Abstract
Background. Infections caused by carbapenem-resistant Acinetobacter baumannii (CR-Ab) have become increasingly prevalent in clinical settings and often result in significant morbidity and mortality due to their multidrug resistance (MDR). Here we present an integrated whole-genome sequencing (WGS) response to a persistent CR-Ab outbreak in a Brisbane hospital between 2016–2018.
Methods. A. baumannii, Klebsiella pneumoniae, Serratia marcescens and Pseudomonas aeruginosa isolates were sequenced using the Illumina platform primarily to establish isolate relationships based on core-genome SNPs, MLST and antimicrobial resistance gene profiles. Representative isolates were selected for PacBio sequencing. Environmental metagenomic sequencing with Illumina was used to detect persistence of the outbreak strain in the hospital.
Results. In response to a suspected polymicrobial outbreak between May to August of 2016, 28 CR-Ab (and 21 other MDR Gram-negative bacilli) were collected from Intensive Care Unit and Burns Unit patients and sent for WGS with a 7 day turn-around time in clinical reporting. All CR-Ab were sequence type (ST)1050 (Pasteur ST2) and within 10 SNPs apart, indicative of an ongoing outbreak, and distinct from historical CR-Ab isolates from the same hospital. Possible transmission routes between patients were identified on the basis of CR-Ab and K. pneumoniae SNP profiles. Continued WGS surveillance between 2016 to 2018 enabled suspected outbreak cases to be refuted, but a resurgence of the outbreak CR-Ab mid-2018 in the Burns Unit prompted additional screening. Environmental metagenomic sequencing identified the hospital plumbing as a potential source. Replacement of the plumbing and routine drain maintenance resulted in rapid resolution of the secondary outbreak and significant risk reduction with no discernable transmission in the Burns Unit since.
Conclusion. We implemented a comprehensive WGS and metagenomics investigation that resolved a persistent CR-Ab outbreak in a critical care setting.
Keywords: carbapenem resistance, Acinetobacter baumannii, CR-Ab, whole-genome sequencing, WGS, intensive care unit, burns ward, metagenomics, genomics, surveillance
Data Summary
The datasets supporting the conclusions of this article are available in the short read archive (SRA) repository, under the following Bioprojects: the complete genomes for MS14413 (GenBank: CP054302.1) and MS14393 (GenBank: CP054303-CP054305) have been deposited under the Bioprojects PRJNA631347 and PRJNA631348, respectively. All isolate Illumina sequencing reads have been deposited under the Bioproject PRJNA631491. All metagenomic Illumina sequencing reads have been deposited under the Bioproject PRJNA631351.
Impact Statement.
Infections with carbapenem-resistant Acinetobacter baumannii (CR-Ab) have a high morbidity and mortality in healthcare settings and can be difficult to treat due to limited susceptibility to available antimicrobials. Rigorous surveillance and intervention methods are necessary to combat its spread within hospitals, particularly among patients in critical care. DNA sequencing has become instrumental in the detection and tracking of bacteria in hospitals, but barriers still exist to its routine implementation. Timely reporting and appropriate communication of findings are important features for the success and integration of genomics in healthcare settings. Here we present a thorough investigation of an ongoing CR-Ab outbreak in a tertiary hospital in Brisbane, Australia, between 2016–2018. Continual analysis and timely communication between the genomics and infection control teams allowed for the eradication of both the initial outbreak and recurrent infections, which were traced back to the hospital plumbing. Continued interventions have resulted in significant risk reduction to patients, with no cases in critical care since 2018. Here we provide examples of our genomics reporting scheme (both the original and the adapted version after ongoing feedback) as well as an interactive visualization of the outbreak using the Healthcare-Associated Infections Visualization Tool (HAIviz; https://haiviz.beatsonlab.com/). We believe this work to be of broad interest to both researchers and clinicians alike.
Introduction
Hospital outbreaks of multidrug-resistant Gram-negative pathogens present great risk to patients and are costly [1, 2]. Whole-genome sequencing (WGS) has been proposed as an effective tool to support infection-control responses to emerging outbreaks within the healthcare environment, but barriers exist to the effective implementation into clinical practice [3].
Acinetobacter baumannii has emerged over recent decades as a major nosocomial pathogen [4]. Its capacity to develop or acquire resistance to multiple antibiotic classes, in addition to intrinsic resistance to desiccation and disinfectants, contributes to persistence of A. baumannii in the hospital environment [5, 6]. It has frequently been a cause of nosocomial outbreaks, particularly in the critical care setting [7–9]. A. baumannii are often resistant to multiple antibiotic classes and the global incidence of extensively-drug-resistant (XDR) or even pan-drug-resistant (PDR) strains have been increasing [10–12]. Carbapenem-resistant A. baumannii (CR-Ab) have been seen at high prevalence in several areas, particularly in the Asian-Pacific region, Latin America and the Mediterranean [13]. Carbapenem resistance in A. baumannii usually arises from the acquisition of genes encoding carbapenemases, particularly OXA-type carbapenemases (e.g. OXA-23), and may be associated with high mortality in vulnerable patients [14].
Here we describe a large outbreak of CR-Ab, and other co-infecting MDR Gram-negative pathogens, occurring within an Intensive Care Unit (ICU) and burns facility. Incorporation of WGS in real-time facilitated rapid characterisation of this complex polymicrobial outbreak, provided a detailed understanding of transmission pathways and helped to direct a successful infection control response.
Methods
Study setting and patient inclusion
Primary isolates were obtained from patients admitted to the Royal Brisbane and Women’s Hospital (RBWH), a tertiary referral hospital with 929 beds in South-East Queensland, Australia. The RBWH has a 36 bed ICU providing highly specialist burns care for all of Queensland. The incidence of CR-Ab is low in Australian hospitals [15]. All new CR-Ab strains are routinely stored in the clinical laboratory for future reference. For the outbreak investigation, any patient admitted to the RBWH who cultured CR-Ab from any clinical or screening specimen from May to August 2016 was identified as a case and included in the primary outbreak analysis. Any CR-Ab cases during the outbreak period were also included to determine if plasmid-mediated resistance and dissemination was relevant, with any MDR Gram-negative bacilli (including ESBL-producing K. pneumoniae , carbapenem-resistant S. marcescens or carbapenem-resistant P. aeruginosa ) prospectively collected for further genomic analysis. Overall these included 28 CR-Ab, three carbapenem-sensitive A. baumannii , ten K . pneumoniae , seven P . aeruginosa , four S . marcescens and three Enterobacter cloacae (the E. cloacae were isolated in relation to a previous outbreak in the same hospital [16]). Stored CR-Ab isolates from a previous outbreak in 2006 [6], as well as other sporadic cases imported from overseas to the RBWH during 2015/2016 (prior to the outbreak) were included for further analysis. These included 17 historical CR-Ab isolates from earlier in 2016 (n=3), 2015 (n=2) and between 2000–2006 (n=12). A. baumannii identified from the outbreak until mid-2018 were also included in the analysis during continued surveillance and infection control monitoring. These included three carbapenem-sensitive A. baumannii and 19 CR-Ab isolates. A complete list of all isolates is provided in File S2 (available in the online version of this article).
Antimicrobial susceptibility testing
All bacterial isolates were identified by MALDI-TOF (Vitek MS; bioMérieux, France). Antimicrobial susceptibility testing was carried out using Vitek 2 automated AST-N426 card (bioMérieux). For the first eight sequential CR-Ab isolates, additional susceptibility testing was undertaken using Etest to determine MICs for meropenem, imipenem, colistin, tigecycline, fosfomycin, amikacin, sulbactam, doxycyline and ceftolozane/tazobactam, with disc diffusion to determine susceptibility to aztreonam and ceftazidime/avibactam. Additional colistin testing was carried out on suspected colistin-resistant isolates using broth microdilution via the MICRONAUT MIC-Strip Colistin (Merlin Diagnostika GmbH). Carbapenemase activity was assessed by the use of the Carba-NP test (RAPIDEC; bioMérieux) and screened for the presence of common carbapenemases found in Enterobacteriaceae using an in-house multiplex real-time PCR (that targets NDM, IMP-4-like, KPC, VIM and OXA-48-like carbapenemases). Once it became clear that all the outbreak strains had an identical antibiogram, susceptibility testing was confined to the Vitek 2 automated AST-N426 panel with MICs to tigecycline, doxycycline and colistin determined by Etest (as the only susceptible agents).
Bacterial culturing and genomic DNA extraction
All isolates were grown on horse blood agar at 37 °C overnight. For all historical and outbreak isolates collected between May–September of 2016, colonies were scraped from plates and resuspended in 5 ml Luria–Bertani (LB) broth. Then, 1.8 ml of resuspension was used for DNA extraction using the UltraClean Microbial DNA Isolation Kit (MO BIO Laboratories) as per the manufacturer’s instructions. All isolates collected after September 2016 were extracted using the DSP DNA Mini Kit on the QIAsymphony SP (Qiagen).
