Abstract
Compartmentalization of cellular functions is at the core of the physiology of eukaryotic cells. Recent evidences indicate that a universal organizing process – phase separation – supports the partitioning of biomolecules in distinct phases from a single homogeneous mixture, a landmark event in both the biogenesis and the maintenance of membrane and non-membrane-bound organelles. In the cell, ‘passive’ (non energy-consuming) mechanisms are flanked by ‘active’ mechanisms of separation into phases of distinct density and stoichiometry, that allow for increased partitioning flexibility and programmability. A convergence of physical and biological approaches is leading to new insights into the inner functioning of this driver of intracellular order, holding promises for future advances in both biological research and biotechnological applications.
1. Introduction
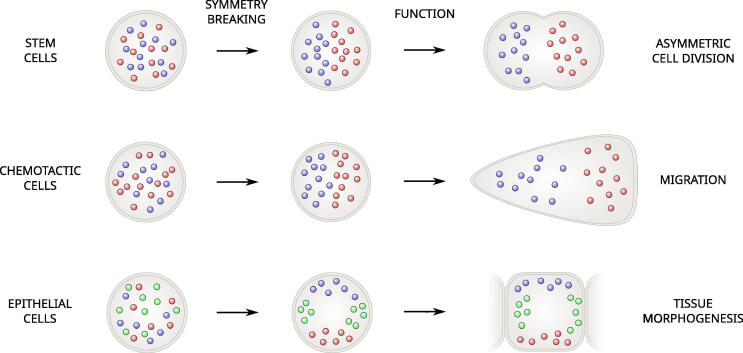
Phase separation is emerging as an overarching organizing principle behind the complex process of compartmentalization of the eukaryotic cell in a multiplicity of specialized subcellular systems. To effectively interact with their environment, cells have to generate inner structures by a sequence of symmetry-breaking events, whereby regions devoted to different vital functions emerge either spontaneously or under the influence of specific driving cues [1]. For instance, stem cells generate differentiated daughter cells by asymmetric cell division, endowing them with specific fate determinants [2] (Figure 1, top). Cells migrating in a chemotactic gradient are initially round but under the influence of a chemotactic factor develop a chemically differentiated front and back allowing them to migrate towards a chemoattractant source [3] (Figure 1, center). Epithelial cells mature chemically differentiated apical, basal and lateral region to integrate into the fine planar architecture of an epithelial tissue [4] (Figure 1, bottom). In these examples, the plasma membrane is subdivided in specialized, chemically differentiated domains by a selforganized phase separation process [5], [6], [7], [8] which in its turn guides the asymmetric positioning of inner structures, such as the cytoskeleton, and membrane-bound organelles, such as the Golgi and endoplasmic reticulum, in the intracellular space [9], [10], [11], [12]. At the same time, the biogenesis of a large class of membrane-less organelles, such as germ granules, stress granules, centrosomes, and nucleoli, takes place by symmetry breaking via a phase separation process [13], [14], [15], [16]. A sophisticated machinery of protein sorting and dispatching contributes to the generation and maintenance of the symmetry-broken, polarized state [17], [18].
Fig. 1.
Symmetry breaking and generation of cellular functions. In asymmetric cell division, fate determinants are distributed asymmetrically between the mother and daughter cell [2]. In chemotactic cells, different molecular factors accumulate in the growing anterior part and in the retracting posterior part of the cell [3]. During epithelial tissue morphogenesis, the apical, lateral and basal regions of the cell acquire different molecular identities [4], [23].
Differently from purely chemical systems that may rely on ‘passive’, non-energy consuming intermolecular interactions for the generation and maintenance of separated phases, biological systems typically employ energy-consuming, non-equilibrium, ‘active’ processes to sustain heterogeneity among the distinct phases that identify its inner compartments [19]. In particular, the ability of biomolecules to separate into phases characterized by distinct states of matter, ranging from liquid to gas or solid, is fundamental to spatially segregate proteins and nucleic acids in cells, thus generating specific compartments with specialized functionalities. Diverse biological processes ranging from RNA metabolism, DNA damage response and signal transduction exploit the fine levels of compartmentalization provided by phase-separated domains for their proper functioning [20], [21], [22].
Here, we review physical mechanism underlying biological phase separation, with a particular attention to the role played by active processes and to their implications. We then highlight representative examples of phase separation processes in the cytosol, nucleosome, and on cell membranes, and discuss some of the functions and mechanisms of regulation of phase separation in cell physiology. Lastly, we expose recent advances in studying and controlling biological phase separation processes in bioengineering and therapeutics.
2. Physics of active and passive phase separation
2.1. Classical theory of phase separation
Mixing-demixing transitions have been thoroughly studied in Physics [24], [25], [26]. There, the ordering of similar molecules by mutual affinity in spatially separated domains is driven by attractive and repulsive interactions, primarily of electrostatic origin, such as those emerging from the interaction of permanent or induced dipoles [27], [28], [29]. The formation of phase-separated domains becomes possible when the demixing tendency of mutually attractive interactions outcompetes the tendency of thermal agitation to mix and homogenize the molecular components of a system. For this reason, in the simplest systems the mixing-demixing transition may take place abruptly when some control parameter (such as the temperature, or the concentration of a particular component) crosses some critical value, and a tipping point is reached.
The initiation of this process of separation into different phases, characterized by distinct physical properties and/or molecular compositions, can occur spontaneously as a consequence of stochastic fluctuations. For instance, the spontaneous nucleation of a germ of a (solid, liquid or gel) condensate phase from a solution starts from the random encounter of two molecules of the solute, resulting in the formation of a (stable or transient) dimer. The growth of a small condensate domain of molecules of the solute is then driven by the balance between the influx of molecules from the solution and the ‘evaporation’ of molecules from the domain [30]. If, after the initial nucleation stage, phase separation takes place sufficiently close to thermodynamic equilibrium, it is possible to describe it in thermodynamical terms as a competition between a bulk and interface free energy. Here, the growth of a spherical domain of the condensate phase allows the system to lower its energy proportionally to the increase in the volume of the domain itself, while at the same time, the free energy of the system is raised by an amount proportional to the increase of interface area, where the energetically favorable contacts are not saturated (the proportionality coefficient is called a ‘surface tension’). A tug of war therefore arises between these two competing effects.
Several observable effects derive from this scenario:
-
1.
Phase separation takes place via a switch-like onset when the concentration of the solute exceeds the threshold concentration that allows a large condensate domain to coexist at equilibrium with the solution.
-
2.
The process tends to minimize the interface area (or perimeter length in two-dimensional systems) between the two phases, leading to the formation of approximately spherical (or circular, in two-dimensional systems) growing domains.
-
3.
The speed of the process is controlled by a degree of metastability, which is approximately proportional to the difference between the concentration of the solute and the threshold concentration.
-
4.
There is a critical size under which domains are unstable and tend to disappear, since for small domains, the energy advantage coming with an increased volume does not repay the cost of an extended droplet boundary; the critical size of approximately spherical or circular domains is inversely proportional to the degree of metastability.
-
5.
Stable domains (i.e., domains larger than the critical size) can be generated either by a large enough random fluctuation (homogeneous nucleation), or by the creation of a large enough nucleation center by some external action (heterogeneous nucleation).
-
6.
When competing for a limited pool of molecules, domains undergo competitive growth, also known as coarsening: larger domains grow at the expense of the molecules that ‘evaporate’ from smaller ones, so that at equilibrium a unique domain survives, in a sort of winner-take-all mechanism; moreover, the growth of domains becomes slower and slower with time, as long as the solute is sequestered by the growing domains, and as a consequence, the degree of metastability decreases and the critical size grows; Lifshits and Slyozov [31], [32] found that under these conditions the average size of domains grows as in both two- and three-dimensional environments. This is ultimately a consequence of the fact that molecules diffuse in a restricted environment without being created or destroyed (in this case it is said that the system exhibits a locally conserved order parameter [25]).
