Abstract
One of the endogenous estrogens, 17β-estradiol (E2) is a female steroid hormone secreted from the ovary. It is well established that E2 causes biochemical and histological changes in the uterus. However, it is not completely understood how E2 regulates the oviductal environment in vivo. In this study, we assessed the effect of E2 on each oviductal cell type, using an ovariectomized-hormone-replacement mouse model, single-cell RNA-sequencing (scRNA-seq), in situ hybridization, and cell-type-specific deletion in mice. We found that each cell type in the oviduct responded to E2 distinctively, especially ciliated and secretory epithelial cells. The treatment of exogenous E2 did not drastically alter the transcriptomic profile from that of endogenous E2 produced during estrus. Moreover, we have identified and validated genes of interest in our datasets that may be used as cell- and region-specific markers in the oviduct. Insulin-like growth factor 1 (Igf1) was characterized as an E2-target gene in the mouse oviduct and was also expressed in human fallopian tubes. Deletion of Igf1 in progesterone receptor (Pgr)-expressing cells resulted in female subfertility, partially due to an embryo developmental defect and embryo retention within the oviduct. In summary, we have shown that oviductal cell types, including epithelial, stromal, and muscle cells, are differentially regulated by E2 and support gene expression changes, such as growth factors that are required for normal embryo development and transport in mouse models. Furthermore, we have identified cell-specific and region-specific gene markers for targeted studies and functional analysis in vivo.
Keywords: embryo development, embryo transport, estrogen, insulin-like growth factor 1, oviduct, scRNA-seq
1 |. INTRODUCTION
The oviduct (or Fallopian tube in humans) is a tube-like structure that connects the ovary to the uterus. The oviduct is divided into four main components known as the infundibulum, ampulla, isthmus, and the uterotubal junction (UTJ, also known as intramural junction in humans). Ovulated eggs enter the oviduct through the infundibulum, the distal most region of the oviduct, and are subsequently fertilized in the ampulla. Embryos undergo divisions and compaction in the isthmus region of the oviduct and transit through the UTJ to reach the uterus. The oviduct provides an optimal microenvironment for gamete fertilization, preimplantation embryo development, and embryo transport.1 The majority of the cells encompassing the epithelial layer of the oviduct are non-ciliated (secretory) cells and ciliated cells. In the bovine oviduct, non-ciliated secretory cells are responsible for secreting a variety of proteins and metabolites to act as growth factors, and metabolic and immune regulators to aid in gamete maturation and fertilization.2 Secretory cells are concentrated mainly in the proximal (isthmus and UTJ) region of the oviduct and are more sparsely dispersed toward the distal (infundibulum and ampulla) region of the oviduct.3 However, ciliated cells are present in the opposing gradient compared to secretory cells: infundibulum > ampulla > isthmus.3 Ciliated cells are responsible for creating a flowing current of oviductal fluid toward the uterus that aids in proper embryo transport during early pregnancy.4 Alteration of secretory function or inflammation of the oviduct, such as hydrosalpinx or salpingitis, negatively impacts the development of preimplantation embryos.5–7 In addition, disruption of ciliary function has been shown to increase the risk for ectopic pregnancy (a condition where the embryo implants outside of the uterus).8 Therefore, the oviduct is crucial for the establishment of a successful pregnancy. Although ectopic pregnancy only occurs in humans, the mouse provides a mammalian model for studying disruptions in embryo transport, a potential contributing factor to ectopic pregnancy in humans.
17β-Estradiol (E2) is mainly produced from the granulosa cells of the ovary. In several mammalian species, elevated levels of E2 alter the histoarchitecture and function of the oviduct.9 It is well-established that E2 is required for ovarian and uterine function. Global knockout of estrogen receptor α (Esr1−/−) in mice leads to female sterility, mainly due to anovulation and a uterine receptivity defect.10 In other female reproductive tissues, such as the uterus, E2 induces the production of growth factors, eg, insulin-like growth factor 1 (IGF1), in the stromal cell layer.11 Subsequently, stromal-derived IGF1 then stimulates uterine epithelial cell production via a paracrine signaling pathway.12 Interestingly, these growth factors (including IGF1, fibroblast growth factors [FGFs], complement component 3 [C3], and demilune cell parotid proteins [DCPPs]) have been identified as embryotrophic factors that facilitate preimplantation embryo development to the blastocyst stage and prevent apoptosis.13–17 However, the mechanistic action and the fundamental understanding of E2 in the oviduct is not completely understood. In the oviduct, several studies showed that E2 exerts its activities via both genomic and non-genomic pathways in rodents.18–22 In this study, we specifically focus on the classical genomic pathway by evaluating gene expression signatures of specific cell types in the oviduct. Our previous work showed that epithelial cell-specific deletion of Esr1 in the oviduct resulted in an alteration of protease and protease inhibitor production, leading to embryo death at 1.5 days post coitus (1.5 dpc).23 However, it is still unclear whether these E2-mediated proteases and protease inhibitors are expressed in specific epithelial cell types of the oviduct.
The use of single cell-RNA sequencing (scRNA-seq) analysis is an increasingly popular tool in the scientific community due to its capability to identify transcriptomes at a single cell resolution as well as to discover the heterogeneity of cell populations within tissues.24 Single cell encapsulation for sequencing pipeline is commercially available.24 In this study we used scRNA-seq analysis to determine the cell types and their response to E2 in the oviduct. Overall, the main objective of this study is to provide fundamental knowledge regarding the roles of E2 on the oviductal cellular response at a single cell resolution. Recent studies published in preprint showed that the epithelial cells from the proximal and distal regions of the oviduct develop from two distinct cell lineages.25 In contrast to these previous studies, our research focuses on the effect of E2 on all cell types present in the oviduct. Lastly, all genes expressed in our scRNA-seq datasets are readily available for the scientific community as a web search format.
2 |. MATERIALS AND METHODS
2.1 |. Animals
All animals were maintained at Washington State University and were handled according to Animal Care and Use Committee guidelines using approved protocols #6147 and 6151. For scRNA-seq experiments, adult female mice (C57B6/J, 8–12 weeks old) were purchased from the Jackson (Jax) laboratory. Animals were ovariectomized and left for 14 days to recover and be rid of ovarian derived-steroid hormones.12 Sesame oil (100 μL) was used as a negative (Vehicle or Veh) control group. 17β-estradiol (E2) at a dose of 0.25 μg was diluted in 100 μL of 10% ethanol and sesame oil. Ovariectomized females were subcutaneously injected with Veh or E2. Twenty-four hours after injection, the oviduct was collected for scRNA-seq analysis. A concentration of 0.25 μg E2 and 24-hour time point were chosen as these were the lowest dose and the least duration to physiologically alter egg transport within the oviduct of the rodents.20 More importantly, 24-hour treatment of 0.25 μg E2 allows direct comparison of the oviductal response to the response documented in the uterus, another E2-responsive tissue.12 Vaginal washes were collected from adult female mice (between 8 and12 weeks old) to determine the stage of the estrous cycle.26 At 11:00 hours, oviducts from females in estrus were collected for subsequent scRNA-seq analysis.
The oviductal and uterine Igf1 conditional knockout mouse model was generated by breeding progesterone receptor-Cre (PgrCre/+)27 with Igf1f/f mice, purchased from Jax.28 The conditional knockout females (PgrCre/+; Igf1f/f) and Igf1f/f control littermates were used in the experiments. Genotyping protocols for PgrCre and Igf1f/f were performed as previously described27 and according to the recommendation from Jax for Igf1f/f. The cell-specific deletion of Igf1 was validated and confirmed using Igf1 in situ hybridization (ISH) analysis using RNA scope, Cat# 443 901: Mm-Igf1 (Advanced Cell Diagnostics (ACD)) according to ACD recommended protocols.
2.2 |. Histological, in situ hybridization (ISH), and immunohistochemistry (IHC) analyses
The oviduct and uterus were collected from female mice for histological analysis. Tissues were fixed in 10% formalin and processed for histological analysis as previously described.26 Formalin-fixed oviductal and uterine tissues were paraffin embedded and sectioned to a 5-μm thickness. The antibodies used were anti-ESR1 antibody (Thermo Fisher Scientific, MA5–13191) at a dilution of 1:200, anti-PGR (Thermo Fisher Scientific, MA5–14505) at 1:400, and Ki67 antibody (BD Pharminogen, 550609) at 1:100. Vectastain ABC system (Vector Laboratories, Burlingame, CA) was used for colorimetric detection. ISH analysis was performed using RNA scope reagent kits and ACD HybEZ II Hybridization System. RNAscope probes #413281Mm-Dcn, #583461Mm-Crabp2, #583471Mm-Serpina1e, and #583481Mm-Pdxk were used for the detection of RNA in the oviductal tissues. Probes #313911Mm-Ppib and #310043 DapB were used as positive and negative controls, respectively. Images were taken using a light microscope (Leica DMi8, Leica Microsystems).
2.3 |. Quantification analysis of Ki67+ and PGR+ cells
Ki67+ and PGR+ cells were quantified using ImageJ software with Cell Counter Plugins. Ki67+ or PGR+ ciliated epithelial cells were counted and calculated as a percentage of total ciliated epithelial cells. A similar counting method was used for secretory and stromal cells (n = 3–6 animals per treatment per group). The average total ciliated cell count for Ki67 analysis was 198 cells/animal. The range for ciliated cell count was 3–292 cells per microscopic field as there were very few ciliated cells present in the isthmus. The average total secretory and stromal cell counts for Ki67 analysis were 141 cells/animal (range: 98–185) and at 255 cells/animal (range: 120–418), respectively. The average total ciliated cell count for PGR analysis was 141 cells/animal (range:0–312 cells per microscopic field; again, as there was a very minimal number of ciliated cells present in the isthmus region). The average total secretory and stromal cell counts for PGR analysis were 160 cells/animal (range: 58–223) and at 271 cells/animal (range: 98–426), respectively.
2.4 |. Oviductal cell sample preparation for scRNA-seq analysis
Oviducts were collected 24 hours after Veh or E2 treatment (n = 5–6 mice/group) or at estrus (n = 5 mice). Oviducts were pooled and collected in Leibovitz-15 media (Gibco, 41 300 070, ThermoFisher Scientific, Carlsbad, CA) supplement with 1% fetal bovine serum (L15 + 1%FBS) and placed on ice. For Veh and E2-treated samples, the oviduct was dissected into distal (infundibulum with ampulla, referred to as InfAmp) or proximal (isthmus with UTJ, referred to as IsthUTJ) regions. To isolate oviductal cells, oviducts were quickly uncoiled by trimming the mesosalpinx in order to locate the ampulla-isthmus junction (AIJ). Then, the tissues were dissected at the AIJ region for subsequent InfAmp and IsthUTJ cell isolation. For estrus samples, the entire length of the oviduct was used. After dissection, 0.25% trypsin-EDTA (Sigma, T4049) was injected into the oviduct using a blunted 30-gauge needle until the trypsin was visibly flushed out from the other end of the oviduct. The tissues were then placed in a tube containing 500 μL of trypsin and incubated on ice for 30 minutes and at 37°C for another 30 minutes. After the incubation, 500 μL of FBS was added to stop the enzymatic activity of trypsin. Oviductal cells were then flushed out from oviductal tubes with 4°C L15 + 10%FBS. At this stage, the epithelial, stromal, and parts of muscle cell layers were visibly flushed out. Only translucent longitudinal muscle cells were left intact. Cell clumps and tissue debris were strained twice through 40-μm cell strainers. Cells were then spun down and resuspended in 0.04% bovine serum albumin AlbuMax (ThermoFisher Scientific, 11020–021) in phosphate-buffered saline. The final cell concentration was targeted for 8,000 cells/run.
scRNA-seq libraries were performed using the manufacturer’s protocol (10X Genomics Inc). Single Cell 3’ v2 chemistry was used. Briefly, individual cells (~8,000 cells/run) were separated into droplets by Gel bead in EMulsion (GEM) technology using 10X Chromium Controller. Emulsion beads were broken and barcoded cDNAs were pooled for amplification. Libraries generated were sequenced using an Illumina HiSeq4000, targeting 400 million reads/run, paired-end, and 100 bp read length.
