Abstract
The microphthalmia-associated transcription factor family (MiT family) proteins are evolutionarily conserved transcription factors that perform many essential biological functions. In mammals, the MiT family consists of MITF (microphthalmia-associated transcription factor or melanocyte-inducing transcription factor), TFEB (transcription factor EB), TFE3 (transcription factor E3), and TFEC (transcription factor EC). These transcriptional factors belong to the basic helix-loop-helix-leucine zipper (bHLH-LZ) transcription factor family and bind the E-box DNA motifs in the promoter regions of target genes to enhance transcription. The best studied functions of MiT proteins include lysosome biogenesis and autophagy induction. In addition, they modulate cellular metabolism, mitochondria dynamics, and various stress responses. The control of nuclear localization via phosphorylation and dephosphorylation serves as the primary regulatory mechanism for MiT family proteins, and several kinases and phosphatases have been identified to directly determine the transcriptional activities of MiT proteins. In different immune cell types, each MiT family member is shown to play distinct or redundant roles and we expect that there is far more to learn about their functions and regulatory mechanisms in host defense and inflammatory responses.
Keywords: autophagy, immune cells, lysosome, metabolism, microphthalmia-associated transcription factor (MITF), MiT family transcription factors, mitochondria, stress response, transcription factor E3 (TFE3), transcription factor EB (TFEB), transcription factor EC (TFEC)
INTRODUCTION
The microphthalmia-associated transcription factor family (MiT family) consists of four transcription factors: MITF (microphthalmia-associated transcription factor or melanocyte-inducing transcription factor), TFEB (transcription factor EB), TFE3 (transcription factor E3), and TFEC (transcription factor EC) (Goding and Arnheiter, 2019; Napolitano and Ballabio, 2016; Oppezzo and Rosselli, 2021). The Mitf gene encoding the first member of the MiT family, MITF, was discovered in 1993 and mutations of Mitf were demonstrated to account for the phenotypes of ‘microphthalmia (mi)’ mice which were originally described in 1942 to have small (microphthalmic) eyes (Hertwig, 1942; Hodgkinson et al., 1993; Krakowsky et al., 1993). In addition to small eyes, mice with a mutant Mitf gene typically display other defects such as deafness and depigmentation (melanocyte defects), and may also show osteopetrosis, mast cell deficiency, heart hypotrophy, and kidney abnormality depending on the type of mutations (Goding and Arnheiter, 2019). For TFE3 and TFEB, corresponding cDNAs were identified in efforts to find immunoglobulin heavy chain enhancer binding proteins (Beckmann et al., 1990; Carr and Sharp, 1990). TFEC was soon afterwards identified in search of a TFE3 homology protein (Zhao et al., 1993). All four MiT family proteins are conserved in vertebrates and a homolog of Mitf is found even in primitive metazoans such as sponges, suggesting that MiT proteins perform important, evolutionary conserved functions (Goding and Arnheiter, 2019; Simionato et al., 2007).
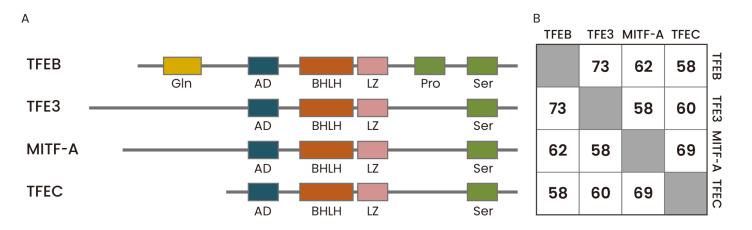
MiT family members share a common basic helix-loop-helix-leucine zipper (bHLH-LZ) domain, which serves as a DNA binding and dimerization domain, and bind DNA as a homodimer or heterodimer (Goding and Arnheiter, 2019). Unlike other bHLH-LZ family transcription factors, MiT family proteins seem to form heterodimers only with transcription factors within the MiT family and not with other bHLH-LZ family proteins (Hemesath et al., 1994). MiT family proteins bind the palindromic, canonical E-box motif (CACGTG) as well as asymmetric M-box sequence (CATGTG) (Aksan and Goding, 1998). The bHLH-LZ domain is well conserved among all members of the MiT family but regulatory regions outside the bHLH-LZ domain show considerable variabilities (Fig. 1). TFEC is the most divergent member of the family and was originally suggested to inhibit TFE3-dependent transcription activation (Zhao et al., 1993).
Fig. 1. Protein domain structures and amino acid sequence similarities of human MiT family proteins.
(A) The four mammalian MiT family members share highly conserved basic helix-loop-helix, leucine zipper (bHLH-ZIP) domain, which serves as a DNA binding and dimerization domain. They also contain an activation domain (or acidic domain, AD) required for transcriptional activation and a serine-rich domain (Ser). In addition, TFEB has a glutamine-rich domain (Gln) and a proline-rich domain (Pro). (B) The amino acid sequence similarities among human MiT family proteins were analyzed using NCBI protein blast. Aligned amino acid sequence ranges are: MITF-A (53-425) vs TFEB (1-349); MITF-A (4-521) vs TFE3 (46-573); MITF-A (221-509) vs TFEC (59-331); TFEB (62-330) vs TFE3 (167-441); TFEB (119-476) vs TFEC (20-347); TFE3 (229-561) vs TFEC (24-331).
TFEB and TFE3 are ubiquitously expressed whereas MITF and TFEC show more tissue-specific expression patterns. Additionally, the Mitf gene has several transcription start sites and its primary RNAs are subjected to various modes of alternative splicing, resulting in at least 12 different protein isoforms (Oppezzo and Rosselli, 2021). Expression of individual MITF isoforms seems to be tissue-specific. For example, the MITF-M isoform is dominantly expressed in melanocytes whereas the MITF-A isoform is more ubiquitously expressed (Goding and Arnheiter, 2019).
Here, we will first briefly summarize diverse functions of MiT family proteins outside the immune system and their common regulatory mechanisms. Then, we will describe the known roles of individual MiT transcription factors in various immune cell types.
BIOLOGICAL FUNCTIONS OF MiT FAMILY TRANSCRIPTION FACTORS OUTSIDE THE IMMUNE SYSTEM
MITF
As mentioned before, mutations of Mitf have been associated with various phenotypes including retinal disorder, deafness, lack of neural crest-derived pigment cells, and defective osteoclast function. Presence of a number of MITF isoforms, due to the multiple transcription start sites in the Mitf gene and alternative splicing events in specific cell types and tissues, may explain the multitude of phenotypes of Mitf mutations (Goding and Arnheiter, 2019). However, it is currently unclear whether each MITF isoform has distinctive roles in the cells they are expressed. Meanwhile, the molecular functions of MITF were most extensively studied in melanocytes and melanoma. Genome-wide analyses of MITF target genes and potential binding sites in melanocytes and melanoma were conducted with multiple approaches (DNA microarray, ChIP-seq, RNA-seq) (Hoek et al., 2008; Strub et al., 2011). The ChIP-seq analysis of genome-wide MITF binding in melanoma cells identified more than 12,000 DNA binding sites and the mRNA expression comparison by RNA-seq analysis between wild type cells and siRNA-mediated MITF knockdown cells showed that several hundred genes are direct targets of MITF (Strub et al., 2011). They include genes controlling cell survival, proliferation, differentiation, senescence, and DNA damage repair. MITF’s roles in cell proliferation seem complex and the ‘rheostat model’ was proposed: low levels of MITF are associated with reduced proliferation, dedifferentiation, and increased invasion, whereas high levels of MITF can be anti-proliferative as well due to cell cycle arrest (Carreira et al., 2005; 2006). In addition, MITF positively regulates peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PGC-1α) that controls mitochondrial biogenesis and oxidative phosphorylation, suggesting the MITF’s role in controlling cellular metabolism (Haq et al., 2013).
MITF, along with TFEB and TFEC, has also been implicated in lysosome biogenesis and autophagy. When MITF-M was over-expressed in melanoma cell lines, the transcription of numerous lysosomal genes and synthesis of lysosomal proteins were enhanced (Ploper et al., 2015). Moreover, microarray analysis of 51 human melanoma cell lines demonstrated that high MITF expression is highly correlated with upregulation of lysosomal genes. MITF also upregulates the transcription of v-ATPase components, which are essential for the acidification of endolysosomes and activation of lysosomal functions (Zhang et al., 2015). Recently, MITF was shown to play an important role in autophagy induction in melanoma (Moller et al., 2019). Autophagosome formation induced by either starvation or mTORC1 inhibition was significantly perturbed and expression of autophagy-related proteins such as LC3B and SQSTM1 was significantly decreased in MITF-deficient cells.
Notably, genomic amplification of Mitf was detected in 5%-20% of melanoma cases (Cancer Genome Atlas Network, 2015). However, critical molecular determinants linking the Mitf amplification and melanoma progression are still ill-defined and poorly understood.
TFEB
Systemic ablation of Tfeb in mice causes embryonic lethality at day E9.5-10.5 due to placental vascularization defects (Steingrimsson et al., 1998). Partly because of the lack of a suitable genetic animal model, TFEB had received relatively little attention until the pioneering works by Ballabio and colleagues, who demonstrated that TFEB transcriptionally induces a network of lysosomal genes (Sardiello et al., 2009). In search for a master transcription factor governing the lysosomal biogenesis, Ballabio and colleagues gathered the promoter sequences of 96 lysosomal genes and found that a palindromic 10 bp sequence (GTCACGTGAC) is highly enriched in the promoter regions of many lysosomal genes. The GTCACGTGAC sequence contains the E-box motif in the middle and is preferentially located about 200 bp upstream of the transcription start sites. They named the sequence as ‘coordinated lysosomal expression and regulation (CLEAR)’ element and demonstrated that TFEB binds the CLEAR elements and upregulates lysosomal gene expression. They further showed that TFEB overexpression results in an expansion of lysosomal compartment and clearance of mutant Huntingtin proteins (Sardiello et al., 2009). Additionally, TFEB was also shown to promote lysosome exocytosis, further contributing to intracellular clearance (Medina et al., 2011). Following these discoveries, many studies have demonstrated that genetically or chemically induced overexpression of TFEB ameliorates lysosomal storage disorders, such as Pompe disease, Gaucher disease, and Batten disease, as well as neurodegenerative diseases, including Huntington’s, Alzheimer’s, and Parkinson’s diseases, in which accumulation of protein aggregates serves as a major pathogenic mechanism (Martini-Stoica et al., 2016; Napolitano and Ballabio, 2016).
Soon after the discovery of TFEB as a master regulator of lysosome biogenesis, its role in autophagy was also revealed (Palmieri et al., 2011; Settembre et al., 2011). Many autophagy-associated genes, such as UVRAG, VPS11, and WIPI, contain the CLEAR element in their promoter regions and are direct targets of TFEB. TFEB is activated by nutrient starvation, a well-known condition for autophagy induction, and TFEB overexpression increases autophagosome formation, autophagosome-lysosome fusion, and substrate degradation. In addition, TFEB facilitates the clearance of damaged mitochondria and lipid droplets via induction of mitophagy and lipophagy (Nezich et al., 2015; Settembre et al., 2013).
