SUMMARY
Topoisomerase II (TOP2) relieves torsional stress by forming transient cleavage complex intermediates (TOP2ccs) that contain TOP2-linked DNA breaks (DSBs). While TOP2ccs are normally reversible, they can be “trapped” by chemotherapeutic drugs such as etoposide and subsequently converted into irreversible TOP2-linked DSBs. Here, we have quantified etoposide-induced trapping of TOP2ccs, their conversion into irreversible TOP2-linked DSBs, and their processing during DNA repair genome-wide, as a function of time. We find that while TOP2 chromatin localization and trapping is independent of transcription, it requires pre-existing binding of cohesin to DNA. In contrast, the conversion of trapped TOP2ccs to irreversible DSBs during DNA repair is accelerated 2-fold at transcribed loci relative to non-transcribed loci. This conversion is dependent on proteasomal degradation and TDP2 phosphodiesterase activity. Quantitative modeling shows that only two features of pre-existing chromatin structure—namely, cohesin binding and transcriptional activity—can be used to predict the kinetics of TOP2-induced DSBs.
INTRODUCTION
TOP2 homodimers induce DNA double-strand breaks (DSBs) by cleaving the DNA phosphodiester backbone and forming protein-DNA adducts consisting of covalent phosphotyrosyl bonds of TOP2 monomers at the 5′ end of the breaks in both strands (Pommier et al., 2016). This structure, termed a TOP2 cleavage complex (TOP2cc), is normally short lived and reversible. After passing another molecule of DNA through the break site, TOP2 re-ligates the ends and subsequently dissociates from DNA without the assistance of additional enzymes. Thus, during a normal catalytic cycle, the transient DSB intermediate is shielded from the canonical DNA damage response. However, TOP2 poisons, such as the anti-cancer agent etoposide (ETO), rapidly stabilize and trap TOP2cc by inhibiting the re-ligation step (Nitiss, 2009). The trapped TOP2ccs can block the progression of RNA and DNA polymerases and in doing so can become abortive TOP2-linked DSBs that are cytotoxic and/or lead to genome instability (Cowell and Austin, 2012b).
Abortive TOP2ccs require removal by cellular DNA repair pathways. This involves, at least in part, hydrolytic removal of TOP2 peptide from the DNA break by tyrosyl-DNA phosphodiesterase 2 (TDP2) (Cortes Ledesma et al., 2009; Zagnoli-Vieira and Caldecott, 2017), thereby generating DSBs with 4-base cohesive overhangs that contain ligatable 5′-phosphate and 3′-hydroxyl termini (Zeng et al., 2011). Since they require no further processing, these protein-free ends are direct substrates for accurate non-homologous end joining (NHEJ) (Gómez-Herreros et al., 2013). TDP2 activity is frequently coupled with proteasomal degradation of TOP2, which exposes and enables access to the TOP2 phosphotyrosyl DNA bond, although proteasome-in-dependent TDP2 activity on unprocessed TOP2ccs has also been reported (Schellenberg et al., 2017).
TDP2 is critical for error-free repair of TOP2-induced breaks arising during transcription (Gómez-Herreros et al., 2017). For example, ETO treatment induces high levels of chromosomal aberrations and translocations in TDP2-deficient cells in a manner that is dependent on transcriptional activity (Gómez-Herreros et al., 2017). DSBs and translocations that arise in the absence of TDP2 are most likely mediated by a mutagenic mechanism of DSB repair that employs endonucleases such as Mre11, which can also remove covalently linked TOP2 from DSBs but which in doing so also remove nucleotides from the DSB termini (Hoa et al., 2016).
In a recent genome-wide study that mapped TOP2 activity following treatment with the TOP2 poison ETO, DNA lesions were found at approximately 25,000 sites (Canela et al., 2017). Most of these were also enriched at Hi-C-defined chromosomal loop anchors demarked by the architectural protein CTCF and the cohesin complex (Canela et al., 2017; Uusküla-Reimand et al., 2016). The repertoire of TOP2 lesions include breakpoint-cluster regions whose rearrangements lead to fusion genes common in human cancer (Canela et al., 2017; Gómez-Herreros et al., 2017). For example, TOP2 colocalized with CTCF/RAD21 binding at recurrent translocation hotspots found in ETO therapy-related acute myeloid leukemia (t-AML) (Ashour et al., 2015; Cowell et al., 2012; Gómez-Herreros et al., 2017).
Translocated genes found in t-AML are frequently transcribed within the same “transcription factory” (Cowell and Austin, 2012a) and can induce AML only from hematopoietic cells or progenitor subsets in which they are highly transcribed (Cowell et al., 2012; Krivtsov et al., 2013). While TOP2 activity and transcription are believed to be critical for translocation in t-AML (Cowell et al., 2012; Gómez-Herreros et al., 2017; Krivtsov et al., 2013), transcription levels do not correlate strongly with TOP2 activity. For example, transcription-dependent etoposide-induced trapping of TOP2ccs occurs only in approximately 1,250 (approximately 5%) of the most transcriptionally active sites (Canela et al., 2017). Moreover, the establishment of architectural loop domains mediated by CTCF and cohesin does not require ongoing transcription (Vian et al., 2018). Thus, the mechanism by which TDP2-dependent NHEJ protects transcriptionally active loci from TOP2-induced DSBs and chromosome aberrations (e.g., translocations) remains unclear (Gómez-Herreros et al., 2017).
END-seq was utilized to comprehensively capture both TOP2ccs and protein-free DSBs and provided a genome-wide representation of all sites of TOP2 action (Canela et al., 2017). Here, we develop a refinement of the original strategy that allows for the discrimination between unprocessed and processed TOP2-induced DSBs. This method allows for the quantification of the probability at which trapped TOP2ccs form at given sites in the genome, the rate of TOP2cc conversion into protein-free DSBs, and the frequency of chromosomal translocations. Based on quantitative modeling, we show that the kinetics of DSB formation can be predicted by only two independent cellular parameters: loop extrusion, which induces TOP2 trapping, and transcription that promotes TOP2cc processing.
RESULTS
Cohesin Is Essential for TOP2 Activity
TOP2 activity is enriched at boundaries of loop domains, usually coincident with the binding of cohesin and CTCF (Canela et al., 2017). To determine how cohesin affects the distribution of TOP2 lesions, we examined mouse embryonic fibroblasts (MEFs) in which SMC3, a cohesin subunit, was depleted by CRE-mediated recombination, resulting in the disruption of nuclear architecture (Busslingeret al., 2017) (Smc3f/f+CRE; Figures 1A–1D). We then mapped sites of TOP2 DNA cleavage by performing END-seq on SMC3-depleted and control MEFs (Smc3+/+) cells after 30 min of ETO treatment. ChlP-seq experiments with antibodies to the cohesin subunit, RAD21, confirmed the depletion of cohesin genome-wide and specifically at CTCF sites, demarcating chromosomal loop anchors (Figures 1B and 1C). As we previously reported in mouse B cells (Canela et al., 2017), TOP2-mediated cleavage in control MEFs colocalized with sites of cohesin binding (Figures 1A and S1A). Upon SMC3 depletion, the END-seq signal dropped precipitously genome-wide (Figures 1A–1D). This drop in END-seq signal was proportional to the decrease in cohesin levels (Figure 1B; 27% and 26% reduction in RAD21 and END-seq levels, respectively). We confirmed these results in human HCT-116 cell lines, in which RAD21 was depleted using an auxin-inducible degron (AID) system (Natsume et al., 2016) (Figures S1B–S1D). Similar to SMC3 depletion in MEFs, RAD21 depletion resulted in a precipitous drop in END-seq signal in HCT-116 genome-wide (Figures S1B–S1D).
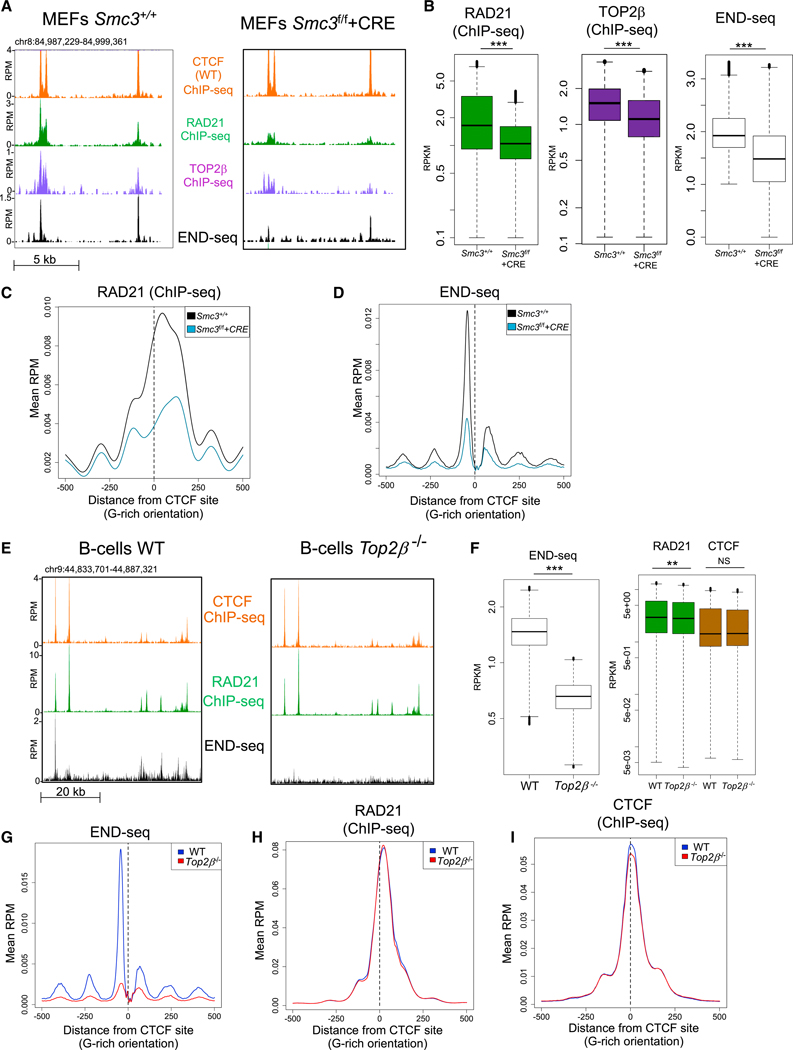
Figure 1. TOP2 Binding and Cleavage Complex (TOP2cc) Formation Is Dependent on Cohesin.
(A) Representative genomic profiles (top to bottom): CTCF, RAD21, TOP2B ChlP-seq, and END-seq. Both CTCF ChlP-seq tracks correspond to Smc3+/+ MEFs. RAD21, TOP2B, and END-seq profiles correspond to Smc3+/+ MEFs (left), and Smc3f/f MEFs in which SMC3 was depleted by CRE-mediated recombination (Smc3f/f +CRE; right). END-seq was performed after 30 min of ETO treatment.
(B) Quantification of the levels of RAD21 and TOP2B binding without ETO, and END-seq upon ETO in Smc3+/+ and Smc3f/f +CRE MEFs. Peaks sets were derived from END-seq signals in Smc3+/+ data. ***p < 1e–20; t test.
(C) Aggregate plot of RAD21 binding in Smc3+/+ (black line) and Smc3f/f +CRE (blue line) MEFs at the double CTCF/RAD21-bound sites ± 500 bp from the CTCF motif (dashed line).
(D) Aggregate plot of END-seq signal upon ETO in Smc3+/+ (black line) and Smc3f/f +CRE (blue line) MEFs at CTCF/RAD21-bound sites ± 500 bp from the CTCF motif (dashed line).
(E) Representative genomic profiles (top to bottom): ChlP-seq for CTCF, ChlP-seq for RAD21, and END-seq in WT (left) and in Top2β−/− (right) B cells. ChlP-seq was performed without ETO treatment.
(F) Left: quantification of the END-seq signal at END-seq peaks in WT and in Top2β−/− B cells. Right: quantification CTCF and RAD21 binding at END-seq peaks in WT and in Top2β−/− B cells. **p < 1e–8; ***p < 1e–30; NS: p > 0.4, t test.
(G-l) Aggregate plots of END-seq signal upon ETO treatment (G), RAD21 binding (H), and CTCF binding (l), in WT (blue line) and Top2β−/− (black line) B cells at CTCF/RAD21-bound sites ± 500 bp from the CTCF motif (dashed line).
The level of chromatin-associated cohesin is balanced by its loading by NIPBL and its removal by WAPL (Tedeschi et al., 2013). To test how elevation of chromatin-bound cohesin impacts TOP2cc formation, we prevented cohesin removal by depleting WAPL from MEFs. In WAPL-deficient cells, chromatin loops are enlarged because of the increased residence time of cohesin on DNA (Haarhuis et al., 2017; Wutz et al., 2017), and we observed a concomitant increase in ETO-induced damage genome-wide (Figures S1E–S1G). Having shown that TOP2 activity at a given site is dependent on and proportional to binding of cohesin, we then wished to determine whether, conversely, cohesin binding was codependent on TOP2. For this purpose, we generated B cells deficient for TOP2B by bone marrow reconstitution with hematopoietic cells from Top2β−/− embryos (Figure 1E). ln G1 cells, in which TOP2B is the major TOP2 isoform (Canela et al., 2017), END-seq signal dropped to background levels in Top2β−/− B cells (Figures 1E and 1F). However, the two genotypes had only minor change in the levels of RAD21, and CTCF levels did not change (Figures 1E–1I). Therefore, TOP2 activity is dependent on cohesin, but not vice versa.
To determine whether cohesin is required for TOP2B occupancy as well as its activity, we performed TOP2B ChlP-seq in WT, SMC3-, and WAPL-deficient cells in the absence of ETO treatment. We found that TOP2B ChlP-seq mirrored its catalytic engagement: TOP2B binding, like TOP2B activity, decreased genome-wide upon depletion of SMC3 (Figures 1A and 1B) and increased upon removal of WAPL (Figure S1E). Thus, both TOP2B binding and activity are downstream of cohesin.
