Abstract
Animal models of drug use have been employed for over 100 years to facilitate the identification of mechanisms governing human substance use and addiction. Most cross-species research on drug use/addiction examines behavioral overlap, but studies assessing neuro-molecular correspondence are lacking. Our study utilized transcriptome-wide data from the hippocampus and ventral tegmental area (VTA)/midbrain from a total of 35 human males with cocaine use disorder/controls and 49 male C57BL/6J cocaine/saline administering/exposed mice. We hypothesized that individual genes (differential expression) and systems of co-expressed genes (gene networks) would demonstrate appreciable overlap across mouse cocaine self-administration and human cocaine use disorder. We found modest, but significant associations between differentially expressed genes associated with cocaine self-administration (short access) and cocaine use disorder within meso-limbic circuitry, but non-robust associations with mouse models of acute cocaine exposure, (cocaine) context re-exposure and cocaine + context re-exposure. Investigating systems of co-expressed genes, we also found several validated gene networks with weak to moderate conservation between cocaine/saline self-administering mice and disordered cocaine users/controls. The most conserved hippocampal and VTA gene networks demonstrated substantial overlap (2,029 common genes) and included novel and previously implicated targets of cocaine use/addiction. Lastly, we conducted expression-based phenome-wide association studies of the nine common hub genes across conserved gene networks and found that they were associated with dopamine/serotonin function, cocaine self-administration and other relevant mouse traits. Overall, our study identified and characterized homologous transcriptional effects between mouse models of cocaine self-administration and human cocaine use disorder that may serve as a benchmark for future research.
Keywords: Cocaine Abuse/Dependence, Cocaine self-administration, Cross-species, Microarray, Multi-Omics, RNA-sequencing
Introduction
For over a hundred years animal models of drug use have provided mechanistic insight into human drug addiction1. Most of the knowledge regarding the neurobiological mechanisms contributing to human drug addiction is derived from extrapolating findings from animal studies. The modus operandi for drug use in animals is the self-administration paradigm (18,865 publications between 2009–2019 via PubMed), in which, animals perform a behavior to consume a drug or saline. Numerous variations of drug self-administration exist to model addiction related behavior, but our study predominately focuses on the classic (short-access) cocaine self-administration paradigm, which is considered a valid model for reinforcement learning and voluntary cocaine use2,3. These features seem important for the development of substance abuse, but controversy remains pertaining to the relationship between classic animal self-administration paradigms and human drug addiction4,5.
Generally the translational relevance for animal models of drug use is investigated through a behavioral lens6,7, whereby researchers assess the overlay of specific animal behaviors with behavioral characteristics or clinical symptomology of human drug use disorder. Such an approach is based almost entirely on face validity of the animal model and the recapitulation of the highly complex aspects of disordered drug use. Transcriptomic methods provide an alternate means of assessing the construct validity of behavioral assays and their relevance to the study of the mechanisms of human conditions. But, despite an expanding body of bioinformatics data, studies investigating cross-species neuro-molecular overlap have been limited – begging the question of whether, or to what degree, gene expression profiles are comparable between animal models of substance use and human drug use disorder.
Human post-mortem brain studies have produced useful information regarding the molecular hallmarks associated with the “end point” of psychiatric disorders8. While useful, these studies have many inherent limitations (for review see9). One pivotal drawback is determining whether human post-mortem findings are specific to a trait or confounded by co-occurring illness, various technical and/or biological biases. Additional complexities exist among drug use disorder research as it is challenging to separate the effects of substance exposure history, drug toxicity and underlying mechanism of addiction. Synchronizing gene expression from precarious post-mortem human data with precise and carefully controlled studies in animal models affords the opportunity to pinpoint and characterize certain neuro-molecular sources of variability; it may also help quantify the pathogenicity of a specific behavior with a diseased state. Such insights will also inform the ongoing dialogue surrounding comparative cross-species approaches in bio-behavioral research10.
The current study queried extant publically available resources to identify transcriptome-wide studies of animal cocaine self-administration and human cocaine use disorder (CUD; DSM-IV abuse and/or dependence). We utilized data from a recent cocaine use/self-administration study that assessed the transcriptomic alterations in multiple reward circuitry regions in mice11. We compared this with two post-mortem studies of individuals diagnosed with CUD versus cocaine-free controls with analogous brain regions – the hippocampus12 and the midbrain/ventral tegmental area (VTA13) Previous cross-species gene expression studies for substance use have been limited to a single brain region and typically have relied on a limited focus on candidate targets or systems14–16. Given the etiology of substance use disorders encompasses many neurological substrates and involves a multitude of molecular systems, research is warranted to examine various tissues and assess global convergence across the transcriptome. Additionally, genes/transcripts tend to act in a broader biological context (e.g., gene co-expression networks), highlighting the importance of investigating cross-species convergence for individual genes and interconnected systems of genes.
We hypothesized that, within meso-limbic reward circuitry 1) individual genes associated with mouse cocaine self-administration would moderately correspond to homologous human genes/transcripts correlated with human CUD and that 2) systems (or networks) of co-expressed genes would demonstrate appreciable transcriptional convergence across mice and men.
Materials and Methods
Human Samples
We obtained male post-mortem human hippocampal (CA4 to CA1 and dentate gyrus) RNA-sequencing (RNA-seq) data from the Sequence-Read-Archive (accession#: SRA029279). After outlier screening16, this sample included seven samples with a diagnosis of CUD and eight drug-free controls (Mage=39.3, s.d.=6.2, range=30–50; 66.67% European American; 33.33% African American). Samples from patients with CUD displayed a chronic pattern of cocaine use before death, died from cocaine toxicity and had prior diagnoses of cocaine abuse or dependence12 (DSM-IV). Note, since DSM-IV cocaine abuse and dependence have been reclassified into a singular DSM-V disorder, we defined these traits as CUD. Control subjects had no history of drug use and died from sudden cardiac or accidental events. All subjects were carefully matched on post-mortem interval (PMI; hours from death to freezer), ethnicity and age. No significant differences were observed for age and PMI across cocaine users and controls |t| < 0.51, all p > 0.623.
Leveraging publically available microarray data from the Gene Expression Omnibus (accession #: GSE54839), we used human post-mortem midbrain samples from twenty males13 (Mage=49.2, s.d.=3.9, range=45–59; 80.0% African American, 20.0% European American). Each individual had three technical replicates, such that, a single person’s sample was run on the HT-12 Bead Chip microarray platform three separate times. Extraction of midbrain samples, which included the ventral tegmental area (VTA) and the substantia nigra (SN), was guided by the presence of neuromelanin and demonstrated enrichment for dopamine transcripts (Bannon et al. 2014). Half of the samples had CUD (a documented history of DSM-IV cocaine abuse) and died from cocaine abuse, toxicity or cocaine-related aortic complications. The other half of midbrain samples had no history of drug or cocaine abuse and died from acute causes such as cardiovascular disease or gunshot wounds. Cases and controls were well matched on ethnicity and did not differ on RNA integrity (degree of RNA degradation), age and brain pH level, all |t| < 1.04, all p > 0.313, but the human midbrain and hippocampal samples did display demographic differences (see Supplementary Table S1 for more information).
