Significance
Using organoids, this study shows that Notch activity and loss of p53 induce a regenerative cell state and recapitulate tumorigenesis. Mutant organoids self-renew and grow independently of essential growth factors and exhibit elevated levels of nuclear Yap, Mll1, and H3K4 trimethylation. These factors are also elevated in human colorectal cancer (CRC) and control viability of patient-derived CRC organoids. Yap interacts with Mll1, and both promote a regenerative cell state that links regenerative processes to tumorigenesis.
Keywords: cancer, Kmt2a, Notch, Yap, regeneration
Abstract
Specified intestinal epithelial cells reprogram and contribute to the regeneration and renewal of the epithelium upon injury. Mutations that deregulate such renewal processes may contribute to tumorigenesis. Using intestinal organoids, we show that concomitant activation of Notch signaling and ablation of p53 induce a highly proliferative and regenerative cell state, which is associated with increased levels of Yap and the histone methyltransferase Mll1. The induced signaling system orchestrates high proliferation, self-renewal, and niche-factor-independent growth, and elevates the trimethylation of histone 3 at lysine 4 (H3K4me3). We demonstrate that Yap and Mll1 are also elevated in patient-derived colorectal cancer (CRC) organoids and control growth and viability. Our data suggest that Notch activation and p53 ablation induce a signaling circuitry involving Yap and the epigenetic regulator Mll1, which locks cells in a proliferative and regenerative state that renders them susceptible for tumorigenesis.
The small intestinal epithelium is continuously renewed by active stem cells that steadily produce absorptive and secretory cells, which enable nutrient supply and protect the epithelium (1). The cellular hierarchy is dynamic and specified cells can reprogram to replenish stem cells after epithelial injury and stem cell loss (2–6). These processes require tight control, as deregulated and persistent repair processes may lead to cancer (7). Regenerative as well as tumorigenic processes in the intestinal epithelium have been linked to the activity of the transcription factor Yap (8–10). Yap mediates the growth of intestinal stem cells and is overexpressed and activated in colon cancer (9, 11–15). Nuclear location controls protein stability and the transcriptional activity of Yap (13, 16). Notch signaling can synergize with Yap in the control of tumor cell proliferation (17, 12). Notch signaling promotes stem cell function, is activated during regeneration, and advances colon cancer progression (18–23). Binding of Notch ligands to Notch receptors located on adjacent cells induces the proteolytic cleavage of the Notch receptor and the released Notch-intracellular-cleaved domain (NICD), which translocate to the nucleus (24). At homeostasis, Notch signaling controls cell specification of absorptive (Notch active) versus secretory (Notch inactive) cells, determined by an autoregulatory circuit called lateral inhibition, in which Notch-activated cells down-regulate the expression of Notch ligands, which results in diminished Notch activation in the adjacent cell (20, 21).
Activation of Notch signaling and loss of p53 in the intestinal epithelium (in NICDEYFPflox/flox/p53flox/flox; VillinCreERT2 mice, here called NICD/p53−/− mice) by inducible Cre-mediated recombination causes the production of the intracellular domain of the Notch receptor (NICD) and ablation of p53 (25). Mice can develop invasive intestinal tumors with metastases, thus recapitulating human tumors to a great extent (25). However, the tumors develop only after months and exhibit accumulation of mutations. The data indicate that Notch activation and loss of p53 generate a stable cell state capable of accumulating mutations, which ultimately result in transformation.
To understand the establishment of a persistent cell state susceptible to oncogenic transformation in NICD/p53−/− mice, we took advantage of studying organoids, which preserve the genomic and cellular complexity of the tissue of origin to a large extent, and consist of stem cells and differentiated cell types (26). Small intestinal organoids form a self-organized cellular hierarchy, regrow over many passages, and require the growth factors R-spondin, Egf, and Noggin for maintenance of stem cells, proliferation, and differentiation control, respectively (27).
Here, we show that activation of Notch and ablation of p53 induce a stable regenerative cell state with a high expression of genes specific for regenerative epithelia such as Clu, Anxa-1, and Trop2, as well as classical Yap target genes such as Ctgf and Cyr61. Such NICD/p53−/− organoids adopt a spheroid shape, self-renew, and grow independently of essential growth factors otherwise supplemented for organoid growth and maintenance, a phenomenon that resembles tumor organoids. NICD/p53−/− organoids exhibit elevated levels of nuclear Yap, Mll1, and H3K4 trimethylation, which we also detected in human CRC tissue. Yap interacts with Mll1 and Wdr5, two essential components of the trithorax-histone methyltransferase complex. We show that the viability of regenerative NICD/p53−/− organoids and human patient-derived CRC organoids depends on both Yap and Mll1. Our data suggest that Notch signaling and loss of p53 induce and maintain a regenerative program with high Yap and Mll1, which link regenerative processes to tumorigenesis.
Results
NICD/p53−/− Mutant Organoids Grow Spheroid Shaped and Niche-Factor Independent.
From the small intestine of NICDEYFPflox/flox/p53flox/flox; VillinCreERT2 mice (25) we established organoids and induced Cre-mediated recombination by 4-hydroxytamoxifen (4-OHT) in culture. The mutagenesis resulted in ablation of p53 and in the production of the NICD (SI Appendix, Fig. 1A). NICD and loss of p53 induced the formation of spheroid-shaped organoids (Fig. 1 A, Upper). The uninduced organoids grew in the known crypt-like organoid structures (27) and served as controls (Fig. 1 A, Lower). Coproduction of EYFP from the Rosa26floxStopfloxNICD-EYFP locus allowed us to trace mutant cells (SI Appendix, Fig. S1B). EYFP tracing, 5 d after induction of mutagenesis, revealed that spheroid formation occurred from a subpopulation of cells (SI Appendix, Fig. S1B). Upon single-cell dissociation, these mutant cells regrew and steadily formed spheroid-shaped organoids (SI Appendix, Fig. S1B and Fig. 1A). NICD/p53−/− organoid cells grew as polarized single epithelial cell layers: electron microscopy showed that epithelial cells had microvilli at the apical surface, facing the inside of the organoid (SI Appendix, Fig. S1C, see enlargements, Upper Right), and the basement membrane was located toward the outside. The cells were joined by lateral tight junctions close to the apical side (see Inset, Below and hatched oval in the enlargement), by desmosomes over the lateral side (marked by yellow asterisks), and by interdigitations in the middle. NICD/p53−/− organoids exhibited an enrichment of actin mesh at the apical surface, shown by whole-mount phalloidin staining (SI Appendix, Fig. S1D).
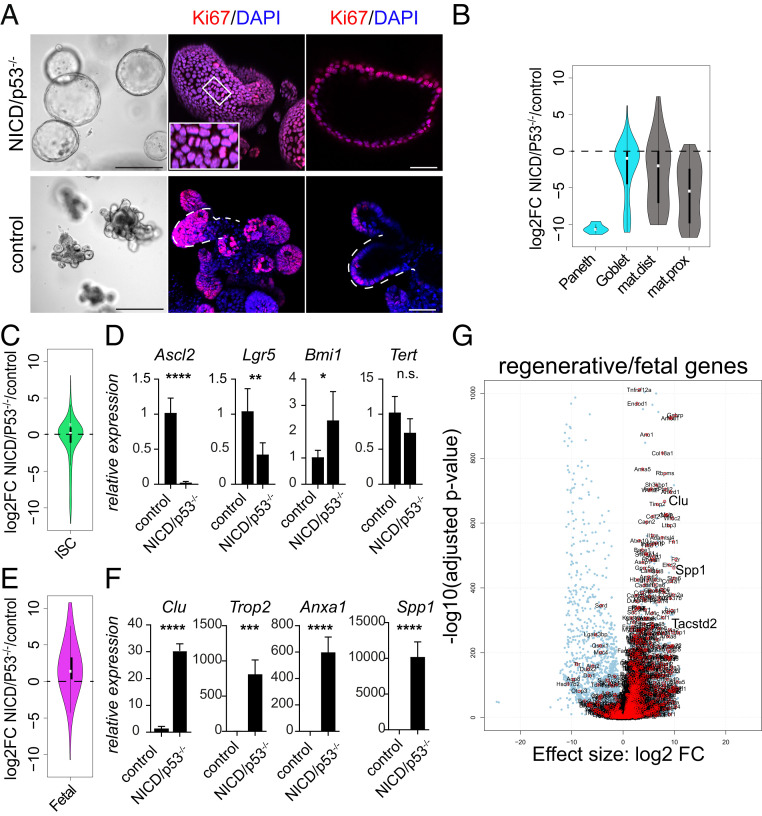
Fig. 1.
