Abstract
Systemic sclerosis is an orphan, systemic autoimmune disease with no FDA-approved treatments. Its heterogeneity and rarity often result in underpowered clinical trials making the analysis and interpretation of associated molecular data challenging. We performed a meta-analysis of gene expression data from skin biopsies of patients with systemic sclerosis treated with five therapies: mycophenolate mofetil, rituximab, abatacept, nilotinib, and fresolimumab. A common clinical improvement criterion of 20% or 5 modified Rodnan skin score was applied to each study. We applied a machine learning approach that captured features beyond differential expression and was better at identifying targets of therapies than the differential expression alone. Regardless of treatment mechanism, abrogation of inflammatory pathways accompanied clinical improvement in multiple studies suggesting that high expression of immune-related genes indicates active and targetable disease. Our framework allowed us to compare different trials and ask if patients who failed one therapy would likely improve on a different therapy, based on changes in gene expression. Genes with high expression at baseline in fresolimumab nonimprovers were downregulated in mycophenolate mofetil improvers, suggesting that immunomodulatory or combination therapy may have benefitted these patients. This approach can be broadly applied to increase tissue specificity and sensitivity of differential expression results.
INTRODUCTION
Systemic sclerosis (SSc) is a rare systemic autoimmune disease characterized by skin fibrosis and lack of FDA-approved therapies. To gain mechanistic insight into the action of experimental therapies, clinical trials in SSc have collected genome-wide gene expression data from skin biopsies before and after treatment (Chakravarty et al., 2015; Chung et al., 2009; Gordon et al., 2015; Hinchcliff et al., 2013; Rice et al., 2015a). However, these studies face limitations. First, because SSc is a rare disease, clinical trials tend to have small sample sizes. Thus, few differentially expressed genes (DEGs) can be detected after multiple hypothesis testing correction (Chakravarty et al., 2015; Gordon et al., 2015). Second, not all therapy- or disease-relevant genes are regulated at the mRNA level. Identifying the functional consequences of treatments for SSc requires analytic methods beyond DEG analysis to infer the full biological impact of a therapy’s action.
In this study, we applied a machine learning method that uses “functional genomic networks” to learn the connectivity patterns of DEGs and extrapolate to the larger functional network in which a therapy is acting. We used our approach to perform comprehensive analyses of gene expression data from five independent therapeutic trials in SSc. We complement our framework with Gene Set Enrichment Analysis (GSEA) (Subramanian et al., 2005) to identify differentially expressed pathways and find broad concordance with our results.
Functional genomic networks are gene-gene interaction networks whose links encode functional relationships between genes, for example, membership in the same pathway. These publicly available, tissue-specific networks were constructed using curated biological information (i.e., process annotations), as well as raw biological data (Greene et al., 2015). In our meta-analyses, we used linear support vector machine (SVM) classifiers to identify the connectivity patterns of the genes modulated during clinically significant treatment response. These patterns were then used to identify relevant non-DEGs. We show that this extrapolated set of genes includes the known therapeutic targets demonstrating the power of our approach.
By examining multiple studies in parallel, we can identify the pathways commonly changed with clinical improvement, regardless of perturbation. We provide a comprehensive description of pathways altered during different treatments and identify molecular signature characteristics of clinical improvement. We show that nonresponders from one trial (fresolimumab) have signatures that suggest possible response to other therapies. These results may be used to guide drug repositioning for patients who do not respond to a given treatment in clinical trials and ultimately for precision medicine in SSc.
RESULTS
We analyzed publicly available gene expression data from clinical trials of five different therapeutics in patients with SSc (detailed information about each study is listed in Supplementary Table S1 online). These included the immunomodulators abatacept (CTLA4-IgG) (Chakravarty et al., 2015), mycophenolate mofetil (MMF) (Hinchcliff et al., 2013; Mahoney et al., 2015), and rituximab (anti-CD20) (Lafyatis et al., 2009; Pendergrass et al., 2012), a tyrosine kinase inhibitor nilotinib (Gordon et al., 2015), and fresolimumab, targeting all isoforms of transforming growth factor-β (TGF-β) (Rice et al., 2015a). All of these trials used skin disease severity measured by the modified Rodnan skin score as one of the outcomes. We applied a common improvement criterion, which was a minimum decrease in the modified Rodnan skin score of 20% or five points (Khanna et al., 2006) after treatment. The proportion of patients with available skin biopsy gene expression data who improved ranged from 27% (rituximab trial) to 83% (abatacept trial) (Table 1). The goal of this study was to understand the molecular processes perturbed by these diverse treatments, analyze those that did not change, and identify the differences between the subjects who did or did not improve.
Table 1.
Overview of improvers and nonimprovers based on the defined common improvement criterion
| Improvers ΔmRSS, ±SD |
Nonimprovers ΔmRSS, ±SD |
||||
|---|---|---|---|---|---|
| Treatment | Improvers:Nonimprovers1 | Post-Base | %(Post-Base) | Post-Base | %(Post-Base) |
| Abatacept | 5:1 | −12.6 ± 3.4 | −49.2 ± 9.0 | N/A | N/A |
| Fresolimumab | 6:4 | −10.2 ± 4.0 | −45.0 ± 12.7 | 3.3 ± 4.8 | 9.1 ± 16.9 |
| Mycophenolate mofetil | 12:6 | −6.4 ± 3.0 | −44.5 ± 14.8 | 0.2 ± 3.1 | 0.7 ± 12.4 |
| Nilotinib | 5:1 | −7.8 ± 2.9 | −31.7 ± 15.6 | N/A | N/A |
| Rituximab | 3:8 | −4.3 ± 0.6 | −27.4 ± 14.9 | 3.0 ± 5.2 | 13.7 ± 19.1 |
The absolute and percent changes in mRSS (± standard deviation) are reported.
