Abstract
The human sliding clamp, Proliferating Cell Nuclear Antigen (hPCNA), interacts with over 200 proteins through a conserved binding motif, the PIP-box, to orchestrate DNA replication and repair. It is not clear how changes to the features of a PIP-box modulate protein binding and thus how they fine-tune downstream processes. Here, we present a systematic study of each position within the PIP-box to reveal how hPCNA-interacting peptides bind with drastically varied affinities. We synthesized a series of 27 peptides derived from the native protein p21 with small PIP-box modifications and another series of 19 peptides containing PIP-box binding motifs from other proteins. The hPCNA-binding affinity of all peptides, characterized as KD values determined by surface plasmon resonance, spanned a 4000-fold range, from 1.83 nM to 7.59 μM. The hPCNA-bound peptide structures determined by X-ray crystallography and modeled computationally revealed intermolecular and intramolecular interaction networks that correlate with high hPCNA affinity. These data informed rational design of three new PIP-box sequences, testing of which revealed the highest affinity hPCNA-binding partner to date, with a KD value of 1.12 nM, from a peptide with PIP-box QTRITEYF. This work showcases the sequence-specific nuances within the PIP-box that are responsible for high-affinity hPCNA binding, which underpins our understanding of how nature tunes hPCNA affinity to regulate DNA replication and repair processes. In addition, these insights will be useful to future design of hPCNA inhibitors.
Keywords: crystallography, peptides, proliferating cell nuclear antigen (PCNA), structural biology, surface plasmon resonance (SPR)
Abbreviations: p21, p21CIP1/WAF1; p21μ, p21 sequence 141 to 155; PIP, hPCNA-interacting protein; SPR, surface plasmon resonance
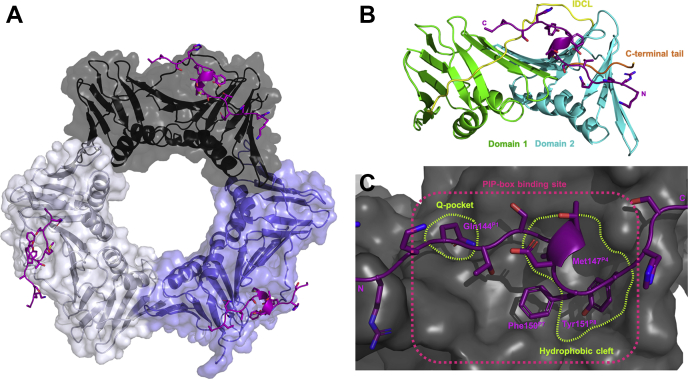
Human Proliferating Cell Nuclear Antigen (hPCNA) is a member of the sliding clamp family of proteins and acts as an essential processivity factor and mediator of DNA replication and repair (1, 2, 3, 4, 5). It is upregulated in the majority of cancers to cope with the increased demand for DNA replication. A hPCNA KO is lethal (6, 7), which reflects its importance in cell cycle progression. hPCNA is a toroidal-shaped homotrimer with 6-fold pseudosymmetry that forms by association of three 27-kDa subunits, with each subunit containing two, nearly symmetrical, domains connected by an unstructured loop termed the interdomain connecting loop (Fig. 1, A and B). The ring-shaped sliding clamp is loaded onto primer–template junctions of DNA and encircles the double strand, sliding with the progressing replication fork, to act as a moving docking platform for enzymes to bind and maintain proximity to the DNA (4, 5, 8, 9).
Figure 1.
Cocrystal structure of p21μ(7KQ1) bound to hPCNA solved by X-ray crystallography. p21μ peptide (purple) shown in cartoon form with side chains as sticks bound to each hPCNA subunit. A, hPCNA shown in cartoon form with transparent surface representation with each subunit indicated in white, gray, and pale purple. B, a hPCNA subunit shown in cartoon ribbons, where the two domains are indicated in green and blue, the IDCL in yellow and the C-terminal tail of hPCNA in orange. C, a zoom-in of a p21μ peptide (purple) bound to hPCNA (gray, surface) where the PIP-box binding site is indicated in pink, the key binding pockets are shown in green, and the conserved PIP-box residues bound are labeled in purple. IDCL, interdomain connecting loop; p21, p21CIP1/WAF1; p21μ, p21 sequence 141 to 155.
More than 200 proteins are known to interact with hPCNA during DNA replication, DNA repair, and cell-cycle regulation. However, factors that control regulation and recruitment of an appropriate sliding clamp binding partner, at the correct time and location, are not well understood. We and others have suggested that a large portion of this control arises from differences in the binding affinity of proteins for hPCNA, which span four orders of magnitude (3, 10, 11, 12). Such proteins compete to bind hPCNA to gain access to the replication fork, where successful binding is ultimately dictated by hPCNA affinity. The cell-cycle regulator protein, p21CIP1/WAF1 (referred to herein as p21), is the highest affinity hPCNA-interacting protein (PIP) known (KD 2.5–90 nM, (13, 14, 15)), and upon binding hPCNA, shuts down replication. This consequently stalls cell-cycle progression to provide a necessary checkpoint for healthy proliferation. p21 requires high affinity for hPCNA to fulfil this role and outcompete other proteins from interacting with hPCNA and hence the replication fork. In contrast, the Y-family translesion polymerases pol λ, pol κ, and pol ι bind hPCNA with micromolar affinity (16), and a peptide derived from the major processive polymerase (pol δp66 452–466) has an affinity for hPCNA of 15.6 μM (14). A fundamental understanding of the molecular-level nuances that dictate hPCNA binding affinity is required to understand the regulation of DNA-replication and DNA-repair processes. Here, we begin to unpack subtle sequence changes that influence hPCNA-binding affinity with an investigation of the structure–activity relationship of peptides that bind hPCNA.
The majority of hPCNA partners bind on the surface of a hPCNA subunit located between the two subunit domains, nestled under the interdomain connecting loop (Fig. 1C). PIPs (or hPCNA-interacting peptides) interact with hPCNA through a consensus sequence aptly named the PIP-box motif. The PIP-box motif is defined as Qxxϕxxψψ, where Q is glutamine, ‘x’ is any amino acid, ‘ϕ’ is a hydrophobic residue, and ‘ψ’ is an aromatic residue (commonly Phe or Tyr). Each of the eight positions of the PIP-box are referred to here as positions 1 through 8, or indicated by a superscript P1 through to P8. It has been argued that the PIP-box definition is part of a larger subset of hPCNA-binding motifs as many noncanonical PIP-box peptides, which are defined by the absence of a glutamine at position 1, bind hPCNA with low micromolar or nanomolar affinity (11, 17). The noncanonical definition will be used here for simplicity of discussion. The highly canonical PIP-box motif of p21, 144QTSMTDFY, was initially defined following an alanine mutation scan from Lys141 to Ser160 and revealed Gln144P1, Met147P4, Phe150P7, and Tyr151P8 as the conserved binding residues (positions 1, 4, 7, and 8 (18, 19)). A 22-mer peptide, derived from p21 (residues 139–160), that contains this PIP-box sequence binds to hPCNA as a single well-defined 310-helical turn that is anchored to the hPCNA surface by a hydrophobic triad (Met, Phe, and Tyr). Despite these insights, the scope and tolerance at each position within the PIP-box is not well understood. A number of point mutations in p21 and modifications in p21-derived peptides have been investigated; however, such modifications have been confined to alanine mutations or simple functional modifications (18, 19, 20, 21, 22). For example, a Tyr151PheP8 modified peptide demonstrated the importance of the tyrosine phenol group that makes a hydrogen bond to Gln131 on hPCNA (22). The nonconserved PIP-box residues (positions 2, 3, 5, and 6) of p21139–160 only participate in three 310-helical stabilizing hydrogen bonds (146–149, 147–150, and 147–151) (10, 15), which has led to the belief that these residues play a limited role in the binding affinity for hPCNA. A thorough investigation is required to develop a nuanced understanding of the key interactions to which the p21 PIP-box sequence owes its high affinity, to begin to understand how affinity for hPCNA plays a role in the regulation of DNA-replication and DNA-repair processes.
Here, a series of short p21-derived peptides is reported, with either one- or two-point modifications made in the PIP-box sequence to study the structure–activity relationship of hPCNA-binding peptides and determine the secondary interactions that fine-tune affinity for hPCNA. The amino acids incorporated at each PIP-box position were chosen to reflect those observed at the respective position in native protein PIP-box sequences. A second series of peptides was prepared with native PIP-box sequences from different hPCNA-binding proteins, inserted within the sequence that flanks the p21 PIP-box sequence. This series of peptides allowed a wider array of sequence combinations to be investigated than point modifications and allows direct comparison of the effect of the PIP-box sequence on hPCNA affinity, as the peptides contain the same sequence flanking the PIP-box. The hPCNA-binding affinity of each peptide was determined by surface plasmon resonance (SPR). The changes in the structure of the peptides bound to hPCNA were studied by X-ray crystallography and computational modeling studies, and correlated to the hPCNA-binding affinity to uncover interactions responsible for the differences in affinity. These studies inform on the design of three new PIP-box sequences to investigate whether cooperative interactions could be predicted from the interactions observed in the high hPCNA affinity modified p21 peptides. This comprehensive and systematic structure–activity investigation advances the ability to predict hPCNA-binding affinity from PIP-box sequence. In addition, it provides insight into how nature has fine-tuned affinity of PIP-box sequences for hPCNA, which can be leveraged to further investigate how hPCNA regulates the DNA-replication and DNA-repair process or develop inhibitors of these interactions.
Results
Fifty-one peptides were synthesized by solid-phase peptide synthesis (see Experimental procedures) to allow systematic study of the structure–activity relationship of peptides that bind hPCNA (Table S1). These peptides comprise two control p21 peptides known to bind hPCNA; a series of 27 point-modified p21 peptides; a second series of 19 peptides, where the PIP-box sequence from a number of hPCNA-binding proteins was included between the sequence that flanks the p21 PIP-box; and a final set of three peptides containing entirely new, rationally designed PIP-box sequences. A short p21 sequence 141 to 155 (referred to herein as p21μ) was chosen as it has been reported the p21139–160 sequence can be shortened without drastically impacting affinity (13). Interestingly, the preparation of this shorter sequence gave less aspartimide formation compared with the longer 22mer, with a M-18 peak in the mass spectrum not apparent for p21μ, in contrast to p21139–160 (unpublished work), thereby improving synthetic yields. All subsequent peptides were based on this shorter p21μ peptide (residues 141–155).
