Abstract
An increasing number of studies have revealed an interaction between gut microbiota and tumors. The enrichment of specific bacteria strains in the intestines has been found to modulate tumor growth and influence the mechanisms of tumor treatment. Various bacteria are involved in modulating the effects of chemotherapeutic drugs currently used to treat patients with cancer, and they affect not only gastrointestinal tract tumors but also distant organ tumors. In addition, changes in the gut microbiota are known to be involved in the antitumor immune response as well as the modulation of the intestinal immune system. As a result, the gut microbiota plays an important role in modulating the efficacy of immune checkpoint inhibitors. Therefore, gut microbiota could be considered as an adjuvant treatment option with other cancer treatment or as another marker for predicting treatment response. In this review, we examine how gut microbiota affects cancer treatments.
Keywords: cancer, cancer therapy, gut microbiota, immune checkpoint inhibitors
INTRODUCTION
Malignant tumors are among the most fatal diseases in modern society (GBD 2015 Disease and Injury Incidence and Prevalence Collaborators, 2016). Carcinogenesis is the result of the stochastic intracellular accumulation of mutations during DNA replication or environmental exposure of various carcinogens, infectious agents, ultraviolet radiation, and toxic substances (Tomasetti and Vogelstein, 2015). Due to the efforts of many researchers, the overall death rate for cancer has been decreasing (Hashim et al., 2016). In addition to the conventional therapy of surgery, chemotherapy and radiation therapy, new treatment therapies including targeted therapy and immunotherapy such as immune checkpoint inhibitors (ICIs) which have recently attracted attention as novel therapeutic drugs, have been emerging for effective treatment modalities (Waldmann, 2003). As a result, the treatment results of patients with cancer are gradually improving (Hashim et al., 2016). However, patients with advanced stage or unfavorable malignant tumors still face a poor prognosis (Coleman et al., 2013; Miller et al., 2019; Nevala-Plagemann et al., 2020). Recently, attention has been drawn to the role of intestinal microbes to influence the therapeutic efficacy of tumor treatments (Louis et al., 2014; Wang et al., 2012; Zhu et al., 2013; Zitvogel et al., 2015).
The intestines of a healthy person contain a complex network of approximately 39 trillion bacteria (Sekirov et al., 2010; Sender et al., 2016). In the last few decades, many studies have been conducted to examine the effects of intestinal microbes on the host (Jandhyala et al., 2015). Intestinal microbes play important roles in normal physiologic development (Ku et al., 2020). In addition, the dysbiosis of intestinal microflora is associated with various diseases ranging from nervous system diseases to metabolic-related and gastrointestinal diseases (Shui et al., 2019). Some evidences indicate that these diseases are associated with tumors (Kuen et al., 2020; Louis et al., 2014; Wang et al., 2012; Zhu et al., 2013; Zitvogel et al., 2015). In the early 20th century, most studies on the relationship between these intestinal microbes and tumors were limited to the pathogenic aspect of microbes (Shui et al., 2019). As a result, many researchers have focused on microorganisms that cause tumors including Helicobacter pylori, hepatitis virus, human papilloma virus and Epstein–Barr virus. However, recent studies have revealed that intestinal microbes are not only involved in the occurrence of tumors but also affect tumor growth and cancer treatment efficacy (Viaud et al., 2013). These surprising discoveries are due to the development of microbial profiling techniques (Panek et al., 2018). With the advancement of technology such as 16S rRNA sequencing, it is possible to discover many microorganisms in the intestines that were previously unidentified (Ji and Nielsen, 2015). Metabolomics and metagenomics studies have described the effects of these intestinal microbes on the human body, and studies of the effects on tumors have been conducted in relation to cancer prevention, tumorigenesis, and anticancer activity (Finlay et al., 2020; Tanoue et al., 2019).
Because antitumor immune responses have been regarded as important in tumor treatment in recent years, attention has also been paid to the effect of intestinal microbes on tumor immunity (Viaud et al., 2013). Compelling evidence indicates that intestinal microbes influence intestinal immune barriers such as intraepithelial lymphocytes development (Jung et al., 2019). Literature has shown that gut microbiota-derived short chain fatty acids play an important role in regulating the adaptive immune response (Kim et al., 2014). Regarding the treatment with ICIs, changes in intestinal microflora affect not only the effects of these ICIs but also their therapeutic toxicity (Choi and Lee, 2020; Viaud et al., 2013). For example, microbial control has been used as a novel therapeutic method to improve the effectiveness of ICIs (Gopalakrishnan et al., 2018; Vetizou et al., 2015). This review discusses the latest studies that have examined the effect of intestinal microbes on tumor growth and antitumor therapy including immunosuppressants.
DEPLETION OF GUT MICROBIOTA AFFECTS TUMOR GROWTH
Antibiotics have saved humanity from infectious diseases and are commonly used drugs in modern medical circumstances (Iizumi et al., 2017). However, they cause dysbiosis of intestinal microbes, which have linked to various disease in recent reports. In addition, gut microbiota depletion due to antibiotics use has various effects on tumor progression and the immune environment.
Recent literature has shown that the loss of intestinal microflora by antibiotics use results in an antitumor effect (Sethi et al., 2018). Several studies have been conducted on gut microbiota depletion using oral antibiotics administration in various mouse tumor models. In one study, the continuous administration of a broad-spectrum antibiotics cocktail of vancomycin, neomycin, metronidazole, ampicillin, and amphotericin B reduced the growth of the tumor burden in pancreatic cancer, colon cancer and melanoma animal models (Sethi et al., 2018). This result was attributed to increased production of interferon-gamma (IFN-γ), interleukin (IL) 17a, and IL-10 in T cells.
