Abstract
A newly-emergent beta-coronavirus, SARS-CoV-2, rapidly has become a pandemic since 2020. It is a serious respiratory disease and caused more than 100 million of deaths in the world. WHO named it COVIA-19 and there is no effective targeted drug for it. The main treatment strategies include chemical medicine, traditional Chinese medicine (TCM) and biologics. Due to SARS-CoV-2 uses the spike proteins (S proteins) on its envelope to infect human cells, monoclonal antibodies that neutralize the S protein have become one of the hot research areas in the current research and treatment of SARS-CoV-2. In this study, we reviewed the antibodies that have been reported to have neutralizing activity against the SARS-CoV-2 infection. According to their different binding epitope regions in RBD or NTD, they are classified, and the mechanism of the representative antibodies in each category is discussed in depth, which provides potential foundation for future antibody and vaccine therapy and the development of antibody cocktails against SARS-CoV-2 mutants.
Keywords: SARS-CoV-2, Antibody, Mechanism, Receptor binding domain (RBD), Epitopes
Abbreviations: SARS-CoV-2, Severe Acute Respiratory Syndrome Coronavirus 2; Acs, antibody cocktails
1. Introduction
A newly-emergent beta-coronavirus disease 2019 (COVID-19), caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), became a global pandemic since its emergence in December 2019 [1]. Protein sequence analysis showed that the virus belongs to SARS-CoV [2]. This is the third pandemic caused by the beta-coronavirus, in addition, SARS-CoV in 2003 and Middle East respiratory syndrome (MERS-CoV) in 2012 [3,4]. Comparison of the SARS-CoV-2 and SARS-CoV sequences indicated that there is 80% of homologue in the sequences [1]. As of April 20, 2021, 141 million people have been infected with the new type of coronary pneumonia and 3.0 million deaths are claimed by SARS-CoV-2 across the globe. (https://covid19.who.int/).
SARS-CoV-2 uses the S proteins on its envelope to infect human cells. The S protein is composed of two subunits, S1 and S2, forming a trimeric spike structures on the envelope of the SARS-CoV-2 [5]. The S1 subunit is responsible for host cell attachment and angiotensin-converting enzyme 2 (ACE2) binding, and the S2 subunit is responsible for the fusion of the virus envelope and host cell membrane [1,[6], [7], [8], [9]]. Studies have shown that by binding to ACE2, the S1 subunit of the spike protein induces conformation changes of S2 subunits from an unstable unfused state to a more stable fused state [[10], [11], [12]]. The hinge-like movement of receptor-binding domain (RBD) produces two different conformational states which are referred to as “up” conformation and “down” conformation. The “up” conformation is the receptor-bound state, which is considered unstable and can bind the receptor and antibody, while the “down” conformation is the receptor-unbound state [5,10,[13], [14], [15]]. This indicates that the hindrance of interaction between the S1 subunit and ACE2 will block the infection of SARS-CoV-2. Neutralizing S1 subunit which composed of RBD and N-terminal domain (NTD) is essential to block the virus from replicating in the body [13]. The high-resolution crystal structures of S protein-antibodies suggest that RBD is responsible for recognizing and binding ACE2, while the function of NTD is not yet clear [16]. But, there is one group speculating that NTD may play an inhibitory role by restraining the conformational changes of the S protein [17]. Therefore, antibodies that bind to RBD or NTD epitopes may become candidates for the treatment of SARS-CoV-2.
Since the genome sequence and three-dimensional (3D) structures of the S protein have been delineated [16], scientists across the world start to work on the development of anti-SARS-CoV-2 drugs to prevent and treat the infections of SARS-CoV-2. Till now, there are still no effective antiviral drugs to treat COVID-19 patients. The current treatment strategies for COVID-19 mainly include chemical medicine, 5,7,3′,4′-tetrahydroxy-2’- (3,3-dimethylallyl) isoflavone [18], desacetylgedunin (DCG) [19], traditional Chinese medicine (TCM), steroid, dexamethasone [20] and various antibodies. At present, the development of vaccines by elucidating host immunity is the fastest and most effective way to solve the COVID-19 pandemic. So far, dozens of antibodies that have neutralizing activity against SARS-CoV-2 have been reported, including monoclonal antibodies (mAbs), synthetic nanobodies (nAbs) and a variety of antibody cocktails (Acs).
In this review, we have summarized the antibodies that have been reported with strong neutralizing activity and high resolution crystal structures, classified the antibodies according to their binding epitopes on RBD, and summarize the mechanism of different types of antibodies. In addition, monoclonal antibodies that bind to NTD, nanobodies and some antibody cocktails are also included. In short, this review tries to provide a comprehensive explanation of the mechanism of SARS-CoV-2 antibodies, and foundation for the development of more effective and safe vaccines.
2. Antibodies against SARS-CoV-2
SARS-CoV-2 antibodies with strong anti-infection and neutralizing activity are one of the research hotspots. Most of them are isolated from COVID-19 convalescent, and have been proved to have neutralizing activity against the virus infection. The structure, function and mechanism of relevant neutralizing antibodies have been reported [[21], [22], [23], [24], [25]].
Most SARS-CoV-2 antibodies block the virus infection by inhibiting the interaction of S protein of SARS-CoV-2 with its receptor, ACE2 [[26], [27], [28], [29], [30], [31], [32]]. Some antibodies bind to non-ACE2 epitopes, NTD, and also have antiviral activity [17,33]. Nanobodies have been reported to bind to RBD and thus hinder ACE2 binding [24,[34], [35], [36], [37]]. Some antibody cocktails which are used in combination of two or more antibodies have super neutralizing activity to SARS-CoV-2 [34,38,39]. In these three types of antibodies, they have a similar mechanism, all of which bind to epitopes on the S protein to inactivate the virus. Among the SARS-CoV-2 target proteins, S protein is the most studied target that can trigger strong host immune response.
