Abstract
Pleistocene climatic oscillations have greatly influenced the evolutionary histories and distribution patterns of most extant species. In this study, the phylogeographic patterns and evolutionary history of Triplophysa stenura were investigated. A total of 359 individuals from 19 locations covering the species’ distribution range were collected, and two mitochondrial genes (COI and Cyt b) and the recombination activating protein 1 gene (Rag1) were analyzed. Two deeply divergent lineages, corresponding geographically to the northern and southern Tanggula Mountain, were observed, and shallow divergences were found within the southern and central Qinghai-Tibetan plateau (QTP). The estimated divergence time between the northern and southern Tanggula lineages was around 1.07 Mya. Within the southern Tanggula Mountain lineages, the Nu and Subansiri rivers populations were split about 0.74 Mya, and the southwestern and central QTP populations were divided with the southeastern QTP ones approximately 0.67 Mya. The divergence times of the lineages were matched with two major glaciations in QTP (the Xixiabangma Glaciation, 1.17–0.80 Mya and Kunlun Glaciation, 0.72–0.62 Mya). Together with demographic history analyses, our results highlighted that major glacial cycles during the mid-Pleistocene played a major role in sculpting the distribution pattern of T. stenura, and led to the gene homogenization crossing the drainage populations in the southwestern and central QTP.
Keywords: The Qinghai-Tibetan plateau, Phylogeography, Climate oscillation, Pleistocene glacial cycles, Triplophysa stenura
BACKGROUND
The Qinghai-Tibetan Plateau (QTP), the highest and largest plateau in the world, is the greatest glaciated track outside the polar region in earth, with a total glacial area of ~100,000 km2 (Lehmkuhl and Owen 2005; Yao et al. 2012). The glaciation throughout eastern QTP is strongly controlled by precipitation changes related to oscillation in the South Asian monsoonal system combined with cooling events (Lehmkuhl and Owen 2005). However, the influence of the South Asian monsoon is weakened significantly across eastern mountain belt, leading to a sharp decrease in precipitation from east to west. Thus, the eastern mountainous margins of the plateau are much more favorable for glacier development compared to the central and western parts (Dong et al. 2010).
The climatic perturbations during the Quaternary have had a profound impact on the distribution pattern and have been ascribed a dominant role in shaping the distribution and genetic attribute of species in the north hemisphere (Hewitt 2000). Species went extinct in situ, survived in refugia or dispersed to new locations during periods of unsuitable climate and then underwent range expansions and colonized new habitations during periods of favorable conditions. Such repeated changes must have left imprints in the evolutionary history of these species (Hewitt 2000 2004; Yang et al. 2008; Wang et al. 2010; He et al. 2016).
Phylogeographic analysis, especially the estimation of divergence time, provides tremendous insight into the patterns and underlying causes of the historical diversification of lineages and is a powerful tool for reconstructing the histories of living species (Funk and Wagner 1995; Avise et al. 2000; Wiens and Donoghue 2004; Lemey et al. 2009; Dincă et al. 2011). Recently, phylogeographic analyses showed that distribution and genetic structure of various species in QTP and its adjacent areas were affected by the climatic oscillations or geological barriers (Xia et al. 2018; Ye et al. 2018; Rana et al. 2019). However, the degrees to which geographic barriers affected for the population structure were depended on several factors such as dispersal ability and habitat preference (Qu et al. 2010; Fan et al. 2012; Xu et al. 2014). The highly migratory species (e.g., birds) were likely to be less affect by geographic barriers compared to less motile species (Zhao et al. 2019). Unlike that of terrestrial animals, the dispersal of freshwater fishes depended on direct connections between drainage basins (Roach et al. 2009; De Macedo‐Soares et al. 2010), and separation among drainage basins caused via the terrestrial environment probably led to the intra-species subdivision of freshwater fishes. However, glacial meltwater during the interglacial period triggered the erosion and incision of river systems which probably prompted species and populations exchanges, while expanded ice during the glacial period may have promoted vicariance due to the form of geographic barriers (e.g., glacier), both of which profoundly influenced the distribution and their genetic structure of cold-adapted fish species. Hence, the freshwater fish are likely serve as good candidates for phylogeographic studies in QTP, allowing examination of how broad-scale historical climatic events have influenced their distribution and genetic structure.
Earth’s climate underwent a fundamental change between c. 1.25 and 0.70 Mya, marked by a shift of previously dominant 41 kya cycles to lately dominant 100 kya cycles. At the same time, the amplitude of glacial-interglacial climate variations became larger (Elderfield et al. 2012). This fundamental shift, known as the mid-Pleistocene climate transition (MPT), is one of an enigma in the Quaternary climate evolution. This climatic transition also arose in QTP, when the plateau entered cryosphere, and caused the appearance of the oldest glaciation (the Xixiabangma Glaciation, 1.17–0.8 Mya) and maximum glaciation (Kunlun Glaciation, 0.72–0.62 Mya) in QTP during the Quaternary.
The Tibetan stone loaches, genus Triplophysa, is one of three principal components of ichthyofauna in QTP, and widely distribute in QTP and its surrounding areas (He et al. 2020). Triplophysa stenura (Nemacheilidae) is a small loach endemic to QTP and adjacent areas (Zhu 1989). It resides in the swift-flowing streams throughout the exorheic rivers in southeast and west QTP and endorheic drainages in north QTP (e.g., tributaries of Kongyu Tso, Tarok Tso and Selin Tso) as well as the Himalayas (e.g., tributaries of Paiku Tso and Dochen Tso). Due to its adaptation to high altitude and widely distribution, T. stenura is tolerant of the complex QTP environment, which makes it an ideal model organism for verifying the effect of QTP’s climate change on the genetic structure. However, prior to this study, genetic surveys of T. stenura was confined to the east QTP and small samples (Ren et al. 2018). In this study, we analyzed the phylogeographic structure and demographic history of T. stenura in QTP and its surrounding areas used three gene segments (COI, Cyt b and Rag1). We collected 359 individuals from 19 locations across the five river basins which almost covered the range of the species (Fig. 1). Thus, the evolutionary history of the species could thoroughly reflect the historic climate events in this region. The aims of this study are to (i) investigate the genetic diversity, phylogeographic structure and demographic history of T. stenura, and (ii) explore the underlying causes of the phylogeographical pattern concerning the mid-Pleistocene climatic oscillation in QTP.
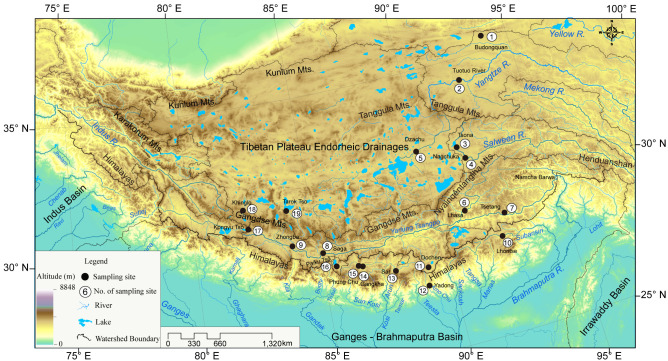
Fig. 1.
Map of the study area and geographical distribution of sampling sites in this study. The sites are numbered according to table 1.
MATERIALS AND METHODS
Ethics statement
The methods involving animals in this study were conducted in accordance with the Laboratory Animal Management Principles of China. All experimental protocols were approved by the Ethics Committee of the Institute of Hydrobiology, Chinese Academy of Sciences.
