1. Introduction
Routine fecal indicator monitoring is used to manage recreational waters throughout the world typically measuring E. coli or enterococci bacteria levels. These fecal indicator bacteria (FIB) are indigenous to the gastrointestinal tract of human and many other animals (APHL 2012, Harwood et al. 2017). The presence of FIB in recreational waters estimates the total level of fecal pollution present and serves as a warning for the potential presence of disease-causing enteric microorganisms. For decades, FIB testing has been a remarkably successful tool for managing recreational water quality. However, there is a growing body of evidence suggesting that viral fecal indicators could further enhance public health protection. Epidemiology, microbial risk assessment, and outbreak surveillance studies report that many illnesses associated with recreating in waters contaminated with sewage result from human enteric virus exposure (Begier et al. 2008, Cabelli et al. 1982, Sinclair et al. 2009, Soller et al. 2010) and that FIB often correlate poorly with these pathogens (Korajkic et al. 2018). As a result, researchers are investigating the potential addition of virus-based somatic and F+ coliphage methodologies to the recreational water monitoring general fecal indicator toolbox.
Coliphage are viruses that infect E. coli and are classified into somatic and F+ (male-specific) groups based on host bacterial cell modes of infection (Cole et al. 2003, Mesquita et al. 2010). There are numerous reliable and inexpensive methods to measure coliphage levels in surface waters (Blanch et al. 2020) and many studies report coliphage in marine waters, rivers, and lakes known to be polluted by fecal waste (Bonadonna et al. 2019, Wanjugi et al. 2018). Furthermore, both coliphage types occur in the gut of humans and are routinely detected in sewage (Korajkic et al. 2020, Nappier et al. 2019). Coliphage are also reported to share similarities in morphology, composition, and persistence with some human DNA and RNA enteric virus pathogens (Gerba 1987, Havelaar et al. 1993, King et al. 2011, Palmateer et al. 1991, Rose et al. 2004). In addition, multiple studies report a likely relationship between coliphage and increased gastroenteritis (Abdelzaher et al. 2010, Griffith et al. 2016, Wiedenmann et al. 2006) suggesting that their presence in recreational waters may indicate a potential public health risk.
Prior to the development of a recreational water quality application, it may be useful to consider the cooccurrence of coliphage and host-associated genetic markers commonly used to characterize fecal pollution sources in recreational waters. Like traditional FIB, coliphage are shed in multiple animal sources, in some instances at similar levels to sewage. For example, somatic coliphages were found in dog, cat, horse, chicken, sheep, cow, and gull feces at similar concentration ranges to sewage (McMinn et al. 2014). The prevalence of these general fecal indicators in multiple animal groups, whether FIB or coliphage, can be limiting for routine recreational water quality monitoring because mitigation strategies often vary by fecal pollution source present. Numerous quantitative fecal source identification methodologies are available to complement routine general fecal indicator recreational water quality monitoring providing the ability to assess in parallel the potential public health risk (general indicator results) and characterize corresponding fecal source pollution trends from key animal groups (fecal source identification results) (Feng et al. 2018, Feng and McLellan 2019, Kildare et al. 2007, McQuaig et al. 2009, Mieszkin et al. 2009, Reischer et al. 2006, Stachler et al. 2017). While there are many applications for quantitative fecal source identification tools such as measuring the influence of rain events (Garcia-Aljaro et al. 2017, Reischer et al. 2008, Shrestha et al. 2020b, Staley et al. 2018), prioritizing sites for mitigation based on human fecal pollution levels (Cao et al. 2018), evaluating the efficacy of source targeted best management practices (Converse et al. 2009, Ervin et al. 2014, McMinn et al. 2019), and identifying seasonal pollutant source trends (Lee et al. 2014, Li et al. 2019, McKee et al. 2020), there also remains a keen interest in the development of fecal source prioritization strategies designed to reduce fecal indicator levels used to regulate local recreational waters (i.e. E. coli, enterococci, etc.) thereby improving public health safety and minimizing the number of recreational water closures. As a result, there is a growing body of research investigating this application in recreational water settings (Li et al. 2019, Shrestha et al. 2020a). However, information on the cooccurrence of coliphage and quantitative fecal source identification targets during routine recreational water monitoring remains scarce.
Our study evaluates paired measurements of host-associated genetic markers (this study) with cultivated viral (somatic and F+ coliphage) and bacterial (E. coli and enterococci) general fecal indicators (Wanjugi et al. 2018) at Great Lakes Basin recreational area beach and river sites. Six recreational area sites including lake beaches and nearby river locations were routinely sampled five days a week over a swimming season (15 weeks) and tested with quantitative real-time PCR (qPCR) targeting host-associated genetic markers indicating human, ruminant, dog, and avian fecal pollution. To prioritize key fecal pollution sources under different fecal indicator routine monitoring schemes, weighted fecal scores for each host-associated genetic marker concentration and indicator combination were estimated using a recently developed Bayesian censored data analysis approach (Cao et al. 2018). Findings demonstrate that the general fecal indicator used for routine monitoring can influence the interpretation of corresponding host-associated genetic marker measurements, in some cases, leading to the prioritization of different pollutant sources for remediation. Different fecal source trends were also observed between Great Lake beach and river sites suggesting disparate management practices may be useful for each water type.
2. Materials and methods
2.1. Site description
Quantitative fecal source identification testing was conducted on the same Great Lakes recreational area surface water samples used in a previous study reporting general fecal indicator levels for E. coli, enterococci, and coliphage (somatic and F+) (Wanjugi et al. 2018). Briefly, recreational areas included Edgewater (Lake Erie, Cuyahoga Watershed, near Cleveland, OH), Grant Park (Lake Michigan, Oak Creek Watershed, south Milwaukee, WI), and Washington Park (Lake Michigan, Trail Creek Watershed, Michigan City, IN). At each recreational area, quantitative fecal source identification testing was conducted at a recreational beach and at a nearby discharging river site (total of 6 sampling sites across all recreational areas). Potential fecal pollution sources at these sites could include combined sewer overflow discharge, urban stormwater runoff, faulty septic systems, local pets (canines), shore birds, ruminants (deer and potential upstream animal feeding operations), as well as nearby wastewater treatment facilities (Grant Park and Edgewater areas only).
2.2. General fecal indicator data sets
This study uses E. coli, enterococci, somatic coliphage, and F+ coliphage general fecal indicator measurements from 365 water samples reported by Wanjugi and collaborators (2018) (Fig. 1). Briefly, E. coli counts (most probable number [MPN] per 100 mL of water sample) were obtained using Colilert Quantitray (Idexx, Westbrook, ME). Enterococci concentrations (colony forming units [CFU] per 100 mL of water sample) were determined by membrane filtration on mEI agar (USEPA 2009). Somatic and F+ coliphage were enumerated (plaque forming units [PFU] per 1 L of water sample) using a dead-end hollow fiber ultrafiltration combined with single agar overlay method (McMinn et al. 2017). Additional details such as positive, negative, and spike recovery controls, as well as sampling site and water type (beach or river) trends are reported elsewhere (Wanjugi et al. 2018).
Fig. 1.
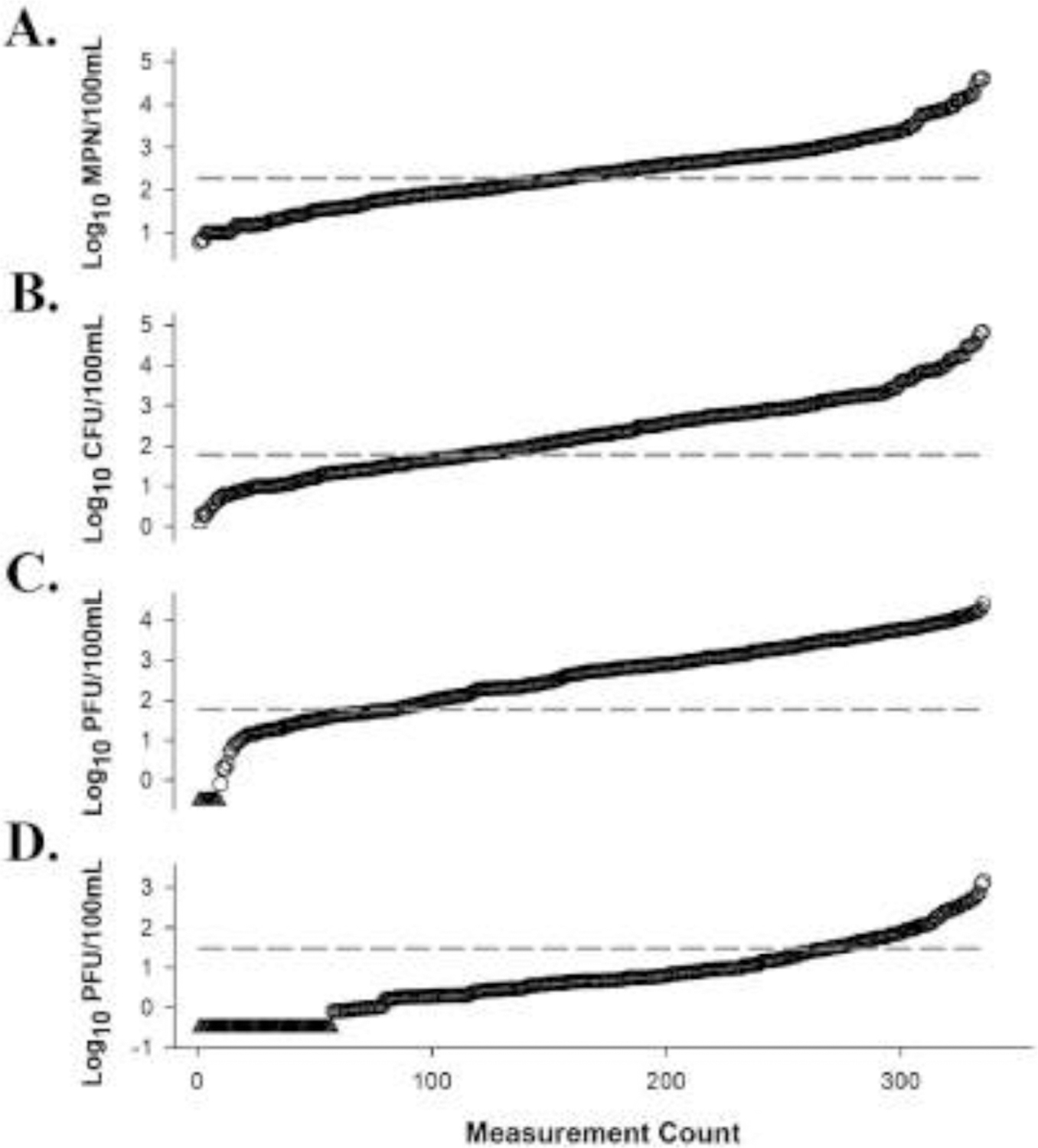
General fecal indicator measurements (n = 365) for E. coli (Panel A), enterococci (Panel B), somatic coliphage (Panel C), and F+ coliphage (Panel D) ranked from lowest to highest quantities (values increase from left to right) (Wanjugi et al. 2018). Data indicated by a triangle denotes a non-detect, while data denoted by circles represents a general fecal indicator measurement. The dashed line represents general fecal indicator data grouping thresholds used in this study for E. coli (190 most probable number (MPN)/100mL), enterococci (60 culture forming unit (CFU)/100mL), somatic coliphage (60 plaque forming unit (PFU)/100mL), and F+ coliphage (30 PFU/100mL).
