Abstract
Background
CD38, a druggable ectoenzyme, is involved in the generation of adenosine, which is implicated in tumour immune evasion. Its expression and role in prostate tumour-infiltrating immune cells (TIICs) have not been elucidated.
Objective
To characterise CD38 expression on prostate cancer (PC) epithelial cells and TIICs, and to associate this expression with clinical outcomes.
Design, setting, and participants
RNAseq from 159 patients with metastatic castration-resistant prostate cancer (mCRPC) in the International Stand Up To Cancer/Prostate Cancer Foundation (SU2C/PCF) cohort and 171 mCRPC samples taken from 63 patients in the Fred Hutchinson Cancer Research Centre cohort were analysed. CD38 expression was immunohistochemically scored by a validated assay on 51 castration-resistant PC (CRPC) and matching, same-patient castration-sensitive PC (CSPC) biopsies obtained between 2016 and 2018, and was associated with retrospectively collected clinical data.
Outcome measurements and statistical analysis
mCRPC transcriptomes were analysed for associations between CD38 expression and gene expression signatures. Multiplex immunofluorescence determined CD38 expression in PC biopsies. Differences in CD38+ TIIC densities between CSPC and CRPC biopsies were analysed using a negative binomial mixed model. Differences in the proportions of CD38+ epithelial cells between non-matched benign prostatic epithelium and PC were compared using Fisher’s exact test. Differences in the proportions of biopsies containing CD38+ tumour epithelial cells between matched CSPC and CRPC biopsies were compared by McNemar’s test. Univariable and multivariable survival analyses were performed using Cox regression models.
Results and limitations
CD38 mRNA expression in mCRPC was most significantly associated with upregulated immune signalling pathways. CD38 mRNA expression was associated with interleukin (IL)-12, IL-23, and IL-27 signalling signatures as well as immunosuppressive adenosine signalling and T cell exhaustion signatures. CD38 protein was frequently expressed on phenotypically diverse TIICs including B cells and myeloid cells, but largely absent from tumour epithelial cells. CD38+ TIIC density increased with progression to CRPC and was independently associated with worse overall survival. Future studies are required to dissect TIIC CD38 function.
Conclusions
CD38+ prostate TIICs associate with worse survival and immunosuppressive mechanisms. The role of CD38 in PC progression warrants investigation as insights into its functions may provide rationale for CD38 targeting in lethal PC.
Patient summary
CD38 is expressed on the surface of white blood cells surrounding PC cells. These cells may impact PC growth and treatment resistance. Patients with PC with more CD38-expressing white blood cells are more likely to die earlier.
Keywords: Adenosine pathway, B lymphocyte, Castration-resistant prostate cancer, CD38, Inflammation, Myeloid cell, Plasmacyte, Tumour microenvironment, T cell exhaustion, Prostate cancer
Take Home Message
CD38 is expressed by phenotypically diverse prostate tumour-infiltrating immune cells (TIICs). CD38+ TIICs are associated with a worse prognosis and may be implicated in prostate cancer immune tolerance. Existing CD38-directed therapies merit evaluation in metastatic prostate cancer clinical trials.
1. Introduction
Metastatic castration-resistant prostate cancer (mCRPC) remains a lethal condition with overall survival (OS) of 2–3 yr [1]. Overcoming immunosuppressive barriers in the prostate tumour microenvironment (TME) is critical to improving prostate cancer (PC) treatment outcomes [1], [2], [3].
CD38 is an ectoenzyme of the ribosyl cyclase family expressed on the surface of immune progenitors, which increases during B lymphocyte maturation into plasmacytes, nonhaematopoietic tissues, and some cancer cells [4], [5], [6], [7], [8], [9], [10], [11], [12], [13]. Both its receptor-ligand and enzymatic functions are implicated in tumorigenesis and tumour progression by modulating immune regulation, metabolism, calcium-mediated signal transduction, cell adhesion, and migration [12], [14], [15], [16]. CD38 catalyses the conversion of nicotinamide adenine dinucleotide (NAD+) to ADP-ribose (ADPR) and cyclic ADPR (cADPR), leading to non-canonical adenosine synthesis [14]. Adenosine has been shown to dampen antitumour immunity through its direct effects on multiple immune cell subsets [12], [14], [17]. Upregulated adenosine synthesis is independently associated with worse PC outcomes [18]. Single-agent adenosine A2A receptor blockade has shown antitumour activity in mCRPC patients [19].
Since CD38 depletes NAD+, it can also impair downstream NAD+-dependent enzymatic and metabolic processes, thereby constraining cellular proliferation, leucocyte differentiation, and function [4], [6], [10], [15]. CD38 upregulation on tumour-infiltrating immune cells (TIICs) has been reported in gastrointestinal malignancies, but not in PC [7], [8]. The growing body of evidence implicating CD38 in tumour immune evasion led us to hypothesise that CD38 expression on prostate TIICs contributes to PC progression and may serve as an important immunotherapeutic target [20]. Indeed, studies in patients with multiple myeloma have shown that anti-CD38 antibodies can deplete immunosuppressive lymphoid and myeloid cells to allow the expansion of effector T cells [21], [22]. Given the availability of several anti-CD38 antibodies, with daratumumab routinely used for the treatment of multiple myeloma, there is an urgent clinical need to elucidate the expression, function, and impact of CD38 in PC [23]. The primary objective of this study was to quantify the expression and clinical impact of CD38+ prostate TIICs as tumours progressed from being castration sensitive to castration resistant, with the long-term goal of repurposing CD38-directed therapies for mCRPC treatment.
2. Patients and methods
2.1. Patients and tissue samples
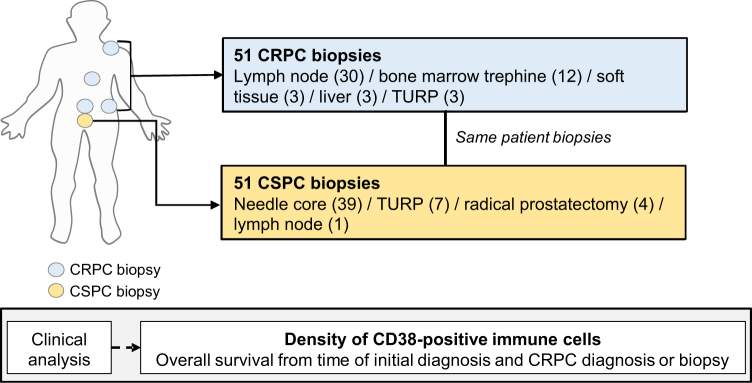
All patients had mCRPC and were treated at The Institute of Cancer Research and Royal Marsden Hospital (ICR/RMH), provided written informed consent, and were enrolled in institutional protocols approved by the RMH ethics review committee (reference no. 04/Q0801/60). Eligible patients (n = 51) had matched histologically confirmed formalin-fixed paraffin-embedded (FFPE) diagnostic (archival) and castration-resistant prostate cancer (CRPC) biopsies. Castration-sensitive PC (CSPC) biopsies were collected from the prostate primary (n = 50) and lymph node (n = 1), and CRPC biopsies were collected from lymph node (n = 30), bone (n = 12), soft tissue (n = 3), liver (n = 3), and prostate (n = 3; Fig. 1). Clinical data were retrospectively collected from electronic hospital medical records (Supplementary Table 1).
