Summary
Propionate and propionyl-CoA accumulation have been associated with the development of mitochondrial dysfunction. In this study, we show that propionate induces intestinal damage in zebrafish when fed a high-fat diet (HFD). The intestinal damage was associated with oxidative stress owing to compromised superoxide dismutase 2 (Sod2) activity. Global lysine propionylation analysis of the intestinal samples showed that Sod2 was propionylated at lysine 132 (K132), and further biochemical assays demonstrated that K132 propionylation suppressed Sod2 activity. In addition, sirtuin 3 (Sirt3) played an important role in regulating Sod2 activity via modulating de-propionylation. Finally, we revealed that intestinal oxidative stress resulting from Sod2 propionylation contributed to compositional change of gut microbiota. Collectively, our results in this study show that there is a link between Sod2 propionylation and oxidative stress in zebrafish intestines and highlight the potential mechanism of intestinal problems associated with high propionate levels.
Subject areas: Gastroenterology, molecular physiology, cell biology
Graphical abstract
Highlights
-
•
Propionate supplementation in high-fat diet induces intestinal damage
-
•
Propionate induces oxidative stress via Sod2 propionylation at 132 lysine site
-
•
Increased Sod2 propionylation is associated with reduced expression of Sirt3
-
•
Intestinal oxidative stress alters gut microbiota composition
Gastroenterology; Molecular physiology; Cell biology
Introduction
Propionate is a ubiquitous short-chain fatty acid (SCFA). It is a common food preservative widely used in the food industry and agriculture including many fish feeds (Rose, 2013). The maximum inclusion level is up to 0.5% (Rose, 2013). Meanwhile, propionate is one of the major fermentation products of the enteric microbiome (Koh et al., 2016). Thus human and animals are exposed to propionate derived from both endogenous and nutritional sources.
As an SCFA, propionate will cross the mitochondrial inner membrane and serve as precursor for generation of propionyl-CoA, which could enter the tricarboxylic acid (TCA) cycle for energy metabolism. Propionyl-CoA can also act as donor for lysine propionylation (Schonfeld and Wojtczak, 2016; Flavin and Ochoa, 1957; Chen et al., 2007; Cheng et al., 2009). Lysine propionylation is a common posttranslational modification (PTM) occurring in histones of eukaryotic cells (Chen et al., 2007; Cheng et al., 2009; Liu et al., 2009; Zhang et al., 2009). Similar to acetylation and butyrylation, histone propionylation is a marker of active chromatin (Kebede et al., 2017). Furthermore, the presence of lysine propionylation in non-histone proteins such as p53, p300, and cAMP response element-binding protein has been reported in 293T cells (Cheng et al., 2009). Although the role of lysine propionylation in non-histone proteins of eukaryotic cells is rarely reported, existing evidences in bacteria suggest a role similar to lysine acetylation in modulating protein function (Starai et al., 2002; Garrity et al., 2007).
Mitochondria are both the source and target of reactive oxygen species (ROS). Impaired mitochondrial respiration will increase ROS production (Bhatti et al., 2017). Excessive ROS accumulation causes oxidative stress, which in turn leads to mitochondrial dysfunction (Bhatti et al., 2017; Wei et al., 1998; Duchen, 2004; Pieczenik and Neustadt, 2007). Propionate may inhibit mitochondrial respiration due to significant propionyl-CoA accumulation (Matsuishi et al., 1991; Schwab et al., 2006). Propionyl-CoA accumulation inhibits the activity of mitochondrial respiration complexes, which implicates the involvement of protein propionylation (Lagerwaard et al., 2020). However, the potential links between protein propionylation and oxidative stress have never been studied.
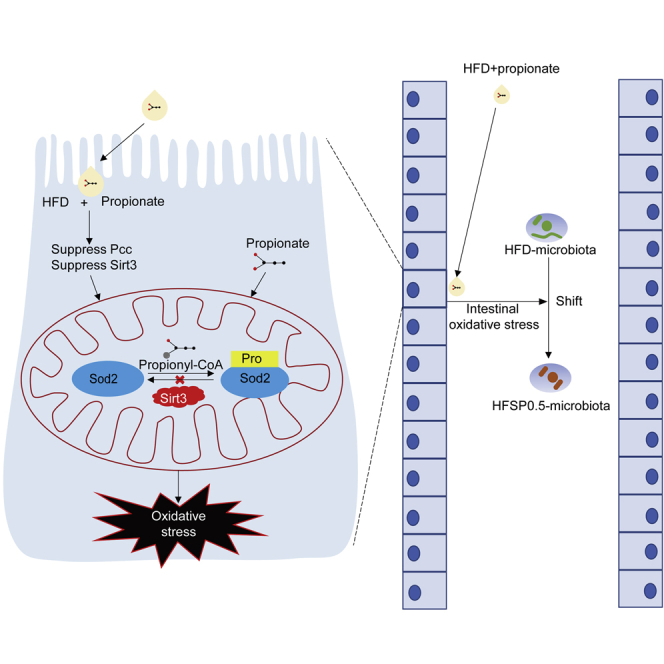
The intestinal epithelium is particularly prone to oxidative damage induced by luminal oxidants because it locates at the interface between the intestinal tissue and luminal environment (Circu and Aw, 2012). In the current study, we observed that propionate induced oxidative damage to zebrafish intestine when fed a high-fat diet (HFD). We established a plausible mechanism underlying propionate-induced intestinal oxidative damage that involved propionylation of superoxide dismutase 2 (Sod2) at the lysine 132 site (K132), which suppressed its activity and resulted in oxidative damage in the intestine. Furthermore, we found that increased propionylation of Sod2 was linked to reduced intestinal expression of sirtuin 3 (Sirt3) in zebrafish fed high fat plus propionate diet.
Results
Propionate supplementation in high-fat diet induces intestine damage in zebrafish
We established a propionate-feeding model by feeding one-month-old zebrafish a series of low- (6% oil) and high (16% oil)-fat diets with 0, 0.25, 0.5, and 1.0% propionate. Both LFD (low fat no propionate) and HFD (high fat no propionate) were tested, as HFD has been a very common basal dietary condition for both human and animals (Du et al., 2014; Zhai et al., 2014; Shapawi et al., 2014). The diets were designated LFD, LFSP0.25, 0.5, and 1.0 for low-fat propionate-supplemented diets and similarly HFD, HFSP0.25, 0.5, and 1.0 for the corresponding HFD. The he zebrafish were fed these diets for 2 weeks (Figure S1; Tables S1 and S2). After 2-week feeding trial, propionate supplementation in LFD showed no effect on zebrafish weight gain and feed efficiency (Figures S1A and S1B), and mortality of LFSP1.0-fed zebrafish was significantly decreased when challenged by Aeromonas veronii (A. veronii Hm091) (Figure S1C). In the context of HFD, propionate supplementation decreased weight gain and feed efficiency at both 0.5% and 1.0% when compared with zebrafish fed the HFD (Figure S1D). Feed efficiency also tended to be reduced, but was only significant for the HFSP0.5 group (Figure S1E). Mortality following challenge with A. veronii Hm091 tended to increase when propionate was added to the diet but was only statistically significant for the HFSP0.5-fed zebrafish (Figure S1F).
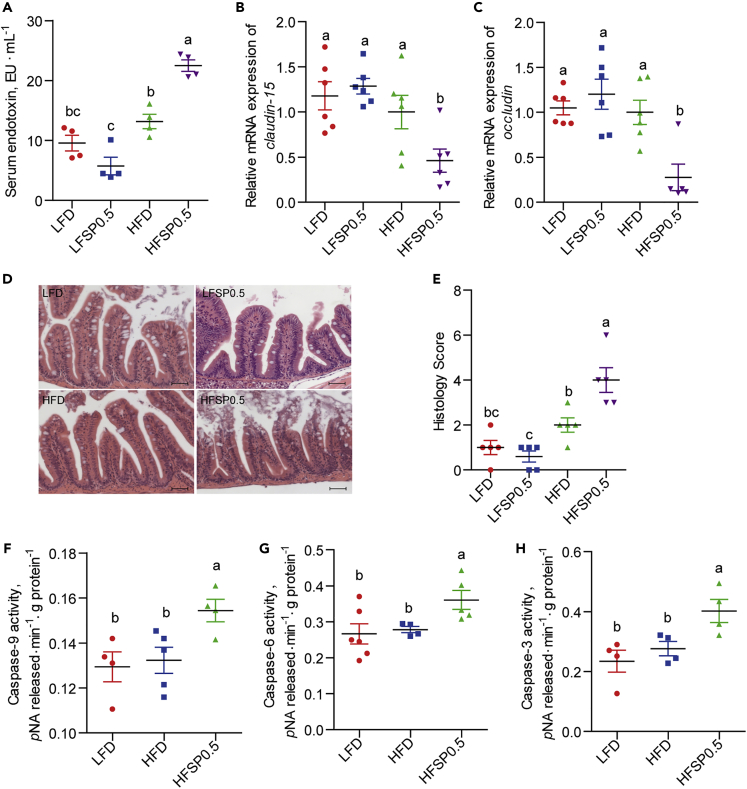
At the end of feeding, serum endotoxin level was increased by 70.8% (Figure 1A), and the relative mRNA expression of claudin-15 and occludin was significantly reduced in HFSP0.5-fed zebrafish as compared with the HFD group (Figures 1B and 1C). This suggests that 0.5% propionate-supplemented HFD might impair intestinal barrier function. Histopathologic examination of intestinal sections showed considerable morphological damage (i.e., breaches in the intestinal epithelium and injury to or loss of intestinal villi) in the HFSP0.5 group compared with those fed HFD (Figures 1D and 1E). These evidences together indicate that dietary propionate induces intestinal damage in the context of HFD. The activity of caspase-9, caspase-6, and caspase-3 were 16.7% (Figure 1F), 29.6% (Figure 1G), and 45.7% (Figure 1H) higher, respectively, in intestines of HFSP0.5-fed zebrafish compared with those fed the HFD, suggesting that a mitochondrial pathway of apoptosis was activated in the HFSP0.5 group. Intestinal caspase-8 and caspase-12 activities were similar in HFSP0.5 and HFD-fed zebrafish (Figures S2A and S2B). In this respect, it is worth noting that 0.5% propionate-supplemented LFD had little or no effect on damage markers such as serum endotoxin content, junctional proteins expression, histological score (Figures 1A–1E), or intestinal caspase-9 and caspase-3 activities (Figure S2C). The results clearly indicate that 0.5% propionate might only cause intestinal damages when accompanied by HFD. From this point, comparisons were only made for the selected groups, mainly LFD, HFD, and HFSP0.5. Thus the following studies aimed to study the mechanisms behind the negative effects of propionate on the intestine of zebrafish fed HFD.
Figure 1.
Propionate induces intestinal damage in the context of high-fat diet
(A) Serum endotoxin in LFD-, LFSP0.5-, HFD-, and HFSP0.5-fed zebrafish at the end of the 2-week feeding trial (n = 4).
(B and C) The relative mRNA expression of (B) claudin-15 and (C) occludin in the intestine of LFD-, LFSP0.5-, HFD-, and HFSP0.5-fed zebrafish at the end of 2-week feeding trial (n = 5–6).
(D) Representative histopathologic image of H&E-stained intestinal sections. The scale bar, 50 μm.
(E) Histological score measuring the severity of the intestinal damage of zebrafish (n = 5).
(F–H) (F) Caspase-9, (G) caspase-6, and (H) caspase-3 activities in the intestine of 1-month-old zebrafish fed the LFD, HFD, or HFSP0.5 for 2 weeks (n = 4–6).
Values are means ± SEM. Means without a common letter are significantly different (p < 0.05). Duncan's test. LFD, low-fat diet; LFSP0.5, 0.5% propionate-supplemented LFD; HFD, high-fat diet; HFSP0.5, 0.5% propionate-supplemented HFD.
See also Figure 1, Figure 2, Figure 3, Figure 4, Figure 5, Figure 6, Figure 7, Figure 8and S2.
