Abstract
Objective
Gut microbiome plays an important role in systemic inflammation and immune response. Microbes can translocate and reside in tumor niches. However, it is unclear how intra-tumor microbiome affects immunity in human cancer. The purpose of this study was to investigate the association between intra-tumor bacteria, infiltrating CD8+ T cells and patient survival in cutaneous melanoma.
Methods:
Using a TCGA cutaneous melanoma RNA-seq data, intra-tumor bacteria and infiltrating CD8+ T cells were determined. Correlation between intra-tumor bacteria and infiltrating CD8+ T cells or chemokine gene expression, and survival analysis of infiltrating CD8+ T cells and Lachnoclostridium in cutaneous melanoma were performed.
Results:
Patients with low levels of CD8+ T cells have significantly shorter survival than those with high one. The adjusted hazard ratio was 1.57 (low vs. high) (95% CI: 1.17–2.10, p = 0.002). Intra-tumor bacteria Lachnoclostridium genus ranked top in a positive association with infiltrating CD8+ T cells (correlation coefficient = 0.38, p = 9.4 x 10−14), followed by Gelidibacter (0.31, p = 1.13x 10−9), Flammeovirga (0.29, p = 1.96 x 10−8), and Acinetobacter (0.28, p = 8.94x 10−8). These intra-tumor genera positively correlated with chemokines CXCL9, CXCL10 and CCL5 expression. High Lachnoclostridium significantly reduced the mortality risk (p = 0.0003). However, no statistically significant correlation was observed between intra-tumor Lachnoclostridium abundance and the levels of either NK, B or CD4+ T cells.
Conclusion:
Intra-tumor-residing gut microbiota could modulate chemokine levels and affect CD8+ T cell infiltration, consequently influencing patient survival in cutaneous melanoma. Manipulating intra-tumor gut microbiome may benefit patient outcome for those with immunotherapy.
Keywords: CD8+ T cells, gut microbiome, intra-tumor bacteria, melanoma, prognosis
Introduction
Immune evasion is a hallmark of human cancer including melanoma, which plays an important role in the development and progression of the diseases. There are a variety of molecular mechanisms underlying this phenomenon. T-cell exhaustion, insufficient lymphocyte infiltration of lymphocytes (“cold” tumors or immune “desert”) and loss of neoantigens are the current main avenues of onco-immunology research under investigation.[1-4] The United States Food and Drug Administration (US FDA)-approved immune checkpoint inhibitors (ICIs), e.g. anti-PD-1/PD-L1 and anti-CTLA4, benefit patients by reinvigorating exhaustive CD8+ T cells residing in tumor tissues. However, not all patients respond to the blockade.[5-8]
It has been reported that a positive association exists between increased T cell infiltration and improved overall survival in advanced melanoma patients. [9, 10] Different strategies have been designed to convert immune ‘desert’ tumors to T cell-inflamed tumors (i.e. ‘hot’ tumors). One strategy is to engineer an oncolytic virus, in which granulocytes-macrophage colony-stimulating factor (GM-CSF) and a PD-L1 inhibitor are co-expressed and could promote the infiltration of T cells into tumors, therefore enhancing the immune response. [11] Talimogene laherparepvec (T-VEC) is a genetically modified type I herpes simplex virus expressing GM-CSF. It can induce the recruitment of T cells into melanoma, via modulating type 1 interferon-related factors and chemokines such as CXCL9 and CXCL10, consequently eliciting systemic anti-tumor specific CD8+ T cell response to shrink tumors.[12] Additionally, patients with advanced melanoma were intratumorally treated with interleukin-12 (IL-12)-expressing Tavokinogene Telseplasmid (TAVO) which caused increased T cell infiltration and tumor shrink.[13] These observations suggest that increasing the recruitment of T cells in tumors may improve patient outcome. Thus, better understanding of factors that affect the abundance of T cells in tumors is critical in improving the efficacy of immunotherapy.
Trillions of microbiota live in the niches of human body, including skin, mouth, gut and vagina. The gut microbiome has been implicated in the initiation and progression of human cancers. [14] Accumulating evidence shows that microbiota are involved in modulating the immune response of melanoma patients to anti-PD-1 immunotherapy.[15, 16] Dysbiosis-induced chronic inflammation can lead to the decline of immunosurveillance and damage of epithelial cell-formed barriers, consequently allowing microbial translocation. Recently, two independent studies have shown the presence of bacteria residing in tumors.[17, 18] By revisiting RNA sequencing (RNA-seq) and/or whole-genome sequencing data archived in The Cancer Genome Alta (TCGA), Poore and colleagues reported unique microbial signatures in tumor tissues and blood within and between most major types of cancer, and the association between the abundance of bacteria and virus infection in tumors.[17] Nejman and colleagues performed RNA-Seq, microscopy and cell culture analysis on 1526 tumors and their adjacent normal tissues for 7 cancer types including melanoma, and demonstrated that intratumor bacteria resided intracellularly in both tumor and immune cells. [18] They also found that patients’ response to immunotherapy had a distinct bacteria patterns with specific bacterial metabolic functions.[18] Previous studies have shown that the activation of Toll-like receptors (TLRs) by bacterial products can drive macrophages to immunosuppressive M2 polarization. [19, 20] However, there are no studies which have explored how intratumor bacteria affect infiltrating effector CD8+ T cell abundance in tumor and prognosis. The purpose of this study was to examine the association between intratumor bacteria and both the abundance of infiltrated CD8+ T cells and movement-related chemokines in cutaneous melanoma.
Materials and Methods
Study subjects and data sources
Upper quartile expectation maximum normalized mRNA levels in Fragments per Kilobase of transcript per Million mapped reads (FPKM) (RNA-seq V2 root-mean-squared error (RSEM)), RNA-seq raw data, and clinical data were retrieved from a The Cancer Genome Alta (TCGA) cutaneous melanoma dataset, which is available at the Genomic Data Commons (GDC) data portal (https://portal.gdc.cancer.gov).
