Abstract
Krüppel‐like factor 5 (KLF5) is a member of the KLF family. Recent studies have suggested that KLF5 regulates the expression of a large number of new target genes and participates in diverse cellular functions, such as stemness, proliferation, apoptosis, autophagy, and migration. In response to multiple signaling pathways, various transcriptional modulation and posttranslational modifications affect the expression level and activity of KLF5. Several transgenic mouse models have revealed the physiological and pathological functions of KLF5 in different cancers. Studies of KLF5 will provide prognostic biomarkers, therapeutic targets, and potential drugs for cancers.
Keywords: posttranslational modification, signaling pathway, targeted therapy, transcription
KLF5 is a key transcription factor in cancers. This article reviews the functions, mechanisms, and regulations of KLF5.
![]()
Abbreviations
- 5FU
5‐Fluorouracil
- ABCG2
ATP‐binding cassette subfamily G member 2
- AGGF1
Angiogenic factor with G‐patch and FHA domains 1
- AKT
AKT serine/threonine kinase
- ALDH
Acetaldehyde dehydrogenase
- ALDH3A1
Aldehyde dehydrogenase 3 family member A1
- AR
Androgen receptor
- ASK1
Apoptotic signal‐regulating kinase 1
- ATXN3L
Ataxin‐3 like
- BAP1
BRCA1 associated protein 1
- BAX
BCL2 associated X
- BECN1
Beclin1
- BET
Bromodomain and extra‐terminal
- BRD4
Bromodomain‐containing 4
- BRG1
Brahma‐related gene 1
- CASC15
Cancer susceptibility candidate 15
- CBP
CREB‐binding protein
- CCL5
C‐C motif chemokine ligand 5
- CCR5
C‐C motif chemokine receptor 5
- ccRCC
Clear cell renal cell carcinoma
- Cdc42
Cell division cycle 42
- CDK7
Cyclin dependent kinase 7
- CDT1
Chromatin licensing and DNA replication Factor 1
- Chk1/2
Checkpoint kinase 1/2
- CINP
Cdk2‐interacting protein
- COX2
Cyclooxygenase 2
- CPD
Cdc4 phosphodegron
- CRC
Colorectal cancer
- CSCs
Cancer stem cells
- CUL4B
Cullin 4B
- CXCR4
Cell‐surface chemokine receptor 4
- DDR
DNA damage repair
- DEX
Dexamethasone
- DII1
Delta like canonical Notch ligand 1
- DNMT1
DNA methyltransferase 1
- DSS
Dextran sodium sulfate
- DUBs
Deubiquitinases
- E2F1/3
E2F transcription factor 1/3
- EFP
Estrogen‐responsive finger protein
- EGFR
Epidermal growth factor receptor
- EGR1
Early growth response 1
- EMT
Epithelial‐mesenchymal transition
- ERCC5
ERCC excision repair 5
- ERα/β
Estrogen receptor α/β
- ESCC
Esophageal squamous cell carcinoma
- ESC
Embryonic stem cell
- FAK
Focal adhesion kinase
- FASN
Fatty acid synthase
- FGF‐BP1
Fibroblast growth factor binding protein 1
- FoxA1
Forkhead box protein A1
- FOXE1
Forkhead box E1
- FOXO1
Forkhead box O1
- FYN
FYN proto‐oncogene kinase
- GADD45A
Growth arrest and DNA damage‐inducible 45 alpha
- GATA4/6
GATA binding protein 4/6
- GCN5
general control non‐derepressible 5
- GDF15
Growth differentiation factor 15
- GlcNAc
O‐linked N‐acetylglucosamine
- GLI1/2
GLI family zinc finger 1 /2
- GLUT‐1
Glucose transporter 1
- GR
Glucocorticoid receptor
- GSK3β
Glycogen synthase kinase 3β
- GSTM1
Glutathione‐S‐transferase M1
- H2AX
H2A.X variant histone
- H3K27ac
Histone H3 lysine 27 acetylation
- HCC
Hepatocellular carcinoma cells
- HCF1
Host cell factor C1
- HDAC1/2/3
Histone deacetylase 1/2/3
- Hes1
Hes family bHLH transcription factor 1
- HIF‐1α
Hypoxia‐inducible factor 1α
- HNF4α
Hepatocyte nuclear factor 4α
- IGF1
Insulin like growth factor 1
- IL1β
Interleukin 1β
- ITGB2
Integrin subunit beta 2
- KLF2
Krüppel‐like factor 2
- KLF4
Krüppel‐like factor 4
- KLF5
Krüppel‐like factor 5
- KLF8
Krüppel‐like factor 8
- KLFs
Krüppel‐like factors
- LIF
Leukemia inhibitory factor
- lncRNAs
Long non‐coding RNAs
- LPA
Lysophosphatidic acid
- MED1
Mediator complex subunit 1
- mESCs
Mouse embryonic stem cells
- miRNAs
MicroRNAs
- MKK4/7
Mitogen‐activated protein kinase 4/7
- MKP‐1
Mitogen‐activated protein kinase phosphatase 1
- mPGES1
microsomal prostaglandin E‐2 synthase 1
- MYC
MYC proto‐oncogene
- NAMPT
Nicotinamide phosphoribosyl transferase
- ncRNAs
Non‐coding RNAs
- NDRG2
N‐myc downregulated gene 2
- NEAT1
Nuclear paraspeckle assembly transcript 1
- NES
Nuclear export signal
- NOTCH1
Notch receptor 1
- NSCLC
Non–small‐cell lung cancer
- OGT1
O‐linked N‐acetylglucosamine transferase
- P300
Adenovirus E1A‐associated 300 kDa protein
- PARP1
Poly ADP‐ribose polymerase 1
- PDGF
Platelet‐derived growth factor
- PDGFA
Platelet‐derived growth factor A
- PKD1
Protein kinase D1
- PMA
Phorbol 12‐myristate 13‐acetate
- PR
Progestogen receptor
- PSA
Prostate‐specific antigen
- PTCH1
Patched 1
- PTEN
Phosphate and tension homology deleted on chromosome ten
- Rac1
Rac family small GTPase 1
- RAD51
RAD51 recombinase
- RARα
Retinoic acid receptor α
- RAS
Rat sarcoma
- RTK
Receptor tyrosine kinase
- SCFFBW7
SCFF‐box and WD repeat domain‐containing 7
- SE
Super enhancer
- SHH
Sonic hedgehog
- SMAD 2/3/4
SMAD family member 2/3/4
- SMURF2
SMAD ubiquitination regulatory factor 2
- SNHG12
Small nucleolar RNA host gene 1
- SOX2/4
SRY‐Box transcription factor 2/4
- SREBP‐1
Sterol‐regulatory‐element‐binding protein‐1
- STAT3
Signal transducer and activator of transcription 3
- TAD
Transactivation domain
- TAZ
Transcriptional co‐activator with PDZ‐binding motif
- TCF3
Transcription factor 3
- TEAD4
TEA domain transcription factor 4
- TGF‐β
Transforming growth factor‐β
- TNBC
Triple‐negative breast cancer
- TNFAIP2
Tumor necrosis factor‐α (TNFα)‐induced protein 2
- TNFRSF11a
Tumor necrosis factor receptor superfamily member 11a
- TNFα/TNFR1
Tumor necrosis factor α/ tumor necrosis factor receptor 1
- TP63
Tumor protein P63
- USP3
Ubiquitin‐specific protease 3
- VEGF
Vascular endothelial growth factor
- WWP1
WW domain‐containing E3 ubiquitin protein ligase 1
- YAP
Yes1 associated transcriptional regulator
- ZEB1/2
Zinc finger E‐box binding homeobox 1/2
- ZF
Zinc finger
1. INTRODUCTION
Krüppel‐like factors (KLFs) belong to a significant transcription factor family that is involved in multiple biological processes and diverse diseases, especially cancers. 1 Seventeen KLFs contain 3 highly conserved and tandem ZF domains at their C‐terminus, which bind to DNA CACC or GC boxes and regulate the transcription of downstream target genes. 2 , 3 To date, several KLFs, such as Krüppel‐like factor 2 (KLF2), Krüppel‐like factor 4 (KLF4), Krüppel‐like factor 8 (KLF8) and KLF5, have been indicated to participate in cancer development and have drawn increasing attention. 1
KLF5 plays vital roles in disease development, especially in cancers and cardiovascular diseases. KLF5 regulates the expression of a wide range of target genes, such as Cyclin D1, 4 p27, 5 Nanog, 6 and Slug. 7 Over the past decade, KLF5 has been reported to participate in various biological functions, such as cell stemness, proliferation, apoptosis, autophagy, and migration. Several outstanding articles have reviewed the roles of KLF5 in cancers in recent years. 8 , 9 , 10 , 11
It is well known that animal models are the best approach to study the roles of KLF5 in physiology and pathology. The study of KLF5 animal models, especially tissue specific, may provide directions for future disease treatments. KLF5 is mainly involved in embryonic development, 12 adipose tissue development, 13 prostate and mammary gland development, 7 , 14 , 15 intestinal crypt morphological maintenance, 16 , 17 , 18 lung morphological development, 19 and energy metabolism 13 etc. The reported mouse models are KLF5‐KO, KLF5‐KI, and KLF5 lineage tracking models, we have summarized these in Table 1.
TABLE 1.
KLF5‐related animal models
| Animal models | Organization types | Phenotypes | References |
|---|---|---|---|
| Klf5 knockout mice | Embryo | Mouse with homozygous knockout of Klf5 died before embryonic day 8.5. | 12 |
| Blood vessel | The medial and adventitial layers of the aortic wall of Klf5 +/− mice are abnormally thinned and dilated; in response to vascular injury, the activation, proliferation, inflammation, and angiogenesis of fibroblasts and smooth muscle cells are impaired. | 12 | |
| Heart | Klf5 +/− mice have reduced heart weight, reduced fibrosis, and thinner heart ventricular walls. | 12 | |
| Gastrointestinal tract | Klf5 +/− mice have malformed gastrointestinal villi, and decrease the number of extracellular matrix and mesenchymal cells. | 12 | |
| Mammary gland | Klf5 mammary gland‐specific knockout mice can be observed to inhibit ductal elongation at 9 wk of age; the lobular alveolar structure is significantly reduced during pregnancy and lactation; the production of whey acidic protein (WAP) in mice during pregnancy and lactation is decrease, and there is a defect in milk secretion. Klf5 knockout decreased the proliferation, survival and stemness of mammary epithelial cells. | 7 | |
| Adipose tissue | The development of white adipose tissue in Klf5 +/− mice is delayed, and lipid droplets in adipocytes are reduced. Klf5 +/− mice can avoid obesity, hypercholesterolemia and impaired glucose tolerance caused by high fat. | 13 | |
| Skeleton | Klf5 deficiency impairs cartilage degradation and calcification in the perinatal period. | 194 | |
| Lung | The fetal lung airway epithelial cells in transgenic mice with specific knockout of Klf5 have inhibited lung maturation during the cystic development stage. Phenotypic abnormalities appear in different components of bronchiolar smooth muscle, pulmonary blood vessels, and respiratory epithelium. Mice with knocked out both alleles of Klf5 died of respiratory distress immediately after birth. Klf5 is essential for lung function and morphogenesis. | 19 | |
| Intestine | Intestinal‐specific deletion of Klf5 using Villin‐Cre showed that Klf5 is required to maintain gut epithelial cell proliferation, differentiation, and positioning along the crypt radial axis; Klf5 deletion in the intestinal epithelium using Shh‐Cre inhibited villus morphogenesis and epithelial differentiation; depletion of Klf5 disrupts the integrity of intestinal stem cells. | 16, 17, 18 | |
| Hematopoietic system | Knockout mice of Klf5 have enlarged spleens and increased peripheral white blood cells; the proportion of eosinophils is significantly increased, while the proportion of neutrophils is downregulated, long‐term hematopoietic progenitor cells are reduced, and the ability to reproduce is reduced. | 195 | |
| Eye | Klf5 was specifically deleted in the ectoderm‐derived structure of the ocular surface of mice, resulting in eyelid defects with malformed meibomian glands, corneal abnormalities, and loss of conjunctival goblet cells; Klf5 contributed to corneal epithelial homeostasis via regulating the expression of desmosomal components. | 196, 197 | |
| Prostate | Prostate‐specific Klf5 heterozygous deletion mice induced hyperplasia with thicker cell layers in the lateral prostate, anterior prostate, and dorsal prostate; Klf5 homozygous deletion caused prostate epithelial cell apoptosis instead of hyperplasia. | 14 | |
| Klf5 transgenic (Tg) mice | Prostate | Knockin of the Klf5 K358R gene in mouse model, the prostate has changed, showing a lighter, smaller and denser tissue morphology; prostate cells were reduced, the acinar area was smaller, and increased differentiation of basal cells into luminal cells. | 15 |
| Skin | Klf5 transgenic mice showed craniofacial defects, extracerebral malformations, persistent abdominal herniation, and ectodermal dysplasia; overexpression of Klf5 in adult mice resulted in hair follicle occlusion, hyperkeratosis and epidermal erosion. | 198 | |
| Esophagus | Esophageal epithelial cells specifically overexpress Klf5 in ED‐L2/Klf5 mouse suprabasal cells without proliferation, but have increased proliferation of basal cells. | 199 |
In the past decade, there has been significant progress in KLF5 studies in cancers. In this review, we summarize the molecular structure, biological functions, transcriptional modulations, posttranslational modifications, signaling pathways, physiological and pathological functions of KLF5, and potential targeting strategies.
2. MOLECULAR STRUCTURE
The KLF5 gene is located at 13q21, and the KLF5 protein consists of 457 amino acid residues. 8 The protein structure of human KLF5 is shown in Figure 1. KLF5 has 3 highly conserved classical C2H2 ZF motifs at the C‐terminus: a proline‐rich transactivation domain (TAD) and a NES. 20 , 21 , 22 The KLF5 TAD contains a PPSY motif and a Cdc4 phosphodegron (CPD) motif (303SPPSS), which recruit the E3 ubiquitin ligases WW domain‐containing E3 ubiquitin protein ligase 1 (WWP1) and SCFFBW7. 22 , 23 Recently, 2 hotspot mutations of KLF5 were identified, 1 in the CPD motif and the other in the DNA binding domain. 24 Mutations at the CPD motif of KLF5 in colorectal cancer (CRC) escape SCFFBW7‐mediated ubiquitination and degradation. 24 Mutations at D418 and E419 within the second zinc finger change the DNA binding properties of KLF5. 24 Interestingly, these mutations are cancer specific. 24
FIGURE 1.
