Abstract
There is currently a strong interest to derive the biological precursor cis,cis-muconic acid from shikimate pathway-branches to develop a biological replacement for adipic acid. Pioneered by the Frost laboratory this concept has regained interest: Recent approaches (Boles, Alper, Yan) however suffer from low product titres. Here an in silico comparison of all strain construction strategies was conducted to highlight stoichiometric optimizations. Using elementary mode analysis new knock-out strategies were determined in Saccharomyces cerevisiae and Escherichia coli. The strain construction strategies are unique to each pathway-branch and organism, allowing significantly different maximum and minimum yields. The maximum theoretical product carbon yields on glucose ranged from 86% (dehydroshikimate-branch) to 69% (anthranilate-branch). In most cases a coupling of product formation to growth was possible. Especially in S. cerevisiae chorismate-routes a minimum yield constraint of 46.9% could be reached. The knock-out targets are non-obvious, and not-transferable, highlighting the importance of tailored strain construction strategies.
Abbreviations: ADH, acetaldehyde dehydrogenase; ANTH, anthranilate; ccMA, cis,cis-muconic acid; cMCs, constrained minimal cut sets; DAHP, 3-deoxy-arabinoheptulosonate 7-phosphate; DHBA, 2,3-dihydroxybenzoate; DHS, 3-dehydroshikimate; E4P, erythrose 4-phosphate; EDP, Entner–Doudoroff pathway; EFM, elementary flux modes; EMA, elementary mode analysis; EPSP, 5-enolpyruvylshikimate-3-phosphate; F6P, fructose-6-phosphate; FRD, fumarate reductase; G6PD, glucose-6-phosphate dehydrogenase; GPDH, glycerol-3-phosphate dehydrogenase; EDA, 2-dehydro-3-deoxy-phosphogluconate aldolase; LDH, lactate dehydrogenase; MAE, malic enzyme; PCA, protocatechuate; PCK, phosphoenolpyruvate carboxykinase; PEP, phosphoenolpyruvate; pHBA, para-hydroxybenzoate; PPP, pentose phosphate pathway; PPS, phosphoenolpyruvate synthase; PTS, phosphotransferase system; PYK, pyruvate kinase; SA, salicylate; WT, wild type; Ymax, maximum theoretical carbon yield; Ymin, minimum theoretical carbon yield
Keywords: Elementary mode analysis, Adipic, Muconic, Knock-out strategies, Carbon yield
Highlights
-
•
Maximum theoretical product carbon yields for biological production of cis,cis-muconic acid from glucose were calculated.
-
•
Existing strain construction strategies were compared in silico and weaknesses were identified.
-
•
New strain construction strategies were determined to allow coupling of product formation to growth for E. coli and S. cerevisiae.
-
•
In particular abolishing one target reaction introduces a minimum yield constraint of 28% in S. cerevisiae.
-
•
Determined knock-out targets are unique to each pathway and organism, highlighting the importance of tailored strain construction strategies.
1. Background
Adipic acid is a main precursor for the production of nylon-6,6 and polyurethanes. To date it is still derived from the precursor benzene in a process of oxidation of cyclohexane to cyclohexanol/cyclohexanone (EPA, 1994, Musser, 2005). While these precursors are all petro-chemistry based and thus non-sustainable, the further oxidation with nitric acid has an additional environmental impact as the greenhouse gas N2O is a large by-product in this process (Alini et al., 2007, Mainhardt and Kruger, 2000). Recently emerging patents (Boussie et al., 2010, Burgard et al., 2012, Picataggio and Beardslee, 2012, Raemakers-Franken et al., 2012), papers and reviews (Chen and Nielsen, 2013, Polen et al., 2012) emphasize the strong scientific and commercial interest to develop a biological replacement for fossil fuel based adipic acid.
A possible replacement precursor is the metabolic intermediate cis,cis-muconic acid (ccMA) which can efficiently be converted to adipic acid via hydrogenation with a yield of 97% (moladipate/molccMA) (Niu et al., 2002). Biotechnological production of ccMA recently experienced a renaissance with the original route established in the late 1990s (Draths and Frost, 1994, Frost and Draths, 1996, Niu et al., 2002) being implemented in yeast (Curran et al., 2013, Weber et al., 2012) and a new route being introduced in E. coli (Lin et al., 2014, Pugh et al., 2014, Sun et al., 2013, Sun et al., 2014). Microbial ccMA production through benzoate degradation in Pseudomonas putida (Choi et al., 1997) has also been explored. Though effective in terms of maximum titre 32 g/L (Bang and Choi, 1995) and yield 91% (Schmidt and Knackmuss, 1984), benzoate, however, can hardly be considered a sustainable feedstock.
1.1. Muconic acid production via the shikimate pathway
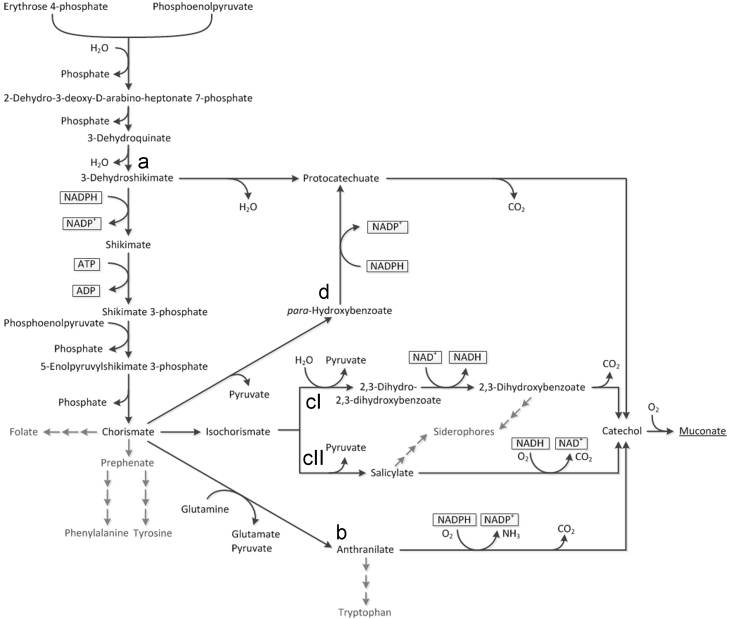
From the shikimate pathway five stoichiometrically unique routes lead to ccMA (Fig. 1). In this study the routes were named according to the key intermediates in the respective pathways: dehydroshikimate=DHS-pathway; anthranilate=ANTH-pathway; 2,3-dihydroxybenzoate=DHBA-pathway; salicylate=SA-pathway and para-hydroxybenzoate=pHBA-pathway. The latter four have in common to proceed via chorismate and are therefore collectively referred to as chorismate derived routes. All routes branch of from shikimate pathway at different intermediates (protocatechuate, anthranilate, isochorismate, para-hydroxybenzoate) and differ in their co-factors and co-metabolites, which makes a significant difference for stoichiometric carbon yields as well as in terms of energy demand and redox equivalents required for ccMA synthesis.
Fig. 1.
Shikimate pathway with routes to muconic acid. Shikimate pathway outgoing from the precursors E4P and PEP with routes to ccMA including co-factors in each step. Adjacent pathways not involved in ccMA synthesis are greyed out. Triple arrows indicate multiple steps. The DHS-pathway (a) proceeds via protocatechuate outgoing from 3-dehydroshikimate, while the ANTH-pathway (b) branches off further downstream from chorismate via anthranilate. The DHBA-pathway (cI) and the SA-pathway (cII) both proceed via isochorismate. A route linking the chorismate derived routes with the DHS-pathway is the pHBA-pathway (d). Although all routes lead to formation of catechol prior to ring opening release of ccMA, the involved co-factors and metabolites to catechol formation are essentially different.
The production of ccMA via shikimate pathway from sugar in E. coli was initially reported by Frost and Draths (1996). The optimized process showed high maximum titres of 36.8 g/L, product yields were comparatively high with 22% (molccMA/molglucose) (Niu et al., 2002). The pathway was established in two steps outgoing from dehydroshikimate (DHS), via protocatechuate and catechol (Fig. 1 branch a) by means of the aroZ gene from Klebsiella pneumoniae encoding dehydroshikimate dehydratase, the aroY gene from K. pneumoniae encoding protocatechuate (PCA) decarboxylase and the catA gene from Acinetobacter calcoaceticus encoding catechol 1,2-dioxygenase. The levels of the entry metabolites to shikimate pathway phosphoenolpyruvate (PEP) and erythrose 4-phosphate (E4P) were increased and a feedback inhibition resistant deoxy-arabino-heptulosonate-7-phosphate (DAHP) synthase was overexpressed. Further, blocking of the pathway below DHS through deletion of the DHS dehydrogenase gene aroE, directed carbon flux to ccMA.