Vancomycin-depleted gut microbiota not only affects the antitumor immune response but also increases the antitumor activity of radiation therapy (Uribe-Herranz et al., 2020). In the melanoma mouse model, the vancomycin-depleted group exhibited increased cross-presentation of tumor-associated antigens to cytotoxic T lymphocytes and IFN-γ production (Uribe-Herranz et al., 2020). However, butyrate secreted from vancomycin-sensitive bacteria plays an important role in enhancing antitumor activity by radiotherapy. As a result, the reduction of specific strains caused by these antibiotics decreases the synergistic action of depleted gut microbiota.
However, antibiotics-induced gut microbiota depletion and dysbiosis have also been shown to increase tumor growth and progression. In this regard, reports indicate that certain microbial metabolites promote tumor growth (Laborda-Illanes et al., 2020). In addition, dysbiosis induced through a broad-spectrum antibiotics cocktail in mouse colon cancer model can lead to increases colon tumor susceptibility via the induction of CD8+ IFN-γ+ T cells (Yu et al., 2020). In this study, increased CD8+ IFN-γ+ T cells in the colon lamina propria induced inflammation in the serous membrane, affecting tumor development. After tumor formation, the antitumor immune response was suppressed by inducing the exhaustion of functional CD8+ T cells (Yu et al., 2020). The use of broad-spectrum antibiotics also affects dendritic cells. The depleted gut microbiota by vancomycin increased the number of systemic CD8a+ dendritic cells, thereby enhancing the effect of adoptive T-cell therapy and increasing IL-12 cytokine in mice (Xu et al., 2017).
The effect of gut microbiota depletion by antibiotics has been reported as both antitumoral and protumoral activity. Although there is still no clear mechanism, this varied tumor response seems to be due to differences in affected bacteria strains by antibiotics and tumor types.
SPECIFIC COMMENSAL GUT BACTERIA MAY MODULATE TUMOR GROWTH AND PROGRESSION
The use of an antibiotics-induced dysbiosis model can confirm a variety of reactions to tumor growth. However, the decrease or change in intestinal microflora by antibiotics has limitations in confirming tumor response to specific bacteria species. Several studies have reported that the enrichment of specific bacteria in animal models is associated with tumor growth.
Some microbial-derived short-chain fatty acids may have anticancer effects. Propionibacterium is known as one of the symbiotic microorganisms in the human intestines (Jan et al., 2002). Among its species, Propionibacterium acidipropionici and freudenreichii were found to induce apoptosis in human colorectal carcinoma cell lines. They produce short-chain fatty acids propionate and acetate, which are known to have cytotoxic effects. These compounds produce reactive oxygen species (ROS) and initiate caspase-3 processing, nuclear chromatin condensation formation, and B cell lymphoma 2 inhibition (Jan et al., 2002), all of which induce apoptosis.
In addition, several studies have shown that Lactobacilli can stimulate host immune cells such as dendritic cells or Th1 responses, leading to the elimination of cancer or precancerous cells in mice rhabdomyosarcoma model (Jacouton et al., 2018; Takagi et al., 2008). However, further studies are necessary to confirm how the bacterial bioproduct acts in inducing these stimulatory effects (Takagi et al., 2008). Lactobacillus casei ATCC334 was found to induce anticancer action through the production of ferrichrome, a bioproduct of the bacteria. The substance was confirmed to have greater anticancer activity than chemotherapy agents such as cisplatin or 5-fluorouracil (5-FU), with less damage to normal cells. This mechanism has been confirmed to function through c-Jun N-terminal kinase activation (Konishi et al., 2016).
Fusobacterium nucleatum is known to be associated with colorectal cancer. F. nucleatum promotes the growth of colorectal carcinoma cells through Fusobacterium adhesin A (FadA), which is involved in beta-catenin signaling activation. The level of FadA gene expression in colon tissue was approximately 10 to 100 times higher in patients with colorectal cancer than in healthy participants. These elevated gene levels are associated with other oncogenic and inflammatory genes (Rubinstein et al., 2013). In addition, F. nucleatum was shown to be associated with colorectal cancer, esophageal cancer, gastric cancer, head and neck cancer and pancreatic cancer (Gaiser et al., 2019; Hsieh et al., 2018; Shin et al., 2017; Yamamura et al., 2016). Bacteroides fragilis also can interact with the epithelial E-cadherin, disrupting intercellular junctions and activating β-catenin signaling by metalloproteinase toxins (Wu et al., 2007).
Peptostreptococcus anaerobius is an anaerobic strain present in the intestines of patients with colorectal cancer. In a colorectal cancer mouse model, this strain was found to accelerate tumor growth (Long et al., 2019). In vitro experiments demonstrated that P. anaerobius directly interacts with the colonic cell line, and as a result, induces cell proliferation by activating the PI3K-Akt pathway of colorectal cancer cells and the NF-κB activation of tumor-associated macrophages. Furthermore, the activation of NF-κB was associated with the proliferation of myeloid-derived suppressor cells and promoted chronic inflammation and tumor progression (Long et al., 2019).
