Abstract
The nuclear envelope is often depicted as a static barrier that regulates access between the nucleus and the cytosol. However, recent research has identified many conditions in cultured cells and in vivo in which nuclear membrane ruptures cause the loss of nuclear compartmentalization. These conditions include some that are commonly associated with human disease, such as migration of cancer cells through small spaces and expression of nuclear lamin disease mutations in both cultured cells and tissues undergoing nuclear migration. Nuclear membrane ruptures are rapidly repaired in the nucleus but persist in nuclear compartments that form around missegregated chromosomes called micronuclei. This review summarizes what is known about the mechanisms of nuclear membrane rupture and repair in both the main nucleus and micronuclei, and highlights recent work connecting the loss of nuclear integrity to genome instability and innate immune signaling. These connections link nuclear membrane rupture to complex chromosome alterations, tumorigenesis, and laminopathy etiologies.
Keywords: membrane dynamics, nuclear lamina, ESCRT-III, chromothripsis, cGAS, TREX1
INTRODUCTION
A major function of the nuclear envelope (NE) is to compartmentalize the genome and regulate access to DNA. In eukaryotes the genome is surrounded by two lipid bilayers, the outer nuclear membrane (ONM) and inner nuclear membrane (INM), that merge at nuclear pore complexes (NPCs) and are contiguous with the endoplasmic reticulum (ER). Assembled on the INM is a complex protein matrix called the nuclear lamina. The main components of the nuclear lamina are the nuclear lamins, which are divided into two types based on their structure: the B-type lamins, lamin B1 and lamin B2, and the A-type lamins, lamin A and its splice variant lamin C in human cells. The lamins are intermediate-filament proteins that form independent meshworks on the INM (Shimi et al. 2015, Turgay et al. 2017). The density of the meshwork can be irregular across the surface of the INM, with gaps for NPCs (Turgay et al. 2017). Lamins localize to the membrane via posttranslational modification (C-terminal farnesylation and methyl esterification, B-type lamins) or interactions with NE transmembrane proteins (NETs) (A-type lamins) and can bind the chromatin, linking the genome to the INM (Gruenbaum & Foisner 2015). The nuclear lamina is also connected to the cytoskeleton by the linker of nucleoskeleton and cytoskeleton (LINC) complex (Jahed & Mofrad 2019). This review refers to the entire INM proteome, including lamins, NETs, INM-localized nucleoplasmic proteins, and the LINC complex, as the nuclear lamina. Together, the three structures of the NE, the membranes, lamina, and pores, establish a barrier between the nucleoplasm and the cytoplasm (Figure 1a). One of the major functions of the nuclear lamina is to resist force on the nuclear membrane and DNA, in part by binding heterochromatin at the nuclear periphery, but nuclear lamina proteins also have critical functions in gene expression and cell cycle regulation (Gruenbaum & Foisner 2015). For this reason, although alterations in nuclear lamina structure have been linked to cancer development and laminopathy etiologies for decades, how changes in NE structure cause disease pathologies is still unclear.
Figure 1.
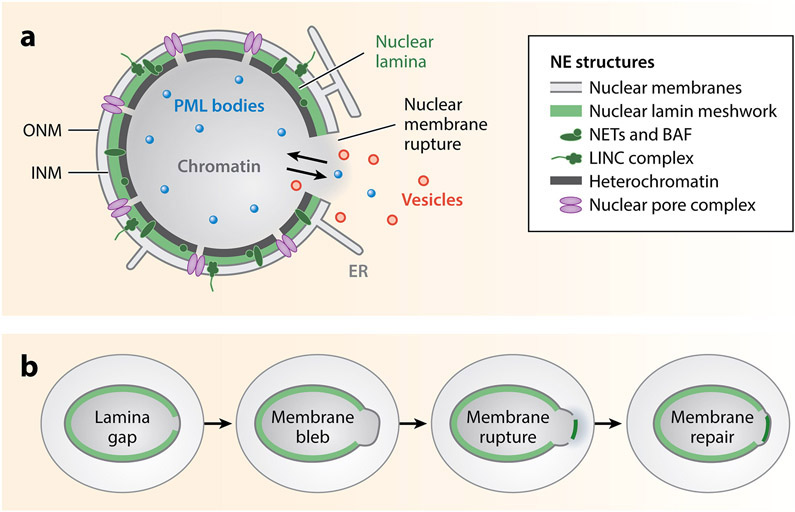
NE structures and membrane rupture. (a) Nucleus undergoing membrane rupture, showing the major NE structures including the membranes, lamina, nuclear pore complexes, chromatin, and peripheral heterochromatin. The nuclear lamina includes the nuclear lamin meshworks, NETs, BAF, and the LINC complex. The nuclear membrane is comprised of the outer and inner nuclear membranes, which are contiguous with each other and the ER. During membrane rupture, a pore is formed in the nuclear membrane that allows the free diffusion of nuclear and cytoplasmic proteins and small organelles, including PML bodies (blue) and vesicles (red) (DeVos et al. 2011, Vargas et al. 2012). (b) Stages of nuclear membrane rupture and repair. Many ruptures begin with the appearance of a gap in the nuclear lamina meshwork, which then becomes the site of membrane blebbing and chromatin herniation followed by membrane rupture when the nucleus is mechanically stressed. A subset of NE proteins (dark green) from the cytoplasm and ER accumulates on the exposed chromatin, along with other cytosolic DNA-binding proteins. These proteins persist at rupture sites even after nuclear compartmentalization is restored in the membrane repair process. This is only one possible order of events. Abbreviations: BAF, barrier-to-autointegration factor; ER, endoplasmic reticulum; INM, inner nuclear membrane; LINC, linker of nucleoskeleton and cytoskeleton; NE, nuclear envelope; NET, NE transmembrane proteins; ONM, outer nuclear membrane; PML, promyelocytic leukemia.
Recent work has highlighted the dynamic properties of the nuclear membrane and demonstrated that misregulation of these dynamics has significant consequences for the cell. One dynamic under investigation is interphase nuclear membrane rupture and repair (Figure 1b). This phenomenon is identified by rapid mislocalization of soluble nuclear and cytoplasmic proteins, frequently fluorescent proteins tagged with a nuclear localization signal (NLS) or nuclear export signal (NES) (de Noronha et al. 2001, De Vos et al. 2011, Vargas et al. 2012), and accumulation of cytosolic proteins on the exposed chromatin, typically BAF (barrier-to-autointegration factor) or cGAS (cyclic GMP-AMP synthase) (Denais et al. 2016, Harding et al. 2017, S. Liu et al. 2018, Mackenzie et al. 2017, Raab et al. 2016). Nuclear membrane ruptures frequently reseal within minutes but can sometimes persist for hours with no loss of cell viability (de Noronha et al. 2001, De Vos et al. 2011, Denais et al. 2016, Vargas et al. 2012), although unrepaired nuclear ruptures eventually lead to death (Kinugasa et al. 2019). In contrast to the primary nucleus, nuclei that form around missegregated chromatin, called micronuclei, rupture at a high frequency and fail to repair their membranes but without triggering cell cycle arrest or death (Crasta et al. 2012, Hatch et al. 2013). In human cells, micronuclei can vary in volume from under 1 μm3 for compartments formed around DNA fragments and small chromosomes to up to several 10s of μm3 for compartments formed around multiple chromosomes. A traditional size cutoff for distinguishing micronuclei from multipolar mitosis products has been that micronuclei must be less than one-third the diameter of the primary nucleus (Fenech 2007). After membrane repair, mislocalized proteins containing a nuclear transport signal can be trafficked into the correct compartment. However, both proteins lacking transport signals and mislocalized organelles remain trapped in the wrong compartment until they are degraded or the cell enters mitosis.
This review highlights the growing interest in how NE disorganization leads to membrane instability, compromised genome stability, and immune activation. These striking consequences have seized the attention of the research community, and we anticipate that further study will continue to yield fundamental insights into the critical barrier function of the NE and the consequences of nuclear decompartmentalization. Although principles are emerging to characterize the biological consequences of nuclear membrane rupture and the cytoplasmic exposure of DNA, there are important distinctions that depend on the causes and positioning of nuclear decompartmentalization (see the sidebar titled Sources of Cytoplasmic DNA).
SOURCES OF CYTOPLASMIC DNA.
Primary Nuclear Membrane Rupture
The primary NE ruptures during interphase in many cancer cell lines, during cell migration through tightly constricted areas, following errors in cell division, and during senescence (Denais et al. 2016, Dou et al. 2015, Maciejowski et al. 2015, Raab et al. 2016, Vargas et al. 2012). Nuclear envelope disruptions can also be induced in nontumorigenic cell lines by lamin B1 deficiency and p53 or Rb depletion (Dou et al. 2015, 2017; Glück et al. 2017; Yang et al. 2017b).
Micronuclei
Micronuclei form when a chromosome or acentric fragment fails to attach appropriately to the mitotic spindle, then lags during anaphase and is not incorporated into one of the two primary nuclei. Nuclear envelope disruption at micronuclei is frequently irreversible and is associated with DNA damage of unknown cause (Hatch et al. 2013).
DNA Bridges
DNA bridges are a distinct type of nuclear aberration that form as a result of errors in cell division caused by telomere fusion or the formation of merotelic kinetochore-microtubule attachments (Maciejowski et al. 2015, Umbreit et al. 2020). Following DNA bridge resolution, stubs of the bridges persist and are usually reincorporated into the primary nucleus after mitotic nuclear envelope breakdown (Maciejowski et al. 2019). DNA bridges are distinct from ultrafine bridges, which are formed by sister chromatid nondisjunction or unresolved recombination intermediates (Chan et al. 2007, 2018).
Extrachromosomal DNA
Circular DNAs are often made up of repetitive sequences such as telomeric or ribosomal DNA (Paulsen et al. 2018). Extrachromosomal DNAs are also found in up to 50% of human cancers and frequently contain oncogenes (Turner et al. 2017).
Mitochondrial Dysfunction
Damage to mitochondria or mitochondrial DNA can cause the accumulation of mitochondrial DNA in the cytosol.
Leaky DNA
So-called leaky DNAs are single- and double-stranded DNA species that escape the nucleus due to endogenous retroelement activation, DNA damage, or DNA replication stress (Coquel et al. 2018, De Cecco et al. 2019, Stetson et al. 2008).
Overview of Primary Nucleus Rupture
Rupture and repair of the nuclear membrane have been observed in primary nuclei in several species both in vitro and in vivo. In vitro, rupture and repair dynamics were first characterized in cells expressing the HIV protein VPR (de Noronha et al. 2001), then validated both in fibroblasts from patients with laminopathies (De Vos et al. 2011), a class of genetic diseases caused by mutations in lamin proteins, and in cancer cells (Vargas et al. 2012). Nontransformed cell lines exhibit little to no membrane rupture (Comaills et al. 2016, De Vos et al. 2011, Takaki et al. 2017, Vargas et al. 2012, Z. Yang et al. 2017), but they can be induced to rupture by parvovirus infection (Cohen et al. 2011), activation of apoptosis (Lindenboim et al. 2014), DNA bridge formation (Maciejowski et al. 2015), external force (Denais et al. 2016, Halfmann et al. 2019, Raab et al. 2016, Zhang et al. 2019), and introduction of oncogenic lesions or signaling (Comaills et al. 2016, Z. Yang et al. 2017). Depletion or mutation of nuclear lamin proteins (Chen et al. 2018, Comaills et al. 2016, De Vos et al. 2011, Hatch & Hetzer 2016, Karoutas et al. 2019, Robijns et al. 2016, Tamiello et al. 2013, Vargas et al. 2012, Xia et al. 2018) or loss of peripheral heterochromatin (Stephens et al. 2018) also induces membrane rupture. In addition, lipid-based transfection increases membrane rupture (Lindenboim et al. 2014). The frequency of membrane rupture is context dependent. Nuclear rupture is rare in cycling cancer cells but occurs several times an hour in migrating cells or cells depleted of lamins (de Vos et al. 2011, Denais et al. 2016, Hatch & Hetzer 2016, Raab et al. 2016).
Nuclear membrane rupture has also been observed in tissues and in vivo. Confined single-cell migration through dense tissue (Raab et al. 2016) or a tumor (Denais et al. 2016, Xia et al. 2018) results in such membrane ruptures, and they are also observed at very low frequencies in chick hearts after transfection (Cho et al. 2019). In addition, nuclear migration causes membrane rupture and repair during muscle development in mice and during pronuclear migration in Caenorhabditis elegans when laminopathy mutations or lamin knockouts are introduced (Chen et al. 2019, Penfield et al. 2018). Nuclear membrane rupture and repair have also been observed in fission yeast upon deletion of multiple NETs (Kinugasa et al. 2019). Collectively, these observations indicate that nuclear membrane rupture is neither a species-specific phenomenon nor a cell culture artifact but a mechanical response to changes in the environment.
Overview of Micronucleus Rupture
Micronuclei are separate nuclear compartments that generally form when chromosomes or chromosome fragments missegregate during mitosis and recruit their own NE. Disruption of mitosis and persistent DNA damage are frequent causes of micronucleation, but micronuclei also accumulate at a low rate in healthy tissue (reviewed in Guo et al. 2019). Micronucleus rupture occurs at high frequencies in many cell types and has been observed in lung cancer patient tissue and early-stage mammalian embryos (Daughtry et al. 2019, Gratia et al. 2019, Harding et al. 2017, Hatch et al. 2013, He et al. 2019, Kneissig et al. 2019, Kort et al. 2016, Ly et al. 2016, Mackenzie et al. 2017, Vázquez-Diez et al. 2016), suggesting that membrane instability is a property of the micronuclei themselves rather than of the cell they are in. Unlike primary nuclei, micronuclei rarely repair their membranes. Prior to rupture, many micronuclei can recruit major NE structures during mitosis, including NPCs, and are able to initiate transcription, replication, DNA damage responses, and centromere specification, although whether these processes function normally is unclear (Crasta et al. 2012, Hatch et al. 2013, He et al. 2019, Soto et al. 2018, Umbreit et al. 2020). In addition, some micronuclei appear to have no nuclear functions due to a lack of NPC assembly in mitosis (S. Liu et al. 2018). After rupture, micronuclei lose nuclear functions and histone acetylation and, if rupture occurs after the initiation of DNA replication, accumulate the DNA damage marker γH2AX (Hatch et al. 2013, Zhang et al. 2015). Chromatin from ruptured micronuclei is generally not lost from the cell and persists until the next round of mitosis (Crasta et al. 2012, Hatch et al. 2013). During this second mitosis, this chromatin can reincorporate into the primary nucleus (Crasta et al. 2012, Hatch et al. 2013), but it frequently missegregates again and becomes a new micronucleus or DNA bridge (He et al. 2019, Soto et al. 2018).
