Abstract
Aging of the nervous system is typified by depressed metabolism, compromised proteostasis, and increased inflammation that results in cognitive impairment. Differential expression analysis is a popular technique for exploring the molecular underpinnings of neural aging, but technical drawbacks of the methodology often obscure larger expression patterns. Co-expression analysis offers a robust alternative that allows for identification of networks of genes and their putative central regulators. In an effort to expand upon previous work exploring neural aging in the marine model Aplysia californica, we used weighted gene correlation network analysis to identify co-expression networks in a targeted set of aging sensory neurons in these animals. We identified twelve modules, six of which were strongly positively or negatively associated with aging. Kyoto Encyclopedia of Genes analysis and investigation of central module transcripts identified signatures of metabolic impairment, increased reactive oxygen species, compromised proteostasis, disrupted signaling, and increased inflammation. Although modules with immune character were identified, there was no correlation between genes in Aplysia that increased in expression with aging and the orthologous genes in oyster displaying long-term increases in expression after a virus-like challenge. This suggests anti-viral response is not a driver of Aplysia sensory neuron aging.
1 Introduction
As an organ composed of long-lived cells, the brain is uniquely susceptible to the deleterious effects of aging, the outcome of which is often cognitive impairment [1,2]. Common hallmarks of brain aging include impaired metabolism, compromised proteostasis, mitochondrial dysfunction, and neuro-inflammation [3–5]. However, what causes these hallmark phenotypes is not well understood and still debated [6–8]. Due to the complexity of mammalian brains, invertebrate models are often employed for the study of aging in the nervous system at the neuronal level.
The marine model Aplysia californica is well suited for study of aging neurons. These mollusks live approximately one year in the wild and mariculture setting and have relatively simple nervous systems of only approximately 10,000 neurons grouped into well mapped circuits [9–11]. Studies on the life history and reflex behaviors of these animals in controlled environments have described weight loss and cognitive impairment in old age similar to that of other animals and humans [12–16]. Investigation of physiology and transcriptomics in easily identifiable neurons and neuron clusters during aging have elucidated neuronal correlates that underpin aging phenotypes at the behavior and organism level [17–23]. Indeed, it is the capacity for vertical integration of investigation, from molecular to behavioral, that makes Aplysia such an effective model for the aging nervous system. Previously, transcriptomic studies in aging sensory neurons of Aplysia identified signatures of common aging hallmarks, namely metabolic and proteostatic impairment [20,24]. However, due to the limitations of differential expression analysis (DEA), including log fold change thresholds and multiple comparison [25], the driving mechanism behind these transcriptional changes could not be identified. A common alternative analysis that can overcome these limitations is weighted gene correlation network analysis (WGCNA).
WGCNA is able to lessen the impact of correction for multiple tests by comparing changes in groups of genes, called modules, as opposed to individual genes and does not employ the strict fold change thresholds used in DEA [25,26]. This method can capture network level changes that integrate the small, coordinated changes of many genes that would otherwise be below the thresholds employed in DEA [27]. Furthermore, WGCNA allows for the identification of putative central driver genes in co-expression networks and inference of putative function for genes that are undescribed or have yet to be experimentally verified via guilt-by-association [26,28,29]. In this study we used WGCNA and eigengene network analysis to identify the central drivers of the transcriptional aging phenotype of Aplysia SN.
2 Methods
2.1 Experimental design
Sequencing data for this study were generated in our previous study [24]. RNA was extracted from A. californica Buccal S Cluster (BSC) and Pleural Ventral Caudal (PVC) sensory neurons across the adult aging spectrum (6–12 months).
2.2 Raw read processing
Raw, 150 base pair, paired end RNA reads from our previous study, available at the NCBI (PRJNA639857), were processed as described previously [24]. Briefly, raw reads were adapter trimmed and quality filtered using the BBDUK software [30] and then quantified by the Salmon software package [31] using the Aplysia californica reference transcriptome from the NCBI ftp site (AplCal3.0 GCF000002075.1).
Salmon derived transcript abundances were imported into the R statistical environment via the tximport R package [32]. Transcripts with a sum total abundance of less than 1 transcript per million (TPM) across all samples were considered not expressed and filtered out. The R package geneFilter was used to remove low variance transcripts using the function varFilter() with the default parameters var.func = IQR and var.cutoff = 0.5 to filter out transcripts with interquartile range (IQR) smaller than the median of all IQR in the expression data [33]. Surrogate variables were identified using the SVA R package [34]. Abundances were then variance stabilized using the vst() function from the DESeq2 R package, with surrogate variables incorporated into the model design and the blind parameter set to FALSE to account of said surrogate variables [35].
2.3 Co-expression analysis
Variance stabilized counts from BSC and PVC samples were used to construct sensory neuron type specific co-expression networks using the WGCNA R package. Briefly, Biweighted midcorrelation in WGCNA was used to construct adjacency matrices for each sensory neuron type independently. A soft power of 16 was used for both PVC and BSC networks to achieve at least 0.8 metric for scale free topology and to minimize mean connectivity (S1 Fig). Adjacency matrices were used to construct signed topological overlap matrices (TOM) for each sensory neuron type. The sensory neuron type-specific TOMs were centered and scaled and combined to make a consensus TOM by taking the minimum of the two modules. Transcripts were hierarchically clustered with the average method using a distance metric of 1-consensus TOM and a minimum module size of 30.
Initial module eigengenes were hierarchically clustered to determine module similarity. Modules with a branch height of 0.25 or less were deemed insufficiently different from their neighbors and merged. Module eigengenes for merged modules were then recalculated (S2 and S3 Figs). Consensus module eigengenes were then correlated to animal chronological age.
Individual transcripts in each module were then correlated to the module eigengene, referred to as module membership, as a measure of the transcript centrality.
Similarly, transcripts were correlated with chronological age, referred to as transcript-age significance (TAS), as a measure of the influence of chronological age on the expression of that transcript. Correlation of module membership with TAS of all transcripts within a module (MM-TAS) was calculated to describe the influence of chronological age on the module as a whole. Only modules with high magnitude (|Pearson cor| ≥ 0.5) and high degree of significance (p ≤ 0.01) of MM-TAS were considered for further downstream analysis. Module hub transcripts were identified using the WGCNA function chooseTopHubInEachModule().
2.4 Module enrichment analysis
Transcript sets of each module were tested for enrichment for Kyoto Encyclopedia of Genes and Genomes (KEGG) canonical pathways using the clusterProfiler R package [36].
All software used and version information is available in Supplementary Table S1 Table and Supplementary Information S1 File. All scripts used for this analysis can be found in the following GitHub repository: [https://github.com/Nicholas-Kron/Kron_Cohort77_CoExpression_Analysis]
2.5 Comparison to immune response in Crassostrea gigas
To assess whether the observed module immune signatures suggested mapping to KEGG orthology and the UNIPROT human proteome represented a bona fide molluscan immune signature, module transcript sets were compared to differential expression resulting from immune challenge in the pacific oyster Crassostrea gigas by Lafont et al (2020) [37].
The Aplysia RefSeq proteome for the current genome build (AplCal3.0, GCF_000002075.1) was downloaded from the RefSeq database. Similarly, the proteome for the C.gigas genome build used by Lafont et al (2020, oyster_v9, GCA_000297859.1) [37] was downloaded from the Ensemble FTP site. The Aplysia proteome was BLASTed against local BLAST database built from said C.gigas proteome using an e value cutoff of less than 0.001 and selecting only the top hit.
The resultant putative protein orthologs between Aplysia and C.gigas were then mapped to their respective gene and transcripts identifiers using the respective genome build gene feature format annotation files (AplCal3.0_genomic.gff version 1.21 and oyster_v9.49.gff3). The new Aplysia to C. gigas mapping file was used to determine the proportion of each co-expression module that mapped to genes differentially expressed in response to immune challenge either due to exposure to poly(I·C) priming, viral challenge, or both [37]. Enrichment for C. gigas orthologs in each module was calculated by Fisher’s exact test with Bonferroni multiple test correction in R using the dhyper function.
3 Results
3.1 Filtering
Removal of transcripts with zero total count and variance filtering with the geneFilter package yielded 11,703 analysis ready transcripts, out of approximately 12,000 transcripts expressed in these sensory neuron types described previously [24]. SVA identified one surrogate variable which was included in the DESeq2 model design.
3.2 Clustering
Hierarchical clustering of transcripts and module merging resulted in 12 consensus co-expression modules, which were assigned arbitrary color names by WGCNA. A thirteenth module wastebasket module was assigned the “grey” designation and not evaluated in downstream analysis. The royalblue, saddlebrown, orange, and pink module eigengenes exhibited significant correlation with chronological age (p ≤ 0.05; Fig 1). Module-trait correlations in both PVC and BSC individually can be found in Supplementary Figure S4 Fig. The hub transcript for each module can be found in Table 1.
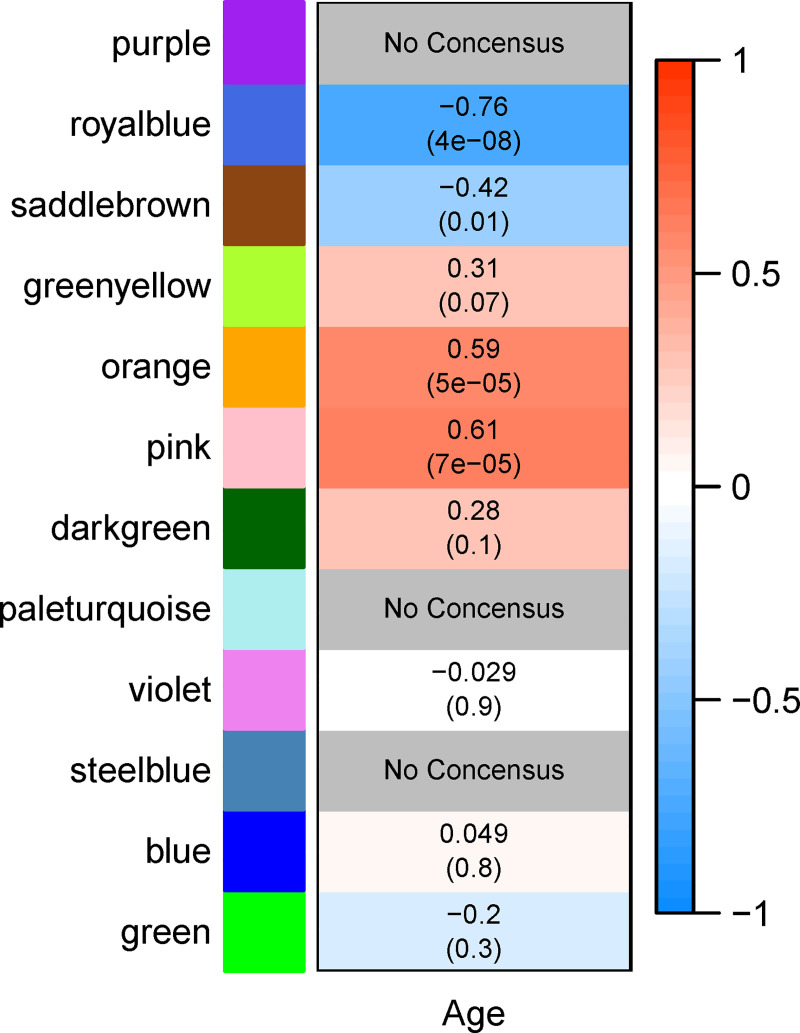
Fig 1. Correlation in Aplysia californica sensory neurons between consensus co-expression modules and animal age.
Each module is arbitrarily assigned a color to assist in reference. This color is denoted on the heatmap row labels of Fig 1 and referred to throughout the figures and tables. Each cell of the heatmap represents the Pearson correlation between a module eigengene (row) and age (column). The upper value within a cell represents the magnitude of correlation. The lower value in parentheses in each cell represents the p-value of the correlation. Cell color denotes direction of correlation (red = positive, blue = negative) and saturation represents magnitude of correlation, with greater magnitude of correlation (top value in each cell) represented by higher saturation. Modules for which sign of eigengene correlation with age between PVC and BSC was inconsistent were colored grey and marked as “No Consensus” due to lack of consensus. Orange and pink modules are significantly correlated with age, while royalblue and saddlebrown are significantly anti-correlated.
Table 1. Co-expression modules identified in Aplysia californica sensory neurons.
| Module | Module n | Hub gene RefSeq ID | Human Ortholog | Ortholog Name |
|---|---|---|---|---|
| blue | 1581 | XM_013088909.1 | MGA | MAX gene-associated protein |
| darkgreen | 225 | XM_013082889.1 | RPA2 | Replication protein A 32 kDa subunit |
| green | 2387 | NM_001204703.1 | GNAO1 | G-protein G(o) subunit alpha |
| greenyellow | 329 | XM_005110768.2 | ZNFX1 | NFX1-type zinc finger-containing protein 1 |
| orange | 166 | XM_005096841.2 | CREB3L3 | Cyclic AMP-responsive element-binding protein 3-like protein 3 |
| paleturquoise | 41 | XM_013087385.1 | - | - |
| pink | 1255 | XM_005111489.2 | NFKBIA | NF-kappa-B inhibitor alpha |
| purple | 3036 | XM_005095177.2 | PSMB7 | Proteasome subunit beta type-7 |
| royalblue | 561 | XM_005101095.2 | - | - |
| saddlebrown | 65 | XM_005089315.2 | - | - |
| steelblue | 64 | XM_013083399.1 | EBF3 | Transcription factor COE3 |
| violet | 36 | XM_013085790.1 | ATXN10 | Ataxin-10 |
Modules were identified using the weighted gene correlation network analysis (WGCNA) R package. The most connected transcript, called the hub gene, is listed by its RefSeq identifier, as well as its BLASTx assigned human ortholog. Hub transcripts with a “-” in the Human Ortholog and Ortholog Name columns could not be mapped to any known human protein, and thus are of unknown function.
3.3 Modules of interest
The expression of each transcript in a module was correlated with the module eigengene, called module membership (MM), and with age, called transcript-age significance (TAS). Modules for which MM and TAS were highly correlated (|Pearson cor| ≥ 0.5, p ≤ 0.01) were investigated further.
Both the royalblue and saddlebrown modules were significantly anti-correlated with age. Eigengene expression trend of both modules was stable until age nine months when expression decreased monotonically (Fig 2). Module membership was highly correlated with the transcript-age significance for age (TAS) in the royalblue module (Pearson cor ≥ 0.8, p ≤ 0.001, Fig 3). However, MM-TAS was not strongly correlated for the saddlebrown module and thus this module was not investigated further (Pearson cor = 0.35, p = 0.004, Fig 3).
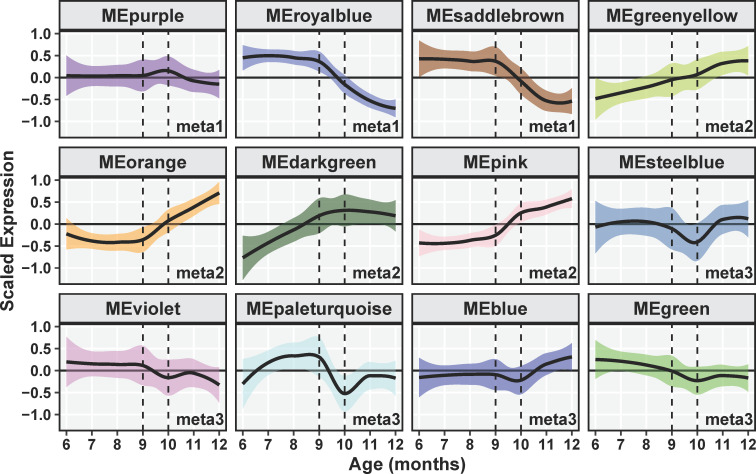
Fig 2. Expression trajectories of consensus co-expression module eigengenes of Aplysia californica sensory neurons over the adult lifespan.
Each cell represents the mean centered and variance scaled expression of a module eigengene, with the solid line the monthly average with colored bounding area representing the standard error. Dotted lines highlight the transition at age 9–10 months, during which most module eigengenes exhibit perturbations of their expression trends.
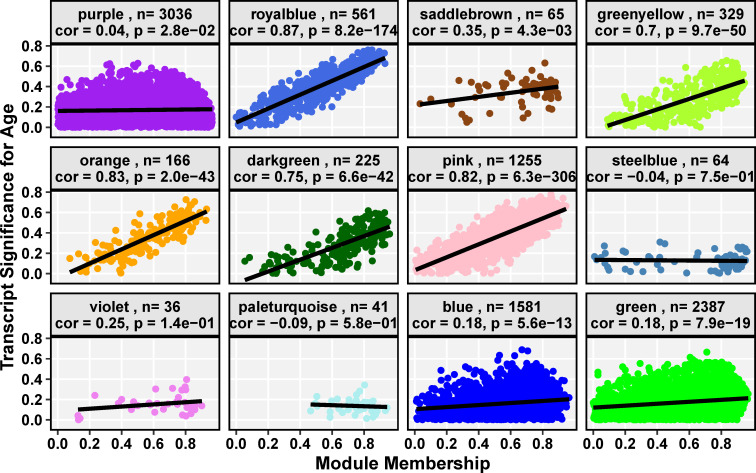
Fig 3. Trait significance–module membership correlation (TS-MM) for each co-expression module of Aplysia californica sensory neurons over the adult lifespan.
The x-axis of each cell is the Pearson correlation of the expression of a transcript and the module eigengene. The y-axis of each cell is the Pearson correlation of the expression of a transcript and chronological age in months. Each module transcript is plotted as a colored point, while the line of best fit, which represents the TS-MM, is rendered in black. Header strips detail the module name, the number of transcripts in that module (n), the TS-MM Pearson correlation value, and the p-value significance of that correlation for each module as calculated by the WGCNA R package. The darkgreen, greenyellow, orange, pink, and royalblue modules have a significant TS-MM ≥ 0.7.
Eigengenes of the pink, orange, darkgreen, and greenyellow modules exhibited increasing expression trends with increasing chronological age for at least some portion of the age span (Fig 2). The pink and orange module eigengenes were significantly correlated with chronological age (p ≤ 0.05, Fig 1) and both modules exhibited high MM-TAS correlation (Pearson cor ≥ 0.7, p ≤ 0.001; Fig 2). Notably, the expression profile of the orange and pink eigengenes resemble the royalblue and saddlebrown eigengene mirrored over the x-axis, exhibiting a linear increase in expression after age 9 months. The greenyellow module exhibited a linear increase in eigengene expression with age across the entire aging span, while darkgreen exhibited increasing expression from ages 6–9 months (Fig 2). While the module eigengenes of these two modules were not significantly correlated with age, both modules exhibited strong MM-TAS correlation (Pearson cor ≥ 0.7, p ≤ 0.001; Fig 3). This suggests aging strongly affects the central regulators of these modules, a notion sufficiently interesting to justify further investigation of these modules.
Many of the age associated modules overlapped with transcriptional trajectories of transcripts differentially expressed in aging in our previous study (Supplementary table S2 Table).
3.4 Enrichment analysis of modules of interest
3.4.1 Royalblue
The royalblue module, which exhibited decreasing eigengene expression trend with age, was enriched for a diverse set of KEGG pathways (Fig 4). Most prominent were the canonical energy metabolism pathways of glycolysis (ko00010), TCA cycle (ko00020), and fatty acid metabolism (ko01212). Amino acid metabolism related pathways such as biosynthesis of amino acids (ko01230) and alanine, aspartate, and glutamate metabolism (ko00250 were also enriched. Another set of enriched KEGG pathways represent elements important to neuronal function such as synaptic vesicle cycle (ko04721), long-term potentiation (ko04720), MAPK signaling (ko04013), and calcium signaling (ko04020). Several enriched pathways represent human neurodegenerative diseases that sit at the nexus of neuronal dysfunction and metabolic failure such as Alzheimer’s disease (ko05010), Huntington’s disease (ko05016), and Parkinson’s disease (ko05012).
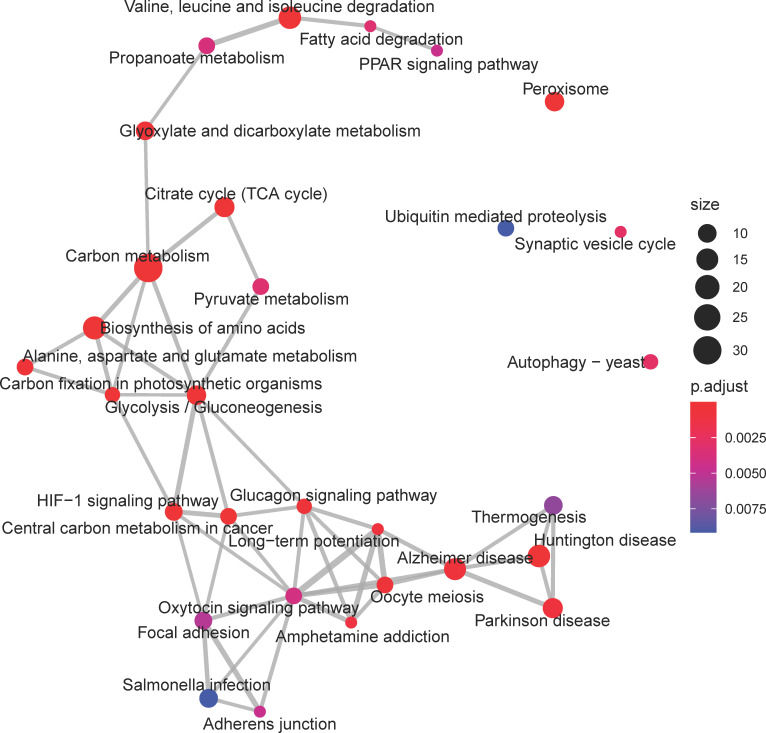
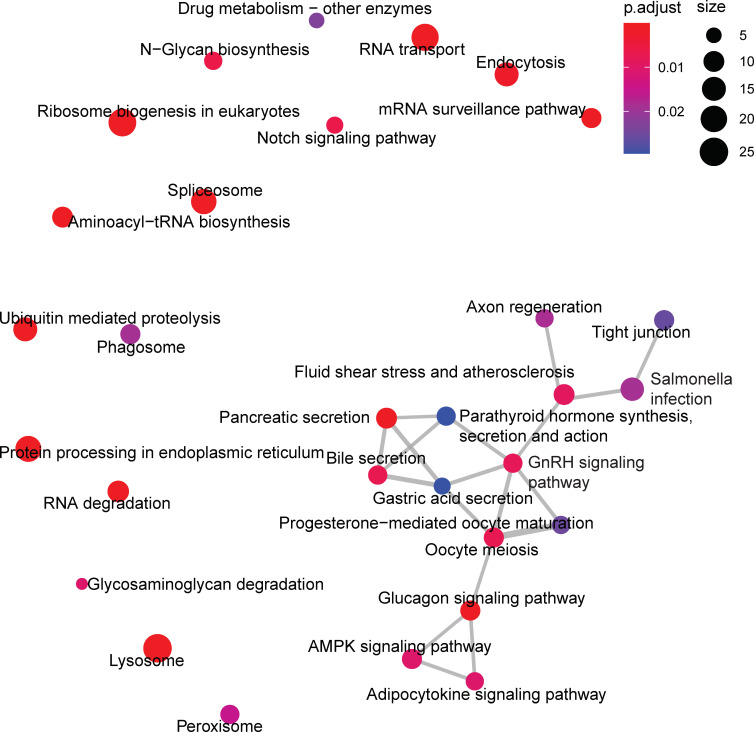
Fig 4. Enrichment map of Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways for the royalblue consensus co-expression module from Aplysia californica sensory neurons.
Each node represents a KEGG pathway, with node size representing the number of transcripts annotated to that pathway, and color denoting the significance of that enrichment (brighter red is most significant). KEGG pathways with overlapping transcripts sets are connected by grey lines, or edges. Edge width is determined by the number of overlapping transcripts. This module exhibits a high degree of gene set overlap between most of the enriched KEGG pathways. Metabolic pathways such as TCA cycle, Glycolysis, and fatty acid degradation are among the largest and most significantly enriched. The module eigengene trend of this module was negatively correlated with age, indicating downregulation of these pathways.
Orthologs also involved in anterograde or retrograde movement of cellular cargo featured prominently among the transcripts with highest module membership in this module. Metabolic enzymes associated with TCA cycle, glycolysis, and mitochondrial fatty acid beta oxidation were also prominent. Reactive oxygen species (ROS) detoxification enzymes and other mitochondrial homeostasis maintenance orthologs were also present (Table 2).
