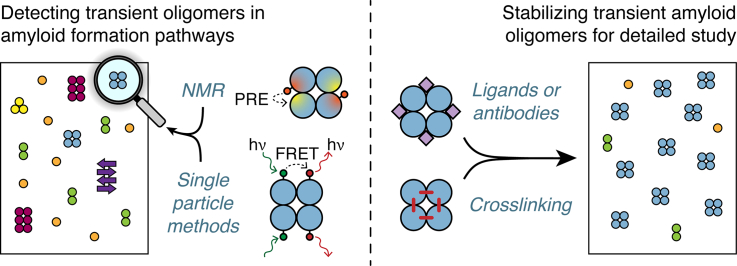
Abstract
Oligomers which form during amyloid fibril assembly are considered to be key contributors towards amyloid disease. However, understanding how such intermediates form, their structure, and mechanisms of toxicity presents significant challenges due to their transient and heterogeneous nature. Here, we discuss two different strategies for addressing these challenges: use of (1) methods capable of detecting lowly-populated species within complex mixtures, such as NMR, single particle methods (including fluorescence and force spectroscopy), and mass spectrometry; and (2) chemical and biological tools to bias the amyloid energy landscape towards specific oligomeric states. While the former methods are well suited to following the kinetics of amyloid assembly and obtaining low-resolution structural information, the latter are capable of producing oligomer samples for high-resolution structural studies and inferring structure-toxicity relationships. Together, these different approaches should enable a clearer picture to be gained of the nature and role of oligomeric intermediates in amyloid formation and disease.
Keywords: Amyloid disease, Transient intermediate, Oligomer stabilization, Chemical tool, NMR, Single particle
Graphical abstract
Highlights
-
•
Methods to study structure, toxicity, and kinetics of transient amyloid oligomers.
-
•
NMR and single particle methods can characterize lowly-populated oligomers.
-
•
Chemical tools/antibodies stabilize oligomers for structural and toxicity studies
-
•
A combination of methods is needed to fully characterize amyloid assembly pathways.
1. Introduction
Amyloid fibril formation is a complex and multifaceted process, characterized by the self-assembly of proteins or peptides into insoluble cross-β deposits [1,2] (Fig. 1A). In healthy individuals, amyloid assembly pathways are highly regulated and accessible only to a small number of proteins, resulting in fibrils which can play important functional roles. These so-called “functional amyloid” fibrils can act as structural scaffolds, protein reservoirs, biofilm components, or be involved in the laying down and storage of long-term memories, amongst other biological roles [3,4]. However, normally soluble proteins and peptides can also be rendered amyloid-competent by specific mutations, changes to post-translational modifications, or changes to the intra- or extracellular conditions [1]. The aberrant and unregulated self-assembly events which subsequently occur are associated with an array of human disorders, ranging from Alzheimer's and Parkinson's diseases, to type II diabetes and dialysis-related amyloidosis [1,2].
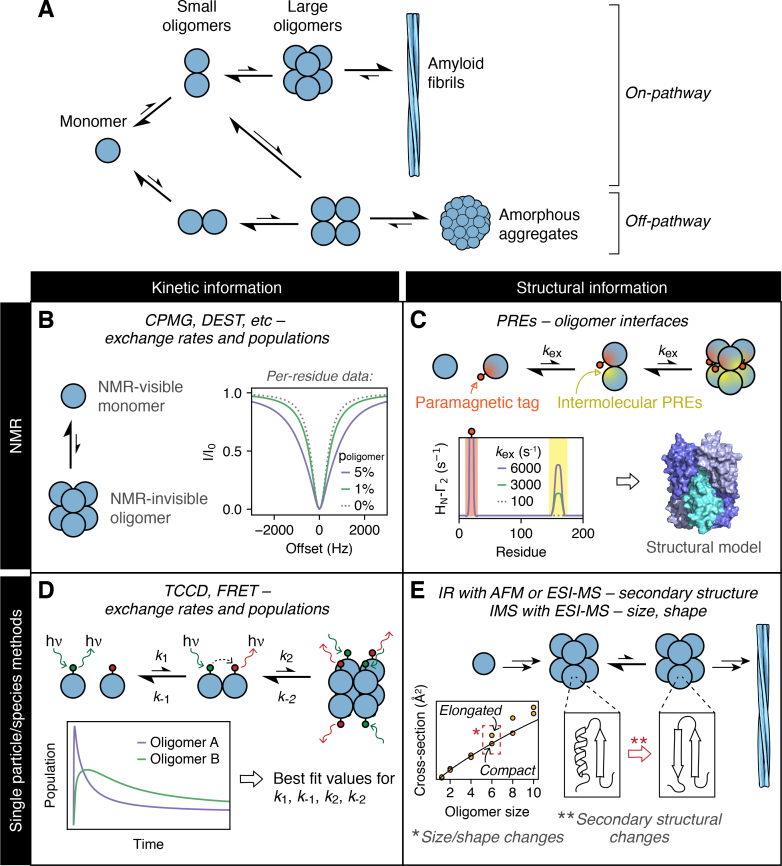
Fig. 1.
Understanding the complex mechanisms of amyloid fibril assembly using “non-perturbing” biophysical methods. A: A schematic of a generic protein self-assembly pathway. The self-association of particular proteins can lead to the formation of cross-β fibrillar structures (amyloid fibrils) but can also lead to the formation of aggregates which lie “off-pathway” from fibril assembly. Assembly intermediates are often lowly-populated, rapidly interconverting, and transiently formed, making them challenging targets for many biophysical techniques, with notable exceptions being NMR (B, C) [[31], [32], [33], [34], [35]] and methods which detect single particles or species (D, E) (e.g. single particle fluorescence - including FRET and 2-color incidence detection, TCCD - [36,37], SPFS [38], and ESI-MS [[39], [40], [41]]). Monomer-oligomer exchange rates and oligomer populations (poligomer) can be obtained through the global fitting of oligomerization models to the observed NMR or single particle data (B, D). The structural information obtained through NMR and single particle/species methods is typically of low resolution but can provide insights into oligomerization interfaces (C), and structural transitions (E). The DEST and PRE data shown in B and C, respectively, were simulated by numerically solving the corresponding McConnell equations using a B1 field of 200 Hz, R2B = 1000 s−1, R2A = 10 s−1, and R1A = 1.5 s−1. The PRE data shown in C were simulated with a distance between residue 160 and the MTSL label (on residue 20) of 1 Å in the excited state (5% population). The black line in the collisional cross-section plot in E represents the expected cross-sections for each oligomer size, assuming isotropic growth, and the dashed red box indicates the point in the self-assembly pathway at which the oligomers of this protein start to undergo structural transitions, as detected by IMS coupled to ESI-MS [17,41]. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
For many disease-associated amyloid proteins, it has been recognized that soluble oligomers which form during, or as a result of, fibril assembly can be the major cytotoxic species associated with cellular dysfunction and disease onset [[5], [6], [7], [8], [9], [10]]. Consequently, there has been intense interest in the study of such assembly intermediates, with the aim to better understand the molecular mechanism(s) by which soluble oligomers form and transition into a cross-β structure, as well as to identify structural features associated with cytotoxicity [11]. Unfortunately, the complexity and heterogeneity of amyloid assembly pathways makes such analyses immensely challenging in vitro, let alone in vivo. Additionally, it is difficult to reconcile the vast range of oligomers observed by different research groups under varying experimental conditions. Oligomeric intermediates are typically metastable, lowly-populated, and interconverting with each other and fibril surfaces within the aggregating mixture [12,13]. As a consequence, the properties of specific oligomers cannot be readily determined in detail using most bulk solution methods, as such approaches typically do not allow information to be extracted about individual species within these diverse populations. The structural and kinetic study of oligomeric intermediates thus requires the use of techniques capable of dissecting the properties of individual species within complex mixtures, coupled with careful data analysis to extract the relevant information about their population, lifetime, and structural features. Some of the most powerful methods include those which can directly detect single particles (such as single particle fluorescence, e.g. fluorescence resonance energy transfer (FRET) [14,15], and single particle force spectroscopy (SPFS) [16]) or single species (such as electrospray ionization mass spectrometry (ESI-MS) [[17], [18], [19], [20]]). Solution-phase nuclear magnetic resonance (NMR) spectroscopy, which is unique amongst ensemble techniques for its ability to extract structural and kinetic information about different protein populations in atom-specific detail, can also be used for the characterization of lowly populated and rapidly interconverting species [21]. Additionally, molecular dynamics simulations using a variety of different force fields and approaches can provide insights into the potential repertoire of oligomers formed, which then require parallel experiments for their verification [[22], [23], [24], [25], [26]]. However, the aforementioned experimental approaches are not without their limitations – in particular, the structural resolution obtained using these techniques is generally insufficient to inform rational drug design approaches. Thus, there remains an on-going need to explore complementary methods for the characterization of amyloid oligomers.
The purposeful manipulation of an amyloidogenic system, with the aim of tuning the self-assembly landscape to favor specific events or species of interest, represents an alternative strategy for the study of amyloid self-assembly. Such an approach can reduce sample heterogeneity and, if used to bias the system towards oligomer formation, may allow mechanistic and structural information to be obtained about these assembly intermediates using a wider range of techniques than would otherwise be possible. Modulation of amyloid assembly can be achieved through the use of small molecule ligands, antibodies, or covalent protein modifications, to generate samples with a range of defined oligomer populations. In combination with methods such as NMR, kinetic modelling, and cell toxicity assays, these samples can provide insights into the role, disease-relevance, and structure of particular oligomeric populations, and validate potential routes for the treatment of amyloid diseases. There are now several examples where the stabilization (or destabilization) of specific species has reduced heterogeneity sufficiently to facilitate detailed structural studies using X-ray crystallography, thereby providing high-resolution insights into these potential therapeutic targets [[27], [28], [29], [30]].
This review provides an overview of the experimental methods and approaches which can be used to study the structure, kinetics, and disease-relevance of transient amyloid oligomers, and provides examples of their use in vitro. Although the ultimate goal of the amyloid field is to perform these experiments in vivo, in vitro studies allow us to explore the energy landscape of amyloid protein self-assembly and identify general trends between certain structural features (e.g. hydrophobicity, secondary structure) and toxicity. We discuss approaches through which observations can be made without intentional perturbation of the self-assembly process (e.g. using solution NMR, single particle methods, and native ESI-MS), as well as those through which the self-assembly equilibrium is deliberately modified to gain information about specific species. These “non-perturbing” and “perturbing” methods are complementary: most non-perturbing techniques are best suited to the determination of kinetic parameters (yielding information about rates of interconversion and population lifetimes), while the resolution of structural information they can obtain is typically lower. By contrast, perturbing methods offer the opportunity to analyze species using high-resolution structural methods, and hence enable detailed structure-toxicity relationships to be determined. The combination of both of these approaches provides an ideal experimental toolkit for generating more complete descriptions of the molecular mechanisms of amyloid fibril formation and their associated cellular toxicity.
2. Non-perturbing methods for the study of amyloid oligomers
2.1. Solution NMR
NMR spectroscopy has the capability to characterize transient and heterogeneous systems in all-atom detail, and thus has been widely utilized in the characterization of amyloid protein assembly [21,42,43]. Many powerful NMR methods are sensitive to the properties of “NMR-invisible” species (i.e. those <1% populated or of high molecular weight) that are in rapid exchange with the NMR-visible state(s) [21,44]. In the context of amyloid assembly, this means that the properties (e.g. size, timescale of formation) of lowly populated (ca. 0.5–15%) amyloid oligomers can be indirectly studied via the monomeric precursor, using experiments such as Carr-Purcell-Meiboom-Gill (CPMG) relaxation dispersion, chemical exchange saturation transfer (CEST), paramagnetic relaxation enhancement (PRE), life-time line broadening, dark exchange saturation transfer (DEST), and off-resonance R1ρ relaxation [42]. Using these methods, the relative populations and exchange rates (100–10,000 s−1) between species can be extracted, providing valuable information about the assembly process. Some experiments (e.g. CPMG relaxation dispersion and CEST) are designed to probe exchange with species whose relaxation properties are not very different to those of the monomer, and thus are best suited to the study of small oligomers [[45], [46], [47]]. By contrast, others, such as DEST, life-time line broadening, and off-resonance R1ρ [33], require exchange of the monomer with a higher molecular weight species, making them ideal for the study of large amyloid oligomers or the exchange between monomer and fibrils [32,48].
The application of these NMR methods, in combination with global fitting of data acquired at a range of protein concentrations and magnetic field strengths, can provide valuable insights into the mechanisms of self-assembly (Fig. 1B). Such an approach has recently been used to interrogate the early stages of aggregation of a minimal peptide from the protein huntingtin (associated with Huntington's disease), providing information about the populations, exchange rates, and secondary structure of oligomers formed by two competing aggregation pathways [31]. Similar strategies have been adopted for other amyloid proteins that form a range of oligomeric states and structures, revealing insights into the binding of amyloid β (Aβ) monomers to fibrils [49], the energy landscape of copper‑zinc superoxide dismutase (SOD1) aggregation [50], and the formation of β2-microglobulin (β2m) oligomers [35,51] – events which are associated with Alzheimer's disease, amyotrophic lateral sclerosis, and dialysis-related amyloidosis, respectively. However, as these methods rely on reversible chemical exchange, this places a limit on the protein concentrations which can be used. There is an apparent irreversibility of the amyloid pathway for samples prepared above their critical protein concentration, and so sample conditions must be carefully selected to minimize the formation of any highly aggregation-prone species, while maintaining a suitable signal-to-noise ratio.
An alternative NMR approach that can be used to study the assembly of amyloid oligomers involves tilting the energy landscape of aggregation in a controlled manner through the use of hydrostatic pressure [34]. Pressure allows the rapid disassembly of oligomeric species before they convert to pressure-resistant fibrils. This technique therefore permits study of the oligomerization equilibria that would not be accessible to the experiments described above under a constant atmospheric pressure. Pressure-jump NMR experiments, in which the pressure inside the NMR cell is rapidly altered (in a matter of milliseconds), have been used recently to study the interconversion of a highly unfolded Aβ monomer (at high pressure) with an oligomeric species (at low pressure), providing residue-specific information about oligomers that show the first signs of conversion into amyloid [34].
In addition to probing the kinetics of protein self-assembly, NMR can also be used to investigate the structural properties of amyloid oligomers [31,[52], [53], [54]]. NMR-derived structural insights into these protein complexes can be achieved through the determination of distance restraints from NMR experiments, which are then used to drive simulated annealing calculations [55] or other molecular dynamics approaches [56]. Due to the transient nature of the complexes present in the early stages of amyloid assembly, traditional nuclear Overhauser effect-based structural investigations typically fail and intermolecular distance restraints tend to instead be derived from PRE studies using paramagnetic spin labels (commonly MTSL; S-(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl methanesulfonothioate) [57] (Fig. 1C). When covalently attached at a specific site on a protein's surface, spin labels can be used to map oligomerization interfaces, even those that are short-lived (< 100 μs) [58]. Data from PRE experiments have been used to generate structural models of β2m dimers and hexamers [35,52] (Fig. 1C), tetramers formed by a minimal huntingtin construct [31], and homo-/heterodimers formed by α- and β-synuclein (the former variant being associated with Parkinson's disease) [53,54].
Due to the size limitations of solution NMR methods, it has remained challenging to investigate large oligomeric assemblies directly using solution NMR-based approaches. However, with the advance of 13C-methyl-TROSY methods as sensitive reporters of protein structure and dynamics, amyloid oligomers can be studied directly (provided that they exist in a sufficiently large population) [59]. Finally, the arsenal of magnetic resonance techniques to study amyloid formation would not be complete without mentioning solid-state NMR and electron paramagnetic resonance (EPR). Both techniques have contributed substantially to our understanding of the structures of amyloid fibrils [60,61], as well as oligomers of Aβ peptides [62,63] and α-synuclein [64], but are frequently geared towards stable, monodisperse samples (e.g. fibrils or stable oligomers), with some notable exceptions [65,66].
