Summary
Background
Alzheimer’s disease is a debilitating and highly heritable neurological condition. As such, genetic studies have sought to understand the genetic architecture of Alzheimer’s disease since the 1990s, with successively larger genome-wide association studies (GWAS) and meta-analyses. These studies started with a small sample size of 1086 individuals in 2007, which was able to identify only the APOE locus. In 2013, the International Genomics of Alzheimer’s Project (IGAP) did a meta-analysis of all existing GWAS using data from 74 046 individuals, which stood as the largest Alzheimer’s disease GWAS until 2018. This meta-analysis discovered 19 susceptibility loci for Alzheimer’s disease in populations of European ancestry.
Recent developments
Three new Alzheimer’s disease GWAS published in 2018 and 2019, which used larger sample sizes and proxy phenotypes from biobanks, have substantially increased the number of known susceptibility loci in Alzheimer’s disease to 40. The first, an updated GWAS from IGAP, included 94 437 individuals and discovered 24 susceptibility loci. Although IGAP sought to increase sample size by recruiting additional clinical cases and controls, the two other studies used parental family history of Alzheimer’s disease to define proxy cases and controls in the UK Biobank for a genome-wide association by proxy, which was meta-analysed with data from GWAS of clinical Alzheimer’s disease to attain sample sizes of 388 324 and 534 403 individuals. These two studies identified 27 and 29 susceptibility loci, respectively. However, the three studies were not independent because of the large overlap in their participants, and interpretation can be challenging because different variants and genes were highlighted by each study, even in the same locus. Furthermore, neither the variant with the strongest Alzheimer’s disease association nor the nearest gene are necessarily causal. This situation presents difficulties for experimental studies, drug development, and other future research.
Where next?
The ultimate goal of understanding the genetic architecture of Alzheimer’s disease is to characterise novel biological pathways that underly Alzheimer’s disease pathogenesis and to identify novel drug targets. GWAS have successfully contributed to the characterisation of the genetic architecture of Alzheimer’s disease, with the identification of 40 susceptibility loci; however, this does not equate to the discovery of 40 Alzheimer’s disease genes. To identify Alzheimer’s disease genes, these loci need to be mapped to variants and genes through functional genomics studies that combine annotation of variants, gene expression, and gene-based or pathway-based analyses. Such studies are ongoing and have validated several genes at Alzheimer’s disease loci, but greater sample sizes and cell-type specific data are needed to map all GWAS loci.
Introduction
Alzheimer’s disease is a neurological condition characterised by progressive decline in cognition, with concomitant functional decline.1 The primary pathological hall mark of Alzheimer’s disease is the aggregation of amyloid β peptides into extracellular plaques and of hyperphosphorylated tau into intracellular neurofibrillary tangles, accompanied by neuro inflammation, gliosis, and neurodegeneration.2
Genetic factors play an important part in the development of Alzheimer’s disease. In autosomal dominant Alzheimer’s disease, highly penetrant mutations in APP, PSEN1, or PSEN2 result in monogenic Alzheimer’s disease, typically with early onset.3 However, most cases of Alzheimer’s disease (99%) involve multiple genetic, environ mental, and lifestyle factors, with genetics accounting for up to 53% of total phenotypic variance.4 Until 2018, the largest genome–wide association study (GWAS) of Alzheimer’s disease had been done in 2013, identifying 19 risk loci.5 Beyond locus identification, characterization of risk loci can implicate functional genetic variants and genes, which can inform mechanistic studies and rational drug development. Compared with drug targets with no evidence of genetic association, drug targets supported by evidence of both genetic association with disease and functional data are twice as likely to progress from phase 1 studies to successful approval.6
In 2018 and 2019, three new GWAS in Alzheimer’s disease have been published, expanding the number of known genome–wide risk loci to 40.7–9 In this Rapid Review, we summarise discovered loci, emphasising that the specific functional or causal gene in each locus is often unknown. To ensure genomic risk loci and lead single nucleotide polymorphisms (SNPs) are consistent across studies, we used the default settings of Functional Mapping and Annotation10 on published GWAS summary statistics, then annotated loci with cytogenetic band using SNPnexus (panel 1).11 Because the lead SNP in each locus varies across studies, we provide a unified list of SNPs associated with Alzheimer’s disease across GWAS, highlighting genetic correlations to emphasise when lead SNPs are equivalent or different. Finally, we discuss the strength of GWAS evidence at different loci for Alzheimer’s disease, and necessary steps to assign a likely functional gene.
Panel 1: Glossary of terms.
Cytogenetic band
Subregion of a chromosome that is visible under a microscope after staining
Exon
Any part of a transcribed gene that is incorporated into the final functional RNA molecule
Genome-wide association study (GWAS)
Observational study of a genome-wide set of genetic variants in different individuals to test whether a variant is associated with a trait
GWAS by proxy
A GWAS in which the phenotype is inferred on the basis of parental phenotype
Locus
A fixed position on the genome where a specific gene or genetic marker is located; in GWAS a locus represents the top single nucleotide polymorphism (SNP) and all SNPs in linkage disequilibrium with it
Linkage disequilibrium
Correlation between genetic variants caused by non-random segregation of alleles located close to one another on a chromosome
Population stratification
Genetic variation that tags population structure instead of the phenotype of interest; stratification can be as subtle as geographical location within a country or as extreme as ethnicity
Quantitative trait locus
A genetic locus that is associated with a quantitative trait such as gene expression, DNA methylation, or protein expression; quantitative trait locus studies are generally done genome-wide at the SNP level
Genome-wide significance
A result that is significant according to the Bonferroni corrected significance threshold (α/n), in which the number of comparisons (n) is the number of independent genetic loci based on linkage disequilibrium; in Europeans, genome-wide significance for an uncorrected α of 0·05 is p less than 5 × 10−8; this threshold differs depending on ethnicity
SNP
A genomic site where a single nucleotide (the basic unit of genetic code) varies between individuals; these variants may or may not affect phenotype
GWAS
In GWAS, millions of common coding and non–coding genetic variants across the genome are tested for association with a trait (panel 2). Functional variants are often not directly genotyped, but can be correlated with genotyped variants due to linkage disequilibrium (in which restricted recombination between loci causes non–random transmission of alleles). Furthermore, functional variants often regulate expression of a nearby gene, rather than changing the coding sequence.16 Thus, GWAS generally do not discover functional variants or genes, but instead identify genetic loci associated with traits. Informatic and functional characterisation is then needed to further identify functional variants and genes.
Panel 2: Rare genetic variant analyses in Alzheimer’s disease.
