Abstract
The mechanisms by which cells adapt to proteotoxic stress are largely unknown, but key to understanding how tumor cells, particularly in vivo, are largely resistant to proteasome inhibitors. Analysis of cancer cell lines, mouse xenografts and patient-derived tumor samples all showed an association between mitochondrial metabolism and proteasome inhibitor sensitivity. When cells were forced to use oxidative phosphorylation rather than glycolysis, they became proteasome inhibitor-resistant. This mitochondrial state, however, creates a unique vulnerability: sensitivity to the small-molecule compound elesclomol. Genome-wide CRISPR/Cas9 screening showed that a single gene, encoding the mitochondrial reductase FDX1, could rescue elesclomol-induced cell death. Enzymatic function and NMR-based analyses further showed that FDX1 is the direct target of elesclomol, which promotes a unique form of copper-dependent cell death. These studies elucidate a fundamental mechanism by which cells adapt to proteotoxic stress and suggests strategies to mitigate proteasome inhibitor-resistance.
Introduction
Cancers often show an increased dependency on protein quality control systems to accommodate the severe proteotoxic stresses that arise from genome instability, biomass accumulation and increased protein turnover 1,2. In such cells, the increased rate of protein synthesis can outstrip proteasome degradation capacity making cancer cells exceedingly vulnerable to proteasome inhibition 3,4. While proteasome inhibitors can effectively kill cancer cells grown in culture without any lineage specificity 5 (Supplementary Fig. 1a and Supplementary Dataset 1), in vivo, cancer cells demonstrate a remarkable ability to adapt to proteasome inhibition 6. In the clinic, proteasome inhibitors have demonstrated limited efficacy, emerging as front-line therapy in only the B-cell malignancies multiple myeloma and mantle cell lymphoma. Yet even these cancers rapidly develop proteasome inhibitor (PI) resistance. The ability of normal cells and most cancer cells to withstand the toxic effects of transient proteasome inhibition suggests that a non-genetic mechanism of resistance exists that is not readily observed in normal cell culture.
Several mechanisms underlying PI resistance have been suggested over the years including constitutive activation of NF-κB 7 and the chaperone machinery 8, alterations in the EGFR/JAK1/STAT3 pathway 9 and mutations in the catalytic sites that are targeted by proteasome inhibitors 10. Recent studies using several genetic technologies have converged on a novel mechanism that involves the down-regulation of one of the many subunits of the 19S regulatory complex of the proteasome which leads to PI resistance in yeast and cancer cell line models 11-14. We refer to this PI-resistant state as the Lo19S state. Notably, multiple myeloma patients that are refractory to proteasome inhibitors often have tumors with spontaneously reduced expression of one or more of the 19S subunits 11,14. These findings suggest that the Lo19S state is a frequent, naturally occurring mechanism in cancer that allows neoplastic cells to resist the toxic effects of proteasome inhibition.
In this study, using a functional genomic approach, we demonstrate that the naturally occurring PI-resistant state is associated with a shift to mitochondrial energy metabolism. In many cases, this shift is sufficient to promote resistance to proteasome inhibitors. However, the metabolic shift creates a new actionable vulnerability: sensitivity to the drug elesclomol (1). We show that elesclomol, previously explored in clinic trials but with unknown mechanism of action, attenuates the ability of cancer cells to cope with proteasome inhibitor-induced toxicity both in culture and in an orthotopic mouse model of multiple myeloma. We further reveal that elesclomol promotes a unique form of copper-dependent cell death that is augmented by the ferredoxin 1 (FDX1) protein, which binds and reduces the elesclomol-Cu(II) complex. This unique mechanism of cell death establishes how a switch in energy metabolism can be exploited to increase the efficacy of a cancer targeting agent and thereby inhibit the adaptation of cancer cells to proteasome inhibition.
Results
Genetic signature of the PI- resistant Lo19S state
To explore the role of the Lo19S state in promoting PI-resistance, we engineered the breast cancer cell line T47D to transiently shift to the Lo19S state by suppressing the expression of the PSMD2 subunit of the 19S complex with an inducible shRNA. Transient induction of the Lo19S state conferred strong resistance to proteasome inhibition (EC90 increased by ~27 fold) (Fig. 1a and Supplementary Fig. 1b). Inducing the Lo19S state by itself did not lead to a significant change in gene expression, suggesting that transcriptional preconditioning was not required for PI resistance in the Lo19S state. However, following proteasome inhibition, gene set enrichment analysis revealed a strong signature of mitochondria-related gene expression specific to the Lo19S state (Supplementary Fig.1c,d and Supplementary Dataset 2).
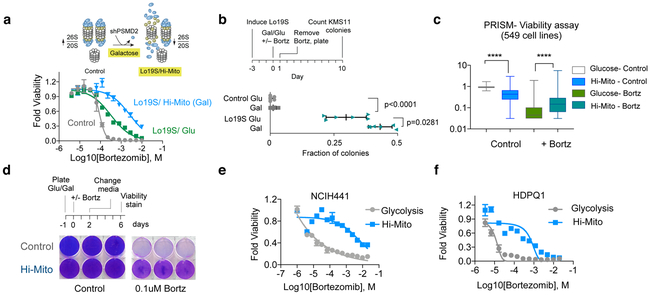
Figure 1. Mitochondrial energy metabolism is associated with the PI-resistant Lo19S state.
a, The Lo19S state was induced by induction of a doxycycline inducible PSMD2 shRNA in T47D breast cancer cells with 0.2 μg/ml doxycycline for 72 hours. Cells were then collected, washed and plated with fresh media without doxycycline. After 24 hours the indicated concentrations of bortezomib were added and the relative viability was measured after 72 hours (mean −/+ SD, n= 4 biologically independent samples). The calculated EC50s and EC90s are also presented. b, Basal oxygen consumption rate (OCR) was measured in control and induced Lo19S state cells (as in a) with and without the additions of 200nM bortezomib (Bortz) for 6 hours (discovery determined using the Two-stage linear step-up procedure of Benjamini, Krieger and Yekutieli, with Q = 1%. Each row was analyzed individually, without assuming a consistent SD, n= 7,8 biologically independent samples). c, MM.1S orthotopic tumors grown out from control and bortezomib-treated mice were analyzed for gene expression with RNA-seq and the gene set enrichment categories (Hallmarks) the mountain plot for Hallmark OXPHOS is plotted. d, Gene set enrichment analysis (GSEA) of genes upregulated in Lo19S but not control tumors was conducted for breast, prostate, thyroid, skin and kidney cancers from the TCGA. The top and bottom 29 categories are plotted. Mitochondrial-associated categories are marked in blue, the rest in orange. The network enrichment of Gene Ontology (GO) terms in the Lo19S state in breast cancer tumors is visualized and color-coded by function (right). Only GO categories showing enrichment with Log2 score >0.5 were used to create the network and visualized was conducted using ClueGO in Cytoscape.
To directly explore the possibility that the Lo19S state results in changes in energy metabolism, we measured oxygen consumption and performed metabolite profiling in Lo19S and control cells. Upon proteasome inhibition, the basal oxygen consumption of control cells dropped, whereas basal oxygen consumption of cells in the PI-resistant, Lo19S state was sustained (Fig. 1b). Metabolite profiling showed no significant change in basal metabolites in control vs. Lo19S cells, but following treatment with proteasome inhibitors, we observed statistically significant changes in tricarboxylic acid cycle (cis-aconitate) and redox (NAD, NADH, GSSG) metabolites (Supplementary Fig. 1e-h and Supplementary Dataset 3). In addition, other metabolites related to the TCA cycle including succinate, alpha-ketoglutarate and glutamate showed a trend toward statistical significance (Supplementary Dataset 3). Thus, proteasome inhibitor treatment in the Lo19S induces mitochondrial gene expression and metabolite changes that are suggestive of a shift in mitochondrial metabolism.
To understand whether increased mitochondrial metabolism is promoted by proteasome inhibitors in vivo, we turned to a multiple myeloma orthotopic xenograft model. We profiled the mRNA using RNA-seq in untreated (control) tumors and tumors that grew out following exposure to bortezomib (bortezomib-resistant tumors). Bortezomib-resistant multiple myeloma samples showed a significant increase in mitochondrial gene expression (oxidative phosphorylation signature) compared to their untreated counterparts (Fig. 1c and Supplementary Dataset 4). Thus, we observed increased levels of mitochondrial genes in two experimental models of PI resistance, both in cell culture and in an orthotopic mouse tumor model.
We further analyzed gene expression data from human tumor tissues in The Cancer Genome Atlas (TCGA), which also revealed a striking enrichment of mitochondrial oxidative phosphorylation (OXPHOS) gene sets among tumors in the Lo19S state (Fig. 1d and Supplementary Dataset 5). Our findings strongly support a model in which the PI-resistant Lo19S state is associated with increased mitochondrial function.
The Hi-Mito state promotes proteotoxic stress tolerance
We reasoned that the mitochondrial gene expression signature associated with the Lo19S state might cooperate to enhance the ability of cells to withstand proteotoxic stress. To test this possibility, we simultaneously induced the Lo19S state and increased the cells’ dependency on mitochondrial metabolism (Hi-Mito state). We induced the Lo19S state by transient suppression of the proteasome subunit protein PSMD2, and induced the Hi-Mito state by replacing glucose with galactose in the cell culture media. This change in the primary carbon source results in the utilization of glutamine, which forces cells into a state of increased mitochondrial dependency 15. Combining the Lo19S and Hi- Mito states had an additive effect on PI resistance, resulting in a 50-fold increase in the EC90 for proteasome inhibitors (Fig. 2a) and promoting the ability of multiple myeloma cells in the Lo19S state to withstand bortezomib treatment (Fig. 2b). In the Hi-Mito state the Lo19S cells exhibited increased maximal oxygen consumption, correlating with their increased PI-resistance (Supplementary Fig. 2a). Interestingly, enhancing mitochondrial metabolism alone caused mild resistance to proteasome inhibition, (Supplementary Fig. 2b), and reduced heat-shock induction in response to bortezomib (Supplementary Fig. 2c) without affecting the overall catalytic activity of cellular proteasomes as determined by measuring the chymotrypsin-like activity of the proteasome (Supplementary Fig. 2d).