NICD/p53−/− mutant organoids grow spheroid shaped and exhibit regenerative properties. (A) Brightfield and immunofluorescence for Ki67 of NICD/p53−/− organoids (Upper) and control organoids (Lower) (Left, Scale bar, 250 µm.); three-dimensional reconstitution of confocal z-stack images (Middle) and optical section (Right) of immunofluorescence for Ki67 (red) and DAPI (blue). (Scale bar, 50 µm.) Surface buds in the controls are marked by dashed lines. The Inset shows dividing cells. (B) Violin plot of differentially regulated genes (DRGs) in NICD/p53−/− compared to control organoids for cell-type-specific gene signatures. (C) Violin plot of stem cell genes differentially regulated in NICD/p53−/− compared to control organoids. (D) Relative mRNA expression of stem cell markers. (E) Violin plot of fetal/regenerative markers differentially regulated in NICD/p53−/− compared to control organoids. (F) Relative mRNA expression of fetal/regenerative genes. (G) Volcano plot of fetal/regenerative makers differentially regulated in NICD/p53−/− organoids. (*P ≤ 0.05, **P ≤ 0.001, ***P ≤ 0.0001.)
NICD/p53−/− organoids grew independently of the growth factors Egf, Noggin, and R-spondin1 (ENR) (SI Appendix, Fig. S1E), which were essential for the growth of control organoids (see cloud of dead cells in SI Appendix, Fig. S1 E, Lower, control −ENR). Growth factor-independent growth was not induced in single-mutant organoids either mutant for p53 or NICD; the culture of both required R-spondin (SI Appendix, Fig. S1F). R-spondin/Wnt signaling is essential for organoid viability and growth (27). However, viability of the NICD/p53−/− organoids was independent of Wnt signaling, as organoid growth was not affected by treatment with the Wnt inhibitor ICG-001 (SI Appendix, Fig. S1G). The results indicate that NICD and p53−/− mutations act in concert to mediate niche-factor-independent growth and self-renewal. Ki67 staining showed that NICD/p53−/− organoid cells were highly proliferative (Fig. 1A, see dividing cells in the Inset, Upper panel), while in control organoids, proliferation was restricted to crypt buds (Fig. 1 A, Lower, marked by white dashed lines).
Notch signaling is known to prevent the differentiation of secretory cells in the intestine by repressing the expression of the pan-secretory marker Math1 (28). Indeed, the expression of the secretory markers Lyz, ChgA, and Gob5 and of Math1 was strongly reduced in NICD/p53−/− organoids compared to controls (SI Appendix, Fig. S1H). NICD/p53−/− organoids did not contain secretory cells, as shown by the absence of Paneth cells (expressing lysozyme, red arrow), goblet cells (expressing the intestinal trefoil factor 3 [ITF], red arrow), and enteroendocrine cells (expressing chromograninA [ChroA], green arrow) (SI Appendix, Fig. S1I).
RNA sequencing of NICD/p53−/− and control organoids confirmed changes in cellular composition when compared to defined cell-type-specific gene signatures generated by single-cell sequencing (29) and from a gene ontology gene set for enteroendocrine cells. Genes specific for secretory cells (blue) as well as the gene signatures of enterocytes (gray) were down-regulated (Fig. 1B). Genes of a stem cell signature (30) revealed a set of up- and down-regulated stem cell genes in NICD/p53−/− organoids, as shown by a violin plot and heat map analysis (Fig. 1C and SI Appendix, Fig. S1J). RT-PCR confirmed that the expression of the classical stem cell markers Lgr5 and Ascl2 was strongly down-regulated, while Bmi1 and Tert levels were elevated or remained unchanged (Fig. 1D). These results show that the NICD/p53−/− organoids are composed of cells that grow niche-factor independently and are in a hyperproliferative, self-renewing cell state, yet have low expression of classical intestinal stem cell genes.
NICD/p53−/− Mutations Induce a Regenerative Program.
Intestinal injury triggers a transient epithelial reprogramming into a highly proliferative state. This epithelium has fetal-like features and can be mimicked in organoids that also display regenerative/fetal-like properties and display a spheroid shape (8), as observed here. We therefore analyzed the expression of fetal/regenerative signature genes in the NICD/p53−/− organoids. NICD/p53−/− organoids exhibited a marked elevation of a fetal/regenerative profile (Fig. 1E). We validated the increased expression of the regenerative genes Clu, Tacstd2 (Trop2), Anxa1, and Spp1 (8, 31, 32) by RT-PCR (Fig. 1F). The strong increase in regenerative gene expression also becomes evident using transcriptome analysis, as displayed in a volcano plot of differentially expressed genes in NICD/p53−/− organoids compared to controls (Fig. 1G). Thus, the NICD and p53−/− mutations induced regenerative-like programs in organoids, which grow niche-factor independently.
NICD/p53−/− Mutations Induce Nuclear Yap That Promotes Growth and Viability of Organoids and Tumors.
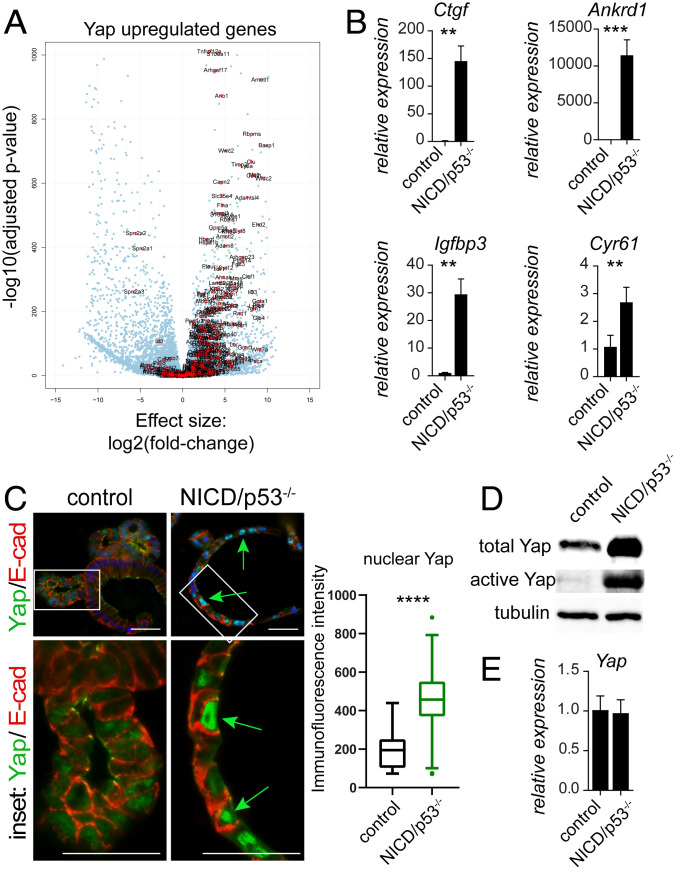
Nuclear translocation and activation of Yap have been implicated in regeneration and tissue repair in the intestinal epithelium (8, 9). Gene expression profiling revealed that genes up-regulated by Yap in intestinal organoids (9) were increased in NICD/p53−/− organoids (Fig. 2A), and genes down-regulated by Yap were decreased, respectively (SI Appendix, Fig. S2A). The increased transcriptional activity of Yap upon expression of NICD and loss of p53 was confirmed by elevated expression of the well-established Yap target genes Ctgf, Cyr61, Ankrd1, and Igfbp3 (33), as assessed by RT-PCR (Fig. 2B). We compared the subcellular location of Yap in NICD/p53−/− and control organoids by immunofluorescence. Strong Yap staining was found in nuclei of NICD/p53−/− organoids (Fig. 2 C, Right, see Inset, Below, marked by green arrows). Yap staining was much weaker in the nuclei of crypt-like cells of control organoids (Fig. 2 C, Left). Western blot confirmed that mutant organoids produced higher levels of total and active Yap (Fig. 2D). However, the levels of Yap mRNA were not changed (Fig. 2E). Tumors from NICD/p53−/− mice (25) also showed nuclear Yap (SI Appendix, Fig. S2B). To address the role of Yap in more detail, we inhibited the transcriptional activity of Yap by the small molecule Verteporfin (VP) (34), which reduced the viability of NICD/p53−/− organoids in time- and concentration-dependent manners (SI Appendix, Fig. S2 C and D). By lentiviral transduction of NICD/p53−/− organoids we introduced a doxycycline-inducible shYap cassette, which upon induction coproduces turboRFP that allows monitoring of shRNA expression (35). Three days of induction with doxycycline reduced the levels of Yap down to 40% (SI Appendix, Fig. S2E) and reduced organoid growth (SI Appendix, Fig. S2F, the control spheroid size is marked by a hatched circle, quantified on the Far Right). These results demonstrate that the growth and viability of NICD/p53−/− organoids is dependent on Yap.