Genome-wide association studies traditionally only capture associations with common genetic variants—those with a minor allele frequency greater than 1%. However, with the release of genotype imputation panels such as the Haplotype Reference Consortium, it is now possible to accurately impute genotypes at allele frequencies as low as 0·1%.12 Furthermore, next-generation sequencing methods such as whole-exome sequencing, whole-genome sequencing, and exome arrays allow for the identification of rare genetic variants (genetic variants with a minor allele frequency less than 1%) associated with disease. The identification of rare genetic variants associated with disease can identify novel loci or, when located within the coding region of a gene, can pinpoint the causal gene in known loci identified in genome-wide association studies. Rare genetic variant analyses in Alzheimer’s disease have identified coding variants located within PLD3,3 TREM2,3 ABI3,13 PLCG2,13 PILRA,14 ABCA7,14 and SORL1.15
GWAS have increased statistical power by adding clinically or pathologically diagnosed cases and controls. This method is time–consuming and expensive due to the extensive efforts needed for recruitment, ascertainment, and genotyping. With the advent of large biobanks such as the UK Biobank (>500 000 participants), large population-based cohorts of genotyped individuals are now available. However, ascertainment of Alzheimer’s disease cases is limited in these biobanks because enrolled participants tend to be too young to have a high probability of developing Alzheimer’s disease; in the UK Biobank, only about 1000 individuals have a diagnosis of Alzheimer’s disease, based on International Classification of Diseases 10 codes. Genome–wide association studies by proxy (GWAX)17 are a novel solution to this problem, using parental history of a trait to identify proxy cases and controls. This approach requires approximately four times as many proxy cases and controls for equivalent power to traditional GWAS. However, GWAX can massively increase statistical power compared with GWAS due to a larger sample size by including younger samples from large biobanks.17 Unknown sample overlap in meta–analysis of cohort studies can lead to false associations due to recruitment of individuals into multiple studies, an issue which large-scale bio banks will exacerbate.18 Funding agencies have begun to address this issue through globally unique identifiers or similar solutions.
An underlying issue for GWAS of Alzheimer’s disease is the use of clinical phenotypes, often in the absence of specific biomarkers or neuropathologically defined phenotypes. Although Alzheimer’s disease is mainly characterised by the presence of amyloid plaques and neurofibrillary tangles, concomitant or alternative neuro degenerative pathologies can lead to clinical phenotypes analogous to Alzheimer’s disease.2 For example, in a community–based autopsy cohort, approximately 60% of patients with clinical diagnoses of Alzheimer’s–type disease were in fact affected by a vascular disease pathology, TDP43, or Lewy body pathology rather than plaques and tangles.19 This problem is further exacerbated in the GWAX framework, because parental history of Alzheimer’s disease is often less precise than clinical diagnoses due to a lack of distinction between Alzheimer’s disease and other dementia subtypes. Phenotypic heterogeneity due to misdiagnosis of Alzheimer’s disease results in genetic heterogeneity and reduced statistical power for GWAS discovery.20 Furthermore, Alzheimer’s disease pathology can be found in cognitively normal individuals, who might develop the clinical manifestations of Alzheimer’s disease if they live long enough.2 Inclusion of young–old (60–69 years) or younger participants as controls in Alzheimer’s disease GWAS might lead to further confounding and reduced statistical power; the genetic aetiology of non–Mendelian early–onset Alzheimer’s disease dementia seems to be the same as in late–onset Alzheimer’s disease dementia,21 but individuals with earlier onset might have higher combined genetic and environmental risk. Rather than using age–matched case–control studies, use of the youngest possible cases with the oldest possible controls might substantially improve the discovery power of GWAS in late–onset diseases such as Alzheimer’s disease.22,23 Existing Alzheimer’s disease GWAS cohorts have generally included a minimum age restriction for cases, and have occasionally used young population controls, both practices that should be avoided in future recruitment if possible.
A potential further confounding factor is population stratification, which causes GWAS associations to tag population differences rather than disease associations.24 To resolve this issue, a homogeneous population is selected in which population outliers are excluded, and principal components are used to reduce overall genetic variation and thus capture and account for remaining stratification due to genetic ancestry.24 In this Rapid Review, we focus on GWAS done in populations of European ancestry due to their large sample sizes and increased statistical power, but GWAS have also been performed in other ethnicities (panel 3). Fine–scale population structure could still be an under lying issue that can be partly accounted for by covarying on more principal components.29 Fully accounting for population structure might require more complex models that adjust for local ancestry of specific regions of DNA to truly be controlled.24
Panel 3: Non-European Alzheimer’s disease genome-wide association studies.
Non-European genome-wide association studies of Alzheimer’s disease have identified genome-wide significant variants in African-American (APOE, 7p12.1 [rs112404845], 13q33.1 [rs16961023], and 19p13.3),25 Chinese (14q22.2 [rs72713460], 21q22.13 [rs928771]),26 and transethnic populations (5q31.3 [rs11168036], 10p14 [rs7920721], and 17q22 [rs2632516]).27 The genomic diversity across populations offers opportunities to discover new loci that might be specific to a particular population, and can improve the identification of functional variants in known loci due to differences in linkage disequilibrium structure.28 As such, it is important to include diverse populations in genetic studies of Alzheimer’s disease.
Advances in Alzheimer’s disease GWAS
The first GWAS for Alzheimer’s disease of 1086 individuals was done in 2007, and only replicated the previous association with APOE.30 Increasing sample sizes from new studies and meta–analysis of existing studies led to the discovery of novel Alzheimer’s disease loci,30 leading to the landmark meta–analysis done by the International Genomics of Alzheimer’s Project (IGAP) in 2013.5 This was the largest Alzheimer’s disease GWAS at the time and was a meta–analysis of earlier GWAS done by the European Alzheimer Disease Initiative, Genetic and Environmental Risk in Alzheimer’s Disease, Cohorts for Heart and Aging Research in Genomic Epidemiology, and the Alzheimer Disease Genetics Consortium. IGAP’s stage 1 discovery phase consisted of 17 008 Alzheimer’s disease cases and 37 154 controls (n=54 162, approximately 8% of cases and controls with pathology confirmed), with stage 2 consisting of a follow–up of the top 11 632 SNPs in an additional 8572 Alzheimer’s disease cases and 11 312 controls (n=74 046). The meta–analysis of stage 1 and stage 2 identified 19 Alzheimer’s disease susceptibility loci.
A GWAX done in 2018 used 314 278 participants from the UK Biobank,8 with 14 338 participants reporting paternal and 27 696 participants reporting maternal family history of Alzheimer’s disease or dementia. Participants were excluded if their parents were younger than 60 years, died before the age of 60 years, or if no age was reported. The GWAX summary statistics were then meta–analysed with stage 1 and 2 of the 2013 IGAP GWAS5 for a total sample of 388 324 individuals. This analysis resulted in the identification of 27 susceptibility loci. Despite concerns that GWAX reflect different underlying genetic architecture than case–control studies due to increased phenotypic heterogeneity, the genetic correlation (the proportion of variance in disease liability shared between two traits) between self–reported parental history of Alzheimer’s disease and clinically diagnosed Alzheimer’s disease was high (maternal rg=0·91; paternal rg=0·66), indicating that this GWAX captured the same genetic architecture as a GWAS of clinical Alzheimer’s disease.8,17
Another GWAX done in 20197 expanded its sample size by meta–analysing the IGAP stage 1 discovery sample,5 a new GWAS from the Psychiatric Genomics Consortium (n=17 477), exome–wide data from the Alzheimer’s Disease Sequencing Project (n=7506), and a GWAX from the UK Biobank (71 880 proxy cases and 383 378 proxy controls) for a total sample of 534 403 individuals. Sample overlap between the Alzheimer’s Disease Sequencing Project and IGAP was accounted for statistically. Instead of meta-analysing paternal and maternal cases and controls, this GWAX used the number of parents with Alzheimer’s disease weighted by the probability of being a case or control on the basis of parental age, rather than excluding participants. This analysis identified 29 susceptibility loci.