2.3. Water sampling
The exact surface water sample grabs used for general fecal indicator monitoring (Wanjugi et al. 2018) were also used for quantitative fecal source identification. Briefly, samples were collected by three local laboratories including Scientific Methods Inc. (Washington Park), City of Racine Health Department (Grant Park), and Northeast Ohio Regional Sewer District (Edgewater). Beach composite (6 separate grabs per day, mixed) and river (single grab per day) water samples were collected over a 15-week beach season between 8am and 10am, five days a week (Sunday-Thursday). For beach composite sampling, transects were arranged in a parallel fashion with each sample location situated 50 m apart with the center sample coinciding with the most heavily used swimming area (data not shown). For each composite sample, equal sample volumes were mixed (total volume = 6L) in a sterilized container prior to fecal indicator testing. River samples were collected 1 m from bank at a depth of 0.3 m below the surface. Fecal indicator bacteria testing was performed by each local laboratory within 4 h of collection. Coliphage samples (2 L each) were transported to the U.S. Environmental Protection Agency research facility (Cincinnati, OH) on ice in sterilized containers. Coliphage samples were eligible for testing only if received within 48 h of sample collection and shipping temperature was maintained at ≤ 4°C. Shipping temperature was monitored with iButton® temperature loggers DS1920 according to manufacturer’s procedures (Maxim Integrated, San Jose, CA).
2.4. Reference DNA preparation
Reference DNA materials consisted of two plasmid constructs (Integrated DNA Technologies, Coralville, IA) and salmon sperm DNA (Sigma-Aldrich, St. Louis, MO). Plasmid constructs for internal amplification controls (IAC) and calibration standards (all DNA targets on single construct) were prepared as previously described (Li et al. 2019). Briefly, plasmid constructs were linearized, quantified, and diluted to generate 102 copies/2µL for IAC reference material and 10, 102, 103, 104, and 105 copies/2µL for calibration standards. A salmon DNA working stock containing 10 µg/mL was prepared by diluting a commercially available 10 mg/mL solution. All reference DNA materials were stored in GeneMate Slick low-adhesion microcentrifuge tubes (ISC BioExpress, Kaysville, UT) at −20°C.
2.5. Water filtration and DNA extraction
A total of 365 water samples were filtered for DNA extraction processing. For each sample, 100 mL was filtered through a 0.45 µm polycarbonate filter (Fisher Scientific, Pittsburg, PA) and placed in a sterile 2 mL screw cap tube containing silica bead mill matrix (GeneRite, North Brunswick, NJ) and stored at −80°C (< 18 months) until DNA extraction. DNA extraction was performed using the DNA-EZ RW02 kit (GeneRite, North Brunswick, NJ) as previously described (Li et al. 2019). Briefly, 600 µL of 0.02 µg/mL salmon sperm DNA (Sigma-Aldrich) was added to each bead mill tube followed by bead milling with a MP FastPrep-24 (MP Biomedicals, LLC Solon, OH) at 6.0 m/s for 30 s. Three method extraction blanks (MEB), with purified water substituted for test sample, were performed with each sample processing batch (12 samples/batch). DNA was eluted with 100 µL elution buffer into GeneMate Slick low-adhesion microcentrifuge tubes (ISC BioExpress, Kaysville, UT). DNA extracts were stored at 4°C prior to qPCR amplification (< 48 h).
2.6. qPCR amplification
Five host-associated qPCR assays were used in this study including two human-associated assays (HF183/BacR287 and HumM2), a ruminant-associated assay (Rum2Bac), a canine-associated assay (DG3), and an avian-associated assay (GFD), as well as a sample processing control (SPC) assay (Sketa22) as previously reported with the following modifications (Green et al., 2012, Green et al., 2014a, Green et al., 2014b, Haugland et al., 2010, Mieszkin et al., 2010, Shanks et al., 2009, USEPA 2019a, USEPA 2019b). All reaction mixtures contained 1X TaqMan Environmental Master Mix (version 2.0; Thermo Fisher Scientific, Grand Island, NY), 0.1X SYBR Green I Dye (GFD assay only; Thermo Fisher Scientific), 0.2 mg/mL bovine serum albumin (Sigma-Aldrich, St. Louis, MO), 1 µM each primer, and 80 nM 6-carboxyfluorescein (FAM)-labeled probe, and 80 nM VIC-labeled probe (multiples reactions only). All reactions contained either 2 µL of DNA sample extract or 10 to 1 × 105 target gene copies of reference DNA calibration standards in a total reaction volume of 25 µL. HF183/BacR287 and HumM2 multiplex reactions also contained 102 copies of IAC template. Triplicate reactions were performed for reference DNA calibration standard testing while all water samples included six replicate reactions. Amplifications were conducted on a QuantStudio 6 Flex Real-Time PCR system (Thermo Fisher Scientific) in MicroAmp optical 96-well reaction plates with MicroAmp 96-well optical adhesive film (Thermo Fisher Scientific). The thermal cycling profile for all assays was 2 min at 95°C followed by 40 cycles of 5 s at 95°C, and 30 s at 60°C (except GFD, 57°C). The threshold was manually set to either 0.03 (HF183/BacR287, DG3, Rum2Bac, and Sketa22) or 0.08 (HumM2 and GFD). Quantification cycle (Cq) values were exported to Microsoft Excel for further analysis. To monitor for potential extraneous DNA contamination during qPCR amplification, six no-template controls (NTC) with purified water substituted for template DNA were performed with each instrument run.
2.7. Data acceptance metrics
A rigorous data acceptance scheme was adopted to ensure high quality data generation in this study. All host-associated qPCR assays were subject to calibration model acceptance criteria [linearity (R2 ≥ 0.980) and amplification efficiency (0.90 to 1.10 where E = 10(−1/slope) − 1)]. A SPC protocol was used to identify suitable DNA recovery from each water sample as previously described (Shanks et al. 2016). Water samples with unacceptable DNA recovery were excluded from the study based on batch-specific (n = 12 samples/batch) criteria derived from repeated MEB spike recovery measurements. SPC proficiency was also determined for each sample batch preparation requiring a standard deviation in Sketa22 qPCR MEB repeated measures of ≤ 0.62 Cq to ensure consistent DNA recovery from one batch to another. The HF183/BacR287 and HumM2 multiplex IAC procedures were used to monitor for amplification inhibition. Any DNA extract indicating evidence of amplification inhibition was discarded. Instrument run-specific IAC proficiency testing (HF183/BacR287 and HumM2 NTC VIC Cq standard deviation ≤ 1.16 or 1.05, respectively) (Shanks et al. 2016) were also conducted to confirm reliable application of amplification inhibition testing from one instrument run to another. For each GFD instrument run, a melt curve analysis with a resolution of 0.3°C was used after thermal cycling to identify spurious amplicons that could confound data interpretation. Reactions yielding a GFD Cq value, but not at the expected melt peak (approximately 82–83°C) or reactions exhibiting a GFD Cq value at the expected melt peak plus an additional non-specific peak were discarded from the study (5.3% reactions discarded; 152 out of 2,881 total reactions; data not shown).
2.8. Calculations and statistics
Master reference DNA calibration models were generated for each qPCR assay from six independent standard curves using a Bayesian Markov Chain Monte Carlo approach (Sivaganesan et al. 2010, Sivaganesan et al. 2008). The lower limit of quantification (LLOQ) was defined as the 95% credible interval upper-bound from repeated measurements (n = 18) of 10 copies per reaction reference DNA standard dilutions. qPCR target concentrations were reported as mean log10 copy number per reaction. Amplification efficiency (E) for each master reference DNA calibration was calculated as follows: E = 10(−1/slope) − 1. Water samples were organized by site to investigate potential variability between sampling locations and by recreational beach (n = 171) and river (n = 171) water type groups based on previous findings reporting key differences in the occurrence of general fecal indicators between these different Great Lakes recreational area water types (Wanjugi et al. 2018). Weighted average fecal score ratios were estimated for each qPCR assay based on defined fecal indicator (E. coli, enterococci, somatic coliphage, and F+ coliphage) data group definitions utilizing all measurements including non-detects, detections below the LLOQ, and measurements within the range of quantification as reported elsewhere (Cao et al. 2018). Bacterial and viral fecal indicator threshold definitions, sample counts, and sample frequencies for beach and river water type groupings are shown in Table 2. Briefly, E. coli samples were organized into two groups for each water type including: 1) samples with E. coli ≥ 190 MPN/100mL [Beach Action Value (BAV) representing estimated illness rate of 32 per 1,000 primary contact recreators (USEPA 2012)] and 2) E. coli < 190 MPN/100mL. For enterococci, samples were organized into two groups for each water type including: 1) enterococci ≥ 60 CFU/100mL [BAV representing estimated illness rate of 32 per 1,000 primary contact recreators (USEPA 2012)] and 2) enterococci < 60 CFU/100mL. There are currently no recommended BAV criteria available for coliphage (somatic and F+). Instead, predicted coliphage BAV estimates were used based on a recent study using quantitative microbial risk assessment to estimate thresholds of 60 PFU/100mL for somatic coliphage and 30 PFU/100mL for F+ coliphage (Boehm 2019). For each individual water sample, 6 replicates were tested to ensure an adequate number of observations for suitable fecal score estimations (Cao et al. 2018). A sample group was eligible for fecal score determination if it had 1) at least 5 samples (5 samples × 6 replicates = 30 data points), and 2) at least one replicate must be a non-detect and one replicate below LLOQ (data groupings consisting of all non-detects and/or all below LLOQ were not eligible). All statistics were conducted with SAS software (Cary, NC), Microsoft Excel, or WinBugs (https://www.mrc-bsu.cam.ac.uk/software/bugs/thebugs-project-winbugs/). Heat maps of host-associated qpCR genetic marker concentrations were generated with Heatmapper with default settings (Babicki et al. 2016).