Fig. 2.
NICD/p53−/− mutations promote nuclear Yap. (A) Volcano plot of DRGs in NICD/p53−/− organoids with indicated genes up-regulated in Yap-expressing organoids. (B) mRNA expression of specific Yap target genes comparing control and NICD/p53−/− organoids. (C) Immunofluorescence staining from control (Left) and NICD/p53−/− organoids (Right) for Yap (green) and E-cadherin (red), nuclei counterstained with DAPI; magnifications of Insets below (Scale bar, 50 µm.); the green arrows point to increased nuclear Yap in the mutants. Quantification of immunofluorescence intensity of nuclear Yap on the Right. (D) Western blot of control and NICD/p53−/− organoids showing increased total and active Yap in the mutants. (E) Similar expression of Yap mRNA in control and NICD/p53−/− organoids, assessed by qRT-PCR. (*P ≤ 0.05, **P ≤ 0.001, ***P ≤ 0.0001.)
A Src, Yap, and Mapk Signaling Cascade in NICD/p53−/− Organoids.
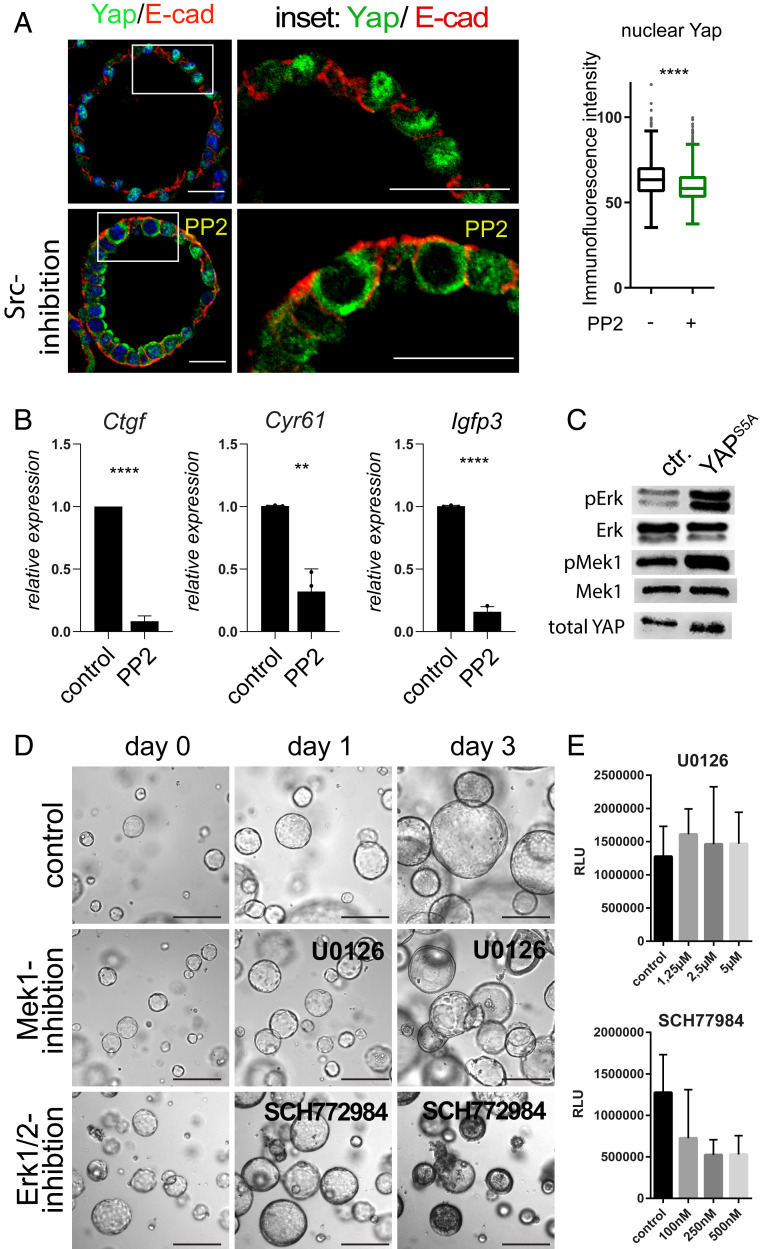
During regeneration, Yap activation is promoted by activity of the tyrosine kinase Src (8), which is activated via Stat3 in p53-ablated cells (36), and Stat3 signaling is also involved in epithelial repair mechanisms (37). RNA sequencing revealed that Stat3 targets (38) were up-regulated in NICD/p53−/− organoids (SI Appendix, Fig. S3A). Strikingly, at 24 h of Src kinase inhibition by PP2, Yap was sequestered in the cytosol (Fig. 3A) and Yap target genes were down-regulated (Fig. 3B). Prolonged inhibition of the Src activity reduced growth and viability of NICD/p53−/− organoids (SI Appendix, Fig. S3B). Western blot analysis of wild-type (WT) small intestinal organoids revealed a decrease of active Yap levels upon inhibition of Src kinase. Although, inhibition of Mst1/2 kinases with XMU-MP1 increased the level of active Yap, this could not rescue the effect of Src kinase inhibition on Yap (SI Appendix, Fig. S3C). In conclusion, Src kinase promotes stability of active Yap downstream of Mst1/2 kinases in wild-type organoids. In contrast, in NICD/p53−/− organoids neither Src, nor Mst1/2 kinase inhibition altered the level of active Yap protein (SI Appendix, Fig. S3C). These data indicate that Src promotes nuclear accumulation but not stability of Yap in NICD/p53−/− organoids to control their growth and viability. Yap has been described to cross-talk with Mapk signaling in regenerative processes of the lung (39), reinforcing the idea that Yap activity mediates Egf-independent growth of NICD/p53−/− organoids. Expression of an active form of Yap (S5A-Yap) in patient-derived colorectal cancer organoids strongly activated Mapk signaling with a pronounced increase in pErk levels (Fig. 3C). Inhibition of Mapk signaling with small molecule inhibitors of the MAP kinases Mek1 (U0126) and Erk1/2 (SCH772984) revealed that growth and viability of NICD/p53−/− organoids were dependent on Erk1/2 activity, whereas inhibition of the upstream kinase Mek1 did not affect organoid growth and viability, as assessed by tracking of organoids and viability assays (Fig. 3 D and E). Inhibition of Yap with VP showed a strong reduction in the level of activated phosphorylated Erk, while the level of activated Mek1 remained unchanged (SI Appendix, Fig. S3D). Erk inhibition down-regulated the expression of the Yap target genes Clu, Egr1, and Ly6a (SI Appendix, Fig. S3E), suggesting a signaling axis of Src-Yap-Erk in promoting the regenerative cell state.
