized to D0 to account for any outliers showing more invasion than typical on D0 (shown in Figure S3A).
Inlet (DMEM with 10% FBS and 1% Pen-Strep for the top channel and fully supplemented EBM-2 media (2% FBS, 1% Pen-Strep, Hydrocortisone, hFGF-B, VEGF, R3-IGF-1, Ascorbic Acid, hEGF, and heparin in proprietary concentrations) for the bottom channel) and effluent media was collected from the media reservoirs at the time of imaging and spun down at 900 rpm (200 rcf) for 5 minutes. The supernatant was collected for mass spectrometry-based metabolomic analysis (see below) and the pelleted cells were resuspended and plated into a 96-well plate. After cell attachment, the cells were stained with 5 μg mL-1 Hoechst 33342 (nuclear dye; Invitrogen, #H1399) and the cells were imaged via the Operetta HCS. The numbers of GFP+ nuclei were quantified via the Harmony software.
Mass spectrometry-based metabolomics of CRC-on-chip
Metabolite Extraction: 100 μL of each sample was extracted with 500 μL extraction solvent (80:20 Methanol:Water) spiked with 10 μL internal standard mix (Cambridge Isotopes Laboratories Inc. Metabolomics QC Kit, #MSK-QC2), vortexed briefly and sonicated for 1 min to precipitate the proteins. After storing for 60 min at −20°C, the samples were centrifuged at 13,000 x g for 30 min at 4°C. 450 μL of supernatant was transferred into a microcentrifuge tube and dried using vacuum centrifugation at room temperature (approximately 2 hours). The dried samples were resuspended in 100 μL resuspension solvent (50:50:0.1 (v/v) Water:Methanol:Formic Acid) with 10 μL internal standard (Cambridge Isotopes Inc. Metabolomics QC Kit, #MSK-QC1) spiked in and vortexed briefly to mix.
Liquid chromatography with tandem mass spectrometry (LC-MS/MS)
Extracted metabolite were analyzed using an ultra high performance liquid chromatography (UHPLC) system 1290 connected to a quadrupole time of flight (Q-TOF 6545) mass spectrometer from Agilent Technologies (Santa Clara, CA, USA) equipped with an orthogonal DUAL AJS-ESI interface. Samples were subjected to both hydrophilic interaction liquid chromatography (HILIC) separation (Agilent Poroshell-HILIC-p 2.1 X 100, 2.7μm; Agilent Technologies, #675775-924) and reverse phase C18 separation (Agilent Zorbax RRHD eclipse plus C18 column 2.1 X 50, 1.8μm; Agilent Technologies, #959757-902) and data were either collected in positive or negative ion mode. Data were acquired from 50 to 1250 m z-1 at 2 spectra s-1. Electrospray ionization (ESI) source conditions were set as follows: gas temperature 290°C, drying gas 9 L min-1, nebulizer 35 psi, fragmentor 125 V, sheath gas temperature 350°C, sheath gas flow 11 L min-1, nozzle voltage 1000V.
For HILIC chromatographic separation, a two-solvent gradient flowing at 0.5 mL min-1 (Mobile Phase A: 10mM Ammonium Acetate in 90:10 Water:Acetonitrile, pH 9.2, 5 μM Agilent InfinityLab Deactivator Additive (Agilent, #5191-4506) per vendor instruction, Mobile Phase B: 10mM Ammonium Acetate in 90:10 Acetonitrile:Water, pH 9.2, 5 μM Agilent InfinityLab Deactivator Additive per vendor instruction) was used. Column was equilibrated at 98% B for 3 minutes and a sample was introduced. The solvent ratio was then reduced from 95% B to 50% B over 10 minutes and then brought back up to 98% B over 5 minutes. For reverse phase C18 chromatographic separation, a two-solvent gradient running at 0.3 mL min-1 (Mobile Phase: A: 100:0.1 Water:Formic Acid, B: 100:0.1 Methanol:Formic Acid) was used. Column was equilibrated at 2% B for 1 minute and a sample was introduced. The solvent ratio was then increased from 2% B to 98% B over 15 minutes and then reduced back to 2% B over 4 minutes. Both HILIC and reverse phase C18 had injection volumes of 2 μL and column temperature of 30°C.