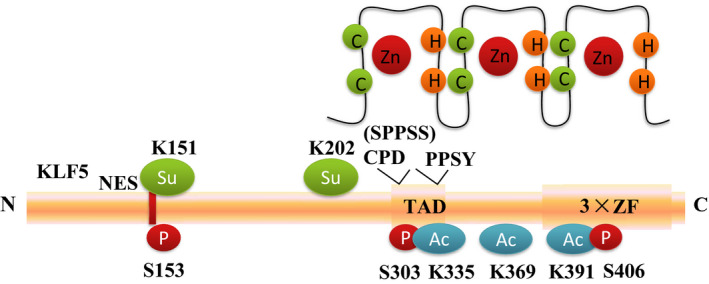
The human KLF5 protein structure. The KLF5 gene is located at 13q21, and the KLF5 protein consists of 457 amino acid residues. KLF5 has 3 highly conserved classical C2H2 ZF motifs at the C‐terminus: a proline‐rich transactivation domain (TAD) and a NES. The KLF5 TAD contains a PPSY motif and a CPD motif (SPPSS), which recruit the E3s WWP1 and SCFFBW7. KLF5 is regulated by multiple post‐transcriptional modifications, including ubiquitination (Ub), phosphorylation (P at S153, S303, S406), acetylation (Ac at K335, K369, K391), and SUMOylation (Su at K151 and K202)
3. TRANSCRIPTIONAL TARGET GENES
As a basic transcription factor, KLF5 predominately promotes the transcription of target genes involved in various cellular functions, such as Cyclin D1, platelet‐derived growth factor A (PDGFA), and fibroblast growth factor binding protein 1 (FGF‐BP1) 4 , 12 , 25 (Table 2). Accumulating evidence from our laboratory has suggested that KLF5 promotes breast cancer by regulating several key target genes, including FGF‐BP1, 25 microsomal prostaglandin E‐2 synthase 1 (mPGES1), 26 tumor necrosis factor‐α (TNFα)‐induced protein 2 (TNFAIP2), 27 Slug, 7 and Cyclin D1. 4 In addition to mPGES1, KLF5 was reported to induce cyclooxygenase 2 (COX2) expression and promote cell proliferation and migration in phosphate and tension homology deleted on chromosome ten (PTEN)‐null mouse embryonic fibroblasts. 28 Consistently, KLF5 also promotes the transcription of Cyclin E1 by binding to its polymorphic enhancer in bladder cancer. 29 KLF5 was reported to promote bladder cancer cell migration by promoting the transcription of the FYN proto‐oncogene kinase (FYN) gene, which thereby activates FAK. 30 Additionally, KLF5 was shown to promote glioblastoma angiogenesis by inducing angiogenic factor with G‐patch and FHA domains 1 (AGGF1) gene transcription in glioma‐associated endothelial cells. 31 In prostate cancer, KLF5 binds to the AR gene promoter to increase its transcription; furthermore, KLF5 interacts with AR to increase MYC proto‐oncogene (MYC), Cyclin D1, and PSA transcription. 32 KLF5 is essential for hematopoietic stem and progenitor cell bone marrow adhesion because KLF5 activates Rab family protein Rab5 transcription. 33 KLF5 interacts with mutant p53 to promote PLA2G16 gene transcription and pancreatic cancer cell glycolysis. 34 KLF5 increases oxidative stress by decreasing glutathione by increasing the transcription of glutathione‐S‐transferase M1 (GSTM1) in B‐cell acute lymphoblastic leukemia cells. 35 Interestingly, the KLF5 E419Q mutant gains new binding sites and activates the transcription of several target genes, including fork head box E1 (FOXE1) and nicotinamide phosphoribosyl transferase (NAMPT). 24 KLF5 can upregulate the transcription of mitogen‐activated protein kinase 7 (MKK7), the upstream kinase of c‐JNK, by binding to the proximal promoter region of the MKK7 gene and promote the apoptosis induced by tumor necrosis factor α/tumor necrosis factor receptor 1 (TNFα/TNFR1). 36 In ESCC, KLF5, tumor protein P63 (TP63), and SRY‐Box transcription factor 2 (SOX2) form a transcription complex that promotes aldehyde dehydrogenase 3 family member A1 (ALDH3A1) gene transcription by binding to its SE. 37 KLF5 inhibits Beclin1 (BECN1) gene transcription and autophagy in prostate cancer cells. 38 KLF5 activates tumor necrosis factor receptor superfamily member 11a (TNFRSF11a) gene transcription by binding to the TNFRSF11a gene promoter in cervical cancer. 39 In non–small‐cell lung cancer (NSCLC), KLF5 promotes the expression of growth differentiation factor 15 (GDF15) and cell proliferation induced by C5a. 40 Moreover, KLF5 activates the transcription of E2F transcription factor 1 (E2F1) and RAD51 recombinase (Rad51) in pancreatic cancer and promotes cell proliferation. 41
TABLE 2.
New identified KLF5 target genes in cancers
| Types | Target genes | Functions | Cancers | References |
|---|---|---|---|---|
| Protein‐encoding genes | TNFAIP2 | Promote cell migration and invasion | Breast cancer | 27 |
| mPGES1 | Promote the production of PGE2 and cell proliferation | Breast cancer | 26 | |
| Slug | Promote stemness | Breast cancer | 7 | |
| GDF15 | Promote cell proliferation | Non–small‐cell lung cancer | 40 | |
| GSTM1 | Increase oxidative stress by reducing glutathione | B‐cell acute lymphoblastic leukemia | 35 | |
| TNFRSF11a | Promote cell proliferation, migration, and invasion | Cervical cancer | 39 | |
| FYN | Promote cell migration | Bladder cancer | 30 | |
| Cyclin E1 | Promote cell cycle | Bladder cancer | 29 | |
| AGGF1 | Promote angiogenesis | Glioblastoma | 31 | |
| HNF4α | Promote cell proliferation | Gastric cancer | 56 | |
| MYC | Promote cell proliferation | Prostate cancer | 32 | |
| BECN1 | Promote autophagy | Prostate cancer | 38 | |
| MKK7 | Promote apoptosis induced by TNFα/TNFR1 | Prostate cancer | 36 | |
| ALDH3A1 | Promote cell proliferation and tumor growth | Esophageal squamous cell carcinoma | 37 | |
| PLA2G16 | Promote glycolysis | Pancreatic cancer | 34 | |
| E2F1, Rad51 | Promote cell proliferation | Pancreatic cancer | 41 | |
| ABCG2 | Reduce the chemotherapy resistance to anthracyclines | Lung cancer | 54 | |
| IGF1 | Inhibit the STAT3 signaling pathway and tumor metastasis | Prostate cancer | 53 | |
| LncRNA | LINC00094 | Promote cell proliferation | Esophageal squamous cell carcinoma | 42 |
| NEAT1 | Promote cell proliferation, and inhibit apoptosis | Gastric cancer | 46 | |
| LINC00346 | Promotes cell growth, migration and invasion | Gastric cancer | 45 | |
| CASC15 | Promotes cell proliferation, invasion, and tumor growth | Breast cancer | 44 | |
| RP1 | Promote tumorigenesis | Breast cancer | 43 | |
| SNHG12 | Promote invasion and metastasis | Colorectal cancer | 47 | |
| miRNA | miR‐27a | Promote cell migration and invasion in response to cholesterol | Clear cell renal cell carcinoma | 48 |
| miR‐200 | Inhibit EMT | Epithelial cells | 50 | |
| miR‐145, miR‐124, miR‐183 | Inhibit cell migration and invasion | Pituitary adenoma | 51 | |
| miR‐192 | Inhibit EMT in the context of p53 loss or mutation | Liver cancer | 49 |
In addition to protein‐encoding genes, KLF5 also regulates the transcription of several long non‐coding RNAs (lncRNAs). LncRNAs are RNA with a length greater than 200 nt. KLF5 and transcription factor 3 (TCF3) activate the transcription of LINC00094 by binding to its SE, thereby promoting the growth of ESCC cells. 42 KLF5 recruits adenovirus E1A‐associated 300 kDa protein (p300) to induce lncRNA RP1 transcription, and RP1 can, in turn, inhibit p27kip1 protein translation to drive breast cancer tumorigenesis. 43 Additionally, KLF5 induces cancer susceptibility candidate 15 (CASC15) to promote breast cancer cell proliferation and migration. 44 KLF5 and MYC induce LINC00346 to promote gastric cancer. 45 The LINC00346 is amplified and overexpressed in gastric cancer. High expression of LINC00346 is correlated with poor pathological stage, large tumor volume, and poor prognosis. 45 LINC00346 functions as a molecular sponge for miR‐34a‐5p and antagonizes its ability to inhibit gastric cancer. 45 Additionally, KLF5 induces nuclear paraspeckle assembly transcript 1 (NEAT1), which recruits Brahma‐related gene 1 (BRG1) to silence growth arrest and DNA damage‐inducible 45 alpha (GADD45A) gene transcription in gastric cancer. 46 KLF5 promotes CRC cell invasion and metastasis by upregulating lncRNA small nucleolar RNA host gene 1 (SNHG12). 47
MicroRNAs (miRNAs) are a group of short (generally 21 to 24 nucleotides in length), non‐coding RNA (ncRNAs) molecules that fine tune gene expression levels. miRNAs play a role in RNA interference, destroying mRNA, suppressing gene expression and controlling translation of target mRNAs, thereby regulating cancer initiation and development. In clear cell renal cell carcinoma (ccRCC), KLF5 positively regulates the transcription of miR‐27a, thereby targeting the E3 ubiquitin ligase FBW7 to promote cell migration and invasion. 48 In the context of p53 loss, KLF5 promotes miR‐192 transcription, which inhibits the expression of zinc finger E‐Box binding homeobox 2 (ZEB2) and EMT in liver cancer cells. 49 Additionally, KLF5 induces the transcription of miR‐200 and inhibits the EMT process of epithelial cells. 50 Furthermore, KLF5 promotes miR‐124, miR‐145, and miR‐183 transcription, therefore inhibiting pituitary adenoma cell migration and invasion. 51
In addition to upregulating target gene transcription, KLF5 can also inhibit the transcription of a few target genes, such as p21, 52 p27 5 , and p16. 41 Additionally, KLF5 and histone deacetylase 1 (HDAC1) cooperate to inhibit the transcription of insulin like growth factor 1 (IGF1) in prostate cancer cells. 53 Recently, KLF5 was shown to suppress the transcription of forkhead box protein A1 (FoxA1), thereby promoting the morphogenesis of intestinal villi. 17 KLF5 also inhibits the transcription of ATP‐binding cassette subfamily G member 2 (ABCG2) and sensitizes lung cancer cells to doxorubicin. 54
4. INTERACTING PROTEINS
Various KLF5‐interacting proteins were identified by immunoprecipitation and mass spectrometry. As shown in Table 3. KLF5 can form a transcription complex with transcription‐related factors or histone modifiers and regulate the transcription of target genes.
TABLE 3.
New identified KLF5 interacting proteins
| Classification | KLF5 interacting proteins |
|---|---|
| Transcription factors | EGR1, 51 GATA4/6, 56 Miz‐1, 60 MYC, 134 TEAD4, 5 SMAD4, 60 ERα/β, 57 , 200 TP63, 37 SOX2, 37 AR, 32 HIF‐1α, 61 HCF1 55 |
| Enzymes | HDAC3, 38 SMURF2, 64 FBW7, 22 BAP1, 55 USP3, 66 ATXN3L, 65 GCN5, 40 GSK3β, 22 EFP, 63 OGT1 55 |
| Others | CNIP, 67 TAZ, 68 YAP 68 |
We reported that KLF5 interacts with the transcription factors host cell factor C1 (HCF1) 55 and TEA domain transcription factor 4 (TEAD4) 5 to regulate FGF‐BP1 and p27 gene transcription in TNBC. Additionally, there is a physical interaction between KLF5 and AR to maintain the transcriptional activity of AR. 32 Moreover, KLF5 cooperates with the transcription factors GATA binding protein 4/6 (GATA4/6) to regulate the transcription of downstream oncogenes (ie, hepatocyte nuclear factor 4α, HNF4α) in gastric cancer. 56 Jiang et al found that KLF5 interacts with TP63 and SOX2 to regulate target gene transcription in ESCC. 37 KLF5 binds to ERα in breast cancer and inhibits the transcriptional activity of estrogen receptor α (ERα). 57 In prostate cancer, estrogen receptor β (ERβ) cooperates with KLF5 to promote forkhead box O1 (FOXO1) transcription and suppress tumor growth. 58 Nuclear early growth response 1 (EGR1) interacts with KLF5 to inhibit the transcription of miR‐124, miR‐145, and miR‐183 and promote the migration and invasion of pituitary adenomas. 51 In response to transforming growth factor‐β (TGF‐β), KLF5 forms a transcription complex with SMAD family member 2/3/4 (SMAD2/3/4) to activate the transcription of p15 and inhibit the transcription of c‐Myc. 59 , 60 Under hypoxia, KLF5 interacts with hypoxia‐inducible factor 1α (HIF‐1α) to promote the transcription of Cyclin B1 and Survivin in lung cancer. 61
KLF5 also binds to enzymes that mediate posttranslational modifications. It has been reported that several E3 ligases, including WWP1, 62 estrogen‐responsive finger protein (EFP), 63 FBW7, 22 and SMAD ubiquitination regulatory factor 2 (SMURF2), 64 interact with KLF5 and promote KLF5 ubiquitination and degradation. Because ubiquitination is reversible, 3 deubiquitinases (DUBs), BRCA1 associated protein‐1 (BAP1), 55 Ataxin‐3 like (ATXN3L), 65 and Ubiquitin‐specific protease 3 (USP3), 66 were identified to mediate the deubiquitination of KLF5. BAP1 has been reported to form a complex with HCF1 and O‐Linked N‐acetylglucosamine transferase (OGT1). 55 OGT1 is an O‐linked N‐acetylglucosamine (GlcNAc) transferase, however whether KLF5 is modified with GlcNAc is unknown. 55 Glycogen synthase kinase 3β (GSK3β) interacts with KLF5 to phosphorylate KLF5 at S303, which promotes FBW7‐mediated KLF5 ubiquitination and degradation. 22 KLF5 and histone acetyltransferase general control non‐derepressible 5 (GCN5) form a transcription complex to activate GDF15 gene transcription in NSCLC cells. 40 Additionally, KLF5 was shown to interact with histone deacetylase 3 (HDAC3) to inhibit the transcription of BECN1. 38
Cdk2‐interacting protein (CINP) was identified as a new KLF5 interacting protein in bladder cancer. 67 CINP knockdown attenuated the transcription of KLF5 target genes, including Cyclin D1 and Caspase 7, and therefore inhibited cell cycle progression and tumorigenesis. 67 Transcriptional co‐activator with PDZ‐binding motif (TAZ) and Yes1 associated transcriptional regulator (YAP), 2 key transcription activators of the Hippo pathway, can competitively antagonize the binding of WWP1 and KLF5 and increase KLF5 protein stability. 68
5. TRANSCRIPTIONAL AND POST‐TRANSCRIPTIONAL MODULATIONS OF KLF5
5.1. Promoter methylation inhibits KLF5 transcription
DNA hypermethylation at gene promoters is a common mechanism that causes transcriptional repression. Accumulating data indicate that KLF5 is regulated by DNA methylation. KLF5 intron 1 is hypermethylated in acute myeloid leukemia, and methylation is associated with poor overall survival. 69 KLF5 expression is also inhibited in ccRCC cells by DNA methyltransferase 1 (DNMT1)‐induced hypermethylation at exon 4. 70 The expression of KLF5 can be recovered by knockdown of DNMT1 or the methylation inhibitor 5‐Aza‐CdR. 70
5.2. Super enhancer
SE consists of clusters of transcriptional enhancers, which are highly enriched for histone H3 lysine 27 acetylation (H3K27ac), bromodomain‐containing 4 (BRD4), mediator complex subunit 1 (MED1) and other transcriptional coactivators. 71 , 72 KLF5 transcription was reported to be regulated by SE in various cancers. 24 KLF5 SE was amplified in head and neck squamous cell carcinoma, lung squamous cell carcinoma, gastric adenocarcinoma, CRC, cervical squamous cell carcinoma, bladder cancer, and esophageal cancer. 24 In tumor cells with SE amplification, the average activation expression of KLF5 was upregulated by 39%. 24 Next, we reported that SE maintains high expression levels of KLF5 in HCC1806 and HCC1937 breast cancer cell lines. 73 The BRD4 inhibitors JQ‐1 and compound 870 and the cyclin dependent kinase 7 (CDK7) inhibitor THZ1 strongly inhibited the transcriptional expression of KLF5 and basal‐like breast cancer cell growth. 73
5.3. miRNAs inhibit KLF5 expression
KLF5 was reported to be targeted by multiple miRNAs, such as miR‐217, 74 miR‐153, 75 miR‐145‐5p, 76 miR‐152‐3p, 77 miR‐5195‐3p, 78 miR‐590‐5p, 79 miR‐4711‐5p, 80 miR‐214‐5p, 81 miR‐21, 82 miR‐211, 31 miR‐320, 83 and miR‐375, 84 in various cancers.