GUT MICROBIOTA MEDIATES CANCER TREATMENT
Chemotherapy
As the link between intestinal microbes and tumor growth has been gradually revealed, studies have examined the synergistic effects of these microbes with antitumor treatments (Roy and Trinchieri, 2017). In addition to existing chemotherapeutic agents, their synergistic effects with radiation therapy and ICIs have been newly discovered (Roy and Trinchieri, 2017). There are some changes in gene expression associated with drug metabolism by the RNA-seq analysis of hepatic drug-processing genes in the germ-free mice (Selwyn et al., 2015). Therefore, intestinal microbes may be involved in the metabolism of antitumor drugs. Continuous exposure to cyclophosphamide-induced changes in intestinal microflora in tumor-bearing mice (Viaud et al., 2013). Table 1 lists the intestinal microbes found to modulate the anticancer effect of chemotherapeutic agents, as described in various papers.
Table 1.
Summary of intestinal microorganism modulating the efficacy of chemotherapeutic drugs
| Bacteria | Chemotherapeutic agent | Interaction mechanism | Reference |
|---|---|---|---|
| Lactobacillus species | Cyclophosphamide | Promoted Th17 and Th1 cell response during cyclophosphamide treatment | Viaud et al. (2013) |
| Segmented filamentous bacteria | |||
| Enterococcus hirae | Cyclophosphamide | Associated with increased CD8/ Treg ratio | Daillere et al. (2016) |
| Barnesiella intestinihominis | Infiltration of interferon-g-producing gd-T cells | ||
| Escherichia coli | 5-FU | Distribution of bacterial deoxynucleotide pools regulates the effect of 5-FU | Scott et al. (2017) |
| Fusobacterium nucleatum | 5-FU | Modulation autophagy pathway and inhibit tumor cell apoptosis | Yu et al. (2017) |
| Oxaliplatin | |||
| Gammaproteobacteria | Gemcitabine | Gemcitabine was converted into an inactivated form by long isoform of cytidine deaminase | Geller et al. (2017) |
| β-glucuronidase producers | Irinotecan | Converting inactivated irinotecan into the active form, SN-38 | Kodawara et al. (2016) |
| Ciprofloxacin | Involved in the activation of ciprofloxacin | Alexander et al. (2017);Wallace et al. (2010) |
The effect of platinum-based chemotherapeutic agents such as oxaliplatin and CpG oligodeoxynucleotides was poor in mice treated with antibiotics (Iida et al., 2013). In the mice administered antibiotics, cytokine secretion and ROS were reduced, resulting in decreased tumor necrosis and cytotoxicity of the antitumor therapy. Correspondingly, in a study of patients with chronic lymphocytic leukemia, tumor progression was observed and the overall survival rate decreased in patients who were prescribed anti-gram-positive antibiotics (Pflug et al., 2016). As a result, anti-gram-positive antibiotics were suggested to negatively affect the cytotoxicity of cyclophosphamide and cisplatin.
As previously shown, continuous exposure of cyclophosphamide-induced changes in the intestinal microflora, and it was confirmed that gram-positive bacteria including Lactobacillus and segmented filamentous bacteria promoted the formation of a pathogenic Th17 and memory Th1 immune responses in mice (Viaud et al., 2013). In the case of the adoptive transfer of these cells, antitumor efficacy was confirmed, suggesting that intestinal microbes play an important role in regulating the antitumor efficacy of cyclophosphamide (Viaud et al., 2013). Enterococcus hirae and Barnesiella intestinihominis were found to be related during cyclophosphamide treatment. In addition, E. hirae was associated with an increase in the CD8/Treg ratio, and B. intestinihominis promoted infiltration of gamma-delta T cells that form IFN-γ in cancer lesions. As a result, clinical results showed that both bacteria can be used as a predictor of long-term progression-free survival when chemoimmunotherapy is administered in patients with ovarian cancer and lung cancer (Daillere et al., 2016).
In a colon cancer model, intratumoral bacteria were found to modulate the effect of chemotherapy. Gemcitabine is converted into an inactivated form by specific gammaproteobacteria present in the tumor, reducing the antitumor effect of the drug. The bacteria contain an enzyme in the form of a long isoform of cytidine deaminase. The suppression of the anticancer effect disappeared when bacteria were killed through antibiotics treatment in colon cancer mouse model (Geller et al., 2017). Moreover, another study, related to gemcitabine, confirmed that the presence of bacteria in an in vivo murine subcutaneous tumor model reduced the cytotoxicity of gemcitabine, thereby affecting the tumor response to chemotherapy (Lehouritis et al., 2015).
In the study of Caenorhabditis elegans model, the effect of fluoropyrimidine was modulated by the composition of Escherichia coli in the intestines. The authors reported that the modulation the effects of fluoropyrimidines was influenced by bacterial vitamins B6, B9 and ribonucleotide metabolism. As a result, the difference in the distribution of bacterial deoxynucleotide pools was found to regulate the effect of 5-FU with nucleoside diphosphate kinase 1 involved in regulating the drug (Scott et al., 2017). Another study with the same model showed that bacterial ribonucleotide metabolism plays an important role in regulating the effects of chemotherapeutic drugs such as 5-FU (Garcia-Gonzalez et al., 2017). Even in an experimental mouse model using 5-FU, the administration of antibiotics was found to reduce the effect of 5-FU treatment in colorectal cancer. In an analysis of 16S rRNA seq, the use of antibiotics resulted in a significant increase in the pathogenic bacteria Escherichia shigella and Enterobacter, and these changes were found to be recovered when taking probiotics in mice model (Yuan et al., 2018). Another intestinal microbe, F. nucleatum, was abundant in patients with colorectal cancer exhibiting poor response to 5-FU and oxaliplatin (Yu et al., 2017). F. nucleatum targets the innate immune signaling TLR4 and MYD88 and contributes to activating the autophagy pathway and colorectal cancer chemoresistance by disrupting apoptosis (Yu et al., 2017).