Table 2. Selection of transcripts with highest correlation to transcript co-expression module eigengene (module membership, MM) in the royalblue consensus module identified in Aplysia californica sensor neurons by WGCNA.
| Refseq ID | MM | MM p value | TAS | TAS p value | Human Ortholog | Ortholog Name | Ortholog Function |
|---|---|---|---|---|---|---|---|
| XM_005096347.2** | 0.93 | 9.9E-33 | -0.63 | 2.95E-09 | DCTN6 | Dynactin subunit 6 | Transport of cellular cargo [38] |
| XM_005105237.1** | 0.93 | 6.4E-32 | -0.73 | 2.46E-13 | GPI | Glucose-6-phosphate isomerase | glycolysis, neurotrophic factor [39] |
| XM_005093202.2** | 0.89 | 2.8E-25 | -0.70 | 7.42E-12 | NAPG | Gamma-soluble NSF attachment protein | Required for vesicular transport between the endoplasmic reticulum and the Golgi apparatus. (UniProt) |
| XM_005101715.2** | 0.88 | 1.1E-24 | -0.70 | 6.98E-12 | EXOC2 | Exocyst complex component 2 | Component of exocyst complex (UniProt) |
| XM_005096859.2** | 0.86 | 1.1E-22 | -0.67 | 1.09E-10 | GPX4 | Phospholipid hydroperoxide glutathione peroxidase | Antioxidant [40] |
| XM_005089329.2** | 0.86 | 3.2E-22 | -0.71 | 1.84E-12 | MAP2K1 | Dual specificity mitogen-activated protein kinase kinase 1 | MEK 1 |
| XM_005105342.2* | 0.85 | 2.2E-21 | -0.76 | 5.45E-15 | SNX30 | Sorting nexin-30 | Possibly intracellular trafficking (UniProt) |
| XM_005103941.2* | 0.84 | 5.9E-21 | -0.55 | 4.21E-07 | SDHAF2 | Succinate dehydrogenase assembly factor 2, mitochondrial | Assembly and function of succinate dehydrogenase complex [41] |
| XM_013079678.1* | 0.84 | 1.0E-20 | -0.66 | 1.71E-10 | VAPA | Vesicle-associated membrane protein-associated protein A | vesicular transport between the endoplasmic reticulum and the Golgi apparatus [42] |
| XM_005106229.2** | 0.84 | 2.9E-20 | -0.57 | 1.83E-07 | ACADS | Short-chain specific acyl-CoA dehydrogenase, mitochondrial | catalyze the first step of mitochondrial fatty acid beta-oxidation [43] |
| XM_005112738.2** | 0.83 | 5.4E-20 | -0.59 | 4.64E-08 | ENO1 | Alpha-enolase | glycolysis |
| XM_013088411.1** | 0.83 | 6.5E-20 | -0.59 | 3.18E-08 | CCDC151 | Coiled-coil domain-containing protein 151 | dynein arm assembly [44] |
| XM_005098999.2** | 0.83 | 8.4E-20 | -0.61 | 7.88E-09 | PARK7 | Protein/nucleic acid deglycase DJ-1 | Antioxidant, neuroprotection [45–47] |
| XM_005096727.2** | 0.83 | 2.7E-19 | -0.61 | 1.13E-08 | GDAP1 | Ganglioside-induced differentiation-associated protein 1 | Regulator of mitochondrial network [48] |
| XM_005111161.2 | 0.79 | 7.9E-17 | -0.56 | 2.47E-07 | |||
| XM_005109966.2** | 0.82 | 3.2E-19 | -0.76 | 6.07E-15 | HADHA | Trifunctional enzyme subunit alpha, mitochondrial | catalyzes the last three of the four reactions of the mitochondrial beta-oxidation pathway [49] |
| XM_005098946.2 | 0.82 | 1.3E-18 | -0.54 | 7.55E-07 | C1orf194 | Uncharacterized protein C1orf194 | Ca2+ homeostasis [50] |
| XM_013081239.1** | 0.82 | 1.3E-18 | -0.53 | 1.23E-06 | PGK1 | Phosphoglycerate kinase 1 | glycolysis |
| XM_005089581.2* | 0.82 | 1.5E-18 | -0.56 | 2.27E-07 | EFCAB1 | EF-hand calcium-binding domain-containing protein 1 | Also called calaxin, binds Ca2+ and dynein [51] |
| XM_005092435.2 | 0.81 | 1.9E-18 | -0.57 | 1.84E-07 | CHMP6 | Charged multivesicular body protein 6 | ESCR-III complex, endosomal cargo sorting [52] |
| NM_001204580.1* | 0.81 | 2.1E-18 | -0.53 | 1.59E-06 | CALM2 | Calmodulin-2 (Fragment) | Ca2+ homeostasis [53] |
| XM_005104646.2** | 0.81 | 2.2E-18 | -0.55 | 4.29E-07 | SOD2 | Superoxide dismutase [Mn], mitochondrial | ROS defense [54] |
| XM_005090003.2 | 0.81 | 3.1E-18 | -0.57 | 1.39E-07 | CAT | Catalase | ROS defense [55] |
| XM_005097336.2* | 0.81 | 3.2E-18 | -0.55 | 3.87E-07 | BLOC1S1 | Biogenesis of lysosome-related organelles complex 1 subunit 1 | anterograde transport [56] |
| XM_005089746.2** | 0.81 | 3.8E-18 | -0.58 | 5.48E-08 | PGAM2 | Phosphoglycerate mutase 2 | glycolysis |
| XM_005108968.2** | 0.81 | 4.0E-18 | -0.64 | 1.09E-09 | GPS2 | G protein pathway suppressor 2 (Fragment) | mitochondrial retrograde signaling, mitochondrial biogenesis, transcriptional activator of nuclear-encoded mitochondrial genes [57] |
| NM_001280826.1** | 0.81 | 5.0E-18 | -0.57 | 1.90E-07 | GAPDH | Glyceraldehyde-3-phosphate dehydrogenase | glycolysis |
| XM_005097828.2** | 0.81 | 5.5E-18 | -0.57 | 1.92E-07 | PDHB | Pyruvate dehydrogenase E1 component subunit beta, mitochondrial | pyruvate dehydrogenase |
| XM_005104734.2** | 0.81 | 7.3E-18 | -0.55 | 3.70E-07 | SDHA | Succinate dehydrogenase [ubiquinone] flavoprotein subunit, mitochondrial | TCA and OXPHOS |
| XM_005100966.2** | 0.80 | 1.9E-17 | -0.62 | 6.83E-09 | DECR2 | Peroxisomal 2,4-dienoyl-CoA reductase | mitochondrial fatty acid beta-oxidation [58] |
| XM_013090573.1* | 0.80 | 2.1E-17 | -0.56 | 2.64E-07 | DLST | Dihydrolipoyllysine-residue succinyltransferase component of 2-oxoglutarate dehydrogenase complex, mitochondrial | TCA |
| XM_005091339.2** | 0.80 | 2.2E-17 | -0.61 | 8.96E-09 | ETFA | Electron transfer flavoprotein subunit alpha, mitochondrial | Electron acceptor, mitochondrial fatty acid beta-oxidation [59] |
| XM_013079116.1* | 0.80 | 2.3E-17 | -0.54 | 6.66E-07 | SYT4 | Synaptotagmin-4 | Retrograde signaling, endocytosis, Ca2+ sensing [60,61] |
| XM_005106740.2** | 0.80 | 2.8E-17 | -0.64 | 8.87E-10 | FUNDC1 | FUN14 domain-containing protein 1 | mitochondrial maintenance [62] |
| XM_005112721.2** | 0.80 | 3.6E-17 | -0.59 | 5.11E-08 | KCNC2 | Potassium voltage-gated channel subfamily C member 2 | ion channel [63] |
| XM_013088705.1 | 0.80 | 4.2E-17 | -0.52 | 2.53E-06 | MFN2 | Mitofusin-2 | mitochondrial fusion [64,65] |
| XM_005105274.2 | 0.79 | 4.8E-17 | -0.63 | 3.04E-09 | CCDC39 | Coiled-coil domain-containing protein 39 | Inner dynein arm assembly [66] |
| XM_013084603.1* | 0.79 | 6.4E-17 | -0.62 | 6.02E-09 | VPS26B | Vacuolar protein sorting-associated protein 26B | Endosome retromer complex [67,68] |
| XM_013079609.1** | 0.79 | 7.4E-17 | -0.56 | 3.10E-07 | IDH3G | Isocitrate dehydrogenase [NAD] subunit gamma, mitochondrial | TCA |
| XM_005091388.2** | 0.79 | 8.9E-17 | -0.68 | 5.00E-11 | NXNL2 | Nucleoredoxin-like protein 2 | ROS defense, neurotrophic factor [69] |
| XM_013087736.1* | 0.79 | 1.0E-16 | -0.52 | 2.89E-06 | NDUFAF2 | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex assembly factor 2 | Mitochondrial complex I assembly and function [70,71] |
| XM_005090280.2** | 0.79 | 1.6E-16 | -0.68 | 2.75E-11 | PDCD6 | Programmed cell death protein 6 | calcium sensor, ER to golgi transport, interacts with ESCRT-III [72] |
| XM_005097549.2* | 0.78 | 2.5E-16 | -0.45 | 5.60E-05 | FMC1 | Protein FMC1 homolog | Plays a role in the assembly/stability of the mitochondrial membrane ATP synthase [73] |
| XM_013084592.1** | 0.78 | 8.4E-16 | -0.56 | 2.79E-07 | ACO2 | Aconitate hydratase, mitochondrial | TCA |
| XM_013086643.1* | 0.76 | 4.3E-15 | -0.55 | 3.79E-07 | KCNAB2 | Voltage-gated potassium channel subunit beta-2 | ion channel subunit, regulates other KCN [74] |
| XM_005110731.2 | 0.76 | 9.3E-15 | -0.50 | 5.63E-06 | SDHC | Succinate dehydrogenase cytochrome b560 subunit, mitochondrial | TCA and OXPHOS |
| XM_005108990.2 | 0.75 | 3.4E-14 | -0.44 | 9.78E-05 | ETFRF1 | Electron transfer flavoprotein regulatory factor 1 | regulator of the electron transfer flavoprotein [75] |
| XM_005104950.2 | 0.74 | 4.2E-14 | -0.53 | 1.70E-06 | KIF3A | Kinesin-like protein | anterograde transport [76] |
| NM_001204727.1* | 0.74 | 9.7E-14 | -0.52 | 2.29E-06 | STAU2 | Double-stranded RNA-binding protein Staufen homolog 2 | transport of neuronal RNA from the cell body to dendrites [77] |
| XM_013089873.1** | 0.74 | 1.3E-13 | -0.61 | 1.08E-08 | ITCH | E3 ubiquitin-protein ligase Itchy homolog | ROS defense [78], Involved in the negative regulation antiviral responses [79] |
| XM_005103012.2** | 0.71 | 3.1E-12 | -0.48 | 2.02E-05 | PRDX5 | Peroxiredoxin-5, mitochondrial | Antioxidant [80] |
The RefSeq ID of each transcript is paired with a human protein ortholog gene symbol and name annotated by BLASTn mapping to the UNIPROT human proteome (UP000005640). The function of each ortholog is detailed in the final column. Correlation of transcript expression and chronological age, transcript-Age correlation (TAS), and significance (TAS p-value) are also listed. Transcripts are marked with “*” if they were among significantly downregulated transcripts in Kron et al 2020 [24] in one neuron type and with “**” if in both. All subsequent tables are organized identically. Common functions include TCA cycle, glycolysis, retrograde and anterograde transport, and calcium homeostasis. The module eigengene trend of this module was negatively correlated with age, indicating downregulation of these processes in aging.
3.4.2 Pink
The pink module, which was positively correlated with age, was significantly enriched for KEGG pathways related to translation, such as ribosome biogenesis in eukaryotes (ko03008) and aminoactyl-tRNA biosynthesis (ko00970), and proteostatic mechanisms, such as lysosome (ko04142), protein processing in the endoplasmic reticulum (ko04141), and ubiquitin mediated proteolysis (ko04120, Fig 5).
Fig 5. Enrichment map of Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways for the pink consensus co-expression module.
Symbol explanation as in Fig 4. The Protein processing in the endoplasmic reticulum, Lysosome, and Ribosome biogenesis in eukaryotes pathways are among the largest and most significantly enriched pathways. This module was positively correlated with age, suggesting these pathways are upregulated in aging.
Investigation of individual transcripts with the highest module membership for the pink module similarly revealed several transcripts annotated to human orthologs involved in transcription, translation, and ribosome biogenesis, as well as modulation of innate immunity and NFkB signaling (Table 3).
Table 3. Selection of transcripts with highest correlation to transcript co-expression module eigengene (module membership, MM) in the pink consensus module identified in Aplysia californica sensory neurons by WGCNA.
| RefSeq ID | MM | MM p value | TAS | TAS p value | Human Ortholog | Ortholog Name | Ortholog Function |
|---|---|---|---|---|---|---|---|
| XM_005111489.2** | 0.95 | 1.8E-36 | 0.66 | 1.4E-10 | NFKBIA | NF-kappa-B inhibitor alpha | NF-kappa-B inhibition [81], anti-inflammatory [82] |
| XM_005111747.2**! | 0.93 | 0.74 | 6.8E-14 | BIRC3 | Baculoviral IAP repeat-containing protein 3 | E3 ubiquitin-protein ligase, NF-kappa-B signaling regulation [83], innate immunity regulation [84] | |
| XM_005106964.2* | 0.84 | 4.1E-32 | 0.56 | 2.3E-20 | |||
| XM_013081148.1 | 0.78 | 3.15E-16 | 0.48 | 2.1E-05 | |||
| XM_005098591.2** | 0.93 | 1.5E-31 | 0.68 | 5.6E-11 | ZUP1 | Zinc finger-containing ubiquitin peptidase 1 | Deubiquitination, DNA damage, replication stress [85] |
| XM_005098861.2 | 0.92 | 1.2E-30 | 0.63 | 2.3E-09 | UBC | Polyubiquitin-C (Fragment) | Ubiquitination [86] |
| XM_005098862.2 | 0.87 | 5.6E-23 | 0.53 | 1.7E-06 | |||
| XM_005105067.2**! | 0.91 | 6.5E-29 | 0.65 | 7.3E-10 | HERC4 | Probable E3 ubiquitin-protein ligase HERC4 | E3 ubiquitin-protein ligase [87] |
| XM_005112342.2** | 0.91 | 6.6E-29 | 0.66 | 1.4E-10 | ZNF343 | Zinc finger protein 343 | transcriptional regulation |
| XM_005108387.2** | 0.91 | 2.1E-28 | 0.63 | 1.7E-09 | ACP7 | Acid phosphatase type 7 | Iron transport, innate immunity, ROS generation [88] |
| XM_005092478.2* | 0.90 | 7.5E-27 | 0.47 | 2.4E-05 | INTS1 | Integrator complex subunit 1 | snRNA [89], eRNA [90], transcriptional attenuation [91,92] |
| XM_005097515.2** | 0.90 | 9.3E-27 | 0.63 | 2.3E-09 | GM2A | Ganglioside GM2 activator | Ganglioside metabolism [93] |
| XM_005111000.2** | 0.90 | 1.3E-26 | 0.61 | 1.1E-08 | GCN1 | eIF-2-alpha kinase activator GCN1 | Global translation repression, gene-specific mRNA translation [94] |
| XM_005090686.2** | 0.89 | 5.2E-26 | 0.60 | 2.4E-08 | SLC16A5 | Monocarboxylate transporter 6 | Glucose and lipid metabolism, possible immune regulation [95] |
| XM_005104555.2** | 0.89 | 6.9E-26 | 0.59 | 3.5E-08 | EFL1 | Elongation factor-like GTPase 1 | Ribosome biogenesis [96] |
| XM_005093568.2** | 0.89 | 7.1E-26 | 0.53 | 1.3E-06 | NAT10 | RNA cytidine acetyltransferase | Ribosome biogenesis [97], E3 ubiquitin-protein ligase, cellular stress sensor [98], translation efficiency [99] |
| XM_005095929.2** | 0.89 | 8.2E-26 | 0.58 | 7.5E-08 | MOS | Proto-oncogene serine/threonine-protein kinase mos | Serine/threonine-protein kinase, MAPK pathway [100] |
| XM_005093424.2*! | 0.89 | 1.8E-25 | 0.66 | 1.5E-10 | BIRC7 | Baculoviral IAP repeat-containing protein 7 | E3 ubiquitin-protein ligase, apoptosis inhibitor [101] |
| XM_005094992.2*! | 0.81 | 2.81E-18 | 0.66 | 1.9E-10 | |||
| XM_005102721.2* | 0.89 | 2.0E-25 | 0.63 | 3.4E-09 | GIMAP4 | GTPase IMAP family member 4 | Apoptosis [102], cytokine signaling [103] |
| XM_005106556.2** | 0.88 | 6.1E-25 | 0.60 | 2.3E-08 | ETF1 | Eukaryotic peptide chain release factor subunit 1 | Translation termination [104] |
| XM_005093041.2 | 0.88 | 1.3E-24 | 0.62 | 5.5E-09 | RNASET2 | Ribonuclease T2 | Innate immunity [105], mtRNA degradation [106] |
| XM_005104568.2** | 0.88 | 3.1E-24 | 0.61 | 1.0E-08 | PLIN2 | Perilipin-2 | Lipid storage, ROS defense [107] |
| XM_013087467.1** | 0.88 | 3.2E-24 | 0.60 | 1.7E-08 | EIF4A2 | Eukaryotic initiation factor 4A-II | Translation initiation [108], Translation inhibition [109] |
| XM_013087273.1** | 0.87 | 7.7E-24 | 0.56 | 2.4E-07 | HEATR1 | HEAT repeat-containing protein 1 | Ribosome biogenesis [110] |
| XM_005101849.2 | 0.87 | 8.5E-24 | 0.57 | 1.3E-07 | EXOSC10 | Exosome component 10 | RNA metabolism [111] |
| XM_005099415.2** | 0.87 | 1.1E-23 | 0.63 | 2.3E-09 | DUSP7 | Dual specificity protein phosphatase 7 | MAPK pathway [112] |
| XM_005088796.2**! | 0.87 | 1.2E-23 | 0.64 | 1.4E-09 | IRF8 | Interferon regulatory factor 8 | Microglia activation and neuroinflammation [113] |
| XM_013084591.1** | 0.87 | 1.6E-23 | 0.64 | 1.5E-09 | PSAP | Prosaposin | Sphingolipid metabolism [114] |
| XM_013087976.1** | 0.87 | 2.9E-23 | 0.60 | 1.9E-08 | EEF2 | Elongation factor 2 | Translation [115] |
| XM_005097092.2** | 0.86 | 6.1E-23 | 0.66 | 2.1E-10 | CTSL | Cathepsin L1 | Lysosomal protease [116], neuropeptide processing [117] |
| XM_005094650.2*! | 0.86 | 6.8E-23 | 0.69 | 2.1E-11 | CYLD | Ubiquitin carboxyl-terminal hydrolase CYLD | NF-kappa-B regulation, deubiquitination [118], Negative regulation of innate immunity [119] |
| XM_005090468.2** | 0.86 | 9.1E-23 | 0.71 | 2.6E-12 | PDE12 | 2’,5’-phosphodiesterase 12 | Negative regulation of innate immunity |
| XM_005099805.2* | 0.86 | 1.3E-22 | 0.49 | 9.2E-06 | CASP3 | Caspase-3 | Apoptosis [120] |
| XM_013087138.1** | 0.86 | 2.6E-22 | 0.50 | 6.1E-06 | GRN | Granulins | Lysosome biogenesis and homeostasis [121] |
| XM_013083177.1** | 0.85 | 6.4E-22 | 0.59 | 5.0E-08 | SIGIRR | Single Ig IL-1-related receptor | Negative regulation of immune signaling [122] |
| XM_005110683.2** | 0.84 | 2.0E-20 | 0.62 | 5.0E-09 | FTH1 | Ferritin heavy chain | Iron storage, ROS defense [123] |
| XM_013089050.1* | 0.81 | 1.8E-18 | 0.56 | 3.2E-07 | MAP3K8 | Mitogen-activated protein kinase kinase kinase 8 | MAPK signaling, NFkB signaling [124] |
| XM_005112843.2**! | 0.81 | 2.2E-18 | 0.50 | 6.3E-06 | RIOK1 | Serine/threonine-protein kinase RIO1 | Ribosome biogenesis [125], p38 MAPK innate immune response suppressor [126] |
| XM_005105539.2* | 0.8 | 2.3E-17 | 0.45 | 6.9E-05 | RIOK3 | Serine/threonine-protein kinase RIO3 | Ribosome biogenesis [127], INF signaling [128], NFkB inhibitor [129], innate immune response [130] |
| XM_005102490.2* | 0.79 | 8.4E-17 | 0.60 | 1.6E-08 | JKAMP | JNK1/MAPK8-associated membrane protein | MAPK signaling, inhibits MAPK8 [131] |
| XM_005092503.2** | 0.78 | 2.3E-16 | 0.66 | 2.3E-10 | TNIP1 | TNFAIP3-interacting protein 1 | NFkB inhibitor [132,133] |
| XM_013087081.2* | 0.78 | 5.3E-16 | 0.48 | 2.15E-05 | DUOX1 | Dual Oxidase 1 | ROS production [134] |
| XM_013088029.1! | 0.69 | 1.3E-11 | 0.54 | 8.6E-07 |
See Table 2 for description of organization. Transcripts are marked with “*” if they were among significantly upregulated transcripts in Kron et al 2020 [24] in one neuron type and with “**” if in both. Transcripts identified as orthologs to genes differentially expressed due to immune challenge in C. gigas are marked with a “!”in the first column. Common categories include ubiquitination, NFkB signaling, innate immunity, ribosome biogenesis, and regulation of transcription and translation. This module was positively correlated with age, suggesting these processes are upregulated in aging.
3.4.3 Orange
The three KEGG pathways significantly enriched in the orange module, which was positively correlated with age, all participate in proteostasis, whether that is proper protein folding localized to the Endoplasmic Reticulum (ER) in the case of protein processing in the endoplasmic reticulum (ko04141) and N-glycan biosynthesis (ko00510), or in protein degradation, in the case of lysosome (ko04142). Transcripts with highest module membership were associated with ER stress or the endoplasmic reticulum associated protein degradation (ERAD) pathway (Table 4).
Table 4. Selection of transcripts with highest correlation to transcript co-expression module eigengene (module membership, MM) in the orange consensus module identified in Aplysia californica sensory neurons by WGCNA.
| Refseq ID | MM | MM p value | TAS | TAS p value | Human Ortholog | Ortholog Name | Ortholog Function |
|---|---|---|---|---|---|---|---|
| XM_005096841.2* | 0.93 | 6.27E-33 | 0.63 | 1.8E-09 | CREB3L3 | Cyclic AMP-responsive element-binding protein 3-like protein 3 | ER stress response transcription factor, acute inflammation [135] |
| NM_001204489.1* | 0.93 | 4.47E-32 | 0.52 | 2.0E-06 | PSEN2 | Presenilin-2 | Endoprotease, Ca2+ homeostasis as ER leak channel [136], ER-Mitochondrial Ca2+ shuttle [137] |
| XM_005101813.2* | 0.89 | 6.49E-26 | 0.56 | 2.5E-07 | LGMN | Legumain | Endopeptidase [138] |
| XM_013080474.1* | 0.89 | 7.70E-26 | 0.60 | 2.0E-08 | EIF2AK3 | Eukaryotic translation initiation factor 2-alpha kinase 3 | Known as PERK, ER stress response [139] |
| XM_005108642.2* | 0.87 | 2.24E-23 | 0.54 | 7.4E-07 | MACO1 | Macoilin | ER localized regulator of neuronal activity [140] |
| XM_013081341.1 | 0.86 | 7.08E-23 | 0.51 | 4.6E-06 | DESI2 | Deubiquitinase DESI2 | Deubiquitinase [141] |
| XM_013088003.1**! | 0.86 | 4.35E-22 | 0.71 | 2.7E-12 | BIRC3 | Baculoviral IAP repeat-containing protein 3 | E3 ubiquitin-protein ligase, NF-kappa-B signaling regulation [83], innate immunity regulation [84] |
| XM_005105526.2** | 0.85 | 1.13E-21 | 0.56 | 2.8E-07 | ABCA3 | ATP-binding cassette sub-family A member 3 | Lipid transporter [142] |
| XM_005113508.1** | 0.85 | 4.97E-21 | 0.57 | 1.0E-07 | PSAP | Prosaposin | Sphingolipid metabolism [114] |
| XM_013086889.1 | 0.84 | 6.78E-21 | 0.45 | 6.2E-05 | SPTLC2 | Serine palmitoyltransferase 2 | Sphingolipid de-novo synthesis |
| XM_013079527.1* | 0.84 | 1.12E-20 | 0.55 | 5.8E-07 | SUN1 | SUN domain-containing protein 1 | Neurogenesis and neuron and glial migration [143], telomere attachment [144] |
| XM_005100110.2* | 0.83 | 6.91E-20 | 0.41 | 3.0E-04 | MBOAT7 | Lysophospholipid acyltransferase 7 | Regulation of free polyunsaturated fatty acids levels [145,146] |
| XM_013085224.1** | 0.83 | 1.34E-19 | 0.68 | 5.7E-11 | VCAN | Versican core protein | Diverse roles [147], including inflammation [148] |
| XM_005096173.2** | 0.83 | 1.71E-19 | 0.58 | 6.3E-08 | BCL3 | B-cell lymphoma 3 protein | Enhances or inhibits NFkB signaling depending on phosphorylation state [149,150] |
| XM_005102600.2* | 0.82 | 1.24E-18 | 0.48 | 1.7E-05 | SLC39A2 | Zinc transporter ZIP2 | Zinc transporter |
| XM_013083287.1* | 0.81 | 4.10E-18 | 0.51 | 4.0E-06 | ADGRG6 | Adhesion G-protein coupled receptor G6 | Schwann cell differentiation and myelination [151] |
| XM_005101146.2* | 0.81 | 4.14E-18 | 0.57 | 1.7E-07 | C16orf62 | UPF0505 protein C16orf62 | Cell surface recycling, including of signaling receptors [152] |
| XM_005095917.2 | 0.79 | 1.10E-16 | 0.36 | 1.8E-03 | STT3B | Dolichyl-diphosphooligosaccharide—protein glycosyltransferase subunit STT3B | N-glycosylates unfolded proteins [153], plays role in ERAD [154] |
| XM_005103010.2 | 0.78 | 3.77E-16 | 0.45 | 7.5E-05 | TXNDC16 | Thioredoxin domain-containing protein 16 | ERAD [155], humoral immune response [156] |
| XM_005100158.2* | 0.78 | 7.92E-16 | 0.43 | 1.4E-04 | UGGT1 | UDP-glucose:glycoprotein glucosyltransferase 1 | ER glycoprotein quality control [157] |
See Table 2 for description of organization. Common functions include Endoplasmic Reticulum (ER) stress response, ER associated protein degradation (ERAD), sphingolipid metabolism, and immune regulation. This module was positively correlated with age, suggesting these process are upregulated in aging.
3.4.4 Darkgreen
For the darkgreen module, which exhibited an increasing eigengene expression trend until age 10 months after which the trend stabilized, KEGG enrichment analysis highlighted processes involved in nucleic acid metabolism, namely DNA replication (ko03030), Nucleotide excision repair (ko03420), and mismatch repair (ko03430) for DNA and RNA transport (ko03013) and RNA degradation (ko03018) for RNA (Fig 6).
