Abstract
The kinetochore is a proteinaceous structure that assembles onto centromeric DNA and mediates chromosome attachment to microtubules during mitosis. This description is deceivingly simple: recent proteomic studies suggest that the diminutive kinetochores of Saccharomyces cerevisiae are comprised of at least 60 proteins organized into as many as 14 different subcomplexes. Many of these proteins, such as the centromeric histone variant CENP-A, and entire subcomplexes, such as the Ndc80Hec1 complex, are conserved from yeast to humans despite the diverse nature of the DNA sequences on which they assemble. There have recently been advances in our understanding of the molecular basis of how kinetochores establish dynamic attachments to spindle microtubules, and how these attachments are correctly oriented to ensure segregation of sister chromatids to daughter cells.
Introduction
The many tasks of the mitotic kinetochore include attaching chromosomes to the mitotic spindle, coupling force production by microtubule polymer dynamics and/or motor proteins to chromosome movement, and inhibiting the anaphase segregation of chromatids until all chromosomes are attached and properly aligned [1,2]. Here, we discuss recent work on the binding of kinetochores to spindle microtubules, on the regulation of the dynamics of kinetochore-attached microtubules, and on the mechanisms that ensure proper orientation of attached chromosomes on the spindle. We will not discuss the spindle checkpoint [2,3] or the specification of kinetochore assembly [4], which have been reviewed recently elsewhere.
Forming and maintaining stable microtubule attachments
The kinetochore forms on centromeric chromatin to generate a microtubule-binding interface that links chromosomes to the mitotic spindle. Numerous proteins and protein subcomplexes have been implicated in proper kinetochore assembly and microtubule attachment [2,5–7], although the precise functions of the majority of these proteins remain unclear. New ideas about how the kinetochore attaches chromosomes to spindle microtubules have emerged from studies of the Ndc80 complex (Ndc80Hec1, Nuf2, Spc24, and Spc25), which is conserved from fungi to humans [8–17,18•,19,20••,21••,22•,23•] (Table 1). The variation in chromosomal architecture and kinetochore-microtubule binding capacity within the animal kingdom [24] makes the conservation of this complex an exciting finding. Immunofluorescence and immuno-electron microscopic analyses of vertebrate cells reveal that Ndc80 complex proteins localize to the outer kinetochore plate [14,15,19,25••], where microtubule plus ends terminate [26]. Ndc80Hec−1 and Nuf2 are stably bound to kinetochores, as assessed by fluorescence recovery after photobleaching [19], and their levels at the kinetochore remain unchanged during the formation of kinetochore-microtubule attachments [15,19,20••,22•, 25••,27]. These attributes make components of the Ndc80 complex good candidates for playing a direct role in mediating interactions between kinetochores and microtubules.
Table 1.
Mitotic defects resulting from inactivation of Ndc80 complex proteins in eukaryotic systems.
| Species | Complex components | Experimental method | Results | References |
|---|---|---|---|---|
| Mitotic progression and morphology | ||||
| H. sapiens | Ndc80Hec1 Nuf2 Spc24 Spc25 | RNAi Nuf2 in HeLa cells | Chromosome alignment and spindle defects. Reduced stability of kinetochore fibers to cold treatment. Reduced inter-kinetochore distances. Poor kinetochore morphology and microtubule binding at EM level [25••]. Active spindle checkpoint. | [15,25••] |
| RNAi Spc24, Spc25, Ndc80Hec1 in HeLas | Chromosome alignment and spindle defects. Active spindle checkpoint. | [22•,23•,27] | ||
| RNAi Nuf2 in HeLa cells | Chromosome alignment and spindle defects. Active spindle checkpoint with 95% depletion. Inactive spindle checkpoint with more penetrant depletions. |
[36•] | ||
| Targeting of kinetochore components | ||||
| RNAi Nuf2, Ndc80Hec1 in HeLa cells | Decreased levels of Mad1 and Mad2 targeting to kinetochores restored by nocodazole depolymerization of microtubules. | [22•,30••] | ||
| RNAi Nuf2 in HeLa cells | For penetrant depletions, Mad1 and Mad2 targeting is abolished. Kinetochore localization is not restored upon nocodazole treatment. | [36•] | ||
| RNAi Spc24, Spc25 in HeLa cells | Loss of outer kinetochore proteins, including Mad1 and Mad2, is mostly restored after nocodazole treatment. | [23•] | ||
| RNAi Nuf2, Spc24, Spc25 in HeLa cells | RNAi of Nuf2 [30••] or Spc25 [22•] abolishes Ndc80 binding at kinetochores. RNAi of Spc24 abolishes kinetochore targeting of Spc25 and vice versa [23•]. Localization is not restored upon nocodazole treatment. | [22•,23•,30••] | ||
| Mitotic progression and morphology | ||||
| X. laevis | Ndc80 Nuf2 Spc24 Spc25 | Injection of a-Nuf2, α-Ndc80 into XTC cells | Failure of chromosome alignment and segregation. Cells exit mitosis prematurely, presumably because spindle checkpoint is inactive. Spindle checkpoint is also inactivated in egg extracts following immunodepletion. |
[20••] |
| Injection of a-Spc24, α-Spc25 into S3 cells | Chromosome alignment and segregation defects. Decreased inter-kinetochore distances. Active spindle checkpoint. | [23•] | ||
| Immunodepletion of Nuf2, Ndc80 from egg extracts | Targeting of kinetochore components Multiple outer kinetochore proteins fail to target in immunodepleted extracts. |
[20••] | ||
| Mitotic progression and morphology | ||||
| C. elegans | Ndc80 Nuf2HIM-10, Spc25KBP-3 | RNAi of Ndc80, Nuf2HIM-10 | Chromosome-microtubule attachments form, but congression and segregation are aberrant. Ndc80, Nuf2, and Spc25 are part of a larger KNL-1/3 complex, which includes Mis12 and 6 novel kinetochore proteins [21••]. | [18•,21 ••] |
| nuf2him-10 mutant | Defective chromosome alignment and disrupted kinetochore structure, as assayed by EM of high pressure frozen embryos. | [11] | ||
| Mitotic progression and morphology | ||||
| S. cerevisiae | Ndc80 Nuf2 Spc24 Spc25 | ndc80, nuf2 mutant and degron alleles | Disrupted kinetochore-microtubule binding. Aberrant centromere movements. Active spindle checkpoint. | [9,10,12,20••,28] |
| spc24, spc25 mutants | Disrupted kinetochore-microtubule binding. Spindle microtubule defects. Inactive spindle checkpoint. | [12,14,16] | ||
| Targeting of kinetochore components: | ||||
| ndc80 mutants | Most kinetochore proteins localize in ChIP assays, except Stu2 and the Dam complex (both require microtubules to target to kinetochores [34,35•]. | [12,28,29•, 31 ••,32] | ||
| spc24, spc25 mutants | Most kinetochore proteins localize in ChIP assays [12]; Bub1 and Mad2 do not in spc25 mutants [32]. | [12,32] | ||
| Mitotic progression and morphology: | ||||
| S. pombe | Ndc80 Nuf2 Spc24 Spc25 | nuf2 mutants | Allele-specific effects on spindle length and spindle checkpoint activity. Defects in centromere motility and chromosome segregation. | [13,17] |
Defects in mitotic progression, spindle and kinetochore morphology, and targeting of kinetochore components are organized above according to experimental system and inhibitory method. H. sapiens, Homo sapiens; S. cerevisiae, Saccharomyces cerevisiae; S. pombe, Schizosaccharomyces pombe; X. laevis, Xenopus laevis. Please see text for comments.
Defects in the Ndc80 complex disrupt kinetochore-microtubule attachments, chromosome congression and chromosome segregation in all systems that have been analyzed, although the underlying cause is controversial (please see Table 1 and all references therein). In addition to technical variations between studies, particularly with respect to mammalian cell RNAi, difficulties in distinguishing between a role in outer kinetochore assembly versus a direct role in kinetochore-microtubule interactions probably underlie many of the apparently contradictory conclusions. Recent findings in tissue culture cells, budding yeast and Caenorhabditis elegans embryos indicate that targeting of most outer kinetochore proteins is largely preserved upon disruption of Ndc80 complex proteins, although some spindle checkpoint protein levels are decreased [12,15,18•,21••,22•,23•,27, 28,29•,30••,31••,32] (Table 1). In contrast, chromosomes assembled in Xenopus egg extracts immunodepleted of Ndc80 and Nuf2 fail to localize multiple outer kinetochore components [20••]. However, as these assembly defects have not yet been rescued using purified proteins, the possibility that they result from the depletion of additional interacting proteins cannot be excluded [20••] (Table 1). Recent work in metazoans has identified a larger network of conserved interacting proteins that includes the Ndc80 complex, making this a likely possibility [21••]. In budding yeast, mutational inactivation of Ndc80 complex subunits results in an inability of microtubule-binding proteins, such as the Dam1 complex and Stu2, to localize to kinetochores as judged by chromatin immunoprecipitations [28,29•,32,33]; however, neither the Dam1 complex nor Stu2 can associate with centromeric DNA in the absence of microtubules [34,35•] (Table 1). Thus, the apparent assembly defects following inhibition of Ndc80 complex function are more likely to be a consequence of defects in forming stable microtubule attachments.
