Abstract
Background
The prognosis of early breast cancer is linked to clinic-pathological stage and the molecular characteristics of intrinsic tumor cells. In some patients, the amount and quality of tumor-infiltrating immune cells appear to affect long term outcome. We aimed to propose a new tool to estimate immune infiltrate, and link these factors to patient prognosis according to breast cancer molecular subtypes.
Methods
We performed in silico analyses in more than 2800 early breast cancer transcriptomes with corresponding clinical annotations. We first developed a new gene expression deconvolution algorithm that accurately estimates the quantity of immune cell populations (tumor immune contexture, TIC) in tumors. Then, we studied associations between these immune profiles and relapse-free and overall survival among the different intrinsic molecular subtypes of breast cancer defined by PAM50 classification.
Results
TIC estimates the abundance of 15 immune cell subsets. Both myeloid and lymphoid subpopulations show different spread among intrinsic molecular breast cancer subtypes. A high abundance of myeloid cells was associated with poor outcome, while lymphoid cells were associated with favorable prognosis. Unsupervised clustering describing the 15 immune cell subsets revealed four subgroups of breast tumors associated with distinct patient survival, but independent from PAM50. Adding this information to clinical stage and PAM50 strongly improves the prediction of relapse or death.
Conclusions
Our findings make it possible to refine the survival stratification of early patients with breast cancer by incorporating TIC in addition to PAM50 and clinical tumor burden in a prognostic model validated in training and validation cohorts.
Keywords: tumor microenvironment, biostatistics, tumor biomarkers, breast neoplasms
Introduction
Breast cancer is the most common cancer in women, and its incidence has markedly increased in recent years. However, most patients are actually diagnosed with early-stage disease, and can thus be cured by locoregional and adjuvant systemic treatments. In recent decades, progress and intensification of adjuvant therapies in early breast cancer has significantly contributed to reducing relapse rates and improving survival.
Beyond classical prognostic factors (eg, age, tumor size, lymph node involvement, histological grade, ER, Ki67, and HER2 expression) used for treatment-related decisions, the development of multigene expression technologies in the early 2000s led to the discovery of transcriptomic heterogeneity among breast cancers, and brought about a refined tumor molecular classification with distinct clinical prognosis.1 Numerous multigene expression assays have been developed to help clinicians to refine the evaluation of the risk of relapse.2–4 Most are based on cancer cell intrinsic biology, with a dominant weight given to proliferation-associated genes.5 Among these assays, the PAM50 assay, which was initially developed to identify the different intrinsic breast cancer subtypes (luminal A/B, HER2 enriched, and basal like),3 6 is able to refine the outcome prediction, not only in ER+, but also in ER− tumors.6
Besides the intrinsic biology of breast cancer cells, recent studies on the tumor microenvironment provide new insights into the prognostic importance of immune response in breast cancer. Breast cancer was largely considered as non-immunogenic for many decades. However, the prognosis of breast cancer now appears to be associated with some features of host immune response, like tumor infiltrating lymphocyte (TIL) density, composition, and activity.7–10 Numerous studies over the past years have highlighted that the presence of high levels of TILs, but also of Th1 immune signatures, is associated with improved survival in early stage breast cancer, especially HER2-overexpressing and triple negative subtypes.11 12 Regarding the latter subtype, it was recently shown that the organization of the immune response within the tumor is also associated with prognosis.13–15 In contrast, a strong tumor infiltration in immunoregulatory cells such as regulatory T lymphocytes (Tregs), M2 macrophages, or a Th2 polarization of the immune response have been associated with a poor prognosis in early breast cancer.12 16 17
Moreover, accumulating data also suggest that the clinical efficacy of cytotoxic therapies commonly used by oncologists (and assessed by pathological complete response after neoadjuvant chemotherapy), may sometimes be positively influenced by local T dependent immune response, here again, mainly in HER2 positive and triple negative subtypes.9 10 18 In the recent context of the arrival of immunotherapy in triple negative breast cancer, a strong tumor infiltration of TILs, but also the presence of an interferon-γ (IFN-γ) inflammatory signature, or a high tumor mutational burden (TMB), have been associated with greater effectiveness of not only immunotherapy but also chemotherapy.19 These results raised the question whether certain breast tumors might benefit more from immune-based interventions and which cancer cell-intrinsic and/or microenvironmental factors define anti-tumor immune response.20
However, it must be noted that all these pioneering studies suffered from technical limitations, only evaluating the global pool of TILs (commonly detected in H&E on stained histological slides via light microscopy), or only one or two particular subtypes of immune cells, detected by specific staining in immunohistochemistry. For instance, with their role as final effectors of immune response, a favorable impact of breast tumor-infiltrating CD8+ T cells on survival has been largely reported.21 22 Nonetheless, all these histological studies offered only a narrow view of the complexity of immune response, immune cell interactions, and immune cell function and diversity in breast tumors. For example, it is still unknown whether the respective proportions of the different subpopulations of CD8+ T cells vary across intrinsic molecular subtypes of breast cancer (based on a transcriptomic definition), and how these different populations could influence early breast cancer prognosis. Moreover, in addition to CD8+ T cells, breast cancers are usually infiltrated by many other lymphoid or myeloid immune cells. This leukocyte complexity, and the respective role of each immune cell population on breast cancer prognosis, is only starting to be described.23 24 Thanks to deconvolution algorithms and public transcriptome datasets, we were able to estimate from tumor bulk mRNA the relative abundance of distinct subsets of immune and non-immune cells, and their relationships with outcome.
In order to reconcile the prognostic information given by both molecular intrinsic subtype (assessed by PAM50), and tumor global immune contexture (TIC) of early breast cancers, we performed complementary in silico analyses in more than 2800 early breast cancer transcriptomes with corresponding clinical annotations. We therefore developed a new gene expression deconvolution algorithm derived from Cibersort,25 and which accurately estimates in tumors not only the respective proportions of immune cells, but also the absolute quantity of distinct immune cell populations in the tumor sample, allowing us to study their statistical associations with relapse-free and overall survival (OS) among the different intrinsic molecular subtypes (defined by PAM50 expression assay). Moreover, this approach gave us the opportunity to study the added value of immune contexture on top of clinical parameters, and tumor molecular intrinsic subtype, for early breast cancer risk stratification.
Material and methods
Deconvolution process for estimating the proportions/quantity of cells
Signature matrices
The signature matrices used for the deconvolution process were built from public data found on Gene Expression Omnibus respository (GEO; https://www.ncbi.nlm.nih.gov/geo/). The building process was (1) to find overexpressed genes in one of the 15 cell types considered, and then (2) to rank the genes for each cell population from most significant to least significant, keeping only the genes with a false discovery rate (FDR)26 p value <0.05. Step (3) was to iteratively add a gene from each cell population of a given level and compute the condition number of the matrix at each step. The matrix kept as the signature matrix for a given level was the one with the smallest condition number as done in.25 Signature matrices can be found in online supplemental file 1.
jitc-2020-002036supp001.XLSX (35.3KB, XLSX)
Optimization problem
The mathematical modeling behind the deconvolution process is based on Cibersort25 except that the constraints of finding a vector of proportions (sum of proportions equal to 1) is directly included in the mathematical model. It yields a constrained version of the well-known support vector regression problem to solve and shows better performance than the Cibersort estimator27 that computes the unconstrained Support Vector Regression (SVR) and then projects it to have a proportion vector (see online supplemental file 2). Figures included in this file shows that our algorithm outperforms Cibersort in terms of better estimation performances, especially for small proportions.
jitc-2020-002036supp002.pdf (341.9KB, pdf)
Absolute versus relative
We chose to work with 15 different types of immune cells (see online supplemental figure S1 for the full list). The choice was based on different factors: purified samples available, the ability to discriminate them, and choice of cells of interest according to their biological relevance in cancer immune response. The list of the different samples used to construct the signature matrices is given in online supplemental file 3. The estimation of proportions of cells was done at each level of cell families; it results in an estimation of the relative proportions of cells inside each level as represented in online supplemental figure S1. However, because our modeling tries to capture all the cells that are present inside a tumor, the absolute quantities can be computed by multiplying the proportion of a given type of cell by its upper parents’ proportions. To be clearer, if the lymphoids represent 10% of the total cells and the T-cells represents 20% of the lymphoids, the absolute quantity of T-cells is obtained by multiplying 20% with 10% which makes 2% of the total number of cells. This can be done up to the fourth level with the CD8 T-cells subtypes.
jitc-2020-002036supp003.pdf (81.8KB, pdf)
jitc-2020-002036supp004.XLSX (77.3KB, XLSX)
There is a lot more information carried out by the absolute quantities. For example, two patients could have a relative T-cells quantity of 10% (ie, 10% of lymphoids are T-cells) but patient A has 10% of lymphoids and patient B only 1%; the absolute quantity distinguishes the two patients: patient A has 1% of T-cells in absolute proportions and patient B only 0.1%.
