Abstract
Combined chemotherapeutic treatment is the method of choice for advanced and metastatic gastric tumors. However, resistance to chemotherapeutic agents is one of the main challenges for the efficient gastric cancer (GC) treatment. Cisplatin (CDDP) is used as an important regimen of chemotherapy for GC which induces cytotoxicity by interfering with DNA replication in cancer cells and inducing their apoptosis. Majority of patients experience cisplatin-resistance which is correlated with tumor metastasis and relapse. Moreover, prolonged and high-dose cisplatin administrations cause serious side effects such as nephrotoxicity, ototoxicity, and anemia. Since, there is a high rate of recurrence after CDDP treatment in GC patients; it is required to clarify the molecular mechanisms associated with CDDP resistance to introduce novel therapeutic methods. There are various cell and molecular processes associated with multidrug resistance (MDR) including drug efflux, detoxification, DNA repair ability, apoptosis alteration, signaling pathways, and epithelial-mesenchymal transition (EMT). MicroRNAs are a class of endogenous non-coding RNAs involved in chemo resistance of GC cells through regulation of all of the MDR mechanisms. In present review we have summarized all of the miRNAs associated with cisplatin resistance based on their target genes and molecular mechanisms in gastric tumor cells. This review paves the way of introducing a miRNA-based panel of prognostic markers to improve the efficacy of chemotherapy and clinical outcomes in GC patients. It was observed that miRNAs are mainly involved in cisplatin response of gastric tumor cells via regulation of signaling pathways, autophagy, and apoptosis.
Keywords: MicroRNA, Cisplatin, Multi drug resistance, Chemo resistance, Cancer
Background
Gastric cancer (GC) is the 6th frequent and 2nd cancer related mortality globally [1]. Surgery is considered as the primary therapy in early stage GC, while majority of the patients are diagnosed in advanced stages of tumor progression that needs chemotherapeutic treatment. The combination of surgery along with chemotherapy is considered a curative approach for treating patients with gastric cancers, leading to higher disease-free survival and reduced risk of tumor relapse and metastasis [2, 3]. Despite the declining trend of GC incidence during recent decades, it still remains as one of the leading cause of cancer related mortalities globally [1]. Apart from fundamental diagnostic and therapeutic advances during recent decade, there is still a high ratio of poor prognosis among GC patients [4]. Surgery and chemotherapy can improve GC survival by 10–15 % [2, 3]. Combined chemotherapeutic treatment is the method of choice for advanced and metastatic gastric tumors. Cisplatin (CDDP) is used as an important regimen of chemotherapy for GC which functions through induction of DNA adducts formation in nucleus and mitochondria [5, 6]. Cisplatin induces cytotoxicity by interfering with DNA replication in cancer cells and inducing their apoptosis. Nevertheless, a majority of patients develop cisplatin-resistance which has been shown to be correlated with cancer metastasis and relapse [7]. Moreover, prolonged and high-dose cisplatin administrations cause serious side effects such as nephrotoxicity, ototoxicity, and anemia [8]. Therefore, it is important to enhance the sensitivity of GC cells to cisplatin in order to maximize the efficacy of chemotherapy for the chemo resistant patients. Since, there is a high rate of recurrence after CDDP treatment in GC patients; it is required to clarify the molecular mechanisms associated with CDDP resistance to introduce novel therapeutic methods. Various mechanisms are involved in drug resistance including drug efflux, reduced apoptosis, increased DNA repair ability, and drug detoxification [9]. Cisplatin accumulates in the mitochondria and causes mitochondrial dysfunctions resulting in the induction of cell apoptosis and oxidative/endoplasmic reticulum stress [10]. Due to the absence of appropriate early detection methods, most GC cases are diagnosed at late stages when the treatment is not effective [11]. About 70 % of GC patients are diagnosed with middle-advanced stage and show a 5-year survival rate of less than 20 % [12].
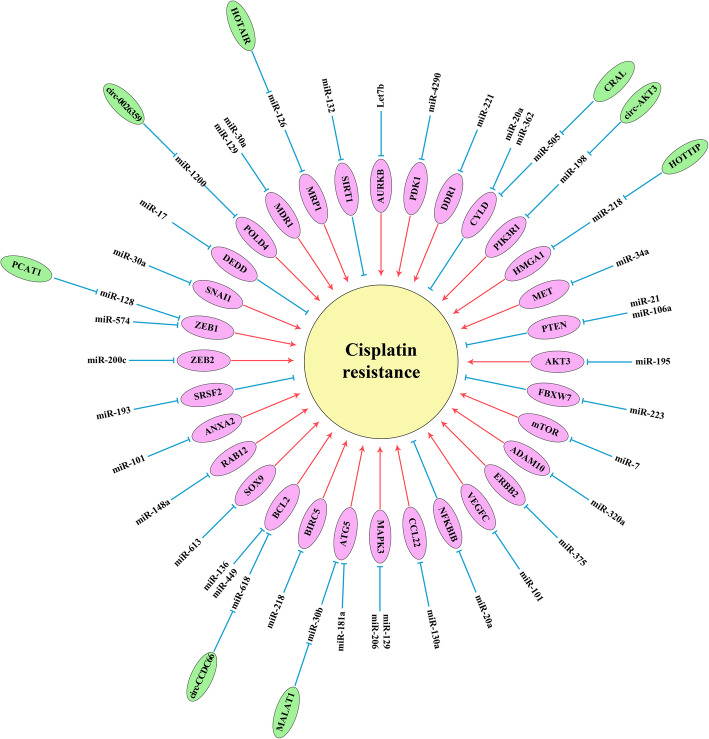
MicroRNAs (miRNAs) are small non-coding RNAs (∼22 nucleotides) involved in regulation of their target mRNAs through translation suppression or degradation [13]. MiRNAs have key roles in regulation of cellular mechanisms such as proliferation, differentiation, and apoptosis. Aberrant expression of miRNAs is associated with the development of chemo resistance [14]. Therefore, in present review we have summarized all of the miRNAs associated with cisplatin resistance to pave the way of introducing a miRNA-based panel of prognostic markers in GC patients (Fig. 1) (Table 1).
Fig. 1.
Molecular mechanisms of miRNAs associated with cisplatin resistance in gastric tumor cells
Table 1.
All of the miRNAs associated with cisplatin (CDDP) resistance in gastric tumor cells
| Gene | Target | Effect on the target | Effect on the tumor cells | Study | Year |
|---|---|---|---|---|---|
| Signaling pathways | |||||
| Circ-AKT3 | miR-198 | Down regulation | Increased CDDP resistance | Huang [6] | 2019 |
| miR-34a | MET | Down regulation | Increased CDDP sensitivity | Zhang [7] | 2016 |
| miR-106a | PTEN | Down regulation | Increased CDDP resistance | Fang [15] | 2013 |
| miR-21 | PTEN | Down regulation | Increased CDDP resistance | Yang [16] | 2013 |
| miR-195 | AKT3 | Down regulation | Increased CDDP sensitivity | Ye [17] | 2017 |
| CRAL | miR-505 | Down regulation | Increased CDDP sensitivity | Wang [18] | 2020 |
| miR-7 | mTOR | Down regulation | Increased CDDP sensitivity | Xu [19] | 2017 |
| miR-375 | ERBB2 | Down regulation | Increased CDDP sensitivity | Zhou [20] | 2016 |
| miR-101 | VEGF-C | Down regulation | Increased CDDP sensitivity | Li [21] | 2016 |
| miR-362 | CYLD | Down regulation | Increased CDDP resistance | Xia [22] | 2014 |
| miR-20a | CYLD | Down regulation | Increased CDDP resistance | Zhu [23] | 2016 |
| miR-20a | NFKBIB | Down regulation | Increased CDDP resistance | Du [24] | 2016 |
| miR-130a | CCL22 | Down regulation | Increased CDDP sensitivity | Fang [25] | 2020 |
| miR-206 | MAPK3 | Down regulation | Increased CDDP sensitivity | Chen [26] | 2019 |
| miR-129 | MAPK3 | Down regulation | Increased CDDP sensitivity | Cao [27] | 2019 |
| Autophagy and apoptosis | |||||
| miR-181a | ATG5 | Down regulation | Increased CDDP sensitivity | Zhao [28] | 2016 |
| MALAT1 | miR-30b | Down regulation | Increased CDDP resistance | Xi [29] | 2019 |
| miR-218 | BIRC5 | Down regulation | Increased CDDP sensitivity | Zhang [30] | 2018 |
| Circ-CCDC66 | miR-618 | Down regulation | Increased CDDP resistance | Zhang [31] | 2020 |
| miR-136 | AEG1, BCL2 | Down regulation | Increased CDDP sensitivity | Yu [32] | 2018 |
| miR-449a | BCL2, CCND1 | Down regulation | Increased CDDP sensitivity | Hu [33] | 2014 |
| miR-143 | IGF1R and BCL2 | Down regulation | Increased CDDP sensitivity | Zhuang [34] | 2015 |
| miR-148a-3p | RAB12, AKAP1 | Down regulation | Increased CDDP sensitivity | Li [35] | 2017 |
| miR-193a-3p | SRSF2 | Down regulation | Increased CDDP resistance | Lee [36] | 2019 |
| Epithelial Mesenchymal Transition (EMT) | |||||
| miR-200c | ZEB2 | Down regulation | Increased CDDP sensitivity | Jiang [37] | 2017 |
| PCAT-1 | miR-128 | Down regulation | Increased CDDP resistance | Guo [38] | 2019 |
| miR-574-3p | ZEB1 | Down regulation | Increased CDDP sensitivity | Wang [39] | 2019 |
| miR-30a | SNAI1, VIM | Down regulation | Increased CDDP sensitivity | Wang [40] | 2016 |
| miR-17 | DEDD | Down regulation | Increased CDDP resistance | Wu [41] | 2018 |
| Drug efflux | |||||
| miR-129 | MDR1 | Down regulation | Increased CDDP sensitivity | Lu [42] | 2017 |
| miR-30a | MDR1 | Down regulation | Increased CDDP sensitivity | Du [43] | 2018 |
| HOTAIR | miR-126 | Down regulation | Increased CDDP resistance | Yan [44] | 2016 |
| HOTAIR | miR-34a | Down regulation | Increased CDDP resistance | Cheng [45] | 2018 |
| miR-132 | SIRT1 | Down regulation | Increased CDDP resistance | Zhang [46] | 2017 |
| Protein kinases | |||||
| Let-7b | AURKB | Down regulation | Increased CDDP sensitivity | Han [47] | 2018 |
| Hsa-Circ-0081143 | miR-646 | Down regulation | Increased CDDP resistance | Xue [48] | 2019 |
| miR-4290 | PDK1 | Down regulation | Increased CDDP sensitivity | Qian [49] | 2020 |
| miR-221-5p | DDR1 | Down regulation | Increased CDDP sensitivity | Jiang [50] | 2020 |
| Transcription factors | |||||
| miR-613 | SOX9 | Down regulation | Increased CDDP sensitivity | Xue [51] | 2019 |
| miR-421 | CASP3 and CDH1 | Down regulation | Increased CDDP resistance | Ge [52] | 2016 |
| Circ-DONSON | miR-802 | Down regulation | Increased CDDP resistance | Liu [53] | 2020 |
| Structural and DNA repair factors | |||||
| miR-101 | ANXA2 | Down regulation | Increased CDDP sensitivity | Bao [54] | 2017 |
| miR-223 | FBXW7 | Down regulation | Increased CDDP resistance | Zhou [55] | 2015 |
| miR-320a | ADAM10 | Down regulation | Increased CDDP sensitivity | Ge [56] | 2017 |
| miR-876-3p | TMED3 | Down regulation | Increased CDDP sensitivity | Peng [57] | 2019 |
| HOTTIP | miR-218 | Down regulation | Increased CDDP resistance | Wang [58] | 2019 |
| miR-138-5p | ERCC1 and ERCC4 | Down regulation | Increased CDDP sensitivity | Ning [59] | 2019 |
| Circ-0026359 | miR-1200 | Down regulation | Increased CDDP resistance | Zhang [60] | 2020 |
Signaling pathways
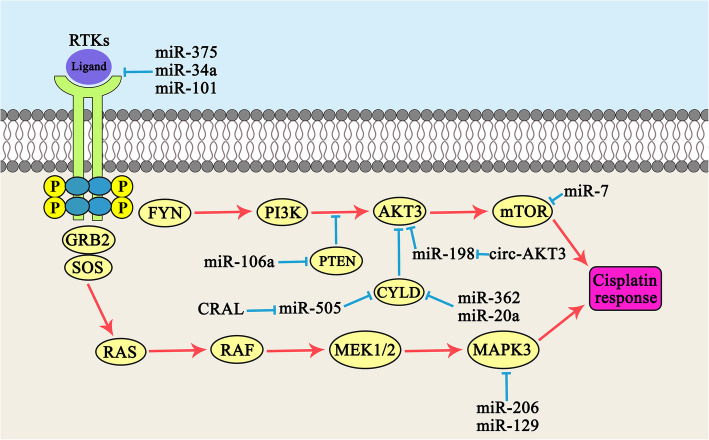