Abbreviations: mRSS, modified Rodnan skin score; SD, standard deviation.
Only subjects with gene expression data were included.
Network-based machine learning captures known treatment targets despite lack of differential expression
We applied a machine learning approach to identify pathways downregulated by treatment (Figure 1). Genes with a nominally significant decrease after treatment (uncorrected P < 0.05, paired t-test) in clinical improvers were supplied as positive examples to an SVM classifier (adapted from Greene et al., 2015; Figure 1); genes that showed no evidence of differential expression were used as negative examples (0.95 < uncorrected P ≤ 1). The classifier learned the connectivity patterns of the DEGs in the Genome-scale Integrated Analysis of gene Networks in Tissues (GIANT) skin network and returned a ranking of all genes in the genome (Greene et al., 2015). Genes with high positive scores are most functionally similar to the nominally significant DEGs from the expression analysis (Greene et al., 2015), but are not necessarily differentially expressed. Thus, top-ranked genes may be unregulated at the mRNA level or “missed” due to small sample sizes, but are highly relevant to response. Similar approaches have been applied to genome-wide association study reanalysis (Greene et al., 2015) and to DEGs for novel viral antagonism mechanism detection (Gorenshteyn et al., 2015).
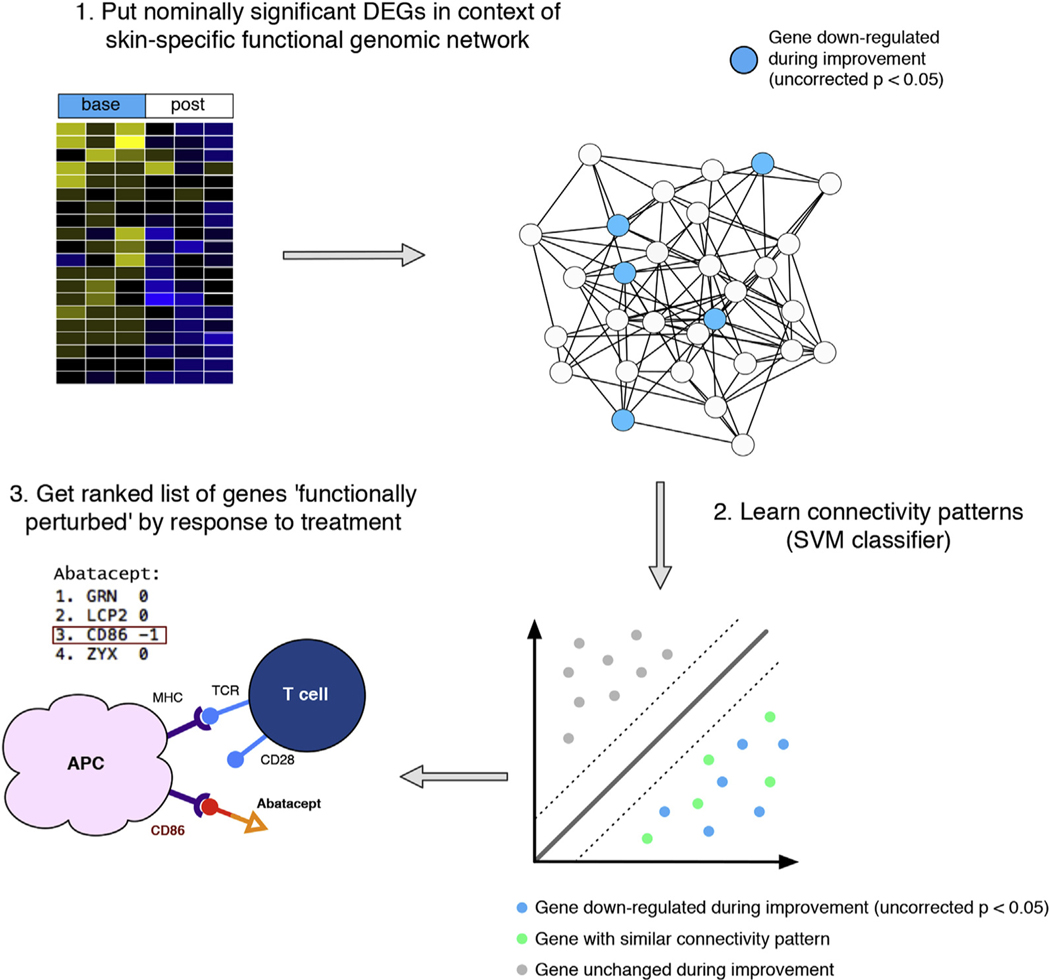
Figure 1. Schematic overview of machine learning approach.
Nominally significant genes that decrease after treatment (uncorrected P < 0.05) in clinically significant improvers are supplied as positive examples to a linear SVM classifier (DEGs); genes that show no evidence of differential expression are used as negative examples. The classifier learns the connectivity patterns of the DEGs in the GIANT skin network and returns a ranked list of gene symbols (highly positive: most like nominally significant DEGs). In the case of abatacept, the classifier returns CD86 as a highly ranked gene—one of the molecules that interacts with this biologic—despite being a negative example. APC, antigen-presenting cell; DEGs, differentially expressed genes; GIANT, Genome-scale Integrated Analysis of gene Networks in Tissues; MHC, major histocompatibility complex; SVM, support vector machine.
