Summary
The makeup of the tumor immune microenvironment may be associated with tumor somatic genomic alterations and plays a key role in tumor progression and response to immunotherapy. We examined the association of tumor-infiltrating T-cell density with TP53 status in surgically treated primary prostate cancer using 3 independent tissue microarray sets, including one set of tumors from grade-matched patients of European American or African American ancestry (n = 391), a retrospective case-cohort of intermediate- and high-risk patients enriched for adverse outcomes (n = 267), and a set of tumors with primary Gleason pattern 5 (n = 77). The presence of TP53 missense mutation, indicated by p53 nuclear accumulation using a genetically validated assay, was significantly associated with increased CD3+ T-cell density (median, 341 versus 231 CD3+ T cells/mm2; P = .004) in the matched European American and African American ancestry patient sets. The same association was present in patients of both ancestries when analyzed separately, despite the fact that p53 nuclear accumulation was less frequent among African American compared with European American tumors (7% versus 3%, P = .2). The validation cohorts of intermediate/high-risk and primary Gleason pattern 5 patients corroborated the association of increased CD3+ T-cell density with presence of p53 nuclear accumulation. In a pooled analysis of all sets, adjusting for clinicopathological variables, CD3+ and CD8+, but not FOXP3+, T-cell densities remained significantly higher in tumors with p53 nuclear accumulation compared with those without. TP53 mutation is associated with higher tumor-infiltrating T-cell density, which may be relevant in future clinical trials of immunotherapy in prostate cancer.
Keywords: Prostatic adenocarcinoma, Tumor-infiltrating lymphocytes, T cells, CD3, CD8, FOXP3, TP53
1. Introduction
The tumor microenvironment plays an important role in tumor progression and response to therapy [1,2]. Until recently, the research focus has been on the cancer cells, with less emphasis on the host’s immune system, which interacts with the cancer cells in a continuous, dynamic, and evolving way. However, the recent success with immunotherapy in multiple tumor types, particularly those with a high burden of tumor-specific neoantigens, has shifted the focus of treatment from the tumor itself to the immune system [3]. These early successes highlight the fact that a deeper understanding of the cross talk between tumor biology and the immune microenvironment is necessary if we wish to predict which patients will receive maximal benefit from these promising but often toxic therapeutic agents.
Compared with other solid tumor types, prostate cancer is generally considered an immunologically “cold” tumor, aside from rare cases with mismatch DNA repair defects, which have a higher tumor-infiltrating lymphocyte density [4,5]. However, some molecular subtypes of mismatch repair proficient prostate cancer may have more lymphocytic infiltration than others, and may thus be potentially better poised for response to immune checkpoint inhibition. We previously reported that ERG gene rearrangements and PTEN deletions are associated with higher densities of T cells within the primary tumor [6]. These findings in human tumors, along with complementary work in preclinical mouse models [7], support the hypothesis that the molecular profile of the tumor is associated with the makeup of the tumor immune microenvironment. A more refined understanding of this interaction between tumor genomics and immune cell infiltrate may help us to better risk stratify and treat prostate cancer patients.
The tumor suppressor TP53 is the most frequently mutated gene in human tumors [8]. In prostate cancer, TP53 mutation is a relatively early, though infrequent event during tumorigenesis. We and others have shown that primary tumors with TP53 mutations have an exceptionally poor prognosis independent of tumor grade and stage and have a shorter time to the development of hormonal resistance [9,10]. Consistent with this, metastatic or castration resistant prostate cancer shows significant enrichment for TP53 alterations compared with primary prostate cancer [11,12]. Studies in other systems have shown that p53 inactivation skews the immune landscape of the tumor microenvironment toward tumor-promoting inflammation [13-16], whereas p53 reactivation may promote antitumor immunity [17-19]. However, in prostate cancer, our understanding of the cellular and molecular processes that link p53 activity to host immune regulation is still incomplete. To address these knowledge gaps, we studied T-lymphocyte infiltration in 3 independent well-defined clinical cohorts with diverse racial ancestry. Having previously validated our method for digital quantification of T-cell density across tissue microarray (TMA) cores [6], we assessed the association of the density of different subsets of T cells (CD3+, CD8+, and FOXP3+) with TP53 missense mutation status in surgically treated prostate cancer patients. We found that TP53 missense mutation is uniformly associated with higher T-cell density within the primary tumor, an observation that has potential implications for future trials of immunotherapy in prostate cancer.