Isolate whole genome sequencing
Illumina WGS of suspected outbreak patient isolates and historical CR-Ab isolates was performed in four batches of between 10 and 18 samples between June and August 2016 at the Australian Centre for Ecogenomics (ACE), The University of Queensland (see Methods in the Supplementary Material). One CR-Ab isolate (MS14413) and one K. pneumoniae isolate (MS14393) were selected for sequencing with Pacific Biosciences (PacBio) Single Molecule Real-Time (SMRT) sequencing on an RSII machine (see Methods in the Supplementary Material). Subsequent Illumina WGS was carried out at Queensland Forensic Scientific Services (QFSS) (see Methods in the Supplementary Material).
Quality control and assembly of WGS data
Illumina raw reads were checked for contamination using Kraken [17] v0.10.5-beta and quality using FastQC v0.11.5 (www.bioinformatics.babraham.ac.uk/projects/). Raw reads were filtered for reads less than 80 bp and quality score less than five using Nesoni clip v0.130 (https://github.com/Victorian-Bioinformatics-Consortium/nesoni). Some reads required further hard trimming with Nesoni clip (10 bp from start, 40 bp from end). Isolates were assembled using SPAdes [18] v3.6.0 at default settings. Contigs less than 10× coverage were removed using a custom script. Assembly metrics were checked for quality using Quast [19] v4.3 (see File S2). Details of the PacBio genome assembly and annotation can be found in Methods in the Supplementary Material (Fig. S1, Table S1).
Genomic analysis and clinical reporting
Between June and August 2016, four reports of detailed bioinformatic analyses were prepared in response to available Illumina data for A. baumannii , K. pneumoniae , P. aeruginosa , S. marcescens and Enterobacter cloacae patient isolates. Comparative genome analysis using variant calling, phylogenetic reconstruction, transmission pathway prediction, MLST resistance gene prediction and plasmid characterization used in the clinical reports are given in Methods in the Supplementary Material. For subsequent analyses of the final genome dataset updated or alternative software was used as described below.
Core SNPs were identified using Snippy [20] (v4.3.6) at default settings and trimmed reads against the complete chromosomes for MS14413 (CR-Ab) and MS14393 ( K. pneumoniae ). Parsnp (v1.2) (at default with ‘-c’ flag) was used to visualize phylogenetic relatedness between the outbreak CR-Ab and the historical A. baumannii isolates. MLST was performed using mlst [21] v2.6 (https://github.com/tseemann/mlst) against the draft assemblies. Both the Oxford [22] and Pasteur [23] MLST schemes were used for the CR-Ab isolates. Resistance genes were identified using Abricate [24] v0.6 against the ResFinder database [25] (accessed 18 August 2017). Abricate was also used to determine plasmid types using the PlasmidFinder database [26] (accessed 18 August 2017). Comparative analyses were completed using the Artemis Genome browser and the Artemic Comparison Tool (ACT). Figures were constructed using EasyFig [27], BRIG [28] and FigTree [29]. The capsular polysaccharide (K) and lipooligosaccharide outer core (OCL) locus for A. baumannii were typed using Kaptive v0.5.1 [30] against the A. baumannii databases provided [31] (accessed 16 Dec 2020).
Metagenomic sequencing and analysis
Metagenomic sequencing of environmental samples and analysis was conducted as described previously [16]. Briefly, swab and water samples from the ICU and Burns Unit were collected in July 2018. DNA was extracted using the Qiagen DNeasy Powersoil extraction kit and sequenced at the Australian Centre for Ecogenomics on an Illumina NextSeq500. Metagenomic sequencing data was used to screen for evidence of the current A. baumannii outbreak strain, as well as a previously identified Enterobacter hormaechei strain responsible for an outbreak in the same ICU in 2015 (described in [16]).
All samples were screened for species using Kraken [17] v1.0 and resistance genes using SRST2 [32] v0.2.0 against the ARG-ANNOT [33] database. Mash [34] v1.1.1 was used at default settings to screen Illumina reads for each samples against our reference CR-Ab sketch (MS14413). Samples that shared ≥90% of hashes were mapped to the reference sequence. Mapped reads were parsed and de novo assembled using SPAdes [18] v3.11.1 for MLST analysis using mlst [21] v2.16.2 and nucleotide comparison using ACT [35] and BRIG [28].
Risk reduction assessment
We aimed to estimate the reduced risk of patient colonization following the identification of ST1050 CR-Ab by environmental metagenomic sequencing and the initiation of enhanced decontamination of hospital plumbing. The incidence rate of CR-Ab was measured pre-intervention and post-intervention. The point of intervention was defined as the targeted initiation of routine plumbing maintenance programme within the Burns and Intensive Care units in August 2018. The intervention was expected to generate immediate results with no lag time. The pre-intervention period was defined as May 2016 to August 2018 and post-intervention period as September 2018 to May 2020. All CR-Ab cases recorded in the hospital during these periods were included. Patients admitted to the Burns and Intensive Care units underwent standard clinical swabbing for surveillance and laboratory method for testing did not change over the study period. Statistical analyses were performed on Rv3.5.1.
Results
Case study
A 25-year-old patient with extensive burn injuries was retrieved from an overseas healthcare facility. As per infection-control protocols, the patient was placed on contact precautions and provided a single room. Initial nasal and rectal screening swabs were negative for MDR pathogens, including CR-Ab. An extended-spectrum beta-lactamase (ESBL)-producing Klebsiella pneumoniae was isolated from the patient’s respiratory secretions on day 4, and within 24 h a similar organism was isolated from blood cultures. Repeated collection of blood cultures demonstrated a polymicrobial culture with ESBL-producing K. pneumoniae , CR-Ab and Pseudomonas aeruginosa on day 6, that tested susceptible to all first-line agents. Over the following days, CR-Ab was also isolated from numerous clinical specimens, including a femoral line tip, endotracheal aspirates, rectal swabs, wound swabs and operative specimens collected from debrided tissue. Blood cultures repeatedly grew CR-Ab, (day 15 and 45 of admission), with the emergence of colistin resistance when tested by Etest (MIC 32 μg ml−1) on day 45. Serratia marcescens was co-cultured in blood on day 15 and was also grown from respiratory secretions and wounds swabs.
Over the next 5 months in 2016, 18 additional patients within the same ICU area were also found to be colonized or infected with phenotypically similar CR-Ab, K. pneumoniae , S. marcescens and/or P. aeruginosa . This included CR-Ab colonized cases identified in patients discharged from the ICU to the Burns Unit or other surgical wards throughout the hospital, and eventually patients admitted to the Burns Unit. The final CR-Ab case was identified several weeks later in a patient discharged from the Burns Unit and transferred to a hospital in a remote part of Queensland. An outbreak investigation team was constituted as soon as it was suspected that an outbreak of CR-Ab had occurred within the ICU and the use of WGS for strain characterization was initiated.
WGS predicted likely transmission pathways and ruled out non-outbreak cases
Between May to August 2016, a total of 55 isolates were recovered from 22 patients (see File S2). These isolates included A. baumannii , K. pneumoniae , S. marcescens, E. cloacae and P. aeruginosa . Species typing and antibiogram analysis alone were insufficient to determine clonal relationships between these isolates. As such, we used WGS to establish the relationship between isolates and predict patient transmission based on SNP accumulation.
We applied WGS in real-time over the course of the outbreak. Four reports aimed at communicating genomic analyses to infection control and other clinical staff at RBWH were delivered during the primary outbreak (22 June, 15 July, 2 August and 29 August). We managed on average a 1 week turn-around time between receiving the isolates and presenting a finalized report, which consisted of (i) a front-page overview of the analysis and key outcomes/interpretations conveyed as short bullet points, (ii) detailed analysis and diagrams on the internal pages, and (iii) method descriptions (see Methods in Supplementary Material). Actual time between receipt of sequencing data and reporting was 8–72 h depending on the complexity of analyses with supplementary interim reports and regular academic-clinical partner meetings necessary to communicate our comparative genomic analyses and help shape the content of the final reports (see File S1 for example reports from 22 June and 29 August, respectively). The Hospital-Acquired Infections Visualization tool (HAIviz) was also developed alongside analysis of this outbreak and was used to interactively display linked cases throughout the hospital wards [File S3 (video)].