Table 2.
Bacterial and viral fecal indicator threshold definitions, sample counts, and frequencies for beach and river site type groupings.
| General Fecal, Indicator | Group Threshold | Beach Type | River Type | ||
|---|---|---|---|---|---|
| Above | Below | Above | Below | ||
| Somatic Coliphage | 60 PFU/100mL | 15 (8.8%) | 156 (91.2%) | 142 (83%) | 29 (17%) |
| F+ Coliphage | 30 PFU/100mL | 0 (0%) | 171 (100%) | 13 (7.6%) | 158 (92.4%) |
| E. coli | 190 MPN/100mL | 42 (24.6%) | 129 (75.4%) | 137 (80.1%) | 34 (19.9%) |
| Enterococci | 60 CFU/100mL | 77 (45%) | 94 (55%) | 142 (83%) | 29 (17%) |
() shows percent of total samples in a given site type (n = 171 total samples per water type).
PFU denotes plaque forming unit.
MPN indicates multiple probable number.
CFU represents culture forming unit.
3. Results
3.1. qPCR quality controls and data acceptance metrics
High-quality data for qPCR fecal source identification experiments were identified through the implementation of a series of quality controls and data acceptance metrics. Calibration model performance parameters are shown in Table 1. Calibration model R2 values were greater than 0.990 and E values ranged from 0.90 (GFD) to 0.96 (HF183/BacR287). Extraneous DNA control reactions indicated 99.6% DNA-free (14 false positives of 3,409 total reactions). False positive Cq values were higher than respective LLOQ for all but two reactions (both from GFD NTC reactions with Cq values of 36.9 and 36.0).
Table 1.
Calibration model performance metrics.
| Assay | Slope | Intercept | LLOQ | R2 | E |
|---|---|---|---|---|---|
| HF183/BacR287 | −3.43 ± 0.03 | 38.4 ± 0.08 | 34.5 | 0.996 | 0.96 |
| HumM2 | −3.45 ± 0.02 | 40.6 ± 0.10 | 37.4 | 0.994 | 0.95 |
| Rum2Bac | −3.52 ± 0.03 | 41.2 ± 0.13 | 37.9 | 0.994 | 0.92 |
| DG3 | −3.48 ± 0.03 | 38.3 ± 0.15 | 35.1 | 0.994 | 0.94 |
| GFD | −3.58 ± 0.04 | 40.5 ± 0.15 | 37.1 | 0.991 | 0.90 |
LLOQ indicates the lower limit of quantification
R2 denotes calibration model linearity
E represents amplification efficiency (E = 10(−1/slope) − 1).
Amplification inhibition was rarely identified in multiplex IAC HF183/BacR287 and HumM2 experiments [3.01%; 11 of 365 DNA extractions]. IAC acceptance thresholds ranged from (HF183/BacR287) 31.41 Cq to 33.39 Cq and 33.05 Cq to 35.29 Cq (HumM2). Competition thresholds were 24.7 Cq for HF183/BacR287 and 26.8 Cq for HumM2. Instrument run-specific IAC proficiency testing yielded a 100% pass rate with NTC VIC Cq standard deviations ranging from 0.16 to 0.52 for HF183/BacR287 (acceptance criteria ≤ 1.16) and 0.09 to 0.50 for HumM2 (acceptance criteria ≤ 1.05) (Shanks et al. 2016).
All 365 water sample DNA extracts passed SPC testing exhibiting negligible matrix interference. SPC acceptance thresholds ranged from 24.1 Cq to 26.5 Cq. A total of 146 DNA extracts were eligible for Cq adjustments ranging from 0.001 Cq to 2.85 Cq. A SPC proficiency test (Shanks et al. 2016) was used to monitor for suitable DNA recovery for each extraction batch. Sketa22 MEB Cq standard deviations ranged from 0.10 to 0.74 across 31 batch preparations resulting in a successful SPC proficiency rate of 96.8% [30 of 31 batches; acceptance criteria ≤ 0.62 (Shanks et al. 2016)]. Twelve DNA extracts from Batch 15 (Sketa22 MEB Cq standard deviation = 0.74) were discarded from the study due to inconsistent DNA recovery.
3.2. Host-Associated qPCR Measurements
Host-associated genetic markers were quantified for eligible water samples (n = 342) using fecal source identification qPCR methods for human (HF183/BacR287 and HumM2), ruminant (Rum2Bac), canine (DG3), and avian (GFD; n = 339 due to melt curve analysis results). Estimated mean log10 copies per reaction concentrations are shown in Fig. 2 organized by respective sample general fecal indicator measurements ranked from lowest to highest levels. A total of 445 samples yielded host-associated qPCR genetic marker measurements within an assay respective range of quantification (26.1% of 1,707 sample measurements) ranging from mean log10 copies per reaction of 0.92 (DG3) to 4.85 (HF183/BacR287). The occurrence of non-detection reactions ranged from 35.3% (GFD) to 86.5% (DG3) and detections below LLOQ spanned 4.5% (Rum2Bac) to 19.5% (HumM2) (Fig. 3).
Fig. 2.
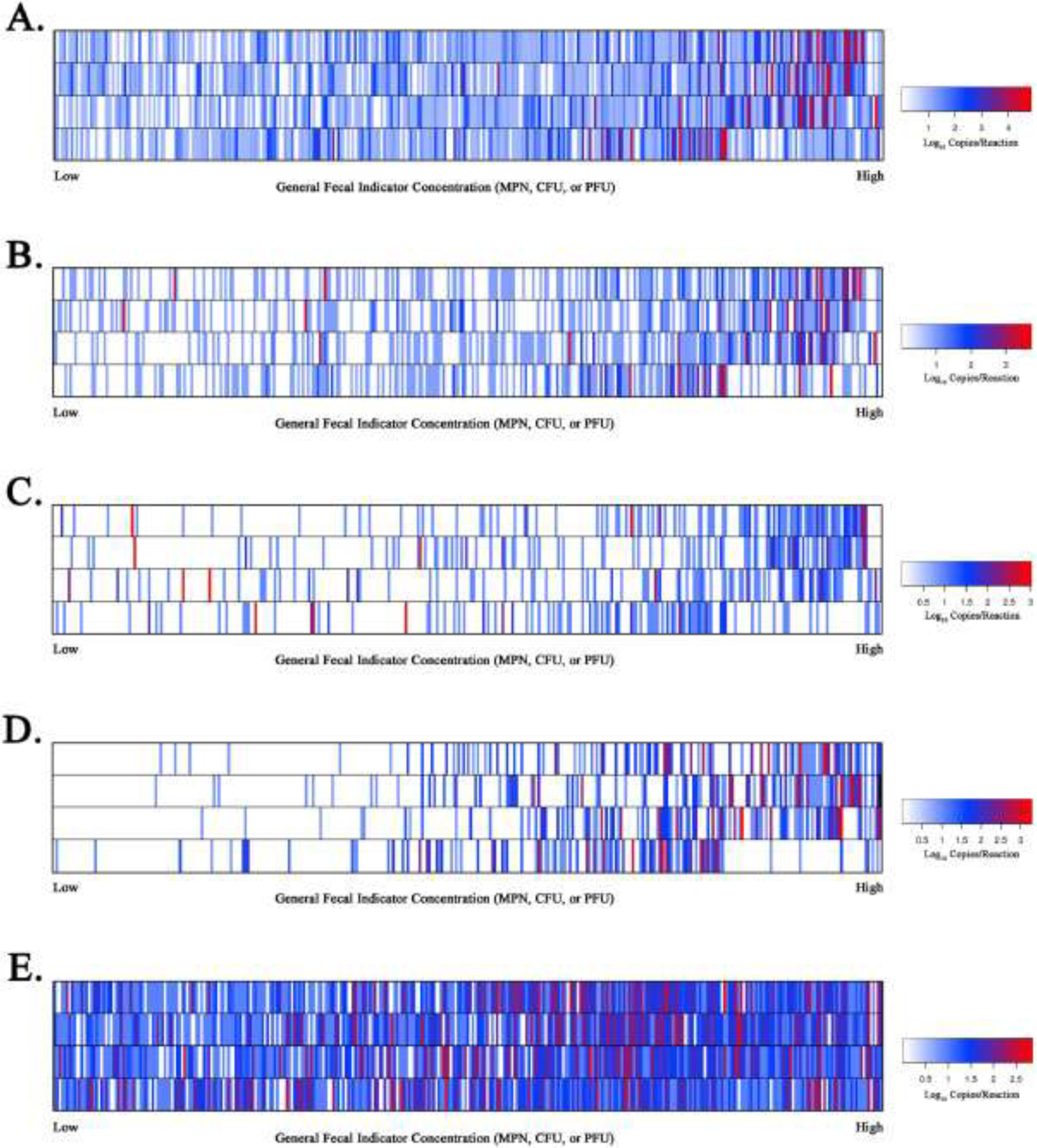
Heat map illustrating measurements of host-associated qPCR genetic marker estimated log10 copies per reaction concentrations for HF183/BacR287 (Panel A), HumM2 (Panel B), DG3 (Panel C), Rum2Bac (Panel D), and GFD (Panel E). Individual water sample estimated log10 copy per reactions concentrations for respective qPCR assays are ranked from lowest to highest (x-axis) based on four different general fecal indicator definitions (y-axis) including, situated from top to bottom, E. coli MPN/100mL, enterococci CFU/100mL, somatic coliphage PFU/100mL, and F+ coliphage PFU/100mL. Heat map keys are shown for each qPCR assay data set reporting estimated log10 copies per reaction color coding information.