MiRNAs are involved in regulation of cisplatin response in gastric tumor cells via MAPK and PI3K/AKT signaling pathways (Fig. 2). The PI3K/AKT is a critical cell survival signaling pathway that promotes apoptosis resistance [61, 62]. PTEN is a tumor suppressor that negatively modulates the activity of PI3K/AKT pathway and is also implicated in tumorigenesis and chemo resistance [63]. PTEN inhibits the activity of PI3K through PIP3 dephosphorylation [64]. An association was observed between the miR-106a ectopic expression and the resultant down regulation of PTEN which suggested the role of aberrant miR-106a expression in cisplatin-resistance of SGC7901/DDP cells. The findings also indicated that cisplatin-resistant SGC7901/DDP cells had significantly higher miR-106a expression levels as compared to parental SGC7901 cells [15]. It has been observed that there was a significant miR-21 up regulation in CDDP-resistant GC cells compared with their parental. MiR-21 increased CDDP resistance via PTEN targeting through the activation of PI3K/AKT pathway [16]. Long noncoding RNAs (lncRNAs) are a group of noncoding RNA longer than 200 bp that can be oncogene or tumor suppressor [65, 66]. They function as decoys for miRNAs or proteins [67]. LncRNAs act as inhibitors of miRNAs via absorbing and suppressing the miRNAs from binding to their target mRNAs in a process that is known as sponging. Cancer susceptibility candidate 2 (CASC2) is a lncRNA that increased CDDP sensitivity by miR-21 sponging and PTEN up regulation [68]. It has been shown that there were significant CASC2 down regulations in CDDP resistant GC tissues and cells. CASC2 increased CDDP sensitivity through miR-19a sponging in GC [69]. AKT is a serine/threonine kinase and the primary downstream effector of PI3K signaling which is involved in tumor progression and disease-free survival [70, 71]. The miR-195 exerts its suppressive effects on proliferation, invasion, and migration of GC cells via AKT3 targeting and suppression of PI3K/AKT pathway. It also increased cisplatin sensitivity in GC cells. Patients with a high miR-195 expression level had significantly longer progression-free survival (PFS) compared with those with miR-195 under expression [17]. MiR-34a enhanced cisplatin-sensitivity in GC cells through modulating PI3K/AKT/BIRC5 pathway [72]. Cylindromatosis (CYLD) functions as a deubiquitinating enzyme and exerts a tumor-suppressive role in multiple malignancies [73, 74]. CYLD is the negative modulator of PI3K/AKT/NF-κB axis and is also implicated in regulation of tumor cell apoptosis [75, 76]. The suppression of PI3K/AKT signaling pathway is effective in attenuating the chemo-resistance of GC cells [77, 78]. It has been observed that Cisplatin Resistance-Associated lncRNA (CRAL) up regulated CYLD and inhibited PI3K/AKT pathway through miR-505 targeting that regulated cisplatin response in GC cells [18]. mTOR belongs to the PIKKs family of serine/threonine protein kinases and is one of the effectors of PI3K/AKT pathway. It has key roles in anabolic cell metabolism through enhancing mRNA translocation and protein synthesis and is also implicated in glucose metabolism and lipid biosynthesis [79, 80]. It was found that gastric cancer tissues had significantly lower levels of miR-7 expressions compared to normal margins. MiR-7 suppressed GC cell proliferation and invasion through targeting mTOR. It also attenuated cisplatin-resistance via suppressing mTOR in GC cells [19].
Fig. 2.
Role of miRNAs in regulation of cisplatin response in gastric tumor cells via MAPK and PI3K/AKT signaling pathways
Circular RNAs (circRNAs) are a group of non-coding RNAs (ncRNAs) forming a covalently closed continuous loop that lacks the 5’ and 3’ terminal nucleotide sequences. CircRNAs are implicated in transcriptional and post-transcriptional regulations [81]. CircRNAs can sponge miRNAs and inhibit their activity through functioning as competitive endogenous RNAs (ceRNAs). As a result, they affects different biological processes that are regulated by miRNAs. The inhibition of miRNAs by circRNAs suggests a novel mechanism of how miRNA’s activity is modulated and extends our understandings of circRNAs mode of action [81]. Activation of the PI3K/AKT signaling pathway enhances cell viability and inhibits apoptosis; therefore, this pathway plays a critical role in the chemo resistance of tumor cells [82, 83]. Induction of PI3K/AKT pathway prevents CASP3 activation by promoting its phosphorylation, thereby suppresses cell apoptosis [84]. It has been observed that there was significant circ-AKT3 up regulation in cisplatin-resistant compared to cisplatin-sensitive GC tissues and cells. Circ-AKT3 facilitated cisplatin-resistance in GC cells via sponging miR-198 and activating the PI3K/AKT signaling pathway [6]. ERBB2 belongs to the epidermal growth factor receptor family and a positive regulator of tumor cell proliferation [85]. Since, PI3K/AKT signaling is implicated in regulation of ERBB2; the induction of ERBB2/PI3K/AKT axis is correlated with chemo resistance [86]. There was a significant miR-375 down regulation in SGC7901/DDP cells compared to parental SGC7901 cells. The protein levels of both ERBB2 and p-AKT were suppressed in response to miR-375 overexpression. Therefore, it was hypothesized that miR-375 enhanced SGC7901/DDP cisplatin-sensitivity via targeting ERBB2/PI3K/AKT axis [20]. VEGF-C is a growth factor belonging to the VEGF family that promotes angiogenesis along with permeability of blood vessels via direct activation of VEGFR2/3 [87]. VEGF-C/VEGFR3 are pivotal regulators of lymphendothelial function by PI3K/AKT pathway [88]. There was significant miR-101 down regulation in GC cell lines in comparison with normal gastric epithelial cells. MiR-101 reduced cell proliferation while increased cisplatin-induced apoptosis in SGC7901/DDP cells via targeting VEGF-C [21].
NF-κB signaling inhibits apoptosis via transcriptional regulation of apoptotic related genes such as TRAF1, TRAF2, c-IAP1, and BCL-2 [89]. NF-κB is a heterodimer protein complex consisting of p65 and p50 subunits which are sequestered by a family of inhibitors called IκBs in the cytoplasm of unstimulated cells. Following the signal-induced degradation of IκB proteins, the NF-κB complex is activated and enters the nucleus to stimulate the transcription of multiple target genes. CYLD is a tumor suppressor deubiquitinating enzyme that functions as an inverse modulator of the NFκB signaling pathway and is implicated in the regulation of tumor cell apoptosis [90]. NFκB inhibition promotes chemo sensitivity in GC cells [22, 91]. BIRC5 and Livin are the members of inhibitor of apoptosis (IAP) family that inhibit caspase activation. It has been shown that there was a significant miR-20a up regulation in cisplatin resistant GC plasma and tissue samples. MiR-30a potentially was contributed to cisplatin resistance in GC through down regulating CYLD which leads to NFκB activation and up regulation of the downstream targets such as BIRC5 and Livin [23]. MiR-362 over expression was observed in gastric tumor tissues and cell lines. The results showed that miR-362 promoted the gastric tumor cell proliferation and resistance to cisplatin-induced apoptosis. Furthermore, miR-362 induced the activity of the NF-κB pathway in GC cells via targeting and down regulating CYLD [22]. NFKB inhibitor beta (NFKBIB) belongs to the NFκB inhibitor family [92]. Persistent activation of NF-κB and over expression of drug resistance-related proteins such as MDR1, MCL-1, BCL-2, and XIAP have critical functions in mediating chemo-resistance in various tumors [93–95]. Survivin and livin are two anti-apoptotic proteins belonging to the inhibitor of apoptosis (IAP) family which block caspase activity and inhibit cell apoptosis [96]. It has been observed that there was significant miR-20a up regulation in GC plasma and tissue samples with cisplatin-resistance. MiR-20a promoted cisplatin-resistance of GC cells through NFκB activation and up regulations of survivin and livin following NFKBIB targeting [24]. Chemokines are small molecules involved in regulation of various cellular processes through binding with G-protein-coupled receptors [97]. CCL22 is a member of the CC chemokine family which is secreted by dendritic cells and macrophages and is linked with tumor immune infiltration via CCR4 receptor [98, 99]. Chemokine receptor CCR4 promotes tumor invasion by activating ERK/NF-κB/MMP13 axis [100]. It decreases anti-tumor immunity in GC, while enhances the immunosuppression of regulatory T lymphocytes, and contributes to the CRC chemo-resistance [101, 102]. It has been reported that miR-130a-5p promoted cisplatin-sensitivity and suppressed malignant progression of the gastric tumor through CCL22 inhibition [25].
The ERK/MAPK signaling pathway is involved in regulation of multiple physiological and pathological processes such as cell proliferation, apoptosis, tumorigenesis, and chemo resistance [103–105]. MAPK is a serine/threonine kinase, which acts in the nuclear translocation of cytosolic proteins through phosphorylation, thereby regulating the functions of nuclear C-fos or C- transcription factors via phosphorylation [106, 107]. There was a significant MAPK3 up regulation in GC tissues compared with normal margins. MiR-206 reduced cisplatin-resistance in GC cells through MAPK3 targeting. It also decreased cell proliferation and increased apoptosis in GC cells [26]. MiR-129 reduced cell proliferation and CDDP resistance via MAPK3 targeting in GC cells [27]. Transforming growth factor beta (TGFB) signaling is involved in embryogenesis, cell differentiation, and apoptosis via activation of SMAD protein complexes that act as transcription factors to modulate the expression of target genes. There was an inverse association between the levels of miR-187 expression and cisplatin resistance in GC cells. The up regulation of miR-187 suppressed the expression of TGF-β1, p-Smad4, ERCC3, and ERCC4 in GC cells implying the role of the TGF-β/Smad axis in miR-187-regulated cisplatin-resistance [108].
Autophagy and apoptosis
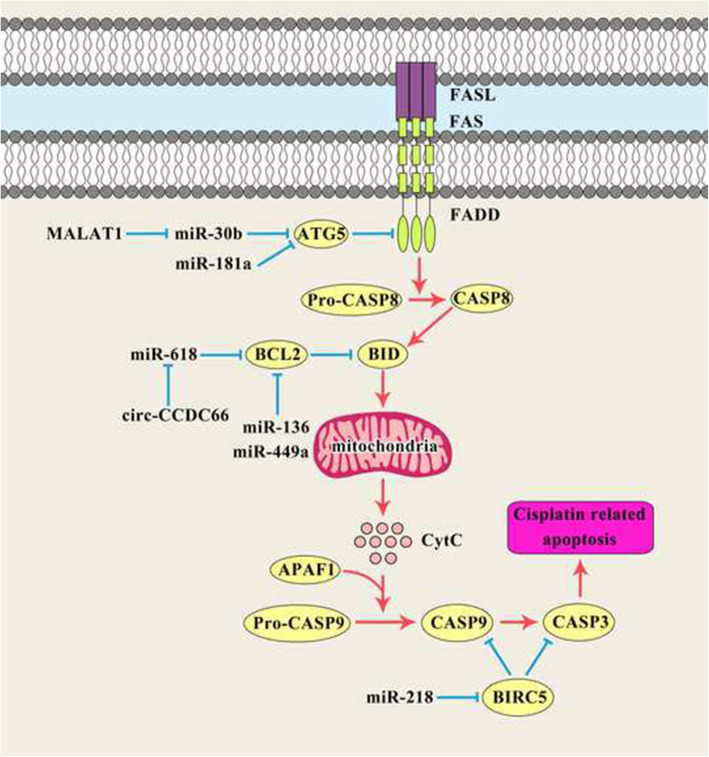
Autophagy is an intracellular self-digesting process for the regulation of cell homeostasis during which malformed or excessive proteins and long-lived or damaged organelles are removed by lysosomes [109]. While the majority of studies indicate that autophagy allows prolonged survival [110, 111], some others show that the autophagy enhances autophagic cell death [112]. Therefore, suppression of autophagy offers an efficient combined therapeutic approach for the regulation of chemo sensitivity in cancer. It has been reported that autophagy exerts oncogenic or tumor-suppressive functions during GC progression [113]. Cisplatin chemotherapy is believed to promote autophagy in some human malignancies [114, 115]. MiRNAs are involved in cisplatin response of gastric tumor cells by regulation of apoptosis and autophagy (Fig. 3). MiR-181a suppresses autophagy in cisplatin-resistant GC cells via ATG5 targeting [28]. MALAT1 and ATG5 improve chemo-resistance of cancer cells through increasing autophagy [116, 117]. It has been observed that there was significant MALAT1 up regulation in cisplatin-resistant GC cells. MALAT1 increased cisplatin-resistance via enhancing autophagic activity in AGS/CDDP gastric tumor cells. MiR-30b attenuated autophagy-related cisplatin-resistance through targeting ATG5 in cisplatin-resistant gastric tumor cells. MALAT1 Up regulated ATG5 in cisplatin-resistant GC cells via functioning as a ceRNA for miR-30b to sequester miR-30b from ATG5 [29].