Figure 2. Increased mitochondrial energy metabolism (Hi-Mito) is sufficient to promote proteotoxic stress tolerance.
a, Control and induced Lo19S T47D cells were plated in media containing either glucose (Glu) or galactose (to induce Hi-Mito state). The viability was measured 72 hours after addition of the indicated concentrations of bortezomib (mean −/+ SD, n= 4 biologically independent samples). Calculated EC90 for control, Lo19S/Glu and Lo19S/Gal is 12.7 nM, 109 nM and 624 nM respectively b, Control and Lo19S state was induced by transient induction of a doxycycline-inducible PSMD2 shRNA in the multiple myeloma KMS11 cell line. Control non-targeting shRNA was used in the control cell lines. Colony formation was assessed after cells were grown in media with either glucose or galactose (Hi-Mito) in the presence or absence of 5nM bortezomib for 24 hours. The relative fraction (compared to untreated glucose grown cells) of colonies formed following 5nM bortezomib treatment is plotted. Statistical analysis was conducted by two-tailed unpaired t-test. Plotted mean −/+ SD, n= 6 biologically independent samples c, The relative viability score of 549 cell lines following growth in glucose (control) or galactose (Hi-Mito) containing media, in the presence or absence of 40nM bortezomib. Data plotted as box (25–75 percentile) and whiskers (min to max), statistical analysis was conducted by two-tailed paired t-test. **** p<0.0001, n= 549 biologically independent samples . d, NCIH2030 cells were grown in media containing glucose (control) or galactose (Hi-Mito) and treated with or without 100nm bortezomib for 2 days. Cell media was changed to control glucose-containing media and presented is the visualization of the cell number after 4 days. Three biologically independent samples presented that are representative of two independent experiments. e-f, NCIH441 (e) and HDPQ1 (f) cells were grown in glucose (Glycolysis) or galactose (Hi-Mito) containing media and cell viability was determined after 72 hours of treatment with indicated concentrations of bortezomib. Non-linear curve fit was performed (mean −/+ SD, n= 4 biologically independent samples).
But is a shift to the Hi-Mito state sufficient to promote PI-resistance? To address this question, we utilized the PRISM pooled cell line barcoding method that allowed us to simultaneously interrogate 549 cancer cell lines16. We grew the PRISM pool of barcoded cells in media containing either glucose or galactose (to induce the Hi-Mito state) and in the presence or absence of bortezomib. After 3 days, we assessed differential viability (Fig. 2c). Overall, the relative viability of cells in the Hi-Mito state was reduced, with some cells exhibiting glucose auxotrophy (Fig. 2c and Supplementary Dataset 6). Despite this, cells in the Hi-Mito state are significantly more resistant to bortezomib across this large panel of genetically diverse cancer cell lines from multiple cell lineages (Fig. 2c and Supplementary Dataset 6). We validated these findings using non-pooled cell lines, and again found that the switch to the Hi-Mito state was sufficient to promote PI-resistance (Fig. 2d-f). Thus, a shift to the Hi-Mito state is sufficient to promote PI resistance.
Elesclomol inhibits adaptation to proteotoxic stress
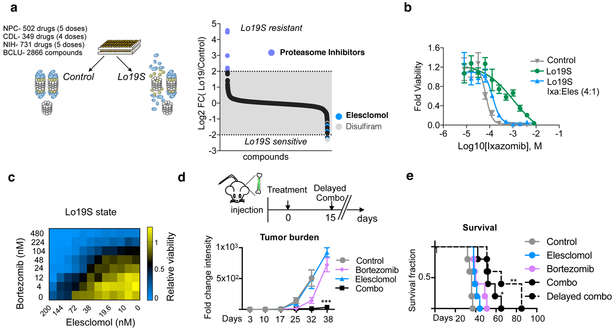
While our findings established an association between mitochondrial metabolism and adaptation to proteotoxic stress, they did not establish a definitive mechanistic link between the two. To probe for mechanism, we undertook a chemical-genomic approach and screened 4,300 small molecules with diverse mechanisms of action (Supplementary Dataset 7) for their ability to kill control or PI-resistant (Lo19S) T47D cells. We found that only proteasome inhibitors showed reduced efficacy in the Lo19S versus the wild-type state, indicating that the Lo19S state does not confer nonspecific drug-resistance (Fig. 3a, Supplementary Fig. 3a and Supplementary Dataset 8). In line with this observed specificity, the Lo19S state sensitized cells to only 3 of the 4,300 compounds (0.07%): the sulfur-containing copper-binders elesclomol and disulfiram and the BCL2/BCLxL inhibitor navitoclax (Fig. 3a, Supplementary Dataset 8). Navitoclax has been previously shown to synergize with proteasome inhibitors 17, a result we confirmed in our system emphasizing the specificity of our experimental system (Supplementary Fig. 3b).
Figure 3. The PI-resistant Lo19S state exhibits increased sensitivity to elesclomol.
a, Viability based small molecule screen was conducted using three distinct libraries (Selleck anti-cancer L3000 drug library (CDL n=349 in 4 doses), the natural product library (NPC n=502 in 5 doses) and the NIH bioactive compound library (n=731 in 5 doses) to determine the compounds that have differential effect on viability between the control versus the induced Lo19S cells. The relative viability of the Lo19S versus control cells was calculated for replica experiments and the log2 of the ratio is plotted. b, The effect on relative cell growth of elesclomol added with the proteasome inhibitor ixazomib at a 1:4 (elesclomol:ixazomib) ratio (the ratio of the EC50s) to Lo19S cells compared to the effect of ixazomib alone added to either Lo19S or control cells. Plotted are the mean −/+ SD n= 4 biologically independent samples. Calculated EC50 for control-Ixazomib, Lo19S-Ixazomib and Lo19S-Ixazomib:elesclomol combo is 69 nM, 1145 nM and 138 nM respectively. c, Heatmap of relative cell growth following addition of different combinations of bortezomib and elesclomol to the Lo19S state cells. d-e, Mice were injected with MM.1S luciferase-expressing cells. Upon tumor formation as judged by bioluminescent signal intensity, treatment with elesclomol (28mg/kg), bortezomib (0.25mg/kg) or the combination (Combo) was initiated (day 0). At day 15, elesclomol was added to the treatment of one of the groups that had received only bortezomib from day 0 to day 15 (combo-delayed group). Tumor burden (d) over time as determined by changes from the baseline radiant flux associated with the BLI signal intensity. Plotted are the mean −/+ SD (multiple unpaired t-test analysis with Holm-Sidak correction on bortezomib alone versus combo group at week 32 and 38 adjusted p-values; p= 0.001 (n=5), p=0.0003 (n=3 bortz n=4 combo) respectively and viability plots (e) are presented as Kaplan Meier survival curve (statistical test: Log-rank of significant difference between the combo treatments over bortezomib alone * p= 0.0244 ** p= 0.0044). n = 5 per group.
Multiple lines of evidence suggest a unique relationship between proteasome inhibitor resistance and elesclomol sensitivity (Fig. 3a and Supplementary Fig. 3c). First, we found that cell lines with naturally occurring Lo19S states were more sensitive to elesclomol than their lineage-matched controls (Supplementary Fig. 3d,e). Second, elesclomol enhanced the effect of PI, specifically in the PI-resistant Lo19S state (Fig. 3b,c and Supplementary Fig. 3f). Third, combination treatment with elesclomol potentiated the response of orthotopic multiple myeloma xenografts to bortezomib in vivo (Fig. 3d,e, Supplementary Fig. 3g,h). These distinct results strongly suggest that elesclomol might impair an adaptive response to PI-induced proteotoxic stress.
FDX1 regulates elesclomol induced toxicity
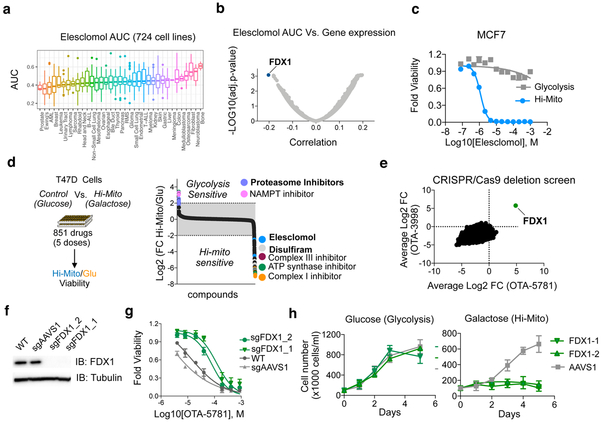
Elesclomol has been previously evaluated as an anti-cancer agent both in pre-clinical models and in patients 18. Elesclomol binds copper, 19-22 and elesclomol-induced cell death has been suggested to be mediated by increased mitochondrial reactive oxygen species (ROS) 23-25. However, the specific mechanism of action of elesclomol and its target in cells have remained unknown. To identify cellular attributes that correlate with the cytotoxic activity of elesclomol, we measured elesclomol sensitivity in dose-response across 724 cancer cell lines using an expanded PRISM screening system (Supplementary Dataset 9). Overall, elesclomol was very potent in inducing cell death across all lineages with most cells showing sensitivity at doses above 100nM (Fig. 4a and Supplementary Dataset 9). To identify genomic correlates of elesclomol sensitivity, analyzed the pre-treatment genomic features of the cell lines (obtained from the Cancer Dependency Map at www.depmap.org) to identify those most correlated with elesclomol AUC estimated by PRISM. The top correlated gene expression feature was the mitochondrial gene FDX1 (Fig. 4b and Supplementary Dataset 9). This correlation is further supportive of a relationship between mitochondrial metabolism and elesclomol sensitivity.