Metabolomics data analysis
The LC-MS/MS data acquired using Agilent Mass Hunter Workstation (“.d” files) were processed in Profinder (Agilent Technologies) for batch recursive extraction of features. Spectral peak extraction was performed with a minimum peak height of 500 counts and charge state of one. Further, retention time and mass alignment corrections were performed on the runs to remove non-reproducible signals. The resulting features were then exported as “.cef” files to Mass Profiler Professional (MPP) software (Agilent Technologies, Santa Clara, CA, USA) for multivariate analysis. Principal Component Analysis (PCA) was performed to check the quality of the samples and then the data containing filtered features were processed by unpaired t-test to find the difference between the groups. Only analytes with p values < 0.05 and fold change > 2 were regarded as statistically significant. Additionally, multiple test correction using Bonferroni was applied to reduce false positives and false negatives in the data. The statistically filtered data was then exported to Mass Hunter for targeted MS/MS analysis. All target metabolites were identified by matching their m/z values and retention times with in-house libraries (PCDL, Agilent technologies) of measured values for the IROA technologies MSMLS compound library. Metabolites that did not match to our internal library were matched against METLIN library or were listed with a predicted molecular formula.
Pathway analysis
To visualize the metabolites within relevant networks of pathways, the detected statistically significant metabolites were mapped to the curated pathways using IPA software (digitalinsights.qiagen.com).
Quantification and statistical analysis
Statistical analysis
Unless otherwise noted, all experiments were performed with 2-3 chips per condition and repeated independently at least 3 times. Analysis of variance (ANOVA) and t-tests were performed using GraphPad Prism 8 software, with the p-value < 0.0001: ∗∗∗∗; p-value < 0.001: ∗∗∗; p-value < 0.01: ∗∗; p-value < 0.05: ∗, as noted in all figure legends. Data is reported as mean with standard error of the mean (SEM) or as boxplots when noted, with boxplots designating the 25th-75th percentiles, median value, minimum value, and maximum value.
Acknowledgments
This research was supported by the NCI Tissue Engineering Consortium R01 CA241137-01A1 grant (SMM), a Stop Cancer Grant (SMM), and the USC Norris Comprehensive Cancer Center Core Grant P30CA014089 (HJL, SMM). We would like to express our deepest gratitude to our philanthropic supporters, particularly the Stephenson family, Emmett, Toni, and Tessa, for their donation of the Operetta HCS platform and the funding support as part of the Stephenson Family Personalized Medicine Center. We are extremely grateful for the expertise, guidance, and support from Emulate, Inc. We would also like to thank: S. Kim for help with image acquisition and organoid images; E. Fong for review of and comments on the manuscript; R. Hill, and D. Ruderman for meaningful discussions; and O. Castellanos, P. Gilmore, and G. Samuel for patient data assistance.
Author contributions
CS designed, performed, analyzed, and interpreted the experiments and wrote the manuscript. SC performed, analyzed, and interpreted mass spectrometry experiments. KG performed experiments, compiled data for figures in the manuscript, and edited the manuscript. RL and ES provided key resources and training on the Emulate platform and performed experiments. NU aided in the visualization of all data, particularly mass spectrometry data, and aided in discussions about experiment design and statistics. DH performed experiments. AYY performed and analyzed mass spectrometry experiments. RS aided in the visualization of cytokine array data. HJL provided clinical expertise. JEK provided conceptualization, experimental design support and interpretation of mass spectrometry data. SMM was responsible for study concept and design, interpretation of data, supervision of the study, and editing and revision of the manuscript. There was no outside writing assistance. All authors had access to the study data and reviewed, edited, and approved the final manuscript.
