Abstract
Constitutive MAPK activation silences genes required for iodide uptake and thyroid hormone biosynthesis in thyroid follicular cells. Accordingly, most BRAFV600E papillary thyroid cancers (PTC) are refractory to radioiodide (RAI) therapy. MAPK pathway inhibitors rescue thyroid-differentiated properties and RAI responsiveness in mice and patient subsets with BRAFV600E-mutant PTC. TGFB1 also impairs thyroid differentiation and has been proposed to mediate the effects of mutant BRAF. We generated a mouse model of BRAFV600E-PTC with thyroid-specific knockout of the Tgfbr1 gene to investigate the role of TGFB1 on thyroid-differentiated gene expression and RAI uptake in vivo. Despite appropriate loss of Tgfbr1, pSMAD levels remained high, indicating that ligands other than TGFB1 were engaging in this pathway. The activin ligand subunits Inhba and Inhbb were found to be overexpressed in BRAFV600E-mutant thyroid cancers. Treatment with follistatin, a potent inhibitor of activin, or vactosertib, which inhibits both TGFBR1 and the activin type I receptor ALK4, induced a profound inhibition of pSMAD in BRAFV600E-PTCs. Blocking SMAD signaling alone was insufficient to enhance iodide uptake in the setting of constitutive MAPK activation. However, combination treatment with either follistatin or vactosertib and the MEK inhibitor CKI increased 124I uptake compared to CKI alone. In summary, activin family ligands converge to induce pSMAD in Braf-mutant PTCs. Dedifferentiation of BRAFV600E-PTCs cannot be ascribed primarily to activation of SMAD. However, targeting TGFβ/activin-induced pSMAD augmented MAPK inhibitor effects on iodine incorporation into BRAF tumor cells, indicating that these two pathways exert interdependent effects on the differentiation state of thyroid cancer cells.
Keywords: thyroid, TGF beta, BRAF, iodide uptake
Introduction
Radioiodine is a key treatment modality for patients with recurrent or metastatic-differentiated thyroid cancer and is also widely applied in the post-operative adjuvant setting. Most papillary thyroid cancers (PTC), which are the most prevalent form of the disease, are associated with clonal non-overlapping activating mutations of genes encoding MAPK pathway signaling effectors, primarily point mutations of BRAF and of the three RAS genes, as well as fusions of receptor tyrosine kinases (Fagin & Wells 2016). There is an inverse correlation between the MAPK signaling flux of PTCs, as measured by its transcriptional output, and the expression of genes required for iodine uptake and thyroid hormone biosynthesis (Cancer Genome Atlas Research Network 2014). BRAFV600E-mutant thyroid cancers have a high MAPK output because this class 1 BRAF-mutant signals as a monomer and is insensitive to the negative feedback effects of ERK on activated RAF dimers (Yao et al. 2015). Accordingly, they also have the most profoundly decreased thyroid differentiation score (TDS) (Cancer Genome Atlas Research Network 2014), a quantitative integrated readout of a set of thyroid differentiation markers, and tend to be more refractory to RAI therapy (Xing et al. 2005).
Treatment of well-differentiated thyroid cells with TGFB1 impairs TSH-induced expression of thyroid-specific genes such as Tg (thyroglobulin) and Slc5a5 (Nis) (Colletta et al. 1989). The Santisteban lab showed that pSMAD2/3 binds to the thyroid lineage transcription factor PAX8 and impairs its transactivation of the sodium iodide symporter (Nis), and that this is reversed by expression of SMAD7 (Costamagna et al. 2004, Riesco-Eizaguirre et al. 2009), which prevents SMAD2/3 activation by promoting degradation of the TGFβ receptor (Kavsak et al. 2000). Moreover, BRAFV600E-induced suppression of Nis expression was shown to be mediated by a TGFB1-driven autocrine loop in PCCL3 (Riesco-Eizaguirre et al. 2009). This group also found that the inhibition of Nis transcription was MEK-independent, implying that redifferentiation is attainable in the setting of high constitutive MAPK activation. This is inconsistent with the evidence that RAF and MEK inhibitors rescue NIS mRNA levels and iodine incorporation in BRAF-mutant cell lines, mouse models and patients (Knauf et al. 2003, Liu et al. 2007, Chakravarty et al. 2011, Ho et al. 2013, Rothenberg et al. 2015, Nagarajah et al. 2016). Nevertheless, BRAFV600E clearly increases TGFB1 expression and pSMAD in cell lines and mouse PTCs(Riesco-Eizaguirre et al. 2009, Knauf et al. 2011),prompting us to investigate the functional role of this pathway in a genetically accurate in vivo context.
By using a combination of genetic and pharmacological approaches, we found that pSMAD activation is increased in BRAF-mutant thyroid cancers, and that this is due to promiscuous engagement of activin and TGFβ family ligands with their corresponding receptors. Inhibition of pSMAD activation in vivo is insufficient to induce the cancer cells to redifferentiate in the context of constitutive MAPK activation. However, suppression of both MAPK and pSMAD pathways leads to enhancement of radioiodine uptake in cancer cells, an effect that could be leveraged for therapeutic benefit.
Materials and methods
Mouse models
Animals used in this study were maintained on a mixed strain background. Animal care and all experimental procedures were approved by the MSKCC Institutional Animal Care and Use Committee. The mouse lines were genotyped by Transnetyx, Inc. (Cordova, TN) using quantitative PCR or by PCR using primers described in Supplementary Table 1 (see section on supplementary materials given at the end of this article).
TPO-Cre mice (Kusakabe et al. 2004) obtained from Dr Shioko Kimura (NCI) were bred with LSL-BrafV600E mice (Mercer et al. 2005) obtained from Dr Catrin Pritchard (University of Leicester, UK) to generate TPO-Cre/LsL-BrafV600E (Braf) mice, resulting in thyroid-specific knock-in of oncogenic Braf (Franco et al. 2011). Tgfbr1f/f (TβR1) mice (Larsson et al. 2003), a generous gift from Dr Yosuke Mukoyama (NIH/NHLBI), were bred with LSL-BrafV600E and TPO-Cre to generate LSL-BrafV600E/Tgfbr1f/f and TPO-Cre/Tgfbr1f/f, respectively. Experimental TPO-Cre/LSL-BrafV600E/Tgfbr1f/f (Braf/TβR1) mice were generated by breeding TPO-Cre/Tgfbr1f/f with LSL-BrafV600E/Tgfbr1f/f.
Generation of Tre-shTgfbr1 mice
The shRNAs to Tgfbr1 and Tgfbr2 were designed using splashRNA (splashrna.mskcc.org) (Pelossof et al. 2017) and cloned into the pMSCV retrovirus expression vector. Retroviral particles expressing the shRNAs were generated as previously described (Dow et al. 2012) and used to infect a TPO-Cre/BrafV600E PTC cell line. Quantitative RT-PCR was used to determine knockdown efficiency and Western blotting for pSMAD to evaluate efficacy for blocking TGFB1-induced SMAD activation (Supplementary Fig. 1D, E and F). Tgfbr1 shRNA #6, the most effective in blocking TGFB1-induced pSMAD (Supplementary Fig. 1D, E and F), was subcloned into the TGMP vector and used to target Tre-shTgfbr1 into the Cola1-homing cassette (CHC) in ESCs derived from TPO-Cre/LSL-BrafV600E/RIK/CHC mice using recombination-mediated cassette exchange (Premsrirut et al. 2011, Dow et al. 2012). The ESC clones confirmed to have single copy of Tre-shTgfbr1 targeted to the CHC were microinjected into blastocysts produced from NCI C57BL/6-cBrd/cBrd/Cr (C57BL/6 albino) mice and implanted into CD-1 pseudopregnant mothers enabling the production of chimeric pups. Chimeric pups were bred to generate TPO-Cre/RIK/Tre-shTgfbr1 mice and experimental TPO-Cre/BrafV600E/RIK/Tre-shTgfbr1 (Braf/shTβR1) animals generated by crossing them with LSL-BrafV600E. Animals were fed doxycycline-impregnated chow (TD01306, Envigo) to induce expression of the Tgfbr1 shRNA and the appropriate knockdown of Tgfbr1 was confirmed by quantitative RT-PCR.
