Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants continue to emerge during the global pandemic and may facilitate escape from current antibody therapies and vaccine protection. Here we showed that the South African variant B.1.351 was the most resistant to current monoclonal antibodies and convalescent plasma from coronavirus disease 2019 (COVID-19)-infected individuals, followed by the Brazilian variant P.1 and the United Kingdom variant B.1.1.7. This resistance hierarchy corresponded with Y144del and 242–244del mutations in the N-terminal domain and K417N/T, E484K, and N501Y mutations in the receptor-binding domain (RBD) of SARS-CoV-2. Crystal structure analysis of the B.1.351 triple mutant (417N-484K-501Y) RBD complexed with the monoclonal antibody P2C-1F11 revealed the molecular basis for antibody neutralization and escape. B.1.351 and P.1 also acquired the ability to use mouse and mink ACE2 receptors for entry. Our results demonstrate major antigenic shifts and potential broadening of the host range for B.1.351 and P.1 variants, which poses serious challenges to current antibody therapies and vaccine protection.
Keywords: SARS-CoV-2, variant of concern, immune escape, antibody, neutralization
Graphical abstract
SARS-CoV-2 variants continue to emerge and spread around the world. Wang et al. conduct comprehensive mutational and crystal structure analyses of the variants and show that variants of concern, and the South African variant B.1.351 in particular, are resistant to many monoclonal antibodies and COVID-19 convalescent plasma and acquire the ability to use mouse and mink ACE2 receptors for infection.
Introduction
Current neutralizing antibody and vaccine strategies against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causative agent of coronavirus disease 2019 (COVID-19), were developed by targeting the prototype SARS-CoV-2 strain identified during the early phase of the pandemic (Cao et al., 2020; Corbett et al., 2020; Hansen et al., 2020; Ju et al., 2020; Liu et al., 2020; Pinto et al., 2020; Robbiani et al., 2020; Vogel et al., 2021; Wang et al., 2020; Wu et al., 2020; Zhou et al., 2020; Zost et al., 2020). Several regulatory agencies recently approved a selection of these antibodies and vaccines for emergency use authorization (EUA), and their rollout to high-risk populations began late in 2020. Eli Lilly and Regeneron developed neutralizing monoclonal antibodies (mAbs) targeting the receptor-binding domain (RBD) of the spike (S) protein that have been shown to reduce the viral loads of affected individuals, COVID-19-related symptoms, and hospitalizations (Chen et al., 2021; Gottlieb et al., 2021; Weinreich et al., 2021). We and others also reported the isolation and characterization several hundred RBD-specific mAbs from SARS-CoV-2-infected individuals (Cao et al., 2020; Hansen et al., 2020; Ju et al., 2020; Liu et al., 2020; Shi et al., 2020; Zost et al., 2020), some of which are under active clinical development. Pfizer/BioNtech and Moderna developed mRNA vaccines that express a stabilized form of the S protein, which demonstrated about 95% efficacy against symptomatic infection and a reduced risk of severe disease (Baden et al., 2021; Polack et al., 2020). Additional vaccine modalities, such as adenovirus-based, protein subunit, and inactivated vaccines, have also demonstrated reasonably good efficacy (Johnson & Johnson, 2021; Novavax, 2021; Voysey et al., 2021; Xia et al., 2021).
However, there is growing concern that the epidemic raging worldwide may be generating new SARS-CoV-2 variants that are antigenically distinct from the prototype strain, rendering current antibody and vaccine strategies ineffective (Callaway, 2021). By May 17, 2021 approximately 1,200,000 SARS-CoV-2 genome sequences were listed in the GISAID database, and more than 6,200 types of amino acid substitutions, deletions, and insertions have been found at least once in the S protein. Of the latter, 1,818 are in the N-terminal domain (NTD), 968 in the RBD, 741 in the subdomains 1 and 2 (SD1-2), and 2,481 in the S2 region (GISAID, 2021). Although these mutations are driven by the intrinsic, error-prone nature of the viral encoded RNA-dependent RNA polymerase (RdRp), their survival and maintenance rely on selective advantage during natural infection, transmission, and host adaptation. For example, the S protein D614G mutation became dominant just a few months into the pandemic and is associated with greater infectivity and transmissibility as well as moderately decreased susceptibility to antibody neutralization (Hou et al., 2020; Korber et al., 2020; Plante et al., 2021; Volz et al., 2021; Weissman et al., 2021; Yurkovetskiy et al., 2020). However, recently identified variants of SARS-CoV-2 (B.1.1.7 in the United Kingdom, B.1.351 in South Africa, and P.1 in Brazil) are raising serious concerns (England, 2020; Fujino et al., 2021; Kupferschmidt, 2021; Maggi et al., 2021; Tegally et al., 2021). They are not only rapidly displacing local SARS-CoV-2 strains but also carry NTD and RBD mutations that are critical for interactions with the ACE2 receptor and neutralizing antibodies (Barnes et al., 2020; Lan et al., 2020; Rambaut et al., 2020; Wang et al., 2021a; Yuan et al., 2020a). Specifically, B.1.1.7, B.1.351, and P.1 share the N501Y mutation, shown previously to enhance binding affinity to ACE2 (Laffeber et al., 2021; Tian et al., 2021). B.1.351 and P.1 each have three mutation sites in common within the RBD—K417N/T, E484K, and N501Y—which may change their antigenic profile. In addition, various deletion mutants are found in the NTD, such as 69–70del and Y144del in B.1.1.7 and 242–244del in B.1.351 (Rambaut et al., 2020; Tegally et al., 2021). A few mutations in the SD1-2 region near the Furin cleavage site are also identified, such as P681H in B.1.1.7 and A701V in B.1.351. Because all of these mutations fall in or are proximal to major S protein antigenic sites, they may adversely affect antibody neutralization induced by natural infection or vaccination.
To test the effect of these mutations on neutralization sensitivity, we generated a panel of 28 SARS-CoV-2 pseudoviruses corresponding to the identified mutations. These included 14 single mutations in the B.1.1.7, B.1.351, or P.1 variants, the K417N-E484K-N501Y mutation, and highly prevalent single mutations in the NTD, RBD, SD1-2, and S2 domains, identified across the entire S protein in the GISAID database (Figure 1 A). The pseudoviruses were then subjected to neutralization tests against 12 human neutralizing mAbs, including some that had received EUA, some isolated initially from SARS-CoV-2-infected individuals in our laboratory, and 23 convalescent plasma samples collected from individuals with SARS-CoV-2 between January and February 2020. Our results show that B.1.351 was the most resistant to mAbs and convalescent plasma neutralization, followed by P.1 and then B.1.1.7. Such a resistance hierarchy was attributed to the extent of mutations identified in the NTD and RBD. Crystal structural analysis of the RBD carrying the triple K417N-E484K-N501Y mutation found in B.1.351 bound with the mAb P2C-1F11 revealed a molecular basis for antibody neutralization and escape. We also show that mutations acquired by B.1.351 and P.1 substantially improve their ability to use mouse and mink ACE2 for entry. These results demonstrate major antigenic shifts and the potential to broaden the host range of B.1.351 and P.1, posing serious challenges to current antibody therapies and vaccine protection.
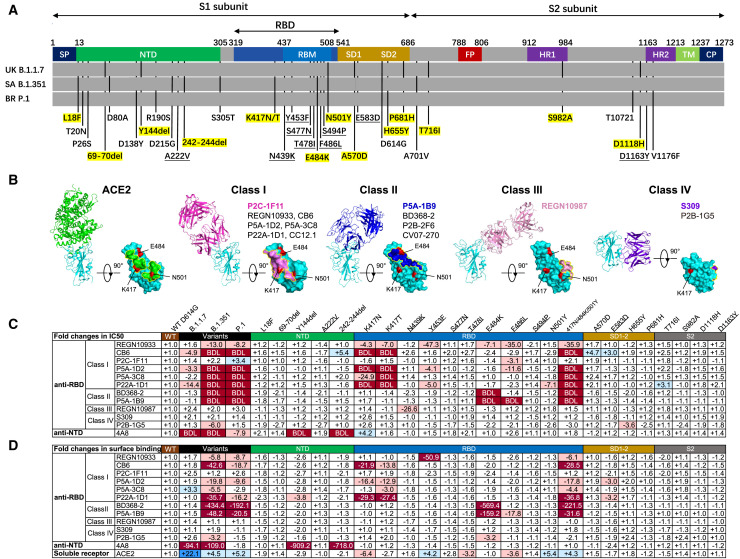
Figure 1.
SARS-CoV-2 variants and single amino acid mutations reduce neutralization and binding of mAbs
(A) Mutant residues along the S protein identified in SARS-CoV-2 variants B.1.1.7, B.1.351, and P.1. Mutant residues in each variant are indicated by black vertical lines. Those highlighted in yellow represent the mutant residues studied here. Nine mutant residues are underlined that were not identified in the three variants but included in the study because of their high representation in the GISAID database. SP, signal peptide; NTD, N-terminal domain; RBD, receptor-binding domain; RBM, receptor binding motif; SD, subdomain; FP, fusion peptide; HR1, heptad repeat 1; HR2, heptad repeat 2; TM, transmembrane domain; CP, cytoplasmic domain.
(B) The binding mode to and footprint of ACE2 and four classes of antibodies on the RBD. ACE2 is colored in green, RBD in cyan, and representative antibodies in four distinct colors. The three mutation residues (K417, E484, and N501) critical for binding to ACE2 and antibodies are shown in red.
(C and D) The fold changes in neutralizing activity, measured by half-maximal inhibitory concentration (IC50) (C), and in binding activity, measured by mean fluorescence intensity (MFI), relative to that of WT D614G. “−” indicates increased resistance and “+” increased sensitivity. IC50 or MFI values highlighted in red indicate that resistance increased at least 3-fold; in blue, sensitivity increased at least threefold; and in white, resistance or sensitivity increased less than 3-fold. BDL (below the detection limit) indicates that the highest concentration of mAbs failed to reach 50% neutralization. Results were calculated from three independent experiments performed in technical duplicates.
See also Figures S1 and S2.