MECHANISM OF NUCLEAR MEMBRANE RUPTURE
A general sequence of events for nuclear membrane rupture is that a gap occurs in the nuclear lamina, creating a weak point in the meshwork that becomes a site of membrane blebbing and/or chromatin herniation when the nucleus is mechanically stressed, ultimately leading to membrane rupture. This model describes the majority of nuclear membrane ruptures that have been visualized in cell culture and in vivo, but exceptions to this pathway suggest that interesting alternative mechanisms exist.
Nuclear Lamina Gaps and Nuclear Membrane Rupture
Nuclear membrane ruptures frequently occur at gaps in the nuclear lamina. Nuclear lamina gaps are holes in the lamin B1 network, also often observable in the NPC and lamin A patterns, that occur singly or in a honeycomb pattern (Vigouroux et al. 2001). Nuclear lamina gaps lack major NE constituents, including NETs, LINC complex proteins, and NPCs (Bercht Pfleghaar et al. 2015, Hale et al. 2008, Hatch & Hetzer 2016,Muchir et al. 2003, Sullivan et al. 1999). For some proteins, such as lamin A, conflicting reports about whether they localize to lamin B1 gaps (Earle et al. 2020, Lindenboim et al. 2014, Muchir et al. 2003, Vigouroux et al. 2001) may highlight either different mechanisms of hole nucleation or the different proteins associated with lamina gaps before and after membrane rupture.
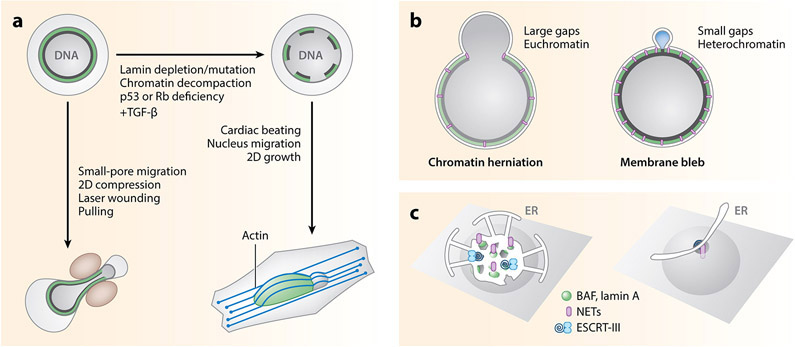
A correlation between nuclear lamina gaps and membrane rupture sites was described in an early nuclear rupture paper (de Noronha et al. 2001) and confirmed in subsequent studies. In support of a causal connection, inhibition of lamina gaps by overexpression of B-type lamins or Lap2β suppresses nuclear rupture (Maciejowski et al. 2015, Robijns et al. 2016, Vargas et al. 2012, Xia et al. 2019) while increasing gap frequency or size induces nuclear rupture (Cho et al. 2019, De Vos et al. 2011, Denais et al. 2016, Earle et al. 2020, Penfield et al. 2018, Raab et al. 2016, Stephens et al. 2018, Tamiello et al. 2013, Z. Yang et al. 2017). Gaps in the nuclear lamina can be observed both in cultured cells and in vivo and arise from several types of perturbations. These disruptions include loss of lamin B1 or lamin A/C expression, expression of lamin mutants, loss of peripheral heterochromatin, or substantial mechanical stress [lamin B1 and A/C loss (Cho et al. 2019; Earle et al. 2020; Lenz-Bohme et al. 1997; Liu et al. 2000; Penfield et al. 2018; Sullivan et al. 1999; Vergnes et al. 2004; Zwerger et al. 2013, 2015), lamin mutant expression (Goldman et al. 2004, Karoutas et al. 2019,Muchir et al. 2004, Vigouroux et al. 2001), peripheral heterochromatin disruption (Furusawa et al. 2015, Jagannathan et al. 2018, Stephens et al. 2018), and mechanical stress (Coffinier et al. 2010, Dahl et al. 2005, Denais et al. 2016, Harada et al. 2014, Hatch & Hetzer 2016, Helfand et al. 2012, Le Berre et al. 2012, Raab et al. 2016, Rowat et al. 2005, Takaki et al. 2017, Tamiello et al. 2013, Thiam et al. 2016, Wiggan et al. 2017)]. In addition, loss of retinoblastoma (Rb) and p53 activity or transforming growth factor (TGF)-β-induced epithelial-to-mesenchymal transition significantly increases nuclear lamina gap formation in cultured cells (Comaills et al. 2016, Z. Yang et al. 2017) (Figure 2a).
Figure 2.
Mechanisms of nuclear membrane rupture and repair. (a) The probability of nuclear membrane rupture depends on the degree of nuclear lamina (green) and peripheral heterochromatin (dark gray) disruption and the degree of mechanical stress. In healthy cells with an intact nuclear lamina, membrane rupture requires a significant amount of force, such as laser-induced rupture or (left) cell migration through a small opening. Disruption of the nuclear lamina by depletion or mutation of lamin proteins, chromatin decompaction, loss of retinoblastoma (Rb) and p53 activity, and activation of EMT genes by TGF-β signaling leads to membrane rupture in less mechanically demanding conditions, such as growth on stiff surfaces where (right) nucleus compression by apical actin bundles drives chromatin herniation and membrane rupture. (b) Nuclear membrane rupture is frequently preceded by or occurs with chromatin herniation and nuclear membrane blebbing. Chromatin herniation, where little to no gap is visible between the membrane and the extruded chromatin, is associated with conditions that cause large lamina gaps and decreased heterochromatin, while membrane blebbing without chromatin is associated with more intact nuclear laminas and significant peripheral heterochromatin. (c) Models of nuclear membrane repair. (Left) Cytosolic BAF binds to exposed chromatin and recruits LEM-domain NETs (purple) embedded in ER sheets and tubules. BAF also recruits lamin A/C and the NET LEMD2, which recruits the ESCRT-III complex. Depletion of BAF, ESCRT-III, or multiple NETs leads to impaired membrane repair. (Right) Alternatively, direct binding of NETs and the ESCRT-III protein Chmp7 to the chromatin and inner nuclear membrane, with or without membrane recruitment, or spreading of the outer nuclear membrane over the membrane gap could recompartmentalize the nucleus in the absence of BAF. Abbreviations: BAF, barrier-to-autointegration factor; Chmp7, charged multivesicular body protein; EMT, epithelial-to-mesenchymal transition; ER, endoplasmic reticulum; ESCRT-III, endosomal sorting complexes required for transport III; LEMD, Lap2, emerin, and Man1 domain; NET, nuclear envelope transmembrane protein; TGF, transforming growth factor.
Although increased lamina strain and high membrane curvature have been proposed as mechanisms for lamina gap formation during mechanical stress (Pfeifer et al. 2018, Sapra et al. 2019, Xia et al. 2019), in many cases the root cause of lamina disruption is unknown. Depletion of lamin B1 or lamin A causes changes in the structure of the lamin meshwork (Shimi et al. 2015) but also causes loss of peripheral heterochromatin (Camps et al. 2014, Sullivan et al. 1999), whose effects on lamina gap formation are also unclear. One possibility is that mechanical stress expands preexisting gaps in the lamina, which could arise stochastically or during NE assembly (Maeshima et al. 2010, Sapra et al. 2019, Turgay et al. 2017). Future analysis of the dynamics of the lamin meshwork in various conditions will likely yield new information about how lamina gaps are nucleated and their stability over time.
Nuclear ruptures and membrane blebs have been observed in the absence of lamina gaps. In cells migrating through small pores, membrane blebs occur over an intact nuclear lamina and are thought to arise when the nuclear lamina peels off the membrane in response to high pressure or membrane curvature (Deviri et al. 2019, Raab et al. 2016, Xia et al. 2019). In addition, chromatin herniations surrounded by an intact lamin B1 meshwork are sometimes observed after chromatin decompaction (Jagannathan et al. 2018, Stephens et al. 2018), but a potential mechanism for this phenomenon has not been identified.
Force and Nuclear Membrane Rupture
Nuclear lamina gaps leave parts of the nuclear membrane unprotected, which become sites of nuclear membrane blebs and membrane rupture when the nucleus is under mechanical stress. Nuclear membrane blebs form when increased intranuclear pressure, often from nucleus deformation, exceeds the capacity of the lamin meshworks and proteins connecting the membrane to the peripheral heterochromatin to buffer the force. Under these conditions, chromatin can flow out through a lamina gap, generating a chromatin herniation (Vargas et al. 2012), or pressure on the membrane at a small lamina gap or at an area of lamina peeling can generate a nucleoplasmic membrane bleb (Denais et al. 2016, Deviri et al. 2017, Le Berre et al. 2012, Raab et al. 2016). Chromatin herniation also likely depends on the availability of free euchromatin, as chromatin hernia are enriched in euchromatin (Shimi et al. 2008) and heterochromatinization prevents chromatin herniation (Figure 2b) (Stephens et al. 2018, 2019b). Protrusion of the nuclear membrane into a bleb releases intranuclear pressure but also increases the area of membrane that is vulnerable to strain and likely to rupture under continued or additional stress (Shah et al. 2017).
Actomyosin contractility is a major source of nuclear deformation and stress in nearly all identified examples of nuclear membrane blebbing and rupture. In cells grown on a stiff substrate, this force likely comes from compression of the nucleus by contractile apical actin bundles that run lengthwise across it (Khatau et al. 2009, Versaevel et al. 2014). Disrupting these bundles by inhibiting myosin activity, disrupting the LINC complex, or moving cells to a soft surface is sufficient to prevent both chromatin herniation and rupture (Hatch & Hetzer 2016, Robijns et al. 2016, Stephens et al. 2018, Tamiello et al. 2013). In contrast, excessive myosin activity in nonmuscle cells causes the formation of nuclear buds and membrane rupture (Takaki et al. 2017). Increased nuclear deformation also increases the size of the membrane bleb and the extent of rupture, defined as the proportion of marker protein mislocalized (Le Berre et al. 2012, Mistriotis et al. 2019, Penfield et al. 2018, Raab et al. 2016, Tamiello et al. 2013, Thiam et al. 2016, Zhang et al. 2019), which is consistent with actin forces directly determining the likelihood of membrane rupture.
Microtubule motors and high membrane curvature can also drive membrane rupture. Inhibition of dynein during pronuclear migration in C. elegans expressing lamin mutants significantly decreases the extent of membrane rupture (Penfield et al. 2018), and inhibition of kinesin prevents membrane rupture during nuclear migration in myofibers lacking lamin A (Earle et al. 2020). Membrane blebs and ruptures are also enriched at areas of high membrane curvature (Denais et al. 2016, Raab et al. 2016, Stephens et al. 2018, Xia et al. 2018), and recent experiments demonstrate that curvature is more predictive of rupture than force (Xia et al. 2018, 2019). However, the relative importance of membrane curvature for mechanical strain versus lamin loss in membrane blebbing and rupture is unclear.
Because the nuclear lamina and heterochromatin buffer mechanical forces on the nucleus (Stephens et al. 2019a), different magnitudes of force are required to rupture the nuclear membrane in cells with a normal NE structure and those in which the lamina or peripheral heterochromatin has been disrupted (Figure 2a). Normal cells in culture rarely experience nuclear membrane ruptures (De Vos et al. 2011, Raab et al. 2016, Z. Yang et al. 2017) and typically have to be subjected to significant force, for example, migration through a small pore, to induce it (Denais et al. 2016, Raab et al. 2016, Thiam et al. 2016, Zhang et al. 2019). In contrast, plating cells on plastic or glass is sufficient to induce rupture when nuclear lamina gaps are present or peripheral heterochromatin is lost (Chen et al. 2018; De Vos et al. 2011; Karoutas et al. 2019; Kinugasa et al. 2019; Maciejowski et al. 2015; Robijns et al. 2016; Stephens et al. 2018, 2019b; Takaki et al. 2017; Tamiello et al. 2013; Vargas et al. 2012; Xia et al. 2018; Z. Yang et al. 2017). Similarly, typical physiological forces, including muscle contraction, nuclear migration, and stiff tissue environments, rarely cause rupture in cells with a normal NE but are sufficient to induce nuclear membrane rupture at a high frequency when nuclear lamina or chromatin organization is disrupted (Chen et al. 2019, Cho et al. 2019, Comaills et al. 2016, Denais et al. 2016, Earle et al. 2020, Furusawa et al. 2015, Helfand et al. 2012, Penfield et al. 2018, Takaki et al. 2017, Xia et al. 2018). A direct test of this hypothesis found that subjecting lamin A–depleted cells to a mechanical strain that resulted in occasional rupture in control cell nuclei was sufficient to rupture nearly all of the nuclei in the depleted cells (Xia et al. 2018). Thus, any conditions that increase chromatin decompaction (Stephens et al. 2018), impair the connections between the membrane and the chromatin (Schreiner et al. 2015), or cause gaps in the lamina (see the section titled Nuclear Lamina Gaps and Nuclear Membrane Rupture) are likely to increase the probability of nuclear membrane rupture.
Although nuclear membrane blebs are frequent sites of membrane rupture, they are not always present before or during these events. Membrane rupture without a membrane bleb has been observed in some cancer cell lines (Denais et al. 2016, Vargas et al. 2012), some cell lines lacking lamin A/C (Chen et al. 2018, Robijns et al. 2016), and some small-pore migration assays (Xia et al. 2019), as well as during pronuclear migration in C. elegans (Penfield et al. 2018) and parvovirus infection (Porwal et al. 2013). It is possible that these experiments missed small or brief membrane blebs at the rupture sites. However, it is equally likely that these examples represent conditions in which membrane rupture is favored over membrane blebbing.
Nuclear Membrane Rupture in Micronuclei
The mechanism of micronucleus rupture follows many of the same patterns as primary nucleus rupture but is less understood. Similar to primary nuclei, micronuclei typically have gaps in the lamina preceding rupture events (Hatch et al. 2013). These gaps are required for membrane rupture, as reducing them by overexpressing either lamin B1 or lamin B2 inhibits micronucleus disruption (Bakhoum et al. 2018, Hatch et al. 2013), but their origin is unclear. One potential explanation is decreased recruitment of lamin proteins to micronuclei (Hatch et al. 2013, Kneissig et al. 2019, S. Liu et al. 2018), perhaps due to defects in NPC insertion and nuclear import (S. Liu et al. 2018). However, gaps in the lamina are observed even in large micronuclei with near-normal recruitment of lamins to the nuclear periphery (S. Liu et al. 2018). An elegant study recently demonstrated that extended contact with midspindle microtubules inhibits NPC and lamin B1 assembly on chromatin, which can leave a large lamina gap if microtubules are touching only part of the micronucleus (S. Liu et al. 2018). Live-cell imaging of lamin B1 in micronuclei showed that lamina gaps visible after mitosis can persist and even expand during interphase (Hatch et al. 2013), suggesting that NE assembly defects could impair membrane stability throughout the cell cycle (Figure 3a).