Generation of mouse papillary thyroid cancer cell lines
PTCs from Braf, Braf/TβR1 and Braf/shTβR1 mice were collected, washed with 1× PBS, minced and placed in digestion medium (MEM with 112 U/mL collagenase type I, 1.2 U/mL dispase and pen/strep). Minced tissues were incubated at 37°C for 45–60 min and then washed twice with Coon’s F12 containing 0.5% bovine brain extract (BBE) (Hammond Cell Tech) and pen/strep/glutamine (PSG) (Gemini Bio Products). Cells were resuspended in Coon’s F12 containing 0.5% BBE and PSG and then plated in CellBind plates (Corning). After culturing for 7–10 days, the culture medium was changed to growth medium (Coon’s F12 containing 5% fetal bovine serum (FBS)(Omega Scientific), 0.5% BBE and PSG). Cells were passaged for at least 1 month and the thyroid cancer origin confirmed by expression of BRAFV600E prior to use.
Reagents
EW7197 (Vactosertib, Catalog No.S7530) and SD208 (Catalog No.S7530) were purchased from SelleckChem. CH5126766 (CKI) was from Chugai Pharmaceutical Co. Follistatin was purchased from Shenandoah Biotechnologies Inc. The TGFB1 neutralizing antibody 1D11 and its isotype control were obtained from the MSKCC MAB Core.
For in vivo experiments, CKI was dissolved in 2 hydroxypropyl-β-cyclo-dextrin and administered once daily by oral gavage at a dose of 3.0 mg/kg. EW7197 was dissolved in artificial gastric fluid formulation (95 mM HCl, 38 mM NaCl and 3.6 mg/mL pepsin) and administered once daily by oral gavage at a dose of 25 mg/kg. Follistatin was dissolved in sterile H2O and administered once daily by i.p. injection at a dose of 25 µg/kg. The TGFB1 neutralizing antibody 1D11 and isotype control were suspended in sterile saline and administrated once every 2 days at a dose of 10 mg/kg.
RNA isolation and quantitative RT-PCR
Thyroid lobes were surgically removed and immediately placed in liquid N2. Cell lines were scraped into PBS, washed once with PBS and the cell pellet used for RNA isolation. RNA was isolated using TRIzol (Invitrogen) or RNAeasy (Affymetrix) and 100–500 ng was reverse transcribed with SuperScript III (Invitrogen) in the presence of random hexamers to generate cDNA. Quantitative PCR using the cDNA as a template was done using Power SYBR Green PCR Master Mix (Applied Biosystems) and primer pairs for mouse β-actin, Tgfbr1, Tgfbr2, Tshr, Tg, Nis, Tpo, Pax8, and Dusp5 (Supplemental Table 1). The cycle threshold values for β-actin and the target genes were determined with a 7500 real-time PCR instrument (Applied Biosystems) and used to calculate the β-actin normalized relative expression using the QGENE program (Tomas et al. 2012).
124I PET imaging
Imaging was performed using an R4 microPET scanner (Concorde Microsystems) as previously described (Chakravarty et al. 2011, Nagarajah et al. 2016). Briefly, mice were gavaged with 2.6–3.5 MBq (70–94 μCi) of Na124I and were imaged 24 and 72 h later under inhalational isoflurane anesthesia at 1.5 L/min. List-mode data were acquired for 5 min using an energy window of 250–750 keV and a coincidence timing window of 6 ns, histogrammed into 2D projected data by Fourier rebinning, and reconstructed by filtered back-projection using a cut-off frequency equal to the Nyquist frequency. Image visualization and analysis were performed using ASIPro VM software (Concorde Microsystems) by drawing a 3D volume of the target lesions to derive the activity concentrations to the two different time points, which in turn was used to calculate a maximum activity concentration by applying an empirical model as previously described (Jentzen et al. 2008). The activity concentration was corrected for partial volume effects using a recovery coefficient derived from previously done phantom studies.
Immunostaining
Mouse thyroids dissected from surrounding tissues were fixed in 4% paraformaldehyde, embedded in paraffin, sectioned, and stained with hematoxylin and eosin (H&E). Sections were also immunostained for CD45. H&E and IHC for CD45 were performed by the MSK Molecular Cytology Core Facility.
Co-immunofluorescent staining for NIS and Pan-cadherin
For immunofluorescence staining, tissue sections deparaffinized and permeabilized with 0.3% Triton were subjected to heat-induced epitope retrieval using citrate-based antigen unmasking solution at pH6 (H-3300; Vector Laboratories). Sections were blocked in PBS containing 3% BSA and 10% normal donkey serum (017-000-121; Jackson Labs) and incubated with rabbit anti-Rat SLC5A5 (kindly provided by Dr Nancy Carrasco, Vanderbilt School of Medicine), followed by A647 labeled donkey anti-rabbit secondary antibodies (A31573; Invitrogen) and sequentially co-stained with A546 labeled pan-cadherin antibody (Sc-515872; Santa Cruz). Z-stack scans of the slides were taken with Pannoramic P250 Flash scanner (3DHistech, Hungary) using 20×/0.8NA objective lens. Regions of interest around the tissues were then drawn and max projection images were exported as .tif files using Caseviewer (3DHistech, Hungary). Images were analyzed using ImageJ/FIJI (NIH, USA) where thresholding and watershedding were used to segment the nuclei in the DAPI channel. NIS expression at the plasma membrane was determined by thresholding the A647/A546 channel and quantifying the co-localization fluorescence intensity at the membrane, and normalizing this to the DAPI/cell counts across the sections.
Western blotting
Mouse thyroid cancer cell lines were plated in CellBind plates in Coon’s F12 with 5% FBS, 0.5% BBE and PSG and incubated for 24–48 h. Medium was then switched to Coon’s F12 with 0.1% BSA and PSG and incubated for 24–48 h. Cells were then washed with ice-cold PBS and harvested by scraping and centrifugation (1000 g for 4 min at 4°C). The cell pellets were resuspended in a lysis buffer consisting of 10 mM Tris-HCl (pH 7.5), 5 mM ETDA, 4 mM EGTA, 1% Triton ×100, protease inhibitor cocktail (Sigma) and phosphatase inhibitor cocktail I and II (Sigma). Cells were placed on ice for 10 min and lysed by passing through a p200 tip. Cell debris was removed by centrifugation (18,000 g for 15 min at 4°C) and the supernatant was collected. Frozen tissues were placed in lysis buffer and then ground with a Polytron homogenizer. Lysates were centrifuged to removed debris (18,000 g for 15 min at 4°C) and supernatant was collected. Protein concentration for all lysates was determined using the MicroBCA kit (Thermo Fisher Scientific). Ten to fifty micrograms of protein lysate was subjected to SDS-PAGE and transferred to PVDF membranes (VWR). The membranes were probed with the indicated antibody and the target protein detected by incubating with species-specific horseradish peroxidase conjugated IgG’s (Santa Cruz) or IRDye fluorophores and then with ECL reagent (Amersham Biosciences). HRP probed blots were developed using the ECL reagent (Amersham Biosciences) and the signal captured using X-ray films or with the KwikQuantTM Imager (http://kindlebio.com/index.php). IRDye-probed blots were imaged using the LICOR Odyssey imaging system (Licor Biosciences). The following antibodies were used: from Cell Signaling pERK (9101), ERK (9102), pSMAD2S465/467 (3101S) and Vinculin (13901S), from Sigma β-actin (A2228), from Santa Cruz SMAD2/3 (sc-8332), from Thermofisher pSMAD2S465/467 (44-244G), from Abcam pSMADT8; from Novus NOX4 (NB110-58849). ImageJ was used to quantify band density.
