Abstract
Clavibacter is an agriculturally important bacterial genus comprising nine host-specific species/subspecies including C. nebraskensis (Cn), which causes Goss's wilt and blight of maize. A robust, simple, and field-deployable method is required to specifically detect Cn in infected plants and distinguish it from other Clavibacter species for quarantine purposes and timely disease management. A multiplex Recombinase Polymerase Amplification (RPA) coupled with a Lateral Flow Device (LFD) was developed for sensitive and rapid detection of Clavibacter and Cn directly from infected host. Unique and conserved genomic regions, the ABC transporter ATP-binding protein CDS/ABC-transporter permease and the MFS transporter gene, were used to design primers/probes for specific detection of genus Clavibacter and Cn, respectively. The assay was evaluated using 52 strains, representing all nine species/subspecies of Clavibacter, other closely related bacterial species, and naturally- and artificially-infected plant samples; no false positives or negatives were detected. The RPA reactions were also incubated in a closed hand at body temperature; results were again specific. The assay does not require DNA isolation and can be directly performed using host sap. The detection limit of 10 pg (~ 3000 copies) and 100 fg (~ 30 copies) was determined for Clavibacter- and Cn-specific primers/probes, respectively. The detection limit for Cn-specific primer/probe set was decreased to 1 pg (~ 300 copies) when 1 µL of host sap was added into the RPA reaction containing tenfold serially diluted genomic DNA; though no effect was observed on Clavibacter-specific primer/probe set. The assay is accurate and has applications at point-of-need diagnostics. This is the first multiplex RPA assay for any plant pathogen.
Subject terms: Plant sciences, Microbiology, Applied microbiology, Bacteriology
Introduction
The Clavibacter genus is a well-known, Gram-positive plant-pathogenic bacterium belonging to the Microbacteriaceae family—contains high GC—and is responsible for several devastating diseases in staple crops worldwide1,2. Previously, this genus consisted of only one species subdivided into nine subspecies, but recently, six subspecies were elevated to the species level-3. The genus now includes C. michiganensis (Cm), C. sepedonicus (Cs), C. insidiosus (Ci), C. tessellarius (Ct), C. capsici (Cc), C. nebraskensis (Cn), affecting tomato, potato, alfalfa, wheat, pepper, and corn, respectively. However, the other three subspecies—C. michiganensis ssp. chiloensis (Cmch) and C. michiganensis ssp. californiensis (Cmca) isolated from tomato, and C. michiganensis ssp. phaseoli (Cmp) from bean—have characteristics that warrant their elevation to the species level as well (D. Arizala and M. Arif; unpublished information).
All six species and three subspecies are host specific, infecting the main host and a few closely related species. Clavibacter species and subspecies are usually disseminated through contaminated seed, propagative materials and soil; secondary infections usually occur through wounds, but in some cases stomata or hydathodes. In the early stages of the infection, all members within the Clavibacter genus reside as biotrophs within xylem vessels eventually leading to systemic disease1,4. Three main factors, including the severity of the disease, the pathogen’s ability to invade seeds, and latent symptomless infections, have necessitated the classification of Cm, Cs, and Ci as quarantine organisms under the European Union Plant Health Legislation, as well as in many other countries2. Despite numerous attempts to breed resistant varieties, no resistant plant cultivars have been commercialized. Due to adverse effects of Clavibacter species on economically important crops, a field-deployable, cost-effective, sensitive and specific detection tool is needed to detect early infections by Clavibacter species at the point-of-need [PON]1,5–8.
Although suitable multiplex molecular techniques are available to detect multiple target pathogens, these techniques are restricted to centralized laboratories and suffer from several other limitations9–11. To address the demand of point-of-need and integrated field-applicable technologies, isothermal nucleic acid amplification methods have been the object of extensive research efforts; many isothermal methods are available, but among them, loop-mediated isothermal amplification [LAMP];12 and recombinase polymerase amplification [RPA];13 are the most commonly used techniques2,14–20. RPA effectiveness is highlighted by rapid detection of multiple targets within a single reaction; however, additional advantages of this technique include the availability of lyophilized reagents, obviation of initial denaturation, high sensitivity, affordability, reduced equipment requirements, and operation at constant low temperature employing recombinase-primer complexes11,14,16,20–23.
RPA possesses an inherent tolerance for sample impurities that may inhibit other platforms based on nucleic acid amplification. Additionally, easy visualization of the results using lateral flow devices can be attained without the involvement of fluorescence readers16,20, which demonstrates the flexibility of RPA for rapid pathogen diagnosis. RPA detection can be achieved as early as in ten minutes, and measurements can be obtained over an extended range of temperatures with a detection limit of 1 fg10,16,20,24,25. It has been reported that even greater optimization of this method can be achieved to prevent false negatives and augment the sensitivity on the multiplex format, although such effects are contingent upon the characteristics of the target sequences, amplicon sizes, primer, and probe design ratios11,24,26,27. While RPA assays are widely used in the detection of animal and human pathogens, its use in plant pathogen detection is limited, but increasing with the availability of commercial kits14,16,20,28–32; yet, no RPA multiplex has been developed for any plant pathogens.
To address the demand for a point-of-need test, we proposed the development of an isothermal multiplex recombinase polymerase amplification assay, not only noted for a high degree of sensitivity and specificity with the potential to be implemented in a laboratory setting, but also for the portable format that enables on-site detection and differentiation of Clavibacter species from C. nebraskensis. This protocol will also serve as a point-of-reference to develop multiplex RPA assays, coupled with a lateral flow device, for other plant pathogens.
Results
In silico specificity validation
We developed two unique primer and probe sets for specific detection of the genus Clavibacter and C. nebraskensis (Fig. 1). The genomic regions, ABC transporter ATP-binding, sugar ABC transporter permease, and MFS transporter, were identified through comparative genomic analyes33. Developed primers/probe sets showed 100% query coverage and 100% identity with only the target species; Clavibacter primer/probe regions were present in all species within the genus Clavibacter, while the Cn primer/probe region was exclusively present in C. nebraskensis. Initial specificity tests were performed with endpoint PCR.
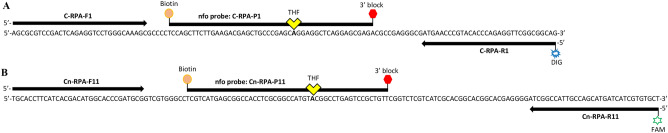
Figure 1.
