Abstract
Cyclocotyla bellones Otto, 1823 (Diclidophoridae) is a monogenean characterised by an exceptional way of life. It is a hyperparasite that attaches itself to the dorsal face of isopods, themselves parasites in the buccal cavity of fishes. In this study, Cy. bellones was found on Ceratothoa parallela (Otto, 1828), a cymothoid isopod parasite of the sparid fish Boops boops off Algeria in the Mediterranean Sea. We provide, for the first time, molecular barcoding information of a hyperparasitic monogenean, the parasitic crustacean host, and the fish host, with COI sequences.
Keywords: Hyperparasite, Cyclocotyla bellones, Polyopisthocotylea, Isopoda, Boops boops, Barcoding
Abstract
Cyclocotyla bellones Otto, 1823 (Diclidophoridae) est un monogène caractérisé par un mode de vie exceptionnel. C’est un hyperparasite qui se fixe sur la face dorsale d’isopodes, eux-mêmes parasites dans la cavité buccale de poissons. Dans cette étude, Cy. bellones a été trouvé sur Ceratothoa parallela (Otto, 1828), un isopode cymothoïdé parasite du poisson sparidé Boops boops au large de l’Algérie, en mer Méditerranée. Nous fournissons, pour la première fois, des informations de codes-barres moléculaires d’un monogène hyperparasite, de l’hôte crustacé parasite et de l’hôte poisson, avec des séquences de COI.
Introduction
Cyclocotyla bellones Otto, 1823 is a diclidophorid Monogenea, hyperparasite of cymothoid isopods of the buccal cavity of the sparid fish Boops boops (Linnaeus, 1758). It was first described in one of the earliest accounts on monogeneans by Otto (1823), who erected the genus Cyclocotyla Otto, 1823 for Cy. bellones collected from the skin of the dorsal face of the garfish Belone belone (Linnaeus, 1760) off Naples, Italy [62]. It was then frequently reported on isopods parasitic of B. boops and of Spicara spp. (see Table 1), with a single record on cymothoids of the carangid Trachurus trachurus (Linnaeus, 1758) [23].
Table 1.
Hosts and localities of Cyclocotyla bellones reported in the literature.
| Host/locality | Reference |
|---|---|
| Belone belone (type host) | |
| Italy, Mediterranean Sea | [62] |
| Ceratothoa oestroides, buccal cavity of Trachurus trachurus | |
| Spain, Atlantic Ocean | [23] |
| Ceratothoa oestroides, buccal cavity of Boops boops | |
| Monaco, Mediterranean Sea | [22] |
| France, Mediterranean Sea | [68] |
| Cymothoids, buccal cavity of Spicara maena | |
| France, Mediterranean Sea | [46] |
| Ceratothoa oestroides, buccal cavity of Spicara maena | |
| Turkey, Mediterranean Sea | [74] |
| France, Mediterranean Sea | [25] |
| Spain, Mediterranean Sea | [14] |
| Algeria, Mediterranean Sea | [5] |
| Emetha audouini, buccal cavity of Spicara maena | |
| Montenegro, Mediterranean Sea | [74] |
| France, Mediterranean Sea | [25] |
| Emetha audouini, buccal cavity of Spicara smaris | |
| Montenegro, Mediterranean Sea | [66] |
| France, Mediterranean Sea | [25] |
| Cymothoidae, Spicara smaris | |
| Montenegro, Mediterranean Sea | [66] |
| Boops boops, Cymothoids | |
| France, Mediterranean Sea | [46, 59] |
| Boops boops, Ceratothoa oestroides | |
| France, Mediterranean Sea | [25] |
| Italy, Mediterranean Sea | [53] |
| Boops boops, E. audouini-Ceratothoa oxyrrhynchaena-Ceratothoa parallela | |
| France, Mediterranean Sea | [25] |
| Boops boops, Cymothoids | |
| France, Mediterranean Sea | [46] |
| Boops boops, Ceratothoa parallela | |
| Turkey, Mediterranean Sea | [74] |
| France, Mediterranean Sea | [68] |
| Algeria, Mediterranean Sea | Present study |
Similarly, a few other diclidophorids were described from parasitic crustaceans: Allodiclidophora squillarum (Parona & Perugia, 1889) [syn. Mesocotyle squillarum Parona & Perugia, 1889] on the bopyrid Bopyrus squillarum Latreille, 1802 off Italy [63]; Allodiclidophora charcoti (Dollfus, 1922) [syn. Cyclobothrium charcoti Dollfus, 1922] on a female Ceratothoa oestroides (Risso, 1816) from the buccal cavity of T. trachurus (Linnaeus, 1758) and of B. boops (Linnaeus, 1758), caught from Spain and Monaco, respectively [22–24]; and Diclidophora merlangi (Kuhn, 1829) Krøyer, 1838 [syn. Dactylocotyle merlangi Kuhn, 1829] collected on Ceratothoa oestroides from the buccal cavity of B. boops from Italy (Taschenberg 1879 in [23]).
The identity of the hyperparasite monogenean, the crustacean parasite-host, or the fish host was not confirmed via DNA sequencing in any of these instances. Hence, as part of an ongoing effort to characterize the parasite biodiversity of fishes off the Southern shores of the Mediterranean Sea [3, 6–12, 15–18, 20, 42], molecular characterization of the three members of the hyperparasite-parasite-host association is provided for the first time.
We do not show detailed illustrations or measurements of monogeneans in this paper because these will be provided in a future article; however, we provide a general illustration and a short description.
Materials and methods
Collection and sampling of fishes
From 2017 to 2019, 624 specimens of B. boops and 45 Pagellus acarne (Risso, 1827) were collected from fish markets off Réghaia on the Algerian coast or directly from local fishermen in Bouharoun (36°37′ N, 2°39′ E). Fish specimens were transferred to the laboratory shortly after capture and identified using keys [26] and examined fresh on the day of purchase. The buccal cavities were carefully examined for isopods. Gill arches were also resected and placed in separate Petri dishes containing filtered sea water. The buccal cavity, isopods and isolated gills were observed under a dissecting microscope for the presence of monogeneans.
Collection of isopods and monogeneans
For the host fish B. boops, all isopods (sometimes with visible diclidophorid monogeneans) were removed from the buccal cavity using dissecting forceps. Monogeneans were isolated from the isopods with a fine dissecting needle. Other diclidophorids (Choricotyle sp.) parasitic on gills of another sparid, Pagellus acarne were removed from the gills using a fine dissecting needle (Table 2). Note that amongst the polyopisthocotyleans collected on P. acarne, only Choricotyle sp. was included as it is remarkably similar in morphology to species of Cyclocotyla; Choricotyle sp. is a parasite of the fish so no isopod was involved in this particular association.
Table 2.
Fishes, Isopoda, Monogenea, and their COI sequences. To ensure full traceability and respect of host-parasite relationships, for Cyclocotyla bellones one monogenean was collected from one parasitic isopod and each fish, isopod and monogenean individuals were sequenced. Choricotyle chrysophryi is a parasite of the fish, so no isopod was involved. All vouchers were deposited in the MNHN.
| Fish species | Fish ID | GenBank fish COI sequence | Isopoda species | Isopoda ID | GenBank Isopoda COI sequence | Voucher | Monogenea species | Monogenea ID | GenBank Monogenea COI sequence | Voucher slide |
|---|---|---|---|---|---|---|---|---|---|---|
| Boops boops | Bobo Br2 | MZ127221 | Ceratothoa parallela (female) | Bobo Br2 Iso1 | MZ127227 | MNHN-IU-2016-9111 | Cyclocotyla bellones | Bobo Br2 Iso1 MO01 | MZ127220 | MNHN HEL1307 |
| Boops boops | Bobo Br5 | MZ127226 | Ceratothoa parallela (female) | Bobo Br5 Iso1 | MZ127219 | MNHN-IU-2016-9112 | Cyclocotyla bellones | Bobo Br5 Iso1 MO01 | MZ127224 | MNHN HEL1308 |
| Boops boops | Bobo Br6 | MZ127217 | Ceratothoa parallela (female) | Bobo Br6 Iso1 | MZ127225 | MNHN-IU-2016-91123 | Cyclocotyla bellones | Bobo Br6 Iso1 MO01 | MZ127218 | MNHN HEL1309 |
| Pagellus acarne | PaacaBr1 | MZ127223 | Choricotyle cf. chrysophryi | Paaca Br1 MO01 | MZ127216 | MNHN HEL1310 | ||||
| Choricotyle cf. chrysophryi | Paaca Br1 MO02 | MZ127222 | MNHN HEL1311 | |||||||
Morphological methods
Monogeneans were preserved in 70% ethanol, stained with acetic carmine, dehydrated in a graded series of alcohol for 15 min each: (70, 96 and 100%), cleared in clove oil, and finally mounted in Canada balsam. Monogeneans were identified on stained whole mounts. Isopods were identified with the help of Prof. Jean-Pierre Trilles (University of Montpellier, France).