Aside from an induction by starvation or deposit of undesirable macromolecules and damaged organelles in the cytoplasm, autophagy can also be induced by an accumulation of unfolded or misfolded proteins in the endoplasmic reticulum (ER) (Rashid et al., 2015). In cells where ER stress was triggered by treatment with brefeldin A and tunicamycin, TFEB and TFE3 become activated and directly induce transcription of ATF4, one of the major transcription factors controlling the unfolded protein response (UPR), and other UPR-associated genes (Martina et al., 2016). TFEB and TFE3 are also activated by genotoxic stress and DNA damage. In TFEB/TFE3 double-knockout cells exposed to DNA damaging conditions, the half-life of p53 is significantly shortened and cell cycle checkpoint gene induction and apoptosis responses are dysregulated (Brady et al., 2018). Therefore, TFEB and TFE3 seem to play critical roles in integrated stress responses in general under various cellular stress conditions such as nutrient deprivation, ER stress, and DNA damage.
TFEB also controls cellular metabolism. Overexpression of TFEB in mouse liver results in major expression changes of genes in cellular lipid metabolic processes via direct transcriptional control of Ppargc1a (encoding PGC-1α) (Settembre et al., 2013). Specifically, genes related to lipid catabolism and oxidation are upregulated by TFEB overexpression whereas genes responsible for lipid biosynthesis such as steroid, fatty acid, and isoprenoid biosynthetic processes are downregulated. During starvation, TFEB expression goes up in the liver, muscle, and kidney, and TFEB controls lipid catabolism in the liver by engaging the autophagic pathway (Settembre et al., 2013). In skeletal muscles, TFEB has been shown to regulate mitochondria biogenesis and control energy balance during exercise (Mansueto et al., 2017). Interestingly, unlike in the liver, the effects of TFEB on cellular metabolism in skeletal muscles do not require the presence of PGC-1α, and neither TFEB activation nor depletion affects autophagy flux in the muscle. Related with the roles of TFEB in metabolic control, the liver-specific overexpression of TFEB in mice prevents development of metabolic syndrome in high fat diet-fed animals and ameliorates ethanol-induced liver injury (Chao et al., 2018; Settembre et al., 2013).
The function of TFEB in cancer has been studied in several cancer types; renal cell carcinoma (RCC) in particular is one of the most studied (Perera et al., 2019; Puertollano et al., 2018). TFEB overexpression, due to chromosomal translocation or genetic amplification, was found in a group of RCC patients, and the TFEB overexpression seems to be associated with poor prognosis (Argani et al., 2016). Moreover, a positive correlation between TFEB and PD-L1 expression has been found in human primary RCC and murine RCC xenograft models and TFEB was suggested to mediate immune evasion and resistance to mTOR inhibition (Zhang et al., 2019). In addition, TFEB has been implicated in specific killing of B cell non-Hodgkin lymphoma (B-NHL) by apilimod, a PIKfyve kinase inhibitor. Apilimod induces B-NHL cell death by destabilization of lysosomes. Interestingly, TFEB is most highly expressed in B-NHL compared to other cancer types and deletion of TFEB by CRISPR-mediated gene knockout gives rise to resistance of B-NHL to apilimod (Gayle et al., 2017). However, functional relevance of TFEB in various cancers and the exact regulatory mechanism in tumorigenesis remain to be elucidated.
TFE3
The tissue expression pattern and known biological functions of TFE3 highly overlap with those of TFEB, even though Tfe3 knockout mice are viable, unlike Tfeb knockout mice. Like TFEB, TFE3 binds the CLEAR motif and induces transcription of lysosomal and autophagy-related genes, and overexpression of TFE3 enhances lysosomal biogenesis (Martina et al., 2014). As mentioned above, TFE3, along with TFEB, also responds to stress signals such as DNA damage (Brady et al., 2018). In terms of metabolic controls, TFE3 overexpression in hepatocytes has been shown to induce transcriptional activation of IRS-2 expression, enhance insulin receptor signaling, and have protective effects in mouse diabetes models by increasing liver glycogen synthesis and decreasing liver triglyceride and blood glucose levels (Nakagawa et al., 2006). Similarly, TFE3 overexpression in skeletal muscles also increases the glycogen synthesis, muscle mass, and insulin sensitivity (Iwasaki et al., 2012). Conversely, Tfe3 knockout mice show defects in mitochondria dynamics, abnormalities in systemic glucose and lipid metabolism, and enhance high fat diet-induced obesity and diabetes (Pastore et al., 2017). Notably, TFEB overexpression in Tfe3 knockout mice as well as TFE3 overexpression in liver-specific Tfeb knockout mice rescues the diet-induced obesity, demonstrating that TFEB and TFE3 can compensate for deficiency of each other. TFE3 has also been implicated in adipose tissue browning (Wada et al., 2016). However, an earlier study showed that adipocyte-specific overexpression of TFE3 suppresses lipolysis and thermogenesis, and therefore the role of TFE3 in adipose tissue browning needs further clarification (Fujimoto et al., 2013).
Like TFEB, TFE3 has been implicated in development and progress of various tumors (Perera et al., 2019; Puertollano et al., 2018). Chromosomal translocation and fusion of TFE3 gene are often found in RCC, alveolar soft part sarcoma, and perivascular epithelioid cell tumor (Ladanyi et al., 2001; Tanaka et al., 2009; Weterman et al., 1996). TFE3, along with MITF and TFEB, also plays an important role in pancreatic cancer (Perera et al., 2015). TFE3 is also implicated in maintaining stem cell state, i.e. preventing differentiation, of embryonic stem cells (Betschinger et al., 2013) and specific point mutations of TFE3, which render the mutant TFE3 proteins hyperactive, are associated with a human developmental disorder (Villegas et al., 2019).
TFEC
TFEC was initially described as lacking the acidic domain (AD) required for transcriptional transactivation and inhibit TFE3-mediated transcriptional activation (Zhao et al., 1993). However, a more recent study found that TFEC functions as a transcriptional activator of the non-muscle myosin II heavy chain A gene in transfected cells (Chung et al., 2001). TFEC expression is highly restricted in macrophages and its physiological roles has been little studied (Rehli et al., 1999).
REGULATION OF MiT FAMILY TRANSCRIPTION FACTORS
The control of intracellular localization serves as a major regulatory mechanism for the function of MiT family transcription factors. Transcriptional factors need to be inside the nucleus to be functional and the nuclear localization of MiT proteins is mainly controlled by phosphorylation and dephosphorylation events (Puertollano et al., 2018). Various kinases and phosphatases have been identified to control the nuclear localization of MITF, TFEB, and TFE3 and phosphorylation target sequences and regulatory mechanisms seem to be well conserved among the three proteins (Fig. 2). Therefore, here we will mainly describe the regulatory mechanisms of TFEB as representative examples.
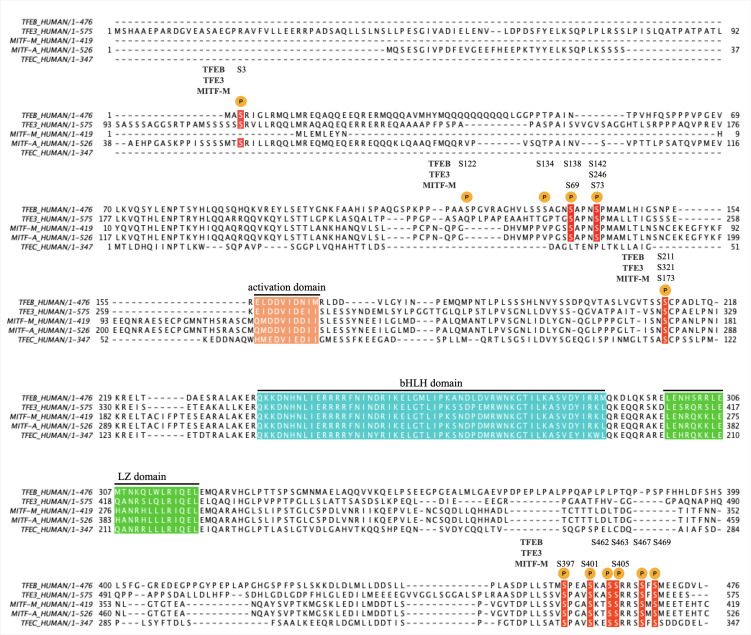
Fig. 2. Amino acid sequence alignment of human MiT family proteins.
A multiple sequence alignment of TFEB (P19484-1), TFE3 (P19532-1), MITF-M (O75030-9), MITF-A (O75030-1), and TFEC (O14948-1) was performed using clustal Omega (ver. 1.2.4). Major phosphorylation sites are denoted as P, and the conserved phosphorylation sites are in red. The activation domain, bHLH (basic-helix-loop-helix) domain, and LZ (leucine zipper) domain are also indicated.
Regulation of TFEB localization by phosphorylation/dephosphorylation was first clearly demonstrated in cells undergoing starvation (Settembre et al., 2011). TFEB is normally phosphorylated at multiple serine residues and stays in the cytoplasm. Upon starvation, it becomes dephosphorylated and translocates into the nucleus to induce autophagy. Among several possible phosphorylation sites, Ser-142 was first identified as a key residue for controlling the nuclear localization. The phospho-mimetic mutation at Ser-142 (S142D) of TFEB inhibits the nuclear translocation and autophagy induction whereas TFEB with the S142A mutation is constitutively localized in the nucleus and induces autophagy even without starvation. Based on a bioinformatics analysis, ERK2 was suggested as the kinase responsible for the phosphorylation of Ser-142 (Settembre et al., 2011). Ser-142 in TFEB corresponds to Ser-73 in MITF-M and the phosphorylation at Ser-73 was also shown to be mediated by ERK2 (Hemesath et al., 1998; Ngeow et al., 2018). Subsequent studies have demonstrated that mTOR kinase couples nutrient conditions to the TFEB subcellular localization (Settembre et al., 2012). In a normal, nutrient-sufficient condition, mTOR is active and phosphorylates TFEB, thereby sequestering it in the cytoplasm. At least three serine residues, Ser-122, 142, and 211, are identified as the phosphorylation target sites of mTOR and inhibition of mTOR induces nuclear translocation of TFEB even in the nutrient-rich conditions (Martina et al., 2012; Roczniak-Ferguson et al., 2012; Vega-Rubin-de-Celis et al., 2017). Moreover, similar to S142A mutation, S211A mutation alone can induce constitutive nuclear localization of TFEB whereas S122A mutation by itself is insufficient in driving nuclear localization of TFEB. Therefore, dephosphorylation at either S142 or S211 seems to be a critical step for nuclear translocation of TFEB. Interestingly, Ser-142 and 211 are conserved in MITF and TFE3, but Ser-122 is only found in TFEB (Fig. 2). Phosphorylation of Ser-211 induces a physical interaction between TFEB and cytosolic 14-3-3 protein, potentially explaining the cytosolic retention of TFEB (Martina et al., 2012; Roczniak-Ferguson et al., 2012). Similarly, phosphorylation of Ser-173 in MITF and Ser-321 in TFE3, each corresponding to Ser-211 in TFEB, facilitates the interaction of MITF and TFE3 with 14-3-3 and retains the proteins in the cytoplasm (Bronisz et al., 2006; Hsu et al., 2018; Martina et al., 2016).
