Abstract
Coronaviruses (CoVs) are a large family of viruses responsible for the severe pathophysiological effects on human health. The most severe outbreak includes Severe Acute Respiratory Syndrome (SARS-CoV), Middle East Respiratory Syndrome (MERS-CoV) and Coronavirus disease 2019 (COVID-19) caused by Severe Acute Respiratory Syndrome coronavirus 2 (SARS-CoV-2). The COVID-19 poses major challenges to clinical management because no specific FDA-approved therapy yet to be available. Thus, the existing therapies are being used for the treatment of COVID-19, which are under clinical trials and compassionate use, based on in vitro and in silico studies. In this review, we summarize the potential therapies utilizing small molecules, bioactive compounds, nucleoside and nucleotide analogs, peptides, antibodies, natural products, and synthetic compounds targeting the complex molecular signaling network involved in COVID-19. In this review>230 natural and chemically synthesized drug therapies are described with their recent advances in research and development being done in terms of their chemical, structural and functional properties. This review focuses on possible targets for viral cells, viral proteins, viral replication, and different molecular pathways for the discovery of novel viral- and host-based therapeutic targets against SARS-CoV-2.
Keywords: Therapeutic drugs, Natural compounds, Viral inhibitors, SARS-CoV-2, COVID-19
Abbreviations: COVID-19, Coronavirus disease 2019; SARS-CoV, severe acute respiratory syndrome coronavirus; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; MERS, Middle East respiratory syndrome; HCoV, human coronavirus; CHIKV, Chikungunya virus; DHODH, dihydroorotate dehydrogenase; HBV, hepatitis B virus; IAV, influenza A virus; HCV, hepatitis C virus; JEV, Japanese encephalitis virus; PEDV, porcine epidemic diarrhea virus; PLpro, papain-like protease; 3CLpro, 3 chymotrypsin-like proteases; RdRp, RNA-dependent RNA polymerase; SAH, S-adenosyl-l-homocysteine; RBD, receptor-binding domain; RSV, respiratory syncytial virus; ZIKV, Zika virus; IMPDH, inosine-monophosphate dehydrogenase; PPIase, peptidyl-prolyl isomerase; IMPTH, inosine-5′-monophosphate dehydrogenase; NS3, non-structural protein 3; VEGF, Vascular Endothelial Growth Factor; ALI, acute lung injury; ARDS, acute respiratory distress syndrome; HTCC, N-(2hydroxypropyl)-3-trimethylammoniumchitosan chloride; SCV, SARS-associated coronavirus; HCMV, Human cytomegalovirus; COX, cyclooxygenase; JAK, Janus-associated kinase; NAK, Numb-associated kinase; HCMV, Human Cytomegalovirus; NS, Not studied; S protein, Spike (S) protein; E, Enveloped protein; ACE2, angiotensin-converting enzyme 2 (ACE2) blockers; TCM, traditional Chinese medicine
Graphical abstract
1. Introduction
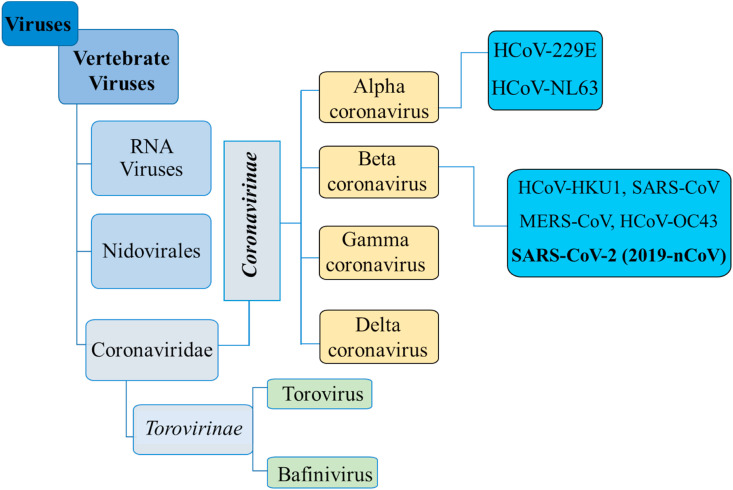
Coronaviruses (CoVs) are enveloped viruses having non-segmented, positive sense single-stranded RNA genome rather than DNA, belonging to the family Coronaviridae and contain the largest genomic RNA among any viruses broadly distributed in humans and other mammals (Pillaiyar et al., 2020; Zumla et al., 2016). CoVs are named from crown-like spikes protruding from their outer surface and grouped in four main sub-groups, mainly alpha, beta, gamma, and delta (Sheahan et al., 2020a, 2020b, 2020b). CoVs were first identified in the mid-1960s, seven of which infect human beings. These are MERS-CoV (the beta coronavirus that causes Middle East Respiratory Syndrome, or MERS), SARS-CoV (the beta coronavirus that causes severe acute respiratory syndrome, or SARS), NL63 (alpha coronavirus), 229E (alpha coronavirus), OC43 (beta coronavirus), HKU1 (beta coronavirus), and severe acute respiratory syndrome-related coronavirus (SARS-CoV-2, novel coronavirus responsible for COVID-19). People around the world commonly get infected by human CoVs like HCoV-229E, HCoV-NL63, HCoV-OC43, and HCoV-HKU1 (B. Chen et al., 2020; Shen et al., 2019). Sometimes CoVs that infect animals can evolve, make people sick and become a new human coronavirus. Recent examples are SARS-CoV, MERS-CoV and SARS-CoV-2 (Pillaiyar et al., 2020; Zumla et al., 2016). The detail taxonomical classification of coronaviruses (according to the International Committee on Taxonomy of Viruses) illustrated in Fig. 1 .
Fig. 1.
Schematic taxonomical classification of coronaviruses (according to the International Committee on Taxonomy of Viruses).
In late December 2019, several cases of unexplained pneumonia have been reported in Wuhan, China. Most of the infected or confirmed patients live near the local Huanan seafood wholesale where live animals are widely sold, where live animals are widely sold. In the early stages of pneumonia, severe acute respiratory infections occur, and some patients develop rapidly into acute respiratory distress syndrome (ARDS), acute respiratory failure and other serious complications (Huang et al., 2020). The Chinese Centers for Disease Control and Prevention identified a new type of coronavirus from a patient's throat swab sample on January 7, 2020. Subsequently, on February 7, 2020, a notice issued by the National Health Committee of China temporarily named the coronavirus-infected pneumonia a New/Novel Coronavirus Pneumonia, referred to as “New Crown Pneumonia” (NCP). On January 13, 2020, the World Health Organization temporarily referred to the coronavirus that caused the disease as 2019 new coronavirus (“2019-nCoV). On January 30, 2020, the disease caused by the virus was temporarily named “2019-nCoV acute respiratory disease” (2019 new type of coronavirus acute respiratory disease). On February 11, 2020, the World Health Organization officially named it “Coronavirus Disease 2019”, abbreviated as “COVID-19". On the same day, the International Viral Classification Commission officially named the disease-causing coronavirus “severe acute respiratory syndrome coronavirus 2”, abbreviated as SARS-CoV-2. According to WHO, the disease caused by Novel Coronavirus (2019-nCoV), or SARS-Cov-2 is now officially called COVID-19 (Huang et al., 2020a; Shen et al., 2019; Zhang and Liu, 2020). By February 25, 2021, more than >120000000 cases of COVID-19 have been confirmed, with an estimated mortality risk of ~3.4%, which was comparatively less than that of major viral outbreaks that occurred in past years (Table 1 ). So far, the infection keeps spreading and more and more exported cases were confirmed in many countries worldwide, posing great pressure on public health security.
Table 1.
Comparative detail on major outbreaks with fatality rate of epi- and pandemics.
| Viral outbreaks | Year identified | Number infected cases | Number of deaths | Number of countries affected | Fatality rate (%) |
|---|---|---|---|---|---|
| Marburg | 1967 | 466 | 373 | 11 | 80 |
| Ebola*** | 1976 | 33577 | 13562 | 9 | 40.4 |
| Hendra | 1994 | 7 | 4 | 1 | 57 |
| H5N1 (Bird flu) | 1997 | 861 | 455 | 18 | 52.8 |
| Nipah | 1998 | 513 | 398 | 2 | 77.6 |
| SARS-CoV | 2002 | 8096 | 774 | 29 | 9.6 |
| H1N1 (Swine flu) ** | 2009 | >762630000 | 284500 | 214 | 17.4 |
| MERS-CoV*** | 2012 | 2494 | 858 | 28 | 34.4 |
| H7N9 (Bird flu) | 2013 | 1568 | 616 | 3 | 39.3 |
| SARS-CoV-2* | 2019 | >120000000 | >2600000 | >219 | ~3.4 |
Regarding COVID-19 treatment and its spread, it is currently unclear; current knowledge is mainly based on known similar coronaviruses. CoVs are a large series of viruses that are common in many different animal species, including camels, cows, cats, and bats. Animal coronaviruses rarely infect people and then spread from person to person, such as the respiratory system related diseases MERS, SARS, and now with SARS-CoV-2 (Pillaiyar et al., 2020; Zumla et al., 2016). The most common case is transmission between close contacts (about 6 feet). Human-to-human transmission is believed to occur mainly through respiratory droplets produced when an infected person coughs or sneezes, similar to the way influenza and other respiratory pathogens spread. These water droplets can land on the mouth or nose of nearby people, or they can be inhaled into the lungs. It is unclear whether a person can contract COVID-19 by touching a surface or object, and then touching their mouth, nose or eyes. Generally, for most respiratory viruses, when patients have the serious symptoms (most sick), they are considered most infectious. It should be noted that how easy it is for the virus to spread from person to person varies depending on the type of virus. Some viruses are highly contagious (such as measles), while others are less common (CDC, 2020; WHO, 2020). There is more to be understood about the transmissibility, severity and other characteristics related to SARS-CoV-2, and the investigation is ongoing.
2. Signs and symptoms
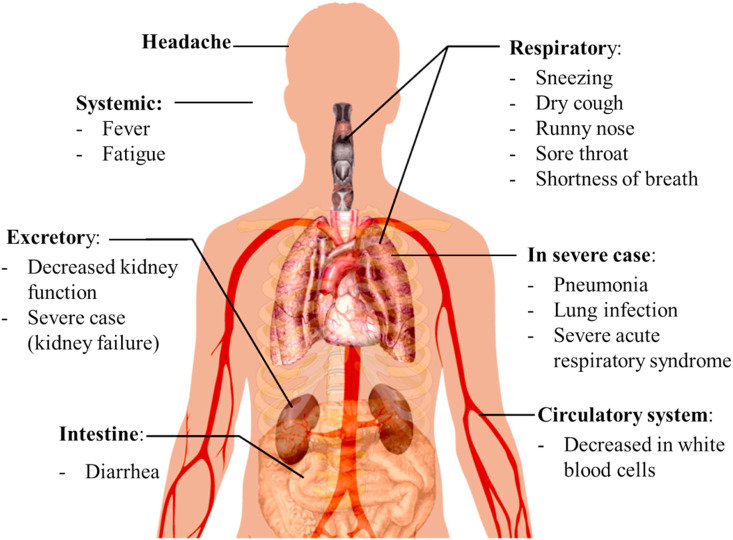
Common symptoms of COVID-19 infection include fever, cough, shortness of breath, and respiratory symptoms (Fig. 2 ). In more severe cases, the infection can cause pneumonia, severe acute respiratory syndrome, kidney failure, and even death. An infected person may be asymptomatic or has symptoms such as fever, cough and shortness of breath, also having diarrhea or upper respiratory symptoms, including sneezing, runny nose and sore throat (CDC, 2020; WHO, 2020). According to WHO, the estimated incubation period for development of symptom after infection ranges from 1 to 14 days, with the median incubation period being 5–6 days. A study found some rare cases with an incubation period of up to 27 days (CDC, 2020; WHO, 2020).
Fig. 2.
Symptoms of COVID-19 caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2).
3. SARS-COV-2 structural details
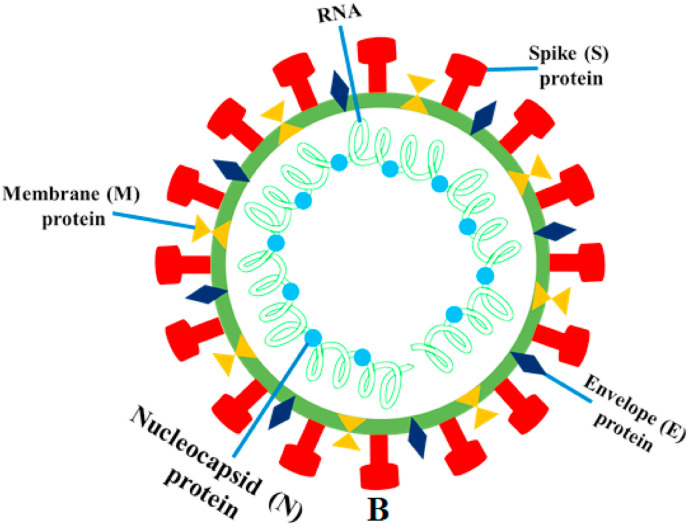
SARS-CoV-2 (2019-nCoV) is an enveloped, single-stranded RNA, positive-sense, β-coronavirus, similar to SARS and MERS. The SARS-CoV-2 genome encodes non-structural proteins, like papain-like protease, helicase, 3-chymotrypsin-like protease, and RNA-dependent RNA polymerase, structural proteins, mainly spike glycoprotein and other accessory proteins (Fig. 3 ) (McKee et al., 2020). From this point of view, the Spike (S), Envelope (E) and Membrane (M) proteins, which are located on the outer surface of the particles are also identified under electron microscope (Dömling and Gao, 2020). A novel type of coronavirus called “Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) was identified as the cause of the respiratory disease outbreak that was first detected in Wuhan, China in 2019. The disease caused by this virus was named as Coronavirus Disease 2019 (COVID-19) (Nile and Kai, 2021).
Fig. 3.
The illustration is created with schematic structural details of the SARS-CoV-2 virion and its major structural proteins. Note that when observed under an electron microscope, the spikes adorned with the outer surface of the virus give rise to the corona like appearance around the virus body.