Several lines of evidence indicate that microtubule attachments can form when Ndc80 complex proteins are depleted, but these attachments are unstable and cannot support chromosome congression and segregation. In vertebrate cells depleted of Nuf2 or Ndc80Hec1, stable fibers of kinetochore microtubules fail to form, but kinetochore checkpoint proteins, such as Mad1 and Mad2, are still significantly depleted from kinetochores [15,19, 20••,22•,23•,25••,27,30••] (Table 1). Interestingly, drug-induced depolymerization of microtubules restores Mad1 and Mad2 targeting to normal levels, indicating that kinetochores assembled in Nuf2- or Ndc80Hec1-depleted cells are fully capable of binding checkpoint proteins [22•,30••]. However, a recent study claims that with more penetrant depletions of Nuf2 and Ndc80Hec1, checkpoint proteins fail to target even after microtubule depolymerization [36•] (Table 1). Although the current data exclude a stoichiometric role for the Ndc80 complex in the targeting of checkpoint proteins, the discrepancy between the different mammalian cell RNAi studies remains to be resolved.
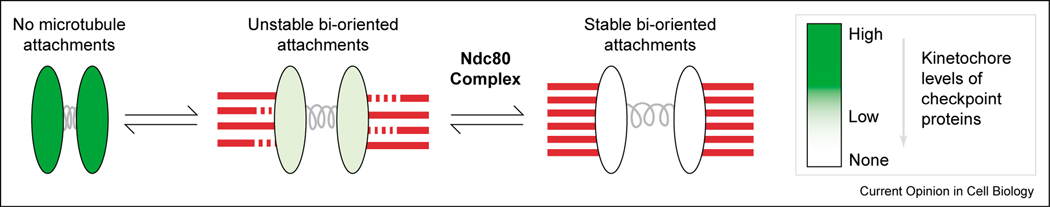
The microtubule-dependent reduction of checkpoint proteins at kinetochores depleted of Ndc80 complex proteins suggests that these kinetochores can form transient, albeit unstable, microtubule attachments (Figure 1). In support of this idea, an electron microscopic analysis of Nuf2-depleted cells indicates that the outer kinetochore morphology is severely perturbed and the number of embedded microtubule plus ends dramatically decreased in these cells [25••]. Furthermore, analyses of C. elegans embryos have demonstrated that partial chromosome alignment and segregation still occur in embryos depleted of Ndc80, Nuf2HIM−10 or Spc25KBP−3, although attachments between chromosomes and the spindle are mechanically unstable [18•,21••] (Table 1). In contrast, a complete failure of chromosome segregation occurs in embryos depleted of the more chromatin-proximal kinetochore components, CENP-AHCP−3 and CENP-CHCP−4 [18•,37].
Figure 1.
Formation of stable kinetochore–microtubule attachments. Microtubules are in red and centromeric chromatin is in gray. Kinetochores are indicated by ovals in various shades of green corresponding to the level of kinetochore-localized spindle checkpoint proteins, which are progressively depleted as microtubules attach to kinetochores. With 95% depletion of Ndc80 complex subunits, significant depletion of checkpoint proteins still occurs, suggesting the existence of some type of attachment between kinetochores and spindle microtubules. However, stable kinetochore fibers do not form, and chromosome alignment and segregation is severely perturbed. Thus, the Ndc80 complex plays a critical role in stabilization of microtubule attachments, allowing the formation of mature kinetochore fibers capable of aligning and segregating chromosomes properly.
Cumulatively, these results suggest that the Ndc80 complex does not act as a targeting scaffold for kinetochore proteins, but rather plays a crucial role in stabilizing microtubule attachments by maintaining the structural integrity of binding sites for microtubule plus ends at the outer kinetochore [25••]. Determining how the Ndc80 complex interacts with other components at the kinetochore should provide a more detailed understanding of how mechanically stable microtubule attachments are formed and maintained. For example, recent studies in C. elegans have identified two novel kinetochore components, KNL-1 and KNL-3, which are required to target the Ndc80 complex to kinetochores [18•,21••]. In addition, both proteins biochemically associate with components of the Ndc80 complex, and this interaction is conserved in human cells [18•,21••]. Coupled in vivo and in vitro analyses of the Ndc80 complex and the larger KNL-1/3 interacting protein network should help decipher how stable kinetochore-microtubule attachments are formed.
Growth of kinetochore-attached microtubules
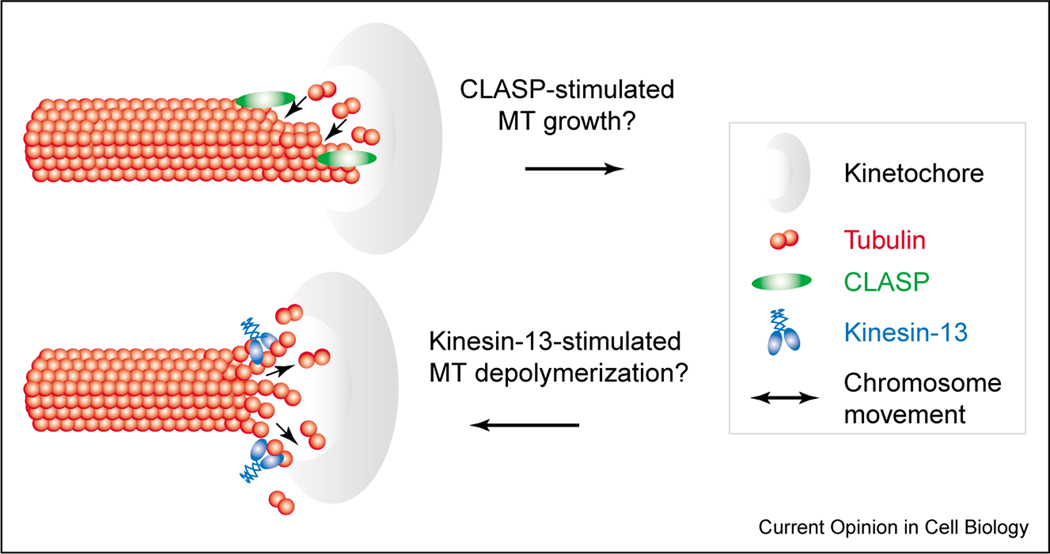
Growth of kinetochore microtubules by the addition of tubulin at the kinetochore is important for chromosome movements prior to anaphase. An example of this type of growth is seen during chromosome congression to the metaphase plate, when microtubule depolymerization at the leading kinetochore is coupled to microtubule polymerization at the lagging kinetochore [1,38]. Kinetochores contain multiple proteins that can promote microtubule growth (for example, see [39–48]), although the physiological contributions of these proteins to kinetochore-microtubule dynamics have not been defined. Recent data have identified the CLASP family of microtubule-associated proteins (MAPs), which includes CLASP1 in humans [49,50•] and MAST/Orbit in Drosophila [51,52], as potential regulators of kinetochore-microtubule dynamics. CLASPs localize near microtubule plus ends and stabilize microtubules when overexpressed as GFP-fusion proteins in interphase cells [49,50•]. CLASPs also localize to kinetochores in a microtubule-independent manner and are the outermost kinetochore proteins identified to date [50•,52]. Inhibition of CLASP activity by antibody microinjection or RNAi does not prevent the formation of kinetochore-microtubule attachments, but rather leads to shortened kinetochore microtubules and suppressed chromosome oscillations [50•,52]. Although exactly how the dynamics of kinetochore fibers are perturbed in these cells remains unclear, current data suggest that CLASPs stimulate the growth of kinetochore-attached microtubules (Figure 2a). This idea is supported by the ability of microtubule-growth-promoting drugs to rescue the short kinetochore microtubule defect in CLASP-inhibited cells [50•,52].
Figure 2.
Regulation of kinetochore–microtubule dynamics. The assembly dynamics of kinetochore-attached microtubules is coupled to chromosome movement on the spindle. Regulators of microtubule dynamics localized to kinetochores, such as the CLASP family of MAPs and the kinesin-13 family of microtubule depolymerases, are likely to play important roles in this process. Kinetochore-localized CLASPs (in green) may stimulate the growth of kinetochore-attached microtubules during anti-poleward movements that align chromosomes at the metaphase plate. Polymerization of kinetochore microtubule plus ends is also necessary for poleward microtubule flux during metaphase. In contrast, the microtubule depolymerase activity of kinesin-13 proteins (in blue) may contribute to poleward chromosome movement, although discrepancies between studies in different systems concerning this putative role need to be resolved. Arrows indicate the predicted direction of chromosome movement. MT, microtubule.