Fig. 3.
Src kinase promotes nuclear Yap, which controls Erk activity. (A) Immunofluorescence staining of untreated NICD/p53−/− (Upper) and 20 µM PP2-treated NICD/p53−/− organoids (Lower) for E-cadherin (red) and Yap (green); Middle, magnifications of Insets. (Scale bar, 50 µm.) Quantification of immunofluorescence intensity of nuclear Yap on the Right. (B) Relative mRNA expression of Yap target genes comparing untreated and PP2-treated NICD/p53−/− organoids. (**P ≤ 0.001, ***P ≤ 0.0001.) (C) Western blot for Mapk activation in organoids with and without doxycycline-induced expression of activated YAP (S5A). (D) Tracking of individual NICD/p53−/− organoids over time treated with U0126 (Mek1/2 inhibitor) and SCH772984 (Erk1/2 inhibitor). (E) Concentration-dependent viability of NICD/p53−/− organoids treated with Mek1/2 and Erk1/2 inhibitors (RLU: relative light units).
Menin-Mll1/Mll2 Interaction Mediates H3K4me3, Cell Proliferation, and Viability of NICD/p53−/− Organoids.
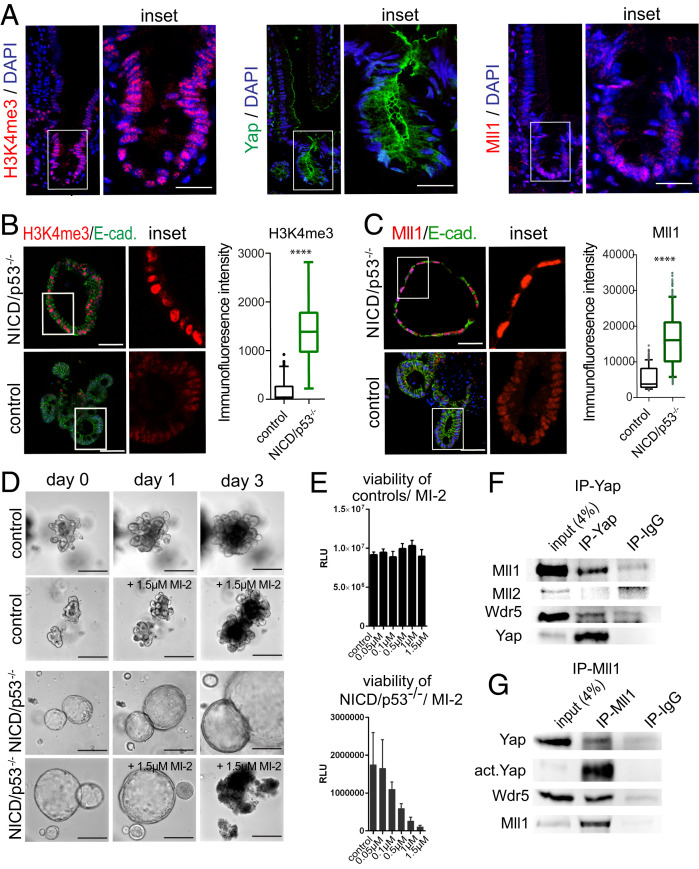
In the intestinal epithelium, Yap promotes transcriptional changes toward a regenerative cell state (8). However, epigenetic factors involved in this process are still to be determined. Elevated H3K4me3 methylation and transcriptional activity has been correlated to Yap (40–42). Since the histone methyltransferase Mll1 has been shown to take part in reprogramming processes (43–45), and we have recently demonstrated a role of Mll1 and H3K4me3 in conferring a cancer stem cell fate in salivary gland and colon tumors (46, 47), we investigated whether Mll1 is involved in the regenerative cell state of NICD/p53−/− organoids. In the small intestinal epithelium we detected pronounced H3K4me3 in the crypt cell compartment (Fig. 4 A, Left). A subset of crypt cells showed nuclear active Yap staining (SI Appendix, Fig. S4A, see arrows), while total Yap levels were increased in all crypt cells, compared to differentiated cells of the villus, in which Yap was predominantly located at the apical cell membrane (Fig. 4 A, Middle). The expression of Mll1 was high in the crypt cell compartment and low in differentiated villus cells (Fig. 4 A, Right). Immunofluorescence for NICD/p53−/− organoids revealed an increase of H3K4me3 levels (Fig. 4 B, Upper pictures, see also enlarged Inset, quantification on the Right) compared to controls (Lower pictures), which suggested an involvement of H3K4 trimethyltransferases in sustaining NICD/p53−/− organoids. We compared Mll1 levels in NICD/p53−/− and control organoids by immunofluorescence and observed a strong increase in Mll1 protein levels in the nuclei of the mutants (Fig. 4 C, Upper) compared to low Mll1 levels in the controls (Fig. 4 C, Lower). Tumors of NICD/p53−/− mice (25) also showed high expression of Mll1 (SI Appendix, Fig. S4B) and high levels of H3K4me3 (SI Appendix, Fig. S4C). Western blotting revealed a strong increase in the protein levels of Mll1 in NICD/p53−/− organoids as well as its homolog Mll2, while Wdr5 and Ash2l, two core components of Mll methyltransferase complexes, were unchanged (SI Appendix, Fig. S4D). However, mRNA levels of Mll1 and Mll2 were not changed in NICD/p53−/− organoids compared to controls (SI Appendix, Fig. S4E). The small molecule MI-2 interferes with the pocket where the scaffold protein Menin binds to Mll1 and Mll2, which impedes their methyltransferase activity (see scheme in SI Appendix, Fig. S4F) (48). MI-2 treatment strongly reduced H3K4me3 in the NICD/p53−/− organoids, and to a much lesser extent H3K4me2 and H3K4me1, as shown by immunofluorescence (SI Appendix, Fig. S4 G–I). Further, MI-2 treatment reduced the proliferation of NICD/p53−/− organoids, as shown by immunofluorescence for Ki67 (SI Appendix, Fig. S4J). Prolonged treatment with MI-2 reduced the viability and induced the collapse of NICD/p53−/− organoids in time- and concentration-dependent manners, while control organoids remained intact (Fig. 4 D and E). These findings indicate that H3K4me3, cell proliferation, and viability of NICD/p53−/− organoids depend on the Menin-Mll1/Mll2 interaction. As the growth and viability of NICD/p53−/− organoids depended on both Yap and Mll1/2, we performed coimmunoprecipitations of endogenous Yap and Mll1, and vice versa, from NICD/p53−/− organoid lysates to address the possibility of interaction. Yap coimmunoprecipitated Mll1 as well as Wdr5, but not Mll2 (Fig. 4F). Mll1 coimmunoprecipitated Yap and its known interacting partner Wdr5 (Fig. 4G) (49). The results reveal that Yap interacts with the histone-methyltransferase complex containing Wdr5 and Mll1.
Fig. 4.