3. The binding modes of neutralizing monoclonal antibodies
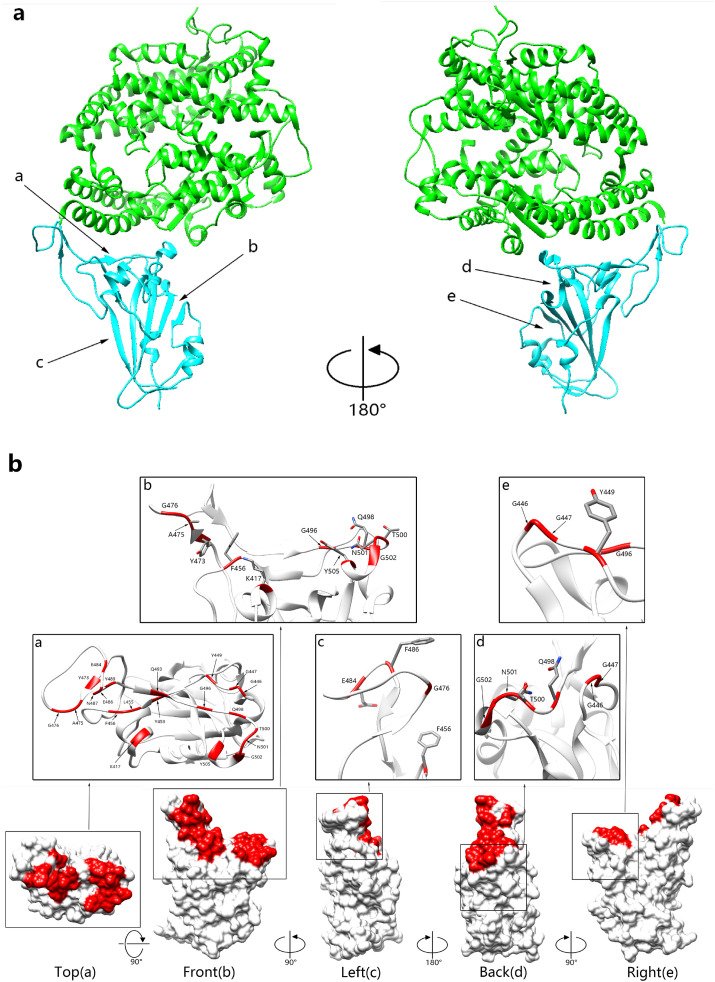
Among 40 reported monoclonal antibodies which are extracted from the COVID-19 convalescent patients, 37 of them compete with ACE2 to bind to RBD and 3 antibodies bind to NTD. Since the epitopes of these antibodies largely overlap with the ACE2-binding sites, they can neutralize SARS-CoV-2 by competing with ACE2 binding at the RBD. Based on the binding sites of ACE2 on RBD, RBD is divided into 5 regions (Fig. 1 ) According to our classification, the main binding sites of ACE2 locate in two regions, a/b, so it can be inferred that antibodies with binding epitopes in these two regions are likely to have strong neutralizing activity. Interestingly, these antibodies bind to RBD at different angles, so the binding epitopes can be partitioned, and antibodies can be classified according to different binding regions.
Fig. 1.
Overall structure of SARS-CoV-2 RBD bound with ACE2. a Structure of SARS-CoV-2 RBD combined with ACE2. ACE2 is colored in green. The SARS-CoV-2 RBD is colored in cyan. b The binding sites of ACE2 on SARS-CoV-2 RBD, define a/b/c/d/e regions on the top, front, left, right, and back sides (from left to right), respectively. The binding sites of ACE2 on SARS-CoV-2 RBD are colored in red. The SARS-CoV-2 RBD is colored in white. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.1. Monoclonal antibodies bind to SARS-CoV-2 RBD
Most of the monoclonal antibodies that bind epitopes on RBD are at a similar angle as ACE2. By comparative analysis, the binding epitopes of S2E12 [32], CV07-250 [40], C1A-B3, C1A-F10, C1A-C2, C1A-B12 [26], BD-236, BD-604, BD-629 [27], CV30 [29], C105 [30], CB6 [31], B38 [41], CC12.1, CC12.3 [42], C002, C102, C119, C121, C144 [43], P17 [44], 2–4 [45], COVA2-04, COVA2-39 [46], CT-P59 [47] are epitopes a/b which are similar to ACE2. There are also some antibodies that are completely dissimilar to ACE2, but they also show strong neutralizing activity: S2M11 [32], S2H13 [48], CV07-270 [40], P2B–2F6 [49], BD-368-2 [27], COVA1-16 [11], H014 [2], EY6A [50], C104, C110 [43], S2A4 [48], C135 [43]. Only a few antibodies bind to other epitopes (Table 1 ).
Table 1.
Summary of monoclonal antibodies that bind to RBD epitopes and their binding epitopes.