Dissecting how kinetochore-bound CLASP regulates microtubule dynamics may provide molecular clues about the mechanism of chromosome alignment, and perhaps about other occurrences of microtubule growth at kinetochores, such as during microtubule flux [53••]. Addition of tubulin at kinetochore-attached microtubule ends may also prove to be an integral part of mitotic spindle assembly. This idea has arisen from recent observations of outward growth of kinetochore microtubules on unattached kinetochores facing away from the spindle. Kinetochore fibers formed in this manner are translocated poleward, connecting the chromosome to the spindle pole [54••]. This phenomenon occurs both in somatic cells recovering from treatment with a drug that prevents separation of spindle poles and in untreated cells, suggesting that cells possess a kinetochore-based pathway for the formation of kinetochore microtubules that is independent of the capture of centrosome-nucleated microtubules [54••]. In addition, laser cutting experiments performed in grasshopper spermatocytes indicate that kinetochore fibers can grow outward from the kinetochore, and must reach the spindle pole before chromosome segregation can occur [55••]. Overall, these studies suggest that kinetochore-mediated growth of microtubules is an intrinsic activity of kinetochores in multiple systems, where it may play a role in attaching chromosomes to the spindle and/or in chromosome movement on the spindle.
Depolymerization of kinetochore-attached microtubules
Depolymerization of microtubules at the kinetochore is thought to drive poleward movements during the alignment and segregation of chromosomes [1,38]. The most logical candidates for this activity are the kinesin-13 family members (formerly called Kin I kinesins) [56], which display microtubule-end-stimulated ATPase activity to induce depolymerization from either end of the microtubule [57–59,60••]. During mitosis, kinesin-13 proteins are localized to kinetochores and to the centromeric region between sister kinetochores [61,62•]. Kinesin-13 proteins are clearly required for proper positioning of chromosomes on the spindle in both vertebrate and invertebrate species [61,62•,63,64••,65], although their precise role at kinetochores remains controversial. Analysis in Drosophila embryos suggests that Klp59C, one of two kinesin-13 proteins in this organism, remains associated with anaphase kinetochores where it may play a role in depolymerizing kinetochore-associated microtubules [64••] (Figure 2b). However, inhibition of MCAK, a kinetochore-localized kinesin-13 in vertebrate cells, does not perturb the rate of poleward chromatid movement during anaphase [62•]. Future work analyzing all kinesin-13 family proteins in vertebrates is necessary to reconcile these observations and to understand how kinesin-13 proteins contribute to poleward chromosome movement.
During mitosis in budding yeast, depletion of the microtubule-binding protein Stu2 suppresses the dynamics of spindle microtubules, including kinetochore microtubules, and results in non-motile centromeres [66•,67]. Kinetochores are not required for the stabilization of spindle microtubules observed in Stu2-depleted cells, suggesting that Stu2 affects microtubule dynamics through a kinetochore-independent mechanism [66•]. These findings imply that Stu2 can destabilize microtubules in vivo, an idea which is supported by in vitro studies demonstrating that recombinant Stu2 can promote microtubule depolymerization [68•] despite its similarity to the XMAP215/TOG family of proteins, which strongly promote microtubule growth in vivo and in vitro [69]. Surprisingly, XMAP215 has also been shown to destabilize microtubules under specific in vitro conditions [70•], highlighting the diversity of the potential interactions between microtubules and members of this MAP family [71]. Whether vertebrate orthologs of Stu2 affect kinetochore-microtubule dynamics is an important question to address in the future. It is also important to note that proteins such as kinesin-13 depolymerases and XMAP215/TOG are prominent global regulators of microtubule dynamics and spindle bipolarity [65,72–79]. Consequently, potential indirect effects of their inhibition must be taken into consideration during phenotypic analysis, and may underlie superficially contradictory results from studies in different systems or studies using different methods of inhibition.
Ensuring proper chromosome orientation through regulation of kinetochore-microtubule attachments
Throughout mitosis, there exists the potential for improper kinetochore-microtubule attachments, which must be resolved to prevent missegregation and aneuploidy [80•]. Proper attachments result in a bi-oriented chromosome, wherein one kinetochore is connected exclusively to microtubules emanating from one spindle pole, while its sister kinetochore is connected to the opposite spindle pole. Such bi-oriented attachments result in tension between the two sister kinetochores [81]. The Aurora B kinase (Ipl1 in budding yeast) has emerged from recent work as a major regulator involved in correcting inappropriate kinetochore-microtubule attachments. Aurora BIpl1 is thought to promote bi-orientation by selectively detaching kinetochore-microtubule attachments that are not under tension [82,83••,84•,85]. Antibody inhibition and small molecule inhibition studies have suggested that this role for Aurora B is conserved in vertebrate cells [86•,87•,88,89,90•]. It is less clear how Aurora BIpl1 recognizes the tension created when proper bi-orientation is achieved, although micromanipulation of spermatocyte kinetochores support the idea that chemical properties of kinetochores, such as phosphorylation, can change in response to tension [91,92]. Biochemical and genetic analyses indicate that the multi-protein Dam1 complex, which is conserved within fungi but not identified elsewhere to date, is the key target of Aurora BIpl1 [93]. Precisely how Aurora BIpl1-mediated phosphorylation of the Daml complex affects kinetochore-microtubule attachments and how this regulation is selectively targeted to attachments that are not under tension remain important future questions.
Chromosome bi-orientation is also promoted by the cohesin protein complex, which connects sister chromatids and sustains inter-kinetochore tension [94,95]. Surprisingly, in budding yeast any type of connection between sister chromatids that is capable of resisting tension can promote bi-orientation [83••]. Similarly, in vertebrate cells, chromosome alignment defects caused by depletion of cohesin are rescued by concomitant inhibition of topoisomerase II to generate a distinct type of physical linkage by catenation of sister DNA strands [96•]. These findings indicate a direct link between the mechanics and biochemistry of the kinetochore-microtubule interface, an idea further strengthened by recent data identifying the kinetochore protein Sgo as a microtubule-stabilizing protein and a protector of the cohesive forces between sister centromeres [97•]. Understanding how centromeric tension is generated and detected will be key to defining how Aurora BIpl1p acts at the kinetochore-microtubule interface to promote detachment (in budding yeast, [83••]) or kinetochore fiber depolymerization (in vertebrate cells, [90•]) as a mechanism to resolve incorrect microtubule attachments.
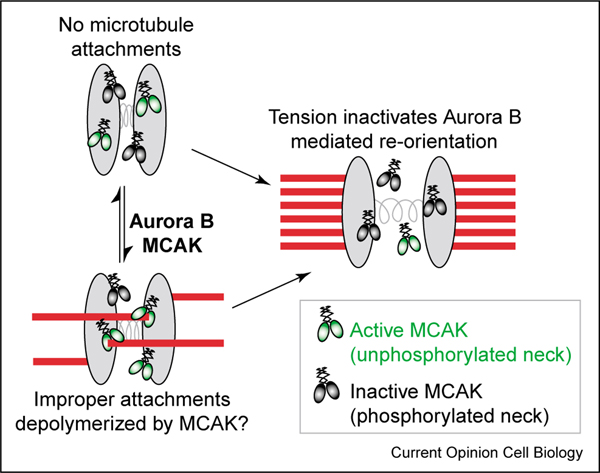
In all eukaryotes, Aurora B is found in a complex with INCENP and Survivin [98]. This highly conserved chromosomal passenger complex exhibits a dynamic localization at the inner centromere early in mitosis and at the spindle midzone following the metaphase-anaphase transition. New components of this complex have been identified in metazoans, termed CSC-1 in C. elegans [99] and Dasra A and Dasra B/Borealin in vertebrates [100•,101•]. Interestingly, recent work suggests that, in addition to its role in promoting bi-orientation, the chromosomal passenger complex also contributes to spindle assembly [89,100•,101•]. In vertebrates, a potential link between Aurora B and both correction of aberrant kinetochore-microtubule attachments and regulation of spindle morphology may be via regulation of the microtubule-depolymerizing kinesin MCAK, a member of the kinesin-13 family. Inactivation of MCAK leads to highly aberrant kinetochore-microtubule attachments [62•] and spindle morphology defects [102]. Aurora B phosphorylates MCAK, promoting its localization to the centromere and inhibiting its microtubule depolymerizing activity (Figure 3) [103••,104••,105••]. In addition to being negatively regulated by Aurora B, MCAK activity is positively regulated in vitro by ICIS, which may work in combination with MCAK to depolymerize microtubules contacting the centromeric domain between sister kinetochores [106•]. Taken together, these recent discoveries indicate that the depolymerizing activity of MCAK is subject to a high degree of local regulation in the vicinity of the kinetochore-microtubule interface (Figure 3). An exciting direction for further research will be to investigate how inactivation of MCAK by Aurora B and stimulation of MCAK by ICIS are dynamically used to stabilize or destabilize kinetochore microtubules during chromosome positioning and attachment correction.
Figure 3.
Error correction mechanisms ensure chromosome bi-orientation on the spindle. Attachment of sister kinetochores to microtubules emanating from opposing spindle poles, a configuration referred to as bi-orientation, is critical to ensure chromosome alignment and segregation. This cartoon depicts how errors in microtubule attachments may be resolved in vertebrate cells by Aurora B kinase and the kinesin-13 protein MCAK, a newly identified substrate of Aurora B. Kinetochores and centromeric heterochromatin are in gray, unphosphorylated MCAK (active microtubule depolymerase) is in green, and MCAK phosphorylated in the neck region (inactive depolymerase) is in black. Throughout mitosis, populations of both phosphorylated and unphosphorylated MCAK exist at the centromere, although the precise localization of active versus inactive MCAK is controversial. One interpretation is that active MCAK is more prevalent when incorrect attachments are present and tension is low. MCAK can then depolymerize inappropriately attached microtubules. Upon attachment of sister kinetochores to opposite spindle poles, the resulting increase in tension prevents Aurora-B-mediated re-orientation, perhaps by affecting local regulation of MCAK activity. In fungi, the Dam1 complex is the key target of Aurora BIpl1 implicated in this error correction process. Such a complex has not been identified in metazoans.