2. Materials and methods
2.1. Patients and tissue samples
With Johns Hopkins institutional review board approval (IRB00089322 and IRB00135395, in accordance with the Declaration of Helsinki), 3 patient sets were included in this study. (1) The first set included a previously described group of 391 patients who underwent radical prostatectomy at Johns Hopkins Hospital, designed to evaluate the association of self-identified European American or African American racial ancestry with patient outcomes [6,20]. TMAs were constructed from matched prostate tumors collected from 100 self-identified African American and 127 self-identified European American patients who underwent radical prostatectomy from 1995 to 2005 (hereafter referred to as matched-race TMA set). An additional TMA set of 85 African American patients with high grade (Gleason 4 + 3 = 7 and higher) disease who underwent radical prostatectomy from 2005 to 2010 (hereafter referred to as high-grade African American set), grade matched with 79 European American patients from this period, were also studied to enrich for adverse oncologic outcomes. (2) The second TMA set included a retrospective case-cohort for metastatic progression, including 267 intermediate- or high-risk patients (predominantly European American) who underwent RP between 1992 and 2010 and received no additional treatment until the time of metastasis [9,21]. (3) The third TMA set included a previously described cohort of radical prostatectomies from 2004 to 2014 with primary Gleason pattern 5 (n = 77) [4]. All TMAs described above included 3 to 4 individual 0.6 mm punches of the dominant tumor nodule from each case (slightly >1 mm2 of tissue for analysis).
2.2. Immunohistochemistry
Immunohistochemistry (IHC) for CD3 and CD8 was performed in a Clinical Laboratory Improvement Amendments–accredited laboratory using a polyclonal rabbit antibody for CD3 (A0452; Dako/Agilent, Santa Clara, CA) and a mouse monoclonal for CD8 (clone C8/C8144B, 760-4250; Cell Marque, Rocklin, CA) on the Ventana Benchmark immunostaining system (Ventana/Roche, Tucson, AZ). IHC for FOXP3 used a rat monoclonal antibody (FJK-16s; Invitrogen/Thermo Fisher, Carlsbad, CA) on the Ventana Discovery Ultra (Ventana/Roche). FOXP3 IHC was unevaluable in a subset of the European American tumors from the matched-race TMA set because of a faulty batch of charged slides, which were incompatible with the Ventana immunostainer. Thus, these data are not included.
p53 IHC was performed on the Ventana Benchmark autostaining system using a mouse monoclonal antibody (BP53-11) after antigen retrieval in CC1 buffer followed by detection with the iView HRP system (Roche/Ventana Medical Systems, Oro Valley, AZ). This protocol was previously validated for detection of TP53 mutations in prostate cancer [9].
2.3. p53 scoring
Each TMA spot containing tumor cells was visually dichotomously scored for presence or absence of nuclear p53 signal by a urologic pathologist blinded to the gene expression data (TLL). As previously genetically validated, a spot was considered to show p53 nuclear accumulation if >10% of tumor nuclei showed p53 positivity [9]. This cutoff value was chosen because it was the most correlated with the presence of wild-type TP53 in a prior genomic validation of this staining protocol in ovarian carcinoma [22] and the most commonly chosen cutoff in a meta-analysis of p53 staining in other tumor types [23]. A tumor was considered to show p53 nuclear accumulation if any sampled spot was scored as p53 positive, and as p53 negative if all sampled spots were scored as p53 negative.
2.4. Image analyses
Automated image analysis for number of cells per millimeter squared was performed for CD3, CD8, and FOXP3 IHC. Stained TMA slides were scanned at ×20 magnification on Nano Zoomer Digital Pathology scanner (Hamamatsu, Shizuoka, Japan). The positively stained cells per millimeter squared tissue were quantitatively scored with the Aperio Digital Pathology software (Leica, Wetzlar, Germany) for each cancer-containing TMA core using a previously validated method [4,6]. In brief, the number of positive cells in each tumor core was calculated by designing a detection mask for brown immunostaining using a modified version of the nuclear analysis parameters in Aperio Image Analysis software. The software-identified count was visually checked by a pathologist in at least 50% of TMA spots selected randomly for evaluation. For the TMA sections, all 4 cores that contained tumor from the dominant tumor nodule were circled and selected for analysis in their entirety; cores were excluded if they did not contain tumor glands, and areas of artifactual staining were manually excluded as well. Parameters were set separately for each stain, and the parameters for each stain were uniformly applied across each TMA set. The cell count in each manually circled area was returned by the Aperio software. The total tumor area analyzed was calculated manually for the TMA spots, assuming that the diameter of each spot was 0.6 mm and multiplying by the total number of spots analyzed for each case. The ratio of positive cells to the total tumor area analyzed was calculated for each case.
2.5. Statistical analysis
For each TMA set, we calculated the median T-cell density stratified by p53 status (in the overall set and within race/ethnicity category), race/ethnicity, stage, and Gleason score. Given nonnormal distributions, median densities were compared by race, stage, Gleason score, and p53 status using the Kruskal-Wallis test. We then assessed whether T-cell density was associated with p53 status independent of clinicopathological factors. We pooled T-cell counts across all sets and modeled the association between T-cell density and p53 status using negative-binomial regression with a natural logarithmic offset term for tissue area at risk, and adjusting for age, race, stage, grade, Prostate Specific Antigen, and TMA set. All statistical analyses were performed in SAS V9.4 (Cary, NC), and tests were 2-sided with a P value of .05 deemed statistically significant.