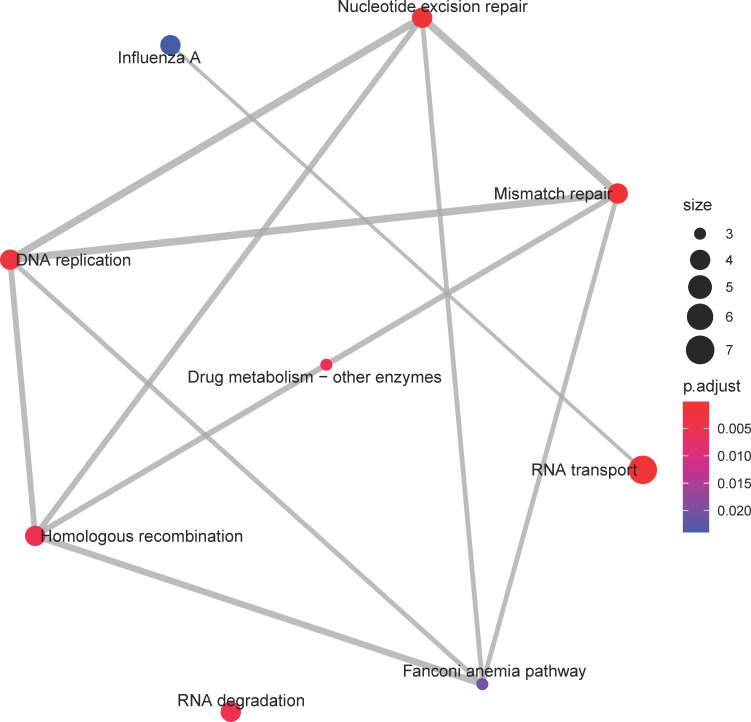
Fig 6. Enrichment map of Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways for the darkgreen consensus co-expression module.
Symbol explanation as in Fig 4. Several pathways dealing with nucleic acid metabolism, such as DNA replication and RNA degradation are significant in this pathway. The expression trend of this module increased linearly until age 10 months after which it stabilized, suggesting increasing activity of these pathways until a stable activity level is reach in old age.
Many of the transcripts with highest module membership were involved with DNA damage response, as well as formation of the nuclear pore, mRNA quality control and export, and immune signaling cascades such as the JAK/STAT cascade, NFkB signaling, and RIG-1 signaling (Table 5).
Table 5. Selection of transcripts with highest correlation to transcript co-expression module eigengene (module membership, MM) in the darkgreen consensus module identified in Aplysia californica sensory neurons by WGCNA.
| Refseq ID | MM | MM p value | TAS | TAS P value | Human Ortholog | Ortholog Name | Ortholog Function |
|---|---|---|---|---|---|---|---|
| XM_013082889.1 | 0.96 | 9.81E-41 | 0.39 | 6.56E-04 | RPA2 | Replication protein A 32 kDa subunit | DNA replication [158], DNA damage repair [159,160] |
| XM_005094195.2 | 0.95 | 3.60E-37 | 0.51 | 4.61E-06 | TRIM3 | Tripartite motif-containing protein 3 | E3 ubiquitin-protein ligase, negative regulator of inflammation [161–163], inhibits synaptic plasticity [164] |
| XM_013086128.1 | 0.94 | 2.71E-34 | 0.47 | 2.42E-05 | TIPARP | Poly [ADP-ribose] polymerase | Inhibitor of AHR-dependent transcription [165], suppressor of INF due to AHR activation [166], activator of INF due to ROS [167] |
| XM_005113357.2! | 0.93 | 7.35E-33 | 0.53 | 1.71E-06 | SOCS2 | Suppressor of cytokine signaling 2[160] | Inhibits JAK/STAT signaling, promotes neurite outgrowth [168], regulates cytokine signaling [169,170] |
| XM_005092855.1! | 0.91 | 3.77E-28 | 0.46 | 3.57E-05 | |||
| XM_005108083.2! | 0.85 | 4.32E-21 | 0.42 | 2.17E-04 | |||
| XM_005092772.2 | 0.93 | 1.77E-32 | 0.29 | 1.39E-02 | ENDOG | Endonuclease G, mitochondrial | mitochondrial biogenenesis and homeostasis [171,172], apoptosis [173,174] |
| XM_005107816.2* | 0.93 | 3.24E-32 | 0.52 | 2.29E-06 | HENMT1 | Small RNA 2’-O-methyltransferase | piRNA biogenesis [175] |
| XM_005095515.2* | 0.93 | 3.35E-32 | 0.60 | 2.12E-08 | ICE2 | Little elongation complex subunit 2 | snRNA transcription [176] |
| XM_005101982.2 | 0.93 | 1.01E-31 | 0.38 | 8.51E-04 | LGALS4 | Galectin-4 | Reduced pro-inflammatory cytokine secretion [177], inhibits myelination [178] |
| XM_013087261.1* | 0.92 | 3.26E-31 | 0.48 | 1.80E-05 | GALC | Galactocerebrosidase | Lysosomal degradation of galactocerebrosides [179] |
| XM_013084530.1 | 0.92 | 1.26E-30 | 0.43 | 1.36E-04 | SMARCD1 | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily D member 1 | Transcription activation and repression via chromatin remodeling as part of SWI/SNF complex [180], immune regulation [181] |
| XM_005102729.2 | 0.92 | 5.39E-30 | 0.28 | 1.59E-02 | WDR53 | WD repeat-containing protein 53 | unknown |
| XM_005105962.2 | 0.92 | 8.43E-30 | 0.46 | 4.99E-05 | EPSTI1 | Epithelial-stromal interaction protein 1 | Macrophage differentiation [182] |
| XM_005111022.2 | 0.92 | 1.03E-29 | 0.52 | 2.03E-06 | RBBP9 | Putative hydrolase RBBP9 | Inhibits TGF-beta growth-inhibition [183] |
| XM_005101042.1 | 0.91 | 1.34E-29 | 0.44 | 9.85E-05 | RPA3 | Replication protein A 14 kDa subunit | DNA damage repair [184] |
| XM_005108381.2 | 0.91 | 1.78E-29 | 0.43 | 1.39E-04 | MOV10L1 | RNA helicase Mov10l1 | piRNA biogenesis [185] |
| XM_013088805.1 | 0.91 | 2.10E-29 | 0.47 | 3.26E-05 | SLC16A14 | Monocarboxylate transporter 14 | Neuronal aromatic-amino-acid transporter [186] |
| XM_005100427.2* | 0.91 | 3.91E-29 | 0.50 | 6.50E-06 | NSMCE1 | Non-structural maintenance of chromosomes element 1 homolog | E3 ubiquitin-protein ligase, DNA damage response, and iron homeostasis [187] |
| XM_013084495.1 | 0.91 | 5.14E-29 | 0.42 | 1.84E-04 | DDX58 | Probable ATP-dependent RNA helicase DDX58 | RIG 1, antiviral immune receptor [188] |
| XM_005091253.2* | 0.91 | 9.96E-29 | 0.56 | 2.70E-07 | TENT4A | Terminal nucleotidyltransferase 4A | mRNA stability and quality control [189,190] |
| XM_013090188.1! | 0.91 | 1.24E-28 | 0.56 | 2.35E-07 | MPEG1 | Macrophage-expressed gene 1 protein | Antibacterial protein, pore-forming protein, innate immunity [191] |
| XM_005088804.2 | 0.91 | 1.75E-28 | 0.44 | 1.00E-04 | PLA2G16 | HRAS-like suppressor 3 | Phospholipid metabolism [192], sensor for sites of viral entry [193] |
| XM_013082099.1 | 0.90 | 1.24E-27 | 0.42 | 1.97E-04 | NUP214 | Nuclear pore complex protein Nup214 | Nuclear pore formation and protein import into nucleus [194,195] |
| XM_013088902.1! | 0.89 | 1.45E-26 | 0.50 | 8.27E-06 | DIS3L2 | DIS3-like exonuclease 2 | Mediates degradation of polyuridylated RNAs [196], mRNA metabolism [197] |
| XM_005089228.2 | 0.89 | 2.04E-26 | 0.48 | 2.03E-05 | SSUH2 | Protein SSUH2 homolog | Odontogenesis, upstream of several transcriptional regulators [163] |
| XM_005107110.2 | 0.89 | 2.47E-26 | 0.43 | 1.52E-04 | PARP14 | Protein mono-ADP-ribosyltransferase PARP14 | Suppresses IFN-gamma response, induces IL-4 response, counteracts PARP9 [198] |
| XM_013081916.1 | 0.89 | 6.38E-26 | 0.53 | 1.16E-06 | NUP98 | Nuclear pore complex protein Nup98-Nup96 | Nuclear pore formation, gene expression regulation [199] |
| XM_005106449.2*! | 0.89 | 1.87E-25 | 0.57 | 1.31E-07 | ZRANB3 | DNA annealing helicase and endonuclease ZRANB3 | Rewinds DNA and maintains genome stability, replication stress response [200,201] |
| XM_005106672.2! | 0.88 | 3.94E-25 | 0.44 | 1.18E-04 | JAK2 | Tyrosine-protein kinase JAK2 | JAK/STAT signaling, interferon gamma response [202] |
| XM_005106450.2 | 0.88 | 5.72E-25 | 0.46 | 3.81E-05 | BANF1 | Barrier-to-autointegration factor | Chromatin organization and gene expression [203], blocks viral DNA replication [204] |
| XM_013085923.1 | 0.87 | 7.99E-24 | 0.42 | 2.17E-04 | TREX2 | Three prime repair exonuclease 2 | DNA repair, mRNA export, transcription [205–207] |
| XM_013080704.1* | 0.87 | 8.51E-24 | 0.52 | 2.95E-06 | VRK1 | Serine/threonine-protein kinase VRK1 | Regulates BANF1 [208], activates ATF2 [209] |
| XM_005107122.2! | 0.87 | 4.34E-23 | 0.35 | 2.44E-03 | GLE1 | Nucleoporin GLE1 | Export of mRNA from nucleus [210] |
| XM_005110865.2 | 0.86 | 5.48E-22 | 0.41 | 2.70E-04 | DXO | Decapping and exoribonuclease protein | Pre-mRNA quality control [211] |
| XM_005113277.2! | 0.85 | 6.73E-22 | 0.28 | 1.77E-02 | NUP88 | Nuclear pore complex protein Nup88 | Nuclear pore formation [212] |
| XM_005099732.2 | 0.85 | 9.14E-22 | 0.35 | 2.59E-03 | MTPAP | Poly(A) RNA polymerase, mitochondrial | mtDNA stabilization [213,214], histone mRNA degradation [215] |
| XM_013087644.1 | 0.77 | 3.02E-15 | 0.39 | 7.51E-04 | RPUSD3 | Mitochondrial mRNA pseudouridine synthase RPUSD3 | mtRNA translation, mitochondrial ribosome biogenesis via pseudouridylation [216,217] |
| XM_005104572.2 | 0.74 | 1.10E-13 | 0.41 | 2.66E-04 | GIMAP1 | GTPase IMAP family member 1 | Immune cell development [218] |
| XM_005109614.2! | 0.73 | 1.84E-13 | 0.30 | 9.16E-03 | XIAP | E3 ubiquitin-protein ligase XIAP | E3 protein-ubiquitin ligase, apoptosis inhibitor [219], NFkB activation [220] |
| XM_005109011.2 | 0.72 | 5.24E-13 | 0.25 | 2.98E-02 | STRA6 | Receptor for retinol uptake STRA6 | Retinol importer [221] |
See Table 2 for description of organization. Common functions include nuclear pore formation, DNA damage repair, immune and inflammation signaling. The expression trend of this module increased linearly until age 10 months after which it stabilized, suggesting increasing activity of these in early age and stable, heightened activity in old age.
3.4.5 Greenyellow
Pathways associated with viral infection and immune signaling dominated the KEGG enrichment results of the greenyellow module, which exhibited an increasing eigengene expression trend with age (Fig 7). Highly significant pathways included classical immune response associated pathways such as NF-kappa-Beta signaling pathway (ko04064), RIG-1-like receptor signaling pathway (ko04622), and NOD-like receptor signaling (ko04621). Transcripts with highest module membership in the greenyellow module mapped to human orthologs involved in the interferon and NFkB signaling pathways (Table 6).
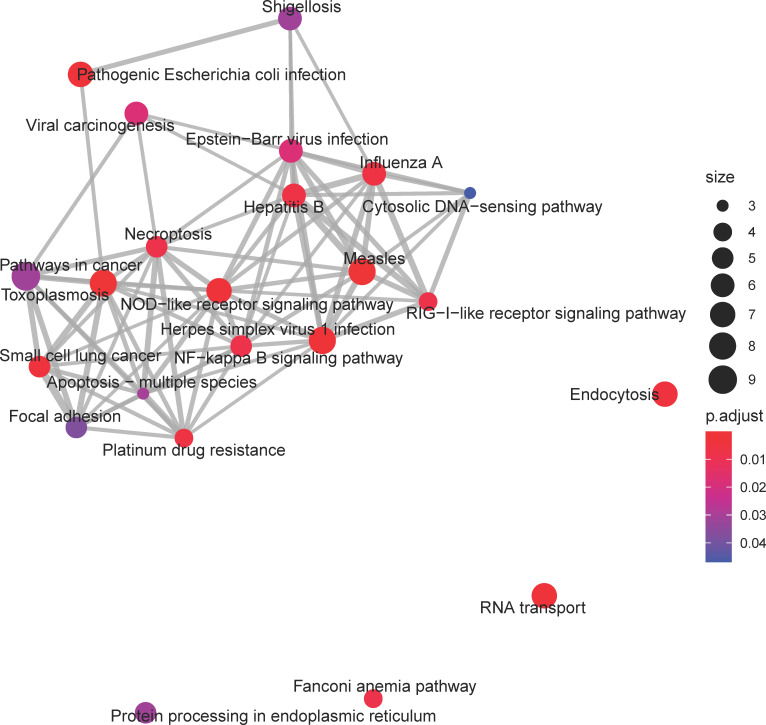
Fig 7. Enrichment map of KEGG pathways for the greenyellow consensus co-expression module.
Symbol explanation as in Fig 4. Many of the significant pathways in this module have overlapping gene sets, many of which are associated with immune activation, such as NOD-like receptor signaling, RIG-I-like receptor signaling, and NF-Kappa-B signaling pathway. The increasing expression trend of this module’s eigengene throughout aging suggests consistently increased activation of these pathways in aging.
Table 6. Selection of transcripts with highest correlation to transcript co-expression module eigengene (module membership, MM) in the greenyellow consensus module identified in Aplysia californica sensory neurons by WGCNA.
| Refseq ID | MM | MM p value | TAS | TAS p value | Human Ortholog | Ortholog Name | Ortholog Function |
|---|---|---|---|---|---|---|---|
| XM_005110768.2! | 0.95 | 1.8E-36 | 0.44 | 1.1E-04 | ZNFX1 | NFX1-type zinc finger-containing protein 1 | Virus detection, IFN response [222], epigenetics [223] |
| XM_005097053.2! | 0.92 | 6.8E-31 | 0.44 | 8.8E-05 | |||
| XM_005105124.2! | 0.86 | 2.9E-22 | 0.53 | 1.4E-06 | |||
| XM_013083625.1 | 0.94 | 4.2E-35 | 0.42 | 1.9E-04 | DTX3L | E3 ubiquitin-protein ligase DTX3L | E3 ubiquitin-protein ligase [224], DNA damage repair [225], INF response [226] |
| XM_005102017.2 | 0.94 | 1.8E-34 | 0.52 | 2.2E-06 | SAMD9 | Sterile alpha motif domain-containing protein 9 | Antiviral stress response [227] |
| XM_005090189.2* | 0.93 | 2.2E-32 | 0.45 | 5.9E-05 | MRE11 | Double-strand break repair protein | DNA damage repair [228], INF response [229] |
| XM_005105514.2 | 0.93 | 3.7E-32 | 0.46 | 4.5E-05 | TRANK1 | TPR and ankyrin repeat-containing protein 1 | Interferon- stimulated gene [230] |
| XM_013082259.1! | 0.93 | 1.1E-31 | 0.48 | 2.1E-05 | TUT4 | Terminal uridylyltransferase 4 | mRNA decay [231], innate immunity [232] |
| XM_005110839.2 | 0.91 | 5.6E-29 | 0.47 | 2.5E-05 | IL17RD | Interleukin-17 receptor D | ERK inhibitor [233], negative regulation of TLR signaling [234] |
| XM_005099718.2 | 0.90 | 7.9E-28 | 0.45 | 7.5E-05 | VCPIP1 | Deubiquitinating protein VCIP135 | Deubiquitination [235] |
| XM_005090454.2 | 0.90 | 1.0E-27 | 0.56 | 3.4E-07 | SACS | Sacsin | Chaperone [236], INF response in oyster |
| XM_005106324.2* | 0.90 | 1.7E-27 | 0.60 | 2.6E-08 | ZNF598 | E3 ubiquitin-protein ligase ZNF598 | E3 ubiquitin-protein ligase and Ribosome quality control [237], translation repression [238], Attenuation of innate immune response [239] |
| XM_005093151.2 | 0.90 | 2.7E-27 | 0.36 | 1.6E-03 | TRIM2 | Tripartite motif-containing protein 2 | E3 ubiquitin-protein ligase [240], antiviral [241] |
| XM_013081468.1* | 0.90 | 3.4E-27 | 0.52 | 2.0E-06 | ASCC3 | Activating signal cointegrator 1 complex subunit 3 | DNA repair [242], NFkB, ATF-1, and SRF signaling [243] |
| XM_005107621.2! | 0.90 | 4.2E-27 | 0.34 | 3.0E-03 | CMTR1 | Cap-specific mRNA (nucleoside-2’-O-)-methyltransferase 1 | IFN signaling, antiviral state establishment [244] |
| XM_013084387.1! | 0.90 | 6.5E-27 | 0.36 | 2.0E-03 | STAT5B | Signal transducer and activator of transcription 5B | GH signaling [245], Il-2 cytokine signaling [246] |
| XM_005095848.2 | 0.90 | 1.3E-26 | 0.25 | 3.4E-02 | ANK1 | Ankyrin-1 | Diverse, including protein localization to membranes [247] |
| XM_005108543.2*! | 0.89 | 1.9E-26 | 0.63 | 2.2E-09 | HERC4 | Probable E3 ubiquitin-protein ligase HERC4 | E3 ubiquitin-protein ligase [87] |
| XM_005094842.2! | 0.89 | 4.7E-26 | 0.47 | 2.9E-05 | IRF1 | Interferon regulatory factor 1 | IFN signaling [248] |
| XM_005096699.2 | 0.89 | 8.4E-26 | 0.31 | 7.1E-03 | PARP12 | Protein mono-ADP-ribosyltransferase PARP12 | Interferon induced gens, negative regulation of translation, NFkB singaling [249] |
| XM_005102776.2* | 0.88 | 4.5E-25 | 0.41 | 2.7E-04 | DDX58 | Probable ATP-dependent RNA helicase DDX58 | RIG 1, antiviral immune receptor [188] |
| XM_005104216.2! | 0.83 | 5.0E-20 | |||||
| XM_005098154.2! | 0.88 | 4.6E-25 | 0.55 | 5.7E-07 | SMG1 | Serine/threonine-protein kinase SMG1 | Nonsense mediated mRNA decay [250], restriction of viral replication [251] |
| XM_005101113.2 | 0.88 | 7.1E-25 | 0.54 | 8.0E-07 | ZC3HAV1L | Zinc finger CCCH-type antiviral protein 1-like | Same family as ZNFX1, unknown function [252] |
| XM_013086154.1 | 0.87 | 1.95E-23 | 0.32 | 5.2E-03 | BIRC2 | Baculoviral IAP repeat-containing protein 2 | E3 ubiquitin-protein ligase, NF-kappa-B signaling regulation [83], innate immunity regulation [84] |
| XM_013086147.1 | 0.84 | 1.57E-20 | |||||
| XM_013089793.1! | 0.86 | 7.8E-23 | 0.43 | 1.7E-04 | CASP8 | Caspase-8 | Apoptosis [253], inflammatory homeostasis via RIPK1 [254] |
| XM_005111821.2 | 0.85 | 1.6E-21 | 0.42 | 2.1E-04 | HIST1H3A | Histone H3.1 | Nucleosome formation, transcription regulation [255] |
| XM_013079365.1 | 0.83 | 6.6E-20 | 0.34 | 2.9E-03 | TLR1 | Toll-like receptor 1 | Innate immune response [256,257] |
| XM_013079178.1! | 0.83 | 7.2E-20 | 0.39 | 6.5E-04 | IRF8 | Interferon regulatory factor 8 | Microglia activation and neuroinflammation [113] |
See Table 2 for description of organization. Common functions include ubiquitination, interferon signaling (IFN), and inflammation. The roughly monotonic increasing trend of this module’s eigengene throughout aging suggests consistently increased activation of these processes in aging.
A full list of KEGG enrichment results can be found in Supplementary Datasheet S1 Dataset. Full transcript sets and their respective module membership values can be found in Supplementary Datasheet S2 Dataset.
3.5 Module enrichment for C. gigas immune response genes
BLAST annotation of Aplysia proteins to the C. gigas proteome and subsequent annotation resulted in 22,715 unique Aplysia transcripts mapped to 10,112 unique C. gigas proteins. Of these 22,715 transcripts, 7,359 were present in one of the identified co-expression modules. Among the 1,547 genes marked as exhibiting coherent regulation profiles following priming with poly(I·C)and viral challenge in Lafont et al (2020) [37], 697 were also present in Aplysia co-expression modules. Enrichment tests for each module revealed that the greenyellow, darkgreen, and pink modules were significantly enriched for C. gigas immune response genes (Fisher’s exact test, p < = 0.00385, Table 7). A full mapping of co-expression module genes to genes DE in Lafont et al (2020) [37] can be found in supplementary file S3 Dataset.
Table 7. Aplysia co-expression module enrichment for orthologs differentially expressed after immune priming and viral challenge in pacific oyster Crassostrea gigas.
| Aplysia transcripts in C. gigas (m+n) | Transcripts in LaFont (m) | Transcripts not in Lafont (n) | signif threshold (ɑ) | Bonferroni (ɑ`= ɑ/14) | |
|---|---|---|---|---|---|
| 7359 | 697 | 6662 | 0.05 | 0.00385 | |
| Module | Transcripts in module (k) | Module transcripts in LaFont (x) | Proportion (x/k) | p-value (p = Σp(x-1:k)) | Sig (p<ɑ`) |
| greenyellow | 224 | 45 | 0.20 | 4.37E-06 | * |
| darkgreen | 156 | 29 | 0.19 | 1.41E-03 | * |
| pink | 906 | 111 | 0.12 | 3.71E-03 | * |
| purple | 2203 | 206 | 0.09 | 6.72E-01 | - |
| blue | 1170 | 109 | 0.09 | 6.78E-01 | - |
| violet | 24 | 2 | 0.08 | 1.00E+00 | - |
| royalblue | 410 | 34 | 0.08 | 9.01E-01 | - |
| grey | 190 | 15 | 0.08 | 9.22E-01 | - |
| green | 1838 | 133 | 0.07 | 1.00E+00 | - |
| orange | 125 | 9 | 0.07 | 9.59E-01 | - |
| steelblue | 39 | 2 | 0.05 | 1.00E+00 | - |
| paleturquoise | 23 | 1 | 0.04 | 1.00E+00 | - |
| saddlebrown | 51 | 1 | 0.02 | 1.00E+00 | - |
Module enrichment for C. gigas immune genes in Aplysia co-expression modules was assessed via Fisher’s exact test. Of the 7359 Aplysia transcripts that were annotated as C. gigas orthologs and present in Aplysia co-expression modules, 697 were present in C. gigas (Lafont et al (2020) [37] m = 697, n = 6662). For each module, the number of transcripts in that module that were marked as C. gigas orthologs (k) and the proportion of those also present in Lafont et al (2020) [37] (x) were used to calculate the hypergeometric distribution in R. The p value was calculated as the sum of all probabilities at least as extreme as k (Σp(x-1:k) which was compared to a significance threshold of 0.05 with Bonferroni multiple test correction for 14 tests. Three modules (greenyellow, darkgreen, and pink) were identified as significantly enriched for C. gigas immune response orthologs.
Of the transcripts in the greenyellow, darkgreen, and pink modules, 17, 7, and 8 respectively exhibited a module membership greater than 0.8 for their respective modules and mapped to a C. gigas gene with a log 2 fold change of greater than or equal to 1 in response to immune priming and/or viral challenge in Lafont et al (2020) [37] (Table 8). Of those, transcripts from the greenyellow and darkgreen modules mapped primarily to C. gigas genes with putative viral function. Furthermore, the greenyellow transcripts mapped to C. gigas genes assigned by Lafont et al (2020) [37] primarily to the interferon-like and RIG-like receptor recognition pathways, while many darkgreen transcripts were assigned to the JAK/STAT signaling.