Propionate induces intestinal oxidative stress in zebrafish fed high-fat diet
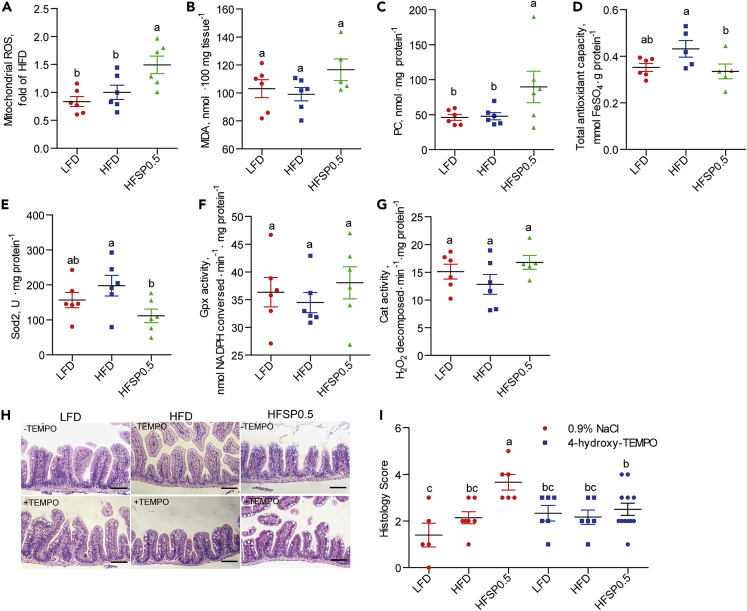
Analysis of intestinal oxidative stress markers showed that mitochondrial ROS (Figure 2A), malonaldehyde (MDA) (Figure 2B), and protein carbonyl (PC) content (Figure 2C) increased by 49.1%, 17.7%, and 87.5%, respectively, in zebrafish fed HFSP0.5 when compared with those maintained on HFD, suggesting the induction of intestinal oxidative stress of dietary propionate in the context of HFD. In contrast, zebrafish fed LFSP0.5 showed similar intestinal ROS to the counterparts fed LFD (Figure S3). The total antioxidant capability (T-AOC) (Figure 2D) and Sod2 activity (Figure 2E) in intestines of HFSP0.5-fed zebrafish were reduced by 22.4% and 43.5%, respectively, as compared with that in HFD-fed zebrafish. However, there were no dietary effects on the activity of other intestinal antioxidant enzymes, such as glutathione peroxidase (Gpx) (Figure 2F) and catalase (Cat) (Figure 2G). Furthermore H&E-stained intestinal sections of HFSP0.5-fed zebrafish administrated 4-hydroxy-TEMPO, an SOD mimic, showed less damage than saline-administered control (Figures 2H and 2I). Taken together, these results indicate that the intestine-damaging effect of HFSP0.5 was due to impaired T-AOC and oxidative stress, which may be at least partially attributable to the inhibition of Sod2 activity.
Figure 2.
Propionate induces intestinal oxidative stress in the context of high-fat diet
(A–C) Intestinal biomarkers for oxidative stress in 1-month-old zebrafish fed the LFD, HFD, or HFSP0.5 for 2 weeks, including (A) mitochondrial ROS, (B) MDA, and (C) PC.
(D) Intestinal total antioxidant capability in 1-month-old zebrafish fed LFD, HFD, or HFSP0.5 for 2 weeks.
(E–G) Intestinal antioxidant enzymes in 1-month-old zebrafish fed the LFD, HFD, or HFSP0.5 for 2 weeks, including (E) Sod2, (F) Gpx, and (G) Cat.
(H) Representative histopathologic images of H&E-stained intestinal sections in zebrafish intraperitoneally injected with 4-hydroxy-TEMPO, a membrane-permeable radical scavenger. The scale bar, 50 μm.
(I) Histological score measuring the severity of the intestinal damage of zebrafish intraperitoneally injected with 4-hydroxy-TEMPO.
Values are means ± SEM (n = 5–12). Means without a common letter are significantly different (p < 0.05). Duncan's test.
See also Figure S3.
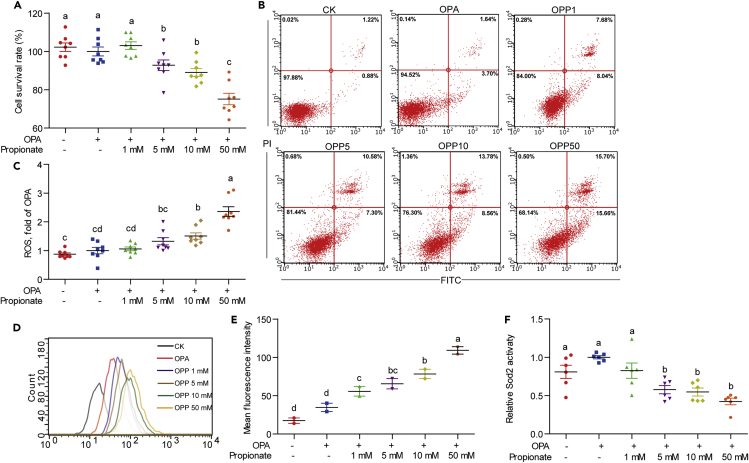
To further validate the effect of propionate on intestinal epithelium, we established an oleic acid-palmitic acid-propionate (OPP) model using the ZF4 cell line where the cells were treated with a mixture of 150 μM oleic acid and 50 μM palmitic acid (OPA) or a mixture of 150 μM oleic acid, 50 μM palmitic acid, and propionate (1, 5, 10 or 50 mM) (OPP1/5/10/50). In this model, OPA treatment acted as the high fat control. In the context of OPA, the effects of propionate on ZF4 cell viability, cell apoptosis, ROS, and Sod2 activity were evaluated. We observed a concentration-dependent decrease of cell survival rate (7.2%–24. 9%) (Figure 3A) and increase of cell apoptotic rate (15.7%–31.4%) (Figure 3B) after OPP treatment for 24 h. The relative cellular ROS levels examined by a fluorescence microplate reader increased by 1.1-, 1.3-, 1.5- and 2.4-fold in the OPP1, OPP5, OPP10, and OPP50 treatments, respectively, as compared with OPA-treated ZF4 cells (Figure 3C). The mean fluorescence intensity of the ROS signal acquired by flow cytometry was 60.3%, 89.0%, 125.5%, and 214.0% higher in OPP1-, OPP5-, OPP10-, and OPP50-treated ZF4 cells, respectively, compared with the OPA controls (Figure 3D and 3E). The Sod2 activity in OPP1-, OPP5-, OPP10-, and OPP50-treated ZF4 cells was inhibited by 17.5%, 42.2%, 45.3%, and 57.7% as compared with OPA controls (Figure 3F).
Figure 3.
ZF4 cell model validating the cytotoxicity of propionate and its role on Sod2 activity
(A) Cell survival rate in ZF4 cells treated with OPA or a mixture of OPA with increasing concentrations of propionate for 24 h (n = 8).
(B) Representative plot of cell apoptotic rate in ZF4 cells treated with OPA or a mixture of OPA with increasing concentrations of propionate (OPP1, OPP5, OPP10, and OPP50) for 24 h.
(C) Relative amount of cellular ROS acquired by a fluorescence microplate reader between ZF4 cells treated with OPA or a mixture of OPA with increasing concentrations of propionate for 24 h (n = 8).
(D) DCFH-DA histogram acquired by flow cytometry between ZF4 cells treated with OPA or mixtures of OPA with increasing concentrations of propionate for 24 h.
(E) Mean fluorescence intensity of DCFH-DA in ZF4 cells treated with OPA or a mixture of OPA with increasing concentrations of propionate for 24 h (n = 2).
(F) Sod2 activity in ZF4 cells treated with OPA or a mixture of OPA with increasing concentrations of propionate for 24 h (n = 6).
Values are means ± SEM. Means without a common letter are significantly different (p < 0.05). Duncan's test.
Propionate induces Sod2 propionylation at 132 lysine site
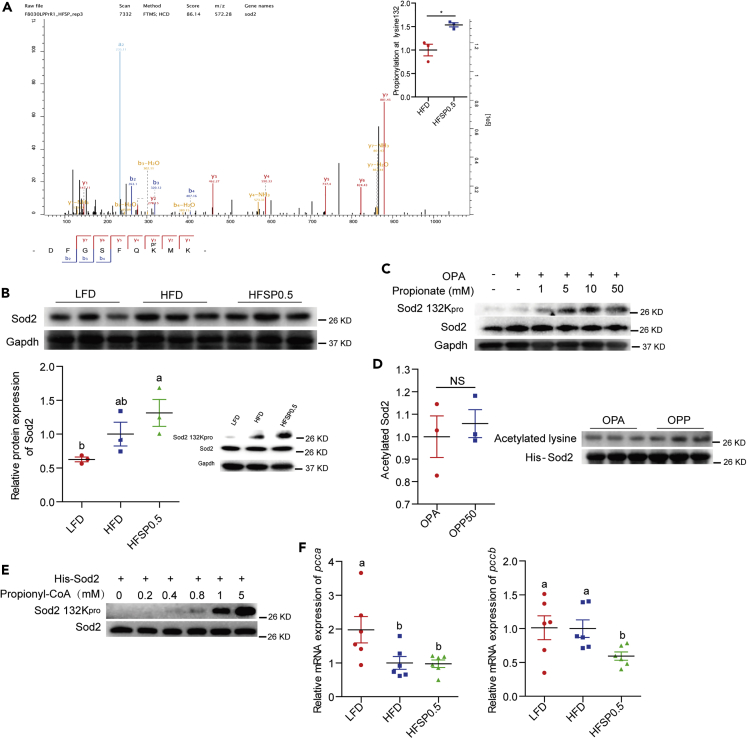
Propionate induced hyper-propionylation of intestinal mitochondrial proteins in zebrafish fed the HFSP0.5 (Figure S4; Table S3). HPLC-MS/MS-based proteomics analysis identified a Sod2 peptide, DFGSFQK+57.0901MN, with a mass shift of +57.0901 Da at the K132 in the intestine of HFSP0.5-fed zebrafish as compared with that in HFD-fed zebrafish (Figure 4A; Table S4). Despite the fact that Sod2 activity was reduced in the HFSP0.5 group when compared with HFD group, the total protein level of intestinal Sod2 was similar in both groups (Figure 4B). Immunoblotting using an anti-propionyl-K132 Sod2 antibody demonstrated increased propionylation of Sod2 K132 in the intestine from HFSP0.5-fed zebrafish as compared with HFD-fed zebrafish (Figure 4B). Consistently, the propionylation of Sod2 K132 was enhanced by exposure to OPP in ZF4 cells (Figure 4C). His-Sod2 overexpressed and immunoprecipitated from HEK293 cells were subject to HPLC-MS/MS analysis. Results showed that his-Sod2 overexpressed in OPA-treated HEK293 cells were acetylated in K70, K77, and K132, whereas that overexpressed in OPP50-treated HEK293 cells showed no significant alteration in acetylation level but acquired extra propionylation at K132 ( Figure 4D and Table 1). Moreover, his-Sod2 overexpressed in HEK293 cells could be readily propionylated at K132 by adding propionyl-CoA in a dose-dependent manner (Figure 4E), confirming that Sod2 K132 could be propionylated. Compared with LFD-fed zebrafish, the mRNA level of the pcca subunit was reduced in the intestines of zebrafish fed HFD and HFSP0.5 (Figure 4F), whereas the expression of the pccb subunit was similar in the LFD and HFD groups but reduced in HFSP0.5 group (Figure 4F).
Figure 4.
Propionate contributes to Sod2 propionylation at 132 lysine site in zebrafish fed high-fat diet
(A) HPLC-MS/MS spectra of an Sod2 peptide bearing propionylation (DFGSFQK+57.0901MN).
(B) A representative western blotting showing patterns of Sod2 expression and Sod2 propionylation at the K132 and quantification of intestinal Sod2 protein level in zebrafish fed LFD, HFD, or HFSP0.5 for 2 weeks (n = 3).
(C) A representative western blotting showing patterns of Sod2 expression and Sod2 propionylation at the K132 in ZF4 cells treated with OPA or OPP.
(D) Western blotting showing the pattern of his-Sod2 acetylation purified from OPA- or OPP-treated HEK293 cells (n = 3).
(E) A representative western blotting showing the pattern of his-Sod2 propionylation at K132 incubated with the indicated concentrations of propionyl-CoA.
(F) The mRNA expression of genes encoding subunits of intestinal Pcc, an enzyme catalyzing the carboxylation of propionyl-CoA, in zebrafish fed HFD or HFSP0.5 for 2 weeks (n = 6).
Values are means ± SEM. A and D were analyzed by Student's t test, ∗, p < 0.05, NS, not significant. B and F were analyzed by Duncan's test and means without a common letter are significantly different (p < 0.05).
See also Figure S4.
Table 1.
Modified lysine site in purified his-Sod2
| Group | Modification type | Peptide modified | Site |
|---|---|---|---|
| OPA | Acetylation | YQEALAKAcGDVTTQVSLQPALK | K77 |
| HHATYVNNLNVTEEKAcYQEALAK | K70 | ||
| DFGSFQKAcMK | K132 | ||
| OPP50 | Acetylation | YQEALAKAcGDVTTQVSLQPALK | K77 |
| HHATYVNNLNVTEEKAcYQEALAK | K70 | ||
| DFGSFQKAcMK | K132 | ||
| Propionylation | DFGSFQKProMK | K132 |
Ac, acetylation site; Pro, propionylation site. K, lysine; OPA, mixture of 150 μM oleic acid and 50 μM palmitic acid; OPP50, mixture of 150 μM oleic acid, 50 μM palmitic acid, and 50 mM propionate.
