Summary
The Southern Cone of South America (SCSA) is a key region for investigations about the peopling of the Americas. However, little is known about the eastern sector, the Argentinian Pampas. We analyzed 18 mitochondrial genomes—7 of which are novel—from human skeletal remains from 3 Early to Late Holocene archaeological sites. The Pampas present a distinctive genetic makeup compared to other Middle to Late Holocene pre-Columbian SCSA populations. We also report the earliest individuals carrying SCSA-specific mitochondrial haplogroups D1j and D1g from Early and Middle Holocene, respectively. Using these deep calibration time points in Bayesian phylogenetic reconstructions, we suggest that the first settlers of the Pampas were part of a single and rapid dispersal ∼15,600 years ago. Finally, we propose that present-day genetic differences between the Pampas and the rest of the SCSA are due to founder effects, genetic drift, and a partial population replacement ∼9,000 years ago.
Subject areas: Genetics, Genomics, Paleogenetics
Graphical abstract
Highlights
-
•
Analysis of 18 ancient human mitochondrial genomes from the Argentinian Pampas.
-
•
Genetic makeup of Early-Mid Holocene Pampas distinct from later neighboring peoples.
-
•
Earliest individuals carrying region-specific mitochondrial haplogroups D1j and D1g.
-
•
First Pampean settlers were part of a single and rapid dispersal ∼15,600 years ago.
Genetics; Genomics; Paleogenetics
Introduction
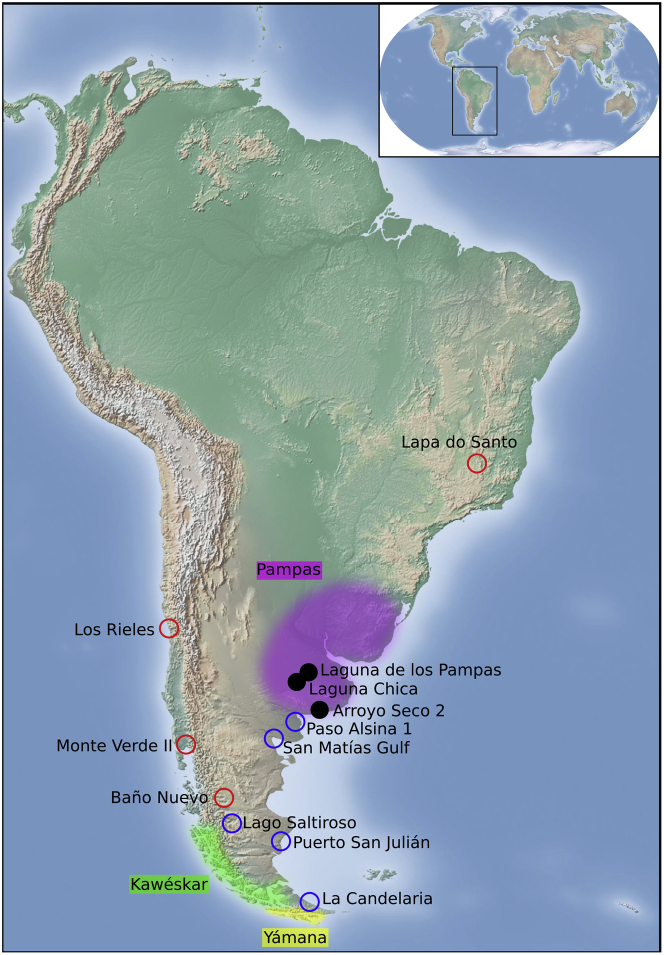
The Southern Cone of South America (SCSA), formed by Argentina, Chile, Uruguay, and Southern Brazil, is a key study area to build a comprehensive picture of the peopling of the Americas. The region is geographically the most distant from Beringia, the entry point of the first settlers of the Americas, and the archaeological record shows that humans arrived in the SCSA roughly 14.3 thousand years ago (kya) (Dillehay et al., 2008; Politis and Prates, 2018), shortly after the initial colonization of the continent (∼16 kya, based on ancient DNA (Llamas et al., 2016)), indicating a rapid spread southwards (Bodner et al., 2012; Prates et al., 2020). A growing body of genetic research addressing the population history of the SCSA has focused mostly on the southernmost archaeological and extant groups from Patagonia and Tierra del Fuego (Lalueza et al., 1997; García-Bour et al., 2004; de Saint Pierre et al., 2012b; de la Fuente et al., 2015, 2018; Crespo et al., 2017, 2018; Nakatsuka et al., 2020b). However, despite its potential importance for the peopling of this region, only a few recent studies included populations from the eastern sector of the SCSA, the Argentinian Pampas (Figure 1) (Perez et al., 2009; Llamas et al., 2016; Posth et al., 2018; Postillone et al., 2020b).
Figure 1.
Map of the Pampas region
The three archaeological sites from which samples for this study derive are highlighted (Laguna de los Pampas, Laguna Chica, and Arroyo Seco 2; black circles), as well as other Early (red circles) and Late Holocene (blue circles) sites mentioned in this study. Los Rieles has some samples that date from the Late Pleistocene. Monte Verde II has signs of human activity dating to the Late Pleistocene. The 19th century territories of the Kawéskar and Yámana populations are shadowed in green and yellow, respectively.
The Argentinian Pampas harbor some of the oldest known archaeological sites in the SCSA, with human presence in the region evident around 14,000 calibrated 14C years before present (calBP) (Politis et al., 2014; Politis et al., 2016) and a rich record of Late Pleistocene sites (∼12.9–11 kya) (Politis, 2008; Mazzanti et al., 2012; Flegenheimer et al., 2013; Martínez et al., 2016). The faunal remains indicate that the subsistence was based on the hunting of camelids, extinct horse, and some extinct megamammals (e.g. giant ground sloths Megatherium and giant glyptodonts Doedicurus) (Martínez et al., 2016; Miotti et al., 2018). The fishtail projectile point (a typical early lithic tool) appeared both in the Pampas and Patagonia at ∼13 kya, a period of time when population size increased in the SCSA (Bodner et al., 2012). However, during the beginning of the Early Holocene (∼11–8 kya), there was a reduction of archaeological sites throughout the Pampas and Patagonia and a disappearance of megafaunal remains as well as fishtail projectile points. This change in the archaeological record precedes a proposed population replacement in parts of South America starting around 9 kya (Posth et al., 2018). While a different local retraction process and a population replacement in the Middle Holocene (∼8–4 kya), concretely in the latest period, have been proposed based on differences in skull morphology as well as a gap in the radiocarbon database (Barrientos and Perez, 2005), genetic evidence and more dates from archaeological sites indicate population continuity in the region since the Early Holocene (Politis, 2008; Mazzanti et al., 2015; Posth et al., 2018; Donadei, 2019). During the Late Holocene (<4 kya), the complexity of human populations in the Pampas increased due to the introduction of pottery most likely from the subtropical lowlands as well as the development of newly derived technologies from the bow and arrow adoption (Politis, 2008). During this period, macro-regional social link and exchange expanded, and, as a result, an individual with Central-Andes-associated genetic ancestry unexpectedly was found at the archaeological site of Laguna Chica, dating to 1700–1565 calBP (Nakatsuka et al., 2020a).
While phylogeographic studies of mitochondrial genomes (mitogenomes) from the past and present Native Americans have revealed that the major founding lineages (A2, B2, C1b, C1c, C1d, D1, and D4h3a) are widespread across the Americas with temporal and spatial variation (Perego et al., 2009; 2010; Bisso-Machado et al., 2012; Llamas et al., 2016), there are specific clades that show a highly restricted geographic distribution (Bodner et al., 2012; de Saint Pierre et al., 2012b; Gómez-Carballa et al., 2018). Thus, their evolutionary history can be highly informative to infer the population history of a particular region. Four clades are found nearly exclusively in people inhabiting the SCSA: B2i2, C1b13, D1g, and D1j (Bodner et al., 2012; de Saint Pierre et al., 2012b). The Pan-American minor founding lineage D4h3a is found at highest frequencies in Patagonia and Tierra del Fuego (Perego et al., 2009; de Saint Pierre et al., 2012a). A2 and B2, which are frequent all over South America, are virtually absent in populations of the extreme South (Lalueza et al., 1997; García-Bour et al., 2004; Crespo et al., 2018).
In particular, the mitochondrial clades D1g and D1j play a prominent role in the discussions surrounding the peopling of the SCSA. The geographical distribution of both clades, identified by Bodner et al. (2012), differs significantly from one another; D1g is found most frequently in Argentinian and Chilean Patagonia, as well as in the Argentinian Pampas (Bodner et al., 2012; de Saint Pierre et al., 2012b; Crespo et al., 2018), while D1j is most frequent in northern and central Argentina (including the Pampas) and Southern Brazil but virtually absent in Chile (Bodner et al., 2012; García et al., 2012; Crespo et al., 2018; de Saint Pierre, 2017). Based on their calculated divergence ages for both clades (D1g: 18.3 ± 2.4 kya; D1j: 13.9 ± 2.9 kya) and the prominence of Monte Verde II in southern Chile as the oldest known site in the SCSA (∼14.3 kya) (Dillehay et al., 2008; Politis and Prates, 2018), Bodner et al. (2012) proposed that the most likely peopling scenario was with one initial founding population arriving via the Pacific coast and subsequently crossing the Andes into Argentina, followed by further diversification of both clades. However, other studies note that the practical non-overlapping distribution of both clades contradicts this hypothesis (de Saint Pierre et al., 2012b; García et al., 2012; de Saint Pierre, 2017). Concretely, de Saint Pierre (2017) proposed much older divergence dates of the two clades (D1g: 22 ± 7 kya; D1j: 16.7 ± 9.4 kya), suggesting that the peopling of South America along the Pacific route might have happened much earlier than suggested by studies that include ancient mitogenomes (Llamas et al., 2016) or genomic data (Moreno-Mayar et al., 2018; Posth et al., 2018). Support for this earlier peopling hypothesis relies on the older dates reported for the Monte Verde locality (Dillehay et al., 2015). However, the association of these older dates with human occupation remains controversial (Politis and Prates, 2018; Prates et al., 2020).
We identify three potential shortcomings of studies addressing the peopling of the SCSA using mitogenomes: (1) Inferences are highly dependent on the prominence of one specific archaeological site; (2) Most of the mitochondrial data derive from either relatively recent archaeological samples or modern extant groups who might not have a local origin; (3) Divergence date estimates are highly dependent on the calibration method applied to the molecular clock when converting estimates of relative rates of molecular evolution into calendar years in the phylogenetic tree. Several studies have shown that calibration points close to the age of the events, such as radiocarbon-dated ancient DNA sequences (so-called tip calibrations), provide the most reliable date inferences (Rieux et al., 2014; Llamas et al., 2016, 2017; Posth et al., 2016).
