Abstract
To increase the repertoire of PCR based laboratory developed tests (LDTs) for the detection of SARS-CoV-2, we describe a new multiplex assay (SORP), targeting the SARS-CoV-2's, Spike and ORF8 genes. The widely used human RNaseP internal control was modified to specifically co-amplify the RNaseP mRNA. The SORP triplex assay was tested on a cohort (n = 372; POS = 144/NEG = 228) of nasopharyngeal flocked swab (NPFS) specimens, previously tested for the presence of SARS-CoV-2 using a PCR assay targeting E and RdRp genes. The overall sensitivity and specificity of the SORP assay was: 99.31% (95% CI: 96.22–99.98%), 100.0% (95% CI: 98.4–100%) respectively. The SORP assay could also detect a panel of variants of concern (VOC) from the B1.1.7 (UK) and B1.351 (SA) lineage. In summary, access to a repertoire of new SARS-CoV-2 LDT's would assist diagnostic laboratories in developing strategies to overcome some of the testing issues encountered during high-throughput SARS-CoV-2 testing.
Keywords: SARS-CoV-2, Spike, ORF8, RNaseP, Variants of concern, LDT
1. Introduction
From the first reports of infections in Wuhan, China leading to its genetic identification and designation as “Wuhan-Hu-1” (GenBank Acc No: MN908947; [1,2], the SARS-CoV-2 virus has spread globally, infecting millions of people worldwide. With the publication of the Wuhan-Hu-1 genome sequence, research groups were able to quickly develop reverse transcription-quantitative polymerase chain reaction (RT-qPCR) protocols [[3], [4], [5], [6], [7]] for specific and sensitive detection of the SARS-CoV-2 RNA. These laboratory-developed tests (LDT's) were openly shared with the World Health Organization (WHO), enabling large scale deployment of SARS-CoV-2 testing [8]. These RT-qPCR based LDT protocols, have become a major part of the global testing SARS-CoV-2 strategy, to quickly detect the virus and develop appropriate containment strategies. Based on recent meta-analyses of current published literature, the most common SARS-CoV-2 genetic targets for these LDT's include, the open reading frame 1a or 1b (ORF1a or 1b) [4,5,9], RNA dependent RNA polymerase (RdRp)/helicase (Hel) [6], Spike [10], envelope (E) [3], 5′-untranslated region (5′-UTR) [11], and nucleocapsid (N) genes [7]. The choice of these gene targets was largely based on previous experience in detecting SARS-CoV-1 [12] and MERS-CoV [13].
Early in the outbreak, the British Columbia Center for Disease Control (BCCDC) public health laboratory, developed an in-house triplex RT-qPCR assay for detecting SARS-CoV-2 [14,15]. This multiplex LDT-, targeted the RdRp and E-genes [3] of the SARS-CoV-2 virus, with the human RNaseP gene [7], co-amplified as an internal control for sample quality. Henceforth referred to as the “BCCDC Triplex”, this assay is now being used in many clinical laboratories in British Columbia, including ours and some other laboratories facilities in Canada [14].
As pandemics evolve, random nucleotide substitution occurs in the viral genome, a genetic event which is increasingly being observed in the SARS-CoV-2's genome [[16], [17], [18]]. From a diagnostic point of view, such nucleotide substitution(s) can negatively affect the binding ability of any validated primer/probe to its target [19,20]. For example, the GISAID data indicates that the last five nucleotides (3′ end) of the RT-qPCR primers developed for SARS-CoV-2 detection commonly contain some form of mutation (range between 0.2% and 3.5%) (www.epicov.org/epi3). Such mutation events can compromise the sensitivity/specificity of any validated RT-qPCR assay, leading to false negative results [[21], [22], [23]] which can compromise both patient care and infection control practices [24]. Such scenarios can be mitigated by (a.) regular genomic surveillance of new SARS-CoV-2 genomes, to verify the binding fidelity of validated primer/probe and (b.) development of new RT-qPCR assays, targeting different conserved genes of the SARS-CoV-2 genome.
Concurrent with the development of SARS-CoV-2 specific LDTs, the global research community also put into place sequencing strategies, to rapidly sequence the SARS-CoV-2 virus and to make those sequences publicly available in near real-time. As a result, as of January 20,20th January 2021, >51,000 and > 400,000 whole genome sequences of SARS-CoV-2, are now available on the NCBI and GISAID databases respectively. The availability of these high-quality whole genome sequences, from a diverse set of geographical locations, have resulted in an enhanced understanding of genetic variation amongst SARS-CoV-2's genomes. The recent reports of a new lineages and potentially highly transmissible variants, commonly referred to as variants of concerns (VOC's) of the SARS-CoV-2 virus, namely the B.1.1.7 and 501Y.V2 in the United Kingdom [25] and Republic of South Africa [26] respectively, is the result of such parallel genomic surveillance programs. Discovery of new strains of SARS-CoV-2 is not surprising as SARS-CoV-2, similar to other RNA respiratory viruses, can mutate quickly [27], and is estimated to accumulate mutations at a rate of about 1–2 mutations per month [28].
To increase the limited repertoire of SARS-CoV-2 specific qRT-PCR assays, we here present a new multiplex SARS-CoV-2 RT-qPCR LDT targeting two conserved regions of the viral genome-the Spike (S) and ORF8 genes. Henceforth, referred to as the SORP (Spike ORF8 mRNaseP) triplex assay, this LDT was validated against the BCCDC reference assay [14] and the US-CDC N1/N2 Nucleocapsid assay [7].