Ironically, the mechanism of attachment regulation by Aurora B could be one reason why it is difficult to elucidate exactly how stable kinetochore-microtubule attachments are formed and maintained. This possibility is illustrated in budding yeast mutants of the conserved kinetochore protein Mis12Mtw1, which contain unattached chromosomes [84•]. This defect is abolished if Aurora BIpl1 is concomitantly inhibited, indicating that active Aurora BIpl1 is detaching defective kinetochore-microtubule attachments in Mis12Mtw1 mutants [84•]. This finding suggests that Aurora BIpl1 inhibition may help to decipher the molecular basis for kinetochore-microtubule attachments. It also highlights the complexity of the regulatory processes at the kinetochore that must be taken into account when analyzing various chromosome segregation defects.
Conclusions and future directions
Dissecting the mechanisms that regulate kinetochore assembly and modulate kinetochore-microtubule attachments is essential to understand chromosome segregation. This will require the combination of precise molecular perturbations with high-resolution assays in living cells. Such efforts will probably be facilitated by recent advances in microscopy [107] and by the emergence of RNA interference and chemical inhibition as new specific methods for disrupting the functions of essential proteins in metazoans. Together, advances in molecular perturbation techniques and the development of increasingly sophisticated assays are gradually bringing the complexities of kinetochore structure and function into focus.
Update
Recent analyses in S.pombe and human cells [108] support work in C. elegans and human cells [21••] indicating the existence of a complex of Mis12Mtw1-interacting proteins required for chromosome segregation. In humans, a complex isolated from interphase nuclear extracts included the heterochromatic proteins HP1a and HP1g, suggesting a molecular link between kinetochore proteins and centromeric heterochromatin [108].
Two recent papers extend analyses of kinetochore-driven kinetochore fiber assembly. Photobleaching and lasermediated kinetochore fiber cutting experiments in Drosophila tissue culture cells indicate that fiber growth can initiate from ‘naked’ kinetochores and connect the kinetochore to the spindle pole via kinetochore-proximal tubulin addition [109]. Interestingly, the microtubule-associated protein CLASP is not required for the initial formation or elongation of kinetochore fibers but is required for polymerization at kinetochores once the fibers reach the pole and initiate flux [110]. This finding indicates the existence of distinct mechanisms that contribute to tubulin addition at kinetochores, and establish CLASPs as integral players in kinetochore fiber dynamics [110].
Acknowledgements
We thank K Oegema for comments on the manuscript. Work in this laboratory is supported by funding from the Ludwig Institute for Cancer Research. SS is supported by an NCI Predoctoral Cancer Cell Biology training grant. AD is a Damon Runyan Scholar supported by the Damon Runyan Cancer Research Foundation (DRS 38-04).
References and recommended reading
Papers of particular interest, published within the annual period of
review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.McIntosh JR, Grishchuk EL, West RR: Chromosome-microtubule interactions during mitosis. Annu Rev Cell Dev Biol 2002, 18:193–219. [DOI] [PubMed] [Google Scholar]
- 2.Cleveland DW, Mao Y, Sullivan KF: Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell 2003, 112:407–421. [DOI] [PubMed] [Google Scholar]
- 3.Lew DJ, Burke DJ: The spindle assembly and spindle position checkpoints. Annu Rev Genet 2003, 37:251–282. [DOI] [PubMed] [Google Scholar]
- 4.Amor DJ, Kalitsis P, Sumer H, ChooKH: Building the centromere: from foundation proteins to 3D organization. Trends Cell Biol 2004, 14:359–368. [DOI] [PubMed] [Google Scholar]
- 5.McAinsh AD, Tytell JD, Sorger PK: Structure, function, and regulation of budding yeast kinetochores. Annu Rev Cell Dev Biol 2003, 19:519–539. [DOI] [PubMed] [Google Scholar]
- 6.Biggins S, Walczak CE: Captivating capture: how microtubules attach to kinetochores. Curr Biol 2003, 13:R449–R460. [DOI] [PubMed] [Google Scholar]
- 7.Fukagawa T: Centromere DNA, proteins and kinetochore assembly in vertebrate cells. Chromosome Res 2004, 12:557–567. [DOI] [PubMed] [Google Scholar]
- 8.Chen Y, Riley DJ, Chen PL, Lee WH: Hec, a novel nuclear protein rich in leucine heptad repeats specifically involved in mitosis. Mol Cell Biol 1997, 17:6049–6056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wigge PA, Jensen ON, Holmes S, Soues S, Mann M, Kilmartin JV: Analysis of the Saccharomyces spindle pole by matrix-assisted laser desorption/ionization (MALDI) mass spectrometry. J Cell Biol 1998, 141:967–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zheng L, Chen Y, Lee WH: Hec1p, an evolutionarily conserved coiled-coil protein, modulates chromosome segregation through interaction with SMC proteins. Mol Cell Biol 1999, 19:5417–5428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Howe M, McDonald KL, Albertson DG, Meyer BJ: HIM-10 is required for kinetochore structure and function on Caenorhabditis elegans holocentric chromosomes. J Cell Biol 2001, 153:1227–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Janke C, Ortiz J, Lechner J, Shevchenko A, Magiera MM, Schramm C, Schiebel E: The budding yeast proteins Spc24p and Spc25p interact with Ndc80p and Nuf2p at the kinetochore and are important for kinetochore clustering and checkpoint control. EMBO J 2001, 20:777–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nabetani A, Koujin T, Tsutsumi C, Haraguchi T, Hiraoka Y: A conserved protein, Nuf2, is implicated in connecting the centromere to the spindle during chromosome segregation: a link between the kinetochore function and the spindle checkpoint. Chromosoma 2001, 110:322–334. [DOI] [PubMed] [Google Scholar]
- 14.Wigge PA, Kilmartin JV: The Ndc80p complex from Saccharomyces cerevisiae contains conserved centromere components and has a function in chromosome segregation. J Cell Biol 2001, 152:349–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DeLuca JG, Moree B, Hickey JM, Kilmartin JV, Salmon ED: hNuf2 inhibition blocks stable kinetochore-microtubule attachment and induces mitotic cell death in HeLa cells. J Cell Biol 2002, 159:549–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Le Masson I, Saveanu C, Chevalier A, Namane A, Gobin R, Fromont-Racine M, Jacquier A, Mann C: Spc24 interacts with Mps2 and is required for chromosome segregation, but is not implicated in spindle pole body duplication. Mol Microbiol 2002, 43:1431–1443. [DOI] [PubMed] [Google Scholar]
- 17.Appelgren H, Kniola B, Ekwall K: Distinct centromere domain structures with separate functions demonstrated in live fission yeast cells. J Cell Sci 2003, 116:4035–4042. [DOI] [PubMed] [Google Scholar]
- 18.Desai A, Rybina S, Muller-Reichert T, Shevchenko A, Hyman A, OegemaK: KNL-1 directs assembly of the microtubule-binding interface of the kinetochore in C. elegans. Genes Dev 2003, 17:2421–2435.• The authors report the discovery of a novel C. elegans kinetochore protein, KNL-1, whose RNAi depletion results in a ‘kinetochore null’ phenotype with dramatic mitotic defects in chromosome alignment and segregation. KNL-1 interacts with CENP-C as well as Ndc80Hec1 and Nuf2HIM−10, both of which require KNL-1 to target to the kinetochore. Thus, KNL-1 acts downstream of CENP-C but upstream of outer kinetochore proteins. For related work describing a mammalian homolog, see [21••].
- 19.Hori T, Haraguchi T, Hiraoka Y, Kimura H, Fukagawa T: Dynamic behavior of Nuf2-Hec1 complex that localizes to the centrosome and centromere and is essential for mitotic progression in vertebrate cells. J CellSci 2003, 116:3347–3362. [DOI] [PubMed] [Google Scholar]
- 20.McCleland ML, Gardner RD, Kallio MJ, Daum JR, Gorbsky GJ, Burke DJ, Stukenberg PT: The highly conserved Ndc80 complex is required for kinetochore assembly, chromosome congression, and spindle checkpoint activity. Genes Dev 2003, 17:101–114.•• The authors use antibody inhibition in Xenopus egg extracts and in Xenopus cultured cells, as well as genetic analysis in budding yeast, to show a conserved role for Ndc80 and Nuf2 in chromosome segregation and spindle checkpoint activity. Immunoprecipitation experiments indicate that Ndc80 and Nuf2 exist in a larger complex. Kinetochore assembly experiments in immunodepleted extracts suggest that this group of proteins is necessary for outer kinetochore assembly. For related work, see [22•,23•,30••].
