Abstract
Mitochondrial DNA (mtDNA) quality and quantity relate to two hallmarks of aging—genomic instability and mitochondrial dysfunction. Physical performance relies on mitochondrial integrity and declines with age, yet the interactions between mtDNA quantity, quality, and physical performance are unclear. Using a validated digital PCR assay specific for mtDNA deletions, we tested the hypothesis that skeletal muscle mtDNA deletion mutation frequency (i.e., a measure of mtDNA quality) or mtDNA copy number predicts physical performance in older adults. Total DNA was isolated from vastus lateralis muscle biopsies and used to quantitate mtDNA copy number and mtDNA deletion frequency by digital PCR. The biopsies were obtained from a cross-sectional cohort of 53 adults aged 50 to 86 years. Before the biopsy procedure, physical performance measurements were collected, including VO2max, modified physical performance test score, 6-min walk distance, gait speed, grip strength, and total lean and leg mass. Linear regression models were used to evaluate the relationships between age, sex, and the outcomes. We found that mtDNA deletion mutation frequency increased exponentially with advancing age. On average from ages 50 to 86, deletion frequency increased from 0.008 to 0.15%, an 18-fold increase. Females may have lower deletion frequencies than males at older ages. We also measured declines in VO2max and mtDNA copy number with age in both sexes. The mtDNA deletion frequency measured from single skeletal muscle biopsies predicted 13.3% of the variation in VO2max. Copy number explained 22.6% of the variation in mtDNA deletion frequency and 10.4% of the lean mass variation. We found predictive relationships between age, mtDNA deletion mutation frequency, mtDNA copy number, and physical performance. These data are consistent with a role for mitochondrial function and genome integrity in maintaining physical performance with age. Analyses of mtDNA quality and quantity in larger cohorts and longitudinal studies could extend our understanding of the importance of mitochondrial DNA in human aging and longevity.
Keywords: Skeletal muscle, Aging, Mitochondria, Mitochondrial DNA, Mutation, Physical performance
Introduction
The molecular alterations that underpin aging are thought to be multifactorial, encompassing at least nine domains of recognized cellular dysfunction [1]. Among these aging hallmarks, two factors—mitochondrial dysfunction and genomic instability—converge on the mitochondrial genome. Age-induced loss of mitochondrial genomes and increasing mtDNA deletion mutations are indicative of genomic instability and result in mitochondrial respiratory dysfunction. There is considerable evidence of age-induced decreases in mitochondrial DNA (mtDNA) integrity and mitochondrial function in many tissues [2, 3]. It is unclear whether mitochondrial DNA alterations manifest as broader physical performance changes with age.
Cells contain multiple copies of mtDNA, and, with advancing age, mtDNA deletion mutations arise from replication errors [4]. In human skeletal muscle, a deletion mutation event typically removes 4–12 kb of the 16.3-kb mtDNA sequence [5]. The resulting mutated mtDNA lacks multiple genes that encode tRNAs and protein subunits necessary for oxidative phosphorylation. With time, the deletion-containing “selfish” genome clonally accumulates within the individual muscle fiber [6]. When the intracellular deletion abundance (i.e., heteroplasmy) exceeds 90% of the total mitochondrial genomes, electron transport chain (ETC) function is disrupted, and cells lose cytochrome c oxidase activity prior to undergoing cell death [7–9]. In 36-month-old rat quadriceps, ~ 80% of the myofibers positive for markers of apoptosis and/or necrosis were ETC-deficient [8]. A similar series of events occurs in humans with mtDNA deletion-containing genetic diseases [9]. The sequential process of mtDNA deletion formation, clonal accumulation, respiratory chain deficiency, and cell death reiterates in an increasing number of muscle fibers with age. mtDNA deletion-containing, ETC-deficient cells have been detected in most aged human tissues including brain, heart, kidney, and skeletal muscle, where they contribute to the cellular phenotypes and tissue degeneration of aging [10–23].
mtDNA quantity, reflected in copy number, declines with age in human skeletal muscle [24, 25]. This decrease occurs in individual muscle fibers [26] and is of greater magnitude in type IIb muscle fibers, which are lost preferentially with age [27]. In mouse quadriceps muscle, mtDNA copy number per square micron of muscle fiber area decreases 1.8-, 1.9-, and 3.2-fold in type I, IIa, and IIb fibers, respectively, between 6 and 24 months of age [26]. The importance of these declines in mtDNA copy number is unclear because mtDNA copy number correlates weakly with mitochondrial function or mitochondrial content [28]. Lower mtDNA copy number is unstable as it may facilitate the segregation of deletion mutants in post-mitotic tissues such as skeletal muscle [29].
Physical performance decreases with age across numerous domains that involve skeletal muscle [30]. For example, whole-body maximum oxygen consumption (VO2max) decreases 3 to 6% per decade for the third and fourth decades of life, but after age 70, the rate accelerates to a > 20% loss per decade [31, 32]. Moreover, physical performance, including VO2max, predicts survival in older adults [33, 34]. VO2max as a performance measure integrates maximal heart rate and stroke volume, blood flow, compliance of heart muscle fibers and arterial walls, peripheral oxygen extraction, as well as mitochondrial oxygen utilization in skeletal muscle [31, 32, 35–37]. Fleg and Lakatta [38] showed that decreased whole-body lean mass contributed to the decline in VO2max with advancing age. Physical performance also is dependent upon mitochondria due to the centrality of this organelle in metabolism, respiration, muscle mass, and oxidative capacity [39, 40]. Individuals with mitochondrial mutations exemplify the link between mitochondria and VO2max [41]. The affected individuals are exercise intolerant and have lower muscle mass [42]. Measurements of mtDNA copy number and deletion frequency have been examined with age in relation to performance measures such as VO2max [25]. In the study by Short et al., mtDNA copy number correlated with age (r = − 0.62) and VO2max (r = 0.48). Deletion mutations were not detected.
The quantitation of deletion mutations in muscle biopsies requires sensitive and specific assays that account for the unique characteristics of these mutations. While heteroplasmy refers to the percent of mutations in single cells, we define deletion frequency as the ratio between deletion-bearing mtDNA molecules and all mtDNA molecules in tissue homogenates. High specificity is required as wild-type mtDNA contains identical PCR priming sites as those present in mtDNA deletions. Moreover, as most mtDNA deletion events contain unique breakpoints, the assay needs to detect a heterogeneous class of molecules. Using a digital PCR assay that accounts for the unique characteristics of mtDNA deletions, we observed, in human skeletal muscle, that mtDNA deletion mutations increase exponentially ~ 98-fold between 20 and 80 years of age [24]. These findings suggest that mtDNA deletion frequency predicts biological age.