Estimation of the refraction of infiltrated lymphocytes
The normalized mRNA expression obtained from the bulk tumor RNA-Seq data was used for the estimation of the fractions of infiltrated lymphocytes in the tumor for each individual, which was deconvoluted by the quanTIseq “Deconvolution” module in immundeconv R package. [21] The quanTIseq is a deconvolution algorithm for estimating the proportions of 10 different immune cell types (B cells, M1 and M2 macrophages, monocytes, neutrophils, natural killer (NK) cells, non-regulatory CD4+ T cells, CD8+ T cells, regulatory T (Treg) cells and myeloid dendritic cells (DC)) in an individual tumor sample under investigation using an immune cell signature gene matrix with the highest specificity and discriminative power. This algorithm uses a constrained least squares regression to ensure no negative values for the cell fractions and their sum not exceeding 1.
Analysis of intra-tumor microbiome
RNA-Seq reads in FASTA format were applied to analyze intra-tumor microbiome using Kraken TCGA microbial detection as previously described elsewhere.[17] Briefly, the pre-processed sequencing reads after the quality examination were aligned to human reference genomes for human transcript identification. The sequences that cannot be mapped to known human reference genomes were then aligned against all known bacterial and archaeal genomes using the ultrafast Kraken algorithm [22], which employs a window search with a default setting of 31-mers to match the k-mer against a database of microbial k-mer for taxonomic identification. With the removal of batch effects, taxonomic count data were normalized into log-count per million (log-cpm) using the Voom algorithm followed by supervised normalization (SNM).
Statistical analysis
Spearman or Pearson Correlation analysis was appropriately used to examine the association between intra-tumor bacteria and both T cell activation score and infiltrated lymphocytes based on the distribution. Overall survival months were calculated as the date from the diagnosis to either the last follow-up or the event (death), whichever one came first. Survival analysis was performed using Kaplan-Meier survival, and multivariate Cox regression model for covariates adjustment to estimate hazard ratios (HRs) and their 95% confidence intervals (95% CIs). All statistical analyses were performed in R (https://www.r-project.org/). Bonferroni correction was used for multiple comparisons. A two-sided p-value less than 0.05 was considered statistically significant.
Results
Patient characteristics
In this study, 447 cutaneous melanoma patients with RNA-seq data available were included, and their characteristics are shown as in supplementary Table 1. Median age was 58 years old (range 15-90), and majority were men (61.7%, n =276). Ninety-seven percent of the patients (n= 424) were non-Hispanic white, 2.8 %, Asian (n =12) and 0.2 % African American (n =1). Over half of patients (n =223) were diagnosed at an early disease stage, and 45.5% were diagnosed with an advanced disease stage. The median of Breslow depth was 3 mm with the range from 0 to 75 mm. Recurrence occurred in some patients at the early disease stage and for those patients, surgical resection was performed for metastatic lesions. For RNA-seq analysis, metastatic samples accounted for 78.3% (n=350), and primary samples 21.7% (n =97). The majority of patients (95.5%) did not receive adjuvant radiotherapy. Samples were provided prior to approval of active adjuvant therapies. The median of follow-up months was 37.6, with the range from 0.07 to 369.6 months. During the follow-up, 47.8% (n=213) patients died, and 52.2% were still alive or missed follow-up with unknown status.
Infiltrated CD8+ T cells and patient survival
Using the algorithm of quanTIseq on the bulk tumor RNA-seq, we estimated the relative abundance of infiltrating CD8+ T cells for each individual. The median of percentage of cellular content constituting CD8+ T cells was 0.79% with the range from 0 to 65.5%. Binarizing by the median, we grouped the patients into two groups, high and low. The association between the infiltrating CD8+ T cells and overall survival was first evaluated using log-rank Kaplan-Meier survival. Patients with high levels of infiltrating CD8+ T cells showed a significantly superior overall survival (log-rank p = 0.0009) (supplementary Figure 1). The median overall survival was 105.0 (95% CI: 66.6 – 167.9) months for patients with high CD8+ T cells, and 65.9 (48.9 – 89.1) months for those with low CD8+ T cells. With the adjustment for patients’ age at diagnosis, disease stage, and sex, a multivariate Cox regression model showed that an increased mortality risk was observed in patients with low infiltrated CD8+ T cells compared to those with high infiltrated CD8+ T cells (Supplementary Table 2). The adjusted HR was 1.57 (95% CI: 1.17 – 2.10, p =0.002). As expected, an elevated mortality risk was also found for those older or with an advanced disease. The adjusted HRs were 1.02 (1.01 – 1.03, p = 0.0002) for patients’ age at diagnosis, and 1.80 (1.34 – 2.42, p < 0.0001) for disease stage. However, there was no association between patients’ sex and mortality risk; the HR was 0.96 (0.71 – 1.30, p = 0.80).
When we examined data from metastatic lesions only (n =340), the statistical significance remained (log-rank p = 0.0014). Patients with high infiltrating CD8+ T cells survived approximately 39 months longer than those with low levels. The median overall survival was 107.0 months (95% CI: 69.0-174.7) for patients with high CD8+ T cells, and 68.0 months (50.7-94.9) for those with low levels (data not shown). Similarly, the adjusted HR was 1.57 (95% CI: 1.17-2.11, p = 0.002) (data not shown).
Similarly, we constructed Kaplan-Meier survival curves for B cells, NK cells and CD4+ T cells. Using the median proportion of either B or NK cells for binary grouping, we found that patients with either high levels of B or NK cells had a better but not statistically significant overall survival compared to those with low ones, respectively (supplementary Figure 2, and 3). The median overall survival was 92.9 (95% CI: 66.6 -148) months for patients with high B cells, and 65.8 (55.5-105) months for those with low B cells (log-rank p value = 0.43). For patients with high NK cells, the median overall survival was 94.9 (95% CI: 63.0-139) months, and 68.1 (61.5-103) months for those with low NK cells (log-rank p value = 0.38). Since the majority of the patients had no infiltrating CD4+ T cells, we used 0 as the cutoff for CD4+ T cells binary grouping (low and high levels). Kaplan-Meier survival curve showed that patients with low levels of CD4+ T cells had improved survival than those with high ones (log-rank p value = 0.00093) (supplementary Figure 4). The median overall survival was 102.0 (95% CI: 69.0-151) months for patients with low CD4+ T cells and 62.8 (49.5-85) months for those with high CD4+ T cells.