In 2019, IGAP did the largest GWAS of clinically diagnosed Alzheimer’s disease to date.9 This analysis increased the IGAP Stage 1 discovery sample to 21 982 cases and 41 944 controls (n=63 926, approximately 9% of cases and controls with confirmed pathology). The meta-analysis with replication samples from stages 2 and 3 produced a final sample size of 35 274 cases and 59 163 controls (n=94 437). As a result, 24 susceptibility loci were discovered.
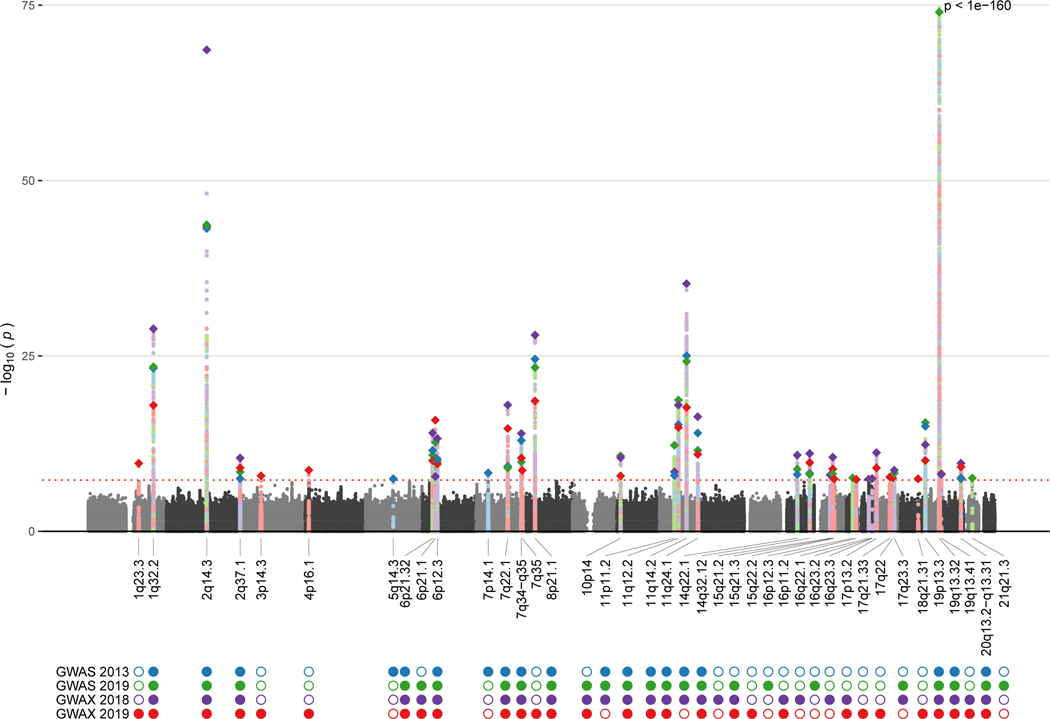
In total, 40 Alzheimer’s disease susceptibility loci reached genome–wide significance in at least one of the four GWAS we discuss (table 1). 15 were replicated across all four GWAS, and nine were significant in two or three studies. Two loci identified by the 2013 GWAS,5 three loci in the 2018 GWAX8 and 2019 GWAS,9 and eight in the 2019 GWAX7 were not replicated at full genome–wide significance in the other studies (table 1, figure). All but four of those loci (7q35, 16q23.3, 17q21.33, and 18q21.31) reached genome–wide suggestive significance (p<1 × 10−5) in at least one of these four studies, suggesting increased sample sizes improved statistical power to detect previous suggestive loci. The direction of effect was also consistent across studies after accounting for linkage disequilibrium in all except six loci (5q14.3, 6p21.1, 7p14.1, 16p12.3, 19p13.3, and 21q21.3), which are among those containing multiple independent variants. However, there was extensive overlap in the sample used between all four Alzheimer’s disease GWAS, so these studies are not independent. Across these 40 susceptibility loci, associations were reported for 78 unique lead SNPs—although most lead SNPs within particular loci were in high linkage disequilibrium, indicating they probably tag the same functional variant or variants (table 1).
Table 1:
Loci associated with Alzheimer’s disease from four GWAS
| Locus | Study | rsID | chr:pos:ref:alt | LD | Nearest Gene (N) | AF | OR (95% CI) | P |
|---|---|---|---|---|---|---|---|---|
| 1q23.3 | X19 | rs4575098 | 1:161155392:G:A | ADAMTS4 (57) | 0.239 | 1.02 (1.01–1.02) | 2.1E-10 | |
| 1q32.2 | S13 | rs6656401 | 1:207692049:A:G | 1 | CR1 (25) | 0.784 | 0.85 (0.82–0.87) | 5.7E-24 |
| X18 | rs6656401 | 1:207692049:A:G | 1 | CR1 (26) | 0.784 | 0.87 (0.85–0.89) | 1.4E-29 | |
| X19 | rs2093760 | 1:207786828:A:G | 1 | CR1 (23) | 0.775 | 0.98 (0.97–0.98) | 1.1E-18 | |
| S19 | rs4844610 | 1:207802552:A:C | 1 | CR1 (25) | 0.780 | 0.85 (0.83–0.88) | 3.6E-24 | |
| 2q14.3 | X19 | rs4663105 | 2:127891427:A:C | 1 | BIN1 (17) | 0.412 | 1.03 (1.03–1.04) | 3.4E-44 |
| S13 | rs6733839 | 2:127892810:C:T | 1 | BIN1 (17) | 0.395 | 1.22 (1.18–1.25) | 6.9E-44 | |
| X18 | rs6733839 | 2:127892810:C:T | 1 | BIN1 (17) | 0.395 | 1.20 (1.17–1.22) | 2.4E-69 | |
| S19 | rs6733839 | 2:127892810:C:T | 1 | BIN1 (17) | 0.395 | 1.20 (1.17–1.23) | 2.1E-44 | |
| 2q37.1 | X19 | rs10933431 | 2:233981912:G:C | 1 | INPP5D (35) | 0.761 | 1.02 (1.01–1.02) | 8.9E-10 |
| S19 | rs10933431 | 2:233981912:G:C | 1 | INPP5D (35) | 0.761 | 1.10 (1.06–1.13) | 3.4E-09 | |
| S13 | rs35349669 | 2:234068476:C:T | 2 | INPP5D (35) | 0.512 | 1.08 (1.05–1.11) | 3.2E-08 | |
| X18 | rs35349669 | 2:234068476:C:T | 2 | INPP5D (35) | 0.512 | 1.07 (1.05–1.09) | 3.6E-11 | |
| 3p14.3 | X19 | rs184384746 | 3:57226150:C:T | HESX1 (27) | 0.002 | 1.21 (1.14–1.30) | 1.2E-08 | |
| 4p16.1 | X19 | rs6448453 | 4:11026028:A:G | CLNK (5) | 0.772 | 0.99 (0.98–0.99) | 1.9E-09 | |
| 5q14.3 | S13 | rs190982 | 5:88223420:G:A | MEF2C (2) | 0.663 | 1.08 (1.05–1.11) | 3.2E-08 | |
| 6p21.32 | X18 | rs34855541 | 6:32559825:A:G | 1 | HLA-DRB1 (116) | 0.135 | 0.90 (0.87–0.92) | 9.5E-15 |
| S19 | rs9271058 | 6:32575406:A:T | 2 | HLA-DRB1 (120) | 0.713 | 0.91 (0.88–0.93) | 1.4E-11 | |
| S13 | rs111418223 | 6:32578530:C:A | 2 | HLA-DQA1 (116) | 0.713 | 0.90 (0.87–0.93) | 2.9E-12 | |
| X19 | rs6931277 | 6:32583357:A:T | 1 | HLA-DQA1 (118) | 0.122 | 0.98 (0.98–0.99) | 8.4E-11 | |
| 6p21.1 | X19 | rs187370608 | 6:40942196:G:A | 1 | UNC5CL (30) | 0.001 | 1.25 (1.19–1.32) | 1.5E-16 |
| S19 | rs114812713 | 6:41034000:G:C | 2 | OARD1 (25) | 0.019 | 1.32 (1.19–1.46) | 2.1E-13 | |
| X18 | rs9381040 | 6:41154650:C:T | 3 | TREML2 (29) | 0.353 | 0.94 (0.92–0.96) | 1.5E-08 | |
| 6p12.3 | S19 | rs9473117 | 6:47431284:A:C | 1 | CD2AP (15) | 0.265 | 1.09 (1.06–1.12) | 1.2E-10 |