NMR currently remains the only ensemble method that can yield atom-specific structural and kinetic information (including rates of interconversion and lifetimes) for lowly populated, transient states, by observing their effect on the main species in solution (usually the monomer). However, rather than inferring such information from the interpretation of NMR data, it is also possible to directly observe transient, oligomeric species using single particle and other non-ensemble averaged methods, as described below.
2.2. Single particle and non-ensemble averaged methods
Single particle techniques are unparalleled in their ability to directly detect individual molecules and populations of interconverting species within heterogeneous mixtures. Single particle fluorescence methods (a term we use here to describe approaches which rely on fluorescent dyes, rather than the measurement of intrinsic protein fluorescence) and SPFS have both played key roles in the amyloid field, allowing kinetic and low-resolution structural information to be obtained for the species populated during fibril assembly [[67], [68], [69]]. Other non-ensemble averaged methods, such as ESI-MS, are also able to directly detect specific oligomer populations during assembly. When coupled to other techniques (e.g. ion mobility spectrometry, hydrogen-deuterium exchange, or infrared spectroscopy), ESI-MS is capable of providing structural information for these individual oligomeric species [18,19].
Single particle fluorescence experiments used for the study of amyloid assembly most commonly require preparation of dual-labelled protein samples, containing an equimolar mixture of protein molecules labelled with one of two distinct fluorescent dyes [67]. From such dual-labelled samples, oligomers can be detected by the observation of simultaneous bursts from the two fluorophores or, in cases where the emission of one fluorophore overlaps with the absorption of the other, by the ability of these molecules to undergo FRET (Fig. 1D) [10,36,37,[70], [71], [72], [73], [74], [75], [76], [77], [78], [79], [80]]. Different oligomer populations can be distinguished from one another based on the intensity of fluorescent bursts [79,80], the FRET efficiency [10,36,37], or the stability of the detected oligomers to particular experimental conditions (e.g. dilution into buffer with high or low ionic strengths, or the presence of proteases) [10,37,75,76,79,81]. By using such measurements to monitor the population of specific oligomers in an aggregating mixture over time, the resulting data can be fitted to self-assembly models to determine the rates of oligomer formation and interconversion, as well as thermodynamic parameters for these protein-protein interactions [36,37,76,78,82] (Fig. 1D).
While single particle fluorescence is immensely powerful, like any technique it relies on certain assumptions and has various limitations. The requirement to perform single particle fluorescence measurements under highly dilute solution conditions (several orders of magnitude less concentrated than the conditions ordinarily used to study protein aggregation in vitro) can limit the types of oligomers which can be observed. Typically, aliquots from a more concentrated aggregating mixture are taken and diluted immediately prior to single particle experiments, and thus unstable and/or highly transient oligomers may dissociate before the data are acquired [79]. However, this issue can be minimized through the use of microfluidic devices to speed up sample analysis [75,83] or by diluting aliquots into a solution of non-fluorescently labelled protein, rather than buffer alone [10]. An additional factor which must be considered is that most single particle fluorescence experiments make use of fluorescently-labelled protein samples, and therefore rely on the assumption that the incorporation of fluorescent dyes, which are often large and hydrophobic, does not perturb the assembly mechanism. This assumption may not always hold true [[84], [85], [86]], and the appropriate selection of fluorophores must be ensured by performing thorough control experiments. It is also possible to perform experiments with fluorescent probes which interact non-covalently with the protein of interest, and which can therefore be added after assembly has taken place [81,82,87,88].
SPFS relies on the use of mechanical force to perturb interatomic interactions and has been extensively used to study the formation of native protein contacts during folding [89]. However, non-native intermolecular contacts that take place during protein misfolding and self-assembly can also be studied through such methods. Atomic force microscopy (AFM) is the primary SPFS technique which has been used to study amyloid assembly, as the high forces accessible in this method are suitable for the study of amyloidogenic oligomerization events [16]; lower forces can be accessed using other, complementary SPFS methods [90], such as optical tweezers [[91], [92], [93], [94]]. Unlike single particle fluorescence measurements, where soluble oligomers formed at any stage during aggregation can be detected within a single experiment, AFM SPFS experiments tend to be designed to focus on specific oligomerization events – commonly dimerization. The lowering and raising of an AFM cantilever to and from a surface controls the formation and then drives dissociation of individual interactions [95]. For the study of dimerization events, one protein is typically attached to the cantilever tip, whilst the partner of interest (often the same protein) is immobilized on a surface on the sample stage [[96], [97], [98], [99], [100], [101], [102], [103], [104], [105], [106], [107], [108]]. Formation of higher oligomers can be studied using tandem repeat oligomers which are tethered between the surface and cantilever tip [109] or, more recently, using the flexible nanoarray (FNA) approach where a number of monomers are immobilized at various points along a flexible polymer [[110], [111], [112], [113]]. In both cases, the site of covalent attachment and the length/flexibility of the covalent linker need to be carefully selected to ensure that steric constraints do not alter the accessibility of key interaction interfaces or change the behavior of the protein. SPFS methods can yield information concerning the lifetime of specific protein-protein interactions and their strength (force resistance). Furthermore, the distance required to move the cantilever away from the surface to cause disruption of the oligomer can be used to infer the site of interaction [95,114].
In addition to its use as a SPFS method, AFM can be used as a surface-imaging technique to directly observe the formation and turnover of oligomers during amyloid assembly [68]. Recently, AFM has been coupled with infrared spectroscopy (AFM-IR) to image the secondary structure content of individual amyloid oligomers at the single aggregate level and to identify structural transitions which occur during the self-assembly of various amyloid proteins, including ataxin-3 (the causative agent of spinocerebellar ataxia type-3 or Machado–Joseph disease) [38], huntingtin exon 1 [115], and α-synuclein [116] (Fig. 1E). Such conformational conversion events are often rate-limiting steps in amyloid formation [117], and thus are key processes to characterize in structural and kinetic detail.
ESI-MS represents an alternative technique for the direct detection of individual monomeric conformers and oligomeric species formed during amyloid assembly. As a soft ionization method, ESI allows non-covalent interactions to be maintained when aggregating proteins are sprayed into a mass spectrometer from a volatile buffer solution [118]. ESI-MS has the advantage over the single particle methods discussed here in that no dye labels or immobilization strategies are required, with the mass accuracy of modern mass spectrometers making it straightforward to detect different oligomeric species that are co-populated [[17], [18], [19], [20]]. Most powerfully, ESI-MS can be directly coupled to other methods, such as ion mobility spectrometry (IMS; which allows oligomers with the same m/z ratio to be separated based on size and shape) [41,[119], [120], [121], [122], [123], [124], [125], [126], [127], [128], [129], [130], [131], [132], [133]], hydrogen-deuterium exchange and related covalent labelling experiments (revealing oligomer interfaces) [[134], [135], [136], [137], [138], [139], [140], [141], [142], [143], [144], [145], [146], [147]], and infrared spectroscopy (IR) [39,40] to yield structural information about the different species present in an aggregating mixture. These techniques have been used to detect structural transitions in amyloid assembly pathways [[39], [40], [41],123,135] (Fig. 1E), produce low-resolution structural models of specific intermediates [144,145,148], and infer the role of different oligomers in amyloid assembly and toxicity [120,130].
The primary limitation of methods which detect single particles or populations is their structural resolution. Unlike NMR, these techniques do not readily reveal residue- or atom-level information about amyloid assembly, and thus cannot facilitate structure-based ligand design approaches. We note, however, that the recent revolution in cryo-electron microscopy (cryo-EM), including advances in image processing algorithms, has allowed individual species within heterogeneous samples to be studied at high resolution [149]. While cryo-EM has already played a key role in the elucidation of fibril structures at near-atomic resolution [150], there are few examples of its application to amyloid oligomers [8]. This technique nonetheless has the potential to be used in the future to reveal high-resolution structural information for oligomeric samples at a “single species” level.
NMR and single particle methods remain invaluable for detecting individual oligomer populations and elucidating amyloid pathways. These approaches, in combination with methods which allow access to higher-resolution information (discussed below), facilitate progress towards more refined descriptions of amyloid assembly pathways.
3. Trapping transient oligomers to facilitate the characterization of amyloid self-assembly
Obtaining high-resolution structural and functional insights into specific amyloid oligomers typically requires the use of samples which predominantly contain a single species. While it is possible to reduce the heterogeneity of oligomer samples through size separation approaches [151], the degree of sample homogeneity achievable through this method is limited, as the self-assembly landscape may start to rapidly re-equilibrate after isolation of individual species. Careful control of sample preparation conditions (e.g. the availability of air-water interfaces, or buffer composition and pH) can bias self-assembly landscapes towards specific oligomeric states (Section 3.1), but the resulting oligomer distributions are often broad. Furthermore, such an approach does not address the experimental challenges which remain concerning how to trap and kinetically/structurally characterize amyloid intermediates in an in vivo setting. Four primary tools have emerged which can favor the production of narrow distributions of oligomers, or sometimes even a specific oligomer state, without requiring specific buffer conditions: oligomer-binding antibodies (Section 3.2), non-covalent small molecule ligands (Section 3.3), covalent ligands or protein modifications (Section 3.4), and crosslinking (Section 3.5).
3.1. Sample preparation strategies
For certain amyloid proteins, different sample preparation strategies have been established which have been shown to favor specific oligomer distributions. Lyophilization, followed by resuspension and incubation at high protein concentrations, has been used to promote the formation of kinetically-trapped α-synuclein oligomers that can then be further enriched by centrifugation, size-exclusion chromatography, or other size separation methods [152]. While oligomer distributions produced by this approach are still broad (predominantly 10-40mers, although species up to 90mers have been detected) [8,153,154], the enrichment of oligomers in these samples has nonetheless facilitated their study via a range of biophysical techniques [8,136,138,[153], [154], [155], [156], [157], [158]]. Notably, lyophilization has been used to produce α-synuclein samples for cryo-EM, leading to two low-resolution (18–19 Å) reconstructions of toxic cylindrical oligomers [8], and solid-state NMR, where structural properties of these same oligomers were compared with those of non-toxic, small molecule-stabilized oligomers [159] to understand the structural determinants of oligomer toxicity [64]. Similarly, samples prepared through incubation of amyloid proteins in carefully-selected buffers have allowed structure-toxicity relationships to be uncovered for oligomers formed by the N-terminal domain of the Escherichia coli HypF protein (prepared in additive-containing solutions) [160,161] and Aβ peptides (prepared in low salt buffers at low temperature [162,163] or in the presence of detergent micelles [26,164]), amongst others. In general, such studies have highlighted that structural features which promote promiscuous interactions with other cellular components (e.g. enhanced surface hydrophobicity or the presence of structured elements which can insert into membranes) are often associated with toxicity [26,64,165,166], while the proportion of β-sheet structure does not generally appear to play a clear role [167].
3.2. Using antibodies to probe the structure and toxicity of oligomers
An emerging therapeutic avenue for the treatment of amyloid diseases is the use of antibodies to capture and neutralize toxic oligomers, due to the ability of these proteins to bind target epitopes with high specificity and affinity [168,169]. Such molecular recognition properties also make antibodies ideal tools for stabilizing specific, transient amyloid intermediates for detailed study [170,171] and for detecting the presence of particular structural features [172]. The A11 antibody, in particular, has played an important role in the amyloid field [6]. Although A11 was raised against an oligomeric mimic of Aβ40 (in the form of gold nanoparticles coated with Aβ40 peptides), this polyclonal antibody recognizes pre-fibrillar, toxic oligomers formed by a range of amyloid proteins with diverse primary sequences and native folds, suggesting that toxicity is associated with a common set of structural features [6]. Understanding precisely what these structural features are, how much they vary between different amyloid proteins, and which oligomeric species within a given self-assembly pathway possess these characteristics is still not understood.
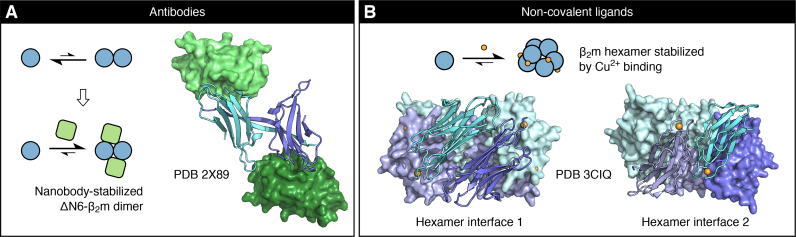
In addition to antibodies which recognize specific structural features in a sequence-independent manner [6], antibodies (and antibody fragments) which bind to, and stabilize, specific oligomers formed by different amyloid proteins have also been developed: this includes antibodies which target oligomers of α-synuclein (SDS-stable dimers/tetramers [173] and SDS-stable trimers/hexamers [174]), Aβ (< 70 kDa oligomers [175,176] and larger aggregates [177]), and a truncated β2m variant (dimers [28]; Fig. 2A). Antibodies have also been developed that interfere with specific stages of amyloid assembly for particular proteins [178]. To guarantee specificity for amyloid oligomers (over monomers or fibrils), antibodies are often generated by screening against samples which have been enriched in oligomers [173,174,177], by screening against cyclic peptides which mimic predicted oligomer epitopes [175,176], by grafting amyloidogenic peptide fragments into the complementary-determining regions of canonical antibody scaffolds [179,180], or through rational in silico design methods [178,181]. The ability of antibodies generated using these approaches to bind oligomers in cell models or in ex vivo samples highlights the physiological- and/or disease-relevance of these antibody-stabilized structures, and in some cases has allowed the roles of specific oligomer sub-classes in disease to be interrogated [[174], [175], [176], [177], [178],181,182]. Oligomer-binding antibodies have also been used to facilitate structural studies of amyloid intermediates – this has been demonstrated by a nanobody-stabilized dimer formed by a truncated β2m variant (ΔN6-β2m; Fig. 2A) [28] and a tetrameric fusion between an antibody and an Aβ peptide fragment [183], both of which yielded high-resolution crystal structures. Thus, in contrast with other approaches discussed thus far in this review, antibody-oligomer complexes have the potential to provide high-resolution insights into the conformational changes which an amyloid protein undergoes during the initiation of self-assembly. Where it is possible to assess the toxicity of such stabilized oligomer complexes, structural features that are associated with disease can be inferred.
Fig. 2.
Non-covalent strategies for the stabilization of particular oligomer populations, exemplified by nanobody- and metal ion-stabilized β2m oligomers. A: Crystal structure of a dimer of a truncated β2m variant (ΔN6-β2m) stabilized by a nanobody (green) [28]. B: Crystal structure of a Cu2+-stabilized hexamer of the H13F β2m variant. The Cu2+ ions are colored in orange [29]. In both A and B, β2m protomers are shown in various shades of blue. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.3. Stabilizing oligomers using non-covalent small molecules
Non-covalent small molecules offer an alternative approach to capture amyloidogenic intermediates. In a similar manner to antibodies [178], such ligands can be invaluable tools to interfere with distinct microscopic events in amyloid self-assembly [[184], [185], [186], [187]], although identification of such compounds is complicated by the poor ligandability of most monomeric amyloid precursors (which are commonly intrinsically disordered or partially folded). The discovery of non-covalent ligands specifically for the oligomeric forms of amyloid proteins presents additional challenges, as protein-protein interactions are inherently difficult to modulate with small molecules [188]. While small molecules have been identified that stabilize natively oligomeric amyloid precursors – e.g. the functional tetramers of transthyretin (the aggregation of which results in familial amyloid polyneuropathy) and immunoglobulin light chain dimers (involved in light chain amyloidosis) [[189], [190], [191], [192]] – the identification of compounds that bind specifically to non-native, oligomeric assembly intermediates is far more challenging: less structural information is available for non-native intermediates to guide ligand design and it can be difficult to detect oligomer-specific interactions due to the low population of these species. Screening methods [193] that are well-suited to the identification of oligomer-binding compounds include kinetics approaches, which can identify the microscopic events that are modulated by a given compound [187], and ESI-MS (sometimes also coupled with IMS), which is sufficiently sensitive to detect lowly-populated intermediates and can identify the oligomeric state of the ligand-bound species [194].