2. Materials & methods
2.1. Clinical specimens
Clinical specimens submitted for routine SARS-CoV-2 testing, at the microbiology & virology laboratories of BC Children's Hospital (BCCH) were used to validate the SORP triplex assay. Nasopharyngeal flocked swabs (NPFS) in universal transport medium (Copan Diagnostics, Murrieta, CA) or NPFS in viral transport medium (YOCON, Beijing, China) were used. The testing population included symptomatic children and adults. Residual total nucleic acid (TNA) extracts, remaining from the primary testing, were anonymized and used for parallel testing on the SORP triplex and US-CDC's N1/N2 Nucleocapsid assay. Because the study involved secondary use of anonymous human biological materials, it was exempted from review by the BC Children's Hospital Research Ethics Board.
2.2. Design of the spike, ORF8, mRNaseP (SORP) triplex assay
SARS-CoV-2 sequences were downloaded from the NCBI GenBank's SARS-CoV-2 data hub on January 20, 2021. Of the original 52,237 downloaded sequences, only 49,812 and 49,426 sequences had the complete sequence information for the ORF8 and S sequences respectively. Truncated and poor-quality sequences were omitted from analysis. These S/ORF8 open reading frames (ORF) were extracted and aligned using the Geneious Prime 2020.2.3 (Biomatters, Auckland, New Zealand) software suite. TaqMan™ primer and probes were designed using the Primer Express 3.0 software (ThermoFisher, USA). The “Wuhan-Hu-1” sequence (RefSeq:NC_045512.2) was used as the reference SARS-CoV-2 sequence in the alignment. BLAST analysis of the candidate primer pairs was done to ensure in silico specificity to SARS-Cov-2 sequences prior to clinical testing. Candidate primers and probes were examined manually against the currently known SARS-Cov-2, Spike and ORF8 variants, deposited in the GISAID (www.gisaid.org) database. If required, degeneracy was incorporated into the primer/probe sequences to account for single nucleotide polymorphisms/deletions.
A modified version of the US CDC's human RNaseP (RNaseP) control (Table 1 ) was designed for the SORP triplex assay (Fig. 1A_Suppl). This modified RNaseP control PCR, henceforth referred to as “mRNaseP”, amplified human RNaseP mRNA and not RNaseP genomic DNA. A new reverse primer (RNaseP-R8) was designed using the Primer Express™ software, on exon 2 of the RNaseP transcript (Ref GenBank Acc: NM_001104546.2) (Fig. 1B_Suppl). Purified human genomic DNA (TaqMan™ Control Genomic DNA human; ThermoFisher, Cat No: 4312660) and purified total nucleic acid (gDNA + mRNA) from human clinical samples, were used as templates to test the performance of the mRNaseP TaqMan™ assay. For multiplexing, Spike, ORF8 and RNaseP probes were labelled with FAM, Cy5 and NED fluorophores respectively. All primer/probes were custom synthesized from IDT (Coralville, IA) and ThermoFisher (Carlsbad, CA).
Table 1.
Sequence of forward and reverse primers and TaqMan™ probes used for the SORP assay. Position of the Spike and ORF 8 primers and probes based on the alignment to the Wuhan-Hu-1 SARS-CoV-2 sequence (RefSeq:NC_045512.2).
| PRIMER/PROBE | SEQUENCE | Nucleotide Position | Ref |
|---|---|---|---|
| Spike-F1 | CCACTAGTCTCTAGTCAGTGTGTTAATY | 21568–21595 | Present work |
| Spike-R1 | AAACTGAGGATCTGAAAACTTTGTC | 21618–21647 | |
| Spike-P1 | FAM-CAACCAGAA/ZEN/CTCAATTACCCCCTGCATACA-IABlkFQ/ | 21690–21716 | |
| ORF8-F1 | GGAGCTAGAAAATCAGCACCTTTAA | 28041–28065 | Present work |
| ORF8-R | TCGATGTACTGAATGGGTGATTTAG | 28093–28117 | |
| ORF8-P | Cy5-TGAATTGTG/TAO/CSTGGATGAGGCTGG-IABlkRQ/ | 28067–28090 | |
| RNaseP-F | AGATTTGGACCTGCGAGCG | US-CDC | |
| RNaseP-P | NED-TTCTGACCTGAAGGCTC-MGBNFQ | BCCDC in house design (Lee) Modified from US-CDC for MGB | |
| RNaseP-R | GAGCGGCTGTCTCCACAAGT | US-CDC | |
| RNaseP-R8 | TCAACGATATGATTGATAGCAACAAC | Present work |
2.3. Total nucleic acid (TNA) extraction
Total nucleic acid (TNA) was extracted from NPFS samples on the QIAsymphony (Qiagen, Hilden, Germany) automated extraction platform, using the DSP Virus/Pathogen kit (Qiagen). The volume of eluate per sample was 80 μL.
2.4. SARS-CoV-2 RT-qPCR assays
All of the RT-qPCR assays were performed using the 4X TaqMan Fast Virus 1-step Master Mix (ThermoFisher: cat No 4444434) on the ABI Fast 7500 real-time PCR system (ThermoFisher, CA). The total reaction volume was 20 μL which included 5 μL of the TNA template. The cycling conditions used were 50 °C for 5 min (Reverse Transcription), 95 °C × 20 s (Enzyme Activation) followed by 45 cycles of 95 °C at 3 s and 60 °C at 30 s, as recommended by the manufacturer.