Fig. 1.
Overview of the clinical cohort. The patient cohort studied included 51 treatment-naïve, castration-sensitive prostate cancer biopsies and 51 same-patient, castration-resistant prostate cancer biopsies. CRPC = castration-resistant prostate cancer; CSPC = castration-sensitive prostate cancer; TURP = transurethral resection of the prostate.
2.2. Immunofluorescence and immunohistochemistry
Immunohistochemistry (IHC) and immunofluorescence (IF) were performed on FFPE tissue sections using an automated staining platform (Bond RX; Leica Biosystems, Buffalo Grove, IL, USA). Antibodies were validated using human tissue and cell lines as controls (Supplementary Fig. 1–3). Optimal antibody concentrations were determined for primary antibodies against CD38, CD33, CD11b, CD15, CD79a, CD3, CD19, CD20, CD138, and EpCAM (Supplementary Table 2).
Multiplex IF using a six-colour panel (CD38, CD33, CD11b, CD15, EpCAM, and DAPI) and a five-colour panel (CD38, CD33, CD79a, CD3 and DAPI) were performed on 4-µm-thick FFPE tissue sections (Supplementary Fig. 4 and 5).
Parallel dual chromogenic IHC staining using the Bond ChromoPlex 1 Dual Detection kit (DS9477; Leica Biosystems) was performed on 4-µm-thick FFPE tissue sections to determine colocalisation of CD38 and CD19, CD38 and CD20, and CD38 and CD138 (Supplementary Fig. 2 and Supplementary material).
2.3. Tissue image acquisition and analysis
Slides were scanned using the Aperio ScanScope (Leica Biosystems) and/or Vectra Automated Multispectral Imaging System (Akoya Biosciences, Marlborough, MA, USA), and analysed using inForm v2.2.1 (Akoya Biosciences) or Halo v3.0 software (Indica Labs, Albuquerque, NM, USA). Tissue segmentation was achieved either by EpCAM positivity (inForm; Supplementary Fig. 4) or a supervised machine learning algorithm to recognise PC foci and surrounding stroma (Halo; Supplementary Fig. 5). Cell segmentation was achieved with nuclear DAPI counterstain. TIICs were phenotypically characterised based on leucocyte cell surface marker staining. Cell densities were algorithmically quantitated. CD38 expression on tumour epithelium was quantitated using the Histo-score (H-score). Positivity was defined as an H-score of >0 (Supplementary material) [24].
2.4. Bioinformatics
A total of 159 mCRPC transcriptomes, generated by the International Stand Up To Cancer/Prostate Cancer Foundation (SU2C/PCF) Prostate Cancer Dream Team, were re-analysed [25]. Transcriptomes were aligned to the human reference genome (GRCh38/hg38) using TopHat2 (version 2.0.7). Gene expression as fragments per kilobase of transcript per million mapped reads (FPKM) was calculated using Cufflinks. Unbiased interrogation of 200 cell signalling pathways utilised gene sets from the Pathway Interaction Database [26]. Data of 171 mCRPC transcriptomes from 63 patients from the Fred Hutchinson CRC cohort were downloaded from cBioportal. Gene profile data were prenormalised with Z score ([raw expression values – mean expression level of the gene]/standard deviation of gene expression level) [27]. Transcript data were analysed with a previously described 14-gene adenosine signature [28], an 18-gene adenosine signature [29], and T cell exhaustion signatures [30], [31].
2.5. Statistical analysis
All statistical analyses were performed using Stata v13.1, GraphPad Prism v6, and R v3.6.2. Two-sided p values of <0.05 were designated as significant. For the unbiased pathway interaction analysis, to adjust for multiple hypothesis testing, false discovery rate (FDR)-adjusted p values (Bonferroni correction) are presented. For analyses of associations in gene expression of specific pathways of interest or CD38+ immune cell density and clinical outcomes, no correction for multiplicity was made. Detailed methods for statistical analysis can be found in the Supplementary material.
3. Results
3.1. Expression of CD38 mRNA on mCRPC biopsy is associated with multiple immune transcripts including T cell exhaustion, IL-23, and adenosine signalling pathway signatures
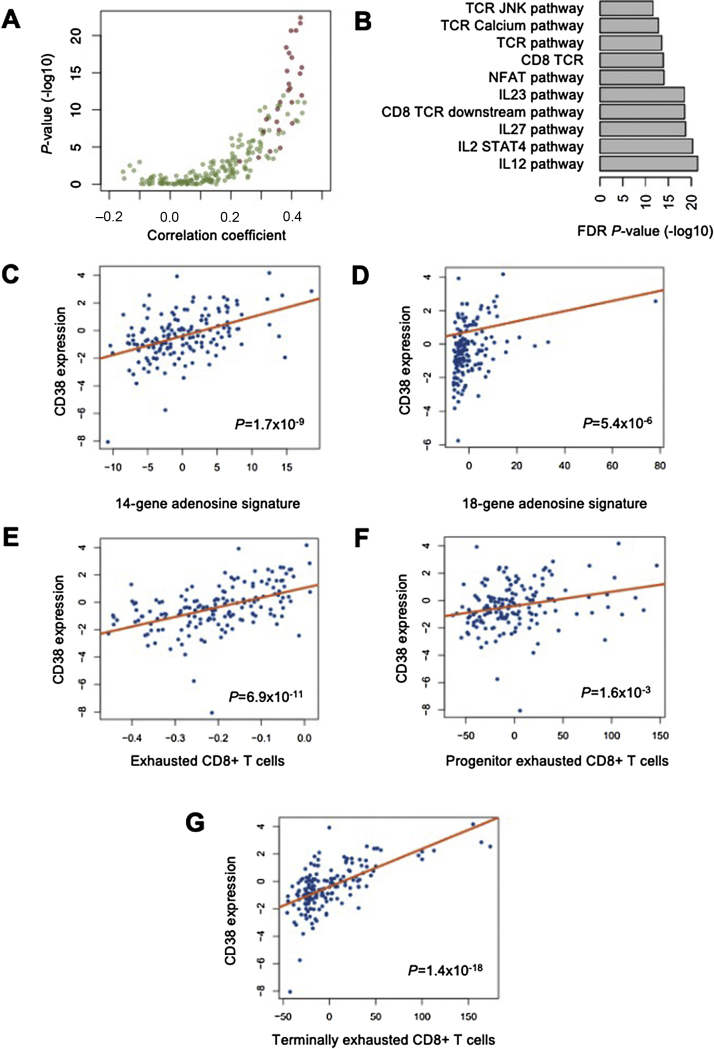
To explore the relevance of CD38 in mCRPC, an unbiased analysis of 200 cellular signalling pathways was performed on 159 mCRPC transcriptomes from the SU2C/PCF mCRPC cohort. CD38 mRNA expression was significantly associated with the upregulation of transcripts from immunoregulatory pathways, and the top ten signalling pathways with the strongest evidence of CD38 association (Bonferroni adjusted p values, all p < 0.001) all being immunomodulatory (Fig. 2A and 2B and Supplementary Table 3). CD38 mRNA expression was associated with interleukin (IL)-23, IL-12, IL-27, transforming growth factor β (TGFβ), IL-10, interferon γ (IFNγ), and tumour necrosis factor α (TNFα) pathway signalling (all p < 0.001). We validated the associations between CD38 and IL-23, TGFβ, IL-10, IFNγ, and TNFα pathway signalling using 171 mCRPC transcriptomes from the publicly available Fred Hutchinson CRC cohort (p < 0.001 for IL-23, TGFb, IL-10; and p < 0.01 for regulation of IFNγ signalling and TNFα signalling); Supplementary Fig. 6 and 7). Since CD38 catalyses immunosuppressive adenosine synthesis, and several CD38-associated cytokines are also associated with adenosine signalling, we evaluated the association between CD38 and two previously reported independent adenosine signatures [14], [28]. Indeed, CD38 mRNA expression was associated with a 14-gene adenosine signature (p < 0.001) [28] and an 18-gene adenosine signature (p < 0.001; Fig. 2C and 2D) [29]. Since adenosine pathway signalling is also associated with impaired T effector cell function [20], [32], we then evaluated whether CD38 is associated with tumour-infiltrating CD8+ T cell exhaustion [30], [31], [32]. CD38 mRNA expression is associated with genes implicated in CD8+ T cell exhaustion and, in particular, that of terminally exhausted CD8+ T cells (p < 0.001; Fig 2E-G) [18], [30], [31]. Taken together, these data suggested that CD38 may contribute to tumour immune tolerance in mCRPC.