In addition to compounds which are identified through screening, small molecules and other ligands which are thought to modulate amyloid assembly in vivo can be used in vitro to gain insight into assembly pathways. In particular, several metal ions with known or proposed roles in amyloid disease have been shown to promote the formation of amyloid oligomers [[195], [196], [197]]. A striking example of this was demonstrated by Calabrese et al., where Cu2+-mediated stabilization of a β2m variant hexamer allowed a high-resolution structure to be obtained of this oligomer by X-ray crystallography, showing a ring-like arrangement of protomers with a native-like fold (Fig. 2B) [29].
For natively disordered amyloid proteins, obtaining discrete oligomer populations through non-covalent binding is immensely challenging, as small molecule or metal ion binding tends to result in a distribution of oligomers [159,[198], [199], [200], [201], [202], [203]]. Nonetheless, through bulk structural measurements in combination with assays in cells, structure-toxicity relationships have been obtained for various natively disordered amyloid proteins, including α-synuclein [159,202,203] and Aβ peptides [159,200,201,204,205].
3.4. Covalent ligands and protein functionalization
In recent years, covalent modification has become an increasingly utilized tool for the modulation of protein-protein interactions [[206], [207], [208], [209], [210], [211], [212], [213], [214], [215]]. A covalently bound small molecule may exert an effect on a protein or protein complex either via (a) non-covalent interactions (which have been reinforced by the covalent bond), or (b) by modifying the protein's surface properties or topography. We will refer to these two categories of modifications as “covalent ligands” and “protein functionalization”, respectively. Covalent ligands can offer improved affinity and selectivity over their non-covalent analogues [213,216,217], thereby overcoming some of the difficulties associated with using small molecules to target the dynamic and poorly structured oligomers formed by many amyloid proteins. Modification of a protein's behavior through functionalization does not require the conjugated chemical moiety to have a high non-covalent affinity for the target; it can instead effectively act as a chemical post-translational modification [218,219], altering the protein's surface properties [27,220] or acting as a steric block [30]. We also note that sequence variation can be considered as a special case of protein functionalization which alters the chemistry and/or steric bulk of protein sidechains without the need for chemical modifications to be performed post-translationally. The effect of sequence variation on the thermodynamic stability of amyloid precursors and oligomers, and on the kinetics of protein self-assembly has been extensively studied and reviewed elsewhere [29,[221], [222], [223], [224], [225], [226], [227], [228]].
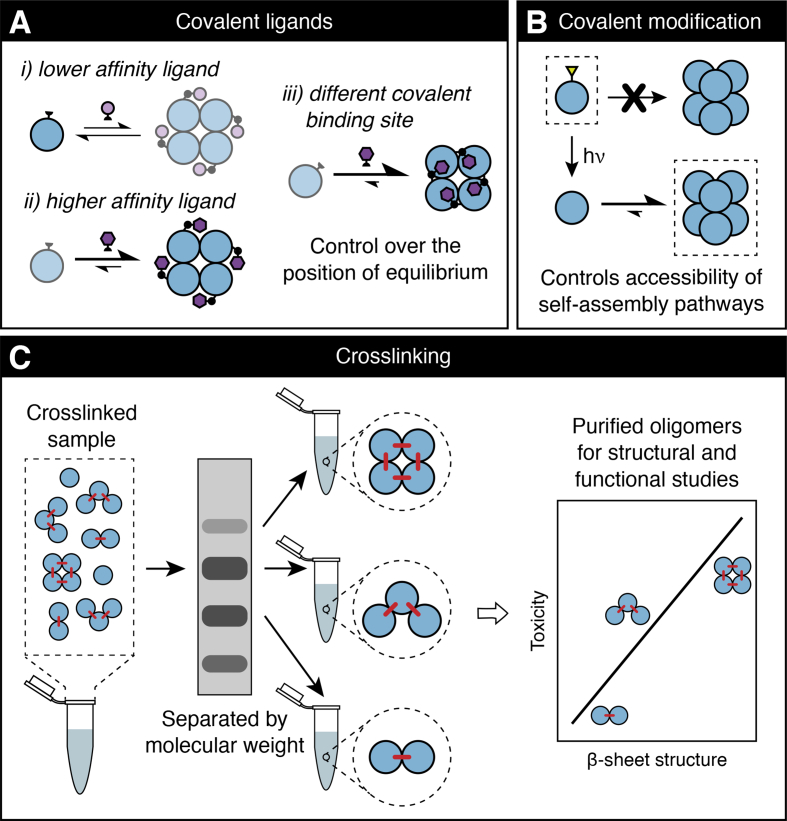
Both categories of covalent modification have shown great promise in dissecting the structure and toxicity of discrete oligomeric species in amyloid assembly, as has recently been demonstrated by a panel of small molecules used to stabilize a tetramer formed by the ΔN6 variant of β2m [27]. These compounds, which were identified through disulfide tethering [229,230], were found to be capable of modulating ΔN6-β2m oligomer populations, primarily by acting as covalent ligands. A series of samples was produced where the tetramer population could be varied (from ca. 5–95%) based on the site of covalent modification and the identity of the small molecule. These samples were used to determine a high-resolution structure of ΔN6-β2m tetramers and demonstrate their role in inhibiting amyloid fibril formation, through the use of X-ray crystallography, solution NMR, and functional assays (Fig. 3A) [27]. The ability of such covalent small molecules to generate specific oligomer populations in a controlled manner makes this a promising platform to study oligomer structures and structure-toxicity relationships for other amyloid proteins.
Fig. 3.
Covalent strategies for the control of oligomer populations in amyloid protein samples. A: Covalent ligands have been shown to be powerful modulators of oligomerization equilibria and can allow oligomer distributions to be tuned based on the affinity and location of the protein-ligand interaction [27]. B: The use of photolabile groups (e.g. N-2-nitrobenzyl) to sterically or chemically prevent amyloid proteins and peptides from accessing particular regions of the self-assembly landscape can allow oligomer populations to be controlled in a UV-dependent manner [30]. C: Covalent crosslinking using the PICUP or diazirine-based approaches represents a means of trapping a distribution of oligomers formed by a self-assembling protein, and the resulting crosslinked species can be separated (e.g. by SDS-PAGE, or using other, higher-throughput size/mass separation approaches) for individual structure and toxicity studies [[239], [240], [241]].
While the strategies discussed above have focused on methods to stabilize particular oligomeric states, sample homogeneity can also be improved by destabilizing or preventing the formation of other species in an amyloid assembly pathway. For example, the formation of inter-peptide backbone hydrogen bonds is essential for fibril formation [231] and so blocking the ability of the backbone to form these non-covalent interactions provides a means to prevent a protein from accessing certain regions of the self-assembly energy landscape. Chemical or steric blocking of backbone hydrogen bonds can be achieved through the incorporation of certain backbone mimics, such as the tripeptide β-strand mimic “Hao” [232] or through amide N-alkylation [233]. These approaches have been shown to restrict the self-assembly of a range of model amyloid peptides, including those derived from Aβ [30,[234], [235], [236]], tau (involved in Alzheimer's disease and other neurodegenerative disorders) [234], β2m [237], and islet amyloid polypeptide (IAPP; associated with type II diabetes) [238]. Recently, it has also been demonstrated that such covalent modifications can be applied in a reversible manner to tune oligomer populations with temporal control. The use of photolabile N-2-nitrobenzyl groups in place of (or in combination with) N-alkylation, for example, can allow precise control over amyloid oligomer populations, and hence cytotoxicity, of model Aβ peptides (Fig. 3B) [30]. The application of this approach to other model peptides and full-length amyloid proteins could thus drive novel insights into the role of different assembly intermediates and structural features in amyloid disease.
3.5. Crosslinking strategies
Crosslinking reactions which are rapid and indiscriminate in their amino acid preferences offer promising opportunities for capturing snapshots of transient oligomers formed by amyloidogenic proteins or peptides [242]. Such reaction characteristics are partially or fully exhibited by both the photo-induced crosslinking of unmodified proteins (PICUP) approach [243] and diazirine-based photochemical crosslinking [244]. The enhancement of oligomer stability resulting from covalent crosslinking renders the assembly intermediates within such samples amenable to purification by size separation approaches, allowing pure oligomer samples to be prepared for detailed structural and functional analysis [239,240,245] (Fig. 3C).
PICUP is a rapid, radical-based crosslinking method which can be used without the need for prior functionalization of the target protein with photoreactive groups [246]. Instead, metal complexes (typically tris-bipyridyl ruthenium(II)) are oxidized in the presence of visible light and subsequently abstract single electrons from amino acid sidechains (commonly tyrosine, cysteine, tryptophan, and methionine), rendering them capable of forming covalent bonds with suitable proximal residues [247]. PICUP has become an established method for the stabilization and study of amyloid oligomers [240,[248], [249], [250]], and the purification of crosslinked samples to yield individual oligomers has been demonstrated using a sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) extraction method [239,240] (Fig. 3C). The ability to obtain individual samples of well-defined (by mass, if not conformation), stabilized oligomers through PICUP has allowed the structure and toxicity of different Aβ oligomers to be probed. Ono et al. successfully isolated crosslinked Aβ40 dimers, trimers, and tetramers (all with at least 94% purity) and were able to individually characterize the secondary structure, amyloidogenicity, and cytotoxicity of each species, ultimately finding a correlation between all these variables [239]. With some modifications [251], the PICUP crosslinking approach, with subsequent oligomer purification, has also been applied to Aβ42 [240].
Despite the advantages of PICUP crosslinking chemistry, the requirement for an amino acid sidechain radical to encounter a radical scavenger or a readily oxidizable sidechain means that intermolecular crosslinking between protein molecules can occur through diffusion-controlled collision events, independently of genuine protein-protein interactions [248]. Such events can be avoided through the use of diazirine-based crosslinking, as the carbenes which form upon irradiation of diazirine groups with UV light have nanosecond lifetimes and are rapidly quenched by reaction with solvent if no crosslinkable residues are in proximity [252]. Although diazirine groups have to be introduced into the target protein or peptide by synthesis [25,241,253], covalent modification (e.g. via cysteine using methanethiosulfonate-functionalized diazirines [254]), or the use of unnatural amino acids during protein expression [255,256], the advantages offered by the short lifetime of the carbene, the high yield of potential crosslinks formed, and the rapidity of their formation when using LED illumination (reducing irradiation times from minutes or hours to seconds) [254] make diazirines attractive tools for capturing transient amyloidogenic interactions [25,241,244,253].
In theory, protein photo-crosslinking is well-suited to trap disease-relevant oligomers in cells or in vivo, as well as to identify the cellular components with which these oligomers interact. However, in practice, such an approach presents many challenges, notably the potential for very low signal-to-noise ratios due to the vast number of species within cells to which an amyloid protein can crosslink, as well as the complexities associated with subsequent data analysis. The use of crosslinking reagents with extremely short lifetimes and which offer the capacity for precise temporal control, such as diazirines, in combination with a method for enrichment of the crosslinked species (e.g. through affinity tags or alkyne-functionalized crosslinkers) addresses some of these challenges. Such an approach, in combination with quantitative proteomics has already been shown to be capable of identifying low-affinity, non-amyloidogenic protein-protein interactions in living cells [257]. Photo-crosslinking thus holds great potential for the study of amyloid assembly in physiologically- and disease-relevant systems.
When attempting oligomer-trapping methods, and particularly crosslinking strategies, it is important to keep in mind that while oligomer stabilization can dramatically improve sample homogeneity and offers advantages for structural characterization, the metastable and dynamic nature of amyloid intermediates can also be an important characteristic, particularly when considering oligomer toxicity. Oligomer dynamics, including protomer dissociation and exchange, can play important roles in toxicity [64,258]. While some crosslinked oligomers have been observed to undergo detectable dynamic motions [250], in other cases, crosslinking has been observed to suppress dissociation events which would otherwise contribute towards cell death [259]. It is therefore vital to employ a range of approaches to unpick the nature and potential cytotoxicity of different oligomers during amyloid assembly, keeping in mind that methods which are best suited to generating samples for high-resolution structure elucidation and those best suited for assessing oligomer toxicity are not necessarily the same.
4. Conclusion
Amyloid assembly intermediates are intimately involved in amyloid diseases, often representing key cytotoxic species which interact aberrantly with each other and other cellular components [11]. While the challenges presented by the transient nature, dynamics, and heterogeneity of these oligomeric intermediates have hindered structural and functional studies, continual advances in biophysical methods and chemical tools have allowed increasingly detailed insights to be gained. In this review, we have explored the role that NMR and single particle/species methods can play in gaining kinetic and low-resolution structural descriptions of lowly populated and transient oligomeric intermediates. In addition, strategies which allow specific oligomer populations to be stabilized (e.g. through the use of antibodies, small molecule ligands, or chemical crosslinking) can facilitate higher-resolution structural studies and investigation of detailed structure-toxicity relationships. It is important, however, to keep a balance between stabilizing a sample sufficiently to make it amenable to analysis through the desired methods and avoiding tipping the oligomerization equilibria to biologically- or disease-irrelevant species. The different strengths and caveats of the aforementioned methods make all these techniques complementary and emphasize the need to employ a wide range of integrated methods and tools from chemistry, biophysics, structural biology, and cellular biology to gain a complete description of these disease-relevant self-assembly pathways and to inform the rational design of much-needed treatments.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgements
We thank members of our laboratories for helpful discussions. This work was supported by The Wellcome Trust (109154/Z/15/Z and 204963), the EPSRC (EP/N035267/1, EP/N013573/1) and the ERC (322408). A.J.W. holds a Royal Society Leverhulme Trust Senior Fellowship (SRF/R1/191087).
Author contributions
EEC and TKK wrote the first draft. All authors curated information, reviewed, and edited the manuscript.
Contributor Information
Andrew J. Wilson, Email: A.J.Wilson@leeds.ac.uk.
Sheena E. Radford, Email: S.E.Radford@leeds.ac.uk.