Figure 4. FDX1 is the primary mediator of elesclomol induced toxicity.
a, The viability of 724 barcoded cell lines was determined for 8 concentrations of elesclomol and the calculated area under the curve (AUC) is presented as box plot for different lineages (Supplementary Data 9). b, Correlation analysis of calculated AUC for elesclomol with the gene expression (obtained from the Cancer Dependency Map at www.depmap.org) in 701 cell lines common to both datasets is plotted with the most significant correlation indicated (FDX1 gene) see Supplementary Data 9. c, MCF7 cells were grown in the presence of either glucose (control, gray) or galactose (Hi-Mito, blue) as the carbon source and the relative cell viability was analyzed 72 hours after addition of the indicated concentrations of elesclomol. Mean −/+ SD n= 4 biologically independent samples is plotted d, T47D cells were grown in the presence of either glucose (Glycolysis) or galactose (Hi-Mito) as the carbon source in the presence of compounds from the Selleck anti-cancer L3000 drug library (CDL) (n= 349 in 4 doses) and the natural product library (NPC) (n=502 in 5 doses). The relative viability of the Hi-Mito versus the glycolytic control cells was calculated for replica experiments and the log2 of the ratio is plotted. Specific compounds are annotated. e, The gene scores of two whole genome CRISPR/Cas9-deletion screens conducted with OTA-5781 (100 nM) and OTA-3998 (1uM) treated K562 cells. The gene score is the mean log2 fold change in abundance of all sgRNAs targeting that gene during the culture period. FDX1 is indicated. f, Western blot analysis of FDX1 and tubulin (loading control) protein expression levels in WT K562 cells (WT) or cells with FDX1 (two distinct sgRNAs) and AAVS1 deletions. Representative of two independent experiments. Full length gels are shown in Supplementary Fig. 7. g, Viability curves of parental K562 cells and cells targeted for either AAVS1 (control) or FDX1 achieved with two sgRNAs using CRISPR/Cas9 were treated with increasing concentrations OTA-5781 and viability was examined after 72 hours. h, The indicated cells were grown in the presence of either glucose or galactose (Hi-Mito) and the relative cell number plotted. For g,h mean −/+ SD n= 4 biologically independent samples.
To further probe the relationship between elesclomol activity and mitochondrial metabolism, we forced cells into the Hi-Mito state and examined their sensitivity to elesclomol. The shift to the Hi-Mito state profoundly enhanced sensitivity to elesclomol across several cell lines, in some cases by as much as 1,000-fold (Fig. 4c and Supplementary Fig. 4a-c). Furthermore, this increased sensitivity to elesclomol in the Hi-Mito state was highly specific to elesclomol. Of 800 drugs tested, only two classes of compounds showed increased killing in the Hi-Mito state: known modulators of mitochondrial function (e.g. Complex I, Complex III and ATP synthase inhibitors) and the copper-binding proteins elesclomol and disulfiram (the two compounds identified in our Lo19S screen) (Fig. 4d Supplementary Dataset 8). In contrast to elesclomol, proteasome inhibitors showed decreased cell killing in the Hi-Mito state, precisely as predicted by our initial findings (Fig. 1). These results again suggest that both elesclomol- and proteasome-induced cytotoxicity are highly dependent on the cell’s primary energy source.
Given that the precise mechanism of action of elesclomol was unknown, its mechanistic relationship to mitochondrial metabolism was not obvious. We thought it unlikely to be acting simply as an inhibitor of the electron transport chain (ETC), because ETC inhibitors, unlike elesclomol, did not score in our Lo19S small molecule screen (Fig. 3a) or synergize with proteasome inhibition in the PI-resistant Lo19S state (Supplementary Fig. 4d). We therefore performed pooled genome-wide CRISPR/Cas9 screens to identify genes whose loss conferred resistance to the elesclomol analogs OTA-5781 (2) and OTA-3998 (3) (Supplementary Fig. 4e,f). In both screens, only a single gene scored: FDX1, encoding the ferredoxin 1 protein (Fig. 4e). FDX1 is the same gene whose basal mRNA expression level was most highly correlated with elesclomol sensitivity (Fig. 4b). We validated the ability of FDX1 knock-out (Fig. 4f) to confer resistance to elesclomol using multiple independent sgRNAs and a non-targeting control (Fig. 4g). The resistance conferred by FDX1 knockout, together with the increased expression that predicted sensitivity to elesclomol, suggested the possibility that elesclomol could act by imparting a toxic gain of function to the protein.
FDX1 is a mitochondrial reductase involved in iron-sulfur (Fe-S) cluster formation 26-28, a process that is essential for mitochondrial function 29. We found that FDX1-null cells were unable to make the metabolic shift that enables growth in the Hi-Mito state (Fig. 4h), despite proliferating similarly to control cells in the presence of glucose. Additionally, FDX1-null cells exhibited reduced levels of basal respiration that was further reduced upon growth in galactose (Supplementary Fig. 4g-i). Analysis of the Cancer Dependency Map showed that, like elesclomol, the genes whose pattern of essentiality most parallels FDX1 loss-of-function were genes involved in mitochondrial function (Supplementary Fig. 4j).
Elesclomol inhibits FDX1-mediated Fe-S cluster biosynthesis
To investigate whether FDX1 might be the direct target of elesclomol, we performed NMR spectroscopy using recombinant FDX1. We found that elesclomol indeed binds the FDX1 α2/α3 helices and β5 strand, but does not bind the paralog protein FDX2 (Fig. 5a and Supplementary Fig. 5a,b). Fe-S clusters are generated from sulfur extracted from cysteine by the iron-sulfur cluster (ISC) core complex (iron plus NFS1-ISD11-ACP-ISCU), and this process requires either FDX1 or FDX2 as a reductant (Fig. 5b). In an in vitro Fe-S cluster assembly reaction, titration of elesclomol or a closely related structural analogue (OTA-5781) caused progressive inhibition of Fe-S cluster production (Fig. 5c and Supplementary Fig. 5c) by directly inhibiting the electron transfer from reduced FDX1 to the NFS1 component of the ISC core complex (Fig. 5d). In contrast, substitution of reduced FDX1 with reduced FDX2 led to much less inhibition of such electron transfer, consistent with the NMR binding data showing that elesclomol specifically binds FDX1 (Supplementary Fig. 5d). Furthermore, elesclomol had no inhibitory effect on cysteine desulfurase activity with DTT as the reductant, further indicating its selectivity for FDX1 (Supplementary Fig. 5e).
Figure 5. Elesclomol inhibits the natural function of FDX1 in the Fe-S cluster biosynthesis, serving as a neo-substrate when bound to copper.
a, Chemical shift perturbation results mapped onto a diagram of the structure of FDX1. Color code: grey, not significantly affected (ΔδNH < 0.01 ppm); blue, significant chemical shift changes (ΔδNH > 0.01 ppm); red, severe line broadening; grey, no assignments. The [2Fe-2S] cluster in FDX1 is indicated by spheres. b, Schematic describing mitochondrial Fe-S cluster biosynthesis. The mitochondrial ISC (iron-sulfur cluster) core complex catalyzes the conversion of cysteine to alanine and generates S0 for iron sulfur cluster assembly. S0 is reduced by FDX1. A [2Fe-2S] cluster is subsequently formed on the scaffold protein ISCU. c, In vitro Fe-S cluster assembly was monitored by following the increase of absorbance at 456 nm. Reduced FDX1 was used as a reducing agent in the presence or absence of either 5X (green) or 10X (blue) elesclomol. Data is representative of two independent experiments d, Electron transfer from reduced FDX1 to the cysteine desulfurase complex was measured upon the addition of cysteine in the presence or absence of elesclomol. Data is representative of two independent experiments e, The molecular structure of elesclomol analogs and their corresponding calculated EC50s (from T47D cells grown in Hi-Mito state). The reactive sulfur substitution with an oxygene is indicated with an asterisk. f, The UV/vis spectra of reduced FDX1 before and after incubation with elesclomol-Cu(II) or Cu(II) alone.
Elesclomol-Cu(II) is an FDX1 neo-substrate
These biochemical results implicate elesclomol as a direct inhibitor of FDX1 activity in the context of Fe-S cluster assembly. However, an important paradox remained: while elesclomol inhibits FDX1, FDX1 knockout did not phenocopy elesclomol activity in cells. Rather, its knockout rescued elesclomol-induced cytotoxicity. Because elesclomol has been shown to complex with copper and shuttle it to the mitochondria 20,21, we hypothesized that cell death induced by elesclomol might best be explained by an interaction between FDX1 and the copper conveyed to it by elesclomol. In support of this idea, substitution of oxygen for just one (OTA-5393 (4)) of the two elesclomol sulfur atoms, critical for binding copper, reduced the activity of the molecule by 300-fold and substitution of both (OTA-5313 (5)) completely inhibited the cytotoxic activity. Other modifications to the molecule, not at the sulfur group (OTA-5781, OTA-3998, OTA-3356 (6), OTA-5673 (7)), did not result in a dramatic change in the cytotoxic activity of the molecule (Fig. 5e and Supplementary Fig. 5f,g).
These observations led us to consider the hypothesis that elesclomol-bound copper might serve as a neo-substrate of FDX1. During physiological Fe-S cluster biosynthesis, natural substrates of FDX1 drive oxidation of the reduced form of FDX1. By measuring the oxidized state of FDX1, we found that neither elesclomol nor Cu(II) alone was capable of oxidizing FDX1, but elesclomol and Cu(II) together led to full oxidation of FDX1 (Fig. 5f). Importantly, FDX2 could not substitute for FDX1 in this reaction, consistent with our NMR and biochemical data showing no binding of elesclomol to FDX2 (Supplementary Fig. 5h). Our findings support a model in which the Cu(II)-elesclomol complex directly binds and inhibits the natural function of FDX1 in the Fe-S cluster biosynthesis pathway, which requires reduced FDX1. This reaction also leads to the production of Cu(I).
