Graphical Abstract
The dense layer of glycans that surrounds each cell serves as an evolvable link between the cell and its environment. Like finch beaks in the Galapagos, this connection to the outside world must be tunable to shifts in selective pressure. Fortunately, glycans have proven optimal as agents of quick and robust adaptation. The enormous yet subtle variation of glycan structures provides Lego-like potential for restructuring the extracellular space. Variation in glycan structure is under the control of hundreds of differentially-regulated biosynthetic and catabolic enzymes (e.g. glycosyltransferases and glycosidases). The actions of these enzymes often occur on the timescale of minutes, allowing for rapid alteration of glycan architecture in response to environmental stimuli. If advantageous, the capability for glycan remodeling can be passed on to subsequent generations. A prime example of these evolutionary benefits lies in host-bacterial interactions. The interface between eukaryotic host cells and bacteria is comprised primarily of glycans that decorate both secreted and membrane-anchored proteins and lipids (1). Under homeostatic conditions, this glycan mix is tuned to foster symbiosis. For example, commensal gut flora can utilize host glycans as a nutrient source and in turn can help train host immune response. But upon pathogen invasion, both host and bacterial sugars are weaponized in an “arms-race” for survival. For instance, host cell immunoreceptors can detect pathogen-associated microbial patterns (PAMPs), that contain sugars (e.g. capsular polysaccharides) to mount an immune response. In defense, bacteria can change their glycan composition to mimic host sugars and subsequently evade detection. An exciting example of such co-evolution has been recently published in Science, where Buscaill et al. detail the role of a host glycosidase in controlling the release of immunogenic flagellin peptides, and bacteria’s defensive maneuvering in response (2).