3. Results
3.1. Baseline clinicopathological characteristics according to p53 status
The presence of TP53 missense mutation was determined by a genetically validated IHC assay for p53 nuclear accumulation in each of 3 previously described TMA sets, one of which (intermediate/high-risk set) had previously published p53 IHC results [9]. The frequency of p53 nuclear accumulation was 5.1% (20/391) in the matched-race TMA set. When analyzed separately according to patient race, the frequency of p53 nuclear accumulation was higher in European American (14/206; 6.8%) compared with African American patients (6/185; 3.3%), although the difference was not statistically significant (P = .2). The frequency of p53 nuclear accumulation was 6.4% (17/267) in the intermediate/high-risk TMA set, comprising mainly European American patients (241/267). The frequency of p53 nuclear accumulation was higher in the third set of primary Gleason pattern 5 patients compared with the other 2 cohorts, at 17.1% (12/70).
Clinicopathological characteristics of the cases by p53 status are shown in Supplementary Tables S1 to S3. Consistent with the much higher prevalence of p53 nuclear accumulation in the in the primary Gleason pattern 5 TMA set compared with the other 2 TMA sets, p53 nuclear accumulation was significantly more common with increasing Gleason score within each set (P = .005 in matched-race set, P = .001 in intermediate/high-risk set). Even within the primary Gleason pattern 5 TMA set (where all tumors were Gleason score 5 + 4 = 9 or 5 + 5 = 10), p53 nuclear accumulation was significantly more common among 5 + 5 = 10 compared with 5 + 4 = 9 tumors (P = .03). P53 nuclear accumulation was only associated with tumor stage within the Gleason pattern 5 TMA set (P = .01), but not within any of the other sets. The prevalence of p53 nuclear accumulation was not significantly associated with patient age or PSA in any of the 3 TMA sets (Supplementary Figure).
3.2. Association of T-cell density with p53 nuclear accumulation
Next, we tested the association of p53 status with tumor-infiltrating T-cell density in each of the 3 TMA sets. The median CD3+ T-cell density was significantly higher among men with p53 nuclear accumulation as compared with the men without p53 nuclear accumulation in all the 3 sets (median, 341 versus 231 CD3+T cells/mm2 [P = .004] in the combined matched-race TMA set;, 416 versus 208 CD3+ T cells/mm2 [P = .006] in the intermediate- and high-risk set;, and 662 versus 344 CD3+ T cells/mm2 [P = .01] in the primary Gleason pattern 5 set; Tables 1-3). Within the matched-race TMA set, the same pattern was seen when each race was analyzed separately (301.5 versus 220.6 CD3+ T cells/mm2 [P = .03] in the European American subset and 486.3 versus 241.5 CD3+ T cells/mm2 [P = .02] in the African American subset). Similar patterns were observed for median CD8+ and FOXP3+ T-cell densities, although not all comparisons remained statistically significant. Although CD8+ T-cell density seemed higher for tumors with p53 nuclear accumulation in the African American subset of the matched-race TMA set (105 versus 57 CD8+ T cells/mm2, P = .05), the difference was not statistically significant in the other sets. The FOXP3+ T-cell density was evaluable in the intermediate- and high-risk set, the primary Gleason pattern 5 set, and the African American subset of the matched-race TMA set. The median FOXP3+ T-cell density was significantly higher among men with p53 nuclear accumulation in the intermediate- and high-risk patients set (51 versus 29 FOXP3+ T cells/mm2, P = .04) and the African American subset of the matched race TMA set (34 versus 16 FOXP3+ T cells/mm2, P = .04), although it did not reach statistical significance in the primary Gleason pattern 5 set (7 versus 4 FOXP3+ T cells/mm2, P = .3).
Table 1.
T-cell densities (cells/mm2) by p53 status in combined matched-race TMA sets
| CD3+ density |
CD8+ density |
FOXP3+ density |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| n (387) | Median | P a | n (388) | Median | P a | n (206) | Median | P a | |
| Overall | |||||||||
| p53 negative b | 367 | 230.8 | .004 * | 368 | 66.3 | .1 | |||
| p53 positive c | 20 | 341.3 | 20 | 90.7 | |||||
| European American | |||||||||
| p53 negative b | 192 | 220.6 | .03 * | 192 | 76.9 | .4 | |||
| p53 positive c | 14 | 301.5 | 14 | 88.4 | |||||
| African American | |||||||||
| p53 negative b | 175 | 241.5 | .02 * | 176 | 57.0 | .05 * | 179 | 15.9 | .04 * |
| p53 positive c | 6 | 486.3 | 6 | 104.8 | 6 | 34.0 | |||
NOTE. FOXP3 immunostaining was unevaluable on a subset of the European American tumors from the matched-race TMA set because of a faulty batch of slides, which were incompatible with Ventana immunostainer. Thus, these data are not included. CD3 and CD8 immunostaining was unevaluable in 4 and 3 African American cases from the race TMA set because of poor staining, respectively.
