Abstract
Objective
In order to isolate and identify bacteria that produce potentially novel bactericidal/bacteriostatic compounds, two ponds on the campus of the Rochester Institute of Technology (RIT) were targeted as part of a bioprospecting effort.
Results
One of the unique isolates, RIT 452 was identified as Exiguobacterium sp. and subjected to whole-genome sequencing. The genome was assembled and in silico analysis was performed to predict the secondary metabolite gene clusters, which suggested the potential of Exiguobacterium RIT452 for producing antibiotic compounds. Extracts of spent growth media of RIT452 were active in disc diffusion assays performed against four reference strains, two Gram-negative (E. coli ATCC 25922 and P. aeruginosa ATCC 27853) and two Gram-positive (B. subtilis BGSC 168 and S. aureus ATCC 25923). Differential extraction and liquid chromatography was used to fractionate the extracts. Efforts to identify and elucidate the structure of the active compound(s) are still ongoing.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13104-021-05644-2.
Keywords: Antibiotic resistance, Exiguobacterium, Whole-genome sequencing, Secondary metabolite, Antibiotics
Introduction
Antibiotic resistance was first discovered in the 1940s, when it was observed that bacteria were become increasingly resistant to measures taken against them [1]. However, this development did not become a global issue until antibiotic abuse in medicine and animal husbandry became more widespread [2–4]. Antibiotic resistance dramatically increased in the 1980s and 1990s due to a discovery void. The progressive increase of antibiotic resistance and the decline in research and development of antibiotics combined to produce the current crisis [5]. Since there have been few developments in the creation of new antibiotic classes for many years, bacteria are becoming increasingly resistant to those that are currently in use [1, 6]. The targets of antibiotics are diverse; protein synthesis, DNA replication, RNA synthesis and the cell membrane are common targets [4]. Antibiotics with novel targets need to be developed for replacing the existing drugs.
The β-lactam core structure is part of a number of antibiotic classes, but β-lactam resistance is widespread and is conferred by over 300 types of β-lactamases, of which 200 are able to hydrolyze extended spectrum cephalosporin antibiotics [7]. Therefore, increasing the structural diversity of antimicrobial compounds becomes important. Estimates of all possible small molecules are pegged at 1063 distinct molecules, of which many may not be accessible by synthesis [8]. Only a small fraction of these molecules are biologically relevant [9]. However, natural compounds offer better chances of finding bioactive molecules with entirely new chemical scaffolds than combinatorial chemistry libraries, since they have evolved to be bioactive, and can often enter cells via transmembrane transporters rather than by passive diffusion [10]. Statistical analysis reveals the continuing trend of chemical novelty in natural products as de-replication tools are increasingly being implemented [11]. In this context, shifting to less known bacterial phyla, rather than over-represented phyla such as the Actinobacteria might increase diversity of the antibiotics identified by prospecting [12]. The goal of our study is to identify novel bacteria that yield potentially novel compounds, which may become starting points to develop more specific and potent antibiotics.
RIT452 was identified during the screening of bacterial isolates from a campus pond, with potential bacteriostatic/bactericidal properties, and its genome was sequenced. The genome was subjected to secondary metabolite analysis via the antibiotics and secondary metabolite analysis shell 5.0 (antiSMASH) [13] and the antibiotic resistance target seeker (ARTS) platforms [14]. Organic compounds extracted from the spent growth media of RIT452 were shown to inhibit the growth of Gram negative and Gram positive reference strains.
The data presented here shows that Exiguobacterium sp. RIT452 produces a broad spectrum antibiotic, which after fractionating by differential extraction and chromatography, was not structurally characterized. The facultative anaerobic Gram-positive genus Exiguobacterium consists of motile, non-spore forming species [15], widely distributed in the environment, including extremophiles growing in high altitude salt plains [16], hot springs [17, 18], oceans, Antarctic dry valleys and permafrost [18], while others tolerate gamma radiation [19], organic solvents [20], chromium and mercury [21–23], arsenic [16, 24–26], pesticides [27], alkaline wastewater [28], and heat [29–32]. All of this is accomplished without the ability to form spores, suggesting other physiological changes help them cope with these stresses.
Main text
Methods
Bacterial growth and characterization
Tryptic soy broth (TSB) cultures were used for DNA extraction, while LB (Lysogeny broth) was used for the antibiotic production cultures. R2A (Reasoner’s 2A) minimal media were used for starvation experiments according to Yang et al. [33]. Tryptic soy agar (TSA)-grown cells were analyzed after weeks 1 and 5 by electron microscopy to examine the morphological changes. RIT452 was identified as an Exiguobacterium sp. based on the 16S rRNA gene sequence. The V3/V4 region was sequenced by Sanger nucleotide sequencing (GeneWiz LLC, South Plainfield, NJ) and was analyzed by the basic local alignment search tool (BLAST) [18].