Table 8. Mapping between Aplysia transcripts with high module membership in the greenyellow, darkgreen, and pink modules and genes differentially expressed as a result of immune priming and viral exposure in C. gigas.
| Module | Aplysia transcript | C. gigas Gene | Antimicrobial activity | Pathways | Human Ortholog | Ortholog Name |
|---|---|---|---|---|---|---|
| greenyellow | XM_005110768.2 | CGI_10023396 | virus | IFN-like pathway and RLR recognition | ZNFX1 | NFX1-type zinc finger-containing protein 1 |
| greenyellow | XM_013082259.1 | CGI_10020126 | V/B | other | TUT4 | Terminal uridylyltransferase 4 |
| greenyellow | XM_005097053.2 | CGI_10003301 | virus | IFN-like pathway and RLR recognition | ZNFX1 | NFX1-type zinc finger-containing protein 1 |
| greenyellow | XM_005107621.2 | CGI_10013977 | other | other | CMTR1 | Cap-specific mRNA (nucleoside-2’-O-)-methyltransferase 1 |
| greenyellow | XM_013084387.1 | CGI_10028719 | virus | JAK/STAT | STAT5B | Signal transducer and activator of transcription 5B |
| greenyellow | XM_005098154.2 | CGI_10024989 | V/B | signaling | SMG1 | Serine/threonine-protein kinase SMG1 (Fragment) |
| greenyellow | XM_005101016.2 | CGI_10021954 | virus | IFN-like pathway and RLR recognition | DHX38 | Pre-mRNA-splicing factor ATP-dependent RNA helicase PRP16 |
| greenyellow | XM_013089793.1 | CGI_10023960 | virus | apoptosis | CASP8 | Caspase-8 |
| greenyellow | XM_005105124.2 | CGI_10023396 | virus | IFN-like pathway and RLR recognition | ZNFX1 | NFX1-type zinc finger-containing protein 1 |
| greenyellow | XM_005104216.2 | CGI_10024392 | virus | IFN-like pathway and RLR recognition | DDX58 | Probable ATP-dependent RNA helicase DDX58 |
| greenyellow | XM_013079178.1 | CGI_10003270 | virus | IFN-like pathway and RLR recognition | IRF8 | Interferon regulatory factor 8 (Fragment) |
| greenyellow | XM_013085543.1 | CGI_10018479 | V/B | signaling | COL21A1 | Collagen alpha-1(XXI) chain |
| greenyellow | XM_013081218.1 | CGI_10021954 | virus | IFN-like pathway and RLR recognition | DHX16 | Pre-mRNA-splicing factor ATP-dependent RNA helicase DHX16 |
| greenyellow | XM_005104310.2 | CGI_10010459 | virus | IFN-like pathway and RLR recognition | DDX58 | Probable ATP-dependent RNA helicase DDX58 |
| greenyellow | XM_005110422.2 | CGI_10003270 | virus | IFN-like pathway and RLR recognition | IRF8 | Interferon regulatory factor 8 |
| greenyellow | XM_005109293.2 | CGI_10002009 | other | other | ELF5 | ETS-related transcription factor Elf-5 |
| greenyellow | XM_005108818.2 | CGI_10020752 | virus | RNAi | DDX58 | Probable ATP-dependent RNA helicase DDX58 |
| darkgreen | XM_005113357.2 | CGI_10019528 | virus | JAK/STAT | SOCS2 | Suppressor of cytokine signaling 2 |
| darkgreen | XM_013090188.1 | CGI_10002181 | bacteria | other | MPEG1 | Macrophage-expressed gene 1 protein |
| darkgreen | XM_005092855.1 | CGI_10019528 | virus | JAK/STAT | SOCS2 | Suppressor of cytokine signaling 2 |
| darkgreen | XM_013088902.1 | CGI_10019733 | virus | other | DIS3L2 | DIS3-like exonuclease 2 |
| darkgreen | XM_005108083.2 | CGI_10019528 | virus | JAK/STAT | SOCS2 | Suppressor of cytokine signaling 2 |
| darkgreen | XM_013091162.1 | CGI_10028125 | virus | other | RANBP2 | E3 SUMO-protein ligase RanBP2 |
| darkgreen | XM_013088005.1 | CGI_10027619 | virus | other | TYMP | Thymidine phosphorylase |
| pink | XM_005088796.2 | CGI_10003270 | virus | IFN-like pathway and RLR recognition | IRF8 | Interferon regulatory factor 8 (Fragment) |
| pink | XM_005097229.2 | CGI_10023430 | other | other | FNIP2 | Folliculin-interacting protein 2 (Fragment) |
| pink | XM_005110283.2 | CGI_10026985 | bacteria | other | PRSS12 | Neurotrypsin |
| pink | XM_013081403.1 | CGI_10026606 | other | other | CBX4 | E3 SUMO-protein ligase CBX4 |
| pink | XM_005099789.2 | CGI_10021954 | virus | IFN-like pathway and RLR recognition | DHX16 | Pre-mRNA-splicing factor ATP-dependent RNA helicase DHX16 |
| pink | XM_013082526.1 | CGI_10025856 | virus | other | MSH2 | DNA mismatch repair protein Msh2 |
| pink | XM_005102215.2 | CGI_10013829 | V/B | other | ANGPTL6 | Angiopoietin-related protein 6 |
| pink | XM_005107413.2 | CGI_10005182 | virus | IFN-like pathway and RLR recognition | ADAR | Double-stranded RNA-specific adenosine deaminase (Fragment) |
Transcripts (column 2) from the greenyellow, darkgreen, and pink (column 1) modules alongside the C. gigas gene to which they mapped (column 3) during BLAST comparison of Aplysia and C. gigas proteomes. Transcripts were selected if they exhibited module membership greater than or equal to 0.8 for their respective module and the C. gigas ortholog exhibited a log 2 fold change of at least 1 in Lafont et al (2020) [37] as a result of immune priming or virus exposure. Columns 4 and 5 list the antimicrobial function and pathway identified by Lafont et al (2020) [37] for each C. gigas gene. Columns 6 and 7 list the gene symbol and gene name for the human ortholog of each Aplysia transcript. A full mapping of Aplysia transcripts to C. gigas genes from Lafont et al (2020) [37] can be found in supplementary file S3 Dataset.
Of those Crassostrea gigas transcripts that exhibited a greater than two-fold increase in expression 10 days after treatment with a viral analog, 84 had clear orthologs in Aplysia. Of those 84 orthologous transcripts in Aplysia, only two exhibited a significant increase in expression in aging sensory neurons in our previous study: an IRF8 ortholog and an uncharacterized protein (S4 Dataset).
4 Discussion
While enrichment analysis and eigengene expression profiles suggested strong overlap with our previous study, several modules and enrichment results identified many facets to the transcriptional dynamics in aging of these sensory neurons not detected in our previous DEA study [24].
Enrichment analysis and eigengene expression trend of the royalblue module strongly resembles that observed in Kron et al (2020) [24] of expression clusters with decreasing expression trends in aging. Key enzymes in glycolysis such as GPI, PGK1, PGAM2, GAPDH, and ENO1, as well as PDHB, which is part of the pyruvate dehydrogenase complex that links glycolysis to the TCA cycle, were among the transcripts with the highest module membership in the royalblue module. Key TCA enzymes DLST, IDH3G, and AOC2, as well as two members of the succinate dehydrogenase complex, SDHA and SDHC, which function as a nexus between the TCA and OXPHOS, were also present. Downregulation of these transcripts suggests decreased TCA cycle activity, and decreased NADH generation for use in OXPHOS. The presence of orthologs to OXPHOS complex assembly proteins SDHAF2, NDUFA2, and FMC1, as well as electron transport chain regulator ETRF1 among transcripts with high module membership may suggest further disruptions of OXPHOS at the complex level [41,71,73,75]. Mitochondria depleted of NADH exhibit impaired antioxidant capacity and increased ROS generation [258].
Several orthologs of major ROS detoxification enzymes, namely GPX4, SOD2, CAT, and PRDX5 are present in the downregulated royalblue module [40,54,55,80]. The further presence of PARK7, NXNL2, and ITCH, which stimulate antioxidant activity, in this downregulated module suggest that the antioxidant system of these neurons is impaired in age [45,47,69,78]. The presence of ortholog mediators of mitochondrial fission-fusion dynamics such as GDAP1 and MFN2, mediator of mitophagy FUNDC1, and promoter of mitochondrial biogenesis GPS2, which plays a role in mitochondrial stress signaling, suggests that maintenance of mitochondrial homeostasis is downregulated in aging [48,57,62,64,65]. Mitochondria act as key Ca2+ reservoirs and dysfunctional mitochondria can lead to perturbed Ca2+ dynamics. This is further suggested by downregulation of orthologs of C1orf194 and CALM2 which act to maintain Ca2+ homeostasis [50,53]. Homeostasis of Ca2+ is critical to the proper function of neurons suggesting that knock-on effects of energy metabolism impairment may have adverse effects on the proper functioning of these sensory neurons with aging. Similarly, the presence of two potassium channels orthologs, KCNC2 and KCNAB2, a delayed rectifier K channel and a subunit of fast-inactivating A-type K channels, respectively, that repolarize neurons during firing and thus play important roles in membrane excitability, further suggests impaired neuronal function [63,74].
A facet of this module that was not captured in the expression clusters is the presence of many downregulated transcripts involved in cellular cargo transport. Orthologs of several proteins involved in retrograde transport, including DCTN6, CCDC151, and EFCAB1, and anterograde transport orthologs like BLOC1S1 and KIF3A, suggest disruptions in communication from soma to synapse and vice versa [38,44,51,56,66,76,259]. Downregulation of STAU2, which plays a key role in transport of RNA from the cell body to dendrites for local translation, suggests disruption of transcription/translation events necessary for long term memory in these sensory neurons [77,260]. Other processes in cellular cargo transport, such as ER to Golgi transport, endocytosis and exocytosis are also represented by downregulated orthologs to NAPG, EXOC2, SNX30, PDCD6, CHMP6, VPS26B, and SYT4 [52,60,68,72]. Proper vesicle transport and recycling are crucial for the signaling function of neurons, and disruptions in timing of these vesicle mediated events can impact neuronal function. The diversity of biological functions present in this module demonstrates the tight coupling of neuronal metabolism, transport of cellular cargo, and signaling.
While the pink module was identified as having an increasing eigengene expression trajectory and a largely a transcriptional and proteostatic character by KEGG enrichment analysis similar to several clusters in Kron et al (2020) [24], the transcripts with the highest module membership, including the hub gene NFKBIA, suggest inflammation plays a central role in this module. Many of these upregulated transcripts mapped to human genes known to be induced by NFkB, including CSTL, BIRC3, FTH1, CYLD, and the hub gene NFKBIA [261–266]. Furthermore, several of these orthologs activate or permit NFkB signaling, including CSTL, BIRC3, GM2A, and NAT10 [83,97,267–270]. The pink module also contains several dampeners of the NFkB signaling cascade and innate immunity, such as NFKBIA, CYLD, PDE12, SIGIRR, RIOK1, RIOK3 JKAMP, and TNIP1, likely to maintain homeostatic control and prevent over-inflammation [118,122,126,129–132,271,272]. While an upregulation of NFKBIA would seem to suggest a decrease in NFkB signaling, the degradation of NKFBIA is a key step in the release of NFkB, allowing translocation to the nucleus [272]. This may suggest that these neurons are upregulating NFKBIA to keep up with growing rates of NKFBIA degradation as a result of increased NFkB signaling [122].
The pink module also contains upregulated translation modulators. This includes genes that promote rRNA maturation and recruitment such as EFL1, NAT10, RIOK1, RIOK3, and HEATR1 [96,97,110,125,127]. Although known for its role in ribosome biogenesis and translation efficiency [97,273,274], NAT10 is also responsible for N4-acetylcytoside (N4A) driven INF and NFkB inflammatory signaling via HMGB1 and NLRP3 inflammasomes, perhaps playing a dual role in this module [270]. Chronic, low grade inflammation as a result of N4A accumulation due to consistent activity of NAT10 may contribute to aging in these neurons.
Translation attenuating orthologs are also present in this module, such as INTS1, GCN1, and EIF4A2 [91,92]. Under amino acid starvation conditions, GCN1 activates GCN2 which in turn phosphorylates EIF4A, which restricts translation [94,275–277]. However, this cascade has also been shown to be part of the antiviral response, specifically to prevent translation of viral RNAs [278,279]. Considering the preponderance of inflammatory genes in the pink module and the methodology that these animals were fed an ad libitum diet, the antiviral function of this cascade is the more likely here. Interestingly, this cascade also modulates synaptic plasticity and memory by inhibiting CREB in the hippocampus. Because CREB is essential for synaptic plasticity, increased CREB inhibition as a result of increased activity of the GCN1-EIF4A cascade during an inflammatory response may have knock-on effects that inhibit synaptic plasticity in these neurons [280].
Like the pink module, the orange module is similar to expression profile clusters found in Kron et al (2020) [24] as it exhibits increasing eigengene expression trajectory in age and classic signatures of ER stress. The hub transcript CREB3L3 (CREBH) and the ortholog with fourth highest module membership EIF2AK3 (PERK) are critical in the ER stress response cascade, while others like UGGT1, STT3B and TNXDC16 are involved in ERAD [135,139,154,155,157]. Interestingly, several of the transcripts in the pink module with high module membership are involved in sphingolipid metabolism, such as PSAP and SPLITC2 [114,281,282]. Sphingolipids, particularly ceramides, play central roles in pro-inflammatory signaling and ER stress, suggesting this module also participates in pro-inflammatory signaling [283]. SPLITC2 in particular has been shown to be upregulated by NFkB, suggesting NFkB signaling also plays a role in the orange module [281].
Several transcripts in the orange module regulate NFkB, such as BCL3 and BIRC3, similar to the pink module [83,149,150]. In addition, upregulation of the orange module hub gene CREB3L3 itself suggests neuro-inflammation as a central component of this module due to the role of this ortholog in the acute inflammatory response [135]. The similarities in the pink and orange modules suggest that each represents a different element of a proteostatic response to inflammation. Interestingly, the darkgreen module has a monotonic increasing eigengene expression trend early in the aging process and then stabilizes when the orange and pink modules enter their monotonic phase.
Many upregulated transcripts in the darkgreen module are orthologous to human genes associated with DNA damage response, including the hub transcript RPA2, RPA3, NSMCE1, ZRANB3, and TREX2 suggesting mounting DNA damage with age in these neurons [160,184,187,201,205]. The presence of several upregulated transcripts orthologous to genes critical to the formation of the nuclear pore such as NUP214, NUP98, and NUP88 as well as the stability and export of mRNA such as TREX2, GLE1, and TENT4A, and transcription and translation regulators like SMARCD1 may suggest this module maintains a particular transcriptional program [180,189,190,206,207,210]. Interestingly, this module contains several upregulated orthologs known to inhibit inflammatory or immune signaling, such as TRIM3, TIPARP, SOCS2, SMARCD1, and LGALS4. This could suggest this module is either acting to suppress or modulate inflammatory signaling to prevent over-inflammation as seen in the pink and orange modules [162,166,170,177,181]. However, several other member orthologs exhibit pro-inflammatory functions, such as the hub transcript RPA2 and XIAP which activate NFkB [220,284], viral RNA sensor and activator of the interferon response pathway DDX58/RIG-1 [285], and genes that support differentiation and maturation of immune cells such as EPSTI1, RBBP9, SSUH2 and GIMAP1 [163,182,183,218]. Aplysia immune hemocyte aggregates are known to play a pivotal role in neuron injury-associated inflammation and perhaps are similarly involved in the inflammatory response suggested by the orthologs present in the pink, orange, and darkgreen modules [286–289]. Curiously, TIPARP and PARP14 function to suppress and activate different cytokine signaling cascades depending on context and could be counted among other previously listed groups [166,167,198]. Upregulation of orthologs to the aforementioned viral RNA sensor RIG-1, sensor of viral entry PLA2G16, antimicrobial peptide MPEG1, and blocker of viral DNA replication BANF1 may suggest a role for this module in the detection of and initial response to viral infection in these neurons [191,193,204]. Interestingly, this may be further supported by the orthologs HENMT1 and MOV10L1 which mediate the formation of PIWI interacting RNAs (piRNA), noncoding RNAs that have antiviral activities in the innate immune systems of insects and possibly all eukaryotes [175,185,290,291]. The final module of interest, the greenyellow module, exhibits a monotonic increase in eigengene expression during the aging process, in parallel with the darkgreen module from age 6–9 months, and then parallel to the pink and orange modules thereafter.
Transcripts with highest module membership in the greenyellow module all represent orthologs to elements of the Interferon (IFN) Mediated response to RNA viruses. Several innate immune mobilized antiviral zinc finger protein orthologs are represented in this module, including PARP12, ZC3HAVIL, TUT4, and three orthologs of ZNFX1, one of which is the hub transcript [249,252,292,293]. Other transcripts are orthologs of interferon-stimulated genes (ISG) that stimulate or facilitate the expression of other ISG, including ZNFX1, MRE11, DTX3L, CMTR1, and IRF1 [222,226,229,244,248]. Among these ISG orthologs are viral dsRNA sensors ZNFX1 and DDX58/RIG1 [222,285]. Other ISG orthologs upregulated in this module are IFN effector genes that have specific antiviral action: SAMD9 triggers anti-viral granule formation, TUT4 uridylates viral RNAs thus tagging them for degradation, SMG1 restricts viral replication, and TRIM2 prevents viral internalization [227,232,241,251]. The upregulation of TLR1 and STAT5b orthologs in the greenyellow module, which cooperate with STRA6 and JAK2 from the darkgreen, capture elements of the pattern recognition signaling cascade needed to mobilize an immune response [246,256,257]. A proper immune response also requires modulators to prevent over-inflammation or to dampen ISG expression once the viral challenge has been dealt with to allow clearance of waste. Several such immune regulator orthologs are also upregulated in the greenyellow module, including IL17RD which exists as part of a larger family of proteins that regulate interleukin (IL) and Toll-like receptor (TLR) signaling, ZNF598, and ASCC3 [234,239,243]. Whereas the pink module contains translation modulators, several of these upregulated transcripts are also orthologs of proteins known to have epigenetic and/or transcription attenuating effects, including PARP12, ZNFX1, and H3.1, possibly to mute global transcription in favor of anti-viral transcriptional programs [223,249,255].
Several of these transcripts also participate in NFkB signaling, promoting inflammation as part of the immune response. These include IRF1 and IRF8 which activate the Type-I Interferon response, CASP8, ASCC3 which is indispensable for NFkB signaling, and PARP12 which activates NFkB [243,249,254,294]. Ubiquitination dynamics are crucial for IFN and NFkB signaling [295], and many of the included upregulated transcripts are orthologs of genes that act as E3 ubiquitin-protein ligases such as DTX3L, ZNF598, TRIM2, and HERC4; as well as the Deubiquitinating enzymes VCPIP1. The multitude of orthologs involved in initiating the NFkB signaling cascade in response to viral infection in the greenyellow module may suggest this module may act upstream of the NFkB stimulated pink and orange modules.
While the discussed results appear to present a convincing picture of the function of these co-expression networks, it is important to approach these inferences with a degree of caution. Genes marked as orthologous by statistical means, such as BLAST or ghostKOALA, do not always perform the same functions in the target species as they do in the annotated species. Very few of these transcripts discussed have been independently investigated in Aplysia and their functions in Aplysia neurons and associated cells are not known. While several orthologs are generally understood to perform conserved roles across genera and their inferred function can be reasonably assumed, such as those involved in glycolysis or the TCA cycle, others involved in more idiosyncratic systems such as the immune response may have a large degree of divergence, especially when comparing the complex immune system of vertebrates such as human to that of evolutionarily distant mollusks.
Although the function of the orthologs discussed were characterized in human, mouse, C. elegans, and/or Drosophila, similar expression patterns to those captured by the pink, orange, darkgreen, and greenyellow modules have also been documented in other mollusks. Both Cathepsin L and MPEG1 orthologs have been shown to take part in the innate immune response of two species of abalone [296–298]. Immune challenge in oysters with viral RNA also resulted in upregulation of common NFkB and JAK/STAT signaling components observed in darkgreen and greenyellow modules, including orthologs to ZNFX1, Sacsin, and MPEG1 along with various IAP, TRIM, caspases, and IRF proteins [37]. To assess to what degree the transcripts inferred to play a role in the neural immune and inflammatory response via annotation to human orthologs represent a bonafide immune response, we further compared our module transcript sets to the transcriptional signature of an acute immune response to immune priming and RNA virus exposure in another mollusk, the Pacific oyster Crassostrea gigas [37]. The pink, greenyellow, and darkgreen modules were significantly enriched for orthologs to putative C. gigas immune response genes, supporting the notion that these modules do indeed represent a component of the Aplysia immune response. Indeed, several DE genes in the C. gigas immune response were orthologs of transcripts with high module membership in the pink, darkgreen, and/or greenyellow modules, such as the aforementioned orthologs of HERK4, BIRC3, IRF1, IRF8, MPEG1, DDX58/RIG-I, and most importantly all three orthologs of ZNFX1, including the greenyellow hub gene. Furthermore, several more transcripts, although not identified as orthologs of genes demonstrated to be DE in Lafont et al (2020) [37], do still map to the same human ortholog as genes identified in Lafont et al (2020) [37], such as sacsin. However, when comparing the list of significantly differentially expressed genes as a result of poly(I·C) in C. gigas to Aplysia orthologous genes significantly upregulated in sensory neuron aging as reported in our previous study, only two orthologs are shared. This suggest that, while the greenyellow and darkreen modules may capture an immune transcriptional program, viral infection and associated immune response are not drivers of transcriptional change in Aplysia sensory neurons. Moreover, this lack of correlation with oyster genes chronically upregulated after exposure to a virus analog indicates the need for caution in interpreting changes in module expression. Even when specific modules that show an increase in expression with aging include transcript members with anti-inflammatory or antiviral functions, these specific genes may not individually exhibit age-related expression changes. On the other hand, a majority of transcripts with high module membership in the royalblue, pink, and orange modules were identified as differentially expressed in our previous study, suggesting inflammation may result in proteostatic stress and mitochondrial dysfunction rather than infection.
Several transcripts from the royalblue module interact with the pink and orange modules, primarily as inhibitors. MTFS2 specifically inhibits the activity of EIF2AK3 from the orange module, which functions to maintain mitochondrial morphology, Ca2+ homeostasis, and limit ROS production [299]. ITCH, via its interaction with TXNIP, promotes an antioxidant state, prevents pro-inflammatory signaling through ROS and NFkB, and prevents TXNIP from inhibiting glycolysis [300,301]. Similarly, PARK7 also inhibits NFkB signaling and the astrocyte inflammatory response [302,303]. Finally, GPS2 also exhibits strong anti-inflammatory activity [304,305]. Conversely, PDE12 from the pink module is known to suppresses mitochondrial translation and contribute to respiratory incompetence [306]. Perhaps the eigengene expression perturbation at 9 months of age common to these expression modules may represent a transition from a state of healthy metabolic activity and antioxidant capacity exemplified by the transcripts of the royalblue module towards an aged state typified by inflammation and protein dyshomeostasis suggested by the transcript sets of the pink and orange modules. The role of ROS represents a potentially interesting link between these modules.
Decreased activity of mitochondrial homeostasis and metabolism mechanism suggested by the royalblue module suggest mitochondrial dysfunction with age, a well-known source of ROS [307]. Furthermore, downregulation of several antioxidant proteins in the royalblue module would suggest decreased capacity for ROS defense. Overexpression of SOD2 and GPX have been demonstrated to inhibit NFkB activation [308,309], thus the downregulation of these and other antioxidants in the royalblue module may in fact potentiate NFkB activation as suggested by the pink and orange modules via increased ROS levels. While the role of ROS in NFkB signaling is complex and cell type specific, prolonged oxidative stress has been shown to increased NFkB signaling and contribute to a pro-inflammatory state [310,311]. Upregulation of FTH by NFkB, as seen in the pink module, has also been suggested to be a major component of the NFkB derived antioxidant response against high H2O2 levels during chronic inflammation and oxidative stress [312]. Furthermore, ER stress resulting from increases in misfolded proteins due to oxidative stress, signatures of which were identified in our previous study and are also suggested by the orange module, has been shown to activate NFkB as well. Specifically, the activity of PERK, which is among the top orthologs in the orange module, plays a crucial role in activation of NFkB during ER stress [313]. In addition to downregulation of antioxidants, co-temporal upregulation of ROS producers like the two DUOX1 orthologs in the pink module suggest an increased in ROS, [88,134].
Indeed, chronic, low-grade inflammation has been suggested to be both cause and consequence of mitochondrial dysfunction and resulting metabolic impairment in neuronal aging [314–316]. These data suggest that chronic inflammation contributes to an increasingly oxidative environment in these neurons, exacerbating mitochondrial dysfunction that is known to occur in aging.
Supporting information
For each cutout, the scale free topology model fit (A), median connectivity (B), mean connectivity (C), and max connectivity (D) of a co-expression matrix (y-axis) at a selected soft threshold power (x-axis) is plotted for both BSC (black) and PVC (Red). A soft power of 16 was selected for both sensory neuron types to achieve a scale free topology fit above 0.9 and to minimize the mean connectivity.
(TIF)
Module eigengene names are arbitrarily assigned. Red horizontal line represents 0.25 branch height threshold for similarity, below which modules are merged.
(TIF)
Colored bars at the bottom represent module assignments for each transcript. Module colors are arbitrarily assigned. The top color set represents the module assignment before merging similar of similar modules, while bottom bar represents module assignment post merge (see S2 Fig). In total, 13 co-expression modules were identified.
(TIF)
Each cell of the heatmap represents the correlation between a module eigengene (row) and phenotype (column). The top number in a cell is the value of Pearson correlation between two eigengenes. The p-value significance of module-trait correlation is the bottom number in each cell in parentheses. The phenotypes are animal weight at sacrifice (weight), latency of two reflex behaviors described in Greer et al 2018: Tail withdrawal reflex time (TWRT) and time to right (TTR), and chronological age of the animal in months at sacrifice (Age). Correlations for age are similar between sensory neuron types for the royalblue, saddlebrown, greenyellow, orange, pink, and darkgreen modules.
(TIF)
All Software and respective versions used for RNA sequencing read quality control and quality assurance, mapping and abundance estimation, and downstream analysis.
(DOCX)
Each cell represents the number of transcripts shared between respective module (row) and cluster (column). The “n” column represents the number of transcripts in a given module, and the “n” row represents the number of transcripts in a given cluster. Values in the “Sum” columns are row sums, e.g. the total number of transcripts in each respective module also present in the clusters. The “% of module” columns represent percentage values calculated by dividing the “Sum” columns by the total number of transcripts in a cluster set (1106 for B clusters, and 1198 for P clusters). Values in the “Sum” row are column sums, e.g. the total number of transcripts in each respective cluster that are also present in the modules. The “cluster %” row represent percentage values calculated by dividing the “Sum” row by the total number of transcripts among all modules (10012).
(DOCX)
All R packages and respective versions used for clustering, analysis, and visualization of RNA sequencing transcript abundances.
(TXT)
Full results for all modules of KEGG enrichment of co-expression modules using clusterProfiler.
(XLSX)
Per module module-membership values and significance for all transcripts assigned to each module.
(XLSX)
This file contains the results of mapping Aplysia californica transcript IDs to Crassostrea gigas gene IDs for transcripts in Aplysia age associated co-expression modules that mapped to C. gigas genes identified as DE in immune priming and viral challenge by Laont et al 2020.