In this study, we used digital PCR quantitation of mtDNA deletions and copy number to examine the relationships between skeletal muscle mtDNA copy number, mtDNA deletion mutation frequency, and physical performance measures in non-diseased older adults. Because physical performance measures reflect a myriad of physiological processes across numerous organ systems, we hypothesized that mtDNA deletion frequency measured from a single muscle biopsy would not correlate with physical performance measures. Here, we show that the converse is true; mtDNA deletion frequency predicts the decline in physical performance with age.
Materials and methods
Personnel analyzing mtDNA quality and quantity were blinded to subject age and performance characteristics. Researchers were unblinded after the molecular analyses were completed and reported.
Human subjects
Adults 50–89 years of age were recruited from the Baltimore, MD, area to participate in studies examining metabolic responses to exercise training. Participants were screened by medical history, physical examination, fasting blood chemistry, and graded maximal exercise test. All participants were current non-smokers (no history of smoking for > 2 years), sedentary (<20 min of aerobic exercise two times per week), and had no history or evidence of cardiovascular, liver, kidney, or lung disease. Participants had asymptomatic graded treadmill exercise tests. The women in the study had not menstruated for at least 1 year. The resultant sample for this study was 53 men and women for whom muscle biopsy samples were available. A subset of 13 participants enrolled in one study did not undergo tests of physical performance; thus, 40 subjects comprise the sample for those variables. The research protocols were approved by the Institutional Review Board at the University of Maryland School of Medicine, and all participants provided written informed consent.
Physical performance measures
Subjects underwent tests of physical performance, including the modified physical performance test (MPPT), 6-min walk distance (6-MWD), fast gait speed, and grip strength. Prior to testing, all individuals were instructed to wear comfortable clothes and shoes and were notified they would be performing walking and balance tests. The MPPT is a nine-item test that includes standing balance, repeated chair stands, gait speed, timed ascent of one flight of stairs, ability to ascend and descend four flights of stairs, donning and doffing a coat, picking up a penny, placing a book on a shelf, and safely turning 360° [43]. The scores range from 0 to 4 for each item, and total scores of < 32 points indicate at least a mild mobility limitation. Chair stands, standing balance tests, and fast gait speed were performed once, with the time for each test recorded and scored. Times for stair ascent–descent were calculated as the average of three attempts; the same standard set of stairs was used for all participants. For 6-MWD, participants were asked to walk as far as possible in a 100-ft corridor for 6 min, and total distance was recorded. Grip strength of the dominant hand was measured with a hydraulic handheld dynamometer (Jamar Inc., Bolingbrook, IL). Participants stood with arms at their side and elbows slightly bent, squeezing the dynamometer with as much force as possible; the average of triplicate measures was recorded for analysis. Gait speed and 6-MWD were not available for one subject due to technical issues (Table 2).
Table 2.
Summary of molecular and performance characteristics
| N | Women | Men | Total | p-value | |
|---|---|---|---|---|---|
| mtDNA deletion frequency (e-04) | 53 | 3.48 (8.02) | 5.83 (13.30) | 3.90 (8.50) | 0.935 |
| mtDNA copy number | 53 | 3853 (797.6) | 3781 (1898) | 3814 (1217) | 0.725 |
| VO2max (ml/kg/min) | 53 | 21.90 ± 0.73 | 25.80 ± 1.10 | 23.45 ± 0.67 | 0.003 |
| MPPT score | 40 | 35.00 (1.50) | 35.00 (1.00) | 35.00 (1.00) | 0.954* |
| 6-min walk distance (ft) | 39 | 1611 ± 39 | 1820 ± 49 | 1691 ± 34 | 0.002 |
| Gait speed (m/s) | 40 | 1.32 ± 0.07 | 1.40 ± 0.08 | 1.35 ± 0.05 | 0.422 |
| Grip strength (kg) | 39 | 23.42 ± 1.16 | 38.46 ± 3.46 | 29.59 ± 1.96 | < 0.001 |
| Total lean mass (kg) | 52 | 45.17 ± 1.09 | 59.83 ± 1.26 | 51.09 ± 1.30 | < 0.001 |
| Right leg lean mass (kg) | 52 | 7.90 ± 0.25 | 10.20 ± 0.28 | 8.83 ± 0.24 | < 0.001 |
Results are presented by means ± SEs or median (IQR)
p-values are for the difference between men and women by using Student’s t-test or Mann–Whitney test, as appropriate
IQR interquartile range, MPPT modified physical performance test, mtDNA mitochondrial DNA, SE standard error, VO2max maximal oxygen uptake
*Continuity correction was applied when running Mann–Whitney test
Maximal oxygen consumption (VO2max)
VO2max was measured by indirect calorimetry (Quark, Cosmed USA, Chicago, IL) during a maximal graded treadmill exercise test. Participants were asked to walk at a constant, comfortable speed through the test with the grade initially set to 0%, increasing every 2 min until VO2max was reached as verified by standard physiological criteria (respiratory exchange ratio > 1.10 or a plateau in VO2 with an increase in workload).
Body mass index (BMI) and body composition
Height was measured with a stadiometer, and weight was measured with a standard laboratory scale. Body mass index (BMI) was calculated as weight (kg) divided by height (m) squared. Total and regional body composition, including total and leg lean mass, were measured by dual-energy X-ray absorptiometry (DXA) (Prodigy or iDXA, LUNAR Radiation, Madison, WI). Data for one subject were not available due to technical issues.
Muscle biopsy
Vastus lateralis muscle biopsies were obtained approximately 12–13 cm above the patella on the anterolateral aspect of the right thigh using a Bergstrom needle (Stille, Solna, Sweden) as previously described [44]. Muscle samples were embedded and rapidly frozen in optimal cutting temperature/gum tragacanth mixture, then stored at −80 °C until analyzed.
Animal tissues
Twenty-two-month-old quadriceps muscle tissue samples from male and female mice were obtained from the NIA Interventions Testing Program. All mouse samples were flash-frozen in liquid nitrogen and stored at – 80 °C until DNA isolation.