| X18 | rs9381563 | 6:47432637:C:T | 2 | CD2AP (15) | 0.656 | 0.93 (0.91–0.95) | 5.8E-14 | |
| X19 | rs9381563 | 6:47432637:C:T | 2 | CD2AP (15) | 0.656 | 0.99 (0.98–0.99) | 2.5E-10 | |
| S13 | rs10948363 | 6:47487762:A:G | 1 | CD2AP (15) | 0.263 | 1.10 (1.07–1.13) | 5.2E-11 | |
| 7p14.1 | S13 | rs2718058 | 7:37841534:A:G | GPR141 (10) | 0.347 | 0.93 (0.90–0.95) | 4.8E-09 | |
| 7q22.1 | X19 | rs1859788 | 7:99971834:A:G | 1 | PILRA (85) | 0.715 | 1.02 (1.01–1.02) | 2.2E-15 |
| S13 | rs1476679 | 7:100004446:C:T | 1 | ZCWPW1 (82) | 0.730 | 1.09 (1.06–1.12) | 5.6E-10 | |
| X18 | rs1476679 | 7:100004446:C:T | 1 | ZCWPW1 (85) | 0.730 | 1.10 (1.07–1.12) | 9.9E-19 | |
| S19 | rs12539172 | 7:100091795:T:C | 1 | NYAP1 (82) | 0.714 | 1.09 (1.06–1.11) | 9.3E-10 | |
| 7q34-q35 | X18 | rs10808026 | 7:143099133:C:A | 1 | EPHA1 (42) | 0.204 | 0.91 (0.89–0.93) | 1.1E-14 |
| S19 | rs10808026 | 7:143099133:C:A | 1 | EPHA1 (42) | 0.204 | 0.90 (0.88–0.93) | 1.3E-10 | |
| X19 | rs7810606 | 7:143108158:T:C | 2 | EPHA1 (44) | 0.425 | 1.01 (1.01–1.02) | 3.6E-11 | |
| S13 | rs11771145 | 7:143110762:G:A | 3 | EPHA1 (42) | 0.396 | 0.90 (0.88–0.93) | 1.1E-13 | |
| 7q35 | X19 | rs114360492 | 7:145950029:C:T | CNTNAP2 (44) | <0.001 | 1.19 (1.12–1.26) | 2.1E-09 | |
| 8p21.1 | X18 | rs4236673 | 8:27464929:A:G | 1 | CLU (24) | 0.611 | 1.12 (1.09–1.14) | 1.1E-28 |
| X19 | rs4236673 | 8:27464929:A:G | 1 | CLU (25) | 0.611 | 1.02 (1.02–1.02) | 2.6E-19 | |
| S13 | rs9331896 | 8:27467686:C:T | 1 | CLU (24) | 0.610 | 1.16 (1.13–1.19) | 2.8E-25 | |
| S19 | rs9331896 | 8:27467686:C:T | 1 | CLU (24) | 0.610 | 1.14 (1.11–1.17) | 4.6E-24 | |
| 10p14 | X19 | rs11257238 | 10:11717397:T:C | 1 | USP6NL (10) | 0.382 | 1.01 (1.01–1.02) | 1.3E-08 |
| X18 | rs7920721 | 10:11720308:A:G | 1 | ECHDC3 (10) | 0.393 | 1.07 (1.05–1.09) | 3.2E-11 | |
| S19 | rs7920721 | 10:11720308:A:G | 1 | ECHDC3 (10) | 0.393 | 1.08 (1.05–1.11) | 1.8E-11 | |
| 11p11.2 | S19 | rs3740688 | 11:47380340:G:T | 1 | SPI1 (43) | 0.526 | 1.09 (1.07–1.12) | 5.5E-13 |
| X18 | rs12292911 | 11:47449072:G:A | 2 | PSMC3 (42) | 0.369 | 1.06 (1.04–1.08) | 3.3E-09 | |
| S13 | rs10838725 | 11:47557871:T:C | 3 | CELF1 (42) | 0.290 | 1.08 (1.05–1.11) | 1.1E-08 | |
| 11q12.2 | S13 | rs983392 | 11:59923508:A:G | 1 | MS4A6A (52) | 0.376 | 0.90 (0.87–0.92) | 6.1E-16 |
| S19 | rs7933202 | 11:59936926:A:C | 1,2 | MS4A6A (52) | 0.341 | 0.89 (0.87–0.92) | 1.9E-19 | |
| X19 | rs2081545 | 11:59958380:C:A | 2 | MS4A6A (52) | 0.342 | 0.98 (0.98–0.99) | 1.6E-15 | |
| X18 | rs1582763 | 11:60021948:G:A | 3 | MS4A4E (52) | 0.328 | 0.92 (0.90–0.93) | 1.0E-18 | |
| 11q14.2 | X19 | rs867611 | 11:85776544:G:A | 1 | PICALM (16) | 0.692 | 1.02 (1.02–1.02) | 2.2E-18 |
| S13 | rs10792832 | 11:85867875:A:G | 2 | PICALM (16) | 0.667 | 1.15 (1.12–1.18) | 9.3E-26 | |
| X18 | rs10792832 | 11:85867875:A:G | 2 | PICALM (16) | 0.667 | 1.13 (1.11–1.15) | 5.1E-36 | |
| S19 | rs3851179 | 11:85868640:T:C | 2 | EED (16) | 0.667 | 1.14 (1.11–1.17) | 6.0E-25 | |
| 11q24.1 | S13 | rs11218343 | 11:121435587:T:C | 1 | SORL1 (6) | 0.034 | 0.77 (0.72–0.82) | 9.7E-15 |
| X18 | rs11218343 | 11:121435587:T:C | 1 | SORL1 (6) | 0.034 | 0.81 (0.77–0.85) | 4.6E-17 | |
| X19 | rs11218343 | 11:121435587:T:C | 1 | SORL1 (6) | 0.034 | 0.97 (0.96–0.98) | 1.1E-11 | |
| S19 | rs11218343 | 11:121435587:T:C | 1 | SORL1 (6) | 0.034 | 0.80 (0.75–0.85) | 2.9E-12 | |
| 14q22.1 | X18 | rs17125924 | 14:53391680:A:G | 1 | FERMT2 (16) | 0.099 | 1.12 (1.08–1.15) | 1.3E-11 |
| S19 | rs17125924 | 14:53391680:A:G | 1 | FERMT2 (16) | 0.099 | 1.14 (1.09–1.18) | 1.4E-09 | |
| S13 | rs17125944 | 14:53400629:T:C | 1 | FERMT2 (16) | 0.097 | 1.14 (1.09–1.19) | 7.9E-09 | |
| 14q32.12 | S13 | rs10498633 | 14:92926952:G:T | 1 | SLC24A4 (23) | 0.228 | 0.91 (0.88–0.94) | 5.5E-09 |
| S19 | rs12881735 | 14:92932828:T:C | 1 | SLC24A4 (23) | 0.238 | 0.92 (0.89–0.94) | 7.4E-09 | |
| X18 | rs12590654 | 14:92938855:G:A | 2 | SLC24A4 (23) | 0.347 | 0.92 (0.90–0.95) | 8.2E-12 | |
| X19 | rs12590654 | 14:92938855:G:A | 2 | SLC24A4 (23) | 0.347 | 0.99 (0.98–0.99) | 1.6E-10 | |
| 15q21.2 | X18 | rs59685680 | 15:51001534:T:G | SPPL2A (19) | 0.247 | 0.93 (0.91–0.96) | 9.2E-09 | |
| 15q21.3 | X19 | rs442495 | 15:59022615:T:C | 1 | ADAM10 (18) | 0.334 | 0.99 (0.98–0.99) | 1.3E-09 |