Five p21μ peptides with a 144P1 point mutation were prepared: p21μ–Q144K, p21μ–Q144M, p21μ–Q144D, p21μ–Q144S, and p21μ–Q144N. Four of these were inspired by the following noncanonical P1 residues: lysine (pol ι, pol κ), methionine (Cdt2, pol η and RNaseH2B), aspartic acid (poly(ADP-ribose) glycohydrolase, PARG), or serine (pol λ) at PIP-box position 1. In addition, asparagine with the amidated, but shorter side chain than glutamine was included (p21μ–Q144N). Five p21μ peptides, with a single amino-acid modification at the conserved position 4 of the PIP-box, were prepared and include valine (p21μ–M147V), isoleucine (p21μ–M147I), or leucine (p21μ–M147L), which are commonly observed in native PIP-box sequences, as well as the smaller but still hydrophobic alanine (p21μ–M147A) and the nonpolar aromatic tryptophan (p21μ–M147W). Aromatic residues, such as phenylalanine and tyrosine, are commonly observed at conserved positions 7 and 8 of the PIP-box. p21μ contains the PheP7TyrP8 combination and consequently three peptides, each with a different permutation of phenylalanine and tyrosine, were prepared (p21μ–Y151F, p21μ–F150Y, and p21μ–FY150/151YF). In addition, a p21μ–F150H peptide was prepared, inspired by the PARG PIP-box. Positively charged residues are commonly observed in native PIP-box sequences at nonconserved positions 2 and 3 (e.g., XPG, FEN1, WRN, PARG, p15, Cdt1). Consequently, eight p21μ peptides with arginine or lysine residues at PIP-box position 2 and/or 3 were prepared (p21μ–T145K, p21μ–T145R, p21μ–S146K, p21μ–S146R, p21μ–TS145/146KK, p21μ–TS145/146KR, p21μ–TS145/146RK, and p21μ–TS145/146RR). Conversely, negatively charged residues are commonly observed at nonconserved positions 5 and 6 of native PIP-box sequences (e.g., pol ι, pol λ, pol κ, p15, Cdt1, XPG, RNaseH2B) and five peptides containing aspartic and/or glutamic acid were prepared (p21μ–T148D, p21μ–T148E, p21μ–TD148/149DE, p21μ–TD148/149EE, and p21μ–D149E). Finally, to investigate a variety of PIP-box sequence combinations, a variety of native PIP-box sequences replaced the p21 PIP-box in a p21μ peptide. Ten canonical PIP-box sequences from hPCNA-binding proteins Cdt1, Pogo, XPG, DNALig1, MCMT, pol δp66, FEN1, p15, WRN, and RecQ5 were chosen, along with eight peptides with noncanonical human PIP-box sequences from pol ι, pol κ, Cdt2, pol η, RNaseH2B, PARG, pol λ, and RFCp14. The PIP-box motif in many of these proteins is located at the protein terminus and does not have a sequence extending C- or N-terminal from the PIP-box (e.g., pol δp66). Consequently, our design allows direct comparison of PIP-box sequence influence on hPCNA affinity and is not influenced by the native flanking sequence.
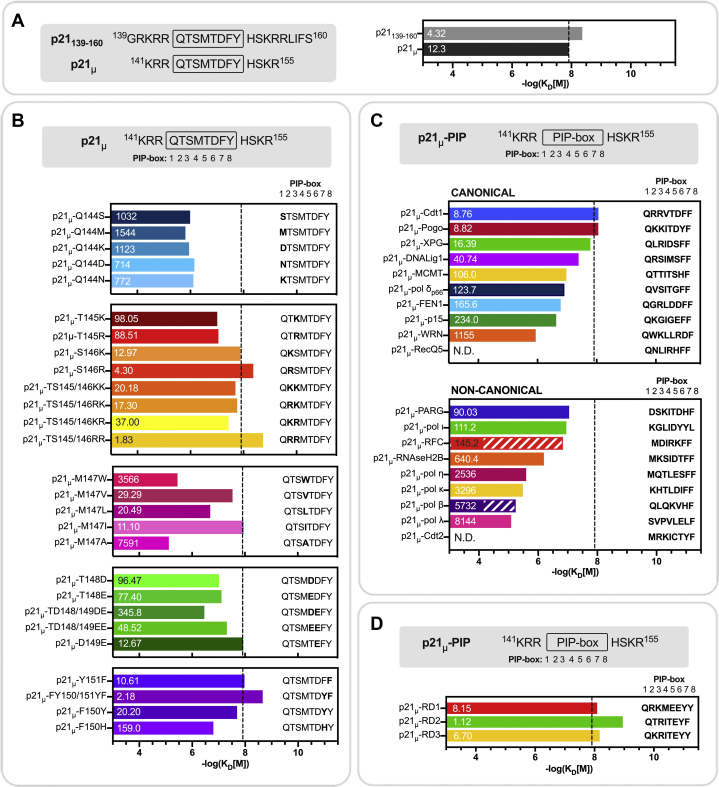
The binding affinity (KD) of each peptide for hPCNA was determined by SPR, and the results are summarized in Figure 2 and Table S2. A representative sample of SPR sensorgrams are included in Fig. S1. p21139–160, prepared as a positive control, bound to hPCNA with a KD value of 4.32 nM (Fig. 2A, Table S2), which agrees with previous literature (13, 14, 15). p21μ (residues 141–155) bound to hPCNA with 12.3 nM affinity, which indicates the truncated sequence was well tolerated, although the peptide is seven amino acids shorter than p21139–160 (Fig. 2A, Table S2). All modified p21 peptides are compared with p21μ throughout the discussion, unless otherwise indicated. The KD value of p21μ is marked as a dashed line on all panels of Figure 2 to provide a benchmark for comparison. The KD values for the modified p21μ peptides spanned four orders of magnitude, with the best binding peptide of these two series having a KD value of 1.83 nM (p21μ–TS145/146RR) and the lowest affinity was 8.14 μM (p21μ–pol λ) (Fig. 2, B and C, Table S2). The binding affinity of p21μ–Cdt2 and p21μ–RecQ5 could not be determined because of nonspecific binding to the sensor chip. Interestingly, the analysis revealed seven peptides with higher affinity than p21μ: p21μ–TS145/146RR (1.83 nM), p21μ–S146R (4.30 nM), p21μ–Cdt1 (8.76 nM), p21μ–Pogo (8.82 nM), p21μ–Y151F (10.6 nM), and p21μ–M147I (11.1 nM) (Fig. 2, B and C). The affinity of canonical PIP-box p21μ peptides (with Gln144P1) here range from 1.83 nM to 3.57 μM (Fig. 2C, ‘canonical’). In general, the noncanonical peptides as a group, unsurprisingly, bound hPCNA with lower affinity than the canonical p21μ peptides (see Fig. 2C).
Figure 2.
Binding affinity (KD) of p21 peptides to PCNA determined by SPR.KD values were calculated using the in-built Biacore Evaluation S200 software and are shown in nanomolar over the respective bar. The bars graphically represent the KD values (in molar, M) as −log(KD[M]). All experiments were repeated to ensure reproducibility, and additional SPR information including fitting errors can be found in Table S2. The dashed line on all panels represents the affinity of p21μ (12.3 nM, −log(KD[M]) = 7.91) to which all modified p21μ peptides are compared throughout the discussion. A, binding affinity of the native p21 peptides. B, rationally mutated p21 peptides with single- or double-point modifications are introduced into the p21 PIP-box. C, binding affinity of peptides containing a native canonical PIP-box from an alternate hPCNA-binding partner (top) or native noncanonical PIP-box from an alternate hPCNA-binding partner (bottom) including two PIP-box sequences which only contain seven amino acids (indicated by the striped bars). The PIP-box sequences are flanked by the same sequence that flanks the p21 PIP-box. D, binding affinity of the rationally designed PIP-box sequences. N.D., could not be determined; p21, p21CIP1/WAF1; SPR, surface plasmon resonance.
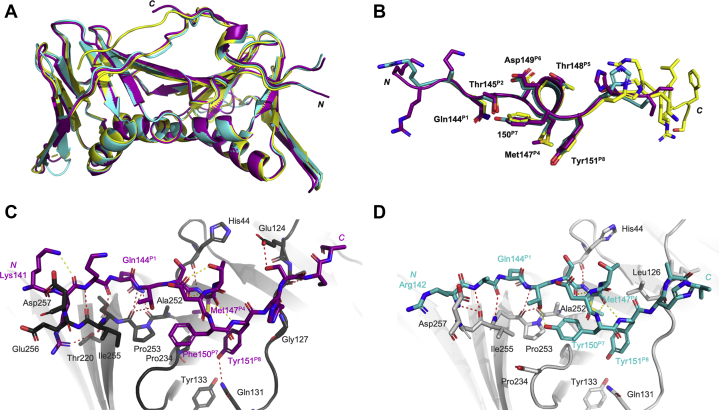
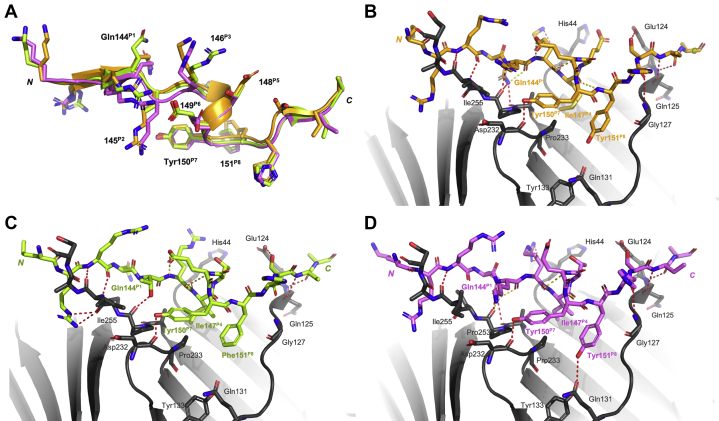
The conformation of the peptides bound to hPCNA was investigated to uncover potential reasons for differences in affinity. Cocrystal structures of p21μ (PDB ID: 7KQ1, 3.30 Å) and p21μ–F150Y bound to hPCNA (PDB ID: 7KQ0, 2.40 Å) were solved. Both served as important controls for our modeling studies: p21μ showed that the shorter peptide binds hPCNA in a similar manner (position on hPCNA surface and secondary structure) as the 22mer p21139–160 peptide (PDB ID: 1AXC), and p21μ–F150Y demonstrated a proof of concept that our docking studies represented the crystal structures accurately. The resulting structures are shown in Figure 3, Figs. S1 and S2, and the data collection and refinement statistics are summarized in Table S3. The cocrystal structure of p21μ bound to hPCNA (Fig. 1) revealed six intermolecular and three intramolecular hydrogen bonds (Fig. 3C, Fig. S2 and Table S4). The PIP-box residues 144 to 151 all retain similar conformations to p21139–160 (1AXC, Fig. 3, A and B), which is represented by an RMSD value of 0.51 Å (Table S18). This is also reflected in a high similarity of the buried surface area of the PIP-box residues of p21μ compared with p21139–160 in 1AXC (Table S19). Gln144P1 of p21μ makes a hydrogen bond with Ala252 and Pro253 within the Q-pocket, but not Ala208, in contrast to Gln144P1 of p21139–160 that formed three hydrogen bonds. The side chain of Met147P4 was packed slightly differently to the analogous Met147P4 in p21139–160 (1AXC). The C-terminal residues of hPCNA (Lys254-Asp257) and N-terminal residues of p21μ (141–143) gave well-defined electron density, in contrast to the p21139–160 structure (1AXC) that revealed an electrostatic interaction between Lys141 and Arg142 of p21μ, and Glu256 and Asp257 of hPCNA (Fig. 3C).
Figure 3.