4. SARS-COV-2 genomic details
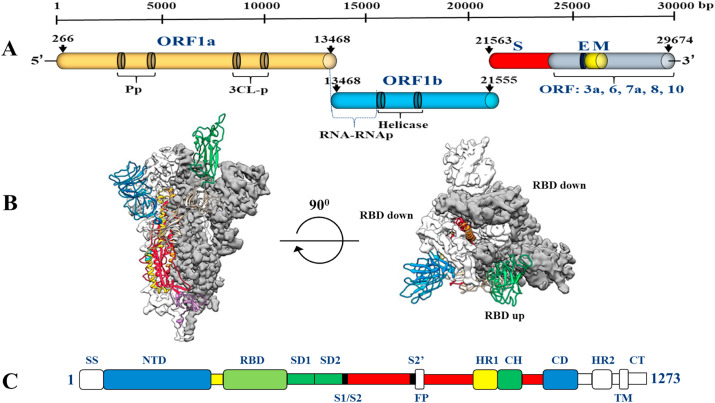
Research evidence shows that SARS-CoV, MERS-CoV and SARS-CoV-2 all originated from bats. The sequence of SARS-CoV-2 is similar to that of the β-coronavirus found in bats, and the virus is genetically different from other coronaviruses, such as severe acute respiratory syndrome-associated coronavirus (SARS), member of Beta-CoV lineage B (that is, the subspecies Sabeco virus) and the Middle East respiratory system Syndrome-associated coronavirus (MERS). The genome of CoVs is a single-stranded sense RNA (+ssRNA) (~30 kb) with a 5′-cap structure and a 3′-poly-A tail. The genome size of CoV (~30 kb) is the largest of all RNA viruses and almost twice the size of the second largest RNA virus. The maintenance of the giant genome size of CoV may be related to the special characteristics of CoV RTC, which contains several RNA processing enzymes, such as the 3′-5′ exoribonuclease enzyme of nsp14, which is unique to CoV among all RNA viruses, and has been proven to be used as a proofreading part of RTC (Chen et al., 2020). Fig. 4 shows the schematic structure of SARS-CoV-2 in the perfusion conformation. Sequence analysis shows that SARS-CoV-2 has a typical genome structure, similar to the β-coronavirus group, including bat-SL ZXC21, bat-SARS (SL)-ZC45, SARS-CoV and MERS-CoV. Based on the phylogenetic tree of CoV, SARS-CoV-2 is more closely related to bat-SL-CoV ZC45 and bat-SL-CoV ZXC21, and is further related to SARS-CoV (Pillaiyar et al., 2020; Zumla et al., 2016).
Fig. 4.
Schematic structures of SARS-CoV-2 S in the prefusion conformation. (A) SARS-CoV-2 genomic structure, with the un-translated region (UTR), open reading frame regions ORF1a and ORF1b, spike (S), envelope (E), membrane (M), and nucleocapsid (N) genes. (B) Select 2D class averages of the particles that were used to calculate the SARS-CoV-2 S reconstruction (left). Side and top views of the prefusion structure of the SARS-CoV-2 S protein with a single RBD in the “up” conformation (right). The two RBD “down” protomers are shown as cryo-EM density in either white or gray and the RBD “up” protomer is shown in ribbons (C) Schematic of SARS-CoV-2 S primary structures, colored by domain. Domains that were excluded from the ectodomain expression construct or could not be visualized in the final map are colored white. SS = signal sequence, NTD= N-terminal domain, RBD = receptor-binding domain, SD1 = subdomain 1, SD2 = subdomain 2, S1/S2= S1/S2 protease cleavage site, S2′ = S2’ protease cleavage site, FP = fusion peptide, HR1 = heptad repeat 1, CH = central helix, CD = connector domain, HR2 = heptad repeat 2, TM = transmembrane domain, CT = cytoplasmic tail. Arrows denote protease cleavage sites (isolate Wuhan-Hu-1, GenBank Acc MN908947).
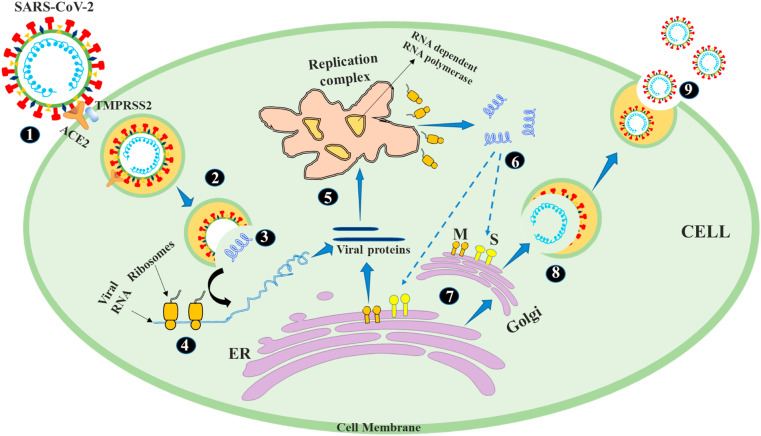
5. SARS-COV-2 infection and life cycle
The spike proteins present on viral outer surface act as a key that allows the virus to enter the cells of a specific host human body. The binding of viral particle to the surface of host human cells through receptors constitutes the first step in the life cycle of coronaviruses. The steps and events involved in the life cycle of SARS-Cov-2 in human cells are shown in Fig. 5 . SARS-CoV-2 virion can enter human cells through endosome or plasma membrane fusion, and the spike protein of SARS-CoV-2 mediates attachment to the host cell membrane and engages angiotensin-converting enzyme 2 (ACE2) as the cellular entry receptor (Shereen et al., 2020). Once the virion enters the complete endosome, cathepsin L activates the spike protein, which is also activated by the cellular serine protease TMPRSS2 in close proximity to the ACE2 receptor, thereby initiating the fusion of the viral membrane and plasma membranes (Hoffmann et al., 2020). Plasma membrane fusion entry is unlikely to trigger host cell anti-viral immunity, so it is more effective for virus replication. Once the virus enters the cell, the gene is translated from the viral genome RNA, and the virus replicates by using viral enzymes such as RNA polymerase. These enzymes are induced by the release of virus from endosomal viral RNA. In addition, the virus hijacks the host machinery, brakes transcription, replicates, and reverse-transcribes its RNA genome for integration into host chromosome, and then reassembles, encapsulates and replicates in infected human cells (Fehr and Perlman, 2015). 5′ end two-thirds of the viral genome encode the polyproteins PP1a and PP1ab, which are cleaved by 3C-like protease (3CLPro) and papain (PLPro) into non-structural protein replicas. An important part of these nonstructural proteins is the RNA-dependent RNA polymerase (RdRp) that forms the replication complex (Fehr and Perlman, 2015; Hoffmann et al., 2020). This replication complex performs transcription of the full-length negative strand. Then, the 3′end of the virus genome encodes four structural proteins, called spike protein (S) envelope (E) protein, nucleocapsid (N) protein and matrix/membrane (M) protein, and a set of accessory proteins (Perlman and Netland, 2009). When the transcription and replication of the viral RNA genome and accessory proteins are completed, the newly synthesized viral protein is trafficked from the endoplasmic reticulum to the Golgi apparatus, and then the mature virion is assembled in the budding vesicles and finally, mature virions are released through the process of exocytosis and release viral replicas outside the host cell and infect nearby cells (Shereen et al., 2020).
Fig. 5.
Life cycle of SARS-Cov-2 in human cell (1. Binding of spike protein to ACE2, 2: TMPRSS2 helps the virion entry, 3: The virion releases its genomic RNA 4: RNA is translated into proteins by the cell's machinery 5: Proteins forms a replication complex to make more RNA 6: Translation and RNA replication 7: Proteins and RNA are assembled into a new virion in the Golgi and released 8: Packaging of RNA synthesized 9: Virion release).
6. Prevention & potential therapies
Currently, there are no any specific drugs or vaccines to prevent or treat 2019 coronavirus disease (COVID-19), as for the majority of other diseases; prevention of the infection by avoiding exposure or close contact to infected persons is the best way in the management of COVID-19. The Centers for Disease Control and Prevention recommended preventive actions to prevent the spread of COVID-19, including; avoiding close contact with infected people, touching eye, nose, mouth, and covering mouth during coughing and sneezing, staying at home in case of illness, cleaning or disinfecting objects and surfaces that are regularly touched. and CDC also recommends people with COVID-19 symptoms should use a mask to prevent the disease from spreading to others (CDC, 2020; WHO, 2020). The use of masks is also important for healthcare professionals and those who take care of infected individuals in a closed environment (home or medical institution). Washing hands with soap and water for at least 20 s after coughing or squeezing, use at least 60% alcohol-containing hand sanitizer. In severe cases, treatment should include care that supports vital organ functions (CDC, 2020; WHO, 2020).
Researchers, clinicians and virologists have been exploring and gaining some experience since the outbreaks of SARS-2003 and MERS-2012. The study of coronaviruses such as SARS and MERS have provided us with several potentially effective drugs. Researchers are using MERS-CoV and SARS-CoV as prototypes to evaluate COVID-19 countermeasures. Broad-spectrum antiviral drugs, such as remdesivir, lopinavir/ritonavir and interferon beta, have shown promising therapeutic effects against MERS-CoV in animal models are currently being used for treatment and prevention of SARS -CoV-2 developed COVID-19 (Pillaiyar et al., 2020; Zumla et al., 2016). Based on previous studies, angiotensin-converting enzyme 2 (ACE2), trans membrane protease serine 2 (TMPRSS2), spike (S) protein, RNA-dependent RNA polymerase (RdRp), angiotensin AT2 receptor, chymotripsin-like protease (3CLpro) and papain-like protease (PLpro) are considered as major targets for development of antiviral drugs against SARS-CoV-2 and another infectious coronavirus (Zumla et al., 2016). Doctors and scientists form different countries, are trying to use different pharmacological strategies to fight COVID-19, which include currently established antiviral drugs, different modes of oxygen therapy or mechanical aeration. Development of vaccines is crucial factor to prevent and control this COVID-19 pandemic as it plays an important role in controlling replication and spread SARS-CoV-2 through production of antibodies against virus and reducing mortality. Currently about 35 vaccine candidates have been entered into a clinical trial, few of them already approved and used against covid treatment and 145 vaccines are in the preclinical phase (Kaur and Gupta, 2020; Rawat et al., 2021). The COVID-19 pandemic requires rapid development of effective treatment strategies in pursuit of three concepts being applied: (1) the first method relies on testing currently known antiviral drugs and verifying their clinical effectiveness. (2) Another model is based on molecular libraries and databases, allowing high computing power and simultaneous verification of millions of potential drugs at the same time. (3) Finally, the third strategy involves targeted treatments aimed at disrupting the genome and function of the virus (Drożdżal et al., 2020; Lu, 2020).
Scientists and physicians around the world have been carrying out an important campaign to understand this emerging disease and its epidemiology to in the context of identifying possible treatment options, finding effective therapeutic agents and developing vaccines. The development of a vaccine may take at least 12–18 months, and the typical schedule for approval of new antiviral therapies may exceed 10 years. Therefore, the reuse of known drugs currently being used for MERS and SARS can significantly accelerate the deployment of new COVID-19 therapies as described in this article. Here are some examples of synthetic (Table 2 ) and natural (Table 3 ) compounds used to treat SARS-CoV and related coronaviruses infection. Their chemical structures details provided in supplementary file (S1).
Table 2.
Commercially available remedies and drugs as possible targets for SARS-CoV-2 and related human coronavirus.
| Name of the therapy | Chemical nature | Molecular formula | Targeted virions | Target virion mechanism | Status as drug | Ref |
|---|---|---|---|---|---|---|
| 2,6-Bis-arylmethyloxy-5-hydroxychromones | Aryl diketoacids | Not available | SARS-Cov, HCV | Inhibits ATPase and helicase activities | Preclinical | Kim et al. (2011) |
| 6′-Fluorinated-Aristeromycin Analogs | Nucleoside analogs | C11H15N5O3 (Aristeromycin) | SARS-CoV, MERS-CoV, CHIKV, ZIKV | RdRp and host cell SAH hydrolase inhibitors | Preclinical studies | Yoon et al. (2019) |
| Abacavir | Nucleoside analog | C14H18N6O | HIV | Reverse transcriptase inhibitor | Approved as HIV drug | Beck et al. (2020) |
| Acyclovir | Doubly flexible synthetic nucleoside analogue | C8H11N5O3 | HSV, HCoV-NL63, MERS-CoV | RNA polymerase inhibitor (RdRp) | Preclinical studies | Beck et al. (2020) |
| Alisporivir | Cyclosporin A-analog | C63H113N11O12 | HCV, HIV, SARS-CoV, MERS-CoV | Non-immunosuppressive, Cyclophilin inhibitor | HCV infection in phase III clinical trial (NCT01860326) | de Wilde et al. (2017) |
| Umifenovir (Arbidol) | Indole derivative | C22H25BrN2O3S | SARS-CoV-2; SARS-CoV, Influenza virus | Block viral fusion and replication | Approved for influenza. Phase 4 for 2019-nCoV, (NCT04260594) | Zhang and Liu (2020) |
| Aryl diketoacids | Enoic acids | C10H8O4 | HIV, SARS-Cov, HCV | NTPase/helicase inhibitors, RdRp inhibitors | Inhibit HIV-1 and HCV Preclinical | Kim et al. (2011) |
| ASC09F | Not available | Not available | HIV, SARS-CoV-2 | Inhibits 3CLpro | Phase 3 for 2019-nCoV, ASC09F/oseltamivir (NCT04261270) | Li and De Clercq (2020) |
| Asunaprevir (BMS-650032) | Oligopeptide | C35H46ClN5O9S | HCV | NS3 protease inhibitor | Approved for HCV, Phase III clinical trials | Beck et al. (2020) |
| Atazanavir | Aza-dipeptide analogue | C38H52N6O7 | HIV, HBV, HCV, SARS-CoV-2 | Protease inhibitor, inhibits 3CLpro | Treat infection of HIV. Preclinical for 2019-nCoV | Beck et al. (2020) |