| Name | PDB | The binding epitopes | Number of residues buried |
|---|---|---|---|
| S2H13 | 7JV2 | a | 18 |
| EY6A | 6ZER | b | 28 |
| S2E12 | 7K45 | a b | 18 |
| CV07-250 | 6XKQ | a b | 20 |
| CV07-270 | 6XKP | a e | 22 |
| S2M11 | 7K43 | a e | 18 |
| P2B–2F6 | 7BWJ | a e | 19 |
| BD-368-2 | 7CHH | a e | 20 |
| C104 | 7K8U | a e | 7 |
| S2A4 | 7JVA | b d | 22 |
| COVA1-16 | 7JMW | b d | 23 |
| H014 | 7CAH | b d | 30 |
| C135 | 7K8Z | d e | 11 |
| COVA2-04 | 7JMO | a b c | 33 |
| C1A-B3 | 7KFW | a b c | 36 |
| C1A-F10 | 7KFY | a b c | 34 |
| C1A-C2 | 7KFX | a b c | 35 |
| C1A-B12 | 7KFV | a b c | 33 |
| BD-236 | 7CHB | a b c | 33 |
| BD-604 | 7CHF | a b c | 33 |
| BD-629 | 7CH5 | a b c | 26 |
| B38 | 7BZ5 | a b c | 38 |
| CV30 | 6XE1 | a b c | 30 |
| C105 | 6XCM | a b c | 29 |
| CB6 | 7C01 | a b c | 32 |
| CC12.1 | 6XC2 | a b c | 40 |
| CC12.3 | 6XC4 | a b c | 24 |
| C002 | 7K8S | a b e | 20 |
| C102 | 7K8M | a b e | 20 |
| C119 | 7K8W | a b e | 24 |
| C121 | 7K8X | a b e | 18 |
| C144 | 7K90 | a b e | 25 |
| P17 | 7CWO | a b e | 20 |
| 2–4 | 6XEY | a b e | 20 |
| COVA2-39 | 7JMP | a b e | 18 |
| CT-P59 | 7CM4 | a b e | 19 |
| C110 | 7K8V | a d e | 23 |
3.1.1. Monoclonal antibody that occupying only one epitope region on RBD
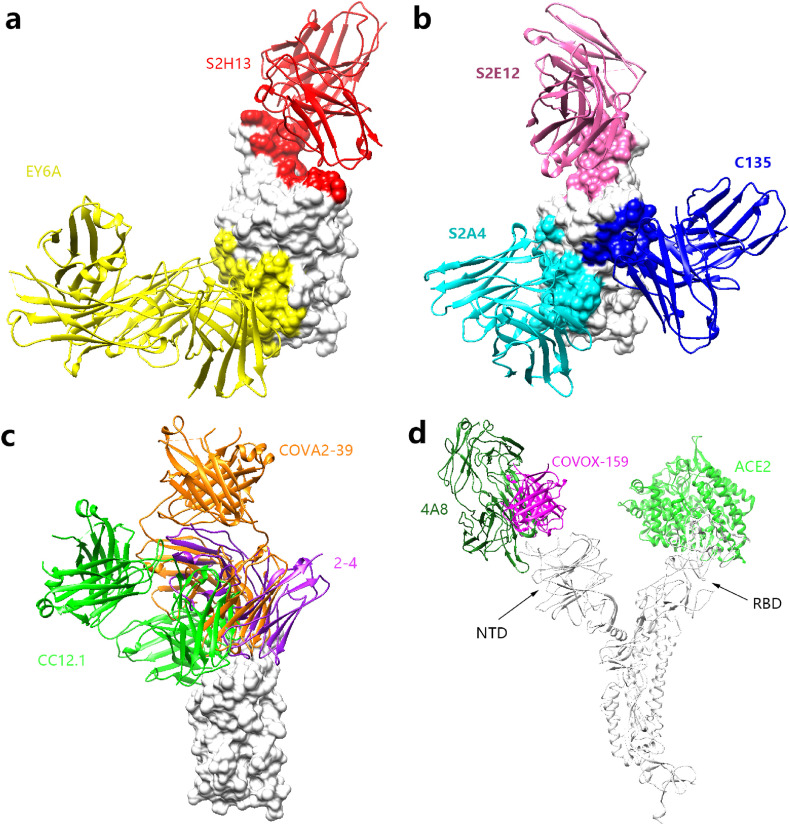
Among these monoclonal antibodies that occupy only one epitope region on RBD are: S2H13, EY6A (Fig. 2 a). S2H13 recognizes the receptor-binding motif (RBM) formed by RBD β-hairpin, and this region can be accessed regardless of whether the S protein is in the up or down conformation. Therefore, S2H13 may neutralize SARS-CoV-2 by recognizing the S protein in the up or down conformation, which is not the case with ACE2 [48]. This explains why S2H13 only binds to one region and also has neutralizing activity. The binding epitopes of EY6A is located at the lower position of region b which is close to the junction of S1 and S2. A protein-protein surface is formed, making the epitope inaccessible to ACE2 [50].
Fig. 2.
Structure comparisons of monoclonal antibodies occupying different epitope regions. a Monoclonal antibodies occupying one epitope region. S2H13 binds to the epitope region a which is colored in red. EY6A binds to region b which is colored in yellow. b Antibodies occupying two epitope regions. S2E12 binds to regions a/b which is colored in hot pink. S2A4 binds to regions b/d which is colored in cyan. C135 binds to regions d/e which is colored in blue. c Antibodies occupying three epitope regions. CC12.1 which is colored in green binds to the side of the ridge, representing this type of antibody that binds to the regions a/b/c. 2–4 which is colored in purple binds to the tip of the ridge, representing this type of antibody that binds to the regions a/b/e. COVA2-39 is colored in orange. d Antibodies that bind SARS-CoV-2 NTD. 4A8 is colored in forest green. COVOX-159 is colored in magenta. ACE2 is colored in greed. S protein is colored in white. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
The antibodies that only bind to one epitope region require multiple antibodies to neutralize SARS-CoV-2. They usually recognize the RBDs of the adjacent S protein in the trimer and keep the S protein in a down conformation, which prevents it access to ACE2.
3.1.2. Monoclonal antibody that occupying two epitope regions on RBD
Similar to the ACE2, these antibody epitopes occupy two regions: epitopes a/b. The binding angle of two antibodies S2E12 and CV07-250 to RBD is very similar to ACE2 (Fig. 2b). The steric hindrance of monoclonal antibodies that block the binding of ACE2 to RBD is one of the most important mechanisms for virus neutralization. The binding mode of CV07-250 to the RBD is unusual which is mainly dominated by light chains, the others are dominated by heavy chains. After interacting with RBD, CV07-250 has a buried surface area 399 Å and 559 Å on the heavy chain and light chains, respectively. But this unique binding mode of CV07-250 does not affect its neutralizing activity. The ELISA experiment showed that the neutralizing activity of CV07-250 was stronger than the CV07-270 reported in the same paper [40].