Cocrystal structures of p21μ(7KQ1, purple) and p21μ–F150Y (7KQ0, blue) bound to hPCNA solved by X-ray crystallography, compared with p21139–160(1AXC, yellow).A, overall binding mode and structure of p21 peptides bound to hPCNA is the same. Peptide and hPCNA shown are cartoons. B, an overlay of peptide structures shows the PIP-box residues (labeled) adopt similar orientations. C and D, intramolecular (yellow dashed lines) and intermolecular (red dashed lines) polar interactions of p21μ (purple, C) and p21μ–F150Y (blue, D) shown as sticks bound to hPCNA in gray. Conserved PIP-box residues, and the N-terminal residue, are labeled in the corresponding peptide color. p21, p21CIP1/WAF1; p21μ, p21 sequence 141 to 155.
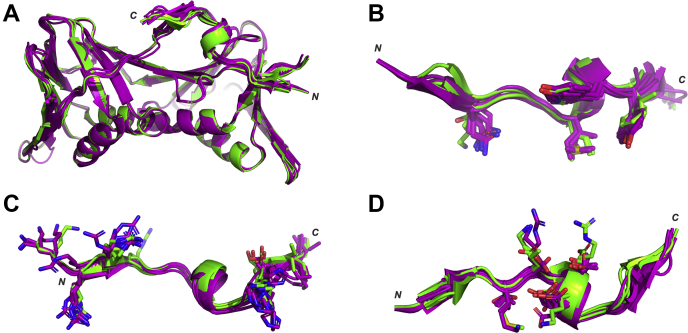
After this, a larger collection of the modified p21μ peptides were selected for further investigation by computational modeling of the hPCNA-bound peptides, to allow high-throughput analysis of more structures. The crystal structure of p21μ (PDB ID: 7KQ1) bound to hPCNA was used as a starting point for modeling as this peptide is the most similar to the modeled peptides, and the structure was overall similar to the p21139–160 structure bound to hPCNA (PDB ID: 1AXC). The peptides chosen for computational analysis were p21μ–S146R, p21μ–M147I, p21μ–D149E, and p21μ–FY150/151YF as these modifications resulted in the largest increase in affinity for the respective PIP-box position. In addition, a representative group containing native PIP-box sequences was also modeled bound to hPCNA: two canonical PIP-box peptides, p21μ–Pogo and p21μ–pol δp66; two noncanonical PIP-box peptides p21μ–pol ι and p21μ–PARG, as well as seven amino-acid PIP-box peptide p21μ–RFC (Figs. S2–S15). The resulting computationally modeled peptide structures bound to hPCNA were compared with the cocrystal structure of p21μ bound to hPCNA, which revealed the complexed structures were similar overall (Fig. 4A), represented by an average RMSD value of 0.223 Å (Table S18). This in turn suggests that the difference in affinity for hPCNA is due to subtle structural changes. The conserved residues all adopted similar conformations between all peptide structures (Fig. 4B); however, the nonconserved residues (positions 2, 3, 5, and 6) and residues flanking the PIP-box were much more varied (Fig. 4, C and D). Intramolecular (peptide-protein) and intermolecular (peptide-peptide) interactions for each hPCNA-bound peptide structure are summarized in Tables S4, S5–S17.
Figure 4.
Superimpositionof cocrystallized or computationally modeled peptide:hPCNA structures. Canonical PIP-box peptides are shown in purple, and noncanonical PIP-box peptide structures are shown in green. Canonical: p21μ, p21μ–F150Y p21μ–S146R, p21μ–M147I, p21μ–D149E, p21μ–FY150/151YF, p21μ–Pogo, and p21μ–pol δp66. Noncanonical: p21μ–pol ι, p21μ–PARG, and p21μ–RFC. A, all peptides adopt a single 310-helical turn upon binding hPCNA. B, conserved PIP-box residues shown with sticks show a high degree of similarity. C, flanking residues shown with sticks are orientated in similar directions but are still flexible. D, nonconserved PIP-box residues shown with sticks and adopt a large variety of orientations. p21, p21CIP1/WAF1; p21μ, p21 sequence 141 to 155.
In addition, the hPCNA-bound peptides p21μ, p21μ–pol δp66, p21μ–pol ι, p21μ–PARG, and p21μ–Pogo were compared with the co-crystal structure reported for the analogous native peptide bound to hPCNA, to investigate how the sequence flanking the PIP-box motif influences binding conformation. These structures are p21139–160 (1AXC, (15)), pol δp66 452–466 (1U76, (14)), pol ι415–437 (2ZVM, (16)), PARG402–420 (5MAV, (23)), and the Pogo PIP-box included in the mutant Pogo-Ligase peptide (1VYJ, (24)). The hPCNA-bound peptide cocrystal structures were overlaid (Fig. S16), and the overall peptide conformation (RMSD values in Table S20) and secondary interactions were compared (Table S21). These data reveal the PIP-box of peptides that contain the p21, pol δp66, PARG, or Pogo PIP-box sequences all adopt similar conformations to the native counterpart on binding hPCNA with RMSD values of 0.51, 0.69, 0.49, and 0.69 Å, respectively (Table S20, Fig. S16). However, the pol ι PIP-box–containing peptides bound to hPCNA adopt very different conformations, represented by an RMSD of 1.13 Å (Fig. S16 and Table S20).
Finally, the affinity and structural information for these two series of peptides was harnessed to design three new PIP-box sequences in an attempt to test our understanding of the requirements for high-affinity hPCNA binding. These new PIP-box sequences were designed to mimic favorable secondary interactions observed for native PIP-box sequences or point-modified peptides (Fig. 5). These sequences were designed with particular primary sequence combinations in mind that correlated with high hPCNA affinity, or particular secondary interactions that appeared important to stabilize the binding conformation which are highlighted throughout the discussion. These sequences were then synthesized within the p21μ scaffold, as before, to give p21μ–RD1–3. The binding affinity to hPCNA was determined by SPR (Fig. 2D) to reveal that all three peptides bind with higher affinity than p21μ and helped inform and refine the interaction map shown in Figure 6. The peptides were then computationally modeled bound to the hPCNA surface (Fig. 7), which indicates that these peptides all bind hPCNA in a similar conformation to p21μ (RMSD 0.181–0.270 Å, Table S18), where a main-chain hydrogen bond between residues 149P6 and 146P3 defines the 310-helical binding conformation (Fig. 7A). The best-performing peptide p21μ–RD2 binds hPCNA with 1.12 nM affinity, which is remarkably the highest affinity hPCNA-binding peptide or protein reported (Fig. 2, Table S2).
Figure 5.
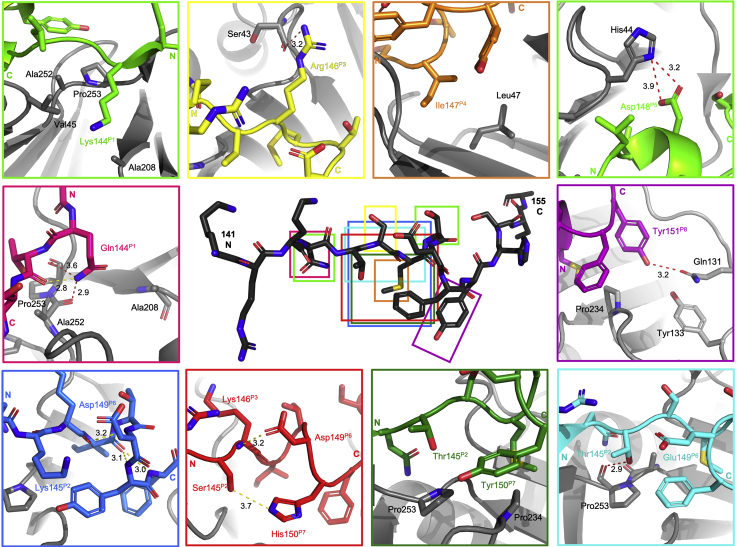
Representative examples of key intermolecular and intramolecular interactions. Structures shown in cartoon format with side chains as sticks. p21μ structure shown in the center in black and the area of key interactions shown in colored rectangles. hPCNA is shown in gray. Computationally modeled peptides bound to hPCNA: p21μ–pol ι, light green; p21μ–S146R, yellow; p21μ–M147I, orange; p21μ–pol δp66, purple; p21μ–PARG, red; p21μ–D149E, light blue; p21μ–Pogo, dark blue; and p21μ–FY150/151YF, dark green. Cocrystal structures of peptides bound to hPCNA: p21μ–F150Y, pink and p21μ, purple. Intramolecular hydrogen bonds are indicated as a yellow dashed line, and intermolecular hydrogen bonds are indicated as a red dashed line. Distances are indicated in angstroms. Elemental coloring: nitrogen, blue; oxygen, red; sulfur, yellow. p21, p21CIP1/WAF1; p21μ, p21 sequence 141 to 155.
Figure 6.
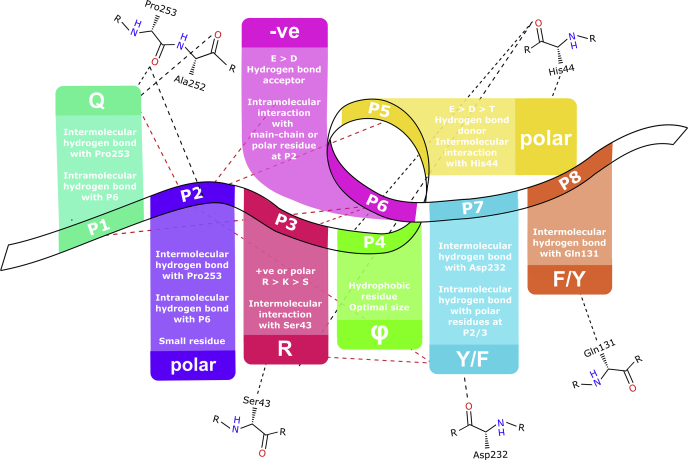
Summary of the guideline to design a high-affinity PIP-box and the interactions each residue participates in when bound to hPCNA. hPCNA residues shown as line structures. Intermolecular interactions to side chain or main chain shown as black dashed lines, intramolecular interactions from side chain or main chain shown as red dashed lines.
Figure 7.
Computational modeling of rationally designed PIP-box peptidesp21μ–RD1, p21μ–RD2, and p21μ–RD3 on hPCNA. hPCNA shown in gray as a cartoon, with interacting amino acids shown as sticks and labeled in gray. Intermolecular polar interactions are shown as red dashed lines and intramolecular polar interactions shown as yellow dashed lines. A, overlay of the structure of the rationally designed PIP-box peptides. B, p21μ–RD1 shown in orange and conserved residues labeled. C, p21μ–RD2 shown in green and conserved residues labeled. D, p21μ–RD3 shown in pink and conserved residues labeled. p21μ, p21 sequence 141 to 155; p21, p21CIP1/WAF1.
Discussion
The binding affinities of the p21μ peptides for hPCNA, and where appropriate, the structure-based information were compared and contrasted to highlight interactions that correlated to changes in hPCNA-binding affinity. Individual modifications at each PIP-box position were first examined for simplicity, then the cooperative interactions that arise were summarized, and finally how these observations pertain to the design of the peptides p21μ–RD1–3 are discussed below.