As a positive control, we tested whether our approach could prioritize known drug targets better than differential expression alone. Abatacept is a fusion protein that binds to CD80 or CD86. Using data from an investigator-initiated trial of abatacept in SSc (Chakravarty et al., 2015), the classifier returned CD86 as the third highest-ranked gene, despite not being differentially expressed (P = 1) (Figure 1). Differential expression analysis missed this gene, but the DEGs were functionally related to CD86 enabling our approach to find it. Beyond single gene targets, we found that our method is better at capturing relevant target gene sets than the t-statistic alone, which is used in many DEG approaches (Figure 2a and b).
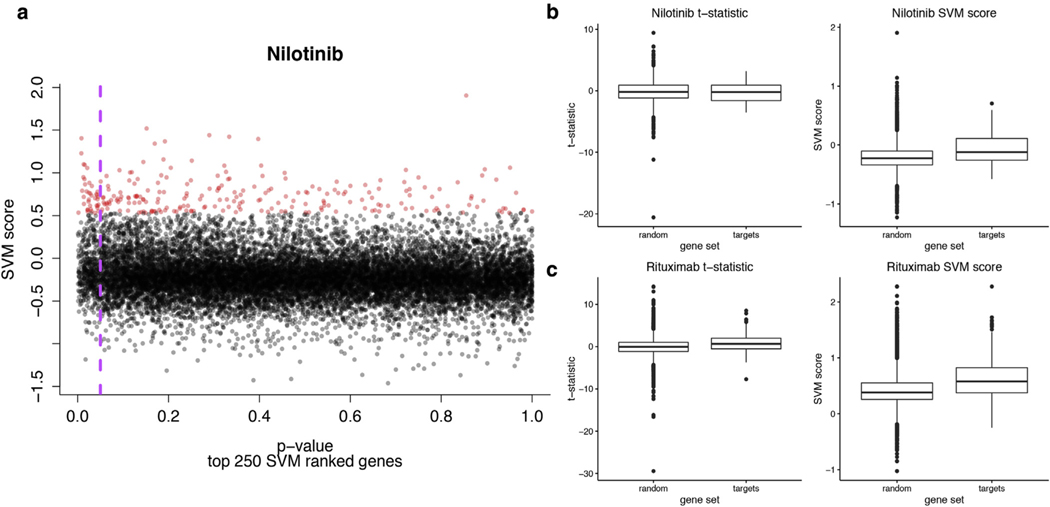
Figure 2. The linear SVM classifier captures features beyond just differential expression and better separates nilotinib targets from random sets of genes than the t-statistic alone.
(a) Scatterplot of SVM scores versus P-values. The genes with the highest SVM scores are highlighted in red. The purple dashed line indicates a P-value = 0.05. (b) Boxplots of SVM scores and t-statistics comparing the target gene set versus 100 gene sets of the same size (“random”). There is no significant difference in the t-statistic distributions (Mann-Whitney-Wilcoxon P = 0.73), whereas the difference in the SVM scores distributions is significant (Mann-Whitney-Wilcoxon P = 0.0004). (c) B-cell gene distributions (as annotated at the protein level using the Human Protein Reference Database) in the rituximab data—t-statistic Mann-Whitney-Wilcoxon P = 1.75 × 10−9, SVM score Mann-Whitney-Wilcoxon P <2.2 × 10−16. SVM, support vector machine.
Nilotinib is a tyrosine kinase inhibitor with a set of known targets (taken from Yoo et al., 2015, [D2]). This target set shows higher SVM scores than random gene sets of the same size (P = 0.0004), but there is no significant difference in t-statistics between target and random genes (P = 0.73) (Figure 2b). Our approach also sheds light on how cell types are perturbed during treatment. Rituximab has been shown to deplete dermal B cells (Lafyatis et al., 2009). B-cell-specific genes, as determined by the immune response in silico study (P = 0.029) (Abbas et al., 2005) and the Human Protein Reference Database (P <2.2 × 10−16) (Prasad et al., 2009), have higher SVM scores than random. In contrast, t-statistics of B-cell genes are either no different than random (immune response in silico; P = 0.38) or the difference is less statistically significant (Human Protein Reference Database; P = 1.75 × 10−9) (Supplementary Figure S1 online, Figure 2c).
Improvement-associated gene signatures indicate common pathways of skin disease resolution
We then investigated if the SVM-generated gene signatures for each therapy were functionally associated using a z-score method (Huttenhower et al., 2009) (Supplementary Figure S2aec online). This approach quantifies whether the top-ranked genes (250) from the SVM are more strongly connected to one another in the skin functional network than random gene sets of the same size. If two gene sets have a z-score >3, that indicates that they are significantly connected and therefore likely functionally related. Every pair of gene signatures from each trial was highly significant (Supplementary Figure S2a–c). Notably, improver signatures are generally more significant than nonimprover signatures (Supplementary Figure S2b) or treatment effect alone (Supplementary Figure S2c), suggesting that there is a core biology underlying the resolution of SSc skin disease.
