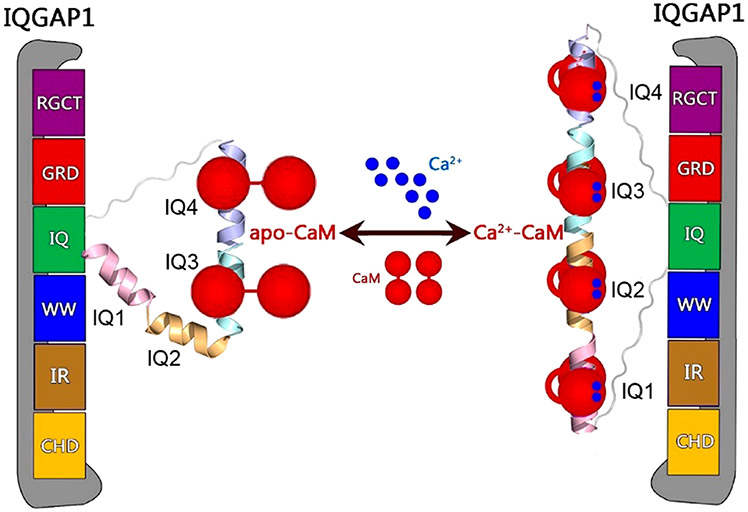
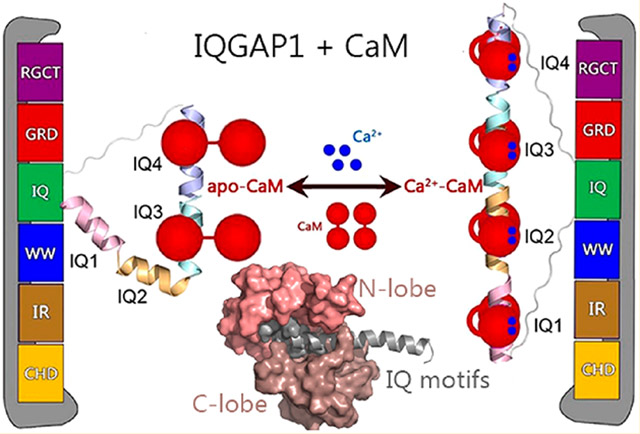
Abstract
IQ domain GTPase-activating scaffolding protein 1 (IQGAP1) mediates cytoskeleton, cell migration, proliferation, and apoptosis events. Calmodulin (CaM) modulates IQGAP1 functions by binding to its four tandem IQ motifs. Exactly how CaM binds the IQ motifs and which functions of IQGAP1 CaM regulates and how are fundamental mechanistic questions. We combine experimental pull-down assays, mutational data, and molecular dynamics simulations to understand the IQ–CaM complexes with and without Ca2+ at the atomic level. Apo-CaM favors the IQ3 and IQ4 motifs but not the IQ1 and IQ2 motifs that lack two hydrophobic residues for interactions with apo-CaM’s hydrophobic pocket. Ca2+-CaM binds all four IQ motifs, with both N- and C-lobes tightly wrapped around each motif. Ca2+ promotes IQ–CaM interactions and increases the amount of IQGAP1-loaded CaM for IQGAP1-mediated signaling. Collectively, we describe IQ–CaM binding in atomistic detail and feature the emergence of Ca2+ as a key modulator of the CaM–IQGAP1 interactions.
Graphical Abstract
IQ domain GTPase-activating protein (IQGAP) is a conserved family of proteins expressed in organisms ranging from yeast, fish, and Xenopus to mammals.1-3 Humans have three IQGAP proteins, IQGAP1–IQGAP3.4,5 IQGAP1 can be detected in almost all cell types and tissues and is the most investigated member of the IQGAP family.6,7 Expression of IQGAP2 and IQGAP3 is limited to specific tissues, i.e., liver for IQGAP28 and brain, lung, testis, and spleen for IQGAP3.4 Human IQGAP proteins are large and multidomain. They comprise an N-terminal calponin homology domain (CHD), a nIQGAP repeat domain (IR), a WW domain, IQ motifs, a Ras GTPase-activating protein (GAP)-related domain (GRD), and a RasGAP C-terminus (RGCT). These domains bind multiple partners to execute diverse cellular functions, including those associated with the cytoskeleton,5 cell migration,9,10 cell proliferation,11-13 and apoptosis.7 Dysfunction of IQGAP1 has been implicated in several neoplasms, including colorectal, breast, gastric, liver, lung, and ovarian cancers.6,14 Approximately 10% of the genes with an increased level of expression in metastatic cells encode IQGAP1 binding partners,6 in line with the significant roles of IQGAP1 in tumor proliferation, invasion, and angiogenesis.15,16
More than 130 proteins have been characterized as IQGAP1 binding partners, and many of these bind to the four tandem IQ motifs or the C-terminal domain.7 IQGAP1 dimerizes, which has been shown to be crucial for its functions.17 IQGAP1 dimerization can follow different pathways. In one, Cdc42 promotes IQGAP1 dimerization through the GRD;18,19 in a second, dimerized N-termini may contribute,20 and in a third, most importantly, IQ motifs mediate dimerization, although exactly how is still unclear.17 IQ motifs with conserved IQ core sequences have been widely identified in human proteins.21 The IQ domain of IQGAP1 contains four tandem IQ motifs, i.e., IQ1–IQ4. IQ motifs are sequences comprised of 20–25 amino acids that contain the consensus sequence [FILV]Qxxx[RK]Gxxx[RK]xx[FILVWY] (where x is any amino acid), which binds to calmodulin (CaM). CaM is the primary calcium (Ca2+) messenger in cells. It has two lobes connected by a flexible linker. Apo-CaM denotes Ca2+-free CaM, which usually adopts the partially bent conformation with the two lobes apart.22 Upon binding to Ca2+, Ca2+-CaM exposes the hydrophobic grooves in the N- and C-lobes to accommodate targets.23-25 IQGAP1 interacts with CaM via four IQ motifs. The interactions are sensitive to Ca2+ and are crucial for IQGAP1 functions.26,27 IQ motifs frequently occur in tandem repeats binding multiple CaM molecules with higher affinity in the absence of Ca2+.28 IQGAP1 differs from most IQ-containing proteins as it binds more Ca2+-CaM than apo-CaM.29,30 Importantly, CaM modulates the binding of IQGAP1 to many of its other binding partners.30-34 Notably, CaM attenuates some of the IQGAP1–target interactions only in the presence of Ca2+.29,35 IQGAP1 binds 2-fold more Ca2+-CaM than apo-CaM.
Despite significant effort, obtaining the experimental structures of IQ–CaM complexes has been challenging, and thus, molecular details for how IQGAP1 interacts with CaM via IQ motifs have been elusive. Insight into the IQGAP1 IQ–CaM structure, its modulation by Ca2+, and how its mechanistic underpinning relates to signaling, foremost by oncogenic KRas, are highly significant aims.