DNA isolation and quality control
De-identified human muscle biopsy samples were obtained from the quadriceps muscle of 53 male and female subjects ranging in age from 50 to 86 years. Muscle samples were powdered under liquid nitrogen using a mortar and pestle. Approximately 25 mg of the powdered muscle was used for DNA isolation, performed by proteinase K and RNase A digestion, phenol/chloroform extraction, and ethanol precipitation, as previously described [24]. Quality control of the DNA was performed via NanoDrop (NanoDrop 2000, Thermo Scientific; Waltham, MA), Qubit (2.0, Invitrogen; Carlsbad, CA), and TapeStation (2200, Agilent; Santa Clara, CA).
mtDNA copy number and deletion frequency quantitation
A 5′ nuclease cleavage assay and droplet digital PCR (ddPCR) were used to quantitate copy numbers for nuclear DNA, total mtDNA, and mtDNA deletions with specific primer/probe sets and cycling conditions for each as previously described [24]. Samples were diluted to the manufacturer’s recommended target range (1 to 5000 copies per microliter) in 25-μl reactions using BioRad ddPCR Supermix for Probes (BioRad; Hercules, CA). Final primer and probe concentrations were 900 nM and 250 nM, respectively. Reactions were partitioned into droplets using a BioRad QX200 droplet generator (BioRad; Hercules, CA) prior to thermocycling. Digital PCR cycling conditions for nDNA and mtDNA copy number included Taq-polymerase activation at 95 °C for 10 min, followed by 40 cycles of denaturation at 94 °C for 30 s and annealing/extension at 60 °C for 2 min. Droplet fluorescence was then read using a BioRad QX200 droplet reader. Target copy numbers per microliter were determined using BioRad QuantaSoft Regulatory Edition Software (version 1.7, BioRad; Hercules, CA). Direct quantitation of mtDNA major arc deletions by ddPCR used cycling conditions of Taq-polymerase activation at 95 °C for 10 min, followed by 60 cycles of denaturation at 94 °C for 15 s, annealing at 55 °C for 1 min, and extension at 72 °C for 6 min. Deletion assays on mouse skeletal muscle DNA used mouse-specific primer/probe sets as previously described [24], with the same cycling conditions as described for the human mtDNA major arc deletion detection.
Statistical analysis
We applied log transformation on mtDNA deletion frequency and Box–Cox transformation [45] on mtDNA copy number to reduce skewness. Both variables were normalized using the min–max method. Participant characteristics were presented as mean ± standard error of the mean (SEM) for continuous variables and counts (proportions) for categorical variables. Student’s t-test for two independent samples, the Mann–Whitney test, or a chi-squared test was used to evaluate the differences in the characteristics between males and females, as appropriate.
Linear regression models were used to evaluate the relationships between age, sex, and the outcomes of interest. The interaction between age and sex was included in the models to evaluate the potential effect of sex on age-related changes. Pearson correlations between age, mtDNA deletion frequency, mtDNA copy number, VO2max, MPPT score, 6-MWD, gait speed, grip strength of the dominant hand, total lean mass, and right lean mass were assessed. The mtDNA deletion frequency (or mtDNA copy number)–related changes in the outcome of interest (i.e., VO2max, MPPT score, 6-MWD, gait speed, grip strength of the dominant hand, total lean mass, or right lean mass) were also evaluated by using linear regression in sequential models. The first model was a model without adjustment for other covariates. The second model was adjusted for age and sex. p-values were two-tailed and were considered statistically significant at values < 0.05. All analyses were performed using R statistical software version 3.6.1 (R Core Team, 2019).
Results
Participant characteristics
We collected quadriceps muscle biopsies and performance measures from 53 subjects from 50 to 86 years of age (Table 1). Of the 53 participants, 60% were women and 40% were men. Average ages were similar between the sexes. There were racial differences by sex in the subject population. Seventy-five percent of the women were African American, while 62% of the men were Caucasian. There were negligible differences in body weight and BMI between women and men.
Table 1.
Participant characteristics
| Women | Men | Total | p-value | |
|---|---|---|---|---|
| N | 32 | 21 | 53 | NA |
| Age (year) | 63.97 ± 1.28 | 65.29 ± 1.69 | 64.49 ± 1.01 | 0.531 |
| Race | 0.007 | |||
| White | 8 | 13 | 21 | |
| African American | 24 | 8 | 32 | |
| Weight (kg) | 83.41 ± 3.31 | 91.03 ± 2.79 | 86.43 ± 2.32 | 0.109 |
| BMI (kg/m2) | 31.37 ± 1.21 | 29.39 ± 0.83 | 30.58 ± 0.81 | 0.233 |
Results are presented by means ± SEs or counts
p-values are for the difference between men and women by using Student’s t-test or chi-squared test, as appropriate
BMI body mass index, NA not applicable, SE standard error
Molecular and physical performance of the study participants
mtDNA deletion frequency ranged from 2.95e-06 to 1.74e-02, and mtDNA copy number ranged from 1381 to 7139 copies per diploid nucleus (Table 2). Without considering age, there was no statistically significant difference in the deletion frequency or copy number between men and women. Lean mass (total and right leg) and physical performance measures (VO2max, 6-MWD, and grip strength) were lower in women.
Relationships between age and molecular or physical performance measures
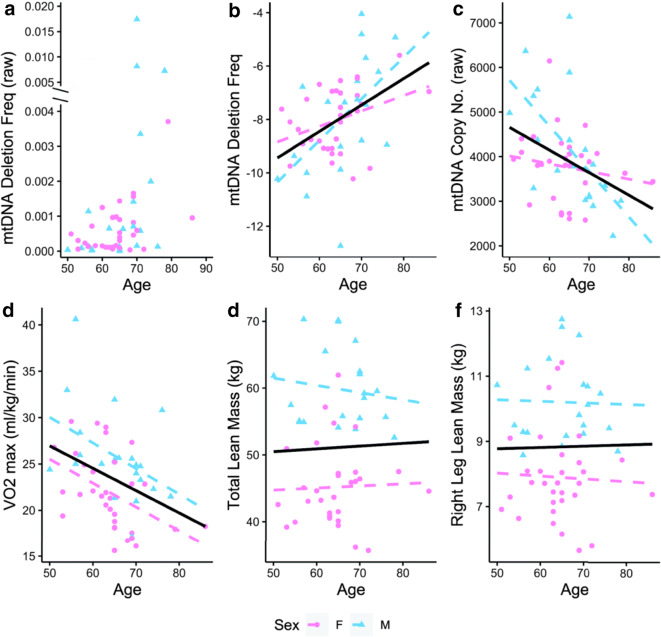
mtDNA deletion frequency increases exponentially with age (Fig. 1a). To account for this non-linear increase and facilitate comparisons with the other linear variables, we log transformed and normalized (min–max) the mtDNA deletion frequency values (Fig. 1b). mtDNA deletion frequency increased with age (r = 0.44, p = 0.001). On average, mtDNA deletion frequency increased 18-fold in individuals from 50 to 86 years of age. mtDNA deletion frequency increased by 10.4% per year on average. The regression equation is loge(mtDNA deletion frequency) = − 14.39 + 0.099 * (age).