Patient dataset
Aggregation of datasets
The cohort of patients with breast cancer comes from different sources but regroup only microarray data coming from the H133a and the H133plus Affymetrix platforms. The CIT cohort comes from the Cartes d’Identité des Tumeurs project from the French Ligue Nationale Contre le Cancer.28 The transcriptomic data are available on ArrayExpress under the accession code E-MTAB-365. The rest of the cohort was downloaded from GEO; part of it comes from a study of KMplot.29 A description of the datasets is given in figure 1, the KMplot and GEO cohorts were used as the training cohort, while the CIT cohort was used for validation purposes. clinical characteristics of patients and tumors are described in online supplemental table 1.
Figure 1.
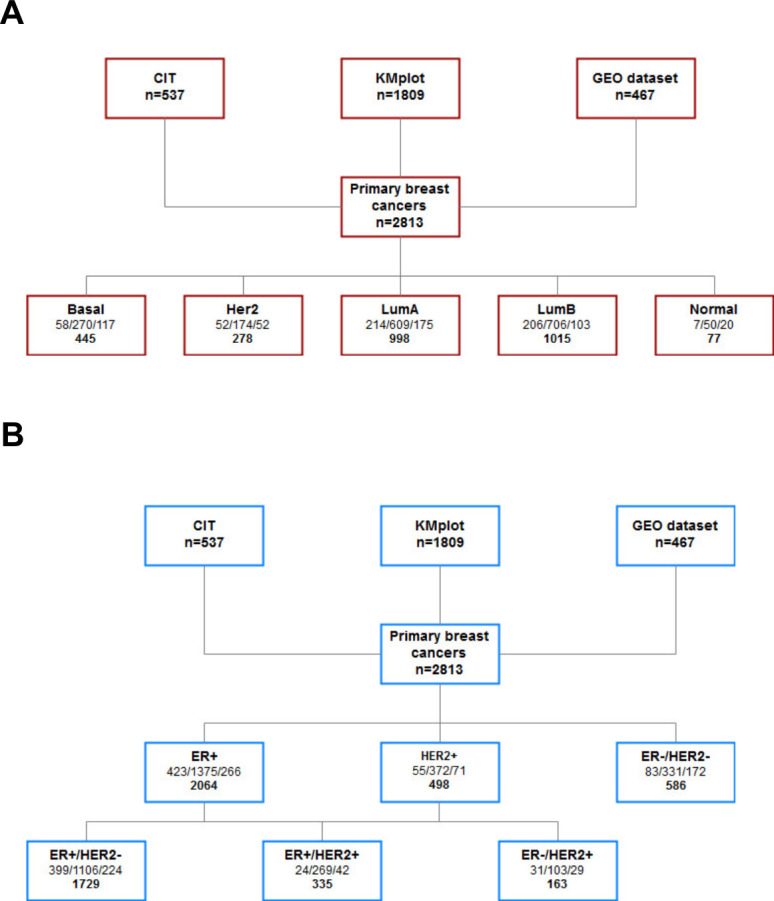
(A–B) Flow chart presenting the different cohorts used in the study and their classification based on PAM50 (A) and on the estrogen receptor (given by IHC when data were available or gene expression otherwise), and on human epidermal growth factor receptor 2 (B). In each box, the first set of numbers corresponds to number of patients broken down by the cohort (respectively, CIT, KMplot, and GEO dataset cohorts), and the bottom number corresponds to total number of patients. IHC, immunohistochemistry; GEO, Gene Expression Omnibus.
jitc-2020-002036supp006.pdf (63KB, pdf)
PAM50 classification from transcriptome
The PAM50 classification was performed on publicly available microarray transcriptomic data using the Genefu30 R package. The summary of the PAM50 classification and the number of patients in each group and each dataset can be found in figure 1A. The normal-like group was not kept for the rest of the analysis. Hereafter, “PAM50 classification” refers to the results of this process.
ER and HER2 status
As there is no complete overlap between the transcriptomic subtypes defined by the PAM50 signature, and the breast cancer subtype assessed by routine pathological techniques (expression of estrogen receptors and HER2),31 we also have evaluated our models within the different breast cancer subtypes, as defined with these routine techniques (namely: luminal non-HER2 (ER+/HER2−), triple negative (ER−/HER2−), and HER2+). When ER or HER status was unknown, the expression of the genes ESR1 and ERBB2 were used to define the patient’s status. A threshold of these two genes was computed using the patients for whom the information was available. The ER threshold was chosen such that it maximizes the correct classification rate in the available patients, and the same was done for HER status (results not shown). This approach has been validated on a test dataset that was not used in the rest of the analysis and (GEO accession number GSE129551). This dataset contains 147 breast cancer samples on which IHC ER and HER2 status are available. Using our cut-off values we obtained 100% well classified rate on the ER status and 97% on the HER2 status (5 misclassified out of 147 samples). These results justify our approach for the rest of this paper. In this way, the ER status was inferred by the described method for 590 patients of the 2736 (21%). The Her2 status was inferred for 1966 patients of the 2736 (72%) which still makes a training for the validation threshold of 770 patients.
Analysis of the estimated immune cell proportions
Testing the differences in cell quantity between molecular subtypes
In order to test whether a given cell population was significantly more abundant in one group of patients compared with another, we performed a Wilcoxon test and used adjusted p values (FDR controlling procedure). An adjusted p value <0.05 was considered as significant.
Clustering based on the proportions of cells
To make sure that the differences observed between patients were not only due to the molecular subtype, we performed clustering of patients based on the absolute quantity of the 15 immune cell types. Hierarchical clustering using the Euclidean distances and Ward’s method was used.
Survival analysis
Univariate survival analysis
Univariate survival analyses were performed using Cox proportional hazards regression models with a log rank test to compare survival distributions. Each variable was taken as a continuous variable. The Kaplan-Meier method was used to calculate survival probabilities. The log-rank test was used for comparison of survival curves. OS was calculated as the time from diagnosis of cancer to the date of death (from any cause). Relapse-free survival (RFS) was computed as the time between start of primary treatment and the time of any breast disease recurrence (local (including second primary breast cancer), regional, or distant). Patients were censored at 10 years for OS and RFS.
FDR-adjusted p values were obtained by Benjamini Hochberg method.
Multivariate survival analysis and models for survival prediction
Multivariate survival analyses were performed with Cox regression models. Patients were censored at 10 years. The “PAM50” model was built using the four classes of the molecular subtype variable. The “TIC” (tumor immune contexture) model includes the estimated absolute quantities of the different cell populations. The “clinical” model was built using the T and N variables considered as factor variables. The variable N is binary and is equal to 0 when there is no lymph node involvement and 1 when there is at least one lymph node involved. The T variable splits the patients in three groups: T1, T2, and T3 (this last category encompasses stages T3 and T4). Patients for whom the clinical information was missing were excluded from the construction and validation of the model. The model combining “PAM50” and “TIC” information models the interactions between the estimated quantities of immune cells and the PAM50 levels. As it was confirmed that each molecular subtype has an impact on the prognostic role of each immune cell population, a LASSO (Least Absolute Shrinkage and Selection Operator) algorithm for variable selection in the Cox model was then used on immune and PAM50 variables and their respective interactions, to select variables that were most strongly related to the outcome. Online supplemental file 4 shows corresponding multivariate Cox models for RFS and OS. The linear predictor of this model was then used to build a final model combining the three types of information (quantitative immune populations, PAM50, and clinical).
jitc-2020-002036supp005.XLSX (24KB, XLSX)
Results
Dataset containing 2813 breast cancer samples from three independent cohorts
A summary of available samples among the three independent cohorts of patients is depicted in figure 1. Samples were classified according to intrinsic molecular subtypes (PAM50) (figure 1A), or classical immunohistochemistry (ER and HER2 status) (figure 1B). Concerning intrinsic molecular subtypes, 998 cases (35.5%) were classified as luminal A subtype, 1015 (36%) as luminal B, 278 (10%) as HER2 enriched, and 445 (16%) as basal-like subtype. Seventy-seven (2.5%) of samples were classified as normal-like subtype, and were thereby excluded from subsequent analyses. A summary of available clinicopathological characteristics is also presented in online supplemental file 4.