To address this challenge and to obtain an atomistic level grasp of IQ–CaM interactions in IQGAP1, we study the interactions of IQ motifs with CaM with and without Ca2+ by both experimental and computational methods, providing mechanistic insight to explain the critical role played by Ca2+. We show that Ca2+ mediates the CaM–IQ interactions, which may further link CaM to IQGAP1-promoted signaling, including the KRas4B/PI3Kα/Akt pathways.12,36-39 Experimental data indicate that IQ motifs in IQGAP1 load more Ca2+-CaM molecules than apo-CaM, and IQ motifs show a preference for the C-lobe versus the N-lobe of CaM. Our modeling and simulations suggest that in the presence of Ca2+, Ca2+-CaM interacts with the IQ motif with both N- and C-lobes tightly wrapped around the motif, forming highly compact complexes, while apo-CaM favors the IQ3 and IQ4 motifs that have three hydrophobic residues interacting with the hydrophobic pockets. Ca2+ enhances the interactions of the IQ motifs with CaM and increases the number of loaded CaM molecules in IQGAP1.
MATERIALS AND METHODS
Plasmids.
IQGAP1 (amino acids 717–916) in the pcDNA3-Myc vector40 was modified by site-directed mutagenesis to generate IQ3R and IQ4R mutations as described previously.41 GST-CaM wild-type, N-half, and C-half were synthesized by polymerase chain reaction (PCR) amplification from pEYFP-mCaM as the template and forward primer 5′-CGGGATCCGCTGATCAGCTGACCGAAGAACAG-3′ and reverse primer 5′-GCTCTAGACTCGAGTCATTTTGCAGTCATCATCTGTACGAATTC-3′ for CaM-F, forward primer 5′-CGGGATCCGCTGATCAGCTGACCGAAGAACAG-3′ and reverse primer 5′-CCGGAATTCTCTAGATCAATCTTTCATTTTTCTAGCCATCATAG-3′ for CaM-N, and forward primer 5′-CGGGATCCACAGATAGTGAAGAAGAAATCCG-3′ and reverse primer 5′-GCTCTAGACTCGAGTCATTTTGCAGTCATCATCTGTACGAATTC-3′ for CaM-C.
PCR products were cut with BamHI and XhoI and inserted into pGEX4T-1 at BamHI-XhoI sites. DNA sequencing was performed to confirm plasmid sequences. All proteins migrated to the expected positions on a sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) gel.
Transcription and Translation (TNT) Product Production and Binding Analysis.
[35S]Methionine-labeled TNT products were synthesized using the TNT Quick Coupled Transcription/Translation system (Promega) as described previously.17,40 Briefly, 1 μg of each pcDNA3-Myc-IQ motif (amino acids 717–916) plasmid was incubated with 40 μL of TNT Quick Master Mix and 20 μCi of [35S]methionine (PerkinElmer Life Sciences) for 90 min at 30 °C. TNT products were confirmed by SDS–PAGE and autoradiography before being used in pull-down assays. GST-CaM (F, N, or C) and GST alone were expressed in Escherichia coli and purified with glutathione Sepharose (GE Healthcare) following the manufacturer’s protocol. The TNT products of the IQ domains of IQGAP1 were incubated with 4 μg of each GST-CaM construct (GST alone was used as the control) in buffer A [50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1% Triton X-100, 1 mM CaCl2, or 1 mM EGTA] with 1% protease inhibitor and 1 mM phenylmethanesulfonyl fluoride for 3 h at 4 °C with rotation. Complexes were washed five times with buffer A and separated by SDS–PAGE. Gels were dried, and autoradiography was performed. Autoradiographs were scanned, and bands quantified with Li-Cor Image Studio Software. Data were graphed and analyzed with Prism.
Modeling of Apo-CaM–IQ and Ca2+-CaM–IQ Complexes.
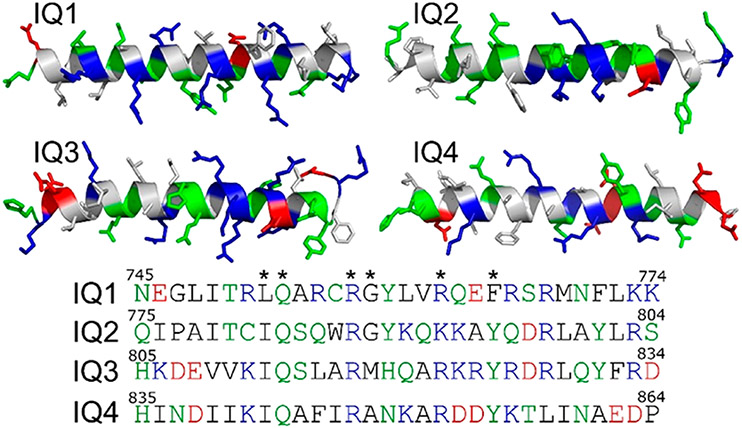
Homology modeling of individual IQ motifs (IQ1, residues 745–774; IQ2, residues 775–804; IQ3, residues 805–834; and IQ4, residues 835–864) (Figure 2) in IQGAP1 was performed by the LOMETS online server that generates template-based protein structure prediction.42,43 The templates used to model IQ1–IQ4 are the segments in Protein Data Bank (PDB) entries 5LJ3, 2DFS, and 3GN4. The predicted secondary structures of IQ1–IQ4 determined by homology modeling (IQ1, CCCHHHHHHHHHHHHHHHHC; IQ2, C C C H H H H H H H H H H H H H H H H C ; I Q 3 , CCHHHHHHHHHHHHHHHHCC; a n d IQ 4 , CCHHHHHHHHHHHHHCCCCC, where C and H represent coiled and helical structures, respectively) show that four IQ motifs in IQGAP1 favored α-helical structures. The apo-CaM–IQ complexes were modeled on the basis of the nuclear magnetic resonance (NMR) structure of the IQ motif in NaV1.5 interacting with apo-CaM (PDB entry 2L53). Modeling of Ca2+-CaM–IQ complexes was performed on the basis of the structural templates of IQ motifs in CaV1.2 interacting with Ca2+-CaM (PDB entry 2F3Y). The IQ motifs in NaV1.5 and CaV1.2 exhibited sequence similarity to IQ motifs in IQGAP1 with the typical IQ core sequence. Four IQ motifs in IQGAP1 were individually superimposed into the apo-CaM and Ca2+-CaM on the basis of the structural alignment of IQ core sequences. The minimization and short MD simulations were performed to relax the generated complexes.
Figure 2.
Characterization of IQ1–IQ4 motifs in IQGAP1. The structures are obtained from homology modeling. The hydrophobic, hydrophilic, basic, and acidic residues are colored white, green, blue, and red, respectively. Asterisks denote the conserved residues in IQ core sequences.
Molecular Simulation Protocols.
The MD simulations were performed by NAMD package44 using the CHARMM allatom additive force field (version C36).45 The NPT simulations were performed at 310 K and 1 atm. The TIP3 water model was used. Na+ and Cl− ions were utilized to neutralize the systems and generated a salt concentration of 0.15 mol/L. Before the production run, the minimizations and short MD simulations were performed to relax the systems. The MD simulations were performed with a time step of 2 fs. Short-range interactions were calculated by the switch function with the twin-range cutoff. Long-rang electrostatic interactions were described by the particle mesh Ewald (PME) algorithm. A set of 1.6 μs simulations were conducted for eight CaM–IQ systems (200 ns for each complex). The analysis was performed using the tools in CHARMM, VMD, and scripts.
Statistical Analysis.