Sampling
Specimens were sampled via cast nets, hand nets and shrimp coops (Table 1). Specimens were stored in 8% formaldehyde solution, and were identified according Zhu (1989). Fin clips were obtained and persevered in 95% ethanol (-20°C).
Table 1.
Collection localities, drainages, sample size (N), numbers of haplotypes (H), and haplotype diversity (h) and nucleotide diversity (π) of the sampled populations of T. stenura in the Qinghai-Tibetan Plateau. Tajima’s D values and Fu’s Fs values shows in six different drainages. The single asterisk (*) for p ≤ 0.05; double asterisks (**) for p ≤ 0.01
| Drainage | Localities | N | H | h | π | Longitude | Latitude | Altitude | Tajima’ D | Fu’Fs | |
| upper Yangtze | 40 | 29 | 0.98 ± 0.00008 | 0.0036 ± 0.00535 | -1.2 | -13.0** | |||||
| 1 | Budongquan | 9 | 8 | 0.97 ± 0.00409 | 0.0033 ± 0.00411 | 93.97 | 35.51 | 4597 | |||
| 2 | Tuotuo River | 31 | 26 | 0.98 ± 0.00012 | 0.0038 ± 0.00539 | 92.43 | 34.21 | 4542 | |||
| Nu River | 47 | 39 | 0.97 ± 0.00021 | 0.0026 ± 0.00443 | -1.2* | -21.7** | |||||
| 3 | Tsona | 16 | 15 | 0.99 ± 0.00008 | 0.0024 ± 0.00347 | 90.87 | 32.03 | 4730 | |||
| 4 | Nagchuka | 14 | 14 | 1.00 ± 0.00073 | 0.0021 ± 0.00238 | 91.55 | 31.5 | 4532 | |||
| 5 | Dza Chu | 17 | 11 | 0.84 ± 0.00785 | 0.0012 ± 0.0021 | 89.84 | 32.13 | 4610 | |||
| Yarlung Tsangpo | 125 | 89 | 0.98 ± 0.00003 | 0.0056 ± 0.00870 | -1.9** | -33.9** | |||||
| 6 | Lhasa | 58 | 41 | 0.97 ± 0.00008 | 0.0030 ± 0.00422 | 91.15 | 29.64 | 3670 | |||
| 7 | Tsetang | 19 | 16 | 0.98 ± 0.00050 | 0.0036 ± 0.00434 | 92.68 | 29.19 | 3538 | |||
| 8 | Saga | 15 | 14 | 0.99 ± 0.00079 | 0.0044 ± 0.00515 | 85.15 | 29.32 | 4474 | |||
| 9 | Zhongba | 33 | 30 | 0.99 ± 0.00011 | 0.0024 ± 0.00550 | 83.98 | 29.76 | 4570 | |||
| Brahmaputra | 53 | 35 | 0.97 ± 0.00016 | 0.0024 ± 0.00439 | -1.54 | -0.09** | |||||
| 11 | Dochen | 22 | 21 | 0.99 ± 0.00023 | 0.0035 ± 0.00427 | 89.16 | 28.03 | 4384 | |||
| 12 | Yadong | 31 | 15 | 0.92 ± 0.00069 | 0.0010 ± 0.00150 | 89.03 | 27.39 | 4111 | |||
| Subansiri River | 33 | 13 | 0.72 ± 0.00633 | 0.0005 ± 0.00128 | -2.1** | -6.9** | |||||
| 10 | Lhontse | 33 | 13 | 0.72 ± 0.00633 | 0.0005 ± 0.00128 | 92.34 | 28.41 | 3940 | |||
| upper Ganges | 26 | 17 | 0.96 ± 0.00047 | 0.0015 ± 0.00282 | -0.53 | -7.3** | |||||
| 13 | Sar | 4 | 3 | 0.83 ± 0.04948 | 0.00040 ± 0.0004 | 87.87 | 28.17 | 4275 | |||
| 14 | Gangkha | 8 | 6 | 0.93 ± 0.00711 | 0.0015 ± 0.00200 | 86.61 | 28.58 | 4300 | |||
| 15 | Phung Chu | 12 | 12 | 1.00 ± 0.00160 | 0.0019 ± 0.00277 | 86.45 | 28.63 | 4362 | |||
| 16 | Paiku Tso | 2 | 2 | 1.00 ± 0.25000 | 0.0008 ± 0.00080 | 85.59 | 28.77 | 4592 | |||
| Endorheic drainages | 35 | 23 | 0.95 ± 0.00044 | 0.0018 ± 0.00310 | -1.84** | -5.5** | |||||
| 17 | Kongyu Tso | 4 | 4 | 1.00 ± 0.03125 | 0.0025 ± 0.00239 | 82.3 | 30.61 | 4924 | |||
| 18 | Khinglo | 28 | 16 | 0.93 ± 0.00091 | 0.0009 ± 0.00133 | 82.21 | 31.31 | 4922 | |||
| 19 | Tarok Tso | 3 | 3 | 1.00 ± 0.07407 | 0.0008 ± 0.00080 | 83.98 | 31.05 | 4572 |
Total DNA extraction and sequencing
Total genomic DNA was extracted using the standard phenol-chloroform method (Maniatis et al. 1982). Fragments of two mitochondrial genes (cytochrome b, Cyt b; cytochrome oxidase subunit 1, COI) and one nuclear gene (recombination activating protein 1, Rag1) were amplified and sequenced based on He and Chen (2006), except that annealing temperature was 57°C for Rag1. The corresponding primer sets are given in table S1.
Molecular diversity and phylogenetic analyses
Sequences were checked with chromatograms using visually in BioEdit 7.0 (Hall et al. 2011) after alignment with Clustal X 2.0 (Larkin et al. 2007). The haplotypes were identified using DnaSP 6.0 (Rozas et al. 2017) and have been deposited in the GenBank library under the Accession Nos MT681283-681640. Haplotype diversity (h), nucleotide diversity (π) were estimated in DnaSP 6.0 (Rozas et al. 2017). The TCS network (Clement et al. 2000) was constructed using PopART based on the haplotypes of Cyt b (Leigh and Bryant 2015).
A spatial analysis of molecular variance was conducted using the spatial analysis of molecular variation (SAMOVA) software with the mitochondrial gene fragments (COI + Cyt b). The SAMOVA takes into consideration geographic locations of sampling, and maximizes the proportion of genetic variance among K groups of populations (Dupanloup et al. 2002). As such, this analysis was useful for statistically differentiating between historically isolated groups in the network. SAMOVA was run with several different group structures to determine the maximum value for FCT (the maximized proportion of total genetic variance due to differences between groups).
Phylogenetic relationships were reconstructed under maximum likelihood (ML) and Bayesian inference (BI) in CIPRES (Miller et al. 2010) using concatenated sequence data and two other Triplophysa species, T. brevicauda and T. stewarti, were chosen as outgroups (GenBank accession nos. MW086881-MW086892). We used PartitionFinder 2.1.1 to identify the appropriate model and partition scheme under the Akaike information criterion (AIC) (Lanfear et al. 2017), which divided our data into two partitions (subset1 = mtDNA; subset2 = Rag1).