Elesclomol mediates copper-dependent cell death
We further probed the role of copper in elesclomol induced cell death by chelating copper with tetrathiomolybdate (TTM). TTM specifically inhibited elesclomol and OTA-5781 induced cell death under the conditions where elesclomol is most potent, both in the Hi-Mito (Fig. 6a, b) and the Lo19S (Supplementary Fig. 6a) states. TTM did not affect the cell death induced by antimycin A or bortezomib (Supplementary Fig. 6b, c). Moreover, the addition of copper to elesclomol at a 1:1 molar ratio prior to treatment significantly reduced cell viability (Fig. 6c) when cells were grown in glycolytic (glucose media) conditions. These results are consistent with the model that copper is both necessary and sufficient for the cytotoxic effects of elesclomol. However, the addition of exogenous copper did not potentiate elesclomol-induced cell death in cells that were forced into the Hi-Mito state (galactose media) where elesclomol is already extremely potent (Fig. 6c), nor was potentiation observed in all cell lines (Supplementary Fig. 6d), suggesting that basal cellular copper homeostasis may also determine elesclomol sensitivity. The elesclomol-mediated, copper-dependent cell-death was not associated with the activation of caspase 3 and 7, which are downstream effectors in the apoptosis pathway (Supplementary Fig. 6e, f). The lack of activation of apoptotic signaling, along with the requirement for copper in elesclomol induced cell death, is reminiscent of iron-dependent cell death (ferroptosis) induced by GPX4 inhibitors 30. Whereas copper chelation rescued elesclomol-induced cell death, neither inhibitors of apoptosis or ferroptosis did so (Fig. 6d and Supplementary Fig. 6g-i). Taken together, our results support a model whereby a state of enhanced mitochondrial metabolism and its associated upregulation of FDX1 enzymatic activity facilitates the cytotoxicity of elesclomol by reducing elesclomol-bound Cu(II), resulting in a unique copper dependent cell death (Fig. 6e). Other copper ionophores such as disulfiram may promote cancer cell death through a similar mechanism.
Figure 6. Elesclomol mediated copper dependent cell death is not inhibited by known apoptosis and ferroptosis inhibitors.
a-b, The viability of T47D cells cultured in the presence of galactose (Hi-Mito state) was examined following the addition of either elesclomol (a) or OTA-5781 (b) in the presence or absence of the copper chelator tetrathiomolybdate (TTM) at 10uM concentration. The mean −/+ SD n= 3 biologically independent samples is plotted. c, NCIH2030 cells were grown in glucose (glycolysis) or galactose (Hi-Mito) containing media and treated with either elesclomol or elesclomol-Cu(II) (at a 1:1 molar ratio). The viability was examined after 48 hours and the mean −/+ SD n= 3 biologically independent samples is plotted. d, The viability of cells after 48 hours of treatment with indicated concentrations of elesclomol in the presence or absence of either TTM (copper chelator at 10uM), Z-VAD (pan-caspase inhibitor at 30uM), ferrostatin-1 or alpha-tocopherol (ferroptosis inhibitors at 10uM and 100uM concentrations respectively). The mean −/+ SD n= 3 biologically independent samples is plotted e, A schematic of the mechanism of FDX1-activated, elesclomol-induced and copper-dependent cell death induction.
Discussion
Our results elucidate a surprising role for cellular metabolism in promoting resistance to cell death induced by proteasome inhibition. We have established that the cellular metabolic shift from glycolysis to high mitochondrial (Hi-Mito) dependency is correlated with, naturally induced by, and in some cases even sufficient to promote PI resistance. This metabolic shift to increased mitochondrial energy metabolism also exposes new druggable dependencies. One such dependency was revealed by the increased sensitivity cancer cells to the compound elesclomol, which preferentially targets both the PI-resistant and Hi-Mito states. We further elucidated a unique mechanism of action of elesclomol by showing that it can both suppress the natural function of FDX1, a critical component in the Fe-S cluster assembly pathway, and when bound to copper can also serve as a neo-substrate to promote a unique form of copper-dependent cell death. Thus, the adaptation to proteotoxic stress entails a cellular metabolic shift resulting in increased sensitivity to copper-mediated cell death.
Long before any of the mechanistic insights reported here, elesclomol underwent clinical development for the treatment of advanced solid tumors in combination with paclitaxel 18. Although the precise mechanism of action for elesclomol was not known, it was shown that elesclomol could bind copper 19,22 and shuttle it to the mitochondria 20,21 generating increased levels of reactive oxygen specifies (ROS) levels eventually resulting in cell death 20,22,24,25. It was hypothesized that the ROS induction by elesclomol is facilitated by reduction of Cu(II) to Cu(I) in the mitochondria 20 a process crucial for inducing cancer cell death by elesclomol. However, the mechanism mediating this reduction was unknown. Here, we describe the interaction of elesclomol with the mitochondrial enzyme FDX1 that can facilitate the elesclomol induced toxicity by two distinct processes. First, direct binding of elesclomol to reduced FDX1 inhibits its role in Fe-S cluster biosynthesis and serves as an upstream regulator of mitochondrial function. Second, the elesclomol-Cu(II) complex serves as a neo-substrate for reduced FDX1 leading to oxidation and the production of Cu(I), which promotes a unique form of copper-dependent cell death that cannot be blocked by apoptosis or ferroptosis inhibitors.
In support of our model that elesclomol is bioactivated by FDX1, we show that FDX1 deletion, was the only gene deletion in two whole genome deletion screens, that enabled increased survival in the presence of two distinct elesclomol analogs and also that elevated FDX1 gene expression is the strongest biomarker for elesclomol induced toxicity across a panel of almost 800 cancer cell lines. Thus, FDX1 activity and increased mitochondria dependent energy metabolism are strong inducers of the cytotoxic effect of elesclomol mediated, copper-dependent cell-death. This type of copper-dependent cell death may not be unique to elesclomol, as other copper binding ionophores 31,32 including disulfiram 33 are also expected to promote a similar mode of cell death that we show is distinct from ferroptosis and is not mediated by the classic apoptosis induced caspase 3 activation. The concept that a shift in core metabolism plays an important role in anticancer drug resistance has recently found support in multiple cancer models 34-38 including in multiple myeloma 39,40. Our findings predict that in some of these models, a similar increased metabolism-regulated sensitivity to copper-dependent cell death may exist.
Our work illuminates a previously described, yet underappreciated link between protein turnover and cellular metabolism that is derived from the requirement of cells to coordinate energy production with the demands of protein homeostasis. We show that altering cell metabolism affects the ability of cells to cope with the inhibition of proteasome function and that perturbation of proteasome function can result in a shift in cellular energy metabolism. This feedback loop might stem from both the energy requirements of protein synthesis 41 and breakdown 42 and from the need to recycle damaged, oxidized and dysfunctional proteins 43 producing amino acids that serve as building blocks for protein synthesis, metabolism and redox pathways 44,45. Moreover, this feedback might be underscored as the cellular metabolic state can also regulate proteasome complex integrity in a post-transcriptional manner by altering the redox sate 46, NADH/NAD+ balance 47,48, and mTOR signaling 49,50. We show that elevated proteasome function is required for biomass expansion of glycolytic cancer cells in glucose-rich culture media and can be readily targeted with proteasome inhibitors. In contrast, oxidative phosphorylation promotes resistance to proteasome inhibition, but reveals increased sensitivity to FDX1 regulated copper-dependent cell death. Combined targeting of these two conserved pathways could provide a feasible, chemotherapeutic strategy as it confronts cancer cells with opposing selective pressures.
Methods Section
Antibodies reagents-
FDX1 (Proteintech Group, Inc (Catalog Number: 12592–1-AP)), tubulin (ab80779 Abcam). Caspase 3/7 cleavage was assessed using the Caspase-Glo® 3/7assay system (Promega) according to manufactures protocol. The signal was normalized to viability as measured by cell titer-Glo.
Compounds- Compounds-
Elesclomol (MedChemexpress Co., LTD. # HY-12040), Ixazomib, disulfiram, antimycin A, ferrostatin-1 and Z-VAD (selleck), Bortezomib (LC-Laboratories) TTM and alpha-tocopherol (Sigma). Elesclomol analogs used (described in US patent #8680100 and #7763658) were received from OnTarget Pharmaceutical Consulting LLC. The purity and identity were determined by LC-UV/MS using an Acquity UPLC system coupled to an SQ Detector 2 (Waters) (purity > 95% see supplementary note for compound structure). ML120 was received from the Schreiber lab (Broad Institute). Drug libraries used were the anti-cancer compound library L3000 (selleck), BML-2865 Natural product library (Enzo), the NIH Clinical Collections (NCC) and the Boston University’s Chemical Methodology and Library Development (CMLD-BU) drug library (http://www.bu.edu/cmd/about-the-bu-cmd/compound-libraries/).
Cell culture methods-
T47D, Lo19S T47D, K562, HeLa, NCIH441, NCIH2030, HDPQ1 and 293T-HS cells were cultured in RPMI-1640 medium supplemented with 10% fetal bovine serum; HEK293, HEK293T and MCF-7 cells were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum. For the glucose, galactose experiments RPMI media lacking glucose was supplemented with dialyzed serum, 1mM pyruvate and either 10mM Glucose or 10mM galactose.
GFP+/Luc+ MM.1S cells were generated by retroviral transduction of the human MM1.S that was purchased from ATCC (Manassas, VA, USA)., using the pGC-GFP/Luc vector. Cells were authenticated by short tandem repeat DNA profiling.
Generation of the Lo19S T47D cell line - For the generation of the T47D Tet-inducible PSMD2 knockdown cell line- the TRIPZ vector with an inducible shRNA targeting PSMD2 was purchased from Dharmacon (clone V3THS_403760). It was introduced to the T47D cells according to manufactures protocol and cells were selected with puromycin 1ug/ml for one week. The cells were exposed to doxycycline for 24 hours and cells were FACS sorted for the top 10% of most RFP expressing cells (highest expression of shRNA). The cells were further cultured in the absence of doxycycline and PSMD2 knockdown was induced as specified in the text.