Declaration of interests
The authors declare no competing interest.
Published: May 21, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.102509.
Supplemental information
References
- Aleman J., Skardal A. A multi-site metastasis-on-a-chip microphysiological system for assessing metastatic preference of cancer cells. Biotechnol. Bioeng. 2019;116:936–944. doi: 10.1002/bit.26871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amos S.E., Choi Y.S. The cancer microenvironment: mechanical challenges of the metastatic cascade. Front. Bioeng. Biotechnol. 2021;9:625859. doi: 10.3389/fbioe.2021.625859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armistead F.J., Gala De Pablo J., Gadêlha H., Peyman S.A., Evans S.D. Physical biomarkers of disease progression: on-chip monitoring of changes in mechanobiology of colorectal cancer cells. Sci. Rep. 2020;10:3254. doi: 10.1038/s41598-020-59952-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai J., Tu T.Y., Kim C., Thiery J.P., Kamm R.D. Identification of drugs as single agents or in combination to prevent carcinoma dissemination in a microfluidic 3D environment. Oncotarget. 2015;6:36603–36614. doi: 10.18632/oncotarget.5464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basson M.D. Effects of repetitive deformation on intestinal epithelial cells. Inflammopharmacology. 2007;15:109–114. doi: 10.1007/s10787-007-1562-8. [DOI] [PubMed] [Google Scholar]
- Basson M.D., Yu C.F., Herden-Kirchoff O., Ellermeier M., Sanders M.A., Merrell R.C., Sumpio B.E. Effects of increased ambient pressure on colon cancer cell adhesion. J. Cell. Biochem. 2000;78:47–61. doi: 10.1002/(sici)1097-4644(20000701)78:1<47::aid-jcb5>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- Batista S., Gregorio A.C., Hanada Otake A., Couto N., Costa-Silva B. The gastrointestinal tumor microenvironment: an updated biological and clinical perspective. J. Oncol. 2019;2019:6240505. doi: 10.1155/2019/6240505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatia S.N., Ingber D.E. Microfluidic organs-on-chips. Nat. Biotechnol. 2014;32:760–772. doi: 10.1038/nbt.2989. [DOI] [PubMed] [Google Scholar]
- Brabletz T., Kalluri R., Nieto M.A., Weinberg R.A. EMT in cancer. Nat. Rev. Cancer. 2018;18:128–134. doi: 10.1038/nrc.2017.118. [DOI] [PubMed] [Google Scholar]
- Bürtin F., Mullins C.S., Linnebacher M. Mouse models of colorectal cancer: past, present and future perspectives. World J. Gastroenterol. 2020;26:1394–1426. doi: 10.3748/wjg.v26.i13.1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caballero D., Kaushik S., Correlo V.M., Oliveira J.M., Reis R.L., Kundu S.C. Organ-on-chip models of cancer metastasis for future personalized medicine: from chip to the patient. Biomaterials. 2017;149:98–115. doi: 10.1016/j.biomaterials.2017.10.005. [DOI] [PubMed] [Google Scholar]
- Carvalho M.R., Barata D., Teixeira L.M., Giselbrecht S., Reis R.L., Oliveira J.M., Truckenmuller R., Habibovic P. Colorectal tumor-on-a-chip system: a 3D tool for precision onco-nanomedicine. Sci. Adv. 2019;5:eaaw1317. doi: 10.1126/sciadv.aaw1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Boutros P.C. VennDiagram: a package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinformatics. 2011;12:35. doi: 10.1186/1471-2105-12-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H., Moon A. Crosstalk between cancer cells and endothelial cells: implications for tumor progression and intervention. Arch. Pharm. Res. 2018;41:711–724. doi: 10.1007/s12272-018-1051-1. [DOI] [PubMed] [Google Scholar]