Sod2 propionylation at 132 lysine site accounts for cellular ROS increase
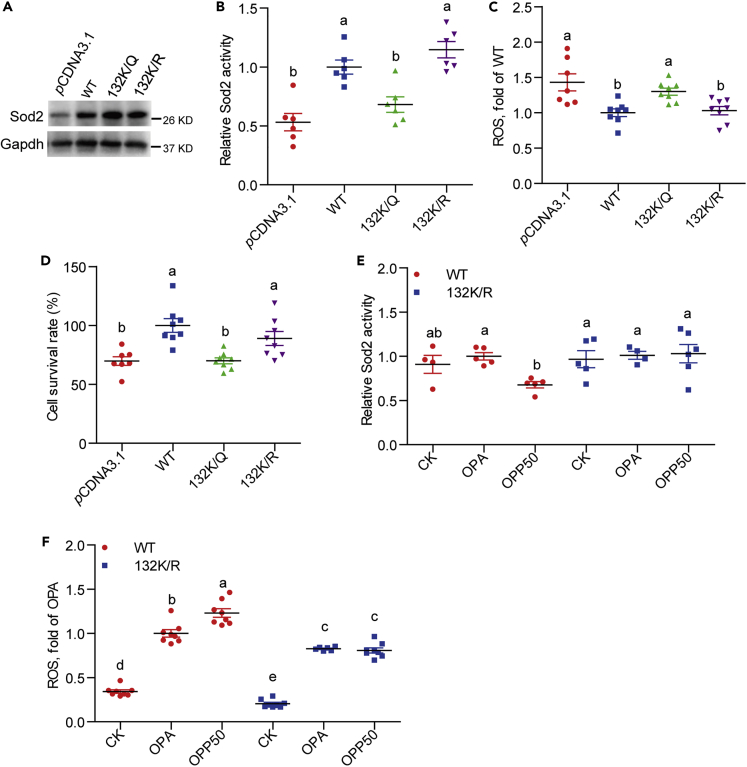
To determine whether propionylation at K132 compromises the activity of Sod2, we generated plasmids expressing mutated zebrafish flag-Sod2 in which K132 was substituted by arginine (R, conserves the positive charge) or glutamine (Q, mimics lysine propionylation) (K132R/Q). The plasmids were then transfected into ZF4 cells with endogenous Sod2 knock-down. SiRNA (mixture of sod2-57 and sod2-412) targeting sod2 was used to reduce its expression in ZF4 cells, and scrambled siRNA was used as a negative control (Figure S5 and Table S5). This resulted in reduced cell viability and elevated ROS (Figure S6). The results showed that overexpression of WT Sod2 and Sod2 K132R/Q compensated the protein content of Sod2 (Figure 5A). Compared with the ZF4 cells transfected with WT Sod2, cells transfected with the Sod2 K132R mutant had similar Sod2 activity, ROS level, and cell viability (Figures 5B–5D), whereas cells transfected with Sod2 K132Q mutant had a 31.9% reduction in Sod2 activity (Figure 5B), 30.0% elevation in ROS level (Figure 5C), and 30.0% inhibition in cell viability (Figure 5D). These results indicated that K132 is a key site for Sod2 activity, and propionylation at K132 compromises the Sod2 activity, leading to enhanced ROS. Moreover, ZF4 cells overexpressed Sod2 K132R (without Sod2 knock-down), maintained the Sod2 activity (Figure 5E), and prevented increase in cellular ROS (Figure 5F) after OPP50 treatment when compared with cells overexpressed with WT Sod2. This supports the finding that Sod2 K132 propionylation reduces Sod2 activity, which leads to enhanced ROS under high fat plus propionate conditions. Collectively, these results indicate that propionylation of Sod2 at K132 inhibits Sod2 activity and accounts for cellular ROS accumulation.
Figure 5.
Sod2 propionylation at 132 lysine site accounts for cellular ROS increase
(A) A representative western blotting showing that overexpression of WT Sod2 and Sod2 K132R/Q compensated Sod2 level in ZF4 cells (with endogenous Sod2 knockdown).
(B–D) (B) Sod2 activity, (C) ROS level, and (D) cell survival rate in ZF4 cells transfecting with WT Sod2 or Sod2 K132R/Q mutants (with endogenous Sod2 knockdown) (n = 6–8).
(E) Sod2 activity and (F) ROS level in ZF4 cells treated with OPA or OPP (50 mM propionate), which were transfected with WT Sod2 and Sod2 K132R in advance (without Sod2 knockdown) (n = 4–8).
Values are means ± SEM. Means without a common letter are significantly different (p < 0.05). Duncan's test. CK, 5% BSA.
See also Figures S5 and S6.
Inhibition of Sirt3 promotes Sod2 propionylation
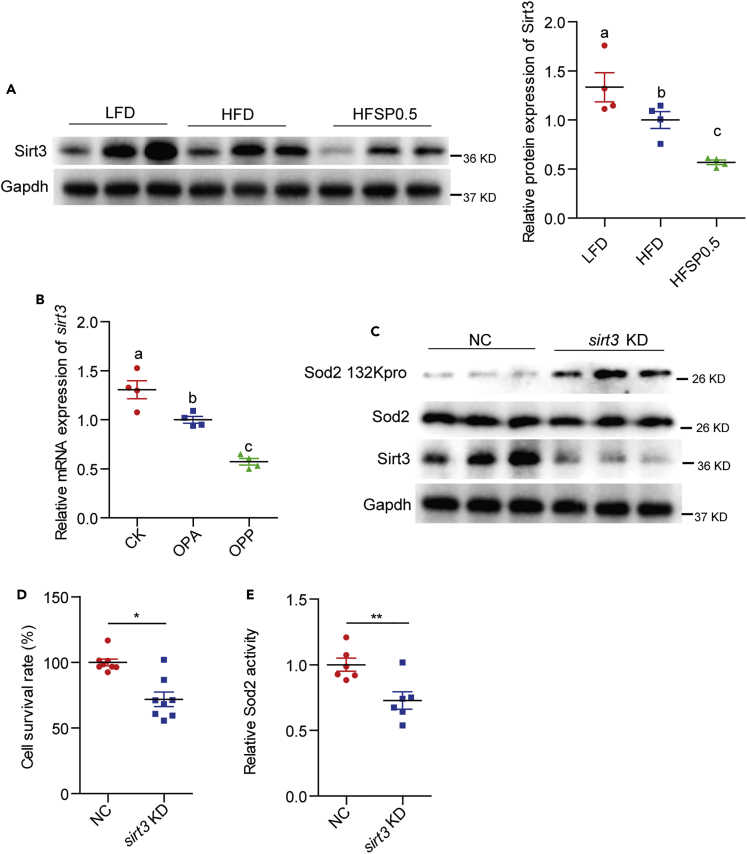
Recent studies have shown that the sirtuin family of deacetylases have de-propionylation activity (Bheda et al., 2011). We evaluated the expression of genes encoding sirtuins in zebrafish intestine among zebrafish fed HFD and HFSP0.5. The relative expression of mRNA encoding intestinal sirt3 was reduced by 48.1% in zebrafish fed HFSP0.5 as compared with those fed HFD (Figure S7A). This observation was confirmed by immunoblotting with an anti-Sirt3 antibody, which showed a clear reduction in Sirt3 protein level in the intestine of HFSP0.5-fed zebrafish as compared with HFD-fed counterparts (Figures 6A and S7B). Moreover, the relative mRNA expression of sirt3 in ZF4 cells treated with OPP50 was 42.8% lower than that in OPA-treated cells (Figure 6B). To identify whether Sirt3 reduction was associated with propionylation of intestinal Sod2 at the K132 site, we knocked down sirt3 with siRNA (mixture of sirt3-391, sirt3-755 and sirt3-885) in ZF4 cells (Figure S8 and Table S5). Immunoblotting with anti-propionyl-K132 Sod2 antibody showed that knockdown of Sirt3 increased propionylation of Sod2 at K132 (Figure 6C). In agreement with increased propionylation of Sod2 at K132 in sirt3-knockdowned ZF4 cells, cell viability and Sod2 activity were decreased by 28.1% and 27.2%, respectively (Figures 6D and 6E). Together, these results indicate that Sirt3 plays an important role in regulating Sod2 activity via modulating propionylation at K132.
Figure 6.
Inhibition of Sirt3 promotes Sod2 propionylation
(A) A representative western blotting showing Sirt3 expression and quantification of intestinal Sirt3 protein level in zebrafish fed LFD, HFD, or HFSP0.5 for 2 weeks (n = 4).
(B) The mRNA expression of sirt3 in ZF4 cells treated with OPA or OPP50 for 24 h (n = 4).
(C) A representative western blotting showing the propionylation of Sod2 at the 132 lysine site in ZF4 cells upon sirt3 knockdown.
(D) Cell survival rate in ZF4 cells with sirt3 knockdown (n = 8).
(E) Sod2 activity in ZF4 cells with sirt3 knockdown (n = 6).
Values are mean ± SEM. A and B were analyzed by Duncan's test, and means without a common letter are significantly different (p < 0.05). D and E were analyzed by Student's t test, ∗, p < 0.05, ∗∗, p < 0.01.
See also Figures S7 and S8.
Intestinal oxidative stress induced by Sod2 propionylation disturbs gut microbiota composition
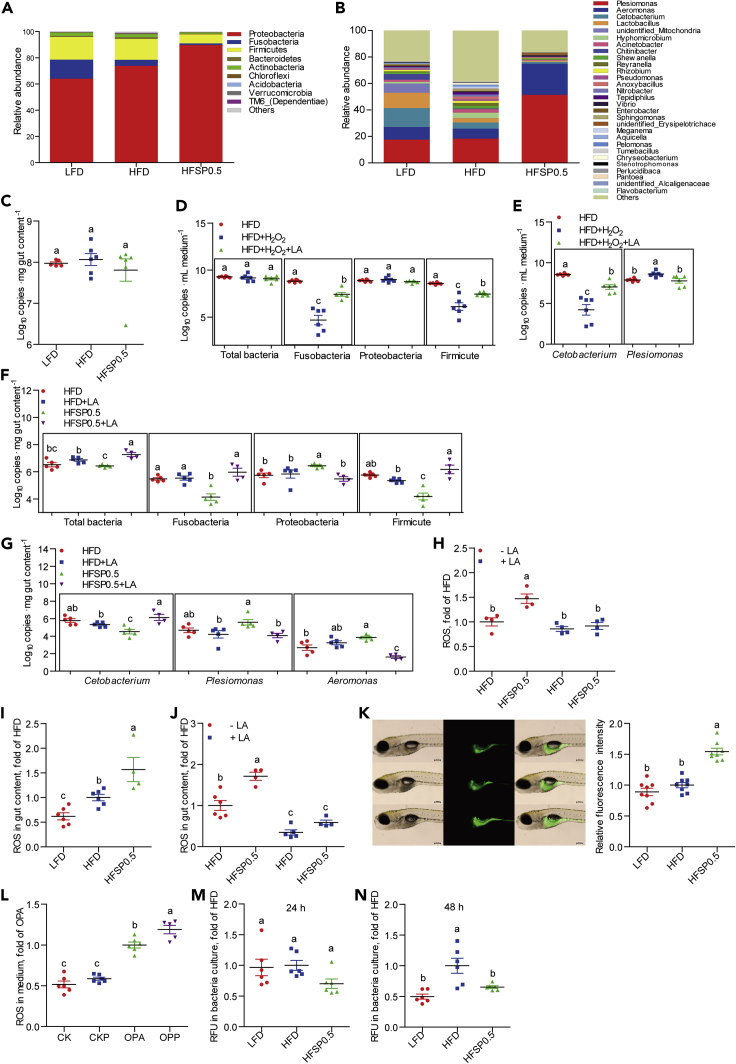
The composition of gut microbiota was assessed via 16S rRNA gene sequencing. In HFD-fed zebrafish, the relative abundance of Proteobacteria, Fusobacteria, and Firmicutes were 73.8%, 4.7%, and 15.9%, respectively (Figure 7A and Table 2), whereas it changed to 89.7%, 1.3%, and 6.8%, respectively, in HFSP0.5-fed zebrafish (Figure 7A and Table 2). The relative abundance of Plesiomonas in HFD-fed zebrafish was 18.3% (Figure 7B and Table 3), which increased to 51.4% in HFSP0.5-fed zebrafish (Figure 7B, Table 3). Although the relative abundance of Cetobacterium was 4.6% in HFD-fed zebrafish, it was reduced to only 1.2% in HFSP0.5-fed zebrafish (Figure 7B, Table 3). The total bacterial counts were comparable in gut contents from zebrafish fed HFD and HFSP0.5 (Figure 7C). These results indicate that Proteobacteria and Plesiomonas are enriched in the intestine of HFSP0.5-fed zebrafish, whereas the abundance of Firmicutes and Cetobacterium are reduced.
Figure 7.
Alteration of gut microbiota is linked to intestinal oxidative stress induced by propionate supplementation in HFD
(A) The composition of gut microbiota at phylum level in 1-month-old zebrafish fed the LFD, HFD, or HFSP0.5 (n = 6).
(B) The composition of gut microbiota at genus level in 1-month-old zebrafish fed the LFD, HFD, or HFSP0.5 (n = 6).
(C) The number of total bacteria in gut content collected from 1-month-old zebrafish fed the LFD, HFD, or HFSP0.5 (Log10 16S rDNA gene copies . mg gut content−1) (n = 6).