Kruskal-Wallis test.
Absence of p53 nuclear accumulation.
Presence of p53 nuclear accumulation.
P < .05.
Table 3.
T-cell densities (cells/mm2) by clinicopathological parameters and p53 status in primary Gleason pattern 5 TMA sets
| CD3+ density |
CD8+ density |
FOXP3+ density |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| n | Median | P a | n | Median | P a | n | Median | P a | |
| Overall | 77 | 379.4 | 77 | 103.5 | 77 | 5.3 | |||
| Race | |||||||||
| White | 64 | 369.7 | .9 | 64 | 107.9 | .9 | 64 | 5.3 | .9 |
| Nonwhite b | 13 | 382.1 | 13 | 96.4 | 13 | 6.2 | |||
| Stage | |||||||||
| T2 N0 | 10 | 413.1 | .04 * | 10 | 100.0 | .02 * | 10 | 6.6 | .6 |
| T3aN0 | 23 | 267.7 | 23 | 74.3 | 23 | 4.4 | |||
| T3bN0 | 27 | 423.7 | 27 | 102.6 | 27 | 4.4 | |||
| T4 N0 | 1 | 270.4 | 1 | 112.3 | 1 | 1.8 | |||
| N0 | 61 | 292.6 | .01 * | 61 | 92.9 | .001 * | 61 | 4.4 | .2 |
| N1 | 16 | 773.5 | 16 | 256.1 | 16 | 6.2 | |||
| Secondary Gleason pattern | |||||||||
| 4 | 72 | 380.8 | .9 | 72 | 107.9 | .9 | 72 | 5.8 | .1 |
| 5 | 5 | 359.9 | 5 | 92.9 | 5 | 3.5 | |||
| p53 negative c | 58 | 344.0 | .01 * | 58 | 107.5 | .1 | 58 | 4.4 | .3 |
| p53 positive d | 12 | 661.8 | 12 | 203.9 | 12 | 7.1 | |||
NOTE. p53 immunostaining was unevaluable in 7 cases from the Gleason pattern 5 TMA set.
From Kruskal-Wallis test.
Including African American, Hispanic, other, and unknown.
Absence of p53 nuclear accumulation.
Presence of p53 nuclear accumulation.
P < .05.
To determine whether T-cell density was associated with p53 status independent of clinicopathological factors, we pooled T-cell counts across all sets and modeled the association between T-cell density and p53 status using negative-binomial regression with a natural logarithmic offset term for tissue area at risk, and adjusting for age, race, stage, grade, PSA, and TMA set. When pooled across sets, adjusted CD3 + T-cell density was significantly higher among tumors with p53 nuclear accumulation (adjusted density, 551.1 cells/mm2) than in those without nuclear accumulation (adjusted density, 352.8 cells/mm2; P = .003). When pooled across sets, adjusted CD8+ T-cell density was significantly higher among tumors with p53 nuclear accumulation (adjusted density, 136.7 cells/mm2) than in those without nuclear accumulation (adjusted density, 98.5 cells/mm2; P = .04). Although the trend was similar, when pooled across sets, adjusted FOXP3+ T-cell density was nonsignificantly higher among tumors with p53 nuclear accumulation (adjusted density, 22.5 cells/mm2) than in tumors without p53 nuclear accumulation (adjusted density, 17.2 cells/mm2; P = .08).
4. Discussion
Accumulating evidence suggests that differing tumor molecular subtypes are associated with distinct tumor immune microenvironments in prostate cancer. We and others have previously reported that ERG gene rearrangements are associated with higher levels of tumor-infiltrating T cells in primary prostate tumors, and our group recently found a similar association with PTEN loss [6,24]. These findings are supported by preclinical studies that have established differing immune microenvironments in transgenic mouse models of prostate cancer with different genomic driver alterations [7]. Importantly, many of these associations between genomic drivers and immune microenvironment are likely highly context specific. For example, in contrast to our recent findings in prostate cancer, a study of melanoma suggested that PTEN loss was associated with fewer tumor-infiltrating CD8+ T cells and a reduced response to checkpoint blockade [25].
To our knowledge, our study is the first to correlate the density of subsets of tumor-infiltrating T cells with presence or absence of TP53 missense mutation, across prostate cancer patients of varying clinical grade and racial ancestry. We found that p53 nuclear accumulation, a biomarker of underlying TP53 missense mutation, is associated with increased tumor-infiltrating lymphocytes independent of clinicopathological factors and racial ancestry. Across 3 independent TMA sets, tumors with p53 nuclear accumulation had higher median CD3+, CD8+, and FOXP3+ T-cell densities, although not all differences were statistically significant. Importantly, this association was significant for CD3+ and CD8+ T cells when all TMA sets were pooled and associations were adjusted for clinicopathological variables including tumor grade and stage. A similar trend, although not statistically significant, was seen for FOXP3+ T cells. Notably, this association remained independent of racial ancestry in the matched-race TMA sets, despite the fact that the prevalence of p53 nuclear accumulation was lower in African American tumors compared with European American tumors. A previous study by Lindquist et al also observed significantly lower TP53 mutation rates by sequencing in patients with African ancestry when compared with the TCGA cohort [26]. These findings in prostate cancer are particularly interesting in light of the fact that TP53 mutations were overall more common in African American patients compared with European American patients when examined across other tumor types [27].