Fig. 3.
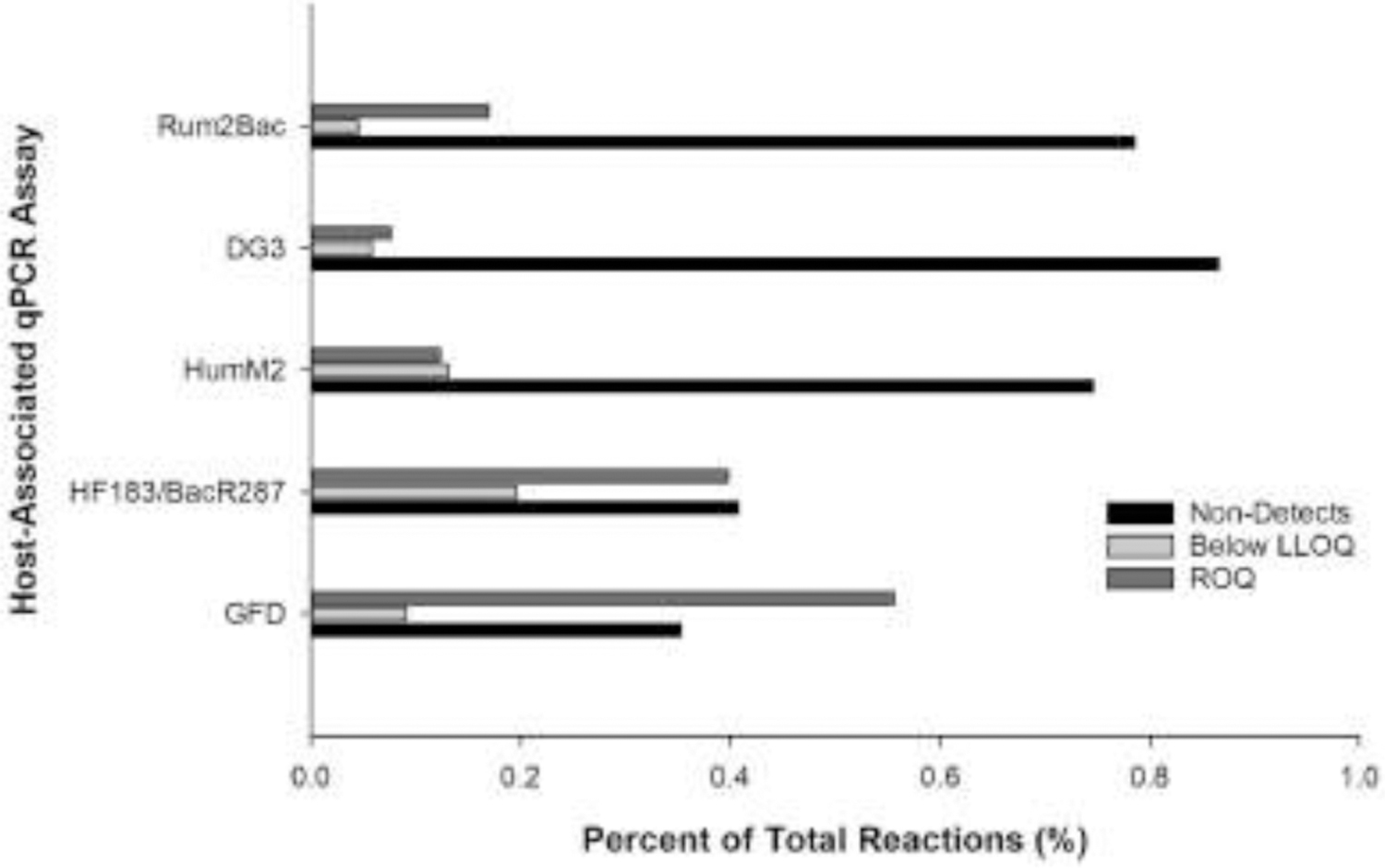
Histogram showing proportion of non-detections, detections below the lower limit of quantification (LLOQ), and measurements within the range of quantification (ROQ) for each host-associated qPCR assay.
3.3. Fecal Score Ratio Determination
Due to the large number of non-detection and detections below LLOQ reactions across all assays and samples (73.8% of data set; Fig. 3), a qPCR censored-data method was used to estimate log10 weighted-average fecal score ratios with 95% BCI to investigate pollution source trends based on four general fecal indicator definitions. By sampling site, only one location yielded a complete set of host-associated genetic marker and general fecal indicator definition combinations eligible for fecal score ratio determination limiting the ability to evaluate site to site variability (Table S1, Fig. 4). In contrast, 87.5% (35 of 40) of all host-associated genetic marker and general fecal indicator definition combinations were eligible for fecal score ratio determination by water type groupings (Fig. 5). A summary of sample counts and frequencies by fecal indicator definition and water type are shown in Table 2. For water type log10 fecal score ratios, ranged from −0.05 (−0.15 to 0.06 95% BCI) (F+ coliphage, river type, GFD) to 2.08 (1.37 to 3.26 95% BCI) (enterococci, river type, Rum2Bac) across water types. A total of 94.3% of eligible assay-general fecal indicator ratios yielded positive values (33 of 35 eligible combinations) with 80% significantly higher in sample groups with elevated general fecal indicator levels (n = 28). Negative ratios were only observed in river type samples for GFD-somatic coliphage and GFD-F+ coliphage groupings. E. coli MPN/100mLwas the only general fecal indicator that was significantly higher regardless of host-associated genetic marker or water type (river or beach types).
Fig. 4.
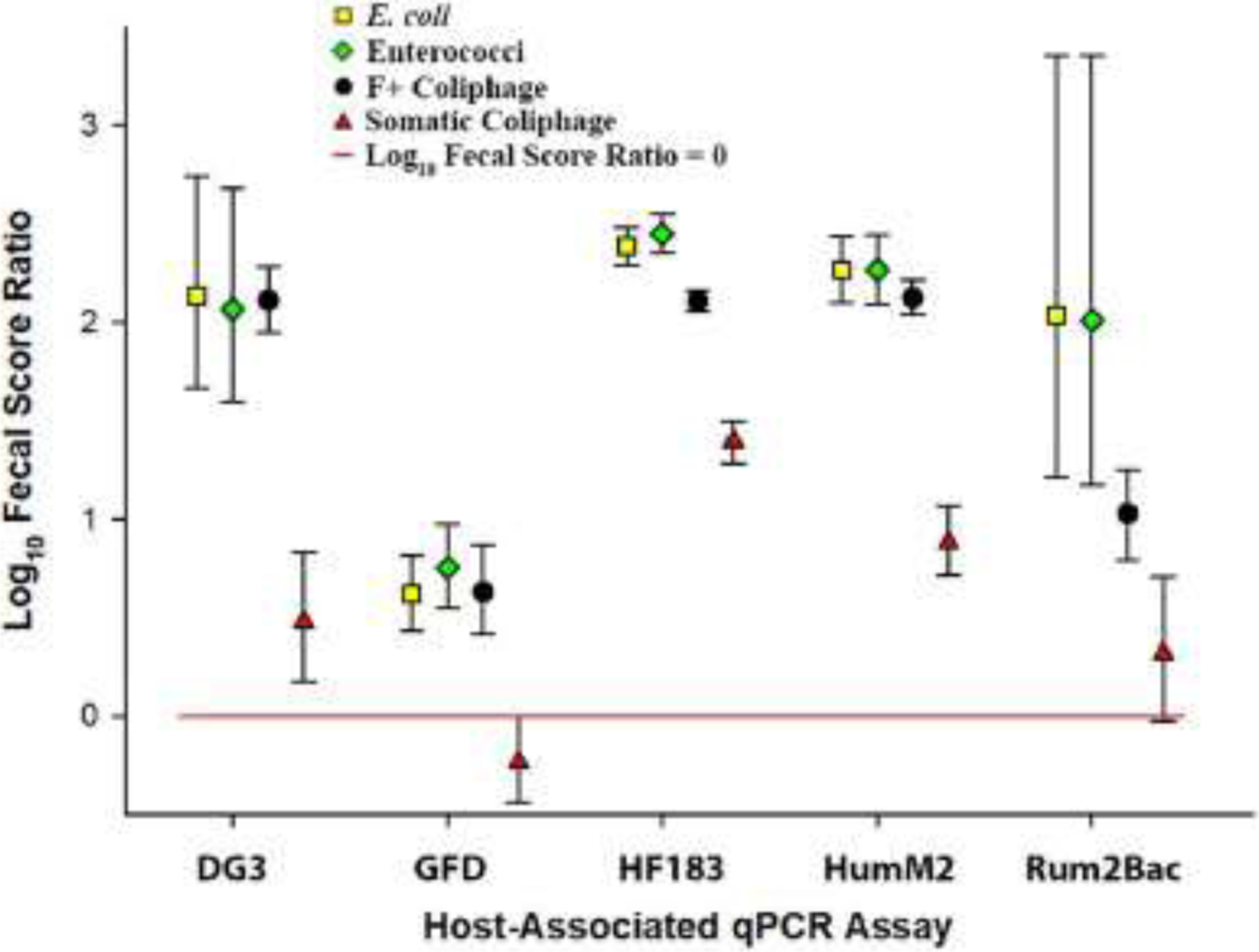
Scatter plot showing the Edgewater river sampling site log10 fecal score ratios with 95% Bayesian credible intervals (BCI) for each fecal indicator definition-host-associated qPCR assay data grouping. Geometric symbols represent respective mean log10 fecal score ratios and error bars depict respective 95% BCI. The horizontal red line depicts a log10 fecal score ratio of zero. Mean log10 fecal score ratio values above the red line with no interval overlap indicate scenarios where the host-associated qPCR genetic marker average log10 concentration is significantly higher when waters are under advisory based on a respective general fecal indicator threshold values used in this study.
Fig. 5.
