Abstract
Nucleic acid nanoparticles (NANPs) represent a highly versatile molecular platform for the targeted delivery of various therapeutics. However, despite their promise, further clinical translation of this innovative technology can be hindered by immunological off-target effects. All human cells are equipped with an arsenal of receptors that recognize molecular patterns specific to foreign nucleic acids and understanding the rules that guide this recognition offer the key rationale for the development of therapeutic NANPs with tunable immune stimulation. Numerous recent studies have provided increasing evidence that in addition to NANPs’ physicochemical properties and therapeutic effects, their interactions with cells of the immune system can be regulated through multiple independently programmable architectural parameters. The results further suggest that defined immunomodulation by NANPs can either support their immunoquiescent delivery or be used for conditional stimulation of beneficial immunological responses.
Graphical Abstract
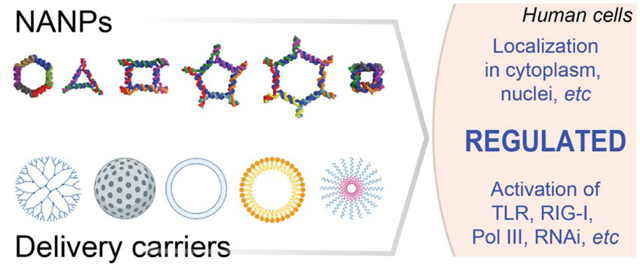
Combinations of different Nucleic Acid Nanoparticles (NANPs) and delivery agents allows to direct desirable and avoid undesirable immunological effects and therapeutic actions.
Versatile Therapeutic Nucleic Acids (TNAs).
In the makeup of a traditional small molecule drug, the dianophore, which is the molecular component determining distribution and delivery, and the pharmacophore, which is the molecular component determining targeted function, are separate activities which are both determined by the drug’s overall chemical structure. Any changes to the makeup in the drug design process can therefore mean for divergent effects between the dianophore and pharmacophore activities, which must be clinically reevaluated for every stage of the drug’s development[1]. Instead, modular therapeutic nucleic acids (TNAs) offer a means of separation between these two facets, because the backbone chemistry and targeting moieties which serve as the dianophore can be used in combination with different nucleotide sequences serving as the pharmacophores with functional independence[1, 2]. Therefore, clinical evaluations of TNAs contribute to a foundation of preliminary bioactivity not only for the individual TNA candidate, but for any TNA with that modular piece in its formulation. Overall, this information and enhanced prediction of biological activity serves to greatly increase the safety profiles of TNA formulations and also has the potential to decrease the overall costs associated with the broad amount of drug candidates entering the clinical pipeline. With 156 TNAs currently undergoing phases I-III clinical trials or awaiting regulatory decisions as of April 2020, this innovative class of therapeutics holds great promise for the regulation of cells to eliminate specific diseases or enhance responses such as for cancer immunotherapy[3, 4].
With the decoding of the human genome and increasing utility in high-throughput sequencing, there has been a growing potential of TNAs to be sequence-specific candidates for targeted gene therapy[5]. A number of TNAs which vary in mechanism have been approved by the U.S. Food and Drug Administration (FDA), including antisense oligonucleotides (ASOs), mRNAs, siRNAs, miRNAs, and aptamers[6] (Fig. 1). ASOs are short sequences designed to bind to specific RNAs to promote their degradation, cleavage, or steric blocking [7]. To date, there are six antisense oligonucleotides which have been approved by the U.S. FDA: three RNase H-competent ASOs (Fomivirsen, Mipomersen, and Inotersen) and three splice-switching ASOs (Eteplirsen, Golodirsen, and Nusinersen)[6]. Two siRNA candidates (Patisiran and Givosiran), which work by undergoing enzyme-mediated RNA interference for post-transcriptional gene silencning, have recently been FDA-approved. There is also one aptamer (Pegaptanib) currently approved[6]. Aptamers are oligonucleotides selected in vitro to specifically recognize and bind a target molecule with high specificity—in this case, as an agonist to prevent angiogenesis[8]. Recently, the versatility of a nucleic acid-based approach has been further expanded by the rapid development and ongoing worldwide distribution of mRNA-based vaccines (BNT162b2 and mRNA-1273) against SARS-CoV-2[9].
Figure 1: Examples of mechanisms of TNA action.
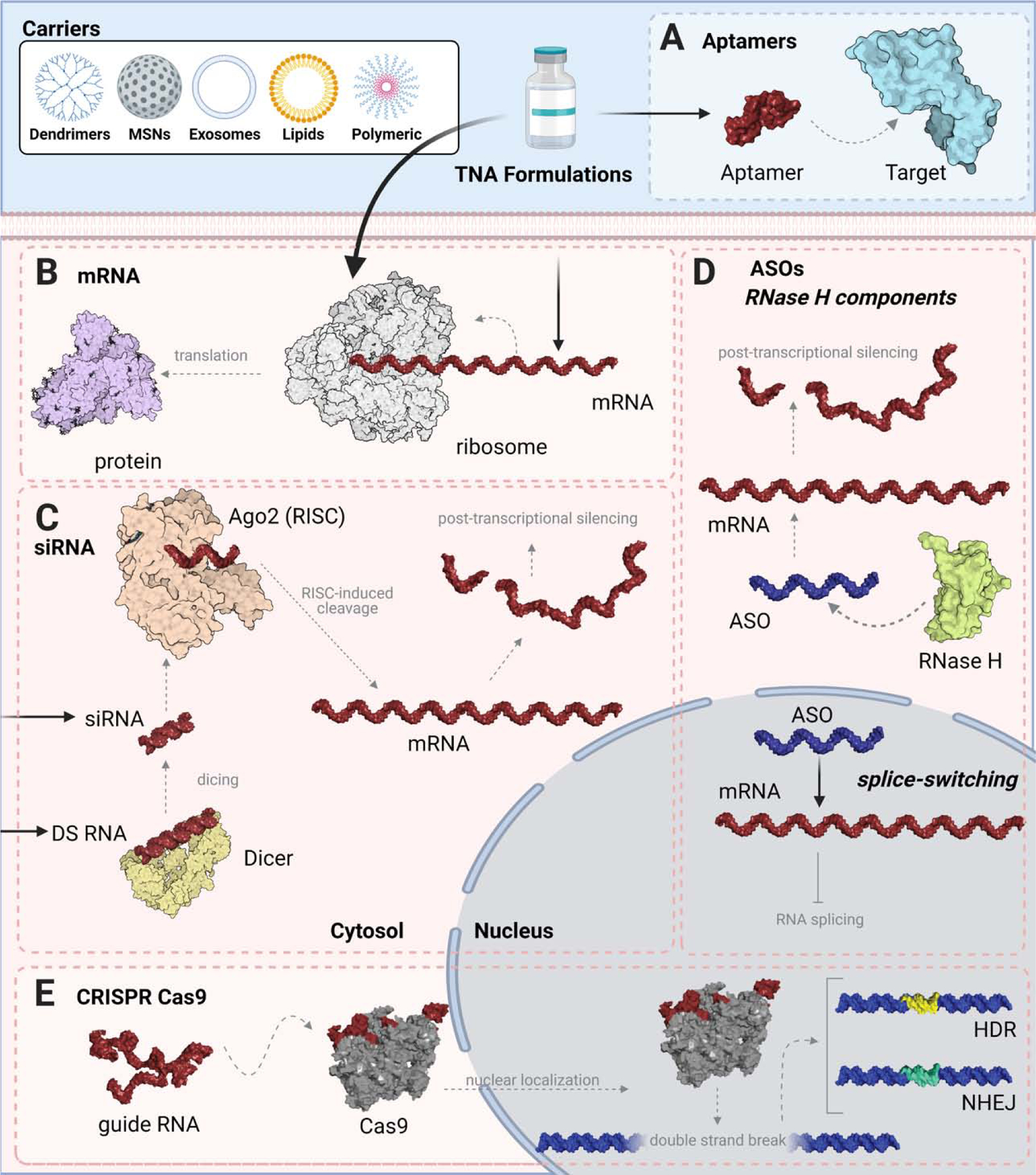
For efficient intracellular delivery, some TNAs require a carrier with several of them exemplified in the upper panel. (A) Aptamers, composed of either DNA, RNA or their chemical analogs, function by binding a specific target molecule. As an example, Pegaptanib[14] (shown using RNAComposer[15, 16]) is schematically shown to bind to VEGF (PDB: 1VPF) for its inhibition to prevent downstream angiogenesis. (B) Delivery of mRNA into the cytoplasm is translated via the ribosome (PDB: 6Y0G) to yield a protein of interest. Spike protein from SARS-CoV-2 (PDB: 6VXX) is shown as a protein product example of mRNA vaccines. (C) For RNAi-induced gene silencing, either Dicer Substrate (DS) RNAs may be introduced for processing by Dicer, or siRNAs may be introduced exogenously. siRNAs are incorporated into the RNA-induced silencing complex (RISC) and guide strands direct sequence-specific mRNA cleavage. For illustration purposes, only the Ago2 component of RISC is shown (PDB: 6CBD). (D) ASOs bind to an endogenous mRNA sequence, where they may act as a steric hinderance for further splicing and translation, or may serve as a target for degradation by RNase H (PDB: 2QK9). (E) CRISPR Cas9 (PDB: 5F9R) utilizes a guide RNA sequence as a template to promote the double strand breakage of a gene. Repair mechanisms including homology directed repair (HDR) or non-homologous end joining (NHEJ) can be implemented for gene editing. Created with Biorender.com
Immunorecognition of TNAs.
Currently available TNAs are often offered as periodic doses, perform a single function, and may present a high recurring cost to patients[10]. The scaled-up production of nucleic acids while maintaining high purity is a challenge[11]. Furthermore, many TNAs require chemical modifications in order to maintain nuclease degradation resistance, cross the plasma membrane, and avoid unwanted side effects from off-target immunostimulation[12]. Pattern recognition receptors (PRRs) are strategically localized to the endosome and cytosol for the targeted recognition of pathogen associated molecular patterns (PAMPs) or damage associated molecular patterns (DAMPs) which allow for the discrimination of self- and non-self-biomaterials. These PRRs include the endosomal Toll-Like Receptors (TLRs)—TLR3, TLR7, TLR8, and TLR9—and the cytosolic PRRs, such as RIG-I-like receptors (RLRs) and DNA sensors. Importantly, activation of these PRRs stimulates downstream signaling cascades that trigger the production of immune mediators. The immune mediators promote cellular recruitment, cellular maturation, and antigen presentation to coordinate adaptive immune responses. While potential TNAs in the translational pipeline may be impeded by the prominence of these nucleic acid receptors, they also have the opportunity to utilize these natural pathways for the favorable activation of cytokine and interferon production. Favorable immunostimulation can be directed to invoke responses for use in immunotherapy, vaccine adjuvants, and antigen delivery[13].
Pattern Recognition Receptor Ligand Characteristics.
TNAs offer a unique platform for the strategic design of immunomodulatory formulations based on the known characteristics of PRR ligands. PRRs discerningly identify nucleic acid ligands due to subcellular localization, composition, nucleoside characteristics, structure, length, and sequence motifs (Table 1). Although PRRs primarily recognize nucleic acids through interaction with their sugar-phosphate backbone in a sequence-independent manner, these sensors are still able to distinguish some characteristic features of nucleic acid ligands.
Table 1.
