Abstract
About 40% of women with infertility and 70% of women with pelvic pain suffer from endometriosis. The pregnancy rate in women undergoing IVF with low endometrial integrin αvβ3 (LEI) expression is significantly lower compared to the women with high endometrial integrin αvβ3 (HEI). Mid-secretory eutopic endometrial biopsies were obtained from healthy controls (C; n=3), and women with HEI (n=4) and LEI (n=4) and endometriosis. Changes in gene expression were assessed using human gene arrays and DNA methylation data were derived using 385 K Two-Array Promoter Arrays. Transcriptional analysis revealed that LEI and C groups clustered separately with 396 differentially expressed genes (DEGs) (P<0.01: 275 up and 121 down) demonstrating that transcriptional and epigenetic changes are distinct in the LEI eutopic endometrium compared to the C and HEI group. In contrast, HEI vs C and HEI vs LEI comparisons only identified 83 and 45 DEGs, respectively. The methylation promoter array identified 1304 differentially methylated regions in the LEI vs C comparison. The overlap of gene and methylation array data identified 14 epigenetically dysregulated genes and quantitative RT–PCR analysis validated the transcriptomic findings. The analysis also revealed that aryl hydrocarbon receptor (AHR) was hypomethylated and significantly overexpressed in LEI samples compared to C. Further analysis validated that AHR transcript and protein expression are significantly (P<0.05) increased in LEI women compared to C. The increase in AHR, together with the altered methylation status of the 14 additional genes, may provide a diagnostic tool to identify the subset of women who have endometriosis-associated infertility.
Keywords: integrin αvβ3, endometriosis, epigenetics, gene expression, infertility, eutopic endometrium, aryl hydrocarbon receptor, methylation
Introduction
Endometriosis is estimated to affect 10–15% of reproductive-aged women. Up to 40% of women with infertility have endometriosis, and over 70% of women with chronic pelvic pain suffer from this condition (Practice Committee of the American Society for Reproductive Medicine, 2006; Bulun, 2009). The predominant mechanism for the development of endometriosis is thought to be the result of retrograde menstruation in susceptible women (Halme et al., 1984). Although 90% of women undergo retrograde menstruation, there is a need to better understand which genetic, epigenetic, and/or environmental factors predispose some women to develop endometriosis. More importantly, endometriosis itself has significant implications for a woman's fertility as well as chronic pelvic pain or dysmenorrhea (Miller et al., 2012, 2017; Tanbo and Fedorcsak, 2017). Assessment of endometrial histology and expression of receptivity markers, such as integrin status (Ceydeli et al., 2006; Chen et al., 2016; Dorostghoal, 2017), may be useful to identify women with defects in uterine receptivity and have the added utility to predict which patients could benefit from medical or surgical intervention (Lessey et al., 1995; Lessey, 2000; Germeyer et al., 2014). Spatial and temporal expression of integrins, which are heterodimeric glycoproteins, have been observed to undergo a dynamic change in the endometrium during the menstrual cycle in women and non-human primates along with extracellular matrix remodeling (Fazleabas et al., 1997; Lessey, 1998). Integrins are a group of cell surface receptors that play critical roles in cell-matrix adhesion and, upon ligand binding, transduces external signals to the cytoskeleton (Giancotti and Ruoslahti, 1999; Salvatori et al., 2014). The pregnancy rate in women undergoing IVF with low αvβ3 integrin expression was significantly lower compared to the women with higher αvβ3 expression (Miller et al., 2012). It is possible that low integrin β3 levels might be related to the histologic delay in the endometrium, as it does not appear until Day 20 of the menstrual cycle. This phenomenon is also referred to as a Type I defect (Lessey et al., 1995; Lessey, 1998). The lower expression of integrin β3 and ‘in phase’ glandular histology (Day 20 and beyond) in the eutopic endometrium of women with endometriosis is referred to as Type II endometrial defect (Lessey et al., 1994; Lessey, 2000).
Epigenetic changes are reversible changes that do not alter the nucleotide sequence but still regulate gene expression and act as a bridge between the environmental signals and genetics (Zhang and Ho, 2011). It is very well established that epigenetic mechanisms regulate female reproductive tract functions and contribute to the non-malignant and malignant pathogenesis of gynecological disorders, including endometriosis (Kim et al., 2007; Izawa et al., 2013; Jones et al., 2013; Dyson et al., 2014; Naqvi et al., 2014; Houshdaran et al., 2016; Kim et al., 2019; Houshdaran et al., 2020). One of the earliest reports related to the aberrant methylation changes in endometriosis focused on the methylation of CpG islands within the 5' untranslated region of the HOXA10 gene (Wu et al., 2005). HOXA10, a homeobox transcription factor, is critical for normal uterine functions and is hypermethylated in the endometrium of baboons and women with endometriosis (Wu et al., 2005; Kim et al., 2007). Subsequent studies have attempted to analyze the epigenetic alterations between: the endometrium of women with and without endometriosis (Houshdaran et al., 2016; Akter et al., 2019); isolated endometrial stromal cells from eutopic and ectopic endometrium (Yamagata et al., 2014); isolated endometrial stromal cells from women with and without endometriosis (Yotova et al., 2017); and in vitro decidualized endometrial stromal cells from normal or diseased endometrium (Logan et al., 2012; Maekawa et al., 2019). These comparisons are very valuable and reveal the extent of epigenetic alterations on phenotypic changes in the endometrium of women with endometriosis (Izawa et al., 2013). However, one of the outstanding questions has been to determine the molecular differences between the subset of women who can get pregnant with endometriosis compared to those who cannot. The opportunity for surgical intervention exists such that women who are operated upon for endometriosis see an improvement in their fertility regardless of the amount of disease at the time of surgery (Marcoux et al., 1997; Joshi et al., 2017). These observations further support the fact that epigenetic changes play a critical role in the pathophysiology of endometriosis and could be reversed by surgical and/or therapeutic interventions. However, there is a significant need to understand the epigenetic regulations in the context of endometriosis-associated infertility.
This study aims to investigate the transcriptomic and epigenetic changes in the eutopic endometrium during the window of implantation in women with endometriosis and low αvβ3 endometrial expression compared to disease-free controls. We hypothesize that women with endometriosis together with low αvβ3 endometrial expression have a distinct transcriptomic signature, which is influenced by epigenetic modification that may contribute to the observed infertility in a subset of women with endometriosis.
Materials and methods
Human sample acquisition
Endometrial biopsies were obtained from women undergoing a workup for infertility at Greenville Health System (Division of Reproductive Endocrinology and Infertility) and the University of North Carolina, per IRB (Pro00013885) protocols approved by both institutions. Samples included timed endometrial biopsies in the mid-secretory phase as well as endometrium obtained in the proliferative phase. None of the subjects had been on hormone therapy for the preceding 3 months. The control samples represent endometrium obtained at laparoscopy or timed to the LH surge in the clinic. In the treatment group, endometriosis was diagnosed at laparoscopy by experienced gynecological surgeons and confirmed by pathology when present. Endometrial biopsies from these patients were saved for immunohistochemistry, and RNA and DNA isolation. The endometrial biopsies were histologically assessed for αvβ3 expression. For protein isolation (western blots), human endometrial samples were obtained through the Michigan State University's Center for Women’s Health Research Female Reproductive Tract Biorepository and Spectrum Health Universal Biorepository (Grand Rapids, MI, USA) (IRB- 2010-085). Participants were 18–45 years of age and had regular menstrual cycles. The details of the samples utilized in this study are provided in the Supplementary Tables SI and SII.
DNA and RNA isolation
Mid-secretory eutopic endometrial biopsies (25 mg) were processed for genomic and DNA and RNA isolation. Genomic DNA was extracted using the QIAGEN DNA isolation kit following the manufacturer’s protocol (QIAGEN, Valencia, CA, USA). Total RNA was isolated using Trizol reagent (Invitrogen, Waltham, MA, USA) as previously described (Joshi et al., 2017), and an RNA quality check was performed using NanoDrop 2000 (Thermo Scientific, Waltham, MA, USA).
DNA methylation analysis
DNA methylation data from low endometrial Integrin αvβ3 (LEI) (n=4), high endometrial integrin αvβ3 (HEI) (n=4), and controls (C, n=3) groups were obtained using the Roche Nimblegen 385 K Two-Array DNA Methylation Promoter Arrays. Using the NimbleGen Dual-Color DNA Labeling Kit, labeling of the immunoprecipitated and input DNA samples with Cy5 and Cy3 was performed. Differential DNA methylation analysis was performed in four steps using statistical software R (http://www.R-project.org/, Vienna, Austria). In the first step, raw data from GPR files were imported and normalized using a LOESS-like method. In the second step, a linear model with Group as the only parameter was fitted for the M value (ratio of IP/Input in log2 scale) of each probe separately using limma (Smyth, 2004). Appropriate contrasts were then used to calculate the difference between HEI vs C, LEI vs C, and LEI vs HEI. In the third step, a sliding window approach was used to identify the enriched differentially methylated regions (DMRs). Specifically, a RandomSet (Newton et al., 2007) analysis was performed on probe scores (−log10 transformed P-value derived in step two) for each sliding window of 750 bps. The sliding windows with <5 probes were ignored. In the last step, a gene-level differential methylation result was generated where each the gene was summarized by the most significant differentially methylated window in the −3.5k to +0.75k promoter region in each comparison.