Human clinical trials
The trials NCT02456701, NCT01947023 and NCT02145143 were performed after approval by the MSKCC Institutional Review Board (IRB numbers 15-046, 13-061 and 14-031, respectively). Written informed consent was obtained from all patients in these studies. Patients received either vemurafenib 960 mg or dabrafenib 150 mg orally twice daily and had sequential biopsies of the same index lesion at baseline (prior to drug) and at 2 weeks while on either vemurafenib or dabrafenib. RNAseq was performed on all samples as described (Dunn et al. 2019).
Datasets and TGFβ family ligand score (TLS)
NCBI GEO human datasets GSE29265, GSE33630 (Dom et al. 2012, Tomas et al. 2012) and GSE65144 (von Roemeling et al. 2015) were downloaded and imported into Partek Genomic suites for normalization and expression analysis. Normalized expression data derived from RNAseq of PTCs in the THCA TCGA dataset (Cancer Genome Atlas Research Network 2014) were obtained from the archive at GDAC Firehose: http://gdac.broadinstitute.org/runs/stddata__2016_01_28/data/THCA/20160128/gdac.broadinstitute.org_THCA.Merge_rnaseqv2__illuminahiseq_rnaseqv2__unc_edu__Level_3__RSEM_genes_normalized__data.Level_3.2016012800.0.0.tar.gz. TGFβ family ligand score is the average log2 fold change of Tgfb1, Tgfb2, Tgfb3, Inhba and Inhbb compared with the median of all samples. The TGFβ transcriptional output score was determined by calculating the average fold change for all mRNAs in the gene set. The TGFβ gene set consisted of genes found regulated by TGFβ (Coulouarn et al. 2008) that were also upregulated in mouse BRAFV600E-PTCs compared to normal thyroid and included the following mRNAs: GNG13, HMGA2, MOSPD3, PCDH1, PDLIM7, PRG4, TGFBR1, TNK2, UBR2 and ZSWIM4.
Statistical analysis
The statistical software GraphPad-Prism (version 8.0; GraphPad Software, Inc.) was used to analyze the data, compute the Pearson correlation coefficients and calculate P-values using unpaired two-tailed Student's t-tests. All data for quantitative RT-PCR, Affy expression array values, immunofluorescence image quantification and Western blots are represented as mean ± s.e.m.
Results
SMAD activation in BRAFV600E-driven PTCs is regulated via MAPK
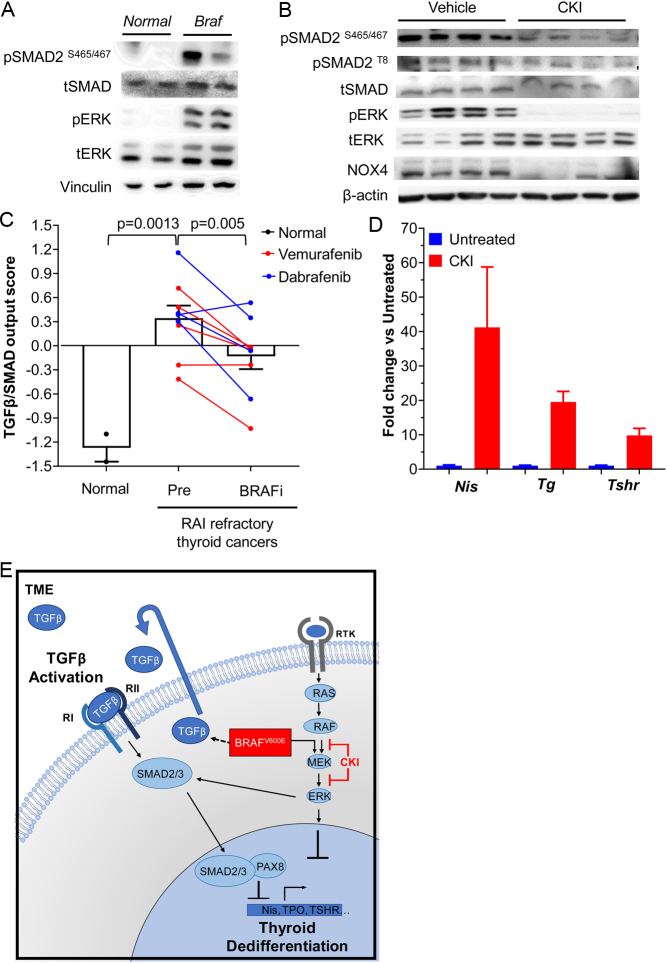
To determine whether the TGFβ/SMAD pathway is activated in BRAFV600E-driven PTC in vivo, we performed pSMAD Western blots in tumor lysates from TPO-Cre/LSL-BrafV600E mice, which develop thyroid cancers with high penetrance by 5 weeks of age (Franco et al. 2011). As shown in Fig. 1A there was a marked increase in pSMAD in BRAFV600E-PTCs compared to normal thyroid. Treatment of mice with 1.5 mg/kg of the MEK inhibitor CH5126766 (CKI), which binds to MEK and places it in an inactive conformation bound to RAF (Ishii et al. 2013), potently blocked MAPK signaling and reduced pSMAD levels as well as expression of the TGFβ downstream target NOX4 (Fig. 1B) (Azouzi et al. 2017). MEK inhibition also reduced phospho-SMAD T8 levels (Fig. 1B), which has been previously shown to be required for optimal TGFβ signaling in mouse thyroid follicular cells (Knauf et al. 2011). In humans, BRAFV600E-driven PTCs had increased SMAD transcriptional output compared to normal thyroid, which was reduced by treatment with RAF inhibitors (Fig. 1C). As previously demonstrated, CKI increased the expression of genes involved in thyroid hormone biosynthesis (Fig. 1D) (Nagarajah et al. 2016). Hence, MAPK and pSMAD signaling are markedly increased in PTC driven by oncogenic BRAF, and the activity of both these pathways is MEK-dependent. Both could converge to impair the thyroid-differentiated function of the cancer cells, but their relative contribution to these effects has not been established (Fig. 1E).
Figure 1.