Both ML and BI were conducted with two partitions scheme and GTR + G + I substitute model. For ML analysis, a majority consensus tree was obtained with 1000 pseudoreplicates in RAxML 8.2.4 (Stamatakis 2014). For BI, two independent runs with four Metropolis-coupled Monte-Carlo Markov Chains (MCMC) were simultaneously conducted with 3 million generations, and 1/1000 sample frequency in MrBayes 3.2 (Ronquist et al. 2012). The first trees with a standard deviation of split frequencies above 0.01 were discarded as the burn-in phase. Nodal support was assessed by calculating the mean posterior probability (BP) values of each node in the resulting consensus tree after burn-in. Phylogenetic tree visualization was performed in FigTree 1.4 (Rambaut 2012).
Divergence time and demographic history analyses
The divergence times (Common Ancestor heights) with 95% highest posterior density (HPD) among lineages were tentatively estimated based the mitochondrial gene fragments using BEAST 2.4.7 (Bouckaert et al. 2014). To our knowledge, there is no well-dated calibration substitute rate for Tibetan stone loaches. The substitute rates of mtDNA protein coding genes for nemacheilian loaches were usually adopted the evolutionary rate of the European cobitids (0.34% to 0.42% substitution rate per million years) (Zhang et al. 2017) or cyprinids (1.05% substitution rate per million years) (Ren et al. 2018). However, the substitute rates may be differences depending on that of substitution models, taxa and molecular markers; thus they may not all be exactly comparable (He and Chen 2007). Here, we first recovered the time-calibrated phylogeny of Triplophysa and other loaches in China: The substitution rate was estimated by three calibration points for Triplophysa and other loaches using three calibration points: (1) the crown group of Cobitidae and Balitoridae were dated to 27.82–100.5 Mya (Chen et al. 2015); (2) the crown group of Botiidae was prior to 5.33–27.82 Mya; and (3) the crown group of genus Triplophysa was corrected to 7.25–27.82 Mya (Prokofiev 2007). We then adopted the lineage substitution rate of T. stenura (0.61% per million years), this substitution rate was slow that of Schizothoracins in QTP, but rapid that of European cobitids (Doadrio and Perdices 2005). The substitution model was set GTR + G + I and a strict clock was used. The simulation was run for 60 million generations and sampling every 1,000 generations, and convergence was evaluated in Tracer 1.7.1 (Rambaut et al. 2018). The time tree was gained after 25% burn-in in TreeAnnotator v 2.4.7. Time tree visualization was performed in FigTree.
Two statistical tests, Tajima’s D (Tajima, 1989) and Fu’s Fs (Fu, 1997), were calculated by two mitochondrial genes (COI, Cyt b) fragments with 10,000 pseudoreplicates in DnaSP 6.0 (Rozas et al. 2017). The significance of both values was calculated from 10000 reted as signatures of population expansion. In addition, we performed a mismatch distribution analysis in DnaSP to investigate population expansion signatures. A multimodal distribution of differences between haplotypes is usually found in samples drawn from populations at demographic equilibrium, whereas the distribution is usually unimodal in populations having undergone recent demographic expansion (Excoffier 2004). To estimate past population sizes, Bayesian Skyline Plot (BSP) was used with a strict molecular clock and 20 million generations in BEAST2.4.7. The substitution model and substitution rate were identical with divergence time analysis. ESS and trace plot were checked ensuring the Skyline parameters for acceptable effective sample sizes values (i.e., > 100). The trend plot of fluctuation of population size were reconstructed using 500 points in Tracer 1.7.1.
RESULTS
Molecular diversity
From 359 individuals, 51 haplotypes of COI (666 bp), 147 haplotypes of Cyt b (1140 bp), 61 haplotypes of Rag1 (701 bp), 168 haplotypes of the mitochondrial genes (1806 bp), and 241 haplotypes of the concatenated gene (2507 bp) were identified. Estimates of nucleotide diversity and haplotype diversity for each sampled site and drainage basins are given in table 1. Haplotype diversity averaged over all populations was 0.99 with a range from 0.72 (0.72 ± 0.00633; Lhontse) to 0.99 (0.99 ± 0.00023; Dochen). The highest nucleotide diversity and the second-highest nucleotide diversity were observed in the Yarlung Tsangpo River (h = 0.98 ± 0.00003; π = 0.00323 ± 0.00731), which might have been caused by the large number of samples from this region.
In spatial analysis (SAMOVA), the highest FCT (FCT = 77.9) was obtained when populations were grouped into six main clusters (Table 2), the biggest of which comprised nine populations including Khinglo, Saga, Zhongba, Tsetang, Yadong, Tarok Tso, Lhasa, Dochen, and Kongyu Tso. The samples from Yangtze were formed a cluster whereas the two different clusters comprised the three populations from Nu River basin (group 3 and group 4; Table 2).
Table 2.
Results from a spatial analysis of molecular variance (SAMOVA) showing F values given different numbers of groupings. Sets of lineages that were combined within the groups are indicated
| Group | |
| 4 groups | |
| 1. Budongquan + Tuotuo River | FSC = 0.43 |
| 2. Lhontse | FST = 0.84 |
| 3. Dza Chu + Nagchuka + Tsona | FCT = 0.72 |
| 4. Phung Chu + Khinglo + Saga + Zhongba + Tsetang + Gangkha + Yadong + Tarok Tso + Lhasa + Dochen | |
| 5 groups | |
| 1.Budongquan + Tuotuo River | FSC = 0.25 |
| 2.Lhontse | FST = 0.83 |
| 3.Dza Chu + Nagchuka + Tsona | FCT = 0.77 |
| 4.Sar + Phung Chu + Gangkha + Paiku Tso | |
| 5.Khinglo + Saga + Zhongba + Tsetang + Yadong + Tarok Tso + Lhasa + Dochen + Kongyu Tso | |
| 6 groups | |
| 1. Budongquan + Tuotuo River | FSC = 0.23 |
| 2. Lhontse | FST = 0.83 |
| 3. Dza Chu | FCT = 0.78 |
| 4. Nagchuka + Tsona | |
| 5. Sar + Phung Chu + Gangkha + Paiku Tso | |
| 6. Khinglo + Saga + Zhongba + Tsetang + Yadong + Tarok Tso + Lhasa + Dochen + Kongyu Tso | |
| 7 groups | |
| 1. Budongquan + Tuotuo River | FSC = 0.16 |
| 2. Lhontse | FST = 0.81 |
| 3. Dza Chu | FCT = 0.77 |
| 4. Nagchuka + Tsona | |
| 5. Yadong | |
| 6. Sar + Phung Chu + Gangkha + Paiku Tso | |
| 7. Khinglo + Saga + Zhongba + Tsetang + Tarok Tso + Lhasa + Dochen + Kongyu Tso |
Phylogenetic relationships and divergence time
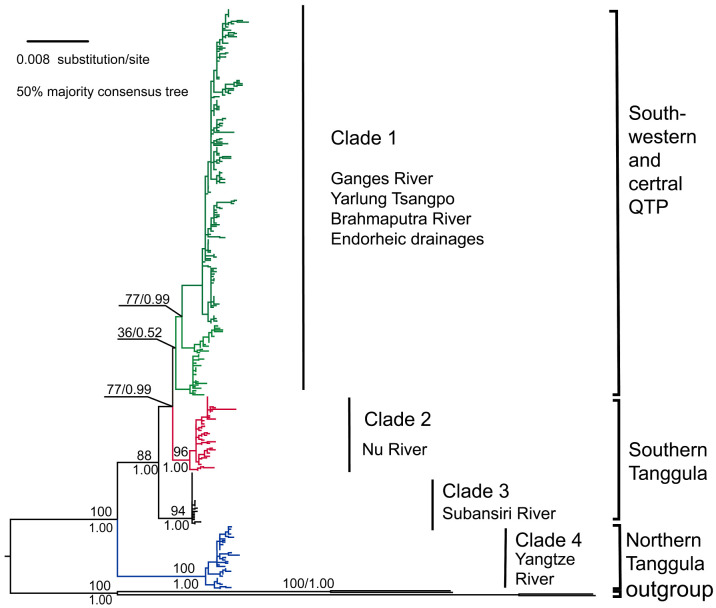
Both ML and BI trees showed a consistency in topology (Fig. 2). Therefore, we only present the ML tree. We found substantial genetic structure within T. stenura, consisting of at least two significantly supported clades (BP = 1, Bootstrap > 90; clades 1–3, clade 4; Fig. 2). The clades 1–3 and clade 4 showed deeply divergent and separated by the intervening Tanggula Mountain. The clade 4 (Bootstrap = 100, BP = 1) geographically was restricted to the upper Yangtze River in the northern of the Tanggula Mountain. In the southern of the Tanggula Mountain, the clade 3 (Bootstrap = 94, BP = 1) composed for haplotypes limited to the Subansiri River (Lhontse) and clade 2 (Bootstrap = 96, BP = 1) consisted of Nu river systems. In the southwestern and central QTP, populations were distributed in two large rivers and three endorheic drainages (Ganges River and the Brahmaputra-Yarlung Tsangpo River; Kongyu Tso, Khinglo and Tarok Tso). The southwestern and central populations QTP were formed clade 1 and displayed a shallow divergence.