Understanding the highly dynamic interplay of cell surface glycans with the extracellular milieu is paramount to understanding immune response as well as many other processes, both physiological and pathological. Unfortunately, progress in the study of cell surface glycans has been slow and difficult. And yet, many of the challenges in glycobiology stem directly from the characteristics that enable the biological activity of glycans. For example, the immense variety and complexity of glycans makes the elucidation of function a daunting task. Similarly, the large number of enzymes involved in glycan biosynthesis and their compensatory mechanisms complicates functional analysis. Recent efforts have therefore centered on developing tools to overcome these obstacles. One such tool is a probe molecule (JJB70, Fig. 1) that reacts with active glycosidases and was utilized by Buscaill et al. to identify a single glycosidase, among the hundreds that convolute the plant apoplast (extracellular space of plants), as an important mediator of flagellin deglycosylation.
Fig. 1. A chemical tool for glycosidase identification.
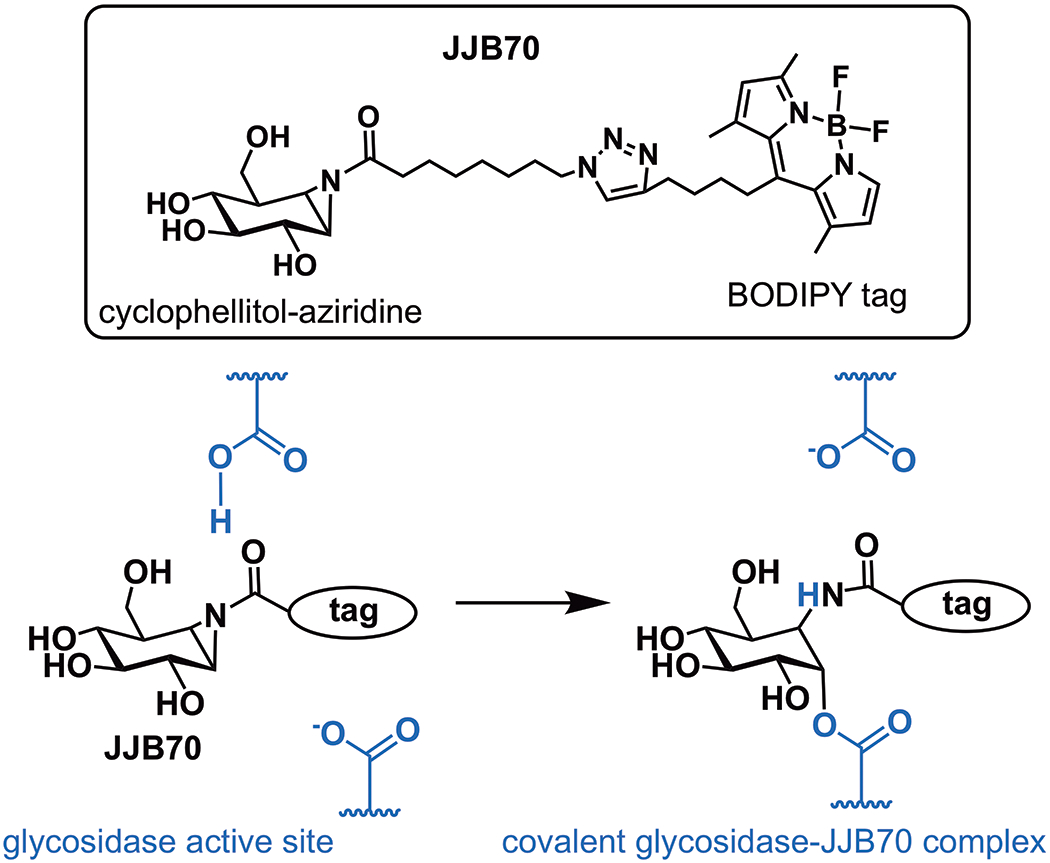
JJB70 is an activity-based probe that fluorescently labels the active site of glycosidases.
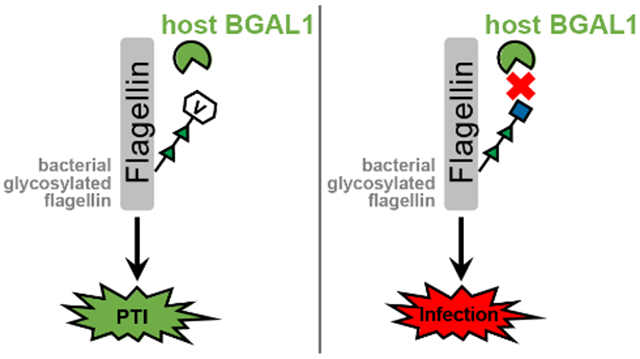
Flagellin is a bacterial protein that constitutes a majority of the flagellar filament necessary for locomotion. Upon bacterial infection of the plant apoplast, flagellin is cleaved by host hydrolases to generate a conserved immunogenic fragment. This elicitor peptide is a PAMP that can be recognized via the conserved receptor-like kinase flagellin sensitive 2 (FLS2). Albeit one of the most studied phenomena in the context of PAMP-triggered immunity (PTI), the events upstream of FLS2-activation have evaded characterization. Mainly, the biochemical mechanism by which flagellin is processed to release the buried elicitor sequence was unknown. Buscaill et al. note that glycosylation of flagellin had been shown to stifle PTI in rice (2); however, the nature of this inhibition also remained ill-defined. The findings presented in the current work, using a relative of tobacco as the model system, shed new light on these long-standing questions, and establish a model in which glycosylation is a key regulator of flagellin-dependent immune response.