Expression analysis
RNA expression data from the same samples used for DNA methylation analysis were derived using Affymetrix Human Gene 1.0 ST Arrays, which covers 32 020 well-established annotated RefSeq coding transcripts. The data were aligned to human reference genome hg18 and analyzed using R, and Bioconductor packages with custom CDF downloaded (Dai et al., 2005). Data were preprocessed using Robust Multiarray Analysis and quality assessed using arrayQualityMetrics. Differential expression analysis was performed using limma (Smyth, 2004). Functional categories from Gene Ontology and Kyoto Encyclopedia of Genes and Genomes (KEGG) associated with differential expression and methylation were discovered using clusterProfiler (Sartor et al., 2009).
The typical limitation of probe-based technologies, such as a microarray, is the small number of observations per gene. This produces low power for statistical tests like Student’s t-test, the most widely used statistical method in genetic studies for comparing two groups, owing to instability in estimating gene-specific variances (Tusher et al., 2001; Smyth et al., 2003). When sample sizes are small, it would be even harder to observe low P-values. In this study, the limited sample size combined with individual genetic variations among patients resulted in low statistical power, which was inflated after correction for multiplicity. As a result, we were unable to identify differentially expressed genes (DEGs) that were significantly changed when applying a false discovery rate (FDR) =0.05. In order not to compromise the biological context of this research and to avoid weakening the biological relevance of the data, unadjusted P-values are reported for the list of the DEGs. However, in this study, we tested and validated most candidate genes by quantitative RT–PCR (qRT–PCR) analysis to confirm changes in expression.
qRT–PCR techniques
qRT–PCR was performed as previously described (Joshi et al., 2017). Briefly, for mRNA analysis, total RNA (1000 ng) was reverse transcribed using a High-Capacity cDNA synthesis kit (ThermoFisher Scientific, Waltham, MA, USA). After cDNA synthesis, the qRT–PCR reaction was carried out for genes using specific primers (Supplementary Table SIII) for nine genes listed in Table I, including aryl hydrocarbon receptor (AHR) and 18S (internal control) with SyBrGn PCR master mix (ThermoFisher Scientific, Waltham, MA, USA). All quantitative real-time PCR (RT–PCR) reactions were run for 40 cycles, and fold change was calculated using ΔΔCt method (Livak and Schmittgen, 2001).
Table I.
Genes that are differentially methylated and differentially expressed in the low endometrial αvβ3 integrin expression vs healthy control comparison.
| Gene symbol | Gene name | Gene expression data |
Methylation array data |
||
|---|---|---|---|---|---|
| log2FC | RNA expression | PEAK | DNA methylation | ||
| RNPC3 | RNA binding region (RNP1, RRM) containing 3 | 0.673 | Up | −0.629 | Decreased |
| SLC18A2 | Solute carrier family 18 member A2 | 1.159 | Up | −0.445 | Decreased |
| SMCHD1 | Structural maintenance of chromosomes flexible hinge domain containing 1 | 0.733 | Up | −0.508 | Decreased |
| SNORD58A | Small nucleolar RNA, C/D box 58A | 1.353 | Up | −0.518 | Decreased |
| PGLYRP2 | Peptidoglycan recognition protein 2 | −0.629 | Down | 0.504 | Increased |
| SNORD82 | Small nucleolar RNA, C/D box 82 | 0.596 | Up | −0.506 | Decreased |
| ANXA3 | Annexin A3 | 0.930 | Up | −0.362 | Decreased |
| HIST1H4F | H4 clustered histone 6 | −0.657 | Down | 0.664 | Increased |
| HIST1H3I | H3 clustered histone 11 | −0.656 | Down | 0.928 | Increased |
| PGK2 | Phosphoglycerate kinase 2 | 1.073 | Up | −0.449 | Decreased |
| AHR | Aryl hydrocarbon receptor | 0.834 | Up | −0.545 | Decreased |
| CCDC146 | Coiled-coil domain containing 146 | 0.628 | Up | −0.487 | Decreased |
| AGPAT5 | 1-acylglycerol-3-phosphate O-acyltransferase 5 | 0.586 | Up | −0.828 | Decreased |
| PLEKHF2 | Pleckstrin homology and FYVE domain containing 2 | 1.117 | Up | −0.507 | Decreased |
The genes highlighted in bold, and italics were validated by quantitative RT–PCR analysis to confirm the gene array results.
Immunohistochemistry and H-SCORE analysis
Immunostaining for beta 3 integrin and AHR proteins was performed on formalin-fixed eutopic endometrial sections from women with and without endometriosis, as previously described (Germeyer et al., 2014). Briefly, paraffin-embedded sections of each sample (5 um) were deparaffinized in xylene and rehydrated. Non-specific binding sites were blocked with 2% normal goat serum for 30 min at room temperature followed by overnight incubation at 4°C with either the mouse monoclonal SSA6 antibody specific to the beta 3 integrin (1:2000) or 1:400 a-AHR receptor (H-211) sc5579 (Santa Cruz Biotechnology Inc., Dallas, TX, USA.). Subsequently, sections were washed with PBS and incubated with biotinylated goat anti-mouse secondary antibody (Vectastain Elite ABC kit, Vector Laboratories Inc., Burlingame, CA, USA) at a dilution of 1:200 for 30 min at room temperature. After rinsing with PBS, 3,3-diaminobenzidine was used as a chromagen, and slides were counterstained with toluidine blue followed by dehydration in a graded series of ethanols, cleared in xylene, and mounted with Permount (Fisher Scientific, Fair Lawn, NJ, USA). The resulting staining was evaluated by a single-blinded observer, and images were captured using the Spot Insight 4 Camera (Diagnostic Instruments, Inc., Sterling Heights, MI, USA) mounted on a Nikon Eclipse E600 Microscope (Nikon Inc., Japan) (Joshi et al., 2017). H-SCORE was calculated using the following equation: H-SCORE=∑ Pi (i+1), where ‘i’ is the intensity of staining with a value of 1, 2, or 3 (weak, moderate, or strong, respectively) and Pi is the percentage of stained epithelial cells for each intensity, varying from 0% to 100%. Low intra-observer (r=0.983; P<0.0001) and inter-observer (r=0.994; P<0.0001) differences for H-SCORE in uterine tissues have been reported previously using this technique (Detre et al., 1995; Jackson et al., 2007). A low integrins H-SCORE was defined as ≤0.7 (Lessey et al., 1994; Franasiak et al., 2014).
Western blot analysis
Endometrial tissues were lysed with lysis buffer (150 mM NaCl, 10 mM Tris–HCl (pH 7.4)), 2.5 mM EDTA, 0.125% Nonidet P-40 (vol/vol)) with both a protease inhibitor cocktail (Roche, Indianapolis, IN, USA) and a phosphatase inhibitor cocktail (Sigma Aldrich, St. Louis, MO, USA). A total of 20 μg of total protein lysates were electrophoresed using SDS-PAGE and transferred onto polyvinylidene difluoride membrane (Millipore Corp., Bedford, MA, USA). Casein (0.5% v/v) was used to block the membrane before exposure to antibodies against AHR and beta-actin overnight. Immunoreactivity was visualized by incubation with a horse-radish peroxidase-linked secondary antibody followed by exposure to enhanced chemiluminescence reagents according to manufacturer's instructions (GE Healthcare Biosciences, Piscataway, NJ, USA).
Statistical analysis
Differences in gene expression were compared following normalization against the 18 s internal control gene. One-way ANOVA was used to test the null hypothesis of group differences, followed by Tukey’s post-hoc test. For qRT–PCR analysis, gene expression was normalized against 18 s followed by the Student’s t-test for pair-wise comparison at a 95% CI (P<0.05) between negative control and endometriosis. The qRT–PCR data were analyzed for the Q-value using the equation: qi=pi×N/I, where pi–ith smallest P-value out of N total P-values of the experiment (Benjamini and Hochberg, 1995) for the correction of multiple testing. The data were analyzed using Graphpad Prism 8.0 statistical software (GraphPad Software, San Diego, CA, USA).
Results
Classification of endometrial samples based on integrin αvβ3 expression
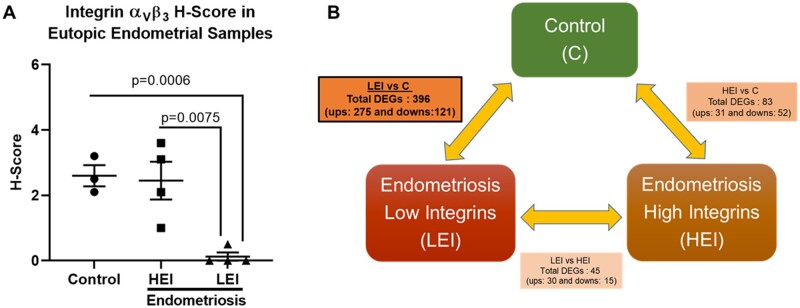
Immunohistochemical evaluation of integrin αvβ3 expression categorized endometrial biopsies into mid-secretory eutopic endometrium from disease-free control (C; n=3), HEI expression and LEI expression women with endometriosis (n=4/group). H-score analysis for αvβ3 expression revealed that LEI expression women with endometriosis had significantly reduced expression of αvβ3 compared to disease-free control (P=0.0006) and HEI expression women with endometriosis (P=0.0075), as shown in Fig. 1A.