Mifepristone induced the tumor suppressor miR‐153 to suppress KLF5 expression and CSCs in TNBC. 75 Interestingly, miR‐153 also targets HIF‐1α 85 and angiopoietin 1 86 in response to hypoxia. miR‐217 targeted KLF5 and suppressed TNBC cell growth, migration, and invasion. 74 The tumor suppressor miR‐145‐5p targeted KLF5 and decreased the proliferation of gastric cancer 76 and cervical carcinoma cells. 87 , 88 The tumor suppressor miR‐152 also targets KLF5 in cervical cancer. 89 miR‐5195‐3p inhibited bladder cancer cell proliferation and invasion by targeting KLF5. 78 miR‐590‐5p targeted KLF5 and inhibited cell proliferation, migration and tumor growth of bladder cancer. 79 Consistently, miR‐4711‐5p suppressed colon cancer cell stemness and cell cycle progression in part by downregulating KLF5. 80 miR‐214‐5p exerted a tumor suppressor function by targeting KLF5 in HCC. 81 Crocin induced the expression of miR‐320 to target KLF5 and inhibited the EMT, migration, and invasion of gastric cancer cells. 83 miR‐375 reduces the expression of KLF5 in oral squamous cell carcinoma. 84
KLF5 was reported to play a context‐dependent role in different cancers. In prostate cancer, highly expressed miR‐21 targeted KLF5 and promoted cancer cell proliferation, survival, migration, and invasion. 82 miR‐21 also plays a similar role in HCC 90 and acute myeloid leukemia. 91 miR‐27a targeted FBW7 and indirectly increased KLF5 expression in ccRCC. 48
5.4. LncRNA
CASC15 promoted KLF5 expression and breast cancer cell proliferation, invasion, and tumor growth by inhibiting miR‐153‐3p. 44 MCM3AP‐AS1 increased KLF5 expression in glioma‐associated endothelial cells by inhibiting miR‐211. 31 In endometrial cancer, UCA1 upregulates the expression of KLF5 by sponging both miR‐1‐3p and miR‐143‐3p. 92 In addition, PVT1 was found to bind to KLF5 and increase KLF5 protein stability by recruiting BAP1. 93 LINC00337 promoted the cancer stem cell‐like properties of cervical cancer cells through the miR‐145/KLF5 axis. 88 LINC00908 promoted the expression of KLF5 by absorbing miR‐143‐3p, thereby promoting the proliferation and survival of CRC cells. 94
6. POSTTRANSLATIONAL MODIFICATIONS
6.1. Phosphorylation
KLF5 has been shown to be phosphorylated by protein kinase C (PKC) at S153, which upregulates its transactivation activities by enhancing the interaction between KLF5 and CBP. 95 KLF5 S406 phosphorylation by extracellular signal regulated kinase 1/2 (ERK1/2) enhanced the interaction between KLF5 and C‐JUN 96 or retinoic acid receptor α (RARα). 97 KLF5 phosphorylation at S303 by GSK3β recruits FBW7 to promote KLF5 ubiquitination and degradation by the proteasome. 22
6.2. Acetylation
KLF5 acetylation at K369 by p300 promoted the transactivation activity of KLF5 and induced p15 transcription in response to TGF‐β. 60 Recently, GCN5 was reported to acetylate KLF5 at K335 and K391 and increase the transcription of the downstream target gene GDF15 and the proliferation of NSCLC cells. 40 It is well known that acetylation is reversible. KLF5 can be deacetylated by histone deacetylase 1/2 (HDAC1/2) and SET, which inhibit the function of KLF5. 98 , 99 A recent study suggested that KLF5 acetylation may regulate KLF5 protein stability. 99 Additionally, acetylated KLF5 is essential to maintain basal progenitor cells and prostate differentiation. 15 Recently, KLF5 was shown to interact with HDAC3 to inhibit the transcription of BECN1. 38
6.3. Ubiquitination
KLF5 has been demonstrated to be ubiquitinated. 100 KLF5 can be ubiquitinated by E3 ligases, such as WWP1, 62 FBW7, 22 SMURF2, 64 and EFP. 63 Interestingly, YAP and TAZ, 2 well known WW domains containing coactivators in the Hippo pathway, competitively bind to the KLF5 PY motif and protect KLF5 from WWP1‐mediated ubiquitinated degradation. 68 , 101 , 102 Curcumin appears to target KLF5 for degradation by downregulating YAP/TAZ in bladder cancer cells. 68
It is well known that ubiquitination is also reversible. In recent years, 3 KLF5 deubiquitinating enzymes, BAP1, ATXN3L, and USP3, have been identified through siRNA library screening. 55 , 65 , 66 BAP1 was shown to promote breast tumor growth and metastasis by stabilizing KLF5. 55 Furthermore, KLF5 forms a complex with BAP1/HCF‐1/OGT1 to regulate the transcription of FGF‐BP1 and p27 and cell cycle progression. 55 In melanoma, the high expression of BAP1 also indicates a poor prognosis for patients, which promotes tumor progression by hindering KLF5 ubiquitination. 103 Although ATXN3L was identified as another KLF5 DUB, the endogenous ATXN3L protein could not be detected because of its low expression or antibody sensitivity. 65 Most recently, USP3 was validated to reduce KLF5 polyubiquitination and increase KLF5 protein stability, therefore promoting breast cancer cell proliferation and tumor growth. 66 Most importantly, the expression levels of USP3 and KLF5 in breast tumors are positively correlated. 66 How these E3s and DUBs are coordinated to regulate KLF5 protein stability and activity in response to signaling requires further investigation.
7. CELLULAR FUNCTIONS
KLF5 is broadly expressed during embryogenesis and in adult tissues. Accumulating evidence suggests that KLF5 is expressed in several tissues, including the colon, 104 intestine, 18 pancreas, 104 stomach, 105 placenta, 106 prostate, 15 testis, 104 breast, 7 bladder, 107 lung, 19 and skeletal muscle. 104 KLF5 has been well documented to regulate multiple cellular processes, including the cell cycle and proliferation, apoptosis and autophagy, migration and invasion, and stemness and differentiation.
7.1. Proliferation and cell cycle progression
Several lines of new evidence support the idea that KLF5 promotes the cell cycle and proliferation. In a mouse model of intestinal‐specific deletion of Klf5, it was found that Klf5 is necessary for gut epithelial cell proliferation. 16 Progesterone and PR promote breast cell proliferation by inducing KLF5 transcription, which in turn upregulates the expression of several genes, including Cyclin A, chromatin licensing and DNA replication factor 1 (CDT1), and E2F transcription factor 3 (E2F3). 108 Consistently, Klf5 knockout in mouse mammary glands showed defects after pregnancy because epithelial cell proliferation was significantly decreased, as examined by Ki67 IHC staining. 7 In addition to FGF‐BP1 and integrin subunit beta 2 (ITGB2), 101 we demonstrated that KLF5 also promoted breast cancer cell proliferation through mPGES1 26 and TNFAIP2. 27 In pancreatic cancer cells, KLF5 promotes the transcription of E2F1, Cyclin D1 and Rad51 while inhibiting the expression of p16. 41 KLF5 is also required for androgen/AR‐dependent prostate cancer cell proliferation. 32
7.2. Stemness and differentiation
KLF5 plays important roles in the self‐renewal of ESCs, tissue‐specific stem cells, and CSCs. Klf5 is induced by leukemia inhibitory factor (LIF)/signal transducer and activator of transcription 3 (STAT3) in mouse ESCs to maintain the undifferentiated state. 109 In mammary glands, KLF5 promotes Slug gene transcription to maintain stemness. 7 Depletion of Klf5 in mouse mammary glands significantly decreased CD24+/CD49fhigh normal breast stem cells. 7 Mammosphere formation and breast regeneration efficiency were suppressed in the absence of Klf5. 7 Knockdown of KLF5 or Slug also reduced the number of acetaldehyde dehydrogenase positive (ALDH+) stem cells in 184B5 and MCF10A breast epithelial cell lines. 7 Consistently, mifepristone, metformin, and miR153 suppress breast CSCs by inhibiting KLF5 expression. 6 , 75 Additionally, Klf5 is essential for maintaining the progenitor cells of prostate basal cells. 15
Several studies have shown that KLF5 plays an important role in the maturation of gut cells. 16 Klf5 depletion destroyed the colonic crypt structure. 16 Klf5 is essential for initiating morphogenesis of the early endoderm into a compartmentalized intestinal epithelium comprised of villi and terminally differentiated cells. 17 miR‐4711‐5p suppressed colon cancer cell stemness by downregulating KLF5. 80 KLF5 depletion also caused the differentiation of gastric cancer cells. 76
7.3. Apoptosis and autophagy
Accumulating evidence suggests that KLF5 is a pro‐survival protein. In pancreatic ductal adenomas, Klf5 blocked Sox4‐induced apoptosis. 110 In breast cancer, we found that KLF5 increases mitogen‐activated protein kinase phosphatase 1 (MKP‐1) protein stability to inhibit apoptosis. 111 Dexamethasone (DEX) increases docetaxel and cisplatin resistance by upregulating KLF5 in TNBC. 112 Consistently, KLF5 downregulation promoted cisplatin‐induced apoptosis by inhibiting the phosphorylation of the DNA damage checkpoint protein checkpoint kinase 1/2(Chk1/2) in NSCLC. 113 KLF5 depletion markedly induced apoptosis of anaplastic thyroid carcinoma cells and increased doxorubicin sensitivity, and likely to be through inhibiting the JNK signaling pathway. 114
In contrast, ectopic KLF5 expression was reported to promote apoptosis in ESCC cells 115 and in LNCaP prostate cancer cells in the presence of phorbol 12‐myristate 13‐acetate (PMA) by activating the JNK signaling pathway. 116 Interestingly, KLF5 inhibited autophagy in castration‐resistant prostate cancer cells by suppressing the transcription of BECN1. 38 KLF5 knockdown decreased the sensitivity of prostate cancer cells to docetaxel. 38 In A2058 melanoma cells, KLF5 knockdown increased autophagosomes and autolysosomes. 103 Therefore, KLF5 plays a context‐dependent role in regulating cell death and drug resistance.
7.4. Migration and invasion
Large numbers of studies suggest that KLF5 promotes cell migration and invasion. KLF5 increased TNBC cell adhesion, migration, and invasion by inducing the transcription of TNFAIP2, which in turn activated Rac family small GTPase 1 (Rac1) and cell division cycle 42 (Cdc42). 27 In agreement with this, miR‐217 inhibits TNBC cell migration and invasion by targeting KLF5. 74 Additionally, depletion of Klf5 in 4T1 mouse breast cancer cells significantly suppressed lung metastasis. 55 KLF5 promoted cervical cancer migration and invasion by inducing TNFRSF11a expression. 39 KLF5 promoted cell migration via the FYN/p‐FAK axis in bladder cancer cells. 30 Cholesterol accelerated ccRCC cell migration and invasion by inducing KLF5. 48 Consistently, miR‐4711‐5p suppressed colon cancer cell migration and invasion by downregulating KLF5. 80 miR‐590‐5p suppressed osteosarcoma cell migration and invasion by targeting KLF5. 79 KLF5 knockdown reduced laryngeal carcinoma Hep‐2 cell migration and invasion and EMT by inhibiting the NF‐κB pathway. 117 Similarly, KLF5 was reported to promote thyroid cancer metastasis by activating the NF‐κB pathway. 118 KLF5 activated LINC00346 transcription and promoted gastric cancer cell migration and invasion. 45 Consistently, crocin inhibited EMT, migration, and invasion of gastric cancer cells partially through increasing the expression of miR‐320, which targets KLF5. 83 Studies have found that overexpression of BAP1 in esophageal cancer cells upregulates the expression of KLF5 and its downstream genes FGF‐BP1 and Cyclin D1, and promotes the proliferation and migration of ECA109 esophageal cancer cells. 119
In sharp contrast, KLF5 knockdown in A549 lung cancer cells induced EMT by downregulating the expression of E‐cadherin and upregulating the expression of Vimentin protein. 120 Consistently, KLF5 is required to maintain epithelial characteristics and prevent EMT induction in epithelial cells by inducing the expression of miR‐200. 50 TTK kinase induced the expression of ZEB1 and EMT in TNBC by inhibiting KLF5‐induced miR‐200 expression. 121 In the absence of p53, KLF5 inhibits EMT in liver cancer cells through the miR‐192/ZEB2 axis. 49 miR‐21 promoted prostate cancer cell migration and invasion by directly targeting KLF5. 82 Similar results were observed in hepatocellular carcinoma. 90 In ESCC, KLF5 loss inhibited notch receptor 1 (NOTCH1) expression and induced invasion when p53 was inactivated. 122 These findings suggest that KLF5 has a context‐dependent role in regulating cell migration and invasion.
8. KLF5 INVOLVED ONCOGENIC SIGNALING PATHWAYS
KLF5 plays important roles in the regulation of multiple cancer‐related signaling pathways (Figure 2).
FIGURE 2.
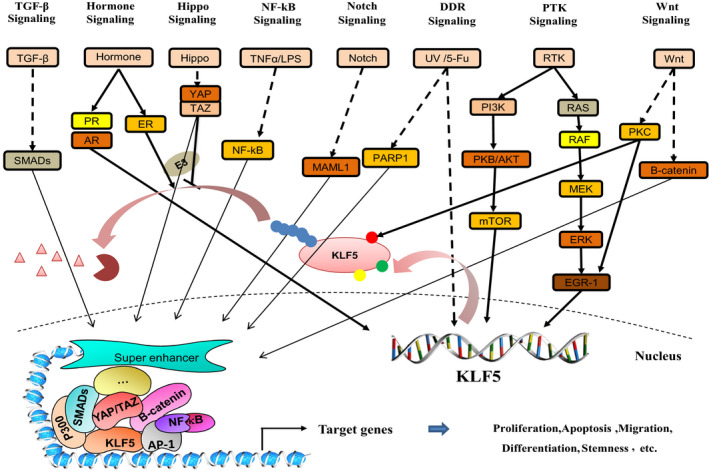
KLF5 is involved in diverse oncogenic signaling pathways. The protein expression and function of KLF5 involve multiple signal pathways, including Wnt, RTK, hormone, TGF‐β, Hippo, NOTCH, NF‐κB and Hedgehog signaling pathways. It can form a transcription complex with a variety of transcription factors to regulate the expression of target genes
8.1. Wnt
The Wnt signaling pathway plays a vital role in various cancers, especially intestinal and CRC. McConnell et al first reported that KLF5 haploinsufficiency inhibited intestinal adenoma formation in ApcMin mice by 96%, with decreased expression of Wnt target genes, including c‐Myc and Cyclin D1. 123 Consistently, KLF5 interacted with β‐catenin to promote β‐catenin nuclear localization. 123 A subsequent study demonstrated that deletion of Klf5 in the gut epithelium decreased epithelial proliferation, differentiation, and cell positioning along the crypt radial axis because of the disruption of canonical Wnt signaling. 16 Nakaya et al further confirmed that inducible deletion of Klf5 in Lgr5+ mouse intestinal stem cells suppressed cell proliferation, survival and production of lethal adenomas and carcinomas induced by an oncogenic mutant of β‐catenin. 18 This conclusion was confirmed by another independent study. Klf5 knockout in Lgr5+ intestinal stem cells inactivated numerous canonical WNT‐responsive and NOTCH‐responsive genes and caused regeneration defects. 124 , 125 In CRC progenitor cells, KLF5 and YAP1 form a complex to induce the transcription of the Ascl2 gene, a Wnt signaling target, to maintain self‐renewability. 126 Taken together, these findings suggest a crucial role of KLF5 in the Wnt signaling pathway and CRC.
8.2. RTK
RTK activates the rat sarcoma/mitogen‐activated protein kinase (RAS/MAPK), phosphatidylinositol 3‐kinase/AKT serine/threonine kinase/mechanistic target of rapamycin kinase (PI3K/AKT/mTOR), and STAT3 pathways. Accumulating evidence suggests that RAS/MAPK/PI3K can increase KLF5 transcription to promote cell proliferation. 127 , 128 Pharmacological inhibition of MEK and PI3K kinases reduced KLF5 protein levels in mouse pancreatic cancer cells. 129 We showed that KLF5 activates ERK in breast cancer by inducing the expression of FGF‐BP1. 25 , 111 Activated ERK phosphorylates and stabilizes the MKP‐1 protein, which decreases MAPK phosphorylation as a negative feedback loop. 111 Interestingly, Klf5 suppressed ERK activity in mESCs by inducing the transcription of Spred1, a negative regulator of ERK signaling. 130
Some studies suggest that KLF5 promotes cancer by activating the PI3K/AKT signaling pathway. For example, KLF5 knockdown inhibited the activation of the PI3K/AKT/mTOR pathway and HIF‐1α‐dependent glycolysis, therefore overcoming hypoxia‐induced cisplatin resistance in NSCLC cells. 131 The latest research shows that knocking down BAP1 in melanoma cells also reduced the phosphorylation of PI3K, AKT and mTOR activated by KLF5. 103 Additionally, KLF5 was reported to promote EMT of HCC by activating the PI3K/AKT/Snail signaling pathway. 132 Furthermore, KLF5 increased the activities of both the PI3K/AKT and ERK1/2 signaling pathways, possibly by upregulating the expression of AGGF1 in glioma‐associated endothelial cells. 31 Consistently, miR‐493‐5p inhibited the proliferation and metastasis of osteosarcoma cells by downregulating KLF5 and inactivating the PI3K/AKT signaling pathway. 133 Resveratrol inhibited the PI3K/protein kinase D1 (PKD1)/AKT pathway and increased KLF5 phosphorylation, which decreased its interaction with c‐Myc in HEK293 cells. 134 However, the opposite conclusion was reached in prostate cancer. Klf5 and PTEN depletion additively increased AKT activity, the expression of HIF‐1α, VEGF, and PDGF, and angiogenesis, although the mechanism by which KLF5 inhibits the PI3K/AKT pathway is unclear. 135 An independent study supported this finding. miR‐21 promoted prostate cancer by directly targeting KLF5, which downregulates the expression of GSK3β and the phosphorylation of AKT. 82
Additionally, KLF5 was reported to activate STAT3. He et al demonstrated that Klf5 knockout decreased Ras‐induced pancreatic tumorigenesis by reducing the activation of STAT3 due to the increase in the expression of N‐myc downregulated gene 2 (NDRG2). 129 ML264, a small molecular inhibitor of KLF5, also inhibited the activation of STAT3 in osteosarcoma. 136 However, KLF5 was reported to inhibit STAT3 activity and prostate cancer metastasis by suppressing IGF1 transcription. 53
8.3. Hormone
In 2013, Liu, R et al summarized the relationships among steroid hormonal signaling (progesterone, estrogen and androgen) and KLF5 in breast cancer. 10 In brief, KLF5 was induced by progesterone in PR‐positive breast cancer cells and promoted cell proliferation and dedifferentiation. 108 Consistently, KLF5 expression is significantly increased in mouse mammary glands after pregnancy, and KLF5 is essential to maintain breast stemness. 7 Mifepristone, a PR antagonist, not only blocked progesterone‐induced KLF5 expression in PR‐positive breast cancer cells 10 , 108 but also inhibited the expression of KLF5 in TNBC by inducing miR‐153. 75
Similarly, androgen also induced KLF5 transcription through AR in prostate cancer cells and KLF5‐mediated androgen‐induced cell‐surface chemokine receptor 4 (CXCR4) expression and migration. 137 This finding was confirmed by an independent study in which androgen induced KLF5 transcription and KLF5 interacted with an interaction with sterol‐regulatory‐element‐binding protein‐1 (SREBP‐1) to induce fatty acid synthase (FASN) gene transcription and in LNCaP prostate cancer cells. 138 In agreement with these results, KLF5 is also an androgen‐responsive gene in human breast carcinomas. 139 Recently, KLF5 was shown to interact with AR to regulate the expression of AR target genes in prostate cancer cells. 32 Additionally, acetylation of KLF5 at K369 is essential for androgen‐mediated organoid organization and prostate regeneration. 15
Glucocorticoids, such as DEX and hydrocortisone, can also induce KLF5 expression through GR and confer docetaxel and cisplatin resistance in TNBC cell lines. 112 This finding was supported by an independent study in which DEX‐bound GR accelerated adipogenesis by directly promoting the expression of KLF5. 140 Interestingly, glucocorticoids also induced the expression of several KLF5 interacting partners, such as TEAD4 141 and YAP, 142 to promote breast cancer growth and metastasis. It is likely that activated GR interacts with the YAP/TEAD4/KLF5 complex to promote cancer cell proliferation, survival, and metastasis.