All data are expressed as means ± the standard deviation (SD). Statistical analysis was performed with GraphPad Prism 7 (GraphPad Software). Radiographs were quantified with Li-Cor Image Studio 5.2 Software. Statistical analysis was performed using one-way analysis of variance (ANOVA), multiple-comparison test, with P ≤ 0.05 considered significant. Significance levels were indicated as follows: n.s. (not significant), p > 0.05; *p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001; ****p ≤ 0.0001.
■ RESULTS
Mutations in the IQ3 and IQ4 Motifs Alter Apo-CaM Association.
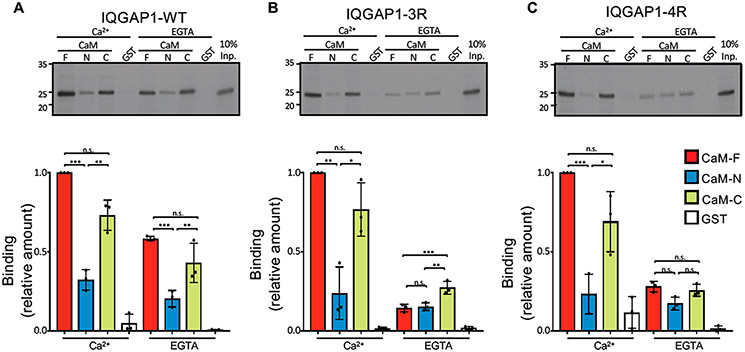
Biochemical binding analyses were performed for the IQ domain (amino acids 717–916) of IQGAP1 interacting with GST-tagged CaM. The IQ domain contains four IQ motifs, IQ1–IQ4. We compared the binding of CaM to the wild-type IQ domain to its binding to two mutant IQ domains, one with mutations in the IQ3 motif (denoted as IQ3R, in which Arg817 and Arg822 are replaced by Gln) and the other with mutations in the IQ4 motif (denoted as IQ4R, in which Arg847 and Arg852 are replaced by Gln).41 In the pull-down assays, CaM was used as the full-length protein (CaM-F, amino acids 1–148), the N-half (CaM-N, residues 1–74), and the C-half (CaM-C, residues 75–148) as described previously.46 In the presence of Ca2+, the wild-type IQ domain stably bound CaM-F (Figure 1A). The level of binding of Ca2+-CaM-N to the wild-type IQ domain is only 32% of the level of Ca2+-CaM-F binding. The level of binding of the wild-type IQ domain to Ca2+-CaM-C was 70% of that seen with Ca2+-CaM-F (Figure 1A). In the absence of Ca2+ (EGTA), the level of binding of apo-CaM-F to the wild-type IQ domain is reduced to 58%. For the mutant IQ3R and IQ4R domains, mutations of the IQ domain had essentially no effect on interactions with Ca2+-CaM. This observation suggests that electrostatic interaction between Ca2+-CaM and the IQ domain is not the major driving force. In contrast, binding of apo-CaM to IQ3R and IQ4R was significantly different from that of apo-CaM to the wild-type IQ domain. Remarkably, the level of binding of apo-CaM-F to IQ3R was reduced to 15% of the level of binding of IQ3R to Ca2+-CaM-F (Figure 1B). The levels of binding of apo-CaM-C and apo-CaM-N to IQ3R were also reduced to 15% and 27%, respectively. Similarly, the level of binding of apo-CaM-F to IQ4R was reduced to 28% (Figure 1C). Interestingly, the extent of binding of apo-CaM-C and apo-CaM-N to IQ4R was very similar to their binding to IQ3R. Binding of the CaM constructs to GST alone was minimal, validating the specificity of the interactions. Coomassie staining revealed that similar amounts of purified CaM and GST protein were present in the binding assays (Figure S1).
Figure 1.
Mutations of IQ3 and IQ4 motifs alter calmodulin binding. In vitro binding analysis of IQ domain constructs of IQGAP1. (A) The wild-type (WT) IQ domain (amino acids 717–916), (B) IQ3R, and (C) IQ4R were labeled with [35S]Met using TNT. Samples were incubated with purified recombinant GST-CaM constructs, namely, full-length (F, amino acids 1–148), N-half (N, amino acids 1–74), and C-half (C, amino acids 75–148) or GST alone. In the top panels, the bound proteins were resolved by SDS–PAGE and imaged by autoradiography. Input (Inp.) is 10% of the IQ domain used for the binding assay. Representative images of three independent experiments are shown. Autoradiographs were quantified with Li-Cor Image Studio Software. The amount of binding of each IQ domain was normalized to CaM-F in Ca2+. The graphs depict binding to CaM-F (red), CaM-N (blue), CaM-C (green), or GST (white). Data are means ± the standard deviation (n = 3). Significance was determined by ANOVA: *p < 0.05; **p < 0.005; ***p < 0.0005; ns, not significant.
Apo-CaM Favors Interaction with IQ3 and IQ4 Motifs.
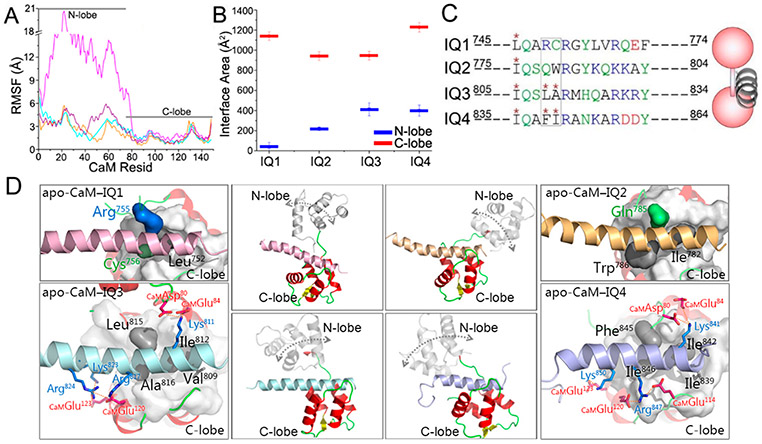
To explore the CaM–IQ interactions and understand the preference of IQ motifs for apo-CaM’s C-lobe in the experiments (Figure 1) at the atomic level, we modeled and simulated apo-CaM in complex with four IQ motifs in IQGAP1. The available structures of IQ peptides from other proteins bound to CaM indicate that the IQ CaM binding motifs adopt α-helical structures (at least when they bind to CaM). The structures of the IQ motifs in IQGAP1 have not been determined. We performed homology modeling to predict the structures of IQ motifs in IQGAP1. The results show that the four IQ motifs in IQGAP1 also favor the α-helical conformation (Figure 2). The structures of apo-CaM bound to IQ motifs have been determined for many CaM targets, including myosin V (PDB entries 2IX7 and 5WSU), NaV1.5 (PDB entry 2L53), NaV1.6 (PDB entry 3WFN), and neuromodulin and neurogranin (PDB entry 4E50).47-51 In the structures, the IQ motifs interact predominantly with the C-lobe of apo-CaM, suggesting a general preference of IQ motifs for the C-lobe of apo-CaM.47-51 This is consistent with our experiments (Figure 1) and indicates that the IQ motifs in IQGAP1 may interact with apo-CaM in a similar way.