These data are perhaps not surprising given that TP53 mutation is associated with increased genomic instability [28], which may be associated with greater immunogenicity in cancer. Studies in other tumor types have also found greater numbers of immune cells in TP53-mutated tumors, although these studies have largely been conducted by examining transcriptomic immune signatures in silico. In breast cancer, significantly higher levels of regulatory T cells (T-reg) were seen by RNA profiling in TP53-mutated tumors compared with those with wild-type TP53, although this association was not seen in other tumor types included in the TCGA [29]. These data are consistent with independent findings in the same TCGA cohort showing that TP53 mutations were enriched in a subset of breast tumors with upregulation of immune-regulatory transcripts, including PDL1, PD1, and FOXP3 [30]. There has been subsequent confirmation of these findings using PD-1 IHC in breast cancers with TP53 mutation [31]. Similar findings have been seen in ovarian and gastric carcinoma as well [32,33].
In addition to the possibility of increased immunogenicity in TP53-mutated tumors due to genomic instability, there are a number of alternative mechanisms that might explain why TP53-mutated prostate cancers have higher levels of tumor-infiltrating T cells. First, it is formally possible that tumor-specific mutations in the p53 protein itself may alter its antigenicity, making it a potential neoantigen. Unfortunately, because of the large number of different p53 hotspot mutations and the distinct potential epitopes for each, vaccines targeting mutant p53 have not yet gained traction. It is also possible that patients may mount an immune response that cross-reacts with wild-type p53 because of stabilization and massive overexpression of p53 with missense mutations in cancer cells. In colorectal cancer patients, anti-p53 immunoreactive T cells can be detected years after tumor resection, which may indicate that there is a lack of immunologic tolerance to the p53 auto-antigen in many patients [34]. Based on this concept, a number of studies have examined the potential of wild-type p53-based vaccines, and these seem to hold some promise [35-38].
Although some studies suggest that TP53 mutation may be associated with antitumor immunity, from our data, it is unclear whether TP53 mutation is associated with an increase in antitumor or protumor immune response. We observed a concomitant increase in total T cells (by CD3+ density) as well as T-reg (FOXP3+) and cytotoxic (CD8+) T cells in prostate tumors with TP53 mutation. This finding seems to be a fairly common one in the prostate. For example, androgen deprivation results in proportional increases in CD8+ and T-reg cells [39]. Our previous observations in primary prostate tumors with ERG gene rearrangement and PTEN loss were similar, where both CD8+ and FOXP3+ T cells increased proportionally [6]. Although we examined the CD8+/FOXP3+ ratio across all cohorts in the current study, we did not see any significant associations with clinicopathological factors or p53 nuclear accumulation (data not shown). Thus, it remains unclear whether the increased immune response with TP53 mutation in prostate cancer is more immunosuppressive or cytotoxic. Given that our group has found that PD-L1 expression is extremely rare in primary prostate cancer [40], we did not look at PD-L1 or PD-1 expression across all of the cohorts in the current study. However, correlation of previously published PD-L1 quantification in the primary Gleason pattern 5 cohort [40] with TP53 status in this cohort did not yield any significant associations (data not shown). Future work examining additional immune checkpoint molecules, such as B7-H3, that are more highly expressed in prostate with underlying molecular subclass may be informative.
The current study has some limitations. First, this study was performed using TMAs, and although we have previously found that T-cell densities measured on TMA spots correlate significantly with T-cell densities measured on standard histological sections (the criterion standard) [6], lymphocyte infiltrates are very heterogeneous in prostate tumors and there can be a sampling bias with TMAs, which may not reflect the true intratumoral lymphocyte density. Similarly, heterogeneity of TP53 mutation and subsequent p53 nuclear accumulation within and between individual tumor foci in primary prostate cancer could be missed in TMA analysis. Another limitation is that our IHC assay for TP53 mutation is sensitive only for missense mutations in TP53 [9,10], comprising approximately two-thirds of the pathogenic TP53 alterations seen in prostate cancer. Because this assay misses TP53 truncation mutations (or very rare homozygous deletions at the TP53 locus), our findings are only representative of the association of TP53 missense mutation with T-cell infiltrate. Additional studies examining a similar association with TP53 truncation mutations will be informative. Finally, and perhaps most importantly, we used a relatively broad definition of tumor-infiltrating lymphocytes, as any lymphocyte in a TMA spot containing tumor cells. We did not distinguish between lymphocytes within the tumor epithelium (quite rare in prostate, see Figure) and those in the stroma. As we continue to learn about the immune microenvironment in primary prostate cancer, we will develop a more refined spatial definition of immune cell compartments that are tumor responsive.