Deposition of specimens
Voucher specimens of monogeneans (hologenophores sensu Pleijel [64]) were deposited in the Muséum National d’Histoire Naturelle, Paris, France (MNHN), under registration numbers MNHN HEL1307–1336. Voucher specimens of isopods (hologenophores) were deposited in the Muséum National d’Histoire Naturelle, Paris, France (MNHN), under registration numbers MNHN-IU-2016-9111–9113. Fish specimens were not deposited.
Molecular methods
For complete traceability of the molecular study, special care was taken to ensure that hosts and monogeneans were labelled with respect to host-parasites relationships [3, 7, 8, 10, 11, 39].
For three individual B. boops, the parasitic female isopod and one monogenean on this individual isopod were extracted (Table 2). A tissue sample from the gill of the fish was taken and a pereopod was detached from each infected isopod and submitted to molecular analysis. For the monogenean, a small lateral part of the body was separated with a scalpel and submitted to molecular analysis, and the rest of the body was mounted on a slide as a voucher for drawing and deposition in a Museum collection. This ensures that the molecular identification of the host fish, the parasitic isopod and their monogenean parasite correspond perfectly at the individual host and parasite levels, and enable morphological assessment of sequenced monogeneans. Slides of monogeneans were deposited in the Muséum National d’Histoire Naturelle, Paris, France (MNHN), under registration numbers MNHN HEL1307–1309.
Molecular barcoding of fish
Total genomic DNA was isolated using a QIAamp DNA Mini Kit (Qiagen, Courtaboeuf, France), according to the manufacturer’s instructions. The 5′ region of the mitochondrial cytochrome c oxidase subunit I (COI) gene was amplified with the primers TelF1 (5′–TCGACTAATCAYAAAGAYATYGGCAC–3′) and TelR1 (5′–ACTTCTGGGTGNCCAAARAATCARAA–3′) [21]. PCR reactions were performed in 20 μL, containing 1 ng of DNA, 1× CoralLoad PCR buffer, 3 mM MgCl2, 66 μM of each dNTP, 0.15 μM of each primer, and 0.5 units of Taq DNA polymerase (Qiagen). The amplification protocol was 4 min at 94 °C, followed by 40 cycles at 94 °C for 30 s, 48 °C for 40 s, and 72 °C for 50 s, with a final extension at 72 °C for 7 min. PCR products were purified (Ampure XP Kit, Beckman Coulter, Brea, CA, USA) and sequenced in both directions on a 3730 × l DNA Analyzer 96-capillary sequencer (Applied Biosystems, Foster City, CA, USA). We used CodonCode Aligner version 3.7.1 software (Codon Code Corporation, Dedham, MA, USA) to edit sequences, compared them to the GenBank database content with BLAST, and deposited them in GenBank (accession numbers in Table 1). Species identification was confirmed with the BOLD identification engine [67].
Molecular barcoding of isopods
Total genomic DNA from a pereopod was isolated using a QIAamp DNA Mini Kit (Qiagen, Courtaboeuf, France), according to the manufacturer’s instructions. The 5′ region of the COI gene was amplified with the “universal” primers LCO1490 (5′–GGTCAACAAATCATAAAGATATTGG–3′) and HCO2198 (5′–TAAACTTCAGGGTGACCAAAAAATCA–3′) [27]. PCR reactions were performed in 20 μL, containing 1 ng of DNA, 1× CoralLoad PCR buffer, 3 mM MgCl2, 66 μM of each dNTP, 0.15 μM of each primer, and 0.5 units of Taq DNA polymerase (Qiagen). The amplification protocol was 4 min at 94 °C, followed by 40 cycles at 94 °C for 30 s, 48 °C for 40 s, and 72 °C for 50 s, with a final extension at 72 °C for 7 min. Sequences were obtained as for fish and were deposited in GenBank (accession numbers in Table 1).
Molecular barcoding of monogeneans
Total genomic DNA was isolated using a QIAmp DNA Micro Kit (Qiagen). The specific primers JB3 (=COIASmit1) (forward 5′–TTTTTTGGGCATCCTGAGGTTTAT–3′) and JB4.5 (=COI-ASmit2) (reverse 5′–TAAAGAAAGAACATAATGAAAATG–3′) were used to amplify a fragment of 424 bp of the COI gene [13, 48]. PCR reactions were performed in 20 μL, containing 1 ng of DNA, 5× iProof HF buffer, 0.25 mM dNTP, 0.15 μM of each primer, and 0.5 units of iProof HF DNA polymerase (Bio-Rad). Thermocycles consisted of an initial denaturation step at 94 °C for 2 min, followed by 37 cycles of denaturation at 94 °C for 30 s, annealing at 48 °C for 40 s, and extension at 72 °C for 50 s. The final extension was conducted at 72 °C for 5 min. Sequences were obtained as for fish and were deposited in GenBank (accession numbers in Table 1).
Trees and distances
For fishes, the phylogenetic analyses included three sequences of B. boops and one Pagellus acarne generated in this study, and other sequences of these fishes available in GenBank (Table 3), whilst Spicara maena (Linnaeus, 1758), a sparid previously reported as a host of Cy. bellones [5] was used as an outgroup.
Table 3.
Accession numbers of COI sequences used in the molecular analysis of fishes. Species previously reported as hosts of Cyclocotyla bellones or as hosts for isopods bearing the latter are in bold. Note that all hosts are sparids. Choricotyle chrysophryi is a parasite of the fish, so no isopod was involved. *, new sequences.
| Host species | Origin | GenBank | Source |
|---|---|---|---|
| Boops boops | Algeria | MZ127221* | Present study |
| Boops boops | Algeria | MZ127226* | Present study |
| Boops boops | Algeria | MZ127217* | Present study |
| Boops boops | Algeria | MK317921 | [8] |
| Boops boops | Algeria | MT666082–MT666086 | [11] |
| Boops boops | Italy | KJ709490 | [47] |
| Boops boops | Malta | KJ709712 | [47] |
| Boops boops | Turkey | KC500341–KC500342 | [40] |
| Boops boops | Turkey | KC500351 | [40] |
| Boops boops | Mediterranean Sea | KJ012295–KJ012296 | [2] |
| Boops boops | Portugal | JQ774986–JQ774987 | [19] |
| Boops boops | Eastern Mediterranean | KJ012383–KJ012385 | [2] |
| Pagellus acarne | Algeria | MZ127223* | Present study |
| Pagellus acarne | Italy | KJ709574 | [47] |
| Pagellus acarne | Portugal | JQ775093–JQ775092 | [19] |
| Pagellus acarne | Spain | FN689213 | [44] |
| Pagellus acarne | Turkey | FN689212 | [44] |
| Spicara maena | Portugal | KJ768312 | [47] |
For isopods, the molecular analysis was based upon mouth dwelling cymothoids, mainly species previously reported as hosts of Cy. bellones (Table 4). The body surface isopod Anilocra clupei Williams & Bunkley-Williams, 1986 was used as an outgroup.
Table 4.