Though no less elaborate, recent approaches employing the same pathway in the yeast S. cerevisiae were so far unable to achieve comparable titres and yields of ccMA (141 mg/L, ≙ 0.9% carbon yield (Curran et al., 2013) and 1.56 mg/L, ≙ 0.01% carbon yield (Weber et al., 2012)).
In Weber et al. (2012) partial deletion of ARO1 (corresponds to the E. coli aroE analogous domain which in yeast is part of a penta-functional enzyme, encoded by ARO1 that mediates stepwise the conversion of DAHP to EPSP via DHS (Weber et al., 2012)) blocked the conversion of 3-dehydroshikimate into shikimate. A DHS dehydratase from Bacillus thuringiensis (aroZ), a PCA decarboxylase composed of three different subunits encoded by the genes B, C and D taken from K. pneumoniae (aroY) and a catechol 1,2-dioxygenases from Acinetobacter radioresistens (catA) composed the pathway. The bottleneck here was the first step: Only very low PCA levels < 7 mg/L could be detected (Weber et al., 2012), which limit the overall ccMA titre.
In Curran et al. (2013) the DHS dehydratase was taken from Podospora anserina and the PCA decarboxylase from Enterobacter cloacae. A catechol 1,2-dioxygenase from Candida albicans completed the DHS-pathway. Knock-out of ARO3 and overexpression of a feedback-resistant ARO4 (DAHP synthase) increased overall flux to aromatics.
Furthermore flux balance analysis was utilized in Curran et al. (2013) to pursue further optimizations: When flux was directed through glucose-6-phosphate dehydrogenase (G6PD) the transketolase would form fructose-6-phosphate (F6P) and glyceraldehyde-3-phosphate from erythrose-4-phosphate and xylulose-5-phosphate. This resulted in a decreased theoretical carbon yield of 60.9% (Curran et al., 2013). Based on these findings pentose phosphate pathway was modified: Deletion of ZWF1, the gene coding for G6PD (R24 in supplementary File 2), would result in carbon flux to enter pentose phosphate (PPP) pathway via transketolase (R31 in supplementary File 2). The gene coding for this (TKL1) was over-expressed in order to favour this. Thus, by blocking the channelling of G6P into PPP and favouring the conversion of pentoses into E4P, flux to shikimate pathway would increase through higher E4P levels derived from F6P.
Also production of ccMA in E. coli via the anthranilate branch of the shikimate pathway (Fig. 1 branch b) could only achieve 390 mg/L ccMA on a glucose and glycerol co-feed (Sun et al., 2013) (equivalent to 4% total carbon product yield). Here the route via the tryptophan precursor anthranilate was established downstream of chorismate. This was done by cloning an anthranilate 1,2-dioxygenase from Pseudomonas aeruginosa (encoded by the antABC gene cluster, cloned as an operon) and a catechol 1,2-dioxygenase (catA1) from Pseudomonas putida in E. coli. Tryptophan biosynthesis was blocked and glutamine synthase was over-expressed in order to strengthen glutamine regeneration, required for anthranilate formation.
The same group recently established two more variations of this route, both partially utilize the siderophore-biosynthesis pathway, outgoing from isochorismate (Fig. 1 branch c): Via 2,3-dihydroxybenzoic acid (cI) a slightly increased titre of 480 mg/L ccMA on a glucose and glycerol co-feed (Sun et al., 2014) (equivalent to 4.95% total carbon product yield) was achieved. Overexpression of entCBA lead to formation of 2,3-dihydroxybenzoic acid, from here a 2,3-dihydroxybenzoate decarboxylase from Klebsiella pneumonia catalysed the formation of catechol. Via salicylic acid (cII) degradation, a greatly increased ccMA titre of 1500 mg/L on a combined glucose/glycerol C-source (Lin et al., 2014) (equivalent to 15.48% total carbon product yield) could be reached. The strain was constructed outgoing from NST 74, a phenylalanine over-producing strain: First aromatic amino acid biosynthesis was blocked, then carbon flux was redirected and production of ccMA established by means of three modules: Phosphoenolpyruvate synthase (ppsA), transketolase 1 (tktA), a feedback inhibition resistant 3-deoxy-d-arabino-heptulosonate-7-phosphate synthase (aroG) and shikimate kinase II (aroL) were overexpressed to improve overall flux to shikimate pathway. Further isochorismate synthase (entC) from E. coli and the isochorismate pyruvate lyase (pchB) from Pseudomonas fluorescens were overexpressed to establish salicylic acid production. Finally a salicylate 1-monoxygenase (nahG) from P. putida DOT-T1E and a catechol 1,2-dioxygenase from P. putida KT-2440 (catA1) were implemented to complete the pathway to ccMA.
Another route to ccMA leads from chorismate via pHBA to PCA, from here the steps coincide with the DHS-pathway (Fig. 1 branch d). Recently it has been shown in E. coli that PCA and catechol can be produced this way, with the highest yield being that of catechol at a titre of 451 mg/L (Pugh et al., 2014) (equivalent to 3.69% total carbon product yield). Engineering was also based on the phenylalanine over-producing strain NST 74. Overexpression of chorismate lyase, by means of ubiC, initiated the pathway, a pHBA hydroxylase (pobA) from Pseudomonas aeruginosa and a protocatechuate decarboxylase from Enterobacter cloacae completed it. To further increase flux to the target compounds the chorismate mutase/prephenate dehydratase (pheA) was disrupted.
While all these routes feature a sophisticated rearrangement of bioconversions on the way to ccMA, they mostly focus on shikimate pathway locally and are based on obvious knock-out targets and modifications to shift flux distribution within the pathway. The only direct approach to incorporate modifications that are not located within the shikimate pathway is reported by Curran et al. (2013) (knock-out of ZWF1 and overexpression of TKL1). Other attempts that can indirectly be seen as more general approaches on metabolic engineering are to strengthen glutamine regeneration in order to favour anthranilate formation (R65 in supplementary File 3) by Sun et al. (2013) and to overexpress the phosphoenolpyruvate synthase and transketolase 1 by Lin et al. (2014) to channel carbon flux to shikimate pathway. Yet the beneficial impact of these modifications cannot be easily foreseen without holistic insight. E.g. transamination is a very common process in many pathways and global effects are unknown.
Given the fact that shikimate pathway is deeply anchored in central metabolism a more rational approach considering the whole metabolism at once could open unknown opportunities to increase production with less effort by e.g. identifying one or two essential knock-out targets in order to significantly minimize the possible metabolic adjustments of the organism and force it into a state of product formation. It will also help to identify crucial elements of the metabolism which serves a better understanding of its peculiarities.
Many computational approaches exist to determine knock-out targets, e.g. the bi-level programming framework OptKnock (Burgard et al., 2003) or the more recent OptFlux (Rocha et al., 2010), which combines strain optimization tasks with pathway analysis. A recent example for model-guided network engineering on yeast shikimate pathway (Brochado and Patil, 2013) shows the potential of these approaches. The issue with these tools is that only one particular solution is computed, resulting in a narrow solution space that draws an incomplete picture of the cells resulting metabolism, thus these tools are considered biased (Lewis et al., 2012).
This can explain the issues of the optimization strategy in Curran et al. (2013): Elimination of a route leading to a reduced maximum yield (deletion of ZWF1) is necessary, but may not be sufficient in order to create a metabolism that operates at high yields. It is still unforeseeable in which other states the cell may be able to operate, potentially evading a high-yield metabolism.
This can be addressed by determining the complete solution space e.g. with elementary mode analysis (EMA) (Schuster and Hilgetag, 1994). In EMA an array of flux distributions can be compared, which makes it possible to assess the reduction of metabolic freedom (minimum/maximum product/biomass yields) in favour of product formation. This delivers a more universal description of the metabolisms degree of freedom after application of the respective deletions. Outgoing from this, a minimum efficiency network may be designed through minimizing the metabolic functionality. That this can be applied in vivo was shown in Trinh et al. (2008).
We used elementary mode analysis (EMA) by means of EFMTool (Terzer and Stelling, 2008), combined with a new method using a holistic approach to compute constrained minimal cut sets (cMCs) (Hadicke and Klamt, 2011). These cut sets represent all available genetic intervention strategies to introduce a constraint that allows a minimum efficiency greater than zero in respect to the desired product. That this concept can be easily transferred and applied to various strain optimization problems shows e.g. the study in Gruchattka et al. (2013). We decided to allow a maximum of five gene deletions to keep the deletion strategy realistic in respect to experimental effort.
