Abstract
Triple-negative breast cancer (TNBC) is a highly aggressive breast cancer (BrC) subtype lacking effective therapeutic targets currently. The development of multi-omics databases facilities the identification of core genes for TNBC. Using TCGA-BRCA and METABRIC datasets, we identified CT83 as the most TNBC-specific gene. By further integrating FUSCC-TNBC, CCLE, TCGA pan-cancer, Expression Atlas, and Human Protein Atlas datasets, we found CT83 is frequently activated in TNBC and many other cancers, while it is always silenced in non-TNBC, 120 types of normal non-testis tissues, and 18 types of blood cells. Notably, according to the TCGA-BRCA methylation data, hypomethylation on chromosome X 116,463,019 to 116,463,039 is significantly correlated with the abnormal activation of CT83 in BrC. Using Kaplan–Meier Plotter, we demonstrated that activated CT83 is significantly associated with unfavorably overall survival in BrC and worse outcomes in some other cancers. Furthermore, GSEA suggested that the abnormal activation of CT83 in BrC is probably oncogenic by triggering the activation of cell cycle signaling. Meanwhile, we also noticed copy number variations and mutations of CT83 are quite rare in any cancer type, and its role in immune infiltration is not significant. In summary, we highlighted the significance of CT83 for TNBC and presented a comprehensive bioinformatics strategy for single-gene analysis in cancer.
Subject terms: Cancer, Breast cancer, Cancer genomics, Epigenetics, Transcriptomics
Introduction
Breast cancer is heterogeneous in molecular features. Triple-negative breast cancer is one of the four classical breast cancer subtypes that account for about 15% to 20% of diagnosed breast cancer1. Molecularly, TNBC is characterized by the lack of estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER2)2. Basal breast cancer is an underlying uncertain subtype3 that shares an expression profile similar to that of the basal-myoepithelial layer of the normal breast and characterized by a strong expression of basal markers such as cytokeratins 5, 6, and 174,5. Despite some discordance, "TNBC" and "basal breast cancer" have been used interchangeably because the majority of basal tumors are also TNBC6. The current clinical management of TNBC is generally challenging7. TNBC is more aggressive compared with other breast cancer subtypes, as patients with TNBC are usually young and more likely to suffer from worse clinical outcomes such as early relapse and visceral metastasis regardless of their higher chemosensitivity. Although the incorporation of targeted agents like poly-ADP-ribose polymerase (PARP)8,9 and immune checkpoint inhibitors10,11 in clinical practice seems promising, TNBC responses to these treatments are still vary considerably12. Moreover, the lack of specific targets is one of the key factors that stymieing the substantial improvement in the targeted therapy of TNBC13. Therefore, the identification of core molecular markers, especially TNBC-specific ones, is still critical at this moment.
Advancement in multi-omics approaches, especially RNA-seq and microarray techniques, have yielded tons of bioinformatics data, greatly driving innovation and progression of scientific studies on cancer markers. The Cancer Genome Atlas (TCGA)14–16 is a landmark cancer genomics program that provided genomic, epigenomic, transcriptomic, and proteomic data in over 20,000 cancer and paired normal samples spanning 33 types of cancer, including breast cancer. The Molecular Taxonomy of Breast Cancer International Consortium (METABRIC) project17,18 documented genomic landscapes of over 2500 breast cancer, including clinical characteristics, gene expression, copy number variations, and mutation profiles of driver genes. Fudan University Shanghai Cancer Center (FUSCC) presented comprehensive clinical, genomic, transcriptomic data of 465 primary TNBC19, which is the largest TNBC genomic project to date. The Cancer Cell Line Encyclopedia (CCLE)20–22 comprehensively characterized over 1000 types of human cancer cell lines and provided open-access bioinformatics data including but not limited to cell phenotypes, RNA-seq, DNA copy number, DNA methylation, and gene mutations. These publicly available bioinformatics data enable the systematic search for TNBC-specific molecular markers and the prediction of their biological functions.
In this study, from over 60,000 genes and 3617 breast cancer samples, we identified a TNBC-specific key gene, CT83, which is significantly overexpressed in TNBC but not in other breast cancer subtypes. Moreover, by analyzing multi-omic bioinformatics data in over 10,000 cancer tissues, 1739 cancer cell lines, 124 types of normal tissues, and 18 types of blood cells, we comprehensively analyzed CT83 in breast cancer and pan-cancer, including its expression patterns (in cancer tissues, normal tissues, cancer cell lines, and blood cells), mutation profiles, copy number variations, methylation status, association with tumor-infiltrating lymphocytes, prognostic significance, and potential biological functions in breast cancer. Briefly, our data not only highlighted the importance of CT83 for TNBC but also presented a comprehensive bioinformatics strategy for the analysis of cancer-related genes.
Methods
Gene screening
The TCGA-BRCA dataset downloaded from the TCGA portal23 (http://tcgaportal.org/download.html) and the METABRIC dataset downloaded from the cBioPortal24,25 (https://www.cbioportal.org/study/summary?id=brca_metabric) were used for gene screening. The integrated file in the TCGA-BRCA dataset (RNA-seq, FPKM) and the mRNA microarray data named “data_mRNA_median_Zscores.txt” in the METABRIC dataset (Microarray, Z-scores) were used for assessing gene expression levels. Data in the “PAM50” column of the TCGA-BRCA dataset and the “Pam50 + claudin-low subtype” column in the file named “data_clinical_patient.txt” of the METABRIC dataset were used for subtype determining. Data were processed with Microsoft Excel, and the t-test was calculated to evaluate statistical significance.
Gene annotations
Gene annotations, mainly including gene synonyms, chromosome locations, transcript lengths, exon information, and protein lengths, were obtained from the Ensembl database (https://www.ensembl.org/index.html)26,27.
mRNA expression
The two datasets used in the screening part and the FUSCC-TNBC dataset19 (RNA-seq, Log2 FPKM) downloaded from the National Omics Data Encyclopedia (NODE; https://www.biosino.org/node/project/detail/OEP000155) were used to analyze the expression of CT83 mRNA in breast cancer tissues of different subtypes. The expression of CT83 mRNA in cancer cell lines was analyzed with data from the Cancer Cell Line Encyclopedia (CCLE) project20. Specifically, the list of breast cancer cell lines was extracted from the file named “Cell_lines_annotations_20181226.txt” downloaded from the CCLE data portal (https://portals.broadinstitute.org/ccle/data), and gene expression was analyzed with the file named “CCLE_RNAseq_genes_rpkm_20180929.gct.gz” The CT83 expression heat maps were generated using the cBioPortal25 (https://www.cbioportal.org) based on data from the TCGA-BRCA14–16 (Pan-cancer Atlas; https://www.cbioportal.org/study/summary?id=brca_tcga_pan_can_atlas_2018), METABRIC17 (https://www.cbioportal.org/study/summary?id=brca_metabric), and the CCLE20 (https://www.cbioportal.org/study/summary?id=ccle_broad_2019) dataset. RNA-seq data in “organism part” of “homo sapiens” obtained from the Expression Atlas database (https://www.ebi.ac.uk/gxa/home) were used to analyze the expression of CT83 in normal human tissues. RNA-seq data based on TCGA and the GTEx28 project obtained from the GEPIA229 database (http://gepia2.cancer-pku.cn) were used to investigate the expression of CT83 in pan-cancer and paired normal tissues.
Copy number variation (CNV)
The CNV profiles named “data_CNA.txt” and the corresponding annotation files named “meta_CNA.txt” of the TCGA-BRCA (Firehose Legacy; https://www.cbioportal.org/study/summary?id=brca_tcga), MATABRIC (https://www.cbioportal.org/study/summary?id=brca_metabric), and CCLE (https://www.cbioportal.org/study/summary?id=ccle_broad_2019) dataset obtained from the cBioPortal database were used to analyze CNV of CT83 in breast cancer tissues and pan-cancer cell lines. The CNV profiles of CT83 in TCGA pan-cancer tissues were obtained from the TIMER2.030 database (http://timer.cistrome.org). The DNA copy number data named “data_linear_CNA.txt” and RNA-seq data named “data_RNA_Seq_v2_expression_median.txt” in the TCGA-BRCA (Firehose Legacy) were used to analyze the correlation between the CT83 DNA copy number and mRNA expression levels. Pearson’s correlation coefficient was calculated using the GraphPad Prism (https://www.graphpad.com) to evaluate their correlation strength. R-squared was calculated to evaluate the DNA-mRNA linear correlation.