- Ciasca G., Papi M., Minelli E., Palmieri V., De Spirito M. Changes in cellular mechanical properties during onset or progression of colorectal cancer. World J. Gastroenterol. 2016;22:7203–7214. doi: 10.3748/wjg.v22.i32.7203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker E., Tanis P.J., Vleugels J.L.A., Kasi P.M., Wallace M.B. Colorectal cancer. Lancet. 2019;394:1467–1480. doi: 10.1016/S0140-6736(19)32319-0. [DOI] [PubMed] [Google Scholar]
- Deryugina E.I., Kiosses W.B. Intratumoral cancer cell intravasation can occur independent of invasion into the adjacent stroma. Cell Rep. 2017;19:601–616. doi: 10.1016/j.celrep.2017.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driehuis E., van Hoeck A., Moore K., Kolders S., Francies H.E., Gulersonmez M.C., Stigter E.C.A., Burgering B., Geurts V., Gracanin A. Pancreatic cancer organoids recapitulate disease and allow personalized drug screening. Proc. Natl. Acad. Sci. U S A. 2019;116:26580–26590. doi: 10.1073/pnas.1911273116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drost J., Clevers H. Organoids in cancer research. Nat. Rev. Cancer. 2018;18:407–418. doi: 10.1038/s41568-018-0007-6. [DOI] [PubMed] [Google Scholar]
- Farshidfar F., Weljie A.M., Kopciuk K.A., Hilsden R., McGregor S.E., Buie W.D., MacLean A., Vogel H.J., Bathe O.F. A validated metabolomic signature for colorectal cancer: exploration of the clinical value of metabolomics. Br. J. Cancer. 2016;115:848–857. doi: 10.1038/bjc.2016.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faubert B., Solmonson A., DeBerardinis R.J. Metabolic reprogramming and cancer progression. Science. 2020;368:eaaw5473. doi: 10.1126/science.aaw5473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Follain G., Herrmann D., Harlepp S., Hyenne V., Osmani N., Warren S.C., Timpson P., Goetz J.G. Fluids and their mechanics in tumour transit: shaping metastasis. Nat. Rev. Cancer. 2020;20:107–124. doi: 10.1038/s41568-019-0221-x. [DOI] [PubMed] [Google Scholar]
- Follain G., Osmani N., Azevedo A.S., Allio G., Mercier L., Karreman M.A., Solecki G., Garcia Leon M.J., Lefebvre O., Fekonja N. Hemodynamic forces tune the arrest, adhesion, and extravasation of circulating tumor cells. Dev. Cell. 2018;45:33–52.e12. doi: 10.1016/j.devcel.2018.02.015. [DOI] [PubMed] [Google Scholar]
- Gaggioli C., Hooper S., Hidalgo-Carcedo C., Grosse R., Marshall J.F., Harrington K., Sahai E. Fibroblast-led collective invasion of carcinoma cells with differing roles for RhoGTPases in leading and following cells. Nat. Cell Biol. 2007;9:1392–1400. doi: 10.1038/ncb1658. [DOI] [PubMed] [Google Scholar]
- Gayer C.P., Basson M.D. The effects of mechanical forces on intestinal physiology and pathology. Cell. Signal. 2009;21:1237–1244. doi: 10.1016/j.cellsig.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geller L.T., Barzily-Rokni M., Danino T., Jonas O.H., Shental N., Nejman D., Gavert N., Zwang Y., Cooper Z.A., Shee K. Potential role of intratumor bacteria in mediating tumor resistance to the chemotherapeutic drug gemcitabine. Science. 2017;357:1156–1160. doi: 10.1126/science.aah5043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez H., Hagerling C., Werb Z. Roles of the immune system in cancer: from tumor initiation to metastatic progression. Genes Dev. 2018;32:1267–1284. doi: 10.1101/gad.314617.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould S.E., Junttila M.R., de Sauvage F.J. Translational value of mouse models in oncology drug development. Nat. Med. 2015;21:431–439. doi: 10.1038/nm.3853. [DOI] [PubMed] [Google Scholar]