Buscaill et al. capitalized on the assumption that pathogenic bacteria would evolve mechanisms to suppress important host immune-enzymes. They employed JJB70, an activity-based protein profiling (ABPP) probe (Fig. 1), in the extracellular space of the plant Nicotiana benthamiana infected with a virulent strain of Pseudomonas syringae. This cyclophellitol-aziridine ABPP probe was developed by the van der Hoorn lab in collaboration with the Overkleeft group and reacts with glutamate/aspartate residues in the active sites of a broad range of glycosidases (3). Covalent ester bonds between the electrophilic acyl-aziridine group of the probe and nucleophilic amino acids are formed only when glycosidases are active. By comparing the labeling profile of infected plants with that of mock-inoculated controls, the authors identified a 45 kDa host protein with 19-fold reduced activity during bacterial invasion. This suppressed glycosidase, β-galactosidase 1 (BGAL1), was shown to contribute to immunity by facilitating release of the flagellin elicitor fragment. Specifically, BGAL1-mediated PTI was dependent upon flagellin production in the bacteria as well as host expression of FLS2. Glycosylation-deficient strains of P. syringae were significantly less pathogenic than wild type irrespective of BGAL1 presence, and cleavage of non-glycosylated flagellin into immunogenic fragments could be blocked using a protease inhibitor cocktail. Together, these data imply that the role of BGAL1 is to deglycosylate flagellin, a step that is imperative for downstream proteolysis to release the elicitor sequence (Fig. 2). Interestingly, two P. syringae strains were shown to evade immune detection by eluding BGAL1. The weapon of one strain is a small molecule that inhibits BGAL1 activity, thereby preventing flagellin processing. A second strain camouflages its flagellin by modifying it with an altered glycan structure that includes a terminal N-acetyl-d-glucosamine (GlcNAc) instead of modified viosamine (mVio). This glycan shield protects the flagellin from BGAL1 digestion. The findings of Buscaill et al. underscore the importance of glycans in mediating host-pathogen interaction and co-evolution, and show that subtle changes in glycan composition and regulation can result in significant evolutionary advantage.
Fig. 2. Mechanism of BGAL1-mediated innate immune response in plant apoplast.
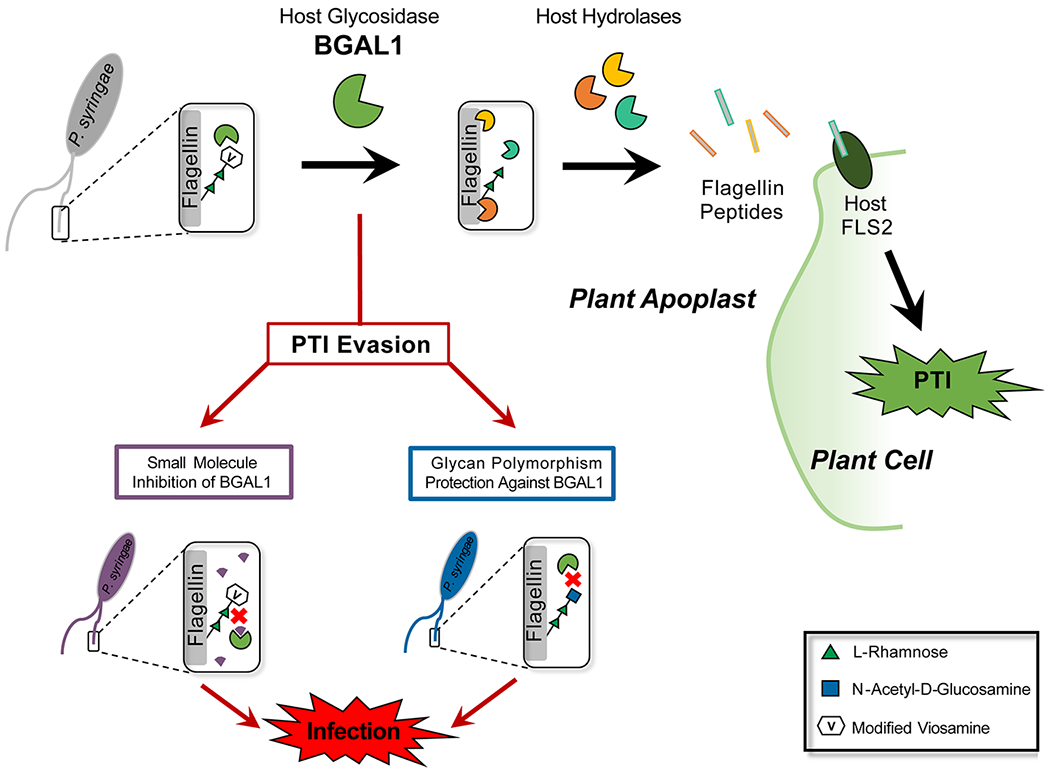
Host BGAL1 removes modified viosamine from P. syringae flagellin, allowing further protease processing of the flagellin to release immunogenic peptides that bind host FL2 and initiate PAMP-triggered immunity (PTI). P. syringae strains can evade PTI by producing a small molecule inhibitor of BGAL1 or by altering the flagellin glycan so that it is no longer a BGAL1 substrate.