Mutations
The TCGA-BRCA (Firehose Legacy; https://bit.ly/36wxqQP; https://bit.ly/2HRYxvb), CCLE-BRCA (https://bit.ly/36xQXjV), TCGA (Firehose Legacy; https://bit.ly/2GtsAZP; https://bit.ly/30AYhrb), and CCLE (https://bit.ly/36wxyzN; https://bit.ly/36x4wzX) dataset in cBioPortal database were used to analyze mutations of CT83 in breast cancer tissues, breast cancer cell lines, pan-cancer tissues, and pan-cancer cell lines, respectively. The correlation analysis between the expression of CT83 and mutations of key genes in breast cancer was performed with the TCGA-BRCA dataset in the TCGA Portal database (http://tcgaportal.org/TCGA/Breast_TCGA_BRCA/index.html). The permutation tests were auto-calculated to evaluate the correlation strength.
Methylation
The correlation between CT83 expression and its methylation in breast cancer tissues was analyzed based on the TCGA-BRCA dataset in the TCGA Portal database (http://tcgaportal.org/TCGA/Breast_TCGA_BRCA/index.html). The methylation status of CT83 was shown as methylation beta-values31. Chromosome locations of involved methylation probes were annotated with the NCBI Genome Data Viewer (https://www.ncbi.nlm.nih.gov/genome/gdv). Pearson’s correlation coefficient was calculated using GraphPad Prism (https://www.graphpad.com) to evaluate the correlation strength31,32. A Pearson’s value of -0.8 or less was considered as a strong correlation.
Immune infiltration
The TIMER2.030 (http://timer.cistrome.org) and TISIDB33 (http://cis.hku.hk/TISIDB) databases were used to analyze the correlation between CT83 mRNA expression and the abundance of tumor-infiltrating lymphocytes in breast cancer tissues and pan-cancer tissues, respectively. CXorf61, one of the aliases of CT83, was used for searching in both database. Spearman’s correlation coefficient was auto-calculated to evaluate the strength of correlation. An absolute Spearman’s value of 0.75 or more was considered as a strong correlation.
Prognosis
Survival analysis was performed using the Kaplan–Meier Plotter34 (https://kmplot.com/analysis) with mRNA data in breast cancer (gene chips; https://kmplot.com/analysis/index.php?p=service&cancer=breast) and pan-cancer (RNA-seq; https://kmplot.com/analysis/index.php?p=service&cancer=pancancer_rnaseq). Hazard ratios (HR), 95% confidence intervals (95%CI), and log-rank test P-values were all auto-calculated by the Kaplan–Meier Plotter. CT83 will be considered as prognostic if a P-value is 0.05 or less.
Gene set enrichment analysis
Gene set enrichment analysis (GSEA)35 was performed according to the official guidelines (https://software.broadinstitute.org/cancer/software/gsea/wiki/index.php/Main_Page). Briefly, the TCGA-BRCA (http://tcgaportal.org/download.html) and METABRIC (https://www.cbioportal.org/study/summary?id=brca_metabric) datasets were downloaded to get gene expression profiles in breast cancer tissues. Next, the “expression dataset” and the “phenotype labels” were prepared based on the required format (https://www.gsea-msigdb.org/gsea/doc/GSEAUserGuideFrame.html?Preparing_Data_Files). Annotated datasets were then analyzed with the GSEA software (v4.1.0, https://www.gsea-msigdb.org/gsea/downloads.jsp) using the the hallmark gene sets (h.all.v7.2.symbols.gmt) and the KEGG gene sets (c2.cp.kegg.v7.2.symbols.gmt). Samples were divided into CT83-positive and CT83-negative cohorts using 1 FPKM (for TCGA-BRCA) and 1 Z-score (for METABRIC) as the cutoffs, respectively. The top 5 positively and negatively enriched gene sets were included for further analysis. Enriched genes that were annotated as “core enrichment” in the Cell Cycle, G2M Checkpoint, and E2F targets were used for the screening of CT83-associated core genes. The expression profiles of CT83 and its correlated genes in breast cancer were obtained using the TCGA-BRCA, METABRIC, and the CCLE-BRCA dataset in the cBioPortal database (https://www.cbioportal.org).
Results
CT83 is significantly overexpressed in TNBC but not in other breast cancer subtypes
To screen for genes that significantly overexpressed in TNBC but not in other breast cancer subtypes, we analyzed the RNA-seq data from the TCGA-BRCA and the mRNA microarray data from the METABRIC datasets. Based on our screening criteria, CT83 is the top-ranked gene in the TCGA-BRCA dataset and second-ranked in the METABRIC dataset (Fig. 1A; Table S1). The median expression of CT83 in TNBC tissues is 10.4 FPKM in the TCGA-BRCA dataset and 2.28 Z-score in the METABRIC dataset, while its maximal median expression levels in non-TNBC tissues are 0 FPKM and -0.18 Z-score, respectively. Cancer/testis antigen 83 (CT83), also known as CXorf61, FLJ20611, FLJ22913, and KK-LC-1, is a protein-coding gene located on the reverse strand of chromosome X (Fig. 1B). It has only one transcript with a length of 517 bp that consisted of 2 exons (Fig. 1C). The protein encoded by CT83 mRNA is 113 aa in length. As the TNBC-specific expression patterns of CT83 were observed in two independent datasets with large sample sizes, we hypothesized that CT83 may play distinct roles in TNBC.
Figure 1.
Screening strategies and basic annotations of CT83. Screening strategies for genes that significantly overexpressed in TNBC but not in non-TNBC subtypes. (B) The location of CT83 on chromosome X. (C) The schematic diagram of the only transcript of CT83.
CT83 is frequently overexpressed in TNBC tissues and cell lines
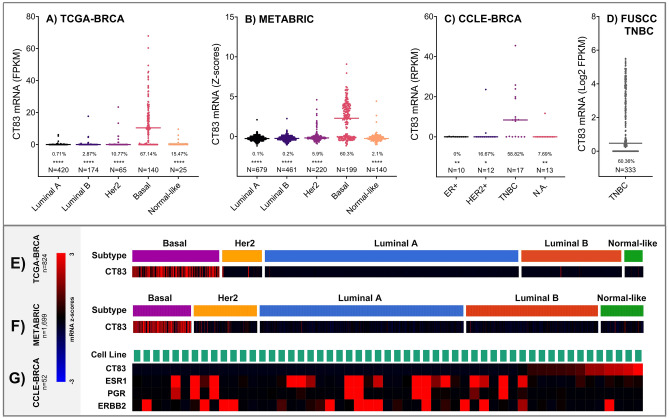
Next, we investigated the RNA-seq and microarray data from the TCGA-BRCA, METABRIC, FUSCC-TNBC, and CCLE-BRCA dataset to get a better understanding of the expression CT83 mRNA in breast cancer tissues and cell lines (Fig. 2). As mentioned in the screening part, the expression of CT83 mRNA is significantly higher in Basal/TNBC tissues compared with other subtypes, and a similar trend was observed in the 57 breast cancer cell lines (including 17 TNBC cell lines) in which the expression of CT83 mRNA was measured (Table S2). Moreover, the positive rate of CT83 mRNA is the highest in either TNBC tissues or cell lines, while its expression is always undetectable in breast cancer tissues or cell lines of other subtypes. The frequency of detectable CT83 mRNA, defined as 1 FPKM/RPKM/Z-score or more, was 67.1% (94/140), 60.3% (125/199), 60.4% (201/333), and 58.8% (10/17) according to data from the TCGA-BRCA, METABRIC, FUSCC-TNBC, and CCLE-BRCA-TNBC dataset, respectively. Although the expression data of CT83 protein was not available in any of the above-mentioned dataset or the Clinical Proteomic Tumor Analysis Consortium (CPTAC) dataset36, data from a previous study suggested that the protein of CT83 is always detectable in TNBC tissues and cell lines37. Briefly, data from 672 TNBC tissues, 2184 non-TNBC breast cancer tissues, 17 TNBC cell lines, and 35 non-TNBC breast cancer cell lines demonstrated that CT83 is distinctively and frequently overexpressed in TNBC.
Figure 2.
The expression of CT83 mRNA in breast cancer. The expression of CT83 mRNA in breast cancer tissues of different subtypes according to data from the TCGA-BRCA (A,E) and METABRIC (B,F) dataset. The expression of CT83 mRNA in TNBC tissues according to data from the FUSCC-TNBC dataset (D). The expression of CT83 mRNA in breast cancer cell lines based on data from the CCLE-BRCA dataset (C,G). The expression of ESR1, PGR, and ERBB2 was plotted for distinguishing the subtype of breast cancer cell lines. The median expression of CT83 in each subtype was labeled as dashes. The positive rate of CT83 mRNA was labeled as percentage numbers in A (> 1 FPKM), 2C (> 1 RPKM), and 2D (> 1 Log2 FPKM), while the positive rate of CT83 in B was not presented because the positive expression cutoff cannot be determined based on the METABRIC raw data. Only samples with available both CT83 expression and subtype information were used for plotting. The asterisks in A–C represent the statistical difference (t-test p values) of CT83 expression between the Basal subtype and other subtypes. *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001.