Fig. 1.
The relationships between age and molecular and performance characteristics. The solid black line denotes the overall linear regression. Dashed lines denote the sex-specific linear regressions. Individual data points are plotted for each subject. a mtDNA deletion frequency (raw values, truncated axis) vs. age. b mtDNA deletion frequency (transformed, normalized) vs. age. c mtDNA copy number (raw values) vs. age. d VO2max vs. age. e Total lean mass vs. age. f Right leg lean mass vs. age
mtDNA copy number decreased with age (r = − 0.39, p = 0.004) (Fig. 1c). Among the physical performance measures, VO2max declined with age (r = − 0.37, p = 0.007) (Fig. 1d). There was no detectable difference in average lean mass in this cohort as a function of age as assessed by DXA (Fig. 1e, f). While sex differences were not a primary aim of the study, the effect of age on mtDNA copy number was different between sexes (p = 0.011) (Fig. 1c). The effect of age on deletion frequency may differ between sexes (p = 0.089) (Fig. 1b), although this analysis is underpowered, as discussed below. There were sex differences in VO2max and lean mass (Table 2); however, these did not interact with age.
Relationships between molecular and physical performance measures
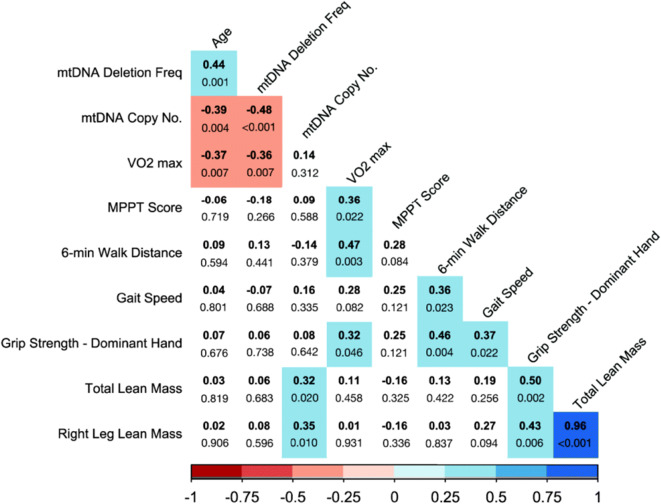
To explore the importance of mtDNA deletion frequency and mtDNA copy number in performance outcomes, we examined Pearson correlations between the variables (Fig. 2). There is a negative correlation between mtDNA deletion frequency and VO2max (r = − 0.36, p = 0.007). Individuals with a higher deletion frequency had a lower VO2max (Fig. 3a). mtDNA deletion frequency was also negatively correlated with mtDNA copy number (r = − 0.48, p < 0.001). Individuals with higher mtDNA copy numbers had lower deletion frequencies (Fig. 3b). There may be a sex dependence to the relationship between deletion frequency and copy number (p = 0.098) (Fig. 3b). mtDNA copy number was positively correlated with both total lean mass (r = 0.32, p = 0.020) (Fig. 3c) and right leg lean mass (r = 0.35, p = 0.010) (data not shown). Total lean mass and right leg lean mass were highly correlated (r = 0.96, p < 0.001).
Fig. 2.
Relationships between molecular and physiologic measures. Pearson correlation coefficients are bolded. Exact p-values are shown for each pairwise comparison. Significant relationships (p < 0.05) are highlighted in color. The strength and direction of significant correlations are indicated by the color and tint
Fig. 3.
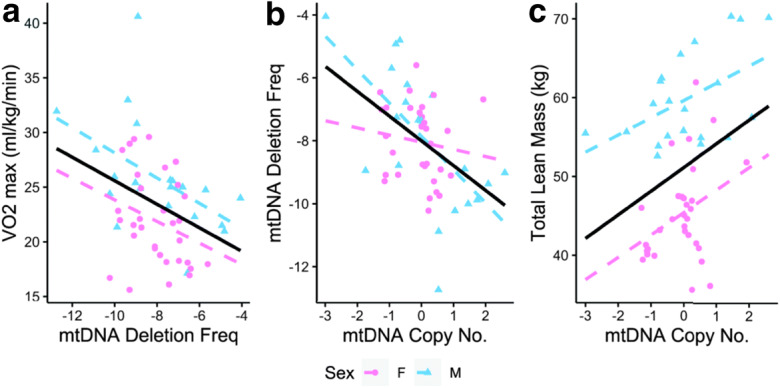
Significant relationships between molecular measures and physical performance measures. The solid black line denotes the overall linear regression. Dashed lines denote the sex-specific linear regressions. Individual data points are plotted for each subject (pink = females, blue = males). a VO2max vs. mtDNA deletion frequency (transformed, normalized). b mtDNA deletion frequency (transformed, normalized) vs. mtDNA copy number (transformed, normalized). c Total lean mass vs. mtDNA copy number (transformed, normalized)
Linear regression modeling of mtDNA deletion frequency and mtDNA copy number
We evaluated the relative contributions of age, sex, and mtDNA deletion frequency to VO2max variation. Together, mtDNA deletion frequency, age, and sex predicted 36.6% of the variation in VO2max. mtDNA deletion frequency alone predicted 13.3% of the variation in VO2max (p = 0.007). However, after accounting for age and sex, the contribution of mtDNA deletion frequency was reduced to 4.7% (p = 0.061).