Fig. 3.
miRNAs are involved in cisplatin response of gastric tumor cell by regulation of apoptosis and autophagy
Survivin belongs to the IAP family and is encoded by the BIRC5 gene [118]. Survivin exerts its pro-survival functions via inhibiting the activity of caspases [119]. It has been shown that survivin form a complex with hepatitis B X-interacting protein to regulate the activation of CASP9 [120]. Studies have also highlighted the role of survivin in anti-cancer chemotherapy resistance and have introduced survivin as a putative biomarker for predicting chemo resistance [120, 121]. It has been reported that there was a significant miR-218 down regulation in cisplatin-resistant SGC7901/DDP gastric tumor cells. MiR-218 promoted cisplatin-sensitivity in SGC7901/DDP cells via BIRC5 inhibition [30]. BCL2 is a negative regulator of apoptosis [122]. It has been reported that there was circCCDC66 up regulation in CDDP-resistant GC patients. Circ-CCDC66 induced CDDC resistance via miR-618 inhibition that resulted in BCL2 up regulation in gastric tumor cells [31]. MiR-136 and miR-449a also increased CDDP sensitivity of gastric tumor cells through BCL2 targeting [32, 33]. There was a significant miR-143 down regulation in cisplatin-resistant GC cell line. MiR-143 regulated cisplatin-resistance through IGF1R and BCL2 targeting. Cisplatin-resistant GC cells transfected with miR-143 mimics had reduced cell proliferation, while increased apoptosis [34]. Mitochondria function in energy production and also play key roles in cancer metabolic homeostasis [123]. Following exposure to anti-cancer drugs, the intrinsic apoptotic pathway is activated and leads to cancer cell death [124]. Mitochondrial fission is a crucial stage in the initiation of mitochondria-dependent apoptosis [125]. Dynamin-Related Protein 1 (DRP1) is considered as an upstream modulator of mitochondrial fission which controls and accomplishes the final part of mitochondrial fission [126]. Mitochondrial fission 1 protein (FIS1) functions as a receptor for recruiting DRP1 to mitochondria that is indirectly involved in mitochondrial fission. It has been demonstrated that FIS1 is implicated in CDDP-sensitivity in tongue squamous cell carcinoma [127]. AKAP1 functions as a scaffold that presents PKA to downstream targets at the mitochondrial membrane to regulate their phosphorylation state [128]. RAB12 belongs to the Ras family of oncogenes and is involved in the induction of autophagy through speeding up autolysosome maturation or suppression of mTORC1 activity [129, 130]. There were miR-148a-3p down regulations in cisplatin-resistant GC tissue and cell lines. MiR-148a-3p enhances cisplatin cytotoxicity in gastric tumor cells through targeted inhibition of RAB12 and AKAP1. In response to cisplatin treatment, miR-148a-3p promoted mitochondrial fission-induced apoptosis via targeting AKAP1 and increasing P53 and DRP1 activation [35]. SRSF2 belongs to the serine/arginine-rich protein family that is critical for splice-site selection during the alternative splicing process of mRNA precursors. CD44 is a GC stem cell marker that plays key roles in modulating tumorigenesis, self-renewal, distant metastasis, and chemo-resistance. It has been reported that miR-193a-3p was positively correlated with cisplatin resistance in CD44 + GC cells. There was inverse correlation between the levels of miR-193a-3p and SRSF2 expressions. CD44 + GC cells had high levels of BCL-2 expression, while low levels of CYCS, BAX, CASP3, and CASP9 expressions compared with CD44- cells. MiR-193a-3p suppressed CDDP-induced mitochondrial cell death in CD44 + GC cells [36].
Epithelial-mesenchymal transition
Epithelial-mesenchymal transition (EMT) is a cellular process in which the epithelial cells lose their cell adhesion and polarity to convert into the invasive mesenchymal cells. It is a pivotal cellular mechanism during tumor growth, metastasis, and chemo resistance [131–134]. The expressions of epithelial genes such as CDH1, ZO-1, and OCLN are suppressed during EMT; while, mesenchymal markers including CDH2, α-SMA, FN, and VIM are overexpressed during this process [135, 136]. The initiation of EMT occurs following the up regulation of its related transcription factors such as SNAI1, TWIST1, and ZEB1 [137]. ZEB2 is a key transcription factor that belongs to the Snail superfamily and is normally expressed in human tissues. ZEB2 suppresses the transcription of CDH1, cytokeratin, MAC1, and mucin proteins through binding to the E-box sequence in the promoter region of target genes. Low expression levels of the above-mentioned proteins are associated with the EMT [138]. ZEB2 expression is also associated with resistance to cisplatin and EGFR inhibitors in ovarian and bladder cancers, respectively [139, 140]. It has been observed that there was significant miR-200c down regulation in GC tissues and SGC7901/DDP cells in comparison with normal margins and parental SGC7901 cells. MiR-200c sensitized GC cells to cisplatin treatment via ZEB2 targeting [37]. There was PCAT-1 up regulation in CDDP-resistant gastric tumor cells. PCAT-1 increased CDDP resistance by of GC cells through miR-128 sponging that resulted in increased levels of ZEB1 expression in gastric tumor cells [38]. It has been observed that miR-574-3p suppressed EMT via ZEB1 targeting. MiR-574-3p also suppressed cisplatin resistance in GC cells both in vivo and in vitro [39]. Another study have reported that there was a significant miR-30a down regulation in cisplatin-resistant GC compared with sensitive tissues. Cisplatin-resistant (SGC7901/DDP) cells also showed significantly lower miR-30a expression levels than the cisplatin-sensitive SGC7901 cells. Morphological features were also different between SGC7901/DDP (extended fibroblastoid-like) and SGC7901 (epithelial-like) cell lines. There were SNAI1 and VIM up regulations in SGC-7901/DDP cells in comparison with SGC-7901 cells. MiR-30a suppressed SNAI1 and VIM expressions and induced morphological changes from an elongated fibroblastoid-like to an epithelial-like morphology in SGC-7901/DDP cells [40]. Death effector domain-containing protein (DEDD) plays important roles in multiple cellular processes such as cell cycle, mitosis, and apoptosis [141]. There is a correlation between EMT and DEDD in various cancers [136, 142]. The findings revealed that miR-17 enhanced EMT and cisplatin-resistance through targeting DEDD in GC cells [41].
Drug efflux
There are various mechanisms involved in development of multidrug resistance (MDR) in tumor cells including DNA repair ability, cell cycle regulation, drug efflux/uptake, and detoxification agents [143–145]. P-glycoprotein (P-gp) is a trans membrane protein encoded by the MDR1 gene that is widely expressed in normal tissues [146]. P-gp is an important ATP-dependent drug efflux transporter playing a critical role in tissue homeostasis, detoxification, and protection against hazardous metabolites. Up regulation of P-gp in tumor cells occurs following exposure to chemotherapy that is observed in almost 30 % of cancer cases [147]. It has been reported that miR-129 down regulated P-gp and activated intrinsic apoptosis pathway via the over expressions of CASP9 and CASP3. It was also found that miR-129 was markedly down regulated in cisplatin-resistant vs. cisplatin-sensitive clinical specimens. MiR-129 reduced cisplatin resistance of GC cells through targeting P-gp [42]. There was a significant miR-30a down regulation in SGC7901/CDDP cells. MiR-30a exerted its inhibitory functions on tumor cell proliferation and cisplatin-resistance via MDR1 targeting [43]. HOTAIR is a lncRNA transcribed from the homeobox C (HOXC) gene [148]. It has been reported that the up regulation of HOTAIR was associated with chemo resistance in various cancers through different mechanisms. HOTAIR promotes chemo resistance of ovarian cancer via WNT pathway activation [149]. It also contributes to tamoxifen resistance in breast cancer through inducing estrogen receptor (ER) signaling [150]. It has been shown that there were HOTAIR up regulations in GC tissues and cisplatin-resistant GC cells compared with normal margins and control cells. HOTAIR promoted resistance to cisplatin treatment in GC cells via miR-126 sponging that resulted in increased activity of PI3K/AKT/MRP1 axis [44]. HOTAIR up regulation was observed in GC tissues and cells compared with normal margins and cell lines. There was also an inverse correlation between the levels of HOTAIR and miR-34a expressions in GC tissues. HOTAIR knockdown suppressed cisplatin-resistance in gastric tumor cells through miR-34a up regulation [45]. Cancer stem cells (CSCs) are a subpopulation of highly oncogenic and chemo resistant tumor cells that have an important role in tumor progression and recurrence [151]. It has been reported that Lgr5 + GC stem cells (GCSCs) had significantly higher levels of miR-132 compared with Lrg5- cells. MiR-132 also increased cisplatin-resistance in Lgr5 + GCSCs both in vitro and in vivo. There was an inverse correlation between the levels of SIRT1 and miR-132 expressions in GC samples. Silencing of SIRT1 led to the up regulation of ABCG2 through the induction of CREB acetylation [46].
Protein kinases
Aurora kinase B (AURKB) belongs to the serine/threonine protein kinases that is implicated in regulation of mitosis and chromosome segregation [152]. AURKB phosphorylates VIM to regulate vimentin filament segregation during cytokinesis. Patients with up regulation of AURKB experience higher overall survival rates compared with those with AURKB down regulation [153]. There was a significant let-7b down regulation in cisplatin-resistant GC cells compared with parental cells. Let-7b induced sensitivity to cisplatin and suppressed tumor growth through targeting AURKB [47]. CDK6 belongs to the cyclin-dependent kinase (CDK) family and is implicated in different biological and pathological processes including cell cycle progression, centrosome stability, apoptosis, angiogenesis, and chemo-resistance [154, 155]. It was found that hsa-circ-0081143 enhanced gastric tumor cell proliferation and chemo-resistance through miR-646/CDK6/KLF5 axis. Hsa-circ-0081143 was significantly overexpressed in GC patients which was associated with higher tumor, nodes, metastases (TNM) stage and poorer prognosis. Inhibition of hsa-circ-0081143 suppressed GC progression, induced apoptosis, and attenuated their cisplatin resistance. There was a significant inverse association between the miR-646 and hsa-circ-0081143 expressions. The hsa-circ-0081143 positively regulated CDK6 expression through targeting miR-646 [48]. In contrast with the normal cells which tend to shift to the aerobic respiration pathway under oxygen availability, tumor cells boost their glucose uptake and continue to undergo glycolysis. Dysregulation of glycolysis is implicated in chemo-resistance [156]. Pyruvate dehydrogenase kinase 1 (PDK1) as a major glycolytic enzyme, inhibits pyruvate oxidation through inactivation of pyruvate dehydrogenase [157]. There was a miR-4290 down regulation in GC tissues which was correlated with advanced TNM stage and poor prognosis. MiR-4290 inhibited glycolysis through directly targeting PDK1 that increased cisplatin sensitivity in GC cells [49]. Receptor tyrosine kinases (RTKs) are activated by growth factors, hormones, and cytokines. They are also key factors during tumor progression and metastasis. Discoidin domain receptor 1 (DDR1) is a RTK which is up regulated in multiple human cancers and is a putative regulator of various biological processes in tumor cells [158, 159]. It functions as a collagen receptor to regulate the ECM remodeling, cell migration, and proliferation via MAPK signaling pathway. DDR1 regulates ECM remodeling by MMPs up regulations to increase cell migration. DDR1 over expression was correlated with unfavorable prognoses [160]. It has been observed that there were significant miR-221-5p down regulations in GC tissues and cells. MiR-221-5p promoted apoptosis and cisplatin-sensitivity, while inhibited gastric tumor cell proliferation and migration through DDR1 targeting [50]. MET is also another RTK that is activated by hepatocyte growth factor (HGF) to regulate cell proliferation and survival. Activated MET interacts with various cytoplasmic downstream proteins such as PLCG1 and SRC which resulted in activation of RAS/ERK and PI3K/AKT signaling pathways. It has been reported that there was significant miR-34a down regulation in cisplatin-resistant GC tissues and cells. MiR-34a reduced cell proliferation, while promoted apoptosis of cisplatin-resistant GC cells by MET targeting [7].