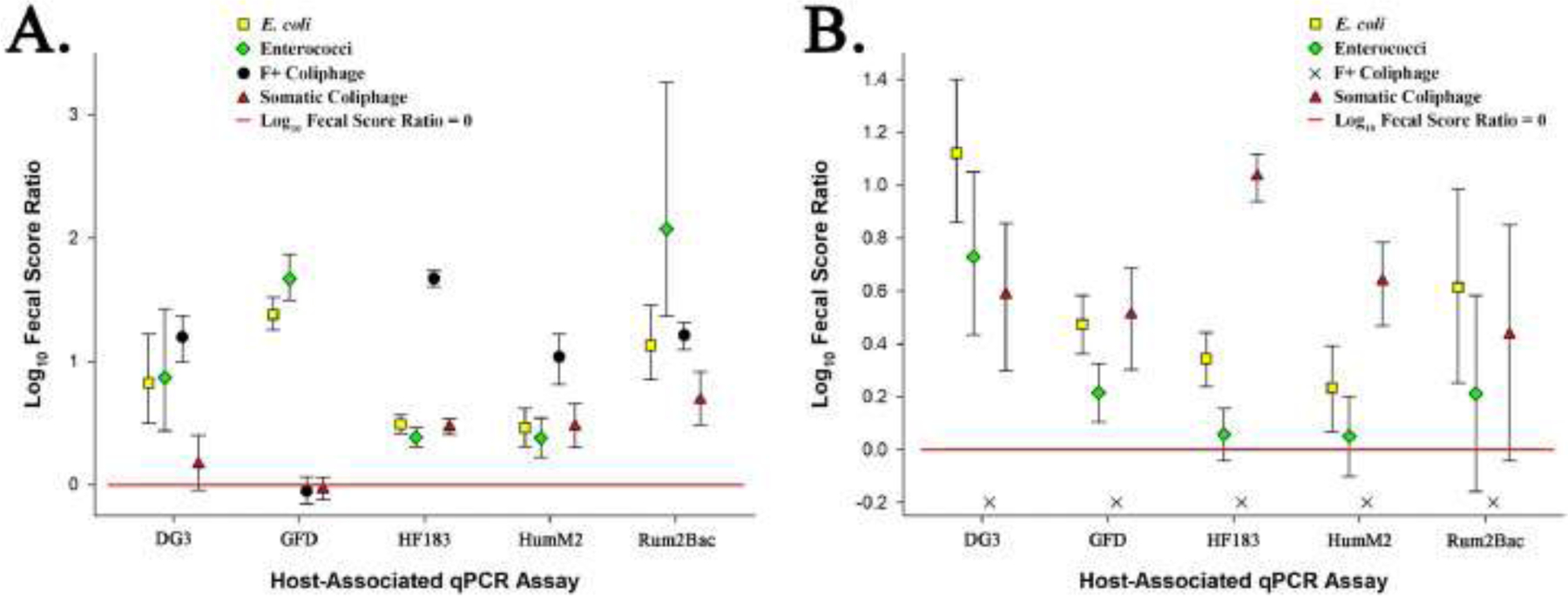
Scatter plot showing river (Panel A) and beach (Panel B) water type log10 fecal score ratios with 95% Bayesian credible intervals (BCI) for each fecal indicator definition-host-associated qPCR assay data grouping. Geometric symbols represent respective mean log10 fecal score ratios and error bars depict respective 95% BCI. The horizontal red line depicts a log10 fecal score ratio of zero. Mean log10 fecal score ratio values above the red line with no interval overlap indicate scenarios where the host-associated qPCR genetic marker average log10 concentration is significantly higher when waters are under advisory based on a respective general fecal indicator threshold values used in this study. An ‘X’ denotes that a general fecal indicator definition – water type data grouping was ineligible for fecal score ratio determination.
4. Discussion
Somatic and F+ coliphage are under consideration as potential surface water quality monitoring tools to identify unsafe levels of fecal pollution in recreational waters (Nappier et al. 2019). However, little is known about the cooccurrence of these virus-based fecal indicators and common host-associated genetic markers used to prioritize pollution source remediation during routine recreational water monitoring. To address this research gap, paired measurements of five host-associated genetic markers indicating human, ruminant, canine, and avian pollution sources were compared with general fecal indicators (E. coli MPN/100mL, enterococci CFU/100mL, somatic coliphage (PFU/100mL) and F+ coliphage (PFU/100mL) in Great Lakes basin water samples collected over a 15-week recreational season. Findings provide important insights on the prioritization of key fecal pollution sources for remediation when monitoring with different fecal indicators and fecal source identification methods, the utility of a recently reported censored data approach for interpreting host-associated genetic marker and fecal indicator measurements, and the application of virus-based fecal indicators for routine recreational water quality monitoring.
4.1. Fecal source pollution trends vary by general fecal indicator
Four fecal pollution sources were investigated in recreational waters from six Great Lakes basin sites including human, ruminant, canine, and avian groups. Host-associated genetic marker measurements were organized into two groups based on a corresponding general indicator threshold definition for E. coli (190 MPN/100mL), enterococci (60 CFU/100mL), somatic coliphage (60 PFU/100mL), or F+ coliphage (30 PFU/100mL) to generate respective log10 average weighted fecal score ratios by sampling site and water type (Fig. 4 and Fig. 5). These ratios identified whether a pollution source, general indicator combination significantly increases, decreases, or does not change when waters are impaired based on a given water quality definition, on average.
4.1.1. Human fecal pollution trends
Human fecal pollution is typically considered the greatest public health risk when present at unsafe levels in recreational waters (Soller et al. 2010, Soller et al. 2014). Human fecal waste can harbor a wide array of public health relevant pathogens. As a result, researchers have spent a considerable amount of time developing, standardizing and validating human-associated fecal source identification technologies. In this study, the recently nationally validated HF183/BacR287 and HumM2 qPCR procedures (Shanks et al., 2016, USEPA 2019a, USEPA 2019b) were used to characterize fecal pollution originating from sewage, septage, and/or human fecal waste. For the Edgewater river sampling site (Fig. 4) and river water type (Fig. 5, Panel A) findings, HF183/BacR287 and HumM2 log10 fecal score ratios were always significantly higher in sample groups with elevated fecal pollution levels regardless of general fecal indicator definition. Agreement amongst all general indicators and human-associated genetic marker log10 fecal score ratio estimates strongly suggests that the management of human fecal pollution sources impacting Great Lakes rivers would substantially reduce the incidence of water impairment. A different trend was observed in Great Lake beach water type samples where human-associated genetic marker log10 fecal score ratios were consistently lower compared to corresponding river values and were only significantly higher in beach samples with E. coli and somatic coliphage general fecal indicator definitions (Fig. 5, Panel B). A lack of agreement between human-associated genetic marker log10 fecal score ratios (HF183/BacR287 and HumM2) results using different general fecal indicator definitions at beach sites (Fig. 5, Panel B) suggests that a manager could identify human waste as a key fecal pollution source when routine monitoring with E. coli or somatic coliphage, but not when using enterococci or F+ coliphage. Additional research is warranted to investigate variable indicator levels and persistence in different human waste types and persistence in freshwater systems.
4.1.2. Canine fecal pollution trends
The extent of recreational water quality contamination attributed to dogs is poorly understood in the Great Lakes basin. Research reports that almost 40% of known human pathogens can infect domestic animal hosts (Cleaveland et al. 2001) and that dogs can harbor antibiotic resistant bacteria (Cinquepalmi et al. 2013, Nam et al. 2013), Campylobacter spp. (Chaban et al. 2010), Giardia duodenalis (Traub et al. 2009), and numerous parasites (Katagiri and Oliveira-Sequieira 2008). There is a growing body of research demonstrating that canine-host associated fecal source identification testing can detect dog waste in surface waters and prompt local community management activities that improve water quality by reducing fecal indicator levels (Ervin et al. 2014, Li et al. 2019, Shrestha et al. 2020b). At Great Lakes basin recreational sites in this study, canine fecal score ratios were significantly higher for both bacterial general fecal indicator definitions regardless of water type (beach or river). However, virus-based general fecal indicators yielded contradictory results. When samples were grouped by the somatic coliphage definition in beach water type samples, the fecal score ratio was significantly higher (95% BCI does not intersect the red line indicative of a log10 fecal score ratio = 0) for somatic coliphage, but a different trend was observed in river water type sample groups (Fig. 5, Panel B). A previous study reports a 2.9 log10 PFU per gram of feces higher level of somatic coliphage compared to F+ coliphage in dog fecal material (McMinn et al. 2014) potentially accounting for beach water type trends. However, an opposite pattern was observed in river water type samples where canine fecal score ratios were significantly higher using the F+ coliphage definition compared to somatic coliphage (Fig. 5, Panel A). This trend was also observed at the Edgewater river site (Fig. 4), but to a different degree suggesting that this discrepancy may vary in magnitude from one site location to another. Additional research is needed to investigate potential explanations for this lack of agreement between coliphage and water types in the Great Lakes basin.
4.1.3. Avian fecal pollution trends
Fecal pollution originating from birds represent a difficult challenge for recreational water quality managers. These animals can harbor public health relevant pathogens and shed antibiotic resistance bacteria (Bonnedahl et al. 2009, Cao et al. 2020, Rodolfo et al. 2020). Birds are common at recreational sites, can easily move from one point to another, and are reported to be a significant source of general fecal indicators via direct contact with water (Brooks et al. 2020, Shrestha et al. 2020b), as well as from surface run-off after rain events (Lee et al. 2020). Like canine pollution trends above, a similar pattern was found for bacterial fecal indicator definitions where log10 fecal score ratios were significantly higher at both beach and river water type sites (Fig. 5). In contrast, avian fecal score ratios were not significantly higher when monitoring with either somatic or F+ coliphage at river water type sites (Fig. 5, Panel A), but exhibited a different trend at beach sites (Fig. 5, Panel B). These conflicting trends across water types could simply be due to lower concentrations of fecal indicators at beach sites potentially introducing more uncertainty into fecal score ratios. However, another plausible explanation could be potential differences in the fate and transport of general fecal indicators originating from bird waste. Lake beaches are typically slow-moving bodies of water likely impacted by birds in the immediate vicinity. In contrast, rivers are often comprised of faster-moving currents flowing downstream composed of waters originating from a broader expanse of tributaries offering a different set of physical and chemical conditions that could influence the composition and persistence of general fecal indicators. This notion is supported by numerous studies reporting differences in the decay of bacterial and viral general indicators under various conditions (Boehm et al. 2019, Kohn and Nelson 2007, Long and Sobsey 2004, Wanjugi et al. 2016, Wu et al. 2016). Another possible explanation is that variable bird species may be contributing coliphage indicators in rivers compared to beach water types. There is limited information on the shedding of somatic and F+ coliphage in many avian species. The Great Lakes basin supports an immense population of resident and migratory birds that could differentially shed these coliphage types. The sole complete sampling site log10 fecal score ratio data set for the Edgewater river location further complicates the interpretation of avian pollution source trends in the context of routine coliphage water quality monitoring. At this site, somatic and F+ coliphage definitions disagreed (Fig. 4), but in an opposite manner compared to beach water type findings (Fig. 5, Panel B) suggesting that patterns can be highly variable on a site by site basis. Additional research is necessary to understand the underlying mechanisms resulting in multiple cooccurrence patterns of the GFD avian-associated genetic marker under different general fecal indicator routine monitoring scenarios in Great Lake basin recreational waters, especially for coliphage-based scenarios.