References
- 1.Chiti F., Dobson C.M. Protein misfolding, amyloid formation, and human disease: a summary of progress over the last decade. Annu. Rev. Biochem. 2017;86:27–68. doi: 10.1146/annurev-biochem-061516-045115. [DOI] [PubMed] [Google Scholar]
- 2.Iadanza M.G., Jackson M.P., Hewitt E.W., Ranson N.A., Radford S.E. A new era for understanding amyloid structures and disease. Nat. Rev. Mol. Cell Biol. 2018;19:755–773. doi: 10.1038/s41580-018-0060-8. [DOI] [PubMed] [Google Scholar]
- 3.Jackson M., Hewitt E. Why are functional amyloids non-toxic in humans? Biomolecules. 2017;7:71. doi: 10.3390/biom7040071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ulamec S.M., Radford S.E. Spot the difference: function versus toxicity in amyloid fibrils. Trends Biochem. Sci. 2020;45:635–636. doi: 10.1016/j.tibs.2020.04.007. [DOI] [PubMed] [Google Scholar]
- 5.Bucciantini M., Giannoni E., Chiti F., Baroni F., Formigli L., Zurdo J., Taddei N., Ramponi G., Dobson C.M., Stefani M. Inherent toxicity of aggregates implies a common mechanism for protein misfolding diseases. Nature. 2002;416:507–511. doi: 10.1038/416507a. [DOI] [PubMed] [Google Scholar]
- 6.Kayed R., Head E., Thompson J.L., McIntire T.M., Milton S.C., Cotman C.W., Glabe C.G. Common structure of soluble amyloid oligomers implies common mechanism of pathogenesis. Science. 2003;300:486–489. doi: 10.1126/science.1079469. [DOI] [PubMed] [Google Scholar]
- 7.Winner B., Jappelli R., Maji S.K., Desplats P.A., Boyer L., Aigner S., Hetzer C., Loher T., Vilar M., Campioni S., Tzitzilonis C., Soragni A., Jessberger S., Mira H., Consiglio A., Pham E., Masliah E., Gage F.H., Riek R. In vivo demonstration that α-synuclein oligomers are toxic. Proc. Natl. Acad. Sci. U. S. A. 2011;108:4194–4199. doi: 10.1073/pnas.1100976108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen S.W., Drakulic S., Deas E., Ouberai M., Aprile F.A., Arranz R., Ness S., Roodveldt C., Guilliams T., De-Genst E.J., Klenerman D., Wood N.W., Knowles T.P.J., Alfonso C., Rivas G., Abramov A.Y., Valpuesta J.M., Dobson C.M., Cremades N. Structural characterization of toxic oligomers that are kinetically trapped during α-synuclein fibril formation. Proc. Natl. Acad. Sci. U. S. A. 2015;112:E1994–E2003. doi: 10.1073/pnas.1421204112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abedini A., Plesner A., Cao P., Ridgway Z., Zhang J., Tu L.-H., Middleton C.T., Chao B., Sartori D.J., Meng F., Wang H., Wong A.G., Zanni M.T., Verchere C.B., Raleigh D.P., Schmidt A.M. Time-resolved studies define the nature of toxic IAPP intermediates, providing insight for anti-amyloidosis therapeutics. Elife. 2016;5 doi: 10.7554/eLife.12977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cremades N., Cohen S.I.A., Deas E., Abramov A.Y., Chen A.Y., Orte A., Sandal M., Clarke R.W., Dunne P., Aprile F.A., Bertoncini C.W., Wood N.W., Knowles T.P.J., Dobson C.M., Klenerman D. Direct observation of the interconversion of normal and toxic forms of α-synuclein. Cell. 2012;149:1048–1059. doi: 10.1016/j.cell.2012.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cremades N., Dobson C.M. The contribution of biophysical and structural studies of protein self-assembly to the design of therapeutic strategies for amyloid diseases. Neurobiol. Dis. 2018;109:178–190. doi: 10.1016/j.nbd.2017.07.009. [DOI] [PubMed] [Google Scholar]
- 12.Breydo L., Uversky V.N. Structural, morphological, and functional diversity of amyloid oligomers. FEBS Lett. 2015;589:2640–2648. doi: 10.1016/j.febslet.2015.07.013. [DOI] [PubMed] [Google Scholar]
- 13.Nagel-Steger L., Owen M.C., Strodel B. An account of amyloid oligomers: facts and figures obtained from experiments and simulations. ChemBioChem. 2016;17:657–676. doi: 10.1002/cbic.201500623. [DOI] [PubMed] [Google Scholar]
- 14.Lerner E., Cordes T., Ingargiola A., Alhadid Y., Chung S., Michalet X., Weiss S. Toward dynamic structural biology: two decades of single-molecule Förster resonance energy transfer. Science. 2018;359 doi: 10.1126/science.aan1133. eaan1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schuler B., Soranno A., Hofmann H., Nettels D. Single-molecule FRET spectroscopy and the polymer physics of unfolded and intrinsically disordered proteins. Annu. Rev. Biophys. 2016;45:207–231. doi: 10.1146/annurev-biophys-062215-010915. [DOI] [PubMed] [Google Scholar]
- 16.Simone Ruggeri F., Habchi J., Cerreta A., Dietler G. AFM-based single molecule techniques: unraveling the amyloid pathogenic species. Curr. Pharm. Des. 2016;22:3950–3970. doi: 10.2174/1381612822666160518141911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Woods L.A., Radford S.E., Ashcroft A.E. Advances in ion mobility spectrometry–mass spectrometry reveal key insights into amyloid assembly. Biochim. Biophys. Acta, Proteins Proteomics. 2013;1834:1257–1268. doi: 10.1016/j.bbapap.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu J., Zheng Q. Applications of mass spectrometry in the onset of amyloid fibril formation: focus on the analysis of early-stage oligomers. Front. Chem. 2020;8:324. doi: 10.3389/fchem.2020.00324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoffmann W., von Helden G., Pagel K. Ion mobility-mass spectrometry and orthogonal gas-phase techniques to study amyloid formation and inhibition. Curr. Opin. Struct. Biol. 2017;46:7–15. doi: 10.1016/j.sbi.2017.03.002. [DOI] [PubMed] [Google Scholar]
- 20.Mitra G. Application of native mass spectrometry in studying intrinsically disordered proteins: a special focus on neurodegenerative diseases. Biochim. Biophys. Acta, Proteins Proteomics. 2019;1867:140260. doi: 10.1016/j.bbapap.2019.07.013. [DOI] [PubMed] [Google Scholar]
- 21.Alderson T.R., Kay L.E. Unveiling invisible protein states with NMR spectroscopy. Curr. Opin. Struct. Biol. 2020;60:39–49. doi: 10.1016/j.sbi.2019.10.008. [DOI] [PubMed] [Google Scholar]
- 22.Strodel B., Lee J.W.L., Whittleston C.S., Wales D.J. Transmembrane structures for Alzheimer’s Aβ1−42 oligomers. J. Am. Chem. Soc. 2010;132:13300–13312. doi: 10.1021/ja103725c. [DOI] [PubMed] [Google Scholar]
- 23.Barz B., Olubiyi O.O., Strodel B. Early amyloid β-protein aggregation precedes conformational change. Chem. Commun. 2014;50:5373–5375. doi: 10.1039/C3CC48704K. [DOI] [PubMed] [Google Scholar]
- 24.Zheng W., Tsai M.-Y., Chen M., Wolynes P.G. Exploring the aggregation free energy landscape of the amyloid-β protein (1–40) Proc. Natl. Acad. Sci. U. S. A. 2016;113:11835–11840. doi: 10.1073/pnas.1612362113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bunce S.J., Wang Y., Stewart K.L., Ashcroft A.E., Radford S.E., Hall C.K., Wilson A.J. Molecular insights into the surface-catalyzed secondary nucleation of amyloid-β40 (Aβ40) by the peptide fragment Aβ16–22. Sci. Adv. 2019;5 doi: 10.1126/sciadv.aav8216. eaav8216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ciudad S., Puig E., Botzanowski T., Meigooni M., Arango A.S., Do J., Mayzel M., Bayoumi M., Chaignepain S., Maglia G., Cianferani S., Orekhov V., Tajkhorshid E., Bardiaux B., Carulla N. Aβ(1-42) tetramer and octamer structures reveal edge conductivity pores as a mechanism for membrane damage. Nat. Commun. 2020;11:3014. doi: 10.1038/s41467-020-16566-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cawood E.E., Guthertz N., Ebo J.S., Karamanos T.K., Radford S.E., Wilson A.J. Modulation of amyloidogenic protein self-assembly using tethered small molecules. J. Am. Chem. Soc. 2020 doi: 10.1021/jacs.0c10629. (In press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Domanska K., Vanderhaegen S., Srinivasan V., Pardon E., Dupeux F., Marquez J.A., Giorgetti S., Stoppini M., Wyns L., Bellotti V., Steyaert J. Atomic structure of a nanobody-trapped domain-swapped dimer of an amyloidogenic β2-microglobulin variant. Proc. Natl. Acad. Sci. U. S. A. 2011;108:1314–1319. doi: 10.1073/pnas.1008560108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Calabrese M.F., Eakin C.M., Wang J.M., Miranker A.D. A regulatable switch mediates self-association in an immunoglobulin fold. Nat. Struct. Mol. Biol. 2008;15:965–971. doi: 10.1038/nsmb.1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Salveson P.J., Haerianardakani S., Thuy-Boun A., Kreutzer A.G., Nowick J.S. Controlling the oligomerization state of Aβ-derived peptides with light. J. Am. Chem. Soc. 2018;140:5842–5852. doi: 10.1021/jacs.8b02658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kotler S.A., Tugarinov V., Schmidt T., Ceccon A., Libich D.S., Ghirlando R., Schwieters C.D., Clore G.M. Probing initial transient oligomerization events facilitating Huntingtin fibril nucleation at atomic resolution by relaxation-based NMR. Proc. Natl. Acad. Sci. U. S. A. 2019;116:3562–3571. doi: 10.1073/pnas.1821216116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fawzi N.L., Ying J., Ghirlando R., Torchia D.A., Clore G.M. Atomic-resolution dynamics on the surface of amyloid-β protofibrils probed by solution NMR. Nature. 2011;480:268–272. doi: 10.1038/nature10577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yuwen T., Brady J.P., Kay L.E. Probing conformational exchange in weakly interacting, slowly exchanging protein systems via off-resonance R1ρ experiments: application to studies of protein phase separation. J. Am. Chem. Soc. 2018;140:2115–2126. doi: 10.1021/jacs.7b09576. [DOI] [PubMed] [Google Scholar]
- 34.Barnes C.A., Robertson A.J., Louis J.M., Anfinrud P., Bax A. Observation of β-amyloid peptide oligomerization by pressure-jump NMR spectroscopy. J. Am. Chem. Soc. 2019;141:13762–13766. doi: 10.1021/jacs.9b06970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Karamanos T.K., Jackson M.P., Calabrese A.N., Goodchild S.C., Cawood E.E., Thompson G.S., Kalverda A.P., Hewitt E.W., Radford S.E. Structural mapping of oligomeric intermediates in an amyloid assembly pathway. Elife. 2019;8 doi: 10.7554/eLife.46574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shammas S.L., Garcia G.A., Kumar S., Kjaergaard M., Horrocks M.H., Shivji N., Mandelkow E., Knowles T.P.J., Mandelkow E., Klenerman D. A mechanistic model of tau amyloid aggregation based on direct observation of oligomers. Nat. Commun. 2015;6:7025. doi: 10.1038/ncomms8025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kjaergaard M., Dear A.J., Kundel F., Qamar S., Meisl G., Knowles T.P.J., Klenerman D. Oligomer diversity during the aggregation of the repeat region of tau. ACS Chem. Neurosci. 2018;9:3060–3071. doi: 10.1021/acschemneuro.8b00250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ruggeri F.S., Longo G., Faggiano S., Lipiec E., Pastore A., Dietler G. Infrared nanospectroscopy characterization of oligomeric and fibrillar aggregates during amyloid formation. Nat. Commun. 2015;6:7831. doi: 10.1038/ncomms8831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seo J., Hoffmann W., Warnke S., Huang X., Gewinner S., Schöllkopf W., Bowers M.T., von Helden G., Pagel K. An infrared spectroscopy approach to follow β-sheet formation in peptide amyloid assemblies. Nat. Chem. 2017;9:39–44. doi: 10.1038/nchem.2615. [DOI] [PubMed] [Google Scholar]
- 40.Hoffmann W., Folmert K., Moschner J., Huang X., von Berlepsch H., Koksch B., Bowers M.T., von Helden G., Pagel K. NFGAIL amyloid oligomers: the onset of beta-sheet formation and the mechanism for fibril formation. J. Am. Chem. Soc. 2018;140:244–249. doi: 10.1021/jacs.7b09510. [DOI] [PubMed] [Google Scholar]
- 41.Bleiholder C., Dupuis N.F., Wyttenbach T., Bowers M.T. Ion mobility–mass spectrometry reveals a conformational conversion from random assembly to β-sheet in amyloid fibril formation. Nat. Chem. 2011;3:172–177. doi: 10.1038/nchem.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Karamanos T.K., Kalverda A.P., Thompson G.S., Radford S.E. Mechanisms of amyloid formation revealed by solution NMR. Prog. Nucl. Magn. Reson. Spectrosc. 2015;88–89:86–104. doi: 10.1016/j.pnmrs.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sekhar A., Kay L.E. An NMR view of protein dynamics in health and disease. Annu. Rev. Biophys. 2019;48:297–319. doi: 10.1146/annurev-biophys-052118-115647. [DOI] [PubMed] [Google Scholar]
- 44.Anthis N.J., Clore G.M. Visualizing transient dark states by NMR spectroscopy. Q. Rev. Biophys. 2015;48:35–116. doi: 10.1017/S0033583514000122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rennella E., Morgan G.J., Kelly J.W., Kay L.E. Role of domain interactions in the aggregation of full-length immunoglobulin light chains. Proc. Natl. Acad. Sci. U. S. A. 2019;116:854–863. doi: 10.1073/pnas.1817538116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Culik R.M., Sekhar A., Nagesh J., Deol H., Rumfeldt J.A.O., Meiering E.M., Kay L.E. Effects of maturation on the conformational free-energy landscape of SOD1. Proc. Natl. Acad. Sci. U. S. A. 2018;115:E2546–E2555. doi: 10.1073/pnas.1721022115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sekhar A., Rumfeldt J.A.O., Broom H.R., Doyle C.M., Sobering R.E., Meiering E.M., Kay L.E. Probing the free energy landscapes of ALS disease mutants of SOD1 by NMR spectroscopy. Proc. Natl. Acad. Sci. U. S. A. 2016;113:E6939–E6945. doi: 10.1073/pnas.1611418113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fusco G., Pape T., Stephens A.D., Mahou P., Costa A.R., Kaminski C.F., Kaminski Schierle G.S., Vendruscolo M., Veglia G., Dobson C.M., De Simone A. Structural basis of synaptic vesicle assembly promoted by α-synuclein. Nat. Commun. 2016;7:12563. doi: 10.1038/ncomms12563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fawzi N.L., Doucleff M., Suh J.-Y., Clore G.M. Mechanistic details of a protein-protein association pathway revealed by paramagnetic relaxation enhancement titration measurements. Proc. Natl. Acad. Sci. U. S. A. 2010;107:1379–1384. doi: 10.1073/pnas.0909370107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sekhar A., Rumfeldt J.A.O., Broom H.R., Doyle C.M., Bouvignies G., Meiering E.M., Kay L.E. Thermal fluctuations of immature SOD1 lead to separate folding and misfolding pathways. Elife. 2015;4 doi: 10.7554/eLife.07296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rennella E., Cutuil T., Schanda P., Ayala I., Gabel F., Forge V., Corazza A., Esposito G., Brutscher B. Oligomeric states along the folding pathways of β2-microglobulin: kinetics, thermodynamics, and structure. J. Mol. Biol. 2013;425:2722–2736. doi: 10.1016/j.jmb.2013.04.028. [DOI] [PubMed] [Google Scholar]
