Summary
The identification of patients with coronavirus disease 2019 and high risk of severe disease is a challenge in routine care. We performed cell phenotypic, serum, and RNA sequencing gene expression analyses in severe hospitalized patients (n = 61). Relative to healthy donors, results showed abnormalities of 27 cell populations and an elevation of 42 cytokines, neutrophil chemo-attractants, and inflammatory components in patients. Supervised and unsupervised analyses revealed a high abundance of CD177, a specific neutrophil activation marker, contributing to the clustering of severe patients. Gene abundance correlated with high serum levels of CD177 in severe patients. Higher levels were confirmed in a second cohort and in intensive care unit (ICU) than non-ICU patients (P < 0.001). Longitudinal measurements discriminated between patients with the worst prognosis, leading to death, and those who recovered (P = 0.01). These results highlight neutrophil activation as a hallmark of severe disease and CD177 assessment as a reliable prognostic marker for routine care.
Keywords: immunology, virology
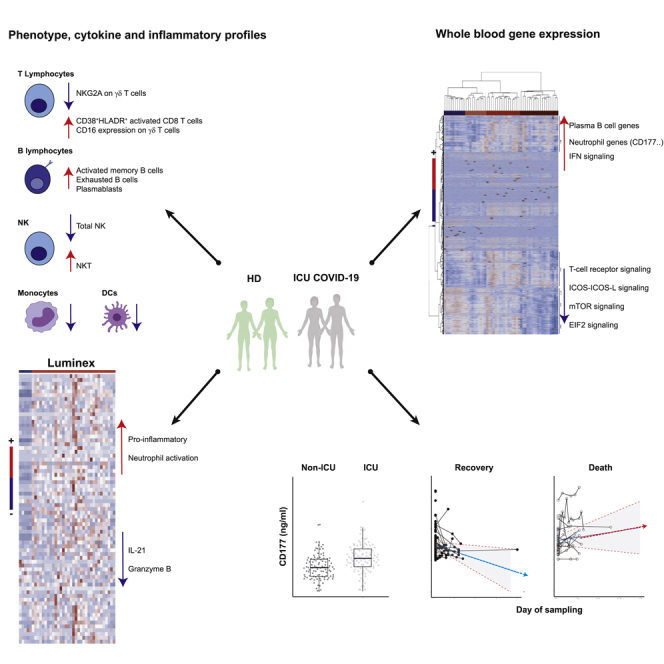
Graphical abstract
Highlights
-
•
Increase in B cells, activated CD8 T cells, NKT, and γδ T NKG2A + cells in severe COVID-19
-
•
Severe COVID-19 is characterized by an increase of neutrophil and inflammatory markers
-
•
Serum CD177 protein levels are increased in patients with COVID-19 in ICU
-
•
Sustained high levels of CD177 discriminated recovery and death of patients with COVID-19
Immunology; Virology
Introduction
The coronavirus disease 2019 (COVID-19) pandemic is caused by a newly described highly pathogenic beta coronavirus, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) (Coronaviridae Study Group of the International Committee on Taxonomy of, 2020; Phelan et al., 2020). COVID-19 consists of a spectrum of clinical symptoms that range from mild upper respiratory tract disease in most cases to severe disease that affects approximately 15% of patients requiring hospitalization (Wu and McGoogan, 2020), some of whom require intensive care because of severe lower respiratory tract illness, acute respiratory distress syndrome, and extrapulmonary manifestations, leading to multiorgan failure and death. Several recent studies have provided important clues about the physiopathology of COVID-19 (Blanco-Melo et al., 2020; Chen et al., 2020a; Kuri-Cervantes et al., 2020; Mehta et al., 2020; Qin et al., 2020). Most compared the immune and inflammatory status of patients at different stages of the disease (Hadjadj et al., 2020; Mathew et al., 2020; Wilk et al., 2020). Thus, several important biomarkers associated with specific phases of the evolution of COVID-19 have thus far been identified (Ponti et al., 2020; Silvin et al., 2020). Inflammation, cytokine storms, and other dysregulated immune responses have been shown to be associated with severe disease pathogenesis (Lucas et al., 2020; Ong et al., 2020). Patients with severe COVID-19 are characterized by elevated numbers of monocytes and neutrophils and lymphopenia (Giamarellos-Bourboulis et al., 2020; Huang et al., 2020; Mathew et al., 2020; Zhou et al., 2020), and a high neutrophil-to-lymphocyte ratio predicts inhospital mortality of critically ill patients (Fu et al., 2020). High levels of proinflammatory cytokines, including interleukin (IL)-6,IL-1β, tumor necrosis factor (TNF), MCP-1, IP-10, and granulocyte colony-stimulating factor (G-CSF), in the plasma (Chen et al., 2020a, 2020b; Giamarellos-Bourboulis et al., 2020; Mathew et al., 2020; Zhou et al., 2020) and a possible defect in type I interferon (IFN) activity have been reported in patients with severe COVID-19 (Arunachalam et al., 2020; Hadjadj et al., 2020; Ong et al., 2020). However, these responses are dynamic, changing rapidly during the clinical course of the disease, which may explain the high variability of the immunological spectrum described (Arunachalam et al., 2020; Bouadma et al., 2020; Lucas et al., 2020). This makes it difficult to deduce a unique profile of the pathophysiology of this infection, which is still undetermined. Furthermore, the high amplitude of the signals generated by the inflammation associated with the disease may hide other pathways that are involved.
From a clinical standpoint, clinicians face the daily challenge of predicting worsening patients owing to the peculiar clinical course of severe COVID-19, characterized by a sudden deterioration of the clinical condition 7 to 8 days after the onset of symptoms. Determination of the onset of the pathological process once infection has been established in a patient with a severe stage of infection is highly imprecise because of the possible paucisymptomatic or asymptomatic phase of the infection, as well as the low specificity of self-limited “flu” illness.
We used a systems immunology approach to identify host factors that were significantly associated with the time to illness onset, severity of the disease (intensive care unit [ICU] or transfer to ICU), and mortality of patients with COVID-19 enrolled in the multicentric French COVID cohort (Yazdanpanah, 2020). In addition to the depletion of T cells and mobilization of B cells, neutrophil activation, and severe inflammation, we show upregulation of CD177 gene expression and protein levels in the blood of patients with COVID-19 in both the COVID-19 cohort and a “confirmatory” cohort, that is, Swiss cohort, relative to healthy subjects. CD177, a neutrophil activation marker, characterized critically ill patients and marked disease progression and death. Our finding highlights the major role of neutrophil activation through CD177 overexpression in the critical clinical transition point in the trajectory of patients with COVID-19.
Results
Overview of the phenotype, cytokine, and inflammatory profiles of patients with COVID-19
Patient characteristics from the French COVID cohort enrolled in this analysis are shown in Table 1. All patients from this cohort were stratified as severe as per criteria of the French COVID cohort (clinicaltrials.gov NCT04262921) (Yazdanpanah, 2020), with 53 (87%) being hospitalized in an ICU (either initially or after clinical worsening or death) and eight were not. The median age was 60 years (interquartile range, 50–69) and 80% were men. Sampling for immunological analyses was performed within three days of entry and after a median of 11 days [7–14] after the onset of symptoms. We first assessed leukocyte profiles by flow cytometry using frozen peripheral blood mononuclear cells (PBMCs) from 50 patients with COVID-19 (with available PBMC samples) and 18 healthy donors (HDs) (14 or 15 HDs were used as controls per immune cell subset).
Table 1.
Patient characteristics of the French COVID cohort (n=61)
| Number of patients | ||
|---|---|---|
| Demographic characteristics | ||
| Age – median (IQR) – years | 61 | 60 (50–69) |
| Male sex – No./total No. (%) | 61 | 49/61 (80) |
| ICU or transfer to ICU or death – No./total No. (%) | 61 | 53/61 (87) |
| Outcome – No./total No. (%) | 61 | |
| Death | 21/61 (34) | |
| Discharge alive | 40/61 (66) | |
| Median interval from first symptoms on admission (IQR) – days | 61 | 11 (7–14) |
| Comorbidities – No./total No. (%) | ||
| Any | 61 | 14/61 (23) |
| Chronic cardiac disease | 61 | 9/61 (15) |
| Hypertension | 61 | 22/61 (36) |
| Chronic pulmonary disease | 61 | 5/61 (8) |
| Asthma | 61 | 4/61 (7) |
| Chronic kidney disease | 61 | 6/61 (10) |
| Chronic neurological disorder | 61 | 2/61 (3) |
| Obesity | 60 | 23/60 (38) |
| Diabetes | 61 | 12/61 (20) |