The mTOR-dependent regulation of TFEB has been elucidated more precisely (Raben and Puertollano, 2016). Lysosomal nutrient sensing mechanisms activate Ragulator, a guanine nucleotide exchange factor for Rag GTPases, located on the lysosomal membrane. Rag GTPases are small GTPases normally present as heterodimers of RagA/B - RagC/D. The activated Ragulator promotes the GDP/GTP exchange of RagA/B to form a RagA/BGTP-RagC/DGDP state, which is an active state of the Rag heterodimer. The activated Rag heterodimer recruits mTORC1 and TFEB onto the lysosomal surface, enabling mTORC1-mediated phosphorylation of TFEB. The first 30 amino acids of TFEB are shown to be essential for Rag-TFEB interaction (Martina and Puertollano, 2013). In starvation conditions, Rag GTPases return to an inactive RagA/BGDP-RagC/DGTP form, which cannot recruit mTORC1 for TFEB phosphorylation, and TFEB is subsequently released from cytosolic 14-3-3 protein and translocates to the nucleus for autophagy induction. A recent study further showed that the interaction of TFEB with the Ragulator/Rag GTPase/mTORC1 complex depends on Ser-3 phosphorylation by MAP4K3 and the MAP4K3-mediated Ser-3 phosphorylation precedes the phosphorylation of Ser-211 by mTORC1 (Hsu et al., 2018). Ser-3 is also conserved in TFE3 and MITF-A, suggesting that both proteins may be subject to the same mode of regulation by MAP4K3 (Fig. 2). In energy-deficient conditions, AMPK becomes activated and promotes nuclear translocation of TFEB and TFE3 without affecting the mTORC1 signaling pathway. However, it is unclear if AMPK directly phosphorylates TFEB and TFE3 and how AMPK facilitates their nuclear translocation (El-Houjeiri et al., 2019).
Several other kinases have been identified to phosphorylate TFEB in a mTOR-independent manner. GSK3β phosphorylates Ser-134 and Ser-138 and inhibits nuclear translocation of TFEB (Li et al., 2016). Accordingly, inhibition of GSK3β results in TFEB nuclear localization and autophagy induction (Marchand et al., 2015; Parr et al., 2012). A recent study revealed that phosphorylation of TFEB at Ser-142 primes for GSK3β-mediated phosphorylation at Ser-138. Phosphorylation of both sites, but neither alone, promotes CRM1 (also called as XPO1)-mediated nuclear export of TFEB (Li et al., 2018). Similarly, Ser-69 in MITF-M, a counterpart of Ser-138 in TFEB, is also phosphorylated by GSK3, contributing to the nuclear export of MITF-M (Ngeow et al., 2018). AKT-mediated phosphorylation of TFEB at Ser-467 also causes the cytoplasmic retention of TFEB while AKT inhibition causes nuclear translocation of TFEB even in the presence of constitutively active mTORC1 (Palmieri et al., 2017). Trehalose, a disaccharide having an autophagy-inducing activity, has been shown to activate TFEB via AKT inhibition and pharmacological inhibition of AKT was suggested as a potential therapeutic strategy for neurodegenerative storage diseases (Palmieri et al., 2017; Sarkar et al., 2007). CDK4 and CDK6, which are located in the nucleus, also phosphorylate TFEB and TFE3 at Ser-142 and Ser-246, respectively and promote their nuclear export (Yin et al., 2020). The CDK4/6-mediated phosphorylation of TFEB and TFE3 is suggested to regulate cell cycle-dependent lysosome biogenesis, and a CDK4/6 inhibitor promotes lysosomal activation and cellular clearance of various substrates.
In contrast to phosphorylation by mTOR, GSK3β, AKT, and CDK4/6, which inhibit nuclear translocation and transcription-activating function of TFEB, PKCβ-mediated phosphorylation of multiple serine residues (Ser-462, Ser-463, Ser-467, Ser-469) in the C-terminal tail of TFEB stabilizes the protein and induces its nuclear localization, resulting in lysosomal biogenesis (Ferron et al., 2013). The C-terminal serine residues are conserved in all four MiT family members. However, Ser-405 in MITF-M (corresponding to Ser-463 in TFEB), along with Ser-397 and Ser-401, seems to be phosphorylated by GSK3 and inhibition of GSK3-mediated phosphorylation promotes stabilization and nuclear translocation of MITF-M, which enhances endolysosomal biogenesis and potentiates Wnt signaling in melanoma cells (Ploper et al., 2015).
As discussed above, dephosphorylation of TFEB at a few key serine residues induces translocation of TFEB into the nucleus where TFEB performs its transcription-activating functions. A siRNA-based screen has identified calcineurin as the major phosphatase for TFEB dephosphorylation (Medina et al., 2015). Similarly, calcineurin also plays a critical role in nuclear localization and activation of TFE3 upon ER stress (Martina et al., 2016). Calcineurin is activated by high intracellular calcium concentration and, therefore, calcium ionophores facilitate nuclear localization of TFEB whereas calcium chelators inhibit it (Medina et al., 2015). In cells under nutrient starvation, the lysosomal calcium channel MCOLN1, also called TRPML1, becomes activated and releases calcium into cytosol from lysosomes (Wang et al., 2015). This results in activation of calcineurin and TFEB translocation. Accordingly, TFEB-mediated enhancement of lysosomal functions is inhibited by blocking MCOLN1 channel expression or activity. PP2A can also dephosphorylate TFEB and TFE3 (Martina and Puertollano, 2018). Activation of TFEB and TFE3 upon acute oxidative stress caused by sodium arsenite treatment involves dephosphorylation of Ser-211 of TFEB and Ser-321 of TFE3. PP2A seems to be able to dephosphorylate TFEB at additional residues, including Ser-109, Ser-114, and Ser-122.
In addition to the localization-based regulation by phosphorylation/dephosphorylation, TFEB activity is subjected to various alternative post-translational regulatory mechanisms. An E3 ubiquitin ligase STUB1 targets phosphorylated, inactive TFEB for degradation via the ubiquitin/proteasome pathway (Sha et al., 2017). In STUB1-deficient cells, phosphorylated TFEB accumulates in the cytoplasm, resulting in reduced TFEB activity. Regulation of protein stability by the ubiquitin/proteasome pathway is also demonstrated for MITF-M, and phosphorylation at Ser-73 and Ser-409 seems to be critical for ubiquitination and degradation of MITF-M (Wu et al., 2000; Xu et al., 2000; Zhao et al., 2011). MITF-M is also regulated by another ubiquitination-like modification, sumoylation, and the sumoylation regulates MITF-M’s transcriptional activity on a subset of target genes (Murakami and Arnheiter, 2005). A histone acetyltransferase GCN5 functions as a TFEB-specific acetyltransferase and acetylation of TFEB by GCN5 decreases TFEB transcriptional activity, thereby inhibiting autophagy induction (Wang et al., 2020). Transcriptional activity of TFEB is also controlled by liquid-liquid phase separation, which promotes the formation of transcriptional condensates for efficient gene expression. Inositol polyphosphate multikinase, IPMK was shown to directly interact with TFEB and inhibit liquid-liquid phase separation of TFEB in the nucleus. Consequently, depletion of IPMK leads to increased TFEB activity and promotion of autophagy and lysosomal functions (Chen et al., 2020).
IMMUNOREGULATORY FUNCTIONS OF MiT FAMILY TRANSCRIPTION FACTORS
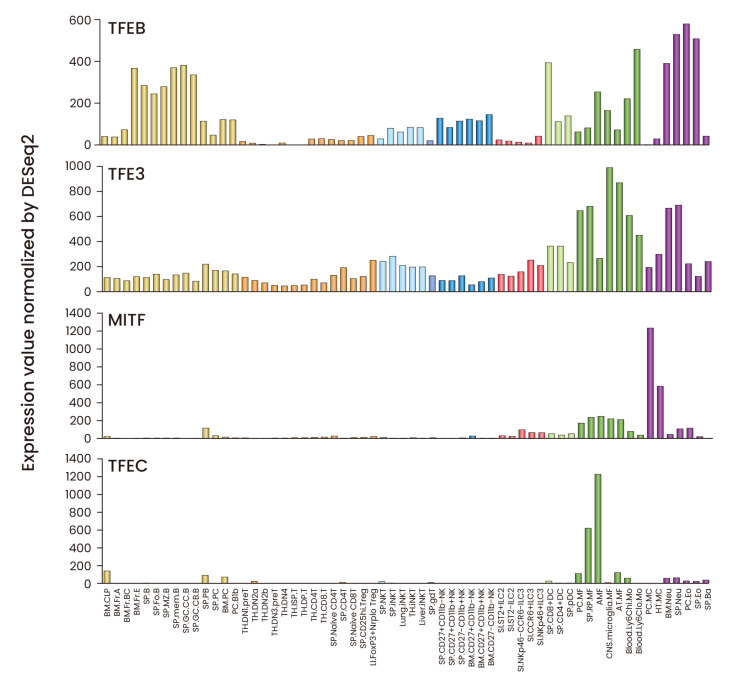
MiT family proteins are expressed in various immune cell types. Analysis of mRNA expression shows that TFEB and TFE3 are expressed in most of immune cell types whereas MITF expression is restricted to myeloid cells with the highest expression in mast cells. TFEC is highly expressed in a subset of macrophages but not in other immune cell types (Fig. 3, Supplementary Table S1). Below we summarize the known function of MiT family proteins in individual immune cell types (Fig. 4).
Fig. 3. Expression patterns of MiT family proteins in mouse immune cells.
The mRNA expression data were downloaded from the ImmGen (Immunological Genome Project) database (https://www.immgen.org) and displayed in bar graphs. The mRNA expression values and the full names of individual cell types can be found in Supplementary Table S1.