- 52.Karamanos T.K., Kalverda A.P., Thompson G.S., Radford S.E. Visualization of transient protein-protein interactions that promote or inhibit amyloid assembly. Mol. Cell. 2014;55:214–226. doi: 10.1016/j.molcel.2014.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Janowska M.K., Wu K.-P., Baum J. Unveiling transient protein-protein interactions that modulate inhibition of alpha-synuclein aggregation by beta-synuclein, a pre-synaptic protein that co-localizes with alpha-synuclein. Sci. Rep. 2015;5:15164. doi: 10.1038/srep15164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu K.-P., Baum J. Detection of transient interchain interactions in the intrinsically disordered protein α-synuclein by NMR paramagnetic relaxation enhancement. J. Am. Chem. Soc. 2010;132:5546–5547. doi: 10.1021/ja9105495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nilges M., Clore G.M., Gronenborn A.M. Determination of three-dimensional structures of proteins from interproton distance data by dynamical simulated annealing from a random array of atoms. FEBS Lett. 1988;239:129–136. doi: 10.1016/0014-5793(88)80559-3. [DOI] [PubMed] [Google Scholar]
- 56.Gurry T., Ullman O., Fisher C.K., Perovic I., Pochapsky T., Stultz C.M. The dynamic structure of α-synuclein multimers. J. Am. Chem. Soc. 2013;135:3865–3872. doi: 10.1021/ja310518p. [DOI] [PubMed] [Google Scholar]
- 57.Clore G.M., Iwahara J. Theory, practice, and applications of paramagnetic relaxation enhancement for the characterization of transient low-population states of biological macromolecules and their complexes. Chem. Rev. 2009;109:4108–4139. doi: 10.1021/cr900033p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tang C., Iwahara J., Clore G.M. Visualization of transient encounter complexes in protein–protein association. Nature. 2006;444:383–386. doi: 10.1038/nature05201. [DOI] [PubMed] [Google Scholar]
- 59.Rosenzweig R., Kay L.E. Bringing dynamic molecular machines into focus by methyl-TROSY NMR. Annu. Rev. Biochem. 2014;83:291–315. doi: 10.1146/annurev-biochem-060713-035829. [DOI] [PubMed] [Google Scholar]
- 60.Tycko R., Wickner R.B. Molecularstructures of amyloid and prion fibrils: consensus versus controversy. Acc. Chem. Res. 2013;46:1487–1496. doi: 10.1021/ar300282r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wu J.W., Breydo L., Isas J.M., Lee J., Kuznetsov Y.G., Langen R., Glabe C. Fibrillar oligomers nucleate the oligomerization of monomeric amyloid β but do not seed fibril formation. J. Biol. Chem. 2010;285:6071–6079. doi: 10.1074/jbc.M109.069542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lendel C., Bjerring M., Dubnovitsky A., Kelly R.T., Filippov A., Antzutkin O.N., Nielsen N.C., Härd T. A hexameric peptide barrel as building block of amyloid-β protofibrils. Angew. Chem. Int. Ed. 2014;53:12756–12760. doi: 10.1002/anie.201406357. [DOI] [PubMed] [Google Scholar]
- 63.Gao Y., Guo C., Watzlawik J.O., Randolph P.S., Lee E.J., Huang D., Stagg S.M., Zhou H.-X., Rosenberry T.L., Paravastu A.K. Out-of-register parallel β-sheets and antiparallel β-sheets coexist in 150-kDa oligomers formed by amyloid-β(1–42) J. Mol. Biol. 2020;432:4388–4407. doi: 10.1016/j.jmb.2020.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fusco G., Chen S.W., Williamson P.T.F., Cascella R., Perni M., Jarvis J.A., Cecchi C., Vendruscolo M., Chiti F., Cremades N., Ying L., Dobson C.M., De Simone A. Structural basis of membrane disruption and cellular toxicity by α-synuclein oligomers. Science. 2017;358:1440–1443. doi: 10.1126/science.aan6160. [DOI] [PubMed] [Google Scholar]
- 65.Varkey J., Langen R. Membrane remodeling by amyloidogenic and non-amyloidogenic proteins studied by EPR. J. Magn. Reson. 2017;280:127–139. doi: 10.1016/j.jmr.2017.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ceccon A., Schmidt T., Tugarinov V., Kotler S.A., Schwieters C.D., Clore G.M. Interaction of huntingtin exon-1 peptides with lipid-based micellar nanoparticles probed by solution NMR and Q-band pulsed EPR. J. Am. Chem. Soc. 2018;140:6199–6202. doi: 10.1021/jacs.8b02619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.De S., Klenerman D. Imaging individual protein aggregates to follow aggregation and determine the role of aggregates in neurodegenerative disease. Biochim. Biophys. Acta, Proteins Proteomics. 2019;1867:870–878. doi: 10.1016/j.bbapap.2018.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ruggeri F.S., Šneideris T., Vendruscolo M., Knowles T.P.J. Atomic force microscopy for single molecule characterisation of protein aggregation. Arch. Biochem. Biophys. 2019;664:134–148. doi: 10.1016/j.abb.2019.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hoffmann A., Neupane K., Woodside M.T. Single-molecule assays for investigating protein misfolding and aggregation. Phys. Chem. Chem. Phys. 2013;15:7934–7948. doi: 10.1039/c3cp44564j. [DOI] [PubMed] [Google Scholar]
- 70.Iljina M., Dear A.J., Garcia G.A., De S., Tosatto L., Flagmeier P., Whiten D.R., Michaels T.C.T., Frenkel D., Dobson C.M., Knowles T.P.J., Klenerman D. Quantifying co-oligomer formation by α-synuclein. ACS Nano. 2018;12:10855–10866. doi: 10.1021/acsnano.8b03575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Iljina M., Garcia G.A., Dear A.J., Flint J., Narayan P., Michaels T.C.T., Dobson C.M., Frenkel D., Knowles T.P.J., Klenerman D. Quantitative analysis of co-oligomer formation by amyloid-beta peptide isoforms. Sci. Rep. 2016;6:28658. doi: 10.1038/srep28658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Castello F., Paredes J.M., Ruedas-Rama M.J., Martin M., Roldan M., Casares S., Orte A. Two-step amyloid aggregation: sequential lag phase intermediates. Sci. Rep. 2017;7:40065. doi: 10.1038/srep40065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Whiten D.R., Cox D., Horrocks M.H., Taylor C.G., De S., Flagmeier P., Tosatto L., Kumita J.R., Ecroyd H., Dobson C.M., Klenerman D., Wilson M.R. Single-molecule characterization of the interactions between extracellular chaperones and toxic α-synuclein oligomers. Cell Rep. 2018;23:3492–3500. doi: 10.1016/j.celrep.2018.05.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Iljina M., Garcia G.A., Horrocks M.H., Tosatto L., Choi M.L., Ganzinger K.A., Abramov A.Y., Gandhi S., Wood N.W., Cremades N., Dobson C.M., Knowles T.P.J., Klenerman D. Kinetic model of the aggregation of alpha-synuclein provides insights into prion-like spreading. Proc. Natl. Acad. Sci. U. S. A. 2016;113:E1206–E1215. doi: 10.1073/pnas.1524128113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Horrocks M.H., Tosatto L., Dear A.J., Garcia G.A., Iljina M., Cremades N., Dalla Serra M., Knowles T.P.J., Dobson C.M., Klenerman D. Fast flow microfluidics and single-molecule fluorescence for the rapid characterization of α-synuclein oligomers. Anal. Chem. 2015;87:8818–8826. doi: 10.1021/acs.analchem.5b01811. [DOI] [PubMed] [Google Scholar]
- 76.Yang J., Dear A.J., Michaels T.C.T., Dobson C.M., Knowles T.P.J., Wu S., Perrett S. Direct observation of oligomerization by single molecule fluorescence reveals a multistep aggregation mechanism for the yeast prion protein Ure2. J. Am. Chem. Soc. 2018;140:2493–2503. doi: 10.1021/jacs.7b10439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tosatto L., Horrocks M.H., Dear A.J., Knowles T.P.J., Dalla Serra M., Cremades N., Dobson C.M., Klenerman D. Single-molecule FRET studies on alpha-synuclein oligomerization of Parkinson’s disease genetically related mutants. Sci. Rep. 2015;5:16696. doi: 10.1038/srep16696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chiou A., Hägglöf P., Orte A., Chen A.Y., Dunne P.D., Belorgey D., Karlsson-Li S., Lomas D.A., Klenerman D. Probing neuroserpin polymerization and interaction with amyloid-β peptides using single molecule fluorescence. Biophys. J. 2009;97:2306–2315. doi: 10.1016/j.bpj.2009.07.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Orte A., Birkett N.R., Clarke R.W., Devlin G.L., Dobson C.M., Klenerman D. Direct characterization of amyloidogenic oligomers by single-molecule fluorescence. Proc. Natl. Acad. Sci. U. S. A. 2008;105:14424–14429. doi: 10.1073/pnas.0803086105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Narayan P., Orte A., Clarke R.W., Bolognesi B., Hook S., Ganzinger K.A., Meehan S., Wilson M.R., Dobson C.M., Klenerman D. The extracellular chaperone clusterin sequesters oligomeric forms of the amyloid-β1−40 peptide. Nat. Struct. Mol. Biol. 2012;19:79–83. doi: 10.1038/nsmb.2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sang J.C., Lee J.-E., Dear A.J., De S., Meisl G., Thackray A.M., Bujdoso R., Knowles T.P.J., Klenerman D. Direct observation of prion protein oligomer formation reveals an aggregation mechanism with multiple conformationally distinct species. Chem. Sci. 2019;10:4588–4597. doi: 10.1039/C8SC05627G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kundel F., Hong L., Falcon B., McEwan W.A., Michaels T.C.T., Meisl G., Esteras N., Abramov A.Y., Knowles T.J.P., Goedert M., Klenerman D. Measurement of tau filament fragmentation provides insights into prion-like spreading. ACS Chem. Neurosci. 2018;9:1276–1282. doi: 10.1021/acschemneuro.8b00094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Horrocks M.H., Rajah L., Jönsson P., Kjaergaard M., Vendruscolo M., Knowles T.P.J., Klenerman D. Single-molecule measurements of transient biomolecular complexes through microfluidic dilution. Anal. Chem. 2013;85:6855–6859. doi: 10.1021/ac4010875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wägele J., De Sio S., Voigt B., Balbach J., Ott M. How fluorescent tags modify oligomer size distributions of the Alzheimer peptide. Biophys. J. 2019;116:227–238. doi: 10.1016/j.bpj.2018.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Anderson V.L., Webb W.W. Transmission electron microscopy characterization of fluorescently labelled amyloid β 1-40 and α-synuclein aggregates. BMC Biotechnol. 2011;11:125. doi: 10.1186/1472-6750-11-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Best R.B. Emerging consensus on the collapse of unfolded and intrinsically disordered proteins in water. Curr. Opin. Struct. Biol. 2020;60:27–38. doi: 10.1016/j.sbi.2019.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bongiovanni M.N., Godet J., Horrocks M.H., Tosatto L., Carr A.R., Wirthensohn D.C., Ranasinghe R.T., Lee J.-E., Ponjavic A., Fritz J.V., Dobson C.M., Klenerman D., Lee S.F. Multi-dimensional super-resolution imaging enables surface hydrophobicity mapping. Nat. Commun. 2016;7:13544. doi: 10.1038/ncomms13544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Horrocks M.H., Lee S.F., Gandhi S., Magdalinou N.K., Chen S.W., Devine M.J., Tosatto L., Kjaergaard M., Beckwith J.S., Zetterberg H., Iljina M., Cremades N., Dobson C.M., Wood N.W., Klenerman D. Single-molecule imaging of individual amyloid protein aggregates in human biofluids. ACS Chem. Neurosci. 2016;7:399–406. doi: 10.1021/acschemneuro.5b00324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Schönfelder J., De Sancho D., Perez-Jimenez R. The power of force: insights into the protein folding process using single-molecule force spectroscopy. J. Mol. Biol. 2016;428:4245–4257. doi: 10.1016/j.jmb.2016.09.006. [DOI] [PubMed] [Google Scholar]
- 90.Neuman K.C., Nagy A. Single-molecule force spectroscopy: optical tweezers, magnetic tweezers and atomic force microscopy. Nat. Methods. 2008;5:491–505. doi: 10.1038/nmeth.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yu H., Dee D.R., Liu X., Brigley A.M., Sosova I., Woodside M.T. Protein misfolding occurs by slow diffusion across multiple barriers in a rough energy landscape. Proc. Natl. Acad. Sci. U. S. A. 2015;112:8308–8313. doi: 10.1073/pnas.1419197112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gupta A.N., Neupane K., Rezajooei N., Cortez L.M., Sim V.L., Woodside M.T. Pharmacological chaperone reshapes the energy landscape for folding and aggregation of the prion protein. Nat. Commun. 2016;7:12058. doi: 10.1038/ncomms12058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Solanki A., Neupane K., Woodside M.T. Single-molecule force spectroscopy of rapidly fluctuating, marginally stable structures in the intrinsically disordered protein α-synuclein. Phys. Rev. Lett. 2014;112:158103. doi: 10.1103/PhysRevLett.112.158103. [DOI] [PubMed] [Google Scholar]
- 94.Churchill C.D.M., Healey M.A., Preto J., Tuszynski J.A., Woodside M.T. Probing the basis of α-synuclein aggregation by comparing simulations to single-molecule experiments. Biophys. J. 2019;117:1125–1135. doi: 10.1016/j.bpj.2019.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lyubchenko Y.L., Kim B., Krasnoslobodtsev A.V., Yu J. Nanoimaging for protein misfolding diseases. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2010;2:526–543. doi: 10.1002/wnan.102. [DOI] [PubMed] [Google Scholar]
- 96.McAllister C., Karymov M.A., Kawano Y., Lushnikov A.Y., Mikheikin A., Uversky V.N., Lyubchenko Y.L. Protein interactions and misfolding analyzed by AFM force spectroscopy. J. Mol. Biol. 2005;354:1028–1042. doi: 10.1016/j.jmb.2005.10.012. [DOI] [PubMed] [Google Scholar]
- 97.Yu J., Malkova S., Lyubchenko Y.L. α-Synuclein misfolding: single molecule AFM force spectroscopy study. J. Mol. Biol. 2008;384:992–1001. doi: 10.1016/j.jmb.2008.10.006. [DOI] [PubMed] [Google Scholar]
- 98.Kim B.-H., Lyubchenko Y.L. Nanoprobing of misfolding and interactions of amyloid β 42 protein. Nanomedicine. 2014;10:871–878. doi: 10.1016/j.nano.2013.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Doherty C.P.A., Young L.M., Karamanos T.K., Smith H.I., Jackson M.P., Radford S.E., Brockwell D.J. A peptide-display protein scaffold to facilitate single molecule force studies of aggregation-prone peptides. Protein Sci. 2018;27:1205–1217. doi: 10.1002/pro.3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Maity S., Lyubchenko Y.L. Force clamp approach for characterization of nano-assembly in amyloid beta 42 dimer. Nanoscale. 2019;11:12259–12265. doi: 10.1039/C9NR01670H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yu J., Lyubchenko Y.L. Early stages for Parkinson’s development: α-synuclein misfolding and aggregation. J. NeuroImmune Pharmacol. 2009;4:10–16. doi: 10.1007/s11481-008-9115-5. [DOI] [PubMed] [Google Scholar]
- 102.Yu J., Warnke J., Lyubchenko Y.L. Nanoprobing of α-synuclein misfolding and aggregation with atomic force microscopy. Nanomedicine. 2011;7:146–152. doi: 10.1016/j.nano.2010.08.001. [DOI] [PubMed] [Google Scholar]