Figure 3.
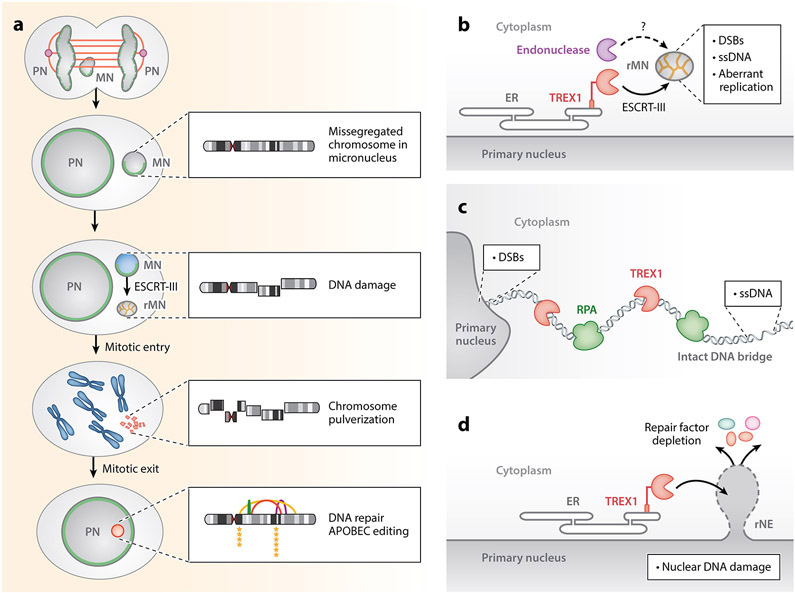
Sources of DNA damage from nuclear rupture. (a) Micronuclei frequently form around lagging chromosomes during mitosis. Association with midspindle microtubules (orange) may inhibit nuclear lamina (green) and NPC assembly. Insets show the progression of DNA damage and repair in the micronucleated chromosome. Chromosome pulverization is apparent in mitosis, and the chromatin from rMNs can missegregate or reincorporate into the nucleus. DNA repair occurs after nuclear reincorporation of the micronucleated chromatin and is primarily driven by nonhomologous end joining. Reassembly of the damaged micronucleated chromatin frequently gives rise to chromothripsis and other structural aberrations, including translocations, insertions, and deletions. APOBEC3B base editing generates clustered C > T and C > G mutations proximal to chromothripsis-associated breakpoints. (b) Micronucleus membrane rupture is associated with DNA damage. Staining with γH2AX, TUNEL, the bacterial protein Gam, RPA32, and native BrdU indicates the presence of DNA DSBs and ssDNA. The ESCRT-III complex localizes to the rMNs and promotes chromatin compaction. ER invasion into rMNs directs TREX1 to resect micronuclear DNA. (c) DNA bridges accumulate ssDNA as marked by RPA32 prior to DNA bridge breakage. ssDNA on DNA bridges is generated by TREX1 resection. DNA bridges are also associated with evidence of DNA DSBs at the connection site to the primary nucleus. (d) Nuclear rupture induces TREX1-dependent DNA DSBs in primary nuclei. Such DSBs are not restricted to the site of NE breakage and may occur throughout the nucleus. DNA damage is associated with the depletion of DNA repair factors from the nucleus. Abbreviations: APOBEC, apolipoprotein B mRNA-editing enzyme, catalytic polypeptide-like; BrdU, bromodeoxyuridine; DSB, double-strand break; ER, endoplasmic reticulum; ESCRT-III, endosomal sorting complexes required for transport III; MN, micronucleus; NPC, nuclear pore complex; PN, primary nucleus; rMN, ruptured micronucleus; rNE, ruptured nuclear envelope; RPA, replication protein A; ssDNA, single-stranded DNA; TREX1, three-prime repair exonuclease 1; TUNEL, terminal deoxynucleotidyl transferase dUTP nick end labeling.
Another unknown in micronucleus rupture is why the membrane ruptures when it does. Lamina gaps are visible at the end of mitosis (Hatch et al. 2013, S. Liu et al. 2018), yet rupture frequently does not occur until many hours later (Hatch et al. 2013). Current data suggest that both high membrane curvature and actin-based compression contribute to membrane rupture in micronuclei. Consistent with a membrane curvature mechanism, small micronuclei tend to rupture sooner than large ones and overall appear less stable (Hatch et al. 2013, Xia et al. 2019). In contrast, rupture of large micronuclei appears to require actin-based compression, as this population of nuclei is stabilized by actin depolymerization (S. Liu et al. 2018). Collectively, these data suggest that the mechanism of membrane rupture is influenced by the size of the micronucleus, but the validity of this hypothesis and the details of the rupture mechanisms remain to be determined.
The identity of the trapped chromosome could also determine micronucleus stability. Recent work presented at the 2019 American Society of Cell Biology meeting (Hatch & Levy 2020) indicated a correlation between gene density and membrane stability in micronuclei. Surprisingly, one of the smallest human chromosomes, chromosome 19, which is also the most gene dense, is also the most stable in micronuclei. Whether this stability is linked to improved nuclear lamina assembly, decreased interactions with midspindle microtubules, or the transcriptional or replication activity of the euchromatin is not yet clear.
Physical Description of Membrane Gaps
Very little is known for sure about how the nuclear membrane ruptures, but recent research suggests several likely scenarios. Electron microscopy images indicate that membrane rupture can occur in either the INM or the ONM, or in both, which leads to formation of a membrane pore (Chen et al. 2018, Karoutas et al. 2019, Kinugasa et al. 2019, Porwal et al. 2013). The frequency of single rupture versus concurrent membrane rupture is unknown, but analysis of green fluorescent protein fused to an NLS (GFP-NLS) localization to the ER lumen during membrane rupture suggests that INM rupture can occasionally precede ONM rupture in triple-lamin knockout cells (Chen et al. 2018). Electron microscopy analysis and superresolution imaging of chromatin-binding proteins suggest a range of membrane pore sizes between 50 nm and 500 nm, consistent with estimates from models of protein diffusion during rupture (Chen et al. 2018, Denais et al. 2016, Karoutas et al. 2019, Porwal et al. 2013, Zhang et al. 2019). These sizes are sufficient to allow mislocalization of large proteins and small organelles during rupture, but the limited amount of available data suggests that the upper and lower bounds of membrane pore sizes are still unknown. Another unknown is how membrane pore expansion is determined. Factors limiting membrane pore size are likely to overlap with those that decrease the extent of rupture, which is defined as the proportion of protein mislocalization, including low force, high lamin A expression, and expression of multiple tethering proteins (Chen et al. 2018, Kinugasa et al. 2019, Le Berre et al. 2012, Raab et al. 2016, Zhang et al. 2019). Future experiments to identify accurate markers of membrane hole size will be critical to advance our understanding of nuclear membrane mechanics and rupture mechanisms.
Summary of Nuclear Membrane Rupture
Nuclear membrane rupture can be viewed as a response to unbalanced pressure across the nuclear membrane. Thus, disruption of the nucleus’s ability to buffer normal forces by increasing nuclear lamina gaps or losing peripheral heterochromatin causes cells in mildly demanding mechanical environments to lose nucleus integrity. However, additional factors, such as the composition and mobility of lipids in the NE (Kinugasa et al. 2019), also are likely to contribute to membrane stability and need to be defined to predict when nuclear deformation will lead to membrane rupture. In addition, the mechanism of lamina gap formation is still unclear in many systems. Improvements in superresolution imaging along with new models of the organization and mechanics of the nuclear periphery promise to provide an unprecedented level of detail about how changes in nuclear lamina structure impact its functions and new insights into how NE dynamics are regulated during membrane rupture as well as normal growth and development.
MEMBRANE REPAIR
One of the remarkable facts to come out of the study of nuclear membrane rupture is that cells have a robust mechanism for repairing the nuclear membrane during interphase. The duration of an individual nuclear membrane rupture is frequently only a few minutes (de Noronha et al. 2001, De Vos et al. 2011, Vargas et al. 2012), but even ruptures lasting hours can eventually be repaired (Chen et al. 2018, Denais et al. 2016, Raab et al. 2016). One explanation is that mitotic NE assembly remains active throughout interphase. Consistent with this model, many of the proteins that recruit and reseal the nuclear membrane at the end of mitosis also accumulate at sites of interphase membrane rupture and facilitate nucleus recompartmentalization. Several types of repair mechanisms have been proposed, including recruitment and attachment of ER sheets to the exposed chromatin, spreading of the existing ONM, plugging through membrane recruitment, and resealing by protein complexes rather than membranes (Figure 2c). For simplicity we refer to all these pathways as membrane repair. Although recruitment of new ER currently has the most experimental support, there is strong evidence that multiple pathways exist.
Membrane Repair via ER Recruitment: BAF, Nuclear Envelope Transmembrane Proteins, and ESCRT-III
The proteins recruited to sites of nuclear membrane rupture largely overlap with those that localize to the core region of telophase chromatin and have critical roles in mitotic NE assembly. These proteins include BAF, lamin A, LEM-domain NE proteins (named for founding members Lap2, emerin, and Man1), and the endosomal sorting complexes required for transport (ESCRT)-III remodeling complex (Denais et al. 2016, Halfmann et al. 2019, Le Berre et al. 2012, Penfield et al. 2018, Raab et al. 2016, Robijns et al. 2016, Young et al. 2020). LEM-domain NETs act redundantly to recruit nuclear membranes to the decondensing chromatin (Anderson et al. 2009, LaJoie & Ullman 2017), and their localization to chromatin during mitosis, although not to the lamina in interphase mammalian cells, is partly dependent on BAF (Haraguchi et al. 2001, 2008; Samwer et al. 2017). BAF is a small (10-kDa) NE protein that homodimerizes to cross-link DNA and bind LEM-domain proteins as well as lamin A or histones (Jamin & Wiebe 2015). BAF is an essential protein, likely because its loss causes extensive multinucleation in mitosis due to a loss of chromatin cross-linking (Samwer et al. 2017). BAF may also be required to recruit ESCRT-III to the midspindle-associated core region. BAF binds LEMD2, which interacts with the ESCRT-III adaptor protein Chmp7 (Gu et al. 2017, Olmos et al. 2016, von Appen et al. 2020, Webster et al. 2016). ESCRT-III recruitment coordinates microtubule removal from the chromatin with the sealing of the resulting membrane holes (Olmos et al. 2015, Vietri et al. 2015). Disrupting LEMD2 binding to BAF prevents ESCRT-III recruitment to the core and results in nuclear morphology defects consistent with delayed membrane sealing (von Appen et al. 2020).
A current model of nuclear membrane repair is that, similar to NE assembly during mitosis, binding of cytosolic BAF to the exposed chromatin initializes recruitment of both new ER membranes to repair the membrane hole and the ESCRT-III complex to reseal the remaining membrane gaps. Cytoplasmic BAF accumulates rapidly at sites of membrane rupture and recruits lamin A/C, Chmp7, and many LEM-domain NETs to this region from the cytoplasm and ER (Denais et al. 2016, Halfmann et al. 2019). The amount of BAF accumulation correlates with the extent of rupture (Denais et al. 2016, Halfmann et al. 2019, Young et al. 2020), suggesting that BAF initially coats the surface of the exposed chromatin. BAF depletion significantly impairs nucleus compartmentalization and may even prevent it in some conditions (Halfmann et al. 2019, Young et al. 2020). Both the DNA- and LEM-binding domains of BAF are required for its function. BAF is recruited to the exposed chromatin via its DNA-binding domain (Halfmann et al. 2019) and loss of the LEM-binding domain does not fully rescue membrane repair defects (Young et al. 2020). Consistent with LEM protein recruitment being critical for membrane repair, NET protein depletion also increases the duration of membrane rupture, and depletion of multiple NETs is additive (Halfmann et al. 2019, Young et al. 2020), suggesting that NETs function redundantly to recruit new membrane. Consistent with this model, BAF depletion decreases ER density at rupture sites (Halfmann et al. 2019).
BAF localization to rupture sites is also required for the recruitment of ESCRT-III (Halfmann et al. 2019), but the function of this complex in membrane repair is unclear. When cells migrate through small pores, loss of ESCRT-III activity substantially increases the duration of nuclear membrane rupture (Denais et al. 2016, Raab et al. 2016), but in cells grown on stiff surfaces, depletion of complex member Chmp4B only increases rupture duration when lamin A is codepleted (Robijns et al. 2016). In addition, Chmp7 depletion had no effect on rupture duration in nontransformed cells ruptured by laser wounding (Halfmann et al. 2019). These data imply that ESCRT-III function is context dependent and, unexpectedly, that its activity is more critical in high-stress situations in which membrane hole sizes are predicted to be larger (Zhang et al. 2019). Because ESCRT-III membrane fusion activity is restricted to pores sized 50 nm or smaller (Gatta & Carlton 2019), these data suggest that ESCRT-III has additional unidentified functions in membrane rupture and repair.
Several experiments suggest that overaccumulation of Chmp7, ESCRT-III, or ER membranes could inhibit membrane repair. Aberrant recruitment of Chmp7 to the nucleus is sufficient to induce nuclear membrane rupture (Vietri et al. 2020), likely due to induction of substantial membrane deformations (Thaller et al. 2019). Similar deformations are observed in ruptured micronuclei and require ESCRT-III activity to occur. However, loss of ESCRT-III is not sufficient to stimulate micronucleus membrane repair (Vietri et al. 2020). In addition, a recent analysis of C. elegans meiosis and mitosis found that overrecruitment or overproduction of ER sheets can impair membrane resealing (Penfield et al. 2020). These studies suggest that the timely release of BAF from rupture sites, potentially by inhibitory phosphorylation (Halfmann et al. 2019), could be as critical for efficient membrane repair as is BAF’s recruitment by limiting membrane accumulation and remodeling.
Alternative Mechanisms of Repair
The ability of nuclei to recompartmentalize after membrane rupture in the absence of BAF, ESCRT-III, or individual NETs strongly suggests that multiple repair mechanisms exist (Figure 2c). Recent studies have identified several potential alternatives. Both BAF and LEMD2 proteins are sufficient to compartmentalize the chromatin (to different degrees) during mitotic exit (Samwer et al. 2017, von Appen et al. 2020). Therefore, these proteins, or others recruited to rupture sites, may have a similar function in interphase. In addition, ER sheet insertion at microtubule-associated holes is sufficient to seal the nucleus during C. elegans meiosis (Penfield et al. 2020), suggesting that membrane recruitment alone could restore nucleus integrity after rupture. Membrane flux from the ER to the ONM could also be increased to promote spreading of the existing nuclear membrane over the gap. Notably, LEMD2 inhibits nuclear membrane flow in fission yeast (Kinugasa et al. 2019, Kume et al. 2019), suggesting that activation of one repair pathway could suppress others. Finally, there may be only one repair pathway with redundant components. For instance, loss of BAF could be compensated for by direct interactions between NETs and the DNA (Dechat et al. 2000, Ulbert et al. 2006), and Chmp7 could be sufficient to reseal membranes in the absence of ESCRT-III (Penfield et al. 2020, Thaller et al. 2019). In the future, it will be interesting to determine which of these potential mechanisms are active in membrane repair and in what contexts.