PRR ligand characteristics and clinical applications.
| Receptor | Ligand Characteristics | Potential Therapeutic Applications | ||||
|---|---|---|---|---|---|---|
| Localization | Nucleic Acid | Sequence | Length (bp) | Agonist | Antagonist | |
| TLR3 | Endosome | dsRNA | Independent | >45 | Antiviral/Adjuvant (e.g., HIV[55, 56], influenza[57, 58], HPV[59–61]), Cancer (e.g., colorectal, melanoma, mammary, prostate[62, 63]) | Inflammatory disease and Autoimmunity (e.g., SLE[63, 64]) |
| TLR7/8 | Endosome | ssRNA, dsRNA, small molecules | GU and AU rich | >19 | Antiviral/Adjuvant (e.g., influenza, hepatitis C[27, 65, 66]), Cancer (e.g., melanoma, colon, lymphoma[67, 68]), Allergy/Asthma[27, 63] | Inflammatory disease and Autoimmunity (e.g., atherosclerosis, SLE ([27, 69, 70]) |
| TLR9 | Endosome | dsDNA; preference for curved DNA | Independent preference: CpG | >15 | Antiviral/Adjuvant (e.g., malaria[71, 72], HIV[73, 74], hepatitis C[65]) Cancer (e.g., melanoma[75], lymphoma[76], colon[77, 78]), Asthma [63] | Inflammatory disease and Autoimmunity (e.g., SLE[64], HIV[79], psoriasis[80]) |
| cGAS | Cytosol | dsDNA, DNA/RNA hybrid, Y-form | Independent | >25 | STING: Adjuvant (e.g., influenza [81, 82], coronaviruses[83, 84]) Cancer (e.g., solid tumors, prostate, lymphoma [85, 86]) | STING: Inflammatory disease and Autoimmunity ([87] |
| IFI16 | Cytosol Nucleus | dsDNA | Independent | >70; optimal 150–200 | STING: Adjuvant (e.g., influenza [81, 82], coronaviruses[83, 84]) Cancer (e.g., solid tumors, prostate, lymphoma [85, 86]) | STING: Inflammatory disease and Autoimmunity ([87] |
| AIM2 | Cytosol | dsDNA | Independent | >80 optimal 280 | Cancer (e.g., colon [88, 89]) | Inflammatory disease and Autoimmunity (e.g. psoriasis, atherosclerosis, neuroinflammation) Cancer (e.g., cutaneous squamous cell carcinoma, melanoma, CAR-T treatment [90]) [88, 89] |
| RNA pol III | Cytosol | dsDNA | AT-rich | >30 | Cancer (colon [91]) | |
| RIG-I | Cytosol | dsRNA | 5’ ppp | >19 | Antiviral/Adjuvant [92] (e.g., influenza [93], ebola[94], rabies[95]) Cancer (e.g., melanoma [75, 96]) | Inflammatory disease and Autoimmunity (e.g., COPD, arthritis [97]) |
| MDA-5 | Cytosol | dsRNA | Independent | >1000 | Adjuvant Cancer [98] | |
RNA ligands are recognized by the sensors TLR3, TLR7, TLR8, MDA-5, and RIG-I. Within the endosome, the recognition of RNA ligands is initiated by their binding to TLRs 3, 7, or 8 which occurs at relatively low pH (<6.5). The activation of TLR3 requires interactions with dsRNA of at least 45 bp long [17] while TLRs 7 and 8 display their preferential recognition for GU and AU-rich ssRNA sequences with a minimal length of 19 bp[18–21]. There is also evidence which suggests that TLR7 can recognize short dsRNAs[22]. In addition to a binding site for RNA, TLRs 7 and 8 uniquely possess binding sites for guanosine and uridine[19, 23–26] and as such, can be activated by small synthetic molecules[27]. In the cytosol, the RLRs MAVS, MDA-5, and RIG-I recognize dsRNA ligands. Despite the fact that both MDA-5 and RIG-I possess two CARD domains, a helicase domain, and a regulatory domain, these two sensors identify distinct RNA ligands[28–30]. RIG-I binds dsRNAs longer than 19 bp through the interaction between the helicase and regulatory domains with the RNA backbone. Additionally, a 5’ di- or triphosphorylation of RNA is required for binding to the regulatory domain of PRRs[29, 31–34]. Similarly, MDA-5 recognizes RNA in a sequence-independent manner; however, it requires long dsRNAs (>1000 bp) for MDA-5 dimers to form filaments[34–36].
DNA ligands are recognized in the endosome by TLR9 and in the cytosol by several DNA sensors including cGAS, IFI16, and AIM2. TLR9 binds DNAs greater than 15 bp in length with unmethylated CpG dinucleotides that are infrequently found in mammalian DNA, but prominent in pathogen DNA[37]. Notably, naturally occurring phosphodiester oligodeoxynucleotides can activate TLR9 via the 2’-deoxyribose backbone independent of CpG motifs, suggesting that subcellular localization of TLR9 to the endosome is essential for discrimination of self and non-self-nucleic acids[37–39]. Additionally, TLR9 activation is enhanced in response to nucleic acid-histone complexes and supercoiled plasmids, indicating preferential binding to curved DNA ligands[37]. Finally, TLR9 is activated in response to DNA-RNA hybrids that are GU-rich and contain CpG motifs[40].
In the cytosol, cGAS binds B-form dsDNA with a minimum of 25 bp in length for efficient activation[41–43]. Recognition of dsDNA is sequence-independent, as electrostatic interactions with the sugar-phosphate backbone facilitate the interaction with two cGAS binding sites[41, 42]. Notably, cGAS can also recognize DNA/RNA hybrids and Y-form DNA, a ssDNA stem loop structure containing guanosines[44, 45]. Similar to cGAS, IFI16 and AIM bind dsDNA in a sequence-independent manner[46–50]. Both of these sensors oligomerize and form filaments along dsDNA which requires a minimum of 70–80 bp. However, optimum activation requires 150–200 and 280 bp for IFI16 and AIM, respectively[47–49, 51]. RNA polymerase III is unique in that it activates an immune response by transcribing AT-rich dsDNA into a ligand for the RNA sensor RIG-I. Importantly, AT-rich regions are required for promoter-independent initiation of transcription to generate a ligand for RIG-I[52–54].
Collectively, these PRRs form the first line of immune defense and are able to survey the cell to identify a wide range of nucleic acid ligands in order to coordinate innate and adaptive immune responses. Based on these known ligand requirements for PRR activation, nucleic acid nanoparticles (NANPs) can be rationally engineered with desired composition, nucleoside characteristics, structure, dimensionality, length of single- or double-stranded regions, and sequence motifs to generate a library of immunomodulatory, biocompatible agents suitable for a broad range of biomedical applications.
Nucleic Acid Nanoparticles (NANPs).
With known base pairing rules and an assortment of resolved naturally occurring motifs, nucleic acids can be designed to assemble into programmed NANPs of desired shapes, sizes, and compositions. An array of such structures have been demonstrated as biocompatible materials for a number of applications, encompassing biosensing[99, 100], drug delivery[101, 102], and as molecular devices[103, 104]. Depending on the design principles and motifs in their composition, NANPs can be assembled entirely from RNA or DNA, or take advantage of hybrid RNA/DNA combinations[105]. Currently, all NANPs can be roughly categorized by two design strategies[106]. The first design strategy, represented by cubes (Fig. 2A), is based solely on intermolecular canonical Watson-Crick interactions and utilizes ssRNAs and/or ssDNAs designed to assemble only with their partner strands and avoid any intramolecular secondary structures[107–109]. These design principles are widely employed in DNA nanotechnology and DNA origami[110, 111] to construct a variety of DNA NANPs. The second strategy, represented by RNA rings (Fig. 2B), is characteristic mainly of RNA nanotechnology and takes advantage of naturally occurring RNA structural and long-range interacting motifs (e.g., RNAI/II inverse RNA kissing loops needed to assemble RNA rings[112]). This design strategy follows empirically rationalized rules to combine, similarly to Lego bricks[113–116], pre-folded RNA motifs (typically formed via intramolecular base-pairing of individual RNA strands) and promote their bottom-up intermolecular assembly with remarkable degree of structural control[117]. In addition to conventional nucleic acids, NANPs can also be composed of chemically modified oligonucleotides which may serve to increase their stability or be modified with fluorophores or small ligands for precise tracking and targeting[118].
Figure 2: Design strategies and functionalization of NANPs.
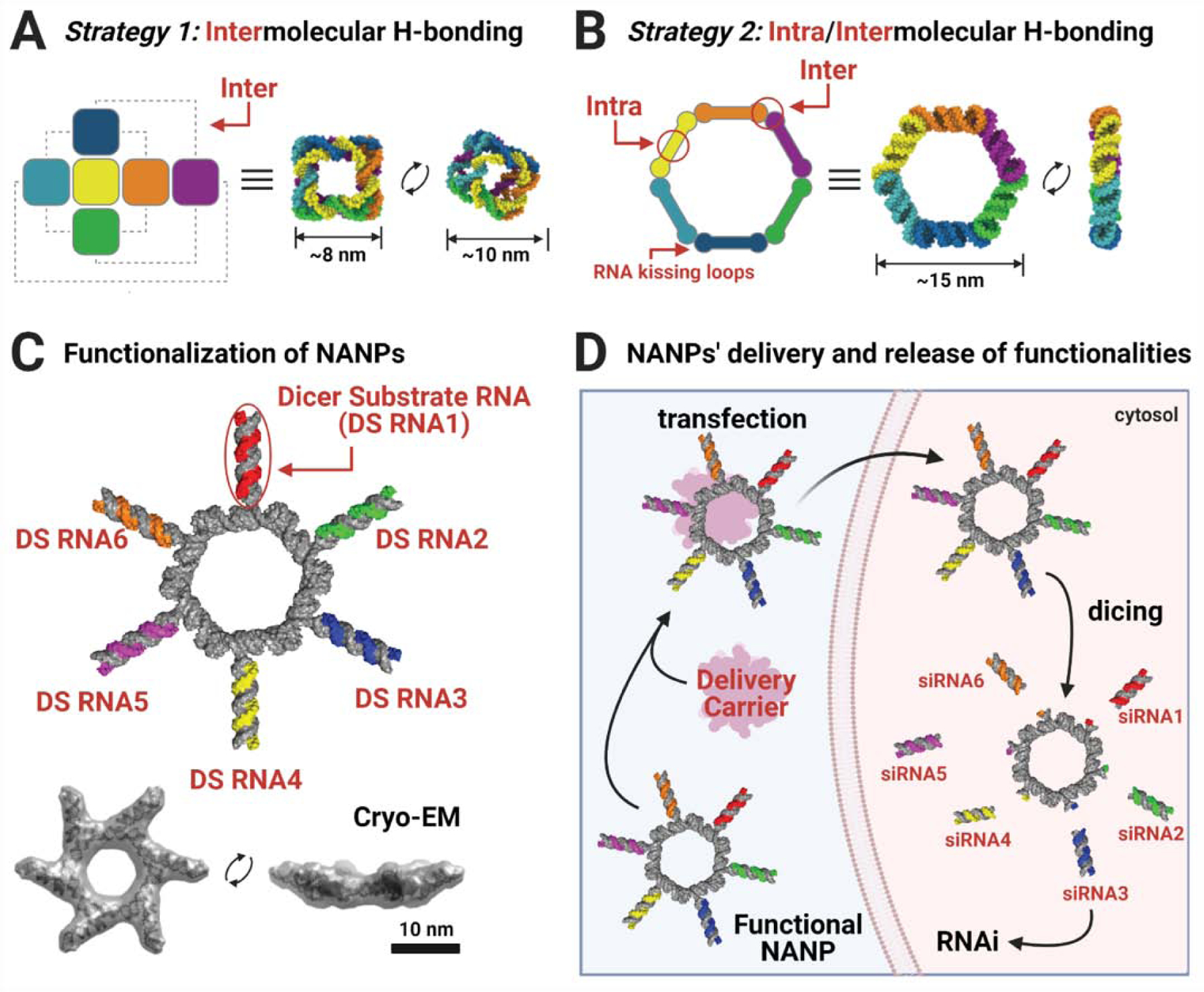
(A) For the formation of cubes, intermolecular hydrogen bonds occur between six oligonucleotides. As these are canonical WC bps, the cubes may be composed of any combination of RNA and/or DNA. (B) For the formation of rings, intramolecular hydrogen bonding first occurs within each strand, exposing single-stranded regions (RNA kissing loop motifs) which can then interact intermolecularly. (C) By extending the sequences in their compositions, NANPs can be functionalized with Dicer Substrate (DS) RNAs which can then enter the RNA interference (RNAi) pathway. Due to their hexameric nature, up to six DS RNAs can be added to each ring for simultaneous knockdown of six different target genes. Cryo-EM (from ref. [119]) demonstrates the structure of the functional RNA rings. (D) Functional NANPs must be combined with a delivery carrier for their transfection into cells, where they may then be processed by Dicer to begin RNAi. Created with Biorender.com.
When multiple TNAs are chosen for simultaneous delivery to the diseased cell (e.g., for combinatorial RNAi[120]), the optimal route of their controllable formulation would be through the their attachment to the individual strands entering the composition of NANPs. Assembly of NANPs would then bring the desired TNAs together with control over their composition and stoichiometry[121], an ability confirmed by several animal trials[105, 119, 122–127]. For example, the functionalization of RNA rings with multiple siRNAs occurs through introduction of Dicer Substrate RNAs (DS RNAs)[128] designed to promote the intracellular Dicer-assisted release of experimentally validated siRNAs[119] (Fig. 2C–D). The functionalization protocols[106] include the extension of the 3’-side of individual monomers with either sense or antisense strands of DS RNA and their further annealing to the complementary strands. Following the same rationale, programmable NANPs also allow for controlled functionalization with different aptamers, fluorescent dyes, proteins, and TNAs[129–131].
Immunomodulation with NANPs.
Stemming from their nucleic acid content, NANPs with biological applications interact with PRRs in the same immune syntax as innate nucleic acids. Therefore, known patterns of NANP recognition can be used to direct their design[11, 132]. Historically, the unknown immunorecognition of nucleic acid constructs has been a major impediment to their further clinical development, with indications that NANPs may need to be classified as a new class of drugs distinct from traditional TNAs[13]. Now, pattern recognition as it relates to NANP design is being thoroughly explored to allow for more control over which interactions with PRRs can occur, which also controls their downstream signaling and cytokine production. While the design of NANPs is already imbued with the ability for innate immunorecognition, NANP scaffolds can also be decorated with known immunostimulatory oligonucleotides[133] or used to present immunogens with spatial precision[134].
Control over the various design aspects allows for NANPs to be tailored for specific immunostimulatory control. For example, one approach (Fig. 3) has been to utilize hybrid NANP fibers which reassociate in the cellular environment for the formation of double-stranded NF-κB decoy DNA duplexes while releasing multiple RNAi inducers[135]. This strategy allows for faster processing of the subsequent downregulated immunostimulation over traditional gene silencing approaches since the products require no additional processing. Various aspects of NANP design can be strategically controlled in this manner for tailored immunomodulation. Working within the cell’s innate nucleic acid recognition capacity to do so allows for NANPs to serve as a molecular language for immune stimulation.