(D) The number of total bacteria, Fusobacteria, Proteobacteria, and Firmicutes after incubation in GAM with or without H2O2 for 48 h (Log10 16S rDNA gene copies . mL medium−1) (n = 6).
(E) The number of Cetobacterium and Plesiomonas after incubation in GAM with or without H2O2 for 48 h (n = 6).
(F) The number of total bacteria, Fusobacteria, Proteobacteria, and Firmicutes in gut content collected from 1-month-old zebrafish fed HFSP0.5 with or without supplementation of LA (Log10 16S rDNA gene copies . mg gut content−1) (n = 4–5).
(G) The numbers of Cetobacterium, Plesiomonas, and Aeromonas in gut content collected from 1-month-old zebrafish fed HFSP0.5 with or without supplementation of LA (Log10 16S rDNA gene copies . mg gut content−1) (n = 4–5).
(H) ROS level in the intestine collected from zebrafish fed the HFD or HFSP0.5 supplemented with LA (n = 4).
(I) ROS level in the gut content collected from zebrafish fed the LFD, HFD, or HFSP0.5 (n = 4–6).
(J) ROS level in gut contents collected from zebrafish fed the HFD or HFSP0.5 supplemented with LA (n = 4–6).
(K) ROS pattern in GF zebrafish fed the LFD, HFD, or HFSP0.5 for 1 week. The scale bar, 200 μm (n = 8).
(L) ROS level in the medium of ZF4 cells treated with OPA or OPP (50 mM propionate) for 24 h (n = 6).
(M and N) ROS generated by in vitro cultured gut microbiota derived from LFD-, HFD-, or HFSP0.5-fed zebrafish at (M) 24 h and (N) 48 h (n = 6).
Values are means ± SEM. Means without a common letter are significantly different (p < 0.05). Duncan's test.
Table 2.
The predominant gut bacterial phylum in zebrafish fed the LFD, HFD, or HFSP0.5 for 4 weeks based on V3–V4 sequences
| Phylum (%) | LFD | HFD | HFSP0.5 |
|---|---|---|---|
| Proteobacteria | 64.1c±5.9b | 73.8 ± 3.1b | 89.7 ± 1.8a |
| Fusobacteria | 14.5 ± 3.8a | 4.7 ± 1.9b | 1.3 ± 0.5b |
| Firmicutes | 17.3 ± 3.5a | 15.9 ± 2.7a | 6.8 ± 1.3b |
| Bacteroidetes | 0.6 ± 0.2a | 1.3 ± 0.6a | 0.6 ± 0.2a |
| Actinobacteria | 2.5 ± 0.6a | 2.3 ± 0.5a | 1.0 ± 0.4a |
Values are expressed as the mean ± SEM, n = 6 biological replicates. Means marked with different letters represent statistically significant results (p < 0.05), whereas the common letter corresponds to results with no statistically significant differences. Duncan's test.
Table 3.
The predominant gut bacterial genus in zebrafish fed the LFD, HFD, or HFSP0.5 for 4 weeks based on V3–V4 sequences
| Genus (%) | LFD | HFD | HFSP0.5 |
|---|---|---|---|
| Plesiomonas | 17.5 ± 6.9b | 18.3 ± 6.9b | 51.4 ± 7.2a |
| Aeromonas | 9.6 ± 2.4a | 7.6 ± 2.5a | 23.5 ± 2.6a |
| Cetobacterium | 14.3 ± 3.8a | 4.6 ± 4.1b | 1.2 ± 4.2b |
| Lactobacillus | 11.5 ± 2.7a | 3.2 ± 3.3b | 0.6 ± 3. 7b |
| Hyphomicrobium | 1.3 ± 0.3b | 4.0 ± 0.3a | 0.4 ± 0.4b |
| Acinetobacter | 1.6 ± 0.9ab | 2.6 ± 0.4a | 0.8 ± 0.1b |
| Chitinibacter | 4.3 ± 4.0a | 0.5 ± 4.0a | 0.1 ± 0.1a |
Values are expressed as the mean ± SEM, n = 6 biological replicates. Means marked with different letters represent statistically significant results (p < 0.05), whereas the common letter corresponds to results with no statistically significant differences. Duncan's test.
To examine the contribution of oxidative stress to the altered composition of gut microbiota, we cultured gut microbiota isolated from HFD-fed zebrafish in GAM containing 2 mmol/L H2O2 for 48 h and evaluated the effect of H2O2 on gut microbiota alteration. The results showed that the number of Fusobacteria, Firmicutes, and Cetobacterium were dramatically reduced (Figures 7D and 7E), whereas the number of Plesiomonas increased (Figures 7D and 7E). Adding 0.5 mg/mL of the antioxidant lipoic acid (LA) to the medium partially recovered the number of Fusobacteria, Firmicutes, and Cetobacterium and restricted the growth of Plesiomonas (Figures 7D and 7E). These results indicate that oxidative stress contributes to the compositional change of gut microbiota in an ex vivo culture system.
To study the transfer value of oxidative stress to gut microbiota in vivo, we performed qPCR analysis to determine the number of Proteobacteria and Plesiomonas in gut content collected from zebrafish fed the HFSP0.5 with or without LA. The results showed that the number of Proteobacteria and Plesiomonas were significantly reduced when zebrafish were fed the HFSP0.5 supplemented LA as compared with the HFSP0.5 only (Figures 7F and 7G). Moreover, the ROS accumulation in intestine was significantly reduced by LA supplementation (Figure 7H). These results indicate that oxidative stress induced by HFSP0.5 may elevate the abundance of Proteobacteria and alter the composition of gut microbiota.
We observed that ROS in gut content from HFSP0.5 zebrafish was 56.7% higher than that in HFD-fed zebrafish (Figure 7I). Consistently, the ROS accumulation in the gut content was significantly reduced when the HFSP0.5 was supplemented with LA (Figure 7J). Here, the probe dichlorofluorescin (DCFH-DA) also gave the same fluorescence pattern in germ-free (GF) zebrafish fed HFD and HFSP0.5 (Figure 7K). Similarly, the ROS level in the medium of ZF4 cells treated with OPP50 for 24 h was 19.0% higher than that in OPA-treated cells (Figure 7L). However, in vitro cultured gut microbiota derived from HFSP0.5-fed zebrafish generated similar ROS level to those fed HFD at 24 h (Figure 7M) and decreased at 48 h (Figure 7N). These results indicate that intestinal oxidative stress is derived from the intestinal tissue rather than from the microbiota. This appears to form an oxidative microenvironment in gut, leading to altered microbiota.
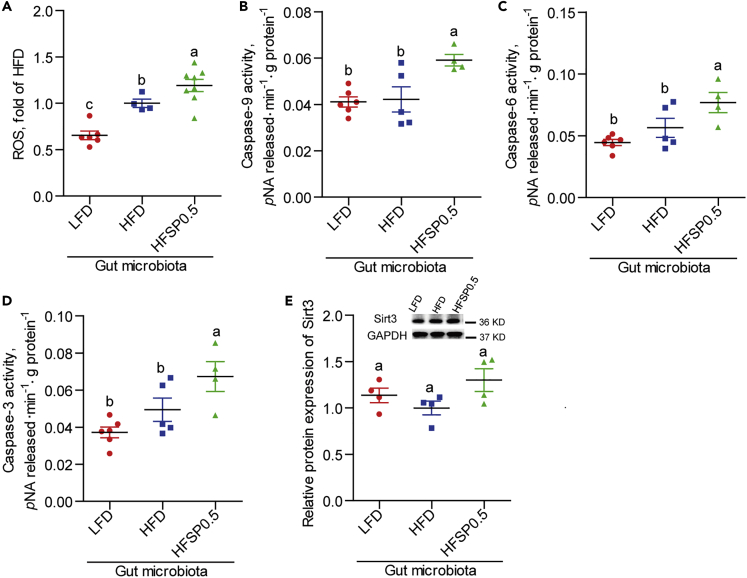
To further investigate the role of microbiota, we transferred gut microbiota from zebrafish fed LFD, HFD, and HFSP0.5 into GF zebrafish and assayed for ROS and caspase activity. The gut microbiota from HFSP0.5-fed zebrafish induced significant elevation of ROS (Figure 8A) and higher caspase-9, caspase-6, and caspase-3 activities as compared with that from HFD-fed zebrafish (Figures 8B–8D). These results indicate that gut microbiota from HFSP0.5-fed zebrafish can activate the mitochondrial death pathway. Gut microbiota from zebrafish fed HFSP0.5 did lead to a moderate, but non-significant, increase in Sirt3 expression (Figure 8E). These results suggest that the role of gut microbiota might be independent of Sirt3.
Figure 8.
Gut microbiota indirectly activate mitochondrial death pathway
(A) The ROS level in GF zebrafish colonized with gut microbiota from 1-month-old zebrafish fed the LFD, HFD, or HFSP0.5 (n = 4–8).
(B–D) (B) Caspase-9, (C) caspase-6, and (D) caspase-3 activities in GF zebrafish colonized with gut microbiota from 1-month-old zebrafish fed the LFD, HFD, or HFSP0.5 (n = 4–6).
(E) Quantification of Sirt3 protein level in GF zebrafish transferred with gut microbiota from 1-month-old zebrafish fed the LFD, HFD, or HFSP0.5 (n = 4). Values are means ± SEM. Means without a common letter are significantly different ( p < 0.05). Duncan's test.
Discussion
In this study, we conclude that dietary propionate induces oxidative stress and consequent intestinal damage in zebrafish when fed HFD. These damages appeared to be attributed to posttranslational propionylation of the mitochondrial antioxidant enzyme Sod2 and reduced expression of Sirt3, which could function as a de-propionylation agent.
Oxidative stress and mitochondrial abnormality are associated with impaired intestinal health. For instance, they have important pathogenetic implications for inflammatory bowel disease (IBD) (Palucka, 2007; Novak and Mollen, 2015). The gastrointestinal (GI ) tract injury effect of nonsteroidal anti-inflammatory drugs (NSAID) is associated with disruption of mitochondrial structure and function (Rafi, 1998; Somasundaram et al., 1997; Kyle, 2014), whereas antioxidant drugs, such as sulfasalazine, are known to have beneficial effects in the treatment of IBD (Bhattacharyya et al., 2014). Oxidative stress is caused by ROS accumulation due to an imbalance in ROS production and removal. Increased ROS production will result in mitochondrial dysfunction (Wei et al., 1998; Duchen, 2004; Pieczenik and Neustadt, 2007). SOD2 is the primary mitochondrial matrix-located enzyme for ROS clearance (Spitz and Oberley, 1989; Zelko et al., 2002). Deficiency of SOD2 activity will impair antioxidant defense and increase oxidative stress, eventually leading to mitochondrial dysfunction (Ramachandran et al., 2011). SOD1, another dismutase, will also remove ROS, but it localizes to the cytoplasm and the intermembrane space of the mitochondria (Fukai and Ushio-Fukai, 2011). SOD2 is more important to prevent oxidative damage than SOD1 (Fukui and Zhu, 2010; Matzuk et al., 1998; Asimakis et al., 2002). In this study, supplementing propionate to HFD increased ROS production and reduced antioxidant capability and Sod2 activity. Moreover, an SOD mimic, 4-hydroxy-TEMPO, relieved the intestinal damage induced by HFSP0.5. These results confirmed that the intestinal damage induced by HFSP0.5 is, at least in part, mediated by oxidative stress resulting from Sod2 inactivation.
Dietary propionate supplementation leads to intestinal Sod2 propionylation at K132. Propionylation of Sod2 was also verified in ZF4 cells treated with OPP and in Sod2 purified from HEK293 cells. In vitro incubation with propionyl-CoA leads to propionylation of his-tagged Sod2, suggesting that Sod2 propionylation was mediated by propionyl-CoA, which can be generated from propionate (Schonfeld and Wojtczak, 2016). Decreased transcription of Pcc subunits in both HFD and HFSP0.5 groups reflects blocked propionyl-CoA metabolism, as PCC is the essential enzyme for catalyzing the carboxylation of propionyl-CoA to methylmalonyl-CoA, which ultimately enters the succinyl-CoA pool and the TCA cycle (Wongkittichote et al., 2017; Xu et al., 2018). Therefore, the inhibitory effect of HFD on pcc subunit expression may interrupt propionyl-CoA metabolism and increase the local concentration of propionyl-CoA in mitochondria, and this could reduce the tolerance of intestine tissue or cells to exogenous propionate.
Recent studies have shown that PTM of SOD2, such as acetylation at 53, 68, 89, 122, and 130 lysine (Qiu et al., 2010; Tao et al., 2010; Lu et al., 2015; Zhang et al., 2016), negatively regulates its antioxidant activity (Qiu et al., 2010; Tao et al., 2010). According to the alignment of amino acid sequences, the 132 lysine of zebrafish Sod2 (EMBL no. AY195857) is aligned to the 130 lysine of SOD2 in mice (AK002534), rats (BC070913), and humans (M36693) (Lin et al., 2009). Lysine propionylation is chemically similar to acetylation, but the extra methylene-group makes the modification bulkier and more hydrophobic (Bheda et al., 2011). The consequence of such modifications was convincingly demonstrated when the Sod2 activity was increased by the exchange of 132 lysine with arginine (to mimic the de-propionylated state) and reduced by the exchange of 132 lysine with glutamine (to mimic propionylation). Sod2 K132/R mutant can also prevent OPP-induced Sod2 inactivation and ROS accumulation. This shows that K132 is the main propionylated site that accounts for reduced Sod2 activity when exposed to high levels of propionyl-CoA concentration derived from exogenous propionate.