Etoposide-Induced Chromosomal Translocations Are Transcription Dependent
Cohesin-mediated loop extrusion and associated TOP2 activity occur largely independently of transcription (Canela et al., 2017; Vian et al., 2018). Given that elevated TOP2-induced translocations in TDP2-defective cells are transcription dependent (Gomez-Herreros et al., 2017), we wished to explore further the relationship between TOP2-cleavage and translocation. To do so, we performed high-throughput genome-wide translocation sequencing (HTGTS), a method which captures “prey” DSBs in the genome by their ability to translocate to a “bait” DSB, generated at a defined location (Frock et al., 2015). To generate a bait DSB (Figure 2A), we expressed a zinc finger nuclease (ZFN) targeting the T cell receptor locus (Tcrβ enhancer on chromosome 6) in Abelson-transformed pre-B cell lines active for V(D)J recombination (Canela et al., 2016; Dorsett et al., 2014).
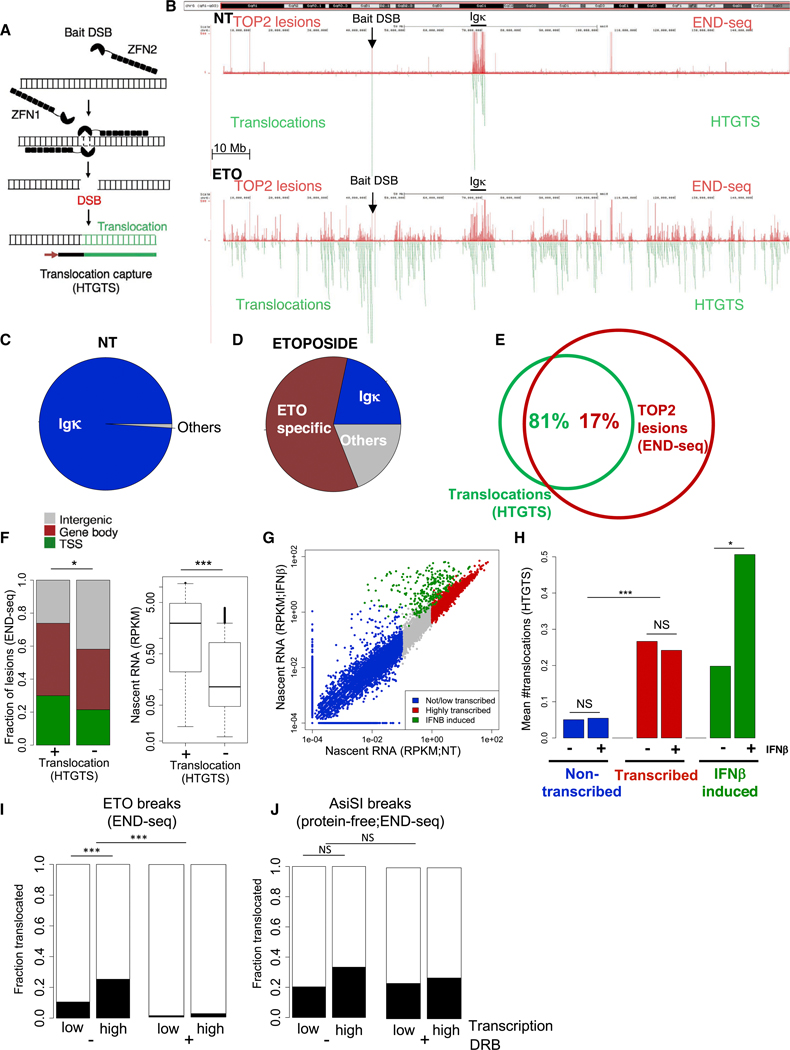
Figure 2. Transcription Promotes Chromosomal Translocations of TOP2cc Sites.
(A) Schematic representation of translocation sequencing by HTGTS. lnduction of a ZFN “bait” break is shown to be translocated to (“prey”) sequences (green). HTGTS uses a primer labeled with biotin to extend from one junction of the bait break to copy the translocation as described (Frock et al., 2015).
(B) Representative genomic profile of chromosome 6 comparing TOP2 lesions, detected by END-seq signal (top, red), and translocations (bottom, green), detected by HTGTS, in pre-B cells not treated (NT, upper) and treated with ETO for 2.5 h (lower). HTGTS was performed 12 h after ETO washout, while END-seq was performed immediately after ETO. Bait DSB position and Igκ locus are indicated. 9 million reads were sequenced for each HTGTS sample.
(C) Pie chart representation of translocations in cis to the bait (chr6) in absence of ETO treatment (NT).
(D) Pie chart representation of translocations in cis to the bait (chr6) upon ETO treatment.
(E) Venn diagram showing the overlap between translocations in cis to the bait (chr6) (green) and TOP2 lesions detected by END-seq upon ETO treatment (chr6) (red). *p < 1e−10, hypergeometric test.
(F) Comparison of the genomic location (left) and nascent RNA levels (right) of TOP2 lesions in chromosome 6 between the lesions that overlap with translocations and those that do not. *p < 1e–4, fisher-test. ***p < 1e–20, t test.
(G) Dotplot comparing of gene transcription levels between NT and lFNβ treatment. Colors represent level of transcription and dependency of lFNβ. *p < 0.01;***p < 1e–35; Wilcoxon test.
(H) Barplot comparing the mean number of translocations detected in the presence or absence of lNFβ in genes that are non-transcribed, transcribed, and lNFβ induced. 6 million reads were sequenced for each sample.
(I) Left: comparison of that fraction of TOP2 lesions that translocate in sites that exhibit low transcription (RPKM < 0.1) versus highly transcribed (RPKM > 1) sites. Right: same comparison upon transcription inhibition with DRB. 6 million reads were sequenced for each sample.
(J) Left: comparison of the fraction of AsiSl-induced protein-free DSBs that translocate to sites that exhibit low transcription (RPKM < 0.1) versus high transcription (RPKM > 1). Right: same comparison upon transcription inhibition with DRB. ***p < 1e–10; NS:p > 0.08. Transcription determined by nascent RNA prior to DRB treatment. 6 million reads were sequenced for each HTGTS sample.
Because canonical or classical NHEJ suppresses chromosome rearrangements (Difilippantonio et al., 2000; Ferguson et al., 2000), we speculated that LlG4 deficiency might increase the levels of DSB processing and translocation. Thus, Lig4−/− cells expressing the ZFN were either non-treated (NT) or treated with ETO for 2.5 h, following which translocations were assessed 12 h after ETO washout (see STAR Methods for details). Since the fusion of two DSBs requires physical juxtaposition, fusions arising in cis with the “bait” DSB were always highly overrepresented in the HTGTS profile (Frock et al., 2015), with 85% of junctions concentrated on chromosome 6 in the NT sample (Figure S2A).
Altogether we found 505 translocations on chromosome 6 in the NT sample, 99% of which were fused to the immunoglobulin kappa (lgκ) locus (Figures 2B, upper panel, and 2C). lgκ is a target of RAG-induced DSBs, which are readily detected by END-seq in pre-B cells undergoing V(D)J recombination (Figure 2B, upper panel) (Canela et al., 2016). Upon ETO treatment, both TOP2 lesions and translocations increased significantly relative to NT samples (2,322 cis translocations; Figure 2B, bottom panel). Peak calling of ETO-induced END-seq peaks, performed relative to the NT sample, revealed approximately 23,000 TOP2 lesions. Although there were considerably more TOP2 lesions (assessed 2.5 h after ETO) than translocations (measured after 12 h), there was a significant overlap between the two (Figures 2D and 2E). Thus, translocations occur largely at sites at which ETO traps TOP2.
TOP2 lesions that exhibited translocation (17% of the total TOP2 lesions; Figure 2E) were more enriched in transcription start sites (TSSs) and gene bodies compared to TOP2 lesions that were not translocated (73% and 58%, respectively; Figure 2F). Moreover, nascent RNA levels at sites associated with TOP2 translocations were higher on average compared to non-translocated sites (Figure 2F, right panel). Highly transcribed genes also translocated more frequently than genes with low or no transcription (Figures 2G and 2H). To directly assess the contribution of transcription to chromosomal translocation, we treated the cells with interferon β (lFNβ) prior to ETO treatment, resulting in the transcriptional induction of approximately 400 genes (Figures 2G and 2H, green). Notably, lFNβ treatment resulted in elevated translocation of these genes, suggesting that transcription facilitates translocation of TOP2-induced DNA breaks.
To test whether transcription is required for TOP2-induced chromosome translocation, we inhibited transcription with 5,6-dichloro-1-beta-D-ribofuranosylbenzimidazole (DRB) prior to ETO treatment (Figure S2B). Without DRB treatment, we observed translocations in 11% and 25% of ETO lesions with low and high levels of transcription, respectively; however, DRB pretreatment abolished translocation of ETO-induced DSBs (Figure 2l; 1% and 2% for ETO lesions with low and high levels of transcription, respectively).
ETO-induced translocations require removal of TOP2 from DNA to generate protein-free DSBs, primarily via TDP2-independent mechanisms involving nucleases (Gómez-Herreros et al., 2017). To test how transcription impacts translocation of protein-free DNA ends, we generated translocations with the restriction enzyme AsiSl, which produces “clean” DSBs at 233 sites in the pre-B cell genome (Canela et al., 2016; lacovoni et al., 2010). AsiSl-induced DSBs also translocated more at transcriptionally active than inactive broken loci (20% and 34% of low and high levels of transcription, respectively), although the differences were not statistically significant (Figure 2J). Moreover, DRB treatment did not decrease translocation of AsiSl-DSBs significantly (Figure 2J). Thus, transcription more profoundly impacts translocations that arise from TOP2ccs compared to those originating from protein-free DNA ends.
Genome-wide Frequency of TOP2-Induced Cleavage Complexes and Protein-free Breaks
To explore how transcription promotes TOP2-mediated translocation, we designed an approach to distinguish between TOP2-induced DSBs before and after TOP2 has been removed from the break. ln our recently developed method END-seq, which maps DSBs at nucleotide resolution (Canela et al., 2016), we ligate a sequencing adaptor to each end of the DNA break inside agarose plugs. lt is important to note that we routinely use, during END-seq, a combination of exonucleases ExoVll and ExoT to ensure that all DSB termini are free of protein so that they can be ligated to sequencing adaptors (Canela et al., 2016, 2017) (Figure 3A). For example, for natural protein-free DSBs such as those induced by RAG endonuclease or AsiSl, ExoT alone is sufficient for DSB detection (Canela et al., 2016). This is illustrated here by the observation that “clean” DSBs induced by AsiSl or RAG are equally detected using either ExoT+ExoVll or ExoT alone (Figure S3A). ln contrast, ETO-induced DSBs are covalently attached to TOP2 via an active site tyrosine at the 5′ termini, requiring the activity of ExoVll in the END-seq protocol to generate ligatable ends (Figures S3B and S3C) (Gao et al., 2012).
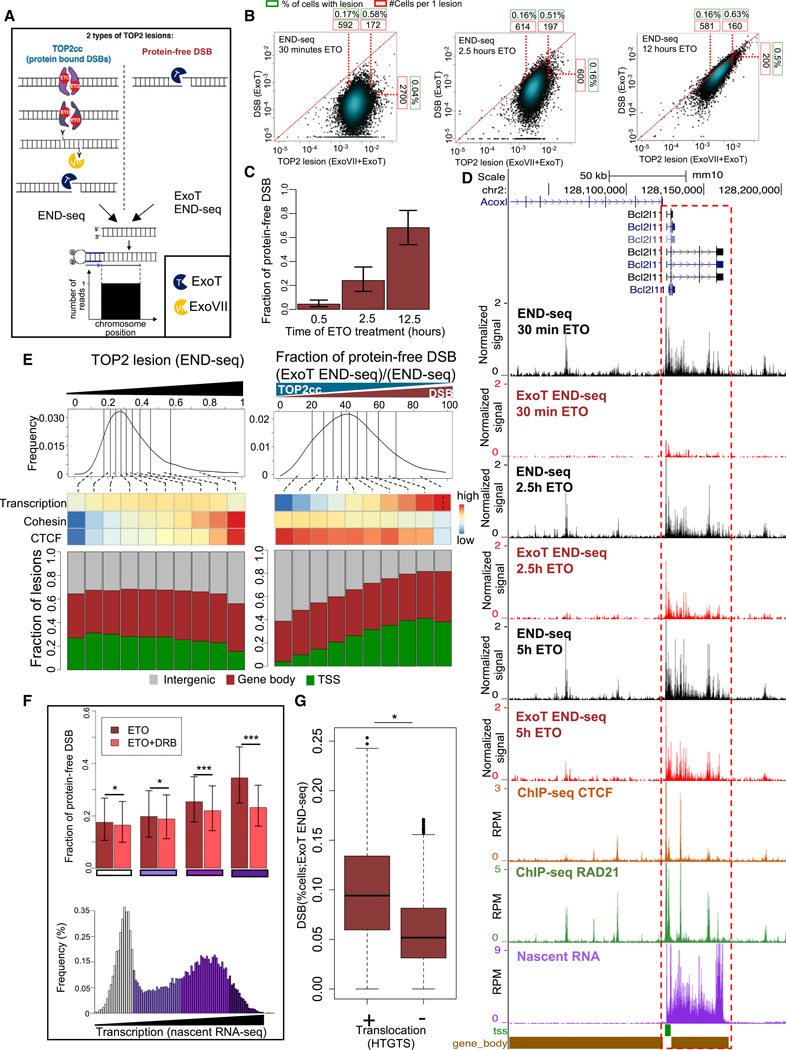
Figure 3. Genome-wide Quantification of the Processing of TOP2cc to Protein-free DSBs Reveals the Facilitating Role of Transcription.