Multiplex RPA primer and probe locations developed to specifically detect genus Clavibacter (A) and C. nebraskensis (B). (A) Target fragment size 129 bp, reverse primer labeled with DIG; (B) Target fragment size 156 bp, reverse primer labeled with FAM. The probe and primer are 49- and 32-bp long, respectively. THF-tetrahydrofuran residue; DIG-digoxigenin; FAM—6-carboxy-fluorescein.
RPA assay optimization
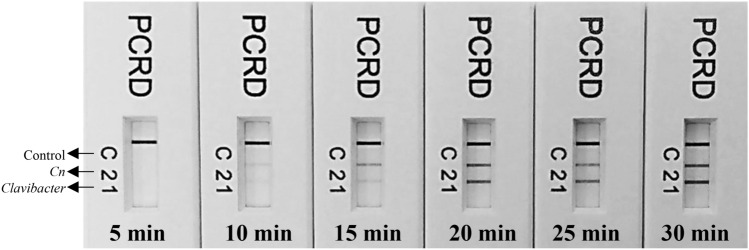
The RPA reaction yielded greater amplification effects (thicker bands) when incubated at 39 °C compared to 37 °C (data not shown). Primer/probe set concentration was also optimized; the results indicated that equal concentrations of each primer (concentration 0.24 µM) and probes (concentration 0.08 µM) in a 50 µL reaction gave the best results (Supplementary Fig. S1). Finally, the effect of RPA incubation time was evaluated from 5 to 30 min. The results indicated that a positive amplification can be obtained in 10 min but with a low band intensity (Fig. 2). However, the 20–30 min incubation time delivered optimum results (Fig. 2). In addition, a brief spin after 4 min once the RPA reaction started showed to have better results compared with no spin.
Figure 2.
Determination of optimum time for successful detection of the genus Clavibacter and C. nebraskensis using the developed multiplex RPA assay. Line 1: detects Clavibacter species/subspecies strains; line 2 detects C. nebraskensis strains, and; line C is a control line. The primer/probe sets for genus Clavibacter and C. nebraskensis are labeled with Biotin/DIG and Biotin/FAM, respectively. The detection can be achieved within 10 min but the optimum line intensity on PCRD was observed in 20–30 min.
RPA assay selectivity and reaction rate
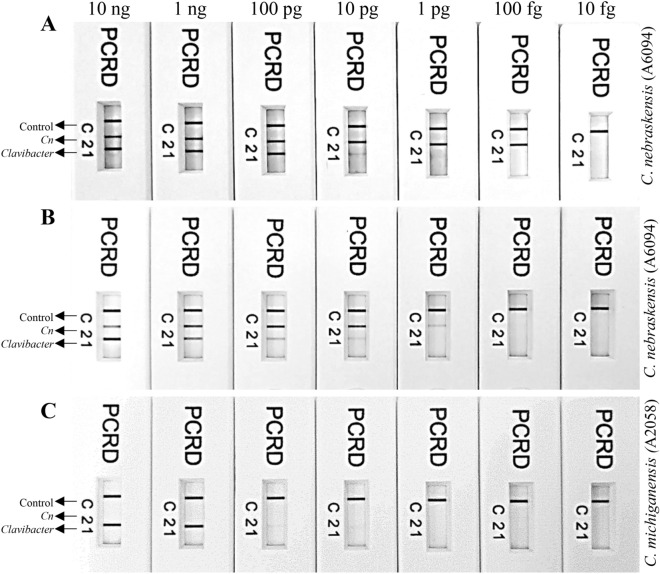
RPA single and multiplex reactions demonstrated high levels of analytical sensitivity and specificity. No false positives were reported using Clavibacter- or Cn-specific primer/probe sets (Fig. 3). All six reported species including Cm, Cs, Ci, Ct, Cc, Cn, and three subspecies Cmch, Cmca, and Cmp within the genus Clavibacter were detected. Clavibacter-specific primer/probe labeled with DIG/Biotin was sufficiently robust to detect all species and subspecies within the genus Clavibacter, whereas, Cn-specific primer/probe labeled with FAM/Biotin showed specific detection of C. nebraskensis strains without displaying cross-detection with other species/subspecies within the genus Clavibacter. Table 1 shows the results of inclusivity and exclusivity tests. All 32 strains representing all species/subspecies within genus Clavibacter were detected using the genus-specific primer/probe set; while a total of 16 samples (14 from pure culture and 2 from naturally infected tissues) were accurately identified as C. nebraskensis using Cn-specific primer/probe set. Results were visible to the unaided eye using the PCRD Nucleic Acid Detector. Lines 1, 2, and C correspond to Clavibacter species, Cn, and control line, respectively (Fig. 3).
Figure 3.
Multiplex RPA assay validation with all known species within the genus Clavibacter. PCRD Nucleic Acid Detector allows the detection of labeled RPA amplicons: line 1- detects the nine DIG/Biotin-labelled Clavibacter species/subspecies amplicons; line 2 -specific detection of FAM/Biotin-labeled C. nebraskensis amplicons, and; line C—control line. NTC (non-template control; water) is included in the last reaction. Strain numbers and species/subspecies names are provided in the figures.
Table 1.
Details of the bacterial strains used in inclusivity and exclusivity panels to validate the multiplex RPA assay for specific detection of genus Clavibacter in general and C. nebraskensis.