CD8 subpopulations according to breast cancer molecular subtypes
Because of their role as effectors in the cytotoxic response against cancer cells, we first investigated whether CD8+ T cell subpopulations were different in ER-positive and ER-negative breast cancers. Figure 2 shows the estimated proportions (figure 2A), and absolute quantities (figure 2B) of CD8 fractions: effector memory CD8 T cells (EM CD8) constitute the majority of CD8 infiltrating breast tumors. Among all CD8 T cells, the estimated proportions of EM CD8 are significantly higher in ER− tumors (p value Wilcoxon test 9e-06), while the proportion of naïve CD8 is higher in ER+ tumors (p value Wilcoxon test 1e-11). There was no difference in proportions as regards central memory (CM) CD8 between ER+ and ER− tumors (p value Wilcoxon test 0.19). Similar trends were observed for the absolute quantities: compared with ER+ tumors, ER− breast cancer tissue contained more CD8 T cells, especially more EM CD8 (p value Wilcoxon test 6e-06), while ER+ tumors contained more naïve CD8+ (p value Wilcoxon test 5.7e-05). Here again, there was no difference in terms of CM CD8 (p value Wilcoxon test 0.54).
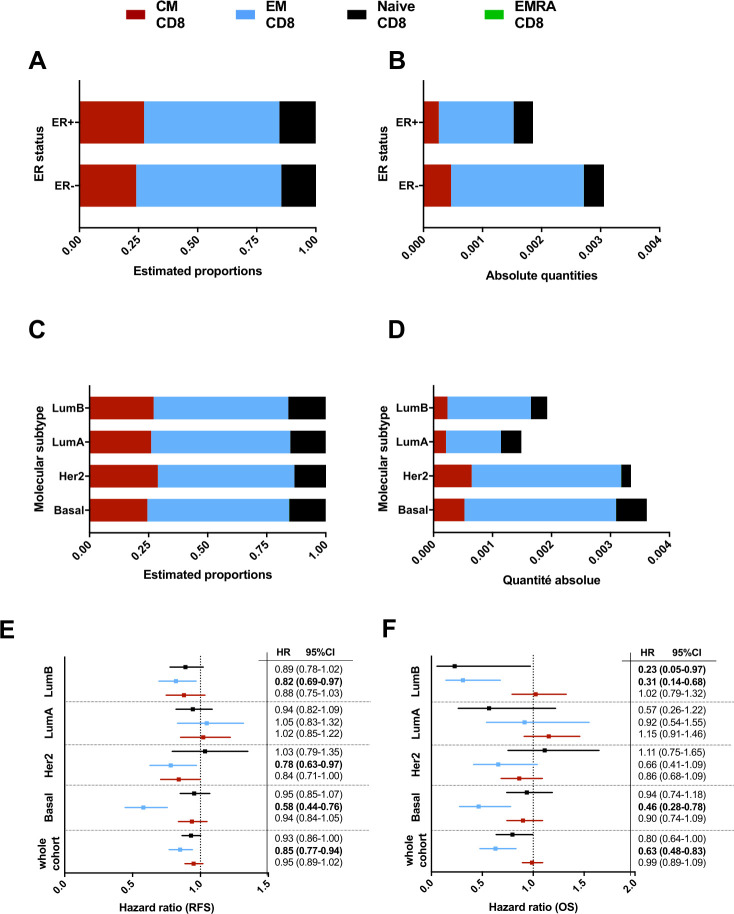
Figure 2.
(A–D) Bar graph representing the estimation of T-CD8 subtype absolute quantities in the different groups based on ER status (A–B) or PAM50 classification (C–D). (E–F) Forest plot representing the HR for relapse free survival (RFS) (E) and overall survival (OS) (F) of the different CD8 subtypes taken as a continuous variable in different PAM50 subgroups.
In view of these differences, we then investigated CD8 T cell subpopulations according to breast cancer molecular subtype, defined by PAM50, and found marked differences (figure 2C, D). HER2-enriched and basal-like molecular subtypes contained higher quantities of EM CD8, but also CM CD8 T cells compared with luminal tumors (Wilcoxon test basal vs luminal, p value=5e-9 and HER2 vs Luminal, p value=2e-4). Among luminal tumors, luminal B subtypes contained more EM CD8 T cells compared with luminal A tumors. Here again, among all CD8 T cells, the estimated proportions of the different CD8 subpopulations show the same results as the absolute quantities between the four molecular subtypes (figure 2D). The largest difference between pam50 subgroups is on the EM CD8 T cells whereas the differences in quantity for the CM CD8 T cells and the naïve CD8 T cells are rather small. According to their specific location in peripheral blood,32 we did not find the significant presence of intratumoral EMRA cells.
Prognostic roles of CD8 subpopulations according to breast cancer molecular subtype
We then examined the relationships between CD8 T cell subpopulations and prognosis of early breast cancer (in terms of RFS and OS) in each tumor molecular subtype (figure 2E, F, respectively). Considering all breast cancer samples, naïve CD8 (HR: 0.930 p=0.12, and HR: 0.796 p=0.13 for RFS and OS, respectively), but mostly EM CD8 (HR: 0.851 p=0.008, and HR: 0.629 p=0.01 for RFS and OS, respectively) are associated with better RFS and OS. Conversely, CM CD8 T cells are not associated with significantly different outcome. The favorable effect of EM CD8 on RFS and OS is particularly marked in the basal-like molecular subtype (HR: 0.580 (0.44 to 0.76), p=3.8e-4, and HR: 0.464 (0.28 to 0.78), p=0.02 for RFS and OS, respectively), and to a lesser extent in HER2 enriched (HR: 0.781 (0.63 to 0.97), p=0.06, and HR: 0.656 (0.41 to 1.04), p=0.18 for RFS and OS, respectively), and luminal B (HR: 0.819 (0.69 to 0.97), p=0.05, and HR: 0.310 (0.14 to 0.68), p=0.02 for RFS and OS, respectively) subtypes. naïve CD8 cells have significant prognostic effect (but numerically modest, considering HRs) in luminal B tumors only. Of note, as for TIL evaluation, CD8 T cell subpopulations are not associated with differences in outcome in luminal A tumors.
TIC is quantitatively different across breast cancer molecular subtypes
In view of these differences between breast cancer molecular subtypes considering only CD8 T cells, we then examined the overall immune cell composition (TIC) obtained by similar deconvolution approaches. Thus, we inferred 15 immune cell subsets among lymphoid and myeloid populations (see top of figure 3).
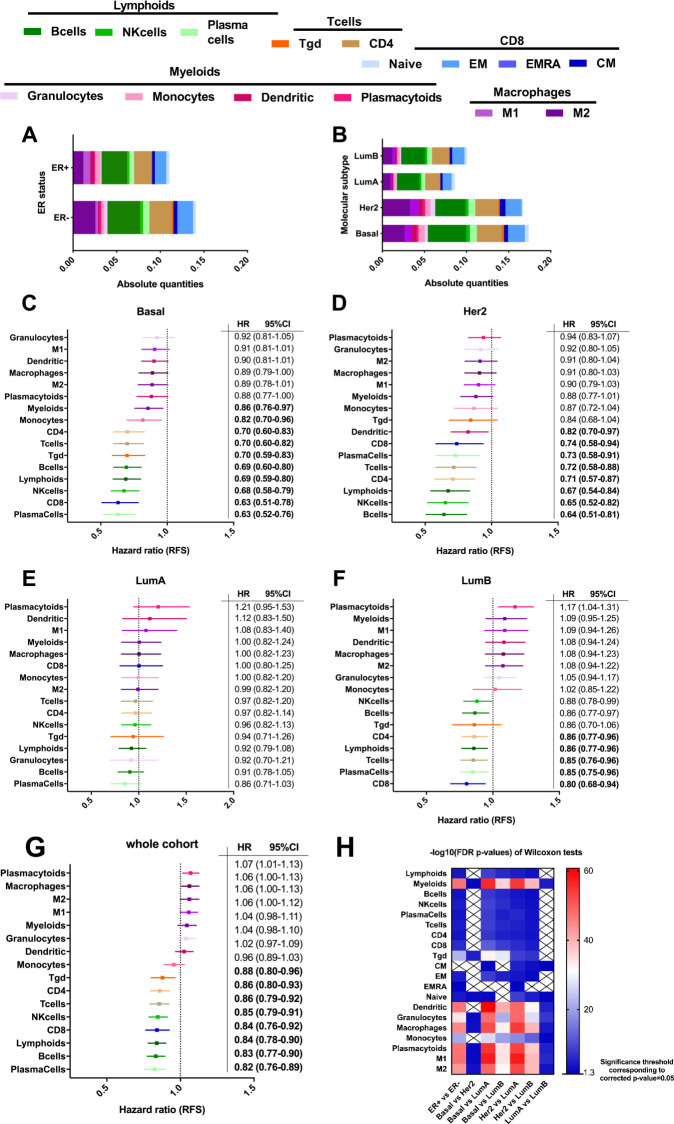
Figure 3.