Figure 1.
Integrin αvβ3 expression is significantly decreased in the subset of women with endometriosis compared to controls. H-score analysis shows that integrin αvβ3 expression is significantly decreased in the subset of women with endometriosis compared to controls (A). Microarray performed on mid-secretory eutopic endometrial samples from three groups of women demonstrates that the highest number of DEGs was observed in the low endometrial integrin αvβ3 (LEI) vs controls (C) comparison (B). Data are displayed as mean±SD and analyzed by One-Way ANOVA followed by Tukey’s multiple comparisons test. HEI, high endometrial integrin αvβ3.
The transcriptomic signature of eutopic endometrium is unique between controls and women with endometriosis
Eutopic endometrial biopsies from all three groups were analyzed for global gene expression profiles and genome-wide changes in promoter methylation patterns in the same samples to identify the influence of an altered methylation signature on gene expression patterns. The data obtained from the gene expression arrays identified 396 DEGs (275 up and 121 down; P<0.05, with +/− 1.5-fold change) in the LEI vs C group comparison. In contrast, 83 DEGs (31 up and 52 down) in the HEI vs C and only 45 (30 up and 15 down) DEGs in the LEI vs HEI comparisons were observed. We observed the highest gene expression changes in the eutopic endometrium between the low integrin endometriosis group vs the control group, suggesting that the gene expression signature of women with low αvβ3 is unique compared to control or women with endometriosis with higher integrin αvβ3 expression (Fig. 1B).
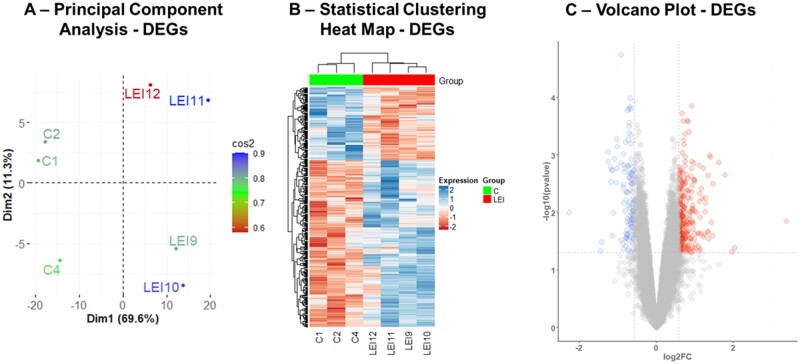
Principal component analysis (PCA) separated the LEI and control endometrial biopsy samples (Fig. 2A). Additionally, the hierarchical clustering of DEGs further supported the PCA analysis, and the LEI and C group samples clustered distinctly differently, as evident by the heatmap in Fig. 2B. The comparative analysis of the eutopic endometrial gene expression between the LEI and C group is represented in the Volcano plot (Fig. 2C). The PCA analysis, statistical clustering heatmap, and volcano plots for HEI vs C and LEI vs HEI comparisons are illustrated in Supplementary Figs S1A–C and S2A–C, respectively. In summary, these data demonstrated that a subset of women with endometriosis has significantly decreased integrin αvβ3 expression along with a higher number of DEGs compared to controls, which might be the primary reason for observed infertility in this subset of women. These observations made a compelling case to primarily focus on the LEI vs C comparison in this study.
Figure 2.
Gene array analysis identified 396 DEGs in the LEI vs control comparisons. PCA for 396 DEGs identified in the LEI vs C comparisons (A). Heatmap of DEGs shows that the two groups (LEI and C) cluster separately. Each column represents an individual sample, and each row represents the individual DEG in LEI vs C comparisons (B). The volcano plot shows the statistical significance (Y-axis) against the fold change (X-axis) between the LEI and C groups (C). Each blue dot represents the significantly decreased expression of an individual gene, and each red dot the significantly increased expression of a particular gene on the Volcano Plot. Dotted horizontal and vertical lines represent the cut-off fold change (±1.5-fold) and statistical significance (P<0.05), respectively.
Functional analysis of DEGs (LEI vs C) in eutopic endometrium of low integrin endometriosis women compared to controls
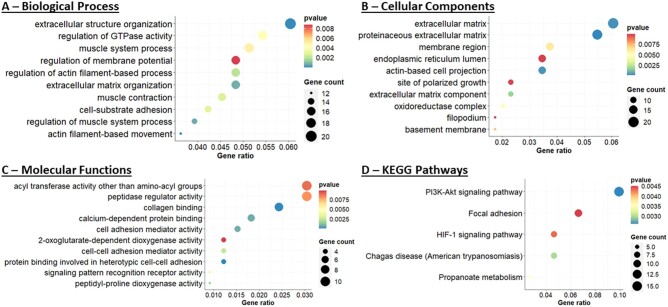
It has been reported that a subset of women with endometriosis and unexplained infertility have low expression of integrin αvβ3 in the eutopic endometrium and have poor pregnancy outcomes following IVF procedures (Miller et al., 2012). However, aberrant molecular functions and pathways associated with this defect are not known in detail. To explore the potentially altered biological functions in the eutopic endometrium of women with endometriosis (LEI) compared to controls (C), we used the R clusterProfiler package to conduct pathway enrichment analysis for 396 DEGs (Yu et al., 2012). This analysis revealed that biological processes (Fig. 3A and Supplementary Table SIV) associated with extracellular structure organization, GTPase activity, actin-filament-based movement, cell-substrate adhesion, and regulation of membrane potential were among the top 10 significantly (P<0.01) altered biological processes in the LEI group compared to controls. Analysis of the cellular component (Fig. 3B) (P<0.01; Supplementary Table SV) affected in the LEI vs C comparison identified extracellular matrix, actin-based cell projection, proteinaceous extracellular matrix, endoplasmic reticulum lumen, oxidoreductase complex, and filopodium as the major affected cellular components. The top 10 significantly (P<0.01) altered molecular function analysis (Fig. 3C and Supplementary Table SVI) resulted in the compromised activity of peptidase regulator, cell adhesion mediator, cell–cell adhesion mediator, protein binding involved in heterotypic cell–cell adhesion, and collagen binding. In addition, both phosphatidylinositol-3-kinase and protein kinase B (PI3K-AKT) and hypoxia-inducible factor signaling pathways are reported to be altered in women with endometriosis (Wu et al., 2011; Yin et al., 2012; Kim et al., 2014, 2015; Matsuzaki and Darcha, 2015; McKinnon et al., 2016; Wu et al., 2019). The KEGG analysis also revealed that the DEGs were mostly enriched in PI3K-AKT signaling, focal adhesion, propanoate metabolism, and HIF-1 signaling were the major affected pathways in the LEI women with endometriosis (Fig. 3D) (P<0.01; Supplementary Table SVII). Together with our comprehensive functional pathway analysis, we reveal that the eutopic endometrium with low integrin expression results in markedly altered biological, cellular and molecular responses that could contribute to the compromised implantation and pregnancy outcomes in the subset of women with endometriosis.
Figure 3.
Downstream molecular and functional analysis of 396 DEGs in the LEI vs control comparisons. Altered biological process (A), molecular functions (B), cellular component (C), and KEGG pathways analysis (D). Color (red to blue) gradient bar represents the degree of P-values for statistical significance, and the size of the circle (smaller to bigger) represents the number of genes involved in each of the molecular or functional pathways.