Results
SARS-CoV-2 variants show decreased susceptibility to mAb neutralization
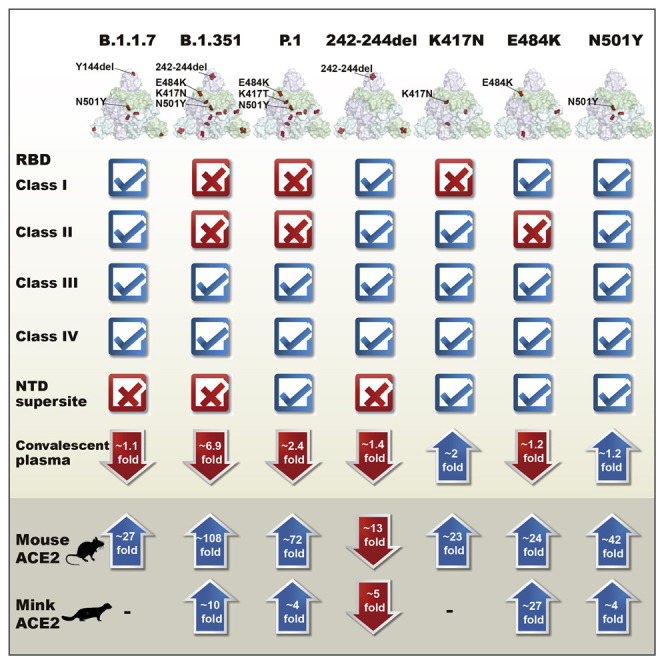
We first evaluated the suspectability of the 28 pseudoviruses to neutralization by 12 mAbs, including 11 anti-RBD mAbs (4 mAbs approved with EUA and 7 other typical antibodies identified from SARS-CoV-2-infected individuals) and 1 epitope, well-defined anti-NTD mAbs 4A8. The anti-RBD mAbs fall into one of four major classes based on structural features affecting their mode of recognition and epitope specificity (Figure 1B; Barnes et al., 2020; Du et al., 2020; Ge et al., 2021; Hansen et al., 2020; Ju et al., 2020; Kreye et al., 2020; Pinto et al., 2020; Shi et al., 2020; Yuan et al., 2020a). These epitope assignments are related to the six major sites put forward by Yuan et al. (2021). The wild-type (WT) pseudovirus used throughout the analysis was the prototype strain with a D614G mutation (WT D614G). As shown in Figure 1C and Figure S1, the B.1.351 and P.1 pseudoviruses were more resistant to mAbs in terms of magnitude and breadth of neutralization relative to B.1.1.7. The B.1.351 and P.1 pseudoviruses were fully resistant to almost all class I and class II anti-RBD mAbs except for P2C-1F11, whereas class III and class IV were largely unaffected. Many mAbs had neutralizing activity below the detection limit (BDL) even when tested at the highest concentration (Figure 1C). The three variants were also resistant to the anti-NTD mAb 4A8, with the B.1.1.7 and B.1.351 pseudoviruses showing greater resistance relative to P.1.
Examination of the resistance patterns across the single- and triple-mutant pseudoviruses showed that the K417N and K417T mutations correlated with resistance to class I mAbs, whereas the E484K mutation correlated with resistance to class II mAbs (Figure 1C). The triple-mutant K417N-E484K-N501Y pseudovirus, like B.1.351 and P.1, was resistant to class I and II mAbs, suggesting that combination of the three RBD mutations is key for conferring complete resistance. Neutralization by the 4A8 mAb was eliminated in two NTD deletion mutant pseudoviruses, Y144del and 242-244del. These deletions were initially identified in B.1.1.7 and B.1.351 and likely contribute to their resistance profiles (Chi et al., 2020; England, 2020; Tegally et al., 2021). Because both deletion mutations largely fall in the NTD supersite, B.1.1.7 and B.1.351 are likely resistant to other members of the same antibody family (Cerutti et al., 2021; Chi et al., 2020; Liu et al., 2020; McCallum et al., 2021; Suryadevara et al., 2021; Wang et al., 2021a).
Of note, the neutralization activities of mAbs approved for EUA (REGN10933, REGN10987, and CB6) or being studied for clinical use (P2C-1F11, BD368-2, S309, and P2B-1G5) were variably affected by pseudoviruses carrying these mutations (Figure 1C; Figure S1). The half-maximal inhibitory concentration (IC50) of REGN10933 dropped 13.0- and 8.2-fold against B.1.351 and P.1, respectively, largely because of the K417N/T and E484K mutations (Figure 1C). Several mutations within the REGN10933 epitope, such as Y453F and F486L, were also associated with a substantial reduction in neutralization. N439K, located in the REGN10987 epitope, showed a 26.6-fold reduction in IC50. The most substantially affected mAb was CB6, one of the paired antibodies developed by Eli Lilly and approved for EUA, for which neutralization against B.1.351 and P.1 pseudoviruses was BDL when the highest concentration (1 μg/mL) was used. The reduction and loss of neutralization were largely attributed to K417N/T (Figure 1C). Despite being a class I or RBS-A antibody, P2C-1F11 was virtually unaffected by the mutations, as were the class III and IV anti-RBD mAbs REGN10987, S309, and P2B-1G5 (Figure 1C).
Next we studied the binding avidity of the 12 mAbs to the 28 mutant S proteins expressed on cell surfaces. There was a strong correlation between neutralization and binding avidity (Figure 1D; Figure S2), indicating that compromised binding avidity is a major escape mechanism. Last, soluble human ACE2 showed improved binding to all three variants as well as pseudoviruses bearing single N501Y and triple K417N-E484K-N501Y mutations (Figure 1D; Figure S2). This finding indicates that N501Y plays a role in enhanced binding, which is consistent with earlier reports of human and mouse ACE2 (Laffeber et al., 2021; Rathnasinghe et al., 2021; Sun et al., 2020; Tian et al., 2021). The single K417N mutation, however, decreased ACE2 binding by about 6.4-fold. These results indicate that the NTD mutations Y144del and 242-244del and the RBD mutations K417N/T, E484K, and N501Y confer substantial mAb resistance. The mAbs studied here with EUA would need to be optimized for the best possible efficacy against new variants.
Structural basis for mAb neutralization and escape
We next studied the structural basis for the mAb P2C-1F11 which is able to maintain neutralization to the variants despite being a class I or RBS-A anti-RBD antibody. We determined the crystal structure of the SARS-CoV-2 RBD carrying K417N-E484K-N501Y mutations (RBD-3M) bound by P2C-1F11 at a resolution of 2.10 Å (PDB: 7E8M; Table S1). Comparing this structure with our previously reported crystal structure of the WT RBD with P2C-1F11 at 3.0 Å (PDB:7CDI), we found that the three mutations did not change the overall binding mode of P2C-1F11 to the RBD (Figure 2 A), as evidenced by a 0.52 Å root-mean-square deviation (RMSD) value for all 581 Cα atoms. However, a couple of subtle changes were identified. One was related to the interactive forces with residue 417 and the other with residue 501. Like other class I and RBS-A antibodies, P2C-1F11 bound to WT K417 through hydrophobic and hydrogen bond interactions. This was largely mediated by its heavy-chain germline residues Y33 and Y52 (Ge et al., 2021). These interactions were diminished by the K417N mutation but replaced by one hydrogen bond between Y52 and mutant N417 (Figure 2B; Table S2). Furthermore, unlike those in class I or RBS-A, P2C-1F11 did not form salt bridge interactions between aspartic acid (D) and K417 (Figures 2B and 2C). The K417N mutation would therefore be less disruptive to P2C-1F11 than to those in class I or RBS-A, such as P5A-1D2, P22A-1D1, and CB6, studied here, and CC12.1, CC12.3, COVA2-04, and COVA2-07, characterized elsewhere (Ge et al., 2021; Yuan et al., 2020a).
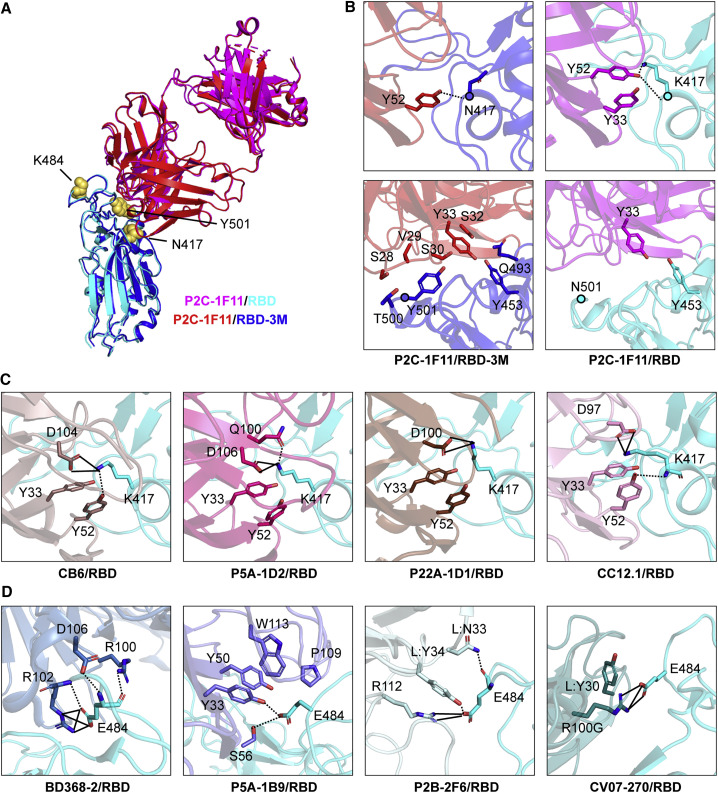
Figure 2.
Structural basis for mAb neutralization and escape
(A) P2C-1F11/RBD-3M crystal structure superimposed onto the P2C-1F11/RBD crystal structure (PDB: 7CDI). P2C-1F11 is colored magenta and RBD is colored cyan in the P2C-1F11/RBD complex. P2C-1F11 and RBD-3M are colored red and blue, respectively, in the P2C-1F11/RBD-3M complex. The three RBD-3M mutated residues (N417, K484, and Y501) are shown as yellow spheres.