SMAD pathway activation in BRAFV600E-induced PTCs. (A) Western blot of normal thyroid and tumor lysates from TPO-Cre/LSL-BrafV600E (Braf) mice probed with antibodies to the indicated proteins. (B) Western blot of tumor tissues from Braf mice treated with vehicle or the MEK inhibitor CKI127 (CKI) probed for the indicated targets. Each lane corresponds to an individual mouse PTC (n = 4 per group). (C) TGFβ-SMAD transcriptional output scores of normal thyroid tissues compared to biopsy samples from patients with RAI-refractory thyroid cancers taken prior to and while on treatment with the RAF kinase inhibitors dabrafenib (blue) or vemurafenib (red) for 2 weeks. Each line depicts paired biopsy results from the same lesion prior to and while on drug. (D) Quantitative RT-PCR of thyroid differentiation markers in tumors from panel B. (E) Interactions between the MAPK pathway and TGFβ signaling in thyroid cancer. Oncogenic BRAF induces dedifferentiation in part by ERK-induced silencing or inactivation of lineage transcription factors and by interfering with TSH-induced cAMP signaling (Mitsutake et al. 2005) (not shown). BRAFV600E also increases tumor cell secretion of TGFB1, leading to SMAD impairment of transactivation of thyroid-specific genes by the lineage transcription factor PAX8. Oncogenic BRAF also induces pERK phosphorylation at the T8 residue of SMAD, promoting its additional phosphorylation and activation by the TGFB1 receptor.
Role of TGFβ/SMAD in BRAFV600E-induced thyroid differentiation in vivo
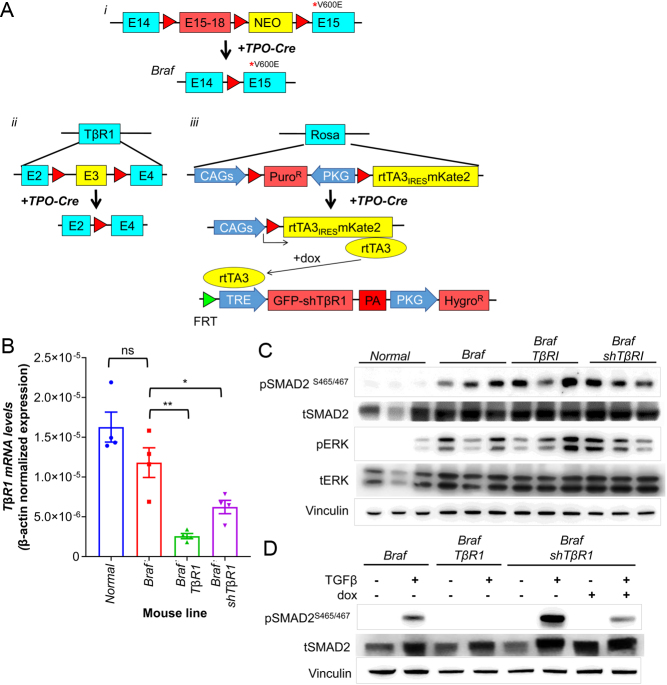
To explore the role of TGFB1 on thyroid-differentiated gene expression in thyroid tumors, we first tested the TGFB1-neutralizing MAB 1D11 and the TGFBR1 (TβR1) kinase inhibitor SD-208 in cell lines derived from mouse BRAFV600E-PTCs, where both effectively blocked the TGFB1-induced SMAD2/3 phosphorylation (Supplementary Fig. 1A). For in vivo experiments, we chose to test the 1D11 antibody, because SD-208 has inhibitory effects on other kinases besides TβR1 (Tandon et al. 2015). Treatment of mice with a 1D11 schedule known to block the TGFB1 activity in vivo (Biswas et al. 2011) did not inhibit pSMAD in mouse BRAFV600E-PTCs (Supplementary Fig. 1B) or enhance the radioiodine uptake (Supplementary Fig. 1C). As this may have been due to the PK of the antibody in the tumor, we generated mouse models of BRAFV600E-PTC with thyroid-specific knockout of the TgfβR1 gene, TPO-Cre/LSL-BrafV600E/TβR1fl/fl (Braf/TβR1) or, to avoid confounding effects of loss of the receptor during development, through post-natal dox-inducible expression of a TβR1 shRNA. We validated a series of TβR1 hairpins in vitro, and selected TβR1 shRNA#6 to generate the mouse model TPO-Cre/LSL-BrafV600E/TetO-GFP_shTβR1/RIK (Braf/shTβR1) (Fig. 2A and Supplementary Fig. 1D, F). The penetrance and histological characteristics of the PTCs in Braf/TβR1 and Braf/shTβR1 was indistinguishable from those of Braf mice (not shown). Despite appropriate knockdown of TβR1 with either model (Fig. 2B), pSMAD levels remained high in thyroid cancer tissues (Fig. 2C), and there was no significant increase in expression of thyroid differentiation markers (Supplementary Fig. 1G). Using cell lines derived from PTCs of Braf/shTβR1 and Braf/TβR1 mice, we found that the loss of TβR1 effectively blocked TGFB1-induced SMAD activation in vitro (Fig. 2D). We concluded that Smad activation in this in vivo context was at least in part TβR1-independent.
Figure 2.
TβR1 knockout in mouse BRAFV600E-PTCs is insufficient to inhibit SMAD activation in vivo. (A) Schematic design of the transgenic lines used in this study and of their recombination when crossed with TPO-Cre mice: (1) Lsl-BrafV600E; (2) TβR1f/f and (3) tetO-shTβR1/RIK. (B) β-actin normalized expression of TβR1 mRNA in thyroid tissues of the indicated mouse lines as determined by quantitative RT-PCR (n = 4/group). *P < 0.05, **P < 0.005, ns = not significant. (C) Western blots for pSMAD and pERK in lysates from normal thyroid or PTCs from Braf, Braf/TβR1 and Braf/shTβR1 mice. (D) Western blots of cell lines derived from PTCs of Braf, Braf/TβR1, and Braf/shTβR1 mice treated with vehicle or 1 ng/mL TGFβ1 for 1 h. Cells treated with dox were incubated for 3 days prior to collection.
Activins induce SMAD in BRAFV600E-induced thyroid cancers
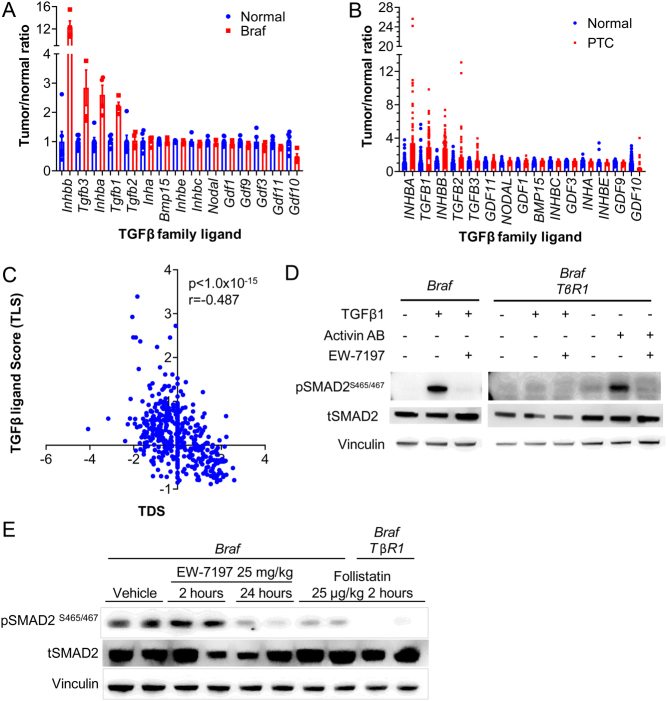
The lack of SMAD inhibition by blocking TGFB1 or TGFBR1 in vivo suggested that other ligands of the TGFβ family might be engaging in this pathway. Activins are disulfide-linked homodimers or heterodimers of primarily two β-subunits, βA and βB, which are transcribed from the Inhba and Inhbb genes, respectively. They activate SMAD2/3 signaling via the activin type 1 receptor ALK4. Expression array data from normal mouse thyroid and BRAFV600E-PTCs showed Inhbb expression to be increased by >15-fold, as compared to a <3-fold increase in Tgfb1 (Fig. 3A). On average human PTCs have a three- to four-fold upregulation of the TGFβ family ligand mRNAs, with INHBA, TGFB1 and INHBB being the most prominent (Fig. 3B and Supplementary Fig. 2A). Analysis of the TCGA dataset of PTC found an inverse correlation between the expression of either INHBA, INHBB, TGFB1, TGFB2 or TGFB3 with the thyroid differentiation score (TDS). The reciprocal relationship with TDS was even stronger when compared with an integrated TGFβ ligand score encompassing expression levels of all five TGFβ family members (Fig. 3C).