To predict the structures of apo-CaM binding to IQ motifs in IQGAP1, we performed molecular modeling, taking the NMR structure of the IQ motif of NaV1.5 in complex with apo-CaM (PDB entry 2L53) as the structural template.52 The IQ motif in NaV1.5 contains the IQ core sequence resembling those of the IQ motifs in IQGAP1. The four IQ motifs in IQGAP1 were individually superimposed onto apo-CaM on the basis of structural alignment of the IQ core residues (Figure S2). This protocol is expected to generate the most likely apo-CaM–IQ interfaces. As in the structure, the IQ motifs in IQGAP1 interacted mainly with the C-lobe of apo-CaM in the simulations. The N-lobe of apo-CaM also interacted with the IQ motifs, but to a lesser extent. The residue-based root-mean-square fluctuation (RMSF) values of apo-CaM show that the interactions between IQ motifs in IQGAP1 and apo-CaM C-lobe were stable, while those with the N-lobe showed larger fluctuations (Figure 3A). The interface areas between IQ motifs and apo-CaM’s C-lobe (~942.1–1228.3 Å2) are larger than those of the N-lobe (~37.7–412.2 Å2) (Figure 3B). In the modeled apo-CaM–IQ complexes, IQ3 and IQ4 individually have three hydrophobic amino acids at positions 1, 4, and 5 (Ile812, Leu815, and Ala816 for IQ3 and Ile842, Phe845, and Ile846 for IQ4) of the IQ core region inserting into the hydrophobic pocket of the apo-CaM’s C-lobe, which is similar to the NMR structure. However, the IQ1 and IQ2 motifs lack two of these residues. Arg755 and Cys756 in the IQ1 motif and Gln785 in the IQ2 motif are hydrophilic, and Trp786 of IQ2 was not fully in the apo-CaM hydrophobic pocket (Figure 3C,D). This suggests that IQ3 and IQ4 favor interacting with the hydrophobic pockets in apo-CaM over IQ1 and IQ2 motifs, in line with the observations that apo-CaM favors IQ3 and IQ4, not IQ1 and IQ2, in IQGAP1.41 In addition, Val809 in IQ3 and Ile839 in IQ4 also formed hydrophobic interactions with CaM’s C-lobe. In the experiments, point mutations of the conserved basic residues at positions 6 and 11 (Arg817 and Arg822 for IQ3 and Arg847 and Arg852 for IQ4, respectively) eliminated the interactions of IQ3 and IQ4 with apo-CaM (Figure 1).41 In the modeled apo-CaM–IQ3 and apo-CaM–IQ4 complexes, the arginine residues at position 6 (Arg817 for IQ3 and Arg847 for IQ4) of the IQ core region contributed to the apo-CaM–IQ interfaces by forming interfacial salt bridges, while the arginine at position 11 (Arg822 for IQ3 and Arg852 for IQ4) appeared to be less important. In the apo-CaM–IQ3 complex, Arg817 consistently interacted with Glu120 at CaM’s C-lobe throughout the trajectory, which also formed interactions with Glu45 at CaM’s N-lobe, indicating its great contributions to the interface. The quantitative analysis also indicates the salt bridges between IQ3 and apo-CaM’ C-lobe with the high contacting probabilities, such as CaMAsp80-IQ3Lys811-CaMGlu84 (contacting probability of 0.84), IQ3Lys823-CaMGlu123 (contacting probability of 0.93), and IQ3Arg824-CaMGlu120 (contacting probability of 1.00). At the interface between the IQ4 motif and CaM’s C-lobe, Arg847 formed dual salt bridges with Glu120 and Glu114 at CaM’s C-lobe with a contacting probability of 1.00, in line with its significance in stabilizing the apo-CaM–IQ4 interface (Figure 3D). Salt bridges with high contacting probabilities at the apo-CaM–IQ4 interface also include CaMAsp80-IQ4Lys841-CaMAsp80 (contacting probability of 0.90) and IQ4Lys850- CaMGlu123 (contacting probability of 1.00).
Figure 3.
Properties of IQ motifs binding to apo-CaM. (A) RMSF values of apo-CaM and (B) interface areas between IQ motifs and CaM in the apo-CaM–IQ complexes. (C and D) Comparison of the sequences and structures of modeled apo-CaM–IQ complexes, with a cartoon depicting the apo-CaM–IQ interaction modes. Color code: red for CaM, pink for the IQ1 motif, yellow for the IQ2 motif, cyan for the IQ3 motif, and purple for the IQ4 motif. The rectangular box in panel C highlights two residues of IQ1–IQ4 in the hydrophobic pockets of apo-CaMs. The double-ended arrows in the middle panels of panel D indicate the motion of the N-lobe of apo-CaMs. Important residue contacts in the apo-CaM–IQ3 and apo-CaM–IQ4 complexes are highlighted.
Ca2+-CaM Accommodates Four IQ Motifs.
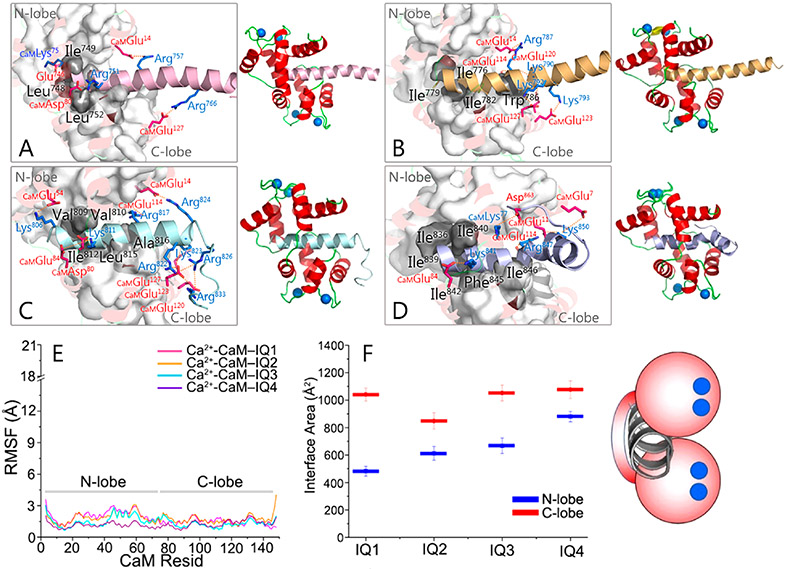
IQ motifs in IQGAP1 load more Ca2+-CaM molecules in the experiments, where hydrophobic interactions play major roles (Figure 1). It has been established that CaM exposes the hydrophobic pockets in two lobes in the presence of Ca2+ and establishes stronger hydrophobic interactions with the targets. Upon binding to partners, particularly peptides, Ca2+-CaM frequently adopts a collapsed conformation with the two lobes wrapped, leading to a compact complex.24,53 The crystal structure of the IQ motifs in CaV1.2 interacting with Ca2+-CaM (PDB entry 2F3Y) provides clues for how IQ motifs bind to Ca2+-CaM.54 We modeled the structures of Ca2+-CaM binding to individual IQ motifs in IQGAP1 by sequence alignment and structural superimposition on this complex (Figure S3).