2.5 |. Human fallopian tube collection
Human fallopian tubes were collected at the Baylor College of Medicine with an Institutional Review Board approval number H-21138. Informed written consent was obtained from the patient prior to undergoing surgery. The tissue was collected from an individual without underlying gynecological diseases who underwent post-partum bilateral salpingectomy for sterilization. To obtain cell singlets, the fallopian tubes were transected so that the luminal portion of the tube was exposed. The mucosal lining of the tubes was gently dissected with a scalpel and enzymatically digested in a solution of 0.05% Trypsin (Sigma, T1426) dissolved in HBSS (Gibco) for 30 minutes, followed by incubation in Collagenase (5 mg/ml, Sigma C5138) and DNAse I (200 μg/ml, Sigma DN25) solution for 20 minutes. The cells were filtered through a 40 μm filter, collected by centrifugation and resuspended in DMEM/F12 (Gibco, 11–330-057) supplemented with 10% FBS (Sigma, F2442) and antibiotics (Gibco, 15–240-096). Live cells were sorted on a BD FACS Aria II at the Cytometry and Cell Sorting Core Laboratory at Baylor College of Medicine (BCM) and submitted for single cell sequencing at the Single Cell Genomics Core Laboratory at BCM. scRNA-seq libraries were also performed using 10X Chromium Controller with Single Cell 3’ v3 chemistry. Libraries generated were sequenced using an Illumina NovaSeq 6000, targeting 300 million reads/run, paired-end, 50 bp read length.
2.6 |. Processing of single cell RNA-seq data
The computer was equipped with AMD Ryzen 9 3900X 12-core, 24-thread and used as a server using Linux operating system. Processing of the raw sequencing data (mkfastq files) was carried out using Cell Ranger v3.1.0 to generate fastq files with default settings. Reference genomes, mm10–3.0.0 and GRCh38–3.0.0, were used for sequence alignment in mouse and human samples, respectively. Web summaries for each sample were shown in Table 1. Fastq files were then processed through Velocyto v0.1.2429–31 to produce loom files with spliced and unspliced mRNA information. Velocyto was run using the command run10x with default setting and a repeat masker gtf file from UCSC Genome Browser for mouse with an assembly for GRCm38/mm10 to mask expressed repeated elements. Generated loom files were then read in scanpy v1.6.032 using JupyterLab33 for analyses and saved in an h5ad format.34 For subsequent data analysis and visualization, a virtual machine (Docker)35 was used to create the analysis environment. Analysis packages including scanpy v1.6.0,32 anndata v0.7.5,36 umap v0.4.6,37 numpy v1.19.4,38 scipy v1.5.3,39 pandas v1.1.4,40 scikit-learn v0.23.2,41 statsmodels v0.12.1,42 python-igraph v0.8.3, 43 louvain v0.7.0,44 and leidenalg v0.8.245 were installed on the Docker.
TABLE 1.
scRNA-seq output for each dataset in this study
| Sample name | Estimate number of cells | Total reads | Means reads/cell | Median gene/cell | Median UMI counts/cell | Total detected gene # | Sequencing saturation |
|---|---|---|---|---|---|---|---|
| Estrus | 6,573 | 331 M | 50,403 | 2,786 | 7,278 | 21,649 | 61.2% |
| V24Am | 4,221 | 54 M | 12,766 | 1,476 | 2,790 | 19,113 | 35.3% |
| V24Is | 1,709 | 77 M | 45,055 | 2,037 | 4,928 | 18,709 | 60.9% |
| E24AM | 2,106 | 62 M | 29,280 | 2,186 | 5,194 | 18,655 | 48.8% |
| E24Is | 1,758 | 59 M | 33,503 | 1,724 | 3,797 | 18,432 | 64.6% |
| Human fallopian tube | 7,318 | 360 M | 49,200 | 1,333 | 3,921 | 24,508 | 74.2% |
| Antibody table | |||||
|---|---|---|---|---|---|
| Peptide/protein target | Name of antibody | Manufacturer, catalog no., and/or name of individual providing the antibody | Species raised in; monoclonal or polyclonal | Dilution used | RRID |
| ESR1 (for IHC analysis) | Estrogen receptor alpha (1D5) | Thermo Fisher Scientific Cat# MA5-13191 | Mouse; monoclonal | 1:200 | AB_10986080 |
| PGR (for IHC analysis) | Progesterone receptor (SP2) | Thermo Fisher Scientific Cat# MA5- 14505 | Rabbit; monoclonal | 1:400 | AB_10980030 |
| Ki67 (for IHC analysis) | Purified mouse anti- Ki67 | BD Pharminogen Cat #550609 | Mouse; monoclonal | 1:100 | AB_393778 |
2.7 |. Quantifications and statistical analysis
Analysis was carried out using scanpy which was designed specifically for scRNA-seq data QC and analysis inspired by Seurat’s clustering tutorial.46 Each sample was filtered out for doublets, genes that were present in less than 3 cells, cells that expressed less than 200 genes and cells that had higher than 5% reads mapped to the mitochondrial genome. Samples were then processed through the default clustering vignette on scanpy including normalization, log1p, and scaling at default settings, “highly variable genes” was set to 5000 genes, and Unique Molecular Identifier (UMI) and percentage mitochondrial were regressed out. Samples were then combined into one file using the concatenate function with batch keys for oviduct region when applicable. principal component analysis (PCA), Uniform Manifold Approximation and Projection (UMAP) clustering analyses were then performed using 10 neighbors and PCs 1–37 based on variance ratio. Clustering was performed using Leiden clustering at 0.06–0.1 resolution based on known gene markers for secretory, ciliated, fibroblast, and muscle cells in the oviduct. Leiden clusters were then subsetted individually for independent analyses. Endothelial (Pecam1+) and immune cells (Cd52+) were filtered out from the dataset. When merging samples using the concatenate feature, there may be technical artifacts from sample collections making it difficult to interconnect different batches or samples. Batch balanced K-Nearest Neighbor (BBKNN)47 corrects these artifacts by using KNN to create connections between batches without interfering with PCA space or UMIs.
Using the generated Loom files from Velocyto, samples were processed following scVelo tutorials31; RNA Velocity Basics, Dynamic Modeling, and Differential Kinetics. The filter and normalize functions were set to 30 minimum shared counts and 10,000 top genes. The remaining functions were kept at default including moments. The RNA velocity map was then projected onto the UMAP and FA (ForceAtlas2)48 plots. FA plots are drawn from the function scanpy.tl.draw_graph() using default settings. Top 500 differentially expressed genes were used for determination of biological processes (BPs) enriched in each sample. These genes were uploaded onto geneontology.org using GO enrichment analysis with PANTHER overrepresentation test49 and Fisher test type with FDR correction when applicable.
2.8 |. Data and code availability
All analyses in this study were saved in JupyterLab and deposited on github in https://github.com/winuthayanon/ovx_ve/ for the ovariectomized Veh- and E2-treated dataset, https://github.com/winuthayanon/estrus/ for the estrus dataset, and https://github.com/winuthayanon/humanFT/ for the human fallopian tube dataset. Raw data as fastq files for the ovariectomized Veh- and E2-treated, the estrus, and the human Fallopian tube datasets were deposited at Gene Expression Omnibus (GSE164291).
2.9 |. Embryo collections
Adult (8–12 weeks old) PgrCre/+; Igf1f/f, and Igf1f/f female mice were bred overnight with C57B6/J male proven breeders. Females with a copulatory plug the next morning were considered pregnant at 0.5 dpc. Embryos were collected from the oviduct and uterus at 3.5 dpc (n = 9–12 mice/genotype). Nonviable, unfertilized, and developmentally delayed embryos were also included in the total number of eggs/embryos. The region (oviduct vs uterus) that the embryos were retrieved from was recorded. Images of embryos collected at 3.5 dpc were taken using a Leica DMi8 microscope.
2.10 |. Statistical analyses
All graphs represent mean ± SEM. Individual value from each mouse was plotted when applicable. Statistical analysis was performed using GraphPad Prism v8.4.0 for Mac OS X (GraphPad Software, Inc, La Jolla, CA). Statistical significance is considered when P < .05 using two-tailed unpaired Student’s t-test with Welch’s correction for simple comparison or two-way ANOVA with Sidak’s multiple comparisons test, unless otherwise indicated.
3 |. RESULTS
3.1 |. E2-targeted gene expression in the oviduct
To determine the biological action of E2 on oviductal cell types, we ovariectomized adult female mice and treated with vehicle control (Veh) or E2 for 24 hours. We found that after treatment with E2, localization of ESR1 in the oviduct decreased in the nucleus but increased in the cytosolic compartment compared to the Veh-treated group (Figure 1A), especially in the epithelial cells of all regions including the infundibulum, ampulla, and isthmus. In our previous study, E2 (0.25 μg) stimulated the proliferative response (increase in Ki67+ cells) in uterine epithelial cells (Figure S1A).12 We therefore investigated whether E2 induced a similar response in the oviduct using the proliferative cell marker, Ki67. Images at a lower magnification are shown in Figure S1. E2 treatment did not significantly change the number of Ki67+ ciliated or secretory cells (Figure 1B,C). However, E2 significantly increased the number of Ki67+ stromal cells in the infundibulum. We also evaluated a known response of E2 treatment in the female reproductive tract, which causes an increase of progesterone receptor (PGR) expression in the stromal cell layer (Figure S1).12 We found that E2 increased overall Pgr transcript levels in the whole oviduct (Figure 1D). However, when we assessed protein levels using semiquantitative IHC analysis, we found that PGR was expressed in all cell types of the oviduct, including secretory epithelial, ciliated epithelial, stromal, and muscle cells (Figure 1E). However, E2 increased PGR expression primarily in the ciliated cell population in the ampulla (insets in Figure 1E,F), while the expression of PGR in other cell types remained comparable to that of Veh-treated. As mentioned above, E2 induces the expression of IGF1 in the uterus.11 Here we found that E2 increased Igf1 transcript levels in the whole oviduct after 24 hours of treatment (Figure 1G). These data suggest that E2 treatment differentially induces cell specific proliferation and PGR expression in the oviduct but in a different fashion compared to the uterus (more detail in the discussion section).