CT83 expression is highly restricted in normal tissues but widely distributed in cancer tissues and cell lines
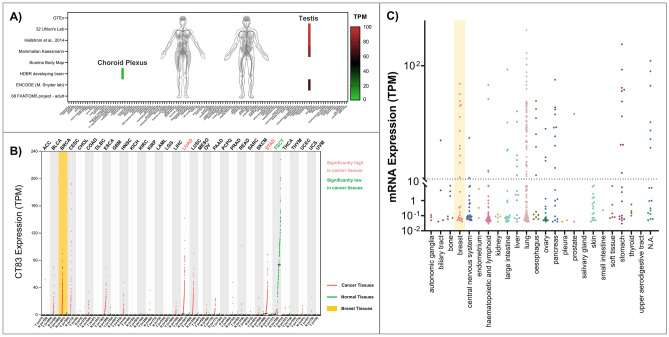
Since the expression of cancer/testis antigens is highly restricted in normal adult tissues and widely distributed in tumor tissues38,39, we examined the expression of CT83 in 120 types of normal human tissues (Fig. 3A), 33 types of cancer tissues (Fig. 3B), 1019 cancer cell lines (Fig. 3C), and 18 types of blood cells (Figure S1). Based on RNA-seq data of 8 datasets in the Expression Atlas database, the expression of CT83 can only be detected in testis tissues in normal adult males and sometimes in brain (choroid plexus) tissues, while no positive CT83 expression is observed in any other normal tissue. Conversely, the abnormal activation of CT83 is often detected in cancer tissues including breast cancer, and its high expression is statistically different from paired normal tissues in lung adenocarcinoma (LUAD) and stomach adenocarcinoma (STAD) tissues. Moreover, the high expression of CT83 mRNA, defined as 10 TPM or more, is also often detected in cancer cell lines, especially in cell lines of breast cancer, liver cancer, lung cancer, pancreas cancer, and stomach cancer. These expression patterns of CT83 are consistent with previous understanding about typical cancer/testis antigens, indicating that CT83 is a potential therapeutic target in cancer immunotherapy40.
Figure 3.
The expression of CT83 in normal tissues, cancer tissues, and cancer cell lines. (A) The expression of CT83 in normal adult tissues based on 8 RNA-seq datasets obtained from the Expression Atlas. (B) The expression of CT83 in pan-cancer and paired normal tissues based on RNA-seq data from the GEPIA2 database. The abbreviations of involved cancers are available on this webpage (https://gdc.cancer.gov/resources-tcga-users/tcga-code-tables/tcga-study-abbreviations). (C) The expression of CT83 in pan-cancer cell lines based on RNA-seq data from the CCLE dataset.
Copy number variation is not the major cause of the abnormal activation of CT83 in cancer
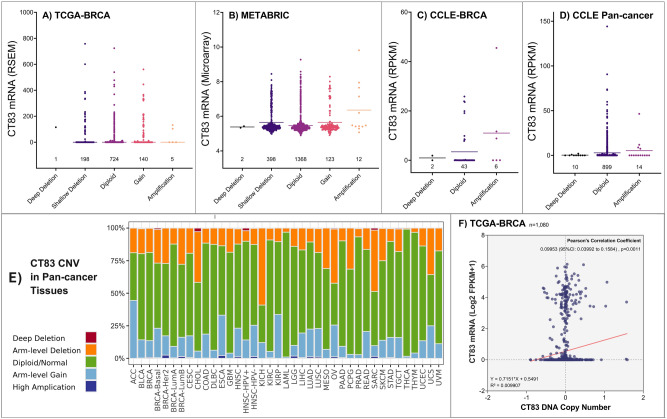
Copy number variation (CNV), especially copy number amplification, is one of the major causes of gene overexpression in cancer41–43. Therefore, we investigated the correlation between the expression level of CT83 mRNA and the copy number status of its DNA to figure out whether the abnormal activation of CT83 in cancer is a consequence of its copy number amplification. According to data from the TCGA-BRCA and the METABRIC dataset, copy number variation (amplification and deep deletion) of CT83 is a rare event in breast cancer tissues (Fig. 4A,B). Among the included 2971 breast cancer tissue samples, CT83 amplification rate was only less than 1% in both datasets. Similarly, CT83 amplification was detected in 6/51 of the CCLE-BRCA cell lines, while its deep deletion was observed in only 2 breast cancer cell lines (Fig. 4C). Apart from breast cancer, copy number variation of CT83 is also uncommon in pan-cancer tissues and cell lines (Fig. 4D,E). Specifically, CT83 amplification was detected in 14/923 (1.52%) cancer cell lines based on CCLE pan-cancer CNV data. In 33 types of TCGA cancer tissues, most of the detected CT83 DNA were diploid, while its DNA amplification or deep deletion was still rare events. Moreover, we also analyzed the linear correlation between its DNA copy number and mRNA expression levels with data from the TCGA-BRCA dataset (Fig. 4F). The R-squared and Pearson’s correlation coefficients of their correlation was 0.009907 and 0.09953 (p = 0.0011), respectively. Taken together, these data showed that copy number variation has no significant correlation with the abnormal activation of CT83 in breast cancer and in other cancers.
Figure 4.
Copy number variations of CT83 in cancer. Copy number variations of CT83 in breast cancer tissues based on data from the TCGA-BRCA (A) and the METABRIC (B) dataset. Copy number variations of CT83 in breast cancer cell lines (C) and pan-cancer cell lines (D) based on data from the CCLE dataset. The median expression levels of CT83 mRNA were labeled as dashes. Copy number variations of CT83 in pan-cancer tissues (E) based on TCGA cancer data. The correlation between CT83 DNA copy number and CT83 mRNA expression levels (F).
CT83 mutation is a rare event in cancer
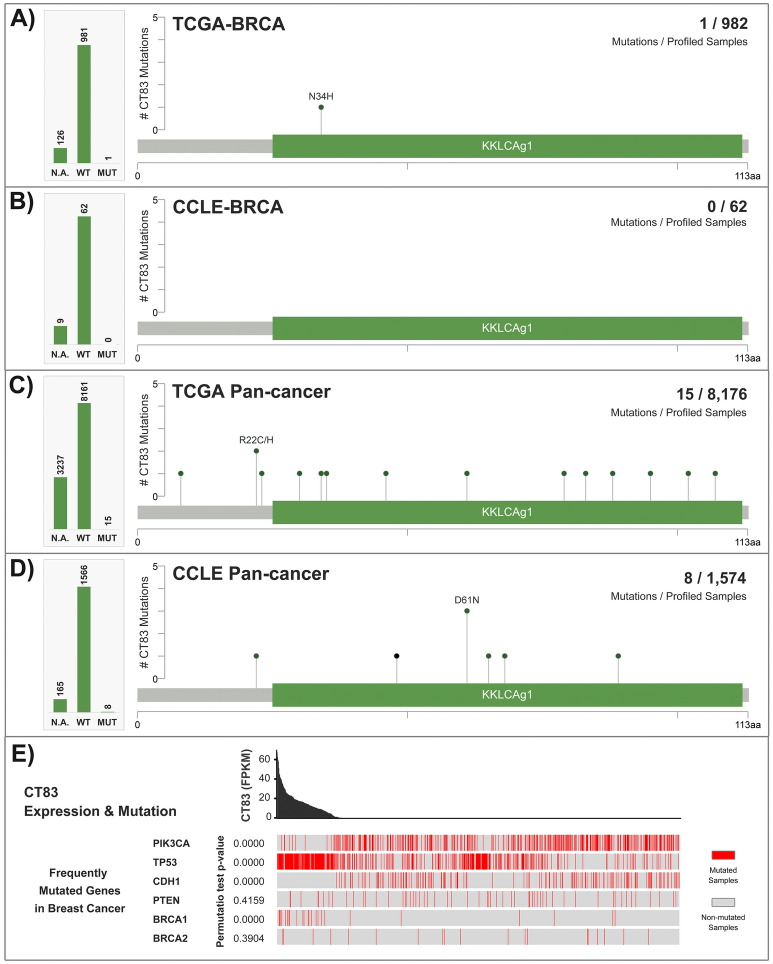
Somatic mutations also contribute to changes in gene expression44, and some driver mutations are critical for cancer development45,46. Hence, we examined mutation profiles of CT83 in cancer. In 982 TCGA-BRCA samples with available CT83 mutation data, only one CT83 mutation was detected, while not any CT83 mutation was observed in the 62 CCLE-BRCA cell lines (Fig. 5A,B). In 8176 TCGA pan-cancer tissues spanning 33 cancer types, mutated CT83 was only detected in 15 samples (Fig. 5C). Similarly, among the 1574 CCLE pan-cancer cell lines, CT83 mutations were only observed in 8 cell lines of 7 mixed cancer types (Fig. 5D). Moreover, CT83 mutations were sporadic that no mutation hotspot was identified (Table S3). In addition, we also assessed the correlation between CT83 mRNA expression levels and mutations of key genes in breast cancer (Fig. 5E). According to the TCGA-BRCA dataset, CT83 expression is statistically higher in TP53-mut (permutation test p = 0.0000) or BRCA1-mut samples (permutation test p = 0.0000), significantly lower in PIK3CA-mut (permutation test p = 0.0000) or CDH1-mut samples (permutation test p = 0.0000), and has no significant correlation with PTEN and BRCA2 mutations. Briefly, data from this part suggested that mutation is not one of the major genomic alternations of CT83 in cancer.