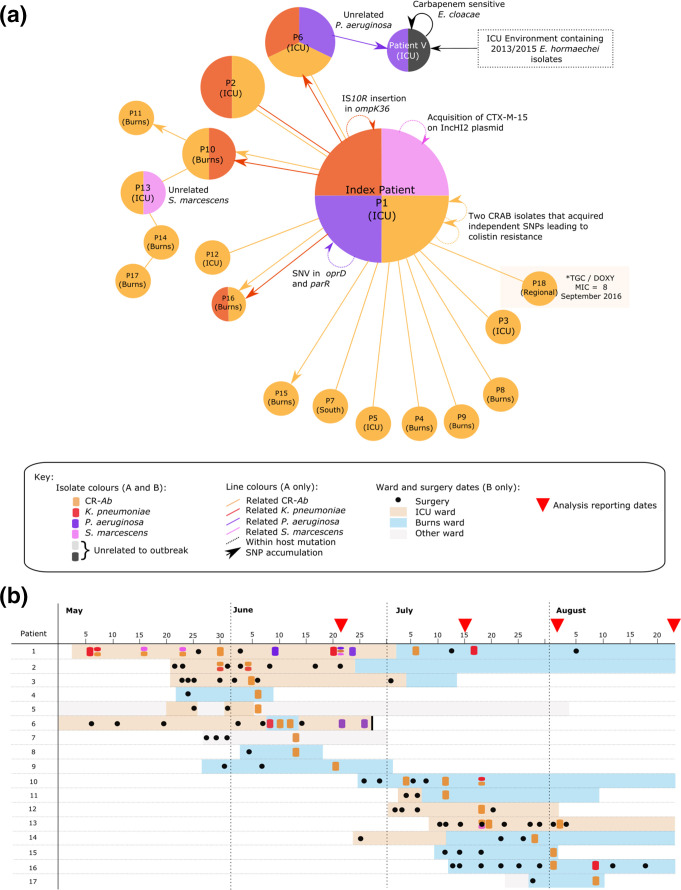
The presumed index patient admitted in early May 2016 was identified with ST1050 (Pasteur ST2) CR-Ab, ST515 K. pneumoniae , ST979 P. aeruginosa and S. marcescens . Using WGS, we found that 16 of the 21 patients admitted following the index patient had bacterial infections related to either the ST1050 CR-Ab or the ST515 K. pneumoniae . Transmission direction based on the accumulation of SNPs was inferred in patients 10, 11, 13, 14, 15, 16 and 17 (Fig. 1a, as indicated by lines with arrows). CR-Ab isolates from the first nine patients (and patient 12) were identical based on core SNPs, making inference of patient transmission impossible using SNPs alone. However, when combined with SNP information from K. pneumoniae isolates, it was possible to infer co-transmission of K. pneumoniae and CR-Ab from the index patient to patient 6 (Fig. 1).
Fig. 1.
Patient relationship matrix describing 2016 outbreak of CR-Ab: (a) Each circle represents a patient, where the size of the circle correlates to the number of isolates from that patient. Colours correspond to bacterial species. Straight lines connecting circles represent patients with identical isolates (with the colour of the line indicating the specific species) at the core-genome level (and as such directionality of transmission cannot be inferred). Lines with arrows (also coloured by species) represent predicted direction of transmission based on the accumulation of SNPs between patients’ isolates. Circular arrows represent changes in individual patient’s isolates, (b) timeline of patient samples, as well as location and surgery dates.
Strains of S. marcescens and P. aeruginosa specific to the index patient were not found in other patients (see Results in Supplementary Material). Two patients had unrelated S. marcescens and P. aeruginosa isolates (patients 13 and 6, respectively). Transmission of the unrelated P. aeruginosa isolate from patient 6 to another patient in the ICU ward (denoted patient V) was detected. Patient V was also found to have an Enterobacter cloacae isolate (later identified as Enterobacter hormaechei by WGS) identical to that identified in a 2015 outbreak from the same hospital [16]. This patient also carried an additional carbapenem-sensitive E. cloacae (blaIMP-4 negative) that was unrelated to the carbapenem-resistant isolate.
Over the course of the outbreak, each species carried by the index patient acquired additional antibiotic resistance mechanisms, via mutations (pmrB mutations in CR-Ab, IS10R insertions in ompK36 in K. pneumoniae, oprD nonsense mutation in P. aeruginosa ) or plasmid gain (IncHI2 in S. marcescens ) (Fig. 1, Results in Supplementary Material and Tables S2 and S3).
An additional CR-Ab was isolated in September 2016 from a patient in a Regional Queensland (QLD) hospital who had previously been admitted to the Brisbane ICU (patient 18, isolate MS14438). Analysis of this isolate found that it was closely related to isolates from the initial outbreak between May to August 2016.
Extensive environmental swabbing throughout the ICU and Burns Unit was conducted on the 16 June 2016, targeting patient bedrooms as well as high-touch areas (e.g. nurse keyboards, trolleys, door handles). However, no bacterial species related to the CR-Ab outbreak were detected in the environment based on traditional culture methods using chromogenic agar.
The outbreak CR-Ab was likely imported into the hospital ICU
In total, 29 CR-Ab isolates related to the ongoing outbreak were collected from 18 patients between May–September 2016. All were found to be ST1050 (Pasteur ST2; global clone [GC] 2) and less than ten SNPs different (Fig. S2). Three carbapenem-sensitive A. baumannii isolated at the same time were found to be different sequence types and unrelated to the outbreak. Comparison of the outbreak ST1050 CR-Ab isolates to historical CR-Ab isolates collected between 2000–2016 from the hospital found no close relationship, indicating that the CR-Ab had likely been introduced into the hospital with the index patient (Fig. S3).
All ST1050 CR-Ab isolates related to the index were found to be extensively resistant to carbapenems, β-lactams, cephalosporins, aminoglycosides and quinolones (Table 1). Resistance to colistin appeared in three isolates from the index patient and was mediated by two independent SNP acquisitions in the sensor kinase gene pmrB (causing the amino acid changes T235I in MS14413 and its descendant MS14402, and R263C in MS14407). These SNP corresponded to MICs of 8, 16 and 64 µg ml−1 for MS14402, MS14413 and MS14407, respectively, by broth microdilution. Antibiotic resistance genes were conserved between all isolates, and included β-lactamases (such as bla OXA-23 and bla OXA-66), streptomycin resistance genes (strA and strB), and aminoglycoside resistance genes [aph(3′)-Ic, aadA1 and the methylase armA]. Finally, a single SNP was found to result in the reversion of a nonsense mutation in a putative type 3 filamentous fimbriae gene (filB). This SNP was identified in the majority of CR-Ab isolates taken after the 4 July 2016 and appears to have arisen independently multiple times across the A. baumannii lineage (Results in Supplementary Material, Fig. S4).
Table 1.