| Bevacizumab (Avastin) | Immunoglobulin G 1 | C6638H10160N1720O2108S44 | SARS-CoV-2 | VEGF inhibitor | Approved in clinical oncotherapy Promising drug for COVID-19. Phase 2/3 trials (NCT04275414) | Pang et al. (2021) |
| Carmofur | Pyrimidine analogue | C11H16FN3O3 | SARS-CoV-2 | Inhibits THE protease (Mpro) | Induce leukoencephalopathy | Jin et al. (2020) |
| Chloroquine | Aminoquinoline | C18H26ClN3 | Broad spectrum: HCoV-229E HCoV-OC43, HIV, Ebola, SARS-CoV, MERS-CoV, SARS-CoV-2 | S protein ACE2 inhibitor, Endosomal acidification | Approved for malaria. Open-label trial for 2019-nCoV (ChiCTR2000029609) | (Zhang and Liu, 2020; Zumla et al., 2016) |
| Chloroquine Phosphate | Phosphate salt of chloroquine | C18H32ClN3O8P2 | SARS-CoV-2 | Inhibits autophagy and toll-like receptors (TLRs) | An antimalarial drug, FDA approved drug for COVID. | (Zhang and Liu, 2020; Zumla et al., 2016) |
| Hydroxychloroquine | Derivative of chloroquine | C18H26ClN3O | SARS-CoV, MERS-CoV, SARS-CoV-2 | Antiparasitic agent | Used to treat autoimmune disease, antimalarial | Dyall et al. (2014) |
| Triflupromazine (1), Fluphenazine (2), Promethazine (3) | Phenothiazine derivative | (1). C18H19F3N2S (2). C22H26F3N3OS (3). C17H20N2S | SARS-CoV, MERS-CoV | Antipsychotic that shows clathrin-mediated endocytosis | First two approved as antipsychotic agents | Li and De Clercq (2020) |
| Chlorpromazine | Phenothiazine | C17H19ClN2S | SARS-CoV, MERS-CoV, HCV | An antipsychotic affects the assembly of clathrin-coated pits at the plasma membrane | Approved as antipsychotic agents | Zumla et al. (2016) |
| Cobicistat (GS-9350) | Monocarboxylic acid amide | C40H53N7O5S2 | HIV, SARS-CoV-2 | Protease inhibitor, inhibits 3CLpro | Approved for HIV and clinical trial at phase 3 for 2019-nCoV | Li and De Clercq (2020) |
| Compound 6 | Pyrimidine derivative | C12H14CIN3O3S | MERS-CoV | Inhibits papain-like protease | Preclinical | Lee et al. (2019) |
| Cyclosporine A | Cyclic non-ribosomal peptide | C62H111N11O12 | SARS-CoV, MERS-CoV, HIV, HCV | Binds to nucleocapsid protein (NP), inhibits viral replication | Approved as immunosuppressive drug in organ transplantation | Zhang and Liu (2020) |
| Darunavir | Furofuran | C27H37N3O7S | HIV, SARS-CoV-2 | Protease inhibitor, inhibits 3CLpro | Approved for HIV and clinical trial at phase 3 for 2019-nCoV | Li and De Clercq (2020) |
| Disulfiram | Carbamoyl derivative | C10H20N2S4 | MERS-CoV, SARS-CoV | Papain-like protease inhibitor | Approved for treat chronic alcoholism | Lin et al. (2018) |
| Dolutegravir | Monocarboxylic acid amide | C20H19F2N3O5 | HIV, SARS-CoV-2 | Second-generation integrase inhibitor | Approved for HIV and Preclinical for 2019-nCoV | Beck et al. (2020) |
| Ebselen (SPI-1005) | Organoselenium compound | C13H9NOSe | HIV, SARS-CoV-2 | Potently inhibits Mpro and viral replication | Used to treat Diabetes Mellitus | Jin et al. (2020) |
| Efavirenz | Non-nucleoside | C14H9ClF3NO2 | HIV, SARS-CoV-2 | Reverse transcriptase (RT) inhibitor, 3CLpro inhibitor | Approved for HIV and Preclinical for 2019-nCoV | Beck et al. (2020) |
| Entecavir | Guanosine nucleoside analogue | C12H15N5O3 | HBV, SARS-CoV-2 | inhibits the reverse transcriptase (RT) viral RNA-dependent HBV DNA polymerase | Approved for HBV and Preclinical for 2019-nCoV | Beck et al. (2020) |
| Favipiravir (T-705) | Pyrazine carboxamide | C5H4FN3O2 | Influenza, SARS-CoV-2 | RNA polymerase inhibitor (RdRp) | Approved as influenza drug in Japan. China approved for 2019-nCoV | Zhang and Liu (2020) |
| Fingolimod (FTY720) | Aminodiol | C19H33NO2 | 2019-nCoV | Sphingosine-1-phosphate receptor agonist and a CB1 receptor antagonist | Approved for treatment of relapsing forms of multiple sclerosis. Phase 2 for 2019-nCoV, NCT04280588. | Wang (2020) |
| Galidesivir (BCX4430) | Adenosine analog | C11H15N5O3 | SARS-CoV, MERS-CoV, IAV, Ebola | RNA polymerase inhibitor (RdRp) | Clinical trials as Phase 1 for yellow fever and Phase 1 for Marburg virus | Warren et al. (2014) |
| GC376 | Bisulfite adduct | C21H30N3NaO6S | TGEV, FIPV and PTV, MERS-CoV, SARS-CoV | Inhibits 3CLpro, Inhibits the replication of viruses | Preclinical studies | Kim et al. (2012) |
| GC813 | Pyrrolidinone based peptide | C22H31ClN3NaO8S | MERS-CoV | Inhibits 3CLpro | Preclinical studies | Pillaiyar et al. (2020) |
| Imatinib | Benzamide | C29H31N7O | SARS-CoV, MERS-CoV | Abelson tyrosine-protein kinase 2 (Abl2) inhibitor | Approved for cancer | Coleman et al. (2016) |
| Trametinib | Pyridopyrimidine | C26H23FIN5O4 | MERS-CoV, SARS-CoV | Inhibits the ERK/MAPK and PI3K/AKT/mTOR signalling pathways | Approved for cancer treatment | Li and De Clercq (2020) |
| Dasatinib | Benzimidazole | C22H26ClN7O2S | MERS-CoV, SARS-CoV | BCR/ABL and Src family tyrosine kinase inhibitor | Approved for cancer treatment | Li and De Clercq (2020) |
| Selumetinib | Benzimidazole | C17H15BrClFN4O3 | MERS-CoV, SARS-CoV | Inhibits the ERK/MAPK and PI3K/AKT/mTOR signaling pathways | Approved for cancer treatment | Li and De Clercq (2020) |
| Rapamycin | Antibiotic | C51H79NO13 | MERS-CoV | Inhibits the ERK/MAPK and PI3K/AKT/mTOR pathways, block early viral entry and/or post-entry | Approved as antifungal agent | Pillaiyar et al. (2020) |
| Laninamivir | Octanoyl ester | C13H22N4O7 | Influenza virus A and B | Neuraminidase inhibitor | Approved as influenza A and B drug | Samson et al. (2014) |
| Loperamide | Phenyl-butanamide | C29H33ClN2O2 | MERS-CoV, SARS-CoV, HCoV-229E | Inhibits viral replication. Opioid receptor binding | Approved as synthetic antidiarrheal agent | de Wilde et al. (2014) |
| Lopinavir | Dicarboxylic acid amide | C37H48N4O5 | HIV, HPV, HCoV-229E, MERS-CoV, SARS-CoV, SARS-CoV-2 | Protease inhibitor, inhibits 3CLpro | Approved for HIV, Phase 3 for 2019-nCoV, Phase 2/3 for MERS | (Chu, 2004; Li and De Clercq, 2020) |
| Methylprednisolone | Corticosteroid | C22H30O5 | MERS-CoV, SARS-CoV | Protease Inhibitor | Treat arthritis and severe allergic reactions. Randomized trial for 2019-nCoV, NCT04323592 | (Huang et al., 2020; Pillaiyar et al., 2020) |
| Mucroporin-M1 | Scorpion venom-derived peptide | Not available | HBV, H5N1, SARS-CoV | Inhibiting viral replication | Drug design to target COVID-19 | Zhang and Liu (2020) |
| Mycophenolic acid | Antibiotic | C17H20O6 | MERS-CoV, HBV, HCV | Inhibits viral replication, Inhibits IMPDH and guanine monophosphate synthesis | Approved as immunosuppressant during organ transplantation | Hart et al. (2014) |
| Nafamostat | Synthetic p-Guanidinobenzoic acid ester | C19H17N5O2 | SARS-CoV-2, MERS-CoV | Serine protease inhibitor, Inhibits spike-mediated membrane fusion | Approved as an anticoagulant therapy | Li and De Clercq (2020) |
| Nelfinavir | Aryl sulfide | C32H45N3O4S | HIV, HBV, HCV, SARS-CoV | Protease inhibitor | Responsible for post-translational in HIV propeptides. Preclinical trials for 2019-nCoV | Zhang and Liu (2020) |
| Neuraminidase inhibitor analogs (compound 3k) | Chlorobenzoic acid derivatives | Not available | SARS-CoV, MERS-CoV | 3CL protease inhibitor | Preclinical | Kumar et al. (2016) |
| Niclosamide | Benzamide | C13H8Cl2N2O4 | SARS-CoV | ACE2 inhibitor, Inhibit replication of virus | Antihelminthic drug Inhibits IFV-A in A549 cells. | Li et al. (2019) |
| Nicotianamine | Metal ligand | C12H21N3O6 | SARS-CoV-2 | S protein and ACE2 inhibitor | Preclinical | Zhang and Liu (2020) |
| Oseltamivir | Ethyl ester of oseltamivir acid | C16H28N2O4 | SARS-CoV-2; Influenza virus | Influenza neuraminidase inhibitor | Approved for influenza, Phase 3 and 4 for 2019-nCoV, NCT04261270 | Lu (2020) |
| Penciclovir | Nucleoside analogue | C10H15N5O3 | HCV, SARS-CoV-2 | RNA polymerase inhibitor (RdRp) | Approved for HSV. Randomized trial for 2019-nCoV | (M. Wang et al., 2020) |
| Peptidomimetic inhibitors (Compound 6) | Aldehyde derivatives | Not available | MERS-CoV, SARS-CoV | 3CL protease inhibitor | Preclinical | Kumar et al. (2016) |
| Peramivir | Cyclopentane derivative | C15H28N4O4 | Influenza A and B | Neuraminidase inhibitor | Approved as influenza A and B drug | (De Clercq and Li, 2016; Lu, 2020) |
| Promazine | Phenothiazine derivative | C17H20N2S | SARS-CoV | Blocking the interaction of S protein and ACE2 | Alternative for the treatment of COVID-19 | Zhang and Liu (2020) |
| Pyrithiobac derivatives (6-5) | Benzoic acids | C13H11ClN2O4S (Pyrithiobac) | SARS-CoV, | 3CL protease inhibitor | Preclinical | Wu et al. (2019) |
| Remdesivir (GS-5734) | Nucleoside analogue | C27H35N6O8P | Ebola, MERS-CoV, SARS-CoV, SARS-CoV-2 | RNA polymerase inhibitor (RdRp) | Randomized trials for SARS-CoV-2 | (Sheahan et al., 2020a; M. Wang et al., 2020) |
| Ribavirin | Nucleoside analogue | C8H12N4O5 | HCV, RSV, MERS-CoV, SARS-CoV, SARS-CoV-2 | Inhibits viral RNA replication and mRNA capping | Approved for HCV and RSV. Randomized trials for SARS and SARS-CoV-2 | (Chan et al., 2013; Lu, 2020) |
| Ritonavir | L-valine derivative | C37H48N6O5S2 | HIV, MERS-CoV, SARS-CoV-2 | Protease inhibitor, inhibits 3CLpro | Approved for HIV, Phase 3 for SARS-CoV-2, Phase 2/3 for MERS | (Chu, 2004; Li and De Clercq, 2020) |
| SK80 | Phenylisoserine derivative | C31H32N2O4 | SARS-CoV | 3CL protease inhibitor | Preclinical | Konno et al. (2017) |
| SSYA10-001 | Triazole derivative | C12H12N4O2S2 | SARS-CoV, MERS-CoV, MHV | Inhibits helicase without affecting ATPase activity | Preclinical | Adedeji et al. (2014) |
| Losartan (Cozaar) | Monopotassium salt | C22H23ClN6O | MERS-CoV, SARS-CoV, SARS-CoV-2 | Angiotensin-receptor blocker | Phase 2 for SARS-CoV-2 (NCT04312009) | (Yan et al., 2020.) |
| Verdinexor (KPT-335) | Synthesized chemical compound | C18H12F6N6O | Influenza A and B virus, Respiratory syncytial virus (RSV) | Blocking XPO1-mediated nuclear export of viral ribonucleoprotein complexes | Inhibitor of Nuclear Export, Under clinical trial FOR Influenza (NCT02431364) | Perwitasari et al. (2014) |
| Zanamivir | Sialic acid-analogue | C12H20N4O7 | Influenza virus | Neuraminidase inhibitor | Approved for influenza virus | Lu (2020) |
| Gemcitabine hydrochloride | Deoxycytidine analog | C9H12ClF2N3O4 | MERS-CoV, SARS-CoV nfluenza virus | DNA metabolism inhibitor, Inhibiting pyrimidine biosynthesis | FDA-approved anticancer agent | (Li et al., 2019; Pillaiyar et al., 2020) |
| Amodiaquine | Quinoline derivative | C20H22ClN3O | MERS-CoV, SARS-CoV, Ebola, ZIKA virus | Targets early events of the viral replication cycle | Approved as antimalarial drug | (Dyall et al., 2014; Li and De Clercq, 2020) |
| Mefloquine | Quinoline derivative | C17H16F6N2O | MERS-CoV, SARS-CoV | Targets early events of the viral replication cycle | Approved as antimalarial drug | (Dyall et al., 2014; Li and De Clercq, 2020) |
| Dihydroartemisinin | Sesquiterpene lactone | C15H24O5 | HIV, HCMV, HBV, influenza virus A | Inhibits replication of virion | Used as antimalarial and anticancer agent | Krishna et al. (2008) |
| E-64-D (Aloxistatin) | L-leucine derivative | C17H30N2O5 | MERS-CoV, SARS-CoV | Cathepsin protease inhibitor | Inhibit calpain activity in intact platelets. | Dyall et al. (2014) |
| Recombinant interferons | Signalling proteins | Not available | SARS-CoV-2; SARS-CoV; MERS-CoV | Interferon response, Inhibiting the viral protein synthesis, disables viral replication | Approved for melanoma (IFN-α2b), metastatic renal cell carcinoma (IFN-α2a), multiple sclerosis (IFN- β1a, 1b), chronic granulomatous disease (IFN-γ) | Li and De Clercq (2020) |
| SAB-301 | Polyclonal antibody | Not available | MERS-CoV | Prevent the virus from infecting and entering cells | Phase 2/3 trial for MERS endemic in Kingdom of Saudi Arabia | Beigel et al. (2018) |
| REGN3048 and REGN 3051 | Monoclonal antibodies | Not available | MERS-CoV | Prevent the virus replication in cell | Phase 1 trial for MERS-CoV (NCT03301090) | de Wit et al. (2018) |
| Nitazoxanide | Thiazolides | C12H9N3O5S | Influenza viruses, HBV, HCV, HIV, SARS-CoV, MERS-CoV, SARS-CoV-2 | Interferon response in host cell | Approved for Diarrhea treatment. Phase III clinical development for Influenza virus -A and B strains | (Li et al., 2019; Pillaiyar et al., 2020) |
| Saracatinib | Anilinoquinazoline | C27H32ClN5O5 | MERS-CoV | Suppression of the SFK signalling pathways, Inhibits viral replication | Approved for treating cancers | Pillaiyar et al. (2020) |