(A–B) Bar graph representing the estimation of cell population absolute quantities in the different groups of ER status (A) and PAM50 (B). (C–G) Forest plot representing the HR for relapse free survival (RFS) of the different immune cell populations taken as a continuous variable in Basal (C), HER2 (D), LumA (E), LumB (F) subgroups, and in the whole cohort (G). (H) Represents the −log10 FDR (false discovery rate) corrected p values of Wilcoxon tests on a heatmap. The Wilcoxon test was used to compare the distributions of absolute quantities of a certain type of cells (in rows) between two subgroups (in columns). The p values were corrected due to the high number of tests using the FDR method. Crosses indicate non-significant tests.
The absolute quantities of different immune subpopulations vary widely between ER+ and ER− breast tumors (figure 3A), notably with significantly higher absolute quantities of M2 macrophages, B cells, CD4 T cells, and CD8 EM T cells in ER− tumors. These differences are even more pronounced when considering molecular intrinsic subtypes (figure 3B). Lymphoid cells were the most represented immune population (especially B cells and CD4 T cells). Briefly, concerning the main differences, among myeloid cells, M2 macrophages are more abundant than M1 in all molecular subtypes. M2 macrophages are more abundant in HER2 enriched, and above all, in basal-like subtypes, compared with luminal tumors (p<0.001). Luminal B tumors contained more macrophages than luminal A tumors (especially more M2) (p<0.001). Similar trends are observed for dendritic cells, with no significant difference between basal-like and HER2 enriched subtypes, but a significantly lower number of this cell subset in luminal subtypes, especially in luminal A tumors. Concerning lymphoid cells, B cells, NK cells, and T cells (both CD8 and CD4) were significantly higher in the basal-like subtype. Here again, there was no significant difference between basal-like and HER2 enriched subtypes, but lymphoid subpopulations were more abundant in these tumors than in luminal tumors. No difference was seen between luminal A and B subtypes concerning lymphoid populations (except for CM CD8 T cells, slightly higher in luminal B tumors). The heatmap of the significance of the differences among PAM50 molecular subtypes of breast cancer for each immune cell population is depicted in figure 3H and a table with average proportions by subgroup is available in online supplemental table 3.
jitc-2020-002036supp008.pdf (50.8KB, pdf)
Prognostic impact of overall TIC among breast cancer molecular subtypes
Figure 3C–F depicts the HRs and 95% CI for RFS for each immune subset taken as a continuous variable, and in the whole population (figure 3G, N=2736), and highlights the complex impact of different immune cells on the outcome of patients with breast cancer. While most myeloid populations are significantly associated with worse RFS (granulocytes, macrophages, especially M2, and dendritic cells), lymphoid subsets (T cells, B cells populations, and NK cells), are generally associated with significantly better outcome.
Figure 3C–F shows the prognostic role of each population according to breast cancer molecular subtype. In the basal-like subtype (figure 3C), all immune cell subtypes tend to be associated with better RFS. However, these associations were mainly statistically significant for lymphoid populations (B cells, T cells, and NK cells), consistent with the high prognostic value of TILs in triple negative breast cancer. In the HER2-enriched subtype (figure 3D), all immune populations also tend to be associated with better prognosis (here again, especially for lymphoid subtypes), but with less marked HRs than in basal-like subtypes. Interestingly, in luminal tumors, the prognostic value of immune cell subsets had a totally different profile, since in luminal A tumors, no immune cell subset was associated with outcome (figure 3E). In luminal B tumors (figure 3F), only B cell (and plasma cells), and T cell subsets (CD4, CD8, and NK cells), were associated with better RFS (but clearly with a lesser impact than in basal-like and HER2 enriched subtypes). In contrast, in this molecular type of breast cancer, most myeloid cells (granulocytes, macrophages both M1 and M2, and in a statistically significant manner, plasmacytoid cells), tended to be associated with worse RFS. These results obtained in luminal tumors reflect the contrasting and complex prognostic role of TILs, and more generally of the immune response, in ER+/HER2− breast cancers. Importantly, similar trends were also obtained for OS, despite many values not statistically significant when considering subgroup analyses (online supplemental figure S2A–E).
jitc-2020-002036supp009.pdf (489.6KB, pdf)
Added value of TIC on top of breast cancer molecular subtype for predicting relapse
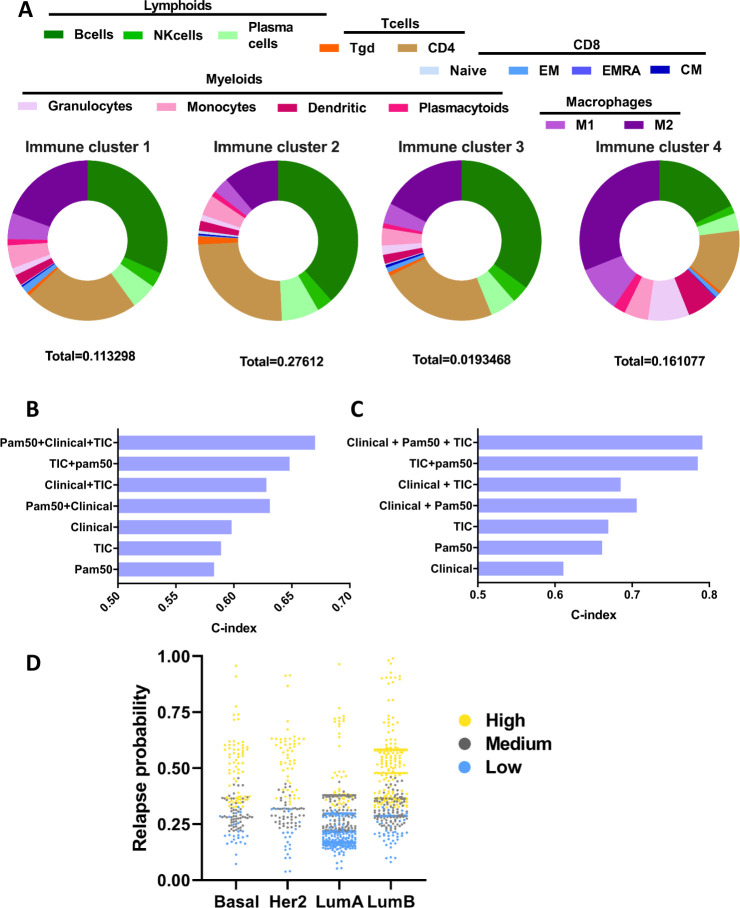
Unsupervised clustering analysis using immune cell relative quantities revealed four subgroups of breast tumors (ImmunoClass 1–4, figure 4A). Cluster 2 is characterized by a high level of myeloids and lymphoids in comparison to the other groups. Overall, this subgroup has a high level of immune cell infiltration. Cluster 4 is characterized by a high proportion of myeloids in comparison to other groups. Lymphoid and more precisely, most T-cell subpopulations are present in small quantities. Cluster 3 is characterized by low infiltration of lymphoids and myeloids, this type of tumor is poor in immune cells and could be considered as “immune desert.” Cluster 1 is the average of the previous cluster, it is not very infiltrated but some immune cells are present in average proportions. As shown in online supplemental figure S3, these immune clusters differ in terms of RFS (online supplemental figure S3A), with cluster 2 harboring better survival than the three others, and cluster 4 associated with the worst RFS. Clusters 1 and 3 have intermediate RFS. These exploratory results are in accordance with the respective prognostic role of lymphoid (favorable) and myeloid cells (unfavorable) in patients with cancer.33 These results have been obtained both in peripheral blood (namely the neutrophil-lymphocyte ratio),34 and in tumor tissue, especially in breast cancer.24 35 In an original, but logical way, patients with mild or low immune infiltration share intermediate prognosis. These results are also in accordance with the intermediate prognosis of moderate TILs infiltration in breast cancer.10 Interestingly, the spread of breast cancer molecular subtypes defined by PAM50 is independent of these TIC clusters (online supplemental figure S3B), suggesting that these two prognostic factors could be additive, to better refine breast cancer patients’ outcomes.
Figure 4.