Fig. 2.
Transcriptome analyses associating CD38 mRNA expression with immunoregulatory signalling pathway transcripts. (A and B) Unbiased transcriptome analyses of RNA-sequencing data from 159 patient biopsies in the SU2C/PCF mCRPC cohort for the association between CD38 and 200 PID signalling pathways. Red dots indicate immune signalling pathways and green dots nonimmunoregulatory pathways. CD38 was most significantly associated with the expression of multiple immune signalling pathway genes, and the ten most significant associations are shown (B). SU2C/PCF mCRPC transcriptome analyses of RNA-sequencing data from 159 patient biopsies for associations between CD38 and (C) a 14-gene adenosine signature, (D) an 18-gene adenosine signature, genes associated with CD8+ T cell exhaustion, (F) progenitor exhausted CD8+ T cell signature, and (G) terminally exhausted CD8+ T cell signature. The p values were calculated using linear regression analysis. mCRPC = metastatic castration-resistant prostate cancer; SU2C/PCF = International Stand Up To Cancer/Prostate Cancer Foundation.
3.2. CD38 is rarely expressed on PC epithelial cells but highly expressed by TIICs
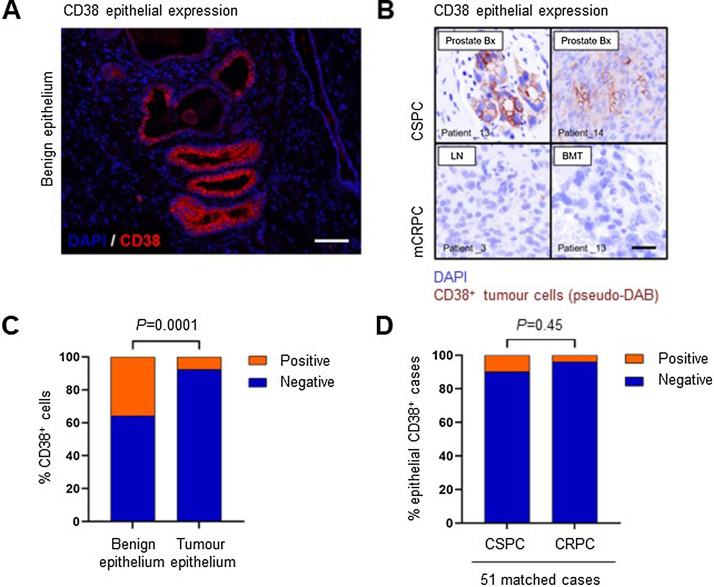
Given the association between CD38 mRNA expression and multiple immune signalling pathway transcripts, we characterised CD38 protein expression on PC cells and TIICs using multicolour IF on 51 same-patient, matched CSPC and CRPC biopsies. CD38 was more frequently expressed on normal prostatic epithelial cells (35.8%) than on tumour epithelium (7.7%; Fisher’s exact test, p < 0.001; Fig 3A–C). There was no significant difference in the proportion of CSPC versus CRPC tumour biopsies that expressed CD38 (5/51 [9.8%] of CSPC biopsies had cytoplasmic and membranous expression; 2/51 [3.9%] of CRPC biopsies had cytoplasmic or membranous expression; McNemar’s test, p = 0.45; Fig 3D). Importantly, CD38 was expressed by phenotypically diverse TIICs, as demonstrated by multicolour IF and dual-colour IHC panels consisting of both lymphoid and myeloid cell surface markers (Fig. 4 and Supplementary Fig. 8).
Fig. 3.
CD38 expression on prostate cancer epithelial cells and benign epithelial cells. (A) Representative immunofluorescence image showing CD38 expression in benign prostatic epithelium (× 200 magnification, scale bar 100 µm). (B) Representative images showing CD38 expression in prostate cancer epithelial cells. Top row shows examples of CD38+ tumours and bottom row shows CD38− tumours (200× magnification, scale bar 50 µm). (C) CD38 expression was more frequent on benign prostatic epithelial cells (35.8%) than on malignant epithelial cells (7.7%). Fisher’s exact test, p < 0.001. (D) Tumour epithelial cell CD38 expression was more frequent in CSPC biopsies (9.8%; 5/51) than in CRPC biopsies (3.9%; 2/51). McNemar’s test, p = 0.45, not significant. BMT = bone marrow trephine; CRPC = castration-resistant prostate cancer; CSPC = castration-sensitive prostate cancer; LN = lymph node; mCRPC = metastatic castration-resistant prostate cancer; Prostate Bx = prostate biopsy.