In contrast, estrogen suppresses KLF5 expression in ERα‐positive breast cancer cells through multiple mechanisms, and KLF5 also suppresses ERα‐mediated gene transcription. 10 It was also shown that 17β‐estradiol suppressed KLF5‐mediated FOXO1 and PDGFA transcription through ERβ in prostate cancer cells. 58
8.4. TGF‐β
The TGF‐β/SMAD pathway plays a dual role in tumorigenesis and development. TGF‐β inhibits epithelial cell proliferation in the early stage of cancer development but promotes metastasis in the late stage of cancer progression. In response to TGF‐β, KLF5 is acetylated by p300, binds to SMADs, and regulates the transcription of p15 and c‐Myc. 60 , 143 Recently, Ras was shown to inhibit TGF‐β‐mediated KLF5 acetylation, block the assembly of the p300‐KLF5‐SMAD complex, and regulate the expression of p15 and c‐Myc. 59
8.5. Hippo
The Hippo pathway regulates cell stemness, proliferation, survival, migration, and organ size. 144 YAP and TAZ are 2 key transcriptional coactivators in the Hippo pathway. Our studies showed that both YAP and TAZ increased KLF5 protein stability by preventing WWP1‐mediated ubiquitination and degradation. 68 , 101 , 102 Consistently, curcumin downregulated the expression of KLF5 by targeting YAP/TAZ and inhibited bladder cancer cell growth. 68 Additionally, TEAD4, a key transcription factor and partner of YAP/TAZ, was found to interact with KLF5 and inhibit p27 gene transcription in breast cancer. 5 In agreement with this, TEAD4, similar to KLF5, promoted breast cancer stemness, cell growth, survival, metastasis, and chemoresistance. 5 , 141 Furthermore, glucocorticoids also induced GR‐dependent TEAD4 transcription, nuclear accumulation, and transcription complex formation. 141 It is reasonable to suspect that KLF5, TEAD4, and GR form a transcription complex to execute the above oncogenic functions in breast cancer.
8.6. NOTCH
KLF5 may be a tumor suppressor in ESCC. KLF5 and p53 maintain the expression of NOTCH1, which suppresses primary human keratinocyte transformation. Loss of both p53 and KLF5 led to the formation of invasive tumors. 122 In the mouse prostate, Klf5 deacetylation activates NOTCH signaling, Hes family bHLH transcription factor 1 (Hes1), Myc, Jagged 1 and delta like canonical NOTCH ligand 1 (Dll1)), which promotes luminal cell proliferation. 15 DAPT, a NOTCH signaling inhibitor, blocked the abnormal phenotype induced by Klf5K358R knockin. 15 Klf5 knockout in Lgr5+ intestinal stem cells led to loss of stem cell identification and premature differentiation because of inactivation of NOTCH and WNT target genes. 125 Interestingly, KLF5, NOTCH, and MYC are substrates of the tumor suppressor SCFFBW7. 145
8.7. NF‐κB
NF‐κB signaling is activated in response to inflammatory stimuli, such as TNFα, interleukin 1β (IL1β), and lipopolysaccharide (LPS). Early studies showed that LPS induced KLF5 expression and that KLF5 was necessary for NF‐κB activation. 146 , 147 TNFα was reported to induce KLF5 by activating p38 in cervical cancer cells. 39 Several studies have suggested that KLF5 activates NF‐κB signaling. For example, KLF5 increased the phosphorylation of IKKβ, IκBα and p65 nuclear translocation in thyroid cancer cells. 118 KLF5 silencing significantly decreased Hep‐2 laryngeal cancer cell proliferation, survival, and migration by reducing the phosphorylation of IκBα and p65. 117 KLF5 also regulates several target genes related to the NF‐κB signaling pathway. We demonstrated that KLF5 and NF‐κB may coordinately induce TNFAIP2 gene transcription in breast cancer cells. 27 , 148 KLF5 was shown to induce the transcription of TNFRSF11a, which promotes cervical cancer cell proliferation and invasion. 39
8.8. Hedgehog
KLF5 also promotes sonic hedgehog (SHH) signaling. KLF5 was highly expressed in esophageal adenocarcinoma, and KLF5 knockdown significantly decreased SHH signaling and cancer cell proliferation and migration. 149 GLI family zinc finger 1 (GLI1), a typical SHH pathway target gene, was significantly downregulated by KLF5 knockdown, however SHH and patched 1 (PTCH1) expression levels were upregulated. 149 Bell et al generated conditional Klf5 knockout mice using SHH‐Cre and found that Klf5 depletion resulted in the inhibition of villus morphogenesis and intestinal epithelial differentiation. 17 Consistently, Klf5‐KO increased the expression of PTCH1 and GLI family zinc finger 2 (GLI2). 17 The mechanism by which KLF5 participates in the SHH signaling pathway remains to be elucidated.
8.9. DNA damage repair
5‐Fluorouracil (5‐FU)‐induced DNA damage activated KLF5 transcription via a p53‐independent mechanism in HCT116 colon cancer cells. KLF5, in turn, induced the expression of Pim1 to promote cell survival. 150 Consistently, irradiation also induced Klf5 expression in mouse intestinal epithelial cells. 151 KLF5 modulates DDR via p21‐mediated growth arrest and BCL2 associated X (BAX)‐mediated apoptosis in response to UV irradiation in TE2 esophageal cancer cells. 152 Klf5 intestinal‐specific heterozygote‐deficient mice had more severe intestinal damage after radiation damage than WT mice. 151 The mechanism was associated with reduced expression of DDR genes, such as ERCC excision repair 5 (ERCC5) and cullin 4B (CUL4B). 151 Additionally, KLF5 was reported to interact with poly ADP‐ribose polymerase 1 (PARP1), an enzyme associated with DDR and apoptosis. 153 Importantly, knockdown of KLF5 sensitized NSCLC cells to cisplatin. 113 KLF5 depletion inhibited the DDR by reducing the activation of Chk1/2 and H2A.X variant histone (H2AX), allowing cells to enter mitosis with damaged DNA. 113 KLF5 knockdown also sensitized TNBC cells to cisplatin and docetaxel. 112
8.10. Hypoxia
KLF5 is involved in hypoxia‐induced cancer behaviors, such as vascular remodeling, 154 cell proliferation, 154 apoptosis, 61 and drug resistance. 131 HIF‐1α is often rapidly activated under hypoxia. 155 In pancreatic cancer, KLF5 is upregulated by hypoxia, and KLF5 interacts with HIF‐1α to induce transcription of some target genes, such as glucose transporter 1 (GLUT‐1). 156 In CRC cells, lysophosphatidic acid (LPA) induces KLF5 expression, which in turn transactivates HIF‐1α gene transcription. 157 Consistently, hypoxia promotes the survival of NSCLC cells by inducing the expression of HIF‐1α in a KLF5‐dependent manner. 61 Gong et al also found that hypoxia upregulated the expression of KLF5 in NSCLC cells and that hypoxia‐induced cisplatin resistance was reversed by KLF5 knockdown because KLF5 knockdown inhibited hypoxia‐induced HIF‐1α expression, PI3K/AKT/mTOR pathway activation, and glycolysis. 131 Crocin inhibits gastric cancer cell migration, invasion and EMT by activating the miR320/KLF5/HIF‐1α axis. 83 In contrast, KLF5 deletion promotes the accumulation of HIF‐1α and angiogenesis in PTEN‐deficient prostate cancer. 135
9. FUNCTIONS OF KLF5 IN VARIOUS CANCERS
The expression of KLF5 is abnormal in a variety of solid tumors, such as breast cancer, prostate cancer, colon cancer, NSCLC, and ESCC. The function of KLF5 is context dependent, although it promotes tumorigenesis in most cancers.
9.1. Breast cancer
Accumulating evidence suggests that KLF5 promotes breast cancer cell stemness, proliferation, survival, adhesion, and migration. In breast cancer, KLF5 promotes cancer cell proliferation and the cell cycle by inducing transcription of FGF‐BP1, 25 mPGES1, 26 TNFAIP2, 27 and Cyclin D1 4 and inhibiting transcription of p27 5 and p21. 26 KLF5 maintains cell stemness by inducing the transcription of Slug 7 and Nanog. 6 KLF5 is essential for progesterone to induce cell proliferation and dedifferentiation. 108 Consistently, breast‐specific Klf5 knockout mice significantly reduced the proliferation, survival, and stemness of breast epithelial cells and inhibited PyMT‐induced tumorigenesis. 7 Klf5 is essential for the formation of milk bubble structures during pregnancy and lactation. 7 Interestingly, KLF5 is also induced by androgen 5α‐dihydrotestosterone and promotes MCF7 cell proliferation. 139 Glucocorticoids also induce KLF5 expression through GR and confer docetaxel and cisplatin resistance to TNBC cells. 112 , 141
PVT1 directly interacts with KLF5 and recruits BAP1 to stabilize the KLF5 protein in breast cancer. 93 CASC15 functions as a competitive endogenous RNA for miR‐153‐3p, which targets KLF5. 44 Interestingly, CASC15 is also a direct target gene of KLF5; therefore, a positive feedback loop is formed to accelerate breast tumor progression. 44 Several drugs or compounds, such as mifepristone 75 and its derivatives, 158 , 159 metformin, 6 and mithramycin A, 160 have been shown to inhibit the expression of KLF5 in TNBC. In addition, the SE inhibitors JQ1, compound 870, and THZ1 potently inhibited the expression of KLF5 in TNBC. 73
9.2. Prostate cancer
Some studies have suggested that KLF5 promotes prostate cancer. KLF5 siRNA delivered by a nanoparticle system inhibited PC‐3 xenograft growth and angiogenesis. 161 Androgen induces KLF5 and CXCR4 expression and promotes prostate cancer cell migration in vitro. 137 This conclusion is supported by a recent study, in which acetylated KLF5 promoted bone metastasis by activating CXCR4. 162 In two androgen‐responsive prostate cancer cell lines, C4‐2B and LNCaP, the expression of KLF5 was upregulated by androgen/AR. 32 At the same time, KLF5 interacts with AR to coordinately increase the expression of AR target genes, including Myc, Cyclin D1 and PSA, which promote cell proliferation and differentiation. 32 Estrogen (17β‐estradiol) promoted prostate tumor angiogenesis through the ERβ/KLF5 pathway. 58
However, a line of evidence also supports that KLF5 is a tumor suppressor in prostate cancer. The KLF5 gene is located at chromosome 13q.21 and is frequently deleted in prostate cells. 163 A low expression level of KLF5 was correlated with poor prognosis in prostate cancer patients. 38 Klf5 deletion promoted PTEN deletion‐initiated luminal‐type mouse prostate tumors. 164 Klf5 inhibited angiogenesis in PTEN‐deficient prostate cancer by attenuating AKT activation and HIF‐1α accumulation. 135 TNFα induced the expression of KLF5 in LNCaP cells and increased the levels of phosphorylation of JNK and apoptosis in response to PMA. 116 Consistently, KLF5 upregulated the transcription of MKK7, the upstream kinase of JNK, and promoted TNFα‐induced apoptosis in LNCaP cells. 36 miR‐21 is highly expressed in prostate cancer. 82 It can directly target KLF5, upregulate GSK3β expression and AKT phosphorylation, and promote prostate cancer cell proliferation, survival, migration, and invasion. 82 Loss and downregulation of KLF5 increased autophagy, thereby decreasing the sensitivity of prostate cancer cells C4‐2 and CWR22RV1 to docetaxel. 38
Therefore, the role of KLF5 in prostate cancer seems controversial. Acetylated KLF5 exhibits antitumor activity; in contrast, deacetylated KLF5 plays a tumor‐promoting effect, which is related to the TGF‐β signaling pathway. 165 KLF5 acetylation in prostate basal cells is essential to maintain the characteristics of basal progenitor cells by inhibiting the NOTCH pathway. 15 KLF5 downregulation promoted the invasion of prostate cancer by increasing the expression of IGF1, which in turn activated the STAT3 signaling pathway. 53 A circular RNA CircCRKL is downregulated in the prostate and can inhibit miR‐141, upregulate the expression of KLF5, and inhibit prostate cancer. 166
9.3. Bladder cancer
KLF5 plays an important role in the normal development of mouse bladder tissue. 107 Our early study indicated that KLF5 promotes bladder cancer cell proliferation. 4 KLF5 can bind to a Cyclin E1 gene enhancer to activate its transcription and increase susceptibility to bladder cancer. 29 In addition, KLF5 promotes bladder cancer cell migration by increasing the expression of FYN. 30 Furthermore, KLF5 promotes angiogenesis in bladder cancer by directly increasing vascular endothelial growth factor A (VEGFA) transcription. 127 Consistently, miR‐5195‐3p targets KLF5 to inhibit the expression of Cyclin D1 and VEGFA and to suppress the proliferation and invasion of bladder cancer cells. 78 Curcumin can inhibit bladder tumor growth by targeting the YAP/TAZ/KLF5/CyclinD1 axis. 68
9.4. Renal cell carcinoma
KLF5 is expressed in kidney and its collecting system. ccRCC is a common subtype of renal cell carcinoma in which KLF5 is highly expressed. 48 KLF5 was shown to increase the expression of miR‐27a, which targets FBW7 and prevents FBW7‐mediated KLF5 ubiquitination and degradation, promoting renal cancer cell migration and invasion. 48 In contrast, DNMT1‐mediated KLF5 promoter hypermethylation inhibits KLF5 expression. 70 KLF5 was shown to inhibit ccRCC xenograft tumor growth and metastasis. 70 The deubiquitinase BAP1 was reported to stabilize KLF5. 55 Interestingly, the BAP1 gene is mutated in ~15% of ccRCC, which also defines a new subtype of renal cell carcinoma. 167 Whether BAP1 mutations cause renal cell carcinoma by promoting KLF5 degradation should be investigated.