FIGURE 1.
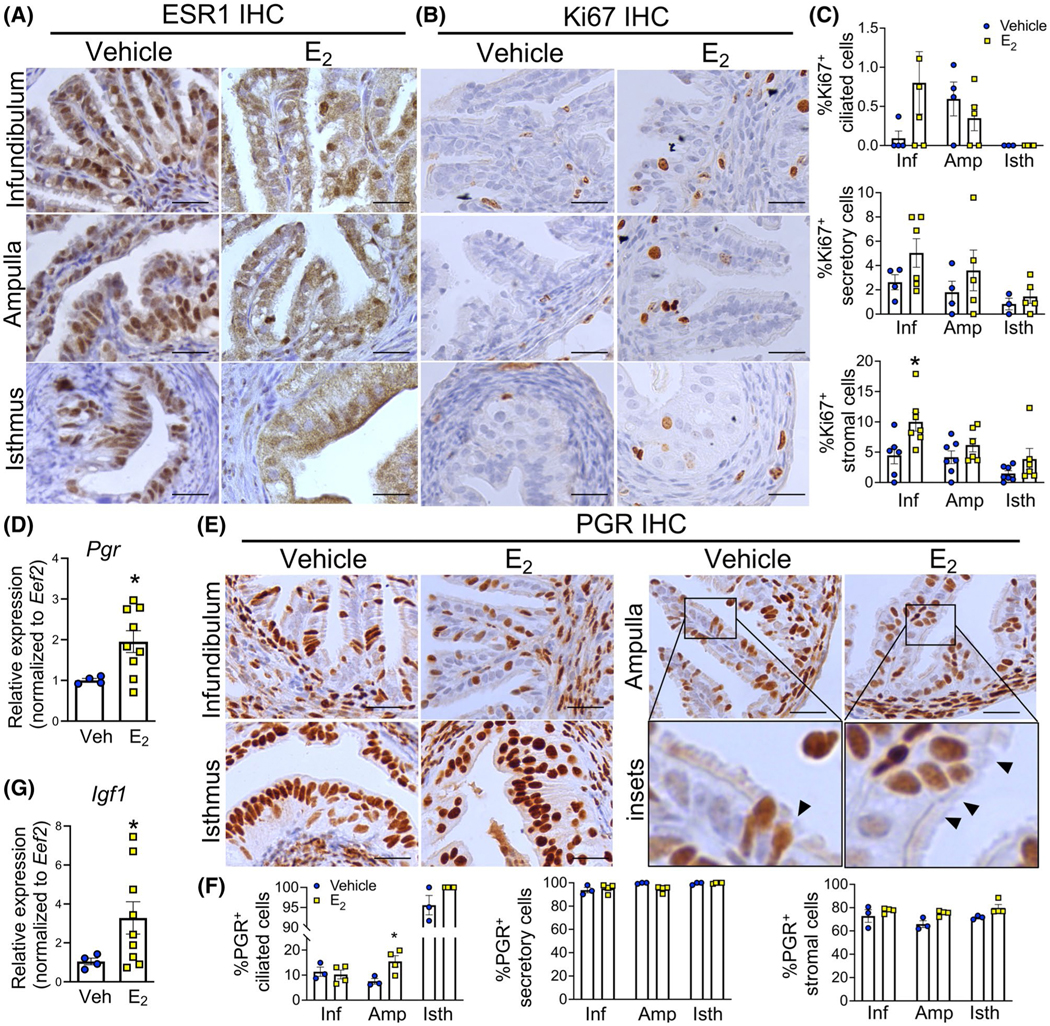
Ki67-positive cells and PGR expression in the mouse oviduct after E2 treatment. A-B, Immunohistochemical (IHC) analyses of ESR1 and Ki67 expression of the infundibulum, ampulla, and the isthmus of the oviduct, respectively. Oviducts were collected from ovariectomized mice after 24 h of treatment with Veh or E2 (0.25 μg/mouse), n = 3–4 mice/group. C, Quantification of Ki67-positive ciliated, secretory and stromal cells. D, Transcript levels of Pgr in the whole oviduct after 24 h of Veh or E2 treatment (n = 4–9 mice/group). E. IHC analysis of PGR expression at the infundibulum, the ampulla and the isthmus after 24 h of treatment with Veh or E2. Insets are enlarged images of area with PGR+ ciliated cells (arrowheads) in the ampulla. F. Quantification of PGR-positive ciliated, secretory and stromal cells. G. Transcript levels of Igf1 in the whole oviduct after 24 h of Veh or E2 treatment (n = 4–9 mice/group). Scale bars = 25 μm. Each dot represents data from each mouse. *P < .05, significantly different when compared to the corresponding Veh, unpaired Student’s t-test with Welch’s correction
3.2 |. E2-mediated transcriptional signatures in specific cell types in the oviduct
As shown above, E2 induced cellular responses in the oviduct in a cell-type- and region-specific manner. To delineate the gene regulatory effect of E2 in a cell-type- and region-specific manner, we employed scRNA-seq analysis. After 24 hours of Veh or E2 treatment, oviducts were isolated into distal (infundibulum and ampulla, referred to as “InfAmp”) or proximal (isthmus and UTJ, referred to as “IsthUTJ”) regions and cell singlets were collected for scRNA-seq analysis. We found that cells from the oviduct were grouped into seven cell clusters (Figure 2A), including ciliated cells (InfAmp) indicated by pink and the expression of Ccdc153 (Figure 2B), secretory cells (InfAmp) indicated by blue and Ovgp1+, secretory cells (IsthUTJ) indicated by green and Fxyd3+, fibroblast (or stromal) Pdgfra-, cells indicated by orange and Igfbp6+; Dcn+; Pdgfra− (Figure S2A), muscle cells indicated by grey and Act2a+, epithelial cells (subset) indicated by maroon and Sprr2f+, and fibroblast Pdgfra+, cells indicated by beige and Dcn+; Pdgfra+. The top 25 genes highly expressed in each cell cluster were shown in Table S1. In addition to cell clusters, we can also distinguish the regional origin (InfAmp vs. IsthUTJ) of each cell population (Figure 2C). Our data showed for the first time that the transcriptional profile in each population was unique to the region. E2 treatment caused a shift of the transcriptome in all cell clusters (Figure 2D). Specific genes in each cluster, region, or treatment in this dataset, can be searched on our laboratory website at www.winuthayanon.com/genes/ovx_ve/.
FIGURE 2.
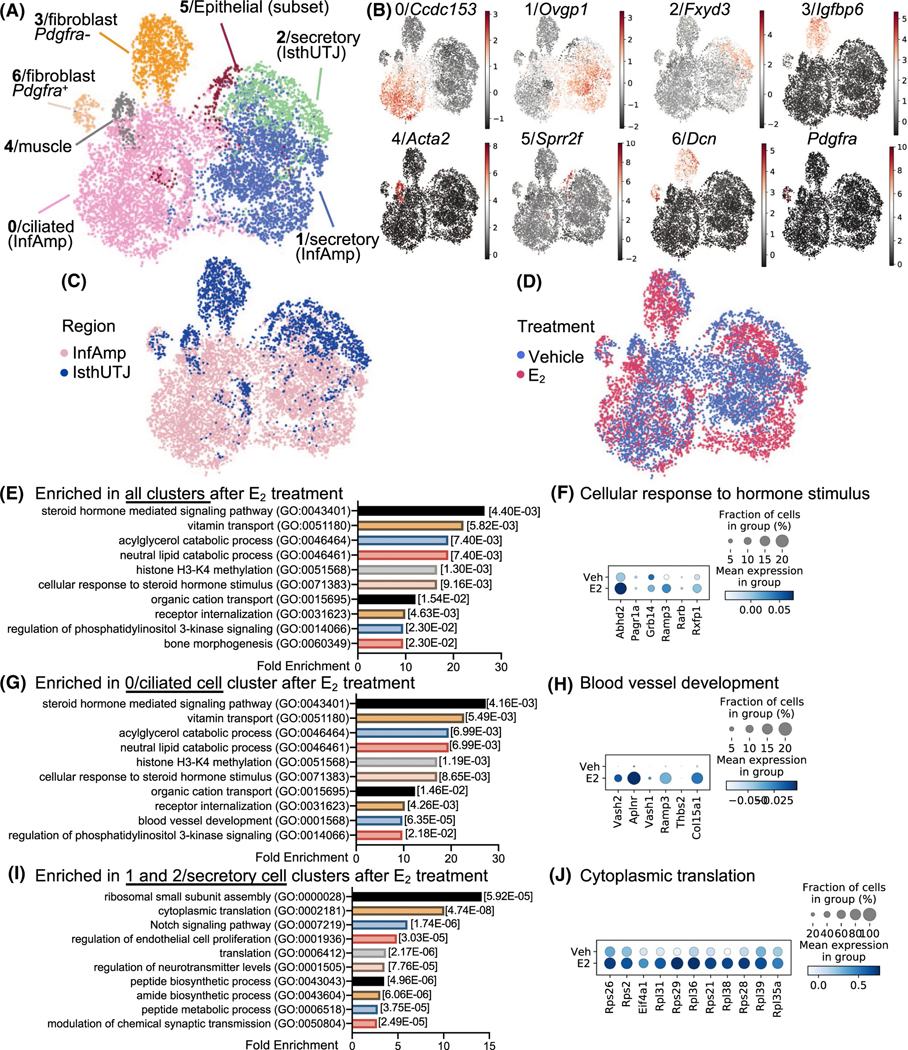
E2 treatment alters the transcriptional profile of the oviduct. A, Cell singlets were isolated from either the “infundibulum + ampulla” (InfAmp) or “isthmus + uterotubal junction (IsthUTJ) and pooled from mice that were ovariectomized and treated with Veh or E2 for 24 h (n = 5–6 mice/treatment). Single cell-RNA sequencing (scRNA-seq) data presented as Uniform Manifold Approximation and Projection (UMAP) plot. Each dot represents one cell. There are seven distinct oviductal cell clusters (#0–6) as indicated by different colors. B, Expression of the top marker gene from each cell cluster. C-D, All cell clusters differentiated by the region (InfAmp vs. IsthUTJ) or the treatment (Veh vs. E2). E, Top biological processes using gene ontology analysis from genes enriched in all clusters after E2 treatment. Numbers in brackets indicate P-values. F, Dot plots of genes enriched in cellular response to steroid hormone stimulus in all clusters. Percentage of cells expressing particular gene(s) in the dataset is visualized by the size of the dot. Mean expression within each category is visualized by color. G, Top biological processes from genes enriched in 0/ciliated cell cluster after E2 treatment. H, Dot plots of genes enriched in blood vessel development in cluster 0/ciliated cells. I, Top biological processes from genes enriched in 1 and 2/secretory cell clusters after E2 treatment. J, Dot plots of genes enriched in cytoplasmic translation in clusters 1 and 2/secretory cells
The effect of E2 and region-specific transcripts in each cell cluster are shown in dot plots (Figure S2B-K and Table S2 for top 500 genes). To determine whether the action of E2 in the oviduct could result from the direct genomic binding of ESR1 on the promoter region of target genes, we integrated the previous ESR1 (chromatin immunoprecipitation sequencing) ChIP-seq dataset from the mouse uterus50,51 by scanning the regulatory region of differentially expressed genes in the oviduct identified from scRNA-seq analysis for the ESR1 binding sites. We found that some of the genes regulated by E2 in all cell types of the oviduct had ESR1 binding sites on their promoter, transcription start sites (TSSs), or at the intragenic regions (Figure S3). This finding indicates that E2 may also mediate its action through the genomic regulatory pathway by the recruitment of ESR1 to the promoters, TSSs, or the intragenic regions of target genes in different cell types in the mouse oviduct, similar to those in the uterus.