Transcription factors
Forkhead box O3a (FOXO3a) is a transcription factor involved in regulation of cell cycle progression, autophagy, and apoptosis [161, 162]. FOXO3a suppresses cell cycle progression via p27Kip1 regulation [163]. There was a significant miR-25 up regulation in cisplatin-resistant GC cells. MiR-25 was associated with reduced sensitivity of SGC-7901 cells to cisplatin treatment by FOXO3a targeting. MiR-25 silencing in cisplatin-resistant SGC-7901/DDP cells led to cell cycle arrest through p27Kip1 down regulation [164]. SOX9 is a developmental transcription factor widely expressed in various embryonic tissues [165, 166]. There were miR-613 down regulations in GC tissues and cell lines. MiR-613 sensitized GC cells to cisplatin, while inhibited cell proliferation, cell cycle progression, and migration through SOX9 targeting [51]. Hypoxia is a frequent process in tumor cells that is associated with pathological processes such as tumorigenesis, angiogenesis, migration, and chemo resistance [167, 168]. HIF-1α is regarded as the main transcription factor involved in regulation of signaling pathways related to cell cycle, apoptosis, and metastasis that results in transformation of cancer cells to a more malignant phenotype under hypoxic conditions [169]. Up regulation of miR-421 in gastric tumor cells suppresses apoptosis, enhances metastasis, and promotes cisplatin-resistance via CASP3 and CDH1 targeting. HIF-1α-induced-miR-421 is implicated in modulation of chemo-resistance and apoptosis suppression. MiR-421 up regulated CDH2, VIM, SNAI1, Twist, SNAI2, MMP-2, while inhibited CDH1 in GC cells [52]. BMI1 is a component of polycomb repressor complex 1 (PRC1) that functions as an oncogene through modulating the cell proliferation, apoptosis, and invasion of tumor cells. Deregulation of BMI1 has been associated with tumorigenesis and chemo resistance in multiple malignancies [170, 171]. It has been reported that there was circDONSON up regulation in cisplatin-resistant GC tissues and cell lines. CircDONSON knockdown enhanced tumor cell apoptosis, reduced cell viability, and attenuated cisplatin-resistance in vitro. CircDONSON indirectly regulated the BMI1 expression levels via sponging miR-802 in GC cells [53].
Structural and DNA repair factors
Annexin A2 (ANXA2) is a calcium-dependent phospholipid-binding membrane protein involved in linkage of protein complexes to the cytoskeletal components [172]. It also regulates cell proliferation, differentiation, and migration in different cancers [173, 174]. It has been shown that there were miR-101 down regulations in GC cells and tissues. MiR-101 increased cisplatin sensitivity of GC cells through ANXA2 targeting [54]. FBXW7 is the substrate detection component of a SCF complex that mediates degradation of various target proteins such as NOTCH1, NOTCH2, and JNK [175, 176]. MiR-223 enhanced cisplatin-resistance of GC by targeting FBXW7 [55]. ADAM metallopeptidase domain 10 (ADAM10) is a metalloproteinase that is frequently up regulated in human tumors which triggers cancer initiation, progression, and metastasis [177, 178]. ADAM10 expression in non-small cell lung cancer and pituitary adenoma contributes to cell migration through activating the NOTCH signaling pathway and regulating the cleavage of CD44, respectively [179, 180]. Moreover, the expression of ADAM10 is correlated with the invasive behavior of cancer cells in pancreatic carcinoma and oral squamous cell carcinoma [181, 182]. It has been reported that there were miR-320 down regulations in GC tissues and cells. MiR-320a enhanced cisplatin-sensitivity in GC cells through ADAM10 suppression [56]. TMED3 is a trans-membrane protein involved in vesicular protein trafficking and Golgi protein modification. There were positive correlations between miR-876-3p down regulation and clinicopathological features including poorer prognoses and shorter overall survival rates of GC patients. MiR-876-3p modulated cisplatin sensitivity and stem cell-like morphology and properties of gastric tumor cells via targeting TMED3. Down regulation of miR-876-3p was more prominent in chemo-resistant than chemo-sensitive GC tissues [57]. HMGA1 is a chromatin-associated protein involved in regulation gene expression. There was HOTTIP up regulation in cisplatin-resistant GC cells. HOTTIP promoted cisplatin resistance through activating HMGA1 via functioning as a ceRNA for miR-218 [58]. ERCC is a pivotal member of nuclear excision repair (NER) that is considered as the principal cause of cisplatin resistance in tumor cells. ERCC1 and ERCC4 heterodimer possesses a potential endonuclease activity to excise the 5’ side of DNA damage during NER process. MiR-138-5p regulates cisplatin-sensitivity of GC cells through ERCC1 and ERCC4 targeting [59]. POLD4 is a component of DNA polymerase delta that has pivotal roles in DNA replication and repair via regulation of POLD1 and 3’ to 5’ proofreading activity [183]. Gastric tumor tissues and cell lines had significant circ_0026359 up regulations compared with normal margins and cell lines. There were significant circ_0026359 up regulations in cisplatin resistant GC tissues and cell lines. Circ_0026359 up regulation was positively correlated with shorter overall and relapse-free survival rates in GC patients. Depletion of circ_0026359 attenuated cisplatin-resistance of gastric tumor cells through inducing miR-1200 activity and down regulating POLD4 [60].
Perspective and hurdles
Direct assessment of tissue biomarkers improves tumor diagnosis, however, the invasive procedures required to obtain tumor biopsies limit their application. An alternative approach is to study miRNAs in body fluids, such as blood, which are also deregulated in cancer [184] and, furthermore, have the distinct advantage of requiring a much less invasive procedure for sample collection. Circulating miRNAs are also stable in different pH conditions, repeated freeze-thawing, and room temperature which support the miRNAs as diagnostic and prognostic molecular biomarkers [185, 186]. Regarding the small size and low concentration of circulating miRNAs in body fluids, accurate quantification should be done by various molecular techniques such as qRT-PCR, Microarray platforms, and Next-generation sequencing [187, 188]. Some of the miRNAs have been reported to affect anticancer drug response [189–191]. Although, single miRNA can be used to predict the chemotherapeutic response, a panel of miRNAs signature is more efficient. A signature of several miRNAs was associated with CDDP and 5-fluorouracil resistances in GC patients [192]. Therefore, drug responses can be affected by the manipulation of miRNA levels. The miRNA-based treatment is the suppression of deregulated miRNAs using miRNA sponges and anti-miRNA oligonucleotides [193]. Since, miRNAs affect the target mRNAs by partial sequence complementarity, the off-target effect results in unfavorable immune response and side effects. They are also sensitive toward the cellular nucleases that prevent the development of efficient miRNA-based therapies. Moreover, antimiRs may also negatively affect physiological functions that are normally regulated by the target miRNAs. Therefore, site specific, lowest optimum concentration, and delivery systems are pivotal factors to obtain the best therapeutic results with the lowest side effects in miRNA based treatment [194]. MiRNA mimics can be delivered by vectors, nanocarriers, and amphiphilic star copolymer [195–197]. Nanoparticle delivery of synthetic anti oncomiRs or synthetic tumor suppressor miRNAs in combination with chemotherapeutic drugs has been reported in cancer therapy [198]. It is required to clarify the molecular interactions of the miRNAs involved in regulation of CDDP response to introduce a prognostic miRNA panel marker for the prediction of CDDP response. For the first time, present review uncovers the molecular interactions of miRNAs during GC progression.
Conclusions
Since, there is a high rate of recurrence after CDDP treatment in GC patients; it is required to clarify the molecular mechanisms associated with CDDP resistance to introduce novel therapeutic methods. MicroRNAs are a class of endogenous non-coding RNAs involved in chemo resistance of GC cells through regulation of all of the MDR mechanisms. In present review we have summarized all of the miRNAs associated with cisplatin resistance based on their target genes and molecular mechanisms in gastric tumor cells. This review paves the way of introducing a miRNA-based panel of prognostic markers to improve the efficacy of chemotherapy and clinical outcomes in GC patients. It was observed that miRNAs are mainly involved in cisplatin response of gastric tumor cells via regulation of signaling pathways, autophagy, and apoptosis.
Acknowledgements
Not applicable.
Abbreviations
- GC
Gastric cancer
- lncRNAs
Long noncoding RNAs
- CASC2
Cancer susceptibility candidate 2
- PFS
Progression-free survival
- circRNAs
Circular RNAs
- ncRNAs
Non-coding RNAs
- ceRNAs
Competitive endogenous RNAs
- CYLD
Cylindromatosis
- NFKBIB
NFKB inhibitor beta
- IAP
Inhibitor of apoptosis
- TGFB
Transforming growth factor beta
- DRP1
Dynamin-Related Protein 1
- EMT
Epithelial-mesenchymal transition
- DEDD
Death effector domain-containing protein
- MDR
Multidrug resistance
- P-gp
P-glycoprotein
- HOXC
Homeobox C
- ER
Estrogen receptor
- CSCs
Cancer stem cell-like cells
- GCSCs
GC stem cells
- AURKB
Aurora kinase B
- CDK
Cyclin-dependent kinase
- PDK1
Pyruvate dehydrogenase kinase 1
- RTKs
Receptor tyrosine kinases
- DDR1
Discoidin domain receptor 1
- HGF
Hepatocyte growth factor
- FOXO3a
Forkhead box O3a
- PRC1
Polycomb repressor complex 1
- ANXA2
Annexin A2
- ADAM10
ADAM metallopeptidase domain 10
- NER
Nuclear excision repair
- CRAL
Cisplatin Resistance-Associated lncRNA
- TNM
Tumor, nodes, metastases
Authors’ contributions
ASZ was involved in search strategy and drafting. MM supervised the project and revised and edited the manuscript. All authors read and approved the final manuscript.
Funding
Not applicable.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Amir Sadra Zangouei, Email: zangoueias961@mums.ac.ir.
Meysam Moghbeli, Email: moghbelim@mums.ac.ir, Email: Meysam_moghbeli@yahoo.com.