Accession numbers of COI sequences used in the molecular analysis of cymothoids. All Ceratothoa species infect the buccal cavity of their fish host, whereas individuals of Anilocra clupei infect the body surface of the fish. Species previously reported as hosts of Cyclocotyla bellones or as hosts for crustaceans bearing the latter are in bold. S, Sparidae. M, Moronidae. C, Carangidae. Cl, Clupeidae. *, new sequences.
| Parasite species | Host species | Origin | GenBank | Source |
|---|---|---|---|---|
| Ceratothoa parallela | Boops boops (S) | Mediterranean Sea (Algeria) | MZ127227* | Present study |
| Ceratothoa parallela | Boops boops (S) | Mediterranean Sea (Algeria) | MZ127219* | Present study |
| Ceratothoa parallela | Boops boops (S) | Mediterranean Sea (Algeria) | MZ127225* | Present study |
| Ceratothoa collaris | Lithognathus mormyrus (S) | Mediterranean Sea (Tyrrhenian Sea) | EF455816 | [41] |
| Ceratothoa sp. | Dentex gibbous (S) | Atlantic Ocean (West Africa) | LC159551 | [33] |
| Ceratothoa oestroides | Sparus aurata (S) | Mediterranean Sea (Adriatic). | GQ240266 | [58] |
| Ceratothoa oestroides | Sparus aurata (S) | Mediterranean Sea (Adriatic) | GQ240267 | [58] |
| Ceratothoa oestroides | Boops boops (S) | Mediterranean Sea (Adriatic) | GQ240272 | [58] |
| Ceratothoa oestroides | Boops boops (S) | Mediterranean Sea (Adriatic) | GQ240273 | [58] |
| Ceratothoa oestroides | Dicentrarchus labrax (M) | Mediterranean Sea (Adriatic) | GQ240276 | [58] |
| Ceratothoa oestroides | Dicentrarchus labrax (M) | Mediterranean Sea (Adriatic) | GQ240277 | [58] |
| Ceratothoa oxyrrhynchaena | Dentex hypselosomus (S) | Pacific Ocean (Japan) | LC159545 | [33] |
| Ceratothoa oxyrrhynchaena | Dentex abei (S) | Pacific Ocean (Japan) | LC160310 | [33] |
| Ceratothoa verrucosa | Pagrus major (S) | Pacific Ocean (Japan) | LC159556 | [33] |
| Ceratothoa verrucosa | Evynnis tumifrons (S) | Pacific Ocean (Japan) | LC160317 | [33] |
| Anilocra clupei | Sardinella zunasi (C) | Pacific Ocean (Japan) | LC159540 | [33] |
| Anilocra clupei | Etrumeus micropus (Cl) | Pacific Ocean (Japan) | LC160309 | [33] |
For monogeneans, most sequences of Diclidophoridae available in GenBank were included in the phylogenetic analysis (Table 5), with three sequences of Cy. bellones and one of Choricotyle cf. chrysophryi obtained in the present study. A sequence of Plectanocotyle gurnardi (Van Beneden & Hesse, 1863), a member of Plectanocotylidae Monticelli, 1903, grouped with the Diclidophoridae in a previous phylogeny of Monogenea [38], was used as an outgroup. Molecular analyses were performed in MEGA, version 7 [45]. The trees were inferred using the neighbour joining (NJ) method [69] and the maximum likelihood (ML) method using MEGA7 [45]. Based on the best model, maximum likelihood was used for the best fitting tree according to the Hasegawa-Kishino-Yano with gamma distribution and invariant sites (HKY + G) for fishes; Tamura 3-parameter with gamma distribution and invariant sites (T92 + I) for isopods, and Tamura 3-parameter with gamma distribution (T92 + G) for monogeneans. The robustness of the inferred analysis was assessed using a bootstrap procedure with 1000 replications. Genetic distances, p-distance and Kimura-2 parameter distance (K-2-P), [43] were estimated with MEGA7 and all codon positions were used.
Table 5.
Accession numbers of COI sequences used in the molecular analysis of diclidophorid monogeneans. *, new sequences.
| Parasite species | Host species | Origin | GenBank | Source |
|---|---|---|---|---|
| Cyclocotyla bellones | Ceratothoa parallela from Boops boops (Cymothoidae/ Sparidae) | Mediterranean Sea (Algeria) | MZ127220* | Present study |
| Cyclocotyla bellones | Ceratothoa parallela from Boops boops | Mediterranean Sea (Algeria) | MZ127224* | Present study |
| Cyclocotyla bellones | Ceratothoa parallela from Boops boops | Mediterranean Sea (Algeria) | MZ127218* | Present study |
| Choricotyle cf. chrysophryi | Pagellus acarne (Sparidae) | Mediterranean Sea (Algeria) | MZ127216* | Present study |
| Choricotyle cf. chrysophryi | Pagellus acarne (Sparidae) | Mediterranean Sea (Algeria) | MZ127222* | Present study |
| Choricotyle anisotremi | Anisotremus scapularis (Haemulidae) | Pacific Ocean (Chile) | KJ794206 | [60] |
| Choricotyle anisotremi | Anisotremus scapularis | Pacific Ocean (Chile) | KJ794207 | [60] |
| Neoheterobothrium affine | Paralichthys dentatus (Paralichthyidae) | Atlantic Ocean (USA) | AB478782 | [75] |
| Neoheterobothrium hirame | Paralichthys olivaceus (Paralichthyidae) | Pacific Ocean (Japan) | AB162615 | [75] |
| Parapedocotyle prolatili | Prolatilus jugularis (Pinguipedidae) | Pacific Ocean (Chile) | KJ794219 | [60] |
| Parapedocotyle prolatili | Prolatilus jugularis | Pacific Ocean (Chile) | KJ794218 | [60] |
| Pedocotyle bravoi | Stellifer minor (Sciaenidae) | Pacific Ocean (Chile) | KJ794210 | [60] |
| Pedocotyle bravoi | Stellifer minor | Pacific Ocean (Chile) | KJ794210 | [60] |
| Plectanocotyle gurnardi | Chelidonichthys lastoviza (Triglidae) | Mediterranean Sea (Algeria) | MK275654 | Ayadi et al. (unpublished) |
BLAST analyses of the COI sequences of monogeneans, isopods and fishes obtained in the present study were performed using NCBI and BOLD databases [67].
Results
Details on parasitism
Boops boops, parasitism by Ceratothoa parallela. Prevalence and intensity: 14% (93 out of 624 fish), up to 2 cymothoids/fish.
Ceratothoa parallela, parasitism by Cyclocotyla bellones. Prevalence and intensity: 11% (10 out of 93 isopods), up to 2 monogeneans/cymothoids.
No Pagellus acarne were infected with cymothoids. All monogeneans were collected from the fish gills.
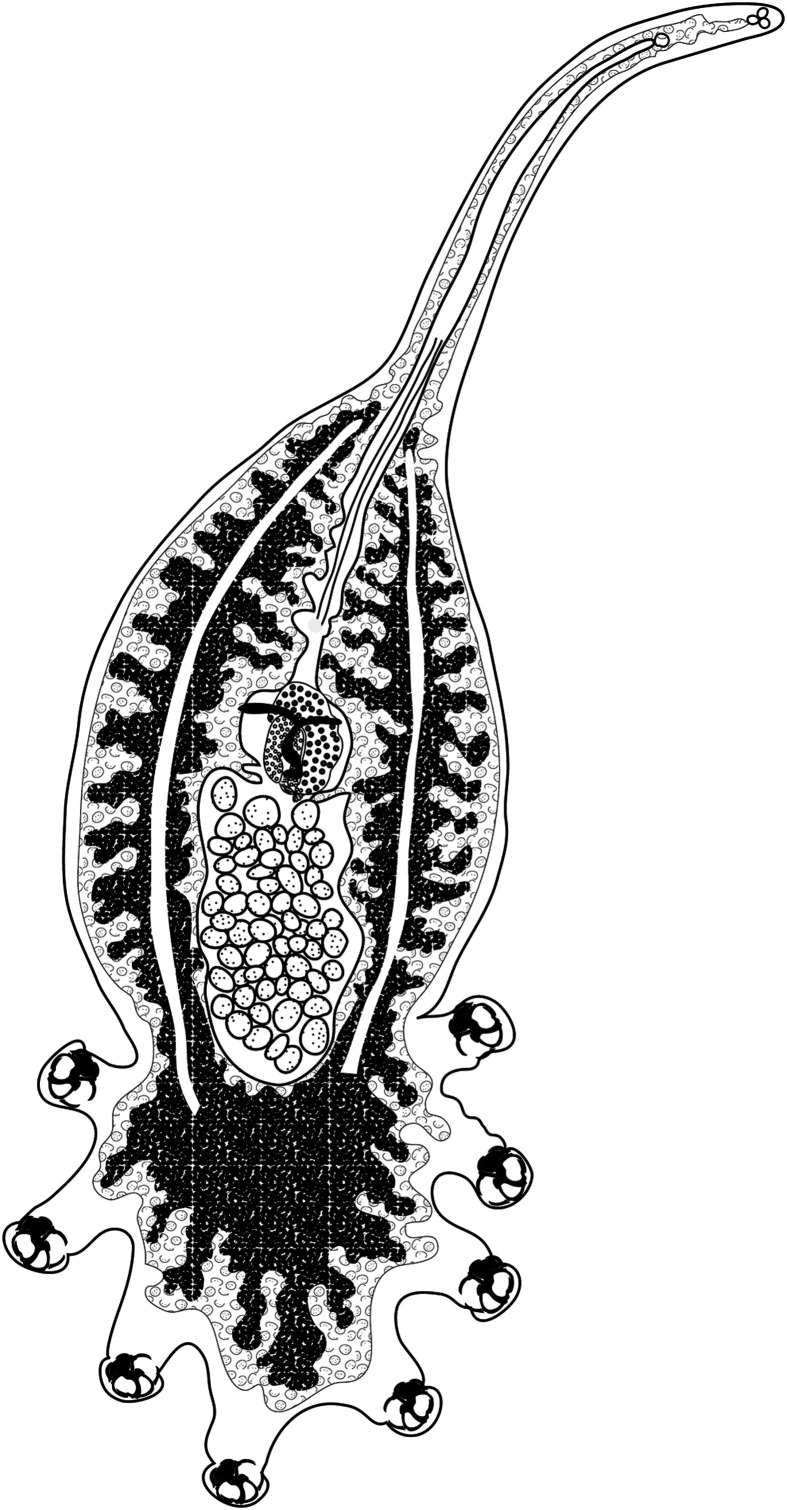
Short description of Cyclocotyla bellones Otto, 1823 (Fig. 1)
Figure 1.