| Camostat | Benzoic acid derivative | C20H22N4O5 | SARS-CoV MERS-CoV HCoV-229E | Cysteine protease inhibitor, blocks endosomal protease mediated cleavage and the endosomal entry pathway | Preclinical | (Pillaiyar et al., 2020; Zumla et al., 2016) |
| K11777 | Piperazine derivative | C32H38N4O4S | SARS-CoV MERS-CoV HCoV-229E Ebola | Cysteine protease inhibitor, targeting endosomal proteases involved in viral entry | Preclinical | Zhou et al. (2015) |
| Nafamostat | Benzoic acids derivative | C19H17N5O2 | SARS-CoV Influnza-A MERS-CoV | Serine protease inhibitor | FDA-approved to treat pancreatitis, approved as an anticoagulant therapy | Li et al. (2019) |
| K22 | Benzamide | C27H25BrN2O3 | SARS-CoV, MERS-CoV, HCoV-229E | Inhibits membrane-bound RNA synthesis and membrane vesicle formation | Preclinical | Lundin et al. (2014) |
| Teicoplanin derivatives | Glycopeptide antibiotic | C80H81Cl2N9O33 | Broad-spectrum (influenza virus, HCoV, Ebola, HIV, HCV) | Inhibits peptidoglycan polymerization | Effective drug against gram-positive infections | (Li and De Clercq, 2020; Szűcs et al., 2018) |
| FA-613 | Carboxylic acid | C18H14BrNO3 | Influenza A and B, RSV, HCoV) SARS-CoV, MERS-CoV | Inhibits DHODH, interferes intracellular pyrimidine synthesis pathways | Preclinical | (Cheung et al., 2017; Li and De Clercq, 2020) |
| Convalescent plasma | Immunoglobulins | Not available | SARS-CoV-2, SARS-CoV, MERS-CoV, influenza | Inhibits virus entry to the target cells | Phase 2 (NCT02190799) | (Chen et al., 2020; Li and De Clercq, 2020) |
| Mycophenolate mofetil | Ester of mycophenolic acid | C23H31NO7 | HCoV-OC43, HCoV-NL63, MERS-CoV MHV-A59 | Inhibits viral replication | Approved as immunosuppressant | Shen et al. (2019) |
| Monensin sodium | Antibiotic salt | C36H61NaO11 | MERS-CoV, HCoV-OC43, and HCov-NL63 | Inhibits viral replication | Antibacterial drug | Shen et al. (2019) |
| Phenazopyridine | Pyridine derivative | C11H12ClN5 | MERS-CoV, HCoV-OC43, and HCov-NL63 | Inhibits viral replication | Urinary tract analgesic, Removed by FDA | Shen et al. (2019) |
| Pyrvinium pamoate | Quinoline derivative | C49H43N3O6 | MERS-CoV, HCoV-OC43, and HCov-NL63 | Inhibits viral replication | DA-approved antihelmintic drug, inhibits WNT pathway signaling. | Shen et al. (2019) |
| Hexamethylene amiloride | Pyrazines | C12H18ClN7O | SARS-CoV, HCoV-229E, and some animal CoVs | Viroporin inhibitor that inhibits the ion channel activity of CoV E | Preclinical | Zumla et al. (2016) |
| Indomethacin | Indole derivative | C19H16ClNO4 | SARS-CoV | COX1 and COX2 inhibitor, Blocking viral RNA synthesis | Approved as anti-Inflammatory, used to treat gout | Amici et al. (2006) |
| Azithromycin | Azalide | C38H72N2O12 | Zika, Ebola, SARS-CoV-2 | Inhibit replication of virus | Approved as antibiotic | Gautret et al. (2020) |
| Tocilizumab | Monoclonal antibody | C6428H9976N1720O2018S42 | SARS-CoV-2 | Treatment of cytokine storms induced by COVID-19 | Phase III clinical development for COVID-19, NCT04361552 | Luo et al. (2020) |
| EIDD-2801 | Prodrug of NHC | C13H19N3O7 | SARS-CoV-2, MERS-CoV, SARS-CoV | Inhibit replication of virus | Preclinical | Sheahan et al. (2020b) |
| β-D-N4 hydroxycytidine (NHC, EIDD-1931) | Ribonucleoside analog | C9H13N3O6 | Influenza, Ebola, SARS-CoV-2, MERS-CoV, SARS-CoV | Inhibit replication of virus | Preclinical | Sheahan et al. (2020b) |
| Bromhexine hydrochloride | Hydrochloride | C14H21Br2ClN2 | Influenza, SARS-CoV-2 | Inhibit transmembrane serine protease 2 | Mucolytic and prophylactic drug | Habtemariam et al. (2020) |
| Triazavirin | Guanine nucleotide | C5H4N6O3S | SARS-CoV-2, H5N1, Ebola | RNA polymerase inhibitor | Antiviral drug | Shahab and Sheikhi (2020) |
| Carfilzomib | Epoxomicin derivate | C40H57N5O7 | SARS-CoV-2 | Protease inhibitor | Approved anticancer drug | Wang (2020) |
| Eravacycline | Antibiotic | C27H31FN4O8 | SARS-CoV-2 | Protease inhibitor | Broad spectrum antibacterial | Wang (2020) |
| Ruxolitinib | Pyrazole | C17H18N6 | SARS-CoV-2 | JAK inhibitor | Anti-arthritic drugs | Stebbing et al. (2020) |
| Fedratinib | Anilinopyrimidine derivative | C27H36N6O3S | SARS-CoV-2 | JAK inhibitor | Anti-arthritic drugs | Stebbing et al. (2020) |
| Baricitinib (Olumiant) | Pyrazole | C16H17N7O2S | SARS-CoV-2 | JAK and NAK inhibitor | Anti-arthritic drugs | Stebbing et al. (2020) |
| Pirfenidone | Pyridinone derivative | C12H11NO | SARS-CoV-2 | Inhibits DNA synthesis | Antifibrotic agent, phase 3 for COVID-19 NCT04282902 | (Su et al., 2020) |
| Nintedanib | Indolinone derivative | C31H33N5O4 | SARS-CoV-2 | Kinase inhibitor | Antifibrotic agent, phase 2 for COVID-19 NCT04338802 | (Su et al., 2020) |
| Sofosbuvir | Nucleoside analogue | C22H29FN3O9P | Hepatitis C SARS-CoV-2 | Bind to RdRp, Inhibits RNA synthesis | Preclinical | Shah et al. (2020) |
| Tenofovir | Acyclic nucleotide analogue of adenosine | C9H14N5O4P | HIV, HBV, SARS-CoV-2 | Bind to RdRp, Inhibits reverse transcriptase | Preclinical | Shah et al. (2020) |
| Tideglusib | Thiadiazolidinone | C19H14N2O2S | SARS-CoV-2 | non-ATP competitive inhibitor of glycogen synthase kinase 3, inhibits Mpro | Potent anti-inflammatory and neuroprotective | Jin et al. (2020) |
| Azvudine | Cystidine analogue | C9H11FN6O4 | HIV, SARS-CoV-2 | Reverse transcriptase inhibitor | Clinical trial for COVID ChiCTR2000029853 | Zhai et al. (2020) |
| Danoprevir (R7227) | Macrocyclic peptidomer | C35H46FN5O9S | HCV, SARS-CoV-2 | Protease inhibitor | Antiviral agent, phase 2 for COVID-19 NCT04338802NCT04291729 | Shah et al. (2020) |
| Baloxavir marboxil | Synthesized compound | C27H23F2N3O7S | Influenza | Inhibits mRNA and protein synthesis | ChiCTR2000029544 | Li and De Clercq (2020) |
| Ciclesonide | Glucocorticoid | C32H44O7 | SARS-CoV-2 | Inhibits virus replication | Treat obstructive airway diseases, under clinical trial for COVID -19 NCT04330586 | Iwabuchi et al. (2020) |
| Paritaprevir (ABT-450) | Synthesized compound | C40H43N7O7S | HCV, SARS-CoV-2 | Protease inhibitor | Preclinical | Shah et al. (2020) |
| Amprenavir | Derivative of hydroxyethylamine sulfonamide | C25H35N3O6S | HIV-1, SARS-CoV-2 | Protease inhibitor | Preclinical | Wu et al. (2020) |
| Adefovir | Acyclic nucleotide analogue of adenosine | C8H12N5O4P | HIV, HBV, SARS- CoV | Reverse transcriptase and Protease inhibitor | Preclinical | Shah et al. (2020) |
| Ivermectin | Macrocyclic lactone | C48H74O14 | Flavivirus, HIV, dengue, influenza, SARS-CoV-2 | Inhibit the non-structural 3 (NS3) helicase | FDA-approved broad-spectrum anti-parasitic drug. | Kumar et al. (2020) |
| Artesunate | Semi-synthetic derivative artemisinin | C19H28O8 | Hepatitis, HCMV, SARS-CoV-2 | Inhibit NF-kB (Nuclear Factor kappa B) | Antimalarial drug | Uzun and Toptas (2020) |
| Dexamethasone | Corticosteroid | C22H29FO5 | SARS-CoV-2 | Potent anti-inflammatory drug treat arthritis | Phase 6 clinical trial for COVID-19, NCT04325061 | Villar et al. (2020) |
| Siltuximab | Monoclonal antibody | C6450H9932N1688O2016S50 | HIV, SARS-CoV-2 | Interleukin-6 Inhibitors | Phase 3 clinical trial for COVID-19 NCT04330638 | Saini et al. (2020) |
| Hydrocortisone | Corticosteroid | C21H30O5 | SARS-CoV-2 | Anti-inflammatory and immunosuppressive, | Phase 3 clinical trials, NCT04348305 | Saini et al. (2020) |
| Boceprevir | Synthetic tripeptide | C27H45N5O5 | HCV, SARS-CoV-2 | Inhibits protease and viral replication | Approved as antiviral agent | Ma et al. (2020) |
| GC-376 | Synthetic compound | C21H30N3NaO8S | SARS, MERS, SARS-CoV-2 | 3C-like protease inhibitor | Treatment for feline infectious peritonitis | Ma et al. (2020) |
| Thalidomide | Synthetic derivative of glutamic acid | C13H10N2O4 | H1N1, SARS-CoV-2 | Inhibits virus replication | Phase 2 clinical trial for COVID-19, NCT04273529 | Saini et al. (2020) |
| Lenalidomide (Revlimid) | Thalidomide analog | C13H13N3O3 | SARS-CoV-2 | Inhibits virus replication | Phase 4 clinical trial for COVID-19, NCT04361643 | Saini et al. (2020) |
| Acalabrutinib | Synthetic compound | C26H23N7O2 | SARS-CoV-2 | Inhibitor of Bruton's tyrosine kinase (BTK), and viral replication | Phase 2 clinical trial for COVID-19, NCT03863184 | Saini et al. (2020) |
| Duvelisib | Synthetic compound | C22H17ClN6O | HIV, hepatitis B, and C SARS-CoV-2 | Inhibitor of phosphatidylinositol 3-kinase (PI3K) and viral replication | Phase 2 clinical trial for COVID-19, NCT04372602 | Saini et al. (2020) |
| ML188 | Acetamide | C₂₆H₃₁N₃O₃ | SARS-CoV, SARS-CoV-2 | 3CLpro inhibitor | Noncovalent small molecule inhibitor | (Loffredo et al., 2021) |
| Famotidine | Propanimidamide | C8H15N7O2S3 | SARS-CoV-2 | Protease inhibitor | Histamine H2-receptor antagonist | (Loffredo et al., 2021) |
| Tilorone | Fluoren-9-ones | C25H34N2O3 | MERS-CoV, Ebola | Inhibit viral replication | Broad-spectrum antiviral and immunomodulator | Ekins and Madrid (2020) |
Table 3.
Different types of natural compounds as possible targets for SARS-CoV-2 and related human coronavirus.
| Name of the compound | Chemical nature | Molecular formula | Targeted virions | Target and inhibition mechanism | Ref |
|---|---|---|---|---|---|
| 229E-HR1P 229E-HR2P | Peptide | Not available | HCoV-229E | Inhibits spike protein-mediated cell-cell fusion | Li and De Clercq (2020) |
| 6-mercaptopurine | Thiopurine analog | C5H4N4S | MERS-CoV, SARS-CoV | Inhibits PLpro | Li and De Clercq (2020) |
| 6-thioguanine | Thiopurine analog | C5H5N5S | MERS-CoV, SARS-CoV | Inhibits PLpro | Li and De Clercq (2020) |
| Aescin | Saponin | C55H86O24 | SARS-CoV | Inhibits glycoprotein | Xian et al. (2020) |
| Arachidonic acid | Fatty acid | C20H32O2 | SARS-CoV-2, SARS and MERS | Supress ACE2 receptor for viral cell entry | Das (2020) |
| Astaxanthin | Carotenoid pigment | C40H52O4 | SARS-CoV-2 | Supress cathepsin L (CatL) and cytokine storm | Liu et al. (2020) |
| Eicosapentaenoic acid | Fatty acid | C20H30O2 | SARS-CoV-2, SARS and MERS | Supress ACE2 receptor for viral cell entry | Das (2020) |
| Docosahexaenoic acid | Fatty acid | C22H32O2 | SARS-CoV-2, SARS and MERS | Supress ACE2 receptor for viral cell entry | Das (2020) |
| Baicalin | Flavone glycoside | C21H18O11 | HIV-1, SARS-CoV, SARS-CoV-2 | Inhibit E-protein, 3CL protease inhibitor | Su et al. (2020) |
| Baicalein | Trihydroxyflavone | C15H10O5 | HIV, SARS-CoV, SARS-CoV-2 | 3CL protease inhibitor | Su et al. (2020) |
| Betulinic acid | Phenolic acid | C30H48O3 | SARS-CoV | Replication, 3CLpro | (D. Zhang et al., 2020) |
| Celastrol | Quinone-methide triterpene | C29H38O4 | SARS-CoV | 3CLpro inhibitory effect | Ryu et al. (2010) |
| Cepharanthine | Alkaloid | C37H38N2O6 | HCoV-OC43, SARS-CoV, SARS-CoV-2 | Protease inhibition | (Islam et al., 2020; McKee et al., 2020) |
| Cinanserin | Cinnamamides | C20H24N2OS | MERS-CoV, SARS-CoV, SARS-CoV-2 | Serotonin receptor antagonist, 3CL protease inhibitor | (Jin et al., 2020; Zhang and Liu, 2020) |
| Chrysin | Dihydroxyflavone | C15H10O4 | SARS-CoV, SARS-CoV-2 | PLpro inhibitor, Inhibits interaction of SARS-CoV (S) Protein and ACE2. | (Islam et al., 2020; Wu et al., 2020) |
| Chlorogenic acid | Polyphenol | C16H18O9 | HCoV-NL63 | Reducing the production of progeny HCoV-NL63 | Weng et al. (2019) |
| Caffeic acid | Polyphenol | C9H8O4 | HCoV-NL63 | Binds to ACE2 receptor, Inhibits viral replication | Weng et al. (2019) |
| Curcumin | Polylphenol | C27H28O12 | SARS-CoV | GSK-3 Inhibitor, Suppress viral replication | Kandeel and Al-Nazawi (2020) |
| Ginkgolide A | Terpenoids | C20H24O9 | SARS-CoV-2 | Protease inhibitor | 99 |
| Gallic acid | Phenolic acid | C7H6O5 | HCoV-NL63 | Inhibits the viral replication | Weng et al. (2019) |
| Cyanidin-3-sambubioside | Flavonoid | C26H29O15+ | Influenza A and B | Neuraminidase inhibitor | Porter and Bode (2017) |
| Dieckol | Phlorotannin | C36H22O18 | SARS-CoV | 3CLpro inhibitor | Park et al. (2013) |
| Dihydrotanshinone I | Lipophilic diterpenes | C18H14O3 | MERS-CoV | 3CLpro and PLpro protease inhibitors | Kim et al. (2018) |
| Emetine | Alkaloid | C29H40N2O4 | MERS-CoV | Inhibits RNA synthesis | Shen et al. (2019) |
| Emodin | Anthraquinone | C15H10O5 | SARS-CoV HCoV-OC43 SARS-CoV-2 | S protein and ACE2 inhibitor | (Ho et al., 2007; Zhang and Liu, 2020) |