(A) Pie charts representing the relative proportions of immune cells in each cluster of patients (ImmunoClass 1–4). The total proportions of these immune cells inside the tumor is indicated for each cluster. (B–C) Bar graphs representing the C-index values obtained by computing different Cox models. The PAM50 model is built using PAM50 classification, the tumor global immune contexture (TIC) is built using immune quantities information and the clinical model is built using T and N clinical variables. The model combining “PAM50” and “TIC” information models the interactions between the estimated quantities of immune cells and the PAM50 levels. As it was confirmed that each molecular subtype has an impact on the prognostic role of each immune cell population, a LASSO algorithm for variable selection in the Cox model was then used on immune and PAM50 variables and their respective interactions, to select variables that were most strongly related to the outcome (overall survival (OS) or relapse-free survival (RFS)). The linear predictor of this model was then used to build a final model combining the three types of information (quantitative immune populations, PAM50, and clinical). The C-index allows comparison between nested Cox models and was computed for the RFS models (B) and for the OS models (C). (D) Scatter plot of the relapse probability at 5 years of each patient of the whole cohort in each of the PAM50 groups. The points were colored based on the classification obtained via our model combining all three types of available information (clinical+PAM50+TIC). The number of patients with low, medium and high risk of relapse according to our final model (using tertiles as the threshold) appears, respectively in blue, gray, and yellow, distinguishing different outcomes in each PAM50 tumor group. LASSO, Least Absolute Shrinkage and Selection Operator.
jitc-2020-002036supp010.pdf (372.3KB, pdf)
Descriptive analysis was performed on both training and validation cohorts and this aggregation of patients is hereafter termed the “whole cohort.” The training cohort comprised the Kmplot and GEO cohorts together, and the validation cohort was the CIT cohort. As the importance and the respective prognostic roles of the different immune cell subsets varied among tumor types, we computed this “immune” prognostic information for each breast cancer molecular subtype with the prognostic information given by clinical stage (T and N status), and PAM50 subtype (figure 4B, C).
Compared with clinical parameters, neither PAM50 alone nor immune TIC cluster alone had better a C-index for the prediction of relapse (figure 4B). Concerning the risk of death, PAM50 alone or TIC cluster alone are slightly better than clinical parameters, but here again with low C-indexes (figure 4C). However, combining these independent parameters substantially increased the prognostic information. Adding immune TIC information to the clinical stage, and mainly to PAM50, strongly increased the C-index for the prediction of relapse or death. Above all, the C-index was best when we added immune TIC information to the model containing both clinical stage and PAM50. These results argue for the complementary prognostic value of these clinical and biological factors.
A scatter plot of the relapse probability at 5 years for each patient from both the training and validation cohorts in each of the PAM50 groups is depicted in figure 4D, according to our classification of risk (low, medium, or high) via our model combining all three types of available information (clinical+PAM50+TIC). This figure highlights the fact that the model can help to stratify patients risk in each molecular subgroup, and can for example identify patients with poor-risk luminal tumors (including luminal A tumors having comparable risk of relapse than HER2 or basal-like tumors).
Together these data underline the capacity of TIC to discriminate patient prognosis in a specific molecular subtype.
Refining survival stratification of patients with breast cancer with the final model incorporating TIC prognostic information
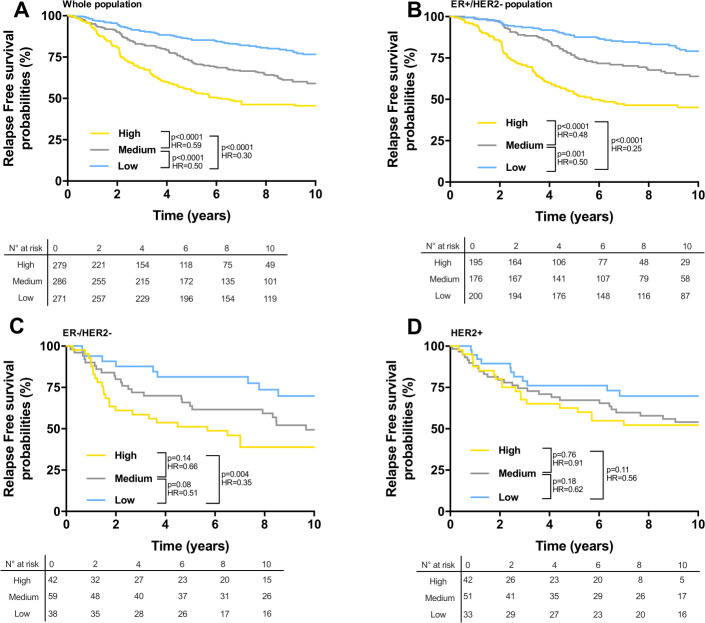
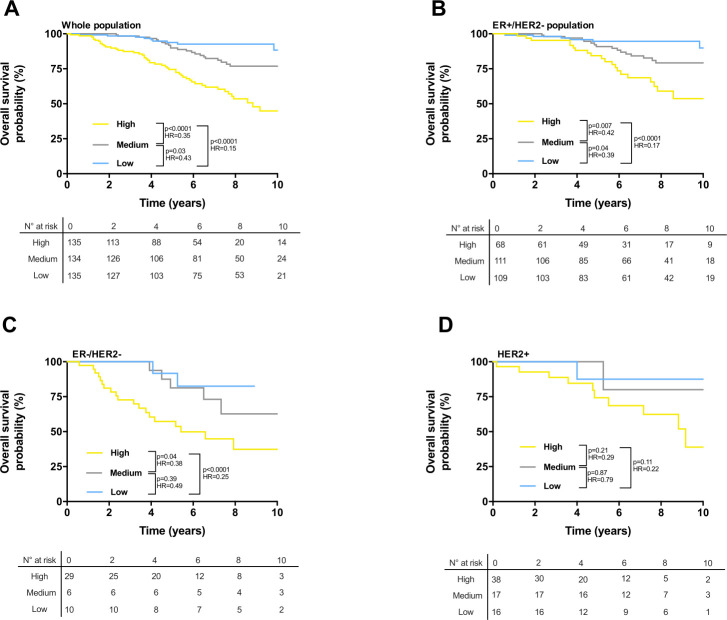
We computed survival of patients with breast cancer according to our final best model, which incorporated clinical stage, PAM50, and immune TIC. A score was obtained as the linear combination between the coefficients estimated by the Cox models (online supplemental table 2) and the variables. This score was then divided in three groups using tertiles (low score, medium, and high) stratifying the patients. In the training cohort (Kmplot and GEO cohorts, n=2276 patients), our final model identified a low-risk group with significantly better RFS compared with medium and high-risk groups in the whole population (figure 5A), the ER+/HER2− population (figure 5B), and the triple negative population (figure 5C). In the population of patients with breast cancer with HER2 positive tumors, there was no significant difference in RFS between the three prognostic groups (figure 5D). The 10-year RFS rates for low-risk groups were 79% (ER+/HER2−), 70% (triple negative), and 70% (HER2), respectively.
Figure 5.
(A–D) Kaplan-Meier estimates for relapse free survival; patients from the training cohort were stratified according to the score obtained from the Cox model combining clinical variables, PAM50 classification, and immune cell estimations, using tertiles as thresholds. Graphs are presented for all patients of the training cohort (A), for ER+/HER2− patients (B), for ER−/HER− patients (C), and for HER2+ patients (D). n.s., not significant, *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
jitc-2020-002036supp007.pdf (38.9KB, pdf)
Similar results were obtained in the training cohort for OS (figure 6A–D). The 10-year OS rates were 90% (ER+/HER2−), 83% (triple negative), and 88% (HER2). OS was not significantly different among the three prognostic groups in triple negative tumor patients (figure 6C), despite a clear poor survival for high-risk group. Overall, regarding the prognostic groups of patients, compared with PAM50 alone, our final classifier enabled better discrimination of the three different populations in terms of outcome, notably a group of patients with nearly 100% OS at 10 years, and in contrast, a group of patients with very poor survival. Note that for ER−/HER2− and HER2+ groups, number of patients is low, results should thus be interpreted with caution.
Figure 6.
(A–D) Kaplan-Meier estimates for overall survival; patients from the training cohort were stratified according to the score obtained from the Cox model combining clinical variables, PAM50 classification and immune cell estimations using tertiles as thresholds. Graphs are presented for all patients of the discovery cohort (A), for ER+/HER2− patients (B), for ER−/HER− patients (C), and for HER2+ patients (D). n.s., not significant, *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
Importantly, similar survival profiles were obtained when testing the same models in the validation cohort (CIT cohort, n=537) (RFS: online supplemental figure S4A–D, OS: online supplemental figure S4E–H).
jitc-2020-002036supp011.pdf (437.9KB, pdf)
Discussion
Thanks to a new gene expression deconvolution algorithm derived from Cibersort, allowing estimation of the relative and absolute quantities of various tumor-infiltrating immune cells, our study highlights the additional prognostic value of overall TIC on top of the breast cancer molecular intrinsic subtypes defined by the PAM50 classifier.