Cyclocotyla bellones Otto, 1823. Whole body, MNHN HEL1312.
Type-host: Belone belone (Linnaeus, 1760), garfish (Belonidae Bonaparte, 1835).
Additional hosts: Bopyrus squillarum Latreille, 1802 (Bopyridae Rafinesque, 1815). Isopods of Spicara maena (Linnaeus, 1758) (Sparidae Rafinesque, 1818), the blotched picarel; of Spicara smaris (Linnaeus, 1758), the picarel; and of B. boops (Linnaeus, 1758) (Sparidae), the bogue. Ceratothoa parallela (Otto, 1828) (Cymothoidae Leach, 1818) from B. boops (this paper).
Type-locality: Italy [62].
Additional localities: Montenegro, France, and Turkey. Off Bouharoun (36° 37′ 24″ N, 2° 39′ 17″ E), off the Algerian coast (this paper).
Specimens from Algeria, from Ceratothoa parallela (Cymothoidae) of buccal cavity. Vouchers deposited in the collection of the Muséum National d’Histoire Naturelle, Paris (MNHN HE1307–HEL1326). Vouchers with molecular information, three specimens mounted on slide, a small lateral part cut off and used for molecular analysis. (MNHN HEL1307, GenBank MZ127220; MNHN HEL1308, GenBank MZ127224; MNHN HEL1309, GenBank MZ127218).
Body elongate, divided into three regions: a tapered anterior region; an enlarged and rounded middle region and a posterior region formed by the haptor. Haptor ovoid, bearing four pairs of pedunculated clamps. Prohaptoral suckers paired, rounded. Caeca ramified medially and laterally. Testicles post-ovarian, follicular, numerous in intercaecal field. Vas deferens sinuous, extending anteriorly to male copulatory organ. Ovary median, folded. Oötype postovarian, surrounded by mass of Mehlis’ glands. Uterus running along body midline and opening into genital atrium. Seminal receptacle present. Genito-intestinal canal originating from left intestinal branch. Vitelline follicles large, coextensive with intestinal caeca and invading haptor. Vitelloducts Y-shaped. Vagina absent.
Molecular identification of fish
The provisional identification of fish species using morphological characteristics was confirmed by the DNA barcoding approach. The obtained sequences were 652 bp long. BLAST analyses of the COI sequences of the present study with NCBI and BOLD databases showed sequence similarity values of 100% for B. boops and 99.85 % for P. acarne.
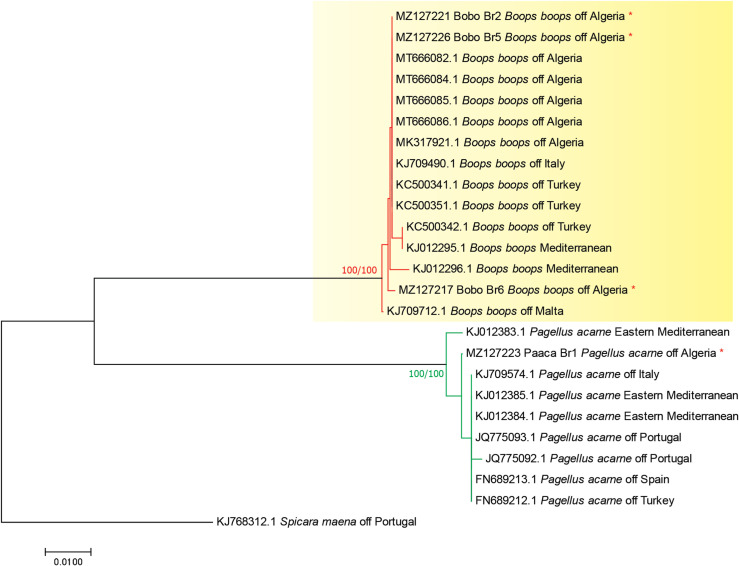
The ML tree is shown in Figure 2. The newly generated sequences of B. boops clustered in a well-supported clade (100% bootstrap). All sequences of P. acarne, including our newly generated sequence, clustered in a single robust clade.
Figure 2.
Molecular phylogenetic analysis based on maximum likelihood, inferred from COI sequences of Boops boops in relation to other hosts of Cyclocotyla bellones. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site.
Distances were computed using Kimura 2-parameter distance and p-distance. Our sequences of B. boops were identical (0% intraspecific variation). All Sequences of B. boops (available in GenBank plus our newly generated sequences) showed little to no variation (0–1%). The divergence among sequences of P. acarne was also low, and ranged between 0 and 1%.
Molecular information on isopods
Isopods were identified as Ceratothoa parallela (Otto, 1828). The newly acquired sequences were 658 bp long. BLAST analyses of the COI sequences with NCBI and BOLD databases showed sequence similarity values of 80.47–80.63% for “Ceratothoa sp.”; however, no sequence identified at the species level was available from the databases.
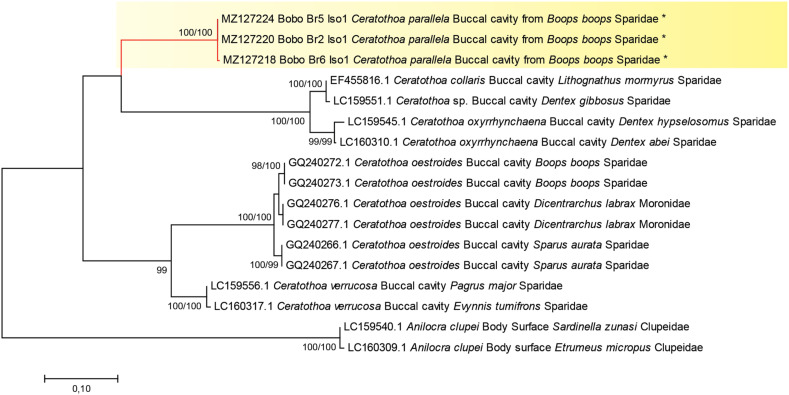
Phylogenetic trees were constructed based on our newly generated COI sequences of Ceratothoa parallela, and combined datasets of Ceratothoa spp., mainly those previously reported as hosts of C. bellones. The analysis involved 13 nucleotide sequences, and there was a total of 409 positions in the final dataset. The topologies were nearly consistent among the ML (Fig. 3) and the NJ trees. The “mouth dwelling cymothoid clade” was well separated from the outgroup, which included body surface isopods. The mouth dwelling cymothoids were separated into two clades, one including C. oestroides and C. verrucosa, and one including C. parallela, C. collaris and C. oxyrrhynchaena.
Figure 3.
Molecular phylogenetic analysis based on maximum likelihood, inferred from COI sequences of Ceratothoa parallela in relation to other cymothoids hosts of Cyclocotyla bellones. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site.
There was no intraspecific variation among our three sequences of Ceratothoa parallela. The mean intraspecies distance of sequences of Ceratothoa oestroides from three hosts, B. boops, Sparus aurata and Dicentrarchus labrax ranged between 0% and 2%, suggesting that the same species is harboured by the three fish species. Distances between individuals of congeneric species were high, the highest being between Ceratothoa oestroides from B. boops and C. parallela, 28%.