9.5. Intestinal and colorectal cancer
Overall, KLF5 is an oncogenic transcription factor in intestinal and CRC. In Klf5+/− mice, DSS induced more severe colitis, suggesting that KLF5 is required for intestinal epithelial repair. 168 Intestinal‐specific deletion of Klf5 using Villin‐Cre showed that Klf5 is required to maintain gut epithelial cell proliferation, differentiation, and positioning along the crypt radial axis. 16 Consistently, Klf5 deletion in the intestinal epithelium using Shh‐Cre inhibited villus morphogenesis and epithelial differentiation. 17 Furthermore, depletion of Klf5 in Lgr5+ stem cells disrupts the integrity and oncogenicity of intestinal stem cells. 18 Klf5 haploinsufficiency rescued the intestinal tumor formation induced by APCMin/+ because KLF5 promotes the nuclear translocation of β‐catenin and upregulates the expression of CyclinD1 and Myc. 123 Similarly, Klf5 haploinsufficiency also decreased intestinal tumor formation in APCMin/+/KRASV12 mice. 169 Klf5 is also essential for intestinal epithelial cell proliferation and transformation by oncogenic KRASG12D. 169 , 170
Zhang et al reported that the second CPD domain of KLF5 and the WD40 domain of FBW7 were frequently mutated, which reduced the degradation of KLF5 protein in CRC. 24 The KLF5 P301S mutation also increased the stability and transcriptional activity of KLF5 in CRC. 171 The tumor suppressors miR‐143 and miR‐145 can downregulate the expression of KLF5 in CRC. 172 LINC00908 is highly expressed in CRC and promotes cell proliferation and survival through the miR‐143‐3p/KLF5 axis. 94 LPA induced HIF‐1α through KLF5 in colon cancer cell lines. 157 Recently, KLF5 was reported to induce lncRNA SNHG12 expression and to promote invasion and metastasis of CRC. 47 KLF5 also induces the transcription of the intestinal differentiation marker gene alkaline phosphatase. 173 ML264 was identified as a KLF5 inhibitor that effectively inhibited the expression of KLF5 and the growth of CRC xenograft tumors. 128 ML264 appears to inhibit the expression of KLF5 by inhibiting its upstream transcription factor EGR1, although the exact mechanism is unknown. 128
9.6. Esophageal squamous cell cancer
Esophageal cancer is a common malignant tumor, and its 5‐y survival rate is less than 20%. Squamous cell carcinoma (51.6%) or adenocarcinoma (41.9%) accounts for more than 90% of esophageal cancers. 174 The function of KLF5 in ESCC is also controversial. KLF5 is expressed in normal esophageal epithelial cells, but its expression is downregulated or absent in ESCC. 115 In abnormally proliferating esophageal squamous epithelial cells, KLF5 promotes NOTCH1 transcription in the context of p53 mutation or loss. 122 KLF5 and NOTCH1 loss is a critical event in squamous cell transformation and invasion. Additionally, KLF5 induces the transcription of apoptotic signal‐regulating kinase 1 (ASK1) and MKK4, which activate the JNK pathway and promote apoptosis. 115
In contrast, KLF5 interacts with TCF3 to induce LINC00094 transcription, thereby promoting the growth of ESCC. 42 In ECA109 cells, BAP1 can promote cell proliferation and migration by upregulating KLF5. 119 Additionally, KLF5, TP63, and SOX2 form a transcription complex to synergistically regulate the expression of target genes, including ALDH3A1, which is highly expressed in ESCC, and ALDH3A1 knockdown inhibits colony formation, cell viability and tumor growth in vivo. 37 This study also reported that the combination of the HDAC inhibitor romidepsin and the BET inhibitor ARV‐771 can synergistically inhibit ESCC. 37
9.7. Gastric cancer
The KLF5, GATA4, and GATA6 genes are independently amplified, and the 3 proteins form a transcription complex to promote the transcription of Hnf4α and tumorigenesis of gastric cancer. 56 Crocin can induce miR320 to target both KLF5 and HIF‐1α to inhibit EMT in gastric cancer cells. 83 Interestingly, miR153 also targets both KLF5 75 and HIF‐1α. 85 Furthermore, miR‐145‐5p targets KLF5 and promotes gastric cancer cell differentiation. 76 In addition, KLF5 and MYC induce the transcription of LINC00346 to promote gastric cancer tumorigenesis. 45 KLF5 can activate the transcription of lncRNA NEAT1, which recruits BRG1 to silence the transcription of GADD45A and promotes gastric cancer cell proliferation and survival. 46 Notably, an early study reported that KLF5 is downregulated in gastric cancer and that KLF5 expression is positively correlated with early, small, and no lymph node metastasis tumors. 175 Zhang et al showed that the expression of KLF5 has no relationship with the prognosis of gastric cancer patients. 176 Interestingly, KLF5 is highly expressed in the tumor‐associated fibroblasts of gastric cancer patients, and its expression is positively correlated with tumor grade, invasion depth, size, metastasis, and poor prognosis. 177 KLF5 knockdown in cancer‐associated fibroblasts not only inhibited the growth of tumor cells but also inhibited their migration and invasion by inhibiting the C‐C motif chemokine ligand 5/C‐C motif chemokine receptor 5 (CCL5/CCR5) axis. 177
9.8. Hepatocellular carcinoma
It has been reported that KLF5 is significantly overexpressed in HCC specimens, and high KLF5 expression predicts a poor prognosis for HCC patients. 132 KLF5 promotes HCC growth and metastasis by activating PI3K/AKT/Snail signaling. 132 Consistently, the tumor suppressor miR‐145‐5p inhibits HCC cell proliferation and migration by targeting KLF5. 178 In contrast, miR‐21 promotes the migration and invasion of HuH7 cells by targeting KLF5. 90 In HCC, the function of KLF5 may depend on the p53 status. 49 When WT p53 is present in HepG2 cells, KLF5 does not regulate cell migration. 49 When p53 is inactivated in Hep3B cells, KLF5 inhibits ZEB2 expression and EMT by promoting miR‐192. 49 The defined role of KLF5 in HCC requires more studies.
9.9. Pancreatic cancer
Pancreatic cancer is one of the most aggressive tumors. KLF5 expression is a poor prognostic marker for pancreatic cancer patients. 41 KLF5 promoted G1/S pancreatic cancer cell cycle progression by inducing the expression of E2F1, Cyclin D1 and Rad51 while inhibiting the expression of p16. 41 In pancreatic cancer, KLF5 is induced by IL‐1β through p38 and hypoxia through HIF‐1α. 156 In pancreatic cancer with p53 mutation, KLF5 induced PLA2G16 expression to promote glycolysis. 34
KLF5 and SMAD4 inhibited the infiltration of T cells in the tumor immune microenvironment and promoted the infiltration of myeloid cells in pancreatic cancer. 179
9.10. Lung cancer
Klf5 is essential for mouse lung development. 19 KLF5 may also have a dual role in lung cancer. KLF5 was shown to promote lung cancer cell proliferation and tumorigenesis through upregulation of Sox4 expression. 180 Recently, KLF5 was reported to be highly expressed in NSCLC, and its expression was significantly higher than that of adjacent tissues, indicating a poor prognosis. 113 KLF5 depletion can overcome cisplatin resistance in NSCLC. 113 Under hypoxic conditions, the expression of KLF5 and HIF‐1α was induced, and KLF5 interacted with HIF‐1α to promote the survival of NSCLC cells. 61 Moreover, KLF5 knockdown inhibited hypoxia‐induced HIF‐1α expression, the PI3K/AKT/mTOR pathway, glycolysis, and cisplatin resistance in NSCLC cells. 131 In A549 cells, KLF5 interacted with GCN5 to induce the expression of GDF15 and promoted cell proliferation and tumor growth. 40 However, it was reported that Klf5 was not necessary in the mouse K‐RasG12D lung tumorigenesis model. 54 In this study, KLF5 expression was positively correlated with better disease‐specific survival of patients with NSCLC. 54
9.11. Leukemia
KLF5 interacted with p53 to induce survivin gene transcription and increase the survival of acute lymphoblastic leukemia cells. 181 However, KLF5 was downregulated in acute myeloid leukemia blast cells because of promoter methylation. 69 , 182 , 183 Low expression of KLF5 was associated with poor overall survival in acute myeloid leukemia patients. 183 In acute myeloid leukemia, miR‐21 targeted KLF5 and promoted the proliferation of acute myeloid leukemia cells in vitro. 91 The expression of KLF5 was also downregulated in BCR‐ABL1+ B‐ALL leukemia. 35 In B‐cell acute lymphoblastic leukemia cells, overexpression of KLF5 can induce imatinib‐resistant cell apoptosis by increasing oxidative stress. 35
9.12. Ovarian cancer
KLF5 was highly expressed in SKOV3 ovarian cancer cells and was positively correlated with high levels of survivin. 184 Knockdown of KLF5 sensitized SKOV3 cells to cisplatin or paclitaxel treatment. 184
9.13. Cervical cancer
KLF5 is an oncogene in cervical cancer. Among 17 KLF members, only KLF5 mRNA was highly expressed in cervical cancer. 185 The KLF5/TNFRSF11a axis promoted the proliferation, migration and invasion of cervical cancer cells. 39 TNFα induced the expression of KLF5 in cervical cancer by activating the p38 signaling pathway. 39 miR‐152 and miR‐145‐5p inhibited cervical cancer cell proliferation by directly inhibiting the expression of KLF5. 87 , 89 Consistently, LINC00337 maintained tumor stem cell‐like characteristics by downregulating miR‐145 and increasing the expression of KLF5. 88
9.14. Head and neck cancer
The KLF5 gene is frequently amplified in salivary adenomas. 186 Liu et al found for the first time that KLF5 can inhibit the proliferation, survival, and migration of Hep‐2 laryngeal cancer cells. 117 Knockdown of KLF5 inhibited EMT by suppressing the NF‐κB signaling pathway. 117
9.15. Melanoma
In melanoma, KLF5 promoted tumor growth. 103 Clinical data showed that KLF5 was highly expressed in melanoma patients, and the WWP1 gene was also downregulated. 103 It was observed in A2058 melanoma cells that KLF5 knockdown decreased cell proliferation, migration, and invasion, but increased autophagy. 103 The transplanted tumor experiments in mice also further verified the cancer‐promoting effect of KLF5 in melanoma. 103 Previous studies have found that KLF5 is downregulated in Ras mutant melanoma cell lines. 187 The function of KLF5 in melanoma needs further exploration.
9.16. Other cancers
In glioblastoma, KLF5 activated the transcription of the AGGF1 gene to promote angiogenesis. 31 In pituitary adenoma cells, KLF5 upregulated the expression levels of miR‐124, miR‐145, and miR‐148 and inhibited cell migration and invasion. 51 In addition, KLF5 is downregulated in nasopharyngeal carcinoma and Ras mutant melanoma cell lines. 188
10. TARGETED THERAPY
Given the important role of KLF5 in tumorigenesis and development, an increasing number of scientists have tried to target this transcription factor for cancer therapy. It is well known that transcription factors are undruggable to date because of their nuclear localization and the lack of small molecular binding pockets. Therefore, more attention has been given to targeting KLF5 upstream positive regulators and downstream effectors. A recent review comprehensively summarized the compounds regulating the expression of KLF5. 11 Here, we listed a few examples of the latest progress, and as shown in Table 4. Recently, several old drugs were reported to inhibit the expression of KLF5. First, mifepristone inhibits KLF5 expression by inducing miR‐153 to suppress TNBC cell proliferation, survival and CSCs. 75 Two mifepristone‐derived compounds, FZU‐00,003 and FZU‐00,004, showed higher efficiency. 158 , 159 Second, metformin suppressed PKA activity, promoted GSK3β‐mediated KLF5 phosphorylation and degradation and decreased TNBC stem cells. 6 Additionally, mithramycin A inhibited TNBC by inhibiting the binding of Sp1 to the KLF5 promoter and the binding of KLF5 to the FGF‐BP1 promoter. 160 Furthermore, curcumin promoted KLF5 degradation in bladder cancer by inhibiting the transcription of YAP/TAZ. 68 Crocin inhibited KLF5 expression by inducing miR‐320 expression in gastric cancer cells. 83 The deacetylase inhibitor sodium butyrate also inhibited KLF5 expression in colon cancer cells. 173
TABLE 4.
KLF5 targeted compounds
| Compounds | Functional mechanism | References |
|---|---|---|
| CID51003603(ML264) | Inhibit the expression of EGR1 and KLF5 | 128, 190 |
| CID5951923, CID46931043, CID46931037 | Inhibit the expression of EGR1 and KLF5 | 189 |
| Metformin | Promote KLF5 phosphorylation and degradation | 6 |
| Mifepristone | Induce miR‐153 to inhibit KLF5 protein translation | 75 |
| Mithramycin A | Inhibit the binding of Sp1 to the KLF5 promoter | 160 |
| Curcumin | Inhibit YAP/TAZ | 68 |
| Crocin | Induce miR‐320 | 83 |
| JQ‐1 and compound 870 | BRD4 inhibitors | 73 |
| THZ1 | CDK7 inhibitor | 73 |
| Sodium butyrate | Histone deacetylase inhibitor | 173 |
Beyond old drugs, we found that the CDK7 inhibitor THZ1 and BRD4 inhibitor JQ‐1 can efficiently downregulate the transcription of KLF5 and inhibit TNBC cell growth in vitro. 73 JQ‐1‐derived compound 870 showed better efficacy. 73 Using ultrahigh‐throughput screening, CID5951923 and ML264 (CID51003603) were identified to inhibit the expression of KLF5 and the proliferation of CRC cells. 128 , 189 We confirmed that ML264 also inhibited KFL5 expression in breast cancer cells. 190 Interestingly, the ML264‐derived compound YD277 lost this capability, although it triggered ER stress in breast cancers. 190
11. CONCLUSION
In general, KLF5 is a critical transcription factor that controls the transcription of multiple downstream target genes. KLF5 can regulate cell stemness and differentiation, proliferation, apoptosis, and autophagy and participate in a variety of cell physiology and pathological processes such as organ development, tissue regeneration, angiogenesis, and disease development. KLF5 is involved in the initiation and development of diverse cancers in a context‐dependent manner. KLF5 promotes several cancers, such as breast cancer, CRC, bladder cancer, and cervical cancer. However, KLF5 may have dual functions in prostate cancer, gastric cancer, lung cancer, and so on. KLF5 undergoes a variety of posttranslational modifications, such as phosphorylation, acetylation, and ubiquitination. KLF5 is regulated by various signaling pathways, including RTK, Hippo, Wnt, etc. Some small molecules have been identified to inhibit KLF5 expression through different mechanisms.
12. PERSPECTIVES
Given the important roles of KLF5 in cancer initiation and development, we should further understand KLF5 biology, including its physiological and pathological functions, downstream effectors, interacting partners, and upstream regulatory mechanism. Eventually, small molecular modulators should be developed to target this key transcription factor for cancer treatment. In this regard, Klf5 transgenic animal models will be very useful in the future. 191
KLF5 participates in a variety of physiological processes by regulating a variety of downstream target genes. Recent studies have found that KLF5 is involved in glycolysis and lipid metabolism. For example, KLF5 promotes glycolysis, inhibits mitochondrial respiration, and promotes pancreatic tumor growth by upregulating the expression of PLA2G16. 34 KLF5 can interact with SREBP‐1 to regulate the expression of FASN, thereby promoting the proliferation of prostate cancer cells. 138 Additionally, KLF5 plays a vital role in inflammation and tumor immunity. Several inflammatory factors, including TNFα 39 and IL1β, 156 induce KLF5 expression. A recent study reported that KLF5 and SMAD4 inhibited the infiltration of T cells in the tumor immune microenvironment and promoted the infiltration of myeloid cells in pancreatic cancer by upregulating the expression of EGFR. 179 Therefore, the combined immunotherapy of EGFR and reshaping of the immune microenvironment may be a promising targeted therapy strategy. 179 Identification of KLF5 target genes, including non‐coding RNAs, that participate in metabolism and affect the immune microenvironment will provide a better idea how to clarify the role of KLF5 in tumor development. High‐throughput analysis methods, such as ChIP‐seq, Hi‐C‐seq, RNA‐seq, and ATAC‐seq, will be useful to identify the functional mechanism and KLF5 direct target genes from the whole genome landscape.
It is important to identify KLF5 interaction proteins and transcriptional complex components. As a transcription factor, KLF5 does not function alone. For example, KLF5‐TP63‐SOX2, KLF5‐GATA4‐GATA6, and KLF5‐SMAD4‐Miz‐1‐p300 transcriptional complexes have been reported in different cancers. 37 , 56 , 60 KLF5 is induced by androgen through AR, and then KLF5 cooperates with AR to promote the transcription of AR downstream target genes and promote cell proliferation. 32 Epigenetic enzymes, such as p300, HDACs, and BAP1, have been reported to participate in gene transcription in addition to modifying the KLF5 protein. It would be interesting to understand how these enzymes function in gene transcription. Disruption of the KLF5 transcriptional complex may be a promising approach to inhibit its oncogenic functions.