PCR amplification and nucleotide sequencing of the 16S V3/V4 regions
Bacteria from ponds at the Rochester Institute of Technology (RIT) were isolated on TSA or R2A media. Each was subjected to PCR using primers, 5′-CCTACGGGNGGCWCGAG-3′ (forward) and 5′-GACTACHVGGGTATCTAATCC-3′ (reverse) designed to amplify the V3/V4 rRNA regions. The following thermal cycler protocol was used: 1 cycle at 95 °C for 2 min, 30 cycles each at 95 °C for 30 s, 52 °C for 30 s and 72 °C for 3 min, 1 cycle at 72 °C for 5 min, and finally infinite hold at 4 °C. The PCR products were separated by gel electrophoresis, followed by Sanger nucleotide sequencing of the amplified sequences (GeneWiz LLC, South Plainfield, NJ) prepared with the V3/V4 forward primer.
Genomic DNA isolation, library preparation, genome sequencing and alignment
RIT452 grown in 3 mL of TSB for 24 h at 30 °C. The DNA isolation and subsequent steps were performed as in Steiner et al. [34].
Predicting secondary metabolite production
The aligned genome sequence of Exiguobacterium sp. RIT 452 was analyzed using the antibiotics and secondary metabolite analysis shell (antiSMASH4.0) webserver [13]. The aligned genome was also analyzed by the antibiotic resistance target seeker (ARTS version 2) webserver [14]. ARTS predicts resistance mechanisms and BGCs from genome sequences [14].
Extraction of organic compounds from culture media
RIT 452 was cultured in 100 mL LB medium at 30 °C shaken at 130 rpm for 24 h. This was inoculated in 900 mL of LB medium and grown for an additional 48 h at 30 °C shaken at 130 rpm. The extraction of organics from the spent media and storage of concentrated extracts were performed as published earlier [34].
Compound fractionation and liquid chromatography (LC)
A five-step extraction and the subsequent separation of the most active extract by liquid chromatography were conducted using known methods [34].
Broth dilution assay vs. clinical pathogens
Three clinical pathogens were used: MRSA USA300-FPR3757 (mecA) [35], E. coli MCR1_NJ [mcr-1, blaNDM-5, strA, strB, aac(6′)-Ib-cr, blaOXA-1, arr-3, sul1, sul2, tet(A)] [36], and P. aeruginosa AR-0230 (aac(3)-Id, aadA2, dfrB5, OXA-4, OXA-50, tet(G), VIM-2) [37]. Briefly, experiments were conducted in cation adjusted Mueller Hinton broth and a starting bacterial inoculum of ~ 106 cfu/mL for each isolate. The minimum inhibitory concentration (MIC) for the crude extract against all three clinical isolates, determined using broth micro-dilution according to the Clinical and Laboratory Standards Institute (CLSI) Guidelines, was 6.25 × (equivalent to metabolites extracted from 6.25 mL of spent media) [38].
Disc diffusion assays
Disc diffusion tests of the organics extracted with different solvents was performed according to Steiner et al. [34].
Disc diffusion assays of LC fractions
The activity was tested against E. coli ATCC 25922, P. aeruginosa ATCC 27853, B. subtilis BGSC 168 and S. aureus ATCC 25923. The activity was better against the Gram positive strains (Additional file 1: Figure S1). Further enrichment of the active compounds was performed with S. aureus ATCC 25923 as the test strain. S. aureus ATCC 25923 was grown overnight in 5 mL LB medium at 30 °C in a 130 rpm shaker incubator. The cells were pelleted and each culture was re-suspended in 2 mL of sterile PBS. 180 μL of the re-suspension was mixed into tubes containing 40 ml of warm LB agar and poured into square petri dishes. The petri plates were cooled for 1 h in a sterile hood. 6 mm sterile blank paper disks (BD Biosciences, USA) were aseptically placed onto each agar plate. Methanol (20 μL), tetracycline stock (10 mg/mL, 22.5 μM, 20 μL), and each fraction (60 μL of each fraction re-suspended in 100 μL methanol) were pipetted onto the disks. The plates were dried aseptically for 1 h and incubated for 16 h at 30 °C. The diameter of each zone of inhibition (ZOI) was measured in mm.
Flow injection analysis (FIA) and liquid chromatography-mass spectrometry (LCMS)
Low resolution mass spectrometry data was obtained on an Agilent LC/MSD VL system by electrospray ionization (ESI) flow injection analysis in the (positive or negative) mode at the Boston University Chemical Instrumentation Center. A reverse-phase C18 Zorbax Eclipse 2.1 × 50 mm column (Agilent) was used, and the mobile phases were water and acetonitrile with 0.1% formic acid. Separation was achieved by a flow rate of 0.15 mL/min and a mobile phase gradient from 5 to 95% acetonitrile in 10 min. The MS settings were: voltage = 3000 V, fragmentor = 70 and chamber temperature = 350 °C.