Using gene ontology (GO) analysis, we found that the top biological processes (BP) that were enriched in all cell clusters (as well as the ciliated cell clusters 0 and 6) after E2 treatment were steroid hormone-mediated signaling pathways (GO:0 043 401, Figure 2E-G). Examples of genes enriched in this cellular response to hormone stimulus included acylglycerol lipase (Abhd2), PAXIP1 associated glutamate rich protein 1 (Pagr1a), growth factor receptor-bound protein 14 (Grb14), receptor activity-modifying protein 3 (Ramp3), retinoic acid receptor beta (Rarb), and relaxin receptor 1 (Rxfp1) (Figure 2F). One of the unique BPs in the ciliated cell clusters was blood vessel development, which included vasohibin 1 and 2 (Vash1 and Vash2), apelin receptor (Aplnr), Ramp3, thrombospondin 2 (Thbs2), and collagen type XV alpha 1 chain (Col15a1) (Figure 2H). The top 3 BPs enriched in the secretory cell clusters included ribosomal small subunit assembly, cytoplasmic translation, and notch signaling pathway (Figure 2I). We showed there that the BP ‘cytoplasmic translation’ enriched in E2-treated samples included ribosomal proteins S21, S26, S28, and S29 (Rps21, Rps26, Rps28, Rps29), ribosomal proteins L31, L35a, L36, L38, L39 (Rpl31, Rpl35a, Rpl36, Rpl38, Rpl39), and eukaryotic translation initiation factor 4A1 (Eif4a1) (Figure 2J). For a complete GOBP analysis for all cell clusters, refer to Table S3.
Based on our previous findings using microarray analysis, we found that proteases and protease inhibitors were regulated by E2- and ESR1-dependent action.23 Here, we found that E2 treatment specifically increased overall expression of kallikrein 8 and 11 (Klk8 and Klk11), prostatin (Prss8), neurotrypsin (Prss12), and serine protease 23 (Prss23) (Figure S4A) in the oviduct. The expression of Klk8 was specific to the secretory (IsthUTJ) cell cluster and Klk11 to the ciliated (InfAmp) cell cluster (Figure S4B). Prss23 showed the highest expression among other proteases and the expression was specific to fibroblast Pdgfra− and muscle cell clusters. E2 increased overall expression of serine protease inhibitors (Serpinb1a, Serping1, and Serpinh1) and WAP four-disulfide core domain protein 18 (Wfdc18 or extracellular peptidase inhibitor) (Figure S4C). Similar to Prss23, protease inhibitors were expressed in fibroblast Pdgfra− and muscle cell clusters, but also in secretory and ciliated cells of the oviduct (Figure S4D). These data suggest that E2 (1) modulates hormone signaling pathways in ciliated cells, (2) increases translation capacity in secretory cells, and (3) regulates the synthesis of proteases and protease inhibitors in multiple cell compartments of the oviduct.
3.3 |. Insights into the effects of exogenous vs. endogenous E2 on oviductal cell responses
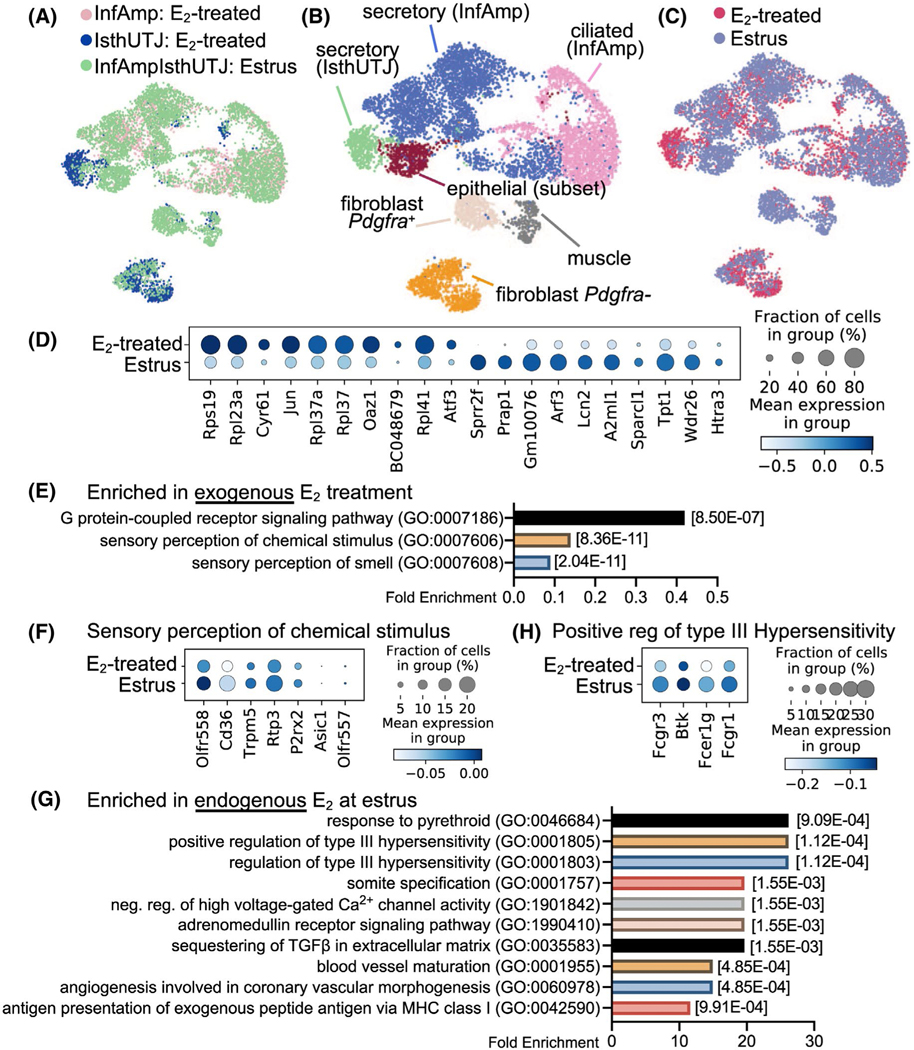
To determine whether exogenous E2 causes differential responses in oviductal cells compared to endogenous E2 at estrus, we compared the scRNA-seq data of oviductal cells collected at estrus to aforementioned E2-treated oviductal cells. Cells isolated during estrus were collected from the entire oviduct (denoted as “InfAmpIsthUTJ:estrus). scRNA-seq data were then combined and distinguished into InfAmp:E2-treated, IsthUTJ:E2-treated, and InfAmpIsthUTJ:estrus (Figure 3A). We found that all seven cell clusters present in the E2-treated group were also detected in the estrus samples (Figure 3B). Colors for each cell cluster were consistent with the dataset from Veh- vs. E2-treated samples (Figure 2A) for ease of comparison. Marker genes for each cell cluster are depicted in Figure S5. Here, we show that endogenous E2 (estrus) had cell populations with similar transcriptome signatures compared to that of exogenous E2 (E2-treated) (Figure 3C) as all cell populations were overlapped between estrus and E2-treated samples. Only one exception was that there were more secretory (IsthUTJ) cell populations in the E2-treated dataset compared to cells from the estrus samples.
FIGURE 3.
Exogenous and endogenous E2 induce differential transcriptional signatures in the oviduct. Endogenous E2 single cell samples were collected from the entire oviduct at the estrus stage (n = 5 mice). scRNA-seq data from estrus (endogenous E2) were combined with data from ovariectomized E2-treated samples (exogenous E2) shown in Figure 2 for comparison analyses. A, Estrus and E2-treated (InfAmp and IsthUTJ) datasets were overlapped. B, After the dataset from E2-treated and estrus samples were combined, scRNA-seq data were analyzed and separated into 7 cell clusters containing the similar cell populations to that in Figure 2. C, Overall similarity and difference between endogenous vs. exogenous E2 in the oviductal cells. D, Dot plot of top 10 differentially expressed genes between exogenous and endogenous E2 in the oviduct. E, Top biological processes using gene ontology analysis of gene enrichment in exogenous E2 treatment. F, Dot plots of genes enriched in sensory perception of chemical stimulus in exogenous E2 treatment. G, Top biological processes of genes enriched in estrus samples. H, Dot plots of genes enriched in positive regulation of type III hypersensitivity in estrus samples
We found that the top 10 genes elevated by E2 treatment were mostly ribosomal proteins including Rpl19, Rpl23a, Rpl37, Rpl37a, and Rpl41 (Figure 3D), indicating a stimulation of translation. Interestingly, common E2-target genes were also expressed at higher levels in the oviduct in E2-treated samples compared to estrus, such as cysteine rich angiogenic inducer 61 (Cyr61, aka IGF-binding protein 10)52 and Jun.53 However, some other known E2-specific genes in the female reproductive tract were unique to estrus, such as small proline-rich protein 2F (Sprr2f) and proline-rich acidic protein 1 (Prap1).54,55 Analysis using gene ontology demonstrated that E2-treated cells showed enrichment of molecules in G-protein-coupled receptor (GPCR) signaling pathways and both sensory perception of chemical stimulus and sensory perception of smell were the subset of GPCR signaling (Figure 3E-F). Specific enriched genes in the sensory perception to chemical stimulus in E2-treated cells included olfactory receptor family 51 subfamily D and E member 1 (Olfr557 and Olfr558), Cd36, transient receptor potential cation channel subfamily M member 5 (Trpm5), receptor transporter protein 3 (Rtp3), purinergic receptor P2X 2 (P2rx2), and acid sensing ion channel subunit 1 (Asic1). Endogenous E2 at estrus showed an enrichment of various BPs (Figure 3G). As an example, transcripts enriched in the estrus sample for the BP of positive regulation of type III hypersensitivity (Figure 3H) were Fc fragment of IgG receptor IIa, IgE receptor Ig, and IgG receptor Ia (Fcgr3, Fcer1g, and Fcgr1), and Bruton tyrosine kinase (Btk). Overall, these data indicate that the oviduct showed a slight difference in gene expression profile when exposed to E2 after ovariectomy compared to the estrus stage, at which time circulating E2 is peaked and progesterone is present at basal levels.
3.4 |. Validation of markers identified from E2-treated InfAmp and E2-treated IsthUTJ samples
To validate the scRNA-seq findings observed from the ovariectomized E2-treated model, we used similar criteria to differentiate cell clusters in the scRNA-seq data from oviductal tissues collected at estrus. We found that all clusters found in E2-treated samples were present in the estrus sample (Figure 4A). The top 10 marker genes for each cell cluster from the estrus dataset shown in the heatmap plot (Figure 4B) were overlapped with the markers identified in Veh- and E2-treated samples (Table S1). Genes expressed in each cluster in this estrus dataset can be searched at www.winuthayanon.com/genes/estrus/.