2.4.1. BCCDC-triplex assay (“reference assay”)
The BCCDC's SARS-CoV-2 triplex assay was used as described earlier [14]. This assay targets the SARS-CoV-2's RdRp and E-gene, with the human RNaseP as the internal control. Henceforth referred to as the “Reference Assay”, this is the primary assay used to test for the presence of SARS-CoV-2 in clinical samples submitted at our BCCH laboratory.
2.4.2. N1/N2/RNaseP US-CDC's assay
The US-CDC's N1 (2019-nCoV_N1)/N2 (2019-nCoV_N2)/RNaseP primer-probe sets were purchased from IDT (Cat No: 10006713) and used as per the US-CDC's testing guidelines [29].
2.4.3. SORP (spike, ORF8, mRNaseP) triplex assay
The SORP RT-qPCR assay reaction consisted of (20 μL) with the following concentrations of the primers and probes: Spike-F1/R1 (0.3 μM), Spike-P1 (0.2 μM), ORF8-F1/ORF8-R (0.4 μM), ORF8-P (0.2 μM), RNaseP-F/R8 (0.05 μM) and RNaseP-P (0.15 μM). The primers and probes used for SORP are listed in Table 1.
2.5. Interpretation algorithm of SARS-CoV-2 assay results & discrepant analysis
2.5.1. BCCDC-triplex assay (“reference assay”)
As per the current clinical reporting guidelines for the use of the BCCDC's triplex assay, a patient sample was considered negative for SARS-CoV-2, if the RdRp and E-gene assays were negative and the RNaseP control gene was positive with a CT ≤40. A negative RNaseP signal indicated improper sample collection or failed extraction, and was reported as “invalid”. Samples positive for both the RdRp and E-gene with CT values =< 40 were considered positive. Single target positive (RdRp or E-gene), were clinically resolved by testing the original patient sample on the Xpert Xpress SARS-CoV-2 (Cepheid, Sunnyvale, CA, USA). The Xpert Xpress's result algorithm was used to guide for the presence/absence of the SARS-CoV-2 in such cases.
2.5.2. SORP triplex assay
The SORP assay was interpreted in the same fashion as described for the BCCDC reference assay, substituting the Spike and ORF8 genes for RdRp and E-gene. Discrepant or weakly positive results for S or ORF8 targets were also resolved on Xpert Xpress SARS-CoV-2 as described above.
2.5.3. US-CDC nucleocapsid singleplex assay
US-CDC's assay Nucleocapsid assay results were interpreted as per the US-CDC's testing guidelines [29].
2.6. Analytical sensitivity and specificity of the SORP triplex assay
Limit of detection (LOD) of the SORP triplex assay was determined using AmpliRun™ Total SARS-CoV-2 control (Vircell, Granada, Spain), an inactivated (swab) SARS-CoV-2 viral control preparation (4.3 × 104 copies/mL). Post-reconstitution in sterile water as per the manufacturer's instructions, the SARS-CoV-2 inactivated virus control was extracted on the QIAsymphony (Qiagen) automated extraction platform, using the DSP Virus/Pathogen kit (Qiagen). Probit regression analysis to determine the 95% detection limit was carried out using the MedCalc Software package (MedCalc Software Ltd, Ostend, Belgium).
The analytical specificity of the SORP assay was evaluated using two methods: in silico analysis using BLAST analysis with common non-target organisms in the NCBI GenBank database (Table. 1_Suppl) and RT-qPCR analysis on a panel of non-target microorganisms. These non-target microorganisms were either positive patient specimens (nasopharyngeal/oropharyngeal swabs, bronchoalveolar lavage fluid, urine, saliva, and nasal washes), type strain cultures or commercial standards (Table. 2_Suppl). Nucleic acid was extracted from each individual organism using the QIAsymphony (Qiagen, Hilden, Germany) automated extraction platform.
3. Results
3.1. SORP primer and probes & genomic alignment
The TaqMan™ primer and probes designed for the Spike and ORF8 gene targets showed 100% homology with the SARS-CoV-2 RefSeq (NC_045512.2; Fig. 1_Suppl). Random mismatches were however observed for both the S and ORF8 amplicons, when it was aligned to ca. 52,237 SARS-CoV-2 sequences, (downloaded on Jan 20, 2021, from the NCBI SARS-CoV-2 hub). These mismatches appeared random in nature suggesting either point mutation or sequencing base call errors. Based on the ca. 52,237 SARS-CoV-2 downloaded sequences, a 99% consensus sequence was generated using the Geneious Software. To incorporate nucleotide variations, degenerate nucleotide(s) was incorporated to account for single nucleotide polymorphism (L18F) detected in the recently reported B1.1.7 and 501Y.V2 SARS-CoV-2 variants (Fig. 2_Suppl). Similarly, the ORF8-F1/-R primers also showed 100% match to the ORF8 consensus sequence except for one C/G substitution in the middle of the ORF-8 probe (Fig. 3_Suppl). To prevent this substitution affecting the probe binding kinetics, a degenerate base (S C/G) was incorporated in the ORF8-probe.