Scanning electron microscopy (SEM)
The microbiological sample preparation followed an open source protocol [39]. Samples were covered with gold–palladium for 2 minutes with an SPI sputter coater to mitigate charging in the electron beam. The SEM was performed at 5 kV using a Mira3Tescan field emission SEM at the Rochester Institute of Technology (RIT) Nano-Imaging Lab.
Results
Strain characterization and phylogeny
The genome was sequenced using an Illumina MiSeq. After assembly, the genome sequence has been deposited in GenBank under accession number QXJB00000000 (BioProject number PRJNA489292; BioSample number SAMN09954399).
Electron microscopy analysis
RIT452 grown on solid media under different conditions and Fig. 1 shows the scanning electron microscopy (SEM) images recorded for each. In minimal media [33], the cells grow longer, closer to 2 µm, and form larger aggregates (Fig. 1A). Healthy cells are around 1 µm in length and around 0.5 µm wide (Fig. 1B); However when healthy cells are grown for prolonged periods in rich broth media, the cell surface alters and appears rougher (Fig. 1C).
Fig. 1.
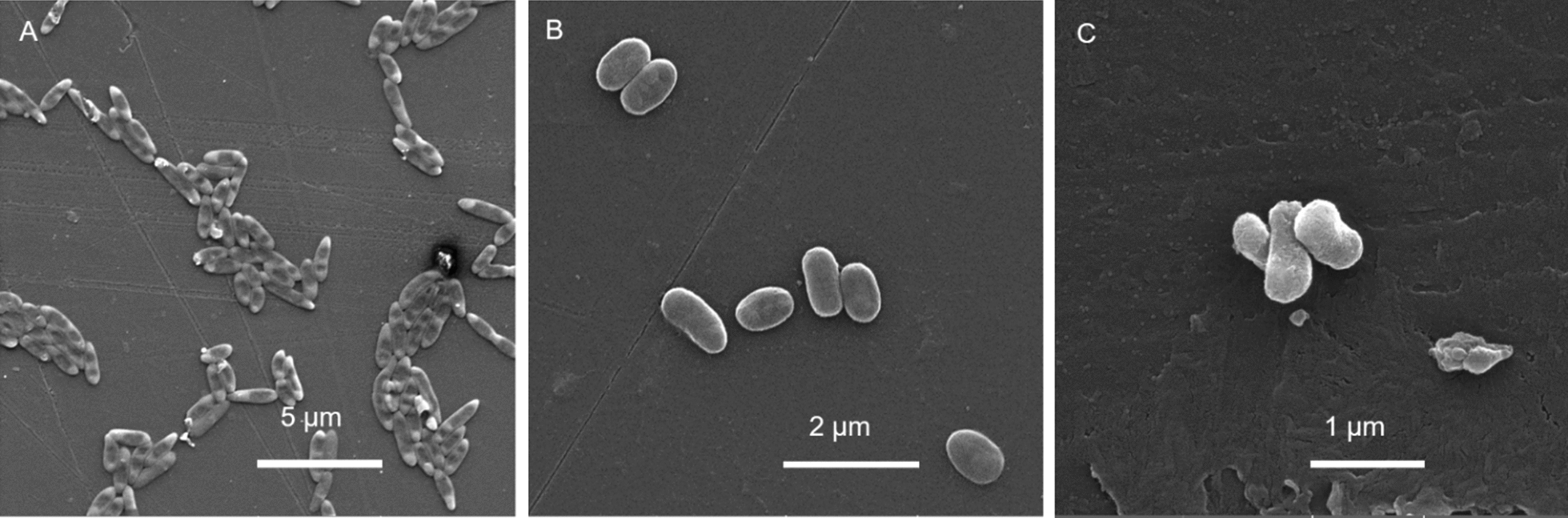
SEM analysis. Exiguobacterium RIT452 under different conditions. A Cell elongation observed due to starvation on R2A (minimal medium); B cells grown on TSA for 1 week and C cells grown on TSA for 5 weeks. The magnifications are 9830 ×, 26,500 ×, and 44,900 ×, respectively
antiSMASH and ARTS results
The antiSMASH 5.0 web tool analyzes genome sequences and predicts the secondary metabolites produced by an organism [13]. antiSMASH predicted that RIT452 contained four biosynthetic gene clusters (BGCs), two terpene clusters, a siderophore cluster, and a putative antibiotic cluster (Table 1). The “putative antibiotic” BGC identified at locus 897206–908755 shares sequence similarity with a Lugdunin locus, an antibiotic first isolated from S. lugdunensis, a commensal of the human nasal cavity [40]. This novel thiazolidine-containing cyclic peptide antimicrobial was shown to be effective against S. aureus. Using the genome sequence of RIT 452, ARTS predicted three BGCs, out of which two show “proximity hits” (Table 1). The “proximity hits” readout in the ARTS program shows scaffold-specific self-resistance [14], which is considered a good way to mine for novel antibiotic BGCs [41].