References
- 1.Bray F, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Cunningham D, et al. Perioperative chemotherapy versus surgery alone for resectable gastroesophageal cancer. N Engl J Med. 2006;355(1):11–20. doi: 10.1056/NEJMoa055531. [DOI] [PubMed] [Google Scholar]
- 3.Bang Y-J, et al. Adjuvant capecitabine and oxaliplatin for gastric cancer after D2 gastrectomy (CLASSIC): a phase 3 open-label, randomised controlled trial. Lancet. 2012;379(9813):315–21. doi: 10.1016/S0140-6736(11)61873-4. [DOI] [PubMed] [Google Scholar]
- 4.De Vita F, et al. Human epidermal growth factor receptor 2 (HER2) in gastric cancer: a new therapeutic target. Cancer Treat Rev. 2010;36:S11–5. doi: 10.1016/S0305-7372(10)70014-1. [DOI] [PubMed] [Google Scholar]
- 5.Wang S, et al. PRDX2 protects against oxidative stress induced by H. pylori and promotes resistance to cisplatin in gastric cancer. Redox Biol. 2020;28:101319. doi: 10.1016/j.redox.2019.101319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang X, et al. Circular RNA AKT3 upregulates PIK3R1 to enhance cisplatin resistance in gastric cancer via miR-198 suppression. Mol Cancer. 2019;18(1):71. doi: 10.1186/s12943-019-0969-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang Z, et al. Upregulation of microRNA-34a enhances the DDP sensitivity of gastric cancer cells by modulating proliferation and apoptosis via targeting MET. Oncol Rep. 2016;36(4):2391–7. doi: 10.3892/or.2016.5016. [DOI] [PubMed] [Google Scholar]
- 8.Kostova IJ, R.p.o.a.-c.d.d. Platinum complexes as anticancer agents. 2006;1(1):1–22. doi: 10.2174/157489206775246458. [DOI] [PubMed] [Google Scholar]
- 9.Gong J, et al., Microparticles and their emerging role in cancer multidrug resistance. 2012; 38(3): p. 226–234. [DOI] [PubMed]
- 10.Wang S-F, et al. Activated integrated stress response induced by salubrinal promotes cisplatin resistance in human gastric cancer cells via enhanced xCT expression and glutathione biosynthesis. Int J Mol Sci. 2018;19(11):3389. doi: 10.3390/ijms19113389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.An Y, et al. miR-23b-3p regulates the chemoresistance of gastric cancer cells by targeting ATG12 and HMGB2. Cell Death Dis. 2015;6(5):e1766–6. doi: 10.1038/cddis.2015.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Calin GA, Croce C.M.J.N.r.c. Croce, MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6(11):857–66. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 13.Tamagawa S, et al. Role of miR-200c/miR-141 in the regulation of epithelial-mesenchymal transition and migration in head and neck squamous cell carcinoma. Int J Mol Med. 2014;33(4):879–86. doi: 10.3892/ijmm.2014.1625. [DOI] [PubMed] [Google Scholar]
- 14.Hummel R, Hussey DJ, Haier J.E.j.o.c. MicroRNAs: predictors and modifiers of chemo-and radiotherapy in different tumour types. Eur J Cancer. 2010;46(2):298–311. doi: 10.1016/j.ejca.2009.10.027. [DOI] [PubMed] [Google Scholar]
- 15.Fang Y, et al. miR-106a confers cisplatin resistance by regulating PTEN/Akt pathway in gastric cancer cells. Acta Biochim Biophys Sin (Shanghai) 2013;45(11):963–72. doi: 10.1093/abbs/gmt106. [DOI] [PubMed] [Google Scholar]
- 16.Yang S-m, et al. miR-21 confers cisplatin resistance in gastric cancer cells by regulating PTEN. Toxicology. 2013;306:162–8. doi: 10.1016/j.tox.2013.02.014. [DOI] [PubMed] [Google Scholar]
- 17.Ye R, Wei B, Li S, Liu W, Liu J, Qiu L, Wu X, Zhao Z, Li J. Expression of miR-195 is associated with chemotherapy sensitivity of cisplatin and clinical prognosis in gastric cancer. Oncotarget. 2017;8(57):97260. [DOI] [PMC free article] [PubMed]
- 18.Wang Z, et al. The long noncoding RNA CRAL reverses cisplatin resistance via the miR-505/CYLD/AKT axis in human gastric cancer cells. RNA Biol. 2020;17(11):1–14. doi: 10.1080/15476286.2020.1850628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu N, Lian YJ, Dai X, Wang YJ. miR-7 increases cisplatin sensitivity of gastric cancer cells through suppressing mTOR. Technol Cancer Res Treat. 2017;16(6):1022-30. [DOI] [PMC free article] [PubMed]
- 20.Zhou N, et al. Upregulation of microRNA–375 increases the cisplatin–sensitivity of human gastric cancer cells by regulating ERBB2. Exp Ther Med. 2016;11(2):625–30. doi: 10.3892/etm.2015.2920. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21.Li G, et al. MicroRNA-101 induces apoptosis in cisplatin-resistant gastric cancer cells by targeting VEGF-C. Mol Med Rep. 2016;13(1):572–8. doi: 10.3892/mmr.2015.4560. [DOI] [PubMed] [Google Scholar]
- 22.Xia JT, Chen LZ, Jian WH, Wang KB, Yang YZ, He WL, He YL, Chen D, Li W. MicroRNA-362 induces cell proliferation and apoptosis resistance in gastric cancer by activation of NF-κB signaling. J Transl Med. 2014;12(1):1-12. [DOI] [PMC free article] [PubMed]
- 23.Zhu M, et al. miR-20a induces cisplatin resistance of a human gastric cancer cell line via targeting CYLD. Mol Med Rep. 2016;14(2):1742–50. doi: 10.3892/mmr.2016.5413. [DOI] [PubMed] [Google Scholar]
- 24.Du Y, et al. miR-20a enhances cisplatin resistance of human gastric cancer cell line by targeting NFKBIB. Tumour Biol. 2016;37(1):1261–9. doi: 10.1007/s13277-015-3921-1. [DOI] [PubMed] [Google Scholar]
- 25.Fang QL, Li KC, Wang L, Gu XL, Song RJ, Lu S. Targeted Inhibition of CCL22 by miR-130a-5p can enhance the sensitivity of cisplatin-resistant gastric cancer cells to chemotherapy. Cancer Manage Res. 2020;12:3865-75. [DOI] [PMC free article] [PubMed]
- 26.Chen Z, et al. MicroRNA-206 facilitates gastric cancer cell apoptosis and suppresses cisplatin resistance by targeting MAPK2 signaling pathway. Eur Rev Med Pharmacol Sci. 2019;23(1):171–80. doi: 10.26355/eurrev_201901_16761. [DOI] [PubMed] [Google Scholar]
- 27.Cao H, et al. MiR-129 reduces CDDP resistance in gastric cancer cells by inhibiting MAPK3. Eur Rev Med Pharmacol Sci. 2019;23(15):6478–85. doi: 10.26355/eurrev_201908_18531. [DOI] [PubMed] [Google Scholar]
- 28.Zhao J, et al. MiR-181a suppresses autophagy and sensitizes gastric cancer cells to cisplatin. Gene. 2016;576(2):828–33. doi: 10.1016/j.gene.2015.11.013. [DOI] [PubMed] [Google Scholar]
- 29.Xi Z, Si J, Nan J. LncRNA MALAT1 potentiates autophagy-associated cisplatin resistance by regulating the microRNA-30b/autophagy-related gene 5 axis in gastric cancer. Intern J Oncolo. 2019;54(1):239-48. [DOI] [PubMed]
- 30.Zhang Z, et al. MicroRNA–218 enhances gastric cancer cell cisplatin sensitivity by targeting survivin. Exp Ther Med. 2018;16(6):4796–802. doi: 10.3892/etm.2018.6802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Q, et al. CircRNACCDC66 regulates cisplatin resistance in gastric cancer via the miR-618/BCL2 axis. Biochem Biophys Res Commun. 2020;526(3):713–20. doi: 10.1016/j.bbrc.2020.03.156. [DOI] [PubMed] [Google Scholar]
- 32.Yu L, Zhou G, D.J.E.R.M.P S, Li MiR-136 triggers apoptosis in human gastric cancer cells by targeting AEG-1 and BCL2. Eur Rev Med Pharmacol Sci. 2018;22(21):7251–6. doi: 10.26355/eurrev_201811_16259. [DOI] [PubMed] [Google Scholar]
- 33.Hu J, et al. miR-449a Regulates proliferation and chemosensitivity to cisplatin by targeting cyclin D1 and BCL2 in SGC7901 cells. Dig Dis Sci. 2014;59(2):336–45. doi: 10.1007/s10620-013-2923-3. [DOI] [PubMed] [Google Scholar]
- 34.Zhuang M, Shi Q, Zhang X, Ding Y, Shan L, Shan X, Qian J, Zhou X, Huang Z, Zhu W, Ding Y. Involvement of miR-143 in cisplatin resistance of gastric cancer cells via targeting IGF1R and BCL2. Tumor Biol. 2015;36(4):2737-45. [DOI] [PubMed]
- 35.Li B, Wang W, Li Z, Chen Z, Zhi X, Xu J, Li Q, Wang L, Huang X, Wang L, Wei S. MicroRNA-148a-3p enhances cisplatin cytotoxicity in gastric cancer through mitochondrial fission induction and cyto-protective autophagy suppression. Cancer Lett. 2017;410:212-27. [DOI] [PMC free article] [PubMed]
- 36.Lee SD, Yu D, Lee DY, Shin HS, Jo JH, Lee YC. Upregulated microRNA-193a-3p is responsible for cisplatin resistance in CD 44 (+) gastric cancer cells. Cancer Sci. 2019;110(2):662-73. [DOI] [PMC free article] [PubMed]
- 37.Jiang T, et al. MicroRNA-200c regulates cisplatin resistance by targeting ZEB2 in human gastric cancer cells. Oncol Rep. 2017;38(1):151–8. doi: 10.3892/or.2017.5659. [DOI] [PubMed] [Google Scholar]
- 38.Guo Y, Yue P, Wang Y, Chen G, Li Y. PCAT-1 contributes to cisplatin resistance in gastric cancer through miR-128/ZEB1 axis. Biomed Pharmacother. 2019;118:109255. [DOI] [PubMed]
- 39.Wang M, et al. MicroRNA-574-3p regulates epithelial mesenchymal transition and cisplatin resistance via targeting ZEB1 in human gastric carcinoma cells. Gene. 2019;700:110–9. doi: 10.1016/j.gene.2019.03.043. [DOI] [PubMed] [Google Scholar]
- 40.Wang LL, Zhang XH, Zhang X, Chu JK. MiR-30a increases cisplatin sensitivity of gastric cancer cells through suppressing epithelial-to-mesenchymal transition (EMT). Eur Rev Med Pharmacol Sci. 2016;20(9):1733-1739. [PubMed]
- 41.Wu DM, Hong XW, Wang LL, Cui XF, Lu J, Chen GQ, Zheng YL. MicroRNA-17 inhibition overcomes chemoresistance and suppresses epithelialmesenchymal transition through a DEDD-dependent mechanism in gastric cancer. Int J Biochem Cell Biol. 2018;102:59-70. [DOI] [PubMed]
- 42.Lu C, et al. MiR-129 regulates cisplatin-resistance in human gastric cancer cells by targeting P-gp. Biomed Pharmacother. 2017;86:450–6. doi: 10.1016/j.biopha.2016.11.139. [DOI] [PubMed] [Google Scholar]
- 43.Du X, et al. miR–30 decreases multidrug resistance in human gastric cancer cells by modulating cell autophagy. Exp Ther Med. 2018;15(1):599–605. doi: 10.3892/etm.2017.5354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yan J, et al. LncRNA HOTAIR promotes cisplatin resistance in gastric cancer by targeting miR-126 to activate the PI3K/AKT/MRP1 genes. Tumour Biol. 2016;37(12):16345–55. doi: 10.1007/s13277-016-5448-5. [DOI] [PubMed] [Google Scholar]
- 45.Cheng C, Qin Y, Zhi Q, Wang J, Qin C. Knockdown of long non-coding RNA HOTAIR inhibits cisplatin resistance of gastric cancer cells through inhibiting the PI3K/Akt and Wnt/β-catenin signaling pathways by up-regulating miR-34a. Int J Biol Macromol. 2018;107:2620-9. [DOI] [PubMed]
- 46.Zhang L, et al. Upregulated miR-132 in Lgr5 + gastric cancer stem cell‐like cells contributes to cisplatin‐resistance via SIRT1/CREB/ABCG2 signaling pathway. Mol Carcinog. 2017;56(9):2022–34. doi: 10.1002/mc.22656. [DOI] [PubMed] [Google Scholar]
- 47.Han X, et al. Let-7b attenuates cisplatin resistance and tumor growth in gastric cancer by targeting AURKB. Cancer Gene Ther. 2018;25(11):300–8. doi: 10.1038/s41417-018-0048-8. [DOI] [PubMed] [Google Scholar]
- 48.Xue M, et al. hsa_circ_0081143 promotes cisplatin resistance in gastric cancer by targeting miR-646/CDK6 pathway. Cancer Cell Int. 2019;19(1):25. doi: 10.1186/s12935-019-0737-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Qian Y, et al. MicroRNA-4290 suppresses PDK1-mediated glycolysis to enhance the sensitivity of gastric cancer cell to cisplatin. Braz J Med Biol Res. 2020;53(5):e9330. doi: 10.1590/1414-431x20209330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jiang X, et al. Promotion of miR-221-5p on the Sensitivity of Gastric Cancer Cells to Cisplatin and Its Effects on Cell Proliferation and Apoptosis by Regulating DDR1. Onco Targets Ther. 2020;13:2333. doi: 10.2147/OTT.S232953. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 51.Xue M, Li G, Sun P, Zhang D, Fang X, Li W. MicroRNA-613 induces the sensitivity of gastric cancer cells to cisplatin through targeting SOX9 expression. Am J Translat Res. 2019;11(2):885. [PMC free article] [PubMed]