Network theory identifies skin-specific functional gene sets
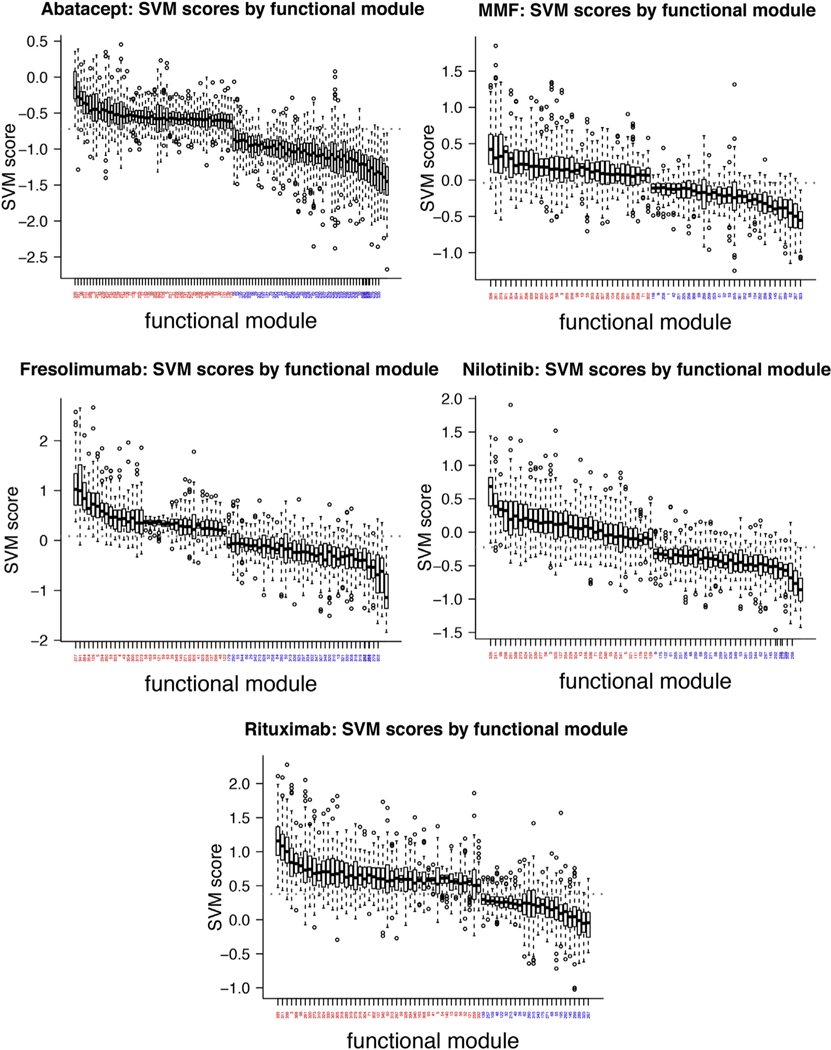
To determine the common pathways underlying skin disease resolution, we used community detection to identify functional modules in the GIANT skin network (Greene et al., 2015) (see the Materials and Methods section). Because of the way these networks were constructed, community detection identifies gene sets that participate in coherent biological processes in a tissue-specific manner. We determined which functional modules had high or low SVM scores to ascertain what pathways were downregulated or unchanged by treatment, respectively. Figure 3 illustrates functional modules with significantly high or low SVM scores as compared with the entire distribution (Wilcoxon test, Bonferroni-adjusted P < 0.001). Below, we report functional modules with high SVM scores for multiple therapies as indicative of “overlapping” modulated biology across trials (we restricted further study to the top 20 modules in each trial for brevity; all met the threshold outlined above). Generally, bottom-ranking functional modules encoded housekeeping processes (e.g., ribosome biogenesis). This is a useful positive control because we do not expect therapies to target such processes.
Figure 3. Boxplots of functional modules with significantly high or low SVM scores (Bonferroni-adjusted P < 0.001).
A red label indicates high SVM scores; blue indicates low scores. The gray dashed line indicates the median SVM score in each case. MMF, mycophenolate mofetil; SVM, support vector machine.
A module (273) enriched for fibrosis-related processes such as response to TGF-β and signaling by platelet-derived growth factor was highly ranked in all studies but abatacept (Table 2). Modules enriched in immune-related processes were highly ranked in all studies except fresolimumab (Table 2). This suggests that fresolimumab, a monoclonal antibody to TGF-β, does not modulate the same processes as other therapeutics, consistent with its mechanism of action. In addition, the perturbation of a module enriched for interleukin-6 signaling (261), specifically targeted by tocilizumab (anti-IL-6R) (faSScinate trial) (Khanna et al., 2015), highlights the central importance of IL-6 in SSc skin disease.
Table 2.
Functional modules predicted to be downregulated in four of five therapeutic trials
| Therapeutics | Functional module | Selected genes | Selected enriched biological processes |
|---|---|---|---|
| Fresolimumab MMF Nilotinib Rituximab |
273 | BMP2, JUN, JUNB, JUND, EDN1, VEGFA, SMAD3, SMAD7 | Response to transforming growth factor beta, response to hypoxia, signaling by PDGF |
| Abatacept MMF Nilotinib Rituximab |
261 | CXCL1, CXCL2, IL6, NFKB2, PLAUR, TNFAIP3 | I-kB kinase/NF-kB signaling, IL-6 signaling, IL-1 signaling |
| 3 | HLA-A, HLA-B, HLA-C, IFITM1-3, CASP1 | Antigen processing and presentation of exogenous peptide antigen via MHC class I, T-cell-mediated cytotoxicity, interleukin-10 secretion | |
| 311 | PTPN22, CD52, HLA-DQB1, IRF4, IRF5, IRF8 | MHC class II protein complex assembly, positive regulation of T-cell proliferation, TCR signaling | |
| 335 | CCL2, CD14, CD163, MS4A6A, TLR2, TLR4, CD86 | Vascular endothelial growth factor production, toll-like receptors cascades, tumor necrosis factor production | |
| 98 | CD3E, CD8A, CD8B, IL2RA, IL2RB, CD247, CD28 | Alpha-beta T-cell activation, lymphocyte costimulation, positive regulation of interleukin-2 production, costimulation by the CD28 family |
Top 20 significant modules only (all modules met Bonferroni-adjusted P < 0.001 threshold). Selected member genes and biological processes are shown. Functional enrichment was performed using gProfileR (Reimand et al., 2011).
Abbreviations: MHC, major histocompatibility complex; MMF, mycophenolate mofetil; PDGF, platelet-derived growth factor.