| X18 | rs593742 | 15:59045774:A:G | 2 | ADAM10 (18) | 0.298 | 0.93 (0.91–0.95) | 2.8E-11 | |
| S19 | rs593742 | 15:59045774:A:G | 2 | ADAM10 (18) | 0.298 | 0.93 (0.91–0.95) | 6.8E-09 | |
| 15q22.2 | X19 | rs117618017 | 15:63569902:C:T | APH1B (18) | 0.107 | 1.02 (1.01–1.02) | 3.3E-08 | |
| 16p12.3 | S19 | rs7185636 | 16:19808163:T:C | IQCK (30) | 0.156 | 0.92 (0.89–0.95) | 2.4E-08 | |
| 16p11.2 | X18 | rs889555 | 16:31122571:C:T | 1 | BCKDK (83) | 0.322 | 0.94 (0.92–0.96) | 4.1E-08 |
| X19 | rs59735493 | 16:31133100:G:A | 1 | KAT8 (83) | 0.324 | 0.99 (0.98–0.99) | 4.0E-08 | |
| 16q22.1 | X18 | rs4985556 | 16:70694000:C:A | IL34 (32) | 0.088 | 1.09 (1.05–1.12) | 3.7E-08 | |
| 16q23.2 | S19 | rs62039712 | 16:79355857:G:A | WWOX (3) | 0.094 | 1.16 (1.09–1.24) | 3.7E-08 | |
| 16q23.3 | X18 | rs12444183 | 16:81773209:A:G | PLCG2 (14) | 0.657 | 1.06 (1.04–1.08) | 3.2E-08 | |
| 17p13.2 | X18 | rs7225151 | 17:5137047:G:A | 1 | SCIMP (51) | 0.118 | 1.10 (1.07–1.13) | 6.1E-12 |
| X19 | rs113260531 | 17:5138980:G:A | 1 | SCIMP (47) | 0.117 | 1.02 (1.01–1.03) | 9.2E-10 | |
| 17q21.33 | X19 | rs28394864 | 17:47450775:G:A | RP11–81K2.1 (46) | 0.471 | 1.01 (1.01–1.02) | 1.9E-08 | |
| 17q22 | X19 | rs2526380 | 17:56398006:C:G | BZRAP1 (31) | 0.449 | 0.97 (0.96–0.98) | 2.6E-08 | |
| 17q23.3 | X18 | rs138190086 | 17:61538148:G:A | 1 | CYB561 (40) | 0.017 | 1.25 (1.16–1.35) | 1.9E-09 |
| S19 | rs138190086 | 17:61538148:G:A | 1 | CYB561 (36) | 0.017 | 1.30 (1.16–1.46) | 5.3E-09 | |
| 18q21.31 | X19 | rs76726049 | 18:56189459:T:C | ALPK2 (13) | 0.011 | 1.06 (1.04–1.08) | 3.3E-08 | |
| 19p13.3 | X19 | rs111278892 | 19:1039323:C:G | 1 | CNN2 (74) | 0.165 | 1.02 (1.01–1.03) | 7.9E-11 |
| X18 | rs3752231 | 19:1043638:C:T | 2 | ABCA7 (75) | 0.239 | 1.09 (1.07–1.12) | 4.4E-13 | |
| S19 | rs3752246 | 19:1056492:G:C | 3 | ABCA7 (75) | 0.838 | 0.87 (0.84–0.90) | 3.1E-16 | |
| S13 | rs4147929 | 19:1063443:A:G | 3 | ABCA7 (75) | 0.840 | 0.87 (0.84–0.90) | 1.1E-15 | |
| 19q13.32 | S13 | rs41289512 | 19:45351516:C:G | 1 | PVRL2* (100) | 0.030 | 5.15 (4.58–5.78) | 2.2E-167 |
| X18 | rs41289512 | 19:45351516:C:G | 1 | PVRL2* (105) | 0.030 | 2.50 (2.37–2.63) | 6.7E-255 | |
| X19 | rs41289512 | 19:45351516:C:G | 1 | PVRL2* (124) | 0.030 | 1.22 (1.21–1.24) | 5.8E-276 | |
| S19 | rs12691088 | 19:45418486:G:A | 2 | APOC1* (101) | 0.016 | 3.39 (3.15–3.65) | 2.7E-238 | |
| 19q13.41 | X19 | rs3865444 | 19:51727962:C:A | 1 | CD33 (75) | 0.336 | 0.99 (0.98–0.99) | 6.3E-09 |
| X18 | rs12459419 | 19:51728477:C:T | 1 | CD33 (75) | 0.336 | 0.94 (0.92–0.96) | 8.0E-09 | |
| 20q13.2-q13.31 | X18 | rs6069736 | 20:54983075:C:T | 1 | CSTF1 (16) | 0.088 | 0.89 (0.86–0.93) | 2.0E-10 |
| S19 | rs6024870 | 20:54997568:G:A | 1,2 | CASS4 (16) | 0.083 | 0.88 (0.85–0.92) | 3.5E-08 | |
| X19 | rs6014724 | 20:54998544:A:G | 1,2 | CASS4 (16) | 0.089 | 0.98 (0.97–0.98) | 6.6E-10 | |
| S13 | rs7274581 | 20:55018260:T:C | 2 | CASS4 (16) | 0.091 | 0.88 (0.84–0.92) | 2.5E-08 | |
| 21q21.3 | S19 | rs2830500 | 21:28156856:C:A | ADAMTS1 (4) | 0.336 | 0.93 (0.91–0.95) | 2.6E-08 |
Nearest protein coding gene to the lead SNP in the locus refers to the nearest protein-coding gene and number of genes within 1 MB of the Functional Mapping and Annotation locus. chr:pos:ref:alt=chromosome, base pair position, reference allele, and alternate allele from human genome build 19. LD=linkage disequilibrium, where numbers are assigned to blocks of highly correlated (R2≥0·8) or identical SNPs in each locus, ordered by the position of the first variant in each block; see appendix (pp 1–2) for further LD information. GWAS=genome-wide association study. OR=odds ratio for alternate allele. S13=2013 GWAS.5 S19=2019 GWAS.9 X18=2018 GWAX.8 X19=2019 GWAX.7 SNP=single nucleotide polymorphism.
These nearest genes are in the APOE locus, where a LD region surrounding the ε4-defining SNP (rs429358) is maximally significant, but the ε2-defining SNP (rs7412) is less significant than the displayed variant, therefore, these signals are likely due to APOE ε4.
Figure: Combined Manhattan plot of four Alzheimer’s disease GWAS, showing loci with genome-wide significance.
Coloured circles represent loci that had genome-wide significance in each study, while empty circles represent non-significant loci for each study. GWAS=genome-wide association studies.