There are eight antibodies occupying other areas of S protein which are different from ACE2. The binding angle of these antibodies including CV07–270, S2M11, P2B–2F6, BD-368-2, S2A4, COVA1-16, H014 and C135 is different from ACE2. This unique binding angle will more or less cause a decrease in neutralizing activity. Among them, S2A4 binds to areas b/d, which is completely different from ACE2. Crystal structure analysis shows, S2A4 will conflict with ACE2 in space after binding with S trimer (Fig. 2b). Another explanation is that after S2A4 binds to RBD, it triggers the S1 subunit to fall off the S protein, thereby losing the ability to bind ACE2 [48]. These indicate the reason why S2A4 has strong neutralizing activity but the binding epitope regions is completely different from ACE2. The other antibody C135 binding epitope regions are d/e, which is in the middle of the S1 subunit. Crystal structure analysis shows C135 does not interfere with ACE2 binding (Fig. 2b), but the structure revealed three C135 bound to an S trimer with two down and one up RBD which may cause spatial conflict with ACE2. And its use in combination with other antibodies as antibody cocktail may increase the chance of the antibody's spatial conflict with ACE2 [43].
3.1.3. Monoclonal antibody that occupying three epitope regions on RBD
Among the antibodies that bind to three RBD epitope regions, most of them bind to epitopes a/b/c which are similar to ACE2: COVA2-04, C1A-B3, C1A-F10, C1A-C2, CA1-B12, BD-236, BD-604, BD-629, CV30, C105, CB6, CC12.1, CC12.3, B38, C144, C002, C121, C119, P17, 2–4, COVA2-39, C102, CT-P59 (Fig. 2c). Interestingly, antibody C110 binds to the epitope regions a/d/e. Crystal structure analysis showed that some of these antibodies bound to the side of the RBD ridge and some bound to the tip of the ridge (Fig. 2c). The binding epitopes of antibody COVA2-39 are between side and tip of the ridge (Fig. 2c). COVA2-39 mainly uses the heavy chain to interact with the epitope, and both the heavy chain and the light chain interact with the ridge. In contrast to this, COVA2-04 was reported at the same time mainly uses the light chain to interact with the ACE2 binding sites, and the heavy chain to interact with the RBD ridge and their binding to RBD is dominated by the heavy chain [46].
3.2. Monoclonal antibodies bind to SARS-CoV-2 NTD
There is another class of antibodies that do not bind to RBD but the NTD on the S protein: 4A8 [17], FC05 [51], COVOX-159 [52]. The structure analysis of 4A8 and COVOX-159 showed that their binding epitope regions do not overlap with ACE2 at all, and they do not compete with ACE2 binding (Fig. 2d). Although it's not clear that the function of NTD in SARS-CoV-2 infection in humans and the specific neutralization mechanism of NTD-binding antibodies, recent studies have shown that this type of antibodies has the effective neutralizing activity on SARS-CoV-2 and MERS-CoV [52,53]. There are also reports showing that the use of such antibodies in combination with RBD antibodies enhances the neutralizing activity of the antibodies [51]. This inspired us that this type of antibody can be used in combination with RBD-binding antibodies as antibody cocktail, which may boost the antiviral therapy as detailed in section 5.
4. Synthetic nanobodies neutralize SARS-CoV-2 infection
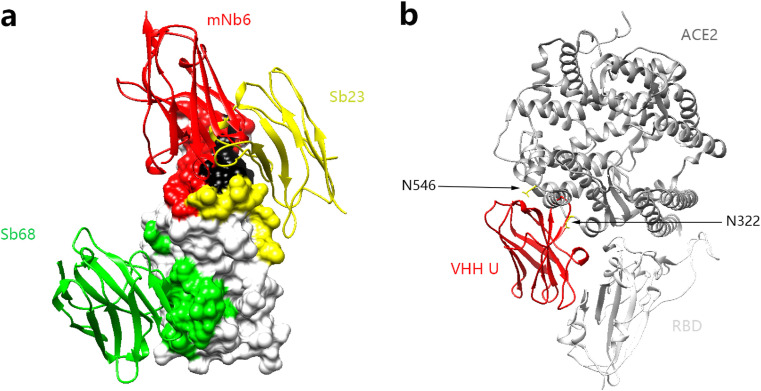
Heavy chain only antibodies that naturally lacks light chains in the peripheral blood of alpaca has structural stability and antigen-binding activity equivalent to that of the conventional antibodies, and is the smallest unit known to bind the target antigen. In 1993, Belgian scientists first discovered and described the characteristics of this type of antibodies, nanobodies [54]. Here, we compared the some nanobodies whose structures are disclosed: mNb6 [24], H11–H4, H11-D4 [34], Sb23 [35], Nb20 [36], Ty1 [37], Sb16, Sb45, Sb68 [55], VHH E, VHH U, VHH V, VHH W [56], VHH-72 [57], SR4, MR17 [58] ( Table 2 Fig. 3 a). Structural analysis shows that most of these nanobodies bound to the ridges of RBD, and therefore have a strong conflict with ACE2, making them has a strong neutralizing activity. However, there are some effective antibodies that bind to regions far away from the ACE2 bind sites, and they bind to the regions b/d of RBD. This is because they only conflicts with the carbohydrate of N322 and N546 on ACE2 (Fig. 3b). This small area of conflict makes it also have a certain neutralizing activity [55,56]. Of note, VHH-72 has cross-neutralizing activity and can block SARS-CoV-1 and SARS-CoV-2 [57]. In general, due to its small size, nanobodies are easy to prepare, access to hidden epitopes, and stable to heat and pH [[59], [60], [61]]. The continuous research of nanobodies will provide a powerful alternative for the prevention and treatment of SARS-CoV-2 infection.