(B) Interactions with P2C-1F11 around RBD-3M N417 and Y501 (left panel) and WT RBD K417 and N501 (right panel).
(C) Interactions between K417 and the representative class I IGHV3-53/3-66 antibodies CB6, P5A-1D2, P22A-1D1, and CC12.1.
(D) Interactions between E484 and the class II antibodies BD368-2, P5A-1B9, P2B-2F6, and CV07-270.
In (C) and (D), antibodies are shown with different colors; hydrogen bond and salt bridges are represented by dashed and black lines, respectively. CB6/RBD (PDB: 7C01), P5A-1D2/RBD (PDB: 7CHO), P22A-1D1/RBD (PDB: 7CHS), CC12.1/RBD (PDB: 6XC2), BD368-2/RBD (PDB: 7CHC), P5A-1B9 (PDB: 7CZX), P2B-2F6 (PDB: 7BWJ), CV07-270 (PDB:6XKP). See also Figure S3 and Tables S1 and S2.
Structural analysis showed that the P2C-1F11 light chain had extensive interactions with RBD-3M residues Y453, Q493, T500, and Y501 around the 501 position but much less interaction with the WT RBD residue asparagine (N) at the 501 position (Figure 2B; Table S2). The extra interactions between RBD-3M and P2C-1F11 around Y501 may also contribute to the retained binding and neutralization of P2C-1F11 against SARS-CoV-2 variants carrying the triple K417N-E484K-N501Y mutation.
Similar to the WT RBD, P2C-1F11 did not directly bind to E484K in RBD-3M (Figure 2B; Table S2). This explains why E484K had no detectable effect on the binding and neutralizing activity of P2C-1F11. However, the E484K mutation resulted in complete loss of BD368-2 and P5A-1B9 neutralization (Figure 1C; Figure S1). Structural analysis showed that the E484 formed salt-bridge and/or hydrogen-bonding interactions with BD368-2, P5A-1B9, P2B-2F6, and CV07-270 (Figure 2D; Du et al., 2020; Ju et al., 2020; Kreye et al., 2020). Therefore, the E484K mutation would diminish such interaction and render this class of antibodies ineffective against B.1.351 and P.1.
Next we measured the equilibrium dissociation constants and calculated KD values of P2C-1F11 binding to the WT RBD and RBD-3M. Consistent with the neutralization results, binding of P2C-1F11 in Fab or full-length immunoglobulin G (IgG) form to RBD-3M was slightly weaker compared with the WT RBD (21.9 versus 14.4 nM and 8.9 versus 2.2 nM, respectively; Figure S3). Furthermore, antibodies that lost their neutralizing activities, including CB6, P5A-1D2, P5A-3C8, P22A-1D1, BD368-2, and P5A-1B9, failed to show detectable levels of binding to RBD-3M (Figure S3). These results indicate that antibody binding activity correlates well with antibody neutralizing activities.
SARS-CoV-2 variants reduced sensitivity to neutralization by convalescent plasma
We next studied the degree to which major variant pseudoviruses conferred resistance to convalescent plasma from SARS-CoV-2-infected individuals. Plasma samples were collected from 23 affected individuals during the early wave of the pandemic between January and February of 2020. Fourteen of these individuals had only mild symptoms, and nine developed severe disease. The average age was 56 with a range of 29–81 years of age. Thirteen were men, and ten were women (Table S3). For each plasma sample, eight serial dilutions were made, starting from 1:60 or 1:200, and neutralization activity was estimated based on half-maximal inhibitory dilution (ID50) and fold changes relative to that against the WT D614G pseudovirus (Figure 3 ; Figure S4). Overall, neutralizing activity was higher in serum from those with severe versus those with mild disease (Figures 3A and 3B). Consistent with the findings for mAbs, the B.1.351 and P.1 pseudoviruses demonstrated more resistance than B.1.1.7 in absolute ID50 (Figures 3A and 3B) and fold changes (Figure 3C) relative to WT D614G. The average reductions in neutralization activity across the 23 plasma samples were at least 6.9-fold against the B.1.351, 2.4-fold against the P.1, and no significant reduction against the B.1.1.7 pseudovirus (Figure 3A). Complete loss of neutralization activity, indicated by BDL in Figure 3C, was observed for 11 plasma samples against pseudovirus B.1.351, four against P.1, and none against B.1.1.7. The remaining plasma demonstrated varying degrees of reduction in neutralization potency.
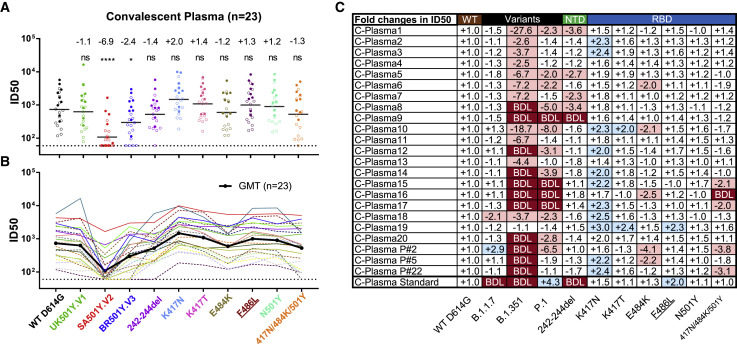
Figure 3.
SARS-CoV-2 variants reduced sensitivity to convalescent plasma neutralization
Reciprocal plasma dilutions (ID50) against SARS-CoV-2 variants are shown by (A) colored dots or (B) colored curves, each of which represents a different convalescent plasma. The geometric mean against each variant is indicated by a black horizontal line in (A) and black curve (B). Plasma samples from individuals with mild and severe disease are indicated by empty or solid circles in (A) and dashed or solid curves in (B). The fold change in ID50 between mutant and WT D614G pseudoviruses is shown by overall average at the top in (A) or individually in (C). “−” indicates an increase in resistance, and “+” indicates an increase in sensitivity. In (C), red highlights indicate a minimum 2-fold increase in resistance; blue a minimum 2-fold increase in sensitivity, and white a less than 2-fold change in resistance or sensitivity. BDL indicates that the highest concentration of plasma (1:60) failed to confer 50% neutralization. Standard plasma was obtained from the NIBSC (code: 20/136). Results of 23 plasma samples and standard plasma were calculated from three independent experiments performed in technical duplicates. ∗p < 0.05; and ∗∗∗∗p < 0.0001; ns, not significant. See also Figure S4.
Examination of the resistance patterns across the single- and triple-mutant pseudoviruses revealed some notable patterns. In particular, the 242–244del mutation in the NTD reduced neutralization activity more than 2-fold for 5 of 23 samples (C-Plasma1, C-Plasma5, C-Plasma7, C-Plasma8, and C-Plasma9), which correlated with the reduction or loss in potency observed against B.1.351 pseudoviruses (Figure 3C; Figure S4). The NTD supersite antibodies were likely to account for such a significant reduction (Cerutti et al., 2021; Chi et al., 2020; McCallum et al., 2021). On the other hand, the single-mutant E484K and triple-mutant K417N-E484K-N501Y pseudoviruses substantially reduced the neutralization activity for 8 of 23 samples (C-Plasma6, C-Plasma10, C-Plasma15, C-Plasma16, C-Plasma17, C-PlasmaP#2, C-PlasmaP#5, and C-PlasmaP#22), which corresponded to their diminished or loss of neutralization against the B.1.351 and P.1 pseudoviruses. Antibodies targeting the receptor binding motif (RBM) likely contributed to such changes (Figure 3C; Figure S4). In contrast, for pseudoviruses carrying only the K417N mutant, neutralization efficacy invariably increased for all plasma sample studied, with an average 2.0-fold improvement. A similar trend was also seen for pseudoviruses carrying only the K417T mutant (Figure 3C; Figure S4). The single mutant N501Y pseudovirus, reported previously to enhance ACE2 binding (Laffeber et al., 2021; Chan et al., 2021; Tian et al., 2021), had limited effects on the neutralizing activity of convalescent plasma.
Last, the plasma standard (code 20/136) obtained from the NIBSC (United Kingdom) failed to neutralize the B.1.1.7 and B.1.351 pseudoviruses, which correlated with loss of efficacy against the 242–244del pseudovirus (Figure 3C; Figure S4). This suggests that anti-NTD antibodies account for a major proportion of neutralizing activity in this sample. These results indicate that B.1.351 was the most resistant pseudovirus against the convalescent plasma tested, followed by P.1 and then B.1.1.7. The loss of plasma neutralizing activities is attributed, to varying degrees, to the 242–244del in the NTD, E484K, and triple K417N-E484K-N501Y mutations in the RBD, depending on individual plasma. Different convalescent plasma appears to respond differently to the mutated pseudoviruses, perhaps reflecting the different compositions and proportions of neutralizing antibodies in each individual generated during natural infection.
Enhanced entry of SARS-CoV-2 variants through mouse and mink ACE2
To study the potential effect of SARS-CoV-2 variants on host range and cross-species transmission, we characterized the ability of ACE2 from nine host species to support entry of 28 SARS-CoV-2 mutant pseudoviruses. HeLa cell lines stably expressing the ACE2 molecules were subjected to infection. The entry efficiency was measured and presented as fold change relative to WT D614G. As shown in Figure 4 , the three variants B.1.1.7, B.1.351, and P.1 gained substantial ability to infect HeLa mouse ACE2, which correlated with pseudoviruses bearing single (K417N, K417T, E484K, and N501Y) and triple (K417N-E484K-N501Y) mutations. This agreed well with recent reports where single N501Y or triple K417N-Q493H-N501Y mutations were found in the mouse-adapted SARS-CoV-2 strains, although the triple mutant causes more severe acute respiratory symptoms and mortality in standard laboratory mice (Rathnasinghe et al., 2021; Sun et al., 2020). Single N501Y mutations found in B.1.1.7 and two of three (K417N and N501Y) found in B.1.351 and P.1 therefore likely enhanced binding to mouse ACE2, improving entry efficiency into HeLa mouse ACE2.