Figure 3: Non-functional RNA/DNA hybrid NANPs can be used for the coordinated activation of RNAi and NF-κB decoys.
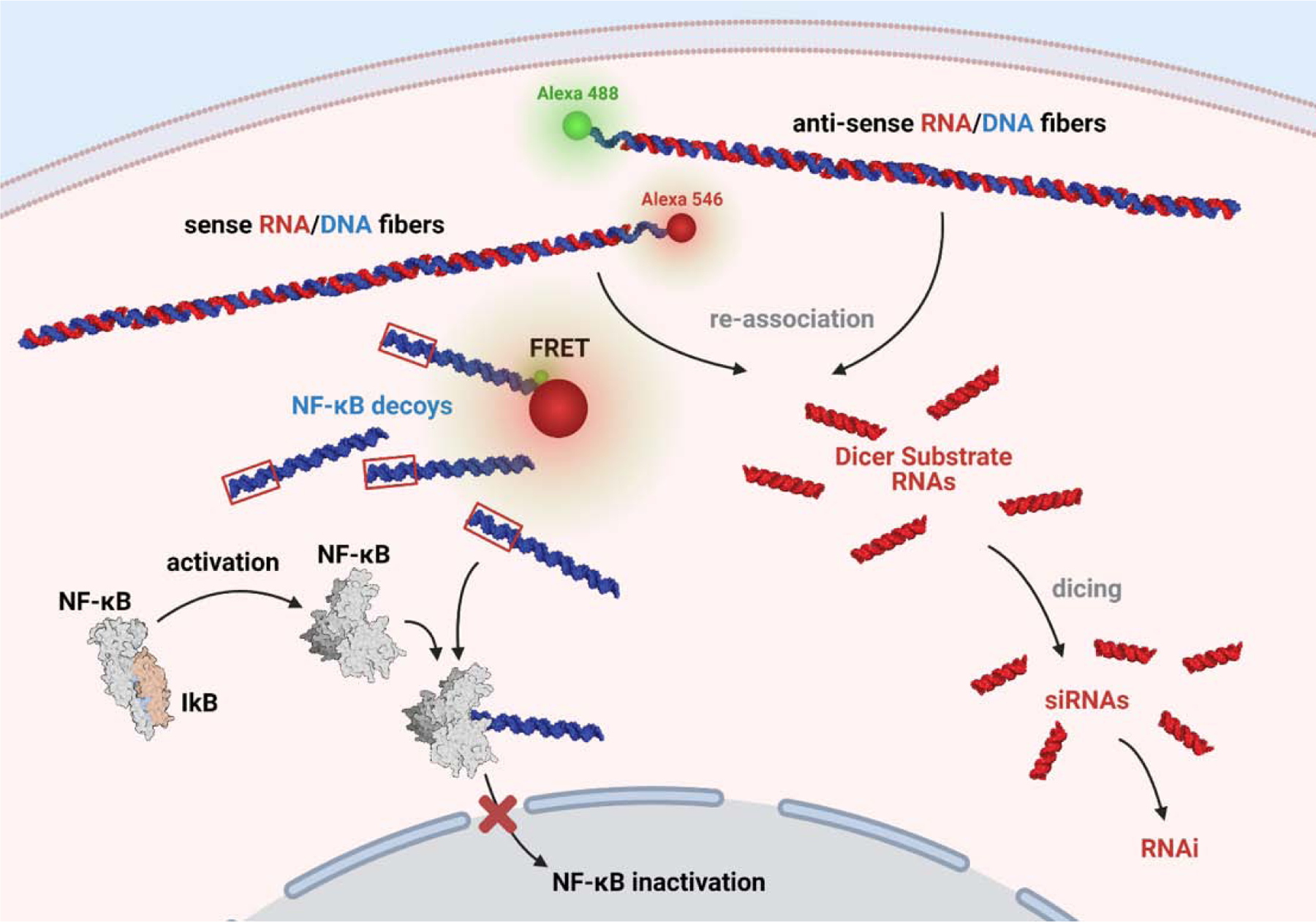
Re-association of hybrid fibers in the cytosol yields two products. First, DS RNAs that are cleaved by Dicer produce functional siRNAs for the silencing of target genes. Second, synthetic dsDNA decoys that readily bind to NF-κB prevent nuclear translocation and activation of NF-κB-induced cytokines. PDBs: 1NFK and 1NFI. Created with Biorender.com
PRRs display cell type-dependent expression and specific localization to subcellular compartments[31, 35, 43, 52, 136–139]. Cell type-dependent differences in receptor expression impact immune mediator production. Data from freshly collected human peripheral blood mononuclear cells (PBMCs) indicates NANPs are primarily identified by pDC that only express TLR7 and TLR9 in the endosome[140, 141]. Additionally, PRRs are localized to specific subcellular compartments, including the endosome and the cytosol[31, 35, 42, 43, 45, 52, 136, 137, 142, 143]. In PBMCs, delivery of NANPs via an endocytic pathway drives the production of IFNs, however, bypassing the endosome using an electroporation method of delivery abrogated any IFN responses[140]. These data suggest subcellular trafficking of NANPs may influence exposure to PRRs and thereby the production of cytokines and interferons or lack thereof. Notably, some NANPs can enter cells in the absence of a carrier, while other NANP formulations require a carrier to overcome the charge repulsion between the phosphate backbone and host cell membranes[102, 144–153]. Recent data indicates carrier-NANP combinations affect both delivery efficiency and enrichment to subcellular compartments[147]. Interestingly, NANPs composed of different nucleic acid compositions but delivered with the same carrier displayed differences in enrichment to the cytosol and endosome[147]. This provides evidence that carrier-NANP combinations are an additional parameter to direct delivery to specific subcellular compartments and either avoid or target specific PRRs.
Chemical composition and immunorecognition of NANPs.
Most PRRs display preferential recognition of RNA vs DNA due to secondary or tertiary structures and nucleoside characteristics of nucleic acids[154, 155]. Nucleic acids are composed of nucleotides that have a phosphate, a sugar (ribose or deoxyribose), and a base (adenine, guanine, cytosine, thymine, uracil). dsDNA is most commonly found in a B-form double helix. In contrast, RNA/DNA hybrids and dsRNAs exist in A-form that is highly compact due to more efficient base-pair stacking. DNA can also exist as a Z-form left-handed helix due to alternating purine-pyrimidine nucleotides (GC)n and high salt concentration[156]. Consistent with PRRs’ preferential recognition of nucleic acid composition, NANP polygons that differ only in composition (RNA vs DNA vs RNA/DNA hybrid) but have the same size, shape, and connectivity induce significant differences in the production of proinflammatory cytokines and interferons[157, 158]. DNA induced minimal production of these immune modulators and immunostimulation increased with incorporation of RNA strands. Polygon NANPs composed of only RNA strands displayed the highest production of proinflammatory cytokines and interferons[157, 158]. To advance the current understanding of properties that contribute to NANP immunomodulation, quantitative structure activity relationship (QSAR) modeling was applied for NANP studies[158] (Fig. 4). Investigation of a focused panel of RNA, DNA, and RNA/DNA polygons demonstrated that NANP physicochemical properties including molecular weight, melting temperature, and half-life predict NANP-stimulated immune responses. Continued characterization of larger NANP panels can be used to expand this predictive model in order to generate a set of design parameters for engineering NANPs with desired immunostimulatory properties. Notably, in vitro and in vivo studies demonstrate polygon and three-dimensional pRNA-based NANPs stimulate minimal production of proinflammatory cytokines, suggesting composition is not the sole factor determining immunostimulatory properties[159–162].
Figure 4: QSAR modeling employs a set of NANPs to predict pro-inflammatory immune responses.
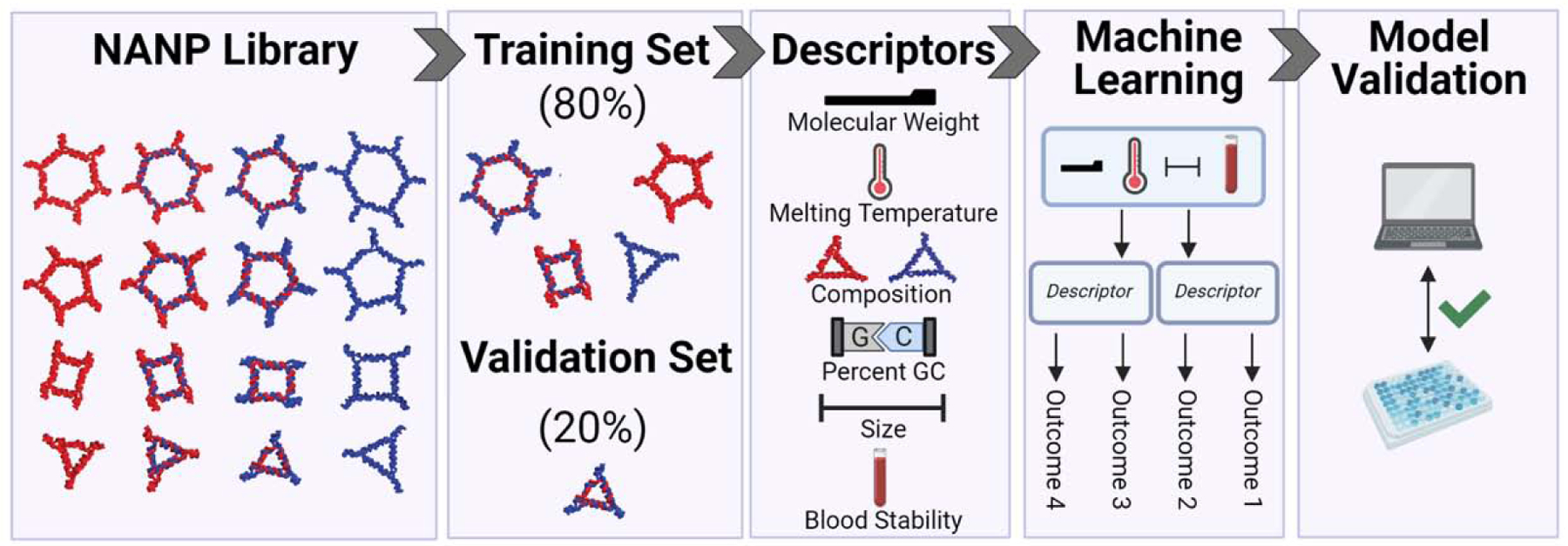
A panel of representative DNA, RNA, and DNA/RNA NANPs was designed with varying descriptors such as molecular weight, melting temperature, size, and stability. A training set composed of 80% of this batch was used for machine learning, where descriptors were matched against outcomes of experimentally found immunostimulations. A validation set was then used to confirm the predicted trends and validate the model. Created with Biorender.com
Due to the sensitivity of RNA to nuclease degradation, chemically modified strands are often incorporated to increase enzymatic stability of TNAs[163–166]. Modification of the 2’-hydroxyl group with 2’-fluoro, 2’-deoxy, or 2’-O-methyl prevents nuclease recognition of RNAs[167]. Additionally, these nucleoside modifications alter PRR receptor identification and have to be considered as an additional descriptor in the NANPs’ and TNAs’ immunomodulation. The 2’-hydroxyl uridine has been demonstrated to be central to endosomal TLR recognition of RNA[167]. TLRs 3 and 7 activation in response to a 140 bp dsRNA ligand was abrogated by the incorporation of 2’-fluoro or 2’-O-methyl pyrimidines[168]. However, RIG-I activation was enhanced by these modifications. Interestingly, 2’-O-methyl modified ligands have also been demonstrated to possess TLR7 antagonistic effects which are able to reduce cytokine and interferon responses to a small molecule TLR7 agonist[169]. Consistent with these findings, 2’-fluoro-modified RNA polygons display RIG-I agonist activity and avoid activation of endosomal TLRs[147]. Additional chemical modifications can include small molecules such as cholesterol to further direct the immune responses. Cholesterol-tagged lipid-DNA nanobarrels have recently been shown to selectively bind white blood cells in the PBMC population in order to suppress the immune response upon interactions with lipopolysaccharides[170].
Architectural parameters and immunorecognition of NANPs.
Although many PRRs recognize nucleic acid ligands in a sequence-independent manner via interaction with the sugar phosphate backbone, several PRRs display preferential binding to specific sequences or sequence motifs. In the endosome, TLR7/8 display preferential recognition for GU- and AU-rich sequences while TLR9 recognizes CpG motifs with unmethylated CG dinucleotides[19–21, 26, 37, 38]. NANPs, hydrogels, polypod structures, and spherical nucleic acids have been developed to deliver agonists with these sequence characteristics to the endosome for eliciting desired TLR-dependent responses (Fig. 5A)[171–173]. In the cytosol, RNA polymerase III requires AT-rich regions for promoter-independent transcription that results in a 5’ triphosphorylated ligand for the RNA sensor, RIG-I[52, 54]. Recent evidence indicates RIG-I-dependent production of pro-inflammatory cytokines and interferons in response to RNA and DNA polygons[147]. NANPs made of in vitro transcribed RNAs strands possess a 5’ triphosphates required for binding RIG-I; therefore, it is not surprising that these NANPs directly trigger RIG-I-dependent production of interferons. In contrast, DNA NANP stimulation of RIG-I-dependent responses is indirect via the involvement of RNA polymerase III dependent mechanism (Fig. 5). Notably, incorporation of RNA strands with 2’-fluoro-modified pyrimidines, known to increase NANP chemical and thermodynamic stability, also enhanced RNA polymerase III-dependent immune responses[147].