Association between intra-tumor bacteria and infiltrated CD8+ T cells
Using the robust algorithm and restricted criteria for bacterial identification, 369 patients were included. In total, 1630 OTUs were identified. The association was analyzed between identified intra-tumor bacteria and infiltrated CD8+ T cell levels. In total, 18 genera were associated with the infiltrated CD8+ T cells with a p value < 0.001. Two of them (Algibacter and Epilithonimonas) had a negative association with CD8+ T cells (correlation coefficients were - 0.19 and – 0.17, respectively, and the p values were 0.0003 and 0.0008, respectively). Sixteen genera had a positive correlation with infiltrated CD8+ cells with a correlation coefficient ranging from 0.17 to 0.38 (p values ranging from 0.0008 to 9.4x10−14) (Figure 1). The top 5 genera were Lachnoclostridium (correlation coefficient = 0.38, p = 9.4 x 10−14), followed by Gelidibacter (correlation coefficient = 0.31, p = 1.13x 10−9), Flammeovirga (correlation coefficient = 0.29, p = 1.96 x 10−8), Acinetobacter (correlation coefficient = 0.28, p = 8.94x 10−8) and Tropicibacter (correlation coefficient = 0.25, p = 1.44 x 10−6).
Figure 1.
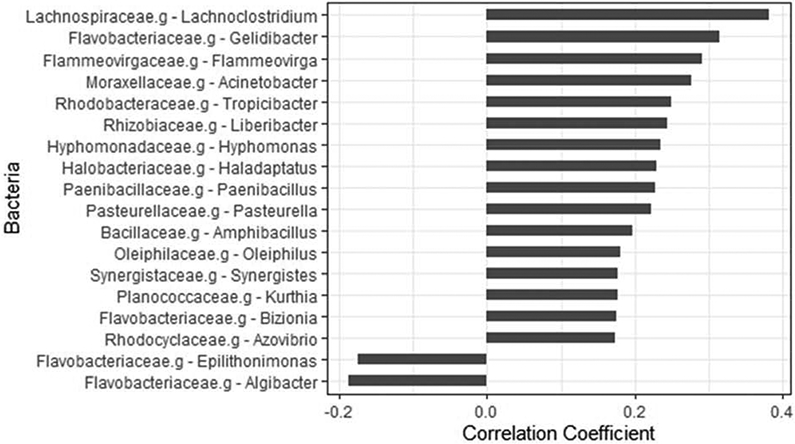
Association between intra-tumor bacteria and infiltrated CD8+ T cells in cutaneous melanoma. Bar plot showing the correlation coefficients for top 18 intra-tumor bacteria genera with a p value < 0.001.
Correlation between intra-tumor bacteria and the chemokines for CD8+ T cells
Given that CXCL9, CXCL10 and CCL5 are important chemokines for CD8+ T cell infiltration, we further examined the correlation between intra-tumor bacteria candidates that were significantly associated with CD8+ T cell infiltration as observed above. Table 1 shows the Spearman correlation coefficients and their corresponding p values. We found that the majority of the bacteria genera was positively associated with infiltrating CD8+ T cells, and had a significantly positive correlation with the chemokines CXCL9, CXCL10 and CCL5, whereas the two genera in negative association with infiltrating CD8+ T cells also had negative correlations with the chemokines. For instance, the Pearson correlation coefficients with Lachnoclostridium were 0.56 (p < 2.2 x10−16) for CXCL9, 0.56 (p < 2.2x 10−16) for CXCL10 and 0.57 (p < 2.2x10−16) for CCL5, respectively. In contrast, the correlation coefficients with Algibacter were −0.13 (p = 0. 009) for CXCL9, −0.09 (p = 0.083) for CXCL10, and −0.14 (p = 0.007) for CCL5, respectively.
Table 1.
Spearman correlation coefficients for the association between intra-tumor bacteria and chemokines for CD8+ T cell infiltration (n=369)
| CXCL9 |
CXCL10 |
CCL5 |
||||
|---|---|---|---|---|---|---|
| Variable | Correlation coefficient |
p-value | Correlation coefficient |
p-value | Correlation coefficient |
p-value |
| Lachnoclostridium | 0.56 | <2.2x10−16 | 0.56 | <2.2x10−16 | 0.57 | <2.2x10−16 |
| Gelidibacter | 0.25 | 7.8x10−7 | 0.23 | 8.4x10−6 | 0.25 | 8.5x10−7 |
| Flammeovirga | 0.45 | <2.2x10−16 | 0.41 | <2.2x10−16 | 0.44 | <2.2x10−16 |
| Acinetobacter | 0.46 | <2.2x10−16 | 0.44 | <2.2x10−16 | 0.46 | <2.2x10−16 |
| Tropicibacter | 0.24 | 3.9x10−6 | 0.21 | 2.2x10−5 | 0.25 | 9.3x10−7 |
| Liberibacter | 0.24 | 1.99x10−6 | 0.20 | 8.4x10−5 | 0.19 | 2.0x10−4 |
| Hyphomonas | 0.27 | 1.2x10−7 | 0.26 | 2.3x10−7 | 0.24 | 2.7x10−6 |
| Paenibacillus | 0.41 | <2.2x10−16 | 0.43 | <2.2x10−16 | 0.31 | 6.5x10−10 |
| Pasteurella | 0.26 | 2.5x10−7 | 0.25 | 1.3x10−6 | 0.17 | 7.9x10−4 |
| Amphibacillus | 0.12 | 0.018 | 0.10 | 0.048 | 0.09 | 0.069 |
| Oleiphilus | 0.06 | 0.278 | 0.03 | 0.529 | 0.12 | 0.024 |
| Synergistes | 0.03 | 0.575 | 0.04 | 0.448 | 0.05 | 0.335 |
| Kurthia | 0.25 | 7.5x10−7 | 0.26 | 3.6x10−7 | 0.13 | 0.009 |
| Bizionia | 0.16 | 0.002 | 0.13 | 0.013 | 0.13 | 0.014 |
| Azovibrio | 0.04 | 0.459 | 0.06 | 0.259 | −0.002 | 0.975 |
| Epilithonionas | −0.18 | 0.0005 | −0.17 | 0.001 | −0.16 | 0.0018 |
| Algibacter | −0.13 | 0.009 | −0.09 | 0.083 | −0.14 | 0.007 |
Association of intra-tumor Lachnoclostridium abundance and mortality