Menin-Mll1/Mll2 interaction mediates H3K4me3, cell proliferation, and viability of NICD/p53−/− organoids. (A) Immunofluorescence staining of small intestinal crypt for H3K4me3 (Left), Yap (Middle), and Mll1 (Right). (Scale bar, 30 µm.) (B) Immunofluorescence staining of NICD/p53−/− (Upper) and control organoids (Lower) for H3K4me3 (red) and E-cadherin (green). (Scale bar, 50 µm.) Magnifications of Insets on the Right. Quantification of staining intensity on the Far Right. (C) Immunofluorescence staining of NICD/p53−/− (Upper) and control organoids (Lower) for Mll1 (red), E-cadherin (green), and DAPI (Scale bar, 50 µm.); Insets are enlarged on the Right. Quantification of staining intensity on the Far Right. (***P ≤ 0.0001.) (D) Brightfield images of individual tracked control (Upper) and NICD/p53−/− organoids (Lower) either untreated or in response to MI-2 treatment. (Scale bar, 250 µm.) (E) Concentration-dependent viability of control (Upper) and NICD/p53−/− organoids (Lower) treated with increasing concentrations of MI-2. (F) Coimmunoprecipitation of endogenous proteins from NICD/p53−/− organoids; Yap coprecipitates with Mll1 and Wdr5, but not with Mll2. (G) Coimmunoprecipitation of endogenous Mll1 with Yap and Wdr5 from lysates of NICD/p53−/− organoids.
YAP and MLL1 Regulate Growth and Viability of Human Patient-Derived Colorectal Cancer Organoids.
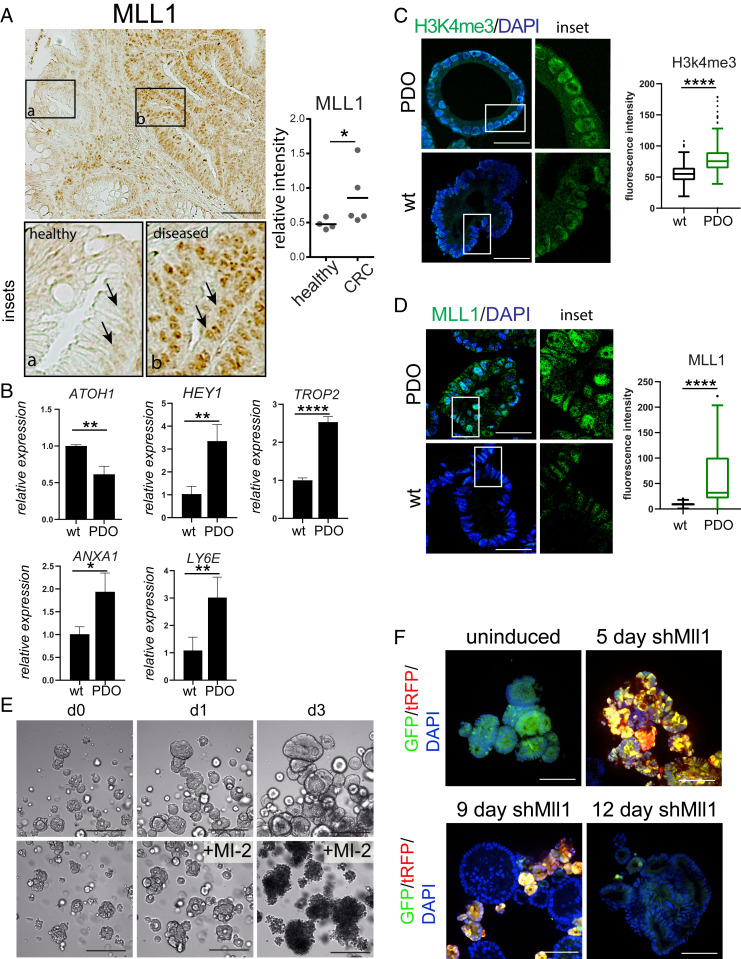
To assess a relevance of YAP and MLL1 in human colon cancer, we analyzed the role of both regulators in tumor biopsies, patient-derived xenografts (PDXs) and patient-derived organoids (PDOs) from human CRC samples (50, 51). We observed nuclear location of YAP and MLL1 in patient-derived xenografts (SI Appendix, Fig. S5 A and B) as well as high levels of MLL1 in tumor biopsies of CRC patients (Fig. 5A, compare Inset b to healthy tissue in Inset a, quantification on the Right). The Cancer Genome Atlas (TCGA) database analysis (at http://www.cbioportal.org) revealed a decreased disease- and progression-free survival of patients with elevated MLL1 levels (SI Appendix, Fig. S5 C and D). Correlation analysis of TCGA colon cancer expression data revealed a coexpression of MLL1 and YAP (SI Appendix, Fig. S5 E and F) as well as of YAP targets (ANKRD1 and CLU), regenerative marker genes (CLU, SPP1, ANXA1, and TROP2/TACSTD2), and Notch targets (HEY1) (SI Appendix, Fig. S5F). We assessed to what extent the patient-derived CRC organoid (PDO) model correlated to the mouse-derived NICD/p53−/− organoids. The analyzed PDOs harbor p53 mutations (52) and indeed remained unresponsive to treatment with Nutlin3a, while the treatment induced cell death in naïve human colon organoids (SI Appendix, Fig. S5 G, Middle). Inhibition of Notch activity with the gamma-secretase inhibitor DAPT did not cause obvious morphological changes (SI Appendix, Fig. S5 G, Right). However, PDOs exhibited higher Notch signaling activity compared to WT organoids, assessed by lower ATOH1 and higher HEY1 expression levels (Fig. 5B). The Notch repressed gene ATOH1 and the Notch suppressed secretory cell-state gene MUC2 (20) were up-regulated in WT organoids but remained unchanged in PDOs upon Notch inhibition (SI Appendix, Fig. S5H). Prolonged inhibition of Notch signaling reduced the growth of the organoids (SI Appendix, Fig. S5I) and reduced the levels of pErk1/2 and activated YAP (SI Appendix, Fig. S5J), demonstrating that Notch activity contributes to YAP activity and promotes MAPK signaling in PDOs. The regenerative genes TROP2 and ANXA1 as well as LY6E also showed higher expression in PDOs (Fig. 5B), which also exhibited higher nuclear YAP (SI Appendix, Fig. S5K) and higher H3K4me3 and MLL1 levels (Fig. 5 C and D), compared to naïve colon organoids. We treated patient-derived organoids with the Yap inhibitor VP and the Menin-Mll1/2 inhibitor MI-2. VP treatment impaired the growth of the human organoids (SI Appendix, Fig. S6A). Treatment with MI-2 for 24 h strongly reduced H3K4me3 (SI Appendix, Fig. S6B), and prolonged MI-2 treatment reduced the growth and caused death of the human organoids (Fig. 5E), while naïve colon organoids survived (SI Appendix, Fig. S6C). We also detected reduced expression of the Yap targets CTGF and CYR61 in PDO upon 24 h of MI-2 treatment (SI Appendix, Fig. S6D). By lentiviral transduction, we introduced a doxycycline-inducible shMLL1 cassette into the human tumor organoids, which upon induction coproduces turboRFP, allowing monitoring of shRNA production (35). Stable integration of the cassette was monitored by GFP expression (Fig. 5F and SI Appendix, Fig. S6E). Noninduced organoids did not express turboRFP (SI Appendix, Fig. S6E). Eight days after MLL1 knockdown organoids producing shMLL1 (green and red) failed to grow (SI Appendix, Fig. S6E). Only organoids incapable of MLL1 knockdown grew (green only) (SI Appendix, Fig. S6E, marked by green arrow). After 12 d of induced shRNA expression, organoid cells with MLL1 knockdown were negatively selected (Fig. 5F, notice the loss of green-red double-positive organoids/cells, SI Appendix, Fig. S6E). Taken together, these results reveal that loss of p53 and activation of Notch signaling promote and maintain a highly proliferative cellular state that resembles the regenerative state and is functionally linked to Yap and MLL1.
Fig. 5.