Figure 5.
Mutations of CT83 in cancer. (A) CT83 mutations in breast cancer tissues, (B) breast cancer cell lines, (C) TCGA pan-cancer tissues, and (D) CCLE pan-cancer cell lines. (E) The correlation between CT83 mRNA expression and mutations of key genes in breast cancer based on data from the TCGA-BRCA dataset. CT83 mutation points were labeled in the corresponding regions in the linear schematic diagram of its protein. Permutation test p-values were calculated by comparing CT83 expression in key-gene-mutated samples with key-gene-unmutated samples. N.A., not available; WT, wild type; MUT, mutated.
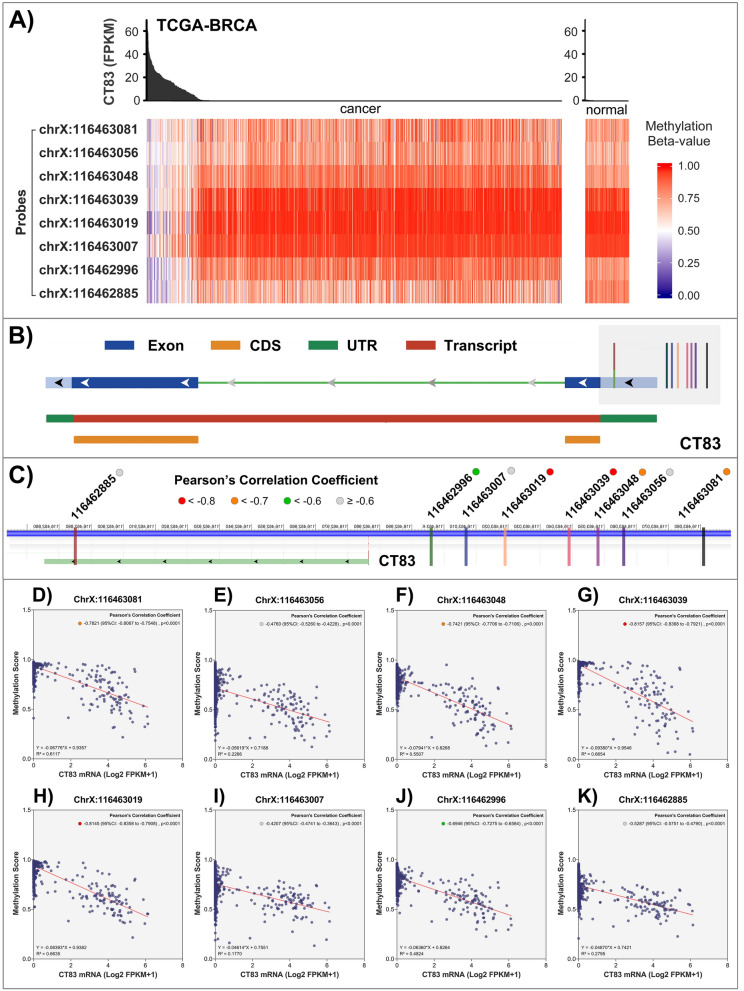
Hypomethylation may induce the abnormal activation of CT83 in cancer
Previous studies have identified that epigenetic modification, especially hypomethylation, is of great importance in the abnormal activation of CT genes in cancer47–49. Therefore, we analyzed the correlation between CT83 mRNA expression levels and its methylation status in breast cancer tissues with the TCGA-BRCA dataset (Fig. 6A). The expression level of CT83 is usually higher when some of its DNA loci are hypomethylated, while CT83 is always silenced when its DNA is hypermethylated. As 8 probes were used to measure CT83 methylation status at different DNA loci, we then assessed these data in detail to find out the DNA locus in which its methylation status is mostly responsible for the expression changes of CT83 (Fig. 6B,C). By evaluating the correlation between the methylation score of a specific probe and the expression level of CT83 mRNA (Fig. 6D–K), we found that methylation of the region between 116,463,019 to 116,463,039 on chromosome X is most closely correlated with CT83 expression. The Pearson’s correlation coefficients for the probe “ChrX:116,463,039” (Fig. 6G) and “ChrX:116,463,019” (Fig. 6H) are − 0.8157 (95% CI − 0.8368 to − 0.7921, p < 0.0001) and − 0.8145 (95% CI − 0.8358 to − 0.7908, p < 0.0001), respectively. These data indicated that hypomethylation of the region 116,463,019 to 116,463,039 on chromosome X may induce the abnormal activation of CT83 in breast cancer tissues.
Figure 6.
CT83 methylation in breast cancer. (A) The correlation between CT83 mRNA expression and its methylation status in breast cancer tissues. (B) Locations of CT83 methylation probes relative to CT83 DNA sequence. It should be noted that CT83 is located on the reverse strand of chromosome X. (C) Enlarged views for locations CT83 methylation probes. Pearson’s correlation coefficients between CT83 mRNA expression levels and methylation beta values were labeled as colorful dots. D-K) Pearson’s correlation coefficients between CT83 mRNA expression levels and methylation beta values of corresponding probes.
CT83 and tumor-infiltrating lymphocytes
The association between cancer/testis antigens and immune components has been widely reported50–52. We analyzed the TIMER2.0 and TISIDB database to assess the correlation between the expression of CT83 and the abundance of tumor-infiltrating lymphocytes (TILs). According to data from the TIMER2.0 database, the expression of CT83 has no significant correlation with any types of TILs in breast cancer tissues or any breast cancer subtypes, with the strongest correlation observed in “T cell CD4 + Th2_XCELL” of the overall cohort (Spearman’s correlation coefficient = 0.4213, p = 0.0000) (Table S4). Moreover, based on data from the TISIDB database, the abundance of TILs in pan-cancer tissues is independent of CT83 expression (Figure S2). For the strongest correlation, CT83 mRNA expression is positively correlated with the abundance of type 17 T helper cells (Th17) in esophageal carcinoma (ESCA) (Spearman’s correlation coefficient = 0.442, p = 0.0000) and negatively correlated with effector memory CD4 T cells (Tem_CD4) in rectum adenocarcinoma (READ) (Spearman’s correlation coefficient = − 0.413, p = 0.0000).
Prognostic analysis
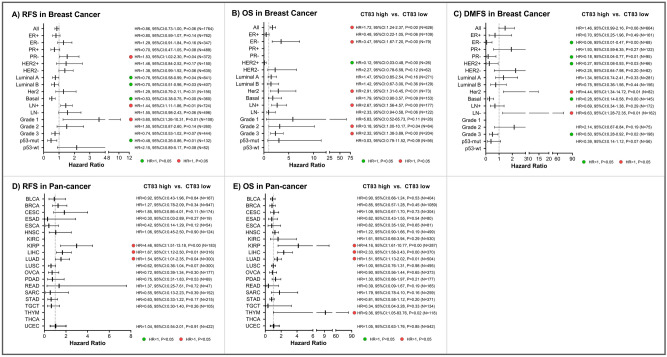
Expression of cancer/testis antigens in cancer is frequently associated with prognosis, both unfavorable prognosis and improved outcomes53–57. Based on this, we assessed the prognostic significance of CT83 in breast cancer as well as in pan-cancer using the KM Plotter database. In breast cancer (Fig. 7A–C), high CT83 expression is statistically associated with poor OS (overall survival; HR = 1.72, 95% CI 1.24–2.37, p = 0.00, N = 626) but not with RFS (relapse-free survival; HR = 0.86, 95% CI 0.73–1.00, p = 0.06, N = 1764) and DMFS (distant metastasis-free survival; HR = 1.46, 95% CI: 0.99–2.16, p = 0.06, N = 664) in the overall analysis. In breast cancer subgroup analysis, high CT83 is considered as an unfavorable risk factor in PR-negative, node-positive, and Grade-1 subgroups for RFS, in ER-negative, HER2 subtype, node-positive, Grade-2, and Grade-3 subgroups for OS, and in HER2 subtype and node-negative subgroups for DMFS. Meanwhile, CT83 high expression is also associated with improved outcomes for RFS in Luminal A, Luminal B, Basal, and p53-mut subgroups, for OS in the HER2-positive subgroup, and for DMFS in ER-negative, PR-negative, HER2-positive, Basal subtype, and Grade-3 subgroups. In pan-cancer analysis (Fig. 7D,E), CT83 high expression is correlated with worse RFS in kidney renal papillary cell carcinoma (KIRP), liver hepatocellular carcinoma (LIHC), and lung adenocarcinoma (LUAD). Similarly, in KIRP, LIHC, LUAD, and Thymoma (THYM), CT83 high expression is associated with shorter OS. In brief, high CT83 mRNA expression is usually unfavorably prognostic for OS in breast cancer, and correlated with worse RFS and OS in KIRP, LIHC, LUAD, and THYM.