Table 2.
Information on synthetic nanobodies that neutralize SARS-CoV-2.
| Name | Source | Target | PDB | The binding epitopes | Number of residues buried |
|---|---|---|---|---|---|
| mNb6 | Yeast surface-displayed library | RBD | 7KKL | a b e | 29 |
| H11–H4 | Llama VHH antibody library | RBD | 6ZBP | a b e | 25 |
| H11-D4 | Llama VHH antibody library | RBD | 6YZ5 | a b e | 20 |
| Nb20 | Llama | RBD | 7JVB | a b e | 21 |
| Ty1 | Alpaca | RBD | 6ZXN | a b e | 28 |
| Sb16 | Plasmids pSb-init | RBD | 7KGK | a b e | 28 |
| Sb45 | Plasmids pSb-init | RBD | 7KGJ | a b e | 29 |
| VHH E | Llama | RBD | 7KSG | a b e | 17 |
| SR4 | High-diversity libraries | RBD | 7C8V | a b e | 28 |
| MR17 | High-diversity libraries | RBD | 7C8W | a b e | 25 |
| Sb23 | Synthetic library, | RBD | 7A25 | a c e | 23 |
| Sb68 | Plasmids pSb-init | RBD | 7KLW | b d | 15 |
| VHH U | Alpaca | RBD | 7KN5 | b d | 17 |
| VHH V | Alpaca | RBD | 7KN6 | b d | 27 |
| VHH W | Alpaca | RBD | 7KN7 | b d | 18 |
| VHH-72 | Llama | RBD | 6WAQ | b d | 22 |
Fig. 3.
Synthetic nanobodies neutralize SARS-CoV-2 RBD. a Antibody mNb6 in red represents antibodies that bind to the epitope regions a/b/e. Sb23 binds to regions a/c/e which is colored in yellow. Sb68 represents antibodies that bind to the epitope regions on b and d which is colored in green. (The overlap sites on RBD between two antibodies are colored in black.) b Overview of conflicting sites of ACE2 and nanobodies which bind to b and d. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
5. Potent antibody cocktails neutralize SARS-CoV-2 infection
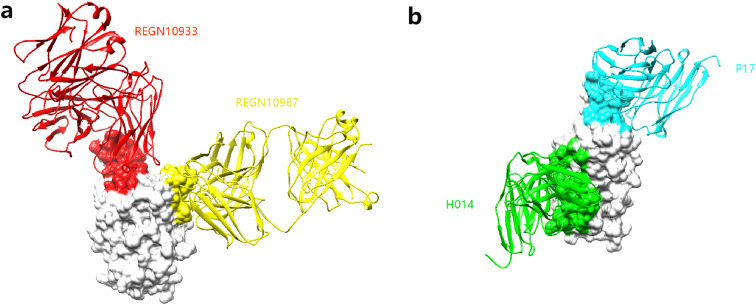
The therapeutic strategy of antibody cocktail is to reduce the potential of the virus from immune evasion, due to viral mutation, which may appear in response to selective pressure from single antibody therapy. Therefore, antibody cocktail has stronger neutralizing activity than a single antibody. Here we compare some antibody cocktails: REGN10933-REGN10987 [38], P17–H014 [62], FC05–H014, FC05-HB27, FC05-HB27, FC05–P17 [63], LY-CoV016 (CB6)-LY-CoV555 [64] (Table 3 ). However, there are some newly reported antibody cocktails: COV2-2196-COV2-2130 is not included due to its crystal structure not yet available [65]. The use of antibody cocktail is achieved under the condition that the two antibodies do not compete with each other to bind to the S protein. However, which part of the S protein each antibody binds to is not strictly required. The crystal structure analysis shows that in most antibody cocktails, one antibody binds to the ridge of the RBD, and the other antibody binds to other parts of the RBD, so the two antibodies do not affect each other (Fig. 4 ). This indicates that in this way of binding, a total of 6 antibodies are bound to one S protein trimer. These six antibodies form a huge barrier in space, preventing any possibility of RBD interacting with host cell receptors and cell surface proteases. The special case is FC05, which is an NTD antibody, used at the same time with RBD antibody, and also shows good neutralizing activity. It is worth noting that FC05 uses a cross-arrangement around the outside of the apex of the S trimer, which completely prevents the domain exchange between prototypes, which cause RBD in a state where it cannot bind to the ACE2 receptor. This explains the simultaneous use of the RBD antibody also shows good neutralizing activity [63]. And due to the good steric hindrance of the antibody cocktail, it may have a better performance in dealing with mutant viruses.
Table 3.
Information on antibody cocktails that neutralize SARS-CoV-2.
| Name | Target | PDB | The binding epitopes | Number of residues buried |
|---|---|---|---|---|
| REGN10933-REGN10987 | RBD | 6XDG | a b d e | 34 |
| P17–H014 | RBD | 7CWN | a b d e | 46 |
| FC05–H014 | RBD + NTD | 7CWS | b d | 30 |
| FC05-HB27 | RBD + NTD | 7CWT | – | – |
| FC05–P17 | RBD + NTD | 7CWU | a e | 20 |
| LY-CoV016(CB6)-LY-CoV555 | RBD | 7C01+7KMG | a b c e | 48 |
Fig. 4.