3.2. Amplification of human mRNA RNaseP (mRNaseP assay)
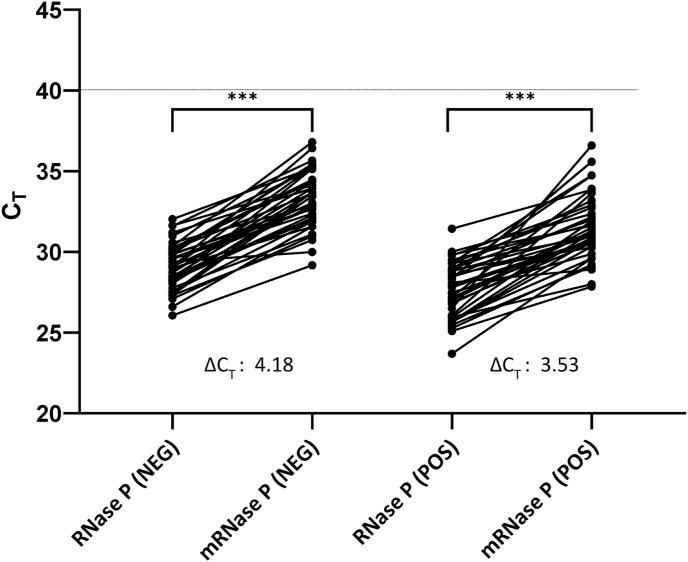
The RNaseP assay, containing the modified reverse primer RNaseP-R8, specifically amplified the human RNaseP mRNA and not RNaseP gDNA (Table 2 ). When the reverse transcription (RT) step was omitted or the template consisted of a purified human gDNA (TaqMan™ Control Genomic DNA human; ThermoFisher, Cat No: 4312660), no positive RNaseP amplification was observed (Table 2). Prior to its implementation as a control in the SORP triplex assay format, the performance of this newly developed mRNA-specific RNaseP TaqMan™ was evaluated on total nucleic acid (mRNA + gDNA), extracted from a cohort (n = 40) of clinical samples that tested negative for SARS-CoV-2. The regular US-CDC's RNaseP which amplified both gDNA and mRNA, was tested on these same samples in parallel. The average CT difference between these two assays (ΔCT = CT mRNaseP - CT RNaseP) was found to be ΔCT = 4.18 (Fig. 1 ). For SARS-CoV-2 positive clinical samples (n = 40), the average CT difference between mRNaseP and RNaseP assay was ΔCT = 3.95. (Fig. 1).
Table 2.
CT values obtained by individual RNAseP and mRNAseP assays, for two different template types-purified human gDNA and total nucleic acid (gDNA + mRNA). The TNA template was used with reverse transcription (+RT) and without reverse transcription (-RT).
| RNaseP TaqMan™ Assay | RT-qPCR Primer/Probes |
Amplicon Size |
TaqMan™ control Human gDNA (0.1 ng/μL) CT |
SARS-CoV-2 NEGATIVE Total Nucleic Acid (mRNA + gDNA) CT |
||||
|---|---|---|---|---|---|---|---|---|
| Forward | Probe | Reverse | mRNA | gDNA | +RT | -RT | ||
| RNaseP | RNaseP-F | RNase-P Probe | RNase-P-R | 65 bp | 65 bp | 32.88 | 27.25 | 28.3 |
| mRNaseP | RNaseP-F | RNase-P Probe | RNase-P-R8 | 105 bp | 2.8 kb | und | 30.14 | und |
Fig. 1.
Distribution of CT's recorded between the RNaseP and mRNaseP TaqMan™ assays for a cohort (n = 40) of SARS-CoV-2 negative and positive NPFS samples. ΔCT = CT (mRNaseP)-CT (RNaseP). A two-sided paired-sample t-test found statistically significant difference between (***P < 0.001) the CT values obtained between RNaseP and mRNaseP TaqMan™ assays, as shown by P values. Dotted line indicates the assay cut-off at CT ≤ 40 as defined in materials and methods.
3.3. Diagnostic performance of BCCDC-Triplex, SORP and N1/N2 assay
3.3.1. BCCDC-triplex assay
Of the total of 372 NPFS samples tested on the BCCDC-RdRp/E/RNaseP triplex assay, 142 were found to be positive for both the RdRp and E gene targets while two samples, S46 (RdRp = undetermined [und]/E = 37.1/mRNaseP = 27.5) and S60 (RdRp = 37.31/E = und/mRNaseP = 31.43) were single gene positives. None of these samples in this cohort tested negative for RNaseP, indicating proper sample collection. To resolve these single gene target positives, the original samples were tested on Xpert Xpress SARS-CoV-2 assay. The S46 sample gave the following results: E = Neg/N2 = 42.3 which as per Xpert Xpress SARS-CoV-2 reporting algorithm, was reported as “presumptive positive”. Interestingly, the S46 sample, when re-tested subsequently on the BIOFIRE® Respiratory Panel 2.1 assay (RP2.1; BioFire Diagnostics, LLC, Salt Lake City, UT) gave a negative result. The other single target patient positive sample S60, when tested on Xpert Xpress SARS-CoV-2, gave the following results: E = 35.4/N2 = 39.0. This as per Xpert Xpress SARS-CoV-2 reporting algorithm, classified the S60 sample as “SARS-CoV-2 positive”.
3.3.2. N1/N2 US-CDC nucleocapsid assay
Parallel testing of the 372 samples previously tested on the BCCDC-Triplex assay, gave the following results-142 positive (N1/N2 positive), 228 negatives (N1/N2 negative). The sample S46 (RdRp = und/E = 37.1/RNaseP = 27.5) which was single gene positive on the reference assay and “presumptive positive” on the Xpert Xpress SARS-CoV-2 assay, was negative for both N1/N2 targets. Sample S60, which was single gene target positive (RdRp = 37.31/E = und/RNaseP = 31.43) on the reference assay, was positive for only N2 positive (N1 = und/N2 = 37.46), classifying it as “Inconclusive” (N1 = und/N2 = 37.46), as per US-CDC's reporting algorithm for the N1/N2 assay. The overall analytical sensitivity and specificity of the N1/N2 assay therefore was: 99.30% (95% CI:96.2–99.9%), 100.0% (95% CI:98.4–100%) respectively (Table 3 ).