Different immunomodulatory treatments modulate distinct functional processes
Intriguingly, none of the top modules were shared between abatacept and MMF only (modules in common across abatacept, MMF, and at least one other treatment were found, Table 2). This is despite the fact that improvers in both studies have been reported to have high “inflammatory” signatures before treatment that were downregulated after treatment (Chakravarty et al., 2015; Hinchcliff et al., 2013). The lack of additional overlapping modules suggests potential heterogeneity within the inflammatory molecular intrinsic subset (Hinchcliff et al., 2013; Milano et al., 2008; Pendergrass et al., 2012; Johnson et al., 2015) that can be targeted by either therapy. To contrast the functional targets of these two therapies, we standardized their SVM scores (z-scores) to make a direct numerical comparison and identified functional modules significantly different between them (Supplementary Figure S3 online). T lymphocyte-, vascular-, and proliferation-related gene sets were likely to be differentially affected by abatacept and MMF (Supplementary Table S2 online). Abatacept had higher scores for vascular- and collagen-related modules (129 and 277), although it is possible that this is due to the greater magnitude of the improvement in the abatacept trial (Table 1). MMF had higher scores for proliferation (modules 285 and 302) and type I interferon modules (module 336). This may be due to the broadly immunosuppressive nature of MMF, which abrogates lymphocyte proliferation, compared with the molecularly precise action of abatacept, which inhibits T-cell costimulation of antigen presenting cells. Overall, there were fewer genes with positive SVM scores for abatacept.
Comparison of network analyses with GSEA results
As a final control for our analyses, we used GSEA to test each study for differential expression of sets of genes (pathways) rather than single genes. GSEA is a method for identifying gene sets that are altered between two phenotypes or timepoints. It is a well-established procedure that is complementary to and provides further validation of our network approach. We used a collection of curated “Hallmark” gene sets (Hallmarks) (Liberzon et al., 2015). The major limitation of GSEA compared with our approach is the requirement that users identify relevant gene sets (in this case Hallmarks) a priori. Nevertheless, the GSEA results were broadly concordant with our network results.
We focused on processes in common between studies, which represent biology relevant to disease resolution either due to intervention or spontaneous improvement. A single Hallmark, epithelial-mesenchymal transition, was significantly decreased after treatment in improvers from all five studies. All therapies except for fresolimumab modulated immune response-related Hallmarks, for example, IL6/JAK/STAT3 signaling and TNFA/NFKB signaling (Supplementary Table S3 online). This agrees with our network-based functional module results (Table 2; modules 261, 3, 311, 335, 98), where we found that immune-related processes were downregulated by all therapies except for fresolimumab.
All therapies except for MMF resulted in a decrease in TGF-β signaling (Supplementary Table S3). This is somewhat in contrast with our network results, where we found that all therapeutics but abatacept downregulated genes implicated in response to transforming growth factor beta (found in module 273). Some differences between the two methods are to be expected. The identification of functional modules is data driven, in contrast to the expert-curated Hallmarks, and we restricted our analysis to top modules only and not all that reached significance. However, there were two common “core enrichment” genes that significantly contributed to the enrichment in TGF-β signaling across all four studies: THBS1 and SERPINE1 (Supplementary Table S3). Both are strongly correlated with modified Rodnan skin score (Farina et al., 2010; Rice et al., 2015b). These genes are in module 304, which is one of the top 20 modules for MMF. This suggests that although MMF did not significantly downregulate TGF-β signaling by GSEA criteria, it is hitting a functionally similar set of genes by our network method. Likewise, although abatacept did not downregulate genes associated with TGF-β signaling, it did downregulate genes enriched for the Reactome pathway Degradation of the extracellular matrix (module 277). Thus, although the pathways that significantly overlap between studies depend somewhat on the method used, the downregulation of collagen or extracellular matrix deposition signaling is commonly found in all studies. In addition, both methods identified differences between the immunomodulatory treatments abatacept and MMF, suggesting that these two therapies may resolve skin disease severity differently. Most importantly, fresolimumab is the only therapy that does not appear to alter immune-related processes as determined by either method.
Fresolimumab nonimprovers may benefit from immunomodulatory treatment
Original studies largely ignored the molecular changes measured in nonimprovers. However, our results suggest that similar processes were downregulated in nonimprovers after treatment across multiple therapies (Supplementary Figure S2b). We hypothesized that the use of nonimprover signatures could help distinguish between processes that were truly downregulated due to improvement and those affected due to treatment alone, thus, allowing us to identify therapies that may be more clinically effective for a particular set of patients.
We used an SVM classifier on the GIANT skin network to classify genes that were uniquely downregulated in improvers after treatment (uncorrected P < 0.05); negative examples were genes uniquely downregulated in nonimprovers. This resulted in genes with highly positive SVM scores being most like genes downregulated in improvers and genes with highly negative SVM scores being most like genes downregulated in nonimprovers. We refer to these as therapeutic “post lists.” We performed a similar analysis to identify genes most like those elevated in improvers (highly positive scores) or in nonimprovers (highly negative scores) before treatment (termed “base lists”; see the Materials and Methods section).
We determined if subjects who failed to respond to fresolimumab (nonimprovers) may have had active inflammatory pathways that could be modulated by one of the other therapies. Functional annotation analyses for the bottom 250 genes in the fresolimumab base list (most like genes elevated in nonimprovers before treatment) showed significant enrichment for lymphocyte aggregation and type I interferon production. We then asked whether fresolimumab nonimprovers were likely to have responded to one of the immunomodulatory treatments analyzed, such as MMF.