(XLSX)
This file contains a list of genes upregulated genes in C. gigas immune respone to viral analogue poly(IC) as reported in Lafont et al. 2020 [37] that have orthologs with evalue of >1E-20 in Aplysia. Aplysia orthologs are annotated with transcript, protein, and gene RefSeq identifiers as well as neuron type-specific adjusted pvalues, expression cluster assignment, and expression cluster direction reported in our previous publication. Only two Aplysia orthologs were identified as significantly upregulated in aging sensory neurons.
(XLSX)
Acknowledgments
We gratefully acknowledge the University of Miami Aplysia Resource staff for their assistance. We are indebted to Dr. Douglas Crawford and Dr. Marjorie Oleksiak for use of their Nanodrop and Bioanalyzer. We thank Dr. Justin Greer for his assistance in RNA extraction and invaluable advice.
Data Availability
The BioProject in which our sequencing data is archived can be accessed here: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA639857. The project has Accession: PRJNA639857, ID: 639857, and title: Aplysia californica Sensory Neuron Time Series Transcriptome.
Funding Statement
This work was funded by the National Institutes of Health Grant (P40OD010952). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. This work was made possible, in part, through access to the Genomics High Throughput Facility Shared Resource of the Cancer Center Support Grant (P30CA-062203) at the University of California, Irvine and NIH shared instrumentation grants 1S10RR025496-01, 1S10OD010794-01, and 1S10OD021718-01.
References
- 1.Grimm A, Eckert A. Brain aging and neurodegeneration: from a mitochondrial point of view. Journal of Neurochemistry. 2017;143(4):418–31. doi: 10.1111/jnc.14037 WOS:000415368800004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Poddar J, Pradhan M, Ganguly G, Chakrabarti S. Biochemical deficits and cognitive decline in brain aging: Intervention by dietary supplements. J Chem Neuroanat. 2019;95:70–80. doi: 10.1016/j.jchemneu.2018.04.002 . [DOI] [PubMed] [Google Scholar]
- 3.Lopez-Otin C, Blasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell. 2013;153(6):1194–217. doi: 10.1016/j.cell.2013.05.039 ; PubMed Central PMCID: PMC3836174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Watts ME, Pocock R, Claudianos C. Brain Energy and Oxygen Metabolism: Emerging Role in Normal Function and Disease. Front Mol Neurosci. 2018;11:216. doi: 10.3389/fnmol.2018.00216 ; PubMed Central PMCID: PMC6023993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mattson MP, Arumugam TV. Hallmarks of Brain Aging: Adaptive and Pathological Modification by doi: 10.1016/j.cmet.2018.05.011 States. Cell Metabolism. 2018;27(6):1176–99. WOS:000434480000007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yankner BA, Lu T, Loerch P. The aging brain. Annu Rev Pathol. 2008;3:41–66. doi: 10.1146/annurev.pathmechdis.2.010506.092044 . [DOI] [PubMed] [Google Scholar]
- 7.Buffenstein R, Edrey YH, Yang T, Mele J. The oxidative stress theory of aging: embattled or invincible? Insights from non-traditional model organisms. Age. 2008;30(2–3):99–109. doi: 10.1007/s11357-008-9058-z WOS:000258881300004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gems D, Partridge L. Genetics of longevity in model organisms: debates and paradigm shifts. Annu Rev Physiol. 2013;75:621–44. doi: 10.1146/annurev-physiol-030212-183712 . [DOI] [PubMed] [Google Scholar]
- 9.Capo TR, Fieber LA, Stommes DL, Walsh PJ. Reproductive output in the hatchery-reared California sea hare at different stocking densities. Contemp Top Lab Anim. 2003;42(5):31–5. WOS:000185698300007. [PubMed] [Google Scholar]
- 10.Moroz LL. Aplysia. Curr Biol. 2011;21(2):R60–1. doi: 10.1016/j.cub.2010.11.028 ; PubMed Central PMCID: PMC4024469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Audesirk TE. A Field Study of Growth and Reproduction in Aplysia Californica. Biol Bull. 1979;157(3):407–21. doi: 10.2307/1541026 . [DOI] [PubMed] [Google Scholar]
- 12.Kempsell AT, Fieber LA. Behavioral aging is associated with reduced sensory neuron excitability in Aplysia californica. Front Aging Neurosci. 2014;6. doi: 10.3389/fnagi.2014.00006 WOS:000339432000001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gerdes R, Fieber LA. Life history and aging of captive-reared California sea hares (Aplysia californica). J Am Assoc Lab Anim. 2006;45(1):40–7. WOS:000236461200006. [PubMed] [Google Scholar]
- 14.Bailey CH, Castellucci VF, Koester J, Chen M. Behavioral changes in aging Aplysia: a model system for studying the cellular basis of age-impaired learning, memory, and arousal. Behav Neural Biol. 1983;38(1):70–81. doi: 10.1016/s0163-1047(83)90399-0 . [DOI] [PubMed] [Google Scholar]
- 15.Rattan KS, Peretz B. Age-dependent behavioral changes and physiological changes in identified neurons in Aplysia californica. J Neurobiol. 1981;12(5):469–78. doi: 10.1002/neu.480120506 . [DOI] [PubMed] [Google Scholar]
- 16.Papka R, Peretz B, Tudor J, Becker J. Age-Dependent Anatomical Changes in an Identified Neuron in the Cns of Aplysia-Californica. Journal of Neurobiology. 1981;12(5):455–68. doi: 10.1002/neu.480120505 WOS:A1981MD97800004. [DOI] [PubMed] [Google Scholar]
- 17.Kempsell AT, Fieber LA. Habituation in the Tail Withdrawal Reflex Circuit is Impaired During Aging in Aplysia californica. Front Aging Neurosci. 2016;8. doi: 10.3389/fnagi.2016.00008 WOS:000369905200001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kempsell AT, Fieber LA. Aging in Sensory and Motor Neurons Results in Learning Failure in Aplysia californica. Plos One. 2015;10(5). doi: 10.1371/journal.pone.0127056 WOS:000354544200147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kempsell AT, Fieber LA. Age-related deficits in synaptic plasticity rescued by activating PKA or PKC in sensory neurons of Aplysia californica. Front Aging Neurosci. 2015;7. doi: 10.3389/fnagi.2015.00007 WOS:000361232000001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Greer JB, Schmale MC, Fieber LA. Whole-transcriptome changes in gene expression accompany aging of sensory neurons in Aplysia californica. BMC genomics. 2018;19(1):529. doi: 10.1186/s12864-018-4909-1 ; PubMed Central PMCID: PMC6042401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moroz LL, Kohn AB. Single-neuron transcriptome and methylome sequencing for epigenomic analysis of aging. Methods Mol Biol. 2013;1048:323–52. doi: 10.1007/978-1-62703-556-9_21 ; PubMed Central PMCID: PMC4045448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moroz LL, Kohn AB. Do different neurons age differently? Direct genome-wide analysis of aging in single identified cholinergic neurons. Front Aging Neurosci. 2010;2. doi: 10.3389/neuro.24.002.2010 ; PubMed Central PMCID: PMC2910937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kadakkuzha BM, Akhmedov K, Capo TR, Carvalloza AC, Fallahi M, Puthanveettil SV. Age-associated bidirectional modulation of gene expression in single identified R15 neuron of Aplysia. BMC genomics. 2013;14:880. doi: 10.1186/1471-2164-14-880 ; PubMed Central PMCID: PMC3909179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kron NS, Schmale MC, Fieber LA. Changes in Metabolism and Proteostasis Drive Aging Phenotype in Aplysia californica Sensory Neurons. Front Aging Neurosci. 2020;12(280).doi: 10.3389/fnagi.2020.573764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Song Y, Pan Y, Liu J. The relevance between the immune response-related gene module and clinical traits in head and neck squamous cell carcinoma. Cancer Manag Res. 2019;11:7455–72. doi: 10.2147/CMAR.S201177 ; PubMed Central PMCID: PMC6689548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. Bmc Bioinformatics. 2008;9. doi: 10.1186/1471-2105-9-9 WOS:000262999900002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nam D, Kim SY. Gene-set approach for expression pattern analysis. Brief Bioinform. 2008;9(3):189–97. doi: 10.1093/bib/bbn001 . [DOI] [PubMed] [Google Scholar]
- 28.Oldham MC, Konopka G, Iwamoto K, Langfelder P, Kato T, Horvath S, et al. Functional organization of the transcriptome in human brain. Nat Neurosci. 2008;11(11):1271–82. doi: 10.1038/nn.2207 ; PubMed Central PMCID: PMC2756411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Serin EAR, Nijveen H, Hilhorst HWM, Ligterink W. Learning from Co-expression Networks: Possibilities and Challenges. Front Plant Sci. 2016;7. doi: 10.3389/fpls.2016.00007 WOS:000373600300001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bushnell B. BBMap. 37.90 ed. sourceforge.net/projects/bbmap/2014.
- 31.Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods. 2017;14(4):417–9. doi: 10.1038/nmeth.4197 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Soneson C, Love MI, Robinson MD. Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Res. 2015;4:1521. doi: 10.12688/f1000research.7563.2 ; PubMed Central PMCID: PMC4712774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gentleman R, Carey V, Huber W, Hahne F. genefilter: genefilter: methods for filtering genes from high-throughput experiments. 1.70.0 ed2020. [Google Scholar]
- 34.Leek JT. svaseq: removing batch effects and other unwanted noise from sequencing data. Nucleic Acids Res. 2014;42(21). WOS:000347914600002. doi: 10.1093/nar/gku864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12). doi: 10.1186/s13059-014-0550-8 WOS:000346609500022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu GC, Wang LG, Han YY, He QY. clusterProfiler: an R Package for Comparing Biological Themes Among Gene Clusters. Omics-a Journal of Integrative Biology. 2012;16(5):284–7. doi: 10.1089/omi.2011.0118 WOS:000303653300007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lafont M, Vergnes A, Vidal-Dupiol J, de Lorgeril J, Gueguen Y, Haffner P, et al. A Sustained Immune Response Supports Long-Term Antiviral Immune Priming in the Pacific Oyster, Crassostrea gigas. mBio. 2020;11(2). doi: 10.1128/mBio.02777-19 ; PubMed Central PMCID: PMC7064767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Deacon SW, Serpinskaya AS, Vaughan PS, Lopez Fanarraga M, Vernos I, Vaughan KT, et al. Dynactin is required for bidirectional organelle transport. J Cell Biol. 2003;160(3):297–301. doi: 10.1083/jcb.200210066 ; PubMed Central PMCID: PMC2172679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Faik P, Walker JI, Redmill AA, Morgan MJ. Mouse glucose-6-phosphate isomerase and neuroleukin have identical 3’ sequences. Nature. 1988;332(6163):455–7. doi: 10.1038/332455a0 . [DOI] [PubMed] [Google Scholar]
- 40.Imai H, Nakagawa Y. Biological significance of phospholipid hydroperoxide glutathione peroxidase (PHGPx, GPx4) in mammalian cells. Free Radic Biol Med. 2003;34(2):145–69. doi: 10.1016/s0891-5849(02)01197-8 . [DOI] [PubMed] [Google Scholar]
- 41.Hao HX, Khalimonchuk O, Schraders M, Dephoure N, Bayley JP, Kunst H, et al. SDH5, a gene required for flavination of succinate dehydrogenase, is mutated in paraganglioma. Science. 2009;325(5944):1139–42. doi: 10.1126/science.1175689 ; PubMed Central PMCID: PMC3881419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weir ML, Xie H, Klip A, Trimble WS. VAP-A binds promiscuously to both v- and tSNAREs. Biochem Biophys Res Commun. 2001;286(3):616–21. doi: 10.1006/bbrc.2001.5437 . [DOI] [PubMed] [Google Scholar]
- 43.Ikeda Y, Okamura-Ikeda K, Tanaka K. Purification and characterization of short-chain, medium-chain, and long-chain acyl-CoA dehydrogenases from rat liver mitochondria. Isolation of the holo- and apoenzymes and conversion of the apoenzyme to the holoenzyme. J Biol Chem. 1985;260(2):1311–25. . [PubMed] [Google Scholar]
- 44.Jerber J, Baas D, Soulavie F, Chhin B, Cortier E, Vesque C, et al. The coiled-coil domain containing protein CCDC151 is required for the function of IFT-dependent motile cilia in animals. Hum Mol Genet. 2014;23(3):563–77. doi: 10.1093/hmg/ddt445 . [DOI] [PubMed] [Google Scholar]
- 45.Taira T, Saito Y, Niki T, Iguchi-Ariga SM, Takahashi K, Ariga H. DJ-1 has a role in antioxidative stress to prevent cell death. Embo Rep. 2004;5(2):213–8. doi: 10.1038/sj.embor.7400074 ; PubMed Central PMCID: PMC1298985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Junn E, Jang WH, Zhao X, Jeong BS, Mouradian MM. Mitochondrial localization of DJ-1 leads to enhanced neuroprotection. J Neurosci Res. 2009;87(1):123–9. doi: 10.1002/jnr.21831 ; PubMed Central PMCID: PMC2752655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Clements CM, McNally RS, Conti BJ, Mak TW, Ting JP. DJ-1, a cancer- and Parkinson’s disease-associated protein, stabilizes the antioxidant transcriptional master regulator Nrf2. Proc Natl Acad Sci U S A. 2006;103(41):15091–6. doi: 10.1073/pnas.0607260103 ; PubMed Central PMCID: PMC1586179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Niemann A, Ruegg M, La Padula V, Schenone A, Suter U. Ganglioside-induced differentiation associated protein 1 is a regulator of the mitochondrial network: new implications for Charcot-Marie-Tooth disease. J Cell Biol. 2005;170(7):1067–78. doi: 10.1083/jcb.200507087 ; PubMed Central PMCID: PMC2171517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Carpenter K, Pollitt RJ, Middleton B. Human liver long-chain 3-hydroxyacyl-coenzyme A dehydrogenase is a multifunctional membrane-bound beta-oxidation enzyme of mitochondria. Biochem Biophys Res Commun. 1992;183(2):443–8. doi: 10.1016/0006-291x(92)90501-b . [DOI] [PubMed] [Google Scholar]
- 50.Sun SC, Ma D, Li MY, Zhang RX, Huang C, Huang HJ, et al. Mutations in C1orf194, encoding a calcium regulator, cause dominant Charcot-Marie-Tooth disease. Brain. 2019;142(8):2215–29. doi: 10.1093/brain/awz151 . [DOI] [PubMed] [Google Scholar]
- 51.Sasaki K, Shiba K, Nakamura A, Kawano N, Satouh Y, Yamaguchi H, et al. Calaxin is required for cilia-driven determination of vertebrate laterality. Commun Biol. 2019;2:226. doi: 10.1038/s42003-019-0462-y ; PubMed Central PMCID: PMC6586612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yorikawa C, Shibata H, Waguri S, Hatta K, Horii M, Katoh K, et al. Human CHMP6, a myristoylated ESCRT-III protein, interacts directly with an ESCRT-II component EAP20 and regulates endosomal cargo sorting. Biochem J. 2005;387(Pt 1):17–26. doi: 10.1042/BJ20041227 ; PubMed Central PMCID: PMC1134928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chin D, Means AR. Calmodulin: a prototypical calcium sensor. Trends Cell Biol. 2000;10(8):322–8. doi: 10.1016/s0962-8924(00)01800-6 WOS:000088382600004. [DOI] [PubMed] [Google Scholar]
- 54.Zelko IN, Mariani TJ, Folz RJ. Superoxide dismutase multigene family: a comparison of the CuZn-SOD (SOD1), Mn-SOD (SOD2), and EC-SOD (SOD3) gene structures, evolution, and expression. Free Radic Biol Med. 2002;33(3):337–49. doi: 10.1016/s0891-5849(02)00905-x . [DOI] [PubMed] [Google Scholar]
- 55.Schriner SE, Linford NJ, Martin GM, Treuting P, Ogburn CE, Emond M, et al. Extension of murine life span by overexpression of catalase targeted to mitochondria. Science. 2005;308(5730):1909–11. doi: 10.1126/science.1106653 WOS:000230120000040. [DOI] [PubMed] [Google Scholar]
- 56.Setty SR, Tenza D, Truschel ST, Chou E, Sviderskaya EV, Theos AC, et al. BLOC-1 is required for cargo-specific sorting from vacuolar early endosomes toward lysosome-related organelles. Mol Biol Cell. 2007;18(3):768–80. doi: 10.1091/mbc.e06-12-1066 ; PubMed Central PMCID: PMC1805088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cardamone MD, Tanasa B, Cederquist CT, Huang J, Mahdaviani K, Li W, et al. Mitochondrial Retrograde Signaling in Mammals Is Mediated by the Transcriptional Cofactor GPS2 via Direct Mitochondria-to-Nucleus Translocation. Mol Cell. 2018;69(5):757–72 e7. doi: 10.1016/j.molcel.2018.01.037 ; PubMed Central PMCID: PMC6022402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hua T, Wu D, Ding W, Wang J, Shaw N, Liu ZJ. Studies of human 2,4-dienoyl CoA reductase shed new light on peroxisomal beta-oxidation of unsaturated fatty acids. J Biol Chem. 2012;287(34):28956–65. doi: 10.1074/jbc.M112.385351 ; PubMed Central PMCID: PMC3436514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Olsen RK, Andresen BS, Christensen E, Bross P, Skovby F, Gregersen N. Clear relationship between ETF/ETFDH genotype and phenotype in patients with multiple acyl-CoA dehydrogenation deficiency. Hum Mutat. 2003;22(1):12–23. doi: 10.1002/humu.10226 . [DOI] [PubMed] [Google Scholar]
- 60.Yoshihara M, Adolfsen B, Galle KT, Littleton JT. Retrograde signaling by Syt 4 induces presynaptic release and synapse-specific growth. Science. 2005;310(5749):858–63. doi: 10.1126/science.1117541 . [DOI] [PubMed] [Google Scholar]
- 61.Korkut C, Li Y, Koles K, Brewer C, Ashley J, Yoshihara M, et al. Regulation of postsynaptic retrograde signaling by presynaptic exosome release. Neuron. 2013;77(6):1039–46. doi: 10.1016/j.neuron.2013.01.013 ; PubMed Central PMCID: PMC3626103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen M, Chen ZH, Wang YY, Tan Z, Zhu CZ, Li YJ, et al. Mitophagy receptor FUNDC1 regulates mitochondrial dynamics and mitophagy. Autophagy. 2016;12(4):689–702. doi: 10.1080/15548627.2016.1151580 WOS:000373983600007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yan L, Herrington J, Goldberg E, Dulski PM, Bugianesi RM, Slaughter RS, et al. Stichodactyla helianthus peptide, a pharmacological tool for studying Kv3.2 channels. Mol Pharmacol. 2005;67(5):1513–21. doi: 10.1124/mol.105.011064 . [DOI] [PubMed] [Google Scholar]
- 64.Cao YL, Meng S, Chen Y, Feng JX, Gu DD, Yu B, et al. MFN1 structures reveal nucleotide-triggered dimerization critical for mitochondrial fusion. Nature. 2017;542(7641):372–6. doi: 10.1038/nature21077 ; PubMed Central PMCID: PMC5319402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Santel A, Fuller MT. Control of mitochondrial morphology by a human mitofusin. J Cell Sci. 2001;114(Pt 5):867–74. 11181170. [DOI] [PubMed] [Google Scholar]
- 66.Merveille AC, Davis EE, Becker-Heck A, Legendre M, Amirav I, Bataille G, et al. CCDC39 is required for assembly of inner dynein arms and the dynein regulatory complex and for normal ciliary motility in humans and dogs. Nat Genet. 2011;43(1):72–8. doi: 10.1038/ng.726 ; PubMed Central PMCID: PMC3509786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bugarcic A, Zhe Y, Kerr MC, Griffin J, Collins BM, Teasdale RD. Vps26A and Vps26B subunits define distinct retromer complexes. Traffic. 2011;12(12):1759–73. doi: 10.1111/j.1600-0854.2011.01284.x . [DOI] [PubMed] [Google Scholar]
- 68.Kim E, Lee Y, Lee HJ, Kim JS, Song BS, Huh JW, et al. Implication of mouse Vps26b-Vps29-Vps35 retromer complex in sortilin trafficking. Biochem Biophys Res Commun. 2010;403(2):167–71. doi: 10.1016/j.bbrc.2010.10.121 . [DOI] [PubMed] [Google Scholar]
- 69.Jaillard C, Mouret A, Niepon ML, Clerin E, Yang Y, Lee-Rivera I, et al. Nxnl2 splicing results in dual functions in neuronal cell survival and maintenance of cell integrity. Hum Mol Genet. 2012;21(10):2298–311. doi: 10.1093/hmg/dds050 ; PubMed Central PMCID: PMC3664437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stroud DA, Surgenor EE, Formosa LE, Reljic B, Frazier AE, Dibley MG, et al. Accessory subunits are integral for assembly and function of human mitochondrial complex I. Nature. 2016;538(7623):123–6. doi: 10.1038/nature19754 . [DOI] [PubMed] [Google Scholar]
- 71.Ogilvie I, Kennaway NG, Shoubridge EA. A molecular chaperone for mitochondrial complex I assembly is mutated in a progressive encephalopathy. J Clin Invest. 2005;115(10):2784–92. doi: 10.1172/JCI26020 ; PubMed Central PMCID: PMC1236688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Inuzuka T, Suzuki H, Kawasaki M, Shibata H, Wakatsuki S, Maki M. Molecular basis for defect in Alix-binding by alternatively spliced isoform of ALG-2 (ALG-2DeltaGF122) and structural roles of F122 in target recognition. BMC Struct Biol. 2010;10:25. doi: 10.1186/1472-6807-10-25 ; PubMed Central PMCID: PMC2927601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li Y, Jourdain AA, Calvo SE, Liu JS, Mootha VK. CLIC, a tool for expanding biological pathways based on co-expression across thousands of datasets. Plos Comput Biol. 2017;13(7):e1005653. doi: 10.1371/journal.pcbi.1005653 ; PubMed Central PMCID: PMC5546725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.McCormack K, McCormack T, Tanouye M, Rudy B, Stuhmer W. Alternative splicing of the human Shaker K+ channel beta 1 gene and functional expression of the beta 2 gene product. FEBS Lett. 1995;370(1–2):32–6. doi: 10.1016/0014-5793(95)00785-8 . [DOI] [PubMed] [Google Scholar]
- 75.Floyd BJ, Wilkerson EM, Veling MT, Minogue CE, Xia C, Beebe ET, et al. Mitochondrial Protein Interaction Mapping Identifies Regulators of Respiratory Chain Function. Mol Cell. 2016;63(4):621–32. doi: 10.1016/j.molcel.2016.06.033 ; PubMed Central PMCID: PMC4992456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kondo S, Sato-Yoshitake R, Noda Y, Aizawa H, Nakata T, Matsuura Y, et al. KIF3A is a new microtubule-based anterograde motor in the nerve axon. J Cell Biol. 1994;125(5):1095–107. doi: 10.1083/jcb.125.5.1095 ; PubMed Central PMCID: PMC2120052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tang SJ, Meulemans D, Vazquez L, Colaco N, Schuman E. A role for a rat homolog of staufen in the transport of RNA to neuronal dendrites. Neuron. 2001;32(3):463–75. doi: 10.1016/s0896-6273(01)00493-7 . [DOI] [PubMed] [Google Scholar]
- 78.Zhang P, Wang C, Gao K, Wang D, Mao J, An J, et al. The ubiquitin ligase itch regulates apoptosis by targeting thioredoxin-interacting protein for ubiquitin-dependent degradation. J Biol Chem. 2010;285(12):8869–79. doi: 10.1074/jbc.M109.063321 ; PubMed Central PMCID: PMC2838308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.You F, Sun H, Zhou X, Sun W, Liang S, Zhai Z, et al. PCBP2 mediates degradation of the adaptor MAVS via the HECT ubiquitin ligase AIP4. Nat Immunol. 2009;10(12):1300–8. doi: 10.1038/ni.1815 . [DOI] [PubMed] [Google Scholar]
- 80.Yamashita H, Avraham S, Jiang S, London R, Van Veldhoven PP, Subramani S, et al. Characterization of human and murine PMP20 peroxisomal proteins that exhibit antioxidant activity in vitro. J Biol Chem. 1999;274(42):29897–904. doi: 10.1074/jbc.274.42.29897 . [DOI] [PubMed] [Google Scholar]
- 81.Scherer DC, Brockman JA, Chen Z, Maniatis T, Ballard DW. Signal-induced degradation of I kappa B alpha requires site-specific ubiquitination. Proc Natl Acad Sci U S A. 1995;92(24):11259–63. doi: 10.1073/pnas.92.24.11259 ; PubMed Central PMCID: PMC40611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Xiao C, Ghosh S. NF-kappaB, an evolutionarily conserved mediator of immune and inflammatory responses. Adv Exp Med Biol. 2005;560:41–5. doi: 10.1007/0-387-24180-9_5 . [DOI] [PubMed] [Google Scholar]