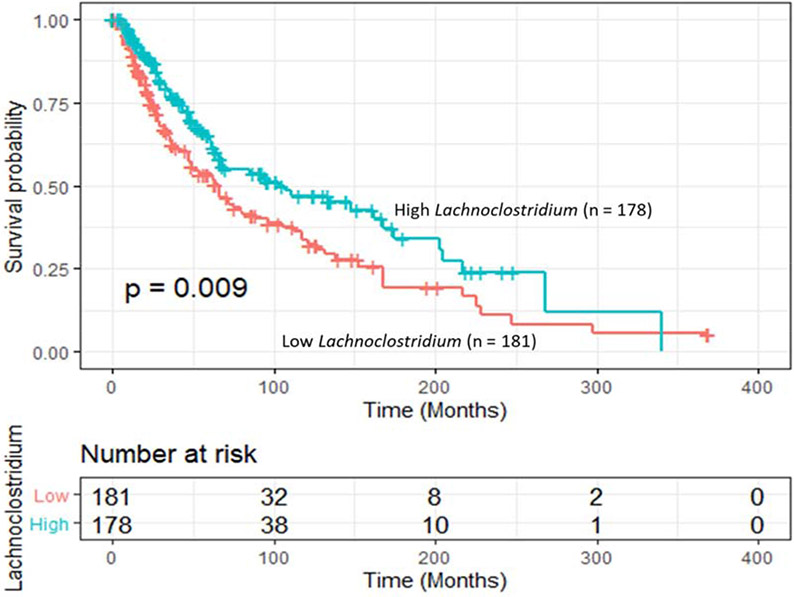
To investigate the association of intra-tumor Lachnoclostridium abundance and overall survival, we first classified the patients into two groups, low or high, based on the median of intra-tumor Lachnoclostridium abundance. Kaplan-Meier survival curves showed that patients with high levels of intra-tumor Lachnoclostridium had statistically significantly improved (appropriately 37.6 months longer) overall survival than those with low ones (log-rank p = 0.009) (Figure 2). The median of overall survival was 103.0 (95% CI: 68.0 – 174.7) months for high levels, and 65.4 (47.3 – 96.2) months for low levels of intra-tumor Lachnoclostridium, respectively.
Figure 2.
Kaplan-Meier overall survival curves stratified by intra-tumor Lachnoclostridium abundance in cutaneous melanoma. Patients with high Lachnoclostridium showed superior overall survival than those with low one (log-rank p value = 0.009).
Table 2 shows the multivariate Cox regression results of intra-tumor Lachnoclostridium abundance and the mortality in cutaneous melanoma, in which the median of Lachnoclostridium abundance was used as a cutoff to classify the patients into high and low groups. In the model 1 with whole group, patients with high abundance showed significantly decreased risk (p = 0.0003). The adjusted HR was 0.55 (95% CI: 0.40 – 0.76). Then we further stratified patients into two subgroups, high or low, based on CD8+ T cells. Among the patients with high levels of CD8+ T cells (model 2), those with a high abundance of Lachnoclostridium had a significantly lower mortality risk than those with a low one (p = 0.046). The adjusted HR was 0.57 (0.33 -0.99). Similarly, among patients with low levels of CD8+ T cells (model 3), a lower but not statistically significantly mortality risk was observed in those with a high abundance of Lachnoclostridium than those with a low one (p = 0.062). The adjusted HR was 0.63 (0.39-1.02). No interaction was found between CD8+ T cells and Lachnoclostridium abundance in patient survival (p value for interaction = 0.92).
Table 2.
Association of intra-tumor Lachnoclostridium abundance with mortality risk in cutaneous melanoma patients
| Variable | HR1 | 95% CI2 | p-value |
|---|---|---|---|
| Model 1 (whole group) | |||
| Lachnoclostridium (high/Low) | 0.55 | 0.40 – 0.76 | 0.0003 |
| Age | 1.02 | 1.01 - 1.03 | 0.0006 |
| Sex (male/female) | 0.91 | 0.66 – 1.29 | 0.634 |
| Disease Stage (III-IV/I-II) | 1.73 | 1.25 – 2.40 | 0.0009 |
| Model 2 (high CD8+ T cells) | |||
| Lachnoclostridium (high/Low) | 0.57 | 0.33 – 0.99 | 0.046 |
| Age | 1.04 | 1.02 – 1.06 | 0.0001 |
| Sex (male/female) | 0.86 | 0.50 – 1.47 | 0.580 |
| Disease Stage (III-IV/I-II) | 1.86 | 1.10 – 3.13 | 0.020 |
| Model 3 (low CD8+ T cells) | |||
| Lachnoclostridium (high/Low) | 0.63 | 0.39 – 1.02 | 0.062 |
| Age | 1.01 | 0.99 – 1.02 | 0.338 |
| Sex (male/female) | 0.92 | 0.60 – 1.43 | 0.717 |
| Disease Stage (III-IV/I-II) | 1.70 | 1.11 – 2.61 | 0.014 |
| p-value for interaction between Lachnoclostridium and CD8+ T cells | 0.92 | ||
HR: adjusted hazard ratio, which was estimated using multivariate Cox regression model.
CI: confidence interval.
Correlation between intra-tumor Lachnoclostridium abundance and B, NK and CD4+ T cells
Spearman correlation analysis showed that a negative but weak correlation between intra-tumor Lachnoclostridium abundance and NK cells, and the correlation coefficient was −0.12 (95% CI: −0.22- −0.01) with a p value of 0.03. A weak positive correlation was observed between intra-tumor Lachnoclostridium abundance and B cells, and the correlation coefficient was 0.09 (−0.01–0.19) with a p value of 0.09. There was a very weak negative correlation between intra-tumor Lachnoclostridium abundance and CD4+ T cells, and the correlation coefficient was −0.04 (−0.15– 0.06) with a p value of 0.42. After multiple comparison correction, none of three lymphocyte subtypes (NK, B and CD4+ T cells) had statistically significant correlation with intra-tumor Lachnoclostridium in cutaneous melanoma.