HFD and propionate-induced Sod2 K132 propionylation is accompanied by reduced Sirt3 expression in the intestine of zebrafish, as well as in ZF4 cells treated with OPP. SIRT3 has been suggested to possess de-propionylation activity. For example, the propionyl-lysine modification in propionyl-CoA synthetase introduced by bacterial Gcn-5-related N-acetyltransferase enzymes can be removed by human SIRT3 (Garrity et al., 2007). Moreover, the absence of SIRT3 will lead to a higher propionyl-lysine levels in mouse lenses (Nahomi et al., 2020). In this study, we showed that sirt3 knockdown increased propionylation of Sod2 at K132 and inhibited its antioxidant activity. This supports the hypothesis that Sirt3 participates in the regulation of Sod2 de-propionylation. Peroxisome-proliferator-activated receptor γ coactivator 1 α (PGC1a) is one of the few known regulators of SIRT3. It will induce SIRT3 expression by binding the ERR-binding element in the promoter region (Kong et al., 2010). The reported transcriptional repressors of SIRT3 include poly (ADP-ribose) polymerase 1 (PARP1) and transcriptional cofactor receptor-interacting protein 140 (RIP140), both of which contribute to oxidative stress and mitochondrial dysfunction (Yoon and Kim, 2016; Kim et al., 2020). In this study, the expression of pgc1α and err in zebrafish intestine and ZF4 cells showed no significant alteration in response to HFSP0.5 and OPP (50 mM) treatment (data not shown), suggesting that the reduction of sirt3 expression might be mediated by its transcriptional repressors. Further studies are needed to clarify these regulations of sirt3 expression in zebrafish fed HFD and propionate.
There is accumulating evidence demonstrating intricate relationships between host oxidative stress and gut microbiota (Qiao et al., 2013; Million et al., 2016). To some extent this probably reflects different tolerances to oxidative stress among anaerobic and aerotolerant bacteria (Liu et al., 2021), although other explanations may also be possible. In this study, we have shown that propionate supplementation to HFD leads to altered gut microbiota with an enrichment of Proteobacteria and Plesiomonas. We also conclusively showed that both conventional and GF zebrafish fed HFSP0.5 had increased luminal ROS production, which was confirmed with observations of elevated ROS level in the medium of ZF4 cells treated with OPP. Furthermore, in vitro and in vivo results showed that the alteration of gut microbiota was attributable to intestinal oxidative stress induced by propionate supplementation to HFD. The detailed mechanisms underlying the relationship between intestinal oxidative stress and gut microbiota remains to be elucidated. It is however quite clear that intestinal oxidative stress induced by exogenous propionate will alter luminal oxidative microenvironment and the gut microbiota.
In the present study, we used gut microbiota transfer to investigate the role of gut microbiota on zebrafish health. We found that transplanting microbiota from HFSP0.5-fed zebrafish to GF zebrafish led to increased ROS accumulation and activation of mitochondrial death pathway. Gut microbiota can stimulate mitochondrial production of ROS via release of various microbiota-associated molecular patterns (MAMPs), including lipopolysaccharides (LPS), flagellin, lipoteichoic acid, lipoprotein, or other toxins (Saint-Georges-Chaumet and Edeas, 2016). These MAMPs are recognized by the pattern recognition receptor (PRR) system in the host cell, which in turn induces ROS production via classical TLR and NLR pathways (Saint-Georges-Chaumet and Edeas, 2016; Emre and Nubel, 2010). Therefore, the alteration in microbiota structure might promote ROS production due to differential microbe-associated molecular patterns (MAMPs)-PRR signaling. Further studies are needed to identify the role of specific bacteria-deprived MAMPs associated with HFSP0.5-microbiota in promoting host ROS production.
Apart from Sod2, lysine propionylation was also observed in other proteins based on the global lysine propionylation analysis (Figure S4; Tables S3 and S4). The first global survey of lysine propionylation has been reported in Cyanobacteria (Yang et al., 2019). To our knowledge, our work was the first global propionylome analysis conducted in animals. The bioinformatics results showed that proteins involved in oxidative phosphorylation (OXPHOS) and ATP synthesis were enriched among the propionylated proteins in the intestine of HFSP0.5-fed zebrafish; this also appeared to be the case with proteins (KEEG pathway) of the TCA cycle. Among the proteins in TCA cycle, malate dehydrogenase 2 (Mdh2) and citrate synthase (Cs) were found to have a complex association with other mitochondrial proteins. This association may indicate other potential regulatory roles of lysine propionylation in mitochondrial metabolism that would act through modulating Mdh2 and Cs activity. These and other roles of global lysine propionylation on mitochondrial energy metabolism deserves further studies.
In propionic acidemia (PA), a human metabolic disorder, the accumulation of propionyl-CoA results in mitochondrial dysfunction and oxidative stress (de Keyzer et al., 2009; Gallego-Villar et al., 2013, 2016). Although studies suggest a role of protein propionylation and oxidative stress in the pathological mechanism of PA, information on possible relationships between mitochondrial protein propionylation and oxidative stress is lacking. Our results identified propionylated Sod2 as one candidate that can cause oxidative stress. It is not unlikely that a similar mechanism may apply to PA pathogenesis. More importantly, propionate is a widely used feed preservative. Our findings clearly indicate potential health hazard of supplementing dietary propionate to fish when combined with HFD formulations, which are very common for fish culture. Moreover, the results also suggest a novel and important aspect of propionate safety in the context of HFD for humans, especially for those with pre-existing intestinal health issues. These issues deserve further investigation in mammalian models.
Limitations of study
In this study, our data demonstrate that propionate supplementation to HFD induces intestinal oxidative stress via Sod2 K132 propionylation. The Sod2 propionylation is associated with decreased expression of Pcc and Sirt3, which leads to increased propionyl-CoA level and reduced depropionylation activity, respectively. However, the underlying mechanisms are not clear and deserves further investigation. Furthermore, our findings suggest potential risk of propionate as feed additive but cannot directly translate to food additive for humans, which requires evaluation in mammal models.
STAR★methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal Anti-Gapdh | Sigma | CAT# SAB2701826; |
| Rabbit polyclonal Anti-Sod2 | GeneTex | CAT#GTX124294; RRID:AB_11174816 |
| Rabbit polyclonal Anti-Sirt3 | Sigma | CAT#AV32388; RRID:AB_1856904 |
| Mouse monoclonal Anti-Mtco1 | Abcam | CAT#ab14705; RRID:AB_2084810 |
| Rabbit polyclonal Acetyl-Lysine antibody | Cell Signaling Technology | CAT#9441; RRID:AB_331805 |
| Rabbit polyclonal Anti-Sod2 K132pro | This paper | N/A |
| Bacterial and virus strains | ||
| Aeromonas veronii Hm091 | Pearl River Fisheries Research Institute | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| 4-Hydroxy-TEMPO | Sigma | CAT#176141 |
| Protease inhibitor cocktail | Cell Signaling Technology | CAT#5871 |
| AlarmaBlue cell viability reagent | Invitrogen | CAT#DAL1025 |
| Sodium oleate | Sigma | CAT#O7501 |
| Palmitic acid | Sigma | CAT#P5585 |
| Sodium propionate | Sigma | CAT#P1880 |
| DCFH-DA | Beyotime | CAT#S0033S |
| Propionyl coenzyme A sodium salt | zzstandard | CAT#ZL-21372 |
| PMSF | Beyotime | CAT#ST506 |
| Lipofectamine 3000 transfection reagent | Invitrogen | CAT#L3000008 |
| Lipofectamine RNAiMAX transfection reagent | Invitrogen | CAT#13778 |
| SYBR Green Supermix | TIANGEN | CAT#FP205 |
| Critical commercial assays | ||
| ToxinSensor Chromogenic LAL Endotoxin Assay Kit | Genscript | CAT#L00350 |
| Caspase-9 Activity Assay Kit | Beyotime | CAT#C1158 |
| Fluorometric Intracellular Ros Kit | Sigma | CAT#MAK143 |
| Lipid Peroxidation (MDA) Assay Kit | Sigma | CAT#MAK085 |
| Protein Carbonyl Content Assay Kit | Sigma | CAT#MAK094 |
| Total Antioxidant Capacity Assay Kit | Beyotime | CAT#S0116 |
| CuZn/Mn-SOD Activity Kit | Beyotime | CAT#S0103 |
| Annexin V-FITC Apoptosis Kit | Sigma | CAT#apoaf |
| QuikChange Site-Directed Mutagenesis Kit | Stratagene | CAT#200523 |
| Deposited data | ||
| Microbiota sequencing data | This paper | BioProject: PRJNA714780 |
| Experimental models: Cell lines | ||
| ZF4: zebrafish embryo | ATCC | CRL-2050 |
| Experimental models: Organisms/strains | ||
| Zebrafish: Tübingen strain | China Zebrafish Resource Center | CZ3 |
| Oligonucleotides | ||
| siRNA | This paper | Table S5 |
| Primers | This paper | Table S6 |
| Recombinant DNA | ||
| Plasmids: pCDHA3.1-flag-Sod2, pCDHA3.1-flag-Sod2132K/Q and pCDHA3.1-flag-Sod2 132K/R | This paper | N/A |
| Plasmid: pCDHA3.1-his-flag-Sod2 | This paper | N/A |
| Software and algorithms | ||
| Guava easyCyte Flow Cytometer software | Merck Millipore, Stafford, VA, USA | N/A |
| GraphPad Prism 5 | GraphPad Software Inc., San Diego, CA, USA | N/A |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Zhigang Zhou (email: zhouzhigang03@caas.cn).
Materials availability
Plasmids generated in this study are available from the lead contact upon reasonable request.
Data and code availability
Microbiota sequencing data in this study are available from the National Center for Biotechnology Information (NCBI) under accession number BioProject: PRJNA714780.
Experimental model and subject details
Animals and diets
All experimental and animal care procedures were approved by the Feed Research Institute of the Chinese Academy of Agricultural Sciences Animal Care Committee under the auspices of the China Council for Animal Care (Assurance No. 2016-AF-FRI-CAAS-001). Healthy, uniformly sized 1-month-old zebrafish (Tübingen strain) were divided into eight groups at random and fed the experimental diets (Tables S1 and S2) at the zebrafish facility of the Feed Research Institute of the Chinese Academy of Agricultural Sciences (Beijing, China) twice a day (9:00, 17:00) to apparent satiation for 2 weeks. For the study both sexes were used. During the feeding period, the rearing temperature was 25–28°C, the dissolved oxygen was >6.0 mg/L, the pH was 7.0–7.2, the nitrogen content was <0.50 mg/L, and the nitrogen content (as NO2) was <0.02 mg/L. All fish were anesthetized with tricaine methanesulfonate (MS222).
ZF4 cell line
The ZF4 cell line was purchased from American Type Culture Collection (Manassas, VA, USA), and cultured according to established protocols (Driever and Rangini, 1993). The medium (DMEM/F12) was obtained from Corning Inc. (New York, USA). Penicillin-Streptomycin solution were purchased from Sigma (St. Louis, MO, USA). Fetal bovine serum was purchased from Corning Inc. (New York, USA). Cells were kept in a cell incubator with 5% CO2 at 28°C.
Method details
Aeromonas veronii challenge assay
The Aeromonas strain used in this study was A. veronii Hm091 isolated by Pearl River Fisheries Research Institute (Guangzhou, China). A. veronii Hm091 was grown for 18 h at 37°C in LB medium. Three replicates of 10 zebrafish from each group were infected with Hm091 at 2 × 107 CFU/mL. For challenge, zebrafish were immersed in water containing the pathogen, and mortality was recorded for the 3 days following infection.
Serum endotoxin assay
Serum endotoxin levels were determined using the ToxinSensor Chromogenic LAL Endotoxin Assay Kit (Genscript, Jiangsu Province, China). In brief, serum samples were dispensed into endotoxin-free vials and then consecutively incubated with LAL and chromogenic substrate consecutively. After the reaction was stopped, absorbance of each sample was read at 545 nm. The serum level of endotoxin in adult zebrafish was expressed as endotoxin units per milliliter serum (EU/mL).
Examination of intestinal histopathology
The intestines of zebrafish were rinsed with sterilized PBS, fixed in 4% formalin solution, and embedded in paraffin. For histological analysis, the liver sections were prepared from the paraffin blocks stained with hematoxylin and eosin (H&E). Images were obtained under a microscope (Carl Zeiss) at a 200× magnification.