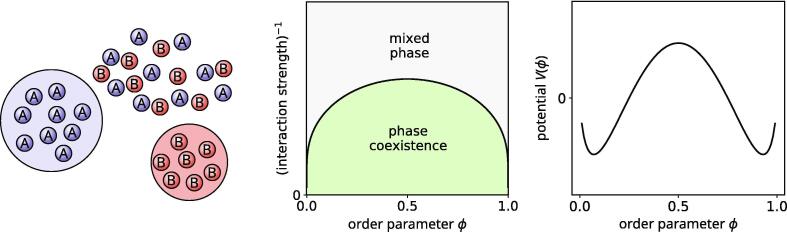
A similar scenario is realized in the case of an initially well-mixed binary mixture of a population of molecules of types A and B (Figure 2, left). characterized by homotypic intermolecular affinity, where islands of pure A- and B-phases can be nucleated and grow in the ‘sea’ of the well-mixed phase.
Fig. 2.
Mixing-demixing transition. Left: schematic representation of the formation of phase-separated domains in a ‘sea’ of the mixed phase. Center: phase diagram of the process. Right: potential part of the free energy in the phase-coexistence region, with the characteristic bistable shape; the two potential wells correspond to the two stable phases.
In both cases, a mathematical description of phase separation can be given by introducing a coarse-grained order parameter (that in the case of the binary mixture can for instance be thought as the difference between the local concentrations of the A- and B-molecules) and a Landau-Ginzburg free-energy density:
| (1) |
where the gradient term penalizes the interfaces between the two phases (K is thus proportional to the surface tension), while is a potential, which depends on the strengths of the intermolecular interactions. There exist a region of parameter space (Figure 2, center), in which the coexistence of competing phases is possible, as develops two minima (Figure 2, right),( corresponding respectively to each of the two physically realizable phases. If is symmetric, both phases are globally stable, otherwise one of the two phases is metastable.
2.2. Active vs. passive phase separation
Classical theories of phase separation rely on concepts of equilibrium statistical mechanics, such as the free energy, that in some cases can be used to approximately describe also non-equilibrium dissipative processes during their relaxation toward an equilibrium state. However, many of the inner workings of living cells are intrinsically nonequilibrium, since they are continuously driven by external and/or internal forces, and can involve transport phenomena and enzymatic processes in which individual molecular components constantly and irreversibly consume and dissipate energy, a defining feature of ‘active’ matter [19], [33], [34]. Such intrinsically out-of-equilibrium processes permit the existence of a larger variety of stationary states than those realizable in close-to-equilibrium conditions [15], [35]. Such processes contribute to generate and maintain the symmetry-broken, structured, compartmentalized state which allows the cell to perform its complex, vital functions. An example of such order-generating, energy-consuming nonequilibrium process is provided by the phase separation of cell membranes into polarized signaling domains driven by autocatalytic loops [5], [36], [37], [38], [39], [40]. In this context, the attractive or repulsive interaction between homotypic molecules is often not direct, but effectively mediated by autocatalytic feedback loops involving auxiliary molecules and sustained by a continuous energy influx, that may be provided for instance by ATP hydrolysis, and is consumed by individual catalytic events.
In the following paragraphs we review two representative models of active phase separation, respectively, in the cytosol and on lipid membranes.
2.3. Active phase separation in the cytosol
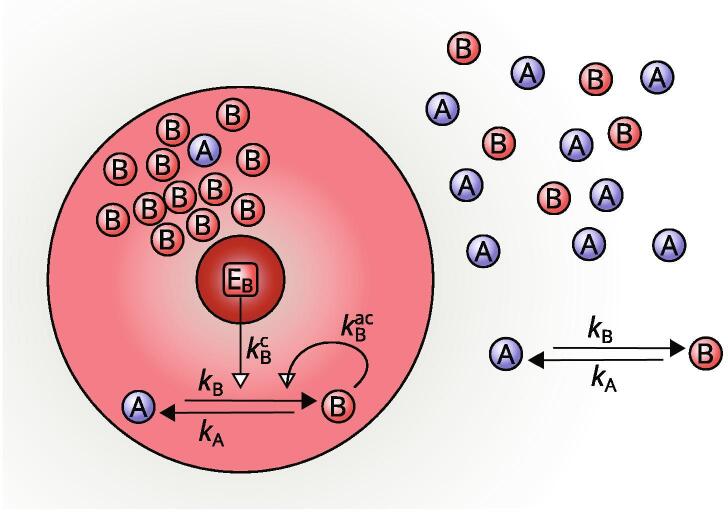
Several organelles generated by phase separation in the cytosol are present at the stationary state in finite numbers and with similar sizes. This cannot be easily explained by classical theories of coarsening kinetics, according to which larger domains grow at the expense of smaller ones, so that in finite systems at the stationary state a single large domain survives. Interestingly, active, catalytic mechanisms may suppress several features of this classical scenario, such as spontaneous nucleation and coarsening, thus allowing the existence of multiple-domain stationary states where the number of domains is controlled [15], [35]. A simple model where these effects may be appreciated describes a population of soluble A-molecules and phase-separating B-molecules, that may be converted into each other, either spontaneously, or by autocatalysis, or by the action of a finite number of spatially localized catalytic cores E (Fig. 3) [15], [35]. In this system, nucleation of new domains is triggered by the catalytic activity of the E-cores, and is suppressed elsewhere, thus allowing for precise control of the number of domains by the upstream regulation of the number of E-cores. Moreover, the nonequilibrium conditions created by the chemical reactions suppress coarsening and stabilize the multiple-domain stationary state. The catalytic activity of a finite number of E-cores irreversibly transforms the A-molecules into phase-separating B-molecules, thus contributing to the growth of B-domains. However, the B-domains are destabilized by the tendency of B-molecules to decay back into soluble A-molecules with a given rate . For small values of , the B-domains grow following a coarsening kinetics that leads the system to a single-domain stationary state. However, if is large enough, larger domains cannot grow indefinitely at the expense of smaller ones, coarsening is suppressed, and a multi-domain stationary state becomes stable.
Fig. 3.
Active phase separation in the cytosol [35]. Soluble A-molecules are converted to phase-separating B-molecules in the proximity of a catalytic core (dark red). An autocatalytic component reinforces the conversion of A into B. Additionally, the interconversion of the A- and B-molecules can also take place by first-order reactions.
2.4. Active phase separation on lipid membranes
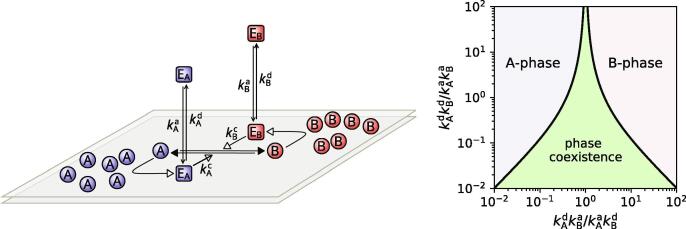
The spontaneous or induced symmetry breaking leading to the partition of lipid membranes into selforganized domains enriched in specific chemical factors is an ineludible feature of many vital functions of eukaryotic cells, such as migration, proliferation, and organogenesis, and is known to imply the action of multiple autocatalytic feedback loops [41], [5], [42], [43], [44], [36], [37], [38], [45], [39], [40]. A simple model of this active phase separation process describes a population of molecules of types A and B, bound to the lipid membrane, where they can laterally diffuse with diffusivity constant D. The two molecular types are interconverted by the action of two enzymes that shuttle between the lipid membrane and the cytosol, where they rapidly diffuse. Each of the two enzyme types binds preferentially to membrane regions enriched in their own product, thus realizing a simple reinforcing feedback loop (Fig. 4) [5], [6], [46], [8], [47]. Based on some of its characteristic properties, this kind of model has been termed ‘mass-conserved’, ‘bistable’, or ‘wave-pinning’ by different authors [48], [49], [50].
Fig. 4.