2. Results
Not only the knock-out targets but also the availability of a feasible strategy depends as much on the pathway as on the organism′s individual metabolism. This resulted in significant differences in the maximum and minimum yields that could be achieved in the different scenarios.
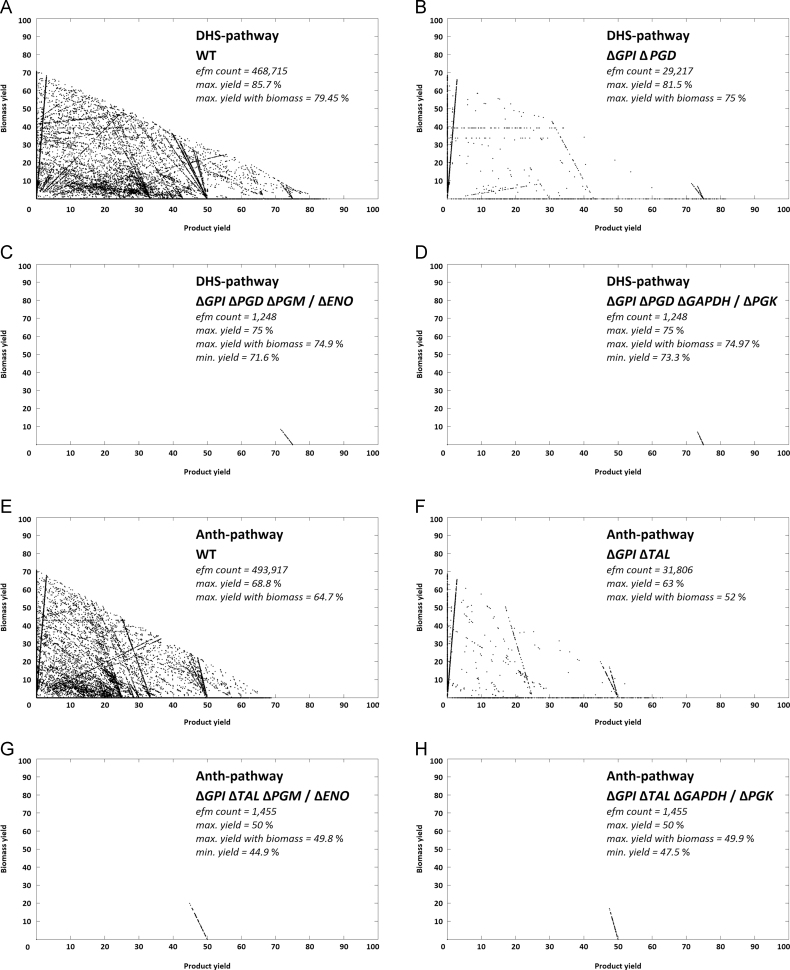
The maximum yields of the pathways were essentially different in both organisms. The highest theoretical carbon yield (Ymax) of the product in respect to the substrate glucose was obtained for the DHS-pathway with 85.7% (0.676 gccMA/gglucose). For chorismate derived pathways most yields were significantly lower and varied between 68.8% (0.5427 gccMA/gglucose) for the ANTH-pathway in E. coli and 80% (0.631 gccMA/gglucose) for the DHBA-pathway in S. cerevisiae. When comparing the two organisms, S. cerevisiae performed generally slightly better than E. coli. This is most significant when comparing the Ymax that also allows growth (formation of biomass), e.g. for the DHS-pathway Ymax with biomass is 5.35% lower in E. coli than in S. cerevisiae (cf. Fig. 3E vs. Fig. 2E). All yields can be found in the figures of the respective networks EFM distribution or in supplementary Table 1.
Fig. 3.
Yield vs. biomass plots of knock-out strategies for E. coli. Product vs. biomass yield plots of the EFM distribution of E. coli networks. For each pathway four scenarios are shown, comparing the wild type with the determined knock-out metabolism, key data as well as respective knock-outs are indicated on the charts. Each point in a chart corresponds to the specific product and biomass yield of the respective elementary flux mode. Yields are carbon yields in %.
Fig. 2.
Yield vs. biomass plots of knock-out strategies for S. cerevisiae. Product vs. biomass yield plots of the EFM distribution of S. cerevisiae DHS- and ANTH-pathway networks. For each pathway four scenarios are shown, comparing the wild type with the determined knock-out metabolism, key data as well as respective knock-outs are indicated on the charts. Each point in a chart corresponds to the specific product and biomass yield of the respective elementary flux mode. Yields are carbon yields in %. A dashed vertical line indicates currently achieved product yields in the respective approaches.
For comparability with the results obtained in Lin et al. (2014) and Sun et al., 2013, Sun et al., 2014) the Ymax′ on glucose and glycerol co-feed were calculated for E. coli. These can also be found in supplementary Table 2. These were slightly higher (e.g. 89.6% for the DHS-pathway and 73.3% for the ANTH pathway). Carbon sources different to glucose were not subject to further analysis.
The respective approaches failed to reach these maxima by a big margin. The dashed green vertical lines in Fig. 2A and B indicate the yields reached in Weber et al. (2012) and Curran et al. (2013) respectively. For the approaches in E. coli the yields are indicated in the same way in suppmentary Fig. 5A for Niu et al. (2002), Fig. 5D for Sun et al. (2013), suppementary Fig. 5F for Sun et al. (2014) and supplementary Fig. 5H for Lin et al. (2014). In order to optimize flux distribution of the metabolisms to favour a higher yield, knock-out targets were determined for each pathway in both organisms. The respective Ymax and Ymin of the knock out networks are indicated on the respective figures or can be found in supplementary Appendix A, Appendix A.
2.1. Two knock-out targets favour high yield metabolism in S. cerevisiae DHS-pathway
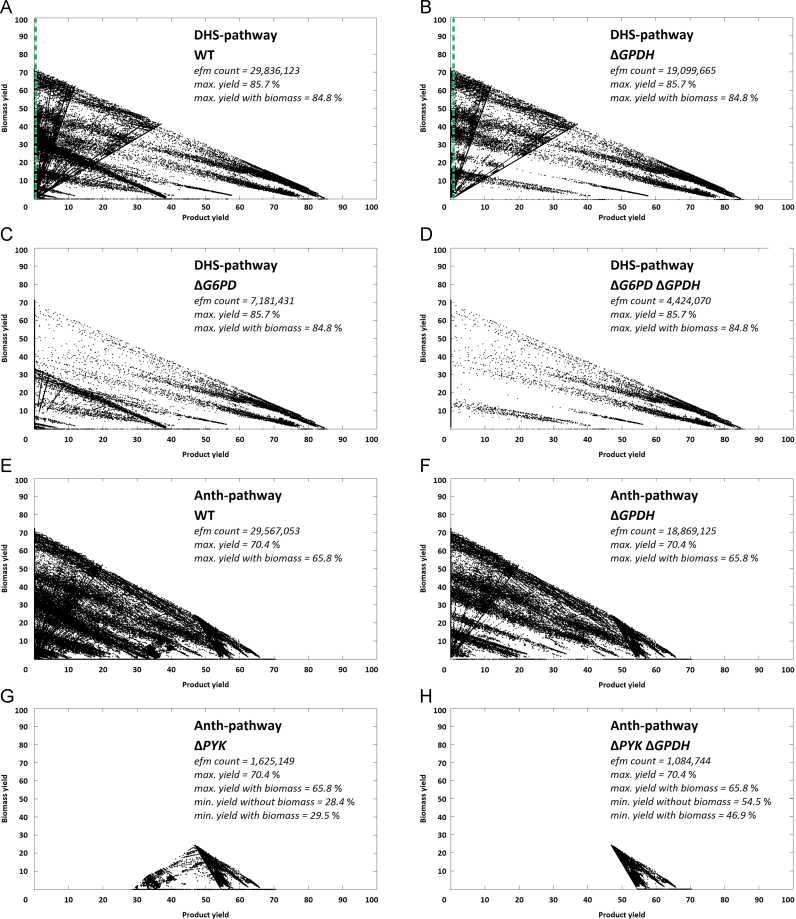
The effect of the deletion of ZWF1, previously applied in vivo (Curran et al., 2013), was investigated. As can be seen from Fig. 2B the deletion of ZWF1, encoding for G6PD (cf. suppementary File 1, R24) eliminates a large number of high biomass+low product yield modes, as desired. This effect can be expanded when combining a G6PD deficiency with a deletion of the glycerol-3-phosphate dehydrogenases (GPDH) (R43) as can be seen in Fig. 2D. Although this removed the bulk of modes with a product yield less than 40%, still a large number of modes with zero product yield remains. No cut sets with five or less knock-out targets could be determined that would result in a minimum efficiency different from zero in respect to the product.