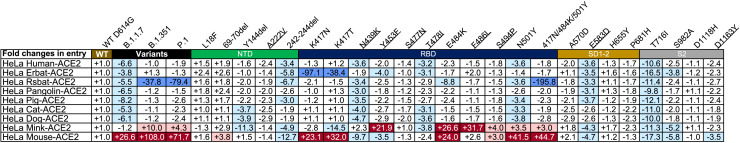
Figure 4.
Entry efficiency of SARS-CoV-2 variants into HeLa cells expressing ACE2 from diverse host species
The values show the fold changes in luciferase activity for each indicated mutant pseudovirus variant compared with WT D614G. “+” indicates an increase in entry efficiency, and “−” indicates a decrease. Red highlights indicate at least a 3-fold increase in efficiency; blue indicates at least a 3-fold decrease in efficiency, and white indicates no change greater than 3-fold. Results were calculated from three independent experiments performed in technical triplicates.
The single-mutant E484K pseudovirus also enhanced entry into HeLa mouse ACE2 and HeLa mink ACE2. Such enhancement occurred only with the E484K-bearing variant B.1.351 and P.1 but not with the E484K-missing variant B.1.1.7, suggesting that the added and/or synergistic effect of E484K with other mutant residues facilitates entry into these two cell lines. Furthermore, two single-mutant pseudoviruses, Y453F and F486L, also substantially improved entry efficiency. These two mutations have been found recently among the mink-associated SARS-CoV-2 circulating in mink farms in Denmark, suggesting their critical role in adaptation and transmission among the mink population (Oude Munnink et al., 2021). In contrast, entry efficiency through ACE2 of the other animal origins was reduced substantially, particularly for B.1.1.7 (Figure 4). For B.1.351 and P1, entry remained largely unchanged, except for Rsbat-ACE2 (Figure 4). These results indicate that the three variants, particularly B.1.351 and P.1, acquired mutations in the RBD that not only facilitated their escape from antibody neutralization but also potentially expanded their host range to mice and mink. Active surveillance of these variants in humans and relevant animal species would be required to minimize potential cross-species transmission.
Discussion
In this study, we evaluated the effect of mutations found in the emerging SARS-CoV-2 variants B.1.1.7, B.1.351, and P.1, as well as some single mutations within or close by the major antigenic sites in the S protein identified in the GISAID database. We identified K417N, E484K, and N501Y mutations in the RBD of the three variants that have profound consequences on antibody neutralization and interaction with mouse and mink ACE2. Mutations in the NTD also substantially reduced neutralizing sensitivity to supersite antibodies as well as convalescent plasma, suggesting that NTD and RBD responses may be contributing jointly to overall protection in polyclonal responses. Because responses to the NTD or RBD can be readily diminished by escape mutations, targeting both regions may be more advantageous. Our entry studies were conducted on ectopically expressed mouse and mink ACE2 and are not necessarily equivalent to natural infection and transmission in the corresponding animals. However, the same K417N and/or N501Y mutations found in mouse-adapted SARS-CoV-2 should raise enough concern about the potential spread of these new variants to mice and beyond. Indeed, recently identified SARS-CoV-2 variants in a mink farm in Denmark have raised another warning sign regarding the complexity of host range and cross-species transmission of SARS-CoV-2 variants. Rigorous and thorough monitoring of relevant animals would be required to better understand such complexity and prevent future outbreaks.
Among the three variants, B.1.351 was the most resistant against mAbs and convalescent plasma, followed by P.1 and then B.1.1.7. This resistance hierarchy corresponded well to genetic mutations in the NTD and RBD that led to major antigenic changes in the S protein. Particularly B.1.351 had the NTD supersite mutation (242–244del) and triple K417N-E484K-N501Y RBD mutation, whereas P.1 had only the latter and B.1.1.7 only the former (Y144del) together with N501Y in the RBD (Figure 1A). The effect of these mutations on conferring antibody resistance clearly indicates that NTD and RBD are major antigenic domains on the S protein. Of note, 5 of 23 plasma samples had a more than 2-fold loss-neutralizing ability resulting from 242–244del alone, and another 5 of 23 plasma samples had a more than 2-fold loss of neutralizing ability resulting from 484K alone, indicating that some people reacted in a more focused way to one epitope and, thus, may be more vulnerable to common circulating variants. Further study is needed to precisely estimate the relative contributions of RBD-directed and NTD-directed neutralizing antibodies to overall plasma neutralizing activity. To date, the most potent mAbs isolated from infected and vaccinated individuals are often dominated by those targeting the RBD, whereas many isolated NTD mAbs failed to reach 100% potency in neutralizing activity (Chi et al., 2020; Hansen et al., 2020; Ju et al., 2020; Liu et al., 2020; Robbiani et al., 2020; Wang et al., 2021b; Zost et al., 2020; McCallum et al., 2021). These results point to a greater effect of RBD antibodies in overall plasma neutralization. Nevertheless, new SARS-CoV-2 variants with increasing numbers of mutations in the NTD and RBD will challenge the efficacy of mAb therapies and vaccine protections.
The effects of these mutations, in terms of mAbs, are clear. B.1.351 and P.1 are resistant to neutralization by many anti-RBD and anti-NTD antibodies, including two (CB6 and REGN10933) already approved for EUA (FDA, 2021; Wang et al., 2021a; Huang et al., 2021; Wang et al., 2021b). Most class I mAbs studied here were disrupted by the K417N/T mutation and those in class II by the E484K mutation. Of note, we observed that the single K417N/T mutant tended to increase rather than decrease the neutralizing activity of non-class I mAbs. Because K417N/T also markedly reduced binding to soluble ACE2, this mutation may shift the balance in favor of binding to an antibody rather than to ACE2. Similar findings were also found for convalescent plasma. More importantly, P2C-1F11, which is currently in therapeutic development, was virtually unaffected by the single K417N/T or the triple K417N-E484K-N501Y mutation. The ability of Y52 in its heavy chain to maintain a hydrogen bond with the mutant N417 while gathering more interactive forces around mutant Y501 provides plausible structural explanations (Figure 2; Table S2). This was verified by similar binding activity of P2C-1F11 to the WT RBD and RBD-3M, indicating that antibody binding activity was well correlated with their neutralizing activities. Considering that most class I antibodies with heavy-chain IGHV3-53/3-66 usage lost efficacy against the K417N mutation, the P2C-1F11 findings show the existence of class I antibodies that were not affected by the mutation. This offers hope for inducing P2C-1F11-like antibodies by vaccine that can maintain strong, broadly neutralizing activity against a wide range of variants, including B.1.351 and P.1.
For convalescent plasma, the most profound effect was observed on the B.1.351 pseudovirus, followed by P.1 and then B.1.1.7. Reduction or complete loss of neutralization activities were variably attributed to the 242–244del mutation in the NTD, E484K, and triple K417N-E484K-N501Y in the RBD, depending on the individual profiles of neutralizing antibodies. Similar reductions in neutralization of B.1.351 have also been reported elsewhere for recovered individuals and individuals vaccinated with mRNA or inactivated vaccines approved under EUA (FDA, 2021; Wang et al., 2021a; Huang et al., 2021; Wang et al., 2021b). Although the specific levels of neutralizing antibody required to confer protection remain uncertain, reductions in antibody titers raise concerns regarding their protective potential against emerging variants, particularly B.1.351. Indeed, reductions in antibody titers have been associated with an increased likelihood of re-infection (Sabino et al., 2021). Recent vaccine trials in South Africa showed reduced efficacy against B.1.351 but maintained efficacy against WT strains (Diamond et al., 2021; Hoffmann et al., 2021; Wu et al., 2021). These results demonstrate that antigenic shifts are occurring among emerging SARS-CoV-2 variants. This calls for immediate reevaluation and updates of mAb therapies and vaccines. In the long run, the goal is to develop universal therapeutic and preventive interventions that maintain efficacy against all strain mutations. Until then, control over the emergence of new variants requires accelerated vaccine rollouts and ardent practice of proven public health measures.