RNA-seq-
For RNA extraction the T47D cells were washed and then collected with Trizol. The MM.1S mouse derived tumors were collected at mice paralysis, by bone marrow flushing from the femurs. Cell pellets were frozen at −80C upon collection of all samples. RNA extraction and purification was conducted with Trizol according to manufactures protocol. For the T47D Lo19S experiment Libraries were prepared for RNA-Seq using Illumina’s TruSeq Stranded mRNA Library Preparation Kit according to manufacturer’s directions. Briefly, total RNA (1ug) is enriched for polyadenylated sequences using oligo dT magnetic bead capture. For the MM.1S tumor analysis Libraries were prepared for RNA-Seq using Illumina’s TruSeq Stranded Total RNA Library Preparation Kit according to manufacturer’s directions. Briefly, total RNA (1ug) is ribo-depleted using biotinylated, target-specific oligos combined with Ribo-Zero rRNA removal beads. The enriched fraction is then fragmented, and first-strand cDNA is generated using random primers. Strand specificity is achieved during second-strand cDNA synthesis by replacing dTTP with dUTP, which quenches the second strand during amplification. The resulting cDNA is A-Tailed and ligated with indexed adapters. Finally, the library is amplified using a DNA Polymerase which cannot incorporate past dUTPs, effectively quenching the second strand during PCR.
RNASeq reads were aligned using STAR (v2.5.4b) 51 using default parameters (except --sjdbOverhang 39) to the human genome (GRCh37) with Ensembl annotation (GRCh37.87) in gtf format. Differential expression was assayed using feature Counts(v1.6.1), with reversely stranded option, and DESeq2(v1.18). K-means clustering, with k=6, was performed using Cluster3 and visualized in Java TreeView. Clustering was performed with only protein-coding genes with at least a median normalized count of 1 in each condition resulting in approximately 13000 genes. Each cluster gene enrichment was analyzed using Gene Ontology (GO) annotations (Extended Table S1). Enrichment analysis presented in Figure 1b was performed using GSEA 52 using HALLMARK gene categories. The full data can be found on GEO (GSE123639)
Disseminated Multiple Myeloma Animal model and survival study-
A total of 25 Female SCID-beige mice at 4–6 weeks of age (Taconic, USA) were utilized for the study. Tumors cells (3 million LUC+/RFP+ MM1S cells/mouse) were introduced into mice via intravenous injection and were allowed to grow until the LUC signal from their tumors reached 1×107 radians (photons/sec/cm2/surface area). Tumor growth was monitored weekly by bioluminescence imaging (BLI), using an IVIS Spectrum-bioluminescent and fluorescent imaging system (Perkins Elmer). Mice were randomly attributed a group of treatment (n=5 per group). Control group was IV injected once a week 200 uL saline solution; Bortezomib (0.25 mg/kg, 100 uL) was IP injected first day of the week, once a week until death of the animal. Elesclomol (25 mg/kg, 100 uL) was subcutaneously injected every 3rd day of the week until death of the animal. Elesclomol+Bortezomib group were treated similarly to previous groups. The delayed group started the Elesclomol treatment 14 days after the inclusion of the animal in the study. Endpoint of the study was dictated by either limb-paralysis of the animal or severe toxicity induced by the treatment as per Dana-Farber Animal policy. Enrichment analysis was performed using GSEA 52 using HALLMARK gene categories.
TCGA analysis-
TCGA data analysis- The Cancer Genome Atlas (TCGA: cancergenome.nih.gov) expression (RNASeq V2) were downloaded using TCGA-assembler 53. RNASeq data were quantified as RSEM. Sigma score was calculated for each primary tumor category separately by calculating a Z-score for every individual proteasome subunit gene and categorizing the tumors as 3-sigma or control as previously described 11. Enrichment analysis was performed using GSEA 52, using H and C2 genesets of MSigDB. GO enrichment was visualized using ClueGO via Cytoscape 54.
The Lo19S T47D drug screen-
Cells with a Dox inducible PSMD2 KD plasmid were incubated in the presence or absence of 1ug/ml of Doxycycline for 48 hours (control versus PSMD2 KD respectively). After 48 hours the cells were collected counted and plated (in the absence of dox) at 1000 cells/well in 384-well opaque, white assay plates (Corning, NY), 50 uL per well, and incubated overnight at 37°C/5% CO2. Compound stocks from the Cancer drug library containing 349 bioactive compounds that were arrayed in dose 10nM, 100nM, 1uM, 10uM (Selleck anti-cancer compound library L3000), the natural products library containing 502 compounds that were utilized in 5 doses (dilution of 1:1000 of 2ug/ml, 0.2ug/ml, 0.002 ug/ml, 0.0002ug/ml), the NIH bioactive library including 731 drugs in dose of 10nM, 100nM, 1uM, 10uM and the Boston University’s CMLD (Chemical Methodology and Library Development) compound deck containing 2866 compounds in one dose (10uM), including novel chemotypes that uniquely probe three-dimensional space by employing stereochemical and positional variation within the molecular framework as diversity elements in library design. 50 nl of compounds were pin-transferred (V&P Scientific, CA, pin tool mounted onto Tecan Freedom Evo 150 MCA96 head, Tecan, CA) into duplicate assay plates and incubated for 72h. The DMSO content was 0.1% within each well. Per plate, there are 32 wells of DMSO vehicle control and 32 wells of positive control compounds. After three days of incubation, 10 uL of CellTiter-Glo (Promega, WI) was added to each well, incubated for 10 minutes and the luminescence output was read on the M1000 Infinite Pro plate reader (Tecan, CA). CellTiter-Glo measures ATP levels in the cell and is used as a surrogate for cell viability.
PRISM assay-
The PRISM assay was conducted as previously described 16 with slight improvements. For the bortezomib (Glucose/galactose) experiment, each condition cells were plated in triplicates. 100,000 cells (of the total pool of 549 barcoded cell lines (Supplementary Dataset 6) in regular media, the next day the day zero control was harvested and media was changed to RPMI with either 10mM Glucose or Galactose. After 24 hours cells were treated with either DMSO or indicated concentrations of bortezomib. Cells were collected and lysed after 3 days. Cells were collected and lysed in DNA buffer (1xPCR buffer Invitrogen, 0.45% NP40, 0.45% Tweem20 and 10% Proteinase K (Qiagen #19133)), incubated at 60°C for 1 hour and then transferred to 384 well plates where proteinase K was inactivated and lysate was taken for genomic DNA analysis.
For the elesclomol experiment, the relative viability of 768 cell lines with elesclomol treatment in RPMI media are measured relative to vehicle control using the PRISM assay 16. The compound is screened in triplicate at 8 doses (4-fold dilutions starting from 10uM) on pools of 20–25 cell lines (Supplementary Dataset 9). After five days, cell lines pools are collapsed into three detection groups and inert control barcodes are added to each group. Genomic DNA from cell lysates was amplified by PCR using primers Biotin pLENTR4 (5′-biotin-CGTCATTACTAACCGGTACGC-3′) and pLENTF1 (5′-GGAATAGAAGAAGAAGGTGG-3′). PCR product was hybridized to Luminex beads with covalently attached antisense barcodes, and streptavidin-phycoerythrin addition, washing and detection on Luminex FlexMap machines was performed as previously described. Median-fluorescence intensity (MFI) values for each cell line barcode in each condition were collected.
To remove low-quality data, cell lines with low separation between log2-MFI(logMFI) values of positive and negative control treatments were removed (strictly standardized mean difference < 2). To correct for non-linear cell-count to MFI relationships, a monotonic spline was used to transform logMFI of the inert control barcodes of each detection group to logMFI of the inert control barcodes for the median DMSO treatment of the same detection group. This transformation was then applied to the remaining, non-control barcodes in each detection group. DMSO-treated cell mixtures were used as reference control for scaling of each cell line signal at the conclusion of each experiment (viability = 100 for each cell line). Thus, the signal from each treated cell line was calculated as 100 × (adjusted median Luminex measurement across replicates)/(median Luminex measurement of DMSO control). Potential batch effects were corrected using ComBat (Johnson et al. 2007). Log2-transformed viability values are corrected independently for each treatment condition. Using the terminology of the original publication, cell lines are treated as probes and batches are defined using cell line pools.
Calculation of AUC and Gene Dependency analysis-
A robust 4-parameter dose response curve is fit using the “drc” R package 55 with the following form:
under the restrictions that l=1and h>0. Subsequently, the area under the log2-dose response curve (AUC) is computed for each cell line 56. The AUC profile obtained from PRISM screen is correlated with the gene expression and genetic dependency data obtained from Cancer Dependency Map (https://depmap.org) in order to detect potential associations. In particular, we computed the Pearson correlation between the PRISM AUC’s (724 cell lines) and each gene’s expression profile (RNAseq measurements of 18,778 protein coding genes over 701 overlapping PRISM cell lines) or genetic dependency profile (results of genome-scale CRISPR/Cas9 knockout viability screens for 17,634 genes over 392 overlapping PRISM cell lines). p-values are computed and corrected using Benjamini-Hochberg (1995) which are available in the Supplementary Dataset 9. The relationship between FDX1 knockout and CRISPR knockouts was assessed across all 391 lines present in the knockout dataset.
The glucose/galactose drug screen-
T47D cells growing in regular media were collected counted and after centrifugation cells were re-suspended in DMEM media without glucose containing 10% dialyzed serum with the addition of either 10mM glucose or 10mM galactose. Cells were then plated at 1000 cells/well in 384-well opaque, white assay plates (Corning, NY), 50 uL per well, and incubated overnight at 37°C/5% CO2. Application of the drug libraries and cell viability was executed as described above for the Lo19S drug screen
Heat-shock reporter-
293T cells (American Type Culture Collection) harboring enhanced GFP fused to firefly luciferase under control of HSP70B′ promoter elements as previously described 57
CRISPR screen –
268M cells were transduced with 22mL human genome-wide cleavage-optimized lentiviral sgRNA library containing Cas9 (http://www.addgene.org/pooled-library/sabatini-crispr-human-high-activity-3-sublibraries) as previously described 58. Cells were allowed to recover for 48h and selected with 3ug/mL puromycin for 72h and transduction efficiency was determined. 50M cells per condition were passaged every 2–3 days throughout the duration of the screen (except where noted), according to the following timeline:
STA5781:
Day 0–7: 4nM
Day 7–26: 6nM
Day 26–34: 100nM
STA3998:
Day 0–7: 25nM
Day 7–23: 44nM
Day 26–31: 1uM
80M cells were collected after puromycin selection, representing the initial cell population, and 5–80M cells were collected at each time point. Genomic DNA was isolated using the QIAmp DNA Blood Maxi, Midi, or Miniprep kit, depending on the cell number of the sample, and high-throughput sequencing libraries were prepared 58, except that the following forward PCR primer was used:
AATGATACGGCGACCACCGAGATCTACACGAATACTGCCATTTGTCTCAAGATCTA
Sequencing reads were aligned to the sgRNA library and the abundance of each sgRNA was calculated. The counts from each sample were normalized for sequencing depth after adding a pseudocount of one. sgRNAs with fewer than 50 reads in the initial reference dataset were omitted from downstream analyses. The log2 fold change in abundance of each sgRNA between the final and initial reference populations was calculated and used to define a CRISPR Score (CS) for each gene. The CS is the average log2 fold change in abundance of all sgRNAs targeting a given gene. Genes represented by fewer than 5 sgRNAs in the initial reference dataset were omitted from downstream analyses.