Table 3.
Sensitivity and Accuracy analysis of 144 Positive and 227 Negatives, tested on SORP Triplex and US-CDC N1/N2 Singleplex assay. * = sample No. S60 classified as “inconclusive” and was excluded from the final analysis.
| SARS-CoV-2 Assay | Positive (n = 144) |
Negative (n = 227) | Sensitivity (95% CI) |
Accuracy |
|---|---|---|---|---|
| SORP Triplex | 143 | 0 | 99.31% (96.19–99.98) |
99.73% (98.51–99.99) |
| US-CDC N1/N2 | 142* | 0 | 99.30% (96.17–99.98) |
99.73% (98.51–99.99) |
3.3.3. SORP triplex assay
Of the 144 SARS-CoV-2 positives detected on the reference BCCDC-Triplex assay, 143 samples were positive for both the S and ORF8 gene targets. The two samples, S46 and S60 which were single gene positives on the reference assay were negative (S/ORF8 = und; mRNaseP = 29.87) and positive (S = 36.09/ORF8 = 37.36/mRNaseP = 33.85) respectively, on the SORP triplex assay. No S/ORF8 positives were detected from the 227 (RdRp = und/E = und) negatives when the same 227 negative NPFS samples, were parallel tested on the SORP Triplex assay. The overall sensitivity and specificity of the SORP triplex assay were: 99.31% (95% CI: 96.22–99.98%) and 100.0% (95% CI: 98.4–100%) respectively (Table 3). In addition, the SORP triplex assay detected both the S and ORF8 targets when tested on a panel of VOC's consisting of SARS-CoV-2 of the B.1.1.7 (UK) and B.1.351 (SA) lineages (Table.3_Suppl).
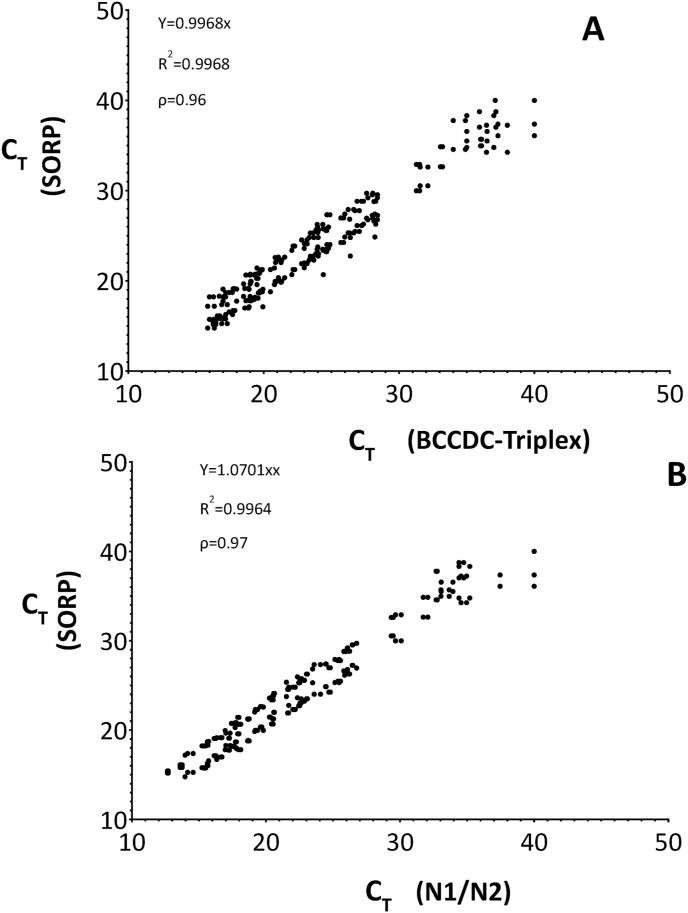
Overall, a high degree of concordance was observed between the SORP triplex assay and the BCCDC triplex assay as seen by the strong correlation in Ct values (R2:0.99; Spearman's ρ = 0.96; P < 0.0001) (Fig. 2 a). Similarly, a strong correlation was also observed between the SORP triplex assay values and the US-CDC's Nucleocapsid (N1/N2) singleplex assay (R2:0.99; Spearman's ρ = 0.97; P < 0.0001). (Fig. 2b).
Fig. 2.
Correlation between CT values obtained between (A.) SORP triplex assay and BCCDC triplex assay (R2:0.99; Spearman's ρ = 0.96; P < 0.0001) (B.) SORP triplex assay and N1/N2 US-CDC Nucleocapsid singleplex assay. (R2:0.99; Spearman's ρ = 0.97; P < 0.0001).
3.4. Analytical sensitivity & specificity
Using a quantified SARS-CoV-2 template, the lowest concentration (95% probability) of the ORF8 and S gene targets detected were: 222.33 c/mL (CI:88.4–1625.9) and 206.69 c/mL (CI:97.14–1294.9) respectively as per Probit regression analysis. On a copies per reaction basis, this was: 1.98 (CI:0.78–14.71) and 1.82 (CI:0.85–11.475) for the ORF8 and S gene targets respectively.