Lamina Organization After Membrane Repair
Nuclear membrane rupture results in the accumulation of many nuclear lamina proteins at a site that previously lacked almost all of them, yet this is insufficient for lamina gap repair. BAF, lamin A, and NETs colocalize at rupture sites for many hours after membrane repair (Denais et al. 2016, Halfmann et al. 2019, Vietri et al. 2020, Young et al. 2020), yet lamin B1, lamin B receptor (LBR), and the nucleoporin Nup58 fail to accumulate at these protein scars (Denais et al. 2016, Deviri et al. 2019, Vietri et al. 2020). Targeting of NPCs and lamin B1 during normal nucleus growth and to the core region during the early G1 phase is an active process requiring cyclin-dependent kinase (CDK)1 and CDK2 activity (Maeshima et al. 2010), but why this pathway would be inactive in cycling cells after nuclear membrane rupture is unclear. Also unknown is whether these noncore proteins can eventually be recruited to membrane rupture sites. Protein scars at these sites are refractory to rerupture, but membrane ruptures at adjacent sites occur frequently (Denais et al. 2016, Young et al. 2020), which is consistent with persistent defects in nuclear lamina organization. Collectively, these observations suggest that membrane rupture does not restore nuclear lamina organization and, conversely, could destabilize the surrounding membrane.
Summary of Membrane Repair
Understanding how the nucleus regains its integrity after membrane rupture is critical both to understanding how cells survive the loss of nucleus compartmentalization and to determining the consequences of nuclear membrane rupture. The current model has many parallels to mitotic NE assembly, including a chromatin scaffold, a requirement for BAF and NET recruitment, and a sensitivity to ESCRT-III complex activity. However, molecular details of the mechanism remain to be elucidated. Of outstanding interest are the function of ESCRT-III in membrane repair and resealing, the physical nature of the membrane openings and how this correlates with repair mechanisms, and why membrane repair fails in micronuclei. Overall, the impressive ability of the nucleus to recompartmentalize after membrane rupture suggests that inhibiting nuclear membrane repair could be an unappreciated component of NE breakdown (Penfield et al. 2018).
CONSEQUENCES OF NUCLEAR MEMBRANE RUPTURE
Nuclear membrane rupture results in the mislocalization of nuclear proteins to the cytosol and the exposure of chromosomal DNA to the cytoplasm. The full impact of this nucleocytoplasmic mixing is likely to be extensive and is not completely understood. Two major consequences of nuclear membrane rupture have come to light in recent years: compromised genomic integrity and the activation of innate immune signaling pathways. These new observations suggest that membrane rupture may play an active role in tumorigenesis, DNA damage, the acquisition of complex chromosome rearrangements, and even metastasis.
NUCLEAR MEMBRANE RUPTURE COMPROMISES GENOMIC INTEGRITY
Nuclear membrane disruption exposes chromosomal DNA to the cytoplasmic environment and threatens the security of the genome. The degree—and possible causes—of DNA damage may depend on context. Nuclear membrane rupture in primary nuclei caused by confined migration of cells is associated with immediate, but transient, induction of nuclear fragmentation and DNA damage (Denais et al. 2016, Pfeifer et al. 2018, Raab et al. 2016). Likewise, intravital imaging of injected melanoma cells that have accumulated in the lung vasculature suggests that similar patterns of nuclear fragmentation occur in vivo (Headley et al. 2016). Notably, DNA damage frequently occurs distal to the site of membrane rupture, suggesting that the entire nucleus is at risk. The extent of DNA damage may be limited by efficient membrane repair (see the section titled Membrane Repair). In contrast, DNA damage at nuclear aberrations, such as micronuclei and DNA bridges, persists following nuclear membrane rupture (Hatch et al. 2013, Maciejowski et al. 2015). These data are consistent with impaired DNA repair in these compartments due to mislocalization of DNA repair proteins but may also indicate ongoing DNA damage associated with compromised membrane repair at these structures (Hatch et al. 2013, Maciejowski et al. 2015). The frequency and types of DNA lesions induced by nuclear membrane rupture have not been comprehensively characterized, but both DNA double-strand breaks (DSBs) and single-stranded (ss)DNA have been documented (Denais et al. 2016; Fouquerel et al. 2019; Hatch et al. 2013; Maciejowski et al. 2015, 2019; Raab et al. 2016; Vietri et al. 2020; Xia et al. 2018, 2019). Damage to chromosomal DNA following nuclear rupture may contribute to the etiology of laminopathies and facilitate genome evolution in tumorigenesis.
Mechanisms of Chromosome Damage After Nuclear Rupture
One prominent source of chromosome damage following nuclear rupture may be the entry of cytoplasmic factors into the nucleus (Maciejowski et al. 2015, 2019) (Figure 3). One prime candidate is TREX1 (three-prime repair exonuclease 1), an abundant, ER-associated 3′-5′ exonuclease that normally degrades cytosolic DNA to suppress inflammation (Stetson et al. 2008). TREX1 localizes to DNA bridges after nuclear rupture, where it generates ssDNA via exonucleolytic resection (Fouquerel et al. 2019; Maciejowski et al. 2015, 2019; Xia et al. 2019). Work in preprint indicates that membrane ruptures at micronuclei and at primary nuclei also enable TREX1 to access and damage DNA (Mohr et al. 2020, Nader et al. 2020).
TREX1-dependent DNA damage after nuclear membrane rupture may depend on recruitment of ER membranes to the rupture site. Electron microscopy data show that ER membranes are present in ruptured micronuclei but are not found in intact micronuclei (Hatch et al. 2013, Vietri et al. 2020). This ER membrane invasion may be a by-product of BAF/ESCRT-III-dependent nuclear membrane repair (Vietri et al. 2020). Deletion of the TREX1 C-terminus disrupts its association with the ER (Stetson et al. 2008) and compromises its ability to localize to micronuclear membrane rupture sites (Mohr et al. 2020). Retethering TREX1 to the ER restores normal TREX1 localization (Mohr et al. 2020). TREX1 colocalizes with Chmp4B at both ruptured nuclei and micronuclei, and Chmp4B depletion interferes with TREX1 localization to ruptured micronuclei (Vietri et al. 2020). In support of a model of ESCRT-III-dependent TREX1 deposition, the ESCRT-III complex was found to be required for ssDNA formation and fragmentation of DNA in micronuclei (Vietri et al. 2020). Human clinical data underline the importance of TREX1 association with the ER: Frameshift mutations that eliminate TREX1’s highly conserved C-terminal extension are associated with systemic lupus erythematosus (Lee-Kirsch et al. 2007). Interestingly, these mutations compromise TREX1 association with the ER but do not affect its catalytic activity (Lee-Kirsch et al. 2007). Collectively, these data suggest that TREX1 localization to nuclear membrane rupture sites and consequent DNA damage depend on its association with the ER and ESCRT-III-dependent membrane remodeling.
One major unknown is how TREX1 gains access to free 3′ ends to initiate resection. A priming endonuclease is likely to facilitate TREX1-mediated resection after nuclear membrane rupture. During granzyme A-mediated cell death, TREX1 collaborates with an endonuclease to fragment DNA (Chowdhury et al. 2006). Candidate endonucleases that may prime TREX1 activity following nuclear membrane rupture include the lysosomal endonuclease DNase II (deoxyribonuclease II), which digests DNA during cell death and phagocytosis (Evans & Aguilera 2003). Biallelic mutations that compromise DNase II endonuclease activity are implicated in type I interferon-mediated autoinflammation (Rodero et al. 2017). DNase II is proposed to play a central role in the clearance of nuclear DNA that accumulates in the cytosol as a result of DNA damage (Lan et al. 2014).
Evidence for Different Rearrangement Architectures
DNA damage caused by nuclear membrane rupture may play a prominent role in facilitating genome evolution in cancer. Nuclear membrane disruption has been implicated in the genesis of chromothripsis, a mutational process that generates highly rearranged chromosomes in an all-at-once event (Stephens et al. 2011). Chromothripsis-associated rearrangements are often numerous, are typically clustered, and affect one or a few chromosomes (Korbel & Campbell 2013). These initial observations introduced a few puzzles: What mechanism(s) could cause the catastrophic damage seen in chromothripsis and how could it be restricted to one chromosome or locus? Micronuclei are one possible solution (Figure 3a,b). Chromosomes missegregated into micronuclei that exhibit nuclear membrane rupture are fragmented and can generate chromothripsis chromosomes in one cell cycle (Zhang et al. 2015). Nonhomologous end joining is essential for the reassembly of chromosomes fragmented in micronuclei (Ly et al. 2016). Chromothripsis may be one extreme outcome: Missegregated chromosomes develop other structural aberrations, including translocations, insertions, and deletions (Ly et al. 2016, 2019). These rearrangements may be stably propagated because micronuclei are often reincorporated into the main nuclear mass during mitosis (Crasta et al. 2012, Hatch et al. 2013).
The molecular mechanisms of micronucleus-associated chromosome shattering are not entirely clear, and there may be additional sources of DNA damage at micronuclei beyond membrane rupture (Crasta et al. 2012). Micronuclei with intact nuclear membranes exhibit defects in DNA replication (Crasta et al. 2012, Zhang et al. 2015). Aberrant replication at micronuclei is followed by mitotic DNA synthesis, DNA damage, and chromothripsis after mitotic NE breakdown (Umbreit et al. 2020).
Chromothripsis has also been observed in several experimental models of telomere crisis (Cleal et al. 2019; Maciejowski et al. 2015, 2019; Mardin et al. 2015). Telomere crisis occurs when depletion of the telomere reserve leads to frequent telomere fusions (Maciejowski & de Lange 2017). The resulting dicentrics drive genomic instability by causing chromosome segregation errors during mitosis. Cancer genomic studies have identified dicentric chromosomes as important precipitants of chromothripsis (Li et al. 2014). Dicentrics are rarely, if ever, broken during mitosis and instead form DNA bridges during the subsequent interphase (Maciejowski et al. 2015, Pampalona et al. 2016) (Figure 3c). Nuclear rupture at DNA bridges formed during telomere crisis is associated with the generation of chromothripsis chromosomes (Maciejowski et al. 2015). Nuclear rupture appears to play a critical role in the acquisition of chromosome rearrangements during telomere crisis. Cells that transit through telomere crisis in the absence of TREX1 exhibit fewer copy number changes and infrequent chromothripsis (Maciejowski et al. 2019). The DNA repair pathways that rejoin chromosomes fragmented during telomere crisis are not completely understood.
In addition to chromosome structural variants, nuclear membrane rupture may also lead to base substitution. Chromosomes isolated in DNA bridges and micronuclei exhibit patterns of hypermutation termed kataegis (Ly et al. 2019; Maciejowski et al. 2015, 2019; Umbreit et al. 2020). Kataegis describes patterns of localized hypermutation that cluster near rearrangement breakpoints and display strand coordination (Nik-Zainal et al. 2012). Most kataegis-associated base substitutions affect cytosines at TpC dinucleotides (Nik-Zainal et al. 2012). The strand coordination and sequence preferences for mutations observed at kataegis are proposed to reflect base editing by the APOBEC3 (apolipoprotein B mRNA-editing enzyme, catalytic polypeptide-like 3) family of cytidine deaminases. The APOBEC3 deaminases play critical roles in viral defense and are proposed to be a major mutagenic force in cancer (Burns et al. 2013a,b; Roberts et al. 2013). Mutational signature analyses have placed the focus on two APOBEC3 paralogs, APOBEC3A and APOBEC3B, as the main candidates for drivers of cancer mutagenesis (Burns et al. 2013a,b; Chan et al. 2015; Landry et al. 2011; Taylor et al. 2013). Kataegis observed at chromothriptic breakpoints in a cell model of telomere crisis is the consequence of cytosine deamination by APOBEC3B (Maciejowski et al. 2019). APOBEC3B base editing during telomere crisis occurs independently of APOBEC3B upregulation and instead depends on ssDNA formation at DNA bridges (Maciejowski et al. 2019). APOBEC3B may also enhance chromothriptic fragmentation during telomere crisis, as clones that transit through telomere crisis in the absence of APOBEC3B exhibit fewer copy number changes per chromothripsis event (Maciejowski et al. 2019). Kataegis can also be recapitulated in yeast by expression of AID/APOBEC family deaminases (Roberts et al. 2012, Sakofsky et al. 2019).
Rupture of the primary nucleus membrane is also associated with heritable genetic changes (Figure 3d). Cell clones isolated after constricted migration, under conditions known to induce nuclear membrane rupture, show considerable genomic variation (Irianto et al. 2017). Likewise, nuclear rupture induced by lamin dysfunction results in chromosome rearrangement patterns including chromothripsis (Karoutas et al. 2019). The cytoplasmic mislocalization of DNA repair factors may also contribute to or exacerbate DNA damage during nuclear rupture (Irianto et al. 2016). DNA repair factors, such as 53BP1, can be retained in the cytoplasm for hours after a membrane rupture event (Xia et al. 2018). Enrichment of the antiviral double-stranded (ds)DNA sensor cGAS at nuclear herniations may also interfere with efficient DNA repair at these sites (Jiang et al. 2019). In the future, it will be interesting to use single-cell approaches to determine if single primary nuclear rupture events are sufficient to fragment chromosomes and generate chromothripsis.
Summary of Genomic Effects of Nuclear Membrane Rupture
Nuclear membrane rupture compromises genomic integrity and is associated with complex chromosome rearrangement architectures. Future work will be necessary to determine the precise causes of DNA damage after nuclear membrane rupture.
NUCLEAR MEMBRANE RUPTURE AND INNATE IMMUNE PATHWAY ACTIVATION
Nuclear membranes may prevent autoimmunity by suppressing innate immune activation (Figure 4). The innate immune system senses and responds to specific molecular patterns that can be indicative of infection or injury. DNA is one such molecular pattern, which can derive from microbial infection or defects in host cell nuclei or mitochondria. In keeping with its importance as an innate immune target, DNA is recognized by multiple sensor proteins, including the well-characterized cytosolic DNA receptor cGAS absent in melanoma 2 (AIM2) and the newly discovered stimulator of interferon genes (STING)-independent DNA-sensing pathway led by DNA-dependent protein kinase (DNA-PK) (Burleigh et al. 2020, Hornung et al. 2009, Sun et al. 2013). Nuclear membrane protection is understood to enable self-DNA to evade detection via physical separation from sensor proteins, which are primarily located in the cytoplasm (Hemmi et al. 2000, Hornung et al. 2009, Sun et al. 2013). Nuclear membrane ruptures may induce an immune response by compromising this separation and facilitating interaction between cytoplasmic DNA sensors and nuclear DNA. However, recent work, including observations that cGAS is present in the nucleus and the discovery of hnRNPA2B1, a nuclear viral DNA sensor, challenges the simplicity of this physical separation model and suggests that other factors may be necessary to help differentiate pathogenic from self-DNA (Gentili et al. 2019, Jiang et al. 2019, H. Liu et al. 2018, Volkman et al. 2019, Wang et al. 2019).