Left: abstract model for active phase separation on lipid membranes [5], [6], [46], [8]. Right: phase diagram. Concentrations are measured in units of ; the graph is symmetric with respect to the vertical axis centered in (here assumed to be unity).
A mathematical description of this process can be given by considering as an order parameter the difference in the surface concentration of the molecules of types A and B [5, Supp. Text]. The evolution of in time can be reduced to the minimization of the effective free energy density:
where is an effective potential, which depends on the chemical reaction rates indicated in Fig. 4. In the region of parameter space depicted in light green in Fig. 4 (right) the coexistence of competing phases is possible, as develops two minima, corresponding respectively to each of the two (A- and B-enriched) phases. This is analogous to the mathematical description of the classical phase separation of a binary mixture, except that now depends on the kinetic rates of a set of non-equilibrium autocatalytic reactions, instead of the equilibrium strength of direct intermolecular interactions. A similar mathematical structure implies similar properties: domains of pure A- and B-phases may nucleate and grow, exhibiting all the ‘classical’ effects, including switch-like onset, minimization of surface tension, existence of critical size, and coarsening [5], [6], [46]. Interestingly, in these conditions coarsening is faster than in the classical Lifshitz-Slyozov prediction, since the order parameter is not conserved: molecules may ‘evaporate’ from any part of a domain, diffuse rapidly in the cytosol, and be captured again in another point of the membrane; correspondingly, the average size of domains is predicted to grow as [6], [46].
These selforganized ‘active’ domains exhibit peculiar features. They cannot exist at the stationary state without a constant influx of energy, and are therefore intrinsically out of equilibrium. Since direct homotypic molecular interactions are here substituted by effective interactions mediated by the autocatalytic loops, the intermolecular distance in such domains can be larger than the molecular size, a characteristic that can be assimilated to that of a ‘gas’ phase. In principle, such domains may be expected to cover large extents of space and to exhibit a high degree of plasticity and fast recycling of their constituents. Interestingly, the size of these membrane domains is limited by the depletion of their constituents from the cytosolic reservoir, and can therefore in principle be controlled by regulating their cytosolic concentrations [5], [6], [46].
It is also worth observing here that actual selforganized active domains are driven by selfreinforcing catalytic loops that usually involve a chain of events where a multiplicity of molecular species participate. Therefore, such domains are expected to be typically multicomponent, and to host a spatially-localized, higher-than-average concentration of the ‘clan’ of all of the molecular species that take part into the relevant autocatalytic feedback loops (the red and blue ‘clans’ in Fig. 4 can be seen as an abstract example).
3. Biological phase separation
3.1. Evidences and functionalities
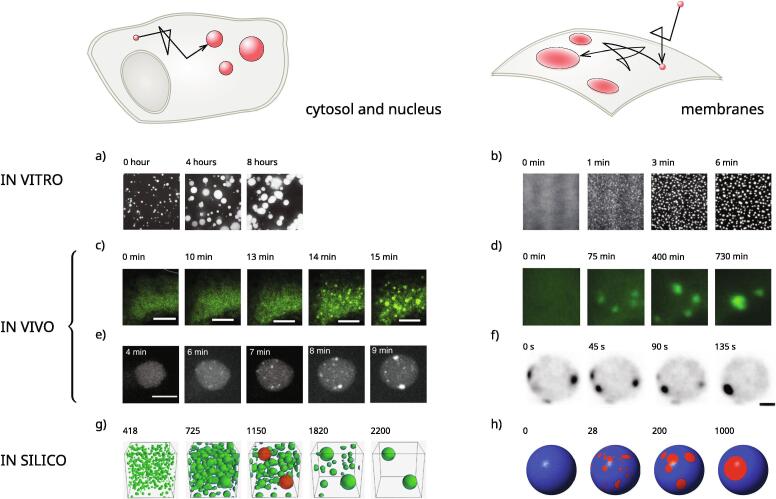
Processes of phase separation have emerged in the recent decade as an ubiquitous feature of eukaryotic cell physiology. Phase separation is a natural process that the cell may exploit to spatially localize biochemical reactions and cellular functions in appropriate subcellular structures and compartments. Two large classes of macromolecules have been identified, that exhibit a particular tendency to phase separate: proteins and RNA molecules containing several repeats of similar binding sites and/or weak interaction regions (multivalent molecules) and proteins (in particular, RNA- and DNA-binding proteins) containing stretches of unstructured, partially unfolded regions enriched in particular polar and charged amino acids (intrinsically disordered, or prion-like, protein regions) [51], [21], [52]. Favored by negligible activation barriers, such feeble interactions promote spontaneous and reversible organization of macromolecules in phase-separated domains, often referred to also as ‘droplets’, or ‘condensates’ [53]. In most cases the state of aggregation of such domains has been reported to be liquid-like, or gel, but occasionally also solid domains have been observed, mainly in association with pathological conditions [21]. On lipid membranes, phase separation into localized domains enriched in specific molecular factors can be also driven by the indirect intermolecular interactions established by networks of autocatalytic loops. Such ‘soft’ domains may have a gas-like structure, as their constituents do not need to be constantly in direct contact [5], [54]. The progressive ‘coarsening’, or ‘ripening’ (see Section 2.1) of phase-separated domains evidenced by time-lapse experiments, which is a signature of phase separation in the presence of a finite pool of molecular factors, has been observed in the cytosol, nucleus, and on lipid membranes, both in vitro (Fig. 5a,b) and in vivo (Fig. 5c–f). Computer simulations of quantitative models of phase separation reproduce the observed coarsening dynamics (Fig. 5g,h).
Fig. 5.
Nucleation and coarsening of selforganized domains in biological phase separation (reproduced from the original with permission, in modified form); a) prion-like FUS protein, associated with the neurodegenerative disease ALS [21]; b) LAT protein, taking part in T cell receptor signal transduction [22]; c) stress granules [14]; d) post-synaptic densities [55]; e) nucleoli and extranucleolar droplets [16]; f) polarity establishment in yeast [45]; g) simulation of the coarsening kinetics of nucleoli and extranucleolar droplets [16]; h) simulation of coarsening kinetics in the establishment of cell polarity [6], [8].
By organizing biochemical reactions in time and space, phase-separated domains allow them to proceed at the right pace by preventing undesired side-reactions in multistage processes [20], [22], storing away biomaterials when the cell comes under stress and releasing them gradually in normal conditions [52], [56], selectively inhibiting or promoting reactions by sequestering [57] or concentrating reactants [58], [59].
Importantly, at the larger, cellular scale, the regulated formation of phase-separated domains allows the right processes to take place at the right time and in the right place. A far from exhaustive list of examples is briefly reviewed here.
The centrosome, which serves as the main microtubule organizing center during mitosis, is nucleated by the centriole and grows by a phase-separation process [15]. Then, by absorbing tubulin from the cytosol and concentrating it in its interior, it favors the multiple nucleation of microtubules from its surface [60].
The nucleolus, which is responsible for ribosome biogenesis, is nucleated around a specific ribosomal DNA region and grows by phase separation, as evidenced by the observed coarsening kinetics (Fig. 5e).
Germ granules, aggregates of protein and RNA that determine the differentiation fate of daughter cells generated by asymmetric cell division, form during mitosis by a phase separation process [13], which is asymmetrically driven by the previous formation of opposite phase-separated domains on the plasma membrane of the mother cell [61].
3.2. Active and passive mechanisms of regulation
The spontaneous aggregation processes described by classical phase separation allow to control properties of subcellular compartments responsible for specific biological functions without any energy expense. A variety of such ‘passive’ control mechanisms is intrinsic in the nature of the process. For example, in many common situations phase separation starts when some parameter of the system crosses a threshold or critical value. This implies that switch-like control over physiological processes may be simply achieved by bringing the concentration levels of some of the relevant chemical factors above or below threshold. For instance, the formation of nucleoli has been shown to require crossing a threshold in the concentration of one of their main constituents, fibrillarin [16].