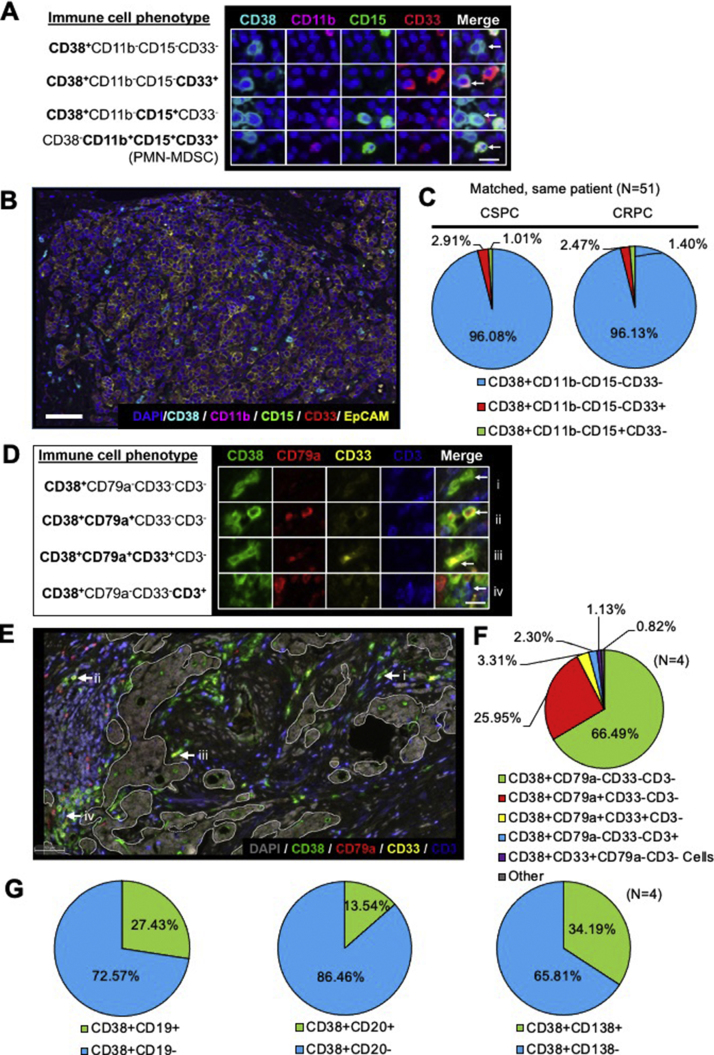
Fig. 4.
CD38 is expressed on a heterogeneous population of TIICs coexpressing myeloid and lymphoid markers. (A) Representative immunofluorescence images showing subpopulations of CD38+ tumour-infiltrating immune cells and PMN-MDSC. White arrows point to a cell with the leucocyte’s phenotype specified on each row (200× magnification, scale bar 100 µm). (B) Representative multispectral six-colour immunofluorescence image of an mCRPC lymph node biopsy stained with DAPI (blue), CD38 (cyan), CD11b (magenta), CD15 (green), CD33 (red), and EpCAM (yellow; 200× magnification, scale bar 100 µm). (C) Proportion of immune cell subpopulations expressing CD38 in CSPC and CRPC biopsies (n = 51). (D) Representative immunofluorescence images of mCRPC biopsy showing subpopulations of CD38+ coexpressing CD79a, both CD33 and CD79a, or CD3. White arrows point to a cell with the leucocyte’s phenotype specified on each row (200× magnification, scale bar 100 µm). (E) Five-colour immunofluorescence micrograph of mCRPC biopsy, with areas of stroma and tumour demarcated by white lines. Image staining for DAPI (grey), CD38 (green), CD79a (red), CD33 (yellow), and CD3 (blue). White arrows indicating representative phenotypes expressing CD38 (200× magnification, scale bar 100 μm. (F, G) Proportion of CD38+ tumour-infiltrating immune cells expressing other immune cell surface markers in 4 CSPC biopsies. CRPC = castration-resistant prostate cancer; CSPC = castration-sensitive prostate cancer; mCRPC = metastatic castration-resistant prostate cancer; PMN-MDSC = polymorphonuclear myeloid-derived suppressor cells; TIIC = tumour-infiltrating immune cell.
3.3. CD38+TIICs show heterogeneous expression of both myeloid and lymphoid cell surface markers
Since both tumour-infiltrating lymphocytes and myelocytes can drive PC growth through paracrine signalling and CD38 is expressed during normal haematopoiesis [2], [3], [11], we hypothesised that CD38 is expressed by diverse TIICs. We evaluated for the coexpression of CD38 with markers expressed by T cells, B cells, and myeloid cells using a multicolour IF panel for CD38, CD33, CD11b, CD15, DAPI (counterstain), and EpCAM (tumour mask) (Fig. 4A and 4B); a multicolour IF panel for CD38, CD79a, CD33, CD3, and DAPI; and dual-colour IHC panels for CD38 coupled with CD19, CD20, or CD138 (Fig. 4D and 4E, and Supplementary Fig. 1–5). CD11b+CD15+CD33+ polymorphonuclear myeloid-derived suppressor cells (PMN-MDSCs) did not express CD38 in any of the CSPC or CRPC biopsies. However, we identified subpopulations of CD38+CD11b–CD15–CD33+ and CD38+CD11b–CD15+CD33– cells, which were likely undergoing myeloid differentiation (Fig. 4A–C). Using the multicolour IF panel for CD38, CD79a, CD33, CD3, and DAPI, we identified subpopulations of CD38+CD79a–CD33–CD3–, CD38+CD79a+CD33–CD3–, and CD38+CD79a+CD33+CD3– TIICs (Fig. 4D–F). Since CD38+CD79a+CD33–CD3– TIICs were a common subset, and both CD38 and CD79a are commonly expressed by B cells [33], [34], we next examined CD38 colocalisation with other B cell surface markers—CD19, CD20, and CD138. A significant proportion of CD38+ TIICs coexpressed CD19+, CD20+, or CD138+, indicating that these were indeed lymphoplasmacytic infiltrates (Fig. 4G). CD38 was rarely coexpressed with the T cell marker CD3. Taken together, we show that CD38 is expressed by heterogeneous populations of TIICs frequently coexpressing B cell and myeloid cell surface markers.
3.4. CD38+ TIICs increase as PC progresses to castration resistance
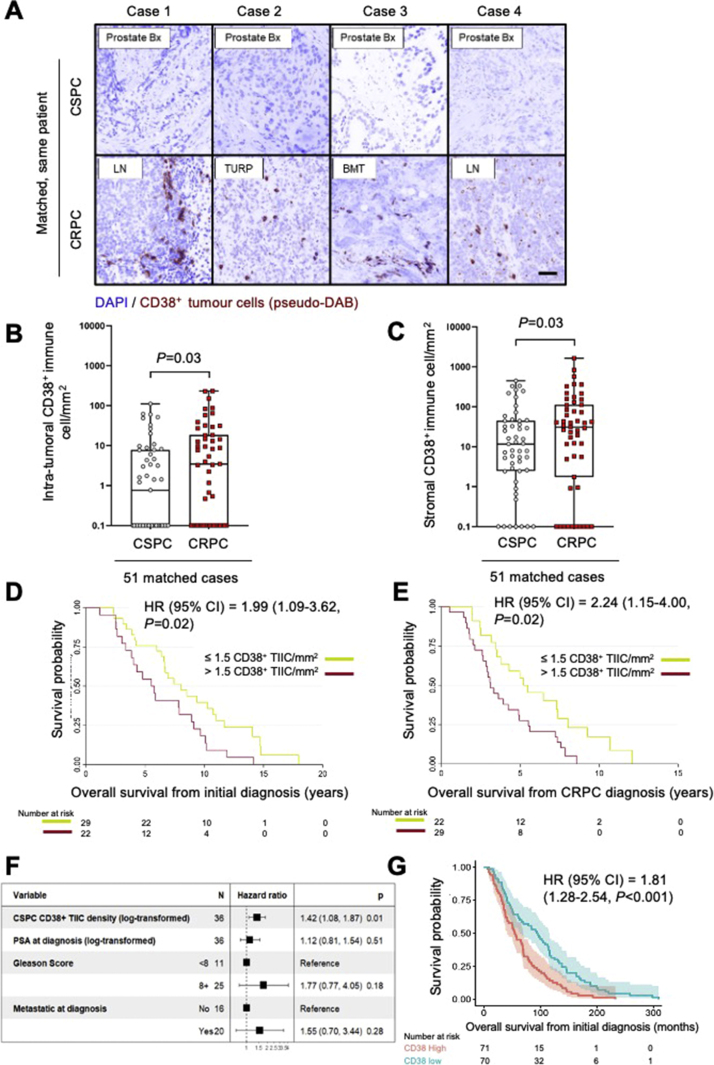
Given that CD38 expression on leucocytes has been associated with tumour progression and is associated with worse prognosis in other solid tumours [7], [8], we evaluated whether an increase in the density of CD38+ TIICs is associated with the emergence of castration resistance. Using multicolour IF and automated quantitative image analyses, we compared CD38+ TIIC density (cells/mm2) in 51 matched CSPC, and CRPC biopsies. CD38+ TIIC density in both the intratumoural and the stromal compartments increased with progression from CSPC to CRPC (negative binomial mixed model; tumour: p = 0.03; stroma: p = 0.03) (Fig. 5A-C). To confirm that the increase in CD38+ TIIC density from CSPC to CRPC was not attributable to differences in background immune cell densities at different biopsy sites, we compared CD38+ TIIC density between biopsies taken from the prostate, lymph nodes, bone, and soft tissue metastases and found no significant difference in CD38+ TIIC density at different CRPC biopsy sites (Kruskal-Wallis test, p = 0.8). Further, there was no difference in CD38+ TIIC density in CRPC between prostatic and nonprostatic metastatic biopsies (Mann-Whitney test, p = 0.6), or between lymph node and non–lymph node metastatic biopsies (Mann-Whitney test, p = 0.3). Overall, these data support the concept that CD38+ TIICs, including lymphoplasmacytoid and myeloid cells, increase with the emergence of CRPC, and suggest that these may be recruited to and/or expand in the CRPC TME to promote treatment-resistant PC growth.