Mouse Samples
We utilized classic cocaine-self administration RNA-seq data from male mice in two brain regions: the ventral hippocampus (n=15), and the VTA11 (n=12; Gene Expression Omnibus accession #: GSE110344). Note this sample consisted of genetically homogenous C57BL/6J mice (6–8 weeks old) were on a 12 hour reverse light-dark cycle, had ad libitum access to food and water, but restricted access to these resources during training and testing. After standard food training, jugular vein catheterization surgery was performed. Mice recovered for 3–5 days and entered single housing before starting self-administration. Mice were randomly assigned to either press a lever for cocaine (0.5 mg/kg/infusion; 20 second time-out; n=6–8 per group/brain-region) or to press a lever for saline (n=6–7 per group/brain-region). Over 10–15 days mice underwent 2-hour self-administration sessions on either a fixed-ratio one (days 5–10) or a fixed-ratio two schedule (for 4–5 days). Twenty-four hours after their last self-administration session mice were euthanized.
In addition to classic cocaine-self administration, the same study used a separate/independent cohort of male C57BL/6J mice to evaluate other cocaine-related behaviors (see11 for more details). Briefly, this cohort experienced classic cocaine or saline self-administration, but after their last trial they experienced a 30-day forced abstinence period before being put in the following test conditions: 1) acute cocaine or saline exposure (among saline self-administering mice; 10 mg/kg intraperitoneal injection of cocaine), 2) re-exposure to only the context of cocaine or saline self-administration (context re-exposure) and 3) re-exposure to cocaine or saline in the context of cocaine/saline self-administration (cocaine re-exposure; 10 mg/kg intraperitoneal injection of cocaine). After removal of two outliers (due to low read counts), we used a total of 10–12, 9–11 and 11 mouse hippocampus and VTA samples for acute cocaine exposure, context re-exposure and cocaine re-exposure groups, respectively. To control for age differences, these mice were euthanized at the same age as the other cohort of mice and all mice experienced cocaine self-administration post-puberty. This independent cohort was primarily used to test the validity of gene networks constructed from the classic cocaine self-administration mice and was also leveraged for post-hoc comparative analyses (see Supplementary Information).
Data Preparation
The current study processed RNA-seq with a unified approach. Specifically, we used Trimmomattic17 to remove Illumina adapters and low quality reads (Phred score < 12). Then, utilizing the Spliced Transcripts Alignment to a Reference tool18 we aligned RNA-seq data to either the human hg19 genome (Homo sapiens) or the mouse mm10 genome (Mus musculus). Our RNA-seq read alignment exceeded recommended thresholds19 for human (M%_reads_aligned=70.60%) and mouse RNA-seq data (M%_reads_aligned =91.89%). In total, we mapped an average of 13,623,098 reads (s.d.=2,889,467) to the human hg19 genome and 26,504,017 reads (s.d.=6,8965,611) to the mouse mm10 genome. Lastly, we transferred mapped RNA-seq reads into discrete RNA-seq counts via HTseq_0.6.120 and utilized Ensembl to annotate genes and transcripts for bioinformatics analyses.
Microarray data from the human midbrain/VTA had three technical replicates per sample/person. Correlations across technical replicates were strong (all Pearson’s r > 0.95) suggesting that gene expression could be collapsed across replicates. Therefore, we averaged microarray feature intensity recordings for individual array ids across technical replicates for each person/individual. Occasionally, multiple array ids exist for one individual gene, so we distilled array ids into a single gene-level expression value via the collapseRow function in R (with default parameters).
To account for gene expression variability among individual datasets, species and brain regions, we normalized data for each specific dataset separately (e.g., within species and within region). Performing normalization of only overlapping genes across species is thought to bias down-stream analyses21. Hence, we normalized transcriptome-wide data using all measured genes specific to each species and brain region, and consequently merged human and mouse datasets to homologous genes listed from the Mouse Genome Informatics website (http://www.informatics.jax.org/). Improperly accounting for differences across RNA-seq and microarray platforms can also be a source of bias for bioinformatic results, thus we harmonized our gene expression normalization using the voom (RNA-seq) and vooma (microarray) commands.
Statistical Analyses
For a schematic overview of our analyses and study design see Fig 1. To identify individual genes associated with both cocaine use/self-administration and CUD, we compared results across human and mouse differential expression analyses (case versus control). These analyses sought to minimize potential biases from analytical techniques by using the same differential expression platform (limma) that conducted empirical Bayes models22. To control for technical noise, all differential expression analyses adjusted for hidden batch effects using the svaseq package, which stabilizes error estimates23. Log fold change estimates from differential expression analyses are still estimated with noise – especially for genes with low baseline expression. To account for this noise, we focused our analyses on differential expression t-statistics, which balance effect size and standard errors for log fold change estimates, and thus, enabled us to (more confidently) characterize comparative gene expression signatures transcriptome-wide. We quantified the association of cross-species gene expression associations using Pearson product-moment correlations, standard linear regressions and linear Gaussian mixture models using FlexMix24. Our mixture modeling approach first estimated how many multivariate normal (Gaussian) distributions best fit the bivariate associations between cross-species differential expression results via the stepFlexMix command (# of replications = 10; # of iterations = 400). Across behaviors and brain regions, we found that two mixture distributions tended to yield the best fit to the data (see Supplementary Tables S2 and S3). Hence, we estimated all mixture models using two finite multivariate normal distributions weighting each data point by their prior probability of belonging to a distribution via the refit command. Our cross-species differential expression analyses included a range of 10,869 to 12,721 homologous genes/transcripts.
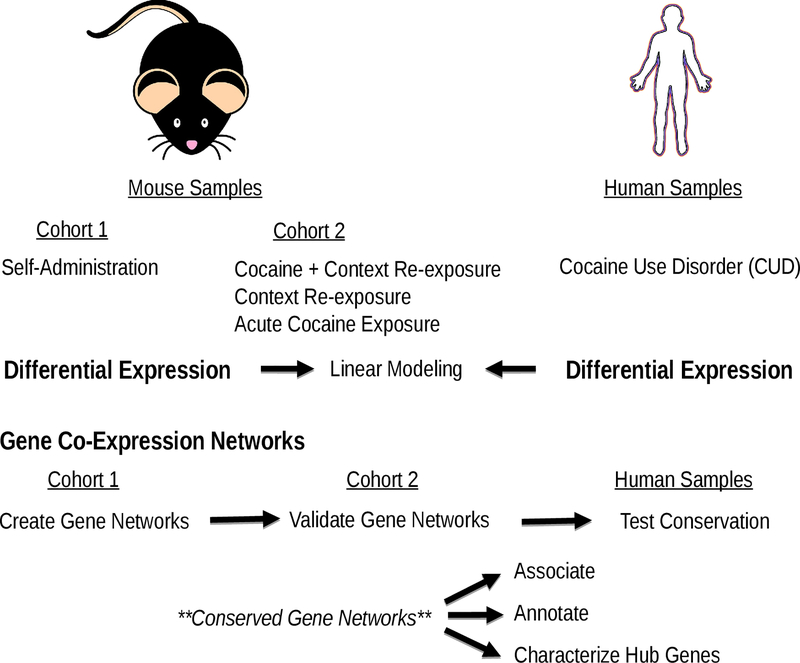
Figure 1:
Schematic overview of our study design. Mouse samples consisted of adult male C57BL/6J from two independent cohorts (from Walker et al. 2018). We also utilized two human samples of adult males diagnosed with cocaine abuse or dependence (Bannon et al. 2014; Zhou et al. 2011), which we defined as cocaine use disorder (CUD). Mouse and human samples used analogous brain regions (hippocampus and ventral tegmental area (VTA)/midbrain).