FIGURE 4.
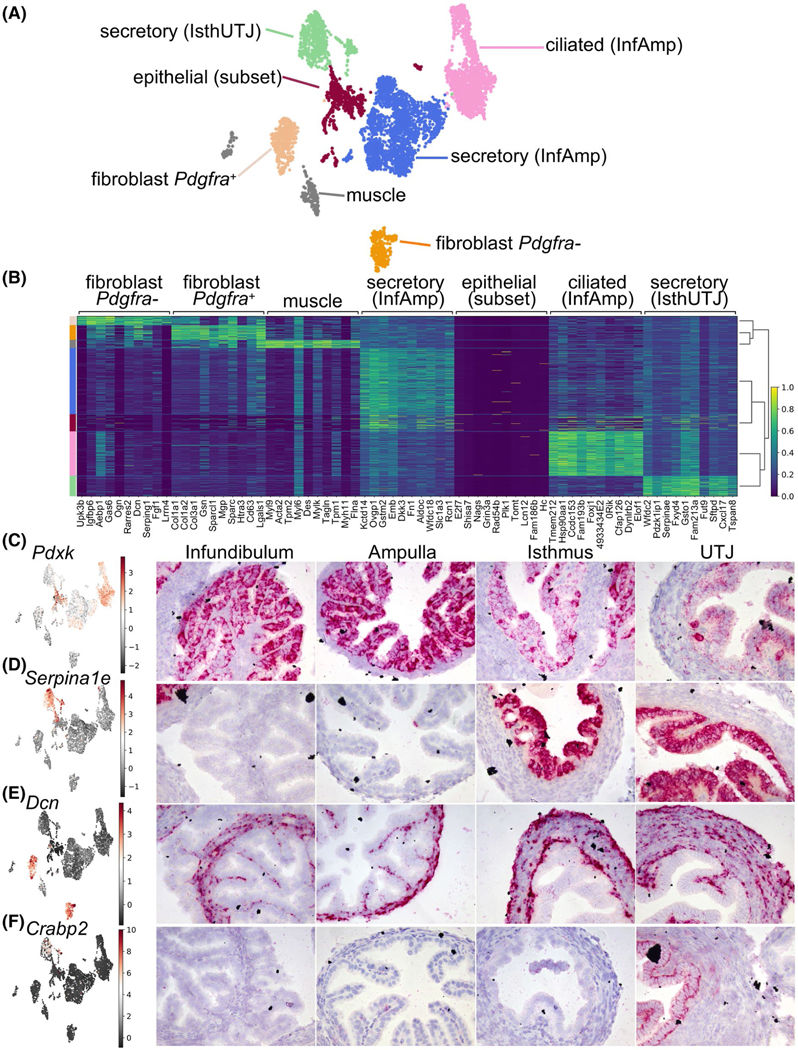
Cells isolated from estrus alone show similarity of cell clusters identified in the E2-treated dataset. A, UMAP plot of scRNA-seq analysis from estrus samples. Cells were separated into 7 cell clusters. B, Heatmap plot of top ten differentially expressed genes in each cell cluster. Color-coded bar on the left is coordinated with the cell clusters in A. In situ hybridization analyses of (C) Pdxk, (D) Serpina1e, (E) Dcn, and (F) Crabp2 transcripts in the oviduct from samples collected randomly at different stages of the estrous cycle from ovarian intact adult female mice
To verify the markers identified from each region from the E2-treated dataset (Table S1), we identified and determined the localization of these marker genes using ISH analysis in oviduct samples randomly collected during various stages of the estrous cycle. We found that pyridoxal kinase (Pdxk, also known as epididymis secretory sperm binding protein) was one of the top genes that was classified as an InfAmp-enriched gene (Table S2). Herein, we show that Pdxk was exclusively present only in the epithelial cells and detected at higher levels in the infundibulum and ampulla compared to isthmus and UTJ (Figure 4C). Additionally, we were able to verify that Serpina1e, one of the top markers identified as IsthUTJ-specific, was only present in the epithelial cells in the isthmus and UTJ (Figure 4D). Decorin (Dcn), a marker for fibroblasts (or stromal cells), was only detected in cells from mesenchymal origin including fibroblast (stroma) and muscle cell layers (Figure 4E). Lastly, cellular retinoic acid binding protein 2 (Crabp2), was found specifically in the secretory (IsthUTJ) cluster and only detected in the epithelial cell layer of the UTJ region (Figure 4F). Together, these data suggest that the markers identified from different populations and regions can be used as region- and cell-specific markers in the mouse oviduct.
3.5 |. Embryotrophic factors, potential E2-target genes in the oviduct
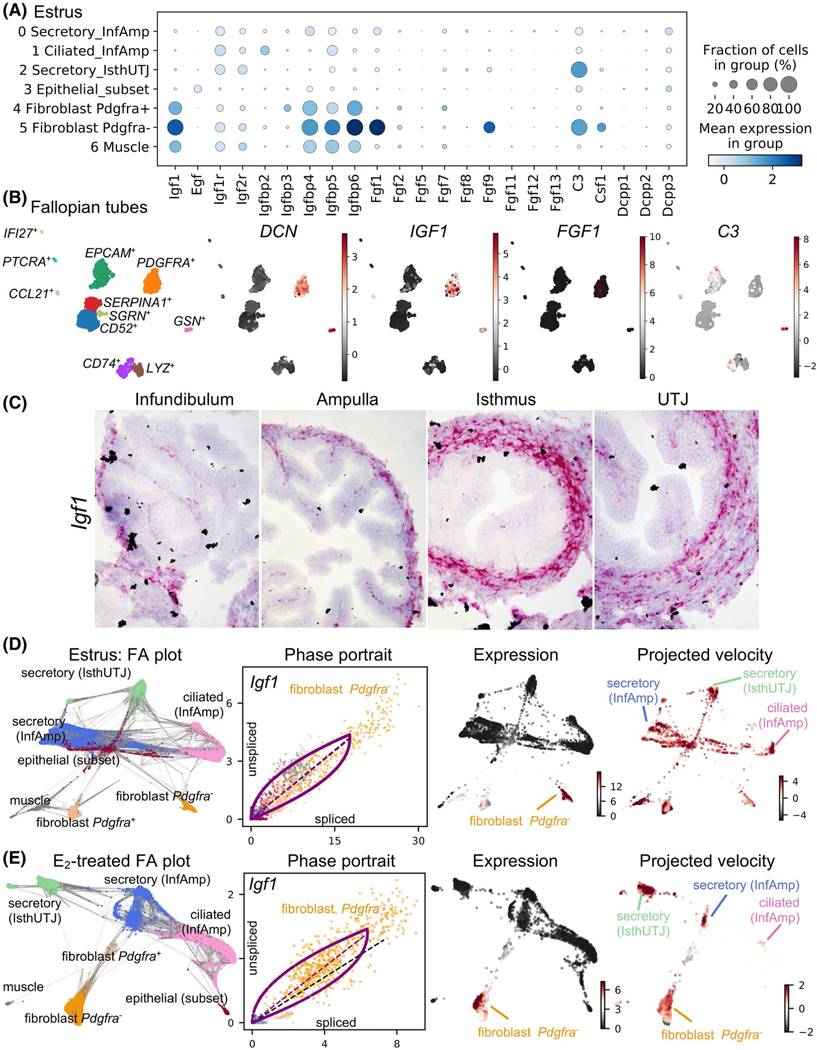
One of the crucial functions of the oviduct is to support preimplantation embryo development. We showed that improper E2 signaling resulted in embryo death.23 Embryotrophic factors were previously shown to improve embryo quality in vitro.13–17 As Igf1, one of the embryotrophic factors identified, was upregulated by E2 (Figure 1G), we looked more closely into the expression of embryotrophic factors in each cell population. We found that Igf1 and its binding proteins (Igfbp4, 5, and 6) were mainly present in the cells from mesenchymal origin including fibroblasts (both Pdgfra− and Pdgfra+) and muscle cells (Figure 5A). Fgf1 and Fgf9 were present in the fibroblast Pdgfra− population. However, C3 was expressed in both fibroblast Pdgfra− and secretory (IsthUTJ) cell clusters. Egf, Csf1, and Dcpp1–3 were expressed at lower levels and in a lower fraction of cells compared to Igf1, Fgf1, and C3. These findings indicate that embryotrophic factors are mainly present in cells originating from the mesenchyme. To determine if IGF1, FGF1, and C3 were also expressed in human Fallopian tube, we isolated cells from one individual as a proof of concept. The heterogeneity of different cell populations within the Fallopian tube is shown in Figure 5B, Figure S6, and Table S5. IGF1 was present in the fibroblast cell population (DCN+ and PDGFRA+). Interestingly, FGF1 was detected only in a subset of fibroblasts. C3 was also expressed in other cell types but at low levels. Specific genes in each cluster in this dataset, can be searched at www.winuthayanon.com/genes/humanFT/.
FIGURE 5.
Unique cell populations in mouse oviducts and human Fallopian tube produce embryotrophic factor(s). A, Dot plot of embryotrophic factors and associated proteins in each cell cluster in the mouse oviduct collected at estrus. Insulin-like growth factor 1 (Igf1), epidermal growth factor (Egf), IGF receptor (Igfr), IGF-binding protein (Igfbp), fibroblast growth factor (Fgf), compliment C3 (C3), colony-stimulating factor 1 (Csf1), and demilune cell parotid proteins 1–3 (Dcpp). Different sizes of the circle represent percentages of the cells with expression of each gene. Color scale represents normalized expression value. B, scRNA-seq analysis of cell clusters from human Fallopian tube isolated from one individual as a proof-of-concept. Marker genes for each cell cluster are indicated in Figure S6. Stromal cells (decorin, DCN+) also express IGF1, FGF1, and C3. C, Expression of Igf1 in the mouse oviduct using ISH analysis. D and E, ForceAtlas2 (FA) plot, phase contrast, expression, and RNA project velocities of Igf1 from mouse oviductal cells at (D) estrus or (E) after E2 treatment. Gray lines in the FA plot represent all velocity-inferred cell-to-cell connections/transitions but do not differentiate the directionality of velocity. Phase portraits depict a spliced vs. unspliced Igf1 mRNA. Black dotted line represents constant transcriptional state. The expression of Igf1 is mainly in the fibroblast cell populations, but the inferred direction of RNA velocity (projected velocity) showed the trajectory towards ciliated (InfAmp), secretory (InfAmp), and secretory (IsthUTJ) cell populations
As Igf1 was present at high levels in both mouse oviducts and human Fallopian tubes, we reasoned that IGF1 was a viable candidate as an E2-target gene in the oviduct and could provide biological potential for oviductal function. First, we validated the expression of Igf1 in the mouse oviduct using ISH analysis, as there was no reliable IGF1 antibody. Here, we showed that Igf1 was indeed expressed in the fibroblasts and muscle cell layers of the oviduct (Figure 5C) and the expression in the epithelial cells was omissible.