Figure 5: PRRs are localized to specific subcellular compartments to screen for PAMPs and DAMPs.
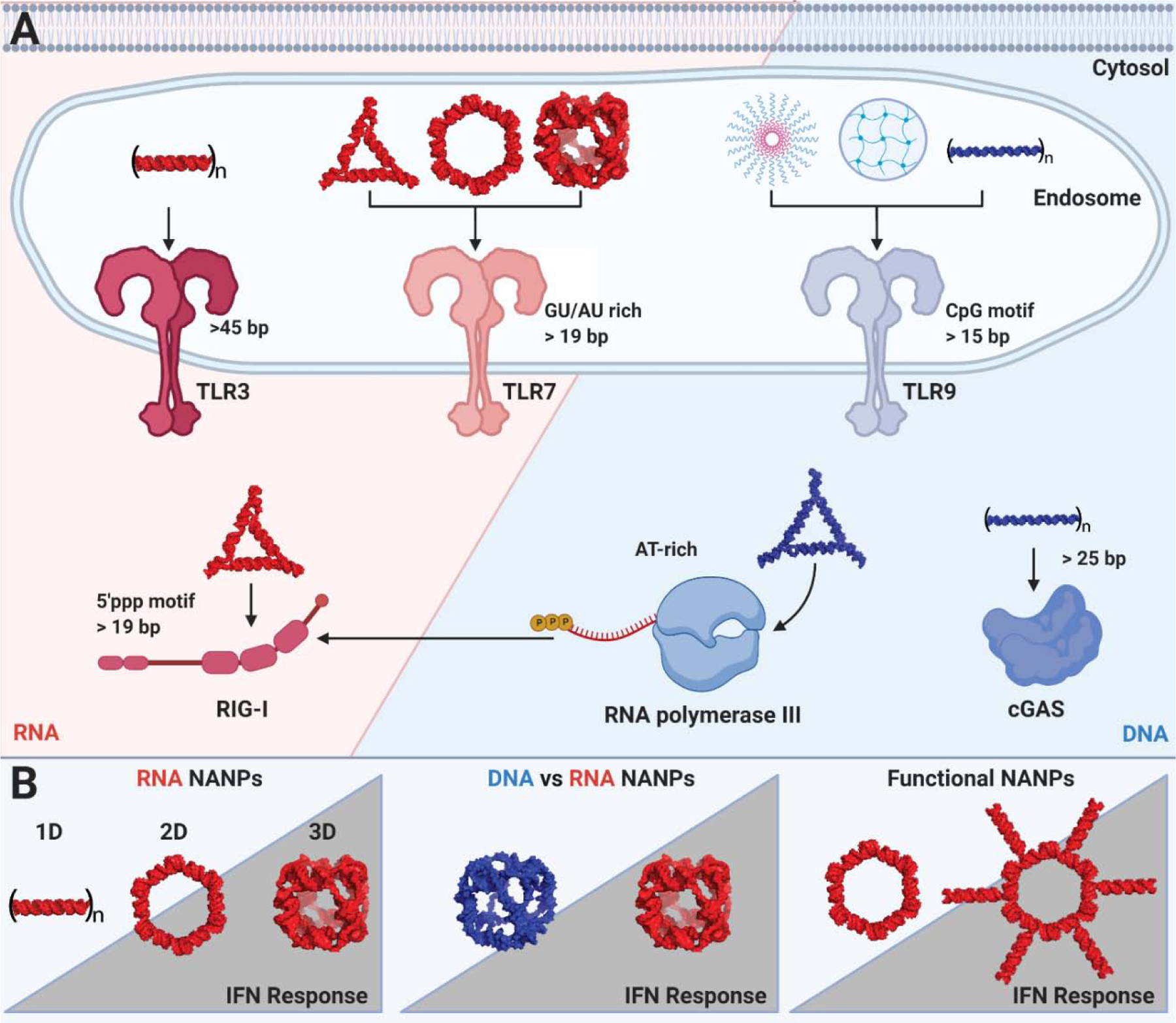
The endosomal and cytosolic sensors display composition, sequence, length, and structure-dependent recognition of nucleic acid ligands. The same ligand preferences determine binding to NANPs. The figure highlights key features of NANPs that meet the necessary ligand characteristics and have been experimentally confirmed to activate nucleic acid PRRs (A) with trends in relative responses in IFN productions across some representative categories of NANPs (B). Created with Biorender.com
NANP size is known to influence half-life and biodistribution in animal models as smaller nanoparticles are more readily cleared via renal clearance while larger nanoparticles accumulate in the liver and spleen[174–176]. Importantly, size is a determining factor in NANP immunostimulatory properties. As NANP size increases for a polygon shape, there are significantly increased levels of proinflammatory cytokines released[140, 160]. One contributing factor is delivery efficiency as larger NANPs may bind more readily in contrast to smaller analogs that can exhibit reduced uptake in the absence of a carrier due to charge repulsion with the negatively charged cell membrane[140, 141, 174]. Additionally, size is an influential factor in predicting NANP immunostimulation as PRRs display length-dependent recognition of nucleic acids[17, 20, 34, 37, 42, 48, 177]. Each of the PRRs has a minimal base-pair requirement for activation. For example, TLR3 requires a minimum of 45 bp for binding and activation[17]. Recent data supports the requirement for nucleic acid strand length as polygon NANPs consisting of dsRNA containing 22 bp per side fail to elicit TLR3 while longer dsRNAs present in nanostructures trigger TLR3 dependent responses (Fig. 5)[147, 178]. In the meantime, RNA polygons stimulate TLR7 and RIG-I-dependent immune responses, which require a minimum of 19 base-pairs for activation[147].
There are several examples of PRRs displaying shape-dependent recognition of nucleic acid ligands. First, in contrast to linearized RNA, circular RNAs (circRNAs) that are ssRNAs found in microbe genomes or produced to drive expression of proteins of interest avoid stimulation of the cellular RNA sensors, RIG-I, TLR3, and TLR7/8, providing an option for an immunoquiescent delivery platform[179]. Importantly, circRNA avoidance of RNA sensor stimulation was independent of nucleoside modifications, indicating a critical role for shape in determining interactions with these PRRs. Similarly, both TLRs 7 and 9 have been demonstrated to recognize the structural features of tRNA and curved DNA ligands, respectively [37, 180, 181]. Consistent with PRR shape-dependent recognition of nucleic acid ligands, NANP immunostimulatory properties are correlated with shape and connectivity. A direct comparison of RNA polygons (triangle, square, pentagon) of identical size demonstrates pentagons are the most potent inducers of proinflammatory cytokines compared to triangles[140]. Additionally, immunostimulation increased with RNA NANP dimensionality from linear fibers to planar rings to globular cube structures (Fig. 5B) with type I IFNs being the key biomarkers produced in response to NANP internalization by phagocytes[141, 182–184]. The same NANPs were also shown to induce type III IFNs, which have never been investigated in detail for traditional TNAs[182]. Globular RNA NANPs, such as RNA cubes, were the most immunostimulatory, when compared to their DNA analogs, inducing enhanced production of cytokines. Furthermore, RNA cubes and RNA rings but not RNA fibers elicit TLR7-dependent responses, suggesting a role for shape in determining the interaction with PRRs [140, 141]. Surprisingly, the magnitude and specificity of the immunostimulation can be additionally regulated by varying the numbers and orientations of TNAs attached to each NANP[183]. Combinations of TNAs can therefore potentially yield a second level of therapeutic activity in which the codelivery of several TNAs into one target cell is met with the synergistic effect of their scaffolding. Several different TNAs may be chosen in order to orchestrate multiple effects in the cellular environment, or the higher concentration of one type of TNA can be utilized for more efficient activity.
Conclusions.
Nucleic acid PRRs identify unique nucleic acid ligands based on cellular localization, composition, nucleoside characteristics, structure, length, and sequence motifs. These PRR ligand characteristics provide a set of guidelines for engineering therapeutic NANPs with defined immunostimulatory properties. Supporting evidence demonstrates that NANPs’ physicochemical properties and architectural parameters can be rationally designed and in turn control immunostimulation. Additionally, QSAR modeling of focused NANP panels indicates the physicochemical properties of NANPs are strong predictors of immunostimulatory properties. Continued research efforts to characterize broader NANP panels with varied sizes, chemical compositions, dimensionalities, and structures, factors demonstrated to influence PRR-dependent responses, will further strengthen these predictive models, thereby providing a molecular language that can be used to generate a vast library of immunomodulatory NANPs. Furthermore, NANPs can be functionalized with RNAi inducers and decoy duplexes to modulate PRRs’ downstream signaling, providing an additional level to control immunostimulation. As such, NANPs are a promising immunotherapeutic platform.
Currently, many small molecules, antibodies, oligonucleotides, and synthetic nucleic acids are being investigated in clinical trials as PRR agonists and antagonists. As discussed above, NANPs can be rationally designed to harness both innate and adaptive immune responses via nucleic acid PRRs. PRR agonists stimulate innate cytokine and interferon responses that can shape adaptive immunity. In particular, the production of interferons promotes cellular antiviral defenses and antigen-specific adaptive immune responses to pathogens and cancer cells. Therefore, immunomodulatory NANPs could be applied as broad-spectrum antivirals, vaccine adjuvants, and cancer immunotherapeutics (Table 1). NANP activation of endosomal PRRs could also be implemented to desensitize allergic responses by directing Th1 responses in contrast to the potent Th2 and IgE-mediated inflammatory response characteristic of allergic diseases. Alternatively, in inflammatory or autoimmune diseases, NANP PRR antagonists or NANPs complexed with functional groups to diminish PRRs’ downstream signaling can be utilized to control overactivation of PRR immune responses (Table 1). Additionally, these immunomodulatory NANP platforms can be accessorized with functional groups targeting disease-related pathways for a combinatorial treatment approach.
Notably, there are additional gaps in the broader adaptation of NANPs. First, in order to cross the plasma membrane for intracellular activity, many NANPs must be complexed with a carrier[141].There are many nanoparticle-based platforms for the delivery of nucleic acids that would add an additional level of tailorability depending on the resultant route of trafficking[185]. The continuous development of novel delivery vehicles has been recommended to overcome this challenge[12]. Second, NANPs must also resist nuclease degradation and traverse complex organ systems to arrive at the target site[6]. While the delivery platform may yield some bioavailability, there are also combinations of chemical modifications which can assist in this regard[147]. Chemical modifications may also serve as an important parameter to regulate NANP-PRR interactions, thereby promoting beneficial or preventing detrimental immune responses[147, 163, 186]. Finally, systemic gaps have been cited as needing to be overcome for further successful clinical translation[187]. Moving past in vitro models and into in vivo demonstrations will assist in obtaining more relevant depictions of the effectiveness of these platforms. There is also greater need for simplicity in design and manufacturing, culminating in more universal nomenclature and protocols that can be adapted. As the summation of these future directions, specific, modular, adjustable, reproducible, and targeted nucleic acid nanoparticles (SMART NANPs) represent the next step in nucleic acid programmability for diverse biomedical applications. The manifestation of tailorable SMART NANPs has a lot to offer for eliciting desirable immunostimulatory and therapeutic profiles.
Acknowledgements.