Altered methylation profile influences the eutopic endometrial gene expression in a subgroup of women with endometriosis
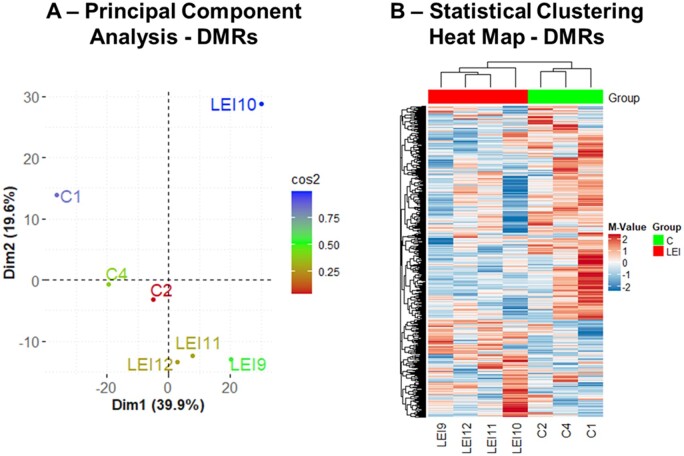
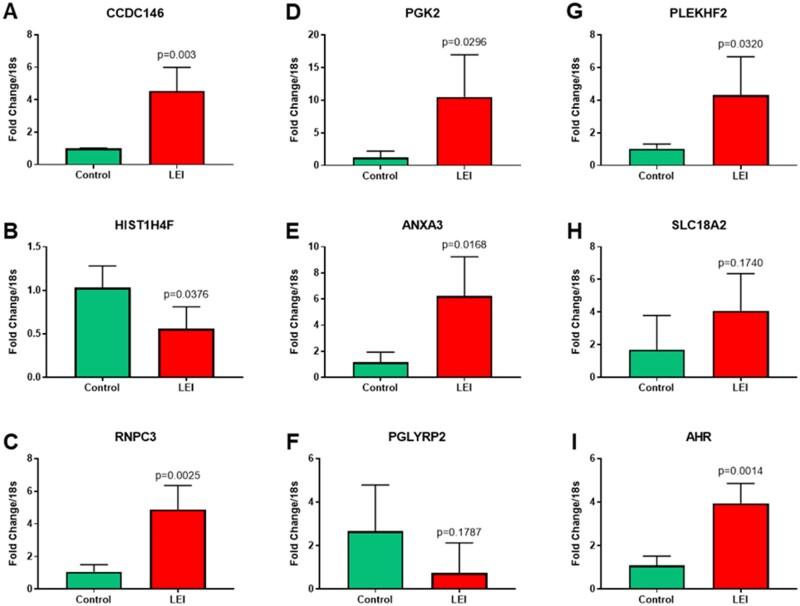
Altered methylation changes in the eutopic endometrium of women with and without endometriosis have been reported previously (Aghajanova and Giudice, 2011; Dyson et al., 2014; Houshdaran et al., 2016, 2020; Liu et al., 2020). However, to the best of our knowledge, aberrant methylation changes specifically in a subset of women with endometriosis-associated infertility and lower eutopic endometrial integrin αvβ3 expression have never been studied. To achieve this objective in this study, the eutopic endometrial samples from LEI and C groups were analyzed on Roche Nimblegen 385 K Two-Array DNA Methylation Promoter Arrays. Differential DNA methylation analysis resulted in substantial changes in global methylation patterns associated with endometriosis. We observed 1304 DMRs, identified by 5 or more probes (FDR<0.006) between LEI compared to controls. PCA and statistical clustering performed on DMRs demonstrated that LEI and C endometrial tissue samples have distinct methylation profiles and segregated the samples from the two groups (Fig. 4A and B). This study also observed 1325 DMRs between HEI vs C (Supplementary Fig. S3A and B) and 1233 DMRs between LEI vs HEI comparisons (Supplementary Fig. S4A and B). However, considering that the highest gene expression changes were observed between LEI and C groups and because of the clinical relevance of endometrial low integrin expression and infertility, we focused on the LEI vs C comparison for further downstream analysis. To identify the changes in the gene expression due to an altered methylome in LEI vs C comparison, we did overlap the DEGs and DMRs data. We created a list of genes that had an opposite pattern in gene expression analysis and methylation status. This analysis identified 14 differentially (Table I) expressed transcripts (P<0.01: 11 up and 3 down), which showed inverse changes in promoter methylation status. The details of these 14 gene symbols and their known function from the Pubmed Gene database are provided in Supplementary Table SVIII. Further, building upon NCBI Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/) search engine, we performed an extensive in silico research to compare and verify the findings of this study with previous similar datasets and explore any possible overlap with reported expression quantitative trait loci (eQTLs) (Mortlock et al., 2020) and/or methylation quantitative trait loci (mQTLs) (Mortlock et al., 2019) datasets (Supplementary Table SVIII). The in silico data obtained from the methylation array and gene array overlap analysis was validated by performing qRT–PCR analysis for nine genes randomly selected from Table I. qRT–PCR data were in agreement with the methylation and gene expression analysis. They suggested that seven genes were significantly (P<0.05) altered in the eutopic endometrium of the LEI group compared to controls (Fig. 5). The expression pattern of the other two genes also matched the array data; however, owing to higher variation, SLC18A2 (P=0.174) and PGLYRP2 (P=0.1787) were not statistically significant (Fig. 5). Q-values analysis for the correction of multiple testing of qRT–PCR data suggested that PGK2, PGLYRP2, and SLC18A2 had Q-values >0.05 threshold (Benjamini and Hochberg, 1995).
Figure 4.
DMRs identified in the LEI vs control comparisons. PCA for 1304 DMRs identified in the LEI vs C comparisons (A). Heatmap of differentially methylated genes demonstrates that the two groups (LEI and C) cluster separately. Each column represents an individual sample, and each row represents the unique DMR in the LEI vs C comparisons (B).
Figure 5.
qRT–PCR validation of genes that are differentially methylated and differentially expressed in the LEI vs control comparison. qRT–PCR validation of genes, which are differentially methylated and differentially expressed in the LEI (n=4) vs C (n=3) comparison. The green bar represents the control samples, and the red bar represents the LEI samples. Data displayed as mean±SD and analyzed by performing an unpaired Student’s t-test. CCDC146 - coiled-coil domain containing 146, HIST1H4F - H4 clustered histone 6, RNPC3 - RNA binding region (RNP1, RRM) containing 3, PGK2 - phosphoglycerate kinase 2, ANXA3 - annexin A3, PGLYRP2 - peptidoglycan recognition protein 2, PLEKHF2 - pleckstrin homology and FYVE domain containing 2, SLC18A2 - solute carrier family 18 member A2, AHR - aryl hydrocarbon receptor.
Taken together, these data suggest that differential methylation of the promoter regulated the subset of DEG in LEI women. We confirmed these previously unreported epigenetically regulated genes by the qRT–PCR analysis, which may serve as a diagnostic marker for observed infertility in a subset of women with endometriosis. Identifying these key transcripts may serve as a valuable tool for appropriate decision making for both clinicians and patients undergoing IVF.
Hypomethylation of the AHR promoter region is associated with increased AHR expression in the eutopic endometrium of LEI women with endometriosis compared to controls
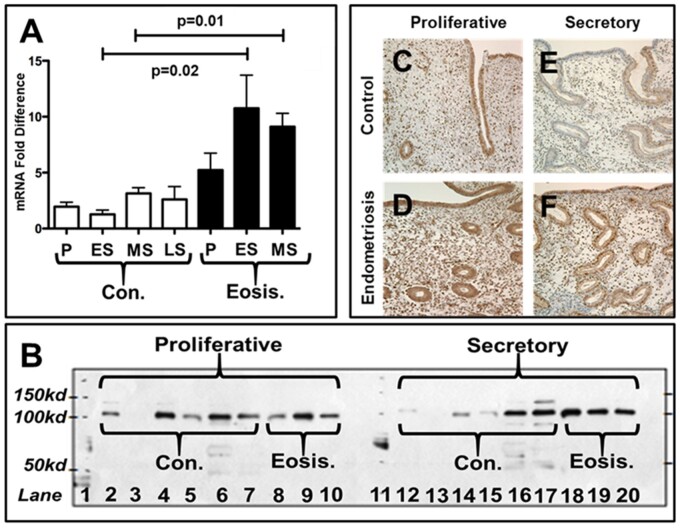
AHR expression is elevated in the eutopic endometrium of LEI women with endometriosis compared to disease-free controls. AHR is reported to be altered in women with endometriosis (Khorram et al., 2002; Mariuzzi et al., 2016); however, the exact mechanism regulating AHR expression in endometriosis is not known. The data from this study for the first time suggested that AHR is epigenetically regulated, and hypomethylation of AHR promoter may be the reason for its increased expression (Table I) in eutopic endometrium of women with endometriosis. In addition, qRT–PCR analysis data strongly suggested that AHR is significantly (P=0.0014) increased in the LEI group compared to controls (Fig. 5I). Further, by qRT–PCR, AHR mRNA expression was noted to be significantly higher in endometriosis samples compared to controls during the early (P=0.02) and mid-secretory phases (P=0.01) (Fig. 6A). Western blots were generated from eutopic endometrial samples from normal controls (n=6) and women with endometriosis (n=3) from the proliferative and secretory phases (Fig. 6B). While proliferative expression was similar in control and endometriosis samples after correcting for beta-actin (not shown), an increased AHR protein level was noted in the secretory phase of women with endometriosis. Immunohistochemistry (Fig. 6C–F) illustrates the appearance and distribution of AHR in normal proliferative and secretory endometrium and differences in AHR staining in the secretory phase of women with endometriosis compared to normal endometrium: the results confirm the qRT–PCR data (Fig. 5I). This difference in AHR appeared more prominent in the nuclei of the epithelial cells. This study has demonstrated that the AHR transcript and protein levels are significantly increased in the mid-secretory eutopic endometrium of women with endometriosis, which may be epigenetically regulated in LEI women with endometriosis.
Figure 6.
AHR transcript and protein expression is significantly increased in women with endometriosis. qRT–PCR analysis of AHR transcript in the eutopic endometrial samples obtained during the menstrual cycle from women with and without endometriosis. Note the significantly increased expression of AHR mRNA during the early and mid-secretory phase of the menstrual cycle in endometrial biopsies obtained from women with endometriosis compared to controls (A). Western blot analysis of endometrial tissue proteins shows the increased AHR expression during the mid-secretory phase in women with endometriosis compared to controls (B). Immunohistochemistry shows an increased signal intensity for AHR protein in both proliferative and mid-secretory endometrial luminal epithelium, glandular epithelium, and stroma of women with endometriosis (D & F) compared to the women without disease (C & E). Data displayed as mean±SD and analyzed by performing an unpaired Student’s t-test (C, control; Eosis, endometriosis, P, proliferative; ES, early secretory, MS, mid secretory, LS, late secretory).