Fig. 5.
CD38+ TIICs increased with the emergence of castration resistance and is associated with worse overall survival. (A) Representative “pseudo-DAB” images showing an increase in CD38+ tumour-infiltrating immune cells as tumours progressed from CSPC to CRPC in matched, same-patient biopsies. Dark-field immunofluorescence images were transformed to pseudo bright-field immunohistochemistry images to facilitate visualisation (200× magnification, scale bar 100 µm). CD38+ tumour-infiltrating immune cell density in the (B) tumour compartment and (C) stromal compartment of 51 matched, same-patient CSPC and CRPC biopsies. The median and interquartile ranges (IQRs) for intratumoural CD38+ immune cell density (cell/mm2) for CSPC (median; IQR: 0.77; 0.00–7.80) and CRPC (median; IQR: 3.49; 0.00–18.33) are shown. The median and IQRs for stromal CD38+ tumour-infiltrating immune cell density (cell/mm2) for CSPC (median; IQR: 11.66; 2.53–44.60) and CRPC (median: 31.13; 3.32–111.06) are shown. The p values were calculated using negative binomial mixed model (CD38+ tumour-infiltrating immune cell density, intratumoural p = 0.03; stromal p = 0.03. (D) Kaplan-Meier curves of overall survival from the time of initial diagnosis for CD38+ tumour-infiltrating immune cell density in CSPC samples. (E) Kaplan-Meier curves of overall survival from time of CRPC diagnosis for CD38+ tumour-infiltrating immune cell density at CRPC biopsy. Hazard ratio (HR) with 95% confidence intervals (CIs) and p values for the univariate Cox survival model are shown. (F) Multivariable analysis of overall survival (n = 36) from the time of initial diagnosis for log-transformed CD38+ immune cell density, log-transformed PSA at the time of diagnosis, Gleason score, and the presence of metastatic disease at the time of diagnosis. HR with 95% CI and p values for multivariable Cox survival models are shown. (G) Kaplan-Meier curves of overall survival from the time of CRPC biopsy for CD38 mRNA expression (n = 159; dichotomised by the median: 0.64 FPKM). HR with 95% CIs and p value for the univariate Cox survival model are shown. BMT = bone marrow trephine; CRPC = castration-resistant prostate cancer; CSPC = castration-sensitive prostate cancer; FPKM = fragments per kilobase of transcript per million mapped reads; LN = lymph node; Prostate Bx = prostate biopsy; PSA = prostate-specific antigen; TIIC = tumour-infiltrating immune cell; TURP = transurethral resection of the prostate.
3.5. An increase in CD38+ TIIC density is associated with worse OS in patients with PC
CD38+ TIIC density increases with progression to CRPC, with CD38 having potential immunosuppressive functions. Next, we determined the association between CD38+ TIIC density and OS. Log-transformed intratumoural CD38+ TIIC density in CSPC biopsies was associated with worse OS from the time of initial diagnosis (hazard ratio [HR]: 1.36, 95% confidence interval [CI]: 1.08–1.70; log-rank test, p = 0.008). Correspondingly, high CD38+ TIIC density (dichotomised based on the median; >1.5 cells/mm2) in the CSPC biopsies was associated with significantly shorter OS from the time of initial diagnosis compared with CSPC biopsies with low CD38+ TIIC density (≤1.5 cells/mm2; HR: 1.99, 95% CI: 1.09–3.62; log-rank test, p = 0.02; Fig. 5D).
Log-transformed intratumoural CD38+ TIIC density in CRPC biopsies was also associated with worse OS from the time of CRPC biopsy (HR: 1.20, 95% CI: 1.00–1.42; log-rank test, p = 0.04) and from the time of CRPC diagnosis (HR: 1.40, 95% CI: 1.14–1.70; log-rank test, p < 0.001). Correspondingly, high CD38+ TIIC density in CRPC biopsies (>1.5 cells/mm2) was associated with significantly shorter OS from the time of CRPC diagnosis than low CD38+ TIIC density in CRPC biopsies (≤1.5 cells/mm2; HR: 2.24, 95% CI: 1.15–4.00; log-rank test, p = 0.02; Fig. 5E).
Multivariable analyses, incorporating available, clinically relevant prognostic factors, including Gleason score, diagnostic prostate-specific antigen (PSA; log transformed), and the presence of metastatic disease at the time of diagnosis, showed that the CD38+ immune cell density (log transformed) in CSPC biopsies was independently associated with worse OS from the time of PC diagnosis (HR: 1.42, 95% CI: 1.08–1.87; log-rank test, p = 0.01; Fig. 5F). Multivariable analysis for OS from CRPC biopsy was not performed, as clinical data from the time of CRPC biopsies were incomplete. Finally, we evaluated the association between CD38 mRNA expression and OS, and showed that patients from the 159-patient SU2C/PCF cohort with high CD38 mRNA expression (dichotomised by the median: 0.64 FPKM) had worse OS than those with low CD38 mRNA expression (HR: 1.81, 95% CI: 1.28–2.54; log-rank test, p < 0.001; Fig. 5G). These findings corroborate the potential role of CD38+ TIICs in PC progression.