Table 1.
Secondary metabolites predicted by antiSMASH 5.0 genome mining
| Cluster no. | Predicted biosynthetic metabolite | Coordinates within the genome | % Similarity to known cluster | ARTS BGC proximity hits |
|---|---|---|---|---|
| 1 | Terpene | 69,899–90,726 | 33 (with carotenoid biosynthetic gene cluster) | – |
| 4 | Siderophore | 5,05,835–5,19,165 | – | Yes |
| 9 | Putative antibiotic | 8,97,206–9,08,755 | 26 (with lugdunin biosynthetic gene cluster) | |
| 20 | Terpene | 1,60,337–1,81,161 | – | Yes |
The compounds shown in this table show four of the 24 compounds antiSMASH predicted, which could potentially exhibit antibiotic characteristics. If core and resistance genes have intersecting locations on the same scaffold as predicted by ARTS, they are marked as BGC proximity hits
Broth dilution assays
The crude extract from 1 Liter of spent RIT 452 media was able to inhibit the pathogens MRSA, E. coli ncr1_NJ, and P. aeruginosa AR-230 at moderate concentrations and the apparent minimum inhibitory concentrations (MIC) in each case is shown in the Additional file 1: Table S1.
Disc diffusion assays
Crude ethyl acetate extract from spent LB media was spotted on sterile discs and plated on LB agar seeded with different species of bacteria which are not clinical pathogens, whereby the extract inhibited the growth of all strains tested (Additional file 1: Figure S1). The zones of inhibition (ZOIs) show a graded increase with increased volume of extract. The dose response curve created using the ZOIs shows that the extract inhibited the growth of S. aureus, E. coli, and P. aeruginosa along roughly the same trend while B. subtilis had a steeper increase in dosage response (Additional file 1: Figure S1). Among the crude extracts obtained by processing with hexanes, toluene, ether, dichloromethane and ethyl acetate, only one fraction (diethyl ether) showed a ZOI.
Enrichment of active metabolites using liquid chromatography
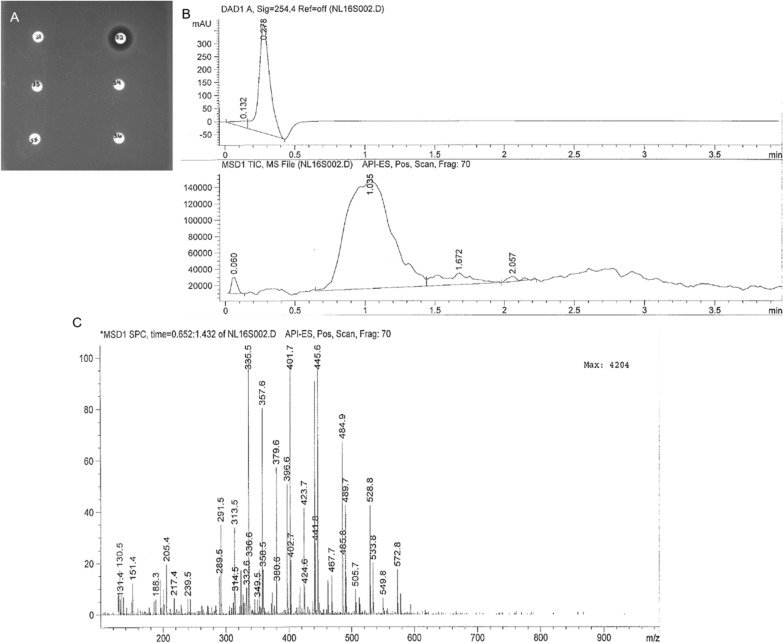
The diethyl ether extract when subjected to liquid chromatography using a C18 column (see methods) with S. aureus as the test strain. The extract when fractionated showed activity only in fraction 32 (ZOI = 15 mm), corresponding to a single peak in the LC channel of the LCMS (Fig. 2A), but at least four ionizable species in the positive scan mode (Fig. 2B). The largest peak corresponding to 0.652 min shows the masses 335.5, 401.7 and 445.6 in the positive scan mode. The active compound(s) were not further characterized.
Fig. 2.
Disc assay using LC fractions 31–36 from diethyl ether extraction and LCMS data. A The LC protocol was run to collect fractions of the RIT452 diethyl ether extract. Each was condensed, fully dried, and re-suspended in 100 µl of methanol. 60 µl of each fraction was added to a sterile diffusion disc and plated on LB agar seeded with S. aureus. The zone of inhibition (ZOI) of fraction 32 is 15 mm after 16 h of growth. B The LC channel in the LCMS shows only one chromatographic peak, while the MS channel shows at least four ionizable species in the positive scan mode. C The MS pattern corresponding to the beginning of the largest MS peak (0.652 min) in B shows the peaks 335.5, 401.7 and 445.6 in the positive scan mode between 100 and 1000 m/z
Discussion
Exiguobacterium strains have been previously reported to nearly triple their cell length after exposure to organic solvents [20], whereas RIT452 also undergoes cell elongation upon starvation. This suggests that cell elongation might be a generic stress response in the Exigobacterium genus. However, it is not known if this response also occurs as a result of interaction with other bacteria.