The individual IQ motifs in IQGAP1 formed stable interactions with Ca2+-CaM in the simulations, in line with the ability of Ca2+-CaM to accommodate four IQ motifs in IQGAP1. Ca2+-CaM maintained a collapsed conformation, with both N- and C-lobes interacting with the IQ motifs (Figure 4A-D). The RMSF values for the N-lobe and C-lobe of Ca2+-CaM are comparable (Figure 4E). However, the interactions of IQ motifs with the C-lobe of Ca2+-CaM (~847.9–1076.5 Å2) were more dominant than in the N-lobe (~482.7–880.4 Å2), as indicated by the larger interface areas (Figure 4F), consistent with the larger number of the IQ motifs bound to the C-lobe than to the N-lobe of Ca2+-CaM in the experiments (Figure 1). Different from apo-CaM, mutations of basic residues in one, two, three, or all four IQ motifs in IQGAP1 cannot fully abolish the Ca2+-CaM–IQ interactions.41 While point mutations of hydrophobic residues distal to the conserved Gln (the “Q” of IQ) of the four IQ motifs attenuated the Ca2+-CaM–IQ interactions, the additional point mutation of Leu in IQ1 and Ile in IQ2–IQ4 (the “I” of IQ) led to complete loss of the Ca2+-CaM interactions.41 This indicates that the hydrophobic interactions are dominant for the Ca2+-CaM–IQ complex. In the modeled Ca2+-CaM–IQ complexes, the hydrophobic residues in IQ motifs interacted with both lobes, which, however, had more contacts with CaM’s C-lobe. The hydrophobic residues of IQ motifs interacting with CaM’s C-lobe (Leu748 and Leu752 for IQ1, Ile779, Ile782, and Trp786 for IQ2, Ile812, Leu815, and Ala816 for IQ3, and Ile842, Phe845, and Ile846 for IQ4) are generally more abundant than those interacting with the N-lobe (Ile749 for IQ1, Ile776 for IQ2, Val809 and Val810 for IQ3, and Ile836, Ile839, and Ile840 for IQ4). The “I” residues of IQ (Leu752 for IQ1, Ile782 for IQ2, Ile812 for IQ3, and Ile842 for IQ4) that are crucial for the Ca2+-CaM–IQ interactions are always inserted into the hydrophobic pocket of the Ca2+-CaM C-lobe. This is consistent with the experiments.41 Quantitative analysis shows that Ca2+-CaM–IQ3 and Ca2+-CaM–IQ4 complexes presented more salt bridges than Ca2+-CaM–IQ1 and Ca2+-CaM–IQ2 complexes. Four salt bridges in the Ca2+-CaM–IQ1 complex [IQ1Glu746-CaMLys75 (contacting probability of 0.99), IQ1Arg751-CaMAsp80 (contacting probability of 0.96), IQ1Arg757-CaMGlu14 (contacting probability of 1.00), and IQ1Arg766-CaMGlu127 (contacting probability of 0.99)], four salt bridges in the Ca2+-CaM–IQ2 complex [IQ2Arg787-CaMGlu14 (contacting probability of 0.97), CaMGlu114-IQ2Lys790-CaMGlu120 (contacting probability of 0.87), IQ2Lys792-CaMGlu127 (contacting probability of 0.78), and IQ2Lys793-CaMGlu123 (contacting probability of 0.91)], eight salt bridges in the Ca2+-CaM–IQ3 complex [IQ3Lys806-CaMGlu54 (contacting probability of 0.90), CaMAsp80-IQ3Lys811-CaMGlu84 (contacting probability of 0.99), IQ3Arg817-CaMGlu114 (contacting probability of 0.99), IQ3Arg822-CaMGlu127 (contacting probability of 1.00), IQ3Lys823-CaMGlu120 (contacting probability of 0.96), IQ3Arg824-CaMGlu14 (contacting probability of 0.95), CaM-Glu123-IQ3Arg826-CaMGlu127 (contacting probability of 1.00), and CaMGlu123-IQ3Arg833-CaMGlu127 (contacting probability of 1.00)], and four salt bridges in the Ca2+-CaM–IQ4 complex [IQ4Lys841-CaMGlu84 (contacting probability of 0.99), CaM-Glu11-IQ4Arg847-CaMGlu114 (contacting probability of 1.00), CaMGlu7-IQ4Lys850-CaMGlu11 (contacting probability of 0.91), and IQ4Asp863-CaMLys77 (contacting probability of 0.88)] were identified, where the conserved arginine residues in the IQ core region consistently showed the contributions.
Figure 4.
Properties of IQ motifs binding to Ca2+-CaM. Snapshots of Ca2+-CaM interacting with (A) IQ1, (B) IQ2, (C) IQ3, and (D) IQ4 motifs, displayed by a cartoon depicting the Ca2+-CaM–IQ interaction mode. (E) RMSF values of Ca2+-CaM and (F) interface areas between IQ motifs and Ca2+-CaM in the Ca2+-CaM–IQ complexes. The figure shows that the conserved basic and hydrophobic residues, especially the “I” of IQ (Leu752 for IQ1, Ile782 for IQ2, Ile812 for IQ3, and Ile842 for IQ4), exhibit significant Ca2+-CaM–IQ interactions. The blue dots denote the Ca2+ in the complexes.
Ca2+ Promotes CaM–IQ Interactions in IQGAP1.
Ca2+ regulates cellular signaling by modulating Ca2+ binding proteins, primarily CaM.55 The IQ motifs in IQGAP1 interact with both apo-CaM and Ca2+-CaM. Apo-CaM favors IQ3 and IQ4, and Ca2+-CaM accommodates four IQ motifs.41 This suggests that Ca2+ may promote IQGAP1 to load more CaM molecules in the IQ region (Figure 1). To further study the CaM–IQ interactions from an energy point of view and quantify the effects of Ca2+ on CaM–IQ interactions, the binding free energies of the CaM–IQ complexes were calculated using molecular mechanics combined with the generalized Born and surface area continuum solvation (MM-GBSA), which is a sum of the enthalpic (gas phase contribution and solvation energy contribution) and entropic contributions.56 In the apo-CaM–IQ complexes, the binding free energies of IQ3 (~−109.0 kcal/mol) and IQ4 (~−106.9 kcal/mol) are comparable (Table 1). Apo-CaM interacts with the IQ3 and IQ4 motifs in IQGAP1 mainly via the C-lobe, while the contributions of the N-lobe are minor. By contrast, both the N-lobe and the C-lobe of Ca2+-CaM established the interactions with the IQ motifs, giving rise to the compact complexes. The interface areas of Ca2+-CaM–IQ complexes are larger than those of apo-CaM–IQ complexes (Figures 3 and 4). As expected, the binding free energies of IQ3 (~−120.8 kcal/mol) and IQ4 (~−129.8 kcal/mol) in complex with Ca2+-CaM were lower than those with apo-CaM (Table 1). This indicates that the presence of Ca2+ enhances the CaM–IQ interfaces in IQGAP1. In the Ca2+-CaM complexes, the binding free energies for the IQ3 and IQ4 motifs are lower than that for the IQ1 (~−106.9 kcal/mol) and IQ2 (~−108.0 kcal/mol) motifs.
Table 1.