As another example of ‘passive’ control mechanism, the size of phase-separated domains is naturally controlled by the limited availability of their molecular components in any given compartment [62], [5].
On the other hand, biological phase separation appears to involve a wide variety of active, energy-consuming processes, raising the natural question about their role in the cell survival strategy. Present evidence suggests that active processes are required both for achieving precise regulation, and for allowing the realization of peculiar out-of-equilibrium stationary states.
As phase separation is driven by the ordering effect of direct or indirect intermolecular interactions, the main control parameter of the process is the intermolecular interaction strength, which may be controlled by several means, such as conformation modifications that hide/expose interaction sites, that may be induced for instance by specific enzymatic actions. For instance, it has been proposed that centrosomes grow by aggregating proteins that can switch between a dephosphorylated, globular, soluble form and a phosphorylated, elongated, phase-separating form [15]. Similarly, the phase separation of germ granules in C. elegans embryos is controlled by a switch between a phosphorylated and unphosphorylated state of the constituent proteins MEG-1 and MEG-3 [63]. Here, active processes may be seen as sparks that ignite the generation, maintenance and remodeling of biological phase-separated domains.
On the other hand, by their irreversible action, ‘active’ enzymatic processes may be essential to sustain out-of-equilibrium stationary states with specific features. For instance, the natural, ‘passive’ coarsening kinetics leading to the formation of a single phase-separated domain may be useful when the establishment of a single-domain stationary state is necessary for survival [45], [64], [65], but it would be harmful when a larger number of phase-separated domains have to exist in the stationary state, as is the case, for instance, of centrosomes, that must be present in exactly two copies. In this case, coarsening must be suppressed [15]. A minimal model of the process assumes that centrosome-forming proteins are actively converted from a soluble to a phase-separating form on the surface of the centriole, and are converted back to the soluble form in the body of the centrosome [15]. The effect of these active transformations is twofold. First, centrosome nucleation is strongly favored around the two centrioles and correspondingly impaired in the cytosol, so that only two centrosomes may be nucleated. Second, the conversion from the phase-separating to the soluble form decreases the ‘greediness’ of the growing centrosomal domains, thus allowing the peaceful coexistence of two centrosomes in the stationary state [15]. This has been proposed as a general mechanism that by suppressing coarsening may allow the generation of stable populations of selforganized domains of controlled size [35].
3.3. Active phase separation in signaling and trafficking
Many signal transduction circuits in the eukaryotic cells organize their activity on the lipid bilayer of membranes in discrete, localized microdomains, enriched in specific signaling molecules. Although the functional implications of this level of spatial organization are still not well understood, it has been proposed that they may be used to ‘digitalize’ incoming signals [66], [67] and to control the spatiotemporal remodeling of the actin cytoskeleton [68], [69].
The formation of signaling microdomains on lipid membranes has been shown to take place by a two-dimensional phase separation process in the signaling pathway upstream of RAS, a small GTPase which plays a pivotal in the transduction of stimuli from the T cell receptor and the Epidermal Growth Factor Receptor [22]. RAS is activated by the associated guanosine exchange factor SOS [70]. The SOS molecule forms a complex with the integral membrane adaptor protein LAT, leading to the recruitment of soluble proteins, such as GRB2, which establish multivalent interactions with LAT. Initially, LAT proteins are uniformly distributed and diffuse freely on the lipid bilayer. By increasing the amount of GRB2, two-dimensional LAT-GRB2-SOS microdomains are formed, controlling SOS membrane dwell time, and ultimately RAS activation kinetics [22]. The coarsening kinetics of LAT microdomains on supported lipid bilayers (Fig. 5b) is a strong evidence of the underlying selforganized phase separation process.
A second example is provided by the assembly of F-actin, which is controlled by the formation of phase-separated two-dimensional membrane-associated microdomains, relying on interactions between phosphorylated Nephrin (membrane receptor), NCK (adaptor protein) and N-WASP (NCK-interacting protein and the activator of the actin nucleation factor Arp2/3 complex) [51], [71]. In this case, N-WASP membrane dwell time is sensitive to NCK concentration. The modulation of multivalent interactions within the condensate fine-tunes dwell times and signaling activity [72].
The selforganized formation of phase-separated signaling domains on the plasma membrane plays a central role in many cellular functions, including chemotaxis, cell division, neuronal growth, immunity, and intracellular protein sorting [40], [44], [5], [6], [61], [39], [73], [74]. In this context, the attractive or repulsive interactions driving the formation of phase-separated domains is often not direct, but effectively mediated by energy-consuming positive and negative feedback loops involving multiple molecular species [40], [41], [42], [44], [5], [43], [36], [37], [38], [45], [39]. A central role is played here by switch-like molecules such as small GTPases proteins [75]. Specific enzymes (GEFs and GAPs) convert the inactive form of small GTPases into the active form, and vice versa, thereby ensuring kinetic control of GTPase activation over time [70], [76]. The two states of small GTPases and the associated GEF and GAP enzymes provide a concrete realization of the scheme of Fig. 4, when for instance an active GTPase recruits its own activator GEF to the lipid membrane in an autocatalytic feedback loop [8].
As an example, in S. cerevisiae the site of budding of a daughter cell is marked by the formation on the cell membrane of a domain enriched in the active form of Cdc42, a member of the Rho family of small GTPases. The formation of this selforganized active domain is driven by an autocatalytic feedback loop, whereby active Cdc42 recruits its own GEF, Cdc24, from the cytosol to the domain [77]. Time-lapse images of the process show that the formation of these domains takes place through successive stages of nucleation, coarsening, and relaxation to a single-domain stationary state (Fig. 5f). The speed of domain formation can be controlled genetically by modifying the effective strength of the intermolecular interaction mediated by the autocatalytic feedback loop [39]. In this system, the formation of a unique domain is fundamental for cell survival: mutations that allow for the formation of multiple domains are lethal [45], [39].
Similarly, the Rab5 small GTPase, which plays a key role in organizing endocytic trafficking, in its active form recruits its own GEF, Rabex5, thus creating a local positive feedback loop that induces the formation of selforganized Rab5-enriched domains on endosomal membranes [78], [40], [79].
Other members of the small GTPase protein family, including both Rac and Rab molecular switches, are likely involved in the generation of selforganized domains driven by complex biochemical circuits including multiple feedback and feedforward loops [80], [70].
Self-organized phase-separated domains generated by direct or mediated intermolecular interactions on lipid membranes have been observed to drive endocytic events [81], [82]. In this regard, it was recently suggested that protein sorting, i.e. the concentration of specific protein cargos in submicrometric lipid vesicles, is enabled by their colocalization in membrane microdomains by means of phase separation [83]. In this scenario, phase separation leads to the spontaneous formation of selforganized active domains enriched, along with the sorted cargo, in specific lipids and in several adaptor, membrane-bending and fission-inducing proteins. Then, the biochemical constituents of this selforganized domain locally induce higher membrane curvature and the consequent nucleation and detachment of a small vesicle. The newly generated vesicle is then constitutively enriched in the biochemical factors of the engulfed domain, thus resulting in a spontaneous distillation process. The main parameter controlling the efficiency of the sorting process is the strength of the effective intermolecular interaction driving phase separation, and optimal sorting is realized for intermediate values of the interaction strength (neither too low nor too high) [83]. A similar picture emerges from recent work, where optimal endocytic efficiency was obtained by tuning the strength of protein assembly at intermediate values using light-inducible oligomerization [107].