(A) Schematic representation of the different species of TOP2 lesions (left from top to bottom: intact TOP2cc, partially degraded TOP2, and completely degraded TOP2; right: protein-free DSB) and their detection by END-seq. “END-seq” utilizes ExoT+ExoVll for detection of total TOP2 lesions, whereas “ExoT END-seq” utilizes ExoT alone for detection of protein-free DSBs.
(B) Scatterplots that show the levels of END-seq signal obtained by utilizing ExoT+ExoVll (xaxis) or ExoT alone (y axis), representing total TOP2 lesion levels and the protein-free DSB fraction, respectively, for each TOP2cc site, after (left to right) 30 min, 2.5 h, and 12.5 h of ETO treatment. Number of cells per single lesion (red boxes) and % cells per lesion (green boxes) are represented for the 10th and 90th percentile of TOP2 lesions and the 90th percentile of DSBs.
(C) The ratio of protein-free DSB to total TOP2 lesions (i.e., “Fraction of protein-free DSB”) for the three time-points of ETO treatment illustrated in (B). Error bars represent the distribution of this fraction over different break sites.
(D) Representative genomic profile ofTOP2 lesions (detected by END-seq using ExoVll+ExoT) and DSBs (detected by END-seq using ExoT alone) after 30 min, 2.5 h, and 5 h ETO; binding ofCTCF, cohesin (RAD21) (detected by ChlP-seq), and nascent RNA-seq in the absence of ETO. Gene body (brown bars) and TSSs (green bars) are indicated. Values in DSB panels are normalized by the corresponding spike-in (see STAR Methods).
(E) Top: TOP2 lesions were divided into deciles with respect to their level ofTOP2 trapping (left), measured at 30 min post ETO treatment, or processing levels (right) (histogram; top row), measured at 2.5 h post ETO treatment. Middle: the average levels of(top to bottom) transcription, cohesin (RAD21), and CTCF binding were plotted as a heatmap for each decile. Correlation with TOP2 trapping: ρ = 0.98, 1, and 0 for CTCF, cohesin and transcription respectively. Correlation with TOP2 processing: ρ = −0.91, −0.98, and 0.51, for CTCF, cohesin, and transcription respectively. Spearman Correlation Coefficient between deciles. Bottom: The distribution of genomic localization (TSSs, gene body, and intergenic) is shown for each decile.
(F) Bottom: histogram of transcription levels. Colors represent the different quartiles, from low and non-transcribed (white) to highly transcribed (dark purple).Top: the average fraction of protein-free DSB was measured for each quartile presented in the histogram below, with or without treatment with transcription inhibitor DRB (red and salmon, respectively). Error bars represent the distribution of this fraction in each quartile over different break sites. *p < 1e–4;***p < 1e–20, t test.
(G) Boxplots show the distribution of DSB levels in translocated and non-translocated TOP2 lesions (represented as % cells). ***p < 1e–74; t test.
Having established an assay to distinguish between unprocessed and processed TOP2-induced DSBs, we compared the rate of processing in transcriptionally active versus inactive loci. To promote the conversion of trapped TOP2ccs into DSBs, we incubated cells with ETO for up to 12.5 h in G1-ar-rested Lig4−/− cells and then measured total ETO-induced TOP2-lesions (trapped TOP2ccs and protein-free DSBs) using a combination of ExoVll+ExoT and the subset of TOP2-induced lesions that are protein-free DSBs using ExoT alone (Figure 3B). We then compared the levels of TOP2 lesions and protein-free DSB over time at the 23,000 TOP2 lesions. To estimate the frequency of cells harboring such lesions, we added to each sample a spike-in control consisting of Lig4−/− cells carrying a single ZFN-induced DSB at the TCRb enhancer (Figure S3D). This site is expected to break in all spike-in cells, which were mixed in at a 2% frequency, allowing us to normalize each of the 23,000 TOP2 lesion peaks using the signal at the TCRβ enhancer (Figure S3D).
Based on this approach, we found that the fraction of Lig4−/− cells with a TOP2 lesion at a given site (measured by ExoT+ ExoVll) remained constant with time (Figure S3E), ranging from 0.17%−0.58% depending on genome location (Figure 3B). ln contrast, protein-free DSBs (detected by ExoT alone) increased with time at all genomic locations (Figure 3B) and comprised 2%−7% of total TOP2 lesions after 30 min of ETO treatment (Figure 3C). This corresponds to the upper value of approximately one protein-free DSB per 2,700 cells (Figure 3B). After 2.5 and 12.5 h of ETO treatment, the fraction of protein-free DSBs was 18%−37% and 55%−82% (Figure 3C), corresponding to the upper values to approximately one and three protein-free DSBs per 600 cells, respectively (Figure 3B). Thus, while both the total level of TOP2 lesions and protein-free DSBs vary across the genome, only the latter accumulates in Lig4−/− cells during prolonged incubation with ETO.
Transcription Increases the Rate of TOP2 Processing
We noticed that the initial level of TOP2cc (detected by ExoVll+ ExoT at 30 min) and the levels of DSBs emerging at early time points did not correlate (Figures 3B and S3F), suggesting that trapped TOP2ccs and their subsequent conversion to DSBs are governed by distinct genomic features. ln agreement with the dependence of TOP2 activity on chromatin-bound cohesin (Figure 1), TOP2 trapping measured at 30 min post-ETO-treatment positively correlated with the binding of cohesin and CTCF (Figures 3D and 3E). ln contrast, genomic regions with the greatest levels of protein-free DSBs were enriched in TSSs, depleted of intergenic regions, and correlated with nascent transcript levels (Figures 3D and 3E). Notably, the negative correlation between protein-free DSBs and CTCF/cohesin occupancy stems from the fact that loop anchors frequently reside within intergenic regions (Ong and Corces, 2014) and hence show low transcription (Figures 3D and S3G). The absolute level of DSBs, being a function of both the initial TOP2 lesion and its processing rate, correlates with CTCF/cohesin binding, as well as with transcription (Figure S3H).
To quantify the impact of transcription on the conversion of trapped TOP2ccs into protein-free DSBs, we grouped genomic locations into quartiles with respect to their transcriptional activity (Figure 3F). For the lowest transcription quartile, approximately 19% of the TOP2 lesions were converted into protein-free DSBs within 2.5 h, whereas in the highest transcription quartile, 35% of these lesions were converted (Figure 3F). ln contrast, there was no increase in TOP2ccs conversion to protein-free DSBs with increased levels of CTCF (Figure S3l) or cohesin (Figure S3J) chromatin binding. Consistent with a causal relationship between transcription and TOP2ccs processing, blocking transcription with DRB 2 h prior to ETO treatment suppressed the enrichment of protein-free DSBs in highly transcribed loci (Figure 3F). Conversely, induction of transcription by lFNβ further increased levels of protein-free DSBs (Figure S3K). Futhermore, we found that translocated regions were associated, on average, with 1.5 times more DSBs than the non-translocated lesions (8.2×10−4 cell−1 and 5.6× 10−4 cell−1, respectively; Figure 3G). Taken together, these results support the model in which transcription increases the rate at which trapped TOP2ccs are converted into protein-free DSBs, which in turn promotes translocation.
Reversible and Irreversible TOP2ccs across the Genome
Since trapped TOP2ccs are intrinsically reversible upon removal of ETO (Long et al., 1985), we reasoned that transcription might have an impact on this reversibility. By measuring levels of TOP2ccs (defined as the difference between total TOP2 lesions detected by ExoVll+ExoT and protein-free DSBs detected by ExoT alone) before and immediately after ETO washout, we defined the fraction of TOP2ccs that are “reversible” and “irreversible” (Figure 4A; light blue versus dark blue areas, respectively). After 2.5 h of ETO treatment, the fraction of total END-seq lesions that were TOP2ccs in Lig4−/− cells was 60% on average (Figure 4A; sum of light blue and dark blue areas), with the remainder being protein-free DSBs. Strikingly, when ETO was washed away and then the cells were immediately processed for END-seq, approximately 88% of the TOP2cc had reversed and so were not detectable by END-seq (Figure 4A; light blue area). However, the level of protein-free DSBs remained the same (Figures 4A, red area, and 4B). lndeed, it is noteworthy that approximately 80% of the TOP2 lesions that had not reversed immediately following ETO washout were already present as protein-free DSBs, indicating that the rate at which irreversible TOP2ccs are converted to a protein-free DSB is rapid (Figures 4A, 4B [ETO peaks], S4A [CTCF sites and TSSs], and S4B).
Figure 4. Genome-wide Quantification of Reversible and Irreversible TOP2cc.
(A) Schematic representation of the different TOP2 lesions. Unprocessed TOP2ccs that disappear immediately upon washout of ETO are referred to as “reversible TOP2ccs” (light blue). Partially processed (or postranslationally modified) TOP2ccs retained upon ETO washout are referred to as “irreversible TOP2ccs” (dark blue), and fully processed TOP2ccs correspond to protein-free DSBs. The fraction of each of theseTOP2 lesions is shown for Lig4−/− cells 2.5 h after ETO treatment.
(B) Scatterplots comparing the levels of TOP2 lesions (measured by ExoVII+ExoT), DSBs (measured by ExoT), and TOP2cc (inferred by substraction of DSBs from the levels of total TOP2 lesions) upon 2.5 h of ETO treatment before (x axis) and immediately after washing out ETO (y axis).
(C-E) Left: TOP2 lesions in Lig4−/− cells (C), Tdp2−/−Lig4−/− cells (D), or Lig4−/− cells treated with the proteasome inhibitor epoxomicin (E), were divided into 200 bins with increasing levels of transcription, and the relative levels of DSBs (red) and reversible and irreversible TOP2cc (light and dark blue, respectively) were measured for each bin. Right: for each genotype, the average levels of transcription is plotted against the fraction of DSBs (red) and irreversible TOP2cc (dark blue) per bin.
To examine whether transcription influences the reversibility of TOP2ccs, we divided all TOP2 lesions into 200 bins with respect to the level of transcription at their individual genomic locations. Because the impact of DNA repair would confound the analyses, we once again examined cells in which LlG4 was absent. Notably, the fraction of TOP2 lesions that were reversible (light blue areas) was inversely correlated with the rate of transcription at the sites of the lesion (Figure 4C, light blue areas). Correspondingly, the fraction of TOP2 lesions that were protein-free DSBs increased with increasing transcriptional activity (Figure 4C; red area). ln contrast, the levels of ETO-induced TOP2 lesions that were neither reversible TOP2ccs nor protein-free DSBs was unaffected by transcriptional activity and consisted of only a very small fraction of the total lesions (Figure 4C, dark blue area). We reasoned that this latter fraction reflected those TOP2ccs that were converted into an irreversible form, but which were not yet processed into a protein-free DSB. Consistent with this hypothesis, additional deletion of TDP2 (Figure S4C), which interferes with the removal of TOP2 peptides from the 5′ termini of TOP2ccs, decreased the fraction of protein-free DSBs while increasing 2.5-fold the fraction of irreversible TOP2ccs (Figures 4C and 4D). This irreversible fraction in Tdp2−/−Lig4−/− cells was relatively insensitive to transcription except at the highest RNA levels (Figure 4D, right panels). We therefore speculate that highly expressed genes are preferentially protected by TDP2 (Gómez-Herreros et al., 2017) because transcriptionally active regions are processed most rapidly to phosphotyrosyl TOP2 peptides.
Proteosome Activity Promotes the Commitment of TOP2ccs to an Irreversible State
The total processed fraction of TOP2ccs (irreversible TOP2ccs [red areas]+DSBs [dark blue areas]) was comparable between Tdp2−/−Lig4−/− and Lig4−/− (Figures 4C, 4D and S4D). We wondered what might be the mechanistic basis for the commitment of reversible TOP2ccs to a processed state. While this was clearly influenced by transcriptional activity, it did not appear to be greatly impacted by TDP2 activity. This in turn suggested that the commitment point lay between the encounter of TOP2ccs by RNA polymerases and the initiation of processing. We thus examined the impact of the proteasome, since activity of the proteasome has been implicated in the early processing of TOP2ccs prior to their subsequent processing by TDP2 or MRE11 nuclease activities (Ban et al., 2013; Mao et al., 2001; Zhang et al., 2006). Indeed, we found that the fraction of TOP2 lesions that were reversible was increased when Lig4−/− cells were co-incubated with the proteasome inhibitor epoxomicin during ETO treatment (Figure 4E). In addition, as expected, the fraction of TOP2ccs that were present as protein-free DSBs was decreased by 5.5-fold, with a small enhancement of DSBs within highly transcribed areas (Figure 4E, right panel). Interestingly, the fraction of irreversible TOP2cc also increased with epoxomicin, suggesting the possibility that post-translationally modified TOP2cc may not be efficiently reversed (Yoshida et al., 2016).
Rates of Protein-free DSB Formation
The above results raise the possibility that the propensity of a genomic site to undergo translocation following ETO treatment depends both on the binding and activity of TOP2 at that site and the rate at which it is processed into a DSB. We therefore sought to quantitatively model the kinetics of DSB formation across the genome.
If TOP2 is active at given sites only in a subset of cells (Figure 5A, purple circles), ETO rapidly traps TOP2 at these sites to generate a long-lived TOP2cc (Figure 5A; blue circles). Assuming that TOP2ccs are processed into DSBs at a rate of kp (Figure 5A, red circles), eventually all lesions will be converted into DSBs (Figure 5A; bottom). As indicated earlier, while DSBs accumulate with time (Figure S5A), the total TOP2 lesions do not change (Figure S3E). Thus, the cellular fraction of DSBs cannot exceed a certain value denoted as Cmax, which corresponds to the total initial lesions per cell at a given site. The uniformity of Cmax from 30 min and onward (Figure S3E) indicates that all the available TOP2 sites are occupied within 30 min of 50-μM ETO treatment.