In silico BLAST analysis of the S and ORF8 amplicon sequence did not show any homology to other coronaviruses including human coronaviruses (229E/HKU1/NL63/NO/OC43), SARS-CoV (ExoN1/Tor2/HSE 1/HKU-39849/Urbani), bat coronaviruses and MERS-CoV (Table. 1_Suppl). Also, no homology was observed for both the S and ORF8 amplicon sequences, when tested in silico on common respiratory viruses like human adenovirus, RSV A/B, influenza virus A/B, human metapneumovirus, human parainfluenza virus and human rhinovirus (Table. 1_Suppl). Empirical testing of the candidate Spike and ORF8 TaqMan™ sets on a panel of clinically positive non-target specimens, which included viral targets like Adenovirus, cytomegalovirus, Epstein Barr virus, human bocavirus and common bacterial targets like Escherichia coli, Bordetella pertussis and Streptococcus pneumoniae, also showed no cross-reactivity, indicating high degree of target specificity (Table. 2_Suppl).
4. Discussion
In this study, we developed a highly sensitive and specific TaqMan™ multiplex assay, to detect SARS-CoV-2, targeting the Spike and ORF8 genes of this virus. Based on the availability of >51,000 high-quality SARS-CoV-2 whole genome sequences, two highly conserved region(s), specific for SARS-CoV-2, were identified for both these genes. Incorporation of nucleotide degeneracy in the Spike forward primer and ORF8 probe, accounted for single nucleotide polymorphisms in the SARS-CoV-2 genomes currently deposited in the NCBI and GISAID databases. In silico analysis of the candidate primer and probes designed for these two genetic targets showed no cross-reactivity with other human coronaviruses, including SARS-CoV-1 and MERS-CoV, and other common respiratory viruses. Empirical testing of the candidate Spike and ORF8 TaqMan™ sets on a panel of clinically positive, non-target specimens, showed no cross-reactivity, confirming a high degree of target specificity (Table. 2_Suppl). The newly developed SORP triplex assay, detected with 99.73% accuracy the 144 positives nasopharyngeal SARS-CoV-2 swab samples, previously tested on the reference BCCDC triplex assay. No single gene target positives were detected amongst the 144 positive samples, nor false positives from the cohort of 227 negative NPFS samples. Overall, a high degree of concordance was observed between the SORP and the BCCDC triplex assays (Spearman's ρ = 0.96) and the widely used FDA licenced, US-CDC's Nucleocapsid assay (Spearman's ρ = 0.97) (Fig. 2). Interestingly, the SORP triplex assay was also able to detect with 100% sensitivity a panel of SARS-CoV-2 VOC's belonging to the B.1.1.7 and B1.135 lineage (Table.3_Suppl).
Getting a single-gene positive can be disruptive to a testing workflow and have to be resolved separately using for example, Xpert Xpress SARS-CoV-2 assay used in our laboratory test setting. Having an assay which gives high rate of dual (S/ORF8) gene positives, results in minimal workflow disruption due to repeat testing and enhances the overall confidence of using it for clinical testing. While no single gene S/ORF8 gene targets were detected in our study, reports of ORF8 deletion in certain SARS-CoV-2 strains [[30], [31], [32]], could potentially result in lack of ORF8 signal when using the SORP triplex assay. Under these circumstances, the end-user would have to resolve it on a separate confirmatory assay which does not target either the S or ORF8 genes for e.g., Xpert Xpress SARS-CoV-2 (Cephid, USA).
In the present work, we attempted to develop a more “contextual” relevance for the human RNaseP internal control widely used in SARS-CoV-2 assays. Alignment of the RNaseP forward (RNaseP-F) and reverse (RNaseP-R) primer sequences, showed that both these primers had binding site on exon 1 (Fig. 1_Suppl_A). This in theory would result in the co-amplification of both mRNA and gDNA, which was also empirically verified in our study (Table 2). The amplification of RNaseP gDNA however, cannot account for the highly sensitive reverse transcription process - a key enzymatic step, required for the amplification of the RNA viral target i.e., SARS-CoV-2 acknowledged to be extremely vulnerable to chemical inhibition [33]. In other words, the lack of US-CDC's defined RNaseP primer/probe set, does not capture the fidelity of the reverse transcription process, which in our opinion, represents a limitation of this highly versatile and widely used control for SARS-CoV-2 testing.
Using the classical exon-spanning strategy, the RNaseP reverse primer (RNaseP-R) was re-designed such that it exclusively amplified RNaseP mRNA (mRNaseP) (Fig. 1_Suppl_B). As a result, amplification of the mRNA-specific RNaseP internal control could now be interpreted to also account for reverse transcription in addition, to improper specimen collection and/or sample nucleic acid extraction failure. In our study, of the >350 samples tested, we did not record any instances of mRNaseP control failure, indicating both proper sample collection and high quality (inhibitor-free) of total nucleic acid extracted on the QIAsymphony extraction platform.
5. Conclusion
In summary, a new SARS-CoV-2 specific LDT assay was designed to improve the resiliency of our COVID-19 testing program. Ready availability of a new SARS-CoV-2 specific LDT, could help diagnostic laboratories, to overcome challenges posed by supply chain limitations [34] and detection of new variants of SARS-CoV-2 [25,26]. Moreover, ready availability of a repertoire of SARS-CoV-2 LDT's, could facilitate the development of a more “panel-centric” testing approach, whereby multiple LDT's are used, to test the presence of SARS-CoV-2 in a clinical sample. This panel-based approach is now being increasingly realized to be more prudent testing approach than the current single LDT approach [[20], [36]].