It is crucial to understand the upstream regulatory mechanisms of KLF5 in cancers to design effective targeting strategies. At the genomic level, gene copy number variations and promoter methylation have been shown to regulate KLF5 expression. Some KLF5 upstream transcription factors and cofactors, including Sp1, 160 AR, 32 PR, 108 GR, 112 and EGR1, 128 have been reported in different cancers.
KLF5 mRNA should be regulated by miRNA, lncRNA, alternative splicing, and even RNA modifications. Finally, KLF5 proteins undergo different posttranslational modifications, including the well known phosphorylation, acetylation, ubiquitination, and SUMOylation. New posttranslational modifications of KLF5 will need further investigation.
It is difficult to target a transcription factor. Alternatively, new downstream effectors may be targeted. For example, KLF5 induces the expression of FGF‐BP1, a secretory protein activating FGF signaling. An anti‐FGF‐BP1 antibody was developed to inhibit growth. 192 , 193 Ma et al showed that blocking the KLF5 target IGF1 with antibodies inhibited the STAT3/ matrix metalloproteinase 9 (MMP9) signaling pathway, thereby reducing the invasion of prostate cancer. 53
Taken together, understanding the functions and functional and regulatory mechanisms of KLF5 will help us to better develop more effective targeted therapies for cancer treatment.
DISCLOSURE
Authors have no conflict of interest.
ACKNOWLEDGMENTS
This work was supported by the National Key Research and Development Program of China (2020YFA0112300 and 2018YFC2000400 to C. Chen), National Natural Science Foundation of China (81830087 and 31771516 to C. Chen),and project of Innovative Research Team of Yunnan Province (2019HC005).
Luo Y, Chen C. The roles and regulation of the KLF5 transcription factor in cancers. Cancer Sci. 2021;112:2097–2117. 10.1111/cas.14910
REFERENCES
- 1. McConnell BB, Yang VW. Mammalian Kruppel‐like factors in health and diseases. Physiol Rev. 2010;90:1337‐1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kaczynski J, Cook T, Urrutia R. Sp1‐ and Kruppel‐like transcription factors. Genome Biol. 2003;4:206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dang DT, Pevsner J, Yang VW. The biology of the mammalian Kruppel‐like family of transcription factors. Int J Biochem Cell Biol. 2000;32:1103‐1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chen C, Benjamin MS, Sun X, et al. KLF5 promotes cell proliferation and tumorigenesis through gene regulation and the TSU‐Pr1 human bladder cancer cell line. Int J Cancer. 2006;118:1346‐1355. [DOI] [PubMed] [Google Scholar]
- 5. Wang C, Nie Z, Zhou Z, et al. The interplay between TEAD4 and KLF5 promotes breast cancer partially through inhibiting the transcription of p27Kip1. Oncotarget. 2015;6:17685‐17697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shi P, Liu W, Tala , et al. Metformin suppresses triple‐negative breast cancer stem cells by targeting KLF5 for degradation. Cell Discov. 2017;3:17010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Liu R, Shi P, Zhou Z, et al. Krüpple‐like factor 5 is essential for mammary gland development and tumorigenesis. J Pathol. 2018;246:497‐507. [DOI] [PubMed] [Google Scholar]
- 8. Dong JT, Chen C. Essential role of KLF5 transcription factor in cell proliferation and differentiation and its implications for human diseases. Cell Mol Life Sci. 2009;66:2691‐2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Diakiw SM, D'Andrea RJ, Brown AL. The double life of KLF5: Opposing roles in regulation of gene‐expression, cellular function, and transformation. IUBMB Life. 2013;65:999‐1011. [DOI] [PubMed] [Google Scholar]
- 10. Liu R, Dong JT, Chen C. Role of KLF5 in hormonal signaling and breast cancer development. Vitam Horm. 2013;93:213‐225. [DOI] [PubMed] [Google Scholar]
- 11. Gao Y, Ding Y, Chen H, Chen H, Zhou J. Targeting Krüppel‐like factor 5 (KLF5) for cancer therapy. Curr Top Med Chem. 2015;15:699‐713. [DOI] [PubMed] [Google Scholar]
- 12. Shindo T, Manabe I, Fukushima Y, et al. Krüppel‐like zinc‐finger transcription factor KLF5/BTEB2 is a target for angiotensin II signaling and an essential regulator of cardiovascular remodeling. Nat Med. 2002;8:856‐863. [DOI] [PubMed] [Google Scholar]
- 13. Oishi Y, Manabe I, Tobe K, et al. Krüppel‐like transcription factor KLF5 is a key regulator of adipocyte differentiation. Cell Metab. 2005;1:27‐39. [DOI] [PubMed] [Google Scholar]
- 14. Xing C, Fu X, Sun X, Guo P, Li M, Dong JT. Different expression patterns and functions of acetylated and unacetylated Klf5 in the proliferation and differentiation of prostatic epithelial cells. PLoS One. 2013;8:e65538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhang B, Ci X, Tao R, et al. Klf5 acetylation regulates luminal differentiation of basal progenitors in prostate development and regeneration. Nat Commun. 2020;11:997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. McConnell B, Kim S, Yu K, et al. Krüppel‐like factor 5 is important for maintenance of crypt architecture and barrier function in mouse intestine. Gastroenterology. 2011;141:1302‐1313, 1313.e1301–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bell SM, Zhang L, Xu Y, et al. Kruppel‐like factor 5 controls villus formation and initiation of cytodifferentiation in the embryonic intestinal epithelium. Dev Biol. 2013;375:128‐139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nakaya T, Ogawa S, Manabe I, et al. KLF5 regulates the integrity and oncogenicity of intestinal stem cells. Can Res. 2014;74:2882‐2891. [DOI] [PubMed] [Google Scholar]
- 19. Wan H, Luo F, Wert SE, et al. Kruppel‐like factor 5 is required for perinatal lung morphogenesis and function. Development. 2008;135:2563‐2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Du JX, Bialkowska AB, McConnell BB, Yang VW. SUMOylation regulates nuclear localization of Krüppel‐like factor 5. J Biol Chem. 2008;283:31991‐32002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shao M, Ge GZ, Liu WJ, et al. Characterization and phylogenetic analysis of Krüppel‐like transcription factor (KLF) gene family in tree shrews (Tupaia belangeri chinensis). Oncotarget. 2017;8:16325‐16339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhao D, Zheng HQ, Zhou Z, Chen C. The Fbw7 tumor suppressor targets KLF5 for ubiquitin‐mediated degradation and suppresses breast cell proliferation. Can Res. 2010;70:4728‐4738. [DOI] [PubMed] [Google Scholar]
- 23. Zhi X, Chen C. WWP1: a versatile ubiquitin E3 ligase in signaling and diseases. Cell Mol Life Sci. 2012;69:1425‐1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhang X, Choi PS, Francis JM, et al. Somatic superenhancer duplications and hotspot mutations lead to oncogenic activation of the KLF5 transcription factor. Cancer Discov. 2018;8:108‐125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zheng HQ, Zhou Z, Huang J, Chaudhury L, Dong JT, Chen C. Kruppel‐like factor 5 promotes breast cell proliferation partially through upregulating the transcription of fibroblast growth factor binding protein 1. Oncogene. 2009;28:3702‐3713. [DOI] [PubMed] [Google Scholar]
- 26. Xia H, Wang C, Chen W, et al. Kruppel‐like factor 5 transcription factor promotes microsomal prostaglandin E2 synthase 1 gene transcription in breast cancer. J Biol Chem. 2013;288:26731‐26740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jia L, Zhou Z, Liang H, et al. KLF5 promotes breast cancer proliferation, migration and invasion in part by upregulating the transcription of TNFAIP2. Oncogene. 2016;35:2040‐2051. [DOI] [PubMed] [Google Scholar]
- 28. Zhang L, Wu Y, Wu J, et al. KLF5‐mediated COX2 upregulation contributes to tumorigenesis driven by PTEN deficiency. Cell Signal. 2020;75:109767. [DOI] [PubMed] [Google Scholar]
- 29. Pattison JM, Posternak V, Cole MD. Transcription factor KLF5 binds a cyclin E1 polymorphic intronic enhancer to confer increased bladder cancer risk. Mol Cancer Res. 2016;14:1078‐1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Du C, Gao Y, Xu S, et al. KLF5 promotes cell migration by up‐regulating FYN in bladder cancer cells. FEBS Lett. 2016;590:408‐418. [DOI] [PubMed] [Google Scholar]
- 31. Yang C, Zheng J, Xue Y, et al. The effect of MCM3AP‐AS1/miR‐211/KLF5/AGGF1 axis regulating glioblastoma angiogenesis. Front Mol Neurosci. 2017;10:437. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 32. Li J, Zhang B, Liu M, et al. KLF5 is crucial for androgen‐ar signaling to transactivate genes and promote cell proliferation in prostate cancer cells. Cancers. 2020;12:748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ishikawa ET, Chang KH, Nayak R, et al. Klf5 controls bone marrow homing of stem cells and progenitors through Rab5‐mediated beta 1/beta 2‐integrin trafficking. Nat Commun. 2013;4:1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Xia W, Bai H, Deng Y, Yang Y. PLA2G16 is a mutant p53/KLF5 transcriptional target and promotes glycolysis of pancreatic cancer. J Cell Mol Med. 2020;24:12642‐12655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhang C, D'Alessandro A, Wellendorf AM, et al. KLF5 controls glutathione metabolism to suppress p190‐BCR‐ABL+ B‐cell lymphoblastic leukemia. Oncotarget. 2018;9:29665‐29679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shi QI, Gao Y, Xu S, et al. Krüppel‐like factor 5 promotes apoptosis triggered by tumor necrosis factor α in LNCaP prostate cancer cells via up‐regulation of mitogen‐activated protein kinase kinase 7. Urol Oncol. 2016;34(58):58.e11‐58.e18. [DOI] [PubMed] [Google Scholar]
- 37. Jiang Y‐Y, Jiang Y, Li C‐Q, et al. TP63, SOX2, and KLF5 establish a core regulatory circuitry that controls epigenetic and transcription patterns in esophageal squamous cell carcinoma cell lines. Gastroenterology. 2020;159:1311‐1327.e1319. [DOI] [PubMed] [Google Scholar]
- 38. Jia J, Zhang H‐B, Shi QI, et al. KLF5 downregulation desensitizes castration‐resistant prostate cancer cells to docetaxel by increasing BECN1 expression and inducing cell autophagy. Theranostics. 2019;9:5464‐5477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ma D, Chang L‐Y, Zhao S, et al. KLF5 promotes cervical cancer proliferation, migration and invasion in a manner partly dependent on TNFRSF11a expression. Sci Rep. 2017;7:15683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhao C, Li Y, Qiu W, et al. C5a induces A549 cell proliferation of non‐small cell lung cancer via GDF15 gene activation mediated by GCN5‐dependent KLF5 acetylation. Oncogene. 2018;37:4821‐4837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Li Y, Kong R, Chen H, et al. Overexpression of KLF5 is associated with poor survival and G1/S progression in pancreatic cancer. Aging. 2019;11:5035‐5057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wang Q‐Y, Peng L, Chen Y, et al. Characterization of super‐enhancer‐associated functional lncRNAs acting as ceRNAs in ESCC. Mol Oncol. 2020;14(9):2203‐2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Jia X, Shi L, Wang X, et al. KLF5 regulated lncRNA RP1 promotes the growth and metastasis of breast cancer via repressing p27kip1 translation. Cell Death Dis. 2019;10:373. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 44. Yu L, Xu Q, Yu W, Duan J, Dai G. LncRNA cancer susceptibility candidate 15 accelerates the breast cancer cells progression via miR‐153‐3p/KLF5 positive feedback loop. Biochem Biophys Res Comm. 2018;506:819‐825. [DOI] [PubMed] [Google Scholar]
- 45. Xu T‐P, Ma P, Wang W‐Y, et al. KLF5 and MYC modulated LINC00346 contributes to gastric cancer progression through acting as a competing endogenous RNA and indicates poor outcome. Cell Death Differ. 2019;26:2179‐2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ma P, Pan Y, Yang F, et al. KLF5‐Modulated lncRNA NEAT1 contributes to tumorigenesis by acting as a scaffold for BRG1 to silence GADD45A in gastric cancer. Mol Ther Nucleic Acids. 2020;22:382‐395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Liao QI, Chen L, Zhang N, et al. Network analysis of KLF5 targets showing the potential oncogenic role of SNHG12 in colorectal cancer. Cancer Cell Int. 2020;20:439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Liu Z, Liu X, Liu S, Cao Q. Cholesterol promotes the migration and invasion of renal carcinoma cells by regulating the KLF5/miR‐27a/FBXW7 pathway. Biochem Biophys Res Comm. 2018;502:69‐75. [DOI] [PubMed] [Google Scholar]
- 49. Sun L, Zhou X, Li Y, et al. KLF5 regulates epithelial‐mesenchymal transition of liver cancer cells in the context of p53 loss through miR‐192 targeting of ZEB2. Cell Adh Migr. 2020;14:182‐194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhang B, Zhang Z, Xia S, et al. KLF5 activates microRNA 200 transcription to maintain epithelial characteristics and prevent induced epithelial‐mesenchymal transition in epithelial cells. Mol Cell Biol. 2013;33:4919‐4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Yang W, Xu T, Qiu P, Xu G. Caveolin‐1 promotes pituitary adenoma cells migration and invasion by regulating the interaction between EGR1 and KLF5. Exp Cell Res. 2018;367:7‐14. [DOI] [PubMed] [Google Scholar]
- 52. Zheng B, Han M, Shu YN, et al. HDAC2 phosphorylation‐dependent Klf5 deacetylation and RAR alpha acetylation induced by RAR agonist switch the transcription regulatory programs of p21 in VSMCs. Cell Res. 2011;21:1487‐1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ma J‐B, Bai J‐Y, Zhang H‐B, et al. KLF5 inhibits STAT3 activity and tumor metastasis in prostate cancer by suppressing IGF1 transcription cooperatively with HDAC1. Cell Death Dis. 2020;11:466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Meyer SE, Hasenstein JR, Baktula A, et al. Kruppel‐like factor 5 is not required for K‐RasG12D lung tumorigenesis, but represses ABCG2 expression and is associated with better disease‐specific survival. Am J Pathol. 2010;177:1503‐1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Qin J, Zhou Z, Chen W, et al. BAP1 promotes breast cancer cell proliferation and metastasis by deubiquitinating KLF5. Nat Commun. 2015;6:8471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Chia NY, Deng N, Das K, et al. Regulatory crosstalk between lineage‐survival oncogenes KLF5, GATA4 and GATA6 cooperatively promotes gastric cancer development. Gut. 2015;64:707‐719. [DOI] [PubMed] [Google Scholar]
- 57. Guo P, Dong XY, Zhao KW, Sun X, Li Q, Dong JT. Estrogen‐induced interaction between KLF5 and estrogen receptor (ER) suppresses the function of ER in ER‐positive breast cancer cells. Int J Cancer. 2010;126:81‐89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Nakajima Y, Osakabe A, Waku T, et al. Estrogen exhibits a biphasic effect on prostate tumor growth through the estrogen receptor beta‐KLF5 pathway. Mol Cell Biol. 2016;36:144‐156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Guo P, Xing C, Fu X, He D, Dong JT. Ras inhibits TGF‐β‐induced KLF5 acetylation and transcriptional complex assembly via regulating SMAD2/3 phosphorylation in epithelial cells. J Cell Biochem. 2020;121:2197‐2208. [DOI] [PubMed] [Google Scholar]
- 60. Guo P, Zhao KW, Dong XY, Sun X, Dong JT. Acetylation of KLF5 alters the assembly of p15 transcription factors in transforming growth factor‐beta‐mediated induction in epithelial cells. J Biol Chem. 2009;284:18184‐18193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Li X, Liu X, Xu Y, et al. KLF5 promotes hypoxia‐induced survival and inhibits apoptosis in non‐small cell lung cancer cells via HIF‐1alpha. Int J Oncol. 2014;45:1507‐1514. [DOI] [PubMed] [Google Scholar]
- 62. Chen C, Sun X, Guo P, et al. Human Kruppel‐like factor 5 is a target of the E3 ubiquitin ligase WWP1 for proteolysis in epithelial cells. J Biol Chem. 2005;280:41553‐41561. [DOI] [PubMed] [Google Scholar]