2.2. Two essential knock-outs enforce yield maximization in S. cerevisiae chorismate derived pathways
For the routes via chorismate a coupling of product formation to central metabolism was possible, allowing the introduction of a minimum yield constraint to the metabolism. In particular deletion of the pyruvate kinases (PYK) (cf. suppementary File 1, R23) leaves no other source for cytosolic pyruvate formation than the release of pyruvate during product formation. The effect can be seen in Fig. 2G exemplary for the ANTH-pathway, where a Ymin of 28.4% was obtained (DHBA-, SA- and pHBA pathway are analogue, cf. suppementary Appendix A, Appendix A. When combined with a deletion of the GPDH (R43) (which eliminated glycerol formation) Ymin increased even further to 46.9% (Fig. 2H). Ymax was unaffected, remaining at 65.8% with/70.4% without formation of biomass (compare Fig. 2E with H).
2.3. Constraining the minimum yield diminishes the maximum yield in E. coli DHS-pathway
Four minimal cut sets could be found that result in two different flux distributions with a minimum efficiency greater than zero. The knock-out of glucose-6-isomerase (GPI) and phosphogluconate dehydrogenase (PGD) (cf. supplementary File 1, R18 and R33) was essential, while the final scenarios could be obtained with the knock-out of either phosphoglycerate mutase (PGM) (R25) or enolase (ENO) (R26) (Fig. 3C) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (R23) or phosphoglycerate kinase (PGK) (R24) (Fig. 3D). This results in Ymins of 71.6% and 73.3% respectively, while Ymax was reduced to just below 75%. No cut set with five or less knock-outs that did not diminish Ymax could be found.
2.4. An alternative knock-out strategy preserves the maximum yield in E. coli chorismate derived pathway
For E. coli ANTH-pathway four cut sets consisting of three knock-out targets analogous to the DHS-pathway could be found: The knock-out of the GPI and the transaldolase (TAL) (cf. supplementary File 1, R18 and R37) is essential, while the same redundant targets PGM or ENO (R25 and R26) and GAPDH or PGK (R23 and R24) as in the DHS-pathway deliver two different flux distributions (Fig. 3G and H). Ymin was 44.9% and 47.5%, respectively. However, enforcing a Ymin came along with a great reduction in number of modes and, above all, Ymax, which was reduced to just below 50%, almost 20% less than in the original metabolism.
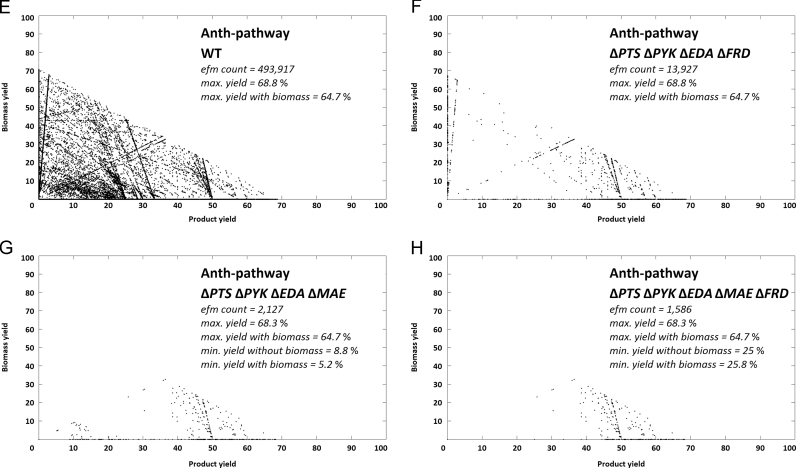
In search for knock-out targets that would preserve Ymax and result in a similar pattern as for S. cerevisiae a second strategy was determined, resulting in a cut set with five knock-out targets. In particular the pyruvate forming reactions of the PTS (cf. supplementary File 1, R16), PYK (R27), 2-dehydro-3-deoxy-phosphogluconate aldolase (EDA) (R32) and the malic enzymes (MAE) (R54 and R55) were identified as compulsory knock-out targets (Fig. 4G). In addition, deletion of the fumarate reductase (FRD) (R47) allowed Ymin to be raised from 5.2% to 25% (Fig. 4H). This knock out strategy was also applicable to the other chorimate derived pathways (cf. supplementary Appendix A, Appendix A).
Fig. 4.
Yield vs. biomass plots of alternative knock-out strategy for E. coli ANTH-pathway. Product vs. biomass yield plots of the EFM distribution of E. coli networks. Four scenarios are shown, comparing the wild type with the determined knock-out metabolism, key data as well as respective knock-outs are indicated on the charts. Each point in a chart corresponds to the specific product and biomass yield of the respective elementary flux mode. Yields are carbon yields in %.
3. Discussion
The advantage of the DHS-pathway over the chorismate derived pathways in terms of higher yield can be explained with the higher energy demand of the latter. In particular the DHS-pathway has no need for co-factors other than O2 and the shikimate-pathway substrates PEP and E4P (cf. Fig. 1). Chorismate derived pathways have direct demand for two NADPH and one ATP (cf. Fig. 1). In addition a second PEP is required, which is equivalent to the demand for two ATP (if regeneration from PYR is assumed, cf. supplementary File 3 R28 and R74). Indirectly the ANTH-pathway needs another ATP, as the transamination step (supplementary File 2 R104b, supplementary File 3 R84b) requires regeneration of glutamate to glutamine (supplementary File 2 R86, supplementary File 3 R65). This also explains the advantage of the pHBA-pathway and the isochorismate derived routes, as these they do not involve a transamination. In addition, in the DHBA-pathway, which features the highest yield of all chorismate derived pathways, an additional redox equivalent in form of NADH is gained.
In order to explain the low and extremely low product yields obtained by Curran et al. (2013) and Weber et al. (2012), distribution of modes has to be considered with the objective of a microorganism in mind: Naturally it will always try to maximize its growth (here the biomass). Looking at the product vs. biomass yield plots (Fig. 2, Fig. 3, Fig. 4) it becomes clear that maximization of growth involves drain of flux from product formation, which is thus minimized: The two extremes high product yield and high biomass yield are mutually exclusive.
The lowest carbon yield is obtained by Weber et al. (0.01%). Looking at the distribution of modes of the respective network (Fig. 2A) this is no surprise, as the applied strategy does not constrain the Ymin. (Curran et al., 2013) apply the ZWF1 knock-out, taking out the G6PD (Fig. 2B) in order to favour a higher carbon yield. This knock-out is also confirmed in this study as an essential requirement to optimize the pathway. When the elementary flux modes (EFMs) for this network are analysed (Fig. 2B) though, it becomes evident that this is not sufficient: Still every distribution of a product carbon yield between 0% and 85% is possible, while the majority features a low yield. From a stoichiometric viewpoint, this can explain why the in vivo product carbon yield of Curran et al. (0.9%), though almost 100-fold higher than in any other study in S. cerevisiae, still remains below the potential of the pathway. To overcome this problem it needs to be approached from a different angle, in order to actually make use of the microorganism׳s goal to maximize its growth, as explained in the following.
3.1. Metabolic peculiarities of S. cerevisiae facilitate knock-out strategies
During the elementary mode analysis (EMA) it became obvious, that out of the two precursors for the shikimate pathway, PEP and E4P, the former is crucial. This has already been realized by Niu et al. (2002) where a difference in theoretical maximum yields of 43% was determined, depending on the availability of PEP. For production of one mole ccMA, one mole of PEP is consumed in the DHS- and two moles in the chorismate derived branches of the shikimate pathway, while only one E4P is needed. In the pathways via chorismate per mole ccMA one mole PYR is released again. If PYR production in central metabolism is taken out, flux to product needs to be proportionally high in order to obtain enough PYR to feed central metabolism. Why is this knock-out strategy now so applicable to and effective in S. cerevisiae? The explanation lies in the nature of the eukaryote׳s metabolism. It is possible because in yeast central metabolism no pyruvate producing reactions other than PYK and MAE exist. Apart from the products of shikimate pathway only the biosynthesis of glycine from glyoxylate, the homocysteine production through transsulfuration and the degradation of alanine and serine also produce pyruvate (Cherry et al., 2012). The anabolic pathways (glycine and homocysteine) are linked to biomass formation themselves and therefore cannot provide additional PYR in order to increase biomass formation. Alanine itself is made from PYR and therefore cannot be regarded an alternative source of PYR while the serine degradation pathway is used for the degradation of extracellular serine as the sole nitrogen source. In addition, it is unique to the shikimate pathway that PEP is a substrate, while all other pyruvate producing reactions start from different metabolites. Also the S. cerevisiae compartmentalization is an advantage, as the malic enzyme is localized in the mitochondria (Boles et al., 1998) it does not have a direct influence on the cytosolic shikimate pathway. Thus coupling of product formation to central metabolism is in silico possible with deletion of only one target (PYK) when using pathways via chorismate for production. Not only the increase in availability of PEP as an educt (which is the main focus in common strain construction strategies), but also the formation of PYR as by-product is essential in order to realize this knock-out strategy. By taking out the conversion of PEP to PYR in central metabolism the shikimate pathway replaces this function, thus enforcing product formation.