CR-Ab MICs and AB resistance genes: table only shows select representative isolates as all CR-Ab were found to have the same AB resistance gene profile and MIC data. Colours represent mechanism of detection: blue, Etest MIC; Green, Disk diffusion zone diameter; Orange, Vitek2; Grey, Resfinder (accessed August 2017). A. baumannii is intrinsically resistant to penicillin and cephalosporins [50]
|
Strain |
MS8413 |
MS8419 |
MS8436 |
MS8442 |
MS8441 |
|
|---|---|---|---|---|---|---|
|
Patient |
4 |
5 |
6 |
7 |
8 |
|
|
Site |
Leg wound |
ETA |
Tissue buttock |
Wound Swab |
Rectal Swab |
|
|
Colistin |
Colistin |
0.125 |
0.25 |
0.25 |
0.125 |
0.5 |
|
Carbapenem |
Mero |
32 |
>32 |
>32 |
>32 |
>32 |
|
Imi |
>32 |
>32 |
n.t |
>32 |
n.t |
|
|
Erta |
>32 |
>32 |
>32 |
>32 |
>32 |
|
|
Beta-lactam and Cephalosporins |
Sulb |
32 |
32 |
64 |
32 |
64 |
|
MER |
R |
R |
R |
R |
R |
|
|
TIM |
R |
R |
R |
R |
R |
|
|
TAZ |
R |
R |
R |
R |
R |
|
|
CRO |
R |
R |
R |
R |
R |
|
|
CAZ |
R |
R |
R |
R |
R |
|
|
FEP |
R |
R |
R |
R |
R |
|
|
KZ |
R |
R |
R |
R |
R |
|
|
Azt |
6 mm R |
6 mm R |
n.t |
6 mm R |
n.t |
|
|
CTZ/TAZ |
>256 |
>256 |
128 |
16 |
96 |
|
|
CAZ/AVI |
16 mm R |
17 mm R |
18 mm R |
15 mm R |
18 mm R |
|
|
blaADC-25 |
+ |
+ |
+ |
+ |
+ |
|
|
blaOXA-23 |
+ |
+ |
+ |
+ |
+ |
|
|
blaOXA-66 |
+ |
+ |
+ |
+ |
+ |
|
|
Aminoglycosides |
Amikacin |
>256 |
>256 |
>256 |
>256 |
>256 |
|
GENT |
R |
R |
R |
R |
R |
|
|
TOB |
R |
R |
R |
R |
R |
|
|
Aph(3′)-Ic-1 |
+ |
+ |
+ |
+ |
+ |
|
|
aadA1 |
+ |
+ |
+ |
+ |
+ |
|
|
armA |
+ |
+ |
+ |
+ |
+ |
|
|
Quinolones |
CIP |
R |
R |
R |
R |
R |
|
NOR |
R |
R |
R |
R |
R |
|
|
Trimmethoprim/ Sulphonamide |
TMP |
R |
R |
R |
R |
R |
|
SXT |
R |
R |
R |
R |
R |
|
|
Sul1 |
+ |
+ |
+ |
+ |
+ |
|
|
Sul2 |
+ |
+ |
+ |
+ |
+ |
|
|
Tigecycline |
Tige |
2 |
2 |
2 |
2 |
4 |
|
Chloramphenicol |
Chloro |
6 mm R |
6 mm R |
n.t |
6 mm R |
n.t |
|
catB8 |
+ |
+ |
+ |
+ |
+ |
|
|
Fosfomycin |
Fosfo |
256 |
512 |
n.t |
128 |
n.t |
|
Tetracycline |
Doxy |
4 |
4 |
2 |
2 |
2 |
|
Macrolides |
mph(E)_3 |
+ |
+ |
+ |
+ |
+ |
|
msr(E)_4 |
+ |
+ |
+ |
+ |
+ |
|
|
Streptomycin |
strA |
+ |
+ |
+ |
+ |
+ |
|
strB |
+ |
+ |
+ |
+ |
+ |
|
n.t, not tested; Mero, Meropenem; Tige, Tigecycline; Sulb, Sulbactam; CTZ/TAZ, Ceftolozane/tazobactam; CAZ/AVI, Ceftazidime/avibactam; Chloro, Chloramphenicol; Fosfo, Fosfomycin; Azt, Aztreonam; Erta, Ertapenem; Doxy, Doxycycline; Imi, Imipenem; KZ, Cephazolin; TMP, Trimethoprim; SXT, Co-trimoxazole; GENT, Gentamicin; TOB, Tobramycin; CRO, Ceftriaxone; CAZ, Ceftazidime; FEP, Cefepime; TAZ, Piperacillin/tazobactam; CIP, Ciprofloxacin; NOR, Norfloxacin; MER, Meropenem; TIM, Ticarcillin/clavulanate.
PacBio sequencing of CR-Ab reveals context of resistance genes and mobile elements
Complete sequencing of a reference CR-Ab isolate (MS14413) from the index patient using long-read sequencing provided a high-quality reference and allowed contextualization of the antibiotic resistance genes (as well as other mobile genetic elements) within the genome. Assembly of the ST1050 CR-Ab reference genome revealed a 4 082 498 bp chromosome with no plasmids. StrA, strB and sul2 resided within a novel AbGRI1 resistance island most closely related to the A. baumannii strain CBA7 (GenBank:NZ_CP020586.1) isolated from Korea in 2017, both of which lacked the tetA-tetR genes commonly found in AbGRI1 (Fig. S5). The CR-Ab isolates also carried Tn6279 (also known as AbGRI3-2), which encompassed a large number of resistance genes including mph(E) and msr(E) (macrolide resistance) and the methylase gene armA (gentamicin resistance) (Fig. S6). Resistance to carbapenems in these CR-Ab isolates was likely driven by the presence of three copies of bla OXA-23 residing in separate Tn2006 transposons within the chromosome (two copies proximal to the capsule region, and the third interrupting a diguanylate cyclase gene, which has previously been implicated in biofilm formation [36]). An ISAba1 insertion sequence upstream of the chromosomal ampC gene was also detected, which has previously been shown to enhance cephalosporin resistance [37]. Additionally, an ISAba125 element was identified upstream of the csu operon, which is a well-characterized chaperone-usher pili assembly system involved in biofilm formation [38].
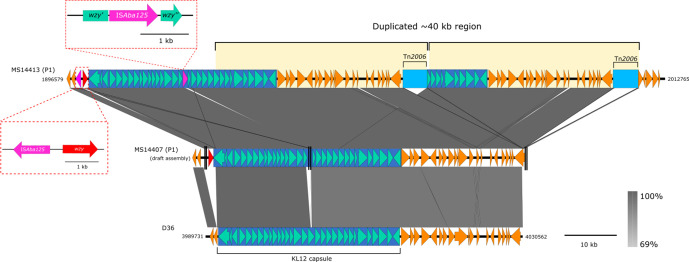
Long-read sequencing revealed an OCL1 oligosaccharide outer core and a KL12 capsule (K) locus, which shares 97% nucleotide identity to the capsule region found in the GC1 A. baumannii strain D36 (GenBank:NZ_CP012952.1) (Fig. S7). However, the wzy gene (a polymerase required for capsular polysaccharide biosynthesis) within the capsule locus was interrupted by an ISAba125 insertion sequence in all CR-Ab isolates. Further comparative analysis found a portion of the capsule locus in MS14413 to share 99% nucleotide identity to the capsule from A. baumannii strain BAL_097 (GenBank: KX712116), which carries a wzy gene at the beginning of the capsule region. This unusual gene placement also appears in MS14413, and likely complements the loss of the internal wzy gene (Fig. 2). The high nucleotide identity at this region also indicates possible recombination.
Fig. 2.
Large ~40 kb tandem duplication found in MS14413: Duplication of part of the capsule (k) region in the MS14413 complete genome (top line), resulting in three chromosomal copies of Tn2006 (third copy at alternate locus). This duplication appears to have arisen in some of the index patient isolates, but not other isolates involved in the outbreak (e.g. MS14407 concatenated draft genome, central line; vertical double black lines represent contig break in the draft assembly, presumed to be caused by the same IS as in MS14413). The wzy gene in the capsule region was found to be interrupted by an ISAba125 element, however a secondary wzy gene was identified at the start of the capsule region. Neither the ISAba125 insertion or secondary wzy gene is found in the KL12 capsule locus of A. baumannii strain D36 (bottom line).
Overlapping the capsule (K) region in MS14413 is a large 41 375 kb tandem duplication, encompassing two copies of Tn2006 (Fig. 2). Analysis of the other CR-Ab isolates using the Illumina de novo assemblies found evidence for this duplication in only one other related colistin-resistant isolates from the index patient (MS14402), suggesting that this duplication arose once and was maintained by a sub-population of CR-Ab within this patient for at least 36 days.
Transmission of K. pneumoniae parallel to CR-Ab transmission
Ten ESBL-producing K. pneumoniae isolates were collected from five patients during the outbreak and were all found to be ST515. Nine of the ten isolates differed by less than ten core SNPs, indicating direct transmission within the ICU ward (Fig. S8). A single isolate from the index patient (MS14418) was found to have an additional 61 core SNPs, consistent with a hypermutator phenotype. Further investigation of this isolate found an in-frame 9 bp deletion in mutH, resulting in the loss of 3 amino acids from this protein (Fig. S9).
All ESBL-positive K. pneumoniae isolates had identical antibiotic resistance gene profiles, including the ESBL gene bla CTX-M-15, other β-lactamases (bla TEM, bla OXA-1) and the aminoglycoside resistance gene aac(6′)Ib-cr (Fig. S6). Two isolates from the index patient (MS14393 and MS14418) developed resistance to carbapenems, which was likely due to an IS10R insertion in the outer membrane porin gene ompK36 (Fig. S8). Isolate MS14433 (from patient 16) also contained an IS10R inserted into ompK36, however the insertion was found to be close to the 5′ boundary of the ompK36 gene and based on in silico analysis there was no evidence that it affected the function of the resulting protein. Isolate MS14393 (from the index patient) also possessed a nonsense mutation in the antibiotic resistance protein repressor gene marR, which could contribute to its overall resistance to antibiotics.
A single K. pneumoniae isolate from the index patient (MS14393) was sequenced using PacBio long-read sequencing to generate a high-quality reference genome, consisting of a 5 492 431 bp chromosome, a 216 803 bp IncF plasmid (pMS14393A), and a 125 232 bp IncA/C plasmid (pMS14393B). Most of the antibiotic resistance genes resided on the IncA/C plasmid in two main loci (Fig. S6). The larger IncF plasmid did not contain any antibiotic resistance genes, but did harbour several heavy metal resistance operons, including resistance to copper, arsenic and mercury (Fig. S10). Comparison of the short-read assemblies to both plasmids confirmed that all ten K . pneumoniae isolates retained both plasmids.