| Smoking History – No./total No. (%) | ||
| Smoking | 61 | 5/61 (8) |
| Laboratory findings on admission - Median (IQR) | ||
| Hemoglobin – g/dL | 57 | 13 (11–14) |
| WBC count – x109/L | 57 | 6 (5–9) |
| Platelet count – x109/L | 57 | 189 (143–270) |
| C-reactive protein (CRP) – mg/L | 57 | 120 (66–195) |
| Blood urea nitrogen (urea) – mmol/L | 57 | 7 (5 – 12) |
| Symptoms on admission – No./total No. (%) | ||
| Fever | 59 | 51/59 (86) |
| Cough | 57 | 40/57 (70) |
| Sore throat | 56 | 4/56 (7) |
| Wheezing | 54 | 6/54 (11) |
| Myalgia | 56 | 21/56 (38) |
| Arthralgia | 55 | 9/55 (16) |
| Fatigue | 57 | 27/57 (47) |
| Dyspnea | 57 | 46/57 (81) |
| Headache | 57 | 11/57 (19) |
| Altered consciousness | 56 | 3/56 (5) |
| Abdominal pain | 53 | 8/53 (15) |
| Vomiting/nausea | 56 | 10/56 (18) |
| Diarrhea | 56 | 11/56 (20) |
| Clinical characteristics on admission – Median (IQR) | ||
| SOFA score (ICU patients) | 34 | 6 (4–8) |
| SAPS2 (ICU patients) | 36 | 32 (27–53) |
| Heart rate – beats per minute | 61 | 87 (76–104) |
| Respiratory rate – breaths per minute | 55 | 24 (20–32) |
| Systolic blood pressure - mmHg | 60 | 130 (109–145) |
| Diastolic blood pressure – mmHg | 60 | 77 (70–87) |
| Oxygen saturation – percent | 61 | 96 (91–98) |
| Oxygen saturation on – No./total No. (%) | 56 | |
| Room air | 17/56 (30) | |
| Oxygen therapy | 39/56 (70) | |
| Treatments – No./total No. (%) | ||
| Antiviral | 60 | 40/60 (66) |
| Antibiotic | 60 | 46/60 (77) |
| Corticosteroids | 60 | 33/60 (55) |
| Antifungal | 60 | 9/60 (15) |
| Hydroxychloroquine | 59 | 8/59 (14) |
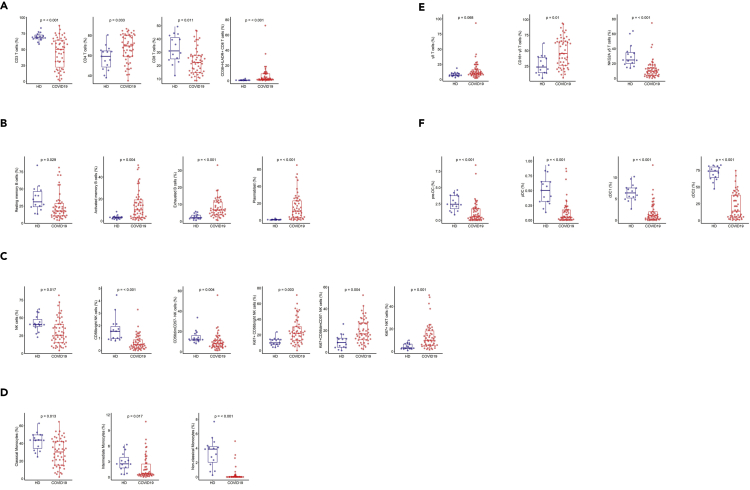
We analyzed 52 immune cell populations (gating strategies are shown in Figure S1); of which, 23 showed significant differences (Wilcoxon test adjusted for multiple comparisons) between patients with COVID-19 and HDs. We not only confirmed previously reported abnormalities but also revealed new immunological features of patients with COVID-19 (Figure S2). Patients with COVID-19 showed a significant reduction in the frequency of total CD3+ T cells and CD8+ T cells relative to HDs, as previously reported (De Biasi et al., 2020; Xu et al., 2020), that expressed an activated phenotype (CD38+HLA-DR+) (Figure 1A). Patients with COVID-19 also showed lower frequencies of resting memory B cells contrasting with higher frequencies of activated memory B cells and exhausted B cells (Figure 1B). As previously reported (Bouadma et al., 2020; Mathew et al., 2020), the proportion of plasmablasts was markedly higher in patients with COVID-19 (median [Q1-Q3]: 10.85% [3–23]) than HDs (0.76% [0.4–0.8]) (P < 0.001). Total natural killer (NK) cell frequencies, more precisely those of the CD56bright and CD56dimCD57- NK cell subpopulations, were lower than in HDs (P = 0.017, P < 0.001, and P = 0.004, respectively) (Figure 1C), whereas a higher proportion of these NK cells, as well as NKT cells, were cycling, expressing Ki67 antigen (CD56bright: 22% [13–30], CD56dimCD57-: 16.8% [11.5–27], and NKT: 10% [5.6–18.2]) (P = 0.003, P = 0.004, and P = 0.001 compared with HD) (Figure 1C). In addition, patients with COVID-19 showed significantly smaller classical (CD14+CD16-), intermediate (CD14+CD16+), and nonclassical (CD14−CD16+) monocyte subpopulations than HDs (P = 0.013, P = 0.017, P < 0.001, respectively) (Figure 1D). Interestingly, patients with COVID-19 tended to exhibit a higher frequency of γδ T cells than HDs (median 10.4% [7.5–16.1] vs 7.3% [6–10] in HDs; P = 0.068) (Figure 1E), with a significant proportion of γδ T cells showing higher expression of the activation marker CD16 (P = 0.01) and lower expression of the inhibitory receptor NKG2A (P < 0.001) than HDs (Figure 1E). Finally, we observed markedly smaller frequencies of dendritic cells (DCs) for all populations studied (pre-DC, plasmacytoid DC (pDC), and conventional DC (cDC1 and cDC2) in patients with COVID-19 than in HDs (P < 0.001, for all comparisons) (Figure 1F). Neutrophils count were available in 44 patients, and the concentration was more elevated in patients with COVID-19 belonging to group 2 and group 3 compared with group 1 (8.109/L vs 3.109/L, p < 0.03). It was also more elevated in patients hospitalized in an ICU (8.109/L vs 2.109/L, p < 0.009).
Figure 1.
Frequency of immune-cell subsets between HDs (n = 18) and patients with COVID-19 (n = 50)
(A) Frequency of total CD3 T cells, CD4 and CD8 T cell subsets, and activated CD38+HLADR+ CD8 T cells.
(B) Frequency of B cell subsets (CD21+CD27+: resting memory, CD21−CD27+: activated memory, CD21−CD27-: exhausted) and plasmablasts (CD38++CD27+) gated on CD19+ B cells.
(C) Frequency of NK cell subsets (gated on CD3- CD14-) CD56Bright: CD56++CD16+, CD56dim: CD56+CD16++CD57+/−, differentiated Ki67+ NK cells (gated on CD56Bright or CD56dimCD57- NK cells) and differentiated Ki67+ NKT cells (gated on CD3+CD56+ cells).
(D) Monocyte subsets (gated on CD3−CD56-) (classical monocytes: CD14+CD16-, intermediate monocytes: CD16+CD14+, non-classical monocytes: CD14−CD16+).
(E) Frequency of γδ T cells (gated on CD3+ T cells) and CD16 and NKG2A expression (gated on γδ CD3 T cells).
(F) Frequency of DC subsets (gated on HLADR+Lin−) (pDC: CD45RA+CD33−CD123+, pre-DC: CD123+CD45RA+, cDC1: CD33+CD123−CD141+CD1clow, cDC2: CD33+CD123−CD14+CD1c+) detected by flow cytometry in PBMCs from n = 50 patients with COVID-19 and n = 18 HDs. The differences between the two groups were evaluated using Wilcoxon rank sum statistical tests. The lower and upper boundaries of the box represent the 25% and 75% percentiles, the whiskers extend to the most extreme data point that is no more than 1.5 times the interquartile range away from the box. Median values (horizontal line in the boxplot) are shown.
See also Figures S1 and S2.
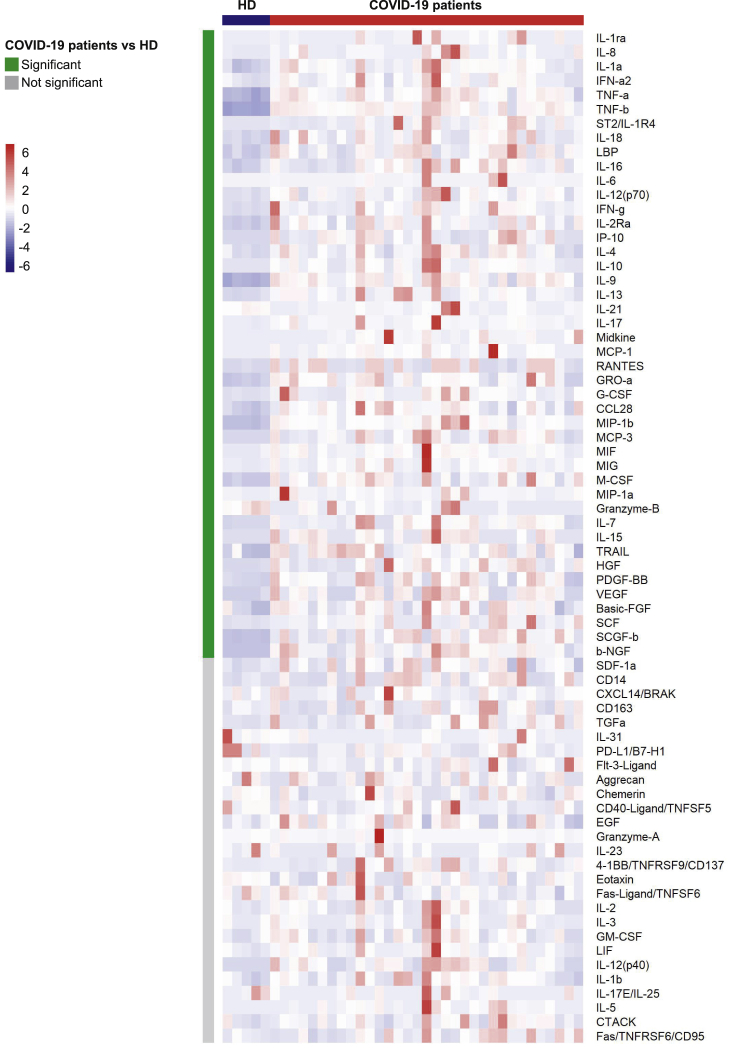
We then evaluated the levels of 71 serum cytokines, chemokines, and inflammatory factors in 33 patients with COVID-19 and 5 HDs. Forty-four analytes differed significantly (Wilcoxon test adjusted for multiple comparisons) between the patients with COVID-19 and HDs (shown in the heatmap in Figure 2 and detailed in Figure S3). The levels of 42 factors were higher, among them, proinflammatory factors (IL-1a, IL-6, IL-18, TNF alpha and β [TNF-α, TNF-β], IL-1ra, ST2/IL-1R4, the acute phase protein lipopolysaccharide-binding protein LBP, IFN-a2); Th1 pathway factors (IL-12 [p70], IFN-γ, IP-10, IL-2Ra); Th2/regulatory cytokines (IL-4, IL-10, IL-13); IL-17, which also promotes G-CSF-mediated granulopoiesis and recruits neutrophils to inflammatory sites; T cell proliferation and activation factors (IL-7, IL-15); growth factors (SCF, SCGF-b, HGF, b-FGF, b-NGF); and a significant number of cytokines and chemokines involved in macrophage and neutrophil activation and chemotaxis (RANTES (CCL5), MIP-1a and b (CCL3 and CCL4), MCP-1 (CCL2), MCP-3 (CCL7), M-CSF, MIF, Gro-a (CXCL1), monokine inducible by γ IFN MIG/CXCL9, IL-8, IL-9). Interestingly, we found higher levels of midkine, a marker usually not detectable in the serum, which enhances the recruitment and migration of inflammatory cells and contributes to tissue damage (Cai et al., 2020). In parallel, granzyme B and IL-21 levels were significantly lower in patients with COVID-19 patients than HDs (P = 0.007 and P = 0.004, respectively) (Figure S3).
Figure 2.
Heatmap of analyte abundance in serum
The colors represent standardized expression values centered around the mean, with variance equal to 1. HD: healthy donors (n = 5), COVID: patients with COVID-19 (n = 33). Each column represents a subject. Each line represents an analyte.
See also Figure S3.