- 103.Kim B.-H., Palermo N.Y., Lovas S., Zaikova T., Keana J.F.W., Lyubchenko Y.L. Single-molecule atomic force microscopy force spectroscopy study of Aβ-40 interactions. Biochemistry. 2011;50:5154–5162. doi: 10.1021/bi200147a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Portillo A.M., Krasnoslobodtsev A.V., Lyubchenko Y.L. Effect of electrostatics on aggregation of prion protein Sup35 peptide. J. Phys. Condens. Matter. 2012;24:164205. doi: 10.1088/0953-8984/24/16/164205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Krasnoslobodtsev A.V., Peng J., Asiago J.M., Hindupur J., Rochet J.-C., Lyubchenko Y.L. Effect of spermidine on misfolding and interactions of alpha-synuclein. PLoS One. 2012;7 doi: 10.1371/journal.pone.0038099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lv Z., Roychaudhuri R., Condron M.M., Teplow D.B., Lyubchenko Y.L. Mechanism of amyloid β−protein dimerization determined using single−molecule AFM force spectroscopy. Sci. Rep. 2013;3:2880. doi: 10.1038/srep02880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lv Z., Condron M.M., Teplow D.B., Lyubchenko Y.L. Nanoprobing of the effect of Cu2+ cations on misfolding, interaction and aggregation of amyloid β peptide. J. NeuroImmune Pharmacol. 2013;8:262–273. doi: 10.1007/s11481-012-9416-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Krasnoslobodtsev A.V., Volkov I.L., Asiago J.M., Hindupur J., Rochet J.-C., Lyubchenko Y.L. α-Synuclein misfolding assessed with single molecule AFM force spectroscopy: effect of pathogenic mutations. Biochemistry. 2013;52:7377–7386. doi: 10.1021/bi401037z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Oberhauser A.F., Marszalek P.E., Carrion-Vazquez M., Fernandez J.M. Single protein misfolding events captured by atomic force microscopy. Nat. Struct. Biol. 1999;6:1025–1028. doi: 10.1038/14907. [DOI] [PubMed] [Google Scholar]
- 110.Krasnoslobodtsev A.V., Zhang Y., Viazovkina E., Gall A., Bertagni C., Lyubchenko Y.L. A flexible nanoarray approach for the assembly and probing of molecular complexes. Biophys. J. 2015;108:2333–2339. doi: 10.1016/j.bpj.2015.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Maity S., Viazovkina E., Gall A., Lyubchenko Y.L. Single-molecule probing of amyloid nano-ensembles using the polymer nanoarray approach. Phys. Chem. Chem. Phys. 2017;19:16387–16394. doi: 10.1039/C7CP02691A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Maity S., Pramanik A., Lyubchenko Y.L. Probing intermolecular interactions within the amyloid β trimer using a tethered polymer nanoarray. Bioconjug. Chem. 2018;29:2755–2762. doi: 10.1021/acs.bioconjchem.8b00387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Maity S., Lyubchenko Y.L. AFM probing of amyloid-beta 42 dimers and trimers. Front. Mol. Biosci. 2020;7:69. doi: 10.3389/fmolb.2020.00069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Farrance O.E., Paci E., Radford S.E., Brockwell D.J. Extraction of accurate biomolecular parameters from single-molecule force spectroscopy experiments. ACS Nano. 2015;9:1315–1324. doi: 10.1021/nn505135d. [DOI] [PubMed] [Google Scholar]
- 115.Ruggeri F.S., Vieweg S., Cendrowska U., Longo G., Chiki A., Lashuel H.A., Dietler G. Nanoscale studies link amyloid maturity with polyglutamine diseases onset. Sci. Rep. 2016;6:31155. doi: 10.1038/srep31155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zhou L., Kurouski D. Structural characterization of individual α-synuclein oligomers formed at different stages of protein aggregation by atomic force microscopy-infrared spectroscopy. Anal. Chem. 2020;92:6806–6810. doi: 10.1021/acs.analchem.0c00593. [DOI] [PubMed] [Google Scholar]
- 117.Serio T.R., Cashikar A.G., Kowal A.S., Sawicki G.J., Moslehi J.J., Serpell L., Arnsdorf M.F., Lindquist S.L. Nucleated conformational conversion and the replication of conformational information by a prion determinant. Science. 2000;289:1317–1321. doi: 10.1126/science.289.5483.1317. [DOI] [PubMed] [Google Scholar]
- 118.Leney A.C., Heck A.J.R. Native mass spectrometry: what is in the name? J. Am. Soc. Mass Spectrom. 2017;28:5–13. doi: 10.1007/s13361-016-1545-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Bernstein S.L., Dupuis N.F., Lazo N.D., Wyttenbach T., Condron M.M., Bitan G., Teplow D.B., Shea J.-E., Ruotolo B.T., Robinson C.V., Bowers M.T. Amyloid-β protein oligomerization and the importance of tetramers and dodecamers in the aetiology of Alzheimer’s disease. Nat. Chem. 2009;1:326–331. doi: 10.1038/nchem.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Illes-Toth E., Ramos M.R., Cappai R., Dalton C., Smith D.P. Distinct higher-order α-synuclein oligomers induce intracellular aggregation. Biochem. J. 2015;468:485–493. doi: 10.1042/BJ20150159. [DOI] [PubMed] [Google Scholar]
- 121.Ilitchev A.I., Giammona M.J., Olivas C., Claud S.L., Lazar Cantrell K.L., Wu C., Buratto S.K., Bowers M.T. Hetero-oligomeric amyloid assembly and mechanism: prion fragment PrP(106–126) catalyzes the islet amyloid polypeptide β-hairpin. J. Am. Chem. Soc. 2018;140:9685–9695. doi: 10.1021/jacs.8b05925. [DOI] [PubMed] [Google Scholar]
- 122.Cole H.L., Kalapothakis J.M.D., Bennett G., Barran P.E., MacPhee C.E. Characterizing early aggregates formed by an amyloidogenic peptide by mass spectrometry. Angew. Chem. Int. Ed. 2010;49:9448–9451. doi: 10.1002/anie.201003373. [DOI] [PubMed] [Google Scholar]
- 123.Smith D.P., Radford S.E., Ashcroft A.E. Elongated oligomers in β2-microglobulin amyloid assembly revealed by ion mobility spectrometry-mass spectrometry. Proc. Natl. Acad. Sci. U. S. A. 2010;107:6794–6798. doi: 10.1073/pnas.0913046107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Leney A.C., Pashley C.L., Scarff C.A., Radford S.E., Ashcroft A.E. Insights into the role of the beta-2 microglobulin D-strand in amyloid propensity revealed by mass spectrometry. Mol. BioSyst. 2014;10:412–420. doi: 10.1039/C3MB70420C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Scarff C.A., Almeida B., Fraga J., Macedo-Ribeiro S., Radford S.E., Ashcroft A.E. Examination of ataxin-3 (atx-3) aggregation by structural mass spectrometry techniques: a rationale for expedited aggregation upon polyglutamine (polyQ) expansion. Mol. Cell. Proteomics. 2015;14:1241–1253. doi: 10.1074/mcp.M114.044610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Do T.D., Economou N.J., LaPointe N.E., Kincannon W.M., Bleiholder C., Feinstein S.C., Teplow D.B., Buratto S.K., Bowers M.T. Factors that drive peptide assembly and fibril formation: experimental and theoretical analysis of Sup35 NNQQNY mutants. J. Phys. Chem. B. 2013;117:8436–8446. doi: 10.1021/jp4046287. [DOI] [PubMed] [Google Scholar]
- 127.Do T.D., LaPointe N.E., Sangwan S., Teplow D.B., Feinstein S.C., Sawaya M.R., Eisenberg D.S., Bowers M.T. Factors that drive peptide assembly from native to amyloid structures: experimental and theoretical analysis of [Leu-5]-Enkephalin mutants. J. Phys. Chem. B. 2014;118:7247–7256. doi: 10.1021/jp502473s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Young L.M., Cao P., Raleigh D.P., Ashcroft A.E., Radford S.E. Ion mobility spectrometry-mass spectrometry defines the oligomeric intermediates in amylin amyloid formation and the mode of action of inhibitors. J. Am. Chem. Soc. 2014;136:660–670. doi: 10.1021/ja406831n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Smith D.P., Woods L.A., Radford S.E., Ashcroft A.E. Structure and dynamics of oligomeric intermediates in β2-microglobulin self-assembly. Biophys. J. 2011;101:1238–1247. doi: 10.1016/j.bpj.2011.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Gessel M.M., Wu C., Li H., Bitan G., Shea J.-E., Bowers M.T. Aβ(39-42) modulates Aβ oligomerization but not fibril formation. Biochemistry. 2012;51:108–117. doi: 10.1021/bi201520b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Gessel M.M., Bernstein S., Kemper M., Teplow D.B., Bowers M.T. Familial Alzheimers disease mutations differentially alter amyloid β-protein oligomerization. ACS Chem. Neurosci. 2012;3:909–918. doi: 10.1021/cn300050d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kłoniecki M., Jabłonowska A., Poznański J., Langridge J., Hughes C., Campuzano I., Giles K., Dadlez M. Ion mobility separation coupled with MS detects two structural states of Alzheimer’s disease Aβ1-40 peptide oligomers. J. Mol. Biol. 2011;407:110–124. doi: 10.1016/j.jmb.2011.01.012. [DOI] [PubMed] [Google Scholar]
- 133.Young L.M., Tu L.-H., Raleigh D.P., Ashcroft A.E., Radford S.E. Understanding co-polymerization in amyloid formation by direct observation of mixed oligomers. Chem. Sci. 2017;8:5030–5040. doi: 10.1039/C7SC00620A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Carulla N., Zhou M., Giralt E., Robinson C.V., Dobson C.M. Structure and intermolecular dynamics of aggregates populated during amyloid fibril formation studied by hydrogen/deuterium exchange. Acc. Chem. Res. 2010;43:1072–1079. doi: 10.1021/ar9002784. [DOI] [PubMed] [Google Scholar]
- 135.Singh J., Udgaonkar J.B. Dissection of conformational conversion events during prion amyloid fibril formation using hydrogen exchange and mass spectrometry. J. Mol. Biol. 2013;425:3510–3521. doi: 10.1016/j.jmb.2013.06.009. [DOI] [PubMed] [Google Scholar]
- 136.Mysling S., Betzer C., Jensen P.H., Jorgensen T.J.D. Characterizing the dynamics of α-synuclein oligomers using hydrogen/deuterium exchange monitored by mass spectrometry. Biochemistry. 2013;52:9097–9103. doi: 10.1021/bi4009193. [DOI] [PubMed] [Google Scholar]
- 137.Przygońska K., Poznański J., Mistarz U.H., Rand K.D., Dadlez M. Side-chain moieties from the N-terminal region of Aβ are involved in an oligomer-stabilizing network of interactions. PLoS One. 2018;13 doi: 10.1371/journal.pone.0201761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Paslawski W., Mysling S., Thomsen K., Jørgensen T.J.D., Otzen D.E. Co-existence of two different α-synuclein oligomers with different core structures determined by hydrogen/deuterium exchange mass spectrometry. Angew. Chem. Int. Ed. 2014;53:7560–7563. doi: 10.1002/anie.201400491. [DOI] [PubMed] [Google Scholar]
- 139.Zhang Y., Rempel D.L., Zhang J., Sharma A.K., Mirica L.M., Gross M.L. Pulsed hydrogen–deuterium exchange mass spectrometry probes conformational changes in amyloid beta (Aβ) peptide aggregation. Proc. Natl. Acad. Sci. U. S. A. 2013;110:14604–14609. doi: 10.1073/pnas.1309175110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Wang H., Shu Q., Rempel D.L., Frieden C., Gross M.L. Continuous and pulsed hydrogen–deuterium exchange and mass spectrometry characterize CsgE oligomerization. Biochemistry. 2015;54:6475–6481. doi: 10.1021/acs.biochem.5b00871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Carulla N., Zhou M., Arimon M., Gairí M., Giralt E., Robinson C.V., Dobson C.M. Experimental characterization of disordered and ordered aggregates populated during the process of amyloid fibril formation. Proc. Natl. Acad. Sci. U. S. A. 2009;106:7828–7833. doi: 10.1073/pnas.0812227106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Pan J., Han J., Borchers C.H., Konermann L. Structure and dynamics of small soluble Aβ(1-40) oligomers studied by top-down hydrogen exchange mass spectrometry. Biochemistry. 2012;51:3694–3703. doi: 10.1021/bi3002049. [DOI] [PubMed] [Google Scholar]
- 143.Li K.S., Rempel D.L., Gross M.L. Conformational-sensitive fast photochemical oxidation of proteins and mass spectrometry characterize amyloid beta 1–42 aggregation. J. Am. Chem. Soc. 2016;138:12090–12098. doi: 10.1021/jacs.6b07543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Mendoza V.L., Antwi K., Barón-Rodríguez M.A., Blanco C., Vachet R.W. Structure of the preamyloid dimer of β-2-microglobulin from covalent labeling and mass spectrometry. Biochemistry. 2010;49:1522–1532. doi: 10.1021/bi901748h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Mendoza V.L., Barón-Rodríguez M.A., Blanco C., Vachet R.W. Structural insights into the pre-amyloid tetramer of β-2-microglobulin from covalent labeling and mass spectrometry. Biochemistry. 2011;50:6711–6722. doi: 10.1021/bi2004894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Borotto N.B., Zhang Z., Dong J., Burant B., Vachet R.W. Increased β-sheet dynamics and D–E loop repositioning are necessary for cu(II)-induced amyloid formation by β-2-microglobulin. Biochemistry. 2017;56:1095–1104. doi: 10.1021/acs.biochem.6b01198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Illes-Toth E., Rempel D.L., Gross M.L. Pulsed hydrogen–deuterium exchange illuminates the aggregation kinetics of α-synuclein, the causative agent for Parkinson’s disease. ACS Chem. Neurosci. 2018;9:1469–1476. doi: 10.1021/acschemneuro.8b00052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Hall Z., Schmidt C., Politis A. Uncovering the early assembly mechanism for amyloidogenic β2-microglobulin using cross-linking and native mass spectrometry. J. Biol. Chem. 2016;291:4626–4637. doi: 10.1074/jbc.M115.691063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Murphy B.J., Klusch N., Langer J., Mills D.J., Yildiz Ö., Kühlbrandt W. Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F1-Fo coupling. Science. 2019;364 doi: 10.1126/science.aaw9128. eaaw9128. [DOI] [PubMed] [Google Scholar]
- 150.Gallardo R., Ranson N.A., Radford S.E. Amyloid structures: much more than just a cross-β fold. Curr. Opin. Struct. Biol. 2020;60:7–16. doi: 10.1016/j.sbi.2019.09.001. [DOI] [PubMed] [Google Scholar]
- 151.Jan A., Hartley D.M., Lashuel H.A. Preparation and characterization of toxic Aβ aggregates for structural and functional studies in Alzheimer’s disease research. Nat. Protoc. 2010;5:1186–1209. doi: 10.1038/nprot.2010.72. [DOI] [PubMed] [Google Scholar]
- 152.Chen S.W., Cremades N. Preparation of α-synuclein amyloid assemblies for toxicity experiments. In: Sigurdsson E.M., Calero M., Gasset M., editors. Amyloid Proteins: Methods and Protocols. Humana Press; New York: 2018. pp. 45–60. [DOI] [Google Scholar]
- 153.Zijlstra N., Blum C., Segers-Nolten I.M.J., Claessens M.M.A.E., Subramaniam V. Molecular composition of sub-stoichiometrically labeled α-synuclein oligomers determined by single-molecule photobleaching. Angew. Chem. Int. Ed. 2012;51:8821–8824. doi: 10.1002/anie.201200813. [DOI] [PubMed] [Google Scholar]
- 154.Lorenzen N., Nielsen S.B., Buell A.K., Kaspersen J.D., Arosio P., Vad B.S., Paslawski W., Christiansen G., Valnickova-Hansen Z., Andreasen M., Enghild J.J., Pedersen J.S., Dobson C.M., Knowles T.P.J., Otzen D.E. The role of stable α-synuclein oligomers in the molecular events underlying amyloid formation. J. Am. Chem. Soc. 2014;136:3859–3868. doi: 10.1021/ja411577t. [DOI] [PubMed] [Google Scholar]
- 155.Celej M.S., Sarroukh R., Goormaghtigh E., Fidelio G.D., Ruysschaert J.-M., Raussens V. Toxic prefibrillar α-synuclein amyloid oligomers adopt a distinctive antiparallel β-sheet structure. Biochem. J. 2012;443:719–726. doi: 10.1042/BJ20111924. [DOI] [PubMed] [Google Scholar]
- 156.Gallea J.I., Celej M.S. Structural insights into amyloid oligomers of the Parkinson disease-related protein α-synuclein. J. Biol. Chem. 2014;289:26733–26742. doi: 10.1074/jbc.M114.566695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.van Rooijen B.D., Claessens M.M.A.E., Subramaniam V. Lipid bilayer disruption by oligomeric α-synuclein depends on bilayer charge and accessibility of the hydrophobic core. Biochim. Biophys. Acta Biomembr. 2009;1788:1271–1278. doi: 10.1016/j.bbamem.2009.03.010. [DOI] [PubMed] [Google Scholar]
- 158.Volles M.J., Lee S.-J., Rochet J.-C., Shtilerman M.D., Ding T.T., Kessler J.C., Lansbury P.T., Jr. Vesicle permeabilization by protofibrillar α-synuclein: implications for the pathogenesis and treatment of Parkinson’s disease. Biochemistry. 2001;40:7812–7819. doi: 10.1021/bi0102398. [DOI] [PubMed] [Google Scholar]