Fig. 2.
ML tree for T. stenura on QTP and in its adjacent drainages based on a combination of genes including two mitochondrial genes (COI, Cyt b) and recombination activating protein 1 gene (Rag1-701 bp). Clade credibility values of major lineages are given for nodes with bootstrap support for ML (above branch) and posterior probability for Bayesian inferences (below branch). Major clades referred to in the text are listed to the right. Different colors were assigned for each clade: Clade 1, green; Clade 2, red; Clade 3, black; Clade 4, blue; Outgroups, black.
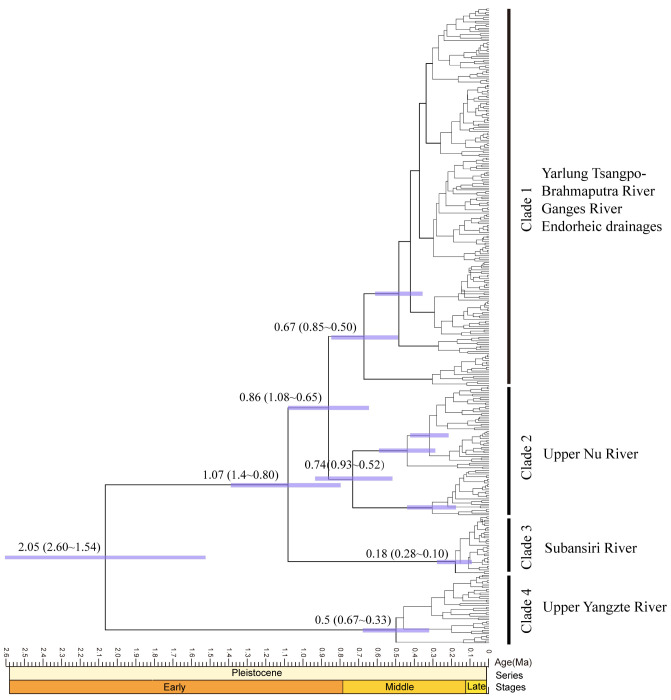
Dated divergence time for the main mtDNA lineages are concentrated in the Pleistocene and shown with 95% HPD in figure 3. The oldest lineages arose at 2.05 Mya (95% HPD: 1.54–2.60 Mya) in the Yangtze River (clade 4), while clade 3 (Lhontse) diverged with the remaining clades at 1.07 Mya (95% CI: 0.80–1.38 Mya). Approximately 0.86 Mya (95% HPD: 0.65–1.08 Mya), clade 1 and clade 2 were separated with other southern Tanggula lineages. The estimated divergence times within clade 2 and clade 1 were tentatively dated back to 0.74 Mya (95% HPD: 0.52–0.93 Mya), 0.67 Mya (95% HPD: 0.50–0.85 Mya) respectively.
Fig. 3.
Bayesian estimates of divergence time for the lineages of T. stenura based on the mitochondrial genes (COI, Cyt b) data set. Dusty purple bars represent 95% highest posterior density for divergence estimates. The numbers on the nodes are million years ago (Mya).
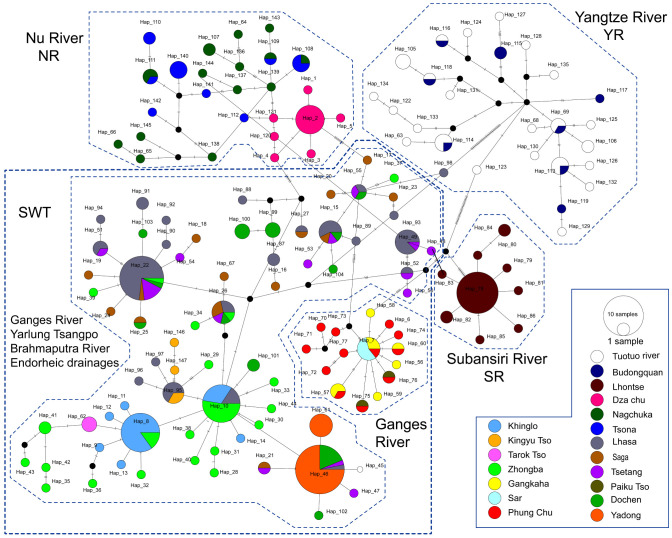
The TCS analysis revealed four distinct separate networks (networks YR, NR, SR, SWT; Fig. 4) clearly corresponded to the four clades recovered by the phylogenetic analyses. Within the network YR, the haplotypes geographically were restricted to the upper Yangtze River, corresponding to phylogenetic clade 4. Haplotypes of population on the Nu River basin (network NR; clade 2) and Subansiri River (network SR; clade 3) formed two separate cluster. Network SWT (clade 1) was restricted among the southwestern and central QTP and showed several star-like topologies, indicative of recent expansions at least. Within network SWT, two regionally defined haplogroups (the Brahmaputra-Yarlung Tsangpo and endorheic drainage basin lineages; Ganges River lineages; Fig. 4) could be distinguished. Moreover, many mixed haplotypes were found to be mixed among the SWT haplogroups, suggesting recent gene flow occurred.
Fig. 4.
TCS network generated by Popart based on cytochrome b haplotypes for the T. stenura in QTP. Numbers in the networks represent haplotype designations and the areas of the circles are proportional to haplotype frequency; Black dots represent missing intermediate haplotypes.