Beta-glucuronidase, an enzyme produced by intestinal microbes, plays an important role in converting inactivated irinotecan into its active form, SN-38. Taking antibiotics consequently inhibits this metabolism, thereby reducing the antitumor efficacy of irinotecan (Kodawara et al., 2016). In addition, beta-glucuronidase is involved in the activation of ciprofloxacin, demonstrating that an important role of intestinal microbes in determining the effectiveness of chemotherapy (Alexander et al., 2017; Wallace et al., 2010).
Immune checkpoint inhibitors
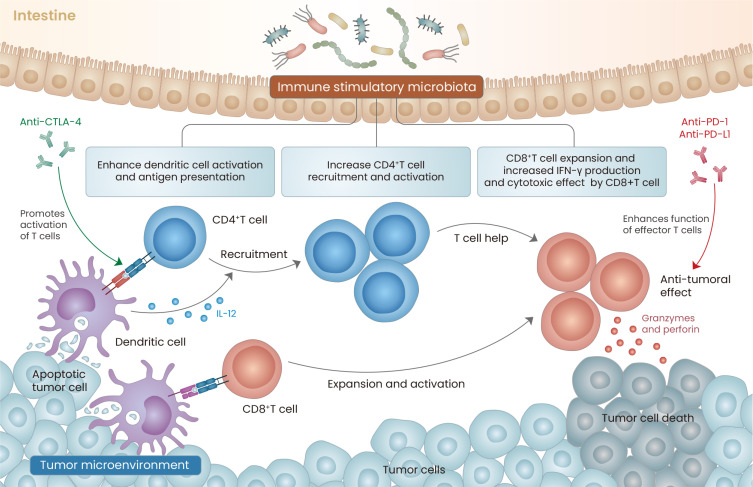
Among the mechanisms reviewed above, some intestinal microbes contribute to the anticancer effect by improving the immune response. With this background, studies have examined the synergistic effects of intestinal microbes with ICIs and found that they can play an important role in increasing the effects of ICIs (Fig. 1).
Fig. 1. Gut microbiota immune modulation and Immune checkpoint inhibitors.
Gut microbiota activates the T cell priming process of dendritic cells. In this process, anti-CTLA 4 antibody has synergy with the Immune stimulatory microbiota (such as Bacteroides species). As a result, it promotes the formation of functional effector T cells. In addition, intestinal microbes affect CD4 T cell recruitment and activation. Enrichment of certain intestinal microbes, such as Akkermansia muciniphila or Bifidobacterium species, contributes to enhancement and expansion of functional CD8 T cells, IFN-γ production, and increased granzyme and perforin secretion. These points contribute to enhancing the effect of the PD-1/PD-L1 blockade, thus contributing to enhancing the antitumor immunity.
Cytotoxic T-lymphocyte-associated protein 4 (CTLA-4) inhibitors are one of the ICIs used in clinical practice currently because of its recognized effectiveness. Bacteroides fragilis of the Bacteroides species among intestinal microbes enhances the antitumor effect of these inhibitors in mice and patients (Vetizou et al., 2015). In the case of using antibiotics in a germ-free mouse model, the effect of the CTLA-4 inhibitor was recovered through the oral gavage of the strain and the adoptive transfer of the bacterial polysaccharides or strain-specific T cells (Vetizou et al., 2015). In the same year, the effect of PD-L1 inhibitor, one of the ICIs, was also found to be affected by gut microbiota in mice (Sivan et al., 2015). After confirming the presence of intestinal microbes common to patients responding to the immunosuppressant, researchers found an enhanced effect of ICI treatment in mice that took Bifidobacterium through oral gavage. They found that this effect is associated with altered dendritic cell activation and an increased degree of CD8+ T-cell activation (Sivan et al., 2015). However, the clinical benefit disappeared in cancer patients with a medical history of taking antibiotics who used ICIs (Routy et al., 2018). Another study transplanted avatar mice with fecal samples from mice treated with ICIs and classified by treatment response (response or nonresponse). Mice treated with fecal microbiota from the response group exhibited a higher effect of ICIs than those with stool from the nonresponse group. In a study of avatar mice transplanted with fecal samples from the respond group or nonrespond group of these ICIs, the mice subjected to fecal microbiota transplantation (FMT) with the stool of the respond group had a higher effect of the ICIs. In the analysis, the degree of this reaction was closely related to the abundance of Akkermansia muciniphila. As a result, the supplementation of A. muciniphila was associated with increased recruitment of CCR9+ CXCR3+ CD4 T cells in an IL-12-dependent manner (Routy et al., 2018).
The development of metagenomics has helped to elucidate the relationship between ICIs and intestinal microbes. In a FMT study of patients with metastatic melanoma, an analysis of 16S rRNA sequencing data performed after transplantation but before ICI treatment showed that patients who responded to ICIs had more strains of Bifidobacterium longum, Collinsella aerofaciens, and Enterococcus faecium. In addition, the respond group showed higher tumor control rate in the germ-free avatar mouse model through FMT (Matson et al., 2018). In a gut microbiome analysis of patients with non-small cell lung cancer who received ICI treatment, Bifidobacteirum bifidum was abundant in the gut of patients who responded to treatment (Lee et al., 2021). Among them, it was confirmed that only specific B. bifidum strains enhance the effect of PD-1 blockade. Moreover, a metabolomic analysis reported that the bacteria strains were associated with increased serum L-tryptophan and IFN-γ secretion of cytotoxic T cells in mice (Lee et al., 2021). In another paper, patients who responded to ICI responders had more abundance of the Ruminococcaceae family and significantly more alpha diversity, and systemic and antitumor immunity were generally elevated, indicating that intestinal microbes were directly related to ICIs (Gopalakrishnan et al., 2018).