Next, we performed cross-species comparisons of gene co-expression patterns. First, we created signed WGCNA gene co-expression networks25 from mouse cocaine self-administration data. For computational limitations, we filtered out lowly expressed genes/transcripts (< 0.1 read count per sample), which resulted in 16,029 and 15,751 genes/transcripts for WGCNA modeling in the hippocampus and VTA, respectively. Using these genes/transcripts, we created a correlation matrix of the expression of all WGCNA genes/transcripts with themselves. We weighted these co-expression matrices by raising the correlations to a power of 12 in the hippocampus and a power of 7 in the VTA, which maximized WGCNA model fit (all scale free topologies > 0.86). Our study transformed weighted correlation matrices into topological overlap matrices using the TOMdist command, which we converted into distance metrics (Euclidean distance) and then further split into clusters of correlated genes using the hclust command. To make discrete WGCNA gene networks we parsed clusters of correlated genes via a standard dynamic tree-cutting algorithm (minimum network size = 50). In total, our WGCNA modeling created 22 hippocampal and 19 VTA gene co-expression networks, which were arbitrarily assigned to a color. Of note, we created gene co-expression networks using 12–15 mice, which is fewer samples than recommended for WGCNA modeling (n > 20). Therefore, we investigated our mouse WGNCA network reproducibility.
To test the stability, validity and conservation of these WGCNA networks, we used a three-step network preservation procedure similar to Vanderlinden et al.26. First, we used a within sample technique to assess the stability of WGCNA networks (e.g., if networks were created by chance). Specifically, we compared the reproducibility of the original gene networks to 100 bootstrapped samples of hippocampal and VTA mouse cocaine self-administration data. Then we quantified the validity of these WGCNA gene networks. We defined validity as whether WGCNA gene networks could be re-created in the same tissue and species but in an independent cohort (of cocaine or saline exposed mice). Lastly, we tested the conservation of WGCNA gene networks within tissue and across species. If mouse WGCNA gene networks were conserved, it would suggest that co-expression patterns from self-administering mice were reproducible in disordered human cocaine users and controls. As recommended by Langfelder et al.27, all WGCNA network preservation analyses used Zsummary statistics, which aggregate across seven measures of gene network reproducibility. Zsummary statistics below a score of 2 indicate that networks/co-expression patterns are not reproducible, whereas scores between 2–10 indicate weak to moderate reproducibility and Zsummary statistics above 10 are thought to be highly robust27.
For the most conserved WGCNA networks, we investigated their associations with mouse cocaine use behaviors and human CUD. If a WGCNA network was enriched for differentially expressed genes (via a Fisher’s Exact test) we considered it to be associated with a trait. Since only a few genes survived a BH-FDR correction for multiple testing (padj < 0.05) for mouse cocaine use behaviors, we used a nominal p-value threshold (p < 0.05) to define differentially expressed genes in mice but a standard BH-FDR threshold in humans (padj < 0.05). To increase power differentially expressed genes were combined across brain regions. Our functional annotation analyses investigated the potential function of conserved cocaine-related genes using the Kyoto Encyclopedia of Genes and Genomes (KEGG; Mouse 2019) pathways in Enrichr28. To control for false positives, we required significant KEGG pathways to survive correction for multiple testing (padj < 0.05) and surpass an enrichment odd’s ratio (OR) > 2.00.
Since hub genes are thought to represent biologically meaningful targets in disease progression29, we focused our analyses on hub genes from the most conserved gene networks. We defined hub genes as the top 1% of most intra-connected network genes (e.g., the most co-expressed with all other genes in the network). To better characterize the functional role of these genes in the brain we performed expression-based phenome-wide association analyses (ePheWASs30; https://systems-genetics.org/). Specifically, these analyses investigated the associations between the expression of a hub gene, in a specific brain region, with 1,250 unique “nervous system” traits – including both molecular and behavioral phenotypes - from the BXD mouse population (consisting of inbred lines derived from the progeny of a cross between C57BL/6J and DBA/2J mice). Significant ePheWAS were required to survive a Bonferroni p-value correction for multiple testing (p=0.05/1,250 or 0.0004).
Results
Differential Expression – Individual Genes
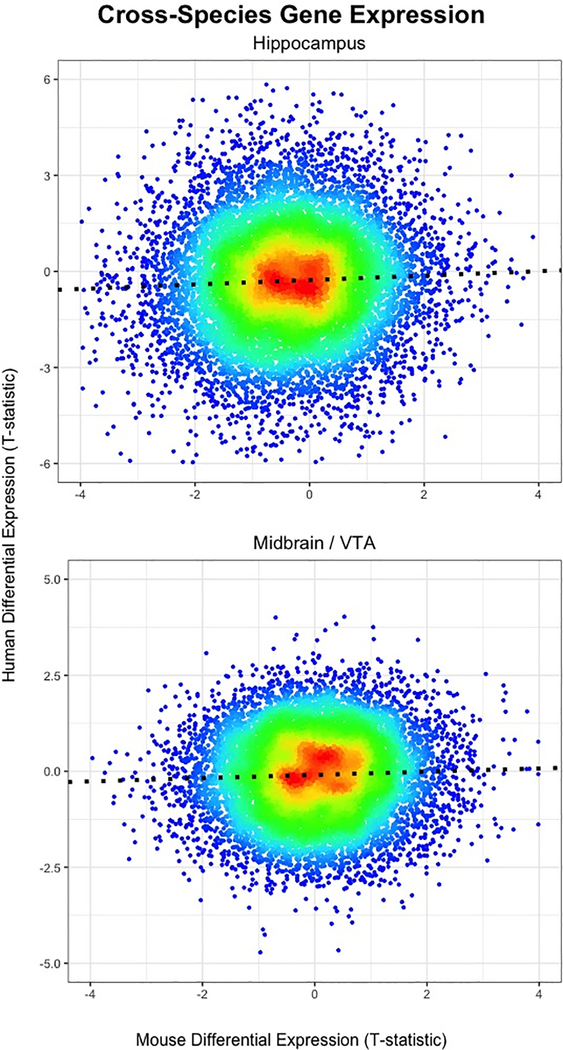
We conducted differential expression analyses within species and tissues to identify specific cocaine-related genes and then quantified the degree of overlap between mice and humans. The genes with increased expression in cocaine self-administering mice tended to also be increased in humans with CUD, in both the hippocampus, B = 0.076, s.e.=0.014, F(1, 12719) = 29.82, p = 4.82e-8, R2 = 0.0023, and VTA/midbrain, B = 0.042, s.e = 0.001, F(1, 11856) = 17.83, p=2.43e-5, R2 = 0.0014 (see Fig 2). While these associations were significant, in the expected direction and reproducible across brain regions, they were modest in magnitude and only accounted for ~0.1%−0.2% of the molecular variance observed in human CUD.
Figure 2:
Heat scatter plots showing overlap of individual genes/transcripts associated with cocaine self-administration in mice (x-axis; n=12–15) and human CUD (y-axis; n=15–20) in the hippocampus (top) and midbrain/VTA (bottom). Note that each dot represents a gene/transcript, which are color coded by their density, with high frequency/density regions shown in red and low frequency genes/transcripts represented in blue.
Perhaps modeling a mixture of distributions may better recapitulate the cross-species gene expression overlap. We fit linear mixture models and found analogous results to our standard linear regression analyses, with a little stronger cross-species relationship in the hippocampus (mixture distribution #2: B= 0.121, p = 4.05e-10 88.8% of genes; mixture distribution #1: B = −0.191, p = 0.074) and nearly identical findings in the VTA/midbrain (mixture distribution #2: B = 0.043, p = 0.001, 93.4% of genes; mixture distribution #1: B = 0.035, p = 0.057; see Supplementary Figure S1). Our study reports these summary data in Supplementary Files S1 and S2 and functionally annotated genes within the differentially expressed genes among the significant mixture distributions (mixture distribution # 2) for each brain region. In the hippocampus, we found enrichment for synaptic plasticity processes, cell death, neurotransmitter, hormonal, and calcium signaling pathways (see Supplementary Table S4), but we detected no significant enrichment in the VTA/midbrain (all padj > 0984).