Discussion
In this study, we demonstrated the association between intra-tumor bacteria and infiltrated CD8+ T cells in cutaneous melanoma using the TCGA cutaneous melanoma RNA-seq database. We first estimated the relative abundance of infiltrated CD8+ T cells, which was shown in a positive associated with mortality risk in the patients as expected. Melanoma patients with a high CD8+ T cell infiltration survived > 3 years longer than those with a low CD8 T-cells. The association remained significant after the adjustment for age, sex and stage. This association held significant for metastatic cases only. This finding is concordance with previous studies.[23, 24]
Cytotoxic CD8+ T cells are a killer lymphocyte, eliminating tumor cells within favorable tumor niches. After the priming, CD8+ T cells, with the guidance of cytokine and chemokine signaling, migrate and infiltrate into tumor sites to execute its anti-tumor function. By engaging T cell receptors (TCR) on cytotoxic T cells with antigenic peptides-MHC-I complex, activated CD8+ T cells release IFN-γ, perforin (PRF2) and granzyme A/B (GZMA/B) to induce the necrosis and pyroptosis of tumor cells. Thus, the abundance of infiltrated CD8+ T cells has been reported in a positive association with better prognosis and response to several types of human cancer.[25] A meta-analysis of 16 studies reported that cancer patients with a high proportion of CD8+ T cells had a significantly reduced mortality risk compared to those with a low one, and the HR was 0.71 (95% CI: 0.62 – 0.82).[26] In the meta-analysis of 7 melanoma prognostic studies, a favorable prognostic significance of CD8+ T cell infiltration was observed with a HR of 0.50 (95% CI: 0.37- 0.69). [27] A recent study demonstrated that high intra-tumor not peri-tumor infiltration of CD8+ T cells particularly with low counts of CD163+ myeloid cells had a significant better survival and response to MAPK inhibitors than low CD8+ T cells and high CD163+ myeloid cells in melanoma patients, and the HR was 0.34 (95% CI: 0.16 – 0.72).[28] In immunotherapy-Naïve melanoma patients, CD8+ T cell infiltration was associated with improved overall survival and significantly expanded with the anti-PD-1 blockade.[29]
In this study, we did not find the statistically significant association of B cells with overall survival in cutaneous melanoma. This is in agreement with the report that there was no difference in infiltrated B cells between responders and non-responders to immunotherapy. [30] For NK cells, we found that patients with high abundance of NK cells survived approximately 26.8 months longer than those with low abundance, although the association was not statistically significant (p = 0.34). However, Cursons and colleagues recently reported a significantly positive association between higher NK cells and better survival in metastatic melanoma patients. [31] Although there is a discrepancy at statistical significance level in the findings between ours and Cursons’ study, the direction of the association in both studies is the same. This discrepancy may be due to different algorithms applied in estimating the abundance of NK cells. Cursons and colleagues developed a gene-set ranking score for NK cells based on the curation of LM22/CIBERSORT for active and resting NK cells and LM7 NK cell gene sets. In contrast, the quanTIseq algorithm was applied for estimation of total abundance NK cells (without further classifying active and resting NK cells) in our study. In addition, it has been reported that there are differences in performance across deconvolution pipelines for transcriptomics data.[32]
Interestingly, we found that patients with low CD4+ T cells had a significantly improved survival than those with high one. This finding seems inconsistent with the previous reports in which the role of CD4+ T cell was demonstrated in driving immune response against tumor. [2, 3, 33] These three previous studies showed that neoantigen vaccines induced CD4+ rather than CD8+ T cell response in either preclinical mouse tumor models or a clinical trial with a very small sample size. In these studies, synthetic peptides were used as vaccines for melanoma treatment, which might have stimulated the differentiation of polyfunctional CD4+ T cells into CD4+ T helper-1 (Th-1) cells, consequently activating CD8+ T cells and secreting effector cytokines, e.g., IFN-γ. In response to the context-dependent microenvironmental signals, versatile CD4+ T cells also can differentiate into other T helper subtypes, e.g., IL-17-releasing Th-17, IL-6- and IL-21-releaseing T-FH, and TGF-β- and IL-10-releasing induced T regulatory cells (iTreg) besides IFN-γ- and TNFα-releasing Th-1 and IL-4- and IL-13-releasing Th-2. [34] Given that immunosuppression is a hallmark of human cancer including melanoma, our finding of the association of high CD4+ T cells and poor survival suggests that CD4+ T cells most likely differentiated into iTreg more than into Th-1 cells in the immunosuppression context of melanoma patients. In agreement with this postulation, it has been reported that expanded unique Th-1-like CD4+ T cells were observed in non-tumor peripheral tissues in an active response to adjuvant therapy.[35] Moreover, in melanoma patients who had received anti-CTLA4 antibody and granulocyte-macrophage colony-stimulating factor (GM-CSF) therapy, circulating CD4+ T cells showed their anti-tumor properties. [35]
Besides their involvement in energy harvest and storage, accumulating evidence shows that gut microbiome also influences systemic inflammation and immunity. By fermenting indigestible fibers, gut microbiome produces short-chain fatty acids (SCFA), such as butyrate, acetate and propionate, which have an important anti-inflammatory activity. These SCFAs also participate intestinal homeostasis in the normal intestine by stimulating cell proliferation and differentiation. [36] Gut dysbiosis leads to the damage of intestinal barriers, allowing microbial translocation from its original niches to other niches where they survive. Recently, two studies used robust approaches to evidence the presence of intra-tumor bacteria.[17, 18] Cancer patients who responded to immunotherapy showed significantly distinct signature of intra-tumor microbiome profile compared to those non-responders.[18] In the extension and support of these studies, we found that intra-tumor bacteria were associated with the abundance of infiltrated CD8+ T cells in cutaneous melanoma. Out of the top 18 bacteria, the majority (n =16) were positively correlated with CD8+ T cell infiltration, with Lachnoclostridum ranking the top, whereas two genera (Elgibacter and Epilithonimonas) were negatively correlated with CD8+ T cell infiltration. Furthermore, survival analysis demonstrated approximately 41% reduction in the mortality risk of patients with the high abundance of intra-tumor Lachnoclostridium compared to those with low one. Our studies suggest that the manipulation of gut microbiomes (particularly Lachnoclostridium) may enhance the abundance of tumor infiltrating CD8+ T cells, thereby improving immunotherapy. Interestingly, no statistically significant correlation was found between intra-tumor Lachnoclostridium abundance and either NK, B or CD4+ T cells. Pre-clinical studies demonstrating translocation of bacteria from the gut to metastatic tumors are also warranted.