3.2. Feasibility of theoretical knock-out targets for practical application
In the strain construction strategy pictured in this study for pathways via chorismate in S. cerevisiae, cytosolic PYR can only be derived in combination with product formation. This is based on current knowledge, assuming that PYR cannot readily cross the mitochondrial membrane back into the cytosol. While the structure responsible for the uptake of pyruvate into the mitochondria has recently been revealed (Bricker et al., 2012, Herzig et al., 2012), the reverse, that is export of PYR from mitochondria into the cytosol, has not been reported to date. Although studies of a PYK knock-out mutant indicate that PYR formed by MAE in the mitochondria can have an influence on cytosolic central metabolism to some extent (Boles et al., 1998), growth of the mutant is only possible on lactate or ethanol and glycerol as carbon source (Boles et al., 1997). Thus, even if transport of mitochondrially formed PYR into the cytosol would be possible, in case of glucose feed, carbon would have to pass through glycolysis and shikimate pathway with product coupling first, before malate would be available to MAE. Only in case of ethanol or lactate feed flux could enter the TCA-cycle downstream of PYR, thus allowing circumvention of product formation.
A possible concern when arguing that product formation is coupled to central metabolism is that formation of PYR does not occur during the final step of ccMA formation. E.g. in case of ANTH-pathway it could be sufficient for the organism to proceed to anthranilate in order to obtain PYR, thus rather accumulating anthranilate than ccMA. From a thermodynamic point of view this is unlikely as the ΔrG′°s for the two subsequent reactions from anthranilate to ccMA are extremely negative (Flamholz et al., 2012), meaning the reaction equilibrium is largely on product side. A high anthranilate concentration would even boost this, favouring the pathway to proceed to the end product ccMA. The same applies for the SA-pathway, while for the DHBA- and the pHBA-pathway, which both proceed via different isomers of dihydroxybenzoic acid, the ΔrG′° of the reaction to catechol is close to 0. This may give preference to the ANTH- and the SA-pathway, although their yields are lower.
Further concerns are related to in vivo application difficulties of the identified knock-out targets in S. cerevisiae. Deletion of the paralogs coding for the two PYK isozymes (see supplementary Table 5 for more details) results in a severe growth defect (Boles et al., 1997). The same applies for the two GPDH isozymes (Hubmann et al., 2011). For GPDH a knock-down strategy exists that retains a viable metabolism while almost completely inhibiting glycerol formation (Hubmann et al., 2011). When combining this with an overexpression of the alternate PYR synthesis route (that is the product pathway) this should enable growth again. In terms of PYK also regulatory factors play a role, as not both isozymes are always expressed to the same level at the same time (Boles et al., 1997), so that deletion of CDC19 only could be sufficient.
3.3. Metabolic peculiarities of E. coli hinder knock-out strategies
The knock-out strategy for E. coli DHS-pathway and the first one for ANTH-pathway similarly introduce a minimum yield constraint: Biomass formation is limited by forcing all flux through the EDP. For this the deletion of GPI is essential in both cases and blocks the entrance (R18) of glycolysis. In DHS-pathway the oxidative part of PPP is blocked through knock-out of PGD (R33) (Fig. 3B), for the ANTH-pathway deletion of TAL (R37) has a similar effect (Fig. 3F). The further four alternative knock-outs PGM, ENO, GAPDH and PGK (R23–26) constrain flux to the biomass precursor 3-PG from either side, resulting in a very narrow distribution of modes. This is not very elegant and results in a massive reduction of Ymax, but also leaves the cell minimal metabolic freedom to adjust to its’ environmental settings. Therefore the value of this strategy for in vivo application is questionable. This extremely constraining knock-out strategy also explains the reduction of Ymax and underlines that not always the shortest intervention strategy is necessarily also the best. That a longer cut set can sometimes also be of advantage is outlined in the following.
An alternate knock-out strategy can preserve Ymax in the ANTH-pathway (and the other chorismate derived pathways) and results in similar coupling of product formation to growth as in S. cerevisiae. However the PEP-pyruvate node in E. coli is significantly different from S. cerevisiae and three additional PYR producing reactions (PTS, EDA, MAE) besides PYK hamper the application of the same knock-out strategy. A knock-out of the phosphotransferase system is known in literature (Escalante et al., 2012) allowing a change of glucose uptake mechanism especially if combined with overexpression of hexose permease (Hernandez-Montalvo et al., 2003). Also combinations of PTS and PYK knock-out (Lee et al., 2005) have been applied successfully, especially in respect to production of aromatic amino acids (Gosset et al., 1996, Meza et al., 2012, Papagianni, 2012). Of the remaining targets (EDA and MAE) each of the single knock-outs is possible (Baba et al., 2006). However more combinations of these knock-out targets have to our knowledge not been described so far. This could be due to the extensive mutations in central metabolism having unpredictable consequences, even though modelling predicts that the resulting metabolism should be viable (see supplementary Table 5 for more details on genes coding for the respective enzymes).
Further, formation of succinate as by-product has been identified as one of the major features in modes with low product yields (Fig. 4G). Succinate, a product of mixed acid fermentation in E. coli, is in these modes formed by R47 (cf. supplementary File 1). It can drain carbon flux from shikimate pathway, as it is obtained from PEP in four steps via oxaloacetate, malate and fumarate (Thakker et al., 2012). The enzyme identified as knock-out target (FRD) is only active under anaerobic conditions while the TCA cycle is inactive (aerobically succinate is only produced if the glyoxylate cycle is active) (Cecchini et al., 2002, Thakker et al., 2012). A knock-out may not be necessary as aerobic fermentation is obligatory for ccMA formation (in the last step oxygen is incorporated in the ring structure of catechol between the two hydroxyl groups, thus releasing ccMA).
One possible approach to facilitate critical knock-outs is the knock-down of the respective target genes. sRNAs in E. coli can be utilized for posttranscriptional repression of a gene (Sharma et al., 2013). Although more difficult in prokaryotes due to the lack of a nucleus, which shortens the timeframe for interactions of the sRNAs with the mRNA between transcription and translation significantly, it is still a promising approach. It results in attenuation rather than full silencing, which is here important to retain a basal activity of critical target genes. This could be combined with an inducible system (a wealth of approaches exists for induced expressions in E. coli (Baneyx, 1999)) in order to further facilitate the cultivation of the strain.
So far it has only been discussed which reactions need to be taken out additionally in respect to yeast in order to also couple product formation to central metabolism. But why is a yield reduction observed in the knock-out strategy for the DHS-pathway and the first ANTH-pathway knock-out strategy in E. coli (Fig. 3)? This is due to limited possibilities to recycle PYR, which exits shikimate pathway, back into central metabolism. Of the replenishing reactions that fill up the PEP-pyruvate-oxaloacetate node in E. coli (Papagianni, 2012, Sauer and Eikmanns, 2005) only the phosphoenolpyruvate carboxykinase (PCK) and the PPS remain. Thus the formation of PEP can either happen from PYR at the expense of two ATP equivalents (PPS) or from oxaloacetate (PCK). In contrast to S. cerevisiae, E. coli does not possess a pyruvate carboxylase which incorporates one CO2 into PYR to form oxaloacetate.
Only two options remain to convert PYR to OAA: One way is to further metabolize the PYR via acetyl-CoA. This leads to decarboxylation. The acetyl-CoA can then only be assimilated via the glyoxylic shunt and one-third of the carbon is lost as CO2. The other possibility for PYR conversion to OAA is the reverse operation of MAE in concert with the malate dehydrogenase and PCK, which would lead to a carbon neutral but ATP dependent conversion of PYR back to PEP. Because of the lower energy demand and the absence of CO2 formation, the final option has a higher carbon yield. The in vivo observation that the MAE is up-regulated in a fully PYK deficient E. coli mutant and the PCK is also highly active (Emmerling et al., 2002) supports this.