- 63. Zhao KW, Sikriwal D, Dong X, Guo P, Sun X, Dong JT. Oestrogen causes degradation of KLF5 by inducing the E3 ubiquitin ligase EFP in ER‐positive breast cancer cells. Biochem J. 2011;437:323‐333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Du JX, Hagos EG, Nandan MO, Bialkowska AB, Yu B, Yang VW. The E3 ubiquitin ligase SMAD ubiquitination regulatory factor 2 negatively regulates Kruppel‐like factor 5 protein. J Biol Chem. 2011;286:40354‐40364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Ge F, Chen W, Qin J, et al. Ataxin‐3 like (ATXN3L), a member of the Josephin family of deubiquitinating enzymes, promotes breast cancer proliferation by deubiquitinating Krüppel‐like factor 5 (KLF5). Oncotarget. 2015;6:21369‐21378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Wu Y, Qin J, Li F, et al. USP3 promotes breast cancer cell proliferation by deubiquitinating KLF5. J Biol Chem. 2019;294:17837‐17847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Wu Q, Fu C, Li M, et al. CINP is a novel cofactor of KLF5 required for its role in the promotion of cell proliferation, survival and tumor growth. Int J Cancer. 2019;144:582‐594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Gao Y, Shi QI, Xu S, et al. Curcumin promotes KLF5 proteasome degradation through downregulating YAP/TAZ in bladder cancer cells. Int J Mol Sci. 2014;15:15173‐15187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Diakiw SM, Kok CH, To LB, Lewis ID, Brown AL, D'Andrea RJ. The granulocyte‐associated transcription factor Kruppel‐like factor 5 is silenced by hypermethylation in acute myeloid leukemia. Leuk Res. 2012;36:110‐116. [DOI] [PubMed] [Google Scholar]
- 70. Fu R‐J, He W, Wang X‐B, et al. DNMT1‐maintained hypermethylation of Krüppel‐like factor 5 involves in the progression of clear cell renal cell carcinoma. Cell Death Dis. 2017;8:e2952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Pott S, Lieb JD. What are super‐enhancers? Nat Genet. 2015;47:8‐12. [DOI] [PubMed] [Google Scholar]
- 72. Hnisz D, Abraham B, Lee T, et al. Super‐enhancers in the control of cell identity and disease. Cell. 2013;155:934‐947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Chen C‐H, Yang N, Zhang Y, et al. Inhibition of super enhancer downregulates the expression of KLF5 in basal‐like breast cancers. Int J Biol Sci. 2019;15:1733‐1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Zhou W, Song F, Wu Q, et al. miR‐217 inhibits triple‐negative breast cancer cell growth, migration, and invasion through targeting KLF5. PLoS One. 2017;12:e0176395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Liu R, Shi P, Nie Z, et al. Mifepristone suppresses basal triple‐negative breast cancer stem cells by down‐regulating KLF5 expression. Theranostics. 2016;6:533‐544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Zhou T, Chen S, Mao X. miR‐145‐5p affects the differentiation of gastric cancer by targeting KLF5 directly. J Cell Physiol. 2019;234:7634‐7644. [DOI] [PubMed] [Google Scholar]
- 77. Luo S, Ding S, Liao J, et al. Excessive miR‐152‐3p results in increased BAFF expression in SLE B‐cells by inhibiting the KLF5 expression. Front Immunol. 2019;10:1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Jiang Z, Zhang Y, Cao R, et al. miR‐5195‐3p inhibits proliferation and invasion of human bladder cancer cells by directly targeting oncogene KLF5. Oncol Res. 2017;25:1081‐1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Cai W, Xu Y, Yin J, Zuo W, Su Z. miR‐590‐5p suppresses osteosarcoma cell proliferation and invasion via targeting KLF5. Mol Med Rep. 2018;18:2328‐2334. [DOI] [PubMed] [Google Scholar]
- 80. Morimoto Y, Mizushima T, Wu X, et al. miR‐4711‐5p regulates cancer stemness and cell cycle progression via KLF5, MDM2 and TFDP1 in colon cancer cells. Br J Cancer. 2020;122:1037‐1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Pang J, Li Z, Wang G, Li N, Gao Y, Wang S. miR‐214‐5p targets KLF5 and suppresses proliferation of human hepatocellular carcinoma cells. J Cell Biochem. 2019;120:1850‐1859. [DOI] [PubMed] [Google Scholar]
- 82. Guan C, Zhang L, Wang S, et al. Upregulation of MicroRNA‐21 promotes tumorigenesis of prostate cancer cells by targeting KLF5. Cancer Biol Ther. 2019;20:1149‐1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Zhou Y, Xu Q, Shang J, Lu L, Chen G. Crocin inhibits the migration, invasion, and epithelial‐mesenchymal transition of gastric cancer cells via miR‐320/KLF5/HIF‐1alpha signaling. J Cell Physiol. 2019;234:17876‐17885. [DOI] [PubMed] [Google Scholar]
- 84. Shi W, Yang J, Li S, et al. Potential involvement of miR‐375 in the premalignant progression of oral squamous cell carcinoma mediated via transcription factor KLF5. Oncotarget. 2015;6:40172‐40185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Gong GH, An FM, Wang Y, Bian M, Wang D, Wei CX. MiR‐153 regulates expression of hypoxia‐inducible factor‐1alpha in refractory epilepsy. Oncotarget. 2018;9:8542‐8547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Liang H, Ge F, Xu Y, et al. miR‐153 inhibits the migration and the tube formation of endothelial cells by blocking the paracrine of angiopoietin 1 in breast cancer cells. Angiogenesis. 2018;21:849‐860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Cao H, Pan G, Tang S, et al. miR‐145‐5p regulates the proliferation, migration and invasion in cervical carcinoma by targeting KLF5. Onco Targets Ther. 2020;13:2369‐2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Han Q, Wu W, Cui Y. LINC00337 regulates KLF5 and maintains stem‐cell like traits of cervical cancer cells by modulating miR‐145. Front Oncol. 2020;10:1433. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 89. Zhang H, Lu Y, Wang S, Sheng X, Zhang S. MicroRNA‐152 acts as a tumor suppressor microRNA by inhibiting Krüppel‐Like Factor 5 in human cervical cancer. Oncol Res. 2019;27:335‐340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Wang J, Chu Y, Xu M, Zhang X, Zhou Y, Xu M. miR‐21 promotes cell migration and invasion of hepatocellular carcinoma by targeting KLF5. Oncol Lett. 2019;17:2221‐2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Li C, Yan H, Yin J, et al. MicroRNA‐21 promotes proliferation in acute myeloid leukemia by targeting Kruppel‐like factor 5. Oncol Lett. 2019;18:3367‐3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Liu T, Wang X, Zhai J, Wang Q, Zhang B. Long noncoding RNA UCA1 facilitates endometrial cancer development by regulating KLF5 and RXFP1 gene expressions. Cancer Biother Radiopharm. 2020. 10.1089/cbr.2019.3278. [DOI] [PubMed] [Google Scholar]
- 93. Tang J, Li Y, Sang Y, et al. LncRNA PVT1 regulates triple‐negative breast cancer through KLF5/beta‐catenin signaling. Oncogene. 2018;37:4723‐4734. [DOI] [PubMed] [Google Scholar]
- 94. Shan T‐D, Tian Z‐B, Li Q, et al. Long intergenic noncoding RNA 00908 promotes proliferation and inhibits apoptosis of colorectal cancer cells by regulating KLF5 expression. J Cell Physiol. 2021;236:889‐899. [DOI] [PubMed] [Google Scholar]
- 95. Zhang Z, Teng CT. Phosphorylation of Kruppel‐like factor 5 (KLF5/IKLF) at the CBP interaction region enhances its transactivation function. Nucleic Acids Res. 2003;31:2196‐2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. He M, Han M, Zheng B, Shu YN, Wen JK. Angiotensin II stimulates KLF5 phosphorylation and its interaction with c‐Jun leading to suppression of p21 expression in vascular smooth muscle cells. J Biochem. 2009;146:683‐691. [DOI] [PubMed] [Google Scholar]
- 97. Zhang XH, Zheng B, Han M, Miao SB, Wen JK. Synthetic retinoid Am 80 inhibits interaction of KLF5 with RAR alpha through inducing KLF5 dephosphorylation mediated by the PI3K/Akt signaling in vascular smooth muscle cells. FEBS Lett. 2009;583:1231‐1236. [DOI] [PubMed] [Google Scholar]
- 98. Nagai R, Suzuki T, Aizawa K, Shindo T, Manabe I. Significance of the transcription factor KLF5 in cardiovascular remodeling. J Thromb Haemost. 2005;3:1569‐1576. [DOI] [PubMed] [Google Scholar]
- 99. Tao R, Zhang B, Li Y, et al. HDAC‐mediated deacetylation of KLF5 associates with its proteasomal degradation. Biochem Biophys Res Comm. 2018;500:777‐782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Chen C, Sun X, Ran Q, et al. Ubiquitin‐proteasome degradation of KLF5 transcription factor in cancer and untransformed epithelial cells. Oncogene. 2005;24:3319‐3327. [DOI] [PubMed] [Google Scholar]
- 101. Zhi X, Zhao D, Zhou Z, Liu R, Chen C. YAP promotes breast cell proliferation and survival partially through stabilizing the KLF5 transcription factor. Am J Pathol. 2012;180:2452‐2461. [DOI] [PubMed] [Google Scholar]
- 102. Zhao D, Zhi X, Zhou Z, Chen C. TAZ antagonizes the WWP1‐mediated KLF5 degradation and promotes breast cell proliferation and tumorigenesis. Carcinogenesis. 2012;33:59‐67. [DOI] [PubMed] [Google Scholar]
- 103. Jia X, Chen H, Ren Y, et al. BAP1 antagonizes WWP1‐mediated transcription factor KLF5 ubiquitination and inhibits autophagy to promote melanoma progression. Exp Cell Res. 2021;402:112506. [DOI] [PubMed] [Google Scholar]
- 104. Shi H, Zhang Z, Wang X, Liu S, Teng CT. Isolation and characterization of a gene encoding human Kruppel‐like factor 5 (IKLF): binding to the CAAT/GT box of the mouse lactoferrin gene promoter. Nucleic Acids Res. 1999;27:4807‐4815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Fujii Y, Yoshihashi K, Suzuki H, et al. CDX1 confers intestinal phenotype on gastric epithelial cells via induction of stemness‐associated reprogramming factors SALL4 and KLF5. Proc Natl Acad Sci U S A. 2012;109:20584‐20589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Sogawa K, Imataka H, Yamasaki Y, Kusume H, Abe H, Fujii‐Kuriyama Y. cDNA cloning and transcriptional properties of a novel GC box‐binding protein, BTEB2. Nucleic Acids Res. 1993;21:1527‐1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Bell SM, Zhang L, Mendell A, et al. Kruppel‐like factor 5 is required for formation and differentiation of the bladder urothelium. Dev Biol. 2011;358:79‐90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Liu R, Zhou Z, Zhao D, Chen C. The induction of KLF5 transcription factor by progesterone contributes to progesterone‐induced breast cancer cell proliferation and dedifferentiation. Mol Endocrinol. 2011;25:1137‐1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Bourillot PY, Savatier P. Krüppel‐like transcription factors and control of pluripotency. BMC Biol. 2010;8:125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. David CJ, Huang YH, Chen M, et al. TGF‐beta tumor suppression through a lethal EMT. Cell. 2016;164:1015‐1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Liu R, Zheng HQ, Zhou Z, Dong JT, Chen C. KLF5 promotes breast cell survival partially through fibroblast growth factor‐binding protein 1‐pERK‐mediated dual specificity MKP‐1 protein phosphorylation and stabilization. J Biol Chem. 2009;284:16791‐16798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Li Z, Dong J, Zou TN, et al. Dexamethasone induces docetaxel and cisplatin resistance‐partially through up‐regulating Kruppel‐like factor 5 in triple‐negative breast cancer. Oncotarget. 2017;8:11555‐11565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Zhang H, Shao F, Guo W, Gao Y, He J. Knockdown of KLF5 promotes cisplatin‐induced cell apoptosis via regulating DNA damage checkpoint proteins in non‐small cell lung cancer. Thorac Cancer. 2019;10:1069‐1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Wang Z, Qiu X, Zhang H, Li W. KLF5 influences cell biological function and chemotherapy sensitivity through the JNK signaling pathway in anaplastic thyroid carcinoma. J Biochem Mol Toxicol. 2020;34:e22469. [DOI] [PubMed] [Google Scholar]
- 115. Tarapore RS, Yang Y, Katz JP. Restoring KLF5 in esophageal squamous cell cancer cells activates the JNK pathway leading to apoptosis and reduced cell survival. Neoplasia. 2013;15:472‐480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Shi QI, Jia J, Hui KE, et al. KLF5 promotes apoptosis induced by phorbol ester as an effector of the autocrine factor TNFα in LNCaP prostate cancer cells. Oncol Lett. 2017;14:1847‐1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Liu FF, Dong L, Yang X, Li DJ, Shen YY, Liu ZL. KLF5 silence attenuates proliferation and epithelial‐mesenchymal transition induction in Hep‐2 cells through NF‐κB signaling pathway. Eur Rev Med Pharmacol Sci. 2019;23:3867‐3875. [DOI] [PubMed] [Google Scholar]
- 118. Ma Y, Wang Q, Liu F, et al. KLF5 promotes the tumorigenesis and metastatic potential of thyroid cancer cells through the NF‐kappaB signaling pathway. Oncol Rep. 2018;40:2608‐2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Wang F, Luo M, Qu H, Cheng Y. BAP1 promotes viability and migration of ECA109 cells through KLF5/CyclinD1/FGF‐BP1. FEBS Open Bio. 2021. 10.1002/2211-5463.13105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Shimamura T, Imoto S, Shimada Y, et al. A novel network profiling analysis reveals system changes in epithelial‐mesenchymal transition. PLoS One. 2011;6:e20804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. King JL, Zhang B, Li Y, et al. TTK promotes mesenchymal signaling via multiple mechanisms in triple negative breast cancer. Oncogenesis. 2018;7:69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Yang Y, Nakagawa H, Tetreault MP, et al. Loss of transcription factor KLF5 in the context of p53 ablation drives invasive progression of human squamous cell cancer. Cancer Res. 2011;71:6475‐6484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. McConnell BB, Bialkowska AB, Nandan MO, Ghaleb AM, Gordon FJ, Yang VW. Haploinsufficiency of Krüppel‐like factor 5 rescues the tumor‐initiating effect of the Apc(Min) mutation in the intestine. Cancer Res. 2009;69:4125‐4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Kim C‐K, Saxena M, Maharjan K, et al. Krüppel‐like Factor 5 regulates stemness, lineage specification, and regeneration of intestinal epithelial stem cells. Cell Mol Gastroenterol Hepatol. 2020;9:587‐609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Goswami S, Gao N. KLF5 governs stemness in the adult intestinal stem cell niche. Cell Mol Gastroenterol Hepatol. 2020;9:705‐706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Wei X, Ye J, Shang Y, et al. Ascl2 activation by YAP1/KLF5 ensures the self‐renewability of colon cancer progenitor cells. Oncotarget. 2017;8:109301‐109318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Gao Y, Wu K, Chen Y, et al. Beyond proliferation: KLF5 promotes angiogenesis of bladder cancer through directly regulating VEGFA transcription. Oncotarget. 2015;6:43791‐43805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Ruiz de Sabando A, Wang C, He Y, et al. ML264, a novel small‐molecule compound that potently inhibits growth of colorectal cancer. Mol Cancer Ther. 2016;15:72‐83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. He P, Yang JW, Yang VW, Bialkowska AB. Kruppel‐like Factor 5, increased in pancreatic ductal adenocarcinoma, promotes proliferation, acinar‐to‐ductal metaplasia, pancreatic intraepithelial neoplasia, and tumor growth in mice. Gastroenterology. 2018;154:1494‐1508.e1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Azami T, Matsumoto K, Jeon H, et al. Klf5 suppresses ERK signaling in mouse pluripotent stem cells. PLoS One. 2018;13:e0207321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Gong T, Cui L, Wang H, Wang H, Han N. Knockdown of KLF5 suppresses hypoxia‐induced resistance to cisplatin in NSCLC cells by regulating HIF‐1α‐dependent glycolysis through inactivation of the PI3K/Akt/mTOR pathway. J Transl Med. 2018;16:164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. An T, Dong T, Zhou H, et al. The transcription factor Kruppel‐like factor 5 promotes cell growth and metastasis via activating PI3K/AKT/Snail signaling in hepatocellular carcinoma. Biochem Biophys Res Comm. 2019;508:159‐168. [DOI] [PubMed] [Google Scholar]