- 21.Cheeseman I, Niessen S, Anderson S: Hyndman F, Yates JR, Oegema K, Desai A: A conserved protein network controls assembly of the outer kinetochore and its ability to sustain tension. Genes Dev 2004, 18:2255–2268.•• By combining RNAi-based functional genomics and mass-spectrometry-based protein identification, the authors identify a network of 10 interacting kinetochore proteins in C. elegans that includes the conserved components Ndc80Hec1, Nuf2HIM10 and Mis12. Functional analysis partitioned this network into three classes and suggested potential roles for each class at the kinetochore. The authors also describe the existence of a related protein network in human cells that includes members of each functional class and four new human kinetochore proteins. For related work, see [18•].
- 22.Bharadwaj R, Qi W, Yu H: Identification of two novel components of the human NDC80 kinetochore complex. J Biol Chem 2004, 279:13076–13085. See annotation to [23•]. [DOI] [PubMed] [Google Scholar]
- 23.McCleland ML, Kallio MJ, Barrett-Wilt GA, Kestner CA, Shabanowitz J, Hunt DF, Gorbsky GJ, Stukenberg PT: The vertebrate Ndc80 complex contains Spc24 and Spc25 homologs, which are required to establish and maintain kinetochore-microtubule attachment. Curr Biol 2004, 14:131–137.• These studies provide further evidence for the conserved nature of the Ndc80 complex by identifying Spc24 and Spc25 orthologs in Xenopus [23•] and humans [22•]. Inhibition of these proteins leads to spindle morphology defects and failure of chromosome alignment. Loss of Mad1 [22•,23•] and Mad2 [22•] at kinetochores in inhibited cells is rescued to a certain extent after depolymerizing spindle microtubules with nocodazole. In contrast, disruption of Ndc80Hec1 kinetochore localization in Spc25-inhibited cells is not rescued by nocodazole treatment [22•]. For related work, see [20••,30••].
- 24.Rieder CL: The formation, structure, and composition of the mammalian kinetochore and kinetochore fibre. Int Rev Cytol 1982, 79:1–58. [DOI] [PubMed] [Google Scholar]
- 25.DeLuca JG, Dong Y, Hergert P, Strauss J, Hickey JM, Salmon ED, McEwen BF: Hec1 and Nuf2 are core components of the kinetochore outer plate essential for organizing microtubule attachment sites. Mol Biol Cell 2004, in press.•• The authors probe the ultrastructural consequences of Nuf2 siRNA in HeLa cells to reveal a severely disrupted outer kinetochore with little microtubule binding. In addition, they confirm localization of Nuf2 at the outer kinetochore plate, but not the corona, by immuno-electron microscopy. On the basis of these findings and others, a model is put forth involving the Ndc80 complex in the formation of structurally stable binding sites for the plus ends of microtubules. Upon depletion of Ndc80 complex components from kinetochores, other microtubule binding factors, such as MAPs and motors, may facilitate a few lateral or end- on kinetochore-microtubule attachments, although these are insufficient to support poleward forces at kinetochores.
- 26.McEwen BF, Heagle AB, Cassels GO, Buttle KF, Rieder CL: Kinetochore fiber maturation in PtK1 cells and its implications for the mechanisms of chromosome congression and anaphase onset. J Cell Biol 1997, 137:1567–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martin-Lluesma S, Stucke VM, Nigg EA: Role of Hec1 in spindle checkpoint signaling and kinetochore recruitment of Mad1/Mad2. Science 2002, 297:2267–2270. [DOI] [PubMed] [Google Scholar]
- 28.He X, Rines DR, Espelin CW, Sorger PK: Molecular analysis of kinetochore-microtubule attachment in budding yeast. Cell 2001, 106:195–206. [DOI] [PubMed] [Google Scholar]
- 29.De Wulf P, McAinsh AD, Sorger PK: Hierarchical assembly of the budding yeast kinetochore from multiple subcomplexes. Genes Dev 2003, 17:2902–2921.• In a biochemical tour de force, the authors develop a model for how ~60 budding yeast kinetochore proteins interact with one another. In addition to identifying six new kinetochore proteins, they propose that the kinetochore is organized into 14 distinct multi-protein complexes that either interact with centromeres or microtubules, or serve as linkers between the two. For related work, see [31••,35•,84•].
- 30.DeLuca JG, Howell BJ, Canman JC, Hickey JM, Fang G, Salmon ED: Nuf2 and Hec1 are required for retention of the checkpoint proteins Mad1 and Mad2to kinetochores. CurrBiol 2003, 13:2103–2109.•• The authors use live analysis of GFP-labeled Mad2 to detect a fivefold decrease in kinetochore levels during mitosis upon RNAi of Nuf2 or Ndc80HEC1 in HeLa cells. In the absence of microtubules, Mad1 and Mad2 localization is normal, suggesting kinetochore assembly is unperturbed and the unstable kinetochore-microtubule attachments that form in the absence of a functional Ndc80HEC1 complex are sufficient to partially deplete kinetochores of spindle checkpoint proteins. In addition, RNAi of Nuf2 reduces NDC80H binding at kinetochores, which is not restored upon depolymerization of spindle microtubules. For related work, see [20••,22•,23•].
- 31.Westermann S, Cheeseman IM, Anderson S, Yates JR III, Drubin DG, Barnes G: Architecture of the budding yeast kinetochore reveals a conserved molecular core. J Cell Biol 2003, 163:215–222.•• Mass spectrometry of affinity-purified kinetochore protein complexes leads to the identification of the Mis12Mtw1 complex, consisting of CENP-H Nnf1, Nsl1, Dsn1 and Mis12Mtw1. Using chromatin immunoprecipitation experiments in wild-type and mutant extracts, the authors test how this complex interacts with other kinetochore components. They propose a conserved molecular core of kinetochore proteins, including CENP-ACse4, CENP-CMif2, CENP-HNnf1 and Ndc80HEC1. For related work, see [29•,35•,84•].
- 32.Gillett ES, Espelin CW, Sorger PK: Spindle checkpoint proteins and chromosome-microtubule attachment in budding yeast. J Cell Biol 2004, 164:535–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Janke C, Ortiz J, Tanaka TU, Lechner J, Schiebel E: Four new subunits of the Dam1-Duo1 complex reveal novel functions in sister kinetochore biorientation. EMBO J 2002, 21:181–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li Y, Bachant J, Alcasabas AA, Wang Y, Qin J, Elledge SJ: The mitotic spindle is required for loading of the DASH complex onto the kinetochore. Genes Dev 2002, 16:183–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scharfenberger M, Ortiz J, Grau N, Janke C, Schiebel E, Lechner J: Nsl1p is essential for the establishment of bipolarity and the localization of the Dam-Duo complex. EMBO J 2003, 22:6584–6597.• These authors also identify the four-protein budding yeast Mis12Mtw1 complex. Focusing in on one of the members, Nsl1, they demonstrate that its presence is required for establishing chromosome biorientation, reminiscent of Aurora BIpl1 activity. However, unlike Aurora BIpl1, Nsl1 appears to promote alignment through the Dam1 complex, which fails to localize in the Nsl1 mutants. For related work, see [29•,31••,84•].
- 36.Meraldi P, Draviam VM, Sorger PK: Timing and checkpoints in the regulation of mitotic progression. Dev Cell 2004, 7:45–60.• This paper reveals that RNAi depletion of Nuf2/Ndc80HEC1 in HeLa cells results in varying levels of Mad1 and Mad2 recruitment to the kinetochore. Partial depletion leads to a slight decrease in Mad1/2 recruitment, while complete depletion abolishes any Mad1/2 targeting. Contrary to other reports [11,12,21••], the authors state that nocodazole treatment does not restore Mad 1/2 localization to kinetochores upon complete disruption of the Ndc80 complex.