Statistical analysis-
The distribution of the log2 fold changes between the final and initial populations for the set of sgRNAs targeting each gene was tested against the distribution for all sgRNAs by the Kolmogorov-Smirnov test. P-values were adjusted for multiple comparisons by the Benjamini-Hochberg (FDR) procedure.
FDX1 gene targeting using single sgRNAs-
plentiCRISPRv2 vector 58 was used as a backbone to insert the gRNAs targeting FDX1 and AAVS1 as control.
BsmBI restriction enzyme digest of the backbone vector was followed by T4 ligation of the digested vector with phosphorylated and annealed oligo pairs:
sgFDX1–1 CACCGGCAGGCCGCTGGATCCAGCG
sgFDX1–1_RC: AAACCGCTGGATCCAGCGGCCTGCC
sgFDX1–2 CACCGTGATTCTCTGCTAGATGTTG
sgFDX1–2_RC: AAACCAACATCTAGCAGAGAATCAC
sgAAVS1: CACCGGGGGCCACTAGGGACAGGAT
sgAAVS1_RC: AAACATCCTGTCCCTAGTGGCCCCC
To generate the lentiviruses, 3 × 106 HEK-293T cells were seeded in a 10 cm plate in DMEM supplemented with 10% IFS. 24 hours later, the cells were transfected with the above sgRNA pLenti-encoding plasmids alongside the Δ VPR envelope and CMV VSV-G packaging plasmids using the XTremeGene 9 Transfection Reagent (Roche). 12 hours after transfection, the medium was aspirated and replaced with 8 ml fresh medium. Virus-containing supernatants were collected 48 hours after the transfection and passed through a 0.45 μm filter to eliminate cells. Transduction of cells was done as described above.
K562 cell were cells were seeded at a density of 300,000 cells/mL in 6-well plates in 2 mL of RPMI containing 8 μg/mL polybrene (EMD Millipore), and then transduced with lentivirus by centrifugation at 2,200 RPM for 90 min at 37° C. After a 24-hour incubation, cells were pelleted to remove virus and then re-seeded into fresh culture medium containing puromycin, and selected for 72 hours. Cells were then single cell FACS sorted into 96 well plates and grown out in the presence of 1ug/ml puromycin.
Viability and cell proliferation assays-
Relative cell number count protocol- The viability of K562 cells was conducted by seeding cells at 250,000 cells/mL in 6-well plates of RPMI. In the presence or absence of different concentrations of compound-1 or −2. Viable cell numbers were counted using countess (Invitrogen) every 2–3 days and cells were reseeded at 250,000 cells/mL in 6-well plates of RPMI to continue growth.
The viability of FDX1 KO K562 cells and AAVS1 KO and WT controls was examined by plating 100,000 cells/mL in 6-well cells in RPMI 1640 media (without glucose or pyruvate containing dialyzed serum) with either 10mM glucose or 10mM galactose. Viable cell numbers were counted every day using countess (Invitrogen). In the indicated experiment 1mM Pyruvate and 100ug/ml of Uridine were added.
Cell viability-
Where indicated relative cell number following different drug treatment was conducted by plating cells at 1000 cells/well in 384 well plates. Indicated concentrations of compounds were added 24 hours after plating at least in triplicate for each condition. viability was measured 72 hours after drug addition, using cell titer-glo (Promega) according to manufactures protocol. Same protocol was applied for the experiments conducted for the Lo19S T47D, MCF-7, T47D, HEK293, HeLa, HDPQ1, NCIH2030 and NCIH441 cells in the context of altered carbon source only the cells were plated at 1000 cells/well in RPMI 1640 media (without glucose and with dialyzed serum) containing either 10mM Glucose or 10mM galactose supplemented with 1mM Pyruvate. The EC50 and EC90 was conducted by performing non-linear fit using Prism software.
Protein Expression and Purification-
Unlabeled samples of ISCU, unlabeled (NFS1-ISD11-Acp)2 (abbreviated as SDA), unlabeled and [U-15N]-FDX1 (truncated first 61 amino acids) and unlabeled FDX2 were produced and purified as described previously 26. FDX1 and FDX2 were reduced by adding a 10-fold excess of sodium dithionite to oxidized FDX1 or FDX2 in an anaerobic chamber (Coy Laboratory). The reduction of FDX1 or FDX2 was monitored by the color change from dark brown to light pink and by recording UV/vis spectra before and after reduction. The sample was dialyzed extensively against anaerobic HN buffer to remove excess sodium dithionite.
NMR Spectroscopy-
NMR spectra were collected at the National Magnetic Resonance Facility at Madison on a 750 MHz (1H) Bruker with a z-gradient cryogenic probe. The buffer used for NMR samples (HNT buffer) contained 20 mM HEPES at pH 7.6, 150 mM NaCl, and 2 mM TCEP. All sample temperatures were regulated at 25 °C. NMRPipe software was used to process the raw NMR data and NMRFAM-SPARKY software was utilized to visualize and analyze the processed NMR data.
To study the interactions of FDX1 with elesclomol, 0.2 mM [U-15N]-FDX1 in HNT buffer were placed in 5 mm Shigemi NMR tubes, and 2D 1H,15N TROSY-HSQC spectra were collected before and after titration of 5 equivalent molar ratio of elesclomol dissolved in HNT buffer.
Chemical shift perturbations (ΔδHN absolute value ppm) were calculated by
| (i) |
where ΔδH and ΔδN are the chemical shift changes in the 1H and 15N dimensions, respectively.
Cysteine desulfurase assay-
The assay was conducted as previously described 59. The protein samples used in the cysteine desulfurase assay and Fe-S cluster assembly experiments were prepared in an anaerobic chamber (Coy Laboratory) with samples buffer-exchanged extensively prior to the experiments with anaerobic buffer containing 20 mM HEPES at pH 7.6 and 150 mM NaCl (HN buffer). The reaction volumes in all the experiments were kept to 1 ml. A Shimadzu UV-1700 uv/visible spectrophotometer with a temperature control unit was used to collect the spectra, and UVProbe 2.21 software (Shimadzu) was used in collecting and analyzing the data.
The cysteine desulfurase assay reactions (300 μL in HN buffer) contained 10 μM SDA. The reductant was 100 μM DTT. 100 μM L-cysteine was added to initiate the reaction. 10 to 200 μM elesclomol (1–20 X relative to SDA) was added to assess their effect on sulfide production. After 20 min of anaerobic incubation at room temperature, the reaction mixture was diluted to 800 μL, and 100 μL of 20 mM N,N-dimethyl-p-phenylenediamine in 7.2 M HCl and 100 μL of 30 mM FeCl3 in 1.2 M HCl were added to quench the reaction and convert sulfide to methylene blue. The quenched reaction was incubated for 15 min at room temperature, and then the absorbance at 670 nm was measured and used to estimate the amount of sulfide by comparison to a standard curve obtained from known concentrations of Na2S.
In vitro Fe-S cluster assembly assay-
The assay was conducted as previously described 60. The in vitro Fe-S cluster assembly assays were carried out as follows. Reaction mixtures (1 mL) prepared in the anaerobic chamber contained 25 μM reduced FDX1 or reduced FDX2 as the reductant, 1 μM SDA, 25 μM ISCU and 100 μM (NH4)2Fe(SO4)2. To test the effect of the drug, reaction mixtures contained 25–250 μM elesclomol (1–20 X relative to FDX1). L-cysteine (at a final concentration 100 μM) was added to initiate each experiment. Samples were then transferred to 1-cm path-length quartz cuvettes, sealed with rubber septa, and uv/vis spectra were collected at 25 °C. The growth of absorbance at 456 nm was used to monitor the assembly of [2Fe-2S] clusters.
Electron transfer assay-
Electron transfer from re-FDX1 SDA complex was monitored as follows. 25 μM reduced FDX1 was mixed with 25 μM SDA, and 125 μM L-cysteine was added to initiate the reaction. To test the effect of the drug, samples contained 25–500 μM elesclomol (1–20 X relative to FDX1). Samples were then transferred to 1 cm path-length quartz cuvettes, sealed with rubber septa, and UV/vis spectra were collected at 25 °C. The growth of absorbance at 456 nm was used to monitor the oxidation of reduced FDX1 as a result of electron transfer to SDA.