To determine gene regulation trajectories, we used RNA velocity analysis to assess the spliced vs. unspliced Igf1 mRNA. RNA velocity of particular genes can be used to predict the future state (inferred direction) of individual cells based on the time derivative of splicing information and gene expression dynamic.29,56 We identified Igf1 as one of the driving genes of the transcriptional dynamic in both estrus and E2-treated datasets (Figure 5D,E). Gray lines in the FA plots represent all velocity-inferred cell-to-cell connections but do not differentiate the directionality of velocity. Phase portraits showed the ratio of spliced vs. unspliced Igf1 mRNA in each cell cluster (color-coded dots) in estrus (Figure 5D) and E2-treated (Figure 5E) samples. Igf1 was strictly expressed in the fibroblast Pdgfra− population but had an inferred direction (projected velocity) upon epithelial cell clusters such as secretory (InfAmp), secretory (IsthUTJ), and ciliated (InfAmp). These data indicate that Igf1 is expressed in the fibroblast population in both mouse and human tissues and Igf1 has a potential impact on gene regulation in other cell types within the mouse oviduct.
3.6 |. Loss of Igf1 expression led to a partial embryo developmental defect and embryo retention in the oviduct
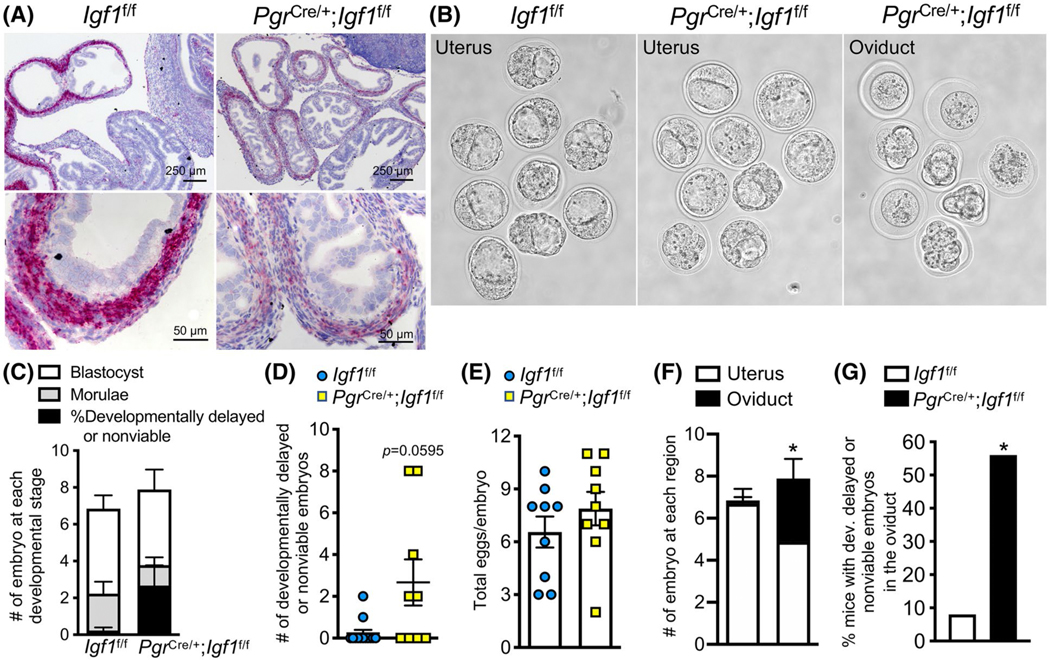
To test whether IGF1 is functionally required for female reproduction, we conditionally ablated Igf1 expression in Pgr expressing cells using the PgrCre/+; Igf1f/f mouse model. We validated that Igf1 transcript levels were significantly reduced in PgrCre/+; Igf1f/f compared to Igf1f/f control oviduct (Figure 6A). The overall histoarchitecture of the oviduct appeared to be similar between PgrCre/+; Igf1f/f and of Igf1f/f mice (Figure 6A). Previous studies showed that PgrCre/+; Igf1f/f females were subfertile.51 However, it is still unclear if the fertility defect was due to a direct disruption in oviductal function. In our study, we showed that morula and blastocyst stage embryos were collected from Igf1f/f females at 3.5 dpc (Figure 6B-D). We found that developmentally delayed or nonviable embryos were present in the PgrCre/+; Igf1f/f females, but not significantly different than those collected from Igf1f/f mice (Figure 6D, P = .0595). The number of total eggs or embryos was similar between PgrCre/+; Igf1f/f, and Igf1f/f females (Figure 6E). Nevertheless, there were significantly more embryos retained in PgrCre/+; Igf1f/f oviduct compared to Igf1f/f females (Figure 6F). Moreover, these embryos retained in the oviduct appeared to be developmentally delayed, unfertilized, or nonviable (Figure 6G). These findings suggest that a loss of IGF1 could lead to an embryo developmental defect and embryo retention in the oviduct.
FIGURE 6.
Oviductal ablation of Igf1 increases developmentally delayed and nonviable embryos in mouse oviducts and inhibits embryo transport. A, Igf1 transcripts in Igf1f/f (control) compared to PgrCre/+; Igf1f/f oviducts. B, Embryos collected from the uterus and/or oviduct from Igf1f/f compared to PgrCre/+; Igf1f/f females at 3.5 days post coitus (dpc). C, Number of embryos at each developmental stage at 3.5 dpc. D, Number of developmentally delayed or nonviable embryos. P = .0595, unpaired Student’s t-test with Welch’s correction. E, Total number of all eggs or embryos present in the oviduct at 3.5 dpc. F, Number of embryos collected from Igf1f/f (left bar) compared to PgrCre/+; Igf1f/f (right bar) at each region of the oviduct at 3.5 dpc. *P < .05, significantly different when compared to the corresponding Igf1f/f mice, unpaired Student’s t-test with Welch’s correction. G, Percentage of mice with developmentally delayed or nonviable embryos in the oviduct. *P < .05, significantly different when compared to the corresponding Igf1f/f group, Fisher’s exact test for contingency plot. n = 9–12 mice/genotype
4 |. DISCUSSION
In the uterus, E2 recruits all epithelial cells to undergo proliferation.52 However, in the oviduct, E2 showed minimal effect on epithelial cell proliferation. It is likely that uterine epithelial cells are more plastic and sensitive to E2 in order to prepare and allow for embryo implantation compared to the oviductal epithelial cells. In contrast to proliferative responses in the uterine tissue, oviductal response to E2 might be through changes in gene expression and cell morphology rather than cell division. Changes in oviductal epithelial cell morphology have been observed in the oviduct at different stages of ovarian cycles in rhesus macaques.57 Moreover, we showed that 24 hours of E2 treatment caused a decreased ESR1 in the nucleus but increased in the cytosolic compartment of oviductal epithelial cells, which was distinct and not observed in the uterus.58 It is established that steroid receptors in subcellular compartments can be altered during different physiological conditions. As such, we reasoned that decreased ESR1 in the nuclear compartment could be due to an overall auto-downregulation of E2 signaling by proteasome-mediated proteolysis, similar to that previously reported in the lactotroph cell line.59 However, to definitively determine whether this is the case for the oviductal cells, future studies need to address the presence of ESR1 in nuclear vs. cytosolic compartments after E2 treatment at different time points using immunoblotting analysis.
The expression pattern of PGR in the oviduct was also distinct from that of the uterus. In the uterus, PGR is mainly present in the epithelial cells in the absence of E2. Upon E2 treatment, uterine expression of PGR is decreased in the epithelial cells but increased in the stromal and muscle cell layers.60 Although developing from the same fetal origin, the Müllerian duct, the oviduct responds to steroid hormones in a distinct manner compared to the uterus. We found that PGR was detected in almost 100% of secretory cells regardless of the oviductal region. The expression of PGR in secretory cells and stromal cells was not altered in the presence or absence of E2. However, E2 significantly increased PGR+ ciliated cells in the ampulla, suggesting that E2-induced PGR expression in the oviduct is exclusively specific to ciliated cells. These data indicate that distinct epithelial cell populations, eg, ciliated and secretory cells, are differentially regulated by steroid hormone signaling, potentially in a region-specific manner, at least in the mouse model.
Using scRNA-seq analysis to determine the transcriptional signature in each cell type, we found that transcriptional signatures of both epithelial cells and fibroblast were altered after E2 treatment. Interestingly, the ciliated cell cluster’s top 1 BP was ‘steroid hormone-mediated signaling pathway’ whereas the secretory cell (from both InfAmp and IsthUTJ) clusters’ top two BPs were ‘ribosomal small subunit assembly’ and ‘cytoplasmic translation’. This further indicates that ciliated and secretory cells of the oviduct respond differentially to E2 in a distinguishable cell-type specific manner. After E2 treatment, secretory cells were enriched for genes responsible for translation. Not surprisingly, exogenous E2 caused a slight shift in the transcriptome of oviductal cell types from that of endogenous E2 during estrus. These differences are likely due to the presence of progesterone at a basal level at the estrus stage in contrast to the absence of progesterone when the mice were ovariectomized and treated with exogenous E2. Moreover, it is unlikely that serum E2 levels are comparable between E2-treated and estrus cycling mice. In addition, adipocyte- and adrenal gland-derived E2 is still present after ovariectomy, which might contribute to the confounding effect on the oviductal cellular response between the E2-treated and estrus groups. However, the majority of the transcriptome signatures remained similar as indicated by overlapping cells when comparing estrus and E2-treated samples.
We showed that E2 exhibited its action through genomic regulation of gene transcription in different cell types. This conclusion was confirmed by the evaluation of ESR1 binding sites on the gene promoters identified from scRNA-seq analysis by the scanning of ESR1 ChIP-seq dataset from the mouse uterus. Nevertheless, it is also likely that E2 regulates nongenomic action in the oviduct. As mentioned earlier, actions of E2 were also shown to be through the activation of nongenomic action of ESR1 by the induction of cAMP and IP3 production in primary epithelial and muscle cells from rat oviducts.21,22 Therefore, this could be one of the reasons we did not observe dramatic changes in the transcription profiles in E2- vs. Veh-treated cells in our study. Another possibility could be due to a suboptimal time point assessed at 24 hours. Therefore, in the follow-up studies, we will evaluate the cell-type-specific response of the oviduct at a shorter time point after E2 treatment.
In addition to previous studies,25 our scRNA-seq analysis from Veh- and E2-treated samples revealed that there are other genes that can be used as region- and cell-type-specific markers. We found that Wt1 was expressed in the infundibulum and ampulla (Table S2), consistent with previous reports.25 Here, we also showed that Pdxk was highly expressed in all epithelial cell types in the infundibulum and ampulla. Pdxk, is also expressed in the bovine oviduct,61 suggesting that its expression in other mammalian species might be region-specific as well. Serpina1e was detected exclusively in the secretory cells of the oviduct in the isthmus and UTJ. Dcn, a small leucine-rich pericellular matrix proteoglycan, was expressed in cells from mesenchymal origin including fibroblast and muscle cells in both mouse oviducts and human Fallopian tubes. We also identified that Crabp2 was expressed in epithelial cells of the UTJ. Mice with biallelic knockout of five Serpina1a–e develop emphysema but showed no reproductive phenotype.62 Dcn−/− and Crabp2−/− mice are also viable and fertile.63,64 These findings suggest that a loss of these genes do not affect female fertility. Therefore, Serpina1e, Dcn, and Crabp2 can be specifically targeted for Cre allele knock-ins as a tool for cell-type- and region-specific deletion of genes of interest in the oviduct.