Figure 4.
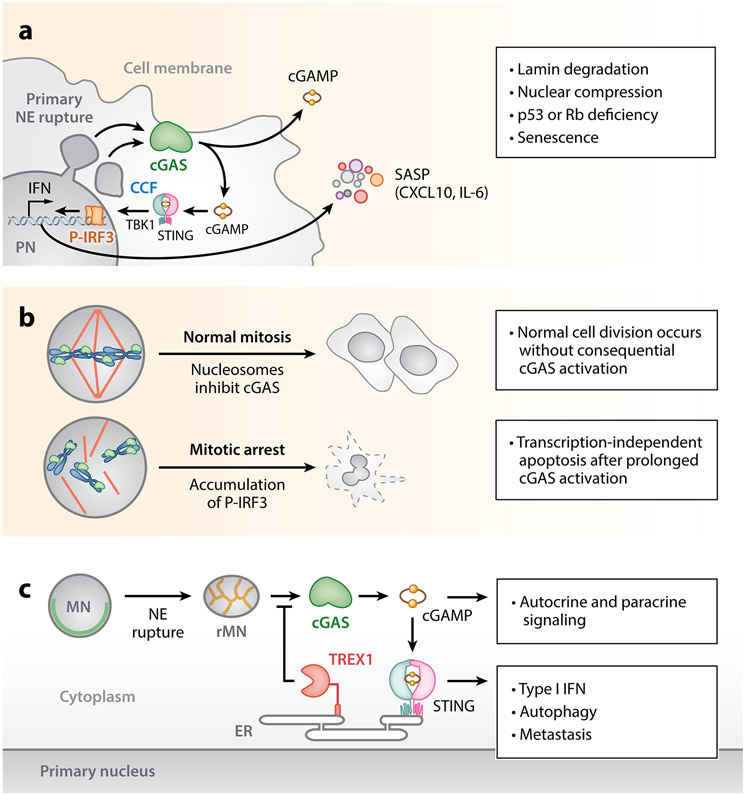
cGAS activation from nuclear rupture. (a) Primary nuclear rupture enables cGAS access to chromosomal DNA. cGAS localizes to chromatin herniations after primary NE rupture. Lamin depletion during senescence can enable cGAS to access chromosomal DNA via the formation of CCFs that bleb and appear to separate from the PN. cGAS detection of CCFs promotes senescence via the STING-dependent production of SASP factors. cGAS-STING activation in this context may promote immune surveillance. (b) cGAS promotes transcription-independent apoptosis following mitotic NE breakdown. cGAS engages with chromatin but is not activated during an unperturbed mitosis. Prolonged mitotic arrest causes a slow buildup of P-IRF3, which eventually causes transcription-independent apoptosis. (c) Nuclear rupture at micronuclei exposes chromosomal DNA to the cytosol and initiates a cGAS-dependent proinflammatory response. cGAS localizes to micronuclei following MN membrane rupture, where it engages chromatin and initiates a STING-dependent proinflammatory transcriptional response. TREX1 resection of micronuclear DNA limits cGAS activation at micronuclei. cGAS activation at micronuclei can drive metastasis through a noncanonical NF-κB signaling response and is associated with the induction of autophagy during telomere crisis. Abbreviations: AMP, adenosine monophosphate; CCF, cytosolic chromatin fragment; cGAMP, cyclic GMP-AMP; cGAS, cyclic GMP-AMP synthase; CXCL10, C-X-C motif chemokine 10; ER, endoplasmic reticulum; GMP, guanosine monophosphate; IFN, interferon; IL-6, interleukin-6; IRF3, interferon regulatory factor 3; MN, micronucleus; NE, nuclear envelope; NF-κB, nuclear factor κ-light chain–enhancer of activated B cells; P-IRF3, phosphorylated IRF3; PN, primary nucleus; rMN, ruptured micronucleus; SASP, senescence-associated secretory phenotype; STING, stimulator of interferon genes; TBK1, TANK-binding kinase 1; TREX1, three-prime repair exonuclease 1.
cGAS Activation by Nuclear Membrane Rupture
The NE may suppress autoimmunity by limiting productive cGAS engagement with self-DNA (Figure 4a). cGAS serves a crucial DNA-sensor role for the cGAS-STING signaling pathway, which induces the expression of the genes for type I interferons as well as other proinflammatory genes in response to the detection of cytosolic DNA (Ablasser & Chen 2019). In principle, cGAS activation following cytoplasmic exposure of genomic DNA due to nuclear membrane rupture is possible, because cGAS binds DNA via sequence-independent interactions and does not require free ends (Civril et al. 2013). Human cGAS is most strongly activated by long (>45 bp) DNA molecules (Kranzusch et al. 2013, Zhou et al. 2018). Engagement with dsDNA elicits a structural switch that stimulates cGAS catalytic activity and the production of cyclic GMP-AMP (cGAMP) (Ablasser et al. 2013, Civril et al. 2013, Gao et al. 2013, Wu et al. 2013). cGAMP directly activates STING, leading to activation of the TBK1 protein kinase, and the phosphorylation and nuclear translocation of its downstream effectors, the transcription factors interferon regulatory transcription factor 3 (IRF3) and nuclear factor κ-light chain–enhancer of activated B cells (NF-κB) (Ishikawa & Barber 2008).
cGAS regulation is critical to organismal health. cGAS hyperactivation is an underlying cause of autoimmune disorders such as Aicardi-Goutières syndrome and lupus (Gao et al. 2015). In principle, the potential for cGAS interaction with self-DNA is an important threat to autoimmunity: Chromatin stimulates cGAMP production, albeit not as effectively as does naked DNA (Mackenzie et al. 2017, Zierhut et al. 2019). cGAS was originally characterized as a predominantly cytoplasmic protein (Sun et al. 2013). Therefore, the NE may limit cGAS hyperactivation by acting as a barrier to prevent unrestrained cGAS engagement with self-DNA. Indeed, nuclear ruptures induced by compression result in a striking localization of transgenic cGAS to chromatin (Denais et al. 2016, Raab et al. 2016). Endogenous cGAS exhibits similar enrichment at sites of chromatin herniation and probable membrane rupture (Xia et al. 2019). In these experiments, cGAS enrichment is specific to sites of membrane rupture and does not extend much further into the chromatin. The reasons for this limited engagement are unclear, but such engagement is similar to dynamics exhibited by the chromatin-binding BAF protein at nuclear rupture sites (Halfmann et al. 2019). Tethering to chromatin may be responsible for inhibiting the spread of cGAS and BAF beyond the site of nuclear rupture. Nuclear blebs also contain increased amounts of acetyl-histone H3, a marker of euchromatin (Xia et al. 2019). One interesting possibility is that altered chromatin marks may facilitate local cGAS access at sites of nuclear disruption. In line with this reasoning, nuclear pressure preferentially pushes euchromatin through gaps in the lamina into nuclear herniations, likely causing the enrichment of euchromatin marks in chromatin herniations (Shimi et al. 2008). Likewise, micronuclei exhibit rapid depletion of histone acetylation following nuclear membrane rupture (Hatch et al. 2013).
Although cGAS exhibits strong enrichment at sites of primary nucleus rupture, cGAS activation is not always apparent and may depend on context. Autophagy-mediated degradation of the nuclear lamina during senescence, a state of permanent cell cycle arrest, results in leakage of chromatin into the cytosol and enables cGAS contact with genomic DNA (Dou et al. 2015, 2017; Glück et al. 2017; H. Yang et al. 2017). cGAS recognition of leaked chromatin induces STING activation and a consequent transcriptional response. Beyond nuclear membrane disruption, other factors may also contribute to cGAS activation during senescence. For example, the cytoplasmic nucleases TREX1 and DNase II exhibit marked reductions during senescence (De Cecco et al. 2019, Takahashi et al. 2018). This downregulation can lead to additional priming of cGAS activation through the accumulation of long interspersed nuclear element (LINE)-1 retrotransposable elements in the cytoplasm (De Cecco et al. 2019). Another possibility is that cytoplasmic nucleases directly suppress cGAS activation at chromatin herniations.
In contrast to nuclear membrane rupture during senescence, primary nuclear rupture triggered by confinement is not sufficient to induce significant cGAS-STING signaling, as measured by the nuclear translocation of IRF3 (Gentili et al. 2019). However, the sensitivity of this assay has not been characterized, and it may not detect modest but physiologically significant responses. Likewise, although cGAS coats chromosomes following mitotic NE breakdown, innate immune signaling pathways are not immediately activated (H. Yang et al. 2017, Zierhut et al. 2019). In this case, the rapid completion of mitosis appears to be an important parameter in the suppression of an immune response (Figure 4b). Mitotic arrest prolongs cGAS-DNA engagement, resulting in gradual IRF3 phosphorylation and eventually apoptosis (Zierhut et al. 2019). The delayed response can be explained by the observations that nucleosomes exhibit a higher affinity for cGAS than naked DNA and can suppress DNA-induced cGAS activity (Zierhut et al. 2019).
The NE was originally proposed to suppress cGAS-driven autoimmunity by acting as a barrier to limit cGAS access to self-DNA. However, new observations that position cGAS inside the nucleus under normal conditions—for at least a portion of the cell cycle—suggest a requirement for additional regulation (Gentili et al. 2019, Jiang et al. 2019, H. Liu et al. 2018, Volkman et al. 2019). Inside the nucleus, cGAS suppresses DNA repair by homologous recombination and is enriched at LINEs and centromere repeats (Gentili et al. 2019, Jiang et al. 2019, H. Liu et al. 2018). Nuclear cGAS can synthesize cGAMP, although at levels 200-fold lower than those following transfection with exogenous DNA (Gentili et al. 2019). Although nucleosomes likely play a role in limiting cGAS activation inside the nucleus, the full extent of regulation is unclear. One possibility is that the balance of free ion concentrations necessary to promote cGAS phase separation is limiting in the nuclear compartment (Du & Chen 2018).
cGAS Activation at Ruptured Micronuclei
In contrast to the unclear connections between nuclear membrane rupture and cGAS activation in primary nuclei, there is stronger evidence for cGAS activation by nuclear membrane disruption in micronuclei (Figure 4c). As discussed in the section titled Nuclear Membrane Rupture in Micronuclei, micronuclei frequently exhibit a catastrophic and persistent nuclear membrane collapse that exposes genomic DNA to the cytoplasm (Gentili et al. 2019, Gratia et al. 2019, Jiang et al. 2019, H. Liu et al. 2018, Volkman et al. 2019, Wang et al. 2019). Similar to observations of nuclear membrane rupture at primary nuclei, rupture at micronuclei results in the rapid and striking accumulation of cGAS at the micronucleus (Harding et al. 2017, Mackenzie et al. 2017). These studies also demonstrate that cGAS localization to micronuclei correlates with the induction of proinflammatory signaling. Consistent with micronucleus-based cGAS activation, blocks in cell cycle progression and consequent loss of micronuclei compromise inflammatory signaling (Harding et al. 2017, Mackenzie et al. 2017).
Under normal circumstances, TREX1 suppresses cGAS hyperactivation by clearing the cytoplasm of DNA (Stetson et al. 2008). TREX1-dependent regulation of cGAS also appears to occur at ruptured micronuclei (Gratia et al. 2019, Mohr et al. 2020). TREX1 deletion enhances cGAS sensing of micronuclei, increases cGAMP production, and stimulates proinflammatory gene expression (Mohr et al. 2020). Rather than clearing DNA, TREX1 appears to regulate cGAS activity by generating ssDNA, which is less capable of stimulating cGAS than is dsDNA (Kranzusch et al. 2013, Mohr et al. 2020).
The differences that can account for cGAS activation following nuclear membrane rupture at micronuclei but not primary nuclei are unclear. One likely explanation for this difference is the persistence of nuclear membrane rupture at micronuclei. Efficient primary nucleus membrane repair may limit cGAS access to genomic DNA and thus inhibit inflammatory signaling. This model is conceptually analogous to findings that a prolonged mitosis is necessary to stimulate cGAS-dependent inflammation (Zierhut et al. 2019). However, other elements may also be at work, because even prolonged primary nuclear membrane rupture is insufficient to stimulate significant cGAS-STING pathway activation (Gentili et al. 2019).
Consequences of Innate Immune Activation Associated with Membrane Rupture
The detection of DNA in the cytosol can trigger diverse responses that depend on context. Activation of the cGAS-STING pathway induces the expression of immune and inflammatory mediators, including type I interferons (Ablasser & Chen 2019). Independent of this transcriptional output, cGAS-STING can promote autophagy, which helps to clear the cytosol of viruses and DNA (Gui et al. 2019). The consequences of cGAS-STING activation are sensitive to cell or tissue background and signaling strength (Gulen et al. 2017).
Nuclear membrane rupture may promote senescence; cGAS is critical for timely cellular senescence (Dou et al. 2017, Glück et al. 2017, H. Yang et al. 2017). Mouse embryonic fibroblasts deficient for cGAS delay senescence and maintain rapid proliferation rates during serial propagation in culture (Glück et al. 2017, H. Yang et al. 2017). Bypass of senescence appears to derive from an inability to initiate the senescence-associated secretory phenotype (SASP) pathway. cGAS also appears to play a critical role in oncogene-induced senescence in vivo, but evidence implicating nuclear membrane rupture in this process is lacking (Glück et al. 2017).
The consequences of cGAS-STING activation after nuclear rupture in cancer are more complex. Recent work indicates that cGAS and STING play pivotal roles in cancer immunity in both cell autonomous and non–cell autonomous manners (Wang et al. 2017). The precise causes of tumor cell–autonomous cGAS activation are not entirely clear but may involve nuclear rupture at micronuclei. Radiotherapy primes antitumor immunity in a STING-dependent manner; however, the sources of immune-stimulatory DNA have not been identified (Harding et al. 2017, Vanpouille-Box et al. 2017). Damage to mitochondria may also play an important role in enhancing immune surveillance in the context of radiotherapy (Tigano et al. 2020). Independent of a potential role in antitumor immunity, nuclear rupture may protect against chromosomal instability by promoting cell death during telomere crisis via cGAS-STING-dependent macroautophagy (Nassour et al. 2019).