An Indian rhizosphere Exiguobacterium strain produces a broad spectrum with an unusual dihydroergotamine-type antibiotic [42], apart from which little is known about bioactive compounds produced by the genus Exiguobacterium. The diversity of microbial metabolites may be partly underestimated, since each BGC does not necessarily produce only one metabolite, and growth conditions may change the metabolite produced by altering the expression of the same BGC [43]. Sequence identity at the level of genes in BGC producing similar compounds were reported to range between 58 and 80% [44]. By these criteria, the similarity to known BGC for our data is not very high, suggesting potential novelty.
The suggestion of metabolite novelty is encouraging since the RIT 452 extract contains compound(s) inhibiting a Gram positive clinical strain (MRSA) at lower concentrations (apparent MIC) than Gram negative clinical strains (Additional file 1: Table S1). Fractionation of the extracts using a non-clinical S. aureus strain shows that the activity is concentrated in the range of 95% acetonitrile, which hints at a relatively non-polar compound. Further “wet lab” experiments are needed to identify the bioactive compound(s) to verify the predictions made by bioinformatics analysis.
Limitations
The active fraction contains more than one ionizable species and the chemical structure/s of the active compound/s are still unknown. MIC values and safety data of the isolated compound are also unknown.
Supplementary Information
Additional file 1: Figure S1. A–D: Disc diffusion assay. RIT452 extracts (on discs) inhibit the growth of (A) E. coli, (B) S. aureus, (C) P. aeruginosa, and (D) B. subtilis. On each plate from top left to right the discs have 20 µl Tetracycline at 10 mg/ml (1), 10 µl extract (2), 20 µl extract (3), 40 µl extract (4), 60 µl extract (5), and 20 µl methanol as a negative control (6). (E) The inhibition zones in each case show a graded increase in the diameter of the zone of inhibition with increasing amounts of extract. Error bars from triplicate readings are shown and it can be seen that the linear range goes approximately up to 20 µL. Table S1. Estimated apparent MICs of the RIT 452 crude extracts computed according to the CLSI guidelines.
Acknowledgements
We thank Prof. Richard Hailstone Center for Imaging Sciences at RIT for help with the SEM analysis, and Norman Lee of the Boston University Chemical Instrumentation Center for running and analyzing the LC-MS samples. We thank Dr. Zachary P. Bulman of the College of Pharmacy, University of Illinois at Chicago, for performing the broth dilution assays with the clinical strains.
Abbreviations
- antiSMASH
Antibiotics and secondary metabolite analysis shell
- ARTS
Antibiotic resistance target seeker
- BGC
Biosynthetic gene cluster
- CLSI
Clinical and Laboratory Standards Institute
- ESI
Electrospray ionization
- LB
Lysogeny broth
- LC
Liquid chromatography
- LCMS
Liquid chromatography-mass spectrometry
- MIC
Minimum inhibitory concentration
- MRSA
Methicillin resistant Staphylococcus aureus
- MS
Mass spectrometry
- R2A
Reasoner’s 2A
- RIT
Rochester Institute of Technology
- SEM
Scanning electron microscopy
- TSA
Tryptic soy agar
- TSB
Tryptic soy broth
- ZOI
Zone of inhibition
Authors’ contributions
Research was conducted by NTC. The manuscript was written by NTC, AP and AOH. NHW helped with whole-genome sequencing and phylogenetic analysis. KKS helped isolate the bacterium and JC helped with liquid chromatography. JA performed assays to facilitate resubmission. The study was designed by AOH and the extraction, chromatography and SEM methods were developed by AP. All authors read and approved the final manuscript.
Funding
This research was funded by a National Institutes of Health award (RG15GM120653) to AOH.
Availability of data and materials
The whole-genome project for Exiguobacterium sp. RIT452 is available in GenBank with the accession number QXJB00000000.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Nicole T. Cavanaugh, Email: cavanaugh.n@northeastern.edu
Anutthaman Parthasarathy, Email: axpsbi@rit.edu.
Narayan H. Wong, Email: narayan.wong@chromamedicine.com
KayLee K. Steiner, Email: kaylee.k.steiner@vanderbilt.edu
Jonathan Chu, Email: jxc9163@rit.edu.
Joseph Adjei, Email: joa9194@g.rit.edu.