Demographic history
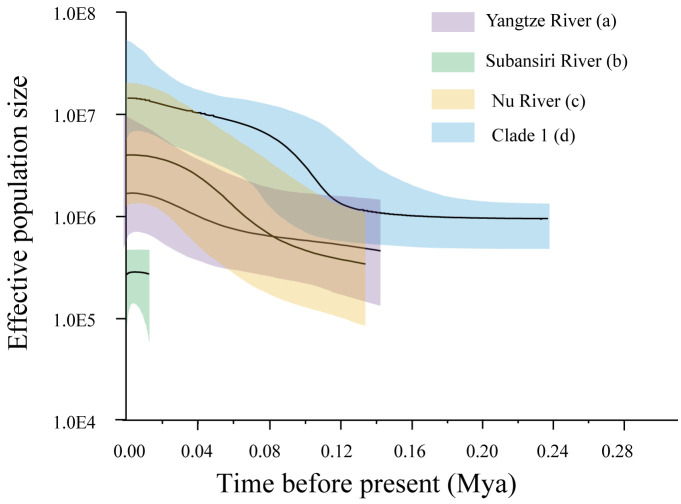
For all of T. stenura populations, Tajima’s D were negative, suggesting that these populations has experienced sudden population expansion, whereas the non-significant value were found in the populations of three different drainages (the Ganges River, Yangtze River and Brahmaputra River; p > 0.05). However, an extremely significant negative values of Fu’s Fs (p < 0.02) indicated tendency of sudden expansion for all of T. stenura populations. These contradiction might have resulted from different sensitiveness levels of statistics. Generally, Tajima’s D can be affected by various factors (e.g., bottleneck effect or founder effect) (Tajima 1989; Nei and Kumar 2000; Schmidt and Pool 2002), but Fu’s Fs is frequently considered as more sensitive to population expansion than Tajima’s D (Fu 1997; Chen et al. 2004). The unimodal distribution showed in the mismatch analysis for four clades indicated a high possibility of historical population expansion. Result of BSP revealed that three clades (clade 1; clade 2; clade 4; Fig. 5) populations showed increment in the effective population size starting approximately 120 thousand years ago (kya), corresponding to the Last Interglacial (Eemian Stage, 130–115 kya). The Subansiri River population (clade 3) had experienced population growth phases since approximately 20 kya. The limited gene flow in low-altitude areas (Lhontse 3910 m; the lowest altitude among the localities we surveyed) may be the cause of the unsynchronized time of population expansion.
Fig. 5.
Bayesian Skyline Plot (BSP) of the Tibetan stone loaches, T. stenura based on mtDNA (COI, Cyt b). The maximum time is set to the upper 95% HPD of the root height. The median estimate (solid line) and 95% HPD limits (Color area) are indicated. (a) Yangtze River populations (clade 4); (b) Subansiri River population (clade 3); (c) Nu River populations (clade 2); (d) the southwestern and central QTP populations (clade 1).
DISCUSSION
Phylogenetic analyses based on the concatenated dataset of three gene fragments (COI + Cyt b + Rag1) showed distinct phylogeographic patterns of Triplophysa stenura. The most obvious geographical pattern exhibited between northern and southern Tanggula Mountain with a deeply lineages divergence, while the southwestern and central QTP populations displayed a shallow divergence (Fig. 2). The previous phylogeographical study of T. stenura based on the two mitochondrial genes (Cyt b; D-loop) revealed that T. stenura is a single lineage with three well supported clades and the divergence occurring at 0.64 Mya (Ren et al. 2018), corresponding to the Kunlun-Huanghe Movement (1.1–0.6 Mya) (Cui et al. 1998). However, the presence of two different phylogeographic patterns in this study suggests that the evolutionary history of T. stenura is more complex than previously assumed. In our study, the southwestern and central QTP populations showed a shallow divergence which did not follow geographic patterns, indicating that other factors may have played a more dominant role than that of tectonic movement in shaping the evolutionary history and genetic divergence of the species.
Interestingly, in most studies, tectonic movement was considered the important factor shaping the genetic structure of QTP species during the Pleistocene. For instance, Ding and Liao (2019) suggested that two major genetic differentiations of Cricetulus kamensis that occurred during the early Pleistocene were influenced by the Qing-Zang tectonic movements. Qu et al. (2010), Cun and Wang (2010), Li et al. (2017) and Wang et al. (2010) emphasized that the uplift of QTP and the subsequently induced climate fluctuations shaped the patterns of genetic diversity observed in extant QTP’s species during the Pleistocene. All these studies proposed that the interaction of historic climate changes and geological events shaped the evolutionary history and genetic divergence of the species in QTP. However, recent review suggested that QTP has been 4,000–5,000 m high since the mid-Eocene, and the Indian summer monsoon, South-east Asian summer monsoon, and Central Asian winter monsoon arose at different times and are unrelated to the uplift of QTP (Renner 2016). Therefore, the explanation that the current phylogeographic structure was influenced by the uplift of QTP during the Pleistocene does not seem plausible. Thus, factors other than distinct orogenic movements may have played the predominant role in shaping the evolutionary history and genetic divergence of the species.
Based on the estimated divergence times obtained from the BEAST analyses, the Yangtze River is likely where T. stenura first arose, approximately at 2.05 Mya (95% HPD: 1.54–2.60 Mya), followed by the southwestward dispersal into the southern Tanggula Mountain (approximately 1.07 Mya). This date well coincides with a cold stage in the Xixiabangma Gaciation (1.17–0.8 Mya) which was the oldest glaciation took place in the late Early Pleistocene. Moreover, the divergences of clades 1 and clade 2 (clade 1, southwestern and central QTP; clade 2, Nu River basin, southern Tanggula Mountain) are approximately 0.68 Mya and 0.74 Mya, respectively. These two dates are well consistent with the largest glaciation (Kunlun Glaciation 0.78–0.62 Mya). During the most extensive glacier stage of Kunlun Glaciation on the plateau, the area covered by ice was found to be approximately 500,000 km2 (Zheng et al. 2002). Following the advancement of glaciers during these glaciations, some glacial refugia (e.g., river valley) might persist on QTP platform, where cold-adapted species (e.g., Triplohysa) could survive to the frigid climate and further promoted the intraspecific divergences. Taberlet and Cheddadi (2002) revealed that the refugia populations were often been accompanied by high levels of genetic diversity and unique haplotypes. In this study, unique haplotypes and high levels of genetic diversity were found in the Yangtze River, Subansiri River and Nu River. Our data implied that the lineages of T. stenura became restricted to at least three different regions that acted as refugia during the mid-Pleistocene glacial period. A logical explanation is that the private haplotypes existed in the glacial refugia of these regions over a long period and survived in situ.
Our data indicated that T. stenura comprises four major clades (clade 1, clade 2, clade 3 and clade 4) and revealed different phylogeographic patterns in QTP. In contrast to northern and southern Tanggula Mountain populations, where populations were strictly nested by drainages (clade 2, 3 and 4 from Nu River, the Subansiri River and Yangtze River, respectively), exhibiting a marked geographical pattern, the southwestern and central QTP populations (clade 1) usually entangled together and lack of a clearly geographic pattern. These different phylogeographic patterns may be due to the asynchronous glacier development in different regions across QTP (Lehmkuhl and Owen 2005; Thompson et al. 2006). The glaciation throughout the northern and southern Tanggula is strongly controlled by precipitation changes related to oscillation in the South Asian monsoonal system combined with cooling events (Lehmkuhl and Owen 2005). However, the influence of the South Asian monsoon is weakens significantly across eastern mountain belt, leading to the precipitation to decrease sharply from south to north, and from east to west. Thus, the eastern mountains of the plateau are much more favorable for glacier development compared to the central and western parts (Dong et al. 2010). The more intensive and extensive glaciers might have been easier to act as a geographical barrier that resulted in restricted gene flow for T. stenura (e.g., northern and southern Tanggula Mountain). Therefore, the restricted gene flow caused by the advancing glaciers is the most likely the reason for the high levels of gene differentiation in the T. stenura populations of northern and southern Tanggula Mountain.