Limitations of study
We note some limitations associated with our study. First, the ACE2 usage data relied on ectopically expressed ACE2 on the surface of human HeLa cells that may not reflect the levels and conformations on their original native cells. Reverse zoonotic infection would need to be verified through future relevant animal studies. Second, because of the limited amount of serum samples, we used 1:60 as our first dilution in the neutralization assay, which may underestimate the effect of variants on neutralization escape. Third, we were only able to study EUA or investigational mAbs with accessible sequences or structure information, and the conclusions may not be applicable to other EUA or investigational mAbs.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| P2C-1F11 | Ju et al., 2020 | N/A |
| P5A-1D2 | Ju et al., 2020 | N/A |
| P5A-3C8 | Ju et al., 2020 | N/A |
| P22A-1D1 | Ju et al., 2020 | N/A |
| P5A-1B9 | Ju et al., 2020 | N/A |
| P2B-1G5 | Ju et al., 2020 | N/A |
| REGN10933 | Hansen et al., 2020 | N/A |
| REGN10987 | Hansen et al., 2020 | N/A |
| CB6 | Shi et al., 2020 | N/A |
| S309 | Pinto et al., 2020 | N/A |
| 4A8 | Chi et al., 2020 | N/A |
| CR3022 | Yuan et al., 2020b | N/A |
| Anti-coronavirus (SARS-CoV-2) spike S2 mouse mAb | MP Biomedicals | Cat#08720401 |
| PE anti-human IgG Fc | Biolegend | Cat#410708 |
| Goat anti-Mouse IgG (H+L) Cross-Adsorbed Secondary Antibody, FITC | Invitrogen | Cat#A16073 |
| PE anti-his tag | Biolegend | Cat#362603 |
| Bacterial and virus strains | ||
| SARS-CoV-2/WH-09/human/2020/CHN | GenBank | MN908947.3 |
| SARS-CoV-2/UK B.1.1.7 | GISAID | EPI_ISL_601443 |
| SARS-CoV-2/SA B.1.351 | GISAID | EPI_ISL_700450 |
| SARS-CoV-2/BR P.1 | GISAID | EPI_ISL_792681 |
| Per site variation v20210517 | GISAID | https://cov.lanl.gov/content/index |
| Biological samples | ||
| International Standard for anti-SARS-CoV-2 immunoglobulin | NISBIC | Cat#20/136 |
| 23 plasma samples from convalescent patients | This paper | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Polyethyleneimine | Polysciences | Cat#24765-1 |
| Endoproteinase Lys-C | Roche | Cat#11047825001 |
| Trypsin | Macgene | Cat#CC017 |
| Fetal bovine serum | GIBCO | Cat#16000-044 |
| ACE2 recombinant protein | This paper | Lan et al., 2020 |
| Recombinant wildtype RBD | This paper | Lan et al., 2020 |
| Recombinant RBD with 417N/484K/501Y mutations | This paper | N/A |
| Cellfectin II Reagents | GIBCO | Cat#10362100 |
| Recombinant protein A | Sino Biological | Cat#10600-P07E |
| His capture kit | Cytiva | Cat#28-9950-56 |
| Critical commercial assays | ||
| QuikChange Lightning Site-Directed Mutagenesis Kit | Agilent | Cat#210519 |
| Bright-Glo™ Luciferase Assay Buffer | Promega | Cat#E264B |
| Bright-Glo™ Luciferase Assay Substrate | Promega | Cat#E263B |
| Deposited data | ||
| P2C-1F11 with mutated RBD structure | This paper | PDB: 7E8M |
| Experimental models: cell lines | ||
| Human: HEK293T, embryo | ATCC | CRL-3216 |
| Human: FreeStyle 293F, embryo | Thermo Fisher | R79007 |
| Human: Huh-7, Male | JCRB | JCRB0403 |
| Human:HeLa, Female | ATCC | CCL-2 |
| HeLa expressing ACE2 from diverse origin, Female | Dr. Qiang Ding’s lab | Liu et al., 2021 |
| Sf9 cells, Female | ATCC | CRL-3357 |
| Recombinant DNA | ||
| pcDNA3.1 | Thermo Fisher | Cat#V79020 |
| pLVX-IRES-zsGreen1 vector | Clontech Laboratories, Inc | Cat#632187 |
| pFastBac-Dual vector | GIBCO | Cat#10712024 |
| pNL4-3- luc -R-E | NIH-AIDS Reagent Program | Cat#3418 |
| Software and algorithms | ||
| Graphpad Prism 7 | GraphPad | www.graphpad.com |
| Microsoft Excel | Microsoft | www.microsoft.com |
| FlowJo 10 software | FlowJo | https://www.flowjo.com/ |
| Biacore Insight Evaluation Software | GE Healthcare | https://www.cytivalifesciences.com/en/us/shop/protein-analysis/spr-label-free-analysis/software/biacore-insight-evaluation-software-p-23528 |
| PHASER (CCP4 Program Suite) | McCoy et al., 2007 | http://www.phaser.cimr.cam.ac.uk/index.php/Phaser_Crystallographic_Software |
| COOT | Emsley et al., 2010 | http://www2.mrc-lmb.cam.ac.uk/Personal/pemsley/coot/ |
| PHENIX | Adams et al., 2010 | http://www.phenix-online.org/ |
| PyMOL | Janson et al., 2017 | https://pymol.org/2/ |
| Chimera | Pettersen et al., 2004 | https://www.cgl.ucsf.edu/chimera/ |
| Other | ||
| Superdex 200 High-Performance column | GE Healthcare | N/A |
| Nanodrop 2000 Spectrophotometer | Thermo Scientific | N/A |
| BD LSRFortassa | BD Biosciences | N/A |
| CM5 sensor chip | GE Healthcare | Cat#BR100530 |
| Biacore 8K | GE Healthcare | N/A |
Resource availability
Lead contact
Further information and requests for resources and primary data should be directed to and will be fulfilled by the Lead Contact, Linqi Zhang (Zhanglinqi@mail.tsinghua.edu.cn).
Materials availability
There are restrictions to the availability of convalescent plasma due to limited stock. We will share these reagents until the stock runs out.
Data and code availability
The coordinates and structure factors files for P2C-1F11 and RBD-3M complex have been deposited to the Protein Data Bank (http://www.rcsb.org) with accession code 7E8M.
Experimental model and subject details
Study approval
This study received approval from the Research Ethics Committee of Beijing Youan Hospital (LL-2020-039-K), Beijing Ditan Hospital (2020-019-01), and Shenzhen Third People’s Hospital (2020-084). The research was conducted in strict accordance with the rules and regulations of the Chinse government for the protection of human subjects. The study subjects agreed and signed the written informed consents for research use of their blood samples.
Human samples
The study enrolled a total of 23 convalescent patients aged from 29 to 81 years old, with an average of 56, infected with SARS-CoV-2 between January to February 2020. SARS-CoV-2 infection status was verified by RT-PCR of nasopharyngeal swab and chest computed tomographic scan. Of the 23 infected individuals, thirteen were males and ten were females. Fourteen individuals developed only mild symptom and the remaining nine developed severe pneumonia. The specific information is provided in Table S3. Convalescent blood samples were collected during hospitalization or follow-up visits in Beijing Youan Hospital, Beijing Ditan Hospital, or Shenzhen Third People’s Hospital, within two months after symptom onset. A blood sample from a healthy control individual was also included. Blood samples were separated into plasma and peripheral blood mononuclear cells (PBMC) by Ficoll-Hypaque gradient (GE Healthcare) centrifugation. All plasma samples were heat-inactivated at 56°C for 1h before being stored at −80°C. PBMCs were maintained in freezing media and stored in liquid nitrogen until use.
Cell lines
HEK293T cells (ATCC, CRL-3216), Huh7 cells (JCRB, JCRB0403), HeLa cells (ATCC, CCL-2) and HeLa cells expressing ACE2 orthologs (kindly provided by Dr. Qiang Ding) were maintained at 37°C in 5% CO2 in Dulbecco’s minimal essential medium (DMEM) containing 10% (v/v) heat-inactivated fetal bovine serum (FBS) and 100 U/mL of penicillin–streptomycin. FreeStyle 293F cells (Thermo Fisher Scientific, R79007) were maintained at 37°C in 8% CO2. Sf9 cells (ATCC, CRL-3357) were maintained at 27°C in Sf-900 II SFM medium.
Method details
Production of mAbs and Fab
P2C-1F11, P5A-1D2, P5A-3C8, P22A-1D2, P5A-1B9 and P2B-1G5 are potent neutralizing mAbs initially isolated from SARS-CoV-2 infected patients by our group and previously published (Ju et al., 2020). BRII-196 and BRII-198 are Fc-modified version of P2C-1F11 and P2B-1G5, respectively, which are being investigated in the phase III clinical trial. Antibodies published by other groups including REGN10933, REGN10987, CB6, S309, 4A8 and CR3022 were synthesized according to the sequences released in Protein Data Bank (PDB) (Chi et al., 2020; Hansen et al., 2020; Pinto et al., 2020; Shi et al., 2020; Yuan et al., 2020b). Antibody production was conducted by co-transfection of the heavy and light chain expression vectors into HEK293F cells using polyethyleneimine (PEI)(Polysciences). After 96h, antibodies secreted into the supernatant were captured by Protein A-Sepharose (GE Healthcare) and eluted by solution buffer Glycine pH 3.0. After further purification by gel-filtration chromatography with Superdex 200 High-Performance column (GE Healthcare), antibody concentration was determined by nanodrop 2000 Spectrophotometer (Thermo Scientific). To produce Fab fragment, purified P2C-1F11 were cleaved using Endoproteinase Lys-C (Roche) with IgG to Lys-C ratio of 4000:1 (w/w) in 10 mM EDTA, 100 mM Tris-HCl, pH 8.5 at 37°C overnight. Fc fragment were removed using Protein A-Sepharose.
Production of SARS-CoV-2 wild-type and variant pseudoviruses
The wild-type pseudovirus used throughout the analysis was the prototype strain (GenBank: MN908947.3) with a D614G mutation (WT D614G). The variant B.1.1.7 (GISAID: EPI_ISL_601443) was constructed with total of 9 mutations including 69-70del, 144del, N501Y, A570D, D614G, P681H, T716I, S982A and D1118H. The variant B.1.351 (GISAID: EPI_ISL_700450) was constructed with 10 mutations including L18F, D80A, D215G, 242-244del, S305T, K417N, E484K, N501Y, D614G and A701V. The variant P.1 (GISAID: EPI_ISL_792681) was constructed with 12 mutations including L18F, T20N, P26S, D138Y, R190S, K417T, E484K, N501Y, D614G, H655Y, T1027I and V1176F. The gene of variants were synthesized In Genwiz, Inc. The single mutations identified from the GISAID database were introduced into the pcDNA3.1 vector encoding WT D614G using QuickChange site-directed mutagenesis (Agilent 210519). SARS-CoV-2 pseudoviruses were generated by co-transfecting HEK293T cells (ATCC) with human immunodeficiency virus backbones expressing firefly luciferase (pNL4-3-R-E-luciferase) and pcDNA3.1 vector encoding either wild-type or mutated S proteins. Viral supernatant was collected 48h or 72h later, centrifuged to remove cell lysis, and stored at −80°C until use.