Our results show that the different CD8 T cell subpopulations, but also more generally, the immune contexture of breast tumors, as assessed by hierarchical clustering based on both myeloid and lymphoid cells, differ between breast cancer molecular intrinsic subtypes. They also have variable and complex associations with different level of clinical impact in terms of patient outcome. Finally, our findings make it possible to refine the survival stratification of patients with early breast cancer by incorporating TIC with PAM50 and clinical tumor burden (T and N stage) in a prognostic model assessed in training and validation cohorts.
The four breast cancer intrinsic subtypes were first described by Perou et al,1 and were initially defined by a large set of genes, subsequently reduced to 50 classifier genes in the PAM50 assay.6 36 Numerous studies have shown that PAM50 provides better estimates of early breast cancer patient outcome than classical clinicopathological factors.3 37 38 Moreover, PAM50 intrinsic subtypes, especially PAM50 proliferation genes, have also been shown to have a predictive value concerning the benefit of chemotherapy.6 38–41 Indeed, among the 50 genes involved in PAM50, the majority are related to proliferation, estrogen, or other growth factor pathways, but are not linked to immune response.6
Breast cancer was previously thought to be a non-immunogenic cancer, but numerous recent studies have suggested that in some cases, the presence of TILs can be a strong indicator of immune response against cancer cells, and thus a favorable prognostic factor.7 9 10 Indeed, in TNBC and HER2-overexpressing breast tumors, retrospective analyses of large adjuvant/neoadjuvant studies have shown that high level of TILs and CD8 T cells, or a Th1 cytotoxic signature, are independently associated with better survival and/or with better response to chemotherapy.7 9–11 18 42 In accordance with these pioneering histological studies, we found that TNBC and HER2-overexpressing cancers are breast tumors with the highest immune cell infiltration, and molecular subtypes in which most, if not all, immune cells are associated, or tend to be associated, with better survival. This was previously demonstrated in transcriptomic analyses from TNBC reported by Denkert et al,43 in which all immune genes were positively correlated among themselves (and with favorable treatment response), even when genes were markers of immunosuppressive populations.
More recent studies, based, like ours, on deconvolution analysis of gene expression data coming from early breast cancer, have highlighted the complexity of tumor-infiltrating immune cell types, and their respective proportions and prognostic roles.23 24 Accordingly, our unsupervised clustering analysis using immune cell quantities revealed four subgroups of breast tumors, largely independent of the PAM50 intrinsic subtype, and with distinct risks of relapse and death. These results indicate that the patterns and prognostic impact of immune infiltrates are not strictly linked to the biological tumor behavior defined by the molecular intrinsic subtypes. Accordingly, we demonstrate here the complementarity of both tumor intrinsic biology and tumor complex immune response, to better assess the prognosis of early breast cancer.
From an immunological point of view, our study demonstrates the prognostic relevance of 15 different subsets of immune cells among myeloid and lymphoid populations. More specifically, we describe here the relative infiltration of different CD8+ T cells. Among TILs, CD8+ T cells are believed to be one of the most important components of cell-mediated immunity, able to directly kill tumor cells presenting tumor-associated antigenic peptides. Accordingly, high tumor-infiltrating CD8+ cell density has been independently associated with improved survival in various types of cancer, including breast cancer.42 In breast cancer, this favorable effect appeared to be restricted to HER2-overexpressing and TNBC, with no significant impact in ER+/HER2− subtype.44
However, many different subpopulations of CD8+ T cells can be found in the tumor immune microenvironment, including naïve, memory, effector, and exhausted lymphocytes. After antigen encounter, naïve CD8+ T cells are activated and differentiate into memory T -cell populations, including effector CD8+ T cells.45 This differentiation is accompanied by the cell’s ability to proliferate and home into lymphoid organs, and to acquire a proinflammatory and cytotoxic phenotype.45 46 Among memory T cell populations, EM T cells are able to rapidly produce IFN-γ after antigenic recall stimulation, while CM T cells express lymph node homing receptors and produce less IFN-γ on stimulation.47
Our analyses revealed that EM CD8+ T cells constitute the predominant population of breast cancer CD8+ infiltrating T cells. These cells were more abundant in basal-like and HER2 molecular subtypes. Such results have been obtained elsewhere by flow cytometry analysis in a small cohort of localized breast cancer.48 Interestingly, in this translational study, and mostly based on luminal breast tumors, these CD8+ TILs expressed checkpoint molecules like programmed cell death protein 1 (PD-1) or TIGIT and are enriched in an exhausted phenotype. However, the authors demonstrated that these CD8+ infiltrating T cells retained the capacity to produce cytokines and to be cytotoxic.48 This could explain the favorable, and preponderant prognostic role of EM CD8+ T cells observed in our study. Indeed, compared with CM or naïve CD8 T cells, our results highlighted the prominent prognostic importance of effector memory cells, among the CD8 T cell population. The presence of CD45RO+ tumor infiltrating memory T cells has been associated with an absence of signs of early metastatic invasion, less advanced pathological stage, and increased survival in pioneering works conducted in localized colorectal cancer samples.49 More recently, a specific favorable role of EM CD8 T cells has been deciphered in ovarian cancer,50 and melanoma.51 In ipilimumab-treated metastatic melanoma patients, peripheral CD8 effector-memory type 1 T-cells have been correlated with more favorable outcome,52 highlighting their potential role as a predictive biomarker for immunotherapy efficacy.
Interestingly, in our breast cancer cohorts, this favorable prognostic effect was observed in basal like and HER2 tumors, but also significantly in luminal B subtypes. This is another means of underlining the complementarity of both immunological and tumor molecular intrinsic information to decipher the complexity of TIL impact in ER+ luminal breast tumor prognosis. Indeed, our results demonstrate a clear and different effect of CD8, and more generally, of the different subsets of immune cells included in TIC, among luminal A and luminal B tumors, with a clear favorable prognostic impact of lymphoid populations (but not myeloid cells, which tend to be associated with worse outcome) for luminal B tumors, whereas no significant effect can be seen for luminal A tumors. These results may partially explain the controversial role of TILs and immune response in luminal breast cancer. In this respect, it should be noted that our final prognostic model incorporating tumor stage, PAM50 and TIC prognostic information was more efficient in ER+ tumors than PAM50 alone or combined with tumor stage, making it possible to discriminate three different populations in terms of outcome, notably a group of patients with nearly 100% OS at 10 years and in contrast, a group of patients with very poor survival considering ER+ localized breast cancer. These results highlight, in our view, the clinical importance of immune response in early breast cancer, even in ER+subtypes.
It should be noted that usually, such correlative studies do not constitute formal proof of a causal relationship, leaving open questions as to whether all immune subsets of TILs cooperate to influence the prognosis of patients, or whether these associations with survival are solely the result of tumor cell intrinsic biological conditions. In this regard, our results show for the first time that different clusters of overall TIC in breast cancer appear to be independent of the intrinsic molecular subtypes usually defined by PAM50. This argues for a specific role of immune response in the prognosis of patients with breast cancer, and for the complementarity of both biological information to refine patient prognosis. This refinement in the prediction of clinical outcome makes it possible to identify, within each subtype of breast cancer, patients with good, intermediate or poor prognosis, thereby opening the door for tailored adjuvant treatment, according to the risk of relapse and death. These results are of particular interest in the recent context of new immunotherapies, since the prognostic and predictive value of antitumor immune response in breast cancer indicates that immunotherapy could constitute a promising option for the treatment of some breast cancers. However, in breast cancer, despite some clinical response observed with anti-PD-1 or anti-programmed death-ligand 1 (PD-L1) monotherapy, the rate of responders remains low compared with other cancer types (such as melanoma, kidney, or non-small cell lung cancer), even in the triple negative subtype, despite higher rates of TILs, PD-L1 expression, and higher TMB.53 A better understanding of tumor-infiltrating immune cell composition among the different breast cancer molecular subtypes could benefit the future design of specific immunotherapeutic clinical trials for patients with breast cancer in each molecular type of disease.