| Ginsenoside Rb1 | Steroid glycosides | C42H72O14 | HIV, SARS-CoV | Prevent viral entry | Li et al. (2005) |
| Glycyrrhetinic acid | Triterpenoids | C30H46O4 | Herpes, HIV, Hepatitis, SARS-CoV | Inhibits viral replication | Wang et al. (2015) |
| Glycyrrhizin | Saponin | C42H62O16 | Herpes, HIV, Hepatitis, SARS-CoV | Inhibits viral replication | Wang et al. (2015) |
| Griffithsin | Algal lectin | Not available | SARS-CoV, MERS-CoV, HCoV-229E, HCoV-OC43, HIV, HCV and Ebola virus | Binds to Spike glycoprotein, inhibiting virus–host cell binding | (Lusvarghi and Bewley, 2016; Zumla et al., 2016) |
| Helichrysetin | Flavonoid | C16H14O5 | SARS-CoV-2, MERS-CoV, | 3CL protease | Zhang and Liu (2020) |
| Herbacetin | Flavonoid | C15H10O7 | SARS-CoV, SARS-CoV-2, MERS-CoV, | 3CL protease | (Jo et al., 2020; Zhang and Liu, 2020) |
| Heparin | Sulfur-rich glycosaminoglycan | C26H42N2O37S5 | SARS-CoV-2 | Anticoagulant, Supress cathepsin L (CatL) | Liu et al. (2020) |
| Homoharringtonine | Alkaloid | C29H39NO9 | SARS-CoV-2 | Inhibits viral replication | Choy et al. (2020) |
| Hesperidin | Dihydroxyflavanone | C28H34O15 | SARS-CoV-2 | ACE2 inhibitor | Wu et al. (2020) |
| Neohesperidin | Flavanone glycoside | C28H34O15 | SARS-CoV-2 | ACE2 inhibitor | Wu et al. (2020) |
| Hesperetin | Trihydroxyflavanone | C16H14O6 | SARS-CoV-2 | Inhibits ACE2 and 3C-like protease | Utomo et al. (2020) |
| HR1P, HR1M, HR1L, HR2L, HR2P, HR2L HR2P-M1, HR2P-M2 | Peptides | Not available | MERS-CoV SARS-CoV-2 | Inhibits replication and spike protein-mediated cell-cell fusion | (Li and De Clercq, 2020; Lu et al., 2014) |
| Iguesterin | Triterpene | C28H36O2 | SARS-CoV | Inhibits 3CLpro | Xian et al. (2020) |
| Kaempferol | Flavonol | C15H10O6 | SARS-CoV, SARS-CoV-2 | PLpro and 3CLpro inhibitor | (D. Zhang et al., 2020) |
| Lignan | Phytonutrients | C25H30O8 | SARS-CoV, SARS-CoV-2 | Inhibition of replication, 3CLpro | (D. Zhang et al., 2020) |
| Luteolin | Flavonoid | C15H10O6 | SARS-CoV | Activation of the NLRP3 inflammasome and modulate inflammatory response | McKee et al. (2020) |
| Lycorine | Alkaloid | C16H17NO4 | HCoV-OC43, HCoV-NL63, MERS-CoV, MHV-A59 | Protein synthesis inhibitor | Li et al. (2005) |
| Apigenin | Flavonoid | C15H10O5 | SARS-CoV | Activation of the NLRP3 inflammasome and modulate inflammatory response to SARS | McKee et al. (2020) |
| Melatonin | Hormone | C13H16N2O2 | SARS-CoV-2 | Regulates ACE2 expression, target papain like protease | (R. Zhang et al., 2020) |
| MERS-5HB | Peptide | Not available | MERS-CoV | Inhibits pseudo typed entry and S protein mediated syncytial formation | Sun et al. (2017) |
| Moupinamide | Alkaloid | C18H19NO4 | SARS-CoV-2 | PLpro inhibitor | (D. Zhang et al., 2020) |
| Myricetin | Flavonoid | C15H10O8 | SARS-CoV | Activation of the NLRP3 inflammasome | McKee et al. (2020) |
| Myricitrin | Glycosyloxyflavone | C21H20O12 | SARS-CoV-2 | Protein kinase inhibitor, 3CLpro receptor inhibitor | Tahir ul Qamar et al. (2020) |
| Methyl rosmarinate | Phenylpropanoids | C19H18O8 | SARS-CoV-2 | 3CLpro receptor inhibitor | Tahir ul Qamar et al. (2020) |
| N-cis-feruloyltyramine | Hydroxycinnamic acid | C18H19NO4 | SARS-CoV-2 | PLpro and 3CLpro inhibitor | (D. Zhang et al., 2020) |
| OC43-HR2P (most promising EK1) | Peptide | Not available | SARS-CoV and MERS-CoV | Spike glycoprotein, inhibits pan-CoV fusion targeting the HR1 domain. | Xia et al. (2019) |
| Oleoylethanolamide | Lipid amide | C20H39NO2 | SARS-CoV-2 | Binds with high affinity to PPAR-a receptors | Ghaffari et al. (2020) |
| Ouabain | ATP1A1-binding cardiotonic steroid | C29H44O12 | MERS-CoV | Inhibit clathrin-mediated endocytosis | Zumla et al. (2016) |
| Oxymatrine | Alkaloid | C15H24N2O2 | HBV | Inhibition of replication | Wang et al. (2011) |
| P21S10 | Peptide | Not available | MERS-CoV | Inhibits spike protein-mediated cell−cell fusion | Li and De Clercq (2020) |
| Pectolinarin | Flavonol | C29H34O15 | SARS-CoV | 3CL protease | Jo et al. (2020) |
| Peptide (P9) | β-defensin derivative | Not available | Broad-spectrum antiviral, SARS-CoV, MERS-CoV, influenza | Inhibits spike protein-mediated cell-cell entry or fusion | Zhao et al. (2016) |
| Pristimerin | Quinone-methide triterpene | C30H40O4 | SARS-CoV | 3CLpro inhibitory effect | Ryu et al. (2010) |
| Quercetin | Flavonoid | C15H10O7 | SARS-CoV | Inhibits 3CLpro and viral replication | Chen et al. (2006) |
| Quercetin-3-β-galactoside | Flavonoid | C21H20O12 | SARS-CoV | 3C-like protease (3CLpro) inhibitor | Chen et al. (2006) |
| Bavachinin | Flavonoid | C21H22O4 | SARS-CoV | Inhibitors of papain-like protease (PLpro). | Islam et al. (2020) |
| Betulonic acid | Pentacyclic triterpenic | C30H46O3 | SARS-CoV | Inhibition of 3CL protease | Islam et al. (2020) |
| Cepharanthine | Alkaloid | C37H38N2O6 | SARS-CoV, HCoV-OC43, SARS-CoV-2 | ACE inhibitor | Xia et al. (2019) |
| Diplacone | Flavonoid | C25H28O6 | SARS-CoV | Inhibition of papain-like protease | Islam et al. (2020) |
| Ferruginol | Diterpenoid | C20H30O | SARS-CoV | Inhibition of viral replication | Islam et al. (2020) |
| Hinokinin | Lignan | C20H18O6 | SARS-CoV | Inhibition of 3CL protease. | Islam et al. (2020) |
| Hirsutenone | Diarylheptanoid | C19H20O5 | SARS-CoV | Inhibits PLpro activity | Xian et al. (2020) |
| Indigo | Organic compound | C16H10N2O2 | SARS-CoV | 3CL protease inhibition. | Islam et al. (2020) |
| Isobavachalcone | Chalcone | C20H20O4 | SARS-CoV | Papain-like protease (PLpro) inhibition | Islam et al. (2020) |
| Juglanin | Cyclic ketone | C20H18O10 | SARS-CoV | Blocks the 3a channel. | Islam et al. (2020) |
| Reserpine | Alkaloid | C33H40N2O9 | SARS-CoV | Inhibits glycoprotein activity | Xian et al. (2020) |
| Rhein | Dihydroxyanthraquinone | C15H8O6 | SARS-CoV | Inhibited interaction (S) protein and ACE2 | Islam et al. (2020) |
| Resveratrol | Polyphenol | C₁₄H₁₂O₃ | MERS-CoV | Inhibits viral replication | Lin et al. (2017) |
| Selamectin | Avermectin | C43H63NO11 | SARS-CoV-2 | Inhibits ACE2 receptor entry | McKee et al. (2020) |
| Rhoifolin | Apigenin derivative | C27H30O14 | SARS-CoV | 3CLpro inhibitor | Jo et al. (2020) |
| Scutellarein | Flavone | C15H10O6 | SARS-CoV-2 | Binds to ACE2 receptor | Chen and Du (2020) |
| Shikonin | Hydroxynaphthoquinones | C16H16O5 | SARS-CoV-2 | Inhibits Mpro | Jin et al. (2020) |
| Silvestrol | Rocaglate derivative | C34H38O13 | MERS-CoV, HCoV-229E, EBOV | Inhibits the DEAD-box RNA helicase eIF4A to affect virus translation | Müller et al. (2018) |
| Sugiol | Diterpenoid | C20H28O2 | SARS-CoV, SARS- CoV-2 | Replication, 3CLpro | (D. Zhang et al., 2020) |
| Tanshinone I | Diterpenoid | C18H12O3 | SARS–CoV | Inhibits PLpro activity | Xian et al. (2020) |
| Tanshinone IIa | Diterpenoid | C19H18O3 | SARS-CoV, SARS- CoV-2 | PLpro and 3CLpro | (D. Zhang et al., 2020) |
| Tingenone | Quinone-methide triterpene | C28H36O3 | SARS-CoV | 3CLpro inhibitory effect | Ryu et al. (2010) |
| Theaflavin | Flavonoid | C29H24O12 | SARS-CoV-2 | Inhibits RdRp activity | Xian et al. (2020) |
| Vitamin C (Ascorbic acid) | Vitamin | C6H8O6 | SARS- CoV-2 | Antioxidant and immunomodulator agent | Boretti and Banik (2020) |
| β-sitosterol | Phytosterol | C₂₉H₅₀O | SARS-CoV | Inhibition of 3CLpro | Mani et al. (2020) |
| Sinigrin | Glucosinolate | C10H17NO9S2 | SARS-CoV | Inhibition of 3CLpro | Mani et al. (2020) |
| α-Helical lipopeptides (e.g. LLS, FFS, IIS, IIK) | Proteins | Not available | MERS-CoV, IAV | Inhibit s protein-mediated cell-cell entry | Wang et al. (2018) |
| Psoralidin | Coumestans | C20H16O5 | SARS-CoV | Inhibits PLpro activity | Mani et al. (2020) |
| Tryptanthrin | Alkaloid | C15H8N2O2 | SARS-CoV | Inhibits PLpro activity | Mani et al. (2020) |
| Amentoflavone | Biflavonoid | C30H18O10 | SARS-CoV | 3CLpro inhibitory effect | Islam et al. (2020) |
| (−)-Catechin gallate | Polyphenol | C22H18O10 | SARS-CoV | Inhibition RNA oligonucleotide | Islam et al. (2020) |
| Savinin | Lignan | C20H16O6 | SARS-CoV | Inhibition of 3CL protease | Islam et al. (2020) |
| Tylophorine | Pentacyclic compound | C24H27NO4 | SARS-CoV | Protease inhibition | Islam et al. (2020) |
6.1. Promising antiviral, antimalarial and anti-HIV agents
Various antiviral, antimalarial and anti-HIV agents are currently being evaluated for use to treat or prevent COVID -19 infections. Currently, several previously available drugs such as Nafamostat, Chloroquine, Hydroxychloroquine, Lopinavir; Ritonavir, Remdesivir, Favipiravir, Lopinavir/Ritonavir, Darunavir/Umifenovir, Nitazoxanide, Ribavirin, Penciclovir, Tocilizumab, Baricitinib, Arbidol, and other antiviral, antimalarial and anti-HIV agents as discussed in Table 1, with structural details provided in supplementary file (Supplementary file S1), Some of these compounds have exhibited promising results in patients and in-vitro clinical studies (Costanzo et al., 2020; Shereen et al., 2020). One of the most common treatments available for SARS-CoV-2 consists of ‘cocktail therapies’ based on various antivirals which are mainly protease inhibitors, the binding of which to the SARS-CoV-1 protease was predicted in silico and in vitro (Costanzo et al., 2020). Various combinational therapies have been used by doctors and researchers for treatment of COVID-19. Thus, the previously approved drugs against MERS, SARS, Malaria and HIV were used as target agent against to block viral protease, clathrin-mediated endocytosis, inhibit the inflammatory cytokine surge, regulate immunity, reduce lung viral loads and improve pulmonary function (Nile et al., 2020). The anti-HIV protease inhibitory drug Kaletra, composed of ritonavir and lopinavir, showed a promising antiviral effect on SARS-CoV and SARS-CoV-2. The other anti-HIV drugs like lopinavir, ritonavir, niclosamide, promazine, and two other HIV inhibitors, PNU and UC2 were also studied as 3CLpro inhibitors of SARS-CoV, demonstrating their potential as templates for designing promising drug against SARS-CoV replication (Ghosh et al., 2020).
Although there have been some preliminary positive reports on use of preexisting antiviral, antimalarial and anti-HIV drugs against treatment of COVID-19 infection, well-designed randomized, controlled clinical trials for evaluating their safety and efficacy will be necessary for the proper treatment of patients diagnosed with COVID-19 in comparison with controls who did not receive the same treatment (Costanzo et al., 2020). The details on potential therapeutic remedies against COVID-19 and related human coronavirus were discussed and presented in Table 1, Table 2, with structural details provided as supplementary file (S1).
6.2. Nucleoside and nucleotide analogs (NAs)
Nucleoside and nucleotide analogs (NAs) are chemically synthesized of purines and pyrimidines analogs having a heterocyclic ring or a sugar moiety. NAs are essential building blocks for nucleic acid biosynthesis and represents as the largest class of anti-inflammatory and antiviral drugs for the treatment of cancer and different viral infections (Pruijssers and Denison, 2019). Some NAs, including amivudine, sofosbuvir, adefovir, telbivudine, entecavir, and tenofovir (Supplementary file S1), have strong antiviral activity and have been used for the treatment of immunodeficiency virus type 1 (HIV-1), hepatitis C (HCV) and hepatitis B (HBV) infection provided proof that these class of compounds used as strong antiviral agents (Fung et al., 2011; Jordheim et al., 2013). Over twenty NAs were approved by US FDA as antiviral drugs for use against various viral infections like; immunodeficiency virus type 1 (HIV-1), hepatitis C (HCV) and hepatitis B (HBV), human cytomegalovirus (HCMV), herpes simplex virus (HSV), varicella zoster virus (VZV)(Mahmoud et al., 2018). These NAs are used to treat both acute and chronic viral infections are delivered as nucleoside and nucleotide precursors or pro-drugs, which are metabolized by host or viral kinases to their active triphosphate once inside the cell and inhibits the viral replication by non-mutually exclusive mechanisms (Pruijssers and Denison, 2019). In this review we summarized the antiviral effects of NAs, mainly remdesivir, lamivudine, amivudine, sofosbuvir, adefovir, entecavir, telbivudine, ribavirin, velpatasvir, and tenofovir against SARS-CoV-2 and related coronaviruses (Table 1, the drug structural details provided as supplementary file (S1).