Figure 7.
Prognostic significance of CT83 in breast cancer and pan-cancer. The hazard ratios of CT83 (high vs. low) for (A) RFS, (B) OS, and (C) DMFS in breast cancer grouped by different clinical characteristics. The hazard ratios of CT83 (high vs. low) for (D) RFS, (E) OS in different cancers. Abbreviations: RFS, relapse-free survival; OS, overall survival; DMFS, distant metastases-free survival; HR, hazard ratio; 95%CI, 95% confidence interval; P, log-rank test P-value; see https://gdc.cancer.gov/resources-tcga-users/tcga-code-tables/tcga-study-abbreviations for TCGA study abbreviations.
Gene set enrichment analysis of CT83 in breast cancer
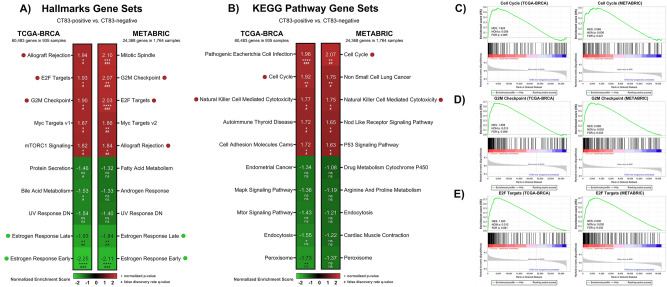
The biological role of CT83 in breast cancer is currently poorly understood. Hence, we performed gene set enrichment analysis (GSEA)35 with the TCGA-BRCA and METABRIC dataset to predict potential functions of CT83 in breast cancer (Fig. 8A,B). By dividing samples into CT83-positive and CT83-negative cohorts, we found that positive CT83 expression is usually correlated with the activation of the Allograft Rejection, E2F Targets, G2M Checkpoint, Cell Cycle, and Natural Killer Cell Mediated Cytotoxicity pathways. Meanwhile, high CT83 expression is also negatively related to the activation of the Estrogen Response Late and Estrogen Response Early pathway. Since deregulation of cell cycle signaling is one of the hallmarks of breast cancer58,59, we then focused on the three cell-cycle-related gene sets that were commonly enriched in both TCGA-BRCA and METABRIC, namely the Cell Cycle, G2M Checkpoint, and E2F Targets pathway (Fig. 8C–E). Based on the enrichment scores of GSEA results, we screened for core enriched genes that were shared in the three gene sets (Figure S3). A total of 16 shared core enriched genes were identified, including CCNB2, CDC20, CDC25A, CDC25B, CDK1, CHEK1, ESPL1, MAD2L1, MCM2, MCM3, MCM5, MCM6, MYC, ORC6, PLK1, and PTTG1. These data suggested that the positive expression of CT83 in breast cancer is correlated with the activation of cell cycle signaling, especially the activation of the 16 shared core enriched genes in Cell Cycle, G2M Checkpoint, and E2F Targets pathways.
Figure 8.
Bioinformatics prediction of CT83 biological functions in breast cancer by GSEA. GSEA of CT83 in breast cancer for (A) hallmark gene sets and (B) KEGG pathway gene sets using the TCGA-BRCA and METABRIC dataset. The red dots represent gene sets positively correlated with CT83 that are common in the TCGA-BRCA and METABRIC dataset with statistical significance, while the green dots were negatively correlated shared gene sets. Details of the three enriched gene sets associated with cell cycle signaling, namely (C) Cell Cycle, (D) G2M Checkpoint, and (E) E2F targets. Abbreviations: NES, normalized enrichment score; NOM p, normalized P-value; FDR q, false discovery rate q-value; *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001; #, q < 0.25; ##, q < 0.05; ###, q < 0.01; ####, q < 0.001.
Next, we examined the expression of the 16 shared core enriched genes in breast cancer using the TCGA-BRCA, METABRIC, and CCLE-BRCA datasets (Figure S4). Compared with other subtypes, expression levels of the 16 genes were usually up-regulated in TNBC samples (Figure S5). In other words, the overexpression of the 16 genes is more frequently observed in CT83-positive breast cancer, while their expression levels are generally low in CT83-negative breast cancer. Furthermore, overexpression of the 16 genes is usually oncogenic by interacting cell cycle signals60–64. Taken together, these data indicated that the aberrant activation of CT83 may interact with one or more of the 16 cell cycle correlated genes to promote tumorigenesis in breast cancer.
Discussion
Cancer/testis (CT) antigens generally refer to a group of proteins with restricted expression in male germ cells in the testis but silenced in other somatic tissues in normal adults38. Extensive data have shown that the abnormal activation of CTAs is frequently observed in cancer tissues65,66, which is one of the driving forces of tumorigenesis67. Currently, more than 200 cancer-related CTAs have been discovered68, and most of them were well documented in the CTDatabase (http://www.cta.lncc.br). Most of CTAs are encoded by genes on chromosome X and thus be classified as CT-X antigens, while those on autosomes are defined as non-X CTAs68. Typically, the expression of CT-X antigens is more testis-restricted than non-X CTAs38. CTAs are of great importance in cancer, especially for cancer immunotherapy65,69. Firstly, CTAs are tumor-specific as their expression is highly restricted in normal tissues and widely distributed in cancer cells65. Secondly, CTAs are immunogenic partly because of the immune-privileged properties of the testis, which can be utilized for the design of anti-cancer vaccines66,70. For example, NY-ESO-1, one of the immunogenic CTAs, can significantly trigger spontaneous, humoral, and cell-mediated immune responses in cancer patients, as can be proved by the presence of serum anti-NY-ESO-1 antibody71 and NY-ESO-1-specific CD8+ T cells69. Moreover, owing to the remarkable resemblances shared by the development of cancer cells and germ cells67, CTAs are considered as markers for cancer stem cells (CSCs)68, and the aberrant expression of CTAs is also associated with the cancer transformation and abnormal differentiation of CSCs72. Lastly, despite the biological functions of the majority of CTAs remain unknown68,73, emerging evidence has highlighted their crucial roles in cellular processes, including cell cycle regulation, cell survival, apoptosis, and signal transduction68,74.
CT83, one of the CT-X antigens, is poorly understood at the current stage. Searching Pubmed with keywords “CT83” and its commonly used synonyms like “CXorf61” and “KK-LC-1,” there were only about 20 studies available regarding CT83 in cancer, mostly in breast cancer, lung cancer, gastric cancer, and hepatocellular cancer. CT83 was first identified as a tumor-specific antigen in a study on lung cancer in 2006, and was then designated as Kita-kyushu lung cancer antigen 1 (KK-LC-1)75. In lung cancer, CT83 is one of the most frequently activated CTAs that its overexpression was often detected in lung cancer tissues and cell lines76–78. Moreover, the abnormal activation of CT83 is associated with advanced tumor stages and unfavorable prognosis in patients with lung cancer77,79, 80. In gastric cancer, CT83 is also frequently overexpressed and related to poor outcomes81,82. Interestingly, the aberrant expression of CT83 in the stomach is associated with Helicobacter pylori infection and atrophic status, and its expression in precancerous gastric lesions was considered as a potential predictor of gastric cancer83–85. Regarding CT83 in breast cancer, several studies have identified and emphasized its TNBC specificity but with a limited sample size37,86,87. Compared with previous findings, our data provided much stronger evidence to support this by analyzing its expression in 3617 breast cancer tissues and 71 breast cancer cell lines. In addition, CT83 was generally considered as an ideal target for immunotherapy in breast cancer40,88 and lung cancer89,90, especially for T-cell-based targeted immunotherapy. Furthermore, there is no risk of intra-family cross-reactivity as CT83 is the only member of its family90. However, apart from its expression patterns, prognostic values, and its significance for immunotherapy, very little is known about the biological functions of CT83, with only one paper available in Pubmed. Chen et al. reported that the abnormal activation of CT83 could promote tumor progression through activating the Notch1/Hes1 signaling in hepatocellular carcinoma91.