Whole-genome shotgun metagenomics detects CR-Ab in hospital environment
Ongoing surveillance was conducted using WGS following the initial outbreak. Despite continual environmental cleaning and routine swabbing, the outbreak CR-Ab strain persisted through to September 2018 (Fig. 3). Swabs collected from surfaces within the ICU and Burns Unit (e.g. handles, tables, shelves, computer equipment) in 2016 and 2017 were unable to detect CR-Ab in the environment and did not yield enough DNA for direct metagenomic sequencing (data not shown).
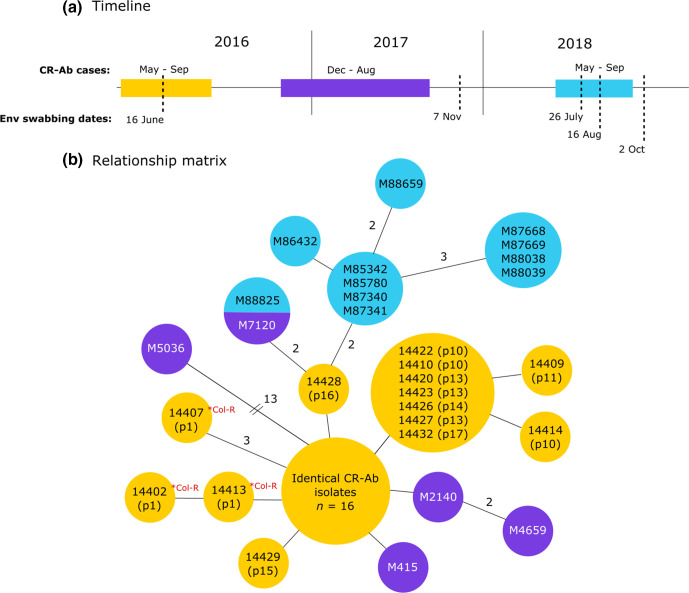
Fig. 3.
Ongoing CR-Ab surveillance from 2016 to 2018: (a) timeline of CR-Ab cases and dates of environmental swabbing between 2016–2018. (b) Relationship matrix of all CR-Ab isolates related to the initial outbreak. Col-R=predicted colistin resistance via mutation in pmrB. Isolates within the same circle are identical at the core genome. Branches represent 1 SNP difference (except where specified). Isolates from the original 2016 outbreak are in yellow. Purple isolates were collected in late 2016–2017. Isolates in blue were collected in 2018. Isolate M88825 was isolated from an Antechamber environment in 2018 and found to be identical at the core SNP level to M7120, isolated in August 2017.
Due to 11 new cases of CR-Ab detected between May to September 2018, additional environmental sampling was carried out in the Burns ward environment. Between July to October 2018, areas of presumed high bacterial load (such as floor drains, plumbing, inside burns bath drains etc) were targeted for environmental sampling (Fig. 4). All samples were subjected to culture using traditional methods (on chromogenic media) and direct DNA extraction and shotgun metagenomic sequencing. Of 50 environmental samples, two were culture positive for CR-Ab (R5666 and R5864), while four were positive based on analysis of the metagenomic sequencing data (R5515, R5510, R5863 and R5864) (Table S4, Fig. 4).
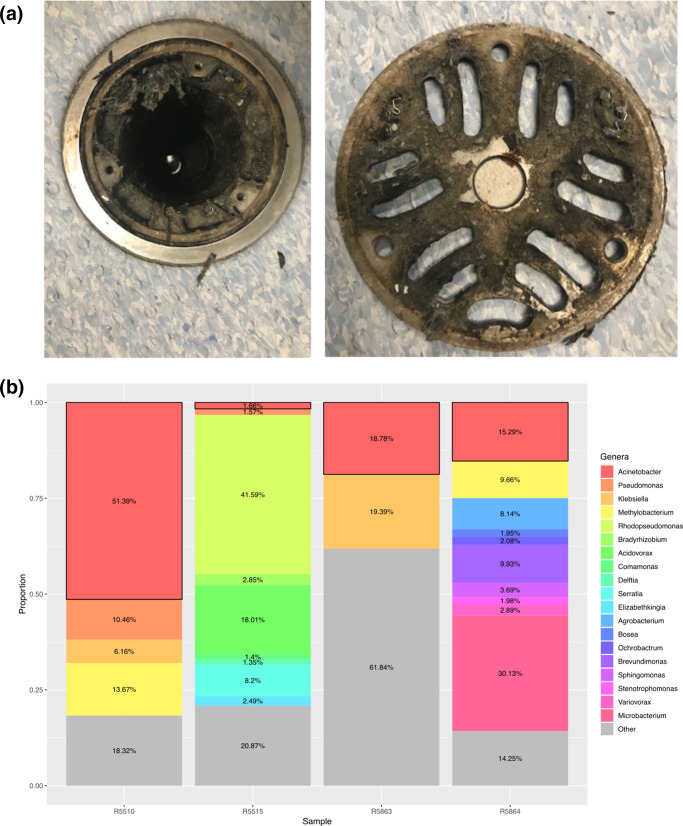
Fig. 4.
Burns bath 3 floor trap and metagenomic read abundance profiles: (a) an example of the biomass uncovered under the floor trap in a Burns Unit bathroom. Areas of high biomass (such as this one) were targeted for environmental screening. (b) Each column shows the relative abundance of paired-end reads for each environmental sample that were classified at a bacterial genus level by comparing against a database of bacterial genomes from RefSeq. Only bacterial genera with a relative abundance >0.5% are shown as distinct. Genera with an abundance of <0.5% are grouped together as ‘Other’ (grey). Boxes outlined in black represent abundance of ‘ Acinetobacter ’.
An ST1050 CR-Ab was cultured using traditional methods from the environmental sample R5666, taken from a crack in a toilet seat being used by a patient colonized with the ST1050 CR-Ab. The depth of sequencing obtained from the same environmental sample, however, was not sensitive enough to be able to confidently detect the presence of the CR-Ab in the metagenomic data. The second positive ST1050 CR-Ab culture came from an environmental sample taken from an Antechamber room connected to patient rooms that had previously been colonized with ST1050 CR-Ab (R5864). Parallel metagenomic sequencing was also able to detect this same ST1050 CR-Ab in the environmental sample (Fig. 5).
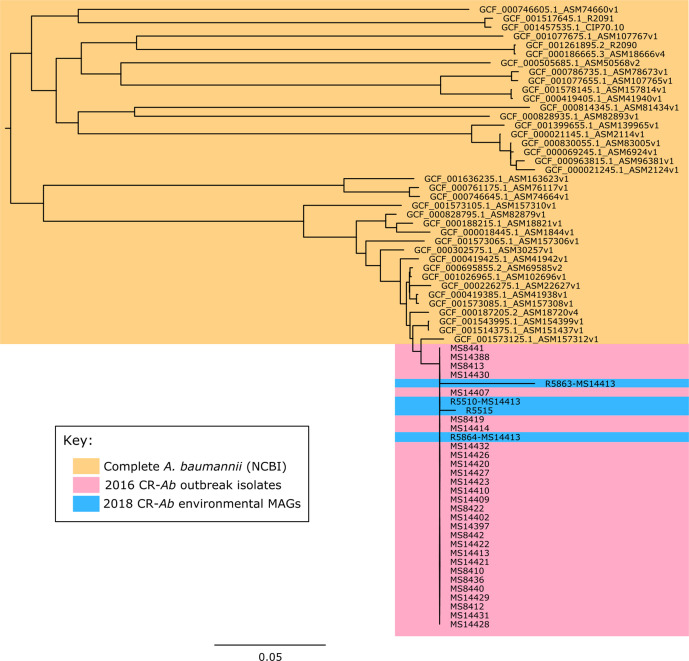
Fig. 5.
Clustering of MAGs with outbreak strains: Mid-point rooted core genome SNP phylogenetic tree contextualizing the metagenome assembled genomes (MAGs) with de novo assemblies of the outbreak strains and publicly available complete A. baumannii genomes (yellow) showing clustering of the MAGs (blue) within the outbreak clade (pink).
Additionally, three other samples were found to have ST1050 CR-Ab based on metagenomic sequencing, despite being culture negative using traditional methods (Fig. 5, Table S4). Samples R5515 (burns bath 2 floor trap water sample) and R5510 (burns bath 2 bath drain hole [interior]) were both positive for ST1050 CR-Ab. Both samples were taken at the same time from proximal locations, and patients colonized with ST1050 CR-Ab were using the burns bath in question. Samples R5863 was also positive for ST1050 CR-Ab, and was taken from the room previously occupied by a patient known to be colonized with ST1050 CR-Ab.