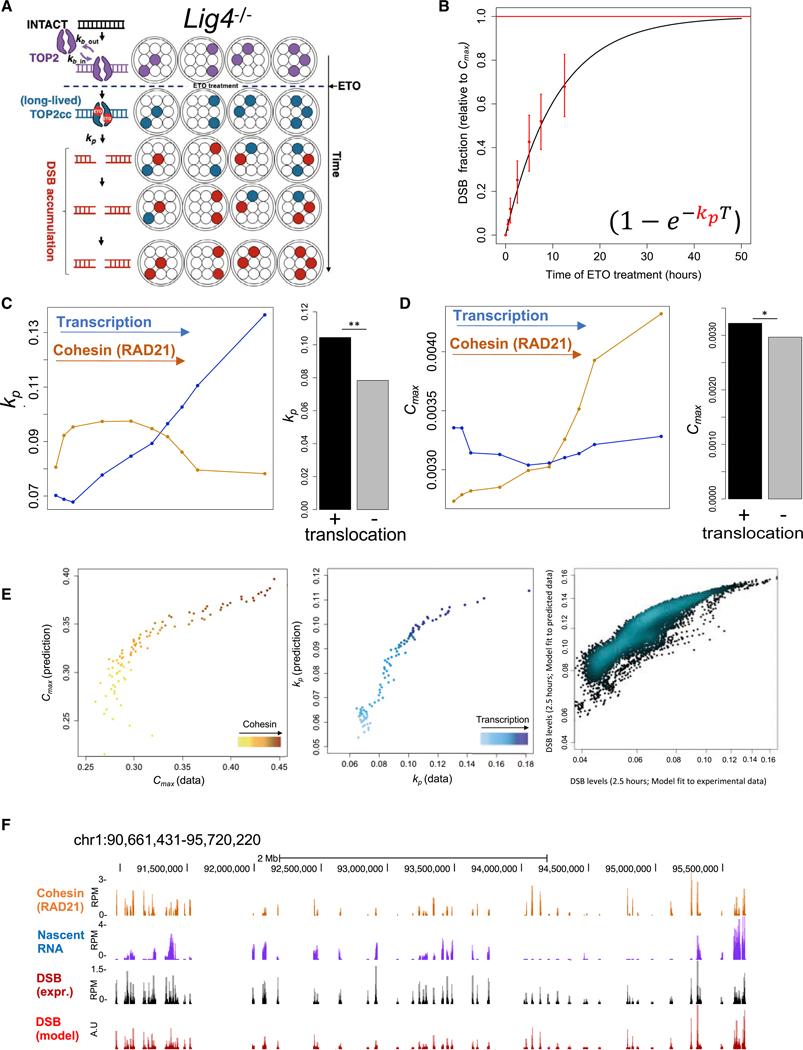
Figure 5. Cohesin Binding and Transcriptional Activity Predict Rates of Protein-free DSB Formation.
(A) Schematic representation of the kinetics of TOP2 lesions before (first row) and after ETO (second row and down) treatment in Lig4−/− cells. Big circles represent different cells, and small circles represent individual genomic sites. TOP2 binding is shown in purple. Long-lived TOP2ccs emerges upon ETO-treatment (blue), which are converted to protein-free DSBs (red). kb_out represents the rate of TOP2cc reversibility, kb_in represents the rate at which TOP2cc are formed (see STAR Methods), and kp represents the rate of TOP2cc processing.
(B) Quantitative modeling the rates of TOP2cc conversion to DSBs. Red points and error bars represent the levels of protein-free DSBs relative to the total TOP2 lesion per cell at all TOP2 lesion sites, both of which were obtained from experimental data. Black line represents the theoretical model after fitting (as described in STAR Methods).
(C and D) Left: lesions were divided into deciles with respect to transcription levels (blue lines) and Cohesin (RAD21) levels (orange lines). The rate of processing (kp) and the levels of TOP2 lesion per cell (Cmax) were obtained at each decile (C and D, respectively), and plotted against transcription/cohesin levels. Cmax: p > 0.2 and p < 1e–20; t test, lowest versus highest decile for transcription and RAD21, respectively.
Right: model was fit to translocated and non-translocated TOP2 lesions and kp (C) and Cmax (D) were derived. *p < 0.05.**p < 0.005, t test.
(E) Left and middle: Cmax and kp parameters respectively, obtained from model fitting to the experimental data (x axis) or predicted by Cmax = α[RAD21]+β and kp = γ[RNA]+δ (y axis; see STAR Methods) for peaks binned by Cohesin and RNA level percentiles. Right: for each pair of Cmax and kp from left and middle panels, DSBs were calculated as where T was set to 2.5 h. Pearson correlations; r = 0.83, r = 0.92, and r = 0.89, for Cmax, kp, and DSB respectively.
(F) Representative genomic profile of Cohesin (RAD21), nascent RNA, experimental DSBs, and predicted DSBs, obtained by integrating Cohesin and transcription into the quantitative model.
In the absence of repair, DSB levels at a given site and time following ETO treatment can be described as where Cmax is the total TOP2 lesion per cell at this site, kp is the processing rate of the site, and T represents time. By knowing the level of DSBs (determined by ExoT, Figure S5A) and Cmax (determined by ExoVII+ExoT, Figure S3E), we could then fit the model to deduce kp (Figure 5B and STAR Methods). We found that the averaged kp is 0.09 h−1, meaning that, on average, 9% of the initial TOP2 lesions are converted to DSBs per h (Figure 5B). TOP2ccs at sites with the highest transcription levels were estimated to be processed at a rate of kp = 0.14 h−1, which is 2-fold higher than sites with the lowest transcription (kp = 0.07 h−1) (Figures 5C and S5B).
While cohesin levels were not strongly predictive of DSB processing (Figures 5C and S5B), they were predictive for initial TOP2cc formation (ie. Cmax, Figures 5D and S5C). For the lesions with the lowest RAD21 level, we estimate Cmax = 0.0026 cell−1, whereas for the lesions with the highest RAD21 levels, it was almost twice as high (Cmax = 0.0043 cell−1). In other words, a TOP2 lesion with high RAD21 levels occurs, on average, once in every 231 cells compared to a TOP2 lesion with low RAD21 levels that occurs once in every 367 cells. Transcription, on the other hand, had very little impact on Cmax (0.0033 cell−1 for both most-highly and most-lowly transcribed sites) (Figures 5D and S5C). We then determined how each of these two parameters, cohesin binding and transcription, influenced translocation. We found that the propensity of TOP2 lesions to undergo translocations was more dependent on kp (kp = 0.0105 and 0.0078 for translocated and non-translocated lesions, respectively) (Figures 5C, right, and S5B) and less so on Cmax (Cmax = 0.32 and 0.3 for translocated and non-translocated lesions, respectively) (Figures 5D, right, and S5B). These results suggest that cohesin facilitates the formation of TOP2 lesions, while TOP2cc processing to DSBs is promoted by transcription. When these two orthogonal factors (high cohesin and transcription) co-exist at a given site, the propensity for translocation increases.
A Molecular Model for Predicting TOP2-Induced Chromosome Breakage Genome-wide
Cmax can be described as a linear function of RAD21 (Figure 5D, orange line), while kp is proportional to transcription (Figure 5C, blue line). We therefore utilized measurements of cohesin binding and transcription at a given site to deduce Cmax and kp, to calculate protein-free DSBs as a function of time after ETO treatment: DSBT = (f[RAD21])(1-e−fiRNA1T). With only these two chromatin features, we were able to predict the DSB levels at a given treatment time and genomic location (Figures 5E and 5F; STAR Methods). Indeed, the model parameters, Cmax and kp (Figure 5E; left and middle panels), and consequently, DSB levels at a given time of ETO treatment (Figure 5E; right panel, and Figure 5F), showed a high correlation between experiment and prediction (see Parameters Prediction section in STAR Methods for full description). Moreover, translocated regions were associated with high levels of predicted DSBs (Figure S5D).
DISCUSSION
Current methods to measure DNA-TOP2 covalent complexes include the ICE (immunocomplex of enzyme) (Nitiss et al., 2012) and RADAR (rapid approach to DNA adduct recovery) assays (Kiianitsa and Maizels, 2013), which rely on the separation of the DNA-protein complexes from bulk cellular proteins and selective quantitation of the DNA bound proteins using anti-TOP2 antibody immunodetection. Other approaches such as alkaline elution (Kohn, 1996) or TARDIS (trapped in agarose DNA immunostaining) (Willmore et al., 1998) and DRT (differential retention of TOP2) (Agostinho et al., 2004) identify TOP2cc as protein-linked DNA breaks or by immunofluorescence after removal of non-covalently bound proteins. These techniques, however, do not provide information on the genomic localization of TOP2cc, nor their rates of conversion to DSBs. Moreover, partially degraded or irreversible TOP2ccs may remain undetectable using antibody-based methods. Here we developed a method to simultaneously measure TOP2ccs and protein-free DSBs genome-wide. As this method is DNA-based, it is not affected by proteolytic degradation of covalently bound protein. Thus, it could be adapted more generally to measure DNA damage generated by repair pathways that require proteolysis of DNA-protein cross-links (Stingele et al., 2017).
Previous studies using short ETO treatments in which TOP2cc conversion to clean DSBs is minimal indicated that TOP2 is active at CTCF-and-cohesin-bound sites, anchoring promoter-enhancer loops and topological domains (Canela et al., 2017; Uusküla-Reimand et al., 2016).
Consistent with these observations, we show here that both TOP2 binding and activity is dependent on cohesin. Among these TOP2 sites are breakpoint cluster regions that are commonly translocated in cancer (Canela et al., 2017). Using the method described in the present study, we can estimate the number of cells required to generate a protein-free DSB at a given location, which is directly proportional to the probability of translocation. The CXCR4 gene, which is highly transcribed and found to be translocated in chronic lymphocytic leukemia (Acunzo et al., 2014), provides an interesting example, since the etiology of this fusion has not previously linked to TOP2 activity. We estimate that it would take 85 cells to generate a TOP2cc within intron 1 of Cxcr4 (which is fused to MAML2 in CLL) (Figure S5E). A protein-free DSB would form at this locus in 1 every 836 cells after 30 min ETO, but after 12.5 h ETO treatment, 1 cell every 98 would have a DSB (Figure S5E). Since loop anchor positions delineated by CTCF/cohesin are well conserved among different cell types, the frequency of TOP2ccs would be similarly preserved (i.e., approximately 1 in 85 cells for CXCR4); however, cell-type differences in transcription or mutations in cohesin (Figures 1A–1D) or its regulatory subunits such as WAPL (Figures S1E and S1F) that either decrease or increase cohesin residence time would be predicted to alter the frequency of TOP2-induced DSBs and translocation.
Here we show that the vast majority of TOPccs that are present during ETO treatment are reversible after washout, especially at sites at which transcriptional activity is low. Only 5%−10% of TOP2ccs at the time of washout are found in an irreversible state, while the remaining lesions have been processed into DSBs. This suggests that TOP2cc processing, once initiated by proteasomal degradation and then completed by TDP2 removal of the phosphotyrosil bond, is exquisitely rapid. Deficiency in TDP2 dramatically decreases TOP2cc conversion into DSBs, resulting in a greater fraction of irreversible TOP2cc; however, the total levels of processed TOP2 (that is, the sum of irreversible TOP2cc and DSBs) are TDP2 independent. This suggests that the bottleneck in DSB formation takes place after TOP2 trapping but before processing initiates. Analogous to the mechanism of transcription-coupled nucleotide excision repair (van Cuijketal., 2014), stalling of RNA polymerase II on transcription-blocking TOP2 lesions could facilitate the recruitment and activation of the proteasome (Mao et al., 2001). However, even at genomic sites that are not actively transcribed, TOP2 lesions are processed, but at a 2-fold slower rate, and translocate approximately 2-fold less frequently than at highly transcribed regions.
Proteasome inhibitors have been widely used in the clinic for treatment of multiple myeloma and mantle cell lymphoma (Manasanch and Orlowski, 2017). We found that inhibition of the proteasome increases the fraction of reversible TOP2ccs while almost completely blocking the conversion of TOP2ccs to DSBs by TDP2 and alternative nucleases. Thus, in the cells employed here, proteasome-independent processing of TOP2ccs is minimal. Since proteasome inhibition increases the levels of reversible TOP2ccs while decreasing DSB formation, one would expect this treatment to be cytoprotective. On the contrary, inhibition of proteasomal activity was shown to potentiate the toxicity of TOP2 poisons (Lee et al., 2016). While DSBs are barely detectable when the proteasome is inhibited, a significant fraction of TOP2ccs remains irreversible (Figure 4E). DSBs can be repaired by several pathways, but TOP2 protein adducts that cannot efficiently be removed by a nucleolytic processing mechanism will block chromatin-based processes such as transcription and DNA replication. Irreversible TOP2cc maybe therefore be even more cytotoxic than DSBs.
Conclusions
While TOP2 activity is largely transcription independent, chromosomal translocation is transcription dependent. We found that etoposide-induced trapping of TOP2cc depends on the pre-existing levels of cohesin bound to DNA. Transcription promotes chromosomal translocation at these sites mainly by accelerating the conversion of TOP2ccs to DSBs, but has less of an impact on stimulating fusions once DNA ends are protein free, as in the case of translocations induced by AsiSI. Based on measured levels of cohesin binding and transcriptional activity at a given location, our quantitative model predicts the level of DSBs as a function of time after the trapping of TOP2 by ETO treatment.