Selective pressures during tumor development may influence innate immune signaling after nuclear rupture via loss of the interferon gene cluster or suppression of cGAS and STING expression by promoter methylation (Bakhoum & Cantley 2018; Konno et al. 2018; Xia et al. 2015, 2016). Persistent nuclear membrane rupture at micronuclei that accumulate in chromosomally unstable cells leads to suppression of type I interferon signaling and upregulation of the alternative NF-κB pathway (Bakhoum et al. 2018, Dou et al. 2017). In this context, nuclear membrane rupture at micronuclei promotes STING-dependent cell invasion and metastasis (Bakhoum et al. 2018). Independent of chromosomal instability, the accumulation of extrachromosomal and mitochondrial DNA as well as the activation of endogenous retroviruses may also place selective pressures on innate immune signaling in cancer cells (Cañadas et al. 2018, Kitajima et al. 2019, Turner et al. 2017, Wu et al. 2019). Likewise, extrachromosomal telomere-repeat DNAs generated as a by-product of alternative telomere-lengthening pathways are associated with compromised type I interferon signaling (Chen et al. 2017).
Summary of Nuclear Membrane Rupture and Innate Immune Activation
cGAS detection of chromosomal DNA serves a crucial role in innate immune sensing of nuclear membrane rupture. Nuclear rupture in micronuclei initiates a cGAS-dependent type I interferon response. In the context of cancer, nuclear membrane rupture in micronuclei and consequent cGAS-STING signaling activates prometastatic pathways. Nucleosomal inhibition of cGAS during mitotic NE breakdown allows normal cell divisions to bypass consequential signaling, but mitotic arrest allows for accumulating cGAS activation and transcription-independent apoptosis. Future work will increase our understanding of the mechanisms that regulate cGAS sensing of nuclear membrane rupture.
CONCLUSIONS AND FUTURE DIRECTIONS
New observations of nuclear membrane rupture have revised the traditional view of the NE as a static barrier. Nuclear membrane rupture occurs in both healthy and disease states, in which it can cause nuclear decompartmentalization and nucleocytoplasmic mixing. Frequent observations of nuclear membrane rupture suggest that it may play a fundamental role in cell health and behavior.
The specific consequences of NE failure have come into focus only recently and are just beginning to be understood. Recent attention has focused on two major consequences of nuclear membrane rupture: compromised genome integrity and innate immune activation. One crucial unanswered question is exactly how nuclear membrane rupture causes DNA DSBs. The current focus is on understanding the role of cytosolic nucleases, such as TREX1, in generating DNA damage following nuclear rupture. Understanding how nuclear membrane rupture affects DNA synthesis may also yield important insights. Comprehensive characterization of the extent and types of DNA damage is also necessary to understand how nuclear membrane ruptures impact genomic integrity. Likewise, a precise molecular understanding of cGAS activation at nuclear rupture sites is necessary to understand innate immune surveillance of nuclear ruptures. Understanding checkpoints that regulate cGAS sensing of nuclear rupture may point toward therapeutic targets to prime antitumor immunity. For instance, inhibition of TREX1 may improve antitumor immune responses by enabling improved cGAS recognition of micronuclei. Another open question is if other DNA sensors, such as AIM2 and DNA-PK, also can sense chromosomal DNA exposed through nuclear membrane rupture.
Finally, it will be important to determine the overall impact of nuclear membrane rupture on cell health. Nuclear membrane rupture appears to be common in cancer-associated nuclear aberrations, with wide-ranging consequences for chromosome rearrangement and metastasis. Nuclear membrane rupture also appears to occur independently of disease, and it is tempting to speculate that nuclear membrane rupture may play an important role in normal cell behavior. We anticipate that future studies will build on the foundational work performed over the last several years and continue to yield exciting advances in understanding the causes of nuclear membrane rupture and the consequences for health and disease.
ACKNOWLEDGMENTS
We would like to thank members of the Maciejowski and Hatch laboratories for critical reading of this manuscript, and Matthieu Piel and Guilherme Nader for sharing unpublished data. Work in J.M.’s laboratory is supported by the National Cancer Institute (R00CA212290), the Pew Charitable Trusts, the V Foundation, the Starr Cancer Consortium, the Geoffrey Beene and Ludwig Centers atMemorial Sloan Kettering Cancer Center (MSKCC), and an MSKCC core grant (P30-CA008748). Work in E.M.H.’s laboratory is supported by a National Institutes of Health grant (R35GM124766-02), the Rita Allen Foundation Scholars program, and a National Institutes of Health/National Cancer Institute Cancer Center Support Grant (P30CA015704).
Glossary
- NE
nuclear envelope
- ONM
outer nuclear membrane
- INM
inner nuclear membrane
- NPC
nuclear pore complex
- NET
nuclear envelope transmembrane protein
- LINC
linker of nucleoskeleton and cytoskeleton; complex composed of Sun-domain proteins in the INM and KASH-domain proteins in the ONM
- BAF
barrier-to-autointegration factor
- cGAS
cyclic GMP-AMP synthase
- LEM
Lap2, emerin, and Man1; domain found in several nuclear envelope transmembrane proteins
- ESCRT-III
endosomal sorting complexes required for transport III; multisubunit complex with membrane remodeling functions
- TREX1
three-prime repair exonuclease 1
- DNase II
deoxyribonuclease II
- APOBEC3
apolipoprotein B mRNA-editing enzyme, catalytic polypeptide-like 3
- STING
stimulator of interferon genes
- cGAMP
cyclic GMP-AMP
- SASP
senescence-associated secretory phenotype
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- Ablasser A, Chen ZJ. 2019. cGAS in action: expanding roles in immunity and inflammation. Science 363(6431):eaat8657. [DOI] [PubMed] [Google Scholar]
- Ablasser A, Goldeck M, Cavlar T, Deimling T, Witte G, et al. 2013. cGAS produces a 2′-5′-linked cyclic dinucleotide second messenger that activates STING. Nature 498(7454):380–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson DJ, Vargas JD, Hsiao JP, Hetzer MW. 2009. Recruitment of functionally distinct membrane proteins to chromatin mediates nuclear envelope formation in vivo. J. Cell Biol 186(2):183–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakhoum SF, Cantley LC. 2018. The multifaceted role of chromosomal instability in cancer and its microenvironment. Cell 174(6):1347–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakhoum SF, Ngo B, Laughney AM, Cavallo J-A, Murphy CJ, et al. 2018. Chromosomal instability drives metastasis through a cytosolic DNA response. Nature 553(7689):467–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bercht Pfleghaar K, Taimen P, Butin-Israeli V, Shimi T, Langer-Freitag S,et al. 2015. Gene-rich chromosomal regions are preferentially localized in the lamin B deficient nuclear blebs of atypical progeria cells. Nucleus 6(1):66–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burleigh K, Maltbaek JH, Cambier S, Green R, Gale M, et al. 2020. Human DNA-PK activates a STING-independent DNA sensing pathway. Sci. Immunol 5(43):eaba4219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns MB, Lackey L, Carpenter MA, Rathore A, Land AM, et al. 2013a. APOBEC3B is an enzymatic source of mutation in breast cancer. Nature 494(7437):366–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns MB, Temiz NA, Harris RS. 2013b. Evidence for APOBEC3B mutagenesis in multiple human cancers. Nat. Genet 45(9):977–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camps J, Wangsa D, Falke M, Brown M, Case CM, et al. 2014. Loss of lamin B1 results in prolongation of S phase and decondensation of chromosome territories. FASEB J. 28(8):3423–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cañadas I, Thummalapalli R, Kim J, Kitajima S, Jenkins R, et al. 2018. Tumor innate immunity primed by specific interferon-stimulated endogenous retroviruses. Nat. Med 24(8):1143–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K, North PS, Hickson ID. 2007. BLM is required for faithful chromosome segregation and its localization defines a class of ultrafine anaphase bridges. EMBO J. 26(14):3397–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K, Roberts SA, Klimczak LJ, Sterling JF, Saini N, et al. 2015. An APOBEC3A hypermutation signature is distinguishable from the signature of background mutagenesis by APOBEC3B in human cancers. Nat. Genet 47(9):1067–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan Y, Fugger K, West SC. 2018. Unresolved recombination intermediates lead to ultra-fine anaphase bridges, chromosome breaks and aberrations. Nat. Cell Biol 20(1):92–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen NY, Kim P, Weston TA, Edillo L, Tu Y, et al. 2018. Fibroblasts lacking nuclear lamins do not have nuclear blebs or protrusions but nevertheless have frequent nuclear membrane ruptures. PNAS 115(40):10100–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen NY, Yang Y, Weston TA, Belling JN, Heizer P, et al. 2019. An absence of lamin B1 in migrating neurons causes nuclear membrane ruptures and cell death. PNAS 116(51):25870–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y-A, Shen Y-L, Hsia H-Y, Tiang Y-P, Sung T-L, Chen L-Y. 2017. Extrachromosomal telomere repeat DNA is linked to ALT development via cGAS-STING DNA sensing pathway. Nat. Struct. Mol. Biol 24(12):1124–31 [DOI] [PubMed] [Google Scholar]
- Cho S, Vashisth M, Abbas A, Majkut S, Vogel K, et al. 2019. Mechanosensing by the lamina protects against nuclear rupture, DNA damage, and cell-cycle arrest. Dev. Cell 49(6):920–35.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury D, Beresford PJ, Zhu P, Zhang D, Sung J-S, et al. 2006. The exonuclease TREX1 is in the SET complex and acts in concert with NM23-H1 to degrade DNA during granzyme A-mediated cell death. Mol. Cell 23(1):133–42 [DOI] [PubMed] [Google Scholar]
- Civril F, Deimling T, de Mann CC, Ablasser A, Moldt M, et al. 2013. Structural mechanism of cytosolic DNA sensing by cGAS. Nature 498(7454):332–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleal K, Jones RE, Grimstead JW, Hendrickson EA, Baird DM. 2019. Chromothripsis during telomere crisis is independent of NHEJ, and consistent with a replicative origin. Genome Res. 29(5):737–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffinier C, Chang S, Nobumori C, Tu Y, Farber EA, et al. 2010. Abnormal development of the cerebral cortex and cerebellum in the setting of lamin B2 deficiency. PNAS 107(11):5076–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S, Marr AK, Garcin P, Panté N. 2011. Nuclear envelope disruption involving host caspases plays a role in the parvovirus replication cycle. J. Virol 85(10):4863–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comaills V, Kabeche L, Morris R, Buisson R, Yu M, et al. 2016. Genomic instability is induced by persistent proliferation of cells undergoing epithelial-to-mesenchymal transition. Cell Rep. 17(10):2632–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coquel F, Silva M-J, Técher H, Zadorozhny K, Sharma S, et al. 2018. SAMHD1 acts at stalled replication forks to prevent interferon induction. Nature 557(7703):57–61 [DOI] [PubMed] [Google Scholar]
- Crasta K, Ganem NJ, Dagher R, Lantermann AB, Ivanova EV, et al. 2012. DNA breaks and chromosome pulverization from errors in mitosis. Nature 482(7383):53–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl KN, Engler AJ, Pajerowski JD, Discher DE. 2005. Power-law rheology of isolated nuclei with deformation mapping of nuclear substructures. Biophys. J 89(4):2855–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daughtry BL, Rosenkrantz JL, Lazar NH, Fei SS, Redmayne N, et al. 2019. Single-cell sequencing of primate preimplantation embryos reveals chromosome elimination via cellular fragmentation and blastomere exclusion. Genome Res. 29(3):367–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Cecco M, Ito T, Petrashen AP, Elias AE, Skvir NJ, et al. 2019. L1 drives IFN in senescent cells and promotes age-associated inflammation. Nature 566(7742):73–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Noronha CM, Sherman MP, Lin HW, Cavrois MV, Moir RD, et al. 2001. Dynamic disruptions in nuclear envelope architecture and integrity induced by HIV-1 Vpr. Science 294(5544):1105–8 [DOI] [PubMed] [Google Scholar]
- De Vos WH, Houben F, Kamps M, Malhas A, Verheyen F, et al. 2011. Repetitive disruptions of the nuclear envelope invoke temporary loss of cellular compartmentalization in laminopathies. Hum. Mol. Genet 20(21):4175–86 [DOI] [PubMed] [Google Scholar]
- Dechat T, Korbei B, Vaughan OA, Vlcek S, Hutchison CJ, Foisner R. 2000. Lamina-associated polypeptide 2alpha binds intranuclear A-type lamins. J. Cell. Sci 113(19):3473–84 [DOI] [PubMed] [Google Scholar]
- Denais CM, Gilbert RM, Isermann P, McGregor AL, te Lindert M,et al. 2016. Nuclear envelope rupture and repair during cancer cell migration. Science 352(6283):353–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deviri D, Discher DE, Safran SA. 2017. Rupture dynamics and chromatin herniation in deformed nuclei. Biophys. J 113(5):1060–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deviri D, Pfeifer CR, Dooling LJ, Ivanovska IL, Discher DE, Safran SA. 2019. Scaling laws indicate distinct nucleation mechanisms of holes in the nuclear lamina. Nat. Phys 15:823–29 [Google Scholar]
- Dou Z, Ghosh K, Vizioli M, Zhu J, Sen P, et al. 2017. Cytoplasmic chromatin triggers inflammation in senescence and cancer. Nature 550(7676):402–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou Z, Xu C, Donahue G, Shimi T, Pan J-A, et al. 2015. Autophagy mediates degradation of nuclear lamina. Nature 527(7576):105–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du M, Chen ZJ. 2018. DNA-induced liquid phase condensation of cGAS activates innate immune signaling. Science 361(6403):eaat1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earle AJ, Kirby TJ, Fedorchak GR, Isermann P, Patel J, et al. 2020. Mutant lamins cause nuclear envelope rupture and DNA damage in skeletal muscle cells. Nat. Mater 19:464–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans CJ, Aguilera RJ. 2003. DNase II: genes, enzymes and function. Gene 322:1–15 [DOI] [PubMed] [Google Scholar]
- Fenech M 2007. Cytokinesis-block micronucleus cytome assay. Nat. Protoc 2(5):1084–104 [DOI] [PubMed] [Google Scholar]