Research reported in this publication was supported by the National Institute of General Medical Sciences of the National Institutes of Health under Award Numbers R01GM120487 and R35GM139587. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Figures were created with Biorender.com.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Khvorova A, Watts JK, The chemical evolution of oligonucleotide therapies of clinical utility, Nat Biotechnol, 35 (2017) 238–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Tai W, Current Aspects of siRNA Bioconjugate for In Vitro and In Vivo Delivery, Molecules, 24 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Wang F, Zuroske T, Watts JK, RNA therapeutics on the rise, Nat Rev Drug Discov, 19 (2020) 441–442. [DOI] [PubMed] [Google Scholar]
- [4].Shen TT, Zhang Y, Zhou SR, Lin SB, Zhang XB, Zhu GZ, Nucleic Acid Immunotherapeutics for Cancer, Acs Applied Bio Materials, 3 (2020) 2838–2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Sridharan K, Gogtay NJ, Therapeutic nucleic acids: current clinical status, Br J Clin Pharmacol, 82 (2016) 659–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Roberts TC, Langer R, Wood MJA, Advances in oligonucleotide drug delivery, Nat Rev Drug Discov, 19 (2020) 673–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Bennett CF, Therapeutic Antisense Oligonucleotides Are Coming of Age, Annu Rev Med, 70 (2019) 307–321. [DOI] [PubMed] [Google Scholar]
- [8].Panigaj M, Johnson MB, Ke W, McMillan J, Goncharova EA, Chandler M, Afonin KA, Aptamers as Modular Components of Therapeutic Nucleic Acid Nanotechnology, ACS Nano, 13 (2019) 12301–12321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Jackson LA, Anderson EJ, Rouphael NG, Roberts PC, Makhene M, Coler RN, McCullough MP, Chappell JD, Denison MR, Stevens LJ, Pruijssers AJ, McDermott A, Flach B, Doria-Rose NA, Corbett KS, Morabito KM, O’Dell S, Schmidt SD, Swanson PA, Padilla M, Mascola JR, Neuzil KM, Bennett H, Sun W, Peters E, Makowski M, Albert J, Cross K, Buchanan W, Pikaart-Tautges R, Ledgerwood JE, Graham BS, Beigel JH, m.-S. Grp, An mRNA Vaccine against SARS-CoV-2-Preliminary Report, New Engl J Med, 383 (2020) 1920–1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Bajan S, Hutvagner G, RNA-Based Therapeutics: From Antisense Oligonucleotides to miRNAs, Cells, 9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Chandler M, Panigaj M, Rolband LA, Afonin KA, Challenges to optimizing RNA nanostructures for large scale production and controlled therapeutic properties, Nanomedicine (Lond), (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Dobrovolskaia MA, McNeil SE, Immunological and hematological toxicities challenging clinical translation of nucleic acid-based therapeutics, Expert Opin Biol Ther, 15 (2015) 1023–1048. [DOI] [PubMed] [Google Scholar]
- [13].Dobrovolskaia MA, Nucleic Acid Nanoparticles at a Crossroads of Vaccines and Immunotherapies, Molecules, 24 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Ng EWM, Shima DT, Calias P, Cunningham ET, Guyer DR, Adamis AP, Pegaptanib, a targeted anti-VEGF aptamer for ocular vascular disease, Nature Reviews Drug Discovery, 5 (2006) 123–132. [DOI] [PubMed] [Google Scholar]
- [15].Antczak M, Popenda M, Zok T, Sarzynska J, Ratajczak T, Tomczyk K, Adamiak RW, Szachniuk M, New functionality of RNAComposer: an application to shape the axis of miR160 precursor structure, Acta biochimica Polonica, 63 (2016) 737–744. [DOI] [PubMed] [Google Scholar]
- [16].Popenda M, Szachniuk M, Antczak M, Purzycka KJ, Lukasiak P, Bartol N, Blazewicz J, Adamiak RW, Automated 3D structure composition for large RNAs, Nucleic Acids Res, 40 (2012) e112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Leonard JN, Ghirlando R, Askins J, Bell JK, Margulies DH, Davies DR, Segal DM, The TLR3 signaling complex forms by cooperative receptor dimerization, Proc Natl Acad Sci U S A, 105 (2008) 258–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Judge AD, Sood V, Shaw JR, Fang D, McClintock K, MacLachlan I, Sequence-dependent stimulation of the mammalian innate immune response by synthetic siRNA, Nat Biotechnol, 23 (2005) 457–462. [DOI] [PubMed] [Google Scholar]
- [19].Diebold SS, Kaisho T, Hemmi H, Akira S, Reis e Sousa C, Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA, Science, 303 (2004) 1529–1531. [DOI] [PubMed] [Google Scholar]
- [20].Heil F, Hemmi H, Hochrein H, Ampenberger F, Kirschning C, Akira S, Lipford G, Wagner H, Bauer S, Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8, Science, 303 (2004) 1526–1529. [DOI] [PubMed] [Google Scholar]
- [21].Hornung V, Guenthner-Biller M, Bourquin C, Ablasser A, Schlee M, Uematsu S, Noronha A, Manoharan M, Akira S, de Fougerolles A, Endres S, Hartmann G, Sequence-specific potent induction of IFN-alpha by short interfering RNA in plasmacytoid dendritic cells through TLR7, Nat Med, 11 (2005) 263–270. [DOI] [PubMed] [Google Scholar]
- [22].Ablasser A, Poeck H, Anz D, Berger M, Schlee M, Kim S, Bourquin C, Goutagny N, Jiang Z, Fitzgerald KA, Rothenfusser S, Endres S, Hartmann G, Hornung V, Selection of molecular structure and delivery of RNA oligonucleotides to activate TLR7 versus TLR8 and to induce high amounts of IL-12p70 in primary human monocytes, J Immunol, 182 (2009) 6824–6833. [DOI] [PubMed] [Google Scholar]
- [23].Gorden KB, Gorski KS, Gibson SJ, Kedl RM, Kieper WC, Qiu XH, Tomai MA, Alkan SS, Vasilakos JP, Synthetic TLR Agonists reveal functional differences between human TLR7 and TLR8, J Immunol, 174 (2005) 1259–1268. [DOI] [PubMed] [Google Scholar]
- [24].Tanji H, Ohto U, Shibata T, Taoka M, Yamauchi Y, Isobe T, Miyake K, Shimizu T, Toll-like receptor 8 senses degradation products of single-stranded RNA, Nat Struct Mol Biol, 22 (2015) 109–115. [DOI] [PubMed] [Google Scholar]
- [25].Shibata T, Ohto U, Nomura S, Kibata K, Motoi Y, Zhang Y, Murakami Y, Fukui R, Ishimoto T, Sano S, Ito T, Shimizu T, Miyake K, Guanosine and its modified derivatives are endogenous ligands for TLR7, Int Immunol, 28 (2016) 211–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Zhang ZK, Ohto U, Shibata T, Krayukhina E, Taoka M, Yamauchi Y, Tanji H, Isobe T, Uchiyama S, Miyake K, Shimizu T, Structural Analysis Reveals that Toll-like Receptor 7 Is a Dual Receptor for Guanosine and Single-Stranded RNA, Immunity, 45 (2016) 737–748. [DOI] [PubMed] [Google Scholar]
- [27].Patinote C, Karroum NB, Moarbess G, Cirnat N, Kassab I, Bonnet PA, Deleuze-Masquefa C, Agonist and antagonist ligands of toll-like receptors 7 and 8: Ingenious tools for therapeutic purposes, Eur J Med Chem, 193 (2020) 112238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, Taira K, Akira S, Fujita T, The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses, Nat Immunol, 5 (2004) 730–737. [DOI] [PubMed] [Google Scholar]
- [29].Yoneyama M, Kikuchi M, Matsumoto K, Imaizumi T, Miyagishi M, Taira K, Foy E, Loo YM, Gale M Jr., Akira S, Yonehara S, Kato A, Fujita T, Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity, J Immunol, 175 (2005) 2851–2858. [DOI] [PubMed] [Google Scholar]
- [30].Saito T, Hirai R, Loo YM, Owen D, Johnson CL, Sinha SC, Akira S, Fujita T, Gale M Jr., Regulation of innate antiviral defenses through a shared repressor domain in RIG-I and LGP2, Proc Natl Acad Sci U S A, 104 (2007) 582–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Hornung V, Ellegast J, Kim S, Brzozka K, Jung A, Kato H, Poeck H, Akira S, Conzelmann KK, Schlee M, Endres S, Hartmann G, 5’-Triphosphate RNA is the ligand for RIG-I, Science, 314 (2006) 994–997. [DOI] [PubMed] [Google Scholar]
- [32].Schlee M, Roth A, Hornung V, Hagmann CA, Wimmenauer V, Barchet W, Coch C, Janke M, Mihailovic A, Wardle G, Juranek S, Kato H, Kawai T, Poeck H, Fitzgerald KA, Takeuchi O, Akira S, Tuschl T, Latz E, Ludwig J, Hartmann G, Recognition of 5’ triphosphate by RIG-I helicase requires short blunt double-stranded RNA as contained in panhandle of negative-strand virus, Immunity, 31 (2009) 25–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, Uematsu S, Jung A, Kawai T, Ishii KJ, Yamaguchi O, Otsu K, Tsujimura T, Koh CS, Reis e Sousa C, Matsuura Y, Fujita T, Akira S, Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses, Nature, 441 (2006) 101–105. [DOI] [PubMed] [Google Scholar]
- [34].Kato H, Takeuchi O, Mikamo-Satoh E, Hirai R, Kawai T, Matsushita K, Hiiragi A, Dermody TS, Fujita T, Akira S, Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5, J Exp Med, 205 (2008) 1601–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Wu B, Peisley A, Richards C, Yao H, Zeng X, Lin C, Chu F, Walz T, Hur S, Structural basis for dsRNA recognition, filament formation, and antiviral signal activation by MDA5, Cell, 152 (2013) 276–289. [DOI] [PubMed] [Google Scholar]
- [36].Pichlmair A, Schulz O, Tan CP, Rehwinkel J, Kato H, Takeuchi O, Akira S, Way M, Schiavo G, Reis e Sousa C, Activation of MDA5 requires higher-order RNA structures generated during virus infection, J Virol, 83 (2009) 10761–10769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Li Y, Berke IC, Modis Y, DNA binding to proteolytically activated TLR9 is sequence-independent and enhanced by DNA curvature, Embo J, 31 (2012) 919–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Haas T, Metzger J, Schmitz F, Heit A, Muller T, Latz E, Wagner H, The DNA sugar backbone 2’ deoxyribose determines toll-like receptor 9 activation, Immunity, 28 (2008) 315–323. [DOI] [PubMed] [Google Scholar]
- [39].Yasuda K, Yu P, Kirschning CJ, Schlatter B, Schmitz F, Heit A, Bauer S, Hochrein H, Wagner H, Endosomal translocation of vertebrate DNA activates dendritic cells via TLR9-dependent and -independent pathways, J Immunol, 174 (2005) 6129–6136. [DOI] [PubMed] [Google Scholar]
- [40].Rigby RE, Webb LM, Mackenzie KJ, Li Y, Leitch A, Reijns MA, Lundie RJ, Revuelta A, Davidson DJ, Diebold S, Modis Y, MacDonald AS, Jackson AP, RNA:DNA hybrids are a novel molecular pattern sensed by TLR9, Embo J, 33 (2014) 542–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Li X, Shu C, Yi G, Chaton CT, Shelton CL, Diao J, Zuo X, Kao CC, Herr AB, Li P, Cyclic GMP-AMP synthase is activated by double-stranded DNA-induced oligomerization, Immunity, 39 (2013) 1019–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Civril F, Deimling T, de Oliveira Mann CC, Ablasser A, Moldt M, Witte G, Hornung V, Hopfner KP, Structural mechanism of cytosolic DNA sensing by cGAS, Nature, 498 (2013) 332–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Sun L, Wu J, Du F, Chen X, Chen ZJ, Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway, Science, 339 (2013) 786–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Herzner AM, Hagmann CA, Goldeck M, Wolter S, Kubler K, Wittmann S, Gramberg T, Andreeva L, Hopfner KP, Mertens C, Zillinger T, Jin T, Xiao TS, Bartok E, Coch C, Ackermann D, Hornung V, Ludwig J, Barchet W, Hartmann G, Schlee M, Sequence-specific activation of the DNA sensor cGAS by Y-form DNA structures as found in primary HIV-1 cDNA, Nat Immunol, 16 (2015) 1025–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Mankan AK, Schmidt T, Chauhan D, Goldeck M, Honing K, Gaidt M, Kubarenko AV, Andreeva L, Hopfner KP, Hornung V, Cytosolic RNA:DNA hybrids activate the cGAS-STING axis, Embo J, 33 (2014) 2937–2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Unterholzner L, Keating SE, Baran M, Horan KA, Jensen SB, Sharma S, Sirois CM, Jin T, Latz E, Xiao TS, Fitzgerald KA, Paludan SR, Bowie AG, IFI16 is an innate immune sensor for intracellular DNA, Nat Immunol, 11 (2010) 997–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Morrone SR, Wang T, Constantoulakis LM, Hooy RM, Delannoy MJ, Sohn J, Cooperative assembly of IFI16 filaments on dsDNA provides insights into host defense strategy, Proc Natl Acad Sci U S A, 111 (2014) E62–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Jin T, Perry A, Jiang J, Smith P, Curry JA, Unterholzner L, Jiang Z, Horvath G, Rathinam VA, Johnstone RW, Hornung V, Latz E, Bowie AG, Fitzgerald KA, Xiao TS, Structures of the HIN domain:DNA complexes reveal ligand binding and activation mechanisms of the AIM2 inflammasome and IFI16 receptor, Immunity, 36 (2012) 561–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Morrone SR, Matyszewski M, Yu X, Delannoy M, Egelman EH, Sohn J, Assembly-driven activation of the AIM2 foreign-dsDNA sensor provides a polymerization template for downstream ASC, Nat Commun, 6 (2015) 7827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Lu A, Li Y, Yin Q, Ruan J, Yu X, Egelman E, Wu H, Plasticity in PYD assembly revealed by cryo-EM structure of the PYD filament of AIM2, Cell Discov, 1 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Stratmann SA, Morrone SR, van Oijen AM, Sohn J, The innate immune sensor IFI16 recognizes foreign DNA in the nucleus by scanning along the duplex, Elife, 4 (2015) e11721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Chiu YH, Macmillan JB, Chen ZJ, RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway, Cell, 138 (2009) 576–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Bauernfeind F, Ablasser A, Kim S, Bartok E, Hornung V, An unexpected role for RNA in the recognition of DNA by the innate immune system, RNA Biol, 7 (2010) 151–157. [DOI] [PubMed] [Google Scholar]