Credit roles of authors
Vijay Gadkar: Conceptualization, Methodology, Validation, Formal analysis, Resources, Project administration, Writing – original draft, Writing – review & editing. David Goldfarb: Formal analysis, Writing – review & editing, Virginia Young: Methodology, Validation. Nicole Watson: Methodology, Validation. Ghada N. Al-Rawahi: Formal analysis, Writing – review & editing. Jocelyn Srigley: Formal analysis, Writing – review & editing. Peter Tilley: Supervision, Formal analysis, Funding acquisition, Project administration, Writing – review & editing.
Declaration of competing interest
The authors report no conflict of interest relevant to this article.
Acknowledgement
This research did not receive any specific grant from funding agencies in the public, commercial or, or not-for-profit sectors. The authors would like to thank Drs. A. Jassem, C. Hogan & L. Huong (BCCDC) for the SARS-CoV-2 VOC panel used in this study.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.mcp.2021.101744.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Wu F., Zhao S., Yu B., et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;580(7803):E7. doi: 10.1038/s41586-020-2202-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu N., Zhang D., Wang W., et al. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2019;382(8):727–733. doi: 10.1056/NEJMoa2001017. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Corman V.M., Landt O., Kaiser M., et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25:2000045. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chan J.F., Yip C.C., To K.K., et al. Improved molecular diagnosis of COVID-19 by the novel, highly sensitive and specific COVID-19-RdRp/Hel real-time reverse transcription-PCR assay validated in vitro and with clinical specimens. J. Clin. Microbiol. 2020;23:58. doi: 10.1128/JCM.00310-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chan W., Ip J.D., Chu A.W., et al. Identification of nsp1 gene as the target of SARS-CoV-2 real-time RT-PCR using nanopore whole-genome sequencing. J. Med. Virol. 2020;92(11):2725–2734. doi: 10.1002/jmv.26140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chan J.F., Yip C.C., To K.K., et al. Improved molecular diagnosis of 1 COVID-19 by the novel, highly sensitive and specific 2 COVID-19-RdRp/Hel real-time reverse transcription-polymerase chain reaction assay validated 3 in vitro and with clinical specimens. J. Clin. Microbiol. 2020;58(5):e00310–e00320. doi: 10.1128/JCM.00310-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Centers for Disease Control and Prevention (Cdc) 2019-novel coronavirus (2019-nCoV) real-time rRT-PCR panel primers and probes. 2020. https://www.cdc.gov/coronavirus/2019-ncov/downloads/rt-pcr-panel-primer-probes.pdf 2020a; Accessed: 09 Apr 2020. Available from:
- 8.Who World Health organization. Coronavirus disease (COVID‐19) technical guidance: laboratory testing for 2019‐nCoV in humans. 2020. https://www.who.int/docs/default-source/coronaviruse/whoinhouseassays.pdf?sfvrsn=de3a76aa_2
- 9.Yip C.C., Ho C.C., Chan J.F., et al. Development of a novel, genome subtraction‐derived, SARS‐CoV‐2‐specific COVID‐19‐nsp2 real‐time RT‐PCR assay and its evaluation using clinical specimens. Int. J. Mol. Sci. 2020;21:2574. doi: 10.3390/ijms21072574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhen W., Berry G.J. Development of a new multiplex real-time RT-PCR assay for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) detection. J Mol Diagnostics. 2020;22(12):1367–1372. doi: 10.1016/j.jmoldx.2020.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vermeiren C., Marchand-Senécal X., Sheldrake E., et al. Comparison of copan E swab and FLOQ swab for COVID-19 diagnosis: working around a supply shortage. J. Clin. Microbiol. 2020;58(6) doi: 10.1128/JCM.00669-20. e00669-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheng V.C., Lau S.K., Woo P.C., et al. Severe acute respiratory syndrome coronavirus as an agent of emerging and re-emerging infection. Clin. Microbiol. Rev. 2007;20(4):660–694. doi: 10.1128/CMR.00023-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chan J.F., Lau S.K., To K.K., et al. Middle East respiratory syndrome coronavirus: another zoonotic Betacoronavirus causing SARS‐like disease. Clin. Microbiol. Rev. 2015;28(2):465–522. doi: 10.1128/CMR.00102-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.LeBlanc J.J., Gubbay J.B., Li Y., et al. Real-time PCR-based SARS-CoV-2 detection in Canadian laboratories. J. Clin. Virol. 2020;128:104433. doi: 10.1016/j.jcv.2020.104433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goldfarb D.M., Tilley P., Al-Rawahi G.N., Srigley J.A., et al. Self-collected saline gargle samples as an alternative to healthcare worker collected nasopharyngeal swabs for COVID-19 diagnosis in outpatients. J. Clin. Microbiol. 2021;59(4):e02427–20. doi: 10.1128/JCM.02427-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Phan T. Genetic diversity and evolution of SARS-CoV-2. Infect. Genet. Evol. 2020;21:104260. doi: 10.1016/j.meegid.2020.104260. 