André O. Hudson, Email: aohsbi@rit.edu
References
- 1.Ventola CL. The antibiotic resistance crisis: part 1: causes and threats. P T. 2015;40:277–83. [PMC free article] [PubMed] [Google Scholar]
- 2.Thornsberry C, Sahm DF, Kelly LJ, Critchley IA, Jones ME, Evangelista AT, Karlowsky JA. Regional trends in antimicrobial resistance among clinical isolates of Streptococcus pneumoniae, Haemophilus influenzae, and Moraxella catarrhalis in the United States: results from the TRUST Surveillance Program, 1999–2000. Clin Infect Dis. 2002;34(Suppl 1):S4–16. doi: 10.1086/324525. [DOI] [PubMed] [Google Scholar]
- 3.Palumbi SR. Humans as the world’s greatest evolutionary force. Science. 2001;293:1786–90. doi: 10.1126/science.293.5536.1786. [DOI] [PubMed] [Google Scholar]
- 4.Walsh C. Antibiotics: actions, origins, resistance. USA: ASM Press; 2003. [Google Scholar]
- 5.Coates A, Hu Y, Bax R, Page C. The future challenges facing the development of new antimicrobial drugs. Nat Rev Drug Discov. 2002;1:895–910. doi: 10.1038/nrd940. [DOI] [PubMed] [Google Scholar]
- 6.Ventola CL. The antibiotic resistance crisis: part 2: management strategies and new agents. P T. 2015;40:344–52. [PMC free article] [PubMed] [Google Scholar]
- 7.Ghafourian S, Sadeghifard N, Soheili S, Sekawi Z. Extended spectrum beta-lactamases: definition, classification and epidemiology. Curr Issues Mol Biol. 2015;17:11–21. [PubMed] [Google Scholar]
- 8.Bohacek RS, McMartin C, Guida WC. The art and practice of structure-based drug design: a molecular modeling perspective. Med Res Rev. 1996;16:3–50. doi: 10.1002/(SICI)1098-1128(199601)16:1<3::AID-MED1>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 9.Lipinski C, Hopkins A. Navigating chemical space for biology and medicine. Nature. 2004;432:855–61. doi: 10.1038/nature03193. [DOI] [PubMed] [Google Scholar]
- 10.Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev. 2001;46:3–26. doi: 10.1016/S0169-409X(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 11.Skinnider MA, Magarvey NA. Statistical reanalysis of natural products reveals increasing chemical diversity. Proc Natl Acad Sci. 2017;114:E6271. doi: 10.1073/pnas.1708560114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smanski MJ, Schlatter DC, Kinkel LL. Leveraging ecological theory to guide natural product discovery. J Ind Microbiol Biotechnol. 2016;43:115–28. doi: 10.1007/s10295-015-1683-9. [DOI] [PubMed] [Google Scholar]
- 13.Blin K, Shaw S, Steinke K, Villebro R, Ziemert N, Lee SY, Medema MH, Weber T. antiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 2019;47:W81–7. doi: 10.1093/nar/gkz310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alanjary M, Kronmiller B, Adamek M, Blin K, Weber T, Huson D, Philmus B, Ziemert N. The antibiotic resistant target seeker (ARTS), an exploration engine for antibiotic cluster prioritization and novel drug target discovery. Nucleic Acids Res. 2017;45:W42–8. doi: 10.1093/nar/gkx360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Collins MD, Lund BM, Farrow JAE, Schleifer KH. Chemotaxonomic study of an alkalophilic bacterium, Exiguobacterium aurantiacum gen. nov., sp. nov. Microbiology. 1983;129:2037–42. doi: 10.1099/00221287-129-7-2037. [DOI] [Google Scholar]
- 16.Castro-Severyn J, Remonsellez F, Valenzuela SL, Salinas C, Fortt J, Aguilar P, Pardo-Esté C, Dorador C, Quatrini R, Molina F, Aguayo D, Castro-Nallar E, Saavedra CP. Comparative genomics analysis of a new Exiguobacterium strain from Salar de Huasco Reveals a repertoire of stress-related genes and arsenic resistance. Front Microbiol. 2017;8:456. doi: 10.3389/fmicb.2017.00456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vishnivetskaya TA, Lucas S, Copeland A, Lapidus A, Glavina del Rio T, Dalin E, Tice H, Bruce DC, Goodwin LA, Pitluck S, Saunders E, Brettin T, Detter C, Han C, Larimer F, Land ML, Hauser LJ, Kyrpides NC, Ovchinnikova G, Kathariou S, Ramaley RF, Rodrigues DF, Hendrix C, Richardson P, Tiedje JM. Complete genome sequence of the thermophilic bacterium Exiguobacterium sp. AT1b. J Bacteriol. 