Molecular characterisation of monogeneans
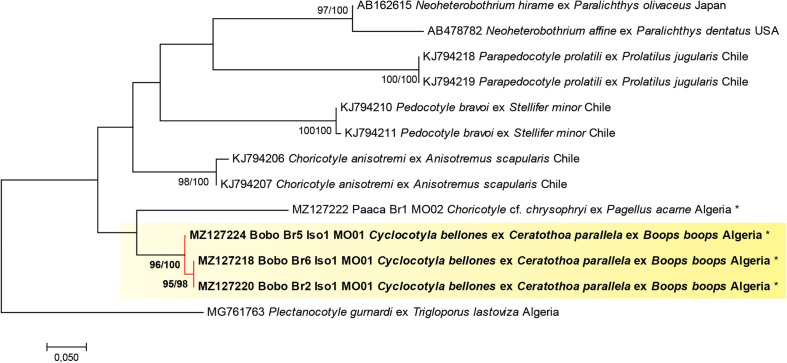
The newly generated sequences were 402 bp long. The COI sequences of Cy. bellones were aligned with other diclidophorid sequences. For trees, the NJ and ML methods led to similar topologies; we show only the ML tree in Figure 4. The analysis involved 13 nucleotide sequences, and there was a total of 337 positions in the final dataset.
Figure 4.
Molecular phylogenetic analysis based on maximum likelihood, inferred from COI sequences of Cyclocotyla bellones in relation to other taxa of diclidophorids; a plectanocotylid was chosen as the outgroup. Values along branches indicate percentage bootstrap support for maximum likelihood and neighbour joining methods (ML/NJ). Nodes without bootstrap values had support lower than 50% and were omitted. There were a total of 389 positions in the final dataset. All specimens of Cyclocotyla bellones were grouped into a single monophylum and showed little variation (≤1%). The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. ex: from.
The three sequences of Cy. bellones reported in the present study formed a well-supported monophyletic lineage (96 bootstraps in ML, 100 in NJ). The phylogenetic analysis therefore supports Cy. bellones as a distinct species from Choricotyle cf. chrysophryi. The sequence of Choricotyle cf. chrysophryi nested within a Cyclocotyla + Choricotyle clade, but Choricotyle was paraphyletic. However, support values for most clades were low and this phylogeny is not discussed further.
Distances were computed using Kimura 2-parameter distance and p-distance. Sequences of Cy. bellones showed little to no variations between them: 0–1%. The differences between sequences of Cy. bellones and Choricotyle cf. chrysophryi from the sparid P. acarne were 18% and 16% in K-2-P and p distance, respectively. Cyclocotyla bellones differed from Neoheterobothrium hirame, and two sequences of Choricotyle anisotremi by 22% and 18%, respectively (p distances were 19% and 16%, respectively). The highest divergence was observed between Cy. bellones and Parapedocotyle prolatili (26–27% in K-2-P, 22–23% in p distance).
Discussion
Hyperparasitic monogeneans
Overall, records of hyperparasitic monogeneans are rather scant. Among the Monopisthocotylea Odhner, 1912, this tripartite association is known for two cases: the Udonellidae Taschenberg, 1879, with Udonella spp. on parasitic copepods of various fishes [73] and the Capsalidae Baird, 1853 with Capsala biparasitica (Goto, 1894) on a copepod on the gills of the tuna Thunnus albacares (Bonnaterre, 1788) [28].
Similarly, hyperparasitism in known in only one family of the Polyopisthocotylea Odhner, 1912, the Diclidophoridae, in four genera: Cyclocotyla [62], Allodiclidophora Yamaguti, 1963 [22, 23, 63, 81], Choricotyle Van Beneden & Hesse, 1863 [28, 32, 51] and Diclidophora Krøyer, 1838 [22, 23]. Except Allodiclidophora squillarum (Parona & Perugia, 1889) reported on the ovigerous lamellae of the bopyrid Bopyrus squillarum, a parasite of shrimp, all the previously mentioned hyperparasites were found on cymothoids of the buccal cavity of marine fishes [22, 23, 28, 32, 51]. However, it is noteworthy that the taxonomic status of the previously mentioned diclidophorids should be further examined as several of them were placed in different genera, based on characters of little generic significance, or considered synonyms with little justification [51, 65, 81]. Hence, molecular markers will be indispensable in future studies for the delimitation of genera and species.
Members of the Diclidophoridae Cerfontaine, 1895 are cosmopolitan polyopisthocotyleans, infecting gills, the operculum, and gill cavity of teleosts and elasmobranchs [54]. To date, 51 genera are assigned to this family (WoRMS, 2020). Even though great progress has been made in generating molecular data for members of this family [4, 38, 49, 50, 52, 56, 59-61, 71, 75, 82], amongst 173 species allocated to this family, molecular markers are available for only 24 species. Internal transcribed spacer (ITS) sequences are known for five species [75, 82]. 18S and 28S RNA ribosomal sequences are available for 18 species across 11 genera [4, 38, 49, 50, 52, 56, 59–61, 71]. COI sequences are the least numerous, known for only 10 species across 6 genera [38, 60, 82].
Cymothoid isopods
The COI divergences observed between the newly generated sequences of Ceratothoa parallela sampled from B. boops ranged from 0 to 2% and did not exceed the 3% and 5% COI thresholds suggested for species-level divergences [34, 35, 70], confirming the morphological identification of the three specimens as conspecific. These findings can further be strengthened by additional COI sequences of this species from various localities and hosts.
Overall, the Cymothoidae are taxonomically challenging as descriptions of many species were originally based only on the morphology of few or sometimes single specimens, thus providing no information on polymorphism [76]. In addition, many misidentifications and incorrect data on species were generated when not considering the morphological variability ascribed to the parasitic lifestyle of cymothoids, polymorphism and sister species, as well as the variations in attachment site and the morphological adaptations [29, 72, 76]. In recent years, numerous genera and species described early were revised, eliminating some of the uncertainties and confusion [30, 72, 76]. Hence, every effort should be made to generate molecular data and deposit specimens in collections, which can be used subsequently.
Ceratothoa is a large genus, with 25 species presently accepted [31]. The species studied here, Ceratothoa parallela, was designated in 2015 as the type-species of the genus [55]. Unfortunately, not many COI sequences are available for members of this genus and of the family of Cymothoidae overall, and almost half are from unpublished works. COI sequences are known for 16 genera [33, 36, 41, 57, 77–80]. 16S sequences are available for 15 genera [33, 37, 41, 78, 80]. 18S sequences are known for 4 genera (all from unpublished works) and 28S sequences are available for only 3 genera [33]. The complete mitochondrial genome is known for 2 genera of Cymothoidae from unpublished works, available only in GenBank: Asotana Schioedte & Meinert, 1881 (Zou et al.) and Ichthyoxenos Herklots, 1870 (Hua et al.) (GenBank MZ127218, MK790137).
Cyclocotyla bellones
Genetic variations among our newly generated sequences of Cy. bellones were very low, 0–1%, a divergence lower than interspecific distances found in other studies of polyopisthocotylean monogeneans (1% vs. 10.2–15.0%; [7]). The divergence between sequences of Cy. bellones and Choricotyle cf. chrysophryi from the sparid P. acarne reached 18% and 16% in K-2-P and p distance, respectively, well above the 3% species threshold often admitted for Monogenea [1], and thus the separation between the two species is confirmed genetically.
In the present study, Cy. bellones was most exclusively on the isopods, mainly on the upper part of the pereon. These results agree with Euzet & Trilles (1961) who found Cy. bellones often attached to the crustacean (telson, pleon, rarely perion) and exceptionally in the buccal cavity of the fish, the palate and the internal edge of the upper lip [25]. We found a single specimen unattached in a Petri dish containing gills and isopods. We carefully examined the gills of B. boops during our four-year survey [6, 8, 11], but we have not observed any Cy. bellones attached to gills nor in the buccal cavity. Hence, we are confident that this monogenean attaches itself to the isopod and not the fish.