Conserved glutamine position 1: Glutamine enhances hPCNA affinity
GlnP1 is known to contribute significantly to hPCNA-binding affinity, where a reported p21 Gln144AlaP1 mutant was not able to effectively inhibit SV40 DNA replication in vitro (18). However, its importance has been disputed because of the prevalence of noncanonical PIP-box sequences (11, 17). Here, the contribution of GlnP1 to hPCNA-binding affinity is quantified in comparison with other common position 1 residues.
Gln144P1 binds in the Q-pocket, and the orientation was unchanged between the canonical computationally modeled or cocrystallized hPCNA-bound peptide structures (Fig. 4B), with hydrogen bonds to hPCNA residues Ala252 and Pro253 within the Q-pocket ranging from 3.0 to 3.6 Å. The Q-pocket of hPCNA went unfilled in the hPCNA-bound noncanonical peptide structures (p21μ–PARG, p21μ–pol ι, p21μ–RFC), where the position 1 residue extended over the Q-pocket to make diverse contacts with the hPCNA surface (see Figs. S10–S12). The inclusion of longer and bulkier side chains at PIP-box position 1, such as lysine (p21μ–Q144K) and methionine (p21μ–Q144M), resulted in peptides with the lowest affinity for hPCNA (KD values of 1.12 and 1.54 μM, respectively). p21μ–pol η, also with Met144P1, bound with similar affinity to p21μ–Q144M at 2.54 μM. p21μ–RNaseH2B, containing a Met144P1, shows 2.4-fold improved affinity compared with p21μ–Q144M (640 nM cf. 1.54 μM). The Lys144P1 containing peptide, p21μ–pol κ, bound hPCNA with 3.30 μM affinity, whereas p21μ–pol ι that also contains a Lys144P1, in contrast, gave a KD value for hPCNA of 111 nM.
Peptides with the point modifications of aspartic acid (p21μ–Q144D), asparagine (p21μ–Q144N), and serine (p21μ–Q144S) at position 1 of the PIP-box displayed significantly lower affinity (50- to 80-fold) than the p21μ peptide, with KD values of 714 and 772 nM and 1.03 μM, respectively (see Fig. 2B, Table S2). p21μ–pol λ, containing Ser144P1, gave a KD value of 8.14 μM, compared to p21μ–PARG with Asp144P1 that binds hPCNA with significantly higher affinity at 90.0 nM. Asp144P1 of p21μ–PARG does not, however, make any clear secondary interactions to explain the higher hPCNA affinity.
p21μ–RFC, with a seven amino-acid PIP-box, was one of the highest affinity noncanonical PIP-box peptides, with a similar KD value to p21μ–PARG and p21μ–pol ι, at 145 nM (Fig. 2C, ‘non-canonical’). It was assumed here that Ile, Phe, and Phe of p21μ–RFC formed a hydrophobic triad (and hence PIP-box positions 4, 7, and 8) to insert into the hydrophobic cleft of hPCNA, which results in an arginine positioned near the Q-pocket (Fig. S12). p21μ–RFC was computationally modeled on the hPCNA surface and indicated that ArgP1 extends over the Q-pocket to make hydrophobic surface contacts with Val45, which is analogous to those made by Lys144P1 in p21μ–pol ι. The other seven amino acid PIP-box in p21μ–pol β, in contrast, was one of the worst performing peptides at 5.73 μM (Fig. 2C, Table S2), where all potential binding modes likely force a polar/charged residue into one of the hydrophobic pockets to create unfavorable interactions and lower binding affinity of p21μ–pol β for hPCNA.
Conserved hydrophobic position 4: Not too big and not too small
The central hydrophobic PIP-box residue plays a key role to stabilize the hPCNA-bound binding structure and anchors the peptide to the hPCNA surface. This is evident by the high number (>5) of interactions the PIP-box position 4 residue makes in all peptides, which are largely hydrophobic interactions (Tables S2, S4–S13). The position 4 PIP-box residue, in all peptides here, either computationally modeled or cocrystallized with hPCNA, is entirely buried within the hydrophobic pocket indicated by a buried surface area of 100% (Table S19).
The hPCNA-bound peptide structures analyzed here all contain a methionine or isoleucine at position 4 of the PIP-box. There was no significant difference between the overall structures of p21μ and p21μ–M147I, confirmed by the RMSD value of 0.191 Å (Table S18). p21μ–M147I has a marginally higher affinity for hPCNA than p21μ, at 11.1 nM (Fig. 2B). The orientation of Met147P4 differed only by slight rotations of the side chain because of its flexibility and large pocket size, and made side-chain interactions with hydrophobic residues Val45, Leu47, and Val48 of hPCNA. A main-chain hydrogen bond interaction between peptide residue Met147P4 and hPCNA residue His44 was evident in all eight of the Met147P4-containing modeled peptides bound to hPCNA. The Ile147P4 peptides p21μ–pol ι and p21μ–PARG similarly make this contact.
Hydrophobic amino-acid substitutions in p21μ–M147V and p21μ–M147L were well tolerated and bound hPCNA with 29.3 nM and 20.5 nM affinity, respectively. p21μ–M147W, with the bulkier Trp147P4, revealed lower affinity for hPCNA at 3.57 μM. However, the peptide containing alanine (p21μ–M147A), with the smallest side chain of this subset, resulted in even weaker affinity for hPCNA, at 7.59 μM. These results reiterate the necessity for a hydrophobic residue at position 4, with an optimal size to fill the hydrophobic pocket and anchor the peptide to the hPCNA surface.
Conserved aromatic positions 7/8: Hydrogen bonds create tighter interactions between peptide and hPCNA
These aromatic residues complete the PIP-box hydrophobic triad that anchors a PIP-box sequence onto the hPCNA surface and stabilizes the single 310-helical turn characteristic of the hPCNA-partner binding conformation (Fig. 4A). The p21μ PIP-box position 8 residue inserts into a hydrophobic pocket on the hPCNA surface formed by Gln131, Ile128, Pro234, Tyr133, and Tyr250 (Fig. 3C). This is exemplified by the Phe150P7Tyr151P8 combination observed in p21μ, where the tyrosine phenol forms a 3.4 Å hydrogen bond to hPCNA Gln131. This interaction is also seen for Tyr151P8 in p21μ–S146R and p21μ–M147I that both hydrogen bond to Gln131 at 2.8 Å with hPCNA affinities of 4.20 and 11.1 nM, respectively.
The p21μ–Y151F peptide has comparable affinity to p21μ (12.3 nM) at 10.6 nM. However, previous literature suggested a Tyr151PheP8–modified p21 peptide resulted in a 3-fold decrease in binding affinity compared with the analogous native peptide (residues 139–160) (22). This may be due to a difference in binding-affinity assay (SPR versus isothermal calorimetry), difference in the assay conditions (e.g., buffer), or a series of small conformational changes that allow p21μ to bind with relatively high affinity, despite being notably shorter than p21139–160. A large number of native PIP-box sequences contain this PheP7PheP8 combination and give KD values that range from 8.76 nM for p21μ–Cdt1 to 3.30 μM for p21μ–pol κ (Fig. 2C).
p21μ–F150Y displayed a slight decrease in affinity for hPCNA, compared with p21μ, with a KD value of 20.2 nM (Fig. 2B, Table S2). The cocrystal structure of p21μ–F150Y bound to hPCNA indicates the Tyr150P7 side chain of p21μ–F150Y forms a 3.7 Å hydrogen bond to Pro253, that is located at the edge of the hydrophobic cleft (Figs. 3D and 5, pink). The phenol group introduced from the Phe150TyrP7 modification participates in a 3.6 Å hydrogen bond with Thr145P2 of the peptide (Fig. 3D), which stabilizes the 310-helical binding conformation of the peptide and may enhance hPCNA binding affinity. TyrP8 often makes a hydrogen bond with Gln131, which is not observed for p21μ–F150Y, suggesting hydrogen bonds between Tyr151P8–Gln131 and Tyr150P7–Thr145P2 cannot both occur at once. Interestingly, there are no human PIP-box proteins reported to contain the TyrP7TyrP8 motif, although one has been identified in the E2F transcription factor from Drosophila melanogaster (11).
The remaining aromatic permutation of TyrP7PheP8 resulted in the greatest improvement in affinity for this subset, with a KD value of 2.18 nM for p21μ–FY150/151YF (Fig. 2B). The computationally modeled structure of p21μ–FY150/151YF bound to hPCNA shows Phe151P8 is lifted slightly out of the pocket, as evidenced by an increased distance from Phe151P8 to Gln131 at 4.8 Å (cf. 3.4 Å in p21μ), and there is also no hydrogen bond evident between Tyr150P7 and Thr145P2. However, a strong intermolecular hydrogen bond from Tyr150P7 to the Ala232 carbonyl may improve hPCNA affinity. The high-affinity p21μ–Pogo (8.82 nM) also displays the TyrP7PheP8 combination.
A sequence modification to include the aromatic histidine, p21μ–F150H, displayed lower affinity for hPCNA at 159 nM, compared with p21μ with a KD value of 4.32 nM (Fig. 2B). The computationally modeled structure of p21μ–PARG bound to hPCNA shows His150P7 positioned similarly to the analogous Phe150P7 in p21μ. His150P7 is angled toward the N terminus of the peptide and makes a secondary interaction with Ser145P2 (Fig. 5, red), which may improve the affinity of the noncanonical PIP-box sequence. Other nonaromatic residues are occasionally observed at PIP-box positions 7 and 8 such as the WRN PIP-box with an aspartic acid at position 7 in place of the conserved aromatic residue. p21μ–WRN was the worst performing native canonical PIP-box peptide with an affinity for hPCNA of 1.15 μM (Fig. 2C), whereas p21μ–pol ι, which contains a Leu151P8, bound hPCNA with 111 nM affinity. The computationally modeled structure of p21μ–pol ι bound to hPCNA suggests Leu151P8 is lifted out of the hydrophobic pocket, compared with Tyr151P8 in p21μ, indicated by an increased distance to Gln131 (3.2 Å in p21μ, to 6.4 Å in p21μ–pol ι). This may explain the 9-fold lower affinity of p21μ–pol ι (111 nM) than p21μ (12.3 nM).