- 37.Oegema K, Desai A, Rybina S, Kirkham M, Hyman AA: Functional analysis of kinetochore assembly in Caenorhabditis elegans. J Cell Biol 2001, 153:1209–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rieder CL, Salmon ED: The vertebrate cell kinetochore and its roles during mitosis. Trends Cell Biol 1998, 8:310–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ligon LA, Shelly SS, Tokito M, Holzbaur EL: The microtubule plus-end proteins EB1 and dynactin have differential effects on microtubule polymerization. Mol Biol Cell 2003, 14:1405–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Faulkner NE, Dujardin DL, Tai CY, Vaughan KT, O’Connell CB, Wang Y, Vallee RB: A role for the lissencephaly gene LIS1 in mitosis and cytoplasmic dynein function. Nat Cell Biol 2000, 2:784–791. [DOI] [PubMed] [Google Scholar]
- 41.Kaplan KB, Burds AA, Swedlow JR, Bekir SS, Sorger PK, Nathke IS: A role for the adenomatous polyposis coli protein in chromosome segregation. Nat Cell Biol 2001, 3:429–432. [DOI] [PubMed] [Google Scholar]
- 42.Nakamura M, Zhou XZ, Lu KP: Critical role for the EB1 and APC interaction in the regulation of microtubule polymerization. CurrBiol 2001, 11:1062–1067. [DOI] [PubMed] [Google Scholar]
- 43.Rogers SL, Rogers GC, Sharp DJ, Vale RD: Drosophila EB1 is important for proper assembly, dynamics, and positioning of the mitotic spindle. J Cell Biol 2002, 158:873–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tai CY, Dujardin DL, Faulkner NE, Vallee RB: Role of dynein, dynactin, and CLIP-170 interactions in LIS1 kinetochore function. J Cell Biol 2002, 156:959–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Coquelle FM, Caspi M, Cordelieres FP, Dompierre JP, Dujardin DL, Koifman C, Martin P, Hoogenraad CC, Akhmanova A, Galjart N et al. : LIS1, CLIP-170’s key to the dynein/dynactin pathway. Mol Cell Biol 2002, 22:3089–3102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Green RA, Kaplan KB: Chromosome instability in colorectal tumor cells is associated with defects in microtubule plus-end attachments caused by a dominant mutation in APC. J Cell Biol 2003, 163:949–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tirnauer JS, Canman JC, Salmon ED, Mitchison TJ: EB1 targets to kinetochores with attached, polymerizing microtubules. Mol Biol Cell 2002, 13:4308–4316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tirnauer JS, Grego S, Salmon ED, Mitchison TJ: EB1-microtubule interactions in Xenopus egg extracts: role of EB1 in microtubule stabilization and mechanisms of targeting to microtubules. Mol Biol Cell 2002, 13:3614–3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Akhmanova A, Hoogenraad CC, Drabek K, Stepanova T, Dortland B, Verkerk T, Vermeulen W, Burgering BM, De Zeeuw CI, Grosveld F et al. : Clasps are CLIP-115 and −170 associating proteins involved in the regional regulation of microtubule dynamics in motile fibroblasts. Cell 2001, 104:923–935. [DOI] [PubMed] [Google Scholar]
- 50.Maiato H, Fairley EA, Rieder CL, Swedlow JR, Sunkel CE, Earnshaw WC: Human CLASP1 is an outer kinetochore component that regulates spindle microtubule dynamics. Cell 2003, 113:891–904.• The authors report localization and functional studies of human CLASP1, which has a more peripheral localization (i.e. is closer to microtubules) than previously studied kinetochore components. CLASP1 inhibition leads to monopolar spindles with centrally localized chromosomes, which exhibit reduced oscillations. Measurements of kinetochore fiber lengths in live cells reveal an overall decrease in kinetochore-microtubule dynamics. The authors suggest that CLASP1 is required to promote polymerization of microtubules at kinetochores.
- 51.Inoue YH, do Carmo Avides M, Shiraki M, Deak P, Yamaguchi M, Nishimoto Y, Matsukage A, Glover DM: Orbit, a novel microtubule-associated protein essential for mitosis in Drosophila melanogaster. J Cell Biol 2000, 149:153–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Maiato H, Sampaio P, Lemos CL, Findlay J, Carmena M, Earnshaw WC, Sunkel CE: MAST/Orbit has a role in microtubule-kinetochore attachment and is essential for chromosome alignment and maintenance of spindle bipolarity. J Cell Biol 2002, 157:749–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Maddox P, Straight A, Coughlin P, Mitchison TJ, Salmon ED: Direct observation of microtubule dynamics at kinetochores in Xenopus extract spindles: implications for spindle mechanics. J Cell Biol 2003, 162:377–382.•• Fluorescent speckle microscopy of fluorescently labeled tubulin in spindles assembled in Xenopus egg extracts enables the authors to analyze in detail the dynamics of kinetochore fibers versus non-kinetochore microtubules during metaphase and anaphase. Although anaphase is dominated by depolymerization of microtubules at the pole, kinetochore-localized depolymerization occasionally occurs, and suggests that tension across the centromere governs kinetochore-mediated alterations in microtubule dynamics.
- 54.Khodjakov A, Copenagle L, Gordon MB, Compton DA, Kapoor TM: Minus-end capture of preformed kinetochore fibres contributes to spindle morphogenesis. J Cell Biol 2003, 160:671–683.•• The authors observe microtubule bundles growing outward from unattached kinetochores facing away from the mitotic spindle in mammalian cells. These bundles are observed looping back towards the spindle and being ‘captured’ and translocated to the spindle pole. This phenomenon occurs in normal cells and in cells recovering from treatment with monastrol (a drug that prevents spindle pole separation by inhibiting a mitotic kinesin and that leads to a monopolar spindle). This work suggests that chromosome attachment to the spindle can occur by capture of preformed kinetochore fibers. For supporting evidence in meiosis, see [55••].
- 55.Chen W, Zhang D: Kinetochore fibre dynamics outside the context of the spindle during anaphase. Nat Cell Biol 2004, 6:227–231.•• By laser-cutting kinetochore fibers of chromosomes in grasshopper spermatocytes, the authors reveal that kinetochore microtubules grow outward from the kinetochore to re-attach to the spindle poles. Only after this reattachment can chromatids undergo poleward movement during anaphase. This suggests that kinetochore-driven polymerization of microtubules may be an inherent property of kinetochores, and that anaphase in these spindles is primarily driven by depolymerization of microtubules at spindle poles. For supporting evidence in mitosis, see [54••].
- 56.Lawrence CJ, Dawe RK, Christie KR, Cleveland DW, Dawson SC, Endow SA, Goldstein LS, Goodson HV, Hirokawa N, Howard J et al. : A standardized kinesin nomenclature. J Cell Biol 2004, 167:19–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Desai A, Verma S, Mitchison TJ, Walczak CE: Kin I kinesins are microtubule-destabilizing enzymes. Cell 1999, 96:69–78. [DOI] [PubMed] [Google Scholar]
- 58.Hertzer KM, Ems-McClung SC, Walczak CE: Kin I kinesins: insights into the mechanism of depolymerization. Crit Rev Biochem Mol Biol 2003, 38:453–469. [DOI] [PubMed] [Google Scholar]
- 59.Ovechkina Y, Wordeman L: Unconventional motoring: an overview of the Kin C and Kin I kinesins. Traffic 2003,4:367–375. [DOI] [PubMed] [Google Scholar]
- 60.Hunter AW, Caplow M, Coy DL, Hancock WO, Diez S, Wordeman L, Howard J: The kinesin-related protein MCAK is a microtubule depolymerase that forms an ATP-hydrolyzing complex at microtubule ends. Mol Cell 2003, 11:445–457.•• Using stabilized microtubules polymerized with GMPCPP (a non-hydro-lysable GTP analog), the authors study the mechanism by which MCAK, a member of the kinesin-13 family, depolymerizes microtubules in vitro. Unlike motile kinesins, MCAK exhibits end-stimulated and tubulin-dimer-stimulated activity. Quantitative analysis of ATPase activity suggests a single binding site for MCAK at the end of each microtubule protofilament. In addition, the authors suggest that one-dimensional diffusion of MCAK along the microtubule lattice facilitates rapid targeting of MCAK to microtubule ends. Once it is at the end, the authors propose that MCAK acts processively to release multiple tubulin subunits per ATP hydrolysis event.
- 61.Maney T, Hunter AW, Wagenbach M, Wordeman L: Mitotic centromere-associated kinesin is important for anaphase chromosome segregation. J Cell Biol 1998, 142:787–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kline-Smith SL, Khodjakov A, Hergert P, Walczak CE: Depletion of centromeric MCAK leads to chromosome congression and segregation defects due to improper kinetochore attachments. Mol Biol Cell 2004, 15:1146–1159.• By disrupting centromeric MCAK with a dominant-negative GFP fusion, the authors observe the consequences on chromosome positioning without perturbing overall spindle morphology. Despite normal rates of chromosome and chromatid movement, chromosome congression and segregation are abnormal; in fact, some chromosomes remain at spindle poles or at the spindle equator even after anaphase has taken place. This is due to the presence of improperly positioned microtubule attachments at kinetochores, which inhibit chromosome movements. These findings imply that MCAK functions to depolymerize improperly attached microtubules at the centromere.
- 63.Walczak CE, Gan EC, Desai A, Mitchison TJ, Kline-Smith SL: The microtubule-destabilizing kinesin XKCM1 is required for chromosome positioning during spindle assembly. CurrBiol 2002, 12:1885–1889. [DOI] [PubMed] [Google Scholar]
- 64.Rogers GC, Rogers SL, Schwimmer TA, Ems-McClung SC, Walczak CE, Vale RD, Scholey JM, Sharp DJ: Two mitotic kinesins cooperate to drive sister chromatid separation during anaphase. Nature 2004, 427:364–370.•• This study explores the activity of the Drosophila kinesin-13 proteins Klp59C and Klp10A, which are shown to depolymerize microtubules in vitro. Antibody-mediated inhibition of KLP59C leads to chromosome congression defects and slowed chromosome segregation. Fluorescent speckle microscopy suggests that this slowed poleward chromatid rate results from a loss of kinetochore-mediated depolymerization of microtubules, causing the chromatids to be moved solely by the depolymerization of kinetochore microtubules at spindle poles, although the presence of improper kinetochore-microtubule attachments cannot be ruled out. The authors also suggest on the basis of similar experiments that KLP10A depolymerizes microtubules at spindle poles during anaphase.