Lymphocyte infiltration is mainly governed by trafficking and adhesion-related molecules, cytokines and chemokines that bind to transmembrane G protein-coupled receptors. Through recognizing microbe-associated molecular patterns (MAMPs), such as bacterial peptidoglycan, flagellin and unmethylated bacterial DNA CpG motifs as antigens), TLRs trigger innate immunity, consequently initiating a complex cascade of signals, and activating a variety of gene expressions, e.g., chemokines, cytokines and other immune response. [37] In vitro cell lines and in vivo tumor-bearing immune-deficient mouse models showed that chemokines, such as CXCL9, CXCL10, CCL5, CCL17, CXCL12, and CXCL13, were expressed by tumor cells on exposure to gut bacteria, and defined chemokines were determinants for the tumor infiltration of distinct T cell subsets.[38] For example, CCL5, CXCL9 and CXCL10 are chemokines for attracting the infiltration of CD8+ T cells and T-helper 1 (Th-1) cells, whereas CCL17 and CXCL17 are for Th-1 and regulatory T cells (Treg). Different species (or genera) showed different capacities to induce the chemokines with distinct signatures. [38] In colorectal cancer (CRC) patients, there was a correlation between the abundance of defined bacteria and high chemokines, enhanced CD8+ T cell infiltration and improved prognosis.[38] For example, Lachnoclostrdium genus was positively correlated with CXCL9, CXCL10, and CD8+ T cells. In addition, microbiome-mediated metabolites may also participate the regulation of tumor infiltrating lymphocytes. For example, in liver cancer, gut microbiome-mediated bile acid metabolites could induce CXCL16 production, which drives the recruitment of NKT cells and consequently immune response against liver cancer.[39] Gut microbiome could promote the recruitment and responses of CD8+ T cells in CRC by altering all-trans-retinoic acid levels.[40, 41] Lachnospiraceae (Lachnoclostridium), a short-chain fatty acid producer, was significantly associated with decreased risk of CRC. [41] Consistent with this finding, previous studies showed that responders to immune checkpoint blockades with -long-term remission had a higher abundance of Lachnospiraceae than non-responders in melanoma and renal clear cell carcinoma and non-small cell lung cancer. In addition, increased CD8+ T cells were observed in responders’ tumors.[42-44] The results of two recently published clinical trials showed that Lachnospiraceae-enriched fecal microbiota transplant (FMT) improved anti-PD-1 immunotherapy, and increased the infiltration of CD8+ T cells in melanoma patients. [45, 46]_ It is interesting that different genera in the same family had opposite directions in association with the abundance of infiltrating CD8+ T cells and its related chemokines CXCL9, CXCL10 and CCL5. For example, in Flavobacteriaceae, Gelidibacter and Bizionia were positively associated with infiltrating CD8+ T cells and the chemokines, whereas Epilithonimonas and Algibacter were negatively associated with infiltrating CD8+ T cells and the chemokines. It is still unknown what component(s) or metabolite(s) determine the modulating effect on infiltration of CD8+ T cells.
One of the limitations of our study is a lack of information regarding systemic therapy, both in the adjuvant setting for advanced disease and for distant metastasis. Of note, the metastatic specimens were collected prior to widespread use of contemporary anti-PD-1-based regimens, and survival was likely not affected by these drugs in the metastatic disease setting. Lack of information on systemic therapy may lead to some bias in survival analysis of infiltrating CD8+ T cells. Thus, we need caution in interpreting the survival analysis results. However, the finding of the prognostic value of infiltrating CD8+ T cells is in agreement with the previous reports in melanoma patients, and is in line with the proof-of-concept of cytotoxic CD8+ T cells in human cancer. However, a strength in this study is a relatively large sample size acquired from multiple sites. In addition, the availability of adjuvant therapy information or not does not change the association of intra-tumor bacteria and infiltrating CD8+ T cells, which is a major question we aimed to address.
In summary, our study demonstrates an association between intra-tumor bacteria and infiltrating CD8+ T cells in cutaneous melanoma. By deep mining RNA-seq data from a large cohort from the TCGA, we found gut microbiomes residing in tumor tissues which are associated with abundance of infiltrating CD8+ T cells and chemokines for CD8+ T cell migration with different intensities and directions. Short-chain fatty acid-producer Lachnoclostridium ranked the top in positive association with infiltrating CD8+ T cells and chemokines CXCL9, CXCL10 and CCL5 in cutaneous melanoma tissue. We also observed that infiltrating CD8+ T cells and high Lachnoclostridium significantly benefit patient survival in cutaneous melanoma. To our knowledge, this is the first study to show that intra-tumor gut microbiomes could affect the abundance of infiltrating CD8+ T cells in cutaneous melanoma. These findings suggest that modulating gut microbiomes may be a novel approach in treating cutaneous melanoma. Particularly, it is warranted to explore whether or not and how Lachnoclostridium can enhance CD8+ T cell infiltration and improve immunotherapy using in vitro 3D organoids and in vivo animal models.
Supplementary Material
Highlight.
High infiltration of cytotoxic CD8+ T cells benefits melanoma patient outcomes.
Intratumor bacteria are associated with the infiltration of cytotoxic CD8+ T cells.
Intratumor bacteria might affect the chemokines for migration of CD8+ T cells.
High Lachnoclostridium load in tumor significantly reduced the mortality risk.
Acknowledgements:
We thank Drs. Rob Knight and Gregory D. Poore at University of California San Diego for providing intra-tumor microbiome data.