In order to address these potential issues and to better understand the population history of the Argentinian Pampas and its implications in the peopling of the SCSA, we analyzed whole mitogenomes from well-dated human skeletal remains from three Early to Late Holocene archaeological sites in the Argentinian Pampas: Arroyo Seco 2 (AS2) (Politis et al., 2014; Politis et al., 2016), Laguna de los Pampas (LLP) (Messineo et al., 2018; Messineo et al., 2019), and Laguna Chica (LCH) (Scheifler et al., 2017; Messineo et al., 2019) (Figures 1 and S1–S3; supplemental information text). We further employed a tip-calibration approach to determine the divergence ages and to better understand the evolutionary history of the SCSA-specific mitochondrial clades D1g and D1j, as well as the Pan-American minor founding lineage D4h3a that has its highest frequencies in Patagonia and Tierra del Fuego.
Results
Ancient DNA authenticity
We reconstructed 18 mitogenomes at an average coverage ranging from 22–419X (Tables 1 and S1). Twelve of these mitogenomes have been reported previously (Llamas et al., 2016; Posth et al., 2018; Nakatsuka et al., 2020a), whereas an additional seven are reported here for the first time. The DNA damage patterns observed at the terminal ends of the sequencing reads are indicative of ancient degraded DNA (Jónsson et al., 2013) and, combined with the low observed contamination estimates (0.5–3%), support the authenticity of the results (Table S1).
Table 1.
Calibrated dates and mitochondrial haplogroups of the 18 Early to Late Holocene individuals from the Argentinian Pampas.
| Site | Sample | Calibrated date | Period | Mitochondrial haplogroup | GenBank ID | Reference |
|---|---|---|---|---|---|---|
| Laguna de los Pampas | LLP.S2.E1 | 10,223–9,764 | Initial Early Holocene | D1j | MW291678 | This study |
| Arroyo Seco 2 | ASO_B27_S36 | 8,960–8,380 | Terminal Early Holocene | C1b | MW291663 | (Posth et al., 2018) |
| ASO_B24_S31 | 8,545–8,188 | C1b+16,311 | MW291669 | This study | ||
| ASO_B13_S20 | 8,545–8,188a | C1c | MW291667 | This study | ||
| ASO_S49 | 8,520–8,200 | C1b | MW291664 | (Posth et al., 2018) | ||
| ASO_B3_S7 | 7,970–7,673 | Middle Holocene | C1b | MW291671 | This study | |
| ASO_B10_S15 | 7,920–7,660 | D1g | MW291661 | (Posth et al., 2018) | ||
| ASO_B10_S17 | 7,920–7,660b | C1b | MW291673 | This study | ||
| ASO_B9_S14a | 7,832–7,573 | D1 | MW291672 | (Llamas et al., 2016) | ||
| ASO_B12_S19 | 7,570–7,300 | A2 | MW291666 | (Llamas et al., 2016) | ||
| ASO_B2_S6 | 7,570–7,290 | A2 | MW291670 | (Posth et al., 2018) | ||
| ASO_B2_S5 | 7,570–7,290c | A2 | MW291665 | This study | ||
| ASO_B1_S3 | 7,330–6,950 | C1c | MW291668 | (Posth et al., 2018) | ||
| ASO_B1_S1 | 7,330–6,950d | C1c | MW291662 | This study | ||
| Laguna Chica | LCH.E1.3 | 7,724–7,589 | A2 | MW291674 | (Posth et al., 2018) | |
| LCH.E2-I2.1 | 6,960–6,790 | B2b | MW291676 | (Posth et al., 2018) | ||
| LCH.E2-I1.2 | 6,780–6,650 | C1b | MW291675 | (Posth et al., 2018) | ||
| LCH.E4.4 | 1,627–1,565 | Late Holocene | D1g5 | MW291677 | (Nakatsuka et al., 2020a) |
Indirect date. Burial 13 (skeleton 20; B13_S20) and burial 24 (skeleton 31; B24_S31) were buried at roughly the same depth and share some characteristics (primary burials with calcrete stones), which indicate they might have the same age (Politis et al., 2014).
Indirect date. Burial 10 was formed by three fully articulated skeletons—1 (B10_S15), 2 (B10_S16), and 3 (B10_S17)—buried at the same time. Date comes from B10_S15 and was extrapolated to B10_S17.
Indirect date. Burial 2 was formed by three fully articulated skeletons—4 (B2_S4), 5 (B2_S5), and 6 (B2_S6)—buried at the same time. The14C date comes from individual B2_S6 and was extrapolated to B2_S5.
Indirect date. Burial 1 was formed by three fully articulated skeletons—1 (B1_S1), 2 (B1_S2), and 3 (B1_S3)—buried at the same time. Date comes from B1_S3 and was extrapolated to B1_S1.
Radiocarbon dating
Radiocarbon dates obtained for some of the successfully sequenced samples show that one newly reported individual from Laguna de los Pampas dates from the initial Early Holocene (LLP.S2.E1; 10,223–9,764 calBP), while four individuals from Arroyo Seco 2 date from the terminal Early Holocene (8,960–8,188 calBP). The majority of samples from Arroyo Seco 2 (n = 9; 7,970–6,950 calBP) and Laguna Chica (n = 3; 7,724–6,650 calBP) date from the Middle Holocene with the exception of one individual from Laguna Chica that dates from the Late Holocene (1,627–1,565 calBP) (Table 1) (Politis et al., 2014; Messineo et al., 2018; Posth et al., 2018; Nakatsuka et al., 2020a). Importantly, a radiocarbon date for a burial in Laguna Chica (LCH.E1.3) is reported for the first time and corresponds to the early Middle Holocene (7,724–7,589 calBP).
Mitochondrial haplogroup diversity
The mitogenomes of the Early to Late Holocene individuals were assigned to 18 distinct haplotypes within known Native American mitochondrial haplogroups (Table 1). The diversity of haplotypes observed in the Early to Middle Holocene Pampas (excluding the Late Holocene individual from Laguna Chica) is characterized by the prevalence of C1 haplotypes (C1b: 35.3% and C1c: 17.6%), a higher frequency of A2 haplotypes (23.5%) compared to D1 haplotypes (17.6%), the detection of a B2b haplotype (6%), and the absence of D4h3a. Both B2b and C1c are absent in other pre-Columbian populations of the Eastern SCSA (de la Fuente et al., 2015; Crespo et al., 2017; Postillone et al., 2020b) but found in modern-day groups of central Argentina (including the Pampas) and northern Patagonia (Bailliet et al., 1994; Merriwether et al., 1995; Bobillo et al., 2010; Perego et al., 2010; Gómez-Carballa et al., 2016; Motti et al., 2020).
The oldest mitogenome obtained in this study from an individual found at the site Laguna de los Pampas (LLP.S2.E1; 10,223–9,764 calBP) exhibits a haplotype basal to D1j, according to the nomenclature suggested by phylotree.org (mtDNA tree Build 17 [18 Feb 2016]) (van Oven, 2015). The haplotype shows the characteristic C16242T and T16311C control region mutation pattern but lacks the T152C substitution (Bodner et al., 2012).
Two D1 mitogenomes, one from the early Middle Holocene Arroyo Seco 2 (ASO_B10_S15; 7,920–7,660 calBP) and the other from the Late Holocene Laguna Chica (LCH.E4.4; 1,627–1,565 calBP), exhibit the mutational pattern A8116G-C16187T characteristic of haplogroup D1g (van Oven and Kayser, 2009; Bodner et al., 2012). The haplotype of the younger LCH.E4.4 shows an additional four substitutions (T55C-A56G-T10595C-T16209C) that are considered part of the mutational motif characterizing the D1g5 subclade, despite lacking three substitutions at nucleotide positions G499A-A3505G-A14693G (Bodner et al., 2012; García et al., 2012). The ASO_B10_S15 D1g mitogenome shares the characteristic substitution T55C, suggesting that the haplotype might be basal to the subclade D1g5.
Time-measured phylogenetic analyses
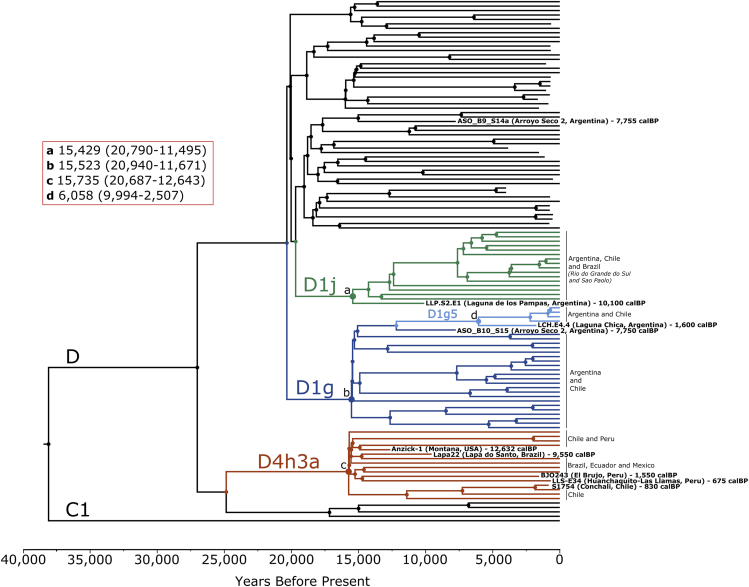
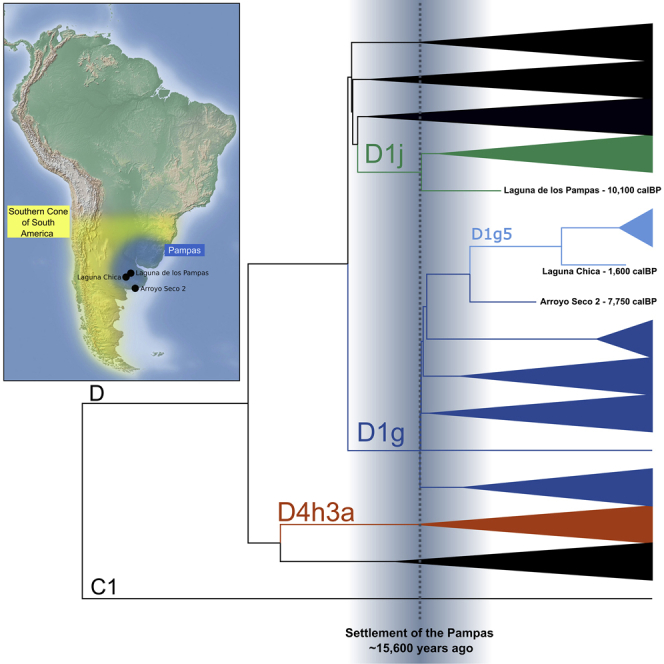
We further investigated the evolutionary relationship between the newly reported Early to Late Holocene D1j and D1g mitogenomes from the Argentinian Pampas and other mitogenomes from the SCSA and the Americas. To achieve this, we constructed a maximum clade credibility tree employing Bayesian phylogenetic inference using BEAST 1.8.0 (Drummond and Rambaut, 2007) (see STAR Methods). A total of 117 pre-Columbian and modern-day haplotypes belonging to haplogroups D1 and D4h3a (Bodner et al., 2012; de Saint Pierre et al., 2012b; Rasmussen et al., 2014; van Oven, 2015; Llamas et al., 2016; Posth et al., 2018) were included to estimate the coalescent times of these haplogroups. The resulting phylogenetic tree (Figures 2 and S4) confirms that the D1j mitogenome from Laguna de los Pampas (LLP.S2.E1) is basal to the entire D1j clade. The Middle Holocene D1g lineage from Arroyo Seco 2 (ASO_B10_S15) is positioned basal to a clade containing the Late Holocene Laguna Chica D1g lineage (LCH.E4.4), which again is ancestral to a number of mitogenomes falling into the subclade D1g5 containing contemporary Pampeans and Patagonians (Figure 2).