6.3. Protein (enzyme) inhibitors
The SARS-CoV-2 containing positive-strand RNA causes severe respiratory syndrome in humans and responsible for COVID-19. This virus contains four structural proteins: Spike (S), Envelope (E), Membrane (M), and Nucleocapsid (N) protein (Fig. 3). S protein plays a role in viral attachment to host cell, E and M proteins are involved in viral assembly, and N protein is needed for RNA synthesis (Dömling and Gao, 2020). Angiotensin-converting enzyme 2 (ACE2), transmembrane protease serine 2 (TMPRSS2), spike (S) protein, RNA-dependent RNA polymerase (RdRp), angiotensin AT2 receptor, chymotripsin-like protease (3CLpro) and papain-like protease (PLpro) are considered as major targets for antiviral drugs against SARS-CoV-2 and another infectious coronavirus (McKee et al., 2020; Zumla et al., 2016). We summarized all the synthetic and natural protein (enzyme) inhibitors used to treat SARS-CoV and related coronaviruses infection in Table 1, Table 2, respectively. The structural details of these synthetic and natural protein (enzyme) inhibitors were provided as supplementary file (S1).
6.4. Corticosteroids
Corticosteroids are a class of drugs used to treat illnesses that result from inflammation and reduces immune system activity by mounting an exaggerated response to something or attacks its own cells (Singh et al., 2020). The study reported by RECOVERY Collaborative Group showed the benefit of dexamethasone for patients with COVID-19 who were receiving mechanical ventilation at the time of randomization. Corticosteroids might be effective in preventing acute respiratory distress syndrome and death for patients having shortness of breath or requires oxygen therapy (The RECOVERY Collaborative Group, 2020). Also, World Health Organization has confirmed that the corticosteroids as a potentially effective for the treatment of COVID-19, and patients' survival rates were improved significantly through the application of dexamethasone and other corticosteroids (Table 1). Interestingly, most of earlier studies conducted on SARS-CoV and MERS-CoV showed adverse outcomes for use of corticosteroid in treatment (Singh et al., 2020). Indeed, the Lancet study also reported that corticosteroids should be avoided for the treatment of COVID19. However, such warnings are mainly based on the experiences in a similar viral illness but not on COVID-19 specifically (Russell et al., 2020). Debates are continuing on potential use of corticosteroids as therapy for the treatment of acute respiratory distress syndrome (ARDS) and COVID -19. Indeed, corticosteroids have been speculated to be used as a potential therapy for ARDS as they have ability to reduce inflammation and fibrosis (Reddy et al., 2020). The various corticosteroids which are used as potential drug candidates were discussed in Table 1 and structural formulas provided as supplementary file (S1).
6.5. Natural products and traditional medicines
Plant based natural products and various traditional medicines have been used as an excellent source for discovery of natural/herbal drugs, as they display great diversity among their chemical structures and wide range of biological activities (Wang et al., 2020). Many natural compounds are widely used as antiviral drugs shown to possess promising antiviral effects against influenza viruses, coronaviruses, herpes simplex virus, human immunodeficiency virus, severe acute respiratory syndrome (SARS), Middle East respiratory syndrome (MERS), and hepatitis B and C viruses (Mani et al., 2020; Xian et al., 2020). Numerous natural compounds have been screened in silico targeting various viral proteins; main protease (3CLpro, also named 3-chymotrypsin-like protease), papain like protease (PLpro), helicase, RNA-dependent RNA polymerase (RdRp), and spike protein(Mani et al., 2020; Wu et al., 2020). Various traditional medications based on indigenous theories and experiences are currently being used in the prevention and treatment various microbial diseases, these medicines mainly includes traditional Chinese medicine (TCM), Indian ayurvedic medicine, ancient Iranian medicine, traditional African medicine and Islamic medicine (Wang et al., 2020). Naturally occurring agents that have potential for prevention of COVID-19 include various alkaloids, anthraquinones, terpenoids, fatty acids, flavonoids, glucosinolates, lignans, peptides, phenolics, proteins, saponins, and vitamins (Wang et al., 2020; Zhang and Liu, 2020). (Table 3 and Supplementary file S1), details about these natural products provided in Table 2 and structural details provided as supplementary file (S1). This comprehensive review provides details insights on some active natural products which are being proposed for COVID-19 drug development and prevention.
6.6. Convalescent plasma
Convalescent (immune) plasma therapy refers to use of antibodies obtained from individual who has been recently recovered from particular resolution of infection and disease (Bloch et al., 2020). Convalescent plasma therapy is a passive immunization used to prevent and manage of infectious diseases and considered to be an emergency intervention in controlling several pandemics like SARS-CoV, West Nile virus, Spanish flu, Ebola virus, and recently emerged COVID-19 (Chen et al., 2020). Food and Drug Administration has recently suggested that administration and study of investigational convalescent plasma therapy may provide effective clinical treatment against COVID-19 (Rajendran et al., 2020). Hence, convalescent plasma transfusion therapy has been the subject of increasing attention, especially in the wake of large-scale epidemics like COVID-19.
7. Conclusions
To date, there is no any approved therapy for prevention or treatment of COVID-19, thus many scientists working on possible drug repurposing by using available different Therefore, therapies preexisting drugs including antivirals, antimalarial, immunosuppressive, antipsychotic, antidiarrheal, antidiabetic, anticancer, antifungal, antibacterial, anticoagulant, and antihelminthic agents have been suggested as potential targets preventives or therapeutics against COVID-19. However, the factors like small sample size, poor quality of drug and long completion period are not allowing obtaining reliable and there is paucity of clinical evidence for the therapeutic efficacy as well as safety of aforementioned agents for COVID-19 treatments. Development of effective therapeutic agents is subordinated to the understanding of molecular mechanisms underlying SARS-CoV-2 replication, pathogenesis and virus-host interaction.
The current available knowledge on the safety and efficacy on various therapies needs proper research, like in vitro studies, animal studies and clinical trials for use as potential drug against COVID-19. Several drugs currently being used and which are under clinical trials are remdesivir, umifenovir, oseltamivir, favipiravir, lopinavir/ritonavir, danoprevir/ritonavir, darunavir/cobicistat, triazavirin, hydroxychloroquine, ASC09F, baloxavir marboxil, azvudine, sofosbuvir/ledipasvir, sofosbuvir/daclatasvir, and emtricitabine/tenofovir (Table 1 2). Also, clinical trials are undergoing for various natural compounds like heparin and vitamin C as therapeutic agents or immune boosters in against COVID-19 infection (Table 2 3). Thus, application of the existing potential candidate therapies may represent an effective strategy for the identification of new pathways and targets for intervention of SARS-CoV-2 infection and pathogenesis. In order to effectively deal with the current strategies, needs further exploration to determine the effective agent/therapies for modifying research conduct for this COVID -19. The safety and efficacy of various suggested COVID-19 therapies needs proper systematic research, coordinated by both preclinical studies and clinical trials.
CRediT authorship contribution statement
Shivraj Hariram Nile: colleted data, Writing – original draft, Writing – review & editing. Arti Nile: collected information on this topic and provided all chemical structural details. Shivkumar Jalde: collected information on this topic and provided all chemical structural details. Guoyin Kai: Writing – review & editing, All authors read and approved the final version of this manuscript.
Funding
This work was supported by National Natural Science Foundation, China (81522049, 31571735, 31270007), Zhejiang Provincial Ten Thousand Program for Leading Talents of Science and Technology Innovation (2018R52050), Zhejiang Natural Science Fund (LY20H280008), Zhejiang Provincial Program for the Cultivation of High-level Innovative Health talents.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We are acknowledged and grateful to Prof. Young-Joon Surh, College of Pharmacy, Seoul National University, Seoul, South Korea for his valuable inputs on English editing and scientific quality improvement.
Handling Editor: Dr. Jose Luis Domingo
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.fct.2021.112333.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Adedeji A.O., Singh K., Kassim A., Coleman C.M., Elliott R., Weiss S.R., Frieman M.B., Sarafianos S.G. Evaluation of SSYA10-001 as a replication inhibitor of severe acute respiratory syndrome, mouse hepatitis, and middle east respiratory syndrome coronaviruses. Antimicrob. Agents Chemother. 2014;58:4894–4898. doi: 10.1128/AAC.02994-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amici C., Coro A., Ciucci A., Chiappa L., Castilletti C., Martella V., Decaro N., Buonavoglia C., Capobianchi M.R., Santoro M. Indomethacin has a potent antiviral activity against SARS coronavirus. Antivir. Ther. 2006;11:1021–1030. [PubMed] [Google Scholar]
- Beck B.R., Shin B., Choi Y., Park S., Kang K. Predicting commercially available antiviral drugs that may act on the novel coronavirus (SARS-CoV-2) through a drug-target interaction deep learning model. Comput. Struct. Biotechnol. J. 2020;18:784–790. doi: 10.1016/j.csbj.2020.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloch E.M., Shoham S., Casadevall A., Sachais B.S., Shaz B., Winters J.L., van Buskirk C., Grossman B.J., Joyner M., Henderson J.P., Pekosz A., Lau B., Wesolowski A., Katz L., Shan H., Auwaerter P.G., Thomas D., Sullivan D.J., Paneth N., et al. Deployment of convalescent plasma for the prevention and treatment of COVID-19. J. Clin. Invest. 2020;130(6):2757–2765. doi: 10.1172/JCI138745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beigel J.H., Voell J., Kumar P., Raviprakash K., Wu H., Jiao J.-A., Sullivan E., Luke T., Davey R.T. Safety and tolerability of a novel, polyclonal human anti-MERS coronavirus antibody produced from transchromosomic cattle: a phase 1 randomised, double-blind, single-dose-escalation study. Lancet Infect. Dis. 2018;18:410–418. doi: 10.1016/S1473-3099(18)30002-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boretti A., Banik B.K. Intravenous vitamin C for reduction of cytokines storm in acute respiratory distress syndrome. PharmaNutrition. 2020;12:100190. doi: 10.1016/j.phanu.2020.100190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC 2020. https://www.cdc.gov/coronavirus/2019-ncov/index.html Retrieved.
- Chan J.F.W., Chan K.-H., Kao R.Y.T., To K.K.W., Zheng B.-J., Li C.P.Y., Li P.T.W., Dai J., Mok F.K.Y., Chen H., Hayden F.G., Yuen K.-Y. Broad-spectrum antivirals for the emerging Middle East respiratory syndrome coronavirus. J. Infect. 2013;67:606–616. doi: 10.1016/j.jinf.2013.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B., Tian E.-K., He B., Tian L., Han R., Wang S., Xiang Q., Zhang S., El Arnaout T., Cheng W. Overview of lethal human coronaviruses. Signal Transduct. Target. Ther. 2020;5:89. doi: 10.1038/s41392-020-0190-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Du Q. Potential natural compounds for preventing 2019-nCoV infection. Preprints. 2020:2020010358. doi: 10.20944/preprints202001.0358.v3. [DOI] [Google Scholar]
- Chen L., Li J., Luo C., Liu H., Xu W., Chen G., Liew O.W., Zhu W., Puah C.M., Shen X., Jiang H. Binding interaction of quercetin-3-β-galactoside and its synthetic derivatives with SARS-CoV 3CLpro: structure–activity relationship studies reveal salient pharmacophore features. Bioorg. Med. Chem. 2006;14:8295–8306. doi: 10.1016/j.bmc.2006.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Xiong J., Bao L., Shi Y. Convalescent plasma as a potential therapy for COVID-19. Lancet Infect. Dis. 2020;20:398–400. doi: 10.1016/S1473-3099(20)30141-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Liu Q., Guo D. Emerging coronaviruses: genome structure, replication, and pathogenesis. J. Med. Virol. 2020;92:418–423. doi: 10.1002/jmv.25681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung N.N., Lai K.K., Dai J., Kok K.H., Chen H., Chan K.-H., Yuen K.-Y., Kao R.Y.T. Broad-spectrum inhibition of common respiratory RNA viruses by a pyrimidine synthesis inhibitor with involvement of the host antiviral response. J. Gen. Virol. 2017;98:946–954. doi: 10.1099/jgv.0.000758. [DOI] [PubMed] [Google Scholar]
- Choy K.-T., Wong A.Y.-L., Kaewpreedee P., Sia S.F., Chen D., Hui K.P.Y., Chu D.K.W., Chan M.C.W., Cheung P.P.-H., Huang X., Peiris M., Yen H.-L. Remdesivir, lopinavir, emetine, and homoharringtonine inhibit SARS-CoV-2 replication in vitro. Antivir. Res. 2020;178:104786. doi: 10.1016/j.antiviral.2020.104786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu C.M. Role of lopinavir/ritonavir in the treatment of SARS: initial virological and clinical findings. Thorax. 2004;59:252–256. doi: 10.1136/thorax.2003.012658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman C.M., Sisk J.M., Mingo R.M., Nelson E.A., White J.M., Frieman M.B. Abelson kinase inhibitors are potent inhibitors of severe acute respiratory syndrome coronavirus and Middle East respiratory syndrome coronavirus fusion. J. Virol. 2016;90:8924–8933. doi: 10.1128/JVI.01429-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo M., De Giglio M.A.R., Roviello G.N. SARS-CoV-2: recent reports on antiviral therapies based on lopinavir/ritonavir, darunavir/umifenovir, hydroxychloroquine, remdesivir, favipiravir and other drugs for the treatment of the new coronavirus. Curr. Med. Chem. 2020;27:4536–4541. doi: 10.2174/0929867327666200416131117. [DOI] [PubMed] [Google Scholar]