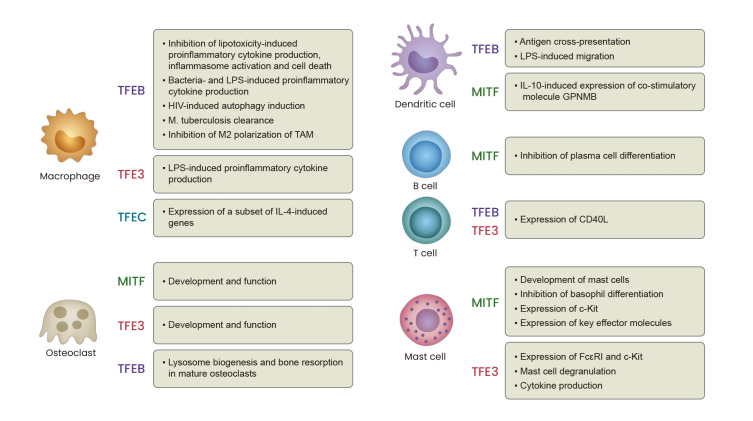
Fig. 4. Diverse functions of MiT family proteins in immune cells.
MiT family transcription factors play diverse roles in various immune cell types. The known roles of each transcription factor in macrophages, osteoclasts, dendritic cells, B cells, T cells, and mast cells are summarized.
Macrophages
Phagocytosis is one of the most important functions of macrophages and MiT family transcription factors regulate lysosome biogenesis, phagosome-lysosome fusion and autophagy in macrophages as in other cell types. In addition, they also regulate cell death and expression of cytokines and chemokines in macrophages. One of the earliest studies on TFEB in macrophages shows that cell death responses induced by co-stimulation with lipopolysaccharide (LPS) and palmitate are preceded by a significant depletion of lysosomes, and TFEB overexpression can rescue the lipotoxicity-induced cell death by preventing lysosome depletion (Schilling et al., 2013). The follow-up study by the same group found that inhibition of mTOR induces nuclear localization of TFEB and protects macrophages from lipotoxic cell death. The protective effect of mTOR inhibition was autophagy-independent and was surprisingly not affected by siRNA-mediated TFEB knockdown (He et al., 2016). However, it is not clear whether a residual presence of TFEB due to an incomplete depletion by the siRNA-mediated approach is enough to mediate the beneficial effect of mTOR inhibition. Treatment of palmitate in LPS-stimulated macrophages also results in increased secretion of proinflammatory cytokines. Ezetimibe, a cholesterol transporter blocker and U.S. Food and Drug Administration (FDA)-approved lipid lowering drug, increases nuclear localization of TFEB, induces autophagy, and reduces both inflammatory cytokine production and NLRP3 inflammasome activation in macrophages co-stimulated with LPS and palmitate (Kim et al., 2017a). These findings suggest that TFEB may exert anti-inflammatory functions in macrophages via autophagy induction in a lipotoxic condition.
In contrast, TFEB and TFE3 are required for production of proinflammatory cytokines and chemokines in responses to pathogen stimulation. Infection of macrophages with Staphylococcus aureus and Escherichia coli promotes nuclear translocation of TFEB, and TFEB depletion using siRNA significantly reduces secretion of proinflammatory cytokines, such as interleukin (IL)-1β, IL-6, and tumor necrosis factor α (TNF-α)) (Visvikis et al., 2014). Similarly, LPS stimulation of macrophages also resulted in rapid nuclear translocation of TFEB and TFE3 (Pastore et al., 2016). Interestingly, the cellular TFEB level goes down after 24 h of LPS stimulation and TFEB is no longer detected in the nucleus, whereas the TFE3 level is constantly maintained and TFE3 stays in the nucleus even after 48 h, suggesting that TFEB and TFE3 are differentially regulated and play possibly non-redundant roles. Genetic ablation of both Tfeb and Tfe3 results in severe impairment in LPS-induced secretion of cytokines and chemokines (Pastore et al., 2016). A more recent study demonstrated that LPS induces TFE3 activation via inhibition of folliculin (FLCN) (Li et al., 2019). FLCN functions as a GTPase-activating protein for RagC/D and thereby activates mTORC1 activity. Depletion of FLCN in macrophages leads to constitutive localization of TFE3 in the nucleus and increased expression of TFE3 target genes. In addition, activated TFE3 induces transcriptional upregulation of RagD and increases mTORC1 activity, forming a feedback loop. The FLCN-depleted macrophages are hyperresponsive to LPS stimulation and secrete significantly larger amounts of proinflammatory cytokines than wild type cells do and this hyperresponsiveness depends on the presence of TFE3. Moreover, loss of FLCN in mouse monocyte lineages results in myeloid cell progenitor expansion in tissues, chronic macrophage activation, and tissue disruptions via TFE3 activation (Li et al., 2019). Therefore, a tight balance of TFE3 activity seems to be critical for homeostatic maintenance of innate immune cells.
HLH-30, a sole MiT protein in Caenorhabditis elegans, is also required for host defense responses against bacterial infection in nematodes. HLH-30 rapidly translocates into the nucleus upon S. aureus infection and almost 80% of genes are upregulated in an HLH-30-dependent manner in C. elegans (Visvikis et al., 2014). These studies demonstrate that MiT family proteins are evolutionarily ancient transcriptional factors performing essential host defense functions.
During infection by various intracellular microorganisms, TFEB plays a critical role in antimicrobial responses by enhancing lysosomal functions and autophagy induction. On the other hand, many pathogens themselves seem to either activate or inhibit TFEB for their own benefit. Interferon (IFN)-γ-stimulated macrophages restrict HIV-1 infection by expressing apolipoprotein L1 (APOL1), which promotes nuclear translocation of TFEB and degradation of viral proteins Gag and Vif. Conversely, HIV-1 Nef protein activates mTOR and sequesters TFEB in the cytoplasm to inhibit lysosomal degradation and autophagy for establishment of permissive infection (Campbell et al., 2015; Taylor et al., 2014).
Mycobacterium tuberculosis (Mtb) replicates inside macrophages by inhibiting autophagy and lysosomal function and by promoting lipid body formation. Upon Mtb infection, expression of miR-33 and its passenger strand miR-33* become upregulated in macrophages and the miRNAs inhibit autophagy by targeting AMPK and TFEB (Ouimet et al., 2016). Accordingly, silencing of miR-33 and miR-33* enhances Mtb clearance via autophagy induction, whereas Tfeb knockdown delays Mtb clearance (Kim et al., 2017b). IFN-γ is a potent activator of macrophages and shows antimycobacterial effects by inducing autophagy. Interestingly, IFN-γ can induce TFEB nuclear translocation through increasing intracellular calcium levels and activating calcineurin, and depletion of calcineurin significantly inhibits antimycobacterial effects of IFN-γ (Singh et al., 2018).
In the case of Coxiella burnetii, the bacteria replicate inside the lysosome-derived Coxiella-containing vacuoles and establishment of this unique bacterial survival niche depends on the nuclear translocation of TFEB and TFE3 and the consequent expression of a network of genes involved in lysosome biogenesis and autophagy (Padmanabhan et al., 2020). Salmonella enterica, another intracellular bacterial pathogen, also significantly increases nuclear localization of TFEB and induces expression of genes associated with immune responses and autophagy (Carey et al., 2020; Najibi et al., 2016). However, another study shows that TFEB expression in bone marrow-derived macrophages decreases upon Salmonella infection and, therefore, the role of TFEB in Salmonella infection needs a further clarification (Rao et al., 2020).
The role of TFEB has also been studied in tumor-associated macrophages (TAMs) in mouse breast cancer models. TAMs isolated from orthotopic breast cancer tissues express lower levels of TFEB and peritoneal macrophages co-cultured with tumor cell-conditioned media (TCM) downregulate expression of TFEB, but not TFE3 and MITF (Fang et al., 2017). Among cancer-derived molecules, transforming growth factor β (TGF-β), but not IL-4 and IL-10, was identified to inhibit TFEB expression and its nuclear translocation. Moreover, TFEB depletion via shRNA led to increased expression of Arg1 and YM1, markers of M2-polarized macrophages, in IL-4- or TCM-treated macrophages, indicating that TFEB potentially inhibits M2-like polarization of macrophages. Indeed, TFEB depletion in macrophages decreased their ability to activate T cells and enhanced cancer cell growth. Conversely, macrophage-specific overexpression of TFEB suppressed breast tumor growth in mice (Fang et al., 2017). A follow-up study by the same group has demonstrated that TFEB activation without TFEB overexpression by trehalose treatment also inhibits tumor growth and TFEB expression is a positive prognostic marker for breast cancer in humans (Li et al., 2020).
TFEC is highly and specifically expressed in macrophages and mice genetically lacking TFEC develop normally and show no obvious defects (Rehli et al., 1999; 2005). TFEC expression in macrophages is further upregulated by IL-4 in a STAT6-dependent manner and a small number of IL-4-induced genes, including Csf3r encoding the G-CSF receptor, are downregulated in TFEC-deficient macrophages (Rehli et al., 2005). In another study, TFEC upregulated by IL-4 was shown to directly bind the promoter region of IL-4Rα and increase its expression, thereby forming the IL-4/TFEC/IL-4R positive feedback loop and facilitating the M2 polarization of macrophages (Wang et al., 2017).
Osteoclasts
Osteoclasts, specialized cells differentiated from monocyte/macrophage precursors in the bone tissues, degrade and reabsorb old bone matrix to maintain bone homeostasis. Osteopetrosis (‘stony bone’ or bone hardening) has long been observed in many but not all of the mice with various Mitf mutant alleles, and it was revealed that only dominant-negative forms of Mitf mutation strongly affect osteoclast development and cause osteopetrosis. Accordingly, TFE3 was identified to bind MITF and has a redundant role in osteoclast development and function (Hershey and Fisher, 2004; Steingrimsson et al., 2002). Mice lacking either Mitf or Tfe3 alone show no bone defects, but Mitf/Tfe3-double knockout mice show the same bone defects as mice having the dominant-negative forms of Mitf mutation. MITF and TFE3 become activated by M-CSF and RANKL signaling pathways during osteoclast differentiation. They are essential for osteoclast proliferation and differentiation and regulate a network of genes associated with bone-degrading function of mature osteoclasts. In addition to MITF and TFE3, TFEB is also activated upon RANKL stimulation via a PKCβ-mediated phosphorylation of C-terminal serine residues and it enhances lysosomal biogenesis in mature osteoclasts. Consequently, osteoclast-specific ablation of TFEB impairs bone resorption processes (Ferron et al., 2013).