Funding:
This work was supported by the US NIH grants P50 CA121974, R01 CA227473, and R01 CA216846 (to Harriet Kluger).
Footnotes
Conflict of Interest: Dr Kluger has received research funding (institutional funding) from Merck, Bristol-Myers Squibb, and Apexigen, and personal fees from Corvus, Nektar, Pfizer, Iovance, Immunocore, Celldex, Array Biopharma, Merck, Bristol-Myers Squibb, Instilbio, Elevate Bio, Clinigen and Shionogi. The other authors have no conflict of interest to declare.
Ethical approval: All procedures performed in this study involving human subjects followed the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. For this type of study format consent is not required. The study presented here complies with the current laws of the United States of America.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Adams S, Gray RJ, Demaria S, Goldstein L, Perez EA, Shulman LN, et al. Prognostic value of tumor-infiltrating lymphocytes in triple-negative breast cancers from two phase III randomized adjuvant breast cancer trials: ECOG 2197 and ECOG 1199. J Clin Oncol 2014;32(27):2959–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sahin U, Derhovanessian E, Miller M, Kloke BP, Simon P, Lower M, et al. Personalized RNA mutanome vaccines mobilize poly-specific therapeutic immunity against cancer. Nature 2017;547(7662):222–226. [DOI] [PubMed] [Google Scholar]
- 3.Ott PA, Hu Z, Keskin DB, Shukla SA, Sun J, Bozym DJ, et al. An immunogenic personal neoantigen vaccine for patients with melanoma. Nature 2017;547(7662):217–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tirosh I, Izar B, Prakadan SM, Wadsworth MH 2nd, Treacy D, Trombetta JJ, et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 2016;352(6282):189–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weber JS, D'Angelo SP, Minor D, Hodi FS, Gutzmer R, Neyns B, et al. Nivolumab versus chemotherapy in patients with advanced melanoma who progressed after anti-CTLA-4 treatment (CheckMate 037): a randomised, controlled, open-label, phase 3 trial. Lancet Oncol 2015;16(4):375–84. [DOI] [PubMed] [Google Scholar]
- 6.Ribas A, Puzanov I, Dummer R, Schadendorf D, Hamid O, Robert C, et al. Pembrolizumab versus investigator-choice chemotherapy for ipilimumab-refractory melanoma (KEYNOTE-002): a randomised, controlled, phase 2 trial. Lancet Oncol 2015;16(8):908–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hodi FS, O'Day SJ, McDermott DF, Weber RW, Sosman JA, Haanen JB, et al. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med 2010;363(8):711–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Robert C, Thomas L, Bondarenko I, O'Day S, Weber J, Garbe C, et al. Ipilimumab plus dacarbazine for previously untreated metastatic melanoma. N Engl J Med 2011;364(26):2517–26. [DOI] [PubMed] [Google Scholar]
- 9.Kluger HM, Zito CR, Barr ML, Baine MK, Chiang VL, Sznol M, et al. Characterization of PD-L1 Expression and Associated T-cell Infiltrates in Metastatic Melanoma Samples from Variable Anatomic Sites. Clin Cancer Res 2015;21(13):3052–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vilain RE, Menzies AM, Wilmott JS, Kakavand H, Madore J, Guminski A, et al. Dynamic Changes in PD-L1 Expression and Immune Infiltrates Early During Treatment Predict Response to PD-1 Blockade in Melanoma. Clin Cancer Res 2017;23(17):5024–5033. [DOI] [PubMed] [Google Scholar]
- 11.Wang G, Kang X, Chen KS, Jehng T, Jones L, Chen J, et al. An engineered oncolytic virus expressing PD-L1 inhibitors activates tumor neoantigen-specific T cell responses. Nat Commun 2020;11(1):1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bommareddy PK, Zloza A, Rabkin SD, Kaufman HL. Oncolytic virus immunotherapy induces immunogenic cell death and overcomes STING deficiency in melanoma. Oncoimmunology 2019;8(7):1591875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Algazi A, Bhatia S, Agarwala S, Molina M, Lewis K, Faries M, et al. Intratumoral delivery of tavokinogene telseplasmid yields systemic immune responses in metastatic melanoma patients. Ann Oncol 2020;31(4):532–540. [DOI] [PubMed] [Google Scholar]
- 14.Farhana L, Banerjee HN, Verma M, Majumdar APN. Role of Microbiome in Carcinogenesis Process and Epigenetic Regulation of Colorectal Cancer. Methods Mol Biol 2018;1856:35–55. [DOI] [PubMed] [Google Scholar]
- 15.Gopalakrishnan V, Helmink BA, Spencer CN, Reuben A, Wargo JA. The Influence of the Gut Microbiome on Cancer, Immunity, and Cancer Immunotherapy. Cancer Cell 2018;33(4):570–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gopalakrishnan V, Spencer CN, Nezi L, Reuben A, Andrews MC, Karpinets TV, et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science 2018;359(6371):97–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Poore GD, Kopylova E, Zhu Q, Carpenter C, Fraraccio S, Wandro S, et al. Microbiome analyses of blood and tissues suggest cancer diagnostic approach. Nature 2020;579(7800):567–574. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Nejman D, Livyatan I, Fuks G, Gavert N, Zwang Y, Geller LT, et al. The human tumor microbiome is composed of tumor type-specific intracellular bacteria. Science 2020;368(6494):973–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liew FY, Xu D, Brint EK, O'Neill LA. Negative regulation of toll-like receptor-mediated immune responses. Nat Rev Immunol 2005;5(6):446–58. [DOI] [PubMed] [Google Scholar]
- 20.Hatziapostolou M, Polytarchou C, Aggelidou E, Drakaki A, Poultsides GA, Jaeger SA, et al. An HNF4alpha-miRNA inflammatory feedback circuit regulates hepatocellular oncogenesis. Cell 2011;147(6):1233–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Finotello F, Mayer C, Plattner C, Laschober G, Rieder D, Hackl H, et al. Molecular and pharmacological modulators of the tumor immune contexture revealed by deconvolution of RNA-seq data. Genome Med 2019;11(1):34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wood DE, Salzberg SL. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol 2014;15(3):R46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gide TN, Quek C, Menzies AM, Tasker AT, Shang P, Holst J, et al. Distinct Immune Cell Populations Define Response to Anti-PD-1 Monotherapy and Anti-PD-1/Anti-CTLA-4 Combined Therapy. Cancer Cell 2019;35(2):238–255 e6. [DOI] [PubMed] [Google Scholar]