- 52.Ge X, et al. MicroRNA-421 regulated by HIF-1α promotes metastasis, inhibits apoptosis, and induces cisplatin resistance by targeting E-cadherin and caspase-3 in. gastric cancer. 2016;7(17):24466. [DOI] [PMC free article] [PubMed]
- 53.Liu Y, et al. CircRNA DONSON contributes to cisplatin resistance in gastric cancer cells by regulating miR-802/BMI1 axis. Cancer Cell Int. 2020;20(1):1–12. doi: 10.1186/s12935-016-0378-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bao J, et al. miR-101 alleviates chemoresistance of gastric cancer cells by targeting ANXA2. Biomed Pharmacother. 2017;92:1030–7. doi: 10.1016/j.biopha.2017.06.011. [DOI] [PubMed] [Google Scholar]
- 55.Zhou X, et al. MiR-223 promotes the cisplatin resistance of human gastric cancer cells via regulating cell cycle by targeting FBXW7. J Exp Clin Cancer Res. 2015;34(1):28. doi: 10.1186/s13046-015-0145-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ge X, et al. miR-320a modulates cell growth and chemosensitivity via regulating ADAM10 in gastric cancer. Mol Med Rep. 2017;16(6):9664–70. doi: 10.3892/mmr.2017.7819. [DOI] [PubMed] [Google Scholar]
- 57.Peng C, Huang K, Liu G, Li Y, Yu C. MiR-876-3p regulates cisplatin resistance and stem cell‐like properties of gastric cancer cells by targeting TMED 3. J Gastroenterol Hepatol. 2019;34(10):1711-9. [DOI] [PubMed]
- 58.Wang J, et al. Exosome-mediated transfer of lncRNA HOTTIP promotes cisplatin resistance in gastric cancer cells by regulating HMGA1/miR-218 axis. Onco Targets Ther. 2019;12:11325. doi: 10.2147/OTT.S231846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ning J, Jiao Y, Xie X, Deng X, Zhang Y, Yang Y, Zhao C, Wang H, Gu K. miR-138-5p modulates the expression of excision repair cross-complementing proteins ERCC1 and ERCC4, and regulates the sensitivity of gastric cancer cells to cisplatin. Oncol Rep. 2019;41(2):1131-9. [DOI] [PubMed]
- 60.Zhang Z, et al., Circular RNA circ_0026359 Enhances Cisplatin Resistance in Gastric Cancer via Targeting miR-1200/POLD4 Pathway. Biomed Res Int, 2020. 2020: p. 5103272. [DOI] [PMC free article] [PubMed]
- 61.Sun Y, Clair DK, Fang F, Warren GW, Rangnekar VM, Crooks PA, Clair WH. The radiosensitization effect of parthenolide in prostate cancer cells is mediated by nuclear factor-κB inhibition and enhanced by the presence of PTEN. Mol Cancer Ther. 2007;6(9):2477-86. [DOI] [PMC free article] [PubMed]
- 62.Jiang Z, Pore N, Cerniglia GJ, Mick R, Georgescu MM, Bernhard EJ, Hahn SM, Gupta AK, Maity A. Phosphatase and tensin homologue deficiency in glioblastoma confers resistance to radiation and temozolomide that is reversed by the protease inhibitor nelfinavir. Cancer Res. 2007;67(9):4467-73. [DOI] [PubMed]
- 63.Cantley LC, Neel BG. New insights into tumor suppression: PTEN suppresses tumor formation by restraining the phosphoinositide 3-kinase/AKT pathway. Proc Nat Acad Sci. 1999;96(8):4240-5. [DOI] [PMC free article] [PubMed]
- 64.Maehama T, Okahara F, Kanaho Y. The tumour suppressor PTEN: involvement of a tumour suppressor candidate protein in PTEN turnover. Biochem Soc Transact. 2004;32(2):343-7. [DOI] [PubMed]
- 65.Zangouei AS, et al. Non coding RNAs as the critical factors in chemo resistance of bladder tumor cells. Diagn Pathol. 2020;15(1):136. doi: 10.1186/s13000-020-01054-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rahmani Z, Mojarrad M, Moghbeli M. Long non-coding RNAs as the critical factors during tumor progressions among Iranian population: an overview. Cell Biosci. 2020;10:6. doi: 10.1186/s13578-020-0373-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mattick JS, Rinn J.L.J.N.s. Discovery and annotation of long noncoding RNAs. Nat Struct Mol Biol. 2015;22(1):5–7. doi: 10.1038/nsmb.2942. [DOI] [PubMed] [Google Scholar]
- 68.Feng Y, et al. Modulation of CASC2/miR-21/PTEN pathway sensitizes cervical cancer to cisplatin. Arch Biochem Biophys. 2017;623:20–30. doi: 10.1016/j.abb.2017.05.001. [DOI] [PubMed] [Google Scholar]
- 69.Zheng L, Lin S, Lv C. MiR-26a-5p regulates cardiac fibroblasts collagen expression by targeting ULK1. Scientific reports. 2018;8(1):2104. doi: 10.1038/s41598-018-20561-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sun M, Wang G, Paciga JE, Feldman RI, Yuan ZQ, Ma XL, Shelley SA, Jove R, Tsichlis PN, Nicosia SV, Cheng JQ. AKT1/PKBα kinase is frequently elevated in human cancers and its constitutive activation is required for oncogenic transformation in NIH3T3 cells. Am J Pathol. 2001;159(2):431-7. [DOI] [PMC free article] [PubMed]
- 71.Zhou X, et al. Activation of the Akt/mammalian target of rapamycin/4E-BP1 pathway by ErbB2 overexpression predicts tumor progression in breast cancers. Clin Cancer Res. 2004;10(20):6779–88. doi: 10.1158/1078-0432.CCR-04-0112. [DOI] [PubMed] [Google Scholar]
- 72.Cao W, et al. miR-34a regulates cisplatin-induce gastric cancer cell death by modulating PI3K/AKT/survivin pathway. Tumour Biol. 2014;35(2):1287–95. [DOI] [PubMed]
- 73.Rajan N, Ashworth A.J.T.l.o. Inherited cylindromas: lessons from a rare tumour. Lancet Oncol. 2015;16(9):e460–9. doi: 10.1016/S1470-2045(15)00245-4. [DOI] [PubMed] [Google Scholar]
- 74.Nikolaou K, et al. Inactivation of the deubiquitinase CYLD in hepatocytes causes apoptosis, inflammation, fibrosis, and cancer. Cancer Cell. 2012;21(6):738–50. doi: 10.1016/j.ccr.2012.04.026. [DOI] [PubMed] [Google Scholar]
- 75.Van Andel H, Kocemba KA, de Haan-Kramer A, Mellink CH, Piwowar M, Broijl A, van Duin M, Sonneveld P, Maurice MM, Kersten MJ, Spaargaren M. Loss of CYLD expression unleashes Wnt signaling in multiple myeloma and is associated with aggressive disease. Oncogene. 2017;36(15):2105-15. [DOI] [PubMed]
- 76.Urbanik T, et al. CYLD deletion triggers nuclear factor-κB-signaling and increases cell death resistance in. murine hepatocytes. 2014;20(45):17049. [DOI] [PMC free article] [PubMed]
- 77.Obenauf AC, et al. Therapy-induced tumour secretomes promote resistance and tumour progression. Nature. 2015;520(7547):368–72. doi: 10.1038/nature14336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mayer IA, Arteaga CL. The PI3K/AKT pathway as a target for cancer treatment. Ann Rev Med. 2016;67:11-28. [DOI] [PubMed]
- 79.Kenerson HL, et al. Activated mammalian target of rapamycin pathway in the pathogenesis of tuberous sclerosis complex renal tumors. Cancer Res. 2002;62(20):5645–50. [PubMed] [Google Scholar]
- 80.Dowling RJ, et al. Dissecting the role of mTOR: lessons from mTOR inhibitors. Biochim Biophys Acta. 2010;1804(3):433–9. doi: 10.1016/j.bbapap.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 81.Kulcheski FR, Christoff AP, Margis R. Circular RNAs are miRNA sponges and can be used as a new class of biomarker. J Biotechnol. 2016;238:42-51. [DOI] [PubMed]
- 82.Yu HG, et al. Phosphoinositide 3-kinase/Akt pathway plays an important role in chemoresistance of gastric cancer cells against etoposide and doxorubicin induced cell death. Int J Cancer. 2008;122(2):433–43. doi: 10.1002/ijc.23049. [DOI] [PubMed] [Google Scholar]
- 83.Lee S, et al. Activation of PI3K/Akt pathway by PTEN reduction and PIK3CA mRNA amplification contributes to cisplatin resistance in an ovarian cancer cell line. Gynecol Oncol. 2005;97(1):26–34. doi: 10.1016/j.ygyno.2004.11.051. [DOI] [PubMed] [Google Scholar]
- 84.Siddik ZHJO. Cisplatin: mode of cytotoxic action and molecular basis of resistance. Oncogene. 2003;22(47):7265–79. doi: 10.1038/sj.onc.1206933. [DOI] [PubMed] [Google Scholar]
- 85.Bang Y, J.J.J.o.c.g. Advances in the management of HER2-positive advanced gastric and gastroesophageal junction cancer. J Clin Gastroenterol. 2012;46(8):637–48. doi: 10.1097/MCG.0b013e3182557307. [DOI] [PubMed] [Google Scholar]
- 86.Knuefermann C, et al. HER2/PI-3K/Akt activation leads to a multidrug resistance in human breast adenocarcinoma cells. Oncogene. 2003;22(21):3205–12. doi: 10.1038/sj.onc.1206394. [DOI] [PubMed] [Google Scholar]
- 87.Liang X, et al. VEGF signal system: the application of antiangiogenesis. Curr Med Chem. 2014;21(7):894–910. doi: 10.2174/09298673113206660264. [DOI] [PubMed] [Google Scholar]
- 88.Makinen T, et al. Isolated lymphatic endothelial cells transduce growth, survival and migratory signals via the VEGF-C/D receptor VEGFR-3. EMBO J. 2001;20(17):4762–73. doi: 10.1093/emboj/20.17.4762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Baldwin A. S.J.T.J.o.c.i. Control of oncogenesis cancer therapy resistance by the transcription factor NF-κB. 2001;107(3):241–6. doi: 10.1172/JCI11991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kovalenko A, Chable-Bessia C, Cantarella G, Israël A, Wallach D, Courtois G. The tumour suppressor CYLD negatively regulates NF-κB signalling by deubiquitination. Nature. 2003;424(6950):801-5. [DOI] [PubMed]
- 91.Manu KA, Shanmugam MK, Ramachandran L, Li F, Fong CW, Kumar AP, Tan P, Sethi G. First evidence that γ-tocotrienol inhibits the growth of human gastric cancer and chemosensitizes it to capecitabine in a xenograft mouse model through the modulation of NF-κB pathway. Clin Cancer Res. 2012;18(8):2220-9. [DOI] [PubMed]
- 92.Hayden MS, Ghosh SJC. Shared principles in NF-κB signaling. 2008;132(3):344–62. doi: 10.1016/j.cell.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 93.Wang Z-H, et al. Enhanced antitumor efficacy by the combination of emodin and gemcitabine against human pancreatic cancer cells via downregulation of the expression of XIAP in vitro and in vivo. Int J Oncol. 2011;39(5):1123–31. doi: 10.3892/ijo.2011.1115. [DOI] [PubMed] [Google Scholar]
- 94.Li F, Sethi G. Targeting transcription factor NF-κB to overcome chemoresistance and radioresistance in cancer therapy. Biochimica et Biophysica Acta (BBA)-Rev Cancer. 2010;1805(2):167-80. [DOI] [PubMed]
- 95.Lin X, Zhang X, Wang Q, Li J, Zhang P, Zhao M, Li X. Perifosine downregulates MDR1 gene expression and reverses multidrug-resistant phenotype by inhibiting PI3K/Akt/NF-κB signaling pathway in a human breast cancer cell line. Neoplasma. 2012;59(3):248-56. [DOI] [PubMed]
- 96.Vucic D, Fairbrother WJ. The inhibitor of apoptosis proteins as therapeutic targets in cancer. Clin Cancer Res. 2007;13(20):5995-6000. [DOI] [PubMed]
- 97.Baggiolini M. Chemokines and leukocyte traffic. Nature. 1998;392(6676):565–8. doi: 10.1038/33340. [DOI] [PubMed] [Google Scholar]
- 98.Yamashita U, Kuroda E. Regulation of macrophage-derived chemokine (MDC/CCL22) production. Crit Rev Immunol. 2002;22(2). [PubMed]
- 99.Wiedemann GM, et al. Peritumoural CCL1 and CCL22 expressing cells in hepatocellular carcinomas shape the tumour immune infiltrate. Pathology. 2019;51(6):586–92. doi: 10.1016/j.pathol.2019.06.001. [DOI] [PubMed] [Google Scholar]
- 100.Ou B, et al. CCR4 promotes metastasis via ERK/NF-kappaB/MMP13 pathway and acts downstream of TNF-alpha in colorectal cancer. Oncotarget. 2016;7(30):47637–49. doi: 10.18632/oncotarget.10256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Mizukami Y, Kono K, Kawaguchi Y, Akaike H, Kamimura K, Sugai H, Fujii H. CCL17 and CCL22 chemokines within tumor microenvironment are related to accumulation of Foxp3+ regulatory T cells in gastric cancer. Int J Cancer. 2008;122(10):2286-93. [DOI] [PubMed]
- 102.Wei C, Yang C, Wang S, Shi D, Zhang C, Lin X, Xiong B. M2 macrophages confer resistance to 5-fluorouracil in colorectal cancer through the activation of CCL22/PI3K/AKT signaling. OncoTargets Ther. 2019;12:3051. [DOI] [PMC free article] [PubMed]