2011;193:2880–1. doi: 10.1128/JB.00303-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vishnivetskaya TA, Chauhan A, Layton AC, Pfiffner SM, Huntemann M, Copeland A, Chen A, Kyrpides NC, Markowitz VM, Palaniappan K, Ivanova N, Mikhailova N, Ovchinnikova G, Andersen EW, Pati A, Stamatis D, Reddy TB, Shapiro N, Nordberg HP, Cantor MN, Hua XS, Woyke T. Draft genome sequences of 10 strains of the genus Exiguobacterium. Genome Announc. 2014 doi: 10.1128/genomeA.01058-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pathak R, Kumar R, Gautam HK. Cross-species induction and enhancement of antimicrobial properties in response to gamma irradiation in Exiguobacterium sp. HKG 126. Indian J Microbiol. 2013;53:130–6. doi: 10.1007/s12088-013-0369-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kongpol A, Kato J, Tajima T, Vangnai AS. Characterization of acetonitrile-tolerant marine bacterium Exiguobacterium sp. SBH81 and its tolerance mechanism. Microbes Environ. 2012;27:30–5. doi: 10.1264/jsme2.ME11228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pattanapipitpaisal P, Mabbett AN, Finlay JA, Beswick AJ, Paterson-Beedle M, Essa A, Wright J, Tolley MR, Badar U, Ahmed N, Hobman JL, Brown NL, Macaskie LE. Reduction of Cr(VI) and bioaccumulation of chromium by gram positive and gram negative microorganisms not previously exposed to Cr-stress. Environ Technol. 2002;23:731–45. doi: 10.1080/09593332308618367. [DOI] [PubMed] [Google Scholar]
- 22.Okeke BC. Bioremoval of hexavalent chromium from water by a salt tolerant bacterium, Exiguobacterium sp. GS1. J Ind Microbiol Biotechnol. 2008;35:1571–9. doi: 10.1007/s10295-008-0399-5. [DOI] [PubMed] [Google Scholar]
- 23.Petrova MA, Mindlin SZ, Gorlenko ZM, Kaliaeva ES, Soina VS, Bogdanova ES. Mercury-resistant bacteria from permafrost sediments and prospects for their use in comparative studies of mercury resistance determinants. Genetika. 2002;38:1569–74. [PubMed] [Google Scholar]
- 24.Anderson CR, Cook GM. Isolation and characterization of arsenate-reducing bacteria from arsenic-contaminated sites in New Zealand. Curr Microbiol. 2004;48:341–7. doi: 10.1007/s00284-003-4205-3. [DOI] [PubMed] [Google Scholar]
- 25.Ordoñez OF, Rasuk MC, Soria MN, Contreras M, Farías ME. Haloarchaea from the Andean Puna: biological role in the energy metabolism of arsenic. Microb Ecol. 2018 doi: 10.1007/s00248-018-1159-3. [DOI] [PubMed] [Google Scholar]
- 26.Belfiore C, Ordoñez OF, Farías ME. Proteomic approach of adaptive response to arsenic stress in Exiguobacterium sp. S17, an extremophile strain isolated from a high-altitude Andean lake stromatolite. Extremophiles. 2013;17:421–31. doi: 10.1007/s00792-013-0523-y. [DOI] [PubMed] [Google Scholar]
- 27.Kumar A, Singh V, Kumar R. Characterization of an alkaliphile, Exiguobacterium sp. and it’s application in bioremediation. p 115. https://citeseerx.ist.psu.edu/viewdoc/download?doi=10.1.1.128.2541&rep=rep1&type=pdf#page=114
- 28.López L, Pozo C, Rodelas B, Calvo C, Juárez B, Martínez-Toledo MV, González-López J. Identification of bacteria isolated from an oligotrophic lake with pesticide removal capacities. Ecotoxicology. 2005;14:299–312. doi: 10.1007/s10646-003-6367-y. [DOI] [PubMed] [Google Scholar]
- 29.Usuda Y, Kawasaki H, Shimaoka M, Utagawa T. Molecular characterization of guanosine kinase gene from a facultative alkalophile, Exiguobacterium aurantiacum ATCC 35652. Biochim Biophys Acta. 1998;1442:373–9. doi: 10.1016/S0167-4781(98)00166-3. [DOI] [PubMed] [Google Scholar]
- 30.Suga S, Koyama N. Purification and properties of a novel azide-sensitive ATPase of Exiguobacterium aurantiacum. Arch Microbiol. 2000;173:200–5. doi: 10.1007/s002039900129. [DOI] [PubMed] [Google Scholar]
- 31.Kasana RC, Pandey CB. Exiguobacterium: an overview of a versatile genus with potential in industry and agriculture. Crit Rev Biotechnol. 2018;38:141–56. doi: 10.1080/07388551.2017.1312273. [DOI] [PubMed] [Google Scholar]
- 32.Kasana RC, Yadav SK. Isolation of a psychrotrophic Exiguobacterium sp. SKPB5 (MTCC 7803) and characterization of its alkaline protease. Curr Microbiol. 2007;54:224–9. doi: 10.1007/s00284-006-0402-1. [DOI] [PubMed] [Google Scholar]