Nonconserved position 2/3: A positively charged residue at P3 increases affinity
No studies to date have explicitly looked at the role of the position 2 and 3 PIP-box residues, as the initial alanine scan that identified the key residues in the PIP-box revealed that mutation of residues Thr145P2 and Ser146P3 in p21 did not significantly impact the hPCNA affinity (18). Ser146P3 in the cocrystal structure of p21μ bound to hPCNA is orientated toward the hPCNA surface and makes a hydrogen bond with the main-chain carbonyl of His44 in 1 of the 3 monomer repeats. Thr145P2 makes a 3.3 Å main-chain hydrogen bond to Pro253 (Fig. 3C) in all three repeats (PDB ID: 7KQ1). Inclusion of lysine or arginine at 145P2 (p21μ–T145K and p21μ–T145R) resulted in 8- and 7-fold decreased affinity for hPCNA, respectively, relative to p21μ (Fig. 2B), whereas p21μ–S146K, with Lys146P3, revealed affinity for hPCNA comparable with p21μ. However, an Arg146P3 instead resulted in a 2.8-fold improvement in the KD value, to 4.30 nM for p21μ–S146R (Fig. 2B). The computationally modeled structure of p21μ–S146R bound to hPCNA indicates Arg146P3 main-chain amide makes a 3.2 Å intramolecular hydrogen bond to the carbonyl of the Asp149P6 side chain (Fig. S6B), and a 2.8 Å intermolecular hydrogen bond to Ser43 on the hPCNA surface (Fig. 5, yellow). These interactions may together enhance the hPCNA-binding affinity of p21μ–S146R. The high-affinity peptides p21μ–XPG and p21μ–FEN1 also contain an Arg146P3 and bind hPCNA with 16.4 and 166 nM affinity, respectively (Fig. 2C). p21μ–Cdt1 binds with even higher affinity at 8.76 nM and contains an ArgP2ArgP3 motif that results in a significant improvement to hPCNA binding affinity when included in the p21μ peptide, p21μ–TS145/146RR, which has a KD value of 1.83 nM.
p21μ-Pogo binds hPCNA with slightly higher affinity (8.82 nM) than p21μ–TS145/146KK, which contains the same LysP2LysP3 motif (20.2 nM, Fig. 2C). The computationally modeled structure of p21μ–Pogo bound to hPCNA shows Lys146P3 makes an analogous 3.2 Å intramolecular hydrogen bond to Arg146P3 in p21μ–S146R, with the 149P6 residue, but no intermolecular interactions (Fig. 5, dark blue). The two other permutations of arginine and lysine as dual modifications at positions 145 and 146 (p21μ–TS145/146RK, p21μ–TS145/146KR) both resulted in lower KD values for hPCNA, than p21μ, at 17.3 and 37.0 nM (Fig. 2B).
Negatively charged residues are not commonly observed at positions 2 and 3; however, Asp146P3 is present in the RFC PIP-box sequence. The computationally modeled p21μ–RFC peptide bound to hPCNA indicates Asp146P3 makes a 3.2 Å intramolecular interaction with the positively charged Lys149P6 (Fig. S12). This complementary interaction is equivalent to the interaction seen for peptides such as p21μ–S146R where Arg146P3 and Asp149P6 interact. Such intramolecular interactions may contribute to the unexpectedly high affinity of p21μ–RFC (145 nM) by stabilizing the 310-helical binding conformation.
The PIP-box sequences of pol ι and pol δp66 contain neutral residues at PIP-box positions 2 and 3, 421GlyLeu and 457ValSer, respectively. Both p21μ–pol ι and p21μ–pol δp66 bind hPCNA with lower affinity than p21μ at 111 and 124 nM, respectively, and lack the ability to make a PIP-box position 2 side-chain hydrogen bond with hPCNA, as in p21μ. This observation reinforces the conclusion that positively charged or polar residues at these nonconserved positions lead to enhanced hPCNA-binding affinity.
Nonconserved position 5/6: Side chains with hydrogen-bond donor and acceptor character increase affinity
The ThrP5AspP6 motif in p21 and Pogo is thought to be responsible, in part, for the high hPCNA affinity observed because of the resultant hydrogen bonds (25). The initial report of an alanine scan of p21 that identified the PIP-box motif noted Asp149P6 as an important residue for inhibition of SV40 DNA replication (18). The cocrystal structure of p21139–160 (PDB ID: 1AXC) highlights two intramolecular interactions of Asp149P6 and one of Thr145P5 (10, 15). The cocrystal structure of p21μ bound to hPCNA (PDB ID: 7KQ1) here shows the Asp149P6 side chain (hydrogen bond acceptor) makes a hydrogen bond with the Thr145P2 side chain in 1 of the 3 monomer repeats. Residues Asp149P6 and Ser146P3 in p21μ participate in an intramolecular main-chain hydrogen bond to define a 310-helix (Fig. 3C). These two hydrogen bonds are the only interactions of the PIP-box position 5 or 6 residues in p21μ. The analogous interaction is made in the computationally modeled structure of p21μ–PARG and p21μ–Pogo bound to hPCNA. The Asp149P6 side chain in p21μ–S146R acts as a hydrogen bond donor to interact with the main chain of Arg145P2.
Incorporation of a negatively charged residue at position 5 of the PIP-box (p21μ–T148E and p21μ–T148D) resulted in ~6- to 8-fold lower affinity for hPCNA, than p21μ. The KD value of p21μ–TD148/149DE was notably worse than p21μ, at 28-fold lower affinity. However, this was recovered to only 4-fold lower than p21μ for a GluP5GluP6 modification (for p21μ–TD148/149EE) (Fig. 2B) and highlights that modifications must be considered in the context of the whole sequence. A number of peptides display an intermolecular hydrogen bond with the His44 side chain of hPCNA where an AspP5 or GluP5 acts as a hydrogen bond donor (e.g., Fig. 5, light green) and is more common in noncanonical than canonical PIP-box sequences. A negatively charged residue at position 6, in contrast, is more likely to form intramolecular interactions, rather than intermolecular. This can be reasoned by the resulting side-chain positions when the 310-helical binding conformation is adopted. When negatively charged residues are not included at positions 5 and 6, high-affinity partners generally contain polar rather than hydrophobic residues at these PIP-box positions (see DNALig1 or MCMT, Fig. 2C). In p21μ–pol ι, the unusual Tyr149P6 points away from the surface in the computationally modeled structure bound to hPCNA, and the 310-helix stabilizing 3.3 Å intramolecular main-chain hydrogen bond is instead made between position 6 and Ile147P4 (Fig. S11).
Individual PIP-box modifications must be considered in the context of the entire PIP-box sequence
All p21μ peptides with greater than 20 nM affinity for hPCNA display charge or hydrogen-bond complementary residues on either side of the hydrophobic residue at position 4, supporting the idea that this configuration aids stabilization of the 310-helical binding conformation and in turn enhances affinity. This is seen in p21μ–Pogo and p21μ–XPG that bind with higher affinity than the p21μ peptide (Fig. 2), and Figure 4D clearly shows that side chains of residues at position 2 and 6 are generally angled toward one another.
Cooperative interactions, in the form of charge-complementary pairs, may be more important in noncanonical PIP-box peptides. The RFC seven amino-acid PIP-box contains a charge complementary pair on either side of the hydrophobic position 4 residue; however, this is in the less-common orientation where the negatively charged residue (Asp) is at position 3 and the positively charged residue (Arg/Lys) is at positions 5/6. Similarly, the divergent, noncanonical PARG PIP-box in the p21μ–PARG peptide displays relatively high hPCNA affinity at 90.0 nM despite the lack of a canonical GlnP1 and contains His150P7 rather than phenylalanine or tyrosine. Interestingly, this peptide binds with higher affinity than p21μ–F150H, suggesting the expected affinity loss due to the absence of the Gln144P1 is overcome by the remaining nonconserved residues, such as the charge complementary Lys146P3 and Asp149P6, and hydrogen bond between His150P7 and Ser145P2 (Fig. 5, red). The PARG PIP-box also contains the ThrP5AspP6 motif observed in the high-affinity hPCNA partners p21 and Pogo-Ligase. The highly divergent PIP-box of the p21μ–pol ι peptide also shows relatively high hPCNA affinity (111 nM), in alignment with the higher-than-expected affinity displayed by the native pol ι415–437 peptide (0.39 μM, Table 1 (16)). This PIP-box lacks a second aromatic residue at position 8; in addition, position 1 is not glutamine. However, the pol ι PIP-box does display a number of interactions that may stabilize the binding conformation and improve affinity in the absence of these traditional elements, for example, a hydrogen bond between Asp425P5 and His44 of hPCNA (cf. Fig. 5, light green) and an intramolecular Tyr427P8 and Lys421 hydrogen bond (Fig. S16F, yellow).
Table 1.
The effect of changing the sequence flanking the PIP-box motif, on the hPCNA-binding affinity
| Name | Sequence | Affinity KD | Native peptides | Affinity KD | Ref. | |
|---|---|---|---|---|---|---|
| p21μ–Pogo | KRRQKKITDYFHSKR | 8.82 nM | < | SAVLQKKITDYFHPKKa | 100 nM | (21, 24) |
| p21μ–Cdt1 | KRRQRRVTDFFHSKR | 8.76 nM | < | 2MEQRRVTDFFARRR15 | 7.20 μM | (26) |
| p21μ–pol δp66 | KRRQVSITGFFHSKR | 124 nM | < | 452KANRQVSITGFFQRK466 | 15.6 μM | (14) |
| p21μ–FEN1 | KRRQGRLDDFFHSKR | 166 nM | < | 331SRQGSTQGRLDDFFKVTGSL350 | 59.9 μM | (14) |
| p21μ–p15 | KRRQKGIGEFFHSKR | 234 nM | < | 41APVCVRPTPKWQKGIGEFFAA72 | 5.56 μM | (46) |
| p21μ–PARG | KRRDSKITDHFHSKR | 90.0 nM | < | 402QHGKKDSKITDHFMRLPKA420 | 3.3 μM | (23) |
| p21μ–pol ι | KRRKGLIDYYLHSKR | 111 nM | < | 415ALNTAKKGLIDYYLMPSLSTTSR437 | 0.39 μM | (16) |
| p21μ–pol η | KRRMQTLESFFHSKR | 2.54 μM | > | 693CKRPRPEGMQTLESFFKPLTH713 | 0.4 μM | (16) |
| p21μ–pol κ | KRRKHTLDIFFHSKR | 3.30 μM | < | 861PKHTLDIFFK874PLTHb | 4.9 μM | (16) |
| p21μ–Cdt2 | KRRMRKICTYFHSKR | NDa | >> | 704SSMRKICTYFHRKS717 | 57 nM | (26) |
| p21μ–RNAseH2B | KRRMKSIDTFFHSKR | 640 nM | < | 290DKSGMKSIDTFFGVKNKKKIGKV312 | 35 μM | (47) |
PL, Pogo-Ligase.
A comparison of p21μ:PIP-box hybrid peptides and the analogous native peptides from which the PIP-box derives.
PL peptide is a mutant hybrid peptide and not an entirely native sequence.
PLTH is not part of the native sequence and was added to the sequence to improve binding affinity.
Some peptides such as p21μ–pol δp66 and p21μ–FEN1 bind with unexpectedly lower affinity than p21μ despite strict canonical sequences. p21μ–FEN1 is particularly interesting as it contains not only charge complementarity by means of residues (Arg146P3 to Asp148P5 and Asp149P6) on either side of the hydrophobic position 4 residue but also an Arg146P3 which considerably enhanced hPCNA binding affinity when introduced as a point modification in p21μ (p21μ–S146R, 4.30 nM). The other sequence differences in p21μ–FEN1 (compared with p21μ) are the PheP7PheP8 motif (seen in p21μ–Y151F, 10.6 nM) and Leu147P4 (seen in p21μ–M147L, 20.5 nM) that did not drastically impact affinity in the point-modified peptides. Perhaps, the lower affinity of p21μ–FEN1 is owed in part to the AspP5AspP6 combination, as the p21μ–T148D peptide showed a notably lower hPCNA affinity relative to p21μ (96.5 nM, cf. 4.32 nM for p21μ). This is an example of how each PIP-box modification must be considered in context of the entire PIP-box sequence and the importance of different sequence combinations. The common interactions that appear to contribute to enhanced hPCNA-binding affinity have been summarized in Figure 6 to provide guidelines to predict cooperative interactions and hence affinity.