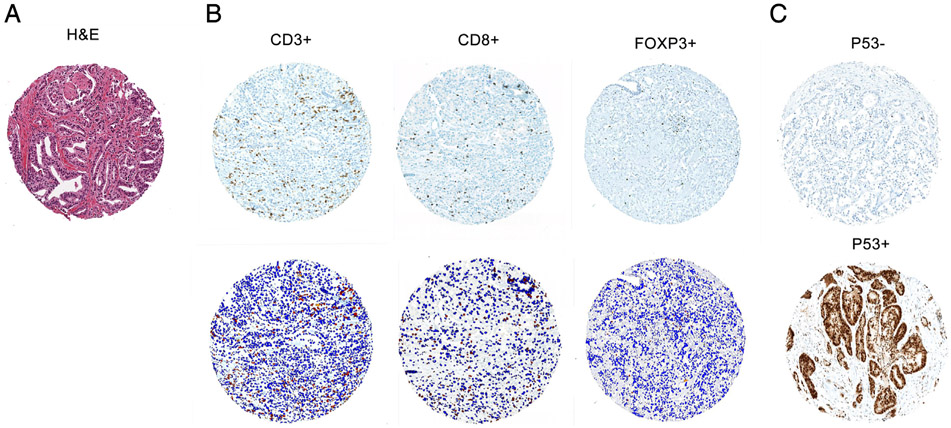
Figure.
Representative E;hematoxylin-eosin staining (A) and lymphocyte immunostaining (B) in prostate tumor on TMA cores. Immunostaining for CD3, CD8, and FOXP3 identifies respective specific subsets of tumor-infiltrating lymphocytes in prostate tumor (top). Aperio image software identifies CD3+ T cells, CD8+ T cells, and FOXP3+ T cells (red) in selected tumor regions and surrounding tumor and stromal nuclei (blue; bottom). C, Representative p53 immunostaining on TMA cores. p53 nuclear accumulation (p53+) signifies TP53 missense mutation. All photo-micrographs are reduced from ×200.
In conclusion, this is the first study in primary prostate cancer to examine associations between digitally quantified tumor-infiltrating T-cell densities and inferred TP53 missense mutation among patients of varying racial ancestry. We found that T-cell density varies significantly with TP53 status in both patients of European American and African American descent. These findings are of particular interest given that our group and others have shown that prostate tumors with TP53 mutations have generally dismal outcomes, with a high frequency of hormone-resistant disease [9,10]. Thus, the possibility that immunotherapies could potentially be efficacious in this aggressive subset is particularly encouraging. More broadly, our findings support the concept that the molecular characteristics of prostate cancer may provide an important framework for patient-targeted immunotherapy. Ultimately, integrating clinical, pathologic, somatic molecular subtyping, and immune microenvironment data is becoming increasingly important in the era of precision medicine, and additional studies of this subject are warranted.
Supplementary Material
Table 2.
T-cell densities (cells/mm2) by clinicopathological parameters and p53 status in intermediate/high-risk TMA sets
| CD3+ density |
CD8+ density |
FOXP3+ density |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| n (257) | Median | P b | n (262) | Median | P b | n (239) | Median | P b | |
| Race | |||||||||
| White | 233 | 215.7 | .3 | 237 | 69.0 | .1 | 218 | 30.5 | .2 |
| Nonwhite a | 24 | 193.6 | 25 | 45.1 | 21 | 22.1 | |||
| Stage | |||||||||
| T2 N0 | 73 | 205.1 | .3 | 74 | 65.9 | 0.2 | 66 | 26.1 | .1 |
| T3aN0 | 110 | 212.2 | 110 | 62.8 | 102 | 30.9 | |||
| T3bN0 | 32 | 208.7 | 35 | 84.0 | 33 | 28.3 | |||
| T4 N0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | |||
| N0 | 215 | 207.8 | .1 | 219 | 67.2 | .9 | 201 | 28.3 | .1 |
| N1 | 39 | 277.6 | 40 | 68.1 | 36 | 34.5 | |||
| Gleason score | |||||||||
| 6 | 1 | 308.6 | .7 | 1 | 247.6 | .5 | 1 | 59.2 | .2 |
| 3 + 4 | 109 | 209.6 | 111 | 67.2 | 99 | 29.2 | |||
| 4 + 3 | 51 | 231.7 | 53 | 71.6 | 50 | 29.2 | |||
| 8 | 29 | 183.9 | 30 | 57.5 | 28 | 22.1 | |||
| 9 | 64 | 233.4 | 64 | 70.0 | 59 | 33.6 | |||
| p53 negative c | 241 | 207.8 | .006 * | 245 | 66.3 | .5 | 222 | 29.2 | .04 * |
| p53 positive d | 16 | 415.9 | 17 | 71.6 | 17 | 51.3 | |||
NOTE. CD3, CD8, and FOXP3 immunostaining was unevaluable in 10, 5, and 28 cases from the intermediate/high-risk TMA set because of poor staining, respectively. Tumor stage and Gleason score information was not available for a few cases.