Binding Free Energies for IQ motifs Binding to Apo-CaM and Ca2+-CaMa
| system | ΔGb | ΔGsol + ΔGgas | −TΔS | system | ΔGb | ΔGsol + ΔGgas | −TΔS |
|---|---|---|---|---|---|---|---|
| apo-CaM–IQ3 | −109.0 ± 9.3 | −133.1 ± 9.3 | 24.1 | apo-CaM–IQ4 | −106.9 ± 10.4 | −131.2 ± 10.4 | 24.3 |
| Ca2+-CaM–IQ1 | −106.9 ± 7.6 | −130.9 ± 7.6 | 24.0 | Ca2+-CaM–IQ2 | −108.0 ± 7.7 | −132.0 ± 7.7 | 24.0 |
| Ca2+-CaM–IQ3 | −120.8 ± 10.4 | −145.0 ± 10.4 | 24.2 | Ca2+-CaM–IQ4 | −129.8 ± 8.9 | −154.0 ± 8.9 | 24.2 |
The average binding free energy ⟨ΔGb⟩ is a sum of the gas phase contribution ⟨ΔGgas⟩, the solvation energy contribution ⟨ΔGsol⟩, and the entropic contribution (−TΔS).
■ DISCUSSION
IQGAP1 is a critical scaffold protein that mediates cellular processes by assembling and regulating diverse binding partners. Partners targeting the same domain in IQGAP1 may accomplish productive crosstalk during cell signaling events.57,58 Thus, surprisingly, CaM has been demonstrated to interact with the IQ motifs of IQGAP1 and mediate its functions,59 either by directly attenuating the interactions of IQGAP1 with other targets, including F-actin and Cdc42,29,60,61 or by being a key component of targets assembled by IQGAP1.26 Ca2+ promotes the interactions of CaM with IQ motifs of IQGAP1, thus mediating cellular events in which IQGAP1 is involved.
The significance of the CaM–IQ interactions, but the absence of experimental molecular structures, likely due to the existence of multiple states, and CaM’s still enigmatic mechanism, led us to first model the CaM–IQ complex. Complicating the mechanistic question was the presence of four tandem IQ motifs in IQGAP1. In addition, experimental data pointed to differences between the presence and absence of Ca2+. The experimental data showed that fewer IQ motifs bound to apo-CaM than to Ca2+-CaM (Figure 1A). While the interaction of Ca2+-CaM with mutant IQ3R and IQ4R domains is highly compatible with the wild-type IQ domain, the level of binding of apo-CaM to IQ3R and IQ4R was less than that to wild-type IQ (Figure 1B,C). The simulations suggest that apo-CaM favors interactions with the IQ3 and IQ4 motifs but not the IQ1 and IQ2 motifs (Figure 3), which is largely due to the absence of the two hydrophobic residues at positions 4 and 5 of the IQ core region in the IQ1 and IQ2 motifs. This finding is consistent with our previous experimental data.41 The models also verify the current experimental data, highlighting the preference of IQ motifs for apo-CaM’s C-lobe and the importance of arginine residues for apo-CaM–IQ interactions (Figures 1B,C and 3D). These molecular details underscore the significance of the experimentally introduced mutations in Ca2+-CaM–IQ interactions.12,41
The interactions of the IQ motifs in IQGAP1 with CaM are sensitive to Ca2+. Many IQGAP1-mediated cellular oncogenic signaling pathways, including KRas4B/PI3Kα/Akt, are sensitive to Ca2+-CaM.59,62-64 Ca2+-CaM,38,65 particularly when phosphorylated at Tyr99,36,37 contributes to full PI3K activation by binding to both nSH2 and cSH2 regulatory domains of PI3K and releasing their autoinhibition.66 IQGAP1 binds PI3Kα via the WW domain.67 The WW domain is adjacent to the IQ domains in IQGAP1. This suggests that IQGAP1 may gather CaM molecules to PI3Kα for its activation. In this scenario, Ca2+ emerges as a key regulator. Without Ca2+, IQGAP1 may load two apo-CaM molecules via the IQ3 and IQ4 motifs. In the presence of Ca2+, four IQ motifs in IQGAP1 interact with four Ca2+-CaM molecules. Figure 5 provides our model for Ca2+-dependent activation of IQGAP1. The level of binding of Ca2+-CaM to IQGAP1 is 2-fold greater than that of apo-CaM.61 This mechanism agrees with the conserved basic and hydrophobic residues in the CaM–IQ interfaces, explains experimental data, and provides molecular details of the dynamic CaM–IQ interactions in IQGAP1.
Figure 5.
Schematic illustration of Ca2+-dependent CaM–IQ interactions in IQGAP1.
Supplementary Material
ACKNOWLEDGMENTS
All simulations were performed using the high-performance computational facilities of the Biowulf PC/Linux cluster at the National Institutes of Health, Bethesda, MD (https://hpc.nih.gov/).
Funding
This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health (NIH), under Contract HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. This research was also supported (in part) by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research, and the Intramural Research Program of the NIH Clinical Center.
Footnotes
The authors declare no competing financial interest.
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.biochem.9b00854.
Coomassie-stained SDS–PAGE gel showing similar GST protein amounts used for the binding assays and the initial and final structures of the apo-CaM–IQ and Ca2+-CaM–IQ complexes (PDF)
REFERENCES
- (1).Hedman AC, Smith JM, and Sacks DB (2015) The biology of IQGAP proteins: beyond the cytoskeleton. EMBO Rep. 16, 427–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Shannon KB (2012) IQGAP Family Members in Yeast, Dictyostelium, and Mammalian Cells. Int. J. Cell Biol 2012, 894817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Choi S, and Anderson RA (2016) IQGAP1 is a phosphoinositide effector and kinase scaffold. Adv. Biol. Regul 60, 29–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Wang S, Watanabe T, Noritake J, Fukata M, Yoshimura T, Itoh N, Harada T, Nakagawa M, Matsuura Y, Arimura N, and Kaibuchi K (2007) IQGAP3, a novel effector of Rac1 and Cdc42, regulates neurite outgrowth. J. Cell Sci 120, 567–577. [DOI] [PubMed] [Google Scholar]
- (5).Briggs MW, and Sacks DB (2003) IQGAP proteins are integral components of cytoskeletal regulation. EMBO Rep. 4, 571–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).White CD, Brown MD, and Sacks DB (2009) IQGAPs in cancer: A family of scaffold proteins underlying tumorigenesis. FEBS Lett. 583, 1817–1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).White CD, Erdemir HH, and Sacks DB (2012) IQGAP1 and its binding proteins control diverse biological functions. Cell. Signalling 24, 826–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Schmidt VA, Chiariello CS, Capilla E, Miller F, and Bahou WF (2008) Development of hepatocellular carcinoma in Iqgap2-deficient mice is IQGAP1 dependent. Mol. Cell. Biol 28, 1489–1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Noritake J, Watanabe T, Sato K, Wang S, and Kaibuchi K (2005) IQGAP1: a key regulator of adhesion and migration. J. Cell Sci 118, 2085–2092. [DOI] [PubMed] [Google Scholar]