The climatic oscillations resulted in the repeated drastic environmental changes during the Quaternary. When climate cooled down and the glacial ice expanded during the glacial period, species retreated to refugia to persist the severe conditions during the glacial period (Yang et al. 2008; Guo et al. 2012; Yu et al. 2013; Liang et al. 2017). Following glacial retreat, the species could be recolonized into the formerly glaciated areas. The geographical barriers formed by glaciers might have been not enough strong to restrict gene flow for less glacier regions on QTP. It is plausible that the advancement of glaciers might have had less influence on the populations in less glacier regions and that glacial meltwater may have driven fish gene exchange among adjacent catchments during the interglacial period. For regions with less ice, such as the southwestern and central QTP, the unrestricted gene flow during the glacial period and driven gene exchange during the interglacial may lead to gene homogenization across the drainage populations.
Within clade 1, the shallow phylogenetic structure and mixed haplotype were found over a widespread geographical distribution. The clade was included two large rivers (Brahmaputra-Yarlung Tsangpo River and Ganges River) and three endorheic lakes (Kongyu Tso, Khinglo, Tarok Tso; Fig. 2; Table 1) on the southwestern and central QTP. The shared haplotypes occurred in each other among 13 populations in clade 1, which suggests that extensive bilateral gene flow existed. This phenomenon could be explained as repeated population expansion/contraction caused by climate oscillation during the late Early Pleistocene, which resulted in the gene homogenization in adjacent catchments by temporary glacial meltwater connections. During the interglacial period, increased rainfall and melting water generated several temporary channels among drainages (Fan et al. 2012). These temporary connections propelled gene flow among drainages, reducing their phylogenetic and genetic differentiation. The neutrality tests were significant, and mismatch distribution (Table 1) were unimodal, revealing that the three drainage populations (the upper Ganges, the Brahmaputra-Yarlung Tsangpo River and endorheic drainage basin) underwent at least once sudden expansion. Furthermore, the result of BSP revealed that three clades (clade 1, clade 2, clade 4; Fig. 5) had experienced population growth phases since approximately 120 kya, corresponding to the last interglacial period. The population expansion may be driven by the warm and wet climate during the last interglacial period (Lv et al. 2018; Zhao et al. 2019). In addition, the endorheic and exorheic drainages were probably connection during the late Pleistocene and Holocene by temporary watercourses. These temporary connections drove the extensive bilateral gene flow among drainages, decreasing the phylogenetic and genetic differentiation on the central and western QTP.
CONCLUSIONS
Our results showed that the current phylogeo-graphic structure of Triplophysa stenura was sculpted by climatic oscillation during the mid-Pleistocene, not just by the tectonic movements. Following the advancement of glaciers during the multiple Pleistocene glaciations, three drainages (Yangtze River, Nu River and Subansiri River) existed refugia in the northern and southern Tanggula Mountain, giving T. stenura a chance to survive in situ. The spatial differences of glacier development in QTP was driven the different phylogeographic patterns. For the regions with more extensive glaciers (e.g., the northern and southern Tanggula Mountain) glacial acted as a geographical barrier, restricted gene flow for T. stenura. It is for the result of deep divergence among the northern and southern Tanggula Mountain drainages. However, the less ice regions like the southwestern and central QTP, T. stenura populations undergo repeated expansion/contraction during the late Pleistocene glacial cycles, which lead to the gene homogenization on the southwestern and central QTP.
Supplementary materials
Primers used in this study. (download)
Acknowledgments
We appreciate Professor Yifeng Chen for this valuable comments and suggestions. We are thankful to Dr Xiaoyun Sui, Xiu Feng, Ren Zhu and Jie Zhao for experiments and field samplings. This research was supported by the Second Tibetan Plateau Scientific Expedition Program (2019QZKK05010102), National Natural Science Foundation of China (32070436, 31900374, 31572248), and Finance Special Fund of Chinese Ministry of Agriculture and Rural Affairs of the People’s Republic of China (Fisheries resources and environment survey in the key water areas of Tibet).
List of abbreviations
- QTP
the Qinghai-Tibetan Plateau.
- Mya
million years ago.
- kya
thousand years ago
- COI
Cytochrome oxidase subunit 1.
- Cyt b
Cytochrome b.
- Rag1
Recombination activating 1.
- SAMOVA
Spatial analysis of molecular variation.
- ML
Maximum likelihood.
- BI
Bayesian inference.
- HPD
highest posterior density.
Footnotes
Authors’ contributions: All authors conceived the ideas; JXH and DKH led the writing and analyzed the data; JXH conducted experiments. DKH conducted the fieldwork sampling.
Competing interests: All authors declare that they have no conflict of interests regarding the publication of this paper.
Availability of data and materials: All the haplotypes were submitted to GenBank with accession numbers MT681383-MT681640. The outgroup sequences were submitted to GenBank with accession numbers MW086881-MW086892.
Consent for publication: Not applicable.
Ethics approval consent to participate: All experimental protocols were approved by the Ethics Committee of the Institute of Hydrobiology, Chinese Academy of Sciences.
References
- Avise JC, Nelson WS, Bowen BW, Walker D. 2000. Phylogeography of colonially nesting seabirds, with special reference to global matrilineal patterns in the sooty tern (Sterna fuscata). Mol Ecol 9(11):1783–1792. doi:10.1046/j.1365-294x.2000.01068.x. [DOI] [PubMed]
- Bouckaert R, Heled J, Kuehnert D, Vaughan T, Wu CH, Xie D, Suchard MA, Rambaut A, Drummond AJ. 2014. BEAST 2: A Software Platform for Bayesian Evolutionary Analysis. PLoS Comput Biol 10(4):e1003537. doi:10.1371/journal. pcbi.1003537. [DOI] [PMC free article] [PubMed]
- Chen CA, Ablan MCA, McManus JW, Bell JD, Tuan VS, Cabanban AS, Shao KT. 2004. Population structure and genetic variability of six bar wrasse (Thallasoma hardwicki) in northern South China sea revealed by mitochondrial control region sequences. Mar Biotechnol 6(4):312–326. doi:10.1007/s10126-003-0028-2. [DOI] [PubMed]
- Chen GJ, Liao W, Lei XQ. 2015. First fossil cobitid (Teleostei: Cypriniformes) from Early-Middle Oligocene deposits of South China. Vertebrat Palasiatic 53(4):299–309.
- Clement M, Posada D, Crandall KA. 2000. TCS: a computer program to estimate gene genealogies. Mol Ecol 9(10):1657–1659. doi:10.1046/j.1365-294x.2000.01020.x. [DOI] [PubMed]
- Cui ZJ, Wu YQ, Liu GN, Ge DK, Pang QQ, Xu QH. 1998. On Kunlun Yellow River tectonic movement. Sci China Ser D Earth Sci 41(6):592–600. doi:10.1007/bf02878741.
- Cun YZ, Wang XQ. 2010. Plant recolonization in the Himalaya from the southeastern Qinghai-Tibetan Plateau: Geographical isolation contributed to high population differentiation. Mol Phylogenet Evol 56(3):972–982. doi:10.1016/j.ympev.2010.05.007. [DOI] [PubMed]
- De Macedo‐Soares PHM, Petry AC, Farjalla VF, Caramaschi EP. 2010. Hydrological connectivity in coastal inland systems: lessons from a Neotropical fish metacommunity. Ecol Freshw Fish 19(1):7–18. doi:10.1111/j.1600-0633.2009.00384.x.