This concept, that the sequence as a whole must be considered to anticipate hPCNA affinity, extends to the sequence that flanks the PIP-box. The p21μ peptides considered here all comprise the same p21-derived PIP-box flanking sequence, so any changes in affinity or structure can be attributed to changes induced by the PIP-box sequence itself. The affinity of p21μ peptides containing a native PIP-box was compared with the reported affinity of the analogous native peptide sequence to reveal an interesting trend: All five of the p21μ native canonical PIP-box hybrid peptides display higher affinity than the native peptide (Table 1). The most significant difference is greater than 820-fold for the Cdt1 PIP-box peptides, where p21μ–Cdt1 binds with 8.76 nM and Cdt12–15 binds with 7.20 μM affinity (26). This may be correlated to the number of positively charged residues in the flanking region, where the native canonical sequences contain four or less positively charged residues compared with the p21μ with four arginine residues. In contrast, four of the six noncanonical PIP-box:p21μ hybrids showed lower affinity relative to the native sequence (Table 1).
Rationally designed PIP-box sequences: Designing the highest affinity hPCNA partner to date
PIP-box sequences were designed to investigate different cooperative amino-acid combinations and were inspired by the binding affinity results of the modified p21μ peptides in conjunction with the structural observations from cocrystal structures, computationally modelled peptides, and native PIP-box structures, which are summarized graphically in Figures 5 and 6. Three PIP-box sequences were synthesized to trial whether amino-acid combinations not previously observed together could give rise to high hPCNA affinity. For example, would a Glu148P5 interact with the positively charged residues at P2/P3 and the Glu149P6 interact with His44 simultaneously to improve hPCNA affinity of p21μ–RD1.
Three peptides were synthesized, the affinity for hPCNA determined by SPR, and the hPCNA-bound structures computationally modelled to reveal, remarkably, that all three peptides bound with higher affinity than the native p21μ (Fig. 2D, Table S2). The highest affinity peptide p21μ–RD2 bound hPCNA with 1.12 nM affinity, which is 11-fold higher affinity than p21μ and 3-fold higher than p21139–160 despite being seven amino acids shorter. This makes p21μ–RD2, a PIP-box peptide rationally designed amino acid by amino acid, the highest affinity hPCNA-binding peptide or protein to date, displaying a significant improvement in affinity for hPCNA. The computationally modeled structures of these peptides bound to hPCNA were analyzed to confirm whether the hypothesized cooperative interactions were observed and contributed to this impressive affinity and allowed us to refine the observations incorporated into the secondary interaction map in Figure 6 that provides guidelines to design high affinity a PIP-box sequence.
In p21μ–RD1 and p21μ–RD3, the bond between Tyr150P7 and Pro253 is stronger than that of p21μ, at distances of 3.5 Å and 3.4 Å, respectively, which may contribute to the improved affinity for hPCNA. The highest affinity peptide p21μ–RD2 (KD 1.12 nM) has four defined intramolecular hydrogen bond interactions (Fig. 7C), which all involve PIP-box residues and may stabilize the 310-helical binding conformation and lead to the enhanced hPCNA affinity. In particular, the Glu149P6 forms a main-chain hydrogen bond to Arg146P3. p21μ–RD2 contains an Arg146P3 that makes a hydrogen bond to Ser43 in the computationally modeled structure bound to hPCNA, which is also observed for p21μ–S146R that bound hPCNA with high affinity (4.30 nM, Fig. 5, yellow). In addition, Thr145P3 side chain of p21μ–RD2 hydrogen bonds to the Pro253 carbonyl, and the Tyr150P7 phenol hydrogen bonds with Asp232 main-chain carbonyl (Fig. 7C). The 146ArgP3 modification resulted in increased affinity in p21μ–S146R and was included in p21μ–RD2 and p21μ–RD3; however, the affinity of p21μ–RD3 was not significantly different to p21μ and suggests the other modifications may be working to oppose an increase in affinity. Interestingly, p21μ–RD1 with the TyrP7TyrP8 motif only makes an intermolecular hydrogen bond between Tyr150P7 and Asp232, as seen in p21μ–F150Y; however, p21μ–RD3 makes an intermolecular interaction through the phenol of both Tyr150P7 (to Asp232) and Tyr151P8 (to Gln131). This is further evidence that sequence modifications work cooperatively to alter structure and affinity.
Conclusions and outlook
This work provides a comprehensive study using a set of hPCNA-binding peptides and highlights a series of interactions that synergistically contribute to high hPCNA affinity, as summarized in Figure 6. The glutamine residue and hydrophobic triad are essential to enhance hPCNA-binding affinity and adhere a peptide to the hPCNA surface. The PIP-box position 1 glutamine residue and its hydrogen bond network contribute notably to hPCNA binding affinity, although glutamine is not essential to confer hPCNA binding. The PIP-box position 4 hydrophobic residue, which fills the hydrophobic cleft, is essential to a high-affinity hPCNA interaction. A variety of combinations of tyrosine and phenylalanine (FY, YF, FF, or YY) are most commonly preferred at positions 7 and 8 of the PIP-box, to stabilize the characteristic 310-helical turn. However, the highest binding affinity was observed for peptides containing either PheP7TyrP8 or TyrP7PheP8 (rather than TyrP7TyrP8 or PheP7PheP8). A tyrosine residue at PIP-box position 7 makes an intramolecular interaction, commonly with the PIP-box position 2 residue, where the phenol is oriented toward the N terminus of the peptide, whereas a position 8 tyrosine generally hydrogen bonds with Gln131 of hPCNA within the hydrophobic cleft. The cocrystal structure of p21μ–F150Y bound to hPCNA is the first TyrP7TyrP8 peptide reported to interact with hPCNA. The PIP-box Tyr151P7 forms a hydrogen bond to Thr145P2, but a hydrogen bond from position 8 to Gln131 is absent, which suggests these two proposed interactions cannot occur at once.
Complementary hydrogen bonding or charge pairs on either side of the hydrophobic residue at position 4 stabilize the 310-helix binding conformation and appear particularly important to increase hPCNA affinity of noncanonical PIP-box sequences. Introduction of a positively charged residue at position 3 is more favorable than at position 2 to increase hPCNA-binding affinity. The nonconserved PIP-box combinations that lead to increased hPCNA-binding affinity include a positively charged residue at position 3 and negatively charged residue at position 6, where both are preceded by a small polar amino-acid that may hydrogen-bond intramolecularly or with the hPCNA surface. Negatively charged residues at position 5 more commonly hydrogen-bond intermolecularly with hPCNA, in particular with His44, whereas position 6 residues often form intramolecular hydrogen bonds. Finally, an increase in positive charge in the sequence that flanks a canonical PIP-box motif appears to further enhance binding affinity. This is the first study that has extensively looked at the effect of nonconserved PIP-box combinations on hPCNA-binding affinity and has identified key modifications that increase affinity such as the PIP-box position 3 arginine.
p21μ–RD2, with PIP-box QTRITEYF, is the highest affinity hPCNA-binding partner reported to date (1.12 nM) and is seven amino acids shorter than the previous title holder in p21139–160 (4.32 nM). This peptide conforms explicitly to the guidelines for secondary interactions that lead to high-affinity hPCNA binding set out in Figure 6. It is important to note that p21μ–RD2 provides just one solution to the puzzle of obtaining high affinity to hPCNA and many other combinations may also lead to high hPCNA binding affinity. This study has highlighted the complexity of the secondary interaction network that gives rise to high-affinity binding of short peptides to hPCNA and has made significant progress to define the rules of high-affinity hPCNA interaction. The importance of these interactions, in particular the intermolecular interactions with hPCNA, may be further probed by mutation of key hPCNA residues such as His44. Furthermore, it emphasizes that to predict hPCNA-binding affinity, the PIP-box sequence must be considered as a whole, where small sequence changes can give rise to large changes in affinity, dependent on the rest of the PIP-box sequence. The insights gained here can be used to inform design of new hPCNA-binding peptides or tune the affinity of hPCNA peptides and proteins to probe interactions important in DNA replication and DNA repair. Consequently, this study provides an insight into how nature has fine-tuned affinity of the native hPCNA-binding proteins to allow tight regulation of DNA-replication and DNA-repair processes. This knowledge may be further leveraged to design inhibitors of human hPCNA interactions for therapeutic applications.
Experimental procedures
Peptide synthesis
All peptides were synthesized using Fmoc/t-Bu Solid-Phase Peptide Synthesis (27). Six peptides were synthesized using Rink Amide resin on a Liberty Blue peptide synthesizer (CEM Corp); 31 peptides were synthesized on a Prelude peptide synthesizer (PTI); five peptides were synthesized manually as detailed in the Supplementary information. The remaining ten peptides were purchased from Shanghai Royobiotech at >95% purity. Synthesized peptides were purified using semipreparatory RP-HPLC using a Phenomenex Luna C18(2) or Phenomenex Aeris Peptide C18 column (10 mm × 250 mm, 5 μm) over a linear gradient of water and acetonitrile, with 0.1% TFA, at 4 ml/min and UV detection at 220 nm. Peptide identity was confirmed by high resolution mass spectrometry using an Agilent 6230 ESI-TOF MS. Peptide purity was characterized on an Agilent 1260 Infinity analytical RP-HPLC equipped with a Phenomenex Luna C18(2) column (4.6 mm × 250 mm, 5 μm) using a linear gradient of 0 to 50% acetonitrile with 0.1% TFA and in water with 0.1% TFA, over 15 min at 1.5 ml/min and visualized at 220 nm. Detailed methods are included in the Electronic Supplementary Information and characterization data are listed in Table S1.
Protein expression and purification
A glycerol stock of Escherichia coli BL21 (λDE3) cells carrying a hPCNA–pMCSG19 plasmid (with no purification tag) were grown in a 50-ml overnight culture. Eight 1-l baffled flasks of LB with 100 μg/ml of ampicillin were inoculated with 6.3 ml of the overnight culture. Cultures were incubated at 37 °C until an absorbance at 600 nm of 0.5 and induced with 0.5 mM IPTG. Cultures were grown overnight at 16 °C with shaking at 200 rpm. Cultures were pelleted at 5000g for 20 min. After removing the supernatant, pellets were resuspended in 30 ml of buffer A (20 mM Tris, pH 7.5, 20 mM NaCl, 2 mM DTT) and then lysed by five rounds of cell disruption by a microfluidics cell disrupter. Lysate was pelleted at 45,000g for 60 min, and the supernatant was collected for purification.
hPCNA was purified at 4 °C by fast protein liquid chromatography, using an anion exchange diethylaminoethyl column (HiTrap diethylaminoethyl FF 5 ml column), equilibrated in buffer A (20 mM Tris pH 7.5, 20 mM NaCl, 2 mM DTT), and protein was eluted using buffer B (20 mM Tris, pH 7.5, 0.7 M NaCl, 2 mM DTT). Fractions were analyzed by SDS-PAGE, and those of interest indicating containing protein at ~28 kDa were selected and pooled and treated with ammonium sulphate to bring the concentration to 1.5 M. Protein was purified again by a hydrophobic column (HiTrap Phenyl FF [high sub] 5 ml column) equilibrated in buffer C (20 mM Tris, pH 7.5, 20 mM NaCl, 2 mM DTT, 0.5 mM EDTA, 1.5 M ammonium sulphate), and protein was eluted using buffer D (20 mM Tris, pH 7.5, 0.5 mM EDTA, 2 mM DTT). Fractions were analyzed by SDS-PAGE, and those of interest were dialyzed overnight in buffer E (20 mM Tris, pH 7.5, 20 mM NaCl, 1 mM DTT).