STAR★METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Andre Nussenzweig (andre_nussenzweig@nih.gov).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell Culture and Mice
Abelson-transformed pre-B cell lines WT(Lig4f/f) and Lig4−/− were generated as described (Bredemeyer et al., 2006). Tdp2−/− pre-B cells were generated from WT(Lig4f/f cells by CRISPR-Cas9 with an sgRNA (target sequence ATGCGGCGGAGCCCGCA GGGCCGGCAGGG) targeting the exon 1 of Tdp2. TDP2 activity was determined as described (Zagnoli-Vieira et al., 2018). Tdp2−/− Lig4−/− cells were generated by retroviral CRE infection of Tdp2−/−Lig4f/f cells. In all experiments, pre-B cells were arrested in G1 with 3 μM imatinibfor48 hr (Bredemeyer etal., 2006) and then treated with etoposide for the times indicated. Etoposide was not washed out during plug preparations for END-seq. For reversibility experiments, after treatment with etoposide for 2.5 h, half of the cells were held in media with etoposide, and half were washed 3 times in cold media and processed at the same time.
For translocations, ZFN was induced by treating (during the last 24 h of imatinib) with doxycycline at 1 μg/mL (Dorsett et al., 2014). For induction of AsiSI, cells were treated in addition with tamoxifen at 1 μM during the last 4 h of treatment of doxycycline and imatinib (Canela et al., 2016). After 2.5 h treatment with 50 μM of etoposide, pre-B cells were washed 3 times in cold media and cultured for 12 h in media without etoposide but with imatinib, doxycycline and tamoxifen present.
For transcription inhibition or stimulation, and proteasome inhibition drug pre-treatment was performed for 2 h with 150 mM DRB (D-ribofuranosylbenzimidazole, Sigma) or 100 U/mL mouse interferon beta (PBL) or 2 μM of epoxomicin (Sigma) prior to etoposide treatment or induction of AsiSI with tamoxifen. Drugs were maintained during the treatment with etoposide or induction of AsiSI and after washout of ETO.
Lymphocyte separation media was used after harvesting the cells in all experiments to separate dead cells from live cells. Mouse embryonic fibroblasts (MEFs) Smc3f/f ERT2-Cre and Waplf/f cells were cultured and floxed alleles deleted as described (Busslinger et al., 2017). After 10 days in confluence and serum starvation (2% charcoal-treated FBS) cells were harvested for ChIP-seq or treated with 50 μM of etoposide during 30 min and processed for END-seq.
HCT-116-CMV-OsTir1 and HCT-116-RAD21-mAID-Clover cells were obtained from (Natsume et al., 2016), arrested in G1 by the treatment with 20 μM of lovastatin for 24 h and treated with auxin for 4 h before 50 μM etoposide treatment for 30 min.
Mature Top2β−/− B cells were generated by bone marrow reconstitution of lethally irradiated recipient mice with fetal liver cells derived from WT and Top2β−/− embryos. Recipient mice were irradiated with 950 rads, 4 h before injection of 3 million donor fetal liver cells. Between 6 to 12 weeks after reconstitution mature resting B cells were isolated from recipient mouse spleen with anti-CD43 MicroBeads (Miltenyi Biotech). B cells were activated with LPS (25 μg/mL; Sigma), IL-4 (5 ng/mL; Sigma) and RP105 (0.5 μg/mL; BD) for 12 hr as described (Callen et al., 2013) and treated for 30 min with 50μM of etoposide. All animal experiments were approved by the NCI Animal Care and Use Committee.
METHOD DETAILS
END-seq
Single cell suspensions of B cells (15 million), pre-B cells (40 million), MEFS (3 million), HCT116 (7 million) were untreated or treated with drugs to alter transcription or inhibit the proteasome and/or etoposide or AsiSI induction as indicated in the ‘Cell Cultures, treatments and Mice’ section. A detailed END-seq protocol can be found in (Canela et al., 2017). Cells were washed twice in cold PBS and resuspended in cold cell suspension buffer (Bio-Rad CHEF Mammalian Genomic DNA plug kit), equilibrated for 5 min at room temperature, mixed with 2% melted CleanCut agarose (Bio-Rad CHEF Mammalian Genomic DNA plug kit) prewarmed at 37°C for a final concentration of 0.75%, and transferred immediately into plug molds and let them solidify at 4°C for 20 min. Embedded cells were lysed and digested using Proteinase K (50°C, 1 hour then 37°C for 7 h). Etoposide and drugs were maintained at the same experimental concentrations in all the steps until digestion with Proteinase K. Plugs were rinsed in TE buffer and treated with RNase A at 37°C, 1 hour. The next enzymatic reactions were performed with the DNA in agarose plugs to prevent shearing. DNA ends were blunted for 1 hour at 37°C with Exonuclease VII followed by Exonuclease T (NEB) for 1 hour at 25°C to detect all TOP2 lesions, or only with Exonuclease T (NEB) 1 hour at 25°C to detect only DSBs. After blunting, A-tailing was performed to attach dA to the free 3′-OH, followed by ligation of “END-seq hairpin adaptor 1,” listed in reagents section, using NEB Quick Ligase. Agarose plugs were then melted and dialyzed and DNA was sonicated to a median shear length of 170bp using Covaris S220 sonicator for 4 min at 10% duty cycle, peak incident power 175, 200 cycles per burst, at 4°C. DNA was ethanol-precipitated and dissolved in 70 μl TE buffer. Biotinylated DNA was isolated using MyOne Streptavidin C1 Beads (ThermoFisher #650–01), followed by end repair (dNTPs, T4 polymerase (NEB), Klenow (NEB), T4 PNK) and dA-tailing (Klenow exo- (NEB), dATP). The second end was ligated to “END-seq hairpin adaptor 2” using NEB Quick Ligase. Hairpins were digested using USER (NEB), and the resulting DNA fragments were PCR amplified using TruSeq barcoded primer p5, AATGATACGGCGACCACCGAGATCTACACNNNNNNNNACACTCTTTCCCTACACGACGCTCTTCCGATC*T and TruSeq barcoded primer p7, CAAGCAGAAGACGGCATACGAGANNNNNNNGTGACTGGAGTTCAGACGTGTGCTCTTCCGATC*T, (NNNNNNNN represents barcode and * a phosphothiorate bond) listed in reagents. PCR fragments were isolated by size selection from agarose gel, selecting 200–500 bp fragments followed by DNA purification using QIAquick Gel Extraction Kit. Libraries were quantified using KAPA Library Quantification Kit and sequenced using Illumina NextSeq 500 or 550.
ChIP-seq
Cells were fixed adding 37% formaldehyde (F1635, Sigma) to a final concentration of 1% and incubating them at 37C for 10 min. Fixation was quenched by addition of 1M glycine (Sigma) in PBS at a final concentration of 125 mM. Twenty million fixed cells were washed twice with cold PBS and pellets were snap frozen in dry ice and stored at 80°C. Fixed cell pellets of 20 million cells were thawed on ice and resuspended in 2 mL of cold RIPA buffer (10 mM TrisHCl pH 7.5, 1 mM EDTA, 0.1% SDS, 0.1% sodium deoxycholate, 1% Triton X-100, 1 Complete Mini EDTA free proteinase inhibitor (Roche)). Sonication was performed using the Covaris S220 sonicator at duty cycle 20%, peak incident power 175, cycle/burst 200 for 30 min at 4°C or using the Branson sonifier at amplitude 35%, 12 cycles of 20 s sonication and 30 s of pause at 4°C. Chromatin were clarified by centrifugation at 21,000 g at 4°C for 10 min and precleared with 80 ul prewashed Dynabeads protein A (ThermoFisher) for 30 min at 4°C. 40 μl prewashed Dynabeads protein A were incubated with 10 μg of each respective antibody in 100 μl of PBS for 10 min at room temperature in continuous mixing, washed twice in PBS for 5 min and added to 1 mL of chromatin followed by overnight incubation at 4°C on a rotator. Beads were then collected in a magnetic separator (DynaMag-2 Invitrogen), washed twice with cold RIPA buffer, twice with RIPA buffer containing 0.3M NaCl, twice with LiCl buffer (0.25 M LiCl, 0.5% Igepal-630, 0.5% sodium deoxycholate), once with TE (10 mM Tris pH 8.0, 1mM EDTA) plus 0.2% Triton X-100, and once with TE. Crosslinking was reversed by incubating the beads at 65°C for 4 hr in the presence of 0.3% SDS and 1mg/mL of Proteinase K (Ambion). DNA was purified using Zymo ChIP DNA clean and concentrator kit (Zymo Research) and eluted in 20 μl. The entire ChIP DNA was used to prepare Illumina sequencing libraries. End-repair was performed in 75 μl of T4 ligase reaction buffer, 0.4 mM of dNTPs, 4 U of T4 DNA polymerase (NEB), 13.5 U of T4 Polynucleotide Kinase (NEB) and 1.5 U of Klenow fragment (NEB) at 24°C for 30 min in a ThermoMixer C at 400 rpm. End-repair reaction was cleaned using 2X Agencourt AMPure XP beads and eluted in 15 μl of EB that was used for A-tailing reaction in 30 μl of NEBNext dA-Tailing reaction buffer (NEB) with 7.5 U of Klenow fragment exo- (NEB) at 37°C for 30 min. The 30 μl of the A-tailing reaction were mixed with Quick Ligase buffer 2X (NEB), 3,000 U of Quick ligase and 5 nM of annealed adaptor (Illumina truncated adaptor) in a volume of 75 μl and incubated at 25C for 20 min. Adaptor was prepared by annealing the following HPLC oligos: 5′-phosphate/GATCGGAAGAGCACACGTCT-3′ and 5′-ACACTCTTTCCCTACACGACGCTCTTCCGATC*T-3′ (*phosphorothioate bond). Ligation was stopped by adding 50mM of EDTA and cleaned with 1.8X Agencourt AM- Pure XP beads and eluted in 15ul of EB that was used for PCR amplification in a 50 uL reaction with 1 mM primers TruSeq barcoded primer p5, AATGATACGGCGACCACCGAGATCTACACNNNNNNNNACACTCTTTCCCTACACGACGCTCTTCCGATC*T, TruSeq barcoded primer p7, CAAGCAGAAGACGGCATACGAGANNNNNNNGTGACTGGAGTTCAGACGTGTGCTCTTCCGATC*T, (NNNNNNNN represents barcode and * a phosphothiorate bond), and 2X Kapa HiFi HotStart Ready mix (Kapa Biosciences). The temperature settings during the PCR amplification were 45 s at 98°C followed by 15 cycles of 15 s at 98°C, 30 s at 63°C, 30 s at 72°C and a final 5 min extension at 72°C. PCR reactions were cleaned with Agencourt AMPure XP beads (Beckman Coulter), run on a 2% agarose gel and a smear of 200–500bp was cut and gel purified using QIAquick Gel Extraction Kit (QIAGEN). Library concentration was determined with KAPA Library Quantification Kit for Illumina Platforms (Kapa Biosystems). Sequencing was performed on the Illumina Nextseq500 (75bp single-end reads). Antibodies for ChiP-seq were: anti-RAD21 (ab992, Abcam), anti-CTCF (07–729, Millipore) and TOP2B Antibody (H-286) (sc13059, SantaCruz).
Nascent RNA-seq
Nascent RNA sequencing was performed as previously described (Canela et al., 2017). Pre-B cells were arrested in G1 by treatment with 3 μM imatinib for 48 h and then pre-treated or not with drugs affecting transcription and treated with etoposide for 2.5 h or untreated. Next, cells were washed 3 times in cold media and cultured back for new media, then 5 million cells were labeled with 0.5 mM 5-ethynyl uridine (EU) during 30 min keeping the transcriptional drugs in the media. Total RNA extraction was performed using TRIzol (Invitrogen) and 2 μg was rRNA depleted using the NEBNext rRNA Depletion kit (human/mouse/rat) (New England Biosciences). rRNA-depleted RNA was used for biotinylation via the Click-IT reaction (Click-iT Nascent RNA Capture Kit, ThermoFisher C10365) using manufacturer’s specification. First-strand cDNA synthesis of the captured nascent RNA was done using SuperScript VILO cDNA synthesis kit (Invitrogen), followed by purification with 1.8X volume of AMPureXP beads (1.8X) and elution in 20 μL. Second-strand synthesis was performed using 0.6 mM dATP, 0.6 mM dCTP, 0.6 mM dGTP, and 1.2 mM dUTP in the presence of 2 U of RNase H (Invitrogen) and 20 Units of E. coli DNA polymerase I (Invitrogen) in a total volume of 30 uL for 2.5 h at 16°C. cDNA was purified using 1.8X volume AMPure XP beads and eluted in 20 μL of EB. Sequencing libraries were then prepared. End-repair was performed in 50 μL of T4 ligase reaction buffer (1X), dNTPs (0.4 nM), T4 DNA polymerase (NEB, 3 Units), T4 Polynucleotide Kinase (NEB, 9 Units) and Klenow fragment (NEB, 1 Unit) at 24°C for 30 min in a ThermoMixer at 400 rpm. End-repair reaction was cleaned using 1.8X volume AMPureXP beads and eluted in 15 μL of EB. A-tailing was performed using NEBNext dA-Tailing reaction buffer (NEB, 1X) with Klenow fragment exo- (NEB, 7.5 U) at 37°C for 30 min. A-tailing reaction was mixed with Quick Ligase Buffer (NEB), Quick ligase (NEB, 3000 Units) and 5 nM of annealed adaptor (Illumina truncated adaptor) in a volume of 75 mL and incubated at 25°C for 20 min. The adaptor was prepared by annealing the following HPLC oligos: 5′-phosphate/GATCGGAAGAGCACACG TCT-3′ and 5′-ACACTCTTTCCCTACACGACGCTCTTCCGATC*T-3′ (*phosphoro-thioate bond). Ligation reaction was stopped by adding 50 mM of EDTA, then DNA was purified with 1.8X volume AMPure XP beads and eluted in 15 ml of EB. 0.5 U of Uracil- DNAglycosylase (ThermoFisher) was added for 15 min at 37°C. PCR amplification was performed in 50 μL reaction with 1 μM primers TruSeq barcoded primer p5, AATGATACGGCGACCACCGAGATCTACACNNNNNNNNACACTCTTTCCCTACACGACGCTCTTCC GATC*T, TruSeq barcoded primer p7, CAAGCAGAAGACGGCATACGAGANNNNNNNGTGACTGGAGTTCAGACGTGTGCTCTTCC GATC*T, (NNNNNNNN represents barcode and * a phosphothiorate bond), and 2X Kapa HiFi HotStart Ready mix (Kapa Biosciences). PCR program: 98°C, 45 s; 15 cycles [98°C, 15 s; 63°C, 30 s; 72°C, 30 s]; 72°C, 5 min. PCR reactions were cleaned with AMPure XP beads, and 200–500 bp fragments were isolated after running on 2% agarose gel. Libraries were purified using QIA-quick Gel Extraction Kit (QIAGEN). Library concentration was determined with KAPA Library Quantification Kit for Illumina Platforms (Kapa Biosystems). Sequencing was performed using Illumina Nextseq 500 (75bp single-end reads).