- Grassart A., Malarde V., Gobaa S., Sartori-Rupp A., Kerns J., Karalis K., Marteyn B., Sansonetti P., Sauvonnet N. Bioengineered human organ-on-chip reveals intestinal microenvironment and mechanical forces impacting Shigella infection. Cell Host Microbe. 2019;26:435–444.e434. doi: 10.1016/j.chom.2019.08.007. [DOI] [PubMed] [Google Scholar]
- Hachey S.J., Movsesyan S., Nguyen Q.H., Burton-Sojo G., Tankazyan A., Wu J., Hoang T., Zhao D., Wang S., Hatch M.M. An in vitro vascularized micro-tumor model of human colorectal cancer recapitulates in vivo responses to standard-of-care therapy. Lab Chip. 2021;21:1333–1351. doi: 10.1039/d0lc01216e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hapach L.A., Mosier J.A., Wang W., Reinhart-King C.A. Engineered models to parse apart the metastatic cascade. NPJ Precis. Oncol. 2019;3:20. doi: 10.1038/s41698-019-0092-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassell B.A., Goyal G., Lee E., Sontheimer-Phelps A., Levy O., Chen C.S., Ingber D.E. Human organ chip models recapitulate orthotopic lung cancer growth, therapeutic responses, and tumor dormancy in vitro. Cell Rep. 2017;21:508–516. doi: 10.1016/j.celrep.2017.09.043. [DOI] [PubMed] [Google Scholar]
- Huang J.W., Pan H.J., Yao W.Y., Tsao Y.W., Liao W.Y., Wu C.W., Tung Y.C., Lee C.H. Interaction between lung cancer cell and myofibroblast influenced by cyclic tensile strain. Lab Chip. 2013;13:1114–1120. doi: 10.1039/c2lc41050h. [DOI] [PubMed] [Google Scholar]
- Huh D., Kim H.J., Fraser J.P., Shea D.E., Khan M., Bahinski A., Hamilton G.A., Ingber D.E. Microfabrication of human organs-on-chips. Nat. Protoc. 2013;8:2135–2157. doi: 10.1038/nprot.2013.137. [DOI] [PubMed] [Google Scholar]
- Humphries A., Wright N.A. Colonic crypt organization and tumorigenesis. Nat. Rev. Cancer. 2008;8:415–424. doi: 10.1038/nrc2392. [DOI] [PubMed] [Google Scholar]
- Jalili-Firoozinezhad S., Prantil-Baun R., Jiang A., Potla R., Mammoto T., Weaver J.C., Ferrante T.C., Kim H.J., Cabral J.M.S., Levy O. Modeling radiation injury-induced cell death and countermeasure drug responses in a human Gut-on-a-Chip. Cell Death Dis. 2018;9:223. doi: 10.1038/s41419-018-0304-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang K.J., Otieno M.A., Ronxhi J., Lim H.K., Ewart L., Kodella K.R., Petropolis D.B., Kulkarni G., Rubins J.E., Conegliano D. Reproducing human and cross-species drug toxicities using a Liver-Chip. Sci. Transl. Med. 2019;11:1–12. doi: 10.1126/scitranslmed.aax5516. [DOI] [PubMed] [Google Scholar]
- Karnoub A.E., Dash A.B., Vo A.P., Sullivan A., Brooks M.W., Bell G.W., Richardson A.L., Polyak K., Tubo R., Weinberg R.A. Mesenchymal stem cells within tumour stroma promote breast cancer metastasis. Nature. 2007;449:557–563. doi: 10.1038/nature06188. [DOI] [PubMed] [Google Scholar]