Protein pool was concentrated using a centrifugal filter unit (30-kDa molecular mass cut-off) to a volume of less than 10 ml and purified using a size-exclusion column (HiPrep 26/60 Sephacryl S-200 HR 300-ml column), equilibrated in buffer F (20 mM Tris, pH 7.5, 50 mM NaCl, 2 mM DTT, 0.5 mM EDTA), and protein was eluted using the same buffer. Fractions were analyzed by SDS-PAGE, and those of interest were pooled and purified using an anion-exchange Q Sepharose column (ENrich Q 10 × 100 mm 8 ml column), equilibrated in buffer G (20 mM Tris, pH 7.5, 20 mM NaCl, 2 mM DTT), protein was eluted using buffer H (20 mM Tris, pH 7.5, 0.7 M NaCl, 2 mM DTT). Fractions were analyzed by SDS-PAGE, and those of interest were pooled and dialyzed overnight against storage buffer I (20 mM Tris, pH 7.5, 10% glycerol, 2 mM DTT, 0.5 mM EDTA). Protein for crystallography was concentrated to ~10 mg/ml using a centrifugal filter unit (50-kDa molecular mass cut-off) and stored at −80 °C.
SPR assays
The running buffer used for ligand attachment and analyte-binding experiments was 10 mM Hepes buffer with 150 mM NaCl, 3 mM EDTA, and 0.05% Tween-20, adjusted to pH 7.4 with 2 M NaOH. A GE CM5 (series S) sensor chip was primed with the running buffer and preconditioned per the manufacturer’s recommendation with successive injections (2 × 50 s, 30 μl/min) of 50 mM NaOH, 10 mM HCl, 0.1% SDS, 0.85% H3PO4, and glycine, pH 9.5, respectively. The surface was then activated with an injection of 0.2 M 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide and 50 mM N-hydroxysuccinimide (600 s, 10 μl/min). hPCNA (5 μl, 12 mg/ml) was diluted into the running buffer (245 μl). Only once the preactivation was complete was the protein further diluted to a final concentration of 25 μg/ml in 10 mM NaAc (~pH 4.6) by addition of hPCNA/Hepes (50 μl) to a solution of 100 mM NaAc (50 μl) and water (400 μl). This solution was immediately injected over only one flow cell (10 μl/min) until ~1500 RU was reached at stabilization. Both flow cells were then blocked with 1.0 M ethanolamine, pH 8.5 (600 s, 10 μl/min). The chip was left to stabilize for 2 h before sample injections commenced.
Peptides (approx. 2 mg) were dissolved in MilliQ water and centrifuged (7800 rpm, 10 min) to remove any particulate. The peptide stock concentration was determined by 205 nm absorbance (A205), where 2 μl of the stock was further diluted in water (10–50 fold) and a measurement taken in triplicate with a NanoDrop2000 and baseline referenced to 750 nm absorbance. The ε205 for each peptide was calculated using an online calculator (http://nickanthis.com/tools/a205.html, (28)); however, an additional glycine residue was added to each peptide sequence to account for the terminal amide of the synthesized peptides. The peptide stock solution concentration was then calculated per c = (A205/ε205 × l) × DF, where the concentration is in molar, A205 is absorbance at 205 nm calculated as an average of three readings, l is the pathlength in centimeter (0.1 cm for Nanodrop), ε205 is the molar absorptivity at 205 nm, and DF is the dilution factor. The stock concentrations are tabulated in Table S2. The peptides were then diluted into the running buffer before further dilution as necessary.
Steady-state affinity experiments were conducted at a flow rate of 30 μl/min, with a starting contact time of 40 s and dissociation of 60 s, and extended if a steady state could not be reached. A 1 in 2 serial dilution, eight times, was performed for each peptide, and the resulting solutions were injected sequentially from the lowest to highest concentration, preceded by a buffer-only blank injection. After each injection, the surface was regenerated with 2 M NaCl (2 × 30 s, 25 μl/min). After an optimal concentration range was found, the series of injections were repeated to ensure reproducibility. The top concentration for the final concentration range for each peptide is listed in Table S2. All data were analyzed using the GE Biosystems Biacore S200 Evaluation Software. All data are summarized in Table S2 and Figure 2.
Protein–peptide cocrystallization experiments
hPCNA was mixed with peptide of interest at 1:1.2 M ratio, and after incubation on ice for 30 min, the sample was pelleted at 16,000g for 10 min to remove aggregates. The supernatant containing peptide-bound protein was stored at −80 °C. Crystals were grown by hanging-drop vapor-diffusion method in 24-well linbro plates containing 500 μl well solution, by mixing 1 μl protein and peptide with an equal volume of the well solution (29, 30, 31). Initial cocrystallization screens with all p21μ-modified peptides and hPCNA were attempted. Diffracting crystals of hPCNA bound to p21μ (residues 141–150) were formed in 8% Tacsimate and 20% PEG at 16 °C after 4 weeks. Diffracting crystals of hPCNA bound to p21μ–F150Y were formed in 0.18 M magnesium acetate and 20% PEG at room temperature after 8 weeks. Crystals were mounted on cryoloops, cryoprotected using paratone-N, and flash-cooled in liquid nitrogen (29, 30, 31). Data were collected at 100 K using the MX1 beamline at the Australian Synchrotron (32). Diffraction data were indexed and integrated using X-ray Detector Software (33). Pointless (CCP4i) (34) was used to create an mtzfile for scaling. Data were scaled using Aimless (CCP4i) (35, 36) to a resolution of 3.30 Å for p21μ and 2.43 Å for p21μ–F150Y. Phasing was solved by molecular replacement using Phaser MR (CCP4i) (37) using a search model for p21μ of (PDB ID: 1AXC, human (15)) and for p21μ–F150Y (PDB ID: 4RJF, human (22)). Solutions were refined in phenix.refine (38) in iterative rounds with manual rebuilding in Coot (39, 40). Data collection and refinement statistics for hPCNA in complex with p21μ or p21μ–F150Y are summarized in Table S3. The final structures are deposited on the RCSB database under accession numbers 7KQ1 and 7KQ0, respectively.
Computational modelling
Models of hPCNA and p21 analogue peptide structures were constructed using the solved structure of hPCNA bound with p21μ peptide as a starting template (PDB ID: 7KQ1). The peptides analyzed were p21μ–F150Y, p21μ–S146R, p21μ–M147I, p21μ–D149E, p21μ–FY150/151YF, p21μ–PARG, p21μ–Pogo, p21μ–pol δp66, p21μ–pol ι, p21μ–RFC, p21μ–RD1, p21μ–RD2, and p21μ–RD3. The residue(s) being investigated were mutated to the amino acid of interest, and unresolved side chains of residues were modeled into the computational structure. Energy minimization/annealing (n = 30) for refinement was carried out in ICM-Pro Molsoft (41, 42). Refined models were analyzed using PyMOL to validate the model by comparing against (p21μ structure) and assess side-chain interactions (43). The resulting structures were visualized in PyMOL and are depicted in Figs. S4–S15. Additional analysis was carried out using the RING server (44) and PoseView (45).
Data availability
Atomic coordinates and structure factors for the reported crystal structures have been deposited with the RCSB Protein Data bank under accession numbers 7KQ1 and 7KQ0.
Supporting information
This article contains supporting information (14, 15, 16, 23, 24, 28, 44, 48).
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
This research was undertaken in part using the MX1 beamline at the Australian Synchrotron, part of Australian Nuclear Science and Technology Organisation. The facilities of the OptoFab node of the Australian National Fabrication Facility and associated Commonwealth and South Australian State Government funding are also gratefully acknowledged.
The research was supported by the Australian Research Council Centre of Excellence for Nanoscale BioPhotonics (CNBP) (CE140100003).
Author contributions
A. J. H. conceptualization, formal analysis, investigation (peptide synthesis, SPR assays), validation, writing–original draft, and writing–review and editing; B. A. V. formal analysis, investigation, (protein synthesis, protein crystallography, computational modeling), validation, writing–original draft, and writing–review and editing; W. K. resources, supervision, and investigation, (peptide synthesis), validation, and writing–review and editing; T. C. investigation, validation, and writing–review and editing; D. B. S.–supervision, investigation, (peptide synthesis), and writing–review and editing; A. D. A. resources, supervision, and writing–review and editing; J. B. B. resources, supervision, validation, and writing–review and editing.
Funding and additional information
A. J. H. and B. A. V. are supported by Australian Government Research Training Program Stipends (RTPS).
Edited by Patrick Sung
Contributor Information
Andrew D. Abell, Email: andrew.abell@adelaide.edu.au.
John B. Bruning, Email: john.bruning@adelaide.edu.au.