In similar analyses, mtDNA copy number, age, and sex predicted 30.6% of the mtDNA deletion frequency variation. mtDNA copy number contributes 22.6% of the variation in mtDNA deletion frequency (p < 0.001). After accounting for age and sex, mtDNA copy number predicted 10.9% (p = 0.008) of the variation in deletion frequency. mtDNA copy number, age, and sex predicted 67.6% of the variation in lean mass. mtDNA copy number contributes 10.4% of the variation of lean mass (p = 0.020). mtDNA copy number predicted 7.2% (p = 0.002) of the variation in lean mass after accounting for age and sex.
Sex differences in mtDNA deletion frequency in 22-month-old mouse skeletal muscle
To examine further the impact of sex on mtDNA deletion frequency, we measured deletion frequency in quadriceps muscle from 22-month UM-HET3 mice that are genetically heterogeneous. In this mouse strain, we observed a significantly lower mtDNA deletion frequency in female (0.0027%) versus male (0.0146%) mice (p < 0.001) (Fig. 4).
Fig. 4.
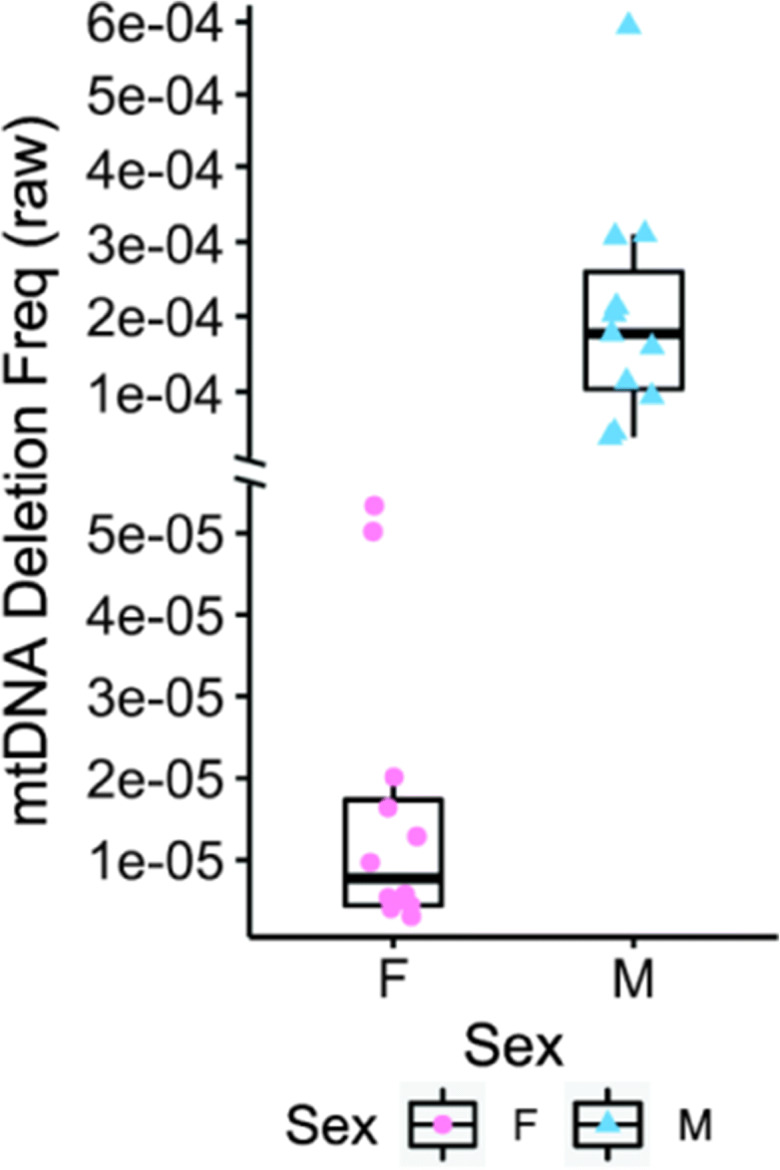
MtDNA deletion frequency is lower in female mice compared to male mice at 22 months of age. Individual data points are plotted for each mouse. The y-axis is discontinuous to facilitate plotting, as there is a large difference between means. Box plots denote median, IQR, and raw values
Discussion
The cellular impact of age-induced mtDNA deletions includes ETC dysfunction and cell death. The data in this study extend the impact of mtDNA deletions to physical performance by showing that increased mtDNA deletion frequency predicts decreased VO2max. Similarly, mtDNA copy number predicts the decline in lean mass and the increase in mtDNA deletion frequency.
mtDNA deletion mutations have been observed to increase with age in many tissues and species [5, 10–23]; however, prior approaches lacked sufficient sensitivity and specificity to enable comparison with physiological outcomes. The increase in deletion frequency found in this study is consistent with the exponential increase we observed (~100-fold) in a separate smaller cohort of adults aged 20 to 80 [24]. The association between age and mtDNA deletion frequency was not statistically different between the 20–80-year-old cohort in our previous study and the cohort in this study. Much of the increase in deletion frequency occurs between 50 and 80 years of age, a finding that helps to target appropriate ages for deploying this assay. A previous study examining physical performance failed to detect deletion mutations using a semi-quantitative gel electrophoresis approach with PCR primer sets that cannot detect age-induced mtDNA deletions [46]. Our validated, quantitative approach is sensitive down to 0.6 deletion-containing mtDNA molecules per million wild-type mtDNA genomes and uses PCR primers and probes explicitly designed to quantitate age-induced deletions independent of PCR efficiency [24].
We found that mtDNA copy number is predictive of lean muscle mass and mtDNA deletion frequency. We did not observe a correlation between mtDNA copy number and VO2max. This contrasts with data from Short et al., where copy number was predictive of VO2max between the ages of 18 and 89 [25]. Our cohort was composed of individuals aged 50 to 86. The finding that a lower mtDNA copy number predicts a higher deletion frequency with age is consistent with the observation that a lower mtDNA copy number leads to genomic instability and facilitates the intracellular accumulation of “selfish” deletion mutants in post-mitotic tissues such as skeletal muscle [29]. In contrast to many other studies, we did not detect the well-characterized age-related decline in lean or total muscle mass. The lack of observed muscle mass loss with age in our study is likely due to the age range of our cohort, the use of DXA to assess lean muscle mass, and the recruitment of only healthy subjects.