- 83.Mahoney DJ, Cheung HH, Mrad RL, Plenchette S, Simard C, Enwere E, et al. Both cIAP1 and cIAP2 regulate TNFalpha-mediated NF-kappaB activation. Proc Natl Acad Sci U S A. 2008;105(33):11778–83. doi: 10.1073/pnas.0711122105 ; PubMed Central PMCID: PMC2575330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bertrand MJ, Doiron K, Labbe K, Korneluk RG, Barker PA, Saleh M. Cellular inhibitors of apoptosis cIAP1 and cIAP2 are required for innate immunity signaling by the pattern recognition receptors NOD1 and NOD2. Immunity. 2009;30(6):789–801. doi: 10.1016/j.immuni.2009.04.011 . [DOI] [PubMed] [Google Scholar]
- 85.Haahr P, Borgermann N, Guo X, Typas D, Achuthankutty D, Hoffmann S, et al. ZUFSP Deubiquitylates K63-Linked Polyubiquitin Chains to Promote Genome Stability. Mol Cell. 2018;70(1):165–74 e6. doi: 10.1016/j.molcel.2018.02.024 . [DOI] [PubMed] [Google Scholar]
- 86.Huang F, Kirkpatrick D, Jiang X, Gygi S, Sorkin A. Differential regulation of EGF receptor internalization and degradation by multiubiquitination within the kinase domain. Mol Cell. 2006;21(6):737–48. doi: 10.1016/j.molcel.2006.02.018 . [DOI] [PubMed] [Google Scholar]
- 87.Rodriguez CI, Stewart CL. Disruption of the ubiquitin ligase HERC4 causes defects in spermatozoon maturation and impaired fertility. Dev Biol. 2007;312(2):501–8. doi: 10.1016/j.ydbio.2007.09.053 . [DOI] [PubMed] [Google Scholar]
- 88.Schenk G, Mitić N, Hanson GR, Comba P. Purple acid phosphatase: A journey into the function and mechanism of a colorful enzyme. Coordination Chemistry Reviews. 2013;257(2):473–82. doi: 10.1016/j.ccr.2012.03.020 [DOI] [Google Scholar]
- 89.Rienzo M, Casamassimi A. Integrator complex and transcription regulation: Recent findings and pathophysiology. Biochim Biophys Acta. 2016;1859(10):1269–80. doi: 10.1016/j.bbagrm.2016.07.008 . [DOI] [PubMed] [Google Scholar]
- 90.Lai F, Gardini A, Zhang A, Shiekhattar R. Integrator mediates the biogenesis of enhancer RNAs. Nature. 2015;525(7569):399–403. doi: 10.1038/nature14906 ; PubMed Central PMCID: PMC4718573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tatomer DC, Elrod ND, Liang D, Xiao MS, Jiang JZ, Jonathan M, et al. The Integrator complex cleaves nascent mRNAs to attenuate transcription. Genes Dev. 2019;33(21–22):1525–38. doi: 10.1101/gad.330167.119 ; PubMed Central PMCID: PMC6824465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Elrod ND, Henriques T, Huang KL, Tatomer DC, Wilusz JE, Wagner EJ, et al. The Integrator Complex Attenuates Promoter-Proximal Transcription at Protein-Coding Genes. Mol Cell. 2019;76(5):738–52 e7. doi: 10.1016/j.molcel.2019.10.034 ; PubMed Central PMCID: PMC6952639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mahuran DJ. The GM2 activator protein, its roles as a co-factor in GM2 hydrolysis and as a general glycolipid transport protein. Biochim Biophys Acta. 1998;1393(1):1–18. doi: 10.1016/s0005-2760(98)00057-5 . [DOI] [PubMed] [Google Scholar]
- 94.Marton MJ, Crouch D, Hinnebusch AG. GCN1, a translational activator of GCN4 in Saccharomyces cerevisiae, is required for phosphorylation of eukaryotic translation initiation factor 2 by protein kinase GCN2. Mol Cell Biol. 1993;13(6):3541–56. doi: 10.1128/mcb.13.6.3541-3556.1993 ; PubMed Central PMCID: PMC359824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Jones RS, Tu C, Zhang M, Qu J, Morris ME. Characterization and Proteomic-Transcriptomic Investigation of Monocarboxylate Transporter 6 Knockout Mice: Evidence of a Potential Role in Glucose and Lipid Metabolism. Mol Pharmacol. 2019;96(3):364–76. doi: 10.1124/mol.119.116731 ; PubMed Central PMCID: PMC6693307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Finch AJ, Hilcenko C, Basse N, Drynan LF, Goyenechea B, Menne TF, et al. Uncoupling of GTP hydrolysis from eIF6 release on the ribosome causes Shwachman-Diamond syndrome. Genes Dev. 2011;25(9):917–29. doi: 10.1101/gad.623011 . PubMed Central PMCID: PMC3084026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ito S, Horikawa S, Suzuki T, Kawauchi H, Tanaka Y, Suzuki T, et al. Human NAT10 is an ATP-dependent RNA acetyltransferase responsible for N4-acetylcytidine formation in 18 S ribosomal RNA (rRNA). J Biol Chem. 2014;289(52):35724–30. doi: 10.1074/jbc.C114.602698 ; PubMed Central PMCID: PMC4276842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Liu X, Tan Y, Zhang C, Zhang Y, Zhang L, Ren P, et al. NAT10 regulates p53 activation through acetylating p53 at K120 and ubiquitinating Mdm2. Embo Rep. 2016;17(3):349–66. doi: 10.15252/embr.201540505 ; PubMed Central PMCID: PMC4772976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Arango D, Sturgill D, Alhusaini N, Dillman AA, Sweet TJ, Hanson G, et al. Acetylation of Cytidine in mRNA Promotes Translation Efficiency. Cell. 2018;175(7):1872–86 e24. doi: 10.1016/j.cell.2018.10.030 ; PubMed Central PMCID: PMC6295233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Singh B, Arlinghaus RB. Mos and the cell cycle. Prog Cell Cycle Res. 1997;3:251–9. doi: 10.1007/978-1-4615-5371-7_20 . [DOI] [PubMed] [Google Scholar]
- 101.Ma L, Huang Y, Song Z, Feng S, Tian X, Du W, et al. Livin promotes Smac/DIABLO degradation by ubiquitin-proteasome pathway. Cell Death Differ. 2006;13(12):2079–88. doi: 10.1038/sj.cdd.4401959 . [DOI] [PubMed] [Google Scholar]
- 102.Schnell S, Demolliere C, van den Berk P, Jacobs H. Gimap4 accelerates T-cell death. Blood. 2006;108(2):591–9. doi: 10.1182/blood-2005-11-4616 . [DOI] [PubMed] [Google Scholar]
- 103.Heinonen MT, Kanduri K, Lahdesmaki HJ, Lahesmaa R, Henttinen TA. Tubulin- and actin-associating GIMAP4 is required for IFN-gamma secretion during Th cell differentiation. Immunol Cell Biol. 2015;93(2):158–66. doi: 10.1038/icb.2014.86 ; PubMed Central PMCID: PMC4355353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Frolova L, Le Goff X, Rasmussen HH, Cheperegin S, Drugeon G, Kress M, et al. A highly conserved eukaryotic protein family possessing properties of polypeptide chain release factor. Nature. 1994;372(6507):701–3. doi: 10.1038/372701a0 . [DOI] [PubMed] [Google Scholar]
- 105.Greulich W, Wagner M, Gaidt MM, Stafford C, Cheng Y, Linder A, et al. TLR8 Is a Sensor of RNase T2 Degradation Products. Cell. 2019;179(6):1264–75 e13. doi: 10.1016/j.cell.2019.11.001 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Liu P, Huang J, Zheng Q, Xie L, Lu X, Jin J, et al. Mammalian mitochondrial RNAs are degraded in the mitochondrial intermembrane space by RNASET2. Protein Cell. 2017;8(10):735–49. doi: 10.1007/s13238-017-0448-9 ; PubMed Central PMCID: PMC5636749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Jin Y, Tan YJ, Chen LP, Liu Y, Ren ZQ. Reactive Oxygen Species Induces Lipid Droplet Accumulation in HepG2 Cells by Increasing Perilipin 2 Expression. Int J Mol Sci. 2018;19(11). doi: 10.3390/ijms19113445 WOS:000451528500161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gingras AC, Raught B, Sonenberg N. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu Rev Biochem. 1999;68:913–63. doi: 10.1146/annurev.biochem.68.1.913 . [DOI] [PubMed] [Google Scholar]
- 109.David R. eIF4A2 helps silence mRNAs. Nat Rev Mol Cell Bio. 2013;14(5):266–. doi: 10.1038/nrm3573 [DOI] [Google Scholar]
- 110.Prieto JL, McStay B. Recruitment of factors linking transcription and processing of pre-rRNA to NOR chromatin is UBF-dependent and occurs independent of transcription in human cells. Genes Dev. 2007;21(16):2041–54. doi: 10.1101/gad.436707 ; PubMed Central PMCID: PMC1948859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Januszyk K, Liu Q, Lima CD. Activities of human RRP6 and structure of the human RRP6 catalytic domain. Rna. 2011;17(8):1566–77. doi: 10.1261/rna.2763111 ; PubMed Central PMCID: PMC3153979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Dowd S, Sneddon AA, Keyse SM. Isolation of the human genes encoding the pyst1 and Pyst2 phosphatases: characterisation of Pyst2 as a cytosolic dual-specificity MAP kinase phosphatase and its catalytic activation by both MAP and SAP kinases. J Cell Sci. 1998;111 (Pt 22):3389–99. . [DOI] [PubMed] [Google Scholar]
- 113.Yoshida Y, Yoshimi R, Yoshii H, Kim D, Dey A, Xiong H, et al. The transcription factor IRF8 activates integrin-mediated TGF-beta signaling and promotes neuroinflammation. Immunity. 2014;40(2):187–98. doi: 10.1016/j.immuni.2013.11.022 ; PubMed Central PMCID: PMC4105266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kishimoto Y, Hiraiwa M, O’Brien JS. Saposins: structure, function, distribution, and molecular genetics. J Lipid Res. 1992;33(9):1255–67. . [PubMed] [Google Scholar]
- 115.Kaul G, Pattan G, Rafeequi T. Eukaryotic elongation factor-2 (eEF2): its regulation and peptide chain elongation. Cell Biochem Funct. 2011;29(3):227–34. doi: 10.1002/cbf.1740 . [DOI] [PubMed] [Google Scholar]
- 116.Brix K, Dunkhorst A, Mayer K, Jordans S. Cysteine cathepsins: cellular roadmap to different functions. Biochimie. 2008;90(2):194–207. doi: 10.1016/j.biochi.2007.07.024 . [DOI] [PubMed] [Google Scholar]
- 117.Funkelstein L, Beinfeld M, Minokadeh A, Zadina J, Hook V. Unique biological function of cathepsin L in secretory vesicles for biosynthesis of neuropeptides. Neuropeptides. 2010;44(6):457–66. doi: 10.1016/j.npep.2010.08.003 ; PubMed Central PMCID: PMC3058267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Trompouki E, Hatzivassiliou E, Tsichritzis T, Farmer H, Ashworth A, Mosialos G. CYLD is a deubiquitinating enzyme that negatively regulates NF-kappaB activation by TNFR family members. Nature. 2003;424(6950):793–6. doi: 10.1038/nature01803 . [DOI] [PubMed] [Google Scholar]
- 119.Friedman CS O’Donnell MA, Legarda-Addison D, Ng A, Cardenas WB, Yount JS, et al. The tumour suppressor CYLD is a negative regulator of RIG-I-mediated antiviral response. Embo Rep. 2008;9(9):930–6. doi: 10.1038/embor.2008.136 ; PubMed Central PMCID: PMC2529351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Nicholson DW, Ali A, Thornberry NA, Vaillancourt JP, Ding CK, Gallant M, et al. Identification and inhibition of the ICE/CED-3 protease necessary for mammalian apoptosis. Nature. 1995;376(6535):37–43. doi: 10.1038/376037a0 . [DOI] [PubMed] [Google Scholar]
- 121.Tanaka Y, Suzuki G, Matsuwaki T, Hosokawa M, Serrano G, Beach TG, et al. Progranulin regulates lysosomal function and biogenesis through acidification of lysosomes. Hum Mol Genet. 2017;26(5):969–88. doi: 10.1093/hmg/ddx011 . [DOI] [PubMed] [Google Scholar]
- 122.Qin J, Qian Y, Yao J, Grace C, Li X. SIGIRR inhibits interleukin-1 receptor- and toll-like receptor 4-mediated signaling through different mechanisms. J Biol Chem. 2005;280(26):25233–41. doi: 10.1074/jbc.M501363200 . [DOI] [PubMed] [Google Scholar]
- 123.Arosio P, Elia L, Poli M. Ferritin, Cellular Iron Storage and Regulation. Iubmb Life. 2017;69(6):414–22. doi: 10.1002/iub.1621 WOS:000403902700005. [DOI] [PubMed] [Google Scholar]
- 124.Waterfield MR, Zhang M, Norman LP, Sun SC. NF-kappaB1/p105 regulates lipopolysaccharide-stimulated MAP kinase signaling by governing the stability and function of the Tpl2 kinase. Mol Cell. 2003;11(3):685–94. doi: 10.1016/s1097-2765(03)00070-4 . [DOI] [PubMed] [Google Scholar]
- 125.Widmann B, Wandrey F, Badertscher L, Wyler E, Pfannstiel J, Zemp I, et al. The kinase activity of human Rio1 is required for final steps of cytoplasmic maturation of 40S subunits. Mol Biol Cell. 2012;23(1):22–35. doi: 10.1091/mbc.E11-07-0639 ; PubMed Central PMCID: PMC3248900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Chen YW, Ko WC, Chen CS, Chen PL. RIOK-1 Is a Suppressor of the p38 MAPK Innate Immune Pathway in Caenorhabditis elegans. Front Immunol. 2018;9:774. doi: 10.3389/fimmu.2018.00774 ; PubMed Central PMCID: PMC5913292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Baumas K, Soudet J, Caizergues-Ferrer M, Faubladier M, Henry Y, Mougin A. Human RioK3 is a novel component of cytoplasmic pre-40S pre-ribosomal particles. Rna Biol. 2012;9(2):162–74. doi: 10.4161/rna.18810 ; PubMed Central PMCID: PMC3346313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Feng J, De Jesus PD, Su V, Han S, Gong D, Wu NC, et al. RIOK3 is an adaptor protein required for IRF3-mediated antiviral type I interferon production. J Virol. 2014;88(14):7987–97. doi: 10.1128/JVI.00643-14 ; PubMed Central PMCID: PMC4097797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Shan J, Wang P, Zhou J, Wu D, Shi H, Huo K. RIOK3 interacts with caspase-10 and negatively regulates the NF-kappaB signaling pathway. Mol Cell Biochem. 2009;332(1–2):113–20. doi: 10.1007/s11010-009-0180-8 . [DOI] [PubMed] [Google Scholar]
- 130.Takashima K, Oshiumi H, Takaki H, Matsumoto M, Seya T. RIOK3-mediated phosphorylation of MDA5 interferes with its assembly and attenuates the innate immune response. Cell Rep. 2015;11(2):192–200. doi: 10.1016/j.celrep.2015.03.027 . [DOI] [PubMed] [Google Scholar]
- 131.Kadoya T, Khurana A, Tcherpakov M, Bromberg KD, Didier C, Broday L, et al. JAMP, a Jun N-terminal kinase 1 (JNK1)-associated membrane protein, regulates duration of JNK activity. Mol Cell Biol. 2005;25(19):8619–30. doi: 10.1128/MCB.25.19.8619-8630.2005 ; PubMed Central PMCID: PMC1265750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Mauro C, Pacifico F, Lavorgna A, Mellone S, Iannetti A, Acquaviva R, et al. ABIN-1 binds to NEMO/IKKgamma and co-operates with A20 in inhibiting NF-kappaB. J Biol Chem. 2006;281(27):18482–8. doi: 10.1074/jbc.M601502200 . [DOI] [PubMed] [Google Scholar]
- 133.Ashida H, Kim M, Schmidt-Supprian M, Ma A, Ogawa M, Sasakawa C. A bacterial E3 ubiquitin ligase IpaH9.8 targets NEMO/IKKgamma to dampen the host NF-kappaB-mediated inflammatory response. Nat Cell Biol. 2010;12(1):66–73; sup pp 1–9. doi: 10.1038/ncb2006 ; PubMed Central PMCID: PMC3107189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.McCallum KC, Garsin DA. The Role of Reactive Oxygen Species in Modulating the Caenorhabditis elegans Immune Response. Plos Pathog. 2016;12(11):e1005923. doi: 10.1371/journal.ppat.1005923 ; PubMed Central PMCID: PMC5104326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Zhang K, Shen X, Wu J, Sakaki K, Saunders T, Rutkowski DT, et al. Endoplasmic reticulum stress activates cleavage of CREBH to induce a systemic inflammatory response. Cell. 2006;124(3):587–99. doi: 10.1016/j.cell.2005.11.040 . [DOI] [PubMed] [Google Scholar]
- 136.Tu H, Nelson O, Bezprozvanny A, Wang Z, Lee SF, Hao YH, et al. Presenilins form ER Ca2+ leak channels, a function disrupted by familial Alzheimer’s disease-linked mutations. Cell. 2006;126(5):981–93. doi: 10.1016/j.cell.2006.06.059 ; PubMed Central PMCID: PMC3241869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Zampese E, Fasolato C, Kipanyula MJ, Bortolozzi M, Pozzan T, Pizzo P. Presenilin 2 modulates endoplasmic reticulum (ER)-mitochondria interactions and Ca2+ cross-talk. Proc Natl Acad Sci U S A. 2011;108(7):2777–82. doi: 10.1073/pnas.1100735108 ; PubMed Central PMCID: PMC3041131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Chen JM, Dando PM, Stevens RA, Fortunato M, Barrett AJ. Cloning and expression of mouse legumain, a lysosomal endopeptidase. Biochem J. 1998;335 (Pt 1):111–7. doi: 10.1042/bj3350111 ; PubMed Central PMCID: PMC1219758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Rozpedek W, Pytel D, Mucha B, Leszczynska H, Diehl JA, Majsterek I. The Role of the PERK/eIF2alpha/ATF4/CHOP Signaling Pathway in Tumor Progression During Endoplasmic Reticulum Stress. Curr Mol Med. 2016;16(6):533–44. doi: 10.2174/1566524016666160523143937 ; PubMed Central PMCID: PMC5008685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Miyara A, Ohta A, Okochi Y, Tsukada Y, Kuhara A, Mori I. Novel and conserved protein macoilin is required for diverse neuronal functions in Caenorhabditis elegans. PLoS Genet. 2011;7(5):e1001384. doi: 10.1371/journal.pgen.1001384 ; PubMed Central PMCID: PMC3093358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Xie X, Wang X, Jiang D, Wang J, Fei R, Cong X, et al. PPPDE1 is a novel deubiquitinase belonging to a cysteine isopeptidase family. Biochem Biophys Res Commun. 2017;488(2):291–6. doi: 10.1016/j.bbrc.2017.04.161 . [DOI] [PubMed] [Google Scholar]
- 142.Ban N, Matsumura Y, Sakai H, Takanezawa Y, Sasaki M, Arai H, et al. ABCA3 as a lipid transporter in pulmonary surfactant biogenesis. J Biol Chem. 2007;282(13):9628–34. doi: 10.1074/jbc.M611767200 . [DOI] [PubMed] [Google Scholar]
- 143.Zhang X, Lei K, Yuan X, Wu X, Zhuang Y, Xu T, et al. SUN1/2 and Syne/Nesprin-1/2 complexes connect centrosome to the nucleus during neurogenesis and neuronal migration in mice. Neuron. 2009;64(2):173–87. doi: 10.1016/j.neuron.2009.08.018 ; PubMed Central PMCID: PMC2788510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Ding X, Xu R, Yu J, Xu T, Zhuang Y, Han M. SUN1 is required for telomere attachment to nuclear envelope and gametogenesis in mice. Dev Cell. 2007;12(6):863–72. doi: 10.1016/j.devcel.2007.03.018 . [DOI] [PubMed] [Google Scholar]
- 145.Gijon MA, Riekhof WR, Zarini S, Murphy RC, Voelker DR. Lysophospholipid acyltransferases and arachidonate recycling in human neutrophils. J Biol Chem. 2008;283(44):30235–45. doi: 10.1074/jbc.M806194200 ; PubMed Central PMCID: PMC2573059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Lee HC, Inoue T, Imae R, Kono N, Shirae S, Matsuda S, et al. Caenorhabditis elegans mboa-7, a member of the MBOAT family, is required for selective incorporation of polyunsaturated fatty acids into phosphatidylinositol. Mol Biol Cell. 2008;19(3):1174–84. doi: 10.1091/mbc.e07-09-0893 ; PubMed Central PMCID: PMC2262980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Wu YJ, La Pierre DP, Wu J, Yee AJ, Yang BB. The interaction of versican with its binding partners. Cell Res. 2005;15(7):483–94. doi: 10.1038/sj.cr.7290318 . [DOI] [PubMed] [Google Scholar]
- 148.Wight TN, Kang I, Merrilees MJ. Versican and the control of inflammation. Matrix Biol. 2014;35:152–61. doi: 10.1016/j.matbio.2014.01.015 ; PubMed Central PMCID: PMC4039577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Zhang Q, Didonato JA, Karin M, McKeithan TW. BCL3 encodes a nuclear protein which can alter the subcellular location of NF-kappa B proteins. Mol Cell Biol. 1994;14(6):3915–26. doi: 10.1128/mcb.14.6.3915-3926.1994 ; PubMed Central PMCID: PMC358758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Bundy DL, McKeithan TW. Diverse effects of BCL3 phosphorylation on its modulation of NF-kappaB p52 homodimer binding to DNA. J Biol Chem. 1997;272(52):33132–9. doi: 10.1074/jbc.272.52.33132 . [DOI] [PubMed] [Google Scholar]
- 151.Mogha A, Benesh AE, Patra C, Engel FB, Schoneberg T, Liebscher I, et al. Gpr126 functions in Schwann cells to control differentiation and myelination via G-protein activation. J Neurosci. 2013;33(46):17976–85. doi: 10.1523/JNEUROSCI.1809-13.2013 ; PubMed Central PMCID: PMC3828454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.McNally KE, Faulkner R, Steinberg F, Gallon M, Ghai R, Pim D, et al. Retriever is a multiprotein complex for retromer-independent endosomal cargo recycling. Nat Cell Biol. 2017;19(10):1214–25. doi: 10.1038/ncb3610 ; PubMed Central PMCID: PMC5790113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Ruiz-Canada C, Kelleher DJ, Gilmore R. Cotranslational and posttranslational N-glycosylation of polypeptides by distinct mammalian OST isoforms. Cell. 2009;136(2):272–83. doi: 10.1016/j.cell.2008.11.047 ; PubMed Central PMCID: PMC2859625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Sato T, Sako Y, Sho M, Momohara M, Suico MA, Shuto T, et al. STT3B-dependent posttranslational N-glycosylation as a surveillance system for secretory protein. Mol Cell. 2012;47(1):99–110. doi: 10.1016/j.molcel.2012.04.015 . [DOI] [PubMed] [Google Scholar]
- 155.Riemer J, Hansen HG, Appenzeller-Herzog C, Johansson L, Ellgaard L. Identification of the PDI-family member ERp90 as an interaction partner of ERFAD. Plos One. 2011;6(2):e17037. doi: 10.1371/journal.pone.0017037 ; PubMed Central PMCID: PMC3040216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Harz C, Ludwig N, Lang S, Werner TV, Galata V, Backes C, et al. Secretion and immunogenicity of the meningioma-associated antigen TXNDC16. J Immunol. 2014;193(6):3146–54. doi: 10.4049/jimmunol.1303098 . [DOI] [PubMed] [Google Scholar]
- 157.Arnold SM, Fessler LI, Fessler JH, Kaufman RJ. Two homologues encoding human UDP-glucose:glycoprotein glucosyltransferase differ in mRNA expression and enzymatic activity. Biochemistry. 2000;39(9):2149–63. doi: 10.1021/bi9916473 . [DOI] [PubMed] [Google Scholar]
- 158.Erdile LF, Wold MS, Kelly TJ. The primary structure of the 32-kDa subunit of human replication protein A. J Biol Chem. 1990;265(6):3177–82. . [PubMed] [Google Scholar]
- 159.Sleeth KM, Sorensen CS, Issaeva N, Dziegielewski J, Bartek J, Helleday T. RPA mediates recombination repair during replication stress and is displaced from DNA by checkpoint signalling in human cells. J Mol Biol. 2007;373(1):38–47. doi: 10.1016/j.jmb.2007.07.068 . [DOI] [PubMed] [Google Scholar]
- 160.He Z, Henricksen LA, Wold MS, Ingles CJ. RPA involvement in the damage-recognition and incision steps of nucleotide excision repair. Nature. 1995;374(6522):566–9. doi: 10.1038/374566a0 . [DOI] [PubMed] [Google Scholar]
- 161.Kawai T, Akira S. Regulation of innate immune signalling pathways by the tripartite motif (TRIM) family proteins. EMBO Mol Med. 2011;3(9):513–27. doi: 10.1002/emmm.201100160 ; PubMed Central PMCID: PMC3377094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Wang M, Wu J, Zhou E, Chang X, Gan J, Cheng T. Forkhead box o3a suppresses lipopolysaccharide-stimulated proliferation and inflammation in fibroblast-like synoviocytes through regulating tripartite motif-containing protein 3. J Cell Physiol. 2019;234(11):20139–48. doi: 10.1002/jcp.28615 . [DOI] [PubMed] [Google Scholar]
- 163.Xiong F, Ji Z, Liu Y, Zhang Y, Hu L, Yang Q, et al. Mutation in SSUH2 Causes Autosomal-Dominant Dentin Dysplasia Type I. Hum Mutat. 2017;38(1):95–104. doi: 10.1002/humu.23130 . [DOI] [PubMed] [Google Scholar]
- 164.Schreiber J, Vegh MJ, Dawitz J, Kroon T, Loos M, Labonte D, et al. Ubiquitin ligase TRIM3 controls hippocampal plasticity and learning by regulating synaptic gamma-actin levels. J Cell Biol. 2015;211(3):569–86. doi: 10.1083/jcb.201506048 . PubMed Central PMCID: PMC4639863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.MacPherson L, Tamblyn L, Rajendra S, Bralha F, McPherson JP, Matthews J. 2,3,7,8-Tetrachlorodibenzo-p-dioxin poly(ADP-ribose) polymerase (TiPARP, ARTD14) is a mono-ADP-ribosyltransferase and repressor of aryl hydrocarbon receptor transactivation. Nucleic Acids Res. 2013;41(3):1604–21. doi: 10.1093/nar/gks1337 ; PubMed Central PMCID: PMC3562000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Yamada T, Horimoto H, Kameyama T, Hayakawa S, Yamato H, Dazai M, et al. Constitutive aryl hydrocarbon receptor signaling constrains type I interferon-mediated antiviral innate defense. Nat Immunol. 2016;17(6):687–94. doi: 10.1038/ni.3422 . [DOI] [PubMed] [Google Scholar]