4. Phase separation in bioengineering and therapeutics
Biomolecular condensates originated by both proteins and nucleic acids are rapidly attracting the interest of biotech industry [84]. The ability of biological polymers to coalesce and quickly disappear from the cell cytoplasm, as well as to speeding up reactions by concentrating reactants, provides a source of inspiration for the development of novel biomedical applications. As a result, four biotech companies (Faze Medicines, Nereid Therapeutics, Transition Bio, Dewpoint Therapeutics) were launched in less than two years (2019–20), with the goal of exploiting biological condensates for human disease treatment [85]. Notably, applications of biological condensates in biotechnologies overlap with their biological function described in cells [22], [56], [86], [87], [52], [88], [89], [90], [91] including: the generation of liquid compartments in cells, the regulation of biochemical catalysis in both space and time, and the partitioning of drugs. As an example, a synthetic condensate (OT organelle) provides customized orthogonal translation and protein engineering in semisynthetic eukaryotic cells [91]. In addition, specific nucleation of phase separation at precise genomic locus induced by a dead Cas9 chimera (CasDROP) could help to understand how phase separation can alter 3D genome organization [92]. Furthermore, tuning and mixing of distinct compartments is forced by fusion proteins containing different combinations of protein modules [93], [94], [95], [96]. Parallelly, selective partitioning into liquid ordered membrane domains is exploited in a wide variety of nanotechnological applications aimed at delivering both small molecules as well as proteins and nucleic acids into cells [97], [98].
Among the distinct phase separation processes, liquid–liquid phase separation is the most promising and capable to originate cell-like structures [99] or bioreactors [100] via interaction among biomolecules [13], [101], [102], [52]. Accordingly, by varying the intra/intermolecular interaction strengths encoded in a biopolymer sequence, the process of condensate formation may be appropriately tuned. As an example, DNA nanostructures phase separate in a DNA-poor and a DNA-rich phase, similarly to coacervate [103], based on both salt concentration and temperature, two physical quantities that control base-pairing of the DNA duplex [104]. These quantities may be tuned to ensure selective cargo capturing and partitioning; such a strategy may be used to compartmentalize distinct biochemical reaction inside the same DNA droplet [105].
In the last years, the principles of liquid–liquid phase separation were used to develop a broad range of drug delivery vehicles. In particular, coacervates generated spontaneously by self-assembly in an aqueous medium are designed to encapsulate labile proteins or drugs, thereby preserving their bioactivity from the surrounding environment. Notably, coacervates do not require organic solvents for their assembly and therefore are defined as the most safe vehicle to deliver pharmacological agents to patients compared to hydrogels and microparticles [106].
Altogether, these evidences point to the utmost relevance of the phase separation process in biotechnological and bioengineering applications.
5. Perspective
Phase separation is emerging as a universal organizing principle, allowing cells to break down their physiological activity in a multitude of functional compartments. Liquid–liquid phase separation has been shown to account for the compartmentalization of biomaterial in many specialized organelles in the cytoplasm and nucleus. On lipid membranes, active phase separation underlies the dynamics of signaling and trafficking domains that organize the complex logistic of molecular sorting. In an exciting convergence of physical and biological intuition, increasing quantitative and theoretical understanding of the principles governing spontaneous and induced phase separation holds promise of gaining control over these exquisite, still partly enigmatic microscopic and submicroscopic mechanisms to achieve new breakthroughs in biotechnology and therapeutics.
Future research is likely to focus on discriminating the active and passive aspects of biological phase separation, highlighting the regulating and promoting role of catalytic events, deepening our understanding about the state of aggregation (solid, liquid, gel, gas-like,…) of the selforganized domains that drive the main physiological activities of the cell. In parallel, the engineering of functionalized biomimetic particles capable of interfering with such activities with therapeutic goals represents a fascinating perspective.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We gratefully acknowledge enlightening discussions with Igor Kolokolov, Vladimir Lebedev, Guido Serini, and Marco Zamparo. This research has received funding from AIRC: IG21875 project (P.I. Emilio Hirsch), MFAG 2020 - ID. 24897 project (P.I. Carlo Cosimo Campa), and from IIGM: Internal Grant Program.
Contributor Information
Luca Dall’Asta, Email: luca.dallasta@polito.it.
Andrea Gamba, Email: andrea.gamba@polito.it.
Emilio Hirsch, Email: hirsch@unito.it.
Carlo C. Campa, Email: carlocosimo.campa@iigm.it.
References
- 1.Li R., Bowerman B. Symmetry breaking in biology. Cold Spring Harb Perspect Biol. 2010;2 doi: 10.1101/cshperspect.a003475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Venkei Z.G., Yamashita Y.M. Emerging mechanisms of asymmetric stem cell division. J Cell Biol. 2018;217:3785–3795. doi: 10.1083/jcb.201807037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ridley A.J., Schwartz M.A., Burridge K., Firtel R.A., Ginsberg M.H., Borisy G., Parsons J.T., Horwitz A.R. Cell migration: integrating signals from front to back. Science. 2003;302:1704–1709. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- 4.Shewan A., Eastburn D.J., Mostov K. Phosphoinositides in cell architecture. Cold Spring Harbor Perspect Biol. 2011;3 doi: 10.1101/cshperspect.a004796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gamba A., de Candia A., Di Talia S., Coniglio A., Bussolino F., Serini G. Diffusion limited phase separation in eukaryotic chemotaxis. Proc Natl Acad Sci U S A. 2005;102:16927–16932. doi: 10.1073/pnas.0503974102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gamba A., Kolokolov I., Lebedev V., Ortenzi G. Patch coalescence as a mechanism for eukaryotic directional sensing. Phys Rev Lett. 2007;99 doi: 10.1103/PhysRevLett.99.158101. [DOI] [PubMed] [Google Scholar]
- 7.Veglio A., Gamba A., Nicodemi M., Bussolino F., Serini G. Symmetry breaking mechanism for epithelial cell polarization. Phys Rev E. 2009;80 doi: 10.1103/PhysRevE.80.031919. [DOI] [PubMed] [Google Scholar]
- 8.Semplice M., Veglio A., Naldi G., Serini G., Gamba A. A bistable model of cell polarity. PLoS One. 2012;7 doi: 10.1371/journal.pone.0030977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bornens M. Organelle positioning and cell polarity. Nat Rev Mol Cell Biol. 2008;9:874–886. doi: 10.1038/nrm2524. [DOI] [PubMed] [Google Scholar]
- 10.Yadav S., Puri S., Linstedt A.D. A primary role for Golgi positioning in directed secretion, cell polarity, and wound healing. Mol Biol Cell. 2009;20:1728–1736. doi: 10.1091/mbc.E08-10-1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rodriguez-Boulan E., Macara I.G. Organization and execution of the epithelial polarity programme. Nat Rev Mol Cell Biol. 2014;15:225–242. doi: 10.1038/nrm3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ravichandran Y., Goud B., Manneville J.-B. The Golgi apparatus and cell polarity: Roles of the cytoskeleton, the Golgi matrix, and Golgi membranes. Curr Opin Cell Biol. 2020;62:104–113. doi: 10.1016/j.ceb.2019.10.003. [DOI] [PubMed] [Google Scholar]