From loci to genes
Discovery of 40 risk loci does not equate to the discovery of 40 risk genes. Many susceptibility loci in Alzheimer’s disease are annotated as the nearest gene to the lead SNP. Furthermore, different studies have identified different lead SNPs and sometimes report different nearest genes within the same loci, such as 7q22.1 for which the closest genes identified across the four studies were PILRA, ZCWPW1, and NYAP1 (table 1). However, only about a third of trait–associated genes are the nearest gene.31 There are 1343 protein–coding genes within the 1 MB cis–regulatory region for gene expression across the 40 risk loci (table 1). As such, mapping of SNPs to the nearest gene can be false, and might result in incorrect assumptions about the relevant molecular pathways underlying disease. Consequently, genetic research is now moving from identification of loci associated with diseases to determination of function and causation.32–34
Several methods prioritise candidate causal variants and genes within loci.32,35 Conditional analysis attempts to identify whether there are multiple independent signals within a locus by iterative use of traditional genetic association methods,35 repeatedly covarying on top variants until no signal remains, but requires raw genetic data and is somewhat stringent; it ignores variants with truly independent effects on genes if they are in strong linkage disequilibrium with lead SNPs. Other statistical fine–mapping methods use either raw data or summary statistics to predict less stringent sets of variants that are likely to be causal on the basis of association statistics and the linkage disequilibrium structure of the locus.35 Genomic annotation can then be used to assign biological function to the variants selected via fine mapping to further prioritise likely functional variants.
Candidate functional SNPs in regulatory and coding regions affect genes differently. Those occurring within the protein coding region can affect protein structure or lead to alternative splicing, potentially resulting in altered function or in some cases loss of function.35 Genetic variants located in non–coding regions often influence phenotypes by altering the expression of nearby genes (expression quantitative trait loci [eQTLs]).35 Genetic variants with evidence of colocalisation between variant associations and gene expression can be used to prioritise functional genes, although this method is limited by availability of gene expression datasets from tissues or cells relevant to the trait.35 Many, but not all, existing methods also assume there is only one functional variant in the locus.35 In addition to these in–silico approaches, candidate causal variants or genes should be experimentally validated in cell–based systems or model organisms to evaluate their biological function.32,35
The investigators of the 2019 GWAS9 and 2019 GWAX7 did a series of functional genomic analyses to prioritise putative risk genes (table 2). In the GWAS, a priority score was constructed for genes located within 500 KB of the linkage disequilibrium region for the risk locus associated with each lead SNP, which comprised the sum of several categories of evidence: exonic functional annotation, expression and eQTLs in all tissues and in those relevant to Alzheimer’s disease, correlation between expression and tau burden, differential expression in Alzheimer’s disease, or evidence based on biological pathways. By contrast, the investigators of the GWAX study used an approach that functionally annotated genome–wide significant SNPs, then mapped them to genes on the basis of localisation within a gene, association with gene expression in any tissue, and the presence of chromatin interactions. They then did a gene–based association analysis that estimated the aggregate effect of all SNPs in a gene on a trait to identify genes that were significantly associated with Alzheimer’s disease. Finally, they further prioritised genes for which the functionally annotated SNPs affected gene expression, methylation, or histone acetylation in brain tissue. In all three of the GWAS and GWAX studies7–9 the investigators also did gene–set analyses to identify biological pathways that were over–represented among genes identified via gene–based associations analysis.