Potent antibody cocktails neutralize SARS-CoV-2 S protein. a Antibody cocktail REGN10933-REGN10987 binds to the RBD. REGN10933 binds to regions a/b which is colored in red. REGN10987 binds to regions d/e which is colored in yellow. b Antibody cocktail P17–H014 binds to the RBD. P17 binds to regions a/e which is colored in cyan. H014 binds to regions a/e which is colored in green. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
6. Discussion
The pandemic of COVID-19 has been going on for more than one year, and it is far from termination in the world. Finding effective treatment methods for COVID-19 is still the driving force of scientists' efforts. At present, many structures of SARS-CoV-2 antibodies have been determined and most of them have potent neutralizing activity. We have summarized three representative strategies of antibodies therapies that have been reported so far: monoclonal antibodies, nanobodies, and antibody cocktails. They have their own binding characteristics, and it is difficult to unify their mechanism of action, but most of the antibodies bind to the RBD regions of the S protein so that the virus no longer interact with its receptor ACE2. Therefore, viruses cannot infect human cells and lose their infective activity.
Most of the monoclonal antibodies have the same binding epitope regions shared with ACE2. Although a few antibodies bind epitopes are different from ACE2, they also have strong neutralizing activity. This is because they still have steric hindrance with ACE2 in space after binding to RBD, thereby blocking the binding of ACE2 to RBD. The most representative antibodies are S2A4 and C135. Interestingly, some monoclonal antibodies combined with NTD also produced strong neutralizing activity, but the role of NTD in virus infection in humans is currently unclear. The crystal structure analysis showed that the S protein trimer bound NTD antibody to make the RBD in “down” state where it could not bind ACE2, and it was found that steric hindrance occurred after overlapping the antibody and ACE2. These explain the neutralizing activity of NTD antibodies. Due to their small size, nanobodies can bind to deep epitope regions, and the reported nanobodies have super neutralizing activity. It is worth noting that due to the far distance between NTD and RBD in space, which provides conditions for the combined use of RBD antibody and NTD antibody as an antibody cocktail. And considering the smaller size of nanobodies, nanobodies and NTD antibodies may be better as antibody cocktail. The use of these antibody cocktails will likely greatly enhance the neutralizing activity of antibodies.
In a recent report, antibody LY-CoV555 can no longer neutralize SARS-CoV-2 mutant B.1.351. Therefore, by analyzing the neutralizing effect of LY-CoV016 (CB6)-LY-CoV555 on the virus infection, Starr et al. explained that the mutation of single amino acid resulted in a new epitope, which may provide novel epitopes for production of antibodies and antibody cocktails [64]. This suggests that the selective pressure of neutralizing antibodies on SARS-CoV-2 drive virus mutations and may increase the probability of inactivation of the current effective antibodies. It may also cause the invalidation of the vaccines currently on the market due to the epitopes of SARS-CoV-2 mutated. Accordingly, the advantages of antibody cocktail will be gradually appreciated and may be more effective strategy for combating SARS-CoV-2. And the development of future vaccines should also take the SARS-CoV-2 mutants into account, in order to prevent another large-scale outbreak of COVID-19 more comprehensively and effectively.
Authors’ contributions
J. Sun and J. Wei conceived and designed the Subject. D. Jin carried out this research. J. Sun and D. Jin analyzed the result. D. Jin and J. Sun wrote the paper. All authors have read and approved the final manuscript to be published
Declaration of competing interest
We declare that we do not have any commercial or associative interest that represents a conflict of interest in connection with the work submitted.
Acknowledgements
This study was supported by the National Natural Science Foundation of China(Grant No. 81273308).
References
- 1.Zhou P., Yang X.L., Wang X.G., Hu B., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deng Y., Lv Z., Ye Q., Cao L., et al. Structural basis for neutralization of SARS-CoV-2 and SARS-CoV by a potent therapeutic antibody. Science. 2020;369:1505–1509. doi: 10.1126/science.abc5881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zaki A.M., van Boheemen S., Bestebroer T.M., et al. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 4.Drosten C., Günther S., Preiser W., et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 5.Gui M., Song W., Zhou H., et al. Cryo-electron microscopy structures of the SARS-CoV spike glycoprotein reveal a prerequisite conformational state for receptor binding. Cell Res. 2017;27:119–129. doi: 10.1038/cr.2016.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoffmann M., Kleine-Weber H., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. e278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Walls A.C., Park Y.J., Tortorici M.A., et al. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292. doi: 10.1016/j.cell.2020.02.058. e286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hofmann H., Pohlmann S. Cellular entry of the SARS coronavirus. Trends Microbiol. 2004;12:466–472. doi: 10.1016/j.tim.2004.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walls A.C., Tortorici M.A., Snijder J., et al. Tectonic conformational changes of a coronavirus spike glycoprotein promote membrane fusion. Proc. Natl. Acad. Sci. U. S. A. 2017;114:11157–11162. doi: 10.1073/pnas.1708727114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yuan Y., Cao D., Zhang Y., et al. Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains. Nat. Commun. 2017;8 doi: 10.1038/ncomms15092. 15092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu H., Wu N.C., Yuan M., S, et al. Cross-neutralization of a SARS-CoV-2 antibody to a functionally conserved site is mediated by avidity. Immunity. 