4. Discussion
This is the first study to characterise the expression and potential clinical impact of CD38, a druggable ectoenzyme, on PC epithelial cells and prostate TIICs, with the goal of evaluating CD38 as a prognostic biomarker in lethal PC and repurposing CD38-directed therapies to abrogate the potential deleterious effects of CD38+ TIICs. We show that CD38 is highly expressed on phenotypically diverse prostate TIICs and these cells are independently associated with worse OS.
The association between CD38 mRNA expression and the adenosine signature supports the notion that CD38 generates adenosine via the non-canonical CD38-CD203 axis in the prostate TME and contributes to tumour immune evasion, at least in part, by suppressing effector T cell function [14], [30], [31]. Another study of primary prostatectomy specimens showed that upregulation of CD73, a key enzyme in canonical adenosine synthesis, in the adjacent normal prostatic epithelium was associated with worse prognosis [18]. Our analyses support such a paracrine relationship, involving components of the adenosine pathway as well as between tumour cells and surrounding immune cells. We also show that the presence of CD38+ TIICs is associated with shorter OS, underscoring the potential importance of the paracrine interactions between immune cells and tumour cells mediated by soluble factors including adenosine and IL-23 [2], [17]. Whilst adenosine signalling can increase infiltration with immune cell subsets implicated in the development of T cell exhaustion, their precise relationship in PC remains unclear. We showed an association between CD38 expression and T cell exhaustion, which is corroborated by a study of PSA-specific T cells from patients with untreated PC, showing higher expression of CD38 and the T cell exhaustion marker TIM3 (HAVCR2) than that of PSA-specific T cells from healthy controls [35]. These findings have significant therapeutic implications, given current drug development efforts to target the adenosine pathways, via CD39, CD73, A2A, and A2B receptors, as well as to reinvigorate T effector cell function [36].
CD38 may also impact tumour-infiltrating lymphocyte function by depleting NAD+, which is critical for enzymatic processes regulating cellular metabolism, and lymphocyte differentiation and function [4], [6], [10], [16]. Consistent with previous studies, CD38 is silenced in tumour cells likely because metabolically active cells rely upon NAD+ and NAD+-dependent enzymes for glycolysis and mitochondrial biogenesis [6], [10]. In leucocytes, upregulation of CD38 inhibits effector function since NAD+-dependent signalling, potentially through the NAD+-SIRT1-FOXO1 axis, promotes effector T cell differentiation and antitumour potential. CD38 blockade can increase intracellular NAD+ and activate T cell–mediated immunity, despite the presence of an immunosuppressive TME [4].
We also found that CD38 is coexpressed with myeloid and lymphoid cell surface markers on diverse prostate TIICs. We have previously shown that PMN-MDSCs infiltrating human PC produce IL-23 to activate androgen-receptor pathway signalling [2]. Interestingly, IL-23 signalling pathway expression was significantly associated with CD38 expression. Unlike circulating MDSCs in patients with advanced colorectal cancer [7], CD38 was not expressed by prostate tumour-infiltrating PMN-MDSCs, but colocalised with the myeloid markers CD33 or CD15; similar immature CD38+CD15hiCD33lo myeloid cells have been observed in patients with head and neck and non–small cell lung cancer, and may confer immunosuppressive capacity [8]. In addition, a subset of CD38+ cells expressed the B cell markers CD19, CD20, CD79a, and CD138. Since CD38 is often expressed by plasmacytes, our data provide further evidence that B cells may play a role in promoting more aggressive PC biology [3]. Interestingly, we also observed CD38 coexpression with CD79a, a pan-B cell marker, and the myeloid cell surface marker CD33, suggesting abnormal differentiation. These data corroborate reports of CD79a expression by circulating myeloid cells in patients with lung and breast cancers in response to tumour-secreted cytokines [37]. Collectively, our findings suggest that CD38+ TIICs, of which a significant proportion likely represent B lymphocytes and myeloid inflammatory cells, may contribute to PC progression.
Limitations of this study are that for the matched, same-patient cohort, the tumour samples were collected at a single UK centre, with the overall sample size being limited to 51 matched cases (102 samples). Furthermore, whilst same-patient biopsies were collected, biopsies were not matched for disease site, although we have shown that there was no significant difference between CD38+ TIIC density in mCRPC biopsies taken from different disease sites. Apart from the unbiased pathway interaction analyses, the threshold for statistical significance was not adjusted for multiple hypothesis testing. These analyses were hypothesis generating and the p values should be interpreted descriptively. Finally, we do not dissect the regulation and functions of CD38 on TIICs. To this end, ongoing studies will characterise the regulation, recruitment, and functions of CD38+ TIIC in lethal PC.
Our findings are critically important for PC biomarker and therapeutic development. CD38 modulation could synergise with and overcome resistance to existing therapies targeting canonical adenosine synthesis. CD38 expression may predict resistance to existing therapies blocking canonical adenosine synthesis (eg, anti-CD73 and/or anti-CD39 antibodies) and aid patient selection for rational combinations simultaneously targeting adenosine synthesis and its receptors. The metabolic and immunomodulatory functions of CD38 may be harnessed to program naïve T cells or reinvigorate exhausted T cells towards antitumour immunity.
5. Conclusions
Our data support CD38 expression on TIICs as a potential prognostic biomarker, future studies of CD38 as a therapeutic target in lethal PC, and the feasibility of testing CD38 as a predictive biomarker.
Author contributions: Johann S. de Bono had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Study concept and design: de Bono, Crespo.
Acquisition of data: Crespo, Guo, Gurel, Sharp, Petremolo, Sumanasuriya, Rodrigues, Ferreira, Pereira, Figueiredo, Mehra, Neeb, Gil, Yuan, PCF/SU2C International Prostate Cancer Dream Team.
Analysis and interpretation of data: Guo, Crespo, Gurel, Dolling, Seed, Yuan.
Drafting of the manuscript: de Bono, Guo, Crespo.
Critical revision of the manuscript for important intellectual content: de Bono, Guo, Crespo, Gurel, Petremolo, Dolling, Rekowski, Rodrigues, Ferreira, Mehra, Lambros, Seed, Yuan, Terstappen, Alimonti, Drake.
Statistical analysis: Dolling, Rekowski, Yuan, Seed.
Obtaining funding: de Bono.
Administrative, technical, or material support: Crespo, Gurel, Rodrigues, Ferreira, Figueiredo, Pereira, Neeb, Gil.
Supervision: de Bono.
Other: None.