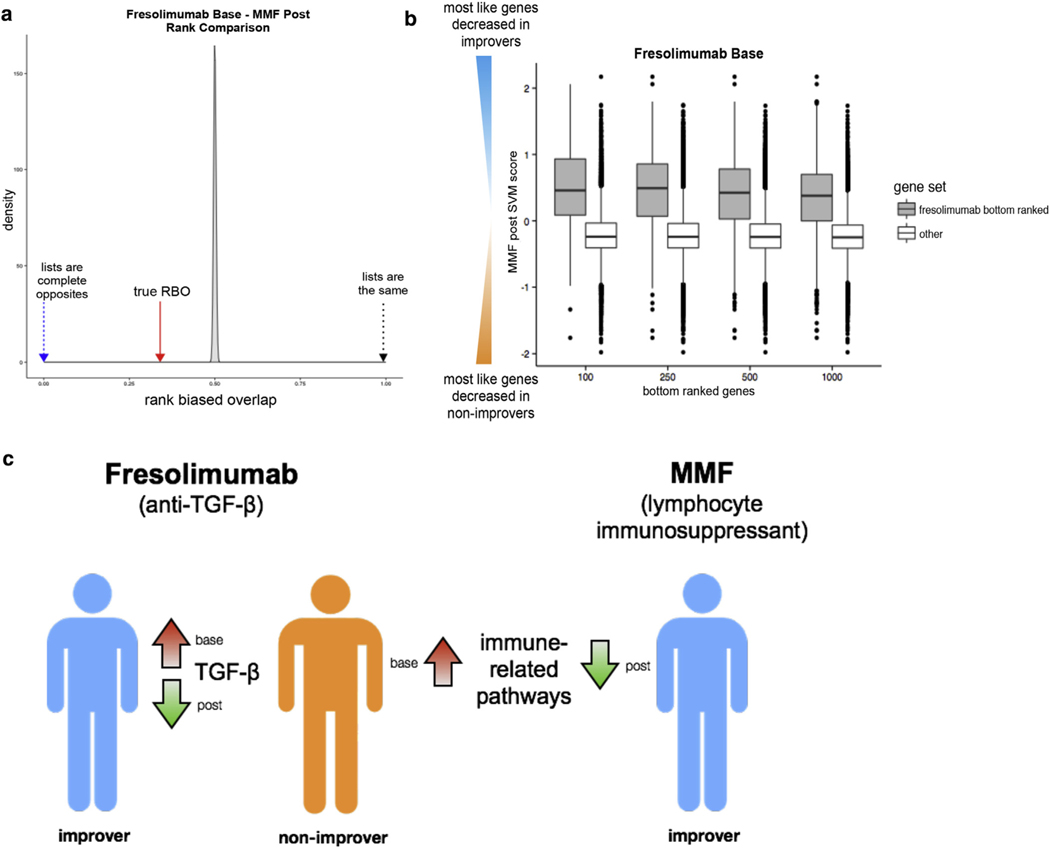
We determined if bottom-ranking genes in the fresolimumab base list were high-ranking genes in the MMF post list—that is, if genes most like those elevated in fresolimumab nonimprovers were also genes most like those downregulated uniquely in MMF improvers. First, we compared the fresolimumab base and MMF post lists using a metric called rank-biased overlap (the extrapolated version; Webber et al., 2010). The rank-biased overlap is a measure of the average agreement between two lists that takes the value 1 if they are exactly the same, 0 if they are exactly opposite, and 0.5 if the rankings are random with respect to each other (Figure 4a). The rank-biased overlap is an analog of rank correlation that is appropriate for lists that do not contain all of the same elements, as the base and post lists in this study (see Supplementary Methods online). The fresolimumab base and MMF post ranked lists have rank-biased overlap = 0.34, suggesting that they are significantly dissimilar. We also asked if the bottom 250 genes from the fresolimumab base list were enriched near the top of the MMF post list using a pattern-matching strategy (Lamb et al., 2006); the Kolmogorov–Smirnov statistic was highly significant (permuted P < 0.001) indicating that was the case. Finally, we showed that bottom-ranked fresolimumab base gene sets of various sizes had significantly highly positive MMF post-SVM scores (Figure 4b). These results suggest that fresolimumab nonimprovers not only had immune-related signatures active before treatment and not modulated by therapy, but also that these pathways may have been modulated by MMF treatment (Figure 4c).
Figure 4. Several complementary methods suggest that fresolimumab nonimprovers may have benefitted from MMF treatment.
(a) Density plot of the rank-biased overlap (RBO) null distribution (permuted SVM scores). The red arrow indicates the true RBO (0.34) between fresolimumab base and MMF post ranked lists. (b) Boxplot illustrating that fresolimumab base bottom-ranked gene sets of increasing size (100, 250, 500, and 1,000 genes) have significantly highly positive MMF post-SVM scores. (c) Schematic overview of findings. Fresolimumab nonimprovers have elevated immune-related genes pretreatment; immune-related genes are uniquely decreased in MMF improvers. MMF, mycophenolate mofetil; SVM, support vector machine; TGF-β, transforming growth factor-β.
DISCUSSION
Successfully treating SSc manifestations requires modulation of many biological pathways, which must occur downstream of a therapeutic molecule binding to its set of targets. Genome-wide gene expression data are now routinely gathered in clinical trials and provide insight into the functional consequence of treatment. A limitation is that most analyses only examine genes with statistically significant changes at the mRNA level, which may not fully capture the clinical impact of treatment.
We applied an approach that leverages functional genomic networks and machine learning to extrapolate from DEGs to the broader functional context in which a therapy is acting. Our strategy goes well beyond differential gene expression to identify the core processes and their critical component genes that change in response to therapy. As a positive control, we found that while a therapeutic’s targets are not always differentially expressed, the DEGs are highly functionally similar to the known therapy targets.