- Kasendra M., Luc R., Yin J., Manatakis D.V., Kulkarni G., Lucchesi C., Sliz J., Apostolou A., Sunuwar L., Obrigewitch J. Duodenum Intestine-Chip for preclinical drug assessment in a human relevant model. Elife. 2020;9:e50135. doi: 10.7554/eLife.50135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasendra M., Tovaglieri A., Sontheimer-Phelps A., Jalili-Firoozinezhad S., Bein A., Chalkiadaki A., Scholl W., Zhang C., Rickner H., Richmond C.A. Development of a primary human Small Intestine-on-a-Chip using biopsy-derived organoids. Sci. Rep. 2018;8:2871. doi: 10.1038/s41598-018-21201-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.J., Huh D., Hamilton G., Ingber D.E. Human gut-on-a-chip inhabited by microbial flora that experiences intestinal peristalsis-like motions and flow. Lab Chip. 2012;12:2165–2174. doi: 10.1039/c2lc40074j. [DOI] [PubMed] [Google Scholar]
- Kim H.J., Li H., Collins J.J., Ingber D.E. Contributions of microbiome and mechanical deformation to intestinal bacterial overgrowth and inflammation in a human gut-on-a-chip. Proc. Natl. Acad. Sci. U S A. 2016;113:E7–E15. doi: 10.1073/pnas.1522193112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labernadie A., Kato T., Brugues A., Serra-Picamal X., Derzsi S., Arwert E., Weston A., Gonzalez-Tarrago V., Elosegui-Artola A., Albertazzi L. A mechanically active heterotypic E-cadherin/N-cadherin adhesion enables fibroblasts to drive cancer cell invasion. Nat. Cell Biol. 2017;19:224–237. doi: 10.1038/ncb3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledford H. Translational research: 4 ways to fix the clinical trial. Nature. 2011;477:526–528. doi: 10.1038/477526a. [DOI] [PubMed] [Google Scholar]
- Lee S.W.L., Adriani G., Ceccarello E., Pavesi A., Tan A.T., Bertoletti A., Kamm R.D., Wong S.C. Characterizing the role of monocytes in T cell cancer immunotherapy using a 3D microfluidic model. Front. Immunol. 2018;9:416. doi: 10.3389/fimmu.2018.00416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak I.W., Evaniew N., Ghert M. Lost in translation: animal models and clinical trials in cancer treatment. Am. J. Transl. Res. 2014;6:114–118. [PMC free article] [PubMed] [Google Scholar]
- McAleer C.W., Long C.J., Elbrecht D., Sasserath T., Bridges L.R., Rumsey J.W., Martin C., Schnepper M., Wang Y., Schuler F. Multi-organ system for the evaluation of efficacy and off-target toxicity of anticancer therapeutics. Sci. Transl. Med. 2019;11:eaav1386. doi: 10.1126/scitranslmed.aav1386. [DOI] [PubMed] [Google Scholar]
- Nguyen D.T., Lee E., Alimperti S., Norgard R.J., Wong A., Lee J.J., Eyckmans J., Stanger B.Z., Chen C.S. A biomimetic pancreatic cancer on-chip reveals endothelial ablation via ALK7 signaling. Sci. Adv. 2019;5:eaav6789. doi: 10.1126/sciadv.aav6789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olejniczak A., Szarynska M., Kmiec Z. In vitro characterization of spheres derived from colorectal cancer cell lines. Int. J. Oncol. 2018;52:599–612. doi: 10.3892/ijo.2017.4206. [DOI] [PubMed] [Google Scholar]
- Ooft S.N., Weeber F., Dijkstra K.K., McLean C.M., Kaing S., van Werkhoven E., Schipper L., Hoes L., Vis D.J., van de Haar J. Patient-derived organoids can predict response to chemotherapy in metastatic colorectal cancer patients. Sci. Transl. Med. 2019;11:eaay2574. doi: 10.1126/scitranslmed.aay2574. [DOI] [PubMed] [Google Scholar]