- Dincă V, Dapporto L, Vila R. 2011. A combined genetic-morphometric analysis unravels the complex biogeographical history of Polyommatus icarus and Polyommatus celina Common Blue butterflies. Mol Ecol 20(18):3921–3935. doi:10.1111/j.1365-294X.2011.05223.x. [DOI] [PubMed]
- Ding L, Liao J. 2019. Phylogeography of the Tibetan hamster Cricetulus kamensis in response to uplift and environmental change in the Qinghai-Tibet Plateau. Ecol Evol 9(12):7291–7306. doi:10.1002/ece3.5301. [DOI] [PMC free article] [PubMed]
- Doadrio I, Perdices A. 2005. Phylogenetic relationships among the Ibero-African cobitids (Cobitis, Cobitidae) based on cytochrome b sequence data. Mol Phylogenet Evol 37(2):484–493. doi:10.1016/j.ympev.2005.07.009. [DOI] [PubMed]
- Dong G, Yi C, Chen L. 2010. An introduction to the physical geography of the Qiangtang Plateau: a frontier for future geoscience research on the Tibetan Plateau. Phys Geogr 31(6):475–492. doi:10.2747/0272-3646.31.6.475.
- Dupanloup I, Schneider S, Excoffier L. 2002. A simulated annealing approach to define the genetic structure of populations. Mol Ecol 11(12):2571–2581. doi:10.1046/j.1365-294X.2002.01650.x. [DOI] [PubMed]
- Elderfield H, Ferretti P, Greaves M, Crowhurst S, McCave IN, Hodell D, Piotrowski AM. 2012. Evolution of ocean temperature and ice volume through the mid-Pleistocene climate transition. Science 337:704–709. doi:10.1126/science.1221294. [DOI] [PubMed]
- Excoffier L. 2004. Patterns of DNA sequence diversity and genetic structure after a range expansion: lessons from the infinite-island model. Mol Ecol 13(4):853–864. doi:10.1046/j.1365-294X.2003.02004.x. [DOI] [PubMed]
- Fan Q, Ma H, Hou G. 2012. Late Pleistocene lake and glaciation evolution on the northeastern Qinghai-Tibetan Plateau: a review. Environ Earth Sci 66(2):625–634. doi:10.1007/s12665-011-1271-x.
- Fu YX. 1997. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics 147(2):915–925. doi:10.1017/S0016672397002966. [DOI] [PMC free article] [PubMed]
- Funk VA, Wagner WL. 1995. Biogeographic patterns in the Hawaiian Islands. In: Hawaiian biogeography: evolution on a hot spot archipelago, Smithsonian Institution Press, Washington, USA, pp. 379–419.
- Guo X, Liu L, Wang Y. 2012. Phylogeography of the Phrynocephalus vlangalii Species Complex in the Upper Reaches of the Yellow River Inferred from mtDNA ND4-tRNALEU Segments. Asian Herpetol Res 3(1):52–68. doi:10.3724/sp.j.1245.2012.00052.
- Hall T, Biosciences I, Carlsbad C. 2011. BioEdit: an important software for molecular biology. GERF Bull Biosci 2(1):60–61.
- He D, Chen Y. 2006. Biogeography and molecular phylogeny of the genus Schizothorax (Teleostei: Cyprinidae) in China inferred from cytochrome b sequences. J Biogeogr 33(8):1448–1460. doi:10.1111/j.1365-2699.2006.01510.x.
- He D, Chen Y. 2007. Molecular phylogeny and biogeography of the highly specialized grade schizothoracine fishes (Teleostei: Cyprinidae) inferred from cytochrome b sequences. Chin Sci Bull 52(6):777–788. doi:10.1007/s11434-007-0123-2.
- He D, Chen Y, Liu C, Tao J, Ding C, Chen Y. 2016. Comparative phylogeography and evolutionary history of schizothoracine fishes in the Changtang Plateau and their implications for the lake level and Pleistocene climate fluctuations. Ecol Evol 6(3):656–674. doi:10.1002/ece3.1890. [DOI] [PMC free article] [PubMed]
- He D, Sui X, Sun H, Tao J, Ding C, Chen Y, Chen Y. 2020. Diversity, pattern and ecological drivers of freshwater fish in China and adjacent areas. Rev Fish Biol Fish 30:387–404. doi:10.1007/s11160-020-09600-4.
- Hewitt G. 2000. The genetic legacy of the Quaternary ice ages. Nature 405(6789):907–913. doi:10.1038/35016000. [DOI] [PubMed]
- Hewitt GM. 2004. Genetic consequences of climatic oscillations in the Quaternary. Philos T Roy Soc B 359(1442):183–195. doi:10.1098/rstb.2003.1388. [DOI] [PMC free article] [PubMed]
- Lanfear R, Frandsen PB, Wright AM, Senfeld T, Calcott B. 2017. PartitionFinder 2: New Methods for Selecting Partitioned Models of Evolution for Molecular and Morphological Phylogenetic Analyses. Mol Biol Evol 34(3):772–773. doi:10.1093/molbev/msw260. [DOI] [PubMed]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. Clustal W and clustal X version 2.0. Bioinformatics 23(21):2947–2948. doi:10.1093/bioinformatics/btm404. [DOI] [PubMed]
- Lehmkuhl F, Owen LA. 2005. Late Quaternary glaciation of Tibet and the bordering mountains: a review. Boreas 34(2):87–100. doi:10.1080/03009480510012908.
- Leigh JW, Bryant D. 2015. POPART: full-feature software for haplotype network construction. Methods Ecol Evol 6(9):1110–1116. doi:10.1111/2041-210x.12410.
- Lemey P, Salemi M, Vandamme AM. 2009. The phylogenetic handbook: a practical approach to phylogenetic analysis and hypothesis testing. Cambridge University Press, New York, USA, pp. 1–723.
- Li Y, Ludwig A, Peng Z. 2017. Geographical differentiation of the Euchiloglanis fish complex (Teleostei: Siluriformes) in the Hengduan Mountain Region, China: Phylogeographic evidence of altered drainage patterns. Ecol Evol 7(3):928–940. doi:10.1002/ece3.2715. [DOI] [PMC free article] [PubMed]
- Liang Y, He D, Jia Y, Sun H, Chen Y. 2017. Phylogeographic studies of Schizothoracine fishes on the central Qinghai-Tibet Plateau reveal the highest known glacial microrefugia. Sci Rep 7(1):10983. doi:10.1038/s41598-017-11198-w. [DOI] [PMC free article] [PubMed]
- Lv X,Cheng J,Meng Y,Chang Y,Xia L,Wen Z,Ge D,Liu S, Yang Q. 2018. Disjunct distribution and distinct intraspecific diversification of Eothenomys melanogaster in South China. BMC Evol Biol 18(1):50. doi:10.1186/s12862-018-1168-3. [DOI] [PMC free article] [PubMed]
- Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In: 2010 gateway computing environments workshop (GCE): Ieee. pp. 1–8. doi:10.1109/GCE.2010.5676129.
- Maniatis T, Fritsch EF, Sambrook J. 1982. Molecular cloning: a laboratory manual: Cold spring harbor laboratory press, New York. pp. 721–722. doi:10.2307/1309366.
- Nei M, Kumar S, Nei M. 2000. Molecular evolution and phylogenetics. Oxford university press, New York, USA, pp. 1–333.