Our study has other limitations: thus, the immunological parameters that we integrated into our models do not take into account the degree of activation or inactivation of the different immune populations (evaluated in part by the degree of expression of the checkpoints of immune response), nor the spatial organization of these immune cells, to which we have not had access. As these two pieces of information are also associated with the prognosis of early breast cancer,12 13 15 54 it could be interesting to study to what extent these parameters could be complementary with our approach to further refine the individual prognosis.
In addition, whether our tool could also help to select patients for standard adjuvant systemic treatment (eg, endocrine treatment or chemotherapy) remains unclear. Additional studies in cohorts of patients treated with neoadjuvant strategies are ongoing, and would help us to understand whether overall TIC, and our prognostic model also enable identification of responding tumors or not (as a predictive biomarker).
In conclusion, our study provides new insights into the complexity of breast cancer immune response, and its clinical relevance, when combined with tumor intrinsic molecular subtypes to refine prognosis of patients with breast cancer in each subtype. We bring together in the context of early breast cancer, the prognostic information given by both molecular intrinsic subtype (assessed by PAM50), and by the overall TIC assessed by a new gene expression deconvolution algorithm. However, the complexity of immune infiltrate interplay, and how they influence long term outcome, remains largely unclear in breast cancer, as in the majority of other solid cancers. In the future, novel biological techniques, like single-cell molecular analyses will certainly help to reveal the profound complexity of cellular subsets, and their molecular and functional properties. Together with an improved understanding of tumor cell intrinsic biology, these techniques of high analytic resolution could be complementary to multiplexed histological analyses55 and high throughput bioinformatic tools in deciphering the complex landscape of immune response against cancer cells, and providing better rationales for the design of future clinical trials in immunotherapy and oncology.
Acknowledgments
We wish to thank Fiona Ecarnot for English correction and helpful comments.
Footnotes
Contributors: Conceptualization: SL and QK. Methodology: QK and CT. Validation: QK, VD, LA, MT, EL, FG, CT, and SL. Formal analysis: QK. Writing: QK, VD, LA, MT, EL, FG, CT, and SL. Supervision: SL.
Funding: The authors have not declared a specific grant for this research from any funding agency in the public, commercial or not-for-profit sectors.
Competing interests: None declared.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
Data sharing not applicable as no datasets generated and/or analyzed for this study. Data sharing not applicable as no datasets were generated for this study. Codes are available upon request.
References
- 1.Perou CM, Sørlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature 2000;406:747–52. 10.1038/35021093 [DOI] [PubMed] [Google Scholar]
- 2.Paik S, Shak S, Tang G, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med 2004;351:2817–26. 10.1056/NEJMoa041588 [DOI] [PubMed] [Google Scholar]
- 3.Nielsen TO, Parker JS, Leung S, et al. A comparison of PAM50 intrinsic subtyping with immunohistochemistry and clinical prognostic factors in tamoxifen-treated estrogen receptor-positive breast cancer. Clin Cancer Res 2010;16:5222–32. 10.1158/1078-0432.CCR-10-1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jerevall P-L, Ma X-J, Li H, Salunga R, et al. Prognostic utility of HOXB13:IL17BR and molecular grade index in early-stage breast cancer patients from the Stockholm trial. Br J Cancer 2011;104:1762–9. 10.1038/bjc.2011.145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sotiriou C, Wirapati P, Loi S, et al. Gene expression profiling in breast cancer: understanding the molecular basis of histologic grade to improve prognosis. J Natl Cancer Inst 2006;98:262–72. 10.1093/jnci/djj052 [DOI] [PubMed] [Google Scholar]
- 6.Parker JS, Mullins M, Cheang MCU, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol 2009;27:1160–7. 10.1200/JCO.2008.18.1370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Criscitiello C, Esposito A, Trapani D, et al. Prognostic and predictive value of tumor infiltrating lymphocytes in early breast cancer. Cancer Treat Rev 2016;50:205–7. 10.1016/j.ctrv.2016.09.019 [DOI] [PubMed] [Google Scholar]
- 8.Stanton SE, Adams S, Disis ML. Variation in the incidence and magnitude of tumor-infiltrating lymphocytes in breast cancer subtypes: a systematic review. JAMA Oncol 2016;2:1354–60. 10.1001/jamaoncol.2016.1061 [DOI] [PubMed] [Google Scholar]
- 9.Savas P, Salgado R, Denkert C, et al. Clinical relevance of host immunity in breast cancer: from TILs to the clinic. Nat Rev Clin Oncol 2016;13:228–41. 10.1038/nrclinonc.2015.215 [DOI] [PubMed] [Google Scholar]
- 10.Denkert C, von Minckwitz G, Darb-Esfahani S, et al. Tumour-infiltrating lymphocytes and prognosis in different subtypes of breast cancer: a pooled analysis of 3771 patients treated with neoadjuvant therapy. Lancet Oncol 2018;19:40–50. 10.1016/S1470-2045(17)30904-X [DOI] [PubMed] [Google Scholar]
- 11.Desmedt C, Haibe-Kains B, Wirapati P, et al. Biological processes associated with breast cancer clinical outcome depend on the molecular subtypes. Clin Cancer Res 2008;14:5158–65. 10.1158/1078-0432.CCR-07-4756 [DOI] [PubMed] [Google Scholar]
- 12.Kristensen VN, Vaske CJ, Ursini-Siegel J, et al. Integrated molecular profiles of invasive breast tumors and ductal carcinoma in situ (DCIS) reveal differential vascular and interleukin signaling. Proc Natl Acad Sci U S A 2012;109:2802–7. 10.1073/pnas.1108781108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Denkert C, Wienert S, Klauschen F. Analyzing the immunological landscape of a Tumor—Heterogeneity of immune infiltrates in breast cancer as a new prognostic indicator. JNCI 2018;110:129–30. 10.1093/jnci/djx188 [DOI] [Google Scholar]
- 14.Heindl A, Sestak I, Naidoo K, et al. Relevance of spatial heterogeneity of immune infiltration for predicting risk of recurrence after endocrine therapy of ER+ breast cancer. J Natl Cancer Inst 2018;110:166–75. 10.1093/jnci/djx137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gruosso T, Gigoux M, Manem VSK, et al. Spatially distinct tumor immune microenvironments stratify triple-negative breast cancers. J Clin Invest 2019;129:1785–800. 10.1172/JCI96313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bates GJ, Fox SB, Han C, et al. Quantification of regulatory T cells enables the identification of high-risk breast cancer patients and those at risk of late relapse. J Clin Oncol 2006;24:5373–80. 10.1200/JCO.2006.05.9584 [DOI] [PubMed] [Google Scholar]
- 17.Mahmoud SMA, Lee AHS, Paish EC, et al. Tumour-infiltrating macrophages and clinical outcome in breast cancer. J Clin Pathol 2012;65:159–63. 10.1136/jclinpath-2011-200355 [DOI] [PubMed] [Google Scholar]
- 18.Ignatiadis M, Singhal SK, Desmedt C, et al. Gene modules and response to neoadjuvant chemotherapy in breast cancer subtypes: a pooled analysis. J Clin Oncol 2012;30:1996–2004. 10.1200/JCO.2011.39.5624 [DOI] [PubMed] [Google Scholar]
- 19.Karn T, Denkert C, Weber KE, et al. Tumor mutational burden and immune infiltration as independent predictors of response to neoadjuvant immune checkpoint inhibition in early TNBC in GeparNuevo. Ann Oncol 2020;31:1216–22. 10.1016/j.annonc.2020.05.015 [DOI] [PubMed] [Google Scholar]
- 20.Thomas R, Al-Khadairi G, Decock J. Immune checkpoint inhibitors in triple negative breast cancer treatment: promising future prospects. Front Oncol 2020;10:600573. 10.3389/fonc.2020.600573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fridman WH, Zitvogel L, Sautès-Fridman C, et al. The immune contexture in cancer prognosis and treatment. Nat Rev Clin Oncol 2017;14:717–34. 10.1038/nrclinonc.2017.101 [DOI] [PubMed] [Google Scholar]
- 22.Fridman WH, Pagès F, Sautès-Fridman C, et al. The immune contexture in human tumours: impact on clinical outcome. Nat Rev Cancer 2012;12:298–306. 10.1038/nrc3245 [DOI] [PubMed] [Google Scholar]
- 23.Ali HR, Chlon L, Pharoah PDP, et al. Patterns of immune infiltration in breast cancer and their clinical implications: a gene-expression-based retrospective study. PLoS Med 2016;13:e1002194. 10.1371/journal.pmed.1002194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bense RD, Sotiriou C, Piccart-Gebhart MJ, et al. Relevance of tumor-infiltrating immune cell composition and functionality for disease outcome in breast cancer. J Natl Cancer Inst 2017;109. 10.1093/jnci/djw192. [Epub ahead of print: 13 Oct 2016]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods 2015;12:453–7. 10.1038/nmeth.3337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Benjamini Y, Drai D, Elmer G, et al. Controlling the false discovery rate in behavior genetics research. Behav Brain Res 2001;125:279–84. 10.1016/s0166-4328(01)00297-2 [DOI] [PubMed] [Google Scholar]
- 27.Klopfenstein Q, Vaiter S. Linear support vector regression with linear constraints. arXiv:191102306 [math, stat] [Internet], 2019. Available: http://arxiv.org/abs/1911.02306 [Accessed 2 Feb 2020].