Dendritic cells
Dendritic cells (DCs) bridge innate and adaptive immune responses. They capture pathogens in the peripheral tissues, migrate into regional draining lymph nodes, and present pathogen-derived peptide antigens loaded on MHC I and MHC II to cognate T cells for initiation of T cell-dependent immune responses. The classical MHC I pathway mediates presentation of intracellular pathogen-derived peptides, that are synthesized inside DCs, to CD8+ T cells. In contrast, the MHC II pathway presents extracellular antigens, which gain entry to the endolysosomal compartments of DCs via endocytosis or phagocytosis, and activates CD4+ T cells. In addition to the classical MHC I and II pathways, a specific subset of DCs—termed cDC1—can also display peptides from extracellular antigens onto MHC I and present them to CD8+ T cells via the cross-presentation pathway. The cross-presentation pathway in cDC1 involves endocytosis/phagocytosis of extracellular antigens and their transport from endolysosomal compartments into the cytoplasm. For efficient cross-presentation, antigenic peptides need to be preserved inside the endolysosomes without too much degradation. Therefore, the lysosomal pH and proteolytic capacity of cDC1 need to be carefully regulated and, in general, a milder pH and a weaker proteolysis in the endolysosomes are preferred for cross-presentation (Joffre et al., 2012). Accordingly, TFEB expression is usually lower in cDC1 compared to other DC subsets. Furthermore, overexpression of TFEB results in decreased lysosomal pH and increased expression of proteases (CatD, CatL, and CatS), leading to an inefficient cross-presentation and a rather enhanced MHC II pathway-mediated presentation of extracellular antigens. Consequently, TFEB overexpression in DCs reduces extracellular antigen-induced CD8+ T cell activation in vivo (Samie and Cresswell, 2015).
TFEB also plays a role in DC migration. After pathogen sensing in the periphery, mature DCs migrate towards lymph nodes in a continuous, directional manner, via activation and positioning of actin-based motor protein myosin II at the cell rear. This event is regulated by lysosomal calcium release through MCOLN1 located on the lysosomal membrane. LPS-activated TFEB induces MCOLN1 expression and is required for fast DC migration (Bretou et al., 2017).
In the case of MITF, it was shown to be phosphorylated and to translocate into the nucleus in monocyte-derived DCs after treatment of IL-10 or an AKT inhibitor, resulting in the expression of a coinhibitory molecule GPNMB (Gutknecht et al., 2015). However, the physiological role of MITF in DCs has not been extensively studied.
B cells
The original mi/mi mice, in which MITF is non-functional due to deletion of a critical arginine residue in the bHLH-LZ domain, display an absence of maturing B cells in the bone marrow and the osteopetrotic phenotype and defective bone marrow environment of the mi/mi mice was suggested to be responsible for the observed B cell development defects (Roundy et al., 1999; 2003). Therefore, when bone marrow cells from the mi/mi mice were transferred into RAG2-deficient recipients, development of both B1 and B2 cells, as well as T cells, were largely normal, confirming that endogenous MITF is dispensable for lymphocyte development (Lin et al., 2004). However, the chimeric mice displayed spontaneous B cell activation, higher IgM antibody titers, and autoantibody production, and it was found that MITF inhibits antibody-secreting plasma cell differentiation by normally repressing expression of IRF4, a transcription factor critical for the terminal differentiation of plasma cells (Lin et al., 2004).
In case of TFE3, it was initially suggested to play a role in B cell activation because chimeric mice generated by injection of embryonic stem cells having a defective TFE3 gene into RAG2-/- blastocysts showed decreased serum antibody levels despite the normal B and T cell development (Merrell et al., 1997). However, detailed analysis of another TFE3-targeted mice showed no B cell phenotypes and the functional role of TFE3 in B cells still needs to be discovered (Steingrimsson et al., 2002).
T cells
In T cells, TFE3 is constitutively expressed while TFEB expression is post-transcriptionally induced by T cell receptor activation (Huan et al., 2006). Mice expressing a dominant negative mutant TFE3 protein, containing the HLH-LZ domain without the DNA-binding basic region and the transcriptional activation domain, in T cells show defective germinal center responses and a hyper-IgM syndrome due to an impaired expression of CD40L in CD4+ T cells. Notably, Cd40lg (encoding CD40L) contains several E-box motifs in the promoter region where both TFE3 and TFEB directly bind to activate the transcription. The dominant negative mutant TFE3 is expected to inhibit functions of both TFE3 and TFEB by forming non-functional heterodimers. Genetic depletions of both TFE3 and TFEB also result in defective CD40L expression, whereas ablation of either alone shows a normal CD40L expression, suggesting that TFE3 and TFEB play a redundant role for CD40L expression in T cells (Huan et al., 2006).
Mast cells
MITF is most highly expressed in mast cells and various mast cell defects were identified in several MITF mutant mouse strains (Kitamura et al., 2002). MITF deficiency in either mast cells themselves or in the surrounding tissue environment has been shown to impact the mast cell development, indicating both intrinsic and extrinsic roles of MITF in mast cell development (Morii et al., 2004). Specifically, MITF has been shown to enhance expression of key molecules for mast cell development and function, including c-Kit (colony-stimulating factor receptor), tryptases, chymases, and tryptophan hydroxylase (Kitamura et al., 2002). Recently, it has been demonstrated that mast cells differentiate from ‘pre-basophil and mast cell progenitors (pre-BMPs)’ while antagonistic regulation by MITF and C/EBPα specifies the mast cell versus basophil fate. MITF directs the differentiation of pre-BMPs into mast cells and represses basophil development by inhibiting expression of C/EBPα. Conversely, C/EBPα promotes basophil development and prevents mast cell development by inhibiting MITF expression. Importantly, overexpression of MITF alone in committed basophil progenitors is sufficient in redirecting the cellular programming into mast cell development (Qi et al., 2013).
TFE3 has also been implicated in mast cell function. TFE3-deficient mice show normal mast cell numbers in various tissues but expression of FcεRI and c-Kit is significantly lower in TFE3-deficient mast cells. Moreover, TFE3-deficient mast cells display defective functionalities including impaired degranulation and cytokine releases in vitro and compromised allergic responses in vivo (Yagil et al., 2012).
PERSPECTIVES
Since the first discovery of MITF almost 30 years ago, the biological roles of the MiT family transcriptional factors and their regulatory mechanisms have been studied by many cell biologists. Thanks to their successful efforts, we now know that MiT family proteins perform several important functions in a variety of cellular processes such as proliferation, differentiation, lysosome biogenesis, autophagy, metabolism, and stress responses. However, despite the fact that MiT family proteins perform essential and evolutionarily conserved host defense responses in lower organisms, our understanding of the roles of MiT family transcriptional factors in mammalian immune cell functions are still limited. Considering the growing attention on the roles of lysosomal functions and cellular metabolism on immune cell regulation, we anticipate that far more will soon be revealed on how MiT family proteins control the immune system and the homeostasis of organisms at large.
Supplemental Materials
Note: Supplementary information is available on the Molecules and Cells website (www.molcells.org).
ACKNOWLEDGMENTS
We apologize for not being able to describe and cite many important works related to biology of the MiT family proteins due to space limitation. We thank Hojune Kwak for English editing. This work was supported by grants from the National Research Foundation of Korea (2016M3A9D3918546, 2020R1A2C2011307) and Korea Advanced Institute of Science and Technology (KAIST).
Footnotes
CONFLICT OF INTEREST
The authors have no potential conflicts of interest to disclose.
REFERENCES
- Aksan I., Goding C.R. Targeting the microphthalmia basic helix-loop-helix-leucine zipper transcription factor to a subset of E-box elements in vitro and in vivo. Mol. Cell. Biol. 1998;18:6930–6938. doi: 10.1128/MCB.18.12.6930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argani P., Reuter V.E., Zhang L., Sung Y.S., Ning Y., Epstein J.I., Netto G.J., Antonescu C.R. TFEB-amplified renal cell carcinomas: an aggressive molecular subset demonstrating variable melanocytic marker expression and morphologic heterogeneity. Am. J. Surg. Pathol. 2016;40:1484–1495. doi: 10.1097/PAS.0000000000000720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckmann H., Su L.K., Kadesch T. TFE3: a helix-loop-helix protein that activates transcription through the immunoglobulin enhancer muE3 motif. Genes Dev. 1990;4:167–179. doi: 10.1101/gad.4.2.167. [DOI] [PubMed] [Google Scholar]
- Betschinger J., Nichols J., Dietmann S., Corrin P.D., Paddison P.J., Smith A. Exit from pluripotency is gated by intracellular redistribution of the bHLH transcription factor Tfe3. Cell. 2013;153:335–347. doi: 10.1016/j.cell.2013.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady O.A., Jeong E., Martina J.A., Pirooznia M., Tunc I., Puertollano R. The transcription factors TFE3 and TFEB amplify p53 dependent transcriptional programs in response to DNA damage. Elife. 2018;7:e40856. doi: 10.7554/eLife.40856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretou M., Saez P.J., Sanseau D., Maurin M., Lankar D., Chabaud M., Spampanato C., Malbec O., Barbier L., Muallem S., et al. Lysosome signaling controls the migration of dendritic cells. Sci. Immunol. 2017;2:eaak9573. doi: 10.1126/sciimmunol.aak9573. [DOI] [PubMed] [Google Scholar]