Oxidation of FDX1 by elesclomol-Cu(II) complex-
The assay was conducted as previously described 26. Elesclomol-Cu(II) complex was made by mixing 1:1 molar ratio of elesclomol with CuCl2 dissolved in H2O. The resulting Elesclomol-Cu(II) water solution developed a dark red color. FDX1 and FDX2 were reduced by adding a 10-fold excess of sodium dithionite to oxidized FDX1 or FDX2 in an anaerobic chamber (Coy Laboratory). The reduction of FDX1 or FDX2 was monitored by the color change from dark brown to light pink and by recording UV/vis spectra before and after reduction. The samples were dialyzed extensively against an anaerobic buffer containing 20 mM HEPES and 150 mM NaCl (HN buffer) to remove excess sodium dithionite. Four 1-mL reactions were prepared under anaerobic conditions, each having following composition: 1) 20 μM reduced FDX1 and 200 μM elesclomol; 2) 20 μM reduced FDX1 and 200 μM CuCl2; 3) 20 μM reduced FDX1 and 200 μM elesclomol-Cu(II) complex; 4) 20 μM reduced FDX2 and 200 μM elesclomol-Cu(II) complex. After mixing, the reactions were immediately transferred to 1-cm path-length quartz cuvettes, sealed with rubber septa, taken out of the anaerobic chamber and placed in a Shimadzu UV-1700 UV/vis spectrophotometer equipped with a temperature control system. Initial and final (3 hours after the reactions were initiated) UV/vis scans were collected at 25 °C.
MM colony formation assay-
KMS11 cell lines transduced with either a doxycycline inducible shPSMD2 or control vector were grown in the presence of doxycycline for 72 hours. The media was replaced with media containing wither glucose or galactose as the carbon source and indicated concentrations 5nM bortezomib. After 24 hours of treatment 1,000 cells were seeded in methylcellulose for colony development. These were monitored and counted after 10 days.
Oxygen consumption rate analysis-
Oxygen consumption rate (OCR) of intact cells was measured using an XF24 Extracellular Flux Analyzer (Seahorse Bioscience) for the K562 cell experiments and the XF96 Extracellular Flux Analyzer (Seahorse Bioscience) for the T47D Lo19S cells. For analysis of WT or FDX1 KO K562 cells, cells were seeded in four replicate wells on Seahorse XF24 cell culture plates coated with CellTak in the indicated growth media and assayed 5h later. Three basal OCR measurements were taken in 8-min intervals, followed by sequential injections of 1 μM oligomycin, 2.5 μM FCCP, and 1 μM antimycin A, taking three measurements at 8-min intervals following each treatment. Basal respiration was determined as the difference between basal OCR and OCR after antimycin A treatment, and maximal respiration as the difference between OCR after FCCP and antimycin A treatments.
Metabolite Extraction and analysis by LCMS-
Cells were washed in ice-cold 0.9% NaCl and metabolites were extracted in ice-cold 80% methanol containing 500 nM isotopically labeled amino acid mix (Cambridge Isotope Laboratories) as internal standards. Samples were vortexed at 4∘C for 10 minutes and cleared by centrifugation for 10 minutes at max speed at 4∘C. Supernatants were transferred to a new tube and samples were evaporated to dryness using a speed vac.
Metabolite profiling was conducted on a QExactive bench top orbitrap mass spectrometer equipped with an Ion Max source and a HESI II probe, which was coupled to a Dionex UltiMate 3000 HPLC system (Thermo Fisher Scientific, San Jose, CA). External mass calibration was performed using the standard calibration mixture every 7 days. Typically, samples were reconstituted in 100 uL water and 2 uL were injected onto a SeQuant® ZIC®-pHILIC 150 × 2.1 mm analytical column equipped with a 2.1 × 20 mm guard column (both 5 mm particle size; EMD Millipore). Buffer A was 20 mM ammonium carbonate, 0.1% ammonium hydroxide; Buffer B was acetonitrile. The column oven and autosampler tray were held at 25∘C and 4∘C, respectively. The chromatographic gradient was run at a flow rate of 0.150 mL/min as follows: 0–20 min: linear gradient from 80–20% B; 20–20.5 min: linear gradient form 20–80% B; 20.5–28 min: hold at 80% B. The mass spectrometer was operated in full-scan, polarity-switching mode, with the spray voltage set to 3.0 kV, the heated capillary held at 275∘C, and the HESI probe held at 350∘C. The sheath gas flow was set to 40 units, the auxiliary gas flow was set to 15 units, and the sweep gas flow was set to 1 unit. MS data acquisition was performed in a range of m/z = 70–1000, with the resolution set at 70,000, the AGC target at 1×106, and the maximum injection time at 20 msec. An additional scan (m/z 220–700) in negative mode only was included to enhance detection of nucleotides. Relative quantitation of polar metabolites was performed with XCalibur QuanBrowser 2.2 (Thermo Fisher Scientific) using a 5 ppm mass tolerance and referencing an in-house library of chemical standards.
Supplementary Material
Acknowledgments
This work is dedicated to the memory of Susan Lindquist who served as a great inspiration as a scientist, mentor and human being. We thank L. Clayton, C. Kayatekin, B. Bevis, N. Kanarek, N. Dharia, V. Viswanathan, J. Eaton, T. Ast, I. Fung, B. Wang and J. McFarland for constructive discussion and comments. Special thanks to D. Sabatini, D. Pincus, J. Rettenmaier and V. Mootha for providing critical comments and reviewing of the manuscript and G. Fink for his supervision. We thank G. Botta, J. Fonseka, J. Roth, S. Bender for help with the PRISM experiments setup and analysis. We thank the Koch Institute Swanson Biotechnology Center for technical support, specifically J. Cheah for her support conducting the chemical drug screen. NMR spectroscopy was carried out at the National Magnetic Resonance Facility at Madison which is supported by National Institutes of Health (NIH) grant P41GM103399; other work at the University of Wisconsin-Madison was supported by funds from the Biochemistry Department. P.T was supported by EMBO Fellowship ALTF 739–2011 and by the Charles A. King Trust Postdoctoral Fellowship Program. A.D was supported by the Multiple Myeloma Research Foundation. T.R.G is an HHMI investigator.
Footnotes
Competing Financial Interests
T.R.G is a consultant in GlaxoSmithKline, Sherlock Biosciences and Foundation medicine; co-founder of Sherlock Biosciences and Foundation medicine. W.Y is employed by Ranok Therapeutics Co. Ltd. and is also the sole proprietor of OnTarget Pharmaceutical Consulting LLC.
Data availability statement
The data that support the findings of this study are available within the paper (and its supplementary information files) or on a public server (GEO-GSE123639). Additional information and reasonable requests for data, resources, sequences, and reagents should be directed to and will be fulfilled by the corresponding author.
References
- 1.Luo J et al. A genome-wide RNAi screen identifies multiple synthetic lethal interactions with the Ras oncogene. Cell 137, 835–848, doi: 10.1016/j.cell.2009.05.006 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Petrocca F et al. A genome-wide siRNA screen identifies proteasome addiction as a vulnerability of basal-like triple-negative breast cancer cells. Cancer cell 24, 182–196, doi: 10.1016/j.ccr.2013.07.008 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adams J et al. Proteasome inhibitors: a novel class of potent and effective antitumor agents. Cancer Res 59, 2615–2622 (1999). [PubMed] [Google Scholar]
- 4.Deshaies RJ Proteotoxic crisis, the ubiquitin-proteasome system, and cancer therapy. BMC biology 12, 94, doi: 10.1186/s12915-014-0094-0 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Holbeck SL, Collins JM & Doroshow JH Analysis of Food and Drug Administration-approved anticancer agents in the NCI60 panel of human tumor cell lines. Mol Cancer Ther 9, 1451–1460, doi: 10.1158/1535-7163.MCT-10-0106 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Orlowski RZ & Kuhn DJ Proteasome inhibitors in cancer therapy: lessons from the first decade. Clin Cancer Res 14, 1649–1657, doi: 10.1158/1078-0432.CCR-07-2218 (2008). [DOI] [PubMed] [Google Scholar]
- 7.Markovina S et al. Bortezomib-resistant nuclear factor-kappaB activity in multiple myeloma cells. Mol Cancer Res 6, 1356–1364, doi: 10.1158/1541-7786.MCR-08-0108 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li B et al. The Nuclear Factor (Erythroid-derived 2)-like 2 and Proteasome Maturation Protein Axis Mediate Bortezomib Resistance in Multiple Myeloma. J Biol Chem 290, 29854–29868, doi: 10.1074/jbc.M115.664953 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang XD et al. Tight Junction Protein 1 Modulates Proteasome Capacity and Proteasome Inhibitor Sensitivity in Multiple Myeloma via EGFR/JAK1/STAT3 Signaling. Cancer cell 29, 639–652, doi: 10.1016/j.ccell.2016.03.026 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kisselev AF, van der Linden WA & Overkleeft HS Proteasome inhibitors: an expanding army attacking a unique target. Chem Biol 19, 99–115, doi: 10.1016/j.chembiol.2012.01.003 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsvetkov P et al. Suppression of 19S proteasome subunits marks emergence of an altered cell state in diverse cancers. Proc Natl Acad Sci U S A 114, 382–387, doi: 10.1073/pnas.1619067114 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsvetkov P et al. Compromising the 19S proteasome complex protects cells from reduced flux through the proteasome. eLife 4, doi: 10.7554/eLife.08467 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shi CX et al. CRISPR genome-wide screening identifies dependence on the proteasome subunit PSMC6 for Bortezomib sensitivity in multiple myeloma. Mol Cancer Ther, doi: 10.1158/1535-7163.MCT-17-0130 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Acosta-Alvear D et al. Paradoxical resistance of multiple myeloma to proteasome inhibitors by decreased levels of 19S proteasomal subunits. eLife 4, e08153, doi: 10.7554/eLife.08153 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gohil VM et al. Nutrient-sensitized screening for drugs that shift energy metabolism from mitochondrial respiration to glycolysis. Nat Biotechnol 28, 249–255, doi: 10.1038/nbt.1606 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu C et al. High-throughput identification of genotype-specific cancer vulnerabilities in mixtures of barcoded tumor cell lines. Nat Biotechnol 34, 419–423, doi: 10.1038/nbt.3460 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ackler S et al. The Bcl-2 inhibitor ABT-263 enhances the response of multiple chemotherapeutic regimens in hematologic tumors in vivo. Cancer chemotherapy and pharmacology 66, 869–880, doi: 10.1007/s00280-009-1232-1 (2010). [DOI] [PubMed] [Google Scholar]