MLL1 in human colorectal cancer. (A) Immunohistochemistry for MLL1 on CRC patient tissue sections (Scale bar, 100 µm.); comparing healthy (Inset a) and tumor (Inset b) epithelia. Quantification of MLL1 expression in healthy and CRC patient tissues on the Right (n = 5). (B) Relative mRNA expression of Notch and regenerative marker genes. (C) Immunofluorescence for H3K4me3 (green) of patient-derived and WT human organoids (Scale bar, 50 µm); quantification is on the Right. (D) Immunofluorescence for MLL1 (green) of patient-derived and WT human organoids (Scale bar, 50 µm); quantification is on the Right. (C and D) Comparison of two individual organoid lines of each condition and a total of 8 to 10 organoids measured. (E) Brightfield images of untreated (Upper) and 2.5 µM MI-2 treated patient-derived CRC organoids over the indicated time points. (Scale bar, 250 µm.) (F) Lentiviral shRNA knockdown of MLL1 in patient-derived organoids 5 d and 12 d after induction with doxycycline. Infected cells capable of shRNA production express GFP (green) and shRNA-producing cells express turboRFP (red) after doxycycline-induction. (Scale bar, 80 µm.) (*P ≤ 0.05, **P ≤ 0.001, ***P ≤ 0.0001.)
Discussion
Our work shows that combination of Notch activation and p53 ablation induces and locks intestinal organoids in a regenerative state. This cell state is persistent and enables self-renewal and growth, independent of otherwise essential niche factors. The regenerative and niche-factor-independent cell state is only acquired upon combination of Notch activation and loss of p53 and involves activation of Yap, which is required for self-renewal and viability. Yap orchestrates high proliferation by promoting Mapk signaling and elevates H3K4me3 in concert with Mll1. We show that Yap interacts with Mll1 and Wdr5, which suggests a connection of Yap with Mll1-mediated histone methylation.
Intestinal repair programs involve transient increases in nuclear Yap (8, 10, 37) and increased proliferation and expression of genes specific for regenerative epithelia, such as Clu, Anxa-1, and Trop2 (8, 31). These processes occur to induce a highly proliferative epithelium, which is essential for wound healing. During regeneration this is a transient process, while our data here suggest that mutational processes may aberrantly induce and maintain such a state, which could predispose cells to carcinogenesis. We demonstrate that NICD/p53−/− organoids exhibit pronounced activation and nuclear translocation of Yap as well as Mll1 that locks cells in a regenerative and highly proliferative cell state. During regeneration, intestinal epithelial cells are reprogrammed into a primitive state, which encompasses the activation of Yap and suppression of classical adult stem cell genes (8). NICD/p53−/− organoids resemble this and exhibit pronounced induction of regenerative genes and a decreased expression of the classical adult intestinal stem cell genes Ascl2 and Lgr5. In organoids and in vivo the regenerative cell state with high Yap expression is induced by altered signaling, which has been linked to changes in the extracellular matrix (8). The two mutations in NICD/p53−/− organoids are sufficient to induce this regenerative cell state, promote Yap target genes, and constitutively elevate the expression of the regenerative genes Clu, Anxa-1, and Trop2. Clu is expressed by unique cells in the intestinal epithelium, which transiently expand in a Yap1-dependent manner upon tissue injury (8, 31). This Clu+ cell population is subsequently able to replenish Lgr5+ stem cells and to regenerate the epithelium (31). Regeneration involves the reformation of crypt-based stem cell niches, which requires the generation of Notch-active stem cells and Notch-inactive secretory Paneth cells. In a process called symmetry breaking, Yap activity causes expression of Notch ligands, which results in Notch activation in adjacent cells. Such Notch-active cells down-regulate the expression of Notch ligands, which in turn results in lower Notch activity in the neighboring, Yap-active cell. The cells with low Notch activity differentiate into secretory Paneth cells that promote reconstitution of the stem cell niche (53). Of note, Notch signaling prevents secretory cell differentiation (21, 28). Accordingly, the constitutive Notch activity in NICD/p53−/− organoids blocks symmetry breaking and secretory cell differentiation, and cells adopt a spheroid shape and are locked in a Yap-active regenerative state. Furthermore, we discovered that Yap activity results in Erk activation, and thus may promote the Egf-independent growth and resistance to Mek1 inhibition of NICD/p53−/− organoids. Mechanisms of growth factor-independent activation of Erk by Yap has been described involving the AXL receptor kinase (54), as well as Mek-independent Erk activation (55). Moreover, it has been shown that Yap transcriptional activity mediates resistance to MEK1/2 inhibitors (56, 57). Together, Yap and Notch signaling activity control organoid growth and maintenance of the regenerative cell state.
A remarkable finding of our study is the crucial role of Mll1 and H3K4 trimethylation in the regenerative NICD/p53−/− organoids. Mll1 has been demonstrated to take part in reprogramming processes (43–45), changes in histone modifications such as H3K4me3 occur during cell reprogramming, and reprogramming factors recruit core components of Mll histone methyltransferase complexes like Wdr5 (58). In addition, the reprogramming factor Yap has been implicated in chromatin remodeling (59). We here find that Yap interacts with Wdr5 and Mll1 in NICD/p53−/− organoids, which suggests a link between Yap-induced reprogramming and Mll1 activity in the regenerative intestinal epithelium. Our data reveal Mll1 as an epigenetic factor that is involved in Yap-dependent reprogramming into a fetal-like cell state. Whether Mll1 participates in the control of symmetry breaking in cell specification is an interesting revelation for future research. We found that the high nuclear levels of Yap in NICD/p53−/− organoids were dependent on active Src, which supports the finding of integrin-regulated and Src-dependent Yap activation in cell reprogramming during regeneration (8). In hematopoiesis, integrin signaling is induced by Mll1 (60). These data support the notion of a signaling cascade of Mll1, integrin/Src, and Yap in NICD/p53−/− cells.
In colorectal tumors, Notch1 has been shown to characterize a subset of cancer stem cells that are undifferentiated, proliferative, and self-renewing, but lack expression of the characterized cancer stem cell markers such as Lgr5 (61). Such Notch1+ cancer stem cells were not analyzed for Yap activity. However, given similar properties of the Notch1+ cancer stem cells and our NICD/p53−/− organoids, coherent combination with our observation that Notch activity promotes Yap in patient-derived organoids supposes that Yap activity might promote the Notch-active cancer stem cell.
While the role of Yap in cancer is well established (15), the implication of Mll1 and histone modifiers in colon cancer is an emerging field of research (47). Recent experiments in cell cultures and in xenografted tumor cells showed that Mll1 is crucial in solid cancer cells (62–64). We had previously shown that genetic and pharmacological inhibition of Mll1 in mouse salivary gland, human head and neck cancer, and a Wnt-dependent intestinal cancer model prevented tumor formation (46, 47). In leukemia, inhibition of Mll1 and other chromatin modifiers is effective as a treatment option (65, 66). We here show that treatment of mouse and human intestinal tumor organoids with the small molecule Menin-Mll1 inhibitor MI-2 strongly decreases H3K4me3, cell proliferation, and organoid viability. Cells of human colon cancer organoids with induced knockdown of Mll1 were negatively selected, which further indicates the crucial role of Mll1 in sustaining these cancer cells. Our study suggests that the Menin-Mll1 complex is a key regulatory unit in intestinal cancer and proposes future investigations into Mll1 as a novel therapeutic target in colorectal cancer.
Altogether, our study points to a crucial role of Notch, Yap, and the Mll1/Wdr5 complex in intestinal tumorigenesis and regeneration. The data suggest that constitutive activation of Notch in p53-deficient cells promotes Yap and Mll1, reprograms cells into a regenerative state, induces niche-factor-independent growth, and—if persistent—renders cells susceptible to tumorigenesis.
Materials and Methods
See SI Appendix, Supplementary Methods for additional details.
Organoid Culture.