- Bronisz A., Sharma S.M., Hu R., Godlewski J., Tzivion G., Mansky K.C., Ostrowski M.C. Microphthalmia-associated transcription factor interactions with 14-3-3 modulate differentiation of committed myeloid precursors. Mol. Biol. Cell. 2006;17:3897–3906. doi: 10.1091/mbc.e06-05-0470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell G.R., Rawat P., Bruckman R.S., Spector S.A. Human immunodeficiency virus type 1 Nef inhibits autophagy through transcription factor EB sequestration. PLoS Pathog. 2015;11:e1005018. doi: 10.1371/journal.ppat.1005018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Network, author. Genomic classification of cutaneous melanoma. Cell. 2015;161:1681–1696. doi: 10.1016/j.cell.2015.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey K.L., Paulus G.L.C., Wang L., Balce D.R., Luo J.W., Bergman P., Ferder I.C., Kong L., Renaud N., Singh S., et al. TFEB transcriptional responses reveal negative feedback by BHLHE40 and BHLHE41. Cell Rep. 2020;33:108371. doi: 10.1016/j.celrep.2020.108371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr C.S., Sharp P.A. A helix-loop-helix protein related to the immunoglobulin E box-binding proteins. Mol. Cell. Biol. 1990;10:4384–4388. doi: 10.1128/mcb.10.8.4384-4388.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carreira S., Goodall J., Aksan I., La Rocca S.A., Galibert M.D., Denat L., Larue L., Goding C.R. Mitf cooperates with Rb1 and activates p21Cip1 expression to regulate cell cycle progression. Nature. 2005;433:764–769. doi: 10.1038/nature03269. [DOI] [PubMed] [Google Scholar]
- Carreira S., Goodall J., Denat L., Rodriguez M., Nuciforo P., Hoek K.S., Testori A., Larue L., Goding C.R. Mitf regulation of Dia1 controls melanoma proliferation and invasiveness. Genes Dev. 2006;20:3426–3439. doi: 10.1101/gad.406406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao X., Wang S., Zhao K., Li Y., Williams J.A., Li T., Chavan H., Krishnamurthy P., He X.C., Li L., et al. Impaired TFEB-mediated lysosome biogenesis and autophagy promote chronic ethanol-induced liver injury and steatosis in mice. Gastroenterology. 2018;155:865–879.e12. doi: 10.1053/j.gastro.2018.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D., Wang Z., Zhao Y.G., Zheng H., Zhao H., Liu N., Zhang H. Inositol polyphosphate multikinase inhibits liquid-liquid phase separation of TFEB to negatively regulate autophagy activity. Dev. Cell. 2020;55:588–602.e7. doi: 10.1016/j.devcel.2020.10.010. [DOI] [PubMed] [Google Scholar]
- Chung M.C., Kim H.K., Kawamoto S. TFEC can function as a transcriptional activator of the nonmuscle myosin II heavy chain-A gene in transfected cells. Biochemistry. 2001;40:8887–8897. doi: 10.1021/bi002847d. [DOI] [PubMed] [Google Scholar]
- El-Houjeiri L., Possik E., Vijayaraghavan T., Paquette M., Martina J.A., Kazan J.M., Ma E.H., Jones R., Blanchette P., Puertollano R., et al. The transcription factors TFEB and TFE3 link the FLCN-AMPK signaling axis to innate immune response and pathogen resistance. Cell Rep. 2019;26:3613–3628.e6. doi: 10.1016/j.celrep.2019.02.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang L., Hodge J., Saaoud F., Wang J., Iwanowycz S., Wang Y., Hui Y., Evans T.D., Razani B., Fan D. Transcriptional factor EB regulates macrophage polarization in the tumor microenvironment. Oncoimmunology. 2017;6:e1312042. doi: 10.1080/2162402X.2017.1312042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferron M., Settembre C., Shimazu J., Lacombe J., Kato S., Rawlings D.J., Ballabio A., Karsenty G. A RANKL-PKCbeta-TFEB signaling cascade is necessary for lysosomal biogenesis in osteoclasts. Genes Dev. 2013;27:955–969. doi: 10.1101/gad.213827.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto Y., Nakagawa Y., Satoh A., Okuda K., Shingyouchi A., Naka A., Matsuzaka T., Iwasaki H., Kobayashi K., Yahagi N., et al. TFE3 controls lipid metabolism in adipose tissue of male mice by suppressing lipolysis and thermogenesis. Endocrinology. 2013;154:3577–3588. doi: 10.1210/en.2013-1203. [DOI] [PubMed] [Google Scholar]
- Gayle S., Landrette S., Beeharry N., Conrad C., Hernandez M., Beckett P., Ferguson S.M., Mandelkern T., Zheng M., Xu T., et al. Identification of apilimod as a first-in-class PIKfyve kinase inhibitor for treatment of B-cell non-Hodgkin lymphoma. Blood. 2017;129:1768–1778. doi: 10.1182/blood-2016-09-736892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goding C.R., Arnheiter H. MITF-the first 25 years. Genes Dev. 2019;33:983–1007. doi: 10.1101/gad.324657.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutknecht M., Geiger J., Joas S., Dorfel D., Salih H.R., Muller M.R., Grunebach F., Rittig S.M. The transcription factor MITF is a critical regulator of GPNMB expression in dendritic cells. Cell Commun. Signal. 2015;13:19. doi: 10.1186/s12964-015-0099-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haq R., Shoag J., Andreu-Perez P., Yokoyama S., Edelman H., Rowe G.C., Frederick D.T., Hurley A.D., Nellore A., Kung A.L., et al. Oncogenic BRAF regulates oxidative metabolism via PGC1alpha and MITF. Cancer Cell. 2013;23:302–315. doi: 10.1016/j.ccr.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L., Weber K.J., Diwan A., Schilling J.D. Inhibition of mTOR reduces lipotoxic cell death in primary macrophages through an autophagy-independent mechanism. J. Leukoc. Biol. 2016;100:1113–1124. doi: 10.1189/jlb.3A1015-463R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemesath T.J., Price E.R., Takemoto C., Badalian T., Fisher D.E. MAP kinase links the transcription factor Microphthalmia to c-Kit signalling in melanocytes. Nature. 1998;391:298–301. doi: 10.1038/34681. [DOI] [PubMed] [Google Scholar]
- Hemesath T.J., Steingrimsson E., McGill G., Hansen M.J., Vaught J., Hodgkinson C.A., Arnheiter H., Copeland N.G., Jenkins N.A., Fisher D.E. Microphthalmia, a critical factor in melanocyte development, defines a discrete transcription factor family. Genes Dev. 1994;8:2770–2780. doi: 10.1101/gad.8.22.2770. [DOI] [PubMed] [Google Scholar]
- Hershey C.L., Fisher D.E. Mitf and Tfe3: members of a b-HLH-ZIP transcription factor family essential for osteoclast development and function. Bone. 2004;34:689–696. doi: 10.1016/j.bone.2003.08.014. [DOI] [PubMed] [Google Scholar]
- Hertwig P. Neue Mutationen und Koppelungsgruppen bei der Hausmaus. Z. Indukt. Abstamm. Vererbungsl. 1942;80:220–246. German. [Google Scholar]
- Hodgkinson C.A., Moore K.J., Nakayama A., Steingrimsson E., Copeland N.G., Jenkins N.A., Arnheiter H. Mutations at the mouse microphthalmia locus are associated with defects in a gene encoding a novel basic-helix-loop-helix-zipper protein. Cell. 1993;74:395–404. doi: 10.1016/0092-8674(93)90429-t. [DOI] [PubMed] [Google Scholar]
- Hoek K.S., Schlegel N.C., Eichhoff O.M., Widmer D.S., Praetorius C., Einarsson S.O., Valgeirsdottir S., Bergsteinsdottir K., Schepsky A., Dummer R., et al. Novel MITF targets identified using a two-step DNA microarray strategy. Pigment Cell Melanoma Res. 2008;21:665–676. doi: 10.1111/j.1755-148X.2008.00505.x. [DOI] [PubMed] [Google Scholar]
- Hsu C.L., Lee E.X., Gordon K.L., Paz E.A., Shen W.C., Ohnishi K., Meisenhelder J., Hunter T., La Spada A.R. MAP4K3 mediates amino acid-dependent regulation of autophagy via phosphorylation of TFEB. Nat. Commun. 2018;9:942. doi: 10.1038/s41467-018-03340-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huan C., Kelly M.L., Steele R., Shapira I., Gottesman S.R., Roman C.A. Transcription factors TFE3 and TFEB are critical for CD40 ligand expression and thymus-dependent humoral immunity. Nat. Immunol. 2006;7:1082–1091. doi: 10.1038/ni1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki H., Naka A., Iida K.T., Nakagawa Y., Matsuzaka T., Ishii K.A., Kobayashi K., Takahashi A., Yatoh S., Yahagi N., et al. TFE3 regulates muscle metabolic gene expression, increases glycogen stores, and enhances insulin sensitivity in mice. Am. J. Physiol. Endocrinol. Metab. 2012;302:E896–E902. doi: 10.1152/ajpendo.00204.2011. [DOI] [PubMed] [Google Scholar]
- Joffre O.P., Segura E., Savina A., Amigorena S. Cross-presentation by dendritic cells. Nat. Rev. Immunol. 2012;12:557–569. doi: 10.1038/nri3254. [DOI] [PubMed] [Google Scholar]
- Kim S.H., Kim G., Han D.H., Lee M., Kim I., Kim B., Kim K.H., Song Y.M., Yoo J.E., Wang H.J., et al. Ezetimibe ameliorates steatohepatitis via AMP activated protein kinase-TFEB-mediated activation of autophagy and NLRP3 inflammasome inhibition. Autophagy. 2017a;13:1767–1781. doi: 10.1080/15548627.2017.1356977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.S., Lee H.M., Kim J.K., Yang C.S., Kim T.S., Jung M., Jin H.S., Kim S., Jang J., Oh G.T., et al. PPAR-alpha activation mediates innate host defense through induction of TFEB and lipid catabolism. J. Immunol. 2017b;198:3283–3295. doi: 10.4049/jimmunol.1601920. [DOI] [PubMed] [Google Scholar]
- Kitamura Y., Morii E., Jippo T., Ito A. Effect of MITF on mast cell differentiation. Mol. Immunol. 2002;38:1173–1176. doi: 10.1016/s0161-5890(02)00058-5. [DOI] [PubMed] [Google Scholar]
- Krakowsky J.M., Boissy R.E., Neumann J.C., Lingrel J.B. A DNA insertional mutation results in microphthalmia in transgenic mice. Transgenic Res. 1993;2:14–20. doi: 10.1007/BF01977676. [DOI] [PubMed] [Google Scholar]
- Ladanyi M., Lui M.Y., Antonescu C.R., Krause-Boehm A., Meindl A., Argani P., Healey J.H., Ueda T., Yoshikawa H., Meloni-Ehrig A., et al. The der(17)t(X;17)(p11;q25) of human alveolar soft part sarcoma fuses the TFE3 transcription factor gene to ASPL, a novel gene at 17q25. Oncogene. 2001;20:48–57. doi: 10.1038/sj.onc.1204074. [DOI] [PubMed] [Google Scholar]
- Li J., Wada S., Weaver L.K., Biswas C., Behrens E.M., Arany Z. Myeloid Folliculin balances mTOR activation to maintain innate immunity homeostasis. JCI Insight. 2019;5:e126939. doi: 10.1172/jci.insight.126939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Friedrichsen H.J., Andrews S., Picaud S., Volpon L., Ngeow K., Berridge G., Fischer R., Borden K.L.B., Filippakopoulos P., et al. A TFEB nuclear export signal integrates amino acid supply and glucose availability. Nat. Commun. 2018;9:2685. doi: 10.1038/s41467-018-04849-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Hodge J., Liu Q., Wang J., Wang Y., Evans T.D., Altomare D., Yao Y., Murphy E.A., Razani B., et al. TFEB is a master regulator of tumor-associated macrophages in breast cancer. J. Immunother. Cancer. 2020;8:e000543. doi: 10.1136/jitc-2020-000543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Xu M., Ding X., Yan C., Song Z., Chen L., Huang X., Wang X., Jian Y., Tang G., et al. Protein kinase C controls lysosome biogenesis independently of mTORC1. Nat. Cell Biol. 2016;18:1065–1077. doi: 10.1038/ncb3407. [DOI] [PubMed] [Google Scholar]