There are several web-based platforms that provide comprehensive strategies for gene analysis in cancer, such as the cBioPortal25. Nevertheless, certain limitations impede the use of a single platform for a highly customized analysis. For example, gene expression data in normal tissues and normal blood cells cannot be obtained from the cBioPortal, but can be easily retrieved from the GEPIA2 and the HPA database, respectively. Moreover, the KM Plotter provided more powerful tools to perform survival analysis, as the sample size in KM Plotter is larger and subgroup analysis is ready-to-use. Similarly, the formatted methylation data in the TCGA Portal is more understandable than the raw TCGA data. Therefore, we presented a comprehensive bioinformatics strategy for the analysis of cancer-related genes, including their expression patterns, genetic alternations, epigenetic modifications, correlation with TILS, prognostic significance, and potential biological functions. Briefly, there are several basic criteria of our strategies: (1) included datasets or platforms should be publicly available; (2) raw data should be easily retrieved if possible; (3) analysis could be performed with code-free methods; (4) the whole workflow should be customizable based on specific purposes. We believe these criteria guarantee the comprehensibility and the repeatability of the associated methods and results.
The reliability of our strategies can be supported by many previous laboratory-confirmed findings, as described in the above review of CT83. Moreover, some of our bioinformatics findings will probably facilitate further laboratory studies on CT83. For example, based on the CCLE-BRCA dataset, the majority of breast cancer cell lines were CT83-negative, even some TNBC cell lines (such as MDA-MB-453, HCC1187, and DU4475), which is quite helpful by guiding our choice of appropriate cell models. Similarly, data from TCGA-BRCA suggested that CNVs and mutations of CT83 in breast cancer or pan-cancer are quite rare, thus the two areas should not be the research priority in future studies. Meanwhile, we predicted that the hypomethylation of the region 116,463,019 to 116,463,039 on chromosome X may induce the abnormal activation of CT83 in breast cancer. Several independent studies support the reliability of this prediction. Firstly, the expression of many CTAs is regulated through epigenetic modifications especially DNA methylation, as strong correlations were observed between the methylation levels of their promoters and the expression levels of CTAs in cancer48. Secondly, hypomethylation of CT83 was often detected in CT83-positive breast cancer samples40, and the hypomethylating agent 5-aza-2′-deoxycytidine can active CT83 expression in CT83-negative cells37. However, data regarding the exact locations of DNA methylation that responsible for the abnormal activation of CT83 is not available in previous publications. We will present further evidence on this in our future studies.
Apart from CT83, the other TNBC-specific genes presented in table S1 may also play critical roles in TNBC, such as HORMAD1 and ART3. For example, HORMAD1 overexpression is associated with homologous recombination deficiency in triple-negative breast cancers92. Moreover, Wang et al. reported that focal hypomethylation is correlated with the frequent overexpression of HORMAD1 in basal-like breast cancer93. They also demonstrated that the epigenetic activation of HORMAD1 may reduce the sensitivity of basal-like tumors to Rucaparib treatment. Similarly, Tan et al. reported that overexpression of ART3 in TNBC cells will increase cell proliferation, invasion, and survival in vitro and in vivo, while inverted phenotypes could be observed when ART3 was knocked down94. However, biological functions as well as molecular mechanisms of these genes in breast cancer should be further investigated, as only 8 and 2 papers were available when PubMed was searched with keywords “breast cancer + HORMAD1” or “breast cancer + ART3,” respectively.
Conclusion
In conclusion, we highlighted the significance of CT83 for TNBC and presented a comprehensive bioinformatics strategy for single-gene analysis in cancer. CT83 is frequently overexpressed in TNBC and many cancers but silenced in normal non-testis tissues and blood cells. CNV and mutation are not the main genetic alternations of CT83 in cancer, and CT83 has no significant correlation with TILs in breast cancer or other cancers. Hypomethylation is probably one of the causes of the abnormal activation of CT83 in breast cancer. CT83 is not prognostic in breast cancer but is valuable for the prognosis prediction in KIRP, LIHC, and LUAD. Overexpression of CT83 in breast cancer may be oncogenic by triggering the activation of signaling associated with the cell cycle.
Supplementary Information
Acknowledgements
This study was supported by the China Scholarship Council (CSC). The results shown here are in whole or part based upon data generated by the TCGA Research Network (https://www.cancer.gov/tcga). We greatly appreciated the Cancer Cell Line Encyclopedia, cBioPortal, Ensembl, Expression Atlas, GEPIA, KM Plotter, National Omics Data Encyclopedia, TCGA-Portal, TIMER2.0, TISIDB, and UALCAN for providing high-quality, ready-to-use, open-access data and platforms.
Author contributions
C.C. conceived of the original idea and wrote the manuscript with input from all authors. D.G. and J.H. performed the calculations. Y.G. verified the analytical methods. R.Q. and X.H. verified the numerical results. L.L. supervised the findings of this work. All authors discussed the results and commented on the manuscript.
Data availability
The availability of all datasets generated or analyzed during the current study is described in corresponding "Methods" sections, and other data are available from the corresponding author on reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-91290-4.
References
- 1.Yin L, Duan J-J, Bian X-W, Yu S-C. Triple-negative breast cancer molecular subtyping and treatment progress. BCR. 2020;22:1–13. doi: 10.1186/s13058-020-01296-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Perou CM, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 3.Foulkes WD, Smith IE, Reis-Filho JS. Triple-negative breast cancer. N. Engl. J. Med. 2010;363:1938–1948. doi: 10.1056/NEJMra1001389. [DOI] [PubMed] [Google Scholar]
- 4.Badve S, et al. Basal-like and triple-negative breast cancers: A critical review with an emphasis on the implications for pathologists and oncologists. Mod. Pathol. 2011;24:157–167. doi: 10.1038/modpathol.2010.200. [DOI] [PubMed] [Google Scholar]
- 5.Alluri P, Newman LA. Basal-like and triple-negative breast cancers: Searching for positives among many negatives. Surg. Oncol. Clin. N. Am. 2014;23:567–577. doi: 10.1016/j.soc.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prat A, et al. Molecular characterization of basal-like and non-basal-like triple-negative breast cancer. Oncologist. 2013;18:123. doi: 10.1634/theoncologist.2012-0397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Park JH, Ahn J-H, Kim S-B. How shall we treat early triple-negative breast cancer (TNBC): From the current standard to upcoming immuno-molecular strategies. ESMO Open. 2018;3:e000357. doi: 10.1136/esmoopen-2018-000357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Geenen JJJ, Linn SC, Beijnen JH, Schellens JHM. PARP inhibitors in the treatment of triple-negative breast cancer. Clin. Pharmacokinet. 2018;57:427–437. doi: 10.1007/s40262-017-0587-4. [DOI] [PubMed] [Google Scholar]
- 9.Papadimitriou M, Mountzios G, Papadimitriou CA. The role of PARP inhibition in triple-negative breast cancer: Unraveling the wide spectrum of synthetic lethality. Cancer Treat. Rev. 2018;67:34–44. doi: 10.1016/j.ctrv.2018.04.010. [DOI] [PubMed] [Google Scholar]
- 10.Heimes A-S, Schmidt M. Atezolizumab for the treatment of triple-negative breast cancer. Expert Opin. Investig. Drugs. 2019;28:1–5. doi: 10.1080/13543784.2019.1552255. [DOI] [PubMed] [Google Scholar]
- 11.Kwa MJ, Adams S. Checkpoint inhibitors in triple-negative breast cancer (TNBC): Where to go from here. Cancer. 2018;124:2086–2103. doi: 10.1002/cncr.31272. [DOI] [PubMed] [Google Scholar]