Organoids were generated from small intestine of VillinCreERT2; NICDflox/flox; p53flox/flox mice (25), cultured in small intestinal organoid media, and treated with the indicated compounds. Mutagenesis was induced in culture 4-OHT (details in SI Appendix, Supplementary Methods).
Colorectal Cancer Samples and Patient-Derived Cancer Organoid.
Analysis of human colon material was approved by the local Institutional Review Board of Charité University Medicine (Charité Ethics, 10117 Berlin, Germany) (EA 1/069/11 and EA2/008/18) and the ethics committee of the Medical University of Graz (Ethics Commission of the Medical University of Graz, 8036 Graz, Austria), confirmed by the ethics committee of the St. John of God Hospital Graz (23-015 ex 10/11). Experiments conformed to the World Medical Association Declaration of Helsinki and the Department of Health and Human Services Belmont Report. Obtained samples were deidentified before preparation and analysis in the laboratory. Cancer organoid and naïve WT colon organoidcultures were established and propagated as described before (51, 67).
Generation of Lentiviral Particles.
For doxycycline-inducible shRNA knockdown of Yap and Mll1 in combination with a fluorescent reporter the pInducer tool kit vectors were used (35); lentiviral particles were produced to infect organoids (details in SI Appendix, Supplementary Methods).
Histology, Immunohistochemistry, and Light Electron Microscopy.
Immunohistochemistry was performed on formaldehyde-fixed and paraffin-embedded sections. Images were taken with a DIM6000 (Leica), LSM710 (Zeiss), and CSU-W1 (Nikon). Images were analyzed with Fiji and Imaris 8 (Bitplane/Andor) software. For electron microscopy, ultrathin sections of fixed organoids were stained with uranyl acetate and lead citrate, and examined at 80 kV with a Morgagni electron microscope (details in SI Appendix, Supplementary Methods).
Western Blots and Coimmunopreciptiation.
See SI Appendix, Supplementary Methods for details.
qRT-PCR and RNA Sequencing.
Total RNA of organoids was isolated using TRIzol extraction (Invitrogen) and purified via phenol/chloroform extraction. RNA was reverse transcribed with random hexamer primers (Invitrogen) and MMLV Reverse Transcriptase (Promega, 200 U/μL), following the manufacturer’s instructions. For quantitative reverse transcription, PCR was performed in a CFX96-C1000T thermal cycler (Bio-Rad) or RNA was further processed for mRNA sequencing (details in SI Appendix, Supplementary Methods).
Supplementary Material
Acknowledgments
We thank Matthias Richter and Konstantin Grohmann from the Advanced Light Microscopy unit of MDC for support with image acquisition and data analysis, Marcel Harrig for great reliability maintaining the mouse colony, and Daniele Franze for uploading the RNA-sequencing data. This work was supported by MDC and Charité University Medicine central resources and DFG grants Si-1983/3-1 and Si-1983/4-1 (to M.S.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2019699118/-/DCSupplemental.
Data Availability
RNA-sequencing data have been deposited in ExpressArray (http://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-6588). All study data are included in the article and/or supporting information.
References
- 1.van der Flier L. G., Clevers H., Stem cells, self-renewal, and differentiation in the intestinal epithelium. Annu. Rev. Physiol. 71, 241–260(2009). [DOI] [PubMed] [Google Scholar]
- 2.Tetteh P. W., et al., Replacement of lost Lgr5-positive stem cells through plasticity of their enterocyte-lineage daughters. Cell Stem Cell 18, 203–213(2016). [DOI] [PubMed] [Google Scholar]
- 3.Buczacki S. J., et al., Intestinal label-retaining cells are secretory precursors expressing Lgr5. Nature 495, 65–69(2013). [DOI] [PubMed] [Google Scholar]
- 4.van Es J. H., et al., Dll1+ secretory progenitor cells revert to stem cells upon crypt damage. Nat. Cell Biol. 14, 1099–1104(2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yan K. S., et al., Intestinal enteroendocrine lineage cells possess homeostatic and injury-inducible stem cell activity. Cell Stem Cell 21, 78–90.e6(2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harnack C., et al., R-spondin 3 promotes stem cell recovery and epithelial regeneration in the colon. Nat. Commun. 10, 4368(2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beachy P. A., Karhadkar S. S., Berman D. M., Tissue repair and stem cell renewal in carcinogenesis. Nature 432, 324–331(2004). [DOI] [PubMed] [Google Scholar]
- 8.Yui S., et al., YAP/TAZ-dependent reprogramming of colonic epithelium links ECM remodeling to tissue regeneration. Cell Stem Cell 22, 35–49.e7(2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gregorieff A., Liu Y., Inanlou M. R., Khomchuk Y., Wrana J. L., Yap-dependent reprogramming of Lgr5(+) stem cells drives intestinal regeneration and cancer. Nature 526, 715–718(2015). [DOI] [PubMed] [Google Scholar]
- 10.Cai J., et al., The Hippo signaling pathway restricts the oncogenic potential of an intestinal regeneration program. Genes Dev. 24, 2383–2388(2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Imajo M., Ebisuya M., Nishida E., Dual role of YAP and TAZ in renewal of the intestinal epithelium. Nat. Cell Biol. 17, 7–19(2015). [DOI] [PubMed] [Google Scholar]
- 12.Zhou D., et al., Mst1 and Mst2 protein kinases restrain intestinal stem cell proliferation and colonic tumorigenesis by inhibition of Yes-associated protein (Yap) overabundance. Proc. Natl. Acad. Sci. U.S.A. 108, E1312–E1320(2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moroishi T., Hansen C. G., Guan K. L., The emerging roles of YAP and TAZ in cancer. Nat. Rev. Cancer 15, 73–79(2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Steinhardt A. A., et al., Expression of Yes-associated protein in common solid tumors. Hum. Pathol. 39, 1582–1589(2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zanconato F., Cordenonsi M., Piccolo S., YAP/TAZ at the roots of cancer. Cancer Cell 29, 783–803(2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Piccolo S., Dupont S., Cordenonsi M., The biology of YAP/TAZ: Hippo signaling and beyond. Physiol. Rev. 94, 1287–1312(2014). [DOI] [PubMed] [Google Scholar]
- 17.Camargo F. D., et al., YAP1 increases organ size and expands undifferentiated progenitor cells. Curr. Biol. 17, 2054–2060(2007). [DOI] [PubMed] [Google Scholar]
- 18.Okamoto R., et al., Requirement of Notch activation during regeneration of the intestinal epithelia. Am. J. Physiol. Gastrointest. Liver Physiol. 296, G23–G35(2009). [DOI] [PubMed] [Google Scholar]
- 19.Carulli A. J., et al., Notch receptor regulation of intestinal stem cell homeostasis and crypt regeneration. Dev. Biol. 402, 98–108(2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Es J. H., et al., Notch/gamma-secretase inhibition turns proliferative cells in intestinal crypts and adenomas into goblet cells. Nature 435, 959–963(2005). [DOI] [PubMed] [Google Scholar]
- 21.Shroyer N. F., et al., Intestine-specific ablation of mouse atonal homolog 1 (Math1) reveals a role in cellular homeostasis. Gastroenterology 132, 2478–2488(2007). [DOI] [PubMed] [Google Scholar]
- 22.Bu P., et al., A microRNA miR-34a-regulated bimodal switch targets Notch in colon cancer stem cells. Cell Stem Cell 12, 602–615(2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sikandar S. S., et al., NOTCH signaling is required for formation and self-renewal of tumor-initiating cells and for repression of secretory cell differentiation in colon cancer. Cancer Res. 70, 1469–1478(2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bray S. J., Notch signalling in context. Nat. Rev. Mol. Cell Biol. 17, 722–735(2016). [DOI] [PubMed] [Google Scholar]
- 25.Chanrion M., et al., Concomitant Notch activation and p53 deletion trigger epithelial-to-mesenchymal transition and metastasis in mouse gut. Nat. Commun. 5, 5005(2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clevers H., Modeling development and disease with organoids. Cell 165, 1586–1597(2016). [DOI] [PubMed] [Google Scholar]