2020;53:1272–1280. doi: 10.1016/j.immuni.2020.10.023. e1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kirchdoerfer R.N., Wang N., Pallesen J., et al. Stabilized coronavirus spikes are resistant to conformational changes induced by receptor recognition or proteolysis. Sci. Rep. 2018;8 doi: 10.1038/s41598-018-34171-7. 15701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wrapp D W.N., Ks Corbett, et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kirchdoerfer R.N., Cottrell C.A., Wang N., et al. Pre-fusion structure of a human coronavirus spike protein. Nature. 2016;531:118–121. doi: 10.1038/nature17200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walls A.C., Xiong X., Park Y.J., et al. Unexpected receptor functional mimicry elucidates activation of coronavirus fusion. Cell. 2019;176:1026–1039. doi: 10.1016/j.cell.2018.12.028. e1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lan J., Ge J., Yu J., et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 17.Remsik Y., Chi X., Zhang J., Zhang G., et al. A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2. Science. 2020;369:650–655. doi: 10.1126/science.abc6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tahir Ul Qamar M., Alqahtani S.M., et al. Structural basis of SARS-CoV-2 3CL(pro) and anti-COVID-19 drug discovery from medicinal plants. Int. J. Pharm. Anal. 2020;10:313–319. doi: 10.1016/j.jpha.2020.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baildya N., Khan A.A., Ghosh N.N., et al. Screening of potential drug from Azadirachta Indica (Neem) extracts for SARS-CoV-2: an insight from molecular docking and MD-simulation studies. J. Mol. Struct. 2021;1227 doi: 10.1016/j.molstruc.2020.129390. 129390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li F., Michelson A.P., Foraker R., et al. Computational analysis to repurpose drugs for COVID-19 based on transcriptional response of host cells to SARS-CoV-2. BMC Med. Inf. Decis. Making. 2021;21 doi: 10.1186/s12911-020-01373-x. 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Y., Wan Y., Liu P., et al. A humanized neutralizing antibody against MERS-CoV targeting the receptor-binding domain of the spike protein. Cell Res. 2015;25:1237–1249. doi: 10.1038/cr.2015.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang C., Li W., Drabek D., et al. A human monoclonal antibody blocking SARS-CoV-2 infection. Nat. Commun. 2020;11 doi: 10.1038/s41467-020-16256-y. 2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wan J., Xing S., Ding Y L., et al. Human-IgG-neutralizing monoclonal antibodies block the SARS-CoV-2 infection. Cell Rep. 2020;32 doi: 10.1016/j.celrep.2020.107918. 107918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Faust B., Schoof M., Saunders R.A., et al. An ultrapotent synthetic nanobody neutralizes SARS-CoV-2 by stabilizing inactive Spike. Science. 2020;370:1473–1479. doi: 10.1126/science.abe3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prabakaran P., Gan J., Feng Y., et al. Structure of severe acute respiratory syndrome coronavirus receptor-binding domain complexed with neutralizing antibody. J. Biol. Chem. 2006;281:15829–15836. doi: 10.1074/jbc.M600697200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clark S.A., Clark L.E., Pan J., et al. Molecular basis for a germline-biased neutralizing antibody response to SARS-CoV-2. bioRxiv. 2020 doi: 10.1101/2020.11.13.381533. [DOI] [Google Scholar]
- 27.Du S., Cao Y., Zhu Q., et al. Structurally resolved SARS-CoV-2 antibody shows high efficacy in severely infected hamsters and provides a potent cocktail pairing strategy. Cell. 2020;183:1013–1023. doi: 10.1016/j.cell.2020.09.035. e1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu H., Yuan M., Wu N.C., Lee C.D., et al. Structural basis of a shared antibody response to SARS-CoV-2. Science. 2020;369:1119–1123. doi: 10.1126/science.abd2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hurlburt N.K., Seydoux E., Y, et al. Structural basis for potent neutralization of SARS-CoV-2 and role of antibody affinity maturation. Nat. Commun. 2020;11 doi: 10.1038/s41467-020-19231-9. 5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barnes C.O., West A.P., et al. Structures of human antibodies bound to SARS-CoV-2 spike reveal common epitopes and recurrent features of antibodies. Cell. 2020;182:828–842. doi: 10.1016/j.cell.2020.06.025. e816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shi R., Shan C., Duan X., et al. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature. 2020;584:120–124. doi: 10.1038/s41586-020-2381-y. [DOI] [PubMed] [Google Scholar]
- 32.Tortorici Ma B.M., Fa Lempp, et al. Ultrapotent human antibodies protect against SARS-CoV-2 challenge via multiple mechanisms. Science. 2020;370:950–957. doi: 10.1126/science.abe3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang L., Cao L., Gao X.S., Zheng B.Y., Zhu F.C. A proof of concept for neutralizing antibody-guided vaccine design against SARS-CoV-2. bioRxiv. 2020 doi: 10.1101/2020.09.23.309294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huo J., Le Bas A., Ruza R.R., et al. Neutralizing nanobodies bind SARS-CoV-2 spike RBD and block interaction with ACE2. Nat. Struct. Mol. Biol. 2020;27:846–854. doi: 10.1038/s41594-020-0469-6. [DOI] [PubMed] [Google Scholar]