HeLa cell lines expressing ACE2 from diverse origin
HeLa cells expressing ACE2 orthologs were kindly provided by Dr. Qiang Ding at Tsinghua University School of Medicine. The species names and accession numbers for the ACE2 orthologs used are as follows: Human, Homo sapiens, NP_001358344.1; Rsbat, Rhinolophus sinicus, KC881004.1; Erbat, Rousettus aegyptiacus, XP_015974412.1; Pangolin, Manis javanica, XP_017505746.1; Pig, Sus scrofa, NP_001116542.1; Cat, Felis catus, XP_023104564.1; Dog, Canis lupus familiaris, NP_001158732.1; Mink, Mustela lutreola, MT560518.1; Mouse, Mus musculus, NP_001123985.1. The cDNAs encoding ACE2 orthologs were synthesized by GenScript and cloned into pLVX-IRES-zsGreen1 vectors (Catalog No. 632187, Clontech Laboratories, Inc) with a C-terminal FLAG tag. VSV-G pseudotyped lentiviruses expressing ACE2 orthologs were produced and used to generate HeLa-ACE2 cells as previously described (Liu et al., 2021). For studying entry efficiency of SARS-CoV-2 variants, HeLa-ACE2 cells were added to 96 well plates, mixed with 50 ul of pseudovirus, and analyzed the luciferase activities 60 h after infection using Bright-Glo Luciferase Assay Vector System (Promega Bioscience). Absolute and fold changes between mutated and WT D614G were used to estimate the entry efficiency of SARS-CoV-2 variants.
mAbs and plasma neutralization using pseudoviruses
Serial dilutions of mAbs were prepared with the highest concentration of 1 μg/ml except for S309 and P2B-1G5 (10 μg/ml). Serial dilutions of convalescent plasma were prepared with the highest dilution of 1:60 except for P#2, P#5, P#22 and International Standard for anti-SARS-CoV-2 immunoglobulin (human) (NIBSC code: 20/136) where 1:200 was the initial dilution. Wild-type or mutated spike pseudovirus were mixed with mAbs or plasma and incubated at 37°C for 1h. HeLa-ACE2 cells (except for S309 where Huh7 cell was used) were then added into the mixture and incubated at 37°C for 60h before cell lysis for measuring luciferase-activity. The percent of neutralization was determined by comparing with the virus control.
Binding of mAb and ACE2 to cell surface-expressed wild-type and mutated S glycoprotein
The entire procedure was conducted as previously published (Ge et al., 2021). Specifically, HEK293T cells were transfected with expression plasmids encoding either wild-type or mutated SARS-CoV-2 S glycoproteins, and incubated at 37°C for 36 h. Cells were digested from the plate with trypsin and distributed onto 96-well plates. Cells were washed twice with 200 μL staining buffer (PBS with 2% heated-inactivated fetal bovine serum (FBS)) between each of the following steps. First, cells were stained with the testing mAb, S2-specific monoclonal antibody (MP Biomedicals, Singapore 08720401), or ACE2 recombinant protein at 4°C for 30 min in 100 μL staining buffer. Then, PE-labeled anti-human IgG Fc (Biolegend 410718), anti-mouse IgG FITC (ThermoFisher Scientific A10673), or anti-his PE secondary antibody (Miltenyi 130120787) was added in 40 μL staining buffer at 4°C for 30 min. After extensive washes, the cells were resuspended and analyzed with BD LSRFortassa (BD Biosciences, USA) and FlowJo 10 software (FlowJo, USA). The serial dilution of concentration of mAbs and ACE2 were tested and the lowest saturated concentrations were used in the assay (2 μg/ml for ACE2, 0.1 μg/ml for all mAbs except for P2B-1G5 and CR3022 where 2 μg/ml was used). HEK293T cells with mock transfection were stained as background control. Antibody binding percentages were calculated by the ratio between mutated over wild-type MFI normalized in relative to that of S2 specific antibody. All MFI values were weighted by multiplying the number of positive cells in the selected gates.
Recombinant RBD and ACE2 protein
Recombinant wild-type RBD, RBD with 417N-484K-501Y mutations, and human receptor ACE2 peptidase domain were expressed using the Bac-to-Bac Baculovirus System (Invitrogen) as previously described (Lan et al., 2020). Specifically, SARS-CoV-2 RBD (residues Arg319 to Lys529) or ACE2 (residues Ser19 to Asp615) containing the gp67 secretion signal peptide and a C-terminal hexahistidine was inserted into pFastBac-Dual vectors (Invitrogen) and transformed into DH10 Bac component cells. The recombinant bacmid was extracted and further transfected into Sf9 cells using Cellfectin II Reagents (Invitrogen). The recombinant viruses were harvested from the transfected supernatant and amplified to generate high-titer virus stock. Viruses were then used to infect Sf9 cells for protein expression. Secreted RBD and ACE2 were harvested from the supernatant, captured by Ni-NTA Sepharose (GE Healthcare) and purified by gel filtration chromatography.
Antibody binding kinetics measured by SPR
The binding kinetics of IgG or Fab form of mAbs to SARS-CoV-2 RBD or mutated RBD were analyzed by SPR (Biacore 8K, GE Healthcare). Specifically, recombinant protein A (Sino Biological) or anti-his antibody (Cytiva) were covalently immobilized to a CM5 sensor chip via amine groups in 10 mM sodium acetate buffer (pH 4.5) for a final RU around 7000. The running buffer was composed of 10mM PBS, pH 7.2 and 0.05% Tween 20. For IgG form, diverse IgG form of mAbs were captured by the sensor chip immobilized with recombinant protein A and then Serial dilutions of wild-type and mutated SARS-CoV-2 RBDs flowed through the sensor chip system. For Fab form, wild-type and mutated SARS-CoV-2 RBDs were captured by the sensor chip immobilized with anti-his antibody and then dilutions of P2C-1F11 Fab flowed through the sensor chip system. The resulting data were fitted to a 1:1 binding model using Biacore Evaluation Software (GE Healthcare).
Crystal analysis and data collection
P2C-1F11 Fab fragments were mixed with SARS-CoV-2 RBD containing K417N-E484K-N501Y mutations at a molar ratio of 1:1.2, incubated on ice for 2 h, and further purified by gel-filtration chromatography. The purified complex was concentrated to 11 mg/mL in HBS buffer (10 mM HEPES, pH 7.2, 150 mM NaCl) for crystallization. Screening trials were performed at 18°C. The sitting drop vapor diffusion method was used by mixing 0.2 μL of protein with 0.2 μL of reservoir solution. Crystals of RBD–Fab complexes were successfully obtained in 0.2 M Ammonium sulfate, 0.1M Tris 8.5, 12% w/v PEG 8000. Diffraction data were collected at the BL17U1 beamline of the Shanghai Synchrotron Research Facility (SSRF) and auto-processed with aquarium pipeline (Yu et al., 2019).
Structural determination and refinement
Structures were determined by the molecular replacement method using PHASER (CCP4 Program Suite) (McCoy et al., 2007). Search models were the SARS-CoV-2 RBD structure (PDB: 6M0J) and the heavy and light chain variable domain structures available in the PDB with the highest sequence identities. Subsequent model building and refinement were performed using COOT and PHENIX, respectively (Adams et al., 2010; Emsley et al., 2010). All structural figures were generated using PyMOL and Chimera (Janson et al., 2017; Pettersen et al., 2004).
Quantification and statistical analysis
The technical and independent experiment replicates were indicated in the figure legends. Half-maximal inhibitory concentration (IC50) of mAb or dilutions (ID50) of convalescent plasmas were calculated by the equation of four-parameter dose inhibition response using Graphpad Prism 7.0. The fold change of mutant S relative to WT D614G in binding or neutralization were calculated by simple division of respective IC50 or ID50 values. The overall fold change of mutant pseudovirus relative to WT D614G in neutralization of convalescent plasma was calculated by the comparison of geometric mean of the ID50 value of the 23 plasma samples. The significance of neutralizing activities of convalescent plasma against each mutant pseudovirus relative to D614G was estimated using the unpaired Mann-Whiteny t test by Graphpad Prism 7 (n = 23).
Acknowledgments
We acknowledge the work and contributions of all health care providers from Beijing Youan Hospital, Beijing Ditan Hospital, and Shenzhen Third People’s Hospital. We also thank affected individuals for their active participation. This study was supported by the National Key Plan for Scientific Research and Development of China (2020YFC0848800 and 2020YFC0849900), the National Natural Science Foundation (81530065, 91442127, and 32000661), the Beijing Municipal Science and Technology Commission (D171100000517 and Z201100005420019), and the Science and Technology Innovation Committee of Shenzhen Municipality (202002073000002). It is also supported by funds from the COVID-19 Science and Technology Project of Beijing Hospitals Authority (YGZX-C1), the Beijing Advanced Innovation Center for Structural Biology, the Tsinghua University Scientific Research Program (20201080053 and 2020Z99CFG004), the Tencent Foundation, the Shuidi Foundation, the TH Capital and the National Science Fund for Distinguished Young Scholars (82025022), and the China Postdoctoral Science Foundation (2020T130062ZX). The funders had no role in study design, data collection, analysis, and interpretation or writing of the report. We thank the SSRF BL17U1 beamline for data collection and processing.
Author contributions
L.Z., X.W., and T.Z. conceived and designed the study. R.W., Q.Z., and J.G. performed most of the experiments with assistance from R.Z., J.L., and P.C. J.G. and J. L. produced the RBD and ACE2 recombinant proteins and solved and analyzed the crystal structure of the antibody-RBD complex. P.C. provided assistance with antibody production. W.R and Q.D. provided HeLa cell lines expressing ACE2 from diverse origins. T.Z., F.Z., Z.Z., and H.L. provided clinical care and management of infected individuals, particularly recruitment and following up of study subjects. T.Z., F.Z., Z.Z., B.J., B.S., F.Y., Y.F., X.L., and X.S. conducted sample collection, processing, and initial characterization. R.W., Q.Z., J.G., T.Z., X.W., and L.Z. had full access to the data in the study, generated figures and tables, and take responsibility for the integrity and accuracy of the data presentation. X.W. and L.Z. wrote the manuscript. All authors reviewed and approved the final version of the manuscript.
Declaration of interests
Patent applications have been filed on monoclonal antibodies targeting SARS-CoV-2 (patent application number: PCT/CN2020/108718; patent applicants: The Third People’s Hospital of Shenzhen and Tsinghua University). Q.Z., B.J., X.S., Z.Z., and L.Z. are the inventors. Q.Z., X.S., and L.Z. are shareholders of TSB Therapeutics.
Published: June 8, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.immuni.2021.06.003.