Detection of caspase activity
The activities of caspase-3, caspase-6, and caspase-9 were determined using an assay kit (Beyotime Biotechnology, Shanghai, China) according to the manufacturer's instructions. The optical density of the reaction product was examined at 405 nm. The enzyme activity is expressed as the rate of p-nitroaniline (pNA) released from the substrate per gram protein (μmol pNA released min−1 · g−1 protein).
ROS detection in intestinal mitochondria
Intestinal mitochondria were isolated by using a tissue mitochondria isolation kit (Beyotime Biotechnology, Shanghai, China). In brief, fresh intestine was collected from zebrafish and homogenized for 10 times in the isolation buffer using a glass homogenizer. The homogenate was then centrifugated at 1000 g for 10 min. The supernatant was then aspirated and further centrifugated at 11,000 g for 10 min. The precipitation was resuspended in the mitochondria storage buffer. The purity of isolated mitochondria was assessed according to Fernandez-Vizarra et al. (2006) (Figures S9A–S9D). Then isolated mitochondria were incubated with a fluorescent probe provided by the ROS kit (Sigma, USA) for 2 h in a 96-well flat-bottom plate. The fluorescence intensity was measured excitation 490 nm and emission 520 nm. The ROS level was expressed as the fold change compared with the HFD group. Rosup was applied as a positive control (Figure S9E).
Detection of oxidative parameters
Fresh intestine was homogenized in ice-cold MDA lysis buffer and centrifuged at 13,000 g for 10 min to remove insoluble material. Lipid peroxidation was determined by the reaction of malondialdehyde (MDA) with thiobarbituric acid using a lipid peroxidation assay kit according to the manufacturer's instructions (Sigma, USA). In brief, the supernatant was collected and incubated with the TBA solution at 95°C for 60 min to generate MDA-TBA adduct. The optical density of the MDA-TBA adduct was examined at 532 nm. Lipid peroxidation was expressed as MDA content per 100 milligram tissue (nmol · 100 mg tissue−1). Oxidation of proteins was determined by the formation of stable dinitrophenyl hydrazine adducts derived from protein carbonyl (PC) groups with 2, 4-dinitrophenylhydrazine using protein carbonyl content assay kit according to the manufacturer's instructions (Sigma, USA). The optical density was examined at 375 nm. Oxidation of proteins was expressed as PC content per milligram protein (nmol · mg protein−1).
Evaluation of total antioxidant capacity
Fresh intestine was homogenized in ice-cold PBS to release the antioxidants. The homogenate was then centrifugated at 12,000 g for 5 min and the supernatant collected for subsequent analysis. Total antioxidant capacity (T-AOC) was measured by the production of blue Ferric-tripyridyltriazine (Fe2+-TPTZ) resulting from the reduction of Fe3+ TPTZ complex under acidic conditions. The optical density was measured at 593 nm. T-AOC was defined as the production of FeSO4 per gram protein (mmol FeSO4 · g protein−1).
Evaluation of antioxidant enzyme activity
Fresh intestine was homogenized in ice-cold PBS, centrifugated at 12,000 g for 5 min, and the supernatant collected for analysis of enzyme activities. The activities of Sod2, glutathione peroxidase (Gpx), and catalase (Cat) were detected using a CuZn/Mn-SOD activity kit, a cellular GPX assay kit, and a catalase (CAT) assay kit (Beyotime Biotechnology, Shanghai, China), respectively. The SOD2 assay included the inhibitors A and B for Cu/Zn SOD (SOD1) (Zeng et al., 2019). Sod2 activity was measured as the inhibition of water soluble tetrazol salt (WST) reduction in a xanthine-xanthine oxidase system. The optical density was measured at 450 nm. Sod2 activity was expressed in units (U) and was the percent inhibition of WST reduction. The Sod2 activity in the intestine was expressed as the inhibition of WST reduction per milligram protein (U · mg protein−1). Relative Sod2 activity in ZF4 cell and zebrafish larva was expressed as fold change of indicated group. The Gpx activity was analyzed by monitoring NADPH consumption at 340nm using the Gpx detection buffer (3.125 mM NADPH, 3.75 mM GSH and glutathione reductase) and peroxide. The conversion of 1 mmol of NADPH to NADP+ by Gpx per 1 min at 25°C and pH8.0 was set as 1 U. The activity of Gpx was expressed as the conversion rate of NADPH to NADP+ per milligram protein (nmol NADPH conversed · min−1 · mg protein−1). The Cat activity was determined by incubating protein in the Cat detection buffer and H2O2. At the end of the reaction, the optical density was measured at 520 nm. One U was defined as the decomposition of 1 mmol H2O2 at 25°C and pH7.0. The activity of Cat was expressed as the rate of H2O2 decomposition per milligram protein (mmol H2O2 decomposed · min−1 · mg protein−1).
Intraperitoneal injection of 4-hydroxy-TEMPO
After 2-week feeding trial, ten zebrafish from LFD, HFD, and HFSP0.5 groups were respectively divided into two groups: (1) control zebrafish injected intraperitonealy (i.p.) with saline (0.9% NaCl); (2) zebrafish treated i.p. with 4-hydroxy TEMPO dissolved in saline (10 mg . kg b.w.−1) every other day. Intestines were collected at the sixth day for H&E staining.
Identification of lysine propionylated sites by HPLC-MS/MS
The intestine was ground by liquid nitrogen into cell powder and then transferred to a 5 mL centrifuge tube. After that, four volumes of lysis buffer (8 M urea, 1% protease inhibitor cocktail, 3 μM trichostation, 50 mM nicotinamide) were added to the cell powder, followed by sonication three times on ice using a high intensity ultrasonic processor. The remaining debris was removed by centrifugation at 12,000 g at 4°C for 10 min. Finally, the supernatant was collected, and the protein concentration was determined with BCA kit according to the manufacturer's instructions. For digestion, the protein solution was reduced with 5 mM dithiothreitol for 30 min at 56°C and alkylated with 11 mM iodoacetamide for 15 min at room temperature in darkness. The protein sample was then diluted by adding 100 mM TEAB to urea concentration less than 2M. Finally, trypsin was added at 1:50 trypsin-to-protein mass ratio for the first digestion overnight and 1:100 trypsin-to-protein mass ratio for a second 4-h digestion. To enrich propionylation-modified peptides, tryptic peptides dissolved in NETN buffer (100 mM NaCl, 1 mM EDTA, 50 mM Tris-HCl, 0.5% NP-40, pH 8.0) were incubated with prewashed antibody beads (PTM202, PTM Bio) at 4°C overnight with gentle shaking. Then the beads were washed four times with NETN buffer and twice with H2O. The bound peptides were eluted from the beads with 0.1% trifluoroacetic acid. Finally, the eluted fractions were combined and vacuum-dried. For HPLC-MS/MS analysis, the resulting peptides were desalted with C18 ZipTips (Millipore) according to the manufacturer's instructions.
The tryptic peptides were dissolved in 0.1% formic acid (solvent A), directly loaded onto a home-made reversed-phase analytical column (15 cm length, 75 μm i.d.). The gradient was comprised of an increase from 6% to 23% solvent B (0.1% formic acid in 98% acetonitrile) over 26 min, 23%–35% in 8 min, and climbing to 80% in 3 min then holding at 80% for the last 3 min, all at a constant flow rate of 400 nL/min on an EASY-nLC 1000 UPLC system. The peptides were subjected to NSI source followed by tandem mass spectrometry (MS/MS) in Q ExactiveTM Plus (Thermo) coupled online to the UPLC. The electrospray voltage applied was 2.0 kV. The m/z scan range was 350–1800 for full scan, and intact peptides were detected. The resulting MS/MS data were processed using Maxquant search engine (v.1.5.2.8). Tandem mass spectra were searched against the Danio rerio database concatenated with a reverse decoy database. Trypsin/P was specified as cleavage enzyme allowing up to four missing cleavages. The mass tolerance for precursor ions was set as 20 ppm in first search and 5 ppm in main search, and the mass tolerance for fragment ions was set as 0.02 Da. Carbamidomethyl on Cys was specified as fixed modification, and propionylation modification and oxidation on Met were specified as variable modifications. FDR was adjusted to <1%, and minimum score for modified peptides was set >40.
Cell viability analysis
ZF4 cell was first seeded on 96-well plates and incubated for 24 h to sub-confluence. Then ZF4 cell was exposed to fresh medium added with a mixture of 150 μM oleic acid and 50 μM palmitic acid (OPA) and a mixture of OPA and increasing concentrations of propionate (OPP). At the end of the exposure period, fresh medium with 10% AlarmaBlue cell viability reagent (Invitrogen, Grand Island, NY, USA) was added. After a 1-h incubation, fluorescence was measured with the SynergyH1 microplate reader (Biotek) at excitation and emission wavelengths of 485 nm and 595 nm, respectively. The ratio of cell viability was calculated using the fluorescence readings of the OPA and OPP treatments.
Cell apoptosis analysis
Cell apoptosis detection was performed with Annexin V-fluorescein isothiocyanate (FITC) kits (Sigma). After exposure to OPA or OPP for 24 h, the cells were collected and incubated with Annexin V-FITC and propidium iodide in binding buffer for 10 min in darkness at room temperature. The analysis was conducted by the Guava easyCyte Flow Cytometer (Merck Millipore, Stafford, VA, USA).
Cellular ROS determination
Relative amount of cellular ROS was determined in ZF4 cells by using a fluorometric intracellular ROS kit (Sigma, USA). After exposure to OPA or OPP for 24 h, 100 μL/well of reaction mix containing ROS fluorogenic sensor was added into the 96-well black plate. After a 2-h incubation, the fluorescence was acquired with excitation and emission wavelengths of 490 nm and 520 nm, respectively. The relative fluorescence of ROS was calibrated by cell viability. ROS level was expressed as the fold change of the OPA group. The mean fluorescence intensity of ROS signal was detected by using a fluorescent probe DCFH-DA. After exposure to OPA or OPP for 24 h, ZF4 cell was incubated with DCFH-DA for 30 min and then harvested for fluorescence detection. The analysis was conducted by the Guava easyCyte Flow Cytometer (Merck Millipore, Stafford, VA, USA). The mean fluorescence intensity of ROS signal was expressed in MFI/5000 cells.
Determination of ROS in gut content and cell medium
Fresh gut contents were homogenized in cold PBS. The homogenates were then centrifuged at 11,000 g for 15 min at 4°C, and the supernatant was used for the measurement of ROS. The process of sample preparation is no more than 30 min. Cell medium was centrifuged at 11,000 g for 15 min at 4°C, and the supernatant was used for the measurement of ROS. Then both gut content and cell medium supernatants were incubated with fluorogenic sensor provided by a commercial kit (Sigma, USA) for 2 h to detect the relative level of ROS. This fluorogenic sensor could react with ROS and result in a fluorometric product, which could be detected by fluorescence microplate reader with excitation and emission wavelengths of 490 nm and 520 nm, respectively. The relative ROS level in gut content was expressed as the fold change of the HFD group. The relative ROS level in cell medium was expressed as the fold of OPA group.
Gene silencing with siRNA
Scrambled siRNA (negative control), sod2, and sirt3 siRNA (Table S5) were synthesized by GenePharma Co. Ltd. (Shanghai, China). Cells were first seeded on 6-well plates (Corning) and incubated for 24 h to sub-confluence. Then the cells were transfected with the appropriate siRNAs using Lipofectamine RNAiMAX Transfection Reagent (Invitrogen). Efficiency of the siRNA was determined by qPCR.
In vitro propionylation assay
In vitro propionylation assay was carried out following established protocols (Liu et al., 2020) with minor modification. The reactions consist of propionylation buffer containing 20 mM HEPEs, 0.1 mg/mL BSA, 1 mM DTT, 3 μg purified Sod2, propionyl-CoA (0, 0.2, 0.4, 0.8, 1, 5 mM), 1 mM PMSF, and 1 × phosphatase inhibitor. Reaction mixture was incubated at 30°C for 2 h.
Plasmid construction and transfection
The Sod2 was cloned into pCDNA3.1 and generated pCDNA3.1-flag-Sod2. Point (Site) mutations of Sod2 (pCDHA3.1-flag-Sod2 132K/Q and pCDHA3.1-flag-Sod2 132K/R) were generated by QuikChange Site-Directed Mutagenesis kit (Stratagene). Both WT Sod2 and Sod2 mutant plasmids were transfected into ZF4 cells using Lipofectamine 3000 Transfection Reagent (Invitrogen). For protein purification, His tag was added to pCDNA3.1-flag-Sod2 to generate pCDNA3.1-his-flag-Sod2 and overexpressed in HEK293 cells.
Gut microbiota analysis
The 16s V3–V4 region was amplified using the primers U341F (5′-CGGCAACGAGCGCAACCC-3′) and U806 (5′-CCATTGTAGCACGTGTGTAGCC-3′). The 16S ribosomal RNA gene sequencing was performed by Novogene Bioinformatics Technology Co. Ltd (Beijing, China) using the Illumina HiSeq platform. Then the raw pair-end readings were subjected to a quality-control procedure using the UPARSE-operational taxonomic unit (OTU) algorithm (Edgar., 2013). The qualified reads were clustered to generate OTUs at the 97% similarity level using the USEARCH sequence analysis tool (Edgar., 2010). A representative sequence of each OTU was assigned to a taxonomic level in the Ribosomal Database Project (RDP) database using the RDP classifier (Maidak et al., 1997).