Whole blood gene expression profiles show a specific signature for patients with COVID-19
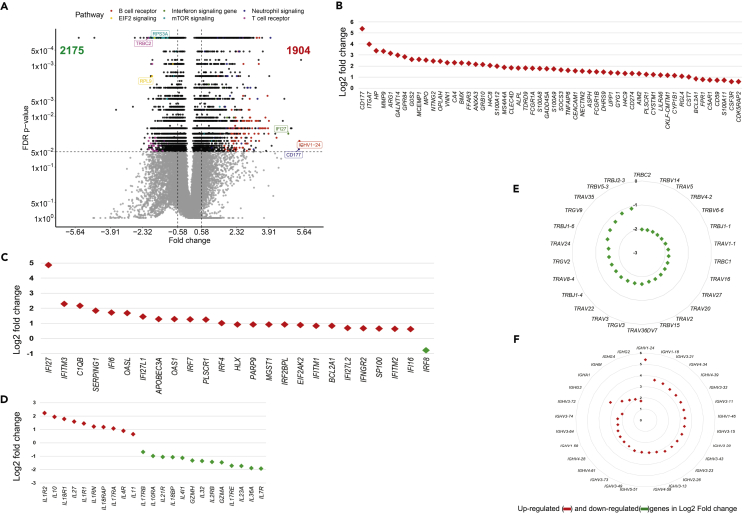
The comparison of gene abundance in whole blood between patients with COVID-19 (n = 44) and HDs (n = 10) showed 4,079 differentially expressed genes (DEGs) with an absolute fold change ≥1.5, including 1,904 that were upregulated and 2,175 that were downregulated (Figure 3A). The main pathways associated with the DEG correspond to the immune response, including neutrophil and IFN signaling, T and B cell receptor responses, metabolism, protein synthesis, and regulators of the eIF2 and mammalian target of rapamycin (mTOR) signaling pathways (Figure 3A). Although several of these pathways involved multiple cell types, analysis of the neutrophil pathway showed higher abundance of genes mainly related to neutrophil activation, their interaction with endothelial cells, and migration (Figure 3B). Among the most highly expressed genes, this signature included CD177, a specific marker of neutrophil adhesion to the endothelium and transmigration (Bayat et al., 2010), HP (haptoglobin), a marker of granulocyte differentiation and released by neutrophils in response to activation (Theilgaard-Monch, 2006), VNN1 (hematopoietic cell trafficking), GPR84 (neutrophil chemotaxis), MMP9 (neutrophil activation and migration), and S100A8 and S100A12 (neutrophil recruitment, chemotaxis, and migration). The S100A12 protein is produced predominantly by neutrophils and is involved in inflammation and the upregulation of vascular endothelial cell adhesion molecules (Roth et al., 2003) (Figure 3B).
Figure 3.
Whole blood gene expression in COVID-19 patients and HDs
(A) Volcano plot showing differentially expressed genes (DEG) as per the log2 fold change (log2 FC) and Benjamini-Hocberg False Discovery Rate (FDR) with thresholds at absolute log2 FC ≥ log2(1.5) and FDR ≤0.05.
(B) Main top DEG related to neutrophils.
(C and D) Main DEG related to IFN and interleukin responses, respectively.
(E) Main TCRV T cell repertoire DEG.
(F) Main B-cell IGHV repertoire DEG. Red symbols represent overabundant genes in COVID-19 relative to HD, green symbols represent underabundant genes.
In parallel, we observed a higher abundance of several IFN-stimulating genes (ISGs) (IFI27, IFITM3, IFITM1, IFITM2, IFI6, IRF7, IRF4) (Figure 3C) and cytokines and cytokine receptors (IL-1R, IL-18R1, IL-18RAP, IL-4R, IL-17R, IL-10) (Figure 3D). Consistent with profound T cell lymphopenia, the expression of several families of T cell receptor genes was lower (Figure 3E). We observed severe dysregulation of T cell function that involved inhibition of serine/threonine kinase PKCθ signaling (Z score = −4.46), as well as the inducible T cell costimulator/ICOSL axis (Z score = −4.5) (Figure S4). In contrast to the results for T cells, the peripheral expansion of memory B cells and plasmablasts was associated with broad expansion of the B cell receptor (BCR) (Figure 3F and Table S2).
We also observed genes belonging to several crucial pathways and biological processes that had not been previously reported to characterize patients with COVID-19 to be underrepresented. These included eIF2 signaling, with many downregulated genes, such as ribosomal proteins and eukaryotic translation initiation factors (Figure S4A), common targets of the integrated stress response (ISR), including antiviral defense (Levin and London, 1978; Pakos-Zebrucka et al., 2016). In addition, we also found genes involved in signaling through mTOR (Figure S4B), a member of the phosphatidylinositol-3-kinase-related kinase family of protein kinases. Prediction analysis using Ingenuity pathways showed both lower eIF2 (Z score = - 6.8) and mTOR (Z score = −2.2) signaling in patients with COVID-19 than HDs.
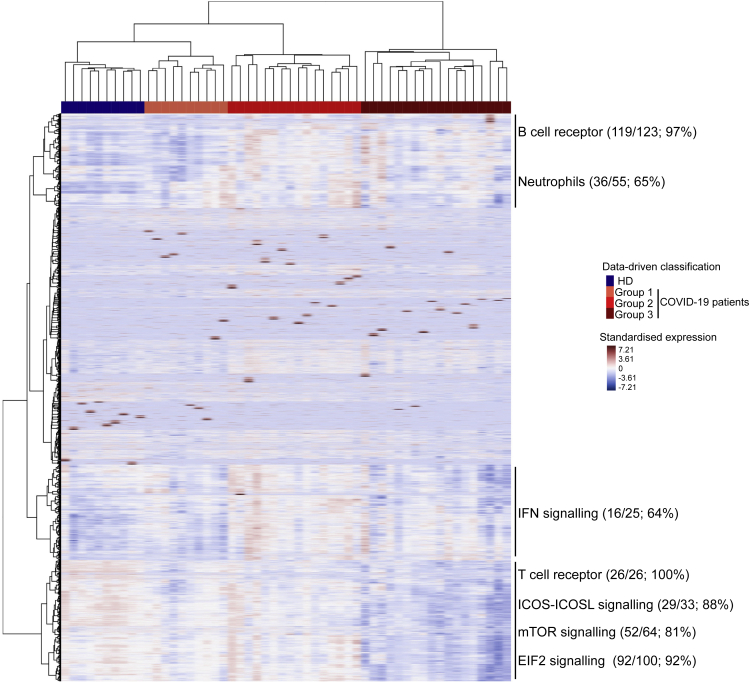
Unsupervised whole blood gene expression profiles reveal distinct features of patients with COVID-19
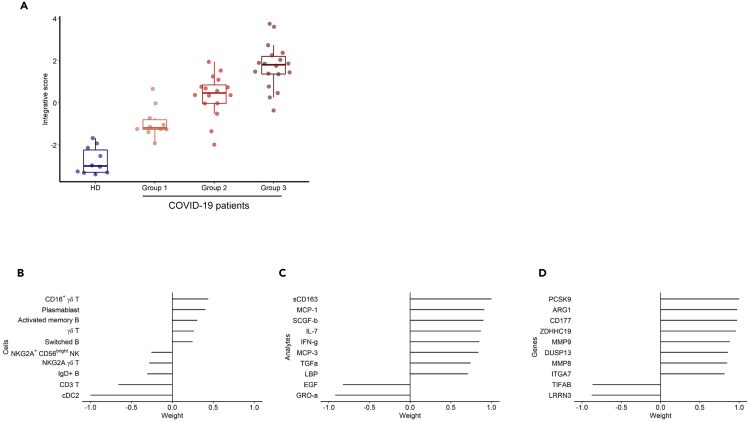
Unsupervised classification of 44 patients with COVID-19 and 10 HDs identified three distinct groups of patients with COVID-19: 10 in group 1, 16 in group 2, and 18 in group 3 (Figure 4). Detailed patient characteristics as per the group are presented in Table S1. Among a large set of clinical and biological characteristics, the analysis showed the differential clustering to not be explained by the severity of the disease. Indeed, the median Sequential Organ Failure Assessment (SOFA) score and Simplified Acute Physiology Score (SAPS2), which include a large number of physiological variables (Le Gall, 1993; Vincent et al., 1998) and evaluate the clinical severity of the disease (a high score is associated with a worse prognosis) of patients with COVID-19, were 6 [4–7] and 36 [28–53], respectively, with no significant differences between groups. Nevertheless, we observed a significant difference from symptoms onset to the admission, which ranged from 7 [6–11] days for patients in group 1 to 11 [10–14] and 13 [9–14] days for patients in groups 2 and 3, respectively (P = 0.04, Kruskal-Wallis test). Finally, group 1 which was the closest to HDs in terms of gene profile consisted of patients in the early days of the disease (Table S1).
Figure 4.
Heatmap of standardized gene expression
The colors represent standardized expression values centered around 0, with variance equal to 1. Each column represents a subject. This heatmap was built by unsupervised hierarchical clustering of log2-counts-per-million RNA-seq transcriptomic data from whole blood (29,302 genes) and subjects (n = 54) using the Euclidean distance and Ward's method. Seven blocks are highlighted as per the features of gene expression across the groups of individuals. Enrichment (number and % of genes of a given pathway selected in the block) of pathways of interest are shown for each block.
See also Table S1.
Analysis of the genes contributing to the differences between groups confirmed and extended the findings described previously (Figures 3 and S4). Several pathways were highly represented in sectors of the heatmap defined as per gene abundance across patient groups. For example, 97% of the genes making up the BCR and 65% of those involved in neutrophil responses were represented among the genes showing a greater abundance in COVID-19 groups 2 and 3 than group 1 and HDs (Figure 4). Other pathways, such as those for IFN (64%), TCR (100%), iCOS-iCOS-L (88%), mTOR (81%), and eIF2 signaling (92%) were also highly represented. The IFN-signaling genes, such as IFI44L, IFIT2, and IRF8, a regulator of type I IFN (α, β), were significantly more abundant at earlier stages (in patients from group 2) and tended to be less abundant in group 3, at more advanced stages of the disease. Finally, the abundance of genes belonging to T cell pathways (TCR, iCOS-iCOSL signaling) or mTOR and eIF2 signaling was lower in group 3, that is to say, those who were analyzed after a longer time from symptom onset to the admission. The findings described previously highlight the heterogeneity of patients with COVID-19.
Integrative analysis of all biomarkers reveals the major contribution of CD177 in the clinical outcome of patients with COVID-19
We performed an integrative analysis using all available data to disentangle the relative contribution of the various markers at the scale of every patient. We thus pooled the data for 29,302 genes from whole blood RNA sequencing (RNA-seq), cell phenotypes (52 types), and cytokines (71 analytes) using the recently described MOFA approach (Argelaguet et al., 2019), which is a statistical framework for dimension reduction adapted to the multiomics context. The data are reduced to components that are linear combinations of variables explaining interpatient variability across the three biological measurement modalities. The first component, that we called our integrative score, discriminated between the three groups of RNA sequencing (initially defined by hierarchical classification based on gene abundance only) and HDs (Figure 5A), although it only explained a portion of the variability within each of the three types of markers (14% of gene expression, 14% of cell phenotypes, and 5% of cytokines). Figures 5B–5D show the contribution of each type of markers to the integrative score. The main contributors for the cell phenotype were the significantly lower frequency of cDC2 and T cells and, marginally, the higher number of plasmablasts and CD16+ γδ T cells in patients with COVID-19 (Figure 5B). The contribution of soluble factors was marked by higher levels of soluble CD163 (sCD163), a marker of polarized M2 macrophages involved in tissue repair (Zhi et al., 2017), in more advanced COVID-19 groups (Figure 5C). Indeed, CD163 gene expression was also significantly higher in patients with COVID-19 than in HDs (log2 fold change = +1.55; FDR = 4.79 10−2). sCD163 has also been reported to be a marker of disease severity in critically ill patients with various inflammatory or infectious conditions (Buechler et al., 2013). Interestingly, the genes that contribute the most to the synthesis of this factor were part of the neutrophil module (CD177, ARG1, MMP9) (Figure 5D). Integrated analysis also revealed higher expression of proprotein convertase subtilisin/kexin type 9 (PCSK9). High plasma PCSK9 protein levels highly correlate with the development and aggravation of subsequent multiple organ failure during sepsis (Boyd et al., 2016; Dwivedi et al., 2016). Of note, high PCSK9 levels have been recently associated with severe Dengue infection (Gan et al., 2020). Finally, the increasing abundance of CD177 gene expression as per the group was again clearly apparent (Figure S4). An additional cell-type-specific significance analysis has been performed to check the robustness of the CD177 differential expression according to the cell-type frequencies (Shen-Orr et al., 2010). We found that CD177 differential expression between patients with COVID-19 and HD was not fully explained by population variations. Indeed, it remained significant after deconvolution in several leukocytes subpopulations (notably FDR of 0.04 within T cells and 0.03 within monocytes).