CONCLUSION
The study of interactions between intestinal microbiota and malignant tumors has a short history. To date, various strains are known to affect tumor treatment, but no studies have clearly described the corresponding mechanism. In addition, methods to use these intestinal microorganisms as therapeutic agents such as probiotics or FMT require further investigation. Studies on the clinical application of gut microbiota are ongoing for the purpose of predicting and diagnosing cancer, as well as the potential for improving the effect or alleviating side effects of antitumor treatment (Wong et al., 2019). In recent studies, FMT can overcomes the resistance to immunotherapy in melanoma patients whose cancers do not respond (Baruch et al., 2021; Davar et al., 2021). These clinical results suggest that gut microbiota modulation through FMT can be effectively used for cancer treatment. Because the intestinal environment is a system in which microorganisms form a huge network, more research needs to be conducted to understand this complex ecosystem and apply it for treatment.
ACKNOWLEDGMENTS
This research was supported by the Bio & Medical Technology Development Program of the National Research Foundation (NRF) & funded by the Korean government (MSIT) (No. 2021M3A9H3015688).
Footnotes
AUTHOR CONTRIBUTIONS
J.K. and H.K.L. conceived and wrote the manuscript. H.K.L. secured funding.
CONFLICT OF INTEREST
The authors have no potential conflicts of interest to disclose.
REFERENCES
- Alexander J.L., Wilson I.D., Teare J., Marchesi J.R., Nicholson J.K., Kinross J.M. Gut microbiota modulation of chemotherapy efficacy and toxicity. Nat. Rev. Gastroenterol. Hepatol. 2017;14:356–365. doi: 10.1038/nrgastro.2017.20. [DOI] [PubMed] [Google Scholar]
- Baruch E.N., Youngster I., Ben-Betzalel G., Ortenberg R., Lahat A., Katz L., Adler K., Dick-Necula D., Raskin S., Bloch N., et al. Fecal microbiota transplant promotes response in immunotherapy-refractory melanoma patients. Science. 2021;371:602–609. doi: 10.1126/science.abb5920. [DOI] [PubMed] [Google Scholar]
- Choi J., Lee S.Y. Clinical characteristics and treatment of immune-related adverse events of immune checkpoint inhibitors. Immune Netw. 2020;20:e9. doi: 10.4110/in.2020.20.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman R.L., Monk B.J., Sood A.K., Herzog T.J. Latest research and treatment of advanced-stage epithelial ovarian cancer. Nat. Rev. Clin. Oncol. 2013;10:211–224. doi: 10.1038/nrclinonc.2013.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daillere R., Vetizou M., Waldschmitt N., Yamazaki T., Isnard C., Poirier-Colame V., Duong C.P.M., Flament C., Lepage P., Roberti M.P., et al. Enterococcus hirae and Barnesiella intestinihominis facilitate cyclophosphamide-induced therapeutic immunomodulatory effects. Immunity. 2016;45:931–943. doi: 10.1016/j.immuni.2016.09.009. [DOI] [PubMed] [Google Scholar]
- Davar D., Dzutsev A.K., McCulloch J.A., Rodrigues R.R., Chauvin J.M., Morrison R.M., Deblasio R.N., Menna C., Ding Q., Pagliano O., et al. Fecal microbiota transplant overcomes resistance to anti-PD-1 therapy in melanoma patients. Science. 2021;371:595–602. doi: 10.1126/science.abf3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlay B.B., Goldszmid R., Honda K., Trinchieri G., Wargo J., Zitvogel L.J.N.R.I. Can we harness the microbiota to enhance the efficacy of cancer immunotherapy? Nat. Rev. Immunol. 2020;20:522–528. doi: 10.1038/s41577-020-0374-6. [DOI] [PubMed] [Google Scholar]
- Gaiser R.A., Halimi A., Alkharaan H., Lu L., Davanian H., Healy K., Hugerth L.W., Ateeb Z., Valente R., Fernandez Moro C., et al. Enrichment of oral microbiota in early cystic precursors to invasive pancreatic cancer. Gut. 2019;68:2186–2194. doi: 10.1136/gutjnl-2018-317458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Gonzalez A.P., Ritter A.D., Shrestha S., Andersen E.C., Yilmaz L.S., Walhout A.J.M. Bacterial metabolism affects the C. elegans response to cancer chemotherapeutics. Cell. 2017;169:431–441.e8. doi: 10.1016/j.cell.2017.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GBD 2015 Disease and Injury Incidence and Prevalence Collaborators, author. Global, regional, and national incidence, prevalence, and years lived with disability for 310 diseases and injuries, 1990-2015: a systematic analysis for the Global Burden of Disease Study 2015. Lancet. 2016;388:1545–1602. doi: 10.1016/S0140-6736(16)31678-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geller L.T., Barzily-Rokni M., Danino T., Jonas O.H., Shental N., Nejman D., Gavert N., Zwang Y., Cooper Z.A., Shee K., et al. Potential role of intratumor bacteria in mediating tumor resistance to the chemotherapeutic drug gemcitabine. Science. 2017;357:1156–1160. doi: 10.1126/science.aah5043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopalakrishnan V., Spencer C.N., Nezi L., Reuben A., Andrews M.C., Karpinets T.V., Prieto P.A., Vicente D., Hoffman K., Wei S.C., et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science. 2018;359:97–103. doi: 10.1126/science.aan4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashim D., Boffetta P., La Vecchia C., Rota M., Bertuccio P., Malvezzi M., Negri E. The global decrease in cancer mortality: trends and disparities. Ann. Oncol. 2016;27:926–933. doi: 10.1093/annonc/mdw027. [DOI] [PubMed] [Google Scholar]