4.1.4. Ruminant fecal pollution trends
Ruminant fecal waste is reported to pose a similar recreational water public health risk to human waste (Soller et al. 2014). These animals can harbor pathogens such as E. coli O157:H7 which requires a minute dose to have potentially lethal effects (Pruimboom-Brees et al. 2000). Many recreational areas in the Great Lakes basin are near agricultural facilities housing domesticated ruminants (i.e. cattle, sheep, goat) or areas inhabited by deer. In this study, the ruminant-associated genetic marker (Rum2Bac) was significantly higher for all general fecal indicator sample definitions at the Edgewater river site (Fig. 4) and across river water type groupings (Fig. 5, Panel A), except for the somatic coliphage definition at the Edgewater river site. In contrast, all but the E. coli definition were either not eligible for fecal score ratio calculation (F+ coliphage) or not significantly different with beach water type groupings (Fig. 5, Panel B). These findings suggest a substantial difference in ruminant fecal pollution impact in rivers compared to beaches in the Great Lakes basin. Unlike beaches, river waters are impacted by pollutants from a broad expanse of upstream land surfaces, collecting and concentrating sediments, nutrients, biological contaminants, and any other potential substances present in the drainage basin, especially after rainfall. Further investigation of potential links between ruminant and local precipitation could help explain differences between river and beach water types observed in this study.
4.2. The fecal score ratio advantage
The fecal score approach employed in this study is a recently developed censored data strategy that estimates a weighted average log10 copies/100mL derived from a collection of samples grouped together to address a specific water quality management question (Cao et al. 2018). For fecal source identification qPCR applications, censored data are usually Cq measurements greater than the respective LLOQ threshold or non-detects (Cq = 40). For these measurements, the exact number of DNA target molecules in a reaction cannot be firmly established. Censored data can present a substantial challenge when interpreting data, especially when a large proportion of measurements fall in this category (> 10%). In fecal source identification applications, censored data values are often assigned an arbitrary fraction of the LLOQ (Kundu et al. 2013, Weidhass et al. 2011) or deleted (Staley et al. 2016, Templar et al. 2016) potentially introducing bias into data interpretations which routinely make up more than 50% of all total measurements (> 70% in this study). The fecal score ratio approach provides an alternative strategy to compare the average concentration of a host-associated genetic marker between two groups of samples using all data measurements without the need to fabricate any Cq values. To date, this censored data tactic has been successfully used to rank recreational water sites based on a specific pollution source (Cao et al. 2018, Li et al. 2019), to investigate potential links between rainfall and host-associated genetic marker measurements, and describe fecal pollution source trends in waters routinely monitored by E. coli and enterococci (Shrestha et al. 2020b). However, the fecal score approach does have limitations such as the requirement to group samples together which prevents higher resolution assessment of temporal or site-specific variability in fecal source identification investigations. Fecal score determination also entails that qPCR measurements from a defined group of samples must contain at least one Cq measurement below LLOQ and one non-detect (data groupings consisting of all non-detects or all below LLOQ are not eligible), potentially limiting utility in waters consistently yielding extremely low levels of the target host-associated genetic marker of interest. Nevertheless, the fecal score ratio approach presented here repeatedly identified clear trends between paired measurements of a host-associated genetic marker and a general fecal indicator leading to useful insights to inform water quality management.
4.3. Implications for routine recreational water quality monitoring
Findings have multiple implications for reducing fecal indicator levels at recreational sites routinely monitored for bacterial (E. coli and enterococci) or viral (somatic and F+ coliphage) indicators and host-associated genetic markers. Most notably, there was not a single instance where all fecal score ratios for a given water type and host-associated genetic marker were in agreement (Fig. 4 and Fig. 5) demonstrating that fecal indicator selection for routine recreational water quality monitoring can lead to the prioritization of different key pollution sources for remediation. For water type comparisons, F+ fecal score ratios were the most variable yielding a significant difference from other indicator fecal score ratios 70% of the time (7 of 10 possible combinations). In contrast, E. coli, enterococci, or somatic coliphage fecal score ratios were in agreement with at least one other indicator more than 80% of the time. The highest level of agreement occurred between bacterial-based E. coli and enterococci fecal score ratios (80%) following a similar trend observed in a recent study of Chicago Great Lake beaches using rapid enterococci qPCR and cultivation-based E. coli general fecal indicator definitions in conjunction with the same host-associated genetic marker panel reported here (Shrestha et al. 2020b). In contrast, virus-based somatic and F+ coliphage fecal score ratios were significantly different from each other or ineligible for calculation (F+ coliphage at beach sites) in 90% (9 of 10) of host-associated genetic marker and water type sample group combinations. These contradictory trends suggest that somatic and F+ coliphage groups, can originate and/or persist in recreational waters in a profoundly different manner compared to traditional E. coli and enterococci general fecal indicators. This study also suggests that different environmental stressors such as solar irradiance, ultraviolet absorbance, and water temperature in riverine compared to lake beach environments may result in different occurrence outcomes. It is also important to recognize that there are currently no recreational water quality thresholds recommend by regulatory authorities for somatic and F+ coliphage monitoring to date. In this study, proposed thresholds derived from a quantitative microbial risk assessment model (Boehm 2019) were used to investigate putative routine water quality monitoring trends. It is possible that different threshold values could result in variable fecal score ratio pollution trends observed here, especially for the F+ coliphage threshold (30 PFU/100mL). This F+ coliphage threshold resulted in no water impairments at Great Lake beach sites in this study compared to 15 events using the somatic coliphage threshold of 60 PFU/100mL (Table 2). Additional research is needed to determine if the predicted F+ coliphage (30 PFU/100mL) and somatic (60 PFU/100mL) thresholds used here are suitable for recreational water quality applications, especially in Great Lake beach environments. Finally, the scope of this study was to characterize the potential impact of recreational water routine monitoring with four general fecal indicator approaches on the prioritization of key pollutant sources using corresponding quantitative fecal source identification measurements. Further investigation of this rich data set will undoubtedly lead to other useful strategies and information further improving water quality management in the Great Lakes region.
5. Conclusion
The potential adoption of virus-based coliphage testing would provide recreational water quality managers with new options for routine general fecal indicator-based water quality monitoring. However, there is limited information on the cooccurrence of coliphage and host-associated genetic markers in recreational waters used to prioritize pollutant sources for remediation. This study characterized trends in fecal pollution originating from human, avian, ruminant, and canine sources at beach lake and river recreational sites in the Great Lake basin under four general fecal indicator routine water quality definitions. Key findings include:
Interpretation of fecal source identification results can vary depending on fecal indicator used for routine water quality monitoring potentially leading to the prioritization of variable pollutant sources for remediation.
Routine monitoring with E. coli (MPN/100mL) or enterococci (CFU/100mL) bacterial fecal indicators almost always led to the identification of the same fecal pollution trends regardless of pollutant source or water type.
Routine monitoring with somatic or F+ coliphage (PFU/100mL) viral fecal indicators often led to contradictory fecal pollution source trends compared to bacterial indicators.
Variability in bacterial and viral indicator defined fecal source trends suggest factors such as fecal shedding, persistence, and transport properties may be responsible for different occurrence patterns.
Lake beach and river water types often exhibited variable fecal indicator and host-associated genetic marker cooccurrence trends suggesting different management practices may be necessary to mitigate pollution.
The censored data fecal score ratio approach used here repeatedly identified trends between paired measurements of a host-associated genetic marker and a general fecal indicator leading to useful insights despite high frequencies of non-detects in final data sets.
Additional research is warranted to characterize animal shedding, the potential for naturalized sources, as well as fate and transport of virus-based coliphage general fecal indicators.