Table 2:
Implicated genes in AD risk loci using functional genomics analyses
| Kunkle et al 2019 | Jansen et al 2019 | Functional Evidence† | |||
|---|---|---|---|---|---|
| Locus | Nearest Gene | Implicated Gene | Nearest Gene | Implicated Gene | |
| 1q32.2 | CR1 | CR1 | CR1 | CR1 | CR1 copy number variation |
| 2q14.3 | BIN1 | BIN1 | BIN1 | BIN1 | BIN1 3bp insertion |
| 2q37.1 | INPP5D | INPP5D | INPP5D | - | |
| 6p21.32 | HLA-DRB1 | HLA-DRB1 PSMB8 | HLA-DQA1 | HLA-DRB1 HLA-DRA | |
| 6p21.1 | OARD1 | TREM2 | UNC5CL | - | TREM2 rare variants |
| 6p12.3 | CD2AP | CD2AP | CD2AP | CD2AP | |
| 7q22.1 | NYAP1 | PILRA AGFG2 | PILRA | PILRA ZCWPW1 STAG3 GATS | PILRA G78R 38 |
| 7q34-q35 | EPHA1 | FAM121B | EPHA1 | EPHA1 ZYX | |
| 8p21.1 | CLU | CLU PTK2B | CLU | CLU PTK2B | CLU rare coding variants & indels |
| 11p11.2 | SPI1 | PSMC3 | - | - | SPI1 myeloid eQTL |
| 11q12.2 | MS4A6A | MS4A6A | MS4A6A | MS4A3 | MS4A4A, MS4A6A myeloid eQTL |
| 11q14.2 | EED | EED PICALM | PICALM | PICALM | |
| 11q24.1 | SORL1 | SORL1 | SORL1 | - | SORL1 rare variants |
| 14q22.1 | FERMT2 | STYX | - | - | |
| 14q32.12 | SLC24A4 | RIN3 | SLC24A4 | SLC24A4 | |
| 15q21.3 | ADAM10 | ADAM10 | ADAM10 | ADAM10 | ADAM10 rare variants |
| 16p12.3 | IQCK | IQCK | - | - | |
| 17p13.2 | - | - | SCIMP | SCIMP | |
| 19p13.3 | ABCA7 | ABCA7 | CNN2 | ABCA7 CNN2 | ABCA7 LoF mutations & deletions |
| 19q13.41 | - | - | CD33 | CD33 | CD33 splicing variants |
| 20q13.2-q13.31 | CASS4 | CASS4 | CASS4 | CASS4 | |
See Pimenova, Raj & Goate 2018 21 for an in-depth review.
The 2019 GWAS9 identified 53 genes across 20 non-APOE loci with a priority score of greater than 5; the 23 genes with the highest score in each locus are presented in table 2. Conditional analysis indicated that there are probably several functional variants in the 8p21.1 and 6p21.1 loci. The 2019 GWAX7 identified 35 genes across 15 non–APOE loci that were implicated in three of the four gene mapping or gene–based analyses used, with 25 genes having further evidence of brain–specific QTL annotations; the 22 genes with the highest evidence in each locus are presented in table 2. Conditional analyses indicated multiple independent association signals in the 2q14.3, 6p21.1, 8p21.1, and 19p13.3 loci. For the APOE locus, eight genes (PVR, TOMM40, BCAM, APOC1, APOC4, CLPTM1, IGSF23, and APOE) were implicated by all four of the gene–mapping or gene–based analyses used. PVR and TOMM40 also have further evidence of brain–specific eQTL and methylation QTLs (mQTLs). Conditional analysis also indicated that there are potentially multiple independent signals in the APOE locus. Pathway analysis in the 2018 and 2019 GWAX7,8 and the 2019 GWAS9 implicated pathways related to lipid traits, tau, and APP or amyloid β. The 2018 GWAX8 and 2019 GWAS9 also implicated immune response pathways. These results remained significant even after removal9 or conditioning of APOE.8
11 genes overlapped between the analyses in the 2019 GWAX7 and in the 2019 GWAS:9 CR1, BIN1, HLA-DRB1, CD2AP, PILRA, CLU, PT2KB, PICALM, ADAM10, ABCA7, and CASS4 (table 2). Independent of these two studies, fine mapping, whole–exome or whole–genome sequencing, targeted resequencing, and in–vitro experimental validation studies have identified common or rare functional variants in APOE, CR1, BIN1, TREM2, CLU, PILRA, SORL1, ADAM10, ABCA7, and CD33 (table 2).37 Gene expression analysis has also identified eQTLs in either myeloid cells or microglia for SPI1, MS4A4A, and MS4A6A (table 2).37 Despite this progress, functional variants or genes have not been identified for most Alzheimer’s disease loci.
A limitation of functional genomics studies in Alzheimer’s disease is the lack of well–powered QTL data for microglia, which are probably the most relevant disease-affected cell type on the basis of SNP heritability.38 No large QTL datasets exist for human microglia, leading investigators to use QTL datasets for peripheral myeloid cells. Additionally, most QTL datasets are generated under baseline conditions, but some functional variants might be dependent on intrinsic or extrinsic factors not present under baseline conditions. As a result, the inability to assign functional variants or causal genes might be due to the absence of annotations in disease–relevant conditions.
The limitations of functional genomics are high lighted in the APOE locus in which the combination of a gene–dense region, complex linkage disequilibrium structure, and non–specific QTL datasets led the investigators of the 2019 GWAX7 to implicate seven other genes in addition to APOE as potential functional genes in this locus.
Conclusions and future directions
GWAS have identified 40 loci that are associated with Alzheimer’s disease in European populations, 24 of which are replicated at genome–wide suggestive significance.5,7–9 Functional genomics studies further suggests APOE, CR1, BIN1, TREM2, CLU, SORL1, ADAM10, ABCA7, CD33, SPI1, and PILRA as the likely causal genes in their respective loci.37 Although GWAS have made substantial progress in characterisation of the genetic architecture of Alzheimer’s disease, much work remains to identify the functional genetic variants and biological mechanisms underlying the observed associations of genetic loci with Alzheimer’s disease. This research will require multi–omic datasets from relevant cell types such as microglia, which have been strongly implicated in Alzheimer’s disease pathogenesis.38 Multiethnic GWAS will aid in mapping of specific variants because of divergent genetic variation (panel 3). Research efforts will be also needed to overcome the challenge of obtaining the necessary sample sizes in cell–type–specific datasets, especially if some associations only apply to specific subsets of cells (eg, activated microglia).38 Until functional mapping of GWAS associations are complete, putative gene annotations should be interpreted cautiously.
Supplementary Material
Search strategy and selection criteria.
We searched PubMed for genome-wide association studies of Alzheimer’s disease published between Jan 1, 2017, and July 1, 2019, using the terms: ((Alzheimer Disease[MeSH Terms]) AND association study, genome wide[MeSH Terms]) AND (“2017/01/01”[Date - Publication] : “2019/07/31”[Date - Publication]) AND English [LA] NOT review[pt]. We included studies in which the outcome was clinically diagnosed late-onset Alzheimer’s disease or a family history of Alzheimer’s disease that were done in participants of European ancestry. The final reference list was generated on the basis of relevance and novelty to this Rapid Review.
Acknowledgments
Declaration of interests
SJA, BFH, and AMG were supported by the JPB Foundation and by the National Institute of Health (U01 AG058635; principal investigator AMG). AMG served on the scientific advisory board for Denali Therapeutics from 2015 to 2018, and has also served as a consultant for Biogen, AbbVie, Pfizer, GlaxoSmithKline, Eisai, and Illumina.