| Strain ID | Other ID | Micro-organism | Host | Geographic origin | GenBank accesion numberc | Multiplex RPA Results | |
|---|---|---|---|---|---|---|---|
| CM | Cmn11 | ||||||
| A6203 | DP104A | Clavibacter nebraskensis | Corn | OK, USA | MH560461 | + | + |
| A6204 | DP114A | C. nebraskensis | Corn | TX, USA | MH560462 | + | + |
| A6205 | DP115 | C. nebraskensis | Corn | IA, USA | MH560463 | + | + |
| A6206b | DP117 | C. nebraskensis | Corn | NE,USA | MH560464 | + | + |
| A6207 | DP121 | C. nebraskensis | Corn | TX, USA | MH560465 | + | + |
| A6208 | DP122A | C. nebraskensis | Corn | NE, USA | MH560466 | + | + |
| A6209 | DP134 | C. nebraskensis | Corn | TX, USA | MH560467 | + | + |
| A6210b | DP137 | C. nebraskensis | Corn | TX, USA | MH560468 | + | + |
| A6211 | DP139B | C. nebraskensis | Corn | TX, USA | MH560469 | + | + |
| A6212 | DP164 | C. nebraskensis | Corn | NE, USA | MH560470 | + | + |
| A6213 | DP165c | C. nebraskensis | Corn | IA, USA | MH560471 | + | + |
| A6094b | NCPPB 2579; LMG 3698 | C. nebraskensis | Corn | NE, USA | MH560472 | + | + |
| A6095 | 20037 | C. nebraskensis | Corn | NE, USA | MH560473 | + | + |
| A6096 | 200800460 | C. nebraskensis | Corn | NE, USA | MH560474 | + | + |
| K-1a | C. nebraskensis | Corn | KA, USA | MH560475 | + | + | |
| K-2a | C. nebraskensis | Corn | KA, USA | MH560476 | + | + | |
| A2058b | H-160 | C. michiganensis | Tomato | ID, USA | MH560477 | + | − |
| A4004 | 47 | C. michiganensis | Tomato | OH, USA | MH560481 | + | − |
| A4763 | N 7388A | C. michiganensis | Tomato | Morocco | MH560485 | + | − |
| A4758b | N 212 | C. michiganensis | Tomato | China | MH560484 | + | − |
| A4598 | Cmm038 | C. michiganensis | Tomato | WA, USA | MH560482 | + | − |
| A4690 | CMM 461 | C. michiganensis | Tomato | Portugal | MH560483 | + | − |
| A2645 | S47 | C. michiganensis | Tomato | CA, USA | MH560480 | + | − |
| A1746 | A 518–1 | C. michiganensis | Tomato | HI, USA | MH560479 | + | − |
| A6262b | C222; A5747 | C. michiganensis | Tomato | OR, USA | MH560488 | + | − |
| A2041 | R8 | C. sepedonicus | Potato | Unkown | MH560493 | + | − |
| A6172 | ATCC 33113 | C. sepedonicus | Potato | Canada | MH560494 | + | − |
| A6101 | ATCCBAA-2690; ZUM3936; CFBP 8217 | C. m. spp. chiloensis | Tomato | Chile | MH560495 | + | − |
| A6112 | 1646, P5005 | C. capsici | Peper | China | MH560497 | + | − |
| A6113 | 1647, P5006 | C. capsici | Peper | China | MH560498 | + | − |
| A6134 | C55 | C. m. spp. californiensis | Tomato | CA, USA | MH560499 | + | − |
| A6135 | LPPA982/LMG 27667 | C. m. spp. phaseoli | Bean | Spain | MH560500 | + | − |
| A1149 | QR-80 | C. insidiosus | Alfalfa | KS, USA | MH560501 | + | − |
| A6109 | ZUM 4260; LMG7294; ATCC33566 | C. tessellarius | Wheat | USA | MH56502 | + | − |
| A6214 | DP101 | Microbacterium sp. | Corn | IA, USA | MH547375 | − | − |
| A1150 | ATCC 12975 | Rhodococcus fascians | Unknown | USA | MH605375 | − | − |
| A1147 | ATCC 9682 | Curtobacterium flaccumfaciens pv. poinsettiae | Poinsettia | USA | MH605380 | − | − |
| A2118 | A602.18 | Xanthomonas axonopodis pv. phaseoli | Bean | Unknown | * | − | − |
| A6187 | CC87 | Paenibacillus validus | Unknown | Unknown | * | − | − |
| A1152 | ATCC 13,659 | Rathayibacter rathayi | Orchard grass | United Kingdom | MH605382 | − | − |
| A5735 | T0012 | Microbacterium oleivorans | Tomato | Guatemala | * | ||
| A6218 | DP122B | Curtobacterium flaccumfaciens pv. flaccumfaciens | Corn | NE, USA | MH547376 | − | − |
| PL22 | GBp1A | D. dianthicola | Potato | HI, USA | MK189269 | − | − |
| A6222 | DP138 | Pantoea agglomerans | Corn | WI, USA | MH517382 | − | − |
| A6223 | DP140 | Klebsiella sp. | Corn | IA, USA | MH547379 | − | − |
| A2961 | C58 | Agrobacterium tumefaciens | Cherry | NY, USA | * | − | − |
| A3465 | 26 | Ralstonia solanacearum | Tomato | GE, USA | * | − | − |
| A5058 | A-4 | Xanthomonas campestris pv. campestris | Cabbage | GE, USA | * | − | − |
| A3453 | 256 | Ralstonia solanacearum | Potato | Costa Rica | * | − | − |
| A6230 | DP162A | Pseudomonas sp. | Corn | SD, USA | MH547378 | − | − |
| A5371 | CC26 | Pectobacterium carotovorum | Aglaonema | HI, USA | MH453528 | − | − |
| A1790 | PSS | Pseudomonas syringae pv. syringae | Rice | South Africa | * | − | − |
| A6178 | CC36 | Pseudomonas syringae pv. tomato | Tomato | GA, USA | MH605378 | − | − |
| A1702 | K618; B-111 | Xanthomonas euvesicatoria | Tomato | CA, USA | MG847407 | − | − |
+ Indicated positive amplification.
− Indicated negative amplification.
aDNA extracted from infected corn tissues; no purified strain. bStrains used to infect corn/tomato plants.
cReference number (GenBank accession numbers) are adopted from Larrea-Sarmiento et al. 2019. Most of the strains included to validate this assays were also previously used by Larrea-Sarmiento et al. 2019.
ATCC American Type Culture Collection, USA.
CFBP Collection Française de Bactéries associées aux plantes (French Collection of Plant associated bacteria),
LMG Collection of Laboratorium voor Microbiologie en Microbiele Genetica, Belgicum.
*Sequences were not reported by Larrea-Sarmiento et al.. 2019.