- 18.O’Day S et al. Phase II, randomized, controlled, double-blinded trial of weekly elesclomol plus paclitaxel versus paclitaxel alone for stage IV metastatic melanoma. J Clin Oncol 27, 5452–5458, doi: 10.1200/JCO.2008.17.1579 (2009). [DOI] [PubMed] [Google Scholar]
- 19.Hasinoff BB, Yadav AA, Patel D & Wu X The cytotoxicity of the anticancer drug elesclomol is due to oxidative stress indirectly mediated through its complex with Cu(II). J Inorg Biochem 137, 22–30, doi: 10.1016/j.jinorgbio.2014.04.004 (2014). [DOI] [PubMed] [Google Scholar]
- 20.Nagai M et al. The oncology drug elesclomol selectively transports copper to the mitochondria to induce oxidative stress in cancer cells. Free Radic Biol Med 52, 2142–2150, doi: 10.1016/j.freeradbiomed.2012.03.017 (2012). [DOI] [PubMed] [Google Scholar]
- 21.Soma S et al. Elesclomol restores mitochondrial function in genetic models of copper deficiency. Proc Natl Acad Sci U S A 115, 8161–8166, doi: 10.1073/pnas.1806296115 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yadav AA, Patel D, Wu X & Hasinoff BB Molecular mechanisms of the biological activity of the anticancer drug elesclomol and its complexes with Cu(II), Ni(II) and Pt(II). J Inorg Biochem 126, 1–6, doi: 10.1016/j.jinorgbio.2013.04.013 (2013). [DOI] [PubMed] [Google Scholar]
- 23.Barbi de Moura M et al. Mitochondrial respiration--an important therapeutic target in melanoma. PLoS One 7, e40690, doi: 10.1371/journal.pone.0040690 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blackman RK et al. Mitochondrial electron transport is the cellular target of the oncology drug elesclomol. PLoS One 7, e29798, doi: 10.1371/journal.pone.0029798 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kirshner JR et al. Elesclomol induces cancer cell apoptosis through oxidative stress. Mol Cancer Ther 7, 2319–2327, doi: 10.1158/1535-7163.MCT-08-0298 (2008). [DOI] [PubMed] [Google Scholar]
- 26.Cai K, Tonelli M, Frederick RO & Markley JL Human Mitochondrial Ferredoxin 1 (FDX1) and Ferredoxin 2 (FDX2) Both Bind Cysteine Desulfurase and Donate Electrons for Iron-Sulfur Cluster Biosynthesis. Biochemistry 56, 487–499, doi: 10.1021/acs.biochem.6b00447 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sheftel AD et al. Humans possess two mitochondrial ferredoxins, Fdx1 and Fdx2, with distinct roles in steroidogenesis, heme, and Fe/S cluster biosynthesis. Proc Natl Acad Sci U S A 107, 11775–11780, doi: 10.1073/pnas.1004250107 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shi Y, Ghosh M, Kovtunovych G, Crooks DR & Rouault TA Both human ferredoxins 1 and 2 and ferredoxin reductase are important for iron-sulfur cluster biogenesis. Biochim Biophys Acta 1823, 484–492, doi: 10.1016/j.bbamcr.2011.11.002 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arroyo JD et al. A Genome-wide CRISPR Death Screen Identifies Genes Essential for Oxidative Phosphorylation. Cell Metab 24, 875–885, doi: 10.1016/j.cmet.2016.08.017 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang WS et al. Regulation of ferroptotic cancer cell death by GPX4. Cell 156, 317–331, doi: 10.1016/j.cell.2013.12.010 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shimada K et al. Copper-Binding Small Molecule Induces Oxidative Stress and Cell-Cycle Arrest in Glioblastoma-Patient-Derived Cells. Cell Chem Biol 25, 585–594 e587, doi: 10.1016/j.chembiol.2018.02.010 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tardito S et al. Copper binding agents acting as copper ionophores lead to caspase inhibition and paraptotic cell death in human cancer cells. J Am Chem Soc 133, 6235–6242, doi: 10.1021/ja109413c (2011). [DOI] [PubMed] [Google Scholar]
- 33.Cen D, Brayton D, Shahandeh B, Meyskens FL Jr. & Farmer PJ Disulfiram facilitates intracellular Cu uptake and induces apoptosis in human melanoma cells. J Med Chem 47, 6914–6920, doi: 10.1021/jm049568z (2004). [DOI] [PubMed] [Google Scholar]
- 34.Kuntz EM et al. Targeting mitochondrial oxidative phosphorylation eradicates therapy-resistant chronic myeloid leukemia stem cells. Nature medicine 23, 1234–1240, doi: 10.1038/nm.4399 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee KM et al. MYC and MCL1 Cooperatively Promote Chemotherapy-Resistant Breast Cancer Stem Cells via Regulation of Mitochondrial Oxidative Phosphorylation. Cell Metab 26, 633–647 e637, doi: 10.1016/j.cmet.2017.09.009 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matassa DS et al. Oxidative metabolism drives inflammation-induced platinum resistance in human ovarian cancer. Cell Death Differ 23, 1542–1554, doi: 10.1038/cdd.2016.39 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vazquez F et al. PGC1alpha expression defines a subset of human melanoma tumors with increased mitochondrial capacity and resistance to oxidative stress. Cancer cell 23, 287–301, doi: 10.1016/j.ccr.2012.11.020 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vellinga TT et al. SIRT1/PGC1alpha-Dependent Increase in Oxidative Phosphorylation Supports Chemotherapy Resistance of Colon Cancer. Clin Cancer Res 21, 2870–2879, doi: 10.1158/1078-0432.CCR-14-2290 (2015). [DOI] [PubMed] [Google Scholar]
- 39.Soriano GP et al. Proteasome inhibitor-adapted myeloma cells are largely independent from proteasome activity and show complex proteomic changes, in particular in redox and energy metabolism. Leukemia 30, 2198–2207, doi: 10.1038/leu.2016.102 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zaal EA et al. Bortezomib resistance in multiple myeloma is associated with increased serine synthesis. Cancer Metab 5, 7, doi: 10.1186/s40170-017-0169-9 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Frumkin I et al. Gene Architectures that Minimize Cost of Gene Expression. Mol Cell 65, 142–153, doi: 10.1016/j.molcel.2016.11.007 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peth A, Nathan JA & Goldberg AL The ATP costs and time required to degrade ubiquitinated proteins by the 26 S proteasome. J Biol Chem 288, 29215–29222, doi: 10.1074/jbc.M113.482570 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Raynes R, Pomatto LC & Davies KJ Degradation of oxidized proteins by the proteasome: Distinguishing between the 20S, 26S, and immunoproteasome proteolytic pathways. Mol Aspects Med 50, 41–55, doi: 10.1016/j.mam.2016.05.001 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vabulas RM & Hartl FU Protein synthesis upon acute nutrient restriction relies on proteasome function. Science 310, 1960–1963, doi: 10.1126/science.1121925 (2005). [DOI] [PubMed] [Google Scholar]
- 45.Suraweera A, Munch C, Hanssum A & Bertolotti A Failure of amino acid homeostasis causes cell death following proteasome inhibition. Mol Cell 48, 242–253, doi: 10.1016/j.molcel.2012.08.003 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang X, Yen J, Kaiser P & Huang L Regulation of the 26S proteasome complex during oxidative stress. Sci Signal 3, ra88, doi: 10.1126/scisignal.2001232 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cho-Park PF & Steller H Proteasome regulation by ADP-ribosylation. Cell 153, 614–627, doi: 10.1016/j.cell.2013.03.040 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tsvetkov P et al. NADH binds and stabilizes the 26S proteasomes independent of ATP. J Biol Chem 289, 11272–11281, doi: 10.1074/jbc.M113.537175 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rousseau A & Bertolotti A An evolutionarily conserved pathway controls proteasome homeostasis. Nature, doi: 10.1038/nature18943 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhao J, Zhai B, Gygi SP & Goldberg AL mTOR inhibition activates overall protein degradation by the ubiquitin proteasome system as well as by autophagy. Proc Natl Acad Sci U S A 112, 15790–15797, doi: 10.1073/pnas.1521919112 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dobin A et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21, doi: 10.1093/bioinformatics/bts635 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Subramanian A et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102, 15545–15550, doi: 10.1073/pnas.0506580102 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhu Y, Qiu P & Ji Y TCGA-assembler: open-source software for retrieving and processing TCGA data. Nat Methods 11, 599–600, doi: 10.1038/nmeth.2956 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shannon P et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13, 2498–2504, doi: 10.1101/gr.1239303 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ritz C, Baty F, Streibig JC & Gerhard D Dose-Response Analysis Using R. PLoS One 10, e0146021, doi: 10.1371/journal.pone.0146021 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Safikhani Z et al. Revisiting inconsistency in large pharmacogenomic studies. F1000Res 5, 2333, doi: 10.12688/f1000research.9611.3 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wijeratne EM et al. Structure-activity relationships for withanolides as inducers of the cellular heat-shock response. J Med Chem 57, 2851–2863, doi: 10.1021/jm401279n (2014). [DOI] [PubMed] [Google Scholar]
- 58.Wang T, Lander ES & Sabatini DM Viral Packaging and Cell Culture for CRISPR-Based Screens. Cold Spring Harb Protoc 2016, pdb prot090811, doi: 10.1101/pdb.prot090811 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cai K, Frederick RO, Tonelli M & Markley JL ISCU(M108I) and ISCU(D39V) Differ from Wild-Type ISCU in Their Failure To Form Cysteine Desulfurase Complexes Containing Both Frataxin and Ferredoxin. Biochemistry 57, 1491–1500, doi: 10.1021/acs.biochem.7b01234 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cai K, Frederick RO, Tonelli M & Markley JL Interactions of iron-bound frataxin with ISCU and ferredoxin on the cysteine desulfurase complex leading to Fe-S cluster assembly. J. Inorg. Biochem. 183, 107–116, doi: 10.1016/j.jinorgbio.2018.03.007 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available within the paper (and its supplementary information files) or on a public server (GEO-GSE123639). Additional information and reasonable requests for data, resources, sequences, and reagents should be directed to and will be fulfilled by the corresponding author.