- (10).Mataraza JM, Briggs MW, Li Z, Entwistle A, Ridley AJ, and Sacks DB (2003) IQGAP1 promotes cell motility and invasion. J. Biol. Chem 278, 41237–41245. [DOI] [PubMed] [Google Scholar]
- (11).Wang JB, Sonn R, Tekletsadik YK, Samorodnitsky D, and Osman MA (2009) IQGAP1 regulates cell proliferation through a novel CDC42-mTOR pathway. J. Cell Sci 122, 2024–2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Brown MD, and Sacks DB (2006) IQGAP1 in cellular signaling: bridging the GAP. Trends Cell Biol. 16, 242–249. [DOI] [PubMed] [Google Scholar]
- (13).Meyer RD, Sacks DB, and Rahimi N (2008) IQGAP1-dependent signaling pathway regulates endothelial cell proliferation and angiogenesis. PLoS One 3, No. e3848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Yin N, Shi J, Wang DP, Tong T, Wang MR, Fan FY, and Zhan QM (2012) IQGAP1 interacts with Aurora-A and enhances its stability and its role in cancer. Biochem. Biophys. Res. Commun 421, 64–69. [DOI] [PubMed] [Google Scholar]
- (15).Jameson KL, Mazur PK, Zehnder AM, Zhang JJ, Zarnegar B, Sage J, and Khavari PA (2013) IQGAP1 scaffold-kinase interaction blockade selectively targets RAS-MAP kinase-driven tumors. Nat. Med 19, 626–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Jadeski L, Mataraza JM, Jeong HW, Li Z, and Sacks DB (2008) IQGAP1 stimulates proliferation and enhances tumorigenesis of human breast epithelial cells. J. Biol. Chem 283, 1008–1017. [DOI] [PubMed] [Google Scholar]
- (17).Ren JG, Li Z, Crimmins DL, and Sacks DB (2005) Self-association of IQGAP1: characterization and functional sequelae. J. Biol. Chem 280, 34548–34557. [DOI] [PubMed] [Google Scholar]
- (18).LeCour L, Boyapati VK, Liu J, Li ZG, Sacks DB, and Worthylake DK (2016) The Structural Basis for Cdc42-Induced Dimerization of IQGAPs. Structure 24, 1499–1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Ozdemir ES, Jang H, Gursoy A, Keskin O, Li Z, Sacks DB, and Nussinov R (2018) Unraveling the molecular mechanism of interactions of the Rho GTPases Cdc42 and Rac1 with the scaffolding protein IQGAP2. J. Biol. Chem 293, 3685–3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Liu J, Kurella VB, LeCour L, Vanagunas T, and Worthylake DK (2016) The IQGAP1 N-Terminus Forms Dimers, and the Dimer Interface Is Required for Binding F-Actin and Calcium-Bound Calmodulin. Biochemistry 55, 6433–6444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Bahler M, and Rhoads A (2002) Calmodulin signaling via the IQ motif. FEBS Lett. 513, 107–113. [DOI] [PubMed] [Google Scholar]
- (22).Ishida H, Nakashima K, Kumaki Y, Nakata M, Hikichi K, and Yazawa M (2002) The solution structure of apocalmodulin from Saccharomyces cerevisiae implies a mechanism for its unique Ca2+ binding property. Biochemistry 41, 15536–15542. [DOI] [PubMed] [Google Scholar]
- (23).Zhang M, Tanaka T, and Ikura M (1995) Calcium-induced conformational transition revealed by the solution structure of apo calmodulin. Nat. Struct. Mol. Biol 2, 758–767. [DOI] [PubMed] [Google Scholar]
- (24).Tidow H, and Nissen P (2013) Structural diversity of calmodulin binding to its target sites. FEBS J. 280, 5551–5565. [DOI] [PubMed] [Google Scholar]
- (25).Zhao L, Lai L, and Zhang Z (2019) How calcium ion binding induces the conformational transition of the calmodulin N-terminal domain-an atomic level characterization. Phys. Chem. Chem. Phys 21, 19795–19804. [DOI] [PubMed] [Google Scholar]
- (26).Ren JG, Li Z, and Sacks DB (2008) IQGAP1 integrates Ca2+/calmodulin and B-Raf signaling. J. Biol. Chem 283, 22972–22982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Briggs MW, Li Z, and Sacks DB (2002) IQGAP1-mediated stimulation of transcriptional co-activation by beta-catenin is modulated by calmodulin. J. Biol. Chem 277, 7453–7465. [DOI] [PubMed] [Google Scholar]
- (28).Wolenski JS (1995) Regulation of calmodulin-binding myosins. Trends Cell Biol. 5, 310–316. [DOI] [PubMed] [Google Scholar]
- (29).Ho YD, Joyal JL, Li Z, and Sacks DB (1999) IQGAP1 integrates Ca2+/calmodulin and Cdc42 signaling. J. Biol. Chem 274, 464–470. [DOI] [PubMed] [Google Scholar]
- (30).Joyal JL, Annan RS, Ho YD, Huddleston ME, Carr SA, Hart MJ, and Sacks DB (1997) Calmodulin modulates the interaction between IQGAP1 and Cdc42. J. Biol. Chem 272, 15419–15425. [DOI] [PubMed] [Google Scholar]
- (31).Li ZG, Kim SH, Higgins JMG, Brenner MB, and Sacks DB (1999) IQGAP1 and calmodulin modulate E-cadherin function. J. Biol. Chem 274, 37885–37892. [DOI] [PubMed] [Google Scholar]
- (32).Briggs MW, Li ZG, and Sacks DB (2002) IQGAP1-mediated stimulation of transcriptional co-activation by beta-catenin is modulated by calmodulin. J. Biol. Chem 277, 7453–7465. [DOI] [PubMed] [Google Scholar]
- (33).Jeong HW, Li Z, Brown MD, and Sacks DB (2007) IQGAP1 binds Rap1 and modulates its activity. J. Biol. Chem 282, 20752–20762. [DOI] [PubMed] [Google Scholar]
- (34).McNulty DE, Li ZG, White CD, Sacks DB, and Annan RS (2011) MAPK Scaffold IQGAP1 Binds the EGF Receptor and Modulates Its Activation. J. Biol. Chem 286, 15010–15021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Mateer SC, McDaniel AE, Nicolas V, Habermacher GM, Lin MJ, Cromer DA, King ME, and Bloom GS (2002) The mechanism for regulation of the F-actin binding activity of IQGAP1 by calcium/calmodulin. J. Biol. Chem 277, 12324–12333. [DOI] [PubMed] [Google Scholar]
- (36).Nussinov R, Zhang M, Tsai CJ, and Jang H (2018) Calmodulin and IQGAP1 activation of PI3Kalpha and Akt in KRAS, HRAS and NRAS-driven cancers. Biochim. Biophys. Acta, Mol. Basis Dis 1864, 2304–2314. [DOI] [PubMed] [Google Scholar]
- (37).Zhang M, Jang H, Gaponenko V, and Nussinov R (2017) Phosphorylated Calmodulin Promotes PI3K Activation by Binding to the SH2 Domains. Biophys. J 113, 1956–1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Nussinov R, Tsai CJ, Muratcioglu S, Jang H, Gursoy A, and Keskin O (2015) Principles of K-Ras effector organization and the role of oncogenic K-Ras in cancer initiation through G1 cell cycle deregulation. Expert Rev. Proteomics 12, 669–682. [DOI] [PubMed] [Google Scholar]
- (39).Nussinov R, Wang G, Tsai CJ, Jang H, Lu S, Banerjee A, Zhang J, and Gaponenko V (2017) Calmodulin and PI3K Signaling in KRAS Cancers. Trends Cancer 3, 214–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Li L, Li Z, and Sacks DB (2005) The transcriptional activity of estrogen receptor-alpha is dependent on Ca2+/calmodulin. J. Biol. Chem 280, 13097–13104. [DOI] [PubMed] [Google Scholar]