RPA limit of detection
Three analytical sensitivity assays were performed to assess the detection limit of the developed multiplex RPA assay (Fig. 4). For each RPA assay, 2 µL of tenfold serially diluted genomic DNA was added as a template. In the first experiment, the limit of detection was determined using the DNA extracted from pure Cn culture; a detection limit of 10 pg (~ 3000 copies) and 100 fg (~ 30 copies) was observed for Clavibacter- and Cn-specific primer/probe sets, respectively. In the second experiment, the inhibitory effect of plant inhibitors was evaluated, 1 µL of corn plant extract was added to each reaction, containing tenfold serially diluted DNA and other RPA reaction components; a detection limit of 10 pg (~ 3000 copies) and 1 pg (~ 300 copies) was observed for Clavibacter- and Cn-specific primer/probe sets, respectively. In a 3rd assay, pure genomic DNA from C. michiganensis was serially diluted and used for sensitivity assay; detection limit of 10 pg (~ 3000 copies) was observed with Clavibacter-specific primer/probe set while, as expected, no amplification with Cn-specific primer/probe primer/probe set (Fig. 4).
Figure 4.
Limit of detection determination of multiplex RPA assays developed for specific detection of genus Clavibacter in general and C. nebraskensis. Line 1: detects Clavibacter species/subspecies strains; line 2 detects C. nebraskensis strains, and; line C is a control line. The primer/probe sets for genus Clavibacter and C. nebraskensis are labeled with Biotin/DIG and Biotin/FAM, respectively. A tenfold genomic DNA serial dilution (10 ng–10 fg) was used to perform the sensitivity assays. (A) Limit of detection determination using pure C. nebraskensis (A6094) DNA; detected until 100 fg and 10 pg using Clavibacter and C. nebraskensis specific primers/probe set, respectively. (B) Spiked multiplex test performed by adding 1 µL of corn plant sap in tenfold serially diluted C. nebraskensis (A6094) genomic DNA; detected until 1000 fg and 10 pg using Clavibacter and C. nebraskensis specific primers/probe set, respectively. (C) Detection using tenfold serially diluted C. michiganensis (A2058) purified genomic DNA; detected until 10 pg by Clavibacter genus-specific primers/probe—no detection with C. nebraskensis specific primers/probe set.
Detection from artificially and naturally infected plant samples and field applicability
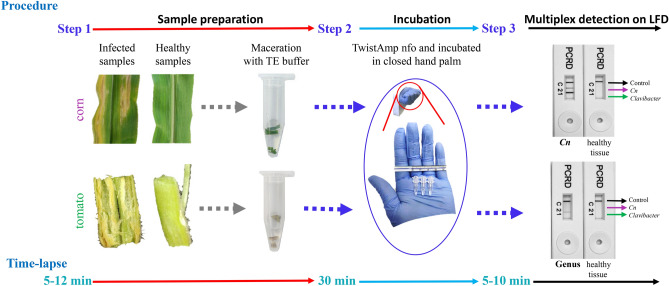
Determination of specificity and detection range of RPA, entailed for point-of-need assays, were carried out using tomato and corn tissues collected from symptomatic 30 dpi plants inoculated with Cm and Cn, respectively. Mock inoculated plant tissues served as a negative control. To evaluate the robustness of the developed RPA assay, no DNA purification was performed—crude sap from infected plants was used directly in the RPA reaction. Testing additional parameters for field viability, temperature conditions, and incubation time were evaluated, and RPA assays were performed in the palm of a closed hand without employing additional equipment. Positive results were observed from tomato and corn tissues infected with Cm (only with Clavibacter-specific assay) and Cn. Presence of pathogens from infected plants was cross-confirmed using end-point PCR (data not shown). Neither positive results nor cross-amplification were observed when mock-inoculated tissues were tested on multiplex reactions (only the control line was observed) (Fig. 5). These results confirmed that the developed RPA assay can specifically detect any species/subspecies within the genus Clavibacter directly from infected plant materials without DNA purification or specialized equipment. Additionally, portable Lateral flow PCD Nucleic Acid Detectors from Abingdon Health help to detect multiple targets without exhibiting false positive or false negative results, producing a robust multi-target RPA assay.
Figure 5.
Schematic steps involve performing multiplex RPA intend for on-filed detection of genus Clavibacter in general and C. nebraskensis. Line 1: detects Clavibacter species/subspecies strains; line 2 detects C. nebraskensis strains, and; line C is a control line. The primer/probe sets for genus Clavibacter and C. nebraskensis are labeled with Biotin/DIG and Biotin/FAM, respectively. Step 1, both infected and healthy plant tissues were processed using TE buffer and a pestle; Step 2, the crude extract was directly used in RPA reactions as a template and incubated using closed hand palm for 30 min; Step 3, visual detection of multiplex RPA results on RPA-PCRD test strips displaying a specific detection of Clavibacter and C. nebraskensis from corn leaves in the upper PCRD, while Clavibacter detection from tomato stem in the lower PCRD. All the reactions were carried out using multiplex assays with TwistAmp-nfo kit. No cross-reactivity or negative results were observed despite using plant extract that contains plant inhibitors.
Discussion
In this study, we reported a reliable, specific, and sensitive multiplex RPA assay for the detection of the widely prevalent plant-pathogenic genus Clavibacter, in general, and more specifically, the re-emerged and increasingly important Goss’ Wilt pathogen, C. nebraskensis. With potential application for point-of-need detection in the field, this proof-of-concept assay based upon the combination of unique genomic regions and multiplex RPA detections coupled with a portable lateral flow device (LFD) may function as a potential tool to rapidly detect numerous targets without specialized equipment. Furthermore, in published literature on current RPA methods, there was no report of simultaneous detection of multiple plant pathogens in laboratory or field settings; thus a number of research studies may benefit from the findings of this study.
The ABC transporter ATP-binding protein CDS/ABC-transporter permease was selected for the detection of all known species and subspecies within the genus Clavibacter, and the MFS transporter gene for the specific detection of C. nebraskensis. These genomic regions were used previously by Dobhal et al. (2019) and Larrea-Sarmiento et. al (2019) to develop TaqMan qPCR and loop-mediated isothermal amplification-based assays and were proven to be unique, conserved, and highly specific. Besides previous genomic analyses, the regions have also been cross-checked with multiple genomes of each species and found to be specific (data not shown)2,33. Previously, RPA assays for plant bacterial pathogens using TwistAmp nfo chemistry coupled with LFD, have shown high specificity and ease of application16,20. The multiplex RPA assay developed here was tested with Clavibacter- and Cn-specific primer/probe sets using a total of 32 strains representing all known species/subspecies within the genus Clavibacter, including 14 strains of C. nebraskensis; the obtained results were specific (Table 1). The identity of these strains was previously confirmed using the dnaA gene region2,33.