- 27.Sato T., et al., Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459, 262–265(2009). [DOI] [PubMed] [Google Scholar]
- 28.Yang Q., Bermingham N. A., Finegold M. J., Zoghbi H. Y., Requirement of Math1 for secretory cell lineage commitment in the mouse intestine. Science 294, 2155–2158(2001). [DOI] [PubMed] [Google Scholar]
- 29.Haber A. L., et al., A single-cell survey of the small intestinal epithelium. Nature 551, 333–339(2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Muñoz J., et al., The Lgr5 intestinal stem cell signature: Robust expression of proposed quiescent '+4′ cell markers. EMBO J. 31, 3079–3091(2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ayyaz A., et al., Single-cell transcriptomes of the regenerating intestine reveal a revival stem cell. Nature 569, 121–125(2019). [DOI] [PubMed] [Google Scholar]
- 32.Mustata R. C., et al., Identification of Lgr5-independent spheroid-generating progenitors of the mouse fetal intestinal epithelium. Cell Rep. 5, 421–432(2013). [DOI] [PubMed] [Google Scholar]
- 33.von Eyss B., et al., A MYC-driven change in mitochondrial dynamics limits YAP/TAZ function in mammary epithelial cells and breast cancer. Cancer Cell 28, 743–757(2015). [DOI] [PubMed] [Google Scholar]
- 34.Liu-Chittenden Y., et al., Genetic and pharmacological disruption of the TEAD-YAP complex suppresses the oncogenic activity of YAP. Genes Dev. 26, 1300–1305(2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meerbrey K. L., et al., The pINDUCER lentiviral toolkit for inducible RNA interference in vitro and in vivo. Proc. Natl. Acad. Sci. U.S.A. 108, 3665–3670(2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wörmann S. M., et al., Loss of P53 function activates JAK2-STAT3 signaling to promote pancreatic tumor growth, stroma modification, and gemcitabine resistance in mice and is associated with patient survival. Gastroenterology 151, 180–193.e12(2016). [DOI] [PubMed] [Google Scholar]
- 37.Taniguchi K., et al., A gp130-Src-YAP module links inflammation to epithelial regeneration. Nature 519, 57–62(2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Azare J., et al., Constitutively activated Stat3 induces tumorigenesis and enhances cell motility of prostate epithelial cells through integrin beta 6. Mol. Cell. Biol. 27, 4444–4453(2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu Z., et al., MAPK-mediated YAP activation controls mechanical-tension-induced pulmonary alveolar regeneration. Cell Rep. 16, 1810–1819(2016). [DOI] [PubMed] [Google Scholar]
- 40.Oh H., et al., Yorkie promotes transcription by recruiting a histone methyltransferase complex. Cell Rep. 8, 449–459(2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Croci O., et al., Transcriptional integration of mitogenic and mechanical signals by Myc and YAP. Genes Dev. 31, 2017–2022(2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oh H., et al., Genome-wide association of Yorkie with chromatin and chromatin-remodeling complexes. Cell Rep. 3, 309–318(2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Muller S., Nayak A., Inhibition of MLL1 histone methyltransferase brings the developmental clock back to naïve pluripotency. Stem Cell Investig. 3, 58(2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang Q., et al., Reprogramming of the epigenome by MLL1 links early-life environmental exposures to prostate cancer risk. Mol. Endocrinol. 30, 856–871(2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu L., et al., Targeting Mll1 H3K4 methyltransferase activity to guide cardiac lineage specific reprogramming of fibroblasts. Cell Discov. 2, 16036(2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhu Q., et al., The Wnt-driven Mll1 epigenome regulates salivary gland and head and neck cancer. Cell Rep. 26, 415–428.e5(2019). [DOI] [PubMed] [Google Scholar]
- 47.Grinat J., et al., The epigenetic regulator Mll1 is required for Wnt-driven intestinal tumorigenesis and cancer stemness. Nat. Commun. 11, 6422(2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grembecka J., et al., Menin-MLL inhibitors reverse oncogenic activity of MLL fusion proteins in leukemia. Nat. Chem. Biol. 8, 277–284(2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Song J. J., Kingston R. E., WDR5 interacts with mixed lineage leukemia (MLL) protein via the histone H3-binding pocket. J. Biol. Chem. 283, 35258–35264(2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Boehnke K., et al., Assay establishment and validation of a high-throughput Screening platform for three-dimensional patient-derived colon cancer organoid cultures. J. Biomol. Screen. 21, 931–941(2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schütte M., et al., Molecular dissection of colorectal cancer in pre-clinical models identifies biomarkers predicting sensitivity to EGFR inhibitors. Nat. Commun. 8, 14262(2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schumacher D., et al., Heterogeneous pathway activation and drug response modelled in colorectal-tumor-derived 3D cultures. PLoS Genet. 15, e1008076(2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Serra D., et al., Self-organization and symmetry breaking in intestinal organoid development. Nature 569, 66–72(2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu M. Z., et al., AXL receptor kinase is a mediator of YAP-dependent oncogenic functions in hepatocellular carcinoma. Oncogene 30, 1229–1240(2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jiang C. C., et al., MEK-independent survival of B-RAFV600E melanoma cells selected for resistance to apoptosis induced by the RAF inhibitor PLX4720. Clin. Cancer Res. 17, 721–730(2011). [DOI] [PubMed] [Google Scholar]
- 56.Lin L., et al., The Hippo effector YAP promotes resistance to RAF- and MEK-targeted cancer therapies. Nat. Genet. 47, 250–256(2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Coggins G. E., et al., YAP1 mediates resistance to MEK1/2 inhibition in neuroblastomas with hyperactivated RAS signaling. Cancer Res. 79, 6204–6214(2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Qin H., Zhao A., Zhang C., Fu X., Epigenetic control of reprogramming and transdifferentiation by histone modifications. Stem Cell Rev. Rep. 12, 708–720(2016). [DOI] [PubMed] [Google Scholar]
- 59.Hillmer R. E., Link B. A., The roles of hippo signaling transducers Yap and Taz in chromatin remodeling. Cells 8, E502(2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang W., et al., Enhancing hematopoiesis from murine embryonic stem cells through MLL1-induced activation of a rac/rho/integrin signaling Axis. Stem Cell Reports 14, 285–299(2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mourao L., et al., Lineage tracing of Notch1-expressing cells in intestinal tumours reveals a distinct population of cancer stem cells. Sci. Rep. 9, 888(2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ansari K. I., Kasiri S., Mandal S. S., Histone methylase MLL1 has critical roles in tumor growth and angiogenesis and its knockdown suppresses tumor growth in vivo. Oncogene 32, 3359–3370(2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gallo M., et al., A tumorigenic MLL-homeobox network in human glioblastoma stem cells. Cancer Res. 73, 417–427(2013). [DOI] [PubMed] [Google Scholar]
- 64.Malik R., et al., Targeting the MLL complex in castration-resistant prostate cancer. Nat. Med. 21, 344–352(2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Borkin D., et al., Pharmacologic inhibition of the Menin-MLL interaction blocks progression of MLL leukemia in vivo. Cancer Cell 27, 589–602(2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dafflon C., et al., Complementary activities of DOT1L and Menin inhibitors in MLL-rearranged leukemia. Leukemia 31, 1269–1277(2017). [DOI] [PubMed] [Google Scholar]
- 67.Heuberger J., et al., Epithelial response to IFN-γ promotes SARS-CoV-2 infection. EMBO Mol. Med. 13, e13191(2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-sequencing data have been deposited in ExpressArray (http://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-6588). All study data are included in the article and/or supporting information.