Evidence showed that supplementation of culture media with embryotrophic factors increases embryo quality and the number of blastocysts produced in vitro.13–17,65 Previous studies showed that deletion of Igf1 affects female fertility in mice.51 The reproductive defect in PgrCre/+; Igf1f/f mice did not seem to emanate from a uterine phenotype.51 However, it was still unclear whether the PgrCre/+; Igf1f/f mice had an oviductal defect. Here, we illustrated that a loss of Igf1 in the oviduct resulted in a trending increase in the number of embryos with delayed development or nonviable embryos. Although the number was not significantly different from the control group, it is astounding that a lack of a single factor, Igf1, could result in a detectable defect in preimplantation embryo development. One possibility could be that liver-derived circulatory IGF1 present in the oviductal fluid could also act as an additional source of IGF1 for the embryos in the PgrCre/+; Igf1f/f mice. Nevertheless, our findings provide evidence that the oviduct provides a finely tuned balance for embryos to develop normally in the oviduct. In addition to an embryo developmental defect, we also found that these developmentally delayed, nonviable embryos, or unfertilized eggs were retained in the oviduct in vivo. This phenomenon has also been observed in mares,66–68 hamsters,69 and rats.70 To functionally determine the role of IGF1 on oviductal function, future studies could address the proliferative response in the epithelial and stromal cells after E2 treatment as well as the transcriptional dynamics in ciliated vs. secretory cells in the PgrCre/+; Igf1f/f oviducts.
As the IGF1 receptor is also present on bovine embryos,71,72 it is possible that IGF1 secreted from the oviduct promotes a positive-feedback loop to the embryo in a paracrine fashion in mammals. We showed that Igf1 was produced in the stromal and muscle cells of the oviduct. In the uterus, stromal-derived IGF1 stimulates epithelial cell proliferation through a paracrine mechanism.11 It was previously shown that secreted factors present in the extracellular vesicles can diffuse through the zona pellucida and bind to the plasma membrane of bovine preimplantation embryos.73 Therefore, it is possible that in the mouse oviduct, secreted IGF1 can be diffused in a similar fashion as in the uterus through epithelial cells and promote embryo development in the oviductal lumen. In IVF settings, human embryos are often cultured in media supplemented with growth factors.74 Therefore, this paracrine signaling of IGF1 likely supports preimplantation embryo development in mammalian species, including humans. Although the presence of developmentally delayed or nonviable embryos in the PgrCre/+; Igf1f/f was not significantly different from Igf1f/f mice, we found that IGF1 provided a non-negligible function in the oviduct for proper embryo development. Future studies could address whether C3 is required for normal embryo development as C3 was another embryotrophic factor identified in both mouse oviducts and human Fallopian tubes. In addition, more human samples are required to characterize the transcriptome and heterogeneity of cell populations within the Fallopian tube.
In summary, we showed the heterogeneity of epithelial cell and fibroblast populations in the oviduct. Each cell type distinctly responded to the presence of the steroid hormone, E2. Not surprisingly, oviductal cells behaved differently from that of the uterus (or the mammary gland75) in response to E2, suggesting a tissue-specific regulation of steroid hormones within the female reproductive tract in mammals. The use of scRNA-seq analysis revealed that supraphysiological treatment of E2 triggers a relatively similar response in oviductal cell populations to that of endogenous E2 during the estrus stage. We found that IGF1 is a potential candidate as an E2-target gene in the oviduct and is necessary for normal embryo development and transport. Therefore, IGF1 could potentially be an endpoint marker for diagnosis of problematic pregnancies in humans. Our findings suggest that E2 regulation of oviductal cell response and function could provide crucial support for preimplantation embryo development and transport in mammals.
Supplementary Material
ACKNOWLEDGMENT
The authors thank Yu Ishikawa for oviductal cell isolation protocol; Anna Willie, Jeffery Erickson and Jalene Velezques for some of the immunohistochemical staining; and Ryan Driskell for initial assistance with scRNA-seq data analysis pipeline.
Funding information
Burroughs Wellcome Fund (BWF); Louis Stokes Alliance for Minority Participation (LSAMP) Research Award; HHS | NIH | National Institute of Environmental Health Sciences (NIEHS), Grant/Award Number: 1ZIAES103311 and 1ZIAES070065; HHS | NIH | Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD), Grant/Award Number: R01HD042311, R01HD097087, R01HD097087-01S1, R01HD097087-02S1 and R00HD096057
Abbreviations:
- E2
17β-estradiol
- ESR1
estrogen receptor α
- IGF1
insulin-like growth factor 1
- IHC
immunohistochemical
- ISH
in situ hybridization
- PGR
progesterone receptor
- scRNA-seq
single-cell RNA-sequencing
- UTJ
uterotubal junction
Footnotes
SUPPORTING INFORMATION
Additional Supporting Information may be found online in the Supporting Information section.
CONFLICT OF INTEREST
The authors have stated explicitly that there are no conflicts of interest in connection with this article.
REFERENCES
- 1.Li S, Winuthayanon W. Oviduct: roles in fertilization and early embryo development. J Endocrinol. 2017;232(1):R1–R26. [DOI] [PubMed] [Google Scholar]
- 2.Pillai VV, Weber DM, Phinney BS, Selvaraj V. Profiling of proteins secreted in the bovine oviduct reveals diverse functions of this luminal microenvironment. PLoS One. 2017;12(11):e0188105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stewart CA, Behringer RR. Mouse oviduct development. Results Probl Cell Differ. 2012;55:247–262. [DOI] [PubMed] [Google Scholar]
- 4.Niwa S, Nakajima K, Miki H, Minato Y, Wang D, Hirokawa N. KIF19A is a microtubule-depolymerizing kinesin for ciliary length control. Dev Cell. 2012;23(6):1167–1175. [DOI] [PubMed] [Google Scholar]
- 5.Ozmen B, Diedrich K, Al-Hasani S. Hydrosalpinx and IVF: assessment of treatments implemented prior to IVF. Reprod BioMed Online. 2007;14(2):235–241. [DOI] [PubMed] [Google Scholar]
- 6.Mukherjee T, Copperman AB, McCaffrey C, Cook CA, Bustillo M, Obasaju MF. Hydrosalpinx fluid has embryotoxic effects on murine embryogenesis: a case for prophylactic salpingectomy. Fertil Steril. 1996;66(5):851–853. [DOI] [PubMed] [Google Scholar]
- 7.Beyler SA, James KP, Fritz MA, Meyer WR. Hydrosalpingeal fluid inhibits in-vitro embryonic development in a murine model. Hum Reprod. 1997;12(12):2724–2728. [DOI] [PubMed] [Google Scholar]
- 8.Saraiya M, Berg CJ, Kendrick JS, Strauss LT, Atrash HK, Ahn YW. Cigarette smoking as a risk factor for ectopic pregnancy. Am J Obstet Gynecol. 1998;178(3):493–498. [DOI] [PubMed] [Google Scholar]
- 9.Barton BE, Herrera GG, Anamthathmakula P, et al. Roles of steroid hormones in oviductal function. Reproduction. 2020;159(3): R125–R137. [PMC free article] [PubMed] [Google Scholar]
- 10.Lubahn DB, Moyer JS, Golding TS, Couse JF, Korach KS, Smithies O. Alteration of reproductive function but not prenatal sexual development after insertional disruption of the mouse estrogen receptor gene. Proc Natl Acad Sci USA. 1993;90(23):11162–11166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu L, Pollard JW. Estradiol-17 beta regulates mouse uterine epithelial cell proliferation through insulin-like growth factor 1 signaling. Proc Natl Acad Sci USA. 2007;104(40):15847–15851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Winuthayanon W, Hewitt SC, Orvis GD, Behringer RR, Korach KS. Uterine epithelial estrogen receptor alpha is dispensable for proliferation but essential for complete biological and biochemical responses. Proc Natl Acad Sci USA. 2010;107(45):19272–19277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harvey MB, Kaye PL. IGF-2 stimulates growth and metabolism of early mouse embryos. Mech Dev. 1992;38(3):169–173. [DOI] [PubMed] [Google Scholar]
- 14.Doherty AS, Temeles GL, Schultz RM. Temporal pattern of IGF-I expression during mouse preimplantation embryogenesis. Mol Reprod Dev. 1994;37(1):21–26. [DOI] [PubMed] [Google Scholar]
- 15.Rappolee DA, Basilico C, Patel Y, Werb Z. Expression and function of FGF-4 in peri-implantation development in mouse embryos. Development. 1994;120(8):2259. [DOI] [PubMed] [Google Scholar]
- 16.Lee Y-L, Lee K-F, Xu J-S, et al. The Embryotrophic Activity of Oviductal Cell-derived Complement C3b and iC3b, a Novel Function of Complement Protein in Reproduction. J Biol Chem. 2004;279(13):12763–12768. [DOI] [PubMed] [Google Scholar]
- 17.Lee K-F, Xu J-S, Lee Y-L, Y eung WSB. Demilune cell and parotid protein from murine oviductal epithelium stimulates preimplantation embryo development. Endocrinology. 2006;147(1):79–87. [DOI] [PubMed] [Google Scholar]
- 18.Orihuela PA, Croxatto HB. Acceleration of oviductal transport of oocytes induced by estradiol in cycling rats is mediated by nongenomic stimulation of protein phosphorylation in the oviduct. Biol Reprod. 2001;65(4):1238–1245. [DOI] [PubMed] [Google Scholar]
- 19.Orihuela PA, Parada-Bus tamante A, Cortes PP, Gatica C, Croxatto HB. Estrogen receptor, cyclic adenosine monophosphate, and protein kinase A are involved in the nongenomic pathway by which estradiol accelerates oviductal oocyte transport in cyclic rats. Biol Reprod. 2003;68(4):1225–1231. [DOI] [PubMed] [Google Scholar]
- 20.Croxatto HB, Ortiz ME, Forcelledo ML, et al. Hormonal control of ovum transport through the rat oviduct. Archi de Biol Med Experiment. 1991;24:403. [Google Scholar]
- 21.Oróstica ML, Lopez J, Rojas I, et al. Estradiol increases cAMP in the oviductal secretory cells through a nongenomic mechanism. Reproduction. 2014;148(3):285–294. [DOI] [PubMed] [Google Scholar]
- 22.Reuquén P, Oróstica ML, Rojas I, Díaz P, Parada-Bus tamante A, Orihuela PA. Estradiol increases IP3 by a nongenomic mechanism in the smooth muscle cells from the rat oviduct. Reproduction. 2015;150(4):331. [DOI] [PubMed] [Google Scholar]
- 23.Winuthayanon W, Bernhardt ML, Padilla-Banks E, et al. Oviductal estrogen receptor alpha signaling prevents protease-mediated embryo death. Elife. 2015;4:e10453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zheng GX, Terry JM, Belgrader P, et al. Massively parallel digital transcriptional profiling of single cells. Nat Commun. 2017;8:14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ford MJ, Harwalkar K, Pacis AS, et al. Oviduct epithelial cells constitute two developmentally distinct lineages that are spatially separated along the distal-proximal axis. bioRxiv. 2020.08.21.261016. 10.1101/2020.08.21.261016 [DOI] [PubMed] [Google Scholar]
- 26.Winuthayanon W, Hewitt SC, Orvis GD, Behringer RR, Korach KS. Uterine epithelial estrogen receptor α is dispensable for proliferation but essential for complete biological and biochemical responses. Proc Natl Acad Sci USA. 2010;107(45):19272–19277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Soyal SM, Mukherjee A, Lee KY, et al. Cre-mediated recombination in cell lineages that express the progesterone receptor. Genesis. 2005;41(2):58–66. [DOI] [PubMed] [Google Scholar]
- 28.Liu JL, Grinberg A, Westphal H, et al. Insulin-like growth factor-I affects perinatal lethality and postnatal development in a gene dosage-dependent manner: manipulation using the Cre/loxP system in transgenic mice. Mol Endocrinol. 1998;12(9):1452–1462. [DOI] [PubMed] [Google Scholar]
- 29.La Manno G, Soldatov R, Zeisel A, et al. RNA velocity of single cells. Nature. 2018;560(7719):494–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.velocyto: Estimating RNA velocity in single cell RNA sequencing datasets. https://velocyto.org. Accessed December 4, 2020.