We also performed post-hoc analyses that evaluated the cross-species differential expression overlap from three additional cocaine behaviors in mice: 1) acute cocaine exposure, 2) context re-exposure and 3) cocaine re-exposure. But, we found no replicable associations with human CUD across brain regions (see Supplementary Figure S1 and Supplementary Table S5).
Gene Co-expression Analyses – Systems of Genes
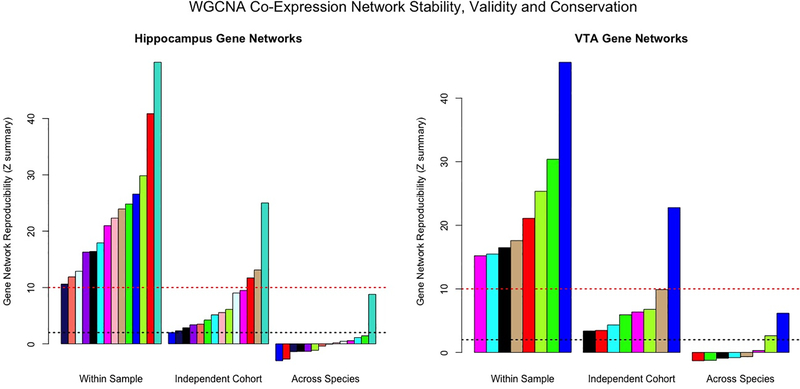
Our next step was to address the cross-species correspondence of biological systems in the hippocampus and VTA/midbrain. Using mice from the classic cocaine self-administration model, we created 22 data-driven WGCNA gene co-expression networks in the hippocampus and 19 WGCNA gene co-expression networks in the VTA. We focused our analyses on the stable (reproducible within sample) and valid (preserved in an independent mouse cohort) gene co-expression networks, which included 14-gene co-expression networks in the hippocampus (all Zsummary > 2.017) and eight in the VTA (all Zsummary > 3.367; see Fig 3 or Supplementary Table S6). We investigated whether these robust WGCNA networks (gene co-expression patterns) were conserved within-tissue and across species. In the hippocampus, only the turquoise WGCNA gene network (3,218 genes) demonstrated moderate conservation in human CUD and controls, Zsummary=8.793. In the mouse VTA, two gene co-expression networks demonstrated weak to moderate conservation in the human midbrain: the green-yellow network (212 genes; Zsummary=2.631) and the blue network (3,726 genes; Zsummary=6.168).
Figure 3:
Barplot displaying the reproducibility/robustness of WGCNA gene co-expression networks derived from the mouse hippocampus (left) and VTA (right). Each bar represents a specific gene network - each arbitrarily assigned to a color - and constructed from cocaine self-administration mouse data. The three sections of the x-axes show whether gene networks were reproducible within sample (100 bootstrapped samples), in a separate group of cocaine/saline-exposed mice (independent cohort/same study; Walker et al. 2018) or conserved in post-mortem human CUD/controls (across species). The y-axes are the Zsummary preservation statistic values. Bars surpassing the dashed red line indicate gene networks that are highly robust and bars that exceed the dashed black line are considered weak to moderately reproducible. Note the turquoise hippocampal gene network and blue VTA gene network are highly stable/valid and are weak to moderately conserved in human tissue.
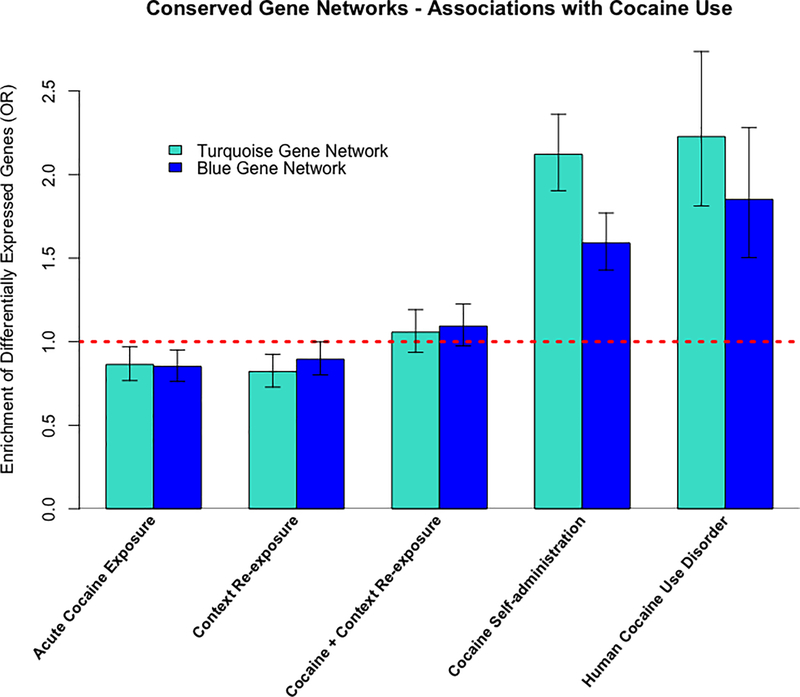
We selected the most conserved WGCNA gene co-expression networks per brain region for follow-up investigation - the turquoise hippocampal network (Supplementary File S3) and blue VTA network (Supplementary File S4). The turquoise and blue gene networks were associated with cocaine self-administration (all p < 2.2e-16) and human CUD (all p < 4.07e-09) but not acute cocaine exposure, context re-exposure or cocaine and context re-exposure (see Fig 4). Our functional annotation analyses identified enrichment for cocaine/drug addiction, neurotransmitter signaling, synaptic plasticity processes and various other KEGG pathways that were both shared and specific among brain regions (see Table 1). We further investigated the overlap between the turquoise and blue gene networks and found over 2,000-shared genes across networks (see Supplementary Figure S2). Despite, the substantial similarities in gene co-expression network structure, we found minimal overlap of differentially expressed genes (p < 0.05) across the hippocampus and VTA from the mouse cocaine self-administration paradigm (see Supplementary Figure S3). We tested whether these networks included just a high baseline expression of genes, but our analyses discovered no significant differences in average baseline expression in blue or turquoise networks than other genes (see Supplementary Tables S7 and S8). Overall, these results suggest substantial transcriptional overlap across brain regions at the systems level but not at the individual gene/transcript level.
Figure 4:
Association of conserved gene networks with cocaine use tested via enrichment of differentially expressed genes from mouse cocaine use (p < 0.05) and human cocaine use disorder (padj < 0.05).
Table 1.