Including African American, Hispanic, other, and unknown.
From Kruskal-Wallis test.
Absence of p53 nuclear accumulation.
Presence of p53 nuclear accumulation.
P < .05.
Acknowledgments
H.K. and T.L. conceptualization, data curation, formal analysis and writing- original draft and review and editing; J. L. and J.B. contributed to formal analysis; L.G., L. M., and L.R. contributed to data curation; C.J. contributed to conceptualization, formal analysis, and writing- original draft; S.T., K. S., A.D.M., M.E., and E.S. contributed to conceptualization, data curation, and writing-original draft.
Funding/Support:
This work was supported by 2 Health Disparity Research Awards from the Congressionally Directed Medical Research Program- Prostate Cancer Research Program (W81XWH-15-1-0661 to S.A.T., E.M.S., and T.L.L. and W81XWH-17-1-0286 to K.S.S., T.L.L., L.B.G., and H.B.K.), the National Institutes of Health/National Cancer Institute Prostate SPORE P50CA58236, and the National Cancer Institute Cancer Center Support Grant 5P30CA006973-52. L.R. and L.M. were supported by fellowships from the CUPID (Cancer in the Under-Privileged Indigent or Disadvantaged) Program.
Footnotes
Competing interests: T.L.L. has received research support from Ventana Medical Systems for other studies.
Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.humpath.2019.02.006.
References
- [1].Hanahan D, Coussens LM. Accessories to the crime: functions of cells recruited to the tumor microenvironment. Cancer Cell 2012;21:309–22 [DOI] [PubMed] [Google Scholar]
- [2].Egeblad M, Nakasone ES, Werb Z. Tumors as organs: complex tissues that interface with the entire organism. Dev Cell 2010;18:884–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Hodi FS, O’Day SJ, McDermott DF, et al. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med 2010;363:711–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Guedes LB, Antonarakis ES, Schweizer MT, et al. MSH2 loss in primary prostate cancer. Clin Cancer Res 2017;23:6863–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Rosty C, Walsh MD, Lindor NM, et al. High prevalence of mismatch repair deficiency in prostate cancers diagnosed in mismatch repair gene mutation carriers from the colon cancer family registry. Fam Cancer 2014;13:573–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Kaur HB, Guedes LB, Lu J, et al. Association of tumor-infiltrating T-cell density with molecular subtype, racial ancestry and clinical outcomes in prostate cancer. Mod Pathol 2018;31:1539–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Bezzi M, Seitzer N, Ishikawa T, et al. Diverse genetic-driven immune landscapes dictate tumor progression through distinct mechanisms. Nat Med 2018;24:165–75. [DOI] [PubMed] [Google Scholar]
- [8].Kandoth C, McLellan MD, Vandin F, et al. Mutational landscape and significance across 12 major cancer types. Nature 2013;502:333–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Guedes LB, Almutairi F, Haffner MC, et al. Analytic, preanalytic, and clinical validation of p53 IHC for detection of TP53 missense mutation in prostate cancer. Clin Cancer Res 2017;23:4693–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Maughan BL, Guedes LB, Boucher K, et al. p53 status in the primary tumor predicts efficacy of subsequent abiraterone and enzalutamide in castration-resistant prostate cancer. Prostate Cancer Prostatic Dis 2018; 21:260–8. [DOI] [PubMed] [Google Scholar]
- [11].Robinson D, Van Allen EM, Wu YM, et al. Integrative clinical genomics of advanced prostate cancer. Cell 2015;161:1215–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Barbieri CE, Baca SC, Lawrence MS, et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat Genet 2012;44:685–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Guo G, Marrero L, Rodriguez P, Del Valle L, Ochoa A, Cui Y. Trp53 inactivation in the tumor microenvironment promotes tumor progression by expanding the immunosuppressive lymphoid-like stromal network. Cancer Res 2013;73:1668–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Cui Y, Guo G. Immunomodulatory function of the tumor suppressor p53 in host immune response and the tumor microenvironment. Int J Mol Sci 2016;17:E1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Raj N, Attardi LD. Tumor suppression: p53 alters immune surveillance to restrain liver cancer. Curr Biol 2013;23:R527–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Menendez D, Shatz M, Resnick MA. Interactions between the tumor suppressor p53 and immune responses. Curr Opin Oncol 2013;25:85–92. [DOI] [PubMed] [Google Scholar]
- [17].Martins CP, Brown-Swigart L, Evan GI. Modeling the therapeutic efficacy of p53 restoration in tumors. Cell 2006;127:1323–34. [DOI] [PubMed] [Google Scholar]
- [18].Ventura A, Kirsch DG, McLaughlin ME, et al. Restoration of p53 function leads to tumour regression in vivo. Nature 2007;445:661–5. [DOI] [PubMed] [Google Scholar]