VO2max is understood to be an integrative measure of physical performance sensitive to multiorgan functional status, including respiratory, cardiac, and skeletal muscle function and metabolism (i.e., systems responsible for the uptake, transportation, and consumption of oxygen) [31, 32, 35–37]. With the large number of possible etiologies for the age-induced decline in VO2max, it is somewhat surprising that some of the variability can be attributed to mtDNA deletion frequency as assessed from a skeletal muscle biopsy. This suggests that either mtDNA deletion frequency reflects a primary deficit in muscle mitochondrial homeostasis or perhaps that skeletal muscle mtDNA deletion frequency correlates with the presence of mtDNA deletions in the other tissues and organs that also modulate VO2max.
While mtDNA deletion frequency increases dramatically across the lifespan, the burden occurring at a specific instant in time is low. This low bulk measure in tissue homogenates contrasts with the exceedingly high abundance of deletion mutations within single cells [7, 10, 17]. In these single cells, mtDNA deletion mutations activate skeletal muscle apoptosis and necrosis and fiber breakage, and result in muscle fiber loss [8]. Therefore, quantitation of mtDNA deletion mutations in homogenates predicts the abundance of ETC-deficient fibers [26] that are destined to undergo apoptosis/necrosis and die. In humans, the instantaneous deletion mutation frequency is similar to the instantaneous rate of fiber loss. In human vastus lateralis muscle, on average, ~ 25 fibers (0.008%) are lost every day, between the ages of 50 and 80 [47]. The rate of fiber loss also accelerates with advancing age. The instantaneous measure of deletion frequency does not integrate the whole of the phenomena that compounds across a lifespan [24]. Thus, mtDNA deletion frequency is a measurement of age-induced mitochondrial DNA damage that predicts the rate of muscle fiber loss with age. Data in this study show that the association of age-induced mtDNA deletions extends to organismal performance, which may exist as a primary muscle deficit, or, as noted above, perhaps that deletion mutation frequency in skeletal muscle is indicative of systemic mtDNA deletion frequency. Cross-tissue comparison of mtDNA deletion frequency within individuals and other measurements of age-induced damage across human lifespan will aid in specifying the molecular, cellular, and tissue origins of the performance deficit.
Our data suggest an effect of sex on mtDNA deletion frequency in aging humans. The age-induced increase in mtDNA deletion frequency in females is slower. If mtDNA deletion frequency is a bona fide determinant of human aging, we would expect females to have longer lifespans, which is well established. In 22-month-old mice, from a strain where female mice live longer, we measured a clear relationship between sex and mtDNA deletion frequency. In our cohort of human subjects, the effect size (Cohen’s f) of age by sex on mtDNA deletion frequency is 0.248 (a medium effect). With the assumption of a two-sided test with an alpha level of 0.05, a sample size of 182 is required to have 80% power of detecting the effect of age by sex interaction on mtDNA deletion frequency. Similarly, we found that mtDNA deletion frequency is a predictor of VO2max. However, after taking other predictors of VO2max (age and sex) into account, the contribution of mtDNA deletion frequency was less clear. Future studies with larger sample sizes are needed to better clarify the independent contribution of mtDNA deletion frequency to physical performance.
mtDNA deletions are an active contributor to the aging process in skeletal muscle. These data suggest that the importance of mtDNA deletions extends beyond muscle to other physical performance measures that, in themselves, are predictors of longevity and healthspan. Measurements of mitochondrial DNA quality are potentially useful predictors for interventions that target the debilitating chronic diseases of aging. mtDNA deletion frequency and mtDNA copy number integrate many aspects of mitochondrial quality control, which may contribute to the mitochondrial dysfunction and genome instability that are recognized as hallmarks of aging.
Funding
This work is supported by the National Institute on Aging at the National Institutes of Health (grant numbers R56AG060880, R01AG055518, K02AG059847, and R21AR072950).
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Allen Herbst and Steven J. Prior have contributed equally to this work.
References
- 1.Lopez-Otin C, Blasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell. 2013;153(6):1194–1217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lanza IR, Sreekumaran Nair K. Regulation of skeletal muscle mitochondrial function: genes to proteins. Acta Physiol (Oxford) 2010;199(4):529–547. doi: 10.1111/j.1748-1716.2010.02124.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sun N, Youle RJ, Finkel T. The mitochondrial basis of aging. Mol Cell. 2016;61(5):654–666. doi: 10.1016/j.molcel.2016.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Persson O, Muthukumar Y, Basu S, Jenninger L, Uhler JP, Berglund AK, McFarland R, Taylor RW, Gustafsson CM, Larsson E, Falkenberg M. Copy-choice recombination during mitochondrial L-strand synthesis causes DNA deletions. Nat Commun. 2019;10(1):759. doi: 10.1038/s41467-019-08673-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bua E, Johnson J, Herbst A, Delong B, McKenzie D, Salamat S, Aiken JM. Mitochondrial DNA-deletion mutations accumulate intracellularly to detrimental levels in aged human skeletal muscle fibers. Am J Hum Genet. 2006;79(3):469–480. doi: 10.1086/507132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gitschlag BL, Tate AT, Patel MR. Nutrient status shapes selfish mitochondrial genome dynamics across different levels of selection. Elife. 2020;9. 10.7554/eLife.56686. [DOI] [PMC free article] [PubMed]