- 24.Van Allen EM, Miao D, Schilling B, Shukla SA, Blank C, Zimmer L, et al. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science 2015;350(6257):207–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tumeh PC, Harview CL, Yearley JH, Shintaku IP, Taylor EJ, Robert L, et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature 2014;515(7528):568–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gooden MJ, de Bock GH, Leffers N, Daemen T, Nijman HW. The prognostic influence of tumour-infiltrating lymphocytes in cancer: a systematic review with meta-analysis. Br J Cancer 2011;105(1):93–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fu Q, Chen N, Ge C, Li R, Li Z, Zeng B, et al. Prognostic value of tumor-infiltrating lymphocytes in melanoma: a systematic review and meta-analysis. Oncoimmunology 2019;8(7):1593806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Massi D, Rulli E, Cossa M, Valeri B, Rodolfo M, Merelli B, et al. The density and spatial tissue distribution of CD8(+) and CD163(+) immune cells predict response and outcome in melanoma patients receiving MAPK inhibitors. J Immunother Cancer 2019;7(1):308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Edwards J, Wilmott JS, Madore J, Gide TN, Quek C, Tasker A, et al. CD103(+) Tumor-Resident CD8(+) T Cells Are Associated with Improved Survival in Immunotherapy-Naive Melanoma Patients and Expand Significantly During Anti-PD-1 Treatment. Clin Cancer Res 2018;24(13):3036–3045. [DOI] [PubMed] [Google Scholar]
- 30.Liu D, Schilling B, Liu D, Sucker A, Livingstone E, Jerby-Arnon L, et al. Integrative molecular and clinical modeling of clinical outcomes to PD1 blockade in patients with metastatic melanoma. Nat Med 2019;25(12):1916–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cursons J, Souza-Fonseca-Guimaraes F, Foroutan M, Anderson A, Hollande F, Hediyeh-Zadeh S, et al. A Gene Signature Predicting Natural Killer Cell Infiltration and Improved Survival in Melanoma Patients. Cancer Immunol Res 2019;7(7):1162–1174. [DOI] [PubMed] [Google Scholar]
- 32.Avila Cobos F, Alquicira-Hernandez J, Powell JE, Mestdagh P, De Preter K. Benchmarking of cell type deconvolution pipelines for transcriptomics data. Nat Commun 2020;11(1):5650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kreiter S, Vormehr M, van de Roemer N, Diken M, Lower M, Diekmann J, et al. Mutant MHC class II epitopes drive therapeutic immune responses to cancer. Nature 2015;520(7549):692–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tay RE, Richardson EK, Toh HC. Revisiting the role of CD4(+) T cells in cancer immunotherapy-new insights into old paradigms. Cancer Gene Ther 2021;28:5–17. doi: 10.1038/s41417-020-0183-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spitzer MH, Carmi Y, Reticker-Flynn NE, Kwek SS, Madhireddy D, Martins MM, et al. Systemic Immunity Is Required for Effective Cancer Immunotherapy. Cell 2017;168(3):487–502 e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parada Venegas D, De la Fuente MK, Landskron G, Gonzalez MJ, Quera R, Dijkstra G, et al. Short Chain Fatty Acids (SCFAs)-Mediated Gut Epithelial and Immune Regulation and Its Relevance for Inflammatory Bowel Diseases. Front Immunol 2019;10:277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Belkaid Y, Hand TW. Role of the microbiota in immunity and inflammation. Cell 2014;157(1):121–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cremonesi E, Governa V, Garzon JFG, Mele V, Amicarella F, Muraro MG, et al. Gut microbiota modulate T cell trafficking into human colorectal cancer. Gut 2018;67(11):1984–1994. [DOI] [PubMed] [Google Scholar]
- 39.Ma C, Han M, Heinrich B, Fu Q, Zhang Q, Sandhu M, et al. Gut microbiome-mediated bile acid metabolism regulates liver cancer via NKT cells. Science 2018;360(6391). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bhattacharya N, Yuan R, Prestwood TR, Penny HL, DiMaio MA, Reticker-Flynn NE, et al. Normalizing Microbiota-Induced Retinoic Acid Deficiency Stimulates Protective CD8(+) T Cell-Mediated Immunity in Colorectal Cancer. Immunity 2016;45(3):641–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu AI, Zhao L, Eaton KA, Ho S, Chen J, Poe S, et al. Gut Microbiota Modulate CD8 T Cell Responses to Influence Colitis-Associated Tumorigenesis. Cell Rep 2020;31(1):107471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Routy B, Le Chatelier E, Derosa L, Duong CPM, Alou MT, Daillere R, et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science 2018;359(6371):91–97. [DOI] [PubMed] [Google Scholar]
- 43.Chaput N, Lepage P, Coutzac C, Soularue E, Le Roux K, Monot C, et al. Baseline gut microbiota predicts clinical response and colitis in metastatic melanoma patients treated with ipilimumab. Ann Oncol 2017;28(6):1368–1379. [DOI] [PubMed] [Google Scholar]
- 44.Frankel AE, Deshmukh S, Reddy A, Lightcap J, Hayes M, McClellan S, et al. Cancer Immune Checkpoint Inhibitor Therapy and the Gut Microbiota. Integr Cancer Ther 2019;18:1534735419846379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baruch EN, Youngster I, Ben-Betzalel G, Ortenberg R, Lahat A, Katz L, et al. Fecal microbiota transplant promotes response in immunotherapy-refractory melanoma patients. Science 2021;371(6529):602–609. [DOI] [PubMed] [Google Scholar]
- 46.Davar D, Dzutsev AK, McCulloch JA, Rodrigues RR, Chauvin JM, Morrison RM, et al. Fecal microbiota transplant overcomes resistance to anti-PD-1 therapy in melanoma patients. Science 2021;371(6529):595–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.