Germ-free zebrafish generation and treatment
Germ-free (GF) zebrafish were derived from normal zebrafish and reared following established protocols (Rawls et al., 2006). Zebrafish larvae hatched from their chorions at 3 days postfertilization (dpf). Each group had six bottles with 20 fish per bottle. At 5 dpf, the yolk was largely absorbed, and the GF-zebrafish was subjected with feeding of LFD, HFD, and HFSP0.5 or transfer of gut microbiota deprived from adult zebrafish fed LFD, HFD, and HFSP0.5. At 12 dpf, GF zebrafish fed LFD, HFD, and HFSP0.5 were collected for living ROS detection. At 8 dpf, zebrafish larvae transferred with LFD, HFD, and HFSP0.5-gut microbiota were collected for ROS detection, enzymes detection, and qPCR.
Detection of ROS in whole zebrafish larvae
The detection of ROS in whole zebrafish larvae was conducted following a Novel procedure (Lackmann et al., 2018). In brief, zebrafish larvae were washed twice with GF-zebrafish medium (GZM) and incubated with 5 μM DCFH-DA for 30 min. After the exposure to DCFH-DA, larvae were washed twice with GZM and placed individually on a drop of 3% methyl cellulose poured onto a microscope glass slide. The texture of methyl cellulose made sure that the larvae kept still. Images of zebrafish larva were obtained with Zeiss stereoscopic microscope (AXIO Zoom. V16). Image analysis and quantification were performed using ImageJ.
In vitro culture of gut microbiota and quantification
Fresh gut content samples pooled from five zebrafish were put on ice and diluted in 5 mL sterile, ice-cold PBS. Within 30 min of sample collection, bacteria were seeded into Gifu anaerobic medium (GAM): GAM supplemented with 2 mmol/L H2O2 (GAM + H2O2); GAM supplemented with 2 mmol/L H2O2 and 0.05% LA (GAM + H2O2+LA). After an incubation period of 48 h at 28°C, the number of total bacteria or a specific phylotype was quantified by qPCR according to Zhang et al. (Zhang et al., 2019). In brief, bacterial DNA were extracted from equal volume of bacterial culture (2 mL) by using a TIANamp Bacteria DNA kit (Tiangen, Beijing, China). Then the extracted DNA were subjected qPCR by using primers set for universal bacteria or specific bacterial groups targeting the 16S rDNA gene (Table S6). 16S rDNA of each bacterial strain was cloned into the pLB vector (Tiangen, Beijing, China) as a copy number standard. For each qPCR standard, the copy number concentration was calculated based on the length of the PCR product and the average mass of a DNA base pair. For the gut microbiota cultured in vitro, results were expressed as Log10 copy numbers of bacterial 16S rDNA per mL medium (Log10 copies . mL medium−1).
Detection of ROS in gut microbiota medium
Gut microbiota from LFD, HFD, and HFSP0.5-fed zebrafish were cultured in GAM. At 24 h and 48 h, bacteria were collected by centrifuging at 12,000 g for 5 min, and supernatants were subjected to ROS detection with fluorogenic sensor provided by a commercial kit (Sigma, USA) for 2 h. The fluorometric product could be detected by fluorescence microplate reader with excitation and emission wavelengths of 490 nm and 520 nm, respectively. The relative ROS level in bacteria cultures was calibrated by bacteria population in each culture.
Western blotting
Zebrafish intestine or larval zebrafish were homogenized in ice-cold HBSS buffer mixed with 1 mM PMSF and phosphatase inhibitors. Equivalent amounts of total protein were loaded into a 12% SDS-PAGE for electrophoresis and then transferred into a PVDF membrane (Millipore, USA). After blocking nonspecific binding with 5% skimmed milk in TBST, the PVDF membrane was incubated with primary antibodies, i.e., antibodies against zebrafish Gapdh (Sigma, SAB2708126, 1:2000), Sod2 (Genetex, GTX124294, 1:1000), Sirt3 (Sigma, AV32388, 1:1000), Mtco1 (Abcam, ab14109, 1:2000), acetylated lysine (Cell Signaling Technology, 9441, 1:1000), and customized Sod2 K132pro (Jingjie PTM BIO, 1:500). The blots were developed using HRP-conjugated secondary antibodies (GE Health, 1:3000) and the ECL-plus system.
Total RNA extraction, reverse transcription, and qPCR
Total RNA was isolated using Trizol reagent and then reverse transcribed to cDNA. The qPCR was performed using SYBRGreen Supermix according to the manufacturer's instructions (Tiangen, Beijing, China). The results were stored, managed, and analyzed via LightCycler 480 software (Roche, Basel, Switzerland). The qPCR primers used are listed in Table S6.
Quantification and statistical analysis
All of the statistical analyses were conducted using GraphPad Prism 5 software (GraphPad Software Inc., San Diego, CA, USA). Results are expressed as the means ± standard errors of the means (SEMs). Comparisons between two groups were analyzed using the Student's t test, and comparisons between multiple groups were analyzed using one-way ANOVA followed by a Duncan's test. The statistical significance was set at p < 0.05. The number of replicates for each experiment are indicated in the figure legends.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (NSFC 31925038, 32061133004) and the National Key R&D Program of China (2018YFD0900400). The authors thank Prof. Nicholas Clarke for linguistic proof reading.
Author contributions
Z.Zhou. designed and supervised the research. Q.D. and C.R. wrote the paper. Z.Zhou. and C.R. gave conceptual advice for the paper. J.L.C., E.R., and R.E.O. reviewed and helped to revise the manuscript. Q.D. performed experiments and acquired data. Z.Zhang. and Y.L. assisted in the qPCR, western blot, gut microbiota analysis, and siRNA knockdown experiments. H.L. and Q.H. participated in zebrafish husbandry and sampling. C.R., Y.Y., and Z.Zhang. co-analyzed and discussed the results. All authors read and approved the final manuscript.
Declaration of interests
The authors declare no competing interests.
Published: June 25, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.102515.
Contributor Information
Chao Ran, Email: ranchao@caas.cn.
Zhigang Zhou, Email: zhouzhigang03@caas.cn.
Supplemental information
References
- Asimakis G.K., Lick S., Patterson C. Postischemic recovery of contractile function is impaired in SOD2(+/-) but not SOD1(+/-) mouse hearts. Circulation. 2002;105:981–986. doi: 10.1161/hc0802.104502. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya A., Chattopadhyay R., Mitra S., Crowe S.E. Oxidative stress: an essential factor in the pathogenesis of gastrointestinal mucosal diseases. Physiol. Rev. 2014;94:329–354. doi: 10.1152/physrev.00040.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatti J.S., Bhatti G.K., Reddy P.H. Mitochondrial dysfunction and oxidative stress in metabolic disorders - a step towards mitochondria based therapeutic strategies. Biochim. Biophys. Acta Mol. Basis Dis. 2017;1863:1066–1077. doi: 10.1016/j.bbadis.2016.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bheda P., Wang J.T., Escalante-Semerena J.C., Wolberger C. Structure of Sir2Tm bound to a propionylated peptide. Protein Sci. 2011;20:131–139. doi: 10.1002/pro.544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Sprung R., Tang Y., Ball H., Sangras B., Kim S.C., Falck J.R., Peng J., Gu W., Zhao Y. Lysine propionylation and butyrylation are novel post-translational modifications in histones. Mol. Cell Proteomics. 2007;6:812–819. doi: 10.1074/mcp.M700021-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z., Tang Y., Chen Y., Kim S., Liu H., Li S.S., Gu W., Zhao Y. Molecular characterization of propionyllysines in non-histone proteins. Mol. Cell Proteomics. 2009;8:45–52. doi: 10.1074/mcp.M800224-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Circu M.L., Aw T.Y. Intestinal redox biology and oxidative stress. Semin. Cell Dev. Biol. 2012;23:729–737. doi: 10.1016/j.semcdb.2012.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Keyzer Y., Valayannopoulos V., Benoist J.F., Batteux F., Lacaille F., Hubert L., Chretien D., Chadefeaux-Vekemans B., Niaudet P., Touati G. Multiple OXPHOS deficiency in the liver, kidney, heart, and skeletal muscle of patients with methylmalonic aciduria and propionic aciduria. Pediatr. Res. 2009;66:91–95. doi: 10.1203/PDR.0b013e3181a7c270. [DOI] [PubMed] [Google Scholar]
- Driever W., Rangini Z. Characterization of a cell line derived from zebrafish (Brachydanio rerio) embryos. In Vitro Cell. Dev. Biol. Anim. 1993;29A:749–754. doi: 10.1007/BF02631432. [DOI] [PubMed] [Google Scholar]
- Du S.F., Wang H.J., Zhang B., Zhai F.Y., Popkin B.M. China in the period of transition from scarcity and extensive undernutrition to emerging nutrition-related non-communicable diseases, 1949-1992. Obes. Rev. 2014;15:8–15. doi: 10.1111/obr.12122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duchen M.R. Mitochondria in health and disease: perspectives on a new mitochondrial biology. Mol. Aspects Med. 2004;25:365–451. doi: 10.1016/j.mam.2004.03.001. [DOI] [PubMed] [Google Scholar]
- Edgar R.C. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods. 2013;10:996–+. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- Edgar R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- Emre Y., Nubel T. Uncoupling protein UCP2: when mitochondrial activity meets immunity. FEBS Lett. 2010;584:1437–1442. doi: 10.1016/j.febslet.2010.03.014. [DOI] [PubMed] [Google Scholar]
- Fernandez-Vizarra E., Fernandez-Silva P., Enriquez J.A. Isolation of mitochondria from mammalian tissues and cultured Cells. In: Celis J.E., editor. Cell Biology: A Laboratory Handbook. Elsevier-Academic Press; 2006. pp. 69–77. [Google Scholar]
- Flavin M., Ochoa S. Metabolism of propionic acid in animal tissues. I. Enzymatic conversion of propionate to succinate. J. Biol. Chem. 1957;229:965–979. [PubMed] [Google Scholar]
- Fukai T., Ushio-Fukai M. Superoxide dismutases: role in redox signaling, vascular function, and diseases. Antioxidants & Redox Signaling. 2011;15:1583–1606. doi: 10.1089/ars.2011.3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukui M., Zhu B.T. Mitochondrial superoxide dismutase SOD2, but not cytosolic SOD1, plays a critical role in protection against glutamate-induced oxidative stress and cell death in HT22 neuronal cells. Free Radic. Biol. Med. 2010;48:821–830. doi: 10.1016/j.freeradbiomed.2009.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego-Villar L., Perez-Cerda C., Perez B., Abia D., Ugarte M., Richard E., Desviat L.R. Functional characterization of novel genotypes and cellular oxidative stress studies in propionic acidemia. J. Inherit. Metab. Dis. 2013;36:731–740. doi: 10.1007/s10545-012-9545-3. [DOI] [PubMed] [Google Scholar]
- Gallego-Villar L., Rivera-Barahona A., Cuevas-Martin C., Guenzel A., Perez B., Barry M.A., Murphy M.P., Logan A., Gonzalez-Quintana A., Martin M.A. In vivo evidence of mitochondrial dysfunction and altered redox homeostasis in a genetic mouse model of propionic acidemia: implications for the pathophysiology of this disorder. Free Radic. Biol. Med. 2016;96:1–12. doi: 10.1016/j.freeradbiomed.2016.04.007. [DOI] [PubMed] [Google Scholar]
- Garrity J., Gardner J.G., Hawse W., Wolberger C., Escalante-Semerena J.C. N-Lysine propionylation controls the activity of propionyl-CoA synthetase. J. Biol. Chem. 2007;282:30239–30245. doi: 10.1074/jbc.M704409200. [DOI] [PubMed] [Google Scholar]
- Kebede A.F., Nieborak A., Shahidian L.Z., Le Gras S., Richter F., Gomez D.A., Baltissen M.P., Meszaros G., Magliarelli H.D., Taudt A. Histone propionylation is a mark of active chromatin. Nat. Struct. Mol. Biol. 2017;24:1048–+. doi: 10.1038/nsmb.3490. [DOI] [PubMed] [Google Scholar]