4. Conclusions
When using pathways via chorismate it was possible to apply a minimum yield constraint with as little as one knock-out in S. cerevisiae without any loss in Ymax. Although the Ymax of the DHS-pathway is higher, only the chorismate derived pathways allow for this strain construction strategy. In E. coli five knock-outs are necessary to achieve a similar picture without drop in Ymax. Therefore yeast seems to be the favourable organism for production of ccMA in terms of knock-out strategies.
This study not only highlights the importance of pathway choice, but also clearly points out how differences in central metabolism of the host organism can affect a synthetic pathway.
5. Methods
5.1. Elementary flux mode analysis
EFMs were calculated using EFMTool 4.7.1 (Terzer and Stelling, 2008) freely available from http://www.csb.ethz.ch/tools/efmtool in MATLAB R2012b (The MathWorks, Natick, USA) on the HPC cluster (Barrine) of the University of Queensland using a node with four eight core X7550 CPUs @ 2 GHz and 1024 GB of 1066 MHz RAM running a PBSPro batch system. Each mode represents a feasible, steady-state, flux distribution of a metabolic network. The maximum carbon yields of the different elementary modes were calculated in MATLAB by drawing carbon balances around the transport reactions into and out of the network. The yield for each product in each mode is defined by:
Fluxproduct and Fluxsubstrate represent the flux rates for products and substrates leaving and entering the balance area, Carbonproduct and Carbonsubstrate are the numbers of carbon atoms in the product and substrate. Yields were converted to per cent; for a graphical depiction of the distribution of modes the biomass yields were plotted against the yields of the desired product for a given network.
5.2. Construction of metabolic networks
Stoichiometric networks of Escherichia coli and Saccharomyces cerevisiae were compiled based on literature (Jol et al., 2012, Krömer et al., 2013, Krömer et al., 2006) and metabolic pathway databases (Caspi et al., 2014, Cherry et al., 2012, Kanehisa and Goto, 2000, Keseler et al., 2013, Kanehisa et al., 2014) and can be found in the supplementary material (supplementary File 1). The two different branches to ccMA via shikimate pathway were compiled from literature (Draths and Frost, 1994, Sun et al., 2013) and named according to its branch off metabolite (DHS or ANTH). These networks contain central carbon metabolism, which is only comprised of glycolysis, EDP (E. coli only), PPP, TCA cycle, glyoxylate cycle, anaplerosis, glutamate/glutamine interconversion, electron transport chain, and biomass formation which was broken down to central metabolism derived precursors (supplementary File 1, R95 in S. cerevisiae network and R76 in E. coli network). Amino acid biosynthesis was not included in the model; this concerns also routes to aromatic amino acids. In case of shikimate pathway this already corresponds to obvious knock-out targets where production of aromatic amino acids and folate precursors is inhibited to direct flux to the sole product ccMA.
5.3. Determination of knock-out targets
For determination of knock-out targets the method of constrained minimal cut sets was used, as described in Hadicke and Klamt (2011). The desired modes where defined depending on the Ymax of each network: S. cerevisiae DHS-pathway modes with a yield > 60%, S. cerevisiae ANTH-pathway modes with a yield > 50%, E. coli DHS-pathway modes with a yield > 70%, E. coli ANTH-pathway modes with a yield > 45%. Of these “desired modes” at least 10% had to remain after application of the determined cut sets. All remaining “target modes” where to be abolished. Cut sets were calculated allowing five or less knock-outs with respect to feasibility of the targets (Ruckerbauer et al., 2014): E.g. reactions that are catalysed by the same enzyme cannot be separate knock-out targets (for instance transketolase), neither can spontaneous reactions be targets (ATP hydrolysis). Biomass formation was also defined to be not a knock-out target, as all sets that abolish growth are infeasible. The reactions that are excluded from the cut set are highlighted in supplementary File 1.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
J.O.K. and N.A.J.H. jointly conceived the study and edited the manuscript. N.A.J.H. designed the metabolic networks, conducted the analysis, and drafted the manuscript. Both authors read and approved the final manuscript.
Acknowledgements
This study was funded by the Australian Research Council (DE120101549).
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.meteno.2014.09.001.
Contributor Information
Nils J.H. Averesch, Email: n.averesch@uq.edu.au.
Jens O. Krömer, Email: j.kromer@uq.edu.au.
Appendix A. Supplementary materials
Excel file containing the reactions of CM of each organism supplemented by the respective pathway branch. Knock-out targets are highlighted in red. The file contains two excel sheets (S. cerevisiae & E. coli), each containing different networks for the respective pathways. The determined knock-out targets are indicated in red for each cut set, potential overexpression targets in green. Reactions that were excluded from identification as knock-out target are highlighted yellow.
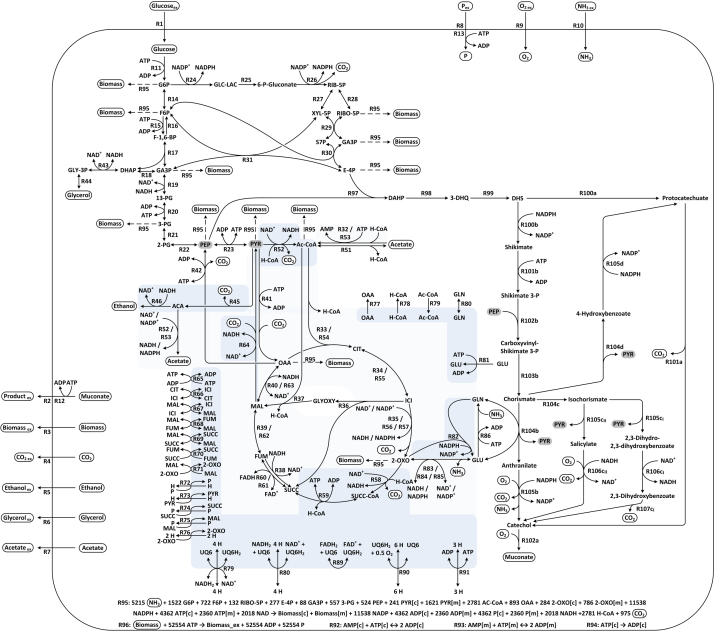
Supplementary file 2.
Map of S. cerevisiae metabolic network. Graphical depiction of the metabolites interactions in the S. cerevisiae network. The reactions are identified by their number according to the network file (supplementary File 1). For better clarity “phosphate” (P) was left out from reactions in the figure unless involved in transport. Contribution to biomass formation (R95) is indicated with dashed arrows for the respective compounds. The two alternative routes to product formation are labelled with “a”–“d” according to Fig. 1. Mitochondrial processes are highlighted in blue (mitochondrial only) and light blue (reactions exist in mitochondria and cytosol).
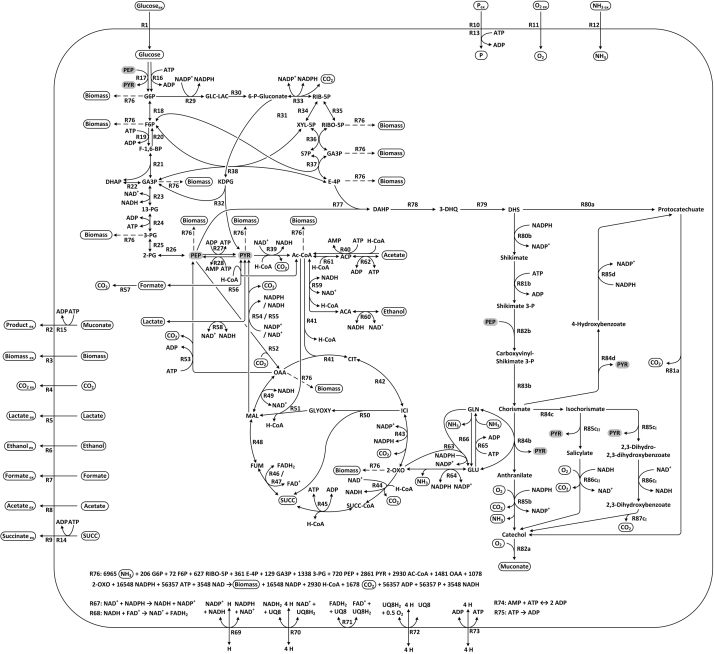
Supplementary file 3.