Supplementary Material
Footnotes
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Appendix. Supplementary materials
References
- Abdelzaher AM, Wright ME, Ortega C, Solo-Gabriele HM, Miller G, Elmir S, Newman X, Shih P, Bonilla JA, Bonilla TD, Plamer CJ, Scott T, Lukasik J, Harwood VJ, McQuaig S, Sinagalliano CD, Gidley ML, Plano LRW, Zhu X, Wang JD, Fleming LE, 2010. Presence of pathogens and indicator microbes at a non-point source subtropical recreational marine beach. Applied and Environmental Microbiology 76 (3), 724–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- APHL, 2012. Standard Methods for the Examination of Water and Wastewater. American Public Health Association, Washington DC. [Google Scholar]
- Babicki S, Arndt D, Marcu A, Liang Y, Grant JR, Maciejewski A, Wishart DS, 2016. Heatmapper: web-enabled heat mapping for all. Nucleic Acids Research 44 W147–W153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begier EM, Obserste MS, Landry ML, Brennan T, Mlynarski D, Mshar PA, Frenette K, Rabatsky-Ehr T, Purviance K, Neapul A, Nix WA, Pallasnsch MA, Perguson D, Cartter ML, Hadler JL, 2008. An outbreak of concurrent echovirus 30and coxsackievirusA1 infections associated with sea swimming among a group of travelers to Mexico. Clinical Infectious Diseases 47, 616–623. [DOI] [PubMed] [Google Scholar]
- Blanch A, Lucena F, Muniesa M, Jofre J, 2020. Fast and easy methods for the detection of coliphages. Journal of Microbiological Methods 173, 105940. [DOI] [PubMed] [Google Scholar]
- Boehm AB, 2019. Risk-based water quality thresholds for coliphages in surface waters: effect of temperature and contamination aging. Environmental Science Processes & Impacts 21, 2031. [DOI] [PubMed] [Google Scholar]
- Boehm AB, Silverman AI, Schriewer A, Goodwin KD, 2019. Systematic review and meta-analysis of decay rates of waterborne mammalian viruses and coliphages in surface waters. Water Research 164, 114898. [DOI] [PubMed] [Google Scholar]
- Bonadonna L, Briancesco R, Suffredini E, Coccia A, Della-Libera S, Carducci A, Verani M, Federigi I, Iaconelli M, Bonanno-Ferraro G, Mancini P, Veneri C, Ferretti E, Lucentini L, Gramaccioni L, La-Rosa G, 2019. Enteric viruses, somatic coliphages and Vibrio species in marine bathing and non-bathing waters in Italy. Marine Pollution Bulletin 149, 110570. [DOI] [PubMed] [Google Scholar]
- Bonnedahl J, Drobni M, Gauthier-Clerc M, Hernandez J, Granholm S, Kayser Y, Melhus A, Kahlmeter G, Waldenstrom J, Johansson A, Olsen B, 2009. Dissemination of Escherichia coli with CTX-M type ESBL between humans and yellow-legged gulls in the south of France. PLoS ONE 4, e5958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks YM, Spirito CM, Bae JS, Hong A, Mosier EM, Sausele DJ, Fernandez-Baca CP, Epstein JL, Shapley DJ, Goodman LB, Anderson RR, Glaser AL, Richardson RE, 2020. Fecal indicator bacteria, fecal source tracking markers, and pathogens detected in two Hudson River tributaries. Water Research 171, 115342. [DOI] [PubMed] [Google Scholar]
- Cabelli VJ, Dufour AP, McCabe LJ, Levin MA, 1982. Swimming-associated gastroenteritis and water quality. American Journal of Epidemiology 115, 606–616 [DOI] [PubMed] [Google Scholar]
- Cao J, Hu Y, Liu F, Wang Y, Bi Y, Lv N, Li J, Zhu B, Gao GF, 2020. Metagenomic analysis reveals the microbiome and resistome in migratory birds. Microbiome 8, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Sivaganesan M, Kelty CA, Wang D, Boehm AB, Griffith JF, Weisberg SB, Shanks OC, 2018. A human fecal contamination score for ranking recreational sites using the HF183/BacR287 quantitative real-time PCR method. Water Research 128, 148–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaban B, Ngeleka M, Hill JE, 2010. Detection and quantification of 14 Campylobacter species in pet dogs reveals an increase in species richness in feces of diarrheic animals. BMC Microbiology 10, 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cinquepalmi V, Monno R, Fumarola L, Ventrella G, Calia C, Greco MF, Vito D, Soleo L, 2013. Environmental contamination by dog faeces: a public health problem? International JOurnal of Environmental Research and Public Health 10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleaveland S, Laurenson MK, Taylor LH, 2001. Diseases of humans and their domestic mammals: pathogen characteristics, host range and risk of emergence. Philosphical Transactions of the Royal Soceity B 356, 991–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole D, Long SC, Sobsey MD, 2003. Evaluation of F-RNA and DNA coliphages as source-specific indicators of fecal contamination in surface waters. Applied and Environmental Microbiology 69, 6507–6514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Converse RR, Blackwood AD, Kirs M, Noble RT, Griffith JF,2009. Rapid QPCR-based assay for fecal Bacteroides spp. as a tool for assessing fecal conamination in recreational waters. Water Research 43, 4 828–4 837 [DOI] [PubMed] [Google Scholar]
- Ervin JC, Van De Werfhorst L, Murray J, Holden P, 2014. Microbial source tracking in a coastal california watershed reveals canines as controllable sources of fecal contamination. Environmental Science and Technology 48, 9043–9052 [DOI] [PubMed] [Google Scholar]
- Feng S, Bootsma MJ, McLellan SL, 2018. Human-associated Lachnospiraceae genetic markers improve detection of fecal pollution sources in urban waters. Applied and Environmental Microbiology 84 e0 0309–0 0318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S, McLellan SL, 2019. Highly specific sewage-derived Bacteroides quantitative PCR assays target sewage-polluted waters. Applied and Environmental Microbiology 85 e02696–02618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Aljaro C, Martin-Diaz J, Vinas-Balada E, W., C.-C., Lucena F, Blanch AR, 2017. Mobilisation of microbial indicators, microbial source tracking markers and pathogens after rainfall events. Water Research 112, 248–253 [DOI] [PubMed] [Google Scholar]
- Gerba CP, 1987. Phage as indicators of fecal pollution. Wiley Interscience, New York, NY. [Google Scholar]
- Green HC, Dick LK, Gilpin B, Samadpour M, Field KG, 2012. Genetic markers for rapid PCR-based identification of gull, Canada goose, duck, and chicken fecal contamination in water. Applied and Environmental Microbiology 78, 503–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green HC, Haugland R, Varma M, Millen HT, Borchardt MA, Field KG, Kelty CA, Sivaganesan M, Shanks OC, 2014a. Improved HF183 quantitative real-time PCR assay for characterization of human fecal pollution in ambient surface water samples. Applied and Environmental Microbiology 80 (10), 3086–3094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green HC, White KM, Kelty CA, Shanks OC, 2014b. Development of rapid canine fecal source identification PCR-based assays. Environmental Science and Technology (48) 11453–11461 [DOI] [PubMed] [Google Scholar]
- Griffith JF, Weisberg SB, Arnold BF, Cao Y, Schiff KC, Colford JM,2016. Epidemiologic evaluation of multiple alternate microbial water quality monitoring indicators at three California beaches. Water Research 94, 371–381 [DOI] [PubMed] [Google Scholar]
- Harwood VJ, Shanks OC, Korajkic A, Verbyla M, Ahmed A, Iriate M, 2017. General and host-associated bacterial indicators of faecal pollution. UNESCO, East Lansing, MI [Google Scholar]
- Haugland RA, Varma M, Kelty CA, Peed L, Sivaganesan M, Shanks OC, 2010. Evaluation of genetic markers from the 16S rRNA gene V2 region for use in quantitative detection of selected Bacteroidales species and human fecal waste by real-time PCR. Systematic and Applied Microbiology 33, 348–357 [DOI] [PubMed] [Google Scholar]
- Havelaar A, van Olphen M, Drost YC, 1993. F-specific RNA bacteriophages are adequate model organisms for enteric viruses in freshwater. Applied and Environmental Microbiology 59, 2956–2962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katagiri S, Oliveira-Sequieira TC, 2008. Prevalence of dog intestinal parasites and risk perception of zoonotic infection by dog owners in Sao Paulo State, Brazil. Zoonoses and Public Health 55, 406–413 [DOI] [PubMed] [Google Scholar]
- Kildare BJ, Leutenegger CM, McSwain BS, Bambic DG, Rajal VB, Wuertz S, 2007. 16S rRNA-based assays for quantitative detection of universal, human-, cow-, and dog-specific fecal Bacteroidales: a Bayesian approach. Water Research 41, 3701–3715 [DOI] [PubMed] [Google Scholar]
- King AMQ, Adams MJ, Carstens EB, Lefkowitz WJ, 2011. Virus taxonomy: classification and nomenclature of viruses. Elsevier Academic Press, London, UK. [Google Scholar]
- Kohn T, Nelson KL, 2007. Sunlight-mediated inactiviation of MS2 coliphage via exogenous singlet oxygen produced by sensitizers in natural waters. Environmental Science & Technology 41, 192–197 [DOI] [PubMed] [Google Scholar]
- Korajkic A, McMinn BR, Harwood VJ, 2018. Relationships between microbial indicators and pathogens in recreational water settings. International Journal of Environmental Research and Public Health 15, 2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korajkic A, McMinn BR, Herrmann MP, Sivaganesan M, Kelty CA, Clinton P, Nash MS, Shanks OC, 2020. Viral and bacterial fecal indicators in untreated wastewater across the contiguous United States exhibit geospatial trends. Applied and Environmental Microbiology 86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundu A, McBride G, Wuertz S, 2013. Adenovirus-associated health risks for recreational activities in a multi-use coastal watershed based on site-specific quantitative microbial risk assessment. Water Research 47, 6309–6325 [DOI] [PubMed] [Google Scholar]
- Lee D, Lee H, Trevors JT, Weir SC, Thomas JL, Habash M,2014. Characterization of sources and loadings of fecal pollutants using microbial source tracking assays in urban and rural areas of Grand River Watershed, southwestern Ontario. Water Research 53, 123–131 [DOI] [PubMed] [Google Scholar]
- Lee S, Suits M, Wituszynski D, Winston R, Martin J, Lee J,2020. Residential urban stormwater runoff: a comprehensive profile of microbiome and antibiotic resistance. Science of the Total Environment 723, 138033. [DOI] [PubMed] [Google Scholar]
- Li X, Sivaganesan M, Kelty CA, Zimmer-Faust A, Clinton P, Reichman JR, Johnson Y, Matthews W, Bailey S, Shanks OC, 2019. Large-scale implementation of standardized quantitative real-time PCR fecal source identification procedures in the Tillamook Bay Watershed. PLoS ONE 14 (6), e0216827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long SC, Sobsey MD, 2004. A comparison of the survival of F + RNA and F-t-DNA coliphages in lake water microcosms. Water Health 2, 15–22 [PubMed] [Google Scholar]