- Orimo A., Gupta P.B., Sgroi D.C., Arenzana-Seisdedos F., Delaunay T., Naeem R., Carey V.J., Richardson A.L., Weinberg R.A. Stromal fibroblasts present in invasive human breast carcinomas promote tumor growth and angiogenesis through elevated SDF-1/CXCL12 secretion. Cell. 2005;121:335–348. doi: 10.1016/j.cell.2005.02.034. [DOI] [PubMed] [Google Scholar]
- Osmani N., Follain G., Garcia Leon M.J., Lefebvre O., Busnelli I., Larnicol A., Harlepp S., Goetz J.G. Metastatic tumor cells exploit their adhesion repertoire to counteract shear forces during intravascular arrest. Cell Rep. 2019;28:2491–2500.e2495. doi: 10.1016/j.celrep.2019.07.102. [DOI] [PubMed] [Google Scholar]
- Pavesi A., Tan A.T., Koh S., Chia A., Colombo M., Antonecchia E., Miccolis C., Ceccarello E., Adriani G., Raimondi M.T. A 3D microfluidic model for preclinical evaluation of TCR-engineered T cells against solid tumors. JCI Insight. 2017;2:e89762. doi: 10.1172/jci.insight.89762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson M.D., Mooseker M.S. Characterization of the enterocyte-like brush border cytoskeleton of the C2BBe clones of the human intestinal cell line, Caco-2. J. Cell Sci. 1992;102(Pt 3):581–600. doi: 10.1242/jcs.102.3.581. [DOI] [PubMed] [Google Scholar]
- Pino M.S., Kikuchi H., Zeng M., Herraiz M.T., Sperduti I., Berger D., Park D.Y., Iafrate A.J., Zukerberg L.R., Chung D.C. Epithelial to mesenchymal transition is impaired in colon cancer cells with microsatellite instability. Gastroenterology. 2010;138:1406–1417. doi: 10.1053/j.gastro.2009.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail D., Joyce J. Microenvironmental regulation of tumor progression and metastasis. Nat. Med. 2013;19:1423–1437. doi: 10.1038/nm.3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramzy G.M., Koessler T., Ducrey E., McKee T., Ris F., Buchs N., Rubbia-Brandt L., Dietrich P.Y., Nowak-Sliwinska P. Patient-derived in vitro models for drug discovery in colorectal carcinoma. Cancers (Basel) 2020;12:1423. doi: 10.3390/cancers12061423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahai E., Astsaturov I., Cukierman E., DeNardo D.G., Egeblad M., Evans R.M., Fearon D., Greten F.R., Hingorani S.R., Hunter T. A framework for advancing our understanding of cancer-associated fibroblasts. Nat. Rev. Cancer. 2020;20:174–186. doi: 10.1038/s41568-019-0238-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarvestani S.K., DeHaan R.K., Miller P.G., Bose S., Shen X., Shuler M.L., Huang E.H. A tissue engineering approach to metastatic colon cancer. iScience. 2020;23:101719. doi: 10.1016/j.isci.2020.101719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T., Stange D.E., Ferrante M., Vries R.G., Van Es J.H., Van den Brink S., Van Houdt W.J., Pronk A., Van Gorp J., Siersema P.D. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett's epithelium. Gastroenterology. 2011;141:1762–1772. doi: 10.1053/j.gastro.2011.07.050. [DOI] [PubMed] [Google Scholar]
- Sato T., Vries R.G., Snippert H.J., van de Wetering M., Barker N., Stange D.E., van Es J.H., Abo A., Kujala P., Peters P.J. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- Siegel R.L., Miller K.D., Fuchs H.E., Jemal A. Cancer statistics, 2021. CA Cancer J. Clin. 2021;71:7–33. doi: 10.3322/caac.21654. [DOI] [PubMed] [Google Scholar]
- Siegel R.L., Miller K.D., Goding Sauer A., Fedewa S.A., Butterly L.F., Anderson J.C., Cercek A., Smith R.A., Jemal A. Colorectal cancer statistics, 2020. CA Cancer J. Clin. 2020;70:145–164. doi: 10.3322/caac.21601. [DOI] [PubMed] [Google Scholar]