- Fouquerel E, Barnes RP, Uttam S, Watkins SC, Bruchez MP, Opresko PL. 2019. Targeted and persistent 8-oxoguanine base damage at telomeres promotes telomere loss and crisis. Mol. Cell 75(1):117–30.e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furusawa T, Rochman M, Taher L, Dimitriadis EK, Nagashima K, et al. 2015. Chromatin decompaction by the nucleosomal binding protein HMGN5 impairs nuclear sturdiness. Nat. Commun 6(1):6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao D, Li T, Li X-D, Chen X, Li Q-Z, et al. 2015. Activation of cyclic GMP-AMP synthase by self-DNA causes autoimmune diseases. PNAS 112(42):E5699–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao P, Ascano M, Wu Y, Barchet W, Gaffney BL, et al. 2013. Cyclic [G(2′,5′)pA(3′,5′)p] is the metazoan second messenger produced by DNA-activated cyclic GMP-AMP synthase. Cell 153(5):1094–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatta AT, Carlton JG. 2019. The ESCRT-machinery: closing holes and expanding roles. Curr. Opin. Cell Biol 59:121–32 [DOI] [PubMed] [Google Scholar]
- Gentili M, Lahaye X, Nadalin F, Nader G, Lombardi E, et al. 2019. The N-terminal domain of cGAS determines preferential association with centromeric DNA and innate immune activation in the nucleus. Cell Rep. 26(9):2377–93.e13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glück S, Guey B, Gulen M, Wolter K, Kang T-W, et al. 2017. Innate immune sensing of cytosolic chromatin fragments through cGAS promotes senescence. Nat. Cell Biol 19(9):1061–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman RD, Shumaker DK, Erdos MR, Eriksson M, Goldman AE, et al. 2004. Accumulation of mutant lamin A causes progressive changes in nuclear architecture in Hutchinson-Gilford progeria syndrome. PNAS 101(24):8963–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratia M, Rodero MP, Conrad C, Bou Samra E, Maurin M, et al. 2019. Bloom syndrome protein restrains innate immune sensing of micronuclei by cGAS. J. Exp. Med 216(5):1199–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruenbaum Y, Foisner R. 2015. Lamins: nuclear intermediate filament proteins with fundamental functions in nuclear mechanics and genome regulation. Annu. Rev. Biochem 84:131–64 [DOI] [PubMed] [Google Scholar]
- Gu M, LaJoie D, Chen OS, von Appen A, Ladinsky MS, et al. 2017. LEM2 recruits CHMP7 for ESCRT-mediated nuclear envelope closure in fission yeast and human cells. PNAS 114(11):E2166–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui X, Yang H, Li T, Tan X, Shi P, et al. 2019. Autophagy induction via STING trafficking is a primordial function of the cGAS pathway. Nature 567(7747):262–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulen MF, Koch U, Haag SM, Schuler F, Apetoh L, et al. 2017. Signalling strength determines proapoptotic functions of STING. Nat Commun. 8(1):427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Ni J, Liang Z, Xue J, Fenech MF, Wang X. 2019. The molecular origins and pathophysiological consequences of micronuclei: new insights into an age-old problem. Mutat. Res 779:1–35 [DOI] [PubMed] [Google Scholar]
- Hale CM, Shrestha AL, Khatau SB, Stewart-Hutchinson PJ, Hernandez L, et al. 2008. Dysfunctional connections between the nucleus and the actin and microtubule networks in laminopathic models. Biophys. J 95(11):5462–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfmann CT, Sears RM, Katiyar A, Busselman BW, Aman LK,et al. 2019. Repair of nuclear ruptures requires barrier-to-autointegration factor. J. Cell Biol 218:2136–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada T, Swift J, Irianto J, Shin J-W, Spinler KR, et al. 2014. Nuclear lamin stiffness is a barrier to 3D migration, but softness can limit survival. J. Cell Biol 204(5):669–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haraguchi T, Kojidani T, Koujin T, Shimi T, Osakada H, et al. 2008. Live cell imaging and electron microscopy reveal dynamic processes of BAF-directed nuclear envelope assembly. J. Cell. Sci 121(Pt 15):2540–54 [DOI] [PubMed] [Google Scholar]
- Haraguchi T, Koujin T, Segura-Totten M, Lee KK, Matsuoka Y, et al. 2001. BAF is required for emerin assembly into the reforming nuclear envelope. J. Cell. Sci 114(Pt 24):4575–85 [DOI] [PubMed] [Google Scholar]
- Harding SM, Benci JL, Irianto J, Discher DE, Minn AJ, Greenberg RA. 2017. Mitotic progression following DNA damage enables pattern recognition within micronuclei. Nature 548(7668):466–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch EM, Fischer AH, Deerinck TJ, Hetzer MW. 2013. Catastrophic nuclear envelope collapse in cancer cell micronuclei. Cell 154(1):47–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch EM, Hetzer MW. 2016. Nuclear envelope rupture is induced by actin-based nucleus confinement. J. Cell Biol 215(1):27–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch EM, Levy DL. 2020. Nucleus structure and dynamics. Mol. Biol. Cell 31(6):397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He B, Gnawali N, Hinman AW, Mattingly AJ, Osimani A, Cimini D. 2019. Chromosomes missegregated into micronuclei contribute to chromosomal instability by missegregating at the next division. Oncotarget 10(28):2660–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Headley MB, Bins A, Nip A, Roberts EW, Looney MR, et al. 2016. Visualization of immediate immune responses to pioneer metastatic cells in the lung. Nature 531(7595):513–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfand BT, Wang Y, Pfleghaar K, Shimi T, Taimen P, Shumaker DK. 2012. Chromosomal regions associated with prostate cancer risk localize to lamin B-deficient microdomains and exhibit reduced gene transcription. J. Pathol 226(5):735–45 [DOI] [PubMed] [Google Scholar]
- Hemmi H, Takeuchi O, Kawai T, Kaisho T, Sato S, et al. 2000. A Toll-like receptor recognizes bacterial DNA. Nature 408(6813):740–45 [DOI] [PubMed] [Google Scholar]
- Hornung V, Ablasser A, Charrel-Dennis M, Bauernfeind F, Horvath G, et al. 2009. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature 458(7237):514–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irianto J, Pfeifer CR, Bennett RR, Xia Y, Ivanovska IL, et al. 2016. Nuclear constriction segregates mobile nuclear proteins away from chromatin. Mol. Biol. Cell 27(25):4011–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irianto J, Xia Y, Pfeifer CR, Athirasala A, Ji J, et al. 2017. DNA damage follows repair factor depletion and portends genome variation in cancer cells after pore migration. Curr. Biol 27(2):210–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H, Barber GN. 2008. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature 455(7213):674–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagannathan M, Cummings R, Yamashita YM. 2018. A conserved function for pericentromeric satellite DNA. eLife 7:e34122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahed Z, Mofrad MR. 2019. The nucleus feels the force, LINCed in or not! Curr. Opin. Cell Biol 58:114–19 [DOI] [PubMed] [Google Scholar]
- Jamin A, Wiebe MS. 2015. Barrier to Autointegration Factor (BANF1): interwoven roles in nuclear structure, genome integrity, innate immunity, stress responses and progeria. Curr. Opin. Cell Biol 34:61–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Xue X, Panda S, Kawale A, Hooy RM, et al. 2019. Chromatin-bound cGAS is an inhibitor of DNA repair and hence accelerates genome destabilization and cell death. EMBO J. 38(21):e102718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karoutas A, Szymanski W, Rausch T, Guhathakurta S, Rog-Zielinska EA, et al. 2019. The NSL complex maintains nuclear architecture stability via lamin A/C acetylation. Nat. Cell Biol 21(10):1248–60 [DOI] [PubMed] [Google Scholar]
- Khatau SB, Hale CM, Stewart-Hutchinson PJ, Patel MS, Stewart CL, et al. 2009. A perinuclear actin cap regulates nuclear shape. PNAS 106(45):19017–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinugasa Y, Hirano Y, Sawai M, Ohno Y, Shindo T, et al. 2019. The very-long-chain fatty acid elongase Elo2 rescues lethal defects associated with loss of the nuclear barrier function in fission yeast cells. J. Cell. Sci 132(10):jcs229021. [DOI] [PubMed] [Google Scholar]
- Kitajima S, Ivanova E, Guo S, Yoshida R, Campisi M, et al. 2019. Suppression of STING associated with LKB1 loss in KRAS-driven lung cancer. Cancer Discov. 9(1):34–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kneissig M, Keuper K, de Pagter MS, van Roosmalen MJ, Martin J, et al. 2019. Micronuclei-based model system reveals functional consequences of chromothripsis in human cells. eLife 8:e50292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konno H, Yamauchi S, Berglund A, Putney RM, Mulé JJ, Barber GN. 2018. Suppression of STING signaling through epigenetic silencing and missense mutation impedes DNA damage mediated cytokine production. Oncogene 37(15):2037–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korbel JO, Campbell PJ. 2013. Criteria for inference of chromothripsis in cancer genomes. Cell 152(6):1226–36 [DOI] [PubMed] [Google Scholar]
- Kort DH, Chia G, Treff NR, Tanaka AJ, Xing T, et al. 2016. Human embryos commonly form abnormal nuclei during development: a mechanism of DNA damage, embryonic aneuploidy, and developmental arrest. Hum. Reprod 31(2):312–23 [DOI] [PubMed] [Google Scholar]
- Kranzusch PJ, Lee A, Berger JM, Doudna JA. 2013. Structure of human cGAS reveals a conserved family of second-messenger enzymes in innate immunity. Cell Rep. 3(5):1362–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kume K, Cantwell H, Burrell A, Nurse P. 2019. Nuclear membrane protein Lem2 regulates nuclear size through membrane flow. Nat. Commun 10(1):1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaJoie D, Ullman KS. 2017. Coordinated events of nuclear assembly. Curr. Opin. Cell Biol 46:39–45 [DOI] [PubMed] [Google Scholar]
- Lan Y, Londoño D, Bouley R, Rooney MS, Hacohen N. 2014. DNase2a deficiency uncovers lysosomal clearance of damaged nuclear DNA via autophagy. Cell Rep. 9(1):180–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry S, Narvaiza I, Linfesty DC, Weitzman MD. 2011. APOBEC3A can activate the DNA damage response and cause cell-cycle arrest. EMBO Rep. 12(5):444–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Berre M, Aubertin J, Piel M. 2012. Fine control of nuclear confinement identifies a threshold deformation leading to lamina rupture and induction of specific genes. Integr. Biol 4(11):1406–14 [DOI] [PubMed] [Google Scholar]
- Lee-Kirsch M, Gong M, Chowdhury D, Senenko L, Engel K, et al. 2007. Mutations in the gene encoding the 3′-5′ DNA exonuclease TREX1 are associated with systemic lupus erythematosus. Nat. Genet 39(9):1065–67 [DOI] [PubMed] [Google Scholar]
- Lenz-Bohme B, Wismar J, Fuchs S, Reifegerste R, Buchner E, et al. 1997. Insertional mutation of the Drosophila nuclear lamin Dm0 gene results in defective nuclear envelopes, clustering of nuclear pore complexes, and accumulation of annulate lamellae. J. Cell Biol 137(5):1001–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Schwab C, Ryan SL, Papaemmanuil E, Robinson HM, et al. 2014. Constitutional and somatic rearrangement of chromosome 21 in acute lymphoblastic leukaemia. Nature 508(7494):98–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindenboim L, Sasson T, Worman HJ, Borner C, Stein R. 2014. Cellular stress induces Bax-regulated nuclear bubble budding and rupture followed by nuclear protein release. Nucleus 5(6):527–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Zhang H, Wu X, Ma D, Wu J, et al. 2018. Nuclear cGAS suppresses DNA repair and promotes tumorigenesis. Nature 563(7729):131–36 [DOI] [PubMed] [Google Scholar]
- Liu J, Rolef Ben-Shahar T, Riemer D, Treinin M, Spann P, et al. 2000. Essential roles for Caenorhabditis elegans lamin gene in nuclear organization, cell cycle progression, and spatial organization of nuclear pore complexes. Mol. Biol. Cell 11(11):3937–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Kwon M, Mannino M, Yang N, Renda F, et al. 2018. Nuclear envelope assembly defects link mitotic errors to chromothripsis. Nature 561(7724):551–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ly P, Brunner SF, Shoshani O, Kim D, Lan W, et al. 2019. Chromosome segregation errors generate a diverse spectrum of simple and complex genomic rearrangements. Nat. Genet 51(4):705–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ly P, Teitz LS, Kim DH, Shoshani O, Skaletsky H, et al. 2016. Selective Y centromere inactivation triggers chromosome shattering in micronuclei and repair by non-homologous end joining. Nat. Cell Biol 19:68–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maciejowski J, Chatzipli A, Dananberg A, de Lange T, Campbell P. 2019. APOBEC3B-dependent kataegis and TREX1-driven chromothripsis in telomere crisis. bioRxiv 725366. 10.1101/725366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maciejowski J, de Lange T. 2017. Telomeres in cancer: tumour suppression and genome instability. Nat. Rev. Mol. Cell Biol 18(3):175–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maciejowski J, Li Y, Bosco N, Campbell PJ, de Lange T. 2015. Chromothripsis and kataegis induced by telomere crisis. Cell 163(7):1641–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie KJ, Carroll P, Martin C-A, Murina O, Fluteau A, et al. 2017. cGAS surveillance of micronuclei links genome instability to innate immunity. Nature 548(7668):461–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeshima K, Iino H, Hihara S, Funakoshi T, Watanabe A, et al. 2010. Nuclear pore formation but not nuclear growth is governed by cyclin-dependent kinases (Cdks) during interphase. Nat. Struct. Mol. Biol 17(9):1065–71 [DOI] [PubMed] [Google Scholar]
- Mardin BR, Drainas AP, Waszak SM, Weischenfeldt J, Isokane M, et al. 2015. A cell-based model system links chromothripsis with hyperploidy. Mol. Syst. Biol 11(9):828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mistriotis P, Wisniewski EO, Bera K, Keys J, Li Y, et al. 2019. Confinement hinders motility by inducing RhoA-mediated nuclear influx, volume expansion, and blebbing. J. Cell Biol 218(12):4093–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr L, Toufektchan E, Chu K, Maciejowski J. 2020. ER-directed TREX1 limits cGAS recognition of micronuclei. bioRxiv 102103. 10.1101/2020.05.18.102103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchir A, Medioni J, Laluc M, Massart C, Arimura T, et al. 2004. Nuclear envelope alterations in fibroblasts from patients with muscular dystrophy, cardiomyopathy, and partial lipodystrophy carrying lamin A/C gene mutations. Muscle Nerve 30(4):444–50 [DOI] [PubMed] [Google Scholar]
- Muchir A, van Engelen BG, Lammens M, Mislow JM, McNally E, et al. 2003. Nuclear envelope alterations in fibroblasts from LGMD1B patients carrying nonsense Y259X heterozygous or homozygous mutation in lamin A/C gene. Exp. Cell Res 291(2):352–62 [DOI] [PubMed] [Google Scholar]
- Nader GPF, Agüera-Gonzalez S, Routet F, Gratia M, Maurin M, et al. 2020. Compromised nuclear envelope integrity drives tumor cell invasion. bioRxiv 110122. 10.1101/2020.05.22.110122 [DOI] [PubMed] [Google Scholar]