- 35.Custodio T.F., Das H., et al. Selection, biophysical and structural analysis of synthetic nanobodies that effectively neutralize SARS-CoV-2. Nat. Commun. 2020;11 doi: 10.1038/s41467-020-19204-y. 5588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xiang Y., Nambulli S., et al. Versatile, multivalent nanobody cocktails efficiently neutralize SARS-CoV-2. bioRxiv. 2020 doi: 10.1101/2020.08.24.264333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hanke L., Vidakovics Perez L., et al. An alpaca nanobody neutralizes SARS-CoV-2 by blocking receptor interaction. Nat. Commun. 2020;11 doi: 10.1038/s41467-020-18174-5. 4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hansen J B.A., Ke Pascal, V Russo, et al. Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Science. 2020;369:1010–1014. doi: 10.1126/science.abd0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang N., Sun Y., Feng R., et al. Structure-based development of human antibody cocktails against SARS-CoV-2. Cell Res. 2021;31:101–103. doi: 10.1038/s41422-020-00446-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kreye J., Reincke S.M., Kornau H.C., et al. A therapeutic non-self-reactive SARS-CoV-2 antibody protects from lung pathology in a COVID-19 hamster model. Cell. 2020;183:1058–1069. doi: 10.1016/j.cell.2020.09.049. e1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu Y., Wang F., Shen C., et al. A noncompeting pair of human neutralizing antibodies block COVID-19 virus binding to its receptor ACE2. Science. 2020;368:1274–1278. doi: 10.1126/science.abc2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yuan M., Liu H., Wu N.C., et al. Structural basis of a shared antibody response to SARS-CoV-2. Science. 2020;369 doi: 10.1126/science.abd2321. eabd2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barnes C.O., Jette C.A., Abernathy M.E., et al. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature. 2020 doi: 10.1038/s41586-020-2852-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yao H., Sun Y., Deng Y.Q., et al. Rational development of a human antibody cocktail that deploys multiple functions to confer Pan-SARS-CoVs protection. Cell Res. 2021;31:25–36. doi: 10.1038/s41422-020-00444-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu L., Wang P., Nair M.S., et al. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature. 2020;584:450–456. doi: 10.1038/s41586-020-2571-7. [DOI] [PubMed] [Google Scholar]
- 46.Wu N.C., Yuan M., Liu H., et al. An alternative binding mode of IGHV3-53 antibodies to the SARS-CoV-2 receptor binding domain. Cell Rep. 2020;33 doi: 10.1016/j.celrep.2020.108274. 108274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim C., Ryu D.K., Lee J., et al. A therapeutic neutralizing antibody targeting receptor binding domain of SARS-CoV-2 spike protein. Nat. Commun. 2021;12 doi: 10.1038/s41467-020-20602-5. 288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Piccoli L., Park Y.J., Tortorici M.A., et al. Mapping neutralizing and immunodominant sites on the SARS-CoV-2 spike receptor-binding domain by structure-guided high-resolution serology. Cell. 2020;183:1024–1042. doi: 10.1016/j.cell.2020.09.037. e1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ju B., Zhang Q., Ge J., et al. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature. 2020;584:115–119. doi: 10.1038/s41586-020-2380-z. [DOI] [PubMed] [Google Scholar]
- 50.Zhou D., Duyvesteyn H.M.E., Chen C.P., et al. Structural basis for the neutralization of SARS-CoV-2 by an antibody from a convalescent patient. Nat. Struct. Mol. Biol. 2020;27:950–958. doi: 10.1038/s41594-020-0480-y. [DOI] [PubMed] [Google Scholar]
- 51.Zhang L., Cao L., Gao X.-S., et al. A proof of concept for neutralizing antibody-guided vaccine design against SARS-CoV-2. bioRxiv. 2020 doi: 10.1101/2020.09.23.309294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dejnirattisai W., Zhou D., Ginn H.M., et al. The antigenic anatomy of SARS-CoV-2 receptor binding domain. Cell. 2021 doi: 10.1016/j.cell.2021.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhou H., Chen Y., Zhang S., P, et al. Structural definition of a neutralization epitope on the N-terminal domain of MERS-CoV spike glycoprotein. Nat. Commun. 2019;10 doi: 10.1038/s41467-019-10897-4. 3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hamers-Casterman C., Atarhouch T., et al. Naturally occurring antibodies devoid of light chains. Nature. 1993;363:446–448. doi: 10.1038/363446a0. [DOI] [PubMed] [Google Scholar]
- 55.Ahmad J., Jiang J., Boyd L.F., et al. Synthetic nanobody-SARS-CoV-2 receptor-binding domain structures identify distinct epitopes. bioRxiv. 2021 doi: 10.1101/2021.01.27.428466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Koenig P.A., Das H., Liu H., et al. Structure-guided multivalent nanobodies block SARS-CoV-2 infection and suppress mutational escape. Science. 2021;371 doi: 10.1126/science.abe6230. eabe6230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wrapp D., De Vlieger D., Corbett K.S., et al. Structural basis for potent neutralization of betacoronaviruses by single-domain camelid antibodies. Cell. 2020;181:1004–1015. doi: 10.1016/j.cell.2020.04.031. e1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li T., Cai H., Yao H., et al. Potent synthetic nanobodies against SARS-CoV-2 and molecular basis for neutralization. bioRxiv. 2020 doi: 10.1101/2020.06.09.143438. [DOI] [Google Scholar]
- 59.van der Linden Rh F.L., Geus B de, et al. Comparison of physical chemical properties of llama VHH antibody fragments and mouse monoclonal antibodies. Biochim. Biophys. Acta. 1999;1431:37–46. doi: 10.1016/s0167-4838(99)00030-8. [DOI] [PubMed] [Google Scholar]
- 60.Pérez Jm R.J., Jj Prompers, et al. Thermal unfolding of a llama antibody fragment: a two-state reversible process. Biochemistry. 2001;40:74–83. doi: 10.1021/bi0009082. [DOI] [PubMed] [Google Scholar]
- 61.Dumoulin M., Conrath K., et al. Single-domain antibody fragments with high conformational stability. Protein Sci. 2002;11:500–515. doi: 10.1110/ps.34602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yao H., Sun Y., Deng Y.Q., et al. Rational development of a human antibody cocktail that deploys multiple functions to confer Pan-SARS-CoVs protection. Cell Res. 2020;31:25–36. doi: 10.1038/s41422-020-00444-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang N., Sun Y., Feng R., et al. Structure-based development of human antibody cocktails against SARS-CoV-2. Cell Res. 2020;31:101–103. doi: 10.1038/s41422-020-00446-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Starr T.N., Greaney A.J., et al. Complete map of SARS-CoV-2 RBD mutations that escape the monoclonal antibody LY-CoV555 and its cocktail with LY-CoV016. bioRxiv. 2021 doi: 10.1101/2021.02.17.431683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dong J., Zost S.J., Greaney A.J., et al. Genetic and structural basis for recognition of SARS-CoV-2 spike protein by a two-antibody cocktail. bioRxiv. 2021 doi: 10.1101/2021.01.27.428529. [DOI] [Google Scholar]