- Hsieh Y.Y., Tung S.Y., Pan H.Y., Yen C.W., Xu H.W., Lin Y.J., Deng Y.F., Hsu W.T., Wu C.S., Li C. Increased abundance of Clostridium and Fusobacterium in gastric microbiota of patients with gastric cancer in Taiwan. Sci. Rep. 2018;8:158. doi: 10.1038/s41598-017-18596-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iida N., Dzutsev A., Stewart C.A., Smith L., Bouladoux N., Weingarten R.A., Molina D.A., Salcedo R., Back T., Cramer S., et al. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science. 2013;342:967–970. doi: 10.1126/science.1240527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iizumi T., Battaglia T., Ruiz V., Perez Perez G.I. Gut microbiome and antibiotics. Arch. Med. Res. 2017;48:727–734. doi: 10.1016/j.arcmed.2017.11.004. [DOI] [PubMed] [Google Scholar]
- Jacouton E., Michel M.L., Torres-Maravilla E., Chain F., Langella P., Bermudez-Humaran L.G. Elucidating the immune-related mechanisms by which probiotic strain Lactobacillus casei BL23 displays anti-tumoral properties. Front. Microbiol. 2018;9:3281. doi: 10.3389/fmicb.2018.03281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan G., Belzacq A.S., Haouzi D., Rouault A., Metivier D., Kroemer G., Brenner C. Propionibacteria induce apoptosis of colorectal carcinoma cells via short-chain fatty acids acting on mitochondria. Cell Death Differ. 2002;9:179–188. doi: 10.1038/sj.cdd.4400935. [DOI] [PubMed] [Google Scholar]
- Jandhyala S.M., Talukdar R., Subramanyam C., Vuyyuru H., Sasikala M., Nageshwar Reddy D. Role of the normal gut microbiota. World J. Gastroenterol. 2015;21:8787–8803. doi: 10.3748/wjg.v21.i29.8787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji B., Nielsen J. From next-generation sequencing to systematic modeling of the gut microbiome. Front. Genet. 2015;6:219. doi: 10.3389/fgene.2015.00219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung J., Surh C.D., Lee Y.J. Microbial colonization at early life promotes the development of diet-induced CD8αβ intraepithelial T cells. Mol. Cells. 2019;42:313–320. doi: 10.14348/molcells.2019.2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C.H., Park J., Kim M. Gut microbiota-derived short-chain Fatty acids, T cells, and inflammation. Immune Netw. 2014;14:277–288. doi: 10.4110/in.2014.14.6.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodawara T., Higashi T., Negoro Y., Kamitani Y., Igarashi T., Watanabe K., Tsukamoto H., Yano R., Masada M., Iwasaki H.J.B., et al. The inhibitory effect of ciprofloxacin on the β‐glucuronidase‐mediated deconjugation of the irinotecan metabolite SN‐38‐G. Basic Clin. Pharmacol. Toxicol. 2016;118:333–337. doi: 10.1111/bcpt.12511. [DOI] [PubMed] [Google Scholar]
- Konishi H., Fujiya M., Tanaka H., Ueno N., Moriichi K., Sasajima J., Ikuta K., Akutsu H., Tanabe H., Kohgo Y. Probiotic-derived ferrichrome inhibits colon cancer progression via JNK-mediated apoptosis. Nat. Commun. 2016;7:12365. doi: 10.1038/ncomms12365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku K., Park I., Kim D., Kim J., Jang S., Choi M., Choe H.K., Kim K. Gut microbial metabolites induce changes in circadian oscillation of clock gene expression in the mouse embryonic fibroblasts. Mol. Cells. 2020;43:276–285. doi: 10.14348/molcells.2020.2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuen D.S., Kim B.S., Chung Y. IL-17-producing cells in tumor immunity: friends or foes? Immune Netw. 2020;20:e6. doi: 10.4110/in.2020.20.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laborda-Illanes A., Sanchez-Alcoholado L., Dominguez-Recio M.E., Jimenez-Rodriguez B., Lavado R., Comino-Mendez I., Alba E., Queipo-Ortuno M.I. Breast and gut microbiota action mechanisms in breast cancer pathogenesis and treatment. Cancers (Basel) 2020;12:2465. doi: 10.3390/cancers12092465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.H., Cho S.Y., Yoon Y., Park C., Sohn J., Jeong J.J., Jeon B.N., Jang M., An C., Lee S., et al. Bifidobacterium bifidum strains synergize with immune checkpoint inhibitors to reduce tumour burden in mice. Nat. Microbiol. 2021;6:277–288. doi: 10.1038/s41564-020-00831-6. [DOI] [PubMed] [Google Scholar]
- Lehouritis P., Cummins J., Stanton M., Murphy C.T., McCarthy F.O., Reid G., Urbaniak C., Byrne W.L., Tangney M. Local bacteria affect the efficacy of chemotherapeutic drugs. Sci. Rep. 2015;5:14554. doi: 10.1038/srep14554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long X., Wong C.C., Tong L., Chu E.S.H., Ho Szeto C., Go M.Y.Y., Coker O.O., Chan A.W.H., Chan F.K.L., Sung J.J.Y., et al. Peptostreptococcus anaerobius promotes colorectal carcinogenesis and modulates tumour immunity. Nat. Microbiol. 2019;4:2319–2330. doi: 10.1038/s41564-019-0541-3. [DOI] [PubMed] [Google Scholar]