- 65.Goshima G, Vale RD: The roles of microtubule-based motor proteins in mitosis: comprehensive RNAi analysis in the Drosophila S2 cell line. J Cell Biol 2003, 162:1003–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pearson CG, Maddox PS, Zarzar TR, Salmon ED, Bloom K: Yeast kinetochores do not stabilize Stu2p-dependent spindle microtubule dynamics. Mol Biol Cell 2003, 14:4181–4195.• The authors explore the relationship between kinetochores and spindle microtubule dynamics using stu2 mutants in budding yeast. The dramatic decrease in spindle (kinetochore and non-kinetochore) microtubule turnover in stu2 mutants is not affected by preventing kinetochore assembly using stringent ndc10 mutants. Surprisingly, a partial loss of function ndc10 mutant rescues the microtubule dynamics defects in the absence of Stu2 activity, suggesting that the kinetochore and Stu2 differentially regulate spindle microtubule turnover in budding yeast.
- 67.Kosco KA, Pearson CG, Maddox PS, Wang PJ, Adams IR, Salmon ED, Bloom K, Huffaker TC: Control of microtubule dynamics by Stu2p is essential for spindle orientation and metaphase chromosome alignment in yeast. Mol Biol Cell 2001, 12:2870–2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.van Breugel M, Drechsel D, Hyman A: Stu2p, the budding yeast member of the conserved Dis1/XMAP215 family of microtubule-associated proteins is a plus end-binding microtubule destabilizer. J Cell Biol 2003, 161:359–369.• The authors analyze Stu2 activity in vitro and find that this MAP specifically binds to microtubule plus ends and induces microtubule catastrophes (transition from microtubule growth to shrinkage) by slowing polymerization rates. For related work, see [70•].
- 69.Kinoshita K, Habermann B, Hyman AA: XMAP215: a key component of the dynamic microtubule cytoskeleton.Trends Cell Biol 2002, 12:267–273. [DOI] [PubMed] [Google Scholar]
- 70.Shirasu-Hiza M, Coughlin P, Mitchison T: Identification of XMAP215 as a microtubule-destabilizing factor in Xenopus egg extract by biochemical purification. J Cell Biol 2003, 161:349–358.• The authors use fractionated Xenopus egg extracts to identify XMAP215 as a strong promoter of microtubule destabilization. This activity is observed under conditions where microtubules are stabilized with GMPCPP (a non-hydrolyzable GTP analog) and little free tubulin is available. Both full-length protein and an N-terminal truncation of XMAP215 can depolymerize microtubules by increasing the rate of microtubule depolymerization, suggesting that the ability of XMAP215 to simultaneously increase microtubule lengths and turnover rates may be separable activities. For related work, see [68•].
- 71.Popov AV, Karsenti E: Stu2p and XMAP215: turncoat microtubule-associated proteins? Trends Cell Biol 2003, 13:547–550. [DOI] [PubMed] [Google Scholar]
- 72.Walczak CE, Mitchison TJ, Desai A: XKCM1: a Xenopus kinesin-related protein that regulates microtubule dynamics during mitotic spindle assembly. Cell 1996, 84:37–47. [DOI] [PubMed] [Google Scholar]
- 73.Tournebize R, Popov A, Kinoshita K, Ashford AJ, Rybina S, Pozniakovsky A, Mayer TU, Walczak CE, Karsenti E, Hyman AA: Control of microtubule dynamics by the antagonistic activities of XMAP215and XKCM1 in Xenopus egg extracts. Nat Cell Biol 2000, 2:13–19. [DOI] [PubMed] [Google Scholar]
- 74.Kinoshita K, Arnal I, Desai A, Drechsel DN, Hyman AA: Reconstitution of physiological microtubule dynamics using purified components. Science 2001, 294:1340–1343. [DOI] [PubMed] [Google Scholar]
- 75.Maney T, Wagenbach M, Wordeman L: Molecular dissection of the microtubule depolymerizing activity of mitotic centromere-associated kinesin. J Biol Chem 2001, 276:34753–34758. [DOI] [PubMed] [Google Scholar]
- 76.Kline-Smith SL, Walczak CE: The microtubule-destabilizing kinesin XKCM1 regulates microtubule dynamic instability in cells. Mol Biol Cell 2002, 13:2718–2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gergely F, Draviam VM, Raff JW: The ch-TOG/XMAP215 protein is essential for spindle pole organization in human somatic cells. Genes Dev 2003, 17:336–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Holmfeldt P, Stenmark S, Gullberg M: Differential functional interplay of TOGp/XMAP215 and the KinI kinesin MCAK during interphase and mitosis. EMBO J 2004, 23:627–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cassimeris L, Morabito J: TOGp, the human homolog of XMAP215/Dis1, is required for centrosome integrity, spindle pole organization, and bipolar spindle assembly. Mol Biol Cell 2004, 15:1580–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cimini D, Moree B, Canman JC, Salmon ED: Merotelic kinetochore orientation occurs frequently during early mitosis in mammalian tissue cells and error correction is achieved by two different mechanisms. J Cell Sci 2003, 116:4213–4225.• The authors have previously shown that cells recovering from nocodazole depolymerization of microtubules exhibit an increased frequency of improper microtubule attachments at kinetochores. Here, they find that improper attachments occur frequently in early mitosis in untreated cells, although virtually all of them are corrected and do not lead to missegregation of chromosomes. High-resolution fluorescence imaging suggests that error detection and correction mechanisms resolve these attachments both before and during anaphase.
- 81.Tanaka TU: Bi-orienting chromosomes on the mitotic spindle. Curr Opin Cell Biol 2002, 14:365–371. [DOI] [PubMed] [Google Scholar]
- 82.Biggins S, Murray AW: The budding yeast protein kinase Ipl1/ Aurora allows the absence of tension to activate the spindle checkpoint. Genes Dev 2001, 15:3118–3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dewar H, Tanaka K, Nasmyth K, Tanaka TU: Tension between two kinetochores suffices for their bi-orientation on the mitotic spindle. Nature 2004, 428:93–97.•• Using unreplicated dicentric and monocentric minichromosomes in budding yeast, the authors demonstrate that the tension created by the physical linkage of bi-oriented sister kinetochores is sufficient to ensure proper chromosome alignment and that Aurora BIpl1 and INCENPSli15 are required for this process. This idea is further supported by the finding that bi-orientation is partially restored in topoisomerase-II-defective cohesion mutants. For supporting evidence in vertebrates, see [96•].
- 84.Pinsky BA, Tatsutani SY, Collins KA, Biggins S: An Mtw1 complex promotes kinetochore biorientation that is monitored by the Ipl1/Aurora protein kinase. Dev Cell 2003, 5:735–745.• Using a Mis12Mtw1 mutant, the authors identify Dsn1 as a suppressor of mtw-1 and show that it is present in the Mis12Mtw1 complex. While the Mis12Mtw1 mutant appears unable to form kinetochore-microtubule attachments to some chromosomes, this defect is abolished when Aurora BIpl1 is also inactivated, suggesting that the Mis12Mtw1 mutant has improper attachments that are being detached by Aurora BIpl1 activity. For related work, see [29•,31••,35•].
- 85.Tanaka TU, Rachidi N, Janke C, Pereira G, Galova M, Schiebel E, Stark MJ, Nasmyth K: Evidence that the Ipl1—Sli15 (Aurora kinase-INCENP) complex promotes chromosome biorientation by altering kinetochore-spindle pole connections. Cell 2002, 108:317–329. [DOI] [PubMed] [Google Scholar]
- 86.Ditchfield C, Johnson VL, Tighe A, Ellston R, Haworth C, Johnson T, Mortlock A, Keen N, Taylor SS: Aurora B couples chromosome alignment with anaphase by targeting BubR1, Mad2 and Cenp-E to kinetochores. J Cell Biol 2003, 161:267–280.See annotation to [87•].
- 87.Hauf S, Cole RW, LaTerra S, Zimmer C, Schnapp G, Walter R, Heckel A, van Meel J, Rieder CL, Peters JM: The small molecule Hesperadin reveals a role for Aurora B in correcting kinetochore-microtubule attachment and in maintaining the spindle assembly checkpoint. J Cell Biol 2003, 161:281–294.• These studies explore the mitotic defects that ensue from small molecule inhibition of Aurora kinase using ZM447439 [86•] (also called AKI for Aurora kinase inhibitor [90•]), or Hesperadin [87•] in HeLa cells. Consistent with a role for Aurora B in the correction of improper kinetochore microtubule attachments, chromosome orientation and congression is defective in these cells. In addition, decreased levels of BubRI, CENP-E and Mad2 are found at kinetochores in ZM447439-treated cells [86•]. For supporting evidence, see [90•].
- 88.Shannon KB, Salmon ED: Chromosome dynamics: new light on Aurora B kinase function. Curr Biol 2002, 12:R458–R460. [DOI] [PubMed] [Google Scholar]
- 89.Kallio MJ, McCleland ML, Stukenberg PT, Gorbsky GJ: Inhibition of aurora B kinase blocks chromosome segregation, overrides the spindle checkpoint, and perturbs microtubule dynamics in mitosis. Curr Biol 2002, 12:900–905. [DOI] [PubMed] [Google Scholar]
- 90.Lampson MA, Renduchitala K, Khodjakov A, Kapoor TM: Correcting improper chromosome-spindle attachments during cell division. Nat Cell Biol 2004, 6:232–237.• Evidence is presented that hints at the mechanism by which vertebrate cells correct mal-oriented chromosomes. The authors first generate incorrect attachments by inhibiting spindle bipolarity using the drug monastrol, then observe how cells recover after monastrol washout. The authors directly observe the correction of mal-attached chromosomes at the spindle poles. This process appears to involve depolymerization of kinetochore fibers. Importantly, Aurora kinases are required for this process, as improper attachments persist upon addition of small molecule inhibitors of Aurora activity. For supporting evidence, see [86•,87•].