- Lin L., Gerth A.J., Peng S.L. Active inhibition of plasma cell development in resting B cells by microphthalmia-associated transcription factor. J. Exp. Med. 2004;200:115–122. doi: 10.1084/jem.20040612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansueto G., Armani A., Viscomi C., D'Orsi L., De Cegli R., Polishchuk E.V., Lamperti C., Di Meo I., Romanello V., Marchet S., et al. Transcription factor EB controls metabolic flexibility during exercise. Cell Metab. 2017;25:182–196. doi: 10.1016/j.cmet.2016.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchand B., Arsenault D., Raymond-Fleury A., Boisvert F.M., Boucher M.J. Glycogen synthase kinase-3 (GSK3) inhibition induces prosurvival autophagic signals in human pancreatic cancer cells. J. Biol. Chem. 2015;290:5592–5605. doi: 10.1074/jbc.M114.616714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martina J.A., Chen Y., Gucek M., Puertollano R. MTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of TFEB. Autophagy. 2012;8:903–914. doi: 10.4161/auto.19653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martina J.A., Diab H.I., Brady O.A., Puertollano R. TFEB and TFE3 are novel components of the integrated stress response. EMBO J. 2016;35:479–495. doi: 10.15252/embj.201593428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martina J.A., Diab H.I., Lishu L., Jeong A.L., Patange S., Raben N., Puertollano R. The nutrient-responsive transcription factor TFE3 promotes autophagy, lysosomal biogenesis, and clearance of cellular debris. Sci. Signal. 2014;7:ra9. doi: 10.1126/scisignal.2004754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martina J.A., Puertollano R. Rag GTPases mediate amino acid-dependent recruitment of TFEB and MITF to lysosomes. J. Cell Biol. 2013;200:475–491. doi: 10.1083/jcb.201209135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martina J.A., Puertollano R. Protein phosphatase 2A stimulates activation of TFEB and TFE3 transcription factors in response to oxidative stress. J. Biol. Chem. 2018;293:12525–12534. doi: 10.1074/jbc.RA118.003471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martini-Stoica H., Xu Y., Ballabio A., Zheng H. The autophagy-lysosomal pathway in neurodegeneration: a TFEB perspective. Trends Neurosci. 2016;39:221–234. doi: 10.1016/j.tins.2016.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina D.L., Di Paola S., Peluso I., Armani A., De Stefani D., Venditti R., Montefusco S., Scotto-Rosato A., Prezioso C., Forrester A., et al. Lysosomal calcium signalling regulates autophagy through calcineurin and TFEB. Nat. Cell Biol. 2015;17:288–299. doi: 10.1038/ncb3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina D.L., Fraldi A., Bouche V., Annunziata F., Mansueto G., Spampanato C., Puri C., Pignata A., Martina J.A., Sardiello M., et al. Transcriptional activation of lysosomal exocytosis promotes cellular clearance. Dev. Cell. 2011;21:421–430. doi: 10.1016/j.devcel.2011.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrell K., Wells S., Henderson A., Gorman J., Alt F., Stall A., Calame K. The absence of the transcription activator TFE3 impairs activation of B cells in vivo. Mol. Cell. Biol. 1997;17:3335–3344. doi: 10.1128/MCB.17.6.3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moller K., Sigurbjornsdottir S., Arnthorsson A.O., Pogenberg V., Dilshat R., Fock V., Brynjolfsdottir S.H., Bindesboll C., Bessadottir M., Ogmundsdottir H.M., et al. MITF has a central role in regulating starvation-induced autophagy in melanoma. Sci. Rep. 2019;9:1055. doi: 10.1038/s41598-018-37522-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morii E., Oboki K., Ishihara K., Jippo T., Hirano T., Kitamura Y. Roles of MITF for development of mast cells in mice: effects on both precursors and tissue environments. Blood. 2004;104:1656–1661. doi: 10.1182/blood-2004-01-0247. [DOI] [PubMed] [Google Scholar]
- Murakami H., Arnheiter H. Sumoylation modulates transcriptional activity of MITF in a promoter-specific manner. Pigment Cell Res. 2005;18:265–277. doi: 10.1111/j.1600-0749.2005.00234.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najibi M., Labed S.A., Visvikis O., Irazoqui J.E. An evolutionarily conserved PLC-PKD-TFEB pathway for host defense. Cell Rep. 2016;15:1728–1742. doi: 10.1016/j.celrep.2016.04.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa Y., Shimano H., Yoshikawa T., Ide T., Tamura M., Furusawa M., Yamamoto T., Inoue N., Matsuzaka T., Takahashi A., et al. TFE3 transcriptionally activates hepatic IRS-2, participates in insulin signaling and ameliorates diabetes. Nat. Med. 2006;12:107–113. doi: 10.1038/nm1334. [DOI] [PubMed] [Google Scholar]
- Napolitano G., Ballabio A. TFEB at a glance. J. Cell Sci. 2016;129:2475–2481. doi: 10.1242/jcs.146365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nezich C.L., Wang C., Fogel A.I., Youle R.J. MiT/TFE transcription factors are activated during mitophagy downstream of Parkin and Atg5. J. Cell Biol. 2015;210:435–450. doi: 10.1083/jcb.201501002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngeow K.C., Friedrichsen H.J., Li L., Zeng Z., Andrews S., Volpon L., Brunsdon H., Berridge G., Picaud S., Fischer R., et al. BRAF/MAPK and GSK3 signaling converges to control MITF nuclear export. Proc. Natl. Acad. Sci. U. S. A. 2018;115:E8668–E8677. doi: 10.1073/pnas.1810498115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppezzo A., Rosselli F. The underestimated role of the microphthalmia-associated transcription factor (MiTF) in normal and pathological haematopoiesis. Cell Biosci. 2021;11:18. doi: 10.1186/s13578-021-00529-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouimet M., Koster S., Sakowski E., Ramkhelawon B., van Solingen C., Oldebeken S., Karunakaran D., Portal-Celhay C., Sheedy F.J., Ray T.D., et al. Mycobacterium tuberculosis induces the miR-33 locus to reprogram autophagy and host lipid metabolism. Nat. Immunol. 2016;17:677–686. doi: 10.1038/ni.3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmanabhan B., Fielden L.F., Hachani A., Newton P., Thomas D.R., Cho H.J., Khoo C.A., Stojanovski D., Roy C.R., Scott N.E., et al. Biogenesis of the spacious Coxiella-containing vacuole depends on host transcription factors TFEB and TFE3. Infect. Immun. 2020;88:e00534–19. doi: 10.1128/IAI.00534-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmieri M., Impey S., Kang H., di Ronza A., Pelz C., Sardiello M., Ballabio A. Characterization of the CLEAR network reveals an integrated control of cellular clearance pathways. Hum. Mol. Genet. 2011;20:3852–3866. doi: 10.1093/hmg/ddr306. [DOI] [PubMed] [Google Scholar]
- Palmieri M., Pal R., Nelvagal H.R., Lotfi P., Stinnett G.R., Seymour M.L., Chaudhury A., Bajaj L., Bondar V.V., Bremner L., et al. mTORC1-independent TFEB activation via Akt inhibition promotes cellular clearance in neurodegenerative storage diseases. Nat. Commun. 2017;8:14338. doi: 10.1038/ncomms14338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parr C., Carzaniga R., Gentleman S.M., Van Leuven F., Walter J., Sastre M. Glycogen synthase kinase 3 inhibition promotes lysosomal biogenesis and autophagic degradation of the amyloid-beta precursor protein. Mol. Cell. Biol. 2012;32:4410–4418. doi: 10.1128/MCB.00930-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastore N., Brady O.A., Diab H.I., Martina J.A., Sun L., Huynh T., Lim J.A., Zare H., Raben N., Ballabio A., et al. TFEB and TFE3 cooperate in the regulation of the innate immune response in activated macrophages. Autophagy. 2016;12:1240–1258. doi: 10.1080/15548627.2016.1179405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastore N., Vainshtein A., Klisch T.J., Armani A., Huynh T., Herz N.J., Polishchuk E.V., Sandri M., Ballabio A. TFE3 regulates whole-body energy metabolism in cooperation with TFEB. EMBO Mol. Med. 2017;9:605–621. doi: 10.15252/emmm.201607204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera R.M., Di Malta C., Ballabio A. MiT/TFE family of transcription factors, lysosomes, and cancer. Annu. Rev. Cancer Biol. 2019;3:203–222. doi: 10.1146/annurev-cancerbio-030518-055835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera R.M., Stoykova S., Nicolay B.N., Ross K.N., Fitamant J., Boukhali M., Lengrand J., Deshpande V., Selig M.K., Ferrone C.R., et al. Transcriptional control of autophagy-lysosome function drives pancreatic cancer metabolism. Nature. 2015;524:361–365. doi: 10.1038/nature14587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ploper D., Taelman V.F., Robert L., Perez B.S., Titz B., Chen H.W., Graeber T.G., von Euw E., Ribas A., De Robertis E.M. MITF drives endolysosomal biogenesis and potentiates Wnt signaling in melanoma cells. Proc. Natl. Acad. Sci. U. S. A. 2015;112:E420–E429. doi: 10.1073/pnas.1424576112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puertollano R., Ferguson S.M., Brugarolas J., Ballabio A. The complex relationship between TFEB transcription factor phosphorylation and subcellular localization. EMBO J. 2018;37:e98804. doi: 10.15252/embj.201798804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi X., Hong J., Chaves L., Zhuang Y., Chen Y., Wang D., Chabon J., Graham B., Ohmori K., Li Y., et al. Antagonistic regulation by the transcription factors C/EBPalpha and MITF specifies basophil and mast cell fates. Immunity. 2013;39:97–110. doi: 10.1016/j.immuni.2013.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raben N., Puertollano R. TFEB and TFE3: linking lysosomes to cellular adaptation to stress. Annu. Rev. Cell Dev. Biol. 2016;32:255–278. doi: 10.1146/annurev-cellbio-111315-125407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao S., Xu T., Xia Y., Zhang H. Salmonella and S. aureus escape from the clearance of macrophages via controlling TFEB. TFEB. Front. Microbiol. 2020;11:573844. doi: 10.3389/fmicb.2020.573844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashid H.O., Yadav R.K., Kim H.R., Chae H.J. ER stress: autophagy induction, inhibition and selection. Autophagy. 2015;11:1956–1977. doi: 10.1080/15548627.2015.1091141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehli M., Lichanska A., Cassady A.I., Ostrowski M.C., Hume D.A. TFEC is a macrophage-restricted member of the microphthalmia-TFE subfamily of basic helix-loop-helix leucine zipper transcription factors. J. Immunol. 1999;162:1559–1565. [PubMed] [Google Scholar]