Plumbing maintenance programme implemented in response to genomic investigation
Shotgun metagenomic detection of the outbreak strain in the hospital plumbing provided the evidence base for implementation of a sustainable infection prevention strategy. Consequently, a routine plumbing maintenance programme was instituted. Every month, pipes were soaked for 30 min in sodium hydroxide, with additional soaking and scrubbing of drain plates. Since the implementation of these measures, no further cases of CR-Ab have been detected in the Burns Unit or Intensive Care Unit (ICU) following 28 September 2018. Periodic environmental surveillance of CR-Ab in drains and plumbing in the Burns unit has been ongoing as of May 2020.
Significant reduction of risk following interventions
A total of 32 CR-Ab cases were recorded over 28 months in the pre-intervention period, compared to four CR-Ab cases over 21 months in the post-intervention period. All cases identified at pre-intervention period were admitted to the Burns or Intensive Care Units during their hospitalization. Conversely, three out of the four CR-Ab cases detected in the post-intervention period had no obvious epidemiological link to exposure in the Burns and Intensive Care Units. The fourth case attended an external wound clinic, which was also attended by RBWH patients, although no specific patient or environmental link was proven. The last CR-Ab case detected in the Burns and Intensive Care unit was in September 2018, and there was no new case detected in the hospital after May 2019 (Fig. S11). The incidence rate post-intervention was two CR-Ab cases per year, significantly reduced from the pre-intervention incidence rate of 13 CR-Ab case per year (P<0.001). The post-intervention CR-Ab incidence rate was reduced by 17 % compared to the pre-intervention period (incidence rate reduction=0.17, 95 % CI: 0.06–0.47).
Discussion
CR-Ab are an increasingly dire threat to global public health. Their proficiency at surviving for long periods of time in environments whilst under antibiotic pressure is largely due to the positive selection of both intrinsic and acquired resistance and survival mechanisms. As such, they present a significant problem in healthcare settings, which typically have high antibiotic use as well as a large cohort of vulnerable patients. Understanding the mechanisms behind their resistance and transmission, as well as their possible environmental reservoirs, is key to combating further colonization and infection in hospital settings. Here we present a comprehensive analysis of an outbreak of CR-Ab using isolate and environmental metagenomic sequencing to fully elucidate transmission, determine new cases rapidly and detect possible environmental reservoirs within the hospital.
Genomics is being rapidly established in clinical settings, particularly in response to outbreaks [39, 40]. This is due not only to the higher discriminatory power that WGS provides, but also the complete picture that WGS captures by yielding the entire genome. The current cost and turnaround time for sequencing and analysis also make this type of investigation more feasible in nosocomial settings. In this study, initial sequencing of the outbreak CR-Ab isolates (and associated bacterial species) confirmed an already suspected outbreak, and so despite providing more insight into possible transmission routes, it did not greatly affect the infection control response. However, genomics superseded traditional methods when it came to (i) contextualizing outbreak isolates with previous CR-Ab strains from the hospital (to determine the likely source), and (ii) contextualizing new CR-Ab isolates as they appeared after the initial outbreak to determine whether there was an ongoing problem in the hospital. While having a slightly faster turnaround time, traditional methods alone would not have been able to confidently assess either of these scenarios. Regular meetings and reporting of the genomic results provided the hospital with actionable information and greater insight into the ongoing outbreak. These cross-disciplinary discussions facilitated the communication of complex genomic data into the clinical setting, providing guiding principles for subsequent WGS reporting of multidrug resistant bacterial pathogens at this hospital, and prompting the development of an interactive online visualization for communicating genomic epidemiology data (see HAIviz; File S3).
In addition to providing evidence for related isolates, WGS was also a valuable tool for discerning unrelated isolates, in many cases preventing ward or operating theatre closures and mitigating the associated financial costs to the hospital [41, 42]. It is plausible that with continued, ongoing sequencing of clinically significant bacteria in high-risk environments (e.g. ICU and Burns Unit) the risk of outbreaks could be reduced if evidence of transmission was detected early. During this study, we were able to detect transmission of an E. hormaechei unrelated to the outbreak at hand, but linked to a blaIMP-4 carbapenemase-producing Enterobacteriaceae (CPE) outbreak from the same hospital the year prior [16]. We were also able to identify transmission of an unrelated meropenem-resistant P. aeruginosa isolate, highlighting how WGS can detect transmission well before it becomes known to staff. Routine WGS can also lead to a reduction in the costs associated with responding to an established outbreak. A study of a similar outbreak in Brisbane determined the cost per patient related to the outbreak to be six-times higher than unrelated patients [43]. However, the feasibility (i.e. access to sequencing facilities and analysis) of routinely sequencing multidrug-resistant organisms is not yet achievable for many hospitals, particularly in low-resource settings. Despite this, recent collaborative efforts in the Philippines [44] have demonstrated how retrospective sequencing and capacity building for prospective sequencing can be achieved. WGS is also becoming increasingly cheap and portable (e.g. Oxford Nanopore Technology), and when coupled with more accessible cloud-based infrastructure, routine sequencing in these settings has greater potential.
Determining relatedness and transmission using genomics has historically relied on the number of core SNP differences between isolates [45–47]. However, this approach has several flaws, including a general lack of consensus on SNP cutoffs and what number defines a related isolate within a particular species, as well as the fact that it largely ignores other genomic differences, such as large insertions, inversion and rearrangements. It also does not account for hypermutators, which we observed in the case of the K. pneumoniae isolate MS14418. More recent methods have explored the use of transmission probabilities by taking into account isolation time and species mutation rate [48], but these methods appear more suited to outbreaks spanning large timeframes. Most studies to date that have used SNP distances have used them retrospectively and under research conditions, thereby avoiding the necessity to conform to standardized metrics and allow case-by-case judgments to be made on isolates. Moving forward, translating this approach into standardized clinical settings will likely present several hurdles. In our study, with the exception of the hypermutator strain MS14418 there was no ambiguity using SNP distances to determine relatedness due to the observed low mutation rate. However, because of this, many isolates were unable to be discriminated, with several identical at the core-genome level. We were surprised that the initial polymicrobial nature of this outbreak enabled deduction of transmission routes by examining SNP differences between their respective companion K. pneumoniae isolates, which appeared to have coinfected with the CR-Ab. However, all of these transmissions were from the index patient and were already recognized by the clinical team. In contrast, the spread of CR-Ab between the ICU and Burns Units in July could be traced to transmission of CR-Ab carrying a discriminatory SNP from the index patient to patient 10 in the Burns Unit with subsequent transmission of CR-Ab to patient 11, 14 and 17 in the Burns Unit and patient 13 in the ICU (Fig. 1). Further work into routinely automating the identification of both SNPs and pan-genome markers (such as gain/loss of regions or movement of mobile elements) could assist in further characterizing this outbreak and others.
Metagenomic sequencing of the environment was able to identify several areas positive for ST1050 CR-Ab. In one case, metagenomic sequencing analysis and traditional culture methods were concordant and both identified the ST1050 CR-Ab. In all other cases, either traditional culture or metagenomic sequencing was able to recover the ST1050 CR-Ab, highlighting the advantage of using both methods during an outbreak. In addition to the ST1050 CR-Ab, we were also able to use the metagenomic sequencing data to search for a previously identified E. hormaechei strain, which caused a small outbreak in 2015 in the same ICU [16] and was found again during this study. This highlights the wider utility of metagenomic sequencing to search for not only a single strain of interest, but potentially several, while also gaining greater insight into the microbial communities within the hospital plumbing.
While metagenomic sequencing was able to recover more positive results than the traditional methods, it has several limitations, including the necessity for high bacterial loads (such that there is sufficient starting DNA to sequence) and the increased costs (in our study, we observed that at least 5 Gigabase pairs of sequencing data is required to get a basic amount of depth and sensitivity when looking for specific strains, roughly equivalent to 5x the required sequencing for a single isolate). In future, initial PCR from the environmental DNA targeting a known marker in the outbreak strain could help narrow the candidates for complete metagenomic sequencing. Further work is required to refine these methods and determine an accurate guideline, particularly as it relates to sequencing depth and sensitivity.
All of the positive sequencing and culture results from the environmental sampling were from areas presently or previously being used by patients colonized with the ST1050 CR-Ab. As such, we cannot be sure that the identified ST1050 CR-Ab was present in these environments prior to colonization, or if it was shed from the patient. Subsequent environmental sampling was carried out after each round of cleaning, and no CR-Ab was detected afterwards. It is most likely that the CR-Ab detected in the environmental reservoirs were shed from the patients, however this result does indicate the ease of transmission of this organism from colonized patients to fomites within the hospital, where they then might transmit to other areas or to hospital staff [49]. Burns baths are a particular risk to patients, as denuded skin is easily colonized and presents a high risk for subsequent infection. ‘Splash-back’ from sinks and/or drains where MDR bacteria, such as CR-Ab, reside could present a hypothetical route for reinfection and ongoing transmission.