- 33.Yang Y, Yang J, Wu WM, Zhao J, Song Y, Gao L, Yang R, Jiang L. Biodegradation and mineralization of polystyrene by plastic-eating mealworms: part 2. role of gut microorganisms. Environ Sci Technol. 2015;49:12087–93. doi: 10.1021/acs.est.5b02663. [DOI] [PubMed] [Google Scholar]
- 34.Steiner KK, Parthasarathy A, Wong NH, Cavanaugh NT, Chu J, Hudson AO. Isolation and whole-genome sequencing of Pseudomonas sp. RIT 623, a slow-growing bacterium endowed with antibiotic properties. BMC Res Notes. 2020;13:370. doi: 10.1186/s13104-020-05216-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Diep BA, Gill SR, Chang RF, Phan TH, Chen JH, Davidson MG, Lin F, Lin J, Carleton HA, Mongodin EF, Sensabaugh GF, Perdreau-Remington F. Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. Lancet. 2006;367:731–9. doi: 10.1016/S0140-6736(06)68231-7. [DOI] [PubMed] [Google Scholar]
- 36.Mediavilla JR, Patrawalla A, Chen L, Chavda KD, Mathema B, Vinnard C, Dever LL, Kreiswirth BN. Colistin- and carbapenem-resistant Escherichia coli harboring mcr-1 and blaNDM-5, causing a complicated urinary tract infection in a patient from the United States. MBio. 2016 doi: 10.1128/mBio.01191-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lutgring JD, Machado MJ, Benahmed FH, Conville P, Shawar RM, Patel J, Brown AC. FDA-CDC Antimicrobial Resistance Isolate Bank: a publicly available resource to support research, development, and regulatory requirements. J Clin Microbiol. 2018 doi: 10.1128/JCM.01415-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wikler MA. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically: approved standard. CLSI. 2015;26:M7–A7. [Google Scholar]
- 39.Parthasarathy A. 2019. Scanning electron microscopy (SEM) for microbes- a simple and inexpensive method for sample preparation. 10.13140/RG.2.2.26222.05449
- 40.Zipperer A, Konnerth MC, Laux C, Berscheid A, Janek D, Weidenmaier C, Burian M, Schilling NA, Slavetinsky C, Marschal M, Willmann M, Kalbacher H, Schittek B, Brötz-Oesterhelt H, Grond S, Peschel A, Krismer B. Human commensals producing a novel antibiotic impair pathogen colonization. Nature. 2016;535:511–6. doi: 10.1038/nature18634. [DOI] [PubMed] [Google Scholar]
- 41.Thaker MN, Waglechner N, Wright GD. Antibiotic resistance-mediated isolation of scaffold-specific natural product producers. Nat Protoc. 2014;9:1469–79. doi: 10.1038/nprot.2014.093. [DOI] [PubMed] [Google Scholar]
- 42.Shanthakumar SP, Duraisamy P, Vishwanath G, Selvanesan BC, Ramaraj V, Vasantharaj DB. Broad spectrum antimicrobial compounds from the bacterium Exiguobacterium mexicanum MSSRFS9. Microbiol Res. 2015;178:59–65. doi: 10.1016/j.micres.2015.06.007. [DOI] [PubMed] [Google Scholar]
- 43.Martinet L, Naômé A, Deflandre B, Maciejewska M, Tellatin D, Tenconi E, Smargiasso N, de Pauw E, van Wezel GP, Rigali S. A single biosynthetic gene cluster is responsible for the production of bagremycin antibiotics and ferroverdin iron chelators. mBio. 2019 doi: 10.1128/mBio.01230-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tran PN, Yen MR, Chiang CY, Lin HC, Chen PY. Detecting and prioritizing biosynthetic gene clusters for bioactive compounds in bacteria and fungi. Appl Microbiol Biotechnol. 2019;103:3277–87. doi: 10.1007/s00253-019-09708-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. A–D: Disc diffusion assay. RIT452 extracts (on discs) inhibit the growth of (A) E. coli, (B) S. aureus, (C) P. aeruginosa, and (D) B. subtilis. On each plate from top left to right the discs have 20 µl Tetracycline at 10 mg/ml (1), 10 µl extract (2), 20 µl extract (3), 40 µl extract (4), 60 µl extract (5), and 20 µl methanol as a negative control (6). (E) The inhibition zones in each case show a graded increase in the diameter of the zone of inhibition with increasing amounts of extract. Error bars from triplicate readings are shown and it can be seen that the linear range goes approximately up to 20 µL. Table S1. Estimated apparent MICs of the RIT 452 crude extracts computed according to the CLSI guidelines.
Data Availability Statement
The whole-genome project for Exiguobacterium sp. RIT452 is available in GenBank with the accession number QXJB00000000.