- Nassour J, Radford R, Correia A, Fusté J, Schoell B, et al. 2019. Autophagic cell death restricts chromosomal instability during replicative crisis. Nature 565(7741):659–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nik-Zainal S, Alexandrov LB, Wedge DC, Loo P, Greenman CD, et al. 2012. Mutational processes molding the genomes of 21 breast cancers. Cell 149(5):979–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmos Y, Hodgson L, Mantell J, Verkade P, Carlton JG. 2015. ESCRT-III controls nuclear envelope reformation. Nature 522(7555):236–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmos Y, Perdrix-Rosell A, Carlton JG. 2016. Membrane binding by CHMP7 coordinates ESCRT-III-dependent nuclear envelope reformation. Curr. Biol 26(19):2635–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pampalona J, Roscioli E, Silkworth WT, Bowden B, Genescà A, et al. 2016. Chromosome bridges maintain kinetochore-microtubule attachment throughout mitosis and rarely break during anaphase. PLOS ONE 11(1):e0147420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen T, Kumar P, Koseoglu MM, Dutta A. 2018. Discoveries of extrachromosomal circles of DNA in normal and tumor cells. Trends Genet. 34(4):270–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfield L, Shankar R, Szentgyörgyi E, Laffitte A, Mauro MS, et al. 2020. Regulated lipid synthesis and LEM2/CHMP7 jointly control nuclear envelope closure. J. Cell Biol 219(5):e201908179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfield L, Wysolmerski B, Mauro M, Farhadifar R, Martinez MA, et al. 2018. Dynein-pulling forces counteract lamin-mediated nuclear stability during nuclear envelope repair. Mol. Biol. Cell 29(7):852–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer CR, Xia Y, Zhu K, Liu D, Irianto J, et al. 2018. Constricted migration increases DNA damage and independently represses cell cycle. Mol. Biol. Cell 29(16):1948–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porwal M, Cohen S, Snoussi K, Popa-Wagner R, Anderson F, et al. 2013. Parvoviruses cause nuclear envelope breakdown by activating key enzymes of mitosis. PLOS Pathog. 9(10):e1003671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raab M, Gentili M, de Belly H, Thiam HR, Vargas P, et al. 2016. ESCRT III repairs nuclear envelope ruptures during cell migration to limit DNA damage and cell death. Science 352(6283):359–62 [DOI] [PubMed] [Google Scholar]
- Roberts SA, Lawrence MS, Klimczak LJ, Grimm SA, Fargo D, et al. 2013. An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nat. Genet 45(9):970–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts SA, Sterling J, Thompson C, Harris S, Mav D, et al. 2012. Clustered mutations in yeast and in human cancers can arise from damaged long single-strand DNA regions. Mol. Cell 46(4):424–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robijns J, Molenberghs F, Sieprath T, Corne TDJ, Verschuuren M, De Vos WH. 2016. In silico synchronization reveals regulators of nuclear ruptures in lamin A/C deficient model cells. Sci. Rep 6:30325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodero MP, Tesser A, Bartok E, Rice GI, Mina E, et al. 2017. Type I interferon-mediated autoinflammation due to DNase II deficiency. Nat. Commun 8:2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowat AC, Foster LJ, Nielsen MM, Weiss M, Ipsen JH. 2005. Characterization of the elastic properties of the nuclear envelope. J. R. Soc. Interface 2(2):63–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakofsky CJ, Saini N, Klimczak LJ, Chan K, Malc EP, et al. 2019. Repair of multiple simultaneous double-strand breaks causes bursts of genome-wide clustered hypermutation. PLOS Biol. 17(9):e3000464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samwer M, Schneider MWG, Hoefler R, Schmalhorst PS, Jude JG, et al. 2017. DNA cross-bridging shapes a single nucleus from a set of mitotic chromosomes. Cell 170(5):956–72.e23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapra KT, Qin Z, Dubrovsky-Gaupp A, Aebi U, Müller DJ, et al. 2019. Nonlinear mechanics of lamin filaments and the meshwork topology build an emergent nuclear lamina. bioRxiv 846550. 10.1101/846550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiner SM, Koo PK, Zhao Y, Mochrie SGJ, King MC. 2015. The tethering of chromatin to the nuclear envelope supports nuclear mechanics. Nat. Commun 6:7159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah P, Wolf K, Lammerding J. 2017. Bursting the bubble—nuclear envelope rupture as a path to genomic instability? Trends Cell Biol. 27(8):546–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimi T, Kittisopikul M, Tran J, Goldman AE, Adam SA, et al. 2015. Structural organization of nuclear lamins A, C, B1, and B2 revealed by superresolution microscopy. Mol. Biol. Cell 26(22):4075–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimi T, Pfleghaar K, Kojima S-I, Pack C-G, Solovei I, et al. 2008. The A- and B-type nuclear lamin networks: microdomains involved in chromatin organization and transcription. Genes Dev. 22(24):3409–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto M, García-Santisteban I, Krenning L, Medema RH, Raaijmakers JA. 2018. Chromosomes trapped in micronuclei are liable to segregation errors. J. Cell. Sci 131(13):jcs214742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens AD, Banigan EJ, Marko JF. 2019a. Chromatin’s physical properties shape the nucleus and its functions. Curr. Opin. Cell Biol 58:76–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens AD, Liu PZ, Banigan EJ, Almassalha LM, Backman V, et al. 2018. Chromatin histone modifications and rigidity affect nuclear morphology independent of lamins. Mol. Biol. Cell 29(2):220–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens AD, Liu PZ, Kandula V, Chen H, Almassalha LM, et al. 2019b. Physicochemical mechanotransduction alters nuclear shape and mechanics via heterochromatin formation. Mol. Biol. Cell 30(17):2320–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens PJ, Greenman CD, Fu B, Yang F, Bignell GR, et al. 2011. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell 144(1):27–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stetson DB, Ko JS, Heidmann T, Medzhitov R. 2008. Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell 134(4):587–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan T, Escalante-Alcalde D, Bhatt H, Anver M, Bhat N, et al. 1999. Loss of A-type lamin expression compromises nuclear envelope integrity leading to muscular dystrophy. J. Cell Biol 147(5):913–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Wu J, Du F, Chen X, Chen ZJ. 2013. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339(6121):786–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi A, Loo T, Okada R, Kamachi F, Watanabe Y, et al. 2018. Downregulation of cytoplasmic DNases is implicated in cytoplasmic DNA accumulation and SASP in senescent cells. Nat. Commun 9:1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaki T, Montagner M, Serres MP, Le Berre M, Russell M, et al. 2017. Actomyosin drives cancer cell nuclear dysmorphia and threatens genome stability. Nat. Commun 8:16013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamiello C, Kamps MAF, van den Wijngaard A, Verstraeten VLRM, Baaijens FPT, et al. 2013. Soft substrates normalize nuclear morphology and prevent nuclear rupture in fibroblasts from a laminopathy patient with compound heterozygous LMNA mutations. Nucleus 4(1):61–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor BJ, Nik-Zainal S, Wu Y, Stebbings LA, Raine K, et al. 2013. DNA deaminases induce break-associated mutation showers with implication of APOBEC3B and 3A in breast cancer kataegis. eLife 2:e00534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaller DJ, Allegretti M, Borah S, Ronchi P, Beck M, Lusk CP. 2019. An ESCRT-LEM protein surveillance system is poised to directly monitor the nuclear envelope and nuclear transport system. eLife 8:e45284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiam H-R, Vargas P, Carpi N, Crespo CL, Raab M, et al. 2016. Perinuclear Arp2/3-driven actin polymerization enables nuclear deformation to facilitate cell migration through complex environments. Nat. Commun 7:10997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tigano M, Vargas DC, Tremblay-Belzile S, Fu Y, Sfeir A. 2020. Nuclear sensing of mitochondrial DNA breaks enhances immune surveillance. bioRxiv 929075. 10.1101/2020.01.31.929075 [DOI] [PubMed] [Google Scholar]
- Turgay Y, Eibauer M, Goldman AE, Shimi T, Khayat M, et al. 2017. The molecular architecture of lamins in somatic cells. Nature 543(7644):261–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner KM, Deshpande V, Beyter D, Koga T, Rusert J, et al. 2017. Extrachromosomal oncogene amplification drives tumour evolution and genetic heterogeneity. Nature 543(7643):122–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulbert S, Platani M, Boue S, Mattaj IW. 2006. Direct membrane protein-DNA interactions required early in nuclear envelope assembly. J. Cell Biol 173(4):469–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umbreit NT, Zhang C-Z, Lynch LD, Blaine LJ, Cheng AM, et al. 2020. Mechanisms generating cancer genome complexity from a single cell division error. Science 368(6488):p.eaba0712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanpouille-Box C, Alard A, Aryankalayil MJ, Sarfraz Y, Diamond JM, et al. 2017. DNA exonuclease Trex1 regulates radiotherapy-induced tumour immunogenicity. Nat. Commun 8:15618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas JD, Hatch EM, Anderson DJ, Hetzer MW. 2012. Transient nuclear envelope rupturing during interphase in human cancer cells. Nucleus 3(1):88–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vázquez-Diez C, Yamagata K, Trivedi S, Haverfield J, FitzHarris G. 2016. Micronucleus formation causes perpetual unilateral chromosome inheritance in mouse embryos. PNAS 113(3):626–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergnes L, Péterfy M, Bergo MO, Young SG, Reue K. 2004. Lamin B1 is required for mouse development and nuclear integrity. PNAS 101(28):10428–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versaevel M, Braquenier J-B, Riaz M, Grevesse T, Lantoine J, Gabriele S. 2014. Super-resolution microscopy reveals LINC complex recruitment at nuclear indentation sites. Sci. Rep 4:7362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vietri M, Schink KO, Campsteijn C, Wegner CS, Schultz SW, et al. 2015. Spastin and ESCRT-III coordinate mitotic spindle disassembly and nuclear envelope sealing. Nature 522(7555):231–35 [DOI] [PubMed] [Google Scholar]
- Vietri M, Schultz SW, Bellanger A, Jones CM. 2020. Unrestrained ESCRT-III drives chromosome fragmentation and micronuclear catastrophe. Nat. Cell Biol 22(7):856–67 [DOI] [PubMed] [Google Scholar]
- Vigouroux C, Auclair M, Dubosclard E, Pouchelet M, Capeau J, et al. 2001. Nuclear envelope disorganization in fibroblasts from lipodystrophic patients with heterozygous R482Q/W mutations in the lamin A/C gene. J. Cell. Sci 114(24):4459–68 [DOI] [PubMed] [Google Scholar]
- Volkman HE, Cambier S, Gray EE, Stetson DB. 2019. Tight nuclear tethering of cGAS is essential for preventing autoreactivity. eLife 8:e47491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Appen A, LaJoie D, Johnson IE, Trnka M, Pick SM, et al. 2020. A role for liquid-liquid phase separation in ESCRT-mediated nuclear envelope reformation. Nature 582(7810):115–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Hu S, Chen X, Shi H, Chen C, et al. 2017. cGAS is essential for the antitumor effect of immune checkpoint blockade. PNAS 114(7):1637–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Wen M, Cao X. 2019. Nuclear hnRNPA2B1 initiates and amplifies the innate immune response to DNA viruses. Science 365(6454):eeav0758. [DOI] [PubMed] [Google Scholar]
- Webster BM, Thaller DJ, Jäger J, Ochmann SE, Borah S, Lusk CP. 2016. Chm7 and Heh1 collaborate to link nuclear pore complex quality control with nuclear envelope sealing. EMBO J. 35(22):2447–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiggan O, Schroder B, Krapf D, Bamburg JR, DeLuca JG. 2017. Cofilin regulates nuclear architecture through a myosin-II dependent mechanotransduction module. Sci. Rep 7:40953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Sun L, Chen X, Du F, Shi H, et al. 2013. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science 339(6121):826–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Turner KM, Nguyen N, Raviram R, Erb M, et al. 2019. Circular ecDNA promotes accessible chromatin and high oncogene expression. Nature 575:699–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia T, Konno H, Ahn J, Barber GN. 2015. Deregulation of STING signaling in colorectal carcinoma constrains DNA damage responses and correlates with tumorigenesis. Cell Rep. 14(2):282–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia T, Konno H, Barber GN. 2016. Recurrent loss of STING signaling in melanoma correlates with susceptibility to viral oncolysis. Cancer Res. 76(22):6747–59 [DOI] [PubMed] [Google Scholar]
- Xia Y, Ivanovska IL, Zhu K, Smith L, Irianto J, et al. 2018. Nuclear rupture at sites of high curvature compromises retention of DNA repair factors. J. Cell Biol 217(11):3796–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Y, Pfeifer CR, Zhu K, Irianto J, Liu D, et al. 2019. Rescue of DNA damage after constricted migration reveals a mechano-regulated threshold for cell cycle. J. Cell Biol 218(8):2545–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Wang H, Ren J, Chen Q, Chen ZJ. 2017. cGAS is essential for cellular senescence. PNAS 114(23):E4612–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Maciejowski J, de Lange T. 2017. Nuclear envelope rupture is enhanced by loss of p53 or Rb. Mol. Cancer Res 15(11):1579–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young AM, Gunn AL, Hatch EM. 2020. BAF facilitates interphase nuclear envelope repair through recruitment of nuclear transmembrane proteins. Mol. Biol. Cell 31(15):1551–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C-Z, Spektor A, Cornils H, Francis JM, Jackson EK, et al. 2015. Chromothripsis from DNA damage in micronuclei. Nature 522(7555):179–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Tamashunas AC, Agrawal A, Torbati M, Katiyar A, et al. 2019. Local, transient tensile stress on the nuclear membrane causes membrane rupture. Mol. Biol. Cell 30(7):899–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Whiteley AT, de Mann CC, Morehouse BR, Nowak RP, et al. 2018. Structure of the human cGAS-DNA complex reveals enhanced control of immune surveillance. Cell 174(2):300–11.e11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zierhut C, Yamaguchi N, Paredes M, Luo J-D, Carroll T, Funabiki H. 2019. The cytoplasmic DNA sensor cGAS promotes mitotic cell death. Cell 178(2):302–15.e23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwerger M, Jaalouk DE, Lombardi ML, Isermann P, Mauermann M, et al. 2013. Myopathic lamin mutations impair nuclear stability in cells and tissue and disrupt nucleo-cytoskeletal coupling. Hum. Mol. Genet 22(12):2335–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwerger M, Roschitzki-Voser H, Zbinden R, Denais C, Herrmann H, et al. 2015. Altering lamina assembly reveals lamina-dependent and -independent functions for A-type lamins. J. Cell. Sci 128(19):3607–20 [DOI] [PMC free article] [PubMed] [Google Scholar]