KEGG Pathway Enrichment for Conserved WGCNA Gene Co-expression Networks
| Potential Functions of Conserved Systems/Networks Across Humans and Mice | ||||||
|---|---|---|---|---|---|---|
| Hippocampus (Turquoise) | VTA (blue) | |||||
| KEGG Pathway | OR | p-value | padj | OR | p-value | padj |
| Synaptic Vesicle Cycle | 3.25 | 3.19E-13 | 3.12E-11 | 3.04 | 1.87E-13 | 2.83E-11 |
| Alzheimer's Disease | 2.46 | 6.92E-14 | 1.05E-11 | 2.18 | 1.63E-11 | 1.23E-09 |
| Oxidative Phosphorylation | 2.47 | 4.62E-11 | 3.50E-09 | 2.16 | 7.23E-09 | 2.19E-07 |
| Parkinson's Disease | 2.30 | 1.03E-09 | 4.46E-08 | 2.31 | 1.90E-11 | 1.15E-09 |
| Long Term Potentiation | 2.70 | 1.09E-07 | 2.53E-06 | 2.60 | 3.70E-08 | 9.33E-07 |
| Dopaminergic Synapse | 2.13 | 1.87E-07 | 3.15E-06 | 2.22 | 1.05E-09 | 4.56E-08 |
| GABAergic synapse | 2.36 | 4.79E-07 | 6.30E-06 | 2.24 | 4.14E-07 | 6.60E-06 |
| Glutamatergic synapse | 2.30 | 5.23E-08 | 1.32E-06 | 2.10 | 3.02E-07 | 5.72E-06 |
| Cardiac muscle Contraction | 2.16 | 4.40E-05 | 3.10E-04 | 2.10 | 2.39E-05 | 2.69E-04 |
| Endocytosis | NS | NS | NS | 2.15 | 3.64E-16 | 1.10E-13 |
| Amphetamine Addiction | NS | NS | NS | 2.57 | 5.76E-08 | 1.34E-06 |
| Ubiquitin Mediated Proteolysis | NS | NS | NS | 2.02 | 1.82E-07 | 3.94E-06 |
| Nicotine Addiction | NS | NS | NS | 2.45 | 9.55E-05 | 7.42E-04 |
| Cocaine Addiction | NS | NS | NS | 2.27 | 1.48E-04 | 9.95E-04 |
| Long Term Depression | NS | NS | NS | 2.06 | 2.91E-04 | 1.80E-03 |
| … Sodium Reabsorption | NS | NS | NS | 2.01 | 5.51E-03 | 2.20E-02 |
| Axon Guidance | 2.15 | 2.96E-10 | 1.79E-08 | NS | NS | NS |
| … Endocannabanoid Signaling | 2.08 | 1.23E-07 | 2.67E-06 | NS | NS | NS |
| Adrenergic Signaling …. | 2.07 | 2.08E-07 | 3.31E-06 | NS | NS | NS |
| Spliceosme | 2.13 | 2.49E-07 | 3.76E-06 | NS | NS | NS |
| ErbB Signaling Pathway | 2.23 | 8.36E-06 | 7.45E-05 | NS | NS | NS |
| Gap Junction | 2.18 | 1.43E-05 | 1.14E-04 | NS | NS | NS |
| SNARE … in Vesicular Transport | 2.27 | 3.63E-03 | 1.36E-02 | NS | NS | NS |
| Circadian Rhythm | 2.08 | 1.50E-02 | 4.46E-02 | NS | NS | NS |
This table shows significantly enriched KEGG pathways (padj < 0.05 & OR > 2.0) common and specific to the turquoise hippocampal and blue VTA gene co-expression networks. Note we highlighted in grey that the blue VTA gene network was enriched for the Cocaine Addiction KEGG pathway.
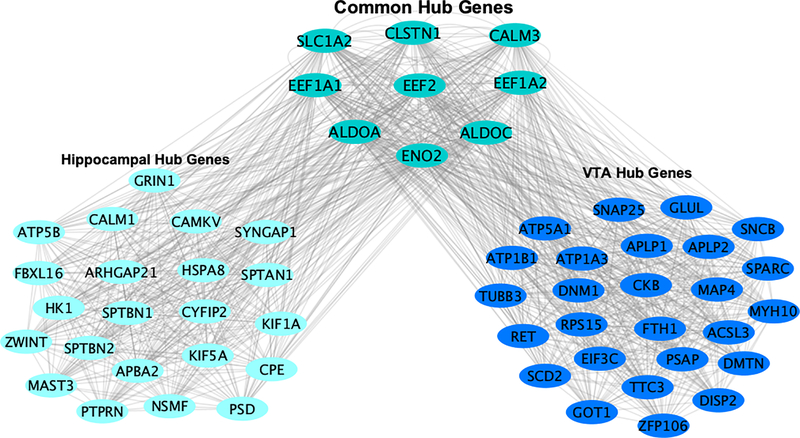
We concentrated on hub genes for the moderately conserved turquoise and blue gene co-expression networks. In total, we identified 32 and 37 hub genes for the turquoise (hippocampus) and blue (VTA) gene networks, respectively. Note that nine hub genes were in the top 99% of gene network intra-connectedness in both the turquoise and blue gene co-expression networks, which we defined as common hub genes. We visualized both the common and specific hub gene structure across networks/brain regions (see Fig 5). Our results suggest that common hub genes could have an important role in multiple brain regions, but the function of these hub genes in the context of cocaine use is not fully understood.
Figure 5:
Hub genes are defined as being in the top 99th %tile of gene network intraconnectedness. Common hub genes are in the top 1% of gene network connectedness in both the turquoise hippocampal gene network (left) and blue VTA gene network (right). Region specific hub genes are in the top 1% of their respective gene network but not in the top 1% of connectedness in the other gene network.
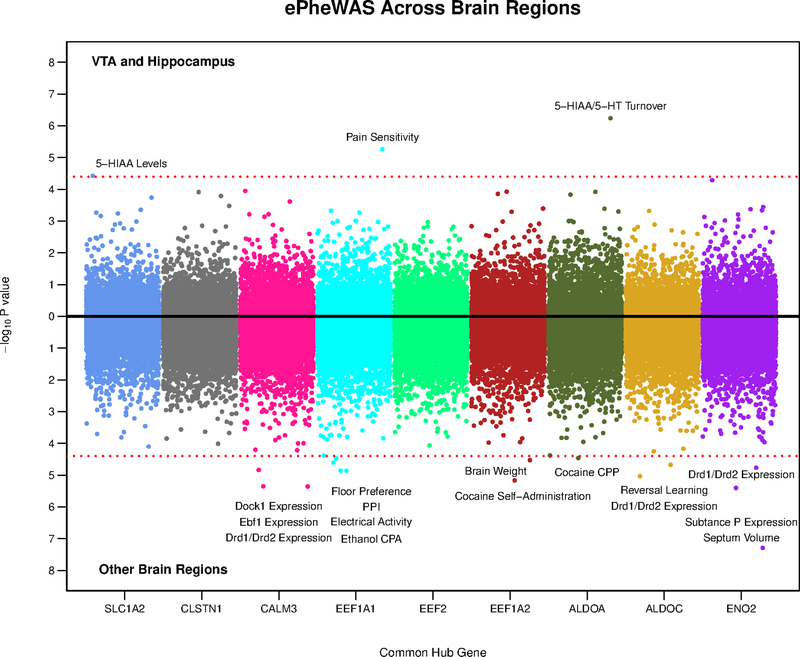
To investigate specific behavioral mechanisms related to the expression of our common hub genes, we conducted ePheWASs using BXD mice30 - first in the VTA and hippocampus and subsequently in eight other brain regions, including: the amygdala, cerebellum, hypothalamus, nucleus accumbens, striatum, pituitary, prefrontal cortex and whole brain. In total, our common hub genes exhibited eighteen significant neuro-transcriptional associations (with thirteen unique mouse traits) in eight different brain regions (see Fig 6). These results suggest that our common hub genes exert a broad functional role spread across the brain (see Supplementary File S5). Most significant ePheWAS associations were molecular. Slc1a2, Calm3, Aldoa, Aldoc and Eno2 were associated with dopamine and serotonin function in the VTA and striatum. Additionally, we found significant ePheWAS associations between Eef1a1, Aldoa and Eef1a2 with alcohol conditioned place aversion31 cocaine conditioned place preference32 and cocaine self-administration33, respectively. Overall, these analyses better characterize the behavioral and neuro-molecular role of our common hub genes within particular brain regions.