HTGTS
HTGTS was performed as described (Hu et al., 2016). Primers are listed in reagents table (ZNF-bio,/5Biosg/CAGAAGCCTTCA GTATGCACCA, I7-ZNF-nested-barcode3,CTCGGCATTCCTGCTGAACCGCTCTTCCGATCTAGGCAGAATTCCACTTAACCCCAACTCCAG, I7-ZNF-nested-barcode4,CTCGGCATTCCTGCTGAACCGCTCTTCCGATCTTCCTGAGCTTCCACTTAACCCCAACTCCAG, Adaptor-upper,GGTACACGACGCTCTTCCGATCTNNNNNN/3AmMO/,Adaptor-lower-mod,/5Phos/AGATCGGAAGAGCGTCGTGTACC/3AmMO/, I5-bridge,AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATC*T, P5-I5c,AATGATACGGCGACCACCGAGATCTACACTCTTT*C, P7-I7c,CAAGCAGAAGACGGCATACGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTT*C). Libraries (fragment size 500 – 1,000 bp) were purified and sequenced using Illumina MiSeq. For translocation analysis the standard LAM-HTGTS bioinformatic pipeline was used (Hu et al., 2016).
Tyrosyl DNA phosphodiesterase activity assay
Tyrosyl DNA phosphodiesterase activity was measured in whole cells-extracts (WCE) as previously described (Zagnoli-Vieira et al., 2018). In summary, WCE was prepared by resuspension of Tdp2+/+ or Tdp2−/− mouse pre-B cells in lysis buffer (40 mM Tris/HCl pH 7.5,100 mM NaCl, 0.1% Tween-20,1 mM DTT, 1 mM PMSF, 1x EDTA free protease cocktail inhibitor), followed by 30 min incubation on ice and mild sonication. The WCE was clarified by centrifugation for 10 min at 4°C at 16000 g in a microfuge and the protein concentration quantified using the bicinchoninicacid (BCA) assay reagent (ThermoFisher). WCE from 1-BR normal human fibroblasts (control) and 850-BR TDP2 mutated patient-derived fibroblasts (Zagnoli-Vieira et al., 2018) was prepared as above as controls. Clarified WCE (0.5 μg total protein from B cells and 2.5 μg total protein from primary human fibroblasts) was incubated with 80 nM of a 19-mer TDP2 single-stranded oligonucleotide substrate harboring a Cy5-tyrosine linked 5′-terminus and a 3′-linked black hole quencher (BHQ) (Midland certified reagent company), oraTDP1 single-stranded oligonucleotide substrate harboring a 3′-linked black hole quencher (BHQ) and a Cy5-tyrosine linked 3′-terminus (Midland certified reagent company), in reaction buffer (50 mM Tris/HCl pH8.0,10 mM MgCl2,80 mM KCl, 1 mM DTT, 0.05% Tween-20) in 10 μl total volume at room temperature. Cy5 fluorescence was measured at 640 nm at the indicated time intervals on a BMG PHERAstar plate reader. Buffer was employed as a negative control for the absence of TDP2 activity and benzonase (500U) (Sigma) was employed for complete hydrolysis of the substrate as a positive control.
ExoVII and ExoT in vitro blunting assay
ExoVII and ExoT blunting assay was performed as described previously for TDP2 in Gao et at 2012. Briefly, recombinant exonucleases from NEB were used at the following concentrations: for ExoVII 0.2 U/μl, 0.1 U/μl and 0.05 U/μl, and for ExoT 0.125 U/μl, 0.062 U/μl and 0.031 U/μl, with 1 nM of 32P-internally-labeled DNA constructs bearing a 5′-tyrosine moiety on a protruding 4-nt overhang or on a blunt end. For the preparation of internally labeled DNA substrates, a 22-nt DNA (5′-GCGCAGCTAGCGGCGGATG*G*C*A-3′ * represents a phosphothiorate bond) was labeled with 32P at the 5′-end with [γ−32P]ATP (PerkinElmer Life Sciences) and T4 polynucleotide kinase (NEB). An 18-nt DNA (5′-TCCGTTGAAGCCTGCTTT-3′) harboring the phosphotyrosine at the 5′-end was mixed with the 5′-labeled 22-nt DNA before annealing to a 36-nt (B36: 5′ P-TGCCATCCGCCGCTAGCTGCGCAAAGCAGGCTTCAA-3′) or a 40-nt (B40: 5′P-TGCCATCCGCCGCTAGCTGCGCAAAGCAGGCTTCAACGGA-3′ DNA with complementary sequence. The nicks were sealed with T4 DNA ligase (NEB). The resulting internally labeled 40-nt and 36-nt product harboring 5′-phosphotyrosine, was then column-purified and eluted for use (named 5′Y40/B40, and 5′Y40/B36 respectively). All oligonucleotides were synthesized by Integrated DNA Technologies. Reactions were performed in 20 ml reaction in buffer NEBuffer4. ExoVII reactions were incubated at 37°C 1 hour and ExoT reactions at 25°C 1 hour, double reactions were performed sequentially first with ExoVII and then with ExoT. As positive control 0.1 mM of TDP2 was used at 25°C for 15 min in reaction buffer containing 80 mM KCl, 5 mM MgCl2, 0.1 mM EDTA, 1 mM DTT, 40 μg/mL bovine serum albumin 50 mM Tris-HCl, pH = 7.5, and 0.01% Tween-20. The reactions were terminated by addition of formamide gel loading buffer and analyzed by denaturing PAGE (16%). Gels were dried and exposed on PhosphorImager screens. Imaging was done using a Typhoon 8600 imager (GE Healthcare, Little Chalfont, United Kingdom).
QUANTIFICATION AND STATISTICAL ANALYSIS
Genome alignment
Tags were aligned to the mouse (GRCm38p2/mm10) or human (GRCm37/hg19) genomes using Bowtie (version 1.1.2)(Langmead et al., 2009) with the options-best-all -strata-l 50. For ChIP-seq, we allow 2 mismatches and discarded tags with multiple alignments (-n 2 -m 1). For Nascent RNA-seq, we allow 3 mismatches (−n 3 −m1). For END-seq, we allowed 3 mismatches and kept the best strata for tags with multiple alignments (-n 3 −k 1).
Peak Calling
Peaks were called using MACS 1.4.3 (Zhang et al., 2008). END-seq peaks were called using the parameters:-nolambda,-nomodel and-keep-dup = all (keep all redundant reads). For ETO-treated END-seq data peak calling, we used the corresponding non-treated samples as control, keeping > 10 fold-enriched peaks. For pre-B cell data (Figures 2, 3, 4, and 5), in order to perform a direct comparison of the levels of breaks across experiments, peak calling was done at the END-seq (ExoT+ExoVII) data presented in Figure 2 (carried out after 2.5 h of ETO), and all the analyses thereafter including the time course, reversibility and kinetics utilized this peak dataset. ChIP-seq peaks were called using default parameters. Nascent RNA-seq peaks were called using the parameters-nolambda-nomodel to account for wide peaks.
CTCF Site Detection
Position Weight Matrix (PWM) of CTCF was taken from JASPAR database (Mathelier et al., 2014). With this PWM, we scanned the mouse (Figures 1C, 1D, 1G–1I, and S1G), or human (Figure S1D) genomes using the FIMO tool (Grant et al., 2011) for significant CTCF sites (p < 1 × 10−4).
END-seq, ChIP-seq, and Nascent RNA-seq Data Analysis
For comparisons between different genotypes, RAD21, CTCF, TOP2B ChIP-seq signal, and END-seq signal were calculated, as RPKM, within END-seq peaks, for the corresponding cell type, and around CTCF sites (both described above). For ChIP-seq and Nascent RNA data, only non-redundant reads were counted. For END-seq data, all reads were counted. For END-seq data from pre-B cells, where cells with ZFN break were spiked-in to the library at a 1:50 (2%) ratio, the RPKM value for each peak was divided by the signal around the spiked-in breaks and then multiplied by 2, to get values as cell-percentage. From this value, X, the # of cells per lesion (Figure 3B) were calculated as 100/X.
For the kinetics model and visualization purposes, outliers were removed from the data as follows: First, the interquartile, iqr, was calculated using the R function IQR. The outlier cutoff was then set to outlier_cutoff = iqr*1.5, and data points that were lower than Q1-outlier_cutoff or greater than Q3+outlier_cutoff (whereby Q1 and Q3 are the first and third quartiles, respectively), were discarded.
Data Reproducibility
END-seq and ExoT END-seq carried out upon 2.5 h of ETO treatment were performed multiple times in this study (three END-seq and two ExoT END-seq), once for each experiment. To assess the reproducibility, we compared the 2.5 hour ETO END-seq experiments (measuring total TOP2 lesion) from Figures 2, 3, and 4 (Figures S6A and S6B; referred to as L#2,L#3 and L#4, respectively), and the 2.5 ETO ExoT END-seq experiments (measuring protein-free DSB) from Figures 3 and 4 (Figure S6C referred to as D#3, and D#4). We found that the Pearson correlation between the TOP2 lesion replicates is greater than 0.99, suggesting high reproducibility (Figure S6A). L#2, L#3,and L#4 showed an average level of 0.44%,0.34% and 0.41% cells that carries the lesion respectively (Figure S6B). Similar to TOP2 lesions, DSB replicates showed high correlation (r = 0.9) (Figure S6C). Fraction of protein-free DSB (DSB/Lesion ratio), showed some variability among experiments, with 0.27 and 0.4 of the lesion that is processed to DSB for Figure 3 and Figure 4, respectively (Figure S6D).
HTGTS Analysis
Sites that underwent chromosomal translocations to the bait site, residing at chr6:41554860-41555099, were identified as described (Hu et al., 2016). For ETO treated and non-treated experiments, data from two experiments were aggregated. To avoid proximity bias, reads that resided within 10 kilobases from the bait site were discarded. The remaining reads were clustered using bedtools merge function with the parameter -d 5000; that is, reads within 5kbfrom each other were merged. To avoid background reads, clusters that contained less than 5 reads were discarded. TOP2 lesions, or AsiSI breaks that overlapped with at least one cluster, were defined to be translocated. For translocations within genes, the total number of reads that overlapped with genes was calculated.
TOP2 Lesion Detection
The fraction of the different TOP2 lesion species (Reversible TOP2cc, Irreversible TOP2cc and protein-free DSB) from the total lesion were calculated as follows: First, ExoT END-seq signal was divided by the END-seq signal (using ExoVII+ExoT)for all theTOP2 lesion sites (determined by peak calling as described above), to obtain the DSB fraction ([DSB]). Similarly, TOP2cc fraction, [TOP2CC], was defined as [TOP2CC] = 1-[DSB] fraction for each lesion. To calculate the reversible TOP2cc fraction ([TOP2CC]R), we subtracted the data obtained from the END-seq ETO washout experiment (see above), from the data obtained from END-seq ETO experiment without washout, to get [TOP2CC]R. Finally, the irreversible TOP2cc fraction ([TOP2CC]I) was defined as [TOP2CC]I= 1-[DSB]-[TOP2CC]R.
Genomic Annotations
Transcription start sites (TSSs) and gene body coordinates were defined using RefSeq genes annotations taken from the UCSC database (Karolchik et al., 2004). For genomic localization analyses, all sites that reside within 1kb of annotated TSSs, were considered as “TSSs.”
Mathematical Modeling
Kinetics Model for TOP2 Processing
[INTACT]: the fraction of cells that are not currently cleaved by TOP2 at the site, but serve as “latent”/potential TOP2 sites. [TOPCC]: the fraction of cells with a TOP2-bound break (TOP2cc). [DSB]: the fraction of cells harboring protein-free DSB.
The model is built under the assumption that each genomic site has a maximal capacity of TOP2 lesion frequency, denoted as Cmax. This assumption is supported by the observation that in conditions in which DNA repair is deficient (L/g4−/−), TOP2 lesion levels, measured by END-seq, are fixed in time.
Cmax is therefore determined as follows:
In the absence of DNA repair (Lig4−/−), the kinetics of these fractions, after introduction of ETO, is determined by the ordinary differential equations:
where kb_out represents the rate of TOP2cc reversibility, kb_in represents the rate at which TOP2cc are formed, and kp is the rate at which TOP2cc is processed into protein-free DSB (Figure 5A).
ETO treatment results in the trapping of TOP2cc, and therefore sets kb_out to be 0. The fact that by 30 min TOP2cc reaches Cmax, indicates that kb_in2.
Note that the processing rate kp is insensitive to the value of kb_in, so long as it is greater than 1.3 (Figure S6E).