- 7.Herbst A, Pak JW, McKenzie D, Bua E, Bassiouni M, Aiken JM. Accumulation of mitochondrial DNA deletion mutations in aged muscle fibers: evidence for a causal role in muscle fiber loss. J Gerontol. 2007;62(3):235–245. doi: 10.1093/gerona/62.3.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheema N, Herbst A, McKenzie D, Aiken JM. Apoptosis and necrosis mediate skeletal muscle fiber loss in age-induced mitochondrial enzymatic abnormalities. Aging Cell. 2015;14(6):1085–1093. doi: 10.1111/acel.12399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aure K, Fayet G, Leroy JP, Lacene E, Romero NB, Lombes A. Apoptosis in mitochondrial myopathies is linked to mitochondrial proliferation. Brain. 2006;129(Pt 5):1249–1259. doi: 10.1093/brain/awl061. [DOI] [PubMed] [Google Scholar]
- 10.Wanagat J, Cao Z, Pathare P, Aiken JM. Mitochondrial DNA deletion mutations colocalize with segmental electron transport system abnormalities, muscle fiber atrophy, fiber splitting, and oxidative damage in sarcopenia. FASEB J. 2001;15(2):322–332. doi: 10.1096/fj.00-0320com. [DOI] [PubMed] [Google Scholar]
- 11.Aspnes LE, Lee CM, Weindruch R, Chung SS, Roecker EB, Aiken JM. Caloric restriction reduces fiber loss and mitochondrial abnormalities in aged rat muscle. FASEB J. 1997;11(7):573–581. doi: 10.1096/fasebj.11.7.9212081. [DOI] [PubMed] [Google Scholar]
- 12.Bua EA, McKiernan SH, Wanagat J, McKenzie D, Aiken JM. Mitochondrial abnormalities are more frequent in muscles undergoing sarcopenia. J Appl Physiol (1985) 2002;92(6):2617–2624. doi: 10.1152/japplphysiol.01102.2001. [DOI] [PubMed] [Google Scholar]
- 13.Chung SS, Weindruch R, Schwarze SR, McKenzie DI, Aiken JM. Multiple age-associated mitochondrial DNA deletions in skeletal muscle of mice. Aging (Milano) 1994;6(3):193–200. doi: 10.1007/BF03324239. [DOI] [PubMed] [Google Scholar]
- 14.Eimon PM, Chung SS, Lee CM, Weindruch R, Aiken JM. Age-associated mitochondrial DNA deletions in mouse skeletal muscle: comparison of different regions of the mitochondrial genome. Dev Genet. 1996;18(2):107–113. doi: 10.1002/(SICI)1520-6408(1996)18:2<107::AID-DVG3>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 15.Herbst A, Wanagat J, Cheema N, Widjaja K, McKenzie D, Aiken JM. Latent mitochondrial DNA deletion mutations drive muscle fiber loss at old age. Aging Cell. 2016;15:1132–1139. doi: 10.1111/acel.12520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee CM, Lopez ME, Weindruch R, Aiken JM. Association of age-related mitochondrial abnormalities with skeletal muscle fiber atrophy. Free Radic Biol Med. 1998;25(8):964–972. doi: 10.1016/S0891-5849(98)00185-3. [DOI] [PubMed] [Google Scholar]
- 17.Schwarze SR, Lee CM, Chung SS, Roecker EB, Weindruch R, Aiken JM. High levels of mitochondrial DNA deletions in skeletal muscle of old rhesus monkeys. Mech Ageing Dev. 1995;83(2):91–101. doi: 10.1016/0047-6374(95)01611-3. [DOI] [PubMed] [Google Scholar]
- 18.Taylor SD, Ericson NG, Burton JN, Prolla TA, Silber JR, Shendure J, Bielas JH. Targeted enrichment and high-resolution digital profiling of mitochondrial DNA deletions in human brain. Aging Cell. 2014;13(1):29–38. doi: 10.1111/acel.12146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kraytsberg Y, Kudryavtseva E, McKee AC, Geula C, Kowall NW, Khrapko K. Mitochondrial DNA deletions are abundant and cause functional impairment in aged human substantia nigra neurons. Nat Genet. 2006;38(5):518–520. doi: 10.1038/ng1778. [DOI] [PubMed] [Google Scholar]
- 20.Parkinson GM, Dayas CV, Smith DW. Increased mitochondrial DNA deletions in substantia nigra dopamine neurons of the aged rat. Curr Aging Sci. 2014;7(3):155–160. doi: 10.2174/1874609808666150122150850. [DOI] [PubMed] [Google Scholar]
- 21.Reeve A, Meagher M, Lax N, Simcox E, Hepplewhite P, Jaros E, Turnbull D. The impact of pathogenic mitochondrial DNA mutations on substantia nigra neurons. J Neurosci. 2013;33(26):10790–10801. doi: 10.1523/JNEUROSCI.3525-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Song L, McMackin M, Nguyen A, Cortopassi G. Parkin deficiency accelerates consequences of mitochondrial DNA deletions and Parkinsonism. Neurobiol Dis. 2017;100:30–38. doi: 10.1016/j.nbd.2016.12.024. [DOI] [PubMed] [Google Scholar]
- 23.McKiernan SH, Tuen VC, Baldwin K, Wanagat J, Djamali A, Aiken JM. Adult-onset calorie restriction delays the accumulation of mitochondrial enzyme abnormalities in aging rat kidney tubular epithelial cells. Am J Physiol Ren Physiol. 2007;292(6):F1751–F1760. doi: 10.1152/ajprenal.00307.2006. [DOI] [PubMed] [Google Scholar]
- 24.Herbst A, Lee CC, Vandiver AR, Aiken JM, McKenzie D, Hoang A, et al. Mitochondrial DNA deletion mutations increase exponentially with age in human skeletal muscle. Aging Clin Exp Res. 2020. 10.1007/s40520-020-01698-7. [DOI] [PMC free article] [PubMed]
- 25.Short KR, Bigelow ML, Kahl J, Singh R, Coenen-Schimke J, Raghavakaimal S, Nair KS. Decline in skeletal muscle mitochondrial function with aging in humans. Proc Natl Acad Sci U S A. 2005;102(15):5618–5623. doi: 10.1073/pnas.0501559102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Herbst A, Widjaja K, Nguy B, Lushaj EB, Moore TM, Hevener AL, McKenzie D, Aiken JM, Wanagat J. Digital PCR quantitation of muscle mitochondrial DNA: age, fiber type, and mutation-induced changes. J Gerontol A Biol Sci Med Sci. 2017;72(10):1327–1333. doi: 10.1093/gerona/glx058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miljkovic N, Lim JY, Miljkovic I, Frontera WR. Aging of skeletal muscle fibers. Ann Rehabil Med. 2015;39(2):155–162. doi: 10.5535/arm.2015.39.2.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Larsen S, Nielsen J, Hansen CN, Nielsen LB, Wibrand F, Stride N, Schroder HD, Boushel R, Helge JW, Dela F, Hey-Mogensen M. Biomarkers of mitochondrial content in skeletal muscle of healthy young human subjects. J Physiol. 2012;590(14):3349–3360. doi: 10.1113/jphysiol.2012.230185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Filograna R, Koolmeister C, Upadhyay M, Pajak A, Clemente P, Wibom R, Simard ML, Wredenberg A, Freyer C, Stewart JB, Larsson NG. Modulation of mtDNA copy number ameliorates the pathological consequences of a heteroplasmic mtDNA mutation in the mouse. Sci Adv. 2019;5(4):eaav9824. doi: 10.1126/sciadv.aav9824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Distefano G, Goodpaster BH. Effects of exercise and aging on skeletal muscle. Cold Spring Harb Perspect Med. 2018;8(3). 10.1101/cshperspect.a029785. [DOI] [PMC free article] [PubMed]