Figure 3.
Blocking both ALK4 and ALK5 is required to completely inhibit SMAD activation in BrafV600E-induced PTCs. (A and B) Expression of the Tgfβ family ligands from Affymetrix expression arrays in (A) mouse and (B) human normal thyroids and PTCs. (C) Pearson correlation coefficient between the integrated TGFβ family ligand score (TLS) and TDS using the TCGA PTC dataset (Cancer Genome Atlas Research Network 2014). (D) Western blots of thyroid tumor lysates from Braf mice treated with EW7197 and of Braf/TβR1 mice treated with follistatin.
Treatment of cell lines derived from Braf and Braf/TβR1 mouse PTCs with activin AB markedly induced SMAD phosphorylation. Hence, SMAD activation in BRAFV600E-PTC cells can be induced via TβR1 (Alk5) or Acvr1b (Alk4) receptors, which is blocked by the dual ALK4 and ALK5 inhibitor EW7197 (vactosertib) (Fig. 3D).
To investigate the roles of activin and TGFB1 on SMAD activation in vivo, tumor-bearing Braf and Braf/TβR1 mice were treated with EW7197 or with follistatin, a potent inhibitor of several TGFβ family ligands (primarily activin and to a lesser extent BMPs, myostatin, and possibly TGFB3 – but not TGFB1 or 2) (Nogai et al. 2008, Sepporta et al. 2013). Treatment of Braf mice with EW7197 or follistatin effectively decreased pSMAD in the PTCs. Follistatin had a greater inhibitory effect on SMAD phosphorylation in PTCs from Braf/TβR1 as compared to Braf mice, indicating that both activin and Tgfβ co-regulate this pathway in these tumors (Fig. 3E).
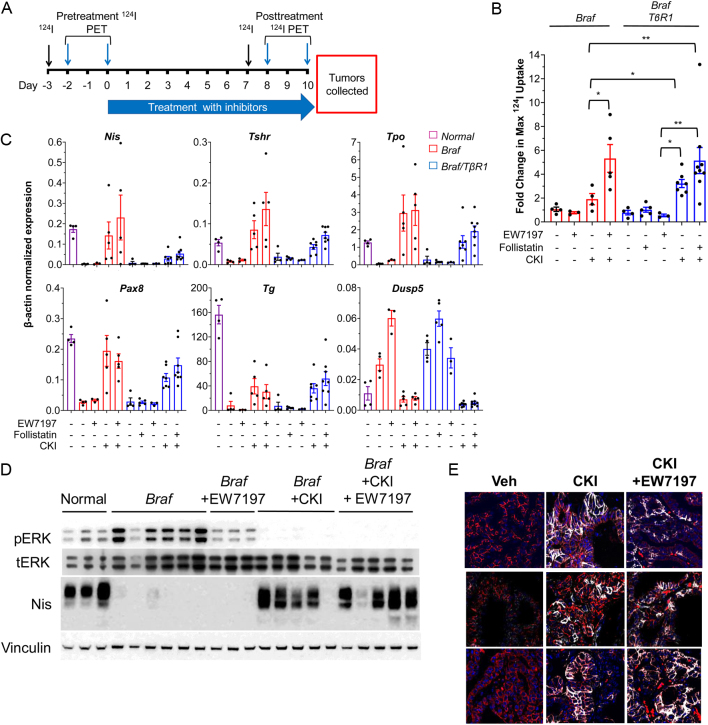
To explore the role of SMAD in BRAFV600E-induced loss of iodide uptake in PTCs, Braf mice were treated with a 10 days course of EW7197 or follistatin. Quantitative 124I-PET scans were performed prior to therapy and at days 7–10 (Schema in Fig. 4A). EW7197 or follistatin monotherapy had no effect on 124I uptake in PTCs of either Braf or Braf/TβR1 mice. Accordingly, these drugs failed to rescue expression of Nis or any other thyroid differentiation marker in PTCs from either mouse model (Fig. 4).
Figure 4.
Co-inhibition of ALK4 and 5 in combination with MAPK blockade increases 124I uptake in Braf mice. (A) Schema for mouse experiments testing effects of CKI and/or EW7197 or follistatin on 124I incorporation into thyroid tumors from the indicated mouse models. (B) Fold-change in maximum 124I uptake in Braf and Braf/TβR1 mice treated with CKI, EW7197 or follistatin or the indicated combinations (*P < 0.05, **P < 0.01). (C and D) Quantitative RT-PCR (C) and Western blots (D) of tumor tissues from B (collected after second 124I PET study). (E) Immunofluorescent staining for NIS and pan-cadherin in tumors from mice treated as indicated in (A).
We next examined whether SMAD inhibition further increased the iodide uptake induced by CKI. 124I PET imaging was performed prior to and while on treatment with CKI alone or in combination with the SMAD pathway inhibitors (Fig. 4A). Treatment with CKI caused a greater increase in 124I uptake in Braf/TβR1 compared to Braf mice (Fig. 4B). EW7197, which inhibits pSMAD profoundly in BRAFV600E-PTCs (Fig. 3C), also augmented the effects of CKI in Braf mice. Accordingly, there was a trend toward higher iodine uptake in Braf/TβR1 mice treated with the combination of follistatin and CKI compared to CKI alone. Taken together, combined blockade of ALK4 and ALK5 by pharmacological targeting alone (EW7197) or in combination with genetic ablation (follistatin + TβR1 knockout) cooperates with MAPK inhibition to augment iodine uptake in Braf-mutant PTC.
Potent inhibition of pSMAD with either EW7197 in Braf mice or follistatin in Braf/TβR1 mice did not increase Nis, Tpo, Tg, or Pax8 mRNA levels, consistent with their lack of effect on iodide uptake (Fig. 4C). By contrast, treatment with CKI markedly increased all four markers. When CKI was combined with EW7197, there was a trend toward higher Nis and Tshr mRNA. CKI restored NIS protein to levels approaching those seen in normal thyroid, but the addition of EW7197 did not result in further augmentation (Fig. 4D and Supplementary Fig. 3A). TGFβ has been shown to block the recruitment of immune cells to the tumor microenvironment (Tauriello et al. 2018). Treatment of Braf mice with EW7197 or CKI + EW7197 did not increase CD45+ cell infiltration (Supplementary Fig. 3B), indicating that decreased tumor cell purity was unlikely to account for the lack of clear increase in thyroid differentiation markers in the tissue extracts. SMAD activation has been shown to reduce plasma membrane localization of NIS via upregulation of NOX4 (Azouzi et al. 2017). We tested this by IHC for NIS in tumors from Braf mice treated with vehicle, CKI or CKI + EW7197. CKI markedly increased membrane-localized NIS compared to vehicle, which was not further augmented with the CKI + EW7197 combination (Fig. 4E and Supplementary Fig. 3C).