Technical expertise along with instrument requirements and infrastructure limitations are common concerns when considering the practicality of diagnostic assays. RPA not only overcomes these limitations but also provides great advantages for on-site detection capability. The preclusion of DNA purification and the elimination of sophisticated equipment provides greater affordability to compensate for technology gaps and allows for the end-user to feasibly conduct multiplex detection tests in the field. TwistDx RPA reagents are available in lyophilized form, and the kit was portable and user-friendly. Molecular analyses that use crude plant extracts as starting materials are often affected by inhibitory sample impurities32. However, the remarkable tolerance to inhibitors offered by RPA led to the development of this rapid sample procedure for RPA assays utilizing tissue ground in TE buffer and applying the crude sap directly as a template11,16,20.
The nature of the recombinase primer complexes used in RPA reactions enables amplification between 37–42 °C13. The low incubation temperature and short reaction time (10–30 min) enhance the efficiency and flexibility of RPA for rapid multiplex pathogen detection (Figs. 2 and 3). Although the intensity of the line on LFD was not intense, a positive reaction can be obtained in as little as 10 min. However, optimum amplification was observed when the RPA reaction was incubated for 20–30 min at 39 °C. Both Gamze et al. (2020) and Ahmed et al. (2018) demonstrated optimum amplification at 30 min with no false positives or negatives. Previously in 2016, Arif and co-workers presented similar optimized test conditions with the Rathayibacter toxicus-specific RPA assay14. The development of RPA based on equipment-free incubation using body temperature was employed previously by Crannell and collaborators for detection of intenstinal protozoa (2014) but this method has not been used for the detection of plant pathogens. This study showed successful, equipment-free incubation of RPA reactions at body temperature in the closed palm.
A thoroughly validated LAMP assay for specific detection of all known subspecies/species in the genus Clavibacter has already been developed by Dobhal and co-workers (2019), but the high-temperature requirement (ca. 65 °C) limits the usefulness of LAMP without portable equipment designed for field applications; moreover, the LAMP assay was not able to discriminate C. nebraskensis from other Clavibacter species.
An optimized diagnostic tool must be sufficiently sensitive to detect the target within latently infected/symptomless samples. The developed RPA assay detected the target sequences as low as 10 pg and 100 fg of genomic DNA of the genus Clavibacter (all species/subspecies) and C. nebraskensis, respectively. However, a tenfold reduction in the sensitivity of the Cn-specific primer/probe set was observed when host sap was added into the tenfold serially diluted genomic DNA (Fig. 4). Previously, Ahmed et al. (2018) reported an RPA assay for specific detection of genus Pectobacterium using TwistDx-nfo coupled with Milenia LFD; the assay showed detection as low as 10 fg from both sensitivity and spiked sensitivity assays. Detection limits of RPA assays are subject to distortion by background noise due to the formation of primer-dimers during the amplification of multiple targets or competition among the primers for the recombinase proteins18,27. Multiplex RPA assays were also optimized by adjusting the primer to probe ratio to maximize signal detection; equal concentrations of Clavibacter- and Cn-specific primer/probe sets displayed similar intensity lines across the strip, with no presence of false positives or false negatives.
Current multiplex RPA methodology is highly specific and sufficiently sensitive to detect multiple targets. The selection of unique targets by genome comparison resulted in specific primer/probe sets enabling the rapid and reliable performance of multiplex RPA. Additionally, efficient sample preparation generates results in about 40–50 min without DNA purification and sophisticated equipment. The developed multiplex RPA assay is ready for point-of-need applications. The developed tools have potential applications in biosecurity, disease surveys, routine diagnostics, seed testing, and epidemiological studies.
Materials and methods
Bacterial isolates and DNA extraction
Strains were obtained from the Pacific Bacterial Collection at the University of Hawai’i at Manoa. Thirty-four bacterial strains representing all species/subspecies within the genus Clavibacter were used in the inclusivity panel. The exclusivity panel was comprised of twenty strains, including Gram-positive and Gram-negative bacterial strains, and characterized as either related or disparate plant pathogens affecting the same crops as the Clavibacter spp. (Table 1). Almost the same strain panels were used by Larrea-Sarmiento et al.33 to validate a multiplex qPCR assay developed for Clavibacter species and C. nebraskensis.
Gram-positive and gram-negative strains were cultured on TZC-S and TZC media, respectively34. Plates were incubated for 2 days at 26 °C (± 2 °C) for DNA extraction or colony PCR. Bacterial genomic DNA was extracted using DNeasy UltraClean Microbial Kit (MO BIO; QIAGEN, Germantown, MD) according to the manufacturer’s instructions. DNA was quantified using the NanoDrop v.2000 spectrophotometer (Thermo Fisher Scientific, Inc., Worcester, MA) for further sensitivity assays. DNA of naturally infected plant samples were received from Kansas State University (Jarred Yasuhara-Bell and James P. Stack)33.
Target selection and primer and probe design
Unique genomic regions were identified using the comparative genomic analyses previously reported33; ABC transporter ATP-binding protein CDS/ABC-transporter permease and MFS transporter were used for genus Clavibacter and C. nebraskensis, respectively. Primers and probes were designed following the instructions provided by TwistDx Ltd. (Maidenhead, UK), with lengths of 32 bp and 49 bp, respectively. Primers and probes specificity was evaluated in silico using the NCBI GenBank BLASTn tool35. The primers and probes were also aligned with Clavibacter species genomes using Geneious version 7.1. Details of primers and probes, synthesized by Integrated DNA Technologies (IDT, Coralville, IA) and LGC Biosearch Technologies (Petaluma, CA), respectively, are provided in Table 2. Reverse primers were labeled with fluorescent 6-FAM (6-Carboxyfluorescein) and DIG (Digoxigenin) while both probes were labeled with biotin.
Table 2.