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shen Z., Xiao Y., Kang L., et al. Genomic diversity of SARS-CoV-2 in coronavirus disease 2019 patients. Clin. Infect. Dis. 2020;4:203. doi: 10.1093/cid/ciaa203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matyášek R., Kovařík A. Mutation patterns of human SARS-CoV-2 and bat RaTG13 coronavirus genomes are strongly biased towards C>U transitions, indicating rapid evolution in their hosts. Genes. 2020;11(7):E761. doi: 10.3390/genes11070761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ziegler K., Steininger P., Ziegler R., et al. SARS-CoV-2 samples may escape detection because of a single point mutation in the N gene. Euro Surveill. 2020;39 doi: 10.2807/1560-7917.ES.2020.25.39.2001650. pii:2001650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bal A., Destras G., Gaymard A., et al. 2020. Screening of the H69 and V70 Deletions in the SARS CoV-2 Spike Protein SARS-CoV-2 Samples May Escape Detection Because of a Single Point Mutation in the N Gene. Medrxiv.https://www.medrxiv.org/content/10.1101/2020.11.10.20228528v1.full [Google Scholar]
- 21.Artesi M., Bontems S., Göbbels P., Franckh M., Maes P., et al. A recurrent mutation at position 26340 of SARS-CoV-2 is associated with failure of the E gene quantitative reverse transcription-PCR utilized in a commercial dual-target diagnostic assay. Journal of Clinical Microbiology Sep. 2020;58(10):e01598–20. doi: 10.1128/JCM.01598-20. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Falasca F., Sciandra I., Di Carlo D., et al. Detection of SARS-COV N2 Gene: very low amounts of viral RNA or false positive? J. Clin. Virol. 2020;133:104660. doi: 10.1016/j.jcv.2020.104660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hasan M.R., Sundararaju, Manickam C., Mirza F., Al-Hail H., et al. A novel point mutation in the N gene of SARS-CoV-2 may affect the detection of the virus by RT-qPCR. J. Clin. Microbiol. 2021;59(4):e03278–-20. doi: 10.1128/JCM.03278-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Woloshin S., Patel N., Kesselheim A.S. False negative tests for SARS-CoV-2 infection- challenges and implications. N. Eng. J. Med. 2020;6(6):e38. doi: 10.1056/NEJMp2015897. 383. [DOI] [PubMed] [Google Scholar]
- 25.Rambaut A., Loman N., Pybus O., et al. Preliminary genomic characterisation of an emergent SARS-CoV-2 lineage in the UK defined by a novel set of spike mutations. Virological.org. 2020 https://virological.org/t/preliminary-genomic-characterisation-of-an-emergent-sars-cov-2-lineage-in-the-uk-defined-by-a-novel-set-of-spike-mutations/563 20 Dec 2020. [Google Scholar]
- 26.Tegally H., Wilkinson E., Giovanetti M., Iranzadeh A., Fonseca V., et al. Medrxiv; 2020. Emergence and Rapid Spread of a New Severe Acute Respiratory Syndrome-Related Coronavirus 2 (SARS-CoV-2) Lineage with Multiple Spike Mutations in South Africa.https://www.medrxiv.org/content/10.1101/2020.12.21.20248640v1 [Google Scholar]
- 27.Li X., Wang W., Zhao X., et al. Transmission dynamics and evolutionary history of 2019‐nCoV. J. Med. Virol. 2020;92(5):501‐511. doi: 10.1002/jmv.25701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Duchene S., Featherstone L., Haritopoulou-Sinanidou M., Rambaut A., Lemey P., Baele G. Temporal signal and the phylodynamic threshold of SARS-CoV-2. Virus Evolution. 2020;6(2):veaa061. doi: 10.1093/ve/veaa061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Centers for Disease Control and Prevention (Cdc) CDC 2019-novel coronavirus (2019-nCoV) real-time RT-PCR diagnostic panel. 2020. https://www.fda.gov/media/134922/download 2020b; Accessed: 09 Apr 2020. Available from:
- 30.Gong Y.N., Tsao K.C., Hsiao M.J., et al. SARS-CoV-2 genomic surveillance in Taiwan revealed novel ORF8-deletion mutant and clade possibly associated with infections in Middle East. Emerg. Microb. Infect. 2020;9(1):1457–1466. doi: 10.1080/22221751.2020.1782271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Su Y.C.F., Anderson D.E., Young B.E., et al. Discovery and genomic characterization of a 382-nucleotide deletion in ORF7b and ORF8 during the early evolution of SARS-CoV-2. mBio. 2020;11(4):e01610–e01620. doi: 10.1128/mBio.01610-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zinzula L. Lost in deletion: the enigmatic ORF8 protein of SARS-CoV-2. Biochem. Biophys. Res. Commun. 2021;538:116–124. doi: 10.1016/j.bbrc.2020.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schrader C., Schielke A., Ellerbroek L., et al. PCR inhibitors – occurrence, properties and removal. J. Appl. Microbiol. 2012;113(5):1014–1026. doi: 10.1111/j.1365-2672.2012.05384.x. [DOI] [PubMed] [Google Scholar]
- 34.Lippi G., Simundic A.M., Plebani M., et al. Potential preanalytical and analytical vulnerabilities in the laboratory diagnosis of 243 coronavirus disease 2019 (COVID-19) Clin. Chem. Lab. Med. 2020;58(7):1070–1076. doi: 10.1515/cclm-2020-0285. [DOI] [PubMed] [Google Scholar]
- 36.Peñarrubia L, Ruiz M, Porco R, Rao SN, Juanola-Falgarona M, Manissero D, López-Fontanals M, Pareja J. Multiple assays in a real-time RT-PCR SARS-CoV-2 panel can mitigate the risk of loss of sensitivity by new genomic variants during the COVID-19 outbreak. Int. J. Infect. Dis. 2020;97:225–229. doi: 10.1016/j.ijid.2020.06.027. https://pubmed.ncbi.nlm.nih.gov/32535302/ [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.