The use of glycan polymorphism and glycan remodeling as defensive strategies is common on both sides of the host-bacteria “arms-race” for survival. As mentioned by Buscaill et al., Pseudomonas aeruginosa and Burkholderia cenocepacia are both cystic fibrosis-related pathogens that, like P. syringae, elicit stronger immune responses when flagellin glycosylation is disrupted (2). Glycan polymorphism occurs on the host side as well: for example, human ABO blood group determinants are glycan structures that have been linked to differences in susceptibility to multiple pathogens. Blood group distributions vary in regions where these pathogens are or are not endemic, implying that human cells also exploit changes in glycan structure to adapt to the selective pressure of pathogen invasion (reviewed in (4)). Further complicating the picture, glycan recognition events are subject to multifactorial evolutionary pressures. For example, a secreted protein toxin from the human gut pathogen Vibrio cholerae has long been known to recognize glycan structures on human cells, but recent data indicate that the bacterial toxin can also recognize chemically-identical glycans found on other bacterial species that occupy the same niche (5). Did cholera toxin evolve this glycan binding specificity to recognize host, competitor, or both? The mechanisms behind these observations remain incompletely understood; however, it is clear that characterizing the roles of glycans in virulence and co-evolution is imperative to evolving our understanding and treatment of infectious diseases.
The findings of Buscaill et al. illustrate just one of many glycan-mediated tactical concealment mechanisms that characterize host-pathogen interactions. By utilizing a pathogen’s evolutionary adaptation of suppressing immune-enzymes as a foothold for identifying glycosidases involved in PTI, the authors establish a paradigm for identifying, characterizing, and potentially combatting more of these ever-evolving glycan-controlled mechanisms of pathogen invasion. As the glycobiology toolbox continues to be expanded and improved, we expect that additional hidden roles for cell surface sugars in the immune response will be revealed.
Acknowledgements
The authors acknowledge support from the National Institutes of Health (R01GM090271).
References
- 1.Hooper LV, Gordon JI (2001) Glycans as legislators of host–microbial interactions: spanning the spectrum from symbiosis to pathogenicity, Glycobiology 11, 1R. [DOI] [PubMed] [Google Scholar]
- 2.Buscaill P, Chandrasekar B, Sanguankiattichai N, Kourelis J, Kaschani F, Thomas EL, Morimoto K, Kaiser M, Preston GM, Ichinose Y, van der Hoorn RAL (2019) Glycosidase and glycan polymorphism control hydrolytic release of immunogenic flagellin peptides, Science 364, eaav0748. [DOI] [PubMed] [Google Scholar]
- 3.Chandrasekar B, Colby T, Emran Khan Emon A, Jiang J, Hong TN, Villamor JG, Harzen A, Overkleeft HS, van der Hoorn RA (2014) Broad-range glycosidase activity profiling. Molecular & Cellular Proteomics 13, 2787–2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cooling L (2015) Blood groups in infection and host susceptibility. Clinical Microbiology Reviews 28, 801–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Patry RT, Stahl M, Perez-Munoz ME, Nothaft H, Wenzel CQ, Sacher JC, Coros C, Walter J, Vallance BA, Szymanski CM (2019) Bacterial AB5 toxins inhibit the growth of gut bacteria by targeting ganglioside-like glycoconjugates. Nature Comm. 10, 1390. [DOI] [PMC free article] [PubMed] [Google Scholar]