Discussion
Endometriosis is a multifactorial disease, and many theories are proposed for its cause, including retrograde menses, genetic, epigenetic, inflammation, environmental contaminant, lifestyle, or idiopathic causes (Sampson, 1927; Burney et al., 2007, 2009; Hickey et al., 2014; Sourial et al., 2014; Houshdaran et al., 2016; Powell et al., 2016; Saare et al., 2016; Fung et al., 2018; Saare et al., 2018; Poli-Neto et al., 2020). Irrespective of the cause, it appears that the presence of ectopic lesions is associated with changes in the eutopic endometrium. Such changes have been noted previously and thought to represent a response to systemic inflammatory changes of the eutopic endometrial transcriptome and epigenome, which may contribute to dysregulated uterine function as evident by the observation that about 50% of the women with endometriosis have compromised fertility (Carter, 1994; Balasch et al., 1996; Burney et al., 2007; Meuleman et al., 2009; Houshdaran et al., 2016; Yoo et al., 2016, 2017; Fung et al., 2018; Saare et al., 2018). This was further validated in our baboon model of induced disease, which demonstrated that the presence of endometriotic lesions directly impacts eutopic gene expression (Hastings, 2006; Afshar, 2013; Joshi, 2015). Hence, it is crucial to understand the molecular and cellular changes that contribute to dysregulated endometrial function in this subset of women with endometriosis. Integrin αvβ3, a cell surface adhesion receptor, expression is increased owing to enhanced expression of the β3 subunit after Day 19 of the menstrual cycle, which is regulated by the transcription factor HOXA10 in response to progesterone (Lessey, 1998; Illera et al., 2000; Daftary et al., 2002; Illera et al., 2003; Singh and Aplin, 2009). We and others have previously shown that integrin αvβ3 expression is decreased in the mid-secretory endometrium of a subset of women with endometriosis with unexplained infertility (Lessey et al., 1994; Lessey, 1998; Lessey and Castelbaum, 2002; Tei et al., 2003; Singh and Aplin, 2009; Young and Lessey, 2010). In this study, we investigated the differential gene expression signature of eutopic endometrium in women with low αvβ3 integrin expression and a confirmed diagnosis of endometriosis compared to the disease-free endometrium from women with higher αvβ3 integrin expression. Further, we compared the global DNA methylation patterns of gene promoter regions in the eutopic endometrial biopsies to investigate the impact of aberrant DNA methylation on the observed gene expression changes in women with endometriosis and LEI.
We observed the highest DEGs (total—396; 275 up and 121 down-regulated genes) in the LEI vs C comparison. In contrast, a recent RNA-seq study conducted without considering the stage of the menstrual cycle reported no DEGs in the eutopic endometrium of infertile women with endometriosis compared to the control group (Da Broi et al., 2019). However, a previous report suggests that 1286 genes were dysregulated in endometrial biopsies of patients with severe endometriosis compared to those with the mild disease during the window of implantation (Aghajanova and Giudice, 2011). These observations further support this study's objective to separate the subgroup of endometriosis patients carefully, and precisely focus on the mid-secretory phase to study the possible causes of infertility associated with endometriosis. In our analyses, there were 83 DEGs (31 up and 52 down-regulated genes) in the HEI vs C comparison and 45 DEGs (30 up and 15 down-regulated genes) in the LEI vs HEI comparison. A three-way comparative analysis for DEGs demonstrated that mid-secretory endometrium of women with endometriosis and LEI has a distinctly different gene expression profile compared to receptive endometrium from disease-free women. Further, downstream bioinformatic functional analysis was performed on 396 DEGs. Interestingly, we observed a significant enrichment of the DEGs involved in biological processes, such as extracellular matrix organization, cell adhesion, GTPase activity, and actin-filament-based movement. These biological processes are critical for embryo movement, attachment, and invasion into the endometrial stroma (Grewal et al., 2008; Margarit et al., 2009; Singh and Aplin, 2009; Grewal et al., 2010). The GTPase binding of small monomeric proteins (RHO–Ras Homology) regulates actin polymerization. Grewal et al. have reported that GTPase-dependent endometrial stromal compartment remodeling is vital for embryo implantation (Grewal et al., 2008, 2010). Cell adhesion molecules are indispensable for successful implantation during uterine receptivity (Donaghay and Lessey, 2007; Margarit et al., 2009; Singh and Aplin, 2009), and our analysis clearly showed that the cell-substrate adhesion process is significantly compromised in LEI women with endometriosis (Merviel et al., 2001). The implantation process comprises three steps—apposition, adhesion, and invasion (Bischof and Campana, 1997; Su-Mi and Jong-Soo, 2017). The cellular component analysis for the DEGs in the LEI vs C comparison identified that most of these DEGs converge to the extracellular matrix, basement membrane, proteinaceous extracellular matrix, and site of polarized growth extracellular matrix component. This finding validates our data and supports the previous studies showing that these cellular components are significantly compromised in women with endometriosis with LEI.
Regulation of gene expression is multifactorial and involves chromatin changes, transcription factor binding, non-coding RNAs, methylation, post-transcriptional modifications, and other mechanisms (Phillips, 2008). This study was extended to specifically investigate the influence of abnormal methylation on the DEGs in LEI endometrial samples compared to controls. To explore this, we performed the methylation array on the same samples utilized for the gene array. We observed 1304 DMRs in LEI compared to controls. PCA and hierarchical clustering distinctly separated the control and LEI samples emphasizing that the methylation signature is unique to both the groups in the mid-secretory phase of the menstrual cycle. Previously, Houshdaran et al. (2016) reported the highest change in the methylome in the mid-secretory samples obtained from women with endometriosis compared to other phases of the menstrual cycle within the disease group and the disease-free controls, suggesting that the presence of ectopic lesions alters the epigenetic signature in eutopic endometrium and affects the uterine receptivity in women with the disease. A study focusing on methylation differences between ectopic lesions and eutopic tissues identified 1753 DMRs in ectopic vs control endometrium, and 2108 DMRs in ectopic vs matched eutopic endometrium (Barjaste et al., 2019). Using the machine-learning approach, Akter et al. (2019) also identified 365 DMRs among the total 2 577 382 DMRs analyzed in endometrial biopsies obtained from women with and without endometriosis.
Following the methylation signature comparison between LEI and C group, we overlapped these data with the gene array to identify the specific genes altered as a result of aberrant methylation. We identified 14 genes (RNPC3, SLC18A2, SMCHD1, SNORD58A, PGLYRP2, SNORD82, ANX3, HIST1H4F, HIST1H3I, PGK2, AHR, CCDC146, AGPAT5, and PLEKHF2), which demonstrated an inverse methylation pattern against their mRNA expression pattern in our tissue samples analyzed from the LEI and control groups. They may serve as novel candidate genes to understand infertility associated with endometriosis. We randomly selected nine genes (RNPC3, SLC18A2, PGLYRP2, ANX3, HIST1H4F, PGK2, AHR, CCDC146, and PLEKHF2) and validated their expression by RT–PCR analysis. Seven out of these nine genes were significantly (P<0.05) altered and matched the expression profile, and two of the genes did follow the expression trend but were not statistically significant. These observations provide a strong basis for the experimental approach of directly comparing gene and epigenetic arrays and using in silico analysis to identify genes that may serve as endometrial biomarkers of endometriosis-associated infertility.
Given the potential role of AHR in the pathogenesis of endometriosis, we investigated the expression differences of AHR in the human endometrium. This is the first study to demonstrate a differential expression in AHR during the menstrual cycle in the eutopic endometrium of women with endometriosis but not in normal controls. Analysis of the AHR transcript reveals cycle stage-specific upregulation only in infertile women with endometriosis. Furthermore, the protein expression is also specifically increased in the secretory phase, as demonstrated by immunohistochemistry and western blot analysis. In contrast to our findings, a previous report indicated little AHR variation throughout the menstrual cycle (Igarashi et al., 2005), which was supported by a study from Khorram et al. (2002); however, the latter study also observed an increased expression of AHR in postmenopausal women on hormone replacement therapy, again suggesting a stimulus by ovarian sex steroids. Recent studies by Mortlock et al. identified eQTLs and mQTLs for AHR in the endometrium, suggesting that this gene’s expression and methylation are also under genomic regulation (Mortlock et al., 2019, 2020). It is not clear whether low αvβ3 expression in women with endometriosis is a cause or effect. Based on protein–protein interaction analysis, we speculate that increased AHR interacts with SP1, which in turn regulates αvβ3 integrin expression. However, more studies are warranted to understand AHR’s role in regulating integrin expression in the eutopic endometrium of women with endometriosis.