Discussion
PTCs driven by BRAFV600E have a greater frequency of nodal recurrences and are usually refractory to RAI treatment (Nikiforova et al. 2003, Elisei et al. 2008, Xing et al. 2013). They are also overrepresented in FDG-PET positive metastatic disease (Ricarte-Filho et al. 2009). The relative resistance to 131I is explained by the disruption in expression of NIS and other thyroid-specific genes that are required for the incorporation of iodine into thyroid cells (1;2). Multiple mechanisms that are not mutually exclusive have been proposed to explain these effects: (1) BRAF activation inhibits PAX8 expression, a paired domain lineage transcription factor that regulates transcription of NIS and other thyroid-specific genes. (2) NKX2-1, a transcription factor essential for thyroid-differentiated gene expression is phosphorylated by ERK, which disrupts its ability to transactivate target genes (Missero et al. 2000). (3) Expression of BRAFV600E increases TGFB1 secretion in rat thyroid PCCL3 cells creating an autocrine loop that activates SMADs (Riesco-Eizaguirre et al. 2009), which in turn bind to PAX8 and impair its transactivation of the Nis gene promoter (Costamagna et al. 2004). This induction of the TGFB1 autocrine loop by BRAFV600E in PCCL3 cells was shown to be MEK-independent (Riesco-Eizaguirre et al. 2009).
The objective of this study was to determine the role of the TGFβ signaling pathway on thyroid differentiation in genetically accurate models of BRAFV600E-induced thyroid cancer in vivo. We found that SMAD phosphorylation was induced in mouse BRAFV600E-PTCs, and that a TGFβ transcriptional output signature was present in advanced RAI-refractory human BRAF-mutant thyroid cancers. We employed pharmacological and genetic approaches to target TGFβ signaling in the mouse models and found that either approach effectively blocked TGFβ signaling in cell lines derived from the murine BRAFV600-PTCs but failed to do so in vivo. We found that other ligands in the TGFβ family were activating SMAD in vivo, which was buttressed by evidence of marked upregulation of Inhbb in BRAFV600E-PTCs. Accordingly, SMAD phosphorylation was inhibited in the PTCs by follistatin, an activin inhibitor. However, follistatin was only able to completely block pSMAD activation in mice with thyrocyte-specific genetic ablation of TβR1, indicating that both activin and TGFβ play a role in SMAD activation, with activin being dominant. We, therefore, chose the ALK4/5 inhibitor EW7197 to explore the role of SMAD activation on iodide uptake in mouse BRAFV600E-driven PTCs, leading to the conclusion that potent inhibition of SMAD alone was insufficient to restore expression of genes involved in thyroid hormone production or iodide uptake.
We previously showed that both BRAFV600E-PTC cells and tumor-associated macrophages contribute TGFB1 to the tumor microenvironment (Knauf et al. 2011). RNAseq of flow-sorted PTC cells from the TPO-Cre/LsL-BrafV600E model used in this paper showed that Tgfb1 and Activin mRNA levels are higher in the BRAFV600E-PTC cells compared to normal thyroid follicular cells (not shown). We do not know which cell types, other than BRAFV600E-PTC cells, contribute to the intratumoral pool of activin.
Potent MAPK pathway blockade promotes redifferentiation and increased iodide uptake in BRAFV600E-driven thyroid cancer (Nagarajah et al. 2016). Here, we show that this is associated with a decrease in SMAD activation, which is likely due to a reduction in ERK-dependent phosphorylation of the threonine 8 residue of SMAD. Phosphorylation at this site by MAPK amplifies the activation of SMAD by ligand-bound TGFβ family receptors (Wrighton et al. 2009, Knauf et al. 2011). These convergent effects on SMAD may explain the increase in iodide uptake in Braf mice treated with the combination of EW7197 and CKI compared to CKI alone. However, MAPK activation suppresses the differentiation program largely through SMAD-independent mechanisms, since potent inhibition of SMAD failed to restore thyroid-differentiated gene expression in the setting of persistent MAPK pathway activation. This likely explains why the treatment combination only resulted in a trend toward increased Nis and Tshr mRNA compared to MEK inhibition alone.
In summary, activation of the MAPK pathway by BRAFV600E promotes SMAD activation in vivo through TGFβ and activin-induced pathways. Increased expression of TGFβ and activin ligands is common in PTCs and this is inversely correlated with an integrated transcriptional output score associated with thyroid differentiation. TGFβ-induced signaling through SMAD has been shown to suppress the expression of NIS and other thyroid differentiation markers in BRAF-WT thyroid cells (Colletta et al. 1989, Nicolussi et al. 2003, Costamagna et al. 2004). However, signaling via SMAD is not the primary mechanism for suppression of thyroid-specific gene expression or loss of iodide uptake in the context of MAPK activation in vivo, as inhibition of SMAD alone did not improve any of these biomarkers. The precise mechanism underpinning the additive effect of SMAD and MEK inhibition on iodide uptake is unclear. It was not associated with clear differences in the expression of NIS. NIS abundance and membrane localization are only some of the variables that impact iodide incorporation (others include iodide organification and iodide efflux, which we did not assess).
The enhancement of iodine uptake may reflect the close interplay between these two pathways, which mutually reinforce overlapping effects on the thyroid-differentiated state during dynamic changes in their respective signaling outputs. The translational implication of these findings is intriguing and has not been explored so far. Any clinical trials looking into synergistic effects of blocking SMAD and MAPK pathways should select agents that target both ALK4 and ALK5 and consider using them for short intervals because of the potential tumor-promoting effects of SMAD inhibition in cancer.
Supplementary Material
Declaration of interest
JAF is a consultant for Loxo Oncology, received grant support from Eisai and is a co-inventor of intellectual property focused on HRAS as a biomarker for treating cancer using tipifarnib which has been licensed by MSK to Kura.
Funding
This work was supported by grants from the US National Institutes of Health RO1-CA50706-23 (JAF), R01-CA 249663-01A1 (JAF, ALH), RO1-CA184724-01A1 (ALH, JAF), P50-CA 172012-01 (JAF) and P30-CA008748/CA/NCI (Craig Thompson PI), Cycle for Survival (ALH), the Jayme and Peter Flowers fund, the Frank D Cohen fund, Cycle for Survival and the Translational Research Oncology Training program 5T32CA160001 (MS).
Acknowlegdements
The authors thank Dr Yosuke Mukoyama (National Heart, Lung and Blood institute, MD, USA) for the Tgfbr1-flox mice, Dr Shioko Kimura (National Cancer Institute, MD, USA) for the Tpo-Cre mice, and Dr Catrin Pritchard (University of Leicester, UK) for the LSL-BrafV600E/+ mice. Also, thank the Research Animal Resource Center and the Molecular Cytology and the Small Animal Imaging Cores of the Sloan Kettering Institute for their support.