- [19].Xue W, Zender L, Miething C, et al. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature 2007; 445:656–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Tosoian JJ, Almutairi F, Morais CL, et al. Prevalence and prognostic significance of PTEN loss in African-American and European-American men undergoing radical prostatectomy. Eur Urol 2017;71:697–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Ross AE, Johnson MH, Yousefi K, et al. Tissue-based genomics augments post-prostatectomy risk stratification in a natural history cohort of intermediate- and high-risk men. Eur Urol 2016;69:157–65. [DOI] [PubMed] [Google Scholar]
- [22].Yemelyanova A, Vang R, Kshirsagar M, et al. Immunohistochemical staining patterns of p53 can serve as a surrogate marker for TP53 mutations in ovarian carcinoma: an immunohistochemical and nucleotide sequencing analysis. Mod Pathol 2011;24:1248–53. [DOI] [PubMed] [Google Scholar]
- [23].Liu J, Li W, Deng M, Liu D, Ma Q, Feng X. Immunohistochemical determination of p53 protein overexpression for predicting p53 gene mutations in hepatocellular carcinoma: a meta-analysis. PLoS One 2016;11:e0159636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Flammiger A, Bayer F, Cirugeda-Kuhnert A, et al. Intratumoral T but not B lymphocytes are related to clinical outcome in prostate cancer. APMIS 2012;120:901–8. [DOI] [PubMed] [Google Scholar]
- [25].Peng W, Chen JQ, Liu C, et al. Loss of PTEN promotes resistance to T cell-mediated immunotherapy. Cancer Discov 2016;6:202–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Lindquist KJ, Paris PL, Hoffmann TJ, et al. Mutational landscape of aggressive prostate tumors in African American men. Cancer Res 2016;76:1860–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Yuan J, Hu Z, Mahal BA, et al. Integrated analysis of genetic ancestry and genomic alterations across cancers. Cancer Cell 2018;34:549–60 [e549]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Song H, Hollstein M, Xu Y. p53 gain-of-function cancer mutants induce genetic instability by inactivating ATM. Nat Cell Biol 2007;9:573–80. [DOI] [PubMed] [Google Scholar]
- [29].Siemers NO, Holloway JL, Chang H, et al. Genome-wide association analysis identifies genetic correlates of immune infiltrates in solid tumors. PLoS One 2017;12:e0179726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Hendrickx W, Simeone I, Anjum S, et al. Identification of genetic determinants of breast cancer immune phenotypes by integrative genome-scale analysis. Oncoimmunology 2017;6:e1253654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Gatalica Z, Snyder C, Maney T, et al. Programmed cell death 1 (PD-1) and its ligand (PD-L1) in common cancers and their correlation with molecular cancer type. Cancer Epidemiol Biomarkers Prev 2014;23:2965–70. [DOI] [PubMed] [Google Scholar]
- [32].Wieser V, Gaugg I, Fleischer M, et al. BRCA1/2 and TP53 mutation status associates with PD-1 and PD-L1 expression in ovarian cancer. Oncotarget 2018;9:17501–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Jiang Z, Liu Z, Li M, Chen C, Wang X. Immunogenomics analysis reveals that TP53 mutations inhibit tumor immunity in gastric cancer. Transl Oncol 2018;11:1171–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].van der Burg SH, de Cock K, Menon AG, et al. Long lasting p53-specific T cell memory responses in the absence of anti-p53 antibodies in patients with resected primary colorectal cancer. Eur J Immunol 2001;31:146–55. [DOI] [PubMed] [Google Scholar]
- [35].Vierboom MP, Nijman HW, Offringa R, et al. Tumor eradication by wild-type p53-specific cytotoxic T lymphocytes. J Exp Med 1997;186:695–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Zwaveling S, Vierboom MP, Ferreira Mota SC, et al. Antitumor efficacy of wild-type p53-specific CD4(+) T-helper cells. Cancer Res 2002;62:6187–93. [PubMed] [Google Scholar]
- [37].Hardwick NR, Frankel P, Ruel C, et al. p53-reactive T cells are associated with clinical benefit in patients with platinum-resistant epithelial ovarian cancer after treatment with a p53 vaccine and gemcitabine chemotherapy. Clin Cancer Res 2018;24:1315–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Chung V, Kos FJ, Hardwick N, et al. Evaluation of safety and efficacy of p53MVA vaccine combined with pembrolizumab in patients with advanced solid cancers. Clin Transl Oncol 2019;21:363–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Sorrentino C, Musiani P, Pompa P, Cipollone G, Di Carlo E. Androgen deprivation boosts prostatic infiltration of cytotoxic and regulatory T lymphocytes and has no effect on disease-free survival in prostate cancer patients. Clin Cancer Res 2011;17:1571–81. [DOI] [PubMed] [Google Scholar]
- [40].Haffner MC, Guner G, Taheri D, et al. Comprehensive evaluation of programmed death-ligand 1 expression in primary and metastatic prostate cancer. Am J Pathol 2018;188:1478–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.