- 13.Brangwynne C.P., Eckmann C.R., Courson D.S., Rybarska A., Hoege C., Gharakhani J., Julicher F., Hyman A.A. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science. 2009;324:1729–1732. doi: 10.1126/science.1172046. [DOI] [PubMed] [Google Scholar]
- 14.J.R. Wheeler, T. Matheny, S. Jain, R. Abrisch, R. Parker, Distinct stages in stress granule assembly and disassembly, eLife 5 (2016). [DOI] [PMC free article] [PubMed]
- 15.Zwicker D., Decker M., Jaensch S., Hyman A.A., Jülicher F. Centrosomes are autocatalytic droplets of pericentriolar material organized by centrioles. Proc Nat Acad Sci. 2014;111:E2636–E2645. doi: 10.1073/pnas.1404855111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berry J., Weber S.C., Vaidya N., Haataja M., Brangwynne C.P. RNA transcription modulates phase transition-driven nuclear body assembly. Proc Nat Acad Sci. 2015;112:E5237–E5245. doi: 10.1073/pnas.1509317112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mellman I., Nelson W.J. Coordinated protein sorting, targeting and distribution in polarized cells. Nat Rev Mol Cell Biol. 2008;9:833–845. doi: 10.1038/nrm2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo Y., Sirkis D.W., Schekman R. Protein sorting at the trans-Golgi network. Ann Rev Cell Devel Biol. 2014;30:169–206. doi: 10.1146/annurev-cellbio-100913-013012. [DOI] [PubMed] [Google Scholar]
- 19.Fang X., Kruse K., Lu T., Wang J. Nonequilibrium physics in biology. Rev Mod Phys. 2019;91 [Google Scholar]
- 20.Feric M., Vaidya N., Harmon T.S., Mitrea D.M., Zhu L., Richardson T.M., Kriwacki R.W., Pappu R.V., Brangwynne C.P. Coexisting liquid phases underlie nucleolar subcompartments. Cell. 2016;165:1686–1697. doi: 10.1016/j.cell.2016.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Patel A., Lee H.O., Jawerth L., Maharana S., Jahnel M., Hein M.Y., Stoynov S., Mahamid J., Saha S., Franzmann T.M. A liquid-to-solid phase transition of the als protein fus accelerated by disease mutation. Cell. 2015;162:1066–1077. doi: 10.1016/j.cell.2015.07.047. [DOI] [PubMed] [Google Scholar]
- 22.Su X., Ditlev J.A., Hui E., Xing W., Banjade S., Okrut J., King D.S., Taunton J., Rosen M.K., Vale R.D. Phase separation of signaling molecules promotes T cell receptor signal transduction. Science. 2016;352:595–599. doi: 10.1126/science.aad9964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cerruti B., Puliafito A., Shewan A.M., Yu W., Combes A.N., Little M.H., Chianale F., Primo L., Serini G., Mostov K.E., Celani A., Gamba A. Polarity, cell division, and out-of-equilibrium dynamics control the growth of epithelial structures. J Cell Biol. 2013;203:359–372. doi: 10.1083/jcb.201305044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Binder K., Stauffer D. Statistical theory of nucleation, condensation and coagulation. Adv Phys. 1976;25:343–396. [Google Scholar]
- 25.Bray A.J. Theory of phase-ordering kinetics. Adv Phys. 2002;51:481–587. [Google Scholar]
- 26.Wadhawan V., Puri S., editors. Kinetics of phase transitions. CRC Press; 2009. [Google Scholar]
- 27.Israelachvili J.N. 3 ed. Academic Press; 2011. Intermolecular and Surface Forces, Third Edition: Revised Third Edition. [Google Scholar]
- 28.Leckband D., Israelachvili J. Intermolecular forces in biology. Q Rev Biophys. 2001;34:105–267. doi: 10.1017/s0033583501003687. [DOI] [PubMed] [Google Scholar]
- 29.Chandler D., Weeks J.D., Andersen H.C. Van der Waals picture of liquids, solids, and phase transformations. Science. 1983;220:787–794. doi: 10.1126/science.220.4599.787. [DOI] [PubMed] [Google Scholar]
- 30.Lifshitz E.M., Pitaevskii L.P. Physical kinetics. Butterworth Heinemann. 1981 [Google Scholar]
- 31.Lifshitz I., Slezov V. Kinetics of diffusive decomposition of supersaturated solid solutions. Soviet Physics JETP. 1959;35:331–339. [Google Scholar]
- 32.Slezov V.V. Wiley-VCH Verlag GmbH & Co. KGaA; Weinheim: 2009. Kinetics of first order phase transitions. [Google Scholar]
- 33.Marenduzzo D. An introduction to the statistical physics of active matter: motility-induced phase separation and the “generic instability” of active gels. Eur Phys J Spec Top. 2016;225:2065–2077. [Google Scholar]
- 34.Lindenberg K., Metzler R., Oshanin G. World Scientific Publishing Europe Ltd; 2019. Chemical kinetics: beyond the textbook. [Google Scholar]
- 35.Zwicker D., Hyman A.A., Juelicher F. Suppression of Ostwald ripening in active emulsions. Phys Rev E. 2015;92 doi: 10.1103/PhysRevE.92.012317. [DOI] [PubMed] [Google Scholar]
- 36.Arai Y., Shibata T., Matsuoka S., Sato M.J., Yanagida T., Ueda M. Self-organization of the phosphatidylinositol lipids signaling system for random cell migration. Proc Nat Acad Sci. 2010;107:12399–12404. doi: 10.1073/pnas.0908278107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goehring N.W., Trong P.K., Bois J.S., Chowdhury D., Nicola E.M., Hyman A.A., Grill S.W. Polarization of PAR proteins by advective triggering of a pattern-forming system. Science. 2011;334:1137–1141. doi: 10.1126/science.1208619. [DOI] [PubMed] [Google Scholar]
- 38.Johnson J.M., Jin M., Lew D.J. Symmetry breaking and the establishment of cell polarity in budding yeast. Curr Opin Genet Develop. 2011;21:740–746. doi: 10.1016/j.gde.2011.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu C.-F., Chiou J.-G., Minakova M., Woods B., Tsygankov D., Zyla T.R., Savage N.S., Elston T.C., Lew D.J. Role of competition between polarity sites in establishing a unique front. Elife. 2015;4 doi: 10.7554/eLife.11611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cezanne A., Lauer J., Solomatina A., Sbalzarini I.F., Zerial M. A non-linear system patterns Rab5 GTPase on the membrane. Elife. 2020;9 doi: 10.7554/eLife.54434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weiner O.D., Neilsen P.O., Prestwich G.D., Kirschner M.W., Cantley L.C., Bourne H.R. A PtdInsP 3-and Rho GTPase-mediated positive feedback loop regulates neutrophil polarity. Nat Cell Biol. 2002;4:509–513. doi: 10.1038/ncb811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wong K., Pertz O., Hahn K., Bourne H. Neutrophil polarization: spatiotemporal dynamics of RhoA activity support a self-organizing mechanism. Proc Nat Acad Sci. 2006;103:3639–3644. doi: 10.1073/pnas.0600092103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Onsum M., Rao C.V. A mathematical model for neutrophil gradient sensing and polarization. PLoS Comput Biol. 2007;3 doi: 10.1371/journal.pcbi.0030036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kölsch V., Charest P.G., Firtel R.A. The regulation of cell motility and chemotaxis by phospholipid signaling. J Cell Sci. 2008;121:551–559. doi: 10.1242/jcs.023333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Howell A.S., Jin M., Wu C.-F., Zyla T.R., Elston T.C., Lew D.J. Negative feedback enhances robustness in the yeast polarity establishment circuit. Cell. 2012;149:322–333. doi: 10.1016/j.cell.2012.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gamba A., Kolokolov I., Lebedev V., Ortenzi G. Universal features of cell polarization processes. J Stat Mech. 2009;P02019 [Google Scholar]
- 47.Zamparo M., Chianale F., Tebaldi C., Cosentino-Lagomarsino M., Nicodemi M., Gamba A. Dynamic membrane patterning, signal localization and polarity in living cells. Soft Matter. 2015;11:838–849. doi: 10.1039/c4sm02157f. [DOI] [PubMed] [Google Scholar]