Figure 2.
Maximum clade credibility tree for haplogroups D1 and D4h3a
The estimated mean coalescent times are shown for clades D1j (a), D1g (b), D4h3a (c), and subhaplogroup D1g5 (d) in years before present (credible intervals in parentheses).
The radiocarbon dates of the ancient sequences from this and previously published studies were used as tip calibrations in the phylogenetic analysis (Table 1; Figure 2). We observe highly synchronous estimates of the times to the most recent common ancestor (TMRCA) for all three haplogroups D1g, D1j, and D4h3a (Figure S5), with an average divergence time of ∼15.6 kya (Figure 2). The TMRCA estimates for D1g (95% highest posterior density interval: 20.9–11.7 kya) and D1j (20.8–11.5 kya) fall well within the divergence age estimated for the founding lineage D1 in our study of 28.6–14.3 kya and previously published estimates of 23.7–12.5 kya (Llamas et al., 2016). Similarly, for D4h3a (21.6–12.6 kya), we were able to reproduce the previous TMRCA estimates of 20.9–12.6 kya (Llamas et al., 2016).
Discussion
Few studies so far have addressed the past genetic diversity of Early to Late Holocene hunter-gatherer populations in the Argentinian Pampas and their genetic relationship with populations of the SCSA. Positioned at the Eastern sector edge of the SCSA and harboring some of the earliest known archaeological sites, the region might have played a crucial role during the peopling of the region.
Our analysis revealed a higher diversity of mitochondrial haplotypes and a different distribution of haplogroup frequencies in the Early to Middle Holocene Pampas compared to other published Middle to Late Holocene pre-Columbian Eastern Patagonian and SCSA populations (Cabana et al., 2006; Marrero et al., 2007; García and Demarchi, 2009; Bobillo et al., 2010; Nores and Demarchi, 2011; de Saint Pierre et al., 2012b; Motti, 2012; Cardoso et al., 2013; de la Fuente et al., 2015; Crespo et al., 2017; Arencibia et al., 2019; Motti et al., 2020; Postillone et al., 2020a, 2020b) (Table S2). These include Late Holocene groups from the geographically near site of Paso Alsina 1, at the eastern Pampa-Patagonia transition (Postillone et al., 2020b), and other sites along the Atlantic coast of Argentinian Patagonia (Motti et al., 2015; Crespo et al., 2017). The difference in haplogroup frequencies between our data set and those previously published is driven by the predominance of haplogroup C1 and the presence of B2b in the Early to Middle Holocene Pampas, while haplogroup D1 is predominant and B2 is mostly absent in the Middle to Late Holocene pre-Columbian Eastern Patagonian and SCSA populations. Even though haplogroup C1 was prevalent in a late 19th and early 20th century indigenous cemetery in Tierra del Fuego (La Candelaria), haplogroup D1 had a high frequency and B2 was not found (Motti et al., 2020). Interestingly, the Late Holocene study in the Córdoba Province (Argentina) presents similar frequencies to our study, as haplogroup C is also predominant and it is followed by B and A, with haplogroup D being the least frequent (Nores and Demarchi, 2011). Moreover, haplogroup B has also been reported at high frequencies in individuals from the Early Holocene site Baño Nuevo (Manríquez et al., 2011) and Late Holocene site Lago Salitroso (Arencibia et al., 2019), both in south-western Patagonia. Since these last studies are based on general identification of mitochondrial haplogroup variants or short, low-resolution hypervariable region 1 sequences, as opposed to complete mitogenomes, it is not possible to determine if those individuals carried the same haplotypes as observed in this study.
Lineages falling into the B2b clade are found throughout South America, most frequently in the Andes, and the clade most likely evolved in the North of Mesoamerica shortly after the initial entry into the Americas (Brandini et al., 2018). Notwithstanding, the most common B2b lineages in the Pampas and central Argentina today are the derived lineages B2b3 or B2b14 (Bobillo et al., 2010; Brandini et al., 2018), while the Middle Holocene Laguna Chica individual (LCH.E2-I2.1) exhibits a less derived B2b haplotype. Gómez-Carballa et al. (2018) performed a phylogeographic analysis of present-day B2 mitogenomes. Their study uses a bifurcation model to explain the presence of the two subclades in the Pampas and central Argentina, proposing that the less frequent B2b14 arrived from the Andes, and B2b3 arrived via an Amazonian/Atlantic route. The presence of the basal B2b haplotype in the Middle Holocene Pampas suggests that individuals carrying an ancestral B2b haplotype spread in the northern SCSA earlier than the B2b3-B2b14 divergence, with the derived clades either arriving in subsequent waves or differentiating later via random genetic drift. These results reinforce the need to include ancient DNA analysis to disentangle past demographic events. Like the B2b haplotype, most A2, C1b, and C1c haplotypes found in the Early and Middle Holocene individuals from the Pampas are relatively basal to the Native American founding haplotypes (Perego et al., 2010). This observation is in agreement with the proposed rapid dispersal of the initial settlers throughout the American continent (Bodner et al., 2012; Llamas et al., 2016; Prates et al., 2020). However, due to the lack of available information for these haplogroups in the SCSA, changes in haplotype frequency through the Holocene period cannot be inferred.
D1g and D1j are the two main mitochondrial haplogroups thought to have a major role in the peopling of the SCSA (Bodner et al., 2012; de Saint Pierre et al., 2012b; García et al., 2012; Crespo et al., 2017; Crespo et al., 2018; de Saint Pierre, 2017). The finding of a D1g haplotype in Arroyo Seco 2 (ASO_B10_S15) supports the idea that D1g was already in the Pampas during the early Middle Holocene. It has been previously suggested that D1g arrived in the SCSA via the Pacific coast with trans-Andean migrations to the East based on its geographical distribution and frequency patterns in modern-day populations in southern Chile, Argentinian Patagonia, and the Pampas (Bodner et al., 2012; de Saint Pierre et al., 2012b; Crespo et al., 2017; de Saint Pierre, 2017). This early Middle Holocene D1g lineage from Arroyo Seco 2 is basal to a subclade containing the Late Holocene D1g5 mitogenome from Laguna Chica (LCH.E4.4), which is itself basal to the modern-day D1g5 lineages. The derived clade D1g5 has been proposed to have differentiated during the early peopling of the SCSA based on its geographically structured internal clades (Motti et al., 2019; Prieto et al., 2020). However, we find no evidence to corroborate this hypothesis as we estimated a divergence time of only ∼6 kya for D1g5 (Figure 2), much later than the peopling of the SCSA, and indicative that D1g5 was present in the Pampas before migrating southwards.
While Bodner et al. (2012) proposed that both D1g and D1j arrived in the SCSA via the Pacific coast, García et al. (2012) and Postillone et al. (2020b) have proposed that D1j probably migrated from central Argentina. This hypothesis was based on the D1j geographic distribution, which extended along the Patagonian Atlantic coast in the Late Holocene groups (Motti et al., 2015; Crespo et al., 2017; Postillone et al., 2020b), and is presently more frequent in the Eastern Pampas, while absent in western Patagonia and the extreme South (Bodner et al., 2012; García et al., 2012). The Early Holocene D1j mitogenome in Laguna de los Pampas is basal in the D1j phylogeny and supports the hypothesis that D1j spread from the Pampas. Interestingly, a cranial morphometric study showed some affinities between the Early Holocene sites of Laguna de los Pampas and Lagoa Santa (Brazil) (Menéndez et al., 2015). These observations, among others, have been used to suggest a possible Atlantic migration route (Miotti, 2006). However, genetic evidence supporting the so-called Paleoamerican morphology has recently been disproven (Raghavan et al., 2015; Moreno-Mayar et al., 2018). Furthermore, the D1j haplotype from Laguna de los Pampas lacks the T152C substitution but has the characteristic C16242T and T16311C substitutions. García et al. (2012) argue that the mutations at T16311C and T152C co-occur in both D1j and other D1 haplotypes found in central Argentina and propose that the substitution at T152C preceded the one at C16242T. Again, the ancient mitogenome from Laguna de los Pampas does not support this hypothesis as our observations indicate that the substitution C16242T preceded T152C. Nevertheless, it should be noted that 152 and 16,311 are mutational hotspots as described by Soares et al. (2009), increasing the odds of a recurrent mutation event.
The divergence ages for D1g and D1j have been estimated several times in previous studies using a range of methods and calibrations of the mitochondrial mutation rate. Bodner et al. (2012) used the HKY85 model and the human-chimpanzee split time (Goodman et al., 1998)—as shown in Mishmar et al. (2003)—and reported 18.3 ± 2.4 kya for D1g and 13.9 ± 2,9 kya for D1j. They also used the corrected rho-based molecular clock proposed by Soares et al. (2009) (also based on the human-chimpanzee split time) and obtained similar time estimates (D1g: 19.7 ± 3 kya; D1j: 14.9 ± 4.7 kya). De Saint Pierre, Bravi, et al. (2012) employed a Bayesian phylogenetic method and multiple mutation rates to propose a divergence date of ∼15 kya for D1g. In that study, they argued that the rate based on internal calibration points in the human mitochondrial tree (Endicott and Ho, 2008) was more concordant with the evolutionary process than when using any rate based on the deep human-chimpanzee split. However, de Saint Pierre (2017) recalculated both dates using a corrected rho-based molecular clock based on the human-chimpanzee split (Soares et al., 2009) and pushed the D1g and D1j divergence dates back to 22 ± 7 kya and 16.7 ± 9.4 kya, respectively. This result led to the proposal that D1g and D1j arrived in the SCSA via two temporally distinct migration pulses (de Saint Pierre, 2017). Here, we adopted a Bayesian phylogenetic method with tip dates calibration (Rieux and Balloux, 2016) and our coalescence time estimates for D1j, D1g, and even D4h3a were synchronous at ∼15.6 kya. A main strength of the present study is the ability to directly calibrate the molecular clock for all 3 clades (D1g, D1j, and D4h3a) via radiocarbon-dated ancient mitochondrial sequences. While using deep nodes as opposed to tips to calibrate the molecular clock does not affect the tree topology in phylogenetic reconstructions, it does impact significantly the evolutionary substitution rate and ultimately the dating of demographic events (Rieux et al., 2014). In fact, tip calibration is a much more reliable method when time-stamped sequences are available (Rieux and Balloux, 2016), such as is the case for recent human mitogenome studies based on ancient DNA (Brotherton et al., 2013; Fu et al., 2013; Rieux et al., 2014; Llamas et al., 2016; Posth et al., 2016; Nieves-Colón et al., 2020).