Details of multiplex RPA primers and probes developed for specific detection of Clavibacter in general and C. nebraskensis.
| Name | Sequences | Length (bp) | Product size (bp) | Target |
|---|---|---|---|---|
| C-RPA-F1 | 5′-AGCGCGTCCGACTCAGAGGTCCTGGGCAAAGC-3′ | 32 | 129 | Clavibacter |
| C-RPA-R1 | 5′/DigN/CTGCCGCCGAACCTCTGGGTGTACGGGTTCAT-3′ | 32 | ||
| C-RPA-P | 5′/Biosg/TCCAGCTTCTTGAAGACGAGCTGCCCGAGC-dSpacer-GGAGGCTCAGGAGCGAGA/Spacer-3′ | 49 | ||
| Cn-RPA-F11 | 5′-TGCACCTTCATCACGACATGGCACCCGATGCG-3′ | 32 | 156 | C. nebraskensis |
| Cn-RPA-R11 | 5′/56-FAM/AGCACACGATGATCATGCTGGCAATGGCCGAT-3′ | 32 | ||
| Cn-RPA-P11 | 5′/Biosg/TCGTCATGAGCGGCCACCTCGCGGCCATGT-dSpacer-CGGCCTGAGTCCGCTGTT/Spacer-3′ | 49 |
Efficiency and specificity of the primers were first evaluated in single end-point PCR reactions with a final volume of 20 µL, containing 1 µL of both sense and anti-sense primers (5 µM), 10 µL of GoTaq Green Master Mix, 7 µL nuclease free-water, and 1 µL of genomic DNA. PCR assays were performed with the following conditions: initial denaturation at 95 °C for 5 min, with 35 cycles of 94 °C for 45 secs, 60 °C for 1 min and 72 °C for 1 min, and a final extension at 72 °C for 5 min.
Single RPA assay design
TwistAmp nfo kits were supplied by TwistDx Ltd., Cambridge, United Kingdom. First, single RPA reactions were performed to validate the specificity of each primer/probe set. The components and conditions were followed as recommended by the manufacturer with slight modifications: each reaction containing an RPA TwistAmp nfo freeze-dried enzyme pellet was rehydrated with 45.5 µL of master-mix consisting of 29.5 µL rehydration buffer, 2.1 µL of each reverse/forward primer set (10 µM), 1.2 µL of probe (5 µM) and 10.6 of nuclease-free water. To each reaction tube, 2 µL of either purified DNA or crude plant extract was added followed by 2.5 µL of Mg-acetate; the tube was mixed by inversion and brief centrifugation.
Multiplex RPA assay optimization
The multiplex RPA assays were optimized in three phases using the TwistAmp nfo kit. For initial optimization, RPA was run at two incubation temperatures, 37 °C and 39 °C, and amplicons resolved in 1.5% agarose gels to identify the optimal amplification temperature. Upon selection of the optimal temperature, primers (10 mM)/probe (5 µM) concentrations were evaluated in three ratios in multi-target assays 1-low (Cn 1.2 µL/1.4 µL = 0.24 µM/0.14 µM; Clavibacter 0.8 µL /0.6 µL = 0.16 µM/0.06 µM), 2-equal (Cn 1.2 µL/0.8 µL = 0.24 µM/0.08 µM; Clavibacter 1.2 µL/0.8 µL = 0.24 µM/0.08 µM) and 3-high (Cn 1.5 µL/0.9 µL = 0.3 µM/0.09 µM; Clavibacter 1.0 µL/0.6 µL = 0.2 µM/0.06 µM). The third phase of assay optimize determined optimum incubation time. Multiplex RPA assays were performed using pure DNA extracted from C. nebraskensis with incubation periods ranging from 5 to 30 min.
Sensitivity and spiked multiplex RPA assays
Two independent assays targeting Clavibacter and Cn were integrated into one multiplex assay. Multiplex RPA assay components were: rehydrating RPA TwistDx-nfo pellet with 45.5 µL of master mix containing 1.2 µL each RPA primer (total 4 primers were added, 10 µM; final concentration 0.24 µM of each primer), 0.8 µL each probe (5 µM—total two probes; final concentration 0.08 µM of each probe), 29.5 µL 1X rehydration buffer and 9.6 µL nuclease-free water. A volume of 2.5 µL of MgOAc applied to the lid, and 2 µL of DNA (10 ng to 100 fg) was added to the final reaction; the reaction was mixed and briefly centrifuged before incubation. For each RPA assay, the relative concentration of purified genomic DNA was measured using the NanoDrop v.2000 spectrophotometer. The limit of detection of RPA multiplex assays was determined using serially diluted genomic DNA from 10 ng to 100 fg (A4757-C. michiganensis and A6094-C. nebraskensis). Effect of plant extract was evaluated by performing a spiked test: one µL of template DNA was used from each tenfold serial dilution of pure bacterial genomic DNA and mixed with 1 µL of crude extract from a healthy corn leaf tissue. Finally, 2 µL of the template was used from each spiked dilution in an RPA reaction with a total volume of 50 µL. Nuclease-free water and/or healthy corn tissue were included in every run as negative controls for the DNA and spiked sensitivity assays, respectively. The copy numbers were calculated using the online tool (http://www.scienceprimer.com/copy-number-calculator-for-realtime-pcr).
Field-applicability
In order to test the specificity of RPA assay as a method for point-of-need detection, a total of six strains, three Cm A2058, A4758, A5747, and three Cn A6206, A6210, A6094, were selected for inoculation of tomato and corn plants, respectively. Strains were grown on YSC medium, and bacterial inoculum was prepared using PBS buffer pH 7 to a final concentration of 108 cells per ml. For Cn, corn plants in the V3 developmental stage were inoculated at the third leaf as described by Ahmad et al.36. For Cm, the stems of three-week-old tomato plants were pierced between the two cotyledons with a needle as described by Kaneshiro et al.37; 10 µL of Cm inoculum was poured directed onto the wound. For both Cn and Cm assays, a negative control was mock-inoculated with 10 uL of PBS alone. Inoculated and control plants were placed into translucent plastic bags for 24 h to maintain humidity. Plant tissues were collected after 30 days-post-inoculation33.
A master mix containing all RPA reagents, except DNA template (2.0 µL) and MgOAc was prepared, as described above and distributed into individual RPA reaction tubes containing a dried enzyme pellet. Field applicability of multiplex RPA reaction was tested by grinding 100 mg each of healthy and infected corn leaf tissue, and healthy and infected tomato stem tissue in 200 µL of TE buffer; the crude sap/extract was used as a template without isolating the DNA. For each reaction, 2 µL of crude extract was added followed by 2.5 µL MgOAc. The contents were thoroughly mixed by shaking and immediately incubated in a closed hand for 30 min.