Cyclocotyla bellones has been recorded on several hosts and from different localities (Table 1). It has been reported on cymothoids of the oral cavity of four fish species, B. boops, Spicara maena (Linnaeus, 1758), S. smaris (Linnaeus, 1758) and T. trachurus. Distributions of the previously mentioned host species overlap as all hosts co-exist in the Mediterranean Sea or in the Eastern Atlantic Ocean. The parasitic isopods have a larger distribution extending to the northern Indo-pacific and coexist with other fish hosts in the Black Sea, Mediterranean and Eastern Atlantic. Therefore, while we took a conservative position and tentatively identified the diclidophorid from cymothoids on B. boops as Cy. bellones, it is not unlikely that this hyperparasite is a species complex.
Conflict of interest
The Editor-in-Chief of Parasite is one of the authors of this manuscript. COPE (Committee on Publication Ethics, http://publicationethics.org), to which Parasite adheres, advises special treatment in these cases. In this case, the peer-review process was handled by an Invited Editor, Jérôme Depaquit.
Acknowledgments
We thank Prof. Jean-Paul Trilles (University of Montpellier, Montpellier, France) for the identification of the isopods. This research was supported by la Direction Générale de la Recherche Scientifique et du Développement Technologique (DGRSDT, Algiers, Algeria), by l’Institut de Systématique, Évolution, Biodiversité (ISYEB, MNHN, Paris, France), and by the DeepBlue Project: Distance Crossborder Traineeship Programme co-funded by “The European Maritime and Fisheries Fund (EMFF)”.
Cite this article as: Bouguerche C, Tazerouti F, Gey D & Justine J-L. 2021. Triple barcoding for a hyperparasite, its parasitic host, and the host itself: a study of Cyclocotyla bellones (Monogenea) on Ceratothoa parallela (Isopoda) on Boops boops (Teleostei). Parasite 28, 49.
References
- 1.Aquaro G, Salogni C, Galli P, Gibelli L, Gelmetti D. 2012. Sciadicleithrum variabilum (Dactylogyridae: Monogenea) infection in Symphysodon discus: a case report. Fish Pathology, 47(1), 23–26. [Google Scholar]
- 2.Armani A, Guardone L, Castigliego L, D’Amico P, Messina A, Malandra R, Gianfaldoni D, Guidi A. 2015. DNA and Mini-DNA barcoding for the identification of Porgies species (family Sparidae) of commercial interest on the international market. Food Control, 50, 589–596. [Google Scholar]
- 3.Ayadi ZEM, Gey D, Justine J-L, Tazerouti F. 2017. A new species of Microcotyle (Monogenea: Microcotylidae) from Scorpaena notata (Teleostei: Scorpaenidae) in the Mediterranean Sea. Parasitology International, 66(2), 37–42. [DOI] [PubMed] [Google Scholar]
- 4.Baverstock PB, Fielke R, Johnson AM, Bray RA, Beveridge I. 1991. Conflicting phylogenetic hypotheses for the parasitic platyhelminths tested by partial sequencing of 18S ribosomal RNA. International Journal for Parasitology, 21, 329–339. [DOI] [PubMed] [Google Scholar]
- 5.Benhamou F, Marzoug D, Boutiba Z, Kostadinova A, Pérez-Del-Olmo A. 2017. Parasite communities in two sparid fishes from the western Mediterranean: a comparative analysis based on samples from three localities off the Algerian coast. Helminthologia, 54(1), 26–35. [Google Scholar]
- 6.Bouguerche C. 2019. Étude taxinomique des Polyopisthocotylea Odhner, 1912 (Monogenea, Plathelminthes) parasites de quelques Téléostéens de la côte algérienne. Université des Sciences et de la Technologie Houari Boumediene: Algiers, Algeria. [Google Scholar]
- 7.Bouguerche C, Gey D, Justine J-L, Tazerouti F. 2019. Microcotyle visa n. sp. (Monogenea: Microcotylidae), a gill parasite of Pagrus caeruleostictus (Valenciennes) (Teleostei: Sparidae) off the Algerian coast, Western Mediterranean. Systematic Parasitology, 96(2), 131–147. [DOI] [PubMed] [Google Scholar]
- 8.Bouguerche C, Gey D, Justine J-L, Tazerouti F. 2019. Towards the resolution of the Microcotyle erythrini species complex: description of Microcotyle isyebi n. sp. (Monogenea, Microcotylidae) from Boops boops (Teleostei, Sparidae) off the Algerian coast. Parasitology Research, 118(5), 1417–1428. [DOI] [PubMed] [Google Scholar]
- 9.Bouguerche C, Justine J-L, Tazerouti F. 2020. Redescription of Flexophora ophidii Prost & Euzet, 1962 (Monogenea, Diclidophoridae) from Ophidion barbatum (Ophidiidae) off the Algerian coast, Mediterranean Sea. Systematic Parasitology, 97, 827–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bouguerche C, Tazerouti F, Gey D, Justine J-L. 2019. Redescription and molecular characterisation of Allogastrocotyle bivaginalis Nasir & Fuentes Zambrano, 1983 (Monogenea: Gastrocotylidae) from Trachurus picturatus (Bowdich) (Perciformes: Carangidae) off the Algerian coast, Mediterranean Sea. Systematic Parasitology, 96(8), 681–694. [DOI] [PubMed] [Google Scholar]
- 11.Bouguerche C, Tazerouti F, Gey D, Justine J-L. 2020. No vagina, one vagina, or multiple vaginae? An integrative study of Pseudaxine trachuri (Monogenea, Gastrocotylidae) leads to a better understanding of the systematics of Pseudaxine and related genera. Parasite, 27, 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bouguerche C, Tazerouti F, Justine J-L. 2021. Four polyopisthocotyleans (Platyhelminthes: Monogenea) from carangid fishes in the Mediterranean, off the Algerian coasts. Current Research in Parasitology & Vector-Borne Diseases, 1, 100026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bowles J, Blair D, McManus DP. 1995. A molecular phylogeny of the human schistosomes. Molecular Phylogenetics and Evolution, 4(2), 103–109. [DOI] [PubMed] [Google Scholar]
- 14.Carreras Aubets M. 2013. Parasites of three fish species of commercial interest from the north-western Mediterranean sea Mullus barbatus, Spicara maena and Trachinus draco (Osteichthyes, Perciformes). Thesis. Universitat Autònoma Barcelona: Barcelona, Spain. [Google Scholar]
- 15.Chaabane A, Justine J-L, Gey D, Bakenhaster MD, Neifar L. 2016. Pseudorhabdosynochus sulamericanus (Monogenea, Diplectanidae), a parasite of deep-sea groupers (Serranidae) occurs transatlantically on three congeneric hosts (Hyporthodus spp.), one from the Mediterranean Sea and two from the western Atlantic. PeerJ, 4, e2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chaabane A, Neifar L, Gey D, Justine J-L. 2016. Species of Pseudorhabdosynochus (Monogenea, Diplectanidae) from groupers (Mycteroperca spp., Epinephelidae) in the Mediterranean and Eastern Atlantic Ocean, with special reference to the “beverleyburtonae group” and description of two new species. PLoS One, 11(8), e0159886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chaabane A, Neifar L, Justine J-L. 2015. Pseudorhabdosynochus regius n. sp. (Monogenea, Diplectanidae) from the mottled grouper Mycteroperca rubra (Teleostei) in the Mediterranean Sea and Eastern Atlantic. Parasite, 22, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chaabane A, Neifar L, Justine J-L. 2017. Diplectanids from Mycteroperca spp. (Epinephelidae) in the Mediterranean Sea: Redescriptions of six species from material collected off Tunisia and Libya, proposal for the ‘Pseudorhabdosynochus riouxi group’, and a taxonomic key. PLoS One, 12(2), e0171392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Costa FO, Landi M, Martins R, Costa MH, Costa ME, Carneiro M, Alves MJ, Steinke D, Carvalho GR. 2012. A ranking system for reference libraries of DNA barcodes: application to marine fish species from Portugal. PLoS One, 7(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Derouiche I, Neifar L, Gey D, Justine J-L, Tazerouti F. 2019. Holocephalocotyle monstrosae n. gen. n. sp. (Monogenea, Monocotylidae) from the olfactory rosette of the rabbit fish, Chimaera monstrosa (Holocephali, Chimaeridae) in deep waters off Algeria. Parasite, 26, 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dettaï A, Adamowizc SJ, Allcock L, Arango CP, Barnes DKA, Barratt I, Chenuil A, Couloux A, Cruaud C, David B, Denis F, Denys G, Diaz A, Eléaume M, Feral J-P, Froger A, Gallut C, Grant R, Griffiths HJ, Held C, Hemery L, Hosie G, Kuklinski P, Lecointre G, Linse K, Lozouet P, Mah C, Monniot F, Norman MD, O’Hara T, Ozouf-Costaz C, Piedallu C, Pierrat B, Poulin E, Puillandre N, Riddle M, Samadi S, Saucède T, Schubart C, Smith PJ, Stevens DW, Steinke D, Strugnell JM, Tarnowska K, Wadley V, Améziane N. 2011. DNA barcoding and molecular systematics of the benthic and demersal organisms of the CEAMARC survey. Polar Science, 5(2), 298–312. [Google Scholar]