Supporting information
References
- 1.De Biasio A., Blanco F.J. Proliferating cell nuclear antigen structure and interactions: Too many partners for one dancer? Adv. Protein Chem. Struct. Biol. 2013;91:1–36. doi: 10.1016/B978-0-12-411637-5.00001-9. [DOI] [PubMed] [Google Scholar]
- 2.Tsurimoto T. PCNA binding proteins. Front. Biosci. 1999;4:D849–D858. doi: 10.2741/tsurimoto. [DOI] [PubMed] [Google Scholar]
- 3.Maga G., Hubscher U. Proliferating cell nuclear antigen (PCNA): A dancer with many partners. J. Cell Sci. 2003;116:3051–3060. doi: 10.1242/jcs.00653. [DOI] [PubMed] [Google Scholar]
- 4.Moldovan G.L., Pfander B., Jentsch S. PCNA, the maestro of the replication fork. Cell. 2007;129:665–679. doi: 10.1016/j.cell.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 5.Boehm E.M., Gildenberg M.S., Washington M.T. The many roles of PCNA in eukaryotic DNA replication. Enzymes. 2016;39:231–254. doi: 10.1016/bs.enz.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stoimenov I., Helleday T. PCNA on the crossroad of cancer. Biochem. Soc. Trans. 2009;37:605–613. doi: 10.1042/BST0370605. [DOI] [PubMed] [Google Scholar]
- 7.Zhongyun D., Wortman M., Tan Z., Dillehay K. World Intellectual Property Organisation, International Bureau; 2012. WO 2012/033938 A2, Identification of PCNA Targeting Compounds for Cancer Therapy and PCNA Function Regulation. [Google Scholar]
- 8.De March M., Merino N., Barrera-Vilarmau S., Crehuet R., Onesti S., Blanco F.J., De Biasio A. Structural basis of human PCNA sliding on DNA. Nat. Commun. 2017;8:13935. doi: 10.1038/ncomms13935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Majka J., Burgers P.M.J. The PCNA–RFC families of DNA clamps and clamp loaders. Prog. Nucleic Acid Res. Mol. Biol. 2004;78:227–260. doi: 10.1016/S0079-6603(04)78006-X. [DOI] [PubMed] [Google Scholar]
- 10.Horsfall A.J., Abell A.D., Bruning J.B. Targeting PCNA with peptide mimetics for therapeutic purposes. ChemBioChem. 2019;21:442–450. doi: 10.1002/cbic.201900275. [DOI] [PubMed] [Google Scholar]
- 11.Prestel A., Wichmann N., Martins J.M., Marabini R., Kassem N., Broendum S.S., Otterlei M., Nielsen O., Willemoes M., Ploug M., Boomsma W., Kragelund B.B. The PCNA interaction motifs revisited: Thinking outside the PIP-box. Cell. Mol. Life Sci. 2019;76:4923–4943. doi: 10.1007/s00018-019-03150-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gonzalez-Magana A., Blanco F.J. Human PCNA structure, function and interactions. Biomolecules. 2020;10:570. doi: 10.3390/biom10040570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wegener K.L., McGrath A.E., Dixon N.E., Oakley A.J., Scanlon D.B., Abell A.D., Bruning J. Rational design of a 310-helical PIP-box mimetic targeting PCNA - the human sliding clamp. Chem. Eur. J. 2018;24:11325–11331. doi: 10.1002/chem.201801734. [DOI] [PubMed] [Google Scholar]
- 14.Bruning J.B., Shamoo Y. Structural and thermodynamic analysis of human PCNA with peptides derived from DNA polymerase-delta p66 subunit and flap endonuclease-1. Structure. 2004;12:2209–2219. doi: 10.1016/j.str.2004.09.018. [DOI] [PubMed] [Google Scholar]
- 15.Gulbis J.M., Kelman Z., Hurwitz J., O’Donnell M., Kuriyan J. Structure of the C-terminal region of p21 WAF1/CIP1 complexed with human PCNA. Cell. 1996;87:297–306. doi: 10.1016/s0092-8674(00)81347-1. [DOI] [PubMed] [Google Scholar]
- 16.Hishiki A., Hashimoto H., Hanafusa T., Kamei K., Ohashi E., Shimizu T., Ohmori H., Sato M. Structural basis for novel interactions between human translesion synthesis polymerases and proliferating cell nuclear antigen. J. Biol. Chem. 2009;284:10552–10560. doi: 10.1074/jbc.M809745200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boehm E.M., Washington M.T. R.I.P. to the PIP: PCNA-binding motif no longer considered specific. Bioessays. 2016;38:1117–1122. doi: 10.1002/bies.201600116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Warbrick E., Lane D.P., Glover D.M., Cox L.S. A small peptide inhibitor of DNA replication defines the site of interaction between the cyclin-dependent kinase inhibitor p21 WAF1 and proliferating cell nuclear antigen. Curr. Biol. 1995;5:275–282. doi: 10.1016/s0960-9822(95)00058-3. [DOI] [PubMed] [Google Scholar]
- 19.Warbrick E. PCNA binding through a conserved motif. Bioessays. 1998;20:195–199. doi: 10.1002/(SICI)1521-1878(199803)20:3<195::AID-BIES2>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 20.Zheleva D.I., Zhelev N.Z., Fischer P.M., Duff S.V., Warbrick E., Blake D.G., Lane D.P. A quantitative study of the in vitro binding of the C-terminal domain of p21 to PCNA: Affinity, stoichiometry, and thermodynamics. Biochemistry. 2000;39:7388–7397. doi: 10.1021/bi992498r. [DOI] [PubMed] [Google Scholar]
- 21.Warbrick E. A functional analysis of PCNA-binding peptides derived from protein sequence, interaction screening and rational design. Oncogene. 2006;25:2850–2859. doi: 10.1038/sj.onc.1209320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kroker A.J., Bruning J.B. p21 exploits residue Tyr151 as a tether for high-affinity PCNA binding. Biochemistry. 2015;54:3483–3493. doi: 10.1021/acs.biochem.5b00241. [DOI] [PubMed] [Google Scholar]
- 23.Kaufmann T., Grishkovskaya I., Polyansky A.A., Kostrhon S., Kukolj E., Olek K.M., Herbert S., Beltzung E., Mechtler K., Peterbauer T., Gotzmann J., Zhang L., Hartl M., Zagrovic B., Elsayad K. A novel non-canonical PIP-box mediates PARG interaction with PCNA. Nucleic Acids Res. 2017;45:9741–9759. doi: 10.1093/nar/gkx604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kontopidis G., Wu S.-Y., Zheleva D.I., Taylor P., McInnes C., Lane D.P., Fischer P.M., Walkinshaw M.D. Structural and biochemical studies of human proliferating cell nuclear antigen complexes provide a rationale for cyclin association and inhibitor design. Proc. Natl. Acad. Sci. U. S. A. 2005;102:1871–1876. doi: 10.1073/pnas.0406540102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Choe K.N., Moldovan G.-L. Forging ahead through darkness: PCNA, still the principal conductor at the replication fork. Mol. Cell. 2016;65:380–392. doi: 10.1016/j.molcel.2016.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hayashi A., Giakoumakis N.N., Heidebrecht T., Ishii T., Panagopoulos A., Caillat C., Takahara M., Hibbert R.G., Suenaga N., Stadnik-Spiewak M., Takahashi T., Shiomi Y., Taraviras S., von Castelmur E., Lygerou Z. Direct binding of Cdt2 to PCNA is important for targeting the CRL4Cdt2 E3 ligase activity to Cdt1. Life Sci. Alliance. 2018;1 doi: 10.26508/lsa.201800238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fields G.B., Noble R.L. Solid phase peptide synthesis utilizing 9-fluorenylmethoxycarbonyl amino acids. Int. J. Pept. Protein Res. 1990;35:161–214. doi: 10.1111/j.1399-3011.1990.tb00939.x. [DOI] [PubMed] [Google Scholar]
- 28.Anthis N.J., Clore G.M. Sequence-specific determination of protein and peptide concentrations by absorbance at 205 nm. Protein Sci. 2013;22:851–858. doi: 10.1002/pro.2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marshall A.C., Kroker A.J., Murray L.A., Gronthos K., Rajapaksha H., Wegener K.L., Bruning J.B. Structure of the sliding clamp from the fungal pathogen Aspergillus fumigatus (AfumPCNA) and interactions with human p21. FEBS J. 2017;284:985–1002. doi: 10.1111/febs.14035. [DOI] [PubMed] [Google Scholar]
- 30.Frkic R.L., Chua B.S., Shin Y., Pascal B.D., Novick S.J., Kamenecka T.M., Griffin P.R., Bruning J.B. Structural and dynamic elucidation of a non-acid PPARγ partial agonist: SR1988. Nucl. Receptors Res. 2018;5:101350. doi: 10.11131/2018/101350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pederick J.L., Thompson A.P., Bell S.G., Bruning J.B. d-Alanine-d-alanine ligase as a model for the activation of ATP-grasp enzymes by monovalent cations. J. Biol. Chem. 2020;295:7894–7904. doi: 10.1074/jbc.RA120.012936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cowieson N.P., Aragao D., Clift M., Ericsson D.J., Gee C., Harrop S.J., Mudie N., Panjikar S., Price J.R., Riboldi-Tunnicliffe A., Williamson R., Caradoc-Davies T. MX1: A bending-magnet crystallography beamline serving both chemical and macromolecular crystallography communities at the Australian Synchrotron. J. Synchrotron Radiat. 2015;22:187–190. doi: 10.1107/S1600577514021717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kabsch W. XDS (X-ray detector software) Acta Crystallogr. D Biol. Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Evans P. Scaling and assessment of data quality. Acta Crystallogr. D Biol. Crystallogr. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 35.Winn M.D., Ballard C.C., Cowtan K.D., Dodson E.J., Emsley P., Evans P.R., Keegan R.M., Krissinel E.B., Leslie A.G.W., McCoy A., McNicholas S.J., Murshudov G.N., Pannu N.S., Potterton E.A., Powell H.R. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Potterton E., Briggs P.J., Turkenburg M.G.W., Dodson E. A graphical user interface to the CCP4 program suite. Acta Crystallogr. D Biol. Crystallogr. 2003;59:1131–1137. doi: 10.1107/s0907444903008126. [DOI] [PubMed] [Google Scholar]
- 37.McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storonia L.C., Reada R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liebschner D., Afonine P.V., Baker M.L., Bunkóczi G., Chen V.B., Croll T.I., Hintze B., Hung L.W., Jain S., McCoy A.J., Moriarty N.W., Oeffner R.D., Poon B.K., Prisant M.G., Read R.J. Phenix: Macromolecular structure determination using X-rays, neutrons and electrons: Recent developments in phenix. Acta Crystallogr. D Biol. Crystallogr. 2019;75:861–877. doi: 10.1107/S2059798319011471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Afonine P.V., Grosse-Kunstleve R.W., Echols N., Headd J.J., Moriarty N.W., Mustyakimov M., Terwilliger T.C., Urzhumtsev A., Zwart P.H., Adams P.D. Towards automated crystallographic structure refinement with phenix.Refine. Acta Crystallogr. D Biol. Crystallogr. 2012;68:352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Emsley P., Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 41.Abagyan R., Totrov M., Kuznetsov D. ICM - new method for protein modeling and design: Applications to docking and structure prediction from the distorted native conformation. J. Comput. Chem. 1994;15:488–506. [Google Scholar]
- 42.Abagyan R., Totrov M. Biased probability Monte Carlo conformational searches and electrostatic calculations for peptides and proteins. J. Mol. Biol. 1994;235:983–1002. doi: 10.1006/jmbi.1994.1052. [DOI] [PubMed] [Google Scholar]
- 43.Schrodinger, LLC . 2015. The PyMOL Molecular Graphics System, Version 1.8. [Google Scholar]
- 44.Piovesan D., Minervini G., Tosatto S.C. The RING 2.0 web server for high quality residue interaction networks. Nucleic Acids Res. 2016;44:W367–W374. doi: 10.1093/nar/gkw315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stierand K., Maass P.C., Rarey M. Molecular complexes at a glance: Automated generation of two-dimensional complex diagrams. Bioinformatics. 2006;22:1710–1716. doi: 10.1093/bioinformatics/btl150. [DOI] [PubMed] [Google Scholar]
- 46.De March M., Barrera-Wilarmau S., Crespan E., Mentegari E., Merino N., Gonzalez-Magana A., Romano-Moreno M., Maga G., Crehuet R., Onesti S., Blanco F., J, De Biasio A. p15PAF binding to PCNA modulates the DNA sliding surface. Nucleic Acids Res. 2018;46:9816–9828. doi: 10.1093/nar/gky723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Duffy C.M., Hilbert B.J., Kelch B.A. A disease-causing variant in PCNA disrupts a promiscuous protein binding site. J. Mol. Biol. 2016;428:1023–1040. doi: 10.1016/j.jmb.2015.11.029. [DOI] [PubMed] [Google Scholar]
- 48.Wlodawer A., Minor W., Dauter Z., Jaskolski M. Protein crystallography for aspiring crystallographers or how to avoid pitfalls and traps in macromolecular structure determination. FEBS J. 2013;280:5705–5736. doi: 10.1111/febs.12495. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atomic coordinates and structure factors for the reported crystal structures have been deposited with the RCSB Protein Data bank under accession numbers 7KQ1 and 7KQ0.