- Louis P., Hold G.L., Flint H.J. The gut microbiota, bacterial metabolites and colorectal cancer. Nat. Rev. Microbiol. 2014;12:661–672. doi: 10.1038/nrmicro3344. [DOI] [PubMed] [Google Scholar]
- Matson V., Fessler J., Bao R., Chongsuwat T., Zha Y., Alegre M.L., Luke J.J., Gajewski T.F.J.S. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science. 2018;359:104–108. doi: 10.1126/science.aao3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller K.D., Nogueira L., Mariotto A.B., Rowland J.H., Yabroff K.R., Alfano C.M., Jemal A., Kramer J.L., Siegel R.L. Cancer treatment and survivorship statistics, 2019. CA Cancer J. Clin. 2019;69:363–385. doi: 10.3322/caac.21565. [DOI] [PubMed] [Google Scholar]
- Nevala-Plagemann C., Hidalgo M., Garrido-Laguna I. From state-of-the-art treatments to novel therapies for advanced-stage pancreatic cancer. Nat. Rev. Clin. Oncol. 2020;17:108–123. doi: 10.1038/s41571-019-0281-6. [DOI] [PubMed] [Google Scholar]
- Panek M., Cipcic Paljetak H., Baresic A., Peric M., Matijasic M., Lojkic I., Vranesic Bender D., Krznaric Z., Verbanac D. Methodology challenges in studying human gut microbiota - effects of collection, storage, DNA extraction and next generation sequencing technologies. Sci. Rep. 2018;8:5143. doi: 10.1038/s41598-018-23296-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pflug N., Kluth S., Vehreschild J.J., Bahlo J., Tacke D., Biehl L., Eichhorst B., Fischer K., Cramer P., Fink A.M., et al. Efficacy of antineoplastic treatment is associated with the use of antibiotics that modulate intestinal microbiota. Oncoimmunology. 2016;5:e1150399. doi: 10.1080/2162402X.2016.1150399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Routy B., Le Chatelier E., Derosa L., Duong C.P.M., Alou M.T., Daillere R., Fluckiger A., Messaoudene M., Rauber C., Roberti M.P., et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science. 2018;359:91–97. doi: 10.1126/science.aan3706. [DOI] [PubMed] [Google Scholar]
- Roy S., Trinchieri G. Microbiota: a key orchestrator of cancer therapy. Nat. Rev. Cancer. 2017;17:271–285. doi: 10.1038/nrc.2017.13. [DOI] [PubMed] [Google Scholar]
- Rubinstein M.R., Wang X., Liu W., Hao Y., Cai G., Han Y.W. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/β-catenin signaling via its FadA adhesin. Cell Host Microbe. 2013;14:195–206. doi: 10.1016/j.chom.2013.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott T.A., Quintaneiro L.M., Norvaisas P., Lui P.P., Wilson M.P., Leung K.Y., Herrera-Dominguez L., Sudiwala S., Pessia A., Clayton P.T., et al. Host-microbe co-metabolism dictates cancer drug efficacy in C. elegans . Cell. 2017;169:442–456.e18. doi: 10.1016/j.cell.2017.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekirov I., Russell S.L., Antunes L.C., Finlay B.B. Gut microbiota in health and disease. Physiol. Rev. 2010;90:859–904. doi: 10.1152/physrev.00045.2009. [DOI] [PubMed] [Google Scholar]
- Selwyn F.P., Cui J.Y., Klaassen C.D. RNA-Seq quantification of hepatic drug processing genes in germ-free mice. Drug Metab. Dispos. 2015;43:1572–1580. doi: 10.1124/dmd.115.063545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sender R., Fuchs S., Milo R. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 2016;14:e1002533. doi: 10.1371/journal.pbio.1002533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethi V., Kurtom S., Tarique M., Lavania S., Malchiodi Z., Hellmund L., Zhang L., Sharma U., Giri B., Garg B., et al. Gut microbiota promotes tumor growth in mice by modulating immune response. Gastroenterology. 2018;155:33–37.e6. doi: 10.1053/j.gastro.2018.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J.M., Luo T., Kamarajan P., Fenno J.C., Rickard A.H., Kapila Y.L. Microbial communities associated with primary and metastatic head and neck squamous cell carcinoma - a high fusobacterial and low streptococcal signature. Sci. Rep. 2017;7:9934. doi: 10.1038/s41598-017-09786-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shui L., Yang X., Li J., Yi C., Sun Q., Zhu H. Gut microbiome as a potential factor for modulating resistance to cancer immunotherapy. Front. Immunol. 2019;10:2989. doi: 10.3389/fimmu.2019.02989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivan A., Corrales L., Hubert N., Williams J.B., Aquino-Michaels K., Earley Z.M., Benyamin F.W., Lei Y.M., Jabri B., Alegre M.L., et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science. 2015;350:1084–1089. doi: 10.1126/science.aac4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi A., Ikemura H., Matsuzaki T., Sato M., Nomoto K., Morotomi M., Yokokura T. Relationship between the in vitro response of dendritic cells to Lactobacillus and prevention of tumorigenesis in the mouse. J. Gastroenterol. 2008;43:661–669. doi: 10.1007/s00535-008-2212-7. [DOI] [PubMed] [Google Scholar]