Beyond mitochondrial DNA results, nuclear genome data from Posth et al. (2018) demonstrated that the ancient individuals from Arroyo Seco 2 have the highest affinity to present-day populations of the SCSA and that Arroyo Seco 2 was an admixed population containing two main genetic ancestries, including the Early Holocene Lapa do Santo (Brazil)-associated ancestry and the Middle-Late Holocene ancestry predominant in present-day South Americans. Furthermore, they observed that ancient populations from central Chile (Los Rieles, 10,900 calBP) do not have a significant Lapa do Santo-associated ancestry, even though the most recent samples (Los Rieles, 5,100 calBP) also have the genetic ancestry related to all modern-day South Americans. These observations led Posth et al. (2018) to postulate a population replacement in South America ∼9 kya. However, the Laguna de los Pampas individual analyzed in this study and carrying the D1j haplogroup predates such putative replacement event, which indicates that D1j was present before the replacement. Considering that Arroyo Seco 2 harbors both genetic ancestries, the proposed replacement event could not have been complete as some mitochondrial lineages persist since the Late Pleistocene until today. A partial population replacement might reasonably explain the high diversity of haplotypes and distinct distribution of haplogroup frequencies observed in the Early and Middle Holocene Pampas compared to the Middle and Late Holocene pre-Columbian SCSA populations.
Hence, the synchronous estimated divergence dates for D1g, D1j, and D4h3a reported in this study (∼15.6 kya) suggest that the three clades might have emerged during the initial peopling of the Americas (∼16 kya) (Llamas et al., 2016) and diversified en route to South America. At ∼9 kya, a partial population replacement (Posth et al., 2018) as well as founder effects and genetic drift might have erased D1g and D1j outside the SCSA but not D4h3a. Alternatively, Prates et al. (2020) suggested that the earliest chronological threshold for the peopling of South America was ∼15.5 kya (16.6–15 kya) using a quantitative analysis of screened radiocarbon databases. Therefore, it could also be possible that the settling of South America led to diversification and D1g and D1j arose in South America. However, this scenario would imply that although D4h3a emerged at the same time, it might have had a different genetic history given its distribution across the entire American Pacific coast both in pre-Columbian and contemporary populations and its high frequency in Patagonia and Tierra del Fuego (Perego et al., 2009; Moraga et al., 2010; de Saint Pierre et al., 2012a).
In conclusion, the most parsimonious explanation for our results is that at least D1g, D1j, and likely B2b arrived in the Pampas during the initial settlement of the region as inferred from archaeological evidence (∼14,000 calBP) (Politis et al., 2014; Politis et al., 2016), molecular dating (∼20–11 kya), and quantitative analysis of radiocarbon dates (16.6–15 kya) (Prates et al., 2020). Despite the caveat that mitochondrial data from the present study do not allow us to infer specific migration routes, it is unlikely that the peopling of the Pampas was through the Pacific coast followed by the crossing of the Southern Andes. The Patagonian Ice Sheet reached the 35th parallel south during the Late Glacial Maximum and it was still in the Le Glacial phase ∼16–14 kya (Dickinson, 2011; Rabassa et al., 2011). Even if trans-cordilleran passes were open during that time, it is unlikely that they were suitable for human transit. Thus, an Atlantic or inland route is more likely for the peopling of the Pampas. Additionally, a partial population replacement took place ∼9 kya in South America and reached both the Pampas and the rest of the SCSA (Posth et al., 2018). Hence, some derived lineages that are currently specific to the Southern Cone could have been brought in by the second wave. For example, the basal B2b haplogroup found in Laguna Chica could have been present during the initial settlement of the Pampas, while present-day derived B2b3 and B2b14 could have come from the Andes and the Atlantic Route, respectively (Gómez-Carballa et al., 2018). Finally, Arroyo Seco 2 has the highest affinity to present-day SCSA indigenous populations (Posth et al., 2018), which suggests that the Pampas and the rest of the SCSA share some of their demographic history. Our results are consistent with the Pampas being one of the sources of the ancient Patagonian gene pool, and contemporary genetic differences observed between both regions could stem from founder effects, genetic drift, and the partial population replacement ∼9 kya.
Limitations of the study
There are limitations associated with the study of mtDNA, as this genetic marker represents the evolutionary history of the female population at a single locus and sample sizes are often small. Moreover, mitochondrial data from the present study do not allow us to test specific migration routes.
STAR★METHODS
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, Peptides, and Recombinant Proteins | ||
| Pfu Turbo Cx Hotstart DNA Polymerase | Agilent Technologies | 600412 |
| Herculase II Fusion DNA Polymerase | Agilent Technologies | 600679 |
| 2x HI-RPM hybridization buffer | Agilent Technologies | 5190-0403 |
| 0.5 M EDTA pH 8.0 | BioExpress | E177 |
| Sera-Mag Magnetic Speed-beads Carboxylate-Modified (1 μm, 3EDAC/PA5) | GE LifeScience | 6.51521E+13 |
| USER enzyme | New England Biolabs | M5505 |
| UGI | New England Biolabs | M0281 |
| Bst DNA Polymerase2.0, large frag. | New England Biolabs | M0537 |
| PE buffer concentrate | QIAGEN | 19065 |
| Proteinase K | Sigma Aldrich | P6556 |
| Guanidine hydrochloride | Sigma Aldrich | G3272 |
| 3M Sodium Acetate (pH 5.2) | Sigma Aldrich | S7899 |
| Water | Sigma Aldrich | W4502 |
| Tween-20 | Sigma Aldrich | P9416 |
| Isopropanol | Sigma Aldrich | 650447 |
| Ethanol | Sigma Aldrich | E7023 |
| 5M NaCl | Sigma Aldrich | S5150 |
| 1M NaOH | Sigma Aldrich | 71463 |
| 20% SDS | Sigma Aldrich | 5030 |
| PEG-8000 | Sigma Aldrich | 89510 |
| 1 M Tris-HCl pH 8.0 | Sigma Aldrich | AM9856 |
| dNTP Mix | Thermo Fisher Scientific | R1121 |
| ATP | Thermo Fisher Scientific | R0441 |
| 10x Buffer Tango | Thermo Fisher Scientific | BY5 |
| T4 Polynucleotide Kinase | Thermo Fisher Scientific | EK0032 |
| T4 DNA Polymerase | Thermo Fisher Scientific | EP0062 |
| T4 DNA Ligase | Thermo Fisher Scientific | EL0011 |
| Maxima SYBR Green kit | Thermo Fisher Scientific | K0251 |
| 50x Denhardt’s solution | Thermo Fisher Scientific | 750018 |
| SSC Buffer (20x) | Thermo Fisher Scientific | AM9770 |
| GeneAmp 10x PCR Gold Buffer | Thermo Fisher Scientific | 4379874 |
| Dynabeads MyOne Streptavidin T1 | Thermo Fisher Scientific | 65602 |
| Salmon sperm DNA | Thermo Fisher Scientific | 15632-011 |
| Human Cot-I DNA | Thermo Fisher Scientific | 15279011 |
| DyNAmo HS SYBR Green qPCR Kit | Thermo Fisher Scientific | F410L |
| Methanol, certified ACS | VWR | EM-MX0485-3 |
| Acetone, certified ACS | VWR | BDH1101-4LP |
| Dichloromethane, certified ACS | VWR | EMD-DX0835-3 |
| Hydrochloric acid, 6N, 0.5N & 0.01N | VWR | EMD-HX0603-3 |
| Critical Commercial Assays | ||
| High Pure Extender from Viral Nucleic Acid Large Volume Kit | Roche | 5114403001 |
| MinElute PCR Purification Kit | QIAGEN | 28006 |
| NextSeq® 500/550 High Output Kit v2 (150 cycles) | Illumina | FC-404-2002 |
| HiSeq® 4000 SBS Kit (50/75 cycles) | Illumina | FC-410-1001/2 |
| Deposited Data | ||
| Sequencing Data | GenBank | MW291661-MW291678 |
| Genotype Data | Reich Lab website | https://reich.hms.harvard.edu/datasets |
| Software and Algorithms | ||
| Samtools v1.11 | Li et al., 2009 | http://samtools.sourceforge.net/ |
| BWA v0.7.17-r1188 | Li and Durbin, 2009 | http://bio-bwa.sourceforge.net/ |
| SeqPrep v2 | https://github.com/jstjohn/SeqPrep | https://github.com/jstjohn/SeqPrep |
| AdapterRemoval v2 | Schubert et al., 2016 | https://github.com/MikkelSchubert/adapterremoval |
| Dedeup v0.12.07 | Peltzer et al., 2016 | https://eager.readthedocs.io/en/latest/ |
| PMDtools v0.60 | Skoglund et al., 2014 | https://github.com/pontussk/PMDtools |
| Haplogrep v2.0 | https://haplogrep.uibk.ac.at/index.html | |
| ContamMix | Fu et al. (2013) | https://github.com/DReichLab/ADNA-Tools |
| MEGA6 | Tamura et al., 2013 | https://www.megasoftware.net |
| mapDamage2.0 | Jónsson et al. (2013) | https://ginolhac.github.io/mapDamage/ |
| MUSCLE v3.8.1551 | Edgar, 2004 | https://www.drive5.com/muscle/ |
| Geneious v9.1.8 | https://www.geneious.com/ | |
| PartitionFinder v2.1.1 | Lanfear et al. (2012) | https://www.robertlanfear.com/partitionfinder/ |
| BEAST 1.8.0 | Drummond and Rambaut (2007) | https://beast.community |
| Tracer v1.7.1 | https://github.com/beast-dev/tracer/releases/tag/v1.7.1 | |
| TreeAnnotator v2.6.0 | https://beast2.blogs.auckland.ac.nz/treeannotator/ | |
| FigTree v1.4.4 | http://tree.bio.ed.ac.uk/software/ | |
Resource availability
Lead contact
Further information and requests should be directed to and will be fulfilled by the lead contact Xavier Roca-Rada (xavier.rocarada@adelaide.edu.au).