- 22.Dollfus R. 1922. Complément à la description de Cyclobothrium charcoti Dollfus, 1922. Bulletin de la Société zoologique de France, 47, 348–352. [Google Scholar]
- 23.Dollfus RP. 1922. Cyclobothrium charcoti, n. sp. Trématode ectoparasite sur Meinertia oestroides (Risso): parasites recueillis pendant la croisière océanographique du “Pourquoi-Pas?” sous le commandement du Dr. J.-B. Charcot, en 1914: 1re note. Bulletin de la Société Zoologique de France, 47, 287–296. [Google Scholar]
- 24.Dollfus RP. 1946. Notes et informations. Annales de Parasitologie Humaine et Comparée, 21(1–2), 93–96. [PubMed] [Google Scholar]
- 25.Euzet L, Trilles J. 1961. Sur l’anatomie et la biologie de Cyclocotyla bellones (Otto, 1821) (Monogenea-Polyopisthocotylea). Revue Suisse de Zoologie, 68(2), 16. [Google Scholar]
- 26.Fischer W, Bauchot M-L, Schneider M. 1987. (Révision 1). Méditerranée et mer Noire. Zone de pêche 37. Volume II. Vertébrés. Publication préparée par la FAO, résultat d’un accord entre la FAO et la Commission des Communautés Européennes (Projet GCP/INT/422/EEC) financée conjointement par ces deux organisations. Fiches FAO d’identification des espèces pour les besoins de la pêche, Vo1.2. FAO: Rome. p. 761–1530. [Google Scholar]
- 27.Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. 1994. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Molecular Marine Biology and Biotechnology, 3(5), 294–299. [PubMed] [Google Scholar]
- 28.Goto S. 1894. Studies on the ectoparasitic Trematodes of Japan. Tokyo pp. [Google Scholar]
- 29.Hadfield KA, Bruce NL, Smit NJ. 2010. Redescription of the monotypic genus Cinusa Schioedte and Meinert, 1884 (Isopoda, Cymothoidae), a buccal-cavity isopod from South Africa. Zootaxa, 2437(1), 51–68. [Google Scholar]
- 30.Hadfield KA, Bruce NL, Smit NJ. 2016. Redescription of poorly known species of Ceratothoa Dana, 1852 (Crustacea, Isopoda, Cymothoidae), based on original type material. ZooKeys, 592, 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hadfield KA, Smit NJ. 2020. Review of the global distribution and hosts of the economically important fish parasitic isopod genus Ceratothoa (Isopoda: Cymothoidae), including the description of Ceratothoa springbok n. sp. from South Africa. International Journal for Parasitology, 50(10–11), 899–919. [DOI] [PubMed] [Google Scholar]
- 32.Hargis WJ. 1955. Monogenetic trematodes of Gulf of Mexico fishes. Part IX. The family Diclidophoridae Fuhrmann, 1928. Transactions of the American Microscopical Society, 74(4), 377–388. [Google Scholar]
- 33.Hata H, Sogabe A, Tada S, Nishimoto R, Nakano R, Kohya N, Takeshima H, Kawanishi R. 2017. Molecular phylogeny of obligate fish parasites of the family Cymothoidae (Isopoda, Crustacea): evolution of the attachment mode to host fish and the habitat shift from saline water to freshwater. Marine Biology, 164(5), 105. [Google Scholar]
- 34.Hebert PDN, Cywinska A, Ball SL, deWaard JR. 2003. Biological identifications through DNA barcodes. Proceedings of the Royal Society of London. Series B: Biological Sciences, 270(1512), 313–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Janssen A, Stuckas H, Vink A, Arbizu PM. 2019. Biogeography and population structure of predominant macrofaunal taxa (Annelida and Isopoda) in abyssal polymetallic nodule fields: implications for conservation and management. Marine Biodiversity, 49(6), 2641–2658. [Google Scholar]
- 36.Joca LK, Leray VL, Zigler KS, Brusca RC. 2015. A new host and reproduction at a small size for the “snapper-choking isopod” Cymothoa excisa (Isopoda: Cymothoidae). Journal of Crustacean Biology, 35(2), 292–294. [Google Scholar]
- 37.Jones CM, Miller TL, Grutter AS, Cribb TH. 2008. Natatory-stage cymothoid isopods: description, molecular identification and evolution of attachment. International Journal for Parasitology, 38(3–4), 477–491. [DOI] [PubMed] [Google Scholar]
- 38.Jovelin R, Justine J-L. 2001. Phylogenetic relationships within the polyopisthocotylean monogeneans (Platyhelminthes) inferred from partial 28S rDNA sequences. International Journal for Parasitology, 31, 393–401. [DOI] [PubMed] [Google Scholar]
- 39.Justine J-L, Rahmouni C, Gey D, Schoelinck C, Hoberg EP. 2013. The monogenean which lost its clamps. PLoS One, 8(11), e79155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Keskİn E, Atar HH. 2013. DNA barcoding commercially important fish species of Turkey. Molecular Ecology Resources, 13(5), 788–797. [DOI] [PubMed] [Google Scholar]
- 41.Ketmaier V, Joyce D, Horton T, Mariani S. 2008. A molecular phylogenetic framework for the evolution of parasitic strategies in cymothoid isopods (Crustacea). Journal of Zoological Systematics and Evolutionary Research, 46(1), 19–23. [Google Scholar]
- 42.Kheddam H, Justine J-L, Tazerouti F. 2016. Hexabothriid monogeneans from the gills of deep-sea sharks off Algeria, with the description of Squalonchocotyle euzeti n. sp. (Hexabothriidae) from the kitefin shark Dalatias licha (Euselachii, Dalatiidae). Helminthologia, 53(4), 354–362. [Google Scholar]
- 43.Kimura M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. Journal of Molecular Evolution, 16(2), 111–120. [DOI] [PubMed] [Google Scholar]
- 44.Kochzius M, Seidel C, Antoniou A, Botla SK, Campo D, Cariani A, Vazquez EG, Hauschild J, Hervet C, Hjörleifsdottir S, Hreggvidsson G, Kappel K, Landi M, Magoulas A, Marteinsson V, Nölte M, Planes S, Tinti F, Turan C, Venugopal MN, Weber H, Blohm D. 2010. Identifying fishes through DNA barcodes and microarrays. PLoS One, 5(9), e12620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Molecular Biology and Evolution, 33, 1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lambert A. 1978. Recherches sur les stades larvaires des Monogènes de Poissons. Annales de Parasitologie Humaine et Comparée, 53(6), 551–559. [PubMed] [Google Scholar]
- 47.Landi M, Dimech M, Arculeo M, Biondo G, Martins R, Carneiro M, Carvalho GR, Brutto SL, Costa FO. 2014. DNA barcoding for species assignment: The case of Mediterranean marine fishes. PLoS One, 9(9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Littlewood DTJ, Rohde K, Clough KA. 1997. Parasite speciation within or between host species? - Phylogenetic evidence from site-specific polystome monogeneans. International Journal for Parasitology, 27, 1289–1297. [DOI] [PubMed] [Google Scholar]
- 49.Littlewood DTJ, Rohde K, Clough KA. 1999. The interrelationships of all major groups of Platyhelminthes: phylogenetic evidence from morphology and molecules. Biological Journal of the Linnean Society, 66, 75–114. [Google Scholar]
- 50.Litvaitis MK, Rohde K. 1999. A molecular test of platyhelminth phylogeny: inferences from partial 28S rDNA sequences. Invertebrate Biology, 118, 42–56. [Google Scholar]