- 31.Ross R, Blair SN, Arena R, Church TS, Despres JP, Franklin BA, Haskell WL, Kaminsky LA, Levine BD, Lavie CJ, Myers J, Niebauer J, Sallis R, Sawada SS, Sui X, Wisloff U, American Heart Association Physical Activity Committee of the Council on L, Cardiometabolic H, Council on Clinical C, Council on E, Prevention, Council on C, Stroke N, Council on Functional G, Translational B, Stroke C Importance of assessing cardiorespiratory fitness in clinical practice: a case for fitness as a clinical vital sign: a scientific statement from the American Heart Association. Circulation. 2016;134(24):e653–e699. doi: 10.1161/CIR.0000000000000461. [DOI] [PubMed] [Google Scholar]
- 32.Kim CH, Wheatley CM, Behnia M, Johnson BD. The effect of aging on relationships between lean body mass and VO2max in rowers. PLoS One. 2016;11(8):e0160275. doi: 10.1371/journal.pone.0160275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Studenski S, Perera S, Patel K, Rosano C, Faulkner K, Inzitari M, Brach J, Chandler J, Cawthon P, Connor EB, Nevitt M, Visser M, Kritchevsky S, Badinelli S, Harris T, Newman AB, Cauley J, Ferrucci L, Guralnik J. Gait speed and survival in older adults. JAMA. 2011;305(1):50–58. doi: 10.1001/jama.2010.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harber MP, Kaminsky LA, Arena R, Blair SN, Franklin BA, Myers J, Ross R. Impact of cardiorespiratory fitness on all-cause and disease-specific mortality: advances since 2009. Prog Cardiovasc Dis. 2017;60(1):11–20. doi: 10.1016/j.pcad.2017.03.001. [DOI] [PubMed] [Google Scholar]
- 35.Rogers MA, Hagberg JM, Martin WH, 3rd, Ehsani AA, Holloszy JO. Decline in VO2max with aging in master athletes and sedentary men. J Appl Physiol (1985) 1990;68(5):2195–2199. doi: 10.1152/jappl.1990.68.5.2195. [DOI] [PubMed] [Google Scholar]
- 36.Coen PM, Jubrias SA, Distefano G, Amati F, Mackey DC, Glynn NW, Manini TM, Wohlgemuth SE, Leeuwenburgh C, Cummings SR, Newman AB, Ferrucci L, Toledo FG, Shankland E, Conley KE, Goodpaster BH. Skeletal muscle mitochondrial energetics are associated with maximal aerobic capacity and walking speed in older adults. J Gerontol A Biol Sci Med Sci. 2013;68(4):447–455. doi: 10.1093/gerona/gls196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Carrick-Ranson G, Hastings JL, Bhella PS, Shibata S, Fujimoto N, Palmer D, Boyd K, Levine BD. The effect of age-related differences in body size and composition on cardiovascular determinants of VO2max. J Gerontol A Biol Sci Med Sci. 2013;68(5):608–616. doi: 10.1093/gerona/gls220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fleg JL, Morrell CH, Bos AG, Brant LJ, Talbot LA, Wright JG, Lakatta EG. Accelerated longitudinal decline of aerobic capacity in healthy older adults. Circulation. 2005;112(5):674–682. doi: 10.1161/CIRCULATIONAHA.105.545459. [DOI] [PubMed] [Google Scholar]
- 39.Standley RA, Distefano G, Trevino MB, Chen E, Narain NR, Greenwood B, Kondakci G, Tolstikov VV, Kiebish MA, Yu G, Qi F, Kelly DP, Vega RB, Coen PM, Goodpaster BH. Skeletal muscle energetics and mitochondrial function are impaired following 10 days of bed rest in older adults. J Gerontol A Biol Sci Med Sci. 2020;75(9):1744–1753. doi: 10.1093/gerona/glaa001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pollock RD, O'Brien KA, Daniels LJ, Nielsen KB, Rowlerson A, Duggal NA, Lazarus NR, Lord JM, Philp A, Harridge SDR. Properties of the vastus lateralis muscle in relation to age and physiological function in master cyclists aged 55-79 years. Aging Cell. 2018;17(2):e12735. doi: 10.1111/acel.12735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.DiMauro S. Mitochondrial myopathies. Curr Opin Rheumatol. 2006;18(6):636–641. doi: 10.1097/01.bor.0000245729.17759.f2. [DOI] [PubMed] [Google Scholar]
- 42.Hou Y, Xie Z, Zhao X, Yuan Y, Dou P, Wang Z. Appendicular skeletal muscle mass: a more sensitive biomarker of disease severity than BMI in adults with mitochondrial diseases. PLoS One. 2019;14(7):e0219628. doi: 10.1371/journal.pone.0219628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brown M, Sinacore DR, Binder EF, Kohrt WM. Physical and performance measures for the identification of mild to moderate frailty. J Gerontol A Biol Sci Med Sci. 2000;55(6):M350–M355. doi: 10.1093/gerona/55.6.m350. [DOI] [PubMed] [Google Scholar]
- 44.Hennessey JV, Chromiak JA, Della Ventura S, Guertin J, MacLean DB. Increase in percutaneous muscle biopsy yield with a suction-enhancement technique. J Appl Physiol (1985) 1997;82(6):1739–1742. doi: 10.1152/jappl.1997.82.6.1739. [DOI] [PubMed] [Google Scholar]
- 45.Box G, Cox D. An analysis of transformations. J R Stat Soc Ser B Methodol. 1964;26(2):211–252. [Google Scholar]
- 46.Hebert SL, Marquet-de Rouge P, Lanza IR, McCrady-Spitzer SK, Levine JA, Middha S, Carter RE, Klaus KA, Therneau TM, Highsmith EW, Nair KS. Mitochondrial aging and physical decline: insights from three generations of women. J Gerontol. 2015;70(11):1409–1417. doi: 10.1093/gerona/glv086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lexell J, Taylor CC, Sjostrom M. What is the cause of the ageing atrophy? Total number, size and proportion of different fiber types studied in whole vastus lateralis muscle from 15- to 83-year-old men. J Neurol Sci. 1988;84(2-3):275–294. doi: 10.1016/0022-510X(88)90132-3. [DOI] [PubMed] [Google Scholar]