- Rehli M., Sulzbacher S., Pape S., Ravasi T., Wells C.A., Heinz S., Sollner L., El Chartouni C., Krause S.W., Steingrimsson E., et al. Transcription factor Tfec contributes to the IL-4-inducible expression of a small group of genes in mouse macrophages including the granulocyte colony-stimulating factor receptor. J. Immunol. 2005;174:7111–7122. doi: 10.4049/jimmunol.174.11.7111. [DOI] [PubMed] [Google Scholar]
- Roczniak-Ferguson A., Petit C.S., Froehlich F., Qian S., Ky J., Angarola B., Walther T.C., Ferguson S.M. The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Sci. Signal. 2012;5:ra42. doi: 10.1126/scisignal.2002790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roundy K., Kollhoff A., Eichwald E.J., Weis J.J., Weis J.H. Microphthalmic mice display a B cell deficiency similar to that seen for mast and NK cells. J. Immunol. 1999;163:6671–6678. [PubMed] [Google Scholar]
- Roundy K., Smith R., Weis J.J., Weis J.H. Overexpression of RANKL implicates IFN-beta-mediated elimination of B-cell precursors in the osteopetrotic bone of microphthalmic mice. J. Bone Miner. Res. 2003;18:278–288. doi: 10.1359/jbmr.2003.18.2.278. [DOI] [PubMed] [Google Scholar]
- Samie M., Cresswell P. The transcription factor TFEB acts as a molecular switch that regulates exogenous antigen-presentation pathways. Nat. Immunol. 2015;16:729–736. doi: 10.1038/ni.3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardiello M., Palmieri M., di Ronza A., Medina D.L., Valenza M., Gennarino V.A., Di Malta C., Donaudy F., Embrione V., Polishchuk R.S., et al. A gene network regulating lysosomal biogenesis and function. Science. 2009;325:473–477. doi: 10.1126/science.1174447. [DOI] [PubMed] [Google Scholar]
- Sarkar S., Davies J.E., Huang Z., Tunnacliffe A., Rubinsztein D.C. Trehalose, a novel mTOR-independent autophagy enhancer, accelerates the clearance of mutant huntingtin and alpha-synuclein. J. Biol. Chem. 2007;282:5641–5652. doi: 10.1074/jbc.M609532200. [DOI] [PubMed] [Google Scholar]
- Schilling J.D., Machkovech H.M., He L., Diwan A., Schaffer J.E. TLR4 activation under lipotoxic conditions leads to synergistic macrophage cell death through a TRIF-dependent pathway. J. Immunol. 2013;190:1285–1296. doi: 10.4049/jimmunol.1202208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settembre C., De Cegli R., Mansueto G., Saha P.K., Vetrini F., Visvikis O., Huynh T., Carissimo A., Palmer D., Klisch T.J., et al. TFEB controls cellular lipid metabolism through a starvation-induced autoregulatory loop. Nat. Cell Biol. 2013;15:647–658. doi: 10.1038/ncb2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settembre C., Di Malta C., Polito V.A., Garcia Arencibia M., Vetrini F., Erdin S., Erdin S.U., Huynh T., Medina D., Colella P., et al. TFEB links autophagy to lysosomal biogenesis. Science. 2011;332:1429–1433. doi: 10.1126/science.1204592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settembre C., Zoncu R., Medina D.L., Vetrini F., Erdin S., Erdin S., Huynh T., Ferron M., Karsenty G., Vellard M.C., et al. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J. 2012;31:1095–1108. doi: 10.1038/emboj.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sha Y., Rao L., Settembre C., Ballabio A., Eissa N.T. STUB1 regulates TFEB-induced autophagy-lysosome pathway. EMBO J. 2017;36:2544–2552. doi: 10.15252/embj.201796699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simionato E., Ledent V., Richards G., Thomas-Chollier M., Kerner P., Coornaert D., Degnan B.M., Vervoort M. Origin and diversification of the basic helix-loop-helix gene family in metazoans: insights from comparative genomics. BMC Evol. Biol. 2007;7:33. doi: 10.1186/1471-2148-7-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh N., Kansal P., Ahmad Z., Baid N., Kushwaha H., Khatri N., Kumar A. Antimycobacterial effect of IFNG (interferon gamma)-induced autophagy depends on HMOX1 (heme oxygenase 1)-mediated increase in intracellular calcium levels and modulation of PPP3/calcineurin-TFEB (transcription factor EB) axis. Autophagy. 2018;14:972–991. doi: 10.1080/15548627.2018.1436936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steingrimsson E., Tessarollo L., Pathak B., Hou L., Arnheiter H., Copeland N.G., Jenkins N.A. Mitf and Tfe3, two members of the Mitf-Tfe family of bHLH-Zip transcription factors, have important but functionally redundant roles in osteoclast development. Proc. Natl. Acad. Sci. U. S. A. 2002;99:4477–4482. doi: 10.1073/pnas.072071099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steingrimsson E., Tessarollo L., Reid S.W., Jenkins N.A., Copeland N.G. The bHLH-Zip transcription factor Tfeb is essential for placental vascularization. Development. 1998;125:4607–4616. doi: 10.1242/dev.125.23.4607. [DOI] [PubMed] [Google Scholar]
- Strub T., Giuliano S., Ye T., Bonet C., Keime C., Kobi D., Le Gras S., Cormont M., Ballotti R., Bertolotto C., et al. Essential role of microphthalmia transcription factor for DNA replication, mitosis and genomic stability in melanoma. Oncogene. 2011;30:2319–2332. doi: 10.1038/onc.2010.612. [DOI] [PubMed] [Google Scholar]
- Tanaka M., Kato K., Gomi K., Matsumoto M., Kudo H., Shinkai M., Ohama Y., Kigasawa H., Tanaka Y. Perivascular epithelioid cell tumor with SFPQ/PSF-TFE3 gene fusion in a patient with advanced neuroblastoma. Am. J. Surg. Pathol. 2009;33:1416–1420. doi: 10.1097/PAS.0b013e3181a9cd6c. [DOI] [PubMed] [Google Scholar]
- Taylor H.E., Khatua A.K., Popik W. The innate immune factor apolipoprotein L1 restricts HIV-1 infection. J. Virol. 2014;88:592–603. doi: 10.1128/JVI.02828-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega-Rubin-de-Celis S., Pena-Llopis S., Konda M., Brugarolas J. Multistep regulation of TFEB by MTORC1. Autophagy. 2017;13:464–472. doi: 10.1080/15548627.2016.1271514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villegas F., Lehalle D., Mayer D., Rittirsch M., Stadler M.B., Zinner M., Olivieri D., Vabres P., Duplomb-Jego L., De Bont E., et al. Lysosomal signaling licenses embryonic stem cell differentiation via inactivation of Tfe3. Cell Stem Cell. 2019;24:257–270.e8. doi: 10.1016/j.stem.2018.11.021. [DOI] [PubMed] [Google Scholar]
- Visvikis O., Ihuegbu N., Labed S.A., Luhachack L.G., Alves A.F., Wollenberg A.C., Stuart L.M., Stormo G.D., Irazoqui J.E. Innate host defense requires TFEB-mediated transcription of cytoprotective and antimicrobial genes. Immunity. 2014;40:896–909. doi: 10.1016/j.immuni.2014.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada S., Neinast M., Jang C., Ibrahim Y.H., Lee G., Babu A., Li J., Hoshino A., Rowe G.C., Rhee J., et al. The tumor suppressor FLCN mediates an alternate mTOR pathway to regulate browning of adipose tissue. Genes Dev. 2016;30:2551–2564. doi: 10.1101/gad.287953.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Gao Q., Yang M., Zhang X., Yu L., Lawas M., Li X., Bryant-Genevier M., Southall N.T., Marugan J., et al. Up-regulation of lysosomal TRPML1 channels is essential for lysosomal adaptation to nutrient starvation. Proc. Natl. Acad. Sci. U. S. A. 2015;112:E1373–E1381. doi: 10.1073/pnas.1419669112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Huang Y., Liu J., Zhang J., Xu M., You Z., Peng C., Gong Z., Liu W. Acetyltransferase GCN5 regulates autophagy and lysosome biogenesis by targeting TFEB. EMBO Rep. 2020;21:e48335. doi: 10.15252/embr.201948335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Zhu J., Zhang L., Zhang Z., He L., Mou Y., Deng Y., Cao Y., Yang P., Su Y., et al. Role of C/EBP homologous protein and endoplasmic reticulum stress in asthma exacerbation by regulating the IL-4/signal transducer and activator of transcription 6/transcription factor EC/IL-4 receptor alpha positive feedback loop in M2 macrophages. J. Allergy Clin. Immunol. 2017;140:1550–1561.e8. doi: 10.1016/j.jaci.2017.01.024. [DOI] [PubMed] [Google Scholar]
- Weterman M.A., Wilbrink M., Geurts, van Kessel A. Fusion of the transcription factor TFE3 gene to a novel gene, PRCC, in t(X;1)(p11;q21)-positive papillary renal cell carcinomas. Proc. Natl. Acad. Sci. U. S. A. 1996;93:15294–15298. doi: 10.1073/pnas.93.26.15294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M., Hemesath T.J., Takemoto C.M., Horstmann M.A., Wells A.G., Price E.R., Fisher D.Z., Fisher D.E. c-Kit triggers dual phosphorylations, which couple activation and degradation of the essential melanocyte factor Mi. Genes Dev. 2000;14:301–312. [PMC free article] [PubMed] [Google Scholar]
- Xu W., Gong L., Haddad M.M., Bischof O., Campisi J., Yeh E.T., Medrano E.E. Regulation of microphthalmia-associated transcription factor MITF protein levels by association with the ubiquitin-conjugating enzyme hUBC9. Exp. Cell Res. 2000;255:135–143. doi: 10.1006/excr.2000.4803. [DOI] [PubMed] [Google Scholar]
- Yagil Z., Hadad Erlich T., Ofir-Birin Y., Tshori S., Kay G., Yekhtin Z., Fisher D.E., Cheng C., Wong W.S., Hartmann K., et al. Transcription factor E3, a major regulator of mast cell-mediated allergic response. J. Allergy Clin. Immunol. 2012;129:1357–1366.e5. doi: 10.1016/j.jaci.2011.11.051. [DOI] [PubMed] [Google Scholar]
- Yin Q., Jian Y., Xu M., Huang X., Wang N., Liu Z., Li Q., Li J., Zhou H., Xu L., et al. CDK4/6 regulate lysosome biogenesis through TFEB/TFE3. J. Cell Biol. 2020;219:e201911036. doi: 10.1083/jcb.201911036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Duan Y., Xia M., Dong Y., Chen Y., Zheng L., Chai S., Zhang Q., Wei Z., Liu N., et al. TFEB mediates immune evasion and resistance to mTOR inhibition of renal cell carcinoma via induction of PD-L1. Clin. Cancer Res. 2019;25:6827–6838. doi: 10.1158/1078-0432.CCR-19-0733. [DOI] [PubMed] [Google Scholar]
- Zhang T., Zhou Q., Ogmundsdottir M.H., Moller K., Siddaway R., Larue L., Hsing M., Kong S.W., Goding C.R., Palsson A., et al. Mitf is a master regulator of the v-ATPase, forming a control module for cellular homeostasis with v-ATPase and TORC1. J. Cell Sci. 2015;128:2938–2950. doi: 10.1242/jcs.173807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao G.Q., Zhao Q., Zhou X., Mattei M.G., de Crombrugghe B. TFEC, a basic helix-loop-helix protein, forms heterodimers with TFE3 and inhibits TFE3-dependent transcription activation. Mol. Cell. Biol. 1993;13:4505–4512. doi: 10.1128/mcb.13.8.4505-4512.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Fiske B., Kawakami A., Li J., Fisher D.E. Regulation of MITF stability by the USP13 deubiquitinase. Nat. Commun. 2011;2:414. doi: 10.1038/ncomms1421. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.