- 51.Llewellyn J. 1941. A revision of the monogenean family Diclidophoridae Fuhrmann, 1928. Parasitology, 33(4), 416–430. [Google Scholar]
- 52.Lockyer AE, Olson PD, Littlewood DTJ. 2003. Utility of complete large and small subunit rRNA genes in resolving the phylogeny of the Neodermata (Platyhelminthes): implications and a review of the cercomer theory. Biological Journal of the Linnean Society, 78, 155–171. [Google Scholar]
- 53.López-Román R, Guevara Pozo D. 1976. Cyclocotyla bellones Otto, 1821 (Monogenea) presente en Meinertia oestreoides en cavidad bucal de Boops boops en aguas del Mar de Alborán. Revista Ibérica de Parasitología, 36, 135–138. [Google Scholar]
- 54.Mamaev YL. 1976. The system and phylogeny of monogeneans of the family Diclidophoridae. Trudy Biologo-Pochvenngo Instituta, Novaya Seriya, 35, 57–80. [Google Scholar]
- 55.Martin MB, Bruce NL, Nowak BF. 2015. Review of the fish-parasitic genus Ceratothoa Dana, 1852 (Crustacea: Isopoda: Cymothoidae) from Australia, with description of two new species. Zootaxa, 3963(3), 251–294. [DOI] [PubMed] [Google Scholar]
- 56.Mendoza-Franco EF, Tun MdCR, Anchevida AdJD, del Rio Rodríguez RE. 2018. Morphological and molecular (28S rRNA) data of monogeneans (Platyhelminthes) infecting the gill lamellae of marine fishes in the Campeche Bank, southwest Gulf of Mexico. ZooKeys, 783, 125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mladineo I, Bott NJ, Nowak BF, Block BA. 2010. Multilocus phylogenetic analyses reveal that habitat selection drives the speciation of Didymozoidae (Digenea) parasitizing Pacific and Atlantic bluefin tunas. Parasitology, 137(6), 1013–1025. [DOI] [PubMed] [Google Scholar]
- 58.Mladineo I, Šegvić T, Grubišić L. 2009. Molecular evidence for the lack of transmission of the monogenean Sparicotyle chrysophrii (Monogenea, Polyopisthocotylea) and isopod Ceratothoa oestroides (Crustacea, Cymothoidae) between wild bogue (Boops boops) and cage-reared sea bream (Sparus aurata) and sea bass (Dicentrarchus labrax). Aquaculture, 295(3–4), 160–167. [Google Scholar]
- 59.Mollaret I, Jamieson BGM, Justine J-L. 2000. Phylogeny of the Monopisthocotylea and Polyopisthocotylea (Platyhelminthes) inferred from 28S rDNA sequences. International Journal for Parasitology, 30, 171–185. [DOI] [PubMed] [Google Scholar]
- 60.Oliva ME, Sepulveda FA, González MT. 2014. Parapedocotyle prolatili gen. n. et sp. n., a representative of a new subfamily of the Diclidophoridae (Monogenea), a gill parasite of Prolatilus jugularis (Teleostei: Pinguipedidae) from Chile. Folia Parasitologica, 61(6), 543. [PubMed] [Google Scholar]
- 61.Olson PD, Littlewood DTJ. 2002. Phylogenetics of the Monogenea – evidence from a medley of molecules. International Journal for Parasitology, 32, 233–244. [DOI] [PubMed] [Google Scholar]
- 62.Otto AW. 1823. Beschreibung einiger neuen Mollusken und Zoophyten. Offprint from Leopoldinisch-Carolinische deutsche Akademie der Naturforscher. Nova acta Leopoldina, v. XI, pp. 273–314. [Google Scholar]
- 63.Parona C, Perugia A. 1889. Mesocotyle squillarum, n. sub. gen., n. sp. di trematode ectoparassita del Bopyrus squillarum. Bollettino Scientifico, 11, 76–80. [Google Scholar]
- 64.Pleijel F, Jondelius U, Norlinder E, Nygren A, Oxelman B, Schander C, Sundberg P, Thollesson M. 2008. Phylogenies without roots? A plea for the use of vouchers in molecular phylogenetic studies. Molecular Phylogenetics and Evolution, 48(1), 369–371. [DOI] [PubMed] [Google Scholar]
- 65.Price EW. 1943. North American monogenetic trematodes: VI. The family Diclidophoridae (Diclidophoroidea). Journal of the Washington Academy of Sciences, 33(2), 44–54. [Google Scholar]
- 66.Radujkovic BM, Euzet L. 1989. Parasites des poissons marins du Monténégro: Monogènes. In: Radujkovic, B. M & Raibaut, A. (Eds) Faune des parasites de poissons marins des côtes du Monténégro (Adriatique Sud). Acta Adriatica, 30(1/2), 51–135. [PubMed] [Google Scholar]
- 67.Ratnasingham S, Hebert PDN. 2007. BOLD: The Barcode of Life Data System (www. barcodinglife. org). Molecular Ecology Notes, 7(3), 355–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Renaud F, Romestand B, Trilles J-P. 1980. Faunistique et écologie des métazoaires parasites de Boops boops Linnaeus (1758) (Téléostéen Sparidae) dans le Golfe du Lion. Annales de Parasitologie Humaine et Comparée, 55(4), 467–476. [DOI] [PubMed] [Google Scholar]
- 69.Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Molecular Biology and Evolution, 4, 406–425. [DOI] [PubMed] [Google Scholar]
- 70.Santamaria CA, Bluemel JK, Bunbury N, Curran M. 2017. Cryptic biodiversity and phylogeographic patterns of Seychellois Ligia isopods. PeerJ, 5, e3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sinnappah ND, Lim L-HS, Rohde K, Tinsley R, Combes R, Verneau O. 2001. A paedomorphic parasite associated with a neotenic amphibian host: phylogenetic evidence suggests a revised systematic position for Sphyranuridae within anuran and turtle polystomatoineans. Molecular Phylogenetics and Evolution, 18, 189–201. [DOI] [PubMed] [Google Scholar]
- 72.Smit NJ, Bruce NL, Hadfield KA. 2014. Global diversity of fish parasitic isopod crustaceans of the family Cymothoidae. International Journal for Parasitology: Parasites and Wildlife, 3(2), 188–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Smit NJ, Bruce NL, Hadfield KA. 2019. Parasitic Crustacea: State of knowledge and future trends. Springer pp: New York, United States. [Google Scholar]
- 74.Solak K, Öktener A, Trilles J-P, Solak CN. 2007. Report on the monogenean Cyclocotyla bellones and three cymothoids parasitizing two fish species from the Aegean Sea coasts of Turkey. Acta Adriatica, 45(2), 145–154. [PubMed] [Google Scholar]
- 75.Tsutsumi N, Yoshinaga T, Kamaishi T, Nakayasu C, Ogawa K. 2003. Effects of water temperature on the development of the monogenean Neoheterobothrium hirame on Japanese flounder Paralichthys olivaceus. Fish Pathology, 38(2), 41–47. [Google Scholar]
- 76.Van der Wal S. 2018. Biodiversity and systematics of branchial cavity inhabiting fish parasitic isopods (Cymothoidae) from sub-Sahara Africa. North-West University, Potchefstroom Campus. [Google Scholar]
- 77.Van der Wal S, Smit NJ, Hadfield KA. 2017. Redescription and molecular characterisation of the fish parasitic isopod Norileca indica (Milne Edwards, 1840)(Crustacea: Isopoda: Cymothoidae) with a key to the genus. African Zoology, 52(3), 163–175. [Google Scholar]
- 78.Welicky RL, Hadfield KA, Sikkel PC, Smit NJ. 2017. Molecular assessment of three species of Anilocra (Isopoda, Cymothoidae) ectoparasites from Caribbean coral reef fishes, with the description of Anilocra brillae sp. n. ZooKeys, 663, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Welicky RL, Smit NJ. 2019. Redescription and molecular characterisation of the fish ectoparasite Anilocra capensis Leach, 1818 (Isopoda: Cymothoidae), with description of six new species of Anilocra Leach, 1818 from Africa. Parasites & Vectors, 12(1), 387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wetzer R. 2001. Hierarchical analysis of mtDNA variation and the use of mtDNA for isopod systematics (Crustacea: Isopoda). Contributions to Zoology, 70(1), 23–39. [Google Scholar]
- 81.Yamaguti S. 1963. Systema Helminthum Volume IV Monogenea and Aspidocotylea. John Wiley & Sons; pp. [Google Scholar]
- 82.Yoshinaga T, Tsutsumi N, Hall KA, Ogawa K. 2009. Origin of the diclidophorid monogenean Neoheterobothrium hirame Ogawa, 1999, the causative agent of anemia in olive flounder Paralichthys olivaceus. Fisheries Science, 75(5), 1167–1176. [Google Scholar]