- Das U.N. Can bioactive lipids inactivate coronavirus (COVID-19)? Arch. Med. Res. 2020;51:282–286. doi: 10.1016/j.arcmed.2020.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Clercq E., Li G. Approved antiviral drugs over the past 50 years. Clin. Microbiol. Rev. 2016;29:695–747. doi: 10.1128/CMR.00102-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wilde A.H., Falzarano D., Zevenhoven-Dobbe J.C., Beugeling C., Fett C., Martellaro C., Posthuma C.C., Feldmann H., Perlman S., Snijder E.J. Alisporivir inhibits MERS- and SARS-coronavirus replication in cell culture, but not SARS-coronavirus infection in a mouse model. Virus Res. 2017;228:7–13. doi: 10.1016/j.virusres.2016.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wilde A.H., Jochmans D., Posthuma C.C., Zevenhoven-Dobbe J.C., van Nieuwkoop S., Bestebroer T.M., van den Hoogen B.G., Neyts J., Snijder E.J. Screening of an FDA-approved compound library identifies four small-molecule inhibitors of Middle East respiratory syndrome coronavirus replication in cell culture. Antimicrob. Agents Chemother. 2014;58:4875–4884. doi: 10.1128/AAC.03011-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit E., Feldmann F., Okumura A., Horne E., Haddock E., Saturday G., Scott D., Erlandson K.J., Stahl N., Lipsich L., Kyratsous C.A., Feldmann H. Prophylactic and therapeutic efficacy of mAb treatment against MERS-CoV in common marmosets. Antivir. Res. 2018;156:64–71. doi: 10.1016/j.antiviral.2018.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dömling A., Gao L. Chemistry and biology of SARS-CoV-2. Chem. 2020;6:1283–1295. doi: 10.1016/j.chempr.2020.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drożdżal S., Rosik J., Lechowicz K., Machaj F., Kotfis K., Ghavami S., Łos M.J. FDA approved drugs with pharmacotherapeutic potential for SARS-CoV-2 (COVID-19) therapy. Drug Resist. Updates. 2020;53:100719. doi: 10.1016/j.drup.2020.100719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyall J., Coleman C.M., Hart B.J., Venkataraman T., Holbrook M.R., Kindrachuk J., Johnson R.F., Olinger G.G., Jahrling P.B., Laidlaw M., Johansen L.M., Lear-Rooney C.M., Glass P.J., Hensley L.E., Frieman M.B. Repurposing of clinically developed drugs for treatment of Middle East respiratory syndrome coronavirus infection. Antimicrob. Agents Chemother. 2014;58:4885–4893. doi: 10.1128/AAC.03036-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekins S., Madrid P.B. Tilorone, a broad-spectrum antiviral for emerging viruses. Antimicrob. Agents Chemother. 2020;64 doi: 10.1128/AAC.00440-20. e00440-20,/aac/64/5/AAC.00440-20.atom. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehr A.R., Perlman S. In: Coronaviruses. Maier H.J., Bickerton E., Britton P., editors. Springer New York; New York, NY: 2015. Coronaviruses: an overview of their replication and pathogenesis; pp. 1–23. [DOI] [Google Scholar]
- Fung J., Lai C.-L., Seto W.-K., Yuen M.-F. Nucleoside/nucleotide analogues in the treatment of chronic hepatitis B. J. Antimicrob. Chemother. 2011;66:2715–2725. doi: 10.1093/jac/dkr388. [DOI] [PubMed] [Google Scholar]
- Gautret P., Lagier J.-C., Parola P., Hoang V.T., Meddeb L., Mailhe M., Doudier B., Courjon J., Giordanengo V., Vieira V.E., Tissot Dupont H., Honoré S., Colson P., Chabrière E., La Scola B., Rolain J.-M., Brouqui P., Raoult D. Hydroxychloroquine and azithromycin as a treatment of COVID-19: results of an open-label non-randomized clinical trial. Int. J. Antimicrob. Agents. 2020;56:105949. doi: 10.1016/j.ijantimicag.2020.105949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaffari S., Roshanravan N., Tutunchi H., Ostadrahimi A., Pouraghaei M., Kafil B. Oleoylethanolamide, A bioactive lipid amide, as A promising treatment strategy for coronavirus/COVID-19. Arch. Med. Res. 2020;51:464–467. doi: 10.1016/j.arcmed.2020.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh A.K., Brindisi M., Shahabi D., Chapman M.E., Mesecar A.D. Drug development and medicinal chemistry efforts toward SARS‐Coronavirus and Covid‐19 therapeutics. ChemMedChem. 2020;15:907–932. doi: 10.1002/cmdc.202000223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habtemariam S., Nabavi S.F., Ghavami S., Cismaru C.A., Berindan-Neagoe I., Nabavi S.M. Possible use of the mucolytic drug, bromhexine hydrochloride, as a prophylactic agent against SARS-CoV-2 infection based on its action on the Transmembrane Serine Protease 2. Pharmacol. Res. 2020;157:104853. doi: 10.1016/j.phrs.2020.104853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart B.J., Dyall J., Postnikova E., Zhou H., Kindrachuk J., Johnson R.F., Olinger G.G., Frieman M.B., Holbrook M.R., Jahrling P.B., Hensley L. Interferon-β and mycophenolic acid are potent inhibitors of Middle East respiratory syndrome coronavirus in cell-based assays. J. Gen. Virol. 2014;95:571–577. doi: 10.1099/vir.0.061911-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho T., Wu S., Chen J., Li C., Hsiang C. Emodin blocks the SARS coronavirus spike protein and angiotensin-converting enzyme 2 interaction. Antivir. Res. 2007;74:92–101. doi: 10.1016/j.antiviral.2006.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.-H., Nitsche A., Müller M.A., Drosten C., Pöhlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam M.T., Sarkar C., El‐Kersh D.M., Jamaddar S., Uddin S.J., Shilpi J.A., Mubarak M.S. Natural products and their derivatives against coronavirus: a review of the non‐clinical and pre‐clinical data. Phytother. Res. ptr. 2020;6700 doi: 10.1002/ptr.6700. [DOI] [PubMed] [Google Scholar]
- Iwabuchi K., Yoshie K., Kurakami Y., Takahashi K., Kato Y., Morishima T. Therapeutic potential of ciclesonide inhalation for COVID-19 pneumonia: report of three cases. J. Infect. Chemother. 2020;26:625–632. doi: 10.1016/j.jiac.2020.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., Zhang B., Li X., Zhang L., Peng C., Duan Y., Yu J., Wang L., Yang K., Liu F., Jiang R., Yang Xinglou, You T., Liu Xiaoce, Yang Xiuna, Bai F., Liu H., Liu Xiang, Guddat L.W., Xu W., Xiao G., Qin C., Shi Z., Jiang H., Rao Z., Yang H. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582:289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- Jo S., Kim S., Shin D.H., Kim M.-S. Inhibition of SARS-CoV 3CL protease by flavonoids. J. Enzym. Inhib. Med. Chem. 2020;35:145–151. doi: 10.1080/14756366.2019.1690480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordheim L.P., Durantel D., Zoulim F., Dumontet C. Advances in the development of nucleoside and nucleotide analogues for cancer and viral diseases. Nat. Rev. Drug Discov. 2013;12:447–464. doi: 10.1038/nrd4010. [DOI] [PubMed] [Google Scholar]
- Kandeel M., Al-Nazawi M. Virtual screening and repurposing of FDA approved drugs against COVID-19 main protease. Life Sci. 2020;251:117627. doi: 10.1016/j.lfs.2020.117627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur S.P., Gupta V. COVID-19 Vaccine: a comprehensive status report. Virus Res. 2020;288:198114. doi: 10.1016/j.virusres.2020.198114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.Y., Kim Y.I., Park S.J., Kim I.K., Choi Y.K., Kim S.-H. Safe, high-throughput screening of natural compounds of MERS-CoV entry inhibitors using a pseudovirus expressing MERS-CoV spike protein. Int. J. Antimicrob. Agents. 2018;52:730–732. doi: 10.1016/j.ijantimicag.2018.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M.K., Yu M.-S., Park H.R., Kim K.B., Lee C., Cho S.Y., Kang J., Yoon H., Kim D.-E., Choo H., Jeong Y.-J., Chong Y. 2,6-Bis-arylmethyloxy-5-hydroxychromones with antiviral activity against both hepatitis C virus (HCV) and SARS-associated coronavirus (SCV) Eur. J. Med. Chem. 2011;46:5698–5704. doi: 10.1016/j.ejmech.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Lovell S., Tiew K.-C., Mandadapu S.R., Alliston K.R., Battaile K.P., Groutas W.C., Chang K.-O. Broad-spectrum antivirals against 3C or 3C-Like proteases of picornaviruses, noroviruses, and coronaviruses. J. Virol. 2012;86:11754–11762. doi: 10.1128/JVI.01348-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konno H., Onuma T., Nitanai I., Wakabayashi M., Yano S., Teruya K., Akaji K. Synthesis and evaluation of phenylisoserine derivatives for the SARS-CoV 3CL protease inhibitor. Bioorg. Med. Chem. Lett. 2017;27:2746–2751. doi: 10.1016/j.bmcl.2017.04.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishna S., Bustamante L., Haynes R.K., Staines H.M. Artemisinins: their growing importance in medicine. Trends Pharmacol. Sci. 2008;29:520–527. doi: 10.1016/j.tips.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R., Gupta N., Kodan P., Mittal A., Soneja M., Wig N. Battling COVID-19: using old weapons for a new enemy. Trop. Dis. Travel Med. Vaccines. 2020;6:6. doi: 10.1186/s40794-020-00107-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar V., Tan K.P., Wang Y.M., Lin S.W., Liang P.H. Identification, synthesis and evaluation of SARS-CoV and MERS-CoV 3C-like protease inhibitors. Bioorg. Med. Chem. 2016;24:3035–3042. doi: 10.1016/j.bmc.2016.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H., Ren J., Pesavento R.P., Ojeda I., Rice A.J., Lv H., Kwon Y., Johnson M.E. Identification and design of novel small molecule inhibitors against MERS-CoV papain-like protease via high-throughput screening and molecular modeling. Bioorg. Med. Chem. 2019;27:1981–1989. doi: 10.1016/j.bmc.2019.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C.-C., Wang X.-J., Wang H.-C.R. Repurposing host-based therapeutics to control coronavirus and influenza virus. Drug Discov. Today. 2019;24:726–736. doi: 10.1016/j.drudis.2019.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., De Clercq E. Therapeutic options for the 2019 novel coronavirus (2019-nCoV) Nat. Rev. Drug Discov. 2020;19:149–150. doi: 10.1038/d41573-020-00016-0. [DOI] [PubMed] [Google Scholar]
- Li S., Chen C., Zhang H., Guo H., Wang H., Wang L., Zhang X., Hua S., Yu J., Xiao P. Identification of natural compounds with antiviral activities against SARS-associated coronavirus. Antivir. Res. 2005;67:18–23. doi: 10.1016/j.antiviral.2005.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin M.-H., Moses D.C., Hsieh C.-H., Cheng S.-C., Chen Y.-H., Sun C.-Y., Chou C.-Y. Disulfiram can inhibit MERS and SARS coronavirus papain-like proteases via different modes. Antivir. Res. 2018;150:155–163. doi: 10.1016/j.antiviral.2017.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S.-C., Ho C.-T., Chuo W.-H., Li S., Wang T.T., Lin C.-C. Effective inhibition of MERS-CoV infection by resveratrol. BMC Infect. Dis. 2017;17:144. doi: 10.1186/s12879-017-2253-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T., Luo S., Libby P., Shi G.-P. Cathepsin L-selective inhibitors: a potentially promising treatment for COVID-19 patients. Pharmacol. Ther. 2020;213:107587. doi: 10.1016/j.pharmthera.2020.107587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loffredo M., Lucero H., Chen D.Y., O'Connell A., Bergqvist S., Munawar A., Bandara A., De Graef S., Weeks S.D., Douam F., Saeed M., Munawar A.H. The in-vitro effect of famotidine on sars-cov-2 proteases and virus replication. Sci. Rep. 2021;11(1):5433. doi: 10.1038/s41598-021-84782-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H. Drug treatment options for the 2019-new coronavirus (2019-nCoV) Biosci Trends. 2020;14(1):69–71. doi: 10.5582/bst.2020.01020. [DOI] [PubMed] [Google Scholar]
- Lu L., Liu Q., Zhu Y., Chan K.-H., Qin L., Li Y., Wang Q., Chan J.F.-W., Du L., Yu F., Ma C., Ye S., Yuen K.-Y., Zhang R., Jiang S. Structure-based discovery of Middle East respiratory syndrome coronavirus fusion inhibitor. Nat. Commun. 2014;5:3067. doi: 10.1038/ncomms4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundin A., Dijkman R., Bergström T., Kann N., Adamiak B., Hannoun C., Kindler E., Jónsdóttir H.R., Muth D., Kint J., Forlenza M., Müller M.A., Drosten C., Thiel V., Trybala E. Targeting membrane-bound viral RNA synthesis reveals potent inhibition of diverse coronaviruses including the middle east respiratory syndrome virus. PLoS Pathog. 2014;10 doi: 10.1371/journal.ppat.1004166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo P., Liu Y., Qiu L., Liu X., Liu D., Li J. Tocilizumab treatment in COVID‐19: a single center experience. J. Med. Virol. 2020;92:814–818. doi: 10.1002/jmv.25801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusvarghi S., Bewley C. Griffithsin: an antiviral lectin with outstanding therapeutic potential. Viruses. 2016;8:296. doi: 10.3390/v8100296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C., Sacco M.D., Hurst B., Townsend J.A., Hu Y., Szeto T., Zhang X., Tarbet B., Marty M.T., Chen Y., Wang J. Boceprevir, GC-376, and calpain inhibitors II, XII inhibit SARS-CoV-2 viral replication by targeting the viral main protease. Cell Res. 2020;30:678–692. doi: 10.1038/s41422-020-0356-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmoud S., Hasabelnaby S., Hammad S., Sakr T. Antiviral nucleoside and nucleotide analogs: a review. J. Adv. Pharm. Res. 2018;2:73–88. doi: 10.21608/aprh.2018.5829. [DOI] [Google Scholar]
- Mani J.S., Johnson J.B., Steel J.C., Broszczak D.A., Neilsen P.M., Walsh K.B., Naiker M. Natural product-derived phytochemicals as potential agents against coronaviruses: a review. Virus Res. 2020;284:197989. doi: 10.1016/j.virusres.2020.197989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKee D.L., Sternberg A., Stange U., Laufer S., Naujokat C. Candidate drugs against SARS-CoV-2 and COVID-19. Pharmacol. Res. 2020;157:104859. doi: 10.1016/j.phrs.2020.104859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller C., Schulte F.W., Lange-Grünweller K., Obermann W., Madhugiri R., Pleschka S., Ziebuhr J., Hartmann R.K., Grünweller A. Broad-spectrum antiviral activity of the eIF4A inhibitor silvestrol against corona- and picornaviruses. Antivir. Res. 2018;150:123–129. doi: 10.1016/j.antiviral.2017.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nile S.H., Kai G. Recent clinical trials on natural products and traditional Chinese medicine combating the COVID-19. Indian J. Microbiol. 2021;61:10–15. doi: 10.1007/s12088-020-00919-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nile S.H., Nile A., Qiu J., Li L., Jia X., Kai G. COVID-19: pathogenesis, cytokine storm and therapeutic potential of interferons. Cytokine Growth Factor Rev. 2020;53:66–70. doi: 10.1016/j.cytogfr.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang J., Xu F., Aondio G., Li Y., Fumagalli A., Lu M., Valmadre G., Wei J., Bian Y., Canesi M., Damiani G., Zhang Y., Yu D., Chen J., Ji X., Sui W., Wang B., Wu S., Kovacs A., Revera M., Wang H., Jing X., Zhang Y., Chen Y., Cao Y. Efficacy and tolerability of bevacizumab in patients with severe Covid-19. Nat. Commun. 2021;12(1):814. doi: 10.1038/s41467-021-21085-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.Y., Kim J.H., Kwon J.M., Kwon H.J., Jeong H.J., Kim Y.M., Kim D., Lee W.S., Ryu Y.B. Dieckol, a SARS-CoV 3CLpro inhibitor, isolated from the edible brown algae Ecklonia cava. Bioorg. Med. Chem. 2013;21:3730–3737. doi: 10.1016/j.bmc.2013.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlman S., Netland J. Coronaviruses post-SARS: update on replication and pathogenesis. Nat. Rev. Microbiol. 2009;7:439–450. doi: 10.1038/nrmicro2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perwitasari O., Johnson S., Yan X., Howerth E., Shacham S., Landesman Y., Baloglu E., McCauley D., Tamir S., Tompkins S.M., Tripp R.A. Verdinexor, a Novel selective inhibitor of nuclear export, reduces influenza A virus replication in vitro and in vivo. J. Virol. 2014;88:10228–10243. doi: 10.1128/JVI.01774-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillaiyar T., Meenakshisundaram S., Manickam M. Recent discovery and development of inhibitors targeting coronaviruses. Drug Discov. Today. 2020;25:668–688. doi: 10.1016/j.drudis.2020.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter R.S., Bode R.F. A review of the antiviral properties of black elder (Sambucus nigra L.) products. Phytother Res. 2017;31:533–554. doi: 10.1002/ptr.5782. [DOI] [PubMed] [Google Scholar]