- 159.Ehrnhoefer D.E., Bieschke J., Boeddrich A., Herbst M., Masino L., Lurz R., Engemann S., Pastore A., Wanker E.E. EGCG redirects amyloidogenic polypeptides into unstructured, off-pathway oligomers. Nat. Struct. Mol. Biol. 2008;15:558–566. doi: 10.1038/nsmb.1437. [DOI] [PubMed] [Google Scholar]
- 160.Campioni S., Mannini B., Zampagni M., Pensalfini A., Parrini C., Evangelisti E., Relini A., Stefani M., Dobson C.M., Cecchi C., Chiti F. A causative link between the structure of aberrant protein oligomers and their toxicity. Nat. Chem. Biol. 2010;6:140–147. doi: 10.1038/nchembio.283. [DOI] [PubMed] [Google Scholar]
- 161.Capitini C., Patel J.R., Natalello A., D’Andrea C., Relini A., Jarvis J.A., Birolo L., Peduzzo A., Vendruscolo M., Matteini P., Dobson C.M., De Simone A., Chiti F. Structural differences between toxic and nontoxic HypF-N oligomers. Chem. Commun. 2018;54:8637–8640. doi: 10.1039/C8CC03446J. [DOI] [PubMed] [Google Scholar]
- 162.Ahmed M., Davis J., Aucoin D., Sato T., Ahuja S., Aimoto S., Elliott J.I., Van Nostrand W.E., Smith S.O. Structural conversion of neurotoxic amyloid-β1–42 oligomers to fibrils. Nat. Struct. Mol. Biol. 2010;17:561–567. doi: 10.1038/nsmb.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Chimon S., Shaibat M.A., Jones C.R., Calero D.C., Aizezi B., Ishii Y. Evidence of fibril-like β-sheet structures in a neurotoxic amyloid intermediate of Alzheimer’s β-amyloid. Nat. Struct. Mol. Biol. 2007;14:1157–1164. doi: 10.1038/nsmb1345. [DOI] [PubMed] [Google Scholar]
- 164.Serra-Batiste M., Ninot-Pedrosa M., Bayoumi M., Gairí M., Maglia G., Carulla N. Aβ42 assembles into specific β-barrel pore-forming oligomers in membrane-mimicking environments. Proc. Natl. Acad. Sci. U. S. A. 2016;113:10866–10871. doi: 10.1073/pnas.1605104113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Evangelisti E., Cascella R., Becatti M., Marrazza G., Dobson C.M., Chiti F., Stefani M., Cecchi C. Binding affinity of amyloid oligomers to cellular membranes is a generic indicator of cellular dysfunction in protein misfolding diseases. Sci. Rep. 2016;6:32721. doi: 10.1038/srep32721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Ladiwala A.R.A., Litt J., Kane R.S., Aucoin D.S., Smith S.O., Ranjan S., Davis J., Van Nostrand W.E., Tessier P.M. Conformational differences between two amyloid β oligomers of similar size and dissimilar toxicity. J. Biol. Chem. 2012;287:24765–24773. doi: 10.1074/jbc.M111.329763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Vivoli Vega M., Cascella R., Chen S.W., Fusco G., De Simone A., Dobson C.M., Cecchi C., Chiti F. The toxicity of misfolded protein oligomers is independent of their secondary structure. ACS Chem. Biol. 2019;14:1593–1600. doi: 10.1021/acschembio.9b00324. [DOI] [PubMed] [Google Scholar]
- 168.Valera E., Spencer B., Masliah E. Immunotherapeutic approaches targeting amyloid-β, α-synuclein, and tau for the treatment of neurodegenerative disorders. Neurotherapeutics. 2016;13:179–189. doi: 10.1007/s13311-015-0397-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Liu J., Yang B., Ke J., Li W., Suen W.-C. Antibody-based drugs and approaches against amyloid-β species for Alzheimer’s disease immunotherapy. Drugs Aging. 2016;33:685–697. doi: 10.1007/s40266-016-0406-x. [DOI] [PubMed] [Google Scholar]
- 170.De Genst E., Messer A., Dobson C.M. Antibodies and protein misfolding: from structural research tools to therapeutic strategies. Biochim. Biophys. Acta, Proteins Proteomics. 2014;1844:1907–1919. doi: 10.1016/j.bbapap.2014.08.016. [DOI] [PubMed] [Google Scholar]
- 171.Ma B., Zhao J., Nussinov R. Conformational selection in amyloid-based immunotherapy: survey of crystal structures of antibody-amyloid complexes. Biochim. Biophys. Acta, Gen. Subj. 2016;1860:2672–2681. doi: 10.1016/j.bbagen.2016.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Glabe C.G. Structural classification of toxic amyloid oligomers. J. Biol. Chem. 2008;283:29639–29643. doi: 10.1074/jbc.R800016200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Emadi S., Barkhordarian H., Wang M.S., Schulz P., Sierks M.R. Isolation of a human single chain antibody fragment against oligomeric α-synuclein that inhibits aggregation and prevents α-synuclein-induced toxicity. J. Mol. Biol. 2007;368:1132–1144. doi: 10.1016/j.jmb.2007.02.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Emadi S., Kasturirangan S., Wang M.S., Schulz P., Sierks M.R. Detecting morphologically distinct oligomeric forms of α-synuclein. J. Biol. Chem. 2009;284:11048–11058. doi: 10.1074/jbc.M806559200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Silverman J.M., Gibbs E., Peng X., Martens K.M., Balducci C., Wang J., Yousefi M., Cowan C.M., Lamour G., Louadi S., Ban Y., Robert J., Stukas S., Forloni G., Hsiung G.-Y.R., Plotkin S.S., Wellington C.L., Cashman N.R. A rational structured epitope defines a distinct subclass of toxic amyloid-beta oligomers. ACS Chem. Neurosci. 2018;9:1591–1606. doi: 10.1021/acschemneuro.7b00469. [DOI] [PubMed] [Google Scholar]
- 176.Gibbs E., Silverman J.M., Zhao B., Peng X., Wang J., Wellington C.L., Mackenzie I.R., Plotkin S.S., Kaplan J.M., Cashman N.R. A rationally designed humanized antibody selective for amyloid beta oligomers in Alzheimer’s disease. Sci. Rep. 2019;9:9870. doi: 10.1038/s41598-019-46306-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Morgado I., Wieligmann K., Bereza M., Rönicke R., Meinhardt K., Annamalai K., Baumann M., Wacker J., Hortschansky P., Malešević M., Parthier C., Mawrin C., Schiene-Fischer C., Reymann K.G., Stubbs M.T., Balbach J., Görlach M., Horn U., Fändrich M. Molecular basis of β-amyloid oligomer recognition with a conformational antibody fragment. Proc. Natl. Acad. Sci. U. S. A. 2012;109:12503–12508. doi: 10.1073/pnas.1206433109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Aprile F.A., Sormanni P., Perni M., Arosio P., Linse S., Knowles T.P.J., Dobson C.M., Vendruscolo M. Selective targeting of primary and secondary nucleation pathways in Aβ42 aggregation using a rational antibody scanning method. Sci. Adv. 2017;3 doi: 10.1126/sciadv.1700488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Perchiacca J.M., Ladiwala A.R.A., Bhattacharya M., Tessier P.M. Structure-based design of conformation- and sequence-specific antibodies against amyloid β. Proc. Natl. Acad. Sci. U. S. A. 2012;109:84–89. doi: 10.1073/pnas.1111232108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Ladiwala A.R.A., Bhattacharya M., Perchiacca J.M., Cao P., Raleigh D.P., Abedini A., Schmidt A.M., Varkey J., Langen R., Tessier P.M. Rational design of potent domain antibody inhibitors of amyloid fibril assembly. Proc. Natl. Acad. Sci. U. S. A. 2012;109:19965–19970. doi: 10.1073/pnas.1208797109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Aprile F.A., Sormanni P., Podpolny M., Chhangur S., Needham L.-M., Ruggeri F.S., Perni M., Limbocker R., Heller G.T., Sneideris T., Scheidt T., Mannini B., Habchi J., Lee S.F., Salinas P.C., Knowles T.P.J., Dobson C.M., Vendruscolo M. Rational design of a conformation-specific antibody for the quantification of Aβ oligomers. Proc. Natl. Acad. Sci. U. S. A. 2020;117:13509–13518. doi: 10.1073/pnas.1919464117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Tian H., Davidowitz E., Lopez P., He P., Schulz P., Moe J., Sierks M.R. Isolation and characterization of antibody fragments selective for toxic oligomeric tau. Neurobiol. Aging. 2015;36:1342–1355. doi: 10.1016/j.neurobiolaging.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Streltsov V.A., Varghese J.N., Masters C.L., Nuttall S.D. Crystal structure of the amyloid-β p3 fragment provides a model for oligomer formation in Alzheimer’s disease. J. Neurosci. 2011;31:1419–1426. doi: 10.1523/JNEUROSCI.4259-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Kumar S., Birol M., Schlamadinger D.E., Wojcik S.P., Rhoades E., Miranker A.D. Foldamer-mediated manipulation of a pre-amyloid toxin. Nat. Commun. 2016;7:11412. doi: 10.1038/ncomms11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Kumar S., Hamilton A.D. α-helix mimetics as modulators of Aβ self-assembly. J. Am. Chem. Soc. 2017;139:5744–5755. doi: 10.1021/jacs.6b09734. [DOI] [PubMed] [Google Scholar]
- 186.Kumar S., Henning-Knechtel A., Chehade I., Magzoub M., Hamilton A.D. Foldamer-mediated structural rearrangement attenuates Aβ oligomerization and cytotoxicity. J. Am. Chem. Soc. 2017;139:17098–17108. doi: 10.1021/jacs.7b08259. [DOI] [PubMed] [Google Scholar]
- 187.Habchi J., Chia S., Limbocker R., Mannini B., Ahn M., Perni M., Hansson O., Arosio P., Kumita J.R., Challa P.K., Cohen S.I.A., Linse S., Dobson C.M., Knowles T.P.J., Vendruscolo M. Systematic development of small molecules to inhibit specific microscopic steps of Aβ42 aggregation in Alzheimer’s disease. Proc. Natl. Acad. Sci. U. S. A. 2017;114:E200–E208. doi: 10.1073/pnas.1615613114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Arkin M.R., Tang Y., Wells J.A. Small-molecule inhibitors of protein-protein interactions: progressing toward the reality. Chem. Biol. 2014;21:1102–1114. doi: 10.1016/j.chembiol.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Bulawa C.E., Connelly S., DeVit M., Wang L., Weigel C., Fleming J.A., Packman J., Powers E.T., Wiseman R.L., Foss T.R., Wilson I.A., Kelly J.W., Labaudinière R. Tafamidis, a potent and selective transthyretin kinetic stabilizer that inhibits the amyloid cascade. Proc. Natl. Acad. Sci. U. S. A. 2012;109:9629–9634. doi: 10.1073/pnas.1121005109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Sant’Anna R., Gallego P., Robinson L.Z., Pereira-Henriques A., Ferreira N., Pinheiro F., Esperante S., Pallares I., Huertas O., Rosário Almeida M., Reixach N., Insa R., Velazquez-Campoy A., Reverter D., Reig N., Ventura S. Repositioning tolcapone as a potent inhibitor of transthyretin amyloidogenesis and associated cellular toxicity. Nat. Commun. 2016;7:10787. doi: 10.1038/ncomms10787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Brumshtein B., Esswein S.R., Salwinski L., Phillips M.L., Ly A.T., Cascio D., Sawaya M.R., Eisenberg D.S. Inhibition by small-molecule ligands of formation of amyloid fibrils of an immunoglobulin light chain variable domain. Elife. 2015;4 doi: 10.7554/eLife.10935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Morgan G.J., Yan N.L., Mortenson D.E., Rennella E., Blundon J.M., Gwin R.M., Lin C.-Y., Stanfield R.L., Brown S.J., Rosen H., Spicer T.P., Fernandez-Vega V., Merlini G., Kay L.E., Wilson I.A., Kelly J.W. Stabilization of amyloidogenic immunoglobulin light chains by small molecules. Proc. Natl. Acad. Sci. U. S. A. 2019;116:8360–8369. doi: 10.1073/pnas.1817567116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Young L.M., Ashcroft A.E., Radford S.E. Small molecule probes of protein aggregation. Curr. Opin. Chem. Biol. 2017;39:90–99. doi: 10.1016/j.cbpa.2017.06.008. [DOI] [PubMed] [Google Scholar]
- 194.Young L.M., Saunders J.C., Mahood R.A., Revill C.H., Foster R.J., Tu L.-H., Raleigh D.P., Radford S.E., Ashcroft A.E. Screening and classifying small-molecule inhibitors of amyloid formation using ion mobility spectrometry–mass spectrometry. Nat. Chem. 2015;7:73–81. doi: 10.1038/nchem.2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Viles J.H. Metal ions and amyloid fiber formation in neurodegenerative diseases. Copper, zinc and iron in Alzheimer’s, Parkinson’s and prion diseases. Coord. Chem. Rev. 2012;256:2271–2284. doi: 10.1016/j.ccr.2012.05.003. [DOI] [Google Scholar]
- 196.Beck M.W., Oh S.B., Kerr R.A., Lee H.J., Kim S.H., Kim S., Jang M., Ruotolo B.T., Lee J.-Y., Lim M.H. A rationally designed small molecule for identifying an in vivo link between metal–amyloid-β complexes and the pathogenesis of Alzheimer’s disease. Chem. Sci. 2015;6:1879–1886. doi: 10.1039/C4SC03239J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Calabrese M.F., Miranker A.D. Metal binding sheds light on mechanisms of amyloid assembly. Prion. 2009;3:1–4. doi: 10.4161/pri.3.1.8601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Fu Z., Aucoin D., Ahmed M., Ziliox M., Van Nostrand W.E., Smith S.O. Capping of Aβ42 oligomers by small molecule inhibitors. Biochemistry. 2014;53:7893–7903. doi: 10.1021/bi500910b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Andrich K., Hegenbart U., Kimmich C., Kedia N., Bergen H.R., 3rd, Schönland S., Wanker E., Bieschke J. Aggregation of full-length immunoglobulin light chains from systemic light chain amyloidosis (AL) patients is remodeled by epigallocatechin-3-gallate. J. Biol. Chem. 2017;292:2328–2344. doi: 10.1074/jbc.M116.750323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Mannini B., Habchi J., Chia S., Ruggeri F.S., Perni M., Knowles T.P.J., Dobson C.M., Vendruscolo M. Stabilization and characterization of cytotoxic Aβ40 oligomers isolated from an aggregation reaction in the presence of zinc ions. ACS Chem. Neurosci. 2018;9:2959–2971. doi: 10.1021/acschemneuro.8b00141. [DOI] [PubMed] [Google Scholar]
- 201.Tew D.J., Bottomley S.P., Smith D.P., Ciccotosto G.D., Babon J., Hinds M.G., Masters C.L., Cappai R., Barnham K.J. Stabilization of neurotoxic soluble β-sheet-rich conformations of the Alzheimer’s disease amyloid-β peptide. Biophys. J. 2008;94:2752–2766. doi: 10.1529/biophysj.107.119909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Fonseca-Ornelas L., Schmidt C., Camacho-Zarco A.R., Fernandez C.O., Becker S., Zweckstetter M. Small-molecule-induced soluble oligomers of α-synuclein with helical structure. Chem. Eur. J. 2017;23:13010–13014. doi: 10.1002/chem.201703001. [DOI] [PubMed] [Google Scholar]
- 203.Hong D.-P., Fink A.L., Uversky V.N. Structural characteristics of α-synuclein oligomers stabilized by the flavonoid baicalein. J. Mol. Biol. 2008;383:214–223. doi: 10.1016/j.jmb.2008.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Lopez del Amo J.M., Fink U., Dasari M., Grelle G., Wanker E.E., Bieschke J., Reif B. Structural properties of EGCG-induced, nontoxic Alzheimer’s disease Aβ oligomers. J. Mol. Biol. 2012;421:517–524. doi: 10.1016/j.jmb.2012.01.013. [DOI] [PubMed] [Google Scholar]