Supplemental information
References
- Adams P.D., Afonine P.V., Bunkóczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W., et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta crystallographica. Section D, Biological crystallography. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baden L.R., El Sahly H.M., Essink B., Kotloff K., Frey S., Novak R., Diemert D., Spector S.A., Rouphael N., Creech C.B., et al. COVE Study Group Efficacy and Safety of the mRNA-1273 SARS-CoV-2 Vaccine. N. Engl. J. Med. 2021;384:403–416. doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes C.O., Jette C.A., Abernathy M.E., Dam K.A., Esswein S.R., Gristick H.B., Malyutin A.G., Sharaf N.G., Huey-Tubman K.E., Lee Y.E., et al. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature. 2020;588:682–687. doi: 10.1038/s41586-020-2852-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callaway E. Fast-spreading COVID variant can elude immune responses. Nature. 2021;589:500–501. doi: 10.1038/d41586-021-00121-z. [DOI] [PubMed] [Google Scholar]
- Cao Y., Su B., Guo X., Sun W., Deng Y., Bao L., Zhu Q., Zhang X., Zheng Y., Geng C., et al. Potent Neutralizing Antibodies against SARS-CoV-2 Identified by High-Throughput Single-Cell Sequencing of Convalescent Patients’ B Cells. Cell. 2020;182:73–84.e16. doi: 10.1016/j.cell.2020.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerutti G., Guo Y., Zhou T., Gorman J., Lee M., Rapp M., Reddem E.R., Yu J., Bahna F., Bimela J., et al. Potent SARS-CoV-2 neutralizing antibodies directed against spike N-terminal domain target a single supersite. Cell Host Microbe. 2021;29:819–833.e7. doi: 10.1016/j.chom.2021.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K.K., Tan T.J.C., Narayanan K.K., Procko E. An engineered decoy receptor for SARS-CoV-2 broadly binds protein S sequence variants. Sci. Adv. 2021 doi: 10.1101/2020.10.18.344622. Published online December 21, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P., Nirula A., Heller B., Gottlieb R.L., Boscia J., Morris J., Huhn G., Cardona J., Mocherla B., Stosor V., et al. BLAZE-1 Investigators SARS-CoV-2 Neutralizing Antibody LY-CoV555 in Outpatients with Covid-19. N. Engl. J. Med. 2021;384:229–237. doi: 10.1056/NEJMoa2029849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi X., Yan R., Zhang J., Zhang G., Zhang Y., Hao M., Zhang Z., Fan P., Dong Y., Yang Y., et al. A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2. Science. 2020;369:650–655. doi: 10.1126/science.abc6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett K.S., Flynn B., Foulds K.E., Francica J.R., Boyoglu-Barnum S., Werner A.P., Flach B., O’Connell S., Bock K.W., Minai M., et al. Evaluation of the mRNA-1273 Vaccine against SARS-CoV-2 in Nonhuman Primates. N. Engl. J. Med. 2020;383:1544–1555. doi: 10.1056/NEJMoa2024671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond M., Chen R., Xie X., Case J., Zhang X., VanBlargan L., Liu Y., Liu J., Errico J., Winkler E., et al. SARS-CoV-2 variants show resistance to neutralization by many monoclonal and serum-derived polyclonal antibodies. Res. Sq. 2021 doi: 10.1038/s41591-021-01294-w. rs.3.rs-228079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du S., Cao Y., Zhu Q., Yu P., Qi F., Wang G., Du X., Bao L., Deng W., Zhu H., et al. Structurally Resolved SARS-CoV-2 Antibody Shows High Efficacy in Severely Infected Hamsters and Provides a Potent Cocktail Pairing Strategy. Cell. 2020;183:1013–1023.e13. doi: 10.1016/j.cell.2020.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of Coot. Acta crystallographica. Section D, Biological crystallography. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FDA . EUA Information; 2021. Coronavirus Disease 2019 (COVID-19) [Google Scholar]
- Fujino T., Nomoto H., Kutsuna S., Ujiie M., Suzuki T., Sato R., Fujimoto T., Kuroda M., Wakita T., Ohmagari N. Novel SARS-CoV-2 Variant Identified in Travelers from Brazil to Japan. Emerg. Infect. Dis. 2021;27:1243–1245. doi: 10.3201/eid2704.210138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge J., Wang R., Ju B., Zhang Q., Sun J., Chen P., Zhang S., Tian Y., Shan S., Cheng L., et al. Antibody neutralization of SARS-CoV-2 through ACE2 receptor mimicry. Nat. Commun. 2021;12:250. doi: 10.1038/s41467-020-20501-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GISAID . 2021. COVID-19 Viral Genome Analysis Pipeline. https://cov.lanl.gov/content/index. [Google Scholar]
- Gottlieb R.L., Nirula A., Chen P., Boscia J., Heller B., Morris J., Huhn G., Cardona J., Mocherla B., Stosor V., et al. Effect of Bamlanivimab as Monotherapy or in Combination With Etesevimab on Viral Load in Patients With Mild to Moderate COVID-19: A Randomized Clinical Trial. JAMA. 2021;325:632–644. doi: 10.1001/jama.2021.0202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen J., Baum A., Pascal K.E., Russo V., Giordano S., Wloga E., Fulton B.O., Yan Y., Koon K., Patel K., et al. Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Science. 2020;369:1010–1014. doi: 10.1126/science.abd0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Arora P., Groß R., Seidel A., Hörnich B.F., Hahn A.S., Krüger N., Graichen L., Hofmann-Winkler H., Kempf A., et al. SARS-CoV-2 variants B.1.351 and P.1 escape from neutralizing antibodies. Cell. 2021;184:2384–2393.e12. doi: 10.1016/j.cell.2021.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y.J., Chiba S., Halfmann P., Ehre C., Kuroda M., Dinnon K.H., 3rd, Leist S.R., Schäfer A., Nakajima N., Takahashi K., et al. SARS-CoV-2 D614G variant exhibits efficient replication ex vivo and transmission in vivo. Science. 2020;370:1464–1468. doi: 10.1126/science.abe8499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B., Dai L., Wang H., Hu Z., Yang X., Tan W., Gao G.F. Serum sample neutralisation of BBIBP-CorV and ZF2001 vaccines to SARS-CoV-2 501Y.V2. Lancet Microbe. 2021 doi: 10.1016/S2666-5247(21)00082-3. Published online April 13, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janson G., Zhang C., Prado M.G., Paiardini A. PyMod 2.0: improvements in protein sequence-structure analysis and homology modeling within PyMOL. Bioinformatics. 2017;33:444–446. doi: 10.1093/bioinformatics/btw638. [DOI] [PubMed] [Google Scholar]
- Johnson & Johnson . 2021. Johnson & Johnson COVID-19 Vaccine Authorized by U.S. FDA For Emergency Use - First Single-Shot Vaccine in Fight Against Global Pandemic.https://www.prnewswire.com/news-releases/johnson--johnson-covid-19-vaccine-authorized-by-us-fda-for-emergency-use-301236878.html [Google Scholar]
- Ju B., Zhang Q., Ge J., Wang R., Sun J., Ge X., Yu J., Shan S., Zhou B., Song S., et al. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature. 2020;584:115–119. doi: 10.1038/s41586-020-2380-z. [DOI] [PubMed] [Google Scholar]
- Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Hengartner N., Giorgi E.E., Bhattacharya T., Foley B., et al. Sheffield COVID-19 Genomics Group Tracking Changes in SARS-CoV-2 Spike: Evidence that D614G Increases Infectivity of the COVID-19 Virus. Cell. 2020;182:812–827.e19. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreye J., Reincke S.M., Kornau H.C., Sánchez-Sendin E., Corman V.M., Liu H., Yuan M., Wu N.C., Zhu X., Lee C.D., et al. A Therapeutic Non-self-reactive SARS-CoV-2 Antibody Protects from Lung Pathology in a COVID-19 Hamster Model. Cell. 2020;183:1058–1069.e19. doi: 10.1016/j.cell.2020.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupferschmidt K. Fast-spreading U.K. virus variant raises alarms. Science. 2021;371:9–10. doi: 10.1126/science.371.6524.9. [DOI] [PubMed] [Google Scholar]
- Laffeber C., de Koning K., Kanaar R., Lebbink J.H.G. Experimental Evidence for Enhanced Receptor Binding by Rapidly Spreading SARS-CoV-2 Variants. Journal of molecular biology. 2021;433:167058. doi: 10.1016/j.jmb.2021.167058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- Liu L., Wang P., Nair M.S., Yu J., Rapp M., Wang Q., Luo Y., Chan J.F., Sahi V., Figueroa A., et al. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature. 2020;584:450–456. doi: 10.1038/s41586-020-2571-7. [DOI] [PubMed] [Google Scholar]
- Liu Y., Hu G., Wang Y., Ren W., Zhao X., Ji F., Zhu Y., Feng F., Gong M., Ju X., et al. Functional and genetic analysis of viral receptor ACE2 orthologs reveals a broad potential host range of SARS-CoV-2. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2025373118. e2025373118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maggi F., Novazzi F., Genoni A., Baj A., Spezia P.G., Focosi D., Zago C., Colombo A., Cassani G., Pasciuta R., et al. Imported SARS-COV-2 Variant P.1 Detected in Traveler Returning from Brazil to Italy. Emerg. Infect. Dis. 2021;27:1249–1251. doi: 10.3201/eid2704.210183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCallum M., De Marco A., Lempp F.A., Tortorici M.A., Pinto D., Walls A.C., Beltramello M., Chen A., Liu Z., Zatta F., et al. N-terminal domain antigenic mapping reveals a site of vulnerability for SARS-CoV-2. Cell. 2021;184:2332–2347.e2316. doi: 10.1016/j.cell.2021.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J. Phaser crystallographic software. J. Appl. Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novavax . 2021. Novavax COVID-19 Vaccine Demonstrates 89.3% Efficacy in UK Phase 3 Trial.https://ir.novavax.com/news-releases/news-release-details/novavax-covid-19-vaccine-demonstrates-893-efficacy-uk-phase-3 [Google Scholar]