- 12.Vagia E, Mahalingam D, Cristofanilli M. The landscape of targeted therapies in TNBC. Cancers. 2020;12:916. doi: 10.3390/cancers12040916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vikas P, Borcherding N, Zhang W. The clinical promise of immunotherapy in triple-negative breast cancer. Cancer Manag. Res. 2018;10:6823. doi: 10.2147/CMAR.S185176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu J, et al. An integrated TCGA pan-cancer clinical data resource to drive high-quality survival outcome analytics. Cell. 2018;173:400–416. doi: 10.1016/j.cell.2018.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sanchez-Vega F, et al. Oncogenic signaling pathways in the cancer genome atlas. Cell. 2018;173:321–337. doi: 10.1016/j.cell.2018.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ding L, et al. Perspective on oncogenic processes at the end of the beginning of cancer genomics. Cell. 2018;173:305–320. doi: 10.1016/j.cell.2018.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Curtis C, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pereira B, et al. The somatic mutation profiles of 2,433 breast cancers refines their genomic and transcriptomic landscapes. Nat. Commun. 2016;7:1–16. doi: 10.1038/ncomms11479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang Y-Z, et al. Genomic and transcriptomic landscape of triple-negative breast cancers: Subtypes and treatment strategies. Cancer Cell. 2019;35:428–440. doi: 10.1016/j.ccell.2019.02.001. [DOI] [PubMed] [Google Scholar]
- 20.Ghandi M, et al. Next-generation characterization of the cancer cell line encyclopedia. Nature. 2019;569:503–508. doi: 10.1038/s41586-019-1186-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barretina J, et al. The cancer cell line encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li H, et al. The landscape of cancer cell line metabolism. Nat. Med. 2019;25:850–860. doi: 10.1038/s41591-019-0404-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu S, Feng Y, Zhao S. Proteins with evolutionarily hypervariable domains are associated with immune response and better survival of basal-like breast cancer patients. Comput. Struct. Biotechnol. J. 2019;17:430–440. doi: 10.1016/j.csbj.2019.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gao J, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cerami E, et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yates AD, et al. Ensembl 2020. Nucleic Acids Res. 2020;48:D682–D688. doi: 10.1093/nar/gkz1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zerbino DR, et al. Ensembl 2018. Nucleic Acids Res. 2018;46:D754–D761. doi: 10.1093/nar/gkx1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Consortium, G. The genotype-tissue expression (GTEx) project. Nat. Genet.45, 580–585 (2013). [DOI] [PMC free article] [PubMed]
- 29.Tang Z, Kang B, Li C, Chen T, Zhang Z. GEPIA2: An enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019;47:W556–W560. doi: 10.1093/nar/gkz430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li T, et al. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020;48:W509–W514. doi: 10.1093/nar/gkaa407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ma X, Wang Y-W, Zhang MQ, Gazdar AF. DNA methylation data analysis and its application to cancer research. Epigenomics. 2013;5:301–316. doi: 10.2217/epi.13.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weisenberger DJ. Characterizing DNA methylation alterations from the cancer genome atlas. J. Clin. Investig. 2014;124:17–23. doi: 10.1172/JCI69740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ru B, et al. TISIDB: An integrated repository portal for tumor-immune system interactions. Bioinformatics (Oxford, England) 2019;35:4200–4202. doi: 10.1093/bioinformatics/btz210. [DOI] [PubMed] [Google Scholar]
- 34.Györffy B, et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res. Treat. 2010;123:725–731. doi: 10.1007/s10549-009-0674-9. [DOI] [PubMed] [Google Scholar]
- 35.Subramanian A, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen F, Chandrashekar DS, Varambally S, Creighton CJ. Pan-cancer molecular subtypes revealed by mass-spectrometry-based proteomic characterization of more than 500 human cancers. Nat. Commun. 2019;10:1–15. doi: 10.1038/s41467-018-07882-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paret C, et al. CXorf61 is a target for t cell based immunotherapy of triple-negative breast cancer. Oncotarget. 2015;6:25356. doi: 10.18632/oncotarget.4516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Simpson AJG, Caballero OL, Jungbluth A, Chen Y-T, Old LJ. Cancer/testis antigens, gametogenesis and cancer. Nat. Rev. Cancer. 2005;5:615–625. doi: 10.1038/nrc1669. [DOI] [PubMed] [Google Scholar]
- 39.Scanlan MJ, Gure AO, Jungbluth AA, Old LJ, Chen Y-T. Cancer/testis antigens: An expanding family of targets for cancer immunotherapy. Immunol. Rev. 2002;188:22–32. doi: 10.1034/j.1600-065X.2002.18803.x. [DOI] [PubMed] [Google Scholar]
- 40.Yao J, et al. Tumor subtype-specific cancer-testis antigens as potential biomarkers and immunotherapeutic targets for cancers. Cancer Immunol. Res. 2014;2:371–379. doi: 10.1158/2326-6066.CIR-13-0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Matsui A, Ihara T, Suda H, Mikami H, Semba K. Gene amplification: Mechanisms and involvement in cancer. Biomol. Concepts. 2013;4:567–582. doi: 10.1515/bmc-2013-0026. [DOI] [PubMed] [Google Scholar]
- 42.Zhang F, Gu W, Hurles ME, Lupski JR. Copy number variation in human health, disease, and evolution. Annu. Rev. Genom. Hum. Genet. 2009;10:451–481. doi: 10.1146/annurev.genom.9.081307.164217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Goh JY, et al. Chromosome 1q21.3 amplification is a trackable biomarker and actionable target for breast cancer recurrence. Nat. Med. 2017;23:1319. doi: 10.1038/nm.4405. [DOI] [PubMed] [Google Scholar]
- 44.Jia P, Zhao Z. Impacts of somatic mutations on gene expression: An association perspective. Brief. Bioinform. 2017;18:413–425. doi: 10.1093/bib/bbw037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Iranzo J, Martincorena I, Koonin EV. Cancer-mutation network and the number and specificity of driver mutations. Proc. Natl. Acad. Sci. USA. 2018;115:E6010–E6019. doi: 10.1073/pnas.1803155115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Martincorena I, Campbell PJ. Somatic mutation in cancer and normal cells. Science (New York, N.Y.) 2015;349:1483–1489. doi: 10.1126/science.aab4082. [DOI] [PubMed] [Google Scholar]
- 47.Kim R, Kulkarni P, Hannenhalli S. Derepression of cancer/testis antigens in cancer is associated with distinct patterns of DNA hypomethylation. BMC Cancer. 2013;13:1–10. doi: 10.1186/1471-2407-13-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.De Smet C, Lurquin C, Lethé B, Martelange V, Boon T. DNA methylation is the primary silencing mechanism for a set of germ line- and tumor-specific genes with a CpG-rich promoter. Mol. Cell. Biol. 1999;19:7327–7335. doi: 10.1128/MCB.19.11.7327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sigalotti L, et al. Intratumor heterogeneity of cancer/testis antigens expression in human cutaneous melanoma is methylation-regulated and functionally reverted by 5-aza-2′-deoxycytidine. Cancer Res. 2004;64:9167–9171. doi: 10.1158/0008-5472.CAN-04-1442. [DOI] [PubMed] [Google Scholar]
- 50.Rooney MS, Shukla SA, Wu CJ, Getz G, Hacohen N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell. 2015;160:48–61. doi: 10.1016/j.cell.2014.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Robbins PF, et al. Tumor regression in patients with metastatic synovial cell sarcoma and melanoma using genetically engineered lymphocytes reactive with NY-ESO-1. J. Clin. Oncol. 2011;29:917. doi: 10.1200/JCO.2010.32.2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li B, et al. Comprehensive analyses of tumor immunity: Implications for cancer immunotherapy. Genome Biol. 2016;17:1–16. doi: 10.1186/s13059-016-1028-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van Duin M, et al. Cancer testis antigens in newly diagnosed and relapse multiple myeloma: Prognostic markers and potential targets for immunotherapy. Haematologica. 2011;96:1662. doi: 10.3324/haematol.2010.037978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Svobodová S, et al. Cancer-testis antigen expression in primary cutaneous melanoma has independent prognostic value comparable to that of breslow thickness, ulceration and mitotic rate. Eur. J. Cancer (Oxford, England : 1990) 2011;47:460–469. doi: 10.1016/j.ejca.2010.09.042. [DOI] [PubMed] [Google Scholar]
- 55.Freitas M, et al. Expression of cancer/testis antigens is correlated with improved survival in glioblastoma. Oncotarget. 2013;4:636. doi: 10.18632/oncotarget.950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhou X, et al. Heterogeneous expression of CT10, CT45 and GAGE7 antigens and their prognostic significance in human breast carcinoma. Jpn. J. Clin. Oncol. 2013;43:243–250. doi: 10.1093/jjco/hys236. [DOI] [PubMed] [Google Scholar]