Map of E. coli metabolic network. Graphical depiction of the metabolites interactions in the E. coli network. The reactions are identified by their number according to the network file (supplementary File 1). For better clarity “phosphate” (P) was left out from reactions in the figure unless involved in transport. Contribution to biomass formation (R78) is indicated with dashed arrows for the respective compounds. The two alternative routes to product formation are labelled with “a” to “d” according to fig. 1. Key metabolites for the knock-out strategy are highlighted in grey.
Supplementary Material
References
- Alini S., Basile F., Blasioli S., Rinaldi C., Vaccari A. Development of new catalysts for N2O-decomposition from adipic acid plant. Appl. Catal. B Environ. 2007;70:323–329. [Google Scholar]
- Baba T., Ara T., Hasegawa M., Takai Y., Okumura Y., Baba M., Datsenko K.A., Tomita M., Wanner B.L., Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2006;2 doi: 10.1038/msb4100050. (Article ID: 2006.0008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baneyx F. Recombinant protein expression in Escherichia coli. Curr. Opin. Biotechnol. 1999;10:411–421. doi: 10.1016/s0958-1669(99)00003-8. [DOI] [PubMed] [Google Scholar]
- Bang S.-G., Choi C.Y. DO-stat fed-batch production of cis,cis-muconic acid from benzoic acid by Pseudomonas putida BM014. J. Ferment. Bioeng. 1995;79:381–383. [Google Scholar]
- Boles E., de Jong-Gubbels P., Pronk J.T. Identification and characterization of MAE1, the Saccharomyces cerevisiae structural gene encoding mitochondrial malic enzyme. J. Bacteriol. 1998;180:2875–2882. doi: 10.1128/jb.180.11.2875-2882.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boles E., Schulte F., Miosga T., Freidel K., Schluter E., Zimmermann F.K., Hollenberg C.P., Heinisch J.J. Characterization of a glucose-repressed pyruvate kinase (Pyk2p) in Saccharomyces cerevisiae that is catalytically insensitive to fructose-1,6-bisphosphate. J. Bacteriol. 1997;179:2987–2993. doi: 10.1128/jb.179.9.2987-2993.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boussie T.R., Dias E.L., Fresco Z.M., Murphy V.J. Production of adipic acid and derivates from carbohydrate-containing materials. 2010 (Patent no: US20100317822A1) [Google Scholar]
- Bricker D.K., Taylor E.B., Schell J.C., Orsak T., Boutron A., Chen Y.C., Cox J.E., Cardon C.M., Van Vranken J.G., Dephoure N., Redin C., Boudina S., Gygi S.P., Brivet M., Thummel C.S., Rutter J. A mitochondrial pyruvate carrier required for pyruvate uptake in yeast, Drosophila, and humans. Science. 2012;337:96–100. doi: 10.1126/science.1218099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brochado A.R., Patil K.R. Overexpression of O-methyltransferase leads to improved vanillin production in baker׳s yeast only when complemented with model-guided network engineering. Biotechnol. Bioeng. 2013;110:656–659. doi: 10.1002/bit.24731. [DOI] [PubMed] [Google Scholar]
- Burgard A.P., Pharkya P., Maranas C.D. Optknock: a bilevel programming framework for identifying gene knockout strategies for microbial strain optimization. Biotechnol. Bioeng. 2003;84:647–657. doi: 10.1002/bit.10803. [DOI] [PubMed] [Google Scholar]
- Burgard A.P., Pharkya P., Osterhout R.E. Microorganisms for the production of adipic acid and other compounds. 2012 (Patent no: US8088607B2) [Google Scholar]
- Caspi R., Altman T., Billington R., Dreher K., Foerster H., Fulcher C.A., Holland T.A., Keseler I.M., Kothari A., Kubo A., Krummenacker M., Latendresse M., Mueller L.A., Ong Q., Paley S., Subhraveti P., Weaver D.S., Weerasinghe D., Zhang P., Karp P.D. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 2014;42:D459–D471. doi: 10.1093/nar/gkt1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecchini G., Schroder I., Gunsalus R.P., Maklashina E. Succinate dehydrogenase and fumarate reductase from Escherichia coli. Biochim. Biophys. Acta. 2002;1553:140–157. doi: 10.1016/s0005-2728(01)00238-9. [DOI] [PubMed] [Google Scholar]
- Chen Y., Nielsen J. Advances in metabolic pathway and strain engineering paving the way for sustainable production of chemical building blocks. Curr. Opin. Biotechnol. 2013 doi: 10.1016/j.copbio.2013.03.008. [DOI] [PubMed] [Google Scholar]
- Cherry J.M., Hong E.L., Amundsen C., Balakrishnan R., Binkley G., Chan E.T., Christie K.R., Costanzo M.C., Dwight S.S., Engel S.R., Fisk D.G., Hirschman J.E., Hitz B.C., Karra K., Krieger C.J., Miyasato S.R., Nash R.S., Park J., Skrzypek M.S., Simison M., Weng S., Wong E.D. Saccharomyces Genome Database: the genomics resource of budding yeast. Nucleic Acids Res. 2012;40:D700–D705. doi: 10.1093/nar/gkr1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi W.J., Lee E.Y., Cho M.H., Choi C.Y. Enhanced production of cis,cis-muconate in a cell-recycle bioreactor. J. Ferment. Bioeng. 1997;84:70–76. [Google Scholar]
- Curran K.A., Leavitt J.M., Karim A.S., Alper H.S. Metabolic engineering of muconic acid production in Saccharomyces cerevisiae. Metab. Eng. 2013;15:55–66. doi: 10.1016/j.ymben.2012.10.003. [DOI] [PubMed] [Google Scholar]
- Draths K.M., Frost J.W. Environmentally compatible synthesis of adipic acid from d-glucose. J. Am. Chem. Soc. 1994;116:399–400. [Google Scholar]
- Emmerling M., Dauner M., Ponti A., Fiaux J., Hochuli M., Szyperski T., Wuthrich K., Bailey J.E., Sauer U. Metabolic flux responses to pyruvate kinase knockout in Escherichia coli. J. Bacteriol. 2002;184:152–164. doi: 10.1128/JB.184.1.152-164.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EPA, Background Report AP-42, Section 6.2, Adipic Acid Production. Compilation of Air Pollutant Emission Factors. U.S. Environmental Protection Agency, OAQPS/TSD/EIB, Research Triangle Park, 1994, p. 6.