Financial disclosures: Johann S. de Bono certifies that all conflicts of interest, including specific financial interests and relationships and affiliations relevant to the subject matter or materials discussed in the manuscript (eg, employment/affiliation, grants or funding, consultancies, honoraria, stock ownership or options, expert testimony, royalties, or patents filed, received, or pending), are the following: Christina Guo, Mateus Crespo, Bora Gurel, David Dolling, Jan Rekowski, Adam Sharp, Antonella Petremolo, Semini Sumanasuriya, Daniel N. Rodrigues, Ana Ferreira, Rita Pereira, Ines Figueiredo, Niven Mehra, Maryou B.K. Lambros, Antje Neeb, Veronica Gil, Wei Yuan, and Johann S. de Bono are all employees of The Institute of Cancer Research (ICR), which has a commercial interest in abiraterone. The ICR operates a Rewards to Inventors scheme through which employees of the ICR may receive financial benefit following commercial licensing. Johann S. de Bono has served on advisory boards and received fees from companies including AstraZeneca, Astellas, Bayer, Bioxcel Therapeutics, Boehringer Ingelheim, Cellcentric, Daiichi Sankyo, Eisai, Genentech/Roche, Genmab, GSK, Janssen, Merck Serono, Merck Sharp & Dohme, Menarini/Silicon Biosystems, Orion, Pfizer, Qiagen, Sanofi Aventis, Sierra Oncology, Taiho, and Vertex Pharmaceuticals. He is an employee of The ICR, which have received funding or other support for his research work from AstraZeneca, Astellas, Bayer, Cellcentric, Daiichi, Genentech, Genmab, GSK, Janssen, Merck Serono, MSD, Menarini/Silicon Biosystems, Orion, Sanofi Aventis, Sierra Oncology, Taiho, Pfizer, and Vertex, and which has a commercial interest in abiraterone, PARP inhibition in DNA repair defective cancers, and PI3K/AKT pathway inhibitors (no personal income). He was named as an inventor, with no financial interest, for patent 8,822,438. Johann S. de Bono is a National Institute for Health Research (NIHR) Senior Investigator. The views expressed in this article are those of the author(s) and not necessarily those of the NHS, the NIHR, or the Department of Health. Charles G. Drake has served on advisory boards for AZ Medimmune, Bayer, BMS, Compugen, Ferring, F-Star, Genocea, Janssen, Kleo, Merck, Merck-Serono, Pfizer, Pierre Fabre, Roche/Genentech, Shattuck Labs, Tizona, Urogen, and Werewolf. He is an employee of Johns Hopkins University, which hold the BMS patent. He has stock or financial interest in the following companies: Compugen, Harpoon, Kleo, Tizona, Urogen, and Werewolf. Adam Sharp has received travel support from Sanofi and Roche-Genentech, and speaker honoraria from Astellas Pharma. Niven Mehra has served on advisory boards (compensated and institutional) for Roche, MSD, BMS, Bayer, Astellas, and Janssen; received research support (institutional) from Astellas, Janssen, Pfizer, Roche, and Sanofi Genzyme; and received travel support from Astellas and MSD. Wei Yuan received travel grant from Jilin Huarui Gene Technology Ltd. No relevant conflicts of interest were disclosed by other authors.
Funding/Support and role of the sponsor: We acknowledge research funding for this work from Cancer Research UK, Prostate Cancer UK, the Movember Foundation through the London Movember Centre of Excellence (CEO13_2-002), the Prostate Cancer Foundation, The V Foundation for Cancer Research (D2016-022), the UK Department of Health through an Experimental Cancer Medicine Centre (ECMC) grant, and Sanofi Aventis. Professor Johann de Bono is a National Institute for Health Research (NIHR) Senior Investigator. The views expressed in this article are those of the author(s) and not necessarily those of the NHS, the NIHR, or the Department of Health.
Associate Editor: Matthew Cooperberg
Statistical Editor: Emily Zambor
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.eururo.2021.01.017.
Contributor Information
Johann S. de Bono, Email: johann.de-bono@icr.ac.uk.
International SU2C PCF Prostate Cancer Dream Team:
Dan Robinson, Eliezer M. Van Allen, Yi-Mi Wu, Nikolaus Schultz, Robert J. Lonigro, Juan-Miguel Mosquera, Bruce Montgomery, Mary-Ellen Taplin, Colin C. Pritchard, Gerhardt Attard, Himisha Beltran, Wassim Abida, Robert K. Bradley, Jake Vinson, Xuhong Cao, Pankaj Vats, Lakshmi P. Kunju, Maha Hussain, Scott A. Tomlins, Kathleen A. Cooney, David C. Smith, Christine Brennan, Javed Siddiqui, Rohit Mehra, Yu Chen, Dana E. Rathkopf, Michael J. Morris, Stephen B. Solomon, Jeremy C. Durack, Victor E. Reuter, Anuradha Gopalan, Jianjiong Gao, Massimo Loda, Rosina T. Lis, Michaela Bowden, Stephen P. Balk, Glenn Gaviola, Carrie Sougnez, Manaswi Gupta, Evan Y. Yu, Elahe A. Mostaghel, Heather H. Cheng, Hyojeong Mulcahy, Lawrence D. True, Stephen R. Plymate, Heidi Dvinge, Roberta Ferraldeschi, Penny Flohr, Susana Miranda, Zafeiris Zafeiriou, Nina Tunariu, Joaquin Mateo, Raquel Perez-Lopez, Francesca Demichelis, Brian D. Robinson, Marc Schiffman, David M. Nanus, Scott T. Tagawa, Alexandros Sigaras, Kenneth W. Eng, Olivier Elemento, Andrea Sboner, Elisabeth I. Heath, Howard I. Scher, Kenneth J. Pienta, Philip Kantoff, Johann S. de Bono, Mark A. Rubin, Peter S. Nelson, Levi A. Garraway, Charles L. Sawyers, and Arul M. Chinnaiyan
Appendix A. Supplementary data
The following are Supplementary data to this article:
References
- 1.Sartor O, de Bono JS. Metastatic prostate cancer. N Engl J Med. 2018;378:1653–1654. doi: 10.1056/NEJMc1803343. [DOI] [PubMed] [Google Scholar]
- 2.Calcinotto A., Spataro C., Zagato E. IL-23 secreted by myeloid cells drives castration-resistant prostate cancer. Nature. 2018;559:363–369. doi: 10.1038/s41586-018-0266-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ammirante M., Luo J.L., Grivennikov S., Nedospasov S., Karin M. B-cell-derived lymphotoxin promotes castration-resistant prostate cancer. Nature. 2010;464:302–305. doi: 10.1038/nature08782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chatterjee S., Daenthanasanmak A., Chakraborty P. CD38-NAD(+) axis regulates immunotherapeutic anti-tumor T cell response. Cell Metab. 2018;27 doi: 10.1016/j.cmet.2017.10.006. 85–100.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen L., Diao L., Yang Y. CD38-mediated immunosuppression as a mechanism of tumor cell escape from PD-1/PD-L1 blockade. Cancer Discov. 2018;8:1156–1175. doi: 10.1158/2159-8290.CD-17-1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chmielewski J.P., Bowlby S.C., Wheeler F.B. CD38 inhibits prostate cancer metabolism and proliferation by reducing cellular NAD(+) pools. Mol Cancer Res. 2018;16:1687–1700. doi: 10.1158/1541-7786.MCR-17-0526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karakasheva T.A., Dominguez G.A., Hashimoto A. CD38+ M-MDSC expansion characterizes a subset of advanced colorectal cancer patients. JCI insight. 2018;3 doi: 10.1172/jci.insight.97022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karakasheva T.A., Waldron T.J., Eruslanov E. CD38-expressing myeloid-derived suppressor cells promote tumor growth in a murine model of esophageal cancer. Cancer Res. 2015;75:4074–4085. doi: 10.1158/0008-5472.CAN-14-3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu X., Grogan T.R., Hieronymus H. Low CD38 identifies progenitor-like inflammation-associated luminal cells that can initiate human prostate cancer and predict poor outcome. Cell Rep. 2016;17:2596–2606. doi: 10.1016/j.celrep.2016.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mottahedeh J., Haffner M.C., Grogan T.R. CD38 is methylated in prostate cancer and regulates extracellular NAD. Cancer Metab. 2018;6:13. doi: 10.1186/s40170-018-0186-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Terstappen L.W., Huang S., Safford M., Lansdorp P.M., Loken M.R. Sequential generations of hematopoietic colonies derived from single nonlineage-committed CD34+CD38– progenitor cells. Blood. 1991;77:1218–1227. [PubMed] [Google Scholar]