- 48.Otsuji M., Ishihara S., Co C., Kaibuchi K., Mochizuki A., Kuroda S. A mass conserved reaction-diffusion system captures properties of cell polarity. PLoS Comput Biol. 2007;3 doi: 10.1371/journal.pcbi.0030108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Beta C., Amselem G., Bodenschatz E. A bistable mechanism for directional sensing. New J Phys. 2008;10 [Google Scholar]
- 50.Mori Y., Jilkine A., Edelstein-Keshet L. Wave-pinning and cell polarity from a bistable reaction-diffusion system. Biophys J. 2008;94:3684. doi: 10.1529/biophysj.107.120824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Banjade S., Rosen M.K. Phase transitions of multivalent proteins can promote clustering of membrane receptors. Elife. 2014;3 doi: 10.7554/eLife.04123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Franzmann T.M., Jahnel M., Pozniakovsky A., Mahamid J., Holehouse A.S., Nüske E., Richter D., Baumeister W., Grill S.W., Pappu R.V. Phase separation of a yeast prion protein promotes cellular fitness. Science. 2018;359 doi: 10.1126/science.aao5654. [DOI] [PubMed] [Google Scholar]
- 53.Banani S.F., Lee H.O., Hyman A.A., Rosen M.K. Biomolecular condensates: organizers of cellular biochemistry. Nat Rev Mol Cell Biol. 2017;18:285–298. doi: 10.1038/nrm.2017.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Matsuoka S., Shibata T., Ueda M. Asymmetric PTEN distribution regulated by spatial heterogeneity in membrane-binding state transitions. PLoS Comput Biol. 2013;9 doi: 10.1371/journal.pcbi.1002862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zeng M., Shang Y., Araki Y., Guo T., Huganir R.L., Zhang M. Phase transition in postsynaptic densities underlies formation of synaptic complexes and synaptic plasticity. Cell. 2016;166:1163–1175. doi: 10.1016/j.cell.2016.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Riback J.A., Katanski C.D., Kear-Scott J.L., Pilipenko E.V., Rojek A.E., Sosnick T.R., Drummond D.A. Stress-triggered phase separation is an adaptive, evolutionarily tuned response. Cell. 2017;168:1028–1040. doi: 10.1016/j.cell.2017.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hirose T., Virnicchi G., Tanigawa A., Naganuma T., Li R., Kimura H., Yokoi T., Nakagawa S., Bénard M., Fox A.H. NEAT1 long noncoding RNA regulates transcription via protein sequestration within subnuclear bodies. Mol Biol Cell. 2014;25:169–183. doi: 10.1091/mbc.E13-09-0558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kaplan A., Reinhold L. CO2 concentrating mechanisms in photosynthetic microorganisms. Ann Rev Plant Biol. 1999;50:539–570. doi: 10.1146/annurev.arplant.50.1.539. [DOI] [PubMed] [Google Scholar]
- 59.Oltrogge L.M., Chaijarasphong T., Chen A.W., Bolin E.R., Marqusee S., Savage D.F. Multivalent interactions between CsoS2 and Rubisco mediate α)carboxysome formation. Nat Struct Mol Biol. 2020;27:281–287. doi: 10.1038/s41594-020-0387-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Woodruff J.B., Gomes B.F., Widlund P.O., Mahamid J., Honigmann A., Hyman A.A. The centrosome is a selective condensate that nucleates microtubules by concentrating tubulin. Cell. 2017;169:1066–1077. doi: 10.1016/j.cell.2017.05.028. [DOI] [PubMed] [Google Scholar]
- 61.Hoege C., Hyman A.A. Principles of PAR polarity in caenorhabditis elegans embryos. Nat Rev Mol Cell Biol. 2013;14:315–322. doi: 10.1038/nrm3558. [DOI] [PubMed] [Google Scholar]
- 62.Goehring N.W., Hyman A.A. Organelle growth control through limiting pools of cytoplasmic components. Curr Biol. 2012;22:R330–R339. doi: 10.1016/j.cub.2012.03.046. [DOI] [PubMed] [Google Scholar]
- 63.Wang J.T., Smith J., Chen B.-C., Schmidt H., Rasoloson D., Paix A., Lambrus B.G., Calidas D., Betzig E., Seydoux G. Regulation of RNA granule dynamics by phosphorylation of serine-rich, intrinsically disordered proteins in C. elegans. Elife. 2014;3 doi: 10.7554/eLife.04591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nicodemi M., Prisco A. Symmetry-breaking model for X-chromosome inactivation. Phys Rev Lett. 2007;98 doi: 10.1103/PhysRevLett.98.108104. [DOI] [PubMed] [Google Scholar]
- 65.Cerase A., Armaos A., Neumayer C., Avner P., Guttman M., Tartaglia G.G. Phase separation drives X-chromosome inactivation: a hypothesis. Nat Struct Mol Biol. 2019;26:331–334. doi: 10.1038/s41594-019-0223-0. [DOI] [PubMed] [Google Scholar]
- 66.Tian T., Harding A., Inder K., Plowman S., Parton R.G., Hancock J.F. Plasma membrane nanoswitches generate high-fidelity Ras signal transduction. Nat Cell Biol. 2007;9:905–914. doi: 10.1038/ncb1615. [DOI] [PubMed] [Google Scholar]
- 67.Villaseñor R., Nonaka H., Del Conte-Zerial P., Kalaidzidis Y., Zerial M. Regulation of EGFR signal transduction by analogue-to-digital conversion in endosomes. Elife. 2015;4 doi: 10.7554/eLife.06156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gomez T.S., Billadeau D.D. A FAM21-containing WASH complex regulates retromer-dependent sorting. Develop Cell. 2009;17:699–711. doi: 10.1016/j.devcel.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Weiner O.D., Marganski W.A., Wu L.F., Altschuler S.J., Kirschner M.W. An actin-based wave generator organizes cell motility. PLoS Biol. 2007;5 doi: 10.1371/journal.pbio.0050221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cherfils J., Zeghouf M. Regulation of small gtpases by gefs, gaps, and gdis. Physiol Rev. 2013;93:269–309. doi: 10.1152/physrev.00003.2012. [DOI] [PubMed] [Google Scholar]
- 71.Kim S., Kalappurakkal J.M., Mayor S., Rosen M.K. Phosphorylation of nephrin induces phase separated domains that move through actomyosin contraction. Mol Biol Cell. 2019;30:2996–3012. doi: 10.1091/mbc.E18-12-0823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Case L.B., Zhang X., Ditlev J.A., Rosen M.K. Stoichiometry controls activity of phase-separated clusters of actin signaling proteins. Science. 2019;363:1093–1097. doi: 10.1126/science.aau6313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schelski M., Bradke F. Neuronal polarization: From spatiotemporal signaling to cytoskeletal dynamics. Mol Cell Neurosci. 2017;84:11–28. doi: 10.1016/j.mcn.2017.03.008. [DOI] [PubMed] [Google Scholar]
- 74.Stone M.B., Shelby S.A., Núñez M.F., Wisser K., Veatch S.L. Protein sorting by lipid phase-like domains supports emergent signaling function in B lymphocyte plasma membranes. Elife. 2017;6 doi: 10.7554/eLife.19891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hutagalung A.H., Novick P.J. Role of Rab GTPases in membrane traffic and cell physiology. Physiol Rev. 2011;91:119–149. doi: 10.1152/physrev.00059.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Campa CC, Germena G, Ciraolo E, Copperi F, Sapienza A, Franco I, et al., Rac signal adaptation controls neutrophil mobilization from the bone marrow. Sci Signal 2016;9:ra124–ra124. [DOI] [PubMed]
- 77.Irazoqui J.E., Gladfelter A.S., Lew D.J. Scaffold-mediated symmetry breaking by Cdc42p. Nat Cell Biol. 2003;5:1062–1070. doi: 10.1038/ncb1068. [DOI] [PubMed] [Google Scholar]
- 78.Zerial M., McBride H. Rab proteins as membrane organizers. Nat Rev Mol Cell Biol. 2001;2:107–117. doi: 10.1038/35052055. [DOI] [PubMed] [Google Scholar]
- 79.Bezeljak U., Loya H., Kaczmarek B., Saunders T.E., Loose M. Stochastic activation and bistability in a Rab GTPase regulatory network. Proc Natl Acad Sci U S A. 2020;117:6540–6549. doi: 10.1073/pnas.1921027117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Campa C.C., Margaria J.P., Derle A., Giudice M.D., Santis M.C.D., Gozzelino L., Copperi F., Bosia C., Hirsch E. Rab11 activity and PtdIns(3)p turnover removes recycling cargo from endosomes. Nat Chem Biol. 2018;14:801–810. doi: 10.1038/s41589-018-0086-4. [DOI] [PubMed] [Google Scholar]
- 81.Bergeron-Sandoval L-P, Heris HK, Chang C, Cornell CE, Keller SL, François P, et al., Endocytosis caused by liquid-liquid phase separation of proteins, bioRxiv (2018).