- Escalante A., Salinas Cervantes A., Gosset G., Bolivar F. Current knowledge of the Escherichia coli phosphoenolpyruvate-carbohydrate phosphotransferase system: peculiarities of regulation and impact on growth and product formation. Appl. Microbiol. Biotechnol. 2012;94:1483–1494. doi: 10.1007/s00253-012-4101-5. [DOI] [PubMed] [Google Scholar]
- Flamholz A., Noor E., Bar-Even A., Milo R. eQuilibrator—the biochemical thermodynamics calculator. Nucleic Acids Res. 2012;40:D770–D775. doi: 10.1093/nar/gkr874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost J.W., Draths K.M. Synthesis of adipic acid from biomass-derived carbon sources. 1996 (Patent no: US005487987A) [Google Scholar]
- Gosset G., Yong-Xiao J., Berry A. A direct comparison of approaches for increasing carbon flow to aromatic biosynthesis in Escherichia coli. J. Ind. Microbiol. 1996;17:47–52. doi: 10.1007/BF01570148. [DOI] [PubMed] [Google Scholar]
- Gruchattka E., Hadicke O., Klamt S., Schutz V., Kayser O. In silico profiling of Escherichia coli and Saccharomyces cerevisiae as terpenoid factories. Microb. Cell Factories. 2013;12:84. doi: 10.1186/1475-2859-12-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadicke O., Klamt S. Computing complex metabolic intervention strategies using constrained minimal cut sets. Metab. Eng. 2011;13:204–213. doi: 10.1016/j.ymben.2010.12.004. [DOI] [PubMed] [Google Scholar]
- Hernandez-Montalvo V., Martinez A., Hernandez-Chavez G., Bolivar F., Valle F., Gosset G. Expression of galP and glk in a Escherichia coli PTS mutant restores glucose transport and increases glycolytic flux to fermentation products. Biotechnol. Bioeng. 2003;83:687–694. doi: 10.1002/bit.10702. [DOI] [PubMed] [Google Scholar]
- Herzig S., Raemy E., Montessuit S., Veuthey J.L., Zamboni N., Westermann B., Kunji E.R., Martinou J.C. Identification and functional expression of the mitochondrial pyruvate carrier. Science. 2012;337:93–96. doi: 10.1126/science.1218530. [DOI] [PubMed] [Google Scholar]
- Hubmann G., Guillouet S., Nevoigt E. Gpd1 and Gpd2 fine-tuning for sustainable reduction of glycerol formation in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2011;77:5857–5867. doi: 10.1128/AEM.05338-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jol S.J., Kummel A., Terzer M., Stelling J., Heinemann M. System-level insights into yeast metabolism by thermodynamic analysis of elementary flux modes. PLoS Comput. Biol. 2012;8:e1002415. doi: 10.1371/journal.pcbi.1002415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M., Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M., Goto S., Sato Y., Kawashima M., Furumichi M., Tanabe M. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res. 2014;42:D199–D205. doi: 10.1093/nar/gkt1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keseler I.M., Mackie A., Peralta-Gil M., Santos-Zavaleta A., Gama-Castro S., Bonavides-Martinez C., Fulcher C., Huerta A.M., Kothari A., Krummenacker M., Latendresse M., Muniz-Rascado L., Ong Q., Paley S., Schroder I., Shearer A.G., Subhraveti P., Travers M., Weerasinghe D., Weiss V., Collado-Vides J., Gunsalus R.P., Paulsen I., Karp P.D. EcoCyc: fusing model organism databases with systems biology. Nucleic Acids Res. 2013;41:D605–D612. doi: 10.1093/nar/gks1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krömer J.O., Nunez-Bernal D., Averesch N.J., Hampe J., Varela J., Varela C. Production of aromatics in Saccharomyces cerevisiae – a feasibility study. J. Biotechnol. 2013;163:184–193. doi: 10.1016/j.jbiotec.2012.04.014. [DOI] [PubMed] [Google Scholar]
- Krömer J.O., Wittmann C., Schroder H., Heinzle E. Metabolic pathway analysis for rational design of l-methionine production by Escherichia coli and Corynebacterium glutamicum. Metab. Eng. 2006;8:353–369. doi: 10.1016/j.ymben.2006.02.001. [DOI] [PubMed] [Google Scholar]
- Lee S.J., Lee D.Y., Kim T.Y., Kim B.H., Lee J., Lee S.Y. Metabolic engineering of Escherichia coli for enhanced production of succinic acid, based on genome comparison and in silico gene knockout simulation. Appl. Environ. Microbiol. 2005;71:7880–7887. doi: 10.1128/AEM.71.12.7880-7887.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis N.E., Nagarajan H., Palsson B.O. Constraining the metabolic genotype–phenotype relationship using a phylogeny of in silico methods. Nat. Rev. Microbiol. 2012;10:291–305. doi: 10.1038/nrmicro2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y., Sun X., Yuan Q., Yan Y. Extending shikimate pathway for the production of muconic acid and its precursor salicylic acid in Escherichia coli. Metab. Eng. 2014;23:62–69. doi: 10.1016/j.ymben.2014.02.009. [DOI] [PubMed] [Google Scholar]
- Mainhardt H., Kruger D. N2O emissions from adipic acid and nitric acid production. 2000 IPCC report. Available at: <http://www.ipcc-nggip.iges.or.jp/public/gp/bgp/3_2_Adipic_Acid_Nitric_Acid_Production.pdf >. [Google Scholar]
- Meza E., Becker J., Bolivar F., Gosset G., Wittmann C. Consequences of phosphoenolpyruvate:sugar phosphotranferase system and pyruvate kinase isozymes inactivation in central carbon metabolism flux distribution in Escherichia coli. Microb. Cell Factories. 2012;11:127. doi: 10.1186/1475-2859-11-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musser M.T. Adipic Acid. In: Wiley-VCH, editor. ULLMANN׳S Encyclopedia of Industrial Chemistry. Wiley-VCH; Weinheim: 2005. [Google Scholar]
- Niu W., Draths K.M., Frost J.W. Benzene-free synthesis of adipic acid. Biotechnol. Prog. 2002;18:201–211. doi: 10.1021/bp010179x. [DOI] [PubMed] [Google Scholar]
- Papagianni M. Recent advances in engineering the central carbon metabolism of industrially important bacteria. Microb. Cell Factories. 2012;11:50. doi: 10.1186/1475-2859-11-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picataggio S., Beardslee T. Biological methods for preparing adipic acid. 2012 (Patent no: US20120021474A1) [Google Scholar]
- Polen T., Spelberg M., Bott M. Toward biotechnological production of adipic acid and precursors from biorenewables. J. Biotechnol. 2013;167(2):75–84. doi: 10.1016/j.jbiotec.2012.07.008. [DOI] [PubMed] [Google Scholar]
- Pugh S., McKenna R., Osman M., Thompson B., Nielsen D.R. Rational engineering of a novel pathway for producing the aromatic compounds p-hydroxybenzoate, protocatechuate, and catechol in Escherichia coli. Process Biochem. 2014 [Google Scholar]
- Raemakers-Franken P.C., Schürmann M., Trefzer A.C., De Wildeman S.M.A. Preparation of adipic acid. 2012 (Patent no: US20120028320A1) [Google Scholar]
- Rocha I., Maia P., Evangelista P., Vilaca P., Soares S., Pinto J.P., Nielsen J., Patil K.R., Ferreira E.C., Rocha M. OptFlux: an open-source software platform for in silico metabolic engineering. BMC Syst. Biol. 2010;4:45. doi: 10.1186/1752-0509-4-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruckerbauer D.E., Jungreuthmayer C., Zanghellini J. Design of optimally constructed metabolic networks of minimal functionality. PLoS One. 2014;9:e92583. doi: 10.1371/journal.pone.0092583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer U., Eikmanns B.J. The PEP–pyruvate–oxaloacetate node as the switch point for carbon flux distribution in bacteria. FEMS Microbiol. Rev. 2005;29:765–794. doi: 10.1016/j.femsre.2004.11.002. [DOI] [PubMed] [Google Scholar]
- Schmidt E., Knackmuss H.-J. Production of cis,cis-muconate from benzoate and 2-fluoro-cis,cis-muconate from 3-fluorobenzoate by 3-chlorobenzoate degrading bacteria. Appl. Microbiol. Biotechnol. 1984;20:351–355. [Google Scholar]
- Schuster S., Hilgetag C. On elementary flux modes in biochemical reaction systems at steady state. J. Biol. Syst. 1994;2:165–182. [Google Scholar]
- Sharma V., Sakai Y., Smythe K.A., Yokobayashi Y. Knockdown of recA gene expression by artificial small RNAs in Escherichia coli. Biochem. Biophys. Res. Commun. 2013;430:256–259. doi: 10.1016/j.bbrc.2012.10.141. [DOI] [PubMed] [Google Scholar]
- Sun X., Lin Y., Huang Q., Yuan Q., Yan Y. A novel muconic acid biosynthetic approach by shunting tryptophan biosynthesis via anthranilate. Appl. Environ. Microbiol. 2013;79:4024–4030. doi: 10.1128/AEM.00859-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X., Lin Y., Yuan Q., Yan Y. Biological production of muconic acid via a prokaryotic 2,3-dihydroxybenzoic acid decarboxylase. ChemSusChem. 2014;7(9):2478–2481. doi: 10.1002/cssc.201402092. [DOI] [PubMed] [Google Scholar]
- Terzer M., Stelling J. Large-scale computation of elementary flux modes with bit pattern trees. Bioinformatics. 2008;24:2229–2235. doi: 10.1093/bioinformatics/btn401. [DOI] [PubMed] [Google Scholar]
- Thakker C., Martinez I., San K.Y., Bennett G.N. Succinate production in Escherichia coli. Biotechnol. J. 2012;7:213–224. doi: 10.1002/biot.201100061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinh C.T., Unrean P., Srienc F. Minimal Escherichia coli cell for the most efficient production of ethanol from hexoses and pentoses. Appl. Environ. Microbiol. 2008;74:3634–3643. doi: 10.1128/AEM.02708-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber C., Bruckner C., Weinreb S., Lehr C., Essl C., Boles E. Biosynthesis of cis,cis-muconic acid and its aromatic precursors, catechol and protocatechuic acid, from renewable feedstocks by Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2012;78:8421–8430. doi: 10.1128/AEM.01983-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Excel file containing the reactions of CM of each organism supplemented by the respective pathway branch. Knock-out targets are highlighted in red. The file contains two excel sheets (S. cerevisiae & E. coli), each containing different networks for the respective pathways. The determined knock-out targets are indicated in red for each cut set, potential overexpression targets in green. Reactions that were excluded from identification as knock-out target are highlighted yellow.
Supplementary Material