- [54].Ablasser A, Bauernfeind F, Hartmann G, Latz E, Fitzgerald KA, Hornung V, RIG-I-dependent sensing of poly(dA:dT) through the induction of an RNA polymerase III-transcribed RNA intermediate, Nat Immunol, 10 (2009) 1065–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Buitendijk M, Eszterhas SK, Howell AL, Toll-like receptor agonists are potent inhibitors of human immunodeficiency virus-type 1 replication in peripheral blood mononuclear cells, AIDS Res Hum Retroviruses, 30 (2014) 457–467. [DOI] [PubMed] [Google Scholar]
- [56].Saxena M, Sabado RL, La Mar M, Mohri H, Salazar AM, Dong H, Correa Da Rosa J, Markowitz M, Bhardwaj N, Miller E, Poly-ICLC, a TLR3 Agonist, Induces Transient Innate Immune Responses in Patients With Treated HIV-Infection: A Randomized Double-Blinded Placebo Controlled Trial, Front Immunol, 10 (2019) 725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Renu S, Feliciano-Ruiz N, Lu F, Ghimire S, Han Y, Schrock J, Dhakal S, Patil V, Krakowka S, HogenEsch H, Renukaradhya GJ, A Nanoparticle-Poly(I:C) Combination Adjuvant Enhances the Breadth of the Immune Response to Inactivated Influenza Virus Vaccine in Pigs, Vaccines (Basel), 8 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Takeda Y, Takaki H, Fukui-Miyazaki A, Yoshida S, Matsumoto M, Seya T, Vaccine adjuvant ARNAX promotes mucosal IgA production in influenza HA vaccination, Biochem Biophys Res Commun, 506 (2018) 1019–1025. [DOI] [PubMed] [Google Scholar]
- [59].Chen S, Ou R, Tang J, Deng X, Wu Y, van Velkinburgh JC, Ni B, Xu Y, Enhanced antitumor effects of HPV16E7(49–57)-based vaccine by combined immunization with poly(I:C) and oxygen-regulated protein 150, Cancer Epidemiol, 37 (2013) 172–178. [DOI] [PubMed] [Google Scholar]
- [60].Domingos-Pereira S, Decrausaz L, Derre L, Bobst M, Romero P, Schiller JT, Jichlinski P, Nardelli-Haefliger D, Intravaginal TLR agonists increase local vaccine-specific CD8 T cells and human papillomavirus-associated genital-tumor regression in mice, Mucosal Immunol, 6 (2013) 393–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Sajadian A, Tabarraei A, Soleimanjahi H, Fotouhi F, Gorji A, Ghaemi A, Comparing the effect of Toll-like receptor agonist adjuvants on the efficiency of a DNA vaccine, Arch Virol, 159 (2014) 1951–1960. [DOI] [PubMed] [Google Scholar]
- [62].Le Naour J, Galluzzi L, Zitvogel L, Kroemer G, Vacchelli E, Trial watch: TLR3 agonists in cancer therapy, Oncoimmunology, 9 (2020) 1771143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Anwar MA, Shah M, Kim J, Choi S, Recent clinical trends in Toll-like receptor targeting therapeutics, Med Res Rev, 39 (2019) 1053–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Patra MC, Achek A, Kim GY, Panneerselvam S, Shin HJ, Baek WY, Lee WH, Sung J, Jeong U, Cho EY, Kim W, Kim E, Suh CH, Choi S, A Novel Small-Molecule Inhibitor of Endosomal TLRs Reduces Inflammation and Alleviates Autoimmune Disease Symptoms in Murine Models, Cells, 9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Dominguez-Molina B, Machmach K, Perales C, Tarancon-Diez L, Gallego I, Sheldon JL, Leal M, Domingo E, Ruiz-Mateos E, Toll-Like Receptor 7 (TLR-7) and TLR-9 Agonists Improve Hepatitis C Virus Replication and Infectivity Inhibition by Plasmacytoid Dendritic Cells, J Virol, 92 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Van Hoeven N, Fox CB, Granger B, Evers T, Joshi SW, Nana GI, Evans SC, Lin S, Liang H, Liang L, Nakajima R, Felgner PL, Bowen RA, Marlenee N, Hartwig A, Baldwin SL, Coler RN, Tomai M, Elvecrog J, Reed SG, Carter D, A Formulated TLR7/8 Agonist is a Flexible, Highly Potent and Effective Adjuvant for Pandemic Influenza Vaccines, Sci Rep, 7 (2017) 46426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Bellmann L, Cappellano G, Schachtl-Riess JF, Prokopi A, Seretis A, Ortner D, Tripp CH, Brinckerhoff CE, Mullins DW, Stoitzner P, A TLR7 agonist strengthens T and NK cell function during BRAF-targeted therapy in a preclinical melanoma model, Int J Cancer, 146 (2020) 1409–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Chi H, Li C, Zhao FS, Zhang L, Ng TB, Jin G, Sha O, Anti-tumor Activity of Toll-Like Receptor 7 Agonists, Front Pharmacol, 8 (2017) 304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Liu CL, Santos MM, Fernandes C, Liao M, Iamarene K, Zhang JY, Sukhova GK, Shi GP, Toll-like receptor 7 deficiency protects apolipoprotein E-deficient mice from diet-induced atherosclerosis, Sci Rep, 7 (2017) 847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Knoepfel T, Nimsgern P, Jacquier S, Bourrel M, Vangrevelinghe E, Glatthar R, Behnke D, Alper PB, Michellys PY, Deane J, Junt T, Zipfel G, Limonta S, Hawtin S, Andre C, Boulay T, Loetscher P, Faller M, Blank J, Feifel R, Betschart C, Target-Based Identification and Optimization of 5-Indazol-5-yl Pyridones as Toll-like Receptor 7 and 8 Antagonists Using a Biochemical TLR8 Antagonist Competition Assay, J Med Chem, 63 (2020) 8276–8295. [DOI] [PubMed] [Google Scholar]
- [71].Gao W, Sun X, Li D, Sun L, He Y, Wei H, Jin F, Cao Y, Toll-like receptor 4, Toll-like receptor 7 and Toll-like receptor 9 agonists enhance immune responses against blood-stage Plasmodium chabaudi infection in BALB/c mice, Int Immunopharmacol, 89 (2020) 107096. [DOI] [PubMed] [Google Scholar]
- [72].Gao W, Sun X, Li D, Sun L, He Y, Wei H, Jin F, Cao Y, Toll-like receptor 7 and Toll-like receptor 9 agonists effectively enhance immunological memory in Plasmodium chabaudi infected BALB/c mice, Int Immunopharmacol, 81 (2020) 106248. [DOI] [PubMed] [Google Scholar]
- [73].Godot V, Tcherakian C, Gil L, Cervera-Marzal I, Li G, Cheng L, Ortonne N, Lelievre JD, Pantaleo G, Fenwick C, Centlivre M, Mouquet H, Cardinaud S, Zurawski SM, Zurawski G, Milpied P, Su L, Levy Y, TLR-9 agonist and CD40-targeting vaccination induces HIV-1 envelope-specific B cells with a diversified immunoglobulin repertoire in humanized mice, PLoS Pathog, 16 (2020) e1009025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Vibholm LK, Konrad CV, Schleimann MH, Frattari G, Winckelmann A, Klastrup V, Jensen NM, Jensen SS, Schmidt M, Wittig B, Zuwala K, Mack K, Olesen R, Hua S, Lichterfeld M, Ostergaard L, Denton PW, Tolstrup M, Sogaard OS, Effects of 24-week Toll-like receptor 9 agonist treatment in HIV type 1+ individuals, AIDS, 33 (2019) 1315–1325. [DOI] [PubMed] [Google Scholar]
- [75].Levy ES, Chang R, Zamecnik CR, Dhariwala MO, Fong L, Desai TA, Multi-Immune Agonist Nanoparticle Therapy Stimulates Type I Interferons to Activate Antigen-Presenting Cells and Induce Antigen-Specific Antitumor Immunity, Mol Pharm, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Frank MJ, Reagan PM, Bartlett NL, Gordon LI, Friedberg JW, Czerwinski DK, Long SR, Hoppe RT, Janssen R, Candia AF, Coffman RL, Levy R, In Situ Vaccination with a TLR9 Agonist and Local Low-Dose Radiation Induces Systemic Responses in Untreated Indolent Lymphoma, Cancer Discov, 8 (2018) 1258–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Scheetz LM, Yu M, Li D, Castro MG, Moon JJ, Schwendeman A, Synthetic HDL Nanoparticles Delivering Docetaxel and CpG for Chemoimmunotherapy of Colon Adenocarcinoma, Int J Mol Sci, 21 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Younes AI, Barsoumian HB, Sezen D, Verma V, Patel R, Wasley M, Hu Y, Dunn JD, He K, Chen D, Menon H, Masrorpour F, Gu M, Yang L, Puebla-Osorio N, Cortez MA, Welsh JW, Addition of TLR9 agonist immunotherapy to radiation improves systemic antitumor activity, Transl Oncol, 14 (2021) 100983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Offersen R, Nissen SK, Rasmussen TA, Ostergaard L, Denton PW, Sogaard OS, Tolstrup M, A Novel Toll-Like Receptor 9 Agonist, MGN1703, Enhances HIV-1 Transcription and NK Cell-Mediated Inhibition of HIV-1-Infected Autologous CD4+ T Cells, J Virol, 90 (2016) 4441–4453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Mussari CP, Dodd DS, Sreekantha RK, Pasunoori L, Wan H, Posy SL, Critton D, Ruepp S, Subramanian M, Watson A, Davies P, Schieven GL, Salter-Cid LM, Srivastava R, Tagore DM, Dudhgaonkar S, Poss MA, Carter PH, Dyckman AJ, Discovery of Potent and Orally Bioavailable Small Molecule Antagonists of Toll-like Receptors 7/8/9 (TLR7/8/9), ACS Med Chem Lett, 11 (2020) 1751–1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Wang J, Li PY, Wu MX, Natural STING Agonist as an “Ideal” Adjuvant for Cutaneous Vaccination, J Invest Dermatol, 136 (2016) 2183–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Lirussi D, Weissmann SF, Ebensen T, Nitsche-Gloy U, Franz HBG, Guzman CA, Cyclic Di-Adenosine Monophosphate: A Promising Adjuvant Candidate for the Development of Neonatal Vaccines, Pharmaceutics, 13 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Wu JJ, Zhao L, Han BB, Hu HG, Zhang BD, Li WH, Chen YX, Li YM, A novel STING agonist for cancer immunotherapy and a SARS-CoV-2 vaccine adjuvant, Chem Commun (Camb), 57 (2021) 504–507. [DOI] [PubMed] [Google Scholar]
- [84].Chattopadhyay S, Hu CJ, Nanomedicinal delivery of stimulator of interferon genes agonists: recent advances in virus vaccination, Nanomedicine (Lond), 15 (2020) 2883–2894. [DOI] [PubMed] [Google Scholar]
- [85].Ding C, Song Z, Shen A, Chen T, Zhang A, Small molecules targeting the innate immune cGASSTINGTBK1 signaling pathway, Acta Pharm Sin B, 10 (2020) 2272–2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Le Naour J, Zitvogel L, Galluzzi L, Vacchelli E, Kroemer G, Trial watch: STING agonists in cancer therapy, Oncoimmunology, 9 (2020) 1777624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Liu Y, Lu X, Qin N, Qiao Y, Xing S, Liu W, Feng F, Liu Z, Sun H, STING, a promising target for small molecular immune modulator: A review, Eur J Med Chem, 211 (2021) 113113. [DOI] [PubMed] [Google Scholar]
- [88].Sharma BR, Karki R, Kanneganti TD, Role of AIM2 inflammasome in inflammatory diseases, cancer and infection, Eur J Immunol, 49 (2019) 1998–2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Xu S, Li X, Liu Y, Xia Y, Chang R, Zhang C, Inflammasome inhibitors: promising therapeutic approaches against cancer, J Hematol Oncol, 12 (2019) 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Liu D, Xu X, Dai Y, Zhao X, Bao S, Ma W, Zha L, Liu S, Liu Y, Zheng J, Shi M, Blockade of AIM2 inflammasome or alpha1-AR ameliorates IL-1beta release and macrophage-mediated immunosuppression induced by CAR-T treatment, J Immunother Cancer, 9 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Liang X, Xie R, Su J, Ye B, Wei S, Liang Z, Bai R, Chen Z, Li Z, Gao X, Inhibition of RNA polymerase III transcription by Triptolide attenuates colorectal tumorigenesis, J Exp Clin Cancer Res, 38 (2019) 217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Yong HY, Luo DH, RIG-I-Like Receptors as Novel Targets for Pan-Antivirals and Vaccine Adjuvants Against Emerging and Re-Emerging Viral Infections, Frontiers in Immunology, 9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Wong PT, Goff PH, Sun RJ, Ruge MJ, Ermler ME, Sebring A, O’Konek JJ, Landers JJ, Janczak KW, Sun W, Baker JR Jr., Combined Intranasal Nanoemulsion and RIG-I Activating RNA Adjuvants Enhance Mucosal, Humoral, and Cellular Immunity to Influenza Virus, Mol Pharm, 18 (2021) 679–698. [DOI] [PubMed] [Google Scholar]