Figure 5.
Integrative analysis. Integrative analysis of the data of RNA-seq (29,302 genes) from 44 patients with COVID-19, cell phenotype (52 types) from 45 patients with COVID-19, and serum analytes (71 analytes) from 33 patients with COVID-19 using a sparse principal component analysis approach, MOFA v2
(A–D)(A) Integrative score as per the patient groups defined by the hierarchical clustering of the RNA-seq data. Top 10 marker contributions (as per the weight from −1 to 1) of the cell phenotypes (B), serum analytes (C), and RNA-seq (D). The integrative score corresponds to the first factor of the analysis and allows the ordering of individuals along an axis centered at 0. Individuals with an opposite sign for the factor therefore have opposite characteristics.
See also Figure S5.
Serum CD177 protein levels are associated with the clinical outcome of patients with COVID-19
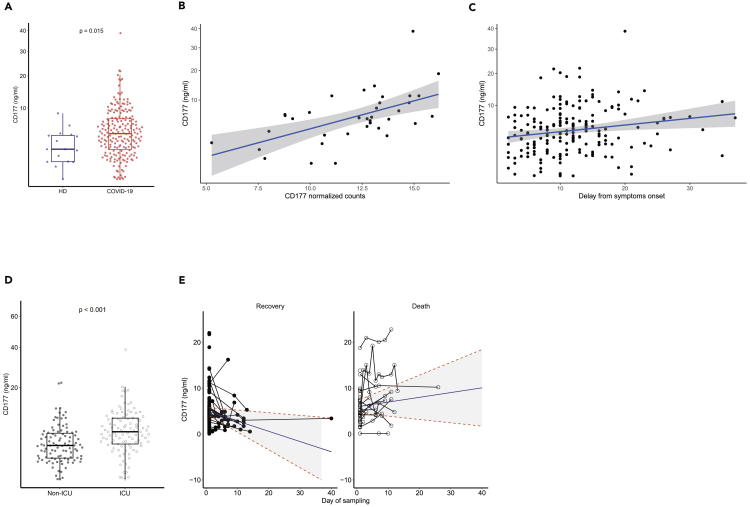
Given the contribution of the neutrophil activation pathway in the clustering of patients with COVID-19, we sought neutrophil-activation features that could act as possible reliable markers of disease evolution. We focused on CD177 because i) it is a neutrophil-specific marker representative of neutrophil activation, ii) it was the most highly differentially expressed gene in patients, and iii) the protein can be measured in the serum, making its use as a marker clinically applicable. Thus, we used an enzyme-linked immunosorbent assay (ELISA) to quantify CD177 in the serum of 203 patients with COVID-19 (115 patients from the French cohort and 88 patients from the Swiss COVID-19 cohort that we used as “a confirmatory” cohort, patient characteristics are described in Table 2), 21% of whom the measurements were repeated (from 2 to 10 measurements per individual). First, we confirmed the significantly higher median serum protein level in the global cohort of patients with COVID-19 (4.5 [2.2–7.4]) relative to that of 16 HDs (2.2 [0.9–4.2]) (P = 0.015, Wilcoxon test) (Figure 6A). Second, we found a robust agreement between CD177 gene expression measured by RNA-seq and CD177 protein levels measured by ELISA (intraclass correlation coefficient 0.88) (Figure 6B).
Table 2.
Characteristics of patients involved in the CD177 analysis
| Number of patients | French cohort (n = 115) | Swiss cohort (n = 88) | |
|---|---|---|---|
| Demographic characteristics | |||
| Age – median (IQR) – years | 200 | 62 (54–72) | 63 (57–74) |
| Male sex – No./total No. (%) | 201 | 82/113 (73) | 56/88 (64) |
| ICU or transfer to ICU or death – No. /total No. (%) | 200 | 61/112 (54) | 40/88 (45) |
| Outcome - No. /total No. (%) | 173 | ||
| Death | 32/107 (30) | 8/66 (12) | |
| Discharge alive | 75/107 (70) | 58/66 (88) | |
| Median interval from first symptoms on admission (IQT) | 192 | 13 (9-18) | 12 (9-17) |
| Comorbidities - No./total No. (%) | |||
| Chronic cardiac disease | 197 | 22/109 (20) | 25/88 (28) |
| Chronic pulmonary disease | 197 | 14/109 (13) | 9/88 (10) |
| Diabetes | 197 | 23/109 (21) | 26/88 (30) |
| Laboratory findings on admission – Median (IQR) | |||
| C-reactive protein (CRP) – mg/L | 34 | 122 (62–196) | |
| Lactate dehydrogenase (LDH) UI/L | 31 | 466 (337–533) | |
| Clinical characteristics on admission – Median (IQR) | |||
| Score SOFA | 41 | 4 (2–7) | |
| Score SAPS2 | 40 | 32 (27–49) |
Figure 6.
Distribution of the CD177 marker and association with clinical outcomes of patients with COVID-19
(A) Measurement of CD177 (ng/mL). HD: Healthy donors (n = 16), patients with COVID-19 (n = 203). The difference between the two groups was evaluated using Wilcoxon rank sum statistical tests. The median values (horizontal line in the boxplot) are shown. The lower and upper boundaries of the box represent the 25% and 75% percentiles.
(B) Correlation between normalized CD177 values of gene expression measured by RNA-seq and CD177 protein by ELISA (ng/mL) from 36 patients with COVID-19. The blue line represents the linear regression line and the gray area the 95% prediction confidence interval.
(C) Association between CD177 serum concentration and time from symptom onset to the admission (n = 192). This association was tested using Spearman correlation tests. The blue line represents the linear regression line and the gray area the 95% confidence interval.
(D) Measurement of CD177 serum concentration in patients hospitalized in an intensive care unit (ICU) or not (n = 196). Wilcoxon rank tests were used. The median values (horizontal line in the boxplot) are shown. The lower and upper boundaries of the box represent the 25% and 75% percentiles.
(E) Change of CD177 concentration over time according to the occurrence of death for 172 patients with COVID-19 and a total of 248 measurements. Predictions were calculated using a mixed effect models for longitudinal data.
See also Figure S6.
Then, we examined the association of clinical characteristics and outcomes with serum CD177 concentration at the time of admission. The serum CD177 concentration was positively associated with the time from symptom onset to the admission (r = 0.22, P = 0.0026) (Figure 6C) and was higher for patients hospitalized in an ICU (6.0 ng/mL [3.5–9.4] vs 3.3 ng/mL [1.5–5.6], P < 0.001) (Figure 6D). The association between serum CD177 levels and hospitalization in an ICU was independent of the usual risk factors, such as age, sex, chronic cardiac or pulmonary diseases, or diabetes (multivariable logistic regression, adjusted odds ratio 1.14 per unit increase, P < 0.001). We observed a trend toward a positive association with the SOFA and SAPS2 risk scores that was not statistically significant (P = 0.17 and P = 0.074, respectively) (Figures S6A and S6B). CD177 levels were not associated with other conditions that contribute to a high risk of severe disease, such as diabetes (P = 0.632), chronic cardiac disease (P = 0.833), chronic pulmonary disease (P = 0.478), or age of the patient (P = 0.83).
We then examined the dynamics of the CD177 concentration in 172 patients with COVID-19, with longitudinal serum samples, using all available measurements (Figure 6E). At the first measurement, the average concentration of CD177 was not significantly different between the patients who died and those who recovered (5.93 vs 5.06, P = 0.26, Wald test). When looking at the change of CD177 concentration over time, it appears clearly that the concentration was decreasing in those who recovered (−0.22 ng/mL/day, 95% CI -0.307; −0.139), whereas it was stable in those who died later on (+0.10 ng/mL/day, 95% confidence interval -0.014; +0.192). These results show that the stability of CD177 protein levels in patients with severe COVID-19 during the course of the disease is a hallmark of a worse prognosis, leading to death.
Discussion
Here, we investigated factors that influence the clinical outcomes of patients with severe COVID-19 involved in a multicentric French cohort combining standardized whole-blood RNA-seq analyses, in-depth phenotypic analysis of immune cells, and measurements of a large panel of serum analytes. An integrated and global overview of host markers revealed several pathways associated with the course of COVID-19 disease, with a prominent role for neutrophil activation. This signature included CD177, a specific neutrophil marker of activation, adhesion to the endothelium, and transmigration. The correlation between CD177 gene abundance and serum protein levels in the blood of patients with COVID-19 underscores the importance of this marker, making the measurement of CD177 protein levels a reliable approach that is largely accessible in routine care. We also demonstrated that the dynamics of serum CD177 levels is strongly associated with the severity of COVID-19 disease, ICU hospitalization, and survival in an additional cohort of patients. During follow-up, the CD177 protein levels decrease in patients who are recovering, while staying high in those who died.
CD177 is a glycosylphosphatidylinositol (GPI)-anchored protein expressed by a variable proportion of human neutrophils. It plays a key role in the regulation of neutrophils by modulating their migration and activation. For example, the CD177 molecule has been identified as the most dysregulated parameter in purified neutrophils from patients with septic shock (Demaret et al., 2016) and in severe influenza (Tang et al., 2019). Clinically, neutrophil chemotaxis, infiltration of endothelial cells, and extravasation into alveolar spaces have been described in lung autopsies from deceased patients with COVID-19 (Fu et al., 2020). Elevated CD177 mRNA expression has also been described for patients with acute Kawasaki disease (Huang et al., 2019) and resistant to IV Ig therapy (Jing et al., 2020; Ko et al., 2019), a syndrome that has been described as a possible complication of SARS-CoV-2 infection in children (Toubiana et al., 2020; Viner and Whittaker, 2020). Our results are also consistent with those obtained using animal models, suggesting an important role for neutrophil activation in the severity of infection with respiratory viruses through their migration toward infected lungs, and in humans infected with influenza (Brandes et al., 2013; Narasaraju et al., 2011; Zaas et al., 2009).