We cannot rule out the possibility that the subject composition of each of the cohorts included in this work influenced our results. However, we show that by putting nominally significant DEGs in the context of functional networks we can identify highly relevant genes (e.g., target kinases for nilotinib, B-cell genes for rituximab) that the original studies did not identify as significant (Figure 2, Supplementary Figure S2). Furthermore, our machine learning approach allows us to take advantage of nonimprovers in some studies and apply conservative cutoffs for gene sets of interest.
Our results add to the growing body of evidence that molecular phenotyping of SSc patients before treatment may increase the likelihood of meaningful clinical response. In particular, we observe the abrogation of inflammatory pathways in multiple trials regardless of a therapy’s mechanism of action, which supports the hypothesis that the high expression of immune-related genes may represent an active disease state that is most clinically actionable.
Across multiple studies, improvers were characterized by post-treatment downregulation of immune and fibrotic signaling. Of particular interest was the modulation of epithelial-mesenchymal transition in improvers from all trials. Genes comprising the epithelial-mesenchymal transition Hallmark showed significant enrichment in extracellular matrix organization and extracellular matrix organization-related functional terms, for example, cell adhesion, vasculature development, and collagen formation. This corroborates our previous work showing that an extracellular matrix organization functional module occupies a central part in the putative network structure of SSc (Mahoney et al., 2015).
However, we also noted subtle differences in the pathways modulated by abatacept and MMF, both immunomodulatory therapies, suggesting that there is no “panacea” for the inflammatory subset as of yet. Conversely, fresolimumab was shown to be less likely to downregulate immune pathways than the other therapeutics, and fresolimumab-treated nonimprovers had elevated inflammatory processes at baseline. This suggests that patients with elevated baseline inflammatory signature may benefit from an immunosuppressant, perhaps in combination with anti-TGF-β therapy.
In this work, we addressed some of the special considerations of pilot studies in a rare disease, and we aimed to be conservative, particularly because multiple trials were under examination. As the field continues to conduct small trials, it will be important not only to amass more molecular data, but also to build new frameworks to analyze and interpret them. Our results show that functional genomic networks are a powerful complement to purely statistical techniques. By extrapolating from a noisy list of DEGs, we have identified common mechanisms of action for multiple therapies, as well as critical differences between them.
MATERIALS AND METHODS
DEG analysis
DEG analysis was performed using the Comparative Marker Selection GenePattern module (Reich et al., 2006) using default parameters. For pre- and post-treatment comparisons, paired t-tests were used; for baseline comparisons, unpaired t-tests were used. The t-statistics and uncorrected P-values used throughout the text are taken from this analysis. Datasets used in this study are available in Gene Expression Omnibus under the following accession numbers: GSE66321, GSE55036, GSE76886, GSE65405, and GSE32413.
Functional genomic network analyses
All SVM classifiers were implemented using the network-guided genome-wide association study analysis from the GIANT webserver (http://giant.princeton.edu) in the context of the GIANT skin network (Greene et al., 2015). The features supplied to the SVM are the GIANT skin network edge weights. For improver signatures, the positive examples were genes downregulated after treatment in improvers (uncorrected P < 0.05) and the negative examples were genes unchanged after treatment (0.95 < uncorrected P ≤ 1); note that where there were enough nonimprovers with gene expression data, DEGs found in both improvers and nonimprovers were filtered out to better identify genes relevant to efficacy rather than solely to treatment. The same approach was used for nonimprover signatures. To generate “base” ranked lists, the positive examples were genes that were higher in improvers before treatment (uncorrected P < 0.05) and the negative examples were genes that were higher in nonimprovers before treatment (uncorrected P < 0.05). To generate “post” ranked lists, the positive examples were genes downregulated in improvers after treatment (uncorrected P < 0.05) and the negative examples were genes downregulated in nonimprovers after treatment (uncorrected P < 0.05). For the boxplots in Figure 2b and c and density plots in Supplementary Figure S1a and b, 100 gene sets of the same size as the target or B-cell gene sets were randomly sampled and one-sided Mann-Whitney-Wilcoxon tests were used to compare the distributions. More information on functional genomic and network methods is in the Supplementary Methods.
Supplementary Material
ACKNOWLEDGMENTS
JNT would like to thank members of the Whitfield Lab, J. K. Gordon, and C. S. Greene for helpful discussion. This work has been supported by grants from the Scleroderma Research Foundation (www.srfcure.org) to MLW, the Dr Ralph and Marian Falk Medical Research Trust Catalyst and Transformational Awards to MLW, and the National Institutes of Health P50 AR060780 and P30AR061271 to MLW. JNT received support from the John H. Copenhaver Jr and William H. Thomas, MD, 1952 Junior Fellowship from Dartmouth Graduate Studies and from the Molecular Cellular Biology at Dartmouth Training Grant (T32GM00870).
Abbreviations:
- DEG
differentially expressed gene
- GIANT
Genome-scale Integrated Analysis of gene Networks in Tissues
- GSEA
Gene Set Enrichment Analysis
- MMF
mycophenolate mofetil
- SSc
systemic sclerosis
- SVM
support vector machine
- TGF-β
transforming growth factor-β
Footnotes
SUPPLEMENTARY MATERIAL
Supplementary material is linked to the online version of the paper at www.jidonline.org, and at http://dx.doi.org/10.1016/j.jid.2016.12.007.
CONFLICT OF INTEREST
MLW is a Scientific Founder of Celdara Medical LLC and has filed patents for biomarkers in systemic sclerosis. JMM has been a paid consultant for Celdara Medical LLC.