- 31.scVelo - RNA velocity generalized through dynamical modeling. https://scvelo.readthedocs.io/. Accessed December 4, 2020.
- 32.Wolf A, Ramirez F, Rybakov S. Scanpy tutorials. https://scanpy-tutorials.readthedocs.io/en/latest/. Accessed December 4, 2020. [Google Scholar]
- 33.JupyterLab Documentation. https://jupyterlab.readthedocs.io/en/stable/. Accessed December 4, 2020.
- 34.LOOMPY. http://loompy.org/. Accessed December 4, 2020.
- 35.Docker. https://www.docker.com/. Accessed December 4, 2020.
- 36.An annotated data matrix. https://anndata.readthedocs.io/en/latest/anndata.AnnData.html. Accessed December 4, 2020.
- 37.McInnes L, Healy J, Melville JUMAP. UMAP: uniform manifold approximation and projection for dimension reduction. https://umap-learn.readthedocs.io/en/latest/. Accessed December 4, 2020. [Google Scholar]
- 38.NumPy: The fundamental package for scientific computing with Python. https://numpy.org/. Accessed December 4, 2020.
- 39.SciPy: a Python-based ecosystem of open-source software for mathematics, science, and engineering. https://www.scipy.org/. Accessed December 4, 2020.
- 40.pandas: Python Data Analysis Library. https://pandas.pydata.org/. Accessed December 4, 2020.
- 41.scikit-learn: Machine Learning in Python. https://scikit-learn.org/stable/. Accessed December 4, 2020.
- 42.statsmodels: statistical models, hypothesis tests, and data exploration. https://www.statsmodels.org/stable/index.html. Accessed December 4, 2020.
- 43.igraph: Python Package Index with pre-compiled wheels for most Python distributions and platforms. https://igraph.org/python/. Accessed December 4, 2020.
- 44.louvain: a general algorithm for methods of community detection in large networks. https://pypi.org/project/louvain/. Accessed December 4, 2020.
- 45.leidenalg: a general algorithm for methods of community detection in large networks. https://pypi.org/project/leidenalg/. Accessed December 4, 2020.
- 46.Stuart T, Butler A, Hoffman P, et al. Comprehensive integration of single-cell data. Cell. 2019;177(7):1888–1902 e1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Polański K, Young MD, Miao Z, Meyer KB, Teichmann SA, Park J-E. BBKNN: fast batch alignment of single cell transcriptomes. Bioinformatics. 2019;36(3):964–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.ForceAtlas2 for Python. https://github.com/bhargavchippada/forceatlas2. Accessed December 4, 2020.
- 49.Thomas PD, Campbell MJ, Kejariwal A, et al. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 2003;13(9):2129–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hewitt SC, Li L, Grimm SA, et al. Research resource: whole-genome estrogen receptor alpha binding in mouse uterine tissue revealed by ChIP-seq. Mol Endocrinol. 2012;26(5):887–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hewitt SC, Lierz SL, Garcia M, et al. A distal super enhancer mediates estrogen-dependent mouse uterine-specific gene transcription of Igf1 (insulin-like growth factor 1). J Biol Chem. 2019;294(25):9746–9759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hewitt SC, Deroo BJ, Hansen K, et al. Estrogen receptor-dependent genomic responses in the uterus mirror the biphasic physiological response to estrogen. Mol Endocrinol. 2003;17(10):2070–2083. [DOI] [PubMed] [Google Scholar]
- 53.Eeckhoute J, Carroll JS, Geistlinger TR, Torres-Arzayus MI, Brown M. A cell-type-specific transcriptional network required for estrogen regulation of cyclin D1 and cell cycle progression in breast cancer. Genes Dev. 2006;20(18):2513–2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hong SH, Nah HY, Lee JY, et al. Estrogen regulates the expression of the small proline-rich 2 gene family in the mouse uterus. Mol Cells. 2004;17(3):477–484. [PubMed] [Google Scholar]
- 55.Diao H, Xiao S, Zhao F, Ye X. Uterine luminal epithelium-specific proline-rich acidic protein 1 (PRAP1) as a marker for successful embryo implantation. Fertil Steril. 2010;94(7):2808–2811.e2801. [DOI] [PubMed] [Google Scholar]
- 56.Bergen V, Lange M, Peidli S, Wolf FA, Theis FJ. Generalizing RNA velocity to transient cell states through dynamical modeling. Nat Biotechnol. 2020. [DOI] [PubMed] [Google Scholar]
- 57.Brenner RM. Renewal of oviduct cilia during the menstrual cycle of the rhesus monkey. Fertil Steril. 1969;20(4):599–611. [DOI] [PubMed] [Google Scholar]
- 58.Winuthayanon W, Lierz SL, Delarosa KC, et al. Juxtacrine activity of estrogen receptor alpha in uterine stromal cells is necessary for estrogen-induced epithelial cell proliferation. Sci Rep. 2017;7(1):8377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Alarid ET, Bakopoulos N, Solodin N. Proteasome-mediated proteolysis of estrogen receptor: a novel component in autologous down-regulation. Mol Endocrinol. 1999;13(9):1522–1534. [DOI] [PubMed] [Google Scholar]
- 60.Tibbetts TA, Mendoza-Meneses M, O’Malley BW, Conneely OM. Mutual and intercompartmental regulation of estrogen receptor and progesterone receptor expression in the mouse uterus1. Biol Reprod. 1998;59(5):1143–1152. [DOI] [PubMed] [Google Scholar]
- 61.Almiñana C, Corbin E, Tsikis G, et al. Oviduct extracellular vesicles protein content and their role during oviduct–embryo cross-talk. Reproduction. 2017;154(3):253–268. [DOI] [PubMed] [Google Scholar]
- 62.Borel F, Sun H, Zieger M, et al. Editing out five Serpina1 paralogs to create a mouse model of genetic emphysema. Proc Natl Acad Sci USA. 2018;115(11):2788–2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Danielson KG, Baribault H, Holmes DF, Graham H, Kadler KE, Iozzo RV. Targeted disruption of decorin leads to abnormal collagen fibril morphology and skin fragility. J Cell Biol. 1997;136(3):729–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lampron C, Rochette-Egly C, Gorry P, et al. Mice deficient in cellular retinoic acid binding protein II (CRABPII) or in both CRABPI and CRABPII are essentially normal. Development. 1995;121(2):539–548. [DOI] [PubMed] [Google Scholar]
- 65.Velazquez MA, Zaraza J, Oropeza A, Webb R, Niemann H. The role of IGF1 in the in vivo production of bovine embryos from superovulated donors. Reproduction. 2009;137(2):161. [DOI] [PubMed] [Google Scholar]
- 66.Weber JA, Freeman DA, Vanderwall DK, Woods GL. Prostaglandin E2 hastens oviductal transport of equine embryos1. Biol Reprod. 1991;45(4):544–546. [DOI] [PubMed] [Google Scholar]
- 67.Weber JA, Freeman DA, Vanderwall DK, Woods GL. Prostaglandin E2 secretion by oviductal transport-stage equine embryos1. Biol Reprod. 1991;45(4):540–543. [DOI] [PubMed] [Google Scholar]
- 68.Freeman DA, Woods GL, Vanderwall DK, Weber JA. Embryo-initiated oviductal transport in mares. Reproduction. 1992;95(2):535–538. [DOI] [PubMed] [Google Scholar]
- 69.Ortiz ME, Bedregal P, Carvajal MI, Croxatto HB. Fertilized and unfertilized ova are transported at different rates by the hamster oviduct1. Biol Reprod. 1986;34(4):777–781. [DOI] [PubMed] [Google Scholar]
- 70.Ortiz ME, Llados C, Croxatto HB. Embryos of different ages transferred to the rat oviduct enter the uterus at different times1. Biol Reprod. 1989;41(3):381–384. [DOI] [PubMed] [Google Scholar]
- 71.Velazquez MA, Hadeler KG, Herrmann D, et al. In vivo oocyte IGF-1 priming increases inner cell mass proliferation of in vitro-formed bovine blastocysts. Theriogenology. 2012;78(3):517–527. [DOI] [PubMed] [Google Scholar]
- 72.Bonilla AQS, Oliveira LJ, Ozawa M, Newsom EM, Lucy MC, Hansen PJ. Developmental changes in thermoprotective actions of insulin-like growth factor-1 on the preimplantation bovine embryo. Mol Cell Endocrinol. 2011;332(1):170–179. [DOI] [PubMed] [Google Scholar]
- 73.Harris EA, Stephens KK, Winuthayanon W. Extracellular vesicles and the oviduct function. Int J Mol Sci. 2020;21(21):8280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chronopoulou E, Harper JC. IVF culture media: past, present and future. Human Reproduct Update. 2014;21(1):39–55. [DOI] [PubMed] [Google Scholar]
- 75.Feng Y, Manka D, Wagner K-U, Khan SA. Estrogen receptor-α expression in the mammary epithelium is required for ductal and alveolar morphogenesis in mice. Proc Natl Acad Sci USA. 2007;104(37):14718–14723. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All analyses in this study were saved in JupyterLab and deposited on github in https://github.com/winuthayanon/ovx_ve/ for the ovariectomized Veh- and E2-treated dataset, https://github.com/winuthayanon/estrus/ for the estrus dataset, and https://github.com/winuthayanon/humanFT/ for the human fallopian tube dataset. Raw data as fastq files for the ovariectomized Veh- and E2-treated, the estrus, and the human Fallopian tube datasets were deposited at Gene Expression Omnibus (GSE164291).