- Tanoue T., Morita S., Plichta D.R., Skelly A.N., Suda W., Sugiura Y., Narushima S., Vlamakis H., Motoo I., Sugita K., et al. A defined commensal consortium elicits CD8 T cells and anti-cancer immunity. Nature. 2019;565:600–605. doi: 10.1038/s41586-019-0878-z. [DOI] [PubMed] [Google Scholar]
- Tomasetti C., Vogelstein B. Cancer etiology. Variation in cancer risk among tissues can be explained by the number of stem cell divisions. Science. 2015;347:78–81. doi: 10.1126/science.1260825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uribe-Herranz M., Rafail S., Beghi S., Gil-de-Gomez L., Verginadis I., Bittinger K., Pustylnikov S., Pierini S., Perales-Linares R., Blair I.A., et al. Gut microbiota modulate dendritic cell antigen presentation and radiotherapy-induced antitumor immune response. J. Clin. Invest. 2020;130:466–479. doi: 10.1172/JCI124332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetizou M., Pitt J.M., Daillere R., Lepage P., Waldschmitt N., Flament C., Rusakiewicz S., Routy B., Roberti M.P., Duong C.P., et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science. 2015;350:1079–1084. doi: 10.1126/science.aad1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viaud S., Saccheri F., Mignot G., Yamazaki T., Daillere R., Hannani D., Enot D.P., Pfirschke C., Engblom C., Pittet M.J., et al. The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide. Science. 2013;342:971–976. doi: 10.1126/science.1240537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldmann T.A. Immunotherapy: past, present and future. Nat. Med. 2003;9:269–277. doi: 10.1038/nm0303-269. [DOI] [PubMed] [Google Scholar]
- Wallace B.D., Wang H., Lane K.T., Scott J.E., Orans J., Koo J.S., Venkatesh M., Jobin C., Yeh L.A., Mani S., et al. Alleviating cancer drug toxicity by inhibiting a bacterial enzyme. Science. 2010;330:831–835. doi: 10.1126/science.1191175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T., Cai G., Qiu Y., Fei N., Zhang M., Pang X., Jia W., Cai S., Zhao L. Structural segregation of gut microbiota between colorectal cancer patients and healthy volunteers. ISME J. 2012;6:320–329. doi: 10.1038/ismej.2011.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong S.H., Kwong T.N.Y., Wu C.Y., Yu J. Clinical applications of gut microbiota in cancer biology. Semin. Cancer Biol. 2019;55:28–36. doi: 10.1016/j.semcancer.2018.05.003. [DOI] [PubMed] [Google Scholar]
- Wu S., Rhee K.J., Zhang M., Franco A., Sears C.L. Bacteroides fragilis toxin stimulates intestinal epithelial cell shedding and gamma-secretase-dependent E-cadherin cleavage. J. Cell Sci. 2007;120(Pt 11):1944–1952. doi: 10.1242/jcs.03455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C., Ruan B., Jiang Y., Xue T., Wang Z., Lu H., Wei M., Wang S., Ye Z., Zhai D.J.B., et al. Antibiotics-induced gut microbiota dysbiosis promotes tumor initiation via affecting APC-Th1 development in mice. Biochem. Biophys. Res. Commun. 2017;488:418–424. doi: 10.1016/j.bbrc.2017.05.071. [DOI] [PubMed] [Google Scholar]
- Yamamura K., Baba Y., Nakagawa S., Mima K., Miyake K., Nakamura K., Sawayama H., Kinoshita K., Ishimoto T., Iwatsuki M., et al. Human microbiome Fusobacterium nucleatum in esophageal cancer tissue is associated with prognosis. Clin. Cancer Res. 2016;22:5574–5581. doi: 10.1158/1078-0432.CCR-16-1786. [DOI] [PubMed] [Google Scholar]
- Yu A.I., Zhao L., Eaton K.A., Ho S., Chen J., Poe S., Becker J., Gonzalez A., McKinstry D., Hasso M., et al. Gut microbiota modulate CD8 T cell responses to influence colitis-associated tumorigenesis. Cell Rep. 2020;31:107471. doi: 10.1016/j.celrep.2020.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu T., Guo F., Yu Y., Sun T., Ma D., Han J., Qian Y., Kryczek I., Sun D., Nagarsheth N., et al. Fusobacterium nucleatum promotes chemoresistance to colorectal cancer by modulating autophagy. Cell. 2017;170:548–563.e16. doi: 10.1016/j.cell.2017.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan L., Zhang S., Li H., Yang F., Mushtaq N., Ullah S., Shi Y., An C., Xu J. The influence of gut microbiota dysbiosis to the efficacy of 5-Fluorouracil treatment on colorectal cancer. Biomed. Pharmacother. 2018;108:184–193. doi: 10.1016/j.biopha.2018.08.165. [DOI] [PubMed] [Google Scholar]
- Zhu Q., Gao R., Wu W., Qin H. The role of gut microbiota in the pathogenesis of colorectal cancer. Tumour Biol. 2013;34:1285–1300. doi: 10.1007/s13277-013-0684-4. [DOI] [PubMed] [Google Scholar]
- Zitvogel L., Galluzzi L., Viaud S., Vetizou M., Daillere R., Merad M., Kroemer G. Cancer and the gut microbiota: an unexpected link. Sci. Transl. Med. 2015;7:271ps1. doi: 10.1126/scitranslmed.3010473. [DOI] [PMC free article] [PubMed] [Google Scholar]