- 103.Jiao S, Wang S, Y.J.E.R.M.P S, Huang LncRNA PRNCR1 promoted the progression of eclampsia by regulating the MAPK signal pathway. Eur Rev Med Pharmacol Sci. 2018;22(12):3635–42. doi: 10.26355/eurrev_201806_15240. [DOI] [PubMed] [Google Scholar]
- 104.McGivern N, et al. Activation of MAPK signalling results in resistance to saracatinib (AZD0530) in ovarian cancer. Oncotarget. 2018;9(4):4722. doi: 10.18632/oncotarget.23524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hew KE, et al. MAPK activation predicts poor outcome and the MEK inhibitor, selumetinib, reverses antiestrogen resistance in ER-positive high-grade serous ovarian cancer. Clin Cancer Res. 2016;22(4):935–47. doi: 10.1158/1078-0432.CCR-15-0534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Xiao P, et al. Correlations of neuronal apoptosis with expressions of c-Fos and c-Jun in rats with post-ischemic reconditioning damage. Eur Rev Med Pharmacol Sci. 2018;22(9):2832–8. doi: 10.26355/eurrev_201805_14984. [DOI] [PubMed] [Google Scholar]
- 107.McGinnis LA, et al. MAPK3/1 (ERK1/2) and myosin light chain kinase in mammalian eggs affect myosin-II function and regulate the metaphase II state in a calcium-and zinc-dependent manner. Biol Reprod. 2015;92(6):p. 146, 1–14. [DOI] [PMC free article] [PubMed]
- 108.Zhu QL, Li Z, Lv CM, Wang W. MiR-187 influences cisplatin-resistance of gastric cancer cells through regulating the TGF-β/Smad signaling pathway. Eur Rev Med Pharmacol Sci. 2019;23(22):9907-14. [DOI] [PubMed]
- 109.Mizushima N, D.J.J.A.R N, Klionsky Protein turnover via autophagy: implications for metabolism. Annu Rev Nutr. 2007;27:19–40. doi: 10.1146/annurev.nutr.27.061406.093749. [DOI] [PubMed] [Google Scholar]
- 110.White EJ, N.r.c. Deconvoluting the context-dependent role for autophagy in cancer. Nat Rev Cancer. 2012;12(6):401–10. doi: 10.1038/nrc3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kuo YZ, Tai YH, Lo HI, Chen YL, Cheng HC, Fang WY, Lin SH, Yang CL, Tsai ST, Wu LW. MiR-99a exerts anti‐metastasis through inhibiting myotubularin‐related protein 3 expression in oral cancer. Oral Dis. 2014;20(3):e65-75. [DOI] [PubMed]
- 112.Das G, Shravage BV, Baehrecke J.C.S.H.p.i.b. Regulation and function of autophagy during cell survival and cell death. Cold Spring Harb Perspect Biol. 2012;4(6):a008813. doi: 10.1101/cshperspect.a008813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Qian HR, Yang Y. Functional role of autophagy in gastric cancer. Oncotarget. 2016;7(14):17641. [DOI] [PMC free article] [PubMed]
- 114.Zou Z, et al. MicroRNA-30a sensitizes tumor cells to cis-platinum via suppressing beclin 1-mediated autophagy. J Biol Chem. 2012;287(6):4148–56. doi: 10.1074/jbc.M111.307405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zhang Y, et al. Regulation of autophagy by miR-30d impacts sensitivity of anaplastic thyroid carcinoma to cisplatin. Biochem Pharmacol. 2014;87(4):562–70. doi: 10.1016/j.bcp.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Sui X, et al. Autophagy and chemotherapy resistance: a promising therapeutic target for cancer treatment. Cell Death Dis. 2013;4(10):e838–8. doi: 10.1038/cddis.2013.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Yuan P, Cao W, Zang Q, Li G, Guo X, Fan J. The HIF-2α-MALAT1-miR-216b axis regulates multi-drug resistance of hepatocellular carcinoma cells via modulating autophagy. Biochem Biophys Res Commun. 2016;478(3):1067-73. [DOI] [PubMed]
- 118.Chen D-Q, et al. HDAC 1/4-mediated silencing of microRNA-200b promotes chemoresistance in human lung adenocarcinoma cells. Oncotarget. 2014;5(10):3333. doi: 10.18632/oncotarget.1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Li F, et al. Control of apoptosis and mitotic spindle checkpoint by survivin. Nature. 1998;396(6711):580–4. doi: 10.1038/25141. [DOI] [PubMed] [Google Scholar]
- 120.Marusawa H, Matsuzawa SI, Welsh K, Zou H, Armstrong R, Tamm I, Reed JC. HBXIP functions as a cofactor of survivin in apoptosis suppression. EMBO J. 2003;22(11):2729-40. [DOI] [PMC free article] [PubMed]
- 121.Singh N, et al. Clinical aspects for survivin: a crucial molecule for targeting drug-resistant cancers. Drug Discov Today. 2015;20(5):578–87. doi: 10.1016/j.drudis.2014.11.013. [DOI] [PubMed] [Google Scholar]
- 122.Radha G, Raghavan S.C.J.B.e.B.A.-R.o.C. BCL2: A promising cancer therapeutic target. Biochim Biophys Acta Rev Cancer. 2017;1868(1):309–14. doi: 10.1016/j.bbcan.2017.06.004. [DOI] [PubMed] [Google Scholar]
- 123.Sarosiek KA, et al. Developmental regulation of mitochondrial apoptosis by c-Myc governs age-and tissue-specific sensitivity to cancer therapeutics. Cancer Cell. 2017;31(1):142–56. doi: 10.1016/j.ccell.2016.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Chonghaile TN, Sarosiek KA, Vo TT, Ryan JA, Tammareddi A, Moore VD, Deng J, Anderson KC, Richardson P, Tai YT, Mitsiades CS. Pretreatment mitochondrial priming correlates with clinical response to cytotoxic chemotherapy. Science. 2011;334(6059):1129-33. [DOI] [PMC free article] [PubMed]
- 125.Suen DF, Norris KL, Youle RJ. Mitochondrial dynamics and apoptosis. Genes Dev. 2008;22(12):1577-90. [DOI] [PMC free article] [PubMed]
- 126.Cerveny KL, et al. Regulation of mitochondrial fusion and division. Trends Cell Biol. 2007;17(11):563–9. doi: 10.1016/j.tcb.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 127.Fan S, et al. miR-483-5p determines mitochondrial fission and cisplatin sensitivity in tongue squamous cell carcinoma by targeting FIS1. Cancer Lett. 2015;362(2):183–91. doi: 10.1016/j.canlet.2015.03.045. [DOI] [PubMed] [Google Scholar]
- 128.Merrill RA, Strack S. Mitochondria: a kinase anchoring protein 1, a signaling platform for mitochondrial form and function. Int J Biochem Cell Biol. 2014;48:92–6. doi: 10.1016/j.biocel.2013.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Sirohi K, et al. M98K-OPTN induces transferrin receptor degradation and RAB12-mediated autophagic death in retinal ganglion cells. Autophagy. 2013;9(4):510–27. doi: 10.4161/auto.23458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Matsui T, Fukuda M. Rab12 regulates mTORC1 activity and autophagy through controlling the degradation of amino-acid transporter PAT4. EMBO Rep. 2013;14(5):450-7. [DOI] [PMC free article] [PubMed]
- 131.Abbaszadegan MR, et al. WNT and NOTCH signaling pathways as activators for epidermal growth factor receptor in esophageal squamous cell carcinoma. Cell Mol Biol Lett. 2018;23:42. doi: 10.1186/s11658-018-0109-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Moghbeli M, et al. Correlation of Wnt and NOTCH pathways in esophageal squamous cell carcinoma. J Cell Commun Signal. 2016;10(2):129–35. doi: 10.1007/s12079-016-0320-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Moghbeli M, et al. Role of Msi1 and MAML1 in Regulation of Notch Signaling Pathway in Patients with Esophageal Squamous Cell Carcinoma. J Gastrointest Cancer. 2015;46(4):365–9. doi: 10.1007/s12029-015-9753-9. [DOI] [PubMed] [Google Scholar]
- 134.Moghbeli M, et al. Role of MAML1 in targeted therapy against the esophageal cancer stem cells. J Transl Med. 2019;17(1):126. doi: 10.1186/s12967-019-1876-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Du B, Shim JS. Targeting epithelial–mesenchymal transition (EMT) to overcome drug resistance in cancer. Molecules. 2016;21(7):965. [DOI] [PMC free article] [PubMed]
- 136.Lv Q, Wang W, Xue J, Hua F, Mu R, Lin H, Yan J, Lv X, Chen X, Hu ZW. DEDD interacts with PI3KC3 to activate autophagy and attenuate epithelial–mesenchymal transition in human breast cancer. Cancer Res. 2012;72(13):3238-50. [DOI] [PubMed]
- 137.Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial–mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15(3):178-96. [DOI] [PMC free article] [PubMed]
- 138.Hill L, Browne G, Tulchinsky E. ZEB/miR‐200 feedback loop: at the crossroads of signal transduction in cancer. Int J Cancer. 2013;132(4):745-54. [DOI] [PubMed]
- 139.Haslehurst AM, et al. EMT transcription factors snail and slug directly contribute to cisplatin resistance in ovarian cancer. BMC Cancer. 2012;12(1):91. doi: 10.1186/1471-2407-12-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Adam L, et al. miR-200 expression regulates epithelial-to-mesenchymal transition in bladder cancer cells and reverses resistance to epidermal growth factor receptor therapy. Clin Cancer Res. 2009;15(16):5060–72. doi: 10.1158/1078-0432.CCR-08-2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Hua F, et al. DEDD decreases Smad3 activity, promotes tumor cell apoptosis and inhibits proliferation. Yao Xue Xue Bao. 2013;48(5):680–5. [PubMed] [Google Scholar]
- 142.Lv Q, Hua F, Hu Z-WJA. DEDD, a novel tumor repressor, reverses epithelial-mesenchymal transition by activating selective autophagy. Autophagy. 2012;8(11):1675–6. doi: 10.4161/auto.21438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Szakács G, et al. Targeting multidrug resistance in cancer. 2006;5(3):219–34. doi: 10.1038/nrd1984. [DOI] [PubMed] [Google Scholar]
- 144.Rabik CA, Dolan J.C.t.r. Molecular mechanisms of resistance and toxicity associated with platinating agents. Cancer Treat Rev. 2007;33(1):9–23. doi: 10.1016/j.ctrv.2006.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Johnstone RW, Ruefli AA, Lowe SWJC. Apoptosis: a link between cancer genetics chemotherapy. 2002;108(2):153–64. doi: 10.1016/s0092-8674(02)00625-6. [DOI] [PubMed] [Google Scholar]
- 146.Borst P, Schinkel A.H.J.T.J.o.c.i. P-glycoprotein ABCB1: a major player in drug handling by mammals. J Clin Invest. 2013;123(10):4131–3. doi: 10.1172/JCI70430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Trock BJ, Leonessa F, Clarke J.J.o.t.N.C.I. Multidrug resistance in breast cancer: a meta-analysis of MDR1/gp170 expression and its possible functional significance. J Natl Cancer Inst. 1997;89(13):917–31. doi: 10.1093/jnci/89.13.917. [DOI] [PubMed] [Google Scholar]
- 148.Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329(5992):689-93. [DOI] [PMC free article] [PubMed]
- 149.Li J, Yang S, Su N, Wang Y, Yu J, Qiu H, He X. Overexpression of long non-coding RNA HOTAIR leads to chemoresistance by activating the Wnt/β-catenin pathway in human ovarian cancer. Tumor Biol. 2016;37(2):2057-65. [DOI] [PubMed]
- 150.Xue X, et al. LncRNA HOTAIR enhances ER signaling and confers tamoxifen resistance in breast cancer. Oncogene. 2016;35(21):2746–55. doi: 10.1038/onc.2015.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Yamashita T, Wang X.W.J.T.J.o.c.i. Cancer stem cells in the development of liver cancer. J Clin Invest. 2013;123(5):1911–8. doi: 10.1172/JCI66024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Vader G, Lens J.B.e.B.A.-R.o.C. The Aurora kinase family in cell division and cancer. Biochim Biophys Acta. 2008;1786(1):60–72. doi: 10.1016/j.bbcan.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 153.Enjoji M, et al. BubR1 and AURKB overexpression are associated with a favorable prognosis in gastric cancer. Mol Med Rep. 2009;2(4):589–96. doi: 10.3892/mmr_00000142. [DOI] [PubMed] [Google Scholar]
- 154.Patnaik A, Rosen LS, Tolaney SM, Tolcher AW, Goldman JW, Gandhi L, Papadopoulos KP, Beeram M, Rasco DW, Hilton JF, Nasir A. Efficacy and safety of abemaciclib, an inhibitor of CDK4 and CDK6, for patients with breast cancer, non–small cell lung cancer, and other solid tumors. Cancer Disc. 2016;6(7):740-53. [DOI] [PubMed]
- 155.Chen-Kiang S, Di M, Liberto, Huang X. Targeting CDK4 and CDK6 in cancer therapy. Google Patents; 2016.