- [94].Suschak JJ, Dupuy LC, Shoemaker CJ, Six C, Kwilas SA, Spik KW, Williams JA, Schmaljohn CS, Nanoplasmid Vectors Co-expressing Innate Immune Agonists Enhance DNA Vaccines for Venezuelan Equine Encephalitis Virus and Ebola Virus, Mol Ther Methods Clin Dev, 17 (2020) 810–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Doener F, Hong HS, Meyer I, Tadjalli-Mehr K, Daehling A, Heidenreich R, Koch SD, Fotin-Mleczek M, Gnad-Vogt U, RNA-based adjuvant CV8102 enhances the immunogenicity of a licensed rabies vaccine in a first-in-human trial, Vaccine, 37 (2019) 1819–1826. [DOI] [PubMed] [Google Scholar]
- [96].Iurescia S, Fioretti D, Rinaldi M, The Innate Immune Signalling Pathways: Turning RIG-I Sensor Activation Against Cancer, Cancers (Basel), 12 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Rawling DC, Jagdmann GE Jr., Potapova O, Pyle AM, Small-Molecule Antagonists of the RIG-I Innate Immune Receptor, ACS Chem Biol, 15 (2020) 311–317. [DOI] [PubMed] [Google Scholar]
- [98].Sultan H, Salazar AM, Celis E, Poly-ICLC, a multi-functional immune modulator for treating cancer, Semin Immunol, 49 (2020) 101414. [DOI] [PubMed] [Google Scholar]
- [99].Karadeema RJ, Stancescu M, Steidl TP, Bertot SC, Kolpashchikov DM, The owl sensor: a ‘fragile’ DNA nanostructure for the analysis of single nucleotide variations, Nanoscale, 10 (2018) 10116–10122. [DOI] [PubMed] [Google Scholar]
- [100].Roark BK, Tan LA, Ivanina A, Chandler M, Castaneda J, Kim HS, Jawahar S, Viard M, Talic S, Wustholz KL, Yingling YG, Jones M, Afonin KA, Fluorescence blinking as an output signal for biosensing, ACS sensors, 1 (2016) 1295–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Li Z, Wang H, Yin H, Bennett C, Zhang HG, Guo P, Arrowtail RNA for Ligand Display on Ginger Exosome-like Nanovesicles to Systemic Deliver siRNA for Cancer Suppression, Sci Rep, 8 (2018) 14644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Halman JR, Kim KT, Gwak SJ, Pace R, Johnson MB, Chandler MR, Rackley L, Viard M, Marriott I, Lee JS, Afonin KA, A cationic amphiphilic co-polymer as a carrier of nucleic acid nanoparticles (Nanps) for controlled gene silencing, immunostimulation, and biodistribution, Nanomedicine, 23 (2020) 102094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Simmel FC, Yurke B, Singh HR, Principles and Applications of Nucleic Acid Strand Displacement Reactions, Chem Rev, 119 (2019) 6326–6369. [DOI] [PubMed] [Google Scholar]
- [104].Afonin KA, Desai R, Viard M, Kireeva ML, Bindewald E, Case CL, Maciag AE, Kasprzak WK, Kim T, Sappe A, Stepler M, Kewalramani VN, Kashlev M, Blumenthal R, Shapiro BA, Co-transcriptional production of RNA-DNA hybrids for simultaneous release of multiple split functionalities, Nucleic Acids Research, 42 (2014) 2085–2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Afonin KA, Viard M, Martins AN, Lockett SJ, Maciag AE, Freed EO, Heldman E, Jaeger L, Blumenthal R, Shapiro BA, Activation of different split functionalities on re-association of RNA-DNA hybrids, Nature nanotechnology, 8 (2013) 296–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Afonin KA, Grabow WW, Walker FM, Bindewald E, Dobrovolskaia MA, Shapiro BA, Jaeger L, Design and self-assembly of siRNA-functionalized RNA nanoparticles for use in automated nanomedicine, Nature Protocols, 6 (2011) 2022–2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Afonin KA, Bindewald E, Yaghoubian AJ, Voss N, Jacovetty E, Shapiro BA, Jaeger L, In vitro assembly of cubic RNA-based scaffolds designed in silico, Nature nanotechnology, 5 (2010) 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Afonin KA, Kasprzak W, Bindewald E, Puppala PS, Diehl AR, Hall KT, Kim TJ, Zimmermann MT, Jernigan RL, Jaeger L, Shapiro BA, Computational and experimental characterization of RNA cubic nanoscaffolds, Methods (San Diego, Calif.), 67 (2014) 256–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Afonin KA, Kasprzak WK, Bindewald E, Kireeva M, Viard M, Kashlev M, Shapiro BA, In silico design and enzymatic synthesis of functional RNA nanoparticles, Accounts of Chemical Research, 47 (2014) 1731–1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Chidchob P, Sleiman HF, Recent advances in DNA nanotechnology, Curr Opin Chem Biol, 46 (2018) 63–70. [DOI] [PubMed] [Google Scholar]
- [111].Pinheiro AV, Han D, Shih WM, Yan H, Challenges and opportunities for structural DNA nanotechnology, Nature nanotechnology, 6 (2011) 763–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Grabow WW, Zakrevsky P, Afonin KA, Chworos A, Shapiro BA, Jaeger L, Self-assembling RNA nanorings based on RNAI/II inverse kissing complexes, Nano letters, 11 (2011) 878–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Westhof E, Masquida B, Jaeger L, RNA tectonics: towards RNA design, Fold Des, 1 (1996) R78–88. [DOI] [PubMed] [Google Scholar]
- [114].Jaeger L, Leontis NB, Tecto-RNA: One-Dimensional Self-Assembly through Tertiary Interactions This work was carried out in Strasbourg with the support of grants to N.B.L. from the NIH (1R15 GM55898) and the NIH Fogarty Institute (1-F06-TW02251–01) and the support of the CNRS to L.J. The authors wish to thank Eric Westhof for his support and encouragement of this work, Angew Chem Int Ed Engl, 39 (2000) 2521–2524. [DOI] [PubMed] [Google Scholar]
- [115].Jaeger L, Westhof E, Leontis NB, TectoRNA: modular assembly units for the construction of RNA nano-objects, Nucleic Acids Res, 29 (2001) 455–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Guo P, Zhang C, Chen C, Garver K, Trottier M, Inter-RNA interaction of phage phi29 pRNA to form a hexameric complex for viral DNA transportation, Mol Cell, 2 (1998) 149–155. [DOI] [PubMed] [Google Scholar]
- [117].Afonin KA, Grabow WW, Walker FM, Bindewald E, Dobrovolskaia MA, Shapiro BA, Jaeger L, Design and self-assembly of siRNA-functionalized RNA nanoparticles for use in automated nanomedicine, Nature protocols, 6 (2011) 2022–2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Jasinski D, Haque F, Binzel DW, Guo PX, Advancement of the Emerging Field of RNA Nanotechnology, Acs Nano, 11 (2017) 1142–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Afonin KA, Viard M, Koyfman AY, Martins AN, Kasprzak WK, Panigaj M, Desai R, Santhanam A, Grabow WW, Jaeger L, Heldman E, Reiser J, Chiu W, Freed EO, Shapiro BA, Multifunctional RNA nanoparticles, Nano Lett, 14 (2014) 5662–5671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Grimm D, Kay MA, Combinatorial RNAi: a winning strategy for the race against evolving targets?, Mol Ther, 15 (2007) 878–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Dao BN, Viard M, Martins AN, Kasprzak WK, Shapiro BA, Afonin KA, Triggering RNAi with multifunctional RNA nanoparticles and their delivery, DNA and RNA Nanotechnology, 1 (2015) 27–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Rychahou P, Haque F, Shu Y, Zaytseva Y, Weiss HL, Lee EY, Mustain W, Valentino J, Guo P, Evers BM, Delivery of RNA nanoparticles into colorectal cancer metastases following systemic administration, ACS Nano, 9 (2015) 1108–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Feng L, Li SK, Liu H, Liu CY, LaSance K, Haque F, Shu D, Guo P, Ocular delivery of pRNA nanoparticles: distribution and clearance after subconjunctival injection, Pharm Res, 31 (2014) 1046–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Li H, Lee T, Dziubla T, Pi F, Guo S, Xu J, Li C, Haque F, Liang XJ, Guo P, RNA as a stable polymer to build controllable and defined nanostructures for material and biomedical applications, Nano Today, 10 (2015) 631–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Shu Y, Pi F, Sharma A, Rajabi M, Haque F, Shu D, Leggas M, Evers BM, Guo P, Stable RNA nanoparticles as potential new generation drugs for cancer therapy, Adv Drug Deliv Rev, 66 (2014) 74–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Binzel DW, Shu Y, Li H, Sun M, Zhang Q, Shu D, Guo B, Guo P, Specific Delivery of MiRNA for High Efficient Inhibition of Prostate Cancer by RNA Nanotechnology, Mol Ther, 24 (2016) 1267–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Lee H, Lytton-Jean AK, Chen Y, Love KT, Park AI, Karagiannis ED, Sehgal A, Querbes W, Zurenko CS, Jayaraman M, Peng CG, Charisse K, Borodovsky A, Manoharan M, Donahoe JS, Truelove J, Nahrendorf M, Langer R, Anderson DG, Molecularly self-assembled nucleic acid nanoparticles for targeted in vivo siRNA delivery, Nat Nanotechnol, 7 (2012) 389–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Rose SD, Kim DH, Amarzguioui M, Heidel JD, Collingwood MA, Davis ME, Rossi JJ, Behlke MA, Functional polarity is introduced by Dicer processing of short substrate RNAs, Nucleic Acids Res, 33 (2005) 4140–4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Afonin KA, Kasprzak WK, Bindewald E, Kireeva M, Viard M, Kashlev M, Shapiro BA, In silico design and enzymatic synthesis of functional RNA nanoparticles, Acc Chem Res, 47 (2014) 1731–1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Guo P, The emerging field of RNA nanotechnology, Nat Nanotechnol, 5 (2010) 833–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Shu D, Shu Y, Haque F, Abdelmawla S, Guo P, Thermodynamically stable RNA three-way junction for constructing multifunctional nanoparticles for delivery of therapeutics, Nat Nanotechnol, 6 (2011) 658–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Chandler M, Johnson MB, Panigaj M, Afonin KA, Innate immune responses triggered by nucleic acids inspire the design of immunomodulatory nucleic acid nanoparticles (NANPs), Curr Opin Biotech, 63 (2020) 8–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Khisamutdinov EF, Li H, Jasinski DL, Chen J, Fu J, Guo P, Enhancing immunomodulation on innate immunity by shape transition among RNA triangle, square and pentagon nanovehicles, Nucleic Acids Res, 42 (2014) 9996–10004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Dobrovoskaia MA, Bathe M, Opportunities and challenges for the clinical translation of structuredDNAassemblies as gene therapeutic delivery and vaccine vectors, Wires Nanomed Nanobi, 13 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Ke W, Hong E, Saito RF, Rangel MC, Wang J, Viard M, Richardson M, Khisamutdinov EF, Panigaj M, Dokholyan NV, Chammas R, Dobrovolskaia MA, Afonin KA, RNA-DNA fibers and polygons with controlled immunorecognition activate RNAi, FRET and transcriptional regulation of NF-κB in human cells, Nucleic acids research, 47 (2019) 1350–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Lee BL, Moon JE, Shu JH, Yuan L, Newman ZR, Schekman R, Barton GM, UNC93B1 mediates differential trafficking of endosomal TLRs, Elife, 2 (2013) e00291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Kim YM, Brinkmann MM, Paquet ME, Ploegh HL, UNC93B1 delivers nucleotide-sensing toll-like receptors to endolysosomes, Nature, 452 (2008) 234–238. [DOI] [PubMed] [Google Scholar]
- [138].Fukui R, Saitoh S, Matsumoto F, Kozuka-Hata H, Oyama M, Tabeta K, Beutler B, Miyake K, Unc93B1 biases Toll-like receptor responses to nucleic acid in dendritic cells toward DNA-but against RNA-sensing, J Exp Med, 206 (2009) 1339–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Hornung V, Rothenfusser S, Britsch S, Krug A, Jahrsdorfer B, Giese T, Endres S, Hartmann G, Quantitative expression of toll-like receptor 1–10 mRNA in cellular subsets of human peripheral blood mononuclear cells and sensitivity to CpG oligodeoxynucleotides, J Immunol, 168 (2002) 4531–4537. [DOI] [PubMed] [Google Scholar]