- 57.von Boehmer L, et al. MAGE-C2/CT10 protein expression is an independent predictor of recurrence in prostate cancer. PLoS ONE. 2011;6:e21366. doi: 10.1371/journal.pone.0021366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thu KL, Soria-Bretones I, Mak TW, Cescon DW. Targeting the cell cycle in breast cancer: Towards the next phase. Cell Cycle (Georgetown, Tex.) 2018;17:1871–1885. doi: 10.1080/15384101.2018.1502567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 60.Shen T, Huang S. The role of Cdc25A in the regulation of cell proliferation and apoptosis. Anticancer Agents Med. Chem. 2012;12:631–639. doi: 10.2174/187152012800617678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xie B, Wang S, Jiang N, Li JJ. Cyclin B1/CDK1-regulated mitochondrial bioenergetics in cell cycle progression and tumor resistance. Cancer Lett. 2019;443:56–66. doi: 10.1016/j.canlet.2018.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang Y, et al. MCM family in gastrointestinal cancer and other malignancies: From functional characterization to clinical implication. Biochim. Biophys. acta. 2020;1874:188415. doi: 10.1016/j.bbcan.2020.188415. [DOI] [PubMed] [Google Scholar]
- 63.Cunningham CE, et al. The CINs of polo-like kinase 1 in cancer. Cancers. 2020;12:2953. doi: 10.3390/cancers12102953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Boutros R, Lobjois V, Ducommun B. CDC25 phosphatases in cancer cells: Key players? Good targets? Nat. Rev. Cancer. 2007;7:495–507. doi: 10.1038/nrc2169. [DOI] [PubMed] [Google Scholar]
- 65.Suri A. Cancer testis antigens—Their importance in immunotherapy and in the early detection of cancer. Expert Opin. Biol. Ther. 2006;6:379–389. doi: 10.1517/14712598.6.4.379. [DOI] [PubMed] [Google Scholar]
- 66.Wei X, et al. Cancer-testis antigen peptide vaccine for cancer immunotherapy: Progress and prospects. Transl. Oncol. 2019;12:733–738. doi: 10.1016/j.tranon.2019.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Old LJ. Cancer/testis (CT) antigens—A new link between gametogenesis and cancer. Cancer Immun. 2001;1:1. [PubMed] [Google Scholar]
- 68.Salmaninejad A, et al. Cancer/testis antigens: Expression, regulation, tumor invasion, and use in immunotherapy of cancers. Immunol. Invest. 2016;45:619–640. doi: 10.1080/08820139.2016.1197241. [DOI] [PubMed] [Google Scholar]
- 69.Nishikawa H, et al. Cancer/testis antigens are novel targets of immunotherapy for adult t-cell leukemia/lymphoma. Blood. 2012;119:3097–3104. doi: 10.1182/blood-2011-09-379982. [DOI] [PubMed] [Google Scholar]
- 70.Meinhardt A, Hedger MP. Immunological, paracrine and endocrine aspects of testicular immune privilege. Mol. Cell. Endocrinol. 2011;335:60–68. doi: 10.1016/j.mce.2010.03.022. [DOI] [PubMed] [Google Scholar]
- 71.Maio M, et al. Analysis of cancer/testis antigens in sporadic medullary thyroid carcinoma: Expression and humoral response to NY-ESO-1. J. Clin. Endocrinol. Metab. 2003;88:748–754. doi: 10.1210/jc.2002-020830. [DOI] [PubMed] [Google Scholar]
- 72.Gordeeva O. Cancer-testis antigens: Unique cancer stem cell biomarkers and targets for cancer therapy. Semin. Cancer Biol. 2018;53:75–89. doi: 10.1016/j.semcancer.2018.08.006. [DOI] [PubMed] [Google Scholar]
- 73.Al-Khadairi G, Decock J. Cancer testis antigens and immunotherapy: Where do we stand in the targeting of PRAME? Cancers. 2019;11:984. doi: 10.3390/cancers11070984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cheng Y-H, Wong EW, Cheng CY. Cancer/testis (CT) antigens, carcinogenesis and spermatogenesis. Spermatogenesis. 2011;1:209–220. doi: 10.4161/spmg.1.3.17990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fukuyama T, et al. Identification of a new cancer/germline gene, KK-LC-1, encoding an antigen recognized by autologous CTL induced on human lung adenocarcinoma. Cancer Res. 2006;66:4922–4928. doi: 10.1158/0008-5472.CAN-05-3840. [DOI] [PubMed] [Google Scholar]
- 76.Jin S, et al. Establishment of cancer/testis antigen profiling based on clinicopathological characteristics in resected pathological stage III non-small cell lung cancer. Cancer Manag. Res. 2018;10:2031. doi: 10.2147/CMAR.S164043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cohen AS, et al. Cell-surface marker discovery for lung cancer. Oncotarget. 2017;8:113373. doi: 10.18632/oncotarget.23009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shigematsu Y, et al. Clinical significance of cancer/testis antigens expression in patients with non-small cell lung cancer. Lung Cancer (Amsterdam, Netherlands) 2010;68:105–110. doi: 10.1016/j.lungcan.2009.05.010. [DOI] [PubMed] [Google Scholar]
- 79.Baba T, et al. Cancer/testis antigen expression as a predictor for epidermal growth factor receptor mutation and prognosis in lung adenocarcinoma. Eur. J. Cardiothorac. Surg. 2013;43:759–764. doi: 10.1093/ejcts/ezs426. [DOI] [PubMed] [Google Scholar]
- 80.Jin S, et al. Cancer/testis antigens (CTAs) expression in resected lung cancer. Onco Targets Ther. 2018;11:4491. doi: 10.2147/OTT.S159491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Shida A, et al. Frequent high expression of Kita-Kyushu lung cancer antigen-1 (KK-LC-1) in gastric cancer. Anticancer Res. 2015;35:3575–3579. [PubMed] [Google Scholar]
- 82.Chen J, et al. An innovative prognostic model based on four genes in Asian patient with gastric cancer. Cancer Res. Treat. 2021;53:148. doi: 10.4143/crt.2020.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fukuyama T, et al. Expression of KK-LC-1, a cancer/testis antigen, at non-tumour sites of the stomach carrying a tumour. Sci. Rep. 2018;8:1–7. doi: 10.1038/s41598-018-24514-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Shida A, et al. Cancer/testis antigen, kita-kyushu lung cancer antigen-1 and ABCD stratification for diagnosing gastric cancers. World J. Gastroenterol. 2020;26:424. doi: 10.3748/wjg.v26.i4.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Futawatari N, et al. Early gastric cancer frequently has high expression of KK-LC-1, a cancer-testis antigen. World J. Gastroenterol. 2017;23:8200. doi: 10.3748/wjg.v23.i46.8200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kondo Y, et al. Detection of KK-LC-1 protein, a cancer/testis antigen, in patients with breast cancer. Anticancer Res. 2018;38:5923–5928. doi: 10.21873/anticanres.12937. [DOI] [PubMed] [Google Scholar]
- 87.Zhong G, Lou W, Shen Q, Yu K, Zheng Y. Identification of key genes as potential biomarkers for triple-negative breast cancer using integrating genomics analysis. Mol. Med. Rep. 2020;21:557–566. doi: 10.3892/mmr.2019.10867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kaufmann J, Wentzensen N, Brinker TJ, Grabe N. Large-scale in-silico identification of a tumor-specific antigen pool for targeted immunotherapy in triple-negative breast cancer. Oncotarget. 2019;10:2515. doi: 10.18632/oncotarget.26808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ichiki Y, et al. Development of adoptive immunotherapy with KK-LC-1-specific TCR-transduced γδt cells against lung cancer cells. Cancer Sci. 2020;111:4021. doi: 10.1111/cas.14612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Marcinkowski B, et al. Cancer targeting by TCR gene-engineered t cells directed against Kita-Kyushu lung cancer antigen-1. J. Immunother. Cancer. 2019;7:1–9. doi: 10.1186/s40425-019-0678-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chen Z, et al. Hypomethylation-mediated activation of cancer/testis antigen KK-LC-1 facilitates hepatocellular carcinoma progression through activating the Notch1/Hes1 signalling. Cell Prolif. 2019;52:e12581. doi: 10.1111/cpr.12581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Watkins J, et al. Genomic complexity profiling reveals that HORMAD1 overexpression contributes to homologous recombination deficiency in triple-negative breast cancers. Cancer Discov. 2015;5:488–505. doi: 10.1158/2159-8290.CD-14-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wang X, et al. Epigenetic activation of HORMAD1 in basal-like breast cancer: Role in rucaparib sensitivity. Oncotarget. 2018;9:30115. doi: 10.18632/oncotarget.25728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Tan L, et al. ART3 regulates triple-negative breast cancer cell function via activation of akt and ERK pathways. Oncotarget. 2016;7:46589. doi: 10.18632/oncotarget.10306. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The availability of all datasets generated or analyzed during the current study is described in corresponding "Methods" sections, and other data are available from the corresponding author on reasonable request.