- 205.Ryan T.M., Roberts B.R., McColl G., Hare D.J., Doble P.A., Li Q.-X., Lind M., Roberts A.M., Mertens H.D.T., Kirby N., Pham C.L.L., Hinds M.G., Adlard P.A., Barnham K.J., Curtain C.C., Masters C.L. Stabilization of nontoxic Aβ-oligomers: insights into the mechanism of action of hydroxyquinolines in Alzheimer’s disease. J. Neurosci. 2015;35:2871–2884. doi: 10.1523/JNEUROSCI.2912-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Wang N., Majmudar C.Y., Pomerantz W.C., Gagnon J.K., Sadowsky J.D., Meagher J.L., Johnson T.K., Stuckey J.A., Brooks C.L., III, Wells J.A., Mapp A.K. Ordering a dynamic protein via a small molecule stabilizer. J. Am. Chem. Soc. 2013;135:3363–3366. doi: 10.1021/ja3122334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Wang N., Lodge J.M., Fierke C.A., Mapp A.K. Dissecting allosteric effects of activator-coactivator complexes using a covalent small molecule ligand. Proc. Natl. Acad. Sci. U. S. A. 2014;111:12061–12066. doi: 10.1073/pnas.1406033111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Lodge J.M., Majmudar C.Y., Clayton J., Mapp A.K. Covalent chemical cochaperones of the p300/CBP GACKIX domain. ChemBioChem. 2018;19:1907–1912. doi: 10.1002/cbic.201800173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Lodge J.M., Rettenmaier T.J., Wells J.A., Pomerantz W.C., Mapp A.K. FP tethering: a screening technique to rapidly identify compounds that disrupt protein–protein interactions. MedChemComm. 2014;5:370–375. doi: 10.1039/C3MD00356F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Sijbesma E., Hallenbeck K.K., Leysen S., de Vink P.J., Skóra L., Jahnke W., Brunsveld L., Arkin M.R., Ottmann C. Site-directed fragment-based screening for the discovery of protein-protein interaction stabilizers. J. Am. Chem. Soc. 2019;141:3524–3531. doi: 10.1021/jacs.8b11658. [DOI] [PubMed] [Google Scholar]
- 211.Akçay G., Belmonte M.A., Aquila B., Chuaqui C., Hird A.W., Lamb M.L., Rawlins P.B., Su N., Tentarelli S., Grimster N.P., Su Q. Inhibition of Mcl-1 through covalent modification of a noncatalytic lysine side chain. Nat. Chem. Biol. 2016;12:931–936. doi: 10.1038/nchembio.2174. [DOI] [PubMed] [Google Scholar]
- 212.Lee S., Wales T.E., Escudero S., Cohen D.T., Luccarelli J., Gallagher C.G., Cohen N.A., Huhn A.J., Bird G.H., Engen J.R., Walensky L.D. Allosteric inhibition of antiapoptotic MCL-1. Nat. Struct. Mol. Biol. 2016;23:600–607. doi: 10.1038/nsmb.3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Choi S., Connelly S., Reixach N., Wilson I.A., Kelly J.W. Chemoselective small molecules that covalently modify one lysine in a non-enzyme protein in plasma. Nat. Chem. Biol. 2010;6:133–139. doi: 10.1038/nchembio.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Harvey E.P., Hauseman Z.J., Cohen D.T., Rettenmaier T.J., Lee S., Huhn A.J., Wales T.E., Seo H.-S., Luccarelli J., Newman C.E., Guerra R.M., Bird G.H., Dhe-Paganon S., Engen J.R., Wells J.A., Walensky L.D. Identification of a covalent molecular inhibitor of anti-apoptotic BFL-1 by disulfide tethering. Cell Chem. Biol. 2020;27:647–656. doi: 10.1016/j.chembiol.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Gambini L., Baggio C., Udompholkul P., Jossart J., Salem A.F., Perry J.J.P., Pellecchia M. Covalent inhibitors of protein–protein interactions targeting lysine, tyrosine, or histidine residues. J. Med. Chem. 2019;62:5616–5627. doi: 10.1021/acs.jmedchem.9b00561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Dal Corso A., Catalano M., Schmid A., Scheuermann J., Neri D. Affinity enhancement of protein ligands by reversible covalent modification of neighboring lysine residues. Angew. Chem. Int. Ed. 2018;57:17178–17182. doi: 10.1002/anie.201811650. [DOI] [PubMed] [Google Scholar]
- 217.Bradshaw J.M., McFarland J.M., Paavilainen V.O., Bisconte A., Tam D., Phan V.T., Romanov S., Finkle D., Shu J., Patel V., Ton T., Li X., Loughhead D.G., Nunn P.A., Karr D.E., Gerritsen M.E., Funk J.O., Owens T.D., Verner E., Brameld K.A., Hill R.J., Goldstein D.M., Taunton J. Prolonged and tunable residence time using reversible covalent kinase inhibitors. Nat. Chem. Biol. 2015;11:525–531. doi: 10.1038/nchembio.1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Krall N., da Cruz F.P., Boutureira O., Bernardes G.J.L. Site-selective protein-modification chemistry for basic biology and drug development. Nat. Chem. 2016;8:103–113. doi: 10.1038/nchem.2393. [DOI] [PubMed] [Google Scholar]
- 219.Wright T.H., Bower B.J., Chalker J.M., Bernardes G.J.L., Wiewiora R., Ng W.L., Raj R., Faulkner S., Vallée M.R.J., Phanumartwiwath A., Coleman O.D., Thézénas M.-L., Khan M., Galan S.R.G., Lercher L., Schombs M.W., Gerstberger S., Palm-Espling M.E., Baldwin A.J., Kessler B.M., Claridge T.D.W., Mohammed S., Davis B.G. Posttranslational mutagenesis: a chemical strategy for exploring protein side-chain diversity. Science. 2016;354 doi: 10.1126/science.aag1465. aag1465. [DOI] [PubMed] [Google Scholar]
- 220.Ruiz J., Boehringer R., Grogg M., Raya J., Schirer A., Crucifix C., Hellwig P., Schultz P., Torbeev V. Covalent tethering and residues with bulky hydrophobic side chains enable self-assembly of distinct amyloid structures. ChemBioChem. 2016;17:2274–2285. doi: 10.1002/cbic.201600440. [DOI] [PubMed] [Google Scholar]
- 221.Sant’Anna R., Almeida M.R., Varejāo N., Gallego P., Esperante S., Ferreira P., Pereira-Henriques A., Palhano F.L., de Carvalho M., Foguel D., Reverter D., Saraiva M.J., Ventura S. Cavity filling mutations at the thyroxine-binding site dramatically increase transthyretin stability and prevent its aggregation. Sci. Rep. 2017;7:44709. doi: 10.1038/srep44709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Wang Y., Truex N.L., Vo N.D.P., Nowick J.S. Effects of charge and hydrophobicity on the oligomerization of peptides derived from IAPP. Bioorg. Med. Chem. 2018;26:1151–1156. doi: 10.1016/j.bmc.2017.10.001. [DOI] [PubMed] [Google Scholar]
- 223.Mannini B., Mulvihill E., Sgromo C., Cascella R., Khodarahmi R., Ramazzotti M., Dobson C.M., Cecchi C., Chiti F. Toxicity of protein oligomers is rationalized by a function combining size and surface hydrophobicity. ACS Chem. Biol. 2014;9:2309–2317. doi: 10.1021/cb500505m. [DOI] [PubMed] [Google Scholar]
- 224.Yang X., Meisl G., Frohm B., Thulin E., Knowles T.P.J., Linse S. On the role of sidechain size and charge in the aggregation of Aβ42 with familial mutations. Proc. Natl. Acad. Sci. U. S. A. 2018;115:E5849–E5858. doi: 10.1073/pnas.1803539115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Hatami A., Monjazeb S., Milton S., Glabe C.G. Familial Alzheimer’s disease mutations within the amyloid precursor protein alter the aggregation and conformation of the amyloid-β peptide. J. Biol. Chem. 2017;292:3172–3185. doi: 10.1074/jbc.M116.755264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Routledge K.E., Tartaglia G.G., Platt G.W., Vendruscolo M., Radford S.E. Competition between intramolecular and intermolecular interactions in an amyloid-forming protein. J. Mol. Biol. 2009;389:776–786. doi: 10.1016/j.jmb.2009.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Yee A.W., Aldeghi M., Blakeley M.P., Ostermann A., Mas P.J., Moulin M., de Sanctis D., Bowler M.W., Mueller-Dieckmann C., Mitchell E.P., Haertlein M., de Groot B.L., Boeri Erba E., Forsyth V.T. A molecular mechanism for transthyretin amyloidogenesis. Nat. Commun. 2019;10:925. doi: 10.1038/s41467-019-08609-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Ebo J.S., Guthertz N., Radford S.E., Brockwell D.J. Using protein engineering to understand and modulate aggregation. Curr. Opin. Struct. Biol. 2020;60:157–166. doi: 10.1016/j.sbi.2020.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Wilson C.G., Arkin M.R. Probing structural adaptivity at PPI interfaces with small molecules. Drug Discov. Today Technol. 2013;10:e501–e508. doi: 10.1016/j.ddtec.2012.10.009. [DOI] [PubMed] [Google Scholar]
- 230.Erlanson D.A., Braisted A.C., Raphael D.R., Randal M., Stroud R.M., Gordon E.M., Wells J.A. Site-directed ligand discovery. Proc. Natl. Acad. Sci. U. S. A. 2000;97:9367–9372. doi: 10.1073/pnas.97.17.9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Fändrich M., Dobson C.M. The behaviour of polyamino acids reveals an inverse side chain effect in amyloid structure formation. EMBO J. 2002;21:5682–5690. doi: 10.1093/emboj/cdf573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Nowick J.S., Chung D.M., Maitra K., Maitra S., Stigers K.D., Sun Y. An unnatural amino acid that mimics a tripeptide β-strand and forms β-sheetlike hydrogen-bonded dimers. J. Am. Chem. Soc. 2000;122:7654–7661. doi: 10.1021/ja001142w. [DOI] [Google Scholar]
- 233.Spencer R., Chen K.H., Manuel G., Nowick J.S. Recipe for β-sheets: foldamers containing amyloidogenic peptide sequences. Eur. J. Org. Chem. 2013;2013:3523–3528. doi: 10.1002/ejoc.201300221. [DOI] [Google Scholar]
- 234.Liu C., Sawaya M.R., Cheng P.-N., Zheng J., Nowick J.S., Eisenberg D. Characteristics of amyloid-related oligomers revealed by crystal structures of macrocyclic β-sheet mimics. J. Am. Chem. Soc. 2011;133:6736–6744. doi: 10.1021/ja200222n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Kreutzer A.G., Spencer R.K., McKnelly K.J., Yoo S., Hamza I.L., Salveson P.J., Nowick J.S. A hexamer of a peptide derived from Aβ 16–36. Biochemistry. 2017;56:6061–6071. doi: 10.1021/acs.biochem.7b00831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Truex N.L., Wang Y., Nowick J.S. Assembly of peptides derived from β-sheet regions of β-amyloid. J. Am. Chem. Soc. 2016;138:13882–13890. doi: 10.1021/jacs.6b06000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Spencer R.K., Kreutzer A.G., Salveson P.J., Li H., Nowick J.S. X-ray crystallographic structures of oligomers of peptides derived from β2-microglobulin. J. Am. Chem. Soc. 2015;137:6304–6311. doi: 10.1021/jacs.5b01673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Wang Y., Kreutzer A.G., Truex N.L., Nowick J.S. A tetramer derived from islet amyloid polypeptide. J. Organomet. Chem. 2017;82:7905–7912. doi: 10.1021/acs.joc.7b01116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Ono K., Condron M.M., Teplow D.B. Structure-neurotoxicity relationships of amyloid β-protein oligomers. Proc. Natl. Acad. Sci. U. S. A. 2009;106:14745–14750. doi: 10.1073/pnas.0905127106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Hayden E.Y., Conovaloff J.L., Mason A., Bitan G., Teplow D.B. Preparation of pure populations of covalently stabilized amyloid β-protein oligomers of specific sizes. Anal. Biochem. 2017;518:78–85. doi: 10.1016/j.ab.2016.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Preston G.W., Radford S.E., Ashcroft A.E., Wilson A.J. Covalent cross-linking within supramolecular peptide structures. Anal. Chem. 2012;84:6790–6797. doi: 10.1021/ac301198c. [DOI] [PubMed] [Google Scholar]
- 242.Preston G.W., Wilson A.J. Photo-induced covalent cross-linking for the analysis of biomolecular interactions. Chem. Soc. Rev. 2013;42:3289–3301. doi: 10.1039/c3cs35459h. [DOI] [PubMed] [Google Scholar]
- 243.Bitan G., Teplow D.B. Rapid photochemical cross-linking - a new tool for studies of metastable, amyloidogenic protein assemblies. Acc. Chem. Res. 2004;37:357–364. doi: 10.1021/ar000214l. [DOI] [PubMed] [Google Scholar]
- 244.Preston G.W., Radford S.E., Ashcroft A.E., Wilson A.J. Analysis of amyloid nanostructures using photo-cross-linking: in situ comparison of three widely used photo-cross-linkers. ACS Chem. Biol. 2014;9:761–768. doi: 10.1021/cb400731s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Hayden E.Y., Teplow D.B. Continuous flow reactor for the production of stable amyloid protein oligomers. Biochemistry. 2012;51:6342–6349. doi: 10.1021/bi3007687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Fancy D.A., Kodadek T. Chemistry for the analysis of protein-protein interactions: rapid and efficient cross-linking triggered by long wavelength light. Proc. Natl. Acad. Sci. U. S. A. 1999;96:6020–6024. doi: 10.1073/pnas.96.11.6020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Fancy D.A., Denison C., Kim K., Xie Y., Holdeman T., Amini F., Kodadek T. Scope, limitations and mechanistic aspects of the photo-induced cross-linking of proteins by water-soluble metal complexes. Chem. Biol. 2000;7:697–708. doi: 10.1016/S1074-5521(00)00020-X. [DOI] [PubMed] [Google Scholar]
- 248.Bitan G., Lomakin A., Teplow D.B. Amyloid β-protein oligomerization: prenucleation interactions revealed by photo-induced cross-linking of unmodified proteins. J. Biol. Chem. 2001;276:35176–35184. doi: 10.1074/jbc.M102223200. [DOI] [PubMed] [Google Scholar]
- 249.Ono K., Ikeda T., Takasaki J., Yamada M. Familial Parkinson disease mutations influence α-synuclein assembly. Neurobiol. Dis. 2011;43:715–724. doi: 10.1016/j.nbd.2011.05.025. [DOI] [PubMed] [Google Scholar]
- 250.Banerjee S., Sun Z., Hayden E.Y., Teplow D.B., Lyubchenko Y.L. Nanoscale dynamics of amyloid β-42 oligomers as revealed by high-speed atomic force microscopy. ACS Nano. 2017;11:12202–12209. doi: 10.1021/acsnano.7b05434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Yamin G., Huynh T.-P.V., Teplow D.B. Design and characterization of chemically stabilized Aβ42 oligomers. Biochemistry. 2015;54:5315–5321. doi: 10.1021/acs.biochem.5b00318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Admasu A., Gudmundsdóttir A.D., Platz M.S., Watt D.S., Kwiatkowski S., Crocker P.J. A laser flash photolysis study of p-tolyl(trifluoromethyl)carbene. J. Chem. Soc. Perkin Trans. 1998;2:1093–1100. doi: 10.1039/a707586c. [DOI] [Google Scholar]
- 253.Smith D.P., Anderson J., Plante J., Ashcroft A.E., Radford S.E., Wilson A.J., Parker M.J. Trifluoromethyldiazirine: an effective photo-induced cross-linking probe for exploring amyloid formation. Chem. Commun. 2008:5728–5730. doi: 10.1039/b813504e. [DOI] [PubMed] [Google Scholar]
- 254.Horne J.E., Walko M., Calabrese A.N., Levenstein M.A., Brockwell D.J., Kapur N., Wilson A.J., Radford S.E. Rapid mapping of protein interactions using tag-transfer photocrosslinkers. Angew. Chem. Int. Ed. 2018;57:16688–16692. doi: 10.1002/anie.201809149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Chou C., Uprety R., Davis L., Chin J.W., Deiters A. Genetically encoding an aliphatic diazirine for protein photocrosslinking. Chem. Sci. 2011;2:480–483. doi: 10.1039/C0SC00373E. [DOI] [Google Scholar]
- 256.Dziuba D., Hoffmann J., Hentze M.W., Schultz C. A genetically encoded diazirine analogue for RNA–protein photo-crosslinking. ChemBioChem. 2020;21:88–93. doi: 10.1002/cbic.201900559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Kleiner R.E., Hang L.E., Molloy K.R., Chait B.T., Kapoor T.M. A chemical proteomics approach to reveal direct protein-protein interactions in living cells. Cell Chem. Biol. 2018;25:110–120. doi: 10.1016/j.chembiol.2017.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Birol M., Kumar S., Rhoades E., Miranker A.D. Conformational switching within dynamic oligomers underpins toxic gain-of-function by diabetes-associated amyloid. Nat. Commun. 2018;9:1312. doi: 10.1038/s41467-018-03651-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Tipping K.W., Karamanos T.K., Jakhria T., Iadanza M.G., Goodchild S.C., Tuma R., Ranson N.A., Hewitt E.W., Radford S.E. pH-induced molecular shedding drives the formation of amyloid fibril-derived oligomers. Proc. Natl. Acad. Sci. U. S. A. 2015;112:5691–5696. doi: 10.1073/pnas.1423174112. [DOI] [PMC free article] [PubMed] [Google Scholar]