Materials availability
This study did not generate new unique reagents.
Data and code availability
The data underlying this article are available in GenBank under accession numbers MW291661–MW291678.
Experimental model and subject details
Archaeological context
We analyzed 18 whole mitochondrial genomes—7 of which are reported in this study for the first time—from human skeletal remains from three Early to Late Holocene archaeological sites in the Argentinian Pampas:
Arroyo Seco 2: 13
Laguna de los Pampas: 1
Laguna Chica: 4
Brief description of archaeological sites
The Arroyo Seco 2 (AS2) site (Figure S1) is located outside the city of Tres Arroyos. It is an open-air archaeological site situated on a low-lying knoll between a small temporary lake and a shallow creek (S 38º36’ and W 60º24’). From 1979 to the most recent excavations in 2019, a total of 77 units (314 m2) were opened in the site, including shovel tests and 3 long trenches. AS2 is a multicomponent site with several occupation episodes and a chronological range from the Late Pleistocene to historical times (Politis et al., 2014; Politis et al., 2016; Rafuse, 2017).
The hunting/scavenging events of the early hunter-gatherers at the AS2 site likely reflect at least two episodes. Temporary campsites were established in the area for the butchering/processing of Megatherium and the extinct horses Equus neogeus (at ∼12,200 14C years BP) and Hippidon (at ca. 11,180 14C years BP). During this period, other species of megafauna (Toxodon, Hemiauchenia and Glossotherium) were at the site, although the evidence of human agency is still inconclusive for these taxa. Lithic artefacts are mainly unifacially retouched quartzite flakes. Formal tools include side and end scrapers as well as bifacially retouched knifes. All raw materials are non-local and have different proveniences (Tandilia Hill Range, Ventania Hill Range, Atlantic seashore, etc.). After the extinction of the megamammals, there is a gap in the human occupation of AS2. During the Early Holocene (∼ 8,500 14C years BP), the site was occupied again by guanaco (Lama guanicoe) hunters, who established several overlapping campsites. Medium and large triangular projectile points, as well as a variety of unifacial quartzite and chert tools, characterize the lithic technology during this period. Around this time, funerary activities are abundant of human skeletons (n = 50) of both sexes and all age categories, dating between 7,819 ± 61 14C years BP and 4,487 ± 45 14C years BP (n = 27 dates). Thirteen of these skeletons were analyzed for this study (Table 1). The burial modalities are varied, including simple and multiple primary burials and simple and multiple funeral packages. The earliest level of burial included five skeletons with projectile points (midsized triangular stemless) stuck between and within the bones. Grave goods consisted of marine shell beads and necklaces of canid canines were recorded in some skeletons, indicating an early and complex treatment of the dead.
The Laguna de los Pampas (LLP) (Figure S2) and Laguna Chica (LC) (Figure S3) archaeological localities are placed in the Central Pampas Dunefields, one of the aeolian unit system from the eastern center of Argentina. These localities are situated in the current margins of shallow lakes, with recurrent hunter-gatherer occupations through the Holocene (Messineo et al., 2019). In LLP, eight burials were recovered in two sectors of a lake’s beach (sector 1: S 35º19’42’’ and W 61º31’50’’; sector 2: S 35º19’56’’ and W 61º31’53’’) due to re-exposition by water erosion. Moreover, isolated human bone remains corresponding to other eight individuals were found. The inhumations dated from the Early and Middle Holocene (Messineo et al., 2019). Burial 1 of LLP includes a multiple primary burial composed of two individuals: one adult female dating from 8,971 ± 77 14C years BP (LLP.S2.E1, AA-90127) and one sub-adult of 2–4 years old dating from 8,835 ± 83 14C years BP (L.LLP.S2.43, AA-93221). Burials 2 and 3 are simple primary and they dated back to 5,688 ± 36 14C years BP (L.LLP.S2.1062, AA-108848) and 5,819 ± 24 14C years BP (L.LLP.S1.E3, MAMS-24770), respectively. Bone remains of burial 4 and 5 were found on the surface and the method of the burials were not determined. They dated back to 5,924 ± 40 14C years BP (L.LLP.S2.E4, AA-106730) and 7,089 ± 37 14C years BP (L.LLP.S1.2706, AA-110832), respectively (Messineo et al., 2019). Burials 6, 7 and 8 are simple primary but they still do not have radiocarbon dates. Only one of these skeletons was analyzed in this study (Table 1).
The Laguna Chica (LCH) archaeological locality is located in the current margins of a small, temporal, shallow lake (S 36º5’8.80’’ and W 62º20’31.44’’) in the southeast of the Hinojo-Las Tunas Shallow Lake System. Seven burials were identified: four in Sector A located in the southern part of the shallow lake and three in Sector B in the western area. Inhumations dated from the Middle to the Late Holocene (Scheifler et al., 2017; Messineo et al., 2019). These burials were partially exposed at the lake’s beach due to water erosion. Only the cranium, the mandible and some remains of the thorax region were recovered from Burial N 1 (sample SC50-L763, LCH.E1.3). The morphological study of this individual’s scarce remains determined a probable male adult (genetically confirmed) and dated back to 6,870 ± 30 14C years BP (PSUAMS-6965). This individual was associated with a decorated pendant made of a canine of a jaguar (yaguareté, Panthera onca). Burial N 2 contained multiple primary burials represented with two individuals who had a dorsal disposition of the bodies with the lower limbs flexed. Individual N 1 (sample SC50-L762, LCH.E2-I1.2) was a female adult that dated back to 5,930 ± 15 14C years BP (UCIAMS-185302). Individual N 2 (sample SC50-L761, LCH.E2-I2.1) was a male adult that dated back to 6,080 ± 15 14C years BP (UCIAMS-185303). Burial N 4 (sample SC50-L764, LCH.E4.4) was an infant of undetermined sex. This individual dated back to 1,750 ± 15 14C years BP (UCIAMS-185301). Burials 3, 5 and 7 are simple primary represented by adult. The individual of the Burial 7 was associated with a bone tool made of a metapodial of guanaco (Lama guanicoe). Bone remains of burial 6 were found on the surface and the method of the burials were not determined. These last burials do not have radiocarbon dates yet.
Stratigraphic excavations have been performed in LLP and LCH locality, and abundant lithic materials and bones have been found. These lithic materials are characterized by a predominance of orthoquartzite and chert, which outcrops in the Tandilia Hill Range, 300 km far from the sites. Other lithic raw materials, such as granite, silicified dolomite, metaquartzite, rhyolite, basalt, silex, micaceous schist, and obsidian, were present in low frequencies. Some of these rocks come from diverse sectors of the Pampas (Ventania Hill Range, xerophytic woodland, and Tehuelche Mantle), and extra-regional areas. A high diversity of lithic tools was found, including side scrapers, end-scrapers, knives, multipurpose tools, triangular projectile points, etc. (Messineo et al., 2019). In addition, a standardize bone technology was also identified in LLP and centered in the production of beveled tools and blunted points made on guanaco tibiae (Messineo et al., 2019). The analysis of the material indicates that these sites were occupied during Holocene times. It might represent a succession of residential camps in the border of the shallow lakes by hunter-gatherers focused on the exploitation of guanaco (Lama guanicoe), although armadillo shell plates and Rheidae eggshells were well represented. In addition, complete eggs of greater rhea with a small perforation at the minor pole were found in both sites. They were possibly employed as flasks for storing water by hunter-gatherer groups in environments with shortage of sources of fresh water (Messineo et al., 2019).
Methods details
Radiocarbon dating and calibration
The new AMS 14C date reported in this article (LCH.E1.3; 7,724–7,589 calBP) has been generated at the Pennsylvania State University AMS Radiocarbon Laboratory, which is part of the Energy and Environmental Sustainability Laboratories (EESL). The Radiocarbon Laboratory is equipped with a National Electronics Corporation compact spectrometer with a 0.5MV accelerator (NEC 1.5SDH-1). The primary modifications impacting analytical measurement error are the use of a spherical ionizer ion source operating at high cathode voltage (7V) to generate intense C-beams, plus injection beam line changes for better ion-optical matching to the accelerator. The injector modifications include a second einzel lens plus an increased ion source voltage running at 47.5 kV combined with a redesigned large-gap injector magnet (DF01319) (Beverly et al., 2010). These alterations allow for analytical error in the 2–3‰ range for near modern samples under currents of up to 200 μA of 12C and routinely generating 100–120 μA of 12C from ∼0.7mg C samples. Radiocarbon ages are δ13C-corrected for mass dependent fractionation with δ13C values measured on the AMS and compared with OXII standards for normalization. Pre-treatment of sample (collagen extraction) was made in Laboratory of Pre-treatment for Isotopic Analysis (LAPREI) which is part of the Instituto de Investigaciones Arqueológicas y Palentolólógicas del Cuaternario Pampeano (INCUAPA), Argentina, following standard protocols.
XAD Amino Acids: Sample was physically cleaned using hand tools and sectioned with disposable Dremel cut-off wheels and then demineralized in 0.5 N HCl for 2–3 days at 5°C. The demineralized collagen pseudomorph was gelatinized at 60°C in 1–2 mL 0.01 N HCl for eight to ten hours. Sample gelatine was pipetted into a pre-cleaned 10 mL disposable syringe with an attached 0.45 mm Millex Durapore PVDF filter (precleaned with methanol and Nanopure H2O) and driven into a thick-walled culture tube. The filtered solution was lyophilized, and percent gelatinization and yield determined by weight. The sample gelatine was then hydrolyzed in 2 mL 6 N HCl for 22 h at 110—C. Supelco ENVI-ChroSPE (Solid Phase Extraction; Sigma-Aldrich) columns were prepped with 2 washes of HCl (2 mL) and rinsed with 10 mL DI H2O. With a 0.45 mm Millex Durapore filter attached, the SPE Column was equilibrated with 50 mL 6 N HCl and the washings discarded. 2 mL collagen hydrolyzate as HCl was pipetted onto the SPE column and driven with an additional 10 mL 6 N HCl dropwise with the syringe into a 20mmculture tube. The hydrolyzate was finally dried into a viscous syrup by passing UHP N2 gas over the sample heated at 50°C for 12 h. All calibrated 14C ages were calculated using OxCal version 4.3 (Bronk Ramsey, 2013).
Ancient DNA laboratory work
All samples were processed in the dedicated clean rooms at UCSC Paleogenomics Lab in Santa Cruz (UC-PL) or at David Reich’s lab at Harvard Medical School (HMS), following strict precautions to minimize contamination (Llamas et al., 2017). DNA was extracted from tooth roots and bone samples (Table S1) using a commonly applied extraction method optimized to retain short DNA fragments (Dabney et al., 2013) with modifications described in Posth et al. (2018). Partially UDG treated internally barcoded double stranded DNA (dsDNA) libraries were generated following the protocol by Rohland et al. (2015). Sequencing libraries were enriched for mitochondrial DNA using in-solution hybridization capture at HMS or UC-PL following the protocols described in Llamas et al. (2016). Enriched libraries were sequenced (2x150) on Illumina NextSeq sequencers at HMS or UC-PL. Raw reads were processed and mapped against hg19 as described in Posth et al. (2018).