The neutrophil activation signature is a specific feature of the homing of activated neutrophils toward infected lung tissue in acute lung injury (Juss et al., 2016), followed by the initiation of aggressive responses and the release of neutrophil extracellular traps (NETs), leading to an oxidative burst and the initiation of thrombus formation (Darbousset et al., 2012). Previous studies have reported elevated levels of circulating NETs in COVID-19 (Barnes et al., 2020a). Consistent with this finding and extending these data, we showed the differential expression of NET-related genes (Brandes et al., 2013; Narasaraju et al., 2011; Tang et al., 2019) (S100A8, S100A9, and S100A12), confirming the recently described elevated expression of calprotectin (heterodimer of S100A8 and A9) in patients with severe COVID-19 (Silvin et al., 2020). The association of neutrophil activation signature with COVID-19 severity has also been described recently with CD177 gene being one of the most differentially expressed gene in advanced disease (Aschenbrenner et al., 2021; Schulte-Schrepping et al., 2020). Likely, we believe that our data extended significantly these recent observations showing that CD177 is increased both at the level of coding RNA and at the protein level. Moreover, we show also that CD177 is not only a marker of severity but also of death as revealed by the longitudinal analysis which was confirmed in a second cohort.
Although, it is difficult to formally conclude whether CD177 is a causal factor of disease progression or a consequence of the severity of the disease, our data strongly show that CD177 is a valid hallmark of the physiopathology of COVID-19. This observation suggests that the activation of neutrophils, triggered by the infection, is a fundamental element of the innate response. However, persistent activation of this pathway may constitute, along with other factors (e.g., “cytokine storm”), fatal harm possibly associated with the critical turning point in the clinical trajectory of patients during the second week of the infection.
Neutrophil activation was associated with significant changes in the level of gene expression of several pathways, some concordantly associated with disturbances in immune-cell populations and cytokine and inflammatory profiles. We reveal a complex picture of the activation of innate immunity, assessed by significant changes in the expression of several genes involved in IFN signaling and the response to stress and the production of inflammatory/activation markers, with a balance between proinflammatory signals (increased expression of IL-1R1, IL-18R1, and its accessory chain IL-18 RAP) and anti-inflammatory cytokines or regulators (increased expression of IL-10, IL-4R, IL-27, IL-1RN) (Kim et al., 2005; Migliorini et al., 2020). The frequency of γδ T cells, a subpopulation of CD3+ T cells that were first described in the lung (Augustin et al., 1989) and that play critical roles in anti-viral immune responses, tissue healing, and epithelial cell maintenance (Cheng and Hu, 2017), was elevated and they expressed an activation marker (CD16) and low levels of the inhibitory receptor NKG2A, suggesting possible killing capacity. In accordance with our observation that eIF2 signaling is significantly inhibited in patients with COVID-19, recent studies have shown that coronaviruses encode ISR antagonists, which act as competitive inhibitors of eIF2 signaling (Rabouw et al., 2016, 2020). Similarly, the inhibition of mTOR signaling that we found in patients with COVID-19 is consistent with the impaired mTOR signaling reported in blood myeloid dendritic cells of patients with COVID-19 (Arunachalam et al., 2020). Interestingly, we observed a markedly smaller proportion of all DC (pre-DC, pDC, cDC1, and cDC2) and monocyte cell populations, including classical, intermediate, and nonclassical subpopulations, in patients with COVID-19 than in HDs. Based on these observations, it can be hypothesized that the impairment in IFN-α production observed in patients with severe COVID-19 may be the result of both a decrease in the number of pDCs, which are natural IFN-producing cells (Ali et al., 2019), and inhibition of mTOR signaling, a regulator of IFN-α production, in these cells (Kaur et al., 2012).
We confirmed the previously reported expansion of B cell populations (Arunachalam et al., 2020; Bouadma et al., 2020) in patients with COVID-19, and our results showed that the anti-SARS-CoV-2 B cell repertoire is commonly mobilized. The marked upregulation of IGV gene families included the IgHV1-24 family, described to be specific for COVID-19 (Brouwer et al., 2020). The expanded VH4-39 family was also recently reported to be the most highly represented in S-specific SARS-CoV-2 sequences (Brouwer et al., 2020). We also found enrichment of VH3-33, previously described in a set of clonally related anti-SARS-CoV-2 receptor-binding domain antibodies (Barnes et al., 2020b).
Globally, these results show that the defense against SARS-CoV-2 after pathogen recognition triggers a fine-tuned program that not only includes the production of antiviral (IFN signaling) and proinflammatory cytokines but also signals the cessation of the response and a strong disturbance of adaptive immunity.
The same pathways (immune and stress responses through eIF2 signaling, neutrophil and IFN signaling, T and B cell receptor responses, and mTOR pathways) contributed to the ability to discriminate between three groups of patients with severe COVID-19 in an unsupervised analysis. One limitation of our study was that we did not repeat the RNA-seq analyses in these specific groups of patients. Nonetheless, it is noteworthy that these groups differed significantly by the time from disease onset. These findings provide clues in our understanding of the wide range of profiles previously described for COVID-19 by showing that these patterns may be mainly related to time-dependent changes in the blood during the course of the infection (Laing et al., 2020; Ong et al., 2020). For example, the observed lower abundance of IFN signaling genes in the last group of patients with COVID-19 may be owing to decreased abundance in more advanced disease and/or patients who constitutively present a lower abundance of ISG, as described in previous studies (Bastard et al., 2020).
Several months after the emergence of this new disease, treatment options for patients with severe disease requiring hospitalization are still limited to corticosteroids, which has emerged as the treatment of choice for critically ill patients (Prescott and Rice, 2020; Sterne et al., 2020). However, interventions that can be administered early during the course of infection to prevent disease progression and longer-term complications are urgently needed. A major obstacle for the design of “adapted” therapies to the various stages of disease evolution is a lack of markers associated with sudden worsening of the disease of patients with moderate to severe disease and markers to predict improvement. Our results show that the measurement of CD177 during the course of the disease may be helpful in following the response to treatment and revision of the prognosis. In addition, they suggest that therapies aiming to control neutrophil activation and chemotaxis should be considered for the treatment of hospitalized patients.
Limitations of the study
HDs were collected from the French Blood Donors Organization (Etablissement Français du sang) before the COVID-19 outbreak. Age to donate blood is limited between 18 and 65 years (with the necessary medical agreement beyond 60 years). Owing to the age of hospitalized patients with COVID-19 (median: 61 years), it was not possible to have age-matched HDs. So we could not exclude that differences observed between HD and patients with COVID-19 could be associated to the age difference. Samples were not available for the entire cohort of 61 patients for every experimentation but clinical characteristics were not different between included and excluded patients for each assay, that is cell phenotype (all p > 0.97), seric markers (all p > 0.58) and gene expression (all p > 0.15). Finally, although we have identified a set of gene characterizing activation of a neutrophil pathway in patients, as well as an association with increased serum CD177, a marker highly specific of neutrophils, we did not study the phenotype of blood neutrophils from patients included in our cohorts for practical reasons.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse Monoclonal anti-CD38 FITC | BD Biosciences | #340909 |
| Mouse Monoclonal anti-HLADR PE | BD Biosciences | #347401 |
| Mouse Monoclonal anti-CD4 BV421 | BD Biosciences | #562424 |
| Mouse Monoclonal anti-CD8 APCH7 | BD Biosciences | #560179 |
| Mouse Monoclonal anti-CD3 Alexa 700 | BD Biosciences | #557943 |
| Rat Monoclonal anti-CCR7 Alexa647 | BD Biosciences | #557734 |
| Mouse Monoclonal anti-CD21 PE | BD Biosciences | #555422 |
| Mouse Monoclonal anti-CD27 APC | BD Biosciences | #337169 |
| Mouse Monoclonal anti-CD45 Alexa 700 | BD Biosciences | #560566 |
| Mouse Monoclonal anti-CD56 PECF594 | BD Biosciences | #564849 |
| Mouse Monoclonal anti-HLADR BV605 | BD Biosciences | #562845 |
| Mouse Monoclonal anti-CD33 BV421 | BD Biosciences | #562854 |
| Mouse Monoclonal anti-CD141 BV711 | BD Biosciences | #563155 |
| Mouse Monoclonal anti-CD45RA PerCpCy5.5 | BD Biosciences | #563429 |
| Mouse Monoclonal anti-HLA ABC BV786 | BD Biosciences | #740982 |
| Mouse Monoclonal anti-CD86 PECF594 | BD Biosciences | #562390 |
| Mouse Monoclonal anti-PD1 BV605 | BD Biosciences | #563245 |
| Mouse Monoclonal anti-TCR gamma delta | BD Biosciences | #559878 |
| Mouse Monoclonal anti-CD45RA PEefluor 610 | ebiosciences | #61-0458-42 |
| Mouse Monoclonal anti-Ki67 PercPe710 | ebiosciences | #46-5698-82 |
| Mouse Monoclonal anti-CD19 PC7 | Beckman Coulter | #IM3628 |
| Mouse Monoclonal anti-CD38 PercpCy5.5 | Biolegend | #303522 |
| Mouse Monoclonal anti-IgM Pacific Blue | Biolegend | #314514 |
| Mouse Monoclonal anti-CD16 APC Cy7 | Biolegend | #302018 |
| Mouse Monoclonal anti-CD14 BV605 | Biolegend | #301834 |
| Mouse Monoclonal anti-CD1c PECy7 | Biolegend | #331516 |
| Mouse Monoclonal anti-CD40 PE | Biolegend | #334308 |
| Mouse Monoclonal Lineage FITC | Biolegend | #348801 |
| Mouse Monoclonal anti-CD57 PercPCy5.5 | Biolegend | #359622 |
| F(ab')2-Goat anti-Human IgD FITC | Invitrogen | #H15501 |
| Mouse Monoclonal anti-CD123 APC | Miltenyi Biotec | #130-113-322 |
| Mouse Monoclonal anti-NKG2A PEVio770 | Miltenyi Biotec | #130-113-567 |
| BD Cytofix/cytoperm fixation/permeabilization kit | BD Biosciences | # 554714 |
| Biological Samples | ||
| French COVID-19 patients | French COVID cohort | clinicaltrials.gov NCT04262921 |
| Swiss COVID-19 patients | Swiss cohort | Swiss ethics protocol ID: 2020-00620 |
| Critical Commercial Assays | ||
| Human Magnetic Luminex Assay (CD163, ST2, and CD14, LBP) | R&D Systems | LXSAHM-2 kits |
| Human XL Cyt Disc Premixed Mag Luminex Perf Assay Kit |
R&D Systems | LXSAHM-19 kit |
| 48-Plex Bio-Plex Pro Human Cytokine | Bio-Rad | #12007283 |
| CD177 ELISA Kit | ThermoFisher Scientific | EH80RBX5 |
| Deposited Data | ||
| Raw and analyzed data | This paper | GEO code: GSE171110 |
| Software and Algorithms | ||
| DIVA v6.2 | BD Bioscences | https://www.bdbiosciences.com/en-us/instruments/research-instruments/research-software/flow-cytometry-acquisition/facsdiva-software |
| Bio-Plex Manager v6.1 | Biorad | https://www.bio-rad.com/fr-fr/product/bio-plex-manager-software-standard-edition?ID=5846e84e-03a7-4599-a8ae-7ba5dd2c7684 |
| hg19 human reference genome | This paper | https://www.ncbi.nlm.nih.gov/assembly/GCF_000001405.13/ |
| STAR - v. 2.5.3ar, and quantified relative to annotation model hg19 - GENCODE Genes - release 19 | N/A | [https://www.gencodegenes.org/human/release_19.html] |
| Sequence Analysis Viewer (SAV) version 2.1.8. | ||
| R (version 3.6) | The R Foundation for Statistical Computing, Vienna, Austria | https://www.r-project.org/ |
| FlowJo v9 | Treestar | https://www.flowjo.com/solutions/flowjo/downloads |
| SPICE v5.22 | https://doi.org/10.1002/cyto.a.21015 | (http://exon.niaid.nih.gov/spice) |
| Ingenuity Pathway software v.51963813. | Qiagen | Ingenuity Pathway Analysis (IPA) - QIAGEN Online Shop |
Resource availability
Lead contact
Further information and requests for resources should be directed to and will be fulfilled by the lead contact, Yves Lévy (yves.levy@aphp.fr).