- 133. Zhang Z, Luo G, Yu C, Yu G, Jiang R, Shi X. MicroRNA‐493‐5p inhibits proliferation and metastasis of osteosarcoma cells by targeting Kruppel‐like factor 5. J Cell Physiol. 2019;234:13525‐13533. [DOI] [PubMed] [Google Scholar]
- 134. Yang H, Chen Q, Sun F, et al. Down‐regulation of the klf5‐c‐Myc interaction due to klf5 phosphorylation mediates resveratrol repressing the caveolin‐1 transcription through the PI3K/PKD1/Akt pathway. PLoS One. 2017;12:e0189156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Ci X, Xing C, Zhang B, et al. KLF5 inhibits angiogenesis in PTEN‐deficient prostate cancer by attenuating AKT activation and subsequent HIF1alpha accumulation. Mol Cancer. 2015;14:91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Huang H, Han Y, Chen Z, et al. ML264 inhibits osteosarcoma growth and metastasis via inhibition of JAK2/STAT3 and WNT/beta‐catenin signalling pathways. J Cell Mol Med. 2020;24:5652‐5664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Frigo DE, Sherk AB, Wittmann BM, et al. Induction of Kruppel‐like factor 5 expression by androgens results in increased CXCR4‐dependent migration of prostate cancer cells in vitro. Mol Endocrinol. 2009;23:1385‐1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Lee MY, Moon JS, Park SW, Koh YK, Ahn YH, Kim KS. KLF5 enhances SREBP‐1 action in androgen‐dependent induction of fatty acid synthase in prostate cancer cells. Biochem J. 2009;417:313‐322. [DOI] [PubMed] [Google Scholar]
- 139. Takagi K, Miki Y, Onodera Y, et al. Kruppel‐like factor 5 in human breast carcinoma: a potent prognostic factor induced by androgens. Endocr Relat Cancer. 2012;19:741‐750. [DOI] [PubMed] [Google Scholar]
- 140. Park YK, Ge K. Glucocorticoid receptor accelerates, but is dispensable for, adipogenesis. Mol Cell Biol. 2017;37:e00260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. He L, Yuan L, Sun Y, et al. Glucocorticoid receptor signaling activates TEAD4 to promote breast cancer progression. Can Res. 2019;79:4399‐4411. [DOI] [PubMed] [Google Scholar]
- 142. Sorrentino G, Ruggeri N, Zannini A, et al. Glucocorticoid receptor signalling activates YAP in breast cancer. Nat Commun. 2017;8:14073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Guo P, Dong XY, Zhao K, Sun X, Li Q, Dong JT. Opposing effects of KLF5 on the transcription of MYC in epithelial proliferation in the context of transforming growth factor beta. J Biol Chem. 2009;284:28243‐28252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Shi P, Feng J, Chen C. Hippo pathway in mammary gland development and breast cancer. Acta Biochim Biophys Sin. 2015;47:53‐59. [DOI] [PubMed] [Google Scholar]
- 145. Yumimoto K, Nakayama KI. Recent insight into the role of FBXW7 as a tumor suppressor. Semin Cancer Biol. 2020;67:1‐15. [DOI] [PubMed] [Google Scholar]
- 146. Chanchevalap S, Nandan MO, McConnell BB, et al. Kruppel‐like factor 5 is an important mediator for lipopolysaccharide‐induced proinflammatory response in intestinal epithelial cells. Nucleic Acids Res. 2006;34:1216‐1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Chen HL, Chong IW, Lee YC, et al. Kruppel‐Like Factor 5 mediates proinflammatory cytokine expression in lipopolysaccharide‐induced acute lung injury through upregulation of nuclear factor‐kappa b phosphorylation in vitro and in vivo. Mediat Inflamm. 2014;2014:281984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Jia L, Shi Y, Wen Y, Li W, Feng J, Chen C. The roles of TNFAIP2 in cancers and infectious diseases. J Cell Mol Med. 2018;22:5188‐5195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Ng CK, Ma K, Cheng Y, Miyashita T, Harmon JW, Meltzer SJ. Kruppel‐like factor 5 promotes sonic hedgehog signaling and neoplasia in Barrett's esophagus and esophageal adenocarcinoma. Transl Oncol. 2019;12:1432‐1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Zhao Y, Hamza MS, Leong HS, et al. Kruppel‐like factor 5 modulates p53‐independent apoptosis through Pim1 survival kinase in cancer cells. Oncogene. 2008;27:1‐8. [DOI] [PubMed] [Google Scholar]
- 151. Li M, Gu Y, Ma Y‐C, et al. Krüppel‐like factor 5 promotes epithelial proliferation and DNA damage repair in the intestine of irradiated mice. Int J Biol Sci. 2015;11:1458‐1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Yang Y, Goldstein BG, Chao HH, Katz JP. KLF4 and KLF5 regulate proliferation, apoptosis and invasion in esophageal cancer cells. Cancer Biol Ther. 2005;4:1216‐1221. [DOI] [PubMed] [Google Scholar]
- 153. Suzuki T, Nishi T, Nagino T, et al. Functional interaction between the transcription factor Kruppel‐like factor 5 and poly(ADP‐ribose) polymerase‐1 in cardiovascular apoptosis. J Biol Chem. 2007;282:9895‐9901. [DOI] [PubMed] [Google Scholar]
- 154. Li X, He Y, Xu Y, et al. KLF5 mediates vascular remodeling via HIF‐1alpha in hypoxic pulmonary hypertension. Am J Physiol Lung Cell Mol Physiol. 2016;310:L299‐310. [DOI] [PubMed] [Google Scholar]
- 155. Harris AL. Hypoxia–a key regulatory factor in tumour growth. Nat Rev Cancer. 2002;2:38‐47. [DOI] [PubMed] [Google Scholar]
- 156. Mori A, Moser C, Lang SA, et al. Up‐regulation of Kruppel‐Like Factor 5 in pancreatic cancer is promoted by interleukin‐1 beta signaling and hypoxia‐inducible factor‐1 alpha. Mol Cancer Res. 2009;7:1390‐1398. [DOI] [PubMed] [Google Scholar]
- 157. Lee SJ, No YR, Dang DT, et al. Regulation of hypoxia‐inducible factor 1alpha (HIF‐1alpha) by lysophosphatidic acid is dependent on interplay between p53 and Kruppel‐like factor 5. J Biol Chem. 2013;288:25244‐25253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Liu R, Chen H, Zhao P, et al. Mifepristone derivative FZU‐00,003 suppresses triple‐negative breast cancer cell growth partially via miR‐153‐KLF5 axis. Int J Biol Sci. 2020;16:611‐619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Lin Y, Liu R, Zhao P, et al. Discovery of novel mifepristone derivatives via suppressing KLF5 expression for the treatment of triple‐negative breast cancer. Eur J Med Chem. 2018;146:354‐367. [DOI] [PubMed] [Google Scholar]
- 160. Liu R, Zhi X, Zhou Z, et al. Mithramycin A suppresses basal triple‐negative breast cancer cell survival partially via down‐regulating Kruppel‐like factor 5 transcription by Sp1. Sci Rep. 2018;8:1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Yagi N, Manabe I, Tottori T, et al. A nanoparticle system specifically designed to deliver short interfering RNA inhibits tumor growth in vivo. Can Res. 2009;69:6531‐6538. [DOI] [PubMed] [Google Scholar]
- 162. Zhang B, Li Y, Wu Q, et al. Acetylation of KLF5 maintains EMT and tumorigenicity to cause chemoresistant bone metastasis in prostate cancer. Nat Commun. 2021;12:1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Chen C, Bhalala HV, Vessella RL, Dong JT. KLF5 is frequently deleted and down‐regulated but rarely mutated in prostate cancer. Prostate. 2003;55:81‐88. [DOI] [PubMed] [Google Scholar]
- 164. Xing C, Ci X, Sun X, et al. Klf5 deletion promotes Pten deletion‐initiated luminal‐type mouse prostate tumors through multiple oncogenic signaling pathways. Neoplasia. 2014;16:883‐899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Li X, Zhang B, Wu Q, et al. Interruption of KLF5 acetylation converts its function from tumor suppressor to tumor promoter in prostate cancer cells. Int J Cancer. 2015;136:536‐546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Nan C, Wang Y, Yang S, Chen Y. circCRKL suppresses the progression of prostate cancer cells by regulating the miR‐141/KLF5 axis. Pathol Res Pract. 2020;216:153182. [DOI] [PubMed] [Google Scholar]
- 167. Peña‐Llopis S, Vega‐Rubín‐de‐Celis S, Liao A, et al. BAP1 loss defines a new class of renal cell carcinoma. Nat Genet. 2012;44:751‐759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. McConnell BB, Kim SS, Bialkowska AB, Yu K, Sitaraman SV, Yang VW. Kruppel‐like factor 5 protects against dextran sulfate sodium‐induced colonic injury in mice by promoting epithelial repair. Gastroenterology. 2011;140:540‐549.e542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Nandan MO, Ghaleb AM, McConnell BB, Patel NV, Robine S, Yang VW. Krüppel‐like factor 5 is a crucial mediator of intestinal tumorigenesis in mice harboring combined ApcMin and KRASV12 mutations. Mol Cancer. 2010;9:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Nandan MO, Bialkowska AB, Yang VW. KLF5 mediates the hyper‐proliferative phenotype of the intestinal epithelium in mice with intestine‐specific endogenous K‐Ras(G12D) expression. Am J Cancer Res. 2018;8:723‐731. [PMC free article] [PubMed] [Google Scholar]
- 171. Bialkowska AB, Liu Y, Nandan MO, Yang VW. A colon cancer‐derived mutant of Krüppel‐like factor 5 (KLF5) is resistant to degradation by glycogen synthase kinase 3β (GSK3β) and the E3 ubiquitin ligase F‐box and WD repeat domain‐containing 7α (FBW7α). J Biol Chem. 2014;289:5997‐6005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Pagliuca A, Valvo C, Fabrizi E, et al. Analysis of the combined action of miR‐143 and miR‐145 on oncogenic pathways in colorectal cancer cells reveals a coordinate program of gene repression. Oncogene. 2013;32:4806‐4813. [DOI] [PubMed] [Google Scholar]
- 173. Shin J, Carr A, Corner GA, et al. The intestinal epithelial cell differentiation marker intestinal alkaline phosphatase (ALPi) is selectively induced by histone deacetylase inhibitors (HDACi) in colon cancer cells in a Kruppel‐like factor 5 (KLF5)‐dependent manner. J Biol Chem. 2014;289:25306‐25316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Daly JM, Fry WA, Little AG, et al. Esophageal cancer: results of an American College of Surgeons Patient Care Evaluation Study. J Am Coll Surg. 2000;190:562‐572; discussion 572–563. [DOI] [PubMed] [Google Scholar]
- 175. Kwak MK, Lee HJ, Hur K, et al. Expression of Kruppel‐like factor 5 in human gastric carcinomas. J Cancer Res Clin Oncol. 2008;134:163‐167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Zhang Z, Zhu X. Clinical significance of Lysophosphatidic Acid Receptor‐2 (LPA2) and Kruppel‐Like Factor 5 (KLF5) protein expression detected by tissue microarray in gastric adenocarcinoma. Med Sci Monit. 2019;25:4705‐4715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Yang T, Chen M, Yang X, et al. Down‐regulation of KLF5 in cancer‐associated fibroblasts inhibit gastric cancer cells progression by CCL5/CCR5 axis. Cancer Biol Ther. 2017;18:806‐815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Liang H, Sun H, Yang J, Yi C. miR‐145‐5p reduces proliferation and migration of hepatocellular carcinoma by targeting KLF5. Mol Med Rep. 2018;17:8332‐8338. [DOI] [PubMed] [Google Scholar]
- 179. Li J, Yuan S, Norgard RJ, et al. Epigenetic and transcriptional control of the epidermal growth factor receptor (EGFR) regulates the tumor immune microenvironment in pancreatic cancer. Cancer Discov. 2021;11(3):736‐753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Li Q, Dong Z, Zhou F, Cai X, Gao Y, Wang LW. Kruppel‐like factor 5 promotes lung tumorigenesis through upregulation of Sox4. Cell Physiol Biochem. 2014;33:1‐10. [DOI] [PubMed] [Google Scholar]
- 181. Zhu N, Gu L, Findley HW, et al. KLF5 Interacts with p53 in regulating survivin expression in acute lymphoblastic leukemia. J Biol Chem. 2006;281:14711‐14718. [DOI] [PubMed] [Google Scholar]
- 182. Humbert M, Halter V, Shan D, et al. Deregulated expression of Kruppel‐like factors in acute myeloid leukemia. Leuk Res. 2011;35:909‐913. [DOI] [PubMed] [Google Scholar]
- 183. Diakiw SM, Perugini M, Kok CH, et al. Methylation of KLF5 contributes to reduced expression in acute myeloid leukaemia and is associated with poor overall survival. Br J Haematol. 2013;161:884‐888. [DOI] [PubMed] [Google Scholar]
- 184. Dong Z, Yang L, Lai D. KLF5 strengthens drug resistance of ovarian cancer stem‐like cells by regulating survivin expression. Cell Prolif. 2013;46:425‐435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Marrero‐Rodriguez D, Taniguchi‐Ponciano K, Jimenez‐Vega F, et al. Kruppel‐like factor 5 as potential molecular marker in cervical cancer and the KLF family profile expression. Tumour Biol. 2014;35:11399‐11407. [DOI] [PubMed] [Google Scholar]
- 186. Giefing M, Wierzbicka M, Rydzanicz M, Cegla R, Kujawski M, Szyfter K. Chromosomal gains and losses indicate oncogene and tumor suppressor gene candidates in salivary gland tumors. Neoplasma. 2008;55:55‐60. [PubMed] [Google Scholar]
- 187. Bloethner S, Chen BW, Hemminki K, et al. Effect of common B‐RAF and N‐RAS mutations on global gene expression in melanoma cell lines. Carcinogenesis. 2005;26:1224‐1232. [DOI] [PubMed] [Google Scholar]
- 188. Fang W, Li X, Jiang Q, et al. Transcriptional patterns, biomarkers and pathways characterizing nasopharyngeal carcinoma of Southern China. J Transl Med. 2008;6:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Bialkowska AB, Crisp M, Bannister T, et al. Identification of small‐molecule inhibitors of the colorectal cancer oncogene Kruppel‐like factor 5 expression by ultrahigh‐throughput screening. Mol Cancer Ther. 2011;10:2043‐2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Chen Z, Wu Q, Ding YE, et al. YD277 suppresses triple‐negative breast cancer partially through activating the endoplasmic reticulum stress pathway. Theranostics. 2017;7:2339‐2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Zeng L, Li W, Chen CS. Breast cancer animal models and applications. Zool Res. 2020;41:477‐494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Tassi E, Wellstein A. Tumor angiogenesis: initiation and targeting ‐ therapeutic targeting of an FGF‐binding protein, an angiogenic switch molecule, and indicator of early stages of gastrointestinal adenocarcinomas. Cancer Res Treat. 2006;38:189‐197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Tassi E, Henke RT, Bowden ET, et al. Expression of a fibroblast growth factor‐binding protein during the development of adenocarcinoma of the pancreas and colon. Cancer Res. 2006;66:1191‐1198. [DOI] [PubMed] [Google Scholar]
- 194. Shinoda Y, Ogata N, Higashikawa A, et al. Kruppel‐like factor 5 causes cartilage degradation through transactivation of matrix metalloproteinase 9. J Biol Chem. 2008;283:24682‐24689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Shahrin NH, Diakiw S, Dent LA, Brown AL, D'Andrea RJ. Conditional knockout mice demonstrate function of Klf5 as a myeloid transcription factor. Blood. 2016;128:55‐59. [DOI] [PubMed] [Google Scholar]
- 196. Kenchegowda D, Swamynathan S, Gupta D, Wan H, Whitsett J, Swamynathan SK. Conditional disruption of mouse Klf5 results in defective eyelids with malformed meibomian glands, abnormal cornea and loss of conjunctival goblet cells. Dev Biol. 2011;356:5‐18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. Kenchegowda D, Harvey SA, Swamynathan S, Lathrop KL, Swamynathan SK. Critical role of Klf5 in regulating gene expression during post‐eyelid opening maturation of mouse corneas. PLoS One. 2012;7:e44771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. Sur I, Rozell B, Jaks V, Bergström A, Toftgård R. Epidermal and craniofacial defects in mice overexpressing Klf5 in the basal layer of the epidermis. J Cell Sci. 2006;119:3593‐3601. [DOI] [PubMed] [Google Scholar]
- 199. Goldstein BG, Chao HH, Yang Y, Yermolina YA, Tobias JW, Katz JP. Overexpression of Kruppel‐like factor 5 in esophageal epithelia in vivo leads to increased proliferation in basal but not suprabasal cells. Am J Physiol Gastrointest Liver Physiol. 2007;292:G1784‐1792. [DOI] [PubMed] [Google Scholar]
- 200. Nakajima Y, Akaogi K, Suzuki T, et al. Estrogen regulates tumor growth through a nonclassical pathway that includes the transcription factors ERβ and KLF5. Sci Signal. 2011;4:ra22. [DOI] [PubMed] [Google Scholar]