REFERENCES
- Abbas AR, Baldwin D, Ma Y, Ouyang W, Gurney A, Martin F, et al. Immune response in silico (IRIS): immune-specific genes identified from a compendium of microarray expression data. Genes Immun 2005;6: 319–31. [DOI] [PubMed] [Google Scholar]
- Chakravarty EF, Martyanov V, Fiorentino D, Wood TA, Haddon DJ, Jarrell JA, et al. Gene expression changes reflect clinical response in a placebo-controlled randomized trial of abatacept in patients with diffuse cutaneous systemic sclerosis. Arthritis Res Ther 2015;17:159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung L, Fiorentino DF, Benbarak MJ, Adler AS, Mariano MM, Paniagua RT, et al. Molecular framework for response to imatinib mesylate in systemic sclerosis. Arthritis Rheum 2009;60:584–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farina G, Lafyatis D, Lemaire R, Lafyatis R. A four-gene biomarker predicts skin disease in patients with diffuse cutaneous systemic sclerosis. Arthritis Rheum 2010;62:580–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon JK, Martyanov V, Magro C, Wildman HF, Wood TA, Huang WT, et al. Nilotinib (Tasigna) in the treatment of early diffuse systemic sclerosis: an open-label, pilot clinical trial. Arthritis Res Ther 2015;17:213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorenshteyn D, Zaslavsky E, Fribourg M, Park CY, Wong AK, Tadych A, et al. Interactive big data resource to elucidate human immune pathways and diseases. Immunity 2015;43:605–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene CS, Krishnan A, Wong AK, Ricciotti E, Zelaya RA, Himmelstein DS, et al. Understanding multicellular function and disease with human tissue-specific networks. Nat Genet 2015;47:569–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinchcliff M, Huang CC, Wood TA, Matthew Mahoney J, Martyanov V, Bhattacharyya S, et al. Molecular signatures in skin associated with clinical improvement during mycophenolate treatment in systemic sclerosis. J Invest Dermatol 2013;133:1979–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttenhower C, Haley EM, Hibbs MA, Dumeaux V, Barrett DR, Coller HA, et al. Exploring the human genome with functional maps. Genome Res 2009;19:1093–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson ME, Mahoney JM, Taroni J, Sargent JL, Marmarelis E, Wu MR, et al. Experimentally-derived fibroblast gene signatures identify molecular pathways associated with distinct subsets of systemic sclerosis patients in three independent cohorts. PLoS One 2015;10:e0114017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna D, Denton CP, Jahreis A, van Laar JM, Cheng S, Spotswood H, et al. OP0054 safety and efficacy of subcutaneous tocilizumab in adults with systemic sclerosis: week 48 data from the fascinate trial. Ann Rheum Dis 2015;74:87–8. [Google Scholar]
- Khanna D, Furst DE, Hays RD, Park GS, Wong WK, Seibold JR, et al. Minimally important difference in diffuse systemic sclerosis: results from the D-penicillamine study. Ann Rheum Dis 2006;65:1325–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafyatis R, Kissin E, York M, Farina G, Viger K, Fritzler MJ, et al. B cell depletion with rituximab in patients with diffuse cutaneous systemic sclerosis. Arthritis Rheum 2009;60:578–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, Wrobel MJ, et al. The connectivity map: using gene-expression signatures to connect small molecules, genes, and disease. Science 2006;313:1929–35. [DOI] [PubMed] [Google Scholar]
- Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst 2015;1:417–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahoney JM, Taroni J, Martyanov V, Wood TA, Greene CS, Pioli PA, et al. Systems level analysis of systemic sclerosis shows a network of immune and profibrotic pathways connected with genetic polymorphisms. PLoS Comput Biol 2015;11:e1004005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milano A, Pendergrass SA, Sargent JL, George LK, McCalmont TH, Connolly MK, et al. Molecular subsets in the gene expression signatures of scleroderma skin. PLoS One 2008;3:e2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pendergrass SA, Lemaire R, Francis IP, Mahoney JM, Lafyatis R, Whitfield ML. Intrinsic gene expression subsets of diffuse cutaneous systemic sclerosis are stable in serial skin biopsies. J Invest Dermatol 2012;132:1363–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad TSK, Goel R, Kandasamy K, Keerthikumar S, Kumar S, Mathivanan S, et al. Human protein reference database—2009 update. Nucleic Acids Res 2009;37:D767–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich M, Liefeld T, Gould J, Lerner J, Tamayo P, Mesirov JP. GenePattern 2.0. Nat Genet 2006;38:500–1. [DOI] [PubMed] [Google Scholar]
- Reimand J, Arak T, Vilo J. g:Profiler—a web server for functional interpretation of gene lists (2011 update). Nucleic Acids Res 2011;39:W307–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice LM, Padilla CM, McLaughlin SR, Mathes A, Ziemek J, Goummih S, et al. Fresolimumab treatment decreases biomarkers and improves clinical symptoms in systemic sclerosis patients. J Clin Invest 2015a;125:2795–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice LM, Ziemek J, Stratton EA, McLaughlin SR, Padilla CM, Mathes AL, et al. A longitudinal biomarker for the extent of skin disease in patients with diffuse cutaneous systemic sclerosis. Arthritis Rheumatol 2015b;67: 3004–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledgesased approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 2005;102:15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webber W, Moffat A, Zobel J. A similarity measure for indefinite rankings. ACM Trans Inform Syst 2010;28:20. [Google Scholar]
- Yoo M, Shin J, Kim J, Ryall KA, Lee K, Lee S, et al. DSigDB: drug signatures database for gene set analysis. Bioinformatics 2015;31:3069–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.