- Oude Munnink B.B., Sikkema R.S., Nieuwenhuijse D.F., Molenaar R.J., Munger E., Molenkamp R., van der Spek A., Tolsma P., Rietveld A., Brouwer M., et al. Transmission of SARS-CoV-2 on mink farms between humans and mink and back to humans. Science. 2021;371:172–177. doi: 10.1126/science.abe5901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Pinto D., Park Y.J., Beltramello M., Walls A.C., Tortorici M.A., Bianchi S., Jaconi S., Culap K., Zatta F., De Marco A., et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature. 2020;583:290–295. doi: 10.1038/s41586-020-2349-y. [DOI] [PubMed] [Google Scholar]
- Plante J.A., Liu Y., Liu J., Xia H., Johnson B.A., Lokugamage K.G., Zhang X., Muruato A.E., Zou J., Fontes-Garfias C.R., et al. Spike mutation D614G alters SARS-CoV-2 fitness. Nature. 2021;592:116–121. doi: 10.1038/s41586-020-2895-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J.L., Pérez Marc G., Moreira E.D., Zerbini C., et al. C4591001 Clinical Trial Group Safety and Efficacy of the BNT162b2 mRNA Covid-19 Vaccine. N. Engl. J. Med. 2020;383:2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Public Health England . 2020. . Investigation of novel SARS-COV-2 variant: Variant of Concern 202012/01: technical briefings.https://www.gov.uk/government/publications/investigation-of-novel-sars-cov-2-variant-variant-of-concern-20201201 [Google Scholar]
- Rambaut A., Loman N., Pybus O., Barclay W., Barrett J., Carabelli A., Connor T., Peacock T., Robertson D.L., Volz E., on behalf of the COVID-19 Genomics Consortium UK (CoG-UK) 2020. Preliminary genomic characterisation of an emergent SARS-CoV-2 lineage in the UK defined by a novel set of spike mutations.https://virological.org/t/preliminary-genomic-characterisation-of-an-emergent-sars-cov-2-lineage-in-the-uk-defined-by-a-novel-set-of-spike-mutations/563 [Google Scholar]
- Rathnasinghe R., Jangra S., Cupic A., Martínez-Romero C., Mulder L.C.F., Kehrer T., Yildiz S., Choi A., Mena I., De Vrieze J., et al. The N501Y mutation in SARS-CoV-2 spike leads to morbidity in obese and aged mice and is neutralized by convalescent and post-vaccination human sera. medRxiv. 2021 doi: 10.1101/2021.01.19.21249592. [DOI] [Google Scholar]
- Robbiani D.F., Gaebler C., Muecksch F., Lorenzi J.C.C., Wang Z., Cho A., Agudelo M., Barnes C.O., Gazumyan A., Finkin S., et al. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature. 2020;584:437–442. doi: 10.1038/s41586-020-2456-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabino E.C., Buss L.F., Carvalho M.P.S., Prete C.A., Jr., Crispim M.A.E., Fraiji N.A., Pereira R.H.M., Parag K.V., da Silva Peixoto P., Kraemer M.U.G., et al. Resurgence of COVID-19 in Manaus, Brazil, despite high seroprevalence. Lancet. 2021;397:452–455. doi: 10.1016/S0140-6736(21)00183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi R., Shan C., Duan X., Chen Z., Liu P., Song J., Song T., Bi X., Han C., Wu L., et al. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature. 2020;584:120–124. doi: 10.1038/s41586-020-2381-y. [DOI] [PubMed] [Google Scholar]
- Sun S., Gu H., Cao L., Chen Q., Yang G., Li R.-T., Fan H., Ye Q., Deng Y.-Q., Song X., et al. Characterization and structural basis of a lethal mouse-adapted SARS-CoV-2. bioRxiv. 2020 doi: 10.1101/2020.11.10.377333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suryadevara N., Shrihari S., Gilchuk P., VanBlargan L.A., Binshtein E., Zost S.J., Nargi R.S., Sutton R.E., Winkler E.S., Chen E.C., et al. Neutralizing and protective human monoclonal antibodies recognizing the N-terminal domain of the SARS-CoV-2 spike protein. Cell. 2021;184:2316–2331.e15. doi: 10.1016/j.cell.2021.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegally H., Wilkinson E., Lessells R.J., Giandhari J., Pillay S., Msomi N., Mlisana K., Bhiman J.N., von Gottberg A., Walaza S., et al. Sixteen novel lineages of SARS-CoV-2 in South Africa. Nat. Med. 2021;27:440–446. doi: 10.1038/s41591-021-01255-3. [DOI] [PubMed] [Google Scholar]
- Tian F., Tong B., Sun L., Shi S., Zheng B., Wang Z., Dong X., Zheng P. Mutation N501Y in RBD of Spike Protein Strengthens the Interaction between COVID-19 and its Receptor ACE2. bioRxiv. 2021 doi: 10.1101/2021.02.14.431117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel A.B., Kanevsky I., Che Y., Swanson K.A., Muik A., Vormehr M., Kranz L.M., Walzer K.C., Hein S., Guler A., et al. BNT162b vaccines protect rhesus macaques from SARS-CoV-2. Nature. 2021;592:283–289. doi: 10.1038/s41586-021-03275-y. [DOI] [PubMed] [Google Scholar]
- Volz E., Hill V., McCrone J.T., Price A., Jorgensen D., O’Toole Á., Southgate J., Johnson R., Jackson B., Nascimento F.F., et al. COG-UK Consortium Evaluating the Effects of SARS-CoV-2 Spike Mutation D614G on Transmissibility and Pathogenicity. Cell. 2021;184:64–75.e11. doi: 10.1016/j.cell.2020.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voysey M., Clemens S.A.C., Madhi S.A., Weckx L.Y., Folegatti P.M., Aley P.K., Angus B., Baillie V.L., Barnabas S.L., Bhorat Q.E., et al. Oxford COVID Vaccine Trial Group Safety and efficacy of the ChAdOx1 nCoV-19 vaccine (AZD1222) against SARS-CoV-2: an interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK. Lancet. 2021;397:99–111. doi: 10.1016/S0140-6736(20)32661-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Zhang Y., Huang B., Deng W., Quan Y., Wang W., Xu W., Zhao Y., Li N., Zhang J., et al. Development of an Inactivated Vaccine Candidate, BBIBP-CorV, with Potent Protection against SARS-CoV-2. Cell. 2020;182:713–721.e9. doi: 10.1016/j.cell.2020.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Nair M.S., Liu L., Iketani S., Luo Y., Guo Y., Wang M., Yu J., Zhang B., Kwong P.D., et al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature. 2021;593:130–135. doi: 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- Wang Z., Schmidt F., Weisblum Y., Muecksch F., Barnes C.O., Finkin S., Schaefer-Babajew D., Cipolla M., Gaebler C., Lieberman J.A., et al. mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. Nature. 2021;592:616–622. doi: 10.1038/s41586-021-03324-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinreich D.M., Sivapalasingam S., Norton T., Ali S., Gao H., Bhore R., Musser B.J., Soo Y., Rofail D., Im J., et al. Trial Investigators REGN-COV2, a Neutralizing Antibody Cocktail, in Outpatients with Covid-19. N. Engl. J. Med. 2021;384:238–251. doi: 10.1056/NEJMoa2035002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman D., Alameh M.G., de Silva T., Collini P., Hornsby H., Brown R., LaBranche C.C., Edwards R.J., Sutherland L., Santra S., et al. D614G Spike Mutation Increases SARS CoV-2 Susceptibility to Neutralization. Cell Host Microbe. 2021;29:23–31.e4. doi: 10.1016/j.chom.2020.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K., Werner A.P., Koch M., Choi A., Narayanan E., Stewart-Jones G.B.E., Colpitts T., Bennett H., Boyoglu-Barnum S., Shi W., et al. Serum Neutralizing Activity Elicited by mRNA-1273 Vaccine. N. Engl. J. Med. 2021;384:1468–1470. doi: 10.1056/NEJMc2102179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S., Zhong G., Zhang J., Shuai L., Zhang Z., Wen Z., Wang B., Zhao Z., Song X., Chen Y., et al. A single dose of an adenovirus-vectored vaccine provides protection against SARS-CoV-2 challenge. Nat. Commun. 2020;11:4081. doi: 10.1038/s41467-020-17972-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia S., Zhang Y., Wang Y., Wang H., Yang Y., Gao G.F., Tan W., Wu G., Xu M., Lou Z., et al. Safety and immunogenicity of an inactivated SARS-CoV-2 vaccine, BBIBP-CorV: a randomised, double-blind, placebo-controlled, phase 1/2 trial. Lancet Infect. Dis. 2021;21:39–51. doi: 10.1016/S1473-3099(20)30831-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F., Wang Q., Li M., Zhou H., Liu K., Zhang K., Wang Z., Xu Q., Xu C., Pan Q., He J. Aquarium: an automatic data-processing and experiment information management system for biological macromolecular crystallography beamlines. J. Appl. Cryst. 2019;52:472–477. [Google Scholar]
- Yuan M., Liu H., Wu N.C., Lee C.D., Zhu X., Zhao F., Huang D., Yu W., Hua Y., Tien H., et al. Structural basis of a shared antibody response to SARS-CoV-2. Science. 2020;369:1119–1123. doi: 10.1126/science.abd2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M., Wu N.C., Zhu X., Lee C.D., So R.T.Y., Lv H., Mok C.K.P., Wilson I.A. A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science. 2020;368:630–633. doi: 10.1126/science.abb7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M., Huang D., Lee C.D., Wu N.C., Jackson A.M., Zhu X., Liu H., Peng L., van Gils M.J., Sanders R.W., et al. Structural and functional ramifications of antigenic drift in recent SARS-CoV-2 variants. Science. 2021:eabh1139. doi: 10.1126/science.abh1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurkovetskiy L., Wang X., Pascal K.E., Tomkins-Tinch C., Nyalile T.P., Wang Y., Baum A., Diehl W.E., Dauphin A., Carbone C., et al. Structural and Functional Analysis of the D614G SARS-CoV-2 Spike Protein Variant. Cell. 2020;183:739–751.e8. doi: 10.1016/j.cell.2020.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zost S.J., Gilchuk P., Case J.B., Binshtein E., Chen R.E., Nkolola J.P., Schäfer A., Reidy J.X., Trivette A., Nargi R.S., et al. Potently neutralizing and protective human antibodies against SARS-CoV-2. Nature. 2020;584:443–449. doi: 10.1038/s41586-020-2548-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The coordinates and structure factors files for P2C-1F11 and RBD-3M complex have been deposited to the Protein Data Bank (http://www.rcsb.org) with accession code 7E8M.