- 91.Li X, Nicklas RB: Tension-sensitive kinetochore phosphorylation and the chromosome distribution checkpoint in praying mantid spermatocytes. J Cell Sci 1997, 110:537–545. [DOI] [PubMed] [Google Scholar]
- 92.Nicklas RB, Ward SC, Gorbsky GJ: Kinetochore chemistry is sensitive to tension and may link mitotic forces to a cell cycle checkpoint. J Cell Biol 1995, 130:929–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cheeseman IM, Anderson S, Jwa M, Green EM, Kang J, Yates JR lll, Chan CS, Drubin DG, Barnes G: Phospho-regulation of kinetochore-microtubule attachments by the Aurora kinase Ipl1p. Cell 2002, 111:163–172. [DOI] [PubMed] [Google Scholar]
- 94.Tanaka T, Fuchs J, Loidl J, Nasmyth K: Cohesin ensures bipolar attachment of microtubules to sister centromeres and resists their precocious separation. Nat Cell Biol 2000, 2:492–499. [DOI] [PubMed] [Google Scholar]
- 95.Sonoda E, Matsusaka T, Morrison C, Vagnarelli P, Hoshi O, Ushiki T, Nojima K, FukagawaT, Waizenegger IC, Peters JM et al. : Scc1/Rad21/Mcd1 is required for sister chromatid cohesion and kinetochore function in vertebrate cells. Dev Cell 2001, 1:759–770. [DOI] [PubMed] [Google Scholar]
- 96.Vagnarelli P, Morrison C, Dodson H, Sonoda E, Takeda S, Earnshaw WC: Analysis of Scc1-deficient cells defines a key metaphase role of vertebrate cohesin in linking sister kinetochores. EMBO Rep 2004, 5:167–171.• The authors explore mitosis in a chicken DT40 cell line that is conditionally deficient in Scc1, a subunit of cohesin. These cells show metaphase alignment defects and high levels of the checkpoint protein BubR1 at kinetochores. Drug-mediated inhibition of topoisomerase II, the enzyme required for chromosome decatenation, rescues the alignment defect in Scc1-deficient cells and reduces BubR1 levels at kinetochores. As cohesion loss could be partially compensated for by catenation, the authors propose that physical linkage between sister chromatids capable of sustaining tension is required for bi-orientation. For supporting evidence in yeast, see [83••].
- 97.Salic A, Waters JC, Mitchison TJ: Vertebrate shugoshin links sister centromere cohesion and kinetochore microtubule stability in mitosis. Cell 2004, 118:567–578.• The authors identify a new microtubule-associated protein in vertebrates with similarities to Drosophila MEI-S332 and fungal Sgo1 (Shugoshin), proteins that previously have only been implicated in centromere cohesion during meiosis. Sgo binds and stabilizes microtubules in Xenopus egg extracts. In addition, Sgo is present at kinetochores in HeLa cells, where it appears to stabilize cohesive forces between sister chromatids and is also required for kinetochore-microtubule stability. Thus, Sgo analysis reveals an exciting potential link between chromatid cohesion and kinetochore-microtubule stability.
- 98.Andrews PD, Knatko E, Moore WJ, Swedlow JR: Mitotic mechanics: the auroras come into view. Curr Opin Cell Biol 2003, 15:672–683. [DOI] [PubMed] [Google Scholar]
- 99.Romano A, Guse A, Krascenicova I, Schnabel H, Schnabel R, Glotzer M: CSC-1: a subunit of the Aurora B kinase complex that binds to the survivin-like protein BIR-1 and the incenp-like protein ICP-1. J Cell Biol 2003, 161:229–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sampath SC, Ohi R, Leismann O, Salic A, Pozniakovski A, Funabiki H: The chromosomal passenger complex is required for chromatin-induced microtubule stabilization and spindle assembly. Cell 2004, 118:187–202. See annotation to [101•]. [DOI] [PubMed] [Google Scholar]
- 101.Gassmann R, Carvalho A, Henzing AJ, Ruchaud S, Hudson DF, Honda R, Nigg EA, Gerloff DL, Earnshaw WC: Borealin: a novel chromosomal passenger required for stability of the bipolar mitotic spindle. J Cell Biol 2004, 166:179–191.• Both groups report the identification of a new member of the chromosomal passenger complex in Xenopus [100•] and in human cells [100•,101•]. The homologous proteins Dasra A [100•] and Dasra B/ Borealin [100•,101•] co-precipitate with Aurora B, Survivin and INCENP, and localize to centromeres pre-anaphase and to the midzone thereafter. RNAi-induced depletion of Dasra B/Borealin in HeLa cells leads to chromosome misalignment and missegregation [100•,101•] as well as extra asters at the spindle poles [101•]. Experiments in frog egg extracts suggest that chromosomal passenger proteins regulate spindle formation through inhibition of the microtubule depolymerase activity of MCAK. The chromosomal enrichment of the passenger complex is proposed to provide a mechanism for microtubule stabilization in the vicinity of chromatin that is independent of the previously described Ran-GTP pathway [100•]. For related work, see [105••].
- 102.Kline-Smith SL, Walczak CE: Mitotic spindle assembly and chromosome segregation: refocusing on microtubule dynamics. Mol Cell 2004, 15:317–327. [DOI] [PubMed] [Google Scholar]
- 103.Andrews PD, Ovechkina Y, Morrice N, Wagenbach M, Duncan K, Wordeman L, Swedlow JR: Aurora B regulates MCAK at the mitotic centromere. Dev Cell 2004, 6:253–268. See annotation to [105••]. [DOI] [PubMed] [Google Scholar]
- 104.Lan W, Zhang X, Kline-Smith SL, Rosasco SE, Barrett-Wilt GA, Shabanowitz J, Hunt DF, Walczak CE, Stukenberg PT: Aurora B phosphorylates centromeric MCAK and regulates its localization and microtubule depolymerization activity. Curr Biol 2004, 14:273–286. See annotation to [105••]. [DOI] [PubMed] [Google Scholar]
- 105.Ohi R, Sapra T, Howard J, Mitchison TJ: Differentiation of cytoplasmic and meiotic spindle assembly MCAK functions by Aurora-B-dependent phosphorylation. Mol Biol Cell 2004, 15:2895–2906.•• These studies show that the kinesin-13 protein MCAK is phosphorylated at multiple sites by Aurora B kinase. This phosphorylation inhibits MCAK depolymerization of microtubules, and may also help load MCAK onto centromeres in mitosis. Surprisingly, phosphorylation of a single site (Ser196) in the kinesin neck region strongly inhibits Xenopus MCAK depolymerization of microtubules in vitro [104••]. Analyses of multi-site phosphorylation mutants and phosphoepitope antibody staining suggest that phosphorylation alters the subcellular location of MCAK at the centromere. This may provide a mechanism for Aurora B to direct MCAK to depolymerize specific subsets of microtubules. In addition, a non-phosphorylatable mutant of MCAK leads to monopolar spindles in Xenopus egg extracts without gross disruptions in microtubule polymer, suggesting that phosphorylation of MCAK is also important for bipolar spindle formation [105••]. For related work, see [100•,101•].
- 106.Ohi R, Coughlin ML, Lane WS, Mitchison TJ: An inner centromere protein that stimulates the microtubule depolymerizing activity of a KinI kinesin. Dev Cell 2003, 5:309–321.• In an effort to identify novel microtubule binding proteins from Xenopus egg extracts, the authors discover ICIS, an F-box domain protein with a novel role in regulating microtubule dynamics. ICIS increases the depolymerization activity of purified MCAK in vitro, immunoprecipitates with MCAK, INCENP and Aurora B from egg extracts, and is required for bipolar spindle formation. By immuno-electron microscopy, ICIS is localized to the inner centromere and on microtubules coming into contact with this region. On the basis of these findings, the authors propose that ICIS may be used at the centromere to enhance the depolymerase activity of MCAK during microtubule attachment correction.
- 107.Rieder CL, Khodjakov A: Mitosis through the microscope: advances in seeing inside live dividing cells. Science 2003, 300:91–96. [DOI] [PubMed] [Google Scholar]
- 108.Obuse C, Iwasaki O, Kiyomitsu T, Goshima G, Toyoda Y, Yanagida M: A conserved Mis12 centromere complex is linked to heterochromatic HP1 and outer kinetochore protein Zwint-1. Nat Cell Biol 2004, 6:1135–1141. [DOI] [PubMed] [Google Scholar]
- 109.Maiato H, Rieder CL and Khodjakov A: Kinetochore-driven formation of kinetochore fibers contributes to spindle assembly during animal mitosis. J Cell Biol 2004, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Maiato H, Khodjakov A and Rieder CL: Drosophila CLASP is required for microtubule subunit incorporation into fluxing kinetochore fibers. Nat Cell Biol 2004, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]