Figure 6:
Miami plot showing the results of our ePheWASs of common hub genes from the turquoise and blue gene networks. Each dot is a nervous system phenotype/trait (1,250 unique traits), which are color coded by hub gene (x-axis). The y-axis represents the –log10 p-value from the association of a hub gene’s expression with a particular trait in a certain brain region. The top part of the plot shows results from the VTA or hippocampus and the bottom includes results from eight other cortical/sub-cortical brain regions. The dashed red line denotes the Bonferonni correction for multiple testing (0.05/1,250) and thus, all dots surpassing this threshold indicate significant associations between the expression of a gene with a trait. Note that our ePheWAS analyses utilized BXD mice, which are derived by crossing C57BL/6J and DBA/2J inbred mice. PPI = pre-pulse inhibition; CPP = conditioned place preference; CPA = conditioned place/taste aversion.
Discussion
The current study identified modest to moderate conservation of transcriptional responses across mouse cocaine self-administration and human CUD within reward circuitry. Specifically, we found convergent gene expression for individual genes/transcripts and systems of co-expressed genes/transcripts in the hippocampus and midbrain/VTA for the ubiquitous short access, intravenous cocaine self-administration model. Our study is the first to quantify the extent to which meso-limbic gene expression findings from mouse models of cocaine self-administration recapitulate human CUD.
While the correlated neuro-transcriptional response between mouse cocaine self-administration and human CUD was significant, and in the expected direction, it was surprisingly modest. We considered various interpretations for these potentially sobering effect sizes. Inbred mice pressing a cocaine lever for two-weeks and individuals with CUD have different degrees of exposure duration, use severity and genetic backgrounds, which may recruit separate dimensions of biological variation. Perhaps the molecular mechanisms governing reward learning are akin to the development of substance use behaviors, but not necessarily tapping into the core pathogenic features of the addictive process, or at least the “end state.” Thus, the neuro-pathophysiology of compulsive human cocaine use may be far more complex than the transcriptional thumbprint of a well-controlled operant behavior. Alternative models of drug use may improve cross-species prediction. Although, acute cocaine exposure, context re-exposure and cocaine re-exposure models tended to be less robust predictors of human CUD than the classic self-administration paradigm (see Supplementary Figure S2).
On the other hand, human post-mortem brain tissue may be responsible for dampening cross-species effect sizes. If only a slim proportion of the observed human gene expression is specific to CUD then our cross-species effects may seem small but could be capturing a fair amount of disordered cocaine use variability and be obfuscated by potential human confounds (co-occurring illness, cocaine toxicity etc.). It is also possible that our cross-species comparisons do not follow a positive linear association. For instance, during drug tolerance, neurotransmission genes may demonstrate expression in one direction from short-term or acute use, but exhibit compensatory expression in the opposite direction through chronic drug exposure, which has been observed in cross-species alcohol research15. The aforementioned interpretations are not mutually exclusive and, in practice, could act in a combinatorial way to mitigate cross-species associations. Regardless, the current study speaks to the difficulty of modeling and capturing the molecular relevance of specific animal systems for complex human diseases. We rely on models as simplifications of complicated traits, to enable examination of specific facets with precise experimental perturbations, but no model in isolation recapitulates the daunting complexity of the human experience.
We extend previous cross-species drug use research34 by investigating the convergence of co-expression patterns across species in the hippocampus and VTA/midbrain. While only a few of the mouse gene co-expression networks surpassed the threshold of (weak to moderate) reproducibility in analogous human tissues, these systems included thousands of genes. Reassuringly, we found that the most conserved gene co-expression networks included numerous candidates from the cocaine addiction pathway and demonstrated associations with mouse cocaine self-administration and human CUD. To identify critical targets of these conserved systems, we honed in on the major players (hub genes), which may regulate the expression and/or function for a multitude of these genes. These genes tended to be involved in broad nervous system functions, such as: synaptic plasticity (Cyfip2, Ret, Tubb3, Sptbn1, Sptbn2, Spatn2), glutamate transmission (Slc1a2, Grin1, Glul) and calcium signaling (Calm1, Calm2, Calm3, Atp1b1, Camkv) potentially indicating a conserved and pleotropic role for more general dimensions of drug use and/or addiction. Accordingly, previous research demonstrates genome-wide associations of these genes with other human substance use disorder traits commonly co-morbid with cocaine abuse/dependence, including: alcohol (Arhgap2135), tobacco (Kif1a36), cannabis (Ret37) and heroin (Myh1038) dependence symptoms, as well as internalizing (Psd39, Tubb340 and Zwint41) and externalizing psychiatric disorders (Dnm142 and Fbxl1643).
From the conserved VTA and hippocampus gene networks, we observed nine common hub genes that may modulate brain function in multiple regions. Notably, over half of these genes (Calm3, Aldoc, Clstn1, Eef1a1 and Slc1a2) were also hub genes in a PFC gene network associated with human CUD that was enriched for analogous KEGG pathways44. It is possible that common hub genes recruit similar biological systems relevant to cocaine addiction across various brain regions. The expression of our common hub genes were also associated with relevant traits from the mouse realm - such as dopamine and serotonin function (Slc1a2, Calm3, Aldoa, Aldoc and Eno2), cocaine reward and/or memory (Aldoa) and cocaine self-administration (Eef1a2) in the striatum, VTA, hypothalamus and cerebellum. In sum, we identify and characterize critical genes from conserved biological systems relevant to cocaine use and rank targets for follow-up investigation within particular tissues.
The current study has various limitations. Our sample sizes were quite small and thus our results may be capturing a fair amount of noise. Ascertainment of brain tissues was performed in similar but not identical neurobiological substrates. That is, mouse tissues were collected from the ventral hippocampus and VTA, whereas human samples used tissue from the entire hippocampus and dopamine enriched regions of the midbrain (VTA and substantia nigra). Another inconsistency regarding our cross-species comparisons was that human samples differed in various technical and demographic variables (Supplementary Table S1) and their findings were a result of heterogeneous genetic predisposition and environmental exposure, whereas the C57BL/6J mice were matched on age, genetics and stemmed from very controlled environmental conditions. We observed the strongest cross-species overlap among gene co-expression networks, but WGCNA networks were derived from a single genetic line of mice exposed to one environmental perturbation. These analyses may be more appropriate among genetically diverse strains of mice (e.g. diversity outbred), which have demonstrated robust neuro-molecular overlap with human CUD45.
In light of the current study, we suggest various directions for future research. Relatively large post-mortem human brain samples are publically available for alcohol46 and opiates47 and could be integrated with a multitude of traits from model systems. Animal models of compulsive drug use (e.g., escalated use, aversion resistant users etc.) have apparent face valid behavioral correspondence to clinical symptomology of drug use disorders48, but whether, or to what extent, these molecular mechanisms recapitulate genuine features of human addiction is not known. Because there are ten times more expression studies for cocaine use in animals than humans49, comparative neuro-transcriptomic work would benefit from selecting human post-mortem brain regions that are supported by the vast literature and bioinformatics database in animals (e.g., striatum/nucleus accumbens). Unfortunately, the largest post-mortem human brain study on cocaine was conducted in dorsal-lateral PFC neurons50, which seems to lack a homologous substrate in the rodent brain. Heterogeneity in cell types across species can pose an additional bias from cross-species comparisons51. Thus, a cleaner signal may emerge from integrating single cell bioinformatics analyses across species. Ideally, cross-species research will aggregate across various layers of the transcriptome and employ global integrative strategies incorporating a multitude of model systems and human traits to illuminate the molecular crossroads among particular models and disease processes.