Portable Lateral flow PCRD Nucleic Acid Detectors containing two different detection lines and one positive control line were coupled to visualize the results. A total of 5 µL of RPA product was added to 70 µL of PCRD extraction buffer, and a total of 75 µL were applied to the receptacle of a portable lateral flow device. Carbon particles at the capture lines of the lateral flow device enabled specific detection of amplicons labeled with either DIG/Biotin and/or FAM/Biotin within 10 min, and the results were visible to the unaided eye.
Conclusions
The multiplex RPA format coupled with LFD, which requires no nucleic acid purification or sophisticated instrumentation, can be performed at a low-temperature, with a short incubation time, and achieves sensitive detection makes the current multiplex RPA assay ideal for in-field and in-lab detection. Until now, multiplex RPA assays capable of accurate and simultaneous detection and differentiation of plant pathogens have not been published. The developed multiplex RPA assay has the potential for rapid and sensitive detection of multiple targets in field settings.
Supplementary Information
Acknowledgements
This work was supported by the USDA National Institute of Food and Agriculture, Hatch project 9038H, managed by the College of Tropical Agriculture and Human Resources. The strains used in this study were revived and further characterized with grant support from the National Science Foundation (NSF-CSBR Grant No. DBI-1561663). The mention of trade names or commercial products in this publication does not imply recommendation or endorsement by the University of Hawaii. The granting agencies had no role in study design, data collection, and analysis, decision to publish, or preparation of the manuscript.
Author contributions
M.A. conceptualization, funding acquisition, project administration, resources; A.L.S. performed the experiments, analyze the data, wrote the first draft; all authors reviewed the manuscript.
Data availability
All sequencing data is available in NCBI GenBank database.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-91336-7.
References
- 1.Yasuhara-Bell J, Alvarez AM. Seed-associated subspecies of the genus Clavibacter are clearly distinguishable from Clavibacter michiganensis subsp. michiganensis. Int. J. Syst. Evol. Microbiol. 2015;65:811–826. doi: 10.1099/ijs.0.000022. [DOI] [PubMed] [Google Scholar]
- 2.Dobhal S, Larrea-Sarmiento A, Alvarez AM, Arif M. Development of a loop-mediated isothermal amplification assay for specific detection of all known subspecies of Clavibacter michiganensis. J. Appl. Microbiol. 2019;126(2):388–401. doi: 10.1111/jam.14128. [DOI] [PubMed] [Google Scholar]
- 3.Li X, Tambong J, Yuan K, Chen W, Xu H, Levesque CA, De Boer SH. Re-classification of Clavibacter michiganensis subspecies on the basis of whole genome and multi-locus sequence analyses. Int. J. Syst. Evol. Microbiol. 2018;68:234–240. doi: 10.1099/ijsem.0.002492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tambong JT. Comparative genomics of Clavibacter michiganensis subspecies, pathogens of important agricultural crops. PLoS ONE. 2017;12(3):e0172295. doi: 10.1371/journal.pone.0172295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Biddle J, McGee D, Braun E. Seed transmission of Clavibacter michiganense subsp. nebraskense in corn. Plant Dis. 1990;74(11):908–911. doi: 10.1094/PD-74-0908. [DOI] [Google Scholar]
- 6.Nemeth J, Laszlo E, Emödy L. Clavibacter michiganensis ssp. insidiosus in lucerne seeds. EPPO Bull. 1991;21(4):713–718. doi: 10.1111/j.1365-2338.1991.tb01305.x. [DOI] [Google Scholar]
- 7.Franc G. Persistence and latency of Clavibacter michiganensis subsp. sepedonicus in field-grown seed potatoes. Plant Dis. 1999;83(3):247–250. doi: 10.1094/PDIS.1999.83.3.247. [DOI] [PubMed] [Google Scholar]
- 8.Eichenlaub R, Gartemann K. The Clavibacter michiganensis subspecies: molecular investigation of Gram-positive bacterial plant pathogens. Annu. Rev. Phytopathol. 2011;49(1):445–464. doi: 10.1146/annurev-phyto-072910-095258. [DOI] [PubMed] [Google Scholar]
- 9.Crannell Z, Castellanos-Gonzalez A, Nair G, Mejia R, White AC, Richards-Kortum R. Multiplexed recombinase polymerase amplification assay to detect intestinal protozoa. Anal. Chem. 2016;88(3):1610–1616. doi: 10.1021/acs.analchem.5b03267. [DOI] [PubMed] [Google Scholar]
- 10.Wu YD, Xu MJ, Wang QQ, Zhou CX, Wang M, Zhu XQ, Zhou DH. Recombinase polymerase amplification (RPA) combined with lateral flow (LF) strip for detection of Toxoplasma gondii in the environment. Vet. Parasitol. 2017;243:199–203. doi: 10.1016/j.vetpar.2017.06.026. [DOI] [PubMed] [Google Scholar]
- 11.Lobato I, O’Sullivan C. Recombinase polymerase amplification: basics, applications and recent advances. TrAC Trends Anal. Chem. 2018;98:19–35. doi: 10.1016/j.trac.2017.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, Hase T. Loop-mediated isothermal amplification of DNA. Nucl. Acids Res. 2000;28(12):E63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Piepenburg O, Williams CH, Stemple DL, Armes NA. DNA detection using recombination proteins. PLoS Biol. 2006;4(7):e204. doi: 10.1371/journal.pbio.0040204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arif M, Busot GY, Mann R, Rodoni B, Stack JP. Detection of the select agent Rathayibacter toxicus using recombinase polymerase amplification coupled with a lateral flow device. (Abstr.) Phytopathology. 2016;106:S4.23. [Google Scholar]