Quantification and statistical analysis
Contamination estimation
We evaluated evidence for ancient DNA authenticity by measuring the rate of damage in the first nucleotide, flagging individuals as potentially contaminated if they had a less than 3% cytosine-to-thymine substitution rate. Additionally, we estimated mtDNA contamination using contamMix version 1.0–12 Fu et al. (2013). The software was run with down-sampling to 50x for samples above that coverage, --trimBases 2, 8 threads, 4 chains, and 2 copies, taking the first one that finished.
Mitochondrial DNA analyses
The sequencing read pileups were visualized in Geneious v9.1.8 (Biomatters; available from http://www.geneious.com/). SNPs were called in Geneious for all polymorphisms with minimum coverage 2 and a minimum variant frequency 0.7. The assembly and the resulting list of SNPs were verified manually and compared to SNPs reported at phylotree.org (mtDNA tree Build 17 [18 Feb 2016] (van Oven, 2015)) as described in Llamas et al. (2016).
Phylogenetic analyses
A total of 117 pre-Columbian and modern-day haplotypes belonging to haplogroups D1 and D4h3a and one C1a outgroup were compiled (Bodner et al., 2012; de Saint Pierre et al., 2012a, 2012b; Rasmussen et al., 2014; van Oven, 2015; Llamas et al., 2016; Posth et al., 2018) and aligned using Geneious v2019.2.1. Common indels and mutational hotspots were excluded (nucleotide positions 309.1C(C), 315.1C, AC indels at 515–522, 16182C, 16183C, 16193.1C(C), and C16519T) (van Oven and Kayser, 2009). PartitionFinder v2.1.1 (Lanfear et al., 2012) was used to select best-fit partitioning schemes and models of molecular evolution. The best model was TRN+G (TN93) for the whole mitogenome. BEAST 1.8.0 (Drummond and Rambaut, 2007) was used with tip date calibrations to reconstruct the phylogeny and constrained monophyly for D, D1g, D1j and D4h3a.
Strict clock was rejected according to a preliminary analysis with uncorrelated lognormal relaxed clock, so a relaxed clock was used. Three Markov Chain Monte Carlo (MCMC) chains of 100 million steps were performed, with sampling of parameters every 10,000 steps. Parameter traces were monitored in Tracer v1.7.1 to ensure convergence of the MCMC chains and effective sample sizes higher than 200. The three chains performed with highly similar results. The initial 10 million steps were discarded as burn-in using TreeAnnotator v2.6.0 and the final tree was visualized using FigTree v1.4.4.
Acknowledgments
The reported investigation of the individuals from Arroyo Seco 2, Laguna Chica, and Laguna de los Pampas has been permitted by the government of the Province of Buenos Aires (Nº de Registro 2014-3-A-125-1 and 2017-3-A-172-1). No indigenous communities in the counties have reclaimed Arroyo Seco 2 and Laguna de los Pampas remains. For Laguna Chica, in addition to obtaining permits from the provincial heritage institutions, the indigenous community living near the site (Comunidad Indígena Mapuche-Tehuelche Cacique Pincen) approved the study after consultation and participation in the rescue excavation (the skeletal remains will be re-buried). The authors want to acknowledge Beatriz Araujo Pincen, one of the leaders of the community as well as the local collaborators Ramon Coria, Juanjo Estevez, Ariel Grub, Marisa Martin, Sonia Finoccio, and Vicente Benito. The Municipality of Tres Arroyos was always very helpful during fieldwork. The authors want to also acknowledge Brendan Culleton for his assistance in the radiocarbon dating and for the summary of the methodology. X.R.-R. is supported by a Molecular Anthropology PhD scholarship from the University of Adelaide. L.F.-S. was supported by a U.S. National Science Foundation (NSF) grant (1515138) and the Wenner-Gren Foundation (SC-14-62). G.P. received support by PUE-CONICET 2015–2021 granted to INCUAPA and PICT 2015-2777 from ANPCYT, Argentina. P.G.M. received additional support by the National Geographic Society (#NGS-50543R-18) and CONICET (PIP Nº 0414). Y.S. is supported by an Australian Research Council Discovery Project (ARC DP190103705). J.C.T. is supported by an ARC Indigenous Discovery Grant (IN180100017). B.L. is supported by an ARC Future Fellowship (FT170100448). K.M.H. was supported from 2015 to 2017 by the National Science Foundation (NSF) SBE postdoctoral fellowship (1513501).
Author contributions
X.R.-R., K.M.H., B.L., and L.F.-S. performed the wet lab and dry lab genetic analyses. G.P., P.G.M., N.S., C.S., and M.G. provided archaeological samples, radiocarbon dates, and contextual information. D.R. provided genetic data. All authors interpreted the genetic results. X.R.-R., G.P., B.L., and L.F.-S. wrote the manuscript with input from all co-authors.
Declaration of interests
The authors declare no competing interests.
Published: June 25, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.102553.
Contributor Information
Bastien Llamas, Email: bastien.llamas@adelaide.edu.au.
Lars Fehren-Schmitz, Email: lfehrens@ucsc.edu.
Supplemental information
References
- Arencibia V., Crespo C., García Guraieb S., Russo M.G., Dejean C.B., Goñi R. Análisis genético poblacional de grupos cazadores recolectores del Holoceno tardío del Lago Salitroso (Santa Cruz, Argentina) RevArgAntropBiol. 2019;21:004. [Google Scholar]
- Bailliet G., Rothhammer F., Carnese F.R., Bravi C.M., Bianchi N.O. Founder mitochondrial haplotypes in Amerindian populations. Am. J. Hum. Genet. 1994;55:27–33. [PMC free article] [PubMed] [Google Scholar]
- Barrientos G., Perez S.I. Was there a population replacement during the Late mid-Holocene in the southeastern Pampas of Argentina? Archaeological evidence and Paleoecological basis. Quat. Int. 2005;132:95–105. [Google Scholar]
- Beverly R.K., Beaumont W., Tauz D., Ormsby K.M., von Reden K.F., Santos G.M., Southon J.R. The keck carbon cycle ams laboratory, university of California, irvine: status report. Radiocarbon. 2010;52:301–309. [Google Scholar]
- Bisso-Machado R., Bortolini M.C., Salzano F.M. Uniparental genetic markers in South amerindians. Genet. Mol. Biol. 2012;35:365–387. doi: 10.1590/S1415-47572012005000027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobillo M.C., Zimmermann B., Sala A., Huber G., Röck A., Bandelt H.J., Corach D., Parson W. Amerindian mitochondrial DNA haplogroups predominate in the population of Argentina: towards a first nationwide forensic mitochondrial DNA sequence database. Int. J. Leg. Med. 2010;124:263–268. doi: 10.1007/s00414-009-0366-3. [DOI] [PubMed] [Google Scholar]
- Bodner M., Perego U.A., Huber G., Fendt L., Röck A.W., Zimmermann B., Olivieri A., Gómez-Carballa A., Lancioni H., Angerhofer N. Rapid coastal spread of First Americans: novel insights from South America’s Southern Cone mitochondrial genomes. Genome Res. 2012;22:811–820. doi: 10.1101/gr.131722.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandini S., Bergamaschi P., Cerna M.F., Gandini F., Bastaroli F., Bertolini E., Cereda C., Ferretti L., Gómez-Carballa A., Battaglia V. The Paleo-Indian entry into South America according to mitogenomes. Mol. Biol. Evol. 2018;35:299–311. doi: 10.1093/molbev/msx267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronk Ramsey C. Recent and planned developments of the program OxCal. Radiocarbon. 2013;55:720–730. [Google Scholar]
- Brotherton P., Haak W., Templeton J., Brandt G., Soubrier J., Jane Adler C., Richards S.M., Der Sarkissian C., Ganslmeier R., Friederich S. Neolithic mitochondrial haplogroup H genomes and the genetic origins of Europeans. Nat. Commun. 2013;4:1764. doi: 10.1038/ncomms2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabana G.S., Merriwether D.A., Hunley K., Demarchi D.A. Is the genetic structure of Gran Chaco populations unique? Interregional perspectives on native South American mitochondrial DNA variation. Am. J. Phys. Anthropol. 2006;131:108–119. doi: 10.1002/ajpa.20410. [DOI] [PubMed] [Google Scholar]
- Cardoso S., Palencia-Madrid L., Valverde L., Alfonso-Sánchez M.A., Gómez-Pérez L., Alfaro E., Bravi C.M., Dipierri J.E., Peña J.A., de Pancorbo M.M. Mitochondrial DNA control region data reveal high prevalence of Native American lineages in Jujuy province, NW Argentina. Forensic Sci. Int. Genet. 2013;7 doi: 10.1016/j.fsigen.2013.01.007. e52–5. [DOI] [PubMed] [Google Scholar]
- Crespo C., Favier Dubois C., Russo M., Lanata J., Dejean C. First analysis of ancient mtDNA genetic diversity in Northern coast of Argentinean Patagonia. J. Archaeol. Sci. Rep. 2017;12:91–98. [Google Scholar]
- Crespo C.M., Lanata J.L., Cardozo D.G., Avena S.A., Dejean C.B. Ancient maternal lineages in hunter-gatherer groups of Argentinean Patagonia. Settlement, population continuity and divergence. J. Archaeol. Sci. Rep. 2018;18:689–695. [Google Scholar]
- Dabney J., Knapp M., Glocke I., Gansauge M.T., Weihmann A., Nickel B., Valdiosera C., García N., Pääbo S., Arsuaga J.L., Meyer M. Complete mitochondrial genome sequence of a Middle Pleistocene cave bear reconstructed from ultrashort DNA fragments. Proc. Natl. Acad. Sci. U S A. 2013;110:15758–15763. doi: 10.1073/pnas.1314445110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson W.R. Geological perspectives on the Monte Verde archeological site in Chile and pre-Clovis coastal migration in the Americas. Quat. Res. 2011;76:201–210. [Google Scholar]
- Dillehay T.D., Ramírez C., Pino M., Collins M.B., Rossen J., Pino-Navarro J.D. Monte Verde: seaweed, food, medicine, and the peopling of South America. Science. 2008;320:784–786. doi: 10.1126/science.1156533. [DOI] [PubMed] [Google Scholar]