In conclusion, this is one of the first studies to test the extent to which neuro-molecular correlates identified from animal models of cocaine use generalize to humans diagnosed with CUD. Our analyses characterize and provided insight into specific targets at the intersection of an established mouse paradigm and a corresponding human trait. The current study, thus, contributes to the burgeoning dialogue surrounding the pre-clinical, theoretical and empirical implications of animal research for human disease. We found that meso-limbic gene expression from a single, granular animal paradigm applied in a single strain of mice recapitulated only a modest fraction of the transcriptional effects observed in human disease. To fully capture the complexity of human addiction a broader spectrum of model organism diversity and experimental breadth needs to be encompassed in cross-species analyses.
Supplementary Material
Acknowledgements
We express our gratitude towards the funding sources that supported our project: P50 DA039841, R01 DA037927 and DP1 DA042103.
Data availability
We obtained human cocaine use disorder data from Sequence-Read-Archive (accession#: SRA029279) and the Gene Expression Omnibus (accession#: (accession #: GSE54839). The RNA-seq data for mouse cocaine use is available on Gene Expression Omnibus accession (accession#: GSE110344).
References
- 1.Biberfeld J (1916). Zur Kenntnis der Kreislaufwirkung einiger Chinaalkaloide und ihres Verhaltens im Organismus. Archiv F. Experiment. Path. u. Pharmakol. 46, 362–382. [Google Scholar]
- 2.Panlilio LV, & Goldberg SR (2007). Self-administration of drugs in animals and humans as a model and an investigative tool. Addiction. 102, 1863–1870. 10.1111/j.1360-0443.2007.02011.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Planeta CS (2013). Animal models of alcohol and drug dependence. Revista Brasileira Psiquiatria. 35, 140–146. 10.1590/1516-4446-2013-1149. [DOI] [PubMed] [Google Scholar]
- 4.Ahmed SH (2012). The science of making drug-addicted animals. Neuroscience. 211, 107–125. 10.1016/j.neuroscience.2011.08.014. [DOI] [PubMed] [Google Scholar]
- 5.Viveros M-P, Mendrek A, Paus T, Belen L-RA, Marco EM, Yehuda R, … Wagner EJ (2012). A comparative, developmental, and clinical perspective of neurobehavioral sexual dimorphisms. Fronteirs in Neuroscience. 6, 1–21. 10.3389/fnins.2012.00084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Katz JL, & Higgins ST (2003). The validity of the reinstatement model of craving and relapse to drug use. Psychopharmacology. 168, 21–30. 10.1007/s00213-003-1441-y. [DOI] [PubMed] [Google Scholar]
- 7.Roberts DCS, Morgan D, & Liu Y (2007). How to make a rat addicted to cocaine. Prog Neuropsychopharmacol Biol Psychiatry. 31(8), 1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Farris SP, Riley BP, Williams RW, Mulligan MK, Miles MJ, Lopez MF, … Mayfield RD (2018). Cross-species molecular dissection across alcohol behavioral domains. Alcohol. 72, 19–31. 10.1016/j.alcohol.2017.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mccullumsmith RE, Hammond JH, Shan D, & Meador-woodruff JH (2013). Postmortem Brain : An Underutilized Substrate for Studying Severe Mental Illness. Neuropsychopharmacology. 39(1), 65–87. 10.1038/npp.2013.239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cates H, Schoenrock SA, Guglielmo G. De, Benca-bachman CE, Shu C, & Kallupi M (2019). National Institute on Drug Abuse genomics consortium white paper : Coordinating efforts between human and animal addiction studies. Genes Brain and Behavior. 18, 1–12. 10.1111/gbb.12577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Walker DM, Cates HM, Loh YHE, Purushothaman I, Ramakrishnan A, Cahill KM, … Nestler EJ (2018). Cocaine Self-administration Alters Transcriptome-wide Responses in the Brain’s Reward Circuitry. Biological Psychiatry. 84(12), 867–880. 10.1016/j.biopsych.2018.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou Z, Yuan Q, Mash DC, & Goldman D (2011). Substance-specific and shared transcription and epigenetic changes in the human hippocampus chronically exposed to cocaine and alcohol. PNAS. 108(16). 10.1073/pnas.1018514108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bannon MJ, Johnson MM, Michelhaugh SK, Hartley ZJ, Halter SD, David JA, … Schmidt CJ (2014). A molecular profile of cocaine abuse includes the differential expression of genes that regulate transcription, chromatin, and dopamine cell phenotype. Neuropsychopharmacology. 39(9), 2191–2199. 10.1038/npp.2014.70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Egervari G, Landry J, Callens J, Fullard JF, Roussos P, Keller E, & Hurd YL (2017). Striatal H3K27 acetylation linked to glutamatergic gene dysregulation in human heroin abusers holds promise as therapeutic target. Biological Psychiatry. 81(7), 585–594. 10.1016/j.biopsych.2016.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Enoch M, Zhou Z, Kimura M, Mash DC, Yuan Q, & Goldman D (2012). GABAergic Gene Expression in Postmortem Hippocampus from Alcoholics and Cocaine Addicts; Corresponding Findings in Alcohol-Naïve P and NP Rats. Plos ONE. 7(1), 1–11. 10.1371/journal.pone.0029369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huggett SB, & Stallings MC (2019). Cocaine’omics: Genome-wide and transcriptome-wide analyses provide biological insight into cocaine use and dependence. Addiction Biology. (January), 1–10. 10.1111/adb.12719. [DOI] [PubMed] [Google Scholar]
- 17.Bolger AM, Lohse M, & Usadel B (2014). Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics, 30(15). 2114–2120. 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, … Gingeras TR (2013). STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 29(1), 15–21. 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Conesa A, Madrigal P, Tarazona S, Gomez-cabrero D, Cervera A, Mcpherson A, … Mortazavi A (2016). A survey of best practices for RNA-seq data analysis. Genome Biology. 17(13), 1–19. 10.1186/s13059-016-0881-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anders S, Pyl PT, & Huber W (2015). Genome analysis HTSeq — a Python framework to work with high-throughput sequencing data. Bioinformatics. 31(2), 166–169. 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liao B, & Zhang J (2004). Low Rates of Expression Profile Divergence in Highly Expressed Genes and Tissue-Specific Genes During Mammalian Evolution. 10.1093/molbev/msj119. [DOI] [PubMed]
- 22.Smyth GK (2005). Limma : Linear Models for Microarray Data. [Google Scholar]
- 23.Leek JT (2014). Svaseq: Removing batch effects and other unwanted noise from sequencing data. Nucleic Acids Research. 42(21), e161. 10.1093/nar/gku864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leisch F FlexMix : A General Framework for Finite Mixture Models and Latent Class Regression in R; 2004. [Google Scholar]