- 15.Song J, Liu C, Mauk MG, Rankin SC, Lok JB, Greenberg RM, Bau HH. Two-stage isothermal enzymatic amplification for concurrent multiplex molecular detection. Clin. Chem. 2017;63(3):714–722. doi: 10.1373/clinchem.2016.263665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ahmed FA, Larrea-Sarmiento A, Alvarez AM, Arif M. Genome-informed diagnostics for specific and rapid detection of Pectobacterium species using recombinase polymerase amplification coupled with a lateral flow device. Sci. Rep. 2018;8:15972. doi: 10.1038/s41598-018-34275-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Larrea-Sarmiento A, et al. Development of a genome-informed loop-mediated isothermal amplification assay for rapid and specific detection of Xanthomonas euvesicatoria. Sci. Rep. 2018;8:14298. doi: 10.1038/s41598-018-32295-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lau, H. Y., & Botella, J. R. Advanced DNA-based point-of-care diagnostic methods for plant diseases detection. Front. Plant Sci.8 (2017). [DOI] [PMC free article] [PubMed]
- 19.Ocenar J, Arizala D, Boluk G, Dhakal U, Gunarathne S, Paudel S, Dobhal S, Arif M. Development of a robust, field-deployable loop-mediated isothermal amplification (LAMP) assay for specific detection of potato pathogen Dickeya dianthicola targeting a unique genomic region. PLoS ONE. 2019;14(6):e0218868. doi: 10.1371/journal.pone.0218868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boluk G, Dobhal S, Crockford AB, Melzer M, Alvarez AM, Arif M. Genome-informed recombinase polymerase amplification assay coupled with a lateral flow device for in-field detection of Dickeya species. Plant Dis. 2020;104(8):2217–2224. doi: 10.1094/PDIS-09-19-1988-RE. [DOI] [PubMed] [Google Scholar]
- 21.De Paz HD, Brotons P, Muñoz-Almagro C. Molecular isothermal techniques for combating infectious diseases: towards low-cost point-of-care diagnostics. Expert Rev. Mol. Diagn. 2014;14(7):827–843. doi: 10.1586/14737159.2014.940319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clancy E, Higgins O, Forrest MS, Boo TW, Cormican M, Barry T, Piepenburg O, Smith TJ. Development of a rapid recombinase polymerase amplification assay for the detection of Streptococcus pneumoniae in whole blood. BMC Infect. Dis. 2015;15:481. doi: 10.1186/s12879-015-1212-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Daher RK, Stewart G, Boissinot M, Bergeron MG. Recombinase polymerase amplification for diagnostic applications. Clin. Chem. 2016;62(7):947–958. doi: 10.1373/clinchem.2015.245829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kersting S, Rausch V, Bier FF, von Nickisch-Rosenegk M. Multiplex isothermal solid-phase recombinase polymerase amplification for the specific and fast DNA-based detection of three bacterial pathogens. Microchim. Acta. 2014;181(13–14):1715–1723. doi: 10.1007/s00604-014-1198-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang Y, Qin X, Sun Y, Cong G, Li Y, Zhang Z. Development of isothermal recombinase polymerase amplification assay for rapid detection of Porcine Circovirus Type 2. Biomed. Res. Int. 2017;2017:8403642. doi: 10.1155/2017/8403642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Crannell ZA, Rohrman B, Richards-Kortum R. Equipment-free incubation of recombinase polymerase amplification reactions using body heat. PLoS ONE. 2014;9(11):e112146. doi: 10.1371/journal.pone.0112146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim JY, Lee JL. Development of a multiplex real-time recombinase polymerase amplification (RPA) assay for rapid quantitative detection of Campylobacter coli and jejuni from eggs and chicken products. Food Control. 2017;73:1247–1255. doi: 10.1016/j.foodcont.2016.10.041. [DOI] [Google Scholar]
- 28.Mekuria TA, Zhang S, Eastwell KC. Rapid and sensitive detection of Little cherry virus 2 using isothermal reverse transcription-recombinase polymerase amplification. J. Virol. Methods. 2014;205:24–30. doi: 10.1016/j.jviromet.2014.04.015. [DOI] [PubMed] [Google Scholar]
- 29.Zhang S, Ravelonandro M, Russell P, McOwen N, Briard P, Bohannon S, Vrient A. Rapid diagnostic detection of plum pox virus in Prunus plants by isothermal AmplifyRP using reverse transcription-recombinase polymerase amplification. J. Virol. Methods. 2014;207:114–120. doi: 10.1016/j.jviromet.2014.06.026. [DOI] [PubMed] [Google Scholar]
- 30.Miles TD, Martin FN, Coffey MD. Development of rapid isothermal amplification assays for detection of Phytophthora spp. in plant tissue. Phytopathology. 2015;105(2):265–278. doi: 10.1094/PHYTO-05-14-0134-R. [DOI] [PubMed] [Google Scholar]
- 31.Londoño MA, Harmon CL, Polston JE. Evaluation of recombinase polymerase amplification for detection of begomoviruses by plant diagnostic clinics. Virol. J. 2016;13(1):48. doi: 10.1186/s12985-016-0504-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kapoor R, Srivastava N, Kumar S, Saritha RK, Sharma SK, Jain RK, Baranwal VK. Development of a recombinase polymerase amplification assay for the diagnosis of banana bunchy top virus in different banana cultivars. Adv. Virol. 2017;162(9):2791–2796. doi: 10.1007/s00705-017-3399-9. [DOI] [PubMed] [Google Scholar]
- 33.Larrea-Sarmiento A, Alvarez AM, Stack JP, Arif M. Synergetic effect of non-complementary 5’ AT-rich sequences on the development of a multiplex TaqMan real-time PCR for specific and robust detection of Clavibacter michiganensis and C. michiganensis subsp. nebraskensis. PLoS ONE. 2019;14(7):e0218530. doi: 10.1371/journal.pone.0218530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Norman D, Alvarez A. A rapid method for presumptive identification of Xanthomonas campestris pv. dieffenbachiae and other Xanthomonas. Plant Dis. 1989;73(8):654–658. doi: 10.1094/PD-73-0654. [DOI] [Google Scholar]
- 35.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 36.Ahmad A, Mbofung GY, Acharya J, Schmidt CL, Robertson AE. Characterization and comparison of Clavibacter michiganensis subsp. nebraskensis strains recovered from epiphytic and symptomatic infections of maize in Iowa. PLoS ONE. 2015;10(11):e0143553. doi: 10.1371/journal.pone.0143553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kaneshiro WS, Mizumoto CY, Alvarez AM. Differentiation of Clavibacter michiganensis subsp. michiganensis from seed-borne saprophytes using ELISA, Biolog and 16S rDNA sequencing. Eur. J. Plant. Pathol. 2006;116:45–56. doi: 10.1007/s10658-006-9036-1. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequencing data is available in NCBI GenBank database.