Materials availability
No materials were newly generated for this paper.
Data and code availability
RNA sequencing data that support the findings of this study have been deposited in Gene Expression Omnibus repository with the accession codes GSE171110. Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact: Yves Lévy (yves.levy@aphp.fr)
Experimental model and subject details
Subjects
We enrolled a subgroup of patients with COVID-19 of the prospective French COVID cohort in this immunological study which is part of the cohort main objectives. The median age of patients with COVID-19 was 60 years [50-69], and 80% were men. Ethics approval was given on February 5 by the French Ethics Committee CPP-Ile-de-France VI (ID RCB: 2020-A00256-33). Eligible patients were those who were hospitalized with virologically confirmed COVID-19. Briefly, nasopharyngeal swabs were performed on the day of inclusion for SARS-CoV-2 testing as per the World Health Organization (WHO) or French National Health Agency guidelines. Viral loads were quantified by real-time semiquantitative reverse transcriptase polymerase chain reactions using either the Charité WHO protocol (testing the E gene and RdRp) or the Pasteur institute assay (testing the E gene and two other RdRp targets, IP2 and IP4). The study was conducted with the understanding and the consent of each participant or its surrogate covering the sampling, storage, and use of biological samples. The time from symptom onset to the admission has been retrospectively collected by the interview of patients enrolled in the national “French COVID-19 cohort.” The Swiss cohort was approved by the ethical commission (CER-VD; Swiss ethics protocol ID: 2020-00620) and all subjects provided written informed consent. Blood from healthy donors (HDs) was collected from the French Blood Donors Organization (Etablissement Français du sang) before the COVID-19 outbreak. HD characteristics are shown in Table S3.
Method details
Quantification of serum analytes
In total, 71 analytes were quantified in heat-inactivated serum samples by multiplex magnetic bead assays or ELISA. Serum samples from five healthy donors were also assayed as controls. The following kits were used as per the manufacturers’ recommendations: LXSAHM-2 kits for CD163,ST2,CD14 and LBP (R&D Systems); the LXSAHM-19 kit for IL-21, IL-23, IL-31, EGF, Flt-3 Ligand, Granzyme B, Granzyme A, IL-25, PD-L1/B7-H1, TGF-α, Aggrecan, 4-1BB/CD137, Fas, FasL,CCL-28, Chemerin, sCD40L, CXCL14, and Midkine (R&D Systems); and the 48-Plex Bio-Plex Pro Human Cytokine screening kit for IL-1β, IL-1rα, IL-2, IL-4, IL-5, IL-6, IL-7, IL-8 / CXCL8, IL-9, IL-10, IL-12 (p70), IL-13, IL-15, IL-17A / CTLA8, Basic FGF (FGF-2), Eotaxin / CCL11, G-CSF, GM-CSF, IFN-γ, IP-10/CXCL10, MCP-1 /CCL2, MIP-1α / CCL3, MIP-1β / CCL4, PDGF-BB (PDGF-AB/BB), RANTES/CCL5, TNF-α, VEFG (VEGF-A), IL-1a, IL-2Ra (IL-2R), IL-3, IL-12 (p40), IL-16, IL-18, CTACK / CCL27, GRO-a /CXCL1 (GRO), HGF, IFN-α2, LIF, MCP-3 / CCL7, M-CSF, MIF, MIG/CXCL9, b-NGF, SCF, SCGF-b ,SDF-1α ,TNF-b/LTA, and TRAIL (Bio-Rad). The data were acquired using a Bio-Plex 200 system. Extrapolated concentrations were used and the out-of-range values were entered at the highest or lowest extrapolated concentration. Values were standardized for each cytokine across all displayed samples (centered around the observed mean, with variance equal to 1). CD177 quantification was performed on non-inactivated serum samples (diluted 1:2 or 1:10) using a Human CD177 ELISA Kit (ThermoFisher Scientific), according to the manufacturer’s instructions.
Cell phenotyping
Immune-cell phenotyping was performed using an LSR Fortessa 4-laser (488, 640, 561, and 405 nm) flow cytometer (BD Biosciences) and Diva software, version 6.2. FlowJo software, version 9.9.6 (Tree Star Inc.), was used for data analysis. CD4+ and CD8+ T cells were analyzed for CD45RA and CCR7 expression to identify the naive, memory, and effector cell subsets for coexpression of activation (HLA-DR and CD38) and exhaustion/senescence (CD57and PD1) markers. CD19+ B cell subsets were analyzed for the markers CD21 and CD27. ASC (plasmablasts) were identified as CD19+ cells expressing CD38 and CD27. We used CD16, CD56, and CD57 to identify NK cell subsets. γδ T cells were identified using an anti-TCR γδ antibody. HLA-DR, CD33, CD45RA, CD123, CD141, and CD1c were used to identify dendritic cell subsets, as previously described (See et al., 2017). Extracellular labeling was performed for all antibodies except for Ki 67 for which an intracellular labeling was performed with the BD cytofix/cytoperm kit (BD Biosciences).
RNA sequencing
Total RNA was purified from whole blood using the Tempus™ Spin RNA Isolation Kit (ThermoFisher Scientific). RNA was quantified using the Quant-iT RiboGreen RNA Assay Kit (Thermo Fisher Scientific) and quality control performed on a Bioanalyzer (Agilent). Globin mRNA was depleted using GLOBINclear Kit (Invitrogen) before mRNA library preparation with the TruSeq® Stranded mRNA Kit, as per the Illumina protocol. Libraries were sequenced on an Illumina HiSeq 2500 V4 system. Sequencing quality control was performed using Sequence Analysis Viewer. FastQ files were generated on the Illumina BaseSpace Sequence Hub. Transcript reads were aligned to the hg18 human reference genome using Salmon v0.8.2 (Patro et al., 2017) and quantified relative to annotation model "hsapiens_gene_ensembl" recovered from the R package biomaRt v2.42.1 (Durinck et al., 2009). Quality control of the alignment was performed via MultiQC v1.4 (Patro et al., 2017). Finally, counts were normalized as counts per million.
Quantification and statistical analysis
Subgroups of patients with COVID-19 were identified from unsupervised hierarchical clustering of log2-counts-per-million RNA-seq transcriptomics from whole blood using the Euclidean distance and Ward’s method. Differential expression analysis was carried out using dearseq (Gauthier et al., 2019) to contribute to the analysis of genes of which the abundance differed across the three subgroups of patient with COVID-19 and healthy subjects. Once the groups were defined by hierarchical clustering, the analysis of the genes contributing to each group was performed by selecting genes with an absolute fold change of ≥ 1.5 in the comparison of interest for which the difference in expression between HDs and patients with COVID-19 was significant (P ≤ 0.05) (to avoid so called “double dipping” [https://arxiv.org/abs/2012.02936]). Pathway analyses of the genes involved in each comparison was performed using Ingenuity Pathway Analysis (IPA ®, Qiagen, Redwood City, California, Version 57662101, 2020). For canonical pathway analysis, a Z-score ≥ 2 was defined as the threshold for significant activation, whereas a Z-score ≤ −2 was defined as the threshold for significant inhibition.
The integrative analysis of the three types of biological data (RNA-seq, cell phenotypes, serum analytes) was performed using MOFA+ (Argelaguet et al., 2019), a sparse factor analysis method. It provides latent variables which are linear combination of the most influential factors for explaining interpatient variability across the three biological measurement modalities. The first component is presented and called integrative score here. The analyses of factors associated with CD177 protein concentration were performed using nonparametric Wilcoxon test or Spearman correlation coefficient when appropriate. To look at the independent association of CD177 with ICU, a logistic regression for the prediction of hospitalization in the ICU adjusted for age, sex, chronic cardiac disease, chronic pulmonary disease, and diabetes was fitted. The analysis of repeated measurements of CD177 over time was performed by using a linear mixed effect model adjusted for time from hospitalization and an interaction with survival outcome (death or recovery). The model included a random intercept and a random slope with an unstructured matrix for variance parameters. Predictions of marginal trajectories were performed. All analyses, if not stated otherwise, were performed using R software, version 3.6.3. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL: https://www.R-project.org/
Additional ressources
A subgroup of patients with COVID-19 of the prospective French COVID cohort was enrolled in this study. French COVID cohort was registered at: https://clinicaltrials.gov/ct2/show/NCT04262921
Acknowledgments
We thank the patients who donated their blood. We thank F. Mentre, S. Tubiana, the French COVID cohort, and REACTing (REsearch & ACtion emergING infectious diseases) for cohort management. We thank the scientific advisory board of the French COVID-19 cohort composed of Dominique Costagliola, Astrid Vabret, Hervé Raoul, and Laurence Weiss. We thank Romain Lévy for fruitful discussions. This work was supported by INSERM and the Investissements d’Avenir program, Vaccine Research Institute (VRI), managed by the ANR under reference ANR-10-LABX-77-01 and the CARE project funded from the Innovative Medicines Initiative 2 Joint Undertaking (JU) under grant agreement No 101005077. The JU receives support from the European Union’s Horizon 2020 research and innovation programme and EFPIA and Bill & Melinda Gates Foundation, Global Health Drug Discovery Institute, University of Dundee. The French COVID Cohort is funded through the Ministry of Health and Social Affairs and Ministry of Higher Education and Research dedicated COVID19 fund and PHRC n°20–0424 and the REACTing consortium. Funding sources were not involved in the study design, data acquisition, data analysis, data interpretation, or writing of the manuscript.