- 82.Kozak M, Kaksonen M. Phase separation of Ede1 promotes the initiation of endocytic events, bioRxiv (2019) 861203.
- 83.Zamparo M., Valdembri D., Serini G., Kolokolov I.V., Lebedev V.V., Dall’Asta L., Gamba A. Optimality in self-organized molecular sorting. Phys Rev Lett. 2021;126 doi: 10.1103/PhysRevLett.126.088101. [DOI] [PubMed] [Google Scholar]
- 84.Strzyz P. Drugs enter a liquid phase. Nat Rev Mol Cell Biol. 2020;21 doi: 10.1038/s41580-020-0268-2. 419–419. [DOI] [PubMed] [Google Scholar]
- 85.Dolgin E. Drug startups coalesce around condensates. Nat Biotechnol. 2021;39:123–125. doi: 10.1038/s41587-021-00828-4. [DOI] [PubMed] [Google Scholar]
- 86.Bakthavachalu B., Huelsmeier J., Sudhakaran I.P., Hillebrand J., Singh A., Petrauskas A., Thiagarajan D., Sankaranarayanan M., Mizoue L., Anderson E.N. RNP-granule assembly via ataxin-2 disordered domains is required for long-term memory and neurodegeneration. Neuron. 2018;98:754–766. doi: 10.1016/j.neuron.2018.04.032. [DOI] [PubMed] [Google Scholar]
- 87.Fang X., Wang L., Ishikawa R., Li Y., Fiedler M., Liu F., Calder G., Rowan B., Weigel D., Li P. Arabidopsis FLL2 promotes liquid–liquid phase separation of polyadenylation complexes. Nature. 2019;569:265–269. doi: 10.1038/s41586-019-1165-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Powers S.K., Holehouse A.S., Korasick D.A., Schreiber K.H., Clark N.M., Jing H., Emenecker R., Han S., Tycksen E., Hwang I. Nucleo-cytoplasmic partitioning of ARF proteins controls auxin responses in Arabidopsis thaliana. Mol Cell. 2019;76:177–190. doi: 10.1016/j.molcel.2019.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Agudo-Canalejo J., Schultz S.W., Chino H., Migliano S.M., Saito C., Koyama-Honda I., Stenmark H., Brech A., May A.I., Mizushima N. Wetting regulates autophagy of phase-separated compartments and the cytosol. Nature. 2020:1–5. doi: 10.1038/s41586-020-2992-3. [DOI] [PubMed] [Google Scholar]
- 90.Klein I.A., Boija A., Afeyan L.K., Hawken S.W., Fan M., Dall’Agnese A., Oksuz O., Henninger J.E., Shrinivas K., Sabari B.R. Partitioning of cancer therapeutics in nuclear condensates. Science. 2020;368:1386–1392. doi: 10.1126/science.aaz4427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Reinkemeier C.D., Girona G.E., Lemke E.A. Designer membraneless organelles enable codon reassignment of selected mRNAs in eukaryotes. Science. 2019;363 doi: 10.1126/science.aaw2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Shin Y., Chang Y.-C., Lee D.S., Berry J., Sanders D.W., Ronceray P., Wingreen N.S., Haataja M., Brangwynne C.P. Liquid nuclear condensates mechanically sense and restructure the genome. Cell. 2018;175:1481–1491.e13. doi: 10.1016/j.cell.2018.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Banani S.F., Rice A.M., Peeples W.B., Lin Y., Jain S., Parker R., Rosen M.K. Compositional control of phase-separated cellular bodies. Cell. 2016;166:651–663. doi: 10.1016/j.cell.2016.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sanders D.W., Kedersha N., Lee D.S., Strom A.R., Drake V., Riback J.A., Bracha D., Eeftens J.M., Iwanicki A., Wang A. Competing protein-RNA interaction networks control multiphase intracellular organization. Cell. 2020;181:306–324. doi: 10.1016/j.cell.2020.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yang P., Mathieu C., Kolaitis R.-M., Zhang P., Messing J., Yurtsever U., Yang Z., Wu J., Li Y., Pan Q. G3BP1 is a tunable switch that triggers phase separation to assemble stress granules. Cell. 2020;181:325–345. doi: 10.1016/j.cell.2020.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Guillén-Boixet J., Kopach A., Holehouse A.S., Wittmann S., Jahnel M., Schlüßler R., Kim K., Trussina I.R., Wang J., Mateju D. RNA-induced conformational switching and clustering of G3BP drive stress granule assembly by condensation. Cell. 2020;181:346–361. doi: 10.1016/j.cell.2020.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Momin N., Lee S., Gadok A.K., Busch D.J., Bachand G.D., Hayden C.C., Stachowiak J.C., Sasaki D.Y. Designing lipids for selective partitioning into liquid ordered membrane domains. Soft Matter. 2015;11:3241–3250. doi: 10.1039/c4sm02856b. [DOI] [PubMed] [Google Scholar]
- 98.Bordovsky S.S., Wong C.S., Bachand G.D., Stachowiak J.C., Sasaki D.Y. Engineering lipid structure for recognition of the liquid ordered membrane phase. Langmuir. 2016;32:12527–12533. doi: 10.1021/acs.langmuir.6b02636. [DOI] [PubMed] [Google Scholar]
- 99.Nakatani N., Sakuta H., Hayashi M., Tanaka S., Takiguchi K., Tsumoto K., Yoshikawa K. Specific spatial localization of actin and DNA in a water/water microdroplet: Self-emergence of a cell-like structure. ChemBioChem. 2018;19:1370–1374. doi: 10.1002/cbic.201800066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Dewey D.C., Strulson C.A., Cacace D.N., Bevilacqua P.C., Keating C.D. Bioreactor droplets from liposome-stabilized all-aqueous emulsions. Nat Commun. 2014;5:1–9. doi: 10.1038/ncomms5670. [DOI] [PubMed] [Google Scholar]
- 101.Chong S., Dugast-Darzacq C., Liu Z., Dong P., Dailey G.M., Cattoglio C., Heckert A., Banala S., Lavis L., Darzacq X. Imaging dynamic and selective low-complexity domain interactions that control gene transcription. Science. 2018;361 doi: 10.1126/science.aar2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Elbaum-Garfinkle S., Kim Y., Szczepaniak K., Chen C.C.-H., Eckmann C.R., Myong S., Brangwynne C.P. The disordered P granule protein LAF-1 drives phase separation into droplets with tunable viscosity and dynamics. Proc Nat Acad Sci. 2015;112:7189–7194. doi: 10.1073/pnas.1504822112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Jeon B.-J., Nguyen D.T., Abraham G.R., Conrad N., Fygenson D.K., Saleh O.A. Salt-dependent properties of a coacervate-like, self-assembled DNA liquid. Soft Matter. 2018;14:7009–7015. doi: 10.1039/c8sm01085d. [DOI] [PubMed] [Google Scholar]
- 104.Nguyen D.T., Saleh O.A. Tuning phase and aging of DNA hydrogels through molecular design. Soft Matter. 2017;13:5421–5427. doi: 10.1039/c7sm00557a. [DOI] [PubMed] [Google Scholar]
- 105.Sato Y., Sakamoto T., Takinoue M. Sequence-based engineering of dynamic functions of micrometer-sized DNA droplets. Sci Adv. 2020;6:eaba3471. doi: 10.1126/sciadv.aba3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Johnson N.R., Wang Y. Coacervate delivery systems for proteins and small molecule drugs. Exp Opin Drug Deliv. 2014;11:1829–1832. doi: 10.1517/17425247.2014.941355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Day Kasey J., Kago Grace, Wang Liping, Richter J. Blair, Hayden Carl C., Lafer Eileen M. Liquid-like protein interactions catalyse assembly of endocytic vesicles. Nat. Cell Biol. 2021;23:366–376. doi: 10.1038/s41556-021-00646-5. [DOI] [PMC free article] [PubMed] [Google Scholar]