- 12.Hogan K.A., Chini C.C.S., Chini E.N. The multi-faceted ecto-enzyme CD38: roles in immunomodulation, cancer, aging, and metabolic diseases. Front Immunol. 2019;10:1187. doi: 10.3389/fimmu.2019.01187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Terstappen L.W., Johnsen S., Segers-Nolten I.M., Loken M.R. Identification and characterization of plasma cells in normal human bone marrow by high-resolution flow cytometry. Blood. 1990;76:1739–1747. [PubMed] [Google Scholar]
- 14.Horenstein A.L., Chillemi A., Zaccarello G. A CD38/CD203a/CD73 ectoenzymatic pathway independent of CD39 drives a novel adenosinergic loop in human T lymphocytes. Oncoimmunology. 2013;2 doi: 10.4161/onci.26246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barbosa M.T., Soares S.M., Novak C.M. The enzyme CD38 (a NAD glycohydrolase, EC 3.2.2.5) is necessary for the development of diet-induced obesity. FASEB J. 2007;21:3629–3639. doi: 10.1096/fj.07-8290com. [DOI] [PubMed] [Google Scholar]
- 16.Camacho-Pereira J., Tarragó M.G., Chini C.C.S. CD38 dictates age-related NAD decline and mitochondrial dysfunction through an SIRT3-dependent mechanism. Cell Metab. 2016;23:1127–1139. doi: 10.1016/j.cmet.2016.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Young A., Mittal D., Stagg J., Smyth M.J. Targeting cancer-derived adenosine: new therapeutic approaches. Cancer Discov. 2014;4:879–888. doi: 10.1158/2159-8290.CD-14-0341. [DOI] [PubMed] [Google Scholar]
- 18.Leclerc B.G., Charlebois R., Chouinard G. CD73 expression is an independent prognostic factor in prostate cancer. Clin Cancer Res. 2016;22:158–166. doi: 10.1158/1078-0432.CCR-15-1181. [DOI] [PubMed] [Google Scholar]
- 19.Bendell J., Bauer T., Patel M. Abstract CT026: Evidence of immune activation in the first-in-human phase Ia dose escalation study of the adenosine 2a receptor antagonist, AZD4635, in patients with advanced solid tumors. Cancer Res. 2019;79 CT026-CT. [Google Scholar]
- 20.Rodrigues D.N., Rescigno P., Liu D. Immunogenomic analyses associate immunological alterations with mismatch repair defects in prostate cancer. J Clin Invest. 2018;128:5185. doi: 10.1172/JCI125184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krejcik J., Frerichs K.A., Nijhof I.S. Monocytes and granulocytes reduce CD38 expression levels on myeloma cells in patients treated with daratumumab. Clin Cancer Res. 2017;23:7498–7511. doi: 10.1158/1078-0432.CCR-17-2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feng X., Zhang L., Acharya C. Targeting CD38 suppresses induction and function of T regulatory cells to mitigate immunosuppression in multiple myeloma. Clin Cancer Res. 2017;23:4290–4300. doi: 10.1158/1078-0432.CCR-16-3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van de Donk N, Usmani SZ. CD38 antibodies in multiple myeloma: mechanisms of action and modes of resistance. Front Immunol. 2018;9:2134. doi: 10.3389/fimmu.2018.02134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Detre S., Saclani Jotti G., Dowsett M. A “quickscore” method for immunohistochemical semiquantitation: validation for oestrogen receptor in breast carcinomas. J Clin Pathol. 1995;48:876–878. doi: 10.1136/jcp.48.9.876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Robinson D., Van Allen E.M., Wu Y.M. Integrative clinical genomics of advanced prostate cancer. Cell. 2015;161:1215–1228. doi: 10.1016/j.cell.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schaefer C.F., Anthony K., Krupa S. PID: the pathway interaction database. Nucleic Acids Res. 2009;37 doi: 10.1093/nar/gkn653. D674–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kumar A., Coleman I., Morrissey C. Substantial interindividual and limited intraindividual genomic diversity among tumors from men with metastatic prostate cancer. Nat Med. 2016;22:369–378. doi: 10.1038/nm.4053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sidders B., Zhang P., Goodwin K. Adenosine signaling is prognostic for cancer outcome and has predictive utility for immunotherapeutic response. Clin Cancer Res. 2020;26:2176–2187. doi: 10.1158/1078-0432.CCR-19-2183. [DOI] [PubMed] [Google Scholar]
- 29.Fong L., Hotson A., Powderly J.D. Adenosine 2A receptor blockade as an immunotherapy for treatment-refractory renal cell cancer. Cancer Discov. 2020;10:40–53. doi: 10.1158/2159-8290.CD-19-0980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wherry E.J., Ha S.J., Kaech S.M. Molecular signature of CD8+ T cell exhaustion during chronic viral infection. Immunity. 2007;27:670–684. doi: 10.1016/j.immuni.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 31.Miller B.C., Sen D.R., Al Abosy R. Subsets of exhausted CD8(+) T cells differentially mediate tumor control and respond to checkpoint blockade. Nat Immunol. 2019;20:326–336. doi: 10.1038/s41590-019-0312-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wherry E.J., Kurachi M. Molecular and cellular insights into T cell exhaustion. Nat Rev Immunol. 2015;15:486–499. doi: 10.1038/nri3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nutt S.L., Hodgkin P.D., Tarlinton D.M., Corcoran L.M. The generation of antibody-secreting plasma cells. Nat Rev Immunol. 2015;15:160–171. doi: 10.1038/nri3795. [DOI] [PubMed] [Google Scholar]
- 34.Chu PG, Arber DA. CD79: a review. Appl Immunohistochem Mol Morphol. 2001;9:97–106. doi: 10.1097/00129039-200106000-00001. [DOI] [PubMed] [Google Scholar]
- 35.Japp A.S., Kursunel M.A., Meier S. Dysfunction of PSA-specific CD8+ T cells in prostate cancer patients correlates with CD38 and Tim-3 expression. Cancer Immunol Immunother. 2015;64:1487–1494. doi: 10.1007/s00262-015-1752-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Allard D., Allard B., Stagg J. On the mechanism of anti-CD39 immune checkpoint therapy. J Immunother Cancer. 2020;8 doi: 10.1136/jitc-2019-000186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luger D., Yang Y.A., Raviv A. Expression of the B-cell receptor component CD79a on immature myeloid cells contributes to their tumor promoting effects. PLoS One. 2013;8 doi: 10.1371/journal.pone.0076115. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.