References
- Azouzi N, Cailloux J, Cazarin JM, Knauf JA, Cracchiolo J, Al GA, Hartl D, Polak M, Carre A, El MMet al. 2017. NADPH oxidase NOX4 is a critical mediator of BRAF(V600E)-induced downregulation of the sodium/iodide symporter in papillary thyroid carcinomas. Antioxidants and Redox Signaling 26 864–877.( 10.1089/ars.2015.6616) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas S, Nyman JS, Alvarez J, Chakrabarti A, Ayres A, Sterling J, Edwards J, Rana T, Johnson R, Perrien DSet al. 2011. Anti-transforming growth factor ss antibody treatment rescues bone loss and prevents breast cancer metastasis to bone. PLoS ONE 6 e27090. ( 10.1371/journal.pone.0027090) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network 2014. Integrated genomic characterization of papillary thyroid carcinoma. Cell 159 676–690. ( 10.1016/j.cell.2014.09.050) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarty D, Santos E, Ryder M, Knauf JA, Liao XH, West BL, Bollag G, Kolesnick R, Thin TH, Rosen Net al. 2011. Small-molecule MAPK inhibitors restore radioiodine incorporation in mouse thyroid cancers with conditional BRAF activation. Journal of Clinical Investigation 121 4700–4711. ( 10.1172/JCI46382) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colletta G, Cirafici AM, Di Carlo A.1989. Dual effect of transforming growth factor beta on rat thyroid cells: inhibition of thyrotropin-induced proliferation and reduction of thyroid-specific differentiation markers. Cancer Research 49 3457–3462. [PubMed] [Google Scholar]
- Costamagna E, Garcia B, Santisteban P.2004. The functional interaction between the paired domain transcription factor Pax8 and Smad3 is involved in transforming growth factor-beta repression of the sodium/iodide symporter gene. Journal of Biological Chemistry 279 3439–3446. ( 10.1074/jbc.M307138200) [DOI] [PubMed] [Google Scholar]
- Coulouarn C, Factor VM, Thorgeirsson SS.2008. Transforming growth factor-beta gene expression signature in mouse hepatocytes predicts clinical outcome in human cancer. Hepatology 47 2059–2067. ( 10.1002/hep.22283) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dom G, Tarabichi M, Unger K, Thomas G, Oczko-Wojciechowska M, Bogdanova T, Jarzab B, Dumont JE, Detours V, Maenhaut C.2012. A gene expression signature distinguishes normal tissues of sporadic and radiation-induced papillary thyroid carcinomas. British Journal of Cancer 107 994–1000. ( 10.1038/bjc.2012.302) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dow LE, Premsrirut PK, Zuber J, Fellmann C, McJunkin K, Miething C, Park Y, Dickins RA, Hannon GJ, Lowe SW.2012. A pipeline for the generation of shRNA transgenic mice. Nature Protocols 7 374–393. ( 10.1038/nprot.2011.446) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn LA, Sherman EJ, Baxi SS, Tchekmedyian V, Grewal RK, Larson SM, Pentlow KS, Haque S, Tuttle RM, Sabra MMet al. 2019. Vemurafenib redifferentiation of BRAF mutant, RAI-refractory thyroid cancers. Journal of Clinical Endocrinology and Metabolism 104 1417–1428. ( 10.1210/jc.2018-01478) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elisei R, Ugolini C, Viola D, Lupi C, Biagini A, Giannini R, Romei C, Miccoli P, Pinchera A, Basolo F.2008. BRAF(V600E) mutation and outcome of patients with papillary thyroid carcinoma: a 15-year median follow-up study. Journal of Clinical Endocrinology and Metabolism 93 3943–3949. ( 10.1210/jc.2008-0607) [DOI] [PubMed] [Google Scholar]
- Fagin JA, Wells Jr SA.2016. Biologic and clinical perspectives on thyroid cancer. New England Journal of Medicine 375 2307. ( 10.1056/NEJMc1613118) [DOI] [PubMed] [Google Scholar]
- Franco AT, Malaguarnera R, Refetoff S, Liao XH, Lundsmith E, Kimura S, Pritchard C, Marais R, Davies TF, Weinstein LSet al. 2011. Thyrotrophin receptor signaling dependence of Braf-induced thyroid tumor initiation in mice. PNAS 108 1615–1620. ( 10.1073/pnas.1015557108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho AL, Grewal RK, Leboeuf R, Sherman EJ, Pfister DG, Deandreis D, Pentlow KS, Zanzonico PB, Haque S, Gavane Set al. 2013. Selumetinib-enhanced radioiodine uptake in advanced thyroid cancer. New England Journal of Medicine 368 623–632. ( 10.1056/NEJMoa1209288) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii N, Harada N, Joseph EW, Ohara K, Miura T, Sakamoto H, Matsuda Y, Tomii Y, Tachibana-Kondo Y, Iikura Het al. 2013. Enhanced inhibition of ERK signaling by a novel allosteric MEK inhibitor, CH5126766, that suppresses feedback reactivation of RAF activity. Cancer Research 73 4050–4060. ( 10.1158/0008-5472.CAN-12-3937) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jentzen W, Freudenberg L, Eising EG, Sonnenschein W, Knust J, Bockisch A.2008. Optimized 124I PET dosimetry protocol for radioiodine therapy of differentiated thyroid cancer. Journal of Nuclear Medicine 49 1017–1023. ( 10.2967/jnumed.107.047159) [DOI] [PubMed] [Google Scholar]
- Kavsak P, Rasmussen RK, Causing CG, Bonni S, Zhu H, Thomsen GH, Wrana JL.2000. Smad7 binds to Smurf2 to form an E3 ubiquitin ligase that targets the TGF beta receptor for degradation. Molecular Cell 6 1365–1375. ( 10.1016/s1097-2765(0000134-9) [DOI] [PubMed] [Google Scholar]
- Knauf JA, Kuroda H, Basu S, Fagin JA.2003. RET/PTC-induced dedifferentiation of thyroid cells is mediated through Y1062 signaling through SHC-RAS-MAP kinase. Oncogene 22 4406–4412. ( 10.1038/sj.onc.1206602) [DOI] [PubMed] [Google Scholar]
- Knauf JA, Sartor MA, Medvedovic M, Lundsmith E, Ryder M, Salzano M, Nikiforov YE, Giordano TJ, Ghossein RA, Fagin JA.2011. Progression of BRAF-induced thyroid cancer is associated with epithelial-mesenchymal transition requiring concomitant MAP kinase and TGFbeta signaling. Oncogene 30 3153–3162. ( 10.1038/onc.2011.44) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusakabe T, Kawaguchi A, Kawaguchi R, Feigenbaum L, Kimura S.2004. Thyrocyte-specific expression of Cre recombinase in transgenic mice. Genesis 39 212–216. ( 10.1002/gene.20043) [DOI] [PubMed] [Google Scholar]
- Larsson J, Blank U, Helgadottir H, Bjornsson JM, Ehinger M, Goumans MJ, Fan X, Leveen P, Karlsson S.2003. TGF-beta signaling-deficient hematopoietic stem cells have normal self-renewal and regenerative ability in vivo despite increased proliferative capacity in vitro. Blood 102 3129–3135. ( 10.1182/blood-2003-04-1300) [DOI] [PubMed] [Google Scholar]
- Liu D, Hu S, Hou P, Jiang D, Condouris S, Xing M.2007. Suppression of BRAF/MEK/MAP kinase pathway restores expression of iodide-metabolizing genes in thyroid cells expressing the V600E BRAF mutant. Clinical Cancer Research 13 1341–1349. ( 10.1158/1078-0432.CCR-06-1753) [DOI] [PubMed] [Google Scholar]