Author contributions
YL and AW conceived and designed the study. MC, JFT, YY, LB, DB, CLa, GP, and MP participated in sample and clinical data collection. EF, MDe, MS, CLe, and PT performed the experiments. MD, MG, BH, and CLe analyzed the data. YL, RT, AW, HH, MS, BJH, and CL analyzed and interpreted the data. YL, RT, AW, and HH drafted the first version and wrote the final version of the manuscript. All authors approved the final version.
Declaration of interests
None of the authors has any conflict of interest to declare.
Published: July 23, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.102711.
Contributor Information
Yves Lévy, Email: yves.levy@aphp.fr.
Rodolphe Thiébaut, Email: rodolphe.thiebaut@u-bordeaux.fr.
Supplemental information
References
- Ali S., Mann-Nüttel R., Schulze A., Richter L., Alferink J., Scheu S. Sources of type I interferons in infectious immunity: plasmacytoid dendritic cells not always in the driver's seat. Front. Immunol. 2019;10:778. doi: 10.3389/fimmu.2019.00778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argelaguet R., Clark S.J., Mohammed H., Stapel L.C., Krueger C., Kapourani C.A., Imaz-Rosshandler I., Lohoff T., Xiang Y.L., Hanna C.W. Multi-omics profiling of mouse gastrulation at single-cell resolution. Nature. 2019;576:487. doi: 10.1038/s41586-019-1825-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arunachalam P.S., Wimmers F., Mok C.K.P., Perera R., Scott M., Hagan T., Sigal N., Feng Y., Bristow L., Tak-Yin Tsang O. Systems biological assessment of immunity to mild versus severe COVID-19 infection in humans. Science. 2020;369:1210–1220. doi: 10.1126/science.abc6261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aschenbrenner A.C., Mouktaroudi M., Krämer B., Oestreich M., Antonakos N., Nuesch-Germano M., Gkizeli K., Bonaguro L., Reusch N., Baßler K. Disease severity-specific neutrophil signatures in blood transcriptomes stratify COVID-19 patients. Genome Med. 2021;13:7. doi: 10.1186/s13073-020-00823-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augustin A., Kubo R.T., Sim G.K. Resident pulmonary lymphocytes expressing the gamma/delta T-cell receptor. Nature. 1989;340:239–241. doi: 10.1038/340239a0. [DOI] [PubMed] [Google Scholar]
- Barnes B.J., Adrover J.M., Baxter-Stoltzfus A., Borczuk A., Cools-Lartigue J., Crawford J.M., Daßler-Plenker J., Guerci P., Huynh C., Knight J.S. Targeting potential drivers of COVID-19: neutrophil extracellular traps. J. Exp. Med. 2020;217:e20200652. doi: 10.1084/jem.20200652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes C.O., West A.P., Jr., Huey-Tubman K.E., Hoffmann M.A.G., Sharaf N.G., Hoffman P.R., Koranda N., Gristick H.B., Gaebler C., Muecksch F. Structures of human antibodies bound to SARS-CoV-2 spike reveal common epitopes and recurrent features of antibodies. Cell. 2020;182:828–842 e816. doi: 10.1016/j.cell.2020.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastard P., Rosen L.B., Zhang Q., Michailidis E., Hoffmann H.-H., Zhang Y., Dorgham K., Philippot Q., Rosain J., Béziat V. Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science. 2020;370:eabd4585. doi: 10.1126/science.abd4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayat B., Werth S., Sachs U.J.H., Newman D.K., Newman P.J., Santoso S. Neutrophil transmigration mediated by the neutrophil-specific antigen CD177 is influenced by the endothelial S536N dimorphism of platelet endothelial cell adhesion molecule-1. J. Immunol. 2010;184:3889–3896. doi: 10.4049/jimmunol.0903136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco-Melo D., Nilsson-Payant B.E., Liu W.C., Uhl S., Hoagland D., Moller R., Jordan T.X., Oishi K., Panis M., Sachs D. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020;181:1036–1045 e1039. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouadma L., Wiedemann A., Patrier J., Surenaud M., Wicky P.H., Foucat E., Diehl J.L., Hejblum B.P., Sinnah F., de Montmollin E. Immune alterations in a patient with SARS-CoV-2-related acute respiratory distress syndrome. J. Clin. Immunol. 2020;40:1082–1092. doi: 10.1007/s10875-020-00839-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd J.H., Fjell C.D., Russell J.A., Sirounis D., Cirstea M.S., Walley K.R. Increased plasma PCSK9 levels are associated with reduced endotoxin clearance and the development of acute organ failures during sepsis. J. Innate Immun. 2016;8:211–220. doi: 10.1159/000442976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandes M., Klauschen F., Kuchen S., Germain R.N. A systems analysis identifies a feedforward inflammatory circuit leading to lethal influenza infection. Cell. 2013;154:197–212. doi: 10.1016/j.cell.2013.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouwer P.J.M., Caniels T.G., van der Straten K., Snitselaar J.L., Aldon Y., Bangaru S., Torres J.L., Okba N.M.A., Claireaux M., Kerster G. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science. 2020;369:643–650. doi: 10.1126/science.abc5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buechler C., Eisinger K., Krautbauer S. Diagnostic and prognostic potential of the macrophage specific receptor CD163 in inflammatory diseases. Inflamm. Allergy Drug Targets. 2013;12:391–402. doi: 10.2174/18715281113126660060. [DOI] [PubMed] [Google Scholar]
- Cai Y.Q., Lv Y., Mo Z.C., Lei J., Zhu J.L., Zhong Q.Q. Multiple pathophysiological roles of midkine in human disease. Cytokine. 2020;135:155242. doi: 10.1016/j.cyto.2020.155242. [DOI] [PubMed] [Google Scholar]
- Chen G., Wu D., Guo W., Cao Y., Huang D., Wang H., Wang T., Zhang X., Chen H., Yu H. Clinical and immunological features of severe and moderate coronavirus disease 2019. J. Clin. Invest. 2020;130:2620–2629. doi: 10.1172/JCI137244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Qiu Y., Wang J., Liu Y., Wei Y. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng M., Hu S. Lung-resident gammadelta T cells and their roles in lung diseases. Immunology. 2017;151:375–384. doi: 10.1111/imm.12764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coronaviridae Study Group of the International Committee on Taxonomy of V. The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darbousset R., Thomas G.M., Mezouar S., Frère C., Bonier R., Mackman N., Renné T., Dignat-George F., Dubois C., Panicot-Dubois L. Tissue factor–positive neutrophils bind to injured endothelial wall and initiate thrombus formation. Blood. 2012;120:2133–2143. doi: 10.1182/blood-2012-06-437772. [DOI] [PubMed] [Google Scholar]
- De Biasi S., Meschiari M., Gibellini L., Bellinazzi C., Borella R., Fidanza L., Gozzi L., Iannone A., Lo Tartaro D., Mattioli M. Marked T cell activation, senescence, exhaustion and skewing towards TH17 in patients with COVID-19 pneumonia. Nat. Commun. 2020;11:3434. doi: 10.1038/s41467-020-17292-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demaret J., Venet F., Plassais J., Cazalis M.-A., Vallin H., Friggeri A., Lepape A., Rimmelé T., Textoris J., Monneret G. Identification of CD177 as the most dysregulated parameter in a microarray study of purified neutrophils from septic shock patients. Immunol. Lett. 2016;178:122–130. doi: 10.1016/j.imlet.2016.08.011. [DOI] [PubMed] [Google Scholar]
- Durinck S., Spellman P.T., Birney E., Huber W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat. Protoc. 2009;4:1184–1191. doi: 10.1038/nprot.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwivedi D.J., Grin P.M., Khan M., Prat A., Zhou J., Fox-Robichaud A.E., Seidah N.G., Liaw P.C. Differential expression of PCSK9 modulates infection, inflammation, and coagulation in a murine model of sepsis. Shock. 2016;46:672–680. doi: 10.1097/SHK.0000000000000682. [DOI] [PubMed] [Google Scholar]
- Fu J., Kong J., Wang W., Wu M., Yao L., Wang Z., Jin J., Wu D., Yu X. The clinical implication of dynamic neutrophil to lymphocyte ratio and D-dimer in COVID-19: a retrospective study in Suzhou China. Thromb. Res. 2020;192:3–8. doi: 10.1016/j.thromres.2020.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan E.S., Tan H.C., Duyen H.L.T., Trieu H.T., Wills B., Ooi E.E., Seidah N.G., Yacoub S. Dengue virus induces PCSK9 expression to alter antiviral responses and disease outcomes. J. Clin. Invest. 2020;130:5223–5234. doi: 10.1172/JCI137536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauthier M., Agniel D., Thiébaut R., Hejblum B.P. dearseq: a variance component score test for RNA-Seq differential analysis that effectively controls the false discovery rate. NAR Genom. Bioinform. 2019;2:lqaa093. doi: 10.1093/nargab/lqaa093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giamarellos-Bourboulis E.J., Netea M.G., Rovina N., Akinosoglou K., Antoniadou A., Antonakos N., Damoraki G., Gkavogianni T., Adami M.E., Katsaounou P. Complex immune dysregulation in COVID-19 patients with severe respiratory failure. Cell Host Microbe. 2020;27:992–1000 e1003. doi: 10.1016/j.chom.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjadj J., Yatim N., Barnabei L., Corneau A., Boussier J., Smith N., Pere H., Charbit B., Bondet V., Chenevier-Gobeaux C. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science. 2020;369:718–724. doi: 10.1126/science.abc6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W.D., Lin Y.T., Tsai Z.Y., Chang L.S., Liu S.F., Lin Y.J., Kuo H.C. Association between maternal age and outcomes in Kawasaki disease patients. Pediatr. Rheumatol. 2019;17:46. doi: 10.1186/s12969-019-0348-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing Y., Ding M., Fu J., Xiao Y., Chen X., Zhang Q. Neutrophil extracellular trap from Kawasaki disease alter the biologic responses of PBMC. Biosci. Rep. 2020;40 doi: 10.1042/BSR20200928. BSR20200928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juss J.K., House D., Amour A., Begg M., Herre J., Storisteanu D.M.L., Hoenderdos K., Bradley G., Lennon M., Summers C. Acute respiratory distress syndrome neutrophils have a distinct phenotype and are resistant to phosphoinositide 3-kinase inhibition. Am. J. Resp Crit. Care. 2016;194:961–973. doi: 10.1164/rccm.201509-1818OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur S., Sassano A., Majchrzak-Kita B., Baker D.P., Su B., Fish E.N., Platanias L.C. Regulatory effects of mTORC2 complexes in type I IFN signaling and in the generation of IFN responses. Proc. Natl. Acad. Sci. 2012;109:7723–7728. doi: 10.1073/pnas.1118122109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.H., Han S.Y., Azam T., Yoon D.Y., Dinarello C.A. Interleukin-32: a cytokine and inducer of TNFalpha. Immunity. 2005;22:131–142. doi: 10.1016/j.immuni.2004.12.003. [DOI] [PubMed] [Google Scholar]