Therefore, for simplicity, we set [INTACT]0 and [TOP2CC]0 (the [INTACT] and [TOP2CC] fractions immediately after ETO introduction) to be 0 and Cmax, respectively.
Under these assumptions, the [DSB] at time T can be denoted as (Figure 5B).
Data Fitting
Cmax associated with a genomic site was obtained directly from the END-seq data, by averaging the TOP2 lesions (after spike-in normalization - see details above) over the different time points (30 min, 1, 2.5, 5, 7.5 and 12 h), at each pre-defined END-seq peak. For the averaged kinetics of all lesions (Figure 5B), or subset of lesions (Figures 5C and 5D), Cmax was taken as the average Cmax over the corresponsing peak set, after removal of outliers, as described above. To obtain the rate of processing, kp, the averaged fraction of protein-free DSB was first calculated for each time point, as described in TOP2 Lesion Detection section above, and the data was fitted to the function using the nls R package.
Parameters Prediction
As Cmax and kp linearly correlate with log(RAD21) and log(RNA) levels, respectively (Figures 5C and 5D), we sought to predict the DSB levels at time T, by incorporating RAD21 ChIP-seq and transcription levels (hereafter reffered to as [RAD21] and [RNA]) into the kinetics model, such that Cmax = α[RAD21]+β and kp, = γ[RNA]+δ. To predict α, β, γ and δ, we divided the peaks set into 1000 quantiles with respect to RAD21 ChIP-seq or transcription levels, and performed a linear regression between RAD21 and transcription, over the END-seq peaks, and the kp at these quantiles, using the R function lm. This regression revealed the value of the above parameters to be: α = 0.037, β = 0.27, γ = 0.01 and δ = 0.1. To test this prediction on independent data (Figure 5E), we divided the peaks into training and validation sets. First, we obtained the α, β, γ and δ coefficients using the training set. Then, using these coefficients, [RAD21] and [RNA], we predicted Cmax and kp of the validation set. We repeated this procedure 10 times with different training and validation sets, and used the mean Cmax and kp to predict the DSB levels at a given time of ETO treatment (Figure 5E).
Data Visualization
Aligned-reads bed files were first converted to bedgraph files using bedtools genomecov (Quinlan and Hall, 2010; PMID: 20110278) following by bedGraphToBigWig to make a bigwig file (Kent et al., 2010; PMID: 20639541). Visualization of genomic profiles was done by the UCSC browser (Kent et al., 2002; PMID:12045153). Genome browser profiles were normalized to present RPM. For the time course END-seq data (Figure 3D) and ETO washout experiments (Figure S4A), data was further normalized using spike-in (see END-seq, ChIP-seq, and Nascent RNA-seq Data Analysis section above).
Heatmaps were produced using the R package pheatmap. For aggregate plots around CTCF sites and TSSs, signal was smoothed using smooth.spline function in R.
Statistical Analysis
Statistical analysis was performed using R version 3.3.1 (http://www.r-project.org). The statistical tests are reported in the figure legend and main text.
DATA AND SOFTWARE AVAILABILITY
Original scans of the radioactive gels (phosphorimager scans) for the ExoVII and ExoT in vitro blunting assay are available in Mendeley Data: https://doi.org/10.17632/2gsyvtr9c4.1. The accession number for the raw and analyzed sequencing data reported in this paper is GEO: GSE129529.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-CTCF, rabbit polyclonal | Millipore | Cat# 107-729; RRID: AB_441965 |
| Anti-Rad21, rabbit polyclonal | Abcam | Cat# ab992; RRID: AB_2176601 |
| Topo IIβ, rabbit polyclonal | Santa Cruz | Cat# sc13059, clone H-286: RRID: AB_2205866 |
| RP105 (Purified Rat Anti-Mouse CD180) | BD Biosciences | Cat# 552128; RRID: AB_394343 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Etoposide | Sigma Aldrich | Cat# E1383 |
| Trizol | Invitrogen | Cat# 15596026 |
| 5,6-Dichlorobenzimidazole 1-β-D-ribofuranoside (DRB) | Sigma Aldrich | Cat# D1916 |
| Mouse IFN Beta | PBL Assay Science | Cat# 12405-1 |
| Imatinib mesylate (STI-571) | Selleckchem | Cat# S1026 |
| Lipopolysaccharide (LPS) | Sigma Aldrich | Cat# L-2630 |
| Doxycycline hyclate | Sigma Aldrich | Cat# D9891 |
| 4-Hydroxytamoxifen | Sigma Aldrich | Cat# H7904 |
| IL-4 from mouse, Interleukin-4, recombinant | Sigma Aldrich | Cat# I-1020 |
| Lovastatin | Abcam | Cat# ab120614 |
| 3-indole acetic acid | Sigma Aldrich | Cat# I3750 |
| Recombinant TDP2 | Gao et al., 2012 | N/A |
| Epoxomicin | Sigma Aldrich | Cat# E3652 |
| Puregene Proteinase K enzyme | QIAGEN | Cat# 158920 |
| Puregene RNase A Solution | QIAGEN | Cat# 158924 |
| T4 DNA Polymerase | NEB | Cat# M0203L |
| T4 Polynucleotide Kinase | NEB | Cat# M0201L |
| DNA Polymerase I, Large (Klenow) Fragment | NEB | Cat# M0210L |
| Exonuclease T (ExoT) | NEB | Cat# M0265L |
| Exonuclease VII (ExoVII) | NEB | Cat# M0379L |
| Klenow Fragment (3′→5′ exo-) | NEB | Cat# M0212L |
| Quick Ligation Kit | NEB | Cat# M2200L |
| USER enzyme | NEB | Cat# M5505L |
| KAPA HiFi HotStart ReadyMix (2X) | KAPA Biosystems | Cat# KK2600 |
| MyOne Streptavidin C1 Beads | ThermoFisher | Cat# 650-01 |
| Agencourt AMPure XP beads | Beckman Coulter | Cat# A63881 |
| TDP2 substrate (Cy5-5′Tyrosine-ssDNA19-BHQ) | Zagnoli-Vieira et al., 2018 | N/A |
| TDP1 substrate (BHQ-ssDNA13-3′Tyrosine-Cy5) | Zagnoli-Vieira et al., 2018 | N/A |
| Critical Commercial Assays | ||
| Click-IT EdU Alexa Fluor 488 Flow Cytometry Assay Kit | ThermoFisher | Cat# C10425 |
| Click-iT RNA Alexa Fluor 594 Imaging kit | ThermoFisher | Cat# C10330 |
| Anti-CD43 (Ly-48) MicroBeads mouse | Miltenyi Biotech | Cat# 130-049-80 |
| NEBNext rRNA Depletion kit | NEB | Cat# E6310S |
| Click-iT Nascent RNA Capture Kit | ThermoFisher | Cat# C10365 |
| KAPA Library Quantification Kit | Kapa Biosciences | Cat# KK4824 |
| CHEF Mammalian Genomic DNA plug kit | Bio-Rad | Cat# 1703591 |
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
| Deposited Data | ||
| Raw and analyzed data | This paper | GSE129529 |
| ChIP-seq CTCF MEF B6 | Canela et al., 2017 | GSM2635593 |
| AsiSI and RAG preB exoT and exoT exoVII | Canela et al., 2016 | PRJNA326246 |
| Scans of the radioactive gels for the ExoVII and ExoT in vitro blunting assay | This paper | https://doi.org/10.17632/2gsyvtr9c4.1 |
| Experimental Models: Cell Lines | ||
| Pre-B cell lines | Bredemeyer et al., 2006 | N/A |
| HCT116 cell lines (gift from Masato Kanemaki) | Natsume et al., 2016 | N/A |
| Primary MEFs WT, Smc3−/F, WaplF/F | Busslinger et al., 2017 | N/A |
| Pre-B cell line Lig4−/−, tet-inducible ZFN, tet-inducible AsiSI | Canela et al., 2016 | N/A |
| Pre-B cells Tdp2−/− and Lig4−/− Tdp2−/− | This paper | N/A |
| Experimental Models: Organisms/Strains | ||
| C57BL/6NCr mice | Charles River | Strain code# 027 |
| Top2β−/− mice | Yang et al., 2000 | N/A |
| Oligonucleotides | ||
| END-seq adaptor 1, 5′-phosphate-GATCGGAAGAGCGTCGTGTAGGGAAAGAGTGUU[Biotin-dT]U[Biotin-dT]UUACACTCTTTCCCTACACGACGCTCTTCCGATCT-3′ | Canela et al., 2016 | N/A |
| END-seq adaptor 2, 5′-phosphate-GATCGGAAGAGCACACGTCUUUUUUUUAGACGTGTGCTCTTCCGATC*T-3′ | Canela et al., 2016 | N/A |
| TruSeq barcoded primer p5, 5′-AATGATACGGCGACCACCGAGATCTACACNNNNNNNNACACTCTTTCCCTACACGACGCTCTTCCGATC*T-3′ (N = barcode) | This paper | N/A |
| TruSeq barcoded primer p7, 5′-CAAGCAGAAGACGGCATACGAGANNNNNNNGTGACTGGAGTTCAGACGTGTGCTCTTCCGATC*T -3′ (N = barcode) | This paper | N/A |
| Illumina truncated adaptor 1, 5′-ACACTCTTTCCCTACACGACGCTCTTCCGATC*T-3′ | Canela et al., 2017 | N/A |
| Illumina truncated adaptor 2, 5′-phosphate-GATCGGAAGAGCACACGTCT-3′ | Canela et al., 2017 | N/A |
| YT18, 5′-tyrosine-TCCGTTGAAGCCTGCTTT-3′ | Gao et al., 2012 | N/A |
| T22 5′-GCGCAGCTAGCGGCGGATG*G*C*A-3′ | This paper (modified from Gao et al., 2012) | N/A |
| B36 5′-phosphate-TGCCATCCGCCGCTAGCTGCGCAAAGCAGGCTTCAA-3′ | Gao et al., 2012 | N/A |
| B40 5′-phosphate-TGCCATCCGCCGCTAGCTGCGCAAAGCAGGCTTCAACGGA-3′ | Gao et al., 2012 | N/A |
| ZNF-bio 5′-/Biosg/CAGAAGCCTTCAGTATGCACCA-3′ | This paper | N/A |
| I7-ZNF-nested-barcode 5′-CTCGGCATTCCTGCTGAACCGCTCTTCCGATCTNNNNNNNNTTCCACTTAACCCCAACTCCAG-3′ (N = barcode) |
This paper (modified from Hu et al., 2016) | N/A |
| Adaptor-upper 5′-GGTACACGACGCTCTTCCGATCTNNNNNN/3AmMO/-3′ | This paper (modified from Hu et al., 2016) | N/A |
| Adaptor-lower 5′-phosphate-AGATCGGAAGAGCGTCGTGTACC/3AmMO/-3′ | This paper (modified from Hu et al., 2016) | N/A |
| I5-bridge 5′-AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATC*T-3′ | This paper (modified from Hu et al., 2016) | N/A |
| P5-I5c 5′-AATGATACGGCGACCACCGAGATCTACACTCTTT*C-3′ | This paper (modified from Huetal., 2016) | N/A |
| P7-I7c 5′-CAAGCAGAAGACGGCATACGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTT*C-3′ | Hu et al., 2016 | N/A |
| Software and Algorithms | ||
| Bowtie 1.1.2 | Langmead et al., 2009 | https://sourceforge.net/projects/bowtie-bio/files/bowtie/1.1.2/ |
| MACS 1.4.3 | Zhang et al., 2008 | https://pypi.org/pypi/MACS/1.4.3 |
| UCSC database | Karolchik et al., 2004 | https://genome.ucsc.edu |
| UCSC Genome Browser | Kent et al., 2002 | https://genome.ucsc.edu |
| Bedtools | Quinlan and Hall, 2010 | https://github.com/arq5x/bedtools2 |
| R 3.3.1 | R Core Team, 2008 | https://www.r-project.org/ |
ACKNOWLEDGMENTS
We thank Sam John for comments on the manuscript; Barry Sleckman for the pre-B cell lines and Masato Kanemaki for HCT116 RAD21-mAID cells; Fred Alt for discussions; Yoram Louzoun for advice on quantitative modeling; Jennifer Wise and Kelly Smith for assistance with animal work; Xuguang Tai for assistance on primary lymphocytes isolation; and David Goldstein and the CCR Genomics core for sequencing support. Y.P. and S.N.H. are supported by the Intramural Program of the NIH, Center for Cancer Research (Z01 BC006161). Research in the laboratory of J.-M.P. is funded by Boehringer Ingelheim and grants by the Austrian Research Promotion Agency (FFG-834223FFG-852936) and the European Research Council H2020 (693949). A.C. is in part supported by the Kyoto University Hakubi Project. The A.N. laboratory is supported by the Intramural Research Program of the NIH, an Ellison Medical Foundation Senior Scholar in Aging Award (AG-SS-2633-11), the Department of Defense Idea Expansion (W81XWH-15-2-006) and Breakthrough (W81XWH-16-1-599) Awards, the Alex Lemonade Stand Foundation Award, and an NIH Intramural FLEX Award.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.molcel.2019.04.030.
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPORTING CITATIONS
The following references appear in the Supplemental Information: Chase and Richardson (1974).