- Pruijssers A.J., Denison M.R. Nucleoside analogues for the treatment of coronavirus infections. Curr. Opin. Virol. 2019;35:57–62. doi: 10.1016/j.coviro.2019.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajendran K., Krishnasamy N., Rangarajan J., Rathinam J., Natarajan M., Ramachandran A. Convalescent plasma transfusion for the treatment of COVID‐19: systematic review. J. Med. Virol. 2020;92:1475–1483. doi: 10.1002/jmv.25961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawat K., Kumari P., Saha L. COVID-19 vaccine: a recent update in pipeline vaccines, their design and development strategies. Eur. J. Pharmacol. 2021;892:173751. doi: 10.1016/j.ejphar.2020.173751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy K., O'Kane C., McAuley D. Corticosteroids in acute respiratory distress syndrome: a step forward, but more evidence is needed. Lancet Respir. Med. 2020;8:220–222. doi: 10.1016/S2213-2600(20)30048-5. [DOI] [PubMed] [Google Scholar]
- Russell C.D., Millar J.E., Baillie J.K. Clinical evidence does not support corticosteroid treatment for 2019-nCoV lung injury. Lancet. 2020;395:473–475. doi: 10.1016/S0140-6736(20)30317-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu Y.B., Park S.-J., Kim Y.M., Lee J.-Y., Seo W.D., Chang J.S., Park K.H., Rho M.-C., Lee W.S. SARS-CoV 3CLpro inhibitory effects of quinone-methide triterpenes from Tripterygium regelii. Bioorg. Med. Chem. Lett. 2010;20:1873–1876. doi: 10.1016/j.bmcl.2010.01.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini K.S., Lanza C., Romano M., de Azambuja E., Cortes J., de las Heras B., de Castro J., Lamba Saini M., Loibl S., Curigliano G., Twelves C., Leone M., Patnaik M.M. Repurposing anticancer drugs for COVID-19-induced inflammation, immune dysfunction, and coagulopathy. Br. J. Canc. 2020;123:694–697. doi: 10.1038/s41416-020-0948-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahab S., Sheikhi M. Triazavirin - potential inhibitor for 2019-nCoV Coronavirus M protease: a DFT study. Curr. Mol. Med. 2020;10 doi: 10.2174/1566524020666200521075848. 2174/1566524020666200521075848. [DOI] [PubMed] [Google Scholar]
- Samson M., Abed Y., Desrochers F.-M., Hamilton S., Luttick A., Tucker S.P., Pryor M.J., Boivin G. Characterization of drug-resistant influenza virus A(H1N1) and A(H3N2) variants selected in vitro with Laninamivir. Antimicrob. Agents Chemother. 2014;58:5220–5228. doi: 10.1128/AAC.03313-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah B., Modi P., Sagar S.R. In silico studies on therapeutic agents for COVID-19: drug repurposing approach. Life Sci. 2020;252:117652. doi: 10.1016/j.lfs.2020.117652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheahan T.P., Sims A.C., Leist S.R., Schäfer A., Won J., Brown A.J., Montgomery S.A., Hogg A., Babusis D., Clarke M.O., Spahn J.E., Bauer L., Sellers S., Porter D., Feng J.Y., Cihlar T., Jordan R., Denison M.R., Baric R.S. Comparative therapeutic efficacy of remdesivir and combination lopinavir, ritonavir, and interferon beta against MERS-CoV. Nat. Commun. 2020;11:222. doi: 10.1038/s41467-019-13940-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheahan T.P., Sims A.C., Zhou S., Graham R.L., Pruijssers A.J., Agostini M.L., Leist S.R., Schäfer A., Dinnon K.H., Stevens L.J., Chappell J.D., Lu X., Hughes T.M., George A.S., Hill C.S., Montgomery S.A., Brown A.J., Bluemling G.R., Natchus M.G., Saindane M., Kolykhalov A.A., Painter G., Harcourt J., Tamin A., Thornburg N.J., Swanstrom R., Denison M.R., Baric R.S. An orally bioavailable broad-spectrum antiviral inhibits SARS-CoV-2 in human airway epithelial cell cultures and multiple coronaviruses in mice. Sci. Transl. Med. 2020;12 doi: 10.1126/scitranslmed.abb5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L., Niu J., Wang C., Huang B., Wang W., Zhu N., Deng Y., Wang H., Ye F., Cen S., Tan W. High-throughput screening and identification of potent broad-spectrum inhibitors of coronaviruses. J. Virol. 2019;93 doi: 10.1128/JVI.00023-19. e00023-19,/jvi/93/12/JVI.00023-19.atom. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shereen M.A., Khan S., Kazmi A., Bashir N., Siddique R. COVID-19 infection: origin, transmission, and characteristics of human coronaviruses. J. Adv. Res. 2020;24:91–98. doi: 10.1016/j.jare.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A.K., Majumdar S., Singh R., Misra A. Role of corticosteroid in the management of COVID-19: a systemic review and a Clinician's perspective. Diabetes Metab. Syndr. Clin. Res. Rev. 2020;14:971–978. doi: 10.1016/j.dsx.2020.06.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbing J., Phelan A., Griffin I., Tucker C., Oechsle O., Smith D., Richardson P. COVID-19: combining antiviral and anti-inflammatory treatments. Lancet Infect. Dis. 2020;20:400–402. doi: 10.1016/S1473-3099(20)30132-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su H., Yao S., Zhao W., Li M., Liu J., Shang W., Xie H., Ke C., Gao M., Yu K., Liu H., Shen J., Tang W., Zhang L., Zuo J., Jiang H., Bai F., Wu Y., Ye Y., Xu Y. Discovery of baicalin and baicalein as novel, natural product inhibitors of SARS-CoV-2 3CL protease in vitro. bioRxiv. 2020 doi: 10.1101/2020.04.13.038687. 2020.04.13.038687. [DOI] [Google Scholar]
- Sun Y., Zhang H., Shi J., Zhang Z., Gong R. Identification of a novel inhibitor against middle east respiratory syndrome coronavirus. Viruses. 2017;9:255. doi: 10.3390/v9090255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szűcs Z., Kelemen V., Le Thai S., Csávás M., Rőth E., Batta G., Stevaert A., Vanderlinden E., Naesens L., Herczegh P., Borbás A. Structure-activity relationship studies of lipophilic teicoplanin pseudoaglycon derivatives as new anti-influenza virus agents. Eur. J. Med. Chem. 2018;157:1017–1030. doi: 10.1016/j.ejmech.2018.08.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahir ul Qamar M., Alqahtani S.M., Alamri M.A., Chen L.-L. Structural basis of SARS-CoV-2 3CLpro and anti-COVID-19 drug discovery from medicinal plants. J. Pharm. Anal. 2020;10:313–319. doi: 10.1016/j.jpha.2020.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The RECOVERY Collaborative Group Dexamethasone in hospitalized patients with Covid-19-Preliminary report. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2021436. NEJMoa2021436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utomo R.Y., Ikawati M., Meiyanto E. Revealing the potency of citrus and galangal constituents to halt SARS-CoV-2 infection. Preprints. 2020;2020:2020030214. doi: 10.20944/preprints202003.0214.v1. [DOI] [Google Scholar]
- Uzun T., Toptas O. Artesunate: could be an alternative drug to chloroquine in COVID-19 treatment? Chin. Med. 2020;15:54. doi: 10.1186/s13020-020-00336-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villar J., Ferrando C., Martínez D., Ambrós A., Muñoz T., Soler J.A., Aguilar G., Alba F., González-Higueras E., Conesa L.A., Martín-Rodríguez C., Díaz-Domínguez F.J., et al. Dexamethasone treatment for the acute respiratory distress syndrome: a multicentre, randomised controlled trial. Lancet Respir. Med. 2020;8:267–276. doi: 10.1016/S2213-2600(19)30417-5. [DOI] [PubMed] [Google Scholar]
- Wang C., Zhao L., Xia S., Zhang T., Cao R., Liang G., Li Y., Meng G., Wang W., Shi W., Zhong W., Jiang S., Liu K. De novo design of α-helical lipopeptides targeting viral fusion proteins: a promising strategy for relatively broad-spectrum antiviral drug discovery. J. Med. Chem. 2018;61:8734–8745. doi: 10.1021/acs.jmedchem.8b00890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Huang J., Yeung A.W.K., Tzvetkov N.T., Horbańczuk J.O., Willschke H., Gai Z., Atanasov A.G. The significance of natural product derivatives and traditional medicine for COVID-19. Processes. 2020;8:937. doi: 10.3390/pr8080937. [DOI] [Google Scholar]
- Wang J. Fast identification of possible drug treatment of coronavirus disease-19 (COVID-19) through computational drug repurposing study. J. Chem. Inf. Model. 2020;60:3277–3286. doi: 10.1021/acs.jcim.0c00179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Yang R., Yuan B., Liu Y., Liu C. The antiviral and antimicrobial activities of licorice, a widely-used Chinese herb. Acta Pharm. Sin. B. 2015;5(4):310–315. doi: 10.1016/j.apsb.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M., Shi Z., Hu Z., Zhong W., Xiao G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30:269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.-P., Zhao W., Xue R., Zhou Z.-X., Liu F., Han Y.-X., Ren G., Peng Z.-G., Cen S., Chen H.-S., Li Y.-H., Jiang J.-D. Oxymatrine inhibits hepatitis B infection with an advantage of overcoming drug-resistance. Antivir. Res. 2011;89:227–231. doi: 10.1016/j.antiviral.2011.01.005. [DOI] [PubMed] [Google Scholar]
- Warren T.K., Wells J., Panchal R.G., Stuthman K.S., Garza N.L., Van Tongeren S.A., Dong L., Retterer C.J., Eaton B.P., Pegoraro G., Honnold S., Bantia S., Kotian P., Chen X., Taubenheim B.R., Welch L.S., Minning D.M., Babu Y.S., Sheridan W.P., Bavari S. Protection against filovirus diseases by a novel broad-spectrum nucleoside analogue BCX4430. Nature. 2014;508:402–405. doi: 10.1038/nature13027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng J.R., Lin C.S., Lai H.C., Lin Y.P., Wang C.Y., Tsai Y.C., Wu K.C., Huang S.H., Lin C.W. Antiviral activity of Sambucus FormosanaNakai ethanol extract and related phenolic acid constituents against human coronavirus NL63. Virus Res. 2019;273:197767. doi: 10.1016/j.virusres.2019.197767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO 2020. https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200121-sitrep-1-2019-ncov.pdf Retrieved.
- Wu C., Liu Y., Yang Y., Zhang P., Zhong W., Wang Y., Wang Q., Xu Y., Li M., Li X., Zheng M., Chen L., Li H. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm. Sin. B. 2020;10:766–788. doi: 10.1016/j.apsb.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu R.-J., Zhou K.-X., Yang Haijin, Song G.-Q., Li Y.-H., Fu J.-X., Zhang X., Yu S.-J., Wang L.-Z., Xiong L.-X., Niu C.-W., Song F.-H., Yang Haitao, Wang J.-G. Chemical synthesis, crystal structure, versatile evaluation of their biological activities and molecular simulations of novel pyrithiobac derivatives. Eur. J. Med. Chem. 2019;167:472–484. doi: 10.1016/j.ejmech.2019.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia S., Yan L., Xu W., Agrawal A.S., Algaissi A., Tseng C.-T.K., Wang Q., Du L., Tan W., Wilson I.A., Jiang S., Yang B., Lu L. A pan-coronavirus fusion inhibitor targeting the HR1 domain of human coronavirus spike. Sci. Adv. 2019;5:eaav4580. doi: 10.1126/sciadv.aav4580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xian Y., Zhang J., Bian Z., Zhou H., Zhang Z., Lin Z., Xu H. Bioactive natural compounds against human coronaviruses: a review and perspective. Acta Pharm. Sin. B. 2020;10:1163–1174. doi: 10.1016/j.apsb.2020.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan F., Huang F., Xu J., Yang P., Qin Y., Lv J., Zhang S., Ye L., Gong M., Liu Z., Wei J., Xie T., Xu K.F., Gao G.F., Wang F.S., Cai L., Jiang C. Antihypertensive drugs are associated with reduced fatal outcomes and improved clinical characteristics in elderly COVID-19 patients. Cell Discov. 2020;6:77. doi: 10.1038/s41421-020-00221-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon J., Kim G., Jarhad D.B., Kim H.-R., Shin Y.-S., Qu S., Sahu P.K., Kim H.O., Lee H.W., Wang S.B., Kong Y.J., Chang T.-S., Ogando N.S., Kovacikova K., Snijder E.J., Posthuma C.C., van Hemert M.J., Jeong L.S. Design, synthesis, and anti-RNA virus activity of 6′-fluorinated-aristeromycin analogues. J. Med. Chem. 2019;62:6346–6362. doi: 10.1021/acs.jmedchem.9b00781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai P., Ding Y., Wu X., Long J., Zhong Y., Li Y. The epidemiology, diagnosis and treatment of COVID-19. Int. J. Antimicrob. Agents. 2020;55:105955. doi: 10.1016/j.ijantimicag.2020.105955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Wu K., Zhang X., Deng S., Peng B. In silico screening of Chinese herbal medicines with the potential to directly inhibit 2019 novel coronavirus. J. Integr. Med. 2020;18:152–158. doi: 10.1016/j.joim.2020.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Liu Y. Potential interventions for novel coronavirus in China: a systematic review. J. Med. Virol. 2020;92:479–490. doi: 10.1002/jmv.25707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., Wang X., Ni L., Di X., Ma B., Niu S., Liu C., Reiter R.J. COVID-19: melatonin as a potential adjuvant treatment. Life Sci. 2020;250:117583. doi: 10.1016/j.lfs.2020.117583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H., Zhou J., Zhang K., Chu H., Liu D., Poon V.K.-M., Chan C.C.-S., Leung H.-C., Fai N., Lin Y.-P., Zhang A.J.-X., Jin D.-Y., Yuen K.-Y., Zheng B.-J. A novel peptide with potent and broad-spectrum antiviral activities against multiple respiratory viruses. Sci. Rep. 2016;6:22008. doi: 10.1038/srep22008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Vedantham P., Lu K., Agudelo J., Carrion R., Nunneley J.W., Barnard D., Pöhlmann S., McKerrow J.H., Renslo A.R., Simmons G. Protease inhibitors targeting coronavirus and filovirus entry. Antivir. Res. 2015;116:76–84. doi: 10.1016/j.antiviral.2015.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zumla A., Chan J.F.W., Azhar E.I., Hui D.S.C., Yuen K.-Y. Coronaviruses — drug discovery and therapeutic options. Nat. Rev. Drug Discov. 2016;15:327–347. doi: 10.1038/nrd.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.