- 167.Kozaki T, Komano J, Kanbayashi D, Takahama M, Misawa T, Satoh T, et al. Mitochondrial damage elicits a TCDD-inducible poly(ADP-ribose) polymerase-mediated antiviral response. Proc Natl Acad Sci U S A. 2017;114(10):2681–6. doi: 10.1073/pnas.1621508114 ; PubMed Central PMCID: PMC5347618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Goldshmit Y, Walters CE, Scott HJ, Greenhalgh CJ, Turnley AM. SOCS2 induces neurite outgrowth by regulation of epidermal growth factor receptor activation. J Biol Chem. 2004;279(16):16349–55. doi: 10.1074/jbc.M312873200 . [DOI] [PubMed] [Google Scholar]
- 169.Letellier E, Haan S. SOCS2: physiological and pathological functions. Front Biosci (Elite Ed). 2016;8:189–204. . [DOI] [PubMed] [Google Scholar]
- 170.Kazi JU, Ronnstrand L. Suppressor of cytokine signaling 2 (SOCS2) associates with FLT3 and negatively regulates downstream signaling. Mol Oncol. 2013;7(3):693–703. doi: 10.1016/j.molonc.2013.02.020 ; PubMed Central PMCID: PMC5528470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Tiranti V, Rossi E, Ruiz-Carrillo A, Rossi G, Rocchi M, DiDonato S, et al. Chromosomal localization of mitochondrial transcription factor A (TCF6), single-stranded DNA-binding protein (SSBP), and endonuclease G (ENDOG), three human housekeeping genes involved in mitochondrial biogenesis. Genomics. 1995;25(2):559–64. doi: 10.1016/0888-7543(95)80058-t . [DOI] [PubMed] [Google Scholar]
- 172.Duguay BA, Smiley JR. Mitochondrial nucleases ENDOG and EXOG participate in mitochondrial DNA depletion initiated by herpes simplex virus 1 UL12.5. J Virol. 2013;87(21):11787–97. doi: 10.1128/JVI.02306-13 ; PubMed Central PMCID: PMC3807374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Zhdanov DD, Fahmi T, Wang X, Apostolov EO, Sokolov NN, Javadov S, et al. Regulation of Apoptotic Endonucleases by EndoG. DNA Cell Biol. 2015;34(5):316–26. doi: 10.1089/dna.2014.2772 ; PubMed Central PMCID: PMC4426297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Kim JS, Lee JH, Jeong WW, Choi DH, Cha HJ, Kim DH, et al. Reactive oxygen species-dependent EndoG release mediates cisplatin-induced caspase-independent apoptosis in human head and neck squamous carcinoma cells. Int J Cancer. 2008;122(3):672–80. doi: 10.1002/ijc.23158 . [DOI] [PubMed] [Google Scholar]
- 175.Kirino Y, Mourelatos Z. 2’-O-methyl modification in mouse piRNAs and its methylase. Nucleic Acids Symp Ser (Oxf). 2007;(51):417–8. doi: 10.1093/nass/nrm209 . [DOI] [PubMed] [Google Scholar]
- 176.Hu D, Smith ER, Garruss AS, Mohaghegh N, Varberg JM, Lin C, et al. The little elongation complex functions at initiation and elongation phases of snRNA gene transcription. Mol Cell. 2013;51(4):493–505. doi: 10.1016/j.molcel.2013.07.003 ; PubMed Central PMCID: PMC4104523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Paclik D, Danese S, Berndt U, Wiedenmann B, Dignass A, Sturm A. Galectin-4 controls intestinal inflammation by selective regulation of peripheral and mucosal T cell apoptosis and cell cycle. Plos One. 2008;3(7):e2629. doi: 10.1371/journal.pone.0002629 ; PubMed Central PMCID: PMC2440804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Stancic M, Slijepcevic D, Nomden A, Vos MJ, de Jonge JC, Sikkema AH, et al. Galectin-4, a novel neuronal regulator of myelination. Glia. 2012;60(6):919–35. doi: 10.1002/glia.22324 . [DOI] [PubMed] [Google Scholar]
- 179.Chen YQ, Rafi MA, de Gala G, Wenger DA. Cloning and expression of cDNA encoding human galactocerebrosidase, the enzyme deficient in globoid cell leukodystrophy. Hum Mol Genet. 1993;2(11):1841–5. doi: 10.1093/hmg/2.11.1841 . [DOI] [PubMed] [Google Scholar]
- 180.Wang W, Xue Y, Zhou S, Kuo A, Cairns BR, Crabtree GR. Diversity and specialization of mammalian SWI/SNF complexes. Genes Dev. 1996;10(17):2117–30. doi: 10.1101/gad.10.17.2117 . [DOI] [PubMed] [Google Scholar]
- 181.Chi T. A BAF-centred view of the immune system. Nat Rev Immunol. 2004;4(12):965–77. doi: 10.1038/nri1501 . [DOI] [PubMed] [Google Scholar]
- 182.Kim YH, Lee JR, Hahn MJ. Regulation of inflammatory gene expression in macrophages by epithelial-stromal interaction 1 (Epsti1). Biochem Biophys Res Commun. 2018;496(2):778–83. doi: 10.1016/j.bbrc.2017.12.014 . [DOI] [PubMed] [Google Scholar]
- 183.Woitach JT, Zhang M, Niu CH, Thorgeirsson SS. A retinoblastoma-binding protein that affects cell-cycle control and confers transforming ability. Nat Genet. 1998;19(4):371–4. doi: 10.1038/1258 . [DOI] [PubMed] [Google Scholar]
- 184.Lin YL, Shivji MK, Chen C, Kolodner R, Wood RD, Dutta A. The evolutionarily conserved zinc finger motif in the largest subunit of human replication protein A is required for DNA replication and mismatch repair but not for nucleotide excision repair. J Biol Chem. 1998;273(3):1453–61. doi: 10.1074/jbc.273.3.1453 . [DOI] [PubMed] [Google Scholar]
- 185.Zheng K, Xiol J, Reuter M, Eckardt S, Leu NA, McLaughlin KJ, et al. Mouse MOV10L1 associates with Piwi proteins and is an essential component of the Piwi-interacting RNA (piRNA) pathway. Proc Natl Acad Sci U S A. 2010;107(26):11841–6. doi: 10.1073/pnas.1003953107 ; PubMed Central PMCID: PMC2900664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Roshanbin S, Lindberg FA, Lekholm E, Eriksson MM, Perland E, Ahlund J, et al. Histological characterization of orphan transporter MCT14 (SLC16A14) shows abundant expression in mouse CNS and kidney. Bmc Neurosci. 2016;17(1):43. doi: 10.1186/s12868-016-0274-7 ; PubMed Central PMCID: PMC4929735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Weon JL, Yang SW, Potts PR. Cytosolic Iron-Sulfur Assembly Is Evolutionarily Tuned by a Cancer-Amplified Ubiquitin Ligase. Mol Cell. 2018;69(1):113–25 e6. doi: 10.1016/j.molcel.2017.11.010 . [DOI] [PubMed] [Google Scholar]
- 188.Shi Y, Yuan B, Zhu W, Zhang R, Li L, Hao X, et al. Ube2D3 and Ube2N are essential for RIG-I-mediated MAVS aggregation in antiviral innate immunity. Nat Commun. 2017;8:15138. doi: 10.1038/ncomms15138 ; PubMed Central PMCID: PMC5418627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Ogami K, Cho R, Hoshino S. Molecular cloning and characterization of a novel isoform of the non-canonical poly(A) polymerase PAPD7. Biochem Biophys Res Commun. 2013;432(1):135–40. doi: 10.1016/j.bbrc.2013.01.072 . [DOI] [PubMed] [Google Scholar]
- 190.Lim J, Kim D, Lee YS, Ha M, Lee M, Yeo J, et al. Mixed tailing by TENT4A and TENT4B shields mRNA from rapid deadenylation. Science. 2018;361(6403):701–4. doi: 10.1126/science.aam5794 . [DOI] [PubMed] [Google Scholar]
- 191.McCormack RM, de Armas LR, Shiratsuchi M, Fiorentino DG, Olsson ML, Lichtenheld MG, et al. Perforin-2 is essential for intracellular defense of parenchymal cells and phagocytes against pathogenic bacteria. Elife. 2015;4. doi: 10.7554/eLife.06508 ; PubMed Central PMCID: PMC4626811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Uyama T, Jin XH, Tsuboi K, Tonai T, Ueda N. Characterization of the human tumor suppressors TIG3 and HRASLS2 as phospholipid-metabolizing enzymes. Biochim Biophys Acta. 2009;1791(12):1114–24. doi: 10.1016/j.bbalip.2009.07.001 . [DOI] [PubMed] [Google Scholar]
- 193.Staring J, von Castelmur E, Blomen VA, van den Hengel LG, Brockmann M, Baggen J, et al. PLA2G16 represents a switch between entry and clearance of Picornaviridae. Nature. 2017;541(7637):412–6. doi: 10.1038/nature21032 . [DOI] [PubMed] [Google Scholar]
- 194.Kraemer D, Wozniak RW, Blobel G, Radu A. The human CAN protein, a putative oncogene product associated with myeloid leukemogenesis, is a nuclear pore complex protein that faces the cytoplasm. Proc Natl Acad Sci U S A. 1994;91(4):1519–23. doi: 10.1073/pnas.91.4.1519 ; PubMed Central PMCID: PMC43191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Fornerod M, van Deursen J, van Baal S, Reynolds A, Davis D, Murti KG, et al. The human homologue of yeast CRM1 is in a dynamic subcomplex with CAN/Nup214 and a novel nuclear pore component Nup88. Embo J. 1997;16(4):807–16. doi: 10.1093/emboj/16.4.807 ; PubMed Central PMCID: PMC1169681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Ustianenko D, Hrossova D, Potesil D, Chalupnikova K, Hrazdilova K, Pachernik J, et al. Mammalian DIS3L2 exoribonuclease targets the uridylated precursors of let-7 miRNAs. Rna. 2013;19(12):1632–8. doi: 10.1261/rna.040055.113 ; PubMed Central PMCID: PMC3884668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Lubas M, Damgaard CK, Tomecki R, Cysewski D, Jensen TH, Dziembowski A. Exonuclease hDIS3L2 specifies an exosome-independent 3’-5’ degradation pathway of human cytoplasmic mRNA. Embo J. 2013;32(13):1855–68. doi: 10.1038/emboj.2013.135 ; PubMed Central PMCID: PMC3981170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Iwata H, Goettsch C, Sharma A, Ricchiuto P, Goh WW, Halu A, et al. PARP9 and PARP14 cross-regulate macrophage activation via STAT1 ADP-ribosylation. Nat Commun. 2016;7:12849. doi: 10.1038/ncomms12849 ; PubMed Central PMCID: PMC5095532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Capitanio JS, Montpetit B, Wozniak RW. Human Nup98 regulates the localization and activity of DExH/D-box helicase DHX9. Elife. 2017;6. doi: 10.7554/eLife.18825 ; PubMed Central PMCID: PMC5338925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Weston R, Peeters H, Ahel D. ZRANB3 is a structure-specific ATP-dependent endonuclease involved in replication stress response. Genes Dev. 2012;26(14):1558–72. doi: 10.1101/gad.193516.112 ; PubMed Central PMCID: PMC3404384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Ciccia A, Nimonkar AV, Hu Y, Hajdu I, Achar YJ, Izhar L, et al. Polyubiquitinated PCNA recruits the ZRANB3 translocase to maintain genomic integrity after replication stress. Mol Cell. 2012;47(3):396–409. doi: 10.1016/j.molcel.2012.05.024 ; PubMed Central PMCID: PMC3613862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Sakatsume M, Igarashi K, Winestock KD, Garotta G, Larner AC, Finbloom DS. The Jak kinases differentially associate with the alpha and beta (accessory factor) chains of the interferon gamma receptor to form a functional receptor unit capable of activating STAT transcription factors. J Biol Chem. 1995;270(29):17528–34. doi: 10.1074/jbc.270.29.17528 . [DOI] [PubMed] [Google Scholar]
- 203.Segura-Totten M, Kowalski AK, Craigie R, Wilson KL. Barrier-to-autointegration factor: major roles in chromatin decondensation and nuclear assembly. J Cell Biol. 2002;158(3):475–85. doi: 10.1083/jcb.200202019 ; PubMed Central PMCID: PMC2173821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Wiebe MS, Traktman P. Poxviral B1 kinase overcomes barrier to autointegration factor, a host defense against virus replication. Cell Host Microbe. 2007;1(3):187–97. doi: 10.1016/j.chom.2007.03.007 ; PubMed Central PMCID: PMC1978190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Mazur DJ, Perrino FW. Excision of 3’ termini by the Trex1 and TREX2 3’—>5’ exonucleases. Characterization of the recombinant proteins. J Biol Chem. 2001;276(20):17022–9. doi: 10.1074/jbc.M100623200 . [DOI] [PubMed] [Google Scholar]
- 206.Schneider M, Hellerschmied D, Schubert T, Amlacher S, Vinayachandran V, Reja R, et al. The Nuclear Pore-Associated TREX-2 Complex Employs Mediator to Regulate Gene Expression. Cell. 2015;162(5):1016–28. doi: 10.1016/j.cell.2015.07.059 ; PubMed Central PMCID: PMC4644235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Garcia-Oliver E, Garcia-Molinero V, Rodriguez-Navarro S. mRNA export and gene expression: the SAGA-TREX-2 connection. Biochim Biophys Acta. 2012;1819(6):555–65. doi: 10.1016/j.bbagrm.2011.11.011 . [DOI] [PubMed] [Google Scholar]
- 208.Nichols RJ, Wiebe MS, Traktman P. The vaccinia-related kinases phosphorylate the N’ terminus of BAF, regulating its interaction with DNA and its retention in the nucleus. Mol Biol Cell. 2006;17(5):2451–64. doi: 10.1091/mbc.e05-12-1179 ; PubMed Central PMCID: PMC1446082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Sevilla A, Santos CR, Vega FM, Lazo PA. Human vaccinia-related kinase 1 (VRK1) activates the ATF2 transcriptional activity by novel phosphorylation on Thr-73 and Ser-62 and cooperates with JNK. J Biol Chem. 2004;279(26):27458–65. doi: 10.1074/jbc.M401009200 . [DOI] [PubMed] [Google Scholar]
- 210.Watkins JL, Murphy R, Emtage JL, Wente SR. The human homologue of Saccharomyces cerevisiae Gle1p is required for poly(A)+ RNA export. Proc Natl Acad Sci U S A. 1998;95(12):6779–84. doi: 10.1073/pnas.95.12.6779 ; PubMed Central PMCID: PMC22633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Jiao X, Chang JH, Kilic T, Tong L, Kiledjian M. A mammalian pre-mRNA 5’ end capping quality control mechanism and an unexpected link of capping to pre-mRNA processing. Mol Cell. 2013;50(1):104–15. doi: 10.1016/j.molcel.2013.02.017 ; PubMed Central PMCID: PMC3630477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Bonnin E, Cabochette P, Filosa A, Juhlen R, Komatsuzaki S, Hezwani M, et al. Biallelic mutations in nucleoporin NUP88 cause lethal fetal akinesia deformation sequence. PLoS Genet. 2018;14(12):e1007845. doi: 10.1371/journal.pgen.1007845 ; PubMed Central PMCID: PMC6307818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Tomecki R, Dmochowska A, Gewartowski K, Dziembowski A, Stepien PP. Identification of a novel human nuclear-encoded mitochondrial poly(A) polymerase. Nucleic Acids Res. 2004;32(20):6001–14. doi: 10.1093/nar/gkh923 ; PubMed Central PMCID: PMC534615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Nagaike T, Suzuki T, Katoh T, Ueda T. Human mitochondrial mRNAs are stabilized with polyadenylation regulated by mitochondria-specific poly(A) polymerase and polynucleotide phosphorylase. J Biol Chem. 2005;280(20):19721–7. doi: 10.1074/jbc.M500804200 . [DOI] [PubMed] [Google Scholar]
- 215.Mullen TE, Marzluff WF. Degradation of histone mRNA requires oligouridylation followed by decapping and simultaneous degradation of the mRNA both 5’ to 3’ and 3’ to 5’. Genes Dev. 2008;22(1):50–65. doi: 10.1101/gad.1622708 ; PubMed Central PMCID: PMC2151014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Antonicka H, Choquet K, Lin ZY, Gingras AC, Kleinman CL, Shoubridge EA. A pseudouridine synthase module is essential for mitochondrial protein synthesis and cell viability. Embo Rep. 2017;18(1):28–38. doi: 10.15252/embr.201643391 ; PubMed Central PMCID: PMC5210091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Arroyo JD, Jourdain AA, Calvo SE, Ballarano CA, Doench JG, Root DE, et al. A Genome-wide CRISPR Death Screen Identifies Genes Essential for Oxidative Phosphorylation. Cell Metab. 2016;24(6):875–85. doi: 10.1016/j.cmet.2016.08.017 ; PubMed Central PMCID: PMC5474757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Saunders A, Webb LM, Janas ML, Hutchings A, Pascall J, Carter C, et al. Putative GTPase GIMAP1 is critical for the development of mature B and T lymphocytes. Blood. 2010;115(16):3249–57. doi: 10.1182/blood-2009-08-237586 . [DOI] [PubMed] [Google Scholar]
- 219.Deveraux QL, Takahashi R, Salvesen GS, Reed JC. X-linked IAP is a direct inhibitor of cell-death proteases. Nature. 1997;388(6639):300–4. doi: 10.1038/40901 . [DOI] [PubMed] [Google Scholar]
- 220.Lu M, Lin SC, Huang Y, Kang YJ, Rich R, Lo YC, et al. XIAP induces NF-kappaB activation via the BIR1/TAB1 interaction and BIR1 dimerization. Mol Cell. 2007;26(5):689–702. doi: 10.1016/j.molcel.2007.05.006 ; PubMed Central PMCID: PMC1991276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Isken A, Golczak M, Oberhauser V, Hunzelmann S, Driever W, Imanishi Y, et al. RBP4 disrupts vitamin A uptake homeostasis in a STRA6-deficient animal model for Matthew-Wood syndrome. Cell Metab. 2008;7(3):258–68. doi: 10.1016/j.cmet.2008.01.009 ; PubMed Central PMCID: PMC2561276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Wang Y, Yuan S, Jia X, Ge Y, Ling T, Nie M, et al. Mitochondria-localised ZNFX1 functions as a dsRNA sensor to initiate antiviral responses through MAVS. Nat Cell Biol. 2019;21(11):1346–56. doi: 10.1038/s41556-019-0416-0 . [DOI] [PubMed] [Google Scholar]
- 223.Ishidate T, Ozturk AR, Durning DJ, Sharma R, Shen EZ, Chen H, et al. ZNFX-1 Functions within Perinuclear Nuage to Balance Epigenetic Signals. Mol Cell. 2018;70(4):639–49 e6. doi: 10.1016/j.molcel.2018.04.009 ; PubMed Central PMCID: PMC5994929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Takeyama K, Aguiar RC, Gu L, He C, Freeman GJ, Kutok JL, et al. The BAL-binding protein BBAP and related Deltex family members exhibit ubiquitin-protein isopeptide ligase activity. J Biol Chem. 2003;278(24):21930–7. doi: 10.1074/jbc.M301157200 . [DOI] [PubMed] [Google Scholar]
- 225.Yan Q, Dutt S, Xu R, Graves K, Juszczynski P, Manis JP, et al. BBAP monoubiquitylates histone H4 at lysine 91 and selectively modulates the DNA damage response. Mol Cell. 2009;36(1):110–20. doi: 10.1016/j.molcel.2009.08.019 ; PubMed Central PMCID: PMC2913878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Zhang Y, Mao D, Roswit WT, Jin X, Patel AC, Patel DA, et al. PARP9-DTX3L ubiquitin ligase targets host histone H2BJ and viral 3C protease to enhance interferon signaling and control viral infection. Nat Immunol. 2015;16(12):1215–27. doi: 10.1038/ni.3279 ; PubMed Central PMCID: PMC4653074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Liu J, McFadden G. SAMD9 is an innate antiviral host factor with stress response properties that can be antagonized by poxviruses. J Virol. 2015;89(3):1925–31. doi: 10.1128/JVI.02262-14 ; PubMed Central PMCID: PMC4300762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Carney JP, Maser RS, Olivares H, Davis EM, Le Beau M, Yates JR, 3rd, et al. The hMre11/hRad50 protein complex and Nijmegen breakage syndrome: linkage of double-strand break repair to the cellular DNA damage response. Cell. 1998;93(3):477–86. doi: 10.1016/s0092-8674(00)81175-7 . [DOI] [PubMed] [Google Scholar]
- 229.Kondo T, Kobayashi J, Saitoh T, Maruyama K, Ishii KJ, Barber GN, et al. DNA damage sensor MRE11 recognizes cytosolic double-stranded DNA and induces type I interferon by regulating STING trafficking. Proc Natl Acad Sci U S A. 2013;110(8):2969–74. doi: 10.1073/pnas.1222694110 ; PubMed Central PMCID: PMC3581880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Irudayam JI, Contreras D, Spurka L, Subramanian A, Allen J, Ren S, et al. Characterization of type I interferon pathway during hepatic differentiation of human pluripotent stem cells and hepatitis C virus infection. Stem Cell Res. 2015;15(2):354–64. doi: 10.1016/j.scr.2015.08.003 ; PubMed Central PMCID: PMC4600668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Lim J, Ha M, Chang H, Kwon SC, Simanshu DK, Patel DJ, et al. Uridylation by TUT4 and TUT7 marks mRNA for degradation. Cell. 2014;159(6):1365–76. doi: 10.1016/j.cell.2014.10.055 ; PubMed Central PMCID: PMC4720960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Le Pen J, Jiang H, Di Domenico T, Kneuss E, Kosalka J, Leung C, et al. Terminal uridylyltransferases target RNA viruses as part of the innate immune system. Nat Struct Mol Biol. 2018;25(9):778–86. doi: 10.1038/s41594-018-0106-9 ; PubMed Central PMCID: PMC6130846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Torii S, Kusakabe M, Yamamoto T, Maekawa M, Nishida E. Sef is a spatial regulator for Ras/MAP kinase signaling. Dev Cell. 2004;7(1):33–44. doi: 10.1016/j.devcel.2004.05.019 . [DOI] [PubMed] [Google Scholar]
- 234.Mellett M, Atzei P, Bergin R, Horgan A, Floss T, Wurst W, et al. Orphan receptor IL-17RD regulates Toll-like receptor signalling via SEFIR/TIR interactions. Nat Commun. 2015;6:6669. doi: 10.1038/ncomms7669 . [DOI] [PubMed] [Google Scholar]
- 235.Wang Y, Satoh A, Warren G, Meyer HH. VCIP135 acts as a deubiquitinating enzyme during p97-p47-mediated reassembly of mitotic Golgi fragments. J Cell Biol. 2004;164(7):973–8. doi: 10.1083/jcb.200401010 ; PubMed Central PMCID: PMC2172062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Anderson JF, Siller E, Barral JM. The neurodegenerative-disease-related protein sacsin is a molecular chaperone. J Mol Biol. 2011;411(4):870–80. doi: 10.1016/j.jmb.2011.06.016 . [DOI] [PubMed] [Google Scholar]
- 237.Sundaramoorthy E, Leonard M, Mak R, Liao J, Fulzele A, Bennett EJ. ZNF598 and RACK1 Regulate Mammalian Ribosome-Associated Quality Control Function by Mediating Regulatory 40S Ribosomal Ubiquitylation. Mol Cell. 2017;65(4):751––60 e4.. doi: 10.1016/j.molcel.2016.12.026 ; PubMed Central PMCID: PMC5321136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Morita M, Ler LW, Fabian MR, Siddiqui N, Mullin M, Henderson VC, et al. A novel 4EHP-GIGYF2 translational repressor complex is essential for mammalian development. Mol Cell Biol. 2012;32(17):3585–93. doi: 10.1128/MCB.00455-12 ; PubMed Central PMCID: PMC3422012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Wang G, Kouwaki T, Okamoto M, Oshiumi H. Attenuation of the Innate Immune Response against Viral Infection Due to ZNF598-Promoted Binding of FAT10 to RIG-I. Cell Rep. 2019;28(8):1961–70 e4. doi: 10.1016/j.celrep.2019.07.081 . [DOI] [PubMed] [Google Scholar]
- 240.Balastik M, Ferraguti F, Pires-da Silva A, Lee TH, Alvarez-Bolado G, Lu KP, et al. Deficiency in ubiquitin ligase TRIM2 causes accumulation of neurofilament light chain and neurodegeneration. Proc Natl Acad Sci U S A. 2008;105(33):12016–21. doi: 10.1073/pnas.0802261105 ; PubMed Central PMCID: PMC2575299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Sarute N, Ibrahim N, Medegan Fagla B, Lavanya M, Cuevas C, Stavrou S, et al. TRIM2, a novel member of the antiviral family, limits New World arenavirus entry. PLoS Biol. 2019;17(2):e3000137. doi: 10.1371/journal.pbio.3000137 ; PubMed Central PMCID: PMC6380604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Dango S, Mosammaparast N, Sowa ME, Xiong LJ, Wu F, Park K, et al. DNA unwinding by ASCC3 helicase is coupled to ALKBH3-dependent DNA alkylation repair and cancer cell proliferation. Mol Cell. 2011;44(3):373–84. doi: 10.1016/j.molcel.2011.08.039 ; PubMed Central PMCID: PMC3258846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Jung DJ, Sung HS, Goo YW, Lee HM, Park OK, Jung SY, et al. Novel transcription coactivator complex containing activating signal cointegrator 1. Mol Cell Biol. 2002;22(14):5203–11. doi: 10.1128/MCB.22.14.5203-5211.2002 ; PubMed Central PMCID: PMC139772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Williams GD, Gokhale NS, Snider DL, Horner SM. The mRNA Cap 2’-O-Methyltransferase CMTR1 Regulates the Expression of Certain Interferon-Stimulated Genes. mSphere. 2020;5(3). doi: 10.1128/mSphere.00202-20 ; PubMed Central PMCID: PMC7227766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Silva CM, Lu H, Day RN. Characterization and cloning of STAT5 from IM-9 cells and its activation by growth hormone. Mol Endocrinol. 1996;10(5):508–18. doi: 10.1210/mend.10.5.8732682 . [DOI] [PubMed] [Google Scholar]
- 246.Lin JX, Leonard WJ. The role of Stat5a and Stat5b in signaling by IL-2 family cytokines. Oncogene. 2000;19(21):2566–76. doi: 10.1038/sj.onc.1203523 . [DOI] [PubMed] [Google Scholar]