Transcriptomic and epigenomic changes are dynamic in the endometrium and are greatly influenced by ovarian steroids in a menstrual stage-specific manner. A study conducted by Saare et al. (2016) found that the methylation signature was very similar between controls and endometriosis patients. They reported that only 28 DMRs were present in endometrial tissues obtained at various stages of the menstrual cycle from women with endometriosis (n = 31) compared to controls (n = 24). Further, in the same study, it was reported that unsupervised hierarchical clustering of DMRs segregated in endometrial tissues based on menstrual cycle stage instead of the presence or absence of endometriosis (Saare et al., 2016). This observation strongly advocates the critical consideration of the menstrual cycle stage while performing disease vs control comparisons for ‘Omic’ studies. The major strength of our study is that all the endometrial tissue biopsies were obtained during the window of uterine receptivity and the stage of the menstrual cycle was dated carefully by the pathologist. Furthermore, endometrial biopsies were screened immunohistochemically for integrin αvβ3 expression, and the samples were categorized based on the integrin αvβ3 H-score into the HEI and LEI groups before they were further processed for the gene and methylation arrays. In addition, we performed an overlay of the transcriptome with the methylome to identify the transcripts specifically influenced by altered methylation status. The PCA segregated the samples in each group (C, LEI, and HEI) among various gene and methylation array comparisons. In silico downstream analysis of transcriptomic data defined the dysregulated cellular and molecular functions in the LEI group compared to controls. The major limitation of this study is the small sample size, which may have influenced our ability to identify DEGs that were significantly changed when applying a FDR=0.05 threshold. Another limitation was that it was not feasible to overcome the heterogeneous nature of the cell population in the samples, which is a drawback with any tissue biopsy. Despite these limitations, the results obtained have identified a previously unreported set of differentially methylated genes expressed in the mid-secretory endometrium of LEI samples from women with endometriosis. In addition, validation of the array data suggested possible epigenetic regulation was responsible for the reported increase in expression of AHR in the context of endometriosis. However, AHR has been reported to be regulated by eQTLs and mQTLs in multiple tissues (Mortlock et al., 2019, 2020). Furthermore, we also identified the cellular and molecular functional pathways contributing to the observed infertility phenotype in a subset of women with endometriosis. Together this study suggests the critical involvement of epigenetic and transcriptomic alterations in the eutopic endometrium of women with endometriosis and highlights the need for future studies involving a bigger sample population to better understand the effect of epigenetic changes on the transcriptomic and proteomic functions of the endometrium during the window of uterine receptivity.
Supplementary data
Supplementary data are available at Molecular Human Reproduction online.
Data availability
The data underlying this article are available in the Dryad Digital Repository, at https://doi.org/10.5061/dryad.5tb2rbp3j.
Supplementary Material
Acknowledgments
The authors thank Ms Samantha Hrbek and Mr Mark Olson (Michigan State University, Grand Rapids, MI, USA) for their excellent technical assistance for DNA and RNA sample preparation and qRT–PCR assays. We also thank the Spectrum Health Universal Biorepository Grand Rapids for provided endometrial samples for this project as a part of the Michigan State University's Center for Women's Health Research Female Reproductive Tract Biorepository. The authors are grateful to Dr. Jing Chen (Gene expression core, University of Cincinnati) for his help in processing and normalizing the gene and methylation array data. The authors also thank Gail Grossman at the University of North Carolina Immunohistochemistry core to help with the immunostaining.
Authors’ roles
N.R.J., B.A.L., and A.T.F. conceived the concept of this study and designed the experimental approach. H.-R.K.-G. performed the genomic, epigenomic data analysis, and downstream bioinformatics analysis. D.S.R. helped with bioinformatics analysis. N.R.J., K.F., E.H., J.Y.Y., and L.Y. performed the experiments. S.-M.H. assisted the epigenetic methylation array experimental design. S.L.Y. and B.A.L. provided the clinical samples, clinical insight into the study design, and edited the manuscript. J.-W.J. provided critical intellectual support, assisted with data analysis, and edited the manuscript. N.R.J., H.-R.K.-G., B.A.L., and A.T.F. wrote the manuscript. All authors contributed to the final editing and reviewed the manuscript.
Funding
This study was funded by NIH Grants (R01 HD042280, HD083273 to ATF).
Conflict of interest
B.A.L. has licensed technology related to alpha V/Beta 3 testing for endometrial receptivity with CiceroDx.
References
- Afshar Y, Hastings J, Roqueiro D, Jeong JW, Gudice LC, Fazieabas AT.. Changes in eutopic endometrial gene expression during the progression of experimental endometriosis in the baboon, Papio anubis. Biology of Reprod 2013;88:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aghajanova L, Giudice LC.. Molecular evidence for differences in endometrium in severe versus mild endometriosis. Reprod Sci 2011;18:229–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akter S, Xu D, Nagel SC, Bromfield JJ, Pelch K, Wilshire GB, Joshi T.. Machine learning classifiers for endometriosis using transcriptomics and methylomics data. Front Genet 2019;10:766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasch J, Creus M, Fabregues F, Carmona F, Ordi J, Martinez-Román S, Vanrell JA.. Visible and non-visible endometriosis at laparoscopy in fertile and infertile women and in patients with chronic pelvic pain: a prospective study. Hum Reprod 1996;11:387–391. [DOI] [PubMed] [Google Scholar]
- Barjaste N, Shahhoseini M, Afsharian P, Sharifi-Zarchi A, Masoudi-Nejad A.. Genome-wide DNA methylation profiling in ectopic and eutopic of endometrial tissues. J Assist Reprod Genet 2019;36:1743–1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y.. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Methodol 1995;57:289–300. [Google Scholar]
- Bischof P, Campana A.. Trophoblast differentiation and invasion: its significance for human embryo implantation. Early Pregnancy 1997;3:81–95. [PubMed] [Google Scholar]
- Bulun SE. Endometriosis. N Engl J Med 2009;360:268–279. [DOI] [PubMed] [Google Scholar]
- Burney RO, Hamilton AE, Aghajanova L, Vo KC, Nezhat CN, Lessey BA, Giudice LC.. MicroRNA expression profiling of eutopic secretory endometrium in women with versus without endometriosis. Mol Hum Reprod 2009;15:625–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burney RO, Talbi S, Hamilton AE, Vo KC, Nyegaard M, Nezhat CR, Lessey BA, Giudice LC.. Gene expression analysis of endometrium reveals progesterone resistance and candidate susceptibility genes in women with endometriosis. Endocrinology 2007;148:3814–3826. [DOI] [PubMed] [Google Scholar]
- Carter JE. Combined hysteroscopic and laparoscopic findings in patients with chronic pelvic pain. J Am Assoc Gynecol Laparosc 1994;2:43–47. [DOI] [PubMed] [Google Scholar]
- Ceydeli N, Kaleli S, Calay Z, Erel CT, Akbas F, Ertungealp E.. Difference in alpha(v)beta3 integrin expression in endometrial stromal cell in subgroups of women with unexplained infertility. Eur J Obstet Gynecol Reprod Biol 2006;126:206–211. [DOI] [PubMed] [Google Scholar]
- Chen G, Xin A, Liu Y, Shi C, Chen J, Tang X, Chen Y, Yu M, Peng X, Li L. et al. Integrins β1 and β3 are biomarkers of uterine condition for embryo transfer. J Transl Med 2016;14:303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Broi MG, Meola J, Plaça JR, Peronni KC, Rocha CV Jr, Silva WA Jr, Ferriani RA., Navarro PA.. Is the profile of transcripts altered in the eutopic endometrium of infertile women with endometriosis during the implantation window? Hum Reprod 2019;34:2381–2390. [DOI] [PubMed] [Google Scholar]
- Daftary GS, Troy PJ, Bagot CN, Young SL, Taylor HS.. Direct regulation of beta3-integrin subunit gene expression by HOXA10 in endometrial cells. Mol Endocrinol 2002;16:571–579. [DOI] [PubMed] [Google Scholar]
- Dai M, Wang P, Boyd AD, Kostov G, Athey B, Jones EG, Bunney WE, Myers RM, Speed TP, Akil H. et al. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res 2005;33:e175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detre S, Saclani Jotti G, Dowsett M.. A "quickscore" method for immunohistochemical semiquantitation: validation for oestrogen receptor in breast carcinomas. J Clin Pathol 1995;48:876–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaghay M, Lessey BA.. Uterine receptivity: alterations associated with benign gynecological disease. Semin Reprod Med 2007;25:461–475. [DOI] [PubMed] [Google Scholar]