- Dillehay T.D., Ocampo C., Saavedra J., Sawakuchi A.O., Vega R.M., Pino M., Collins M.B., Scott Cummings L., Arregui I., Villagran X.S. New archaeological evidence for an early human presence at Monte Verde, Chile. PLoS One. 2015;10 doi: 10.1371/journal.pone.0141923. e0141923–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donadei J.P. Local and nonlocal rocks: technological strategies and raw material management. Hunter-gatherer mobility for mid-Holocene groups of eastern Tandilia range (Argentina) J. Archaeol. Sci. Rep. 2019;24:264–275. [Google Scholar]
- Drummond A.J., Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 2007;7:214–218. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endicott P., Ho S.Y. A bayesian evaluation of human mitochondrial substitution rates. Am. J. Hum. Genet. 2008;82:895–902. doi: 10.1016/j.ajhg.2008.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flegenheimer N., Miotti L., Mazzia N. Rethinking early objects and landscape in the Southern Cone: fishtail point concentrations in the Pampas and Northern Patagonia. In: Graf K., Ketron C., Waters M., editors. Paleoamerican Odyssey. Center for the Study of First Americans; 2013. pp. 359–376. [Google Scholar]
- Fu Q., Mittnik A., Johnson P.L.F., Bos K., Lari M., Bollongino R., Sun C., Giemsch L., Schmitz R., Burger J. A revised timescale for human evolution based on ancient mitochondrial genomes. Curr. Biol. 2013;23:553–559. doi: 10.1016/j.cub.2013.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Fuente C., Galimany J., Kemp B.M., Judd K., Reyes O., Moraga M. Ancient marine hunter-gatherers from Patagonia and Tierra Del Fuego: diversity and differentiation using uniparentally inherited genetic markers. Am. J. Phys. Anthropol. 2015;158:719–729. doi: 10.1002/ajpa.22815. [DOI] [PubMed] [Google Scholar]
- de la Fuente C., Ávila-Arcos M.C., Galimany J., Carpenter M.L., Homburger J.R., Blanco A., Contreras P., Cruz Dávalos D., Reyes O., San Roman M. Genomic insights into the origin and diversification of late maritime hunter-gatherers from the Chilean Patagonia. Proc. Natl. Acad. Sci. U S A. 2018;115:E4006–E4012. doi: 10.1073/pnas.1715688115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García A., Demarchi D.A. Incidence and distribution of native American mtDNA haplogroups in central Argentina. Hum. Biol. 2009;81:59–69. doi: 10.3378/027.081.0105. [DOI] [PubMed] [Google Scholar]
- García A., Pauro M., Nores R., Bravi C.M., Demarchi D.A. Phylogeography of mitochondrial haplogroup D1: an early spread of subhaplogroup D1j from Central Argentina. Am. J. Phys. Anthropol. 2012;149:583–590. doi: 10.1002/ajpa.22174. [DOI] [PubMed] [Google Scholar]
- García-Bour J., Pérez-Pérez A., Álvarez S., Fernández E., López-Parra A.M., Arroyo-Pardo E., Turbón D. Early Population Differentiation in Extinct Aborigines from Tierra del Fuego-Patagonia: ancient mtDNA Sequences and Y-Chromosome STR Characterization. Am. J. Phys. Anthropol. 2004;123:361–370. doi: 10.1002/ajpa.10337. [DOI] [PubMed] [Google Scholar]
- Gómez-Carballa A., Moreno F., Álvarez-Iglesias V., Martinón-Torres F., García-Magariños M., Pantoja-Astudillo J.A., Aguirre-Morales E., Bustos P., Salas A. Revealing latitudinal patterns of mitochondrial DNA diversity in Chileans. Forensic Sci. Int. Genet. 2016;20:81–88. doi: 10.1016/j.fsigen.2015.10.002. [DOI] [PubMed] [Google Scholar]
- Gómez-Carballa A., Pardo-Seco J., Brandini S., Achilli A., Perego U.A., Coble M.D., Diegoli T.M., Álvarez-Iglesias V., Martinón-Torres F., Olivieri A. The peopling of South America and the trans-Andean gene flow of the first settlers. Genome Res. 2018;28:767–779. doi: 10.1101/gr.234674.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman M., Porter C.A., Czelusniak J., Page S.L., Schneider H., Shoshani J., Gunnell G., Groves C.P. Toward a phylogenetic classification of primates based on DNA evidence complemented by fossil evidence. Mol. Phylogenet. Evol. 1998;9:585–598. doi: 10.1006/mpev.1998.0495. [DOI] [PubMed] [Google Scholar]
- Jónsson H., Ginolhac A., Schubert M., Johnson P.L.F., Orlando L. MapDamage2.0: fast approximate Bayesian estimates of ancient DNA damage parameters. Bioinformatics. 2013;29:1682–1684. doi: 10.1093/bioinformatics/btt193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalueza C., Pérez-Pérez A., Prats E., Cornudella L., Turbón D. ‘Lack of founding Amerindian mitochondrial DNA lineages in extinct Aborigines from Tierra del Fuego–Patagonia’. Hum. Mol. Genet. 1997;6:41–46. doi: 10.1093/hmg/6.1.41. [DOI] [PubMed] [Google Scholar]
- Lanfear R., Calcott B., Ho S.Y., Guindon S. PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol. Biol. Evol. 2012;29:1695–1701. doi: 10.1093/molbev/mss020. [DOI] [PubMed] [Google Scholar]
- Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G.R. Durbin. 1000 Genome Project Data Processing Subgroup, The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llamas B., Fehren-Schmitz L., Valverde G., Soubrier J., Mallick S., Rohland N., Nordenfelt S., Valdiosera C., Richards S.M., Rohrlach A. Ancient mitochondrial DNA provides high-resolution time scale of the peopling of the Americas. Sci. Adv. 2016;2:e1501385. doi: 10.1126/sciadv.1501385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llamas B., Willerslev E., Orlando L. Human evolution: a tale from ancient genomes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2017;372:20150484. doi: 10.1098/rstb.2015.0484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manríquez G., Moraga M., Santoro C., Aspillaga E., Arriaza B.T., Rothhammer F. Morphometric and mtDNA analyses of archaic skeletal remains from Southwestern South America. Chungará (Arica) 2011;43:283–292. [Google Scholar]
- Marrero A.R., Bravi C., Stuart S., Long J.C., Pereira das Neves Leite F., Kommers T., Carvalho C.M., Pena S.D., Ruiz-Linares A., Salzano F.M., Cátira Bortolini M. Pre- and post-Columbian gene and cultural continuity: the case of the Gaucho from southern Brazil. Hum. Hered. 2007;64:160–171. doi: 10.1159/000102989. [DOI] [PubMed] [Google Scholar]
- Martínez G., Gutiérrez M.A., Messineo P.G., Kaufmann C.A., Rafuse D.J. Subsistence strategies in Argentina during the late Pleistocene and early Holocene. Quat. Sci. Rev. 2016;144:51–65. [Google Scholar]
- Mazzanti D.L., Martínez G.A., Quintana C. Early settlements in eastern Tandilia, Buenos Aires Province, Argentina: archaeological contexts and site formation processes. In: Miotti L., Salemme M., Flegenheimer N., Goebel T., editors. Special Edition: Southbound. Late Pleistocene Peopling of Latin America. Texas University; 2012. pp. 115–119. [Google Scholar]
- Mazzanti D.L., Martínez G.A., Quintana C. Asentamientos del Holoceno medio en Tandilia oriental. Aportes para el conocimiento de la dinámica poblacional de la región pampeana, Argentina. Relaciones de la Sociedad Argentina de Antropología. 2015;XL:209–231. [Google Scholar]
- Menéndez L.P., Perez S.I., Pucciarelli H.M., Bonomo M., Messineo P.G., Gonzalez M.E., Politis G.G. Early holocene human remains from the Argentinean pampas: cranial variation in South America and the American peopling. PaleoAmerica. 2015;1:251–265. [Google Scholar]
- Merriwether D.A., Rothhammer F., Ferrell R.E. Distribution of the four founding lineage haplotypes in native Americans suggests a single wave of migration for the New World. Am. J. Phys. Anthropol. 1995;98:411–430. doi: 10.1002/ajpa.1330980404. [DOI] [PubMed] [Google Scholar]
- Messineo P.G., González M.E., Álvarez M.C., Pal N. Las ocupaciones humanas en la localidad Arqueológica Laguna de Los Pampas (Campo De Dunas Del Centro Pampeano, Argentina) Durante El Holoceno. Latin Am. Antiq. 2018;29:736–753. [Google Scholar]
- Messineo P.G., Barros M.P., Pal N., Scheifler N.A. Transporting rocks to an empty environment of lithic raw materials. The case of the Central Pampean Dunefields (Argentina) J. Archaeol. Sci. Rep. 2019;25:433–446. [Google Scholar]
- Messineo P.G., Scheifler N.A., Álvarez M.C., González M., Pal N., Barros P., Politis G. A model of human occupation in the Central Pampean Dunefields of Argentina. PaleoAmerica: A J. Early Hum. Migration Dispersal. 2019;5:378–391. [Google Scholar]
- Miotti L.L. 2° Simposio Internacional el Hombre Temprano en América. 2006. La fachada atlántica, como puerta de ingreso alternativa de la colonización humana de América del Sur durante la transición Pleistoceno/Holoceno; pp. 155–188. [Google Scholar]
- Miotti L., Tonni E., Marchionni L. What happened when the Pleistocene megafauna became extinct? Quat. Int. 2018;473:173–189. [Google Scholar]
- Mishmar D., Ruiz-Pesini E., Golik P., Macaulay V., Clark A.G., Hosseini S., Brandon M., Easley K., Chen E., Brown M.D. Natural selection shaped regional mtDNA variation in humans. Proc. Natl. Acad. Sci. U S A. 2003;100:171–176. doi: 10.1073/pnas.0136972100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moraga M., de Sanit Pierre M., de Saint Torres F., Ríos J. Kinship by maternal via between the last descendants of kawéskar ethnicity and burials in the patagonian channels: evidence from the study of mitochondrial lineages. Magallania. 2010;38:103–114. [Google Scholar]
- Moreno-Mayar J.V., Vinner L., de Barros Damgaard P., de la Fuente C., Chan J., Spence J.P., Allentoft M.E., Vimala T., Racimo F., Pinotti T. Early human dispersals within the Americas. Science. 2018;362:eaav2621. doi: 10.1126/science.aav2621. [DOI] [PubMed] [Google Scholar]
- Motti J.M.B. Universidad Nacional de La Plata; 2012. Caracterización de linajes maternos en la población actual del Noroeste y Centro-oeste argentinos. [Google Scholar]
- Motti J.M.B., Hagelberg E., Lindo J., Malhi R.S., Bravi C.M., Guichón R.A. Primer genoma mitocondrial en restos humanos de la costa de Santa Cruz, Argentina. Magallania. 2015;43:119–131. [Google Scholar]
- Motti J.M.B., Muñoz A.S., Cruz I., D’Angelo del Campo M.D., Borrero L.A., Bravi C.M., Guichón R.A. Arqueología de la Patagonia: El pasado en las arenas; 2019. Análisis de ADN mitocondrial en restos humanos del Holoceno tardío del sur de Santa Cruz; pp. 493–503. [Google Scholar]