References
- 1.2019 Alzheimer’s disease facts and figures. Alzheimers Dement 2019; 15: 321–87. [Google Scholar]
- 2.DeTure MA, Dickson DW. The neuropathological diagnosis of Alzheimer’s disease. Mol Neurodegener 2019; 14: 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kamboh IM. A brief synopsis on the genetics of Alzheimer’s disease. Curr Genet Med Rep 2018; 6: 133–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ridge PG, Hoyt KB, Boehme K, et al. Assessment of the genetic variance of late-onset Alzheimer’s disease. Neurobiol Aging 2016; 41: 200.e13–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lambert JC, Ibrahim–Verbaas CA, Harold D, et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet 2013; 45: 1452–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.King EA, Davis WJ, Degner JF. Are drug targets with genetic support twice as likely to be approved? Revised estimates of the impact of genetic support for drug mechanisms on the probability of drug approval. Biorxiv 2019. 10.1101/513945 (accessed Oct 23, 2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jansen IE, Savage JE, Watanabe K, et al. Genome-wide meta-analysis identifies new loci and functional pathways influencing Alzheimer’s disease risk. Nat Genet 2019; 51: 404–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marioni RE, Harris SE, Zhang Q, et al. GWAS on family history of Alzheimer’s disease. Transl Psychiat 2018; 8: 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kunkle BW, Grenier–Boley B, Sims R, et al. Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates Aβ, tau, immunity and lipid processing. Nat Genet 2019; 51: 414–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Watanabe K, Taskesen E, van Bochoven A, Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat Commun 2017; 8: 1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dayem Ullah AZ, Oscanoa J, Wang J, Nagano A, Lemoine NR, Chelala C. SNPnexus: assessing the functional relevance of genetic variation to facilitate the promise of precision medicine. Nucleic Acids Res 2018; 46: W109–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Consortium H, McCarthy S, Das S, et al. A reference panel of 64,976 haplotypes for genotype imputation. Nat Genet 2016; 48: ng.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sims R, van der Lee SJ, Naj AC, et al. Rare coding variants in PLCG2, ABI3, and TREM2 implicate microglial-mediated innate immunity in Alzheimer’s disease. Nat Genet 2017; 49: 1373–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bis JC, Jian X, Kunkle BW, et al. Whole exome sequencing study identifies novel rare and common Alzheimer’s-associated variants involved in immune response and transcriptional regulation. Mol Psychiatr 2018; published online Aug 14. DOI: 10.1038/s41380-018-0112-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Campion D, Charbonnier C, Nicolas G. SORL1 genetic variants and Alzheimer disease risk: a literature review and meta-analysis of sequencing data. Acta Neuropathol 2019; 138: 173–86. [DOI] [PubMed] [Google Scholar]
- 16.Tam V, Patel N, Turcotte M, Bossé Y, Paré G, Meyre D. Benefits and limitations of genome-wide association studies. Nat Rev Genet 2019; 20: 467–84. [DOI] [PubMed] [Google Scholar]
- 17.Liu JZ, Erlich Y, Pickrell JK. Case-control association mapping by proxy using family history of disease. Nat Genet 2017; 49: 325–31. [DOI] [PubMed] [Google Scholar]
- 18.Lin DY, Sullivan PF. Meta-analysis of genome-wide association studies with overlapping subjects. Am J Hum Genetics 2009; 85: 862–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nelson PT, Dickson DW, Trojanowski JQ, et al. Limbic-predominant age-related TDP-43 encephalopathy (LATE): consensus working group report. Brain 2019; 142: 1503–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de Vlaming R, Okbay A, Rietveld CA, et al. Meta-GWAS Accuracy and Power (MetaGAP) calculator shows that hiding heritability is partially due to imperfect genetic correlations across studies. PLoS Genet 2017; 13: e1006495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cruchaga C, Del–Aguila JL, Saef B, et al. Polygenic risk score of sporadic late-onset Alzheimer’s disease reveals a shared architecture with the familial and early-onset forms. Alzheimer’s Dementia 2018; 14: 205–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oliynyk R. Evaluating the potential of younger cases and older controls cohorts to improve discovery power in genome-wide association studies of late-onset diseases. J Personalized Medicine 2019; 9: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tesi N, van der Lee SJ, Hulsman M, et al. Centenarian controls increase variant effect sizes by an average twofold in an extreme case–extreme control analysis of Alzheimer’s disease. Eur J Hum Genet 2019; 27: 244–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hellwege JN, Keaton JM, Giri A, Gao X, Edwards DR, Edwards TL. Population stratification in genetic association studies. Curr Protoc Hum Genetics 2017; 95: 1.22.1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mez J, Chung J, Jun G, et al. Two novel loci, COBL and SLC10A2, for Alzheimer’s disease in African Americans. Alzheimer’s Dementia 2017; 13: 119–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou X, Chen Y, Mok KY, et al. Identification of genetic risk factors in the Chinese population implicates a role of immune system in Alzheimer’s disease pathogenesis. Proc National Acad Sci 2018; 115: 201715554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jun GR, Chung J, Mez J, et al. Transethnic genome-wide scan identifies novel Alzheimer’s disease loci. Alzheimer’s Dementia 2017; 13: 727–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gurdasani D, Barroso I, Zeggini E, Sandhu MS. Genomics of disease risk in globally diverse populations. Nat Rev Genet 2019; 20: 520–35. [DOI] [PubMed] [Google Scholar]
- 29.Haworth S, Mitchell R, Corbin L, et al. Apparent latent structure within the UK Biobank sample has implications for epidemiological analysis. Nat Commun 2019; 10: 333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Raghavan N, Tosto G. Genetics of Alzheimer’s Disease: the importance of polygenic and epistatic components. Curr Neurol Neurosci 2017; 17: 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Porcu E, Rüeger S, Lepik K, et al. Mendelian randomization integrating GWAS and eQTL data reveals genetic determinants of complex and clinical traits. Nat Commun 2019; 10: 3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gallagher MD, Chen–Plotkin AS. The post-GWAS era: from association to function. Am J Hum Genetics 2018; 102: 717–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Novikova G, Kapoor M, TCW J, et al. Integration of Alzheimer’s disease genetics and myeloid genomics reveals novel disease risk mechanisms. Biorxiv 2019. 10.1101/694281 (accessed Oct 23, 2019). [DOI] [Google Scholar]
- 34.Nott A, Holtman IR, Coufal NG, et al. Brain cell type–specific enhancer–promoter interactome maps and disease-risk association. Science 2019; 366: 1134–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cannon ME, Mohlke KL. Deciphering the emerging complexities of molecular mechanisms at GWAS loci. Am J Hum Genetics 2018; 103: 637–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rathore N, Ramani SR, Pantua H, et al. Paired immunoglobulin-like type 2 receptor alpha G78R variant alters ligand binding and confers protection to Alzheimer’s disease. PLoS Gen 2018; 14: e1007427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pimenova AA, Raj T, Goate AM. Untangling genetic risk for Alzheimer’s disease. Biol Psychiat 2018; 83: 300–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Verheijen J, Sleegers K. Understanding Alzheimer disease at the interface between genetics and transcriptomics. Trends Genet 2018; 34: 434–47. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.