REFERENCES
- Acunzo M, Romano G, Wernicke D, Balatti V, Rassenti LZ, dell’Aquila M, Kipps TJ, Pekarsky Y, and Croce CM (2014). Translocation t(2;11) in CLL cells results in CXCR4/MAML2 fusion oncogene. Blood 124, 259–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agostinho M, Rino J, Braga J, Ferreira F, Steffensen S, and Ferreira J (2004). Human topoisomerase IIalpha: targeting to subchromosomal sites of activity during interphase and mitosis. Mol. Biol. Cell 15, 2388–2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashour ME, Atteya R, and El-Khamisy SF (2015). Topoisomerase-mediated chromosomal break repair: an emerging player in many games. Nat. Rev. Cancer 15, 137–151. [DOI] [PubMed] [Google Scholar]
- Ban Y, Ho CW, Lin RK, Lyu YL, and Liu LF (2013). Activation of a novel ubiquitin-independent proteasome pathway when RNA polymerase II encounters a protein roadblock. Mol. Cell. Biol 33, 4008–4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredemeyer AL, Sharma GG, Huang CY, Helmink BA, Walker LM, Khor KC, Nuskey B, Sullivan KE, Pandita TK, Bassing CH, and Sleckman BP (2006). ATM stabilizes DNA double-strand-break complexes during V(D)J recombination. Nature 442, 466–470. [DOI] [PubMed] [Google Scholar]
- Busslinger GA, Stocsits RR, van der Lelij P, Axelsson E, Tedeschi A, Galjart N, and Peters JM (2017). Cohesin is positioned in mammalian genomes by transcription, CTCF and Wapl. Nature 544, 503–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callen E, Di Virgilio M, Kruhlak MJ, Nieto-Soler M, Wong N, Chen HT, Faryabi RB, Polato F, Santos M, Starnes LM, et al. (2013). 53BP1 mediates productive and mutagenic DNA repair through distinct phosphoprotein interactions. Cell 153, 1266–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canela A, Sridharan S, Sciascia N, Tubbs A, Meltzer P, Sleckman BP, and Nussenzweig A (2016). DNA breaks and end resection measured genome-wide by end sequencing. Mol. Cell 63, 898–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canela A, Maman Y, Jung S, Wong N, Callen E, Day A, Kieffer-Kwon KR, Pekowska A, Zhang H, Rao SSP, et al. (2017). Genome organization drives chromosome fragility. Cell 170, 507–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chase JW, and Richardson CC (1974). Exonuclease VII of Escherichia coli. Mechanism of action. J. Biol. Chem 249, 4553–4561. [PubMed] [Google Scholar]
- Cortes Ledesma F, El Khamisy SF, Zuma MC, Osborn K, and Caldecott KW (2009). A human 5′-tyrosyl DNA phosphodiesterase that repairs topo- isomerase-mediated DNA damage. Nature 461, 674–678. [DOI] [PubMed] [Google Scholar]
- Cowell IG, and Austin CA (2012a). Do transcription factories and TOP2B provide a recipe for chromosome translocations in therapy-related leukemia? Cell Cycle 11, 3143–3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowell IG, and Austin CA (2012b). Mechanism of generation of therapy related leukemia in response to anti-topoisomerase II agents. Int. J. Environ. Res. Public Health 9, 2075–2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowell IG, Sondka Z, Smith K, Lee KC, Manville CM, Sidorczuk-Lesthuruge M, Rance HA, Padget K, Jackson GH, Adachi N, and Austin CA (2012). Model for MLL translocations in therapy-related leukemia involving topoisomerase IIβ-mediated DNA strand breaks and gene proximity. Proc. Natl. Acad. Sci. USA 109, 8989–8994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Difilippantonio MJ, Zhu J, Chen HT, Meffre E, Nussenzweig MC, Max EE, Ried T, and Nussenzweig A (2000). DNA repair protein Ku80 suppresses chromosomal aberrations and malignant transformation. Nature 404,510–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsett Y, Zhou Y, Tubbs AT, Chen BR, Purman C, Lee BS, George R, Bredemeyer AL, Zhao JY, Sodergen E, et al. (2014). HCoDES reveals chromosomal DNA end structures with single-nucleotide resolution. Mol. Cell 56, 808–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson DO, Sekiguchi JM, Chang S, Frank KM, Gao Y, DePinho RA, and Alt FW (2000). The nonhomologous end-joining pathway of DNA repair is required for genomic stability and the suppression of translocations. Proc. Natl. Acad. Sci. USA 97, 6630–6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frock RL, Hu J, Meyers RM, Ho YJ, Kii E, and Alt FW (2015). Genome-wide detection of DNA double-stranded breaks induced by engineered nucleases. Nat. Biotechnol 33, 179–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao R, Huang SY, Marchand C, and Pommier Y (2012). Biochemical characterization of human tyrosyl-DNA phosphodiesterase 2 (TDP2/TTRAP): a Mg(2+)/Mn(2+)-dependent phosphodiesterase specific for the repair of topoisomerase cleavage complexes. J. Biol. Chem 287, 30842–30852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Herreros F, Romero-Granados R, Zeng Z, Alvarez-Quilón A, Quintero C, Ju L, Umans L, Vermeire L, Huylebroeck D, Caldecott KW, and Córtes-Ledesma F (2013). TDP2-dependent non-homologous end-joining protects against topoisomerase II-induced DNA breaks and genome instability in cells and in vivo. PLoS Genet. 9, e1003226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gùmez-Herreros F, Zagnoli-Vieira G, Ntai I, Martínez-Macías MI, Anderson RM, Herrero-Ruíz A, and Caldecott KW (2017). TDP2 suppresses chromosomal translocations induced by DNA topoisomerase II during gene transcription. Nat. Commun 8, 233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant CE, Bailey TL, and Noble WS (2011). FIMO: scanning for occurrences of a given motif. Bioinformatics 27, 1017–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haarhuis JHI, van der Weide RH, Blomen VA, Yanez-Cuna JO, Amendola M, van Ruiten MS, Krijger PHL, Teunissen H, Medema RH, van Steensel B, et al. (2017). The cohesin release factor WAPL restricts chromatin loop extension. Cell 169, 693–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoa NN, Shimizu T, Zhou ZW, Wang ZQ, Deshpande RA, Paull TT, Akter S, Tsuda M, Furuta R, Tsutsui K, et al. (2016). Mre11 is essential for the removal of lethal topoisomerase 2 covalent cleavage complexes. Mol. Cell 64, 580–592. [DOI] [PubMed] [Google Scholar]
- Hu J, Meyers RM, Dong J, Panchakshari RA, Alt FW, and Frock RL (2016). Detecting DNA double-stranded breaks in mammalian genomes by linear amplification-mediated high-throughput genome-wide translocation sequencing. Nat. Protoc 11, 853–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- lacovoni JS, Caron P, Lassadi I, Nicolas E, Massip L, Trouche D, and Legube G (2010). High-resolution profiling of gammaH2AX around DNA double strand breaks in the mammalian genome. EMBO J. 29, 1446–1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolchik D, Hinrichs AS, Furey TS, Roskin KM, Sugnet CW, Haussler D, and Kent WJ (2004). The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 32, D493–D496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, and Haussler D (2002). The human genome browser at UCSC. Genome Res. 12, 996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Zweig AS, Barber G, Hinrichs AS, and Karolchik D (2010). BigWig and BigBed: enabling browsing of large distributed data sets. Bioinformatics 26, 2204–2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiianitsa K, and Maizels N (2013). A rapid and sensitive assay for DNA-protein covalent complexes in living cells. Nucleic Acids Res. 41, e104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohn KW (1996). Beyond DNA cross-linking: history and prospects of DNA-targeted cancer treatment-fifteenth Bruce F. Cain Memorial Award Lecture. Cancer Res. 56, 5533–5546. [PubMed] [Google Scholar]
- Krivtsov AV, Figueroa ME, Sinha AU, Stubbs MC, Feng Z, Valk PJ, Delwel R, Döhner K, Bullinger L, Kung AL, et al. (2013). Cell of origin determines clinically relevant subtypes of MLL-rearranged AML. Leukemia 27, 852–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, and Salzberg SL (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KC, Bramley RL, Cowell IG, Jackson GH, and Austin CA (2016). Proteasomal inhibition potentiates drugs targeting DNA topoisomerase ll. Biochem. Pharmacol 103, 29–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long BH, Musial ST, and Brattain MG (1985). Single- and double-strand DNA breakage and repair in human lung adenocarcinoma cells exposed to etoposide and teniposide. Cancer Res. 45, 3106–3112. [PubMed] [Google Scholar]
- Manasanch EE, and Orlowski RZ (2017). Proteasome inhibitors in cancer therapy. Nat. Rev. Clin. Oncol 14, 417–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y, Desai SD, Ting CY, Hwang J, and Liu LF (2001). 26 S proteasome-mediated degradation of topoisomerase II cleavable complexes. J. Biol. Chem 276, 40652–40658. [DOI] [PubMed] [Google Scholar]
- Mathelier A, Zhao X, Zhang AW, Parcy F, Worsley-Hunt R, Arenillas DJ, Buchman S, Chen CY, Chou A, lenasescu H, et al. (2014). JASPAR 2014: an extensively expanded and updated open-access database of transcription factor binding profiles. Nucleic Acids Res. 42, D142–D147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natsume T, Kiyomitsu T, Saga Y, and Kanemaki MT (2016). Rapid protein depletion in human cells by auxin-inducible degron tagging with short homology donors. Cell Rep. 15, 210–218. [DOI] [PubMed] [Google Scholar]
- Nitiss JL (2009). DNA topoisomerase II and its growing repertoire of biological functions. Nat. Rev. Cancer 9, 327–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitiss JL, Soans E, Rogojina A, Seth A, and Mishina M (2012). Topoisomerase assays. Curr Protoc Pharmacol Chapter 3. Unit 3.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong CT, and Corces VG (2014). CTCF: an architectural protein bridging genome topology and function. Nat. Rev. Genet 15, 234–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pommier Y, Sun Y, Huang SN, and Nitiss JL (2016). Roles of eukaryotic topoisomerases in transcription, replication and genomic stability. Nat. Rev. Mol. Cell Biol 17, 703–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, and Hall IM (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team (2008). R: A language and environment for statistical computing. R Foundation for Statistical Computing; http://www.R-project.org/. [Google Scholar]
- Schellenberg MJ, Lieberman JA, Herrero-Rulz A, Butler LR, Williams JG, Muñoz-Cabello AM, Mueller GA, London RE, Cortés-Ledesma F, and Williams RS (2017). ZATT (ZNF451)-mediated resolution of topoisomerase 2 DNA-protein cross-links. Science 357, 1412–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stingele J, Bellelli R, and Boulton SJ (2017). Mechanisms of DNA-protein crosslink repair. Nat. Rev. Mol. Cell Biol 18, 563–573. [DOI] [PubMed] [Google Scholar]
- Tedeschi A, Wutz G, Huet S, Jaritz M, Wuensche A, Schirghuber E, Davidson IF, Tang W, Cisneros DA, Bhaskara V, et al. (2013). Wapl is an essential regulator of chromatin structure and chromosome segregation. Nature 501, 564–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uusküla-Reimand L, Hou H, Samavarchi-Tehrani P, Rudan MV, Liang M, Medina-Rivera A, Mohammed H, Schmidt D, Schwalie P, Young EJ, et al. (2016). Topoisomerase II beta interacts with cohesin and CTCF at topological domain borders. Genome Biol. 17, 182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Cuijk L, Vermeulen W, and Marteijn JA (2014). Ubiquitin at work: the ubiquitous regulation of the damage recognition step of NER. Exp. Cell Res 329, 101–109. [DOI] [PubMed] [Google Scholar]
- Vian L, Pekowska A, Rao SSP, Kieffer-Kwon KR, Jung S, Baranello L, Huang SC, El Khattabi L, Dose M, Pruett N, et al. (2018). The energetics and physiological impact of cohesin extrusion. Cell 173, 1165–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willmore E, Frank AJ, Padget K, Tilby MJ, and Austin CA (1998). Etoposide targets topoisomerase IIalpha and IIbeta in leukemic cells: isoform-specific cleavable complexes visualized and quantified in situ by a novel immunofluorescence technique. Mol. Pharmacol 54, 78–85. [DOI] [PubMed] [Google Scholar]
- Wutz G, Varnai C, Nagasaka K, Cisneros DA, Stocsits RR, Tang W, Schoenfelder S, Jessberger G, Muhar M, Hossain MJ, et al. (2017). Topologically associating domains and chromatin loops depend on cohesin and are regulated by CTCF, WAPL, and PDS5 proteins. EMBO J. 36, 3573–3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Li W, Prescott ED, Burden SJ, and Wang JC (2000). DNA Topoisomerase IIβ and Neural Development. Science 287, 131–134. [DOI] [PubMed] [Google Scholar]
- Yoshida MM, Ting L, Gygi SP, and Azuma Y (2016). SUMOylation of DNA topoisomerase IIα regulates histone H3 kinase Haspin and H3 phosphorylation in mitosis. J. Cell Biol 273, 665–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zagnoli-Vieira G, and Caldecott KW (2017). TDP2, TOP2, and SUMO: what is ZATT about? Cell Res. 27, 1405–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zagnoli-Vieira G, Bruni F, Thompson K, He L, Walker S, de Brouwer APM, Taylor R, Niyazov D, and Caldecott KW (2018). Confirming TDP2 mutation in spinocerebellar ataxia autosomal recessive 23 (SCAR23). Neurol Genet 4, e262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Z, Cortés-Ledesma F, El Khamisy SF, and Caldecott KW (2011). TDP2/TTRAP is the major 5′-tyrosyl DNA phosphodiesterase activity in vertebrate cells and is critical for cellular resistance to topoisomerase II-induced DNA damage. J. Biol. Chem 286, 403–09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A, Lyu YL, Lin CP, Zhou N, Azarova AM, Wood LM, and Liu LF (2006). A protease pathway for the repair of topoisomerase II-DNA covalent complexes. J. Biol. Chem 287, 35997–36003. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, and Liu XS (2008). Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Original scans of the radioactive gels (phosphorimager scans) for the ExoVII and ExoT in vitro blunting assay are available in Mendeley Data: https://doi.org/10.17632/2gsyvtr9c4.1. The accession number for the raw and analyzed sequencing data reported in this paper is GEO: GSE129529.