Conclusion
By using WGS to assist in a large outbreak of CR-Ab (and other MDR Gram-negative bacilli) we show how genomics can be used to improve rapid respond measures and outbreak management, as well as provide in-depth characterization of the outbreak strains to establish a historical database that can be used to guide responses to future outbreaks. We also show how direct sequencing of environmental samples was able to detect evidence of the outbreak strain leading to key changes in infection control policy.
Supplementary Data
Funding information
L.W.R. was supported by an Australian Government Research Training Programme (RTP) Scholarship. S.A.B., P.N.A.H. and M.A.S. were supported by fellowships from the Australian National Health and Medical Research Council (GNT1090456, GNT1157530 and GNT1106930, respectively). The work was supported by VC Strategic Intitiative Funding from The University of Queensland (2016–2018) and grants from the Queensland Genomics Health Alliance (now Queensland Genomics), Queensland Health, Queensland Government. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Acknowledgements
We acknowledge all staff at the Royal Brisbane and Women’s Hospital and Pathology Queensland who were involved in patient care and the clinical response to the outbreak described in this study. We acknowledge the facilities, and the scientific and technical assistance of staff at the Australian Centre for Ecogenomics Sequencing Facility [The University of Queensland (UQ)] and the Public Health Microbiology Laboratory at Queensland Forensic and Scientific Services (Queensland Health). We thank Thomas Cuddihy (QFAB Bioinformatics and Research Computing Centre, UQ) for high-performance computing support. We thank Kate Peters and Minh-Duy Phan (Schembri lab, UQ) For technical assistance preparing genomic DNA. We also thank Nguyen Thi Khanh Nhu (Schembri lab, UQ) and Chelsie Smith [The University of Queensland Centre for Clinical Research (UQCCR)] for their assistance with additional colistin testing of A. baumannii isolates.
Author contributions
L.W.R., P.N.A.H. and S.A.B. designed study. P.N.A.H., G.R.N., N.G., J.M. and J.L, coordinated patient inclusion and isolate collection from the hospital. L.W.R. and T.H. collected environmental samples. T.H. and K.H. coordinated infection control response. L.W.R., B.M.F. and W.L. performed experiments. L.W.R., S.A.B., B.M.F. and P.N.A.H. prepared clinical reports. All authors contributed to the interpretation of results. S.A.B., P.N.A.H., M.A.S. and D.P. supervised aspects of the project and provided essential expert analysis. L.W.R., P.N.A.H., S.A.B., W.L. and T.H. wrote the manuscript. All authors read and approved the final manuscript.
Conflicts of interest
P.N.A.H. has received research grants from MSD and Shionogi Ltd, outside of the submitted work, and speaker’s fees from Pfizer paid to The University of Queensland. D.L.P. reports receiving grants and personal fees from Shionogi and Merck Sharp and Dohme and personal fees from Pfizer, Achaogen, AstraZeneca, Leo Pharmaceuticals, Bayer, GlaxoSmithKline, Cubist, Venatorx, and Accelerate. J.L. has received personal fees from Pfizer and MSD and grants from MSD paid to The University of Queensland. The other authors have no conflicts of interest to declare.
Ethical statement
Ethics approval was provided by the RBWH HREC as a low-risk study with waiver of consent (HREC/16/QRBW/581).
Footnotes
Abbreviations: CPE, carbapenemase-producing Enterobacteriaceae; CR-Ab, carbapenem-resistant Acinetobacter baumannii; ESBL, extended-spectrum beta-lactamase; GC, global clone; ICU, intensive care unit; LB, Luria-Bertani; MAG, metagenome assembled genome; MALDI-TOF, matrix assisted laser desorption ionization-time of flight; MDR, multidrug-resistant; MIC, minimum inhibitory concentration; MLST, multilocus sequence typing; PacBio SMRT, pacific biosciences single molecule real-time; PCR, polymerase chain reaction; PDR, pan-drug-resistant; SNP, single nucleotide polymorphism; ST, sequence type; WGS, whole genome sequencing; XDR, extensively-drug-resistant.
All supporting data, code and protocols have been provided within the article or through supplementary data files. Four supplementary tables, 11 supplementary figures and supplementary video are available with the online version of this article.
References
- 1.Otter JA, Burgess P, Davies F, Mookerjee S, Singleton J, et al. Counting the cost of an outbreak of carbapenemase-producing Enterobacteriaceae: an economic evaluation from a hospital perspective. Clin Microbiol Infect. 2017;23:188–196. doi: 10.1016/j.cmi.2016.10.005. [DOI] [PubMed] [Google Scholar]
- 2.Cruickshank MMC. Reducing Harm to Patients from Healthcare Associated Infections: an Australian Infection Prevention and Control Model for Acute Hospitals. Sydney: Australian Commission on Safety and Quality in Health Care; 2009. [Google Scholar]
- 3.Gilchrist CA, Turner SD, Riley MF, Petri WA, Hewlett EL. Whole-genome sequencing in outbreak analysis. Clin Microbiol Rev. 2015;28:541–563. doi: 10.1128/CMR.00075-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peleg AY, Seifert H, Paterson DL. Acinetobacter baumannii: emergence of a successful pathogen. Clin Microbiol Rev. 2008;21:538–582. doi: 10.1128/CMR.00058-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Denton M, Wilcox MH, Parnell P, Green D, Keer V, et al. Role of environmental cleaning in controlling an outbreak of Acinetobacter baumannii on a neurosurgical intensive care unit. J Hosp Infect. 2004;56:106–110. doi: 10.1016/j.jhin.2003.10.017. [DOI] [PubMed] [Google Scholar]
- 6.Doidge M, Allworth AM, Woods M, Marshall P, Terry M, et al. Control of an outbreak of carbapenem-resistant Acinetobacter baumannii in Australia after introduction of environmental cleaning with a commercial oxidizing disinfectant. Infect Control Hosp Epidemiol. 2010;31:418–420. doi: 10.1086/651312. [DOI] [PubMed] [Google Scholar]
- 7.Wang SH, Sheng WH, Chang YY, Wang LH, Lin HC, et al. ealthcare-associated outbreak due to pan-drug resistant Acinetobacter baumannii in a surgical intensive care unit. J Hosp Infect. 2003;53:97–102. doi: 10.1053/jhin.2002.1348. [DOI] [PubMed] [Google Scholar]
- 8.Valencia R, Arroyo LA, Conde M, Aldana JM, Torres M-J, et al. Nosocomial outbreak of infection with pan-drug-resistant Acinetobacter baumannii in a tertiary care university hospital. Infect Control Hosp Epidemiol. 2009;30:257–263. doi: 10.1086/595977. [DOI] [PubMed] [Google Scholar]
- 9.del Mar Tomas M, Cartelle M, Pertega S, Beceiro A, Llinares P, et al. Hospital outbreak caused by a carbapenem-resistant strain of Acinetobacter baumannii: patient prognosis and risk-factors for colonisation and infection. Clin Microbiol Infect. 2005;11:540–546. doi: 10.1111/j.1469-0691.2005.01184.x. [DOI] [PubMed] [Google Scholar]
- 10.Nowak J, Zander E, Stefanik D, Higgins PG, Roca I, et al. High incidence of pandrug-resistant Acinetobacter baumannii isolates collected from patients with ventilator-associated pneumonia in Greece, Italy and Spain as part of the MagicBullet clinical trial. J Antimicrob Chemother. 2017;72:3277–3282. doi: 10.1093/jac/dkx322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xie R, Zhang XD, Zhao Q, Peng B, Zheng J. Analysis of global prevalence of antibiotic resistance in Acinetobacter baumannii infections disclosed a faster increase in OECD countries. Emerg Microbes Infect. 2018;7:31. doi: 10.1038/s41426-018-0038-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jones CL, Clancy M, Honnold C, Singh S, Snesrud E, et al. Fatal outbreak of an emerging clone of extensively drug-resistant Acinetobacter baumannii with enhanced virulence. Clin Infect Dis. 2015;61:145–154. doi: 10.1093/cid/civ225. [DOI] [PubMed] [Google Scholar]
- 13.Kamolvit W, Sidjabat HE, Paterson DL. Molecular epidemiology and mechanisms of carbapenem resistance of Acinetobacter spp. in Asia and Oceania. Microb Drug Resist. 2015;21:424–434. doi: 10.1089/mdr.2014.0234. [DOI] [PubMed] [Google Scholar]
- 14.Brown S, Amyes S. OXA (beta)-lactamases in Acinetobacter: the story so far. J Antimicrob Chemother. 2006;57:1–3. doi: 10.1093/jac/dki425. [DOI] [PubMed] [Google Scholar]
- 15.ACSQHC) ACoSaQiHC Aura 2019: third Australian report on antimicrobial use and resistance in human health. in. Sydney. 2019.