- 247.Bennett V, Baines AJ. Spectrin and ankyrin-based pathways: metazoan inventions for integrating cells into tissues. Physiol Rev. 2001;81(3):1353–92. doi: 10.1152/physrev.2001.81.3.1353 . [DOI] [PubMed] [Google Scholar]
- 248.Pine R. Constitutive expression of an ISGF2/IRF1 transgene leads to interferon-independent activation of interferon-inducible genes and resistance to virus infection. J Virol. 1992;66(7):4470–8. doi: 10.1128/JVI.66.7.4470-4478.1992 ; PubMed Central PMCID: PMC241256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Welsby I, Hutin D, Gueydan C, Kruys V, Rongvaux A, Leo O. PARP12, an interferon-stimulated gene involved in the control of protein translation and inflammation. J Biol Chem. 2014;289(38):26642–57. doi: 10.1074/jbc.M114.589515 ; PubMed Central PMCID: PMC4176246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Yamashita A. Role of SMG-1-mediated Upf1 phosphorylation in mammalian nonsense-mediated mRNA decay. Genes Cells. 2013;18(3):161–75. doi: 10.1111/gtc.12033 . [DOI] [PubMed] [Google Scholar]
- 251.Balistreri G, Horvath P, Schweingruber C, Zund D, McInerney G, Merits A, et al. The host nonsense-mediated mRNA decay pathway restricts Mammalian RNA virus replication. Cell Host Microbe. 2014;16(3):403–11. doi: 10.1016/j.chom.2014.08.007 . [DOI] [PubMed] [Google Scholar]
- 252.Fu M, Blackshear PJ. RNA-binding proteins in immune regulation: a focus on CCCH zinc finger proteins. Nat Rev Immunol. 2017;17(2):130–43. doi: 10.1038/nri.2016.129 ; PubMed Central PMCID: PMC5556700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Boldin MP, Goncharov TM, Goltsev YV, Wallach D. Involvement of MACH, a novel MORT1/FADD-interacting protease, in Fas/APO-1- and TNF receptor-induced cell death. Cell. 1996;85(6):803–15. doi: 10.1016/s0092-8674(00)81265-9 . [DOI] [PubMed] [Google Scholar]
- 254.Lalaoui N, Boyden SE, Oda H, Wood GM, Stone DL, Chau D, et al. Mutations that prevent caspase cleavage of RIPK1 cause autoinflammatory disease. Nature. doi: 10.1038/s41586-019-1828-5 ;577(7788):103–8. PubMed Central PMCID: PMC6930849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Tagami H, Ray-Gallet D, Almouzni G, Nakatani Y. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell. 2004;116(1):51–61. doi: 10.1016/s0092-8674(03)01064-x . [DOI] [PubMed] [Google Scholar]
- 256.Medzhitov R. Toll-like receptors and innate immunity. Nat Rev Immunol. 2001;1(2):135–45. doi: 10.1038/35100529 . [DOI] [PubMed] [Google Scholar]
- 257.Kaisho T, Akira S. Toll-like receptors and their signaling mechanism in innate immunity. Acta Odontol Scand. 2001;59(3):124–30. doi: 10.1080/000163501750266701 . [DOI] [PubMed] [Google Scholar]
- 258.Mitacek RM, Ke YL, Prenni JE, Jadeja R, VanOverbeke DL, Mafi GG, et al. Mitochondrial Degeneration, Depletion of NADH, and Oxidative Stress Decrease Color Stability of Wet-Aged Beef Longissimus Steaks. J Food Sci. 2019;84(1):38–50. doi: 10.1111/1750-3841.14396 WOS:000455527800006. [DOI] [PubMed] [Google Scholar]
- 259.Ross JL, Wallace K, Shuman H, Goldman YE, Holzbaur EL. Processive bidirectional motion of dynein-dynactin complexes in vitro. Nat Cell Biol. 2006;8(6):562–70. doi: 10.1038/ncb1421 . [DOI] [PubMed] [Google Scholar]
- 260.Heraud-Farlow JE, Sharangdhar T, Li X, Pfeifer P, Tauber S, Orozco D, et al. Staufen2 regulates neuronal target RNAs. Cell Rep. 2013;5(6):1511–8. doi: 10.1016/j.celrep.2013.11.039 . [DOI] [PubMed] [Google Scholar]
- 261.Hay RT, Vuillard L, Desterro JM, Rodriguez MS. Control of NF-kappa B transcriptional activation by signal induced proteolysis of I kappa B alpha. Philos Trans R Soc Lond B Biol Sci. 1999;354(1389):1601–9. doi: 10.1098/rstb.1999.0504 ; PubMed Central PMCID: PMC1692667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Gaur U, Aggarwal BB. Regulation of proliferation, survival and apoptosis by members of the TNF superfamily. Biochem Pharmacol. 2003;66(8):1403–8. doi: 10.1016/s0006-2952(03)00490-8 . [DOI] [PubMed] [Google Scholar]
- 263.James MA, Lee JH, Klingelhutz AJ. Human papillomavirus type 16 E6 activates NF-kappaB, induces cIAP-2 expression, and protects against apoptosis in a PDZ binding motif-dependent manner. J Virol. 2006;80(11):5301–7. doi: 10.1128/JVI.01942-05 ; PubMed Central PMCID: PMC1472131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Kwak EL, Larochelle DA, Beaumont C, Torti SV, Torti FM. Role for NF-kappa B in the regulation of ferritin H by tumor necrosis factor-alpha. J Biol Chem. 1995;270(25):15285–93. doi: 10.1074/jbc.270.25.15285 . [DOI] [PubMed] [Google Scholar]
- 265.Biswas G, Guha M, Avadhani NG. Mitochondria-to-nucleus stress signaling in mammalian cells: nature of nuclear gene targets, transcription regulation, and induced resistance to apoptosis. Gene. 2005;354:132–9. doi: 10.1016/j.gene.2005.03.028 ; PubMed Central PMCID: PMC3800739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Jono H, Lim JH, Chen LF, Xu H, Trompouki E, Pan ZK, et al. NF-kappaB is essential for induction of CYLD, the negative regulator of NF-kappaB: evidence for a novel inducible autoregulatory feedback pathway. J Biol Chem. 2004;279(35):36171–4. doi: 10.1074/jbc.M406638200 . [DOI] [PubMed] [Google Scholar]
- 267.Xu SQ, Zhang H, Yang XD, Qian YW, Xiao Q. Inhibition of cathepsin L alleviates the microglia-mediated neuroinflammatory responses through caspase-8 and NF-kappa B pathways. Neurobiol Aging. 2018;62:159–67. doi: 10.1016/j.neurobiolaging.2017.09.030 WOS:000418478600014. [DOI] [PubMed] [Google Scholar]
- 268.Wang YR, Qin S, Han R, Wu JC, Liang ZQ, Qin ZH, et al. Cathepsin L plays a role in quinolinic acid-induced NF-Kappab activation and excitotoxicity in rat striatal neurons. Plos One. 2013;8(9):e75702. doi: 10.1371/journal.pone.0075702 ; PubMed Central PMCID: PMC3779166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Wendeler M, Hoernschemeyer J, Hoffmann D, Kolter T, Schwarzmann G, Sandhoff K. Photoaffinity labelling of the human GM2-activator protein. Mechanistic insight into ganglioside GM2 degradation. Eur J Biochem. 2004;271(3):614–27. doi: 10.1111/j.1432-1033.2003.03964.x . [DOI] [PubMed] [Google Scholar]
- 270.Duan J, Zhang Q, Hu X, Lu D, Yu W, Bai H. N(4)-acetylcytidine is required for sustained NLRP3 inflammasome activation via HMGB1 pathway in microglia. Cell Signal. 2019;58:44–52. doi: 10.1016/j.cellsig.2019.03.007 . [DOI] [PubMed] [Google Scholar]
- 271.Wood ER, Bledsoe R, Chai J, Daka P, Deng H, Ding Y, et al. The Role of Phosphodiesterase 12 (PDE12) as a Negative Regulator of the Innate Immune Response and the Discovery of Antiviral Inhibitors. J Biol Chem. 2015;290(32):19681–96. doi: 10.1074/jbc.M115.653113 ; PubMed Central PMCID: PMC4528132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Krappmann D, Scheidereit C. Regulation of NF-kappa B activity by I kappa B alpha and I kappa B beta stability. Immunobiology. 1997;198(1–3):3–13. doi: 10.1016/s0171-2985(97)80022-8 . [DOI] [PubMed] [Google Scholar]
- 273.Dominissini D, Rechavi G. N(4)-acetylation of Cytidine in mRNA by NAT10 Regulates Stability and Translation. Cell. 2018;175(7):1725–7. doi: 10.1016/j.cell.2018.11.037 . [DOI] [PubMed] [Google Scholar]
- 274.Cai S, Liu X, Zhang C, Xing B, Du X. Autoacetylation of NAT10 is critical for its function in rRNA transcription activation. Biochem Biophys Res Commun. 2017;483(1):624–9. doi: 10.1016/j.bbrc.2016.12.092 . [DOI] [PubMed] [Google Scholar]
- 275.Cambiaghi TD, Pereira CM, Shanmugam R, Bolech M, Wek RC, Sattlegger E, et al. Evolutionarily conserved IMPACT impairs various stress responses that require GCN1 for activating the eIF2 kinase GCN2. Biochem Biophys Res Commun. 2014;443(2):592–7. doi: 10.1016/j.bbrc.2013.12.021 . [DOI] [PubMed] [Google Scholar]
- 276.Pereira CM, Sattlegger E, Jiang HY, Longo BM, Jaqueta CB, Hinnebusch AG, et al. IMPACT, a protein preferentially expressed in the mouse brain, binds GCN1 and inhibits GCN2 activation. J Biol Chem. 2005;280(31):28316–23. doi: 10.1074/jbc.M408571200 . [DOI] [PubMed] [Google Scholar]
- 277.Roobol A, Roobol J, Bastide A, Knight JR, Willis AE, Smales CM. p58IPK is an inhibitor of the eIF2alpha kinase GCN2 and its localization and expression underpin protein synthesis and ER processing capacity. Biochem J. 2015;465(2):213–25. doi: 10.1042/BJ20140852 . [DOI] [PubMed] [Google Scholar]
- 278.del Pino J, Jimenez JL, Ventoso I, Castello A, Munoz-Fernandez MA, de Haro C, et al. GCN2 has inhibitory effect on human immunodeficiency virus-1 protein synthesis and is cleaved upon viral infection. Plos One. 2012;7(10):e47272. doi: 10.1371/journal.pone.0047272 ; PubMed Central PMCID: PMC3479103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Berlanga JJ, Ventoso I, Harding HP, Deng J, Ron D, Sonenberg N, et al. Antiviral effect of the mammalian translation initiation factor 2alpha kinase GCN2 against RNA viruses. Embo J. 2006;25(8):1730–40. doi: 10.1038/sj.emboj.7601073 ; PubMed Central PMCID: PMC1440839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Costa-Mattioli M, Gobert D, Harding H, Herdy B, Azzi M, Bruno M, et al. Translational control of hippocampal synaptic plasticity and memory by the eIF2alpha kinase GCN2. Nature. 2005;436(7054):1166–73. doi: 10.1038/nature03897 ; PubMed Central PMCID: PMC1464117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Chang ZQ, Lee SY, Kim HJ, Kim JR, Kim SJ, Hong IK, et al. Endotoxin activates de novo sphingolipid biosynthesis via nuclear factor kappa B-mediated upregulation of Sptlc2. Prostaglandins Other Lipid Mediat. 2011;94(1–2):44–52. doi: 10.1016/j.prostaglandins.2010.12.003 ; PubMed Central PMCID: PMC3366150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Hornemann T, Penno A, Rutti MF, Ernst D, Kivrak-Pfiffner F, Rohrer L, et al. The SPTLC3 subunit of serine palmitoyltransferase generates short chain sphingoid bases. J Biol Chem. 2009;284(39):26322–30. doi: 10.1074/jbc.M109.023192 ; PubMed Central PMCID: PMC2785320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Pralhada Rao R, Vaidyanathan N, Rengasamy M, Mammen Oommen A, Somaiya N, Jagannath MR. Sphingolipid metabolic pathway: an overview of major roles played in human diseases. J Lipids. 2013;2013:178910. doi: 10.1155/2013/178910 ; PubMed Central PMCID: PMC3747619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Chen CC, Juan CW, Chen KY, Chang YC, Lee JC, Chang MC. Upregulation of RPA2 promotes NF-kappaB activation in breast cancer by relieving the antagonistic function of menin on NF-kappaB-regulated transcription. Carcinogenesis. 2017;38(2):196–206. doi: 10.1093/carcin/bgw123 . [DOI] [PubMed] [Google Scholar]
- 285.Hagele H, Allam R, Pawar RD, Anders HJ. Double-stranded RNA activates type I interferon secretion in glomerular endothelial cells via retinoic acid-inducible gene (RIG)-1. Nephrol Dial Transplant. 2009;24(11):3312–8. doi: 10.1093/ndt/gfp339 . [DOI] [PubMed] [Google Scholar]
- 286.Farr M, Zhu D-F, Povelones M, Valcich D, Ambron RT. Direct interactions between immunocytes and neurons after axotomy in Aplysia. Journal of Neurobiology. 2001;46(2):89–96. . [DOI] [PubMed] [Google Scholar]
- 287.Farr M, Mathews J, Zhu DF, Ambron RT. Inflammation causes a long-term hyperexcitability in the nociceptive sensory neurons of Aplysia. Learn Mem. 1999;6(3):331–40. ; PubMed Central PMCID: PMC311296. [PMC free article] [PubMed] [Google Scholar]
- 288.Abrams TW. Studies on Aplysia neurons suggest treatments for chronic human disorders. Curr Biol. 2012;22(17):R705–11. doi: 10.1016/j.cub.2012.08.011 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Clatworthy A, Castro G, Budelmann B, Walters E. Induction of a cellular defense reaction is accompanied by an increase in sensory neuron excitability in Aplysia. The Journal of Neuroscience. 1994;14(5):3263–70. doi: 10.1523/jneurosci.14-05-03263.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Aravin AA, Hannon GJ, Brennecke J. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science. 2007;318(5851):761–4. doi: 10.1126/science.1146484 . [DOI] [PubMed] [Google Scholar]
- 291.Parrish NF, Fujino K, Shiromoto Y, Iwasaki YW, Ha H, Xing J, et al. piRNAs derived from ancient viral processed pseudogenes as transgenerational sequence-specific immune memory in mammals. Rna. 2015;21(10):1691–703. doi: 10.1261/rna.052092.115 ; PubMed Central PMCID: PMC4574747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Hamada H, Yamamura M, Ohi H, Kobayashi Y, Niwa K, Oyama T, et al. Characterization of the human zinc finger nfx1type containing 1 encoding ZNFX1 gene and its response to 12Otetradecanoyl13acetate in HL60 cells. Int J Oncol. 2019;55(4):896–904. doi: 10.3892/ijo.2019.4860 . [DOI] [PubMed] [Google Scholar]
- 293.Minoda Y, Saeki K, Aki D, Takaki H, Sanada T, Koga K, et al. A novel Zinc finger protein, ZCCHC11, interacts with TIFA and modulates TLR signaling. Biochem Biophys Res Commun. 2006;344(3):1023–30. doi: 10.1016/j.bbrc.2006.04.006 . [DOI] [PubMed] [Google Scholar]
- 294.Simon PS, Sharman SK, Lu C, Yang D, Paschall AV, Tulachan SS, et al. The NF-kappaB p65 and p50 homodimer cooperate with IRF8 to activate iNOS transcription. BMC Cancer. 2015;15:770. doi: 10.1186/s12885-015-1808-6 ; PubMed Central PMCID: PMC4619452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Chen J, Chen ZJ. Regulation of NF-kappaB by ubiquitination. Curr Opin Immunol. 2013;25(1):4–12. doi: 10.1016/j.coi.2012.12.005 ; PubMed Central PMCID: PMC3594545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Kemp IK, Coyne VE. Identification and characterisation of the Mpeg1 homologue in the South African abalone, Haliotis midae. Fish Shellfish Immunol. 2011;31(6):754–64. doi: 10.1016/j.fsi.2011.07.010 . [DOI] [PubMed] [Google Scholar]
- 297.Bathige SD, Umasuthan N, Whang I, Lim BS, Won SH, Lee J. Antibacterial activity and immune responses of a molluscan macrophage expressed gene-1 from disk abalone, Haliotis discus discus. Fish Shellfish Immunol. 2014;39(2):263–72. doi: 10.1016/j.fsi.2014.05.012 . [DOI] [PubMed] [Google Scholar]
- 298.Shen JD, Cai QF, Yan LJ, Du CH, Liu GM, Su WJ, et al. Cathepsin L is an immune-related protein in Pacific abalone (Haliotis discus hannai)—Purification and characterization. Fish Shellfish Immunol. 2015;47(2):986–95. doi: 10.1016/j.fsi.2015.11.004 . [DOI] [PubMed] [Google Scholar]
- 299.Munoz JP, Ivanova S, Sanchez-Wandelmer J, Martinez-Cristobal P, Noguera E, Sancho A, et al. Mfn2 modulates the UPR and mitochondrial function via repression of PERK. Embo J. 2013;32(17):2348–61. doi: 10.1038/emboj.2013.168 ; PubMed Central PMCID: PMC3770335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Muoio DM. TXNIP links redox circuitry to glucose control. Cell Metabolism. 2007;5(6):412–4. doi: 10.1016/j.cmet.2007.05.011 WOS:000247115400004. [DOI] [PubMed] [Google Scholar]
- 301.Shembade N, Parvatiyar K, Harhaj NS, Harhaj EW. The ubiquitin-editing enzyme A20 requires RNF11 to downregulate NF-kappaB signalling. Embo J. 2009;28(5):513–22. doi: 10.1038/emboj.2008.285 ; PubMed Central PMCID: PMC2657574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.McNally RS, Davis BK, Clements CM, Accavitti-Loper MA, Mak TW, Ting JP. DJ-1 enhances cell survival through the binding of Cezanne, a negative regulator of NF-kappaB. J Biol Chem. 2011;286(6):4098–106. doi: 10.1074/jbc.M110.147371 PubMed Central PMCID: PMC3039338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Kim KS, Kim JS, Park JY, Suh YH, Jou I, Joe EH, et al. DJ-1 associates with lipid rafts by palmitoylation and regulates lipid rafts-dependent endocytosis in astrocytes. Hum Mol Genet. 2013;22(23):4805–17. doi: 10.1093/hmg/ddt332 . [DOI] [PubMed] [Google Scholar]
- 304.Cardamone MD, Krones A, Tanasa B, Taylor H, Ricci L, Ohgi KA, et al. A protective strategy against hyperinflammatory responses requiring the nontranscriptional actions of GPS2. Mol Cell. 2012;46(1):91–104. doi: 10.1016/j.molcel.2012.01.025 ; PubMed Central PMCID: PMC3327812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Fan R, Toubal A, Goni S, Drareni K, Huang Z, Alzaid F, et al. Loss of the co-repressor GPS2 sensitizes macrophage activation upon metabolic stress induced by obesity and type 2 diabetes. Nat Med. 2016;22(7):780–91. doi: 10.1038/nm.4114 . [DOI] [PubMed] [Google Scholar]
- 306.Rorbach J, Nicholls TJ, Minczuk M. PDE12 removes mitochondrial RNA poly(A) tails and controls translation in human mitochondria. Nucleic Acids Res. 2011;39(17):7750–63. doi: 10.1093/nar/gkr470 ; PubMed Central PMCID: PMC3177208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Boveris A, Navarro A. Brain mitochondrial dysfunction in aging. Iubmb Life. 2008;60(5):308–14. doi: 10.1002/iub.46 WOS:000255598500014. [DOI] [PubMed] [Google Scholar]
- 308.Manna SK, Zhang HJ, Yan T, Oberley LW, Aggarwal BB. Overexpression of manganese superoxide dismutase suppresses tumor necrosis factor-induced apoptosis and activation of nuclear transcription factor-kappaB and activated protein-1. J Biol Chem. 1998;273(21):13245–54. doi: 10.1074/jbc.273.21.13245 . [DOI] [PubMed] [Google Scholar]
- 309.Kretz-Remy C, Mehlen P, Mirault ME, Arrigo AP. Inhibition of I kappa B-alpha phosphorylation and degradation and subsequent NF-kappa B activation by glutathione peroxidase overexpression. J Cell Biol. 1996;133(5):1083–93. doi: 10.1083/jcb.133.5.1083 ; PubMed Central PMCID: PMC2120847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Lingappan K. NF-kappaB in Oxidative Stress. Curr Opin Toxicol. 2018;7:81–6. doi: 10.1016/j.cotox.2017.11.002 ; PubMed Central PMCID: PMC5978768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Li N, Karin M. Is NF-κB the sensor of oxidative stress? The FASEB Journal. 1999;13(10):1137–43. 10.1096/fasebj.13.10.1137. [DOI] [PubMed] [Google Scholar]
- 312.Papa S, Bubici C, Pham CG, Zazzeroni F, Franzoso G. NF-kappaB meets ROS: an ’iron-ic’ encounter. Cell Death Differ. 2005;12(10):1259–62. doi: 10.1038/sj.cdd.4401694 . [DOI] [PubMed] [Google Scholar]
- 313.Deng J, Lu PD, Zhang Y, Scheuner D, Kaufman RJ, Sonenberg N, et al. Translational repression mediates activation of nuclear factor kappa B by phosphorylated translation initiation factor 2. Mol Cell Biol. 2004;24(23):10161–8. doi: 10.1128/MCB.24.23.10161-10168.2004 ; PubMed Central PMCID: PMC529034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Deleidi M, Jaggle M, Rubino G. Immune aging, dysmetabolism, and inflammation in neurological diseases. Front Neurosci. 2015;9:172. doi: 10.3389/fnins.2015.00172 ; PubMed Central PMCID: PMC4453474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Currais A. Ageing and inflammation—A central role for mitochondria in brain health and disease. Ageing Res Rev. 2015;21:30–42. doi: 10.1016/j.arr.2015.02.001 WOS:000355037900003. [DOI] [PubMed] [Google Scholar]
- 316.Garaschuk O, Semchyshyn HM, Lushchak VI. Healthy brain aging: Interplay between reactive species, inflammation and energy supply. Ageing Res Rev. 2018;43:26–45. doi: 10.1016/j.arr.2018.02.003 WOS:000433016300004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
For each cutout, the scale free topology model fit (A), median connectivity (B), mean connectivity (C), and max connectivity (D) of a co-expression matrix (y-axis) at a selected soft threshold power (x-axis) is plotted for both BSC (black) and PVC (Red). A soft power of 16 was selected for both sensory neuron types to achieve a scale free topology fit above 0.9 and to minimize the mean connectivity.
(TIF)
Module eigengene names are arbitrarily assigned. Red horizontal line represents 0.25 branch height threshold for similarity, below which modules are merged.
(TIF)
Colored bars at the bottom represent module assignments for each transcript. Module colors are arbitrarily assigned. The top color set represents the module assignment before merging similar of similar modules, while bottom bar represents module assignment post merge (see S2 Fig). In total, 13 co-expression modules were identified.
(TIF)
Each cell of the heatmap represents the correlation between a module eigengene (row) and phenotype (column). The top number in a cell is the value of Pearson correlation between two eigengenes. The p-value significance of module-trait correlation is the bottom number in each cell in parentheses. The phenotypes are animal weight at sacrifice (weight), latency of two reflex behaviors described in Greer et al 2018: Tail withdrawal reflex time (TWRT) and time to right (TTR), and chronological age of the animal in months at sacrifice (Age). Correlations for age are similar between sensory neuron types for the royalblue, saddlebrown, greenyellow, orange, pink, and darkgreen modules.
(TIF)
All Software and respective versions used for RNA sequencing read quality control and quality assurance, mapping and abundance estimation, and downstream analysis.
(DOCX)
Each cell represents the number of transcripts shared between respective module (row) and cluster (column). The “n” column represents the number of transcripts in a given module, and the “n” row represents the number of transcripts in a given cluster. Values in the “Sum” columns are row sums, e.g. the total number of transcripts in each respective module also present in the clusters. The “% of module” columns represent percentage values calculated by dividing the “Sum” columns by the total number of transcripts in a cluster set (1106 for B clusters, and 1198 for P clusters). Values in the “Sum” row are column sums, e.g. the total number of transcripts in each respective cluster that are also present in the modules. The “cluster %” row represent percentage values calculated by dividing the “Sum” row by the total number of transcripts among all modules (10012).
(DOCX)
All R packages and respective versions used for clustering, analysis, and visualization of RNA sequencing transcript abundances.
(TXT)
Full results for all modules of KEGG enrichment of co-expression modules using clusterProfiler.
(XLSX)
Per module module-membership values and significance for all transcripts assigned to each module.
(XLSX)
This file contains the results of mapping Aplysia californica transcript IDs to Crassostrea gigas gene IDs for transcripts in Aplysia age associated co-expression modules that mapped to C. gigas genes identified as DE in immune priming and viral challenge by Laont et al 2020.
(XLSX)
This file contains a list of genes upregulated genes in C. gigas immune respone to viral analogue poly(IC) as reported in Lafont et al. 2020 [37] that have orthologs with evalue of >1E-20 in Aplysia. Aplysia orthologs are annotated with transcript, protein, and gene RefSeq identifiers as well as neuron type-specific adjusted pvalues, expression cluster assignment, and expression cluster direction reported in our previous publication. Only two Aplysia orthologs were identified as significantly upregulated in aging sensory neurons.
(XLSX)
Data Availability Statement
The BioProject in which our sequencing data is archived can be accessed here: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA639857. The project has Accession: PRJNA639857, ID: 639857, and title: Aplysia californica Sensory Neuron Time Series Transcriptome.