- 28.Guedj M, Marisa L, de Reynies A, et al. A refined molecular taxonomy of breast cancer. Oncogene 2012;31:1196–206. 10.1038/onc.2011.301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Györffy B, Lanczky A, Eklund AC, et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat 2010;123:725–31. 10.1007/s10549-009-0674-9 [DOI] [PubMed] [Google Scholar]
- 30.Gendoo DMA, Ratanasirigulchai N, Schröder MS, et al. Genefu: an R/Bioconductor package for computation of gene expression-based signatures in breast cancer. Bioinformatics 2016;32:1097–9. 10.1093/bioinformatics/btv693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cejalvo JM, Pascual T, Fernández-Martínez A, et al. Distribution of the PAM50 breast cancer subtypes within each pathology-based group: a combined analysis of 15,339 patients across 29 studies. Ann Oncol 2017;28:v603. 10.1093/annonc/mdx391.026 [DOI] [Google Scholar]
- 32.Yu X, Zhang L, Chaudhry A, et al. Unravelling the heterogeneity and dynamic relationships of tumor-infiltrating T cells by single-cell RNA sequencing analysis. J Leukoc Biol 2020;107:917–32. 10.1002/JLB.6MR0320-234R [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thorsson V, Gibbs DL, Brown SD, et al. The immune landscape of cancer. Immunity 2018;48:812–30. 10.1016/j.immuni.2018.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gago-Dominguez M, Matabuena M, Redondo CM, et al. Neutrophil to lymphocyte ratio and breast cancer risk: analysis by subtype and potential interactions. Sci Rep 2020;10:13203. 10.1038/s41598-020-70077-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gentles AJ, Newman AM, Liu CL, et al. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat Med 2015;21:938–45. 10.1038/nm.3909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Prat A, Parker JS, Fan C, et al. PAM50 assay and the three-gene model for identifying the major and clinically relevant molecular subtypes of breast cancer. Breast Cancer Res Treat 2012;135:301–6. 10.1007/s10549-012-2143-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chia SK, Bramwell VH, Tu D, et al. A 50-gene intrinsic subtype classifier for prognosis and prediction of benefit from adjuvant tamoxifen. Clin Cancer Res 2012;18:4465–72. 10.1158/1078-0432.CCR-12-0286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martín M, Prat A, Rodríguez-Lescure A, et al. PAM50 proliferation score as a predictor of weekly paclitaxel benefit in breast cancer. Breast Cancer Res Treat 2013;138:457–66. 10.1007/s10549-013-2416-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cheang MCU, Voduc KD, Tu D, et al. Responsiveness of intrinsic subtypes to adjuvant anthracycline substitution in the NCIC.CTG MA.5 randomized trial. Clin Cancer Res 2012;18:2402–12. 10.1158/1078-0432.CCR-11-2956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Prat A, Bianchini G, Thomas M, et al. Research-based PAM50 subtype predictor identifies higher responses and improved survival outcomes in HER2-positive breast cancer in the NOAH study. Clin Cancer Res 2014;20:511–21. 10.1158/1078-0432.CCR-13-0239 [DOI] [PubMed] [Google Scholar]
- 41.Jørgensen CLT, Nielsen TO, Bjerre KD, et al. PAM50 breast cancer intrinsic subtypes and effect of gemcitabine in advanced breast cancer patients. Acta Oncol 2014;53:776–87. 10.3109/0284186X.2013.865076 [DOI] [PubMed] [Google Scholar]
- 42.Mahmoud SMA, Paish EC, Powe DG, et al. Tumor-infiltrating CD8+ lymphocytes predict clinical outcome in breast cancer. J Clin Oncol 2011;29:1949–55. 10.1200/JCO.2010.30.5037 [DOI] [PubMed] [Google Scholar]
- 43.Denkert C, von Minckwitz G, Brase JC, et al. Tumor-infiltrating lymphocytes and response to neoadjuvant chemotherapy with or without carboplatin in human epidermal growth factor receptor 2-positive and triple-negative primary breast cancers. J Clin Oncol 2015;33:983–91. 10.1200/JCO.2014.58.1967 [DOI] [PubMed] [Google Scholar]
- 44.Ali HR, Provenzano E, Dawson S-J, et al. Association between CD8+ T-cell infiltration and breast cancer survival in 12,439 patients. Ann Oncol 2014;25:1536–43. 10.1093/annonc/mdu191 [DOI] [PubMed] [Google Scholar]
- 45.Gattinoni L, Speiser DE, Lichterfeld M, et al. T memory stem cells in health and disease. Nat Med 2017;23:06:18–27. 10.1038/nm.4241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hamann D, Baars PA, Rep MH, et al. Phenotypic and functional separation of memory and effector human CD8+ T cells. J Exp Med 1997;186:1407–18. 10.1084/jem.186.9.1407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sallusto F, Lenig D, Förster R, et al. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature 1999;401:708–12. 10.1038/44385 [DOI] [PubMed] [Google Scholar]
- 48.Egelston CA, Avalos C, Tu TY, et al. Human breast tumor-infiltrating CD8+ T cells retain polyfunctionality despite PD-1 expression. Nat Commun 2018;9:4297. 10.1038/s41467-018-06653-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pagès F, Berger A, Camus M, et al. Effector memory T cells, early metastasis, and survival in colorectal cancer. N Engl J Med 2005;353:2654–66. 10.1056/NEJMoa051424 [DOI] [PubMed] [Google Scholar]
- 50.Lieber S, Reinartz S, Raifer H, et al. Prognosis of ovarian cancer is associated with effector memory CD8+ T cell accumulation in ascites, CXCL9 levels and activation-triggered signal transduction in T cells. Oncoimmunology 2018;7:e1424672. 10.1080/2162402X.2018.1424672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wistuba-Hamprecht K, Di Benedetto S, Schilling B, et al. Phenotypic characterization and prognostic impact of circulating γδ and αβ T-cells in metastatic malignant melanoma. Int J Cancer 2016;138:698–704. 10.1002/ijc.29818 [DOI] [PubMed] [Google Scholar]
- 52.Wistuba-Hamprecht K, Martens A, Heubach F, et al. Peripheral CD8 effector-memory type 1 T-cells correlate with outcome in ipilimumab-treated stage IV melanoma patients. Eur J Cancer 2017;73:61–70. 10.1016/j.ejca.2016.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Emens LA. Breast cancer immunotherapy: facts and hopes. Clin Cancer Res 2018;24:511–20. 10.1158/1078-0432.CCR-16-3001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gaynor N, Crown J, Collins DM. Immune checkpoint inhibitors: key trials and an emerging role in breast cancer. Semin Cancer Biol 2020. 10.1016/j.semcancer.2020.06.016. [Epub ahead of print: 02 Jul 2020]. [DOI] [PubMed] [Google Scholar]
- 55.Carstens JL, Correa de Sampaio P, Yang D, et al. Spatial computation of intratumoral T cells correlates with survival of patients with pancreatic cancer. Nat Commun 2017;8:15095. 10.1038/ncomms15095 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jitc-2020-002036supp001.XLSX (35.3KB, XLSX)
jitc-2020-002036supp002.pdf (341.9KB, pdf)
jitc-2020-002036supp003.pdf (81.8KB, pdf)
jitc-2020-002036supp004.XLSX (77.3KB, XLSX)
jitc-2020-002036supp006.pdf (63KB, pdf)
jitc-2020-002036supp005.XLSX (24KB, XLSX)
jitc-2020-002036supp008.pdf (50.8KB, pdf)
jitc-2020-002036supp009.pdf (489.6KB, pdf)
jitc-2020-002036supp010.pdf (372.3KB, pdf)
jitc-2020-002036supp007.pdf (38.9KB, pdf)
jitc-2020-002036supp011.pdf (437.9KB, pdf)
Data Availability Statement
Data sharing not applicable as no datasets generated and/or analyzed for this study. Data sharing not applicable as no datasets were generated for this study. Codes are available upon request.