- Prokofiev A. 2007. Redescription of a fossil loach Triplophysa opinata (Yakowlew, 1959) from the Miocene of Kirgizia (Balitoridae: Nemacheilinae). J Ichthyol 47(1):26–31. doi:10.1134/S0032945207010031.
- Qu Y, Lei F, Zhang R, Lu X. 2010. Comparative phylogeography of five avian species: implications for Pleistocene evolutionary history in the Qinghai-Tibetan plateau. Mol Ecol 19(2):338–351. doi:10.1111/j.1365-294X.2009.04445.x. [DOI] [PubMed]
- Rambaut A. 2012. FigTree v1. 4. 3. Available at http://tree.bio.ed.ac. uk/software/figtree/.
- Rambaut A, Drummond AJ, Xie D, Baele G, Suchard MA. 2018. Posterior Summarization in Bayesian Phylogenetics Using Tracer 1.7. Syst Biol 67(5):901–904. doi:10.1093/sysbio/syy032. [DOI] [PMC free article] [PubMed]
- Rana SK, Luo D, Rana HK, O'Neill AR, Sun H. 2019. Geoclimatic factors influence the population genetic connectivity of Incarvillea arguta (Bignoniaceae) in the Himalaya-Hengduan Mountains biodiversity hotspot. J Syst Evol (Published online). doi:10.1111/jse.12521.
- Ren Q, Yang JX, Chen XY. 2018. Phylogeographical and morphological analyses of Triplophysa stenura (Cypriniformes: Nemacheilidae) from the three Parallel Rivers region, China. Zool Stud 57:26. doi:10.6620/zs.2018.57-26. [DOI] [PMC free article] [PubMed]
- Renner SS. 2016. Available data point to a 4-km-high Tibetan Plateau by 40 Ma, but 100 molecular-clock papers have linked supposed recent uplift to young node ages. J Biogeogr 43(8):1479–1487. doi:10.1111/jbi.12755.
- Roach KA, Thorp JH, Delong MD. 2009. Influence of lateral gradients of hydrologic connectivity on trophic positions of fishes in the Upper Mississippi River. Freshw Biol 54(3):607–620. doi:10.1111/j.1365-2427.2008.02137.x.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Hohna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice Across a Large Model Space. Syst Biol 61(3):539–542. doi:10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed]
- Rozas J, Ferrer MA, Carlos SDJ, Guirao RS, Librado P, Ramos OSE, Sanchez GA. 2017. DnaSP 6: DNA Sequence Polymorphism Analysis of Large Data Sets. Mol Biol Evol 34(12):3299–3302. doi:10.1093/molbev/msx248. [DOI] [PubMed]
- Schmidt D, Pool J. 2002. The effect of population history on the distribution of the Tajima’s D statistic. Population English Edition. pp. 1–8. doi:10.1677/joe.0.0330211.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30(9):1312–1313. doi:10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed]
- Taberlet P, Cheddadi R. 2002. Quaternary refugia and persistence of biodiversity. Science 297(5589):2009–2010. doi:10.1126/science.297.5589.2009. [DOI] [PubMed]
- Tajima F. 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123(3):585–595. [DOI] [PMC free article] [PubMed]
- Thompson LG, Mosley TE, Davis ME, Mashiotta TA, Henderson KA, Lin PN, Yao T. 2006. Ice core evidence for asynchronous glaciation on the Tibetan Plateau. Quat Int 154:3–10. doi:10.1016/j.quaint.2006.02.001.
- Wang H, Qiong L, Sun K, Lu F, Wang Y, Song Z, Wu Q, Chen J, Zhang W. 2010. Phylogeographic structure of Hippophae tibetana (Elaeagnaceae) highlights the highest microrefugia and the rapid uplift of the Qinghai-Tibetan Plateau. Mol Ecol 19(14):2964–2979. doi:10.1111/j.1365-294X.2010.04729.x. [DOI] [PubMed]
- Wiens JJ, Donoghue MJ. 2004. Historical biogeography, ecology and species richness. Trends Ecol Evol 19(12):639–644. doi:10.1016/j.tree.2004.09.011. [DOI] [PubMed]
- Xia M, Tian Z, Zhang F, Khan G, Gao Q, Xing R, Zhang Y, Yu J, Chen S. 2018. Deep Intraspecific Divergence in the Endemic Herb Lancea tibetica (Mazaceae) Distributed Over the Qinghai-Tibetan Plateau. Front Genet 9:492. doi:10.3389/fgene.2018.00492. [DOI] [PMC free article] [PubMed]
- Xu W, Yin W, Chen A, Li J, Lei G, Fu C. 2014. Phylogeographical analysis of a cold-temperate freshwater fish, the Amur sleeper (Perccottus glenii) in the Amur and Liaohe River basins of Northeast Asia Zoolog Sci 31(10):671–679. doi:10.2108/zs130046. [DOI] [PubMed]
- Yang FS, Li YF, Ding X, Wang XQ. 2008. Extensive population expansion of Pedicularis longiflora (Orobanchaceae) on the Qinghai-Tibetan Plateau and its correlation with the Quaternary climate change. Mol Ecol 17(23):5135–5145. doi:10.1111/j.1365-294X.2008.03976.x. [DOI] [PubMed]
- Yao T, Thompson L, Yang W, Yu W, Gao Y, Guo X, Yang X, Duan K, Zhao H, Xu B, Pu J, Lu A, Xiang Y, Kattel DB, Joswiak D. 2012. Different glacier status with atmospheric circulations in Tibetan Plateau and surroundings. Nat Clim Chang 2(9):663–5667. doi:10.1038/nclimate1580.
- Ye Z,Yuan J,Li M,Damgaard J,Chen P,Zheng C,Yu H,Fu S, Bu W. 2018. Geological effects influence population genetic connectivity more than Pleistocene glaciations in the water strider Metrocoris sichuanensis (Insecta: Hemiptera: Gerridae). J Biogeogr 45(3):690–701. doi:10.1111/jbi.13148.
- Yu G, Zhang M, Rao D, Yang J. 2013. Effect of Pleistocene climatic oscillations on the phylogeography and demography of red knobby newt (Tylototriton shanjing) from southwestern China. PLoS ONE 8(2):e56066. doi:10.1371/journal.pone.0056066. [DOI] [PMC free article] [PubMed]
- Zhang F, Zhu L, Zhang L, Wang W, Sun G. 2017. Phylogeography of freshwater fishes of the Qilian Mountains area (Triplophysa leptosoma, Cobitidae: Cypriniformes). Environ Biol Fishes 100(11):1383–1396. doi:10.1007/s10641-017-0650-x.
- Zhao M, Chang Y, Kimball RT, Zhao J, Lei F, Qu Y. 2019. Pleistocene glaciation explains the disjunct distribution of the Chestnut-vented Nuthatch (Aves, Sittidae). Zool Scr 48(1):33–45. doi:10.1111/zsc.12327.
- Zheng BX, Xu QQ, Shen YP. 2002. The relationship between climate change and Quaternary glacial cycles on the Qinghai-Tibetan Plateau: review and speculation. Quat Int 97:93–101. doi:10.1016/s1040-6182(02)00054-x.
- Zhu S. 1989. The loaches of the subfamily Nemacheilinae in China (Cypriniformes: Cobitidae). Jiangsu Science and Technology Publishing House, Nanjing, China.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primers used in this study. (download)