- Kim S., Piao S., Lee I., Nagar H., Choi S.J., Shin N., Kim D.W., Shong M., Jeon B.H., Kim C.S. CR6 interacting factor 1 deficiency induces premature senescence via SIRT3 inhibition in endothelial cells. Free Radic. Biol. Med. 2020;150:161–171. doi: 10.1016/j.freeradbiomed.2020.02.017. [DOI] [PubMed] [Google Scholar]
- Koh A., De Vadder F., Kovatcheva-Datchary P., Backhed F. From dietary fiber to host physiology: short-chain fatty acids as key bacterial metabolites. Cell. 2016;165:1332–1345. doi: 10.1016/j.cell.2016.05.041. [DOI] [PubMed] [Google Scholar]
- Kyle S.S. Mechanisms of NSAID-induced small intestinal injury: role of bacterial beta-glucuronidase, the microbiome, and mitochondria. Microbiol. Immunol. 2014;28:975–986. [Google Scholar]
- Kong X.X., Wang R., Xue Y., Liu X.J., Zhang H.B., Chen Y., Fang F., Chang Y.S. Sirtuin 3, a new target of PGC-1 alpha, plays an important role in the suppression of ROS and mitochondrial biogenesis. PLoS One. 2010;5:e11707. doi: 10.1371/journal.pone.0011707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackmann C., Santos M., Rainieri S., Barranco A., Hollert H., Spirhanzlova P., Velki M., Seiler T.B. Novel procedures for whole organism detection and quantification of fluorescence as a measurement for oxidative stress in zebrafish (Danio rerio) larvae. Chemosphere. 2018;197:200–209. doi: 10.1016/j.chemosphere.2018.01.045. [DOI] [PubMed] [Google Scholar]
- Lagerwaard B., Pougovkina O., Bekebrede A.F., te Brinke H., Wanders R.J.A., Nieuwenhuizen A.G., Keijer J., de Boer V.C.J. Increased protein propionylation contributes to mitochondrial dysfunction in liver cells and fibroblasts, but not in myotubes. J. Inherit. Metab. Dis. 2020;44:438–449. doi: 10.1002/jimd.12296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C.T., Tseng W.C., Hsiao N.W., Chang H.H., Ken C.F. Characterization, molecular modelling and developmental expression of zebrafish manganese superoxide dismutase. Fish Shellfish Immunol. 2009;27:318–324. doi: 10.1016/j.fsi.2009.05.015. [DOI] [PubMed] [Google Scholar]
- Liu B., Lin Y.H., Darwanto A., Song X.H., Xu G.L., Zhang K.L. Identification and characterization of propionylation at histone H3 lysine 23 in mammalian cells. J. Biol. Chem. 2009;284:32288–32295. doi: 10.1074/jbc.M109.045856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Dai J., Lan X., Fan B., Dong T., Zhang Y., Han M. Intestinal bacteria are potential biomarkers and therapeutic targets for gastric cancer. Microb. Pathog. 2021;151:104747. doi: 10.1016/j.micpath.2021.104747. [DOI] [PubMed] [Google Scholar]
- Liu X., Zhu C.C., Zha H.Y., Tang J.H., Rong F.J., Chen X.Y., Fan S.J., Xu C.X., Du J., Zhu J.J. SIRT5 impairs aggregation and activation of the signaling adaptor MAVS through catalyzing lysine desuccinylation. EMBO J. 2020;39:e103285. doi: 10.15252/embj.2019103285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J.Q., Cheng K.Y., Zhang B., Xu H., Cao Y.Z., Guo F., Feng X.D., Xia Q. Novel mechanisms for superoxide-scavenging activity of human manganese superoxide dismutase determined by the K68 key acetylation site. Free Radic. Biol. Med. 2015;85:114–126. doi: 10.1016/j.freeradbiomed.2015.04.011. [DOI] [PubMed] [Google Scholar]
- Maidak B.L., Olsen G.J., Larsen N., Overbeek R., McCaughey M.J., Woese C.R. The RDP (ribosomal database Project) Nucleic Acids Res. 1997;25:109–110. doi: 10.1093/nar/25.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuishi T., Stumpf D.A., Seliem M., Eguren L.A., Chrislip K. Propionate mitochondrial toxicity in liver and skeletal muscle: acyl CoA levels. Biochem. Med. Metab. Biol. 1991;45:244–253. doi: 10.1016/0885-4505(91)90027-i. [DOI] [PubMed] [Google Scholar]
- Matzuk M.M., Dionne L., Guo Q.X., Kumar T.R., Lebovitz R.M. Ovarian function in superoxide dismutase 1 and 2 knockout mice. Endocrinology. 1998;139:4008–4011. doi: 10.1210/endo.139.9.6289. [DOI] [PubMed] [Google Scholar]
- Million M., Tidjani Alou M., Khelaifia S., Bachar D., Lagier J.C., Dione N., Brah S., Hugon P., Lombard V., Armougom F. Increased gut redox and Depletion of anaerobic and methanogenic prokaryotes in severe acute malnutrition. Sci. Rep. 2016;6:26051. doi: 10.1038/srep26051. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Nahomi R.B., Nandi S.K., Rakete S., Michel C., Fritz K.S., Nagaraj R.H. Lysine malonylation and propionylation are prevalent in human lens proteins. Exp. Eye Res. 2020;190:107864. doi: 10.1016/j.exer.2019.107864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak E.A., Mollen K.P. Mitochondrial dysfunction in inflammatory bowel disease. Front. Cell. Dev. Biol. 2015;3:62. doi: 10.3389/fcell.2015.00062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palucka T. Toying with a ridiculous material. Mrs Bull. 2007;32:283. [Google Scholar]
- Pieczenik S.R., Neustadt J. Mitochondrial dysfunction and molecular pathways of disease. Exp. Mol. Pathol. 2007;83:84–92. doi: 10.1016/j.yexmp.2006.09.008. [DOI] [PubMed] [Google Scholar]
- Qiao Y., Sun J., Ding Y.Y., Le G.W., Shi Y.H. Alterations of the gut microbiota in high-fat diet mice is strongly linked to oxidative stress. Appl. Microbiol. Biotechnol. 2013;97:1689–1697. doi: 10.1007/s00253-012-4323-6. [DOI] [PubMed] [Google Scholar]
- Qiu X.L., Brown K., Hirschey M.D., Verdin E., Chen D. Calorie restriction reduces oxidative stress by SIRT3-mediated SOD2 activation. Cell Metab. 2010;12:662–667. doi: 10.1016/j.cmet.2010.11.015. [DOI] [PubMed] [Google Scholar]
- Rafi S.S. ETH; 1998. Studies on the Pathogenesis of NSAID-Induced Damage to the Gastrointestinal Tract with Special Reference to the Mitochondria; p. 326317. [Google Scholar]
- Ramachandran A., Lebofsky M., Weinman S.A., Jaeschke H. The impact of partial manganese superoxide dismutase (SOD2)-deficiency on mitochondrial oxidant stress, DNA fragmentation and liver injury during acetaminophen hepatotoxicity. Toxicol. Appl. Pharmacol. 2011;251:226–233. doi: 10.1016/j.taap.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawls J.F., Mahowald M.A., Ley R.E., Gordon J.I. Reciprocal gut microbiota transplants from zebrafish and mice to germ-free recipients reveal host habitat selection. Cell. 2006;127:423–433. doi: 10.1016/j.cell.2006.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose M.D. Efsa panel on food additives and nutrient sources added to food (ans) EFSA J. 2013;11:3234. [Google Scholar]
- Saint-Georges-Chaumet Y., Edeas M. Microbiota-mitochondria inter-talk: consequence for microbiota-host interaction. Pathog. Dis. 2016;74:ftv096. doi: 10.1093/femspd/ftv096. [DOI] [PubMed] [Google Scholar]
- Schonfeld P., Wojtczak L. Short- and medium-chain fatty acids in energy metabolism: the cellular perspective. J. Lipid Res. 2016;57:943–954. doi: 10.1194/jlr.R067629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab M.A., Sauer S.W., Okun J.G., Nijtmans L.G.J., Rodenburg R.J.T., van den Heuvel L.P., Drose S., Brandt U., Hoffmann G.F., Ter Laak H. Secondary mitochondrial dysfunction in propionic aciduria: a pathogenic role for endogenous mitochondrial toxins. Biochem. J. 2006;398:107–112. doi: 10.1042/BJ20060221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapawi R., Ebi I., Yong A.S.K., Ng W.K. Optimizing the growth performance of brown-marbled grouper, Epinephelus fuscoguttatus (Forskal), by varying the proportion of dietary protein and lipid levels. Anim. Feed Sci. Technol. 2014;191:98–105. [Google Scholar]
- Somasundaram S., Rafi S., Hayllar J., Sigthorsson G., Jacob M., Price A.B., Macpherson A., Mahmod T., Scott D., Wrigglesworth J.M., Bjarnason I. Mitochondrial damage: a possible mechanism of the "topical" phase of NSAID induced injury to the rat intestine. Gut. 1997;41:344–353. doi: 10.1136/gut.41.3.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitz D.R., Oberley L.W. An assay for superoxide dismutase activity in mammalian tissue homogenates. Anal Biochem. 1989;179:8–18. doi: 10.1016/0003-2697(89)90192-9. [DOI] [PubMed] [Google Scholar]
- Starai V.J., Celic I., Cole R.N., Boeke J.D., Escalante-Semerena J.C. Sir2-dependent activation of acetyl-CoA synthetase by deacetylation of active lysine. Science. 2002;298:2390–2392. doi: 10.1126/science.1077650. [DOI] [PubMed] [Google Scholar]
- Tao R.D., Coleman M.C., Pennington J.D., Ozden O., Park S.H., Jiang H.Y., Kim H.S., Flynn C.R., Hill S., McDonald W.H. Sirt3-Mediated deacetylation of evolutionarily conserved lysine 122 regulates MnSOD activity in response to stress. Mol. Cell. 2010;40:893–904. doi: 10.1016/j.molcel.2010.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y.H., Lu C.Y., Lee H.C., Pang C.Y., Ma Y.S. Oxidative damage and mutation to mitochondrial DNA and age-dependent decline of mitochondrial respiratory function. Ann. N Y Acad. Sci. 1998;854:155–170. doi: 10.1111/j.1749-6632.1998.tb09899.x. [DOI] [PubMed] [Google Scholar]
- Wongkittichote P., Mew N.A., Chapman K.A. Propionyl-CoA carboxylase - a review. Mol. Genet. Metab. 2017;122:145–152. doi: 10.1016/j.ymgme.2017.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J.Y., Xu Y., Xu Z., Zhai L.H., Ye Y., Zhao Y.M., Chu X.H., Tan M.J., Ye B.C. Protein acylation is a general regulatory mechanism in biosynthetic pathway of acyl-CoA-derived natural products. Cell Chem. Biol. 2018;25:984–+. doi: 10.1016/j.chembiol.2018.05.005. [DOI] [PubMed] [Google Scholar]
- Yang M.K., Huang H., Ge F. Lysine propionylation is a widespread post-translational modification involved in regulation of photosynthesis and metabolism in Cyanobacteria. Int. J. Mol. Sci. 2019;20:4792. doi: 10.3390/ijms20194792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon S.P., Kim J. Poly(ADP-ribose) polymerase 1 contributes to oxidative stress through downregulation of sirtuin 3 during cisplatin nephrotoxicity. Anat. Cell Biol. 2016;49:165–176. doi: 10.5115/acb.2016.49.3.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelko I.N., Mariani T.J., Folz R.J. Superoxide dismutase multigene family: a comparison of the CuZn-SOD (SOD1), Mn-SOD (SOD2), and EC-SOD (SOD3) gene structures, evolution, and expression. Free Radic. Biol. Med. 2002;33:337–349. doi: 10.1016/s0891-5849(02)00905-x. [DOI] [PubMed] [Google Scholar]
- Zeng X.L., Yang J.N., Hu O., Huang J., Ran L., Chen M.T., Zhang Y., Zhou X., Zhu J.D., Zhang Q.Y. Dihydromyricetin ameliorates nonalcoholic fatty liver disease by improving mitochondrial respiratory capacity and redox homeostasis through modulation of SIRT3 signaling. Antioxid. Redox Signal. 2019;30:163–183. doi: 10.1089/ars.2017.7172. [DOI] [PubMed] [Google Scholar]
- Zhai F.Y., Du S.F., Wang Z.H., Zhang J.G., Du W.W., Popkin B.M. Dynamics of the Chinese diet and the role of urbanicity, 1991-2011. Obes. Rev. 2014;15:16–26. doi: 10.1111/obr.12124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K., Chen Y., Mang Z.H., Zhao Y.M. Identification and verification of lysine propionylation and butyrylation in yeast core histones using PTMap software. J. Proteome Res. 2009;8:900–906. doi: 10.1021/pr8005155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H.H., Ma X.J., Wu L.N., Zhao Y.Y., Zhang P.Y., Zhang Y.H., Shao M.W., Liu F., Li F., Qin G.J. Sirtuin-3 (SIRT3) protects pancreatic beta-cells from endoplasmic reticulum (ER) stress-induced apoptosis and dysfunction. Mol. Cell Biochem. 2016;420:95–106. doi: 10.1007/s11010-016-2771-5. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Ran C., Ding Q.W., Liu H.L., Xie M.X., Yang Y.L., Xie Y.D., Gao C.C., Zhang H.L., Zhou Z.G. Ability of prebiotic polysaccharides to activate a HIF1 alpha-antimicrobial peptide axis determines liver injury risk in zebrafish. Commun. Biol. 2019;2:274. doi: 10.1038/s42003-019-0526-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Microbiota sequencing data in this study are available from the National Center for Biotechnology Information (NCBI) under accession number BioProject: PRJNA714780.