- 16.Roberts LW, Harris PNA, Forde BM, Ben Zakour NL, Catchpoole E, et al. Integrating multiple genomic technologies to investigate an outbreak of carbapenemase-producing Enterobacter hormaechei . Nat Commun. 2020;11:466. doi: 10.1038/s41467-019-14139-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wood DE, Salzberg SL. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 2014;15:R46. doi: 10.1186/gb-2014-15-3-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gurevich A, Saveliev V, Vyahhi N, Tesler G. QUAST: quality assessment tool for genome assemblies. Bioinformatics. 2013;29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seemann T. Snippy: fast bacterial variant calling from NGS reads. 2015.
- 21.Seemann T. mlst: https://github.com/tseemann/mlst. "This publication made use of the PubMLST website ( https://pubmlst.org/) developed by Keith Jolley (Jolley & Maiden 2010, BMC Bioinformatics, 11:595) and sited at the University of Oxford. The development of that website was funded by the Wellcome Trust"
- 22.Bartual SG, Seifert H, Hippler C, Luzon MAD, Wisplinghoff H, et al. Development of a multilocus sequence typing scheme for characterization of clinical isolates of Acinetobacter baumannii . J Clin Microbiol. 2005;43:4382–4390. doi: 10.1128/JCM.43.9.4382-4390.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Diancourt L, Passet V, Nemec A, Dijkshoorn L, Brisse S. The population structure of Acinetobacter baumannii: expanding multiresistant clones from an ancestral susceptible genetic pool. PLoS One. 2010;5:e10034. doi: 10.1371/journal.pone.0010034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seemann T. Abricate, Github https://github.com/tseemann/abricate
- 25.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, et al. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother. 2012;67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carattoli A, Zankari E, García-Fernández A, Voldby Larsen M, Lund O, et al. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother. 2014;58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sullivan MJ, Petty NK, Beatson SA. Easyfig: a genome comparison visualizer. Bioinformatics. 2011;27:1009–1010. doi: 10.1093/bioinformatics/btr039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alikhan N-F, Petty NK, Ben Zakour NL, Beatson SA. blast ring image generator (BRIG): simple prokaryote genome comparisons. BMC Genomics. 2011;12:402. doi: 10.1186/1471-2164-12-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rambaut A. FigTree. 2009.
- 30.Wyres KL, Wick RR, Gorrie C, Jenney A, Follador R, et al. Identification of Klebsiella capsule synthesis loci from whole genome data. Microb Genom. 2016;2:e000102. doi: 10.1099/mgen.0.000102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wyres KL, Cahill SM, Holt KE, Hall RM, Kenyon JJ. Identification of Acinetobacter baumannii loci for capsular polysaccharide (KL) and lipooligosaccharide outer core (OCL) synthesis in genome assemblies using curated reference databases compatible with Kaptive . Microb Genom. 2020;6 doi: 10.1099/mgen.0.000339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Inouye M, Dashnow H, Raven L-A, Schultz MB, Pope BJ, et al. SRST2: rapid genomic surveillance for public health and hospital microbiology Labs. Genome Med. 2014;6:90. doi: 10.1186/s13073-014-0090-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gupta SK, Padmanabhan BR, Diene SM, Lopez-Rojas R, Kempf M, et al. ARG-ANNOT, a new bioinformatic tool to discover antibiotic resistance genes in bacterial genomes. Antimicrob Agents Chemother. 2014;58:212–220. doi: 10.1128/AAC.01310-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ondov BD, Treangen TJ, Melsted P, Mallonee AB, Bergman NH, et al. Mash: fast genome and metagenome distance estimation using MinHash. Genome Biol. 2016;17:132. doi: 10.1186/s13059-016-0997-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carver TJ, Rutherford KM, Berriman M, Rajandream MA, Barrell BG. Parkhill J: ACT: the Artemis Comparison Tool. Bioinformatics. 2005;21:3422–3423. doi: 10.1093/bioinformatics/bti553. [DOI] [PubMed] [Google Scholar]
- 36.Whiteley CG, Lee D-J. Bacterial diguanylate cyclases: structure, function and mechanism in exopolysaccharide biofilm development. Biotechnol Adv. 2015;33:124–141. doi: 10.1016/j.biotechadv.2014.11.010. [DOI] [PubMed] [Google Scholar]
- 37.Corvec S, Caroff N, Espaze E, Giraudeau C, Drugeon H, et al. AmpC cephalosporinase hyperproduction in Acinetobacter baumannii clinical strains. J Antimicrob Chemother. 2003;52:629–635. doi: 10.1093/jac/dkg407. [DOI] [PubMed] [Google Scholar]
- 38.Tomaras AP, Dorsey CW, Edelmann RE, Actis LA. Attachment to and biofilm formation on abiotic surfaces by Acinetobacter baumannii: involvement of a novel chaperone-usher pili assembly system. Microbiology. 2003;149:3473–3484. doi: 10.1099/mic.0.26541-0. [DOI] [PubMed] [Google Scholar]
- 39.Quainoo S, Coolen JPM, van Hijum SAFT, Huynen MA, Melchers WJG, et al. Whole-genome sequencing of bacterial pathogens: the future of nosocomial outbreak analysis. Clin Microbiol Rev. 2017;30:1015–1063. doi: 10.1128/CMR.00016-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kwong JC, Lane CR, Romanes F, Gonçalves da Silva A, Easton M, et al. Translating genomics into practice for real-time surveillance and response to carbapenemase-producing Enterobacteriaceae: evidence from a complex multi-institutional KPC outbreak. PeerJ. 2018;6:e4210. doi: 10.7717/peerj.4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jiang Y, Resch S, Liu X, Rogers SO, Askari R, et al. The cost of responding to an Acinetobacter outbreak in critically ill surgical patients. Surg Infect. 2016;17:58–64. doi: 10.1089/sur.2015.036. [DOI] [PubMed] [Google Scholar]
- 42.Sadique Z, Lopman B, Cooper BS, Edmunds WJ. Cost-effectiveness of ward closure to control outbreaks of norovirus infection in United Kingdom National health service hospitals. J Infect Dis. 2016;213 Suppl 1:S19–26. doi: 10.1093/infdis/jiv410. [DOI] [PubMed] [Google Scholar]
- 43.Rodriguez-Acevedo AJ, Lee XJ, Elliott TM, Gordon LG. Hospitalization costs for patients colonized with carbapenemase-producing Enterobacterales during an Australian outbreak. J Hosp Infect. 2020;105:146–153. doi: 10.1016/j.jhin.2020.03.009. [DOI] [PubMed] [Google Scholar]
- 44.Argimón S, Masim MAL, Gayeta JM, Lagrada ML, Macaranas PKV, et al. Integrating whole-genome sequencing within the National Antimicrobial Resistance Surveillance Program in the Philippines. Nat Commun. 2020;11:2719. doi: 10.1038/s41467-020-16322-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Harris SR, Feil EJ, Holden MTG, Quail MA, Nickerson EK, et al. Evolution of MRSA during hospital transmission and intercontinental spread. Science. 2010;327:469–474. doi: 10.1126/science.1182395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schürch AC, Arredondo-Alonso S, Willems RJL, Goering RV. Whole genome sequencing options for bacterial strain typing and epidemiologic analysis based on single nucleotide polymorphism versus gene-by-gene-based approaches. Clin Microbiol Infect. 2018;24:350–354. doi: 10.1016/j.cmi.2017.12.016. [DOI] [PubMed] [Google Scholar]
- 47.Peacock SJ, Parkhill J, Brown NM. Changing the paradigm for hospital outbreak detection by leading with genomic surveillance of nosocomial pathogens. Microbiology. 2018;164:1213–1219. doi: 10.1099/mic.0.000700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stimson J, Gardy J, Mathema B, Crudu V, Cohen T, et al. Beyond the SNP threshold:identifying outbreak clusters using inferred transmissions. Mol Biol Evol. 2019;36:587–603. doi: 10.1093/molbev/msy242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kramer A, Schwebke I, Kampf G. How long do nosocomial pathogens persist on inanimate surfaces? A systematic review. BMC Infect Dis. 2006;6:130. doi: 10.1186/1471-2334-6-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sugawara E, Nikaido H. Ompa is the principal nonspecific slow porin of Acinetobacter baumannii . J Bacteriol. 2012;194:4089–4096. doi: 10.1128/JB.00435-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.