- Dorostghoal M, Ghaffari H-O-A, Shahbazian N, Mirani M.. Endometrial expression of β3 integrin, calcitonin and plexin-B1 in the window of implantation in women with unexplained infertility. Int J Reprod Biomed 2017;15:33–40. [PMC free article] [PubMed] [Google Scholar]
- Dyson MT, Roqueiro D, Monsivais D, Ercan CM, Pavone ME, Brooks DC, Kakinuma T, Ono M, Jafari N, Dai Y. et al. Genome-wide DNA methylation analysis predicts an epigenetic switch for GATA factor expression in endometriosis. PLoS Genet 2014;10:e1004158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazleabas AT, Bell SC, Fleming S, Sun J, Lessey BA.. Distribution of integrins and the extracellular matrix proteins in the baboon endometrium during the menstrual cycle and early pregnancy1. Biol Reprod 1997;56:348–356. [DOI] [PubMed] [Google Scholar]
- Franasiak JM, Holoch KJ, Yuan L, Schammel DP, Young SL, Lessey BA.. Prospective assessment of midsecretory endometrial leukemia inhibitor factor expression versus alphanubeta3 testing in women with unexplained infertility. Fertil Steril 2014;101:1724–1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung JN, Mortlock S, Girling JE, Holdsworth-Carson SJ, Teh WT, Zhu Z, Lukowski SW, McKinnon BD, McRae A, Yang J. et al. Genetic regulation of disease risk and endometrial gene expression highlights potential target genes for endometriosis and polycystic ovarian syndrome. Sci Rep 2018;8:11424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germeyer A, Savaris RF, Jauckus J, Lessey B.. Endometrial beta3 integrin profile reflects endometrial receptivity defects in women with unexplained recurrent pregnancy loss. Reprod Biol Endocrinol 2014;12:53–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giancotti FG, Ruoslahti E.. Integrin signaling. Science 1999;285:1028–1033. [DOI] [PubMed] [Google Scholar]
- Grewal S, Carver J, Ridley AJ, Mardon HJ.. Human endometrial stromal cell rho GTPases have opposing roles in regulating focal adhesion turnover and embryo invasion in vitro. Biol Reprod 2010;83:75–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal S, Carver JG, Ridley AJ, Mardon HJ.. Implantation of the human embryo requires Rac1-dependent endometrial stromal cell migration. Proc Natl Acad Sci USA 2008;105:16189–16194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halme J, Hammond MG, Hulka JF, Raj SG, Talbert LM.. Retrograde menstruation in healthy women and in patients with endometriosis. Obstet Gynecol 1984;64:151–154. [PubMed] [Google Scholar]
- Hastings JM, Fazleabas AT.. A baboon model for endometriosis: implications for fertility. Reprod Biol Endocrinol 2006;4(Suppl 1):S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hever A, Roth RB, Hevezi P, Marin ME, Acosta JA, Acosta H, Rojas J, Herrera R, Grigoriadis D, White E. et al. Human endometriosis is associated with plasma cells and overexpression of B lymphocyte stimulator. Proc Natl Acad Sci USA 2007;104:12451–12456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickey M, Ballard K, Farquhar C.. Endometriosis. BMJ 2014;348:g1752. [DOI] [PubMed] [Google Scholar]
- Houshdaran S, Nezhat CR, Vo KC, Zelenko Z, Irwin JC, Giudice LC.. Aberrant endometrial DNA methylome and associated gene expression in women with endometriosis. Biol Reprod 2016;95:93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houshdaran S, Oke AB, Fung JC, Vo KC, Nezhat C, Giudice LC.. Steroid hormones regulate genome-wide epigenetic programming and gene transcription in human endometrial cells with marked aberrancies in endometriosis. PLoS Genet 2020;16:e1008601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull ML, Escareno CR, Godsland JM, Doig JR, Johnson CM, Phillips SC, Smith SK, Tavaré S, Print CG, Charnock-Jones DS.. Endometrial-peritoneal interactions during endometriotic lesion establishment. Am J Pathol 2008;173:700–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igarashi TM, Bruner-Tran KL, Yeaman GR, Lessey BA, Edwards DP, Eisenberg E, Osteen KG.. Reduced expression of progesterone receptor-B in the endometrium of women with endometriosis and in cocultures of endometrial cells exposed to 2,3,7,8-tetrachlorodibenzo-p-dioxin. Fertil Steril 2005;84:67–74. [DOI] [PubMed] [Google Scholar]
- Illera MJ, Cullinan E, Gui Y, Yuan L, Beyler SA, Lessey BA.. Blockade of the alpha(v)beta(3) integrin adversely affects implantation in the mouse. Biol Reprod 2000;62:1285–1290. [DOI] [PubMed] [Google Scholar]
- Illera MJ, Lorenzo PL, Gui YT, Beyler SA, Apparao KB, Lessey BA.. A role for alphavbeta3 integrin during implantation in the rabbit model. Biol Reprod 2003;68:766–771. [DOI] [PubMed] [Google Scholar]
- Izawa M, Taniguchi F, Terakawa N, Harada T.. Epigenetic aberration of gene expression in endometriosis. Front Biosci (Elite Ed) 2013;5:900–910. [DOI] [PubMed] [Google Scholar]
- Jackson KS, Brudney A, Hastings JM, Mavrogianis PA, Kim JJ, Fazleabas AT.. The altered distribution of the steroid hormone receptors and the chaperone immunophilin FKBP52 in a baboon model of endometriosis is associated with progesterone resistance during the window of uterine receptivity. Reprod Sci 2007;14:137–150. [DOI] [PubMed] [Google Scholar]
- Jones A, Teschendorff AE, Li Q, Hayward JD, Kannan A, Mould T, West J, Zikan M, Cibula D, Fiegl H. et al. Role of DNA methylation and epigenetic silencing of HAND2 in endometrial cancer development. PLoS Med 2013;10:e1001551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi NR, Su RW, Chandramouli GVR, Kahoo SK, Jeong JW, Young SL, Lessey, Fazleabas AT.. Altered expression of microRNA-451 in eutopic endometrium of baboons (Papio anubis) with endometriosis. Hum Reprod 2015;30:2881–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi NR, Miyadahira EH, Afshar Y, Jeong J-W, Young SL, Lessey BA, Serafini PC, Fazleabas AT.. Progesterone resistance in endometriosis is modulated by the altered expression of microRNA-29c and FKBP4. J Clin Endocrinol Metab 2017;102:141–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khorram O, Garthwaite M, Golos T.. Uterine and ovarian aryl hydrocarbon receptor (AHR) and aryl hydrocarbon receptor nuclear translocator (ARNT) mRNA expression in benign and malignant gynaecological conditions. Mol Hum Reprod 2002;8:75–80. [DOI] [PubMed] [Google Scholar]
- Kim BG, Yoo JY, Kim TH, Shin JH, Langenheim JF, Ferguson SD, Fazleabas AT, Young SL, Lessey BA, Jeong JW.. Aberrant activation of signal transducer and activator of transcription-3 (STAT3) signaling in endometriosis. Hum Reprod 2015;30:1069–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JJ, Taylor HS, Lu Z, Ladhani O, Hastings JM, Jackson KS, Wu Y, Guo SW, Fazleabas AT.. Altered expression of HOXA10 in endometriosis: potential role in decidualization. Mol Hum Reprod 2007;13:323–332. [DOI] [PubMed] [Google Scholar]
- Kim TH, Yoo J-Y, Choi K-C, Shin J-H, Leach RE, Fazleabas AT, Young SL, Lessey BA, Yoon H-G, Jeong J-W.. Loss of HDAC3 results in nonreceptive endometrium and female infertility. Sci Transl Med 2019;11:eaaf7533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TH, Yu Y, Luo L, Lydon JP, Jeong JW, Kim JJ.. Activated AKT pathway promotes establishment of endometriosis. Endocrinology 2014;155:1921–1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessey BA. Endometrial integrins and the establishment of uterine receptivity. Hum Reprod 1998;13:247–258. [DOI] [PubMed] [Google Scholar]
- Lessey BA. Medical management of endometriosis and infertility. Fertil Steril 2000;73:1089–1096. [DOI] [PubMed] [Google Scholar]
- Lessey BA, Castelbaum AJ.. Integrins and implantation in the human. Rev Endocr Metab Disord 2002;3:107–117. [DOI] [PubMed] [Google Scholar]
- Lessey BA, Castelbaum AJ, Sawin SW, Buck CA, Schinnar R, Bilker W, Strom BL.. Aberrant integrin expression in the endometrium of women with endometriosis. J Clin Endocrinol Metab 1994;79:643–649. [DOI] [PubMed] [Google Scholar]
- Lessey BA, Castelbaum AJ, Sawin SW, Sun J.. Integrins as markers of uterine receptivity in women with primary unexplained infertility. Fertil Steril 1995;63:535–542. [PubMed] [Google Scholar]
- Liu H, Huang X, Mor G, Liao A.. Epigenetic modifications working in the decidualization and endometrial receptivity. Cell Mol Life Sci 2020;77:2091–2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD.. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001;25:402–408. [DOI] [PubMed] [Google Scholar]
- Logan PC, Steiner M, Ponnampalam AP, Mitchell MD.. Cell cycle regulation of human endometrial stromal cells during decidualization. Reprod Sci 2012;19:883–894. [DOI] [PubMed] [Google Scholar]
- Maekawa R, Tamura I, Shinagawa M, Mihara Y, Sato S, Okada M, Taketani T, Tamura H, Sugino N.. Genome-wide DNA methylation analysis revealed stable DNA methylation status during decidualization in human endometrial stromal cells. BMC Genomics 2019;20:324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcoux S, Maheux R, Berube S.. Laparoscopic surgery in infertile women with minimal or mild endometriosis. Canadian Collaborative Group on Endometriosis. N Engl J Med 1997;337:217–222. [DOI] [PubMed] [Google Scholar]