- Mercer K, Giblett S, Green S, Lloyd D, DaRocha Dias S, Plumb M, Marais R, Pritchard C.2005. Expression of endogenous oncogenic V600EB-raf induces proliferation and developmental defects in mice and transformation of primary fibroblasts. Cancer Research 65 11493–11500. ( 10.1158/0008-5472.CAN-05-2211) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Missero C, Pirro MT, Di Lauro R.2000. Multiple ras downstream pathways mediate functional repression of the homeobox gene product TTF-1. Molecular and Cellular Biology 20 2783–2793. ( 10.1128/mcb.20.8.2783-2793.2000) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsutake N, Knauf JA, Mitsutake S, Mesa Jr C, Zhang L, Fagin JA.2005. Conditional BRAFV600E expression induces DNA synthesis, apoptosis, dedifferentiation, and chromosomal instability in thyroid PCCL3 cells. Cancer Research 65 2465–2473. ( 10.1158/0008-5472.CAN-04-3314) [DOI] [PubMed] [Google Scholar]
- Nagarajah J, Le M, Knauf JA, Ferrandino G, Montero-Conde C, Pillarsetty N, Bolaender A, Irwin C, Krishnamoorthy GP, Saqcena Met al. 2016. Sustained ERK inhibition maximizes responses of BrafV600E thyroid cancers to radioiodine. Journal of Clinical Investigation 126 4119–4124. ( 10.1172/JCI89067) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolussi A, D'Inzeo S, Santulli M, Colletta G, Coppa A.2003. TGF-beta control of rat thyroid follicular cells differentiation. Molecular and Cellular Endocrinology 207 1–11. ( 10.1016/s0303-7207(0300238-7) [DOI] [PubMed] [Google Scholar]
- Nikiforova MN, Kimura ET, Gandhi M, Biddinger PW, Knauf JA, Basolo F, Zhu Z, Giannini R, Salvatore G, Fusco Aet al. 2003. BRAF mutations in thyroid tumors are restricted to papillary carcinomas and anaplastic or poorly differentiated carcinomas arising from papillary carcinomas. Journal of Clinical Endocrinology and Metabolism 88 5399–5404. ( 10.1210/jc.2003-030838) [DOI] [PubMed] [Google Scholar]
- Nogai H, Rosowski M, Grun J, Rietz A, Debus N, Schmidt G, Lauster C, Janitz M, Vortkamp A, Lauster R.2008. Follistatin antagonizes transforming growth factor-beta3-induced epithelial-mesenchymal transition in vitro: implications for murine palatal development supported by microarray analysis. Differentiation: Research in Biological Diversity 76 404–416. ( 10.1111/j.1432-0436.2007.00223.x) [DOI] [PubMed] [Google Scholar]
- Pelossof R, Fairchild L, Huang CH, Widmer C, Sreedharan VT, Sinha N, Lai DY, Guan Y, Premsrirut PK, Tschaharganeh DFet al. 2017. Prediction of potent shRNAs with a sequential classification algorithm. Nature Biotechnology 35 350–353. ( 10.1038/nbt.3807) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Premsrirut PK, Dow LE, Kim SY, Camiolo M, Malone CD, Miething C, Scuoppo C, Zuber J, Dickins RA, Kogan SCet al. 2011. A rapid and scalable system for studying gene function in mice using conditional RNA interference. Cell 145 145–158. ( 10.1016/j.cell.2011.03.012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricarte-Filho JC, Ryder M, Chitale DA, Rivera M, Heguy A, Ladanyi M, Janakiraman M, Solit D, Knauf JA, Tuttle RMet al. 2009. Mutational profile of advanced primary and metastatic radioactive iodine-refractory thyroid cancers reveals distinct pathogenetic roles for BRAF, PIK3CA, and AKT1. Cancer Research 69 4885–4893. ( 10.1158/0008-5472.CAN-09-0727) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riesco-Eizaguirre G, Rodríguez I, De la Vieja A, Costamagna E, Carrasco N, Nistal M, Santisteban P.2009. The BRAFV600E oncogene induces transforming growth factor beta secretion leading to sodium iodide symporter repression and increased malignancy in thyroid cancer. Cancer Research 69 8317–8325. ( 10.1158/0008-5472.CAN-09-1248) [DOI] [PubMed] [Google Scholar]
- Rothenberg SM, Daniels GH, Wirth LJ.2015. Redifferentiation of iodine-refractory BRAF V600E-mutant metastatic papillary thyroid cancer with Dabrafenib-response. Clinical Cancer Research 21 5640–5641. ( 10.1158/1078-0432.CCR-15-2298) [DOI] [PubMed] [Google Scholar]
- Sepporta MV, Tumminello FM, Flandina C, Crescimanno M, Giammanco M, La Guardia M, di Majo D, Leto G.2013. Follistatin as potential therapeutic target in prostate cancer. Targeted Oncology 8 215–223. ( 10.1007/s11523-013-0268-7) [DOI] [PubMed] [Google Scholar]
- Tandon M, Salamoun JM, Carder EJ, Farber E, Xu S, Deng F, Tang H, Wipf P, Wang QJ.2015. SD-208, a novel protein kinase D inhibitor, blocks prostate cancer cell proliferation and tumor growth in vivo by inducing G2/M cell cycle arrest. PLoS ONE 10 e0119346. ( 10.1371/journal.pone.0119346) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tauriello DVF, Palomo-Ponce S, Stork D, Berenguer-Llergo A, Badia-Ramentol J, Iglesias M, Sevillano M, Ibiza S, Canellas A, Hernando-Momblona Xet al. 2018. TGFbeta drives immune evasion in genetically reconstituted colon cancer metastasis. Nature 554 538–543. ( 10.1038/nature25492) [DOI] [PubMed] [Google Scholar]
- Tomas G, Tarabichi M, Gacquer D, Hebrant A, Dom G, Dumont JE, Keutgen X, Fahey TJ, Maenhaut C, Detours V.2012. A general method to derive robust organ-specific gene expression-based differentiation indices: application to thyroid cancer diagnostic. Oncogene 31 4490–4498. ( 10.1038/onc.2011.626) [DOI] [PubMed] [Google Scholar]
- von Roemeling CA, Marlow LA, Pinkerton AB, Crist A, Miller J, Tun HW, Smallridge RC, Copland JA.2015. Aberrant lipid metabolism in anaplastic thyroid carcinoma reveals stearoyl CoA desaturase 1 as a novel therapeutic target. Journal of Clinical Endocrinology and Metabolism 100 E697–E709. ( 10.1210/jc.2014-2764) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrighton KH, Lin X, Feng XH.2009. Phospho-control of TGF-beta superfamily signaling. Cell Research 19 8–20. ( 10.1038/cr.2008.327) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing M, Westra WH, Tufano RP, Cohen Y, Rosenbaum E, Rhoden KJ, Carson KA, Vasko V, Larin A, Tallini Get al. 2005. BRAF mutation predicts a poorer clinical prognosis for papillary thyroid cancer. Journal of Clinical Endocrinology and Metabolism 90 6373–6379. ( 10.1210/jc.2005-0987) [DOI] [PubMed] [Google Scholar]
- Xing M, Alzahrani AS, Carson KA, Viola D, Elisei R, Bendlova B, Yip L, Mian C, Vianello F, Tuttle RMet al. 2013. Association between BRAF V600E mutation and mortality in patients with papillary thyroid cancer. JAMA 309 1493–1501. ( 10.1001/jama.2013.3190) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Z, Torres NM, Tao A, Gao Y, Luo L, Li Q, de Stanchina E, Abdel-Wahab O, Solit DB, Poulikakos PIet al. 2015. BRAF mutants evade ERK-dependent feedback by different mechanisms that determine their sensitivity to pharmacologic inhibition. Cancer Cell 28 370–383. ( 10.1016/j.ccell.2015.08.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



 This work is licensed under a
This work is licensed under a