- 156.Wang L, Yang Q, Peng S, Liu X. The combination of the glycolysis inhibitor 2-DG and sorafenib can be effective against sorafenib-tolerant persister cancer cells. OncoTargets Ther. 2019;12:5359. [DOI] [PMC free article] [PubMed]
- 157.Roche TE, Baker JC, Yan X, Hiromasa Y, Gong X, Peng T, Dong J, Turkan A, Kasten SA. Distinct regulatory properties of pyruvate dehydrogenase kinase and phosphatase isoforms. Progr Nucleic Acid Res Mol Biol. 2001;70:33-75. [DOI] [PubMed]
- 158.Yuge R, et al. Silencing of Discoidin Domain Receptor-1 (DDR1) concurrently inhibits multiple steps of metastasis cascade in gastric cancer. Transl Oncol. 2018;11(3):575–84. doi: 10.1016/j.tranon.2018.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Zangouei AS, et al. Role of tyrosine kinases in bladder cancer progression: an overview. Cell Commun Signal. 2020;18(1):127. doi: 10.1186/s12964-020-00625-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Hur H, et al. Discoidin domain receptor 1 activity drives an aggressive phenotype in gastric carcinoma. BMC Cancer. 2017;17(1):87. doi: 10.1186/s12885-017-3051-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Huang H, Tindall DJ. FOXO factors: a matter of life and death. Future Oncol. 2006;2(1):83–9. doi: 10.2217/14796694.2.1.83. [DOI] [PubMed] [Google Scholar]
- 162.Nho RS, Hergert P.J.W.j.o.b.c. FoxO3a and disease progression. World J Biol Chem. 2014;5(3):346. doi: 10.4331/wjbc.v5.i3.346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Rathbone CR, Booth FW, Lees SJ. FoxO3a preferentially induces p27Kip1 expression while impairing muscle precursor cell-cycle progression. Muscle Nerve. 2008;37(1):84-9. [DOI] [PubMed]
- 164.Yang L, et al. Downregulation of microRNA-196a inhibits human liver cancer cell proliferation and invasion by targeting FOXO1. Oncol Rep. 2017;38(4):2148–54. doi: 10.3892/or.2017.5873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Huang C-Z, et al. Sox9 transcriptionally regulates Wnt signaling in intestinal epithelial stem cells in hypomethylated crypts in the diabetic state. Stem Cell Res Ther. 2017;8(1):1–15. doi: 10.1186/s13287-016-0461-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Lefebvre V, Dvir-Ginzberg M.J.C.t.r. SOX9 and the many facets of its regulation in the chondrocyte lineage. Connect Tissue Res. 2017;58(1):2–14. doi: 10.1080/03008207.2016.1183667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Semenza G.L.J.A.R.o.P.M.o.D. Oxygen sensing, hypoxia-inducible factors, and disease pathophysiology. Annu Rev Pathol. 2014;9:47–71. doi: 10.1146/annurev-pathol-012513-104720. [DOI] [PubMed] [Google Scholar]
- 168.Xiao H, Tong R, Ding C, Lv Z, Du C, Peng C, Cheng S, Xie H, Zhou L, Wu J, Zheng S. γ-H2AX promotes hepatocellular carcinoma angiogenesis via EGFR/HIF-1α/VEGF pathways under hypoxic condition. Oncotarget. 2015;6(4):2180. [DOI] [PMC free article] [PubMed]
- 169.De Francesco EM, et al. HIF-1α/GPER signaling mediates the expression of VEGF induced by hypoxia in breast cancer associated fibroblasts (CAFs) Breast Cancer Res. 2013;15(4):R64. doi: 10.1186/bcr3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Kim M, et al. Silencing Bmi1 expression suppresses cancer stemness and enhances chemosensitivity in endometrial cancer cells. Biomed Pharmacother. 2018;108:584–9. doi: 10.1016/j.biopha.2018.09.041. [DOI] [PubMed] [Google Scholar]
- 171.Ojo D, et al. Polycomb complex protein BMI1 confers resistance to tamoxifen in estrogen receptor positive breast cancer. Cancer Lett. 2018;426:4–13. doi: 10.1016/j.canlet.2018.03.048. [DOI] [PubMed] [Google Scholar]
- 172.Zhuang H, et al. Human epididymis protein 4 in association with Annexin II promotes invasion and metastasis of ovarian cancer cells. Mol Cancer. 2014;13(1):243. doi: 10.1186/1476-4598-13-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Zhang ZD, Li YO, Fan Q, Zhao B, Tan B, Zhao XF. Annexin A2 is implicated in multi-drug-resistance in gastric cancer through p38MAPK and AKT pathway. Neoplasma. 2014;61(6):627-37. [DOI] [PubMed]
- 174.Zhai H, et al. Annexin A2 promotes glioma cell invasion and tumor progression. J Neurosci. 2011;31(40):14346–60. doi: 10.1523/JNEUROSCI.3299-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Nateri AS, et al. The ubiquitin ligase SCFFbw7 antagonizes apoptotic JNK signaling. Science. 2004;303(5662):1374–8. doi: 10.1126/science.1092880. [DOI] [PubMed] [Google Scholar]
- 176.Wu G, et al. SEL-10 is an inhibitor of notch signaling that targets notch for ubiquitin-mediated protein degradation. Mol Cell Biol. 2001;21(21):7403–15. doi: 10.1128/MCB.21.21.7403-7415.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.You B, Shan Y, Shi S, Li X, You Y. Effects of ADAM 10 upregulation on progression, migration, and prognosis of nasopharyngeal carcinoma. Cancer Sci. 2015;106(11):1506-14. [DOI] [PMC free article] [PubMed]
- 178.Arima T, et al. Nuclear translocation of ADAM-10 contributes to the pathogenesis and progression of human prostate cancer. Cancer Sci. 2007;98(11):1720–6. doi: 10.1111/j.1349-7006.2007.00601.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Pan Y, et al. ADAM10 promotes pituitary adenoma cell migration by regulating cleavage of CD44 and L1. J Mol Endocrinol. 2012;49(1):21. doi: 10.1530/JME-11-0174. [DOI] [PubMed] [Google Scholar]
- 180.Guo J, He L, Yuan P, Wang P, Lu Y, Tong F, Wang Y, Yin Y, Tian J, Sun J. ADAM10 overexpression in human non-small cell lung cancer correlates with cell migration and invasion through the activation of the Notch1 signaling pathway. Oncol Rep. 2012;28(5):1709-18. [DOI] [PubMed]
- 181.Jones AV, Lambert DW, Speight PM, Whawell SA. ADAM 10 is over expressed in oral squamous cell carcinoma and contributes to invasive behaviour through a functional association with αvβ6 integrin. FEBS Lett. 2013;587(21):3529-34. [DOI] [PubMed]
- 182.Gaida MM, et al. Expression of A disintegrin and metalloprotease 10 in pancreatic carcinoma. Int J Mol Med. 2010;26(2):281–8. doi: 10.3892/ijmm_00000463. [DOI] [PubMed] [Google Scholar]
- 183.Meng X, et al. DNA damage alters DNA polymerase delta to a form that exhibits increased discrimination against modified template bases and mismatched primers. Nucleic Acids Res. 2009;37(2):647–57. doi: 10.1093/nar/gkn1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Bertoli G, Cava C, Castiglioni I. MicroRNAs: New Biomarkers for Diagnosis, Prognosis, Therapy Prediction and Therapeutic Tools for Breast Cancer. Theranostics. 2015;5(10):1122–43. doi: 10.7150/thno.11543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Cortez MA, et al. MicroRNAs in body fluids–the mix of hormones and biomarkers. Nat Rev Clin Oncol. 2011;8(8):467–77. doi: 10.1038/nrclinonc.2011.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Mo MH, et al. Cell-free Circulating miRNA Biomarkers in Cancer. J Cancer. 2012;3:432–48. doi: 10.7150/jca.4919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Kroh EM, et al. Analysis of circulating microRNA biomarkers in plasma and serum using quantitative reverse transcription-PCR (qRT-PCR) Methods. 2010;50(4):298–301. doi: 10.1016/j.ymeth.2010.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Wu X, et al. De novo sequencing of circulating miRNAs identifies novel markers predicting clinical outcome of locally advanced breast cancer. J Transl Med. 2012;10:42. doi: 10.1186/1479-5876-10-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Hummel R, et al. Mir-148a improves response to chemotherapy in sensitive and resistant oesophageal adenocarcinoma and squamous cell carcinoma cells. J Gastrointest Surg. 2011;15(3):429–38. doi: 10.1007/s11605-011-1418-9. [DOI] [PubMed] [Google Scholar]
- 190.Kovalchuk O, et al. Involvement of microRNA-451 in resistance of the MCF-7 breast cancer cells to chemotherapeutic drug doxorubicin. Mol Cancer Ther. 2008;7(7):2152–9. doi: 10.1158/1535-7163.MCT-08-0021. [DOI] [PubMed] [Google Scholar]
- 191.Xia L, et al. miR-15b and miR-16 modulate multidrug resistance by targeting BCL2 in human gastric cancer cells. Int J Cancer. 2008;123(2):372–9. doi: 10.1002/ijc.23501. [DOI] [PubMed] [Google Scholar]
- 192.Kim CH, et al. miRNA signature associated with outcome of gastric cancer patients following chemotherapy. BMC Med Genomics. 2011;4:79. doi: 10.1186/1755-8794-4-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Weiler J, Hunziker J, Hall J. Anti-miRNA oligonucleotides (AMOs): ammunition to target miRNAs implicated in human disease? Gene Ther. 2006;13(6):496–502. doi: 10.1038/sj.gt.3302654. [DOI] [PubMed] [Google Scholar]
- 194.To KK, et al. MicroRNAs in the prognosis and therapy of colorectal cancer: From bench to bedside. World J Gastroenterol. 2018;24(27):2949–73. doi: 10.3748/wjg.v24.i27.2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Brown BD, et al. Endogenous microRNA regulation suppresses transgene expression in hematopoietic lineages and enables stable gene transfer. Nat Med. 2006;12(5):585–91. doi: 10.1038/nm1398. [DOI] [PubMed] [Google Scholar]
- 196.McDermott AM, et al. The therapeutic potential of microRNAs: disease modulators and drug targets. Pharm Res. 2011;28(12):3016–29. doi: 10.1007/s11095-011-0550-2. [DOI] [PubMed] [Google Scholar]
- 197.Wang H, et al. Recent progress in microRNA delivery for cancer therapy by non-viral synthetic vectors. Adv Drug Deliv Rev. 2015;81:142–60. doi: 10.1016/j.addr.2014.10.031. [DOI] [PubMed] [Google Scholar]
- 198.Wei L, et al. The emerging role of microRNAs and long noncoding RNAs in drug resistance of hepatocellular carcinoma. Mol Cancer. 2019;18(1):147. doi: 10.1186/s12943-019-1086-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.