- Margarit L, Gonzalez D, Lewis PD, Hopkins L, Davies C, Conlan RS, Joels L, White JO.. L-selectin ligands in human endometrium: comparison of fertile and infertile subjects. Hum Reprod 2009;24:2767–2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariuzzi L, Domenis R, Orsaria M, Marzinotto S, Londero AP, Bulfoni M, Candotti V, Zanello A, Ballico M, Mimmi MC. et al. Functional expression of aryl hydrocarbon receptor on mast cells populating human endometriotic tissues. Lab Invest 2016;96:959–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki S, Darcha C.. Co-operation between the AKT and ERK signaling pathways may support growth of deep endometriosis in a fibrotic microenvironment in vitro. Hum Reprod 2015;30:1606–1616. [DOI] [PubMed] [Google Scholar]
- McKinnon BD, Kocbek V, Nirgianakis K, Bersinger NA, Mueller MD.. Kinase signalling pathways in endometriosis: potential targets for non-hormonal therapeutics. Hum Reprod Update 2016;22:382–403. [DOI] [PubMed] [Google Scholar]
- Merviel P, Challier JC, Carbillon L, Foidart JM, Uzan S.. The role of integrins in human embryo implantation. Fetal Diagn Ther 2001;16:364–371. [DOI] [PubMed] [Google Scholar]
- Meuleman C, Vandenabeele B, Fieuws S, Spiessens C, Timmerman D, D'Hooghe T.. High prevalence of endometriosis in infertile women with normal ovulation and normospermic partners. Fertil Steril 2009;92:68–74. [DOI] [PubMed] [Google Scholar]
- Newton MA, Quintana FA, den Boon JA, Sengupta S, Ahlquist P.. Random-set methods identify distinct aspects of the enrichment signal in gene-set analysis. Ann Appl Stat 2007;1:85–106. [Google Scholar]
- Miller JE, Ahn SH, Monsanto SP, Khalaj K, Koti M, Tayade C.. Implications of immune dysfunction on endometriosis associated infertility. Oncotarget 2017;8:7138–7147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller PB, Parnell BA, Bushnell G, Tallman N, Forstein DA, Higdon HL III, Kitawaki J, Lessey BA.. Endometrial receptivity defects during IVF cycles with and without letrozole. Hum Reprod 2012;27:881–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortlock S, Kendarsari RI, Fung JN, Gibson G, Yang F, Restuadi R, Girling JE, Holdsworth-Carson SJ, Teh WT, Lukowski SW. et al. specific regulation of transcription in endometrium and association with disease. Hum Reprod 2020;35:377–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortlock S, Restuadi R, Levien R, Girling JE, Holdsworth-Carson SJ, Healey M, Zhu Z, Qi T, Wu Y, Lukowski SW. et al. Genetic regulation of methylation in human endometrium and blood and gene targets for reproductive diseases. Clin Epigenet 2019;11:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naqvi H, Ilagan Y, Krikun G, Taylor HS.. Altered genome-wide methylation in endometriosis. Reprod Sci 2014;21:1237–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips T. Regulation of transcription and gene expression in eukaryotes. Nature Educ 2008;1:199. [Google Scholar]
- Poli-Neto OB, Meola J, Rosa-e-Silva JC, Tiezzi D.. Transcriptome meta-analysis reveals differences of immune profile between eutopic endometrium from stage I-II and III-IV endometriosis independently of hormonal milieu. Sci Rep 2020;10:313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell JE, Fung JN, Shakhbazov K, Sapkota Y, Cloonan N, Hemani G, Hillman KM, Kaufmann S, Luong HT, Bowdler L, Painter JN. et al. Endometriosis risk alleles at 1p36.12 act through inverse regulation of CDC42 and LINC00339. Hum Mol Genet 2016;25:5046–5058. [DOI] [PubMed] [Google Scholar]
- Practice Committee of the American Society for Reproductive Medicine. Endometriosis and infertility. Fertil Steril 2006;86:S156–S160. [DOI] [PubMed] [Google Scholar]
- Predeus AV, Vashukova ES, Glotov AS, Danilova MM, Osinovskaya NS, Malysheva OV, Shved NY, Ganbarli N, Yarmolinskaya MI, Ivashchenko TE, Baranov VS.. Next-generation sequencing of matched ectopic and eutopic endometrium identifies novel endometriosis-related genes. Russ J Genet 2018;54:1358–1365. [Google Scholar]
- Saare M, Krigul KL, Laisk-Podar T, Ponandai-Srinivasan S, Rahmioglu N, Lalit Kumar PG, Zondervan K, Salumets A, Peters M.. DNA methylation alterations—potential cause of endometriosis pathogenesis or a reflection of tissue heterogeneity? Biol Reprod 2018;99:273–282. [DOI] [PubMed] [Google Scholar]
- Saare M, Modhukur V, Suhorutshenko M, Rajashekar B, Rekker K, Sõritsa D, Karro H, Soplepmann P, Sõritsa A, Lindgren CM. et al. The influence of menstrual cycle and endometriosis on endometrial methylome. Clin Epigenetics 2016;8:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvatori M, Peloso A, Patel T, Hemal S, Zambon JP, Katari R, Orlando G.. Chapter 17 - Renal regeneration: the bioengineering approach. In: Orlando G, Lerut J, Soker S, Stratta RJ (eds). Regenerative Medicine Applications in Organ Transplantation. Boston: Academic Press, 2014,227–239. [Google Scholar]
- Sampson JA. Metastatic or embolic endometriosis, due to the menstrual dissemination of endometrial tissue into the venous circulation. Am J Pathol 1927;3:93–110. [PMC free article] [PubMed] [Google Scholar]
- Sartor MA, Leikauf GD, Medvedovic M.. LRpath: a logistic regression approach for identifying enriched biological groups in gene expression data. Bioinformatics 2009;25:211–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sha G, Wu D, Zhang L, Chen X, Lei M, Sun H, Lin S, Lang J.. Differentially expressed genes in human endometrial endothelial cells derived from eutopic endometrium of patients with endometriosis compared with those from patients without endometriosis. Hum Reprod 2007;22:3159–3169. [DOI] [PubMed] [Google Scholar]
- Singh H, Aplin JD.. Adhesion molecules in endometrial epithelium: tissue integrity and embryo implantation. J Anat 2009;215:3–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 2004;3:Article3. [DOI] [PubMed] [Google Scholar]
- Smyth GK, Yang YH, Speed T.. Statistical issues in cDNA microarray data analysis. Methods Mol Biol 2003;224:111–136. [DOI] [PubMed] [Google Scholar]
- Sourial S, Tempest N, Hapangama DK.. Theories on the pathogenesis of endometriosis. Int J Reprod Med 2014;2014:179515–179515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su-Mi K, Jong-Soo K.. A review of mechanisms of implantation. Dev Reprod 2017;21:351–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanbo T, Fedorcsak P.. Endometriosis-associated infertility: aspects of pathophysiological mechanisms and treatment options. Acta Obstet Gynecol Scand 2017;96:659–667. [DOI] [PubMed] [Google Scholar]
- Tei C, Maruyama T, Kuji N, Miyazaki T, Mikami M, Yoshimura Y.. Reduced expression of alphavbeta3 integrin in the endometrium of unexplained infertility patients with recurrent IVF-ET failures: improvement by danazol treatment. J Assist Reprod Genet 2003;20:13–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G.. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA 2001;98:5116–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu MH, Hsiao KY, Tsai SJ.. Hypoxia: the force of endometriosis. J Obstet Gynaecol Res 2019;45:532–541. [DOI] [PubMed] [Google Scholar]
- Wu MH, Lin SC, Hsiao KY, Tsai SJ.. Hypoxia-inhibited dual-specificity phosphatase-2 expression in endometriotic cells regulates cyclooxygenase-2 expression. J Pathol 2011;225:390–400. [DOI] [PubMed] [Google Scholar]
- Wu Y, Halverson G, Basir Z, Strawn E, Yan P, Guo SW.. Aberrant methylation at HOXA10 may be responsible for its aberrant expression in the endometrium of patients with endometriosis. Am J Obstet Gynecol 2005;193:371–380. [DOI] [PubMed] [Google Scholar]
- Yamagata Y, Nishino K, Takaki E, Sato S, Maekawa R, Nakai A, Sugino N.. Genome-wide DNA methylation profiling in cultured eutopic and ectopic endometrial stromal cells. PLoS One 2014;9:e83612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin X, Pavone ME, Lu Z, Wei J, Kim JJ.. Increased activation of the PI3K/AKT pathway compromises decidualization of stromal cells from endometriosis. J Clin Endocrinol Metab 2012;97:E35–E43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo J-Y, Jeong J-W, Fazleabas AT, Tayade C, Young SL, Lessey BA.. Protein inhibitor of activated STAT3 (PIAS3) is down-regulated in eutopic endometrium of women with endometriosis1. Biol Reprod 2016;95:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo JY, Kim TH, Fazleabas AT, Palomino WA, Ahn SH, Tayade C, Schammel DP, Young SL, Jeong JW, Lessey BA.. KRAS activation and over-expression of SIRT1/BCL6 contributes to the pathogenesis of endometriosis and progesterone resistance. Sci Rep 2017;7:6765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yotova I, Hsu E, Do C, Gaba A, Sczabolcs M, Dekan S, Kenner L, Wenzl R, Tycko B.. Epigenetic alterations affecting transcription factors and signaling pathways in stromal cells of endometriosis. PLoS One 2017;12:e0170859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young SL, Lessey BA.. Progesterone function in human endometrium: clinical perspectives. Semin Reprod Med 2010;28:5–16. [DOI] [PubMed] [Google Scholar]
- Yu G, Wang L-G, Han Y, He Q-Y.. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 2012;16:284–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Ho SM.. Epigenetics meets endocrinology. J Mol Endocrinol 2011;46:R11–R32. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article are available in the Dryad Digital Repository, at https://doi.org/10.5061/dryad.5tb2rbp3j.