- (41).Li Z, and Sacks DB (2003) Elucidation of the interaction of calmodulin with the IQ motifs of IQGAP1. J. Biol. Chem 278, 4347–4352. [DOI] [PubMed] [Google Scholar]
- (42).Wu S, and Zhang Y (2007) LOMETS: a local meta-threading-server for protein structure prediction. Nucleic Acids Res. 35, 3375–3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Zheng W, Zhang C, Wuyun Q, Pearce R, Li Y, and Zhang Y (2019) LOMETS2: improved meta-threading server for fold-recognition and structure-based function annotation for distant-homology proteins. Nucleic Acids Res. 47, W429–W436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, and Schulten K (2005) Scalable molecular dynamics with NAMD. J. Comput. Chem 26, 1781–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Brooks BR, Brooks CL, Mackerell AD, Nilsson L, Petrella RJ, Roux B, Won Y, Archontis G, Bartels C, Boresch S, Caflisch A, Caves L, Cui Q, Dinner AR, Feig M, Fischer S, Gao J, Hodoscek M, Im W, Kuczera K, Lazaridis T, Ma J, Ovchinnikov V, Paci E, Pastor RW, Post CB, Pu JZ, Schaefer M, Tidor B, Venable RM, Woodcock HL, Wu X, Yang W, York DM, and Karplus M (2009) CHARMM: The Biomolecular Simulation Program. J. Comput. Chem 30, 1545–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Li Z, Zhang Y, Hedman AC, Ames JB, and Sacks DB (2017) Calmodulin Lobes Facilitate Dimerization and Activation of Estrogen Receptor-alpha. J. Biol. Chem 292, 4614–4622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Houdusse A, Gaucher JF, Krementsova E, Mui S, Trybus KM, and Cohen C (2006) Crystal structure of apo-calmodulin bound to the first two IQ motifs of myosin V reveals essential recognition features. Proc. Natl. Acad. Sci U. S. A 103, 19326–19331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Baek K, Brown RS, Birrane G, and Ladias JA (2007) Crystal structure of human cyclin K, a positive regulator of cyclin-dependent kinase 9. J. Mol. Biol 366, 563–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Chichili VPR, Xiao Y, Seetharaman J, Cummins TR, and Sivaraman J (2013) Structural basis for the modulation of the neuronal voltage-gated sodium channel NaV1.6 by calmodulin. Sci. Rep 3, 2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Kumar V, Chichili VP, Zhong L, Tang X, Velazquez-Campoy A, Sheu FS, Seetharaman J, Gerges NZ, and Sivaraman J (2013) Structural basis for the interaction of unstructured neuron specific substrates neuromodulin and neurogranin with Calmodulin. Sci. Rep 3, 1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Li J, Chen Y, Deng Y, Unarta IC, Lu Q Huang X, and Zhang M (2017) Ca(2+)-Induced Rigidity Change of the Myosin VIIa IQ Motif-Single alpha Helix Lever Arm Extension. Structure 25, 579–591.e574. [DOI] [PubMed] [Google Scholar]
- (52).Chagot B, and Chazin WJ (2011) Solution NMR structure of Apo-calmodulin in complex with the IQ motif of human cardiac sodium channel NaV1.5. J. Mol. Biol 406, 106–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Meador WE, Means AR, and Quiocho FA (1992) Target enzyme recognition by calmodulin: 2.4 A structure of a calmodulin-peptide complex. Science 257, 1251–1255. [DOI] [PubMed] [Google Scholar]
- (54).Fallon JL, Halling DB, Hamilton SL, and Quiocho FA (2005) Structure of calmodulin bound to the hydrophobic IQ domain of the cardiac Ca(v)1.2 calcium channel. Structure 13, 1881–1886. [DOI] [PubMed] [Google Scholar]
- (55).Trybus KM, Gushchin MI, Lui H, Hazelwood L, Krementsova EB, Volkmann N, and Hanein D (2007) Effect of calcium on calmodulin bound to the IQ motifs of myosin V. J. Biol. Chem 282, 23316–23325. [DOI] [PubMed] [Google Scholar]
- (56).Jang H, Banerjee A, Chavan T, Gaponenko V, and Nussinov R (2017) Flexible-body motions of calmodulin and the farnesylated hypervariable region yield a high-affinity interaction enabling K-Ras4B membrane extraction. J. Biol. Chem 292, 12544–12559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Lui WY, Mruk DD, and Cheng CY (2005) Interactions among IQGAP1, Cdc42, and the cadherin/catenin protein complex regulate sertoli-germ cell adherens junction dynamics in the testis. J. Cell. Physiol 202, 49–66. [DOI] [PubMed] [Google Scholar]
- (58).Wallrabe H, Cai Y, Sun Y, Periasamy A, Luzes R, Fang X, Kan HM, Cameron LC, Schafer DA, and Bloom GS (2013) IQGAP1 interactome analysis by in vitro reconstitution and live cell 3-color FRET microscopy. Cytoskeleton 70, 819–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Brown MD, Bry L, Li ZG, and Sacks DB (2008) Actin Pedestal Formation by Enteropathogenic Escherichia coli Is Regulated by IQGAP1, Calcium, and Calmodulin. J. Biol. Chem 283, 35212–35222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Briggs MW, and Sacks DB (2003) IQGAP1 as signal integrator: Ca2+, calmodulin, Cdc42 and the cytoskeleton. FEBS Lett. 542, 7–11. [DOI] [PubMed] [Google Scholar]
- (61).Joyal JL, Annan RS, Ho YD, Huddleston ME, Carr SA, Hart MJ, and Sacks DB (1997) Calmodulin modulates the interaction between IQGAP1 and Cdc42. Identification of IQGAP1 by nanoelectrospray tandem mass spectrometry. J. Biol. Chem 272, 15419–15425. [DOI] [PubMed] [Google Scholar]
- (62).Andrews WJ, Bradley CA, Hamilton E, Daly C, Mallon T, and Timson DJ (2012) A calcium-dependent interaction between calmodulin and the calponin homology domain of human IQGAP1. Mol. Cell. Biochem 371, 217–223. [DOI] [PubMed] [Google Scholar]
- (63).Kholmanskikh SS, Koeller HB, Wynshaw-Boris A, Gomez T, Letourneau PC, and Ross ME (2006) Calcium-dependent interaction of Lis1 with IQGAP1 and Cdc42 promotes neuronal motility. Nat. Neurosci 9, 50–57. [DOI] [PubMed] [Google Scholar]
- (64).Chen F, Zhu HH, Zhou LF, Wu SS, Wang J, and Chen Z (2010) IQGAP1 is overexpressed in hepatocellular carcinoma and promotes cell proliferation by Akt activation. Exp Mol. Med 42, 477–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Joyal JL, Burks DJ, Pons S, Matter WF, Vlahos CJ, White MF, and Sacks DB (1997) Calmodulin activates phosphatidylinositol 3-kinase. J. Biol. Chem 272, 28183–28186. [DOI] [PubMed] [Google Scholar]
- (66).Zhang M, Jang H, and Nussinov R (2019) The mechanism of PI3Kα activation at the atomic level. Chem. Sci 10, 3671–3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Choi S, Hedman AC, Sayedyahossein S, Thapa N, Sacks DB, and Anderson RA (2016) Agonist-stimulated phosphatidylinositol-3,4,5-trisphosphate generation by scaffolded phosphoinositide kinases. Nat. Cell Biol 18, 1324–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.