- Ko T.M., Chang J.S., Chen S.P., Liu Y.M., Chang C.J., Tsai F.J., Lee Y.C., Chen C.H., Chen Y.T., Wu J.Y. Genome-wide transcriptome analysis to further understand neutrophil activation and lncRNA transcript profiles in Kawasaki disease. Sci. Rep. 2019;9:328. doi: 10.1038/s41598-018-36520-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuri-Cervantes L., Pampena M.B., Meng W., Rosenfeld A.M., Ittner C.A.G., Weisman A.R., Agyekum R., Mathew D., Baxter A.E., Vella L. bioRxiv; 2020. Immunologic Perturbations in Severe COVID-19/sars-CoV-2 Infection. [Google Scholar]
- Laing A.G., Lorenc A., Del Molino Del Barrio I., Das A., Fish M., Monin L., Munoz-Ruiz M., McKenzie D.R., Hayday T.S., Francos-Quijorna I. A dynamic COVID-19 immune signature includes associations with poor prognosis. Nat. Med. 2020;26:1623–1635. doi: 10.1038/s41591-020-1038-6. [DOI] [PubMed] [Google Scholar]
- Le Gall J.R. A new Simplified Acute Physiology Score (SAPS II) based on a European/North American multicenter study. JAMA. 1993;270:2957–2963. doi: 10.1001/jama.270.24.2957. [DOI] [PubMed] [Google Scholar]
- Levin D., London I.M. Regulation of protein synthesis: activation by double-stranded RNA of a protein kinase that phosphorylates eukaryotic initiation factor 2. Proc. Natl. Acad. Sci. U S A. 1978;75:1121–1125. doi: 10.1073/pnas.75.3.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas C., Wong P., Klein J., Castro T.B.R., Silva J., Sundaram M., Ellingson M.K., Mao T., Oh J.E., Israelow B. Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature. 2020;584:463–469. doi: 10.1038/s41586-020-2588-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew D., Giles J.R., Baxter A.E., Oldridge D.A., Greenplate A.R., Wu J.E., Alanio C., Kuri-Cervantes L., Pampena M.B., D’Andrea K. Deep immune profiling of COVID-19 patients reveals distinct immunotypes with therapeutic implications. Science. 2020;369:eabc8511. doi: 10.1126/science.abc8511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta P., McAuley D.F., Brown M., Sanchez E., Tattersall R.S., Manson J.J. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet. 2020;395:1033–1034. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migliorini P., Italiani P., Pratesi F., Puxeddu I., Boraschi D. The IL-1 family cytokines and receptors in autoimmune diseases. Autoimmun. Rev. 2020;19:102617. doi: 10.1016/j.autrev.2020.102617. [DOI] [PubMed] [Google Scholar]
- Narasaraju T., Yang E., Samy R.P., Ng H.H., Poh W.P., Liew A.A., Phoon M.C., van Rooijen N., Chow V.T. Excessive neutrophils and neutrophil extracellular traps contribute to acute lung injury of influenza pneumonitis. Am. J. Pathol. 2011;179:199–210. doi: 10.1016/j.ajpath.2011.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong E.Z., Chan Y.F.Z., Leong W.Y., Lee N.M.Y., Kalimuddin S., Haja Mohideen S.M., Chan K.S., Tan A.T., Bertoletti A., Ooi E.E. A dynamic immune response shapes COVID-19 progression. Cell Host Microbe. 2020;27:879–882 e872. doi: 10.1016/j.chom.2020.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pakos-Zebrucka K., Koryga I., Mnich K., Ljujic M., Samali A., Gorman A.M. The integrated stress response. EMBO Rep. 2016;17:1374–1395. doi: 10.15252/embr.201642195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patro R., Duggal G., Love M.I., Irizarry R.A., Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods. 2017;14:417–419. doi: 10.1038/nmeth.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelan A.L., Katz R., Gostin L.O. The novel coronavirus originating in wuhan, China: challenges for global Health governance. JAMA. 2020;323:709–710. doi: 10.1001/jama.2020.1097. [DOI] [PubMed] [Google Scholar]
- Ponti G., Maccaferri M., Ruini C., Tomasi A., Ozben T. Biomarkers associated with COVID-19 disease progression. Crit. Rev. Clin. Lab. Sci. 2020;57:389–399. doi: 10.1080/10408363.2020.1770685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott H.C., Rice T.W. Corticosteroids in COVID-19 ARDS. JAMA. 2020;324:1292. doi: 10.1001/jama.2020.16747. [DOI] [PubMed] [Google Scholar]
- Qin C., Zhou L., Hu Z., Zhang S., Yang S., Tao Y., Xie C., Ma K., Shang K., Wang W. Dysregulation of immune response in patients with COVID-19 in Wuhan, China. Clin. Infect. Dis. 2020;71:762–768. doi: 10.1093/cid/ciaa248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabouw H.H., Langereis M.A., Knaap R.C., Dalebout T.J., Canton J., Sola I., Enjuanes L., Bredenbeek P.J., Kikkert M., de Groot R.J. Middle East respiratory coronavirus accessory protein 4a inhibits PKR-mediated antiviral stress responses. PLoS Pathog. 2016;12:e1005982. doi: 10.1371/journal.ppat.1005982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabouw H.H., Visser L.J., Passchier T.C., Langereis M.A., Liu F., Giansanti P., van Vliet A.L.W., Dekker J.G., van der Grein S.G., Saucedo J.G. Inhibition of the integrated stress response by viral proteins that block p-eIF2-eIF2B association. Nat. Microbiol. 2020;5:1361–1373. doi: 10.1038/s41564-020-0759-0. [DOI] [PubMed] [Google Scholar]
- Roth J., Vogl T., Sorg C., Sunderkötter C. Phagocyte-specific S100 proteins: a novel group of proinflammatory molecules. Trends Immunol. 2003;24:155–158. doi: 10.1016/s1471-4906(03)00062-0. [DOI] [PubMed] [Google Scholar]
- Schulte-Schrepping J., Reusch N., Paclik D., Baßler K., Schlickeiser S., Zhang B., Krämer B., Krammer T., Brumhard S., Bonaguro L. Severe COVID-19 is marked by a dysregulated myeloid cell compartment. Cell. 2020;182:1419–1440.e1423. doi: 10.1016/j.cell.2020.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- See P., Dutertre C.A., Chen J., Gunther P., McGovern N., Irac S.E., Gunawan M., Beyer M., Handler K., Duan K. Mapping the human DC lineage through the integration of high-dimensional techniques. Science. 2017;356:eaag3009. doi: 10.1126/science.aag3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen-Orr S.S., Tibshirani R., Khatri P., Bodian D.L., Staedtler F., Perry N.M., Hastie T., Sarwal M.M., Davis M.M., Butte A.J. Cell type–specific gene expression differences in complex tissues. Nat. Methods. 2010;7:287–289. doi: 10.1038/nmeth.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvin A., Chapuis N., Dunsmore G., Goubet A.G., Dubuisson A., Derosa L., Almire C., Henon C., Kosmider O., Droin N. Elevated Calprotectin and abnormal myeloid cell subsets discriminate severe from mild COVID-19. Cell. 2020;182:1401–1418.e18. doi: 10.1016/j.cell.2020.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterne J.A.C., Murthy S., Diaz J.V., Slutsky A.S., Villar J., Angus D.C., Annane D., Azevedo L.C.P., Berwanger O., Cavalcanti A.B. Association between administration of systemic corticosteroids and mortality among critically ill patients with COVID-19. JAMA. 2020;324:1330. doi: 10.1001/jama.2020.17023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang B.M., Shojaei M., Teoh S., Meyers A., Ho J., Ball T.B., Keynan Y., Pisipati A., Kumar A., Eisen D.P. Neutrophils-related host factors associated with severe disease and fatality in patients with influenza infection. Nat. Commun. 2019;10:3422. doi: 10.1038/s41467-019-11249-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theilgaard-Monch K. Haptoglobin is synthesized during granulocyte differentiation, stored in specific granules, and released by neutrophils in response to activation. Blood. 2006;108:353–361. doi: 10.1182/blood-2005-09-3890. [DOI] [PubMed] [Google Scholar]
- Toubiana J., Poirault C., Corsia A., Bajolle F., Fourgeaud J., Angoulvant F., Debray A., Basmaci R., Salvador E., Biscardi S. Kawasaki-like multisystem inflammatory syndrome in children during the covid-19 pandemic in Paris, France: prospective observational study. BMJ. 2020;369:m2094. doi: 10.1136/bmj.m2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent J.-L., de Mendonca A., Cantraine F., Moreno R., Takala J., Suter P.M., Sprung C.L., Colardyn F., Blecher S. Use of the SOFA score to assess the incidence of organ dysfunction/failure in intensive care units. Crit. Care Med. 1998;26:1793–1800. doi: 10.1097/00003246-199811000-00016. [DOI] [PubMed] [Google Scholar]
- Viner R.M., Whittaker E. Kawasaki-like disease: emerging complication during the COVID-19 pandemic. Lancet. 2020;395:1741–1743. doi: 10.1016/S0140-6736(20)31129-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilk A.J., Rustagi A., Zhao N.Q., Roque J., Martinez-Colon G.J., McKechnie J.L., Ivison G.T., Ranganath T., Vergara R., Hollis T. A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nat. Med. 2020;26:1070–1076. doi: 10.1038/s41591-020-0944-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z.Y., McGoogan J.M. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China summary of a report of 72 314 cases from the Chinese center for disease control and prevention. JAMA. 2020;323:1239–1242. doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- Xu B., Fan C.Y., Wang A.L., Zou Y.L., Yu Y.H., He C., Xia W.G., Zhang J.X., Miao Q. Suppressed T cell-mediated immunity in patients with COVID-19: a clinical retrospective study in Wuhan, China. J. Infect. 2020;81:e51–e60. doi: 10.1016/j.jinf.2020.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazdanpanah Y. Impact on disease mortality of clinical, biological, and virological characteristics at hospital admission and overtime in COVID-19 patients. J. Med. Virol. 2020;93:2149–2159. doi: 10.1002/jmv.26601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaas A.K., Chen M., Varkey J., Veldman T., Hero A.O., Lucas J., Huang Y., Turner R., Gilbert A., Lambkin-Williams R. Gene expression signatures diagnose influenza and other symptomatic respiratory viral infections in humans. Cell Host Microbe. 2009;6:207–217. doi: 10.1016/j.chom.2009.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhi Y., Gao P., Xin X., Li W., Ji L., Zhang L., Zhang X., Zhang J. Clinical significance of sCD163 and its possible role in asthma (Review) Mol. Med. Rep. 2017;15:2931–2939. doi: 10.3892/mmr.2017.6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z., Ren L., Zhang L., Zhong J., Xiao Y., Jia Z., Guo L., Yang J., Wang C., Jiang S. Heightened innate immune responses in the respiratory tract of COVID-19 patients. Cell Host Microbe. 2020;27:883–890 e882. doi: 10.1016/j.chom.2020.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA sequencing data that support the findings of this study have been deposited in Gene Expression Omnibus repository with the accession codes GSE171110. Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact: Yves Lévy (yves.levy@aphp.fr)