- Motti J.M.B., Winingear S., Valenzuela L.O., Nieves-Colón M.A., Harkins K.M., García Laborde P., Bravi C.M., Guichón R.A., Stone A.C. Identification of the geographic origins of people buried in the cemetery of the Salesian Mission of Tierra del Fuego through the analyses of mtDNA and stable isotopes. J. Archaeol. Sci. Rep. 2020;33:102559. [Google Scholar]
- Nakatsuka N., Lazaridis I., Barbieri C., Skoglund P., Rohland N., Mallick S., Posth C., Harkins-Kinkaid K., Ferry M., Harney É. A paleogenomic reconstruction of the deep population history of the Andes. Cell. 2020;181:1131–1131.e21. doi: 10.1016/j.cell.2020.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsuka N., Luisi P., Motti J.M.B., Salemme M., Santiago F., D'Angelo Del Campo M.D., Vecchi R.J., Espinosa-Parrilla Y., Prieto A., Adamski N. Ancient genomes in South Patagonia reveal population movements associated with technological shifts and geography. Nat. Commun. 2020;11:3868. doi: 10.1038/s41467-020-17656-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieves-Colón M.A., Pestle W.J., Reynolds A.W., Llamas B., de la Fuente C., Fowler K., Skerry K.M., Crespo-Torres E., Bustamante C.D., Stone A.C. Ancient DNA reconstructs the genetic legacies of precontact Puerto Rico communities. Mol. Biol. Evol. 2020;37:611–626. doi: 10.1093/molbev/msz267. [DOI] [PubMed] [Google Scholar]
- Nores R., Demarchi D.A. Análisis de haplogrupos mitocondriales en restos humanos de sitios arqueológicos de la provincia de Córdoba. Revista Argentina de Antropología Biológica. 2011;13:43–54. [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A.S. Kumar. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Oven M. PhyloTree Build 17: growing the human mitochondrial DNA tree. Forensic Sci. Int. Genet. Suppl. Ser. 2015;5:e392–e394. [Google Scholar]
- van Oven M., Kayser M. Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation. Hum. Mutat. 2009;30:E386–E394. doi: 10.1002/humu.20921. [DOI] [PubMed] [Google Scholar]
- Peltzer A., Jäger G., Herbig A., Seitz A., Kniep C., Krause J. EAGER: efficient ancient genome reconstruction. Genome Biol. 2016;17:60. doi: 10.1186/s13059-016-0918-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perego U.A., Achilli A., Angerhofer N., Accetturo M., Pala M., Olivieri A., Hooshiar Kashani B., Ritchie K.H., Scozzari R., Kong Q.P. Distinctive paleo-Indian migration routes from Beringia marked by two rare mtDNA haplogroups. Curr. Biol. 2009;19:1–8. doi: 10.1016/j.cub.2008.11.058. [DOI] [PubMed] [Google Scholar]
- Perego U.A., Angerhofer N., Pala M., Olivieri A., Lancioni H., Hooshiar Kashani B., Carossa V., Ekins J.E., Gómez-Carballa A., Huber G. The initial peopling of the Americas: a growing number of founding mitochondrial genomes from Beringia. Genome Res. 2010;20:1174–1179. doi: 10.1101/gr.109231.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez S.I., Bernal V., Gonzalez P.N., Sardi M., Politis G.G. Discrepancy between cranial and DNA data of early Americans: implications for American peopling. PLoS One. 2009;4:e5746. doi: 10.1371/journal.pone.0005746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Politis G. Handbook of South American Archaeology. Springer; 2008. The pampas and campos of South America. pp. 235–261. [Google Scholar]
- Politis G.G., Gutiérrez M.A., Scabuzzo C. In: Estado actual de las investigaciones en el sitio arqueológico Arroyo Seco 2: (partido de tres arroyos, provincia de Buenos Aires, Argentina) Politis G.G., Gutiérrez M.A., Scabuzzo C., editors. INCUAPA-CONICET, UNICEN; 2014. pp. 329–370. [Google Scholar]
- Politis G.G., Prates L. Clocking the arrival of Homo sapiens in the southern cone of South America. In: Harvati K., Jäger G., Reyes-Centeno H., editors. New Perspectives on the Peopling of the Americas. Words, Bones, Genes, Tools. DFG Center for Advanced Studies Series; 2018. pp. 79–106. [Google Scholar]
- Politis G.G., Gutiérrez M.A., Rafuse D.J., Blasi A. The arrival of Homo sapiens into the Southern Cone at 14,000 years ago. PLoS One. 2016;11:e0162870. doi: 10.1371/journal.pone.0162870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posth C., Renaud G., Mittnik A., Drucker D.G., Rougier H., Cupillard C., Valentin F., Thevenet C., Furtwängler A., Wißing C. Pleistocene mitochondrial genomes suggest a single major dispersal of non-africans and a late glacial population turnover in Europe. Curr. Biol. 2016;26:827–833. doi: 10.1016/j.cub.2016.01.037. [DOI] [PubMed] [Google Scholar]
- Posth C., Nakatsuka N., Lazaridis I., Skoglund P., Mallick S., Lamnidis T.C., Rohland N., Nägele K., Adamski N., Bertolini E. Reconstructing the deep population history of central and South America. Cell. 2018;175:1185–1185.e22. doi: 10.1016/j.cell.2018.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postillone M.B., Cobos V.A., Urrutia C., Dejean C., Gonzalez P.N., Perez S.I., Bernal V. Mitochondrial DNA diversity and evolutionary history of native human populations of Argentinean Northwest Patagonia (Argentina) Hum. Biol. 2020;91:57–79. doi: 10.13110/humanbiology.91.2.01. [DOI] [PubMed] [Google Scholar]
- Postillone M.B., Martínez G., Flensborg G., Dejean C.B. First analysis of mitochondrial lineages from the eastern Pampa–Patagonia transition during the final late Holocene. Am. J. Phys. Anthropol. 2020;171:659–670. doi: 10.1002/ajpa.24016. [DOI] [PubMed] [Google Scholar]
- Prates L., Politis G.G., Perez S.I. Rapid radiation of humans in South America after the last glacial maximum: a radiocarbon-based study. PLoS one. 2020;15:e0236023. doi: 10.1371/journal.pone.0236023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieto A., Morano S., Cárdenas P., Sierpe V., Calas E., Christensen M., Lefevre C., Laroulandie V., Espinosa-Parrilla Y., Ramirez O. A Novel Child Burial from Tierra del Fuego: a Preliminary Report. J. Isl. Coastal Archaeol. 2020;15:436–454. [Google Scholar]
- Rabassa J., Coronato A., Martínez O. Late Cenozoic glaciations in Patagonia and Tierra del Fuego: an updated review. Biol. J. Linn. Soc. 2011;103:316–335. [Google Scholar]
- Rafuse D.J. Early to Middle holocene subsistence strategies in the pampas region: evidence from the Arroyo Seco 2 site. J. Archaeol. Sci. Rep. 2017;12:673–683. [Google Scholar]
- Raghavan M., Steinrücken M., Harris K., Schiffels S., Rasmussen S., DeGiorgio M., Albrechtsen A., Valdiosera C., Ávila-Arcos M.C., Malaspinas A.S. Genomic evidence for the Pleistocene and recent population history of Native Americans. Science. 2015;349:aab3884. doi: 10.1126/science.aab3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen M., Anzick S.L., Waters M.R., Skoglund P., DeGiorgio M., Stafford T.W., Rasmussen S., Moltke I., Albrechtsen A., Doyle S.M. The genome of a Late Pleistocene human from a Clovis burial site in western Montana. Nature. 2014;506:225–229. doi: 10.1038/nature13025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieux A., Balloux F. Inferences from tip-calibrated phylogenies: a review and a practical guide. Mol. Ecol. 2016;25:1911–1924. doi: 10.1111/mec.13586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieux A., Eriksson A., Li M., Sobkowiak B., Weinert L.A., Warmuth V., Ruiz-Linares A., Manica A., Balloux F. Improved calibration of the human mitochondrial clock using ancient genomes. Mol. Biol. Evol. 2014;31:2780–2792. doi: 10.1093/molbev/msu222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohland N., Harney E., Mallick S., Nordenfelt S., Reich D. Partial uracil-DNA-glycosylase treatment for screening of ancient DNA. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2015;370:20130624. doi: 10.1098/rstb.2013.0624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Saint Pierre M. Antiquity of mtDNA lineage D1g from the southern cone of South America supports pre-Clovis migration. Quat. Int. 2017;444:19–25. [Google Scholar]
- de Saint Pierre M., Bravi C.M., Motti J.M.B., Fuku N., Tanaka M., Llop E., Bonatto S.L., Moraga M. An alternative model for the early peopling of Southern South America revealed by analyses of three mitochondrial DNA haplogroups. PLoS One. 2012;7:e43486. doi: 10.1371/journal.pone.0043486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Saint Pierre M., Gandini F., Perego U.A., Bodner M., Gómez-Carballa A., Corach D., Angerhofer N., Woodward S.R., Semino O., Salas A. Arrival of paleo-Indians to the southern cone of South America: new clues from mitogenomes. PLoS One. 2012;7 doi: 10.1371/journal.pone.0051311. e51311–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheifler N.A., Messineo P.G., Antiñir A. Cazadores-recolectores en el sistema lagunar Hinojo-Las Tunas (región pampeana, área Oeste) durante la transición Holoceno temprano-medio y tardío. Primeros resultados de las investigaciones arqueológicas. Comechingonia, Revista de Arqueología. 2017;21:287–314. [Google Scholar]
- Schubert M., Lindgreen S. Adapter removal v2: rapid adapter trimming, identification, and read merging. BMC Res. Notes. 2016;9:88. doi: 10.1186/s13104-016-1900-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoglund P., Northoff B.H., Shunkov M.V., Derevianko A.P., Pääbo S., Krause J.M.J. Separating endogenous ancient DNA from modern day contamination in a Siberian Neandertal. Proc. Natl. Acad. Sci. USA. 2014;111:2229–2234. doi: 10.1073/pnas.1318934111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares P., Ermini L., Thomson N., Mormina M., Rito T., Röhl A., Salas A., Oppenheimer S., Macaulay V., Richards M.B. Correcting for purifying selection: an improved human mitochondrial molecular clock. Am. J. Hum. Genet. 2009;84:740–759. doi: 10.1016/j.ajhg.2009.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article are available in GenBank under accession numbers MW291661–MW291678.