- [140].Hong EP, Halman JR, Shah AB, Khisamutdinov EF, Dobrovolskaia MA, Afonin KA, Structure and Composition Define Immunorecognition of Nucleic Acid Nanoparticles, Nano Lett, 18 (2018) 4309–4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Hong E, Halman JR, Shah A, Cedrone E, Truong N, Afonin KA, Dobrovolskaia MA, Toll-Like Receptor-Mediated Recognition of Nucleic Acid Nanoparticles (NANPs) in Human Primary Blood Cells, Molecules, 24 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Choi MK, Wang ZC, Ban T, Yanai H, Lu Y, Koshiba R, Nakaima Y, Hangai S, Savitsky D, Nakasato M, Negishi H, Takeuchi O, Honda K, Akira S, Tamura T, Taniguchi T, A selective contribution of the RIG-I-like receptor pathway to type I interferon responses activated by cytosolic DNA, P Natl Acad Sci USA, 106 (2009) 17870–17875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Hornung V, Ablasser A, Charrel-Dennis M, Bauernfeind F, Horvath G, Caffrey DR, Latz E, Fitzgerald KA, AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC, Nature, 458 (2009) 514–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Nordmeier S, Ke W, Afonin KA, Portnoy V, Exosome mediated delivery of functional nucleic acid nanoparticles (NANPs), Nanomedicine, 30 (2020) 102285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Kim T, Viard M, Afonin KA, Gupta K, Popov M, Salotti J, Johnson PF, Linder C, Heldman E, Shapiro BA, Characterization of Cationic Bolaamphiphile Vesicles for siRNA Delivery into Tumors and Brain, Mol Ther Nucleic Acids, 20 (2020) 359–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Juneja R, Vaderevu H, Halman JR, Tarannum M, Rackley L, Dobbs J, Marquez J, Chandler M, Afonin KA, Vivero-Escoto JL, Combination of nucleic acid and mesoporous silica nanoparticles: optimization and therapeutic performance in vitro, ACS Appl Mater Interfaces, 12 (2020) 38873–38886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Johnson MB, Halman JR, Miller DK, Cooper JS, Khisamutdinov EF, Marriott I, Afonin KA, The immunorecognition, subcellular compartmentalization, and physicochemical properties of nucleic acid nanoparticles can be controlled by composition modification, Nucleic Acids Res, 48 (2020) 11785–11798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Juneja R, Lyles Z, Vadarevu H, Afonin KA, Vivero-Escoto JL, Multimodal Polysilsesquioxane Nanoparticles for Combinatorial Therapy and Gene Delivery in Triple-Negative Breast Cancer, ACS Appl Mater Interfaces, 11 (2019) 12308–12320. [DOI] [PubMed] [Google Scholar]
- [149].Cruz-Acuna M, Halman JR, Afonin KA, Dobson J, Rinaldi C, Magnetic nanoparticles loaded with functional RNA nanoparticles, Nanoscale, 10 (2018) 17761–17770. [DOI] [PubMed] [Google Scholar]
- [150].Parlea L, Puri A, Kasprzak W, Bindewald E, Zakrevsky P, Satterwhite E, Joseph K, Afonin KA, Shapiro BA, Cellular Delivery of RNA Nanoparticles, ACS Comb Sci, 18 (2016) 527–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Gupta K, Mattingly SJ, Knipp RJ, Afonin KA, Viard M, Bergman JT, Stepler M, Nantz MH, Puri A, Shapiro BA, Oxime ether lipids containing hydroxylated head groups are more superior siRNA delivery agents than their nonhydroxylated counterparts, Nanomedicine (Lond), (2015) 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Gupta K, Afonin KA, Viard M, Herrero V, Kasprzak W, Kagiampakis I, Kim T, Koyfman AY, Puri A, Stepler M, Sappe A, KewalRamani VN, Grinberg S, Linder C, Heldman E, Blumenthal R, Shapiro BA, Bolaamphiphiles as carriers for siRNA delivery: From chemical syntheses to practical applications, J Control Release, 213 (2015) 142–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Kim T, Afonin KA, Viard M, Koyfman AY, Sparks S, Heldman E, Grinberg S, Linder C, Blumenthal RP, Shapiro BA, In Silico, In Vitro, and In Vivo Studies Indicate the Potential Use of Bolaamphiphiles for Therapeutic siRNAs Delivery, Mol Ther Nucleic Acids, 2 (2013) e80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Wu JX, Chen ZJ, Innate Immune Sensing and Signaling of Cytosolic Nucleic Acids, Annu Rev Immunol, 32 (2014) 461–488. [DOI] [PubMed] [Google Scholar]
- [155].Kawasaki T, Kawai T, Toll-like receptor signaling pathways, Front Immunol, 5 (2014) 461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Minchin S, Lodge J, Understanding biochemistry: structure and function of nucleic acids, Essays Biochem, 63 (2019) 433–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Bui MN, Brittany Johnson M, Viard M, Satterwhite E, Martins AN, Li Z, Marriott I, Afonin KA, Khisamutdinov EF, Versatile RNA tetra-U helix linking motif as a toolkit for nucleic acid nanotechnology, Nanomedicine, 13 (2017) 1137–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Johnson MB, Halman JR, Satterwhite E, Zakharov AV, Bui MN, Benkato K, Goldsworthy V, Kim T, Hong E, Dobrovolskaia MA, Khisamutdinov EF, Marriott I, Afonin KA, Programmable Nucleic Acid Based Polygons with Controlled Neuroimmunomodulatory Properties for Predictive QSAR Modeling, Small, 13 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Abdelmawla S, Guo S, Zhang L, Pulukuri SM, Patankar P, Conley P, Trebley J, Guo P, Li QX, Pharmacological characterization of chemically synthesized monomeric phi29 pRNA nanoparticles for systemic delivery, Mol Ther, 19 (2011) 1312–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Guo S, Li H, Ma M, Fu J, Dong Y, Guo P, Size, Shape, and Sequence-Dependent Immunogenicity of RNA Nanoparticles, Mol Ther Nucleic Acids, 9 (2017) 399–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Khisamutdinov EF, Li H, Jasinski DL, Chen J, Fu J, Guo PX, Enhancing immunomodulation on innate immunity by shape transition among RNA triangle, square and pentagon nanovehicles, Nucleic Acids Res, 42 (2014) 9996–10004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Yin H, Wang H, Li Z, Shu D, Guo P, RNA Micelles for the Systemic Delivery of Anti-miRNA for Cancer Targeting and Inhibition without Ligand, ACS Nano, 13 (2019) 706–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].Choung S, Kim YJ, Kim S, Park HO, Choi YC, Chemical modification of siRNAs to improve serum stability without loss of efficacy, Biochem Bioph Res Co, 342 (2006) 919–927. [DOI] [PubMed] [Google Scholar]
- [164].Allerson CR, Sioufi N, Jarres R, Prakash TP, Naik N, Berdeja A, Wanders L, Griffey RH, Swayze EE, Bhat B, Fully 2’-modified oligonucleotide duplexes with improved in vitro potency and stability compared to unmodified small interfering RNA, J Med Chem, 48 (2005) 901–904. [DOI] [PubMed] [Google Scholar]
- [165].Kratschmer C, Levy M, Effect of Chemical Modifications on Aptamer Stability in Serum, Nucleic Acid Ther, 27 (2017) 335–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [166].Hassler MR, Turanov AA, Alterman JF, Haraszti RA, Coles AH, Osborn MF, Echeverria D, Nikan M, Salomon WE, Roux L, Godinho B, Davis SM, Morrissey DV, Zamore PD, Karumanchi SA, Moore MJ, Aronin N, Khvorova A, Comparison of partially and fully chemically-modified siRNA in conjugate-mediated delivery in vivo, Nucleic Acids Res, 46 (2018) 2185–2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [167].Sioud M, Single-stranded small interfering RNA are more immunostimulatory than their double-stranded counterparts: A central role for 2 ‘-hydroxyl uridines in immune responses, Eur J Immunol, 36 (2006) 1222–1230. [DOI] [PubMed] [Google Scholar]
- [168].Lee Y, Urban JH, Xu L, Sullenger BA, Lee J, 2 ‘ Fluoro Modification Differentially Modulates the Ability of RNAs to Activate Pattern Recognition Receptors, Nucleic Acid Ther, 26 (2016) 173–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [169].Robbins M, Judge A, Liang L, McClintock K, Yaworski E, MacLachlan I, 2’-O-methyl-modified RNAs act as TLR7 antagonists, Mol Ther, 15 (2007) 1663–1669. [DOI] [PubMed] [Google Scholar]
- [170].Arulkumaran N, Lanphere C, Gaupp C, Burns JR, Singer M, Howorka S, DNA Nanodevices with Selective Immune Cell Interaction and Function, ACS Nano, (2021). [DOI] [PubMed] [Google Scholar]
- [171].Komura F, Okuzumi K, Takahashi Y, Takakura Y, Nishikawa M, Development of RNA/DNA Hydrogel Targeting Toll-Like Receptor 7/8 for Sustained RNA Release and Potent Immune Activation, Molecules, 25 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [172].Guan CX, Chernyak N, Dominguez D, Cole L, Zhang B, Mirkin CA, RNA-Based Immunostimulatory Liposomal Spherical Nucleic Acids as Potent TLR7/8 Modulators, Small, 14 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [173].Toy R, Keenum MC, Pradhan P, Phang K, Chen P, Chukwu C, Nguyen LAH, Liu J, Jain S, Kozlowski G, Hosten J, Suthar MS, Roy K, TLR7 and RIG-I dual-adjuvant loaded nanoparticles drive broadened and synergistic responses in dendritic cells in vitro and generate unique cellular immune responses in influenza vaccination, J Control Release, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [174].Gustafson HH, Holt-Casper D, Grainger DW, Ghandehari H, Nanoparticle Uptake: The Phagocyte Problem, Nano Today, 10 (2015) 487–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [175].Jasinski DL, Li H, Guo P, The Effect of Size and Shape of RNA Nanoparticles on Biodistribution, Mol Ther, 26 (2018) 784–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [176].Ghimire C, Wang HZ, Li H, Vieweger M, Xu CC, Guo PX, RNA Nanoparticles as Rubber for Compelling Vessel Extravasation to Enhance Tumor Targeting and for Fast Renal Excretion to Reduce Toxicity, Acs Nano, 14 (2020) 13180–13191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [177].Shu C, Li X, Li P, The mechanism of double-stranded DNA sensing through the cGAS-STING pathway, Cytokine Growth Factor Rev, 25 (2014) 641–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [178].Qi X, Liu X, Matiski L, Rodriguez Del Villar R, Yip T, Zhang F, Sokalingam S, Jiang S, Liu L, Yan H, Chang Y, RNA Origami Nanostructures for Potent and Safe Anticancer Immunotherapy, ACS Nano, 14 (2020) 4727–4740. [DOI] [PubMed] [Google Scholar]
- [179].Wesselhoeft RA, Kowalski PS, Parker-Hale FC, Huang YX, Bisaria N, Anderson DG, RNA Circularization Diminishes Immunogenicity and Can Extend Translation Duration In Vivo, Mol Cell, 74 (2019) 508-+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [180].Jockel S, Nees G, Sommer R, Zhao Y, Cherkasov D, Hori H, Ehm G, Schnare M, Nain M, Kaufmann A, Bauer S, The 2’-O-methylation status of a single guanosine controls transfer RNA-mediated Toll-like receptor 7 activation or inhibition, J Exp Med, 209 (2012) 235–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [181].Gehrig S, Eberle ME, Botschen F, Rimbach K, Eberle F, Eigenbrod T, Kaiser S, Holmes WM, Erdmann VA, Sprinzl M, Bec G, Keith G, Dalpke AH, Helm M, Identification of modifications in microbial, native tRNA that suppress immunostimulatory activity, Journal of Experimental Medicine, 209 (2012) 225–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [182].Hong E, Halman JR, Shah AB, Khisamutdinov EF, Dobrovolskaia MA, Afonin KA, Structure and Composition Define Immunorecognition of Nucleic Acid Nanoparticles, Nano Lett, 18 (2018) 4309–4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [183].Rackley L, Stewart JM, Salotti J, Krokhotin A, Shah A, Halman JR, Juneja R, Smollett J, Lee L, Roark K, Viard M, Tarannum M, Vivero-Escoto J, Johnson PF, Dobrovolskaia MA, Dokholyan NV, Franco E, Afonin KA, RNA Fibers as Optimized Nanoscaffolds for siRNA Coordination and Reduced Immunological Recognition, Adv Funct Mater, 28 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [184].Ke W, Hong E, Saito RF, Rangel MC, Wang J, Viard M, Richardson M, Khisamutdinov EF, Panigaj M, Dokholyan NV, Chammas R, Dobrovolskaia MA, Afonin KA, RNA-DNA fibers and polygons with controlled immunorecognition activate RNAi, FRET and transcriptional regulation of NF-kappaB in human cells, Nucleic Acids Res, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [185].Halman JR, Kim KT, Gwak SJ, Pace R, Johnson MB, Chandler MR, Rackley L, Viard M, Marriott I, Lee JS, Afonin KA, A cationic amphiphilic co-polymer as a carrier of nucleic acid nanoparticles (Nanps) for controlled gene silencing, immunostimulation, and biodistribution, Nanomed-Nanotechnol, 23 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [186].Broering R, Real CI, John MJ, Jahn-Hofmann K, Ickenstein LM, Kleinehr K, Paul A, Gibbert K, Dittmer U, Gerken G, Schlaak JF, Chemical modifications on siRNAs avoid Toll-like-receptor-mediated activation of the hepatic immune system in vivo and in vitro, Int Immunol, 26 (2014) 35–46. [DOI] [PubMed] [Google Scholar]
- [187].Afonin KA, Dobrovolskaia MA, Church G, Bathe M, Opportunities, Barriers, and a Strategy for Overcoming Translational Challenges to Therapeutic Nucleic Acid Nanotechnology, ACS Nano, 14 (2020) 9221–9227. [DOI] [PMC free article] [PubMed] [Google Scholar]