- 25.Langfelder P, & Horvath S (2008). WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 9, 559. 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vanderlinden LA, Saba LM, Kechris K, Miles MF, Hoffman PL, & Tabakoff B (2013). Whole Brain and Brain Regional Coexpression Network Interactions Associated with Predisposition to Alcohol Consumption. PLoS ONE. 8(7). 10.1371/journal.pone.0068878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Langfelder P, Luo R, Oldham MC, & Horvath S (2011). Is my network module preserved and reproducible? PLoS Computational Biology. 7(1). 10.1371/journal.pcbi.1001057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, … Ma’ayan A (2016). Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Research. 44(W1), W90–7. 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Langfelder P, Mischel PS, & Horvath S (2013). When Is Hub Gene Selection Better than Standard Meta-Analysis? PLoS ONE. 8(4). 10.1371/journal.pone.0061505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li H, Wang X, Rukina D, Morgenthaler S, Williams RW, Auwerx J, … Auwerx J (2018). An Integrated Systems Genetics and Omics Toolkit to Probe Gene Function Article An Integrated Systems Genetics and Omics Toolkit to Probe Gene Function. Cell Systems. 6, 90–102. [DOI] [PubMed] [Google Scholar]
- 31.Risinger FO, & Cunningham CL (2006). Ethanol-Induced Taste Aversion in BXD Recombinant Inbred Mice. Alcoholism Clinical & Experimental Research. 22, 1234–1244. [PubMed] [Google Scholar]
- 32.Philip VM, Duvvuru S, Gomero B, Ansah TA, Blaha CD, Cook MN, … Chesler EJ (2010). High-throughput behavioral phenotyping in the expanded panel of BXD recombinant inbred strains. Genes, Brain and Behavior. 9, 129–159. 10.1111/j.1601-183X.2009.00540.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dickson PE, Miller MM, Calton MA, Bubier JA, Cook MN, Goldowitz D, … Mittleman G (2016). Systems genetics of intravenous cocaine self-administration in the BXD recombinant inbred mouse panel. Psychopharmacology. 5(6), 1–8. 10.4172/2157-7633.1000305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bogenpohl JW, Smith ML, Farris SP, Dumur CI, Lopez MF, Becker HC, … Miles MF (2019). Cross-Species Co-analysis of Prefrontal Cortex Chronic Ethanol Transcriptome Responses in Mice and Monkeys. Fronteirs in Molecular Neuroscience. 12, 1–18. 10.3389/fnmol.2019.00197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gelernter J, Zhou H, Nunez YZ, Mutirangura A, Malison RT, & Kalayasiri R (2019). Genomewide association study of alcohol dependence and related traits in a Thai population. Alcohol Clin Exp Res. 42(5), 861–868. 10.1111/acer.13614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hällfors J, Palviainen T, Surakka I, Gupta R, Buchwald J, Raevuori A, … Loukola A (2018). Genome-wide association study in Finnish twins highlights the connection between nicotine addiction and neurotrophin signaling pathway. Addiction Biology. 24, 549–561. 10.1111/adb.12618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sanchez-roige S, Fontanillas P, Elson SL, Gray JC, Wit H. De, Davis LK, … Palmer AA (2017). Genome-wide association study of alcohol use disorder identification test ( AUDIT ) scores in 20 328 research participants of European ancestry. Addiction Biology. 24, 121–131. 10.1111/adb.12574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kalsi G, Euesden J, Coleman JRI, Ducci F, Aliev F, Newhouse SJ, … Breen G (2016). Genome-Wide Association of Heroin Dependence in Han Chinese. PLoS ONE. 11(12), 1–18. 10.1371/journal.pone.0167388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wei K, Ho D, Han S, Nielsen JV, Jancic D, Hing B, … Potash JB (2018). Genome-wide association study of seasonal affective disorder. Translational Psychiatry, 8, 1–8. 10.1038/s41398-018-0246-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guo W, Samuels JF, Wang Y, Cao H, Ritter M, Nestadt PS, … Shugart YY (2017). Polygenic risk score and heritability estimates reveals a genetic relationship between ASD and OCD. European Neuropsychopharmacology, 27(7), 657–666. 10.1016/j.euroneuro.2017.03.011 [DOI] [PubMed] [Google Scholar]
- 41.Hall LS, Adams MJ, Arnau-soler A, Clarke T, Howard DM, Zeng Y, … Mcintosh AM (2018). Genome-wide meta-analyses of strati fi ed depression in Generation Scotland and UK Biobank. Translational Psychiatry, 1–12. 10.1038/s41398-017-0034-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martin J, Walters RK, Demontis D, Mattheisen M, Lee SH, Robinson E, … Neale BM (2018). Archival Report A Genetic Investigation of Sex Bias in the Prevalence of Attention-De fi cit / Hyperactivity Disorder. Biological Psychiatry, 83, 1044–1053. 10.1016/j.biopsych.2017.11.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lesch K, Tobias T, Rebecca JR, Christoph H, Nguyen TT, Craig DW, … Jacob C (2008). Molecular genetics of adult ADHD : converging evidence from genome-wide association and extended pedigree linkage studies. J Neural Transm, 115, 1573–1585. 10.1007/s00702-008-0119-3 [DOI] [PubMed] [Google Scholar]
- 44.Huggett SB, & Stallings MC (2020). Genetic Architecture and Molecular Neuropathology of Human Cocaine Addiction: Results from Multi-Omic Multi-Ancestry Analyses. The Journal of Neuroscience. 10.1523/JNEUROSCI.2879-19.2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saul MC, Bagley J,R, Bailey LS, Datta U, Dickson PE, Dodd R, Gagnon LH, Kimble VM, Leonardo M, Kim S, Olson A, Roy T, Schoenrock SA, Wilcox T, Jentsch D, Logan RW, McClung CA, Philip VM, Reinholdt LG, Sukoff SJ, Tarantino LM, & Chesler EJ (2020). Widespread genetic effects and sex differences play a crucial role in addiction. bioRxiv. [Google Scholar]
- 46.Rao X, Thapa KS, Chen AB, Lin H, Gao H, Reiter JL, … Liu Y (2019). Allele-specific expression and high-throughput reporter assay reveal functional genetic variants associated with alcohol use disorders. Molecular Psychiatry 10.1038/s41380-019-0508-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saad MH, Rumschlag M, Guerra MH, Savonen CL, Jaster AM, Olson PD, … Bannon MJ (2019). Differentially expressed gene networks, biomarkers, long noncoding RNAs, and shared responses with cocaine identified in the midbrains of human opioid abusers. Scientific Reports. 9(1), 1–9. 10.1038/s41598-018-38209-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hopf FW, Lesscher HMB, & Clinic G (2014). Rodent models for compulsive alcohol intake. Alcohol. 48(3), 253–264. 10.1016/j.alcohol.2014.03.001.Rodent [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lull ME, Freeman WM, Vrana KE, & Mash DC (2008). Correlating Human and Animal Studies of Cocaine Abuse and Gene Expression. Ann. N.Y. Acad. Sci. 1141, 58–75. 10.1196/annals.1441.013 [DOI] [PubMed] [Google Scholar]
- 50.Ribeiro EA, Scarpa JR, Garamszegi SP, Kasarskis A, Mash DC, & Nestler EJ (2017). Gene Network Dysregulation in Dorsolateral Prefrontal Cortex Neurons of Humans with Cocaine Use Disorder. Scientific Reports. 7(1), 1–10. 10.1038/s41598-017-05720-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Breschi A, Gingeras TR, & Guigó R (2017). Comparative transcriptomics in human and mouse. Nature Reviews Genetics. 18(7), 425–440. 10.1038/nrg.2017.19 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
We obtained human cocaine use disorder data from Sequence-Read-Archive (accession#: SRA029279) and the Gene Expression Omnibus (accession#: (accession #: GSE54839). The RNA-seq data for mouse cocaine use is available on Gene Expression Omnibus accession (accession#: GSE110344).