- McKee A, Molina M, Cyterski M, Couch A, 2020. Microbial source tracking (MST) in Chattahoochee River national recreational area: seasonal and precipitation trends in MST marker concentrations, and associations with E. coli leves, pathogentic presence, and land use. Water Research 171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMinn BR, Ashbolt NJ, Korajkic A, 2017. Bacteriophages as indicators of faecal pollution and enteric virus removal. Letters in Applied Microbiology 65, 11–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMinn BR, Klemm S, Korajkic A, Wyatt KM, Herrmann MP, Haugland RA, Lu J, Villegas EN, Frye C, 2019. A constructed wetland for treatment of an impacted waterway and the influence of native waterfowl on its perceived effectiveness. Ecological Engineering 128, 48–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMinn BR, Korajkic A, Ashbolt NJ, 2014. Evaluation of Bacteroides fragilis GB-124 bacteriophages as novel human-associated faecal indicators in the United States. Letters in Applied Microbiology 59, 115–121 [DOI] [PubMed] [Google Scholar]
- McQuaig S, Scott TM, Harwood VJ, Farrah SR, Lukaskik JO,2009. Quantification of human polyomaviruses JC Virus and BK Virus by TaqMan quantitative PCR and comparison to other water quality indicators in water and fecal samples. Applied and Environmental Microbiology 75, 3379–3388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesquita MMF, Stimson J, Chae GT, Tufenkji N, Ptacek CJ, Blowes DW, Emelko MB, 2010. Optimal preparation and purification of PRD1-like bacteriophages for use in environmental fate and transport studies. Water Research 44, 1114–1125 [DOI] [PubMed] [Google Scholar]
- Mieszkin S, Furet JP, Corthier G, Gourmelon M, 2009. Estimation of pig fecal contamination in a river catchment by real-time PCR using two pig-specific Bacteroidales 16S rRNA genetic markers. Applied and Environmental Microbiology 75, 3045–3054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mieszkin S, Yala JF, Joubrel R, Gourmelon M, 2010. Phylogenetic analysis of Bacteroidales 16S rRNA gene sequences from human and animal effluents and assessment of rumaint faecal pollution by real-time PCR. Journal of Applied Microbiology 108, 974–984 [DOI] [PubMed] [Google Scholar]
- Nam EK, Ko S, Chae JS, Hwang CY, 2013. Characterization and zoonotic potential of uropathogenic Escherichia coli isolated from dogs. Journal of Microbiology and Biotechnology 23 (3), 422–429 [DOI] [PubMed] [Google Scholar]
- Nappier SP, Hong T, Ichida A, Goldstone A, Eftime SE, 2019. Occurrence of coliphage in raw wastewater and in ambient water: a meta-analysis. Water Research 153, 263–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmateer GA, Dutka BJ, Janzen EM, Meissner SM, Sakeelaries MG, 1991. Coliphage and bacteriophage as indicators of recreational water quality. Water Research 25, 355–357 [Google Scholar]
- Pruimboom-Brees IM, Morgan TW, Ackermann MR, Nystrom ED, Samuel JE, Cornick NA, Moon HW, 2000. Cattle lack vascular receptors for Escherichia coli O157:H7 Shiga toxins. In: Proceedings of the National Academy of Sciences of the United States of America, 97, pp. 10325–10329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reischer GH, Haider JM, Sommer R, Stadler H, Keiblinger KM, Hornek R, Zerobin W, Mach RL, Farnleitner AH, 2008. Quantitative microbial faecal source tracking with sampling guided by hydrological catchment dynamics. Environmental Microbiology 10 (10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reischer GH, Kasper DC, Steinborn R, Mach RL, Farnleitner AH,2006. Quantitative PCR Method for Sensitive Detection of Ruminant Fecal Pollution in Freshwater and Evaluation of This Method in Alpine Karstic Regions. Applied and Environmental Microbiology 72 (8), 5610–5614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodolfo T, Rivera D, Duenas F, Sallaberry-Pincheira N, Hamilton-West C, Adell AD, Moreno-Switt AI, 2020. Salmonella in raptors and aquatic wild birds in Chile. Journal of Wildlife Diseases 10.7589/2019-08–198 [DOI] [PubMed]
- Rose JB, Farrah SR, Harwood VJ, Levine AD, Lukaskik J, Menendez P and Scott TM (2004) Reduction of pathogens, indicator bacteria, and alternative indicators by wastewater treatment and reclamation processes
- Shanks OC, Kelty CA, Oshiro R, Haugland RA, Madi T, Brooks L, Field KG, Sivaganesan M, 2016. Data acceptance criteria for standardized human-associated fecal source identification quantitative real-time PCR methods. Applied and Environmental Microbiology 82 (9), 2773–2782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanks OC, Kelty CA, Sivaganesan M, Varma M, Haugland RA, 2009. Quantitative PCR for genetic markers of human fecal pollution. Applied and Environmental Microbiology 75, 5507–5513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrestha A, Kelty CA, Sivaganesan M, Shanks OC, Dorevitch S, 2020a. Fecal pollution source characterization at non-point source impacted beaches under dry and wet weather conditions. Water Research In Press [DOI] [PMC free article] [PubMed]
- Shrestha A, Kelty CA, Sivaganesan M, Shanks OC, Dorevitch S, 2020b. Fecal pollution source characterization at non-point source impacted beaches under dry and wet weather conditions. Water Research, 116014. [DOI] [PMC free article] [PubMed]
- Sinclair RG, Jones EL, Gerba CP, 2009. Viruses in recreational water-borne disease outbreaks: a review. Journal of Applied Microbiology 107, 1769–1780 [DOI] [PubMed] [Google Scholar]
- Sivaganesan M, Haugland RA, Chern EC, Shanks OC, 2010. Improved strategies and optimization of calibration models for real-time PCR absolute quantification. Water Research 44, 4726–4735 [DOI] [PubMed] [Google Scholar]
- Sivaganesan M, Seifring S, Varma M, Haugland RA, Shanks OC, 2008. A Bayesian method for calculating real-time quantitative PCR calibration curves using absolute plasmid DNA standards. BMC Bioinformatics 9, 120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soller JA, Schoen ME, Bartrand T, Ravenscroft JE, Ashbolt NJ, 2010. Estimated human health risks from exposure to recreational waters impacted by human and non-human sources of faecal contamination. Water Research 44, 4674–4691 [DOI] [PubMed] [Google Scholar]
- Soller JA, Schoen ME, Varghese A, Ichida AM, Boehm AB, Efitm S, Ashbolt NJ, Ravenscroft JE, 2014. Human health risk implications of multiple sources of faecal indicator bacteria in a recreational water body. Water Research 66, 254–264 [DOI] [PubMed] [Google Scholar]
- Stachler E, Kelty CA, Sivaganesan M, Li X, Bibby K, Shanks OC, 2017. Development of CrAssphage quantitative real-time PCR assays for human fecal pollution measurement. Environmental Science & Technology 51, 9146–9154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staley ZR, Chuong JD, Hill SJ, Brabuski J, Shokralla S, Edge TA,2018. Fecal source tracking and eDNA profiling in an urban creek following an extreme rain event. Scientific Reports 8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staley ZR, Grabuski J, Sverko E, Edge TA, 2016. Comparison of microbial and chemical source tracking markers to identify fecal contamination sources in the Humber River and associated stormwater outfalls. Applied and Environmental Microbiology 10.1128/AEM.01675-01616 [DOI] [PMC free article] [PubMed]
- Templar HA, Dila DK, Bootsma MJ, Corsi SR, McLellan SL, 2016. Quantification of human-associated fecal indicators reveal sewage from urban watersheds as source of pollution in Lake Michigan. Water Research 100, 556–567 [DOI] [PubMed] [Google Scholar]
- Traub RJ, Inpankaew T, Reid SA, Sutthikornchai C, Sukthana Y, Roberston ID, Thompson RC, 2009. Transmission cycles of Giardia duodenalis in dogs and humans in Temple communities in Bangkok - A critical evaluation of its prevalence using three diagnostic tests in the field in the absence of a gold standard. Acta Tropica 111, 125–132 [DOI] [PubMed] [Google Scholar]
- USEPA (2009) Method 160 0: Enterococci in water by membrane filtration using membrane-Enterococcus indoxyl-B-D-glucoside agar (mEI). Oshiro R (ed), Washington, DC [Google Scholar]
- USEPA, 2012. Recreational Water Quality Criteria. Office of Water [Google Scholar]
- USEPA, 2019a. In: Water, E.O.o. (Ed.), Method 1696: Characterization of human fecal pollution in water by HF183/BacR287 TaqMan quantitative polymerase chain reaction (qPCR) assay, ed. United States Environmental Protection Agency, Washington DC [Google Scholar]
- USEPA, 2019b. In: Water, E.O.o. (Ed.), Method 1697: Characterization of human fecal pollution in water by HumM2 TaqMan quantitative polymerase chain reaction (qPCR) assay, ed. United States Environmental Protection Agency, Washington DC [Google Scholar]
- Wanjugi P, Sivaganesan M, Korajkic A, Kelty CA, McMinn BR, Ulrich R, Harwood VJ, Shanks OC, 2016. Differential decomposition of bacterial and viral fecal indicators in common human pollution types. Water Research 105, 591–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanjugi P, Sivaganesan M, Korajkic A, McMinn BR, Kelty CA, Rhodes ER, Cyterski M, Zepp R, Oshima K, Stachler E, Kinzelman J, Kurdas SR, Citriglia M, Fu-Chih H, Shanks OC, 2018. Incidence of somatic and F + coliphage in Great Lake Basin recreational waters. Water Research 140, 200–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidhass JL, Macbeth TW, Olsen RL, Harwood VJ, 2011. Quantitative PCR for poultry-specific Brevibacterium marker gene correlates with bacterial and chemical indicators of water pollution in a watershed impacted by land application of poultry litter. Applied and Environmental Microbiology 10.1128/AEM.02555-02510 [DOI] [PMC free article] [PubMed]
- Wiedenmann A, Kruger P, Dietz K, Lopez-Pila JM, Szewzyk R, Botzenhart K, 2006. A randomized controlled trial assessing infectious disease risks from bathing in fresh recreational waters in relation tot he concentration ofEscherichia coli, intestinal enterococci, Clostridium perfringens, and somatic coliphages. Environmental Health Perspectives 114, 4130–4141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Cao Y, Young B, Yuen Y, Jiang S, Melendez D, Griffith JF, Stewart JR, 2016. Decay of coliphages in sewage-contaminated freshwater: uncertainty and seasonal effects. Environmental Science & Technology 50, 11593–11601 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