- Skardal A., Devarasetty M., Forsythe S., Atala A., Soker S. A reductionist metastasis-on-a-chip platform for in vitro tumor progression modeling and drug screening. Biotechnol. Bioeng. 2016;113:2020–2032. doi: 10.1002/bit.25950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sontheimer-Phelps A., Chou D.B., Tovaglieri A., Ferrante T.C., Duckworth T., Fadel C., Frismantas V., Sutherland A.D., Jalili-Firoozinezhad S., Kasendra M. Human colon-on-a-chip enables continuous in vitro analysis of colon mucus layer accumulation and physiology. Cell. Mol. Gastroenterol. Hepatol. 2020;9:507–526. doi: 10.1016/j.jcmgh.2019.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sontheimer-Phelps A., Hassell B.A., Ingber D.E. Modelling cancer in microfluidic human organs-on-chips. Nat. Rev. Cancer. 2019;19:65–81. doi: 10.1038/s41568-018-0104-6. [DOI] [PubMed] [Google Scholar]
- Tan B., Qiu Y., Zou X., Chen T., Xie G., Cheng Y., Dong T., Zhao L., Feng B., Hu X. Metabonomics identifies serum metabolite markers of colorectal cancer. J. Proteome Res. 2013;12:3000–3009. doi: 10.1021/pr400337b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thodeti C.K., Matthews B., Ravi A., Mammoto A., Ghosh K., Bracha A.L., Ingber D.E. TRPV4 channels mediate cyclic strain-induced endothelial cell reorientation through integrin-to-integrin signaling. Circ. Res. 2009;104:1123–1130. doi: 10.1161/CIRCRESAHA.108.192930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlachogiannis G., Hedayat S., Vatsiou A., Jamin Y., Fernandez-Mateos J., Khan K., Lampis A., Eason K., Huntingford I., Burke R. Patient-derived organoids model treatment response of metastatic gastrointestinal cancers. Science. 2018;359:920–926. doi: 10.1126/science.aao2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walrath J.C., Hawes J.J., Van Dyke T., Reilly K.M. Genetically engineered mouse models in cancer research. Adv. Cancer Res. 2010;106:113–164. doi: 10.1016/S0065-230X(10)06004-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyckoff J.B., Jones J.G., Condeelis J.S., Segall J.E. A critical step in metastasis: in vivo analysis of intravasation at the primary tumor. Cancer Res. 2000;60:2504–2511. [PubMed] [Google Scholar]
- Yamashiro Y., Yanagisawa H. The molecular mechanism of mechanotransduction in vascular homeostasis and disease. Clin. Sci. (Lond) 2020;134:2399–2418. doi: 10.1042/CS20190488. [DOI] [PubMed] [Google Scholar]
- Yao Y., Xu X., Yang L., Zhu J., Wan J., Shen L., Xia F., Fu G., Deng Y., Pan M. Patient-derived organoids predict chemoradiation responses of locally advanced rectal cancer. Cell Stem Cell. 2019;26:17–26.e6. doi: 10.1016/j.stem.2019.10.010. [DOI] [PubMed] [Google Scholar]
- Ying L., Zhu Z., Xu Z., He T., Li E., Guo Z., Liu F., Jiang C., Wang Q. Cancer associated fibroblast-derived hepatocyte growth factor inhibits the paclitaxel-induced apoptosis of lung cancer A549 cells by up-regulating the PI3K/akt and GRP78 signaling on a microfluidic platform. PLoS One. 2015;10:e0129593. doi: 10.1371/journal.pone.0129593. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All relevant data are available from the Lead Contact upon request. The published article includes all datasets generated or analyzed during this study.